Abstract
Maternal education was strongly correlated with adolescent brain morphology, cognitive performances, and mental health. However, the molecular basis for the effects of maternal education on the structural neurodevelopment remains unknown. Here, we conducted gene-environment–wide interaction study using the Adolescent Brain Cognitive Development cohort. Seven genomic loci with significant gene-environment interactions (G×E) on regional gray matter volumes were identified, with enriched biological functions related to metabolic process, inflammatory process, and synaptic plasticity. Additionally, genetic overlapping results with behavioral and disease-related phenotypes indicated shared biological mechanism between maternal education modified neurodevelopment and related behavioral traits. Finally, by decomposing the multidimensional components of maternal education, we found that socioeconomic status, rather than family environment, played a more important role in modifying the genetic effects on neurodevelopment. In summary, our study provided analytical evidence for G×E effects regarding adolescent neurodevelopment and explored potential biological mechanisms as well as social mechanisms through which maternal education could modify the genetic effects on regional brain development.
The interplay between gene and maternal education on adolescent brain volumes was studied in both biological and social contexts.
INTRODUCTION
Substantial brain development occurs during childhood, which continues up to early adolescence. While previous studies revealed high heritability regarding structural adolescent brain (1), the role of variable environmental factors, especially family socioeconomic status (SES), during adolescent neurodevelopment remains one of the most important questions in neuroscience. Neuroimaging studies have demonstrated that maternal education (ME), a central aspect of SES, is positively correlated with cortical surface areas in regions (middle and inferior temporal gyrus, superior frontal, inferior parietal, and postcentral) related to language, reading, various executive functions, and spatial skills (2), gray matter volume (GMV) of amygdala and hippocampal (3), and variability of regional growth patterns (4). Studies examining composite measures of SES and its different components suggested ME as one of the strongest predictors of children’s cognitive development (5–7), mental/physical health (8–11), and educational outcomes (7, 12).
From the social perspective, ME is strongly associated with human capital and household wealth (13, 14), stable family structure (15), and supportive parenting behaviors (16), including cognitive stimulation, selection of academically advantageous childcare arrangements, and high-quality child-directed speech (17, 18). However, from a biological perspective, it remains unknown how ME could interplay with genetic components in structuring adolescent neurodevelopment.
Gene-environment–wide interaction study (GEWIS) is a widely used approach for identifying genetic loci with differential effects on the phenotype stratified by the levels of environmental exposure. It has been previously used to characterize the molecular basis of posttraumatic stress on the risk of suicidal behaviors, and identified extracellular matrix biology and synaptic plasticity as biological interactors on the genetic risk of suicidality (19). Here, by leveraging the genetic and neuroimaging data from a prospective multicenter adolescent cohort, we aim to investigate the interaction effects between ME and single-nucleotide polymorphisms (SNPs) on adolescent neurodevelopment by conducting GEWIS using additive genetic model. Multiple variants were identified to achieve significant main and interaction effects on neurodevelopment, with enriched biological functions relating to metabolic process, inflammatory process, and synaptic plasticity. Results were validated using an independent adolescent cohort and via gene-based and gene-set analyses. Additionally, genetic correlation analysis was conducted between genetic variants with significant gene environment interactions and those associated with multiple behavioral and disease-related phenotypes to examine potentially shared biological mechanism. Finally, conditional analyses indicated that, compared to family environment, ME is more likely to modify the genetic effects on neurodevelopment through socioeconomic status.
RESULTS
ME exhibited strong correlation with regional GMV
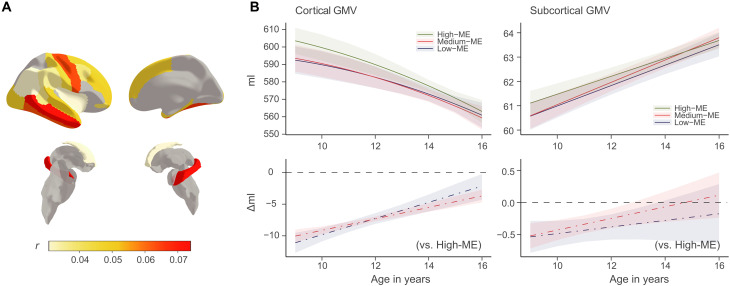
To facilitate comparisons across different educational systems, adolescents from the Adolescent Brain Cognitive Development (ABCD) Study and IMAGEN study were grouped into three categories based on their ME level: mother completed a university education level, the equivalent, or above (High-ME); mother completed a high school education and the equivalent (Medium-ME); and mother completed less than a high school education (Low-ME). Baseline characteristics of these 13,862 adolescents stratified by ME levels are shown in Table 1. Neither study showed significant sex differences in the distributions of ME (X2 = 4.08, P = 0.130 for ABCD; X2 = 0.35, P = 0.840 for IMAGEN). Higher ME was significantly associated with higher total GMV, adjusting for age, site, handedness, sex, and estimated intracranial volume (r = 0.09, P < 0.001). Brain regions exhibiting the strongest correlation with ME include middle temporal (r = 0.07, Padj < 0.001), fusiform (r = 0.07, Padj < 0.001), precentral (r = 0.06, Padj < 0.001), inferior temporal (r = 0.06, Padj < 0.001), and frontal pole (r = 0.05, Padj < 0.001) [Fig. 1A and table S1; Benjamini-Yekutieli false discovery rate (BY-FDR) method]. We also examined the longitudinal associations between ME and structural neurodevelopment, and found that compared to High-ME, the effects of Medium-ME and Low-ME on total GMV remained significant until mid-adolescence, although weakening over time (Fig. 1B and table S2).
Table 1. Baseline characteristics for adolescents in the ABCD and IMAGEN cohorts, stratified by ME.
Continuous characteristics were described using mean and SD, and categorical characteristics are presented as frequency and percentages per category. P values were obtained by one-way analysis of variance (ANOVA) test for continuous variables and chi-square test for categorical variables. ns: P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. High-ME, high maternal education, defined as mother completed a university education level, the equivalent, or above; Medium-ME, medium maternal education, defined as mother completed a high school education and the equivalent; Low-ME, low maternal education, defined as mother completed less than a high school education.
| ABCD study | |||||
|---|---|---|---|---|---|
| All participants (N = 11,780) | High-ME (N = 9567) | Medium-ME (N = 1234) | Low-ME (N = 763) | P value | |
| Age (years), mean (SD) | 9.93 ± 0.64 | 9.93 ± 0.65 | 9.90 ± 0.64 | 9.89 ± 0.63 | ns |
| Male, N (%) | 6147 (52.2%) | 5000 (52.3%) | 646 (52.4%) | 370 (48.5%) | ns |
| Ethnic, N (%) | *** | ||||
| White | 7459 (63.3%) | 6571 (68.7%) | 517 (41.9%) | 276 (36.2%) | |
| Black | 1840 (15.6%) | 1156 (12.1%) | 431 (34.9%) | 199 (26.1%) | |
| Other | 2480 (21.1%) | 1840 (19.2%) | 286 (23.2%) | 288 (37.7%) | |
| Paternal education, N (%) | *** | ||||
| University, the equivalent, or above | 7313 (77.1%) | 6841 (85.6%) | 275 (33.6%) | 83 (16.5%) | |
| High school and the equivalent | 1418 (14.9%) | 879 (11.0%) | 383 (46.8%) | 122 (24.2%) | |
| Less than high school | 754 (7.9%) | 271 (3.4%) | 160 (19.6%) | 299 (59.3%) | |
| Socioeconomic condition, N (%) | *** | ||||
| ≥$100K | 4529 (42.0%) | 4414 (49.3%) | 73 (7.0%) | 7 (1.2%) | |
| $50K–$100K | 3049 (28.3%) | 2682 (30.0%) | 257 (24.6%) | 65 (11.0%) | |
| <$50K | 3193 (29.6%) | 1858 (20.8%) | 714 (68.4%) | 519 (87.8%) | |
| Intracranial volume (liters), mean (SD) | 1.49 ± 0.14 | 1.50 ± 0.14 | 1.46 ± 0.14 | 1.43 ± 0.14 | *** |
| Total brain volume (liters), mean (SD) | 0.66 ± 0.06 | 0.66 ± 0.06 | 0.64 ± 0.06 | 0.63 ± 0.06 | *** |
| IMAGEN study | |||||
| All participants (N = 2082) | High-ME (N = 1056) | Medium-ME (N = 692) | Low-ME (N = 294) | P value | |
| Age (years), mean (SD) | 14.39 ± 0.40 | 14.39 ± 0.40 | 14.40 ± 0.41 | 14.40 ± 0.40 | ns |
| Male, N (%) | 1021 (49.0%) | 522 (49.4%) | 335 (48.4%) | 148 (50.3%) | ns |
| Ethnic, N (%) | ns | ||||
| White | 1859 (89.3%) | 943 (89.3%) | 626 (90.5%) | 260 (88.4%) | |
| Black | 24 (1.2%) | 14 (1.3%) | 4 (0.6%) | 4 (1.4%) | |
| Other | 199 (9.6%) | 99 (9.4%) | 62 (9.0%) | 30 (10.2%) | |
| Paternal education, N (%) | *** | ||||
| University, the equivalent, or above | 1062 (52.3%) | 808 (77.2%) | 197 (28.9%) | 51 (18.0%) | |
| High school and the equivalent | 557 (27.4%) | 155 (14.8%) | 339 (49.8%) | 57 (20.1%) | |
| Less than high school | 411 (20.2%) | 83 (7.9%) | 145 (21.3%) | 175 (61.8%) | |
| Socioeconomic condition | 0.72 ± 1.03 | 0.58 ± 0.95 | 0.81 ± 1.07 | 0.96 ± 1.12 | *** |
| Intracranial volume (liters), mean (SD) | 1.54 ± 0.15 | 1.55 ± 0.15 | 1.53 ± 0.14 | 1.51 ± 0.15 | *** |
| Total brain volume (liters), mean (SD) | 0.60 ± 0.06 | 0.61 ± 0.06 | 0.60 ± 0.06 | 0.59 ± 0.06 | *** |
Fig. 1. The influence of ME on regional GMVs.
(A) Spearman correlations between ME and regional GMVs, adjusting for age, site, handedness, sex, and estimated intracranial volume. Regions in gray indicate nonsignificant correlations after BH-FDR correction. (B) Longitudinal effects of ME on cortical (left) and subcortical brain GMVs. Both GMVs (top) and GMV change rate (bottom) for participants with high, medium, and low levels of ME are estimated. The bands indicate 95% confidence intervals for predicted GMV at the top and relative regression coefficients at the bottom.
ME modified the genetic effects on neurodevelopment via metabolic process, inflammatory process, and synaptic plasticity
To investigate the role of ME on the genetic effects on structural neurodevelopment, we conducted GEWIS using 7662 adolescents in the ABCD study. To reduce the false-positive rate, Bonferroni-corrected genome-wide significance levels considering the number of independent traits were applied (Materials and Methods). Eleven independent loci were identified with genome-wide significant main or interaction effects (fig. S1 and Fig. 2). Among the seven loci with significant interaction effects where SNP effects vary by levels of ME, three of them also showed significant main effects on regional brain GMV. Absolute effect sizes of these loci on regional GMVs were relatively larger and more significant in adolescents with Low-ME, and considerably weaker among those with Medium-ME and High-ME (Table 2). These results, especially those found in loci with significant main effects and effects in adolescents with Low-ME, suggested a possible role of the genotype as a diathesis and low ME as a stressor, while the results found in loci with nonsignificant main effects were more likely to indicate the differential susceptibility to ME levels. Detailed annotations for all 11 significant loci are listed in table S3.
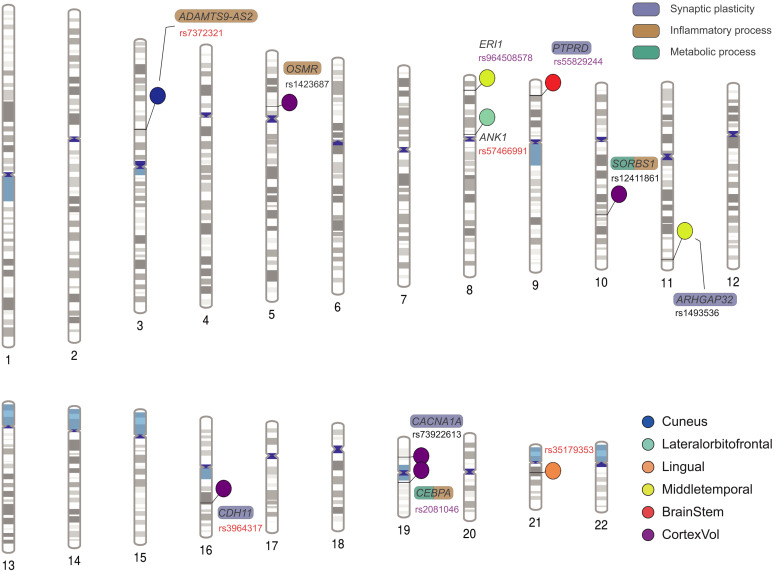
Fig. 2. Genomic loci with genome-wide significant main and interaction effects with ME on regional brain GMVs.
SNPs in red indicate interaction effects only, while those in purple indicate both main and interaction effects. These SNPs are then mapped to functional genes using position, molecular phenotype quantitative trait loci, chromatin interaction, and in silico functional prediction mapping methods. Genes involved in synaptic plasticity, inflammatory process, and metabolic process are highlighted in purple, brown, and green, respectively. Plot created using PhenoGram (http://visualization.ritchielab.org).
Table 2. Genomic loci showing genome-wide significant main or interaction effects with ME on brain GMVs.
SNPs with significant main or interaction effects were clumped using PLINK, and only those with the smallest P values in each locus are shown in the table. The position of SNP was built upon GRCh38. BETA indicates the effect of each SNP on regional GMV among participants with high, medium, and low levels of ME. As ME was treated as a continuous variable, the genetic effects were estimated using a horizontal shift of ME. MAF, minor allele frequency.
| Phenotype | SNP (Chr:BP) | Allele | MAF | High-ME | Medium-ME | Low-ME | G×ME effect | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BETA | P | BETA | P | BETA | P | BETA | P | ||||
| Cuneus | rs7372321 (3:64728139) | C/T | 0.09 | 0.10 | 0.001 | −0.28 | 1.07 × 10−5 | −0.65 | 1.84 × 10−7 | 0.38 | 9.22 × 10−9 |
| Lateral orbitofrontal | rs57466991 (8:41762594) | A/G | 0.17 | 0.03 | 0.112 | −0.20 | 2.29 × 10−7 | −0.42 | 2.05 × 10−8 | 0.22 | 1.02 × 10−8 |
| Lingual | rs35179353 (21:19854073) | C/T | 0.11 | 0.06 | 0.016 | −0.27 | 1.13 × 10−6 | −0.59 | 5.27 × 10−8 | 0.32 | 9.14 × 10−9 |
| Middle temporal | rs964508578 (8:9081668) | C/T | 0.53 | 0.03 | 0.018 | −0.13 | 3.81 × 10−7 | −0.29 | 2.13 × 10−9 | 0.16 | 1.68 × 10−10 |
| rs1493536 (11:129307277) | C/T | 0.44 | 0.00 | 0.897 | 0.14 | 8.71 × 10−9 | 0.29 | 5.42 × 10−9 | −0.14 | 3.87 × 10−8 | |
| Brain stem | rs55829244 (9:9533520) | T/G | 0.12 | 0.02 | 0.310 | −0.22 | 4.91 × 10−8 | −0.46 | 1.01 × 10−8 | 0.24 | 1.10 × 10−8 |
| Cortex | rs1423687 (5:39103314) | A/G | 0.35 | 1.2 × 10−3 | 0.913 | 0.12 | 1.11 × 10−8 | 0.23 | 7.18 × 10−9 | −0.12 | 5.21 × 10−8 |
| rs12411861 (10:95539609) | G/A | 0.18 | −3.0 × 10−3 | 0.824 | 0.12 | 2.36 × 10−8 | 0.25 | 7.56 × 10−9 | −0.13 | 4.62 × 10−8 | |
| rs3964317 (16:65497034) | A/G | 0.38 | −0.02 | 0.164 | 0.10 | 2.48 × 10−7 | 0.22 | 1.50 × 10−8 | −0.12 | 1.03 × 10−8 | |
| rs73922613 (19:13549883) | A/G | 0.05 | 0.01 | 0.617 | −0.28 | 1.68 × 10−8 | −0.57 | 5.54 × 10−9 | 0.29 | 1.26 × 10−8 | |
| rs2081046 (19:33262430) | T/C | 0.28 | −0.04 | 0.003 | 0.10 | 6.27 × 10−7 | 0.23 | 9.34 × 10−10 | −0.13 | 2.87 × 10−11 | |
One intergenic SNP, rs2081046 on chromosome 19, was found to be functionally associated with CCAAT enhancer binding protein α (CEBPA), a transcription factor involved in glucose and lipid metabolism (20). This gene was also found to have immunomodulatory effects, such as regulating proinflammatory cytokines (21), and its deficiency or overexpression could abrogate granulocyte differentiation (22) or induce monocytic differentiation in mixed lineage leukemia (MLL) fusion protein-mediated leukemias (23). Three other SNPs were mapped to genes involved in glucose/lipid metabolic and inflammatory processes as well. Specifically, rs12411861 was an intron variant of SORBS1, which encodes a Casitas B-lineage lymphoma (CBL)–associated protein involved in the insulin signaling, insulin-stimulated glucose transport (24, 25), and lipid biosynthetic process (26), while a few studies have also suggested a potential regulatory role of SORBS1 in immune system through the nuclear factor κB (NF-κB) pathway (27). For another two SNPs, rs1423687 was functionally mapped to OSMR involved in cytokine signaling, especially the interleukin-6 (IL-6) family (28, 29), and rs7372321 was an intro variant of ADAMTS9-AS2, which was widely studied in human cancers due to its relationship with the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway (30, 31) implicated in inflammation (32). In addition, the remaining SNPs showed evidence in synaptic activity of the brain, including an intron variant of CACNA1A (rs73922613), which encoded a subunit of neuronal calcium channel (33) and was involved in a broad phenotypic spectrum of early developmental delay (34) and neuropsychiatric disorders (35), an intron variant of PTPRD (rs55829244) encoding a neuronal cell adhesion molecule and synaptic specifier, and a variant mapped to CDH11 (rs3964317) that correlated with altered dendritic complexity and neuronal/synaptic activity (36).
Note that, although loci identified with significant SNP×ME effects exhibit more significant effects on brain development in population with Low-ME, this is not the case using a whole-genome evaluation (fig. S2). On the basis of external genome-wide association study (GWAS) results for regional brain GMV (37, 38), we found an increase of heritability among participants with High-ME. Moreover, following previous studies to control for potential confounding (39), we included both the covariate-by-environment and the covariate-by-gene interaction term in the GEWIS model as a sensitive analysis. Sixteen of 17 SNPs identified with significant interaction effects exhibited Benjamini–Hochberg false discovery rate (BH-FDR)–corrected significance (Padj < 0.05) (table S4), although none of them passed the stringent genome-wide significance due to small sample size and increased model complexity. The inclusion of multiple interaction effects also complicates the interpretation of the G×E results, making it challenging to compare with the original GEWIS findings.
GEWIS findings were successfully validated
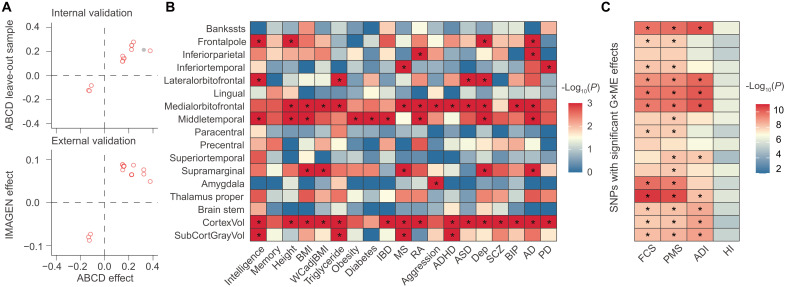
Both internal and external validation were performed (Fig. 3A). First, the leave-out ABCD samples with their siblings were used as an internal validation set, where 15 of 17 SNPs (88%) identified with significant interaction effects were also found to have BH-FDR–adjusted significant interaction effects (fig. S3), although not reaching the genome-wide significance. Next, an independent external sample, IMAGEN cohort (n = 1982), was adopted to replicate the GEWIS findings. Because of the small sample size of IMAGEN and possible population heterogeneity due to age differences, study sites, etc., we examined the sign concordance of the SNP×ME effects between the two studies and reduction of P values after meta-analysis with ABCD (40). All significant SNP×ME effects identified in ABCD had the same effect direction with IMAGEN and smaller P values after meta-analysis with IMAGEN (table S5).
Fig. 3. Validation of G×ME and the interpretation of the influence of ME on neurodevelopment.
