Abstract
Our lab has identified that transcripts and proteins of the cyclooxygenase (COX-1 and COX-2) isoenzymes are expressed during the early stages of vertebrate embryonic development, and that global COX-1/2 inhibition disrupts neural crest (NC) cell maturation in Ambystoma mexicanum (axolotl) embryos, with intriguing implications for tissue regeneration and healing. NC cells are embryonic stem cells that differentiate into various adult tissues including craniofacial cartilage, bone, and neurons in the peripheral and enteric nervous systems. Naproxen (NPX), a common non-steroidal anti-inflammatory drug (NSAID) used to relieve pain and inflammation, exerts its effects through COX-1 and COX-2 inhibition. Embryonic exposures to NSAIDs have been linked to preterm birth, neural tube closure defects, abnormal enteric innervation, and craniofacial malformations, potentially due to disrupted neural tube or NC cell development. To investigate the phenotypic and molecular effects of NPX exposure on NC development and differentiation, we exposed late neurula and early tailbud stage axolotl embryos to various concentrations of NPX and performed immunohistochemistry (IHC) for markers of migratory and differentiating NC cells. Our results reveal that NPX exposure impairs the migration of SOX9+ NC cells, leading to abnormal development of craniofacial cartilage structures, including Meckel’s cartilage in the jaw. NPX exposure also alters the expression of markers associated with peripheral and central nervous system (PNS and CNS) development, suggesting concurrent neurodevelopmental changes.
Keywords: cyclooxygenase, COX-1, COX-2, neural crest, axolotl, naproxen, NSAID
I. Introduction
Non-steroidal anti-inflammatory drugs (NSAIDs) such as ibuprofen, acetaminophen, and naproxen (NPX) are drugs that mediate pain and inflammation via inhibition of the cyclooxygenase (COX) isoenzymes. There are two major COX isoforms, COX-1 and COX-2, which produce hormone-like signaling molecules called prostaglandins (PGs) from arachidonic acid isolated from membrane phospholipids [1]. Among other functions, PGs are responsible for generating inflammation and the fever, pain, redness, and swelling that follow inflammatory processes [2]. Despite being well-studied in normal tissue homeostasis [3–7], pathophysiologic inflammation [8–13], and tissue healing contexts [14–18], much less is known about the role of COX signaling during embryonic development.
In mammalian models, in utero exposure to NSAIDs causes developmental defects in craniofacial, cardiovascular, and gastrointestinal tissues [19, 20]. Work in zebrafish embryos identified that genes encoding COX signaling pathway factors are expressed at the earliest stages of cell specification and that COX signaling is necessary for major developmental processes, including gastrulation [21, 22]. In zebrafish, mouse, and chicken embryo models, COX inhibition via ibuprofen exposure during early development caused abnormal colonization of the gut by enteric neural crest (NC)-derived cells, which become the cells of the enteric nervous system (ENS). In these studies, exposure to ibuprofen inhibited normal migration and proliferation of embryonic NC stem cells [23]. Additionally, COX signaling inhibition in chicken embryos altered the expression of factors involved in key pathways for NC cell development and migration including the Wnt/β-catenin and TGF-β pathways [24]. Conversely, induction of inflammation in chicken embryos also appeared to affect the expression of TGF-β signaling members and ultimately caused abnormal NC migration and development [25]. Clearly, there is a delicate balance between inflammatory and anti-inflammatory signaling that must be maintained for normal development to occur, but we still lack an understanding of the role of the COX signaling pathway within this balance.
Considering the types of developmental abnormalities described above, COX signaling appears to play a role in the normal development of NC and neural tube cells. NC cells are multipotent stem cells in the developing embryo that form at the neural plate border adjacent to the neural plate and subsequently delaminate from the neural tube and undergo an epithelial-to-mesenchymal transition (EMT) to then migrate out into peripheral tissues and give rise to a diverse array of derivatives [26, 27]. NC derivatives range from melanocytes and cells of the peripheral and enteric nervous systems (PNS and ENS) to adipocytes, and craniofacial bone and cartilage [26, 27]. Consequently, a diverse range of developmental anomalies can be attributed to abnormal NC cell development — termed ‘neurocristopathies’—including cleft palate, heart defects, and enteric neuropathies [28].
To delaminate from the developing neural tube, migrate into peripheral tissues, and differentiate into the correct cell type derivative, NC cells alter their gene expression, cell-cell and cell-matrix adhesion, and cytoskeletal arrangements, and become migratory and invasive [27, 29]. Normal NC cell development, therefore, requires precise timing and a complex network of signaling pathways that drive the expression of key factors within their gene regulatory network (GRN). The complexity of these processes makes NC cells vulnerable to assaults including exposure to a variety of potential toxicants and teratogens that can disrupt these cellular processes [30].
We investigated the effects of globally inhibiting COX signaling during development by NPX exposure in externally developing, vertebrate Ambystoma mexicanum (axolotl) embryos. Here, we demonstrate that embryonic expression of COX pathway genes is conserved across species. We additionally identify possible etiologies of NSAID-mediated developmental defects, specifically by molecularly characterizing the onset of developmental anomalies after NPX exposure by analyzing changes in the expression of key factors marking NC cell development. We show that exposing axolotl embryos to biologically relevant concentrations of NPX causes abnormal NC migration marked by a reduction in SOX9 protein expression. This reduced migration further manifests as stage-dependent changes in axial and craniofacial development marked by abnormal localization and patterning of COL2A1-positive craniofacial cartilage structures. Specifically, NPX exposure inhibits normal migration of NC cells fated to become craniofacial chondrocytes, leading to a hypoplastic, immature phenotype of Meckel’s cartilage. We also demonstrate that NPX exposure causes changes in the expression of markers of peripheral and central nervous system (PNS and CNS) development that may indicate an abnormal neurodevelopmental phenotype accompanying the changes in craniofacial chondrogenesis.
II. Methods/Experimental Design
Animal use and NPX exposures
Adult axolotls were housed and mated in accordance with the UC Davis Institutional Animal Care and Use Committee protocol #23047. Embryos were collected and staged according to the axolotl developmental staging chart [31]. The axolotls were cultured between 18°C - 22°C in Holtfreter’s (HF) solution (15 mM NaCl, 0.6 mM NaHCO3, 0.2 mM KCl, 0.2 mM CaCl2, and 0.2 mM MgSO4•7H20) for approximately 72 hours to reach stage 19 (early tailbud stage). Embryos were then either maintained in 100% HF solution or were exposed to various biologically relevant concentrations (5, 10, or 20 μg/mL) of NPX (C14H14O3) dissolved in HF solution [32]. Solutions were replaced once daily until the embryos reached the stage of interest for collection. Embryos remained in their jelly coats during exposure and incubation. Embryos were then cultured at room temperature (RT, approximately 21°C – 22°C) and were collected for immunohistochemistry (IHC) at one of three stages relating to NC cell development: stage 28 (specification and early migration), stage 36 (migration), and stage 45 (differentiation). Embryos were staged based on two factors: their developmental age and their gross morphology compared to control embryos from the same clutch. Control embryos were monitored to determine the time it took to reach the developmental stages of interest and these timepoints were used to track developmental progress of the treated embryos. The development of key morphological characteristics such as the eye, gills, dorsal fin, and the degree of axial elongation were also weighed alongside developmental age to determine the stage of treated embryos. Once the embryos reached stages of interest, they were manually dechorionated through surgical dissection with forceps prior to fixation of the embryos in 4% paraformaldehyde (PFA).
Immunohistochemistry
IHC was performed as previously described with some modifications [33]. Briefly, embryos were fixed in 4% PFA in phosphate buffer for 1 hour then quickly washed three times in 1x TBS (500 mM Tris-HCl pH 7.4, 1.5 M NaCl, and 10 mM CaCl2) containing 0.1% Triton X-100 (TBST + Ca2+) to remove remaining PFA. They were then incubated in a blocking buffer solution (10% donkey serum in 1x TBST + Ca2+) overnight at 4°C. Table 1 lists the primary antibodies used for IHC in this study. Antibodies were diluted (1:2–1:500) in blocking buffer with the optimized dilutions for each antibody (Table 1). Embryos were incubated in primary antibodies and 4’,6-diamidino-2-phenylindole (DAPI) stain at 4°C for 48 hours. Next, embryos were washed in TBST buffer and secondary Alexa Fluor antibodies were added. Embryos were kept in the secondary antibody mixes overnight at 4°C, post-fixed in 4% PFA for 1 hour at RT, then washed in 1x TBST + Ca2+ six times for 15 minutes each wash prior to whole mount imaging.
Table 1.
Primary antibodies used for IHC.
| Antibody Target | Application | Dilution | Antigen Species | Isotype | Source |
|---|---|---|---|---|---|
| COL2A1 | IHC | 1:250 | Chicken | mouse IgG1 | DSHB (AB_528165) |
| ECAD | IHC | 1:500 | Human | mouse IgG2a | BD Biosciences (61081) |
| GFAP | IHC | 1:250 | Human | rabbit IgG | Proteintech (16825-1-AP) |
| PAX7 | IHC | 1:5 | Chicken | mouse IgG1 | DSHB (AB_528428) |
| SOX9 | IHC | 1:500 | Human | rabbit IgG | Millipore (AB5535) |
| TUBB3 | IHC | 1:250 | Chicken | mouse IgG2a | R&D Systems (MAB1195) |
Fluorescence microscopy
Embryos were imaged both in whole mount and after cryosectioning using the Zeiss Axio Imager.M2 microscope. Wholemount stage 28 and 36 embryos were imaged at 10X magnification, focused on the cranial region of the embryo while stage 45 embryos were imaged at 5X magnification. Embryos were washed in sucrose and embedded in gelatin for cryosectioning (see below). They were then cryosectioned at 16–25 μm thick sections. Cryosections were imaged using the ZEISS Apotome.2 at 10X-63X magnification. Images were obtained using ZEN Blue 3.0 optical processing software.
Embedding and cryosectioning
Embryos were incubated in 5% sucrose at 4°C overnight, and then in 15% sucrose at 4°C overnight. They were then incubated in 10% gelatin overnight at 37°C. Embryos were then embedded in 10% gelatin, flash-frozen in liquid nitrogen, and stored at −80° C until cryosectioning. Embryos were cryosectioned with a Microm NX70 cryostat.
Image analysis
Bright field images were evaluated to determine changes in gross morphology of the developing embryos. Head length, dorsal fin height, eye diameter, and pre-optic length were all measured using Adobe Illustrator. For these measurements, line segments were converted to μm using in-image scale bars and the length was compared between treatments. Cell counts for SOX9 and PAX7+ cells were performed using the Adobe Photoshop counter tool as previously described by our lab [33]. Dorsoventral (DV) displacement was calculated as a measurement of cell migration; the methods for these calculations were adapted from a previously described statistical shape analysis technique [34]. For DV displacement, Illustrator was used to draw three lines overlying the transverse section of the embryo: the first was drawn from the dorsal-most point on the section down to the ventral-most point (the total length of the DV axis); the second was drawn from the same starting point until the furthest migrated SOX9 or PAX7+ cell in the section; the last was drawn starting from that same cell and ending at the ventral point of the DV axis. The DV displacement was calculated using these measurements and trigonometric equations to represent the migration distance of cranial NC cells in each transverse section (Supplemental Figure 7) to account for differences in embryo sizes.
