Abstract
The ability to discriminate between deleterious and neutral amino acid substitutions in the genes of patients remains a significant challenge in human genetics. The increasing availability of genomic sequence data from multiple vertebrate species allows inclusion of sequence conservation and physicochemical properties of residues to be used for functional prediction. In this study, the RET receptor tyrosine kinase serves as a model disease gene in which a broad spectrum (≥116) of disease-associated mutations has been identified among patients with Hirschsprung disease and multiple endocrine neoplasia type 2. We report the alignment of the human RET protein sequence with the orthologous sequences of 12 non-human vertebrates (eight mammalian, one avian, and three teleost species), their comparative analysis, the evolutionary topology of the RET protein, and predicted tolerance for all published missense mutations. We show that, although evolutionary conservation alone provides significant information to predict the effect of a RET mutation, a model that combines comparative sequence data with analysis of physiochemical properties in a quantitative framework provides far greater accuracy. Although the ability to discern the impact of a mutation is imperfect, our analyses permit substantial discrimination between predicted functional classes of RET mutations and disease severity even for a multigenic disease such as Hirschsprung disease.
Keywords: deleterious substitutions, evolutionary conservation, physicochemical properties
The capacity to predict the functional consequence of any sequence variation in a given coding (or noncoding) sequence represents the Holy Grail in genetics. There is clearly a need to more rapidly connect DNA sequence variants to phenotype (disease). The availability and extensive analysis of mRNAs and cDNAs have resulted in an established vocabulary for coding sequences, permitting their identification from sequence alone with great accuracy. However, prediction of the deleterious or neutral nature of variants often requires knowledge of protein function and the chemical nature of its amino acid constituents. These parameters can now be estimated through analysis of evolutionary sequence conservation and the physicochemical properties of proteins. Several algorithms are available to elucidate the nature of substitutions, taking advantage of predictions from the neutral theory of evolution (1, 2), the conservation of key residues among protein family members (3), and the chemical nature of individual amino acid residues (4, 5). Although these approaches have increased our confidence in identifying deleterious substitutions, such predictions are not facile and are often fraught with problems resulting from our limited understanding of the underlying biology. It is also abundantly clear that not all sequences, and consequently not all variants, are created equal with respect to their conservation, role in the activity of the protein, tolerance for change, and/or potential impact on the phenotype.
The recent availability of a completed genome sequence of multiple vertebrate species is revolutionizing how we search for, and discriminate between, deleterious and neutral nucleotide and amino acid substitutions. Systematic comparison of genomic sequences from different organisms now represents a central focus of contemporary genome analysis. However, the development and examination of relevant tools require the identification of model genes in which a spectrum of mutations has been independently identified in a disease process.
One such gene encodes the RET receptor tyrosine kinase (TK) (6, 7). Mutations in RET play a central role in two common heritable disorders: Hirschsprung disease [(HSCR), On-Line Mendelian Inheritance in Man (OMIM) no. 142623], a multigenic developmental defect resulting in aganglionosis and functional intestinal obstruction in neonates, and multiple endocrine neoplasia type 2 (MEN2, OMIM no. 171400), a dominantly inherited cancer predisposition. More than 116 mutations in RET have been reported in HSCR and MEN2 patients (6). RET encodes a receptor TK with a signal peptide, a cysteine-rich domain (CYS), a transmembrane region, a conserved intracellular TK catalytic domain, a calcium-binding domain (Ca2+), and four extracellular cadherin-like ligand-binding domains 1–4 (CLD1–4).
RET mutations have been divided into four subclasses (8–10) based on detailed functional analyses of ≈20 mutations. Class I mutations lie within CLD1–4 and are proposed to interfere with RET maturation and its translocation to the plasma membrane. Class II mutations lie within the CYS and are shared with MEN2A/familial medullary thyroid carcinoma, replacing Cys-609, -611, -618, or -620 with another residue, and result in constitutive activation of the receptor and reduction in the number of mature receptors at the cell surface (11). Class III mutations occur within the TK domain reducing the catalytic activity of the receptor (8). Class IV mutations are found in the region around RETY1062 and compromise the efficiency with which RET binds to its effector molecules (6). Importantly, functional validation of these mutational classes is incomplete, and most RET mutations have not been functionally tested. Thus, it is unclear which substitutions are truly deleterious and which are neutral. This is a general problem in human genetics where most mutations cannot be functionally tested.
