Summary
Loss of proteostasis is a hallmark of aging that underlies many age-related diseases. Different cell compartments experience distinctive challenges in maintaining protein quality control, but how aging regulates subcellular proteostasis remains underexplored. Here, by targeting the misfolding-prone FlucDM luciferase to the cytoplasm, mitochondria, and nucleus, we established transgenic sensors to examine subcellular proteostasis in Drosophila. Analysis of detergent-insoluble and -soluble levels of compartment-targeted FlucDM variants indicates that thermal stress, cold shock, and pro-longevity inter-organ signaling differentially affect subcellular proteostasis during aging. Moreover, aggregation-prone proteins that cause different neurodegenerative diseases induce a diverse range of outcomes on FlucDM insolubility, suggesting that subcellular proteostasis is impaired in a disease-specific manner. Further analyses with FlucDM and mass spectrometry indicate that pathogenic tauV337M produces an unexpectedly complex regulation of solubility for different FlucDM variants and protein subsets. Altogether, compartment-targeted FlucDM sensors pinpoint a diverse modulation of subcellular proteostasis by aging regulators.
Keywords: subcellular proteostasis, cell compartments, organelles, tools for aging research, inter-tissue signaling, myokines, protein quality control
Graphical abstract
Highlights
-
•
Establishes transgenic sensors of subcellular proteostasis in Drosophila
-
•
Demonstrates their use to monitor proteostasis in different cell compartments
-
•
Examines how inter-organ signaling impacts subcellular proteostasis
-
•
Shows that pathogenic tauV337M differentially remodels proteome solubility
Motivation
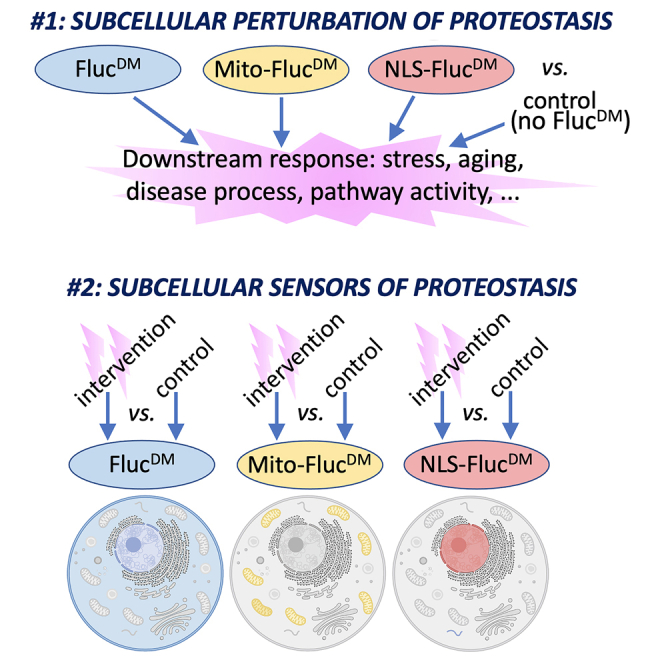
Eukaryotic cells have evolved complex functions via their compartmentalization into organelles and subcellular microenvironments. Such specialization of the cellular space enabled optimal function but also resulted in compartment-specific challenges to proteostasis. However, due to the paucity of tools, how subcellular proteostasis is regulated in physiological and pathological conditions remains largely uncharted. In this study, we generated Drosophila strains that ubiquitously express misfolding-prone FlucDM protein variants that have been engineered to localize to the nucleus and mitochondria, along with an untargeted variant that localizes primarily to the cytoplasm. By validating these tools in several contexts, we demonstrate that compartment-targeted FlucDM variants can be used to either perturb or to monitor subcellular proteostasis, depending on the experimental design.
Curley et al. develop transgenic sensors of subcellular proteostasis in Drosophila based on misfolding-prone FlucDM protein variants that localize to the cytoplasm, nucleus, and mitochondria. They demonstrate that these tools can be used to monitor how cellular stress, inter-organ signaling, and aggregation-prone proteins impact subcellular proteostasis during aging in Drosophila.
Introduction
Age-related diseases arise from interconnected, degenerative events that occur at multiple levels in organs, tissues, cells, and subcellular compartments.1,2,3,4,5 Although the decline of proteostasis is a defining feature of many age-related diseases,6,7,8 little is known about how protein quality control is regulated in different cellular organelles and compartments during aging.1,2,3 Because each cellular compartment/organelle is characterized by its folding environment and proteolytic capacities, disease processes and environmental stressors that challenge protein quality control may impact each organelle differently.1,2
The previous development of tools in C. elegans and cell culture has provided remarkable insight into this subject. For example, redox reporters targeted to the endoplasmic reticulum (ER) and the cytoplasm indicate that the redox state of these cell compartments is profoundly remodeled during aging in C. elegans. Specifically, the ER is oxidizing in young animals but shifts toward reducing conditions during aging, whereas the cytosol becomes more oxidizing.9 Another study used a general (i.e., cytoplasmic) sensor of proteostasis based on a structurally destabilized firefly luciferase mutant (FlucDM) and found that aging leads to an increase in sensor insolubility in C. elegans.10 Organelle-targeted sensors can also be used to identify organelle-specific components of proteostasis. For example, by using model substrates targeted to the nucleus and the cytoplasm, studies in yeast have unveiled that proteostasis in these compartments is regulated by distinct sets of proteins and that the ubiquilin Dsk2 is required to clear nuclearly misfolded proteins.11 Moreover, compartment-specific sensors of proteostasis may also be used to monitor inter-organelle crosstalk, which is a component of age-related processes.1,2,12,13,14 In addition to working as sensors, previous studies have found that misfolding-prone proteins15,16 and organelle-targeted reporters such as mito-GFP17 can impact the specific compartment where they are targeted and induce adaptive and maladaptive stress responses.18,19,20,21 On this basis, depending on the experimental design, organelle-targeted reporter proteins may be utilized both as sensors of proteostasis or as probes to perturb proteostasis in a specific subcellular compartment.
