Abstract
Morphological features of organismal body plans are often highly conserved within large taxa. For example, segmentation is a shared and defining feature of all insects. Screens in Drosophila identified genes responsible for the development of body segments, including the “pair-rule” genes (PRGs), which subdivide embryos into double-segment units in a previously unexpected pre-patterning step. Here we show that the milkweed bug Oncopeltus fasciatus also uses a pair rule for embryo subdivision but Oncopeltus employs different genes for this process. We identified the gene Blimp1 as an Oncopeltus PRG based on its expression pattern, tested its function with RNA interference and CRISPR-Cas9, and generated the first PR mutant in this species. Although it does not have PR function in Drosophila, like Drosophila PRGs, Blimp1 encodes a transcription factor required for embryonic viability. Thus, pair-rule subdivision of the insect body plan is more highly conserved than the factors mediating this process, suggesting a developmental constraint on this pre-patterning step.
Variable segmentation gene cohorts give rise to a conserved insect body plan.
INTRODUCTION
While phenotypic diversity relies on genotypic changes, the reverse is not always true: The same phenotype can be derived from different developmental processes and/or driven by different genes in different species, a phenomenon named developmental systems drift [DSD, (1, 2)]. A classic example of DSD comes from observations of vulval development in two different species of nematodes, where mutants for the homeotic gene lin-39 lack a vulva in both species. In one species, this phenotype arises through cell death, while in the other species, it arises through a redirection of cell fate (3). The extensive turnover of genes required for proper segment formation documented in insects (reviewed below) presents another clear example of underlying evolutionary change despite conservation of phenotype.
Although they are incredibly diverse, arthropods share an embryonic stage that looks remarkably similar across taxa—the segmented germ band (4). At this stage, the embryo is fully elongated and the anterior-posterior (AP) axis is divided into discrete units—segments—by intersegmental grooves. Observations that the gene engrailed (en) is expressed in the posterior of each segment in all species examined (5) provided further evidence for the high degree of conservation of this stage across taxa. Diversification in form and function of each segment later in development occurs after the basic body plan is established at the segmented germ-band stage and is evident by the incredible morphological diversity seen across insect lineages. Less obvious but also long appreciated is the diversity of developmental modes used across lineages to subdivide the AP axis and reach the segmented germ-band stage. In some insects, the segments are patterned around the same time at the blastoderm stage, the so-called simultaneous or long-germ mode of segmentation. In others, segment specification occurs sequentially, with anterior segments patterned first (in some lineages at blastoderm stage), and the remaining segments added sequentially from a posterior segment addition zone (SAZ), the so-called sequential or short-germ mode of segmentation (6). These classifications were initially made experimentally by observing which segments form after pinching or cauterizing embryos (4) but more recently have been based on gene expression patterns (7).
Our knowledge of how segmenting the AP axis is achieved at the molecular level comes mainly from the model organism Drosophila melanogaster, which forms segments simultaneously. In screens for embryonic patterning mutants, a class of mutant phenotypes was found in which an equivalent portion of every other body segment was missing (8). These phenotypes suggested that the first periodic pattern to be defined along the AP axis of the embryo has double-segment periodicity and follows what was termed a “pair rule” (8). The existence of this pre-pattern with two-segment periodicity that is established before morphological segments form was later supported by the expression patterns of the cloned genes: Most of the nine Drosophila pair-rule genes (PRGs) are expressed in the primordia of every other segment, the regions missing in the corresponding mutants. These expression patterns provided a direct visualization of the double-segment periodicity of these genes’ activity across the body axis. For example, the expression patterns of even-skipped (eve) and fushi tarazu (ftz) were found to be roughly complementary to each other, each corresponding to the primordia of one segment-wide unit, or half of the double-segment subdivision, with the entire unit (eve + ftz) repeated multiple times along the entire AP axis (9). Another five PRGs were found to be expressed in other complementary or staggered striped patterns (10, 11). All of the PRGs encode transcription factors that are required for proper establishment of en stripes in the posterior of every segment where it helps to maintain segment boundaries. The existence of PRGs and this intermediate pre-patterning step defining alternate segment primordia was unexpected, and thus the question of how widely such pair-rule (PR) patterning occurs outside of Drosophila has interested researchers since its discovery.
The conservation of PR patterning has been approached by analyzing the expression, and, in some cases, the function, of orthologs of the Drosophila PRGs in arthropod species for which these assays are tractable. Early studies using this comparative molecular genetics approach suggested that PR patterning was limited to simultaneously segmenting insects. For instance, in a sequentially segmenting grasshopper, Eve was found to localize broadly in the posterior SAZ during germ-band elongation rather than in stripes (12). Similarly, grasshopper Ftz was observed in the posterior germ band and the presumptive central nervous system but not in the primordia of alternate segments as in Drosophila (13). However, the notion that the presence or absence of PR patterning correlates with segmentation mode was abandoned after PR-like alternate-segment expression of eve was observed in several species of beetles displaying different segmentation modes (14). Since that time, the sequentially segmenting beetle Tribolium castaneum has become the leading model for sequentially segmenting insects, and an unbiased genetic screen identified two PR mutants (itchy and scratchy) in this species (15).
While patterning segments by a pair rule may be conserved between Drosophila and Tribolium, is the same PRG cohort used in each case? This question has been evaluated across many in-depth studies of the expression and function of the Tribolium orthologs of the Drosophila PRGs. Tribolium orthologs of paired (prd) and sloppy paired (slp) displayed clear PR alternate-segment deletions when knocked down using RNA interference (RNAi) (16), and these were later found to be the genes mutated in the scratchy and itchy mutants, respectively (17). RNAi targeting the Tribolium orthologs of eve, odd-skipped (odd), and runt (run) yielded embryos with axial elongation defects displaying only head structures (16), suggesting that roles for these genes in germ-band elongation may preclude detection of roles in segment delineation. However, milder RNAi knockdown of eve and odd produced partial PR-like segmentation defects in both Tribolium and another beetle Dermestes maculatus (18–20). Yet, for orthologs of Drosophila hairy and ftz, no PR-like segmentation defects have been reported following depletion in Tribolium (16, 21). This body of work (i) demonstrated that a pre-patterning pair rule exists outside of Drosophila and that (ii) a pair rule also acts to subdivide the embryo for species that add segments sequentially. It further suggests that (iii) several of Drosophila PRGs have retained PR function since the divergence of Coleoptera and Diptera over 300 million years ago (22) and (iv) highlights the difficulty in assessing gene function due to the pleiotropic roles of some genes of interest in sequentially segmenting species as well as (v) the challenges of comparing mutant Drosophila phenotypes to much more variable RNAi knockdown phenotypes.
In several other non-model insects, many of the Drosophila PRG orthologs have likewise been found to be part of the PRG cohort. In the lepidopteran Bombyx mori, the Drosophila PRG orthologs eve, odd, and run are expressed in PR patterns (23, 24). In Hymenoptera, PR expression was demonstrated for four PRG orthologs in the honeybee (25) and six PR orthologs in a wasp (26, 27). Outside of Holometabola, PR-like expression of fushi tarazu factor 1 (ftz-f1) was observed in a cricket (28). Even in some more distantly related arthropods, such as centipedes and a spider mite, some Drosophila PRG orthologs were reported to display PR-like expression (29, 30).
However, other studies have shown that individual PRG orthologs have been gained, lost, or changed function during insect radiations. For example, the Drosophila PRG ftz likely arose as a duplication of the Hox gene Antennapedia and in some extant lineages maintains a Hox-like expression pattern. In the lineage leading to Drosophila, however, a switch in cofactor interaction motifs and expression pattern likely allowed ftz to enter the PRG network (31). In mosquitoes, an ortholog of the Drosophila PRG prd has not been found, while it exists in more basally branching lineages, suggesting that this gene was lost entirely in this group. Its function was replaced by family member gooseberry (gsb), as gsb mutants display PR segmentation defects in Anopheles (32). In some insect species much more distantly related to Drosophila, lack of PR function for several PRG orthologs has been observed. In the cockroach Periplaneta, hairy is expressed segmentally and in the SAZ rather than in a PR manner, and its knockdown did not result in PR defects (33). Similarly, segmental expression of slp, run, and eve, was observed in this species, and RNAi did not produce clear PR defects for any gene (34).
In the sequentially segmenting milkweed bug Oncopeltus fasciatus, orthologs of the Drosophila PRGs are expressed segmentally rather than in a PR pattern and RNAi knockdown did not produce PR defects for any of these orthologs (35–37). Does a pair rule exist in Oncopeltus? The transcription factor–encoding gene E75A is expressed in alternate segment primordia in Oncopeltus, and RNAi knockdown produced PR-like defects (38). Oncopeltus is therefore a particularly interesting species in which to study PR patterning, as the only PR regulator identified to date (E75A) is not involved in segmentation in Drosophila. The combinatorial inputs of nine PRGs in Drosophila regulate downstream genes to spatially define segment formation; it therefore seems unlikely that this complex process could be accomplished by a single gene, and thus it is likely that additional PRGs remain to be characterized in Oncopeltus.
Seeking an unbiased approach for Oncopeltus segmentation gene discovery, we produced transcriptomes from three stages during Oncopeltus embryogenesis and began an in situ hybridization–based screen of transcription factor–encoding genes coexpressed with E75A. Here, we show that one gene examined during this screen, B lymphocyte–induced maturation protein 1 (Of-Blimp1, also known as prdm1 in vertebrates), displays PR-like expression in the blastoderm and during much of germ-band elongation. Blimp1 is a member of the PRDM gene family; like other members of this family, it encodes a transcription factor with a SET-like domain. Blimp1 orthologs sampled across Metazoa additionally share five C-terminal zinc fingers (39). We have investigated the function of Of-Blimp1 using RNAi, somatic CRISPR, and used a co-CRISPR strategy to obtain germline CRISPR mutants. Phenotypes derived from all three functional perturbations are consistent with Blimp1 being a PRG in Oncopeltus. Thus, while the PRG cohort may vary between Drosophila and Oncopeltus, the pair rule per se is highly conserved.
RESULTS
First-pass expression pattern screen identifies several Of-E75A-coexpressed, spatially regulated genes
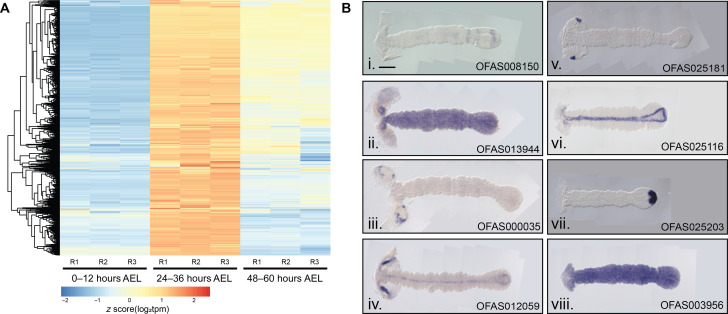
Since Of-E75A was the only gene shown to have PR function in Oncopeltus, we reasoned that other Oncopeltus PRGs would likely be expressed in a similar temporal manner. RNA extracted from embryos 0 to 12, 24 to 36, and 48 to 60 hours after egg laying (AEL) was sequenced. These developmental stages were chosen because they displayed clear changes in expression level of Of-E75A, with no expression detected at 0 to 12 and 48 to 60 hours AEL and high expression detected at 24 to 36 hours AEL, as determined by reverse transcription polymerase chain reaction (RT-PCR) (fig. S1). mRNA was sequenced and edgeR was used to identify differentially expressed genes across the three time points (40). In total, we identified 13,376 differentially expressed gene models that were clustered into coexpression modules using weighted gene coexpression network analysis (WGCNA) (41). Of-E75A was identified in a 2363-gene coexpression module up-regulated specifically at 24 to 36 hours (Materials and Methods and Fig. 1A). Of these, 135 gene models, including Of-E75A, were predicted to encode transcription factors (42) and were thus considered candidate PRGs.
Fig. 1. Genes coexpressed with Of-E75A were screened by expression pattern.
(A) Heatmap displaying expression levels of genes coexpressed with E75A as determined by WGCNA clustering. All genes in this cluster show elevated expression levels at 24 to 36 hours AEL relative to the two other time points. Three replicates (R) were sequenced per time point. Each row represents a single gene, each column a different RNA sample. (B) Expression patterns of Of-E75A–coexpressed genes. Expression was visualized by in situ hybridization with digoxygenin-labeled probes in 24- to 48-hour AEL embryos. A subset of screened genes is shown here and the rest in fig. S2. (i) OFAS008150 is expressed in stripes in the anterior SAZ and broadly in the posterior SAZ, suggestive of a role in segmentation. (ii) OFAS013944 expression was observed ubiquitously in germ-band–stage embryos, (iii) OFAS000035 (annotated as H6-like-homeobox) expression was observed in specific domains of the head lobes, (iv) OFAS012059 expression was observed in the head lobes and along the midline, (v) OFAS025181 (annotated as tailup) expression was observed in a single dot in each head lobe, (vi) OFAS025116 (annotated as single-minded) expression was observed along the midline, (vii) OFAS025203 (annotated as Brachyury) expression was observed in the posterior tip of the SAZ, and (viii) OFAS003956 expression was observed ubiquitously. Genes are named on the basis of published annotations (42).