The majority of RET mutations have been identified in association with HSCR. Consequently, discrimination between neutral and deleterious RET substitutions may be complicated by contributions of mutations at additional loci (12–14). Similarly, common noncoding RET susceptibility variants may result in the identification of a rare coding sequence polymorphism as a causative mutation (7, 15). We propose that the publicly reported RET missense mutations (n ≥ 96) represent an ideal substrate with which to examine the power of methods that combine comparative sequence analysis and physicochemical properties to predict deleterious amino acid substitutions.
We have predicted orthologous RET protein sequences from the genomic sequences of 12 non-human vertebrate species, including two primates, two artiodactyls, two carnivores, two rodents, one bird, and three teleost species, successfully aligning all but one teleost (Tetraodon) to the human reference. We set out to use these data to test three hypotheses. First, conservation at the RET protein sequence level is a reliable predictor of functional constraint. Second, functional constraint on RET, the physical distribution of mutations within the protein, and clinical severity are correlated. Finally, we examined whether the severity of missense mutations may be predicted from their evolutionary and physicochemical characteristics.
Methods
Annotation of the Human RET Protein. GenBank accession no. NP_000314 (www.ncbi.nlm.nih.gov) was used for the human RET protein reference. Exon positions were calculated from annotation at the University of California, Santa Cruz, Genome Browser (http://genome.ucsc.edu). Functional domains were identified by using the following prediction algorithms: CLD1–4, Anders et al. (16); transmembrane region, SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html) (17); TK, Pfam (http://pfam.wustl.edu) (18).
Prediction of RET Orthologs and Alignment with Human RET Sequence. The corresponding proteins for each of the 12 non-human species (chimpanzee, baboon, cow, pig, cat, dog, rat, mouse, chicken, zebrafish, Fugu, and Tetraodon) used in this study were obtained as follows: first, the genomic sequence was obtained from all species as described by Emison et al. (15); second, protein predictions were obtained for each DNA sequence, by using the genscan software (http://bioweb.pasteur.fr/seqanal/interfaces/genscan-simple.html) (19) and default parameters; third, results from genscan were compared with the NP_000314 reference sequence by using blast (www.ncbi.nlm.nih.gov) (20) to identify RET protein orthologs; fourth, RET protein orthologs were aligned to the human reference by using clustalw (http://clustalw.genome.ad.jp) (21). The predicted Tetraodon protein sequence could be aligned with no more than 31% (339/1,114) of residues within the human RET reference (NP_000314), predominantly within the TK domain, and was not used in subsequent alignments. Accession numbers corresponding to all sequences used in this study are provided in Table 1, which is published as supporting information on the PNAS web site.
Calculating Relative Rates of RET Protein Evolution. The relative rate of protein evolution was estimated as reported by Simon et al. (5). A maximum likelihood tree was inferred with the program protml (ftp://ftp.ism.ac.jp/pub/ISMLIB/MOLPHY). For every window of 19 amino acids in the alignment, the total length of a likelihood tree inferred by codeml (part of paml, http://abacus.gene.ucl.ac.uk/software/paml) was recorded. Each value was divided by the average value over all windows to obtain a relative rate.
Estimation of Mutational Tolerance. The impact of an amino acid substitution on RET protein function was predicted as described by Stone and Sidow (22). Briefly, we built a phylogeny from the RET protein alignment of orthologs using the program semphy (23), and each sequence i was assigned a tree-based weight wi for use in subsequent calculations. In parallel, we chose six physicochemical property scales to consider, namely hydropathy (24), polarity (25), charge (25), volume (26), and free energy in both α helix and β strand conformations (27). We chose a threshold of P = 0.01 to reject the null hypothesis and declare an amino acid incompatible, that is, substitutions for which P < 0.01 were predicted to be deleterious.
Results
Annotation of the Human RET Protein Sequence. Predicted and known functional domains within the human RET protein sequence (NP_000314) were identified as follows: a signal peptide is predicted within residues 1–27 (http://pfam.wustl.edu); four CLDs are predicted within residues 28–516 (Anders et al., ref. 16), the second of which is also predicted by pfam (http://pfam.wustl.edu); residue 272 is predicted to be a calcium-binding site and is located between the second and third cadherin-like domains; residues 637–659 comprise a predicted transmembrane region (SOSUI); and, residues 724-1009 are predicted to harbor a TK domain (pfam and the Protein Kinase Resource, http://pkr.sdsc.edu). This region contains two common protein kinase motifs (GEGEFGKV, residues 731–738; and HRDLAARN, residues 872–879) separated by a domain of ≈50 amino acids, that is predicted to be solely structural. These predictions are consistent with previous reports (8, 11).