(A) Validation of significant GEWIS results obtained from ABCD using the leave-out samples as an internal validation (top) and independent adolescent study IMAGEN as an external validation (bottom). (B) Genetic overlap between SNPs showing G×ME effects on regional brain GMVs and those showing strong associations with multiple diseases and related traits. * indicates a significant overlap after BY-FDR correction. BMI, body mass index; WCadjBMI, waist circumference–adjusted BMI; IBD, inflammatory bowel disease; MS, multiple sclerosis; RA, rheumatoid arthritis; ADHD, attention-deficit/hyperactivity disorder; ASD, autism; Dep, depression; SCZ, schizophrenia; BIP, bipolar disorder; AD, Alzheimer’s disease; PD, Parkinson’s disease. (C) Significant G×ME effects after adjusting for family or social environmental factors. * indicates remained genome-wide significant G×ME effects. FCS, family conflict score; PMS, parenting monitoring score; ADI, area deprivation index; HI, household income.
Gene-based and gene-set analyses indicated similar mechanisms underlying the G×ME interactions on neurodevelopment
To reveal the mechanisms of ME on neurodevelopment at a higher biological level, we performed gene-based analysis using summary statistics from our GEWIS in MAGMA. Eleven genes were identified to achieve Bonferroni-corrected genome-wide significant interactions with ME (G×ME) on regional GMVs (table S6). Notably, we observed significant G×ME interactions for CSMD1 on the whole cortical (P = 3.84 × 10−12), subcortical (P = 2.36 × 10−7), and middle temporal (P = 5.39 × 10−7) GMV, with suggestive effects on lateral orbitofrontal (P = 1.10 × 10−5), medial orbitofrontal (P = 3.48 × 10−6), and thalamus (P = 2.40 × 10−6) GMV. CSMD1 encodes a complement pathway inhibitor in regulating C3/CR3-dependent axonal pruning (41) involved in the development of the central nervous system. Abnormal axonal pruning may induce both neurodegenerative and psychiatric disorders (42–44), and CSMD1 has been reported to be correlated with cognitive functions including working memory and episodic memory (45, 46). Other significant genes included CTNNA2, which encodes a cell adhesion protein related to dendritic spine and synaptic connection stability (47–49), and ANK2, which encodes a major ankyrin-B polypeptides required for normal structural connectivity in the central nervous system (CNS) (50). In addition, SORBS1 (P = 5.32 × 10−6), with suggestive gene-based interaction effects, was also implicated in the SNP-based GEWIS.
To examine whether the identified genes with significant G×ME interaction effects converged on functional gene sets and pathways, we conducted gene-set analysis using MAGMA. Eight significant gene sets (table S7) were identified to be involved in neurodevelopment, including four sets related to inflammation regulation and synaptogenesis: GOBP_regulation_of_mast_cell_activation_involved_in_immune_response, GOBP_tachykinin_receptor_signaling_pathway, GOBP_chemorepulsion_of_axon, and Curated_gene_sets_gery_cebp_targets. These gene sets were found to have differential effects on accumbens (P = 1.21 × 10−6), lateral occipital (P = 9.47 × 10−7), caudal middle frontal (P = 1.03 × 10−6), and posterior cingulate (P = 3.15 × 10−7) GMVs separately, stratified by the levels of ME.
Significant genetic overlaps were observed between G×ME effects on regional neurodevelopment and related traits
Independent SNP effect concordance analyses (iSECAs) were conducted to investigate the overlap between our GEWIS summary statistics and related traits, including neuropsychiatric and neurological disorders, and physical, psychological and disease-related phenotypes. Significant genomic overlaps (P < 1.0 × 10−3) were observed between variants with significant SNP×ME effects on medial orbitofrontal/middle temporal and related traits (Fig. 3B). These variants were also reported to be related to height, body mass index (BMI), rheumatoid arthritis (RA), depression, and Alzheimer’s disease (AD). These results indicated that the biological mechanisms through which ME could modify the genetic effects on specific brain regions are likely to overlap with behavioral and disease-related phenotypes. While significant pleiotropy was observed between G×ME effects on regional brain GMVs and GWAS of related traits, we did not find significant evidence for concordance or discordance effect directions (fig. S4).
ME is more likely to modify the genetic effects on neurodevelopment through socioeconomic status rather than family environment
To investigate whether the influence of ME on neurodevelopment could be explained by social/family environments or genetic pathways (51), we first examined the association between SNPs with significant SNP×ME effects and educational attainment/ME in a large meta-analysis (52). Neither significant associations were observed between these SNPs and educational attainment (table S8) nor ME as well as other related environmental variables, adjusting for age, gender, and population stratification (table S9). We also found no differences in both the SNP×ME effects and the SNP-ME correlations between genetically related and unrelated mother and child dyads (fig. S5). These results suggested that ME is more likely to modify the genetic effects on neurodevelopment through environmental inheritance rather than genetic inheritance. To decompose the multidimensional components of ME, we separately adjusted for socioeconomic and family environmental factors in our GEWIS analysis. We found that most$ of SNPs with significant SNP×ME effects remained significant when adjusting for family environmental factors including family conflict score and parenting monitoring score, while few SNPs remained significant when adjusting for household (household income) and neighborhood socioeconomic status [area deprivation index (ADI)] (Fig. 3C). These results jointly suggested that, compared to micro-family environment, macro-SES may play a more manifested role in modifying the genetic effects on neurodevelopment.
DISCUSSION
Here, we investigated the interaction effects between SNP/gene and ME on structural brain development using large-scale adolescent cohorts. Eleven independent SNPs with genome-wide significant main or interaction effects were identified, with middle temporal most susceptible to the effect modification by ME. Most loci showing significant interactions with ME also achieved genome-wide significance for main effects on neurodevelopment. Considering that the impacts of these loci on regional GMVs were larger and more significant with lower levels of ME, the diathesis-stress model (53, 54) may be considered in interpreting these G×ME interaction results, especially for those loci with significant main effects. Specific genotypes may work as diatheses by increasing the vulnerability of individuals’ brain development in low ME environments, while higher ME may be interpreted as a protective factor for child’s brain development. These loci were mapped to functional genes involved in synaptic plasticity (i.e., CACNA1A and PTPRD), metabolic process (i.e., SORBS1), and immune process (i.e., CEBPA and OSMR). Synaptic plasticity refers to the dynamic modulation of the strength of synaptic connections, enabling neural activities generated by the external environment to shape brain morphology and functions (55). Previous studies have reported that metabolism played an important role in neuronal development and neural activities. Specifically, glucoses work as a main energy source for neuronal differentiation, morphogenesis, and synaptic function (56), while lipids, such as myelin, work as components of cellular structural machinery (57) and bioactive molecules (58) implicated in neuronal signaling processes (59). Meanwhile, immune system is crucial to the production of brain-derived neurotrophic factor (60, 61), survival of multiple neural cells (62, 63), regulation of myelination (64, 65), and formation and pruning of the synapses (66, 67) via diverse pathways including phagocytosis and complement cascade. Other mapped genes were reported to be associated with related phenotypes, including CNS development, neurocognition (such as math capability and memory), psychiatric disorders (such as bipolar and depression), and neurodegenerative disorders (such as AD and Parkinson’s disease).
The GEWIS findings obtained from the ABCD cohort were further validated in both the leave-out sample of ABCD and IMAGEN, an independent and large-scale cohort of adolescents. Eighty-eight percent of the SNPs with significant interaction effects were validated in the internal validation using the leave-out ABCD sample. Because of limited sample size of IMAGEN and potential heterogeneity between the two studies, we adopted a robust validation approach by checking the concordance of effect directions and reduction of P values after meta-analyzing both cohorts (40). The external validation yielded concordant directions for all significant SNP×ME effects between ABCD and IMAGEN, suggesting replicability of the original GEWIS results. It is important to note that age is crucial for the reproducibility of the GEWIS results: Adolescent GMV changes during this critical period follow a quadratic pattern (68); in addition, moderation effects of age in the study of both genetic and environmental influences were highlighted in previous studies (69). Besides, gene-based analysis also confirmed the role of SORBS1 in interacting with ME during brain development. Previous studies suggested that CSMD1 play critical roles in regulating axonal pruning through phagocytosis (41), and consistently, we observed differential effects of CSMD1 on cortical and regional GMVs (including middle temporal, thalamus, lateral orbitofrontal, and medial orbitofrontal) among adolescents with high ME compared to those with low ME. Notably, these brain regions form the default mode network (DMN) (70) and are largely activated in tasks requiring participants to understand and interact with others (71). Our results confirmed previous findings that environmental factors, including high levels of income (72), ME (72), and supportive parental practices (73, 74), could interact with genes in inducing high within-DMN connectivity in children and adolescents (75). Further, gene-set analysis strengthened the findings of metabolic and immune alterations as potential interactive pathways where ME could modify structural brain development.
Next, inspired by the Scarr-Rowe hypothesis (76), which referred to a better realization of children’s genetic predisposition in resource-rich family environments, we conducted whole-genome analysis to calculate the heritability across different ME levels and found an increase of heritability in high compared to low ME. As previous work has stressed out the importance of using the entire genome to entangle the contribution of gene-environment interactions (54), it should be cautious that significant results found in participants with Low-ME cannot be interpreted as higher heritability.
Considerable overlap was observed between genetic variants interacting with ME and those associated with neuropsychiatric/neurological disorders and physical/psychological traits. This indicated that ME could affect neuropsychiatric/neurological disorders and related traits via similar pathways as in the neurodevelopment of corresponding brain regions, highlighting the consideration of environmental background when studying genetic relationships between phenotypes. It has been called genetic correlation–by–environment interactions in previous studies (77), referred to changes in pleiotropic effects across different environments. Although marginal associations have been reported between ME and neuropsychiatric/neurological disorders (10, 78–82), focusing on brain regions with significant interactions revealed consistent findings with previous studies that these common genetic variants are linked to pathways involved in metabolic and inflammatory disorders, neuropsychiatric disorders, and intelligence.
From the social context, ME reflects multiple environmental dimensions, summarized by family environment and SES. Higher levels of ME demonstrate significant associations with higher family income (14, 83), maternal psychological well-being (84), stable family structure, as well as better parenting patterns (18). Results from our analysis suggested that SES (household and neighborhood conditions), rather than micro-family environment (such as family conflict and parenting patterns), played a more important role in modifying the genetic effects on neurodevelopment. This finding has important implications for welfare state policy and interventions aimed at addressing the negative effects of social inequality during adolescent brain development. Specifically, adequate efforts could be allocated in improving family income and social housing assistance. However, it did not mean that the role of parent-child communication should be negated. The decomposition of the interaction effects between gene and ME into different environment pathways could be much more complex as shown in previous studies (51, 85, 86).
There are several limitations associated with our study. First, due to limited availability of adolescent cohorts, sample sizes in both the discovery and validation step of our study remained relatively small compared to existing GWASs, which could lead to decreased statistical power in detecting the true gene-by-environment interaction effects. Moreover, when ME was treated as a continuous variable to increase statistical power, we may overlook the potential departure from linearity. Second, due to our research aim of discovering biological processes that may be influenced by ME, we mainly focused on the SNP-level annotations in this research regardless of the whole-genome information. It should be particularly cautious when interpreting this result in terms of heritability. Third, only genes with the strongest evidence of SNP association were considered in the annotation and interpretation of GEWIS results. Those with moderate or low SNP associations were omitted since the Variant-to-Gene (V2G) evidence score from OpenTarget was mostly derived from a single mapping method, which lacks reliability. Fourth, the overlap between genetic variants that significantly interact with ME on brain development and those significantly correlated with neuropsychiatric/neurological disorders and related traits only demonstrated pleiotropy instead of causal relationships. Future research is needed to explore the causal relationship between these variants and confirm the role of ME on modifying the genetic effects on diseases and related traits. Further, although we did not find any evidence to support correlations between genotypes and ME, the potential existence of gene-environment interactions cannot be ignored (87). Trio-based analysis was needed to further elucidate different sources of gene-environment interactions, which was out of our scope. When decomposing components of ME, environment factors in different pathways were adjusted separately in the GEWIS model, ignoring the complex relationships between socioeconomic factors and family environment. More complex covariate adjustment method should be developed to solve this problem.
MATERIALS AND METHODS
Data sources
Data from two adolescent cohorts were used throughout this study. The ABCD study was used to identify SNP/gene × ME interaction effects on structural brain development, and the IMAGEN study was used for replication. Detailed descriptions about these two cohorts are described elsewhere (88, 89). Demographic and environmental factors and neuroimaging data from the curated annual ABCD release 2.0 (age 9 to 10 years, N = 11,811) and IMAGEN (age at 14 years, N = 2082) were used in the analyses. Quality-controlled T1-weighted neuroimaging data were processed using FreeSurfer v6, and regional GMVs were extracted using aparc and aseg atlases. Details on the preprocessing of neuroimaging data can be found in (90) for ABCD and at https://github.com/imagen2/imagen_mri for IMAGEN. Participants with regional GMVs beyond 4 Interquartile Ranges (IQRs) were regarded as outliers and excluded from the analyses. Genotype data were quality controlled using PLINK 1.90, where SNPs with call rates <95%, minor allele frequency <1%, and deviation from the Hardy-Weinberg equilibrium with P < 1 × 10−10 were excluded from the analysis. Because of genetic diversity (91) and low linkage disequilibrium (LD) levels of African populations (92), a total of 2387 ABCD subjects self-reporting ancestral origins as Black or African American were excluded. Considering that ABCD is oversampled for siblings and twins, and thereby has a nested structure, we randomly selected one participant within a family (the kinship relationship between participants was decided by genetically inferred zygosity status in acspsw03 file). The excluded participants in this step together with their siblings were further used as an internal validation set. Details on preprocessing of the genotype data can be found in (93, 94, 95). After stringent quality control, a total of 5,020,358 SNPs and 7662 participants in ABCD, and 5,966,316 SNPs and 1982 participants in IMAGEN were included in the final analyses.
Ethics statement
All the cohort data used in this study comply with relevant ethical regulations. The ABCD study was supported by the National Institutes of Health (NIH), and the IMAGEN study was approved by local ethnical research committees at each research site: King’s College London, University of Nottingham, Trinity College Dublin, University of Heidelberg, Technische Universitat Dresden, Commissariat a l’Energie Atomique et aux Energies Alternatives, and University Medical Center. Informed consent was sought from all participants and a parent/guardian of each participant if under 18 years in all studies.
Measures of mother-child relationship, SES, and family environment
Mother-child relationship
The kinships between mother-child dyads were self-reported. Genetically related mothers were defined by those reported as the child’s biological mother, while non–genetically related mothers were defined by those reported as the child’s adoptive mother, custodial mother, or others.
Maternal education
ME was categorized according to the highest degree attained by one’s biological mother. Adolescents whose mother completed a higher professional programs or university programs were coded as High-ME, those whose mother completed a general higher secondary education or equivalent were coded as Medium-ME, and those whose mother was only involved in primary education/lower secondary education were coded as Low-ME. For ABCD, High-ME: some college/associate degree/bachelor’s degree/master’s degree/professional school degree/doctoral degree; Medium-ME: high school graduate/general educational development (GED) or equivalent diploma general; Low-ME: 1st to 12th grade. For IMAGEN, High-ME: professional qualification/bachelor’s degree/advanced diploma; Medium-ME: A levels or a BTEC (Business and Technology Education Council) national diploma/NVQ (National Vocational Qualification) or GNVQ (General National Vocational Qualification); Low-ME: O levels, GCSE (General Certificate of Secondary Education) or CSE (Certificate of Secondary Education)/less than primary school education.
Household and neighborhood SES
Household income was defined as the total combined family income for the past 12 months before the investigation. It is recoded as an ordinal variable: 0 for less than 50,000 US dollars, 1 for 50,000 to 100,000 US dollars, and 2 for more than 100,000 US dollars. Neighborhood SES was measured using the ADI, which was calculated based on the participant’s primary residential address. The ADI is based on census data on 17 different factors including income, education, employment, and housing quality and provides rankings of neighborhoods as a national percentile (96). Higher ADI reflects greater disadvantages in terms of SES.
Family environment
Family conflict score was estimated as the average of nine questions from the ABCD Parent Family Environment Scale-Family Conflict Subscale Modified from PhenX, which assesses conflict between family members, including parents and children (97). Participants with more than five missing answers were excluded. Missing values were imputed as the average score across all answered questions. Parental monitoring score was calculated as the average of the five questions from the ABCD Parental Monitoring Survey, which reflects overall high parental monitoring behaviors (98).
Statistical analysis
Correlation analysis
To examine the overall impact of ME on brain development, we combined data from ABCD and IMAGEN, and performed linear regression of ME on regional brain GMVs adjusting for age, site, handedness, sex, and estimated intracranial volume. To assess the longitudinal impact of ME on regional GMVs, we adopted a two-stage model fitting approach, where estimated intracranial volume was first estimated using a model linear in age adjusting for sex, site, and handedness, and regional GMVs were then fitted using quadratic term of age and linear age by ME interaction. Model selection was performed using likelihood-ratio test (table S2), and BY-FDR was used to correct for multiple testing.
Gene-environment–wide interaction study
To investigate the interactions between genetic variants and ME on brain development, we assumed an additive genetic model where the number of risk alleles was treated as a continuous variable (coded as 0, 1, and 2), and regional brain GMVs were used as the phenotype. The primary analytic model included SNP, ME, and SNP×ME interaction term as variables of interest, and sex, baseline age, estimated intracranial volume, site, handedness, and top m principal components of genomic marker variations (m = 20) as covariates using PLINK 2 (99). To save degrees of freedom and increase the statistical power, ME was also treated as a continuous variable (coded as 0, 1, and 2 for Low-ME, Medium-ME, and High-ME, respectively). Since the genetic effects on brain GMV at Medium-ME (relative to Low-ME) were approximately half of those at High-ME (fig. S6), it is reasonable to treat ME as a continuous variable. A sensitivity analysis for treating ME as a nominal categorical variable was further conducted to test the linearity of genetic effects with varying levels of ME. The main effect of SNP referred to the genetic effects in the population with low-ME (ME = 0). A horizontal shift of ME was applied to estimate the corresponding genetic effects for population with different ME levels. While previous studies have suggested potential confounding effects for G×E terms (39), we also included both the covariate-by-environment and the covariate-by-gene interaction term in the GEWIS model as a sensitive analysis. However, the inclusion of multiple interaction effects would make it challenging to interpret both the main genetic effect and G×E results, which cannot be compared with the GEWIS results directly.
Additionally, due to the emphasis on the utilization of whole-genome information (54), we calculated the polygenetic score (PGS) using external GWAS (37, 38) [PRSice-2 (100)] for ABCD participants. Considering the possible genetic contribution difference in the brain morphology between adolescents and adults, we selected GWAS summary statistics, which was also conducted in adolescents. Thus, only those for subcortical regions found with significant G×ME results and intracranial volume that was used as a measurement for the total brain development were used. The heritability was estimated by the correlation analysis between PGS and the corresponding GMV phenotype adjusted for sex, baseline age, site, handedness, and top 20 principal components of genomic marker variations.
To replicate the GEWIS results, we first conducted an internal validation using sibling pairs in ABCD, where the shared family environment within siblings was modeled as the random effect (lmer 1.1-34 package). The BH-FDR method was used for multiple testing. Next, we performed the same analytic steps in IMAGEN (m = 20) as an external replication set and check for the effect concordance on significant interaction effects between ABCD and IMAGEN. Inverse variance weighted meta-analysis was used to detect the concordance of interaction effects between GEWISABCD and GEWISIMAGEN. Because the presence of passive gene-environment correlations may influence the estimated G×ME interactions, that is, the child inherits both genotypes and environments from their parents (87), Pearson’s correlation analysis was used to test the independence between identified loci with significant G×ME interactions and ME. We also tested for other family environmental factors and socioeconomic factors due to their correlations with ME. The BH-FDR method was used for multiple testing correction.
Functional mapping
Significant SNPs in GEWIS were identified based on their P values for the main or interaction effects and clumped by LD for independence [r2 < 0.6 in the 1000 Genomes phase 3 reference (101)] within a 250-kb window using PLINK 1.9. SNPs within genes are mapped to genes based on physical positions. For SNPs in intergenic regions, it is challenging to identify possible causal genes underlying association signals requiring a profound understanding of how they alter gene expressions, instead of solely relying on proximity-based approaches. Thus, V2G pipeline from Open Targets Genetics (www.opentargets.org) were used to map them to genes with evidence scores. The V2G model integrates evidence from molecular phenotype quantitative trait loci, chromatin interaction, in silico functional predictions, and distance between the variant and the canonical transcript start site of genes (102). Only mapped genes with the strongest evidence score were reported in the interpretation of GEWIS results.
Gene-based and gene-set analyses
Gene-based association analyses were performed in MAGMA v1.08 (103) in Functional Mapping and Annotation (FUMA) platform (104) using G×ME summary statistics from the GEWIS conducted in ABCD. Associations were tested using the SNP-wise mean model, in which the sum of −log (SNP P values) for SNPs located within the transcript region was used as the test statistic. LD correction was estimated from the 1000 Genomes phase 3 reference. P values from the gene-based association analyses were then used to test whether candidate gene sets belong to specific biological pathways or processes.
