Abstract
NF-κB activation is reciprocally regulated by RelA/p65 acetylation and deacetylation, which are mediated by histone acetyltransferases (HATs) and deacetylases (HDACs). Here we demonstrate that in leukemia cells, NF-κB activation by the HDAC inhibitors (HDACIs) MS-275 and suberoylanilide hydroxamic acid was associated with hyperacetylation and nuclear translocation of RelA/p65. The latter events, as well as the association of RelA/p65 with IκBα, were strikingly diminished by either coadministration of the IκBα phosphorylation inhibitor Bay 11-7082 (Bay) or transfection with an IκBα superrepressor. Inhibition of NF-κB by pharmacological inhibitors or genetic strategies markedly potentiated apoptosis induced by HDACIs, and this was accompanied by enhanced reactive oxygen species (ROS) generation, downregulation of Mn-superoxide dismutase and XIAP, and c-Jun N-terminal kinase 1 (JNK1) activation. Conversely, N-acetyl l-cysteine blocked apoptosis induced by Bay/HDACIs by abrogating ROS generation. Inhibition of JNK1 activation attenuated Bay/HDACI lethality without affecting NF-κB inactivation and ROS generation. Finally, XIAP overexpression dramatically protected cells against the Bay/HDACI regimen but failed to prevent ROS production and JNK1 activation. Together, these data suggest that HDACIs promote the accumulation of acetylated RelA/p65 in the nucleus, leading to NF-κB activation. Moreover, interference with these events by either pharmacological or genetic means leads to a dramatic increase in HDACI-mediated lethality through enhanced oxidative damage, downregulation of NF-κB-dependent antiapoptotic proteins, and stress-related JNK1 activation.
Histone deacetylase inhibitors represent a diverse group of compounds which interfere with the function of histone deacetylases (HDACs), enzymes which regulate the acetylation status of histones (40). Gene transcription is regulated by multiple processes, including modifications in chromatin, the scaffolding around which DNA strands are attached. The function of histones can be modified by multiple processes, including phosphorylation, ADP-ribosylation, methylation, ubiquitination, sumoylation, and acetylation (6, 52). Acetylation is reciprocally regulated by histone acetyltransferases (HATs) and HDACs (40). In general, deacetylation of chromatin, e.g., by HDACs, results in a less-relaxed, closed structure which inhibits gene transcription (15). In addition, HDACs can act as direct transcriptional repressors through their association with corepressor complexes (59). In leukemia, the identification of HDAC/corepressor complexes (i.e., those associated with AML-1/ETO) which block leukemic cell maturation (2) has prompted the development of pharmacological HDAC inhibitors (32). In human leukemia cells, short-chain fatty acid HDAC inhibitors, such as sodium butyrate or phenylbutyrate, have long been known to induce differentiation when administered at mM concentrations (25). More recently, newer HDAC inhibitors which are approximately 1,000-fold more potent on a molar basis than butyrate derivatives in triggering leukemic cell maturation have been developed. The HDAC inhibitor group includes benzamide derivatives, such as MS-275 (50), and hydroxamic acid derivatives, such as suberoylanilide hydroxamic acid (SAHA) (23). In addition to reactivating the leukemic cell differentiation program through induction of the endogenous cyclin-dependent kinase inhibitor p21CIP1 (32, 47), HDAC inhibitors, when administered at above-threshold concentrations, induce apoptosis (25, 47, 48). Apoptosis is regulated by multiple events, including the production of reactive oxygen species (ROS) (47, 48) and the generation of ceramide (39), among others.
The NF-κB (nuclear factor kappa B) family of transcription factors consists of five members in mammalian cells: RelA (also referred to as p65), RelB, c-Rel, p50/p105, and p52/p100. These factors are frequently found in heterodimeric or homodimeric complexes (11). NF-κB, corresponding to a heterodimer consisting of two subunits, RelA/p65 and p50, is involved in the regulation of a variety of physiologic processes, including differentiation, proliferation, inflammation, and survival, among others (11). Ordinarily, NF-κB is sequestered in the cytoplasm in an inactive form due to binding to the IκBα protein (30). Various stimuli, including DNA damage, activate kinases referred to as IκB kinases (IKKs), which phosphorylate IκBα, targeting it for proteasomal degradation (31). Once released from the complex involving IκBα, NF-κB translocates to the nucleus, where it binds to DNA and promotes the transcription of a number of genes that oppose apoptosis, including XIAP and Bcl-xL (9, 54). Recently, it has been noted that acetylation and phosphorylation of RelA/p65 play important roles in regulation of NF-κB activation (12). In the former case, RelA/p65 is reversibly acetylated by HATs p300 and CBP at lysine residues 218, 221, and 310. Acetylation of RelA/p65, particularly at lysine 310 and to a lesser extent at lysine 221, enhances NF-κB/DNA binding and attenuates its interaction with IκBα (13). Acetylated RelA/p65 is subsequently deacetylated by HDACs, most notably, HDAC3 (12, 33), which promotes binding to IκBα, leading in turn to IκBα-dependent nuclear export of the complex (12, 13). Thus, it has been proposed that deacetylation of RelA/p65 by HDAC3 represents an intracellular switch that controls the activation status of NF-κB and translocation of RelA/p65 (12).
There is accumulating evidence that NF-κB activation status plays a critical role in regulating the response of cells, including those of neoplastic origin, to HDAC inhibitors. For example, HDAC inhibitors such as trichostatin A (TSA) have been shown to activate NF-κB (3, 41), possibly as a consequence of RelA/p65 acetylation resulting from HDAC3 inhibition, and in so doing, to diminish the lethality of HDAC inhibitors (41), e.g., by promoting transcription of antiapoptotic target genes, such as XIAP and Bcl-xL, among others (9, 54). A corollary of this model is that interference with HDAC inhibitor-mediated NF-κB activation would permit the proapoptotic actions of HDAC inhibitors to predominate.
Although the observations that HDAC inhibitors induce NF-κB activation and that NF-κB activation status influences the cellular response to HDAC inhibitors are well established, relatively little is known about the relationship between these events and RelA/p65 acetylation, particularly in the case of newer, expanded-spectrum HDAC inhibitors, such as SAHA and MS-275. To address these issues, we have employed both genetic and pharmacological approaches to assess the functional role of RelA/p65 acetylation and NF-κB activation on the response of leukemia cells to HDAC inhibitors. For the latter, we have employed Bay 11-7082, an inhibitor of IκBα phosphorylation which disrupts NF-κB function by sparing IκBα from proteasomal degradation, thereby permitting it to bind and inactivate NF-κB (44). Here we report that both SAHA and MS-275 potently increase the acetylation and nuclear accumulation of RelA/p65 in leukemia cells and that these actions are prevented by both Bay 11-7082 and transfection with an IκBα superrepressor. This leads to reduced IκBα proteasomal degradation, increased association of RelA/p65 with IκBα, diminished nuclear translocation of RelA/p65, inhibition of NF-κB-dependent superoxide dismutase 2 (SOD2) expression accompanied by an increase in generation of ROS, downregulation of XIAP, activation of the stress-related c-Jun N-terminal kinase (JNK) pathway, and a dramatic potentiation of mitochondrial dysfunction and apoptosis. Collectively, these findings suggest that acetylation of RelA/p65 by HDAC inhibitors may play a key role in the cytoprotective NF-κB response to such agents and that interruption of this process by pharmacological or genetic means can dramatically increase HDAC inhibitor-mediated antileukemic potential.
MATERIALS AND METHODS
Cells and reagents.
U937, HL-60, Jurkat, and Raji cells were provided by the American Type Culture Collection and maintained in RPMI 1640 medium containing 10% fetal bovine serum as previously reported (17). U937 cells were stably transfected with Ser32/Ser36-mutated IκBα cDNA (encoding a protein referred to as IκBα superrepressor [IκBα-SR]) or an empty vector (pcDNA3.1 [3.1]) and clones selected with G418 (18). U937 cells overexpressing Bcl-2 were obtained as described previously (19). U937 cells with stable overexpression of XIAP and TAM67 were kindly provided by Donald Kufe (Dana-Farber Cancer Institute, Boston, MA) (20) and Michael J. Birrer (National Cancer Institute, Rockville, MD) (7), respectively. TAM67 is mutated form of c-Jun in which the transactivation domain is deleted.
Two cDNA oligonucleotides (5′-gatccGATCAATGGCTACACAGGAttcaagagaTCCTGTGTAGCCATTGATCttttttggaaa-3′ and 3′-gCTAGTTACCGATGTGTCCTaagttctctAGGACACATCGGTAACTAGaaaaaaccttttcga-5′ [uppercase indicates sequences specifically targeting the RelA/p65 gene, and lowercase indicates sequences for cloning into the vector]), which were designed by using the siRNA Target Finder tool (Ambion, Austin, TX), encode a hairpin small interfering RNA (siRNA) targeting the coding region between nucleotides 186 and 204 downstream of the first nucleotide of the start codon of the human RelA/p65 gene. The oligonucleotides were synthesized, annealed, and cloned into BamHI/HindIII sites of pSilencer 3.1-H1 hygro, an RNA polymerase III-based expression vector (Ambion, Austin, TX). U937 cells were transfected with this construct, and clones were selected by hygromycin and screened for stably downregulated RelA/p65 expression.
In some cases, U937 cells were transiently transfected with 1 μg JNK1 (mitogen-activated protein [MAP] kinase 8) annealed double-stranded RNA interference oligonucleotide (Orbigen, San Diego, CA) (18) by using the Amaxa Nucleofector device (program V-01) with Cell Line Nucleofector Kit V (Amaxa GmbH, Cologne, Germany) as per the manufacturer's instructions. The transfected cells were recovered for 6 h at 37°C, after which cells were treated with drugs for 24 h and subjected to analysis of cell viability by using ViaCount reagent (Guava Technologies, Hayward, CA) and the Guava Personal Cytometer as per the manufacturer's instructions.
The HDAC inhibitor MS-275 was kindly provided by Osamu Nakanishi (Shering, AG), and SAHA was purchased from Biovision (Mountain View, CA), dissolved in dimethyl sulfoxide (DMSO), stored at −80°C, and subsequently diluted with serum-free RPMI medium prior to use. Bay 11-7082 (an inhibitor of IκBα phosphorylation; Alexis, San Diego, CA), SP600125 (a selective JNK inhibitor; Calbiochem, San Diego, CA), anisomycin (a JNK/stress-activated protein kinase [SAPK] activator; Biomol, Plymouth Meeting, PA), Etoposide (VP-16; Sigma, St. Louis, MO), and resveratrol (an activator of SIRT1; Biomol) were dissolved in sterile DMSO and stored at −20°C under light-protected conditions. SN50 (a specific NF-κB-inhibitory peptide; Alexis) and its control inactive peptide SN50M were dissolved in sterile phosphate-buffered saline, aliquoted, and stored at −20°C. The free-radical scavenger N-acetyl l-cysteine (L-NAC; Calbiochem) and a cell-permeable SOD mimetic and peroxynitrite scavenger, MnTBAP [Mn(III) tetrakis(4-benzoic acid)porphyrin chloride; Biomol], were prepared in sterile water immediately before use. In all experiments, the final concentration of DMSO did not exceed 0.1%.