Physical Distribution of RET Mutations. Although 96 missense mutations (Table 2, which is published as supporting information on the PNAS web site), among ≥116 total mutations, have been detected among patients with HSCR and MEN2, there has thus far been no report of phenotype–genotype correlation that satisfies both clinical phenotypes. In large part, this failure is HSCR-specific, because RET mutations are both necessary and sufficient for MEN2. However, HSCR largely results from interaction between mutations in RET and those in yet unknown genes. Furthermore, noncoding mutations at RET are also important in HSCR (7, 14, 15). We hypothesized that a comparative sequence-based approach may have more power to detect correlation between RET mutations and HSCR. Many of the previously reported RET mutations lie in domains of known or predicted function. We hypothesize that disease-associated substitutions would predominate within predicted functional domains. To test this hypothesis, we plotted the distribution of known missense mutations along the RET protein. Although one-third (32/96) of mutant sites lie within the largest functional domain (TK, 286/1,114 residues), RET mutations are found throughout the protein and do not associate exclusively with one or more functional domains (Supporting Text and Fig. 4, which are published as supporting information on the PNAS web site). Interestingly, this observation suggests that some regions of the RET protein are functional, although the basis for such constraint is not yet apparent. We reasoned that comparative sequence analysis can identify other functional units within the RET protein and uncover previously nonobvious correlations.
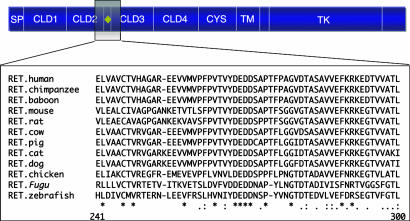
Prediction and Alignment of RET Protein Orthologs. The ORFs of 12 non-human vertebrate RET orthologs were identified and aligned as described in Fig. 1, which illustrates a 60-residue window encompassing residues 237–296 of CLD2–3 of the RET amino acid multisequence alignment. The entire alignment is available in Supporting Text and Fig. 5, which is published as supporting information on the PNAS web site. As expected, maximal identity is observed between the reference (human) and the most recently diverged species, the non-human primates. Similarity decreases as one progresses through the mammalian radiation to greater evolutionary distances and is lowest as one reaches the avian and teleost species. Importantly, because our study was directed at the identification of disease-associated mutations in the human population, we excluded all residues lacking orthologous positions in the human reference from subsequent analyses, including novel exons (data not shown). We successfully aligned 11 of 12 RET orthologs with the human reference (see Methods) and built a phylogeny from their coding sequences using semphy (Friedman et al., ref. 23). We observed 1.3-aa substitutions per position across 100 million years in the alignment, marked conservation consistent with the critical biological function of RET signaling in vertebrate development (McCallion and Chakravarti, ref. 6). Although the average variation among orthologs was low, we hypothesized that the fluctuations in the relative rate of evolution across the length of primary sequence would be sufficient to uncover important features of the protein.
Fig. 1.
Eleven of 12 RET orthologs may be successfully aligned with the human reference RET protein. Schematic representation of RET protein functional domains and example of their amino acid multiple alignment (residues 237–296, inclusive). SP, signal peptide; yellow diamond, Ca2+-binding site; TM, transmembrane domain. The complete multiple alignment is provided in Supporting Text.
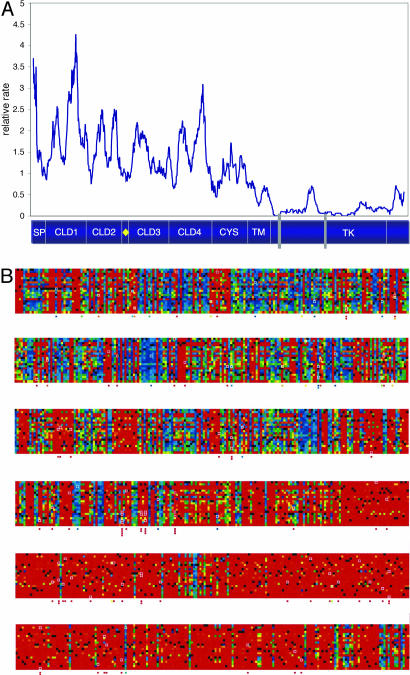
Relative Rate of RET Protein Evolution. To assess whether other functional domains exist, we set out to establish the relative rates of conservation across the protein using the algorithm reported by Simon et al. (5). We posited that critical functional domains evolve more slowly than less functional or purely structural regions. Such regions include, but are not restricted to, the known functional domains of RET. To test this hypothesis, we have placed the known functional domains of the RET protein on the relative rate plot (Fig. 2A). A distinct correlation exists between lower local rates and critical features of RET like the TK, a domain essential for RET signaling activity. Interestingly, the relative rate increases around the predicted structural domain separating TK1 and TK2. Locally reduced rates of evolution are also observed at the CYS/transmembrane domains. Consistent with this observation, CYS is known to be critical for establishment of the secondary structure of the receptor (16). The N terminus half of the protein demonstrates much greater variation in the local rates of evolution, although certain features may also be discerned. Notably, the functionally critical calcium-binding site falls within a region with a low relative rate of evolution (Fig. 2 A). Furthermore, the four CLD domains demonstrate great variability in their relative rates of evolution, suggesting that motifs essential to protein function may be embedded in a less-constrained sequence within these domains. Importantly, although mutational analyses may be significantly enhanced by deep evolutionary sequence comparisons, they may not account for all sources of functional constraint. To thoroughly assess potential correlation between the location of each mutation, functional constraint, and phenotypic impact, we incorporated an additional metric accounting for physicochemical constraint.
