Abstract
The carboxy-terminal domain (CTD) of RNA polymerase II large subunit acts as a platform to assemble the RNA processing machinery in a controlled way throughout the transcription cycle. In yeast, recent findings revealed a physical connection between phospho-CTD, generated by the Ctk1p kinase, and protein factors having a function in small nucleolar RNA (snoRNA) biogenesis. The snoRNAs represent a large family of polymerase II noncoding transcripts that are associated with highly conserved polypeptides to form stable ribonucleoprotein particles (snoRNPs). In this work, we have studied the biogenesis of the snoRNPs belonging to the box H/ACA class. We report that the assembly factor Naf1p and the core components Cbf5p and Nhp2p are recruited on H/ACA snoRNA genes very early during transcription. We also show that the cotranscriptional recruitment of Naf1p and Cbf5p is Ctk1p dependent and that Ctk1p and Cbf5p are required for preventing the readthrough into the snoRNA downstream genes. All these data suggest that proper cotranscriptional snoRNP assembly controls 3′-end formation of snoRNAs and, consequently, the release of a functional particle.
Great advances have been made in determining the architecture of the “gene expression factory” (36), which is characterized by a surprising number of factors associated together to ensure an efficient and regulated RNA biogenesis. In all processes allowing a mature and functional mRNA, a central role is played by the carboxy terminal domain (CTD) of the RNA polymerase II (Pol II) large subunit (17, 39, 43) that in vivo undergoes an extensive phosphorylation and dephosphorylation throughout the transcription cycle (4, 50). It is well known that a number of trans-acting factors required for pre-mRNA processing directly or indirectly bind to differently modified forms of CTD acting at different stages during transcription (25, 42). Four cyclin-dependent CTD kinases (CTDKs) have been described in Saccharomyces cerevisiae which are important for CTD function: Kin28p, Srb10p, Bur1p, and CTDK-1. CTDK-1 is a nonessential complex involved in the regulation of mRNA synthesis at the level of transcriptional elongation, pre-mRNA 3′-end formation, and nuclear export. Ctk1p is the CTDK-1 subunit (29) required for the bulk of Ser2 phosphorylation during elongation (3). Buratowski and colleagues have found that this modification is required for coupling 3′-end processing and transcription of mRNAs (1). Nevertheless, a growing number of evidence suggest that in yeast the role of CTD is not confined to the recruitment of factors involved in mRNA maturation. RNA Pol II transcribes mRNAs as well as noncoding RNAs, such as snRNAs, small nucleolar RNAs (snoRNAs), and, in higher eukaryotes, micro-RNAs. snoRNAs are small RNA molecules localized in the nucleolus, where they mainly participate in the modification and processing of rRNA. They include the so-called box C/D and box H/ACA families that catalyze, on substrate RNAs, the methylation of the 2′-hydroxyl residue of the ribose moiety (2′-O methylation) and the isomerization of uracil residues to pseudouridine (Ψ) (24). Only a subset of snoRNAs is required for different cleavage events during pre-rRNA maturation: the box C/D snoRNAs U3 and U14 (20, 30) and the box H/ACA snR10 and snR30 (34, 45).
The two classes of snoRNAs have conserved secondary structures, and the transcripts are associated with a specific set of proteins forming ribonucleoparticles (snoRNPs). The conserved C and D box elements, together with the internal stem (stem II), form a specific structural core motif (kink-turn) required for the binding of the conserved box C/D snoRNP protein factors: Snu13p, Nop1p, Nop58p, and Nop56p (27, 28, 49). Nop1p carries the enzymatic activity of the box C/D snoRNPs (10, 37) and, together with Snu13p and Nop58p, is a core assembly factor crucial for snoRNA stability, subcellular localization, and function of the particle (9). Nop1p was previously shown to assemble very early on the nascent snoRNA-containing transcript, and its binding to the transcript was demonstrated to be required both for processing of intron-encoded snoRNAs and for efficient 3′-end formation of independently transcribed snoRNAs (14, 33).
