Summary
Hepatocellular carcinoma (HCC) remains a leading cause of cancer-related deaths worldwide, and a large proportion is attributable to viral causes, including hepatitis B (HBV) and C viruses (HCV). The pathogenesis of viral-mediated HCC can differ between HBV and HCV, but it is unclear how much these differences influence the tumors’ final molecular and immune profiles. Additionally, there are known sex differences in the molecular etiology of HCC, but sex differences have not been explored in the context of viral-mediated HCC. To determine the extent to which the viral status and sex impact the molecular and immune profiles of HCC, we performed differential expression and immune cell deconvolution analyses. We identified a large number of differentially expressed genes unique to the HBV or HCV tumor:tumor-adjacent comparison. Pathway enrichment analyses demonstrated that changes unique to the HCV tumor:tumor-adjacent tissue were dominated by changes in immune pathways. Immune cell deconvolution demonstrated that HCV tumor-adjacent tissue had the largest immune cell infiltrate, with no difference in the immune profiles within HBV and HCV tumor samples. Overall, this work demonstrates the convergence of HBV- and HCV-mediated HCC on a similar transcriptomic landscape and immune profile despite differences in the surrounding tissue.
Keywords: hepatocellular carcinoma, liver cancer, immune infiltration, transcriptomics, sex differences, immune system, gene expression profiling, hepatitis
This study explores the differences in viral-mediated forms of hepatocellular carcinoma (HCC). We found that HCC tumors, regardless of hepatitis B (HBV) or C virus (HCV) etiology, appear to be more biologically similar than the corresponding tumor-adjacent tissue. This suggests a convergence on common pathways toward cancers even when the origin differs.
Introduction
Hepatocellular carcinoma (HCC [MIM: 114550]) remains a critical health challenge worldwide, leading to over 600,000 deaths annually.1 Risk factors for HCC include hepatitis B, C, and D viruses (HBV [MIM: 610424], HCV [MIM: 609532], and HDV); alcoholic liver disease; and non-alcoholic fatty liver disease (MIM: 613282).2 Approximately 50% of HCC is attributable to HBV infection.2 The risk of HCC with HCV has been reduced with the introduction of antiviral therapies that have led to a sustained virological response to HCV.3 However, antiviral therapies do not reduce the risk of HCC in individuals who have already progressed to cirrhosis, and 30% of HCC remains attributable to HCV infection.1
The pathogenesis of HCC from HBV and HCV can be attributable to multiple underlying mechanisms. Both viruses can be associated with persistent inflammation, immune-mediated oxidative stress from chronic infection, and abnormal regulation of signaling pathways.1,4 HBV can integrate into the host genome, causing insertional mutagenesis, which may be carcinogenic.5 Chronic HCV has also been associated with steatosis and subsequent progression to fibrosis and cirrhosis.6 Additionally, in endemic areas, HBV is primarily transmitted perinatally,7 leading to lifelong infections that may elicit significantly different immune responses compared to HCV, which is primarily transmitted through direct contact with blood later in life.8
Despite the differences in the underlying pathogenesis of viral-mediated HCC, the majority of treatment guidelines do not discriminate based on etiology.9,10,11 Within the last few years, immune checkpoint inhibitors, including anti-PD1, anti-PDL1, and anti-CTLA-4 treatments, have been approved by the US Food and Drug Administration for the treatment of advanced HCC.12,13,14 Studies to date have been underpowered to detect differences in the response of HBV and HCV to immune checkpoint inhibitors.15,16 Previous studies have demonstrated differences in the immune microenvironment in HBV- and HCV-mediated HCC,17 suggesting the importance of understanding the tumor microenvironment in viral-mediated liver cancer.
Sex differences in the incidence, mortality, and genetic profile of HCC have been documented,18,19 and previous work from our lab has documented sex differences in the molecular etiologies of HCC.20 However, there is less known about the sex differences in viral-mediated HCC. Here, we performed differential expression analyses on a cohort of viral-mediated HCC cases with paired tumor-adjacent tissue. We segregated our analyses first by viral etiology and then by the combination of viral etiology and sex to illuminate the underlying molecular profiles and immune landscapes in these tumors and the adjacent tissue. We discuss the challenges stemming from sampling biases and the need for increased sampling of female tumors to fully probe the biological mechanisms leading to the differences in incidence and mortality from HCC in males and females. Furthermore, while we identified several genes with differential expression in HBV- and HCV-mediated liver cancer, we demonstrated that the tumor tissue appears to converge on a more similar transcriptional landscape and immune profile compared to the tumor-adjacent tissue. Together, these results highlight the importance of considering sex and etiology in defining the transcriptional and immune profiles of HCC.
Material and methods
Data acquisition and processing
Whole transcriptome data (RNA sequencing [RNA-seq]) from tumor and tumor-adjacent viral-mediated HCC samples were obtained from the International Cancer Genome Consortium LIRI-JP dataset.21 Tumor-adjacent samples were taken from adjacent liver tissue. Healthy liver samples from unaffected individuals were analyzed from data collected from the GTEx consortium.22 All data analyses were performed with Arizona State University High-Performance Computing resources.23 The LIRI-JP RNA-seq FASTA files were visualized for quality using FASTQC.24 Data were trimmed using Trimmomatic with parameters of 2 seed mismatches, palindrome clip threshold 30, simple clip threshold 10, leading quality value 3, trailing quality value 3, sliding window size 4, minimum window quality 30, and minimum read length of 50.25 Samples were revisualized using FASTQC after trimming. Samples from one individual (RK023) were excluded due to a mean quality score dropping below 30 for a large portion of the sequence length, whereas all other samples had mean quality scores above 30 for the full length (Figure S1). Transcript expression levels were quantified using Salmon.26 The Salmon index was built based on the Gencode HG38 version 29 genome. Pseudoalignment was carried out using automatic library-type detection. A total of 147 (53.3%) of the 276 samples were detected by Salmon to be stranded, and 129 (46.7%) were unstranded. To verify this, we additionally used GuessMyLT27 to infer the library type based on the FASTQ files. The results of this analysis were concordant with the library types detected by Salmon. No differences were identified in the total sequencing depth, total number of reads mapped, or the proportion of unstranded/stranded library types when segregated by viral etiology, sex, and tumor status (Figure S2).
Sample quality control
We utilized multidimensional scaling as implemented in the plotMDS function of the R package limma version 3.58.1 to calculate distances between samples based on gene expression levels.28 The sequencing library type was identified as a key source of variation in the data (stranded/non-stranded; Figure S3A). After removal of the sequencing library type with the removeBatchEffect function from limma, the majority of samples clustered strongly by tumor/tumor adjacent on principal component 1 and sex on principal component 2 (Figure S3B). However, a small number of samples were found in clusters that did not match their annotation. For all samples, we plotted the gene expression of XIST [MIM: 314670] and DDX3Y [MIM: 400010] (Figures S3C and S3D), and six samples were excluded from subsequent analysis where the inferred sex chromosome complement from gene expression did not match the reported sex of the individual or the tumor and tumor-adjacent samples clustered together, suggesting possible contamination. Two paired samples had tumor and tumor-adjacent samples that clustered in the opposite groups (e.g., the sample marked as tumor grouped with tumor adjacent, and the matched tumor adjacent sample grouped with the tumor samples), and the annotations for these were changed to match the appropriate group. The distribution of the final cohort by tumor/tumor adjacent, sex, and etiology included 276 samples (Table 1; Figure S3E). The age and stage of the tumors were compared between the groups segregated by viral etiology, sex, and tumor status (Figures S4A and S4B). HCV samples were taken from significantly older individuals compared to the HBV samples, but no difference was found within HBV or HCV samples. No difference in the distribution of stage was identified between any of the groups. The sex distribution was compared between the groups segregated by viral etiology and tumor status, and no difference in the proportion of samples from males and females was identified (Figure S4C).
