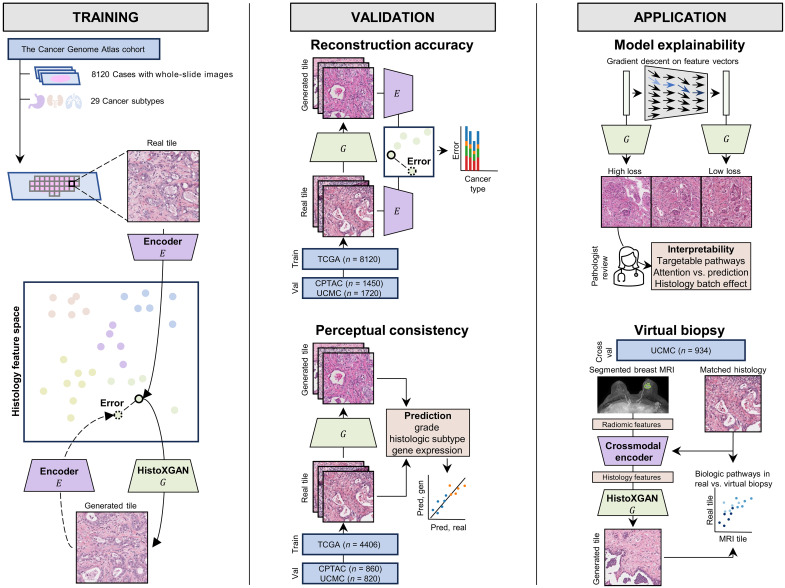
Abstract
Artificial intelligence models have been increasingly used in the analysis of tumor histology to perform tasks ranging from routine classification to identification of molecular features. These approaches distill cancer histologic images into high-level features, which are used in predictions, but understanding the biologic meaning of such features remains challenging. We present and validate a custom generative adversarial network—HistoXGAN—capable of reconstructing representative histology using feature vectors produced by common feature extractors. We evaluate HistoXGAN across 29 cancer subtypes and demonstrate that reconstructed images retain information regarding tumor grade, histologic subtype, and gene expression patterns. We leverage HistoXGAN to illustrate the underlying histologic features for deep learning models for actionable mutations, identify model reliance on histologic batch effect in predictions, and demonstrate accurate reconstruction of tumor histology from radiographic imaging for a “virtual biopsy.”
A generative adversarial network can accurately reconstruct pan-cancer histology from a generalized latent space.
INTRODUCTION
Histopathologic analysis of tumors is an essential step in the diagnosis and treatment of cancer in modern clinical oncology. The initial diagnosis of cancers depends on morphological assessment of biopsy samples, and molecular profiling now informs prognosis and clinical therapeutic decisions in almost every cancer subtype. Machine learning and, more specifically, deep learning has been successfully applied to most standard steps of pathologic image analysis and can segment (1), diagnose (2), grade (3), and even predict recurrence or treatment response for tumors (4). As the field has evolved, studies have moved beyond basic pattern recognition toward identifying deeper disease traits and complex morphological features, including the identification of genomic and transcriptomic profiles directly from histology (5–7). Conceptually, deep learning models often condense complex visual information from histopathology into a small number of higher-order features for prediction, often using pretraining from large image datasets such as ImageNet (8) or feature extractors trained with self-supervised learning (SSL) (9–11). However, the opacity of these high-level features limits adoption and deployment due to concerns about model trustworthiness (12), and lack of interpretability limits the ability to gain new insight from the histologic patterns recognized by models.
A range of techniques exist for explaining machine learning model predictions in medical imaging, including saliency mapping, attention mechanisms, and perturbation-based approaches (13, 14). However, these approaches may identify regions important for prediction, such as tumor epithelium, but may not identify which characteristics of these regions have led to a positive or negative prediction (14). This is critically important in validation studies as results may be confounded by batch effects that are hard to characterize without thoroughly evaluating the features underpinning a model’s prediction (15). Generative adversarial networks (GANs) provide an attractive alternative framework for explainability. GAN frameworks like StyleGAN2 train a generator to produce realistic synthetic images able to fool a discriminator network, and the resulting generator latent space captures semantic concepts and can be manipulated for intuitive image editing (13). Conditional GANs can be used to interpolate between two histologic classes, but training is time-consuming, and such an approach can only embed a limited number of classes (5).
We consider an alternative approach to synthetic image generation—If histology could be reconstructed from high-level features derived from SSL-based extractors, the visual meaning of these features (or models trained from these features) can be deciphered. In addition, reconstruction of histology from base features enables the development of accurate cross-modal autoencoders to reconstruct histology from other forms of data (16, 17), such as sequencing or magnetic resonance imaging (MRI), enabling a “virtual biopsy.” Approaches like Encoder4Editing and pix2style2pix allow the embedding of images into the latent space of a GAN but cannot successfully reconstruct histologic images in their base configurations (18, 19). To address this, we present HistoXGAN (Histology feature eXplainability Generative Adversarial Network), a custom architecture that uses features from highly effective SSL-based feature extractors to accurately reconstruct histology.
RESULTS
Accurate reconstruction of histologic structures
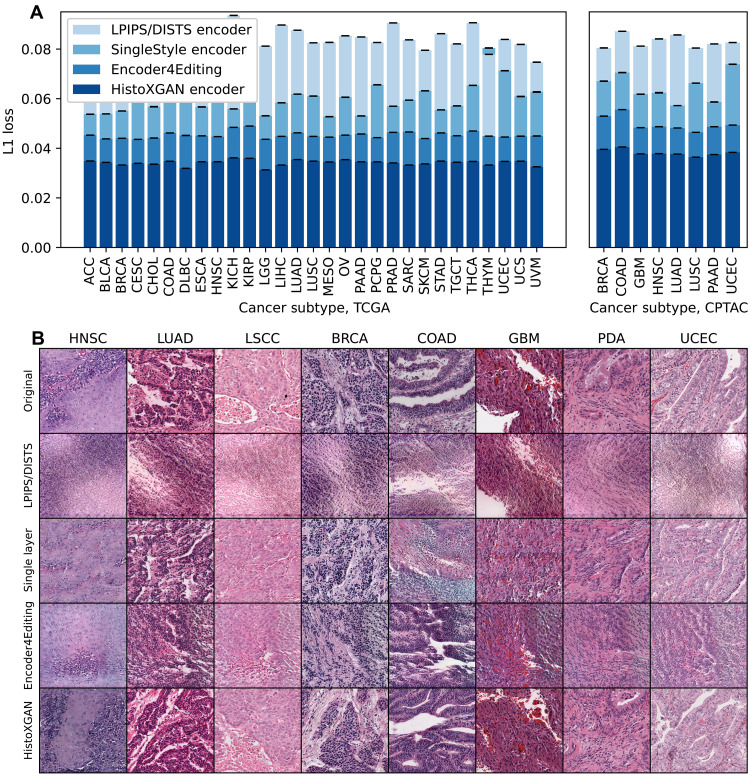
The HistoXGAN architecture is a GAN (13, 20) that ensures consistency of key image features extracted by SSL feature extractors during image generation (Fig. 1, left, and fig. S1) (11, 21, 22). In this way, generated images are structurally similar, but the location of image structures is allowed to vary between images. With this approach, images can be generated by providing a feature vector directly as the input latent vector without requiring a separate encoder to project features into the StyleGAN latent space. This model was trained using 8120 cases (8232 slides with 5,733,871 image tiles) from 29 cancer types in The Cancer Genome Atlas (TCGA). We compared L1 loss/mean absolute error (Fig. 1, middle) between extracted features from the input and reconstructed images generated by HistoXGAN and alternative encoders across 8120 cases in the training TCGA dataset and an additional n = 1328 cases from the Clinical Proteomics Tumor Analysis Consortium (CPTAC) dataset. HistoXGAN achieved the lowest reconstruction error, with a mean error of 0.034 [95% confidence interval (CI), 0.034 to 0.034] across TCGA and 0.038 (95% CI, 0.038 to 0.038) across CPTAC for reconstruction of CTransPath features (Fig. 2 and Table 1), a mean error of 0.010 (95% CI, 0.010 to 0.010) across TCGA and 0.011 (95% CI, 0.011 to 0.011) across CPTAC for reconstruction of RetCCL features (fig. S2 and table S1), and a mean error of 0.689 (95% CI, 0.689 to 0.689) across TCGA and 0.731 (95% CI, 0.731 to 0.731) in CPTAC for reconstruction of UNI features (fig. S3 and table S2). This represented a 22, 17, and 28% improvement over the second-best model, Encoder4Editing, for reconstruction of CTransPath, RetCCL, and UNI features, respectively, in CPTAC. This improved feature reconstruction was reflected in pathologist review of images generated by HistoXGAN: eight tiles, one from each cancer subtype in the validation cohort, were regenerated using the four encoders, and four pathologists with over 50 years of combined experience were presented with the generated images in random order. The HistoXGAN reconstructions were judged as the most similar to input images in 75% (24 of 32) of cases when using CTransPath features, 84% (27 of 32) of cases using RetCCL features, and 94% (31 of 32) of cases using UNI features (Fig. 2B and figs. S2B and S3B).
Fig. 1. Overview of HistoXGAN training, validation, and application.
As illustrated on the (left), the HistoXGAN generator G was trained using 8120 cases across 29 cancer types in TCGA. HistoXGAN takes as an input a histologic feature vector derived from any self-supervised feature extractor E and generates a histology tile with near-identical features with respect to the same feature extractor. As shown in the (middle), in this study, we demonstrate that this architecture accurately reconstructs the encoded features from multiple feature extractors in both the training TCGA dataset and external datasets of 3201 slides from 1450 cases from CPTAC and 2656 slides from 1720 cases from University of Chicago Medical Center (UCMC). In addition, we demonstrate that the real and reconstructed images carry near-identical information of interpretable pathologic features such as grade, histologic subtype, and gene expression data. As illustrated to the (right), we showcase the applications of this architecture for model interpretability using gradient descent to illustrate features used in deep learning model predictions. Through systematic review of these features with expert pathologists, we identify characteristics of cancers with targetable pathways, such as homologous recombination deficiency (HRD) and PIK3CA in breast cancer, and illustrate application for attention based models. Last, we train a crossmodal encoder to translate MRI radiomic features into histology features using paired breast MRIs and histology from 934 breast cancer cases from the UCMC and apply HistoXGAN to generate representative histology directly from MRI.
Fig. 2. Reconstruction accuracy in training and validation datasets for CTransPath encoders.
We compare reconstruction accuracy from the real and reconstructed images for HistoXGAN and other architectures for embedding images in GAN latent space. For comparison, we use encoders designed to recreate images from a StyleGAN2 model trained identically to the HistoXGAN model. The Learned Perceptual Image Patch Similarity (LPIPS)/Deep-Image Structure and Texture Similarity (DISTS) encoder uses an equal ratio of LPIPS/DISTS loss between the real and reconstructed images to train the encoder. The Single-Layer and Encoder4Editing encoders are trained to minimize L1 loss between CTransPath feature vector of the real and reconstructed images. (A) HistoXGAN provides more accurate reconstruction of CTransPath features across the TCGA dataset used for GAN training (n = 8120) and solid tumor CPTAC validation (n = 1328) dataset, achieving an average of 30% improvement in L1 loss over the Encoder4Editing encodings in the validation dataset. (B) HistoXGAN reconstructed images consistently provided more accurate representations of features from the input image across cancer types in the CPTAC validation dataset.
Table 1. Reconstruction accuracy in training and validation datasets for CTransPath encoders.
