Highlights
-
•
Mesocorticolimbic and opioid pathways are key in substance use disorders (SUD).
-
•
Specific gene polymorphisms like DRD2, GABRA, COMT, and DAT may relate to SUD relapse.
-
•
Epigenetics play a role in SUD, such as CpG hypermethylation.
-
•
HTR2A gene methylation links to impulsivity and attentional bias in cocaine users.
Keywords: Substance use disorder, Relapse, Genetic, Genome-wide association-studies (GWAS), Epigenetic, Methylation, Single nucleotide polymorphism (SNP)
Abstract
Several genetic and epigenetic factors contribute to the elevated substance use disorder (SUD) relapse vulnerability, yet a comprehensive investigation into these factors is lacking. This review aims to delve into current literature to highlight key genomic factors associated with SUD relapse.
Focusing on genetic predisposition and epigenetic modifications the review synthesized research findings of several genetic polymorphisms, histone modifications and DNA methylation patterns contributing to the initiation of SUD and the elevated relapse susceptibility. Notably, specific gene polymorphisms, such as Dopamine Receptor D2 gene (DRD2), Gamma-Aminobutyric Acid Receptor Alpha gene (GABRA2), Catechol-O-methyltransferase (COMT) gene, Dopamine Transporter (DAT1) gene and others were identified to be connected to various patterns of SUD relapse. Furthermore, SUD initiation and relapse has been shown to be influenced by epigenetics. Specifically, CpG hypermethylation has been associated with severe alcohol use disorder in the 5′ untranslated region of the Bladder Cancer Associated Protein gene (BLCAP) and the upstream region of the Active BCR Related gene (ABR). Co-users of cannabis and tobacco showed notable variations in CpG site methylation, especially at the Aryl Hydrocarbon Receptor Repressor (AHRR), and factor II receptor-like 3 gene sites (F2RL3).
In conclusion, there is good evidence of certain associations between genomic factors and relapse to SUD. However, further research is needed to ascertain causality effects of these factors and develop novel interventions for effective treatment and relapse prevention.
1. Introduction
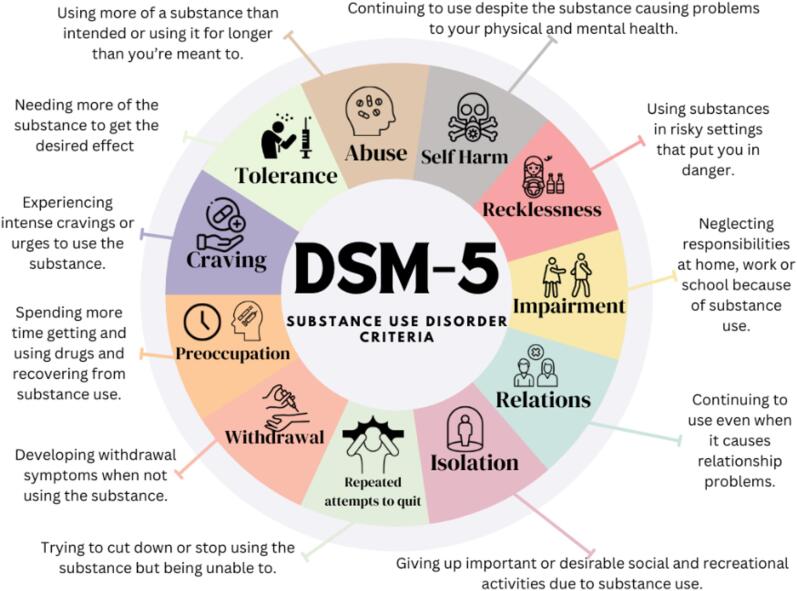
Substance Use Disorder (SUD) is a complex, chronic, and often relapsing disorder manifested by compulsive drug-seeking behavior, despite harmful consequences on the individual’s health, occupation, and social life (National Institute on Drug Abuse, 2020). While understanding the primary factors that predispose and trigger the onset of SUD is necessary, a comprehensive examination of recovery process is as also crucial to overcome the high relapse incidence (Aszalos et al., 1999, Pasareanu et al., 2015). Relapse refers to the reoccurrence of two or more of the SUD criteria due to returning to substance use, during the recovery phase, after a period of remission Fig. 1 (Diagnostic and statistical manual of mental disorders : DSM-5™, 2013; Moe et al., 2022). It indicates a setback in the treatment process, hence identifying reliable factors associated with relapse to SUD can guide the development of innovative treatment modalities to improve SUD outcomes and prevent relapse.
Fig. 1.
DSM-5: substance use disorder criteria. Note. The content of the figure is adapted from Diagnostic and Statistical Manual of Mental Disorders: DSM-5™ (2013) and designed using Canva. Own work. (Diagnostic and statistical manual of mental disorders: DSM-5™ 2013).
Biological genetic biomarkers are thought to have a significant role in predicting relapse vulnerability, although research is yet to provide sufficient evidence of specific genetic predictors of relapse. Genome-wide association-studies (GWAS), and Epigenome-wide association-studies (EWAS) have been utilized to detect genetic polymorphisms and epigenetic modifications that are associated with increased relapse rates (Alblooshi et al., 2019, Cozzoli et al., 2021). Genes that regulate reward pathways have been studied to explore the association between specific genetic polymorphism or epigenetic modifications and the susceptibility to substance abuse and potential relapse. These variations and modifications can disrupt crucial biological processes associated with reward process, leading to substance abuse onset or recurrence (Di Chiara, G. & Imperato, 1988; Willuhn et al., 2010). It is important to note that while this study focuses primarily on genomic factors associated with relapse to SUD, initial diagnoses were also taken into consideration, due to the overlap seen among certain genetic polymorphisms contributing to SUD onset and those influencing relapse, such as the GABRA 2 gene (Bauer et al., 2012, Fehr et al., 2006, Sun and Wolf, 2009). This approach allows for a comprehensive understanding of the genetic landscape that impact SUD recurrence, providing the researchers with a valuable foundation for investigating genes associated with SUD initiation in the context of relapse. While genomic factors associated with relapse to SUD may overlap with those involved in the initial onset, it is unclear if they are identical, since psychological, physiological, and environmental triggers may differ at these two disease stages. Understanding genomic relapse-specific factors offers insights into intervention, enabling the development of targeted treatment plans, reducing relapse likelihood and improving treatment outcomes.
Studies on genomic risk factors for SUD relapse reveal significant knowledge gaps, particularly due to the narrow focus on a limited set of genes. A more comprehensive approach utilizing GWAS remains largely underexplored in the context of SUD relapse. Furthermore, existing research typically focuses on one particular component genetic polymorphisms and epigenetic modifications without comprehensively addressing the complex interplay among these factors. Our review aims to bridge this gap by critically appraising the available evidence and identifying areas where further research is needed to improve our understanding and provide clinically relevant methods to reduce relapse risk.
2. Neural pathways in the brain associated with reward activation
The induction of neuroadaptive changes resulting from chronic substance exposure has been the focus of recent studies. It is believed that these changes may subsequently contribute to the development and reinforcement of addictive behavior (Nestler & Lüscher, 2019). Studies of the addictive nature of substances primarily identified two key pathways: the mesocorticolimbic dopaminergic pathway and endogenous opioid pathway(Blum et al., 2012, von Zastrow et al., 2003).
The mesocorticolimbic dopaminergic pathway consists of dopamine neurons projecting from the ventral tegmental area to the limbic system (mesolimbic pathway) and cortical system (mesocortical pathway). These pathways have been shown to be activated by addictive substances at different levels (Camí & Farré, 2003). The mesolimbic dopaminergic pathway is responsible for the pleasurable and rewarding properties in the human brain (Arias-Carrión et al., 2010). It establishes a neural link between VTA and ventral striatum which encompasses the nucleus accumbens (NA) (Hyman and Malenka, 2001, Pezze and Feldon, 2004). By stimulating the dopamine neurons in the VTA area, dopamine release is increased in the NA leading to the activation of the reward system that basically mediates feeling of desire and pleasure due to the wanting and liking, respectively. Therefore, positive reinforcement effect is facilitated (Berridge, 2012, Di Chiara et al., 2004).
The mesolimbic pathway is activated, either directly or indirectly, by addictive drugs through the release, inhibition of degradation, or inhibition of reuptake of dopamine neurotransmitter (Engert & Pruessner, 2008). This causes disturbance in the dopamine homeostasis which in turn leads to neuroadaptation and hypodopaminergic state (Febo et al., 2017). For example, it has been shown that alcohol directly stimulates the release of dopamine in the NA (Di Chiara and Imperato, 1988). Psychostimulants consumption affects the reward pathway by directly increasing dopamine concentration in the shell of the NA, blocking dopamine reuptake from the synapse, enhancing the release of Glutamate, the excitatory neurotransmitter, and indirectly activating N-methyl-D-aspartate (NMDA) receptors (Aragona et al., 2008, Febo et al., 2017). Mesolimbic pathway can be influenced by the ventral pallidum and amygdala that also receive dopamine neurons from the VTA. Ventral pallidum enhances the primacy positive reinforcement associated with addictive substances (Volkow, Nora D. et al., 2003), while the amygdala is associated with conditional learning that leads to the formation of discrete stimulus–reward association. This process links the drug-related cues to the rewarding effects of addictive substances, hence the drug-related cues become emotionally salient stimuli, increasing the craving to addictive substances (Mahler and Berridge, 2009, See, 2005).
Along with mesolimbic area, VTA also extends its dopaminergic neurons into the various regions of the mesocortical area, including the prefrontal cortex, orbitofrontal cortex and anterior cingulate, with prefrontal cortex being the key target of the pathway (Belin et al., 2013; Volkow et al., 1993). It has been established that the prefrontal cortex is responsible for regulating executive function, decision making, emotional control, impulsive control, and cognitive function (Belin et al., 2013, Volkow et al., 1993). Interestingly, dysregulation of mesocortical pathway has been reported in patients with SUD (Belin et al., 2013, Volkow et al., 2016). This dysregulation manifests as down regulation of dopamine neurotransmitter in the prefrontal cortex, resulting in morphological and functional changes, consequently impairing the decision-making ability, and dysregulating emotional and craving control that leads to an elevated relapse risk (Belin et al., 2013, Volkow et al., 2016).
