Abstract
Bacterial stem and root rot (BSRR) in sweetpotato caused by Dickeya dadantii is one of the ten major diseases of sweetpotatoes in China. However, the molecular mechanism underlying the resistance of sweetpotato to D. dadantii remains unclear. This study adopted a resistance identification assay that conformed Guangshu87 (GS87) as BSRR-resistant and Xinxiang (XX) as susceptible. Compared to XX, GS87 effectively prevented the invasion and dissemination of D. dadantii in planta. An RNA sequencing (RNA-seq) analysis identified 54,844 expressed unigenes between GS87 and XX at four different stages. Further, it revealed that GS87 was more able to regulate the expressions of more unigenes after the inoculation with D. dadantii, including resistance (R) and transcription factors (TF) genes. Moreover, content measurements of disease resistance-related phytohormones showed that both jasmonic acids (JAs) and salicylic acids (SAs) accumulated in D. dadantii-inoculated sweetpotatoes, and JAs may negatively regulate sweetpotato resistance against D. dadantii and accumulated faster than SAs. Meanwhile, determinations of ROS production rate and relevant enzymatic/non-enzymatic activity highlighted the vital roles of reactive oxygen species (ROS) and superoxide dismutase (SOD) in confering GS87 resistance against D. dadantii. Additionally, several hub genes with high connectivity were highlighted through Protein–Protein interaction (PPI) network analysis. In summary, the findings in this study contribute to the understanding of the different responses of resistant and susceptible sweetpotato cultivars to D. dadantii infection, and it also provide the first insight into the relevant candidate genes and phytohormones involved in the resistance of sweetpotato to D. dadantii.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05774-2.
Keywords: Sweetpotato, Dickeya dadantii, RNA sequencing, Phytohormone, Reactive oxygen species, Superoxide dismutase
Background
Sweetpotato (Ipomoea batatas (L.) Lam) is an important crop plant with extensive adaptability and abundant nutriments, ranking eighth in the global annual production (https://www.fao.org/home/en). China is the biggest global sweetpotato producer and is indispensable in ensuring food security worldwide. However, several diseases have severely limited the sweetpotato production in China. For instance, bacterial stem and root rot (BSRR) of sweetpotato caused by Dickeya dadantii (former synonyms: Erwinia chrysanthemi or Pectobacterium chrysanthemi) is among the ten major sweetpotato diseases in China. Severe BSRR infection causes 50–100% yield losses and quality degradation in sweetpotato [1]. The first BSRR record in China was from several sweetpotato plantations in Guangdong, a southern province. In fact, this disease occurs more commonly and seriously in southern than in northern China as the south is warmer, more humid, and conducive to BSRR [2].
D. dadantii is a broad-spectrum plant necrotrophic enterobacterium that infects through wounds or natural openings in planta, causing typical soft rot symptoms by secreting enzymes, effectors, and other virulence factors that degrade plant cell walls [1, 3, 4]. The dicotyledonous model plant Arabidopsis thaliana is susceptible to D. dadantii, and studies of A. thaliana–D. dadantii pathosystem have been conducted extensively. For example, iron homeostasis is critical for regulating Arabidopsis defense against D. dadantii and is closely connected with the plant cell wall integrity, and D. dadantii development is limited effectively in iron-deficient Arabidopsis plants [5, 6]. Moreover, AtFER1 (as ferritin) and AtNRAMP3/4 (as iron transporters) have been proven to be involved in A. thaliana resistance against D. dadantii [7, 8].
Plants have developed a sophisticated innate immune system involving two levels of immunity: pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) for protection from pathogen attacks [9]. As the first guard line, PTI is mediated by plant membrane-localized pattern-recognition receptors (PRRs) cooperated with the co-receptors through the recognition of pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs). Consequently, PTI triggers specific downstream immune responses, including phyhormones metabolism, reactive oxygen species (ROS) burst, cascade signaling pathways of mitogen-activated protein kinases (MAPKs), and gene expression regulation. Leucine-rich repeat receptor-like kinases (LRR-RLKs) are the biggest RLKs subfamily in plants and play important roles in regulating PTI. For instance, two typical Arabidopsis LRR-RLKs, FLAGELLIN SENSITIVE2 (FLS2) and Elongation factor Thermo unstable (EF-Tu) receptor (EFR), perceive the bacterial flagellin (flg22) and EF-Tu, respectively, thus triggering the downstream immunity response [10, 11]. When the pathogens overcome PTI, a more intense defense response—ETI is triggered. In ETI, the intracellular resistance (R) proteins recognize the pathogen effectors also through phyhormones metabolism, ROS burst, and cascade pathways of MAPKs to inhibit pathogen intrusions [12, 13]. Most reported R proteins contain nucleotide binding site-LRR (NBS-LRR or NLR) domains, such as PigmR and Pm21, which conferred broad-spectrum resistances for crops [14–17]. Whether PTI and ETI function independently or interact mutually against pathogen attacks remains unclear. Until recently, studies demonstrated that PTI and ETI in Arabidopsis interact synergistically when challenged with Pseudomonas syringae [18, 19]. Specifically, PTI membrane receptors are essential for activating ETI, which potentiates PTI responses. More evidence demonstrated that PTI and ETI integrity and interaction ensure comprehensive immunity in plants [20–22].
To further elucidate the intricate dynamics of plant-pathogen interactions, it is crucial to examine the role of phytohormones in modulating these responses. Jasmonic acid (JA) and salicylic acid (SA) are two classical biotic stress-relevant phytohormones that were involved in plant disease resistance against D. dadantii. However, the roles of JA- and SA-mediated pathways on plant resistance could be antagonistic or synergistic [13, 23–26]. JA exerts chemoattraction for D. dadantii to assist this bacterium to invade into the host, and it also induces the expressions of a few pathogenesis-associated genes in D. dadantii. Moreover, an Arabidopsis aso1 mutant with JA synthesis deficiency was more resistant to D. dadantii than the wild type. Hence, the role of JA in helping plants to resist against D. dadantii remains controversial. JA was synthesized from an important precursor—12-oxo-phytodienoic acid (OPDA) through sequential catalyses governed by several key enzymes, including lipoxygenase (LOX), allene oxide synthase (AOS) and allene oxide cyclase (AOC). After being released from the peroxisome into the cytosol, JA is further derived into methyl JA (MeJA) and other amino acid conjugates like jasmonoyl-isoleucine (JA-Ile) and jasmonoyl-phenalanine (JA-Phe) [27, 28]. The most known bioactive JA-derivative, JA-Ile, can ubiquitinate jasmonate ZIM-domain (JAZ) proteins with the assistance of coronatine insensitive1 (COI1) proteins, thus removing the repressions of MYC transcription factors (TFs) and triggering the transcriptional reprogramming of downstream JA-responsive genes. Unlike JA, which mediates plant resistance against necrotrophs, SA regulates plant defense responses towards biotrophs or hemibiotrophs [29]. Pathogenic infection may trigger the biosynthesis and modification of SA by a cascade of enzymes, and then SA and its methylated product, MeSA, carry immune signal from the infection site to the unwounded tissues. This reaction enables plants to establish a systemic acquired resistance (SAR), followed by a series of plant defense responses, including the accumulation of ROS while inhibiting JA biosynthesis [13, 30]. Fagard et al. [25] showed that the JA rather than the SA pathway contributes to the Arabidopsis counterattack against D. dadantii, although the bacterial infection enhances the expressions of several marker genes in the JA and SA pathways.
Fundamental studies and applications of the sweetpotato-D. dadantii system in the past few decades include analyses of pathogenicity divergence and genetic diversity of D. dadantii populations, establishments of BSRR-disease resistance assessment systems, and screenings for resistant and susceptible materials [31–33]. These efforts can advance the molecular breeding of BSRR-resistant sweetpotato cultivar. Introducing R genes into disease-susceptible plants is an economically effective strategy for breeding disease-resistant plants. For instance, applications of the broad-spectrum R gene Rpi-vnt1.1 effectively stifled potato late-blight (the most devastating and notorious potato disease worldwide) [34, 35]. Sm, an NBS-LRR gene from wild tomato, confers significant resistance against gray leaf spot disease for the tomato cultivar Motelle [36]. However, there is no report on a BSRR-resistance gene in sweetpotato so far, let alone the interpretation of the relevant genes function or the molecular mechanisms. Unlike other crop plants, identifying genes in sweetpotato by using traditional positional cloning methods is challenging since the sweetpotato has a highly heterozygous genome and strong self-/cross-incompatibility [37, 38]. Thus, RNA sequencing (RNA-seq) technology is appropriate to overcome these limitations and enable high-throughput analysis of massive functional genes regulating yield, quality, and stress tolerance in sweetpotato [39–42]. In current study, the disease developments of two elite sweetpotatoes, Guangshu 87 and Xinxiang were analyzed followed by RNA-seq, phytohormone detection, and ROS and relevant antioxidants determinations. This study confirmed the resistance differences between GS87 and XX against D. dadantii and identified key genes and phytohormones that respond to sweetpotato resistance against BSRR. Furthermore, GS87 activated ROS production more rapidly and superoxide dismutase (SOD) enzyme may play a dominant role in regulating GS87 resistance to D. dadantii in contrast to XX. In conclusion, this study provides novel gene resources for molecular resistance breeding of sweetpotato and a fundamental mechanism for BSRR disease management.
Methods
Bacterial strain and inocula preparation
The D. dadantii strain Ech36 used in this study was isolated from a BSRR-infected sweetpotato plant. The streak-culture of strain Ech36 on NA medium was incubated in darkness at 30 ℃ for 36 h. Next, a single fresh bacterial colony was mixed with 400 μL of NA liquid medium and cultured overnight at 30 ℃ with shaking at 200 rpm. The bacterial suspension was transferred into a flask containing 10 mL of NA liquid medium and incubated overnight at 30 ℃ with shaking (200 rpm). Then, 3 mL of the overnight culture was diluted with 50 mL of NA liquid medium to adjust the initial OD620 to 0.15, which was subsequently incubated at 30 ℃ for 4 h with shaking (200 rpm) to reach the final OD620 = 1 (approximately 109 cfu/mL) [43]. Then the inoculum was diluted to 108 cfu /mL and 107 cfu /mL using NA liquid medium.
Plant materials and bacterial inoculation
Two elite sweetpotato cultivars, GS87 and XX, were planted at the Baiyun experimental station of Guangdong Academy of Agricultural Sciences, Guangzhou, China. Next, 25 cm-long stems with 6 leaves were trimmed from healthy sweetpotato plants and cultivated in sterile water for 4 days to facilitate the formation and growth of adventitious roots. The ends of newly grown plants were cut carefully to make fresh wounds (without damaging roots) and inoculated immediately with 50 mL of inoculum of 108 cfu/mL and 107 cfu /mL for 30 min, respectively, and then they were rinsed with sterile water. Control plants (CK) were inoculated with 50 mL of NA liquid medium. All the inoculated plants were incubated in sterile water for a week at 30 ℃ and 90% relative humidity under a 13-h light/11-h dark cycle. Afterward, the length of longitudinal lesions along the stems and the number of symptomatic leaves were measured to determine the disease index.