We have examined the spatial expression patterns of 46 E75A-coexpressed genes so far (Fig. 1B and fig. S2). Genes that we have previously examined [prd, odd, slp, and run, (37)] that were identified in this module were not reexamined, but several others with published expression patterns were. The expression pattern of Of-single-minded was previously reported (43), where it was shown to be expressed in a triangle-like linear pattern in blastoderms bordering the mesoderm and along the midline in germ bands; the germ-band–stage expression we observed continues to border the mesoderm in the SAZ, and midline expression in the segmented region was likewise observed (Fig. 1B, vi, OFAS025116). The expression patterns we observed for Of-caudal (fig. S2, OFAS025096) and Of-hunchback (fig. S2, OFAS018348) are consistent with previously published reports (7, 44). The expression pattern we observed for Of-giant (gt) (fig. S2, OFAS025035) is similar to that previously reported (45) in early germ bands and in the head region of later germ bands; however, we did not observe stripes in the anterior SAZ. This discrepancy is not likely explained by differences in gt isoforms as our probe should detect both annotated isoforms but may reflect a poor sample of embryos in our screen that did not capture this stage or highly dynamic gt expression.
Several genes were expressed in clearly defined regions suggesting specific functions that remain to be determined (Fig. 1B, iii to vii), while many were found to be expressed ubiquitously (Fig. 1B, ii and viii). Ubiquitously expressed genes could potentially have PR function [e.g., odd-paired (opa) and ftz-f1 in Drosophila, (10, 11)], which will be tested in the future. Several genes previously annotated as orthologs of Drosophila genes (42) displayed expression patterns reminiscent of their Drosophila and/or Tribolium counterparts. For instance, in addition to expression in the anterior SAZ, Of-SoxN expression was observed in paired spots along the midline, likely the primordia of the central nervous system, like Dmel-SoxN (fig. S2, OFAS008078) (46). Of-cubitus interruptus was found to be expressed in stripes in every segment of the germ band, consistent with a role in hedgehog signaling (fig. S2, OFAS009156). Expression of brachyury at the posterior of the germ band (Fig. 1B, vii, OFAS025203) is nearly identical to the expression of its orthologs in Drosophila, Tribolium, and Locusta (47). Like its Tribolium ortholog (48), expression of Of-Dichaete was observed broadly in the SAZ and in a punctate pattern along the midline of the segmented germ band (fig. S2, OFAS018531). So far, the expression pattern of one gene (represented by gene model OFAS008150) stood out as suggestive of involvement in segmentation as it was expressed in stripes in and around the SAZ (Fig. 1B, i).
The OFAS008150 gene model corresponds to the Oncopeltus ortholog of Blimp1
The OFAS008150 gene model was previously annotated as Of-Blimp1 (42), a gene with a highly conserved structure encoding a transcription factor with an N-terminal SET-like domain (named the PR domain after the proteins PRD-BF1 and RIZ1, henceforth called the PR/SET domain) and five zinc fingers at the C terminus (49–51). The OFAS008150 gene model, however, encodes a partial PR/SET domain and only three zinc fingers which, along with several assembly gaps in the region, suggested that portions of the coding sequence may not be present in the genome assembly. To resolve this, we isolated and sequenced a 3.5 kb Of-Blimp1 cDNA (the predicted OFAS008150 transcript is ~1.3 kb) and compared it to the genome assembly and a PCR-amplified genomic fragment (see Materials and Methods and data S1). The sequenced cDNA encodes a complete PR/SET domain and five zinc fingers, supporting the initial annotation of this gene as Blimp1.
To verify the annotation of this gene model as Of-Blimp1, we searched the Oncopeltus genome assembly using the newly assembled complete PR/SET domain of putative Of-Blimp1 as query, yielding three significant (e value <1 × 10−6) alignments corresponding to three separate gene models: OFAS008150, OFAS008959, and OFAS019593. OFAS008959 encodes nine C2H2-type zinc fingers (InterPro ID 013087), while OFAS019593 encodes only two such zinc fingers. OFAS019593 is located at the end of a scaffold and likely does not represent a full coding sequence; we therefore cannot be certain how many zinc fingers are encoded by this gene. Only three PRDM family genes were found in the D. melanogaster and D. pseudoscura genomes: prdm1/Blimp1, prdm5, and hamlet, which encode five, six, and nine zinc fingers, respectively (39). On the basis of the number of zinc fingers, and if Oncopeltus has the same complement of PRDM family members as Drosophila, OFAS008150 would correspond to Blimp1, OFAS008959 to hamlet, and OFAS019593 to prdm5, which would be expected to encode six zinc fingers.
Of-Blimp1 is expressed in the primordia of alternate segments through much of Oncopeltus germ-band elongation
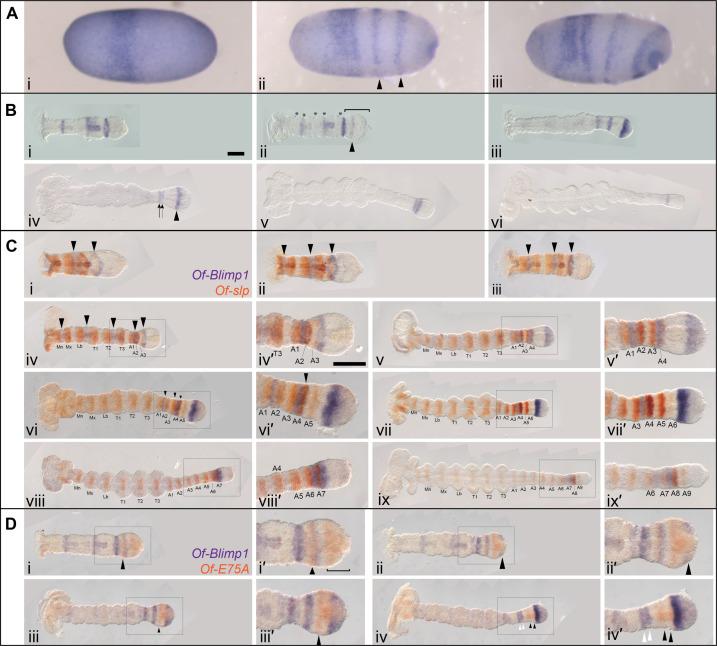
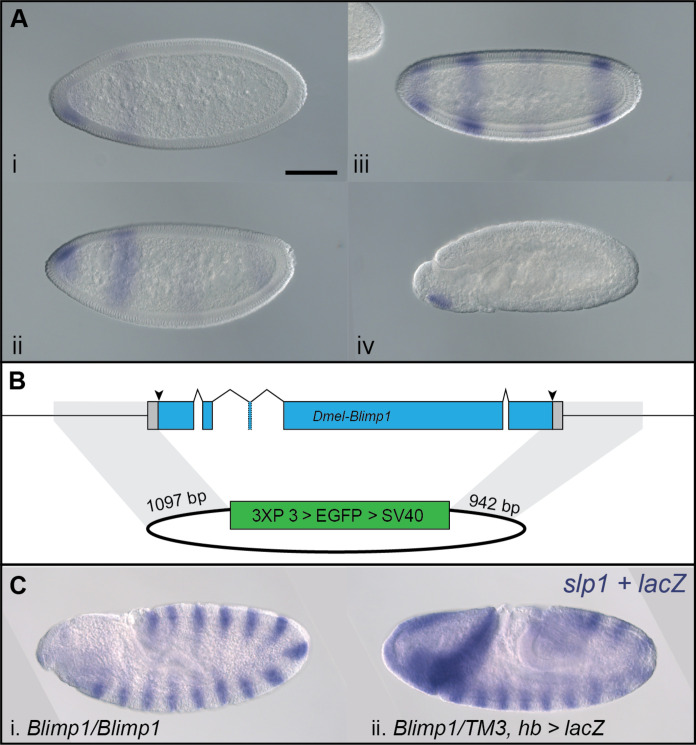
After initially detecting Of-Blimp1 striped expression in and around the SAZ in 24- to 48-hour AEL embryos, we analyzed its expression in greater detail using embryos fixed every 8 hours during blastoderm formation and germ-band extension. Of-Blimp1 expression was first observed in blastoderm-stage embryos at 24 to 32 hours AEL in a broad band positioned at ~one-third the length of the embryo (Fig. 2A, i). In 32- to 40-hour AEL embryos that were starting to gastrulate, as evidenced by an invagination pore at the posterior pole, this broad band remained and two additional stripes had appeared posterior to it (Fig. 2A, ii, arrowheads). In later blastoderm-stage embryos, the broad anterior stripe of Of-Blimp1 expression had split in two, and the two posteriormost stripes had begun to move toward the germ-band invagination site (Fig. 2A, iii). The spacing and number of the Of-Blimp1 stripes in these late blastoderm-stage embryos resemble those of Of-E75A (37, 38) in embryos of comparable age.
Fig. 2. Of-Blimp1 is expressed in alternate segment primordia.
(A) Expression of Of-Blimp1 in blastoderm stage. Digoxygenin-labeled probes were used for Of-Blimp1 in situ hybridization at successive time points in 24- to 32-hour AEL embryos. Expression was first observed in a single thick stripe (i), later two additional stripes appeared (arrowheads) (ii), and as gastrulation proceeded, the broad anterior stripe split into two discrete regions, while the posterior two stripes had begun to move toward the germ-band invagination pore (iii). (B) Expression of Of-Blimp1 through germ-band elongation (i to vi). (C) Double in situ hybridization of segmental Of-slp expression (orange) and Of-Blimp1 (purple). One Of-Blimp1 stripe (arrowheads) was observed posterior to every other Of-slp stripe as the germ band invaginated (i to v). Later, during germ-band elongation, the pattern of Of-Blimp1 coexpression with alternate slp stripes was not regular (vi to ix). (D) Double in situ hybridization of Of-E75A (orange) and Of-Blimp1 (purple) during germ-band elongation. Of-E75A was observed in a stripe anterior to a stripe of Of-Blimp1 in the anterior SAZ (i, arrowhead) and more broadly in the posterior SAZ (i′, bracket). As the broad expression domain of Of-E75A in the SAZ faded at the posterior, Of-Blimp1 expression took its place (ii and ii′, arrowhead). Of-Blimp1 expression in the posterior SAZ later intensified, while the E75A expression just anterior resolved into a stripe (iii and iii′, arrowhead). Two stripes each of Of-Blimp1 (iv and iv′, white arrowheads) and Of-E75A (iv to iv′, black arrowheads) were then observed anterior to an intense Of-Blimp1 stripe in the SAZ. (C, iv′) and (D, iv′) are insets of (C, iv) and (D, iv). Anterior is to the left in all panels. Scale bar, [(B, i) and (C, iv′)] 200 μm.
In early germ bands that lacked clear morphological segments, three stripes of Of-Blimp1 were observed along the length of the embryo (Fig. 2B, i). In slightly older germ bands, a fourth stripe (Fig. 2B, ii, arrowhead) was observed in the SAZ (Fig. 2B, ii, bracket). Also at this stage, morphological segments had begun to appear, and the alignment of segmental furrows (Fig. 2B, ii, asterisks) with Of-Blimp1 stripes suggests that Of-Blimp1 expression coincides with alternate segmental units. In later germ bands in which all gnathal and thoracic segments were morphologically segmented (Fig. 2B, iii), the earlier striped expression faded and additional Of-Blimp1 expression was observed in two stripes—one in the segmented germ band and one around the boundary between segmented germ band and SAZ—as well as in a broad domain in the posterior SAZ. The stripe in the anterior SAZ may split in two, as two stripes were observed in this region in a slightly older embryo (Fig. 2B, iv, arrows). At this time, the posterior SAZ expression domain has resolved into a stripe (Fig. 2B, iv, arrowhead). Two stripes in the anterior SAZ were apparent in later embryos (Fig. 2B, v), and later, only one stripe of Of-Blimp1 expression was detected (Fig. 2B, vi).
While the spacing of the Of-Blimp1 stripes throughout germ-band elongation is suggestive of expression in alternate segment primordia, we sought to better characterize the register of Of-Blimp1 expression by costaining with Of-slp. We and others previously showed that Of-slp is expressed in every segment in the anterior SAZ and the segmented germ band (36, 37) and that this expression persists and is anterior to stripes of invected (inv) expression in every segment, making it a good segmental marker. Of-Blimp1 expression was observed between alternate pairs of Of-slp stripes in early germ bands (Fig. 2C, i to iv). By the time morphological segmentation allows clear identification of slp stripe identity (Fig. 2C, iv), Of-Blimp1 expression was observed in the posterior region of the mandibular (Mn), labial (Lb), and second thoracic (T2) segments, which likely correspond to the stripes seen in blastoderm-stage embryos (Fig. 2A, ii and iii). This alternate segment expression continued as the abdominal segments were added from the posterior during germ-band elongation; Of-Blimp1 stripes were posterior to the slp stripes in A1 and A3 (Fig. 2C, iv and v). Unlike earlier Of-Blimp1 stripes, the Of-Blimp1 stripe posterior to slp stripe A3 seems to extend posteriorly, possibly overlapping the slp A4 stripe, as evidenced by the darker staining of this stripe (Fig. 2C, v and vi). It is unclear whether this expanded Of-Blimp1 expression is a wider stripe or a stripe doublet, possibly corresponding to the pair of stripes seen in Fig. 2B (iv, arrow). As slp stripe A5 (Fig. 2C, vi and vi′) and stripe A6 (Fig. 2C, vii and vii′) appeared, Of-Blimp1 expression remained a single stripe in the SAZ. By the time slp stripe A7 appeared, Of-Blimp1 was observed in stripes posterior to slp stripes A6 and A7 (Fig. 2C, viii and viii′). After slp stripe A8 and A9 had formed, Of-Blimp1 was no longer observed in the SAZ (Fig. 2C, ix and ix′).