Table 1.
International Cancer Genome Consortium LIRI-JP dataset samples, segregated by sex and etiology
| Reported male individual |
Reported female individual |
|||
|---|---|---|---|---|
| Tumor tissue | Tumor-adjacent tissue | Tumor tissue | Tumor-adjacent tissue | |
| HBV | 37 | 31 | 8 | 7 |
| HCV | 70 | 59 | 32 | 32 |
| Total | 107 | 90 | 40 | 39 |
Samples included are those that passed all quality control steps.
Filtering and processing
FPKM (fragments per kilobase of exon per million fragments mapped) and trimmed mean of M-values were obtained using the R package EdgeR, version 4.0.16.29 We retained genes with a mean FPKM value of ≥0.5 in any group segregated by sex and viral status and read count of >6 in at least 10 samples across all samples under investigation, resulting in 12,466 genes.
Differential expression
To detect differentially expressed genes between the sexes, tissues, and viral infection cases, we used linear regression as implemented in limma. Filtered raw counts per million reads were log2 normalized and adjusted for quality using the voomWithQualityWeights function in the limma R package.30 Differential expression analyses were performed for the following comparisons: (1) all tumor vs. all tumor adjacent, (2) HBV tumor vs. HBV tumor adjacent, (3) HCV tumor vs. HCV tumor adjacent, (4) HBV tumor vs. HCV tumor, (5) HBV tumor adjacent vs. HCV tumor adjacent, (6) male HBV tumor vs. male HBV tumor adjacent, (7) male HCV tumor vs. male HCV tumor adjacent, (8) female HBV tumor vs. female HBV tumor adjacent, (9) female HCV tumor vs. female HCV tumor adjacent, (10) male tumor vs. female tumor, and (11) male tumor adjacent vs. female tumor adjacent. For the male tumor:tumor adjacent and female tumor:tumor adjacent comparisons, differential expression analysis was repeated with a randomly down-sampled set of samples to make the number of male and female samples equivalent. Library type was added as a covariate for each model. Correlation between measurements between tumor and tumor-adjacent samples from the same individual was accounted for in the linear modeling using the duplicateCorrelation function.28 Differentially expressed genes were identified using the limma/voom pipeline with empirical Bayes statistics. Upset plots were generated using the R package UpSetR version 1.4.0.31
Pathway enrichment analysis
Differentially expressed genes were then analyzed for overrepresentation of biological processes. Hypergeometric testing was performed using the R package clusterProfiler version 2.1.632 to identify significantly overrepresented Gene Ontology (GO) terms. p values were adjusted for false discovery rate (FDR), and GO terms with an adjusted p value of less than 0.05 were considered to be significantly overrepresented pathways. GO enrichment analyses were visualized as treemaps using ReviGo.33
Immune cell deconvolution
Immune cell deconvolution was performed across all samples with transcripts per million (TPM) normalized expression data using xCell34 and quanTIseq35 through the R package immunedeconv version 2.1.0.36
Results
Segregation by viral etiology enables the identification of distinct sets of differentially expressed genes
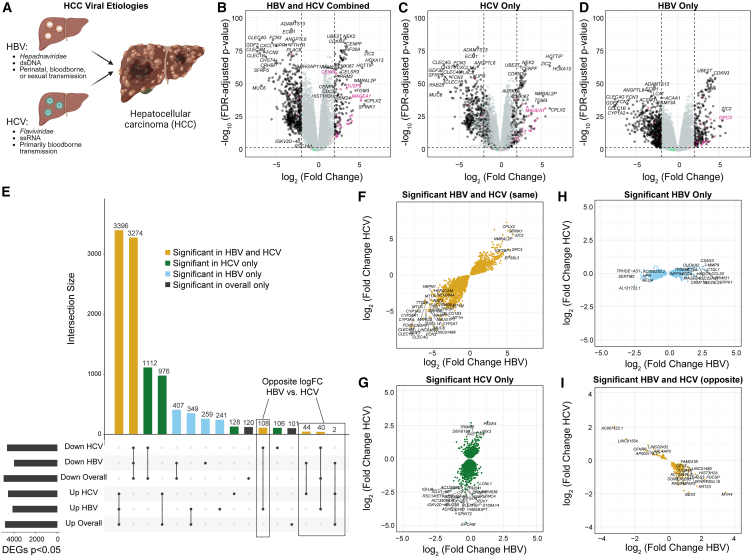
HBV and HCV are both known etiologies for HCC, despite their differences in viral class, genome type, and transmission route (Figure 1A). To probe the molecular phenotypes of HCC arising from HBV or HCV, we performed differential expression analyses across all tumor vs. tumor-adjacent samples regardless of etiology (147 tumor vs. 129 tumor adjacent; Figure 1B), HCV tumor vs. tumor-adjacent samples only (102 tumor vs. 91 tumor adjacent; Figure 1C), and HBV tumor vs. tumor-adjacent samples only (45 tumor vs. 38 tumor adjacent; Figure 1D). Genes were considered differentially expressed if they had a false discovery rate of less than 0.05.
Figure 1.
Segregation by viral etiology increases the identification of differentially expressed genes
(A) Overview of hepatitis B (HBV) and hepatitis C (HCV) viruses. HBV is a hepadnaviridae with a double-stranded DNA (dsDNA) genome and is primarily transmitted perinatally but can also be bloodborne or sexually transmitted. HCV is a flaviviridae with a single-stranded RNA (ssRNA) genome and has primarily bloodborne transmission.
(B–D) Volcano plots of differentially expressed genes from (B) HBV and HCV combined, (C) HCV only, and (D) HBV only. X-linked genes are indicated in pink, Y-linked in green, and autosomal in black.
(E) Upset plot of differentially expressed genes from each of the comparisons. Genes shared in all comparisons are colored yellow, genes unique to the HCV subset are green, and genes unique to the HBV subset are light blue.
(F–I) Comparison of the logFC in HBV and HCV for (F) genes significant in both HBV and HCV tumors in the same direction, (G) genes significant only in the HCV subset, (H) genes significant only in the HBV subset, and (I) genes significant in both HBV and HCV but in opposite directions.
We then evaluated the degree of overlap between the lists of differentially expressed genes from each comparison (Figure 1E). Across all comparisons, we detected 10,663 unique differentially expressed genes. Of these, 6,670 (62.6%, 3,396 upregulated in tumors and 3,274 downregulated in tumors) were significant across all samples, HCV samples, and HBV samples. A total of 2,088 genes (19.6%, 1,112 downregulated in tumors, 976 upregulated in tumors) were identified as significant in the comparison across all samples as well as HCV samples alone, but not in the analysis of HBV samples alone. Similarly, 756 genes (7.1%, 407 downregulated in tumors, 349 upregulated in tumors) were significant in the analysis of all samples and HBV samples alone, but not in HCV samples alone. Moreover, 500 genes not identified as differentially expressed in the overall comparison were identified as differentially expressed in HBV samples only (259 downregulated in tumors and 241 upregulated), while 234 genes not identified as differentially expressed in the overall comparison were identified as differentially expressed in HCV samples (128 upregulated in tumors and 106 downregulated). A total of 221 genes identified in the overall comparison were not detected as significant in either HBV or HCV samples when segregated.
