Abstract
Background
The tumor immune microenvironment (TIME) plays a pivotal role in determining ovarian cancer (OC) prognosis. Long non-coding RNAs (lncRNAs) are key regulators of immune response and tumor progression in OC. Among these, tumor-infiltrating B cells represent an emerging target in immune response pathways. However, the specific involvement of B cell-related lncRNAs (BCRLs) in OC remains unclarified.
Methods
Leveraging single-cell and bulk RNA-sequencing data, correlation analysis identified BCRLs in ovarian serous cystadenocarcinoma (OV) from the TCGA database. Subsequently, BCRLIs were filtered through COX survival analysis and the LASSO algorithm, leading to the development of a B cell-related lncRNA scoring system (BCRLss). The predictive accuracy of BCRLss for prognosis in TCGA-OV was assessed and externally validated in an independent cohort. Functional enrichment analyses were conducted to elucidate biological pathways associated with risk subgroups. Additionally, the relationship between BCRLss and TIME was investigated through multiple algorithms and consensus clustering, uncovering potential immune response targets. Drug sensitivity analyses further identified potential therapeutic options tailored to risk subgroups. The highest risk score lncRNA was selected for in vitro validation.
Results
The BCRLss was constructed using six BCRLIs. Survival analysis revealed an improved prognosis in the low-risk group, with results corroborated by external validation in the ICGC-OV cohort. ROC analysis and nomogram construction confirmed the strong prognostic accuracy of BCRLss. Enrichment analysis highlighted associations between risk subgroups and tumor immune pathways, with the low-risk group demonstrating a more robust immune response and elevated expression of immune checkpoint-related genes. Drug sensitivity tests revealed notable differences across risk subgroups. In vitro experiments confirmed elevated LINC01150 expression in OC cells, and LINC01150 knockdown significantly inhibited the proliferation, invasion, and migration of SKOV3 cells.
Conclusions
In conclusion, BCRLss provides a reliable prognostic tool for predicting clinical outcomes and the immune landscape of patients with OC, offering valuable guidance for immunotherapy target selection and personalized treatment strategies.
Keywords: Tumor immune microenvironment, Immunotherapy, Tumor-infiltrating B cell, LncRNA, Ovarian cancer, Prognosis
1. Introduction
Ovarian cancer (OC) ranks among the most prevalent malignancies in women, with incidence rates surpassed only by cervical and endometrial cancer [1]. OC exhibits diverse pathological subtypes, with epithelial ovarian cancer accounting for 90 %. The remaining 10 % are primarily composed of germ cell tumors, sex cord-stromal tumors—common in individuals under 30—and rarer tumors such as small cell carcinomas, which, despite rapid growth, often have a favorable prognosis [2]. Serous cystadenocarcinoma represents the most frequent epithelial subtype [3], typically diagnosed at advanced stages (III-IV) [4]. While surgery and chemotherapy remain the primary treatment strategies, effective options for recurrent or metastatic OC post-chemotherapy are limited. Although research is actively exploring novel therapies, such as targeting the PI3K/AKT/mTOR pathway, these approaches are still in experimental and clinical trial stages [5,6]. Previous studies have highlighted the significant role of immune-infiltrating cells within the tumor microenvironment (TME) in modulating tumor cell behavior [7]. Immunotherapies and targeted treatments focusing on the tumor immune microenvironment (TIME) hold considerable potential for OC [8,9]. A multicenter study revealed that differences in the TIME landscape are closely linked to overall survival (OS), with long-term survivors showing higher levels of pro-inflammatory immune cell infiltration [10]. This highlights the importance of assessing TIME status in patients with OC to better predict prognosis [11].
The prognostic value of tumor-infiltrating T cells is well established, with immune checkpoint inhibitors (ICIs) driving breakthroughs in antitumor therapy by enhancing T cell activity. However, these studies have also underscored the limitations of T cell responses alone [12,13]. Emerging evidence points to the role of tumor-infiltrating B cells in mediating tumor immunity and immune evasion via antibody secretion and antigen presentation pathways [14,15]. Previous research has linked B cell dysfunction in OC to poor prognosis [16,17]. B cells are also crucial for immune regulation in OC. For example, in OC with omental metastasis, the production of cytokines and chemokines (such as IFNg and CXCL10) activates CD20+ B cells, driving their differentiation, promoting the formation of tertiary lymphoid structures linked to favorable outcomes, and enhancing the anti-tumor immune response [18]. In summary, targeting B cells has the potential to boost OC immunogenicity, representing a promising direction for future research.
Long non-coding RNAs (lncRNAs) are ubiquitously present in the nucleus, cytoplasm, microcirculation, and various body fluids [19]. Notably, due to advancements in detection methods, the number of identified lncRNAs now surpasses the total number of protein-coding genes [20]. These molecules play critical roles in mRNA splicing, stability, transcription, and post-transcriptional modifications [[21], [22], [23]], contributing to processes such as tumor metastasis, invasion, drug resistance, prognosis, and regulation of the TIME [24,25]. Additionally, lncRNAs are involved in B cell growth, activation, epigenetic modification, and immune responses [26,27]. For instance, knocking down lncRNA 2900052N01Rik disrupts the LPS/TLR4 pathway, inhibiting B cell proliferation, activation, and differentiation [28]. In OC, lncRNAs play pivotal roles as well. For example, lncRNA TLR8-AS1 is upregulated in OC and acts as an oncogene, promoting metastasis and chemotherapy resistance through activation of the NF-kB signaling pathway [29]. Research by Hanyuan Liu et al. demonstrated that lncRNA PLADE enhances cisplatin sensitivity in OC by downregulating heterogeneous nuclear ribonucleoprotein D (HNRNPD) via VHL-mediated ubiquitination [30]. Another study revealed that lncRNA SNHG12 facilitates immune escape in OC by modulating IL-6 secretion from M2 macrophages, thereby reducing PD-L1 expression and miR-21 stability [31]. Moreover, prior studies have confirmed that tumor-infiltrating lymphocyte-related lncRNAs and their associated risk signatures can serve as reliable prognostic markers in OC, and are predictive of immune infiltration and activity [32,33]. However, the role of B cell-related lncRNAs (BCRLs) in OC remains unexplored.
This study aimed to identify BCRLs as potential biomarkers for OC. By constructing and validating a B cell-related lncRNA scoring system (BCRLss), this research examined the prognostic features in OC and the landscape of TIME.
All pathological types included in this study were ovarian serous cystadenocarcinoma (OV). Using single-cell RNA sequencing (scRNA-seq) and extensive transcriptome data, BCRLs were identified, and BCRLss was developed to predict the prognosis of patients with OV, with validation in an independent external cohort. Additionally, the relationship between BCRLss and TIME was investigated, providing insights into immune activity and drug sensitivity, which could inform personalized clinical treatments. The oncogenic potential of the high-risk lncRNA LINC01150 in OC was further confirmed through in vitro experiments.
2. Method
2.1. Data capture
The transcriptome and clinical data for constructing the BCRLss in this study were sourced from the OV cohort in The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Patients selected for analysis had pathologic stage III-IV disease and a survival time exceeding 30 days. Somatic mutation data from the TCGA-OV cohort were also incorporated. Transcriptomic data from normal ovarian tissue were retrieved from the Genotype-Tissue Expression (GTEx) database (https://commonfund.nih.gov/GTex), and the external validation cohort was sourced from the International Cancer Genome Consortium (ICGC) database, specifically the OV cohort from Australia (https://dcc.icgc.org/). Immune-related genes were acquired from the IMMPORT database (https://www.immport.org/home) and InnateDB (https://www.innatedb.ca/). Tumor-infiltrating B cell-related genes were sourced from the GSE147082 single-cell OC cohort available through the Tumor Immune Single-cell Hub (TISCH) database (http://tisch.comp-genomics.org/).
2.2. Identifying hub genes in B cell
Differentially expressed genes (DEGs) between the B cell cluster and other cell clusters were extracted from the GSE147082 cohort, using a fold change threshold of 1.5. These DEGs were then intersected with immune-related genes to narrow the selection. The intersecting genes were submitted to the String database (https://cn.string-db.org/) to construct a protein-protein interaction (PPI) network. Gene importance within the PPI network was determined by calculating the degree value (the number of neighboring nodes for each gene). Genes with a degree value ≥ 20 were considered B cell hub genes (BCHGs) in the TCGA-OV cohort for further investigation.
2.3. Identification of B cell-related lncRNA
Next, the expression levels of BCHGs were extracted from the TCGA-OV dataset, distinguishing between mRNAs and lncRNAs within the TCGA-OV transcriptomic matrix. Pearson correlation analysis was then performed to identify BCRLs in TCGA-OV.
2.4. Establishment of BCRLss
These BCRLs were merged with the survival data of TCGA-OV patients. Patients were subsequently divided into training and validation groups in a 1:1 ratio through random allocation. COX regression analysis was employed to identify BCRLs with significant prognostic relevance. Risk lncRNAs and their respective risk coefficients were then mapped using the LASSO algorithm to construct the BCRLss [34]. Based on the median risk score in the training cohort, patients were stratified into high-risk and low-risk groups. The risk score is calculated as:
In this equation, Exp(i) represents the expression of the lncRNA in the sample, Coef(i) represents the risk coefficient of the lncRNA, and Riskscore represents the risk score of the lncRNA used to construct BCRLss in the sample.
2.5. Verification of BCRLss
The clinical predictive performance of BCRLss was validated from several perspectives. Initially, survival analysis was conducted to confirm the prognostic relevance of the identified risk lncRNAs. To examine differential expression, transcriptome data from 88 normal ovarian tissue samples from the GTEx database and TCGA-OV were extracted and converted to TPM format. After applying log2 transformation, expression differences between normal ovarian samples and TCGA-OV were analyzed. The diagnostic ability of these risk lncRNAs was evaluated using receiver operating characteristic (ROC) curves, where a larger Area Under the Curve (AUC) indicates better diagnostic performance.
Principal component analysis (PCA) was then applied to TCGA-OV data to reduce dimensionality and assess patient distribution based on the primary features of all genes, BCHGs, BCRLs, and BCRLss. For enhanced validation accuracy, the ICGC-OV dataset, consisting of 81 patients, was introduced as an external validation cohort. Several visualizations were generated, including distribution heatmaps, survival-related risk curves, risk heatmaps, and Kaplan-Meier (K-M) survival curves for the training cohort, testing cohort, and ICGC-OV cohort. Additionally, the prognostic significance of BCRLss in high- and low-risk groups was explored across various clinical subgroups, such as age, tumor grade, stage, and tumor residual status.