As Of-Blimp1 was found to be expressed in alternate segments across much of the AP axis, it is possible it could be spatially coexpressed with Of-E75A, so far the only gene for which a role in PR patterning had been described in Oncopeltus (38). Double in situ hybridization revealed that while the expression patterns of Of-E75A and Of-Blimp1 appear very similar, their expression domains do not overlap during germ-band elongation. In early embryos, Of-E75A was seen anterior to a stripe of Of-Blimp in the anterior SAZ (Fig. 2D, i and i′, arrowhead), with additional Of-E75A expression broadly in the posterior SAZ. Later, as Of-E75A expression cleared from the posterior SAZ, Of-Blimp1 expression took its place (Fig. 2D, ii and ii′). Of-E75A expression was observed in the SAZ anterior to a broad domain of Of-Blimp1 expression in the posterior SAZ (Fig. 2D, iii and iii′). In an even later embryo, when the posterior SAZ domain of Of-Blimp1 expression had narrowed to a stripe and a stripe of expression in the segmented germ band had appeared to split into two stripes (Fig. 2D, iv and iv′, white arrowheads), Of-E75A expression was distinct and nonoverlapping in a doublet of stripes just posterior (Fig. 2D, iv and iv′, black arrowheads).
In sum, Of-Blimp1 is expressed in stripes in the blastoderm in a register similar to that of Of-E75A and in every other segment primordium through much of germ-band elongation. This alternate segment expression is characteristic of most of the Drosophila PRGs. This PR expression pattern does not overlap that of Of-E75A, the other known Oncopeltus PRG, suggesting that the two genes are active in different sets of cells.
eRNAi suggests that Of-Blimp1 is a key regulator of segmentation
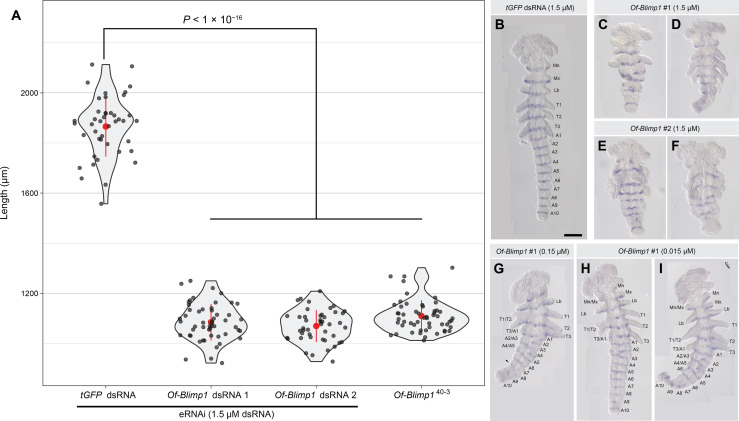
To probe the function of Of-Blimp1 during embryogenesis, we conducted embryonic RNAi (eRNAi), separately using two nonoverlapping double-stranded RNAs (dsRNAs) for knockdown (Fig. 3). Of-Blimp1 dsRNA-1 was injected at three concentrations (0.015, 0.15, and 1.5 μM), while Of-Blimp1 dsRNA-2 was injected at 1.5 μM. Knockdown by either Of-Blimp1 dsRNA at the highest concentration (1.5 μM) produced significantly shorter 67- to 71.5-hour AEL embryos compared with controls (Fig. 3A). Injection of dsRNA-1 produced a mean embryo length of 1084.6 ± 10 μm (SEM) (n = 53) and injection of dsRNA-2 produced embryos with a mean length of 1069.3 ± 9.5 μm (n = 45), compared to a mean length of 1865.6 ± 18.3 μm (n = 43) seen in tGFP dsRNA–injected control embryos (Fig. 3). Thus, Of-Blimp1 knockdown embryos were thus reduced to ~60% wild-type–like length, similar to the ~half-sized embryos seen for Drosophila PR-mutants.
Fig. 3. Knockdown of Of-Blimp1 results in shortened embryos and segmental fusions suggesting PR-like function.
(A) Violin plots displaying the distribution of embryonic lengths in control (tGFP dsRNA-injected), Of-Blimp1 eRNAi embryos (67 to 71.5 hours AEL), and Of-Blimp140–3 presumptive homozygotes. The means and SD are shown in red. The data are plotted in gray dots. (B) tGFP dsRNA–injected embryo with one inv stripe in each of the six appendage bearing segments and each of the 10 abdominal segments. (C and D) Embryos injected with 1.5 μM Of-Blimp1 dsRNA-1 and (E and F) embryos injected with 1.5 μM Of-Blimp1 dsRNA-2 displaying reduced segment number and concomitant reduced inv stripe number. (G) Embryo injected with 0.15 μM Of-Blimp1 dsRNA-1. The right half of the embryo is mostly wild type, while segments are fused in the left half. Arrowhead marks a possible fusion of A7 and A8. (H and I) Embryos injected with 0.015 μM Of-Blimp1 dsRNA-1 displaying partial segment loss. The right half of both embryos exhibits wild-type segment number, while segment fusions are seen on the left side. All eRNAi embryos fixed at 67 to 71.5 hours AEL; Of-Blimp140-3 embryos fixed at 48-72 h AEL. Scale bar, (B) 200 μm.
In the abdomen, wild-type individuals express inv in 10 stripes—one for every abdominal segment (Fig. 3B), but Of-Blimp1 knockdown individuals displayed only 4 to 5 inv stripes in this region (Fig. 3, C to F). When injected at the highest concentration, injection of either Of-Blimp1 dsRNA resulted in embryos displaying at most four appendage-bearing segments in the trunk region (Fig. 3, C to F) compared to six in wild-type individuals (Mn through T3; Fig. 3B). Injecting lower amounts of dsRNA produced weaker and more variable phenotypes, reflecting partial gene knockdown (Fig. 3, G to I). Many of these embryos displayed bilateral phenotypic differences, with one side displaying wild-type segments and the other side displaying segment fusions (Fig. 3, G to I), allowing us to identify the patterns of segment loss. Fusions between the Mn and Mx segments (Fig. 3, H and I), the first and second thoracic segments (T1 and T2; Fig. 3, G to I), the third thoracic and first abdominal segments (T3 and A1; Fig. 3, G to I), the second and third abdominal segments (A2 and A3; Fig. 3, G and I), and the fourth and fifth abdominal segments (A4 and A5; Fig. 3, G and I) were all observed. The observed alternating pattern of segment fusions is similar to phenotypes seen for hypomorphic alleles of Drosophila PRGs (8). Together, these results are suggestive of PR-like function for Of-Blimp1.
Of-v co-mutation expedites screening of Of-Blimp1 mutations
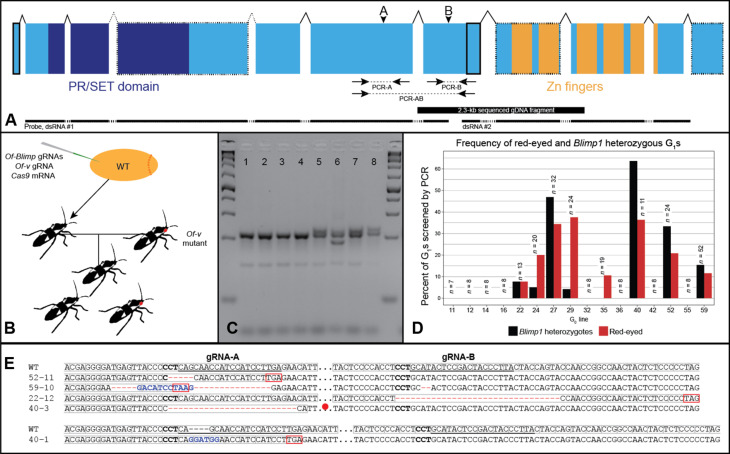
Given the segmental abnormalities and reduced embryonic length observed after eRNAi, we next sought to generate Of-Blimp1 germline mutations to more precisely determine its role in segmentation. We reasoned that introducing indels upstream of the zinc finger–encoding region would likely result in loss-of-function alleles by inducing a frameshift and precluding any mutant protein from binding DNA. We further reasoned that two guide RNAs (gRNAs) spaced apart by several hundred base pairs (bp) might result in large deletions of the intervening sequence, thus producing alleles that could be clearly identified by PCR. Two gRNA target sites (A and B) met these criteria and displayed no substantial alignments to off-target sites in the genome. gRNA-A targets a site two exons upstream of the zinc finger–encoding region, while gRNA-B’s target site falls in the exon directly upstream of this region (Fig. 4A, arrowheads).
Fig. 4. CRISPR-Cas9–generated germline Of-Blimp1 mutations.
(A) Gene structure for the sequenced Of-Blimp1 CDS. Of-Blimp1 encodes a PR/SET domain (dark blue) and five zinc fingers (orange). Gene regions missing from the genome assembly are outlined in dotted lines. Regions missing from the gene model but present in the genome assembly are outlined in solid lines. Dotted introns indicate unknown intron-exon boundaries due to the lack of genomic sequence in this region. Arrowheads indicate gRNA target sites. PCR screening amplicons indicated beneath structure by dotted lines. (B) CRISPR-Cas9 injections and crossing scheme. (C) Heteroduplex mobility assay example. All eight samples from siblings from same G0 cross. Samples 1 to 4 were genotyped as wild type due to the lack of heteroduplex bands. Samples 5 to 8 were genotyped as heterozygous. (D) The presence of red-eyed G1s is correlated with the presence of G1 Of-Blimp1 heterozygotes. Of-Blimp1 heterozygotes were only recovered from G0 crosses yielding Of-v red-eyed progeny. The number of G1s screened from each G0 cross, above bars. (E) Alignments between Of-Blimp1 wild-type allele and five Of-Blimp1 loss-of-function mutations used in this study. gRNA target sites underlined in wild-type sequences. Gray boxes, reading frame; complement of protospacer-adjacent motif (PAM) site, bold; premature stop codons, boxed red or red circle. Deleted nucleotides, red dashes; inserted nucleotides, blue.
Of-Blimp1 gRNAs A and B were coinjected with a previously used Of-vermilion (Of-v) gRNA (52) and Cas9 mRNA into wild-type embryos (Fig. 4B). Of-v mutation results in red eyes in contrast to wild-type black eyes; the Of-v gRNA was therefore injected to provide a visible marker for CRISPR-Cas9 co-mutation, which has proven to be a successful approach in many model systems (53). Of 1304 embryos injected, only 160 (12.3%) hatched. Among the unhatched embryos, we observed clear segmental abnormalities, suggesting that the low hatch rate was a consequence of lethal somatic mutations since the survival rate for embryo injection in our hands is usually >50%. Furthermore, all 82 G0s that survived to adulthood (51.3% of hatchlings) had wild-type eyes; in contrast, when the Of-v gRNA is injected without Blimp1 gRNAs, ~70% of G0s had at least one red eye (52). This result suggests that somatic mutation at the Of-v locus was accompanied by mutation at the Of-Blimp1 locus and that the only G0s to survive had a low frequency of somatic biallelic mutations at both loci. We hypothesize that red-eyed G0s did not hatch because of an accompanying lethal degree of somatic biallelic Of-Blimp1 mutation.
All surviving G0s were crossed to Of-v virgins, and of these, 68 G0s produced G1 offspring (Fig. 4B). G1 offspring were first screened by eye color to determine which G0s had undergone germline mutation of Of-v. Because of Of-v X-linkage (52), G0 male crosses were only considered v-yielding if any female progeny had red eyes, while G0 female crosses were considered v-yielding if any G1 progeny had red eyes. Overall, 10 crosses were v-yielding. We reasoned that five outcomes at the Of-Blimp1 locus were possible: no mutation at either gRNA target site, indel at site A only, indel at site B only, indels at both sites, or deletion between sites A and B. As removal of a single leg does not impede a bug’s ability to mate and reproduce, we screened G1 individuals by performing at least one of the following PCRs with genomic DNA extracted from a single dissected leg: PCR-A, which amplifies a 211-bp region around site A; PCR-B, which amplifies a 171-bp region around site B; and PCR-AB, which amplifies a 751-bp region around both sites (Fig. 4A, arrows). PCR-A and PCR-B were used to identify indels at sites A and B, respectively, and PCR-AB was used to identify large deletions between the two sites. PCR products from PCR-A and PCR-B were subjected to a heteroduplex mobility assay (see Materials and Methods) to identify heterozygotes by the presence of heteroduplex bands (Fig. 4C).
In total, 258 G1 individuals (195 G1s from 8 v-yielding G0 lines and 63 G1s from 8 non–v-yielding G0s) were screened by PCR, and 41 of these were identified as Blimp1 heterozygotes (Fig. 4D). Notably, all Blimp1 heterozygotes came from v-yielding G0 lines (Fig. 4D). Thus, about 21% of all G1s from v-yielding G0s were found to be heterozygotes; a similar rate among non–v-yielding G0s would be expected to yield at least 10 heterozygotes, and yet none were found in our screen, demonstrating the usefulness of our co-CRISPR strategy with Of-v as a visible marker. Among the heterozygotes, 38 had indels at site A as determined by the heteroduplex mobility assay, 2 at site B, and 1 at both sites. Since different alleles produce different patterns of homoduplex and heteroduplex bands, we selected individuals with unique banding patterns to propagate and maintain as lines (compare samples 5, 7, and 8 with sample 6 in Fig. 4C). After sequencing alleles from several lines, we selected five lines with frameshift mutations to maintain (Fig. 4E). Line Blimp152–11 has a 5-bp deletion at site A; line Blimp159–10 has an 18-bp deletion and substitution of 11-bp at site A and a 2-bp deletion at site B (this latter deletion was not identified by the heteroduplex mobility assay but was found when the allele was sequenced); line Blimp122–12 has a 32-bp deletion at site B; line Blimp140–3 has a 25-bp deletion at site A, and line Blimp140–1 has a 4-bp insertion at site A. The frameshifts present in each of these five alleles result in premature stop codons upstream of the zinc finger–encoding region (Fig. 4E, red boxes, circle). Allele sequences are available in data S1.