Finally, 194 genes were identified as differentially expressed in opposite directions in HBV and HCV samples. Of these, 152 were not identified in the overall comparison but were differentially expressed in opposite directions in HBV and HCV when segregated (108 downregulated in HCV tumor samples and upregulated in HBV tumor samples, and 44 downregulated in HBV tumor samples and upregulated in HCV tumor samples). A total of 42 genes were identified as differentially expressed in the overall comparison and then identified in opposite directions in HBV and HCV when segregated. Of the 42, 40 were originally identified as downregulated in all tumor samples and then identified as downregulated in HCV tumor samples and upregulated in HBV tumor samples when segregated, and 2 were originally identified as upregulated across all tumor samples and then upregulated in HCV tumor samples and downregulated in HBV tumor samples when segregated.
We then evaluated whether the genes identified in the segregated analyses showed similar expression changes from tumor-adjacent to tumor tissue in HBV and HCV samples, despite not passing multiple testing corrections in both etiologies. We plotted the log fold change (logFC) of the tumor:tumor-adjacent change in HBV compared to HCV for each subset of genes (Figures 1F–1I). Across the genes with shared differential expression in HBV and HCV, we fit a linear model and found that our model had an R2 value of 0.833, suggesting that 83.3% of the variability in the data is explained through the model (Figure 1F). By contrast, for the differentially expressed genes expressed in HCV only, the R2 value is 0.200 (Figure 1G) and for the genes differentially expressed in HBV only, the R2 value is only 0.197 (Figure 1G). This finding supports that the differences observed are more likely to be biological differences in the transcriptional profiles of HBV- and HCV-mediated HCC rather than artifacts of the differences in sample size. A majority (95.4%) of the differentially expressed genes with opposite directions in HBV and HCV had an absolute logFC of less than 1.5 in both HBV and HCV. However, a small number show large opposite-FCs, including BEX2 (MIM: 300691),37,38 AP1M2 (MIM: 607309),39 and KRT23 (MIM: 606194),40,41 all of which have been associated with the pathogenesis of HCC (Figure 1I).
Genes unique to HCV are enriched for immune pathways
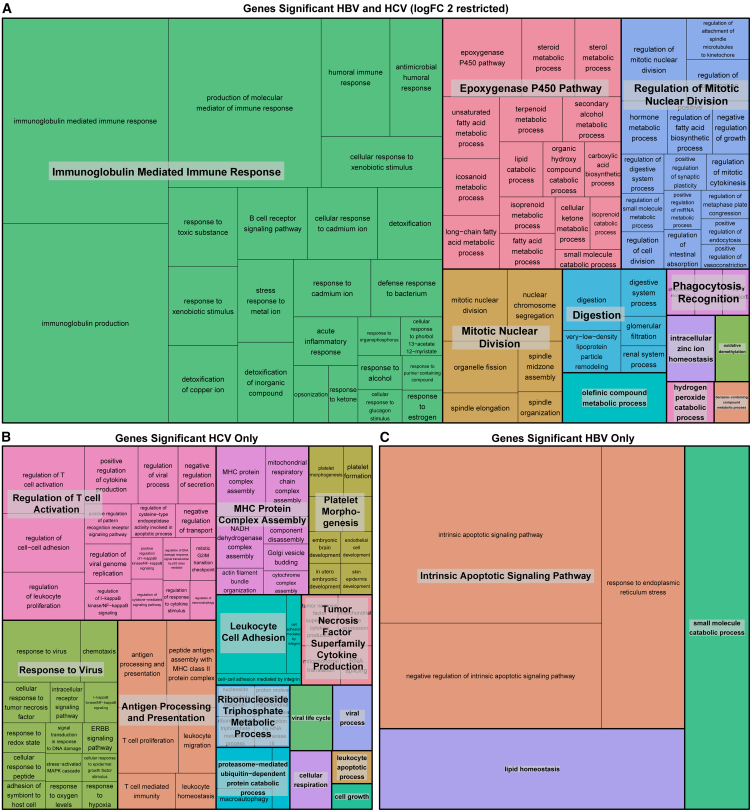
To identify pathways enriched in the differentially expressed genes, we performed GO enrichment analyses on the shared and unique differentially expressed genes for HBV and HCV (Figure 2). The pathways enriched in the shared differentially expressed genes are consistent with the hallmarks of cancer. These pathways include the immune response (“immunoglobulin-mediated immune response”), cell division (“mitotic nuclear division” and “regulation of mitotic nuclear division”), and the “epoxygenase P450 pathway,” which is known to regulate the hepatic inflammatory response (Figure 2A).42 Within the genes uniquely differentially expressed in HCV, we identified a predominance of immune-related pathways, including “regulation of T cell activation,” “response to virus,” “antigen processing and presentation,” “MHC [major histocompatibility complex] protein complex assembly,” and “leukocyte cell adhesion” (Figure 2B). Finally, in the genes significant to HBV only, a few pathways were identified, predominantly the “intrinsic apoptotic signaling pathways” (Figure 2C).
Figure 2.
Genes unique to HCV tumor:tumor-adjacent comparison are enriched for immune pathways
Treemap visualization of GO enrichment analysis from (A) overall tumor vs. tumor-adjacent comparison (restricted by a logFC >2 due to the large number of significant genes), (B) genes unique to HCV tumor vs. tumor-adjacent comparison, and (C) genes unique to HBV tumor vs. tumor-adjacent comparison. The sizes of the boxes reflect the magnitude of the false discovery adjusted p value for the GO enrichment term.
Tumor-adjacent tissue is more distinct than tumor tissue based on viral etiology
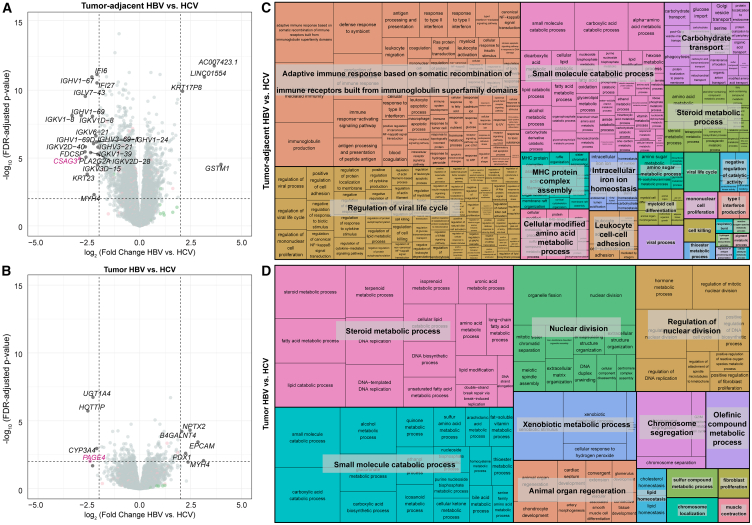
While the previous analyses identified genes that change expression from tumor-adjacent to tumor tissue, stratified by etiology, we were interested in directly examining etiology-based differences in tumor-adjacent and tumor tissue. We therefore performed differential expression analyses on HBV tumor vs. HCV tumor and HBV tumor-adjacent vs. HCV tumor-adjacent samples. When differentially expressed genes are defined as genes with an FDR <0.05, 2,363 genes were identified as upregulated in HCV tumor-adjacent tissue and 1,817 upregulated in HBV tumor-adjacent tissue. By contrast, a smaller number of genes are found to be differentially expressed in the tumor tissue, with 341 upregulated in HCV and 364 upregulated in HBV (Figures 3A and 3B). Of note, when restricting to the genes with the largest logFCs (>2), 14/21 genes upregulated in HCV tumor-adjacent tissue were immunoglobulin genes and 2/21 were interferon-inducible genes, indicating a strong predominance of immune-related genes (Figures S5A–S5U). To establish whether these genes are normally expressed in healthy liver tissue, we probed the expression of each across the normal liver GTEx samples. The majority of genes upregulated in HCV tumor-adjacent tissue, except IFI27 (MIM: 600009), IFI6 (MIM: 147572), and PLA2G2A (MIM: 172411), show average expression below 1 TPM in GTEx normal liver samples, suggesting that the upregulation of these immune-related genes in HCV tumor-adjacent tissue is likely mediated by the viral infection (Figure S5V).