We compare reconstruction accuracy from the real and reconstructed images for HistoXGAN and other architectures for embedding images in GAN latent space. For comparison, we use encoders designed to recreate images from a StyleGAN2 model trained identically to the HistoXGAN model. The LPIPS/DISTS encoder uses an equal ratio of LPIPS/DISTS loss between the real and reconstructed images to train the encoder. The Single Layer and Encoder4Editing encoders are trained to minimize L1 loss between CTransPath feature vector of the real and reconstructed images.
| Source | n | n tiles | LPIP/DISTSL1 loss | Single layer L1 loss | Encoder4Editing L1 loss | HistoXGAN L1 loss | |
|---|---|---|---|---|---|---|---|
| TCGA | ACC | 56 | 78789 | 0.086 (0.010) | 0.054 (0.010) | 0.045 (0.006) | 0.035 (0.004) |
| TCGA | BLCA | 378 | 342,463 | 0.082 (0.009) | 0.054 (0.015) | 0.044 (0.006) | 0.034 (0.004) |
| TCGA | BRCA | 943 | 454,985 | 0.087 (0.010) | 0.055 (0.017) | 0.044 (0.006) | 0.033 (0.004) |
| TCGA | CESC | 267 | 165,088 | 0.083 (0.010) | 0.063 (0.021) | 0.044 (0.006) | 0.034 (0.004) |
| TCGA | CHOL | 38 | 51,414 | 0.087 (0.009) | 0.057 (0.019) | 0.044 (0.006) | 0.034 (0.004) |
| TCGA | COADREAD | 428 | 195,493 | 0.092 (0.013) | 0.071 (0.023) | 0.046 (0.007) | 0.035 (0.004) |
| TCGA | DLBC | 43 | 32,073 | 0.076 (0.011) | 0.063 (0.022) | 0.045 (0.010) | 0.032 (0.005) |
| TCGA | ESCA | 147 | 99,625 | 0.085 (0.010) | 0.057 (0.017) | 0.045 (0.007) | 0.035 (0.005) |
| TCGA | HNSC | 401 | 166,061 | 0.083 (0.009) | 0.060 (0.019) | 0.045 (0.006) | 0.035 (0.004) |
| TCGA | KICH | 101 | 105,461 | 0.094 (0.010) | 0.056 (0.009) | 0.048 (0.006) | 0.036 (0.005) |
| TCGA | KIRP | 270 | 234,740 | 0.091 (0.013) | 0.070 (0.022) | 0.049 (0.007) | 0.036 (0.005) |
| TCGA | LGG | 464 | 155,579 | 0.081 (0.009) | 0.053 (0.014) | 0.044 (0.006) | 0.031 (0.005) |
| TCGA | LIHC | 359 | 331,769 | 0.090 (0.013) | 0.058 (0.019) | 0.045 (0.006) | 0.033 (0.004) |
| TCGA | LUAD | 467 | 335,499 | 0.088 (0.010) | 0.062 (0.019) | 0.046 (0.007) | 0.035 (0.004) |
| TCGA | LUSC | 474 | 370,542 | 0.083 (0.009) | 0.061 (0.019) | 0.045 (0.006) | 0.035 (0.004) |
| TCGA | MESO | 73 | 42,242 | 0.083 (0.009) | 0.053 (0.011) | 0.044 (0.006) | 0.034 (0.004) |
| TCGA | OV | 104 | 108,620 | 0.085 (0.010) | 0.061 (0.019) | 0.045 (0.007) | 0.035 (0.005) |
| TCGA | PAAD | 168 | 119,144 | 0.085 (0.009) | 0.053 (0.012) | 0.046 (0.007) | 0.035 (0.005) |
| TCGA | PCPG | 173 | 207,803 | 0.083 (0.009) | 0.066 (0.022) | 0.044 (0.006) | 0.035 (0.004) |
| TCGA | PRAD | 394 | 202,439 | 0.091 (0.010) | 0.057 (0.015) | 0.046 (0.007) | 0.034 (0.004) |
| TCGA | SARC | 250 | 326,262 | 0.084 (0.011) | 0.059 (0.019) | 0.047 (0.007) | 0.033 (0.005) |
| TCGA | SKCM | 418 | 322,527 | 0.080 (0.009) | 0.063 (0.023) | 0.044 (0.006) | 0.034 (0.004) |
| TCGA | STAD | 371 | 283,734 | 0.086 (0.011) | 0.055 (0.014) | 0.046 (0.008) | 0.035 (0.005) |
| TCGA | TGCT | 129 | 109,128 | 0.082 (0.008) | 0.057 (0.015) | 0.045 (0.007) | 0.034 (0.005) |
| TCGA | THCA | 480 | 279,362 | 0.091 (0.010) | 0.065 (0.022) | 0.047 (0.007) | 0.035 (0.005) |
| TCGA | THYM | 114 | 157,626 | 0.078 (0.010) | 0.080 (0.028) | 0.045 (0.008) | 0.033 (0.006) |
| TCGA | UCEC | 477 | 351,784 | 0.084 (0.010) | 0.071 (0.025) | 0.045 (0.006) | 0.035 (0.004) |
| TCGA | UCS | 53 | 67,853 | 0.082 (0.009) | 0.061 (0.019) | 0.045 (0.006) | 0.035 (0.005) |
| TCGA | UVM | 80 | 35,766 | 0.075 (0.009) | 0.063 (0.020) | 0.045 (0.007) | 0.033 (0.005) |
| CPTAC | BRCA | 105 | 56,318 | 0.084 (0.008) | 0.067 (0.016) | 0.053 (0.008) | 0.040 (0.005) |
| CPTAC | COADREAD | 104 | 76,915 | 0.086 (0.009) | 0.071 (0.015) | 0.056 (0.007) | 0.041 (0.005) |
| CPTAC | GBM | 177 | 143,858 | 0.080 (0.010) | 0.062 (0.016) | 0.048 (0.007) | 0.038 (0.006) |
| CPTAC | HNSC | 108 | 35,064 | 0.080 (0.009) | 0.062 (0.018) | 0.049 (0.007) | 0.038 (0.005) |
| CPTAC | LUAD | 221 | 425,473 | 0.087 (0.009) | 0.057 (0.012) | 0.048 (0.006) | 0.038 (0.004) |
| CPTAC | LUSC | 202 | 359,394 | 0.081 (0.008) | 0.066 (0.021) | 0.046 (0.006) | 0.036 (0.004) |
| CPTAC | PAAD | 164 | 99,093 | 0.082 (0.009) | 0.059 (0.016) | 0.049 (0.008) | 0.037 (0.006) |
| CPTAC | UCEC | 247 | 184,767 | 0.083 (0.010) | 0.074 (0.021) | 0.049 (0.008) | 0.038 (0.006) |
To determine the dependency of HistoXGAN performance on training dataset composition, the CTransPath model trained from 29 cancer types as above was compared to models trained from TCGA Lung Squamous Cell Carcinoma (TCGA-LUSC), TCGA Lung Adenocarcinoma (TCGA-LUAD), and from the eight TCGA sites corresponding to the eight CPTAC validation cohorts (fig. S4 and table S3). Models trained from single tumor sites had worse performance on validation (TCGA-LUSC model mean error 0.0411 in CPTAC; TCGA-LUAD model mean error 0.0430), whereas minimal difference was seen between the 8 tumor–type model (mean error, 0.0380) and the 29 tumor–type model (mean error, 0.0377). The TCGA-LUSC model outperformed the TCGA-LUAD model in reconstruction of squamous cancers [including TCGA Head and Neck Squamous Cell Carcinoma (TCGA-HNSC), CPTAC-HNSC, and CPTAC-LUSC], whereas the TCGA-LUAD model performed better in adenocarcinoma reconstruction [including in TCGA Colorectal Adenocarcinoma (TCGA-COADREAD)]. Visual comparison of model reconstructions demonstrated that the TCGA-LUSC model failed to reconstruct some glandular structures in adenocarcinoma cases (fig. S4B), indicating that lack of exposure to specific histologic features during training may lead to poor reconstruction of those features on application.
Reconstruction of rare tumors and histologic subtypes
We also sought to evaluate HistoXGAN in cases that were poorly represented or unrepresented in the training dataset to better assess limitations of this approach. First, we assessed accuracy of predictions by histologic subtype in breast, lung, and colorectal cancers in the TCGA training (n = 2303), CPTAC (n = 527), and University of Chicago Medical Center (UCMC, n = 1113) validation datasets (fig. S5A and data S1). Performance was robust across subtypes that were rare or absent from the TCGA training subset, including mucinous, metaplastic, and tubular breast cancers; signet ring, enteric, and neuroendocrine lung adenocarcinoma; solid-type lung squamous cell carcinoma; and mucinous and signet ring colorectal adenocarcinoma. Reconstruction error was generally under 0.040 for these rare subtypes, although slightly higher errors were seen in mucinous breast in CPTAC (mean error, 0.043) and UCMC (mean error, 0.057), mucinous lung adenocarcinoma in CPTAC (mean error, 0.042), and mucinous colorectal adenocarcinoma in CPTAC (mean error, 0.041). Visualization of image tiles and reconstruction for breast cancer subtypes using cases from UCMC (fig. S5B) demonstrated key pathologic features of breast cancer subtypes were well represented by HistoXGAN, including tubule formation in tubular cancer, squamous differentiation and keratinization in metaplastic cancer, and mixed ductal and single-file lines of cells in mixed ductal/lobular cancer. Examination of the mucinous cases from UCMC demonstrated that one slide image was out of focus, explaining the higher error seen in UCMC mucinous tumor reconstruction. The in-focus slide had pathologically identifiable mucinous areas, although the mucinous areas were less homogenous than the ground truth source image. Given the consistently lower performance in mucinous tumor reconstruction, this may be one notable limitation of the HistoXGAN architecture when trained across the entire TCGA dataset where mucinous tumors are relatively rare.
To further characterize the limits of HistoXGAN reconstruction, we curated a set of 768 cases from UCMC representing a wide array of 176 OncoTree diagnoses, with samples obtained from 29 different anatomic sites. Average accuracy was consistent with the CPTAC validation cohort and was generally robust across diagnosis and anatomic site (data S2 and S3). Lower accuracy for reconstruction was seen for one slide each of nasopharyngeal cancer (mean error, 0.087), bladder urothelial carcinoma (mean error, 0.060), and low-grade glioma not otherwise specified (mean error, 0.058). However, visualization of reconstructions across these three cases demonstrated that low reconstruction accuracy was largely due to artifacts such as pen markings or oil in slide images (fig. S5C). Last, we assessed HistoXGAN in a cohort of n = 88 cases of acute myeloid leukemia (AML) from CPTAC, revealing a mean error of 0.066. This error was not artifactual: The high reconstruction loss was due to errors in reconstruction, with excessive stellate cytoplasm generated along with stroma more reminiscent of a solid tumor, demonstrating that this approach is not extensible to reconstruction of blood smear images (fig. S5C).