While the mesocorticolimbic pathway is involved in the cognitive and rewards aspects associated with addictive substances use, the nigrostriatal pathway adds complexity to the pathophysiology of SUD. This is due to its role of movement-related functions (regulated by the nigrostriatal pathway) in enhancing the reward circuit, thereby contributing to the development of addictive behavior (Quik et al., 2011). Both nigrostriatal and mesolimbic systems contain neuronal nicotinic acetylcholine receptors that regulate dopamine release, consequently affecting positive reinforcement to addictive substances and motor behaviors (Quik et al., 2011). Moreover, alterations in the nigrostriatal system in habit formation (related to drug use), resulting in behaviors characterized by stereotypic and rigid addictive-substance use pattern, which may contribute to elevated relapse rate (Spanagel & Heilig, 2005).
The reward property can also be experienced through the endogenous opioid system by direct activation of the dopaminergic rewarding system through interfering with the inhibitory neurotransmitter Gamma-Aminobutyric Acid (GABA) receptors which in turn leads to increased dopamine concentrations in the neuronal synapse of the NA (Le Merrer et al., 2009). Additionally, it can activate the reward property indirectly by creating a sensation of reset, sedation, and blissfulness through stimulating the opioid receptors (Trigo et al., 2010). It has been suggested that the reward, reinforcement, and dependence of some addictive substances are in part mediated by the endogenous opioid system (Le Merrer et al., 2009, Trigo et al., 2010). For example, the reinforcing properties of alcohol and tetrahydrocannabinol have been shown to be blocked by the opioid receptor antagonists naltrexone and naloxone, respectively (Braida et al., 2004, Thorsell, 2013).
The chronic consumption of addictive substances is associated with dysregulation and impairment of the mesocorticolimbic dopamine reward pathway, known as reward deficiency syndrome. This occurs due to alterations in the neurotransmission systems in the brain and downregulation of dopaminergic and opioid receptors (Blum et al., 2018). Moreover, reward deficiency syndrome leads to alteration in the brain’s sensitivity towards both natural rewards and addictive substances. This prompts the patient to increase the dose to get the desired effect, which is accompanied by a pattern of compulsive substance use and can lead to abstinence failure during treatment (Christie, 2008, Fraser, 1957, Padmanabhan et al., 2011, World Health Organization, 1994).
Drug-induced changes in the reward system, along with genetic predisposition and epigenetic modifications in genes that regulate reward processes, may collectively induce compulsive drug-seeking, drug-consumption, and relapse to SUD. These genomic factors will be addressed in the following discussion.
3. Genetic factors contributing to relapse to SUD
SUD is a polygenic condition that is affected by many physiological systems in the body which are regulated through multitude of genes responsible for the production and regulation of neurotransmitters, receptors, and other elements in those systems (Deak & Johnson, 2021). Ongoing research on the genetics of SUD relapse has mainly aimed to differentiate individuals with addictive and relapse tendencies from healthy subjects by investigating several polymorphisms within genes involved in the reward pathway, stress response, and morphology and functions of the brain. Genome-wide association studies have been primarily employed to understand the molecular bases of individuals with SUD and highlight the most reliable polymorphisms to SUD initiation and relapse (Alblooshi et al., 2019, Cozzoli et al., 2021) Table 1.
Table 1.
Gene polymorphisms and their association with SUD initiation and relapse.
| Genes Involved | Polymorphism | Association | Reference |
|---|---|---|---|
| Dopamine receptor D2 (DRD2) gene | rs1076560 | Higher relapse rates than non-carriers following alcohol-dependence treatment completion | (Dahlgren et al., 2011) |
| Gamma-Aminobutyric Acid Receptor Alpha (GABRA 2) gene | rs279858 | Increased substance positive reinforcement effect in patients with SUD. Increased susceptibility to different substance dependence (such as opioid, alcohol and cocaine) Increased relapse Increased onset of relapse |
(Fehr et al., 2006, Sun et al., 2018) (Fehr et al., 2006, Sun et al., 2018) (Bauer et al., 2012) |
| Catechol-O-methyltransferase (COMT) gene | rs4680 | Synthesis of enzymes with 3–4 folds lower enzymatic catabolism Increased relapse rate to heroin |
(Owusu Obeng et al., 2017) (Su et al., 2015) |
| Dopamine transporter (DAT1) gene |
9-repeat allele of DAT1 VNTR |
9-repeat carriers had shown higher dopamine transporter protein production than 10-repeat High relapse vulnerability to addictive substance especially during detoxification 9-repeat variation was not associated with alcohol use disorder overall, but with alcoholic subgroup with alcohol withdrawal seizure or delirium tremens |
(Giessen et al., 2009) (Moeller et al., 2013) (YanLei et al., 2011). |
| Opioid Receptor Mu 1 (OPRM1) gene | Rs1799971 |
Risk factor for substance abuse among: Jordanian males Asian population Caucasian males Lack of association with drug addiction within the UAE population. Lack of association with opioid addiction in USA population |
(Al-Eitan et al., 2021) (Rouvinen-Lagerström et al., 2013) (Woodcock et al., 2015) (Alblooshi et al., 2018) (Crowley et al., 2003) |
| Serotonin Transporter (SLC6A4) gene | Short allele of Serotonin Transporter Linked-Polymorphic Region (5-HTTLPR) * | Increased susceptibility to alcohol dependence. Associated with relapse to alcohol use disorder |
(Sander et al., 1997) (Pinto et al., 2008) |
| Ankyrin Repeat and Kinase Domain Containing 1 (ANKK1) gene | rs1800497 | Associated with alcohol addiction. Failed to exhibit an association with alcohol addiction in another study. |
(Wang et al., 2013) (Grzywacz et al., 2019) |
| CACNB2 GRIN2B PLXDC2 PKNOX2 |
Associated with the Alcohol use behavior disorder, Nicotine use behavior disorder and Drug use behavior disorder | (Chang et al., 2022) |
Note. Data summarized by the author.
* This polymorphism is characterized by the number of short sequences repeated in tandem. So, no reference SNP identification.
3.1. Single nucleotide polymorphism (SNP) meaning and general insights
SNP is a common genetic polymorphism at the single-nucleotide level (point mutation) that conventionally occurs in more than 1 % of the population. SNPs can occur in non-coding regions, or coding (exonic) regions (being either synonymous or nonsynonymous). While most of the SNPs are not biologically significant, others, known as Functional SNPs, have demonstrated varying degrees of effect on the gene function and expression (Albert, 2011, Chorley et al., 2008). These variations can either lead to gain or loss of gene function depending on the location of the polymorphism throughout the different genomic regions and its impact on the encoded protein's structure. In the coding region the Nonsynonymous SNP, where nucleotide change leads to substitution of an amino acid by another amino acid, results in alteration in the protein structure and function, either by stabilizing its structure and enhancing its function or destabilizing the protein’s structure and causing a loss of function (due to its degradation) (Albert, 2011, Shastry, 2009). Intronic SNPs used to be considered as non-functional variation, however researchers have revealed that they regulate gene transcription through several mechanisms. They can regulate protein synthesis through influencing the process of mRNA splicing, leading to variation in the gene expression (mRNA sequence), ultimately affecting the quantity and functionality of the synthesized protein (Deng et al., 2017). SNPs within introns may also influence gene expression of imprinted regions due to variations in epigenetic marks (Deng et al., 2017). Moreover, SNPs within introns, which are responsible for transcribing the long non-coding RNA (lncRNA), can impact the structure and function of the resulting lncRNA, thereby influencing its regulatory process (Deng et al., 2017).
3.2. Genetic variants associated with elevated relapse vulnerability
Reward deficiency syndrome, that involves reduction in dopamine or its receptors level (hypodopaminergic state) in synapses within areas of the mesolimbic reward circuit, has been shown to be associated with increased relapse risk, possibly implicating the polymorphisms in genes that affect the brain reward cascade (Blum et al., 2018). A study focusing on the initiation of SUD identified the variation A1 (rs1076560) located in the intron 6 of the Dopamine receptor D2 (DRD2) gene (Sasabe et al., 2007). This SNP has been shown to impact the reward process by reducing the dopamine receptor numbers (Noble et al., 1991) hence reducing the normal brain dopaminergic function (Ritchie & Noble, 1996). This leads to diminished sense of reward from typical stimuli in the mesocorticolimbic dopamine reward system (Blum et al., 2018). A study by (Dahlgren et al., 2011), conducted on 375 alcohol-dependent individuals, aimed to investigate whether A1 allele of the DRD2 gene is associated with increased relapse rate in alcohol-dependent individuals. Specifically, this study demonstrated that carrier of the A1 allele variant exhibited higher relapse rates (89 % of carriers) compared to non-carriers (53 % of non-carrier) following alcohol-dependence treatment completion (Dahlgren et al., 2011).
Transitioning to an analysis of genes pertaining to dopamine synaptic concentration, distinct genes have been shown to be associated with inhibiting dopamine production, catabolizing, and clearing from the synapse. Among those genes, Gamma-Aminobutyric Acid Receptor Alpha gene encodes the GABAA α-2 subunit, which, upon activation by GABA, inhibits dopamine release in the NA leading to reward regulation. Several studies have shown that GABRA2 rs279858 SNP (in exon 5, K132K) (Villafuerte et al., 2012) is significantly associated with increased substance positive reinforcement effect in patients with SUD, (Arias et al., 2014, Roh et al., 2011, Uhart et al., 2013) despite some variations the findings and methodologies. (Arias et al., 2014) study, with a cohort of 28 light drinkers and 24 heavy drinkers European-Americans, found that carriers of the rs279858 C allele experienced greater stimulation and euphoria from alcohol after drinking low-dose or high-dose alcohol (Arias et al., 2014). Similarly, (Roh et al., 2011) study, which involved 110 healthy social drinkers undergoing alcohol clamps, demonstrated that the rs279869, rs279858, and rs279837 SNPs in the GABRA2 gene were significantly associated with alcohol subjective effects, mainly the physiologic responses, stimulant and sedative effects, which known to increase the risk to alcohol use disorders (Roh et al., 2011). Moreover, the rs279858 polymorphism was associated with attenuation of negative response, as found by (Uhart et al., 2013) and can further increase the vulnerability to alcohol use disorder as discussed by (Fehr et al., 2006).
Additionally, the rs279858 SNP has been associated with increased susceptibility to other substance dependence such as heroin. According to (Sun, Y. et al., 2018) this SNP increases heroin vulnerability by affecting the reward network. Heroin addiction, in turn, can disrupt the gene expression of the GABRA2 rs279858 variant, leading to disruption in the reward network and impairment of cognitive function. Ultimately, exacerbating heroin addictive behavior.
