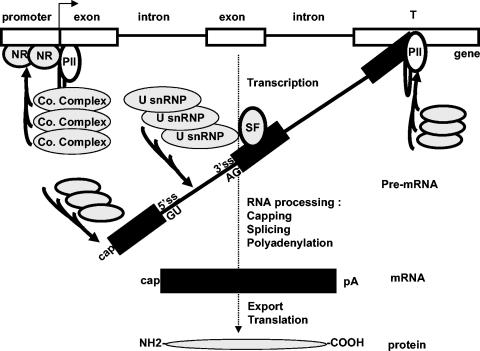
Gene expression is a multistep process starting in the cell nucleus with the synthesis of the primary transcripts that undergo several modifications (including capping, splicing, and polyadenylation) leading to the export of the mature mRNAs into the cytoplasm for translation into proteins. Although an emerging view is that all the steps from transcription to translation are mechanically and functionally coupled, the proteins that are involved in this coupled process are still poorly characterized. In recent years, a growing list of proteins known to control gene expression at the transcriptional level, named transcriptional coregulators, have been independently shown to play additional roles in other steps of gene expression. In this review we compile these emerging data suggesting that a subset of transcriptional coregulators play a major role in the coordination of the individual steps of the gene expression process.
GENE EXPRESSION REGULATION AND TRANSCRIPTIONAL COREGULATORS
Transcriptional stimuli, such as steroid hormones (i.e., androgens, progestins, estrogen, etc.), change the expression of their target genes by binding and modulating the activity of their nuclear receptors (NRs), which recognize and bind specific sequences within target gene promoters. NRs share a signature modular structure consisting of a C-terminal ligand-dependent transcriptional activation domain (AF-2), a central DNA binding domain, and an N-terminal ligand-independent transcriptional activation domain (AF-1) (55, 60). When bound to their target promoters, and like other transcription factors, NRs recruit coregulatory proteins termed coactivators or corepressors that activate or inhibit transcription. Since their discovery in the mid-1990s, the number of transcriptional coregulators has rapidly increased to more than 150. An exhaustive list of NR coregulators is available on the Nuclear Receptor Signaling Atlas website, http://www.nursa.org/index.cfm. Several reviews concerning coregulators have been recently published (5, 14, 29, 42, 60), and we will only briefly describe the background knowledge of their known roles in transcription.
NR coregulators, which were identified as proteins interacting with different NR domains and that can contain specific NR-interacting motifs such as LxxLL or the FxxLF motifs, are often present within dynamic and heterogeneous steady-state complexes (for example, the SRC, TRAP/SMCC/Mediator/SRB, CRSP, DRIP, and ASCOM complexes). Many coregulators are most likely recruited at the promoter level as part of these preformed complexes (14, 60, 62, 91). When present on target promoters, transcriptional coregulators play different roles either due to their specific enzymatic activities (e.g., kinase, acetyl- or methyltransferase, or ubiquitin- or sumo-ligase activities) or due to their ability to recruit other regulator proteins. Certain coregulators play a crucial role in remodeling chromatin structure by modifying histone tails or/and by promoting nucleosome remodeling, in turn facilitating the access of other proteins to the promoter. Finally, transcriptional coregulators recruit and stabilize the basal transcriptional machinery at the promoter, including RNA polymerase II (pol II), leading to the formation of the transcriptional preinitiation and initiation complexes (5, 14, 29, 42, 60). A few reports also have suggested that activated NRs modulate transcriptional elongation, although the potential coregulators involved are not well defined (41, 48, 96). Steroid hormones lead to the relocation of NRs to target promoters; the NRs serve as anchors for the subsequent sequential recruitment of different sets of coregulators, which are generally present in preformed but dynamic steady-state complexes. These coregulator complexes affect various rate-limiting transcriptional steps and lead to the assembly of the complete processive transcriptional machinery, including the general transcriptional factors and pol II. As described below, several transcriptional coregulators also have been implicated in steps that have been termed “posttranscriptional” but that are rather now considered “cotranscriptional” steps.
NUCLEAR RECEPTOR COREGULATORS RELATED TO PROTEINS INVOLVED IN SPLICING
Most eukaryotic genes contain protein coding information within exons separated by noncoding sequences or introns (Fig. 1). Prior to translation, the intronic sequences must be efficiently and accurately removed from the primary transcript or precursor of mRNA (pre-mRNA). The exons have to be joined together to form the translatable mRNA. This process, called splicing, is performed by a large and complex machinery, termed the spliceosome, containing 100 to 300 proteins (Fig. 1 and 2). Several reviews have described the structure of this machinery and the mechanism of splicing (28, 43, 100). Briefly, the spliceosome is made of five small nuclear ribonucleoprotein particles (snRNPs) called U1, U2, U4, U5, and U6, which are very stable and which have been purified (27, 53, 68, 83, 108). Each of the U snRNPs contains a small structured RNA (U1, U2, U4, U5, and U6 snRNAs) bound by 10 to 20 proteins, seven of which (the Sm proteins) are shared by different snRNPs. Numerous other, less stably associated splicing factors (SFs) participate in the formation of the spliceosome and in the splicing reaction, which is performed through sequential steps by virtue of the dynamic assembly and disassembly of complexes on the pre-mRNA substrate.
FIG. 1.
Gene expression is a multistep process, and each step is made by the sequential recruitment of small preformed complexes performing successive reactions. Transcription factors like NRs recognize response elements within target promoters and recruit transcriptional coregulators that exist in preformed small complexes (Co. Complex). The sequential recruitment of different sets of coregulators leads to the formation of the transcriptosome, which ultimately allows the RNA pol II (PII) to be fully processive. After the synthesis of 20 to 40 nt, the 5′ end of the transcript is methylated (cap) through the sequential action of three enzymes. This cap structure plays a role in stability and translation of the mRNA. Similarly, after recognition of sequences at the 3′ end of the gene, transcription stops (T, termination) and the transcript is cleaved upstream of the termination site. The 3′ end is modified by the addition of a polyadenylated tail (pA) that also plays a role in the stability and translation of the mRNA. This step is made by the sequential recruitment of small preformed complexes. As soon as the primary transcript emerges from the RNA pol II CTD, splicing regulatory sequences within exons or introns are recognized by SFs that recruit preformed small complexes (U snRNPs) on the pre-mRNA, which leads to the formation of the spliceosome. The splicing process is in turn coupled to the export of the mRNA translated in the cytosol.
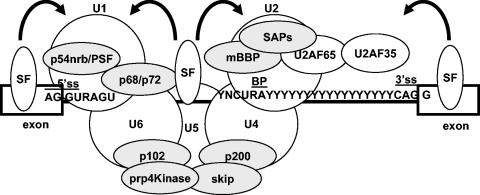
FIG. 2.