Figure 3.
Tumor-adjacent tissue is more distinct than tumor tissue based on viral etiology
(A and B) Volcano plots of differentially expressed genes from (A) tumor-adjacent HBV vs. HCV and (B) tumor HBV vs. HCV samples. In each comparison, genes with higher expression in HCV are on the left side of the plot with negative logFCs, and genes with higher expression in HBV are on the right side of the plot with positive FCs. Genes with an FDR <0.05 and a logFC >2 are labeled on the plot. X-linked genes are indicated in pink, Y-linked in green, and autosomal in black.
(C and D) Treemap visualization of GO enrichment analysis from (C) tumor-adjacent HBV vs. HCV and (D) tumor HBV vs. HCV. The sizes of the boxes reflect the magnitude of the false discovery adjusted p value for the GO enrichment term.
We then performed pathway enrichment analysis on all differentially expressed genes between HBV and HCV in both the tumor and tumor-adjacent samples. Within the HBV tumor-adjacent:HCV tumor-adjacent comparison, pathway enrichment analysis demonstrated enrichment of immune-related pathways including “adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains,” “regulation of viral life cycle,” and “MHC protein complex assembly” (Figure 3C). By contrast, within the smaller number of differentially expressed genes in the tumor tissue, the primary differences were found in metabolism (“steroid metabolic process,” “small molecule catabolic process,” and “xenobiotic metabolic process”) and nuclear division (“nuclear division,” “regulation of nuclear division,” and “chromosome segregation”) (Figure 3D).
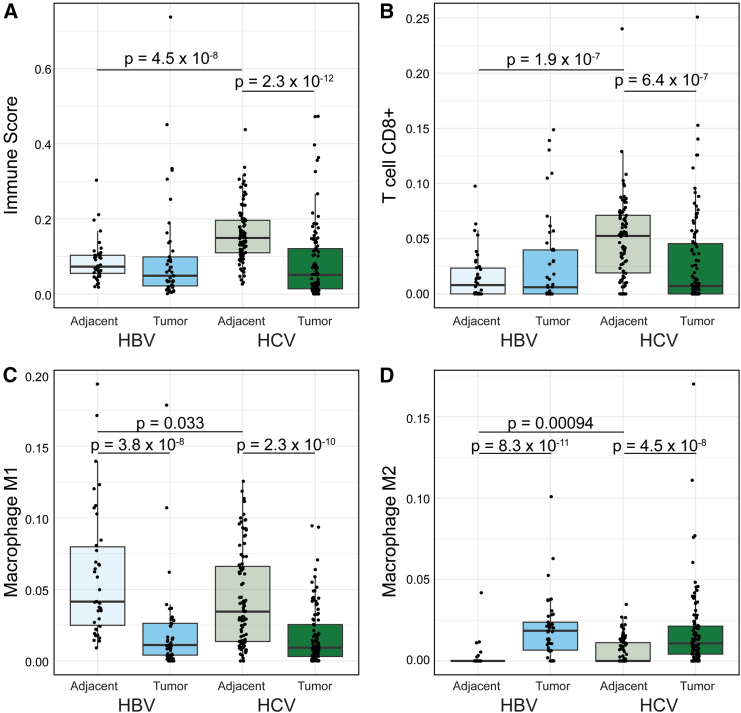
Given the predominance of immune-related pathways identified in the (1) HCV tumor:HCV tumor-adjacent and the (2) HBV tumor-adjacent:HCV tumor-adjacent comparisons, we performed immune cell deconvolution across all samples. Immune cell deconvolution was first performed with xCell34 and demonstrated a significant increase in the immune cell infiltration in HCV tumor-adjacent samples compared to all other samples. This increase is seen in the overall immune score, accounting for all immune cell populations (Figures 4A and S6A). Given the difference in age between HBV and HCV samples, we also assessed for any association of immune infiltration and age within the tumor and tumor-adjacent samples from HBV and HCV and found no significant association (Figures S7A–S7D). Since the enriched pathways suggested changes in the adaptive immune response specifically, we examined the CD8+ T cell population and demonstrated the highest infiltration of CD8+ T cells in HCV tumor-adjacent samples (Figures 4B and S6A). Despite the increased immune infiltration in HBV and HCV tumor-adjacent samples, no differences were observed between HBV and HCV tumor samples.
Figure 4.
Greater immune infiltration in HCV tumor-adjacent tissue
(A and B) xCell immune deconvolution. (A) Comparison of the overall immune infiltration score in tumor and tumor-adjacent tissue segregated by etiology. (B) Comparison of CD8+ T cells in tumor and tumor-adjacent tissue segregated by etiology.
(C and D) quanTIseq immune deconvolution. (C) Comparison of M1 macrophages in tumor and tumor-adjacent tissue segregated by etiology, and (D) comparison of M2 macrophages in tumor and tumor-adjacent tissue segregated by etiology. For all boxplots, the bold line indicates the median, and the upper and lower limits of the boxes indicate the 75th and 25th percentiles, respectively. The lower and upper whiskers indicate the minimum and maximum. Dots outside of the box and whiskers indicate outliers. Significance tested between samples from the same etiology (HBV tumor:HBV tumor-adjacent and HCV tumor:HCV tumor-adjacent) or between samples from the same tissue (HBV tumor:HCV tumor and HBV tumor-adjacent:HCV tumor-adjacent) with Wilcoxon rank-sum tests.