Morphometric analysis of the developing cartilage was performed using Photoshop and FIJI (ImageJ). The total area and circularity of each cross-section through Meckel’s cartilage was measured using the built-in image analysis tools in Photoshop. Cell density was calculated using the measured total area of the cross-sections and cell counts of the DAPI+ cells within the COL2A1+ region which were obtained using the lasso selection tool and counter tool, respectively. Then, using FIJI, the cross sectioned images of Meckel’s cartilage were thresholded into two images for separate analysis: (1) perichondral COL2A1+ band and (2) intra-articular COL2A1+ bundles. For the collagen band, the ImageJ plug-in MorphJ was utilized to determine local thickness and area of staining. For the intra-articular bundles, thresholded images were watershed and internal function Analyze particles was utilized to count and determine the area of each bundle.
GraphPad Prism 10 was used to generate violin plots for the cell count, DV displacement, and morphometric data obtained from bright field and wide field fluorescence microscopy image analysis. Statistical analysis using nonparametric, unpaired Mann-Whitney tests was performed using Prism. The total sample size per IHC marker used in this study are as follows: Stage 28 PAX7 and SOX9: n=13, n=21, n=29, and n=16 for control, 5 μg/mL NPX, 10 μg/mL NPX, and 20 μg/mL NPX, respectively; Stage 45 COL2A1: n=18, n=29, n=21, and n=9; Stage 36 GFAP and TUBB3: n=6, n=15, and n=26; Stage 45 ECAD: n=15, n=14, and n=13. The relevant sample sizes used for quantitative analyses are listed in the individual figure captions. For each embryo, multiple transverse sections were analyzed. Z-scores for Figure 5 describe the axial level at which the image of the transverse section was taken and were determined by identifying the presence of ocular structures within the section included the developing cornea, retina, and lens. Based on the prescence or absence of these structrues, the following z-scores were defined: lens, post-lens, and post-optic.
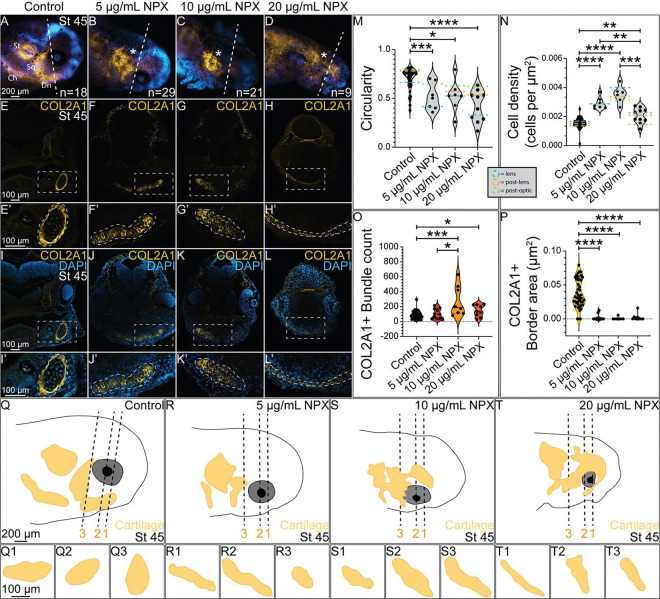
Figure 5. COX inhibition causes abnormal formation of cranial cartilage structures.
IHC for COL2A1 in (A-D) wholemount and (E-L) transverse sections from stage 45 axolotls. (A-D) Dashed line indicates axial level of sections in (E-L). Asterisks mark abnormal COL2A1 localization. (E-L) Dashed boxes indicate region of zoom in. (E’-L’) Enlarged images of Meckel’s cartilage show abnormal tissue morphology and COL2A1 localization following NPX exposure. Dashed outlines highlight the developing chondrocytes within Meckel’s cartilage expressing COL2A1. (M-P) Quantification of circularity and cell density of Meckel’s cartilage, along with quantification of the number of COL2A1+ bundle structures within Meckel’s cartilage and the total area of the COL2A1+ band surrounding Meckel’s cartilage in cross section. Green, orange, and blue dashed lines on the violin plots in M-N indicate the average circularity or cell density values at each axial level (lens= blue, post-lens= orange, and post-optic= green) across the 4 treatment groups. For analysis of COL2A1, the sample sizes analyzed were n=4, n=4, n=2, and n=2 for the respective treatment groups. (Q-T) Drawings of wholemount stage 45 axolotl embryos in A-D with the developing COL2A1+ cranial cartilages drawn in the yellow structures. Dashed lines indicate the axial levels that were analyzed in the circularity and cell density analysis of Meckel’s cartilage in M-N (1 = lens, 2 = post-lens, and 3 = post-optic). (Q1-Q3, R1-R3, S1-S3, and T1-T3) Drawings of transverse sections through Meckel’s cartilage at each axial level across treatment groups. St= supratemporal cartilage, Ch= ceratohyal, Sq= squamate, Dn= dentate (Meckel’s cartilage).
Single-cell sequencing analysis
Publicly available embryonic single cell sequencing data sets were obtained and variably processed according to the organism corresponding to each dataset. Detailed information is described in Supplementary Table 1. All plots were created using built-in Seurat functions [35] or ggplot [36] and were organized in BioRender.
III. Results
Analysis of COX signaling pathway transcript expression during embryogenesis across species
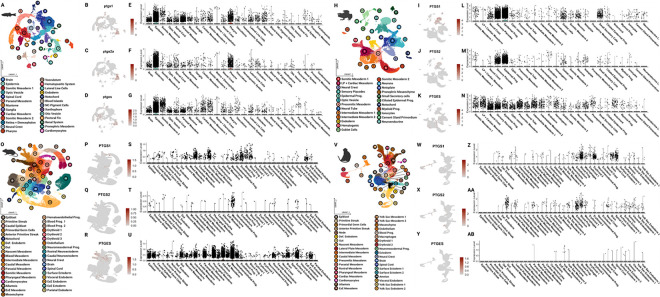
To confirm the conservation, and relevance, of the COX pathway factors during vertebrate embryogenesis, we utilized publicly available whole embryo single cell RNA-sequencing datasets from zebrafish [37–40], frog [41], mouse [42], macaque [43], and axolotls [44]. We selected datasets which incorporated all three germ layers and were staged around early organogenesis. Specifically, the zebrafish dataset represented 12 hours post fertilization through 2 days post fertilization, the African clawed frog was from stage 18 through stage 22, the house mouse was from mixed late gastrulation through embryonic day 8.5, and the cynomolgus macaque dataset ranged from embryonic day 20 through 29. Across species, cells are clustered into roughly 25–35 cell types (Figure 1A, H, O, V) with variability in expression of genes encoding major COX pathway enzymes, particularly COX-1, COX-2 and PTGES (Figure 1B–G, I–N, P–U, W–AB). The enzymes were found to be expressed in epidermal derivatives in the anamniotes (Figure 1B–G, I–N) but were more prevalent in hemato-vascular cell types in the amniotes (Figure 1P–U, W–AB) at the represented stages. Other genes encoding COX pathway elements were identified across species in various stages (Supplemental Figures 1–6). In particular, PTGES3 was found to be ubiquitously expressed across most cell types in all species.
Figure 1. The COX pathway is active in embryonic cell types across vertebrate species.
Analysis was performed of publicly available scRNA-seq data from whole embryos, staged around early organogenesis, of 4 chordate species — Zebrafish (Danio rerio), African clawed frog (Xenopus laevis), House mouse (Mus musculus) and Cynomolgus macaque (Macaca fascicularis). (A, H, O, V) UMAP demonstrating clustering results of whole embryos from vertebrate species. (B-D, I-K, P-R, W-Y) Feature UMAPs demonstrating gene expression of select enzymes across species. (E-G, L-N, S-U, Z-AB) Violin plots demonstrating expression of select enzymes across the major cell types identified by species. NC= neural crest, LP= lateral plate, Prog.= progenitor, Def.= definitive, ExE= extra embryonic.
Previous studies using zebrafish embryos have demonstrated that the expression of COX enzymes during early embryonic development is conserved across mammalian and aquatic vertebrate species [21, 22]. Additionally, the COX signaling pathway is proposed to have a role in several key developmental processes including gastrulation based on COX activity and the effects of NSAIDs during early development [45–47]. Although open-source single cell datasets are limited during the stages of axolotl embryogenesis, we reanalyzed publicly available single cell RNA-sequencing data from axolotl embryonic and regenerating limb buds to confirm that these genes are expressed in our organism of interest. In embryonic axolotls, we identified strong expression of genes encoding the COX enzymes in both early limb field and late limb bud tissue (stages 25–44) (Supplemental Figure 5).
Characterization of relevant markers in axolotl embryos
Axolotls are commonly used research models in the field of regenerative biology [31, 48]; however, there is a gap in knowledge regarding early NC development and differentiation in axolotls. NC cell migration appears to be the most well-studied process within NC development, with previous research in the field focusing on the developmental origins of NC cells [49, 50] and the factors that influence migration of NC cells out from the neural tube to differentiate in the peripheral tissues [51–53]. Researchers have also characterized the role of the extracellular matrix (ECM) in NC migration [54–57], but there is still a gap in our understanding of the timing of NC migration and differentiation. Based on these gaps in knowledge, we first characterized expression of proteins with established roles in the development of NC and neural tube-derived cells in other model systems.