Of-Blimp1 homozygous mutants display approximately half as many segments as wild type
Wild-type Oncopeltus pre-hatchlings have three thoracic segments, each bearing a pair of legs, and 10 abdominal segments, although often only 7 to 8 abdominal segments are clearly visible (Fig. 5A, i and ii). In contrast, presumptive Of-Blimp140–3 homozygotes (Fig. 5A, iii, iv) have many fewer segments than their wild-type siblings at 6 to 7 days AEL. These individuals usually have only one pair of legs [Fig. 5, A (iii) and B (i), black arrowheads]; a second pair of appendages appears as small nubs rather than fully formed legs (Fig. 5B, i, transparent arrowhead). Dissection of the thoracic appendages from a presumptive Blimp1 homozygote shows a slightly enlarged leg (fig. S3D, i) relative to legs from a wild-type–like individual (fig. S3C, i to iii), followed posteriorly by a nub of tissue (fig. S3D, ii). Presumptive homozygotes from all five lines display a very similar phenotype (Fig. 5B and fig. S3A). The mean length of presumptive Of-Blimp140–3 homozygous 48- to 72-hour AEL embryos was 1109 ± 8.6 μm (n = 54), very similar to the embryo lengths observed after Of-Blimp1 eRNAi (Fig. 3A), and 59% the length of wild-type–like (tGFP dsRNA–injected control) embryos.
Fig. 5. Of-Blimp1 homozygotes have about half as many segments as wild type.
(A) Offspring of Of-Blimp140–3 self-cross at the pre-hatchling stage, 6 to 7 days AEL. (i and ii) Wild type–like segmentation. (iii and iv) Presumptive Blimp140–3 homozygote displaying segment loss (i and iii, lateral view; ii and iv, dorsal view). (B) Presumptive Of-Blimp122–12 homozygote (i, lateral view; ii, dorsal view). (C) Of-inv is expressed in the posterior of the gnathal (Mn, Mx, and Lb), thoracic (T1 to T3) segments, and abdominal segments (A1 to A10) in wild-type individuals (i), but only three to four inv stripes were observed in the gnathal and thoracic segments, and abdominal expression of inv was highly variable and less clearly defined, in Of-Blimp140–3 homozygotes (ii to iv). (D to G) Altered Hox gene expression in Of-Blimp1 mutants. (D) While the Mn and Mx segments express Dfd in wild type (i), only one remained in Blimp1 mutants (ii). (E) Wild-type Scr expression, Lb segment and dots on T1 appendages (i); Lb expression unaffected in Blimp140–3 homozygotes (ii), but often lost in T1/T2 fused appendage (arrowhead). (F) Wild-type Ubx expression, A1 and pair of dots in A2 (i); expression restricted to last appendage-bearing segment in Of-Blimp140–3 homozygotes (ii). (G) AbdB expression in posterior abdomen in wild-type (i) and Of-Blimp40–3 homozygotes’ shortened abdomens (ii). inv-stained embryos are 72 to 96 hours AEL; Ubx-stained embryos are 56 to 81 hours AEL; all other stained embryos are 48 to 72 hours AEL. Anterior, left; black arrowheads, T1/T2 fused appendage. Scale bar, (C, i) 200 μm and applies to (C) to (G).
To visualize segment boundaries in Blimp1 mutants, we examined the expression of the segment polarity gene inv in fully extended germ-band–stage embryos. These embryo preparations also allowed clear visualization of the morphology of Of-Blimp1 homozygotes. Of-inv is expressed in the posterior of each of the three mouthpart segments (Mn through Lb), the three thoracic segments (T1 through T3), and the 10 abdominal segments (A1 through A10) (Fig. 5C, i). In presumptive Blimp140–3 homozygotes, there are consistently three to four inv stripes in the mouthpart plus thoracic region (Fig. 5C, ii to iv). Only one pair of leg-like appendages is evident (Fig. 5C, ii to iv, black arrowhead), which often appears distally forked (Fig. 5C, iii), suggesting that this may be a fusion of T1 and T2. Often, a much smaller appendage is seen on the first abdominal segment (Fig. 5C, ii to iv, transparent arrowhead), suggesting that this may be a fusion of T3 and A1. Abdominal inv expression is more disorganized compared to the wild-type pattern; often, this expression takes the form of dots or partial stripes rather than the clearly defined transverse stripes seen in the anterior region of the body. However, the segmental furrows seen along the lateral sides of the abdomen suggest that the abdomen has been reduced to about four to five segments.
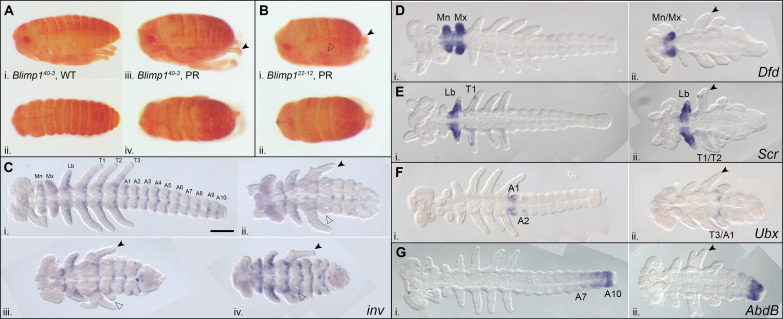
While segment number is clearly reduced in Of-Blimp1 homozygotes, the identities of the remaining segments are unclear. We reasoned that comparing the expression patterns of Hox genes in wild-type and mutant embryos could help illuminate segment identities, as these genes are expressed in specific regions along the AP axis and their expression patterns have been previously characterized in this species (54). In wild-type embryos, Of-Deformed (Dfd) is expressed in the Mn and Mx segments, two appendage-bearing segments in the trunk region that will form part of the mouthparts later in development (Fig. 5D, i). In presumptive mutants, only one appendage-bearing segment expressed Of-Dfd, suggesting that either the Mn or Mx segment is lost (Fig. 5D, ii). This phenotype is consistent across all five Of-Blimp1 lines (fig. S3B).
Of-Sex combs reduced (Scr) is expressed mainly in the Lb segment and in dots on the T1 appendage (Fig. 5E, i). In presumptive Of-Blimp140–3 homozygous mutants, Lb expression of Scr appeared unaffected (Fig. 5E, ii), and the T1 dot of Scr expression was sometimes present. The segment posterior to the Lb segment displays a high degree of variability in the extent of T1 versus T2 identity in the appendages both across embryos and within individual embryos. For instance, in Fig. 5E (ii), one side of the embryo displays the dot of Scr expression on the appendage of this segment, suggesting T1 identity, while the other side does not, suggesting T2 identity. Across embryos, this segment varies in the extent of T1 or T2 appendage loss, often displaying a forked appendage [compare Fig. 5, D (ii), E (ii), F (ii), and G (ii), arrowheads].
In wild-type embryos, Of-Ultrabithorax (Ubx) expression is most pronounced in the first abdominal segment (A1), and two small dots of expression are observed in the A2 segment (Fig. 5F, i). In Of-Blimp40–3 embryos, Ubx is most clearly expressed in the posteriormost appendage-bearing segment (Fig. 5F, ii). This shift in expression domain, along with the observation that this T3 appendage usually degenerates through development leading to only a small nub in pre-hatchling mutants (Fig. 5B, i, and fig. S3D, ii), suggests that the segment remaining displays shared T3/A1 identity. Of-Abdominal B (AbdB) is expressed in a gradient in the posterior abdominal segments in wild-type embryos; the anterior boundary of this expression domain is somewhat variable, but expression is evident in abdominal segments A9 and A10 and often continues anteriorly to A7 (Fig. 5G, i). In Of-Blimp140–3 mutant embryos, AbdB was likewise expressed in the posteriormost abdominal segment or two. This suggests that within the abdomen, segment loss is interspersed along the AP axis, i.e., neither the anterior half nor the posterior half of the abdomen is lost (Fig. 5G, ii). Together, the expression patterns of Hox genes in Blimp1 mutants are consistent with PR defects. Dfd and Ubx expression patterns are most diagnostic and clearly suggest that Of-Blimp1 mutants display Mn/Mx and T3/A1 fusions, and Scr expression is somewhat suggestive of T1/T2 segment fusion.
Given the evidence that loss of Blimp1 function results in PR-like defects, embryos obtained after self-crossing heterozygotes from each of the five lines were phenotypically scored after fixing at 5 days AEL. While most embryos from each line displayed the expected wild-type phenotype, the percentage of embryos displaying a PR-like phenotype was less than the expected 25% (fig. S4A); this percentage ranged from 11.8% (line Of-Blimp140–3) to 20.5% (line Of-Blimp122–12). Aside from the wild-type and PR-like phenotypes, some embryos (6.8 to 10.5%) developed into only a small mass of tissue (“tissue mass phenotype”) (fig. S4C, 6 to 8, and 13). Sometimes, clear structures were present in these tissue masses, such as a compound eye (see fig. S4C6), but, very often, no discernible structures were visible. To understand whether this phenotype is associated with Of-Blimp1 mutation, we genotyped individual embryos from lines Of-Blimp122–12, Of-Blimp140–3, and Of-Blimp159–10. These lines were selected because for each, the indel is large enough for homozygous wild-type and homozygous mutant bands to be distinguished (fig. S4D). All embryos phenotyped as wild type (n = 12) were found to be homozygous for the wild-type allele or heterozygous, and all embryos phenotyped as PR (n = 19) were found to be homozygous for the mutant allele. A total of 74 tissue mass embryos were genotyped, and the majority (78%) were found to be Blimp1 homozygotes, but 20% were either heterozygous or homozygous wild type (fig. S4B). Of this latter group, 80% came from line Of-Blimp122–12. Together, these data suggest that the tissue mass phenotype is caused by an off-target mutation closely linked to Of-Blimp1, and one or more recombination events occurred between these two loci early enough in the Of-Blimp122–12 lineage to separate the alleles.
Somatic CRISPR-Cas9 mutation supports the PR function of Of-Blimp1
The results thus far indicated that Of-Blimp1 homozygotes display PR-patterning defects. However, given the difficulty of identifying specific segments morphologically, we sought to further characterize defects in an unbiased fashion. Since we had observed segmental defects in unhatched G0 embryos following Of-Blimp1 CRISPR-Cas9 injection, we reasoned that somatic mutants displaying mosaic phenotypes—both wild-type and mutant phenotypes in the same sets of segments—might facilitate clear identification of the segments affected in the mutants. We injected wild-type embryos with either Of-Blimp1 gRNA-A and Cas9 mRNA, Of-Blimp1 gRNA-B and Cas9 mRNA, or Cas9 mRNA alone as a negative control. Only embryos that displayed partial segmental fusions, such that the affected segments could be reliably identified by the corresponding unaffected regions, were scored. Thus, embryos were first broadly scored as displaying one of the following phenotypes: specific partial segment fusions, nonspecific segmental defects (embryos displaying defects in segments that could not be reliably identified), undeveloped, and wild type (fig. S5). Specific segmental fusions were recorded for all embryos displaying clear partial segment fusions. As several of the posteriormost segments telescope into each other and are too compact to be reliably scored, only segments anterior to segment A7 were analyzed.
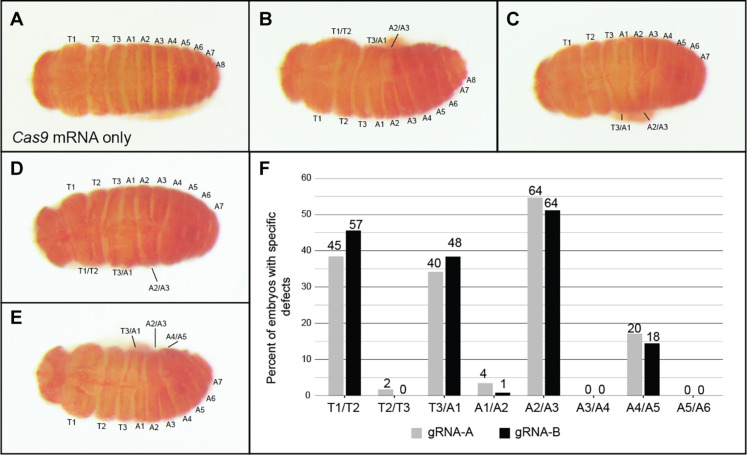
Of the 259 embryos injected with Cas9 mRNA alone, only 1.5% displayed any segmental defects and 79% appeared wild type (Fig. 6A and fig. S5). In contrast, 34% (n = 698) of embryos injected with Cas9 mRNA and Of-Blimp1 gRNA-A and 29% (n = 639) injected with Cas9 mRNA and Of-Blimp1 gRNA-B displayed segmental defects. For the gRNA-A group, 16.8% displayed partial segmental defects that could be scored reliably. Among these, specific pairs of segments displayed fusions consistently along the AP axis: 38.5% displayed fusions of segments T1/T2 (Fig. 6, B and D), while only 1.7% displayed fusions of T2/T3; T3/A1 fusions were observed in 34.2% (Fig. 6, B to E), whereas A1/A2 fusions were observed in only 3.4%; A2/A3 fusions were seen in 54.7% (Fig. 6, B to E) but A3/A4 in none; A4/A5 fusions were observed in 17.1% (Fig. 6E) but A5/A6 in none. Embryos injected with Cas9 mRNA and Of-Blimp1 gRNA-B that displayed specific partial segmental fusions (18.3%) yielded remarkably similar patterns of segmental defects (Fig. 6F), suggesting that the segment fusions observed were due to specific mutation of Of-Blimp1 and not off-target loci.