Paraffin sectioning
The 1 cm-long samples were harvested from the basal stems of plants inoculated with strain Ech36 and fixed in formalin-acetic acid-alcohol solution overnight to better recognize the tissue structure damages after D. dadantii infection. The sectioning process was conducted as described previously with minor modifications [44]. Finally, 10 μm-thick longitudinal sections of samples were sliced by a microtome (Thermo Fisher Scientific, MA, USA) and observed with an optical microscope (Carl Zeiss, Oberkochen, Germany).
Sample collection
Healthy sweetpotato plants with adventitious roots were inoculated with 108 cfu/mL bacterial inoculum for 30 min as described in Plant materials and bacterial inoculation. Afterwards, the 1 cm-long basal stems of the inoculated plants were harvested at 0, 18, and 30 h post inoculation (hpi), immediately frozen in liquid nitrogen, and stored at –80 ℃ prior to analyses of RNA-seq, phytohormone content, ROS levels, enzymatic/non-enzymatic assays, and qRT-PCR. CK was treated in NA liquid medium for 30 min. Each treatment contained three biological replicates.
RNA-seq and data analysis
Total RNA was extracted from the samples using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek, GA, USA) following the manufacturer’s instructions. The cDNA library was constructed, and the fragments were ligated with adapters as described by Li et al. [45] and sequenced on the Illumina NovaSeq 6000 (Illumina, CA, USA) at Gene Denovo Biotechnology Co. (Guangzhou, China). The raw reads containing adapters and low-quality sequences (with > 10% N bases or 50% of the bases having ≤ 20 quality score) were filtered using fastp v0.18.0 [46] to ensure the accuracy of the RNA-seq results. The resultant clean reads were sequentially subjected to analysis of bases composition and de novo assembly using the Trinity software package [47].
The BLASTx tool (http://www.ncbi.nlm.nih.gov/BLAST/) was used for basic annotation of unigenes (E-value threshold of 1e-5) in Nr, Swiss-Prot, KEGG, and COG/KOG databases. GO enrichment was performed based on the Nr annotation from the Blast2GO software [48]. Advanced annotations for protein domain and function were conducted in the Pfam and SAMRT databases using Pfam_Scan (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/Tools/) and HMMER servers (http://hmmer.org/), respectively. Further, predictions of R proteins and TFs were performed against the PRGdb and Plant TFdb databases using Blastp search. RSEM [49] was applied to qualify and normalize the unigene expression as FPKM. Gene differential expression between two groups was determined using the DESeq2 software [50]. The criterion for defining DEGs with statistically significant differences in group comparisons is |absolute log2 fold-change (log2FC) > 1 and FDR < 0.05|. PCA was performed using R package (http://www.r-project.org/) to establish sample correlations.
The interacting relationship among proteins encoded by the DEGs acquired in RNA-seq was analyzed using the STRING database (https://cn.string-db.org/). However, the resulting interaction pairs were numerous. Thus, the interaction pairs with ≥ 500 combined scores were retrieved for further study.
Measurements of phytohormones and data analysis
Here, 1 cm-long stem samples were ground into a fine powder under liquid nitrogen before measuring SA, JA, and their derivatives. Next, the phytohormones were extracted and concentrated using previous protocols [51, 52]. Briefly, 50 mg of the stem powder was mixed with 1 mL of methanol/water/formic acid (15:4:1, V/V/V) and 10 μL of an internal standard solution (100 ng/mL) and vortexed for 10 min. The solution was centrifuged at 12,000 rpm for 5 min at 4 ℃; its supernatant was transferred to a new microcentrifuge tube, dried using nitrogen purging, reconstituted in 100 μL of 80% methanol (V/V), and filtrated using a 0.22 μm membrane filter. The qualitative and quantitative analysis of phytohormones in the samples were then conducted using Analyst 1.6.3 and MultiQuant 3.0.3 softwares upon an ExionLC™ AD ultra-performance liquid chromatography-electrospray tandem QTRAP® 6500 + mass spectrometry (UPLC-ESI–MS/MS, SCIEX, MA, USA) system at MetWare Biotechnology Co. Ltd. (Wuhan, China). The R package pheatmap was used for hierarchical cluster analysis (HCA) of the samples. Statistically significant differences in the phytohormone contents were determined by the threshold |absolute log2FC ≥ 1 and P-value ≤ 1|.
Determinations of ROS levels and relevant enzymes/non-enzymes activity
ROS levels, enzymes (SOD, CAT, and APX) and non-enzymes (AsA and GSH) activity were tested respectively using detection kits following the manufacturer’s instructions (Suzhou Comin, Suzhou,China). Statistics was analyzed using Analysis of variance (ANOVA) methods upon the GraphPad Prism 8 software (https://www.graphpad.com/).
qRT-PCR (quantitative Real-Time PCR) analysis
The RNA extracted from the 1 cm-long basal stems were used for qRT-PCR analysis. The first-strand cDNA was produced from 500 ng of the extracted RNA using the FastKing gDNA Dispelling RT SuperMix kit (TIANGEN, Beijing, China) and diluted 10 times before qRT-PCR. The primers (Table S1) were designed for amplifying the non-conserved sequences of the unigene coding regions using Primer3Plus (https://www.primer3plus.com/index.html). Next, the qRT-PCR was conducted using diluted cDNA as the template in 10 μL of the SsoFast EvaGreen Supermix (Bio-Rad, CA, USA) on a CFX96 Touch real-time PCR detection system (Bio-Rad, USA). Each set was repeated three times. The constitutive sweetpotato β-actin gene (GenBank accession no. AY905538) was chosen as the internal reference, and the unigene expression was measured using the 2–ΔΔCt method [53]. Differences between gene expression levels were determined using the two-tailed Student’s t-test, and the linear regression model was built using the GraphPad Prism 8 software (https://www.graphpad.com/).
Results
Differential resistibility and pathogenicity of D. dadantii in sweetpotato cultivars
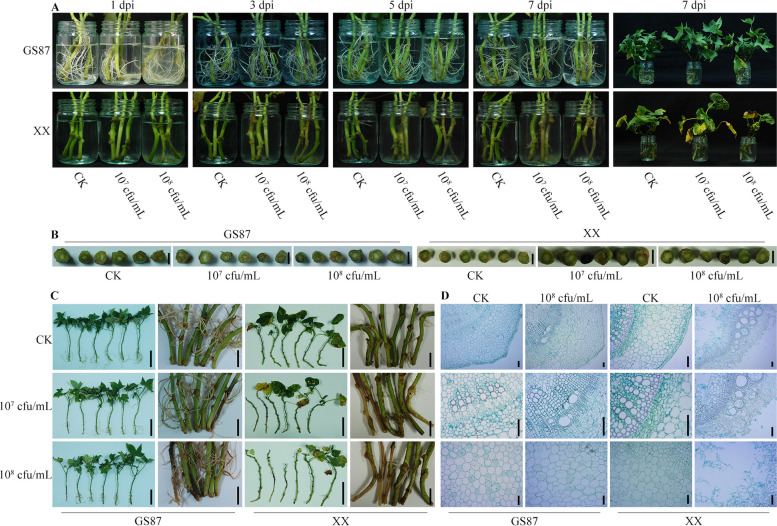
Compared to CK-XX, D. dadantii-infected stems of XX developed water-soaked brown symptoms rapidly within one day either at 107 or at 108 cfu/mL, and the lesions spread along the stems to the top during the next few days (Fig. 1A). Besides, the leaves of the inoculated XX plants gradually became chlorotic and wilted, resulting in plant death (Fig. 1A, C). Different inoculum concentrations caused insignificant differences in the average length of the rotten XX stems (unpublished data). However, a higher concentration significantly increased the disease severity on the leaves and roots of the inoculated XX plants; most leaves became chlorotic, wilted, and abscised without new leaf development, and the roots decayed (Fig. 1C). D. dadantii caused similar concentration effects on GS87 plants under the same conditions, but the overall disease severity was milder than for XX plants (Fig. 1A–C). Despite the minor rotten stem, the GS87 plants inoculated at 107 and 108 cfu/mL of D. dadantii were mostly as normal as CK-87 during the seven days post-inoculation (dpi). Furthermore, the average length of longitudinal rotten stems at 107 and 108 cfu/mL was not significantly different, save for the one or two chlorotic leaves observed at 108 cfu/mL (Fig. 1A–C). All the results above indicated that a higher D. dadantii concentration promote more invasion of sweetpotato. The disease evaluation results revealed that 108 cfu/mL is appropriate for the following studies, and GS87 and XX were identified as resistant and highly susceptible to D. dadantii, respectively.
Fig. 1.
Symptom developments in GS87 and XX plants inoculated with D. dadantii strain E36. A Disease developments in GS87 and XX plants one week after inoculation with 107 cfu/mL and 108 cfu/mL. B Cross sections of the inoculated stems at 7 dpi. Bars = 0.5 cm. C Whole plants and local enlargements of stems and adventitious roots at 7 dpi. Bars = 10 cm. D Histological analysis showed the stem architectures in cross sections at 7 dpi after inoculation with 108 cfu/mL. Bars = 100 μm. CK plants were inoculated with an NA liquid medium
The histological results showed that the cell structure of GS87 stems inoculated with 108 cfu/mL of D. dadantii roughly remained normal with intact epidermis, cortex, and vascular bundles in contrast with CK-87. However, the parenchyma cells of the pith of inoculated GS87 were partially and slightly degraded, comparable to the tiny light-brown lesions observed in the cross-section of infected stems (Fig. 1B, D). Contrarily, the D. dadantii-inoculated XX stems decomposed severely with loosened and fragmentary parenchyma cells in the broken epidermis, cortex, and pith (Fig. 1D). These severe injuries in XX weakened its mechanical support for stems and damaged the absorption, storage, and transportation of water and nutrients, leading to plant maceration and lethality. Intriguingly, the xylem and phloem tissues of the inoculated GS87 and XX stems kept a relatively integrated composition similar to CK. Altogether, GS87 developed a stronger defense strategy to prohibit D. dadantii invasion and dissemination compared to XX.