Here, we expanded the toolkit to study subcellular proteostasis by generating transgenic Drosophila strains that express misfolding-prone firefly luciferase FlucDM variants that localize to the mitochondria, the nucleus, and the cytoplasm. FlucDM was chosen as the starting point for generating sensors of subcellular proteostasis because of the previous studies demonstrating its utility in other model organisms10,22 and because it is an exogenous protein that may be less likely to perturb cellular functions compared to the transgenic expression of an endogenous misfolding-prone protein. By comparing the impact of experimental versus control interventions in the presence of each FlucDM, we find that FlucDM variants can be utilized as sensors of proteostasis. By monitoring the levels of detergent-soluble and -insoluble FlucDM, we find that subcellular proteostasis is differentially regulated by environmental stress, endocrine signaling, and aggregation-prone proteins (such as microtubule-associated protein tau [MAPT]) during aging in Drosophila. Moreover, we find that these tools can be utilized to perturb proteostasis in a compartment-specific manner when a FlucDM variant is compared to an isogenic control with no FlucDM expression. Altogether, these studies indicate that organelle-targeted misfolding-prone proteins may find several experimental applications and that such tools may provide insight into how subcellular proteostasis is differentially cross-regulated in distinct compartments by age-related diseases and anti-aging interventions.
Results
Establishment of transgenic, compartment-targeted, misfolding-prone FlucDM proteins to sense and perturb subcellular proteostasis in Drosophila
FlucDM is a misfolding-prone firefly luciferase protein variant that has been used as a reporter of cytoplasmic proteostasis in cell culture and in C. elegans10 and, more recently, in vivo in mice.22 Detergent-soluble FlucDM is functional, whereas sequestration of FlucDM into detergent-insoluble fractions (which typically correspond to protein aggregates) indicates the occurrence of misfolding and the consequent loss of solubility. In addition to increased misfolding, retention of FlucDM into detergent-insoluble fractions can also indicate deficits in the degradation of misfolded proteins. In line with this model, aging increases FlucDM insolubility in C. elegans,10 confirming that cytoplasmic proteostasis declines with aging.6,7,10,23
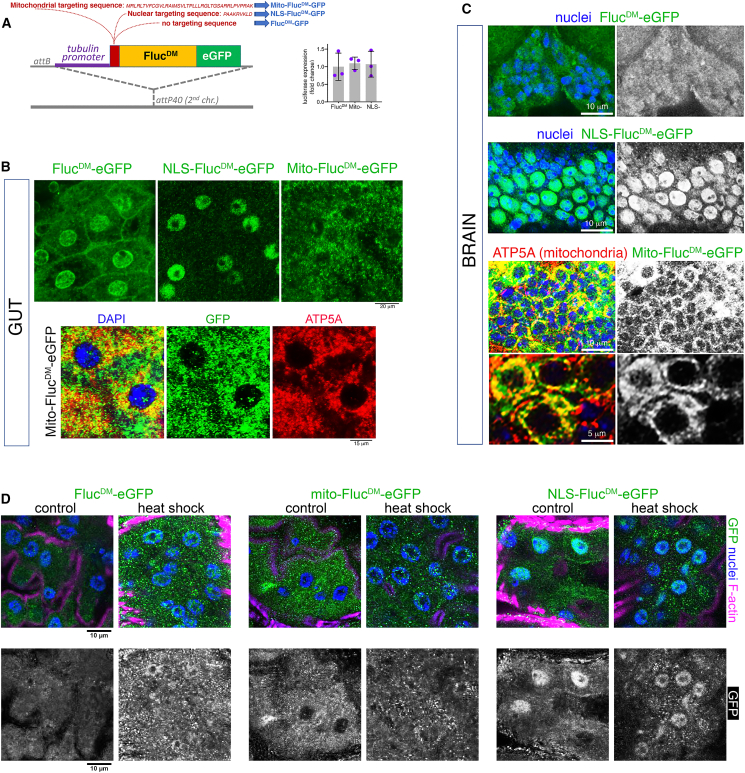
Based on the previously characterized misfolding-prone luciferase,10 we generated compartment-targeted FlucDM variants tagged with enhanced green fluorescent protein (EGFP) for in vivo use in Drosophila (Figure 1A). To generate a FlucDM variant targeted to mitochondria (mito-FlucDM-EGFP), the mitochondrial import sequence from the human mitochondrial COX VIII protein (cytochrome c oxidase subunit 8, a component of the respiratory chain located in the inner mitochondrial membrane)24,25 was fused to the N terminus of FlucDM-EGFP. A standard nuclear targeting sequence (PAAKRVKLD)26 was fused to the FlucDM N terminus to generate a FlucDM-EGFP variant with a nuclear localization signal (NLS-FlucDM-EGFP). Lastly, transgenic flies were also generated for untargeted FlucDM-EGFP (Figure 1A), which has been found previously to display diffuse cytoplasmic localization in C. elegans and cell culture.10
Figure 1.