2.6. Prognostic predictive value of BCRLss
To identify independent prognostic factors affecting advanced OC, univariate and multivariate COX regression analyses were performed by integrating clinical pathological parameters and the risk score of TCGA-OV patients. The predictive value of the risk score in TCGA-OV was further evaluated using ROC analysis based on the AUC. Subsequently, a survival-related nomogram was constructed from the COX analysis results. A calibration curve was also constructed to assess the predictive accuracy of the nomogram.
2.7. Enrichment analysis
To further elucidate the biological functions associated with BCRLss, DEGs between high- and low-risk groups were identified, including both upregulated and downregulated genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were then performed to explore the biological pathways involved [35,36]. Additionally, Gene Set Variation Analysis (GSVA), a non-parametric, unsupervised algorithm, was used to investigate the correlation between risk lncRNAs, risk scores, and tumor-related pathways [37]. Gene Set Enrichment Analysis (GSEA) was also employed to identify pathways enriched in the high- and low-risk groups [38].
2.8. Immunocorrelation analysis of BCRLss
Immune cell expression in TCGA-OV was obtained from the TIMER2.0 platform (http://timer.cistrome.org/), followed by an exploration of the correlation between the BCRLss risk score and immune cell populations using various algorithms, including XCELL, TIMER, QUANTISEQ, MCPCOUNTER, EPIC, and CIBERSORT. To assess immune cell infiltration, the single sample gene set enrichment analysis (ssGSEA) was employed for each TCGA-OV sample, comparing immune cell expression and immune function between the BCRLss risk subgroups. Differences in immune checkpoint gene expression between risk groups were also analyzed. The Immunophenoscore (IPS) for CTLA-4 and PD-1 in TCGA-OV was retrieved from the TCIA database (https://tcia.at/home) to predict variations in response to immunotherapy between the BCRLss risk subgroups. Additionally, the ESTIMATE algorithm (Estimation of Stromal and Immune Cells in Malignant Tumor Tissues Using Expression Data) was applied to evaluate differences in the TME between these subgroups [39].
2.9. Consensus clustering analysis
Consensus unsupervised clustering analysis was performed using the R package 'ConsensusClusterPlus' to determine the optimal number of clusters based on the similarity between risk scores and the proportion of fuzzy clustering measurements. The correlation between different clusters and TIME was then examined.
2.10. Analysis of drug sensitivity
The half-maximal inhibitory concentration (IC50) values for 198 drugs in TCGA-OV samples were derived using the R package 'oncoPredict.' A higher IC50 value indicates increased sensitivity to the drug. Differential analysis identified drugs that may be beneficial for patients in the BCRLss risk subgroups, with commonly used OC drugs highlighted.
2.11. Cell culture and transfection
In the experimental validation, OC cell lines SKOV3, A2780, OVCAR-3, and the normal ovarian cell line IOSE80 were utilized. The cells were cultured at 37 °C with 5 % CO2 in a complete DMEM medium (Gibco, China) containing 10 % fetal bovine serum (FBS) and 1 % penicillin-streptomycin. LINC01150 was transfected with an intervention vector according to NanoTrans 40™ (Biomedical, China) instructions. The siLINC01150 sequence is provided in Supplementary Table S1.
2.12. Quantitative real-time PCR
Total RNA was extracted following the Trizol kit protocol (Thermo Fisher Scientific, USA) and purified using the RNeasy Mini Kit (50) kit (Qiagen, Germany). Complementary DNA (cDNA) was synthesized using the PrimeScript™ RT Master Mix kit (Takara Bio, Japan). Quantitative real-time PCR was performed using the 2X SYBR Green Pro Taq HS Premix kit (Accurate Biology, China), with expression levels normalized to GAPDH. Relative expression was assessed using the 2-ΔΔCt method, and the primer sequences for GAPDH and LINC01150 are listed in Supplementary Table S1.
2.13. Cell proliferation assay based on CCK8 and EDU
After 48 h of transfection, cells were seeded at a low density (2 × 104 cells/ml) into 96-well plates at a volume of 100 μl per well. Cell proliferation was measured using a CCK-8 kit (Yeasen, China) at 24-h intervals for five days. At each time point, 100 μl of CCK-8 solution was added to each well, and the cells were incubated at 37 °C for 2 h. Absorbance was measured at 450 nm using a microplate reader. In parallel, cell proliferation was also assessed using the EDU kit (50 μM) (Guangzhou RiboBio, China) following the manufacturer's instructions, with cells incubated in the Click reaction solution and protected from light. After fixation with 4 % paraformaldehyde, nuclei were stained with Hoechst33342. Fluorescent images of proliferating cells were captured, and positive cells were counted under a fluorescence microscope.
2.14. Colony formation assay
Colony formation assays were performed to evaluate the proliferative capacity of treated cells. Cells (600 cells/well) were seeded into 6-well plates and cultured for two weeks in an incubator at 37 °C with 5 % CO2. Cell growth was monitored daily. After two weeks, cells were fixed with 4 % paraformaldehyde for 30 min and stained with crystal violet for 10 min. Colonies were photographed under a microscope and counted.
2.15. Transwell
For migration and invasion assays, transfected and control cells were suspended in a serum-free medium and seeded into the upper chamber of Transwell inserts (Corning, USA) at a density of 6 × 104 cells per well. For the invasion assay, 50 μL of Matrigel was pre-coated in the upper chamber. A total of 600 μL of 10 % FBS-containing medium was added to the lower chamber. After 24 h of incubation at 37 °C, cells were fixed with 4 % paraformaldehyde for 20 min and stained with crystal violet for 10 min. Cell migration and invasion were photographed under an inverted microscope, and the percentage of migrating cells was calculated.
2.16. Statistical analysis
For statistical analysis, Strawberry Perl (version 5.32.1.1), Cytoscape (version 3.8.1), GraphPad Prism (version 8.0), R software (version 4.1.2), and relevant R packages were used. The Wilcoxon test and Kruskal-Wallis test were employed for comparisons between nonparametric and multiple samples, respectively. Pearson correlation coefficient was calculated to evaluate correlations between variables. KM and COX regression analyses were performed for survival and independent prognostic analyses. Unless otherwise noted, all statistical tests were two-sided, and a P-value of <0.05 was considered statistically significant.
3. Results
3.1. Identification of B cell-related lncRNA in TCGA-OV
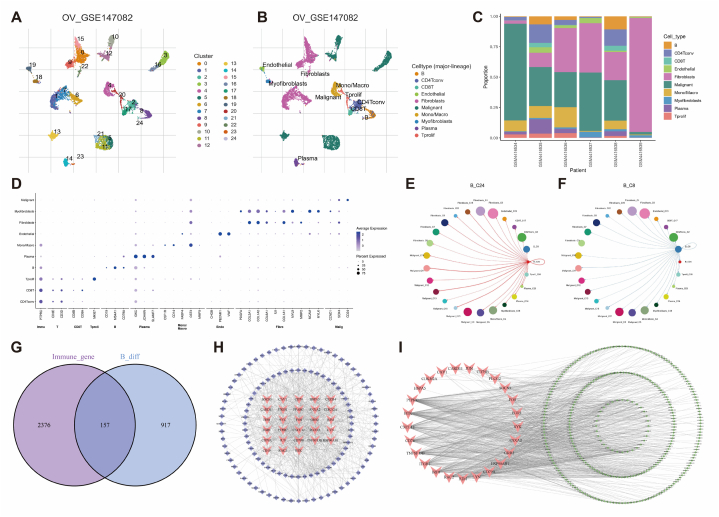
The flowchart (Fig. 1) provides a visual representation of the study's design and methodology. First, the cell cluster types of the GSE147082 cohort were identified using the TISCH database (Fig. 2A and B), and Fig. 2C illustrates the distribution of different cell types within each sample. Fig. 2D shows the correlation between specific biomarkers and single-cell clusters, highlighting the significant heterogeneity among these clusters. Cellular communication analysis indicates a strong interaction between B cells and various cell types in the TIME (Fig. 2E and F). Through differential analysis, 1087 B cell-related DEGs were identified. Intersecting these DEGs with 2533 immune-related genes resulted in 157 overlapping genes (Fig. 2G). The PPI network of these intersecting genes is visualized in Fig. 2H. A total of, 28 BCHGs with a degree ≥20 were selected for further analysis. Pearson Correlation Analysis mapped these 28 BCHGs to 313 BCRLs (Fig. 2I).
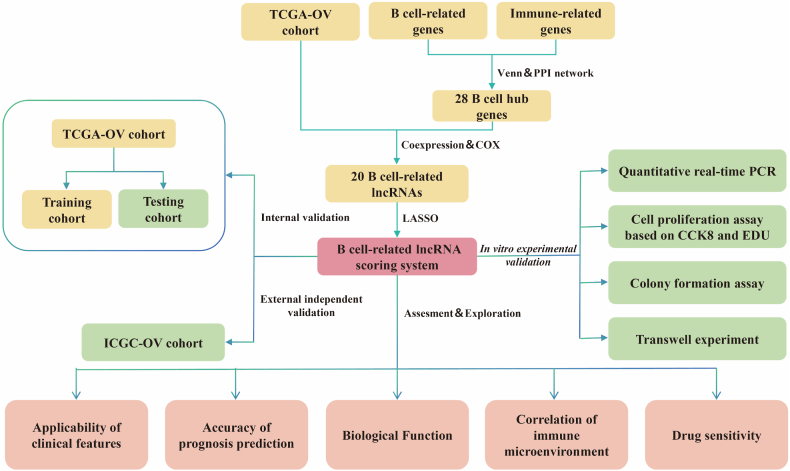
Fig. 1.
Flowchart of the study design.
Fig. 2.