Fig. 2.
Comparative sequence analysis facilitates estimation of the relative rate of RET evolution. (A) Plot of the relative rate of RET evolution using the method of Stone and coworkers (5). Functional domains are annotated as in Fig. 1 with the addition of two gray rectangles corresponding to highly conserved protein kinase motifs (GEGEFGKV and HRDLAARN) at residues 731–738 and 872–879, respectively. (B) Heat plot of the RET protein. The RET protein is displayed in rows, each of 186 residues. The vertical axis of each row represents all 20 amino acids (a–y, top to bottom). Tolerance for any change from wild type to another residue is indicated by color (red, orange, yellow, green, and blue). Red, least most likely to be tolerated; blue, most likely to be tolerated; black, wild-type residue; white box, mutant residue (also shown below the corresponding position). B is enlarged in Supporting Text (See Fig. 6, which is published as supporting information on the PNAS web site).
Predicting Mutational Tolerance. We sought to establish a connection between the evolutionary constraints on the RET protein and the physical distribution of its known disease-causing mutations. As a preliminary analysis, we examined the extent to which causative missense mutations were associated with invariant positions in the alignment. To that end, we computed the proportion of mutations in Table 2 affecting an invariant position (60 of 96) and compared that value with the proportion expected under a null hypothesis that all missense variants of human RET are equally likely to be discovered as contributing to disease. This framework adopts the alignment as an internal control; the expected proportion of causative missense mutations affecting invariant positions is simply the fraction of all potential missense mutations in human RET that change invariant positions in the alignment (2,990 of 6,536). By chance, we would expect on average 44 of the 96 variants in Table 2 to affect invariant positions in the alignment; consequently, the observation of 60 such variants of 96 is highly significant (binomial test, P = 0.0007). These calculations simplify the incidence and discovery of RET mutations by considering causative substitutions that affect the same position in the protein (e.g., R475Q and R475W) to be independent. This overestimates the contribution of invariant positions in the alignment whose identity is crucial to protein structure and function, such as the cysteine residues in CYS that, when substituted, result in MEN2A and familial medullary thyroid carcinoma. We can account for this by grouping causative mutations in Table 2 by position and repeating the earlier analysis. Doing so yields 82 affected positions, 48 of which are invariant in the multiple sequence alignment, as compared with the 509 invariant positions of 1,114 overall (P = 0.01). By either calculation, we observe a significant association between causative missense mutations and invariant positions in the alignment; yet, importantly, strict conservation fails to explain 36 of the 96 missense changes in Table 2. To account for these, we turned to a more sophisticated approach that evaluates the physicochemical suitability of amino acid substitutions relative to the spectrum of variation in the alignment.
We examined the evolutionary constraints on RET by applying Multivariate Analysis of Protein Polymorphism (MAPP), a sequence-based method that can predict the phenotypic impact of coding changes in the human protein (22). The pattern of allowed substitutions at each position was analyzed with respect to six physicochemical properties: hydropathy, polarity, charge, volume, and free energy in both α helix and β strand conformations. The culmination of our analysis is one score for each potential amino acid substitution reflecting its potential for compromising protein function (see Methods). Fig. 2B provides a graphical illustration of these values across the RET protein. This figure, termed a “heat plot,” represents every position along the RET protein (horizontal) and every possible amino acid (vertical). These data are consistent with our previous estimates of relative evolutionary rates (Supporting Text and Fig. 7, which is published as supporting information on the PNAS web site).