The pseudouridylation guide snoRNAs belonging to the H/ACA class are defined by an evolutionary conserved H/ACA motif consisting of a “hairpin-hinge-hairpin-tail” secondary structure (13). Four proteins form a stable complex with H/ACA snoRNAs: Gar1p, Nhp2p, Nop10p, and Cbf5p. In yeast, all these proteins are essential for viability and, with the exception of Gar1p, are required for the stability of H/ACA snoRNAs (2, 5, 15, 26, 48). Cbf5p, initially isolated as a low-affinity centromere binding protein in vitro (22), is believed to be the enzyme that catalyzes the isomerization of uridines to pseudouridines (19, 46). Moreover, it was established that, in vivo, the early assembly factors Naf1p and Shq1p are required to assemble the mature H/ACA snoRNPs (6, 8, 51). It was suggested that Naf1p could directly recruit snoRNPs proteins since it was shown to physically interact with Cbf5p and Nhp2p (8, 21).
In this study, we have analyzed the timing of H/ACA snoRNP assembly by chromatin immunoprecipitation (ChIP) technique and found that Naf1p colocalizes with snoRNA transcription sites together with Cbf5p and Nhp2p. In addition, we show that the presence of both nascent RNA and Ctk1-phospho-CTD is required for the cotranscriptional recruitment of Naf1p and Cbf5p.
We have also investigated the role played by Cbf5p in independently transcribed snoRNAs biosynthesis, and we report that Cbf5p, like Nop1p in the case of the box C/D snoRNAs, is required to obtain proper 3′-end formation/termination of the snoRNAs, preventing transcriptional readthrough into downstream genes.
MATERIALS AND METHODS
Strains and microbiological techniques.
The strains used in this study are listed in Table 1. Standard techniques were used for growth and handling of S. cerevisiae. Epitope TAP tagging of Naf1p and Cbf5p in strain YSB854 was performed as previously described (40).
TABLE 1.
List of strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| YSB854 | MATaura3-52 leu2Δ1 trpΔ63 his3Δ200 lys2Δ202 ctk1Δ::HIS3 | 1 |
| YFP10 | MATaura3-52 leu2Δ1 trpΔ63, his3Δ200 lys2Δ202 ctk1Δ::HIS3 NAF1-TAP::TRP1 | This study |
| YAF10 | MATaade2 his3 leu2 trp1 ura3 NAF1-TAP::TRP1 | 8 |
| YAF60 | MATaade2 arg4 leu2-3,112 trp1-289 ura3-52 CBF5::TAP::URA3 | EUROSCARF |
| YFP60 | MATaura3-52 leu2Δ1 trpΔ63 his3Δ200 lys2Δ202 ctk1Δ::HIS3 CBF5-TAP::TRP1 | This study |
| YAF61 | MATaade2 arg4 leu2-3,112 trp1-289 ura3-52 NHP2::TAP::URA3 | EUROSCARF |
| YAF62 | MATaade2 arg4 leu2-3,112 trp1-289 ura3-52 GAR1::TAP::URA3 | EUROSCARF |
| R1158 | MATahis3-1 leu2-0 met15-0 URA3::CMV-tTA | Open Biosystem |
| YSC1180 | MATahis3-1 leu2-0 met15-0 URA3::CMV-tTA kanR-tetO7-TATA::CBF5 | Open Biosystem |
| ssu72-2 | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 ssu72-2 | 11, 35 |
The ssu72-2 strain, containing conditional temperature-sensitive alleles, was grown at 25°C up to mid-log phase and then was shifted to 37°C for 2 h prior to RNA extraction. The yeast Tet-promoter Hughes strain was grown in YPD medium with 10 μg/ml doxycycline for 24 h to obtain the down regulation of the gene of interest (32).
Chromatin immunoprecipitations.
Cells were grown in YPD to an A600 of 0.6 to 0.8 and processed as previously described (23). Chromatin solution was incubated overnight at 4°C with rabbit immunoglobulin G-agarose beads (Sigma) prewashed in Tris-EDTA buffer. The immunoprecipitated material was washed with 275 to 400 mM NaCl, and the recovered chromatin and the input chromatin were de-cross-linked and analyzed by PCR. Various segments of a gene were amplified after ChIP as depicted in Fig. 1. Their locations cover the following positions indicated with respect to the transcriptional initiation site: −152 to +49, SNR10 (a); +26 to +216, SNR10 (b); −196 to +1, SNR30 (a); +396 to +580, SNR30 (b); and −52 to +141, SNR13. [α-32P]dATP was added to the PCR (1 μCi/12.5 μl). ChIP results were quantified as described by Nedea and colleagues (35): PCR signals were analyzed by PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA) and normalized for amplification efficiency and subtraction of background. Each value in the histogram is the average of the signals of five PCRs performed on five different preparations of immunoprecipitated DNA.