Fig. 6. Mosaic segmentation defects observed after Of-Blimp1 CRISPR-Cas9 somatic mutation.
Somatic mosaics with fusions on only one side of the embryo after injection of (A) Cas9 mRNA only, [(B) and (C)] Cas9 mRNA and Of-Blimp1 gRNA-A, or [(D) and (E)] Cas9 mRNA and Of-Blimp1 gRNA-B. (A) Control embryos displayed wild-type segmentation; [(B) to (F)] embryos with fusions on only one side of the embryo were selected, allowing for identification of affected segments. (B) Partial fusion of segment T1 and T2, T3 and A1, and A2 and A3; (C) partial fusion of segments T3 and A1, A2, and A3; (D) partial fusion of segments T1 and T2, T3 and A1, and A2 and A3; (E) partial fusion of segments T3 and A1, A2 and A3, and A4 and A5. Dorsal, anterior-left views. (F) Frequencies of partial segment fusions observed among embryos displaying scorable segment fusions after injection with Cas9 mRNA and Of-Blimp1 gRNA-A (gray) or gRNA-B (black). A clear trend of alternate segment pair fusions was observed independent of gRNA used. The number of individuals displaying each type of segmental fusion is displayed above the bars.
Although we could not properly score the posteriormost segments in such late-stage embryos, patterns of inv expression in Of-Blimp1 presumptive homozygotes (Fig. 5C) and embryos treated with lower concentrations of Of-Blimp1 dsRNA (Fig. 3, G to I) suggested that segment fusions may be present in the posterior abdomen following Of-Blimp1 depletion. We therefore repeated the injections described above, fixed embryos at 67 to 71 hours AEL, and visualized Of-inv expression by in situ hybridization to directly observe the posterior end of the fully developed germ band (fig. S6). None of the embryos injected with Cas9 mRNA alone displayed segmental defects. A total of 90.3% (n = 72) displayed wild-type inv expression patterns and segment morphology (fig. S6A); the rest were too damaged to score. Of those injected with Cas9 mRNA and gRNA-A, 40% of stained embryos (n = 203) displayed specific defects that could be scored, compared to 36.8% (n = 136) injected with gRNA-B. In addition to observing the same segment fusion pairs seen in the pre-hatchling stage somatic mutants (e.g., A2/A3 fusions; fig. S6B, i), we observed the fusion of the Mn and Mx segments (fig. S6C, v), consistent with our interpretation of the Dfd expression pattern in homozygous germline mutants (Fig. 5D). In the posterior abdomen, segment A6 did not appear to fuse with either neighboring segment, but fusions of A7 and A8 were fairly common [fig. S6, B (i to iii) and C (i and iv)]. A small number of embryos displayed fusions of segments A9 and A10 [fig. S6, B (i and iii) and C (iv)], but many more displayed posterior fusions that could not be scored [“Nonspecific posterior fusions”; fig. S6, C (iii) and D]. Embryos with this latter defect showed reduced inv expression in the A7 through A10 region often accompanied by curling of the posterior abdomen due to differences in length of the left and right halves of the embryo, suggestive of partial segment loss on one side.
In sum, CRISPR-Cas9–mediated somatic mutation produced consistent mosaic defects that facilitated careful identification of the segment pairs that fused after loss of Of-Blimp1. These fusions displayed a PR register with fusion of alternate pairs of segments: T1/T2, T3/A1, A2/A3, and A4/A5 in the most directly scorable regions of the embryo. These alternate segmental fusions are similar to the types of fusion seen for Drosophila PR mutants (8).
Dm-Blimp1 is expressed in stripes but appears to be dispensable for segmentation
Given the clear role of Of-Blimp1 in segmentation, we reexamined the conclusion (55) that the Drosophila ortholog of Blimp1 is not required for wild-type segmentation. Dm-Blimp1 is expressed in the late blastoderm when segmentation of the AP axis is underway. As reported previously (55), Dm-Blimp1 transcripts were first detected in an anterior cap and a stripe about one-third the length of the embryo (Fig. 7A, i), followed by a stripe in the center of the embryo and another at the posterior end (Fig. 7A, ii). Later, the central stripe of Dm-Blimp1 expression resolved into two weak stripes, while the posteriormost stripe became more intense (Fig. 7A, iii). By the end of germ-band extension, Dm-Blimp1 transcripts were only evident in the head region.
Fig. 7. Dm-Blimp1 is not required to establish segments in Drosophila.
(A) Dm-Blimp1 is expressed in early Drosophila embryos. Expression begins in the head followed by formation of a single anterior (i) and additional of two weak posterior stripes (ii). Most Dm-Blimp1 expression then intensified, but the central stripe appeared as two weak stripes (iii). After germ-band formation, Dm-Blimp1 expression was detected only in the head region (iv). (B) Null Dm-Blimp1 allele generated with CRISPR-Cas9. Schematic showing the Dm-Blimp1 gene, which includes five exons. gRNA target sites (arrowheads) were designed to remove the entire coding region. The homology-directed repair plasmid template included ~1-kb homology arms on either side of a 3XP3 > EGFP expression cassette for creation of the Dm-Blimp13XP3 > EGFP allele, in which the entire Dm-Blimp1 CDS is replaced by the 3XP3 > EGFP dominant marker. Homology arms are highlighted in gray. (C) Blimp13XP3 > EGFP homozygous mutants do not show segmentation defects. Blimp13XP3 > EGFP homozygotes and (i) Dm-Blimp13XP3 > EGFP heterozygotes (ii) displayed indistinguishable, wild-type expression of Dm-slp, a segmental marker. Note that heterozygotes also express lacZ in an hb-like pattern due to a transgene on the balancer chromosome. Scale bar, (A, i) 100 μm.
Previous studies (55, 56) have established that Dm-Blimp1 is required for proper tracheal development, but any that considered the effect of Dm-Blimp1 mutation on segmentation used an allele in which a transposable element had been inserted in one of Dm-Blimp1’s introns, which may not result in a total loss of Dm-Blimp1 function. To definitively address the role of Dm-Blimp1 in segmentation, we used CRISPR-Cas9 to create a Dm-Blimp1 allele in which the entire coding DNA sequence (CDS) is replaced by the 3XP3 > enhanced green fluorescent protein (EGFP) dominant fluorescent eye marker (Fig. 7B). Both homozygous (Fig. 7C, i) and heterozygous (Fig. 7C, ii) Dm-Blimp13XP3 > EGFP mutants displayed wild-type–like expression of Dm-slp1 in 14 stripes in germ-band–stage embryos, demonstrating that Dm-Blimp1 is not required for segment establishment.
DISCUSSION
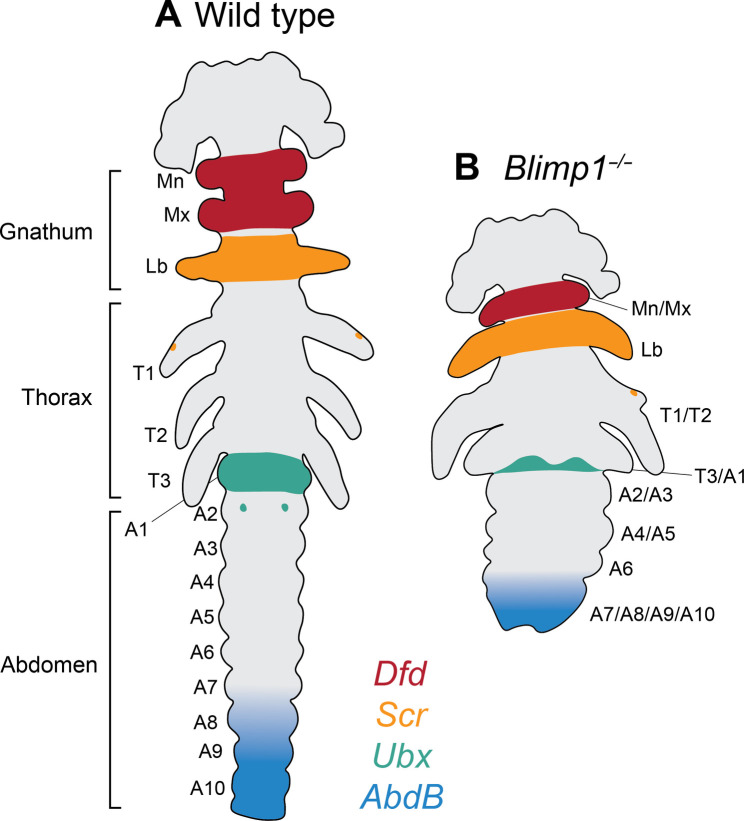
Here, we took an unbiased approach to ask how Oncopeltus, an emerging model for hemimetabolous insects, patterns early embryos according to a pair rule, without the deployment of any Drosophila PRG ortholog for this process. We selected the temporal expression profile (fig. S1) of the only previously identified Of-PRG—E75A—and used in situ hybridization to find additional genes expressed in a PR pattern, identifying Of-Blimp1 (Fig. 1 and 2 and fig. S2). We then carried out functional studies using eRNAi (Fig. 3) and CRISPR-Cas9 genome editing to generate five Of-Blimp1 loss-of-function mutant lines (Figs. 4 and 5 and fig. S3). These experiments support the existence of a pair rule in Oncopeltus and Of-Blimp1’s role in mediating it: (i) Of-Blimp1 is expressed in a manner consistent with two-segment periodicity through much of germ-band elongation; (ii) presumptive homozygotes and eRNAi knockdown embryos display a PR-like shortening of body axis and loss of segments across all tagmata; (iii) where individual segment identities can be clearly discerned, a consistent pattern of segment pair fusions is observed after Blimp1 mutation: Mn/Mx, T1/T2, T3/A1, A2/A3, and A4/A5 (Fig. 8). Last, we verified that Drosophila Blimp1 is not required for PR patterning by generating a definitive null allele (Fig. 7). Thus, the two genes with PR function in Oncopeltus are not PRGs in Drosophila.
Fig. 8. Schematics of wild-type and Blimp1 mutant embryos.
(A) Schematic of a wild-type extended germ-band–stage embryo displaying the full complement of segments and wild-type patterns of Hox gene expression (Fig. 5). (B) Schematic of a Blimp1 homozygous mutant embryo, which displays about half as many segments as wild type and has altered Hox gene expression patterns. Identities of fused segments are based on Hox expression patterns (Fig. 5), somatic CRISPR (Fig. 6 and fig. S6), and eRNAi (Fig. 3). Gnathal segments: Mn, mandibular; Mx, maxillary; Lb, labial. Thoracic segments: T1, prothoracic; T2, mesothoracic; T3, metathoracic; 10 abdominal segments (A1 through A10) are indicated.
While the pattern of segment fusions we observed is mostly in agreement with loss of every other segment, the unperturbed Lb segment following Of-Blimp1 depletion is an exception to this trend. As the pre-gnathal segments (not examined here) are likely patterned differently compared to the gnathal, thoracic, and abdominal segments (57), it is possible that the gnathal segments do not strictly adhere to a pair rule. Most descriptions of the Drosophila PR mutant phenotypes span the limited region of T1 to A8, as the anterior- and posteriormost segments are obscured in such late-stage embryos (8). However, the original description of the Drosophila ftz mutant used scanning electron microscopy to describe defects before head involution and showed that the Mn and Mx segments are present, while the Lb segment is missing (58). The presence of the Mn segment in Drosophila ftz mutants, similar to the presence of the Lb segment in Of-Blimp1 mutants, appears to be an exception to a strictly interpreted pair rule.
This study also contributed to technical advances for the development of Oncopeltus as a model system for hemimetabolous insects (59), and in particular for the order Hemiptera, which includes many pest insects as well as vectors of human disease. While RNAi remains a useful tool to study gene function and was indeed helpful in our study of Of-Blimp1 (Fig. 3), the variability of phenotypes observed after RNAi can sometimes yield ambiguous results. Comparatively, we found that generating a loss-of-function mutation using CRISPR-Cas9 produced phenotypes that, while not entirely devoid of embryo-to-embryo variation, were remarkably similar across lines (fig. S3, A and B). We standardized our protocol for CRISPR-Cas9 genome editing (52, 60) and found that performing co-CRISPR with a visible marker facilitated the isolation of alleles that affect embryonic viability (Fig. 4). This allowed us to generate and maintain the first segmentation mutant in a hemipteran. Together with methods for insertional mutagenesis and transgenesis, these advances in genome editing in Oncopeltus and other non-model insects will allow more researchers to expand the taxonomic sampling of insects in studies of segmentation mechanisms and evo-devo studies more broadly.