Each component of the MAPP score reflects the marginal risk of a substitution due to a specific physicochemical property. Consequently, we can quantify the potential violation of side-chain requirements for any mutation. Each disease-associated RET variant was considered to assess how the protein might be affected. RET missense mutations are bordered in white in Fig. 2B and are also displayed below their corresponding position in the heat plot. Disease-associated RET mutations would be expected to score as deleterious under this framework. Consistent with this hypothesis, functionally tested mutations in classes II–IV (n = 18) all scored as highly deleterious by MAPP (P ≤ 10–5). Interestingly, those previously described as weaker alleles (class I) scored as less deleterious than the others; two of six did not reach significance, two of six had P ≤10–3, and only two had P ≤10–5. Examination of all reported deleterious substitutions further suggests that ≥81% (78 of 96) of the reported RET mutations in HSCR/MEN2 patients are likely to be deleterious (Supporting Text). We hypothesized a direct relationship between the magnitude of predicted MAPP scores and the spectrum of clinical presentations associated with RET mutations.
Although attempts at establishing a clear genotype–phenotype correlation for RET have been restricted to MEN2, known variants have been segregated into experimentally defined categories (8). We hypothesized that these classes, labeled I–IV, were informative of disease severity and, to this end, we partitioned all clinical presentations into two groups, mild and severe. Clinically mild cases comprised the more common short-segment form of HSCR (≥87% of HSCR) (6) as an isolated presentation. Severe presentations included long-segment HSCR (≤13% of HSCR), MEN2, and/or the copresentation of HSCR with MEN2. We built a two-way contingency table of disease severity and class membership and tested for dependence between the two categorizations; however, the χ2 statistic [χ2 (3) = 6.51, P = 0.09] was not large enough to claim that significant association exists. The table reveals that the variants in functional classes II and III tended to result in severe presentation, which is consistent with the putative mechanism of protein impairment in those cases (8); by contrast, the presentation of class I variants was less predictive, consistent with a need for mutations at other loci acting in concert with RET in the genesis of disease (13). We reasoned that class membership was predictive of severity only insofar as it reflects a variant's degree of functional impairment and looked to conservation as an alternative. Because the functional importance of an amino acid residue is known to correlate inversely with evolutionary variation, we propose that genotypic information can now be correlated with both functional class and disease severity.
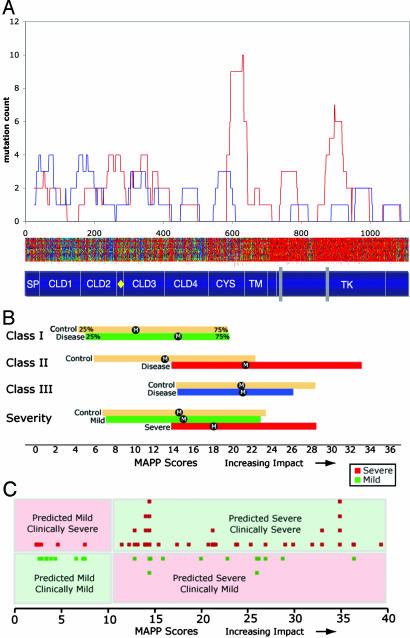
To examine this hypothesis, we used MAPP to quantify the physicochemical constraints in each column of the alignment of RET orthologs; missense variants were then scored relative to the estimated constraints, with higher values indicating greater physicochemical dissimilarity from the observed evolutionary variation in the alignment (Supporting Text). Using this approach, we first asked whether a correlation exists among clinical severity, evolutionary constraint, and physical distribution. We then plotted the frequency distribution of mutations of each class (Fig. 3A, y axis) by their position along the RET protein (x axis) (red, severe; blue, mild). Interestingly, the mutations associated with severe clinical presentations are primarily localized within the functionally critical regions of the RET protein, residues flanking the Ca2+-binding site within the transmembrane domain and the TK domain (Fig. 3A). By contrast, mutations associated with clinically mild presentations are more evenly distributed but are the predominant mutations in areas with higher relative rates. These data show that the patterns of substitution rates across the RET protein are consistent with its known protein landmarks (Ca2+-binding site, CYS and TK domains) but go further to suggest distinct correlations between clinical severity and the physical distribution of mutations.
Fig. 3.
MAPP score estimates identify phenotype:genotype correlations for RET. (A) Distributions of mutations associated with clinically severe (red, long-segment HSCR, MEN2, or copresentation of HSCR and MEN2) and less clinically severe (blue, short-segment HSCR). Also displayed are a schematic representation of RET protein functional domains (Fig. 2A) and a MAPP heat plot (Fig. 2B). (B) Distribution of RET variant MAPP scores corresponding to mutation class (Class, Top) and clinical severity (Severity, Bottom). The interquartile range (25–75%) of scores for each set is shown, with the median denoted by “M.” Interquartile ranges of control distributions are in tan. (C) Prediction of disease severity. Severe (red squares, Upper) and mild (green squares, Lower) variants are plotted against their MAPP scores. Correct predictions (Upper Right and Lower Left) are shown in relief on a green background; incorrect predictions (Upper Left and Lower Right) have a pink background.