A large number of genes were functionally identified in RNA-seq and analyzed
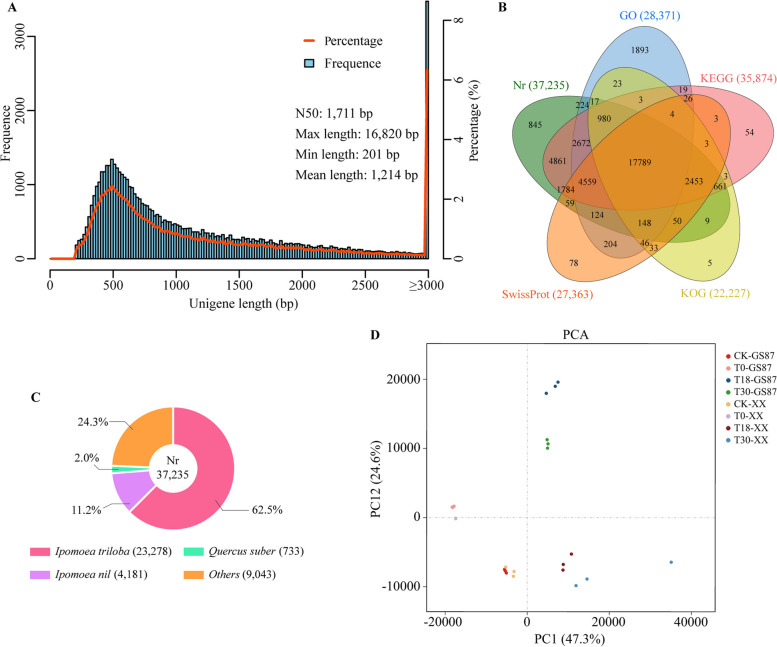
The RNA-seq analysis generated 960.99 million high-quality clean reads consisting of 142.58 Gb clean bases with 92.37–93.53% Q30 and 45.67–47.97% GC contents, and each sample had a relatively low unknown base (N) content (Table 1). This data is similar to another RNA-seq-based study of sweetpotato saccharification mechanism33. Accordingly, de novo assembly yielded 54,844 unigenes, including 2,604 R genes and 1,517 transcription factor (TF) genes (after deleting unigenes mapped to the bacterial transcriptome) with an N50 of 1,711 bp and an average unigene length of 1,214 bp (Fig. 2A). GS87 and XX samples had 52,774 (including 2,520 R genes and 1,507 TF genes) and 45,385 (including 2,405 R genes and 1,403 TF genes) unigenes, respectively (unpublished data). Of 54,844 unigenes, 72.26% were functionally annotated in Nr, GO, KEGG, KOG, and SwissProt databases (Fig. 2B). Most Nr-annotated unigenes were aligned to the genomes of I. triloba and I. nil, two wild relatives of cultivated sweetpotato (Fig. 2C). Furthermore, 28,731 unigenes were classified into three GO terms (level 1), of which 487 unigenes were involved in the immune system process (level 2) (Table S2). Most of the annotated unigenes enriched the KEGG pathways of “Metabolic pathways” (ko01100, 4092), “Biosynthesis of secondary metabolites” (ko01110, 2266) and “Ribosome” (ko03010, 1574), suggesting that the sweetpotato defense against D. dadantii is an energy-cost process. Additionally, three disease-resistant-relevant pathways—“Plant-pathogen interaction (ko04626, 509)”, “Plant hormone signal transduction (ko04075, 365)” and “MAPK signaling pathway-plant (ko04016, 314)” ranked among the top 20 KEGG pathways. Overall, the KOG classifications were consistent with the KEGG enrichment (Table S3 and S4).
Table 1.
Quality assessment of RNA-seq data
| Sample | Raw reads | Clean reads (%) | Raw bases (bp) | Clean bases (bp) | Q20 (%)a | Q30 (%)a | N (%)b | GC content (%) |
|---|---|---|---|---|---|---|---|---|
| CK-GS87-1 | 43,114,924 | 42,951,518 (99.62%) | 6,467,238,600 | 6,395,389,296 | 97.42 | 92.89 | 61,750 (0.00%) | 45.90 |
| CK-GS87-2 | 36,879,782 | 36,752,492 (99.65%) | 5,531,967,300 | 5,473,859,184 | 97.59 | 93.25 | 55,467 (0.00%) | 45.88 |
| CK-GS87-3 | 43,008,784 | 42,862,430 (99.66%) | 6,451,317,600 | 6,392,419,919 | 97.51 | 93.05 | 61,976 (0.00%) | 45.92 |
| T0-GS87-1 | 36,391,086 | 36,264,542 (99.65%) | 5,458,662,900 | 5,403,918,078 | 97.62 | 93.28 | 53,816 (0.00%) | 45.79 |
| T0-GS87-2 | 36,122,112 | 35,986,806 (99.63%) | 5,418,316,800 | 5,359,328,453 | 97.72 | 93.48 | 52,333 (0.00%) | 45.89 |
| T0-GS87-3 | 56,593,696 | 56,380,228 (99.62%) | 8,489,054,400 | 8,415,741,169 | 97.59 | 93.03 | 100,395 (0.00%) | 45.91 |
| T18-GS87-1 | 38,768,196 | 38,517,124 (99.35%) | 5,815,229,400 | 5,735,936,202 | 97.61 | 93.29 | 56,506 (0.00%) | 46.78 |
| T18-GS87-2 | 42,014,168 | 41,715,254 (99.29%) | 6,302,125,200 | 6,212,025,055 | 97.38 | 92.75 | 61,194 (0.00%) | 46.75 |
| T18-GS87-3 | 40,547,828 | 40,356,808 (99.53%) | 6,082,174,200 | 6,011,710,657 | 97.22 | 92.37 | 58,554 (0.00%) | 46.07 |
| T30-GS87-1 | 10,758,112 | 10,668,618 (99.17%) | 1,613,716,800 | 1,588,339,234 | 97.78 | 93.53 | 12,509 (0.00%) | 46.67 |
| T30-GS87-2 | 6,989,218 | 6,948,076 (99.41%) | 1,048,382,700 | 1,035,599,680 | 97.55 | 93.16 | 9,880 (0.00%) | 46.43 |
| T30-GS87-3 | 21,210,984 | 21,061,104 (99.29%) | 3,181,647,600 | 3,137,285,694 | 97.49 | 92.95 | 31,182 (0.00%) | 46.33 |
| CK-XX-1 | 44,562,922 | 44,405,210 (99.65%) | 6,684,438,300 | 6,607,465,046 | 97.66 | 93.35 | 58,734 (0.00%) | 45.67 |
| CK-XX-2 | 45,092,216 | 44,933,498 (99.65%) | 6,763,832,400 | 6,699,461,852 | 97.43 | 92.90 | 63,353 (0.00%) | 45.89 |
| CK-XX-3 | 46,015,234 | 45,856,814 (99.66%) | 6,902,285,100 | 6,835,834,650 | 97.46 | 92.92 | 67,132 (0.00%) | 45.84 |
| T0-XX-1 | 42,266,740 | 42,130,070 (99.68%) | 6,340,011,000 | 6,282,012,599 | 97.57 | 93.11 | 60,722 (0.00%) | 46.07 |
| T0-XX-2 | 44,319,074 | 44,176,452 (99.68%) | 6,647,861,100 | 6,581,362,528 | 97.49 | 93.00 | 64,607 (0.00%) | 46.02 |
| T0-XX-3 | 53,988,786 | 53,808,906 (99.67%) | 8,098,317,900 | 8,032,722,347 | 97.52 | 93.07 | 78,884 (0.00%) | 46.05 |
| T18-XX-1 | 44,720,934 | 44,550,410 (99.62%) | 6,708,140,100 | 6,630,661,698 | 97.49 | 93.06 | 65,353 (0.00%) | 46.26 |
| T18-XX-2 | 49,942,008 | 49,740,896 (99.60%) | 7,491,301,200 | 7,407,888,880 | 97.53 | 93.10 | 72,033 (0.00%) | 46.45 |
| T18-XX-3 | 44,820,040 | 44,626,308 (99.57%) | 6,723,006,000 | 6,632,432,101 | 97.55 | 93.17 | 65,223 (0.00%) | 46.21 |
| T30-XX-1 | 42,620,586 | 42,441,138 (99.58%) | 6,393,087,900 | 6,321,825,263 | 97.60 | 93.30 | 60,808 (0.00%) | 46.20 |
| T30-XX-2 | 49,055,330 | 48,825,962 (99.53%) | 7,358,299,500 | 7,271,071,351 | 97.61 | 93.31 | 71,380 (0.00%) | 47.97 |
| T30-XX-3 | 41,183,068 | 41,021,230 (99.61%) | 6,177,460,200 | 6,112,173,464 | 97.48 | 93.04 | 59,443 (0.00%) | 46.33 |
| Total | 960,985,828 | 956,981,894 | 144,147,874,200 | 142,576,464,400 | – | – | – | – |
aThe ratio of clean bases having quality score over 20 and 30 to all clean bases
bThe ratio of data with unknown (N) bases to all clean bases
Fig. 2.
De novo assembly and analysis of RNA-seq data. A Unigene length distribution. B Venn diagram showing the number of unigenes annotated by five databases. C Best alignments of unigenes to the reference genome against the Nr database. D The principal component analysis model indicated the relationships between cultivars and time points after inoculation with D. dadantii
The principal component analysis (PCA) model showed a small dispersion of the biological replicates of each sweetpotato sample, and all samples were distributed in the four quadrants of two principal components (Fig. 2D). The 0 hpi and CK samples were distinguished from those at 18 and 30 hpi in the first principal component (PC1, 47.3%), indicating that sweetpotato plants respond differently to short- and long-term inoculation with D. dadantii. Besides, GS87 and XX plants showed various responses to the D. dadantii attack, as their corresponding samples were divided into PC2 (24.6%). qRT-PCR using specific primers showed that the expression profiles of nine unigenes were generally consistent with those in the RNA-seq (Fig. S1). Collectively, the results of the PCA model and qRT-PCR demonstrated that the RNA-seq data was reliable.
Key differentially expressed genes (DEGs) encoding R proteins and TFs were uncovered
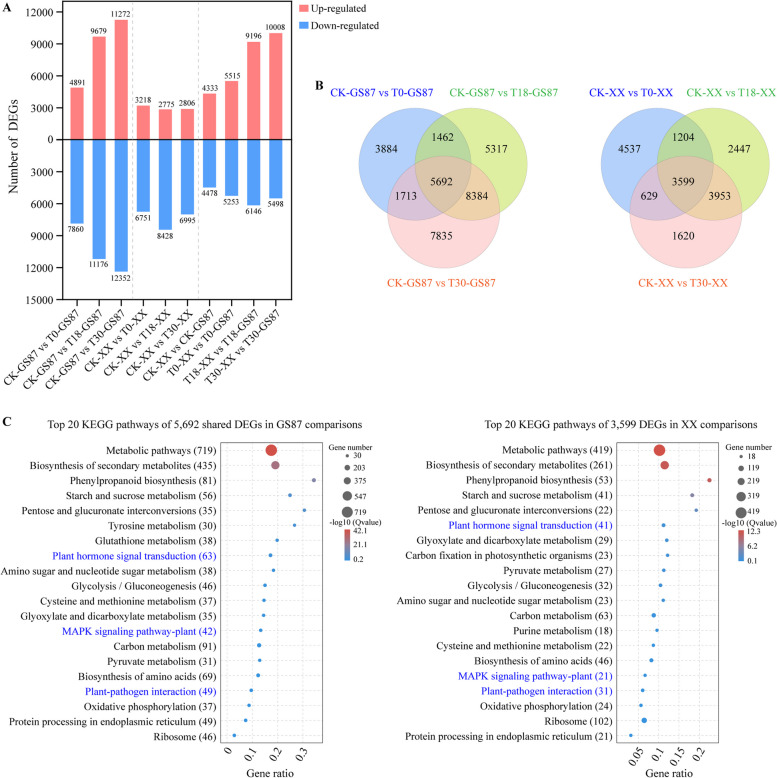
The number of DEGs increased over the treatment time in CK-GS87 vs T0-GS87, CK-GS87 vs T18-GS87, and CK-GS87 vs T30-GS87 comparisons (CK-GS87 as control). More unigenes were down-regulated in D. dadantii-inoculated GS87 samples (T0-GS87, T18-GS87, T30-GS87) compared to CK-GS87. A similar trend was observed in XX comparisons (CK-XX vs T0-XX, CK-XX vs T18-XX, and CK-XX vs T30-XX) (Fig. 3A). The GS87 comparisons had more DEGs (34,287) than XX comparisons (17,989), implying that GS87 manipulated more genes (especially R genes) transcriptionally for defense against D. dadantii infection (Fig. 3A, Table S5 and S6). Moreover, 5,692 common DEGs from GS87 comparisons shared the same top 20 KEGG pathways with 3,599 common DEGs from XX comparisons, including three disease resistance-related pathways “Plant hormone signal transduction (ko04075)”, “MAPK signaling pathway-plant (ko04016)” and “Plant-pathogen interaction (ko04626)”, but the numbers of DEGs in these pathways differed (Fig. 3C).