Generation of compartment-targeted, misfolding-prone FlucDM variants to perturb and sense subcellular proteostasis in Drosophila
(A) Generation of transgenic organelle-targeted sensors of protein quality control based on a misfolding-prone mutant firefly luciferase (FlucDM) fused to EGFP. The mitochondrial targeting sequence from the human mitochondrial COX VIII protein was utilized to generate the mito-FlucDM variant, whereas a standard nuclear targeting sequence was used to generate the NLS-FlucDM. General (untargeted) sensors and reporters for mitochondria and the nucleus were site integrated and are expressed ubiquitously (downstream of a tubulin promoter) and at similar levels, as indicated by qRT-PCR with 3 batches of flies and the mean ± SD (no significant changes, one-way ANOVA).
(B) Immunostaining and confocal microscopy of enterocytes indicate that FlucDM variants exhibit the expected specificity in subcellular localization. General (untargeted) FlucDM is detected in the cytoplasm (but also in the nucleus and plasma membrane), mito-FlucDM is detected in ATP5A-stained mitochondria, and NLS-FlucDM is found in the nucleus. Scale bars represent 20 and 15 μm, as indicated.
(C) Immunostaining and confocal microscopy of brain cells from the antennal lobe indicates a similar localization. The untargeted FlucDM is detected in the cytoplasm, mito-FlucDM is detected in ATP5A-positive mitochondria, and NLS-FlucDM is found in the nucleus. Scale bars represent 10 and 5 μm.
(D) Immunostaining of enterocytes from heat-shocked and control flies identifies FlucDM-GFP aggregates that accumulate in the cytoplasm in response to thermal stress compared to non-heat-shocked controls. Similar heat-induced cytoplasmic aggregates are also found in heat-shocked NLS-FlucDM-EGFP and mito-FlucDM-EGFP cells. In the case of mito-FlucDM-EGFP, these aggregates are recognizable because they produce larger puncta than the staining that corresponds to mito-FlucDM-EGFP-positive mitochondria (B). Scale bar, 10 μm.
To ensure expression in the physiological range, all FlucDM variants were engineered to be expressed downstream of a ubiquitous tubulin promoter and to be site integrated at the same position into the genome via the phiC31 integrase system,27 leading to similar expression levels, as assessed by qRT-PCR (Figure 1A).
Next, immunostaining and confocal microscopy were utilized to determine the subcellular localization of FlucDM protein variants. Considering that the tubulin promoter drives the expression of FlucDM transgenes ubiquitously, FlucDM intracellular localization was monitored in enterocytes, which are a convenient system for these analyses given their large cell size, and in brain cells because of the importance of proteostasis for neurodegeneration.7 Confocal microscopy indicates that FlucDM variants exhibit distinct subcellular localization (Figures 1B and 1C): general (untargeted) FlucDM is detected in the cytoplasm, nucleus, and plasma membrane of enterocytes (Figure 1B) but only in the cytoplasm of brain cells (Figure 1C); NLS-FlucDM is detected in the nucleus of both enterocytes and brain cells (Figures 1B and 1C); and mito-FlucDM is detected in mitochondria, defined by co-staining for the mitochondrial marker ATP5a (ATP synthase F1 subunit alpha) (Figures 1B and 1C). However, not all mito-FlucDM is detected in mitochondria, presumably because mito-FlucDM is synthesized in the cytosol, and its import into mitochondria is suboptimal. Altogether, we established transgenic Drosophila strains that express general (i.e., cytosolic) and compartment-targeted FlucDM variants.
It has been found previously that heat shock results in the accumulation of cytoplasmic aggregates of FlucDM in cell culture.10 On this basis, we tested whether the intracellular localization of compartment-targeted FlucDM variants is remodeled by thermal stress. To this purpose, we examined the subcellular localization of the EGFP-tagged FlucDM variants by immunostaining the enterocytes of heat-shocked flies and controls with anti-GFP antibodies. In agreement with previous findings in cell culture,10 the diffuse localization of the general FlucDM-EGFP sensor shifted to a spotty pattern upon heat shock, indicative of the sequestration of a fraction of the FlucDM-EGFP into cytoplasmic aggregates (Figure 1D). Similar cytoplasmic aggregates were also found in the heat-shocked NLS-FlucDM-EGFP and mito-FlucDM-EGFP cells (Figure 1D). In the case of mito-FlucDM-EGFP, these aggregates were recognized because they were bigger than the speckles that correspond to mitochondria with mito-FlucDM-EGFP (Figure 1D), which largely co-localizes with the mitochondrial marker ATP5a (Figures 1B and 1C). Altogether, these findings indicate that thermal stress induces the sequestration of a fraction of the FlucDM variants into cytoplasmic aggregates (Figure 1D).