Of-Blimp1 and Of-E75A are PRGs expressed in complementary patterns
Of-Blimp1 and Of-E75A are expressed in complementary patterns (Fig. 2D), very similar to sets of Drosophila PRGs, such as eve + ftz or runt + hairy, that are expressed in complementary stripes (61). How does the Of-Blimp1 mutant phenotype compare to phenotypes observed after knockdown of Of-E75A? Within the gnathal region, where Of-Blimp1 loss results in Mn/Mx fusion and no change in Lb segment morphology or expression of Scr, the inverse was observed after E75A RNAi: The Mn and Mx segments were unaffected, while the Lb segment was fused to T1 or lost altogether (38). In the thoracic region, E75A knockdown most often resulted in fusion of T2/T3, although T3/A1 fusions were also observed; in contrast, T2/T3 fusions were not observed in Of-Blimp1 mutants or knockdown embryos. In the abdomen, Of-E75A knockdown produced A3/A4 and A5/A6 fusions, compared to the A2/A3 and A4/A5 fusions shown here after Blimp1 mutation. Thus, these two genes appear to direct PR subdivision of the embryo across different segment boundaries in a nonredundant manner, similar to sets of Drosophila PRGs. The expression of many of the Drosophila PRGs is regulated by gap factors; in Oncopeltus, several of the gap gene orthologs are expressed in broad domains in the blastoderm (7, 45, 62) and thus have the potential to regulate the expression of Of-Blimp1 and/or Of-E75A, but more work is needed to determine whether this is indeed the case.
Striped expression of Blimp1 and E75A has been observed in additional insect species. E75A is also expressed in PR stripes in the harlequin bug Murgantia histrionica, another hemipteran (63), as well as in the jewel wasp Nasonia vitripennis (27), although functional studies have not yet been carried out in either species. E75A was also found to be expressed in a PR pattern in the cricket Gryllus bimaculatus and in possible PR stripes in the cockroach Blattella germanica, but Blattella E75A knockdown did not yield PR defects (28). In Tribolium, Blimp1 was found to be expressed in stripes during germ-band elongation (64), suggesting possible use of this gene in segmentation. Future studies will reveal the extent to which an Oncopeltus-like PRG cohort is used for this process in other species.
Blimp1 is a transcriptional repressor involved in many different developmental processes across Metazoa
Blimp1 is conserved across Metazoa (39) and has been shown to be involved in numerous developmental contexts in insects (55, 56, 65–69). First found for its role as a transcription factor binding to the positive regulatory domain 1 (PRD1) of the human beta-interferon promoter (49), Blimp1 has been most extensively studied for its role in the vertebrate immune system (70). Across diverse developmental contexts, Blimp1 has been found to function as a transcriptional repressor [e.g., (71), including in Drosophila (65)]. The SET domain is the catalytic domain of histone methyltransferases; despite the presence of a SET-like PR domain encoded by Blimp1, it is not thought to have intrinsic methyltransferase activity (51). Rather, Blimp1 has been found to recruit histone-modifying corepressors to the genomic loci to which it binds [e.g., (72)]. Although it is not known how Blimp1 regulates transcription in insects, it likely relies on chromatin-remodeling cofactors, a feature not yet demonstrated for any of the Drosophila PRGs. As E75A similarly appears to function as a transcriptional repressor (73), the juxtaposition of Blimp1 and E75A stripes bears at least superficial similarity to the juxtaposition of Drosophila hairy and runt, which are expressed in largely complementary patterns and encode repressors.
Pair rule patterning per se is more conserved than the genes responsible for this process
As mentioned above, most studies of PR patterning outside Drosophila have analyzed the expression patterns and functions of orthologs of Drosophila PRGs, thus biasing our collective understanding of arthropod segmentation to one that centers this group of genes. However, even within this limited group of genes, many changes in expression and function have been documented across the insect phylogeny. In Anopheles mosquitoes, an ortholog of the Drosophila PRG prd is absent from the genome, with its PR function replaced by Pax-3/7 family member gsb (32). In Drosophila, Ftz-F1 is an obligate partner of Ftz during PR patterning and is expressed ubiquitously (11). However, although ftz-f1 PR function is conserved in Tribolium, ftz-f1 is expressed in PR stripes (74). In the honeybee, several PRG orthologs have taken on maternal roles, in addition to their roles in segmentation (25). For example, ftz is expressed in the anterior tip of embryos and RNAi knockdown resulted in head-patterning defects, along with defects in anterior segmentation (25). These studies demonstrated that PR patterning remained stable despite the gain, loss, or functional change of individual PRGs. The situation for Oncopeltus is particularly interesting, with the only genes having PR function being ones without PR function in Drosophila.
The PRGs, like all the segmentation genes identified to date in any species, are required not only for segment patterning but for viability. Mutations that lead to alterations in their expression and/or function are likely to lead to severe defects if not death, making embryos carrying such mutations unfit. How can these vital regulatory genes change during evolution? The studies cited above provide some insight into how PRG-cohort variation is possible. Entry of a new PRG into the cohort is possible if it partners with a preexisting PRG without changing the original PRG’s function. For example, Ftz may have entered the PRG cohort in lineages leading to Drosophila without activating nontarget genes because of its dependence on Ftz-F1, whose role in PR patterning likely predated that of Ftz, and whose DNA binding properties are dominant for the Ftz/Ftz-F1 partnership (75). Conversely, loss of a gene from the PRG cohort is possible if another gene acts redundantly and is thus primed to replace it. For example, the replacement of prd with gsb in Anopheles was likely possible because gsb had already taken on a PR-expression pattern and because each of these transcription factors bind the same DNA sequences (32). Thus, when expressed in the same pattern, each would likely regulate the same set of target genes without the ectopic activation of nontargets. The studies in Oncopeltus suggest the existence of a wholly different cohort of genes regulating PR patterning. Thus, multiple changes in gene expression and function have occurred since the divergence of lineages leading to extant species, all the while, conserving the patterning of embryos by a pair rule. This is an example of DSD (1) or phenotypic stability (37): The overt process or phenotype remains stable despite evolutionary variation in the genes controlling the process. Future work will identify the full “Oncopeltus PRG cohort” and determine how conserved it is in other insects. In the longer term, this approach will provide insight into the developmental constraints that have preserved a pair rule for insect embryos despite differences in the mode of segment addition and differential utilization of regulatory genes.
MATERIALS AND METHODS
Insect rearing
Laboratory populations of Oncopeltus are maintained on a diet of water and raw organic sunflower seeds in 15.25 inch–by–11 inch–by–11 inch (38.73 cm–by–27.94 cm–by–27.94 cm) (wild type or Of-v lines) or 11 inch–by–7.5 inch–by–5.5 inch (27.94 cm–by–19.05 cm–by–13.95 cm) (individual Of-Blimp1 lines) plastic containers topped with mesh. Lines are regularly maintained at room temperature (RT, ~22°C), but all embryo staging for in situ hybridization or phenotypic analysis was performed at 25°C. Embryo staging for RNA sequencing (RNA-seq) was done at 26°C. Drosophila were reared on a standard cornmeal, molasses, and yeast diet at 25°C or RT.
Oncopeltus embryonic transcriptomes, bioinformatics, and WGCNA clustering
Embryos for RNA-seq were collected over 12-hour periods at 26°C and aged to 0 to 12, 24 to 36, or 48 to 60 hours AEL. These developmental stages were chosen because they previously displayed clear changes in expression levels of Of-E75A. Three replicates were collected per time point. Embryos (~100 μl per replicate) were frozen at −80°C in 150 μl of TRIzol until RNA extraction. To extract RNA, embryos were crushed in TRIzol, followed by the addition of 450 μl of TRIzol and 120 μl of chloroform. After centrifugation, the aqueous phase was moved to a new tube, and an equal volume of chloroform was added. The RNA-containing aqueous layer was again transferred to a new tube and precipitated with isopropanol. RNA pellets were washed with 70% ethanol, dried in a SpeedVac, and then resuspended in 50 μl of water. RNA quality was assessed using an Agilent 2100 bioanalyzer. We verified E75A expression in each sample used for RNA-seq using RT-PCR, which confirmed that E75A was expressed in samples from the second time point but was not detectable in the first and third time points (fig. S1). Primers Of-E75a-F-T7-2 and Of-E75a-3-RT7 were used to amplify E75A, and primers F_Of_actin and R_Of_actin were used to amplify Of-actin as a positive control.
Libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) sequencing. One hundred and fifty–base pair paired-end transcriptomes were sequenced using the Illumina HiSeq4000 system at the University of Maryland School of Medicine’s Institute for Genome Sciences. The Oncopeltus genome assembly was downloaded from the i5k workspace, which was indexed using Bowtie 2v2.2.9 (76). Transcriptome reads were aligned to the assembly using Tophat v2.1.1 (77). HTSeq (78) was used to quantify reads overlapping with gene models in the Oncopeltus official gene set (OGS) v1.2 (42).
Raw counts were normalized as transcripts per million values using the gene model length and sequencing depth. edgeR was used to identify differentially expressed genes using a quasi-likelihood F test with significant genes having false discovery rate <0.05 (40). Differentially expressed genes were sorted into clusters with highly correlated expression profiles using WGCNA v1.69 (41). As an unsigned network was generated, negatively and positively correlated genes clustered together. Each cluster is represented by a different color by WGCNA (“module” in data S2). To generate subclusters informed by the direction of the correlation, the clusters were split by degree of correlation with the eigengene; genes positively correlated with the eigengene are marked inverted = FALSE, and those negatively correlated with the eigengene are marked inverted = TRUE in data S2. Last, transcription factor–encoding genes were selected from the subcluster containing E75A (brown_FALSE; data S2) using the list of gene models previously determined to encode transcription factors (42).
Embryo fixation and in situ hybridization
Oncopeltus embryos were fixed as previously described (38). Briefly, embryos were covered with ≥1.5 ml of water, submerged in a boiling water bath for 3 min, and then transferred to ice until cool. The water was replaced with 1 ml of 1:1 12% paraformaldehyde (PFA):heptane. The embryos were shaken on a rotating platform for 20 min at 300 rpm. The liquid was replaced with 1 ml of 1:1 methanol:heptane, and the embryos were shaken manually for ~30 s. The embryos were then transferred to methanol and stored at −20°C in methanol. After rehydration to phosphate-buffered saline with Tween-20 (PBST), chorions were manually removed. We found that germ bands must be free of yolk to stain with iodophenyl-nitrophenyl-phenyltetrazolium chloride (INT)/bromochloroindolyl phosphate (BCIP), so germ-band–stage embryos were dissected away from the yolk at this time when performing a double in situ hybridization. Embryos were transferred back to methanol and stored at −20°C. Drosophila embryos were fixed according to established protocols.
Probe templates for E75A-coexpressed genes screened by in situ hybridization were PCR amplified from embryonic cDNA using the primers listed in table S1. All other primers are listed in table S2.
Antisense RNA probes for in situ hybridization were generated in vitro using ~200 ng of a PCR-amplified probe template, digoxigenin or biotin RNA labeling mix (Roche), T7 RNA polymerase and buffer, and ribonuclease inhibitor. The reactions were incubated for ≥2 hours at 37°C, LiCl and ethanol precipitated, washed with 70% ethanol, and resuspended in water. For in situ hybridization, the embryos were rehydrated and rinsed several times in PBST. Oncopeltus embryos were washed in 4% PFA for 1.5 hours, Drosophila embryos for 30 min, followed by several PBST rinses. For Drosophila embryos, most of the PBST was removed and embryos were incubated at 95°C for 5 min; this step was not performed for Oncopeltus embryos. All the remaining steps were the same for both species. The embryos were rinsed once in hybridization buffer [5× saline sodium citrate buffer (SSC), 50% (v/v) formamide, heparin (0.05 mg/ml), yeast tRNA (0.05 mg/ml), and 0.1% (v/v) Tween-20] and then incubated in hybridization buffer for at least 30 min at 65°C. The buffer was replaced with the probe diluted in hybridization buffer for overnight incubation at 65°C. A volume of diluted probe was used that ensured that the embryos were completely covered. After probe removal, the following washes were performed for ≥20 min each: two washes in prewarmed hybridization buffer at 65°C, one wash in prewarmed 2× SSC at 65°C, another in 2× SSC at RT, and one wash in 0.2× SSC at RT. All the remaining washes were performed at RT. The embryos were rinsed several times in PBST and then incubated in 10% sheep serum diluted in PBST for ≥30 min. Embryos were incubated in anti-digoxigenin antibody conjugated to alkaline phosphatase (Roche) diluted 1:1600 to 1:2000 in 10% sheep serum for ≥1 hour. Embryos were rinsed several times in PBST and occasionally left in PBST overnight at 4°C. Embryos were washed five times in PBST for ≥10 min each and then three times in fresh alkaline phosphatase staining buffer [0.1 M NaCl, 50 mM MgCl2, 0.1 M tris-HCl (pH 9.5), and 0.1% (v/v) Tween-20] for ≥5 min each. Embryos were stained in the dark with nitro blue tetrazolium (NBT)/BCIP (Roche, 4.5 μl of NBT and 3.5 μl of BCIP diluted in 1 ml of alkaline phosphatase staining buffer). Oncopeltus embryos were often left overnight at RT to stain. After adequate stain had developed, the embryos were rinsed several times in PBST, then in methanol and ethanol, and again in PBST.