To characterize expression of markers across multiple stages of axolotl development we used IHC and fluorescence microscopy. First, we analyzed expression of the transcription factor, PAX7, which is necessary for NC specification and induction of downstream effectors in the NC GRN during early development [58, 59]. At stage 28, we identified expression of PAX7 in NC cells in the dorsal neural tube where specification occurs, as well as in migratory NCs immediately lateral to the neural tube (Figure 2A, A’, pink arrows). At stage 45, PAX7 was maintained in intermittent cells underlying the skin that will develop into pigment cells in the adult axolotl [60]. PAX7 expression in stage 45 embryos was also identified throughout the dorsal neural tube in cells that will develop into interneurons of the motor system in the CNS (Figure 2B, B’, pink dashed outlines) [61–63]. Next, we evaluated expression of the SRY-box transcription factor, SOX9, which is downstream of PAX7 in the NC GRN and essential to numerous developmental processes such as skeletogenesis, sex determination, and gonadogenesis [64–67]. Similar to PAX7, SOX9 is frequently used as a marker of NC cells during NC specification and migration [68]. At stage 28, SOX9 was upregulated in a small number of premigratory NC cells, and several migrating NC cells immediately lateral to the neural tube and in the ventral region of the face (Figure 2C, C’, green arrows). SOX9 expression was maintained in stage 45 embryos with SOX9-expressing cells surrounding the neural tube and colonizing the region behind the eye where they will contribute to the development of the cranial cartilage structures of the skull (Figure 2D, D’, green dashed outlines). We identified that a known downstream target of SOX9 in mammals and the primary ECM component in developing cartilage, Collagen type II apha 1 chain (COL2A1) [69, 70], is expressed following the upregulation of SOX9 expression in early migratory NC cells, with expression of COL2A1 visible in migratory NC cells at stage 36 (Figure 2E, E’, yellow arrows). We observed a similar temporal expression pattern between SOX9 and COL2A1 at stage 45 where COL2A1 is expressed immediately surrounding the neural tube to form the cartilages of the skull, and in the ventral region of the face where SOX9+ cells have initiated the chondrogenic program to promote formation of the cartilages that will form the jaw and palate (Figure 2F, F’, yellow dashed outlines). We then examined the expression of glial fibrillary acidic protein (GFAP), which is expressed by neural progenitors in early development, and later is expressed in mature astrocytes in the CNS [71, 72]. At stage 28, GFAP expression was visible throughout the developing CNS and appeared polarized, with strongest expression at the basal side of the developing neural tube (Figure 2G, G’). At stage 45, the polarized expression of GFAP in the basal side of the neural tube was maintained, but GFAP expression could also be seen astrocyte cell bodies within the neural tube (Figure 2H, H’, orange dashed outlines). Lastly, we evaluated expression of the cell-cell adhesion molecule, E-cadherin (ECAD). In stage 28 embryos, we observed ECAD expression in the developing non-neural ectoderm (NNE) and in the apical region of the neural tube as expected based on prior analysis in other organisms (Figure 2I, I’). In stage 45 embryos, we observed robust expression of ECAD in the developing skin but at these stages it was no longer detectable in the developing brain (Figure 2J, J’, red dashed outlines). Diagrams of the stages and axial levels analyzed in this manuscript are depicted in Figure 2K–M). Following our characterization of the expression of key proteins known to regulate NC development in various vertebrate models, we then investigated how global inhibition of COX signaling alters the expression and localization of these proteins.
Figure 2. Characterization of protein expression during axolotl embryogenesis.
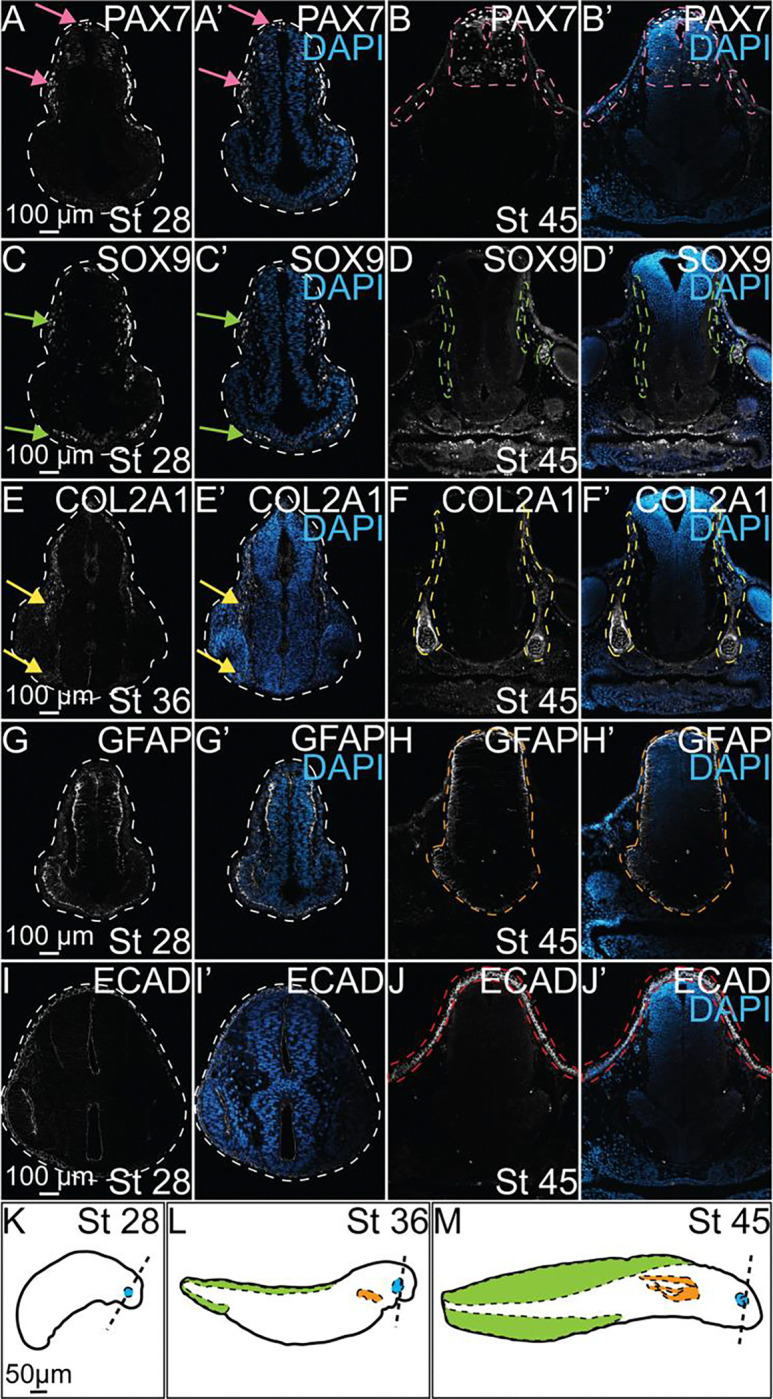
IHC for PAX7 (A-B, A’-B’), SOX9 (C-D, C’-D’), COL2A1 (E-F, E’-F’), GFAP (G-H, G’-H’), and ECAD (I-J, I’-J’) in control axolotl embryos across several stages of development. (A’-J’) IHC for DAPI overlaid with each individual marker to highlight the present anatomical structures at each stage. (A, A’) Pink arrows identify expression of PAX7 in specified NC in the dorsal neural tube and migratory NC. (B, B’) Pink dashed outlines highlight superficial expression of PAX7 in skin and expression in developing interneurons in the NT. (C, C’) Green arrows highight SOX9+ NC, including those beginning to colonize the ventral face. (D, D’) Green dashed outlines identify SOX9 expression surrounding the neural tube and behind the developing eye. (E, E’) Yellow arrows indicate expression of COL2A1 in late migratory NC that follows expression of SOX9 in earlier stages. (F, F’) Yellow dashed outlines highlight COL2A1 expression in the early formation of the cranial cartilages. (H, H’) The orange dashed outline identifies the developing neural tube and expression of GFAP in glial progenitors. (J, J’) The red dashed outlines highlights expression of ECAD in epithelial cells of the developing skin. (K-M) Drawings of axolotl embryos at stages 28, 36, and 45 in development. Dashed lines approximate the axial level in the head at which transverse section images were taken. Developing ocular structures, gills, and dorsal fin are highlighted in blue, orange, and green, respectively.
NPX exposure causes morphological defects
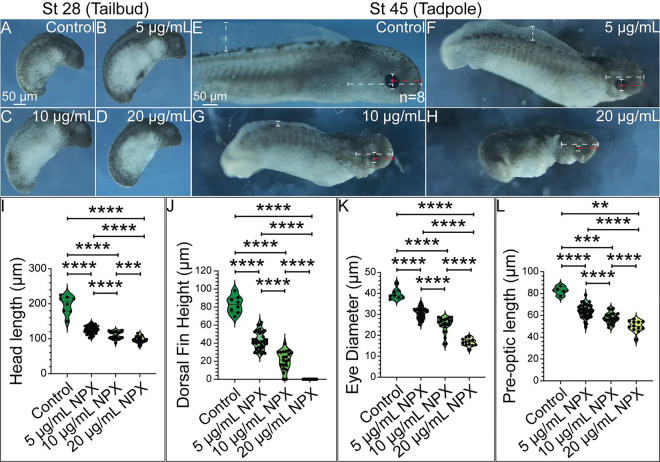
Prior research showed that COX inhibition through NSAID exposure during embryonic development does in fact cause significant developmental defects in NC-derived structures including cardiac malformations, abnormal innervation of the PNS and ENS, and craniofacial abnormalities [19, 20, 73]. Based on this information, we hypothesized that there would be observable morphological changes in axolotl embryo development following NSAID exposure. To test this hypothesis, we exposed axolotl embryos to varying concentrations of NPX and performed morphological analysis to evaluate the incidence and severity of developmental anomalies that occur following embryonic NSAID exposure. Embryos were exposed to NPX at late neurula and early tailbud stages to focus on NC formation and migration stages and to avoid germ layer malformation during gastrulation. The range of concentrations used for NPX exposure experiments were derived from a study evaluating the placental transfer of NPX in human embryos to mimic mammalian biological exposures in the aquatic, externally-developing axolotl [32]. We found that inhibition of COX signaling through NSAID exposure during embryonic development caused a significant reduction in the survival of axolotl embryos in a dose-dependent manner (Supplemental Figure 6). In stage 28 axolotl embryos (tailbud), there were no overt effects on morphological development observed independent of the degree of COX inhibition via NPX exposure (Figure 3A–D). However, in stage 45 axolotl tadpoles, we identified obvious defects in growth and morphology (Figure 3E–H). Specifically, compared to control tadpoles, all those exposed to any concentration of NPX had defects in their head length (Figure 3E–H, I), dorsal fin height (Figure 3E–H, J) eye diameter (Figure 3E–H, K), and pre-optic length (Figure 3E–H, L). Combined with the low rates of survival and concentration-dependent scale of defects, it is clear that global inhibition of COX signaling altered axolotl growth and development. Given their importance in regulating craniofacial development, we then investigated if NPX exposure had an effect on expression of key regulators of NC development that could constitute a molecular mechanism for the development of the craniofacial defects observed.
Figure 3. The degree of NPX exposure corresponds to the severity of morphological effects observed in late-stage axolotl embryos.
(A-D) In stage 28 embryos, NPX appears to have no effect on development demonstrated by the lack of observable defects. (E-H) In stage 45 embryos, increasing concentration of NPX corresponds with the development of progressively more severe morphological defects. Dashed bars demonstrate how morphologic measurements were performed. (I-L) All 3 treatment concentrations produce statistically significant decreases in head length, dorsal fin height, eye diameter, and pre-optic length in stage 45 embryos. Sample sizes for morphologic analysis were n=8, n=40, n=22, and n=19 for the control, 5 μg/mL NPX, 10 μg/mL NPX, and 20 μg/mL NPX treatment groups, respectively. Embryos that did not have grossly appreciable development of their eyes or gills were excluded from the eye diameter, pre-optic length, and head length measurements. Non-parametric, unpaired Mann-Whitney t-tests were performed to analyze these results. p-value= *, **, ***, **** = p-value < 0.05, < 0.01, < 0.001, and <0.0001, respectively.
