Abstract
Undernutrition in Bangladeshi children is associated with disruption of postnatal gut microbiota assembly; compared to standard therapy, a microbiota-directed complementary food (MDCF) significantly improved their ponderal and linear growth. Here, we characterize a fatty acid amide hydrolase (FAAH) from a growth-associated intestinal strain of Faecalibacterium prausnitzii cultured from these children. This enzyme, expressed and purified from E. coli, hydrolyzes a variety of N-acylamides, including oleoylethanolamide (OEA), neurotransmitters and quorum-sensing N-acyl homoserine lactones, and synthesizes a range of N-acylamides, notably N-acyl amino acids. Treating germ-free mice with N-oleoylarginine and N-oleolyhistidine, major products of FAAH OEA metabolism, markedly affected expression of intestinal immune function pathways. Administering MDCF to Bangladeshi children significantly reduced fecal OEA, a satiety factor whose levels were negatively correlated with the abundance and expression of their F. prausnitzii FAAH. This enzyme, structurally and catalytically distinct from mammalian FAAH, is positioned to regulate levels of a variety of bioactive molecules.
Keywords: microbiota-directed therapeutics, childhood undernutrition, gut microbial metabolism, fatty acid amide hydrolase, N-acyl amides, signal transduction
Introduction
A central question in the field of human microbiome research is how microbial communities communicate with their hosts. The pursuit of answers has revealed several examples where the community affects the production or modifies the nature of host-derived signaling molecules or produces pharmacologically active mimetics of host metabolites that affect various facets of host systems physiology. These molecules include neurotransmitters, as well as regulators of metabolic, hormonal and immune function (1–8).
Answering this central question is pertinent to understanding the contributions of the developing gut microbiome to human postnatal development. Recent work has provided evidence that co-development of the gut microbiome and host during the first two years of life is an important determinant of healthy growth. Studies of healthy and undernourished Bangladeshi children have revealed that undernutrition is associated with disruption of a program of microbial community assembly that is normally largely completed during the first 2–3 postnatal years (9). This disrupted development leaves undernourished children with gut microbial communities that appear younger (more immature) than those of their healthy, age-matched counterparts. Preclinical evidence that this immaturity is causally related to the pathogenesis of undernutrition came from transplanting intact uncultured fecal communities from age-matched children with normal anthropometry or with moderate or severe acute malnutrition (MAM or SAM), into germ-free mice fed a diet representative of that consumed by these children. Impaired growth phenotypes were transmitted by gut communities from undernourished donors compared to those from healthy donors. Machine-learning algorithms applied to these transplanted communities identified growth-discriminatory bacterial taxa that were underrepresented in the immature communities (10). These organisms served as therapeutic targets for development of combinations of culturally acceptable, affordable foods that could increase their fitness and express beneficial functions in the gut microbiota in gnotobiotic mouse and piglet models (11).
Based on the results of these preclinical studies, a microbiota-directed complementary food (MDCF-2) was developed. ‘MDCF-2’; contains chickpea, soy flour, peanut paste and green banana plus sugar, vegetable oil, and micronutrients. It was tested in a randomized, controlled feeding study of 12–18-month-old Bangladeshi children with MAM (11, 12). Compared to a conventional, ready-to-use supplementary food (RUSF), MDCF-2 (i) improved the rate of weight gain (increases in weight-for-length Z-score; WLZ), (ii) improved linear growth (length-for-age Z-score; LAZ), (iii) produced greater increases in the abundances of growth-discriminatory taxa and (iv) led to greater changes in the levels of 75 plasma protein biomarkers and mediators of musculoskeletal and CNS development, metabolic regulation and immune function (12, 13). Analyses of fecal bacterial metagenome-assembled genomes (MAGs) from the study population revealed an MDCF-2-dependent increase in the expression of pathways involved in the metabolism of MDCF-2 polysaccharides in a subset of MAGs that were positively correlated with WLZ (14).
These findings set the stage for efforts to identify the nature of signaling processes that occur between WLZ-associated bacteria and the host that underpin the growth-promoting effects of MDCFs. In this study, we identify and characterize a fatty acid amide hydrolase (FAAH) from a strain of Faecalibacterium prausnitzii cultured from a healthy Bangladeshi child who lived in the community where the MDCF clinical trial was conducted. The enzyme is encoded by a strain-restricted gene. Expression of this gene in E. coli and purification of its protein product (FAAH) disclosed that it possesses bidirectional catalytic activity; it can hydrolyze a wide variety of N-acyl amides, with oleoylethanolamide (OEA) a preferred substrate, as well as quorum sensing N-acyl homoserine lactones (AHLs). The enzyme is also able to synthesize N-acyl amides via condensation of fatty acids with amine groups from a broad range of substrates, with arginine the preferred amine donor. Combining in vitro studies with in vivo assays in gnotobiotic mice, and quantification of OEA and oleoylarginine in the feces of children enrolled in the clinical trial, revealed that this strain of F. prausnitzii and its encoded FAAH regulate the levels of a pool of diverse, pharmacologically active signaling molecules in the gut.
Results
Strain-specific F. prausnitzii degradation of N-acylethanolamides
Four- to 5-week-old germ-free C57Bl/6J mice were colonized with either 13 or 14 genome-sequenced human gut bacterial taxa; they included bacterial species previously linked with healthy microbiota maturation in 6 to 24-month-old Bangladeshi children (11, 12, 14, 15). The only difference between the two bacterial consortia was the presence or absence of a strain of F. prausnitzii (Bg7063). F. prausnitzii is a butyrate-producing bacterial species whose abundance and metabolic activity are promoted in undernourished Bangladeshi children treated with an MDCF that significantly accelerates their ponderal and linear growth (12,13). Both groups of animals were fed MDCF for 5 weeks (Methods). After euthanasia, we performed shotgun sequencing of DNA isolated from cecal contents to quantify the absolute abundances of bacterial community members (Fig. 1A), as well as untargeted liquid chromatography-quadrupole time-of-flight mass spectrometry (LC-Qtof-MS) to profile cecal metabolites.
Figure 1. F. prausnitzii strain-specific degradation of N-acylethanolamines.
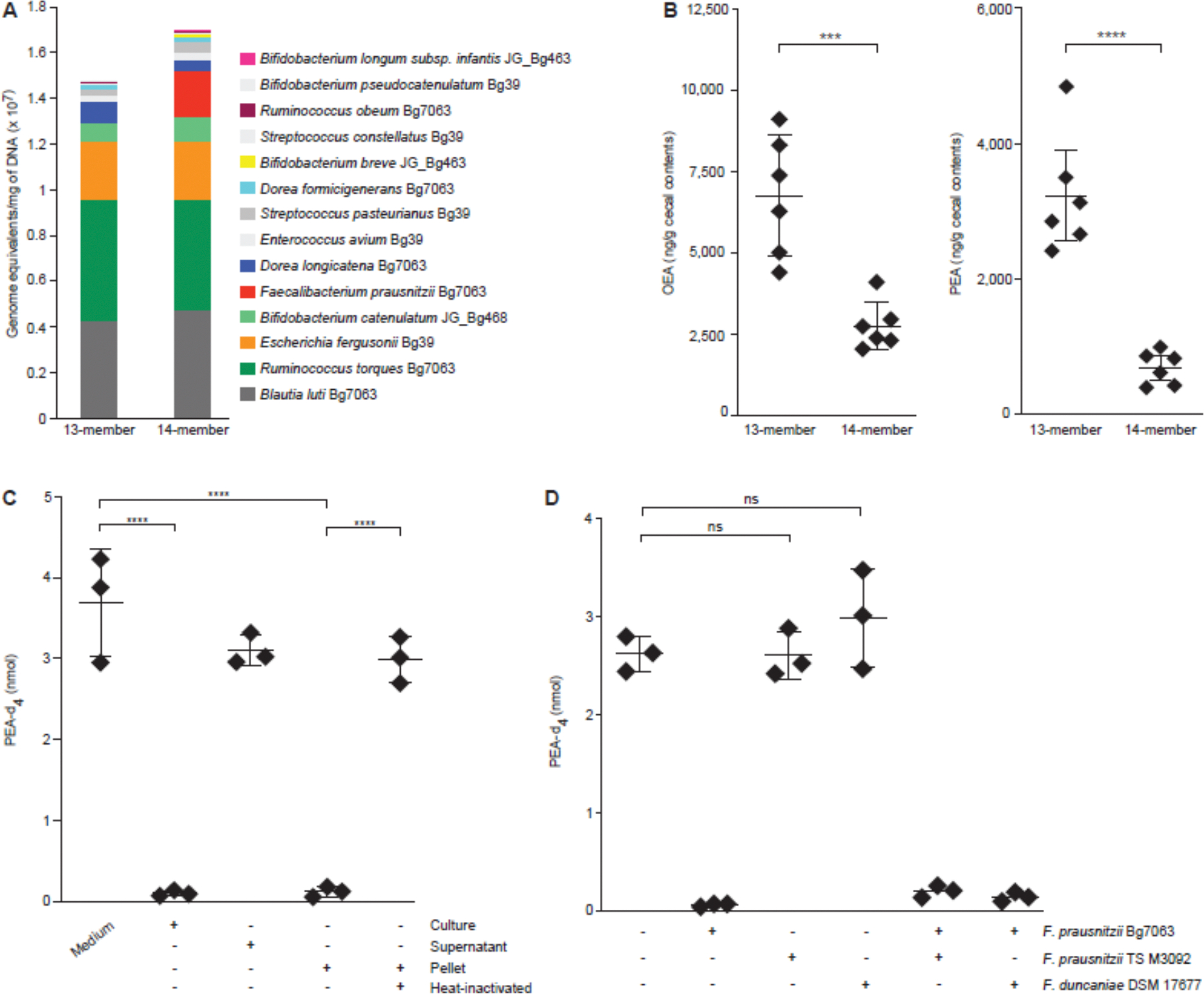
(A) Absolute abundances of organisms comprising the bacterial consortium in cecal contents collected at the time of euthanasia. (B) Oleoylethanolamide (OEA) and palmitoylethanolamide (PEA) levels in the cecal contents of animals from each of the two treatment groups. Each dot represents a sample from a mouse. Mean values ± SD are shown. ****P < 0.0001; ***P < 0.001 (t-test followed by Bonferroni correction). (C) Quantification of PEA-d4 levels using targeted UHPLC-QqQ-MS of medium and cell pellets prepared from F. prausnitzii Bg7063 monocultures after incubation with PEA-d4 for 1 h. Each dot represents a biologic replicate in the indicated treatment group. Mean values ± SD are shown. ****P < 0.0001; ***P < 0.001 (1-way ANOVA followed by post-hoc Tukey’s test). (D) Quantification of PEA-d4 levels in monocultures of the indicated F. prausnitzii strains treated with PEA-d4 for 1 h. NS, not significant (1-way ANOVA followed by post-hoc Tukey’s test).