Moreover, findings from (Bauer et al., 2012) study indicated that GABRA2 and KIBRA genotypes were associated with increased vulnerability to relapse and rapid onset of relapse (Bauer et al., 2012). This study included a reasonable sample size (n = 146), yet an important limitation is related to defining relapse as “any use” rather than the problematic return to substance use.
Catechol-O-methyltransferase (COMT) gene is another important player in the reward system. It encodes the COMT enzyme that has a notable role in dopamine catabolism and the regulation of dopamine concentration in the synaptic cleft (Owusu Obeng et al., 2017). COMT enzyme has been shown to be influenced by genetic polymorphism. For example, the rs4680 SNP (Val 158 Met) located within exon 4 of the COMT gene (Oberacher et al., 2006) produces an enzyme that exhibits three-to-four-time lower enzymatic activity in dopamine catabolism when contrasted with the wild-type allele (Owusu Obeng et al., 2017). This effect can influence reward processing and impulsivity leading to increased relapse to addictive substance use such as heroin and alcohol (Su et al., 2015, Voisey et al., 2011). In the (Su et al., 2015) study with 564 heroin-dependent patients in the abstinent stage, the rs4680 SNP of the COMT gene was found to be associated with relapse to heroin abuse. However, (Voisey et al., 2011) reported that rs165774 and rs4680 were bothassociated with alcohol dependence but not nicotine or opiate dependence. In both studies, limitations necessitate further research. In the study the main limitation was incomplete follow-up data leading to missed relapse cases. In (Voisey et al., 2011) P-values were not adjusted for multiple testing and the opiate and nicotine-dependent groups’ sample sizes were modest, limiting the generalizability of its findings.
Another important gene in the reward pathway is the Dopamine transporter (DAT1) gene (SLC6A3), since it encodes the dopamine transporter protein that plays a vital role in the reuptake of dopamine from the synaptic cleft (Giessen et al., 2009). The DAT1 VNTR (Variable Number Tandem Repeat) genetic variation affects the dopamine transporter expression and is associated with multiple conditions including the SUD (Giessen et al., 2009). The number of repeats in this gene directly affects the quantity of transporter protein produced. Specifically, the 9-repeat carriers had shown higher dopamine transporter protein production than 10-repeat carriers, resulting in an increased dopamine reuptake from the synaptic cleft subsequently resulting in a decreased dopamine concentration within the synapse (Giessen and v. d., Win, M. M. L. d., Tanck, M. W. T., Brink, W. v. d., Baas, F., & Booij, J. , 2009, Pineau et al., 2019). This phenomenon has been shown to significantly correlate with high relapse vulnerability to cocaine among 9-R carries, especially during detoxification treatment phase, due to increased response to drug-related cues as revealed by (Moeller et al., 2013). In contrast, a meta-analysis across various ethnicities, including Mexican Americans, Caucasian and Asian, did not find significant statistical differences in DAT1 gene polymorphism and its allele distribution between alcoholics and controls indicating that SLC6A3 VNTR A9 variation may not be associated with alcohol use disorder overall, but found an association between the variation and alcoholic subgroup with alcohol withdrawal seizure or delirium tremens (YanLei et al., 2011). However, several limitations should be considered including variations in factors, such as diagnostic criteria and gender ratios, the small sample size of the included studies, limited numbers of overall studies included in the meta-analysis, and the presence of addictive and psychiatric disorders (YanLei et al., 2011). Further research is needed to validate and better understand the association between SLC6A3 VNTR 9-Repeat Allele and SUD initiation and relapse.
In addition to the gene polymorphisms associated with increased relapse vulnerability, other gene polymorphisms were examined for their role in increasing SUD susceptibility that might also contribute to relapse vulnerability. A pivotal reward genetic variant is the exonic A118G SNP (rs1799971) of the OPRM1 gene that encodes for the mu-opioid receptor. This nonsynonymous OPRM1variant, which eliminates the N-glycosylation site, is associated with decreased OPRM1 receptor expression and level (Haerian and Haerian, 2013, Ray et al., 2011) hence affects the way neurotransmitters, especially endogenous opioids and dopamine, function in people of different ancestry (Peciña et al., 2015). It has been proposed that this variant is considered a risk factor for substance abuse among Jordanian males (Al-Eitan et al., 2021, Peciña et al., 2015), Asian population (Haerian and Haerian, 2013, Rouvinen-Lagerström et al., 2013), and Caucasian males (Woodcock et al., 2015) and as a risk factor of specifically developing opioid and alcohol use disorders (Clarke et al., 2013, Koller et al., 2012). Yet it was determined that neither the DRD2 rs1076560 SNP nor the OPRM1 rs1799971 SNP demonstrated significant associations with drug addiction within the UAE population (Alblooshi et al., 2018). Of note, this study included subjects who used different types of substances including alcohol, opioids, and non-opioids with no stratification of the disorder phenotype. Older studies have also failed to exhibit an association between the exonic OPRM1 rs1799971 SNP and the opioid addiction (Crowley et al., 2003). However, more recent, large-scale, and well-powered GWAS have confirmed the association with the rs1799971 variant (Deak et al., 2022, Kember et al., 2022, Zhou et al., 2020). The evidence supporting this association is primarily derived from European ancestry samples, possibly due to differences in minor allele frequencies across populations. Overall, the current body of evidence strongly supports a link between opioid use disorder and rs1799971, despite variations among individuals from diverse ancestral backgrounds.
Another important gene polymorphism associated with SUD vulnerability is the Serotonin Transporter Linked Polymorphic Region (5-HTTLPR) genetic variation of the promoter region of the serotonin transporter gene (SLC6A4). The short (S) allele of 5-HTTLPR is associated with reduced serotonin reuptake (Gorwood et al., 2000) and carriers of this allele have been more susceptible to alcohol dependence (Sander et al., 1997). Additionally, findings from (Pinto et al., 2008) study, which included 48 alcohol-dependent male patients in the abstinence phase, indicated that there is a significant association between S allele of the 5-HTTLPR and relapse to alcohol use disorder. However, there is a limitation to the study's generalizability due to its small sample size (Pinto et al., 2008).
A comprehensive meta-analysis demonstrated that the Taq 1A (rs1800497) polymorphism located in exon 8 of ANKK1 gene is associated with alcohol addiction (Palacios et al., 2018; Wang, F. et al., 2013). In contrast, other studies failed to find a significant association between ANKK1 Taq1A genotype and alcohol addiction (Grzywacz et al., 2019). Other comprehensive genetic analysis on 2910 genes associated with SUD from 75 GWAS revealed four genes (CACNB2, GRIN2B, PLXDC2 and PKNOX2) to be associated with the Alcohol use behavior disorder, Nicotine use behavior disorder and Drug use behavior disorder (Chang et al., 2022).
The detection of SNPs related to brain function can be through analyzing DNA samples obtained from brain tissues as well as circulating bodily fluids including saliva and blood. Genetic testing for the analysis of several SNPs, including DRD2 gene rs1076560, GABRA2 rs279858, COMT rs4680 and OPRM1 rs1799971, were performed using blood and saliva samples in several studies. Blood sample is preferred since it has higher DNA concentration, more stable than saliva sample, and less invasive than brain tissue samples (Boyd et al., 2016, Drogou et al., 2020, Suchanecka et al., 2020; Wang et al., 2019).
A major concern in the literature on SUD relapse is the reliance on small candidate gene studies. Complex behavioral phenotypes like SUD relapse are polygenic, and findings from such studies may not hold up over time, as seen with other polygenic psychiatric phenotypes like depression (Border et al., 2019) and schizophrenia (Farrell et al., 2015). In these cases, common genetic variants and genes typically have very small effect sizes, requiring large sample sizes for reliable detection. Similarly, many epigenetic studies in this area focus on only a few genes, which limits their scope. The scarcity of GWAS and EWAS on SUD relapse indicates the urgent need for such comprehensive studies. While small candidate gene studies can serve as hypothesis generators (Jorgensen et al., 2009) and provide foundational insights into gene polymorphisms and epigenetic modifications potentially linked to relapse, their limitations remain a concern. Despite offering limited insights, they fall short of capturing the full complexity of the genomic factors associated with SUD relapse.
Understanding the genomic factors associated with SUD relapse rates can lead to the development of personalized treatment approaches. The identification of genetic and epigenetic risk factors can help in the development of diagnostic kits that can detect individuals with increased susceptibility to relapse and treat them accordingly (Alchakee et al.,2022). Moreover, an individual's genomic profile can guide the behavioral intervention, for example, carriers of the short allele of the 5-HTTLPR genetic variation showed altered mood regulation, therefore the behavioral intervention could focus on this area (Sander et al., 1997). Additionally, identifying genomic risk factors can significantly contribute to the development of novel therapeutic interventions; for example, developing a treatment that targets the OPRM1 gene, which encodes for the mu-opioid receptor, can reduce the risk of opioid addiction, potentially by restoring opioid’s normal binding to their receptors hence improve the treatment outcomes (Haerian and Haerian, 2013, Ray et al., 2011).
Furthermore, regarding epigenetic modifications, studies suggest that HDAC inhibitors could significantly reverse these changes and restore the normal gene function. If confirmed, this and similar agents could be promising treatment approaches (Nestler, 2014).
In summary, by identifying the genomic risk factors of relapse to SUD, determining the patient’s genomic profile, and integrating them with other clinical data, we can develop a precise treatment plan for each patient hence improve treatment outcomes.
4. Epigenetic factors contributing to relapse to SUD
Epigenetic modifications serve as a mechanism through which most of the risk factors interact with the individual’s DNA. Hence, they either activate or suppress the expression of key genes associated with substance addiction and relapse without altering the underlying DNA sequence. Furthermore, epigenetic changes occurring in several brain regions, particularly those associated with dopamine reward system and opioid system, oppose the drug’s effect due to alteration in cellular dynamics, neuroadaptation, and changes in the brain’s reward areas (Nestler & Lüscher, 2019). Additionally, they manifest in various neurocognitive and behavioral consequences, ultimately leading to tolerance, withdrawal effects and impairment in hedonic function (Gipson and Beckmann, 2023, Sinha, 2008). The altered cellular dynamics and neuroadaptation persist while dopaminergic neuron continue to be stimulated by addictive drugs, and even for weeks after complete drug abstinence (Nestler, 2016). Hence, neuroadaptation may contribute to long lasting craving and relapse to drug seeking, especially to psychostimulants, opioids, nicotine, and alcohol (Werner et al., 2021).