Multiple testing corrections
As suggested by Vrieze et al. (105), G×E interactions in association studies were much more difficult to identify than main effects, requiring a sample size of more than 50,000 participants to reach 80% power at the significance threshold of 5 × 10−8. Thus, we adopted a two-step approach to increase the statistical power. First, we conducted GEWIS studies in all the brain phenotypes. Then, we applied multiple testing method to only those phenotypes identified with significant G×E effects (P < 5 × 10−8). As these phenotypes were correlated to some extent, we estimated the effective number of independent variables based on matrix spectral decomposition, using Li and Ji’s method (106). Thus, the Bonferroni-adjusted significant thresholds were set at P < 1.25 × 10−8 (5.0 × 10−8/4). For gene-based and gene-set significance, we applied adjusted significance of P < 1.25 × 10−6 (5.0 × 10−6/4) and suggestive significance of P < 2.50 × 10−5 (1.0 × 10−4/4).
Genetic overlap
Several important neuropsychiatric and neurological disorders, and physical, psychological, and disease-related phenotypes involved in neurodevelopment were selected according to Brouwer et al. (107). Summary statistics were obtained from public GWAS for intelligence (108), memory (109), height (110), BMI (110), waist circumference–adjusted BMI (111), triglyceride (110), obesity (112), diabetes (113), inflammatory bowel disease (114), multiple sclerosis (114), RA (115), aggression (116), attention-deficit/hyperactivity disorder (117), autism (118), depression (119), schizophrenia (120), bipolar disorder (121), AD (122), and Parkinson’s disease (110). To investigate the genetic overlap between SNPs with significant G×ME effects on regional GMVs and those with strong associations with these traits, we performed iSECA (123), which examines pleiotropy and concordance of effect directions between two phenotypes by comparing expected and observed overlap in sets of SNPs with different P value thresholds. Binomial exact test was adopted to test for pleiotropy, and Fisher’s exact test was used to test for effect direction concordance. Empirical P values were generated through permutation testing over 1000 times, and P values for pleiotropy and effect concordance were corrected using the BY-FDR method.
Decomposition of ME
As ME could reflect different social and family advantages, we adjusted resource-related factors (ADI and household income) and family relationship–related factors (family conflict score and parenting monitoring score) in the GEWIS model by adding one specific environment factor and the interaction effect with genotype identified in GEWISABCD. Genome-wide nonsignificance of the G×ME effect after the adjustment of other environmental factors suggests that the moderating effect of ME on genetically influenced brain development could be mediated by the corresponding environmental factor.
Acknowledgments
We thank C. Li for support and recognition of our paper.
Funding: This work received support from the following sources: General Projects of Shanghai Science and Technology Commission (21ZR1405000 to X.L.), National Nature Science Foundation of China (no. 82304241 to X.L.), National Key R&D Program of China (no. 2018YFC1312904 to J.F., no. 2019YFA0709502 to J.F., no. 2019YFA0709500 to J.F., and no. 2023YFE0199700 to X.C.), National Natural Science Foundation of China (82102138), ZJ Lab to J.F., Shanghai Center for Brain Science and Brain-Inspired Technology to J.F., 111 Project (no. B18015 to J.F.), European Union-funded FP6 Integrated Project IMAGEN (Reinforcement-related behaviour in normal brain function and psychopathology) (LSHM-CT-2007-037286 to G.S.), UK Research and Innovation (UKRI) funded UK government’s Horizon Europe funding guarantee (10041392 and 10038599 to G.S.), Horizon 2020 funded ERC Advanced Grant “STRATIFY’ (Brain network based stratification of reinforcement-related disorders) (695313 to G.S.), Human Brain Project (HBP SGA 2, 785907, and HBP SGA 3, 945539 to G.S.), Medical Research Council Grant “c-VEDA” (Consortium on Vulnerability Externalizing Disorders and Addictions) (MR/N000390/1 to G.S.), National Institutes of Health (NIH) (R01DA049238 to G.S., A decentralized macro and micro gene-by-environment interaction analysis of substance use behavior and its brain biomarkers), National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, Bundesministeriumfür Bildung und Forschung (BMBF grants 01GS08152; 01EV0711 to G.S.; Forschungsnetz AERIAL 01EE1406A, 01EE1406B; Forschungsnetz IMAC-Mind 01GL1745B to G.S.), Deutsche Forschungsgemeinschaft (DFG grants SM 80/7-2, SFB 940, TRR 265, NE 1383/14-1 to G.S.), Medical Research Foundation and Medical Research Council (grants MR/R00465X/1 and MR/S020306/1 to S.D.), NIH-funded ENIGMA (grants 5U54EB020403-05 and 1R56AG058854-01 to S.D.), and NSFC grant 82150710554 and environMENTAL grant 101057429. Further support was provided by grants from the ANR (ANR-12-SAMA-0004, AAPG2019—GeBra to J.-L.M.), the Eranet Neuron (AF12-NEUR0008-01—WM2NA and ANR-18-NEUR00002-01—ADORe to J.-L.M.), the Fondation de France (00081242 to J.-L.M.), the Fondation pour la Recherche Médicale (DPA20140629802 to J.-L.M.), the Mission Interministérielle de Lutte-contre-les-Drogues-et-les-Conduites-Addictives (MILDECA to J.-L.M.), the Assistance-Publique-Hôpitaux-de-Paris and INSERM (interface grant to M.-L.P.M.), Paris Sud University IDEX 2012 to J.-L.M., the Fondation de l’Avenir (grant AP-RM-17-013 to M.-L.P.M.), the Fédération pour la Recherche sur le Cerveau; the NIH, Science Foundation Ireland (16/ERCD/3797 to R.W.); and NIH Consortium grant U54 EB020403 to S.D., supported by a cross-NIH alliance that funds Big Data Knowledge Centres of Excellence. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: R.S., X.L., and J.F. conceptualized the study. R.S., X.C., and X.L. designed the analytic approach. R.S. analyzed the data. R.S. and X.C. visualized the results. R.S. and X.L. wrote the manuscript. X.C., T.B., and J.F. helped in interpreting the results. T.B., G.J.B., A.L.W.B., S.D., H.F., A.G., H.G., P.G., A.H., R.B., J.-L.M., M.-L.P.M., E.A., F.N., D.P.O., L.P., S.H., N.H., M.N.S., N.V., H.W., R.W., and G.S. were the principal investigators of IMAGEN Consortium. Imaging, genetic, and behavioral data in the IMAGEN project were acquired and provided by the IMAGEN Consortium. All authors critically reviewed and edited the manuscript.
Competing interests: T.B. served in an advisory or consultancy role for Lundbeck, Medice, Neurim Pharmaceuticals, Oberberg GmbH, and Shire. He received conference support or speaker’s fee by Lilly, Medice, Novartis, and Shire. He has been involved in clinical trials conducted by Shire and Viforpharma. He received royalties from Hogrefe, Kohlhammer, CIP Medien, and Oxford University Press. The present work is unrelated to the above grants and relationships. G.J.B. has received honoraria from General Electric Healthcare for teaching on scanner programming courses. L.P. served in an advisory or consultancy role for Roche and Viforpharma and received speaker’s fee by Shire. She received royalties from Hogrefe, Kohlhammer, and Schattauer. The present work is unrelated to the above grants and relationships. The other authors declare no other competing interests.
Data and materials availability: Data from the ABCD study are available from a dedicated database: https://abcdstudy.org/ by application; data from the IMAGEN study are available upon application: https://imagen2.cea.fr. Summary statistics from published GWASs were described in Materials and Methods in detail and are available at https://atlas.ctglab.nl/. All the summary statistics data for G×ME results could be found at https://zenodo.org/records/13323757. Primary analyses were conducted in R v4.2.2. PLINK 2.0 was used to perform GEWIS. Open Targets Genetics and FUMA were used to perform downstream analysis (gene-based and gene-set analysis, functional mapping). iSECA was used to examine genetic overlaps between summary statistics. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S9
List of IMAGEN Consortium members
References
REFERENCES AND NOTES
- 1.Peper J. S., Schnack H. G., Brouwer R. M., Van Baal G. C., Pjetri E., Szekely E., van Leeuwen M., van den Berg S. M., Collins D. L., Evans A. C., Boomsma D. I., Kahn R. S., Hulshoff Pol H. E., Heritability of regional and global brain structure at the onset of puberty: A magnetic resonance imaging study in 9-year-old twin pairs. Hum. Brain Mapp. 30, 2184–2196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noble K. G., Houston S. M., Brito N. H., Bartsch H., Kan E., Kuperman J. M., Akshoomoff N., Amaral D. G., Bloss C. S., Libiger O., Schork N. J., Murray S. S., Casey B. J., Chang L., Ernst T. M., Frazier J. A., Gruen J. R., Kennedy D. N., Van Zijl P., Mostofsky S., Kaufmann W. E., Kenet T., Dale A. M., Jernigan T. L., Sowell E. R., Family income, parental education and brain structure in children and adolescents. Nat. Neurosci. 18, 773–778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noble K. G., Houston S. M., Kan E., Sowell E. R., Neural correlates of socioeconomic status in the developing human brain. Dev. Sci. 15, 516–527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Muller H. G., Zhu C., Chen Y., Wang J. L., O’Muircheartaigh J., Bruchhage M., Deoni S.; RESONANCE Consortium , Network evolution of regional brain volumes in young children reflects neurocognitive scores and mother’s education. Sci. Rep. 13, 2984 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patra K., Greene M. M., Patel A. L., Meier P., Maternal education level predicts cognitive, language, and motor outcome in preterm infants in the second year of life. Am. J. Perinatol. 33, 738–744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lean R. E., Smyser C. D., Brady R. G., Triplett R. L., Kaplan S., Kenley J. K., Shimony J. S., Smyser T. A., Miller J. P., Barch D. M., Luby J. L., Warner B. B., Rogers C. E., Prenatal exposure to maternal social disadvantage and psychosocial stress and neonatal white matter connectivity at birth. Proc. Natl. Acad. Sci. U.S.A. 119, e2204135119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernald L. C. H., Kariger P., Hidrobo M., Gertler P. J., Socioeconomic gradients in child development in very young children: Evidence from India, Indonesia, Peru, and Senegal. Proc. Natl. Acad. Sci. U.S.A. 109, 17273–17280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speyer L. G., Hall H. A., Hang Y., Hughes C., Murray A. L., Within-family relations of mental health problems across childhood and adolescence. J. Child Psychol. Psychiatry 63, 1288–1296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherar L. B., Griffin T. P., Ekelund U., Cooper A. R., Esliger D. W., van Sluijs E. M., Bo Andersen L., Cardon G., Davey R., Froberg K., Hallal P. C., Janz K. F., Kordas K., Kriemler S., Pate R. R., Puder J. J., Sardinha L. B., Timperio A. F., Page A. S., Association between maternal education and objectively measured physical activity and sedentary time in adolescents. J. Epidemiol. Community Health 70, 541–548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fakhrunnisak D., Patria B., The positive effects of parents’ education level on children’s mental health in Indonesia: A result of longitudinal survey. BMC Public Health 22, 949 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyrose A. K., Klasen F., Otto C., Gniewosz G., Lampert T., Ravens-Sieberer U., Benefits of maternal education for mental health trajectories across childhood and adolescence. Soc. Sci. Med. 202, 170–178 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Lundborg P., Nilsson A., Rooth D.-O., Parental education and offspring outcomes: Evidence from the swedish compulsory school reform. Am. Econ. J. Appl. Econ. 6, 253–278 (2014). [Google Scholar]
- 13.G. S. Becker, Human Capital: A Theoretical and Empirical Analysis, with Special Reference to Education (National Bureau of Economic Research, 1964).
- 14.Krieger N., Williams D. R., Moss N. E., Measuring social class in US public health research: Concepts, methodologies, and guidelines. Annu. Rev. Public Health 18, 341–378 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Jackson M., Kiernan K., McLanahan S., Maternal education, changing family circumstances, and children’s skill development in the United States and UK. Ann. Am. Acad. Pol. Soc. Sci. 674, 59–84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bornstein M., Hahn C. S., Suwalsky J., Haynes O., Socioeconomic status, parenting and child development: The hollingshead fourfactor index of social status and the socioeconomic index of occupations. Mahwah N J, 29–82 (2003). [Google Scholar]
- 17.Hosokawa R., Katsura T., A longitudinal study of socioeconomic status, family processes, and child adjustment from preschool until early elementary school: The role of social competence. Child Adolesc. Psychiatry Ment. Health 11, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belsky J., Bell B., Bradley R. H., Stallard N., Stewart-Brown S. L., Socioeconomic risk, parenting during the preschool years and child health age 6 years. Eur. J. Public Health 17, 508–513 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Wendt F. R., Pathak G. A., Levey D. F., Nunez Y. Z., Overstreet C., Tyrrell C., Adhikari K., De Angelis F., Tylee D. S., Goswami A., Krystal J. H., Abdallah C. G., Stein M. B., Kranzler H. R., Gelernter J., Polimanti R., Sex-stratified gene-by-environment genome-wide interaction study of trauma, posttraumatic-stress, and suicidality. Neurobiol. Stress 14, 100309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olofsson L. E., Orho-Melander M., William-Olsson L., Sjoholm K., Sjostrom L., Groop L., Carlsson B., Carlsson L. M., Olsson B., CCAAT/enhancer binding protein alpha (C/EBPalpha) in adipose tissue regulates genes in lipid and glucose metabolism and a genetic variation in C/EBPalpha is associated with serum levels of triglycerides. J. Clin. Endocrinol. Metab. 93, 4880–4886 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Ramji D. P., Foka P., CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem. J. 365, 561–575 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D. E., Zhang P., Wang N. D., Hetherington C. J., Darlington G. J., Tenen D. G., Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 569–574 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita H., Nakajima H., Nakamura Y., Tsukamoto H., Tanaka Y., Jin G., Yabe M., Asai S., Ono R., Nosaka T., Sugita K., Morimoto A., Hayashi Y., Hotta T., Ando K., Miyachi H., C/EBPalpha and C/EBPvarepsilon induce the monocytic differentiation of myelomonocytic cells with the MLL-chimeric fusion gene. Oncogene 27, 6749–6760 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Chang T.-J., Wang W.-C., Hsiung C. A., He C.-T., Lin M. W., Sheu W. H.-H., Chang Y.-C., Quertermous T., Chen Y.-I., Rotter J. I., Chuang L.-M.; SAPPHIRe Study Group , Genetic variation of SORBS1 gene is associated with glucose homeostasis and age at onset of diabetes: A SAPPHIRe Cohort Study. Sci. Rep. 8, 10574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irvin M. R., Zhi D., Aslibekyan S., Claas S. A., Absher D. M., Ordovas J. M., Tiwari H. K., Watkins S., Arnett D. K., Genomics of post-prandial lipidomic phenotypes in the Genetics of Lipid lowering Drugs and Diet Network (GOLDN) study. PLOS ONE 9, e99509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N., Gu Y., Li L., Chi J., Liu X., Xiong Y., Zhong C., Development and validation of a prognostic classifier based on lipid metabolism-related genes for breast cancer. J. Inflamm. Res. 15, 3477–3499 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vdovenko D., Bachmann M., Wijnen W. J., Hottiger M. O., Eriksson U., Valaperti A., The adaptor protein c-Cbl-associated protein (CAP) limits pro-inflammatory cytokine expression by inhibiting the NF-κB pathway. Int. Immunopharmacol. 87, 106822 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Lantieri F., Bachetti T., OSM/OSMR and interleukin 6 family cytokines in physiological and pathological condition. Int. J. Mol. Sci. 23, 11096 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey G., Radhakrishnan A., Syed N., Thomas J. K., Nadig A., Srikumar K., Mathur P. P., Pandey A., Lin S.-K., Raju R., Prasad T. S. K., Signaling network of Oncostatin M pathway. J. Cell Commun. Signal. 7, 103–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie S., Yu X., Li Y., Ma H., Fan S., Chen W., Pan G., Wang W., Zhang H., Li J., Lin Z., Upregulation of lncRNA ADAMTS9-AS2 promotes salivary adenoid cystic carcinoma metastasis via PI3K/Akt and MEK/Erk signaling. Mol. Ther. 26, 2766–2778 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W., Wang B., Cai Y., Chen J., Lv X., Guo C., Yuan C., ADAMTS9-AS2: A functional long non-coding rna in tumorigenesis. Curr. Pharm. Des. 27, 2722–2727 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Hawkins P. T., Stephens L. R., PI3K signalling in inflammation. Biochim. Biophys. Acta 1851, 882–897 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Catterall W. A., Voltage-gated sodium channels at 60: Structure, function and pathophysiology. J. Physiol. 590, 2577–2589 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerin A. A., Feigenbaum A., Donner E. J., Yoon G., Stepwise developmental regression associated with novel CACNA1A mutation. Pediatr. Neurol. 39, 363–364 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Indelicato E., Nachbauer W., Karner E., Eigentler A., Wagner M., Unterberger I., Poewe W., Delazer M., Boesch S., The neuropsychiatric phenotype in CACNA1A mutations: A retrospective single center study and review of the literature. Eur. J. Neurol. 26, 66-e7 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Frei J. A., Niescier R. F., Bridi M. S., Durens M., Nestor J. E., Kilander M. B. C., Yuan X., Dykxhoorn D. M., Nestor M. W., Huang S., Blatt G. J., Lin Y.-C., Regulation of neural circuit development by Cadherin-11 provides implications for autism. eNeuro 8, ENEURO.0066-21.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hibar D. P., Stein J. L., Renteria M. E., Arias-Vasquez A., Desrivieres S., Jahanshad N., Toro R., Wittfeld K., Abramovic L., Andersson M., Aribisala B. S., Armstrong N. J., Bernard M., Bohlken M. M., Boks M. P., Bralten J., Brown A. A., Chakravarty M. M., Chen Q., Ching C. R. K., Cuellar-Partida G., den Braber A., Giddaluru S., Goldman A. L., Grimm O., Guadalupe T., Hass J., Woldehawariat G., Holmes A. J., Hoogman M., Janowitz D., Jia T., Kim S., Klein M., Kraemer B., Lee P. H., Olde Loohuis L. M., Luciano M., Macare C., Mather K. A., Mattheisen M., Milaneschi Y., Nho K., Papmeyer M., Ramasamy A., Risacher S. L., Roiz-Santianez R., Rose E. J., Salami A., Samann P. G., Schmaal L., Schork A. J., Shin J., Strike L. T., Teumer A., van Donkelaar M. M. J., van Eijk K. R., Walters R. K., Westlye L. T., Whelan C. D., Winkler A. M., Zwiers M. P., Alhusaini S., Athanasiu L., Ehrlich S., Hakobjan M. M. H., Hartberg C. B., Haukvik U. K., Heister A. J. G. A. M., Hoehn D., Kasperaviciute D., Liewald D. C. M., Lopez L. M., Makkinje R. R. R., Matarin M., Naber M. A. M., McKay D. R., Needham M., Nugent A. C., Putz B., Royle N. A., Shen L., Sprooten E., Trabzuni D., van der Marel S. S. L., van Hulzen K. J. E., Walton E., Wolf C., Almasy L., Ames D., Arepalli S., Assareh A. A., Bastin M. E., Brodaty H., Bulayeva K. B., Carless M. A., Cichon S., Corvin A., Curran J. E., Czisch M., de Zubicaray G. I., Dillman A., Duggirala R., Dyer T. D., Erk S., Fedko I. O., Ferrucci L., Foroud T. M., Fox P. T., Fukunaga M., Gibbs J. R., Goring H. H. H., Green R. C., Guelfi S., Hansell N. K., Hartman C. A., Hegenscheid K., Heinz A., Hernandez D. G., Heslenfeld D. J., Hoekstra P. J., Holsboer F., Homuth G., Hottenga J.