Experimental format.
All experiments were performed utilizing logarithmically growing cells (4 to 6 × 105 cells/ml). Cells were pretreated with Bay 11-7082, SN50, or SN50M for 1 h, after which HDAC inhibitors (MS-275 or SAHA) were added and incubated for various intervals, generally 24 h. In some studies, L-NAC, MnTBAP, or SP600125 was added prior to NF-κB inhibitors. After drug treatment, cells were harvested and subjected to further analysis as described below.
Flow cytometry.
Apoptosis, mitochondrial membrane potential (ΔΨm), and ROS production were analyzed by flow cytometry after a staining with annexin V-fluorescein isothiocyanate (FITC) (BD PharMingen, San Diego, CA), dihexyloxacarbocyanine (DiOC6) (Molecular Probes Inc., Eugene, OR), and acetooxymethyl ester of dihydro-dichlorodihydrofluorescein (DCF) (Molecular Probes Inc.), respectively, as described previously (17, 18, 36). In some cases, the extent of apoptosis was also assessed by morphological assessment (17).
Kinase assays.
SAPK/JNK kinase activity was analyzed by using a SAPK/JNK assay kit (Cell Signaling, Beverly, MA) as per the manufacturer's instructions (18). For the IKK kinase assay, 200 μg/condition of protein was incubated with 1 μg of IKK-β antibody (T-20; Santa Cruz Biotech, Santa Cruz, CA) for 1 h at 4°C and then incubated with protein A/G-agarose at 4°C overnight. After four washes were performed with radioimmunoprecipitation assay (RIPA) buffer, the kinase assay was performed by using an IKK-β kinase assay kit (Cell Signaling) and [γ-32P]ATP (3,000 Ci/mmol; ICN Biomedicals, Irvine, CA). The amount (cpm) of γ-32P incorporated into IκBα (Ser32) peptide substrate was determined by scintillation counting.
Western blot analysis.
Samples from either whole-cell pellets or S-100 cytosolic fractions were prepared, and 30 μg/condition of proteins was subjected to Western blot analysis as previously described in detail (17). For analysis of protein phosphorylation, 1 mM (each) Na vanadate and Na pyrophosphate were added to sample buffer, no sodium dodecyl sulfate was included in the transfer buffer, and Tris-buffered saline was used instead of phosphate-buffered saline throughout. The blots were probed with the appropriate dilutions of primary antibody as follows. Where indicated, the blots were reprobed with β-actin antibody (BD PharMingen) to ensure equal loading and transfer of proteins. In some case, the density of blots was quantified using the FluoChem 8800 imaging system (Alpha Innotech, San Leandro, CA) and VideoTesT-Master software (VideoTesT, Ltd., St. Petersburg, Russia). The primary antibodies included the following: anti-acetylated lysine and anti-IκBα antibodies (Upstate Biotech, Lake Placid, NY); anti-IKK-β, anti-RelA/p65, anti-SOD1, anti-SOD2, anti-histone H3, anti-histone H4, anti-phospho-JNK (Thr183/Tyr185), anti-Mcl-1, anti-cytochrome c, and anti-Bcl-xS/L antibodies (Santa Cruz Biotech); anti-phospho-p44/42 MAP kinase (ERK1/2, Thr202/Tyr204), anti-SAPK/JNK, anti-phospho-p38 MAP kinase (Thr180/Tyr182), anti-phospho-IKKα (Ser180)/IKKβ (Ser181), and anti-cleaved caspase 9 antibodies (Cell Signaling); anti-XIAP antibody (Transduction Lab, Lexington, KY); anti-human Bcl-2 oncoprotein (Dako, Carpinteria, CA); anti-poly(ADP-ribose) polymerase (PARP) antibody (Biomol); and anti-caspase 8 antibody (Alexis).
Immunoprecipitation.
The status of RelA/p65 acetylation and the extent of IκBα bound to RelA/p65 were evaluated by immunoprecipitation/Western blot analysis as described previously (17). Briefly, 200 μg protein per condition was incubated under continuous shaking with 1 μg of anti-RelA/p65 antibody (Santa Cruz Biotech) overnight at 4°C. A total of 20 μl/condition of Dynabeads (goat anti-mouse immunoglobulin; Dynal, Oslo, Norway) was added and incubated for an additional 4 h. After being washed three times with RIPA buffer, the bead-bound protein was eluted by being vortexed and being boiled in 1× sample buffer. The samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blot analysis by using anti-acetylated lysine and anti-IκBα antibodies.
Analysis of RelA/p65 translocation.
Nuclear and cytoplasmic fractions were prepared by using the NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL) as per the manufacturer's instructions. The protein amount of each fraction was quantified and 30 μg protein per condition subjected to Western blot analysis by using anti-RelA/p65 antibody.
Electrophoretic mobility shift assay (EMSA).
Consensus oligonucleotides corresponding to the sites of the immunoglobulin κ light-chain enhancer and AP2 binding were purchased from Promega (Madison, WI) and labeled with [γ-32P]ATP. Nuclear extracts were prepared by using NE-PER nuclear and cytoplasmic extraction reagents as described above. A total of 5 μg/condition of nuclear proteins was subjected to EMSA for NF-κB/DNA binding as described previously in detail (18). As a loading control, the same amount (5 μg) of nuclear protein was used for EMSA of AP2/DNA binding.
NF-κB/luciferase reporter assay.
NF-κB TransLucent reporter vector (NF-κB/Luc), which is designed to monitor transcription factor binding activity of NF-κB through the use of a standard luciferase assay, was purchased from Panomics (Redwood City, CA). A total of 4 × 106 cells were transiently transfected with 4 μg of NF-κB/Luc by using the Amaxa Nucleofector device (program V-01) with Nucleofector Kit V (Amaxa GmbH). Cells were incubated for 1 h and subsequently treated with the indicated agents, after which cells were harvested and subjected to a luciferase assay by using a luciferase reporter assay kit (BD Clontech, Palo Alto, CA) as per the manufacturer's instructions. Relative luciferase activity was determined to reflect transcriptional activity of NF-κB, expressed as the fold increase relative to the activity of untreated controls.
Statistical analysis.
For analysis of apoptosis (morphology and annexin V), ΔΨm, ROS production, NF-κB/luciferase reporter assays, and the IKK kinase assay, values represent the means ± standard deviations (SD) for at least three separate experiments performed in triplicate. The levels of significance of the differences between experimental variables were determined by using Student's t test.
RESULTS
Bay 11-7082 blocks NF-κB activation induced by SAHA and MS-275.
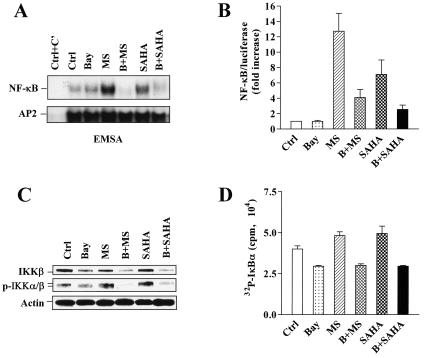
EMSA analysis, as well as the luciferase reporter assay, revealed that 24-h exposure of U937 cells to either 2 μM MS-275 or 1 μM SAHA increased NF-κB/DNA binding and NF-κB/luciferase reporter activity, whereas coadministration of the IκBα phosphorylation inhibitor Bay 11-7082 (3 μM) largely blocked this action (Fig. 1A and B). Similar results for NF-κB/luciferase reporter activity were obtained by transfection with an NF-κB-driven IL-8 promoter/luciferase reporter construct but not with an NF-κB binding site-mutated interleukin-8 promoter/luciferase reporter construct (data not shown) (both constructs kindly provided by Xianjun Feng, Virginia Commonwealth University). Recently, it has been reported that the HDAC inhibitor TSA by itself modestly induces IKK activity while markedly prolonging tumor necrosis factor alpha (TNF-α)-induced IKK activation (1). On the other hand, Bay 11-7082 has been found to reduce IKK-β expression in a dose-dependent manner (35). To test the effects of HDAC inhibitors (e.g., MS-275 and SAHA) and Bay 11-7082 on the levels of total and phosphorylated IKK-β, as well as the activity of IKKs, Western blot analysis and an IKK kinase assay were performed in parallel. Bay 11-7082 reduced expression of both IKK-β and phospho-IKK-α/β (Fig. 1C) as well as IKK activity (Fig. 1D) in both untreated and MS-275- and SAHA-treated U937 cells. On the other hand, MS-275 and SAHA by themselves modestly induced phosphorylation and activation of IKKs and had no effect on total levels of IKK-β. Thus, both SAHA and MS-275 induced various events associated with NF-κB activation in U937 cells, and these effects were antagonized by Bay 11-7082.
FIG. 1.
Bay 11-7082 blocks NF-κB activation induced by HDAC inhibitors, accompanied by downregulation/inactivation of IKK. (A) U937 cells were treated for 24 h with 3 μM Bay 11-7082 with (B) or without (Bay) 2 μM MS-275 (MS) or 1 μM SAHA, after which nuclear extracts were prepared and subjected to EMSA as described in Materials and Methods. Activity of NF-κB/DNA binding is reflected by the extent of binding to 32P-labeled NF-κB consensus oligonucleotide (upper panel). AP2/DNA binding was assessed by using an AP2 consensus oligonucleotide for equal loading controls (lower panel). Ctrl+C', nuclear extract of untreated U937 cells preincubated for 20 min at room temperature with unlabeled oligonucleotides as specific competitor (designed C') controls. The results are representative of three separate experiments. Two additional studies yielded equivalent results. (B) U937 cells were transfected with NF-κB/Luc as described in Materials and Methods. After a 1-h recovery, cells were divided into equivalent aliquots, treated for 24 h with 3 μM Bay 11-7082 with (B) or without (Bay) 2 μM MS-275 (MS) or 1 μM SAHA and then subjected to luciferase assays. Relative luciferase activities were determined by normalizing to total protein. The value for untreated U937 cells was arbitrarily set to 1, and NF-κB/luciferase activity is expressed as the increase relative to the activity of the untreated control (Ctrl). The results represent the means ± SD for three separate experiments performed in triplicate. (C) U937 cells were treated as described for panel A, after which Western blot analysis was performed to assess levels of expression of IKK-β and phosphorylated IKK-α/β. Each lane was loaded with 30 μg of protein; blots were subsequently stripped and reprobed with anti-β-actin to ensure equivalent loading and transfer of protein. The results are representative of three separate experiments. Abbreviations are the same as those for panel B. (D) Alternatively, cells were lysed and subjected to an IKK kinase assay as described in Materials and Methods. IKK activity is reflected by the extent of [32P]ATP incorporated into an IκBα peptide substrate. The results represent the means ± SD for three separate experiments performed in triplicate. Abbreviations are the same as those for panel B.