FIG. 1.
Naf1p is recruited to actively transcribed H/ACA snoRNA genes. (A) ChIP analysis was performed on the YAF10 yeast strain expressing a TAP-tagged version of NAF1 (panel RNase−). The cross-linked chromatin was amplified before (Input) and after (Naf1p) immunoprecipitation. An RNase treatment step was added before immunoprecipitation (panel RNase+). A nontranscribed intergenic region from chromosome V (band *) was used as an internal control by coamplification with each of the gene-specific primers. The diagram on the side shows the oligonucleotides specific for the 5′ end (“a”) and the coding region (“b”) of SNR10, SNR30, and SNR13 genes. (B) Histogram displaying the degree of Naf1p cross-linking on the snoRNA genes. The data are presented as the average (standard deviation, <20%) of signals of five PCRs performed on five preparations of immunoprecipitated DNA, normalized against the controls.
For RNase treatment, RNase-DNasefree (Roche) was added to the whole-cell extract to a final concentration of 5 μg/ml. The whole-cell extract then was incubated for 30 min at 37°C for 30 min and subjected to immunoprecipitation at 4°C overnight.
RT-PCR and Northern blot.
Total RNA was extracted by the hot-phenol method (41). For the reverse transcription-PCR (RT-PCR) experiment, a first-strand cDNA preparation was made using Superscript II reverse transcriptase (Invitrogen) and the primers snR30b and snR30RT, snR33b and snR33RT, snR10b and snR10RT, and snR13b and snR13RT. PCR amplification was with Taq DNA polymerase (Amersham). The primer sets used for PCR analysis were snR30a, snR30b, and snR30RT; snR33a, snR33b, and snR33RT; snR10a, snR10b, and snR10RT; and snR13a, snR13b, and snR13RT. PCRs were performed for 25 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C (for 1 min). For Northern analysis, total RNA was isolated as described above from the indicated strains. Total RNA (5 μg) was separated on 6% polyacrylamide-7 M urea gels and electrotransferred on Hybond-N nylon membrane (Amersham Pharmacia) in 0.5× Tris-borate-EDTA, and small RNA was detected by hybridization with antisense oligonucleotide probes.
To determine the Tet::Cbf5 time course, RNA was extracted from the Tet::Cbf5 strain following growth on doxycycline medium and separated on 1% agarose gel containing formaldehyde. To detect the CBF5 mRNA, a fragment spanning the whole open reading frame of CBF5 was generated by PCR (Cbf5a and Cbf5b oligonucleotides) and labeled by use of the Roche random priming kit. For the detection of 35S and 18S rRNAs, oligonucleotides 033 and 007 were used, respectively.
Oligonucleotides.
The following oligonucleotides were used for RNA analyses by Northern hybridization or RT-PCR (5′-3′): Cbf5-a, CGGAATTCTCAAAGGAGGATTTCGTTATTAAG; Cbf5-b, GAGGCCCGGGAACAAAAGCTGGGTAC; snR30a, GGACGCATGATCTTGAGCTC; snR30b, CAGTATGGTTTTACCCAAAT; snR30RT, TATTCCACCACTAAGTAGGG; snR33a, CCTCTTTGTACGATGGTGTC; snR33b, TGTCCACACACTTCTATATC; snR33RT, GCAATGGTGCAGATTGTGTC; snR10a, CACGTACAGTATCTCCGTCGAGGTT; snR10b, TCATCCGGGCACACGAAGGTAAAC; snR10RT, CTTAGAAAGGAAATGGCAACC; snR13a, GCTCTAGAAGGAAGTTTTTTCCTTTTTAT; snR13b, CGGGATCCGGTCAGATAAAAGTAAAAAAAGGTAGC; snR13RT, CGCTTGCTTAGGCCCAACAG; αsnR10, TCATCCGGGCACACGAAGG; αsnR30, GAGCTCAAGATCATGCGTCC; αsnR13, TTCCACACCGTTACTGATTT; αsnR5, CTCTCGAGCAAGGTCTATTTTAC; αsnR189, ATCTTGCACGTCGTAGAAAC; αU6, GCAGGGGAACTGCTGATCATC; 033, CGCTGCTCACCAATGG; and 007, CTCCGCTTATTGATATGC.
RESULTS
Naf1p is recruited to actively transcribed H/ACA snoRNA genes.