-J., Ikeda M., C. R. Jack, Jr., Jenkinson M., Johnson R., Kanai R., Keil M., Kent J. W. Jr., Kochunov P., Kwok J. B., Lawrie S. M., Liu X., Longo D. L., McMahon K. L., Meisenzahl E., Melle I., Mohnke S., Montgomery G. W., Mostert J. C., Muhleisen T. W., Nalls M. A., Nichols T. E., Nilsson L. G., Nothen M. M., Ohi K., Olvera R. L., Perez-Iglesias R., Pike G. B., Potkin S. G., Reinvang I., Reppermund S., Rietschel M., Romanczuk-Seiferth N., Rosen G. D., Rujescu D., Schnell K., Schofield P. R., Smith C., Steen V. M., Sussmann J. E., Thalamuthu A., Toga A. W., Traynor B. J., Troncoso J., Turner J. A., Valdés Hernández M. C., Ent D., van der Brug M., van der Wee N. J., van Tol M. J., Veltman D. J., Wassink T. H., Westman E., Zielke R. H., Zonderman A. B., Ashbrook D. G., Hager R., Lu L., McMahon F. J., Morris D. W., Williams R. W., Brunner H. G., Buckner R. L., Buitelaar J. K., Cahn W., Calhoun V. D., Cavalleri G. L., Crespo-Facorro B., Dale A. M., Davies G. E., Delanty N., Depondt C., Djurovic S., Drevets W. C., Espeseth T., Gollub R. L., Ho B. C., Hoffmann W., Hosten N., Kahn R. S., Le Hellard S., Meyer-Lindenberg A., Muller-Myhsok B., Nauck M., Nyberg L., Pandolfo M., Penninx B. W., Roffman J. L., Sisodiya S. M., Smoller J. W., van Bokhoven H., van Haren N. E., Volzke H., Walter H., Weiner M. W., Wen W., White T., Agartz I., Andreassen O. A., Blangero J., Boomsma D. I., Brouwer R. M., Cannon D. M., Cookson M. R., de Geus E. J., Deary I. J., Donohoe G., Fernandez G., Fisher S. E., Francks C., Glahn D. C., Grabe H. J., Gruber O., Hardy J., Hashimoto R., Hulshoff Pol H. E., Jonsson E. G., Kloszewska I., Lovestone S., Mattay V. S., Mecocci P., McDonald C., McIntosh A. M., Ophoff R. A., Paus T., Pausova Z., Ryten M., Sachdev P. S., Saykin A. J., Simmons A., Singleton A., Soininen H., Wardlaw J. M., Weale M. E., Weinberger D. R., Adams H. H., Launer L. J., Seiler S., Schmidt R., Chauhan G., Satizabal C. L., Becker J. T., Yanek L., van der Lee S. J., Ebling M., Fischl B., W. T. Longstreth, Jr., Greve D., Schmidt H., Nyquist P., Vinke L. N., van Duijn C. M., Xue L., Mazoyer B., Bis J. C., Gudnason V., Seshadri S., Ikram M. A.; Alzheimer’s Disease Neuroimaging Initiative; CHARGE Consortium; EPIGEN; IMAGEN; SYS, Martin N. G., Wright M. J., Schumann G., Franke B., Thompson P. M., Medland S. E., Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams H. H., Hibar D. P., Chouraki V., Stein J. L., Nyquist P. A., Renteria M. E., Trompet S., Arias-Vasquez A., Seshadri S., Desrivieres S., Beecham A. H., Jahanshad N., Wittfeld K., Van der Lee S. J., Abramovic L., Alhusaini S., Amin N., Andersson M., Arfanakis K., Aribisala B. S., Armstrong N. J., Athanasiu L., Axelsson T., Beiser A., Bernard M., Bis J. C., Blanken L. M., Blanton S. H., Bohlken M. M., Boks M. P., Bralten J., Brickman A. M., Carmichael O., Chakravarty M. M., Chauhan G., Chen Q., Ching C. R., Cuellar-Partida G., Braber A. D., Doan N. T., Ehrlich S., Filippi I., Ge T., Giddaluru S., Goldman A. L., Gottesman R. F., Greven C. U., Grimm O., Griswold M. E., Guadalupe T., Hass J., Haukvik U. K., Hilal S., Hofer E., Hoehn D., Holmes A. J., Hoogman M., Janowitz D., Jia T., Kasperaviciute D., Kim S., Klein M., Kraemer B., Lee P. H., Liao J., Liewald D. C., Lopez L. M., Luciano M., Macare C., Marquand A., Matarin M., Mather K. A., Mattheisen M., Mazoyer B., McKay D. R., McWhirter R., Milaneschi Y., Mirza-Schreiber N., Muetzel R. L., Maniega S. M., Nho K., Nugent A. C., Loohuis L. M., Oosterlaan J., Papmeyer M., Pappa I., Pirpamer L., Pudas S., Putz B., Rajan K. B., Ramasamy A., Richards J. S., Risacher S. L., Roiz-Santianez R., Rommelse N., Rose E. J., Royle N. A., Rundek T., Samann P. G., Satizabal C. L., Schmaal L., Schork A. J., Shen L., Shin J., Shumskaya E., Smith A. V., Sprooten E., Strike L. T., Teumer A., Thomson R., Tordesillas-Gutierrez D., Toro R., Trabzuni D., Vaidya D., Van der Grond J., Van der Meer D., Van Donkelaar M. M., Van Eijk K. R., Van Erp T. G., Van Rooij D., Walton E., Westlye L. T., Whelan C. D., Windham B. G., Winkler A. M., Woldehawariat G., Wolf C., Wolfers T., Xu B., Yanek L. R., Yang J., Zijdenbos A., Zwiers M. P., Agartz I., Aggarwal N. T., Almasy L., Ames D., Amouyel P., Andreassen O. A., Arepalli S., Assareh A. A., Barral S., Bastin M. E., Becker D. M., Becker J. T., Bennett D. A., Blangero J., van Bokhoven H., Boomsma D. I., Brodaty H., Brouwer R. M., Brunner H. G., Buckner R. L., Buitelaar J. K., Bulayeva K. B., Cahn W., Calhoun V. D., Cannon D. M., Cavalleri G. L., Chen C., Cheng C. Y., Cichon S., Cookson M. R., Corvin A., Crespo-Facorro B., Curran J. E., Czisch M., Dale A. M., Davies G. E., De Geus E. J., De Jager P. L., de Zubicaray G. I., Delanty N., Depondt C., DeStefano A. L., Dillman A., Djurovic S., Donohoe G., Drevets W. C., Duggirala R., Dyer T. D., Erk S., Espeseth T., Evans D. A., Fedko I. O., Fernandez G., Ferrucci L., Fisher S. E., Fleischman D. A., Ford I., Foroud T. M., Fox P. T., Francks C., Fukunaga M., Gibbs J. R., Glahn D. C., Gollub R. L., Goring H. H., Grabe H. J., Green R. C., Gruber O., Gudnason V., Guelfi S., Hansell N. K., Hardy J., Hartman C. A., Hashimoto R., Hegenscheid K., Heinz A., Le Hellard S., Hernandez D. G., Heslenfeld D. J., Ho B. C., Hoekstra P. J., Hoffmann W., Hofman A., Holsboer F., Homuth G., Hosten N., Hottenga J. J., Hulshoff Pol H. E., Ikeda M., Ikram M. K., Jack C. R. Jr., Jenkinson M., Johnson R., Jonsson E. G., Jukema J. W., Kahn R. S., Kanai R., Kloszewska I., Knopman D. S., Kochunov P., Kwok J. B., Lawrie S. M., Lemaitre H., Liu X., Longo D. L., Longstreth W. T. Jr., Lopez O. L., Lovestone S., Martinez O., Martinot J. L., Mattay V. S., McDonald C., McIntosh A. M., McMahon K. L., McMahon F. J., Mecocci P., Melle I., Meyer-Lindenberg A., Mohnke S., Montgomery G. W., Morris D. W., Mosley T. H., Muhleisen T. W., Muller-Myhsok B., Nalls M. A., Nauck M., Nichols T. E., Niessen W. J., Nothen M. M., Nyberg L., Ohi K., Olvera R. L., Ophoff R. A., Pandolfo M., Paus T., Pausova Z., Penninx B. W., Pike G. B., Potkin S. G., Psaty B. M., Reppermund S., Rietschel M., Roffman J. L., Romanczuk-Seiferth N., Rotter J. I., Ryten M., Sacco R. L., Sachdev P. S., Saykin A. J., Schmidt R., Schofield P. R., Sigurdsson S., Simmons A., Singleton A., Sisodiya S. M., Smith C., Smoller J. W., Soininen H., Srikanth V., Steen V. M., Stott D. J., Sussmann J. E., Thalamuthu A., Tiemeier H., Toga A. W., Traynor B. J., Troncoso J., Turner J. A., Tzourio C., Uitterlinden A. G., Hernandez M. C., Van der Brug M., Van der Lugt A., Van der Wee N. J., Van Duijn C. M., Van Haren N. E., Van T. E. D., Van Tol M. J., Vardarajan B. N., Veltman D. J., Vernooij M. W., Volzke H., Walter H., Wardlaw J. M., Wassink T. H., Weale M. E., Weinberger D. R., Weiner M. W., Wen W., Westman E., White T., Wong T. Y., Wright C. B., Zielke H. R., Zonderman A. B., Deary I. J., DeCarli C., Schmidt H., Martin N. G., De Craen A. J., Wright M. J., Launer L. J., Schumann G., Fornage M., Franke B., Debette S., Medland S. E., Ikram M. A., Thompson P. M., Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 19, 1569–1582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller M. C., Gene × environment interaction studies have not properly controlled for potential confounders: The problem and the (simple) solution. Biol. Psychiatry 75, 18–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skol A. D., Scott L. J., Abecasis G. R., Boehnke M., Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Lim T. K., Ruthazer E. S., Microglial trogocytosis and the complement system regulate axonal pruning in vivo. eLife 10, e62167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steen V. M., Nepal C., Ersland K. M., Holdhus R., Naevdal M., Ratvik S. M., Skrede S., Havik B., Neuropsychological deficits in mice depleted of the schizophrenia susceptibility gene CSMD1. PLOS ONE 8, e79501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W., Cohen-Woods S., Chen Q., Noor A., Knight J., Hosang G., Parikh S. V., De Luca V., Tozzi F., Muglia P., Forte J., McQuillin A., Hu P., Gurling H. M., Kennedy J. L., McGuffin P., Farmer A., Strauss J., Vincent J. B., Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med. Genet. 15, 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel M., Parkinson disease: CSMD1 gene mutations can lead to familial Parkinson disease. Nat. Rev. Neurol. 13, 641 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Hatzimanolis A., Foteli S., Stefanatou P., Ntigrintaki A. A., Ralli I., Kollias K., Nikolaou C., Gazouli M., Stefanis N. C., Deregulation of complement components C4A and CSMD1 peripheral expression in first-episode psychosis and links to cognitive ability. Eur. Arch. Psychiatry Clin. Neurosci. 272, 1219–1228 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Athanasiu L., Giddaluru S., Fernandes C., Christoforou A., Reinvang I., Lundervold A. J., Nilsson L. G., Kauppi K., Adolfsson R., Eriksson E., Sundet K., Djurovic S., Espeseth T., Nyberg L., Steen V. M., Andreassen O. A., Le Hellard S., A genetic association study of CSMD1 and CSMD2 with cognitive function. Brain Behav. Immun. 61, 209–216 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Abe K., Chisaka O., Van Roy F., Takeichi M., Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat. Neurosci. 7, 357–363 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Park C., Falls W., Finger J. H., Longo-Guess C. M., Ackerman S. L., Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat. Genet. 31, 279–284 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Smith A., Bourdeau I., Wang J., Bondy C. A., Expression of Catenin family members CTNNA1, CTNNA2, CTNNB1 and JUP in the primate prefrontal cortex and hippocampus. Brain Res. Mol. Brain Res. 135, 225–231 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Yang R., Walder-Christensen K. K., Kim N., Wu D., Lorenzo D. N., Badea A., Jiang Y. H., Yin H. H., Wetsel W. C., Bennett V., ANK2 autism mutation targeting giant ankyrin-B promotes axon branching and ectopic connectivity. Proc. Natl. Acad. Sci. U.S.A. 116, 15262–15271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isungset M. A., Conley D., Zachrisson H. D., Ystrom E., Havdahl A., Njolstad P. R., Lyngstad T. H., Social and genetic associations with educational performance in a Scandinavian welfare state. Proc. Natl. Acad. Sci. U.S.A. 119, e2201869119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J. J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T. A., Bowers P., Sidorenko J., Linner R. K., Fontana M. A., Kundu T., Lee C., Li H., Li R., Royer R., Timshel P. N., Walters R. K., Willoughby E. A., Yengo L.; 23andMe Research Team; COGENT (Cognitive Genomics Consortium); Social Science Genetic Association Consortium, Alver M., Bao Y., Clark D. W., Day F. R., Furlotte N. A., Joshi P. K., Kemper K. E., Kleinman A., Langenberg C., Magi R., Trampush J. W., Verma S. S., Wu Y., Lam M., Zhao J. H., Zheng Z., Boardman J. D., Campbell H., Freese J., Harris K. M., Hayward C., Herd P., Kumari M., Lencz T., Luan J., Malhotra A. K., Metspalu A., Milani L., Ong K. K., Perry J. R. B., Porteous D. J., Ritchie M. D., Smart M. C., Smith B. H., Tung J. Y., Wareham N. J., Wilson J. F., Beauchamp J. P., Conley D. C., Esko T., Lehrer S. F., Magnusson P. K. E., Oskarsson S., Pers T. H., Robinson M. R., Thom K., Watson C., Chabris C. F., Meyer M. N., Laibson D. I., Yang J., Johannesson M., Koellinger P. D., Turley P., Visscher P. M., Benjamin D. J., Cesarini D., Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rokholm B., Silventoinen K., Tynelius P., Gamborg M., Sorensen T. I. A., Rasmussen F., Increasing genetic variance of body mass index during the Swedish obesity epidemic. PLOS ONE 6, e27135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boardman J. D., Domingue B. W., Blalock C. L., Haberstick B. C., Harris K. M., McQueen M. B., Is the gene-environment interaction paradigm relevant to genome-wide studies? The case of education and body mass index. Demography 51, 119–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Citri A., Malenka R. C., Synaptic plasticity: Multiple forms, functions, and mechanisms. Neuropsychopharmacology 33, 18–41 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Rumpf S., Sanal N., Marzano M., Energy metabolic pathways in neuronal development and function. Oxf. Open Neurosci. 2, kvad004 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tracey T. J., Steyn F. J., Wolvetang E. J., Ngo S. T., Neuronal lipid metabolism: Multiple pathways driving functional outcomes in health and disease. Front. Mol. Neurosci. 11, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bazinet R. P., Layé S., Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 15, 771–785 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Kirschen G. W., Ge S., Editorial: Changes in metabolic processes affecting brain development. Front. Neurosci. 16, 1002010 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziv Y., Ron N., Butovsky O., Landa G., Sudai E., Greenberg N., Cohen H., Kipnis J., Schwartz M., Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat. Neurosci. 9, 268–275 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Radjavi A., Smirnov I., Kipnis J., Brain antigen-reactive CD4+ T cells are sufficient to support learning behavior in mice with limited T cell repertoire. Brain Behav. Immun. 35, 58–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cunningham C. L., Martinez-Cerdeno V., Noctor S. C., Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 33, 4216–4233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueno M., Fujita Y., Tanaka T., Nakamura Y., Kikuta J., Ishii M., Yamashita T., Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 16, 543–551 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Pang Y., Fan L.-W., Tien L.-T., Dai X., Zheng B., Cai Z., Lin R. C. S., Bhatt A., Differential roles of astrocyte and microglia in supporting oligodendrocyte development and myelination in vitro. Brain Behav. 3, 503–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shigemoto-Mogami Y., Hoshikawa K., Goldman J. E., Sekino Y., Sato K., Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34, 2231–2243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stevens B., Allen N. J., Vazquez L. E., Howell G. R., Christopherson K. S., Nouri N., Micheva K. D., Mehalow A. K., Huberman A. D., Stafford B., Sher A., Litke A. M., Lambris J. D., Smith S. J., John S. W., Barres B. A., The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Miyamoto A., Wake H., Ishikawa A. W., Eto K., Shibata K., Murakoshi H., Koizumi S., Moorhouse A. J., Yoshimura Y., Nabekura J., Microglia contact induces synapse formation in developing somatosensory cortex. Nat. Commun. 7, 12540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bethlehem R. A. I., Seidlitz J., White S. R., Vogel J. W., Anderson K. M., Adamson C., Adler S., Alexopoulos G. S., Anagnostou E., Areces-Gonzalez A., Astle D. E., Auyeung B., Ayub M., Bae J., Ball G., Baron-Cohen S., Beare R., Bedford S. A., Benegal V., Beyer F., Blangero J., Cabez M. B., Boardman J. P., Borzage M., Bosch-Bayard J. F., Bourke N., Calhoun V. D., Chakravarty M. M., Chen C., Chertavian C., Chetelat G., Chong Y. S., Cole J. H., Corvin A., Costantino M., Courchesne E., Crivello F., Cropley V. L., Crosbie J., Crossley N., Delarue M., Delorme R., Desrivieres S., Devenyi G. A., Di Biase M. A., Dolan R., Donald K. A., Donohoe G., Dunlop K., Edwards A. D., Elison J. T., Ellis C. T., Elman J. A., Eyler L., Fair D. A., Feczko E., Fletcher P. C., Fonagy P., Franz C. E., Galan-Garcia L., Gholipour A., Giedd J., Gilmore J. H., Glahn D. C., Goodyer I. M., Grant P. E., Groenewold N. A., Gunning F. M., Gur R. E., Gur R. C., Hammill C. F., Hansson O., Hedden T., Heinz A., Henson R. N., Heuer K., Hoare J., Holla B., Holmes A. J., Holt R., Huang H., Im K., Ipser J., Jack C. R. Jr., Jackowski A. P., Jia T., Johnson K. A., Jones P. B., Jones D. T., Kahn R. S., Karlsson H., Karlsson L., Kawashima R., Kelley E. A., Kern S., Kim K. W., Kitzbichler M. G., Kremen W. S., Lalonde F., Landeau B., Lee S., Lerch J., Lewis J. D., Li J., Liao W., Liston C., Lombardo M. V., Lv J., Lynch C., Mallard T. T., Marcelis M., Markello R. D., Mathias S. R., Mazoyer B., McGuire P., Meaney M. J., Mechelli A., Medic N., Misic B., Morgan S. E., Mothersill D., Nigg J., Ong M. Q. W., Ortinau C., Ossenkoppele R., Ouyang M., Palaniyappan L., Paly L., Pan P. M., Pantelis C., Park M. M., Paus T., Pausova Z., Paz-Linares D., Binette A. P., Pierce K., Qian X., Qiu J., Qiu A., Raznahan A., Rittman T., Rodrigue A., Rollins C. K., Romero-Garcia R., Ronan L., Rosenberg M. D., Rowitch D. H., Salum G. A., Satterthwaite T. D., Schaare H. L., Schachar R. J., Schultz A. P., Schumann G., Scholl M., Sharp D., Shinohara R. T., Skoog I., Smyser C. D., Sperling R. A., Stein D. J., Stolicyn A., Suckling J., Sullivan G., Taki Y., Thyreau B., Toro R., Traut N., Tsvetanov K. A., Turk-Browne N. B., Tuulari J. J., Tzourio C., Vachon-Presseau E., Valdes-Sosa M. J., Valdes-Sosa P. A., Valk S. L., van Amelsvoort T., Vandekar S. N., Vasung L., Victoria L. W., Villeneuve S., Villringer A., Vertes P. E., Wagstyl K., Wang Y. S., Warfield S. K., Warrier V., Westman E., Westwater M. L., Whalley H. C., Witte A. V., Yang N., Yeo B., Yun H., Zalesky A., Zar H. J., Zettergren A., Zhou J. H., Ziauddeen H., Zugman A., Zuo X. N.; 3R-BRAIN; AIBL; Alzheimer’s Disease Neuroimaging Initiative; Alzheimer’s Disease Repository Without Borders Investigators; CALM Team; Cam-CAN; CCNP; COBRE; cVEDA; ENIGMA Developmental Brain Age Working Group; Developing Human Connectome Project; FinnBrain; Harvard Aging Brain Study; IMAGEN; KNE96; Mayo Clinic Study of Aging; NSPN; POND; PREVENT-AD Research Group; VETSA, Bullmore E. T., Alexander-Bloch A. F., Brain charts for the human lifespan. Nature 604, 525–533 (2022).35388223 [Google Scholar]
- 69.Boyce W. T., Sokolowski M. B., Robinson G. E., Genes and environments, development and time. Proc. Natl. Acad. Sci. U.S.A. 117, 23235–23241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alves P. N., Foulon C., Karolis V., Bzdok D., Margulies D. S., Volle E., Thiebaut de Schotten M., An improved neuroanatomical model of the default-mode network reconciles previous neuroimaging and neuropathological findings. Commun. Biol. 2, 370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W., Mai X., Liu C., The default mode network and social understanding of others: What do brain connectivity studies tell us. Front. Hum. Neurosci. 8, 74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao W., Alcauter S., Elton A., Hernandez-Castillo C. R., Smith J. K., Ramirez J., Lin W., Functional network development during the first year: Relative sequence and socioeconomic correlations. Cereb. Cortex 25, 2919–2928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whittle S., Vijayakumar N., Simmons J. G., Dennison M., Schwartz O., Pantelis C., Sheeber L., Byrne M. L., Allen N. B., Role of positive parenting in the association between neighborhood social disadvantage and brain development across adolescence. JAMA Psychiatry 74, 824–832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Degeilh F., Bernier A., Leblanc E., Daneault V., Beauchamp M. H., Quality of maternal behaviour during infancy predicts functional connectivity between default mode network and salience network 9 years later. Dev. Cogn. Neurosci. 34, 53–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Meer D., Hartman C. A., Pruim R. H. R., Mennes M., Heslenfeld D., Oosterlaan J., Faraone S. V., Franke B., Buitelaar J. K., Hoekstra P. J., The interaction between 5-HTTLPR and stress exposure influences connectivity of the executive control and default mode brain networks. Brain Imaging Behav. 11, 1486–1496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scarr-Salapatek S., Race, social class, and IQ. Science 174, 1285–1295 (1971). [DOI] [PubMed] [Google Scholar]
- 77.Wedow R., Zacher M., Huibregtse B. M., Harris K. M., Domingue B. W., Boardman J. D., Education, smoking, and cohort change: Forwarding a multidimensional theory of the environmental moderation of genetic effects. Am. Sociol. Rev. 83, 802–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flemmen H. O., Simonsen C. S., Broch L., Brunborg C., Berg-Hansen P., Moen S. M., Kersten H., Celius E. G., Maternal education has significant influence on progression in multiple sclerosis. Mult. Scler. Relat. Disord. 53, 103052 (2021). [DOI] [PubMed] [Google Scholar]
- 79.Rogers M. A., Plassman B. L., Kabeto M., Fisher G. G., McArdle J. J., Llewellyn D. J., Potter G. G., Langa K. M., Parental education and late-life dementia in the United States. J. Geriatr. Psychiatry Neurol. 22, 71–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cermakova P., Chlapecka A., Csajbok Z., Andryskova L., Brazdil M., Mareckova K., Parental education, cognition and functional connectivity of the salience network. Sci. Rep. 13, 2761 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jarosz E., Gugushvili A., Parental education, health literacy and children’s adult body height. J. Biosoc. Sci. 52, 696–718 (2020). [DOI] [PubMed] [Google Scholar]
- 82.White P. A., Awad Y. A., Gauvin L., Spencer N. J., McGrath J. J., Clifford S. A., Nikiema B., Yang-Huang J., Goldhaber-Fiebert J. D., Markham W., Mensah F. K., van Grieken A., Raat H., Jaddoe V. W. V., Ludvigsson J., Faresjo T.; EPOCH Collaborative Group , Household income and maternal education in early childhood and risk of overweight and obesity in late childhood: Findings from seven birth cohort studies in six high-income countries. Int. J. Obes. 46, 1703–1711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang C., Li H., Liu P. W., Zhang J., Why does spousal education matter for earnings? Assortative mating and cross-productivity. J. Labor Econ. 27, 633–652 (2009). [Google Scholar]
- 84.Berlin S., Social causes of psychological distress. Soc. Serv. Rev. 65, 331–333 (1991). [Google Scholar]
- 85.Tucker-Drob E. M., Bates T. C., Large cross-national differences in gene × socioeconomic status interaction on intelligence. Psychol. Sci. 27, 138–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Figlio D. N., Freese J., Karbownik K., Roth J., Socioeconomic status and genetic influences on cognitive development. Proc. Natl. Acad. Sci. U.S.A. 114, 13441–13446 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boardman J. D., Daw J., Freese J., Defining the environment in gene-environment research: Lessons from social epidemiology. Am. J. Public Health 103 (Suppl. 1), S64–S72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karcher N. R., Barch D. M., The ABCD study: Understanding the development of risk for mental and physical health outcomes. Neuropsychopharmacology 46, 131–142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mascarell Maricic L., Walter H., Rosenthal A., Ripke S., Quinlan E. B., Banaschewski T., Barker G. J., Bokde A. L. W., Bromberg U., Buchel C., Desrivieres S., Flor H., Frouin V., Garavan H., Itterman B., Martinot J. L., Martinot M. P., Nees F., Orfanos D. P., Paus T., Poustka L., Hohmann S., Smolka M. N., Frohner J. H., Whelan R., Kaminski J., Schumann G., Heinz A.; IMAGEN consortium , The IMAGEN study: A decade of imaging genetics in adolescents. Mol. Psychiatry 25, 2648–2671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Casey B. J., Cannonier T., Conley M. I., Cohen A. O., Barch D. M., Heitzeg M. M., Soules M. E., Teslovich T., Dellarco D. V., Garavan H., Orr C. A., Wager T. D., Banich M. T., Speer N. K., Sutherland M. T., Riedel M. C., Dick A. S., Bjork J. M., Thomas K. M., Chaarani B., Mejia M. H., Hagler D. J. Jr., Daniela Cornejo M., Sicat C. S., Harms M. P., Dosenbach N. U. F., Rosenberg M., Earl E., Bartsch H., Watts R., Polimeni J. R., Kuperman J. M., Fair D. A., Dale A. M.; ABCD Imaging Acquisition Workgroup , The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 32, 43–54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teo Y. Y., Small K. S., Kwiatkowski D. P., Methodological challenges of genome-wide association analysis in Africa. Nat. Rev. Genet. 11, 149–160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rosenberg N. A., Huang L., Jewett E. M., Szpiech Z. A., Jankovic I., Boehnke M., Genome-wide association studies in diverse populations. Nat. Rev. Genet. 11, 356–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.the Haplotype Reference Consortium , A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo Q., Chen Q., Wang W., Desrivieres S., Quinlan E. B., Jia T., Macare C., Robert G. H., Cui J., Guedj M., Palaniyappan L., Kherif F., Banaschewski T., Bokde A. L. W., Buchel C., Flor H., Frouin V., Garavan H., Gowland P., Heinz A., Ittermann B., Martinot J. L., Artiges E., Paillere-Martinot M. L., Nees F., Orfanos D. P., Poustka L., Frohner J. H., Smolka M. N., Walter H., Whelan R., Callicott J. H., Mattay V. S., Pausova Z., Dartigues J. F., Tzourio C., Crivello F., Berman K. F., Li F., Paus T., Weinberger D. R., Murray R. M., Schumann G., Feng J.; IMAGEN consortium , Association of a schizophrenia-risk nonsynonymous variant with putamen volume in adolescents: A voxelwise and genome-wide association study. JAMA Psychiatry 76, 435–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.The International HapMap 3 Consortium , Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh G. K., Area deprivation and widening inequalities in US mortality, 1969-1998. Am. J. Public Health 93, 1137–1143 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.R. H. Moos, B. S. Moos, Family Environment Scale Manual: Development, Applications, Research (Social Climate Scale, Center for Health Care Evaluation, Department of Veterans Affairs, Stanford University Medical Center, ed. 4, 2009).
