Abstract
Background
The targeted application of cutting-edge high-throughput molecular data technologies provides an enormous opportunity to address key health, economic and environmental issues in the tropics within the One Health framework. The Earth’s tropical regions are projected to contain > 50% of the world’s population by 2050 coupled with 80% of its biodiversity however these regions are relatively less developed economically, with agricultural productivity substantially lower than temperate zones, a large percentage of its population having limited health care options and much of its biodiversity understudied and undescribed. The generation of high-throughput molecular data and bespoke bioinformatics capability to address these unique challenges offers an enormous opportunity for people living in the tropics.
Main
In this review we discuss in depth solutions to challenges to populations living in tropical zones across three critical One Health areas: human health, biodiversity and food production. This review will examine how some of the challenges in the tropics can be addressed through the targeted application of advanced omics and bioinformatics and will discuss how local populations can embrace these technologies through strategic outreach and education ensuring the benefits of the One Health approach is fully realised through local engagement.
Conclusion
Within the context of the One Health framework, we will demonstrate how genomic technologies can be utilised to improve the overall quality of life for half the world’s population.
Keywords: Genomics, Bioinformatics, One health, Biodiversity, Human health, Food production
Background
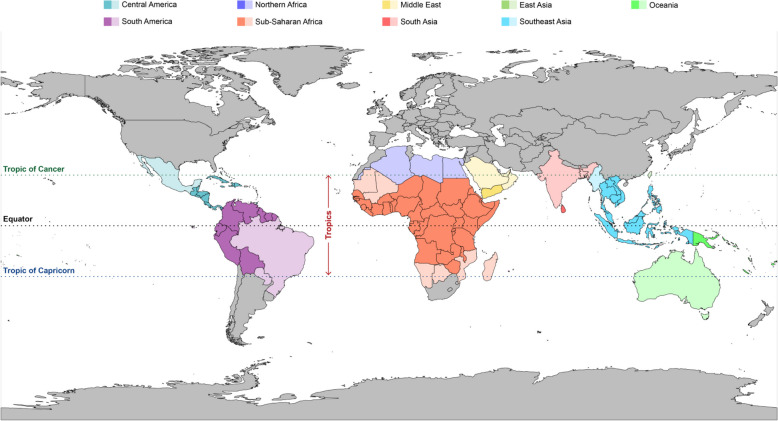
The tropics occupy all land mass between the Tropic of Cancer and the Tropic of Capricorn and contain over 80% of earth’s biodiversity [1] (Fig. 1). About 40% of all people reside in the tropics with current trends predicting this number to reach > 50% by 2050 [1]. There are many challenges unique to the tropics, and genomic technologies and their associated bioinformatic methodologies can play a critical role in addressing these within the One Health framework.
Fig. 1.
Tropical regions of the world. Within each region , countries that lie entirely within the tropics are coloured a darker shade while those lying partially within the tropics are coloured a lighter shade. A minimum of 15% of a country’s landmass lying within the tropical zone was set as a requirement for it to be considered tropical. The tropics are estimated to contain 43% of the world’s population , 80% of the worlds biodiversity and 85% of the poorest people [1].
Historic inequitable distribution of resources means residents of the tropics are more likely to live in extreme poverty or have limited access to healthcare resulting in an average life expectancy of just 60 years across the region. There are also many diseases either unique to or significantly over-represented in tropical regions. For example, over 97% of ~ 400,000 malaria cases and ~ 95% of the annual ~ 1.4 million tuberculosis deaths occurred in the tropics [2] as well as the vast majority of the one billion + cases of neglected tropical diseases (NTDs), a family which includes dengue, leishmania and schistosomiasis amongst others. While NTDs are not generally fatal, they represent a huge burden of disease resulting in wide-spread disability accounting for 22 million disability-adjusted life years (DALY) globally [3].
Globalization has significantly affected the impact of genomic technologies regarding biodiversity, food production and health in both positive and negative ways. Considering biodiversity, it is thought that by most measures, globalization has had a negative impact overall [4]. Through the transfer of technology to regions of the globe formerly outside the reach of major global powers, natural resource exploitation in tropical regions has increased in efficiency, often at the expense of local environments. While biodiversity has suffered as a consequence of the global reach of many modern technologies, innovations that seek to mitigate some of these damages are also being transferred across borders using the same globalised political and economic rails as those more extractive technologies. For example, using technology developed largely in Europe and North America, a network of more than 100 African scientists launched the Africa BioGenome Project (AfricaBP), a project which aims to sequence over 100,000 indigenous species in the interest of safeguarding their genetic biodiversity [5]. One of AfricaBP’s stated aims is to “Improve sharing of data and benefits” which will involve adapting and streamlining the Nagoya Protocol on Access and Benefits Sharing to accommodate the specific cultural sensitivities of the diverse peoples across the African continent. Regarding food production, globalization has had an enormous impact in many areas including expansion of food availability, altered food practices, food security and environmental impact [6]. While globalization can offer greater food security during times of scarcity for underdeveloped nations due to access to new markets (coupled with improved intensive agricultural practices and storage techniques), this also creates new vulnerabilities in terms of global price fluctuations. In some cases, local food sources are being replaced with higher yielding commercial crops like cocoa and coffee [7] which further compounds the risk however the potential to develop genetic breeding programs for under-developed orphan crops (e.g. sorghum, finger millet and cassava) represents a potential path forward for improving local food production, particularly if the program is able to incorporate local farming knowledge [8]. Lastly, human health has come to the fore with the clear disparities observed during the COVID-19 pandemic leading to the proposed Pandemic Accord. The pandemic made clear the urgent need for countries to develop robust synchronized strategies regarding genomic surveillance, pathogen detection and outbreak response. In addition to pandemic preparedness, the development of community based genetics programs to systematically address genetic diseases at the population level is critical in order to ultimately provide customized genetics services including diagnosis and counselling [9]. Regardless of outcomes of the Pandemic Accord, it is clear globalization will continue to influence the widespread uptake and usage of these technologies. It should be noted that while this review focuses collectively across all regions within the tropics, we acknowledge how historic global inequities and current globalization have been a driving force in creating unique challenges specific to individual countries and regions located in the tropics. As such, we attempt to highlight such disparities throughout.
Despite the clear opportunities molecular technologies offer in human health, biodiversity and food production, numerous challenges specific regions of the tropics are currently limiting wide-scale rollout of such applications. For example, the supporting infrastructure to extract, store and sequence samples is not always readily available which requires either local development of such infrastructure or embracing portable solutions such as those offered by Oxford Nanopore Technologies. Another issue is the lack of existing sequence information available, for both the native flora / fauna and the diverse human populations living in the tropics. With the majority of plants and animals lacking a reference genome, many projects must first construct a reference as a costly first step or may be forced to work with the genome of a close relative if available. For the people living in the tropics, an additional challenge is their underrepresentation in the rich genetic variant databases required for applications like precision medicine [10]. The ability to discern population level variation from pathogenic disease-causing variants is critical to any successful precision medicine program and without this ability the potential of such programs in the tropics cannot be fully realised. Lastly, critical to the success of any such programs is the engagement with populations to ensure local uptake of these technologies. Uptake requires harnessing local networks to demonstrate the utility of specific applications via a combination of outreach and education.
Today an increasing number of research institutes located both outside and within the tropics are studying how best to apply high-throughput molecular techniques to address its specific challenges. For example, the Institut Pasteur was founded in 1887 and today is a global network of 33 foreign institutes devoted to solving medical problems in foreign countries with a large focus on the tropics. Similarly, the London School of Health and Hygiene was founded in 1899 and is the constituent college of the University of London that specialises in tropical medicine. Both institutes run comprehensive research programs which try to incorporate new technologies to address issues in tropical regions. Encouragingly, newer institutes and initiatives are increasingly located within the tropics and operating at either a national or even continental level. For example, the Centre for Tropical Bioinformatics and Molecular Biology (CTBMB) operates within tropical Australia, whereas numerous ambitious continent-wide level initiatives include the Institute for Pathogen Genomics (Africa), the African BioGenome Project and the Latin American Genomics Consortium.
In this review we describe and categorise the use of targeted high-throughput molecular data to address challenges within the One Health framework aiming to improve outcomes for populations living in tropical regions. Examples are broadly grouped in health, agricultural and biodiversity applications with relevant domain experts within the author team performing exhaustive literature searches across their respective areas. Applications in these three areas are highly relevant to people living in the tropics with molecular data and bioinformatics already improving outcomes both locally and in the broader global context. These techniques have revolutionised research and clinical practices however they remain under-utilised in many under-developed environments. Lastly, we discuss how to engage with local populations through education, outreach and training. Leveraging genomic technologies within the One Health framework will generate solutions to improve the quality of life for almost half the world’s population.
Main text
Human health in the tropics
Tropical and infectious diseases represent a significant health burden to people living in the tropics. However, increasingly affordable and portable high-throughput molecular technologies are already being trialled to address issues in tropical and infectious disease. For example, an increasing number of clinical metagenomics programs are being trialled for pathogen identification both in hospitals and in the field. Such programs typically yield whole genome sequences which offer improvements over existing assay-based methods in that they are unbiased and can detect novel pathogens as well as identify AMR genes and be used in phylodynamic studies. Vaccine development programs are also increasingly incorporating high-throughput data to better understand the underlying genetic mechanisms which is important in the tropics with only 8 of the 20 WHO defined tropical diseases offering commercially licensed vaccines [11]. Molecular epidemiology is another growing application with advances in portable and rapid long-read sequencing in particular changing how we detect and track infectious diseases. The potential to develop near real-time diagnostics using these technologies represents a huge opportunity to development in-house capabilities in infectious disease detection and treatment.