HDAC inhibitor-induced NF-κB activation is associated with increased acetylation and nuclear localization of RelA/p65, events interrupted by Bay 11-7082.
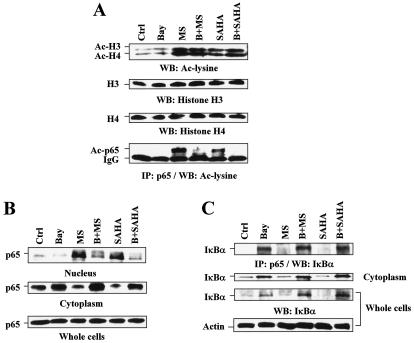
Studies were then undertaken to determine whether increased NF-κB activity triggered by HDAC inhibitors might be related to RelA/p65 acetylation, a process regulated by both HATs (e.g., p300 and CBP) and HDACs (e.g., HDAC3). To this end, an anti-RelA/p65 antibody was employed to immunoprecipitate RelA/p65 protein, which was then subjected to Western blot analysis by using an anti-acetylated lysine antibody. As shown in Fig. 2A, treatment with either 2 μM MS-275 or 1 μM SAHA markedly increased levels of acetylated histones H3 and H4, as well as RelA/p65, which may reflect inhibition of HDAC3 by SAHA and MS-275 (37, 46). Notably, coadministration of Bay 11-7082 had little effect on H3 and H4 acetylation but dramatically diminished RelA/p65 acetylation induced by SAHA and MS-275.
FIG. 2.
HDAC inhibitors induce hyperacetylation and nuclear translocation of RelA/p65, which is reversed by Bay 11-7082 through an increasing association of IκBα with RelA/p65. U937 cells were treated for 24 h with 3 μM Bay 11-7082 with (B) or without (Bay) 2 μM MS-275 (MS) or 1 μM SAHA, after which Western blot and immunoprecipitation analyses were performed. Experiments were conducted at least three times, with similar results. Ctrl, control. (A) Western blot analysis was performed to monitor the acetylation status of histones and total levels of histones H3 and H4 (upper panels). Alternatively, to evaluate the extent of acetylated (Ac) RelA/p65, cells were lysed in RIPA buffer, and 200 μg of protein was immunoprecipitated (IP) with anti-RelA/p65 antibody and subsequently subjected to Western blot (WB) analysis using anti-acetylated lysine antibody (lower panel). (B) Whole-cell lysates, as well as nuclear and cytoplasmic fractions, were prepared as described in Materials and Methods and subjected to Western blot analysis to determine the subcellular distribution of RelA/p65. (C) To assess the association of RelA/p65 with IκBα, immunoprecipitation (IP) was performed using anti-RelA/p65 antibody as described in Materials and Methods. Immunoprecipitates were subjected to Western blot (WB) analysis using anti-IκBα antibodies (upper panel). In parallel, samples of whole-cell lysates and cytoplasmic fractions were subjected to Western blot analysis to monitor the total and cytoplasmic levels of IκBα (lower panels). For Western blot analysis, each lane was loaded with 30 μg of protein; blots were subsequently stripped and reprobed with anti-β-actin, as indicated, to ensure equivalent loading and transfer of protein.
It has been reported that TSA prolongs TNF-α-induced NF-κB/DNA binding activity and the accumulation of NF-κB in the nucleus, events associated with delayed cytoplasmic reappearance of IκBα (1). Because deacetylation of RelA/p65 by HDCA3 promotes IκBα/RelA assembly and IκBα-dependent translocation of nuclear RelA/p65 to the cytoplasm (14), the subcellular distribution of RelA/p65 was then examined. Treatment of cells with MS-275 and SAHA resulted in the accumulation of RelA/p65 in the nuclear fractions while reciprocally decreasing expression of RelA/p65 in the cytoplasmic fractions (Fig. 2B). No alterations in total RelA/p65 levels were noted. Thus, NF-κB activation by SAHA and MS-275 was closely associated with the increase of acetylated RelA/p65 and a shift in distribution of NF-κB from the cytoplasm to the nucleus, where it is presumably acetylated by HATs and deacetylated by HDAC3 (14). Significantly, coadministration of Bay 11-7082 with either MS-275 or SAHA markedly diminished nuclear localization and reciprocally increased cytoplasmic localization of RelA/p65 (Fig. 2B). Finally, Western blot and immunoprecipitation analyses demonstrated that exposure to Bay 11-7082 alone or in conjunction with MS-275 or SAHA resulted in a clear increase in total and cytoplasmic levels of IκBα, as well as RelA/p65-associated IκBα (Fig. 2C). Collectively, these findings indicate that Bay 11-7082 increases the association of RelA/p65 with IκBα and opposes the nuclear accumulation and acetylation of RelA/p65 in HDAC inhibitor-treated cells.
HDAC inhibitors fail to activate NF-κB in U937 cells transfected with an IκBα superrepressor.
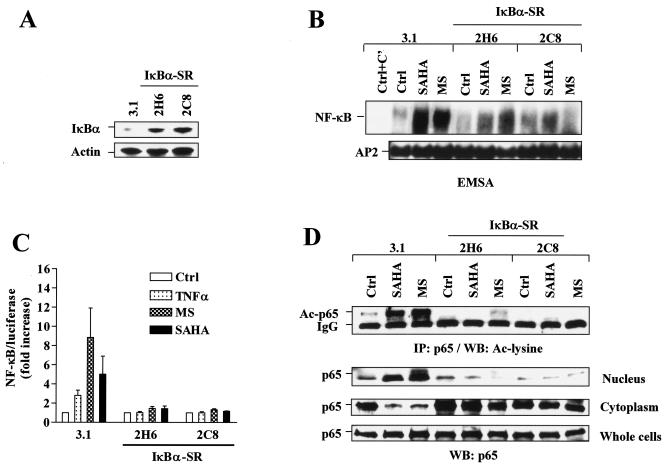
To confirm the role of interference with NF-κB function in the actions of Bay 11-7082, U937 cells were stably transfected with a mutated IκBα construct in which serines 32 and 36 are replaced with alanines. This construct encodes a protein (IκBα-SR) which can no longer be phosphorylated by IKKs and degraded by the proteasome (49). As anticipated, stable transfection of two separate clones with IκBα-SR (2H6 and 2C8) (Fig. 3A) blocked NF-κB/DNA binding (Fig. 3B) as well as NF-κB/luciferase reporter activity (Fig. 3C) induced by 2 μM of either MS-275 or SAHA. Similar results were obtained for NF-κB/luciferase reporter activity by using an NF-κB-driven IL-8 promoter/luciferase construct but not an NF-κB binding site-mutated IL-8 promoter/luciferase reporter construct (data not shown). Consistent with results obtained with Bay 11-7082, both MS-275 and SAHA failed to increase RelA/p65 acetylation and nuclear translocation in IκBα-SR-transfected cells in contrast to empty-vector controls (Fig. 3D). Together, these findings raise the possibility that Bay 11-7082 or stable transfection with an IκBα-SR blocks HDAC inhibitor-induced NF-κB activation by inhibiting IκBα degradation and thus sequestering NF-κB in the cytoplasm. These phenomena could also account for the observed diminution in NF-κB acetylation, which is known to be a nuclear event (12).
FIG. 3.
HDAC inhibitors fail to induce NF-κB activation, as well as RelA/p65 acetylation and nuclear translocation in IκBα-SR-transfected cells. (A) U937 cells were stably transfected with Ser32/Ser36-mutated IκBα cDNA (IκBα-SR) or an empty vector (pcDNA3.1). A Western blot analysis was performed to monitor expression of IκBα. (B) Two separate clones (2H6 and 2C8) of IκBα-SR/U937 cells as well as control 3.1/U937 cells were treated for 24 h with 2 μM of either MS-275 or SAHA, after which nuclear extracts were prepared and subjected to EMSA. AP2/DNA binding was assessed by using an AP2 consensus oligonucleotide for equal loading controls. Ctrl+C', nuclear extract of untreated 3.1/U937 cells preincubated with unlabeled oligonucleotides as specific competitor (C′) controls. Two additional studies yielded equivalent results. (C) IκBα-SR/U937 and 3.1/U937 cells were transfected with NF-κB/Luc and then exposed to 2 μM of MS-275 or SAHA for 24 h, after which a luciferase assay was performed to monitor NF-κB activity. In parallel, cells were treated with 10 ng/ml TNF-α for 30 min as a control. Relative luciferase activities were determined by normalizing to total protein. For each cell line, values for untreated controls were arbitrarily set to a value of 1, and NF-κB/luciferase activity is expressed as the fold increase relative to untreated controls (ctrl). The results represent the means ± SD for three separate experiments performed in triplicate. (D) IκBα-SR/U937 and 3.1/U937 cells were treated as described for panel B, after which immunoprecipitation (IP) and a Western blot (WB) analysis were performed to evaluate the acetylation (Ac) status (upper panel) and subcellular distribution of RelA/p65 (lower panels), respectively. Ctrl, control. The results are representative of three separate experiments. For Western blot analyses in panels A and D, each lane was loaded with 30 μg of protein; blots were reprobed with anti-β-actin, as indicated, to ensure equivalent loading and transfer of protein.
NF-κB inhibition by Bay 11-7082 potentiates mitochondrial dysfunction and apoptosis mediated by HDAC inhibitors.
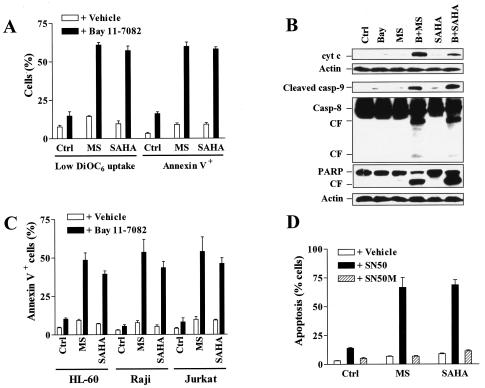
To determine what effect, if any, pharmacological interruption of the NF-κB pathway would exert on the lethality of HDAC inhibitors MS-275 and SAHA, U937 cells were exposed for 24 h to a low, marginally toxic concentration of Bay 11-7082 (3 μM) in conjunction with minimally toxic concentrations of MS-275 (2 μΜ) or SAHA (1 μΜ), after which the loss of ΔΨm, reflected by diminished (low) DiOC6 uptake, and apoptosis were monitored. In each case, coadministration of Bay 11-7082 with MS-275 or SAHA resulted in a dramatic potentiation of loss of ΔΨm, accompanied by an increase in the percentage of annexin V+ cells (Fig. 4A). Consistent with these results, the exposure of U937 cells to 2 μM MS-275 or 1 μM SAHA individually had little effect on cytosolic redistribution of mitochondrial cytochrome c, whereas coexposure to Bay 11-7082 markedly increased translocation of cytochrome c into the cytosolic S-100 fraction (Fig. 4B). These events were associated with a clear increase in cleavage/activation of procaspase-9 and procaspase-8 and PARP degradation. Similar increases in lethality in other human leukemia cell lines, e.g., Jurkat T-lymphoblastic, HL-60 promyelocytic, and Raji B-lymphoblastic leukemia cells (Fig. 4C), were observed. To confirm the role of NF-κB in Bay 11-7082 actions, we utilized the NF-κB inhibitory peptide SN50, which contains the nuclear localization sequence (NLS; residues 360 through 369) of NF-κB p50 linked to the hydrophobic region of the signal peptide of Kaposi fibroblast growth factor and inhibits translocation of NF-κB complex into the nucleus (38). As shown in Fig. 4D, coadministration of SN50, but not of its inactive-mutation control peptide (SN50M), markedly enhanced the lethalities of both MS-275 and SAHA.