When performing a double in situ hybridization, most of the PBST was removed from the tubes and embryos were incubated at 70°C for 30 min to inactivate the alkaline phosphatase conjugated to the first antibody. Embryos were washed in 4% PFA for 20 min, in 10% sheep serum/PBST for ≥30 min, and in anti–biotin-alkaline phosphatase antibody (Sigma-Aldrich) diluted 1:2000 in 10% sheep serum for ≥1 hour. Embryos were washed five times in PBST for ≥10 min each and then three times in alkaline phosphatase staining buffer for ≥5 min each. Embryos were stained overnight with INT/BCIP (Roche, 7.5 μl of stock solution in 1 ml of alkaline phosphatase staining buffer). Germ bands dissected away from yolk were mounted on slides in 70% glycerol and photographed using a Zeiss AxioImager M1 microscope. Multiple images were often required to capture the length of germ bands; these images were stitched together in Photoshop.
Embryonic RNAi
dsRNA was synthesized using the Megascript T7 RNA transcription kit (Ambion). For Of-Blimp1 dsRNA-1, the template was PCR amplified from cDNA using primers ofas008150-FT7 and ofas008150-RT7. For Of-Blimp1 dsRNA-2, the template was PCR amplified from cDNA using primers Of-Blimp-dsRNA2-FT7 and Of-Blimp-dsRNA2-RT7. For tGFP dsRNA, the template was PCR amplified from plasmid pBac[3xP3-DsRed; UAS-Tc′hsp_p-tGFP-SV40] (Addgene, #86453) using primers turbo GFP 1R_T7_2 and T7-tGFP-F. Sense and antisense strands were synthesized simultaneously according to kit instructions, and strands were annealed by denaturing the RNA for 3 min at 95°C and then slowly cooling to ≤40°C. The RNA was LiCl and ethanol precipitated, washed with 70% ethanol, and resuspended in water.
Embryos were injected with dsRNA as previously described (60). Embryos were fixed at 67 to 71.5 hours AEL. For length measurement, the embryos were mounted on slides and photographed under 40.5× magnification using a Zeiss Discovery.V12 microscope, and the length of each embryo was measured using the ZEN software. A Kruskal-Wallis test was used to compare the means between treatment groups, and a post hoc pairwise Wilcoxon rank sum test was used to determine where significant differences lie between treatment groups. The Bonferroni method was used to adjust P values for multiple comparisons.
Of-Blimp1 genomic and cDNA sequence gap filling
Blimp1 orthologs typically encode an intact PR/SET domain and five zinc fingers; however, the OFAS008150 gene model was found to encode only a partial PR/SET domain and three zinc fingers. Furthermore, there are several large gaps in the genome assembly around OFAS008150, suggesting that several portions of the Of-Blimp1 CDS were not assembled. We observed several regions of high RNA-seq read coverage upstream and downstream of the gene model, and individual mapped reads suggested that these were contiguous with portions of the gene model. We amplified 24 to 36 hour AEL cDNA using primers Of-Blimp1-upstream-2 and Of-Blimp1-downstream-2, yielding a 3.5-kb band, which was inserted into pGEM-T-Easy (Promega). Although this fragment is likely not the full-length Of-Blimp1 cDNA (data S1), sequencing revealed a 2-kb open reading frame, encoding a complete PR/SET domain and five zinc fingers, and 1.4 kb of 3′ untranslated region (3′UTR). One exon annotated in the gene model immediately downstream of the partial PR/SET-domain–encoding region was not found in the product amplified. The exon preceding the first zinc finger–encoding region was longer than annotated in the gene model (Fig. 4A, solid-outlined exons). Several exons are missing from the genome assembly altogether, which encode the C-terminal portion of the PR/SET domain, the first two zinc fingers, and the C terminus (Fig. 4A, dashed-outlined exons).
Since we planned to design gRNAs upstream of the zinc finger–encoding region, we wanted to be certain of the exon boundaries and genomic sequence in this region. We PCR amplified Oncopeltus gDNA using primers Of-Blimp-gDNAseq-F1 and Of-Blimp-gDNAseq-R2 with Taq polymerase (NEB), which yielded a 2.3-kb band, which was inserted into pGEM-T-Easy (Promega) and sequenced (data S1).
Design and in vitro synthesis of Of-Blimp1 gRNAs and Cas9 mRNA
CHOPCHOP (79) was used to find potential gRNA target sites within the two exons preceding the zinc finger–encoding region; suggested target sites were assessed for off-target genomic matches by aligning them to the Oncopeltus genome assembly using BLASTn. One target site in each of the two exons with 40 to 60% GC content, low self-complementarity scores, and no predicted off-target matches was selected. The gRNA target sites were named A and B (Fig. 4A, arrowheads). gRNAs were in vitro transcribed: Oligonucleotides were designed for each gRNA sequence, which incorporated a T7 RNA polymerase promoter sequence at the 5′ end and a region annealing to the sgRNA scaffold sequence at the 3′ end (Of-Blimp-gRNA-A, Of-Blimp-gRNA-B). Each oligo was used independently with gRNArev to PCR amplify the gRNA scaffold sequence from plasmid pCFD3 (Addgene, #49410) (80). One hundred nanograms of PCR product was supplied as template for in vitro RNA transcription using T7 polymerase (Megascript T7 kit). Each transcription reaction was incubated at 37°C for 3 hours and then for 15 min after the addition of 2 μl of TURBO DNase I. The products were purified using the Monarch RNA Cleanup kit (NEB) and stored at −80°C.
The template for Cas9 RNA synthesis was PCR amplified from plasmid MLM3613 (Addgene, #42251), which contains the Streptococcus pyogenes Cas9 coding sequence, using Q5 polymerase (NEB) with primers Cas9-mRNA-R and Cas9-mRNA-FT7, the latter of which has a T7 RNA polymerase promoter at the 5′ end. PCR product (250 ng) was used as template in an in vitro RNA synthesis reaction using the HiScribe T7 ARCA mRNA with tailing kit (NEB). The RNA synthesis and tailing reactions were performed following the manufacturer’s instructions. The RNA pre- and post-polyadenylation was checked on an agarose gel to ensure that the transcript was lengthened by the tailing reaction. The final polyadenylated mRNA was lithium chloride–precipitated, washed with 70% ethanol, resuspended in water, and stored at −80°C.
Of-Blimp1 CRISPR-Cas9 injections for germline editing, genotyping, and maintenance of lines
About 1 to 6 hours AEL, wild-type embryos were injected with Cas9 mRNA, Of-Blimp1 gRNA A, and Of-Blimp1 gRNA B (100 ng/μl of each) diluted in injection buffer [5 mM KCl and 0.1 mM phosphate buffer (pH 6.8)]. A single Of-vermilion gRNA (100 ng/μl) (52) was included in the injection mix to allow for a co-CRISPR screening strategy. Embryo injection and post-injection care were performed as previously described (60).
To genotype adults, genomic DNA samples were prepared from G1s by clipping a single mesothoracic leg and then immersing and crushing it in squish buffer [10 mM tris-HCl (pH 8.2), 1 mM EDTA, 25 mM NaCl, and proteinase K (0.2 mg/ml)], followed by incubation at 37°C for 30 min and 95°C for 2 min. One microliter of this gDNA preparation was used in one of three PCR reactions: PCR-A used primers Of-Blimp-screening-F1 and Of-Blimp-screening-R1; PCR-B used primers Of-Blimp-exon8-F and Of-Blimp-screening-R2; and PCR-AB used primers Of-Blimp-screening-F1 and Of-Blimp-screening-R2. Products from PCR-A and -B were subjected to a heteroduplex mobility assay (81) using a 4 to 5% agarose gel, and heterozygotes were identified by the presence of heteroduplex bands. Once the lines were established, primers Of-Blimp-screening-F1 and Of-Blimp-screening-R2 were used to amplify both gRNA target sites; PCR products were inserted into pGEM-T-Easy, and DNA from several colonies per line was sequenced. Nine independent mutant lines were verified by Sanger sequencing, and five lines with frameshifting mutations have been maintained for >10 generations by genotyping virgin females in this manner and outcrossing ~10 heterozygotes from each line to wild-type males each generation.
To genotype individual embryos, 5 days AEL embryos were photographed and phenotyped, squished in 8 μl of squish buffer, and the DNA was prepared and amplified as described above. Only individual embryos from lines Blimp140–3, Blimp122–12, and Blimp159–10 were genotyped in this manner as these Blimp1 alleles (Fig. 4E) contain deletions large enough to allow the PCR bands derived from the Blimp1 mutant and wild-type alleles to be easily distinguished (fig. S4D, samples 13 versus 14).
Of-Blimp1 CRISPR-Cas9 for somatic mutation
Embryos were injected as described above with Of-Blimp1 gRNA A and Cas9 mRNA, Of-Blimp1 gRNA B and Cas9 mRNA, or Cas9 mRNA alone (100 ng/μl of each). To score segment fusions of pre-hatchling stage embryos, the embryos were heat fixed after 7 days at 25°C by submerging in a boiling water bath for 3 min and chorions were manually removed. To score segment fusions in germ bands, the embryos were incubated at 25°C until they reached 67 to 71 hours AEL, when they were fixed according to the method described above (“Embryo fixation”).
D. melanogaster–Blimp1 CRISPR-Cas9 mutagenesis
CRISPR-Cas9 was used to create a null Dmel-Blimp1 allele. The Dmel-Blimp1 genomic region was obtained from Flybase (82). Regions of 200 bp surrounding the start codon (A region) and stop codon (B region) were independently submitted to CHOPCHOP (79) to find gRNA target sites. The gRNA template sequences were inserted into pCFD4 (Addgene, #49411) (80) using primers Dmel-Blimp-gRNA-F and Dmel-Blimp-gRNA-R following the recommendations of CRISPR Fly Design (https://crisprflydesign.org/protocols/). Briefly, the pCFD4 plasmid was amplified using the aforementioned primers. The PCR product and BbsI-cut pCFD4 were gel extracted and Gibson assembled using the HiFi Master Mix (NEB). The resulting plasmid encodes gRNA-A downstream of a U6-1 promoter and gRNA-B downstream of a U6-3 promoter. A repair template to replace the Dmel-Blimp1–coding region with 3XP3 > EGFP was constructed as follows: A 1097-bp homology arm upstream of the gRNA-A cut site was amplified from Drosophila gDNA using primers Dmel-Blimp-5-HDR-F and Dmel-Blimp-5-HDR-R, and a 942-bp homology arm downstream of the gRNA-B cut site was amplified using primers Dmel-Blimp-3-HDR-F and Dmel-Blimp-3-HDR-F. The 3XP3>EGFP construct was amplified from plasmid pbac[3xP3-EGFP;Tc′hsp5′-Gal4Delta-3′UTR] (Addgene, #86449) using primers Dmel-Blimp-HDR-insert-F and Dmel-Blimp-HDR-insert-R. These three PCR products were Gibson assembled into SmaI-digested pUC19 using the HiFi Master Mix (NEB).
The repair plasmid and gRNA expression vector described above were injected (Rainbow Transgenics) into yw;nanos > Cas9/CyO flies, derived from BL#78781 (y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = nanos-Cas9.R}attP40) and BL#22664 (y[1] w[67c23]; Mi{GFP[E.3xP3] = ET1}kkv[MB00004]). Sixty-two G0 flies were outcrossed to yw flies, and G1s expressing GFP in the eyes were selected. GFP+ G1 progeny were identified from 10 G0crosses, and at least two such G1s were selected to found lines by crossing to yw. Their y offspring were selected to eliminate the y rescue marker associated with the nanos > Cas9 element from the population. For two more generations, virgin GFP+ females were outcrossed to yw males to reduce the likelihood of maintaining off-target mutations. The mutant chromosomes were then balanced over the TM3, Ser, hb > lacZ chromosome. The hb > lacZ transgene on this balancer chromosome allowed us to differentiate heterozygous and homozygous embryos by the hb-like lacZ expression domain (Fig. 7D, head staining). Twenty-six balanced lines have been maintained. The genotype of the line used in this study was verified by sequencing both sides of the gene replacement site (data S1).
Acknowledgments
We thank the University of Maryland Evo-Devo-Gen group for many helpful conversations, particularly, T. Kocher for suggesting the RNA-seq approach. We thank J. Wexler, X. Gutierrez Ramos, and A. Cheatle Jarvela for feedback on this manuscript.
Funding: This work was supported by the National Institutes of Health grant R01GM113230 (L.P.) and National Sciences Foundation Graduate Research Program Fellowship (K.R.).