Epigenetic changes primarily include histone modifications, DNA methylation, non-coding RNA activity modification and chromatin remodeler–associated modifications (Nestler & Lüscher, 2019). Among these, DNA methylation and histone modifications have probably received the most extensive focus in research (Cadet, 2016). These modifications influence the activity of genes, which are specialized stretches of DNA determining specific phenotypes within an organism. Each gene carries a genetic code that is transcribed into an mRNA and then translated into a protein. The complex structure of the gene plays an important role in regulating its expression. The promotor region is a DNA sequence located upstream of the gene and provides binding sites for transcription factors that, upon binding to the promotor region, recruits RNA polymerase to start transcribing that specific gene (Polyak & Meyerson, 2003). Promotors often encompass CpG islands that help in regulating gene expression. These islands are rich in cytosine: Guanine dinucleotides. They are typically not methylated during cellular differentiation, with exceptions such as in gene silencing during the inactivation of x chromosome in female. However, some CpG sites become susceptible to methylation when influenced by several environmental factors, such as the chronic consumption of specific addictive substances, where methyl group is added to the cytosine residues preventing gene transcription. The methylation of CpG site leads to gene silencing by both inhibiting the recruitment of factors associated with opening chromatin structure (ZF-CxxC domain proteins) and facilitating the recruitment of complexes that repress gene transcription (Histone deacetylase (HDACs) complexes). This epigenetic modification is highly stable; hence reactivation of the impacted gene becomes extremely challenging (Blackledge & Klose, 2011).
Epigenetic modifications cannot be directly detected by GWAS, but through several other methods that can then be integrated with GWAS to better understanding the genetic base of the epigenetic modification and their role in SUD initiation and relapse. Methods, including Quantitative Trait Loci (QTL) Mapping and EWAS, generate Epigenetic modifications data that can be overlapped with GWAS data to identify genetic basis of the variant that may contribute to the disease or trait studies (Gibbs et al., 2010, Relton and Davey Smith, 2010).
4.1. DNA methylation
DNA methylation is a frequently studied epigenetic modification in the context of SUD, it may occur following the chronic drug abuse influencing gene expression by either increasing or decreasing gene transcription to mRNA, hence affecting cellular functions. Measuring the methylation level of promoter CpG sites indicates the affected-gene’s expression level: hypermethylation is associated with gene silencing, while hypomethylation is with increased gene activity (Blackledge and Klose, 2011, Lea et al., 2018).
Chronic alcohol use has been shown to trigger epigenetic modifications in various genes, and these changes were associated with the severity of the alcohol use disorder, where severe cases led to CpG hypermethylation involving methylation of multiple regions. This included upstream of ABR gene, 5′ untranslated region of BLCAP gene (Philibert et al., 2012) and various CpG sites of the Monoamine oxidase A gene in women (Philibert et al., 2008). Moreover, cannabis with tobacco users have shown significant difference in numerous CPG sites methylation compared to control, with notable emphasis on three sites: AHRR, Alpha-2-Lipoprotein (ALPG) and F2RL3 (Osborne et al., 2020). AHRR methylation was also associated with marijuana use in a diverse population (Nannini et al., 2023). Moreover, (Nielsen et al., 2010) demonstrated that former heroin addicts showed significantly higher methylation level than controls. Specifically, this study showed that opioid use triggered differential hypermethylation in promoter region of the OPRM1 gene, blocking the binding of several transcription factors, ultimately resulting in reduced the gene expression and Mu opioid receptor levels (Nielsen et al., 2010). While understanding the influence of epigenetic modification on the SUD relapse vulnerability has valuable insights, the human study in this area is limited. (Land et al., 2020) conducted a noteworthy study in this area examining how methylation pattern contributes to relapse behaviors in cocaine use disorder (CUD). This study, which involved 48 controls and 53 patients with CUD recruited from two major centers in Texas and Virginia, USA, revealed the significant association between DNA methylation pattern in different sites in the promoter region of the serotonin receptor (HTR2A) gene and personality traits related to relapse in individuals with cocaine use disorder (CUD). Percent methylation at sites −1224 and −253 of HTR2A gene were positively correlated with impulsivity traits and increased attentional bias toward cocaine-related cues in individuals with CUD, respectively. While percent methylation at site −1420 was negatively correlated with the tendency to prefer smaller immediate rewards over larger delayed reward (delay discounting) in control group but not individuals with CUD. These findings indicate that methylation influences relapse-related behaviors, hence impacts the vulnerability to relapse to CUD (Land et al., 2020).
4.2. Histone modifications
Histones, proteins that compress the DNA into nucleosomes, undergo epigenetic modifications that further regulate gene expression by either activating or suppressing gene transcription to mRNA. Histone H3 and H4 types of the core histone possess an N-terminal that is susceptible to several chemical modifications. This is due to the abundant availability of the easily modified Lysine and Arginine amino acids in the N terminal of these subtypes (Shepard & Nugent, 2020).
While histone is susceptible to various chemical modifications, acetylation and methylation are most extensively studied in SUD. Histone acetylation by histone acetyl transferase (HAT) leads to unwrapping the DNA including the gene resulting in the activation of its transcription to mRNA. Conversely, histone methylation by histone methyltransferases can lead to either unwrapping or rewrapping the DNA depending on the methylation site leading to activation or repression of gene transcription to mRNA, respectively (Browne et al., 2020).
Histone acetylation and DNA methylation interact to control gene expression. Promoter CpG hypermethylation was associated with deacetylation of adjacent histone hence leading to gene silencing (Vaissière et al., 2008). Moreover, histone methylation at promoter region can contribute to gene silencing by repressing chromatin structure and restricting transcription factors access to the gene, ultimately inhibiting gene expression (Miller & Grant, 2013).
Evidence from human studies has shown significant correlations between histone modification and SUD. A study has revealed that regulators of histone acetylation, including both HAT and HDACs, are affected by chronic cocaine consumption, that induces the disengagement of HDAC3 from Nuclear receptor 4A2 (NR4A2) resulting in prolonged acetylation of the gene and upregulation of its function. Remarkably, individuals experiencing NR4A2 gene dysfunction in the medial habenula have shown significant association with decreased relapse behavior (López et al., 2019). Moreover, the chronic use of cocaine has also been found to elevate acetylation of H3Ac/H4Ac in 1651 gene promoters and decrease acetylation to 206 gene promoters within the NA (Renthal et al., 2009). This dual impact on acetylation has affected the reward circuit. For instance, chronic and acute cocaine use has been linked to a reduction in CBP, a type of HAT enzyme, consequently decreasing the H3K14Ac and H2BK12Ac in the NA, and may lead to an attenuation of cocaine rewarding effect (Malvaez et al., 2011).
In addition to cocaine, substances like methamphetamine, and opioids, among others, have been shown to induce histone acetylation changes in different brain regions, contributing to the subsequent addictive behavior and potential relapse of SUD (Cheng et al., 2023).
Histone methylation in relation to SUD relapse has been studied in animal models, yet to the best of our knowledge, no studies have been conducted in human subjects. Alonge side acetylation and methylation, phosphorylation and ubiquitination have detrimental role in drug addiction and behavior.
Neuroplasticity involving the brain's ability to change and reorganize synapses has been implicated in SUD (Weerasinghe-Mudiyanselage et al., 2022) . It is thought that the ubiquitin proteasome system plays a critical role in the protein degradation, which affects synaptic plasticity and memory (Fioravante & Byrne, 2011) . Drug addiction appears to share common mechanisms with memory formation processes hence studying the overlap pathway is detrimental (Hyman et al., 2006, Milton and Everitt, 2012). The NA is key in the reinforcing effects of drugs (Cardinal & Everitt, 2004), and changes in NA plasticity are thought to contribute to escalation in drug use (i.e., from recreational, temporary to long term compulsive) (Belin et al., 2009).
The UPS has been linked to synaptic plasticity in cultured NA neurons (Sun, X. & Wolf, 2009), and studies show that drug exposure triggers the degradation of specific proteins in the NA, crucial for synaptic plasticity in addiction development. Targets of the UPS, including proteins like Shank, GKAP, CREB, and deltaFosB, are likely regulated by this process(Carle et al., 2007, Dong et al., 2008, Lee et al., 2008).
On the molecular level, there are 3 steps in the protein synthesis pathway which are initiation, elongation, and termination. Initiation is a rate limiting step and there are several mechanisms involved in controlling this phase (Buffington et al., 2014, Sonenberg and Hinnebusch, 2009). One of the mechanisms is by controlling the compilation of ternary complexes by phosphorylating the translation initiation factor eIF2α. The phosphorylation of eIF2α inhibits overall translation, yet it leads to an increase in translation for a specific group of mRNAs that carry upstream open reading frames in their 5′ untranslated region.(Buffington et al., 2014, Sonenberg and Hinnebusch, 2009). According to (Huang et al., 2016) upon injecting small amount of cocaine to adolescent mice, a reduction in the IF2α phosphorylation was observed in the VTA, along with enhancement in the synaptic inputs to dopaminergic neurons in the VTA leading to drug-related behavior induction. Hence, manipulating eIF2α phosphorylation-mediated translational control in brain reward areas could be a promising approach for treating a wide range of addictive behaviors influenced by various substances.
In exploring the epigenic modifications occurring due to chronic drug consumption, it becomes clear that these molecular changes lead to dysregulation in the neural gene transcription by modifying the genetic information accessibility. One important example is the histone deacetylation process by HDACs that results in condensed chromatin structure and transcriptional repression (Romieu et al., 2011). HDACs have also been associated with DNA methylation and gene silencing (Kennedy et al., 2013). The inhibition of this chromatin remodeling enzyme by HDAC inhibitors has shown promising results in the context of SUD. HDAC inhibitors have been shown to induce the reactivation of genes that had been silenced due to DNA methylation (Ng & Bird, 1999). Additionally, Kennedy and colleagues (2013) findings showed that extended HDAC enzyme inhibition in the NA by HDAC inhibitor led to heightened histone acetylation and reduced DNA methylation. These modifications resulted in decreasing the NA neuron inhibitory level, ultimately opposing the behavioral effects induced by cocaine (Kennedy et al., 2013). Another study investigating HDAC inhibitor’s impact on cocaine users showed that pretreatment with HDAC inhibitor was associated with diminished cocaine reinforcement and motivation for its use in rats (Romieu et al., 2008). A related study further supported these observations, where it demonstrated that HDAC inhibitors had substantially decreased the recurrence of cocaine seeking behavior in rats (Romieu et al., 2011). Collectively, the data suggest that HDAC inhibitor is a promising candidate in the field of SUD, exhibiting the potential to improve treatment outcome and decrease the relapse rate.