Fig. 3.
Analysis of DEGs between GS87 and XX at different time points. A Distribution of DEGs at different time points. XX samples were used as control groups in the four distinct comparisons. B Venn diagram showing DEGs at 0, 18, and 30 hpi with CK as a control in the same sweetpotato cultivar (left, GS87, right, XX). C Top 20 KEGG pathways enriched by 5,692 common DEGs in the GS87 comparisons (left) and 3,599 common DEGs in the XX comparisons (right). Gene ratio indicated the number of unigenes enriched in specific KEGG pathway divided by the total number of all unigenes annotated in the KEGG database
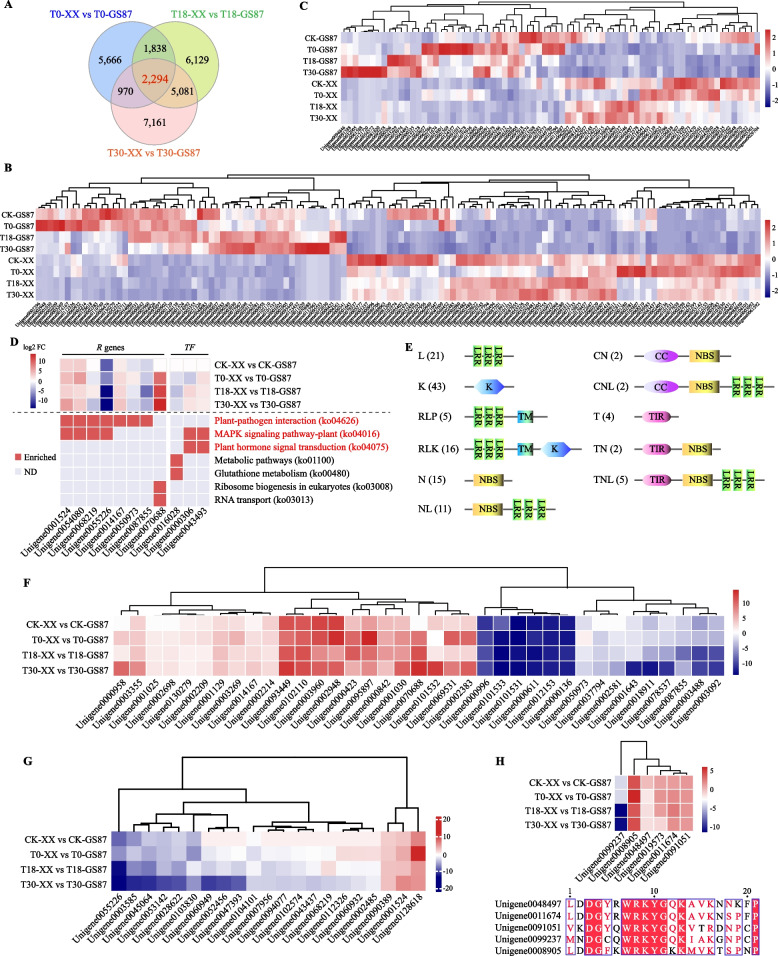
Furthermore, 2,294 shared DEGs were obtained from the overlap of the the Venn diagram of T0-XX vs T0-GS87, T18-XX vs T18-GS87, and T30-XX vs T30-GS87 comparisons (XX samples as controls) (Fig. 4A). Thus, these shared DEGs may be core genes regulating sweetpotato resistance to D. dadantii. This result prompted a more detailed analysis of 2,294 shared DEGs. First, functional annotation analysis showed that over 86% of these shared DEGs enriched 55 GO terms at level 2, and the three most enriched terms are “metabolic process” (848), “cellular process” (833), and “catalytic activity (809)” (Fig. S2). Additionally, 451 and 1,015 DEGs were annotated through KEGG and KOG databases, respectively (Fig. S2). The majority of the DEGs enriched the pathways of metabolisms, transcription, translation and signal transduction, implying that the sweetpotato made a global response to defend against D. dadantii. The putative protein sequences of 2,294 shared DEGs were aligned to PRGdb and plant TFdb databases, identifying 126 R genes and 73 TF genes (Table S7 and S8). The heatmap showed clear divisions in the gene expressions between GS87 and XX (Fig. 4B and C). Moreover, only eight R genes and three TF genes significantly enriched seven KEGG pathways, including “Plant-pathogen interaction (ko04626)”, “MAPK signaling pathway-plant (ko04016)”, and “Plant hormone signal transduction (ko04075)” as the three highly abundant pathways. Additionally, most of the 11 DEGs were highly expressed in inoculated GS87 samples than inoculated XX samples (Fig. 4D). These results suggested that the 11 genes and the relevant pathways may be important for regulating sweetpotato resistance to BSRR.
Fig. 4.
Analysis of the 2,294 DEGs shared by GS87 and XX comparisons at 0, 18, and 30 hpi. A Venn diagram showing the numbers of DEGs from GS87 and XX comparisons at 0, 18, and 30 hpi. XX samples were used as controls at each time point. B and C Expression heatmaps of the shared DEGs encoding 126 R proteins and 73 TFs. D Expression heatmap of 11 DEGs enriched in seven KEGG pathways. ND, not detectable. E Schematic structure of 126 R proteins within 11 families based on the disease resistance-related domain type. L, leucine-rich repeat receptor (LRR); K, kinase; RLP, receptor-like protein with transmembrane (TM) domain; RLK, receptor-like kinase with TM domain; N, nucleotide binding site (NBS); NL, NBS-LRR; CN, coiled-coil (CC)-NBS; CNL, CC-NBS-LRR; T, Toll/interleukin receptor (TIR); TN, TIR-NBS; TNL, TIR-NBS-LRR. F and G Clustering analysis of 37 R genes containing the NBS domain and 21 PRR (including RLP and RLK) genes based on log2FC values. H Clustering analysis of six WRKY genes within TF family log2FC values (top) and multiple amino acid sequence alignments of the core domain in the six WRKY proteins (bottom)
Furthermore, more than half of 126 R genes increased their expressions in XX relative to GS87 (Fig. 4B), suggesting that these genes may negatively respond to the D. dadantii hazard. The PRGdb 4.0 and SMART database searches indicated that these 126 R proteins were functionally divided into 11 distinct families based on the presence of specific disease resistance-relevant domains. Kinase, LRR, and NBS were the most frequent R protein domains (Fig. 4), suggesting that the corresponding domains confer host resistance against pathogens. Compared to XX samples, most DEGs encoding PRRs (including RLPs and RLKs) exhibited a reducing expression level in GS87 samples (Fig. 4G). Moreover, 37 DEGs comprised six NBS-LRR gene families, including N (NBS, 15), NL (NBS-LRR, 11), CN (CC-NBS, 2), CNL (CC-NBS-LRR, 2), TN (TIR-NBS, 2), and TNL (TIR-NBS-LRR, 5) (Fig. 4E, F). Over 50% of the NBS-LRR genes showed higher expression levels in GS87 than XX. Additionally, 73 TF DEGs were classified into 26 families, including one WRKY family with six members (Fig. 4H). Three of the WRKY unigenes (unigene0008905, unigene0011674, and unigene0091051) had higher expression levels in GS87 than XX. After excluding unigene0019573 (with an incomplete WRKY domain), the amino acid sequence alignment showed that the other five WRKY proteins shared a core domain characterized by highly conserved WRKYGQK residue and a common variant of WRKYGKK.
Jasmonic Acids (JAs) and Salicylic Acids (SAs) exhibited diverse dynamic response patterns
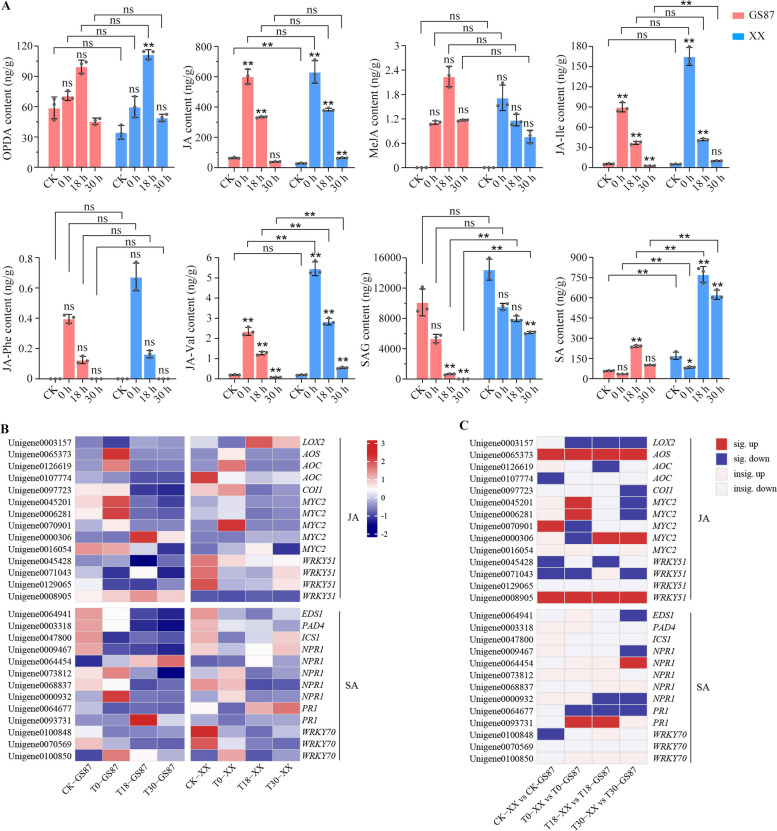
The qualitative and quantitative analysis of phytohormones in the sweetpotato samples showed a high sensitivity and accuracy in detecting JAs and SAs (Fig. S3 and S4).The levels of JA, methyl JA (MeJA), jasmonoyl-isoleucine (JA-Ile), jasmonoyl-phenalanine (JA-Phe), and jasmonoyl-valine (JA-Val) significantly peaked at 0 hpi (except OPDA) and then decreased gradually in the lowermost stem of GS87 and XX plants post-inoculation (Fig. 5A). Furthermore, the content of salicylic acid 2-O-b-D-glucose (SAG, a primary SA glucose conjugate mainly distributed in the vacuole) reduced significantly in the middle and later stage. Conversely, the SA content declined slightly and rose dramatically in the medium term before decreasing at the final time-point (Fig. 5A). Altogether, the JA content accumulated more quickly in both cultivars over the course of time than SA did.