Identification of B cell-related lncRNAs in OV. (A, B) Single-cell sequencing landscape based on UMAP in the GSE147082 cohort. (C) Distribution of different cell clusters in the GSE147082 cohort. (D) Correlation between different cell clusters and their characteristic genes. (E, F) Cell communication between B cells and other cell clusters. (G) Venn diagram showing the intersection of B cell-related and immune-related genes. (H) Protein-protein interaction network of 157 B cell-related genes (genes with degree ≥20 shown in red). (I) Mapping network of 28 B cell hub genes and 313 B cell-related lncRNAs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Construction of BCRLss
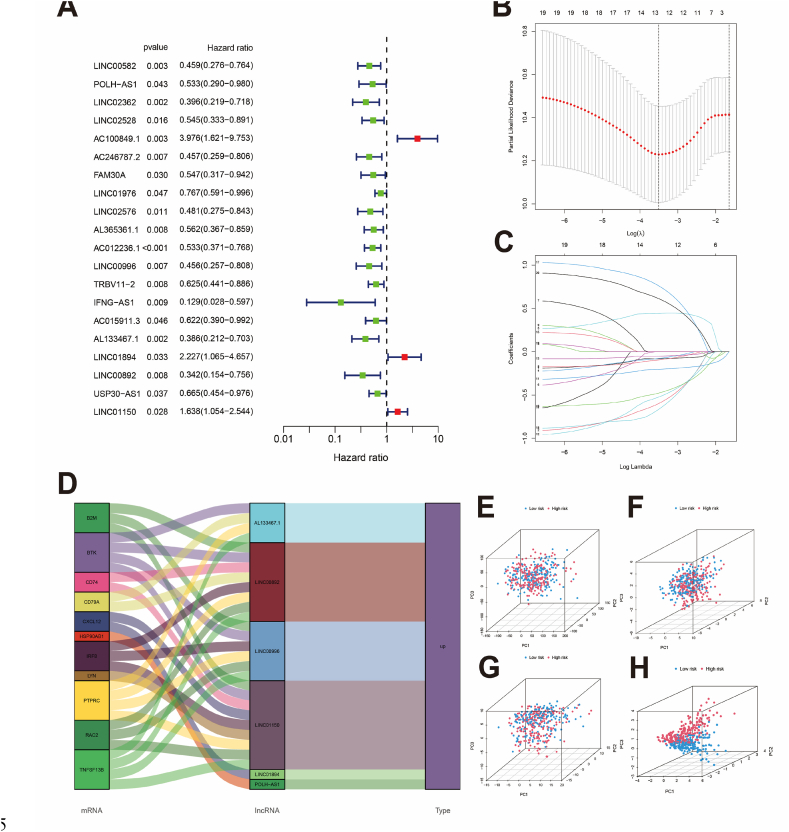
BCRL expression data in TCGA-OV were extracted and merged with survival information. The patients were randomly divided into training and testing cohorts. The training cohort was used to construct the BCRLss, while the testing cohort served as the internal validation set. Statistical analysis indicated that the clinical characteristics of both groups were comparable (Table 1). COX survival analysis identified 20 prognostic BCRLs in the training cohort (Fig. 3A), and 6 risk lncRNAs (POLH-AS1, LINC00996, AL133467.1, LINC01894, LINC00892, and LINC01150) were selected using the LASSO algorithm for BCRLss construction (Fig. 3B and C). The risk lncRNAs and their coefficients are detailed in Table 2. Fig. 3D shows the mapping of BCHGs to risk lncRNAs. Principal component analysis (PCA) demonstrated that BCRLss, as the primary feature, effectively distinguished between patients compared to all genes, BCHGs, and BCRLs (Fig. 3E–H).
Table 1.
Comparison of clinical characteristics between the training and testing cohorts.
| Covariates | Type | Entire cohort | Testing cohort | Training cohort | P-value |
|---|---|---|---|---|---|
| Age | ≤60 | 213 (55.61 %) | 113 (59.16 %) | 100 (52.08 %) | 0.1966 |
| >60 | 170 (44.39 %) | 78 (40.84 %) | 92 (47.92 %) | ||
| Grade | G1 | 1 (0.26 %) | 0 (0 %) | 1 (0.52 %) | 0.5687 |
| G2 | 39 (10.18 %) | 19 (9.95 %) | 20 (10.42 %) | ||
| G3 | 335 (87.47 %) | 167 (87.43 %) | 168 (87.5 %) | ||
| G4 | 1 (0.26 %) | 1 (0.52 %) | 0 (0 %) | ||
| Unknown | 7 (1.83 %) | 4 (2.09 %) | 3 (1.56 %) | ||
| Stage | IIIA | 7 (1.83 %) | 5 (2.62 %) | 2 (1.04 %) | 0.2586 |
| IIIB | 16 (4.18 %) | 11 (5.76 %) | 5 (2.6 %) | ||
| IIIC | 299 (78.07 %) | 147 (76.96 %) | 152 (79.17 %) | ||
| IV | 61 (15.93 %) | 28 (14.66 %) | 33 (17.19 %) | ||
| tumor Residual | >20 mm | 72 (18.8 %) | 42 (21.99 %) | 30 (15.62 %) | 0.3589 |
| 1–10 mm | 185 (48.3 %) | 85 (44.5 %) | 100 (52.08 %) | ||
| 11–20 mm | 26 (6.79 %) | 13 (6.81 %) | 13 (6.77 %) | ||
| No macroscopic disease | 65 (16.97 %) | 33 (17.28 %) | 32 (16.67 %) | ||
| Unknown | 35 (9.14 %) | 18 (9.42 %) | 17 (8.85 %) |
Fig. 3.
Construction of the BCRLss. (A) Identification of survival-related lncRNAs in the training cohort based on Cox analysis. (B, C) LASSO coefficients and partial likelihood deviance of the scoring system. (D) Mapping of risk lncRNAs and B cell hub genes. (E–H) PCA of all genes, B cell hub genes, B cell-related lncRNAs, and risk lncRNAs.
Table 2.
Risk lncRNAs and their corresponding risk coefficients used in the construction of BCRLss.
| BCRlncRNA | Coef | HR | HR (95%CI) | P-value |
|---|---|---|---|---|
| POLH-AS1 | −0.893962038 | 0.533 | 0.290–0.980 | 0.043 |
| LINC00996 | −0.842387676 | 0.456 | 0.257–0.808 | 0.007 |
| AL133467.1 | −0.916867881 | 0.386 | 0.212–0.703 | 0.002 |
| LINC01894 | 0.950234241 | 2.227 | 1.065–4.657 | 0.033 |
| LINC00892 | −1.093890464 | 0.342 | 0.154–0.756 | 0.008 |
| LINC01150 | 1.025415275 | 1.638 | 1.054–2.544 | 0.028 |
HR, hazard ratio; CI, confidence interval.
3.3. Validation of BCRLss
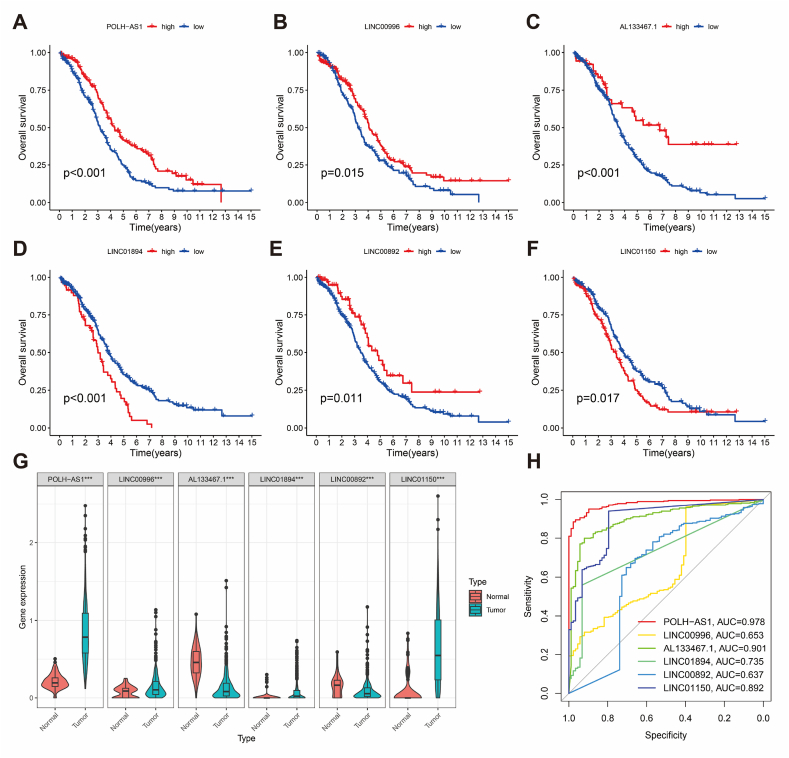
The prognostic performance of BCRLss was validated from three angles. First, K-M survival analysis confirmed the prognostic relevance of the 6 risk lncRNAs in TCGA-OV (Fig. 4A–F). Differential expression analysis revealed that POLH-AS1, LINC00996, LINC01894, and LINC01150 were highly expressed in tumor tissues, while LINC00892 and AL133467.1 were predominantly expressed in normal tissues (Fig. 4G). ROC analysis showed that these risk lncRNAs exhibit strong diagnostic performance, with the potential for use as early diagnostic biomarkers for TCGA-OV (Fig. 4H). POLH-AS1 displayed the highest diagnostic accuracy (AUC = 0.978), whereas LINC00892 had the lowest (AUC = 0.653).
Fig. 4.
Clinical significance of risk lncRNAs in OV. (A–F) Kaplan-Meier survival analysis of risk lncRNAs. (G) Differential expression of risk lncRNAs in normal ovarian tissues and ovarian cancer. (H) ROC analysis for diagnostic performance of risk lncRNAs.