In an ever-increasing number of applications, high-throughput sequencing data is being used to alleviate this burden, largely in the areas of pathogen identification/characterisation, molecular epidemiology, vaccine development, host-pathogen interactions and precision medicine programs (Table 1).
Table 1.
Applications of Omics Technologies to address Health in the tropics
| Category | Omics Technologies | Examples (references) |
|---|---|---|
| Pathogen identification and characterisation |
● Metagenomics (16 S and shotgun) ● Genomics ● Proteomics |
● Neuroleptospirosis discovery [12] ● Aedes aegypti virome [13] ● Klebsiella pneumoniae outbreak [14] ● Necator americanus[15] ● Scabies [16] |
| Molecular epidemiology |
● Metagenomics ● Genomics ● Transcriptomics |
● Bacterial phylodynamics [17] ● Drug resistant Klebsiella pneumoniae clonal grouping [18, 19] ● Drug resistance Aedes albopictus[20] |
| Vaccine development |
● Genomics ● Transcriptomics ● Epigenomics |
● Antibody discovery for diagnosing schistosomiasis[21] ● Host response [22] |
| Host-pathogen interaction |
● Genomics ● Transcriptomics |
● T-cell Receptor (TCR) sequencing [23] |
| Precision medicine |
● Genomics ● Exomes ● Transcriptomics |
● Detect pathogenic variants [26] ● Allele frequencies for underrepresented populations [27] |
-
i)
Pathogen identification and characterisation
The ability to detect causative pathogens driving human disease is an essential precursor to diagnosis and effective treatment. Pathogens causing disease are more common in the tropics and challenging to detect, especially in regional and remote health care settings. Current pathogen identification techniques can be time consuming and imprecise, and critically are unable to detect novel pathogens, rendering them unsuitable for emerging pathogen surveillance that is required in any outbreak scenario. These limitations mean patients are often treated prior to diagnosis with long-duration and broad-spectrum antimicrobials, which contributes to the spread of drug-resistant microbes. Metagenomic next-generation sequencing (mNGS) offers an emerging solution to this problem as it has been successfully used in a research environment to diagnose complex clinical cases where traditional diagnostic tests have failed using both short-read [12] and long-read sequencing [28]. An advantage of this approach is ability to completely reconstruct the genome with high-resolution sequence data enabling strain identification [29], virulence gene detection [19] and phylodynamic studies [17]. While unlikely to replace isolate sequencing (due to host and commensal bacteria contamination), mNGS offers an exciting opportunity to simultaneously detect viruses, bacteria, parasites and fungi in a single test in situations where isolates cannot be obtained, or where health care infrastructure is limited.
Pathogen discovery is also an important tool in predicting future outbreaks and pandemics with the majority of viruses still unknown. The number of known viruses has grown by orders of magnitude since the advent of metagenomic-based high-throughput sequencing. These discoveries are occurring across the tree of life with a recent study identifying 1445 new RNA viruses in invertebrates, including some that are sufficiently divergent to comprise new families [30]. These studies are not limited to understudied organisms however with 140,000 viruses just characterised across ~ 28,000 human microbiome samples, over half of which have never been seen before [31]. Knowledge of the wider viral landscape is critical for monitoring potential outbreak scenarios, with COVID-19 demonstrating how devastating such events can be. The problem is especially acute in the tropics where the virome of most tropical species is largely uncharacterised [13].
-
ii)
molecular epidemiology
New sequencing technologies, especially portable and rapid long-read sequencing is driving important improvements to the way molecular epidemiology is applied in the tropics. Recent outbreaks of Ebola and COVID-19 have demonstrated how near real-time sequencing of pathogens is critical to help better understand both the evolution and transmission of viruses [32]. Recent work by Faust et al. [33] describes their experience utilising long-read sequencing to perform molecular field research on schistosomiasis, trypanosomiasis and rabies for the benefit of the local communities in Uganda. With long reads we can improve diagnostics, better understand disease transmission dynamics and ideally provide feedback to endemic communities regarding clear actionable timelines. Whole-genome sequencing (WGS) is also being used to inform about antimicrobial resistance in near real-time. A recent program in the Philippines incorporated WGS within the established Antimicrobial Resistance Surveillance Program to better understand resistance mechanisms and local transmission patterns [18]. Their work linking resistance phenotypes to WGS data revealed the mixing of genetic strains and subsequent AMR mechanisms which identified the AMR vehicles responsible for driving the expansion of increasing carbapenem resistance rates within the country.
-
iii)
vaccine development
The development of effective vaccines to combat the wide range of tropical and infectious diseases is increasingly relying on sequence information to inform design strategies. With only 8 of 41 NTDs offering commercially licensed vaccines, work is needed to incorporate sequence information to enable rational systems-level vaccine design [34]. A perfect vaccine offers ongoing protection from a pathogen by eliciting both innate and adaptive immunity, however we typically have little understanding of how the host and pathogen interact at a systems level. High-throughput sequencing data offer us insight into how both pathogen and host respond to vaccination and infection [35]. These datasets provide insight into the underlying mechanisms driving immunological memory, protection offered by the vaccine and the efficiency of both antigens and adjuvants. Numerous programs are successfully employing this approach for diseases such as tuberculosis [36, 37], malaria [38] and dengue fever [39].
-
iv)
host-pathogen interactions
The interaction between host and pathogen is recognised as critical to how both populations and individuals respond to infectious disease. Genome sequencing of host and pathogen offers insight into the genomic determinants of adaptive processes driving parasite virulence and host resistance (e.g. malaria [40]). Using this approach, typically genetic variants are identified across both host and pathogen (within populations or even individuals) and the association of these variants with the observed phenotype used to reduce the genomic search space by identifying the driving genetic determinants [41]. Another important application of host-pathogen interactions is detecting proof of horizontal gene transfer, a process whereby an organism acquires genetic material from distantly related species in order to gain new functional capabilities [42]. While common in prokaryotes [43], there are examples of eukaryotes acquiring new functionality from parasites [44]. While much remains to be discovered regarding host-pathogen interactions in the tropics, detailed genomic sequence data offer unprecedented insight into this process.
-
xxii)
Precision medicine
Precision medicine programs are increasingly being rolled out worldwide using a combination of targeted-, exome- and whole-genome sequencing to identify clinically actionable variants in patients [45, 46]. While challenges exist [47], such programs have led to increasingly accurate diagnosis and management of genetic disease. Critical to successful precision medicine programs is the ability to incorporate patient genetic information to develop custom treatment options. There are several challenges that need to be addressed to roll out such programs in tropical regions, largely the need for supporting sequencing infrastructure / analysis capability and the lack of detailed population-specific variant information. The lack of infrastructure requires investments in both lab and analytical capability within health care settings, with most precision medicine programs currently using lab-intensive short-read sequencing capabilities.
Long-read sequencing shows great promise in this space as it requires less infrastructure and can be used in remote locations. Long-read sequencing has now been used to identify disease causing variants [48] and recently researchers at Stanford were able to successfully diagnose a patient in a matter of hours in a critical care setting [49]. Thus, the future of long-read sequencing for disease diagnosis is promising. Another challenge for the tropical population is the lack of ethnically matched allele frequencies within the variant repositories for screening, with this information being critical in prioritising candidate disease causing variants [50]. Without using ethnic matching, uncatalogued population-specific genetic variants may be incorrectly characterized as novel largely due to under-representation of the patient’s ethnic group within the repositories. Despite efforts to increase the representation of peoples from tropical regions, more work is needed to incorporate genetic information from all groups including Indigenous populations [27]. Sequencing efforts such as the Centre for Indigenous Genomics in Australia will begin to address this disparity.
Biodiversity
The biodiversity of the tropics is unparalleled in terms of biomass and numbers of species; however, this diversity is likely underestimated as many tropical species remain undescribed. Not only do we need to know and understand our tropical biodiversity in order to protect it for its own intrinsic value but cataloguing and ultimately preserving tropical biodiversity is critical to maintaining clean water, air and soil and to regulate the climate, recycle nutrients and provide food. Tropical forests currently absorb 15% of the anthropogenic carbon emissions and are critically important for mitigating the effects of climate change [51]. Maintaining biodiversity also represents a huge opportunity with a large ecological reservoir of natural products including antibiotics, many of which have been used as traditional medicines for millennia. Despite traditional usage however, in most cases the active product remains unknown, and more work is needed to identify the active molecules and their mode of action. Antibiotics are of particular interest to address the global challenge of antimicrobial resistance (AMR), which is projected to kill 10 million people annually by 2050 [52, 53].
The tropics are also home to the world’s most diverse marine (coral reefs) and terrestrial (rainforest) ecosystems. In addition to the relational and cultural values that are associated with biodiversity [54], reefs and rainforests help sustain both tropical and non-tropical human populations through the provision of direct ecological services such as food, freshwater and erosion mitigation, and through global-scale effects such as carbon sequestration, climate modulation and as habitat for long-ranging migratory species [55]. Biodiversity is directly linked to ecosystem health and robustness which in turn are the major determinants of an ecosystem’s capacity to support human populations [56, 57]. Current threats to tropical biodiversity include land clearing, over exploitation, climate change, ocean acidification, habitat fragmentation and invasive species [58–60], however modern sequencing technologies provide us with new tools to document, monitor, analyse and advise upon the best strategies to mitigate the worst effects of these threats (Table 2).