Oleoylethanolamide (OEA) and palmitoylethanolamide (PEA) exhibited the greatest differences (reductions) in their levels in animals colonized with the 14-member compared to the 13-member consortium (table S1). OEA and PEA are N-acylethanolamides (NAEs) involved in modulating appetite and energy balance, inflammation and pain (16–19). Follow-up targeted ultra-high performance liquid chromatography-triple quadrupole mass spectrometry (UHPLC-QqQ-MS) of cecal contents confirmed that PEA and OEA levels were significantly lower in animals colonized with the F. prausnitzii-containing 14-member consortium (Bonferroni corrected t test; p=0.0005 and p=0.0012 for PEA and OEA respectively, Fig. 1B). Moreover, a statistically significant negative correlation was found between the abundance F. prausnitzii and cecal levels of OEA and PEA in animals containing the 14-member consortium (Spearman’s ρ = −0.9 and −0.8 respectively; FDR-adjusted p-value < 0.001). Together, these findings suggested that F. prausnitzii Bg7063 was involved in utilizing or transforming these NAEs in vivo.
Given that NAEs can be synthesized by the host in the small intestine, we hypothesized that pharmacologic inhibition of N-acyl phosphatidylethanolamine-specific phospholipase D (NAPE-PLD), the principal mammalian NAE biosynthetic enzyme (20, 21), might reduce gut luminal OEA and PEA levels. However, when gnotobiotic mice were colonized with the consortium of 14 bacterial strains, fed MDCF and then gavaged with a potent and selective NAPE-PLD inhibitor (LEI-401, 60 mg/kg), UHPLC-QqQ-MS disclosed that levels of OEA and PEA in serially collected fecal samples were unaltered compared to similarly colonized animals that received a sham gavage (n=5 mice/treatment group; 1-way ANOVA followed by post-hoc Tukey’s test; table S2A). Together, these results suggest that diet and/or members of the microbiota are sources of these NAEs in intestinal contents.
Supplementing monocultures of F. prausnitzii Bg7063 during the exponential phase of growth at 37 °C under anaerobic conditions with an isotope tracer (PEA-d4) followed by UHPLC-QqQ-MS, disclosed that PEA-d4 levels fell to below the limits of detection within 1 h but were unaltered in control incubations that lacked the Bg7063 strain (Fig. 1C). Follow-up experiments revealed that this activity was present in the cell pellet, but not in the medium and was inactivated by heat-treatment of the cell pellets (Fig. 1C). An isolate cultured from a Malawian child’s fecal sample in our biobank (F. prausnitzii TS M3092; (10)) and a closely related organism (F. duncaniae) had no activity in this enzymatic assay (Fig. 1D). Moreover, there were no ‘dominant negative’ effects observed when the two inactive strains were co-cultured with F. prausnitzii Bg7063 (Fig. 1D).
These results raised the possibility that F. prausnitzii possesses an enzymatic activity analogous to mammalian fatty acid amide hydrolase (FAAH) (22) which confers the ability to hydrolyze NAEs to their corresponding long-chain fatty acids. However, we found that the activity was resistant to two potent and selective inhibitors of mammalian FAAH - PF-3845 and URB597 (table S2B). PF-3845 is piperidine/piperazine urea-based inhibitor that acts via carbamylation of FAAH’s catalytic S241 nucleophile (23). URB597 is a carbamate-based irreversible inhibitor of mammalian FAAH, albeit with lower selectivity than PF-3845 (24). Taken together, our results suggested that F. prausnitzii Bg7063 encodes an enzyme that hydrolyzes NAEs but whose properties are distinct from human FAAH.
We subsequently employed untargeted LC-Qtof-MS coupled with tandem mass fragmentation to analyze product formation at multiple time points following treatment of monocultures of F. prausnitzii Bg7063 with unlabeled PEA. The most abundant reaction product had a mass-to-charge ratio [m/z] of 413.3483 and accumulated in the culture medium within an hour (fig. S1A). To confirm that this species was derived from PEA, we supplemented F. prausnitzii monocultures with PEA, labeled with 13C at the carbonyl carbon of the acyl chain, and observed that the peak shifted by exactly 1.003 Da (m/z=414.3522; Fig. 2A). The high-resolution fragmentation pattern showed large daughter ions that were also shifted by 1.003 Da and with fragments corresponding to arginine that were unchanged (Fig. 2A); therefore, we deduced that the product was an N-acyl amino acid (NAAA) - N-palmitoylarginine (C16:0-Arg). Verification of the molecular structure was obtained by (i) matching the fragmentation pattern with a synthetic C16:0-Arg standard (Fig. 2A), and (ii) purification of the product (m/z=413.3483) from larger scale monocultures using preparative HPLC, followed by NMR spectroscopy (see Methods). Using tandem mass spectrometry, we also confirmed that other NAAAs were present in the PEA treated monocultures, including N-palmitoyllysine (C16:0-Lys) and N-palmitoylhistidine (C16:0-His), albeit at lower levels than C16:0-Arg (fig. S1B).
Figure 2. Characterization and identification of F. prausnitzii FAAH.
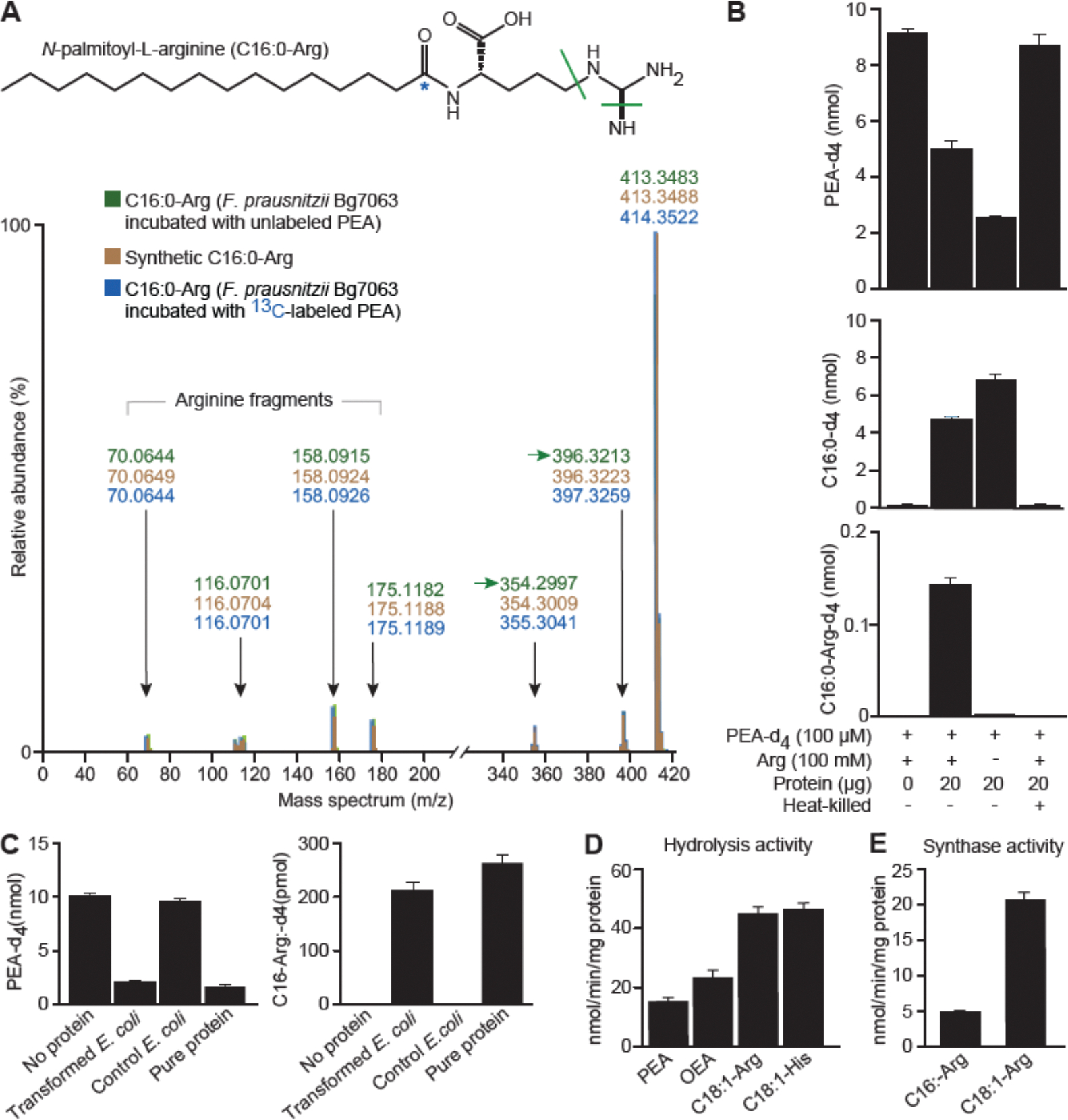
(A) Tandem mass spectrometry fragmentation profile of N-palmitoylarginine (C16:0-Arg) in F. prausnitzii Bg7063 monocultures incubated with (i) unlabeled PEA (indicated in green), C16:0-Arg produced from PEA labelled with 13C (blue asterisk) at the carbonyl carbon (highlighted in blue) and a chemically synthesized unlabeled C16:0-Arg standard (indicated in red). (B) Quantification of labeled PEA (PEA-d4), palmitic acid-d4 (C16:0-d4) and N-palmitoyl-L-arginine-d4 (C16:0-Arg-d4) in reactions containing F. prausnitzii Bg7063 cell lysates. (C) Quantification of PEA-d4 and C16:0-Arg-d4 levels in reaction mixtures containing a cell lysate from E. coli transformed with a CGOBPECO_01956 expression vector, a lysate from E. coli containing the empty vector, purified protein (2 μg) or a no addition (water) control. (D) Reaction rates for purified F. prausnitzii FAAH catalyzed hydrolysis of PEA, OEA, C18:1-Arg, and C18:1-His at 100 μM (see Methods). (E) Reaction rates for purified F. prausnitzii FAAH catalyzed condensation of C16:0 or C18:1 (100 μM) with arginine (100 mM). Mean values ± SD are plotted in panels B-D. All assays involved 3 replicates per condition.