Organism-wide perturbation of subcellular proteostasis with compartment-targeted FlucDM variants extends lifespan and delays neuromuscular aging
Protein misfolding restricted to a specific cell compartment has been found previously to induce adaptive and maladaptive responses locally and even systemically.16,18,19,20,21 For example, the expression of a metastable sarcomeric protein in C. elegans skeletal muscle induces a local and systemic stress response characterized by transcriptional induction of the chaperone Hsp90.15 Moreover, it was recently appreciated that expression of mito-GFP, a GFP with a mitochondrial import sequence that is utilized to identify mitochondria by microscopy, leads to perturbation of mitochondrial proteostasis and induction of the mitochondrial unfolded protein response.17 On this basis, with an appropriate experimental design, FlucDM variants may be utilized to perturb proteostasis in a subcellular compartment and determine the corresponding stress responses induced by such compartment-restricted disruption of proteostasis compared to controls with no FlucDM.
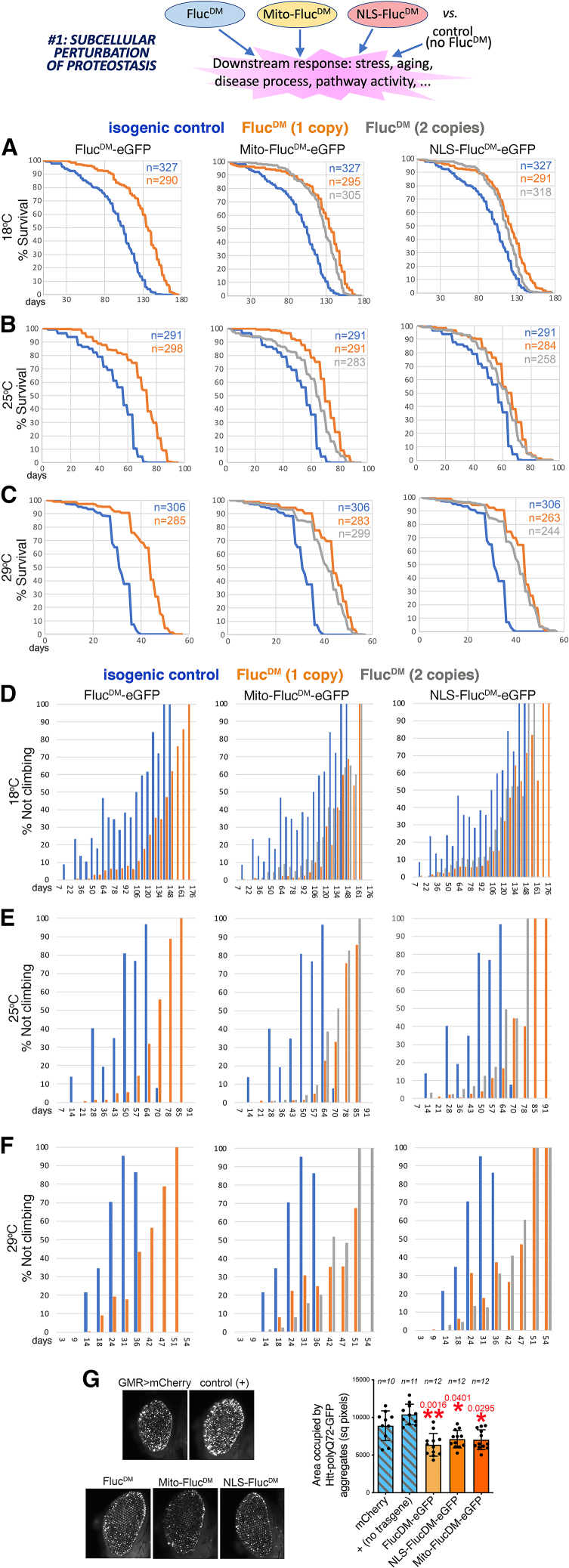
To determine the overarching results of perturbing proteostasis in distinct subcellular compartments at the organism level, we examined whether FlucDM variants regulate lifespan and neuromuscular function during aging compared to isogenic controls with no FlucDM expression. Survival analyses indicate that FlucDM variants extend lifespan, although this occurs differently for each compartment-targeted FlucDM: at 18°C and 25°C, the lifespan extension is more pronounced for the untargeted (i.e., cytoplasmic) FlucDM and for mito-FlucDM compared to NLS-FlucDM (Figures 2A and 2B). However, all FlucDM variants similarly extend lifespan at 29°C (Figure 2C), a temperature that corresponds to mild chronic thermal stress for Drosophila. Interestingly, 2 copies of the mito-FlucDM and NLS-FlucDM transgenes were not more effective in extending lifespan than single copies, suggesting that an optimal adaptive response is already reached with 1 copy and that additional copies of the transgene may not further induce the response or even be detrimental (Figures 2A–2C).