Reconstructed histology retains meaningful representations of tumor biology
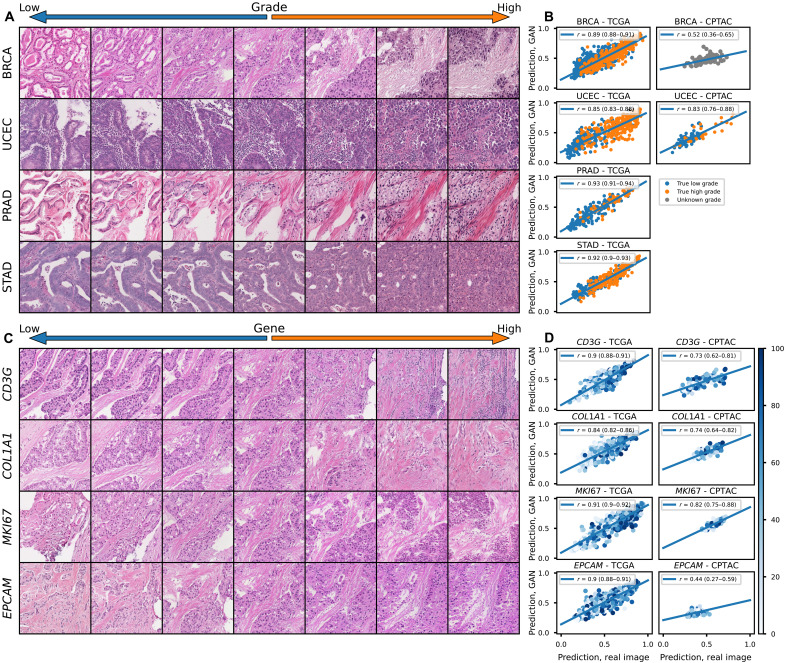
A meaningful synthetic reconstruction of tumor histology from features in a shared latent space should retain key elements of tumor biology that are reflected in pathology – for example, the tumor grade of the reconstructed histology should be identical to the original. To test these aspects of our approach to reconstruction in a systematic and quantitative fashion, we trained deep learning models for grade (TCGA/CPTAC n = 943/100 breast, 168/139 pancreas, 391/107 head and neck, 227/none prostate, 477/99 uterus, 378/none bladder, and 371/none stomach), histologic subtype (743/92 breast with an additional 820 cases included from UCMC, 941/415 lung, 147/none esophageal, and 363/none kidney), and gene expression (941/97 breast), and compared predictions made from whole-slide images to those made from the same set of tiles reconstructed with HistoXGAN (Fig. 1, middle). Models for these tasks were trained with a non-SSL–based architecture, so the predictions are not based on the same image features used to train HistoXGAN. For prediction of high tumor grade (defined as grade 3 for breast, pancreatic, uterine, stomach, and bladder; grade 3 or 4 for head and neck; or Gleason grade 9 or 10 for prostate; Fig. 3 and Table 2), the predictions from real slides and from reconstructed tiles were highly correlated with correlation coefficients ranging from 0.52 (95% CI, 0.36 to 0.65; P = 2.33 × 10−8) in CPTAC Breast Carcinoma (CPTAC-BRCA) to 0.85 (95% CI, 0.79 to 0.89; P = 1.01 × 10−29) in CPTAC Pancreatic Adenocarcinoma (CPTAC-PAAD). Similar findings were seen for the prediction of tumor histologic subtype in TCGA/CPTAC/UCMC breast, lung, esophageal, and renal cancers (fig. S6 and Table 3) as well as for the prediction of gene expression of CD3G, COL1A1, MKI67, and EPCAM in TCGA/CPTAC breast cancer cohorts (Fig. 3 and Table 4). Transition between states of grade, histology, and gene expression can all be readily visualized using HistoXGAN reconstructions (Fig. 3, A and C, and fig. S6A).
Fig. 3. Perceptual consistency of tumor grade and gene expression in reconstructed images.
(A) Illustration of transition between low and high grade (defined as grade 3 for breast, uterine, or stomach, and Gleason grades 9 or 10 for prostate) across a single image from four cancer types. A vector representing high grade is derived from the coefficients of a logistic regression predicting grade from CTransPath features. This vector is subtracted from the base image to visualize lower grade and added to the base image to visualize higher grade. (B) Correlation between predictions of grade from real and reconstructed tiles, averaged per patient, across cancer types, demonstrating a high perceptual similarity of the grade of the real and generated images. For the TCGA datasets, a deep learning model was trained to predict grade from real tiles for each cancer type using threefold cross-validation. The correlation between predictions for real/generated images is aggregated for the three held-out validation sets. For the CPTAC validation, a deep learning model trained across the entire corresponding TCGA dataset was used to generate predictions. True pathologist-determined high versus low grade is annotated on the images when available. (C) Illustration of transition between expression of select genes across a single image from TCGA-BRCA. (D) Correlation between predictions of gene expression from real and reconstructed tiles, averaged per patient, demonstrating a high perceptual similarity of the gene expression of the real and generated images. True gene expression as a percentile from 0 to 100 is indicated by the color of each data point.
Table 2. Perceptual consistency of tumor grade in reconstructed images across cancer types.
Correlation between predictions of grade from real and reconstructed tiles, averaged per patient, across cancer types, demonstrating a high perceptual similarity of the grade of the real and generated images. For the TCGA datasets, a deep learning model was trained to predict grade from real tiles for each cancer type using threefold cross-validation. The correlation between predictions for real/generated images is aggregated for the three held-out validation sets. For the CPTAC validation, a deep learning model trained across the entire corresponding TCGA dataset was used to generate predictions. Average area under the receiver operating characteristic (AUROC) and average precision (AP) are listed for prediction of grade using the above models, as well as when predictions from these models are made with reconstructed (Gen) versions of tiles. The similar AUROC/AP from real tiles and reconstructed tiles illustrates the reconstructed tiles retain informative data with regard to grade.
| Source | n | High grade (%) | Pearson r | P value, correlation | AUROC | AUROC, Gen. | AP | AP, Gen. | |
|---|---|---|---|---|---|---|---|---|---|
| TCGA | BRCA | 943 | 36.5 | 0.89 (0.88–0.91) | < 1 × 10−99 | 0.81 | 0.74 | 0.69 | 0.60 |
| TCGA | PAAD | 168 | 29.8 | 0.85 (0.81–0.89) | 3.90 × 10−49 | 0.52 | 0.58 | 0.32 | 0.40 |
| TCGA | PRAD | 227 | 22.9 | 0.93 (0.91–0.94) | 5.30 × 10−99 | 0.84 | 0.82 | 0.57 | 0.53 |
| TCGA | HNSC | 391 | 25.6 | 0.9 (0.88–0.92) | 9.78 × 10−142 | 0.64 | 0.62 | 0.41 | 0.39 |
| TCGA | UCEC | 477 | 57.0 | 0.85 (0.83–0.88) | 3.07 × 10−137 | 0.92 | 0.94 | 0.85 | 0.88 |
| TCGA | BLCA | 378 | 93.9 | 0.94 (0.93–0.95) | 4.19 × 10−178 | 0.9 | 0.99 | 0.87 | 0.98 |
| TCGA | STAD | 371 | 60.1 | 0.92 (0.9–0.93) | 7.89 × 10−152 | 0.77 | 0.82 | 0.75 | 0.81 |
| CPTAC | BRCA | 100 | N/A | 0.52 (0.36–0.65) | 2.33 × 10−8 | N/A | N/A | N/A | N/A |
| CPTAC | PAAD | 139 | 22.3 | 0.85 (0.79–0.89) | 1.65 × 10−39 | 0.65 | 0.63 | 0.39 | 0.33 |
| CPTAC | HNSC | 107 | N/A | 0.84 (0.77–0.89) | 1.01 × 10−29 | N/A | N/A | N/A | N/A |
| CPTAC | UCEC | 99 | 26.3 | 0.83 (0.76–0.88) | 2.02 × 10−26 | 0.83 | 0.79 | 0.76 | 0.62 |
Table 3. Perceptual consistency of histologic subtype in reconstructed images across cancer types.
Correlation between predictions of histologic subtype from real and reconstructed tiles, averaged per patient, across cancer types, demonstrating a high perceptual similarity of the histologic subtype of the real and generated images. For the TCGA datasets, a deep learning model was trained to predict subtype from real tiles for each cancer type using threefold cross-validation. The correlation between predictions for real/generated images is aggregated for the three held-out validation sets. For CPTAC and UCMC validation, a deep learning model trained across the entire corresponding TCGA dataset was used to generate predictions. AUROC and AP are listed for prediction of histologic subtype using the above models, as well as when predictions from these models are made with reconstructed (Gen) versions of tiles. The similar AUROC/AP from real tiles and reconstructed tiles illustrates the reconstructed tiles retain informative data with regard to histologic subtype.
| Source | n | Histology (%) | Pearson r | P value, correlation | AUROC | AUROC, Gen. | AP | AP, Gen. | |
|---|---|---|---|---|---|---|---|---|---|
| TCGA | BRCA | 734 | Ductal (77.2) Lobular (22.8) | 0.94 (0.93–0.95) | <1 × 10−99 | 0.96 | 0.96 | 0.92 | 0.90 |
| TCGA | LUNG | 941 | Adeno (49.6) Squamous (51.4) | 0.94 (0.92–0.96) | 9.57 × 10−72 | 0.97 | 0.98 | 0.94 | 0.95 |
| TCGA | ESCA | 147 | Adeno (44.9) Squamous (55.1) | 0.95 (0.93–0.96) | <1 × 10−99 | 0.99 | 0.99 | 0.96 | 0.94 |
| TCGA | KIDNEY | 363 | Clear (74.4) Papillary (25.6) | 0.91 (0.9–0.91) | <1 × 10−99 | 0.95 | 0.95 | 0.91 | 0.89 |
| CPTAC | BRCA | 92 | Ductal (92.4) Lobular (7.6) | 0.64 (0.5–0.75) | 5.80 × 10−12 | 0.69 | 0.39 | 0.57 | 0.13 |
| CPTAC | LUNG | 415 | Adeno (49.3) Squamous (50.7) | 0.94 (0.93–0.95) | <1 × 10−99 | 0.96 | 0.96 | 0.92 | 0.90 |
| UCMC | BRCA | 820 | Ductal (85.5) Lobular (14.5) | 0.94 (0.93–0.95) | <1 × 10−99 | 0.88 | 0.79 | 0.59 | 0.46 |
Table 4. Perceptual consistency of gene expression in reconstructed images.