Since immune infiltration is known to impact the response to immunotherapy,43 we were interested in quantifying the relationship between immune cell infiltration in matched tumor-adjacent and tumor samples. We therefore classified each sample as either being above or below the median in terms of immune infiltration. We then classified paired samples into one of four groups: (1) groups with high tumor-adjacent immune infiltration that stayed high in tumor tissue, (2) groups with high tumor-adjacent immune infiltration that dropped to low infiltration in tumor tissue, (3) groups with low tumor-adjacent immune infiltration that stayed low in the tumor tissue, and (4) groups with low tumor-adjacent immune infiltration that changed to high in the tumor tissue (Figure S8). Consistent with the high immune infiltration in HCV tumor-adjacent samples, the majority of HCV samples fell into the first two groups, with 29.5% (26/88) of samples having immune infiltration above the median in both tumor-adjacent and tumor samples and 52.3% (46/88) having immune infiltration above the median in only the tumor-adjacent samples. By contrast, HBV had only 10.5% (4/38) of samples with immune infiltration above the median in both tumor-adjacent and tumor samples and 34.2% (13/38) of samples having immune infiltration above the median in only the tumor-adjacent samples. Both HBV and HCV samples showed very low rates of samples where only the tumor sample had immune infiltration above the median (4.5%, 4/88 for HCV and 7.9%, 3/38 for HBV). The majority of HBV samples had low immune infiltration in both the tumor and tumor-adjacent samples (47.4%, 18/38) compared to a lower proportion for HCV samples (13.6%, 12/88).
We also performed immune cell deconvolution using quanTIseq35 as a second approach. The results from quanTIseq confirmed increased CD8+ T cell infiltration in the HCV tumor-adjacent samples compared to all other samples (Figures S6B and S6C). Additionally, deconvolution with quanTIseq demonstrated a decrease in M1 polarized macrophages (Figures 4C and S6B) and a corresponding increase in M2 polarized macrophages (Figures 4D and S6B) in tumor samples compared to tumor-adjacent samples. Notably, the predominant M2 polarization was consistent across both HBV- and HCV-mediated liver cancer. The difference in macrophage polarization was not detected in the results from xCell, which may be attributable to the overlap in the M1 and M2 gene signatures for xCell. In the development of quanTIseq, marker genes were selected as a subset of the gene sets proposed for xCell, with the removal of any signature genes included in the marker genes for a different cell type. Therefore, quanTIseq may be better equipped to identify differences in subpopulations of cells from a single progenitor cell that may share large numbers of marker genes. Overall, immune cell deconvolution demonstrated a strong immune cell infiltrate in the HCV tumor-adjacent tissue and a consistent immune profile between HBV and HCV tumor samples.
Sex differential expression analysis highlights the need for increased sample size for female HCC
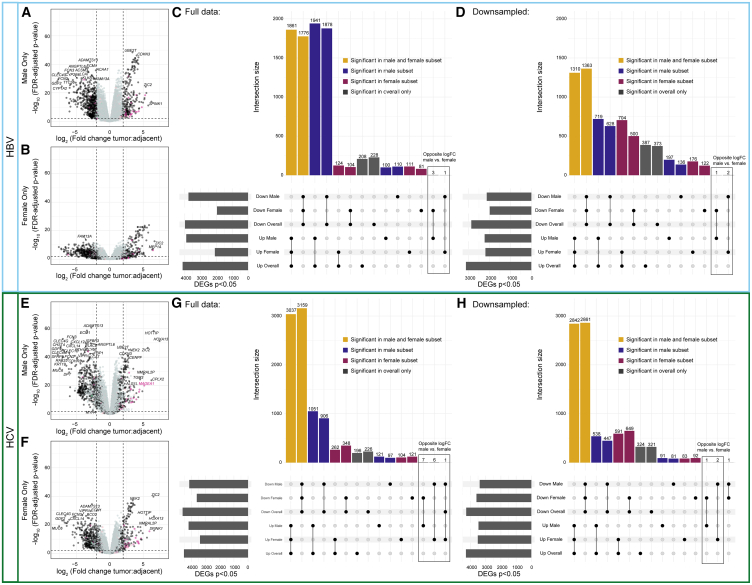
We next explored whether we could identify patterns of sex-differentially expressed genes within HBV and HCV-mediated liver cancer. We performed tumor:tumor-adjacent differential expression analyses, subset by sex and etiology (Figure 5). In HBV, we noticed a striking excess in the number of differentially expressed genes identified in the tumor:tumor-adjacent comparison in male individuals that were not identified in the female subset (Figures 5A–5C). We, therefore, repeated this analysis with random down-sampling of the male individuals and demonstrated that the number of unique differentially expressed genes in males and females was approximately equivalent (Figure 5D). We repeated this approach to study sex differences in HCV-mediated liver cancer, and similarly observed a higher number of differentially expressed genes in the male tumor:tumor-adjacent analysis when sample sizes were unequal (Figures 5E–5G) but not when we down-sampled to have equal representation among samples from males and females (Figure 5H).
Figure 5.
Tumor:tumor-adjacent differential expression in HBV and HCV samples is dominated by male differentially expressed genes
(A and B) Volcano plots of differentially expressed genes from (A) male HBV samples and (B) female HBV samples. X-linked genes are indicated in pink, Y-linked in green, and autosomal in black.
(C) Upset plot of differentially expressed genes from each of the comparisons with the full sample size.
(D) Upset plot of differentially expressed genes from each of the comparisons with down-sampling of male samples to be equal to the number of female samples. Genes shared in all comparisons are colored yellow, genes unique to males are blue, and genes unique to females are maroon.
(E and F) Volcano plots of differentially expressed genes from (E) male HCV samples and (F) female HCV samples.
(G) Upset plot of differentially expressed genes from each of the comparisons with the full sample size.
(H) Upset plot of differentially expressed genes from each of the comparisons with down-sampling of male samples to be equal to the number of female samples.
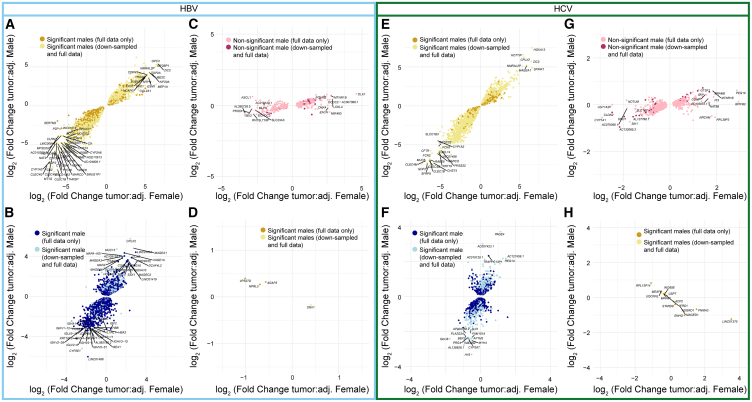
We then evaluated whether the genes identified in the segregated analyses showed similar expression changes from tumor-adjacent to tumor tissue in male and female samples, despite not passing multiple testing corrections in both sexes (Figures 6A–6H). Within HBV, we fit a linear model on all genes shared in the tumor:tumor-adjacent comparison from males and females and found that our model had an R2 value of 0.914, suggesting that 91.4% of the variability in the data is explained through the model (Figure 6A). Of note, within the genes unique to the male tumor:tumor-adjacent comparison, the R2 value is still 0.667, suggesting that there is still a high degree of similarity between the overall logFCs in males and females (Figure 6B). Within the genes significant with down-sampling, the R2 value remains high at 0.755. Within the genes unique to the female tumor:tumor-adjacent comparison, the R2 value drops to 0.446 and is 0.432 among the genes significant with down-sampling (Figure 6C). Of the genes differentially expressed in opposite directions in male tumor:tumor-adjacent vs. female tumor:tumor-adjacent, 4/4 had an absolute logFC of less than 1.5, with 3/4 having an absolute logFC less than 1, suggesting that the biological impact of these differences may be low (Figure 6D).
Figure 6.