Based on these findings, we postulated that F. prausnitzii Bg7063 contains an enzyme or enzymes capable of hydrolyzing NEAs into their respective fatty acids, as well as condensing liberated fatty acids with amino acids present in the culture medium to form NAAAs. To test this hypothesis, F. prausnitzii Bg7063 was grown in a rich medium (LYBHI) to mid-log phase; cells were subsequently pelleted by centrifugation and a cell lysate was prepared by sonication. Both the lysate and the culture supernatant were tested in a reaction mixture that contained PEA-d4 without added arginine (see Methods). 76±3% of the PEA-d4 was degraded by the lysate in one hour; this was accompanied by the appearance of an equimolar amount of palmitic acid-d4. In contrast, the cell-free supernatant obtained from the centrifugation step exhibited no detectable hydrolytic activity (fig. S1C). Addition of both arginine and PEA-d4 to the lysate at the start of the reaction yielded C16:0-Arg (Fig. 2B). Heat treatment inactivated both the degradative and putative synthetic activities of the lysate (Fig. 2B). These observations are consistent with a two-step mechanism for production of NAAAs from NAEs by the F. prausnitzii Bg7063 cell-associated enzymatic activity (see fig. S1D).
Identification of a F. prausnitzii fatty acid amide hydrolase
We employed a combination of experimental and bioinformatic approaches to identify the protein or proteins driving the proposed two-step reaction. Assays using proteins extracted from Bg7063 with SDS, CHAPS, Triton X-100, sodium deoxycholate, NP-40, or Tween-20 disclosed that NP-40 extracts contained enzymatic activity that was significantly greater than any of the other detergents tested (fig. S2A). We fractionated the NP-40 extract using ion-exchange chromatography; fractions obtained at NaCl concentrations between 400 mM and 500 mM displayed the highest PEA degradative and C16:0-Arg-d4 synthetic activities. Analysis of tryptic peptides generated from this fraction revealed that they mapped to 499 proteins in the known/predicted F. prausnitzii Bg7063 proteome (see Methods). To identify candidates for the observed enzymatic activity among these proteins, we applied two criteria: the protein(s) should (i) belong to a ceramidase, amidase, peptidase, hydrolase or ligase family and (ii) be encoded by F. prausnitzii Bg7063 but not by F. prausnitzii TS M3092 and F. duncaniae. Searches of the NCBI and UniProt databases revealed that 14 of the 499 proteins were classified as either ceramidases, amidases, peptidases, hydrolases or ligases, but only one of these was unique in F. prausnitzii Bg7063 (table S3A). This 43,355 Da protein product of the CGOBPECO_01956 locus contains 399 amino acids (table S3B), including a 30-residue N-terminal signal peptide and at least one predicted transmembrane domain (InterPro domain analysis (25)).
We next expressed the protein encoded by CGOBPECO_01956 in E. coli and purified it using His-tag affinity chromatography (fig. S2B). The purified protein, as well as a NP-40 extracts of cell pellets of (i) the E. coli strain containing the vector with the inserted CGOBPECO_01956 ORF or (ii) a control E. coli strain containing the expression vector without the insert, were tested for N-acyl amide hydrolase/synthase activity (see Methods). The results demonstrated that the lysate from CGOBPECO_01956-transfected E. coli, and the His-tagged purified protein from these cells both exhibited bidirectional enzymatic activity similar to that observed in the F. prausnitzii Bg7063 lysate assays when PEA-d4 and arginine were used as substrates. No such activity was present in control lysates from E. coli without the inserted CGOBPECO_01956 open reading frame (Fig. 2C), confirming that CGOBPECO_01956 is the likely F. prausnitzii FAAH.
Comparison of CGOBPECO_01956 and human FAAH revealed no primary sequence similarity (blastp and EMBOSS Needle (26, 27)). The conserved amidase domain present in all known FAAHs (28) is absent in CGOBPECO_01956 based on InterPro domain analysis. We then employed AlphaFold 2 (29) to predict the structure of CGOBPECO_01956. Scoring metrics based on FATCAT (30) and TM-align (31) comparisons of the resulting F. prausnitzii FAAH structure and the known structure of human FAAH (32) highlighted the minimal structural similarity between the two enzymes (TM score = 0.31886; fig. S2C). As with the earlier studies conducted in monocultures, we tested the effects of inhibitors of human FAAH on the activity of the purified F. prausnitzii enzyme. In addition to URB597 and PF-3845, we examined the effects of (i) PF-750, another highly selective, piperidine/piperazine urea-based inhibitor that acts via carbamylation of FAAH’s catalytic S241 nucleophile (34) and (ii) CAY10435 (35, 36), which contains an acyl chain similar to those present in PEA and OEA. Notably, none of these inhibitors had any discernible effects on PEA degradation by the purified protein at a concentration 2.5 μM, which is well above their IC50s for the human enzyme (fig. S2D).
We analyzed the strain specificity of CGOBPECO_01956 by examining all publicly available, fully sequenced genomes of F. prausnitzii in the NCBI genome database. Of 430 genomes, 16 encode this protein (table S3C). A BLASTP search for orthologs of CGOBPECO_01956 in the NCBI non-redundant database using strict thresholds (Score≥400, Identity≥50%, e-value≤10−6) revealed that the closest 37 orthologs are all found in strains belonging to the genus Faecalibacterium (scores: 544–817; identities: 68%-100%) (fig. S3; table S4). CGOBPECO_01956 possesses lower amino acid sequence similarity with proteins with similar annotations in other Firmicutes, e.g., those present in Christensenellaceae sp. (68% identity), Oscillospiraceae sp. (64%), Flavonifractor sp. (62%), Parablautia sp. (58%), Lachnospiraceae sp. (57%), Subdoligranulum variabile (56%), Amedibacterium intestinale (55%), and Butyrivibrio sp. (50%) (table S4). These putative orthologs are annotated as ‘C45 family peptidases’, ‘linear amide C-N hydrolases’, ‘acyl-CoA-6-aminopenicillanic acid acyltransferases’, and ‘carcinine hydrolase/isopenicillin-N N-acyltransferase family proteins’. It remains to be determined whether these predicted proteins share NAE hydrolytic activity with CGOBPECO_01956. However, E-64, a compound that covalently binds to catalytic cysteine nucleophiles (36) of the type found in enzymes in the C45 peptidase clan, had no impact on the catalytic activity of the FAAH (fig. S2D).
Catalytic properties of the purified enzyme
We further assessed the enzymatic activities of the purified recombinant FAAH towards (i) PEA-d4 and (ii) palmitic acid-d31 in the presence of arginine. The results revealed that enzyme’s hydrolytic and synthetic activities peaked at pH 7 (fig. S4A) and were not dependent upon the presence of metal ions (fig. S4B). We subsequently quantified reaction rates for the hydrolytic activity of FAAH against PEA, OEA, C18:1-Arg, and C18:1-Lys (Fig. 2D), and the rate of its condensation of palmitic and oleic acids with arginine (Fig. 2E). The results demonstrated that both the in vitro hydrolytic and synthetic activities of this bacterial FAAH exceed 4 nmol/min/mg. However, akin to human FAAH, F. prausnitzii FAAH exhibits a preference for the hydrolytic reaction, e.g., for C18:1-Arg, hydrolytic activity is significantly higher compared to synthetic activity (48.2±3.4 versus 21.1±0.9 nmol/min/mg enzyme, respectively; p<0.0001; Mann–Whitney U test) (fig. S1D).
Two key questions emerged from these findings; which fatty acids can act as substrates for N-acyl amide synthesis and what amine-containing compounds can serve as donors for condensation with a given acyl chain? To address these questions, we first performed assays using the purified enzyme, arginine and a panel of 25 saturated and unsaturated fatty acids whose chain lengths ranged from C2-C24 (table S5A). Our results indicated that C10-C20 fatty acids are preferred substrates for this FAAH synthetic activity (Fig. 3A). F. prausnitzii is a notable producer of butyrate, a C4 fatty acid that functions as a histone deacetylase inhibitor, is a ligand for several GPCRs, and benefits both intestinal epithelial energy metabolism and gut barrier function (37). However, butyric acid did not serve as a substrate in the in vitro assay. Moreover, the presence of double bonds in the acyl chain affects activity, e.g., while stearic acid (C18:0) was not a substrate, oleic acid (C18:1), which contains a single C9 cis double bond, was the preferred fatty acid substrate among those tested (Fig. 3A). Finally, we examined whether F. prausnitzii FAAH could synthesize two major mammalian endocannabinoids, anandamide and 2-arachidonoyl glycerol. No synthetic activity was detected when arachidonic acid was co-incubated with ethanolamine or glycerol in the presence of the enzyme (fig. S4E).
Figure 3. F. prausnitzii FAAH synthetic and hydrolytic activity.