Correlation between predictions of gene expression from real and reconstructed tiles, averaged per patient, demonstrating a high perceptual similarity of the gene expression of the real and generated images. For TCGA, a deep learning model was trained to predict gene expression from real tiles from TCGA-BRCA using threefold cross-validation. The correlation between predictions for real/generated images is aggregated for the three held-out validation sets. For the CPTAC-BRCA validation, a deep learning model trained across the entire TCGA-BRCA dataset was used to generate predictions. Also listed are the correlations between model predictions and true gene expression, as well as the same correlations made from the reconstructed (Gen) tiles. The similar correlation coefficients from real tiles and reconstructed tiles illustrates the reconstructed tiles retain informative data with regard to histologic subtype.
| Source | Gene | n | Pearson r, real versus gen. | P value | Pearson r, real versus expression | P value | Pearson r, gen. versus expression | P value |
|---|---|---|---|---|---|---|---|---|
| TCGA | CD3G | 941 | 0.9 (0.88–0.91) | <1 × 10−100 | 0.45 (0.39–0.49) | <1 × 10−100 | 0.38 (0.32–0.43) | <1 × 10−100 |
| TCGA | COL1A1 | 941 | 0.84 (0.82–0.86) | 6.88 × 10−269 | 0.45 (0.4–0.5) | <1 × 10−100 | 0.31 (0.26–0.37) | <1 × 10−100 |
| TCGA | MKI67 | 941 | 0.91 (0.9–0.92) | 0.00 × 100 | 0.43 (0.38–0.48) | <1 × 10−100 | 0.35 (0.3–0.41) | <1 × 10−100 |
| TCGA | EPCAM | 941 | 0.9 (0.88–0.91) | 0.00 × 100 | 0.32 (0.26–0.37) | <1 × 10−100 | 0.25 (0.19–0.31) | <1 × 10−100 |
| CPTAC | CD3G | 97 | 0.73 (0.62–0.81) | 3.08 × 10−17 | 0.3 (0.1–0.47) | <1 × 10−100 | 0.21 (0.02–0.4) | 0.04 |
| CPTAC | COL1A1 | 97 | 0.74 (0.64–0.82) | 2.89 × 10−18 | 0.56 (0.4–0.68) | <1 × 10−100 | 0.55 (0.39–0.67) | <1 × 10−100 |
| CPTAC | MKI67 | 97 | 0.82 (0.75–0.88) | 5.34 × 10−25 | 0.04 (−0.16–0.24) | 0.67 | 0.07 (−0.13–0.26) | 0.51 |
| CPTAC | EPCAM | 97 | 0.44 (0.27–0.59) | 5.21 × 10−6 | 0.1 (−0.1–0.29) | 0.33 | −0.04 (−0.24–0.16) | 0.69 |
Identifying models influenced by histologic batch effect
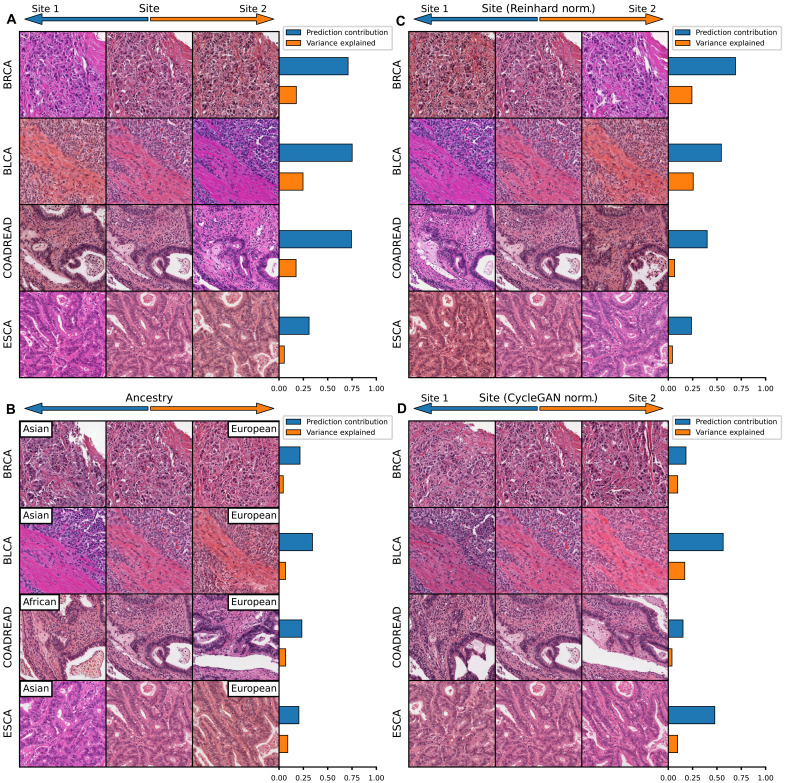
Some features predictable from histology using deep learning can be attributable to batch effect or nonbiologic differences that arise because of slide staining, tissue processing, image resolution, or other differences between batches of cases from model training and evaluation. Notably, differences in slide staining are evident in some of the HistoXGAN image transitions of “standard” pathologic features such as histologic subtype (fig. S5), perhaps most notable in the transition from lung adenocarcinoma to squamous carcinoma. To discern whether these differences in staining are artifactual and introduced by HistoXGAN or reflective of underlying staining differences in lung adenocarcinoma and squamous cell carcinoma in the source dataset, we compare this same transition as calculated from data across all of TCGA versus as calculated from a single TCGA submitting site (Asterand Bioscience; fig. S7A). As expected, the transition derived from a single site features minimal staining differences, suggesting that the staining differences illustrated by HistoXGAN from the full TCGA dataset are due to batch effect between the TCGA adenoacarcinoma and squamous cell carcinoma cases. Furthermore, we assess the impact of stain normalization on model prediction for adenocarcinoma versus squamous cell carcinoma (using the same model trained to verify the similarity of predictions from real versus reconstructed images, as described above). Normalizing image tiles to match the staining of the “adenocarcinoma” image versus the “squamous cell carcinoma” image results in a higher and lower model prediction for likelihood of adenocarcinoma (fig. S7, B and C).
To deconvolute the features used in model prediction, we can apply principal components analysis to the gradients generated from deep learning models for a large set of input images to generate orthogonal feature vectors representing the directions traveled to increase/decrease model prediction. We first apply this approach to models trained to predict grade (true biologic feature) and contributing site (attributable entirely to batch effect) in TCGA-BRCA (n = 934). Principal components are sorted by the relative contribution to the difference in gradients toward an increased/decreased prediction. Whereas a number of distinct components comprise the prediction of higher grade, 69% of the difference in gradient for site prediction is composed of a single component representing a change in tissue stain pattern, and the highest contribution from a single component for grade is 20% (fig. S8 and table S4). We demonstrate that this effect is consistent for site prediction across tumor types with an average of 54% of site prediction attributable to a single component with a strong color variation (Fig. 4A), with a similar visual pattern seen for ancestry prediction (Fig. 4B). Reinhard normalization does not eliminate the dependence on a single-color pattern with 37% of predictions remaining attributable to a single component, although the color pattern of prediction is inverted, likely due to the overcorrection/introducing color changes of image background elements (Fig. 4C). CycleGAN normalization results in an improvement in the dependence of prediction on stain color (Fig. 4D).
Fig. 4. Visualizing histology batch effect and mitigation with normalization.
Tile-based weakly supervised models were trained to predict tissue source site and patient ancestry class (a batch confounded outcome) across select cancer subtypes in TCGA. The gradient with respect to a prediction of these outcomes was calculated for the average feature vector across each slide in the dataset. Principal components analysis was applied to these gradients, and components were sorted by the magnitude of difference of the component between gradients toward each outcome class. The results are then illustrated for this first principal component (i.e., the component contributing most to model prediction). (A) Model predictions for source site are highly homogenous, with an average 54% of the difference in gradients due to the first principal component. Perturbation of images along this component illustrate that it largely represents change in the staining pattern of slides. (B) Slide stain patterns also contribute to prediction of ancestry, although this first principal component constitutes a smaller proportion of gradients. (C) Reinhard normalization does not eliminate the impact of stain pattern on prediction of site, although it leads to an inversion of the stain detected by the model, perhaps due to overcorrection during normalization. (D) CycleGAN normalization reduces the dependence of predictions on a single principal component, and this most predictive component is no longer clearly indicative of staining differences.
Interpretability of models and applications to understand tumor biology
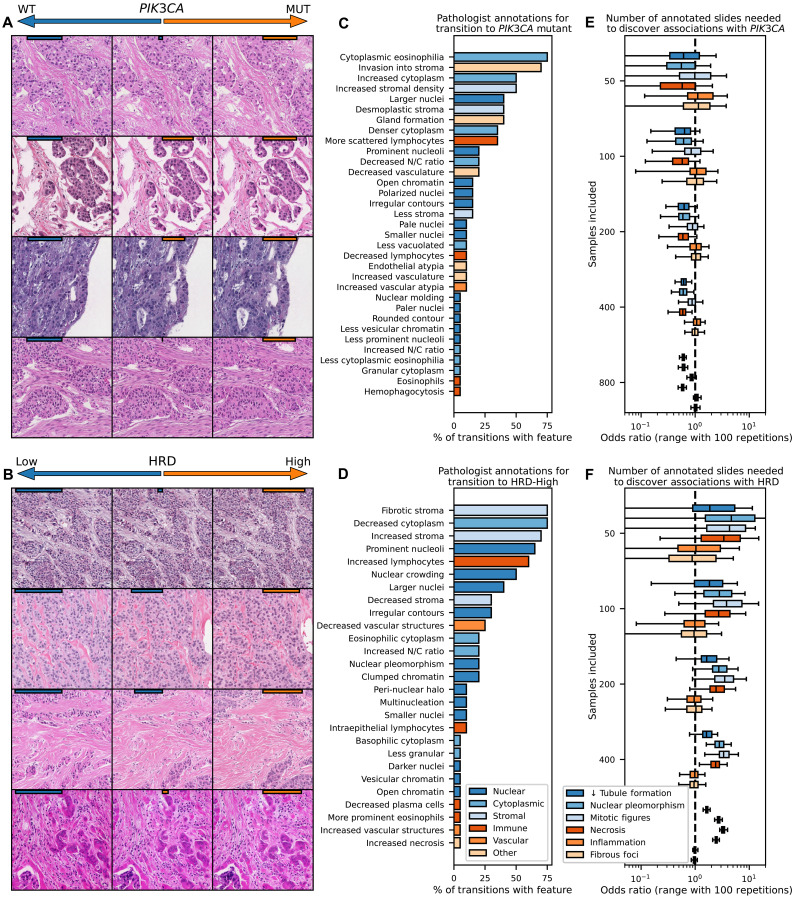
We demonstrate the utility of HistoXGAN to leverage deep learning models to characterize the histologic manifestations of tumor biology. Given the increasing number of targetable molecular alterations predicable from histology (9), we evaluated the explainability of pathways with targeted therapies in breast cancer that have not been thoroughly explored, namely, PIK3CA alterations and homologous recombination deficiency (HRD). Models were trained to predict these alterations in TCGA-BRCA, achieving an average area under the receiver operating characteristic (AUROC) of 0.61 (n = 901; range, 0.58 to 0.63) for PIK3CA alteration and 0.71 (n = 820; range, 0.65 to 0.76) for high HRD score, respectively, on threefold cross-validation, similar to previously published models. Image tiles were altered through gradient descent (Fig. 1, right) to maximize the predictions for PIK3CA mutation and HRD (Fig. 5, A and B), and a set of 22 transitions was reviewed for qualitative analysis of nuclear, cytoplasmic, stromal, immune, and vascular features by four pathologists specializing in breast pathology. Transition to PIK3CA mutation (Fig. 5C) was morphologically associated with increase in abundance (in 45% of transitions) and eosinophilic appearance of cytoplasm (in 68%), increased tubule formation (in 36%), increased invasion into stroma (in 63%), and decreased nuclear to cytoplasmic ratio (in 18%) with variable changes in nuclear size. Transition to high HRD score (Fig. 5D) was associated with prominent nucleoli (in 59%), nuclear crowding/increased nuclear density (45%) with larger (36%) pleomorphic nuclei (18%) with occasional multinucleated cells (9%), increased lymphocytosis (54%), and tumor cell necrosis (5%).
Fig. 5. Illustration of model predictions for targetable alterations in breast cancer.
(A and B) Models were trained to predict PIK3CA mutations and HRD across the TCGA-BRCA dataset (n = 963 for PIK3CA, n = 871 for HRD), as these pathways are common and have Food and Drug Administration–approved therapies. Gradient descent was used to adjust images to maximize/minimize model prediction of PIK3CA/HRD status (with model prediction strength illustrated with orange/blue bars on top of images). Transition to PIK3CA alteration was morphologically associated with increase in abundance and eosinophilic appearance of cytoplasm and stroma, increased tubule formation, and decreased nuclear to cytoplasmic ratio. Transition to high HRD score was associated with nuclear crowding and pleomorphism with occasional multinucleated cells, an increased nuclear/cytoplasmic ratio, increased lymphocytosis, and tumor cell necrosis. (C and D) Structured pathologist review of 20 transitions from low to high model prediction highlight features associated with the selected genomic alterations. (E and F) To determine how many histology slides would be needed to be annotated through traditional methods to uncover these same associations, adjusted odds ratio (estimated through 100 iterations of sampling of listed number of slides) of the association of histologic features with PIK3CA and HRD status are shown as a function of available slides. Tubule formation in PIK3CA and tumor necrosis in HRD that were evident on review of 20 image transitions would require annotation of 400 slides with traditional histologic review to uncover significant associations.