COX inhibition impedes NC migration
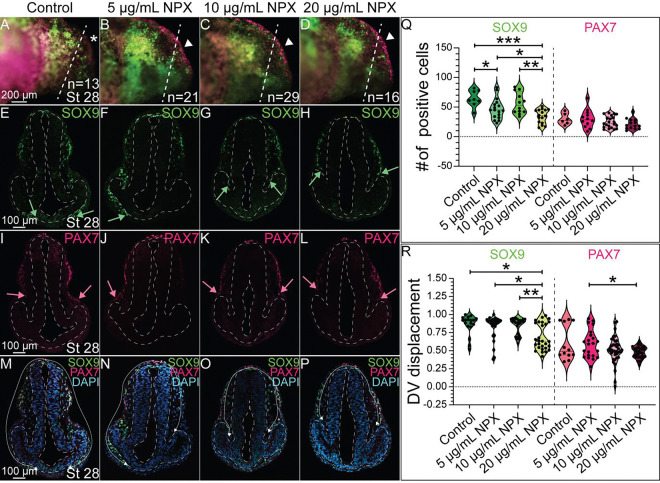
Previous research demonstrated that NSAID exposure affects the migration, proliferation, and survival of NC-derived cells [23]. Our prior work and the gross morphological changes observed in our embryos at stages 36 and 45 following NPX exposure (Figure 3) suggested that COX signaling is necessary for NC development, and therefore we investigated the expression of molecular markers for NC cells and their tissue derivatives. To identify if COX signaling is necessary for NC cell specification and migration, we exposed embryos at late neurula and early tailbud stage to various concentrations of NPX and then used IHC to evaluate changes in expression of two key transcription factors that regulate early NC cell development, SOX9 and PAX7. We identified that although the tailbud embryos appeared morphologically normal in wholemount (Figure 3A–D), compared to control embryos, COX inhibition via NPX exposure at 5 μg/mL and 20 μg/mL reduced the numbers of SOX9+ cranial NC cells (Figure 4A–H, Q). In contrast, there were no significant changes in the number of PAX7+ cells in NPX-exposed versus control embryos, and the ability of PAX7− positive cells to migrate out of the dorsal neural tube following COX inhibition was not significantly reduced compared to control embryos (Figure 4I–L, Q–R). In addition to reducing the number of SOX9+ NC cells, NPX exposure causes a visible reduction in the migratory capacity of these cells (Supplemental Figure 7). When this trend was quantified, we determined that NPX exposure significantly reduced the migration distance of SOX9+ NC cells at the 20 μg/mL NPX concentration compared to control embryos (Figure 4M–P, R).
Figure 4. NPX exposure reduces the migration of SOX9+ cells into the developing cranial region.
IHC for SOX9 and PAX7 in (A-D) whole mount and (E-P) transverse sections of stage 28 axolotl embryos. (A-D) Dashed lines show the axial level in the head that corresponds to the transverse sections. Arrowheads highlight superficial PAX7 expression which appears increased in (B-D) treated embryos compared to PAX7 expression in the (A) control embryos marked by the asterisk. (E-P) Dashed lines outline the neural tube and developing eyes in transverse section. (E-H) Green arrows indicate the extent of migration of SOX9+ NC cells in the cranial region. (I-L) Pink arrows indicate the migration of PAX7+ NC cells into the ventrolateral region of the developing head (M-P) The white arrows demonstrate the migratory path of NC cells into the ventral face. (Q) Quantification of the number of SOX9+ or PAX7+ cranial NC cells based on degree of NPX exposure. (R) Quantification of the migration distance of SOX9+ or PAX7+ NC cells, measured as DV displacement, in transverse section. For SOX9 analysis, the following sample sizes were used: n=4, n=5, n=5, and n=6 embryos for the control, 5 μg/mL NPX, 10 μg/mL NPX, and 20 μg/mL NPX groups, respectively, with an average of 3 transverse sections analyzed per embryo. Similarly, for PAX7 analysis, the sample sizes were n=2, n=5, n=6, and n=6 embryos for the respective treatments, and an average of 3 transverse sections were analyzed per embryo.
Global COX inhibition by NPX alters cranial cartilage formation
Based on the reduction in the number and migratory capacity of SOX9+ NC cells following NPX exposure shown in Figure 4, we hypothesized that there may be downstream effects on NC cell differentiation into cranial chondrocytes. We exposed embryos to the same concentrations of NPX, performed IHC for COL2A1, and then evaluated changes in the development of cranial cartilage structures using morphometric analysis (Figure 5). Alterations in COL2A1 expression were apparent in wholemount stage 45 tadpoles (compare Figure 5A to 5B–D and 5Q to 5R–T). Analysis of COL2A1 in transverse cryosections revealed that in comparison to control tadpoles (Figure 5E, E’, I, I’), which showed normal morphology of the mandibular cartilage (Meckel’s cartilage), embryos exposed to all concentrations of NPX had abnormal development of Meckel’s cartilage demonstrated by abnormal localization of COL2A1 in the lower jaw (Figure 5F–H, J–L). During the process of maturation, chondrocytes begin to secrete specific molecules that define their identity with respect to their stage in the maturation process. Early maturation chondrocytes express high levels of collagen types II and IV and proteoglycans like aggrecan which help draw water into the cartilage tissue, giving the tissue its shape [74]. We indirectly evaluated chondrocyte maturation and cartilage tissue morphogenesis by quantifying 1) the circularity of Meckel’s cartilage in cross-section, 2) the cell density within transverse sections of Meckel’s cartilage, 3) the amount of COL2A1+ signal (bundles) within each cross-section, and 4) the total area of the COL2A1+ border band (perichondrium) that surrounds Meckel’s cartilage in control embryos.
We identified abnormal morphology of Meckel’s cartilage in treated embryos based on the reduction in cross-sectional circularity and elongation of Meckel’s cartilage in cross-section compared to control embryos (Figure 5E’–L’, M). The increase in cell density within Meckel’s cartilage in treated embryos compared to control embryos (Figure 5I’–L’, N) suggests that the chondrocytes in the treated embryos are not developing normally, further indicating that exposure to NPX inhibits normal chondrocyte maturation and cartilage morphogenesis. Although all tadpoles expressed COL2A1 to some extent, the localization of the signal within the cross-sections of Meckel’s cartilage varied. In control embryos, chondrocytes in cross-sections of Meckel’s cartilage show few and small bundles of COL2A1 expression with a discrete border of COL2A1 expression around the outside of Meckel’s cartilage (Figure 5E’, I’, O–P). In contrast, Meckel’s cartilage in treated embryos lacked the distinct COL2A1+ perichondrial border tissue present in control embryos, and Meckel’s cartilage of treated embryos contained more bundles of COL2A1 than in control embryos (Figure 5F’–H’, O–P). Drawings of COL2A1+ cranial cartilages in wholemount stage 45 axolotl embryos in Figure 5A–D demonstrate the consequences of reduced migration of SOX9+ NC cells on patterns of COL2A1 expression in the developing head (Figure 5Q–T). Focusing then on cross-sections of Meckel’s cartilage across several axial levels within the head, the morphology of Meckel’s cartilage is irregular following NSAID exposure across different regions of Meckel’s cartilage (Figure 5Q1–Q3, R1–R3, S1–S3, and T1–T3). Meckel’s cartilage typically has distinct domains across the anterior-posterior axis which uniquely contribute to development of the adult mandible (jaw bone), supporting structures like the mandibular symphysis, and bones of the middle ear, so disruption of its formation throughout the cranial region could has significant consequences for the establishment of these domains and ultimately their contributions to the developed mandibular structural components [75, 76]. These results demonstrate that there are clear craniofacial defects associated with the abnormal NC cells in NPX-exposed embryos at earlier stages and that the cells do not recover.
COX signaling inhibition alters ectodermal derivative formation
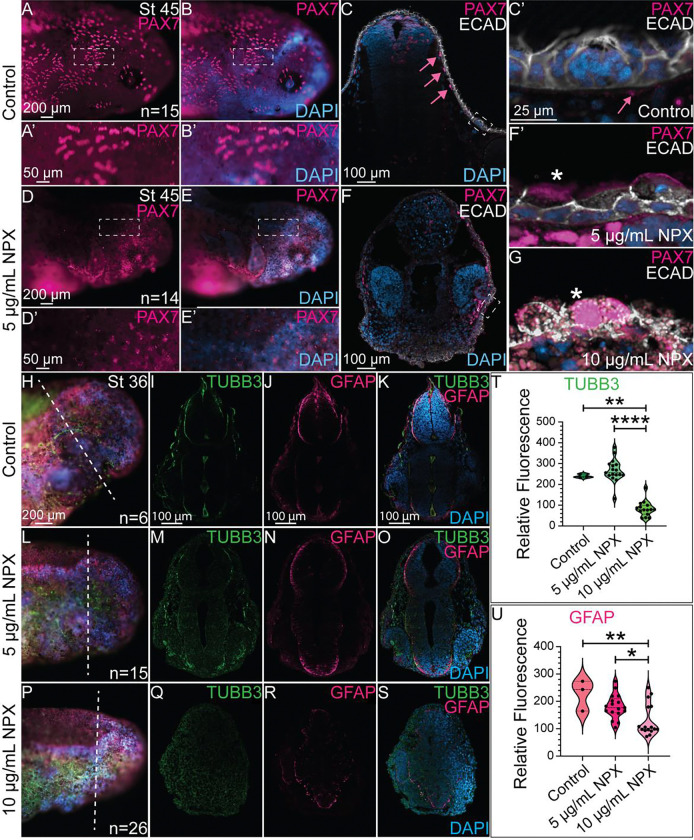
As we observed a reduction in the number of SOX9+ NC progenitors following NPX exposure and subsequent changes in COL2A1 expression and cartilage development, we hypothesized that other NC derivatives may also be affected by COX inhibition following NPX exposure. These drivatives include pigment cells, glia, and sensory neurons of the PNS. The pigment cell types present in axolotls— including melanophores, xanthophores, and iridophores— are also NC-derived and their abundance and distribution in the skin of developing axolotls has been previously described [77]. Previous work in zebrafish showed that PAX7 is an important regulator of melanophore and xanthophore differentiation, so we evaluated PAX7 expression in stage 45 embryos to identify possible effects of NPX exposure on NC-derived pigment cells [60].
In whole mount images, we identified that the normal patterning of pigment cell progenitors marked by PAX7 expression in the cranial region (Figure 6A–A’, B-B’) is abnormal in the NPX-treated embryos (Figure 6D–D’, E-E’). In transverse sections from control embryos, the PAX7+ pigment cells were restricted to the region underlying the ECAD+ epidermis (Figure 6C, C’). In NPX treated tissue sections, the localization of PAX7 was altered. Specifically, PAX7+ cells in the NPX-treated embryos colocalized in cells expressing ECAD (Figure 6F, F’, G). We evaluated expression of ECAD in transverse sections to identify changes in the formation and structure of epidermal sensory organs. These organs of the lateral line sensory system in axolotls are composed of NC-derived glial cells and placodally-derived sensory neurons, and they have well-characterized morphology in the axolotl that can be evaluated through histology [78, 79]. IHC for ECAD revealed that the complex architecture of sensory organs in the skin of control embryos (Figure 6C, C’) was completely lost following exposure to NPX (Figure 6F, F’, G). These data suggest that the development of multiple major lineages of cranial NC cells are affected by early exposure to NPX. In consideration with the data shown in previous figures, there is a strong correlation between exposure to NPX and abnormal patterning, differentiation, and morphology of all ectodermally derived tissue structures.