FIG. 4.
NF-κB inhibitors potentiate mitochondrial dysfunction and apoptosis mediated by HDAC inhibitors. (A) U937 cells were exposed for 24 h to 2 μM MS-275 (MS) or 1 μM SAHA in the absence or presence of 3 μM Bay 11-7082, after which the percentages of cells exhibiting decreased ΔΨm, reflected by low DiOC6 uptake, and annexin V positivity were determined by flow cytometry as described in Materials and Methods. (B) Alternatively, a Western blot analysis was performed to assess expression of cytosolic cytochrome c (cyt c) and cleavage of caspases and PARP. CF, cleavage fragment; Casp, caspase; Ctrl, control; Bay, Bay 11-7082; MS, MS-275; B+MS, Bay 11-7082 plus MS-275; B+SAHA, Bay 11-7082 plus SAHA. Each lane was loaded with 30 μg of protein; blots were subsequently stripped and reprobed with anti-β-actin to ensure equivalent loading and transfer of protein. The results are representative of three separate experiments. (C) HL-60, Jurkat, and Raji cells were exposed to 2 μM MS-275 (MS) or 1 μM SAHA for 24 h in the absence or presence of Bay 11-7082 (HL-60, 3 μM; Jurkat, 2 μM; Raji, 5 μM), after which the percentage of annexin V-positive cells was determined. (D) U937 cells were incubated for 24 h with 2 μM MS-275 (MS) or 1 μM SAHA in the absence and presence of 150 ng/ml NF-κB inhibitory peptide (SN50) or control negative peptide (SN50M), respectively, after which the percentage of cells exhibiting apoptotic morphology was determined as described in Materials and Methods. For panels A, C, and D, results represent the means ± SD for three separate experiments performed in triplicate.
Genetic interruption of the NF-κB pathway sensitizes cells to HDAC inhibitor-mediated lethality.
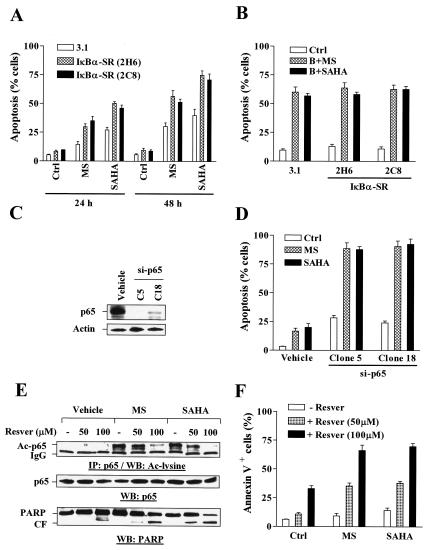
Consistent with the previous results involving pharmacological inhibition of NF-κB, U937 cells stably transfected with IκBα-SR were significantly more sensitive to MS-275- and SAHA-induced apoptosis than those transfected with an empty vector, particularly after 48 h of drug exposure (P < 0.02) (Fig. 5A). Notably, coadministration of Bay 11-7082 did not potentiate HDAC inhibitor-mediated lethality further in IκBα-SR cells (P was >0.05 relative to controls) (Fig. 5B), presumably because NF-κB was already disabled in these cells. Parallel studies with U937 cells stably transfected with a RelA/p65 siRNA construct (si-p65) were performed. Transfection with si-p65 dramatically downregulated the expression of RelA/p65 (clones 5 and 18) (Fig. 5C) and markedly sensitized U937 cells to apoptosis induced by 2 μM of either MS-275 or SAHA (P was <0.01 relative to controls) (Fig. 5D), as well as by lower concentrations (1 μM) of these HDAC inhibitors (data not shown). Taken together, these findings suggest that HDAC inhibitor-induced NF-κB activation provides a protective mechanism against the lethal effects of HDAC inhibitors and that interruption of the NF-κB pathway by either genetic or pharmacological means markedly potentiates HDAC inhibitor-mediated mitochondrial dysfunction and apoptosis.
FIG. 5.
Stable transfection with either IκBα-SR or RelA/p65 siRNA sensitizes U937 cells to the lethalities of HDAC inhibitors, a phenomenon mimicked by coadministration of the SIRT1 activator resveratrol. (A) IκBα-SR/U937 cells as well as 3.1/U937 cells were treated for 24 to 48 h with 2 μM of either MS-275 (MS) or SAHA, after which the percentage of cells exhibiting apoptotic morphology was determined by evaluating Wright-Giemsa-stained cytospin slides as described in Materials and Methods. Ctrl, control. (B) IκBα-SR/U937 and 3.1/U937 cells were treated for 24 h with 2 μM MS-275 (MS) or 1 μM SAHA in the absence or presence (B) of 3 μM Bay 11-7082, after which the percentage of cells exhibiting apoptotic morphology was determined as described for panel A. Ctrl, control. (C) U937 cells were stably transfected with a construct encoding RelA/p65 siRNA as described in Materials and Methods, and a Western blot analysis was performed to monitor expression of RelA/p65. (D) Two clones (clone 5 and 18) of si-RelA/p65-transfected cells were then treated with 2 μM of either MS-275 (MS) or SAHA for 24 h, after which the percentage of cells exhibiting apoptotic morphology was determined as described for panel A. Ctrl, control. (E) U937 cells were incubated for 24 h with 2 μM MS-275 (MS) or 1 μM SAHA in the absence (−) or presence of 50 to 100 μM resveratrol (Resver), after which immunoprecipitation (IP) and Western blot (WB) analyses were performed to assess acetylation and total levels of p65 as well as PARP cleavage. Ac, acetylated; IgG, immunoglobulin G; CF, cleavage fragment. (F) Alternatively, annexin V-FITC/flow cytometric analysis was performed to determine the percentage of apoptotic cells. For panels A, B, D, and F, results represent the means ± SD for three separate experiments performed in triplicate. Ctrl, control; MS, MS-275; Resver, resveratrol. For Western blots shown in panels C and E, each lane was loaded with 30 μg of protein; blots were reprobed with anti-β-actin, as indicated, to ensure equivalent loading and transfer of protein. Two additional studies yielded equivalent results.
The SIRT1 activator resveratrol diminishes HDAC inhibitor-mediated RelA/p65 acetylation and enhances the lethality of HDAC inhibitors.
Very recently, it has been reported that SIRT1, an NAD-dependent class III HDAC of the Sirtuin family, physically interacts with RelA/p65 and inhibits transcriptional activity of NF-κB by deacetylating RelA/p65 at lysine 310. Activation of SIRT1 by resveratrol, a small-molecule agonist of Sirtuin activity, enhances TNF-α-induced apoptosis by modulating the acetylation status of RelA/p65 (60). As SIRT1 is an HDAC which is not sensitive to known inhibitors (e.g., TSA) of class I and II histone deacetylases (5, 40), the possibility that activation of SIRT1 may oppose RelA/p65 acetylation mediated by MS-275 and SAHA and, in so doing, potentiate their lethality arose. To investigate further whether HDAC inhibitor-mediated RelA/p65 acetylation is linked to cytoprotective effects against HDAC inhibitors, studies employing resveratrol were carried out. Coadministration of 50 μM resveratrol modestly diminished MS-275- or SAHA-induced RelA/p65 acetylation, an event accompanied by increased PARP degradation (Fig. 5E) and apoptosis (Fig. 5F), whereas 100 μM markedly diminished this. These findings provide further support for the notion that HDAC inhibitor-mediated RelA/p65 acetylation plays an important cytoprotective role in cells exposed to HDAC inhibitors and that these events can be reversed by activating the SIRT1 class of HDACs.
Potentiation of HDAC inhibitor-mediated lethality by NF-κB inhibition is associated with enhanced ROS generation as well as downregulation of mitochondrial Mn-SOD (SOD2) expression.
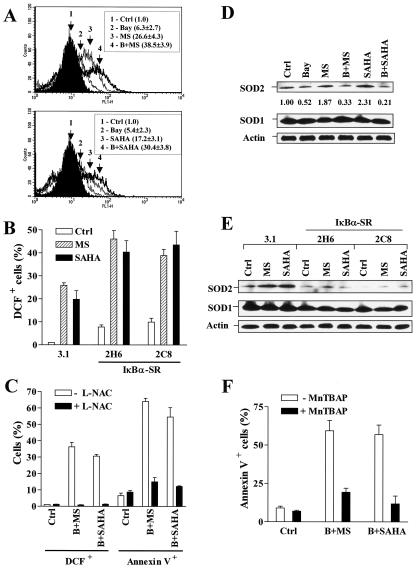
The lethalities of MS-275 and SAHA have been associated with ROS production (47, 48), while ROS generation is closely linked with NF-κB activation in a cell type-specific manner (8). Therefore, attempts to determine whether inhibition of NF-κB modified ROS generation triggered by HDAC inhibitors were made. To this end, U937 cells were exposed to 2 μM MS-275 or 1 μM SAHA in the conjunction with 3 μM Bay 11-7082, after which cellular ROS production was monitored by flow cytometric analysis of the oxidation of dichlorodihydroflurescein. As shown in Fig. 6A, marginally active concentrations of Bay 11-7082 significantly enhanced MS-275- or SAHA-mediated ROS production (P was <0.02 relative to values for HDAC inhibitors alone in both cases). Consistent with these results, stable transfection with IκBα-SR sensitized U937 cells to HDAC inhibitor-induced ROS generation (P was <0.01 relative to empty vector controls) (Fig. 6B). Lastly, the free-radical scavenger L-NAC blocked Bay 11-7082/HDAC inhibitor-induced ROS generation and cytotoxicity (Fig. 6C), indicating a significant functional role for oxidative damage in the lethality of these regimens.
FIG. 6.