Table 2.
Tropical Biodiversity Techniques and applications
| Category | Omics Technologies | Examples (references) |
|---|---|---|
| Biodiversity discovery |
● Metagenomics ● Metabarcoding |
● BIOSCAN project [61] |
| Conservation Genomics | ● Genomics |
● Earth BioGenome Project [62, 63] ● Africa BioGenome Project [5] ● Plant Genomes [64] |
| Threatened species management |
● SNP genotyping ● Target capture sequencing ● qPCR |
● Sea Turtle [67] ● Dugong [68] ● Kuranda Treefrog [69] ● Armoured Mistfrog [70] |
| Invasive species management |
● RADseq ● eDNA |
● Cane Toad [71] ● Golden Mussel [72] ● Yellow Crazy Ant [73] |
| Drug discovery |
● Genome mining ● Metagenomics ● Metabolomics ● Proteomics |
● Streptomyces [74] ● Clostridium [75] ● Dipylidium caninum[76] |
-
i)
Biodiversity Discovery
With much of tropical biodiversity understudied and under described, large scale efforts to describe as many species as efficiently as possible are key. Metagenomics and DNA metabarcoding are two technologies that can find unique genomes and unique species from environmental samples. These techniques are particularly useful because traditional taxonomy is unable to detect cryptic species, which are particularly common in the tropics. The International Barcode of Life Consortium (iBOL) has renewed its efforts to describe all of Earth’s biodiversity, with the 7-year BIOSCAN project (2019–2026) [61] promising to analyse more than 10 million specimens.
-
ii)
Conservation genomics
The advent and refinement of low cost long-read sequencing technologies has emboldened researchers to adopt increasingly ambitious goals regarding the application of bioinformatic technologies to the fields of biodiversity research and conservation. The past decade has seen a proliferation of large consortia that aim to provide high-quality genome assemblies for selected cohorts of species with the goal of providing critical resources for species’ conservation and management. Most major initiatives currently underway can be categorised as either region specific (e.g. 1,000 Chilean Genomes, Darwin Tree of Life [77], European Reference Genome Atlas [78]) or taxon specific (e.g. 10,000 Plant Genomes [64], Global Invertebrate Genomics Alliance [79], Genome 10 K [80]).
At a higher level, in 2018 the Earth BioGenome Project (EBP) was proposed with the explicit aim of sequencing all eukaryotic life on earth [62, 63]. This well-resourced initiative adopted an umbrella-style organisational model through which they aim to facilitate sequencing projects undertaken by member organisations. At the time of writing, over forty consortia are listed as partner members to the EBP, however only four regional initiatives (Africa BioGenome Project [5], AusARG, Oz Mammals Genomics Initiative [81], Taiwan BioGenome Project) cover tropical areas. While most of the taxon-specific initiatives include tropical representatives, in practice tropical species remain underrepresented in the rapidly growing list of sequenced genomes. The current enthusiasm for large-scale genome sequencing projects coupled with the relative lack of sequenced tropical species represents a huge opportunity for future tropical biodiversity-focused sequencing efforts.
-
iii)
threatened species management
The value of genome sequencing to conservation efforts ranges from the assessment of the genetic health of species and the planning of breeding programs to the establishment of naturally occurring variant databases for the implementation of genetic rescue plans of fragmented populations [78, 82, 83]. Recent conservation efforts focusing on the Tasmanian Devil [84] and the kākāpō [85] highlight the value of leveraging a high quality reference genome for planning, implementing and managing conservation strategies.
For species as yet without genomic resources, the use of SNP sequencing and eDNA has been able to advance conservation measures for many tropical species. For example, SNP sequencing has been used to understand the effective population size and genetic connectivity between breeding sites for the critically-endangered Kuranda Treefrog [69] and thereby inform management priorities for this species. Environmental DNA (eDNA) is being used to investigate the true range of threatened species such as the critically-endangered Armoured Mistfrog in north-east Australia [70] and the endangered big-headed turtle Platysternon megacephalum in Hong Kong [86]. With current projections of tropical habitat fragmentation and species loss set to continue in tropical regions [87–89], genome-centric management strategies will likely garner increasing adoption over the coming century.
-
iv)
invasive species management
Invasive species also pose severe risks to tropical biodiversity. In 1991 the Golden Mussel (Limnoperna fortunei (Dunker, 1857)), native to East Asia, was introduced to the Rio Del Plata in Argentina through the expulsion of ship ballast water [90]. This invasive freshwater bivalve mollusc, which was first sequenced in 2018 [91], spread rapidly throughout Argentina, Uruguay, Paraguay and Brazil from where it now risks entering the Amazon river basin [92]. Traditional methods of monitoring of the golden mussel across such an expansive territory would be prohibitively resource intensive, however a recent effort to utilise eDNA to detect traces of Limnoperna DNA from water samples has proven to be an effective and cost-efficient means of monitoring [72].
The use of eDNA for indirect detection of terrestrial tropical species is now well-established [93] and is currently being implemented for monitoring the expansion of invasive species such as the cane toad Rhinella marina in Australia [94, 95] and the detection of Yellow Crazy Ants (Anoplolepis gracilipes) invading the tropical rainforests and streams of north-east Australia [73] through sampling freshwater streams.
-
xxii)
Drug discovery
Tropical marine and rainforest species are exceptionally rich sources of bioactive compounds. Historically, most new drug discoveries came through the targeting of natural compounds present in traditional medicinal plant species [96]. This ‘ethnobotanical’ approach to drug discovery remains a leading source of new bioactive compounds with up to 50% of all natural product-derived drugs having plant-based origins [96].
Bioprospecting for new drug candidates has long involved the targeting of plant, fungi and microbe species but starting in the 1950s, attention turned to marine invertebrates following the discovery of spongothymidine and spongouridine from the tropical marine sponge Tethya crypta. Modern derivatives of these compounds are now powerful antiviral and anticancer drugs [97, 98]. Today sponges and cnidarians account for up to 80% of all invertebrate-based drug discoveries with the remaining 20% coming from a wide range of other phyla [99]. This ‘ecological’ approach to drug discovery, in combination with the previously described ‘ethnobotanical’ approach, constitute the two main components of the ‘biorational’ drug discovery strategy [34].
Although these candidate-based approaches are undoubtedly effective, many of the steps involved in traditional drug discovery and validation pipelines are laborious and costly. While laboratory-based validation of potential drug candidates cannot be replaced, in silico modelling and data mining offer the potential of substantial efficiency gains by narrowing the focus of wet-lab assays and experiments [100, 101]. Following the isolation of a candidate compound, 3D molecular modelling, determination of target affinity, prediction of off-target effects and reverse engineering of biosynthetic pathways are all steps that today benefit from the implementation of bioinformatic pipelines to supplement lab-based experiments and assays. By coupling indigenous knowledge and domain-specific ecological knowledge with modern high-throughput biotechnological and computational approaches, the resulting efficiency gains are beginning to have a positive impact on the enormous costs associated with bringing drugs to market [102–104].
Food production
The tropical climate is ideal for agriculture (crops, livestock, aquaculture) with its high temperature all year round, high rainfall and sun exposure [97]. Unfortunately, these benefits are often counter balanced in many tropical areas due to poor soil fertility, inadequate water supply for irrigation, lack of mechanization and the high prevalence of diseases [7]. Historically, food production in the tropics has been characterised by lower yields and higher variability than in temperate regions despite favourable growing conditions [105]. There are many historical reasons for this disparity, however a major contributing factor is the underdevelopment of breeding programs for tropical cultivars and animal species relative to their temperate equivalents. Advanced genomic breeding techniques specific to tropical species would improve this as they offer the potential to achieve rapid gains in performance through targeted selective practices. The development of rapid and cost-effective genotyping techniques for tropical species would enable powerful techniques for stock improvement such as genomic selection or parentage assignment in circumstances where it would otherwise have been impossible such as mixed spawning aquaculture. Moreover, climate change will require ongoing selective breeding informed by genomic technologies to ensure that production species adapt as the world becomes hotter and less predictable. For example, within the context of meeting the UN sustainable development goals, there is a desire to develop tropical agricultural practices that preserve biodiversity while maintaining high productivity and reliability. Such sustainable farming practices will be reliant on genomic technologies in order to both identify genetic variation driving growth and to understand and monitor their respective microbiomes, which are increasingly being implicated in overall health [106]. Finally, the cultural diversity of tropical peoples and relatively low intensity of food production in these regions means that the range of target species is very large. Technologies for rapid and cost-effective development of genomic resources such as reference genomes, and genotyping arrays will therefore be especially important to the region.
Overcoming these challenges is a huge task on the road to increase food production to feed people living in the tropics. One way to boost productivity in the tropics is to adopt a range of novel molecular technologies to improve breeding of important crops, livestock and aquaculture species as well as providing tools for pathogen detection, food safety, food quality and verification (Table 3).
Table 3.