Transcriptional coregulators present within the spliceosome. SFs recognize exonic or intronic splicing regulatory sequences and help the recognition of neighboring splice sites (5′ SS and 3′ SS) by recruiting the spliceosome subcomplexes or snRNPs (U1, U2, U4, U5, and U6). Several proteins (shown within grey circles) that form part of the spliceosome are also transcriptional coregulators, which suggests that they could be recruited through the transcriptional machinery.
Transcriptional coregulators and the spliceosome.
One of the initial steps in the splicing process is the recognition of the 5′ splice site (SS), which is accomplished in part by the U1 snRNP and the base pairing of the U1 snRNA with the 5′ SS sequence (Fig. 2). This early U1-5′ SS duplex aids the assembly of the “prespliceosome” on the transcript and is then unwound to allow the formation of the U6-5′ SS duplex, leading to the creation of the “active” spliceosome. Interestingly, these two events require proteins that also can act as transcriptional coregulators.
p54nrb (p54 nuclear RNA binding protein) and PSF (polypyrimidine tract-binding protein-associated splicing factor) are two highly homologous RNA binding proteins that form a heterodimer and that likely play similar roles. These two proteins have been suspected to be involved in both transcription and splicing (92). It was reported that p54nrb and PSF (which contains an FXXLF motif) can directly interact with the androgen receptor (AR) and/or with other transcriptional coregulators. We and others have shown that they modulate NR transcriptional activity (3, 35, 58). Recently, it was reported that p54nrb and PSF directly interact with the 5′ SS and that this interaction occurs within large complexes that contain pol II and snRNPs (39). Since p54nrb and PSF directly interact with the pol II carboxy-terminal domain (CTD), these coregulators could serve as a “molecular link” between the transcriptional machinery, the 5′ SS recognition on the transcript, and the splicing machinery (18, 39).
p68 and p72 are two highly homologous proteins that form a heterodimer and that contain a DEAD-box domain characteristic of RNA helicase activities. The RNA helicase activity is required for unwinding RNA duplexes between two different RNAs or within RNA secondary structures (87). Many studies identified p68 and p72 as components of the spliceosome, although their function was only recently clarified (27, 53, 68, 83, 108). Indeed, p68 and p72 were shown to interact with the U1-5′ SS duplex to unwind it, allowing the U6-5′ SS association and the formation of the active spliceosome (51). The essential role of p68 in this process is underlined by the fact that depletion of this protein leads to accumulation of a prespliceosome complex (51). In addition, many studies have demonstrated the importance of p68 and p72 in transcription; in particular, both proteins contain an LXXLL motif and directly interact with NRs, and p68 was shown by chromatin immunoprecipitation to be recruited to a steroid-regulated promoter (19, 62, 89). These data, together with the fact that p68 directly interacts with the pol II CTD (89), suggest that p68 (and probably p72) also could be a “molecular link” between the transcriptional machinery and the recognition and eventual processing of the transcript in the active spliceosome at the 5′ SS, reinforcing the observations that the splicing events occurring at the 5′ SS are strongly connected to transcription (39, 46, 51, 95).
Another early stage in spliceosome assembly involves the 3′ SS which depends on the recognition of the branch point sequence (BPS) by the mammalian branch point binding protein (mBBP) and of the polypyrimidine tract (PY) by the U2 auxiliary factors U2AF65 and U2AF35 (Fig. 2). Once bound to the transcript, these proteins facilitate the association of the U2 snRNP at the 3′ end of the intron through the base pairing of the U2 snRNA on the BPS. Interestingly, several proteins participating in the recognition of the 3′ end of the intron also have been identified within transcriptional complexes.
For instance, mBBP was identified as a potential transcriptional regulator that interacts with the transcriptional elongation factor CA150 and the TET family proteins (24, 106). It was proposed that the function of mBBP in splicing could be coupled to transcriptional elongation (24). In this respect, it is important to underline that the recognition of the BPS by mBBP is facilitated by the simultaneous recognition of the PY tract by U2AF65 (Fig. 2). Interestingly, several proteins structurally related to U2AF65 have been recently implicated in transcription. For instance, PUF60, first identified based on its functional and structural homology with U2AF65 (78), was later identified as “FBP-interacting repressor,” a protein that modulates TFIIH factor activity (50). Moreover, CAPER was reported to be a protein that interacts with the estrogen receptor (ER) and is able to coactivate its transcriptional activity (37). This protein, also known as CC1.3, is highly homologous to the SF U2AF65 and was purified recently as a spliceosome component capable of affecting the splicing reaction (2, 27, 83). We recently identified another U2AF65-related protein, CAPERβ, and we showed that both CAPER and CAPERβ are NR coactivators that enter the splicing process via recruitment to the transcriptional machinery by NRs (16).
Finally, several proteins of the SF3 complex, which is a multisubunit component of the U2 snRNP, have been identified within transcriptional complexes. SF3a120 and SF3b130 were purified as part of several transcriptional complexes (9, 57, 97), and the SF3b14b protein was recently cloned as an ER coregulator (74, 75). Altogether, these observations suggest that several proteins could be potential molecular links between transcription, 3′ SS recognition, and U2 snRNP recruitment.
The U5 snRNP enters the spliceosome after the U1 and U2 snRNPs as part of a tri-snRNP (U4/U6.U5 snRNP) preassembled complex and plays a critical role in the spliceosome rearrangement which leads to the catalytic steps of splicing. One of the U5 snRNP components, the U5 snRNP p102 protein containing LXXLL motifs, was recently identified as an AR-interacting protein that also is a transcriptional coregulator (26, 107). Interestingly, PRP4 kinase, another LXXLL motif-containing protein, has been found in a complex containing the U5 snRNP p102 and the transcriptional coregulators N-CoR and BRG1 (13). Finally, the U5 snRNP p200 helicase, one of the U5 snRNP helicases driving the structural spliceosome rearrangement, shares extensive homology to the NR coregulator ASC-1 p200, containing LXXLL motifs (38).
Spliceosome rearrangement leads to the release of some proteins and the recruitment of new ones, such as SKIP (ski oncogene-interacting protein). SKIP is associated with the spliceosome, and SKIP-interacting proteins are identified as U5 snRNP components (27, 52, 53, 68, 83, 108). SKIP also interacts with and modulates the transcriptional activity of transcription factors such as VDR, the vitamin D receptor (6, 104). Interestingly, a SKIP mutant results in the accumulation of unspliced transcripts generated from a VDR-responsive minigene, supporting the hypothesis that the transcriptional coactivator SKIP could play a role in the transformation of the mature spliceosome into the active spliceosome (52, 53, 65, 104).
The examples provided in the above section clearly demonstrate that several transcriptional coregulators are found within the spliceosome in which they are thought to play specific roles. Prior to discussing the functional aspects of this connection, we will describe a series of transcriptional coregulators that are functionally or structurally related to the splicing factors of the SR family and the hnRNP family of proteins and which appear to participate in spliceosome assembly and in the regulation of RNA splicing.
