Abstract
Photochemically induced dynamic nuclear polarization NMR pulse-labeling techniques have been used to obtain detailed information about side-chain surface accessibilities in the partially folded (molten globule) states of bovine and human α-lactalbumin prepared under a variety of well defined conditions. Pulse labeling involves generating nuclear polarization in the partially folded state, rapidly refolding the protein within the NMR sample tube, then detecting the polarization in the well dispersed native-state spectrum. Differences in the solvent accessibility of specific side chains in the various molten globule states indicate that the hydrophobic clusters involved in stabilizing the α-lactalbumin fold can be formed from interactions between a variety of different hydrophobic residues in both native and nonnative environments. The multiple subsets of hydrophobic clusters are likely to result from the existence of distinct but closely related local minima on the free-energy landscape of the protein and show that the fold and topology of a given protein may be formed from degenerate groups of side chains.
Keywords: hydrophobic cluster, protein folding, pulse labeling, chemically induced dynamic nuclear polarization, molten globule
Very significant advances, both theoretical and experimental, have been made in recent years in elucidating the principles that govern the folding of proteins (1, 2). In particular, the concept of an energy landscape has provided a general framework for interpreting the thermodynamics and kinetics of the folding process through which a polypeptide chain converts from a disorganized unfolded state into the tightly packed native state (N state) at the global energy minimum (3, 4). Recent approaches, involving a combination of computer-simulation techniques with experimental constraints, have enabled three-dimensional structural ensembles of various partially folded species and folding intermediates to be defined and provide a coarse-grained description of the development of interresidue interactions and chain topologies involved in attaining the final native structure (5–7). Of particular importance in this context are compact denatured states, often known as molten globules (MGs), which are commonly characterized by the presence of native-like secondary structure and hydrodynamic radii but lack both native-like packing of the internal amino acid side chains and exclusion of solvent molecules from the hydrophobic core of the protein (8, 9). The detailed experimental description of such states will undoubtedly provide important information about the specific factors stabilizing the overall chain topology at a residue-specific side-chain level (10). However, MG states are very difficult to characterize by using conventional x-ray crystallography or NMR techniques because of their heterogeneous character (11, 12).
We have recently developed NMR techniques to characterize aromatic side chains in transient, kinetic intermediate species present in real-time protein folding experiments (13, 14) or in partially folded states that are stable under equilibrium conditions (15, 16). To circumvent the difficulties in studying such states directly, we have shown that magnetization transfer from the partially folded state to the N state by rapidly refolding the protein can, in principle, yield structural information on the former from the well resolved spectrum of the latter (14–16). However, to carry out such experiments successfully, it is essential that refolding takes place faster than nuclear spin-lattice relaxation. To this end, we use two key methodological procedures. The first is an in situ injection device that permits rapid homogeneous mixing of solutions in the NMR magnet within 50 ms to initiate the refolding reaction (14). The second is photochemically induced dynamic nuclear polarization (photo-CIDNP), a technique involving the reaction of laser-induced photoexcited triplet molecules with surface-accessible tryptophan, tyrosine, and histidine residues to provide information on the accessibilities of specific side chains in the partially folded MG state (17, 18). CIDNP is not observed for residues buried deep within a protein molecule, in contrast to residues with significant exposure to the solvent, thereby permitting the environment of the side chains of specific residues in the protein to be defined.
A wide range of experimental studies has been undertaken to gain insight into the MG state of α-lactalbumin (123 residues, Mr = 14,200; see Table 1, which is published as supporting information on the PNAS web site) (8, 9), a species that has recently gained considerable attention through successful clinical trials as a key component of an antitumor compound (19). Most previous studies have focused on the low pH, acidic MG state (A state) (20). However, it is well established that MG states of α-lactalbumin can also be generated at neutral pH by removal of the bound Ca2+ ion (the “apo state”) or the addition of chemical denaturants at moderate concentrations [sometimes termed the “partly denatured” (P) form] (21). In all forms of the MG species, near-UV CD spectra suggest the substantial weakening of native tertiary interactions, whereas far-UV CD spectra show the persistence of large elements of secondary structure (see Fig. 5, which is published as supporting information on the PNAS web site) (8, 9, 21). Various methods including small-angle x-ray scattering indicate that the hydrodynamic radii of the MG states, regardless of how they are generated, are only slightly larger than that of the N state (21, 22). Together with intrinsic fluorescence spectra and the binding properties of the hydrophobic probe 8-anilino-1-naphthalene sulfonate (ANS), these results lead to the conclusion that the MGs produced under different conditions have broadly similar overall properties (8, 9).
In this article, we describe pulse-labeled photo-CIDNP studies designed to examine the characteristics of the different MG states of bovine and human α-lactalbumin at the level of individual amino acid side chains. The side chains detected by CIDNP (tryptophan, tyrosine, and, less importantly, histidine) are fairly evenly distributed in the α-lactalbumin sequence, allowing the polypeptide chain to be probed throughout its length. Moreover, many of the tryptophan and tyrosine residues are buried in the hydrophobic core of the native proteins. Analysis of the solvent accessibilities of these side chains in the various MG states allows one to compare their respective environments within the ensemble of conformations with those in the native proteins.
Materials and Methods
Proteins and Reagents. Human α-lactalbumin (HLA) and bovine α-lactalbumin (BLA) were purchased from commercial sources, and their various MG states were prepared as described (21). Details of the protein sample preparation and the procedure for rapid refolding are given in Supporting Text, which is published as supporting information on the PNAS web site.
