Abstract
Defective function of the von Hippel-Lindau (VHL) tumor suppressor ablates proteolytic regulation of hypoxia-inducible factor α subunits (HIF-1α and HIF-2α), leading to constitutive activation of hypoxia pathways in renal cell carcinoma (RCC). Here we report a comparative analysis of the functions of HIF-1α and HIF-2α in RCC and non-RCC cells. We demonstrate common patterns of HIF-α isoform transcriptional selectivity in VHL-defective RCC that show consistent and striking differences from patterns in other cell types. We also show that HIF-α isoforms display unexpected suppressive interactions in RCC cells, with enhanced expression of HIF-2α suppressing HIF-1α and vice-versa. In VHL-defective RCC cells, we demonstrate that the protumorigenic genes encoding cyclin D1, transforming growth factor alpha, and vascular endothelial growth factor respond specifically to HIF-2α and that the proapoptotic gene encoding BNip3 responds positively to HIF-1α and negatively to HIF-2α, indicating that HIF-1α and HIF-2α have contrasting properties in the biology of RCC. In keeping with this, HIF-α isoform-specific transcriptional selectivity was matched by differential effects on the growth of RCC as tumor xenografts, with HIF-1α retarding and HIF-2α enhancing tumor growth. These findings indicate that therapeutic approaches to targeting of the HIF system, at least in this setting, will need to take account of HIF isoform-specific functions.
Associations between microenvironmental hypoxia, activation of hypoxia pathways, and aggressively malignant phenotypes are observed across a range of cancers, focusing attention on molecular dissection of these pathways and how they contribute to tumor biology (8, 9). Important insights have been gained through the definition of hypoxia-inducible factor (HIF) as a key transcription factor regulating oxygen-dependent gene expression. HIF is an α/β heterodimeric DNA binding complex that directs an extensive transcriptional response involving the induction of genes with important roles in several aspects of tumor biology such as angiogenesis, glucose/energy metabolism, cellular growth, metastasis, and apoptosis (37). The HIF system is regulated through the activity and abundance of HIF-α subunits. To date, three HIF-α isoforms have been described, with the best characterized being HIF-1α and HIF-2α. In cancer, the HIF system is upregulated both by microenvironmental hypoxia and by genetic events that lead to enhanced translation or stability of HIF-α (37). Activation of HIF in cancer has been shown to contribute to the classical tumor phenotypes of upregulated glycolysis and angiogenesis (23, 28, 34, 36), leading to widespread interest in the HIF system as a target in cancer therapeutics (37). However, though most studies have indicated that HIF activation contributes positively to tumor growth (37), studies of experimental tumors derived from cells with genetic defects in the HIF system have not universally supported this (5). Moreover, HIF target genes encode a wide range of products, some of which, such as the Bcl-2-related proapoptotic protein BNip3, might be predicted to have antitumorigenic properties (3, 15).
The most direct link between genetic events that predispose to cancer and activation of the HIF pathway is observed in tumors associated with inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene, particularly VHL-associated clear-cell renal cell carcinoma (RCC) (29). In hereditary RCC, associated with VHL disease, affected individuals bear a germ line mutation in VHL, with somatic inactivation of the second allele occurring as an early event in RCC development. In sporadic RCC, both VHL alleles are commonly subject to somatic mutation. Though several functions have been proposed for VHL, the best understood is in the regulation of HIF, where VHL acts as the recognition component of an E3 ubiquitin ligase complex that targets HIF-α subunits to the ubiquitin/proteasome degradation pathway following oxygen-dependent prolyl hydroxylation (16). In VHL-defective cells, HIF-α accumulates irrespective of hydroxylation, the HIF system is activated, and a constitutively hypoxic pattern of gene expression is observed. Interestingly, VHL-defective RCC cells show an unusual bias toward HIF-2α rather than HIF-1α expression (22, 29). Though transfection studies have indicated that HIF-1α and HIF-2α activate hypoxia response element-linked reporter genes in a similar manner (42, 45), studies of genetic inactivation have indicated differences, with most studies emphasizing the importance of HIF-1α in directing the transcriptional response to hypoxia (11, 13, 32, 35, 39).
Physiologically, the VHL E3 ubiquitin ligase plays a similar role in the regulation of both HIF-1α and HIF-2α. Both proteins contain two conserved sites of prolyl hydroxylation that are independently targeted by a single hydroxyproline binding site within the β domain of VHL (10, 30). This process is disrupted by all RCC-associated VHL mutations tested to date (17), and overexpression of a HIF-2α gene that escapes VHL-mediated destruction due to mutation of one or both prolyl hydroxylation sites blocks the tumor suppressor action of VHL in experimental tumors grown from transplanted RCC cells (18, 19). In addition, small interfering RNA (siRNA)-mediated downregulation of HIF-2α itself suppresses tumor formation by VHL-defective RCC cells (18, 49). These findings have strongly suggested that upregulation of the HIF system plays a functional role in VHL-associated RCC. However, in apparent contrast, other investigators have found that overexpression of a HIF-1α gene that is mutated at one of the prolyl hydroxylation sites did not block the tumor suppressor action of VHL in RCC cells (27), raising the possibility that despite their similarities, and common upregulation in many types of cancer, HIF-1α and HIF-2α may have nonequivalent effects in the pathogenesis of VHL-associated RCC.
To better understand the role of the HIF pathway in VHL-associated RCC, we have examined the transcriptional selectivity for the HIF-1α and HIF-2α isoforms among a group of genes that are expressed in VHL-defective RCC and have compared responses in VHL-defective RCC with those in other cell lines. We show that HIF target genes show remarkable selectivity for HIF-1α and HIF-2α, that patterns observed in VHL-defective RCC differ significantly from those in other cells, and that the transcriptional selectivity observed in RCC for HIF-2α versus HIF-1α correlates at least in part with predicted positive or negative effects on tumor growth. In keeping with this, we show that effects on RCC tumor formation are nonequivalent, with HIF-2α overexpression promoting and HIF-1α expression retarding tumor growth. In addition, we demonstrate that HIF-α isoforms display reciprocal suppressive interactions in VHL-defective cells that may contribute to the unusual expression pattern of low HIF-1α and high HIF-2α in RCC.
MATERIALS AND METHODS
Cell culture.
All cell lines were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 50 IU/ml penicillin, and 50 μg/ml streptomycin, except for the human kidney 2 (HK-2) cell line, which was maintained in Keratinocyte-SFM supplemented with 5 ng/ml recombinant epithelial growth factor and 0.05 mg/ml bovine pituitary extract (GIBCO-BRL 17005-042). The 786-O cells stably transfected with either an empty vector (PRC3) or VHL (WT-8), the A498 cells stably transfected with VHL (all gifts from William G. Kaelin, Jr.), and the RCC4 cells stably transfected with VHL (RCC4/VHL) (29) were maintained in G418 (1 mg/ml). Cells were grown in normoxia (21% O2) or hypoxia (1% O2) in a Napco 7001 incubator (Precision Scientific). Desferrioxamine and cycloheximide were obtained from Sigma, and dimethyloxalylglycine (MMOG) was a gift from Christopher J. Schofield. For determination of proliferation in standard monolayer culture, approximately 15,000 cells per well were seeded in six-well plates. Each day, cells from three wells of each cell line were harvested and passed through a Coulter counter, with gating between 14 μm and 42 μm.
RNA analysis.
Total RNA was extracted in RNAzol B (Biogenesis) and dissolved in hybridization buffer [80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 400 mM sodium chloride, 1 mM EDTA (pH 8)]. Forty-five micrograms of total RNA was assayed by RNase protection as previously described, with U6 small nuclear RNA serving as an internal control (45). The cyclin D1 (CCND1) riboprobe protected a fragment between bases 3902 and 4060 of the 3′ untranslated region (accession no. NM_053056).
siRNA preparation and transfection.
RNA duplexes targeting HIF-1α and HIF-2α were used as previously described (39). Silencer Negative Control siRNA (Ambion) and Oligofectamine Reagent alone (Invitrogen) were used in control experiments. Cells were plated at 30% confluence in six-well plates in antibiotic-free medium 24 h before transfection. siRNA duplexes (20 nM) were transfected twice at 24-h intervals using Oligofectamine Reagent.
Retroviruses.