Bay 11-7082 and stable transfection with IκBα-SR enhance HDAC inhibitor-mediated ROS generation, accompanied by the downregulation of SOD2. Ctrl, control. (A) U937 cells were exposed for 24 h to 3 μM Bay 11-7082 with (B) or without (Bay) 2 μM MS-275 (MS) (upper panel) or 1 μM SAHA (lower panel), after which the ROS production was determined by a staining with the acetooxymethyl ester of dihydro-dichlorodihydrofluorescein and flow cytometry as described in Materials and Methods. DCF is nonfluorescent in its dihydro form but becomes highly fluorescent upon reaction with ROS. The percentage of cells with increased DCF fluorescence for each condition was determined. (B) Two clones (2H6 and 2C8) of IκBα-SR/U937 cells as well as 3.1/U937 cells were treated for 24 h with 2 μM of either MS-275 (MS) or SAHA, after which the percentage of cells exhibiting positive DCF fluorescence was determined as described for panel A. (C) U937 cells were pretreated with 10 mM L-NAC for 3 h and then exposed to 3 μM Bay 11-7082 (B) plus 2 μM MS-275 (MS) or 1 μM SAHA for additional 24 h, after which the percentages of cells exhibiting positive DCF fluorescence and annexin V-FITC were determined by flow cytometry. (D) U937 cells were treated as described for panel A, after which a Western blot analysis was performed to evaluate levels of SOD1 and SOD2. Density of SOD2 protein bands was quantified by using an imaging system as described in Materials and Methods. Values reflect the ratio of integrated densitometric determinations for each condition relative to untreated controls. Abbreviations are the same as those for panel A. (E) IκBα-SR/U937 cells and 3.1/U937 cells were treated as described for panel B, after which levels of expression of SOD1 and SOD2 were evaluated by Western blot analysis. MS, MS-275. (F) U937 cells were preincubated with 150 μM MnTBAP for 2 h and then treated with 3 μM Bay 11-7082 plus 2 μM MS-275 (B+MS) or 1 μM SAHA (B+SAHA) for additional 24 h. At the end of the incubation interval, cells were harvested and subjected to annexin V/flow cytometric analysis. For panels A through C and F, values represent the means ± SD for three separate experiments performed in triplicate. For Western blot analyses in panels D and E, each lane was loaded with 30 μg of protein; blots were subsequently stripped and reprobed for the expression of β-actin to ensure equivalent loading and transfer of protein. Two additional studies yielded equivalent results.
To investigate further the mechanism of NF-κB inhibition in promotion of HDAC inhibitor-induced ROS generation, expression of superoxide dismutases, which play a key role in protecting cells from oxidative damage, was evaluated. In U937 cells, treatment with MS-275 or SAHA alone resulted in a clearly discernible increase in SOD2 levels (Fig. 6D), consistent with reports that the HDAC inhibitor TSA enhances SOD2 expression (26). Notably, Bay 11-7082 prevented the increase in SOD2 expression, which is known to be an NF-κB-dependent phenomenon (21, 34), in cells exposed to MS-275 or SAHA (Fig. 6D). In contrast, levels of SOD1 did not change. Consistent with these findings, MS-275 and SAHA failed to induce SOD2 expression in IκBα-SR cells relative to their empty-vector controls (Fig. 6E). Lastly, the cell-permeable SOD mimetic MnTBAP, which blocks oxidative damage-mediated cell death by scavenging ROS but not nitric oxide (27, 28), significantly protected cells from Bay 11-7082/HDAC inhibitor lethality (Fig. 6F). Collectively, these findings raise the possibility that interruption of the NF-κB pathway lowers the threshold for HDAC inhibitor-mediated ROS generation and lethality, at least in part, by inhibiting expression of SOD2.
Apoptosis induced by HDAC inhibitors in conjunction with NF-κB inhibition is accompanied by XIAP downregulation and JNK1 activation.
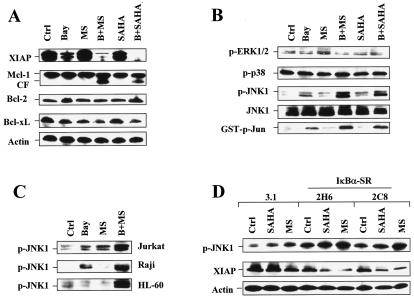
To investigate further mechanisms underlying potentiation of HDAC inhibitor-mediated lethality by NF-κB disruptions, effects on NF-κB-related antiapoptotic and stress-associated proteins were monitored. The expression of XIAP, a well-characterized NF-κB downstream antiapoptotic molecule (54), was inhibited following treatment with Bay 11-7082 alone and was inhibited even more when Bay 11-7082 was coadministered with MS-275 or SAHA (Fig. 7A). Expression of Bcl-xL, another NF-κB downstream target (18), was also modestly decreased after combined treatment (Fig. 7A). In contrast, exposure to Bay 11-7082 and SAHA or MS-275 did not alter expression of Bcl-2 and Mcl-1, although the presence of an Mcl-1 cleavage product was noted following combined treatment (Fig. 7A).
FIG. 7.
NF-κB inhibition results in JNK1 activation and XIAP downregulation in HDAC inhibitor-treated cells. Ctrl, control. (A and B) U937 cells were exposed for 24 h to 3 μM Bay 11-7082 with (B) or without (Bay) 2 μM MS-275 (MS) or 1 μM SAHA, after which a Western blot analysis was performed to evaluate alterations in expression of antiapoptotic proteins (A) and phosphorylation (activation) of ERK1/2, p38, and JNK (B). CF, cleavage fragment. (B) Alternatively, kinase assays were performed, as described in Materials and Methods, to assess SAPK/JNK activity, reflected by phosphorylated (p) GST-c-Jun (bottom panel). (C) Jurkat, Raji, and HL-60 cells were exposed to 2 μM MS-275 with (B+MS) or without (MS) Bay 11-7082 (Jurkat, 2 μM; Raji, 5 μM; HL-60, 3 μM), after which cells were lysed and subjected to Western blot analysis to monitor phosphorylation of JNK. Bay, Bay 11-7082; p, phosphorylated. (D) IκBα-SR/U937 cells, as well as 3.1/U937 cells, were treated for 24 h with 2 μM of either MS-275 (MS) or SAHA, after which a Western blot analysis was performed to assess JNK phosphorylation and XIAP expression. p, phosphorylated. For Western blot analysis in panels A through D, each lane was loaded with 30 μg of protein; blots were subsequently stripped and reprobed for the expression of β-actin, as indicated, to ensure equivalent loading and transfer of protein. The results are representative of three separate experiments.
Activation of the stress-related kinase JNK has been linked to cell death mediated by both inhibition of NF-κB (45, 55) and exposure to HDAC inhibitors (64). As shown in Fig. 7B, treatment with either MS-275 or SAHA in combination with Bay 11-7082 resulted in pronounced increases in JNK1 phosphorylation and JNK activity, manifested by increased levels of glutathione S-transferase-phospho-Jun, whereas individual agents had minimal effects. Comparable increases in JNK1 phosphorylation were also observed for Jurkat, Raji, and HL-60 leukemia cells treated with Bay-11-7082 in combination with MS-275 (Fig. 7C) or SAHA (data not shown). Treatment with these agents individually or in combination did not significantly alter phosphorylation/activation of ERK1/2 and p38 mitogen-activated protein kinase (Fig. 7B).
To verify the role of NF-κB inhibition in XIAP downregulation and JNK1 activation in cells exposed to Bay 11-7082 in combination with HDAC inhibitors, XIAP expression and JNK phosphorylation in U937 cells stably transfected with IκBα (IκBα-SR/U937 cells) were monitored following HDAC inhibitor treatment. As shown in Fig. 7D, U937 cells stably transfected with IκBα-SR displayed a more pronounced downregulation of XIAP and activation of JNK following MS-275 or SAHA treatment than empty-vector controls, consistent with the results obtained for cells coexposed to HDAC inhibitors and Bay 11-7082.
JNK activation plays a functional role in Bay 11-7082/HDAC inhibitor-mediated apoptosis but acts downstream of ROS generation and NF-κB inactivation.
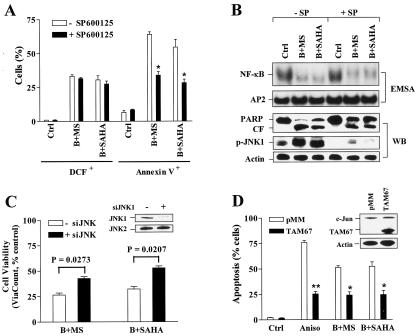
The functional significance of JNK activation in the potentiation of HDAC inhibitor-mediated lethality by Bay 11-7082 was then investigated. SP600125, a selective JNK inhibitor (4), significantly, albeit partially, reduced Bay 11-7082/SAHA- and Bay 11-7082/MS-275-mediated lethalities (P was <0.02 in each case) but failed to diminish ROS generation induced by these regimens (Fig. 8A). EMSA analysis revealed that SP600125 did not prevent inhibition of HDAC inhibitor-mediated NF-κB activation by Bay 11-7082 (Fig. 8B). Western blot analysis confirmed that SP600125 blocked JNK1 activation in cells exposed to these regimens and attenuated PARP degradation (Fig. 8B). Parallel studies employing JNK1 siRNA oligonucleotides demonstrated that downregulation of JNK1 (inset) was associated with significant attenuation of Bay 11-7082/HDAC inhibitor-mediated lethality (P < 0.05) (Fig. 8C). Finally, U937 cells stably expressing TAM67 (Fig. 8D, inset), a dominant-negative c-Jun mutant protein in which the transactivation domain has been deleted (7, 58), were significantly resistant to the lethalities of Bay 11-7082/SAHA and Bay 11-7082/MS-275 regimens, as well as the JNK/SAPK activator anisomycin (Fig. 8D). Thus, these and earlier findings (e.g., results shown in Fig. 7B) indicate that JNK activation plays a functional role in the potentiation of HDAC inhibitor-mediated lethality by Bay 11-7082. Finally, JNK inhibition by SP600125 did not prevent ROS generation or NF-κB inactivation in cells exposed to the Bay 11-7082/HDAC inhibitor regimen, but the blockade of ROS generation by L-NAC did block JNK1 activation (data not shown), indicating that JNK activations lies downstream of oxidative damage and NF-κB inhibition.
FIG. 8.