The H/ACA snoRNP assembly factor Naf1p, besides binding snoRNAs (6, 8), was reported to interact with Cbf5p and Nhp2p core-proteins in a two-hybrid screen and in vitro assays (8, 21); in addition, it was shown to copurify with overexpressed FLAG-tagged Cbf5p in a large-scale proteomic investigation (18). Due to its association with the CTD of Pol II, it was suggested that Naf1p could help the recruitment of H/ACA factors allowing their cotranscriptional assembly (8). To test this possibility, ChIP assays were performed with a strain carrying a C-terminal TAP-tagged version of the Naf1p protein (8). The immunoprecipitated DNA was analyzed by PCR amplification for the presence of the box H/ACA snoRNAs genes SNR10 and SNR30 and of a box C/D snoRNA as a control (SNR13). Each gene was analyzed with two different couples of primers (see schematic representation of Fig. 1): the first couple (probes “a”) recognizes promoter sequences, while the second one (probes “b”) is specific for the transcribed regions. The histogram in Fig. 1 (black boxes) indicates the presence of high levels of Naf1p on the coding regions of box H/ACA snoRNA genes, while the amount of protein recruited on the promoter regions remains close to background. As a further control, a set of middle primers was also utilized. The results show that they give an intermediate level of Naf1p reactivity (see Fig. S1 in the supplemental material). The difference in Naf1p reactivity between the promoter and the transcribed region is higher in SNR30, very likely due to the larger size of this gene that allows a better resolution between two regions (see schematic representation on the side). Since Naf1p binds to RNA in vitro (6, 8), its association with chromatin could be mediated by the interaction with the RNA transcript. To investigate this possibility, Naf1-TAP extracts were treated with RNase before ChIP analysis. The histogram of Fig. 1 (white boxes) shows that, under these conditions, the association of Naf1p with chromatin is strongly reduced, indicating that Naf1p recruitment depends on ongoing transcription.
Cbf5p and Nhp2p are associated with the sites of H/ACA snoRNA transcription.
Since Naf1p was shown to interact with Nhp2p and Cbf5p (8, 21), we asked whether these factors are also delivered early during transcription onto the nascent transcript. Yeast strains carrying chromosomal TAP-tagged versions of one of the CBF5, NHP2, or GAR1 genes were utilized to study the association of H/ACA snoRNP factors with box H/ACA snoRNA transcriptional units. Immunoprecipitated DNA was analyzed by PCR amplification with the same primers as in Fig. 1. Figure 2 indicates that, in comparison with a control box C/D gene (lanes SNR13), the H/ACA genes show a good level of reactivity to the Cbf5 protein (panel A) and to a lower, but still significant, extent to Nhp2p (panel B). On the contrary, signals close to background levels are obtained with the Gar1p factor (panel C). Interestingly, as in the case of Naf1p, also Cbf5p and Nhp2p are detected only on the transcribed region (see preferential reactivity with probes b). These results indicate a cotranscriptional recruitment of Cbf5p and Nhp2p on H/ACA snoRNA precursors and suggest a later association of Gar1p.
FIG. 2.
Cbf5p and Nhp2p are associated with the sites of H/ACA snoRNA transcription. Strains containing TAP-tagged versions of either Cbf5p (A), Nhp2p (B), or Gar1p (C) were analyzed by ChIP of snoRNA genes. Cross-linked chromatin was used for PCR amplification before (panel Input) and after (panel Ip) immunoprecipitation with primers specific for SNR10, SNR30, and SNR13 genes (see diagram in Fig. 1). A nontranscribed intergenic region from chromosome V (band *) was used as an internal control by coamplification with each of the gene-specific primers. The data are presented as the average (standard deviation, <20%) of signals of the PCRs performed on five preparations of immunoprecipitated DNA, normalized against the controls.
Deletion of Ctk1p affects Naf1p and Cbf5p cotranscriptional recruitment on H/ACA transcription units in vivo.
Naf1p was described to interact with the carboxy-terminal domain of the large subunit of RNA polymerase II (8, 18, 21); in addition, it interacts with Cbf5p, which recently has been found in the group of phospho-CTD-associating proteins (PCAPs) (38). These data, together with our experiments with ChIP analysis, suggest a mechanism by which H/ACA assembling factors are recruited to the sites where they are needed by the phospho-CTD of Pol II. To define whether the CTD-serine 2 phosphorylation by Ctk1p is a prerequisite for snoRNP recruitment on snoRNA transcription units, ChIP experiments were carried out with wild-type (CTK1) and ctk1Δ cells.