- 98.Karoly H. C., Callahan T., Schmiege S. J., Ewing S. W., Evaluating the hispanic paradox in the context of adolescent risky sexual behavior: The role of parent monitoring. J. Pediatr. Psychol. 41, 429–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., Lee J. J., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi S. W., O’Reilly P. F., PRSice-2: Polygenic risk score software for biobank-scale data. Gigascience 8, giz082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.The 1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ghoussaini M., Mountjoy E., Carmona M., Peat G., Schmidt E. M., Hercules A., Fumis L., Miranda A., Carvalho-Silva D., Buniello A., Burdett T., Hayhurst J., Baker J., Ferrer J., Gonzalez-Uriarte A., Jupp S., Karim M. A., Koscielny G., Machlitt-Northen S., Malangone C., Pendlington Z. M., Roncaglia P., Suveges D., Wright D., Vrousgou O., Papa E., Parkinson H., MacArthur J. A. L., Todd J. A., Barrett J. C., Schwartzentruber J., Hulcoop D. G., Ochoa D., McDonagh E. M., Dunham I., Open targets genetics: Systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Leeuw C. A., Mooij J. M., Heskes T., Posthuma D., MAGMA: Generalized gene-set analysis of GWAS data. PLOS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Watanabe K., Taskesen E., van Bochoven A., Posthuma D., Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vrieze S. I., Iacono W. G., McGue M., Confluence of genes, environment, development, and behavior in a post genome-wide association study world. Dev. Psychopathol. 24, 1195–1214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li J., Ji L., Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Brouwer R. M., Klein M., Grasby K. L., Schnack H. G., Jahanshad N., Teeuw J., Thomopoulos S. I., Sprooten E., Franz C. E., Gogtay N., Kremen W. S., Panizzon M. S., Loohuis L. M. O., Whelan C. D., Aghajani M., Alloza C., Alnaes D., Artiges E., Ayesa-Arriola R., Barker G. J., Bastin M. E., Blok E., Boen E., Breukelaar I. A., Bright J. K., Buimer E. E. L., Bulow R., Cannon D. M., Ciufolini S., Crossley N. A., Damatac C. G., Dazzan P., de Mol C. L., de Zwarte S. M. C., Desrivieres S., Diaz-Caneja C. M., Doan N. T., Dohm K., Frohner J. H., Goltermann J., Grigis A., Grotegerd D., Han L. K. M., Harris M. A., Hartman C. A., Heany S. J., Heindel W., Heslenfeld D. J., Hohmann S., Ittermann B., Jansen P. R., Janssen J., Jia T., Jiang J., Jockwitz C., Karali T., Keeser D., Koevoets M., Lenroot R. K., Malchow B., Mandl R. C. W., Medel V., Meinert S., Morgan C. A., Muhleisen T. W., Nabulsi L., Opel N., de la Foz V. O., Overs B. J., Martinot M. L. P., Redlich R., Marques T. R., Repple J., Roberts G., Roshchupkin G. V., Setiaman N., Shumskaya E., Stein F., Sudre G., Takahashi S., Thalamuthu A., Tordesillas-Gutierrez D., van der Lugt A., van Haren N. E. M., Wardlaw J. M., Wen W., Westeneng H. J., Wittfeld K., Zhu A. H., Zugman A., Armstrong N. J., Bonfiglio G., Bralten J., Dalvie S., Davies G., Di Forti M., Ding L., Donohoe G., Forstner A. J., Gonzalez-Penas J., Guimaraes J., Homuth G., Hottenga J. J., Knol M. J., Kwok J. B. J., Le Hellard S., Mather K. A., Milaneschi Y., Morris D. W., Nothen M. M., Papiol S., Rietschel M., Santoro M. L., Steen V. M., Stein J. L., Streit F., Tankard R. M., Teumer A., van Ent D., van der Meer D., van Eijk K. R., Vassos E., Vazquez-Bourgon J., Witt S. H.; the IMAGEN Consortium, Adams H. H. H., Agartz I., Ames D., Amunts K., Andreassen O. A., Arango C., Banaschewski T., Baune B. T., Belangero S. I., Bokde A. L. W., Boomsma D. I., Bressan R. A., Brodaty H., Buitelaar J. K., Cahn W., Caspers S., Cichon S., Crespo-Facorro B., Cox S. R., Dannlowski U., Elvsashagen T., Espeseth T., Falkai P. G., Fisher S. E., Flor H., Fullerton J. M., Garavan H., Gowland P. A., Grabe H. J., Hahn T., Heinz A., Hillegers M., Hoare J., Hoekstra P. J., Ikram M. A., Jackowski A. P., Jansen A., Jonsson E. G., Kahn R. S., Kircher T., Korgaonkar M. S., Krug A., Lemaitre H., Malt U. F., Martinot J. L., McDonald C., Mitchell P. B., Muetzel R. L., Murray R. M., Nees F., Nenadic I., Oosterlaan J., Ophoff R. A., Pan P. M., Penninx B., Poustka L., Sachdev P. S., Salum G. A., Schofield P. R., Schumann G., Shaw P., Sim K., Smolka M. N., Stein D. J., Trollor J. N., van den Berg L. H., Veldink J. H., Walter H., Westlye L. T., Whelan R., White T., Wright M. J., Medland S. E., Franke B., Thompson P. M., Hulshoff Pol H. E., Genetic variants associated with longitudinal changes in brain structure across the lifespan. Nat. Neurosci. 25, 421–432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Savage J. E., Jansen P. R., Stringer S., Watanabe K., Bryois J., de Leeuw C. A., Nagel M., Awasthi S., Barr P. B., Coleman J. R. I., Grasby K. L., Hammerschlag A. R., Kaminski J. A., Karlsson R., Krapohl E., Lam M., Nygaard M., Reynolds C. A., Trampush J. W., Young H., Zabaneh D., Hagg S., Hansell N. K., Karlsson I. K., Linnarsson S., Montgomery G. W., Munoz-Manchado A. B., Quinlan E. B., Schumann G., Skene N. G., Webb B. T., White T., Arking D. E., Avramopoulos D., Bilder R. M., Bitsios P., Burdick K. E., Cannon T. D., Chiba-Falek O., Christoforou A., Cirulli E. T., Congdon E., Corvin A., Davies G., Deary I. J., DeRosse P., Dickinson D., Djurovic S., Donohoe G., Conley E. D., Eriksson J. G., Espeseth T., Freimer N. A., Giakoumaki S., Giegling I., Gill M., Glahn D. C., Hariri A. R., Hatzimanolis A., Keller M. C., Knowles E., Koltai D., Konte B., Lahti J., Le Hellard S., Lencz T., Liewald D. C., London E., Lundervold A. J., Malhotra A. K., Melle I., Morris D., Need A. C., Ollier W., Palotie A., Payton A., Pendleton N., Poldrack R. A., Raikkonen K., Reinvang I., Roussos P., Rujescu D., Sabb F. W., Scult M. A., Smeland O. B., Smyrnis N., Starr J. M., Steen V. M., Stefanis N. C., Straub R. E., Sundet K., Tiemeier H., Voineskos A. N., Weinberger D. R., Widen E., Yu J., Abecasis G., Andreassen O. A., Breen G., Christiansen L., Debrabant B., Dick D. M., Heinz A., Hjerling-Leffler J., Ikram M. A., Kendler K. S., Martin N. G., Medland S. E., Pedersen N. L., Plomin R., Polderman T. J. C., Ripke S., van der Sluis S., Sullivan P. F., Vrieze S. I., Wright M. J., Posthuma D., Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davies G., Marioni R. E., Liewald D. C., Hill W. D., Hagenaars S. P., Harris S. E., Ritchie S. J., Luciano M., Fawns-Ritchie C., Lyall D., Cullen B., Cox S. R., Hayward C., Porteous D. J., Evans J., McIntosh A. M., Gallacher J., Craddock N., Pell J. P., Smith D. J., Gale C. R., Deary I. J., Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Mol. Psychiatry 21, 758–767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sakaue S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., Narita A., Konuma T., Yamamoto K., Akiyama M., Ishigaki K., Suzuki A., Suzuki K., Obara W., Yamaji K., Takahashi K., Asai S., Takahashi Y., Suzuki T., Shinozaki N., Yamaguchi H., Minami S., Murayama S., Yoshimori K., Nagayama S., Obata D., Higashiyama M., Masumoto A., Koretsune Y., Gen F., Ito K., Terao C., Yamauchi T., Komuro I., Kadowaki T., Tamiya G., Yamamoto M., Nakamura Y., Kubo M., Murakami Y., Yamamoto K., Kamatani Y., Palotie A., Rivas M. A., Daly M. J., Matsuda K., Okada Y., A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021). [DOI] [PubMed] [Google Scholar]
- 111.Shungin D., Winkler T. W., Croteau-Chonka D. C., Ferreira T., Locke A. E., Magi R., Strawbridge R. J., Pers T. H., Fischer K., Justice A. E., Workalemahu T., Wu J. M. W., Buchkovich M. L., Heard-Costa N. L., Roman T. S., Drong A. W., Song C., Gustafsson S., Day F. R., Esko T., Fall T., Kutalik Z., Luan J., Randall J. C., Scherag A., Vedantam S., Wood A. R., Chen J., Fehrmann R., Karjalainen J., Kahali B., Liu C. T., Schmidt E. M., Absher D., Amin N., Anderson D., Beekman M., Bragg-Gresham J. L., Buyske S., Demirkan A., Ehret G. B., Feitosa M. F., Goel A., Jackson A. U., Johnson T., Kleber M. E., Kristiansson K., Mangino M., Leach I. M., Medina-Gomez C., Palmer C. D., Pasko D., Pechlivanis S., Peters M. J., Prokopenko I., Stancakova A., Sung Y. J., Tanaka T., Teumer A., Van Vliet-Ostaptchouk J. V., Yengo L., Zhang W., Albrecht E., Arnlov J., Arscott G. M., Bandinelli S., Barrett A., Bellis C., Bennett A. J., Berne C., Bluher M., Bohringer S., Bonnet F., Bottcher Y., Bruinenberg M., Carba D. B., Caspersen I. H., Clarke R., Daw E. W., Deelen J., Deelman E., Delgado G., Doney A. S., Eklund N., Erdos M. R., Estrada K., Eury E., Friedrich N., Garcia M. E., Giedraitis V., Gigante B., Go A. S., Golay A., Grallert H., Grammer T. B., Grassler J., Grewal J., Groves C. J., Haller T., Hallmans G., Hartman C. A., Hassinen M., Hayward C., Heikkila K., Herzig K. H., Helmer Q., Hillege H. L., Holmen O., Hunt S. C., Isaacs A., Ittermann T., James A. L., Johansson I., Juliusdottir T., Kalafati I. P., Kinnunen L., Koenig W., Kooner I. K., Kratzer W., Lamina C., Leander K., Lee N. R., Lichtner P., Lind L., Lindstrom J., Lobbens S., Lorentzon M., Mach F., Magnusson P. K., Mahajan A., McArdle W. L., Menni C., Merger S., Mihailov E., Milani L., Mills R., Moayyeri A., Monda K. L., Mooijaart S. P., Muhleisen T. W., Mulas A., Muller G., Muller-Nurasyid M., Nagaraja R., Nalls M. A., Narisu N., Glorioso N., Nolte I. M., Olden M., Rayner N. W., Renstrom F., Ried J. S., Robertson N. R., Rose L. M., Sanna S., Scharnagl H., Scholtens S., Sennblad B., Seufferlein T., Sitlani C. M., Smith A. V., Stirrups K., Stringham H. M., Sundstrom J., Swertz M. A., Swift A. J., Syvanen A. C., Tayo B. O., Thorand B., Thorleifsson G., Tomaschitz A., Troffa C., van Oort F. V., Verweij N., Vonk J. M., Waite L. L., Wennauer R., Wilsgaard T., Wojczynski M. K., Wong A., Zhang Q., Zhao J. H., Brennan E. P., Choi M., Eriksson P., Folkersen L., Franco-Cereceda A., Gharavi A. G., Hedman A. K., Hivert M. F., Huang J., Kanoni S., Karpe F., Keildson S., Kiryluk K., Liang L., Lifton R. P., Ma B., McKnight A. J., McPherson R., Metspalu A., Min J. L., Moffatt M. F., Montgomery G. W., Murabito J. M., Nicholson G., Nyholt D. R., Olsson C., Perry J. R., Reinmaa E., Salem R. M., Sandholm N., Schadt E. E., Scott R. A., Stolk L., Vallejo E. E., Westra H. J., Zondervan K. T.; The ADIPOGen Consortium; The CARDIOGRAMplusC4D Consortium; The CKDGen Consortium; The GEFOS Consortium; The GENIE Consortium; The GLGC; The ICBP; The International Endogene Consortium; The LifeLines Cohort Study; The MAGIC Investigators; The MuTHER Consortium; The PAGE Consortium; The ReproGen Consortium, Amouyel P., Arveiler D., Bakker S. J., Beilby J., Bergman R. N., Blangero J., Brown M. J., Burnier M., Campbell H., Chakravarti A., Chines P. S., Claudi-Boehm S., Collins F. S., Crawford D. C., Danesh J., de Faire U., de Geus E. J., Dorr M., Erbel R., Eriksson J. G., Farrall M., Ferrannini E., Ferrieres J., Forouhi N. G., Forrester T., Franco O. H., Gansevoort R. T., Gieger C., Gudnason V., Haiman C. A., Harris T. B., Hattersley A. T., Heliovaara M., Hicks A. A., Hingorani A. D., Hoffmann W., Hofman A., Homuth G., Humphries S. E., Hypponen E., Illig T., Jarvelin M. R., Johansen B., Jousilahti P., Jula A. M., Kaprio J., Kee F., Keinanen-Kiukaanniemi S. M., Kooner J. S., Kooperberg C., Kovacs P., Kraja A. T., Kumari M., Kuulasmaa K., Kuusisto J., Lakka T. A., Langenberg C., Le Marchand L., Lehtimaki T., Lyssenko V., Mannisto S., Marette A., Matise T. C., McKenzie C. A., McKnight B., Musk A. W., Mohlenkamp S., Morris A. D., Nelis M., Ohlsson C., Oldehinkel A. J., Ong K. K., Palmer L. J., Penninx B. W., Peters A., Pramstaller P. P., Raitakari O. T., Rankinen T., Rao D. C., Rice T. K., Ridker P. M., Ritchie M. D., Rudan I., Salomaa V., Samani N. J., Saramies J., Sarzynski M. A., Schwarz P. E., Shuldiner A. R., Staessen J. A., Steinthorsdottir V., Stolk R. P., Strauch K., Tonjes A., Tremblay A., Tremoli E., Vohl M. C., Volker U., Vollenweider P., Wilson J. F., Witteman J. C., Adair L. S., Bochud M., Boehm B. O., Bornstein S. R., Bouchard C., Cauchi S., Caulfield M. J., Chambers J. C., Chasman D. I., Cooper R. S., Dedoussis G., Ferrucci L., Froguel P., Grabe H. J., Hamsten A., Hui J., Hveem K., Jockel K. H., Kivimaki M., Kuh D., Laakso M., Liu Y., Marz W., Munroe P. B., Njolstad I., Oostra B. A., Palmer C. N., Pedersen N. L., Perola M., Perusse L., Peters U., Power C., Quertermous T., Rauramaa R., Rivadeneira F., Saaristo T. E., Saleheen D., Sinisalo J., Slagboom P. E., Snieder H., Spector T. D., Stefansson K., Stumvoll M., Tuomilehto J., Uitterlinden A. G., Uusitupa M., van der Harst P., Veronesi G., Walker M., Wareham N. J., Watkins H., Wichmann H. E., Abecasis G. R., Assimes T. L., Berndt S. I., Boehnke M., Borecki I. B., Deloukas P., Franke L., Frayling T. M., Groop L. C., Hunter D. J., Kaplan R. C., O’Connell J. R., Qi L., Schlessinger D., Strachan D. P., Thorsteinsdottir U., van Duijn C. M., Willer C. J., Visscher P. M., Yang J., Hirschhorn J. N., Zillikens M. C., McCarthy M. I., Speliotes E. K., North K. E., Fox C. S., Barroso I., Franks P. W., Ingelsson E., Heid I. M., Loos R. J., Cupples L. A., Morris A. P., Lindgren C. M., Mohlke K. L., New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang L., Zheng Z., Fang H., Yang J., A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 53, 1616–1621 (2021). [DOI] [PubMed] [Google Scholar]
- 113.Scott R. A., Scott L. J., Magi R., Marullo L., Gaulton K. J., Kaakinen M., Pervjakova N., Pers T. H., Johnson A. D., Eicher J. D., Jackson A. U., Ferreira T., Lee Y., Ma C., Steinthorsdottir V., Thorleifsson G., Qi L., Van Zuydam N. R., Mahajan A., Chen H., Almgren P., Voight B. F., Grallert H., Muller-Nurasyid M., Ried J. S., Rayner N. W., Robertson N., Karssen L. C., van Leeuwen E. M., Willems S. M., Fuchsberger C., Kwan P., Teslovich T. M., Chanda P., Li M., Lu Y., Dina C., Thuillier D., Yengo L., Jiang L., Sparso T., Kestler H. A., Chheda H., Eisele L., Gustafsson S., Franberg M., Strawbridge R. J., Benediktsson R., Hreidarsson A. B., Kong A., Sigurethsson G., Kerrison N. D., Luan J., Liang L., Meitinger T., Roden M., Thorand B., Esko T., Mihailov E., Fox C., Liu C. T., Rybin D., Isomaa B., Lyssenko V., Tuomi T., Couper D. J., Pankow J. S., Grarup N., Have C. T., Jorgensen M. E., Jorgensen T., Linneberg A., Cornelis M. C., van Dam R. M., Hunter D. J., Kraft P., Sun Q., Edkins S., Owen K. R., Perry J. R. B., Wood A. R., Zeggini E., Tajes-Fernandes J., Abecasis G. R., Bonnycastle L. L., Chines P. S., Stringham H. M., Koistinen H. A., Kinnunen L., Sennblad B., Muhleisen T. W., Nothen M. M., Pechlivanis S., Baldassarre D., Gertow K., Humphries S. E., Tremoli E., Klopp N., Meyer J., Steinbach G., Wennauer R., Eriksson J. G., Mӓnnisto S., Peltonen L., Tikkanen E., Charpentier G., Eury E., Lobbens S., Gigante B., Leander K., McLeod O., Bottinger E. P., Gottesman O., Ruderfer D., Bluher M., Kovacs P., Tonjes A., Maruthur N. M., Scapoli C., Erbel R., Jockel K. H., Moebus S., de Faire U., Hamsten A., Stumvoll M., Deloukas P., Donnelly P. J., Frayling T. M., Hattersley A. T., Ripatti S., Salomaa V., Pedersen N. L., Boehm B. O., Bergman R. N., Collins F. S., Mohlke K. L., Tuomilehto J., Hansen T., Pedersen O., Barroso I., Lannfelt L., Ingelsson E., Lind L., Lindgren C. M., Cauchi S., Froguel P., Loos R. J. F., Balkau B., Boeing H., Franks P. W., Gurrea A. B., Palli D., van der Schouw Y. T., Altshuler D., Groop L. C., Langenberg C., Wareham N. J., Sijbrands E., van Duijn C. M., Florez J. C., Meigs J. B., Boerwinkle E., Gieger C., Strauch K., Metspalu A., Morris A. D., Palmer C. N. A., Hu F. B., Thorsteinsdottir U., Stefansson K., Dupuis J., Morris A. P., Boehnke M., McCarthy M. I., Prokopenko I.