Taken together, the findings from studies of epigenetic modifications offer insights into understanding the neurobiological mechanisms beyond the persistent seeking of the addictive substance, heightened sensitivity to environmental trigger, substance craving, and most importantly relapse even after prolonged period of abstinence (Cozzoli et al., 2021).
5. Conclusion and gap of knowledge
Relapse prevention is an important and challenging goal in the treatment of SUD since patients remain at high risk for many years. Genomic factors are thought to play an important role in the dynamic of relapse to SUD. In the brain, specific gene polymorphisms affect the reward system by influencing the respond to addictive substances through increasing sensitivity to their effect and vulnerability to their craving. Additionally, gene polymorphism can influence the regulation of coping mechanism. Moreover, the reward system can be significantly affected by the chronic use of addictive substances leading to dysregulation in many reward pathways. The dysregulation is primarily caused by epigenetic modifications that may be detected through EWAS. These studies, conducted to identify both genetic polymorphisms and epigenetic modifications, are critical for guiding our understanding of the genomic factors associated with increased relapse rates.
The cumulative evidence presented in this review highlights the significance of understanding the genomic factors that contribute to an elevated SUD relapse rate. However, research is yet to provide sufficient evidence of specific predictors of relapse and validate their viability to reduce relapse rate in patients with SUD. Literature also lacks information regarding the dynamic and plastic nature of epigenetic modification throughout the treatment journey, during abuse, withdrawal, and long-term abstinence, and whether these modifications are reversible with time of abstinence and/or biopsychosocial interventions. Moreover, most GWAS studies have focused on the genetic variants contributing to the development of SUD but not to its relapse. An increased understanding regarding these risk factors will empower organizations and institutes to develop effective, and science-based programs that can help in reducing relapse rates.
Studies on genomic risk factors associated with relapse to SUD revealed notable gaps in knowledge, with several points needing further clarification. Current studies focus on a limited number of genes contributing to SUD relapse, a broader genome-wide analysis is needed to map the genetic architecture of relapse to SUD and potential interaction between candidate genes involved in relapse. Moreover, most GWAS studies have focused on the genetic variants contributing to the development of SUD not to its relapse. In general, the genetic landscape influencing relapse has not been thoroughly explored.
The exact epigenetic modifications and mechanisms by which they influence relapse is not understood yet and worth further exploration. EWAS are necessary to identify these modifications and integrating them with GWAS will help in identifying genetic basis of the variant that may contribute to relapse to SUD. The current literature lacks information regarding the dynamic and plastic nature of epigenetic modification throughout the treatment journey, during abuse, withdrawal, and long-term abstinence, and whether these modifications are reversible with time of abstinence and/or biopsychosocial interventions.
The interplay between various genetic and epigenetic factors needs in-depth exploration to obtain a comprehensive understanding of how they influence each other and various biological systems, ultimately increasing the relapse rate. Also integrating genomic data with other omics data can help in developing biomarkers for relapse to SUD. There is also a lack of population diversity in literature, which limits the generalizability of the findings. This field needs a large-scale and diverse population sample size to ensure that research findings are representative and applicable to various ethnicities. Longitudinal studies are also needed to elucidate the dynamic nature of relapse and its interaction with genomic factors over extended duration.
To gain a more comprehensive understanding of relapse risk, future research should examine the relation between biological, psychological, sociodemographic, and genetic and epigenetic factors. This information can be used to develop personalized interventions for SUD patients to reduce their relapse risk and improve their treatment outcomes.
Author Contributions
NAM: Conceptualization, Methodology, Resources, Writing – original draft, Writing − Review & Editing. HAA: Conceptualization, Methodology, Data Curation, Supervision, validation, Writing − Review & Editing. HAS: Resources, Writing − Review & Editing, Visualization. MOA: Resources, Writing − Review & Editing, Visualization.
Declaration of AI-assisted technologies in the writing process.
During the preparation of this work the authors used ChatGPT in order to improve the language of certain paragraphs. After using this tool, the authors reviewed and further edited the content as needed and take full responsibility for the content of the publication.
CRediT authorship contribution statement
Noora Al-Marzooqi: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Hanan Al-Suhail: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation. Mohammad O. AlRefai: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis, Data curation. Hamid A Alhaj: Writing – review & editing, Writing – original draft, Validation, Supervision, Methodology, Formal analysis, Data curation, Conceptualization.
Funding
This research did not receive any specific funding from any agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Alchakee A., Ahmed M., Eldohaji L., Alhaj H., Saber-Ayad M. Pharmacogenomics in psychiatry practice: The value and the challenges. International Journal of Molecular Sciences. 2022;23(21):13485. doi: 10.3390/ijms232113485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P.R. What is a functional genetic polymorphism? Defining classes of functionality. Journal of Psychiatry & Neuroscience : JPN. 2011;36(6):363–365. doi: 10.1503/jpn.110137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alblooshi, H., Al Safar, H., Fisher, H. F., Cordell, H. J., El Kashef, A., Al Ghaferi, H., Shawky, M., Reece, S., Hulse, G. K., & Tay, G. K. (2019). A case-control genome wide association study of substance use disorder (SUD) identifies novel variants on chromosome 7p14.1 in patients from the United Arab Emirates (UAE). American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 180(1), 68-79. 10.1002/ajmg.b.32708. [DOI] [PubMed]
- Alblooshi H., Hulse G., Osman W., El Kashef A., Shawky M., Al Ghaferi H., Al Safar H., Tay G.K. The frequency of DRD2 rs1076560 and OPRM1 rs1799971 in substance use disorder patients from the United Arab Emirates. Annals of General Psychiatry. 2018;17:22. doi: 10.1186/s12991-018-0192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eitan L.N., Rababa'h D.M., Alghamdi M.A. Genetic susceptibility of opioid receptor genes polymorphism to drug addiction: A candidate-gene association study. BMC Psychiatry. 2021;21(1):5. doi: 10.1186/s12888-020-03006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona B.J., Cleaveland N.A., Stuber G.D., Day J.J., Carelli R.M., Wightman R.M. Preferential enhancement of dopamine transmission within the nucleus accumbens shell by cocaine is attributable to a direct increase in phasic dopamine release events. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(35):8821–8831. doi: 10.1523/JNEUROSCI.2225-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A.J., Covault J., Feinn R., Pond T., Yang B., Ge W., Oncken C., Kranzler H.R. A GABRA2 Variant Is Associated with Increased Stimulation and ‘High’ Following Alcohol Administration. Alcohol and Alcoholism (Oxford, Oxfordshire) 2014;49(1):1–9. doi: 10.1093/alcalc/agt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Carrión O., Stamelou M., Murillo-Rodríguez E., Menéndez-González M., Pöppel E. Dopaminergic reward system: A short integrative review. International Archives of Medicine. 2010;3:24. doi: 10.1186/1755-7682-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszalos R., McDuff D.R., Weintraub E., Montoya I., Schwartz R. Engaging hospitalized heroin-dependent patients into substance abuse treatment. Journal of Substance Abuse Treatment. 1999;17(1–2):149–158. doi: 10.1016/s0740-5472(98)00075-0. [DOI] [PubMed] [Google Scholar]
- Bauer L.O., Covault J., Gelernter J. GABRA2 and KIBRA genotypes predict early relapse to substance use. Drug and Alcohol Dependence. 2012;123(1–3):154–159. doi: 10.1016/j.drugalcdep.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin D., Belin-Rauscent A., Murray J.E., Everitt B.J. Addiction: Failure of control over maladaptive incentive habits. Current Opinion in Neurobiology. 2013;23(4):564–572. doi: 10.1016/j.conb.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Belin D., Jonkman S., Dickinson A., Robbins T.W., Everitt B.J. Parallel and interactive learning processes within the basal ganglia: Relevance for the understanding of addiction. Behavioural Brain Research. 2009;199(1):89–102. doi: 10.1016/j.bbr.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Berridge K.C. From prediction error to incentive salience: Mesolimbic computation of reward motivation. The European Journal of Neuroscience. 2012;35(7):1124–1143. doi: 10.1111/j.1460-9568.2012.07990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N.P., Klose R. CpG island chromatin: A platform for gene regulation. Epigenetics. 2011;6(2):147–152. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K., Modestino E.J., Badgaiyan R.D., Baron D., Thanos P.K., Elman I., Siwicki D., Febo M., Gold M.S. Analysis of Evidence for the Combination of Pro-dopamine Regulator (KB220PAM) and Naltrexone to Prevent Opioid Use Disorder Relapse. EC Psychology and Psychiatry. 2018;7(8):564–579. [PMC free article] [PubMed] [Google Scholar]