HIF-1α, HIF-2α, and control retroviruses were produced using the bicistronic expression plasmids pLZRS-IRES-GFP (14) and pBMN-Z-IRES-Neo (gifts from Maarten van Lohuizen and Garry Nolan, respectively), and Phoenix Amphotropic packaging cells (gift from Garry Nolan). HIF-1α and HIF-2α coding sequences were inserted into the expression plasmids (with removal of the LacZ coding sequence from pBMN-Z-IRES-Neo) using standard recombinant methods that will be provided on request. The HIF-2α DNA binding domain mutant (amino acid residues 24 to 29, RCRRSK to ACAASA) (19) was created in pCDNA3-HIF-2α and subcloned into pLZRS-IRES-GFP following verification of the mutant sequence. Stable polyclonal pools of 786-O cells, infected with retrovirus produced from the pBMN-Z-IRES-Neo plasmids with HIF-1α, HIF-2α, or no insert were selected in G418 (1 mg/ml).
For retrovirus production, Phoenix cells were plated at 50% confluence in a 6-cm dish, incubated for 24 h, transfected with 10 μg of the appropriate retroviral vector using Fugene 6 (Roche), and then incubated at 37°C. After 24 h, cells were replated in a 75-cm2 flask and grown for another 24 h at 32°C. Viral supernatant was collected and filtered (0.45-μm pore size; Millipore) and then supplemented with 4 μg/ml Polybrene (Sigma). Target cells for infection were plated at 30% confluence in six-well plates for 24 h, and medium was then replaced with 0.5 ml of the appropriate viral supernatant with Polybrene for 6 h, followed by supernatant diluted 1:3 with standard growth medium for another 42 h at 37°C. The infection protocol was repeated with fresh retrovirus over a further 48 h. Cells were then harvested or selected in G418 as appropriate. Infection efficiencies between 75% and 95% were routinely achieved in unselected cells using pLZRS-IRES-GFP, as detected by FACS analysis for green fluorescent protein (GFP).
Immunoblotting.
Cells were rinsed in ice-cold phosphate-buffered saline (PBS) and then lysed in urea-sodium dodecyl sulfate (SDS) buffer (6.7 M urea, 10 mM Tris-Cl [pH 6.8], 1 mM dithiothreitol, 10% glycerol, 1% SDS) supplemented with Complete protease inhibitor cocktail (Roche). Protein concentrations were determined using the DC protein assay kit (Bio-Rad), and extracts were normalized for protein content. Whole-cell extracts were resolved by SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane (Millipore). Primary antibodies were mouse monoclonal antibodies against HIF-1α (54; Transduction Labs), HIF-2α (190b) (45), cyclin D1 (ab3; Oncogene Research Products), carbonic anhydrase IX (M75) (48), BNip3 (ANa40; Sigma), and β-tubulin (2-28-33; Sigma) and rabbit polyclonal antibody against glucose transporter-1 (ab652; abcam). Detection was with the ECL Plus system (Amersham Biosciences) and horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibodies (DAKO). Membranes were later stained with Coomassie blue to verify equal loading.
Vascular endothelial growth factor (VEGF) and transforming growth factor alpha (TGF-α) enzyme-linked immunosorbent assays.
Secreted levels of VEGF were measured using a Quantikine human VEGF immunoassay kit (R&D Systems) and normalized to cell number. For measurement of TGF-α in cell lysates, cells were harvested in NP-40 lysis buffer, lightly vortexed on ice for 30 min, and then used in the Quantikine human TGF-α immunoassay kit (R&D Systems). TGF-α levels were normalized to total cellular protein content.
Clinical material and immunohistochemistry.
Kidney specimens obtained at nephrectomy from seven VHL patients were formalin fixed and embedded in paraffin wax. Sections (3 μm) were mounted, dewaxed, rehydrated, and stained with primary antibodies (using antigen retrieval for HIF-1α, HIF-2α, and cyclin D1); then, following antigen-antibody complex detection, sections were counterstained with hematoxylin, dehydrated, and visualized as previously described (26). Cyclin D1 detection was with rabbit anti-human cyclin D1 (SP4; Neomarkers) at 1/200, and BNip3 detection was with mouse monoclonal anti-human BNip3 (ANa40; Sigma) at 1/100.
Xenograft assays in nude mice.
Unselected 786-O cells infected with the pLZRS-IRES-GFP-based HIF-1α, HIF-2α, or control virus (experiment 1) or G418 selected polyclonal pools of 786-O cells infected with the pBMN-Z-IRES-Neo based HIF-1α, HIF-2α, or control virus (experiments 2 and 3) were released by trypsinization and resuspended in PBS, and 107 cells in 100 μl PBS were injected, with or without 100 μl of Matrigel (BD Biosciences), subcutaneously into the dorsal flanks of nu/nu mice. Tumor size was measured twice weekly using calipers, and the xenografts were excised when they reached the maximum permitted size (1.44-cm2 surface area) in the first two experiments, where three sets of five mice each were injected in parallel. In the third set of xenografts, cells were injected into both flanks of three sets of seven mice each and the tumors from all of the groups were excised and weighed at a single time point specified by the first tumor reaching the maximum permitted size.
RESULTS
HIF-α isoform-specific regulation of cyclin D1 in RCC cells.
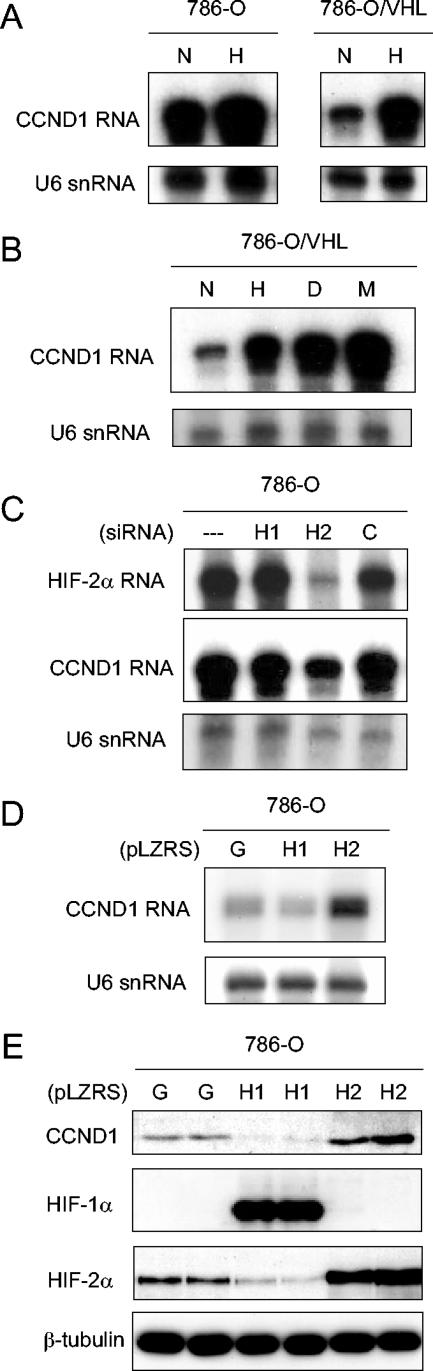
We first focused our analysis on the regulation of cyclin D1, a protein with a well-established role in oncogenesis that has recently been identified as a VHL-regulated gene product (2, 47). This work has demonstrated that cyclin D1 is strongly upregulated in RCC cell lines and tumor specimens and is down regulated by reintroduction of wild-type VHL into VHL-defective RCC cells. Interestingly, several groups, including our own, have shown that cyclin D1 displays hypoxia-inducible behavior in RCC cells re-expressing wild-type VHL but not in other cell types (1, 2, 46). This is illustrated in Fig. 1A for cyclin D1 mRNA in 786-O (PRC3) cells and in the stable transfectant 786-O/VHL (WT-8) expressing wild-type VHL.
FIG. 1.