CIDNP Pulse Labeling. A recently developed rapid-mixing device was used to perform all experiments (14). Briefly, 50 μl of a 1.5-mM solution of α-lactalbumin in its MG state containing 0.15 mM flavin mononucleotide was polarized by using an Ar+ laser (Stabilite 2016-05, 5-W multiline output principally at 488 and 514 nm; Spectra-Physics) for a specified time period (50–500 ms), then pneumatically injected into 280 μl of refolding buffer contained in the NMR sample tube. Complete mixing could be accomplished within 50 ms of injection (14). A free induction decay was acquired after a specified postinjection delay (normally set at 100, 150, or 200 ms) during which time the MG state refolded to the N state. For steady-state CIDNP spectra, the laser irradiation was at 4 W for 100 ms, and difference spectra were generated by subtraction of “dark” spectra (no laser irradiation) from “light” (with laser irradiation) spectra, each being an average of eight transients.
NMR Spectroscopy. All 1H NMR experiments were performed on a spectrometer operating at 600.1 MHz. Experiments on the A state were performed at 25°C. In contrast to the low-pH conditions under which the A state is formed, at neutral pH in the presence of moderate concentrations of denaturant (e.g., 2.0 M guanidinium chloride) or after the removal of the bound Ca2+ ion, there is an equilibrium partitioning of the population of protein molecules into N, MG, and unfolded states (8, 23). Therefore, care was taken to ensure that the MG state was essentially the only species present before the refolding event (for example, the temperature was increased for the P- and apo-MG pulse-labeling experiments; for details, see Fig. 6, which is published as supporting information on the PNAS web site, and Supporting Text). A solvent-suppression method [CHEmical Shift Selective (CHESS) imaging] (24), designed originally for in vivo NMR, was implemented to counteract the effects of bulk solvent motion after injection on the refocusing of the solute magnetization. Each spectrum was the difference between the sum of eight light and eight dark experiments.
Data Analysis. The side-chain 1H resonance assignments of the N-state spectra of BLA and HLA were used to identify peaks in the pulse-labeled spectra (25). No ambiguities arose from minor deviations of chemical shifts resulting from the presence of residual denaturant or temperature differences. Calculations of side-chain static solvent accessibilities were performed with naccess (26) by using the crystallographic structures of native BLA and HLA (27, 28). Residue numbering was identical in both proteins; residue positions of different amino acids are denoted with a superscript to identify the species (e.g., Trp-26BLA and Leu-26HLA).
Results
The pulse-labeling experiment involves initial in situ generation of CIDNP in the MG state, followed by rapid injection of the polarized protein solution into a buffer solution containing excess Ca2+ ions. A delay of 100–200 ms after injection allows the partially folded state to refold, transferring the polarization enhancements to the N state so that the solvent-accessible side chains of the MG state can be identified from the well resolved spectrum of the N state (16). The experiment permits observation of the exchangeable tryptophan indole protons (expected between 9.5 and 12.0 ppm) as well as the aromatic resonances of tryptophan, tyrosine, and histidine (6.5 and 8.0 ppm), greatly aiding the assignment of the CIDNP spectra. Side chains (particularly of tryptophan residues) that have solvent accessibilities as low as ≈15% can be detected under the present conditions (17, 18).
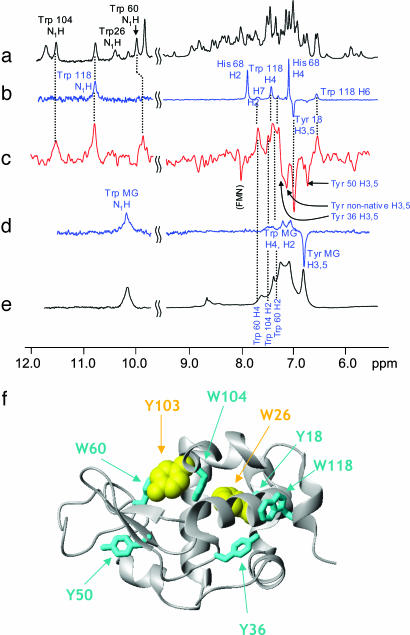
The conventional CIDNP spectrum of the N state of BLA is well resolved, containing side-chain resonances corresponding to one of each of the four tryptophan, four tyrosine, and three histidine residues in the protein (Trp-118, Tyr-18, and His-68; Fig. 1b) (16, 25, 29). In contrast, and consistent with the conventional 1H NMR spectrum (Fig. 1e), the photo-CIDNP spectrum of the A state of BLA (Fig. 1d) exhibits broad lines and poor chemical-shift dispersion because of conformational exchange occurring on the millisecond time scale (16, 29–31). In the A state of BLA, the characteristic emissive signals from the H3,5 protons of the tyrosine residues appear as an intense broad signal at ≈6.8 ppm, whereas the absorptive signals from the aromatic protons of the tryptophan residues (H2, H4, H6, and, at lower intensity, H5 and H7) are seen as broad signals between 7.0 and 7.3 ppm. The overlapping tryptophan NH indole proton resonances appear as a single broad peak at ≈10.2 ppm (Fig. 1 d and e).
Fig. 1.