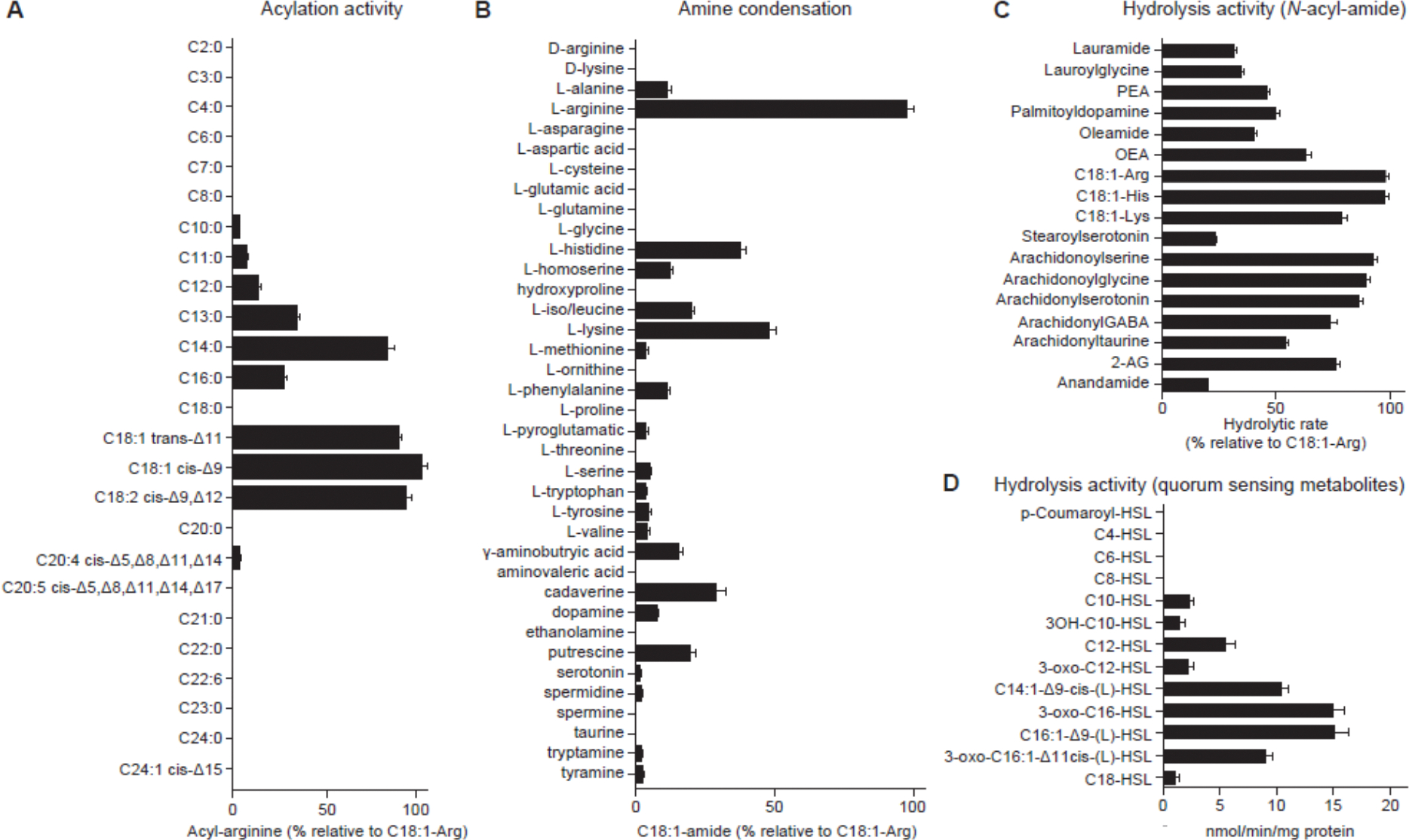
(A) Acylation activity determined in reactions containing the indicated fatty acid (100 μM), arginine (100 mM) and purified F. prausnitzii FAAH (20 μg/ml) (see Methods). The mass spectrometry ion intensity peak area of the most prominent acylation product, oleoylarginine (C18:1-Arg), was set as 100%. The relative utilization preference of the indicated fatty acid as a substrate for FAAH was calculated by dividing the peak area of the corresponding acylarginine by the peak area of C18:1-Arg. (B) Amine donor condensation activity measured by incubating each amine (10 mM) with the oleic acid (100 μM), in the presence of F. prausnitzii FAAH (20 μg/mL). The mass spectrometry ion intensity peak area of the most favored condensation product, C18:1-Arg, was set as 100%. The relative condensation activity of each amine was then calculated by dividing the peak area of the corresponding C18:1-amine by the peak area of C18:1-Arg. (C) FAAH hydrolytic activity for a variety of N-acyl amides with different acyl chains and amine donors. Results are expressed as a percentage of the hydrolytic rate of the indicated N-acyl amide divided by the hydrolytic rate of C18:1-Arg. (D) Reaction rates for F. prausnitzii FAAH catalyzed hydrolysis of quorum sensing N-acyl homoserine lactones. Mean values ± SD are plotted in panels A-D. All assays involved 3 replicates per condition.
We also tested 39 amines (table S5B) in reactions containing the purified enzyme and oleic acid; these amines included 20 L-amino acids, several D-isomers of these amino acids, neuroactive compounds (dopamine, gamma-aminobutyric acid and its methylene homolog, aminovaleric acid) as well as tyramine, taurine, spermidine, spermine, putrescine, cadaverine and ethanolamine. Twenty-one of these amines served as substrates under the in vitro reaction conditions employed (Fig. 3B). Arginine emerged as the preferred substrate followed by lysine and histidine. Only the naturally occurring L-isomers of arginine and lysine served as substrates. Incubating purified F. prausnitzii FAAH with oleic acid and arginine at concentrations typically found in the intestinal lumen (100 μM and 10 μM, respectively; refs. 11, 38, 39) produced levels of C18:1-Arg of 74.33 ± 5.6 nM after 1 h (fig. S4C); this concentration is comparable to the binding affinity of the NAE, anandamide, to the cannabinoid CB1 receptor (40, 41). To determine if C18:1-Arg was present in the gut, we conducted solid phase extraction of cecal contents from mice colonized by 14-member and 13-member communities followed by UHPLC-qTOF-MS. Notably, C18:1-Arg was only detected in samples from mice harboring the 14-member community (37.2 ± 20.8 nM) (fig. S4D), suggesting that F. prausnitzii Bg7063 utilized luminal OEA or oleic acid and arginine to synthesize C18:1-Arg.
We hypothesized that F. prausnitzii FAAH might be able to hydrolyze a broad spectrum of N-acyl amides, including but perhaps not limited to its own synthetic products. This hypothesis was tested using a range of N-acyl amides and the purified enzyme. The enzyme exhibited broad activity against a range of substrates with acyl chains ranging from C12-C20, including PEA, OEA and C18:1-Arg, C18:1-Lys and C18:1-His (Fig. 3C). Interestingly, while glycine and taurine were not substrates for the synthesis reaction in vitro, their acylated forms, lauroylglycine, arachidonoylglycine, and arachidonoyltaurine, were efficiently hydrolyzed. Similarly, just as stearic acid (C18:0) was not a substrate for the condensation reaction, its serotonin conjugate, stearoylserotonin, was a substrate for hydrolysis, albeit at low efficiency compared to arachidonylserotonin (Fig. 3C).
N-acyl homoserine lactones (AHLs) are bacterial lipids crucial for intercellular communication via quorum sensing; they influence biofilm formation and recognition of pathogenic bacteria. Analysis of 12 AHLs (table S5C) that encompass key quorum sensing molecules with acyl chain lengths ranging from C4 to C18, indicated that the purified F. prausnitzii FAAH could hydrolyze several medium and long-chain AHLs (Fig. 3D). Substrates included N-3-oxo-dodecanoyl-L-homoserine lactone (3-oxo-C12-HSL) which is produced by Pseudomonas aeruginosa and (i) impacts various aspects of the innate and adaptive immune system (42, 43), (ii) disrupts the integrity of intestinal barrier by altering tight junctions (44, 45) and (iii) has proapoptotic effects on neutrophils and modulates macrophage function to decrease inflammatory responses (46, 47). Taken together, these results indicate that this FAAH has dual synthetic and hydrolytic capabilities, with the hydrolytic activity being particularly notable for the broad range of substrates that can be utilized.
In vitro assays of the biologic activities of F. prausnitzi FAAH-derived NAAAs
Endogenous fatty acid amides have long been recognized for their ability to serve as ligands for of G-protein coupled receptors (GPCRs) and nuclear hormone receptors (NHRs), such as peroxisome proliferator-activated receptors (PPARs), that are involved in regulating a diverse range of cellular processes (48). In addition, several gut bacteria-derived N-acyl amides have been described that modulate the activity of mammalian GPCRs (49, 50). Therefore, we examined whether the conversion of OEA to other N-acyl amides could have potential pharmacologic implications through effects GPCR- and/or NHR-mediated signaling. To do so, we employed the DiscoverX cell-based reporter assay platform (see Methods) to compare OEA with three of the most prominent N-acyl amino acids produced by FAAH in our in vitro assays (i.e., C18:1-Arg, C18:1-His and C16:0-Arg), for their ability to act as agonists of 73 orphan GPCRs and 18 NHRs. All compounds were chemically synthesized, purified to >99%, and were assayed at 10 μM (a concentration chosen based on their levels document in the intestines of gnotobiotic mice and humans; see below). The results (table S6) disclosed that these compounds were agonists of various GPCRs, albeit with differing specificities and potencies. Signaling through six distinct GPCRs (GPCRs 6, 31, 39, 84, 132 and 139) was activated by at least one of the compounds (threshold, >50% activation). GPR84, which was strongly activated by OEA, has been implicated in NLRP3 inflammasome activation in macrophages in the colonic mucosa of patients with active ulcerative colitis (51). The N-acyl amino acid products of OEA hydrolysis by F. prausnitzii FAAH were much weaker agonists of this GPCR, falling below the 50% activation threshold. Three of the four compounds (C18:1-His, C16:0-Arg and OEA) were agonists of GPR 132, an orphan GPCR recognized for its response to various fatty acid amides (49, 50) and its roles in immune regulation, cell migration, macrophage activation, and hematopoiesis (52). GPR31, which enhances immune responses by inducing dendrite protrusion of intestinal CX3CR1+ cells (53), was activated by C16:0-Arg. In addition, OEA was a strong agonist of PPARδ and to a lesser extent PPARα, whereas the three N-acyl amino acids lacked this activity. Mice with an intestinal epithelial cell-specific deletion of PPARδ have enhanced susceptibility to diet-induced obesity (54); in the context of undernutrition, the hydrolysis of OEA by F. prausnitzii FAAH and potentially other NAEs in the gut might dampen signaling through PPARδ and promote weight gain.
In vivo effects of F. prausnitzi FAAH-derived N-acylamino acids
The results of these in vitro reporter assays provided evidence that substrates and products of F. prausnitzii FAAH activity have the potential to affect host physiology via a variety of signaling pathways. As an initial evaluation of their potential to elicit biological responses in vivo, we tested the effects of the two prominent NAAA products of the F. prausnitzii FAAH synthetic activity in vitro, C18:1-Arg and C18:1-His, as well as OEA in separate groups of 4–5-week-old C57Bl/6J germ-free mice fed MDCF (Fig. 4A). A dose of 8 mg/kg/day of each compound was administered as a single gavage, on experimental days 1, 2, 3, and 4. This dose was selected to achieve NAAA levels in feces in the range of 100 ng/g based on our earlier findings that (i) the level of OEA was >1000 ng/g in cecal samples harvested from gnotobiotic mice colonized with the 14-member defined community that contained F. prausnitzii (Fig. 1B) and (ii) the measured maximum conversion of OEA to C18:1-Arg with the purified FAAH was 11.2 ± 1.6%; fig. S4E). The concentration of C18:1-Arg, C18:1-His, and OEA was quantified by LC-QqQ-MS of cecal contents collected on experimental day 5. (n=5 animals/treatment group). A control group of animals was gavaged with vehicle alone (see Methods for details about the development of the dosing protocol).