The histograms in Fig. 3 show that the recruitment of Naf1p (panel A) and Cbf5p (panel B) to the snR30 coding region is abolished in ctk1Δ strains containing TAP-tagged versions of either of the two proteins. These data indicate that the cotranscriptional assembly of H/ACA snoRNA requires phosphorylation of CTD on Ser2.
FIG. 3.
Ctk1 is required for cotranscriptional recruitment of H/ACA snoRNP in vivo. ChIP analyses were carried out with snoRNP TAP-tagged proteins in both wild-type (black bars) and Ctk1p deletion (gray bars) strains. PCR analysis of immunoprecipitated chromatin was performed on the SN30 (“a” and “b” as diagrammed in Fig. 1) and SNR13 genes. (A) ChIP analysis of Naf1p in CTK1 and ctk1Δ strains. (B) ChIP analysis of Cbf5p in CTK1 and ctk1Δ strains. The upper panels show the amplification products of the snoRNA regions specified above each lane. A nontranscribed intergenic region from chromosome V (band *) was used as an internal control by coamplification with each of the gene-specific primers. The data are presented as the average (standard deviation, <20%) of signals of the PCRs performed on three preparations of immunoprecipitated DNA, normalized against the controls.
The H/ACA snoRNP core protein Cbf5p is required for proper snoRNA 3′-end formation.
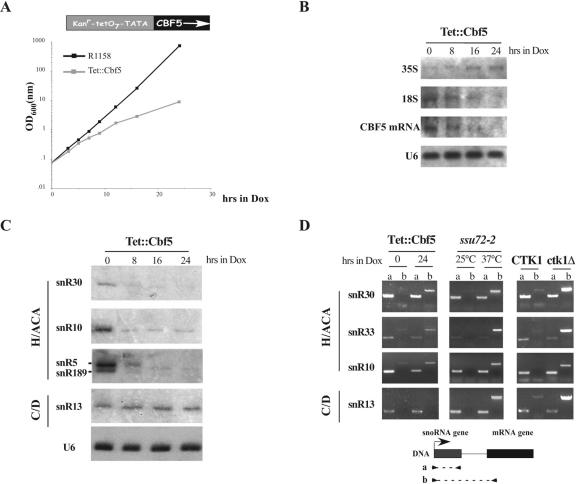
As a next step, we asked whether, as for box C/D snoRNAs, also for box H/ACA the cotranscriptional assembly of the snoRNP operates a quality control on the biosynthesis of the snoRNA by directing proper 3′-end formation of the primary transcript. Therefore, we investigated whether transcriptional readthrough products occurred upon depletion of an early assembly snoRNP factor; such products are diagnostic of alteration in proper 3′-end formation/termination (1). We asked whether Cbf5p, similarly to Nop1p, could be the trans-acting factor responsible for the control of 3′-end formation of box H/ACA snoRNAs. Since the CBF5 gene is an essential gene (22), we used a conditional allele in which the expression of CBF5 is under the control of a shut-off Tet-promoter (see schematic representation in Fig. 4A) (32). In the absence of doxycycline, the Tet promoter is fully active and the growth rate of the Tet::Cbf5 strain is the same as that of its isogenic strain. Addition of doxycycline in a titratable manner allows for downregulation of the promoter until the gene of interest is no longer expressed. Figure 4A shows that the growth rate of the Tet::Cbf5 strain starts to decrease 8 h after doxycycline treatment and stops between 12 and 16 h. In parallel, CBF5 mRNA begins to decrease at 8 h and at 24 h is undetectable (Fig. 4B). Since Western analysis of Cbf5p could not be performed due to the lack of antibodies, we made indirect controls on the depletion of this factor. First of all, we analyzed the levels of H/ACA snoRNA accumulation. Figure 4C shows that already after 8 h a drastic decrease is observed and that at 24 h H/ACA snoRNAs are almost undetectable. As a control, both the snR13 box C/D snoRNA and the U6 snRNA levels are unaffected. We subsequently analyzed rRNA synthesis: panel B shows the progressive depletion of the mature 18S rRNA and the 35S rRNA accumulation following transfer to doxycycline-containing medium. These data are good indicators of the effective depletion of Cbf5p.
FIG. 4.