Regulation of cyclin D1 (CCND1) in 786-O cells. (A and B) RNase protection assay of CCND1 mRNA and U6 snRNA (loading control) in 786-O (PRC3) and 786-O/VHL (WT-8) after exposure to normoxia (N), hypoxia (H), 100 μM desferrioxamine (D), or 1 mM MMOG (M) for 18 h. (C) RNase protection assay of HIF-2α mRNA and CCND1 mRNA in 786-O/VHL (WT-8) after treatment for 48 h with Oligofectamine alone (−), HIF-1α-directed siRNA (H1), HIF-2α-directed siRNA (H2), or control siRNA (C). (D) RNase protection assay of CCND1 mRNA and U6 snRNA in 786-O cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (E) Immunoblots, with the indicated antibodies, of whole-cell lysates from 786-O cells that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). Two independent experiments for each condition are shown in panel E.
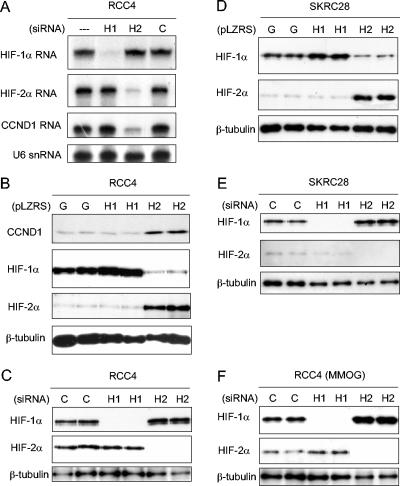
The unusual cell specificity of this response raised questions as to the nature of the hypoxia pathways involved. To address this, 786-O/VHL (WT-8) cells were exposed to the iron chelator desferrioxamine and the 2-oxoglutarate analog MMOG, both of which are powerful inhibitors of HIF hydroxylases. As shown in Fig. 1B, both compounds strongly induced cyclin D1 mRNA. 786-O cells were next transfected with siRNAs that target HIF-1α and HIF-2α specifically. Cyclin D1 mRNA levels were clearly suppressed by siRNA directed against HIF-2α, but not HIF-1α (Fig. 1C). Despite the cell type specificity, these results indicate that hypoxia-inducible expression of cyclin D1 is mediated by the HIF system. They are consistent with the unusual pattern of HIF-α isoform expression in 786-O cells that express HIF-2α alone and correlate with data published during the course of this work demonstrating that HIF-2α overexpression can enhance cyclin D1 expression in RCC cells (1). To analyze this further, 786-O cells were infected with retrovirus, produced using the pLZRS-IRES-GFP vector, expressing (in addition to GFP) either HIF-1α, HIF-2α, or no additional sequence. As anticipated, infection with retrovirus expressing HIF-2α further induced cyclin D1 at both the mRNA (Fig. 1D) and protein (Fig. 1E) levels. However, in contrast, infection of 786-O cells with virus expressing HIF-1α downregulated cyclin D1 (Fig. 1D and E, upper part), a result that was paralleled by downregulation of HIF-2α (Fig. 1E, lower part). Further experiments were performed with RCC4, a VHL-defective cell line that expresses HIF-1α in addition to HIF-2α. Transfection of RCC4 cells with siRNAs targeting HIF-1α or HIF-2α demonstrated clear specificity of cyclin D1 for HIF-2α with no discernible downregulation or even slight upregulation by HIF-1α siRNA (Fig. 2A). Infection of RCC4 cells with retroviruses expressing HIF-α isoforms demonstrated induction by HIF-2α, but not HIF-1α (Fig. 2B).
FIG. 2.
Interaction between HIF-1α and HIF-2α and effects on cyclin D1 expression. (A) RNase protection assay of HIF-1α, HIF-2α, and CCND1 mRNAs (U6 snRNA, loading control) in RCC4 cells after treatment for 48 h with Oligofectamine alone (−), HIF-1α-directed siRNA (H1), HIF-2α-directed siRNA (H2), or control siRNA (C). (B and D) Immunoblots, with the indicated antibodies, of whole-cell lysates from RCC4 and SKRC28 cells that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (C and E) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of RCC4 and SKRC28 whole-cell lysates after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). (F) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of RCC4 whole-cell lysates treated with the previously indicated siRNAs and then exposed to 1 mM MMOG for 18 h. Two independent experiments for each condition are shown in panels B, C, D, E, and F.
Reciprocal interaction between HIF-1α and HIF-2α levels in RCC.
Overall, these findings indicate that cyclin D1 is a specific HIF-2α target in RCC cells. Unexpected down regulation of both cyclin D1 and HIF-2α when 786-O cells were infected with retrovirus expressing HIF-1α also suggested that HIF-α isoforms display an unanticipated interaction in this cell type. Further analysis of this in RCC4 cells indicated that overexpression of HIF-2α strikingly downregulated HIF-1α, suggesting that this interaction is reciprocal (Fig. 2B, middle part). To test whether interactions occur between the endogenous HIF-α proteins, we re-examined the effects of siRNA. Basal levels of HIF-2α were not affected by HIF-1α siRNA under these conditions; however, upregulation of HIF-1α protein was observed following suppression of HIF-2α (Fig. 2C). To determine how general these interactions are in RCC cells, further experiments were performed with SKRC28 cells (7), another VHL-defective RCC cell line expressing both HIF-1α and HIF-2α. Similar results were obtained, with clear suppression of HIF-1α following overexpression of HIF-2α (Fig. 2D) and induction of HIF-1α following siRNA-based suppression of HIF-2α (Fig. 2E). Though the reciprocal effects of HIF-1α on HIF-2α that were observed in 786-O cells were not seen in these experiments, such effects did occur under other conditions. For instance, RCC4 cells retain low-level regulation of HIF-2α by HIF hydroxylase inhibition. When examined following exposure to the hydroxylase inhibitor MMOG, modest but reproducible increases in HIF-2α were observed following siRNA-mediated suppression of HIF-1α levels (Fig. 2F). Densitometric analysis showed that in comparison to a constitutive species, β-tubulin, the increase in HIF-2α following HIF-1α siRNA was 2.1-fold (standard deviation [SD], ±0.3-fold; n = 3). Altogether, therefore, reciprocal interactions between HIF-1α and HIF-2α were apparent across a range of RCC cells.
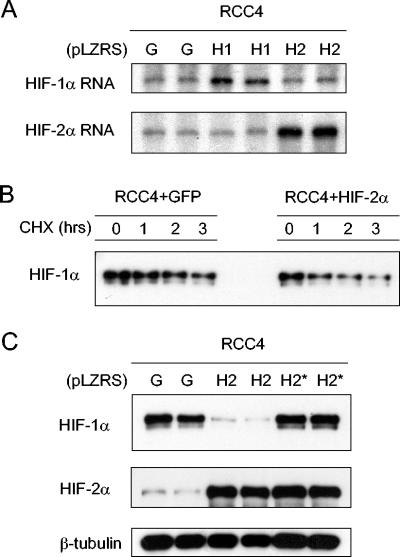
The interactive behavior of HIF-α proteins was in apparent contrast with that of HIF-α mRNAs, which had not demonstrated interactions in siRNA experiments (Fig. 2A, upper parts). To pursue this, we analyzed changes in HIF-α mRNA in RCC4 cells, which exhibit large suppressive effects of HIF-2α overexpression on HIF-1α protein. No such effects were observed on HIF-1α mRNA levels (Fig. 3A), confirming interaction at the protein level. Since this interaction has not been reported in non-RCC cells, we considered whether it might represent effects on an alternative protein degradation pathway(s) that becomes an important determinant(s) of HIF-α protein levels in VHL-defective cells. However, cycloheximide chase experiments indicated that despite an approximately threefold difference in steady-state HIF-1α protein levels in RCC4 cells that did or did not overexpress HIF-2α, the HIF-1α protein half-life was similar (Fig. 3B). Thus, it is most likely that the interaction is mediated through effects on HIF-α protein translation. To determine whether the effector mechanism requires HIF-mediated transcription, a HIF-2α molecule containing a five-amino-acid substitution in its DNA binding domain, that ablates HIF-2α transcriptional activity (19), was created in pLZRS-IRES-GFP and used to infect RCC4 cells. Results are shown in Fig. 3C and indicate that DNA binding is necessary, suggesting, though not formally proving, that HIF-2α transcriptional activity is required for suppression of HIF-1α.
FIG. 3.