- Blum K., Werner T., Carnes S., Carnes P., Bowirrat A., Giordano J., Gold M. Sex, Drugs, and Rock ‘N’ Roll: Hypothesizing Common Mesolimbic Activation as a Function of Reward Gene Polymorphisms. Journal of Psychoactive Drugs. 2012;44(1):38–55. doi: 10.1080/02791072.2012.662112. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4040958/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border R., Johnson E.C., Evans L.M., Smolen A., Berley N., Sullivan P.F., Keller M.C. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. American Journal of Psychiatry. 2019;176(5):376–387. doi: 10.1176/appi.ajp.2018.18070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S.J., Schacht J.P., Prisciandaro J.J., Voronin K., Anton R.F. Alcohol-Induced Stimulation Mediates the Effect of a GABRA2 SNP on Alcohol Self-Administrated among Alcohol-Dependent Individuals. Alcohol and Alcoholism (Oxford, Oxfordshire) 2016;51(5):549–554. doi: 10.1093/alcalc/agw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braida D., Iosuè S., Pegorini S., Sala M. Delta9-tetrahydrocannabinol-induced conditioned place preference and intracerebroventricular self-administration in rats. European Journal of Pharmacology. 2004;506(1):63–69. doi: 10.1016/j.ejphar.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Browne C.J., Godino A., Salery M., Nestler E.J. Epigenetic Mechanisms of Opioid Addiction. Biological Psychiatry. 2020;87(1):22–33. doi: 10.1016/j.biopsych.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington S.A., Huang W., Costa-Mattioli M. Translational control in synaptic plasticity and cognitive dysfunction. Annual Review of Neuroscience. 2014;37:17–38. doi: 10.1146/annurev-neuro-071013-014100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J.L. Epigenetics of Stress, Addiction, and Resilience: Therapeutic Implications. Molecular Neurobiology. 2016;53(1):545–560. doi: 10.1007/s12035-014-9040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camí J., Farré M. Drug addiction. The New England Journal of Medicine. 2003;349(10):975–986. doi: 10.1056/NEJMra023160. [DOI] [PubMed] [Google Scholar]
- Cardinal R.N., Everitt B.J. Neural and psychological mechanisms underlying appetitive learning: Links to drug addiction. Current Opinion in Neurobiology. 2004;14(2):156–162. doi: 10.1016/j.conb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Carle T.L., Ohnishi Y.N., Ohnishi Y.H., Alibhai I.N., Wilkinson M.B., Kumar A., Nestler E.J. Proteasome-dependent and -independent mechanisms for FosB destabilization: Identification of FosB degron domains and implications for DeltaFosB stability. The European Journal of Neuroscience. 2007;25(10):3009–3019. doi: 10.1111/j.1460-9568.2007.05575.x. [DOI] [PubMed] [Google Scholar]
- Chang X., Sun Y., Muhai J., Li Y., Chen Y., Lu L., Chang S., Shi J. Common and distinguishing genetic factors for substance use behavior and disorder: An integrated analysis of genomic and transcriptomic studies from both human and animal studies. Addiction (Abingdon, England) 2022;117(9):2515–2529. doi: 10.1111/add.15908. [DOI] [PubMed] [Google Scholar]
- Cheng J., He Z., Chen Q., Lin J., Peng Y., Zhang J., Yan X., Yan J., Niu S. Histone modifications in cocaine, methamphetamine and opioids. Heliyon. 2023;9(6):e16407. doi: 10.1016/j.heliyon.2023.e16407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley B.N., Wang X., Campbell M.R., Pittman G.S., Noureddine M.A., Bell D.A. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: Current and developing technologies. Mutation Research. 2008;659(1–2):147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M.J. Cellular neuroadaptations to chronic opioids: Tolerance, withdrawal and addiction. British Journal of Pharmacology. 2008;154(2):384–396. doi: 10.1038/bjp.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T., Crist R.C., Kampman K.M., Dackis C.A., Pettinati H.M., O’Brien C.P., Oslin D.W., Ferraro T.N., Lohoff F.W., Berrettini W.H. Low frequency genetic variants in the mu-opioid receptor (OPRM1) affect risk for addiction to heroin and cocaine. Neuroscience Letters. 2013;542:71–75. doi: 10.1016/j.neulet.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzoli D., Daponte A., Fazio S., Ariano V., Quaranta M., Leone V., Ostuni A., Casanova M., Catacchio C.R., Ventura M., Montinaro F. Biomedicines | Free Full-Text | Genomic and Personalized Medicine Approaches for Substance Use Disorders (SUDs) Looking at Genome-Wide Association Studies. Biomedicines. 2021;9(12):1799. doi: 10.3390/biomedicines9121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J.J., Oslin D.W., Patkar A.A., Gottheil E., DeMaria P.A., O'Brien C.P., Berrettini W.H., Grice D.E. A genetic association study of the mu opioid receptor and severe opioid dependence. Psychiatric Genetics. 2003;13(3):169–173. doi: 10.1097/00041444-200309000-00006. [DOI] [PubMed] [Google Scholar]
- Dahlgren A., Wargelius H., Berglund K.J., Fahlke C., Blennow K., Zetterberg H., Oreland L., Berggren U., Balldin J. Do alcohol-dependent individuals with DRD2 A1 allele have an increased risk of relapse? A pilot study. Alcohol and Alcoholism (Oxford, Oxfordshire) 2011;46(5):509–513. doi: 10.1093/alcalc/agr045. [DOI] [PubMed] [Google Scholar]
- Deak J.D., Johnson E.C. Genetics of substance use disorders: A review | Psychological Medicine | Cambridge Core. Psychological Medicine. 2021;51(13):2189–2200. doi: 10.1017/S0033291721000969. https://www.cambridge.org/core/journals/psychological-medicine/article/genetics-of-substance-use-disorders-a-review/B3BAE9D2DCF78C7C4833A8DB4420F5B2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak J.D., Zhou H., Galimberti M., Levey D.F., Wendt F.R., Sanchez-Roige S., Hatoum A.S., Johnson E.C., Nunez Y.Z., Demontis D., Børglum A.D., Rajagopal V.M., Jennings M.V., Kember R.L., Justice A.C., Edenberg H.J., Agrawal A., Polimanti R., Kranzler H.R., Gelernter J. Genome-wide association study in individuals of European and African ancestry and multi-trait analysis of opioid use disorder identifies 19 independent genome-wide significant risk loci. Molecular Psychiatry. 2022;27(10):3970–3979. doi: 10.1038/s41380-022-01709-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N., Zhou H., Fan H., Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8(66):110635–110649. doi: 10.18632/oncotarget.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G., Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(14):5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G., Bassareo V., Fenu S., De Luca M.A., Spina L., Cadoni C., Acquas E., Carboni E., Valentini V., Lecca D. Dopamine and drug addiction: The nucleus accumbens shell connection. Neuropharmacology. 2004;47(Suppl 1):227–241. doi: 10.1016/j.neuropharm.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Diagnostic and statistical manual of mental disorders : DSM-5™ (2013). (5th edition. ed.). American Psychiatric Publishing, a division of American Psychiatric Association.
- Dong C., Upadhya S.C., Ding L., Smith T.K., Hegde A.N. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learning & Memory. 2008;15(5):335–347. doi: 10.1101/lm.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogou C., Sauvet F., Erblang M., Detemmerman L., Derbois C., Erkel M.C., Boland A., Deleuze J.F., Gomez-Merino D., Chennaoui M. Genotyping on blood and buccal cells using loop-mediated isothermal amplification in healthy humans. Biotechnology Reports. 2020;26:e00468. doi: 10.1016/j.btre.2020.e00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert V., Pruessner J.C. Dopaminergic and Noradrenergic Contributions to Functionality in ADHD: The Role of Methylphenidate. Current Neuropharmacology. 2008;6(4):322–328. doi: 10.2174/157015908787386069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell M.S., Werge T., Sklar P., Owen M.J., Ophoff R.A., O'Donovan M.C., Sullivan P.F. Evaluating historical candidate genes for schizophrenia. Molecular psychiatry. 2015;20(5):555–562. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febo M., Blum K., Badgaiyan R.D., Baron D., Thanos P.K., Colon-Perez L.M., Demortrovics Z., Gold M.S. Dopamine homeostasis: Brain functional connectivity in reward deficiency syndrome. Frontiers in Bioscience (Landmark Edition) 2017;22(4):669–691. doi: 10.2741/4509. [DOI] [PubMed] [Google Scholar]
- Fehr C., Sander T., Tadic A., Lenzen K.P., Anghelescu I., Klawe C., Dahmen N., Schmidt L.G., Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatric Genetics. 2006;16(1):9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Fioravante D., Byrne J.H. Protein degradation and memory formation. Brain Research Bulletin. 2011;85(1–2):14–20. doi: 10.1016/j.brainresbull.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.F. Tolerance to and physical dependence on opiates, barbiturates, and alcohol. Annual Review of Medicine. 1957;8:427–440. doi: 10.1146/annurev.me.08.020157.002235. [DOI] [PubMed] [Google Scholar]
- Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., Johnson R., Zielke H.R., Ferrucci L., Longo D.L., Cookson M.R., Singleton A.B. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genetics. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giessen E.M., v. d., M.M.L.D. M.W.T., Win Tanck, Brink W.V.D., Baas F., Booij J. Striatal Dopamine Transporter Availability Associated with Polymorphisms in the Dopamine Transporter Gene SLC6A3. Journal of Nuclear Medicine. 2009;50(1):45–52. doi: 10.2967/jnumed.108.053652. [DOI] [PubMed] [Google Scholar]
- Gipson C.D., Beckmann J.S. Compulsion and substance use disorder: Potential importance of boundary conditions. Neuropsychopharmacology: Official Publication of the American College of. Neuropsychopharmacology. 2023;48(3):432–433. doi: 10.1038/s41386-022-01462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorwood P., Batel P., Adès J., Hamon M., Boni C. Serotonin transporter gene polymorphisms, alcoholism, and suicidal behavior. Biological Psychiatry. 2000;48(4):259–264. doi: 10.1016/s0006-3223(00)00840-4. [DOI] [PubMed] [Google Scholar]
- Grzywacz A., Chmielowiec J., Chmielowiec K., Mroczek B., Masiak J., Suchanecka A., Sipak-Szmigiel O., Szumilas K., Trybek G. The Ankyrin Repeat and Kinase Domain Containing 1 Gene Polymorphism (ANKK1Taq1A) and Personality Traits in Addicted Subjects. International Journal of Environmental Research and Public Health. 2019;16(15):2687. doi: 10.3390/ijerph16152687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerian B.S., Haerian M.S. OPRM1 rs1799971 polymorphism and opioid dependence: Evidence from a meta-analysis. Pharmacogenomics. 2013;14(7):813–824. doi: 10.2217/pgs.13.57. [DOI] [PubMed] [Google Scholar]