Figure 2.
Use of compartment-targeted FlucDM variants to perturb subcellular proteostasis
FlucDM are misfolding-prone proteins and, therefore, they may challenge proteostasis in the subcellular compartment to which they are targeted. On this basis, FlucDM variants can be used as tools to induce a moderate perturbation of subcellular proteostasis when compared to an isogenic control with no FlucDM expression.
(A–C) The untargeted (cytoplasmic) FlucDM, the mitochondrially targeted mito-FlucDM, and the nucleus-targeted NLS-FlucDM extend lifespan (p < 0.001, log -rank test, with n indicated) when compared to isogenic controls with no FlucDM. Distinct FlucDM variants have effects of different magnitudes; the untargeted FlucDM is more effective in extending lifespan at 18°C (A) and at 25°C (B) compared to NLS-FlucDM, whereas similar lifespan extension is seen at 29°C (C) for all FlucDM variants. These findings suggest that moderate perturbation of subcellular proteostasis by FlucDM variants induces a stress response that extends lifespan. A single (orange) or 2 copies (gray) of the FlucDM transgenes similarly extend lifespan compared to the isogenic controls with no FlucDM (blue).
(D–F) Negative geotaxis assays indicate that expression of misfolding-prone FlucDM proteins targeted to distinct subcellular compartments reduces age-related neuromuscular dysfunction during aging compared to isogenic controls that do not express FlucDM. A similar protection is found at 18°C (D), 25°C (E), and 29°C (F). These findings suggest that moderate perturbation of subcellular proteostasis by compartment-targeted FlucDM variants induces an adaptive stress response that improves neuromuscular function; p < 0.001 (log -rank test) with n indicated (A–C).
(G) Aggregates of GFP-tagged pathogenic huntingtin-polyQ can be seen in the retina of GMR>Htt-polyQ72-GFP flies at 30 days of age, but the total area of such aggregates is higher in controls (mCherry and no transgene, +) compared to flies that express FlucDM variants. This suggests that moderate stress induced by FlucDM can induce a hormetic stress response that improves proteostasis. The n (biological replicates) and the mean ± SD are indicated, with ∗p < 0.05 and ∗∗p < 0.01 (one-way ANOVA).
We next examined negative geotaxis; i.e., the startle-induced escape response from gravity.28,29,30 This is an innate behavior that depends on the function of the brain and skeletal muscle and that senesces with aging in Drosophila.28,29,30 These studies indicate that the percentage of flies able to climb progressively decreases with aging in control flies but less so in isogenic flies that express untargeted FlucDM, mito-FlucDM, and NLS-FlucDM at 18°C, 25°C, and 29°C (Figures 2D–2F). Altogether, these findings indicate that organism-wide expression of a misfolding-prone protein targeted to distinct cell compartments induces, to varying degrees, a hormetic response that extends lifespan and preserves neuromuscular function during aging in Drosophila.
It has been shown previously that metastable proteins can trigger an adaptive response that partially preserves or even restores protein quality control.18,19,20,21 On this basis, we next tested whether FlucDM variants regulate proteostasis in a Huntington’s disease model based on the expression of GFP-tagged pathogenic huntingtin in the retina and in which Htt-polyQ72-GFP aggregates progressively form with aging.31,32,33,34 Compared with control mCherry and with no transgene expression, all FlucDM variants significantly reduced Htt-polyQ72-GFP aggregates (Figure 2G), suggesting that the stress response induced by FlucDM improves proteostasis compared to control flies with no FlucDM expression.
In summary, these studies indicate that compartment-targeted FlucDM variants can be utilized to perturb subcellular proteostasis and to identify the consequent hormetic stress responses that are induced compared to isogenic controls with no FlucDM expression.
Transcriptional stress responses induced by organelle-targeted perturbation of proteostasis with FlucDM variants
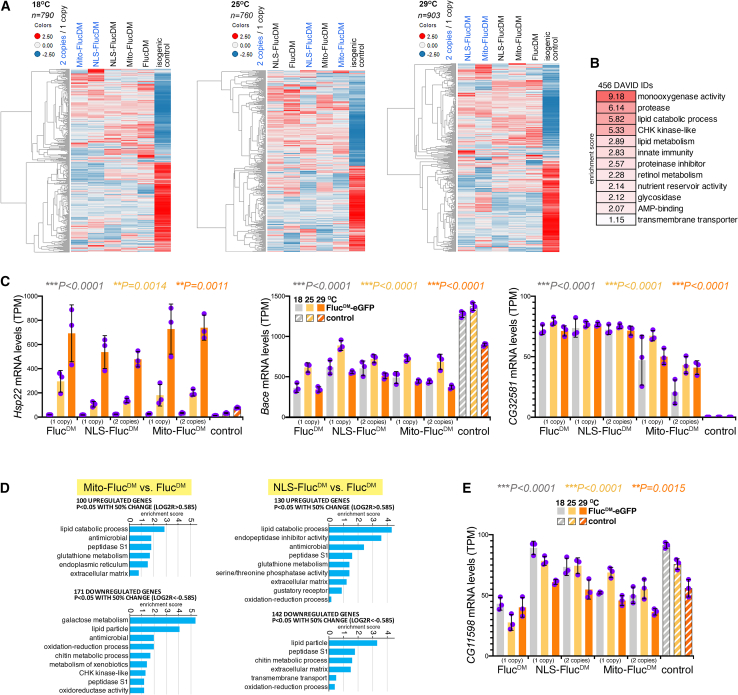
Having established that FlucDM variants induce a non-canonical stress response at the organismal and cellular levels (Figure 2), we next sought to define the underlying transcriptional responses. To this purpose, RNA sequencing (RNA-seq) was done from FlucDM variants and isogenic control flies reared at 18°C, 25°C, and 29°C (Table S1).