Analysis of suppression of HIF-1α by HIF-2α in RCC4 cells. (A) RNase protection assay of HIF-1α and HIF-2α mRNAs in RCC4 cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (B) Representative immunoblot for HIF-1α of RCC4 whole-cell lysates which were infected with retroviral supernatants made from pLZRS containing GFP alone or HIF-2α and then treated with 100 μM cycloheximide (CHX) for 0, 1, 2, or 3 h. Note that to aid densitometric analysis of signal decay, more extract was loaded from cells infected with retroviruses expressing HIF-2α than GFP alone so as to obtain sufficient signal intensity at t = 0. (C) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of whole-cell lysates from RCC4 cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-2α (H2), or a HIF-2α DNA binding domain mutant (H2*). Two independent experiments for each condition are shown in panels A and C.
Transcriptional selectivity for HIF-1α and HIF-2α among genes expressed in RCC cells.
Given these interactions between HIF-α subunits, the unusual ratio of HIF-2α to HIF-1α expression in RCC cells and tumors, and the complete selectivity for HIF-2α manifested by cyclin D1, we next sought to analyze the transcriptional selectivity for HIF-1α or HIF-2α in RCC cells among a wider group of genes. Five gene products of potential relevance to different aspects of tumor biology, BNip3 (a proapoptotic member of the Bcl-2 family), carbonic anhydrase 9 (CAIX), glucose transporter 1 (GLUT-1), VEGF, and TGF-α, were studied.
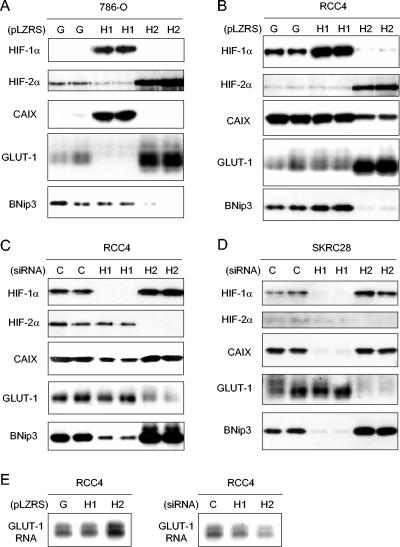
BNip3, CAIX, and GLUT-1 were analyzed by immunoblotting of whole-cell extracts. In the first set of experiments, 786-O cells were infected with retroviruses expressing HIF-1α, HIF-2α, or GFP alone as described above. As before, expression of HIF-1α in 786-O cells suppressed HIF-2α (Fig. 4A, upper two parts). Strikingly, for each of the HIF target gene products, distinct HIF-α isoform-specific responses were observed. CAIX was strongly induced by infection with virus expressing HIF-1α, but not virus expressing HIF-2α (Fig. 4A, third part). GLUT-1 displayed the reverse pattern, with striking induction by HIF-2α, and, like cyclin D1, was downregulated following infection with virus expressing HIF-1α, paralleling the downregulation of HIF-2α (Fig. 4A, fourth part). Results for BNip3 were of particular interest. BNip3 was unresponsive to HIF-1α but clearly downregulated by increased expression of HIF-2α (Fig. 4A, bottom part).
FIG. 4.
HIF-α isoform transcriptional specificity in 786-O, RCC4, and SKRC28 cells. (A and B) Immunoblots for HIF-1α, HIF-2α, CAIX, GLUT-1, and BNip3 of 786-O and RCC4 whole-cell lysates that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (C and D) Immunoblots, with the indicated antibodies, of whole-cell lysates from RCC4 and SKCR28 cells after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). (E) RNase protection assay of GLUT-1 mRNA in RCC4 cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2) and after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). Two independent experiments for each condition are shown in panels A, B, C, and D.
Further experiments were performed with RCC4 and SKRC28 cells (Fig. 4B to D). CAIX was again specifically responsive to HIF-1α, with downregulation in response to HIF-1α- but not HIF-2α-directed RNA interference (RNAi). Interestingly, downregulation was also observed in response to HIF-2α overexpression, due to either direct suppression or indirect effects via suppression of HIF-1α (Fig. 4B). The inability of HIF-1α overexpression in RCC4 to further induce CAIX levels may indicate that endogenous expression of this gene product was already maximal in this cell type (Fig. 4B). GLUT-1 again responded specifically to HIF-2α, being downregulated by HIF-2α RNAi (Fig. 4C and D) and upregulated by HIF-2α overexpression (Fig. 4B). These effects on GLUT-1 protein levels were also observed at the mRNA level (Fig. 4E). Though the findings were not identical, BNip3 also responded similarly to the pattern in 786-O cells. In keeping with downregulation by HIF-2α, BNip3 levels were strikingly reduced in RCC4 cells infected with retroviruses expressing HIF-2α and increased by HIF-2α-directed siRNA. However, in RCC4 and SKRC28 cells BNip3 retained a positive response to HIF-1α, which has been described in non-RCC cell types but was not observed in 786-O cells (Fig. 4B to D).
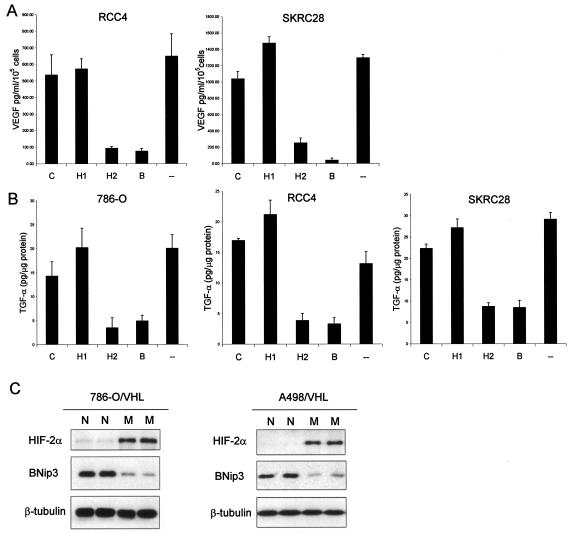
Effects on VEGF and TGF-α expression were assayed by enzyme-linked immunosorbent assay (ELISA) using tissue culture supernatant or cell extract, respectively. Following previous analyses of 786-O cells (39), effects of siRNA-mediated HIF-α suppression on VEGF secretion were examined in RCC4 and SKRC28 cells, whereas effects on TGF-α levels were assessed in 786-O, RCC4, and SKRC28 cells. In each cell line, VEGF and TGF-α production was specifically reduced by siRNA-mediated suppression of HIF-2α but not suppression of HIF-1α (Fig. 5A and B).
FIG. 5.
Unusual patterns of HIF-α-dependent gene expression in RCC cells. (A) VEGF secretion by RCC4 and SKRC28 cells as determined by ELISA of medium supernatant. Cells were treated with siRNAs directed against a control sequence (C), HIF-1α (H1), HIF-2α (H2), both HIF-1α and HIF-2α (B), or Oligofectamine alone (−). VEGF levels were normalized to cell number. Experiments were performed in triplicate at least three times, and error bars correspond to 1 standard deviation. (B) Expression levels of TGF-α in 786-O, RCC4, and SKRC28 cell lysates as determined by ELISA. Cells were treated with siRNAs directed against a control sequence (C), HIF-1α (H1), HIF-2α (H2), both HIF-1α and HIF-2α (B), or Oligofectamine alone (−). TGF-α expression levels in the cell lysates were normalized to total cellular protein content. Experiments were performed in triplicate at least three times, and error bars correspond to 1 standard deviation. (C) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of whole-cell lysates from 786-O/VHL and A498/VHL cells after exposure to normoxia (N) or 1 mM MMOG (M) for 18 h. Two independent experiments for each condition are shown in panel C.
HIF-α isoform-specific patterns of gene expression differ between RCC and non-RCC cell lines.
The above findings indicate that each of six HIF target genes displays a characteristic HIF-α isoform-specific response in a range of VHL-defective RCC cells. These results predict that the unusual bias toward HIF-2α expression in RCC cells would itself lead to unusual patterns of HIF target gene expression. For instance, negative regulation by HIF-2α, and positive regulation by HIF-1α, of BNip3 predicts that in VHL-defective cells displaying a strong bias to HIF-2α isoform expression, the response of BNip3 to activation of the HIF pathway would be inverted. To test this, we examined the effect of the HIF hydroxylase inhibitor MMOG on BNip3 expression in stable transfectants of 786-O and A498 cells re-expressing wild-type VHL. Both of these cells express functional HIF-2α but not HIF-1α. In keeping with predictions, but in contrast with the results of studies in other cell lines (3, 21), BNip3 was markedly downregulated by MMOG in these cells (Fig. 5C). There were no effects on the expression of BNip3 or HIF-2α in response to MMOG treatment in the parental 786-O cell line (data not shown).