Transcriptional coregulators and splicing factors.
The splicing sequences described above (5′ SS, 3′ SS, BPS, and PY) are short degenerate consensus sequences as shown in Fig. 2. Therefore, “false” or “cryptic” SSs are found particularly in long primary transcripts. An important question is how the splicing machinery distinguishes the true SSs from the cryptic ones. One explanation resides in the action of specific SFs able to distinguish regulatory sequences within and around genuine and “false” exons. When bound to such sequences, SFs act either as splicing enhancers, reinforcing the recognition of neighboring SSs (Fig. 2), or as splicing silencers, masking neighboring SSs. As discussed below, one action of SFs is to modulate the selection of SSs (or splicing decisions), a mechanism known as alternative splicing and which leads to the production of multiple different mRNAs from one pre-mRNA. In the following section we discuss the transcriptional coregulators that are structurally or functionally related to the SFs of the SR and the hnRNP families and which play an important role in the SS selection and regulate splicing decisions (Table 1).
TABLE 1.
Transcriptional coregulators structurally or functionally related to splicing factors
| Protein family | RNA binding domain | Name | Comment |
|---|---|---|---|
| hnRNP | KH | mBBP | The essential splicing factor mBBP/SF1 is also a transcriptional corepressor. |
| ASC-1 p50 | Component of the transcriptional ASC-1 complex and affects splicing decisions. | ||
| RGG | hnRNP U/SAFA | Associates and modulates the activity of NR and binds to the nuclear matrix. | |
| 1 RRM | HET/SAFB | Links transcriptional and splicing machineries; binds to nuclear matrix. | |
| TLS | Belongs to the TET family involved in transcription and splicing. | ||
| EWS | Belongs to the TET family. | ||
| TAFII68 | Belongs to the TET family. | ||
| RTA | RNA binding protein interacting with and modulating the ER. | ||
| PERC/PGC-1β | Selective transcriptional coregulator of ER; related to PGC-1α (see below). | ||
| 2 RRMs | CoAA | Affects the activity of several transcriptional factors and splicing decisions. | |
| PSF | Splicing factor involved in modulation of NR activity. | ||
| p54nrb | Related to PSF. | ||
| 3 RRMs | SHARP | Transcriptional coregulator purified in the spliceosome. | |
| SR | TRAP150 | Associates with the TRAP complex and colocalizes with splicing factors. | |
| 1 RRM | PGC-1α | NR coactivator; affects splicing decisions. | |
| PRC | PGC-1-related coactivator. | ||
| 2 RRMs | CAPERα | Affects NR transcriptional activity and splicing decisions. | |
| CAPERβ | Belongs with CAPERα to a U2AF65-related protein family. | ||
| RNA helicase | p68 | Interacts with and modulates ER activity; it is also an essential splicing factor. | |
| p72 | Related to p68. | ||
| ASC-1 p200 | ASC-1 complex component; similar to the p200 helicase of the U5 snRNP. | ||
| DP103 | Coregulator of the steroidogenic factor 1, present in the SMN complex. | ||
| DP97 | ER coregulator. | ||
| Others | SAPs | Some spliceosome-associated proteins are present in transcriptional complexes. | |
| p102 U5 snRNP | Interacts with the AR. | ||
| SKIP | Vitamin D receptor transcriptional coregulator purified in the spliceosome. | ||
| PRP4kinase | Associates with U5 snRNP proteins and transcriptional coregulators. | ||
| MEP50 | Within the methylosome complex, acts also as a transcriptional coregulator. |
Pre-mRNAs, also called heterogeneous nuclear RNAs (hnRNAs) due to their size heterogeneity and cellular location, form densely packed ribonucleoprotein complexes known as heterogeneous nuclear ribonucleoprotein particles (hnRNPs) by associating with hnRNP proteins. hnRNP proteins are involved in all aspects of mRNA metabolism from transcription to translation and even to degradation (44). There are about 20 major hnRNP proteins named from A1 to U and many others that are less abundant. A common feature revealed by primary sequence analysis of multiple hnRNP proteins is a typical modular structure in which one or more RNA binding domains are associated with other (auxiliary) domains containing still poorly defined functions. Three major types of RNA binding domains have been identified to date: (i) the RNA recognition motif (RRM) consisting of a 90-amino-acid sequence, (ii) the RGG box characterized by closely spaced Arg-Gly-Gly repeats, and (iii) the K-homology (KH) motif, a stretch of about 45 amino acids first identified in hnRNP K.
Several KH domain-containing proteins, like hnRNP K, are involved in both splicing and transcription (8). For instance, the KH domain protein Sam68, involved in several “posttranscriptional” steps, was recently identified as interacting with the transcriptional coregulator CBP/p300 and shown to have potent transcriptional repression activity (30). The KH domain protein mBBP, which plays an important role in the recognition of the BPS, was identified as a potential transcriptional corepressor (24, 105). Finally, a KH domain-containing protein, p50 ASC-1, was recently identified in the ASC-1 transcriptional complex; we showed that this protein can affect splicing decisions (2, 38).
Two functionally related hnRNP proteins, hnRNPU/SAF-A, containing an RGG box and an LXXLL motif, and HET/SAF-B, containing an RRM, interact with and modulate the activity of NRs (17, 72). Because these proteins bind the nuclear matrix, DNA, RNA, and pol II CTD, they have been proposed to serve as structural links between the nuclear architecture and transcript synthesis and maturation (see below). A similar role could be played by the proteins of the “TET” family, composed of the highly homologous RRM-containing proteins EWS, TLS, and TAFII68, which act at both the transcriptional and the splicing levels (7, 61, 79).
While CoAA contains two N-terminal RRMs and shows similarities with the hnRNP-like proteins PSF and p54nrb described above, it was identified as a protein interacting with the transcriptional coregulator TRBP/ASC-2. CoAA enhances the transcriptional activity of several transcription factors, and we reported elsewhere that CoAA also affects splicing decisions (2-4, 36). Recently, other RRM-containing proteins have been cloned as NR-interacting proteins. For instance, RTA, which is homologous to the splicing factor FOX-1, was identified as an ER-interacting protein (71). PERC/PGC1-β/ERRL1 was independently cloned by two laboratories showing that this RRM-containing protein interacts with and modulates the transcriptional activity of NRs (45, 49). Finally, SHARP, containing several RRMs, acts as a transcriptional coregulator and copurifies with splicing complexes (93, 108).