Figure 6. NPX alters PAX7 expression and the formation of ectodermally-derived structures..
IHC for PAX7 and ECAD in (A, B, D, and E) whole mount and (C, F) transverse sections. (A’, B’, D’, and E’) Zoom in from A, B, D, and E (dashed boxes). (C) In control embryos, PAX7 and ECAD are localized to pigment cells and superficial sensory organs. Pink arrows highlight subepidermal localization of PAX7+ pigment cells. Dashed box identifies an ECAD+ sensory organ. (C’, F’) Zoom in from sections in C and F (dashed boxes). The pink arrows in panel C’ again highlights the position of PAX7 expression relative to the sensory structures in the skin. (F’, G) Sensory organs from exposed embryos develop abnormally by stage 45. Asterisks indicate mislocalization of PAX7 expression. IHC for TUBB3 and GFAP shown in (H, L, P) wholemount and (I-K, M-O, and Q-S) transverse sections in (H-K) control embryos and (L-S) NPX exposed embryos. (H, L, P) Dashed lines show the axial level of transverse sections. (I-K, M-O, and Q-S) With exposure to increasing concentrations of NPX, there is corresponding reductions in expression of TUBB3 (neurons) and GFAP (astrocytes/glia), in the neural tube. (T, U) Quantification of TUBB3 and GFAP expression in cross-section. The sample sizes used for quantification of both TUBB3 and GFAP expression were n=1, n=2, and n=2 embryos for the control, 5 μg/mL, and 10 μg/mL NPX groups. An average of 7 transverse sections per embryo were quantified for either TUBB3 or GFAP expression.
Abnormal craniofacial development has an increased association with neurodevelopmental disorders [80]. Additionally, previous work identified that in embryos with abnormal NC cell development, there are often defects in other ectodermally-derived structures [81]. The development of NC and neural tube cells are tightly associated due to their physical proximity in the embryo, which means they share a very similar signaling landscape and receive similar mechanical cues while developing [82]. Prior work from our lab has also identified expression of COX pathway factors in the developing CNS of chicken embryos [83]. Based on this information, we hypothesized that NPX exposure may have effects on CNS development in addition to the described effects on NC cells and NC-derived tissues. To test this hypothesis, we exposed embryos to NPX as in prior experiments and subsequently performed IHC for β-III tubulin (TUBB3) and GFAP. At the stages assessed, TUBB3, which marks neuronal microtubules, is expressed in developing neurons in the PNS and CNS [84]. To determine if NPX exposure altered the TUBB3 signal in the CNS or PNS, we performed fluorescence intensity analyses on transverse cryosections and identified that the signal was significantly reduced in embryos exposed to NPX at the 10 μg/mL NPX concentration (Figure 6I, K, T compared to Figure 6Q, S, T). Similarly, the GFAP fluorescence signal in the forming CNS was also reduced in 10 μg/mL NPX-exposed embryos compared to control embryos (compare Figure 6J, K, U to Figure 6R, S, U). These results, along with those shown in previous figures, suggest that there is an overall decline in the development of both NC and CNS derivatives after NPX exposure.
IV. Discussion
Summary
Prior research demonstrated that exposure to NSAIDs during early development caused defects in NC-derived tissues, but we lacked an understanding of the mechanism through which NSAIDs alter NC development and the developmental window during which NSAID-specific effects occur. Our data shows that genes encoding the COX enzymes and other key effector molecules in the signaling pathway appear to be expressed at the right place and time to play a role in early NC development, including NC specification and migration. We found that global COX signlaing inhibition through exposure to the NSAID, NPX, has significant effects on the gross morphological development of axolotl embryos at late stages which can be attributed to abnormal development of NC cells (Figure 3, 7). Although we did not identify any significant changes in the number or migratory capacity of PAX7+ NC cells in tailbud embryos, our data suggests a role for COX signaling during early NC cell migration, specifically in SOX9+ cells fated to become chondrocytes in the developing craniofacial skeleton (Figure 5, 7). In disrupting normal SOX9 signaling, we found that COL2A-expressing cells are capable of populating the developing lower jaw, but cartilage structures with normal morphology at the cellular and tissue level do not form (Figure 5, 7). Interestingly, in the NC cell GRN, SOX9 is not only an important regulator of chondrogenic differentiation in cranial NC cells, but rather it is also important for initiating the expression of SOX10 and the developmental program for the glial and pigment cell lineages of NC cells [69, 70, 85, 86]. We hypothesize that these regulatory relationships explain our data showing that cartilage is not the only NC derivative that appears to be affected by NPX exposure, evidenced by abnormalities seen in ectodermal derivatives of the PNS and CNS. The NC-derived sensory structures of the axolotl skin also develop abnormally after exposure to NPX and show abnormal expression of ECAD and PAX7 demonstrating potential changes in cell potential and fate as a result of COX inhibition. Significantly reduced expression of TUBB3 and GFAP indicate to us that the effects of NPX on development may extend beyond NC cells to include neurodevelopmental effects as well.
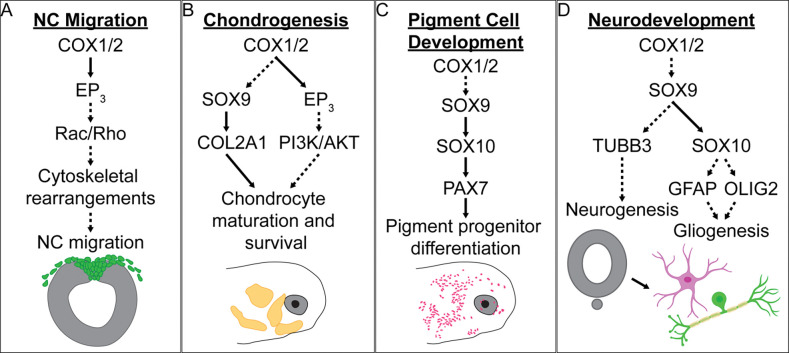
Figure 7. Graphical summary of the proposed mechanisms of developmental NSAID toxicity.
Overview of the proposed mechanisms of NSAID toxicity with respect to (A) NC migration, (B) chondrogenesis, (C) pigment cell development, and (D) neurodevelopment. Solid black arrows indicate established signaling pathways or protein interactions. Dashed arrows highlight possible interactions that could serve as the basis for future studies. Astrocyte and neuron graphical icons were created using BioRender.
COX signaling and SOX9-dependent cartilage and bone development
Our data suggests that COX signaling is necessary for the formation of definitive NC cells and for NC cell migration based on altered SOX9 expression after NPX exposure (Figure 4). These results are in alignment with previous studies which demonstrated that exposure to NSAIDs inhibits NC cell migration, leading to abnormal development of NC-derived tissues, but they suggest a homeostatic role for COX signaling during development in controlling SOX9 expression and cell migration rather than the classical role of the COX signaling pathway in adults as a mediator of inflammation. As SOX9 is expressed in many tissues throughout the developing embryo and is necessary for several key developmental processes including skeletogenesis, nephrogenesis, and gonadogenesis, it is reasonable to conclude that disruption in SOX9 signaling following NSAID exposure may be the cause of other morphological defects that we did not observe [64–67].
In NPX-exposed embryos with altered SOX9 signaling, COL2A1-expressing cells are incapable of populating the developing lower jaw, and create cartilage structures with abnormal morphology at the cellular and tissue level (Figure 5). This reaffirms that the change in SOX9 expression is sufficient to alter expression of COL2A1 in early development of the cranial cartilages [69, 70]. In addition to regulating COL2A1 expression, SOX9 has previously been linked to several steps in cartilage development including mesenchymal condensation, chondrocyte differentiation and maturation [74, 87]. Prior studies identified that SOX9 is essential for maturation and survival of chondrocytes through its interactions with the PI3K/Akt pathway— a cononical regulatory pathway of cell growth, survival, and proliferation [88]. Cross-talk between SOX9 signaling and the PI3K/Akt pathway has previously been studied in various contexts including cancer cell survival and proliferation, and tissue regeneration and fibrosis, which may provide useful insight into future research in the context of embryonic development [89, 90]. So far, our data recapitulates abnormal craniofacial phenotypes following NSAID exposure in early development with similarity to the mandibular hypoplasia phenotype often observed in disorders associated with mutations in the SOX9 gene such as campomelic dysplasia and Pierre Robin sequence [91–93]. These links highlight the need for further investigation into the dysregulation of SOX9 signaling in NSAID-mediated developmental toxicity, because other characteristics of these disorders include abnormal limb development, sex determination, and gonadogenesis, which are all SOX9-dependent processes [64–67].
With respect to the potential role of COX signaling in NC cell migration, there is exisiting evidence to support the hypothesis that NPX-mediated COX inhibition alters directional NC cell migration through disruption of Rac and Rho activation and signaling. During the epithelial to mesenchymal transition (EMT) which preceedes early NC migration, NC cell polarization changes as they delaminate, collectively migrate, and become mesenchymal and invasive. Prior work from our lab and others identified that during NC EMT, there are rapid changes in cell adhesion proteins and actin polymerization, which establishes an expression gradient of small G protiens like Rac1 and RhoA which are involved in essential downstream signaling pathways necessary for normal cell migration and tissue morphogenesis [94–98]. Previous research also identified that prostaglandins can induce activation of Rac1 via siganling through the GPCR, prostaglandin E2 receptor 3 (PTGER3, EP3) [99, 100]. Based on this information, inhibition of COX signaling via NSAID exposure may prevent the necessary cell adhesion and cytoskeletal arrangement changes downstream of SOX9 activation that facilitate establishment of Rac/Rho gradients within the cell, while also preventing activation of Rac/Rho through prostaglandin receptor signaling, ultimately causing reduced cell migration.
COX signaling and PAX7-mediated pigment development
As stated above, SOX9 initiates the expression of SOX10 in the NC GRN, which drives the developmental program for the pigment cell, glial and neuronal lineages of NC cells [69, 70, 85, 86]. Based on our studies, the NC-derived sensory structures and pigment cells in the axolotl skin develop abnormally after exposure to NPX and exhibit abnormal expression of ECAD and PAX7 demonstrating possible changes in cell potential and fate as a result of COX inhibition (Figure 6). PAX7 expression is essential in regulating formation of the melanophore and xanthophore lineages of pigment cells [60, 101], so altered PAX7 expression downstream of the effects on SOX9 could result in abnormal development of these pigment cell types. Specifically, abnormal pigment cell development could occur due to changes in expression of known PAX7 targets including SOX10 in early NC migration, and later, MITF [60, 101, 102]. However, although historical studies on xanthophore development in axolotl exist [103, 104], there are many gaps in our understanding of the genetic and molecular regulation of pigment cell development beyond chromatophores in axolotl morphs outside of the melanoid color variant [105]. Future research should focus on identifying the key regulators of this process and the exact role of PAX7 in the decision between melanophore and xanthophore formation from NC progenitors. Beyond its role in pigment cell development, PAX7 is also a key regulator of myogenesis during embryonic development [106–108]. The dysregulation of PAX7 expression following NSAID-mediated SOX9 reduction could therefore further explain the changes in axial development that we observed (Figure 3).