Analysis of readthrough products in a Cbf5p-depleted strain. (A) Schematic representation of the Tet::Cbf5 allele (top of the panel) and growth of the Tet::Cbf5 (gray squares) and isogenic (black squares) strains following transfer to doxycycline (Dox)-containing medium (bottom of the panel). Cell density (optical density at 600 nm [OD600]) was measured at the indicated time, and the cultures were periodically diluted to be continuously kept in exponential growth. (B) Northern hybridization for the detection of CBF5 mRNA and 35S and 18S rRNAs from the Tet::Cbf5 strain (lanes 0 to 24 h); U6 was used as a normalization control. (C) Northern analysis of snoRNAs (box H/ACA, snR30, snR10, snR5, and snR189; box C/D, snR13) and of the control U6 snRNA. (D) RT-PCR analysis was carried out with set of primers described below the panel on total RNA extracted from following strain: Tet::Cbf5, grown in a complete medium (lanes 0) and then shifted in doxycycline-containing medium for 24 h (lanes 24); ssu72-2, grown at the permissive (lanes 25°C) and restrictive temperatures for 2 h (lanes 37°C); and CTK1 and ctk1Δ (lanes CTK1 and ctk1Δ). No products were observed when reverse transcriptase was omitted during cDNA synthesis (data not shown).
In order to identify readthrough fusion transcripts, RT-PCR analysis was carried out using two couples of primers: the first internal to the snoRNA coding region, while the second covering the region across the snoRNA and the downstream mRNA gene (see schematic representation at the bottom of Fig. 4D). Three different H/ACA snoRNA loci were analyzed: SNR10, SNR30, and SNR33. For all of them, readthrough products were visualized only when the Tet::Cbf5 strain was grown for 24 h in doxycycline (Fig. 4D, lanes 24 h). No such products were visualized with control RNA from the same strain grown in the absence of doxycycline (Fig. 4D, lanes 0 h) and with the control box C/D SNR13 locus. As a positive control, RT-PCR analyses were performed on RNA extracted from the ssu72-2 strain (Fig. 4D, lanes ssu72-2), grown at restrictive temperature and known to have a very strong readthrough phenotype (11, 35). Figure 4D shows that a readthrough phenotype is obtained also in ctk1Δ cells (lanes ctk1Δ), similar to those observed in Cbf5p-depleted cells and as previously described for C/D box snoRNAs (44).
DISCUSSION
Despite the detailed knowledge of the individual protein components of H/ACA snoRNPs, little is known regarding the correlation between transcription and snoRNP assembly. In this study, we have found evidence of a cotranscriptional recruitment of some of the H/ACA snoRNP components and their involvement in the control of snoRNA biosynthesis.
It has been previously suggested that, in S. cerevisiae, Naf1p and Shq1p play a key role in the metabolism of box H/ACA snoRNAs, aiding early steps of ribonucleoparticle assembly (6, 8, 51). The yeast hnRNP-like protein Naf1p localizes to the nucleus, and even if it is not a stable component of the H/ACA snoRNPs, it is required for the accumulation of box H/ACA snoRNAs (6, 8, 51). Moreover, Naf1p was shown to interact with the C-terminal domain of RNA Pol II and with the snoRNP core components Cbf5p and Nhp2p (8, 18, 21).
In this article, ChIP experiments have shown that Naf1p is localized specifically on the coding region of H/ACA snoRNA transcription units while no interaction was observed on the promoter. This behavior, together with the evidence that Naf1p binds to RNA in vitro (8), suggested that its recruitment could be triggered by the appearance of the appropriate RNA target sequences. In line with this hypothesis, an RNase treatment of the extract prior to immunoprecipitation produces a strong reduction of Naf1p association with H/ACA genes. Nevertheless, it cannot be excluded that Naf1p recruitment occurs already at the level of the promoter through the interaction with specific transcriptional factors and that it is subsequently mobilized to the CTD. In this case, this interaction could have been underestimated due to the detection sensitivity of the ChIP technique.
Chromatin immunoprecipitation analysis was carried out to monitor as well the recruitment of three H/ACA proteins, integral components of the mature snoRNP: Cbf5p, Nhp2p, and Gar1p. In analogy with Naf1p, Cbf5p and Nhp2p also show specific immunoprecipitation with the H/ACA genes and only with the transcribed regions. On the contrary, Gar1p does not show any significant reactivity with the H/ACA genes, suggesting that its association to the snoRNP occurs at later posttranscriptional phases. This is in agreement with previous results indicating that Gar1p is not essential for snoRNA accumulation, while it is required for the functionality of the mature particle (2, 16).