Tropical Food Production Techniques and applications
| Category | Omics Technologies | Examples (references) |
|---|---|---|
| Genome editing |
● ZFN ● TALEN ● CRISPR |
● Banana [107] ● Cattle [108] |
| Selective breeding programs |
● GWAS ● Genomics |
● Maize [109] ● Cattle [110] ● Cocoa [111] ● Nile tilapia [114] |
| Pathogen identification and characterisation |
● qPCR ● Metagenomics (16 S and shotgun) ● Genomics |
● Prawn pathogens [115] |
| Food omics (i.e. Food safety, food tracking, foodborne disease) |
● Transcriptomics ● Proteomics ● Metabolomics ● Metabarcoding |
● Coffee bean [116] ● Chlorella [117] ● Bivalve identification [118] |
-
i)
genome editing
Genome editing began in 1985 with the discovery of the Zinc Finger Nucleases (ZFNs), which were able to recognize target and remove sequences using a cutting enzyme [119]. Later, transcription activator-like effector nucleases (TALENs) were favoured over ZFN as they could bind to a single nucleotide [120]. Following this, the CRISPR/CAS9 technology was developed in 2014 [121] as a genome editing tool. CRISPR (clustered regularly interspaced short palindromic repeats) is used by the CRISPR-associated protein 9 (CAS9), an enzyme that can cleave DNA, as a guide that binds to the targeted sequence. This technology has become an essential tool for many applications in tropical agriculture such as disease resistance (banana (Musa spp) [107]), parasite resistance and heat tolerance in livestock (cattle [108]). While promising, CRISPR/CAS9 requires significant infrastructure and expertise to implement. In addition, issues with public acceptance remain a challenge [122]. Despite this, the benefits of gene editing technology could be immense for tropical agriculture by reducing loss due to disease and by increasing production rates.
-
ii)
selective breeding
Molecular data approaches such as genome-wide association studies (GWAS) and genomic selection (GS) are being used in selective breeding programs to improve important production traits. Typically, thousands of biomarkers (generally single-nucleotide polymorphisms, SNPs) are probed and markers are identified that associate with a trait that confers desirable characteristics. The use of GWAS for tropical species can further help to pinpoint specific SNPs associated with traits of economic importance. GWAS analyses are also very useful for understanding the genetic basis of complex traits that are specific to the tropical climate such as heat tolerance or tolerance of poor-quality soil. For example, Ertiro et al. [109] performed GWAS on tropical maize growing in a nitrogen deficient environment to identify a number of markers associated with genes that, if selected for, could enhance production in such nitrogen depleted areas. Similarly with livestock, Otto et al. [110] used GWAS to identify 6 SNPs associated with heat tolerance in Gir X Holstein cattle breeds in the tropics.
Traditional selection methods for genetic improvement of crops and livestock rely solely on identifying desired phenotypes (mass selection) or else using information from established pedigrees to account for theoretical known relationships (Best Linear Unbiased Prediction selection) in order to generate breeding values to drive informed breeding decisions. However, the accuracy of such selection methods can be hindered by the lack of information on relationships among selection candidates as demonstrated by Nayfa et al. [123] for the Nile tilapia (Oreochromis niloticus), where a manually recorded pedigree was often incorrect. Increasingly, GS experiments are incorporating larger numbers of SNPs to improve the accuracy of breeding values, as well as allowing selection at an earlier stage for difficult to measure traits [124]. GS is of particular interest for tropical perennial crop species with a long breeding cycle with high expected genetic gain and has been applied to a number of tropical crops including for disease resistance in cocoa [111] and for fruit quality in citrus [125] (see review [126]). In tropical aquaculture, GS also shows promise for target species with a sufficient level of domestication and for which the required genomic resources are available. Tropical aquaculture species for which GS has already been applied include the prawns Litopenaeus vannamei and Penaeus monodon, the pearl oyster Pinctada maxima [127] the barramundi Lates calcarifer [112, 113] and Nile tilapia Oreochomis notilus [114].
-
iii)
Pathogen detection
Tropical agriculture systems are exposed to a large number of pathogens that can affect the production of crops or decimate entire aquaculture brood stock (e.g. Juvenile Pearl Oyster Mortality Syndrome [128]). Timely identification of pathogens can be undertaken using genomics in combination with quantitative PCR (qPCR): for example Arbon et al. [115] detected 6 known pathogens in wild tropical shrimp P. monodon. Pathogen detection was also used to identify various strains of ticks known to affect humans and livestock in the sub-Saharan region [129]. Metagenomics is a more general technique used to identify all microorganisms in a sample and can be useful in the detection of novel pathogens that affect brood stock and crops, where the actual causative agent is not determined or poorly understood.
-
iv)
Food omics
Food safety is an active area of research that encompasses food verification and tracking as well as the detection and tracking of food-borne disease. Increasingly, consumers want to know the origin and composition of the product that they eat [130], as well as the certainty that the product is safe. Globalization has made this increasingly challenging, however the use of genomics can provide detailed information about products.
Many omics technologies are being actively employed in food safety including transcriptomics, proteomics, metabolomics and metagenomics. Transcriptomics can be very useful in tropical agriculture to elucidate response mechanisms to various conditions. For example, Liu et al. [131] used transcriptomics to discover the molecular pathways of Stylo, a tropical plant, involved with the impact of soil salinity. Understanding such complex mechanisms can be tremendously useful for tropical agriculture in addressing challenges such as poor soil quality and low yield. Proteomics is another approach that studies the collection of proteins produced and modified by an organism at any time. The set of proteins produced at a time is variable and can change as a result of specific circumstances such as stress or infection. For example, coffee beans dried using a different method exhibited a decrease in abundance of proteins linked to heat stress and sugar absorption which significantly impacted the quality of the product [116]. Metabolomics measures metabolites in a cell which informs about the biochemical activities within a cell. These measurements can help understand the reaction of organisms to stressors. For example, metabolomics was used to measure the change of lipid production of tropical Chlorella in nitrogen depleted environments [117]. Finally, metabarcoding is an approach where molecular barcoding is used to identify all the species present in a sample. Metabarcoding has many applications including the identification of the origin of a product. For example, Gense et al. [118] developed a metabarcoding technique to identify bivalve species suitable for human consumption.
Overall, the application of molecular techniques is, and will continue to be beneficial for tropical agriculture by accelerating genetic improvement through genome editing and genomic selection, by limiting losses through pathogen detection and by increasing food safety.
Local education / technology access
-
i)
Bioinformatics education and infrastructure
The previous sections have highlighted the importance of bioinformatics tool and method development focusing on their application in the unique context of the tropics. While many applications have progressed to the proof-of-principle stage, significant challenges in wide-spread delivery exist. Overcoming these challenges requires concerted, cross-disciplinary efforts that are both timely and targeted. While delivery is challenging in any context, bioinformatics literacy and digital infrastructure are poorly developed in vast areas of the tropics exacerbating the issues. The latter had been the focus of the 2021 State of the Tropics Report [1], which highlights the “digital divide” that implies disadvantages for low- and middle-income countries of the tropics.
-
ii)
education and research
There is an unmet need for more bioinformatics training in many universities of low- and middle-income countries, including numerous tropical nations [132]. Likewise, bioinformaticians are underrepresented in many tropical research institutions, which leads to backlogs in high-throughput data analysis. In other cases, large-scale genome projects are not attempted, or put on hold due to the lack of bioinformatics expertise [15]. However, an increasing number of initiatives are attempting to increase bioinformatics capacity in the tropics. One promising example is the National Institute of Health (NIH)-funded Human Heredity and Health in Africa (H3Africa) Bioinformatics Network (H3ABioNet), which was founded to conduct research into genetic and environmental factors involved in the pathology of human diseases [133, 134]. One of H3ABioNet’s goals is to provide bioinformatics training in 16 African countries. H3ABioNet developed a free-of-charge “Introduction to Bioinformatics” course taking into account the particular logistical challenges in Africa [135, 136].
An equivalent to the H3ABioNet initiative is the Asia Pacific Bioinformatics Network (APBioNET), which set out 25 years ago (founded in 1998) to foster the growth of bioinformatics in the Asia-Pacific region (www.apbionet.org) [137]. APBioNET promotes bioinformatics awareness, training, infrastructure, and research among member countries. APBioNet has members from industry, academia, research, government, and international organizations [137]. Another similar initiative is the CABANA project (www.cabana.online.), which aims to build bioinformatics capacity in Latin America. CABANA is supported by the EMBL-EBI and involves nine partners in the higher education and research sectors. CABANA is supported by the Global Challenges Research Fund (GCRF) and focuses on three challenges affecting the tropics: (i) communicable disease, (ii) sustainable food production, and (iii) protection of biodiversity.
Developing and expanding bioinformatics capacity in the tropics will require structured efforts both in educational and research institutions. Aron et al. developed a ten rule-guideline outlining how to develop bioinformatics capacity at an academic institution [132]. A framework for the development of bioinformatics core competencies at higher education or research institutions was designed by The Curriculum Task Force of the International Society of Computational Biology (ISCB) Education Committee [138].
-
iii)
infrastructure
The lack of suitable computational infrastructure in many universities of the tropics could be one of the reasons for the underrepresentation of tropical bioinformatics research labs. One increasingly popular solution for institutions lacking compute infrastructure, is to use the services of centralised data centres, high-performance computing clusters, or cloud-based services, such as the Galaxy bioinformatics infrastructure project. The Galaxy cloud-computing platform for bioinformatics analyses has nodes and support groups serving tropical countries in Africa, Australia, and Asia [139, 140] and covers an increasing number of applications [141, 142].