To validate these findings, we compared annotations for epithelial, nuclear, and mitotic grade as well as annotations for necrosis, lymphocytosis, and fibrous foci across TCGA-BRCA between PIK3CA mutant and wild type and HRD-high and -low tumors, generally yielding consistent findings (tables S5 and S6). To illustrate the benefit of synthetic histology to power discovery, we determined the number of samples required to identify significant associations of mutational status with these annotations. Even the most strongly associated pathologic features, such as increased tubule formation in PIK3CA mutant tumors or nuclear pleomorphism in HRD-high tumors would require annotation of 200 to 400 whole-slide images annotated to demonstrate a significant association, whereas pathologic review of just 22 image transitions clearly uncovered these associations in this study (Fig. 5, E and F).
Furthermore, the emergence of attention-based multiple-instance learning (MIL) necessitates approaches that can disentangle features used for attention versus outcome prediction to truly facilitate interpretability. We demonstrate an application of HistoXGAN to attention-MIL models: Using gradient descent, feature vectors can be perturbed to increase or decrease model attention and model prediction separately, allowing independent visualization along these two axes (fig. S9). Applying this approach to models trained to predict grade and cancer subtype illustrates that low attention for these models is associated with benign-appearing fibrous tissue.
Applying generative histology to enable a virtual tumor biopsy
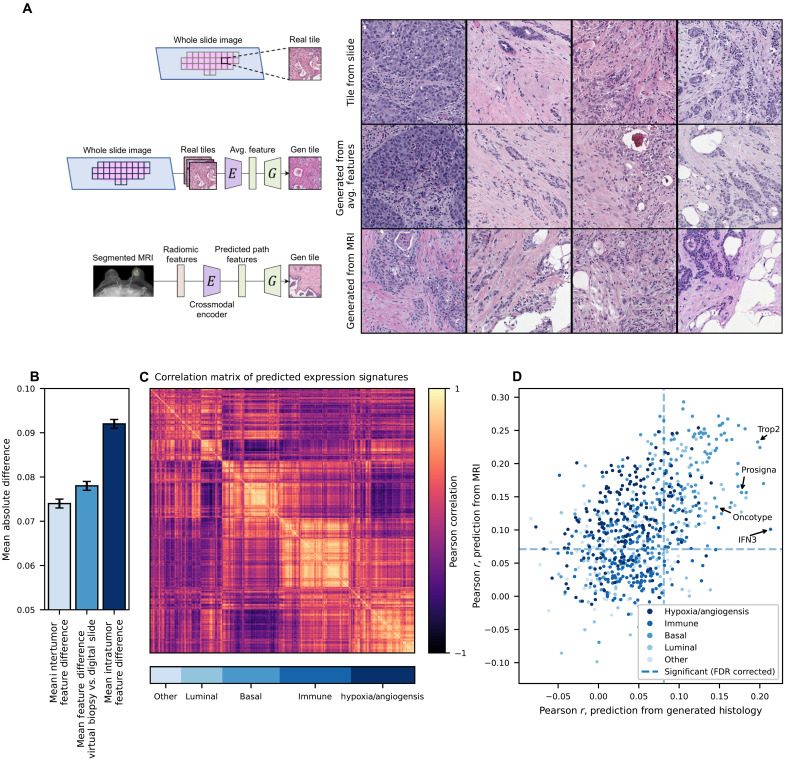
Radiomic analysis of MRI images has been applied to predict key histologic features such as tumor grade and immune infiltrates. However, by predicting histologic SSL feature vectors from radiomic features, a representative tumor image can be reconstructed for downstream analysis, representing a “virtual tumor biopsy.” With fivefold cross-validation across 934 cases with paired MRI and histology, we trained encoders to predict the SSL pathology feature vectors from radiomic features and pooled the predicted features from the held-out test sets for analysis (Fig. 6). Across these test sets, a mean L1 error of 0.078 (95% CI, 0.077 to 0.079) in reconstruction of the histologic feature vectors was observed, comparable to the mean L1 error between pairs of tile images within the tumor across slides (0.074, 95% CI: 0.073 to 0.075), and lower than the average inter-patient difference between average feature vectors of 0.092 (95% CI, 0.092 to 0.093; Fig. 6B). To understand how accurately the recreated images represent true histology across a wide range of meaningful biologic features, we used models pretrained in TCGA to predict grade, histologic subtype, and 775 putatively important gene expression signatures in breast cancer, which can be accurately predicted from histology (average Pearson r between true gene signature and histology prediction of 0.45, range from 0.09 to 0.74; average false discovery rate (FDR)–corrected P value for correlation of 1.10 × 10−5; fig. S10 and data S4). Of note, predictions from these models largely fell into a smaller number of orthogonal categories (Fig. 6C). We found significant correlation between predictions from real histology slide and predictions from virtual biopsy tiles for 213 of these 777 features after FDR correction. Accurate predictions were seen for an IFNγ3 signature (23) (Pearson r, 0.21; 95% CI, 0.15 to 0.28; corrected P = 4.5 × 10−11), a p53 expression module (24) (Pearson r, 0.18; 95% CI, 0.11 to 0.24; corrected P = 4.3 × 10−6), as well as multiple breast cancer prognostic signatures (25, 26), including a histologic grade signature (27) (r, 0.14; 95% CI, 0.07 to 0.20; P = 5.1 × 10−4) research-based version of OncotypeDX recurrence score (r, 0.15; 95% CI, 0.08 to 0.21; P = 6.9 × 10−6; data S5).
Fig. 6. A virtual biopsy reconstructing histology from radiomic features.
An encoder was trained to predict a slide-level average SSL histology feature vector, using 16,379 radiomic features extracted systematically from 934 breast tumors with paired MRI and digital histology available. Fivefold cross-validation was performed, with predictions pooled from the held-out test set. (A) Representative images from the scanned histology slide, reconstruction of the image from an average SSL histology feature vector, and reconstruction directly from radiomic features (predictions made using cases from the test set for each fold). (B) The mean difference between slide-level average features and features from MRI virtual biopsy reconstruction (middle column) was close to the mean difference between tile features within tumors (left) and much less than the mean difference of image tile features between tumors (right). (C) To explore the biologic accuracy of generated images, we used 775 models trained in TCGA to predict RNA signatures as well as models to predict pathologist annotations of grade and histologic subtype. Predictions from these models largely fell into five orthogonal categories as shown in a correlation matrix. (D) Accuracy of predictions for these 777 RNA/histologic features were similar from generated histology and directly from MRI radiomics (without the intermediary of generated histology); in other words, features could only be predicted from generated histology if they were predictable from MRI radiomics. A number of clinically relevant prognostic signatures such as Prosigna and Oncotype were predictable from generated histology.
Although the correlation coefficients for these predictions are not high, the number of positively correlated signatures suggests that some elements of true tumor biology are present in these virtual biopsies. In addition, we found that accurate prediction was largely limited by the accuracy of the radiomic features themselves—a logistic regression trained to predict the result of these 777 features directly from radiomic features performed similarly to reconstructed histology (Fig. 6D). Given that histology images are often available for cases undergoing gene expression profiling, it may be easier to train artificial intelligence models to predict molecular features from histology than from radiographic imaging. This virtual biopsy approach allows for the application of deep learning histology–derived gene expression signatures without the need for a biopsy, which can be used to inform prognosis. By applying pretrained models to virtual biopsy histology in our cohort, we found that intact p53 module expression (24) [hazard ratio (HR) for recurrence free interval 0.83 95% CI, 0.70 to 0.98; P = 0.02] and high histologic grade signature (27) (HR 1.12; 95% CI, 0.95 to 1.13; P = 0.17) identified cases with good/poor prognosis, respectively, although the latter did not reach statistical significance (fig. S11).
DISCUSSION
This study presents HistoXGAN, a GAN architecture for digital pathology image reconstruction that preserves interpretable disease traits. By integrating recent SSL pathology feature extractors (11, 21, 22) with a modified StyleGAN2 generator, HistoXGAN allows interactive manipulation of tissue morphology while maintaining crucial architectural elements indicative of underlying tumor biology. Quantitative analysis across more than 11,000 images validates that HistoXGAN reconstructions accurately recapitulate pathological grade, subtype, and gene expression patterns. Expert pathologist review further corroborates that generated images exhibit superior perceptual similarity to the original histology compared to other modern encoders (18, 28). This approach was robust across a wide array of solid tumors and tissue source sites even beyond those incorporated in model training.
In recent years, deep learning has been applied to predict molecular features of cancers directly from histology with varying degrees of accuracy (6, 9, 29), but understanding the basis of these predictions remains challenging. Conditional GANs have been used to understand clear-cut histologic features but must be retrained for each class comparison and cannot demonstrate multiple transitions simultaneously (5). As HistoXGAN accurately recapitulates histologic features that are easily interpreted by pathologists like cancer grade and subtype, it can likely be applied for discovery of histologic patterns associated with molecular pathways. We applied HistoXGAN to prediction of predictable, actionable alterations in breast cancer, including PIK3CA mutation (30, 31) and HRD status (as defined by high HRD score) (32, 33). Prior studies evaluating histologic features of PIK3CA mutations have described conflicting findings, with one study reporting low grade (34) and others describing sarcomatoid features with areas of high-grade carcinoma (35). We demonstrate that deep learning prediction of PIK3CA was associated with more well-differentiated tubule formation and increased cytoplasm to nuclear ratio but also increased nuclear size/pleomorphism, which may explain the conflicting findings regarding grade in prior studies. Similarly, HRD status and BRCA alteration have been associated with high tumor cell density, with a high nucleus/cytoplasm ratio and conspicuous nucleoli, laminated fibrosis, and high lymphocyte content as well as regions of hemorrhagic suffusion associated with necrotic tissue (36, 37). Review of HistoXGAN images by specialized breast pathologists revealed associated visual features, including nuclear crowding and pleomorphism, an increased nuclear to cytoplasmic ratio, tumor-infiltrating lymphocytes, and areas of tumor necrosis, all of which are consistent with the aforementioned published results. Overall, these findings add confidence that there are true biologic features identified by deep learning models for these alterations, and these features can be used to identify patients at higher likelihood of molecular alterations in standard histologic analysis of tumors. With the rapid growth studies using AI for pathologic image analysis, HistoXGAN may be an important tool to ensure that model predictions are based on rational biologically relevant features.
In addition, the emergence of attention-based MIL necessitates approaches that can disentangle features used for attention versus outcome prediction to facilitate true model interpretability. In general, publications have presented heatmaps of model attention or selected high/low prediction tiles to illustrate potential features used in prediction (10, 38). We demonstrate an elegant application of HistoXGAN to attention-MIL models; using gradient descent, feature vectors can be perturbed to increase or decrease model attention and predictions separately. This enables independent visualization along these two axes, which is critical for understanding whether predictions are driven by meaningful morphology versus dataset biases that attract model attention. Applying this methodology to grade and subtype prediction models illustrates that low model attention correlates with benign fibrous tissue, rather than malignant elements, verifying that attention is applied to tumor regions. Overall, this approach can discern whether attention mechanisms highlight biologically salient regions or whether predictions are partially confounded by irrelevant features that draw attention. Furthermore, we demonstrate that models that are highly confounded by site-specific factors such as ancestry can be quickly identified with HistoXGAN (15, 39). Standard stain normalization such as the Reinhard (40) method demonstrate “overcorrection” of color, as HistoXGAN illustrates that models trained after Reinhard normalization identify the inverse color transition as associated with site, whereas CycleGAN normalization (41) was much more effective at eliminating learned staining patterns of tissue-submitting sites. However, this is also a limitation of HistoXGAN, as any biases in the dataset (such as staining differences between histologic subtypes) will be recapitulated in HistoXGAN visualizations and could lead to false conclusions about the histologic features that distinguish two classes of data.