Comparisons of the logFC values between males and females demonstrate consistency in direction and magnitude
Comparison of the logFC in males and females for (A) genes significant in both male and female tumors in the same direction in HBV, (B) genes significant only in male individuals in HBV, (C) genes significant only in female individuals in HBV, (D) genes significant in both male and female tumors in the opposite directions in HBV, (E) genes significant in both male and female tumors in the same direction in HCV, (F) genes significant only in male individuals in HCV, (G) genes significant only in female individuals in HCV, and (H) genes significant in both male and female tumors in the opposite directions in HCV. For all plots, the lighter-color genes indicate genes that were no longer significant (or gained significance for C/G) in males in the down-sampled differential expression analysis.
Within HCV, the genes shared in the tumor:tumor-adjacent comparison from males and females had an R2 value of 0.923 (Figure 6E). Consistent with the larger sample sizes available for HCV, the genes unique to the tumor:tumor-adjacent comparison in males had an R2 value of 0.490 that increased to 0.621 with down-sampling (Figure 6F). The genes unique to the tumor:tumor-adjacent comparison in females had an R2 value of 0.447 that lowered slightly to 0.403 with down-sampling (Figure 6G). Finally, among the genes differentially expressed in opposite directions for the male tumor:tumor-adjacent and female tumor:tumor-adjacent comparisons, the majority still had logFCs less than 1.5, with only LINC01370 (MIM: 617038) and PNMA3 (MIM: 300675) demonstrating logFCs greater than 1.5 in females and none in males.
Finally, we performed a direct differential expression analysis between male tumor-adjacent and female tumor-adjacent tissue as well as between male tumor and female tumor tissue, subset by etiology (Figure S9). Across all comparisons, the majority of the differentially expressed genes identified were on sex chromosomes, consistent with the expected sex differences. Of note, a larger number of autosomal genes were identified as differentially expressed between male HCV tumors and female HCV tumors, suggesting that there may be an increase in sex differences in these samples (Figure S9). Pathway enrichment analysis did not identify significantly enriched pathways in the male HBV tumor-adjacent:female HBV tumor-adjacent, male HBV tumor:female HBV tumor, or HCV:tumor-adjacent:female HCV tumor-adjacent comparisons. Within the HCV tumor:HCV tumor comparisons, several pathways were identified as enriched, including “small molecule catabolic process,” “diterpenoid metabolic process,” “xenobiotic metabolic process,” and “hormone metabolic process” (Figure S10).
Discussion
Understanding the transcriptional and immune profile differences in HCC segregated by etiology and sex is critical to drive individualized therapeutic decision-making and rational design of therapeutic interventions in HCC. Toward this end, we performed differential expression and pathway analyses on HBV- and HCV-mediated HCC to probe the underlying etiology and sex differences.
Within the tumor:tumor-adjacent differential expression analysis segregated by viral etiology, we first demonstrated large changes in the genes identified in the overall tumor:tumor-adjacent comparison versus the tumor:tumor-adjacent comparisons subset by etiology. In particular, we noted the effect of sample size on the differential expression analyses. Our sample size for HCV was more than double that of HBV, and this was reflected in a much higher consensus between the differential expression results for the overall tumor:tumor-adjacent comparison and the HCV-specific results compared to the HBV-specific results. This manifested as both a smaller number of genes shared between the overall and HBV-specific differential expression results and a larger number of differentially expressed genes identified only in the HBV-specific subset. These results highlight the importance of considering the composition of the samples when identifying differentially expressed genes in a mixed-etiology group.
Of particular interest, a small number of genes demonstrated significant logFCs in opposite directions in the tumor:tumor-adjacent comparison for HBV compared to that for HCV. While the majority of these genes were identified at low FCs, suggesting limited biological effects, a small number demonstrated large opposite FCs. Among these candidates, several have previous associations with HCC pathogenesis in the literature, including BEX2,37,38 AP1M2,39 and KRT23.40,41 BEX2 was identified as upregulated in HBV tumor tissue (logFC = 1.3, p = 0.0026) and downregulated in HCV tumor tissue (logFC = −1.9, p = 7.6 × 10−11). High BEX2 expression has been implicated in the maintenance of cancer stem cells and poor prognosis in HCC.37 Additionally, in mouse models of HCC, BEX2 has been implicated in the oncogenic pathways mediating cell proliferation and metastasis.38 AP1M2 was identified as upregulated in HBV tumor tissue (logFC = 0.95, p = 0.048) and downregulated in HCV tumor tissue (logFC = −1.92, p = 3.24 × 10−8). Upregulation in HBV tumor tissue is consistent with previous studies showing that AP1M2 upregulation by HBV is implicated in the proliferation of liver cancer cells.39 KRT23 was upregulated in HBV tumor tissue (logFC = 1.74, p = 0.022) and downregulated in HCV tumor tissue (logFC = −1.54, p = 0.00095). Previous work has suggested that knockdown of KRT23 reduced HCC cell line proliferation and metastasis.40 Overall, this demonstrates that there are genes with possible biological implications that are differentially regulated in HBV and HCV tumors.
Pathway enrichment analysis on the differentially expressed genes in the HBV tumor:tumor-adjacent and HCV tumor:tumor-adjacent comparisons demonstrate that genes uniquely differentially expressed in HCV tumor:tumor-adjacent are enriched in immune pathways. By contrast, genes uniquely differentially expressed in the HBV tumor:tumor-adjacent genes are enriched in apoptotic pathways, consistent with literature suggesting that HBV plays a significant role in modifying apoptotic pathways in HCC.44 When we further probed the differences in HBV tumor:HCV tumor and HBV tumor-adjacent:HCV tumor-adjacent comparisons, we found that the tumor-adjacent samples had more differentially expressed genes than the tumor comparison. Pathway enrichment analysis on the tumor-adjacent comparison showed significant enrichment across multiple immune pathways.
Due to the repeated observation of enriched immune pathways, we probed the immune infiltration using immune deconvolution analyses. We demonstrated that the HCV tumor-adjacent samples had the highest immune infiltration, with higher immune infiltration than any of the other groups. Of note, despite the high immune infiltration in HCV tumor-adjacent compared to HCV tumor samples, no difference was observed in the immune infiltration in the HCV tumor compared to HBV tumor samples. This is consistent with prior work17 and suggests that the tumor microenvironment converges despite differences in the surrounding regions. We also demonstrated that HBV and HCV tumors had more M2 macrophages and fewer M1 macrophages than tumor-adjacent tissue, suggesting a more immunosuppressive and pro-tumorigenic immune environment, consistent with previous work.45,46 Of note, despite known sex differences in the immune function,47,48,49 there were no clear differences in the immune infiltration in tumor-adjacent or tumor samples between males and females in this dataset.
To probe how the immune infiltration in the tumor-adjacent tissue impacts the immune infiltration in the tumor tissue, we compared immune infiltration in paired samples. We demonstrated that over half of the HCV tumors have high immune infiltration in tumor-adjacent tissue that drops down in the tumor tissue. By contrast, the majority of HBV tumors have low immune infiltration in both the tumor-adjacent and tumor tissue.17 Immune infiltration has previously been identified as a marker of response to immunotherapy.43,50 However, this work raises an interesting question of how the immune infiltration in the tumor-adjacent tissue influences immune infiltration in the emerging tumor and how this impacts treatment efficacy, especially in the context of immunotherapy.