The SR protein family is defined as RNA binding proteins that contain arginine-serine-rich (RS) domains. The presence of RS domains is a marker for RNA splicing function, since proteins containing this domain usually are involved in splicing (90). SR proteins act early in the splicing pathway and aid in the recognition of the 5′ and 3′ SSs. Interestingly, several transcriptional coregulators contain SR domains. For instance, TRAP150, a component of the TRAP transcriptional complex, contains an RS domain and colocalizes with other SR splicing factors within characteristic speckles (94). The NR coregulator PGC-1, containing an LXXLL motif and an RS domain, was identified early as a PPARγ coactivator and was shown to be involved in splicing (63). A new NR transcriptional coregulator, PRC (PGC-1-related coactivator), containing LXXLL and RS motifs, was recently identified, although it has not yet been shown to function in RNA splicing (1). As mentioned above, the two transcriptional coregulators CAPER and CAPERβ, related to the U2AF65 SF, contain RS domains and affect splicing decisions (16). Finally, the general p52 coregulator, a component of the PC4 transcriptional complex, was shown to interact with the SR splicing factor ASF/SF2 and to impact splicing decisions (23).
Another interesting class of proteins associated with the splicing process is the family of ATP-dependent RNA helicases. These proteins use the energy from ATP hydrolysis either to rearrange inter- or intramolecular RNA structures or to dissociate RNA-protein complexes. Such dynamic rearrangements are fundamental for many steps in the life of RNA molecules (87). For instance, multiple helicases have been implicated in correct recognition of pre-mRNA sequences during spliceosome formation, during the rearrangement of the spliceosome, or in regeneration of snRNPs between rounds of splicing. Interestingly, several of the NR transcriptional coregulators mentioned above belong to the RNA helicase family, including p68, p72, and ASC-1 p200 (see above). Other RNA helicases, including DP97 and DP103, interact with NRs and modulate their transcriptional activity (76, 82). The RNA helicase A mediates the association of the transcriptional coregulator CBP with RNA pol II (66).
TRANSCRIPTIONAL COREGULATORS AND THE COORDINATION OF STEPS IN THE GENE EXPRESSION PROCESS
The preceding section, summarized in Table 1, illustrates the large number of important transcriptional coregulators structurally and/or functionally related to proteins involved in “posttranscriptional” steps. This could reflect the fact that a given protein, often multifunctional, plays unrelated roles in the cell. In the following section, we will review results that suggest that different steps of the gene expression process should be considered to be “cotranscriptional” (rather than “posttranscriptional”); several of these steps impact each other. In particular, we will summarize data demonstrating that the implication of some transcriptional coregulators (defined in part as functional proteins recruited to promoters by DNA-bound transcription factors) in splicing depends directly on their implication in transcription, suggesting that the same molecules are engaged in sequential transcriptional and RNA processing events and act as “coupling” proteins.
Cotranscriptional maturation of transcripts.
The synthesis of an mRNA is an immensely complicated process. Each step in the pathway, including transcription, capping, splicing, and polyadenylation, is carried out by complicated molecular machineries (Fig. 1). Because experiments using in vitro systems indicated that each of the major steps can be carried out in isolation, it was initially assumed that the machineries responsible for each step are distinct and function independently in distinct nuclear compartments. However, many recent structural and cytological studies provided numerous pieces of evidence that these steps take place simultaneously and are intimately coupled and, in particular, that SS selection and intron removal can take place on the nascent transcript (33, 99).
Biochemical experiments with cotranscriptional mRNA processing have focused on the role of the RNA pol II CTD. It is now widely accepted that the pol II CTD can act as a platform for nascent transcript maturation. This conclusion is based on the fact that the CTD associates with RNA 5′ capping enzymes, 3′-end-processing factors, and various splicing factors. Moreover, truncation of the CTD inhibits in vivo capping and 3′-end processing and leads to a severe impairment of splicing; other experiments demonstrate that pol II CTD enhances RNA processing reactions. It is believed that the reversible interactions between the pol II CTD and RNA processing factors localize those factors close to their RNA substrates (25, 40, 54, 59, 84, 103). In this regard, it is important to underline that several coregulators that we discussed above have been shown to directly interact with the pol II CTD. These include SAF-A, SAF-B, TLS, EWS, PSF, p54nrb, p68, p72, and PGC-1 (see above). Such examples could provide clues as to how the transcriptional and splicing machineries could communicate through the pol II CTD.
As summarized above, various factors purified in transcriptional complexes seem to play a role in splicing and, inversely, factors first described as involved in splicing appear to play a role in transcription, indicating that these two temporally and spatially linked events share common factors. Another demonstration of the interconnection between these two processes came from Kornblihtt and coworkers, who demonstrated that the promoter identity can impact splicing decisions (11). Kornblihtt and coworkers proposed a model to explain how transcriptional processes can impact splicing decisions based on the concept that the time required to make a transcript can affect the ability of the splicing machinery to recognize splice sites and exons. The demonstration that the speed of pol II in synthesis of a transcript impacts the way in which the transcript is matured clearly reinforces the “cotranscriptional” nature of RNA processing (12, 25, 70, 85, 86).
Functional and biological roles of “coupling” proteins.
On average, a typical human gene contains eight introns and nine exons, which together average about 30,000 nucleotides (nt) in length. An average intron is over 3,000 nt, and an internal exon is only ∼150 nt. Therefore, on average, more than 80% of the pre-mRNA will be removed during the splicing process by elimination of the introns (25, 34, 110). In some cases, introns comprise up to 99% of the pre-mRNA. For example, the dystrophin gene encodes a ∼3-million-nt-long pre-mRNA that gives rise to only 12,000 nt of mature mRNA after the removal of 78 introns (80). The nebulin gene is comprised of 183 exons spanning 249 kb of genomic DNA. Since the start codon is in exon 3 and the stop codon is in exon 183, about 360 SSs must be correctly defined to allow the removal of 180 introns with not a single nucleotide error that would alter the open reading frame (15). Since the SSs are short degenerate consensus sequences, it is a challenge to understand how the splicing machinery can recognize exons that are sometimes very small and awash in huge introns. The fact that the splicing decisions are made coordinately with the synthesis of the transcript adds an additional level of complexity to this process. pol II elongates transcripts with an average rate of 1 to 2 kb/min, which means that one average intron can be synthesized in 1 or 2 minutes and the next one can be made less than 2 minutes later. In this context, the first-appearing 5′ SS easily could be erroneously assembled with the second-appearing 3′ SS of the adjacent intron (25, 69, 110). Obviously, a series of highly structured and coordinated events are required to precisely remove each of the introns during the synthesis of the nascent transcripts. This process may be aided by the action of specific transcriptional coregulators that play an additional nuclear “structural” role.
Indeed, the nucleus is a membrane-bounded organelle that contains the machineries for gene expression steps. It is believed that the nucleus has a kind of skeleton (nuclear matrix), a complex molecular scaffold that binds genomic DNA, RNAs, and specific proteins. It has been proposed that a subset of proteins like hnRNPU/SAF-A or HET/SAF-B that are recruited by transcriptional factors to promoters, that are attached to the nuclear matrix, and that are involved in splicing could play a role in the structural assembly of the different gene expression machineries (10, 67). An increasing number of specialized domains and subnuclear organelles enriched in specific factors have been identified within the nucleus (109). Since several nuclear compartments are defined by their enrichment in certain specific factors (e.g., speckles containing splicing factors), a function of a subset of transcriptional coregulators could be to facilitate communication and exchange between such compartments; this has been proposed for the AR-interacting protein ANT-1, also known as p102 U5 snRNP, described in a preceding section (26, 107). Therefore, a subset of transcriptional coregulators could permit the assembly and the communication of machineries and/or subnuclear domains within the nucleus, while others could facilitate the kinetics of the various transcriptional steps.