Heatmaps of the genes with the highest Z scores indicate that FlucDM flies display substantial gene expression changes compared to isogenic controls with no FlucDM (Figure 3A). Analysis of the Gene Ontology (GO) terms that are modulated by FlucDM include several categories involved in proteostasis and aging, such as proteases, lipid metabolism, and innate immunity (Figure 3B). Examples of differentially regulated genes include the mitochondrial heat shock protein 22 (Hsp22),35 which is among the most highly induced genes in all FlucDM strains compared to isogenic controls: while Hsp22 expression normally rises with heat from 18°C to 29°C (Figure 3C), FlucDM further significantly increases its levels both at 25°C and 29°C. Because previous studies have found a key role for Hsp22 in lifespan regulation,35 these findings suggest that Hsp22 is part of the FlucDM-induced stress response that extends lifespan and improves neuromuscular function (Figure 2). Consistent with this model, NLS-FlucDM, which improved climbing and lifespan less than FlucDM and mito-FlucDM (Figure 2), induced Hsp22 expression less than the other FlucDM variants (Figure 3C). Additional FlucDM-regulated genes include the APP (amyloid precursor protein)-cleaving beta-secretase Bace (which is downregulated by FlucDM) and the ubiquitin ligase CG32581/sordd2 (suppression of retinal degeneration disease 1 upon overexpression 2), which is homologous to human RNF185 (ring finger protein 185) and is highly induced by all FlucDM variants (Figure 3C).
Figure 3.
Compartment-targeted FlucDM variants induce a transcriptional stress response compared to isogenic controls with no FlucDM
(A) Heatmap of the genes that are most differentially regulated indicate that FlucDM variants induce transcriptional adaptive responses that differentiate them from isogenic controls with no FlucDM expression. The average Z scores are color coded; upregulated genes are shown in red and downregulated genes in blue.
(B) Several gene categories are commonly modulated by compartment-targeted FlucDM variants, including genes that encode for proteins with monooxygenase activity, proteases, and proteins involved in lipid catabolism.
(C) Differentially modulated genes such as Hsp22, Bace, and CG32581/sordd2 may provide a mechanistic explanation for how stress responses induced by perturbation of subcellular proteostasis are protective during aging (Figure 2).
(D) Apart from transcriptional stress responses that are commonly induced by all FlucDM variants (B and C), there are also gene categories that are differentially regulated by mito-FlucDM and by NLS-FlucDM versus the untargeted FlucDM.
(E) An example of a gene that is differentially modulated by distinct FlucDM variants is the triacylglycerol lipase CG11598, which is not regulated by NLS-FlucDM, is downregulated by mito-FlucDM, and even more downregulated by the untargeted FlucDM compared to isogenic controls with no FlucDM expression.
In (C) and (E), n = 3 (biological replicates) with the mean ± SD indicated; for each genotype, the mRNA levels (transcripts per million [TPM]) at 18°C (gray), 25°C (yellow), and 29°C (orange) are shown. For the genes in (C) and (E), the color-coded p value summary (one-way ANOVA) is reported for the comparison of FlucDM variants to the control at each temperature. Additional statistical cross-comparisons of each FlucDM to the control at each temperature are reported in the source data file (Table S4).
Further analysis of the transcriptional responses induced by mito-FlucDM and NLS-FlucDM compared to the untargeted FlucDM revealed several gene categories that are differentially regulated by distinct FlucDM proteins: lipid metabolism and antimicrobial peptides were commonly induced by mito-FlucDM and NLS-FlucDM compared to the untargeted FlucDM, whereas components of the ER and gustatory receptors were enriched categories among the genes regulated by mito-FlucDM and by NLS-FlucDM, respectively (Figure 3D). An example of gene that is differentially regulated by distinct FlucDM variants is the triacylglycerol lipase CG11598, which is homologous to human LIPN (lipase family member N) and related lipases: CG11598 is more highly expressed in NLS-FlucDM versus mito-FlucDM and even more so compared to the untargeted FlucDM (Figure 3E). Altogether, transcriptional profiling identifies differentially expressed genes that are induced consistently or distinctly by FlucDM variants compared to isogenic controls with no FlucDM expression.