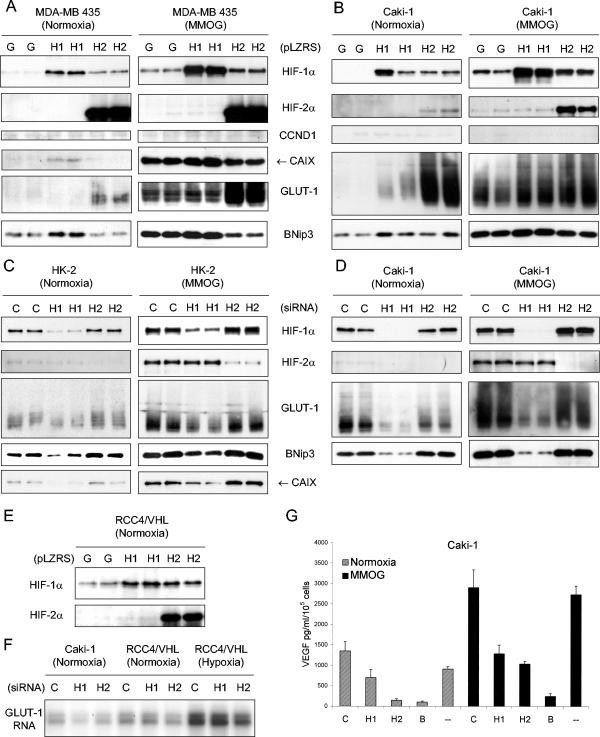
We next wanted to determine if the specific connections between HIF-α isoforms and particular target genes, and the suppressive interactions between HIF-α isoforms in VHL-defective RCC cell lines, differ from those in cells not derived from VHL-associated cancer. To this end, studies were performed with Caki-1 cells (an RCC cell line), HK-2 cells (a human kidney proximal-tubule cell line), and MDA-MB 435 cells (a melanoma cell line, often considered to be breast carcinoma), all of which are wild type for VHL.
Studies using siRNA revealed evidence for a pattern of reciprocal suppression between HIF-1α and HIF-2α similar to that defined in VHL-defective RCC cells; for instance, siRNA-mediated suppression of HIF-2α was associated with upregulation of HIF-1α in MMOG-treated cells, consistent with enhanced synthesis of the HIF-1α protein (Fig. 6C and D, right parts). However, overexpression experiments demonstrated a clear difference; for instance, overexpression of HIF-2α in VHL-competent cells led to enhanced expression of HIF-1α (Fig. 6A and B). Similar results were obtained in stable transfectants of RCC4 cells re-expressing wild-type VHL (Fig. 6E). The positive interaction observed with overexpression in RCC4/VHL but not RCC4 cells indicates that the change in behavior is determined not by cell background but by the presence of an intact VHL degradation pathway and is consistent with previous data indicating that this pathway can be readily saturated, leading to stabilization of HIF-α subunits following overexpression (31, 40). A likely explanation is therefore that in VHL wild-type cells there are two different types of HIF-α interaction taking place, competition in a saturable VHL-mediated degradation pathway and the mutually suppressive (negative) interaction defined above. Indeed, in siRNA experiments, this negative interaction can be observed between HIF-1α and HIF-2α in RCC4/VHL cells (data not shown). Following VHL inactivation, the first process (competition for degradation) no longer operates so that the dominating pattern of HIF-α interaction apparently switches to one of negative interaction.
FIG. 6.
HIF-α isoform transcriptional selectivity in MDA-MB 435, Caki-1, and HK-2 cells. (A and B) Immunoblots, with the indicated antibodies, of MDA-MB 435 and Caki-1 whole-cell lysates that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2) and then exposed to normoxia or 1 mM MMOG for the final 18 h. (C and D) Immunoblots, with the indicated antibodies, of whole-cell lysates from HK-2 and Caki-1 cells after treatment for 48 h with control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2) and then exposed to normoxia or 1 mM MMOG for the final 18 h. (E) Immunoblots for HIF-1α and HIF-2α of RCC4/VHL whole-cell lysates that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). Two independent experiments for each condition are shown in panels A, B, C, D, and E. (F) RNase protection assay of GLUT-1 mRNA in Caki-1, RCC4/VHL (Normoxia), and RCC4/VHL (Hypoxia) cells after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). (G) Secreted levels of VEGF in Caki-1 as determined by ELISA of medium supernatant. Cells were treated with siRNAs directed against a control sequence (C), HIF-1α (H1), HIF-2α (H2), both HIF-1α and HIF-2α (B), or Oligofectamine alone (−) and then exposed to normoxia or 1 mM MMOG for the final 18 h. VEGF levels were normalized to cell number. Experiments were performed in triplicate at least three times, and error bars correspond to 1 standard deviation.
Analysis of gene expression in Caki-1 and non-RCC cells also revealed apparent differences in the transcriptional connections of the HIF pathway. Some genes showed differences in HIF-α isoform specificity in cells derived from VHL-defective RCC versus other cell types, whereas other genes only responded to the HIF pathway in cells derived from VHL-defective RCC.
CAIX was specifically responsive to HIF-1α in all cell backgrounds. Likewise, BNip3 appeared to behave similarly across all cells, demonstrating positive regulation by HIF-1α and either no clear regulation or negative regulation (Fig. 6A) by HIF-2α. Consistent with reports of cell type-restricted responses to hypoxia (2, 46), cyclin D1 was unresponsive to RNAi-based suppression or retrovirus-mediated overexpression of either HIF-α isoform in any cell other than those derived from VHL-defective RCC. Of particular interest were the responses of GLUT-1 and VEGF. In overexpression studies, GLUT-1 responded to both HIF-1α and HIF-2α (Caki-1) or even dominantly to HIF-2α (MDA-MB 435), whereas studies of suppression by RNAi (Caki-1 and HK-2) indicated that endogenous levels were largely maintained by HIF-1α with minimal contribution from HIF-2α (Fig. 6A to D). Similar results were observed when GLUT-1 was assayed in Caki-1 cells at the mRNA level (Fig. 6F, lanes 1 to 3). This pattern is in marked contrast to that in the VHL-defective RCC cells, where the contribution from HIF-1α was minimal, even in RCC4 and SKRC28 cells, where this isoform was expressed at substantial levels (compare Fig. 6C and D with Fig. 4C and D). Interestingly, the switch to HIF-2α dependence of GLUT-1 was not reversed by VHL reconstitution in RCC4 (Fig. 6F). This is consistent with observations in 786-O cells, where dependence of GLUT-1 and certain other target genes on HIF-2α rather than HIF-1α is not reversed by VHL reconstitution (11, 39), suggesting that this is a cell background effect observed in VHL-defective RCC but not directly attributable to VHL itself. The response of VEGF also showed differences between the VHL wild-type and VHL-defective cells (compare Fig. 6G and 5A). For instance, in distinction from results in VHL-defective cells, VEGF production was clearly downregulated by HIF-1α-directed RNAi in Caki-1 cells.
Gene expression patterns in VHL-associated renal lesions.
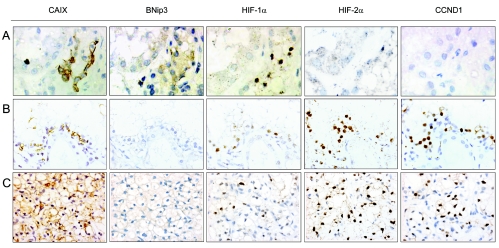
To pursue the clinical relevance of the HIF-α isoform transcriptional selectivity manifest in cultured RCC cells, we next examined immunohistochemical staining patterns in nephrectomy specimens obtained from VHL patients. In such kidneys we have previously demonstrated that activation of the HIF system is an early feature of disease, with HIF-1α being manifest in very early lesions detectable even as single tubular cells, whereas HIF-2α upregulation appeared to occur later with progressively stronger staining in larger and more dysplastic lesions, and overt carcinoma (26). This is illustrated in Fig. 7, where it can be seen that HIF-1α staining is strong in the small, presumably early, lesion (row A) and maintained in the larger cystic (row B) and overtly cancerous lesions (row C), whereas HIF-2α is much more strongly expressed in the cystic and cancerous lesions. In association with this, staining of serial sections for CAIX, BNip3, and cyclin D1 shows that while CAIX is expressed (like HIF-1α) in all lesions, BNip3 and cyclin D1 show strikingly different patterns, with BNip3 strongest in early lesions that express little or no HIF-2α and cyclin D1 being essentially confined to later lesions that do express HIF-2α.