Furthermore, studies have applied cross-modal autoencoders to understand common “latent spaces” between multiple forms of data, but these approaches have not been performed with digital histology (16, 17). In particular, models to predict the histologic diagnosis or cancer phenotypes from imaging have been described as virtual biopsies (42–45) without the intermediate step of tissue histology reconstruction. We demonstrate here that HistoXGAN can be used to create realistic representations of tumor histology directly from imaging radiomic features and that biologic elements of tumor aggressiveness can be identified from the recreated pathology images. As opposed to prior virtual biopsy approaches, generating a representative section of hematoxylin and eosin histology theoretically allows for characterization of any pathologic feature that could be performed from a true biopsy rather than restricting analysis to a limited set of outcomes used during training. In the presented model, this approach is currently most useful for deriving markers of tumor aggressiveness such as grade or other genomic markers of recurrence, which were predictable from generated histology, and could be used to aid in treatment decisions, such as the use of neoadjuvant chemotherapy versus endocrine therapy in hormone receptor–positive breast cancer. This approach could also be used for explainability of radiomic predictions; for example, if a radiomic feature is predictive of response to therapy, histology could be generated as a function of this feature to determine whether the radiomic feature is highly correlated with known pathologic predictors of response such as tumor grade. With a larger training dataset across multiple cancer types, this tool could theoretically be applied as a first step to cancer lesion diagnosis in areas that are inaccessible, or as a quality control check, whereas if biopsy results are discordant with the predicted histology, it may suggest inadequate sampling and need for rebiopsy.
Several opportunities exist to build upon this study’s limitations. HistoXGAN was developed using histology across the TCGA dataset with predominantly solid tumors, and although performance was accurate across a wide array of OncoTree diagnoses, this approach did not generalize to hematologic malignancies such as AML (as indicated by the poor performance in reconstructing the CPTAC AML dataset). Similarly, this approach may not reconstruct representative histology for benign neoplasms or lesions not represented in TCGA. Using generative approaches for discovery of biologic pathways requires a high degree of confidence in the accuracy of the generative model; for example, the identified pathologic characteristics associated with PIK3CA alteration and HRD status are relatively subtle, and it can be difficult to verify the accuracy of features derived from generative images although they are consistent with prior reports. However, the fact that known pathologic features such as grade and histologic subtype are accurately encoded by HistoXGAN provides some confidence that this approach can be used to describe histologic associations with rare mutations that have not yet been fully characterized.
In particular, our virtual biopsy approach as formulated is currently very limited—Only a single histopathology tile is generated, and this histopathology tile represents the averaged features across the entire tumor and thus does not capture heterogeneity; generation is performed at a single magnification; and training was only performed on invasive breast carcinoma without inclusion of benign lesions. Different magnifications may be needed to predict different histologic features; for example, lobular versus ductal subtype was not predictable from MRI-generated histology, but grade was predictable, and the former may require a lower resolution to accurately assess subtype. Although HistoXGAN could be retrained at other resolutions, current SSL-based feature extractors were trained at a 20× resolution, which may limit the accuracy of lower-resolution reconstruction, and feature extractors optimized at lower resolutions may be needed (21, 22). In our current dataset, the exact alignment of tumor pathology within the full MRI image is not available, which limits our ability to translate regional radiomic features into separate histology images. Curation of a dataset with multiple histology images representative of different tumor regions could allow a virtual biopsy to be performed from regional radiomic features to fully capture tumor heterogeneity and generate multiple discrete tumor images per patient. Our virtual biopsy approach focused on characterizing cases with known breast tumors, and similar encoders would need to be trained with cases of benign disease if such a tool was used to distinguish malignant potential of lesions.
In summary, this work presents HistoXGAN, an architecture integrating SSL and GANs to facilitate interpretable manipulation of digital pathology images while maintaining important disease-specific morphological traits. Evaluations in more than 11,000 images demonstrate quantitatively accurate reconstructions as well as qualitative expert pathologist endorsements of similarity. This technology can greatly aid in the interpretability of artificial intelligence models, find novel biologic insights into targetable pathways to accelerate biomarker development, and even be leveraged to noninvasively sample cancer histology for a true virtual biopsy.
MATERIALS AND METHODS
Data sources and image extraction
Patient data and whole-slide images were selected from 29 tumor–type datasets from TCGA (n = 8120) and 8 corresponding tumor types from CPTAC (n = 1327) along with CPTAC AML cases (n = 88) were used for model validation (table S7). Slides and associated clinical data were accessed through the Genomic Data Commons Portal (https://portal.gdc.cancer.gov/). Ancestry was determined using genomic ancestry calls from the work published by Carrot-Zhang and colleagues (46). Annotations for HRD score were taken from Knijnenburg et al. (47) and binarized at a score of ≥42 for training of HRD models. The cohort of 768 cases collected from University of Chicago with a wide array of OncoTree diagnoses was collected from Institutional Review Board (IRB)-approved protocol 20-0238 from patients diagnosed from 2007 to 2020; this cohort was exempted from consent requirements due to the retrospective/deidentified nature of this cohort (data S2 and S3). The cohort of 934 matched pairs of tumor histology/MRI images was collected from University of Chicago under IRB-approved protocol 22-0707 from patients diagnosed from 2006 to 2021, who prospectively consented to a biospecimen repository (table S8). All samples in the above cohorts with image tiles extractable with our Slideflow pipeline were included in the analysis, which was a preestablished inclusion criterion due to the necessity of extracted image tiles in downstream analytic steps. Slide images were extracted using the Slideflow pipeline with a tile size of 51 pixels per 400 μm and filtering to remove tiles with >60% gray space (48). For GAN training and for applications with weakly supervised models without an attention component, the slides were only extracted within pathologist-annotated tumor regions of interest. For attention-based MIL models, the tiles were extracted from unannotated slides.
GAN and encoder training
The HistoXGAN architecture is a custom version of StyleGAN2, composed of a generator G, discriminator D, and an encoder E (fig. S1). Model training consists of two important modifications to the StyleGAN2 architecture. First, the latent vector z used for each batch during the generator training is replaced with the feature vector extracted by the SSL encoder. Second, a weighted L1 loss comparing the SSL-extracted image features from the real image to those extracted from the generated image is added to the generator loss.
| (1) |
The HistoXGAN and StyleGAN2 networks used in this study were trained with 25,000,000 images across the entire TCGA dataset with a batch size of 256, with a lambda weight of 100 for the above L1 loss. Models were trained with CTransPath (21) and RetCCL (22) encoders. For a naïve comparator encoder, an Encoder4Editing architecture was trained to minimize an equal ratio of the Learned Perceptual Image Patch Similarity and Deep-image Structure and Texture Similarity metrics between the real and generated images from the unmodified StyleGAN2 network, as these yielded the most accurate image representation compared to other non-SSL–based comparisons such as L1 loss, L2 loss, and structural similarity. To demonstrate the necessity of the HistoXGAN architecture (which enables direct projection of SSL features into the StyleGAN latent space), we evaluated both an Encoder4Editing and SingleStyle encoder, trained to minimize the same L1 loss from (1). Encoders were trained for 200,000 epochs with a batch size of 8.
Deep learning model training for quantitative assessment
All deep learning models for outcome prediction were trained using the SlideFlow platform. For comparison of predictions of grade, histologic subtype, single gene expression between real/generated images (Fig. 3 and fig. S6), as well as for illustration of PIK3CA/HRD (Fig. 5), and for prediction of tissue source site and ancestry (Fig. 4), we used Xception-based (49) weakly supervised models with ImageNet (8) pretraining, trained for between one and three epochs with batch size of 32. For separate visualization of attention and model predictions (fig. S9), and for prediction of image features for reconstructed histology from MRI (Fig. 6), we used an attention-MIL architecture trained for 20 epochs. All models were trained with a learning rate of 0.0001 and weight decay of 0.00001. Models for grade, histologic subtype, and single gene expression were trained/evaluated with cross-validation for TCGA datasets and retrained across all of TCGA for application to external datasets; other models were trained across the entire TCGA cohort.
Visual representation of transition between histology states
Several approaches are used to demonstrate the robustness of traversing histology feature space within HistoXGAN images. For visualization of grade, histologic subtype, and gene expression patterns (Fig. 3 and fig. S6), a simple logistic regression model was trained to predict these outcomes using averaged SSL-extracted feature vectors across the annotated tumor region from each slide to obtain a coefficient vector vcoef. Using a randomly selected baseline feature vector from the corresponding cancer dataset, interpolation is performed between vbase – vcoef to vbase + vcoef, with images generated at fixed intervals along the interpolation. For visualization along the gradient of a pretrained model M (as in Figs. 4 and 5), we apply gradient descent to iteratively update a base vector to minimize the loss (2) between the model prediction and the target prediction
| (2) |
For visualization of principal components of model predictions (Fig. 4 and fig. S8), a single gradient from the loss as per (2) is generated across a sample of base vectors across the entire dataset. Principal components analysis is used on the resulting gradients to generate 20 orthogonal components c1,2,…n of gradients, and then interpolation is performed between vbase – ci to vbase + ci.
Reconstruction of histology from MRI images
Dynamic contrast enhanced MRI images acquired on 1.5- or 3-T magnet strength scanners and digital histology images scanned at 40× with an Aperio AT2 scanner were obtained for 934 patients. Radiomic features were extracted from the region of each dynamic contrast-enhanced (DCE)-MRI defined by a tumor mask. To generate the tumor masks and visualize results, we used the previously validated TumorSight Viz platform (50). Briefly, TumorSight Viz implements a fully automated segmentation approach consisting of a series of convolutional neural networks trained on pre-contrast and post-contrast DCE-MRIs to obtain an initial tumor mask. Following the tumor segmentation process, radiomic features were extracted from the pre-contrast and post-contrast DCE-MRIs along with mathematically computed subtraction and percent enhancement volume maps. Features were also extracted from the peritumoral regions by eroding or dilating the tumor region by approximately 3 mm. Before feature extraction, each volume map was transformed into eight additional maps using three-dimensional wavelet filters. Wavelet filters, using high- or low-pass filters in each dimension, enhance various frequency components of the volumes and are capable of capturing important textural information (51). The Pyradiomics library was used to generate the wavelet-transformed volume maps (52). Once all volume maps were generated, the features were extracted from a set of standard feature classes, resulting in a total of 16,379 generated features (table S9)3.
Histologic features were extracted from tumor tiles from the matched histology samples using an SSL feature extractor, and the average feature vector in the tumor region was calculated for each case. An encoder was trained for five epochs with a single leaky rectified linear unit hidden layer to convert MRI radiomic features to SSL histologic features. The MRI encoder was trained with a two-component loss, an L1 loss between the encoder prediction and mean SSL histologic feature vector, and an L1 loss between features extracted from an image generated from encoder prediction and the mean SSL feature vector. This was performed across five cross-folds of the University of Chicago dataset, such that a predicted feature vector was generated for each patient in the dataset. Accuracy of reconstruction was assessed by comparing predictions for grade, tumor histologic subtype, and 775 clinically relevant gene expression features that can be identified from histology for reconstructed images to predictions from the same models applied to the original whole-slide images. Predictive models for these clinically relevant gene expression signatures were trained first using threefold cross-validation in TCGA to verify accuracy of predictions (fig. S10), with a composite model trained across TCGA for use in assessment of reconstruction from MRI. For comparison, logistic regression models were trained from MRI features using the same cross-folds to predict these clinical and gene expression features directly from MRI features.