We propose that the observed differences in immune profiles in the tumor-adjacent samples in HBV- versus HCV-mediated HCC are due to the likely timing of infection. HCV infection, which occurs later in life, is recognized as a foreign entity and therefore activates the immune system. In contrast, HBV is often inherited in a neonatal state from the mother, and may not be recognized as a foreign entity. However, we then observe that once a tumor forms, the immune infiltration is similar in both HBV- and HCV-mediated tumor samples, suggesting that tumorigenesis may result in a convergence of immune phenotypes. The convergence of immune phenotypes during tumorigenesis may be attributable to mechanical exclusion or a local immunosuppressive environment, as previously suggested.17
Finally, differential expression analyses in tumor:tumor-adjacent tissue segregated by sex were substantially limited by the low availability of female samples. Specifically, in HBV, the initial segregated analysis suggested a predominance of genes significant in only males, but down-sampling of the male samples demonstrated that this observation was a feature of sample size rather than biology. Additionally, comparisons of the logFCs in males and females suggested that there was still a strong degree of association between the logFCs of genes that were only identified as significant in males, again reaffirming that the differences are likely attributable to sample size. While the differences in sample size between males and females prohibited extensive analysis of underlying biological differences in HCC between males and females, this analysis demonstrates the importance of increasing the available sequencing data in female HCC to enable a more extensive analysis of sex-specific differentially expressed genes.
Overall, in this work, we demonstrated the importance of considering sex and viral etiology in future studies of HCC. HBV- and HCV-mediated HCC appear to converge on similar transcriptomic and immune profiles relative to the surrounding tissue. However, the impact of the differences in the tumor-adjacent tissue on treatment efficacy, particularly of immunotherapies, warrants further research. Additionally, understanding the profiles of the adjacent tissues may further illuminate the pathogenesis and immune evasion pathways in viral-mediated liver cancer. The preliminary data offered here demonstrate that there are sex differences in viral-mediated HCC that cannot be fully characterized with the limited sample sizes available. This finding highlights the importance of increased sampling of female tumors. Finally, the work on both viral- and sex-based differences in HCC underscores that the interpretation of differential expression analyses should be cautious in cases of unequal representation across groups.
Data and code availability
Whole transcriptome data from tumor and tumor-adjacent viral-mediated HCC samples were obtained from the International Cancer Genome Consortium LIRI-JP dataset (controlled access permission to K.H.B. via project DACO-1938). Tumor-adjacent samples were taken from adjacent liver tissue. Healthy liver samples from unaffected individuals were analyzed from data collected from the GTEx consortium (controlled access permission to M.A.W. via project no. 36761: “Assessing shared and divergent sex differences across disease and healthy tissues”). The code for all analyses and visualizations performed is available from https://github.com/SexChrLab/Viral_HCC_Sex_Diff.
Acknowledgments
The authors acknowledge Research Computing at Arizona State University for providing high-performance computing resources that have contributed to the research results reported within this paper. We thank Dr. Tanya Phung for her assistance with data processing. This publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM124827 to M.A.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. E.S.B. was supported by an F30 fellowship, F30CA281056.
Author contributions
Conceptualization, E.S.B., K.H.B., and M.A.W. Methodology, E.S.B., K.H.B., and M.A.W. Formal analysis, E.S.B., A.J., and H.M.N. Investigation, E.S.B., A.J., and H.M.N. Resources, K.T.H., K.H.B., and M.A.W. Writing – original draft, E.S.B., K.H.B., and M.A.W. Writing – review & editing, E.S.B., H.M.N., K.T.H., K.H.B., and M.A.W. Visualization, E.S.B. and A.J. Supervision, K.H.B. and M.A.W.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100373.
Contributor Information
Kenneth H. Buetow, Email: kbuetow@asu.edu.
Melissa A. Wilson, Email: mwilsons@asu.edu.
Web resources
Online Mendelian Inheritance in Man: https://urldefense.com/v3/__http://www.omim.org__;!!IKRxdwAv5BmarQ!a00kXVviVS1IKJltY1ENZdkuAzJiYugyqHI5EzXgklph6ey8wRmkzcqgg25p-M9qVZVuWwtFxL3K3t3HXSTnhhs$
ReviGo: http://revigo.irb.hr/
FastQC: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Supplemental information
References
- 1.Shen C., Jiang X., Li M., Luo Y. Hepatitis Virus and Hepatocellular Carcinoma: Recent Advances. Cancers. 2023;15 doi: 10.3390/cancers15020533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 3.Kanwal F., Kramer J., Asch S.M., Chayanupatkul M., Cao Y., El-Serag H.B. Risk of Hepatocellular Cancer in HCV Patients Treated With Direct-Acting Antiviral Agents. Gastroenterology. 2017;153:996–1005.e1. doi: 10.1053/j.gastro.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 4.D’souza S., Lau K.C., Coffin C.S., Patel T.R. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J. Gastroenterol. 2020;26:5759–5783. doi: 10.3748/wjg.v26.i38.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Péneau C., Imbeaud S., La Bella T., Hirsch T.Z., Caruso S., Calderaro J., Paradis V., Blanc J.-F., Letouzé E., Nault J.-C., et al. Original research: Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2022;71:616. doi: 10.1136/gutjnl-2020-323153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khatun M., Ray R.B. Mechanisms Underlying Hepatitis C Virus-Associated Hepatic Fibrosis. Cells. 2019;8 doi: 10.3390/cells8101249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.di Filippo Villa D., Navas M.-C. Vertical Transmission of Hepatitis B Virus—An Update. Microorganisms. 2023;11:1140. doi: 10.3390/microorganisms11051140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thursz M., Fontanet A. HCV transmission in industrialized countries and resource-constrained areas. Nat. Rev. Gastroenterol. Hepatol. 2014;11:28–35. doi: 10.1038/nrgastro.2013.179. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Sun H., Wang Z., Cong W., Wang J., Zeng M., Zhou W., Bie P., Liu L., Wen T., et al. Guidelines for the Diagnosis and Treatment of Hepatocellular Carcinoma (2019 Edition) Liver Cancer. 2020;9:682–720. doi: 10.1159/000509424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ESMO eUpdate: Hepatocellular Carcinoma Treatment Recommendations. https://www.esmo.org/guidelines/guidelines-by-topic/esmo-clinical-practice-guidelines-gastrointestinal-cancers/hepatocellular-carcinoma-esmo-clinical-practice-guidelines-for-diagnosis-treatment-and-follow-up/eupdate-hepatocellular-carcinoma-treatment-recommendations