Compartment-targeted FlucDM sensors indicate that heat and cold shock distinctly impact subcellular proteostasis
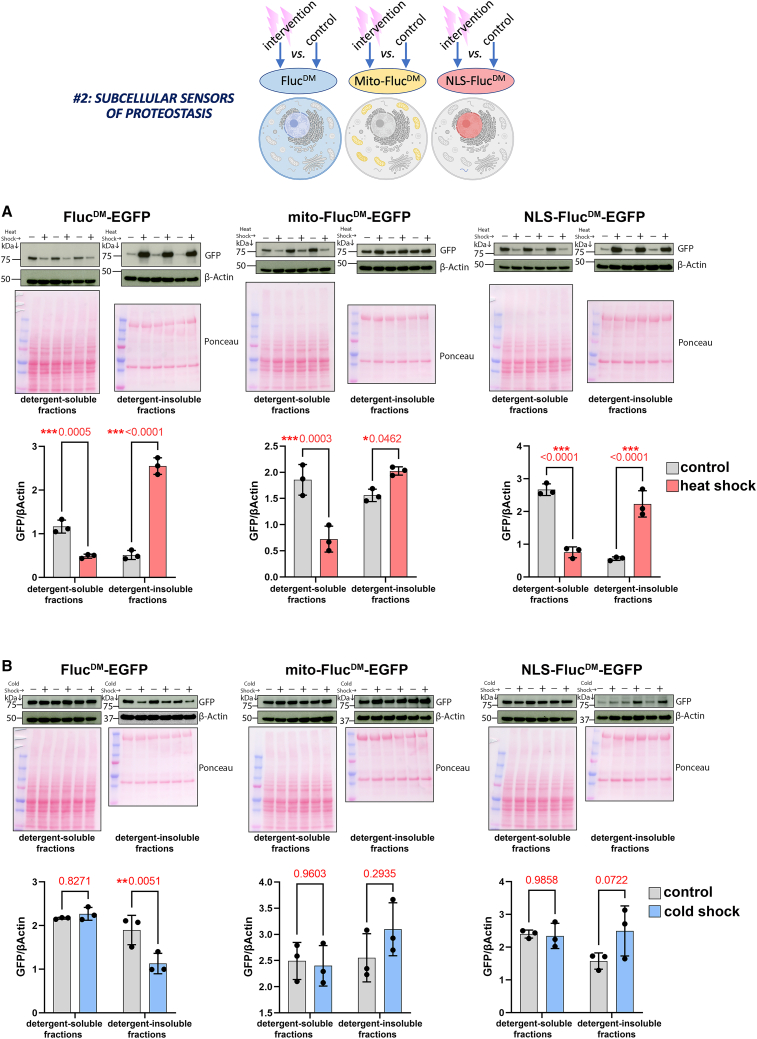
In addition to perturbing proteostasis in subcellular compartments (Figures 2 and 3), FlucDM variants are primarily utilized as sensors of proteostasis, as found previously for a general (i.e., cytoplasmic) FlucDM in C. elegans10 and for compartment-targeted FlucDM in mice.22 To test whether this is also possible in Drosophila, two environmental stressors (cold and heat shock) were utilized to determine how they impact subcellular proteostasis in vivo. While thermal stress (heat shock) is a well-known challenge to proteostasis because of heat-induced protein unfolding,36,37,38 cold shock has been conversely found to improve proteostasis and delay aging.39,40,41,42,43 However, the impact of cold and heat shock on subcellular proteostasis remains largely unexplored. To address this question, FlucDM strains were exposed to heat stress (36°C for 3 h) or cold shock (4°C for 2 h) and compared to isogenic controls kept at the standard temperature of 25°C. Subsequently, the solubility of the untargeted (i.e., cytoplasmic) FlucDM, the mitochondrial mito-FlucDM, and the nuclear NLS-FlucDM was analyzed via western blot of detergent-soluble and -insoluble fractions. As expected based on its capacity to cause protein unfolding,36,37 thermal stress induced profound changes in FlucDM solubility that were, however, of different magnitudes in distinct subcellular compartments. In general, there was a decrease in the soluble levels of all FlucDM variants and a corresponding increase in their detergent-insoluble levels; these heat-induced changes in protein insolubility were the highest for the untargeted (i.e., cytoplasmic) FlucDM, intermediate for the nuclear NLS-FlucDM, and minimal for the mitochondrial mito-FlucDM (Figure 4A). Altogether, these findings suggest that certain cellular compartments (e.g., mitochondria) may be relatively resilient to heat-induced protein unfolding compared to the nucleus and the cytoplasm.
Figure 4.
Use of compartment-targeted FlucDM as sensors of subcellular proteostasis
FlucDM are misfolding-prone proteins and, therefore, can be used to determine whether an experimental intervention modulates proteostasis compared to a control intervention. Specifically, FlucDM protein misfolding leads to its detergent insolubility, which indicates defects in protein quality control.