JNK inhibition attenuates the lethality of the Bay 11-7082/HDAC inhibitor regimen without affecting ROS production or NF-κB inactivation. (A) U937 cells were exposed for 24 h to 3 μM Bay 11-7082 (B) plus 2 μM MS-275 (MS) or 1 μM SAHA in the absence or presence of 10 μM SP600125 (SP), after which the percentages of cells exhibiting positive DCF fluorescence and annexin V-FITC were determined by flow cytometry. Values represent the means ± SD for three separate experiments performed in triplicate. *, significantly lower than the value for treatment with Bay 11-7082 with MS-275 or SAHA in the absence of SP600125 (P < 0.02). Ctrl, control. (B) Alternatively, cells were lysed and subjected to either EMSA (upper panels) or Western blot analysis (lower panels). Abbreviations are the same as those for panel A. (C) U937 cells were transiently transfected with JNK1 siRNA oligonucleotides as described in Materials and Methods. After 24 h, the levels of JNK1 and JNK2 were monitored by Western blot analysis (inset). Following a 6-h posttransfection recovery interval, JNK1 siRNA-transfected cells were exposed to 3 μM Bay 11-7082 plus 2 μM MS-275 (B+MS) or 1 μM SAHA (B+SAHA) for 24 h, after which the percentage of viable cells was determined by ViaCount assay. (D) U937 cells stably transfected with a TAM67 construct (inset, Western blot) or its empty vector (pMM) were treated with 3 μM Bay 11-7082 plus 2 μM MS-275 (B+MS) or 1 μM SAHA (B+SAHA), as well as 50 ng/ml anisomycin (Aniso) for 24 h, after which the percentage of cells exhibiting apoptotic morphology was determined by evaluating Wright-Giemsa-stained cytospin slides. The results represent the means ± SD for three separate experiments performed in triplicate. Ctrl, control. Asterisks signify values that are significantly lower than the values for cells transfected with empty vector (pMM) (**, P < 0.01; *, P < 0.02). For Western blot analyses in panels B through D, 30 μg of protein was loaded in each lane; blots were subsequently stripped and reprobed for the expression of β-actin, as indicated, to ensure equivalent loading and transfer of protein. CF, cleavage fragment. Two additional studies yielded equivalent results.
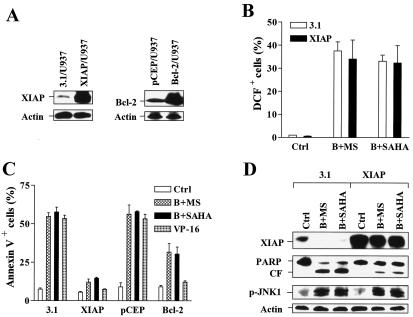
Overexpression of XIAP diminishes the lethality of the Bay 11-7082/HDAC inhibitor regimen but does not affect ROS generation and JNK1 activation.
Lastly, to assess the functional role of XIAP downregulation in these events, U937 cells ectopically overexpressing XIAP (Fig. 9A, left panels) were employed. XIAP/U937 cells exhibited increases in ROS level similar to those of empty-vector controls following coexposure to Bay 11-7082 and either SAHA or MS-275 (Fig. 9B). In striking contrast, XIAP overexpression substantially protected cells from Bay 11-7082/SAHA- or Bay 11-7082/MS-275-mediated apoptosis (Fig. 9C). Comparative studies utilizing U937 cells ectopically overexpressing Bcl-2 were performed (Fig. 9A, right panels). Overexpression of XIAP and overexpression of Bcl-2 protected cells from the topoisomerase inhibitor VP-16 to comparable extents (Fig. 9C). However, whereas Bcl-2 overexpression afforded only partial protection from Bay 11-7082/HDAC inhibitor-mediated lethality, XIAP overexpression conferred virtually complete resistance to these regimens (Fig. 9C). Western blot analysis revealed that Bay 11-7082/HDAC inhibitor regimens failed to reduce XIAP expression in XIAP/U937 cells, events accompanied by diminished PARP cleavage. Moreover, ectopic expression of XIAP did not prevent JNK1 phosphorylation/activation following exposure to Bay 11-7082/HDAC inhibitor regimens (Fig. 9D). Together, these findings suggest that downregulation of XIAP by the Bay 11-7082/HDAC inhibitor regimen plays a significant functional role in the enhanced lethality of this regimen. They also indicate that this event operates downstream or independently of ROS generation and JNK1 activation.
FIG. 9.
Ectopic overexpression of XIAP attenuates apoptosis but not ROS production or JNK1 activation in U937 cells coexposed to Bay 11-7082 and HDAC inhibitors. (A) Western blot analysis was performed to monitor expression of XIAP and Bcl-2 in U937 cells stably transfected with either XIAP (XIAP/U937; left panels) or Bcl-2 (Bcl-2/U937; right panels) construct as well as their corresponding empty-vector controls (pcDNA3.1 [3.1/U937] and pCEP [pCEP/U937]). (B) XIAP/U937 and 3.1/U937 cells were exposed to 3 μM Bay 11-7082 plus 2 μM MS-275 (B+MS) or 1 μM SAHA (B+SAHA) for 24 h, after which the percentage of cells exhibiting DCF fluorescence positivity was evaluated by flow cytometry. (C) XIAP/U937 and Bcl-2/U937 cells, as well as their empty-vector controls, were treated with 3 μM Bay 11-7082 plus 2 μM MS-275 (B+MS) or 1 μM SAHA (B+SAHA) for 24 h, after which the percentage of cells exhibiting positive annexin V-FITC was determined by flow cytometry. In parallel, all cell lines were incubated with 50 μM VP-16 for 5 h as controls. (D) XIAP/U937 and 3.1/U937 cells were treated as described for panel B; a Western blot analysis was performed to monitor expression of XIAP and phospho-JNK (p-JNK1) and cleavage of PARP. CF, cleavage fragment. Other abbreviations are the same as those for panel C. For Western blot analyses in panels A and D, each lane was loaded with 30 μg of protein; blots were subsequently stripped and reprobed for the expression of β-actin to ensure equivalent loading and transfer of protein. The results are representative of three separate experiments. For panels B and C, values represent the means ± SD for three separate experiments performed in triplicate.
DISCUSSION
Although in some cells (e.g., colonic epithelial cells), HDAC inhibitor exposure has been associated with NF-κB inactivation (61), in most cases, activation of NF-κB has been reported (3, 41). While the mechanism by which HDAC inhibitors activate NF-κB remains to be fully elucidated, there is accumulating evidence that this process involves, among other factors, acetylation-related events. Although HDAC inhibitor-induced acetylation of histones (e.g., histone H3 and H4) has been the primary focus of interest, it is clear that HDACs deacetylate a variety of nonhistone proteins. For example, acetylation of heat shock proteins (e.g., Hsp 90) has been implicated in the lethal action of certain expanded-spectrum HDAC inhibitors (24). Based on the findings reported by Chen et al. (12, 13), it has been proposed that acetylation of RelA/p65 represents a molecular switch regulating the duration of NF-κB transcriptional activity and simultaneously replenishing cytoplasmic pools of IκBα/NF-κB (14).
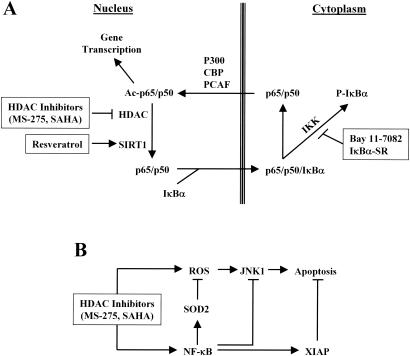
Because both HAT-mediated acetylation and HDAC-mediated deacetylation of RelA/p65 occur exclusively in the nucleus (12), interference with NF-κB nuclear translocation would be expected to diminish acetylation of this transcription factor. Consistent with this model, the exposure of leukemia cells to SAHA or MS-275 markedly increased acetylated RelA/p65, enhanced the nuclear accumulation of RelA/p65, and diminished the association of RelA/p65 with IκBα. Significantly, dysregulation of the IκBα axis, by either pharmacological (i.e., Bay 11-7082) or genetic (i.e., IκBα-SR) means, led to diminished HDAC inhibitor-mediated hyperacetylation of RelA/p65 by the sequestering of NF-κB in cytoplasm. The NF-κB sequestration prevents NF-κB translocation to the nucleus, where HATs and HDACs primarily act (12). It should be noted, however, that activation of NF-κB by RelA/p65 acetylation is not a universal finding. For example, Kiernan et al. reported that acetylation of RelA/p65 diminished NF-κB/DNA binding without altering associations with IκBα, leading to a net export of the complex to the cytoplasmic compartment (33). Based on these observations, it is likely that the net effect of acetylation on the intracellular distribution and transactivation potential of NF-κB may be cell type-specific or, alternatively, may depend upon the specific lysine sites that are acetylated (e.g., K221 and K310 versus K122 and K123) (13, 33). A hypothetical model in which HDAC inhibitors such as MS-275 and SAHA activate NF-κB, which is blocked by pharmacological (Bay 11-7082) or genetic (IκBα-SR) inhibition of IκBα phosphorylation/degradation, is illustrated in Fig. 10A.
FIG. 10.
(A) Schematic diagram of NF-κB activation by HDAC inhibitors. NF-κB activity is regulated by acetylation and deacetylation of RelA/p65 in the nucleus, which are mediated by HATs (e.g., P300, CBP, and PCAF) and HDACs (e.g., HDAC3 and SIRT1), respectively. Acetylation of RelA/p65 blocks the association of p65/p50 with IκBα which transports the NF-κB complex from the nucleus to the cytoplasm and, in so doing, inactivates NF-κB. HDAC inhibitors cause nuclear accumulation of RelA/p65 and NF-κB activation by blocking HDAC-mediated deacetylation of RelA/p65. These events are inhibited by either Bay 11-7082 or IκBα-SR, which blocks phosphorylation and proteasomal degradation of IκBα, leading to sequestration of the NF-κB complex in the cytoplasm. Alternatively, the SIRT1 (a class III HDAC which is not sensitive to known inhibitors of class I and II HDAC) activator resveratrol, by opposing RelA/p65 acetylation, promotes RelA/p65 binding to IκBα, resulting in the same effect. Ac, acetylated. (B) Schematic diagram of possible NF-κB-dependent cytoprotective pathways acting to attenuate the lethalities of HDAC inhibitors. HDAC inhibitors induce apoptosis by multiple mechanisms including induction of ROS and activation of stress-related SAPK/JNK pathways. On the other hand, HDAC inhibitors also trigger cytoprotective mechanisms involving the activation of NF-κB through RelA/p65 acetylation, and in so doing, induction of mitochondrial MnSOD (SOD2) which limits ROS generation, prevention of JNK1 activation, and maintenance of XIAP expression. The lethalities of HDAC inhibitors are determined by a balance between these opposing mechanisms. Blockade of the NF-κB pathway by either pharmacological or genetic means disrupts this balance, resulting in potentiation of HDAC inhibitor-mediated lethality.