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium , An expanded genome-wide association study of type 2 diabetes in europeans. Diabetes 66, 2888–2902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Glanville K. P., Coleman J. R. I., O’Reilly P. F., Galloway J., Lewis C. M., Investigating pleiotropy between depression and autoimmune diseases using the UK biobank. Biol. Psychiatry Glob. Open Sci. 1, 48–58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R. R., Manoharan A., Ortmann W., Bhangale T., Denny J. C., Carroll R. J., Eyler A. E., Greenberg J. D., Kremer J. M., Pappas D. A., Jiang L., Yin J., Ye L., Su D. F., Yang J., Xie G., Keystone E., Westra H. J., Esko T., Metspalu A., Zhou X., Gupta N., Mirel D., Stahl E. A., Diogo D., Cui J., Liao K., Guo M. H., Myouzen K., Kawaguchi T., Coenen M. J., van Riel P. L., van de Laar M. A., Guchelaar H. J., Huizinga T. W., Dieude P., Mariette X., Bridges S. L. Jr., Zhernakova A., Toes R. E., Tak P. P., Miceli-Richard C., Bang S. Y., Lee H. S., Martin J., Gonzalez-Gay M. A., Rodriguez-Rodriguez L., Rantapaa-Dahlqvist S., Arlestig L., Choi H. K., Kamatani Y., Galan P., Lathrop M.; the RACI consortium; the GARNET consortium, Eyre S., Bowes J., Barton A., de Vries N., Moreland L. W., Criswell L. A., Karlson E. W., Taniguchi A., Yamada R., Kubo M., Liu J. S., Bae S. C., Worthington J., Padyukov L., Klareskog L., Gregersen P. K., Raychaudhuri S., Stranger B. E., De Jager P. L., Franke L., Visscher P. M., Brown M. A., Yamanaka H., Mimori T., Takahashi A., Xu H., Behrens T. W., Siminovitch K. A., Momohara S., Matsuda F., Yamamoto K., Plenge R. M., Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pappa I., Pourcain B. S., Benke K., Cavadino A., Hakulinen C., Nivard M. G., Nolte I. M., Tiesler C. M., Bakermans-Kranenburg M. J., Davies G. E., Evans D. M., Geoffroy M. C., Grallert H., Groen-Blokhuis M. M., Hudziak J. J., Kemp J. P., Keltikangas-Jarvinen L., McMahon G., Mileva-Seitz V. R., Motazedi E., Power C., Raitakari O. T., Ring S. M., Rivadeneira F., Rodriguez A., Scheet P. A., Seppala I., Snieder H., Standl M., Thiering E., Timpson N. J., Veenstra R., Velders F. P., Whitehouse A. J., Smith G. D., Heinrich J., Hypponen E., Lehtimaki T., Middeldorp C. M., Oldehinkel A. J., Pennell C. E., Boomsma D. I., Tiemeier H., A genome-wide approach to children’s aggressive behavior: The EAGLE consortium. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 562–572 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Demontis D., Walters R. K., Martin J., Mattheisen M., Als T. D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Baekvad-Hansen M., Cerrato F., Chambert K., Churchhouse C., Dumont A., Eriksson N., Gandal M., Goldstein J. I., Grasby K. L., Grove J., Gudmundsson O. O., Hansen C. S., Hauberg M. E., Hollegaard M. V., Howrigan D. P., Huang H., Maller J. B., Martin A. R., Martin N. G., Moran J., Pallesen J., Palmer D. S., Pedersen C. B., Pedersen M. G., Poterba T., Poulsen J. B., Ripke S., Robinson E. B., Satterstrom F. K., Stefansson H., Stevens C., Turley P., Walters G. B., Won H., Wright M. J.; ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team, Andreassen O. A., Asherson P., Burton C. L., Boomsma D. I., Cormand B., Dalsgaard S., Franke B., Gelernter J., Geschwind D., Hakonarson H., Haavik J., Kranzler H. R., Kuntsi J., Langley K., Lesch K. P., Middeldorp C., Reif A., Rohde L. A., Roussos P., Schachar R., Sklar P., Sonuga-Barke E. J. S., Sullivan P. F., Thapar A., Tung J. Y., Waldman I. D., Medland S. E., Stefansson K., Nordentoft M., Hougaard D. M., Werge T., Mors O., Mortensen P. B., Daly M. J., Faraone S. V., Borglum A. D., Neale B. M., Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grove J., Ripke S., Als T. D., Mattheisen M., Walters R. K., Won H., Pallesen J., Agerbo E., Andreassen O. A., Anney R., Awashti S., Belliveau R., Bettella F., Buxbaum J. D., Bybjerg-Grauholm J., Baekvad-Hansen M., Cerrato F., Chambert K., Christensen J. H., Churchhouse C., Dellenvall K., Demontis D., De Rubeis S., Devlin B., Djurovic S., Dumont A. L., Goldstein J. I., Hansen C. S., Hauberg M. E., Hollegaard M. V., Hope S., Howrigan D. P., Huang H., Hultman C. M., Klei L., Maller J., Martin J., Martin A. R., Moran J. L., Nyegaard M., Naerland T., Palmer D. S., Palotie A., Pedersen C. B., Pedersen M. G., dPoterba T., Poulsen J. B., Pourcain B. S., Qvist P., Rehnstrom K., Reichenberg A., Reichert J., Robinson E. B., Roeder K., Roussos P., Saemundsen E., Sandin S., Satterstrom F. K., Smith G. D., Stefansson H., Steinberg S., Stevens C. R., Sullivan P. F., Turley P., Walters G. B., Xu X.; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team, Stefansson K., Geschwind D. H., Nordentoft M., Hougaard D. M., Werge T., Mors O., Mortensen P. B., Neale B. M., Daly M. J., Borglum A. D., Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Howard D. M., Adams M. J., Clarke T. K., Hafferty J. D., Gibson J., Shirali M., Coleman J. R. I., Hagenaars S. P., Ward J., Wigmore E. M., Alloza C., Shen X., Barbu M. C., Xu E. Y., Whalley H. C., Marioni R. E., Porteous D. J., Davies G., Deary I. J., Hemani G., Berger K., Teismann H., Rawal R., Arolt V., Baune B. T., Dannlowski U., Domschke K., Tian C., Hinds D. A.; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Trzaskowski M., Byrne E. M., Ripke S., Smith D. J., Sullivan P. F., Wray N. R., Breen G., Lewis C. M., McIntosh A. M., Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Trubetskoy V., Pardinas A. F., Qi T., Panagiotaropoulou G., Awasthi S., Bigdeli T. B., Bryois J., Chen C. Y., Dennison C. A., Hall L. S., Lam M., Watanabe K., Frei O., Ge T., Harwood J. C., Koopmans F., Magnusson S., Richards A. L., Sidorenko J., Wu Y., Zeng J., Grove J., Kim M., Li Z., Voloudakis G., Zhang W., Adams M., Agartz I., Atkinson E. G., Agerbo E., Al Eissa M., Albus M., Alexander M., Alizadeh B. Z., Alptekin K., Als T. D., Amin F., Arolt V., Arrojo M., Athanasiu L., Azevedo M. H., Bacanu S. A., Bass N. J., Begemann M., Belliveau R. A., Bene J., Benyamin B., Bergen S. E., Blasi G., Bobes J., Bonassi S., Braun A., Bressan R. A., Bromet E. J., Bruggeman R., Buckley P. F., Buckner R. L., Bybjerg-Grauholm J., Cahn W., Cairns M. J., Calkins M. E., Carr V. J., Castle D., Catts S. V., Chambert K. D., Chan R. C. K., Chaumette B., Cheng W., Cheung E. F. C., Chong S. A., Cohen D., Consoli A., Cordeiro Q., Costas J., Curtis C., Davidson M., Davis K. L., de Haan L., Degenhardt F., DeLisi L. E., Demontis D., Dickerson F., Dikeos D., Dinan T., Djurovic S., Duan J., Ducci G., Dudbridge F., Eriksson J. G., Fananas L., Faraone S. V., Fiorentino A., Forstner A., Frank J., Freimer N. B., Fromer M., Frustaci A., Gadelha A., Genovese G., Gershon E. S., Giannitelli M., Giegling I., Giusti-Rodriguez P., Godard S., Goldstein J. I., Penas J. G., Gonzalez-Pinto A., Gopal S., Gratten J., Green M. F., Greenwood T. A., Guillin O., Guloksuz S., Gur R. E., Gur R. C., Gutierrez B., Hahn E., Hakonarson H., Haroutunian V., Hartmann A. M., Harvey C., Hayward C., Henskens F. A., Herms S., Hoffmann P., Howrigan D. P., Ikeda M., Iyegbe C., Joa I., Julia A., Kahler A. K., Kam-Thong T., Kamatani Y., Karachanak-Yankova S., Kebir O., Keller M. C., Kelly B. J., Khrunin A., Kim S. W., Klovins J., Kondratiev N., Konte B., Kraft J., Kubo M., Kucinskas V., Kucinskiene Z. A., Kusumawardhani A., Kuzelova-Ptackova H., Landi S., Lazzeroni L. C., Lee P. H., Legge S. E., Lehrer D. S., Lencer R., Lerer B., Li M., Lieberman J., Light G. A., Limborska S., Liu C. M., Lonnqvist J., Loughland C. M., Lubinski J., Luykx J. J., Lynham A., Macek M. Jr., Mackinnon A., Magnusson P. K. E., Maher B. S., Maier W., Malaspina D., Mallet J., Marder S. R., Marsal S., Martin A. R., Martorell L., Mattheisen M., McCarley R. W., McDonald C., McGrath J. J., Medeiros H., Meier S., Melegh B., Melle I., Mesholam-Gately R. I., Metspalu A., Michie P. T., Milani L., Milanova V., Mitjans M., Molden E., Molina E., Molto M. D., Mondelli V., Moreno C., Morley C. P., Muntane G., Murphy K. C., Myin-Germeys I., Nenadic I., Nestadt G., Nikitina-Zake L., Noto C., Nuechterlein K. H., O’Brien N. L., O’Neill F. A., Oh S. Y., Olincy A., Ota V. K., Pantelis C., Papadimitriou G. N., Parellada M., Paunio T., Pellegrino R., Periyasamy S., Perkins D. O., Pfuhlmann B., Pietilainen O., Pimm J., Porteous D., Powell J., Quattrone D., Quested D., Radant A. D., Rampino A., Rapaport M. H., Rautanen A., Reichenberg A., Roe C., Roffman J. L., Roth J., Rothermundt M., Rutten B. P. F., Saker-Delye S., Salomaa V., Sanjuan J., Santoro M. L., Savitz A., Schall U., Scott R. J., Seidman L. J., Sharp S. I., Shi J., Siever L. J., Sigurdsson E., Sim K., Skarabis N., Slominsky P., So H. C., Sobell J. L., Soderman E., Stain H. J., Steen N. E., Steixner-Kumar A. A., Stogmann E., Stone W. S., Straub R. E., Streit F., Strengman E., Stroup T. S., Subramaniam M., Sugar C. A., Suvisaari J., Svrakic D. M., Swerdlow N. R., Szatkiewicz J. P., Ta T. M. T., Takahashi A., Terao C., Thibaut F., Toncheva D., Tooney P. A., Torretta S., Tosato S., Tura G. B., Turetsky B. I., Ucok A., Vaaler A., van Amelsvoort T., van Winkel R., Veijola J., Waddington J., Walter H., Waterreus A., Webb B. T., Weiser M., Williams N. M., Witt S. H., Wormley B. K., Wu J. Q., Xu Z., Yolken R., Zai C. C., Zhou W., Zhu F., Zimprich F., Atbasoglu E. C., Ayub M., Benner C., Bertolino A., Black D. W., Bray N. J., Breen G., Buccola N. G., Byerley W. F., Chen W. J., Cloninger C. R., Crespo-Facorro B., Donohoe G., Freedman R., Galletly C., Gandal M. J., Gennarelli M., Hougaard D. M., Hwu H. G., Jablensky A. V., McCarroll S. A., Moran J. L., Mors O., Mortensen P. B., Muller-Myhsok B., Neil A. L., Nordentoft M., Pato M. T., Petryshen T. L., Pirinen M., Pulver A. E., Schulze T. G., Silverman J. M., Smoller J. W., Stahl E. A., Tsuang D. W., Vilella E., Wang S. H., Xu S.; Indonesia Schizophrenia Consortium; PsychENCODE; Psychosis Endophenotypes International Consortium; SynGO Consortium, Adolfsson R., Arango C., Baune B. T., Belangero S. I., Borglum A. D., Braff D., Bramon E., Buxbaum J. D., Campion D., Cervilla J. A., Cichon S., Collier D. A., Corvin A., Curtis D., Forti M. D., Domenici E., Ehrenreich H., Escott-Price V., Esko T., Fanous A. H., Gareeva A., Gawlik M., Gejman P. V., Gill M., Glatt S. J., Golimbet V., Hong K. S., Hultman C. M., Hyman S. E., Iwata N., Jonsson E. G., Kahn R. S., Kennedy J. L., Khusnutdinova E., Kirov G., Knowles J. A., Krebs M. O., Laurent-Levinson C., Lee J., Lencz T., Levinson D. F., Li Q. S., Liu J., Malhotra A. K., Malhotra D., McIntosh A., McQuillin A., Menezes P. R., Morgan V. A., Morris D. W., Mowry B. J., Murray R. M., Nimgaonkar V., Nothen M. M., Ophoff R. A., Paciga S. A., Palotie A., Pato C. N., Qin S., Rietschel M., Riley B. P., Rivera M., Rujescu D., Saka M. C., Sanders A. R., Schwab S. G., Serretti A., Sham P. C., Shi Y., Clair D. S., Stefansson H., Stefansson K., Tsuang M. T., van Os J., Vawter M. P., Weinberger D. R., Werge T., Wildenauer D. B., Yu X., Yue W., Holmans P. A., Pocklington A. J., Roussos P., Vassos E., Verhage M., Visscher P. M., Yang J., Posthuma D., Andreassen O. A., Kendler K. S., Owen M. J., Wray N. R., Daly M. J., Huang H., Neale B. M., Sullivan P. F., Ripke S., Walters J. T. R., O’Donovan M. C.; Schizophrenia Working Group of the Psychiatric Genomics Consortium , Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stahl E. A., Breen G., Forstner A. J., McQuillin A., Ripke S., Trubetskoy V., Mattheisen M., Wang Y., Coleman J. R. I., Gaspar H. A., de Leeuw C. A., Steinberg S., Pavlides J. M. W., Trzaskowski M., Byrne E. M., Pers T. H., Holmans P. A., Richards A. L., Abbott L., Agerbo E., Akil H., Albani D., Alliey-Rodriguez N., Als T. D., Anjorin A., Antilla V., Awasthi S., Badner J. A., Baekvad-Hansen M., Barchas J. D., Bass N., Bauer M., Belliveau R., Bergen S. E., Pedersen C. B., Boen E., Boks M. P., Boocock J., Budde M., Bunney W., Burmeister M., Bybjerg-Grauholm J., Byerley W., Casas M., Cerrato F., Cervantes P., Chambert K., Charney A. W., Chen D., Churchhouse C., Clarke T. K., Coryell W., Craig D. W., Cruceanu C., Curtis D., Czerski P. M., Dale A. M., de Jong S., Degenhardt F., Del-Favero J., DePaulo J. R., Djurovic S., Dobbyn A. L., Dumont A., Elvsashagen T., Escott-Price V., Fan C. C., Fischer S. B., Flickinger M., Foroud T. M., Forty L., Frank J., Fraser C., Freimer N. B., Frisen L., Gade K., Gage D., Garnham J., Giambartolomei C., Pedersen M. G., Goldstein J., Gordon S. D., Gordon-Smith K., Green E. K., Green M. J., Greenwood T. A., Grove J., Guan W., Guzman-Parra J., Hamshere M. L., Hautzinger M., Heilbronner U., Herms S., Hipolito M., Hoffmann P., Holland D., Huckins L., Jamain S., Johnson J. S., Jureus A., Kandaswamy R., Karlsson R., Kennedy J. L., Kittel-Schneider S., Knowles J. A., Kogevinas M., Koller A. C., Kupka R., Lavebratt C., Lawrence J., Lawson W. B., Leber M., Lee P. H., Levy S. E., Li J. Z., Liu C., Lucae S., Maaser A., MacIntyre D. J., Mahon P. B., Maier W., Martinsson L., McCarroll S., McGuffin P., McInnis M. G., McKay J. D., Medeiros H., Medland S. E., Meng F., Milani L., Montgomery G. W., Morris D. W., Muhleisen T. W., Mullins N., Nguyen H., Nievergelt C. M., Adolfsson A. N., Nwulia E. A., O’Donovan C., Loohuis L. M. O., Ori A. P. S., Oruc L., Osby U., Perlis R. H., Perry A., Pfennig A., Potash J. B., Purcell S. M., Regeer E. J., Reif A., Reinbold C. S., Rice J. P., Rivas F., Rivera M., Roussos P., Ruderfer D. M., Ryu E., Sanchez-Mora C., Schatzberg A. F., Scheftner W. A., Schork N. J., Weickert C. S., Shehktman T., Shilling P. D., Sigurdsson E., Slaney C., Smeland O. B., Sobell J. L., Hansen C. S., Spijker A. T., Clair D. S., Steffens M., Strauss J. S., Streit F., Strohmaier J., Szelinger S., Thompson R. C., Thorgeirsson T. E., Treutlein J., Vedder H., Wang W., Watson S. J., Weickert T. W., Witt S. H., Xi S., Xu W., Young A. H., Zandi P., Zhang P., Zollner S.; eQTLGen Consortium; BIOS Consortium, Adolfsson R., Agartz I., Alda M., Backlund L., Baune B. T., Bellivier F., Berrettini W. H., Biernacka J. M., Blackwood D. H. R., Boehnke M., Borglum A. D., Corvin A., Craddock N., Daly M. J., Dannlowski U., Esko T., Etain B., Frye M., Fullerton J. M., Gershon E. S., Gill M., Goes F., Grigoroiu-Serbanescu M., Hauser J., Hougaard D. M., Hultman C. M., Jones I., Jones L. A., Kahn R. S., Kirov G., Landen M., Leboyer M., Lewis C. M., Li Q. S., Lissowska J., Martin N. G., Mayoral F., McElroy S. L., McIntosh A. M., McMahon F. J., Melle I., Metspalu A., Mitchell P. B., Morken G., Mors O., Mortensen P. B., Muller-Myhsok B., Myers R. M., Neale B. M., Nimgaonkar V., Nordentoft M., Nothen M. M., O’Donovan M. C., Oedegaard K. J., Owen M. J., Paciga S. A., Pato C., Pato M. T., Posthuma D., Ramos-Quiroga J. A., Ribases M., Rietschel M., Rouleau G. A., Schalling M., Schofield P. R., Schulze T. G., Serretti A., Smoller J. W., Stefansson H., Stefansson K., Stordal E., Sullivan P. F., Turecki G., Vaaler A. E., Vieta E., Vincent J. B., Werge T., Nurnberger J. I., Wray N. R., Di Florio A., Edenberg H. J., Cichon S., Ophoff R. A., Scott L. J., Andreassen O. A., Kelsoe J., Sklar P.; the Bipolar Disorder Working Group of the Psychiatric Genomics Consortium , Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bellenguez C., Kucukali F., Jansen I. E., Kleineidam L., Moreno-Grau S., Amin N., Naj A. C., Campos-Martin R., Grenier-Boley B., Andrade V., Holmans P. A., Boland A., Damotte V., van der Lee S. J., Costa M. R., Kuulasmaa T., Yang Q., de Rojas I., Bis J. C., Yaqub A., Prokic I., Chapuis J., Ahmad S., Giedraitis V., Aarsland D., Garcia-Gonzalez P., Abdelnour C., Alarcon-Martin E., Alcolea D., Alegret M., Alvarez I., Alvarez