FIG. 7.
Immunohistochemical analysis for HIF-α and transcriptional targets in sections from the kidney of a patient with VHL disease. Immunohistochemical analysis, with the indicated antibodies, of serial sections of representative lesions. Panels: A, an early multicellular lesion; B, a renal cyst; C, an overt clear-cell RCC. Final magnifications: A, ×360; B and C, ×240.
Effects of HIF-1α and HIF-2α overexpression on growth of RCC tumor xenografts.
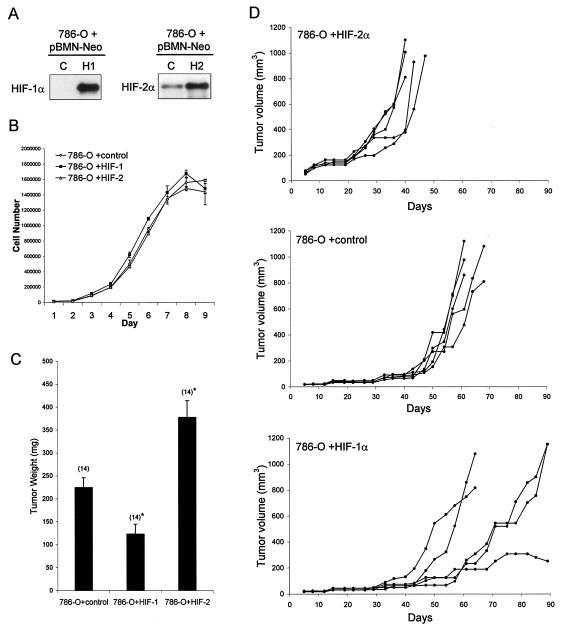
Finally, we tested the effects of enhancing expression of HIF-1α or HIF-2α on the growth of RCC cells as tumor xenografts. In a first set of experiments, 786-O cells were infected with retroviruses expressing HIF-1α, HIF-2α, or GFP alone with the same protocol that was used for the gene expression experiments. Unselected pools of infected cells were implanted subcutaneously into nude mice. Whereas four of five injections of 786-O cells bearing control retrovirus (expressing GFP alone) formed tumors, none of the injections of 786-O infected with retrovirus expressing HIF-1α formed tumors. In contrast, 786-O cells infected with retrovirus expressing HIF-2α formed tumors that grew at an enhanced rate compared to the cells infected with control virus, suggesting disparate effects of HIF-1α and HIF-2α on tumor growth. To pursue this, we made a further set of retroviruses expressing HIF-1α or HIF-2α as part of a bicistronic mRNA also expressing a neomycin resistance gene in pBMN-Z-IRES-Neo. These viruses were used to infect 786-O cells and select pools of cells overexpressing HIF-1α, HIF-2α, or empty vector. Aliquots of these cells were then analyzed for growth as monolayers under standard tissue culture conditions or for growth as tumors after subcutaneous injection into nude mice. In these experiments, injected cells were mixed with Matrigel with the aim of enhancing the tumor take so as to permit better comparison of growth rates. Immunoblotting revealed that moderate overexpression of HIF-1α and HIF-2α was achieved in these cells (Fig. 8A). However, when they were grown as monolayers under standard tissue culture conditions, no differences in the growth rate or maximum cell number were observed (Fig. 8B). In contrast, marked differences were again observed in tumor growth. Two sets of experiments, using separately derived pools of cells were performed. Whereas infection with retrovirus expressing HIF-2α clearly enhanced tumor growth in these experiments, infection with retrovirus expressing HIF-1α had the reverse effect (Fig. 8C and D). Though some variability was seen in growth curves of individual tumors expressing HIF-1α (Fig. 8D), the largest experiment showed significant retardation of growth for this group of tumors (Fig. 8C).
FIG. 8.
Effect of overexpression of HIF-1α or HIF-2α on growth of 786-O cells as monolayers or tumor xenografts. (A) Immunoblots for HIF-1α and HIF-2α of whole-cell lysates from 786-O polyclonal pools of cells that were selected with G418 after infection with retroviral supernatants made from pBMN-Neo containing an empty cassette (LacZ spliced out) as a control (C), HIF-1α (H1), or HIF-2α (H2). (B) Growth of selected polyclonal pools of 786-O cells stably infected with the indicated retroviruses. Growth was measured under standard tissue culture conditions for 9 days, and there was no significant difference in the proliferation rate between the groups. Error bars indicate standard deviations. (C) Tumor weights approximately 5.5 weeks after subcutaneous injection of polyclonal pools of 786-O cells stably infected with the indicated retroviruses. There were 14 tumors analyzed for each group, and error bars indicate 1 standard error. Two-tailed, unpaired Student t tests comparing each HIF-α overexpressing group to the control group were performed, and statistically significant difference is indicated by asterisks for P < 0.005. (D) Growth curves indicated as tumor volume over a 90-day period after subcutaneous injection of polyclonal pools of 786-O cells stably infected with the indicated retroviruses. Five mice with single tumors were analyzed for each group.
DISCUSSION
This study demonstrates that, despite close similarities between the HIF-α isoforms, differential activation of HIF-1α or HIF-2α pathways in VHL-defective RCC cells has nonequivalent or even opposing effects on gene expression and experimental tumor growth. Our investigations differ somewhat from previous studies of the role of HIF in RCC tumor growth, in which two different groups used either mutated HIF-1α (27) or mutated HIF-2α molecules (18, 19), which escape VHL recognition, to test for opposition of the tumor-suppressive effect of reintroducing wild-type VHL into 786-O cells. We used wild-type HIF-α isoforms in assays for effects on tumor growth and directly compared the effects of augmenting HIF-1α or HIF-2α on the growth of 786-O cells as subcutaneous tumors in nude mice. Our data indicate that the apparent discrepancy between the two previous studies arises from real differences between the actions of HIF-1α and HIF-2α, with HIF-2α having positive effects on tumor growth and HIF-1α having negative effects on tumor growth in this setting. Since we also demonstrate suppressive interaction between HIF-α isoforms, we cannot as yet distinguish whether suppression of tumor growth by HIF-1α arises from direct effects, indirect effects due to downregulation of HIF-2α, or a combination of both possibilities. Interestingly, in contrast to effects on tumor growth, no effects were observed on monolayer growth under standard tissue culture conditions, as has been observed in assays of VHL tumor suppressor function based on re-introduction of wild-type VHL into VHL-defective RCC cells (12, 19, 49).
Consistent with differential effects on tumor growth were differential effects on the expression of specific genes with putative pro- and antitumorigenic effects. In particular, HIF-1α positively regulated BNip3 but had no effect on cyclin D1, TGF-α, and VEGF, whereas HIF-2α negatively regulated BNip3 and positively regulated cyclin D1, TGF-α, and VEGF. BNip3 is a member of the Bcl-2 family of apoptosis-regulating proteins and activates caspase-independent necrosis-like cell death by opening the mitochondrial permeability transition pore (43). In most cells, the protein is strongly induced by hypoxia, and an involvement in hypoxic tumor necrosis has been postulated (3, 21). Cyclin D1 is one of the main G1-phase cyclins. Its expression is associated with G1-to-S transition in the cell cycle and upregulated in many types of cancer by a variety of mechanisms (38). TGF-α, on the other hand, is a potent renal cell mitogen that activates the epidermal growth factor receptor pathway and has been proposed to initiate an autocrine loop with this receptor when VHL is inactivated in renal cells (6). CAIX and GLUT-1 are both upregulated in many forms of cancer, but their contribution to tumor growth is less clear. GLUT-1 was found to be a specific HIF-2α target in VHL-defective RCC but not in other cells, whereas CAIX was found to be a specific HIF-1α target in both RCC and non-RCC cells, as has been reported for genes encoding a range of glycolytic enzymes (11). Interestingly, though upregulation of CAIX is commonly observed in RCC (25), reduced staining has been correlated with poorer prognosis in RCC in a recent clinical series (4).