Figure 4. Host effects of F. prausnitzii FAAH synthetic products and correlation between F. prausnitzii FAAH abundance and fecal levels of N-acyl amides in undernourished children.
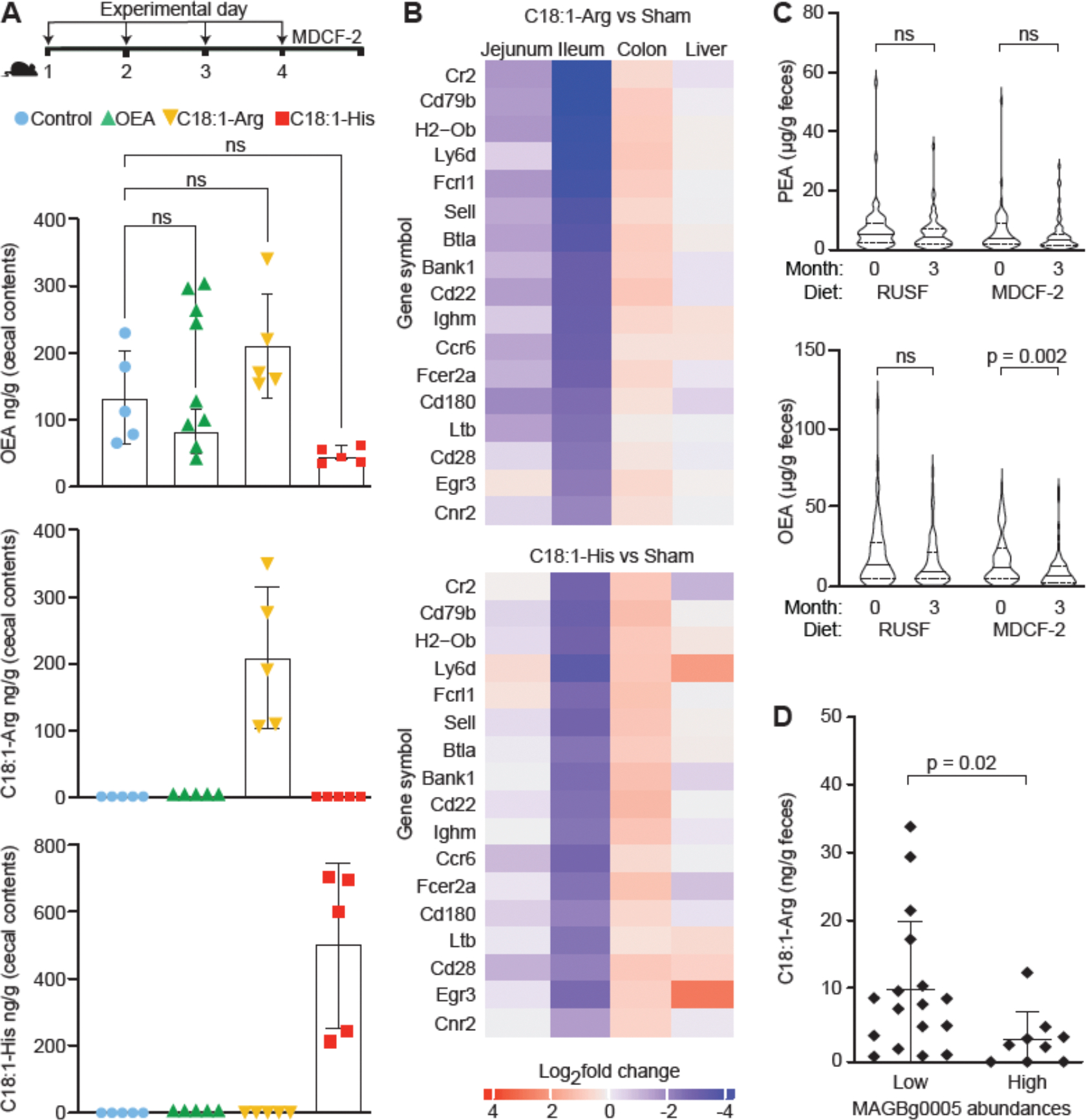
(A) Experimental design. Groups of 4–5-week-old male germ-free animals received the indicated metabolite by oral gavage on experimental days 1, 2, 3, and 4 (n=5 mice/treatment group). Cecal levels of OEA, C18:1-Arg, and C18:1-His in germ-free mice were measured using LC-QqQ-MS. Each dot represents a sample from a mouse in the indicated treatment group. Mean values ± SD are plotted. NS, not significant (1-way ANOVA followed by post-hoc Tukey’s test). (B) Heatmap of log2-fold change in levels of significantly differentially expressed transcripts (rows) that contribute to the 46 pathways (table S8C) whose expression was significantly enriched in both the ileum and jejunum upon treatment with C18:1-Arg and C18:1-His. (C) Fecal levels of OEA and PEA in children (n=117) prior to- and 3-months after initiation of RUSF or MDCF-2 treatment. Data are visualized using violin plots; median values for each analyte are shown. p-values shown were based on Wilcoxon matched-pairs signed rank test. NS, not significant. (D) Fecal levels of C18:1-Arg in samples obtained from the ‘low MAGBg0005 group’ and ‘high MAGBg0005 group’. The p-value was calculated using a Mann-Whitney test.
Oral administration of OEA did not elevate levels of OEA in cecal contents above those in control mice (p>0.18; t-test; Fig. 4B), presumably due to small intestinal absorption/metabolism. However, administration of C18:1-Arg and C18:1-His resulted in significantly higher levels of these molecules in cecal contents (to 208.49 ± 47.44 ng/g and 504.81 ± 110.21 ng/g, respectively); their levels were below the limit of detection in control animals (p<0.0001;1-way ANOVA followed by post-hoc Tukey’s test) (Fig. 4B). Neither C18:1-Arg or C18:1-His were detected in portal vein blood sampled at the time of euthanasia. Since OEA, C18:1-Arg and C18:1-His were administered by gavage at equivalent doses, these results indicate that C18:1-Arg and C18:1-His are either absorbed in the small intestine less efficiently and/or are more resistant than OEA to degradation in the gut.
The effects of C18:1-Arg and C18:1-His on intestinal gene expression in these mice was ascertained by performing RNA-Seq on jejunum, ileum, colon, and liver samples collected at the time of euthanasia (see table S7 for log2-fold change and FDR adjusted p-values). It is challenging to relate changes in gene expression using bulk RNA-Seq to activation of specific orphan GPCRs in the intestine of germ free animals in response to previously uncharacterized ligands due to limited understanding of the downstream target genes of these receptors. With this caveat in mind, we turned to Gene Set Enrichment Analysis (GSEA) to identify Gene Ontology (GO) Biological Pathway terms that were significantly enriched for differentially expressed genes; the results, summarized in table S8A,B, identified 46 pathways that were significantly enriched by treatment with both NAAAs in the jejunum and ileum, but not in the colon or liver (Fig. 4C). All these pathways are related to immune responses (table S8C) and included significant decreases in the expression of genes involved in (i) various facets of B-cell regulation (CD22, a critical modulator of B cell receptor (BCR) signaling, Cd180, Ly6d, Ighm, Bank1, Fcrl1, and Cd79b) (51–53), (ii) T cell activation/regulation (BTLA, Cd28 and Egr3) (54–56), (iii) formation and maintenance of lymphoid tissue (CCR6, Ltb) (57, 58) and (iv) the MHC class II complex (H2-O) (59).
Effects of MDCF-2 administration on fecal levels of NAEs in undernourished children
To examine the relevance of these in vitro and in vivo results to the human gut microbiota, we turned to fecal samples obtained from 12–18-month-old Bangladeshi children with moderate acute malnutrition who were enrolled in the clinical trial that compared the efficacy of dietary supplementation with MDCF-2 versus a standard ready-to-use-supplementary food (RUSF). This 3-month study revealed that compared to RUSF, MDCF-2 treatment increased the rate of weight gain in recipient children and increased the abundance of several bacterial taxa, including F. prausnitzii, in their gut communities (12). LC-MS measurements of NAE levels in fecal samples collected before and after treatment revealed that levels of OEA were significantly reduced by MDCF-2 but not by RUSF (p=0.002 and 0.17 respectively; Wilcoxon matched-pairs signed rank test; n=234 samples from 117 children) (Fig. 4D). PEA levels were not significantly affected by either dietary intervention (Fig. 4D).
Shotgun sequencing of serially collected fecal samples from trial participants had previously allowed us to identify 75 metagenome assembled genomes (MAGs) that were positively correlated with WLZ, including those assigned to F. prausnitzii (14). Among the 13 F. prausnitzii MAGs in this dataset, only MAG Bg0005 had a complete ORF matching that of F. prausnitzii FAAH (CGOBPECO_01956) (table S9). A statistically significant inverse correlation was observed between fecal OEA levels and the abundance of MAG Bg0005 in feces from MDCF-2-treated children (Spearman’s ρ = −0.228, p-value = 0.014). Analysis of microbial RNA-Seq data generated from the same fecal samples (14) disclosed a significant negative correlation between expression of the F. prausnitzii FAAH and fecal OEA levels (Spearman’s ρ = −0.31, p-value = 0.007) (table S10).
We subsequently applied a solid-phase extraction protocol (see Methods) to 26 fecal samples from children in the MDCF-2 arm of the clinical trial. Samples were selected based on the abundance of MAG Bg0005 (n=17 samples comprising a ‘low MAGBg0005 group’ where it represented ≤0.1% of total MAGs, and n= 9 in a ‘high MAG Bg0005 group’ where it comprised ≥15% of all MAGs). UHPLC-qTOF-MS analysis disclosed that (i) C18:1-Arg was the most consistently detected N-acyl amino acid (23 of 26 samples) (table S11), and (ii) fecal levels of this NAAA were significantly lower in the ‘high’ compared to the ‘low’ MAG Bg0005 group (p=0.02; Mann-Whitney test; Fig. 4E). Furthermore, the negative correlation between MAG Bg0005 abundance and levels of C18:1-Arg was statistically significant (Spearman’s ρ = −0.4, p-value = 0.04). Together, these results provide support for the notion that F. prausnitzii FAAH modulates gut luminal levels of N-acylamides.