V., Armstrong N. J., Tsolaki A., Antunez C., Appollonio I., Arcaro M., Archetti S., Pastor A. A., Arosio B., Athanasiu L., Bailly H., Banaj N., Baquero M., Barral S., Beiser A., Pastor A. B., Below J. E., Benchek P., Benussi L., Berr C., Besse C., Bessi V., Binetti G., Bizarro A., Blesa R., Boada M., Boerwinkle E., Borroni B., Boschi S., Bossu P., Brathen G., Bressler J., Bresner C., Brodaty H., Brookes K. J., Brusco L. I., Buiza-Rueda D., Burger K., Burholt V., Bush W. S., Calero M., Cantwell L. B., Chene G., Chung J., Cuccaro M. L., Carracedo A., Cecchetti R., Cervera-Carles L., Charbonnier C., Chen H. H., Chillotti C., Ciccone S., Claassen J., Clark C., Conti E., Corma-Gomez A., Costantini E., Custodero C., Daian D., Dalmasso M. C., Daniele A., Dardiotis E., Dartigues J. F., de Deyn P. P., de Paiva Lopes K., de Witte L. D., Debette S., Deckert J., Del Ser T., Denning N., DeStefano A., Dichgans M., Diehl-Schmid J., Diez-Fairen M., Rossi P. D., Djurovic S., Duron E., Duzel E., Dufouil C., Eiriksdottir G., Engelborghs S., Escott-Price V., Espinosa A., Ewers M., Faber K. M., Fabrizio T., Nielsen S. F., Fardo D. W., Farotti L., Fenoglio C., Fernandez-Fuertes M., Ferrari R., Ferreira C. B., Ferri E., Fin B., Fischer P., Fladby T., Fliessbach K., Fongang B., Fornage M., Fortea J., Foroud T. M., Fostinelli S., Fox N. C., Franco-Macias E., Bullido M. J., Frank-Garcia A., Froelich L., Fulton-Howard B., Galimberti D., Garcia-Alberca J. M., Garcia-Gonzalez P., Garcia-Madrona S., Garcia-Ribas G., Ghidoni R., Giegling I., Giorgio G., Goate A. M., Goldhardt O., Gomez-Fonseca D., Gonzalez-Perez A., Graff C., Grande G., Green E., Grimmer T., Grunblatt E., Grunin M., Gudnason V., Guetta-Baranes T., Haapasalo A., Hadjigeorgiou G., Haines J. L., Hamilton-Nelson K. L., Hampel H., Hanon O., Hardy J., Hartmann A. M., Hausner L., Harwood J., Heilmann-Heimbach S., Helisalmi S., Heneka M. T., Hernandez I., Herrmann M. J., Hoffmann P., Holmes C., Holstege H., Vilas R. H., Hulsman M., Humphrey J., Biessels G. J., Jian X., Johansson C., Jun G. R., Kastumata Y., Kauwe J., Kehoe P. G., Kilander L., Stahlbom A. K., Kivipelto M., Koivisto A., Kornhuber J., Kosmidis M. H., Kukull W. A., Kuksa P. P., Kunkle B. W., Kuzma A. B., Lage C., Laukka E. J., Launer L., Lauria A., Lee C. Y., Lehtisalo J., Lerch O., Lleo A., Longstreth W. Jr., Lopez O., de Munain A. L., Love S., Lowemark M., Luckcuck L., Lunetta K. L., Ma Y., Macias J., MacLeod C. A., Maier W., Mangialasche F., Spallazzi M., Marquie M., Marshall R., Martin E. R., Montes A. M., Rodriguez C. M., Masullo C., Mayeux R., Mead S., Mecocci P., Medina M., Meggy A., Mehrabian S., Mendoza S., Menendez-Gonzalez M., Mir P., Moebus S., Mol M., Molina-Porcel L., Montrreal L., Morelli L., Moreno F., Morgan K., Mosley T., Nothen M. M., Muchnik C., Mukherjee S., Nacmias B., Ngandu T., Nicolas G., Nordestgaard B. G., Olaso R., Orellana A., Orsini M., Ortega G., Padovani A., Paolo C., Papenberg G., Parnetti L., Pasquier F., Pastor P., Peloso G., Perez-Cordon A., Perez-Tur J., Pericard P., Peters O., Pijnenburg Y. A. L., Pineda J. A., Pinol-Ripoll G., Pisanu C., Polak T., Popp J., Posthuma D., Priller J., Puerta R., Quenez O., Quintela I., Thomassen J. Q., Rabano A., Rainero I., Rajabli F., Ramakers I., Real L. M., Reinders M. J. T., Reitz C., Reyes-Dumeyer D., Ridge P., Riedel-Heller S., Riederer P., Roberto N., Rodriguez-Rodriguez E., Rongve A., Allende I. R., Rosende-Roca M., Royo J. L., Rubino E., Rujescu D., Saez M. E., Sakka P., Saltvedt I., Sanabria A., Sanchez-Arjona M. B., Sanchez-Garcia F., Juan P. S., Sanchez-Valle R., Sando S. B., Sarnowski C., Satizabal C. L., Scamosci M., Scarmeas N., Scarpini E., Scheltens P., Scherbaum N., Scherer M., Schmid M., Schneider A., Schott J. M., Selbaek G., Seripa D., Serrano M., Sha J., Shadrin A. A., Skrobot O., Slifer S., Snijders G. J. L., Soininen H., Solfrizzi V., Solomon A., Song Y., Sorbi S., Sotolongo-Grau O., Spalletta G., Spottke A., Squassina A., Stordal E., Tartan J. P., Tarraga L., Tesi N., Thalamuthu A., Thomas T., Tosto G., Traykov L., Tremolizzo L., Tybjaerg-Hansen A., Uitterlinden A., Ullgren A., Ulstein I., Valero S., Valladares O., Broeckhoven C. V., Vance J., Vardarajan B. N., van der Lugt A., Dongen J. V., van Rooij J., van Swieten J., Vandenberghe R., Verhey F., Vidal J. S., Vogelgsang J., Vyhnalek M., Wagner M., Wallon D., Wang L. S., Wang R., Weinhold L., Wiltfang J., Windle G., Woods B., Yannakoulia M., Zare H., Zhao Y., Zhang X., Zhu C., Zulaica M.; EADB; GR@ACE; DEGESCO; EADI; GERAD; Demgene; FinnGen; ADGC; CHARGE, Farrer L. A., Psaty B. M., Ghanbari M., Raj T., Sachdev P., Mather K., Jessen F., Ikram M. A., de Mendonca A., Hort J., Tsolaki M., Pericak-Vance M. A., Amouyel P., Williams J., Frikke-Schmidt R., Clarimon J., Deleuze J. F., Rossi G., Seshadri S., Andreassen O. A., Ingelsson M., Hiltunen M., Sleegers K., Schellenberg G. D., van Duijn C. M., Sims R., van der Flier W. M., Ruiz A., Ramirez A., Lambert J. C., New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ortuno-Sierra J., Fonseca-Pedrero E., Paino M., i Riba S. S., Muniz J., Screening mental health problems during adolescence: Psychometric properties of the Spanish version of the strengths and difficulties questionnaire. J. Adolesc. 38, 49–56 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Ni K., Huang Z., Zhu Y., Xue D., Jin Q., Zhang C., Gu C., The lncRNA ADAMTS9-AS2 regulates RPL22 to modulate TNBC progression via controlling the TGF-β signaling pathway. Front. Oncol. 11, 654472 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martin S., Cule M., Basty N., Tyrrell J., Beaumont R. N., Wood A. R., Frayling T. M., Sorokin E., Whitcher B., Liu Y., Bell J. D., Thomas E. L., Yaghootkar H., Genetic evidence for different adiposity phenotypes and their opposing influences on ectopic fat and risk of cardiometabolic disease. Diabetes 70, 1843–1856 (2021). [DOI] [PubMed] [Google Scholar]
- 126.van der Meer D., Shadrin A. A., O’Connell K., Bettella F., Djurovic S., Wolfers T., Alnaes D., Agartz I., Smeland O. B., Melle I., Sanchez J. M., Linden D. E. J., Dale A. M., Westlye L. T., Andreassen O. A., Frei O., Kaufmann T., Boosting schizophrenia genetics by utilizing genetic overlap with brain morphology. Biol. Psychiatry 92, 291–298 (2022). [DOI] [PubMed] [Google Scholar]
- 127.Kichaev G., Bhatia G., Loh P. R., Gazal S., Burch K., Freund M. K., Schoech A., Pasaniuc B., Price A. L., Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hindley G., Shadrin A. A., van der Meer D., Parker N., Cheng W., O’Connell K. S., Bahrami S., Lin A., Karadag N., Holen B., Bjella T., Deary I. J., Davies G., Hill W. D., Bressler J., Seshadri S., Fan C. C., Ueland T., Djurovic S., Smeland O. B., Frei O., Dale A. M., Andreassen O. A., Multivariate genetic analysis of personality and cognitive traits reveals abundant pleiotropy. Nat. Hum. Behav. 7, 1584–1600 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith A. R., Smith R. G., Burrage J., Troakes C., Al-Sarraj S., Kalaria R. N., Sloan C., Robinson A. C., Mill J., Lunnon K., A cross-brain regions study of ANK1 DNA methylation in different neurodegenerative diseases. Neurobiol. Aging 74, 70–76 (2019). [DOI] [PubMed] [Google Scholar]
- 130.Vujkovic M., Keaton J. M., Lynch J. A., Miller D. R., Zhou J., Tcheandjieu C., Huffman J. E., Assimes T. L., Lorenz K., Zhu X., Hilliard A. T., Judy R. L., Huang J., Lee K. M., Klarin D., Pyarajan S., Danesh J., Melander O., Rasheed A., Mallick N. H., Hameed S., Qureshi I. H., Afzal M. N., Malik U., Jalal A., Abbas S., Sheng X., Gao L., Kaestner K. H., Susztak K., Sun Y. V., DuVall S. L., Cho K., Lee J. S., Gaziano J. M., Phillips L. S., Meigs J. B., Reaven P. D., Wilson P. W., Edwards T. L., Rader D. J., Damrauer S. M., O’Donnell C. J., Tsao P. S.; The HPAP Consortium; Regeneron Genetics Center; VA Million Veteran Program, Chang K. M., Voight B. F., Saleheen D., Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 52, 680–691 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen M. H., Raffield L. M., Mousas A., Sakaue S., Huffman J. E., Moscati A., Trivedi B., Jiang T., Akbari P., Vuckovic D., Bao E. L., Zhong X., Manansala R., Laplante V., Chen M., Lo K. S., Qian H., Lareau C. A., Beaudoin M., Hunt K. A., Akiyama M., Bartz T. M., Ben-Shlomo Y., Beswick A., Bork-Jensen J., Bottinger E. P., Brody J. A., van Rooij F. J. A., Chitrala K., Cho K., Choquet H., Correa A., Danesh J., Di Angelantonio E., Dimou N., Ding J., Elliott P., Esko T., Evans M. K., Floyd J. S., Broer L., Grarup N., Guo M. H., Greinacher A., Haessler J., Hansen T., Howson J. M. M., Huang Q. Q., Huang W., Jorgenson E., Kacprowski T., Kahonen M., Kamatani Y., Kanai M., Karthikeyan S., Koskeridis F., Lange L. A., Lehtimaki T., Lerch M. M., Linneberg A., Liu Y., Lyytikainen L. P., Manichaikul A., Martin H. C., Matsuda K., Mohlke K. L., Mononen N., Murakami Y., Nadkarni G. N., Nauck M., Nikus K., Ouwehand W. H., Pankratz N., Pedersen O., Preuss M., Psaty B. M., Raitakari O. T., Roberts D. J., Rich S. S., Rodriguez B. A. T., Rosen J. D., Rotter J. I., Schubert P., Spracklen C. N., Surendran P., Tang H., Tardif J. C., Trembath R. C., Ghanbari M., Volker U., Volzke H., Watkins N. A., Zonderman A. B., Program V. A. M. V., Wilson P. W. F., Li Y., Butterworth A. S., Gauchat J. F., Chiang C. W. K., Li B., Loos R. J. F., Astle W. J., Evangelou E., van Heel D. A., Sankaran V. G., Okada Y., Soranzo N., Johnson A. D., Reiner A. P., Auer P. L., Lettre G., Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Christakoudi S., Evangelou E., Riboli E., Tsilidis K. K., GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci. Rep. 11, 10688 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Koskeridis F., Evangelou E., Said S., Boyle J. J., Elliott P., Dehghan A., Tzoulaki I., Pleiotropic genetic architecture and novel loci for C-reactive protein levels. Nat. Commun. 13, 6939 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luciano M., Hagenaars S. P., Davies G., Hill W. D., Clarke T. K., Shirali M., Harris S. E., Marioni R. E., Liewald D. C., Fawns-Ritchie C., Adams M. J., Howard D. M., Lewis C. M., Gale C. R., McIntosh A. M., Deary I. J., Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat. Genet. 50, 6–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Okabe T., Nakamura T., Nishimura Y. N., Kohu K., Ohwada S., Morishita Y., Akiyama T., RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-D-aspartate receptor signaling. J. Biol. Chem. 278, 9920–9927 (2003). [DOI] [PubMed] [Google Scholar]
- 136.Yengo L., Vedantam S., Marouli E., Sidorenko J., Bartell E., Sakaue S., Graff M., Eliasen A. U., Jiang Y., Raghavan S., Miao J., Arias J. D., Graham S. E., Mukamel R. E., Spracklen C. N., Yin X., Chen S. H., Ferreira T., Highland H. H., Ji Y., Karaderi T., Lin K., Lull K., Malden D. E., Medina-Gomez C., Machado M., Moore A., Rueger S., Sim X., Vrieze S., Ahluwalia T. S., Akiyama M., Allison M. A., Alvarez M., Andersen M. K., Ani A., Appadurai V., Arbeeva L., Bhaskar S., Bielak L. F., Bollepalli S., Bonnycastle L. L., Bork-Jensen J., Bradfield J. P., Bradford Y., Braund P. S., Brody J. A., Burgdorf K. S., Cade B. E., Cai H., Cai Q., Campbell A., Canadas-Garre M., Catamo E., Chai J. F., Chai X., Chang L. C., Chang Y. C., Chen C. H., Chesi A., Choi S. H., Chung R. H., Cocca M., Concas M. P., Couture C., Cuellar-Partida G., Danning R., Daw E. W., Degenhard F., Delgado G. E., Delitala A., Demirkan A., Deng X., Devineni P., Dietl A., Dimitriou M., Dimitrov L., Dorajoo R., Ekici A. B., Engmann J. E., Fairhurst-Hunter Z., Farmaki A. E., Faul J. D., Fernandez-Lopez J. C., Forer L., Francescatto M., Freitag-Wolf S., Fuchsberger C., Galesloot T. E., Gao Y., Gao Z., Geller F., Giannakopoulou O., Giulianini F., Gjesing A. P., Goel A., Gordon S. D., Gorski M., Grove J., Guo X., Gustafsson S., Haessler J., Hansen T. F., Havulinna A. S., Haworth S. J., He J., Heard-Costa N., Hebbar P., Hindy G., Ho Y. A., Hofer E., Holliday E., Horn K., Hornsby W. E., Hottenga J. J., Huang H., Huang J., Huerta-Chagoya A., Huffman J. E., Hung Y. J., Huo S., Hwang M. Y., Iha H., Ikeda D. D., Isono M., Jackson A. U., Jager S., Jansen I. E., Johansson I., Jonas J. B., Jonsson A., Jorgensen T., Kalafati I. P., Kanai M., Kanoni S., Karhus L. L., Kasturiratne A., Katsuya T., Kawaguchi T., Kember R. L., Kentistou K. A., Kim H. N., Kim Y. J., Kleber M. E., Knol M. J., Kurbasic A., Lauzon M., Le P., Lea R., Lee J. Y., Leonard H. L., Li S. A., Li X., Li X., Liang J., Lin H., Lin S. Y., Liu J., Liu X., Lo K. S., Long J., Lores-Motta L., Luan J., Lyssenko V., Lyytikainen L. P., Mahajan A., Mamakou V., Mangino M., Manichaikul A., Marten J., Mattheisen M., Mavarani L., McDaid A. F., Meidtner K., Melendez T. L., Mercader J. M., Milaneschi Y., Miller J. E., Millwood I. Y., Mishra P. P., Mitchell R. E., Mollehave L. T., Morgan A., Mucha S., Munz M., Nakatochi M., Nelson C. P., Nethander M., Nho C. W., Nielsen A. A., Nolte I. M., Nongmaithem S. S., Noordam R., Ntalla I., Nutile T., Pandit A., Christofidou P., Parna K., Pauper M., Petersen E. R. B., Petersen L. V., Pitkanen N., Polasek O., Poveda A., Preuss M. H., Pyarajan S., Raffield L. M., Rakugi H., Ramirez J., Rasheed A., Raven D., Rayner N. W., Riveros C., Rohde R., Ruggiero D., Ruotsalainen S. E., Ryan K. A., Sabater-Lleal M., Saxena R., Scholz M., Sendamarai A., Shen B., Shi J., Shin J. H., Sidore C., Sitlani C. M., Slieker R. C., Smit R. A. J., Smith A. V., Smith J. A., Smyth L. J., Southam L., Steinthorsdottir V., Sun L., Takeuchi F., Tallapragada D. S. P., Taylor K. D., Tayo B. O., Tcheandjieu C., Terzikhan N., Tesolin P., Teumer A., Theusch E., Thompson D. J., Thorleifsson G., Timmers P., Trompet S., Turman C., Vaccargiu S., van der Laan S. W., van der Most P. J., van Klinken J. B., van Setten J., Verma S. S., Verweij N., Veturi Y., Wang C. A., Wang C., Wang L., Wang Z., Warren H. R., Wei W. B., Wickremasinghe A. R., Wielscher M., Wiggins K. L., Winsvold B. S., Wong A., Wu Y., Wuttke M., Xia R., Xie T., Yamamoto K., Yang J., Yao J., Young H., Yousri N. A., Yu L., Zeng L., Zhang W., Zhang X., Zhao J. H., Zhao W., Zhou W., Zimmermann M. E., Zoledziewska M., Adair L. S., Adams H. H. H., Aguilar-Salinas C. A., Al-Mulla F., Arnett D. K., Asselbergs F. W., Asvold B. O., Attia J., Banas B., Bandinelli S., Bennett D. A., Bergler T., Bharadwaj D., Biino G., Bisgaard H., Boerwinkle E., Boger C. A., Bonnelykke K., Boomsma D. I., Borglum A. D., Borja J. B., Bouchard C., Bowden D. W., Brandslund I., Brumpton B., Buring J. E., Caulfield M. J., Chambers J. C., Chandak G. R., Chanock S. J., Chaturvedi N., Chen Y. I., Chen Z., Cheng C. Y., Christophersen I. E., Ciullo M., Cole J. W., Collins F. S., Cooper R. S., Cruz M., Cucca F., Cupples L. A., Cutler M. J., Damrauer S. M., Dantoft T. M., de Borst G. J., de Groot L., De Jager P. L., de Kleijn D. P. V., de Silva H. J., Dedoussis G. V., den Hollander A. I., Du S., Easton D. F., Elders P. J. M., Eliassen A. H., Ellinor P. T., Elmstahl S., Erdmann J., Evans M. K., Fatkin D., Feenstra B., Feitosa M. F., Ferrucci L., Ford I., Fornage M., Franke A., Franks P. W., Freedman B. I., Gasparini P., Gieger C., Girotto G., Goddard M. E., Golightly Y. M., Gonzalez-Villalpando C., Gordon-Larsen P., Grallert H., Grant S. F. A., Grarup N., Griffiths L., Gudnason V., Haiman C., Hakonarson H., Hansen T., Hartman C. A., Hattersley A. T., Hayward C., Heckbert S. R., Heng C. K., Hengstenberg C., Hewitt A. W., Hishigaki H., Hoyng C. B., Huang P. L., Huang W., Hunt S. C., Hveem K., Hypponen E., Iacono W. G., Ichihara S., Ikram M. A., Isasi C. R., Jackson