Fig. 5.
Analysis of JA, SA, and their corresponding derivatives. A Histograms of phytohormone contents. OPDA, 12-oxo-phytodienoic acid; JA, jasmonic acid; JA-Phe, jasmonoyl-phenalanine; JA-Ile, jasmonoyl-isoleucine; JA-Val, jasmonoyl-valine; MeJA, methyl jasmonate; SAG, salicylic acid 2-O-β-D-glucose; SA, salicylic acid. B Expression patterns of JA- and SA-relevant unigenes in GS87 and XX. LOX, lipoxygenase, AOS, allene oxide synthase, AOC, allene oxide cyclase, ICS1, isochorismate synthase 1. C Unigene expression comparisons between GS87 and XX. Dark red and dark blue rectangles represent significantly up-regulated and down-regulated unigenes with FDR values of < 0.05 between groups. Light red and light blue rectangles represent up-regulated and down-regulated unigenes with FDR values of ≥ 0.05 between groups
Further, the study revealed that key genes involved in JA and SA biosynthesis and signal transduction pathways against D. dadantii. For JA pathway, the expressions of four MYC2 unigenes (Unigene0045201, Unigene0006281, Unigene0070901, and Unigene0016054) and several other unigenes (i.e., Unigene0065373 (AOS), Unigene0126619 (AOC), and Unigene0097723 (COI1)) were immediately up-regulated in GS87 and XX at 0 hpi, but they decreased over the time course compared to CK. The expression patterns of the abovementioned genes matched with the reduced content of JA and its derivatives at the same time point (Fig. 5A, B). Furthermore, unigene0003157 (LOX2) exhibited a distinct content pattern, which first reduced and later rose. Four other JA-relative WRKY-encoding unigenes were differentially expressed in GS87 and XX. In general, D. dadantii inoculation down-regulated the expression of most SA-relevant genes in GS87 and XX. However, most of the SA-related unigenes mentioned in this study were not statistically different between GS87 and XX, unlike the JA-related unigenes (Fig. 5C).
ROS and the relevant antioxidants of sweetpotato responded to D. dadantii infection
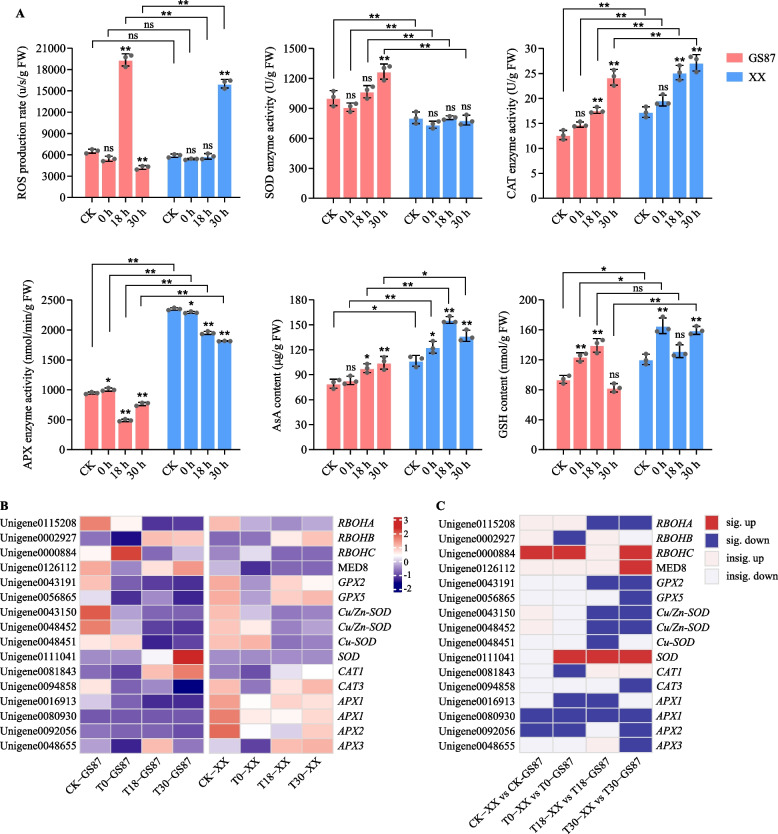
In consideration of the important role of ROS in plant disease resistance, ROS production rate, which reflects ROS content, in GS87 and XX after inoculation with D. dadantii were detected. The results showed that, compared to CK-GS87, ROS production rate in GS87 remained unchanged statistically at 0 hpi until GS87 dramatically increased ROS production rate at 18 hpi and then decreased it at 30 hpi. However, XX had a more delayed ROS production — ROS content peak appeared at 30 hpi (Fig. 6). Furthermore, no significant difference in ROS production rate at CK and 0 hpi between GS87 and XX were observed. These results indicated that GS87 was able to produce ROS more rapidly than XX. Subsequently, five ROS-relevant antioxidants were further analyzed. First of all, it was interesting that there was no obvious difference in SOD enzyme activities between infected XX at 0, 18, 30 hpi and CK-XX. For GS87, SOD enzyme activity remained a similar and low level at 0 and 18 hpi compared to CK-87,whereas it was statistically increased at 30 hpi, which may result from ROS burst. Additionally, except that SOD enzyme activity at 30 hpi in GS87was greater than that in XX, catalase (CAT) and ascorbate peroxidase (APX) enzyme activity, and ascorbic acid (AsA) and glutathione (GSH) contents in GS87 were lower than those in XX. These results implied that SOD in GS87 may play a dominate role in eliminating ROS in comparison to XX.
Fig. 6.
Analysis of ROS and the relevant enzymatic and non-enzymatic antioxidants. A Histograms of ROS production rate, enzymes (SOD, CAT, and APX) activity, and non-enzyme (AsA and GSH) content. ROS, reactive oxygen species; SOD, superoxide dismutase; CAT, catalase; APX, ascorbate peroxidase; AsA, ascorbic acid; GSH, glutathione. B Expression patterns of ROS and antioxidants-relevant unigenes in GS87 and XX, respectively. C Unigene expression comparisons between GS87 and XX. Dark red and dark blue rectangles represent significantly up-regulated and down-regulated unigenes with FDR values of < 0.05 between groups. Light red and light blue rectangles represent up-regulated and down-regulated unigenes with FDR values of ≥ 0.05 between groups
Subsequently, the dynamic expressed patterns of several key DEGs relevant to the biosynthesis or signal transduction of ROS and other antioxidants were analyzed based on the RNA-seq (Fig. 6). Unigene000088 (RBOHC as an ROS producer) and Unigene0126112 (MED8 as an SA enhancer) were significantly induced in infected GS87 than XX, and Unigene0126112 (MED8) exhibited later responses. In additional, the expression levels of Unigene0111041 encoding SOD at all treatment in GS87 were up-regulated compared to those in XX. The expressions of two CAT-encoding unigenes, Unigene0081843 and Unigene0094858, were also induced by D. dadantii attack, of which in GS87 were lower than those in XX. In consistent with the APX enzyme activity, AsA and GSH contents, all APXs and GPXs-encoding genes were down-regulated in GS87 compared to XX.
Hub proteins were focused through Protein–Protein interaction (PPI) network
After removing the interacting protein pairs with combined scores of < 500, a PPI network consisting of 2,338 proteins (including 429 R proteins and 180 TFs) was generated using 10,627 interacting protein pairs (Fig. S5). All four treatments shared 57 strictly hub R proteins, and the remaining 26 were detected at 0, 18, and 30 hpi. Furthermore, the R proteins were correlated to diverse TFs, such as WRKYs, bZIPs, and bHLHs. In some cases, an individual R protein interacted with more than one TF, and vice versa. The number of R proteins and TFs at 0 hpi exceeded those in the CK plants, but the numbers reduced at 18 and 30 hpi; meanwhile, the number of non-TFs or non-R proteins increased. These dynamic changes reflected a tradeoff of the sweetpotato disease resistance and growth, considering that the defense signals had been passed to the downstream proteins. In conclusion, these results demonstrated that sweetpotato rapidly builds a tremendous protein interaction network and increases the number of R proteins to deal with D. dadantii attack.
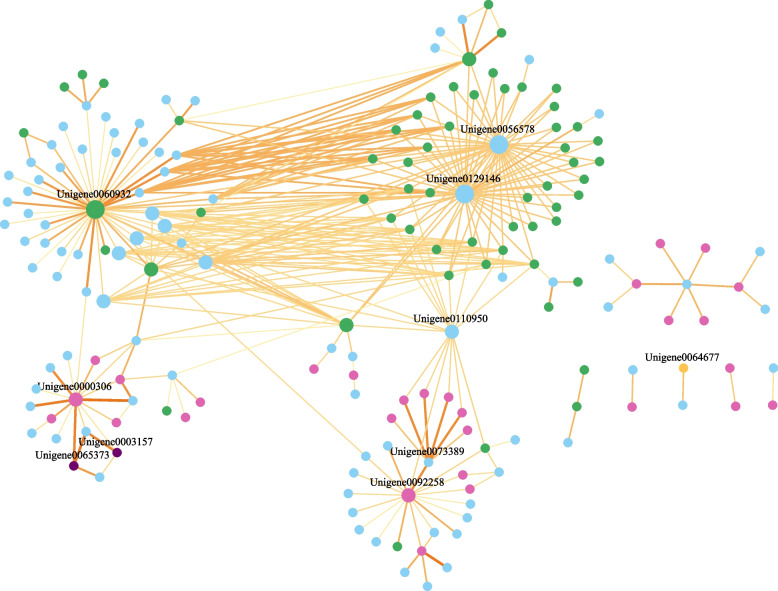
Furthermore, 180 proteins (R, TFs, JA-/SA-pathway-related proteins, and their corresponding interactors) generated a small-scale PPI network shared by GS87 and XX at 0, 18, and 30 hpi (Fig. 7). The small PPI network composed of 349 protein interaction pairs with two sub-networks. The dominant one had 158 proteins connected with others directly or indirectly, and another contained 22 proteins with low interconnectivity and being independent from the main network, probably due to the elimination of protein interactions with < 500 combined score. In the network, Unigene0060932 from RLK family and Unigene0129146 encoding acetyl-CoA carboxylase 1 (ACCase 1) showed the strongest connectivity degree since they directly linked with 45 other proteins, respectively (Table S5, S6 and S7). Thus, the two proteins may have crucial roles in response to BSRR. Unigene0129146 (ACCase 1) showed an interesting phenomenon by connecting directly to almost all R proteins, as did unigene0056578 (thiamin pyrophosphokinase 1, TPK1). In addition, unigene0110950 (glycerol-3-phosphate dehydrogenase3, GPD3) was another hub protein that formed a bridge between TFs and R proteins or other proteins. Unigene0000306 (MYC2) had a relatively high linkage with other proteins, including unigene0003157 (LOX2) and unigene0065373 (AOS).