Discussion
The initial motivation for our study was to determine if a microbiota-directed complementary food (MDCF) that improves the growth of children with moderate acute malnutrition could induce chemical changes in the gut luminal metabolic environment through effects mediated by one or more of the bacterial targets of the MDCF. We have identified a microbial FAAH in a human gut bacterium, F. prausnitzii that exhibits both hydrolytic and synthetic activities towards a broad range of N-acylated amides. The presence of a predicted transmembrane domain and rapid hydrolysis of labelled PEA administered to monocultures of F. prausnitzii with appearance of labeled products in the culture medium suggest that this FAAH is an extracellular facing enzyme. While further structural and mechanistic studies are required for a comprehensive characterization, its discovery underscores how regulation of N-acylamides in the gut luminal environment has a microbial dimension and defines a potentially manipulatable pool of diverse metabolites with various functions.
Human FAAH has received much attention due to its central role in regulating endocannabinoids - compounds that are crucial in controlling various physiological functions including appetite, mood, memory, and pain (16). FAAH, which is highly conserved across eukaryotes, regulates levels of these N-acylamides through a conserved amidase domain; this enzyme has also recently been shown to have the capacity to synthesize and hydrolyze N-acyl amino acids (60). The F. prausnitzii FAAH, while also possessing dual hydrolase/synthase activity, lacks the signature amidase domain of mammalian FAAH. Moreover, it is not sensitive to existing FAAH inhibitors and lacks sequence similarity to its eukaryotic counterparts. Further, the bacterial enzyme has limited phylogenetic distribution, being restricted to a small number of published genomes that belong to members of the genus Feacalibacterium, with lower similarity to predicted proteins in other gut Firmicutes, such as Christensenellaceae sp., Oscillospiraceae sp. and Flavonifractor sp. It remains to be determined whether putative FAAH orthologs encoded by these microbiota members are expressed in vitro and in vivo and if enzymatically active, what their substrate preferences and products are. This knowledge is important given that F. prausnitzii does not exist in isolation and it is possible that hydrolytic and/or synthetic activities of FAAH orthologs encoded by other community members contribute to the host effects of FAAH-derived metabolites.
Human gut-derived F. prausnitzii FAAH has a broad range of substrates, with the ability to irreversibly hydrolyze N-acylethanolamines (NAEs) while being capable of reversible hydrolysis and synthesis of variety of bioactive N-acylamides, including N-acylated amino acids and N-acylated forms of neuroactive compounds (dopamine, GABA). Our results provide a rationale for a comprehensive, albeit ambitious effort to characterize the potential of these newly identified F. prausnitzii FAAH products to act at physiologically relevant levels as agonists and/or antagonists of different receptors, including GPCRs and NHRs.
OEA, whose levels increase in the intestinal epithelium levels after food intake, signals via peroxisome proliferator activated receptors (PPARs) to control the expression of genes involved in absorption and metabolism of fatty acids and activates hypothalamic oxytocinergic neurons to induce satiety (61, 62). Given that undernutrition is associated with poor appetite (63), it is intriguing that in a randomly controlled clinical trial, MDCF-2 treatment of Bangladeshi children with acute malnutrition improved their growth and reduced fecal OEA levels compared to standard therapy. Future studies are needed to assess the relationship between intestinal OEA levels, FAAH abundance/activity and appetite; this includes application of validated tools to assess appetite and satiety in children in low-income settings (64), as well as measurements of circulating levels of key regulators of appetite such as GLP-1, leptin and ghrelin.
While there is already considerable interest in developing F. prausnitzii as a next-generation probiotic for its butyrate producing and anti-inflammatory activities (65), the substrate flexibility of its FAAH offers the potential for hydrolysis and synthesis of a multitude of compounds with a variety of pharmacological activities. For example, administration of an FAAH containing F. prausnitzii strain could shift the equilibrium in the pool of gut N-acyl amides by converting these compounds into other N-acyl amine derivatives. This might be advantageous in certain immunoinflammatory states: e.g., in patients with inflammatory bowel diseases, where elevated levels of PEA and OEA have been associated with reduced levels of F. prausnitzii (66, 67). An additional therapeutic approach could be to administer selected dietary ingredients in a synbiotic formulation with FAAH-containing F. prausnitzii to enable the synthesis of specific, functionally active N-acyl amides in the gut. Finally, the potent hydrolyzing capability of F. prausnitzii FAAH against medium to long chain N-acyl homoserine lactones, also has therapeutic implications. AHLs, used in quorum sensing by Gram-negative bacteria, are crucial for the virulence of pathogens such as Salmonella enteritidis and Pseudomonas aeruginosa. Notably, 3-oxo-C12-HSL and C12-HSL, known for their toxicity to innate and adaptive immune systems (68), can be degraded by F. prausnitzii FAAH, potentially offering host protection.
In summary, the identification of a gut bacterial FAAH with broad substrate specificity and the ability to regulate levels of a variety of bioactive metabolites at the gut mucosa has the potential to provide new insights into how members of our gut microbial community control the levels of signaling molecules with direct effects of host and gut community functions. Moreover, the discovery of this enzyme opens the door to future development of small molecule regulators of the enzyme’s synthetic versus hydrolytic activities that have potential therapeutic applications.
Materials and methods summary
Supplementary Materials include details of methods employed for gnotobiotic mouse husbandry, F. prausnitzii Bg7063 monoculture experiments, purification of palmitoylarginine and NMR analysis, targeted LC-QqQ-MS untargeted LC-Qtof-MS analyses, purification of FAAH activity from F. prausnitzii Bg7063, expression and purification of F. prausnitzii FAAH from E. coli. In addition, we provide descriptions of methods used for measurement of N-acyl amide hydrolase/synthase activities, FAAH inhibitor studies, the GPCR/NHR screen of NAAAs, administration of NAAAs to germ-free mice, host RNA-Seq, solid phase extraction of N-acyl amides from human fecal samples and phylogenetic analysis of F. prausnitzii FAAH orthologs.
Supplementary Material
Acknowledgments
We are indebted to David O’Donnell and Maria Karlsson for their assistance with mouse husbandry. All NMR experiments were performed at the CCIC NMR facility at The Ohio State University. Protein identification analyses were performed by the Mass Spectrometry Technology Access Center (MTAC@MGI) at the McDonnell Genome Institute at Washington University School of Medicine. LEI-401 was a gift from Dr. Mario van der Stelt (Leiden University, The Netherlands). This work was made possible by a long-standing collaborative research program involving members of International Center for Diarrhoeal Disease Research, Bangladesh (icddr,b), and led by Dr. Tahmeed Ahmed.
Funding
This work was supported by National Institutes of Health grant DK30292.
Footnotes
Competing interests: The authors declare no competing interests. J.C., S.V., M.J.B and J.I.G. are inventors on a patent application (application number to be added at galley proof stage) submitted by Washington University in St. Louis that covers therapeutic applications of F. prausnitzii FAAH.
Ethics statement
Fecal samples were collected with parental informed consent from Bangladeshi children with moderate acute malnutrition who were enrolled in a previously reported clinical trial (12; ClinicalTrials.gov identifier: NCT04015999) with approval from the International Center for Diarrhoeal Disease Research, Bangladesh (icddr,b) Ethical Review Committee (Protocol PR-18073). F. prausznitzii Bg7063 was cultured from a fecal sample from a healthy Bangladeshi child collected with informed consent as previously reported (11; NCT02734264). Collection and use of the biospecimens was approved by the Washington University Human Research Protection Office (IRB ID# 201111065). Experiments using gnotobiotic mice were performed with approval from the Washington University Animal Studies Committee (Protocol# 23–0271).
Data deposition: DNA shotgun sequencing and RNA-Seq datasets generated in this study have been deposited at the European Nucleotide Archive under accession number PRJEB72659. Long-read genome sequences for F. prausnitzii TS M3092 and Bg7063 have been deposited in the National Center for Biotechnology Information public reference database. The raw mass spectrometry data files have been deposited in the Dryad repository under DOI: 10.5061/dryad.98sf7m0rt. The Nexus-formatted file for generating the phylogenetic tree in Fig. S3 and its associated FASTA file are available at Dryad under https://doi.org/10.5061/dryad.z34tmpgp8.
Code Availability: All software used was from publicly available sources.
Data and materials availability
Material Transfer Agreements between the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) and Washington University in St. Louis were established for use of the human biospecimens described in this study.