COX signaling and PNS and CNS development
There is much to be discovered about the mechanisms driving development of lateral line sensory organs in the axolotl, but based on previous studies in zebrafish, we know their development requires precise patterns of cell migration, proliferation, and specific cues to differentiate in order to develop normally [109–111]. Examples of necessary cues include interactions with developing glial cells in peripheral tissues and normal innervation by placodally- and NC-derived sensory neurons [110, 112–115]. A potential reduction of SOX10 expression due to reduced SOX9 expression following NSAID exposure would likely have effects on the development of glial and neuronal cells in the periphery, which we observed as reduced peripheral TUBB3 and GFAP expression in embryos exposed to NPX (Figure 6). This putative perturbation of SOX10 expression may impair interactions between developing glial cells and the migrating sensory cell precursors, resulting in abnormal lateral line sensory organ development.
We also identified concentration-dependent abnormalitiesin expression of markers of neurons and glia in the ectodermally-derived CNS. Significantly reduced expression of TUBB3 and GFAP indicate to us that the effects of NPX exposure on development may extend beyond NC cells to include the developing CNS as well (Figure 6). Previous studies have proposed a role for SOX9 in the processes of glial cell fate determination in neuroepithelial progenitor populations and cell proliferation during neocortical expansion which may explain this observed phenotype, but further analyses would be required to definitively link NPX exposure to this phenoytpe in axolotls [116, 117].
Implications for COX signaling in healing and regeneration
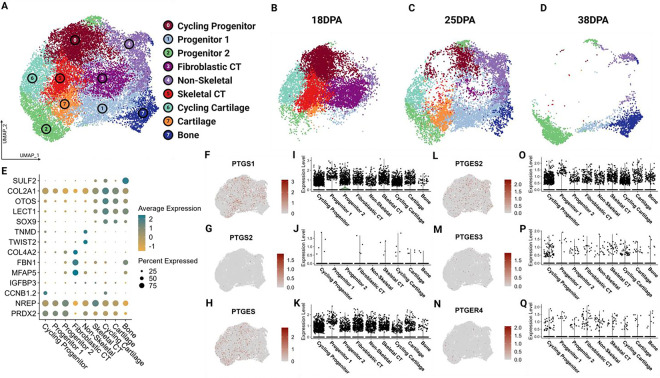
In addition to validating the conserved expression of genes encoding COX pathway factors among vertebrates (Figure 1), we reanalyzed publicly available single cell RNA-sequencing data from axolotl embryonic and regenerating limb buds to see if these genes are also expressed after injury during healing and limb regeneration [44]. In embryonic axolotl, we identified strong expression of COX enzymes in both early limb field and late limb bud tissue (stages 25–44, Supplemental Figure 5). We also able to identified expression of COX pathway factors in the regenerating axolotl limb blastema (Figure 8, Supplemental Figure 8). We further characterized expression of COX pathway elements across cell types by stage and found higher levels of PTGS1, PTGS2 and PTGES in later stages, particularly in cycling progenitors and cartilage.
Figure 8. The COX pathway is active in the regenerating axolotl limb.
Analysis was performed of publicly available scRNA-seq data for 18dpa, 25dpa, and 38dpa regenerating axolotl limb. (A) UMAP demonstrating clustering results of regenerating axolotl limb tissue. (B-D) UMAP split by stage. (E) Dot plot showing expression of major cell type markers. (F-H, L-N) Feature UMAPs demonstrating gene expression of select enzymes. (I-K, O-Q) Violin plots demonstrating expression of select enzymes across clusters.
Prior research in the field has identified an essential role for canonical inflammatory signaling pathways during the early phases of tissue regeneration, and a negative relationship between sustained inflammatory signaling and regenerative capacity/success [118–122]. Our data shows that there is significant expression of COX pathway factors such as PTGS1, PTGS2 and PTGES in local progenitor cell populations and cartilage tissue in the regenerating limb. These results are consistent with previous studies which have identified significant upregulation of COX-2 (PTGS2) expression in various contexts of tissue regeneration and healing [123–125]. Activity of the COX pathway in cartilage tissue in the context of regeneration could suggest that this pathway plays an important role in regulating healing and regeneration of cartilage tissue from progenitors in a similar manner to the regulatory role of COX signaling in SOX9-dependent embryonic development proposed here, rather than its canonical role as a mediator of inflammation. The direct mechanistic link between COX signaling and SOX9 expression, and the potential role of this mechanism in both developmental and tissue regeneration contexts, should be investigated further as this may yield answers that could enhance our understanding of cartilage development and expand our knowledge regarding cartilage tissue healing and regeneration following injury. Any insight gained into the mechanisms downstream of COX signaling that regulate normal cartilage and bone development, healing, and regeneration could help inform advancements in stem cell therapeutics and tissue engineering for use in contexts of musculoskeletal injuries. The time-dependent relationship between inflammation and tissue regeneration success also warrants further investigation and may help explain the mixed results reported in previous studies about the effects of anti-inflammatory medications on tissue healing and regeneration [126–129].
Broader implications of study
Despite the established link between NSAID use and developmental abnormalities, it is reported that one in four women take NSAIDs during pregnancy with 5% of these women using over-the-counter NPX for pain relief [130]. Additionally, NSAID use is not explicitly contraindicated in pregnant women until after week twenty of pregnancy, once significant NC development is already complete [131]. Although pain and inflammation are common complications of pregnancy, we still have a poor understanding of the potential risks of NSAID treatment during prenancy, and as such, it is imperative to understand the effects of NSAID exposure so that safe and effective treatment plans using these therapeutics can be devised for use in medicine. Here, we have established a direct link between inhibition of COX signaling through NSAID exposure and abnormal NC cell development which will serve as the premise for future mechanistic research into the developmental necessity of COX signaling and potential teratogenicity of NSAIDs, which will ultimately inform decision-making regarding pain management during pregnancy. Overall, further research into the developmental toxicity of NSAIDs will allow clinicians to perform more accurate risk assessment when recommending NSAID use to pregnant patients.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the following funding sources: NSF CAREER award 2143217 and NIH R03DE032047-01 to CDR. Funding for EJM was provided by the UC Davis eMCDB T32 (GM-153586) and Research Corporation for Science Advancement (Scialog ABI), and funding for RR was provided by the UC Davis Vision Sciene T32 (NEI-EY015387). We would like to thank the members of the Rogers Lab at UC Davis and the UC Davis community for their discussions and input on this project.
References
- 1.Garavito R.M. and DeWitt D.L., The cyclooxygenase isoforms: structural insights into the conversion of arachidonic acid to prostaglandins. Biochim Biophys Acta, 1999. 1441(2–3): p. 278–87. [DOI] [PubMed] [Google Scholar]
- 2.Ricciotti E. and FitzGerald G.A., Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol, 2011. 31(5): p. 986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez D.E. and Ballaz S.J., The Role of Brain Cyclooxygenase-2 (Cox-2) Beyond Neuroinflammation: Neuronal Homeostasis in Memory and Anxiety. Mol Neurobiol, 2020. 57(12): p. 5167–5176. [DOI] [PubMed] [Google Scholar]
- 4.Sellers R.S., Radi Z.A., and Khan N.K., Pathophysiology of cyclooxygenases in cardiovascular homeostasis. Vet Pathol, 2010. 47(4): p. 601–13. [DOI] [PubMed] [Google Scholar]
- 5.Vegiopoulos A., et al. , Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science, 2010. 328(5982): p. 1158–61. [DOI] [PubMed] [Google Scholar]
- 6.Goldyne M.E., Cyclooxygenase isoforms in human skin. Prostaglandins Other Lipid Mediat, 2000. 63(1–2): p. 15–23. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., et al. , Prostaglandin E2 mediates sensory nerve regulation of bone homeostasis. Nat Commun, 2019. 10(1): p. 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coon D., et al. , The role of cyclooxygenase-2 (COX-2) in inflammatory bone resorption. J Endod, 2007. 33(4): p. 432–6. [DOI] [PubMed] [Google Scholar]
- 9.Hoozemans J.J., et al. , Cyclooxygenase-1 and -2 in the different stages of Alzheimer's disease pathology. Curr Pharm Des, 2008. 14(14): p. 1419–27. [DOI] [PubMed] [Google Scholar]
- 10.Sales K.J. and Jabbour H.N., Cyclooxygenase enzymes and prostaglandins in pathology of the endometrium. Reproduction, 2003. 126(5): p. 559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park G.Y. and Christman J.W., Involvement of cyclooxygenase-2 and prostaglandins in the molecular pathogenesis of inflammatory lung diseases. Am J Physiol Lung Cell Mol Physiol, 2006. 290(5): p. L797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Bergh A., and Damber J.E., Cyclooxygenase-2 expression correlates with local chronic inflammation and tumor neovascularization in human prostate cancer. Clin Cancer Res, 2005. 11(9): p. 3250–6. [DOI] [PubMed] [Google Scholar]
- 13.Jackson L.M., et al. , Cyclooxygenase (COX) 1 and 2 in normal, inflamed, and ulcerated human gastric mucosa. Gut, 2000. 47(6): p. 762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radi Z.A. and Khan N.K., Effects of cyclooxygenase inhibition on bone, tendon, and ligament healing. Inflamm Res, 2005. 54(9): p. 358–66. [DOI] [PubMed] [Google Scholar]
- 15.Futagami A., et al. , Wound healing involves induction of cyclooxygenase-2 expression in rat skin. Lab Invest, 2002. 82(11): p. 1503–13. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi K. and Amagase K., Roles of Cyclooxygenase, Prostaglandin E2 and EP Receptors in Mucosal Protection and Ulcer Healing in the Gastrointestinal Tract. Curr Pharm Des, 2018. 24(18): p. 2002–2011. [DOI] [PubMed] [Google Scholar]
- 17.Jugdutt B.I., Cyclooxygenase inhibition and adverse remodeling during healing after myocardial infarction. Circulation, 2007. 115(3): p. 288–91. [DOI] [PubMed] [Google Scholar]
- 18.Amico C., et al. , Differential expression of cyclooxygenase-1 and cyclooxygenase-2 in the cornea during wound healing. Tissue Cell, 2004. 36(1): p. 1–12. [DOI] [PubMed] [Google Scholar]
- 19.Nakhai-Pour H.R. and Berard A., Major malformations after first trimester exposure to aspirin and NSAIDs. Expert Rev Clin Pharmacol, 2008. 1(5): p. 605–16. [DOI] [PubMed] [Google Scholar]
- 20.Hultzsch S., et al. , First trimester naproxen exposure and outcome of pregnancy - A German case series. Reprod Toxicol, 2021. 103: p. 51–57. [DOI] [PubMed] [Google Scholar]