R. D., Jarvelin M. R., Jin Z. B., Jockel K. H., Joshi P. K., Jousilahti P., Jukema J. W., Kahonen M., Kamatani Y., Kang K. D., Kaprio J., Kardia S. L. R., Karpe F., Kato N., Kee F., Kessler T., Khera A. V., Khor C. C., Kiemeney L., Kim B. J., Kim E. K., Kim H. L., Kirchhof P., Kivimaki M., Koh W. P., Koistinen H. A., Kolovou G. D., Kooner J. S., Kooperberg C., Kottgen A., Kovacs P., Kraaijeveld A., Kraft P., Krauss R. M., Kumari M., Kutalik Z., Laakso M., Lange L. A., Langenberg C., Launer L. J., Le Marchand L., Lee H., Lee N. R., Lehtimaki T., Li H., Li L., Lieb W., Lin X., Lind L., Linneberg A., Liu C. T., Liu J., Loeffler M., London B., Lubitz S. A., Lye S. J., Mackey D. A., Magi R., Magnusson P. K. E., Marcus G. M., Vidal P. M., Martin N. G., Marz W., Matsuda F., McGarrah R. W., McGue M., McKnight A. J., Medland S. E., Mellstrom D., Metspalu A., Mitchell B. D., Mitchell P., Mook-Kanamori D. O., Morris A. D., Mucci L. A., Munroe P. B., Nalls M. A., Nazarian S., Nelson A. E., Neville M. J., Newton-Cheh C., Nielsen C. S., Nothen M. M., Ohlsson C., Oldehinkel A. J., Orozco L., Pahkala K., Pajukanta P., Palmer C. N. A., Parra E. J., Pattaro C., Pedersen O., Pennell C. E., Penninx B., Perusse L., Peters A., Peyser P. A., Porteous D. J., Posthuma D., Power C., Pramstaller P. P., Province M. A., Qi Q., Qu J., Rader D. J., Raitakari O. T., Ralhan S., Rallidis L. S., Rao D. C., Redline S., Reilly D. F., Reiner A. P., Rhee S. Y., Ridker P. M., Rienstra M., Ripatti S., Ritchie M. D., Roden D. M., Rosendaal F. R., Rotter J. I., Rudan I., Rutters F., Sabanayagam C., Saleheen D., Salomaa V., Samani N. J., Sanghera D. K., Sattar N., Schmidt B., Schmidt H., Schmidt R., Schulze M. B., Schunkert H., Scott L. J., Scott R. J., Sever P., Shiroma E. J., Shoemaker M. B., Shu X. O., Simonsick E. M., Sims M., Singh J. R., Singleton A. B., Sinner M. F., Smith J. G., Snieder H., Spector T. D., Stampfer M. J., Stark K. J., Strachan D. P., Hart L. M., Tabara Y., Tang H., Tardif J. C., Thanaraj T. A., Timpson N. J., Tonjes A., Tremblay A., Tuomi T., Tuomilehto J., Tusie-Luna M. T., Uitterlinden A. G., van Dam R. M., van der Harst P., Van der Velde N., van Duijn C. M., van Schoor N. M., Vitart V., Volker U., Vollenweider P., Volzke H., Wacher-Rodarte N. H., Walker M., Wang Y. X., Wareham N. J., Watanabe R. M., Watkins H., Weir D. R., Werge T. M., Widen E., Wilkens L. R., Willemsen G., Willett W. C., Wilson J. F., Wong T. Y., Woo J. T., Wright A. F., Wu J. Y., Xu H., Yajnik C. S., Yokota M., Yuan J. M., Zeggini E., Zemel B. S., Zheng W., Zhu X., Zmuda J. M., Zonderman A. B., Zwart J. A.; 23andMe Research Team; VA Million Veteran Program; DiscovEHR (DiscovEHR and MyCode Community Health Initiative); eMERGE (Electronic Medical Records and Genomics Network); Lifelines Cohort Study; The PRACTICAL Consortium; Understanding Society Scientific Group, Chasman D. I., Cho Y. S., Heid I. M., McCarthy M. I., Ng M. C. Y., O’Donnell C. J., Rivadeneira F., Thorsteinsdottir U., Sun Y. V., Tai E. S., Boehnke M., Deloukas P., Justice A. E., Lindgren C. M., Loos R. J. F., Mohlke K. L., North K. E., Stefansson K., Walters R. G., Winkler T. W., Young K. L., Loh P. R., Yang J., Esko T., Assimes T. L., Auton A., Abecasis G. R., Willer C. J., Locke A. E., Berndt S. I., Lettre G., Frayling T. M., Okada Y., Wood A. R., Visscher P. M., Hirschhorn J. N., A saturated map of common genetic variants associated with human height. Nature 610, 704–712 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Uhl G. R., Martinez M. J., PTPRD: Neurobiology, genetics, and initial pharmacology of a pleiotropic contributor to brain phenotypes. Ann. N. Y. Acad. Sci. 1451, 112–129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rajakulendran S., Kaski D., Hanna M. G., Neuronal P/Q-type calcium channel dysfunction in inherited disorders of the CNS. Nat. Rev. Neurol. 8, 86–96 (2012). [DOI] [PubMed] [Google Scholar]
- 139.Naqvi S., Sleyp Y., Hoskens H., Indencleef K., Spence J. P., Bruffaerts R., Radwan A., Eller R. J., Richmond S., Shriver M. D., Shaffer J. R., Weinberg S. M., Walsh S., Thompson J., Pritchard J. K., Sunaert S., Peeters H., Wysocka J., Claes P., Shared heritability of human face and brain shape. Nat. Genet. 53, 830–839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao B., Luo T., Li T., Li Y., Zhang J., Shan Y., Wang X., Yang L., Zhou F., Zhu Z.; Alzheimer’s Disease Neuroimaging Initiative; Pediatric Imaging, Neurocognition and Genetics, Zhu H., Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co- architecture with cognitive and mental health traits. Nat. Genet. 51, 1637–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lauc G., Huffman J. E., Pucic M., Zgaga L., Adamczyk B., Muzinic A., Novokmet M., Polasek O., Gornik O., Kristic J., Keser T., Vitart V., Scheijen B., Uh H. W., Molokhia M., Patrick A. L., McKeigue P., Kolcic I., Lukic I. K., Swann O., van Leeuwen F. N., Ruhaak L. R., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J., Deelder A. M., Zeng Q., Wang W., Hastie N. D., Gyllensten U., Wilson J. F., Wuhrer M., Wright A. F., Rudd P. M., Hayward C., Aulchenko Y., Campbell H., Rudan I., Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLOS Genet. 9, e1003225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang J., Huffman J. E., Huang Y., Do Valle I., Assimes T. L., Raghavan S., Voight B. F., Liu C., Barabasi A. L., Huang R. D. L., Hui Q., Nguyen X. T., Ho Y. L., Djousse L., Lynch J. A., Vujkovic M., Tcheandjieu C., Tang H., Damrauer S. M., Reaven P. D., Miller D., Phillips L. S., Ng M. C. Y., Graff M., Haiman C. A., Loos R. J. F., North K. E., Yengo L., Smith G. D., Saleheen D., Gaziano J. M., Rader D. J., Tsao P. S., Cho K., Chang K. M., Wilson P. W. F., Program V. A. M. V., Sun Y. V., O’Donnell C. J., Genomics and phenomics of body mass index reveals a complex disease network. Nat. Commun. 13, 7973 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grasby K. L., Jahanshad N., Painter J. N., Colodro-Conde L., Bralten J., Hibar D. P., Lind P. A., Pizzagalli F., Ching C. R. K., McMahon M. A. B., Shatokhina N., Zsembik L. C. P., Thomopoulos S. I., Zhu A. H., Strike L. T., Agartz I., Alhusaini S., Almeida M. A. A., Alnaes D., Amlien I. K., Andersson M., Ard T., Armstrong N. J., Ashley-Koch A., Atkins J. R., Bernard M., Brouwer R. M., Buimer E. E. L., Bulow R., Burger C., Cannon D. M., Chakravarty M., Chen Q., Cheung J. W., Couvy-Duchesne B., Dale A. M., Dalvie S., de Araujo T. K., de Zubicaray G. I., de Zwarte S. M. C., den Braber A., Doan N. T., Dohm K., Ehrlich S., Engelbrecht H. R., Erk S., Fan C. C., Fedko I. O., Foley S. F., Ford J. M., Fukunaga M., Garrett M. E., Ge T., Giddaluru S., Goldman A. L., Green M. J., Groenewold N. A., Grotegerd D., Gurholt T. P., Gutman B. A., Hansell N. K., Harris M. A., Harrison M. B., Haswell C. C., Hauser M., Herms S., Heslenfeld D. J., Ho N. F., Hoehn D., Hoffmann P., Holleran L., Hoogman M., Hottenga J. J., Ikeda M., Janowitz D., Jansen I. E., Jia T., Jockwitz C., Kanai R., Karama S., Kasperaviciute D., Kaufmann T., Kelly S., Kikuchi M., Klein M., Knapp M., Knodt A. R., Kramer B., Lam M., Lancaster T. M., Lee P. H., Lett T. A., Lewis L. B., Lopes-Cendes I., Luciano M., Macciardi F., Marquand A. F., Mathias S. R., Melzer T. R., Milaneschi Y., Mirza-Schreiber N., Moreira J. C. V., Muhleisen T. W., Muller-Myhsok B., Najt P., Nakahara S., Nho K., Loohuis L. M. O., Orfanos D. P., Pearson J. F., Pitcher T. L., Putz B., Quide Y., Ragothaman A., Rashid F. M., Reay W. R., Redlich R., Reinbold C. S., Repple J., Richard G., Riedel B. C., Risacher S. L., Rocha C. S., Mota N. R., Salminen L., Saremi A., Saykin A. J., Schlag F., Schmaal L., Schofield P. R., Secolin R., Shapland C. Y., Shen L., Shin J., Shumskaya E., Sonderby I. E., Sprooten E., Tansey K. E., Teumer A., Thalamuthu A., Tordesillas-Gutierrez D., Turner J. A., Uhlmann A., Vallerga C. L., van der Meer D., van Donkelaar M. M. J., van Eijk L., van Erp T. G. M., van Haren N. E. M., van Rooij D., van Tol M. J., Veldink J. H., Verhoef E., Walton E., Wang M., Wang Y., Wardlaw J. M., Wen W., Westlye L. T., Whelan C. D., Witt S. H., Wittfeld K., Wolf C., Wolfers T., Wu J. Q., Yasuda C. L., Zaremba D., Zhang Z., Zwiers M. P., Artiges E., Assareh A. A., Ayesa-Arriola R., Belger A., Brandt C. L., Brown G. G., Cichon S., Curran J. E., Davies G. E., Degenhardt F., Dennis M. F., Dietsche B., Djurovic S., Doherty C. P., Espiritu R., Garijo D., Gil Y., Gowland P. A., Green R. C., Hausler A. N., Heindel W., Ho B. C., Hoffmann W. U., Holsboer F., Homuth G., Hosten N., Jack C. R. Jr., Jang M., Jansen A., Kimbrel N. A., Kolskar K., Koops S., Krug A., Lim K. O., Luykx J. J., Mathalon D. H., Mather K. A., Mattay V. S., Matthews S., Van Son J. M., McEwen S. C., Melle I., Morris D. W., Mueller B. A., Nauck M., Nordvik J. E., Nothen M. M., O’Leary D. S., Opel N., Martinot M. P., Pike G. B., Preda A., Quinlan E. B., Rasser P. E., Ratnakar V., Reppermund S., Steen V. M., Tooney P. A., Torres F. R., Veltman D. J., Voyvodic J. T., Whelan R., White T., Yamamori H., Adams H. H. H., Bis J. C., Debette S., Decarli C., Fornage M., Gudnason V., Hofer E., Ikram M. A., Launer L., Longstreth W. T., Lopez O. L., Mazoyer B., Mosley T. H., Roshchupkin G. V., Satizabal C. L., Schmidt R., Seshadri S., Yang Q.; Alzheimer’s Disease Neuroimaging Initiative; CHARGE Consortium; EPIGEN Consortium; IMAGEN Consortium; SYS Consortium; Parkinson’s Progression Markers Initiative, Alvim M. K. M., Ames D., Anderson T. J., Andreassen O. A., Arias-Vasquez A., Bastin M. E., Baune B. T., Beckham J. C., Blangero J., Boomsma D. I., Brodaty H., Brunner H. G., Buckner R. L., Buitelaar J. K., Bustillo J. R., Cahn W., Cairns M. J., Calhoun V., Carr V. J., Caseras X., Caspers S., Cavalleri G. L., Cendes F., Corvin A., Crespo-Facorro B., Dalrymple-Alford J. C., Dannlowski U., de Geus E. J. C., Deary I. J., Delanty N., Depondt C., Desrivieres S., Donohoe G., Espeseth T., Fernandez G., Fisher S. E., Flor H., Forstner A. J., Francks C., Franke B., Glahn D. C., Gollub R. L., Grabe H. J., Gruber O., Haberg A. K., Hariri A. R., Hartman C. A., Hashimoto R., Heinz A., Henskens F. A., Hillegers M. H. J., Hoekstra P. J., Holmes A. J., Hong L. E., Hopkins W. D., Pol H. E. H., Jernigan T. L., Jonsson E. G., Kahn R. S., Kennedy M. A., Kircher T. T. J., Kochunov P., Kwok J. B. J., Le Hellard S., Loughland C. M., Martin N. G., Martinot J. L., McDonald C., McMahon K. L., Meyer-Lindenberg A., Michie P. T., Morey R. A., Mowry B., Nyberg L., Oosterlaan J., Ophoff R. A., Pantelis C., Paus T., Pausova Z., Penninx B., Polderman T. J. C., Posthuma D., Rietschel M., Roffman J. L., Rowland L. M., Sachdev P. S., Samann P. G., Schall U., Schumann G., Scott R. J., Sim K., Sisodiya S. M., Smoller J. W., Sommer I. E., Pourcain B. S., Stein D. J., Toga A. W., Trollor J. N., Van der Wee N. J. A., van’t Ent D., Volzke H., Walter H., Weber B., Weinberger D. R., Wright M. J., Zhou J., Stein J. L., Thompson P. M., Medland S. E.; Enhancing NeuroImaging Genetics through Meta-Analysis Consortium (ENIGMA)—Genetics working group , The genetic architecture of the human cerebral cortex. Science 367, eaay6690 (2020).32193296 [Google Scholar]
- 144.Shadrin A. A., Kaufmann T., van der Meer D., Palmer C. E., Makowski C., Loughnan R., Jernigan T. L., Seibert T. M., Hagler D. J., Smeland O. B., Motazedi E., Chu Y., Lin A., Cheng W., Hindley G., Thompson W. K., Fan C. C., Holland D., Westlye L. T., Frei O., Andreassen O. A., Dale A. M., Vertex-wise multivariate genome-wide association study identifies 780 unique genetic loci associated with cortical morphology. Neuroimage 244, 118603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alam T., An M. R., Papaconstantinou J., Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267, 5021–5024 (1992). [PubMed] [Google Scholar]
- 146.de Lange K. M., Moutsianas L., Lee J. C., Lamb C. A., Luo Y., Kennedy N. A., Jostins L., Rice D. L., Gutierrez-Achury J., Ji S. G., Heap G., Nimmo E. R., Edwards C., Henderson P., Mowat C., Sanderson J., Satsangi J., Simmons A., Wilson D. C., Tremelling M., Hart A., Mathew C. G., Newman W. G., Parkes M., Lees C. W., Uhlig H., Hawkey C., Prescott N. J., Ahmad T., Mansfield J. C., Anderson C. A., Barrett J. C., Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Javierre B. M., Burren O. S., Wilder S. P., Kreuzhuber R., Hill S. M., Sewitz S., Cairns J., Wingett S. W., Varnai C., Thiecke M. J., Burden F., Farrow S., Cutler A. J., Rehnstrom K., Downes K., Grassi L., Kostadima M., Freire-Pritchett P., Wang F.; BLUEPRINT Consortium, Stunnenberg H. G., Todd J. A., Zerbino D. R., Stegle O., Ouwehand W. H., Frontini M., Wallace C., Spivakov M., Fraser P., Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lesch K. P., Timmesfeld N., Renner T. J., Halperin R., Roser C., Nguyen T. T., Craig D. W., Romanos J., Heine M., Meyer J., Freitag C., Warnke A., Romanos M., Schafer H., Walitza S., Reif A., Stephan D. A., Jacob C., Molecular genetics of adult ADHD: Converging evidence from genome-wide association and extended pedigree linkage studies. J. Neural Transm. 115, 1573–1585 (2008). [DOI] [PubMed] [Google Scholar]
- 149.Chu T. T., Liu Y., An integrated genomic analysis of gene-function correlation on schizophrenia susceptibility genes. J. Hum. Genet. 55, 285–292 (2010). [DOI] [PubMed] [Google Scholar]
- 150.Scott L. J., Muglia P., Kong X. Q., Guan W., Flickinger M., Upmanyu R., Tozzi F., Li J. Z., Burmeister M., Absher D., Thompson R. C., Francks C., Meng F., Antoniades A., Southwick A. M., Schatzberg A. F., Bunney W. E., Barchas J. D., Jones E. G., Day R., Matthews K., McGuffin P., Strauss J. S., Kennedy J. L., Middleton L., Roses A. D., Watson S. J., Vincent J. B., Myers R. M., Farmer A. E., Akil H., Burns D. K., Boehnke M., Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc. Natl. Acad. Sci. U.S.A. 106, 7501–7506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium , Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Muhleisen T. W., Leber M., Schulze T. G., Strohmaier J., Degenhardt F., Treutlein J., Mattheisen M., Forstner A. J., Schumacher J., Breuer R., Meier S., Herms S., Hoffmann P., Lacour A., Witt S. H., Reif A., Muller-Myhsok B., Lucae S., Maier W., Schwarz M., Vedder H., Kammerer-Ciernioch J., Pfennig A., Bauer M., Hautzinger M., Moebus S., Priebe L., Czerski P. M., Hauser J., Lissowska J., Szeszenia-Dabrowska N., Brennan P., McKay J. D., Wright A., Mitchell P. B., Fullerton J. M., Schofield P. R., Montgomery G. W., Medland S. E., Gordon S. D., Martin N. G., Krasnow V., Chuchalin A., Babadjanova G., Pantelejeva G., Abramova L. I., Tiganov A. S., Polonikov A., Khusnutdinova E., Alda M., Grof P., Rouleau G. A., Turecki G., Laprise C., Rivas F., Mayoral F., Kogevinas M., Grigoroiu-Serbanescu M., Propping P., Becker T., Rietschel M., Nothen M. M., Cichon S., Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 5, 3339 (2014). [DOI] [PubMed] [Google Scholar]
- 153.Cross-Disorder Group of the Psychiatric Genomics Consortium , Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 381, 1371–1379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Smith S. M., Douaud G., Chen W., Hanayik T., Alfaro-Almagro F., Sharp K., Elliott L. T., An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat. Neurosci. 24, 737–745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Comuzzie A. G., Cole S. A., Laston S. L., Voruganti V. S., Haack K., Gibbs R. A., Butte N. F., Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLOS ONE 7, e51954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S9
List of IMAGEN Consortium members
References