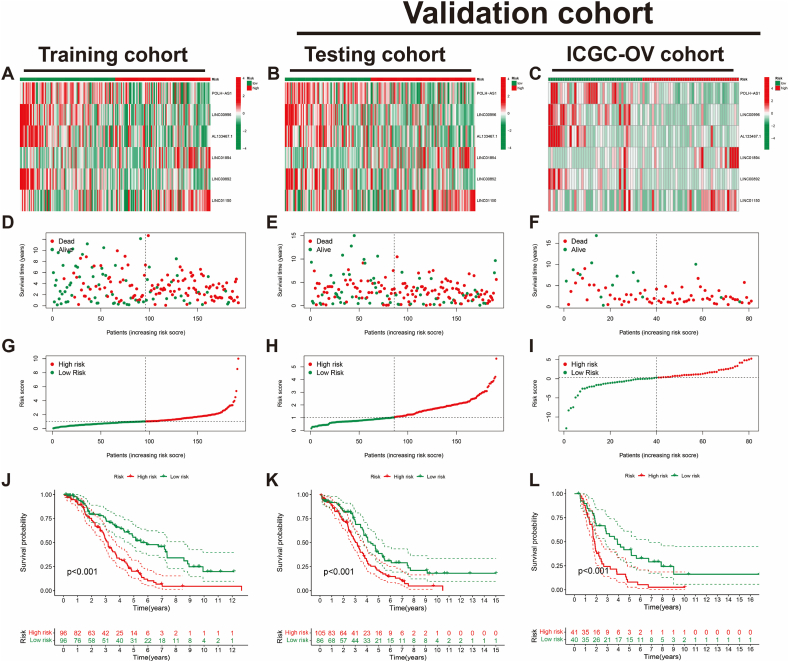
Subsequently, the BCRLss was validated in both the internal testing cohort and the external independent ICGC-OV cohort. The ICGC-OV cohort was stratified into high- and low-risk groups based on the median risk score. The distribution heatmaps (Fig. 5A–C), survival dependency scatter plots (Fig. 5D–F), and risk curves (Fig. 5G–I) demonstrated consistent trends across the training, testing, and ICGC-OV cohorts. Furthermore, Kaplan-Meier survival curves (Fig. 5J–L) indicated that patients in the low-risk group of BCRLss had significantly better prognoses compared to those in the high-risk group across all three cohorts.
Fig. 5.
Validation of BCRLss prognostic performance in OV. (A–C) Heatmap of risk lncRNA expression in the Training cohort, Testing cohort, and ICGC-OV cohort. (D–F) Survival status and time in the Training, Testing, and ICGC-OV cohorts. (G–I) Risk score distribution in the Training, Testing, and ICGC-OV cohorts. (J–L) Kaplan-Meier curves for overall survival in the Training, Testing, and ICGC-OV cohorts.
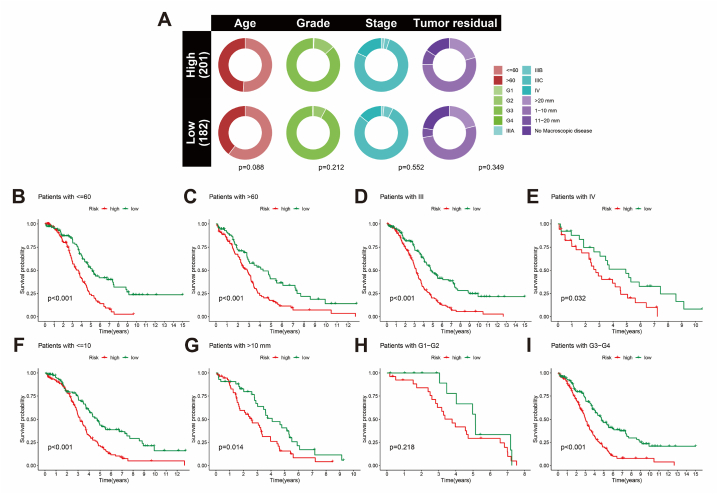
The survival validation of BCRLss across various clinical features is outlined. Fig. 6A illustrates the distribution of patients with different clinical characteristics within the risk subgroups. Notably, patients in the low-risk BCRLss group demonstrated a better prognosis across clinical subgroups, including age (threshold of 60 years) (Fig. 6B and C), stage (III-IV) (Fig. 6D and E), and tumor residual (threshold of 10 mm) (Fig. 6F and G). For tumor grade, survival analysis in G3-G4 remained consistent with this trend. However, no significant difference between high- and low-risk groups was observed in G1-G2, likely due to the limited sample size in this subgroup within TCGA-OV (Fig. 6H and I).
Fig. 6.
Survival analysis of BCRLss across clinical feature subgroups in OV. (A) Heatmap of clinical features in the scoring system. (B, C) Kaplan-Meier curves for age in high- and low-risk groups. (D, E) Kaplan-Meier curves for stage in high- and low-risk groups. (F, G) Kaplan-Meier curves for tumor residual in high- and low-risk groups. (H, I) Kaplan-Meier curves for grade in high- and low-risk groups.
3.4. Prognostic prediction of BCRLss
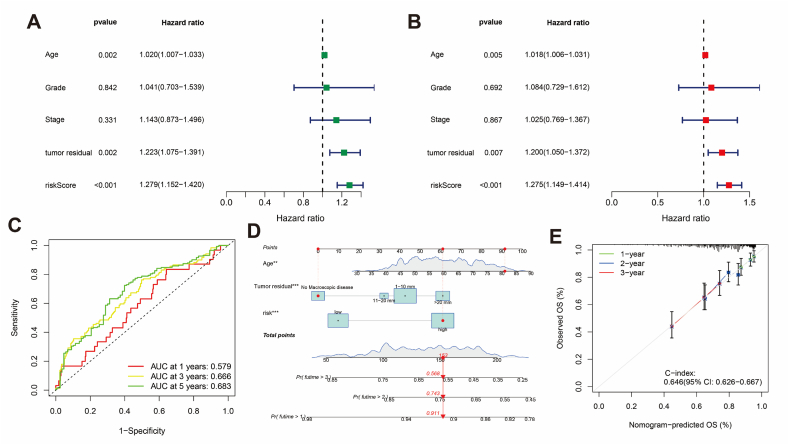
Cox regression analysis identified age, tumor residual, and risk score as independent prognostic factors for TCGA-OV patients (Fig. 7A and B). The AUC values under the ROC survival curve at 1, 3, and 5 years were 0.579, 0.666, and 0.683, respectively (Fig. 7C). A nomogram was constructed to predict patient prognosis, with total points at 1, 2, and 3 years estimated at 0.911, 0.74, and 0.568, respectively (Fig. 7D). The calibration curve indicated good predictive accuracy (C-index = 0.646 [95 % CI: 0.626–0.667]) (Fig. 7E).
Fig. 7.
Prognostic value of BCRLss. (A) Univariate Cox analysis forest plot. (B) Multivariate Cox analysis forest plot. (C) ROC survival curve for OV. (D) Nomogram for survival prediction in OV. (E) Calibration curve of the nomogram.
3.5. Enrichment analysis
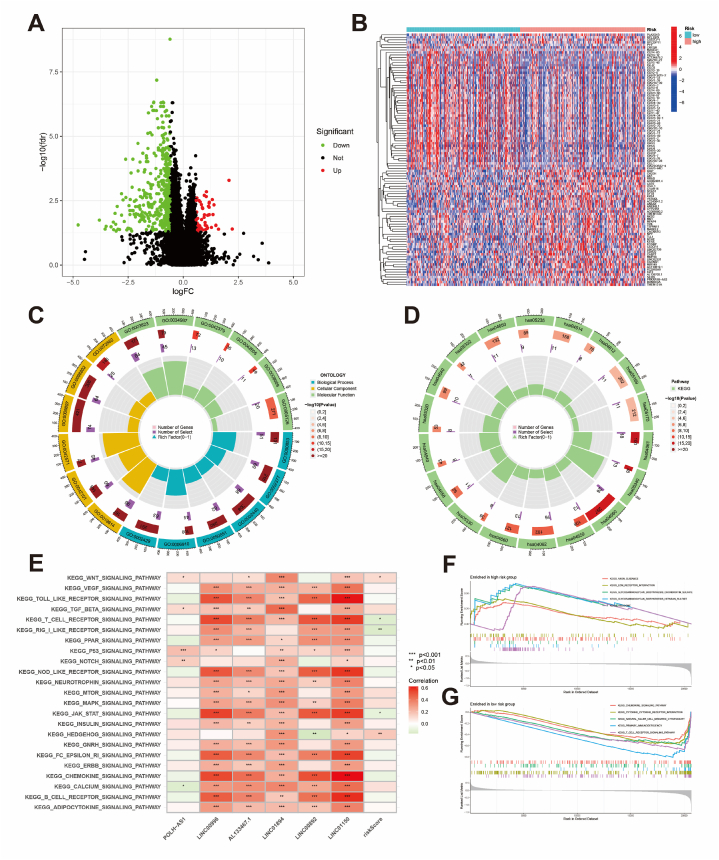
Differential analysis between high- and low-risk groups of BCRLss identified 58 upregulated and 455 downregulated genes (Fig. 8A). Fig. 8B visualizes the top 50 differentially expressed genes in TCGA-OV. GO and KEGG enrichment analyses revealed the biological functions of these DEGs (Fig. 8C and D). GSVA highlighted associations between risk lncRNAs, risk scores, and various tumor immune and metabolic pathways (Fig. 8E). GSEA demonstrated the enrichment of axon guidance and ECM receptor interaction in the high-risk group, while the low-risk group was associated with the chemokine signaling pathway, cytokine-cytokine receptor interaction, and T cell receptor signaling pathway (Fig. 8F and G). Collectively, BCRLss is significantly linked to OC progression and TIME regulation.
Fig. 8.
Enrichment analysis. (A) Identification of DEGs in high- and low-risk groups. (B) Heatmap of DEGs. (C) GO enrichment analysis of DEGs. (D) KEGG enrichment analysis of DEGs. (E) GSVA of risk lncRNA and risk scores. (F, G) GSEA of pathways in high- and low-risk groups.