The 600 MHz 1H NMR and photo-CIDNP spectra of BLA in its N and A states, and the CIDNP pulse-labeled spectrum of the A state at pH 2.0. The 1H NMR (a) and photo-CIDNP spectra (b) of BLA in the N state are shown. CIDNP-active tryptophan, tyrosine, and histidine residues are labeled. (c) Pulse-labeled spectrum recorded 150 ms after initiation of the rapid refolding of the A state to the N state. “Tyr MG” indicates the tyrosine signal from the residual spectrum of the A state. “Tyr nonnative” identifies an emissive tyrosine H3,5 peak whose chemical shift does not correlate with any tyrosine residue in the N state (see text). Photo-CIDNP (d) and 1H NMR (e) spectra of BLA in the A state. (f) Ribbon representation of the three-dimensional structure of BLA (27). The CIDNP-active (cyan) and CIDNP-silent (yellow) residues identified from analysis of the pulse-labeled CIDNP spectra are shown. All three-dimensional structure figures were prepared with the program molmol (version, 2K.1) (56).
The pulse-labeled CIDNP spectrum of the BLA A state (Fig. 1c) is markedly different from the conventional CIDNP spectra of both the initial MG state (Fig. 1d) and the N state (Fig. 1b) (16). Because the half-time of refolding is ≈30 ms (32), and the delay after initiating mixing is 100–200 ms, most of the A-state molecules are expected to be completely refolded at the time the pulse-labeled spectrum is recorded. The most readily assigned peaks are in the range of 10–12 ppm and belong to the exchangeable indole protons of three of the four tryptophan residues (Trp-60, Trp-104, and Trp-118) (Fig. 1c). Confirming these assignments, aromatic proton resonances (H2, H4, and H6) attributable to these three residues are observable in the range of 6.5–8.0 ppm, albeit with some degree of overlap. However, peaks attributable to Trp-26 (e.g., the indole proton at 10.4 ppm) are conspicuously absent from the spectrum, suggesting that this residue is fully shielded from the solvent in the A state. Similarly, characteristic H3,5 emissive peaks for tyrosine residues can observed for Tyr-18, Tyr-36, and Tyr-50, whereas the corresponding resonance of Tyr-103 (at ≈6.5 ppm) is not visible in the spectrum, indicating that the Tyr-103 chain is not solvent accessible in the A state.
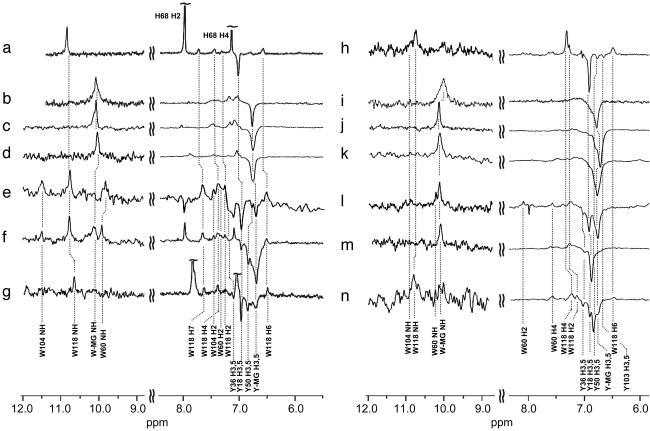
The spectra of the other MG states of BLA and HLA also exhibit characteristically broad peaks and poor chemical-shift dispersion (Fig. 2 b–d and i–k). Pulse-labeled CIDNP spectra recorded under various conditions corresponding to the A, P, and apo states of both proteins are shown in Fig. 2 e–g and l–n. As with the A state of BLA, the spectra contain well resolved resonances enabling side chains that are accessible to solvent in the partially folded states to be identified (for details, see Fig. 3, Supporting Text, and Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
The 600 MHz photo-CIDNP pulse-labeled NMR spectra of the MG states of BLA and HLA. Steady-state photo-CIDNP spectra of the N states of BLA (a) and HLA (h); the steady-state photo-CIDNP spectra of the A state of BLA (b) and HLA (i); the P state (2.0 M guanidinium chloride) of BLA (c) and HLA (j); and the Ca2+-removed (apo-MG) state of BLA (d) and HLA (k) are shown. Below these images, pulse-labeled spectra of the A state of BLA (e) and HLA (l); the P state of BLA (f) and HLA (m); and the apo-MG state of BLA (g) and HLA (n) are shown. Slight differences in chemical shifts result from variations in the solvent and temperature conditions.
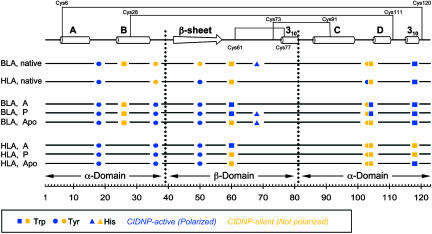
Fig. 3.
Schematic representation of pulse-labeling results for BLA and HLA. The side chains of tryptophan (▪), tyrosine (•), and histidine (▴) are indicated at their positions in the sequence. Blue symbols correspond to CIDNP-active (solvent-accessible) residues, and yellow symbols correspond to CIDNP-silent (solvent-excluded) residues. Secondary-structure elements in the N state (27, 28) and the locations of the disulfide bonds in native α-lactalbumin are shown at the top. The domain boundaries are shown as dotted lines.