Fig. 7.
Protein–Protein interaction (PPI) network analysis. Green dot, R protein; pink dot, TF; dark dot, protein involved in the JA pathway; yellow dot, protein involved in the SA pathway; blue dot, protein other than those mentioned above. The color shades and widths of the lines connecting any two proteins are positively correlated with the combined score of the interacting pairs. The dot size represents the protein connectivity. Unigene0060932, RLK protein; Unigene0056578, thiamine pyrophosphokinase 1-like (TPK1) protein; Unigene0129146, acetyl-CoA carboxylase 1 (ACCase 1) protein; Unigene0000306, MYC2; Unigene0003157, lipoxygenase2 (LOX2); Unigene0065373, allene oxide synthase (AOS); Unigene0110950, glycerol-3-phosphate dehydrogenase3 (GPD3); Unigene0073389, probable LRR receptor-like serine/threonine-protein kinase; Unigene0092258, MYB20; Unigene0064677, pathogenesis-related protein 1 (PR1)
Discussion
Retardation of D. dadantii infection made GS87 more resistant than XX
GS87 is an elite high-yield sweetpotato cultivar widely planted in southern China and is multi-resistant to BSRR, bacterial wilt, and Fusarium wilt. In contrast, XX is another elite cultivar with good taste but susceptible to BSRR. Thus, GS87 and XX are valuable germplasms for studying the molecular mechanism of sweetpotato resistance against BSRR, and the response of GS87 and XX to BSRR is considered for designing and adopting appropriate inoculation methods to reduce disease escape or tolerance resulting from below-threshold inoculation concentrations. In this study, GS87 and XX were inoculated with D. dadantii (the causal agent of BSRR) at concentrations of 107 cfu/mL and 108 cfu/mL. GS87 was more resistant to D. dadantii at 107 cfu/mL and 108 cfu/mL than XX, where D. dadantii caused severe disease with increasing inoculum concentrations. The results implied that 108 cfu/mL of the inoculum can distinguish resistance levels of sweetpotato plants to BSRR. Hence, the following experiments were performed using 108 cfu/mL.
The plant cell wall is the first guard line against phytopathogen intrusions. As one of the most crucial components of plant cell walls, pectin is closely involved in plant immunity [54, 55]. For survival and reproduction, phytopathogens have various strategies for penetrating plant cell walls. For example, D. dadantii secreted plant cell wall-degrading enzymes (especially pectinases) to counteract the pectic-based defense responses from host plants [56]. In this study, microscopic analysis showed a highly intact structure in the stems of GS87 infected with 108 cfu/mL of D. dadantii, a sharp contrast to the bacteria-inoculated XX plants, which exhibited severe cell decomposition and disorganization in the epidermis, cortex, and pith. However, the vascular bundles of GS87 and XX stayed robust. The above findings are similar to the previous observations of D. dadantii infection in A. thaliana leaves, where the bacteria thrived in the decomposed parenchyma cells at the symptomatic regions or colonized intercellular spaces and cell walls around the macerated areas. However, the bacterium was undetectable in the vascular tissues with no observable symptoms [25]. The pectin and lignin contents and corresponding regulator genes should be determined in further studies to understand better why D. dadantii caused different responses between GS87 and XX.
Several R proteins and TFs are involved in resistance to BSRR
RNA-seq is an easy tool for high-throughput analysis of gene expressions at the transcription level [57]. Over 55,000 literatures about RNA-seq from the past ten years can be retrieved from NCBI PubMed sub-database, indicating the wide application and significance of this technology. For sweetpotatoes, RNA-seq has been used to study gene expression patterns in different developmental stages and organs, the mechanisms of plant resistance to biotic and abiotic stress, and the regulation of quality [38, 41, 45, 58]. However, most RNA-seq studies of the D. dadantii–A. thaliana pathosystem were focused on D. dadantii rather than the host [4, 59–61]. Thus, advancing the understanding of plant resistance mechanisms against D. dadantii is of great significance. This study generated 54,844 expressed unigenes using a de no RNA-seq methods to investigate the transcription profiles of GS87 and XX plants challenged with D. dadantii. Though the number of up-/down-regulated genes in GS87 and XX increased over the time course, GS87 had more DEGs (including R genes and TF genes) at each time-point than XX. Altogether, GS87 and XX differ in their defense responses against D. dadantii, and GS87 deployed more genes from an early stage to increasingly induce its resistance.
A total of 2,294 DEGs were shared in three comparisons between GS87 and XX (T0-XX vs T0-GS87, T18-XX vs T18-GS87, T30-XX vs T30-GS87). Most DEGs-encoding proteins functionally enriched metabolic pathways, indicating that the immune response was an energy- and metabolic substance-cost process. Among the 2,294 DEGs, 126 R genes and 73 TF genes were annotated. Moreover, over 50% of the R genes were up-regulated in D. dadantii-challenged GS87 than XX, suggesting the ETI rather than the PTI system was mainly induced in GS87. This study provides the first insight into the global transcriptional responses of sweetpotatoes of different resistance after D. dadantii infection. Furthermore, it advances the understanding of the molecular mechanisms of sweetpotato defenses against D. dadantii.
Phytohormones, ROS, and relevant antioxidants are important for regulating resistance to D. dadantii in sweetpotatoes
Jasmonic acid and SA are vital phytohormones in plant immune reactions. Previous studies showed that JA performed a more dominant role in regulating the pathogenicity of D. dadantii [23, 25]. This study determined the contents of JA, SA, and their corresponding derivatives in D. dadantii-challenged GS87 and XX plants based on the uPLC-MS/MS platform. The contents of JA and its derivatives in GS87 and XX increased quickly and dramatically after infection with D. dadantii and were reduced over time. This phenomenon was consistent with the rapid expression of JA biosynthesis and signal transduction genes at the initial stage, indicating that both cultivars launch a quick JA-based response to D. dadantii. Surprisingly, the contents of JA and its derivatives (OPDA, MeJA, and JA-Phe) were not significantly different between GS87 and XX, except that JA-Ile and JA-Val levels were substantially higher in XX than in GS87. These results agree with the previous findings that JA has a chemoattractant effect on D. dadantii and assists the bacteria in penetrating through wounded plant stems [23]. Similarly, Taurino et al. [62] revealed a rapid induction of JA and JA-Ile contents in potato leaves caused by D. dadantii at the earliest time, followed by a continual reduction at the later stages. However, the potato StAOS1/2 transgenic plants deficient in JA biosynthesis showed a weak resistance to D. dadantii, suggesting a complicated regulatory role of JAs. In fact, many researchers have demonstrated that phytopathogens of various species have evolved various strategies to activate or repress JA biosynthesis and signaling transduction pathways for pathogenesis [63–67].
In terms of the SA pathway, the content patterns of SA and SAG differed from those of JAs. A delayed SA accumulation peaked at the final stages, and the content of SAG was constantly reducing over time, suggesting that SAG is converted to SA after infection by D. dadantii. In this regard, D. dadantii triggered the SA pathway in sweetpotato as in the previous Arabidopsis-D. dadantii study, which also discovered that SA was not likely involved in defense against the bacterial, although the abundance of the PR1 gene increased after bacterial inoculation [25]. In this study, the contents of JAs (except OPDA) and SA in GS87 and XX increased after D. dadantii inoculation. However, GS87 had lower contents of JA-Ile, JA-Val, SA, and SAG than XX at some time points, indicating that these four phytohormones may negatively regulate sweetpotato resistance to D. dadantii. Moreover, the expression trends of some genes involved in JA and SA biosynthesis and signal transduction pathways did not closely follow the changes in JAs and SAs content, implying that JAs and SAs contents in planta were correlated with other factors.
ROS burst in plants is an important sign for plant defense system activation when facing pathogen attacks, and its connections with JA and SA and functions of defense against pathogens were widely studied before. Myers et al. [68] demonstrated that JA hindered activated ROS wave in Arabidopsis responses to local wounding or strong light stress, while SA had the opposite effect. In this study, ROS was produced more quickly and earlier in GS87 than XX, and ROS burst pattern of GS87 seem to keep pace with SA level change mode. SOD, CAT, and APX are important enzymes in scavenging ROS, and AsA-GSH cycle was responsible for reducing H2O2 to H2O via APX, in which APX enzyme activity was negatively related to AsA content [69]. In this study, the difference in boosting SOD enzyme activity as well as in increasing ROS between GS87 and XX may play a key role in confering them different disease-resistance to D. dadantii, as the CAT and APX enzyme ability of GS87 were lower than those of XX. Accordingly, a contrary relationship between APX and AsA (or GSH) in GS87 and XX was observed in either GS87 and XX after pathogenic infection. It was possible that GS87 may adopt other strategies like allocating more metabolic resources to other defense mechanisms rather than relying on CAT or APX.
Significant hub proteins uncovered in the PPI network may be utilized in the resistance study
Three hub proteins, Unigene0060932 (RLK), Unigene0056578 (TPK1), and Unigene0129146 (ACCase 1), were highly connected with other proteins in the PPI network, suggesting their vital functions in regulating resistance to D. dadantii. Previous studies showed that TPK1 is the key enzyme responsible for the thiamine (namely vitamin B1) pyrophosphorylation to thiamine pyrophosphate (TPP) in the cytosol. Thiamine reportedly induces plant disease resistance through an SA-dependent pathway [70, 71]. Futhermore, TPP serves as a coenzyme that assists the production of acetyl-CoA, and ACCase 1 is a restriction enzyme involved in fatty acid biosynthesis and participates in the first step of acetyl-CoA catalysis into malonyl-CoA [72]; hence there is an indirect link between Unigene0056578 (TPK1) and Unigene0129146 (ACCase 1). Moreover, malonyl-CoA is a biosynthesis substrate of fatty acids and many secondary metabolites, including flavonoid, which are involved in defence against pathogens [73]. A previous study in common bean showed that ACCase accumulation after pathogen infection and JA application induces defense [74]. Unexpectedly, the average level of Unigene0129146 in GS87 was 2.5 times lower than XX at 0, 18, and 30 hpi. Moreover, the fragments per kilobase of transcript per million mapped fragments (FPKM) values were low in both cultivars (unpublished data), hence the role of ACCase in regulating R proteins in sweetpotato remains unclear.