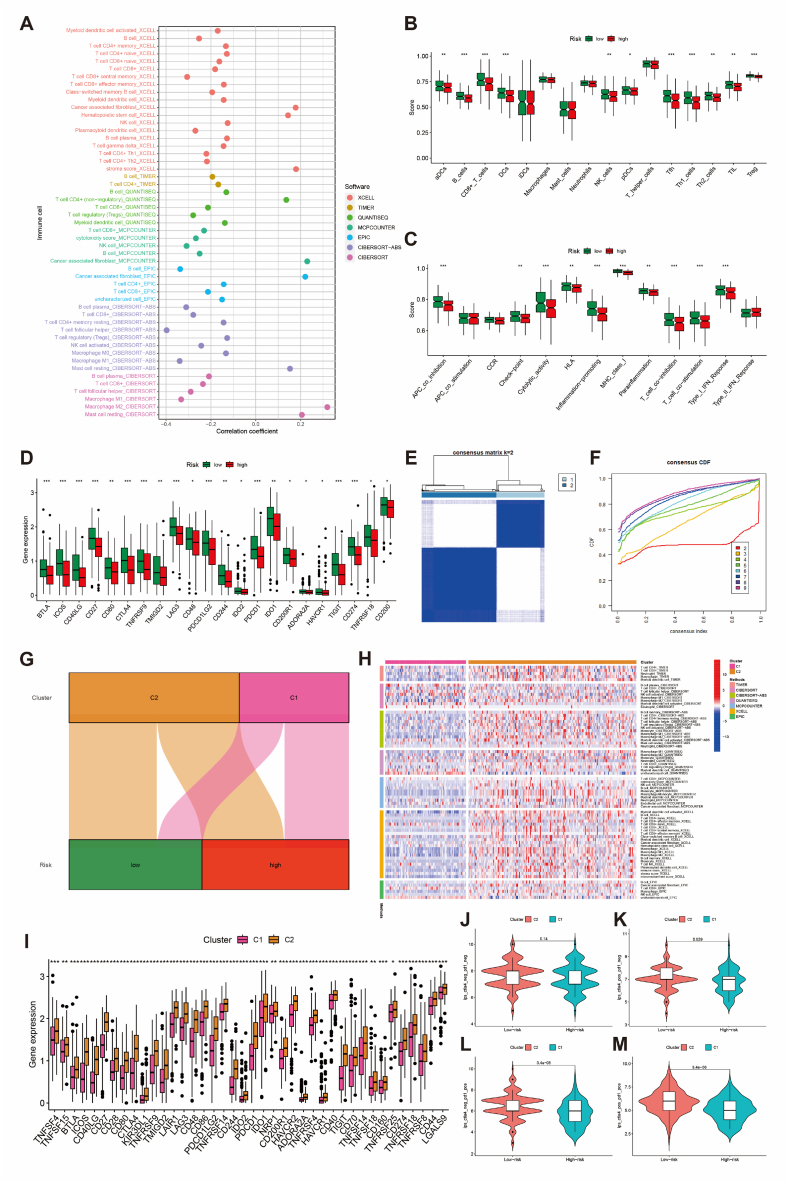
3.6. Landscape of BCRLss in tumor immune microenvironment
Using multiple algorithms from the TIMER2.0 platform, the correlation between cells in the TIME and the risk score was analyzed (Supplementary Fig. 1). Notably, anti-inflammatory cells, such as tumor-associated fibroblasts and M2 macrophages, showed a positive correlation with the risk score, whereas pro-inflammatory cells, including CD8+ T cells and M1 macrophages, were negatively correlated (Fig. 9A). Immune correlation analysis using the ssGSEA algorithm further indicated a higher degree of immune cell infiltration in the low-risk group (Fig. 9B and C). Additionally, immune checkpoint-related genes were more highly expressed in the low-risk group (Fig. 9D).
Fig. 9.
Correlation analysis of BCRLss and the tumor immune microenvironment in OV. (A) Bubble chart of correlation coefficients between immune cells and risk scores. (B) Differential analysis of immune cells between high- and low-risk groups. (C) Differential analysis of immune function between high- and low-risk groups. (D) Differential expression of immune checkpoint-related genes in high- and low-risk groups. (E, F) Consensus clustering analysis based on BCRLss. (G) Sankey diagram of clusters and BCRLss groups. (H) Heatmap of immune cell correlations across clusters. (I) Expression of immune checkpoint-related genes in clusters. (J–M) Predicted immunotherapy response in clusters based on the TICA database. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
An unsupervised consensus clustering analysis based on risk scores (Supplementary Fig. 2) revealed that categorizing patients into two clusters was the most appropriate (Fig. 9E and F). Fig. 9G shows the correlation between the clusters and risk groups, and Fig. 9H illustrates significant differences in cell type expression in TIME across the clusters. Cluster C2 exhibited higher expression of immune checkpoint-related genes than cluster C1. TCIA analysis further suggested that patients in cluster C2 might be more responsive to immune checkpoint inhibitors, specifically anti-CTLA4 and anti-PD1 therapies (Fig. 9J–M). Although StromalScore, ImmuneScore, and ESTIMATEScore did not show significant differences between high- and low-risk groups, the tumor microenvironment scores were significantly higher in the C2 cluster, based on the ESTIMATE algorithm (Supplementary Fig. 3). These findings collectively demonstrate that BCRLss can effectively distinguish patient populations with distinct TIME characteristics.
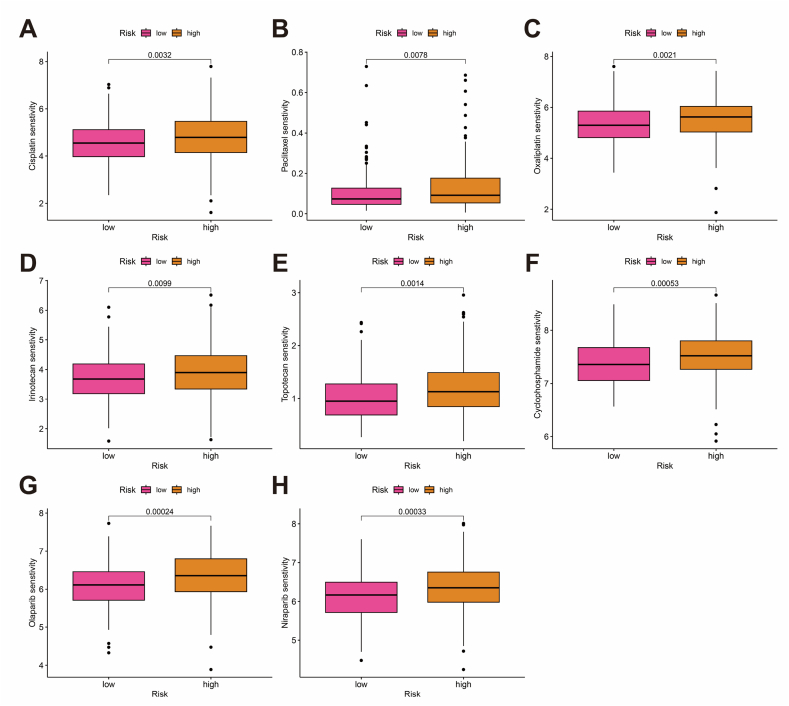
3.7. Screening of drugs for potential clinical benefit
Drug sensitivity predictions were conducted using IC50 values obtained from the GDSC database (Supplementary Fig. 4). Fig. 10A–H displays commonly used OC drugs across different treatment stages, including Cisplatin, Paclitaxel, Cyclophosphamide, Topotecan, Irinotecan, Niraparib, Olaparib, and Oxaliplatin. The data suggest that patients in the low-risk group may benefit more from these drugs.
Fig. 10.
Drug sensitivity analysis. (A–H) Box plot comparison of IC50 values for common clinical drugs between high- and low-risk groups.
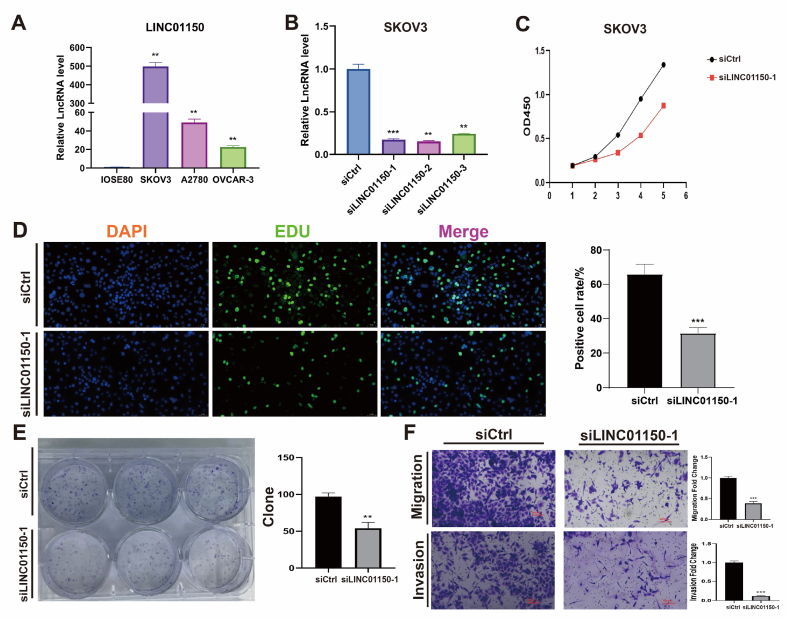
3.8. In vitro validation
The oncogenic potential of the highest risk coefficient lncRNA, LINC01150, in OC, was further investigated through in vitro experiments. Fig. 11A demonstrates that LINC01150 is highly expressed in OC cell lines (SKOV3, A2780, OVCAR-3) compared to the normal ovarian cell line IOSE80. SKOV3, which showed the highest differential expression, was selected for LINC01150 knockdown experiments. The siLINC01150-1 vector, with the highest knockdown efficiency, was chosen for subsequent experiments (Fig. 11B). Functional assays revealed that LINC01150 knockdown significantly inhibited the proliferation of SKOV3 cells, as shown by CCK8, EDU, and colony formation assays (Fig. 11C–E). Moreover, the migration and invasion abilities of the siLINC01150-1 group were markedly reduced (Fig. 11F).
Fig. 11.