Last, an additional emissive peak whose chemical shift (7.1 ppm) does not correspond to the resonances of any tyrosine residue in the N state is observed in the pulse-labeled spectrum of one of the partially folded species, the A state of BLA (Fig. 1c). A signal at this chemical shift has previously been detected in a real-time CIDNP refolding experiment and was attributed to the H3,5 protons of a tyrosine residue in a nonnative environment, suggesting the presence of an intermediate in the folding reaction (31). Its appearance in the pulse-labeled spectrum indicates that the species giving rise to this peak is present before the laser flash; therefore, we suggest that the peak originates from a tyrosine residue in a relatively solvent accessible and significantly nonnative environment in the equilibrium A state. The population of molecules possessing this nonnative tyrosine environment appears to refold more slowly than most of the A-state molecules; if the entire population of MG molecules had refolded to completion within the time after the mixing step, such nonnative peaks would be absent in the pulse-labeled spectrum. Fourier transform infrared (FTIR) studies on the identical A-to-N refolding reaction have also shown the transient existence of nonnative secondary structure (33), supporting the presence of ordered nonnative structures during the folding process. (The presence of nonnative conformers is addressed further below.)
Discussion
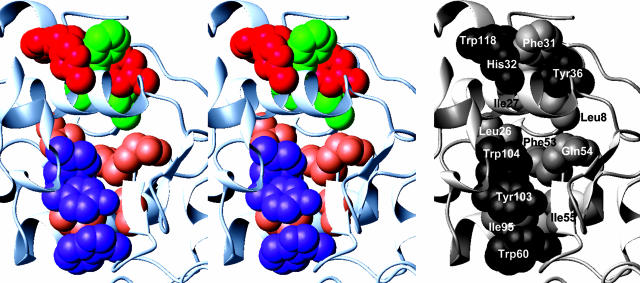
Overview of the Pulse-Labeling Strategy. The structure of native α-lactalbumin consists of an α-domain (residues 1–39 and 81–123), comprising four α-helices and one 310 helix, and a β-domain (residues 40–80), comprising a region of β-sheet structure and another 310 helix (Fig. 3). Two distinct hydrophobic cores are present; one hydrophobic core is formed by residues found within the A and B helices and the C-terminal 310 helix (“aromatic cluster I”), and the other hydrophobic core consists of residues from the C and D helices and part of the β-domain (“aromatic cluster II,” sometimes called the “hydrophobic box”) (Fig. 4) (25, 27, 34). Potentially CIDNP-responsive residues are present in both cluster I (Tyr-36 and Trp-118) and cluster II (Trp-26BLA, Trp-60, Trp-104, and Tyr-103), all of which are largely sequestered from solvent (defined here as <15% accessibility) in the N state. As shown in Fig. 4, these aromatic chains constitute approximately half of all of the residues involved in the packing of the two hydrophobic clusters, demonstrating that tryptophan and tyrosine are well suited to serve as accessibility reporter groups in studying the internal regions of the various MGs. The other three reporter residues (Tyr-18, Tyr-50, and His-68BLA) are situated outside these clusters and are accessible to solvent in the N states.
Fig. 4.
Stereo diagram depicting the side chains of the amino acid residues (in space-filling representation) comprising the two aromatic clusters (25, 27, 34) in HLA (28). (Left) CIDNP-responsive side chains are shown in red (aromatic cluster I) and purple (aromatic cluster II), whereas other hydrophobic residues are shown in green (aromatic cluster I) and coral (aromatic cluster II). (Right) Specific identification of the side chains is shown.
Fig. 1f shows the three-dimensional structure of native BLA on which the CIDNP-active residues are mapped according to their solvent accessibility in the A state. In this state, Trp-26BLA and Tyr-103, both part of cluster II (25, 27, 34), are the only solvent-inaccessible Tyr/Trp side chains; the other six Tyr/Trp residues display clear accessibility to the flavin mononucleotide triplet molecules generated by the laser pulse and, therefore, to solvent. One of the solvent-accessible residues in the A state is Trp-104, whose indole side chain in the native structure is tightly buried and spatially positioned between the two side chains of Trp-26BLA and Tyr-103. Therefore, structure in this region of the protein in the BLA A state must differ significantly from that of the N state. Whereas the side chains of other aromatic residues lining the hydrophobic cluster in the N state (Trp-60, Trp-104, Trp-118, and Tyr-36) demonstrate significant solvent exposure in the A state, it is notable that progressive denaturation experiments monitored by 15N-1H HSQC spectroscopy show that the main chain in the regions of these residues is relatively highly structured (11, 35, 36). Progressive denaturation experiments probe the relative stabilities of the polypeptide main chain in different regions of the protein by determining the concentration of chemical denaturant and/or increase in temperature that is required for them to become unfolded (11).