Generally, NF-κB activation is thought to play an important cytoprotective role against cell death induced by diverse noxious stimuli, including HDAC inhibitors (41). The present studies indicate that in leukemia cells, interruption of the NF-κB pathway by Bay 11-7082 or IκBα-SR dramatically increases the lethal effects of HDAC inhibitors (e.g., SAHA and MS-275). The current findings also raise the possibility that perturbations in HDAC inhibitor-mediated acetylation of RelA/p65 may be critically involved in this phenomenon. In this context, coadministration of resveratrol (60), an activator of SIRT1, which is a class III HDAC insensitive to known inhibitors of class I and II HDACs (5, 40), enhanced the lethality of HDAC inhibitors in association with diminished acetylation of RelA/p65. Thus, the activation of NF-κB associated with p65 acetylation may serve to protect cells from the lethal consequences of certain HDAC inhibitor-mediated actions, particularly, ROS production and induction of mitochondrial dysfunction (47, 48). Conversely, disruption of p65 acetylation by the targeting of IκBα phosphorylation/degradation (e.g., by Bay 11-7082 or IκBα-SR) or more directly through stimulation of HDAC-mediated deacetylation (e.g., by resveratrol) may permit the proapoptotic actions of HDAC inhibitors to predominate, leading to a striking increase in cell death.
The present evidence suggests that potentiation of oxidative damage (e.g., ROS generation) plays a critical role in interactions between Bay 11-7082 and HDAC inhibitors. Involvement of ROS generation in the lethalities of SAHA and MS-275 has previously been reported (47, 48), although the mechanism(s) by which the HDAC inhibitor(s) triggers oxidative damage is (are) not known. Notably, NF-κB activation represents one of the major cellular responses to H2O2 and other mediators of oxidative stress (51). In the present study, coadministration of Bay 11-7082 significantly increased ROS generation by SAHA or MS-275. Similarly, genetic interruption of the NF-κB pathway by IκBα-SR also potentiated HDAC inhibitor-mediated oxidative damage, consistent with the previous considerations (10). Importantly, coadministration of the free-radical scavenger L-NAC blocked ROS generation and enhanced lethality of the Bay 11-7082/HDAC inhibitor regimens, indicating that oxidative damage plays a primary role in synergistic antileukemic effects.
SODs constitute a critical cellular antioxidant enzyme defense system that protects cells from ROS-mediated injury. Of three SOD isoforms, cytosolic CuZn-SOD (SOD1) and mitochondrial Mn-SOD are present in most human normal and neoplastic cells, whereas the expression pattern of extracellular SOD (SOD3) is highly cell type specific and tissue specific (63). Notably, the enhancer element of SOD2 genes contains binding motifs for NF-κB; moreover, activation of NF-κB is essential for induction of SOD2 by multiple stimuli (21, 34). Consistent with these observations, MS-275 and SAHA induced a modest increase in SOD2 expression, whereas inhibition of NF-κB by Bay 11-7082 or IκBα-SR downregulated basal levels of SOD2 and blocked HDAC inhibitor-mediated SOD2 induction. It has also been reported that ROS generation transcriptionally induces SOD expression as a cellular feedback response (29). Thus, NF-κB-dependent SOD2 upregulation induced by HDAC inhibitors may provide a protective feedback against oxidative damage; conversely, interruption of NF-κB activation may disrupt this cellular response by blocking SOD2 expression and enhancing ROS-mediated lethality, consistent with results obtained with the SOD mimetic MnTBAP. Because SOD2 is a mitochondrial Mn-SOD (63), the present findings also suggest, albeit indirectly, that mitochondria represent a major source of ROS generation triggered by HDAC inhibitors. Lastly, recent studies suggest that HDAC inhibitors selectively trigger oxidative injury in transformed cells as a consequence of differential induction of thioredoxin (57), a protein that functions as a hydrogen donor for many protein targets and as an ROS scavenger. Whether HDAC inhibitors exhibit similar selectivities in the induction of SOD2 remains to be determined.
Genetic and pharmacological interruption of the NF-κB pathway in cells exposed to HDAC inhibitors also resulted in perturbations in expression of XIAP, a member of the inhibitor of apoptosis family that acts by inhibiting caspases, particularly caspases 3, 7, and 9 (53). Administration of Bay 11-7082 resulted in a modest decrease in basal XIAP levels and diminished XIAP expression in HDAC inhibitor-treated cells. The finding that ectopic expression of XIAP protected cells from Bay 11-7082/HDAC inhibitor-mediated cell death argues that downregulation of this antiapoptotic protein plays a significant functional role in the lethality of this regimen. It is also noteworthy that enforced expression of XIAP failed to modify ROS production and JNK1 activation in Bay 11-7082/HDAC inhibitor-treated cells. These findings suggest that diminished expression of XIAP operates downstream or independently of other critical events (e.g., NF-κB inactivation, ROS generation, and JNK1 activation) in the cell death hierarchy.
It has been reported that inhibition of NF-κB leads to JNK activation and potentiates the lethality of certain apoptotic stimuli (e.g., TNF-α) (45, 55). JNK activation mediates apoptosis in response to diverse events, possibly through direct effects on release of mitochondrial proapoptotic proteins, such as cytochrome c (56). In the present study, combined inhibition of NF-κB and HDACs resulted in a very pronounced JNK1 activation. Moreover, the observation that blockade of the JNK pathway by pharmacological (e.g., SP600125) or genetic means (transfection with JNK1 siRNA or TAM67) protected cells from this regimen argues strongly for a functional role for JNK1 activation in enhanced cell death. In this context, oxidative stress is a well-known activator of the SEK/JNK cascade, and the interruption of this pathway has been shown to limit damage associated with ROS generation (22). The finding that attenuation of ROS generation in Bay 11-7082/HDAC inhibitor-treated cells (e.g., by L-NAC) blocked JNK activation indicates that under these circumstances, engagement of the stress-related JNK pathway most likely represents a response to oxidative damage. It should be also noted that NF-κB activation, at least induced by TNF-α, has been shown to block JNK activation through an XIAP-related mechanism (55). However, the observation that ectopic expression of XIAP failed to modify Bay 11-7082/HDAC inhibitor-mediated JNK1 phosphorylation argues against an association between XIAP downregulation and JNK activation in the present model system.
In summary, the present findings indicate that interruption of the NF-κB pathway in leukemia cells exposed to HDAC inhibitors results in a cascade of events, including the generation of ROS, downregulation of SOD2 and XIAP, and activation of JNK1, culminating in a striking increase in mitochondrial dysfunction and apoptosis. A hypothetical model summarizing interactions between these factors is illustrated in Fig. 10B. Notably, these events are closely associated with, and very likely related to, a marked increased association of RelA/p65 with IκBα, reduced nuclear translocation of NF-κB, and diminished HDAC inhibitor-mediated acetylation of RelA/p65. Collectively, these findings argue that RelA/p65 acetylation and resulting NF-κB activation by HDAC inhibitors serve to protect cells from what would otherwise be the lethal consequences of oxidative damage. A corollary of this hypothetical model is that interventions that block HDAC inhibitor-induced RelA/p65 acetylation and/or NF-κB activation would be expected to promote the antitumor potential of this class of agents. In this regard, the clinical relevant proteasome inhibitor Bortezomib, which inactivates NF-κB by sparing IκBα from proteasomal degradation (42), has recently been shown to promote HDAC inhibitor-related antitumor effects in malignant hematopoietic cells (43, 62). Currently, the development of pharmacological inhibitors of the NF-κB pathway (e.g., IKK inhibitors) for the treatment of inflammatory or neoplastic diseases has become the subject of considerable interest (16). In view of the present findings, it would be logical to explore antineoplastic interactions between such agents and clinically relevant HDAC inhibitors, particularly those targeting HDAC3. Accordingly, studies addressing these issues are now under way.
Acknowledgments
This work was supported by Public Health Service grants CA-63753, CA-93738, CA-100866, and CA88906 from the National Cancer Institute, grant DK52825 from the National Institutes of Health, award 6045-03 from the Leukemia and Lymphoma Society of America, and a Translational Research award from the V-Foundation.
REFERENCES
- 1.Adam, E., V. Quivy, F. Bex, A. Chariot, Y. Collette, C. Vanhulle, S. Schoonbroodt, V. Goffin, T. L. Nguyen, G. Gloire, G. Carrard, B. Friguet, Y. De Launoit, A. Burny, V. Bours, J. Piette, and C. Van Lint. 2003. Potentiation of tumor necrosis factor-induced NF-κB activation by deacetylase inhibitors is associated with a delayed cytoplasmic reappearance of IκBα. Mol. Cell. Biol. 23:6200-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner, B. P., S. D. Westerheide, and A. S. Baldwin, Jr. 2001. The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. H., T. K. Chen, and M. J. Birrer. 1994. Mechanism of action of a dominant-negative mutant of c-Jun. Oncogene 9:791-799. [PubMed] [Google Scholar]
- 8.Byun, M. S., K. I. Jeon, J. W. Choi, J. Y. Shim, and D. M. Jue. 2002. Dual effect of oxidative stress on NF-kappakB activation in HeLa cells. Exp. Mol. Med. 34:332-339. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., L. C. Edelstein, and C. Gelinas. 2000. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol. Cell. Biol. 20:2687-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, F., V. Castranova, Z. Li, M. Karin, and X. Shi. 2003. Inhibitor of nuclear factor kappaB kinase deficiency enhances oxidative stress and prolongs c-Jun NH2-terminal kinase activation induced by arsenic. Cancer Res. 63:7689-7693. [PubMed] [Google Scholar]
- 11.Chen, L. F., and W. C. Greene. 2004. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 5:392-401. [DOI] [PubMed] [Google Scholar]
- 12.Chen, L. F., W. Fischle, E. Verdin, and W. C. Greene. 2001. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293:1653-1657. [DOI] [PubMed] [Google Scholar]
- 13.Chen, L. F., Y. Mu, and W. C. Greene. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21:6539-6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, L. F., and W. C. Greene. 2003. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J. Mol. Med. 81:549-557. [DOI] [PubMed] [Google Scholar]