Author contributions: Conceptualization: L.P. and K.R. Investigation: K.R., A.H., and L.P. Methodology: K.R. and L.P. Resources: K.R. and L.P. Validation: K.R., A.H., and L.P. Formal analysis: K.R., A.H., and M.C. Software: M.C. Visualization: K.R. and A.H. Supervision: J.D.H. and L.P. Project administration: J.D.H. and L.P. Writing—original draft: K.R. and L.P. Writing—review and editing: K.R., L.P., and A.H. Funding acquisition: L.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw Illumina RNA-seq reads can be found in NCBI BioProject PRJNA1171382. All data needed to evaluate the conclusions of the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Figs. S1 to S6
Tables S1 and S2
Legends for data S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
REFERENCES AND NOTES
- 1.True J. R., Haag E. S., Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3, 109–119 (2001). [DOI] [PubMed] [Google Scholar]
- 2.E. S. Haag, J. R. True, Developmental system drift, in Evolutionary Developmental Biology: A Reference Guide, L. Nuño de la Rosa, G. B. Müller, Eds. (Springer International Publishing, 2021), pp. 99–110. [Google Scholar]
- 3.Eizinger A., Sommer R. J., The homeotic gene lin-39 and the evolution of nematode epidermal cell fates. Science 278, 452–455 (1997). [DOI] [PubMed] [Google Scholar]
- 4.K. Sander, Specification of the basic body pattern in insect embryogenesis, in Advances in Insect Physiology, J. E. Treherne, M. J. Berridge, V. B. Wigglesworth, Eds. (Academic Press, 1976), vol. 12, pp. 125–238. [Google Scholar]
- 5.Patel N. H., Martin-Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S., Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, 955–968 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Davis G. K., Patel N. H., Short, long, and beyond: Molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 47, 669–699 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Liu P. Z., Kaufman T. C., hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 131, 1515–1527 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Nüsslein-Volhard C., Wieschaus E., Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). [DOI] [PubMed] [Google Scholar]
- 9.Lawrence P. A., Johnston P., Macdonald P., Struhl G., Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skipped genes. Nature 328, 440–442 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Benedyk M. J., Mullen J. R., DiNardo S., odd-paired: A zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 8, 105–117 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Yu Y., Li W., Su K., Yussa M., Han W., Perrimon N., Pick L., The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature 385, 552–555 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Patel N. H., Ball E. E., Goodman C. S., Changing role of even-skipped during the evolution of insect pattern formation. Nature 357, 339–342 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Dawes R., Dawson I., Falciani F., Tear G., Akam M., Dax, a locust Hox gene related to fushi-tarazu but showing no pair-rule expression. Development 120, 1561–1572 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Patel N. H., Condron B. G., Zinn K., Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature 367, 429–434 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Maderspacher F., Bucher G., Klingler M., Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev. Genes Evol. 208, 558–568 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Choe C. P., Miller S. C., Brown S. J., A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. U.S.A. 103, 6560–6564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe C. P., Brown S. J., Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev. Biol. 302, 281–294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang J., Reding K., Heffer A., Pick L., Conservation and variation in pair-rule gene expression and function in the intermediate-germ beetle Dermestes maculatus. Development 114, 4625–4636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiyun O., Choe C. P., even-skipped acts as a pair-rule gene in germ band stages of Tribolium development. Dev. Biol. 462, 1–6 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Jeon H., Gim S., Na H., Choe C. P., A pair-rule function of odd-skipped in germband stages of Tribolium development. Dev. Biol. 465, 58–65 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Stuart J. J., Brown S. J., Beeman R. W., Denell R. E., A deficiency of the homeotic complex of the beetle Tribolium. Nature 350, 72–74 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Kumar S., Suleski M., Craig J. M., Kasprowicz A. E., Sanderford M., Li M., Stecher G., Hedges S. B., TimeTree 5: An expanded resource for species divergence times. Mol. Biol. Evol. 39, msac174 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakao H., Characterization of Bombyx embryo segmentation process: Expression profiles of engrailed, even-skipped, caudal, and wnt1/wingless homologues. J. Exp. Zool. B Mol. Dev. Evol. 314B, 224–231 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Nakao H., Analyses of interactions among pair-rule genes and the gap gene Krüppel in Bombyx segmentation. Dev. Biol. 405, 149–157 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Wilson M. J., Dearden P. K., Pair-rule gene orthologues have unexpected maternal roles in the honeybee (Apis mellifera). PLOS ONE 7, e46490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg M. I., Brent A. E., Payre F., Desplan C., Dual mode of embryonic development is highlighted by expression and function of Nasonia pair-rule genes. eLife 3, e01440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor S. E., Dearden P. K., The Nasonia pair-rule gene regulatory network retains its function over 300 million years of evolution. Development 149, dev199632 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wexler J., Pick L., Chipman A., Segmental expression of two ecdysone pathway genes during embryogenesis of hemimetabolous insects. Dev. Biol. 498, 87–96 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dearden P. K., Donly C., Grbić M., Expression of pair-rule gene homologues in a chelicerate: Early patterning of the two-spotted spider mite Tetranychus urticae. Development 129, 5461–5472 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Chipman A. D., Akam M., The segmentation cascade in the centipede Strigamia maritima: Involvement of the Notch pathway and pair-rule gene homologues. Dev. Biol. 319, 160–169 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Heffer A., Shultz J. W., Pick L., Surprising flexibility in a conserved Hox transcription factor over 550 million years of evolution. Proc. Natl. Acad. Sci. U.S.A. 107, 18040–18045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheatle Jarvela A. M., Trelstad C. S., Pick L., Regulatory gene function handoff allows essential gene loss in mosquitoes. Commun. Biol. 3, 540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pueyo J. I., Lanfear R., Couso J. P., Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc. Natl. Acad. Sci. U.S.A. 105, 16614–16619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. Chesebro, Mechanisms of Segmentation in the American Cockroach, Periplaneta americana, thesis, University of Sussex, Brighton, UK (2012). [Google Scholar]
- 35.Liu P. Z., Kaufman T. C., even-skipped is not a pair-rule gene but has segmental and gap-like functions in Oncopeltus fasciatus, an intermediate germband insect. Development 132, 2081–2092 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Auman T., Chipman A. D., Growth zone segmentation in the milkweed bug Oncopeltus fasciatus sheds light on the evolution of insect segmentation. BMC Evol. Biol. 18, 178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reding K., Chen M., Lu Y., Cheatle Jarvela A. M., Pick L., Shifting roles of Drosophila pair-rule gene orthologs: Segmental expression and function in the milkweed bug Oncopeltus fasciatus. Development 146, dev181453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erezyilmaz D. F., Kelstrup H. C., Riddiford L. M., The nuclear receptor E75A has a novel pair-rule-like function in patterning the milkweed bug, Oncopeltus fasciatus. Dev. Biol. 334, 300–310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fumasoni I., Meani N., Rambaldi D., Scafetta G., Alcalay M., Ciccarelli F. D., Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol. Biol. 7, 187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panfilio K. A., Vargas Jentzsch I. M., Benoit J. B., Erezyilmaz D., Suzuki Y., Colella S., Robertson H. M., Poelchau M. F., Waterhouse R. M., Ioannidis P., Weirauch M. T., Hughes D. S. T., Murali S. C., Werren J. H., Jacobs C. G. C., Duncan E. J., Armisén D., Vreede B. M. I., Baa-Puyoulet P., Berger C. S., Chang C.-C., Chao H., Chen M.-J. M., Chen Y.-T., Childers C. P., Chipman A. D., Cridge A. G., Crumière A. J. J., Dearden P. K., Didion E. M., Dinh H., Doddapaneni H. V., Dolan A., Dugan S., Extavour C. G., Febvay G., Friedrich M., Ginzburg N., Han Y., Heger P., Holmes C. J., Horn T., Hsiao Y.-M., Jennings E. C., Johnston J. S., Jones T. E., Jones J. W., Khila A., Koelzer S., Kovacova V., Leask M., Lee S. L., Lee C.-Y., Lovegrove M. R., Lu H.-L., Lu Y., Moore P. J., Munoz-Torres M. C., Muzny D. M., Palli S. R., Parisot N., Pick L., Porter M. L., Qu J., Refki P. N., Richter R., Rivera-Pomar R., Rosendale A. J., Roth S., Sachs L., Santos M. E., Seibert J., Sghaier E., Shukla J. N., Stancliffe R. J., Tidswell O., Traverso L., van der Zee M., Viala S., Worley K. C., Zdobnov E. M., Gibbs R. A., Richards S., Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Biol. 20, 64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachs L., Chen Y.-T., Drechsler A., Lynch J. A., Panfilio K. A., Lässig M., Berg J., Roth S., Dynamic BMP signaling polarized by Toll patterns the dorsoventral axis in a hemimetabolous insect. eLife 4, e05502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novikova A. V., Auman T., Cohen M., Oleynik O., Stahi-Hitin R., Gil E., Weisbrod A., Chipman A. D., The multiple roles of caudal in early development of the milkweed bug Oncopeltus fasciatus. Dev. Biol. 467, 66–76 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Liu P. Z., Patel N. H., giant is a bona fide gap gene in the intermediate germband insect, Oncopeltus fasciatus. Development 137, 835–844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crémazy F., Berta P., Girard F., Sox Neuro, a new Drosophila Sox gene expressed in the developing central nervous system. Mech. Dev. 93, 215–219 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Kispert A., Herrmann B. G., Leptin M., Reuter R., Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev. 8, 2137–2150 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Clark E., Peel A. D., Evidence for the temporal regulation of insect segmentation by a conserved sequence of transcription factors. Development 145, dev155580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller A. D., Maniatis T., Identification and characterization of a novel repressor of β-interferon gene expression. Genes Dev. 5, 868–879 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Huang S., Shao G., Liu L., The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 273, 15933–15939 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Kouzarides T., Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12, 198–209 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Reding K., Lê M., Pick L., Genome editing of the vermilion locus generates a visible eye color marker for Oncopeltus fasciatus. Sci. Rep. 13, 4188 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., Shirayama M., Mello C. C., A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Angelini D. R., Kaufman T. C., Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev. Biol. 283, 409–423 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Ng T., Yu F., Roy S., A homologue of the vertebrate SET domain and zinc finger protein Blimp-1 regulates terminal differentiation of the tracheal system in the Drosophila embryo. Dev. Genes Evol. 216, 243–252 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Öztürk-Çolak A., Stephan-Otto Attolini C., Casanova J., Araújo S. J., Blimp-1 mediates tracheal lumen maturation in Drosophila melanogaster. Genetics 210, 653–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lev O., Chipman A. D., Development of the Pre-gnathal segments in the milkweed bug Oncopeltus fasciatus suggests they are not serial homologs of trunk segments. Front. Cell Dev. Biol. 9, 695135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakimoto B. T., Turner F. R., Kaufman T. C., Defects in embryogenesis in mutants associated with the antennapedia gene complex of Drosophila melanogaster. Dev. Biol. 102, 147–172 (1984). [DOI] [PubMed] [Google Scholar]
- 59.Chipman A. D., Oncopeltus fasciatus as an evo-devo research organism. Genesis 55, e23020 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Reding K., Pick L., High-efficiency CRISPR/Cas9 mutagenesis of the white gene in the milkweed bug Oncopeltus fasciatus. Genetics 215, 1027–1037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark E., Akam M., Odd-paired controls frequency doubling in Drosophila segmentation by altering the pair-rule gene regulatory network. eLife 5, e18215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu P. Z., Kaufman T. C., Krüppel is a gap gene in the intermediate germband insect Oncopeltus fasciatus and is required for development of both blastoderm and germband-derived segments. Development 131, 4567–4579 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Hernandez J., Pick L., Reding K., Oncopeltus-like gene expression patterns in Murgantia histrionica, a new hemipteran model system, suggest ancient regulatory network divergence. EvoDevo 11, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.A. D. Economou, Phylogenetic and Developmental Studies into the Evolution of an Insect Novelty, thesis, University of London, University College London (United Kingdom), England (2008). [Google Scholar]
- 65.Agawa Y., Sarhan M., Kageyama Y., Akagi K., Takai M., Hashiyama K., Wada T., Handa H., Iwamatsu A., Hirose S., Ueda H., Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol. Cell. Biol. 27, 8739–8747 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li C., Yun X., Li B., Dusky-like is required for epidermal pigmentation and metamorphosis in Tribolium castaneum. Sci. Rep. 6, 20102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bunker J., Bashir M., Bailey S., Boodram P., Perry A., Delaney R., Tsachaki M., Sprecher S. G., Nelson E., Call G. B., Rister J., Blimp-1/PRDM1 and Hr3/RORβ specify the blue-sensitive photoreceptor subtype in Drosophila by repressing the hippo pathway. Front. Cell Dev. Biol. 11, 1058961 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu S.-Y., Tong X.-L., Li C.-L., Ding X., Zhang Z.-L., Fang C.-Y., Tan D., Hu H., Liu H., Dai F.-Y., BmBlimp-1 gene encoding a C2H2 zinc finger protein is required for wing development in the silkworm Bombyx mori. Int. J. Biol. Sci. 15, 2664–2675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakamura T., Extavour C. G., The transcriptional repressor Blimp-1 acts downstream of BMP signaling to generate primordial germ cells in the cricket Gryllus bimaculatus. Development 143, 255–263 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Turner C. A., Mack D. H., Davis M. M., Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77, 297–306 (1994). [DOI] [PubMed] [Google Scholar]
- 71.Ren B., Chee K. J., Kim T. H., Maniatis T., PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13, 125–137 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Győry I., Wu J., Fejér G., Seto E., Wright K. L., PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5, 299–308 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Johnston D. M., Sedkov Y., Petruk S., Riley K. M., Fujioka M., Jaynes J. B., Mazo A., Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol. Cell 44, 51–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heffer A., Grubbs N., Mahaffey J., Pick L., The evolving role of the orphan nuclear receptor ftz-f1, a pair-rule segmentation gene. Evol. Dev. 15, 406–417 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Fitzpatrick V. D., Percival-Smith A., Ingles C. J., Krause H. M., Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature 356, 610–612 (1992). [DOI] [PubMed] [Google Scholar]
- 76.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Labun K., Montague T. G., Krause M., Cleuren Y. N. T., Tjeldnes H., Valen E., CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Port F., Chen H.-M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhattacharya D., Van Meir E. G., A simple genotyping method to detect small CRISPR-Cas9 induced indels by agarose gel electrophoresis. Sci. Rep. 9, 4437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Öztürk-Çolak A., Marygold S. J., Antonazzo G., Attrill H., Goutte-Gattat D., Jenkins V. K., Matthews B. B., Millburn G., dos Santos G., Tabone C. J.; Flybase Consortium , FlyBase: Updates to the Drosophila genes and genomes database. Genetics 227, iyad211 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2
Legends for data S1 and S2
Data S1 and S2