The results obtained by the pulse labeling of the A state provide a basis on which a wide range of previous studies can be put into context. Side-chain protons of Tyr-103 show significant 1H NMR chemical shift deviations from random coil values in the A state and give rise to nonnative nuclear Overhauser enhancement (NOE) effects, implying that the structure of this region significantly differs from that in the N state (34). In the A state of guinea pig α-lactalbumin, the Trp-26GPLA indole 1H exchange protection factor is one of the highest (≈103) of all values reported for this state, supporting our conclusion that this side chain is involved in a tightly packed structure (37). Long-range hydrophobic interactions involving core residues such as Tyr-103 and Trp-26BLA have been shown to bring the B- and D-helices into close proximity, leading to ready formation of the Cys-28–Cys-111 disulfide bond during oxidative folding and generation of the native fold (38). Similarly, a disulfide-bridged synthetic peptide construct (residues 1–38 and 101–120) containing these two residues displays native-like signatures in far-UV CD and fluorescence spectra, highlighting the importance of the long-range interactions between Trp-26BLA and Tyr-103 in stabilizing native-like structure (39). Concerning the protein backbone, well resolved 15N-1H HSQC peaks for Tyr-103 and Trp-26BLA NH protons are observed at only the highest urea concentrations (10 M) in progressive denaturation experiments (35), indicating that the backbone regions of these residues are highly recalcitrant to complete unfolding. Hence, in the BLA A state, it appears that only a relatively small number of side chains need to be completely sequestered to form a hydrophobic network for stabilization of the fold.
Comparison of BLA and HLA MGs. Despite the strong homology between BLA and HLA (76% sequence identity), the reporter side chains in the C-terminal region of the MG states of HLA are largely sequestered from solvent, whereas these same side chains exhibit significant exposure to solvent in the MG states of BLA (Fig. 3). For example, the solvent accessibilities of Trp-118 are very different in both the A and P states of BLA and HLA (exposed in BLA; nonnatively sequestered in HLA) (Fig. 3), correlating well with the less strongly denaturing conditions required to unfold the C-terminal region of the main chain of BLA than that of HLA in progressive denaturation experiments (35, 40). Another example is Trp-104, located in the CD loop, where the pulse-labeled spectra of the HLA A and P states indicate that it is highly buried, whereas the BLA spectra indicate solvent exposure (Fig. 3). As a result, in the A and P states of HLA, residues belonging to both aromatic cluster I (Trp-118 and to a lesser extent Tyr-36) and aromatic cluster II (Trp-60, Tyr-103, and Trp-104) are substantially excluded from solvent. By contrast, only residues belonging to aromatic cluster II (including Trp-26BLA and Tyr-103) are solvent-inaccessible in BLA. The relative free energy of unfolding of the MG state (ΔGMG→U) of HLA is significantly greater than that of BLA (23, 35), and therefore, it appears that the differences in stability between the MG states of the two proteins can be attributed, at least in part, to the greater burial of the hydrophobic core of the protein in HLA compared to BLA. These conclusions agree well with and support the α-domain level stability differences between BLA and HLA observed by using peptide constructs (41).
In contrast to the A and P states, the side-chain solvent accessibilities of the Ca2+-depleted, apo-MG states formed at neutral pH for BLA and HLA are similar (Fig. 3). Indeed, except for the sequence substitutions in the two proteins (Trp-26BLA/Leu-26HLA and His-68BLA/Gln-68HLA), the profile of accessible side chains is found to be identical (Fig. 3). In addition to Tyr-36, the exposure of the Trp-118 side chain in both apo-MG species, as found for the A and P states of BLA but not HLA (Fig. 3), suggests that aromatic cluster I is not fully formed in apo-HLA. Therefore, we speculate that both apo-MG states have closely similar hydrophobic cores centered around aromatic cluster II and, if so, may exhibit similar thermodynamic stabilities in contrast to the differences in ΔGMG→U found in the A states.
The Occurrence of Nonnative Interactions. In the N states of HLA and BLA, the backbone amide protons of residues in the D-helix and the C-terminal 310 helix exchange too rapidly to be measured, precluding the use of hydrogen exchange methods to probe this region (11). However, the CIDNP results reveal a high degree of solvent exclusion for the side chains of Tyr-103, Trp-104, and Trp-118 in the A state of HLA, suggesting the formation of stable hydrophobic clusters in this region, a finding that confirms previous suggestions using single proline-substituted mutants (42). Another example is Tyr-36, which, by virtue of being situated in the interdomain loop between the α-domain B-helix and β-domain β-sheet, lacks backbone hydrogen-exchange information. Although excluded from solvent and contributing to aromatic cluster I in the N states of both HLA and BLA (Fig. 4), Tyr-36 is accessible to solvent in all MG states of HLA and BLA (Fig. 3), indicating that side chains involved in the stabilization of the N state may exist in nonnative environments in the MG states.
Other examples of differences between the partially folded states and the N states include the side chain of Tyr-103, which is inaccessible in all MG states of HLA, but accessible in the N state (Fig. 3). Likewise, in agreement with previous 19F NMR and fluorescence experiments (43, 44), Trp-118 is sequestered from solvent in the A and P states of HLA but accessible in the N state, indicating that the partially folded states are not simply associated with the loss of native interactions. Therefore, in addition to the nonnative tyrosine peak observed for the BLA A state, it appears that, in contrast to the situation for the main chain, there are significant nonnative environments for the side chains of aromatic residues of both BLA and HLA in their MG states.