Indeed, if some of the proteins required for the splicing process are recruited prior to or during transcription and not after it has been initiated, full spliceosome assembly will occur more rapidly on the nascent RNA chain. Thus, when a transcriptional signal stimulates its target gene promoter, the ability of such a signal to promote the recruitment of “coupling” proteins would improve the kinetics of full mature transcript production. Such a function could be possible for coregulators like p54nrb, PSF, p68, p72, SAPs, and U2AF65-related proteins that play a role in the recognition of SSs (see above). Nevertheless, it is hard to conceive how a transcription factor could recruit simultaneously the many proteins necessary for transcription as well as the proteins required for splicing of an RNA transcript. Rather, our current concept is that different transcription factor molecules would be recruited in a sequential manner on their target promoter, so that different sets of proteins and “coupling” proteins, once locally enriched, could play a role in the recycling of other proteins or of subcomplexes; this could be the case for SKIP (see above). Supporting this possibility, it is believed that no more than one spliceosome can assemble on a transcript at a given time and that the spliceosome subcomplexes undergo extensive recycling (53, 99). How spliceosome recycling is achieved between successive introns, either in a given transcript or between successive transcripts, remains an unanswered question. Interestingly, activated NRs have been reported to interact with proteins that play a role not only in the building of the spliceosome as described above but also with proteins that play a role in the formation of spliceosome snRNP subcomplexes. For instance, two proteins, PRMT5 and MEP50, components of the methylosome complex that methylates the Sm proteins of the snRNP complexes, were shown to modulate androgen-dependent transcription (32). Moreover, DP103 and the RNA helicase A interact with the survival motor neuron protein SMN involved in the assembly and regeneration of snRNPs and were shown to regulate transcription as described above. Although the initial steps of snRNP assembly take place in the cytosol, it is tempting to speculate that the presence of some snRNP assembly proteins within the nucleus plays a role in recycling spliceosome subcomplexes and that the recruitment of such proteins by transcription factors would improve the kinetics and the coordination of the recycling process in the vicinity of the regulated gene.
The splicing machinery must remove several introns (182 in the case of the nebulin primary transcripts). Importantly, a splicing error that adds or removes even one single nucleotide disrupts the open reading frame of the mRNA. It is now clear that strong quality control steps exist to ensure that mRNA is appropriately processed (98). An emerging view of this quality control process is that early steps of the pathway allow the recruitment of proteins that are crucial for downstream events. Therefore, if an early step is not performed correctly and some of the proteins necessary for the next step are missing, this will lead to the arrest of the process and the destruction of the nascent transcript. Such a mechanism is particularly well illustrated by the molecular connection between transcription elongation and either capping or 3′-end formation. If transcription initiation occurs correctly, RNA pol II allows the recruitment of the capping complex involved in the 5′-end maturation of the transcript (Fig. 1). In turn, this complex “retains” pol II after the synthesis of a 20- to 24-nt-long transcript until the capping complex has accomplished its task (40, 54). In other words, a correct transcriptional initiation process allows capping that in turn allows elongation of transcription. Similarly, a strong interdependency exists between transcription termination and 3′-end processing of the transcript, since pol II allows the recruitment of 3′-end-processing factors that in turn control termination of transcription (81). Similarly, the implication of transcriptional coregulators in splicing could add another quality control checkpoint, since splicing would occur only if some of the proteins described in Fig. 2 were correctly recruited. Supporting the existence of such a checkpoint, it has recently been demonstrated that splicing signals can be transmitted to transcription either in an early stage of transcriptional initiation or during elongation. It is proposed that sampling of the nascent RNA by the U1 snRNA enhances transcription; the recruitment of snRNPs to the transcriptional elongation complex somehow increases its efficiency (21, 22, 46, 56, 95, 110). Therefore, it can be postulated that the appropriate assembly of the transcriptional machinery allows proper construction of the spliceosome, so that when it is correctly assembled on the transcript, it in turn allows the transcriptional machinery to proceed. In addition to this quality control, proteins regulating transcription could also participate in splicing regulation.
In mammalian lineage, the abundance of introns (seven to eight per gene on average) coupled with poor conservation of SSs allows an increasing number of alternative splicing events. Alternative splicing that results in the ligation of differential 5′ and 3′ SSs allows a single gene to produce multiple mRNAs that encode proteins with different functions. Between 30% and 70% of the human genes generate multiple mRNAs by alternative splicing of their primary transcripts, and ∼80% of alternative splicing results in changes in the encoded protein, making this mechanism one of the important sources of human proteome diversity (34). The biological importance of this mechanism is illustrated by the genes involved in cell death, since, from a single gene involved in this cellular program, alternative splicing leads to the production of protein isoforms having either pro- or antiapoptotic effects (102).
Considering the strong impact of splicing decisions on the biological functions of the gene products, alternative splicing must be finely regulated. As mentioned above, it is known that SFs, like SR and hnRNP proteins, can recognize sequences within transcripts (Fig. 1 and 2) and modulate splicing choices. When bound to these sequences, SFs can either mask or aid the recognition of weak neighboring SSs. Increasing evidence suggests a role for transcriptional regulators in alternative splicing regulation. Indeed, the nature of the spliced variants produced by a reporter gene depends on the nature of the promoter driving the gene; the splicing decisions also can be affected by the transcription elongation speed (11, 12, 77, 85, 86, 88). As discussed above, elongation speed can affect the time left for the recognition of splice sites. Moreover, exons are not only recognized by splice site recognition. Instead, exons are defined by pairing either across introns or across exons, multiple sequences recognized by splicing factors in a concerted set of weak protein-RNA and protein-protein interactions that either do or do not result in exon recognition and inclusion. By slowing polymerase in the region of a transcript with an alternative exon, these weak interactions may be favored, leading to inclusion. Finally, in transcripts with large introns where exons are found initially, there is a second step of exon juxtapositioning that must occur, presumably through protein-protein interactions. The CTD becomes an ideal docking platform for these interactions, and again this could be regulated through the elongation speed.