Pathologist image interpretation
To assess accuracy of reconstructed images from HistoXGAN and the three comparator encoders, four pathologists were presented with one image from each cancer in the CPTAC validation set, along with reconstructed images from HistoXGAN and alternative encoders presented in a random order. Study pathologists were asked to select the generated image most similar to the original image (i.e., which image would most likely represent a nearby section of the same tumor). This process was repeated with the CTransPath, RetCCL, and UNI feature extractors.
To characterize the histologic features identified by deep learning models that were predictive of PIK3CA alterations and HRD status, 20 random images were selected from the TCGA-BRCA dataset, and gradient descent was used to alter the base feature vector to produce a high/low likelihood of predicted PIK3CA alteration or high/low HRD score. Images were generated at fixed steps during this transition, and study pathologists were asked to qualitatively describe tumor, cytoplasmic, stromal, immune, and vascular changes that occur during this transition. Features that were consistently identified by most of the pathologists were selected to identify a consensus for pathologic features representing each of these image transitions.
To verify the veracity of these associations, previously reported annotations (53) for epithelial, nuclear, and mitotic grade, as well as for necrosis, inflammation, and fibrous foci were compared among cases with or without PIK3CA alteration or high HRD score. In addition, to determine the minimum number of annotations that would be needed to confirm these associations with traditional whole-slide image review, we repeated these comparisons using 50, 100, 200, 400, or 800 cases of the total TCGA-BRCA cohort, sampled randomly for 100 iterations.
Statistical analysis
For analysis of the perceptual accuracy of reconstruction of grade, histologic subtype, and single gene expression (Tables 2, 3, and 4), the Pearson correlation coefficient was computed between the averaged model prediction across all tiles from whole-slide images and the averaged prediction across regenerated tiles from extracted features from all tiles across these images. For TCGA cohorts, the predictions were grouped from held-out test sets with threefold cross-validation, whereas for CPTAC cohorts, the predictions were all made from the same model. For comparison of previously annotated histologic features (such as tubule formation or nuclear pleomorphism) between PIK3CA-altered/non-altered and HRD high/low cases, adjusted odds ratios and corresponding Wald statistics were computed for each histologic feature using a multivariable logistic regression for all features. For comparison of predictions from MRI-generated images versus real whole-slide images, Pearson correlation coefficient was computed, and FDR correction was performed with Benjamini Hochberg (54) method with false discovery/family wide error rate of 5%. Identical analysis was performed for predictions from logistic regression trained from MRI features to directly predict model outputs. All statistical testing performed was two-sided at an α = 0.05 level. Key analyses, in particular, accuracy of histologic image reconstruction, were performed in duplicate (representing a technical replicate) with identical results. Error bars on bar graphs in figures represent SE.
Acknowledgments
Funding: This work was supported from the following: National Cancer Institute grant K08CA283261 (F.M.H.), National Cancer Institute grant R01CA276652 (A.T.P.), Cancer Research Foundation grant (F.M.H.), Lynn Sage Breast Cancer Foundation (F.M.H.), National Institute of Dental and Craniofacial Research grant R56DE030958 (A.T.P.), European Commission Horizon grant 2021-SC1-BHC (A.T.P.), Adenoid Cystic Carcinoma Research Foundation grant (A.T.P.), Cancer Research Foundation grant (A.T.P.), American Cancer Society grant (A.T.P.), Department of Defense grant BC211095P1 (F.M.H. and A.T.P.), National Cancer Institute grant P50CA058223 (C.M.P.), Breast Cancer Research Foundation BCRF-23-127 (C.M.P.), and Stand Up To Cancer grant (A.T.P.).
Author contributions: Conceptualization: F.M.H., J.D., G.K., and A.T.P. Data curation: F.M.H., S.K., C.F., M.S., C.M.P., G.K., and A.T.P. Resources: F.M.H., S.K., M.S., and C.M.P. Methodology: F.M.H., H.M.H., G.K., and A.T.P. Formal analysis: F.M.H., H.M.H., J.D., Q.Z., J.P., C.F., C.M.P., J.V., K.C., G.K., and A.T.P. Validation: F.M.H., H.M.H., S.R., M.S., and G.K. Investigation: F.M.H., H.M.H., J.V., S.K., Q.Z., J.P., B.F., K.C., and G.K. Software: F.M.H., J.D., and C.M.P. Writing—original draft: F.M.H. and S.R. Writing—review and editing: F.M.H., H.M.H., S.R., J.D., S.K., Q.Z., J.P., C.F., C.M.P., J.V., M.S., K.C., and A.T.P. Visualization: F.M.H. and C.M.P. Supervision: F.M.H. and C.F. Project administration: F.M.H. and S.K. Funding acquisition: F.M.H., C.M.P., and A.T.P.
Competing interests: F.M.H. reports consulting fees from Novartis and Leica Biosystems. S.R. and J.D. report equity ownership in Slideflow Labs. A.T.P. reports consulting fees from Prelude Biotherapeutics LLC, Ayala Pharmaceuticals, Elvar Therapeutics, Abbvie, and Privo, and contracted research with Kura Oncology, Abbvie, and EMD Serono. C.M.P. is an equity stockholder and consultant of BioClassifier LLC. C.M.P. is also listed as an inventor on patent applications for the Breast PAM50 Subtyping assay. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data from TCGA including digital histology and most of the clinical annotations used are available from https://portal.gdc.cancer.gov/ and https://cbioportal.org, and CPTAC images are available from https://wiki.cancerimagingarchive.net/display/Public/CPTAC+Pathology+Slide+Downloads. Annotations for HRD status are available in the published work of Knijnenburg et al. (47), and annotations for genomic ancestry were obtained from Carrot-Zhang et al. (46). Codes used for this analysis, trained models, and matched radiomic features/histology features from the UCMC validation dataset to replicate this analysis are available at https://doi.org/10.5281/zenodo.13785423; code is also available at https://github.com/fmhoward/HistoXGAN. Additional digital images can also be provided pending scientific review and a completed data use agreement. Requests for digital images should be submitted to F.M.H. (frederick.howard@uchospitals.edu). Licensing of code and data is through CC BY-NC 4.0.
Supplementary Materials
The PDF file includes:
Figs. S1 to S11
Tables S1 to S9
Legends for data S1 to S5
Other Supplementary Material for this manuscript includes the following:
Data S1 to S5
REFERENCES AND NOTES
- 1.Qaiser T., Tsang Y.-W., Taniyama D., Sakamoto N., Nakane K., Epstein D., Rajpoot N., Fast and accurate tumor segmentation of histology images using persistent homology and deep convolutional features. Med. Image Anal. 55, 1–14 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Shmatko A., Ghaffari Laleh N., Gerstung M., Kather J. N., Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat. Cancer 3, 1026–1038 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Ramesh S., Dolezal J. M., Pearson A. T., Applications of deep learning in endocrine neoplasms. Surg. Pathol. Clin. 16, 167–176 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Howard F. M., Dolezal J., Kochanny S., Khramtsova G., Vickery J., Srisuwananukorn A., Woodard A., Chen N., Nanda R., Perou C. M., Olopade O. I., Huo D., Pearson A. T., Integration of clinical features and deep learning on pathology for the prediction of breast cancer recurrence assays and risk of recurrence. npj Breast Cancer 9, 25 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolezal J. M., Wolk R., Hieromnimon H. M., Howard F. M., Srisuwananukorn A., Karpeyev D., Ramesh S., Kochanny S., Kwon J. W., Agni M., Simon R. C., Desai C., Kherallah R., Nguyen T. D., Schulte J. J., Cole K., Khramtsova G., Garassino M. C., Husain A. N., Li H., Grossman R., Cipriani N. A., Pearson A. T., Deep learning generates synthetic cancer histology for explainability and education. npj Precis. Oncol. 7, 49 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kather J. N., Heij L. R., Grabsch H. I., Loeffler C., Echle A., Muti H. S., Krause J., Niehues J. M., Sommer K. A. J., Bankhead P., Kooreman L. F. S., Schulte J. J., Cipriani N. A., Buelow R. D., Boor P., Ortiz-Brüchle N., Hanby A. M., Speirs V., Kochanny S., Patnaik A., Srisuwananukorn A., Brenner H., Hoffmeister M., van den Brandt P. A., Jäger D., Trautwein C., Pearson A. T., Luedde T., Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng Y., Wei Z., Yu W., Yin R., Yuan Y., Li B., Tang Z., Lu Y., Yang Y., Spatial transcriptomics prediction from histology jointly through Transformer and graph neural networks. Brief. Bioinform. 23, bbac297 (2022). [DOI] [PubMed] [Google Scholar]
- 8.J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, L. Fei-Fei, ImageNet: A large-scale hierarchical image database in 2009 IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2009), pp. 248–255. [Google Scholar]
- 9.Saldanha O. L., Loeffler C. M. L., Niehues J. M., van Treeck M., Seraphin T. P., Hewitt K. J., Cifci D., Veldhuizen G. P., Ramesh S., Pearson A. T., Kather J. N., Self-supervised attention-based deep learning for pan-cancer mutation prediction from histopathology. npj Precis. Oncol. 7, 35 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner S. J., Reisenbüchler D., West N. P., Niehues J. M., Zhu J., Foersch S., Veldhuizen G. P., Quirke P., Grabsch H. I., van den Brandt P. A., Hutchins G. G. A., Richman S. D., Yuan T., Langer R., Jenniskens J. C. A., Offermans K., Mueller W., Gray R., Gruber S. B., Greenson J. K., Rennert G., Bonner J. D., Schmolze D., Jonnagaddala J., Hawkins N. J., Ward R. L., Morton D., Seymour M., Magill L., Nowak M., Hay J., Koelzer V. H., Church D. N., Matek C., Geppert C., Peng C., Zhi C., Ouyang X., James J. A., Loughrey M. B., Salto-Tellez M., Brenner H., Hoffmeister M., Truhn D., Schnabel J. A., Boxberg M., Peng T., Kather J. N., Transformer-based biomarker prediction from colorectal cancer histology: A large-scale multicentric study. Cancer Cell 41, 1650–1661.e4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R. J., Ding T., Lu M. Y., Williamson D. F. K., Jaume G., Song A. H., Chen B., Zhang A., Shao D., Shaban M., Williams M., Oldenburg L., Weishaupt L. L., Wang J. J., Vaidya A., Le L. P., Gerber G., Sahai S., Williams W., Mahmood F., Towards a general-purpose foundation model for computational pathology. Nat. Med. 30, 850–862 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann J., Blasimme A., Vayena E., Frey D., Madai V. I.; Precise4Q consortium , Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Medi. Inform. Decis. Mak. 20, 310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.T. Karras, S. Laine, M. Aittala, J. Hellsten, J. Lehtinen, T. Aila, Analyzing and improving the image quality of StyleGAN. arXiv:1912.04958 [cs.CV] (2020).