To address challenges with diseases of the tropics, countries are establishing large consortia such as H3Africa, a consortium that facilitates research, resources, and training to study diseases in the African continent [134]. H3Africa supports population-based studies on genomic and environmental risk factors for cardiometabolic disease risk (AWI-GEN) [143], type 2 diabetes (T2D) [144], kidney disease (H3AKDRN) [145], stroke (SIREN) [146], and more. Broader, population-based genome sequencing efforts are underway in India with the GenomeINDIA project, an initiative involving 20 partner institutions in an effort to sequence 10k Indian genomes and identify genetic variants explaining some of India’s high-burden genetic disorders (www.genomeindia.org). GenomeINDIA, along with other national genome projects, feeds into the GenomeAsia 100k (GA100K) project, an effort to counter the underrepresentation of non-European individuals in human genetic studies [147]. Similar national genome sequencing initiatives in the tropics include the Saudi Human Genome Program (SHGP) [148], Genomics Thailand (genomicsthailand.com), ChileGenomico (chilegenomico.med.uchile.cl), and Australian Genomics (australiangenomics.org.au). Most of these projects aim to develop infrastructure and personnel for data management and data analysis [15].
Globalization processes have played a significant role in the development and distribution of sequencing technologies. With the advent of high throughput 2nd generation sequencing-by-synthesis technologies like Illumina it was apparent that purchasing and running such a technology locally represented huge challenges for many tropical countries. With initial price tags near one million USD, expensive proprietary reagents and kits coupled with the need for laboratories capable of DNA/RNA extraction and reliable cold storage many tropical countries lacked the infrastructure and resources to work with these technologies. In lieu of local infrastructure most samples to be sequenced were subsequently shipped overseas at considerable expense; to illustrate only 11 of the 385 African animals sequenced to date were performed locally [5]. While the initial market was dominated by Illumina, globalization contributed to increased competition that has reduced costs with newer companies like BGI Genomics recently setting up the first clinical sequencing facility in Africa. While encouraging, challenges persist in rolling out 2nd generation technologies in under-developed locations with limited supply chains, few regional sales representatives plus insufficient on-site staff with dedicated expertise representing significant hurdles [149]. The emergence of single molecule sequencing technologies such as Oxford Nanopore (3rd generation) represents an exciting opportunity for countries in the tropics with their reduced costs, increased portability, real time processing and reduced technical and computation requirements [150]. Further, with no costly machine purchase required up front and simplified sample prep such technologies are already making significant inroads in under-developed locations with applications in disease monitoring, animal/human population genetics and agriculture. While encouraging, challenges with Nanopore persist including power outages, data transfer, reagent cost, transport and storage as well as insufficient local training [150]. Possible solutions to facilitate wider uptake include further simplification of the DNA/RNA extraction and customized training aimed at under-developed environments [151]. Additionally, while Nanopore currently dominates the portable sequencing space, increased competition will likely improve the options and feasibility over time to enable widespread uptake in the tropics.
Discussion
This review summarises the current impact high-throughput molecular techniques are having for people living in tropical regions across health, biodiversity and food production. Encouragingly, the number of meaningful examples achieving impact is growing in all these areas however challenges remain regarding sequencing cost, required infrastructure and data sharing agreements. Costs will continue to decrease with ongoing improvements in per base sequencing costs while infrastructure requirements are decreasing with the increased availability and maturity of relatively easy to use portable sequencing systems. Data sharing remains a challenge however significant movement is occurring in this space around developing a Pandemic Accord. The recent debates around developing such an accord continue to highlight concerns around sharing of genomic material and information between countries, especially by the High-income countries (often referred to as the Global North) with low and middle income countries (Often referred to as the Global South). Such an accord is a global legally binding instruments to protect and promote people’s health, created by Member States to foster and secure collaboration in areas that impact on the health and well-being of people at local, national and global levels. These international instruments represent a commitment by countries of the world to address the health needs of their citizens to advance their health status and strengthen the socio-economic status of their communities at large.
The final instrument, treaty, accord, agreement or convention (the final name and contents are still under negotiation by the Member states of the World Health Assembly) will aim to establish an equitable and comprehensive system of access and benefit-sharing of effective products to be used in pandemic responses – such as diagnostic tools, medicines and vaccines – often products developed and/or produced through genomic technologies. However the sharing of “pathogens with pandemic potential” and “the genomic sequence of such pathogens” remains one of the contentious issues that has forced the delay of finalisation of the Treaty at the 77th World Health Assembly, in May 2024. Global South countries remain concerned that when they do share genomic data (e.g. South Africa and Omicron [152]), they were immediately faced with sanctions against their country (trade and travel) as well as delays in receiving adequate vaccines developed derived from this information to manage the pandemic in their own country. The Global North is often concerned about recuperating the costs of research and development for these products as well as ensuring stockpiles of their own citizens. Although conventions such as the United Nations (UN) 1992 Convention on Biological Diversity (CBD) which “reaffirmed that States have sovereign rights over their genetic resources, and the authority to determine the rules about accessing these resources” and the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization (2010) which extended further the need for global multilateral benefit-sharing mechanism for transboundary situations such as pandemics and the WHO’s Pandemic Influenza Preparedness Framework provides mechanisms to ensure sovereignty but also equitable access and benefit sharing, however these mechanism have not been fully tested and the present debates highlight that they remain contentious [153].
Concerns have also been raised about the reality of the well-intentioned inclusion of the One Health Approach into the “Pandemic Treaty” related discussions as a means to equitable access integrated human, animal environmental and other ecosystem system such as surveillance data (including genomic data) [154]. Firstly it needs political commitment to implement coordinated transdisciplinary approaches into practice as well as finances, capacity building, and improved exchanges of data and information, which is not comprehensively present in all nation states, and has not been proven scalable at global levels as of yet. Secondly based on a review of the Quadripartite surveillance capacities especially in the wildlife and environment sectors concern has been raised about whether “an expanded Global Early Warning System (GLEWS+) could include all four organisations” [155]. However the authors express optimism that this can occur noting that “Strong and clear jurisdictional legislative and regulatory frameworks are needed” although they make take time to be developed but should not impede implementation of some aspects whilst waiting. Further countries of the Global South, like China has expressed concerns about the need for “concrete mechanisms to build developing countries’ capacities such as technology transfer and financing” to ensure the equity of access and comprehensive implementation of the underlying objective of pandemic preparedness [156]. The Global South in this and similar debates has often felt sidelined by Global North. It reflects, according to Huang et al., “a common wariness among these developing countries about the treaty’s potential for political exploitation and stigmatization. In effect, these shared perspectives are rooted in historical challenges, socio-economic disparities, and a desire for more equitable participation in global health governance” (page 5) [156]. Although this has happened in predominantly the human health realm, such issues may similarly arise in the animal or plant health realm for transboundary biosecurity risks.
Conclusion
Advances in technologies such as DNA sequencing, mass spectrometry and other high-throughput analytical methods continue to increase data generation rates for all omics fields including genomics, transcriptomics, proteomics, metagenomics, and metabolomics. These improvements output higher quality data at lower per-unit cost and offer an unprecedented opportunity to generate large volumes of high-quality molecular data. While massively parallel short-read technologies largely drove the early advances, new developments in technologies including long-read and single-cell sequencing represent a new opportunity to generate increasingly high-resolution data able to answer a whole new class of questions. The analysis of such complex, inter-connected data and their practical application to real-world problems however requires both bioinformatics expertise and domain-specific knowledge of the complex systems. By embracing the One Health approach, we can identify targeted applications to address some of the biggest challenges in the tropics across health, food production and biodiversity and improve the lives of populations living in tropical regions.
Authors’ contributions
I.C, P.C, C.M, U.S, M.H and M.F conceived of the manuscript. All authors wrote and reviewed the manuscript.
Funding
MF is funded by National Health and Medical Research Council Fellowship APP5121190.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Andrew Calcino, Ira Cooke, Pete Cowman, Megan Higgie, Cecile Massault, Ulf Schmitz and Matt A Field contributed equally to this work.
References
- 1.Ann Penny ST, McKenzie M. Daniela Tello Toral and & Hunt., E. State of the Tropics. Australia: James Cook University, Townsville; 2020. [Google Scholar]
- 2.Zammarchi L, Bartalesi F, Bartoloni A. Tuberculosis in tropical areas and immigrants. Mediterr J Hematol Infect Dis. 2014;6:e2014043. 10.4084/MJHID.2014.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzpatrick C, Nwankwo U, Lenk E, de Vlas SJ, Bundy DAP et al. in Major Infectious Diseases (eds rd (2017).
- 4.Ehrenfeld D. Globalisation: effects on biodiversity, environment and society. Conservation and Soc. 2003;1:99–111. [Google Scholar]
- 5.Ebenezer TE, et al. Africa: sequence 100,000 species to safeguard biodiversity. Nature. 2022;603:388–92. 10.1038/d41586-022-00712-4. [DOI] [PubMed] [Google Scholar]
- 6.Reardon, T. & Timmer, C. P. in Handbook of Agricultural Economics Vol. 3 (eds R. Evenson & P. Pingali) 2807–2855 (Elsevier, 2007).