A subset of transcriptional coregulators may also have evolved to regulate the quantitative enhancement of mRNA production as well as in determining the eventual alternative splicing decisions. Using alternatively spliced reporter genes controlled by promoters responsive to steroid hormones, we demonstrated that these hormonal transcriptional stimuli simultaneously control the transcriptional rate of their target genes and the nature of the spliced variants produced by these genes (2-4). We and others have also demonstrated that many of the transcriptional coregulators described in Table 1 affect splicing decisions (2-4, 23, 31, 63, 65, 104). Moreover, these splicing effects mediated by transcriptional coregulators were in some cases shown to be promoter dependent (2-4, 63, 104). For instance, the PPARγ coregulator PGC-1 affects splicing decisions selectively on products synthesized from a PPARγ-regulated promoter (63). We recently demonstrated that the estrogen receptor coregulator CAPER selectively affects the splicing of products synthesized from estrogen-regulated promoters (16). Finally, we demonstrated that the coregulator CoAA affects splicing decisions in a promoter-dependent manner and that the engagement of CoAA in transcription has consequences in splicing (2, 3). Altogether, these results demonstrate that select proteins important for RNA splicing decisions appear to depend on their recruitment by the transcriptional machinery. Certain “classical” SFs could be recruited “directly” onto the nascent RNA transcript whereas others might be incorporated earlier within the transcriptional machinery. Therefore, we propose that transcriptional coactivators not only participate in mRNA syntheses but also provide a prime mechanism by which transcriptional signals influence the nature (exon content) of their target gene products.
CONCLUSIONS
In this review we have compiled data that shows that NR transcriptional coregulators are structurally or functionally related to proteins involved in splicing and that some of these coregulators are actually part of the spliceosome. These observations suggest that a subset of NR coregulators act as dual-function “coupling” proteins between transcription and splicing. These coupling proteins allow a spatial and temporal coordination between transcription and splicing, providing the molecular bases for a quality control checkpoint and for transcriptional stimuli to control not only the quantity, but also the “nature” (exon content), of their target gene products.
Steps other than transcription, capping, polyadenylation, and splicing also are coupled in the gene expression process. Indeed, it is now thought that all these steps even could impact mRNA export and translation. The strong interconnection among the different machineries involved in the gene expression process is illustrated by the results of Wolffe and Meric, who demonstrated that the history of a transcript in the nucleus affects its fate in the cytoplasm (101). In this regard, it is tempting to speculate that the initial transcriptional stimuli allow the recruitment of coregulator proteins that will follow and control the fate of their target gene products until their degradation.
Although we focused our review on NR coregulators, it is important to underline that most of these coregulators participate with other types of DNA binding transcription factors, and of course, many other transcription factors have been shown to affect splicing (20, 47, 64, 73). It is likely that the results of studies of the NR signaling pathway can and will be extended to other transcriptional signaling pathways.
Acknowledgments
This work is supported by NIH grants NICHD 08818 NIH-NIDDK Atlas Program (to B.W.O.), by the Welch Foundation, by grants GM38526 and GM58019 (to S.M.B.), by the INSERM AVENIR program (to D.A. and N.M.), and by the LNCC (to M.D.).
REFERENCES
- 1.Andersson, U., and R. C. Scarpulla. 2001. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 21:3738-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auboeuf, D., D. H. Dowhan, Y. K. Kang, K. Larkin, J. W. Lee, S. M. Berget, and B. W. O'Malley. 2004. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl. Acad. Sci. USA 101:2270-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auboeuf, D., D. H. Dowhan, X. Li, K. Larkin, L. Ko, S. M. Berget, and B. W. O'Malley. 2004. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 24:442-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auboeuf, D., A. Honig, S. M. Berget, and B. W. O'Malley. 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298:416-419. [DOI] [PubMed] [Google Scholar]
- 5.Bastien, J., and C. Rochette-Egly. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1-16. [DOI] [PubMed] [Google Scholar]
- 6.Baudino, T. A., D. M. Kraichely, S. C. Jefcoat, Jr., S. K. Winchester, N. C. Partridge, and P. N. MacDonald. 1998. Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription. J. Biol. Chem. 273:16434-16441. [DOI] [PubMed] [Google Scholar]
- 7.Bertolotti, A., Y. Lutz, D. J. Heard, P. Chambon, and L. Tora. 1996. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 15:5022-5031. [PMC free article] [PubMed] [Google Scholar]
- 8.Bomsztyk, K., O. Denisenko, and J. Ostrowski. 2004. hnRNP K: one protein, multiple processes. Bioessays 26:629-638. [DOI] [PubMed] [Google Scholar]
- 9.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carty, S. M., and A. L. Greenleaf. 2002. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol. Cell. Proteomics 1:598-610. [DOI] [PubMed] [Google Scholar]
- 11.Cramer, P., C. G. Pesce, F. E. Baralle, and A. R. Kornblihtt. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Mata, M., C. R. Alonso, S. Kadener, J. P. Fededa, M. Blaustein, F. Pelisch, P. Cramer, D. Bentley, and A. R. Kornblihtt. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12:525-532. [DOI] [PubMed] [Google Scholar]
- 13.Dellaire, G., E. M. Makarov, J. J. Cowger, D. Longman, H. G. Sutherland, R. Luhrmann, J. Torchia, and W. A. Bickmore. 2002. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 22:5141-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilworth, F. J., and P. Chambon. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20:3047-3054. [DOI] [PubMed] [Google Scholar]
- 15.Donner, K., M. Sandbacka, V. L. Lehtokari, C. Wallgren-Pettersson, and K. Pelin. 2004. Complete genomic structure of the human nebulin gene and identification of alternatively spliced transcripts. Eur. J. Hum. Genet. 12:744-751. [DOI] [PubMed] [Google Scholar]
- 16.Dowhan, D. H., E. P. Hong, D. Auboeuf, A. P. Dennis, M. M. Wilson, S. M. Berget, and B. W. O'Malley. 2005. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell 17:429-439. [DOI] [PubMed] [Google Scholar]
- 17.Eggert, H., M. Schulz, F. O. Fackelmayer, R. Renkawitz, and M. Eggert. 2001. Effects of the heterogeneous nuclear ribonucleoprotein U (hnRNP U/SAF-A) on glucocorticoid-dependent transcription in vivo. J. Steroid Biochem. Mol. Biol. 78:59-65. [DOI] [PubMed] [Google Scholar]