References
- 1.Donia MS, Fischbach MA, Small molecules from the human microbiota. Science 349, 1254766 (2015). doi: 10.1126/science.1254766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silpe JE, Balskus EP, Deciphering Human Microbiota–Host Chemical Interactions. ACS Cent. Sci. 7, 20–29 (2021). doi: 10.1021/acscentsci.0c01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shine EE, Crawford JM, Molecules from the Microbiome. Annu. Rev. Biochem. 90, 789–815 (2021). doi: 10.1146/annurev-biochem-080320-115307 [DOI] [PubMed] [Google Scholar]
- 4.Cohen LJ, Esterhazy D, Kim S-H, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, Chu J, Vila-Farres X, Kaplitt J, Rogoz A, Calle PY, Hunter C, Bitok JK, Brady SF, Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53 (2017). doi: 10.1038/nature23874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colosimo DA, Kohn JA, Luo PM, Piscotta FJ, Han SM, Pickard AJ, Rao A, Cross JR, Cohen LJ, Brady SF, Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors. Cell Host Microbe 26, 273–282 (2019). doi: 10.1016/j.chom.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agus A, Clément K, Sokol H, Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 70, 1174–1182 (2021). doi: 10.1136/gutjnl-2020-323071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massey WJ, Brown JM, Microbial regulation of cholesterol homeostasis. Nat. Microbiol. 7, 1327–1328 (2022). doi: 10.1038/s41564-022-01186-w [DOI] [PubMed] [Google Scholar]
- 8.Yang W, Cong Y, Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 18, 866–877 (2021). doi: 10.1038/s41423-021-00661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA Jr, Ahmed T, Gordon JI, Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014). doi: 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan Y, Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, Gordon JI, Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016). doi: 10.1126/science.aad3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehrig JL, Venkatesh S, Chang H-W, Hibberd MC, Kung VL, Cheng J, Chen RY, Subramanian S, Cowardin CA, Meier MF, O’Donnell D, Talcott M, Spears LD, Semenkovich CF, Henrissat B, Giannone RJ, Hettich RL, Ilkayeva O, Muehlbauer M, Newgard CB, Sawyer C, Head RD, Rodionov DA, Arzamasov AA, Leyn SA, Osterman AL, Hossain MI, Islam M, Choudhury N, Sarker SA, Huq S, Mahmud I, Mostafa I, Mahfuz M, Barratt MJ, Ahmed T, Gordon JI, Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science 365, eaau4732 (2019). doi: 10.1126/science.aau4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen RY, Mostafa I, Hibberd MC, Das S, Mahfuz M, Naila NN, Islam MM, Huq S, Alam MA, Zaman MU, Raman AS, Webber D, Zhou C, Sundaresan V, Ahsan K, Meier MF, Barratt MJ, Ahmed T, Gordon JI, A Microbiota-Directed Food Intervention for Undernourished Children. N. Engl. J. Med. 384, 1517–1528 (2021). doi: 10.1056/NEJMoa2023294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mostafa I, Hibberd MC, Hartman SJ, Rahman MHH, Mahfuz M, Hasan SMT, Ashorn P, Barratt MJ, Ahmed T, Gordon JI A microbiota-directed complementary food intervention in 12–18-month-old Bangladeshi children improves linear growth. EBioMedicine 104, 105166 (2024) doi: 10.1016/j.ebiom.2024.105166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hibberd MC, Webber DM, Rodionov DA, Henrissat S, Chen RY, Zhou C, Lynn HM, Wang Y, Chang H-W, Lee EM, Lelwala-Guruge J, Kazanov MD, Arzamasov AA, Leyn SA, Lombard V, Terrapon N, Henrissat B, Castillo JJ, Couture G, Bacalzo NP, Chen Y, Lebrilla CB, Mostafa I, Das S, Mahfuz M, Barratt MJ, Osterman AL, Ahmed T, Gordon JI, Bioactive glycans in a microbiome-directed food for malnourished children. Nature 625, 157–165 (2024). doi: 10.1038/s41586-023-06838-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang HW, Lee EM, Wang Y, Zhou C, Pruss KM, Henrissat S, Chen RY, Kao C, Hibberd MC, Lynn HM, Webber DM, Crane M, Cheng J, Rodionov DA, A Arzamasov A, Castillo JJ, Couture G, Chen Y, Balcazo NP, Lebrilla CB Jr, Terrapon N, Henrissat B, Ilkayeva O, Muehlbauer MJ, Newgard CB, Mostafa I, Das S, Mahfuz M, Osterman AL, Barratt MJ, Ahmed T, Gordon JI, Prevotella copri and microbiota members mediate the beneficial effects of a therapeutic food for malnutrition. Nat. Microbiol. 9, 922–937 (2024). doi: 10.1038/s41564-024-01628-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn K, Johnson DS, Cravatt BF, Fatty acid amide hydrolase as a potential therapeutic target for the treatment of pain and CNS disorders. Expert Opin. Drug Discov. 4, 763–784 (2009). doi: 10.1517/17460440903018857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cani PD, Plovier H, Van Hul M, Geurts L, Delzenne NM, Druart C, Everard A, Endocannabinoids — at the crossroads between the gut microbiota and host metabolism. Nat. Rev. Endocrinol. 12, 133–143 (2016). doi: 10.1038/nrendo.2015.211 [DOI] [PubMed] [Google Scholar]
- 18.Hansen HS, Role of anorectic N-acylethanolamines in intestinal physiology and satiety control with respect to dietary fat. Pharmacol. Res. 86, 18–25 (2014). doi: 10.1016/j.phrs.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 19.Leung D, Saghatelian A, Simon GM, Cravatt BF, Inactivation of N -Acyl Phosphatidylethanolamine Phospholipase D Reveals Multiple Mechanisms for the Biosynthesis of Endocannabinoids. Biochemistry 45, 4720–4726 (2006). doi: 10.1021/bi060163l [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N, Molecular Characterization of a Phospholipase D Generating Anandamide and Its Congeners. J. Biol. Chem. 279, 5298–5305 (2004). doi: 10.1074/jbc.M306642200 [DOI] [PubMed] [Google Scholar]
- 21.Mock ED, Mustafa M, Gunduz-Cinar O, Cinar R, Petrie GN, Kantae V, Di X, Ogasawara D, Varga ZV, Paloczi J, Miliano C, Donvito G, Van Esbroeck ACM, Van Der Gracht AMF, Kotsogianni I, Park JK, Martella A, Van Der Wel T, Soethoudt M, Jiang M, Wendel TJ, Janssen APA, Bakker AT, Donovan CM, Castillo LI, Florea BI, Wat J, Van Den Hurk H, Wittwer M, Grether U, Holmes A, Van Boeckel CAA, Hankemeier T, Cravatt BF, Buczynski MW, Hill MN, Pacher P, Lichtman AH, Van Der Stelt M, Discovery of a NAPE-PLD inhibitor that modulates emotional behavior in mice. Nat. Chem. Biol. 16, 667–675 (2020). doi: 10.1038/s41589-020-0528-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB, Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 384, 83–87 (1996). doi: 10.1038/384083a0 [DOI] [PubMed] [Google Scholar]
- 23.Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF, Discovery and Characterization of a Highly Selective FAAH Inhibitor that Reduces Inflammatory Pain. Chem. Biol. 16, 411–420 (2009). doi: 10.1016/j.chembiol.2009.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathuria S, Gaetani S, Fegley D, Valiño F, Duranti A, Tontini A, Mor M, Tarzia G, Rana GL, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D, Modulation of anxiety through blockade of anandamide hydrolysis. Nat. Med. 9, 76–81 (2003). doi: 10.1038/nm803 [DOI] [PubMed] [Google Scholar]
- 25.Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, Gough J, Haft DH, Letunić I, Marchler-Bauer A, Mi H, Natale DA, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, InterPro in 2022. Nucleic Acids Res. 51, D418–D427 (2023). doi: 10.1093/nar/gkac993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). doi: 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice P, Longden I, Bleasby A, EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000). doi: 10.1016/s0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 28.Aziz M, Chapman KD, Fatty Acid Amide Hydrolases: An Expanded Capacity for Chemical Communication? Trends Plant Sci. 25, 236–249 (2020). doi: 10.1016/j.tplants.2019.11.002 [DOI] [PubMed] [Google Scholar]
- 29.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y, Godzik A, FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32, W582–W585 (2004). doi: 10.1093/nar/gkh430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005). doi: 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mileni M, Johnson DS, Wang Z, Everdeen DS, Liimatta M, Pabst B, Bhattacharya K, Nugent RA, Kamtekar S, Cravatt BF, Ahn K, Stevens RC, Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc. Natl. Acad. Sci. USA 105, 12820–12824 (2008). doi: 10.1073/pnas.0806121105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn K, Johnson DS, Fitzgerald LR, Liimatta M, Arendse A, Stevenson T, Lund Eric. T., Nugent RA, Nomanbhoy TK, Alexander JP, Cravatt BF, Novel Mechanistic Class of Fatty Acid Amide Hydrolase Inhibitors with Remarkable Selectivity. Biochemistry 46, 13019–13030 (2007). doi: 10.1021/bi701378g [DOI] [PubMed] [Google Scholar]
- 34.Boger DL, Sato H, Lerner AE, Hedrick MP, Fecik RA, Miyauchi H, Wilkie GD, Austin BJ, Patricelli MP, Cravatt BF, Exceptionally potent inhibitors of fatty acid amide hydrolase: The enzyme responsible for degradation of endogenous oleamide and anandamide. Proc. Natl. Acad. Sci. USA 97, 5044–5049 (2000). doi: 10.1073/pnas.97.10.5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glaser ST, Gatley SJ, Gifford AN, Ex Vivo Imaging of Fatty Acid Amide Hydrolase Activity and Its Inhibition in the Mouse Brain. J. Pharmacol. Exp. Ther. 316, 1088–1097 (2006). doi: 10.1124/jpet.105.094748 [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto K, Mizoue K, Kitamura K, Tse WC, Huber CP, Ishida T, Structural basis of inhibition of cysteine proteases by E-64 and its derivatives. Biopolymers 51, 99–107 (1999). doi: [DOI] [PubMed] [Google Scholar]
- 37.Hodgkinson K, El Abbar F, Dobranowski P, Manoogian J, Butcher J, Figeys D, Mack D, Stintzi A, Butyrate’s role in human health and the current progress towards its clinical application to treat gastrointestinal disease. Clin. Nutr. 42, 61–75 (2023). doi: 10.1016/j.clnu.2022.10.024 [DOI] [PubMed] [Google Scholar]
- 38.Song EM, Byeon J-S, Lee SM, Yoo HJ, Kim SJ, Lee S-H, Chang K, Hwang SW, Yang D-H, Jeong J-Y, Fecal Fatty Acid Profiling as a Potential New Screening Biomarker in Patients with Colorectal Cancer. Dig. Dis. Sci. 63, 1229–1236 (2018). doi: 10.1007/s10620-018-4982-y [DOI] [PubMed] [Google Scholar]
- 39.Lüneburg N, Xanthakis V, Schwedhelm E, Sullivan LM, Maas R, Anderssohn M, Riederer U, Glazer NL, Vasan RS, Böger RH, Reference Intervals for Plasma L-Arginine and the L-Arginine:Asymmetric Dimethylarginine Ratio in the Framingham Offspring Cohort. J. Nutr. 141, 2186–2190 (2011). doi: 10.3945/jn.111.148197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thieme U, Schelling G, Hauer D, Greif R, Dame T, Laubender RP, Bernhard W, Thieme D, Campolongo P, Theiler L, Quantification of anandamide and 2-arachidonoylglycerol plasma levels to examine potential influences of tetrahydrocannabinol application on the endocannabinoid system in humans. Drug Test. Anal. 6, 17–23 (2014). doi: 10.1002/dta.1561 [DOI] [PubMed] [Google Scholar]