Conclusions
This study analyzed the comparative time-series RNA-seq profiles and plant disease-relevant phytohormones patterns in resistant (GS87) and susceptible (XX) sweetpotato cultivars challenged by D. dadantii (the causal agent of BSSR). First, D. dadantii-challenged GS87 maintains a stable resistance and overcomes BSRR infection even at higher inoculum concentrations by retaining a highly intact structure in the stem and regulating more genes (especially R genes) than XX. Moreover, the JAs and SAs of sweetpotato are involved in response to D. dadantii infection, and they may have potentially negative effects on sweetpotato resistance to D. dadantii. Lastly, ROS and SOD play vital roles in confering GS87 resistance against D. dadantii. In sum, these findings advance the understanding of molecular differences between sweetpotatoes with variant resistance to D. dadantii and provide core R genes and other hub DEGs as candidates for molecular BSSR-resistance breeding.
Supplementary Information
Supplementary Material 1: Table S1. The primer sequences of nine unigenes. Table S2. GO classifications for 28,731 unigenes Table S3. KEGG pathway enrichments of 10,019 unigenes. a: The number of unigenes enriched in the KEGG B-class and corresponding pathway were listed, respectively. Table S4. KOG classifications for 22,227 unigenes. Table S5. Functional annotation analysis of 34,287 DEGs detected within GS87 samples. a: Classifications of resistance (R) proteins based on PRGdb 4.0 database blast. b: TF: transcription factor. Table S6. Functional annotation analysis of 17,989 DEGs detected within XX samples a: Classifications of resistance (R) proteins based on PRGdb 4.0 database blast. b: TF: transcription factor. Table S7. Information of 126 Ra genes a: Classifications of resistance (R) proteins based on PRGdb 4.0 database blast. b: FC: fold change. XX samples were used as control groups in four distinct comparisons. c: NA: no significant difference. Table S8. Information of 73 TFa genes. a: TF: transcription factor. Classifications of TFs based on plant TFdb database blast. b: FC: fold change. XX samples were used as control groups in four distinct comparisons. c: NA: no significant difference.
Supplementary Material 2: Fig. S1. Validation of RNA-seq data using qRT-PCR. A The relative expressions of nine unigenes in the GS87 and XX samples at CK, 0, 18, and 30 hpi. R genes, unigene0095897, unigene0110749, unigene0101533, and unigene0001418; Pathogenesis-related (PR) genes, unigene0064677 and unigene0000450; WRKY genes, unigene0008905 and unigene0048497, Phenylalanine ammonia-lyase-like (PAL) gene, unigene0047476. The β-actin gene of sweetpotato served as the internal control gene. Data is the mean ± SD (n = 3). **P < 0.01; 0.01 < *P < 0.05; ns: statistically nonsignificant. B The linear regression model described the linear relationship between results of RNA-seq and qRT-PCR, with log2FC determined in RNA-seq and by qRT-PCR in x- and y- axes respectively. Fig. S2. Functional annotation of the 2294 DEGs shared between GS87 and XX at 0, 18, and 30 hpi. A A total of 1,990 DEGs endowed with 55 GO terms (level 2). B A total of 451 DEGs enriched in 19 KEGG pathways. c A total of 1,015 DEGs classified into 25 KOG classes. Fig. S3. The total ion chromatograms (TICs) of each replicates of GS87 and XX. Fig. S4. The regression equation and extracted ion chromatograms (EICs) of JAs and SAs detected in each replicates of GS87 and XX. Fig. S5. Protein-Protein interaction (PPI) network lined by 2,338 proteins collected from four comparisons. Red dots: R proteins shared at four treatments. Green dots: R proteins shared at 0, 18, and 30 hpi. Light blue dots: R proteins unique at each treatment. Triangles: TFs. Squares: proteins of other types. Pink lines highlighted the interactions of R and WRKY proteins, and the grey lines highlighted the other type of interactions.
Acknowledgements
Not applicable.
Abbreviations
- ACCase 1
Acetyl-CoA carboxylase 1
- AOC
Allene oxide cyclase
- AOS
Allene oxide synthase
- APX
Ascorbate peroxidase
- AsA
Ascorbic acid
- BSRR
Bacterial stem and root rot
- COI1
Coronatine insensitive1
- DAMP
Damage-associated molecular patterns
- CAT
Catalase
- DEG
Differentially expressed genes
- ETI
Effector-triggered immunity
- FPKM
Fragments per kilobase of transcript per million mapped fragments
- GPD3
Glycerol-3-phosphate dehydrogenase3
- GSH
Glutathione
- JA
Jasmonic acid
- JA-Ile
Jasmonoyl-isoleucine
- JA-Phe
Jasmonoyl-phenalanine
- JA-Val
Jasmonoyl-valine
- JAZ
Jasmonate ZIM-domain
- LOX
Lipoxygenase
- LRR-RLK
Leucine-rich repeat receptor-like kinase
- MAPK
Mitogen-activated protein kinases
- MeJA
Methyl JA
- MS
Mass spectrometry
- NBS-LRR/NLR
Nucleotide binding site-LRR
- OPDA
12-Oxo-phytodienoic acid
- PAMP
Pathogen-associated molecular patterns
- PPI
Protein-Protein interaction
- PRR
Pattern-recognition receptor
- PTI
Pattern-triggered immunity
- RNA-seq
RNA sequencing
- ROS
Reactive oxygen species
- qRT-PCR
Quantitative Real-time PCR
- SA
Salicylic acid
- SAG
Salicylic acid 2-O-b-D-glucose
- SAR
Systemic acquired resistance
- SOD
Superoxide dismutase
- TPK1
Thiamin pyrophosphokinase 1
- TPP
Thiamine pyrophosphate
Author’s contributions
LH supervised the project. LH, BF, SYX, and JC designed the experiments. SYX, NZ, SL, and TM conducted plant material preparation, sampling, and bacterial inoculation. SYX conducted all the other experiments, analyzed the data and wrote the manuscript; LH, BF and JC revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the earmarked fund from CARS-10-Sweetpotato, and the Sweetpotato Potato Innovation Team of the Modern Agricultural Industry Technology System in Guangdong Province (2023KJ111).
Data availability
The datasets generated and/or analysed during the current study are available in NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/?term =) with BioProject ID (PRJNA1162560).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang L, Fang B, Chen J, Zhang X, Luo Z, Yang Y, et al. Sweetpotato disease identification and control guide. Beijing, China: China Agriculture Press; 2020. [Google Scholar]
- 2.Huang LF, Fang BP, Luo ZX, Chen JY, Zhang XJ, Wang ZY. First report of bacterial stem and root rot of sweetpotato caused by a Dickeya sp (Erwinia chrysanthemi) in China. Plant Dis. 2010;94:1503. [DOI] [PubMed] [Google Scholar]
- 3.Chapelle E, Alunni B, Malfatti P, Solier L, Pédron J, Kraepiel Y, et al. A straightforward and reliable method for bacterial in planta transcriptomics: application to the Dickeya dadantii/Arabidopsis thaliana pathosystem. Plant J. 2015;82:352–62. [DOI] [PubMed] [Google Scholar]
- 4.Pédron J, Chapelle E, Alunni B, Van Gijsegem F. Transcriptome analysis of the Dickeya dadantii PecS regulon during the early stages of interaction with Arabidopsis thaliana. Mol Plant Pathol. 2018;19:647–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aznar A, Patrit O, Berger A, Dellagi A. Alterations of iron distribution in Arabidopsis tissues infected by Dickeya dadantii. Mol Plant Pathol. 2015;16:521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieu NP, Aznar A, Segond D, Rigault M, Simond-Côte E, Kunz C, et al. Iron deficiency affects plant defence responses and confers resistance to Dickeya dadantii and Botrytis cinerea. Mol Plant Pathol. 2012;13:816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dellagi A, Rigault M, Segond D, Roux C, Kraepiel Y, Cellier F, et al. Siderophore-mediated upregulation of Arabidopsis ferritin expression in response to Erwinia chrysanthemi infection. Plant J. 2005;43:262–72. [DOI] [PubMed] [Google Scholar]
- 8.Segond D, Dellagi A, Lanquar V, Rigault M, Patrit O, Thomine S, et al. NRAMP genes function in Arabidopsis thaliana resistance to Erwinia chrysanthemi infection. Plant J. 2009;58:195–207. [DOI] [PubMed] [Google Scholar]
- 9.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9. [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–11. [DOI] [PubMed] [Google Scholar]
- 11.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–60. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Claus LAN, Leslie ME, Tao K, Wu Z, Liu J, et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature. 2020;581:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. [DOI] [PubMed] [Google Scholar]
- 14.Deng Y, Zhai K, Xie Z, Yang D, Zhu X, Liu J, et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017;355:962–5. [DOI] [PubMed] [Google Scholar]
- 15.He H, Zhu S, Zhao R, Jiang Z, Ji Y, Ji J, et al. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol Plant. 2018;11:879–82. [DOI] [PubMed] [Google Scholar]
- 16.Michelmore RW, Christopoulou M, Caldwell KS. Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol. 2013;51:291–319. [DOI] [PubMed] [Google Scholar]
- 17.Xing L, Hu P, Liu J, Witek K, Zhou S, Xu J, et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol Plant. 2018;11:874–8. [DOI] [PubMed] [Google Scholar]
- 18.Ngou BPM, Ahn H-K, Ding P, Jones JD. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 2021;592:110–5. [DOI] [PubMed] [Google Scholar]
- 19.Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021;592:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature. 2021;598:495–9. [DOI] [PubMed] [Google Scholar]
- 21.Tian H, Wu Z, Chen S, Ao K, Huang W, Yaghmaiean H, et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature. 2021;598:500–3. [DOI] [PubMed] [Google Scholar]
- 22.Zhai K, Liang D, Li H, Jiao F, Yan B, Liu J, et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature. 2022;601:245–51. [DOI] [PubMed] [Google Scholar]
- 23.Antunez-Lamas M, Cabrera E, Lopez-Solanilla E, Solano R, González-Melendi P, Chico JM, et al. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol Microbiol. 2009;74:662–71. [DOI] [PubMed] [Google Scholar]
- 24.Denancé N, Sánchez-Vallet A, Goffner D, Molina A. Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci. 2013;4:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagard M, Dellagi A, Roux C, Périno C, Rigault M, Boucher V, et al. Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol Plant Microbe Interact. 2007;20:794–805. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Li Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021;17:e1009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D, Wang J, Xie D. Jasmonate action and biotic stress response in plants. Biotechnol Bull. 2018;34:14–23. [Google Scholar]
- 28.Zhang L, Zhang F, Melotto M, Yao J, He SY. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot. 2017;68:1371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CL, Yuan F, Li XY, Ma RC, Xie H. Jasmonic acid and ethylene signaling pathways participate in the defense response of Chinese cabbage to Pectobacterium carotovorum infection. J Integr Agric. 2021;20:1314–26. [Google Scholar]
- 30.Waszczak C, Carmody M, Kangasjarvi J. Reactive oxygen species in plant signaling. Annu Rev Plant Biol. 2018;69:209–36. [DOI] [PubMed] [Google Scholar]