In vitro experiments. (A) LINC01150 expression in ovarian and ovarian cancer cell lines. (B) Relative expression of LINC01150 after transfection with siRNA. (C) Proliferation of ovarian cancer cells after LINC01150 knockdown based on CCK8 assay. (D) Proliferation of ovarian cancer cells after LINC01150 knockdown based on EDU assay. (E) Proliferation of ovarian cancer cells after LINC01150 knockdown based on colony formation assay. (F) Migration and invasion of ovarian cancer cells after LINC01150 knockdown based on Transwell assay. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
4. Discussion
High-throughput sequencing technology has greatly advanced our understanding of tumor heterogeneity and the exploration of the TIME from a molecular perspective [40]. Constructing reliable predictive factors is essential for achieving precise anti-tumor therapies [41]. Although the combination of CA125 and HE4 provides higher specificity and sensitivity in detecting OC, it remains inadequate for screening and early diagnosis. miRNAs, while promising as clinical biomarkers, face challenges due to complex sample standardization procedures and specific detection platforms [42]. Thus, there is an urgent demand for novel biomarkers to improve molecular diagnostics and prognosis prediction in OC. Furthermore, tumor immune-infiltrating cells in OC exhibit high heterogeneity, which correlates with patient prognosis and response to immunotherapy [8]. Evidence suggests that combining PARP inhibitors with immunotherapy enhances treatment response in patients with BRCA mutations or homologous recombination deficiencies. This effect may arise from BRCA defects triggering STING-mediated innate immune responses, leading to increased production of type I interferons and pro-inflammatory cytokines [43]. Unfortunately, immunotherapy based on ICIs remains limited in OC due to its low immunogenicity [44]. Therefore, identifying novel molecular features that characterize the TIME landscape in OC is critical.
Increasing evidence has demonstrated the significant role of B cells and lncRNAs in the progression and immune activity of OC. Moreover, lncRNAs can regulate the development, proliferation, and differentiation of B cells [45]. B cell-related lncRNAs and their molecular signatures have shown strong clinical potential in predicting prognosis, immune cell infiltration patterns, and immunotherapy responses in solid tumors, such as bladder and liver cancers [46,47]. Additionally, lncRNAs are increasingly recognized as valuable tumor biomarkers [48,49]. However, the potential of B cell-related lncRNAs as biomarkers in OC remains largely unexplored.
scRNA-seq is an advanced gene sequencing technology that enables the exploration of cell subpopulation heterogeneity within the TIME, facilitating the analysis of dynamic cellular transformations and interactions [50,51]. In this study, single-cell transcriptomic data from distinct tumor-infiltrating immune cell subpopulations were obtained from the OC cohort GSE147082. Through differential analysis and PPI network analysis, BCHGs were identified, leading to the construction of a BCRLss composed of 6 risk lncRNAs (POLH-AS1, LINC00996, AL133467.1, LINC01894, LINC00892, LINC01150) using various algorithms. KM survival analysis and ROC analysis collectively demonstrated that these 6 lncRNAs could serve as potential biomarkers for early diagnosis and prognosis evaluation in patients with OV. LINC00996 has previously been reported as a prognostic factor in colorectal cancer and head and neck squamous cell carcinoma (HNSCC) [52,53], with additional studies confirming that its overexpression significantly inhibits lung cancer cell proliferation, migration, and invasion [54]. LINC00892, specifically expressed by CD4+ T cell subsets, is associated with the T cell receptor pathway and may play a role in communication between helper T cells and B cells [55]. POLH-AS1 has been linked to HNSCC prognosis, and AL133467.1 has shown prognostic value in OV by contributing to the construction of an immune-related signature [56]. LINC01150 has been identified as a prognostic marker in lung adenocarcinoma, with potential immune relevance [57]. Despite these insights, the oncogenic role of these risk lncRNAs in OC remains unclear. Therefore, LINC01150, with the highest risk score in BCRLss, was selected for in vitro validation. The results showed that LINC01150 was highly expressed in OC, and its knockdown in the SKOV3 cell line significantly inhibited proliferation, invasion, and metastasis, suggesting that targeting LINC01150 could offer new therapeutic strategies for OC.
Survival analysis indicated that BCRLss was applicable across different clinical subgroups of TCGA-OV patients. Notably, survival variability between high- and low-risk groups was not observed in G1-G2 patients, likely due to the limited sample size. However, a trend toward better prognosis in the low-risk group was noted. The ICGC-OV cohort was introduced as an external validation set, which demonstrated consistency with TCGA-OV in terms of survival-dependent risk profiles and Kaplan-Meier survival analysis. Univariate and multivariate Cox analyses identified age, tumor residual, and risk score as independent prognostic factors for OV. Previous studies have shown that age and tumor residuals (threshold >10 mm) are strongly correlated with disease-free survival (DFS) and OS [58], with further confirmation from a Norwegian study [59]. However, the role of age as an independent prognostic factor in OC remains debated, with a Japanese study suggesting that treatment strategies should focus on physical status rather than age [60]. In addition, ROC survival analysis and the construction of a nomogram further validated the reliable predictive performance of BCRLss in OV.
Increasing evidence underscores the prognostic significance of tumor-infiltrating immune cells in OC. For instance, CD8+ T cells and natural killer cells directly kill cancer cells by producing IFN-γ, a process closely linked to long-term survival in OC [61,62]. Conversely, immune-suppressive cells such as regulatory T cells, fibroblasts, and M2 macrophages facilitate immune evasion, indicating a poorer prognosis [[63], [64], [65]]. In this study, the correlation between risk score and immune cell infiltration in OV was analyzed using multiple algorithms. Results revealed a negative correlation between risk score and immune-promoting cells, including CD8+ T cells, M1 macrophages, plasma cells, and CD4+ memory T cells, which are favorable prognostic markers. Conversely, risk score was positively correlated with immune-suppressive cells such as fibroblasts, regulatory CD4+ T cells, and M2 macrophages, which are associated with poor prognosis [10]. These results suggest that patients in the low-risk group may benefit from enhanced immunotherapy response and have better prognostic outcomes. Additionally, the ssGSEA algorithm showed that patients in the low-risk group exhibited higher immune cell infiltration and more active immune-related functions. Overall, BCRLI effectively captures the immune landscape of patients with OV.
Although ICIs targeting CD274, PDCD1, and CTLA-4 have revolutionized cancer treatment, their efficacy in OC remains limited. A phase III clinical trial in Japan comparing the efficacy of Nivolumab with chemotherapy (gemcitabine or liposomal doxorubicin) in platinum-resistant epithelial OC revealed that, despite good tolerability, no significant improvement in survival was observed [66]. Therefore, identifying new targets to enhance immunotherapy response in OC remains critical. In this study, immune checkpoint-related gene expression was higher in the low-risk group, further suggesting their potential immunotherapy benefits. Moreover, a strong relationship between several key genes and B cells was observed. For instance, IDO1 and IDO2 play roles in B cell-mediated adaptive immunity and inflammation regulation, though with opposing effects. IDO1 suppresses B cell-mediated inflammation, while IDO2 promotes it [67]. The exact pathways by which IDO1 and IDO2 regulate B cell immune responses remain unclear and warrant further investigation [68]. Another noteworthy gene, HAVCR1, emerged as a potential target. Recent studies have shown that the deletion of the protein TIM-1, encoded by HAVCR1, enhances type I interferon (IFN-1) responses in B cells, promoting antigen presentation and co-stimulation, which leads to the expansion of tumor-specific effector T cells. This positions TIM-1 as a promising target for modulating B cell-mediated immune responses [69]. In conclusion, this study provides valuable insights into target selection for immune checkpoint blockade therapy in OV, offering new avenues for improving treatment response.
A drug sensitivity analysis indicated that commonly used OC treatments, including Cisplatin, Paclitaxel, Oxaliplatin, Irinotecan, Topotecan, Cyclophosphamide, Olaparib, and Niraparib, may be more effective for patients in the low-risk group of BCRLss. Platinum-based chemotherapy, such as cisplatin or carboplatin combined with paclitaxel, remains the first-line treatment for advanced OV. However, resistance to platinum-based therapies often leads to reduced OS. While monotherapies (e.g., paclitaxel, irinotecan, topotecan, cyclophosphamide) or combinations with anti-angiogenic drugs are frequently employed, their efficacy is limited. Some studies suggest that patients with platinum-resistant OC may still benefit from re-treatment with platinum-based drugs [70]. In recent years, the focus has shifted toward maintenance therapy. PARP inhibitors have demonstrated the ability to prolong patient survival by targeting DNA double-strand break repair through homologous recombination (S/G2 phase) and non-homologous end joining (G1 phase). These inhibitors act on the mitotic cell cycle and involve several key proteins, including Ku70/80, XLF, MRI/CYREN (bidirectional regulatory), IFFO1, ERCC6L2, and RNaseH2 [71]. The NCT03705156 trial showed that niraparib maintenance therapy reduced disease progression by 68 % in patients with platinum-sensitive OC [72]. Furthermore, olaparib has been shown to benefit patients with platinum-sensitive OC in maintenance therapy, regardless of BRCA mutation status [73]. Overall, this study offers a valuable reference for the development of personalized treatment strategies for patients with OV, although further rigorous clinical trials are required to confirm these findings.
Despite the systematic validation and comprehensive assessment of the constructed scoring system, several limitations remain. Firstly, due to the current accessibility of open data, we were able to validate the scoring system for ovarian cancer only within the TCGA-OV validation cohort and the ICGC-OV cohort. We are unable to extend validation to additional independent cohorts at this time. Future research should aim to further evaluate the predictive value of the scoring system in prospective large-sample clinical studies.Moreover, while our study has demonstrated that targeting LINCO1150 can inhibit the malignant biological behavior of SKOV3 ovarian cancer cells, further investigation is needed to elucidate its specific mechanism of action and its clinical prognostic value in real-world settings.
This study has several limitations. First, all patients included had OV, the most common pathological subtype of OC. Therefore, additional research is necessary to determine the prognostic value of BCRLss in other OC subtypes. Furthermore, while the carcinogenic potential of LINC01150 in OC was confirmed through in vitro experiments, its underlying mechanism remains unclear. Given its potential role in early diagnosis and prognosis evaluation, further investigation into the mechanism of LINC01150 in OC is warranted.
5. Conclusion
In conclusion, BCRLs effectively predicted clinical outcomes in patients with OV and were validated using an external independent cohort. Additionally, BCRLss delineated the immune cell infiltration landscape within the TIME and identified potential targets for immunotherapy, along with identifying patient populations likely to benefit. Drug sensitivity analysis further informed the development of clinical dosing strategies for OC treatment. Lastly, in vitro experiments validated the oncogenic role of BCRL LINC01150, which exhibited the highest risk coefficient, in promoting OC progression.