Nonnative interactions have been well documented in the unfolded state of hen lysozyme, in which natively accessible tryptophan residues were observed to be inaccessible in the early stages of kinetic refolding experiments (13). Backbone relaxation measurements on the unfolded state have also shown that Trp-123 is involved in hydrophobic clusters containing long-range, nonnative interactions that stabilize a native-like core (45). The sequence homology of the c-type lysozymes with the α-lactalbumins (35–40% sequence identity) suggests that similar nonnative hydrophobic clusters involving aromatic residues may be involved in stabilizing a native-like fold in the partially folded forms of HLA and BLA as well. Other similar examples of nonnative interactions have been found for various proteins, indicating that they could be a common feature of partially folded proteins (12, 46–48). One interesting example is the unfolded state of the N-terminal SH3 domain of the Drosophila drk protein, where a natively accessible indole side chain of a tryptophan residue was found buried within a nonnative cluster of various aliphatic and aromatic residues (47). One explanation for this type of phenomenon is that the efficient burial of hydrophobic residues during folding is a mechanism for increasing the stability of the compact denatured species and minimizing the potential for aggregation (2).
Comparisons Between Different MGs of the Same Species. As well as the species-dependence (BLA vs. HLA) of the solvent accessibility profiles, differences are found in the various MG states of the same protein (Fig. 3). In all cases, a variable subset of a small number of side chains appears to remain highly solvent-inaccessible within the core of the MG state, presumably stabilizing the overall fold. Mutagenesis studies on the A state of HLA to discern which residues are most important in generating the native topology have indicated that a relatively small number of residues is sufficient for stabilizing the fold (49). And, as determined previously with a stable HLA variant containing a multiple leucine-substituted α-domain (50), the hydrophobic residues involved in the stabilization of the fold need not be the native ones. The experimental findings described here demonstrate that, at the level of individual side chains and depending on the conditions imposed, a general native-like topology can be stabilized by exploiting a variety of different interactions that include multiple choices in the sequestration of core residues.
Distinct Local Energy Minima on Free-Energy Landscapes. The present finding of heterogeneity in partially folded states supports conclusions from the analysis of structural ensembles generated from computer simulated MG states of HLA (6, 51). In these structures, significant nonnative contacts are found in addition to native-like interactions and native-like secondary structure. Moreover, in the ensemble of structures obtained from Monte Carlo simulations constrained by data from progressive denaturation NMR experiments, key structural cores were found to persist and define native-like topologies even in the absence of a significant number of native-like interactions. Of particular interest is the identification of Leu-26HLA (Trp-26BLA), Tyr-104, and Trp-60 as residues that are involved in long-range contacts within these structural cores (6). In the present study, some or all of the side chains of these four residues have been found to be solvent-inaccessible in the MG states of HLA and BLA.
The simulated free-energy landscape constructed for the A state of HLA contains deep valleys representing ensembles of partially folded conformers surrounded by large energy barriers, suggesting that the MG state is relatively robust against changes in external conditions (6). Spanning different species (BLA vs. HLA) and different conditions (A, P, and apo-MG states), the partially folded forms described here may represent multiple but distinct local minima within these deep valleys, as predicted for the “rough” energy surfaces observed in folding simulations (4). Although clearly distinguishable by using pulse-labeling CIDNP NMR, the similarity of the energy of these minima levels may explain the apparent lack of hydrodynamic and UV/fluorescence/CD spectral differentiation between the various HLA or BLA MGs prepared under different conditions or observed during stopped-flow kinetic studies (8, 9, 32). Moreover, depending on which particular side chains are solvent exposed in a given local minimum, there may be alternative folding, misfolding, and aggregation steps associated with conformational transitions between different regions of the folding energy landscape (2, 52). Particularly interesting examples include the conversion of the apo-MG state of HLA in the presence of oleic acid into a partially folded variant that stimulates apoptosis in tumor cells (19, 53) and the formation of amyloid fibrils from the A state of BLA in the presence of a high concentration of salt (54).
In summary, pulse-labeled photo-CIDNP studies of the various equilibrium partially folded states of α-lactalbumins have enabled us to differentiate patterns of hydrophobic-core surface accessibilities. We propose that these differences reflect different local minima on a generalized free-energy landscape and that multiple subsets of buried residues may be able to encode a particular type of fold. The fact that the amino acid sequence dictates a unique, native three-dimensional structure has long been part of the dogma of protein science. Experimental data, together with computer simulations, are increasingly demonstrating that the complex process of attaining a unique structure may involve degenerate routes involving both native and nonnative interactions. Indeed, the hydrophobic core ensemble of even the native states of proteins can encompass some variations in side-chain conformations (55). The elucidation of such conformational variability will undoubtedly contribute significantly to our understanding of the manner in which the sequence of a protein encodes a highly specific and unique fold, giving rise to distinctive biological function.
Supplementary Material
Acknowledgments
We thank Kiminori Maeda, Jakob J. Lopez, Akihito Matsuyama, and Kevin Hankyu Kim for their assistance during various stages of this project. We especially thank Christina Redfield, Lorna Smith, Valentina E. Bychkova, and Michele Vendruscolo for their advice and comments on the manuscript. This work was supported by a U.K. Foreign and Commonwealth Office Chevening Scholarship (to K.H.M.), the Oxford Centre for Molecular Sciences (K.H.M.), the Biotechnology and Biological Sciences Research Council (K.H.M. and P.J.H.), the European Union Research and Technological Development Project (T.N. and P.J.H.), INTAS [The International Association for the Promotion of Cooperation with Scientists from the New Independent States (NIS) of the Former Soviet Union] Project 02-2126 (to P.J.H.), the Royal Society (P.J.H.), and program grants from the Wellcome Trust and the Lever-hulme Trust (to C.M.D.). The Oxford Centre for Molecular Sciences is supported by the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council.