- 7.Schroth G, Laderach P, Martinez-Valle AI, Bunn C. From site-level to regional adaptation planning for tropical commodities: cocoa in West Africa. Mitig Adapt Strateg Glob Chang. 2017;22:903–27. 10.1007/s11027-016-9707-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gesesse CA, et al. Genomics-driven breeding for local adaptation of durum wheat is enhanced by farmers’ traditional knowledge. Proc Natl Acad Sci USA. 2023;120: e2205774119. 10.1073/pnas.2205774119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kingsmore SF, et al. Next-generation community genetics for low- and middle-income countries. Genome Med. 2012;4:25. 10.1186/gm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley AR, Callier S, Rotimi CN. Diversity and inclusion in genomic research: why the uneven progress? J Community Genet. 2017;8:255–66. 10.1007/s12687-017-0316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engels D, Zhou XN. Neglected tropical diseases: an effective global response to local poverty-related disease priorities. Infect Dis Poverty. 2020;9:10. 10.1186/s40249-020-0630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson MR, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–17. 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakrzewski M, et al. Mapping the virome in wild-caught Aedes aegypti from Cairns and Bangkok. Sci Rep. 2018;8:4690. 10.1038/s41598-018-22945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148ra116. 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovanda A, Zimani AN, Peterlin B. How to design a national genomic project—a systematic review of active projects. Hum Genomics. 2021;15:20. 10.1186/s40246-021-00315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chng L, et al. Molecular diagnosis of scabies using a novel probe-based polymerase chain reaction assay targeting high-copy number repetitive sequences in the Sarcoptes scabiei genome. PLoS Negl Trop Dis. 2021;15: e0009149. 10.1371/journal.pntd.0009149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingle DJ, Howden BP, Duchene S. Development of phylodynamic methods for bacterial pathogens. Trends Microbiol. 2021;29:788–97. 10.1016/j.tim.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Argimon S, et al. Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nat Commun. 2020;11:2719. 10.1038/s41467-020-16322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bialek-Davenet S, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20:1812–20. 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt TL, et al. Spatial population genomics of a recent mosquito invasion. Mol Ecol. 2021. 10.1111/mec.15792. [DOI] [PubMed] [Google Scholar]
- 21.Pearson MS, et al. Immunomics-guided discovery of serum and urine antibodies for diagnosing urogenital schistosomiasis: a biomarker identification study. Lancet Microbe. 2021. 10.1016/s2666-5247(21)00150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luciani F, Bull RA, Lloyd AR. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol. 2012;30:443–52. 10.1016/j.tibtech.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol. 2012;13:121–8. 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- 24.Loughland JR, et al. Plasmacytoid dendritic cells appear inactive during sub-microscopic Plasmodium falciparum blood-stage infection, yet retain their ability to respond to TLR stimulation. Sci Rep. 2017;7:2596. 10.1038/s41598-017-02096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loughland JR, et al. Transcriptional profiling and immunophenotyping show sustained activation of blood monocytes in subpatent Plasmodium falciparum infection. Clin Transl Immunol. 2020;9: e1144. 10.1002/cti2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Field MA. Detecting pathogenic variants in autoimmune diseases using high-throughput sequencing. Immunol Cell Biol. 2020. 10.1111/imcb.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caron NR, et al. Indigenous genomic databases: pragmatic considerations and cultural contexts. Front Public Health. 2020;8: 111. 10.3389/fpubh.2020.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charalampous T, et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783–92. 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 29.Bertelli C, Greub G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin Microbiol Infect. 2013;19:803–13. 10.1111/1469-0691.12217. [DOI] [PubMed] [Google Scholar]
- 30.Shi M, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–43. 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 31.Camarillo-Guerrero LF, Almeida A, Rangel-Pineros G, Finn RD, Lawley TD. Massive expansion of human gut bacteriophage diversity. Cell. 2021;184:1098–109. 10.1016/j.cell.2021.01.029. e1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–32. 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faust CL, et al. Harnessing technology and portability to conduct molecular epidemiology of endemic pathogens in resource-limited settings. Trans R Soc Trop Med Hyg. 2021;115:3–5. 10.1093/trstmh/traa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adegboye O, et al. Natural-product-based solutions for Tropical Infectious diseases. Clin Microbiol Rev. 2021;34: e0034820. 10.1128/CMR.00348-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma M, Krammer F, Garcia-Sastre A, Tripathi S. Moving from empirical to rational vaccine design in the ‘Omics’ Era. Vaccines (Basel). 2019;7:7. 10.3390/vaccines7030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heijmenberg I, et al. ESX-5-targeted export of ESAT-6 in BCG combines enhanced immunogenicity & efficacy against murine tuberculosis with low virulence and reduced persistence. Vaccine. 2021. 10.1016/j.vaccine.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 37.Sathkumara HD, et al. Mucosal delivery of ESX-1-expressing BCG strains provides superior immunity against tuberculosis in murine type 2 diabetes. Proc Natl Acad Sci USA. 2020;117:20848–59. 10.1073/pnas.2003235117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Draper SJ, et al. Malaria vaccines: recent advances and new horizons. Cell Host Microbe. 2018;24:43–56. 10.1016/j.chom.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim SP, et al. Ten years of dengue drug discovery: progress and prospects. Antiviral Res. 2013;100:500–19. 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Garrido-Cardenas JA, Gonzalez-Ceron L, Manzano-Agugliaro F, Mesa-Valle C. Plasmodium genomics: an approach for learning about and ending human malaria. Parasitol Res. 2019;118:1–27. 10.1007/s00436-018-6127-9. [DOI] [PubMed] [Google Scholar]
- 41.Biedrzycka A, Popiolek M, Zalewski A. Host-parasite interactions in non-native invasive species are dependent on the levels of standing genetic variation at the immune locus. BMC Evol Biol. 2020;20:43. 10.1186/s12862-020-01610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulte RD, Makus C, Schulenburg H. Host-parasite coevolution favours parasite genetic diversity and horizontal gene transfer. J Evol Biol. 2013;26:1836–40. 10.1111/jeb.12174. [DOI] [PubMed] [Google Scholar]
- 43.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472–82. 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 44.Gasmi L, et al. Recurrent domestication by Lepidoptera of genes from their parasites mediated by Bracoviruses. PLoS Genet. 2015;11: e1005470. 10.1371/journal.pgen.1005470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunkerton S, et al. A de novo mutation in KMT2A (MLL) in monozygotic twins with Wiedemann-Steiner syndrome. Am J Med Genet A. 2015. 10.1002/ajmg.a.37130. [DOI] [PubMed] [Google Scholar]
- 46.Hamzeh AR, Andrews TD, Field MA. Detecting causal variants in mendelian disorders using whole-genome sequencing. Methods Mol Biol. 2021;2243:1–25. 10.1007/978-1-0716-1103-6_1. [DOI] [PubMed] [Google Scholar]
- 47.Field MA. Bioinformatic challenges detecting genetic variation in precision medicine programs. Front Medicine. 2022;9:806696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merker JD, et al. Long-read genome sequencing identifies causal structural variation in a mendelian disease. Genet Med. 2018;20:159–63. 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goenka SD, et al. Accelerated identification of disease-causing variants with ultra-rapid nanopore genome sequencing. Nat Biotechnol. 2022;40:1035–41. 10.1038/s41587-022-01221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panduro A, Roman S. Personalized medicine in Latin America. Per Med. 2020;17:339–43. 10.2217/pme-2020-0049. [DOI] [PubMed] [Google Scholar]
- 51.Rammig A, Lapola DM. The declining tropical carbon sink. Nat Clim Change. 2021;11:727–8. 10.1038/s41558-021-01135-1. [Google Scholar]
- 52.de Kraker ME, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13: e1002184. 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Travin DY, et al. Structure of ribosome-bound azole-modified peptide phazolicin rationalizes its species-specific mode of bacterial translation inhibition. Nat Commun. 2019;10:4563. 10.1038/s41467-019-12589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan KM, et al. Opinion: why protect nature? Rethinking values and the environment. proceedings of the national academy of sciences of the United States of America. 2016;113:1462–1465. 10.1073/pnas.1525002113. [DOI] [PMC free article] [PubMed]
- 55.Brandon K. Ecosystem Services from tropical forests: review of current science. SSRN Electron J. 2014. 10.2139/ssrn.2622749. [Google Scholar]
- 56.Marselle MR, et al. Pathways linking biodiversity to human health: a conceptual framework. Environ Int. 2021;150: 106420. 10.1016/j.envint.2021.106420. [DOI] [PubMed] [Google Scholar]
- 57.Ross SRPJ, et al. Universal scaling of robustness of ecosystem services to species loss. Nat Commun. 2021;12:5167. 10.1038/s41467-021-25507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Groot R, et al. Global estimates of the value of ecosystems and their services in monetary units. Ecosyst Serv. 2012;1:50–61. 10.1016/j.ecoser.2012.07.005. [Google Scholar]
- 59.Lotze HK. Marine biodiversity conservation. Curr Biol. 2021;31:R1190-5. 10.1016/j.cub.2021.06.084. [DOI] [PubMed] [Google Scholar]
- 60.Morris RJ. Anthropogenic impacts on tropical forest biodiversity: a network structure and ecosystem functioning perspective. Philos Trans R Soc Lond B Biol Sci. 2010;365:3709–18. 10.1098/rstb.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hobern D, BIOSCAN. DNA barcoding to accelerate taxonomy and biogeography for conservation and sustainability. Genome. 2021;64:161–4. 10.1139/gen-2020-0009. [DOI] [PubMed] [Google Scholar]
- 62.Lewin HA, et al. The Earth BioGenome Project 2020: starting the clock. Proc Natl Acad Sci USA. 2022;119:119. 10.1073/pnas.2115635118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewin HA, et al. Earth BioGenome Project: sequencing life for the future of life. Proc Natl Acad Sci USA. 2018;115:4325–33. 10.1073/pnas.1720115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng S, et al. 10KP: a phylodiverse genome sequencing plan. GigaScience. 2018;7:7. 10.1093/gigascience/giy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Field MA, et al. The Australian dingo is an early offshoot of modern breed dogs. Sci Adv. 2022;8: eabm5944. 10.1126/sciadv.abm5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ballard JWO, et al. The Australasian dingo archetype: de novo chromosome-length genome assembly, DNA methylome, and cranial morphology. GigaScience. 2023;12. 10.1093/gigascience/giad018. [DOI] [PMC free article] [PubMed]
- 67.FitzSimmons NN, et al. Phylogeography, genetic stocks, and conservation implications for an Australian endemic marine turtle. Aquat Conserv: Mar Freshw Ecosyst. 2020;30:440–60. 10.1002/aqc.3270. [Google Scholar]
- 68.Plon S, Thakur V, Parr L, Lavery SD. Phylogeography of the dugong (Dugong dugon) based on historical samples identifies vulnerable Indian Ocean populations. PLoS ONE. 2019;14: e0219350. 10.1371/journal.pone.0219350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bertola LV, Higgie M, Zenger KR, Hoskin CJ. Conservation genomics reveals fine-scale population structuring and recent declines in the critically endangered Australian Kuranda Treefrog. Conserv Genet. 2023;24:249–64. 10.1007/s10592-022-01499-7. [Google Scholar]
- 70.Villacorta-Rath C, Hoskin CJ, Strugnell JM, Burrows D. Long distance (> 20 km) downstream detection of endangered stream frogs suggests an important role for eDNA in surveying for remnant amphibian populations. PeerJ. 2021;9: e12013. 10.7717/peerj.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Resh CA, et al. Using Genomics to Link populations of an invasive species to its potential sources. Front Ecol Evol. 2021;9. 10.3389/fevo.2021.575599.