A biochemical investigation of the yeast proteome recognized a large number of factors physically linked to the phospho-CTD generated by the CTDK-1. These proteins are representative of a wide range of functions (e.g., transcription, RNA processing, chromatin structure, DNA metabolism, noncoding RNA biogenesis). The widespread nature of PCAPs points to a complex network of connections between Pol II elongation and other processes, expanding the role played by CTD phosphorylation in functional organization of the nucleus. Intriguingly, recent findings reveal a physical connection between phospho-CTD and protein factors having a function in snRNA modification and snoRNA biogenesis (8, 38). In line with this evidence, Cbf5p was copurified with the Spt5p elongation factor (31) and Naf1p was shown to interact with the carboxy-terminal domain of the large subunit of RNA polymerase II (8, 18, 21). The ChIP experiments performed in this work reveal that phosphorylation of serine 2 by Ctk1p contributes to the cotranscriptional recruitment of box H/ACA snoRNP proteins Naf1p and Cbf5p to the elongating RNA polymerase II, providing another link between snoRNP biogenesis and transcription elongation.
Recently, we showed that the methyltransferase Nop1p associates with box C/D snoRNA genes during transcription and, through its interaction with 3′-end termination machinery, provides a quality control for snoRNA synthesis profoundly influencing both transcription and 3′-end processing (33). The cotranscriptional recruitment of Naf1p and Cbf5p proteins on the chromosome region of H/ACA genes makes them good candidates to control the 3′-end formation of the primary transcripts. As previously observed for C/D box snoRNAs, here we show that in a strain lacking the Cbf5p pseudouridylase, readthrough products extending to the downstream snoRNAs genes are detected. Thus, as for the C/D box, it is an integral component of the mature H/ACA particle that is the sensor controlling correct 3′-end formation.
Recent reports indicated that several subunits of the APT complex are required for 3′-end formation of snoRNAs (7, 11, 35). In particular, Pti1p and Ref2p are required to prevent transcriptional readthrough into downstream genes and were suggested to function in uncoupling cleavage and polyadenylation (7). The finding that Nop1p interacts with Ref2p and enhances its association with the C/D box snoRNA genes (33) suggested a mechanism to mediate polyadenylation-independent 3′-end formation on the specific subpopulation of box C/D snoRNAs. Differently from the C/D box, no interaction was found between GST-Cbf5p, GST-Naf1p, and GST-Nhp2p fusion proteins and any of the APT (Ssu72p, Ref2p, Pti1p, Swd2p, Pta1p) and CFIA (Pcf11p, Clp1p, Rna14p, Rna15p) factors (data not shown) that were previously shown to be involved in snoRNA 3′-end formation (7, 11, 35). Therefore, other still unidentified interactions are likely to mediate the observed coupling between snoRNP assembly and 3′-end formation of H/ACA snoRNAs.
In conclusion, we suggest that, also in the case of H/ACA snoRNA genes, a specific RNA factory is loaded on the nascent transcripts. Naf1p could be transferred during transcription elongation from the chromatin to the newly synthesize snoRNAs and in turn help the recruitment of Nhp2p and Cbf5p. Alternatively, Naf1p could interact with a preassembled complex as suggested by work of Henras and Wang (16, 47). In either case, the cotranscriptional recruitment allows the correct assembly of the H/ACA particle and, at the same time, provides a signal of quality control that allows correct and efficient 3′-end formation (Fig. 5). Further support for the role played by Ser2 phosphorylation in this network is given by the finding that recruitment of Naf1p and Cbf5p on H/ACA snoRNA genes in ctk1Δ cells is severely reduced. The strong readthrough phenotype observed on snoRNA genes in the absence of the Ctk1p kinase suggests that, besides recruiting H/ACA components, Ser2 phosphorylation brings in place 3′-end processing/termination factors, in analogy to what already shown for mRNA genes (1). Future work will be required to address which ones are the factors of the 3′-end formation apparatus that are the sensors of successful H/ACA snoRNP assembly and to elucidate this complex network of interactions.
FIG. 5.