Based on known functions, it seems likely that the HIF-α isoform transcriptional selectivity manifest by one or more of the genes analyzed could contribute to differential effects of the HIF-α isoforms on VHL-associated RCC growth. However, the exact contribution of individual genes was not defined. Given the complexity of the HIF transcriptional cascade and the large number of direct and indirect HIF targets identified in gene expression arrays (11, 15, 47), such an analysis remains a considerable task. Nor can it be deduced that other potentially protumorigenic and antitumorigenic HIF target genes will fall into the above pattern, particularly as in non-RCC settings the balance of evidence favors a positive action of HIF-1α on tumor growth (37). Presumably, the set of HIF-1α and HIF-2α targets in part reflects differences in the physiological role of these proteins in the response to hypoxia that are as yet unclear. Interestingly, HIF-1α has recently been demonstrated to antagonize Myc and induce cell cycle arrest by complexing Myc in a transcriptionally inactive form (20). In future studies it will be interesting to determine if this property is shared by HIF-2α and whether it contributes to effects on RCC tumor growth.
Though HIF-α transcriptional selectivity was observed in all cells, there were important differences between what were essentially two distinct patterns, that observed in VHL-defective RCC cells and that observed in all other cells, including the VHL-competent RCC line Caki-1. Differences in transcriptional targeting were of two types. First, some genes such as cyclin D1 appeared to be transcriptional targets of HIF-2α only in VHL-defective cells. TGF-α may also fall into this category but has been less extensively studied in VHL-competent cells. Second, some genes, such as those for GLUT-1 and VEGF, appeared to be specific HIF-2α targets in VHL-defective RCC cells whereas in VHL-competent Caki-1 and non-RCC cells they appear potentially responsive to both HIF-α isoforms. For instance, for GLUT-1, HIF-α suppression by siRNA in Caki-1 and non-RCC cells indicates clear dependence on HIF-1α, whereas overexpression studies show that enhanced expression of both isoforms, but particularly HIF-2α, can clearly drive GLUT-1 expression. These findings correlate well with published work with other non-RCC cells that has shown that GLUT-1 is induced by overexpression of both HIF-α isoforms in VHL wild-type HEK 293 cells (11) but predominantly dependent on endogenous HIF-1α, as revealed by genetic inactivation studies (39). In contrast, in VHL-defective RCC cells, neither HIF-1α siRNA nor HIF-1α overexpression had any effect on GLUT-1, even in cells retaining substantial levels of HIF-1α that was transcriptionally active on other target genes. Though the mechanistic basis of this difference is unclear, the simplest explanation is that a specific transcriptional connection between HIF-1α and these genes is missing in VHL-defective RCC.
It is difficult to distinguish whether these unusual properties of the HIF system are newly acquired during RCC development as further events following VHL inactivation or whether they reflect unusual intrinsic properties of the cells giving rise to VHL-associated RCC. The clear differences from the related renal cell line Caki-1 favor the occurrence of additional events, though concordance across a number of RCC lines suggests that such events must occur at high probability following VHL inactivation.
Also of interest in understanding the unusual properties of the HIF system in VHL-defective RCC cells is the demonstration of suppressive interactions between HIF-α isoforms. Though a natural antisense to the 3′ untranslated region of HIF-1α has been described (aHIF) (41) that may suppress HIF-1α mRNA in RCC cells, the suppressive interaction we observed was not manifest on the mRNA level, indicating that it represents a different process. Our results are consistent with an action on HIF-α protein translation, though we have not yet defined the mechanism. Whether such a process extends to other components of hypoxia pathways such as the translation of HIF target genes is unclear, though in general we found that changes in expression of HIF target gene transcripts and protein products were concordant. Whatever the mechanism, our analysis suggests that the interaction we describe is not a distinct property of the HIF system in VHL-defective cells; rather, it is uncovered as a direct consequence of VHL inactivation blocking the HIF degradation pathway, which is saturable, leading to competitive interactions between HIF-1α and HIF-2α in the opposite direction of that seen in VHL-competent cells. Nevertheless, this phenomenon could well contribute to the unusual dominant expression of HIF-2α over HIF-1α expression observed in VHL-defective RCC, since upregulation of HIF-2α would now be anticipated to downregulate HIF-1α and vice versa. Such a scenario begs the following question: is upregulation of HIF-2α or downregulation of HIF-1α the primary event in driving the observed bias? Currently, we cannot answer this question, though it is of interest that a recent report has described a shortened HIF-1α transcript in 786-O cells (44) and in previous work we have observed a shortened HIF-1α protein species in A498 cells (M. E. Cockman, Ph.D. thesis, Oxon, 2003), suggesting that at least in some cases inactivation of HIF-1α may be a discrete event.
Overall, our data indicate that HIF-1α and HIF-2α have functionally distinct roles in the biology of VHL-defective RCC, with HIF-2α promoting and HIF-1α retarding tumor growth, and that the HIF system behaves unusually in this setting in a number of ways. There is growing interest in the development of HIF inhibitors as anticancer agents (24, 33, 37). Our findings clearly have relevance to targeting the HIF system in cancer therapy and emphasize the importance of considering isoform-specific effects.
Acknowledgments
This work was funded by the Wellcome Trust, Cancer Research UK, the Medical Research Council, the Urology Research Fund from Guy's Hospital, and a 6th Framework Programme at the E.U. (Euroxy Project).
We thank J. Pastorek for providing the M75 antibody, K. Kranc and S. Bhattacharya for advice on retroviral work, D. Jones for help with ELISAs, and R. Peat, S. Peak, S. Johnson, and D. Watling for assistance in xenograft experiments. K.W.L. was supported by a scholarship from the Agency for Science, Technology, and Research (Singapore). R.R.R. was supported by a Rhodes scholarship.