- 21.Cha Y.I., et al. , Cyclooxygenase-1 signaling is required for vascular tube formation during development. Dev Biol, 2005. 282(1): p. 274–83. [DOI] [PubMed] [Google Scholar]
- 22.Cha Y.I., Solnica-Krezel L., and DuBois R.N., Fishing for prostanoids: deciphering the developmental functions of cyclooxygenase-derived prostaglandins. Dev Biol, 2006. 289(2): p. 263–72. [DOI] [PubMed] [Google Scholar]
- 23.Schill E.M., et al. , Ibuprofen slows migration and inhibits bowel colonization by enteric nervous system precursors in zebrafish, chick and mouse. Dev Biol, 2016. 409(2): p. 473–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parmar B., et al. , Inhibition of Cyclooxygenase-2 Alters Craniofacial Patterning during Early Embryonic Development of Chick. J Dev Biol, 2021. 9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., et al. , The double-edged sword role of TGF-beta signaling pathway between intrauterine inflammation and cranial neural crest development. FASEB J, 2022. 36(1): p. e22113. [DOI] [PubMed] [Google Scholar]
- 26.Achilleos A. and Trainor P.A., Neural crest stem cells: discovery, properties and potential for therapy. Cell Res, 2012. 22(2): p. 288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pla P. and Monsoro-Burq A.H., The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev Biol, 2018. 444 Suppl 1: p. S36–S46. [DOI] [PubMed] [Google Scholar]
- 28.Pilon N., Treatment and Prevention of Neurocristopathies. Trends Mol Med, 2021. 27(5): p. 451–468. [DOI] [PubMed] [Google Scholar]
- 29.Szabo A. and Mayor R., Mechanisms of Neural Crest Migration. Annu Rev Genet, 2018. 52: p. 43–63. [DOI] [PubMed] [Google Scholar]
- 30.Bhagirath A.Y., et al. , Role of Maternal Infections and Inflammatory Responses on Craniofacial Development. Front Oral Health, 2021. 2: p. 735634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamson C.J., Morrison-Welch N., and Rogers C.D., The amazing and anomalous axolotls as scientific models. Dev Dyn, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siu S.S., Yeung J.H., and Lau T.K., An in-vivo study on placental transfer of naproxen in early human pregnancy. Hum Reprod, 2002. 17(4): p. 1056–9. [DOI] [PubMed] [Google Scholar]
- 33.Monroy B.Y., et al. , Expression atlas of avian neural crest proteins: Neurulation to migration. Dev Biol, 2022. 483: p. 39–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobhani F., et al. , Morphometric analysis of the inter-mastoid triangle for sex determination: Application of statistical shape analysis. Imaging Sci Dent, 2021. 51(2): p. 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafemeister C. and Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol, 2019. 20(1): p. 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickham H., ggplot2 : Elegant Graphics for Data Analysis, in Use R!,. 2016, Springer International Publishing : Imprint: Springer,: Cham. p. 1 online resource (XVI, 260 pages 232 illustrations, 140 illustrations in color. [Google Scholar]
- 37.Raj B., et al. , Emergence of Neuronal Diversity during Vertebrate Brain Development. Neuron, 2020. 108(6): p. 1058–1074 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farnsworth D.R., Saunders L.M., and Miller A.C., A single-cell transcriptome atlas for zebrafish development. Dev Biol, 2020. 459(2): p. 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner D.E., et al. , Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science, 2018. 360(6392): p. 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farrell J.A., et al. , Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science, 2018. 360(6392). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrova K., et al. , A new atlas to study embryonic cell types in Xenopus. Dev Biol, 2024. 511: p. 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pijuan-Sala B., et al. , A single-cell molecular map of mouse gastrulation and early organogenesis. Nature, 2019. 566(7745): p. 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai J., et al. , Primate gastrulation and early organogenesis at single-cell resolution. Nature, 2022. 612(7941): p. 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerber T., et al. , Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science, 2018. 362(6413). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha Y.I., et al. , Cyclooxygenase-1-derived PGE2 promotes cell motility via the G-protein-coupled EP4 receptor during vertebrate gastrulation. Genes Dev, 2006. 20(1): p. 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishikawa T.O., et al. , The zebrafish genome contains two inducible, functional cyclooxygenase-2 genes. Biochem Biophys Res Commun, 2007. 352(1): p. 181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosser T., et al. , Developmental expression of functional cyclooxygenases in zebrafish. Proc Natl Acad Sci U S A, 2002. 99(12): p. 8418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voss S.R., Epperlein H.H., and Tanaka E.M., Ambystoma mexicanum, the axolotl: a versatile amphibian model for regeneration, development, and evolution studies. Cold Spring Harb Protoc, 2009. 2009(8): p. pdb emo128. [DOI] [PubMed] [Google Scholar]
- 49.Moury J.D. and Jacobson A.G., The origins of neural crest cells in the axolotl. Dev Biol, 1990. 141(2): p. 243–53. [DOI] [PubMed] [Google Scholar]
- 50.Moury J.D. and Jacobson A.G., Neural fold formation at newly created boundaries between neural plate and epidermis in the axolotl. Dev Biol, 1989. 133(1): p. 44–57. [DOI] [PubMed] [Google Scholar]
- 51.Graveson A.C. and Armstrong J.B., Differentiation of cartilage from cranial neural crest in the axolotl (Ambystoma mexicanum). Differentiation, 1987. 35(1): p. 16–20. [DOI] [PubMed] [Google Scholar]
- 52.Cerny R., et al. , Combined intrinsic and extrinsic influences pattern cranial neural crest migration and pharyngeal arch morphogenesis in axolotl. Dev Biol, 2004. 266(2): p. 252–69. [DOI] [PubMed] [Google Scholar]
- 53.Epperlein H., et al. , Analysis of cranial neural crest migratory pathways in axolotl using cell markers and transplantation. Development, 2000. 127(12): p. 2751–61. [DOI] [PubMed] [Google Scholar]
- 54.Olsson L., et al. , Distribution of keratan sulphate and chondroitin sulphate in wild type and white mutant axolotl embryos during neural crest cell migration. Pigment Cell Res, 1996. 9(1): p. 5–17. [DOI] [PubMed] [Google Scholar]
- 55.Epperlein H.H., et al. , Immunohistochemical demonstration of hyaluronan and its possible involvement in axolotl neural crest cell migration. J Struct Biol, 2000. 132(1): p. 19–32. [DOI] [PubMed] [Google Scholar]
- 56.Lofberg J., Perris R., and Epperlein H.H., Timing in the regulation of neural crest cell migration: retarded "maturation" of regional extracellular matrix inhibits pigment cell migration in embryos of the white axolotl mutant. Dev Biol, 1989. 131(1): p. 168–81. [DOI] [PubMed] [Google Scholar]
- 57.Olsson L., Svensson K., and Perris R., Effects of extracellular matrix molecules on subepidermal neural crest cell migration in wild type and white mutant (dd) axolotl embryos. Pigment Cell Res, 1996. 9(1): p. 18–27. [DOI] [PubMed] [Google Scholar]
- 58.Basch M.L., Bronner-Fraser M., and Garcia-Castro M.I., Specification of the neural crest occurs during gastrulation and requires Pax7. Nature, 2006. 441(7090): p. 218–22. [DOI] [PubMed] [Google Scholar]
- 59.Monsoro-Burq A.H., PAX transcription factors in neural crest development. Semin Cell Dev Biol, 2015. 44: p. 87–96. [DOI] [PubMed] [Google Scholar]
- 60.Nord H., et al. , Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol Biol Cell, 2016. 27(11): p. 1853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vallstedt A. and Kullander K., Dorsally derived spinal interneurons in locomotor circuits. Ann N Y Acad Sci, 2013. 1279: p. 32–42. [DOI] [PubMed] [Google Scholar]
- 62.Gard C., et al. , Pax3- and Pax7-mediated Dbx1 regulation orchestrates the patterning of intermediate spinal interneurons. Dev Biol, 2017. 432(1): p. 24–33. [DOI] [PubMed] [Google Scholar]
- 63.Bandin S., et al. , Immunohistochemical analysis of Pax6 and Pax7 expression in the CNS of adult Xenopus laevis. J Chem Neuroanat, 2014. 57–58: p. 24–41. [DOI] [PubMed] [Google Scholar]
- 64.Cheung M. and Briscoe J., Neural crest development is regulated by the transcription factor Sox9. Development, 2003. 130(23): p. 5681–93. [DOI] [PubMed] [Google Scholar]
- 65.Mori-Akiyama Y., et al. , Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci U S A, 2003. 100(16): p. 9360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jo A., et al. , The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis, 2014. 1(2): p. 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrionuevo F. and Scherer G., SOX E genes: SOX9 and SOX8 in mammalian testis development. Int J Biochem Cell Biol, 2010. 42(3): p. 433–6. [DOI] [PubMed] [Google Scholar]
- 68.Haldin C.E. and LaBonne C., SoxE factors as multifunctional neural crest regulatory factors. Int J Biochem Cell Biol, 2010. 42(3): p. 441–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Q., et al. , Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn, 1997. 209(4): p. 377–86. [DOI] [PubMed] [Google Scholar]
- 70.Lefebvre V., et al. , SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol, 1997. 17(4): p. 2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y., et al. , Glial fibrillary acidic protein-expressing neural progenitors give rise to immature neurons via early intermediate progenitors expressing both glial fibrillary acidic protein and neuronal markers in the adult hippocampus. Neuroscience, 2010. 166(1): p. 241–51. [DOI] [PubMed] [Google Scholar]
- 72.Casper K.B. and McCarthy K.D., GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol Cell Neurosci, 2006. 31(4): p. 676–84. [DOI] [PubMed] [Google Scholar]
- 73.Yoon Y.H., et al. , Evaluation of the toxic effects of celecoxib on Xenopus embryo development. Biochem Biophys Res Commun, 2018. 501(2): p. 329–335. [DOI] [PubMed] [Google Scholar]
- 74.Zheng K., et al. , Chapter 1 - Basic knowledge and research methods, in Bone Cell Biomechanics, Mechanobiology and Bone Diseases, Qian A. and Hu L., Editors. 2024, Academic Press. p. 3–29. [Google Scholar]
- 75.Pitirri M.K., et al. , Meckel's Cartilage in Mandibular Development and Dysmorphogenesis. Front Genet, 2022. 13: p. 871927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Svandova E., et al. , Diverse Fate of an Enigmatic Structure: 200 Years of Meckel's Cartilage. Front Cell Dev Biol, 2020. 8: p. 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frost S.K., Epp L.G., and Robinson S.J., The pigmentary system of developing axolotls. I. A biochemical and structural analysis of chromatophores in wild-type axolotls. J Embryol Exp Morphol, 1984. 81: p. 105–25. [PubMed] [Google Scholar]