- Huang W., Placzek A.N., Viana Di Prisco G., Khatiwada S., Sidrauski C., Krnjević K., Walter P., Dani J.A., Costa-Mattioli M. Translational control by eIF2α phosphorylation regulates vulnerability to the synaptic and behavioral effects of cocaine. 2016;eLife, 5:e12052. doi: 10.7554/eLife.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman S.E., Malenka R.C. Addiction and the brain: The neurobiology of compulsion and its persistence. Nature Reviews Neuroscience. 2001;2(10):695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- Hyman S.E., Malenka R.C., Nestler E.J. Neural mechanisms of addiction: The role of reward-related learning and memory. Annual Review of Neuroscience. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Jorgensen T.J., Ruczinski I., Kessing B., Smith M.W., Shugart Y.Y., Alberg A.J. Hypothesis-Driven Candidate Gene Association Studies: Practical Design and Analytical Considerations. American Journal of Epidemiology. 2009;170(8):986–993. doi: 10.1093/aje/kwp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kember R.L., Vickers-Smith R., Xu H., Toikumo S., Niarchou M., Zhou H., Hartwell E.E., Crist R.C., Rentsch C.T., Davis L.K., Justice A.C., Sanchez-Roige S., Kampman K.M., Gelernter J., Kranzler H.R. Cross-ancestry meta-analysis of opioid use disorder uncovers novel loci with predominant effects in brain regions associated with addiction. Nature Neuroscience. 2022;25(10):1279–1287. doi: 10.1038/s41593-022-01160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P.J., Feng J., Robison A.J., Maze I., Badimon A., Mouzon E., Chaudhury D., Damez-Werno D.M., Haggarty S.J., Han M., Bassel-Duby R., Olson E.N., Nestler E.J. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nature Neuroscience. 2013;16(4):434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller G., Zill P., Rujescu D., Ridinger M., Pogarell O., Fehr C., Wodarz N., Bondy B., Soyka M., Preuss U.W. Possible association between OPRM1 genetic variance at the 118 locus and alcohol dependence in a large treatment sample: Relationship to alcohol dependence symptoms. Alcoholism, Clinical and Experimental Research. 2012;36(7):1230–1236. doi: 10.1111/j.1530-0277.2011.01714.x. [DOI] [PubMed] [Google Scholar]
- Land M.A., Ramesh D., Miller A.L., Pyles R.B., Cunningham K.A., Moeller F.G., Anastasio N.C. Methylation Patterns of the HTR2A Associate With Relapse-Related Behaviors in Cocaine-Dependent Participants. Frontiers in Psychiatry. 2020;11:532. doi: 10.3389/fpsyt.2020.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J., Becker J.A., Befort K., Kieffer B.L. Reward Processing by the Opioid System in the Brain | Physiological Reviews. Physiological Reviews. 2009;89(4) doi: 10.1152/physrev.00005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea A.J., Vockley C.M., Johnston R.A., Del Carpio C.A., Barreiro L.B., Reddy T.E., Tung J. Genome-wide quantification of the effects of DNA methylation on human gene regulation. 2018;eLife doi: 10.7554/eLife.37513. 710.7554/eLife.37513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Choi J., Lee N., Lee H., Kim J., Yu N., Choi S., Lee S., Kim H., Kaang B. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science (New York, N.Y.) 2008;319(5867):1253–1256. doi: 10.1126/science.1150541. [DOI] [PubMed] [Google Scholar]
- López A.J., Hemstedt T.J., Jia Y., Hwang P.H., Campbell R.R., Kwapis J.L., White A.O., Chitnis O., Scarfone V.M., Matheos D.P., Lynch G., Wood M.A. Epigenetic regulation of immediate-early gene Nr4a2/Nurr1 in the medial habenula during reinstatement of cocaine-associated behavior. Neuropharmacology. 2019;153:13–19. doi: 10.1016/j.neuropharm.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler S.V., Berridge K.C. Which Cue to “Want?” Central Amygdala Opioid Activation Enhances and Focuses Incentive Salience on a Prepotent Reward Cue. The Journal of Neuroscience. 2009;29(20):6500–6513. doi: 10.1523/JNEUROSCI.3875-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M., Mhillaj E., Matheos D.P., Palmery M., Wood M.A. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2011;31(47):16941–16948. doi: 10.1523/JNEUROSCI.2747-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.L., Grant P.A. The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. Sub-Cellular Biochemistry. 2013;61:289–317. doi: 10.1007/978-94-007-4525-4_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton A.L., Everitt B.J. The persistence of maladaptive memory: Addiction, drug memories and anti-relapse treatments. Neuroscience and Biobehavioral Reviews. 2012;36(4):1119–1139. doi: 10.1016/j.neubiorev.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Moe F.D., Moltu C., McKay J.R., Nesvåg S., Bjornestad J. Is the relapse concept in studies of substance use disorders a 'one size fits all' concept? A systematic review of relapse operationalisations. Drug and Alcohol Review. 2022;41(4):743–758. doi: 10.1111/dar.13401. [DOI] [PubMed] [Google Scholar]
- Moeller S.J., Parvaz M.A., Shumay E., Beebe-Wang N., Konova A.B., Alia-Klein N., Volkow N.D., Goldstein R.Z. Gene × Abstinence Effects on Drug Cue Reactivity in Addiction: Multimodal Evidence. The Journal of Neuroscience. 2013;33(24):10027–10036. doi: 10.1523/JNEUROSCI.0695-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannini D.R., Zheng Y., Joyce B.T., Kim K., Gao T., Wang J., Jacobs D.R., Schreiner P.J., Yaffe K., Greenland P., Lloyd-Jones D.M., Hou L. Genome-wide DNA methylation association study of recent and cumulative marijuana use in middle aged adults. Molecular Psychiatry. 2023;1–11 doi: 10.1038/s41380-023-02106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute On Drug Abuse. (2020, Drug Misuse and Addiction. National Institute on Drug Abuse. Retrieved Oct 19, 2023, from https://nida.nih.gov/publications/drugs-brains-behavior-science-addiction/drug-misuse-addiction.
- Nestler E.J. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76 Pt B(0 0):259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler E.J. Transgenerational Epigenetic Contributions to Stress Responses: Fact or Fiction? PLoS Biology. 2016;14(3):e1002426. doi: 10.1371/journal.pbio.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler E.J., Lüscher C. The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron. 2019;102(1):48–59. doi: 10.1016/j.neuron.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Bird A. DNA methylation and chromatin modification. Current Opinion in Genetics & Development. 1999;9(2):158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Nielsen D.A., Hamon S., Yuferov V., Jackson C., Ho A., Ott J., Kreek M.J. Ethnic diversity of DNA methylation in the OPRM1 promoter region in lymphocytes of heroin addicts. Human Genetics. 2010;127(6):639–649. doi: 10.1007/s00439-010-0807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble E.P., Blum K., Ritchie T., Montgomery A., Sheridan P.J. Allelic association of the D2 dopamine receptor gene with receptor-binding characteristics in alcoholism. Archives of General Psychiatry. 1991;48(7):648–654. doi: 10.1001/archpsyc.1991.01810310066012. [DOI] [PubMed] [Google Scholar]
- Oberacher H., Pitterl F., Niederstätter H., Weiss E.M., Stadelmann E., Marksteiner J., Parson W. Direct molecular haplotyping of multiple polymorphisms within exon 4 of the human catechol-O-methyltransferase gene by liquid chromatography-electrospray ionization time-of-flight mass spectrometry. Analytical and Bioanalytical Chemistry. 2006;386(1):83–91. doi: 10.1007/s00216-006-0589-9. [DOI] [PubMed] [Google Scholar]
- Osborne A.J., Pearson J.F., Noble A.J., Gemmell N.J., Horwood L.J., Boden J.M., Benton M.C., Macartney-Coxson D.P., Kennedy M.A. Genome-wide DNA methylation analysis of heavy cannabis exposure in a New Zealand longitudinal cohort. Translational Psychiatry. 2020;10(1):114. doi: 10.1038/s41398-020-0800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu Obeng A., Hamadeh I., Smith M. Review of Opioid Pharmacogenetics and Considerations for Pain Management. Pharmacotherapy. 2017;37(9):1105–1121. doi: 10.1002/phar.1986. [DOI] [PubMed] [Google Scholar]
- Padmanabhan A., Geier C.F., Ordaz S.J., Teslovich T., Luna B. Developmental changes in brain function underlying the influence of reward processing on inhibitory control. Developmental Cognitive Neuroscience. 2011;1(4):517–529. doi: 10.1016/j.dcn.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios A., Canto P., Tejeda M.E., Stephano S., Luján H., García-García E., Rojano-Mejía D., Méndez J.P. Complete sequence of the ANKK1 gene in Mexican-Mestizo individuals with obesity, with or without binge eating disorder. European Psychiatry: The Journal of the Association of European Psychiatrists. 2018;54:59–64. doi: 10.1016/j.eurpsy.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Pasareanu A.R., Opsal A., Vederhus J., Kristensen Ø., Clausen T. Quality of life improved following in-patient substance use disorder treatment. Health and Quality of Life Outcomes. 2015;13:35. doi: 10.1186/s12955-015-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peciña M., Love T., Stohler C.S., Goldman D., Zubieta J. Effects of the Mu Opioid Receptor Polymorphism (OPRM1 A118G) on Pain Regulation, Placebo Effects and Associated Personality Trait Measures. Neuropsychopharmacology. 2015;40(4):957–965. doi: 10.1038/npp.2014.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezze M.A., Feldon J. Mesolimbic dopaminergic pathways in fear conditioning. Progress in Neurobiology. 2004;74(5):301–320. doi: 10.1016/j.pneurobio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Philibert R.A., Gunter T.D., Beach S.R., Brody G., Madan A. MAOA Methylation is Associated with Nicotine and Alcohol Dependence in Women. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics : The Official Publication of the International Society of. Psychiatric Genetics. 2008;(5):565–570. doi: 10.1002/ajmg.b.30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert R.A., Plume J.M., Gibbons F.X., Brody G.H., Beach S.R.H. The impact of recent alcohol use on genome wide DNA methylation signatures. Frontiers in Genetics. 2012;3:54. doi: 10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau G., Villemonteix T., Slama H., Kavec M., Balériaux D., Metens T., Baijot S., Mary A., Ramoz N., Gorwood P., Peigneux P., Massat I. Dopamine transporter genotype modulates brain activity during a working memory task in children with ADHD. Research in Developmental Disabilities. 2019;92 doi: 10.1016/j.ridd.2019.103430. [DOI] [PubMed] [Google Scholar]
- Pinto E., Reggers J., Gorwood P., Boni C., Scantamburlo G., Pitchot W., Ansseau M. The Short Allele of the Serotonin Transporter Promoter Polymorphism Influences Relapse in Alcohol Dependence. Alcohol & Alcoholism. 2008;43(4):398–400. doi: 10.1093/alcalc/agn015. [DOI] [PubMed] [Google Scholar]
- Polyak, K., & Meyerson, M. (2003). Holland-Frei Cancer Medicine (6th ed.). BC Decker.