CRediT authorship contribution statement
Yi Huang: Writing – original draft, Formal analysis, Data curation. Zhongxuan Gui: Validation, Conceptualization. Muyun Wu: Project administration, Methodology. Mengmeng Zhang: Software, Formal analysis. Yue Jiang: Visualization. Qiaoqiao Ding: Software. Jinping Yang: Validation, Methodology. Yingquan Ye: Writing – review & editing. Mei Zhang: Resources, Project administration.
Ethics approval and consent to participate
The study's data were culled solely from public databases, which held no personally identifiable information. As such, ethical approval and individual consent were unnecessary for this research.
Data availability
All original data in this study can be obtained upon request from the corresponding author.
Funding
This research received no external funding.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We appreciate the platforms provided by the TISCH2 database, TCGA database, the ICGC database, and the GTEx database, and thank the contributors for their assistance in this collaborative study.In addition, We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39496.
Contributor Information
Jinping Yang, Email: m18895673206@163.com.
Yingquan Ye, Email: onyyq20@163.com.
Mei Zhang, Email: zhangmei@ahmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Penny S.M. Ovarian cancer: an overview. Radiol. Technol. 2020;91(6):561–575. [PubMed] [Google Scholar]
- 2.Saani I., Raj N., Sood R., Ansari S., Mandviwala H.A., Sanchez E., Boussios S. Clinical challenges in the management of malignant ovarian germ cell tumours. Int. J. Environ. Res. Publ. Health. 2023;20(12) doi: 10.3390/ijerph20126089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor E.C., Irshaid L., Mathur M. Multimodality imaging approach to ovarian neoplasms with pathologic correlation. Radiographics : a review publication of the Radiological Society of North America, Inc. 2021;41(1):289–315. doi: 10.1148/rg.2021200086. [DOI] [PubMed] [Google Scholar]
- 4.Sun H., Cao D., Ma X., Yang J., Peng P., Yu M., Zhou H., Zhang Y., Li L., Huo X., Shen K. Identification of a prognostic signature associated with DNA repair genes in ovarian cancer. Front. Genet. 2019;10:839. doi: 10.3389/fgene.2019.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich M., Friedrich D., Kraft C., Rogmans C. Multimodal treatment of primary advanced ovarian cancer. Anticancer Res. 2021;41(7):3253–3260. doi: 10.21873/anticanres.15111. [DOI] [PubMed] [Google Scholar]
- 6.Aliyuda F., Moschetta M., Ghose A., Sofia Rallis K., Sheriff M., Sanchez E., Rassy E., Boussios S. Advances in ovarian cancer treatment beyond PARP inhibitors. Curr. Cancer Drug Targets. 2023;23(6):433–446. doi: 10.2174/1568009623666230209121732. [DOI] [PubMed] [Google Scholar]
- 7.Zhang A., Miao K., Sun H., Deng C.X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022;18(7):3019–3033. doi: 10.7150/ijbs.72534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baci D., Bosi A., Gallazzi M., Rizzi M., Noonan D.M., Poggi A., Bruno A., Mortara L. The ovarian cancer tumor immune microenvironment (time) as target for therapy: a focus on innate immunity cells as therapeutic effectors. Int. J. Mol. Sci. 2020;21(9) doi: 10.3390/ijms21093125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Zhang L., Bai Y., Wang L., Ma X. Therapeutic implications of the tumor microenvironment in ovarian cancer patients receiving PD-1/PD-L1 therapy. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1036298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garsed D.W., Pandey A., Fereday S., Kennedy C.J., Takahashi K., Alsop K., Hamilton P.T., Hendley J., Chiew Y.E., Traficante N., Provan P., Ariyaratne D., Au-Yeung G., et al. The genomic and immune landscape of long-term survivors of high-grade serous ovarian cancer. Nat. Genet. 2022;54(12):1853–1864. doi: 10.1038/s41588-022-01230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Y., Zhang S., Jiang Y., Huang Y., Wang G., Zhang M., Gui Z., Wu Y., Bian G., Li P., Zhang M. Identification of a cancer associated fibroblasts-related index to predict prognosis and immune landscape in ovarian cancer. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-48653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thommen D.S., Koelzer V.H., Herzig P., Roller A., Trefny M., Dimeloe S., Kiialainen A., Hanhart J., Schill C., Hess C., Savic Prince S., Wiese M., Lardinois D., et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018;24(7):994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taefehshokr S., Parhizkar A., Hayati S., Mousapour M., Mahmoudpour A., Eleid L., Rahmanpour D., Fattahi S., Shabani H., Taefehshokr N. Cancer immunotherapy: challenges and limitations. Pathol. Res. Pract. 2022;229 doi: 10.1016/j.prp.2021.153723. [DOI] [PubMed] [Google Scholar]
- 14.Kim S.S., Sumner W.A., Miyauchi S., Cohen E.E.W., Califano J.A., Sharabi A.B. Role of B Cells in responses to checkpoint blockade immunotherapy and overall survival of cancer patients. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2021;27(22):6075–6082. doi: 10.1158/1078-0432.CCR-21-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laumont C.M., Banville A.C., Gilardi M., Hollern D.P., Nelson B.H. Tumour-infiltrating B cells: immunological mechanisms, clinical impact and therapeutic opportunities. Nat. Rev. Cancer. 2022;22(7):414–430. doi: 10.1038/s41568-022-00466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandell G.L., Fisher R.I., Bostick F., Young R.C. Ovarian cancer: a solid tumor with evidence of normal cellular immune function but abnormal B cell function. Am. J. Med. 1979;66(4):621–624. doi: 10.1016/0002-9343(79)91172-0. [DOI] [PubMed] [Google Scholar]
- 17.Dong H.P., Elstrand M.B., Holth A., Silins I., Berner A., Trope C.G., Davidson B., Risberg B. NK- and B-cell infiltration correlates with worse outcome in metastatic ovarian carcinoma. Am. J. Clin. Pathol. 2006;125(3):451–458. [PubMed] [Google Scholar]
- 18.Montfort A., Pearce O., Maniati E., Vincent B.G., Bixby L., Böhm S., Dowe T., Wilkes E.H., Chakravarty P., Thompson R., Topping J., Cutillas P.R., Lockley M., et al. A strong B-cell response is part of the immune landscape in human high-grade serous ovarian metastases. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2017;23(1):250–262. doi: 10.1158/1078-0432.CCR-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kołat D., Hammouz R., Bednarek A.K., Płuciennik E. Exosomes as carriers transporting long non-coding RNAs: molecular characteristics and their function in cancer. Mol. Med. Rep. 2019;20(2):851–862. doi: 10.3892/mmr.2019.10340. (Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokorná M., Černá M., Boussios S., Ovsepian S.V., O'Leary V.B. lncRNA biomarkers of glioblastoma multiforme. Biomedicines. 2024;12(5) doi: 10.3390/biomedicines12050932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell Mol. Med. 2018;22(12):5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan H., Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65(4):625–639. doi: 10.1042/EBC20200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha J.H., Radhakrishnan R., Nadhan R., Gomathinayagam R., Jayaraman M., Yan M., Kashyap S., Fung K.M., Xu C., Bhattacharya R., Mukherjee P., Isidoro C., Song Y.S., et al. Deciphering a GPCR-lncrna-miRNA nexus: Identification of an aberrant therapeutic target in ovarian cancer. Cancer letters. 2024;591 doi: 10.1016/j.canlet.2024.216891. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Wang W., Fan S. Emerging roles of lncRNA in Nasopharyngeal Carcinoma and therapeutic opportunities. Int. J. Biol. Sci. 2022;18(7):2714–2728. doi: 10.7150/ijbs.70292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y.T., Lin J.F., Li T., Li J.J., Xu R.H., Ju H.Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021;41(2):109–120. doi: 10.1002/cac2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zan H., Casali P. Epigenetics of peripheral B-cell differentiation and the antibody response. Front. Immunol. 2015;6:631. doi: 10.3389/fimmu.2015.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brazão T.F., Johnson J.S., Müller J., Heger A., Ponting C.P., Tybulewicz V.L. Long noncoding RNAs in B-cell development and activation. Blood. 2016;128(7):e10–e19. doi: 10.1182/blood-2015-11-680843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., Luo Y., Zhang L., Younis M., Yuan L. Down-regulation of LncRNA 2900052N01Rik inhibits LPS-induced B cell function in vitro. Cell. Immunol. 2021;363 doi: 10.1016/j.cellimm.2021.104321. [DOI] [PubMed] [Google Scholar]
- 29.Xu Q., Lin Y.B., Li L., Liu J. LncRNA TLR8-AS1 promotes metastasis and chemoresistance of ovarian cancer through enhancing TLR8 mRNA stability. Biochemical and biophysical research communications. 2020;526(4):857–864. doi: 10.1016/j.bbrc.2020.03.087. [DOI] [PubMed] [Google Scholar]
- 30.Liu H., Deng S., Yao X., Liu Y., Qian L., Wang Y., Zhang T., Shan G., Chen L., Zhou Y. Ascites exosomal lncRNA PLADE enhances platinum sensitivity by inducing R-loops in ovarian cancer. Oncogene. 2024;43(10):714–728. doi: 10.1038/s41388-024-02940-6. [DOI] [PubMed] [Google Scholar]