- 78.Northcutt R.G., Distribution and innervation of lateral line organs in the axolotl. J Comp Neurol, 1992. 325(1): p. 95–123. [DOI] [PubMed] [Google Scholar]
- 79.Northcutt R.G., Catania K.C., and Criley B.B., Development of lateral line organs in the axolotl. J Comp Neurol, 1994. 340(4): p. 480–514. [DOI] [PubMed] [Google Scholar]
- 80.Tillman K.K., et al. , Increased Risk for Neurodevelopmental Disorders in Children With Orofacial Clefts. J Am Acad Child Adolesc Psychiatry, 2018. 57(11): p. 876–883. [DOI] [PubMed] [Google Scholar]
- 81.Trainor P.A., Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A, 2010. 152A(12): p. 2984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crane J.F. and Trainor P.A., Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol, 2006. 22: p. 267–86. [DOI] [PubMed] [Google Scholar]
- 83.Leathers T.A., Ramarapu R., and Rogers C.D., Spatiotemporal Characterization of Cyclooxygenase Pathway Enzymes During Vertebrate Embryonic Development. bioRxiv, 2024: p. 2024.08.02.606390. [DOI] [PubMed] [Google Scholar]
- 84.Chacon J. and Rogers C.D., Early expression of Tubulin Beta-III in avian cranial neural crest cells. Gene Expr Patterns, 2019. 34: p. 119067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Betancur P., Bronner-Fraser M., and Sauka-Spengler T., Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci U S A, 2010. 107(8): p. 3570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carney T.J., et al. , A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development, 2006. 133(23): p. 4619–30. [DOI] [PubMed] [Google Scholar]
- 87.Akiyama H., et al. , The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev, 2002. 16(21): p. 2813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ikegami D., et al. , Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development, 2011. 138(8): p. 1507–19. [DOI] [PubMed] [Google Scholar]
- 89.Chen S., et al. , Sox9-expressing cells promote regeneration after radiation-induced lung injury via the PI3K/AKT pathway. Stem Cell Res Ther, 2021. 12(1): p. 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Z., et al. , Sox9 promotes renal tubular epithelial-mesenchymal transition and extracellular matrix aggregation via the PI3K/AKT signaling pathway. Mol Med Rep, 2020. 22(5): p. 4017–4030. [DOI] [PubMed] [Google Scholar]
- 91.Al-Qattan M.M. and Almohrij S.A., The Pathogenesis of Pierre Robin Sequence through a Review of SOX9 and Its Interactions. Plast Reconstr Surg Glob Open, 2022. 10(4): p. e4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hossain N., et al. , Phenotype-genotype relationships in Xenopus sox9 crispants provide insights into campomelic dysplasia and vertebrate jaw evolution. Dev Growth Differ, 2023. 65(8): p. 481–497. [DOI] [PubMed] [Google Scholar]
- 93.Csukasi F., et al. , Dominant-negative SOX9 mutations in campomelic dysplasia. Hum Mutat, 2019. 40(12): p. 2344–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sasaki N., Kurisu J., and Kengaku M., Sonic hedgehog signaling regulates actin cytoskeleton via Tiam1-Rac1 cascade during spine formation. Mol Cell Neurosci, 2010. 45(4): p. 335–44. [DOI] [PubMed] [Google Scholar]
- 95.Suzuki D., et al. , Essential mesenchymal role of small GTPase Rac1 in interdigital programmed cell death during limb development. Dev Biol, 2009. 335(2): p. 396–406. [DOI] [PubMed] [Google Scholar]
- 96.Tang C., et al. , Hedgehog signaling is controlled by Rac1 activity. Theranostics, 2022. 12(3): p. 1303–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yalovsky S., et al. , Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol, 2008. 147(4): p. 1527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mayor R. and Theveneau E., The role of the non-canonical Wnt-planar cell polarity pathway in neural crest migration. Biochem J, 2014. 457(1): p. 19–26. [DOI] [PubMed] [Google Scholar]
- 99.Katoh H., Negishi M., and Ichikawa A., Prostaglandin E receptor EP3 subtype induces neurite retraction via small GTPase Rho. J Biol Chem, 1996. 271(47): p. 29780–4. [DOI] [PubMed] [Google Scholar]
- 100.Hatae N., Sugimoto Y., and Ichikawa A., Prostaglandin receptors: advances in the study of EP3 receptor signaling. J Biochem, 2002. 131(6): p. 781–4. [DOI] [PubMed] [Google Scholar]
- 101.Miyadai M., et al. , Pax3 and Pax7 function in combination with Mitf to generate melanophores and xanthophores in medaka and zebrafish. bioRxiv, 2023: p. 2023.06.22.546052. [Google Scholar]
- 102.Miyadai M., et al. , A gene regulatory network combining Pax3/7, Sox10 and Mitf generates diverse pigment cell types in medaka and zebrafish. Development, 2023. 150(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thibaudeau G. and Holder S., Cellular plasticity among axolotl neural crest-derived pigment cell lineages. Pigment Cell Res, 1998. 11(1): p. 38–44. [DOI] [PubMed] [Google Scholar]
- 104.Thibaudeau G., Holder S., and Gerard P., Anterior/posterior influences on neural crest-derived pigment cell differentiation. Pigment Cell Res, 1998. 11(4): p. 189–97. [DOI] [PubMed] [Google Scholar]
- 105.Kabangu M., et al. , Leukocyte Tyrosine Kinase (Ltk) Is the Mendelian Determinant of the Axolotl Melanoid Color Variant. Genes (Basel), 2023. 14(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Buckingham M. and Relaix F., PAX3 and PAX7 as upstream regulators of myogenesis. Semin Cell Dev Biol, 2015. 44: p. 115–25. [DOI] [PubMed] [Google Scholar]
- 107.Musumeci G., et al. , Somitogenesis: From somite to skeletal muscle. Acta Histochem, 2015. 117(4–5): p. 313–28. [DOI] [PubMed] [Google Scholar]
- 108.Otto A., Schmidt C., and Patel K., Pax3 and Pax7 expression and regulation in the avian embryo. Anat Embryol (Berl), 2006. 211(4): p. 293–310. [DOI] [PubMed] [Google Scholar]
- 109.Thomas E.D., et al. , There and back again: development and regeneration of the zebrafish lateral line system. Wiley Interdiscip Rev Dev Biol, 2015. 4(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wada H., et al. , Innervation is required for sense organ development in the lateral line system of adult zebrafish. Proc Natl Acad Sci U S A, 2013. 110(14): p. 5659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laguerre L., et al. , Cell proliferation in the developing lateral line system of zebrafish embryos. Dev Dyn, 2005. 233(2): p. 466–72. [DOI] [PubMed] [Google Scholar]
- 112.Grant K.A., Raible D.W., and Piotrowski T., Regulation of latent sensory hair cell precursors by glia in the zebrafish lateral line. Neuron, 2005. 45(1): p. 69–80. [DOI] [PubMed] [Google Scholar]
- 113.Schuster K., Dambly-Chaudiere C., and Ghysen A., Glial cell line-derived neurotrophic factor defines the path of developing and regenerating axons in the lateral line system of zebrafish. Proc Natl Acad Sci U S A, 2010. 107(45): p. 19531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schlosser G. and Northcutt R.G., Lateral line placodes are induced during neurulation in the axolotl. Dev Biol, 2001. 234(1): p. 55–71. [DOI] [PubMed] [Google Scholar]
- 115.Northcutt R.G. and Brandle K., Development of branchiomeric and lateral line nerves in the axolotl. J Comp Neurol, 1995. 355(3): p. 427–54. [DOI] [PubMed] [Google Scholar]
- 116.Guven A., et al. , Extracellular matrix-inducing Sox9 promotes both basal progenitor proliferation and gliogenesis in developing neocortex. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vogel J.K. and Wegner M., Sox9 in the developing central nervous system: a jack of all trades? Neural Regen Res, 2021. 16(4): p. 676–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng H., et al. , Role of prostaglandin E2 in tissue repair and regeneration. Theranostics, 2021. 11(18): p. 8836–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu M., et al. , PGE2 facilitates tail regeneration via activation of Wnt signaling in Gekko japonicus. J Mol Histol, 2019. 50(6): p. 551–562. [DOI] [PubMed] [Google Scholar]
- 120.Gao X., et al. , Cyclooxygenase-2 deficiency impairs muscle-derived stem cell-mediated bone regeneration via cellular autonomous and non-autonomous mechanisms. Hum Mol Genet, 2016. 25(15): p. 3216–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ho A.T.V., et al. , Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc Natl Acad Sci U S A, 2017. 114(26): p. 6675–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.King M.W., Neff A.W., and Mescher A.L., The developing Xenopus limb as a model for studies on the balance between inflammation and regeneration. Anat Rec (Hoboken), 2012. 295(10): p. 1552–61. [DOI] [PubMed] [Google Scholar]
- 123.Pearl E.J., et al. , Identification of genes associated with regenerative success of Xenopus laevis hindlimbs. BMC Dev Biol, 2008. 8: p. 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bondesen B.A., et al. , The COX-2 pathway is essential during early stages of skeletal muscle regeneration. Am J Physiol Cell Physiol, 2004. 287(2): p. C475–83. [DOI] [PubMed] [Google Scholar]
- 125.Bondesen B.A., Mills S.T., and Pavlath G.K., The COX-2 pathway regulates growth of atrophied muscle via multiple mechanisms. Am J Physiol Cell Physiol, 2006. 290(6): p. C1651–9. [DOI] [PubMed] [Google Scholar]
- 126.Lisowska B., Kosson D., and Domaracka K., Positives and negatives of nonsteroidal anti-inflammatory drugs in bone healing: the effects of these drugs on bone repair. Drug Des Devel Ther, 2018. 12: p. 1809–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Duchesne E., Dufresne S.S., and Dumont N.A., Impact of Inflammation and Anti-inflammatory Modalities on Skeletal Muscle Healing: From Fundamental Research to the Clinic. Phys Ther, 2017. 97(8): p. 807–817. [DOI] [PubMed] [Google Scholar]
- 128.Wilgus T.A., et al. , Reduction of scar formation in full-thickness wounds with topical celecoxib treatment. Wound Repair Regen, 2003. 11(1): p. 25–34. [DOI] [PubMed] [Google Scholar]
- 129.Mescher A.L., Neff A.W., and King M.W., Changes in the inflammatory response to injury and its resolution during the loss of regenerative capacity in developing Xenopus limbs. PLoS One, 2013. 8(11): p. e80477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Werler M.M., et al. , Use of over-the-counter medications during pregnancy. Am J Obstet Gynecol, 2005. 193(3 Pt 1): p. 771–7. [DOI] [PubMed] [Google Scholar]
- 131.O'Rahilly R. and Muller F., The development of the neural crest in the human. J Anat, 2007. 211(3): p. 335–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Korsunsky I., et al. , Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods, 2019. 16(12): p. 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hao Y., et al. , Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol, 2024. 42(2): p. 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.