Author contributions: K.H.M., T.N., I.J.D., P.J.H., and C.M.D. designed research; K.H.M., T.N., and I.J.D. performed research; K.H.M., T.N., I.J.D., P.J.H., and C.M.D. contributed new reagents/analytic tools; K.H.M., T.N., I.J.D., P.J.H., and C.M.D. analyzed data; and K.H.M., P.J.H., and C.M.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CIDNP, chemically induced dynamic nuclear polarization; MG, molten globule; N state, native state; A state, acidic MG state; HLA, human α-lactalbumin; BLA, bovine α-lactalbumin; P, partly denatured.
References
- 1.Fersht, A. R. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Freeman, New York).
- 2.Dobson, C. M. (2003) Nature 426, 884–890. [DOI] [PubMed] [Google Scholar]
- 3.Bryngelson, J. D., Onuchic, J. N., Socci, N. D. & Wolynes, P. G. (1995) Proteins Struct. Funct. Genet. 21, 167–195. [DOI] [PubMed] [Google Scholar]
- 4.Dobson, C. M., Sali, A. & Karplus, M. (1998) Angew. Chem. Int. Ed. 37, 868–893. [DOI] [PubMed] [Google Scholar]
- 5.Vendruscolo, M., Paci, E., Dobson, C. M. & Karplus, M. (2001) Nature 409, 641–645. [DOI] [PubMed] [Google Scholar]
- 6.Vendruscolo, M., Paci, E., Karplus, M. & Dobson, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 14817–14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindorff-Larsen, K., Vendruscolo, M., Paci, E. & Dobson, C. M. (2004) Nat. Struct. Mol. Biol. 11, 443–449. [DOI] [PubMed] [Google Scholar]
- 8.Ptitsyn, O. B. (1995) Adv. Protein Chem. 47, 83–229. [DOI] [PubMed] [Google Scholar]
- 9.Arai, M. & Kuwajima, K. (2000) Adv. Protein Chem. 53, 209–282. [DOI] [PubMed] [Google Scholar]
- 10.Frieden, C. (2003) Biochemistry 42, 12439–12446. [DOI] [PubMed] [Google Scholar]
- 11.Redfield, C. (2004) Methods 34, 121–132. [DOI] [PubMed] [Google Scholar]
- 12.Dyson, H. J. & Wright, P. E. (2004) Chem. Rev. 104, 3607–3622. [DOI] [PubMed] [Google Scholar]
- 13.Hore, P. J., Winder, S. L., Roberts, C. H. & Dobson, C. M. (1997) J. Am. Chem. Soc. 119, 5049–5050. [Google Scholar]
- 14.Mok, K. H., Nagashima, T., Day, I. J., Jones, J. A., Jones, C. J. V., Dobson, C. M. & Hore, P. J. (2003) J. Am. Chem. Soc. 125, 12484–12492. [DOI] [PubMed] [Google Scholar]
- 15.Balbach, J., Forge, V., Lau, W. S., Jones, J. A., van Nuland, N. A. J. & Dobson, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 7182–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyon, C. E., Suh, E.-S., Dobson, C. M. & Hore, P. J. (2002) J. Am. Chem. Soc. 124, 13018–13024. [DOI] [PubMed] [Google Scholar]
- 17.Hore, P. J. & Broadhurst, R. W. (1993) Prog. NMR Spectrosc. 25, 345–402. [Google Scholar]
- 18.Mok, K. H. & Hore, P. J. (2004) Methods 34, 75–87. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson, L., Leijonhufvud, I., Aronsson, A., Mossberg, A.-K. & Svanborg, C. (2004) N. Engl. J. Med. 350, 2663–2672. [DOI] [PubMed] [Google Scholar]
- 20.Kuwajima, K., Nitta, K., Yoneyama, M. & Sugai, S. (1976) J. Mol. Biol. 106, 359–373. [DOI] [PubMed] [Google Scholar]
- 21.Dolgikh, D. A., Gilmanshin, R. I., Brazhnikov, E. V., Bychkova, V. E., Semisotnov, G. V., Venyaminov, S. Y. & Ptitsyn, O. B. (1981) FEBS Lett. 136, 311–315. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka, M., Kuwajima, K., Tokunaga, F. & Goto, Y. (1997) Protein Sci. 6, 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri, T. K., Arai, M., Terada, T. P., Ikura, T. & Kuwajima, K. (2000) Biochemistry 39, 15643–15651. [DOI] [PubMed] [Google Scholar]
- 24.Haase, A., Frahm, J., Hänicke, W. & Matthaei, D. (1985) Phys. Med. Biol. 30, 341–344. [DOI] [PubMed] [Google Scholar]
- 25.Alexandrescu, A. T., Broadhurst, R. W., Wormald, C., Chyan, C.-L., Baum, J. & Dobson, C. M. (1992) Eur. J. Biochem. 210, 699–709. [DOI] [PubMed] [Google Scholar]
- 26.Hubbard, S. J., Campbell, S. F. & Thornton, J. M. (1991) J. Mol. Biol. 220, 507–530. [DOI] [PubMed] [Google Scholar]
- 27.Chrysina, E. D., Brew, K. & Acharya, K. R. (2000) J. Biol. Chem. 275, 37021–37029. [DOI] [PubMed] [Google Scholar]