- Quik M., Perez X.A., Grady S.R. Role of alpha6 nicotinic receptors in CNS dopaminergic function; relevance to addiction and neurological disorders. Biochemical Pharmacology. 2011;82(8):873–882. doi: 10.1016/j.bcp.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R., Ruparel K., Newberg A., Wileyto E.P., Loughead J.W., Divgi C., Blendy J.A., Logan J., Zubieta J., Lerman C. Human Mu Opioid Receptor (OPRM1 A118G) polymorphism is associated with brain mu-opioid receptor binding potential in smokers. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):9268–9273. doi: 10.1073/pnas.1018699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton C.L., Davey Smith G. Epigenetic Epidemiology of Common Complex Disease: Prospects for Prediction, Prevention, and Treatment. PLoS Medicine. 2010;7(10):e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W., Kumar A., Xiao G., Wilkinson M., Covington H.E., Maze I., Sikder D., Robison A.J., LaPlant Q., Dietz D.M., Russo S.J., Vialou V., Chakravarty S., Kodadek T.J., Stack A., Kabbaj M., Nestler E.J. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62(3):335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie T., Noble E.P. [3H]naloxone binding in the human brain: Alcoholism and the TaqI A D2 dopamine receptor polymorphism. Brain Research. 1996;718(1–2):193–197. doi: 10.1016/0006-8993(96)00068-6. [DOI] [PubMed] [Google Scholar]
- Roh S., Matsushita S., Hara S., Maesato H., Matsui T., Suzuki G., Miyakawa T., Ramchandani V.A., Li T., Higuchi S. Role of GABRA2 in Moderating Subjective Responses to Alcohol. Alcoholism, Clinical and Experimental Research. 2011;35(3):400–407. doi: 10.1111/j.1530-0277.2010.01357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu P., Deschatrettes E., Host L., Gobaille S., Sandner G., Zwiller J. The Inhibition of Histone Deacetylases Reduces the Reinstatement of Cocaine-Seeking Behavior in Rats. Current Neuropharmacology. 2011;9(1):21–25. doi: 10.2174/157015911795017317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu P., Host L., Gobaille S., Sandner G., Aunis D., Zwiller J. Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(38):9342–9348. doi: 10.1523/JNEUROSCI.0379-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinen-Lagerström N., Lahti J., Alho H., Kovanen L., Aalto M., Partonen T., Silander K., Sinclair D., Räikkönen K., Eriksson J.G., Palotie A., Koskinen S., Saarikoski S.T. µ-Opioid Receptor Gene (OPRM1) Polymorphism A118G: Lack of Association in Finnish Populations with Alcohol Dependence or Alcohol Consumption. Alcohol and Alcoholism (Oxford, Oxfordshire) 2013;48(5):519–525. doi: 10.1093/alcalc/agt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander T., Harms H., Lesch K.P., Dufeu P., Kuhn S., Hoehe M., Rommelspacher H., Schmidt L.G. Association analysis of a regulatory variation of the serotonin transporter gene with severe alcohol dependence. Alcoholism, Clinical and Experimental Research. 1997;21(8):1356–1359. [PubMed] [Google Scholar]
- Sasabe T., Furukawa A., Matsusita S., Higuchi S., Ishiura S. Association analysis of the dopamine receptor D2 (DRD2) SNP rs1076560 in alcoholic patients. Neuroscience Letters. 2007;412(2):139–142. doi: 10.1016/j.neulet.2006.10.064. [DOI] [PubMed] [Google Scholar]
- See R.E. Neural substrates of cocaine-cue associations that trigger relapse. European Journal of Pharmacology. 2005;526(1–3):140–146. doi: 10.1016/j.ejphar.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Shastry B.S. SNPs: Impact on gene function and phenotype. Methods in Molecular Biology (Clifton. N.J.) 2009;578:3–22. doi: 10.1007/978-1-60327-411-1_1. [DOI] [PubMed] [Google Scholar]
- Shepard R.D., Nugent F.S. Early Life Stress- and Drug-Induced Histone Modifications Within the Ventral Tegmental Area. Frontiers in Cell and Developmental Biology. 2020;8 doi: 10.3389/fcell.2020.588476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R. Chronic Stress, Drug Use, and Vulnerability to Addiction. Annals of the New York Academy of Sciences. 2008;1141:105–130. doi: 10.1196/annals.1441.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R., Heilig M. Addiction and its brain science. Addiction (Abingdon, England) 2005;100(12):1813–1822. doi: 10.1111/j.1360-0443.2005.01260.x. [DOI] [PubMed] [Google Scholar]
- Su H., Li Z., Du J., Jiang H., Chen Z., Sun H., Zhao M. Predictors of heroin relapse: Personality traits, impulsivity, COMT gene val158met polymorphism in a 5-year prospective study in Shanghai, China. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2015;168(8):712–719. doi: 10.1002/ajmg.b.32376. [DOI] [PubMed] [Google Scholar]
- Suchanecka A., Chmielowiec J., Chmielowiec K., Masiak J., Sipak-Szmigiel O., Sznabowicz M., Czarny W., Michałowska-Sawczyn M., Trybek G., Grzywacz A. Dopamine Receptor DRD2 Gene rs1076560, Personality Traits and Anxiety in the Polysubstance Use Disorder. Brain Sciences. 2020;10(5):262. doi: 10.3390/brainsci10050262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wolf M.E. Nucleus accumbens neurons exhibit synaptic scaling that is occluded by repeated dopamine pre-exposure. The European Journal of Neuroscience. 2009;30(4):539–550. doi: 10.1111/j.1460-9568.2009.06852.x. [DOI] [PubMed] [Google Scholar]
- Sun Y., Zhang Y., Zhang D., Chang S., Jing R., Yue W., Lu L., Chen D., Sun Y., Fan Y., Shi J. GABRA2 rs279858-linked variants are associated with disrupted structural connectome of reward circuits in heroin abusers. Translational Psychiatry. 2018;8(1):138. doi: 10.1038/s41398-018-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsell A. The μ-opioid receptor and treatment response to naltrexone. Alcohol and Alcoholism (Oxford, Oxfordshire) 2013;48(4):402–408. doi: 10.1093/alcalc/agt030. [DOI] [PubMed] [Google Scholar]
- Trigo J.M., Martin-García E., Berrendero F., Robledo P., Maldonado R. The endogenous opioid system: A common substrate in drug addiction. Drug and Alcohol Dependence. 2010;108(3):183–194. doi: 10.1016/j.drugalcdep.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Uhart M., Weerts E.M., McCaul M.E., Guo X., Yan X., Kranzler H.R., Li N., Wand G.S. GABRA2 markers moderate the subjective effects of alcohol. Addiction Biology. 2013;18(2):357–369. doi: 10.1111/j.1369-1600.2012.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaissière T., Sawan C., Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutation Research. 2008;659(1–2):40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Villafuerte S., Heitzeg M.M., Foley S., Yau W.-W., Majczenko K., Zubieta J., Zucker R.A., Burmeist M. Impulsiveness and insula activation during reward anticipation are associated with genetic variants in GABRA2 in a family sample enriched for alcoholism. Molecular Psychiatry. 2012;17(5):511–519. doi: 10.1038/mp.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey, J., Swagell, C. D., Hughes, I. P., Lawford, B. R., Young, R. M., & Morris, C. P. (2011). A novel SNP in COMT is associated with alcohol dependence but not opiate or nicotine dependence: a case control study | Behavioral and Brain Functions. Morris, C. P., 7, 1-6. https://link.springer.com/article/10.1186/1744-9081-7-51. [DOI] [PMC free article] [PubMed]
- Volkow N.D., Fowler J.S., Wang G.J., Hitzemann R., Logan J., Schlyer D.J., Dewey S.L., Wolf A.P. Decreased dopamine D2 receptor availability is associated with reduced frontal metabolism in cocaine abusers. Synapse (New York, N.Y.) 1993;14(2):169–177. doi: 10.1002/syn.890140210. [DOI] [PubMed] [Google Scholar]
- Volkow N.D., Fowler J.S., Wang G. The addicted human brain: Insights from imaging studies. Journal of Clinical Investigation. 2003;111(10):1444–1451. doi: 10.1172/JCI200318533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Koob G.F., McLellan A.T. Neurobiologic Advances from the Brain Disease Model of Addiction. The New England Journal of Medicine. 2016;374(4):363–371. doi: 10.1056/NEJMra1511480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zastrow M., Svingos A., Haberstock-Debic H., Evans C. Regulated endocytosis of opioid receptors: Cellular mechanisms and proposed roles in physiological adaptation to opiate drugs. Current Opinion in Neurobiology. 2003;13(3):348–353. doi: 10.1016/s0959-4388(03)00069-2. [DOI] [PubMed] [Google Scholar]
- Wang F., Simen A., Arias A., Lu Q., Zhang H. A large-scale meta-analysis of the association between the ANKK1/DRD2 Taq1A polymorphism and alcohol dependence. Human Genetics. 2013;132(3):347–358. doi: 10.1007/s00439-012-1251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wei C., Xiao F., Chang X., Zhang Y. Influences of COMT rs4680 and OPRM1 rs1799971 Polymorphisms on Chronic Postsurgical Pain, Acute Pain, and Analgesic Consumption After Elective Cesarean Delivery. The Clinical Journal of Pain. 2019;35(1):31–36. doi: 10.1097/AJP.0000000000000654. [DOI] [PubMed] [Google Scholar]
- Weerasinghe-Mudiyanselage P.D.E., Ang M.J., Kang S., Kim J., Moon C. Structural Plasticity of the Hippocampus in Neurodegenerative Diseases. International Journal of Molecular Sciences. 2022;23(6):3349. doi: 10.3390/ijms23063349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner C.T., Altshuler R.D., Shaham Y., Li X. Epigenetic Mechanisms in Drug Relapse. Biological Psychiatry. 2021;89(4):331–338. doi: 10.1016/j.biopsych.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willuhn I., Wanat M.J., Clark J.J., Phillips P.E.M. Dopamine signaling in the nucleus accumbens of animals self-administering drugs of abuse. Current Topics in Behavioral Neurosciences. 2010;3:29–71. doi: 10.1007/7854_2009_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock E.A., Lundahl L.H., Burmeister M., Greenwald M.K. Functional Mu Opioid Receptor Polymorphism (OPRM1 A118G) Associated With Heroin Use Outcomes in Caucasian Males: A Pilot Study. The American Journal on Addictions. 2015;24(4):329–335. doi: 10.1111/ajad.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Lexicon of alcohol and drug terms. World Health. 1994 Organization. [Google Scholar]
- YanLei D., YuQiang N., YuYuan L., Wan Y.J.Y. The association between the SLC6A3 VNTR 9-repeat allele and alcoholism—A meta-analysis. Alcoholism: Clinical and Experimental Research. 2011;35(9):1625–1634. doi: 10.1111/j.1530-0277.2011.01509.x. https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1530-0277.2011.01509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Rentsch C.T., Cheng Z., Kember R.L., Nunez Y.Z., Sherva R.M., Tate J.P., Dao C., Xu K., Polimanti R., Farrer L.A., Justice A.C., Kranzler H.R., Gelernter J. Association of OPRM1 Functional Coding Variant With Opioid Use Disorder. JAMA Psychiatry. 2020;77(10):1072. doi: 10.1001/jamapsychiatry.2020.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.