- 15.Chen, W. Y., and T. M. Townes. 2000. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl. Acad. Sci. USA 97:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai, S., T. Hirayama, S. Abbas, and Y. Abu-Amer. 2004. The IkappaB kinase (IKK) inhibitor, NEMO-binding domain peptide, blocks osteoclastogenesis and bone erosion in inflammatory arthritis. J. Biol. Chem. 279:37219-37222. [DOI] [PubMed] [Google Scholar]
- 17.Dai, Y., C. Yu, V. Singh, L. Tang, Z. Wang, R. McInistry, P. Dent, and S. Grant. 2001. Pharmacological inhibitors of the mitogen-activated protein kinase (MAPK) kinase/MAPK cascade interact synergistically with UCN-01 to induce mitochondrial dysfunction and apoptosis in human leukemia cells. Cancer Res. 61:5106-5115. [PubMed] [Google Scholar]
- 18.Dai, Y., M. Rahmani, and S. Grant. 2003. Proteasome inhibitors potentiate leukemic cell apoptosis induced by the cyclin-dependent kinase inhibitor flavopiridol through a SAPK/JNK- and NF-kappaB-dependent process. Oncogene 22:7108-7122. [DOI] [PubMed] [Google Scholar]
- 19.Dai, Y., P. Dent, and S. Grant. 2003. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) promotes mitochondrial dysfunction and apoptosis induced by 7-hydroxystaurosporine and mitogen-activated protein kinase kinase inhibitors in human leukemia cells that ectopically express Bcl-2 and Bcl-xL. Mol. Pharmacol. 64:1402-1409. [DOI] [PubMed] [Google Scholar]
- 20.Datta, R, E. Oki, K. Endo, V. Biedermann, J. Ren, and D. Kufe. 2000. XIAP regulates DNA damage-induced apoptosis downstream of caspase-9 cleavage. J. Biol. Chem. 275:31733-31738. [DOI] [PubMed] [Google Scholar]
- 21.Dhar, S. K., B. C. Lynn, C. Daosukho, and D. K. St. Clair. 2004. Identification of nucleophosmin as an NF-kappaB co-activator for the induction of the human SOD2 gene. J. Biol. Chem. 279:28209-28219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filomeni, G., K. Aquilano, G. Rotilio, and M. R. Ciriolo. 2003. Reactive oxygen species-dependent c-Jun NH2-terminal kinase/c-Jun signaling cascade mediates neuroblastoma cell death induced by diallyl disulfide. Cancer Res. 63:5940-5949. [PubMed] [Google Scholar]
- 23.Finnin, M. S., J. R. Donigian, A. Cohen, V. M. Richon, R. A. Rifkind, P. A. Marks, R. Breslow, and N. P. Pavletich. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188-193. [DOI] [PubMed] [Google Scholar]
- 24.Fuino, L., P. Bali, S. Wittmann, S. Donapaty, F. Guo, H. Yamaguchi, H. G. Wang, P. Atadja, and K. Bhalla. 2003. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol. Cancer Ther. 2:971-984. [PubMed] [Google Scholar]
- 25.Garcia-Bermejo, L., N. E. Vilaboa, C. Perez, A. Galan, E. De Blas, and P. Aller. 1997. Modulation of heat-shock protein 70 (HSP70) gene expression by sodium butyrate in U-937 promonocytic cells: relationships with differentiation and apoptosis. Exp. Cell Res. 236:268-274. [DOI] [PubMed] [Google Scholar]
- 26.Guo, Z., G. H. Boekhoudt, and J. M. Boss. 2003. Role of the intronic enhancer in tumor necrosis factor-mediated induction of manganous superoxide dismutase. J. Biol. Chem. 278:23570-23578. [DOI] [PubMed] [Google Scholar]
- 27.Hildeman, D. A., T. Mitchell, B. Aronow, S. Wojciechowski, J. Kappler, and P. Marrack. 2003. Control of Bcl-2 expression by reactive oxygen species. Proc. Natl. Acad. Sci. USA 100:15035-15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, H. L., L. W. Fang, S. P. Lu, C. K. Chou, T. Y. Luh, and M. Z. Lai. 2003. DNA-damaging reagents induce apoptosis through reactive oxygen species-dependent Fas aggregation. Oncogene 22:8168-8177. [DOI] [PubMed] [Google Scholar]
- 29.Huang, P., L. Feng, E. A. Oldham, M. J. Keating, and W. Plunkett. 2000. Superoxide dismutase as a target for the selective killing of cancer cells. Nature 407:390-395. [DOI] [PubMed] [Google Scholar]
- 30.Huang, T. T., N. Kudo, M. Yoshida, and S. Miyamoto. 2000. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl. Acad. Sci. USA 97:1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, T. T., S. M. Wuerzberger-Davis, Z. H. Wu, and S. Miyamoto. 2003. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115:565-576. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone, R. W. 2002. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 1:287-299. [DOI] [PubMed] [Google Scholar]
- 33.Kiernan, R., V. Bres, R. W. Ng, M. P. Coudart, S. El Messaoudi, C. Sardet, D. Y. Jin, S. Emiliani, and M. Benkirane. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758-2766. [DOI] [PubMed] [Google Scholar]
- 34.Kiningham, K. K., C. Daosukho, and D. K. St. Clair. 2004. IkappaB-alpha identified as labile repressor of manganese superoxide dismutase (MnSOD) expression. Biochem J. 384:543-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lappas, M., K. Yee, M. Permezel, and G. E. Rice. 2005. Sulfasalazine and BAY 11-7082 interfere with the nuclear factor-kappa B and I kappa B kinase pathway to regulate the release of proinflammatory cytokines from human adipose tissue and skeletal muscle in vitro. Endocrinology 146:1491-1497. [DOI] [PubMed] [Google Scholar]
- 36.Leach, J. K., G. V. Tuyle, P. S. Lin, R. Schmidt-Ullrich, and R. B. Mikkelsen. 2001. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res. 61:3894-3901. [PubMed] [Google Scholar]
- 37.Li, J., M. J. Staver, M. L., Curtin, J. H. Holms, R. R. Frey, R. Edalji, R. Smith, M. R. Michaelides, S. K. Davidsen, and K. B. Glaser. 2004. Expression and functional characterization of recombinant human HDAC1 and HDAC3. Life Sci. 74:2693-2705. [DOI] [PubMed] [Google Scholar]
- 38.Lin, Y. Z., S. Y. Yao, R. A. Veach, T. R. Torgerson, and J. Hawiger. 1995. Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J. Biol. Chem. 270:14255-14258. [DOI] [PubMed] [Google Scholar]
- 39.Maggio, S. C., R. R. Rosato, L. B. Kramer, Y. Dai, M. Rahmani, D. S. Paik, A. C. Czarnik, S. G. Payne, S. Spiegel, and S. Grant. 2004. The histone deacetylase inhibitor MS-275 interacts synergistically with fludarabine to induce apoptosis in human leukemia cells. Cancer Res. 64:2590-2600. [DOI] [PubMed] [Google Scholar]
- 40.Marks, P., R. A. Rifkind, V. M. Richon, R. Breslow, T. Miller, and W. K. Kelly. 2001. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1:194-202. [DOI] [PubMed] [Google Scholar]
- 41.Mayo, M. W., C. E. Denlinger, R. M. Broad, F. Yeung, E. T. Reilly, Y. Shi, and D. R. Jones. 2003. Ineffectiveness of histone deacetylase inhibitors to induce apoptosis involves the transcriptional activation of NF-kappa B through the Akt pathway. J. Biol. Chem. 278:18980-18989. [DOI] [PubMed] [Google Scholar]
- 42.Palombella, V. J., E. M. Conner, J. W. Fuseler, A. Destree, J. M. Davis, F. S. Laroux, R. E. Wolf, J. Huang, S. Brand, P. J. Elliott, D. Lazarus, T. McCormack, L. Parent, R. Stein, J. Adams, and M. B. Grisham. 1998. Role of the proteasome and NF-kappaB in streptococcal cell wall-induced polyarthritis. Proc. Natl. Acad. Sci. USA 95:15671-15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei, X. Y., Y. Dai, and S. Grant. 2004. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor Bortezomib and histone deacetylase inhibitors. Clin. Cancer Res. 10:3839-3852. [DOI] [PubMed] [Google Scholar]
- 44.Pierce, J. W., R. Schoenleber, G. Jesmok, J. Best, S. A. Moore, T. Collins, and M. E. Gerritsen. 1997. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J. Biol. Chem. 272:21096-21103. [DOI] [PubMed] [Google Scholar]
- 45.Reuther-Madrid, J. Y., D. Kashatus, S. Chen, X. Li, J. Westwick, R. J. Davis, H. S. Earp, C. Y. Wang, and A. S. Baldwin, Jr. 2002. The p65/RelA subunit of NF-κB suppresses the sustained, antiapoptotic activity of Jun kinase induced by tumor necrosis factor. Mol. Cell. Biol. 22:8175-8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richon, V. M., S. Emiliani, E. Verdin, Y. Webb, R. Breslow, R. A. Rifkind, and P. A. Marks. 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. USA 95:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosato, R. R., J. A. Almenara, and S. Grant. 2003. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 Cancer Res. 63:3637-3645. [PubMed]
- 48.Ruefli, A. A., M. J. Ausserlechner, D. Bernhard, V. R. Sutton, K. M. Tainton, R. Kofler, M. J. Smyth, and R. W. Johnstone. 2001. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc. Natl. Acad. Sci. USA 98:10833-10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-kappaB in p53-mediated programmed cell death. Nature 404:892-897. [DOI] [PubMed] [Google Scholar]
- 50.Saito, A., T. Yamashita, Y. Mariko, Y. Nosaka, K. Tsuchiya, T. Ando, T. Suzuki, T. Tsuruo, and O. Nakanishi. 1999. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc. Natl. Acad. Sci. USA 96:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreck, R., P. Rieber, and P. A. Baeuerle. 1991. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 10:2247-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasula, S. M., R. Hegde, A. Saleh, P. Datta, E. Shiozaki, J. Chai, R. A. Lee, P. D. Robbins, T. Fernandes-Alnemri, Y. Shi, and E. S. Alnemri. 2001. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410:112-116. [DOI] [PubMed] [Google Scholar]
- 54.Stehlik, C., R. de Martin, I. Kumabashiri, J. A. Schmid, B. R. Binder, and J. Lipp. 1998. Nuclear factor (NF)-kappaB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha-induced apoptosis. J. Exp. Med. 188:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, G., Y. Minemoto, B. Dibling, N. H. Purcell, Z. Li, M. Karin, and A. Lin. 2001. Inhibition of JNK activation through NF-kappaB target genes. Nature 414:313-317. [DOI] [PubMed] [Google Scholar]
- 56.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 57.Ungerstedt, J. S., Y. Sowa, W. S. Xu, Y. Shao, M. Dokmanovic, G. Perez, L. Ngo, A. Holmgren, X. Jiang, and P. A. Marks. 2005. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 102:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verheij, M., R. Bose, X. H. Lin, B. Yao, W. D. Jarvis, S. Grant, M. J. Birrer, E. Szabo, L. I. Zon, J. M. Kyriakis, A. Haimovitz-Friedman, Z. Fuks, and R. N. Kolesnick. 1996. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature 380:75-79. [DOI] [PubMed] [Google Scholar]
- 59.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeung, F., J. E. Hoberg, C. S. Ramsey, M. D. Keller, D. R. Jones, R. A. Frye, and M. W. Mayo. 2004. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin, L., G. Laevsky, and C. Giardina. 2001. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J. Biol. Chem. 276:44641-44646. [DOI] [PubMed] [Google Scholar]
- 62.Yu, C., M. Rahmani, D. Conrad, M. Subler, P. Dent, and S. Grant. 2003. The proteasome inhibitor bortezomib interacts synergistically with histone deacetylase inhibitors to induce apoptosis in Bcr/Abl+ cells sensitive and resistant to STI571. Blood 102:3765-3774. [DOI] [PubMed] [Google Scholar]
- 63.Zelko, I. N., T. J. Mariani, and R. J. Folz. 2002. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33:337-349. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]