- 72.Andrade P, Razzolini E, Baggio RI. See golden mussel! They are everywhere! Environmental DNA supports widespread dissemination of Limnoperna Fortunei in Hydrographic basins in the Paraná State, Brazil. Braz Arch Biol Technol. 2021;64. 10.1590/1678-4324-75years-2021210149.
- 73.Villacorta-Rath C, et al. Invasive terrestrial invertebrate detection in water and soil using a targeted eDNA approach. NeoBiota. 2023;83:71–89. [Google Scholar]
- 74.Iqbal HA, Low-Beinart L, Obiajulu JU, Brady SF. Natural product discovery through improved functional metagenomics in streptomyces. J Am Chem Soc. 2016;138:9341–4. 10.1021/jacs.6b02921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pidot S, Ishida K, Cyrulies M, Hertweck C. Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew Chem Int Ed Engl. 2014;53:7856–9. 10.1002/anie.201402632. [DOI] [PubMed] [Google Scholar]
- 76.Wangchuk P, Constantinoiu C, Eichenberger RM, Field M, Loukas A. Characterization of tapeworm metabolites and their reported biological activities. Molecules. 2019;24: 1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Darwin Tree of Life Project. Sequence locally, think globally: the Darwin Tree of Life Project. Proc Natl Acad Sci USA. 2022;119. 10.1073/pnas.2115642118. [DOI] [PMC free article] [PubMed]
- 78.Formenti G, et al. The era of reference genomes in conservation genomics. Trends Ecol Evol. 2022;37:197–202. 10.1016/j.tree.2021.11.008. [DOI] [PubMed] [Google Scholar]
- 79.Voolstra CR, Scientists GCO, Worheide G, Lopez JV. Advancing genomics through the Global Invertebrate Genomics Alliance (GIGA). Invertebr Syst. 2017;31:1–7. 10.1071/is16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rhie A, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592:737–46. 10.1038/s41586-021-03451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eldridge MDB, et al. The Oz mammals Genomics (OMG) initiative: developing genomic resources for mammal conservation at a continental scale. Aust Zool. 2020;40:505–9. 10.7882/az.2020.003. [Google Scholar]
- 82.Brandies P, Peel E, Hogg CJ, Belov K. The value of reference genomes in the conservation of threatened species. Genes (Basel). 2019;10. 10.3390/genes10110846. [DOI] [PMC free article] [PubMed]
- 83.Paez S, et al. Reference genomes for conservation. Science. 2022;377:364–6. 10.1126/science.abm8127. [DOI] [PubMed] [Google Scholar]
- 84.Farquharson KA, et al. Restoring faith in conservation action: maintaining wild genetic diversity through the tasmanian devil insurance program. iScience. 2022;25: 104474. 10.1016/j.isci.2022.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dussex N, et al. Population genomics of the critically endangered kākāpō. Cell Genomics. 2021;1: 100002. 10.1016/j.xgen.2021.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lam IPY, Sung Y-H, Fong JJ. Using eDNA techniques to find the endangered big-headed turtle (Platysternon megacephalum). PLoS ONE. 2022;17: e0262015. 10.1371/journal.pone.0262015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dubos N, et al. Are narrow-ranging species doomed to extinction? Projected dramatic decline in future climate suitability of two highly threatened species. Perspect Ecol Conserv. 2022;20:18–28. 10.1016/j.pecon.2021.10.002. [Google Scholar]
- 88.Fischer R, et al. Accelerated forest fragmentation leads to critical increase in tropical forest edge area. Sci Adv. 2021;7: eabg7012. 10.1126/sciadv.abg7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tewksbury JJ, Huey RB, Deutsch CA. Ecology. Putting the heat on tropical animals. Science. 2008;320:1296–7. 10.1126/science.1159328. [DOI] [PubMed] [Google Scholar]
- 90.Pastorino G, Darrigran GA, Lunaschi L, Martín SM. Limnoperna fortunei (Dunker, 1857)(Mytilidae), nuevo bivalvo invasor en aguas del Río de la Plata. 1993.
- 91.Uliano-Silva M, et al. A hybrid-hierarchical genome assembly strategy to sequence the invasive golden mussel, Limnoperna Fortunei. GigaScience. 2018;7:gix128. 10.1093/gigascience/gix128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uliano-Silva M, Fernandes FFCF, Holanda IBB, Rebelo M. Invasive species as a threat to biodiversity: the golden mussel Limnoperna fortune approaching the Amazon River basin. Explor Themes Aquat Toxicol Kerala. 2013;1:135–48.
- 93.Huerlimann R, et al. Enhancing tropical conservation and ecology research with aquatic environmental DNA methods: an introduction for non-environmental DNA specialists. Anim Conserv. 2020;23:632–45. 10.1111/acv.12583. [Google Scholar]
- 94.Macgregor LF, Greenlees M, de Bruyn M, Shine R. An invasion in slow motion: the spread of invasive cane toads (Rhinella marina) into cooler climates in southern Australia. Biol Invasions. 2021;23:3565–81. 10.1007/s10530-021-02597-2. [Google Scholar]
- 95.Tingley R, Greenlees M, Oertel S, van Rooyen AR, Weeks AR. Environmental DNA sampling as a surveillance tool for cane toad Rhinella marina introductions on offshore islands. Biol Invasions. 2019;21:1–6. 10.1007/s10530-018-1810-4. [Google Scholar]
- 96.Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13:894–901. 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 97.Bergmann W, Burke DC. Contributions to the study of marine products. XXXIX. The nucleosides of sponges. III. 1 spongothymidine and spongouridine2. J Org Chem. 1955;20:1501–7. [Google Scholar]
- 98.Bergmann W, Feeney RJ. Contributions to the study of marine products. XXXII. The nucleosides of sponges. I. J Org Chem. 1951;16:981–7. [Google Scholar]
- 99.Lindequist U. Marine-Derived pharmaceuticals - challenges and opportunities. Biomol Ther (Seoul). 2016;24:561–71. 10.4062/biomolther.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang J, Liu C, Wang Q, Lin P, Cheng F. In silico polypharmacology of natural products. Brief Bioinform. 2017;19:1153–71. 10.1093/bib/bbx045. [DOI] [PubMed] [Google Scholar]
- 101.Romano JD, Tatonetti NP. Informatics and computational methods in natural product drug Discovery: a review and perspectives. Front Genet. 2019;10. 10.3389/fgene.2019.00368. [DOI] [PMC free article] [PubMed]
- 102.Atanasov AG, et al. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 2021;20:200–16. 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cox PA. The ethnobotanical approach to drug discovery: strengths and limitations. Ciba Found Symp. 1994;185:25–36 discussion 36–41. [PubMed] [Google Scholar]
- 104.Molimau-Samasoni S, et al. Functional genomics and metabolomics advance the ethnobotany of the Samoan traditional medicine matalafi. Proc Natl Acad Sci USA. 2021;118: 118. 10.1073/pnas.2100880118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.West PC, et al. Trading carbon for food: global comparison of carbon stocks vs. crop yields on agricultural land. Proc Natl Acad Sci USA. 2010;107:19645–8. 10.1073/pnas.1011078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tripathi L, Ntui VO, Tripathi JN, Kumar PL. Application of CRISPR/Cas for diagnosis and management of viral diseases of Banana. Front Microbiol. 2020;11: 609784. 10.3389/fmicb.2020.609784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Almeida Camargo LS, Pereira JF. Genome-editing opportunities to enhance cattle productivity in the tropics. CABI Agric Biosci. 2022;3:8. 10.1186/s43170-022-00075-w. [Google Scholar]
- 109.Ertiro BT, et al. Genetic dissection of nitrogen use efficiency in tropical maize through genome-wide association and genomic prediction. Front Plant Sci. 2020;11: 474. 10.3389/fpls.2020.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Otto PI, et al. Genome-wide association studies for tick resistance in Bos taurus x Bos indicus crossbred cattle: a deeper look into this intricate mechanism. J Dairy Sci. 2018;101:11020–32. 10.3168/jds.2017-14223. [DOI] [PubMed] [Google Scholar]
- 111.McElroy MS, et al. Prediction of Cacao (Theobroma cacao) resistance to Moniliophthora spp. Diseases via Genome-Wide Association Analysis and genomic selection. Front Plant Sci. 2018;9: 343. 10.3389/fpls.2018.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jerry DR, et al. Predicted strong genetic gains from the application of genomic selection to improve growth related traits in barramundi (Lates calcarifer). Aquaculture. 2022;549: 737761. 10.1016/j.aquaculture.2021.737761. [Google Scholar]
- 113.Orban L, Shen X, Phua N, Varga L. Toward genome-based selection in Asian seabass: what can we learn from other Food fishes and Farm animals? Front Genet. 2021;12: 506754. 10.3389/fgene.2021.506754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joshi R, Skaarud A, de Vera M, Alvarez AT, Ødegård J. Genomic prediction for commercial traits using univariate and multivariate approaches in Nile tilapia (Oreochromis niloticus). Aquaculture. 2020;516:734641. 10.1016/j.aquaculture.2019.734641. [Google Scholar]