To investigate this relationship, we first considered the extent to which the causative mutations in each class violated the physicochemical constraints implied by the multiple sequence alignment. We began by dividing RET variants according to their class as determined by location of the change in the primary sequence (Fig. 3B). We then compared the MAPP scores of variants in each class, totaling 41 from Class I, 19 from Class II, and 32 from Class III (the 4 from Class IV were omitted). The scores of variants from Classes II and III were predominantly large (Fig. 3B), reflecting extensive conservation of the corresponding sequence and physicochemical dissimilarity between wild-type and mutant residues; as expected, both classes of variants scored significantly higher than those in Class I (Wilcoxon test, I vs. II, P = 0.017; I vs. III, P = 0.001). These results were consistent with the initial analysis of severity by class, so we proceeded to estimate the contribution of sequence location to the range of causative RET variants observed. We partitioned the primary sequence of the protein into intervals defined by the four classes of variants and asked whether the scores of variants identified as causative in each interval were atypically large. To address this, we compared the distribution of scores from each class with a null distribution formed from the scores of all variants achievable by a single missense change within the corresponding sequence interval of the human protein (Fig. 3B). As with the simple conservation analysis, this procedure creates an internal control; under the hypothesis that every missense variant within a given class is equally likely to be discovered as associated with disease, statistics of the causative variants within a class can be evaluated against the corresponding null statistic for significance. We were interested in whether the scores of causative variants for any class appeared atypically large and tested the median score of each class against the median of its respective control distribution. The median scores of causative variants in each class were uniformly larger than the medians of their respective controls, although only the difference for Class II was significant (Wilcoxon test; I vs. control, P = 0.51; II vs. control, P = 0.002; III vs. control, P = 0.22). These data suggest that conservation was more informative of disease potential than class membership and reject the assumption that variation within class boundaries is uniformly deleterious or neutral. Consequently, we reasoned that disease severity might also be inferred from positional variation in the RET alignment.
MAPP scores have been shown to correlate with a variant protein's degree of functional impairment, and in several important cases, with the severity of human disease (Stone and Sidow, ref. 22). We compared the scores of clinically mild (short-segment HSCR; see above) and severe (long-segment HSCR and/or MEN2) variants here in an attempt to establish a similar result for RET (Fig. 3B). We first compared the scores of mild variants with a control formed from the scores of all RET variants achievable by a single missense change in the human protein and observed remarkable similarity between the two distributions (Kolmogorov–Smirnov test, P = 0.92). This supports the hypothesis that the typical missense variant is mildly deleterious (28) and stands in contrast to what we observed for clinically severe cases (Kolmogorov–Smirnov test, P = 0.004). We found that severe scores were significantly larger (Wilcoxon test, P = 0.03), indicating that the changes involved in clinically severe disease tend to radically violate positional constraints, but in interpreting the strength of this observation, we considered that the extensive conservation at positions affected in MEN2 had the potential to skew our results. To disentangle the impact of this set of variants, we reconsidered the previous analysis by restricting attention solely to forms of HSCR. Data arising from the comparison of variants segregated by length of aganglionosis recapitulated our previous findings on severity, although they did not reach statistical significance. However, taken together, our data were consistent with the hypothesis that the clinical severity of diseases of RET might be resolved by comparing the scores of their causative substitutions.
To explore this hypothesis further, we used high MAPP scores in a predictive scheme as an indicator for severe clinical presentation. Adopting a simple threshold for classification, we correctly predicted from genotype the severity of two-thirds of the 75 variants for which data were available (Fig. 3C). The misclassification rate (one in three) can be explained, at least in part, by confounding factors. MAPP assigns the identical score to variants whose relevant alignment columns and substituting amino acids are the same; however, for several pairs of RET mutants satisfying those conditions, we found that the clinical severities differed. Importantly, additional mutations, whether at other loci (12, 13) or noncoding variation at RET (7, 14, 15), may explain the cases in which ostensibly the same variant leads to multiple forms of disease in different patients. Importantly, it may be possible to obtain a stronger comparative signal through the use of additional orthologous sequences, contributing variation to the RET family of orthologs. Consequently, the former may be readily resolved through the comparison of additional orthologs. Quantifying the contributions of noncoding mutations at RET and/or mutations at additional loci should help mitigate the latter.