The snoRNA factory. The snoRNP proteins are required for a quality control mechanism in which the functional assembly of box C/D and H/ACA snoRNAs is monitored during transcription. The snoRNA primary transcript attracts the 3′-end formation complex through snoRNP components. The Nop1p/Cbf5p-3′-end machinery interaction may contribute to negatively regulate poly(A) polymerase (Pap1p) at snoRNA cleavage sites. In the case of box C/D snoRNAs, the specific partner has been identified in the Ref2p factor while for H/ACA it is still unidentified.
Supplementary Material
Acknowledgments
We are grateful to Stephen Buratowski for generously providing the YSB854 yeast strain and for helpful suggestions on ChIP analysis. We thank M. Arceci and G. Ricci for skillful technical help.
This work was partially supported by grants from MURST (FIRB-p.n. RBNE015MPB and RBNE01KXC9-, PRIN-Cofin, and Centro di Eccellenza BEMM).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Bousquet-Antonelli, C., Y. Henry, J.-P. Gèlugne, M. Caizergues-Ferrer, and T. Kiss. 1997. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 16:4770-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387. [DOI] [PubMed] [Google Scholar]
- 5.Dez, C., A. Henras, B. Faucon, D. Lafontaine, M. Caizergues-Ferrer, and Y. Henry. 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dez, C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and Y. Henry. 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dheur, S., L. T. A. Vo, F. Voisinet-Hakil, M. Minet, J. M. Schmitter, F. Lacroute, F. Wyers, and L. Minvielle-Sebastia. 2003. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 22:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatica, A., M. Dlakic, and D. Tollervey. 2002. Naf1p is a box H/ACA snoRNP assembly factor. RNA 8:1502-1514. [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 10.Galardi, S., A. Fatica, A. Bachi, A. Scaloni, C. Presutti, and I. Bozzoni. 2002. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 19:6663-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganem, C., F. Devaux, C. Torchet, C. Jacq, S. Quevillon-Cheruel, G. Labesse, C. Facca, and G. Faye. 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22:1588-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 13.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box H/ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 14.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henras, A. K., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J. P. Gélugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henras, A. K., R. Capeyrou, Y. Henry, and M. Caizergues-Ferrer. 2004. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA 10:1704-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 18.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 19.Hoang, C., and A. R. Ferre-D'Amare. 2001. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107:929-939. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, J. M., and M. Ares, Jr. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, W., K. Middleton, H.-J. Yoon, C. Fouquet, and J. Carbon. 1993. An essential yeast protein, Cbf5p, binds in vitro to centromeres and microtubules. Mol. Cell. Biol. 13:4884-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keogh, M.-C., and S. Buratowski. 2004. Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods Mol. Biol. 257:1-16. [DOI] [PubMed] [Google Scholar]
- 24.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 25.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafontaine, D. L., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H+ACA snoRNAs carry Cbf5p, the putative pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafontaine, D. L., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafontaine, D. L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H. D., J. Zagorski, and M. J. Fournier. 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31-44. [DOI] [PubMed] [Google Scholar]
- 33.Morlando, M., M. Ballarino, P. Greco, E. Caffarelli, B. Dichtl, and I. Bozzoni. 2004. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 23:2392-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 13:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nedea, E., X. He, M. Kim, J. Pootoolal, G. Zhong, V. Canadien, T. Hughes, S. Buratowski, C. L. Moore, and J. Greenblatt. 2003. Organization and function of APT, a sub-complex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and snoRNA 3′ ends. J. Biol. Chem. 278:33000-33010. [DOI] [PubMed] [Google Scholar]
- 36.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115:3865-3871. [DOI] [PubMed] [Google Scholar]
- 37.Omer, A. D., S. Ziesche, H. Ebhardt, and P. P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 99:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phatnani, H. P., J. C. Jones, and A. L. Greenleaf. 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43:15702-15719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 40.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shatkin, A. J., and J. L. Manley. 2000. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 7:838-842. [DOI] [PubMed] [Google Scholar]
- 44.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327-331. [DOI] [PubMed] [Google Scholar]
- 45.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, C., C. C. Query, and U. T. Meier. 2002. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 22:8457-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, C., and U. T. Meier. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23:1857-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Lührmann. 1998. Cbf5p, a potential peudouridine synthase, and Nhp2p, a putative RNA binding protein, are present together with Gar1p in all H box/ACA motif snoRNPs and constitute a common bi-partite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Lührmann. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 50.West, M. L., and J. L. Corden. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, P. K., G. Rotondo, T. Porras, P. Legrain, and G. Chanfreau. 2002. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 277:45235-45242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.