REFERENCES
- 1.Baba, M., S. Hirai, H. Yamada-Okabe, K. Hamada, H. Tabuchi, K. Kobayashi, K. Kondo, M. Yoshida, A. Yamashita, T. Kishida, N. Nakaigawa, Y. Nagashima, Y. Kubota, M. Yao, and S. Ohno. 2003. Loss of von Hippel-Lindau protein causes cell density dependent deregulation of cyclin D1 expression through hypoxia-inducible factor. Oncogene 22:2728-2738. [DOI] [PubMed] [Google Scholar]
- 2.Bindra, R. S., J. R. Vasselli, R. Stearman, W. M. Linehan, and R. D. Klausner. 2002. VHL-mediated hypoxia regulation of cyclin D1 in renal carcinoma cells. Cancer Res. 62:3014-3019. [PubMed] [Google Scholar]
- 3.Bruick, R. K. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 97:9082-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui, M. H., D. Seligson, K. R. Han, A. J. Pantuck, F. J. Dorey, Y. Huang, S. Horvath, B. C. Leibovich, S. Chopra, S. Y. Liao, E. Stanbridge, M. I. Lerman, A. Palotie, R. A. Figlin, and A. S. Belldegrun. 2003. Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: implications for prognosis and therapy. Clin. Cancer Res. 9:802-811. [PubMed] [Google Scholar]
- 5.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, E. Keshert, and E. Keshet. 1998. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 6.de Paulsen, N., A. Brychzy, M. C. Fournier, R. D. Klausner, J. R. Gnarra, A. Pause, and S. Lee. 2001. Role of transforming growth factor-α in von Hippel-Lindau (VHL)−/− clear cell renal carcinoma cell proliferation: a possible mechanism coupling VHL tumor suppressor inactivation and tumorigenesis. Proc. Natl. Acad. Sci. USA 98:1387-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert, T., N. H. Bander, C. L. Finstad, R. D. Ramsawak, and L. J. Old. 1990. Establishment and characterization of human renal cancer and normal kidney cell lines. Cancer Res. 50:5531-5536. [PubMed] [Google Scholar]
- 8.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 9.Hockel, M., and P. Vaupel. 2001. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 93:266-276. [DOI] [PubMed] [Google Scholar]
- 10.Hon, W. C., M. I. Wilson, K. Harlos, T. D. Claridge, C. J. Schofield, C. W. Pugh, P. H. Maxwell, P. J. Ratcliffe, D. I. Stuart, and E. Y. Jones. 2002. Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature 417:975-978. [DOI] [PubMed] [Google Scholar]
- 11.Hu, C. J., L. Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23:9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin, Jr. 1995. Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-826. [DOI] [PubMed] [Google Scholar]
- 13.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, C., H. Lu, K. A. Vincent, S. Shankara, A. J. Belanger, S. H. Cheng, G. Y. Akita, R. A. Kelly, M. A. Goldberg, and R. J. Gregory. 2002. Gene expression profiles in human cardiac cells subjected to hypoxia or expressing a hybrid form of HIF-1α. Physiol. Genomics 8:23-32. [DOI] [PubMed] [Google Scholar]
- 16.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 17.Kaelin, W. G., Jr. 2003. The von Hippel-Lindau gene, kidney cancer, and oxygen sensing. J. Am. Soc. Nephrol. 14:2703-2711. [DOI] [PubMed] [Google Scholar]
- 18.Kondo, K., W. Y. Kim, M. Lechpammer, and W. G. Kaelin, Jr. 2003. Inhibition of HIF-2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1:E83. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, K., J. Klco, E. Nakamura, M. Lechpammer, and W. G. Kaelin, Jr. 2002. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1:237-246. [DOI] [PubMed] [Google Scholar]
- 20.Koshiji, M., Y. Kageyama, E. A. Pete, I. Horikawa, J. C. Barrett, and L. E. Huang. 2004. HIF-1α induces cell cycle arrest by functionally counteracting Myc. EMBO J. 23:1949-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kothari, S., J. Cizeau, E. McMillan-Ward, S. J. Israels, M. Bailes, K. Ens, L. A. Kirshenbaum, and S. B. Gibson. 2003. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 22:4734-4744. [DOI] [PubMed] [Google Scholar]
- 22.Krieg, M., R. Haas, H. Brauch, T. Acker, I. Flamme, and K. H. Plate. 2000. Up-regulation of hypoxia-inducible factors HIF-1α and HIF-2α under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene 19:5435-5443. [DOI] [PubMed] [Google Scholar]
- 23.Kung, A. L., S. Wang, J. M. Klco, W. G. Kaelin, and D. M. Livingston. 2000. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 6:1335-1340. [DOI] [PubMed] [Google Scholar]
- 24.Kung, A. L., S. D. Zabludoff, D. S. France, S. J. Freedman, E. A. Tanner, A. Vieira, S. Cornell-Kennon, J. Lee, B. Wang, J. Wang, K. Memmert, H. U. Naegeli, F. Petersen, M. J. Eck, K. W. Bair, A. W. Wood, and D. M. Livingston. 2004. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 6:33-43. [DOI] [PubMed] [Google Scholar]
- 25.Liao, S. Y., O. N. Aurelio, K. Jan, J. Zavada, and E. J. Stanbridge. 1997. Identification of the MN/CA9 protein as a reliable diagnostic biomarker of clear cell carcinoma of the kidney. Cancer Res. 57:2827-2831. [PubMed] [Google Scholar]
- 26.Mandriota, S. J., K. J. Turner, D. R. Davies, P. G. Murray, N. V. Morgan, H. M. Sowter, C. C. Wykoff, E. R. Maher, A. L. Harris, P. J. Ratcliffe, and P. H. Maxwell. 2002. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell 1:459-468. [DOI] [PubMed] [Google Scholar]
- 27.Maranchie, J. K., J. R. Vasselli, J. Riss, J. S. Bonifacino, W. M. Linehan, and R. D. Klausner. 2002. The contribution of VHL substrate binding and HIF-1α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 1:247-255. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell, P. H., G. U. Dachs, J. M. Gleadle, L. G. Nicholls, A. L. Harris, I. J. Stratford, O. Hankinson, C. W. Pugh, and P. J. Ratcliffe. 1997. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. USA 94:8104-8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 30.Min, J. H., H. Yang, M. Ivan, F. Gertler, W. G. Kaelin, Jr., and N. P. Pavletich. 2002. Structure of an HIF-1α-pVHL complex: hydroxyproline recognition in signaling. Science 296:1886-1889. [DOI] [PubMed] [Google Scholar]
- 31.O'Rourke, J. F., Y. M. Tian, P. J. Ratcliffe, and C. W. Pugh. 1999. Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1α. J. Biol. Chem. 274:2060-2071. [DOI] [PubMed] [Google Scholar]
- 32.Park, S. K., A. M. Dadak, V. H. Haase, L. Fontana, A. J. Giaccia, and R. S. Johnson. 2003. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1α (HIF-1α): role of cytoplasmic trapping of HIF-2α. Mol. Cell. Biol. 23:4959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapisarda, A., B. Uranchimeg, D. A. Scudiero, M. Selby, E. A. Sausville, R. H. Shoemaker, and G. Melillo. 2002. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 62:4316-4324. [PubMed] [Google Scholar]
- 34.Ravi, R., B. Mookerjee, Z. M. Bhujwalla, C. H. Sutter, D. Artemov, Q. Zeng, L. E. Dillehay, A. Madan, G. L. Semenza, and A. Bedi. 2000. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genes Dev. 14:34-44. [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seagroves, T. N., H. E. Ryan, H. Lu, B. G. Wouters, M. Knapp, P. Thibault, K. Laderoute, and R. S. Johnson. 2001. Transcription factor HIF-1 is a necessary mediator of the Pasteur effect in mammalian cells. Mol. Cell. Biol. 21:3436-3444.11313469 [Google Scholar]
- 37.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3:721-732. [DOI] [PubMed] [Google Scholar]
- 38.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 39.Sowter, H. M., R. R. Raval, J. W. Moore, P. J. Ratcliffe, and A. L. Harris. 2003. Predominant role of hypoxia-inducible transcription factor (Hif)-α versus Hif-2α in regulation of the transcriptional response to hypoxia. Cancer Res. 63:6130-6134. [PubMed] [Google Scholar]
- 40.Tanimoto, K., Y. Makino, T. Pereira, and L. Poellinger. 2000. Mechanism of regulation of the hypoxia-inducible factor-1α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19:4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thrash-Bingham, C. A., and K. D. Tartof. 1999. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J. Natl. Cancer Inst. 91:143-151. [DOI] [PubMed] [Google Scholar]
- 42.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 43.Vande Velde, C., J. Cizeau, D. Dubik, J. Alimonti, T. Brown, S. Israels, R. Hakem, and A. H. Greenberg. 2000. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20:5454-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warnecke, C., Z. Zaborowska, J. Kurreck, V. A. Erdmann, U. Frei, M. Wiesener, and K. U. Eckardt. 2004. Differentiating the functional role of hypoxia-inducible factor (HIF)-1α and HIF-2α (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2α target gene in Hep3B and Kelly cells. FASEB J. 18:1462-1464. [DOI] [PubMed] [Google Scholar]
- 45.Wiesener, M. S., H. Turley, W. E. Allen, C. Willam, K. U. Eckardt, K. L. Talks, S. M. Wood, K. C. Gatter, A. L. Harris, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 1998. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1α. Blood 92:2260-2268. [PubMed] [Google Scholar]
- 46.Wykoff, C. C., C. Sotiriou, M. E. Cockman, P. J. Ratcliffe, P. Maxwell, E. Liu, and A. L. Harris. 2004. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br. J. Cancer 90:1235-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zatyka, M., N. F. da Silva, S. C. Clifford, M. R. Morris, M. S. Wiesener, K. U. Eckardt, R. S. Houlston, F. M. Richards, F. Latif, and E. R. Maher. 2002. Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 62:3803-3811. [PubMed] [Google Scholar]
- 48.Zavada, J., Z. Zavadova, S. Pastorekova, F. Ciampor, J. Pastorek, and V. Zelnik. 1993. Expression of MaTu-MN protein in human tumor cultures and in clinical specimens. Int. J. Cancer 54:268-274. [DOI] [PubMed] [Google Scholar]
- 49.Zimmer, M., D. Doucette, N. Siddiqui, and O. Iliopoulos. 2004. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2:89-95. [PubMed] [Google Scholar]