- 31.Huang L, Chen J, Fang B, Luo Z, Zhang X, Wang Z. Pathogenic differentiation and genetic diversity of Dickeya dadantii causing bacterial stem and root rot of sweetpotato. J Plant Prot. 2018;45:1227–34. [Google Scholar]
- 32.Huang L, Luo Z, Fang B, Chen J, Zhang X, Wang Z. A new bacterial stem and root rot disease of sweetpotato in Guangdong. China Acta Phytopathol Sin. 2011;41:18–23. [Google Scholar]
- 33.Lou B, Shen X, Lu G, Lin X, Li Y, Zhang Y, et al. Resistance determination of sweet potato germplasm resources to sweet potato rot. J Zhejiang Agric Sci. 2018;59:2204–7. [Google Scholar]
- 34.Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, et al. Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact. 2009;22:589–600. [DOI] [PubMed] [Google Scholar]
- 35.Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, et al. Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol J. 2019;17:1119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Wang H, Jiang J, Liu M, Liu Z, Tan Y, et al. The Sm gene conferring resistance to gray leaf spot disease encodes an NBS-LRR (nucleotide-binding site-leucine-rich repeat) plant resistance protein in tomato. Theor Appl Genet. 2022;135:1467–76. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Zhang Q, Wang YN, Li Y, ZHai H, Liu QC, et al. Characterization of salt tolerance and Fusarium wilt resistance of a sweetpotato mutant. J Integr Agric. 2017;16:1946–55. [Google Scholar]
- 38.Zhang L, Yu Y, Shi T, Kou M, Sun J, Xu T, et al. Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals the regulatory architecture of gene expression variation in the storage roots of sweet potato. Hortic. 2020;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C, Yao W, Wang J, Wang J, Ai Y, Ma H, et al. A novel effect of glycine on the growth and starch biosynthesis of storage root in sweetpotato (Ipomoea batatas Lam). Plant Physiol Biochem. 2019;144:395–403. [DOI] [PubMed] [Google Scholar]
- 40.Li R, Zhai H, Kang C, Liu D, He S, Liu Q. De novo transcriptome sequencing of the orange-fleshed sweet potato and analysis of differentially expressed genes related to carotenoid biosynthesis. Int J Genomics. 2015;2015:843902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi J, Zhao L, Yan B, Zhu Y, Ma H, Chen W, et al. Comparative transcriptome analysis reveals the transcriptional alterations in growth- and development-related genes in sweet potato plants infected and non-infected by SPFMV, SPV2, and SPVG. Int J Mol Sci. 2019;20:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Zhang Q, Zhai H, Li Y, Wang X, Liu Q, et al. Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci Rep. 2017;7:40819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark CA, Wilder-Ayers JA, Duarte V. Resistance of sweet potato to bacterial root and stem rot caused by Erwinia Chrysanthemi. Plant Dis. 1989;73:984–7. [Google Scholar]
- 44.Wu Y, Zhao S, Li X, Zhang B, Jiang L, Tang Y, et al. Deletions linked to PROG1 gene participate in plant architecture domestication in Asian and African rice. Nat Commun. 2018;9:4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Kou M, Arisha MH, Tang W, Ma M, Yan H, et al. Transcriptomic and metabolic profiling of high-temperature treated storage roots reveals the mechanism of saccharification in sweetpotato (Ipomoea batatas (L) Lam). Int J Mol Sci. 2021;22:6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. [DOI] [PubMed] [Google Scholar]
- 49.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Zhou C, Yan X, Zhang J, Xu J. Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J Sep Sci. 2016;39:1804–13. [DOI] [PubMed] [Google Scholar]
- 52.Floková K, Tarkowská D, Miersch O, Strnad M, Wasternack C, Novák O. UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry. 2014;105:147–57. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 54.Wan J, He M, Hou Q, Zou L, Yang Y, Wei Y, et al. Cell wall associated immunity in plants. Stress Biol. 2021;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D, Kanyuka K, Papp-Rupar M. Pectin: a critical component in cell-wall-mediated immunity. Trends Plant Sci. 2023;28:10–3. [DOI] [PubMed] [Google Scholar]
- 56.Martis BS, Droux M, Nasser W, Reverchon S, Meyer S. Carbon catabolite repression in pectin digestion by the phytopathogen Dickeya dadantii. J Biol Chem. 2022;298:101446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi YX, Liu YB, Rong WH. RNA-seq and its applications: a new technology for transcriptomics. Hereditas. 2011;33:1191–202. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, Zhang Q, Zhai H, Gao S, Yang L, Wang Z, et al. IbBBX24 promotes the jasmonic acid pathway and enhances Fusarium wilt resistance in sweet potato. Plant Cell. 2020;32:1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costechareyre D, Chich JF, Strub JM, Rahbé Y, Condemine G. Transcriptome of Dickeya dadantii infecting Acyrthosiphon pisum reveals a strong defense against antimicrobial peptides. PLoS ONE. 2013;8:e54118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X, Zghidi-Abouzid O, Oger-Desfeux C, Hommais F, Greliche N, Muskhelishvili G, et al. Global transcriptional response of Dickeya dadantii to environmental stimuli relevant to the plant infection. Environ Microbiol. 2016;18:3651–72. [DOI] [PubMed] [Google Scholar]
- 61.Forquet R, Jiang X, Nasser W, Hommais F, Reverchon S, Meyer S. Mapping the complex transcriptional landscape of the phytopathogenic bacterium Dickeya dadantii. Bio. 2022;13:e0052422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taurino M, Abelenda JA, Río-Alvarez I, Navarro C, Vicedo B, Farmaki T, et al. Jasmonate-dependent modifications of the pectin matrix during potato development function as a defense mechanism targeted by Dickeya dadantii virulence factors. Plant J. 2014;77:418–29. [DOI] [PubMed] [Google Scholar]
- 63.Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD. A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PloS Biol. 2013;11:e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patkar RN, Benke PI, Qu Z, Constance Chen YY, Yang F, Swarup S, et al. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat Chem Biol. 2015;11:733–40. [DOI] [PubMed] [Google Scholar]
- 65.Thatcher LF, Gardiner DM, Kazan K, Manners JM. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol Plant Microbe Interact. 2012;25:180–90. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L, Yao J, Withers J, Xin XF, Banerjee R, Fariduddin Q, et al. Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proc Natl Acad Sci U S A. 2015;112:14354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Z, Wu Y, Yang Y, Du M, Zhang X, Guo Y, et al. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell. 2015;27:2032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Myers RJ, Fichman Y, Zandalinas SI, Mittler R. Jasmonic acid and salicylic acid modulate systemic reactive oxygen species signaling during stress responses. Plant Physiol. 2023;191:862–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S. Novel insight into functions of ascorbate peroxidase in higher plants: More than a simple antioxidant enzyme. Redox Biol. 2023;64:102789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahn IP, Kim S, Lee YH. Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol. 2005;138:1505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahn IP, Kim S, Lee YH, Suh SC. Vitamin B1-induced priming is dependent on hydrogen peroxide and the NPR1 gene in Arabidopsis. Plant Physiol. 2007;143:838–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye Y, Nikovics K, To A, Lepiniec L, Fedosejevs ET, Van Doren SR, et al. Docking of acetyl-CoA carboxylase to the plastid envelope membrane attenuates fatty acid production in plants. Nat Commun. 2020;11:6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol. 2021;63:180–209. [DOI] [PubMed] [Google Scholar]
- 74.García-Ponce B, Rocha-Sosa M. The octadecanoid pathway is required for pathogen-induced multi-functional acetyl-CoA carboxylase accumulation in common bean (Phaseolus vulgaris L). Plant Sci. 2000;157:181–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Table S1. The primer sequences of nine unigenes. Table S2. GO classifications for 28,731 unigenes Table S3. KEGG pathway enrichments of 10,019 unigenes. a: The number of unigenes enriched in the KEGG B-class and corresponding pathway were listed, respectively. Table S4. KOG classifications for 22,227 unigenes. Table S5. Functional annotation analysis of 34,287 DEGs detected within GS87 samples. a: Classifications of resistance (R) proteins based on PRGdb 4.0 database blast. b: TF: transcription factor. Table S6. Functional annotation analysis of 17,989 DEGs detected within XX samples a: Classifications of resistance (R) proteins based on PRGdb 4.0 database blast. b: TF: transcription factor. Table S7. Information of 126 Ra genes a: Classifications of resistance (R) proteins based on PRGdb 4.0 database blast. b: FC: fold change. XX samples were used as control groups in four distinct comparisons. c: NA: no significant difference. Table S8. Information of 73 TFa genes. a: TF: transcription factor. Classifications of TFs based on plant TFdb database blast. b: FC: fold change. XX samples were used as control groups in four distinct comparisons. c: NA: no significant difference.
Supplementary Material 2: Fig. S1. Validation of RNA-seq data using qRT-PCR. A The relative expressions of nine unigenes in the GS87 and XX samples at CK, 0, 18, and 30 hpi. R genes, unigene0095897, unigene0110749, unigene0101533, and unigene0001418; Pathogenesis-related (PR) genes, unigene0064677 and unigene0000450; WRKY genes, unigene0008905 and unigene0048497, Phenylalanine ammonia-lyase-like (PAL) gene, unigene0047476. The β-actin gene of sweetpotato served as the internal control gene. Data is the mean ± SD (n = 3). **P < 0.01; 0.01 < *P < 0.05; ns: statistically nonsignificant. B The linear regression model described the linear relationship between results of RNA-seq and qRT-PCR, with log2FC determined in RNA-seq and by qRT-PCR in x- and y- axes respectively. Fig. S2. Functional annotation of the 2294 DEGs shared between GS87 and XX at 0, 18, and 30 hpi. A A total of 1,990 DEGs endowed with 55 GO terms (level 2). B A total of 451 DEGs enriched in 19 KEGG pathways. c A total of 1,015 DEGs classified into 25 KOG classes. Fig. S3. The total ion chromatograms (TICs) of each replicates of GS87 and XX. Fig. S4. The regression equation and extracted ion chromatograms (EICs) of JAs and SAs detected in each replicates of GS87 and XX. Fig. S5. Protein-Protein interaction (PPI) network lined by 2,338 proteins collected from four comparisons. Red dots: R proteins shared at four treatments. Green dots: R proteins shared at 0, 18, and 30 hpi. Light blue dots: R proteins unique at each treatment. Triangles: TFs. Squares: proteins of other types. Pink lines highlighted the interactions of R and WRKY proteins, and the grey lines highlighted the other type of interactions.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/?term =) with BioProject ID (PRJNA1162560).