- 31.Qian M., Ling W., Ruan Z. Long non-coding RNA SNHG12 promotes immune escape of ovarian cancer cells through their crosstalk with M2 macrophages. Aging. 2020;12(17):17122–17136. doi: 10.18632/aging.103653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su R., Jin C., Zhou L., Cao Y., Kuang M., Li L., Xiang J. Construction of a ceRNA network of hub genes affecting immune infiltration in ovarian cancer identified by WGCNA. BMC Cancer. 2021;21(1):970. doi: 10.1186/s12885-021-08711-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M., Shang X., Sun Y., Wu J., Liu G. Integrated analysis of lymphocyte infiltration-associated lncRNA for ovarian cancer via TCGA, GTEx and GEO datasets. PeerJ. 2020;8 doi: 10.7717/peerj.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibshirani R. The lasso method for variable selection in the Cox model. Stat. Med. 1997;16(4):385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canzler S., Hackermüller J. multiGSEA: a GSEA-based pathway enrichment analysis for multi-omics data. BMC Bioinf. 2020;21(1):561. doi: 10.1186/s12859-020-03910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W.H., de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome biology. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa H., Fujita M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018;109(3):513–522. doi: 10.1111/cas.13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghose A., McCann L., Makker S., Mukherjee U., Gullapalli S.V.N., Erekkath J., Shih S., Mahajan I., Sanchez E., Uccello M., Moschetta M., Adeleke S., Boussios S. Diagnostic biomarkers in ovarian cancer: advances beyond CA125 and HE4. Therapeutic advances in medical oncology. 2024;16 doi: 10.1177/17588359241233225. 17588359241233225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boussios S., Karathanasi A., Cooke D., Neille C., Sadauskaite A., Moschetta M., Zakynthinakis-Kyriakou N., Pavlidis N. PARP inhibitors in ovarian cancer: the route to "ithaca". Diagnostics. 2019;9(2) doi: 10.3390/diagnostics9020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anadon C.M., Yu X., Hänggi K., Biswas S., Chaurio R.A., Martin A., Payne K.K., Mandal G., Innamarato P., Harro C.M., Mine J.A., Sprenger K.B., Cortina C., et al. Ovarian cancer immunogenicity is governed by a narrow subset of progenitor tissue-resident memory T cells. Cancer Cell. 2022;40(5):545–557.e513. doi: 10.1016/j.ccell.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkle M., Kluiver J.L., Diepstra A., van den Berg A. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit. Rev. Oncol.-Hematol. 2017;120:77–85. doi: 10.1016/j.critrevonc.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Zhou M., Zhang Z., Bao S., Hou P., Yan C., Su J., Sun J. Computational recognition of lncRNA signature of tumor-infiltrating B lymphocytes with potential implications in prognosis and immunotherapy of bladder cancer. Briefings Bioinf. 2021;22(3) doi: 10.1093/bib/bbaa047. [DOI] [PubMed] [Google Scholar]
- 47.Song H., Huang X.F., Hu S.Y., Lu L.L., Yang X.Y. The LINC00261/MiR105-5p/SELL axis is involved in dysfunction of B cell and is associated with overall survival in hepatocellular carcinoma. PeerJ. 2022;10 doi: 10.7717/peerj.12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao L., Ye Y., Zhang X., Xu X., Wang W., Jiang B. Identification and verification of a PANoptosis-related long noncoding ribonucleic acid signature for predicting the clinical outcomes and immune landscape in lung adenocarcinoma. Heliyon. 2024;10(8) doi: 10.1016/j.heliyon.2024.e29869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Q., Ye Y., Zhang Q., Wu Y., Wang G., Gui Z., Zhang M. PANoptosis-related long non-coding RNA signature to predict the prognosis and immune landscapes of pancreatic adenocarcinoma. Biochemistry and biophysics reports. 2024;37 doi: 10.1016/j.bbrep.2023.101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren X., Zhang L., Zhang Y., Li Z., Siemers N., Zhang Z. Insights gained from single-cell analysis of immune cells in the tumor microenvironment. Annu. Rev. Immunol. 2021;39:583–609. doi: 10.1146/annurev-immunol-110519-071134. [DOI] [PubMed] [Google Scholar]
- 51.Li P.H., Kong X.Y., He Y.Z., Liu Y., Peng X., Li Z.H., Xu H., Luo H., Park J. Recent developments in application of single-cell RNA sequencing in the tumour immune microenvironment and cancer therapy. Military Medical Research. 2022;9(1):52. doi: 10.1186/s40779-022-00414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luan L., Dai Y., Shen T., Yang C., Chen Z., Liu S., Jia J., Li Z., Fang S., Qiu H., Cheng X., Yang Z. Development of a novel hypoxia-immune-related LncRNA risk signature for predicting the prognosis and immunotherapy response of colorectal cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.951455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai Z., Tang B., Chen L., Lei W. Mast cell marker gene signature in head and neck squamous cell carcinoma. BMC Cancer. 2022;22(1):577. doi: 10.1186/s12885-022-09673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Z., Li X., Hu Z., Yang Y., Yang Z., Li S., Zhou Y., Ma J., Li H., Liu X., Cai J., Pu L., Wang X., et al. Linc00996 is a favorable prognostic factor in LUAD: results from bioinformatics analysis and experimental validation. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.932973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iaccarino I., Mourtada F., Reinke S., Patil P., Doose G., Monaco G., Hoffmann S., Siebert R., Klapper W. LINC00892 is an lncRNA induced by T cell activation and expressed by follicular lymphoma-resident T helper cells. Non-coding RNA. 2022;8(3) doi: 10.3390/ncrna8030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y., Huo F.F., Wen Y.Y., Jiang M. Screening and identification of an immune-associated lncRNA prognostic signature in ovarian carcinoma: evidence from bioinformatic analysis. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/6680036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He C., Yin H., Zheng J., Tang J., Fu Y., Zhao X. Identification of immune-associated lncRNAs as a prognostic marker for lung adenocarcinoma. Transl. Cancer Res. 2021;10(2):998–1012. doi: 10.21037/tcr-20-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sait K.H., Alam M.Z., Haque A., Sait H.K., Sait M.K., Anfinan N.M. Survival and prognostic factors in women treated for epithelial ovarian cancer in western region of Saudi Arabia. Saudi Med. J. 2022;43(2):146–155. doi: 10.15537/smj.2022.43.2.20210626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tingulstad S., Skjeldestad F.E., Halvorsen T.B., Hagen B. Survival and prognostic factors in patients with ovarian cancer. Obstet. Gynecol. 2003;101(5 Pt 1):885–891. doi: 10.1016/s0029-7844(03)00123-6. [DOI] [PubMed] [Google Scholar]
- 60.Yoshikawa K., Fukuda T., Uemura R., Matsubara H., Wada T., Kawanishi M., Tasaka R., Kasai M., Hashiguchi Y., Ichimura T., Yasui T., Sumi T. Age-related differences in prognosis and prognostic factors among patients with epithelial ovarian cancer. Molecular and clinical oncology. 2018;9(3):329–334. doi: 10.3892/mco.2018.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato E., Olson S.H., Ahn J., Bundy B., Nishikawa H., Qian F., Jungbluth A.A., Frosina D., Gnjatic S., Ambrosone C., Kepner J., Odunsi T., Ritter G., et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(51):18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nersesian S., Glazebrook H., Toulany J., Grantham S.R., Boudreau J.E. Naturally killing the silent killer: NK cell-based immunotherapy for ovarian cancer. Front. Immunol. 2019;10:1782. doi: 10.3389/fimmu.2019.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamanishi J., Mandai M., Iwasaki M., Okazaki T., Tanaka Y., Yamaguchi K., Higuchi T., Yagi H., Takakura K., Minato N., Honjo T., Fujii S. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y., Mei S., Huang Y., Chen J., Zhang X., Zhang P. Integrative analysis deciphers the heterogeneity of cancer-associated fibroblast and implications on clinical outcomes in ovarian cancers. Comput. Struct. Biotechnol. J. 2022;20:6403–6411. doi: 10.1016/j.csbj.2022.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baek S.H., Lee H.W., Gangadaran P., Oh J.M., Zhu L., Rajendran R.L., Lee J., Ahn B.C. Role of M2-like macrophages in the progression of ovarian cancer. Experimental cell research. 2020;395(2) doi: 10.1016/j.yexcr.2020.112211. [DOI] [PubMed] [Google Scholar]
- 66.Hamanishi J., Takeshima N., Katsumata N., Ushijima K., Kimura T., Takeuchi S., Matsumoto K., Ito K., Mandai M., Nakai H., Sakuragi N., Watari H., Takahashi N., et al. Nivolumab versus gemcitabine or pegylated liposomal doxorubicin for patients with platinum-resistant ovarian cancer: open-label, randomized trial in Japan (NINJA) J. Clin. Oncol. : official journal of the American Society of Clinical Oncology. 2021;39(33):3671–3681. doi: 10.1200/JCO.21.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merlo L.M.F., DuHadaway J.B., Montgomery J.D., Peng W.D., Murray P.J., Prendergast G.C., Caton A.J., Muller A.J., Mandik-Nayak L. Differential roles of Ido1 and Ido2 in T and B cell inflammatory immune responses. Front. Immunol. 2020;11:1861. doi: 10.3389/fimmu.2020.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merlo L.M.F., Peng W., Mandik-Nayak L. Impact of Ido1 and Ido2 on the B cell immune response. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.886225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bod L., Kye Y.C., Shi J., Torlai Triglia E., Schnell A., Fessler J., Ostrowski S.M., Von-Franque M.Y., Kuchroo J.R., Barilla R.M., Zaghouani S., Christian E., Delorey T.M., et al. B-cell-specific checkpoint molecules that regulate anti-tumour immunity. Nature. 2023;619(7969):348–356. doi: 10.1038/s41586-023-06231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bogliolo S., Cassani C., Gardella B., Musacchi V., Babilonti L., Venturini P.L., Ferrero S., Spinillo A. Oxaliplatin for the treatment of ovarian cancer. Expet Opin. Invest. Drugs. 2015;24(9):1275–1286. doi: 10.1517/13543784.2015.1062874. [DOI] [PubMed] [Google Scholar]
- 71.Shah S., Cheung A., Kutka M., Sheriff M., Boussios S. Epithelial ovarian cancer: providing evidence of predisposition genes. Int. J. Environ. Res. Publ. Health. 2022;19(13) doi: 10.3390/ijerph19138113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X.H., Zhu J.Q., Yin R.T., Yang J.X., Liu J.H., Wang J., Wu L.Y., Liu Z.L., Gao Y.N., Wang D.B., Lou G., Yang H.Y., Zhou Q., et al. Niraparib maintenance therapy in patients with platinum-sensitive recurrent ovarian cancer using an individualized starting dose (NORA): a randomized, double-blind, placebo-controlled phase III trial(☆) Ann. Oncol. : official journal of the European Society for Medical Oncology. 2021;32(4):512–521. doi: 10.1016/j.annonc.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 73.Gao Q., Zhu J., Zhao W., Huang Y., An R., Zheng H., Qu P., Wang L., Zhou Q., Wang D., Lou G., Wang J., Wang K., et al. Olaparib maintenance monotherapy in asian patients with platinum-sensitive relapsed ovarian cancer: phase III trial (L-MOCA) Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2022;28(11):2278–2285. doi: 10.1158/1078-0432.CCR-21-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original data in this study can be obtained upon request from the corresponding author.