- 115.Arbon PM, Condon K, Andrade Martinez M, Jerry DR. Molecular detection of six viral pathogens from Australian wild sourced giant black tiger shrimp (Penaeus monodon) broodstock. Aquaculture. 2022;548: 737651. 10.1016/j.aquaculture.2021.737651. [Google Scholar]
- 116.Livramento KGD, et al. Proteomic analysis of coffee grains exposed to different drying process. Food Chem. 2017;221:1874–82. 10.1016/j.foodchem.2016.10.069. [DOI] [PubMed] [Google Scholar]
- 117.Vello V, et al. Metabolomic profiles of tropical Chlorella and parachlorella species in response to physiological changes during exponential and stationary growth phase. Algal Res. 2018;35:61–75. 10.1016/j.algal.2018.08.014. [Google Scholar]
- 118.Gense K, et al. Development of a DNA metabarcoding method for the identification of Bivalve species in Seafood products. Foods. 2021;10. 10.3390/foods10112618. [DOI] [PMC free article] [PubMed]
- 119.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46. 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 120.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346: 1258096. 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 122.Savadori L, et al. Expert and public perception of risk from biotechnology. Risk Anal. 2004;24:1289–99. 10.1111/j.0272-4332.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 123.Nayfa MG, et al. Pipette and paper: combining molecular and genealogical methods to assess a Nile tilapia (Oreochromis niloticus) breeding program. Aquaculture. 2020;523: 735171. 10.1016/j.aquaculture.2020.735171. [Google Scholar]
- 124.Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–29. 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Minamikawa MF, et al. Genome-wide association study and genomic prediction in citrus: potential of genomics-assisted breeding for fruit quality traits. Sci Rep. 2017;7:4721. 10.1038/s41598-017-05100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seyum EG, et al. Genomic selection in tropical perennial crops and plantation trees: a review. Mol Breeding. 2022;42:58. 10.1007/s11032-022-01326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zenger KR, et al. Genomic selection in aquaculture: application, limitations and opportunities with special reference to marine shrimp and pearl oysters. Front Genet. 2018;9: 693. 10.3389/fgene.2018.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Massault C, et al. Association for the Advancement of Animal Breeding and Genetics, vol 1. Armidale: Curran Associates, Inc.; 2019. p. 406–9.
- 129.Nimo-Paintsil SC, et al. Ticks and prevalence of tick-borne pathogens from domestic animals in Ghana. Parasit Vectors. 2022;15:86. 10.1186/s13071-022-05208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Braconi D, Millucci L, Parisi ML, Spiga O, Santucci A. Food authentication and traceability. In: Galanakis CM, editor. Academic Press, Elsevier; 2021. p. 215–45. 10.1016/B978-0-12-821104-5.00003-9.
- 131.Liu Y, et al. A transcriptomic analysis of stylo [Stylosanthes guianensis (Aubl.) Sw.] provides novel insights into the basis of salinity tolerance. Front Sustain Food Syst. 2022;6: 6. 10.3389/fsufs.2022.725656. [Google Scholar]
- 132.Aron S, et al. Ten simple rules for developing bioinformatics capacity at an academic institution. PLoS Comput Biol. 2021;17: e1009592. 10.1371/journal.pcbi.1009592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aron S, et al. The development of a sustainable bioinformatics training environment within the H3Africa Bioinformatics Network (H3ABioNet). Front Educ. 2021;6:6. 10.3389/feduc.2021.725702. [Google Scholar]
- 134.Mulder N, et al. H3Africa: current perspectives. Pharmgenomics Pers Med. 2018;11:59–66. 10.2147/PGPM.S141546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gurwitz KT, et al. Designing a course model for distance-based online bioinformatics training in Africa: the H3ABioNet experience. PLoS Comput Biol. 2017;13: e1005715. 10.1371/journal.pcbi.1005715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mulder NJ, et al. H3ABioNet, a sustainable pan-african bioinformatics network for human heredity and health in Africa. Genome Res. 2016;26:271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Khan AM, Tan TW, Schönbach C, Ranganathan S. APBioNet—Transforming bioinformatics in the Asia-Pacific Region. PLoS Comput Biol. 2013;9: e1003317. 10.1371/journal.pcbi.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mulder N, et al. The development and application of bioinformatics core competencies to improve bioinformatics training and education. PLoS Comput Biol. 2018;14: e1005772. 10.1371/journal.pcbi.1005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537-544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jalili V, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020;48:W395-402. 10.1093/nar/gkaa434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boekel J, et al. Multi-omic data analysis using Galaxy. Nat Biotechnol. 2015;33:137–9. 10.1038/nbt.3134. [DOI] [PubMed] [Google Scholar]
- 142.Thang MWC, Chua XY, Price G, Gorse D, Field MA, MetaDEGalaxy. Galaxy workflow for differential abundance analysis of 16s metagenomic data. F1000Res. 2019;8:726. 10.12688/f1000research.18866.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramsay M, et al. H3Africa AWI-gen collaborative centre: a resource to study the interplay between genomic and environmental risk factors for cardiometabolic diseases in four sub-saharan African countries. Glob Health Epidemiol Genomics. 2016;1:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ekoru K, et al. H3Africa multi-centre study of the prevalence and environmental and genetic determinants of type 2 diabetes in sub-saharan Africa: study protocol. Glob Health Epidemiol Genom. 2016;1:e5. 10.1017/gheg.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Osafo C, et al. Genomic approaches to the burden of kidney disease in sub-saharan Africa: the human heredity and health in Africa (H3Africa) kidney disease research network. Kidney Int. 2016;90:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Akpalu A, et al. Phenotyping stroke in sub-saharan Africa: stroke investigative research and education network (SIREN) phenomics protocol. Neuroepidemiology. 2015;45:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wall JD, et al. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature. 2019;576:106–11. 10.1038/s41586-019-1793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Team SGP. The Saudi Human Genome Program: an oasis in the desert of arab medicine is providing clues to genetic disease. IEEE Pulse. 2015;6:22–6. 10.1109/MPUL.2015.2476541. [DOI] [PubMed] [Google Scholar]
- 149.Helmy M, Awad M, Mosa KA. Limited resources of genome sequencing in developing countries: challenges and solutions. Appl Transl Genom. 2016;9:15–9. 10.1016/j.atg.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wasswa FB, Kassaza K, Nielsen K, Bazira J. MinION whole-genome sequencing in resource-limited settings: challenges and opportunities. Curr Clin Microbiol Rep. 2022;9:52–9. 10.1007/s40588-022-00183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Salazar AN, et al. An educational guide for nanopore sequencing in the classroom. PLoS Comput Biol. 2020;16: e1007314. 10.1371/journal.pcbi.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meier BM, et al. Travel restrictions and variants of concern: global health laws need to reflect evidence. Bull World Health Organ. 2022;100:178-A178. 10.2471/BLT.21.287735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hampton AR, Eccleston-Turner M, Rourke M, Switzer S. Equity in the Pandemic Treaty: access and benefit-sharing as a policy device or a rhetorical device? J Law Med Ethics. 2023;51:217–20. 10.1017/jme.2023.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Michael Bayerlein PAV. "One Health” and Global Health Governance Design and implementation at the international, European, and German levels. 2023. https://www.who.int/news/item/10-12-2023-the-quadripartite-launches-a-guide-to-support-countries-implement-one-health-approach.
- 155.One Health High-Level, Expert P, et al. Developing one health surveillance systems. One Health. 2023;17: 100617. 10.1016/j.onehlt.2023.100617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang Y, Jiang S, Kumah E. China and the WHO pandemic treaty: a dive into stance, underpinnings, and implications. Front Public Health. 2024;12: 1335751. 10.3389/fpubh.2024.1335751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.