Discussion
Our efforts to establish satisfactory phenotype–genotype correlations for RET with MEN2 and HSCR exemplify the challenge to predict function from primary sequence. This task is made more difficult when analyzing coding variation in genes associated with non-Mendelian inheritance. Importantly, RET mutations are central to the genesis of both MEN2 (autosomal dominant) and HSCR (Mendelian and non-Mendelian forms). However, although RET mutations are both necessary and sufficient for MEN2, they are necessary but not sufficient for HSCR (6, 7). Most HSCR cases result from mutations at multiple loci, most frequently coding mutations in RET acting in concert with second site noncomplementing mutation (7, 12–14) and/or with noncoding allelic variation at RET (15). Importantly, noncoding mutations may exist as common (≥10%) disease susceptibility alleles in the general population. We have recently identified one such noncoding mutation, which lies within an enhancer element in the first intron of RET and underlies HSCR susceptibility in the general population (15).
Consistent with the hypotheses that conservation is a reliable indicator of function, we demonstrate that critical TK and transmembrane domains and the Ca2+-binding site are subject to the greatest evolutionary constraint. Importantly, we see a marked association among the physical distribution of mutations, disease severity, and the relative rate of evolution, uncovering a previously nonobvious indication of relationship between RET genotype and both primary related pathologies. These analyses also permitted discrimination among substitutions identified within the same domain boundaries, suggesting that conservation was more informative regarding the nature of a mutation than the domain. These data prompted us to pursue more quantitative measures of evolutionary constraint. Using MAPP (22), we quantified the physicochemical properties at each position along the RET protein given the available data regarding its evolutionary variation. Importantly, MAPP scores have been shown to be consistent with experimental data and known principles of protein evolution, structure, and function (22). Interestingly, these analyses also emphasize previously undescribed features within known functional domains that may represent regions of greater/lesser functional importance. Likewise, our analyses permitted discrimination among experimentally validated classes of disease-associated RET mutations.
We demonstrate that the distributions of RET mutational classes are predictive of clinical presentation and/or severity and report previously uncharacterized phenotype–genotype correlations for RET satisfying both MEN2 and HSCR. This is an important step in the analysis of coding variation underlying both Mendelian and non-Mendelian disease. Interestingly, the distribution of mutation MAPP scores associated with short-segment HSCR and clinically mild presentations approximate control distributions. This observation has several possible explanations. The corresponding variants may comprise weakly deleterious alleles, indicating a potential role for allelic/nonallelic non-complementation in clinical expression, or they may be truly neutral substitutions associated with disease due to the existence of tightly linked causative mutations. We have already demonstrated that some RET mutations may require interaction with variation at additional loci to explain the genesis of HSCR (12–14). However, noncoding RET mutations also contribute to HSCR susceptibility, even in the context of identified RET coding sequence mutations (7, 15), underscoring the potential impact of genetic background in this type of analysis. Importantly, the accuracy of classification schemes like MAPP will further improve when predictions for major loci can be combined with those for additional susceptibility genes and/or with identified cis-acting variation. Similar improvements will be possible with increasing size of genotype collections and depth of evolutionary coverage. Importantly, disease outcome is not a product of amino acid substitution alone. Consequently, prediction of HSCR recurrence risk remains inappropriate even in families where a RET mutation is known.
Our results establish that comparative sequence analyses, when evaluated in the context of the physicochemical properties of variation, are highly predictive of their impact on protein function. More importantly, disease severity is similarly predictable. This approach may now permit novel genetic mapping strategies, grouping cohorts of families based on novel schemes of mutation classification and refined phenotypes, directed toward the identification of additional HSCR susceptibility genes.
Supplementary Material
Acknowledgments
We thank members of the McCallion and Chakravarti laboratories for helpful discussions on this manuscript. We also acknowledge the participants of the National Information Services Corporation Comparative Sequencing Program. This work was supported by grants from the U.S. National Institute of Child Health and Development.
Author contributions: A.C. and A.S.M. designed research; C.S.K. and E.A.G. performed research; C.S.K., E.A.S., M.E.P., E.D.G., and A.S. contributed new reagents/analytic tools; C.S.K., E.A.S., A.C., and A.S.M. analyzed data; and E.A.S. and A.S.M. wrote the paper.
Abbreviations: HSCR, Hirschsprung disease; CLD, cadherin-like ligand-binding domain; CYS, cysteine-rich domain; TK, tyrosine kinase; MEN2, multiple endocrine neoplasia type 2; MAPP, multivariate analysis of protein polymorphism.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AC130189, AC130190, AC140964, AC125509, AC125512, AC125513, AC124163–AC124166, AC122156, AC145014, AC123972, AC123973, AC114881, AC114884, AC138567, AC124155, AC138550, AC124911, AC125500, AC116947, and AC135546).