- 11.Yu S.J. A concise review of updated guidelines regarding the management of hepatocellular carcinoma around the world: 2010-2016. Clin. Mol. Hepatol. 2016;22:7–17. doi: 10.3350/cmh.2016.22.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang T., Kan X., Zheng C. Immune Checkpoint Inhibitors for Advanced Hepatocellular Carcinoma: Monotherapies and Combined Therapies. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.898964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donisi C., Puzzoni M., Ziranu P., Lai E., Mariani S., Saba G., Impera V., Dubois M., Persano M., Migliari M., et al. Immune Checkpoint Inhibitors in the Treatment of HCC. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.601240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sangro B., Sarobe P., Hervás-Stubbs S., Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021;18:525–543. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Khoueiry A.B., Sangro B., Yau T., Crocenzi T.S., Kudo M., Hsu C., Kim T.-Y., Choo S.-P., Trojan J., Welling T.H., et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu A.X., Finn R.S., Edeline J., Cattan S., Ogasawara S., Palmer D., Verslype C., Zagonel V., Fartoux L., Vogel A., et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 17.De Battista D., Zamboni F., Gerstein H., Sato S., Markowitz T.E., Lack J., Engle R.E., Farci P. Molecular Signature and Immune Landscape of HCV-Associated Hepatocellular Carcinoma (HCC): Differences and Similarities with HBV-HCC. J. Hepatocell. Carcinoma. 2021;8:1399–1413. doi: 10.2147/JHC.S325959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 19.Nevola R., Tortorella G., Rosato V., Rinaldi L., Imbriani S., Perillo P., Mastrocinque D., La Montagna M., Russo A., Di Lorenzo G., et al. Gender Differences in the Pathogenesis and Risk Factors of Hepatocellular Carcinoma. Biology. 2023;12 doi: 10.3390/biology12070984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natri H.M., Wilson M.A., Buetow K.H. Distinct molecular etiologies of male and female hepatocellular carcinoma. BMC Cancer. 2019;19:951. doi: 10.1186/s12885-019-6167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Bajari R., Andric D., Gerthoffert F., Lepsa A., Nahal-Bose H., Stein L.D., Ferretti V. The International Cancer Genome Consortium Data Portal. Nat. Biotechnol. 2019;37:367–369. doi: 10.1038/s41587-019-0055-9. [DOI] [PubMed] [Google Scholar]
- 22.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jennewein D.M., Lee J., Kurtz C., Dizon W., Shaeffer I., Chapman A., Chiquete A., Burks J., Carlson A., Mason N., et al. 2023. The Sol Supercomputer at Arizona State University. [DOI] [Google Scholar]
- 24.Babraham bioinformatics - FastQC A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv. 2015 doi: 10.1101/021592. Preprint at. [DOI] [Google Scholar]
- 27.Wik E., Olin H., Haughey C., Klasson L., Dainat J. GUESSmyLT: Software to guess the RNA-Seq library type of paired and single end read files. J. Open Source Softw. 2019;4:1344. doi: 10.21105/joss.01344. [DOI] [Google Scholar]
- 28.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plattner C., Finotello F., Rieder D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol. 2020;636:261–285. doi: 10.1016/bs.mie.2019.05.056. [DOI] [PubMed] [Google Scholar]
- 36.Sturm G., Finotello F., List M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol. Biol. 2020;2120:223–232. doi: 10.1007/978-1-0716-0327-7_16. [DOI] [PubMed] [Google Scholar]
- 37.Fukushi D., Shibuya-Takahashi R., Mochizuki M., Fujimori H., Kogure T., Sugai T., Iwai W., Wakui Y., Abue M., Murakami K., et al. BEX2 is required for maintaining dormant cancer stem cell in hepatocellular carcinoma. Cancer Sci. 2021;112:4580–4592. doi: 10.1111/cas.15115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y.-D., Liu X.-Y., Fang L., Yu H.-Q., Zhang Y.-J., Chen M., Zhang L.-D., Xie C.-M. Mutant Kras and mTOR crosstalk drives hepatocellular carcinoma development via PEG3/STAT3/BEX2 signaling. Theranostics. 2022;12:7903–7919. doi: 10.7150/thno.76873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kou Y., Yan X., Liu Q., Wei X., Zhang B., Li X., Pan W., Kong F., Wang Y., Zheng K., Tang R. HBV upregulates AP-1 complex subunit mu-1 expression via the JNK pathway to promote proliferation of liver cancer cells. Oncol. Lett. 2019;18:456–464. doi: 10.3892/ol.2019.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo D., Ma W., Wang R., Li Y., Chowdhury A.T.m.M., Ren M., Lu X., He S., Lu G. 2020. KRT23 Acts as an Oncogene in Hepatocellular Carcinoma by Regulating P21 via PI3K/AKT/GSK3β Pathway. [DOI] [Google Scholar]
- 41.Kim D., Brocker C.N., Takahashi S., Yagai T., Kim T., Xie G., Wang H., Qu A., Gonzalez F.J. Keratin 23 Is a Peroxisome Proliferator-Activated Receptor Alpha–Dependent, MYC-Amplified Oncogene That Promotes Hepatocyte Proliferation. Hepatology. 2019;70:154–167. doi: 10.1002/hep.30530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuck R.N., Zha W., Edin M.L., Gruzdev A., Vendrov K.C., Miller T.M., Xu Z., Lih F.B., DeGraff L.M., Tomer K.B., et al. The cytochrome P450 epoxygenase pathway regulates the hepatic inflammatory response in fatty liver disease. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu A.X., Abbas A.R., de Galarreta M.R., Guan Y., Lu S., Koeppen H., Zhang W., Hsu C.-H., He A.R., Ryoo B.-Y., et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat. Med. 2022;28:1599–1611. doi: 10.1038/s41591-022-01868-2. [DOI] [PubMed] [Google Scholar]
- 44.Lin S., Zhang Y.-J. Interference of Apoptosis by Hepatitis B Virus. Viruses. 2017;9 doi: 10.3390/v9080230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y., Ge W., Zhou J., Gao B., Qian X., Wang W. The Role of Tumor Associated Macrophages in Hepatocellular Carcinoma. J. Cancer. 2021;12:1284–1294. doi: 10.7150/jca.51346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arvanitakis K., Koletsa T., Mitroulis I., Germanidis G. Tumor-Associated Macrophages in Hepatocellular Carcinoma Pathogenesis, Prognosis and Therapy. Cancers. 2022;14 doi: 10.3390/cancers14010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natri H., Garcia A.R., Buetow K.H., Trumble B.C., Wilson M.A. The Pregnancy Pickle: Evolved immune compensation due to pregnancy underlies sex differences in human diseases. Trends Genet. 2019;35:478–488. doi: 10.1016/j.tig.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S., Cowley L.A., Liu X.-S. Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers, and Therapeutic Strategy. Molecules. 2019;24 doi: 10.3390/molecules24183214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He F., Furones A.R., Landegren N., Fuxe J., Sarhan D. Sex dimorphism in the tumor microenvironment - From bench to bedside and back. Semin. Cancer Biol. 2022;86:166–179. doi: 10.1016/j.semcancer.2022.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Kelley R.K., Sangro B., Harris W., Ikeda M., Okusaka T., Kang Y.-K., Qin S., Tai D.W.-M., Lim H.Y., Yau T., et al. Safety, Efficacy, and Pharmacodynamics of Tremelimumab Plus Durvalumab for Patients With Unresectable Hepatocellular Carcinoma: Randomized Expansion of a Phase I/II Study. J. Clin. Oncol. 2021;39:2991–3001. doi: 10.1200/JCO.20.03555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole transcriptome data from tumor and tumor-adjacent viral-mediated HCC samples were obtained from the International Cancer Genome Consortium LIRI-JP dataset (controlled access permission to K.H.B. via project DACO-1938). Tumor-adjacent samples were taken from adjacent liver tissue. Healthy liver samples from unaffected individuals were analyzed from data collected from the GTEx consortium (controlled access permission to M.A.W. via project no. 36761: “Assessing shared and divergent sex differences across disease and healthy tissues”). The code for all analyses and visualizations performed is available from https://github.com/SexChrLab/Viral_HCC_Sex_Diff.