- 14.Selvaraju R. R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D., Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2020). [Google Scholar]
- 15.Howard F. M., Dolezal J., Kochanny S., Schulte J., Chen H., Heij L., Huo D., Nanda R., Olopade O. I., Kather J. N., Cipriani N., Grossman R. L., Pearson A. T., The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat. Commun. 12, 4423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radhakrishnan A., Friedman S. F., Khurshid S., Ng K., Batra P., Lubitz S. A., Philippakis A. A., Uhler C., Cross-modal autoencoder framework learns holistic representations of cardiovascular state. Nat. Commun. 14, 2436 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang K. D., Belyaeva A., Venkatachalapathy S., Damodaran K., Katcoff A., Radhakrishnan A., Shivashankar G. V., Uhler C., Multi-domain translation between single-cell imaging and sequencing data using autoencoders. Nat. Commun. 12, 31 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O. Tov, Y. Alaluf, Y. Nitzan, O. Patashnik, D. Cohen-Or, Designing an encoder for StyleGAN image manipulation. arXiv:2102.02766 [cs.CV] (2021).
- 19.P. Isola, J.-Y. Zhu, T. Zhou, A. A. Efros, Image-to-image translation with conditional adversarial networks. arXiv:1611.07004 [cs.CV] (2018).
- 20.O. Lang, Y. Gandelsman, M. Yarom, Y. Wald, G. Elidan, A. Hassidim, W. T. Freeman, P. Isola, A. Globerson, M. Irani, I. Mosseri, Explaining in style: Training a GAN to explain a classifier in StyleSpace. arXiv:2104.13369 [cs.CV] (2021).
- 21.Wang X., Yang S., Zhang J., Wang M., Zhang J., Yang W., Huang J., Han X., Transformer-based unsupervised contrastive learning for histopathological image classification. Med. Image Anal. 81, 102559 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Du Y., Yang S., Zhang J., Wang M., Zhang J., Yang W., Huang J., Han X., RetCCL: Clustering-guided contrastive learning for whole-slide image retrieval. Med. Image Anal. 83, 102645 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Wolf D. M., Lenburg M. E., Yau C., Boudreau A., Veer L. J., Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLOS ONE 9, e88309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatza M. L., Lucas J. E., Barry W. T., Kim J. W., Wang Q., Crawford M. D., Datto M. B., Kelley M., Mathey-Prevot B., Potti A., Nevins J. R., A pathway-based classification of human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 107, 6994–6999 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan C., Prat A., Parker J. S., Liu Y., Carey L. A., Troester M. A., Perou C. M., Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med. Genomics 4, 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker J. S., Mullins M., Cheang M. C. U., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J. F., Stijleman I. J., Palazzo J., Marron J. S., Nobel A. B., Mardis E., Nielsen T. O., Ellis M. J., Perou C. M., Bernard P. S., Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heng Y. J., Lester S. C., Tse G. M., Factor R. E., Allison K. H., Collins L. C., Chen Y.-Y., Jensen K. C., Johnson N. B., Jeong J. C., Punjabi R., Shin S. J., Singh K., Krings G., Eberhard D. A., Tan P. H., Korski K., Waldman F. M., Gutman D. A., Sanders M., Reis-Filho J. S., Flanagan S. R., Gendoo D. M., Chen G. M., Haibe-Kains B., Ciriello G., Hoadley K. A., Perou C. M., Beck A. H., The molecular basis of breast cancer pathological phenotypes. J. Pathol. 241, 375–391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. Richardson, Y. Alaluf, O. Patashnik, Y. Nitzan, Y. Azar, S. Shapiro, D. Cohen-Or, Encoding in style: A StyleGAN encoder for image-to-image translation. arXiv:2008.00951 [cs.CV] (2021).
- 29.Fu Y., Jung A. W., Torne R. V., Gonzalez S., Vöhringer H., Shmatko A., Yates L. R., Jimenez-Linan M., Moore L., Gerstung M., Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020). [DOI] [PubMed] [Google Scholar]
- 30.André F., Ciruelos E., Rubovszky G., Campone M., Loibl S., Rugo H. S., Iwata H., Conte P., Mayer I. A., Kaufman B., Yamashita T., Lu Y.-S., Inoue K., Takahashi M., Pápai Z., Longin A.-S., Mills D., Wilke C., Hirawat S., Juric D., Alpelisib for PIK3CA-Mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Turner N. C., Oliveira M., Howell S. J., Dalenc F., Cortes J., Gomez Moreno H. L., Hu X., Jhaveri K., Krivorotko P., Loibl S., Morales Murillo S., Okera M., Park Y. H., Sohn J., Toi M., Tokunaga E., Yousef S., Zhukova L., de Bruin E. C., Grinsted L., Schiavon G., Foxley A., Rugo H. S.; CAPItello-291 Study Group , Capivasertib in hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 388, 2058–2070 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson M., Im S.-A., Senkus E., Xu B., Domchek S. M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., Wu W., Goessl C., Runswick S., Conte P., Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 377, 523–533 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Tutt A. N. J., Garber J. E., Kaufman B., Viale G., Fumagalli D., Rastogi P., Gelber R. D., de Azambuja E., Fielding A., Balmaña J., Domchek S. M., Gelmon K. A., Hollingsworth S. J., Korde L. A., Linderholm B., Bandos H., Senkus E., Suga J. M., Shao Z., Pippas A. W., Nowecki Z., Huzarski T., Ganz P. A., Lucas P. C., Baker N., Loibl S., McConnell R., Piccart M., Schmutzler R., Steger G. G., Costantino J. P., Arahmani A., Wolmark N., McFadden E., Karantza V., Lakhani S. R., Yothers G., Campbell C., Geyer C. E. Jr.; OlympiA Clinical Trial Steering Committee and Investigators , Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N. Engl. J. Med. 384, 2394–2405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zardavas D., Phillips W. A., Loi S., PIK3CA mutations in breast cancer: Reconciling findings from preclinical and clinical data. Breast Cancer Res. 16, 201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheen M. R., Marotti J. D., Allegrezza M. J., Rutkowski M., Conejo-Garcia J. R., Fiering S., Constitutively activated PI3K accelerates tumor initiation and modifies histopathology of breast cancer. Oncogenesis 5, e267–e267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazard T., Bataillon G., Naylor P., Popova T., Bidard F.-C., Stoppa-Lyonnet D., Stern M.-H., Decencière E., Walter T., Vincent-Salomon A., Deep learning identifies morphological patterns of homologous recombination deficiency in luminal breast cancers from whole slide images. Cell Rep. Med. 3, 100872 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soslow R. A., Han G., Park K. J., Garg K., Olvera N., Spriggs D. R., Kauff N. D., Levine D. A., Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod. Pathol. 25, 625–636 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Lu M. Y., Williamson D. F. K., Chen T. Y., Chen R. J., Barbieri M., Mahmood F., Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howard F. M., Kather J. N., Pearson A. T., Multimodal deep learning: An improvement in prognostication or a reflection of batch effect? Cancer Cell 41, 5–6 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Boschman J., Farahani H., Darbandsari A., Ahmadvand P., Van Spankeren A., Farnell D., Levine A. B., Naso J. R., Churg A., Jones S. J., Yip S., Köbel M., Huntsman D. G., Gilks C. B., Bashashati A., The utility of color normalization for AI-based diagnosis of hematoxylin and eosin-stained pathology images. J. Pathol. 256, 15–24 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Runz M., Rusche D., Schmidt S., Weihrauch M. R., Hesser J., Weis C.-A., Normalization of HE-stained histological images using cycle consistent generative adversarial networks. Diagn. Pathol. 16, 71 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aerts H. J. W. L., Velazquez E. R., Leijenaar R. T. H., Parmar C., Grossmann P., Carvalho S., Bussink J., Monshouwer R., Haibe-Kains B., Rietveld D., Hoebers F., Rietbergen M. M., Leemans C. R., Dekker A., Quackenbush J., Gillies R. J., Lambin P., Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R. G. P. M., Granton P., Zegers C. M. L., Gillies R., Boellard R., Dekker A., Aerts H. J. W. L., Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48, 441–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo D., Divard G., Raynaud M., Cohen A., Mone T. D., Rosenthal J. T., Bentall A. J., Stegall M. D., Naesens M., Zhang H., Wang C., Gueguen J., Kamar N., Bouquegneau A., Batal I., Coley S. M., Gill J. S., Oppenheimer F., De Sousa-Amorim E., Kuypers D. R. J., Durrbach A., Seron D., Rabant M., Van Huyen J.-P. D., Campbell P., Shojai S., Mengel M., Bestard O., Basic-Jukic N., Jurić I., Boor P., Cornell L. D., Alexander M. P., Toby Coates P., Legendre C., Reese P. P., Lefaucheur C., Aubert O., Loupy A., A machine learning-driven virtual biopsy system for kidney transplant patients. Nat. Commun. 15, 554 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barros V., Tlusty T., Barkan E., Hexter E., Gruen D., Guindy M., Rosen-Zvi M., Virtual biopsy by using artificial intelligence–based multimodal modeling of binational mammography data. Radiology 306, e220027 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Carrot-Zhang J., Chambwe N., Damrauer J. S., Knijnenburg T. A., Robertson A. G., Yau C., Zhou W., Berger A. C., Huang K.-L., Newberg J. Y., Mashl R. J., Romanel A., Sayaman R. W., Demichelis F., Felau I., Frampton G. M., Han S., Hoadley K. A., Kemal A., Laird P. W., Lazar A. J., Le X., Oak N., Shen H., Wong C. K., Zenklusen J. C., Ziv E.; Cancer Genome Atlas Analysis Network, Cherniack A. D., Beroukhim R., Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell 37, 639–654.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knijnenburg T. A., Wang L., Zimmermann M. T., Chambwe N., Gao G. F., Cherniack A. D., Fan H., Shen H., Way G. P., Greene C. S., Liu Y., Akbani R., Feng B., Donehower L. A., Miller C., Shen Y., Karimi M., Chen H., Kim P., Jia P., Shinbrot E., Zhang S., Liu J., Hu H., Bailey M. H., Yau C., Wolf D., Zhao Z., Weinstein J. N., Li L., Ding L., Mills G. B., Laird P. W., Wheeler D. A., Shmulevich I.; Cancer Genome Atlas Research Network, Monnat R. J., Xiao Y., Wang C., Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 23, 239–254.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolezal J. M., Kochanny S., Dyer E., Ramesh S., Srisuwananukorn A., Sacco M., Howard F. M., Li A., Mohan P., Pearson A. T., Slideflow: deep learning for digital histopathology with real-time whole-slide visualization. BMC Bioinformatics 25, 134 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.F. Chollet, Xception: Deep learning with depthwise separable convolutions. arXiv:1610.02357 [cs.CV] (2017).
- 50.A. Pekis, V. Kannan, E. Kaklamanos, A. Antony, S. Patel, T. Earnest, Seeing beyond cancer: Multi-institutional validation of object localization and 3D semantic segmentation using deep learning for breast MRI. arXiv:2311.16213 [eess.IV] (2023).
- 51.Zhou J., Lu J., Gao C., Zeng J., Zhou C., Lai X., Cai W., Xu M., Predicting the response to neoadjuvant chemotherapy for breast cancer: Wavelet transforming radiomics in MRI. BMC Cancer 20, 100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Griethuysen J. J. M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R. G. H., Fillion-Robin J.-C., Pieper S., Aerts H. J. W. L., Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thennavan A., Beca F., Xia Y., Recio S. G., Allison K., Collins L. C., Tse G. M., Chen Y.-Y., Schnitt S. J., Hoadley K. A., Beck A., Perou C. M., Molecular analysis of TCGA breast cancer histologic types. Cell Genom. 1, 100067 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 to S9
Legends for data S1 to S5
Data S1 to S5