- 28.Ren, J., Stuart, D. I. & Acharya, K. R. (1993) J. Biol. Chem. 268, 19292–19298. [PubMed] [Google Scholar]
- 29.Maeda, K., Lyon, C. E., Lopez, J. J., Cemazar, M., Dobson, C. M. & Hore, P. J. (2000) J. Biomol. NMR 16, 235–244. [DOI] [PubMed] [Google Scholar]
- 30.Improta, S., Molinari, H., Pastore, A., Consonni, R. & Zetta, L. (1995) Eur. J. Biochem. 227, 87–96. [DOI] [PubMed] [Google Scholar]
- 31.Wirmer, J., Kühn, T. & Schwalbe, H. (2001) Angew. Chem. Int. Ed. 40, 4248–4251. [DOI] [PubMed] [Google Scholar]
- 32.Ikeguchi, M., Kuwajima, K., Mitani, M. & Sugai, S. (1986) Biochemistry 25, 6965–6972. [DOI] [PubMed] [Google Scholar]
- 33.Troullier, A., Reinstädler, D., Dupont, Y., Naumann, D. & Forge, V. (2000) Nat. Struct. Biol. 7, 78–86. [DOI] [PubMed] [Google Scholar]
- 34.Alexandrescu, A. T., Evans, P. A., Pitkeathly, M., Baum, J. & Dobson, C. M. (1993) Biochemistry 32, 1707–1718. [DOI] [PubMed] [Google Scholar]
- 35.Wijesinha-Bettoni, R., Dobson, C. M. & Redfield, C. (2001) J. Mol. Biol. 312, 261–273. [DOI] [PubMed] [Google Scholar]
- 36.Schulman, B. A., Kim, P. S., Dobson, C. M. & Redfield, C. (1997) Nat. Struct. Biol. 4, 630–634. [DOI] [PubMed] [Google Scholar]
- 37.Chyan, C.-L., Wormald, C., Dobson, C. M., Evans, P. A. & Baum, J. (1993) Biochemistry 32, 5681–5691. [DOI] [PubMed] [Google Scholar]
- 38.Peng, Z.-y., Wu, L. C. & Kim, P. S. (1995) Biochemistry 34, 3248–3252. [DOI] [PubMed] [Google Scholar]
- 39.Demarest, S. J., Horng, J.-C. & Raleigh, D. P. (2001) Proteins Struct. Funct. Genet. 42, 237–242. [PubMed] [Google Scholar]
- 40.Redfield, C., Schulman, B. A., Milhollen, M. A., Kim, P. S. & Dobson, C. M. (1999) Nat. Struct. Biol. 6, 948–952. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury, F. A. & Raleigh, D. P. (2005) Protein Sci. 14, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quezada, C. M., Schulman, B. A., Froggatt, J. J., Dobson, C. M. & Redfield, C. (2004) J. Mol. Biol. 338, 149–158. [DOI] [PubMed] [Google Scholar]
- 43.Bai, P., Luo, L. & Peng, Z.-y. (2000) Biochemistry 39, 372–380. [DOI] [PubMed] [Google Scholar]
- 44.Chakraborty, S., Ittah, V., Bai, P., Luo, L., Haas, E. & Peng, Z.-y. (2001) Biochemistry 40, 7228–7238. [DOI] [PubMed] [Google Scholar]
- 45.Klein-Seetharaman, J., Oikawa, M., Grimshaw, S. B., Wirmer, J., Duchardt, E., Ueda, T., Imoto, T., Smith, L. J., Dobson, C. M. & Schwalbe, H. (2002) Science 295, 1719–1722. [DOI] [PubMed] [Google Scholar]
- 46.Mok, Y.-K., Alonso, L. G., Lima, L. M. T. R., Bycroft, M. & de Prat-Gay, G. (2000) Protein Sci. 9, 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crowhurst, K. A. & Forman-Kay, J. D. (2003) Biochemistry 42, 8687–8695. [DOI] [PubMed] [Google Scholar]
- 48.Cho, J.-H., Sato, S. & Raleigh, D. P. (2004) J. Mol. Biol. 338, 827–837. [DOI] [PubMed] [Google Scholar]
- 49.Song, J., Bai, P., Luo, L. & Peng, Z.-y. (1998) J. Mol. Biol. 280, 167–174. [DOI] [PubMed] [Google Scholar]
- 50.Wu, L. C. & Kim, P. S. (1997) Proc. Natl. Acad. Sci. USA 94, 14314–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paci, E., Smith, L. J., Dobson, C. M. & Karplus, M. (2001) J. Mol. Biol. 306, 329–347. [DOI] [PubMed] [Google Scholar]
- 52.Chiti, F., Taddei, N., Baroni, F., Capanni, C., Stefani, M., Ramponi, G. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 137–143. [DOI] [PubMed] [Google Scholar]
- 53.Svensson, M., Hakansson, A., Mossberg, A.-K., Linse, S. & Svanborg, C. (2000) Proc. Natl. Acad. Sci. USA 97, 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goers, J., Permyakov, S. E., Permyakov, E. A., Uversky, V. N. & Fink, A. L. (2002) Biochemistry 41, 12456–12551. [DOI] [PubMed] [Google Scholar]
- 55.Lindorff-Larsen, K., Best, R. B., DePristo, M. A., Dobson, C. M. & Vendruscolo, M. (2005) Nature 433, 128–132. [DOI] [PubMed] [Google Scholar]
- 56.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.