- 18.Emili, A., M. Shales, S. McCracken, W. Xie, P. W. Tucker, R. Kobayashi, B. J. Blencowe, and C. J. Ingles. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 19:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Fomenkov, A., Y. P. Huang, O. Topaloglu, A. Brechman, M. Osada, T. Fomenkova, E. Yuriditsky, B. Trink, D. Sidransky, and E. Ratovitski. 2003. P63 alpha mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J. Biol. Chem. 278:23906-23914. [DOI] [PubMed] [Google Scholar]
- 21.Fong, Y., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414:929-933. [DOI] [PubMed] [Google Scholar]
- 22.Furger, A., J. M. O'Sullivan, A. Binnie, B. A. Lee, and N. J. Proudfoot. 2002. Promoter proximal splice sites enhance transcription. Genes Dev. 16:2792-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 24.Goldstrohm, A. C., T. R. Albrecht, C. Sune, M. T. Bedford, and M. A. Garcia-Blanco. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21:7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstrohm, A. C., A. L. Greenleaf, and M. A. Garcia-Blanco. 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31-47. [DOI] [PubMed] [Google Scholar]
- 26.Goto, K., Y. Zhao, M. Saito, A. Tomura, H. Morinaga, M. Nomura, T. Okabe, T. Yanase, R. Takayanagi, and H. Nawata. 2003. Activation function-1 domain of androgen receptor contributes to the interaction between two distinct subnuclear compartments. J. Steroid Biochem. Mol. Biol. 85:201-208. [DOI] [PubMed] [Google Scholar]
- 27.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 29.Hermanson, O., C. K. Glass, and M. G. Rosenfeld. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 30.Hong, W., R. J. Resnick, C. Rakowski, D. Shalloway, S. J. Taylor, and G. A. Blobel. 2002. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol. Cancer Res. 1:48-55. [PubMed] [Google Scholar]
- 31.Honig, A., D. Auboeuf, M. M. Parker, B. W. O'Malley, and S. M. Berget. 2002. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 22:5698-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosohata, K., P. Li, Y. Hosohata, J. Qin, R. G. Roeder, and Z. Wang. 2003. Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol. Cell. Biol. 23:7019-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, S., and D. L. Spector. 1996. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 133:719-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 35.Ishitani, K., T. Yoshida, H. Kitagawa, H. Ohta, S. Nozawa, and S. Kato. 2003. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 306:660-665. [DOI] [PubMed] [Google Scholar]
- 36.Iwasaki, T., W. W. Chin, and L. Ko. 2001. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375-33383. [DOI] [PubMed] [Google Scholar]
- 37.Jung, D. J., S. Y. Na, D. S. Na, and J. W. Lee. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277:1229-1234. [DOI] [PubMed] [Google Scholar]
- 38.Jung, D. J., H. S. Sung, Y. W. Goo, H. M. Lee, O. K. Park, S. Y. Jung, J. Lim, H. J. Kim, S. K. Lee, T. S. Kim, J. W. Lee, and Y. C. Lee. 2002. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol. Cell. Biol. 22:5203-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kameoka, S., P. Duque, and M. M. Konarska. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, H. J., S. H. Jeong, J. H. Heo, S. J. Jeong, S. T. Kim, H. D. Youn, J. W. Han, H. W. Lee, and E. J. Cho. 2004. mRNA capping enzyme activity is coupled to an early transcription elongation. Mol. Cell. Biol. 24:6184-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kino, T., O. Slobodskaya, G. N. Pavlakis, and G. P. Chrousos. 2002. Nuclear receptor coactivator p160 proteins enhance the human immunodeficiency virus (HIV) type 1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J. Biol. Chem. 277:2396-2405. [DOI] [PubMed] [Google Scholar]
- 42.Kinyamu, H. K., and T. K. Archer. 2004. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim. Biophys. Acta 1677:30-45. [DOI] [PubMed] [Google Scholar]
- 43.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 44.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 45.Kressler, D., S. N. Schreiber, D. Knutti, and A. Kralli. 2002. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 277:13918-13925. [DOI] [PubMed] [Google Scholar]
- 46.Kwek, K. Y., S. Murphy, A. Furger, B. Thomas, W. O'Gorman, H. Kimura, N. J. Proudfoot, and A. Akoulitchev. 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 9:800-805. [DOI] [PubMed] [Google Scholar]
- 47.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274:11832-11841. [DOI] [PubMed] [Google Scholar]
- 48.Lee, D. K., and C. Chang. 2003. Molecular communication between androgen receptor and general transcription machinery. J. Steroid Biochem. Mol. Biol. 84:41-49. [DOI] [PubMed] [Google Scholar]
- 49.Lin, J., P. Puigserver, J. Donovan, P. Tarr, and B. M. Spiegelman. 2002. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 277:1645-1648. [DOI] [PubMed] [Google Scholar]
- 50.Liu, J., L. He, I. Collins, H. Ge, D. Libutti, J. Li, J. M. Egly, and D. Levens. 2000. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell 5:331-341. [DOI] [PubMed] [Google Scholar]
- 51.Liu, Z. R. 2002. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacDonald, P. N., D. R. Dowd, C. Zhang, and C. Gu. 2004. Emerging insights into the coactivator role of NCoA62/SKIP in vitamin D-mediated transcription. J. Steroid Biochem. Mol. Biol. 90:179-186. [DOI] [PubMed] [Google Scholar]
- 53.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Luhrmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 54.Mandal, S. S., C. Chu, T. Wada, H. Handa, A. J. Shatkin, and D. Reinberg. 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. USA 101:7572-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manley, J. L. 2002. Nuclear coupling: RNA processing reaches back to transcription. Nat. Struct. Biol. 9:790-791. [DOI] [PubMed] [Google Scholar]
- 57.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathur, M., P. W. Tucker, and H. H. Samuels. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 60.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 61.Meissner, M., S. Lopato, J. Gotzmann, G. Sauermann, and A. Barta. 2003. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell Res. 283:184-195. [DOI] [PubMed] [Google Scholar]
- 62.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 63.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 64.Moyret-Lalle, C., C. Duriez, J. Van Kerckhove, C. Gilbert, Q. Wang, and A. Puisieux. 2001. p53 induction prevents accumulation of aberrant transcripts in cancer cells. Cancer Res. 61:486-488. [PubMed] [Google Scholar]
- 65.Nagai, K., T. Yamaguchi, T. Takami, A. Kawasumi, M. Aizawa, N. Masuda, M. Shimizu, S. Tominaga, T. Ito, T. Tsukamoto, and T. Osumi. 2004. SKIP modifies gene expression by affecting both transcription and splicing. Biochem. Biophys. Res. Commun. 316:512-517. [DOI] [PubMed] [Google Scholar]
- 66.Nakajima, T., C. Uchida, S. F. Anderson, C. G. Lee, J. Hurwitz, J. D. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 67.Nayler, O., W. Stratling, J. P. Bourquin, I. Stagljar, L. Lindemann, H. Jasper, A. M. Hartmann, F. O. Fackelmayer, A. Ullrich, and S. Stamm. 1998. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26:3542-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 69.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115:3865-3871. [DOI] [PubMed] [Google Scholar]