References
- 1.Dayhoff, M. O. (1979) Atlas of Protein Sequence and Structure (National Biomedical Research Foundation, Washington, DC), Vol. 5, Suppl. 3.
- 2.Majewski, J. & Ott, J. (2003) Gene 305, 167–173. [DOI] [PubMed] [Google Scholar]
- 3.Ng, P. C. & Henikoff, S. (2003) Nucleic Acids Res. 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grantham, R. (1974) Science 185, 862–864. [DOI] [PubMed] [Google Scholar]
- 5.Simon, A. L., Stone, E. A. & Sidow, A. (2002) Proc. Natl. Acad. Sci. USA 99, 2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCallion, A. S. & Chakravarti, A. (2004) in Inborn Errors of Development, eds. Epstein, C., Erickson, R. & Wynshaw-Boris, A. (Oxford Univ. Press, San Francisco), Vol. 1, pp. 421–432. [Google Scholar]
- 7.McCallion, A. S., Emison, E. S., Kashuk, C. S., Bush, R. T., Kenton, M., Carrasquillo, M. M., Jones, K. W., Kennedy, G. C., Portnoy, M. E., Green, E. D., et al. (2003) Cold Spring Harbor Symp. Quant. Biol. 68, 373–381. [DOI] [PubMed] [Google Scholar]
- 8.Iwashita, T., Kurokawa, K., Qiao, S., Murakami, H., Asai, N., Kawai, K., Hashimoto, M., Watanabe, T., Ichihara, M. & Takahashi, M. (2001) Gastroenterology 121, 24–33. [DOI] [PubMed] [Google Scholar]
- 9.Manié, S., Santoro, M., Fusco, A. & Billaud, M. (2001) Trends Genet. 17, 580–589. [DOI] [PubMed] [Google Scholar]
- 10.Pelet, A., Geneste, O., Edery, P., Pasini, A., Chappuis, S., Atti, T., Munnich, A., Lenoir, G., Lyonnet, S. & Billaud, M. (1998) J. Clin. Invest. 101, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, M., Iwashita, T., Santoro, M., Lyonnet, S., Lenoir, G. M. & Billaud, M. (1999) Hum. Mutat. 13, 331–336. [DOI] [PubMed] [Google Scholar]
- 12.Bolk, S., Pelet, A., Hofstra, R. M., Angrist, M., Salomon, R., Croaker, D., Buys, C. H., Lyonnet, S. & Chakravarti, A. (2000) Proc. Natl. Acad. Sci. USA 97, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel, S. B., Salomon, R., Pelet, A., Angrist, M., Amiel, J., Fornage, M., Attie-Bitach, T., Olson, J. M., Hofstra, R., Buys, C., et al. (2002) Nat. Genet. 31, 89–93. [DOI] [PubMed] [Google Scholar]
- 14.Carrasquillo, M., McCallion, A. S., Puffenberger, E. G., Kashuk, C. S., Nouri, N. & Chakravarti, A. (2002) Nat. Genet. 32, 237–244. [DOI] [PubMed] [Google Scholar]
- 15.Emison, E. S., McCallion, A. S., Kashuk, C. S., Bush, R. T., Grice, E., Lin, S., Portnoy, M. E., Cutler, D. J., Green, E. D. & Chakravarti, A. (2005) Nature 434, 857–863. [DOI] [PubMed] [Google Scholar]
- 16.Anders, J., Kjar, S. & Ibanez, C. F. (2001) J. Biol. Chem. 276, 35808–35817. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa, T., Boon-Chieng, S. & Mitaku, S. (1998) Bioinformatics 14, 378–379. [DOI] [PubMed] [Google Scholar]
- 18.Sonnhammer, E. L., Eddy, S. R. & Durbin, R. (1997) Proteins 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 19.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 20.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stone, E. A. & Sidow, A. (2005) Genome. Res., in press. [DOI] [PMC free article] [PubMed]
- 23.Friedman, N., Ninio, M., Pe'er, I. & Pupko, T. (2002) J. Comput. Biol. 9, 331–353. [DOI] [PubMed] [Google Scholar]
- 24.Kyte, J. & Doolittle, R. F. (1982) J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 25.Stryer, L. (1995) Biochemistry (Freeman, New York), 4th Ed.
- 26.Zamyatin, A. A. (1972) Prog. Biophys. Mol. Biol. 24, 107–123. [DOI] [PubMed] [Google Scholar]
- 27.Munoz, V. & Serrano, L. (1994) Proteins 20, 301–311. [DOI] [PubMed] [Google Scholar]
- 28.Li, W.-H. (1997) Molecular Evolution (Sinauer, Sunderland, MA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.