- 41.Redlich C, Dlugos A, Hill MN, Patel S, Korn D, Enneking V, Foerster K, Arolt V, Domschke K, Dannlowski U, Redlich R, The endocannabinoid system in humans: significant associations between anandamide, brain function during reward feedback and a personality measure of reward dependence. Neuropsychopharmacology 46, 1020–1027 (2021). doi: 10.1038/s41386-020-00870-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coquant G, Aguanno D, Pham S, Grellier N, Thenet S, Carrière V, Grill J-P, Seksik P, Gossip in the gut: Quorum sensing, a new player in the host-microbiota interactions. World J. Gastroenterol. 27, 7247–7270 (2021). doi: 10.3748/wjg.v27.i42.7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao Y, Zou H, Li J, Song T, Lv W, Wang W, Wang Z, Tao S, Impact of quorum sensing signaling molecules in gram-negative bacteria on host cells: current understanding and future perspectives. Gut Microbes 14, 2039048 (2022). doi: 10.1080/19490976.2022.2039048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vikström E, Bui L, Konradsson P, Magnusson K-E, Role of calcium signalling and phosphorylations in disruption of the epithelial junctions by Pseudomonas aeruginosa quorum sensing molecule. Eur. J. Cell Biol. 89, 584–597 (2010). doi: 10.1016/j.ejcb.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 45.Yadav VK, Singh PK, Kalia M, Sharma D, Singh SK, Agarwal V, Pseudomonas aeruginosa quorum sensing molecule N-3-oxo-dodecanoyl-l-homoserine lactone activates human platelets through intracellular calcium-mediated ROS generation. Int. J. Med. Microbiol. 308, 858–864 (2018). doi: 10.1016/j.ijmm.2018.07.009 [DOI] [PubMed] [Google Scholar]
- 46.Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, Standiford TJ, Ishiguro M, Yamaguchi K, The Pseudomonas aeruginosa Autoinducer N -3-Oxododecanoyl Homoserine Lactone Accelerates Apoptosis in Macrophages and Neutrophils. Infect. Immun. 71, 5785–5793 (2003). doi: 10.1128/IAI.71.10.5785-5793.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coquant G, Grill JP, Seksik P, Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 11, 1827 (2020). doi: 10.3389/fimmu.2020.01827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farrell EK, Merkler DJ, Biosynthesis, degradation and pharmacological importance of the fatty acid amides. Drug Discov. Today 13, 558–568 (2008). doi: 10.1016/j.drudis.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen LJ, Kang HS, Chu J, Huang Y-H, Gordon EA, Reddy BVB, Ternei MA, Craig JW, Brady SF, Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. USA 112, 4825–4834 (2015). doi: 10.1073/pnas.1508737112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang FY, Siuti P, Laurent S, Williams T, Glassey E, Sailer AW, Gordon DB, Hemmerle H, Voigt CA, Gut-inhabiting Clostridia build human GPCR ligands by conjugating neurotransmitters with diet- and human-derived fatty acids. Nat. Microbiol. 6, 792–805 (2021). doi: 10.1038/s41564-021-00887-y [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q, Chen L, Yang H, Fang Y, Wang S, Wang M, Yuan Q, Wu W, Zhang Y, Liu Z, Nan F, Xie X, GPR84 signaling promotes intestinal mucosal inflammation via enhancing NLRP3 inflammasome activation in macrophages. Acta Pharmacol Sin 43, 2042–2054 (2022). doi: 10.1038/s41401-021-00825-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi C, He J, Huang D, Zhao Y, Zhang C, Ye X, Huang Y, Nussinov R, Zheng J, Liu M, Lu W, Activation of orphan receptor GPR132 induces cell differentiation in acute myeloid leukemia. Cell Death Dis. 13, 1004 (2022). doi: 10.1038/s41419-022-05434-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morita N, Umemoto E, Fujita S, Hayashi A, Kikuta J, Kimura I, Haneda T, Imai T, Inoue A, Mimuro H, Maeda Y, Kayama H, Okumura R, Aoki J, Okada N, Kida T, Ishii M, Nabeshima R, Takeda K, GPR31-dependent dendrite protrusion of intestinal CX3CR1+ cells by bacterial metabolites. Nature 566, 110–114 (2019). doi: 10.1038/s41586-019-0884-1 [DOI] [PubMed] [Google Scholar]
- 54.Doktorova M, Zwarts I, Zutphen TV, Dijk THV, Bloks VW, Harkema L, Bruin A, Downes M, Evans RM, Verkade HJ, Jonker JW Intestinal PPARδ protects against diet-induced obesity, insulin resistance and dyslipidemia. Sci Rep. 7, 846 (2017). doi: 10.1038/s41598-017-00889-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark EA, Giltiay NV, CD22: A Regulator of Innate and Adaptive B Cell Responses and Autoimmunity. Front. Immunol. 9, 2235 (2018). doi: 10.3389/fimmu.2018.02235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva P, Justicia A, Regueiro A, Fariña S, Couselo JM, Loidi L, Autosomal recessive agammaglobulinemia due to defect in μ heavy chain caused by a novel mutation in the IGHM gene. Genes Immun. 18, 197–199 (2017). doi: 10.1038/gene.2017.14 [DOI] [PubMed] [Google Scholar]
- 57.Porakishvili N, Memon A, Vispute K, Kulikova N, Clark EA, Rai KR, Nathwani A, Damle RN, Chiorazzi N, Lydyard PM, CD180 functions in activation, survival and cycling of B chronic lymphocytic leukaemia cells. Br. J. Haematol. 153, 486–498 (2011). doi: 10.1111/j.1365-2141.2011.08605.x [DOI] [PubMed] [Google Scholar]
- 58.Ning Z, Liu K, Xiong H, Roles of BTLA in Immunity and Immune Disorders. Front. Immunol. 12, 654960 (2021). doi: 10.3389/fimmu.2021.654960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alegre ML, Frauwirth KA, Thompson CB, T-cell regulation by CD28 and CTLA-4. Nat. Rev. Immunol. 1, 220–228 (2001). doi: 10.1038/35105024 [DOI] [PubMed] [Google Scholar]
- 60.Baron VT, Pio R, Jia Z, Mercola D, Early Growth Response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br. J. Cancer 112, 755–764 (2015). doi: 10.1038/bjc.2014.622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Remouchamps C, Boutaffala L, Ganeff C, Dejardin E, Biology and signal transduction pathways of the Lymphotoxin-αβ/LTβR system. Cytokine Growth Factor Rev. 22, 301–310 (2011). doi: 10.1016/j.cytogfr.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 62.Isaikina P, Tsai CJ, Dietz N, Pamula F, Grahl A, Goldie KN, Guixà-González R, Branco C, Paolini-Bertrand M, Calo N, Cerini F, Schertler GFX, Hartley O, Stahlberg H, Maier T, Deupi X, Grzesiek S, Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist. Sci. Adv. 7, eabg8685 (2021). doi: 10.1126/sciadv.abg8685; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cullum E, Graves AM, Tarakanova VL, Denzin LK, Golovkina T, MHC Class II Presentation Is Affected by Polymorphism in the H2-Ob Gene and Additional Loci. J. Immunol. 207, 5–14 (2021). doi: 10.4049/jimmunol.2100061; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JT, Terrell SM, Li VL, Wei W W, Fischer CR, Long JZ, Cooperative enzymatic control of N-acyl amino acids by PM20D1 and FAAH. Elife 9, e55211 (2020). doi: 10.7554/eLife.55211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartz GJ, Fu J, Astarita G, Li X, Gaetani S, Campolongo P, Cuomo V, Piomelli D, The Lipid Messenger OEA Links Dietary Fat Intake to Satiety. Cell Metabolism 8, 281–288 (2008). doi: 10.1016/j.cmet.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodríguez De Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, Piomelli D, Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-α. Nature 425, 90–93 (2003). doi: 10.1038/nature01921 [DOI] [PubMed] [Google Scholar]
- 67.Brown KH, Peerson JM, Lopez de Romaña G, de Kanashiro HC, Black RE, Validity and epidemiology of reported poor appetite among Peruvian infants from a low-income, periurban community. The American journal of clinical nutrition, 61, 26–32 (1995). doi: 10.1093/ajcn/61.1.26 [DOI] [PubMed] [Google Scholar]
- 68.Nahar B, Hossain M, Ickes SB, N Naila N, Mahfuz M, Hossain D, Denno DM, Walson J, Ahmed T, Development and validation of a tool to assess appetite of children in low income settings. Appetite 134,182–192 (2019). doi: 10.1016/j.appet.2018.12.032 [DOI] [PubMed] [Google Scholar]
- 69.Khan MT, Dwibedi C, Sundh D, Pradhan M, Kraft JD, Caesar R, Tremaroli V, Lorentzon M, Bäckhed F, Synergy and oxygen adaptation for development of next-generation probiotics. Nature 620, 381–385 (2023). doi: 10.1038/s41586-023-06378-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hryhorowicz S, Kaczmarek-Ryś M, Zielińska A, Scott RJ, Słomski R, Pławski A, Endocannabinoid System as a Promising Therapeutic Target in Inflammatory Bowel Disease – A Systematic Review. Front. Immunol. 12, 790803 (2021). doi: 10.3389/fimmu.2021.790803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao Y, Shen J, Ran ZH, Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol. Res. Pract. 2014, 1–7 (2014). doi: 10.1155/2014/872725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sviridova T, Deryabin D, Cyganok O, Chereshnev V, Cytotoxicity of N-dodecanoyl-L-homoserine lactone and 5-N-dodecyl resorcinol to human granulocytes and monocytes: a comparative study. Cent. Eur. J. Immunol. 3, 310–316 (2013). doi: 10.5114/ceji.2013.37752 [DOI] [Google Scholar]
- 73.Wick RR, Judd LM, Gorrie CL, Holt KE, Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13, e1005595 (2017). doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gurevich A, Saveliev V, Vyahhi N, Tesler G, QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013). doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seemann T, Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 76.Sharma J, Mulherkar S, Chen UI, Xiong Y, Bajaj L, Cho BK, Goo YA, Leung HCE, Tolias KF, Sardiello M, Calpain activity is negatively regulated by a KCTD7–Cullin-3 complex via non-degradative ubiquitination. Cell Discov. 9, 32 (2023). doi: 10.1038/s41421-023-00533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krueger F, James F, Ewels P, Afyounian E, Schuster-Boeckler B, FelixKrueger/TrimGalore: v0.6.7 - DOI via Zenodo, version 0.6.7, Zenodo (2021); doi: 10.5281/ZENODO.5127899 [DOI]
- 78.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 80.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A, BioMart – biological queries made easy. BMC Genomics 10, 22 (2009). doi: 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 16, 284–287 (2012). doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797 (2004). doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saitou N, Nei M The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution, 4, 406–425 (1987). doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 85.Felsenstein J, Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985). doi: 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- 86.Paradis E, Schliep K, ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics, 35, 526–528 (2019). doi: 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Material Transfer Agreements between the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) and Washington University in St. Louis were established for use of the human biospecimens described in this study.