- 70.Nogues, G., S. Kadener, P. Cramer, D. Bentley, and A. R. Kornblihtt. 2002. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 277:43110-43114. [DOI] [PubMed] [Google Scholar]
- 71.Norris, J. D., D. Fan, A. Sherk, and D. P. McDonnell. 2002. A negative coregulator for the human ER. Mol. Endocrinol. 16:459-468. [DOI] [PubMed] [Google Scholar]
- 72.Oesterreich, S., A. V. Lee, T. M. Sullivan, S. K. Samuel, J. R. Davie, and S. A. Fuqua. 1997. Novel nuclear matrix protein HET binds to and influences activity of the HSP27 promoter in human breast cancer cells. J. Cell Biochem. 67:275-286. [PubMed] [Google Scholar]
- 73.Ohe, K., E. Lalli, and P. Sassone-Corsi. 2002. A direct role of SRY and SOX proteins in pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 99:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oltra, E., I. Pfeifer, and R. Werner. 2003. Ini, a small nuclear protein that enhances the response of the connexin43 gene to estrogen. Endocrinology 144:3148-3158. [DOI] [PubMed] [Google Scholar]
- 75.Oltra, E., F. Verde, R. Werner, and G. D'Urso. 2004. A novel RING-finger-like protein Ini1 is essential for cell cycle progression in fission yeast. J. Cell Sci. 117:967-974. [DOI] [PubMed] [Google Scholar]
- 76.Ou, Q., J. F. Mouillet, X. Yan, C. Dorn, P. A. Crawford, and Y. Sadovsky. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 15:69-79. [DOI] [PubMed] [Google Scholar]
- 77.Pagani, F., C. Stuani, E. Zuccato, A. R. Kornblihtt, and F. E. Baralle. 2003. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 278:1511-1517. [DOI] [PubMed] [Google Scholar]
- 78.Page-McCaw, P. S., K. Amonlirdviman, and P. A. Sharp. 1999. PUF60: a novel U2AF65-related splicing activity. RNA 5:1548-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Powers, C. A., M. Mathur, B. M. Raaka, D. Ron, and H. H. Samuels. 1998. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol. Endocrinol. 12:4-18. [DOI] [PubMed] [Google Scholar]
- 80.Pozzoli, U., G. Elgar, R. Cagliani, L. Riva, G. P. Comi, N. Bresolin, A. Bardoni, and M. Sironi. 2003. Comparative analysis of vertebrate dystrophin loci indicates intron gigantism as a common feature. Genome Res. 13:764-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16:272-278. [DOI] [PubMed] [Google Scholar]
- 82.Rajendran, R. R., A. C. Nye, J. Frasor, R. D. Balsara, P. G. Martini, and B. S. Katzenellenbogen. 2003. Regulation of nuclear receptor transcriptional activity by a novel DEAD box RNA helicase (DP97). J. Biol. Chem. 278:4628-4638. [DOI] [PubMed] [Google Scholar]
- 83.Rappsilber, J., U. Ryder, A. I. Lamond, and M. Mann. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robert, F., M. Blanchette, O. Maes, B. Chabot, and B. Coulombe. 2002. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 277:9302-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roberts, G. C., C. Gooding, H. Y. Mak, N. J. Proudfoot, and C. W. Smith. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robson-Dixon, N. D., and M. A. Garcia-Blanco. 2004. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 279:29075-29084. [DOI] [PubMed] [Google Scholar]
- 87.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5:232-241. [DOI] [PubMed] [Google Scholar]
- 88.Rosonina, E., M. A. Bakowski, S. McCracken, and B. J. Blencowe. 2003. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 278:43034-43040. [DOI] [PubMed] [Google Scholar]
- 89.Rossow, K. L., and R. Janknecht. 2003. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene 22:151-156. [DOI] [PubMed] [Google Scholar]
- 90.Sanford, J. R., D. Longman, and J. F. Caceres. 2003. Multiple roles of the SR protein family in splicing regulation. Prog. Mol. Subcell. Biol. 31:33-58. [DOI] [PubMed] [Google Scholar]
- 91.Sharma, D., and J. D. Fondell. 2002. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shav-Tal, Y. 2002. PSF and p54(nrb)/NonO-multi-functional nuclear proteins. FEBS Lett. 531:109-114. [DOI] [PubMed] [Google Scholar]
- 93.Shi, Y., M. Downes, W. Xie, H. Y. Kao, P. Ordentlich, C. C. Tsai, M. Hon, and R. M. Evans. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15:1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sutherland, H. G., G. K. Mumford, K. Newton, L. V. Ford, R. Farrall, G. Dellaire, J. F. Caceres, and W. A. Bickmore. 2001. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 10:1995-2011. [DOI] [PubMed] [Google Scholar]
- 95.Tian, H. 2001. RNA ligands generated against complex nuclear targets indicate a role for U1 snRNP in co-ordinating transcription and RNA splicing. FEBS Lett. 509:282-286. [DOI] [PubMed] [Google Scholar]
- 96.Tian, Y., S. Ke, M. Chen, and T. Sheng. 2003. Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J. Biol. Chem. 278:44041-44048. [DOI] [PubMed] [Google Scholar]
- 97.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 98.Vasudevan, S., and S. W. Peltz. 2003. Nuclear mRNA surveillance. Curr. Opin. Cell Biol. 15:332-337. [DOI] [PubMed] [Google Scholar]
- 99.Wetterberg, I., J. Zhao, S. Masich, L. Wieslander, and U. Skoglund. 2001. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 20:2564-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 101.Wolffe, A. P., and F. Meric. 1996. Coupling transcription to translation: a novel site for the regulation of eukaryotic gene expression. Int. J. Biochem. Cell Biol. 28:247-257. [DOI] [PubMed] [Google Scholar]
- 102.Wu, J. Y., H. Tang, and N. Havlioglu. 2003. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog. Mol. Subcell. Biol. 31:153-185. [DOI] [PubMed] [Google Scholar]
- 103.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang, C., D. R. Dowd, A. Staal, C. Gu, J. B. Lian, A. J. van Wijnen, G. S. Stein, and P. N. MacDonald. 2003. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278:35325-35336. [DOI] [PubMed] [Google Scholar]
- 105.Zhang, D., and G. Childs. 1998. Human ZFM1 protein is a transcriptional repressor that interacts with the transcription activation domain of stage-specific activator protein. J. Biol. Chem. 273:6868-6877. [DOI] [PubMed] [Google Scholar]
- 106.Zhang, D., A. J. Paley, and G. Childs. 1998. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J. Biol. Chem. 273:18086-18091. [DOI] [PubMed] [Google Scholar]
- 107.Zhao, Y., K. Goto, M. Saitoh, T. Yanase, M. Nomura, T. Okabe, R. Takayanagi, and H. Nawata. 2002. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment; Identification of the p102 U5 snRNP binding protein as a coactivator for the receptor. J. Biol. Chem. 277:30031-30039. [DOI] [PubMed] [Google Scholar]
- 108.Zhou, Z., J. Sim, J. Griffith, and R. Reed. 2002. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl. Acad. Sci. USA 99:12203-12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zimber, A., Q. D. Nguyen, and C. Gespach. 2004. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell. Signal. 16:1085-1104. [DOI] [PubMed] [Google Scholar]
- 110.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296:91-97. [DOI] [PubMed] [Google Scholar]