(A) Testing compartment-targeted FlucDM with thermal stress (red) versus controls (gray). Western blots of detergent-soluble and -insoluble fractions from whole flies with anti-GFP antibodies detect the EGFP-tagged FlucDM variants targeted to the mitochondria (mito-FlucDM) and the nucleus (NLS-FlucDM) and the untargeted FlucDM that localizes primarily to the cytoplasm. Ponceau staining and β-actin are shown as normalization controls, and the graphs refer to the GFP levels normalized by β-actin. Heat shock significantly decreases the detergent-soluble levels of all compartment-targeted FlucDM. This corresponds to an increase in the detergent-insoluble levels of the untargeted (i.e., cytoplasmic) FlucDM, NLS-FlucDM, and, to a lower extent, mito-FlucDM. Altogether, these findings indicate that thermal stress compromises proteostasis across the cell but more prominently in the cytoplasm and nucleus compared to the mitochondria. n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05 and ∗∗∗p < 0.001 (unpaired two-tailed t test).
(B) Testing compartment-targeted FlucDM with cold shock (blue) versus controls (gray). Shown are western blots of detergent-soluble and -insoluble fractions from whole flies with anti-GFP and anti-β-actin antibodies. Cold shock reduces the detergent-insoluble levels of the untargeted (i.e., cytoplasmic) FlucDM, whereas there are no changes in the detergent-soluble levels and in the detergent-insoluble levels of mito-FlucDM and NLS-FlucDM. Altogether, these findings indicate that cold shock improves cytoplasmic proteostasis but does not impact protein quality control in the nucleus and mitochondria. n = 3 (biological replicates) with the mean ± SD indicated; ∗∗p < 0.01 (unpaired two-tailed t test).
Cold shock impacts the cell differently than thermal stress.39,40,41,42,43 Consistent with this scenario, western blots indicated that there were no changes in the soluble levels of any FlucDM variants, whereas the detergent-insoluble levels were conversely regulated depending on the compartment (Figure 4B): detergent-insoluble levels of the (cytoplasmic) FlucDM were reduced by cold shock but trended toward increasing for NLS-FlucDM, whereas minimal changes were found for mito-FlucDM (Figure 4B). Altogether, the folding status of compartment-targeted FlucDM variants indicates that proteostasis is affected differentially by heat and cold shock in distinct subcellular compartments.
The solubility of compartment-targeted FlucDM proteins is correspondingly regulated by cytoplasmic and mitochondrial chaperones
Proteostasis in distinct subcellular compartments critically relies on dedicated proteolytic systems and chaperones, such as the mitochondrial chaperone Hsp22 and the cytoplasmic chaperone Hsp70.35,44,45,46 On this basis, we tested whether modulation of Hsp22 and Hsp70 impacts the folding status of FlucDM variants. Knockdown of Hsp70 resulted in an ∼70% decline in Hsp70 mRNA levels (Figure S1A) and in a significant decline in the detergent-soluble levels of the untargeted (i.e., cytoplasmic) FlucDM (Figure S1B), whereas there were no changes in the detergent-soluble and -insoluble levels of mito-FlucDM and NLS-FlucDM (Figures S1C and S1D). Moderate (∼3-fold) overexpression of the mitochondrial chaperone Hsp22 (Figure S1A) did not significantly affect the detergent-soluble and -insoluble levels of FlucDM and NLS- FlucDM (Figures S2A and S2C) but significantly reduced the levels of detergent-insoluble mito-FlucDM (Figure S2B), consistent with the known role of Hsp22 in preserving mitochondrial proteostasis.35 Altogether, these findings indicate that chaperones with specific subcellular localizations correspondingly impact the folding status of the FlucDM sensors that are targeted to the same subcellular compartment.
Previous studies in C. elegans have found that aging leads to an increase in FlucDM sensor insolubility.10 On this basis, we examined whether the detergent-soluble and -insoluble levels of FlucDM variants are modulated by aging in extracts from whole flies, heads (enriched for tissues of the central nervous system), and thoraces (enriched for skeletal muscle). In whole flies, there was a significant increase in the detergent-insoluble levels of the untargeted (i.e., cytoplasmic) FlucDM sensor with aging, whereas mito-FlucDM, and NLS-FlucDM were not modulated (Figure S3). Analysis of extracts from heads and thoraces indicates that there are minimal or no changes in the insoluble levels of FlucDM, mito-FlucDM, and NLS-FlucDM during aging (Figure S3).
Similar results were also found by monitoring the folding status of the firefly luciferase FlucDM-GFP sensors with luciferase assays: the signal for FlucDM does not significantly change with aging (10, 30, and 60 days) in fly thoraces. Moreover, the luciferase signal (normalized by total protein content) was higher for the general FlucDM compared to NLS-FlucDM and mito-FlucDM, suggesting that there could be detection limits that can impair the use of luciferase assays for monitoring the folding status of FlucDM variants (Figure S4).
Altogether, these findings indicate that the folding status of FlucDM variants is relatively well preserved in the cytoplasm, nucleus, and mitochondria of aging cells in Drosophila in the absence of environmental shocks or genetic perturbations.
Modulation of subcellular proteostasis by endocrine signaling during aging
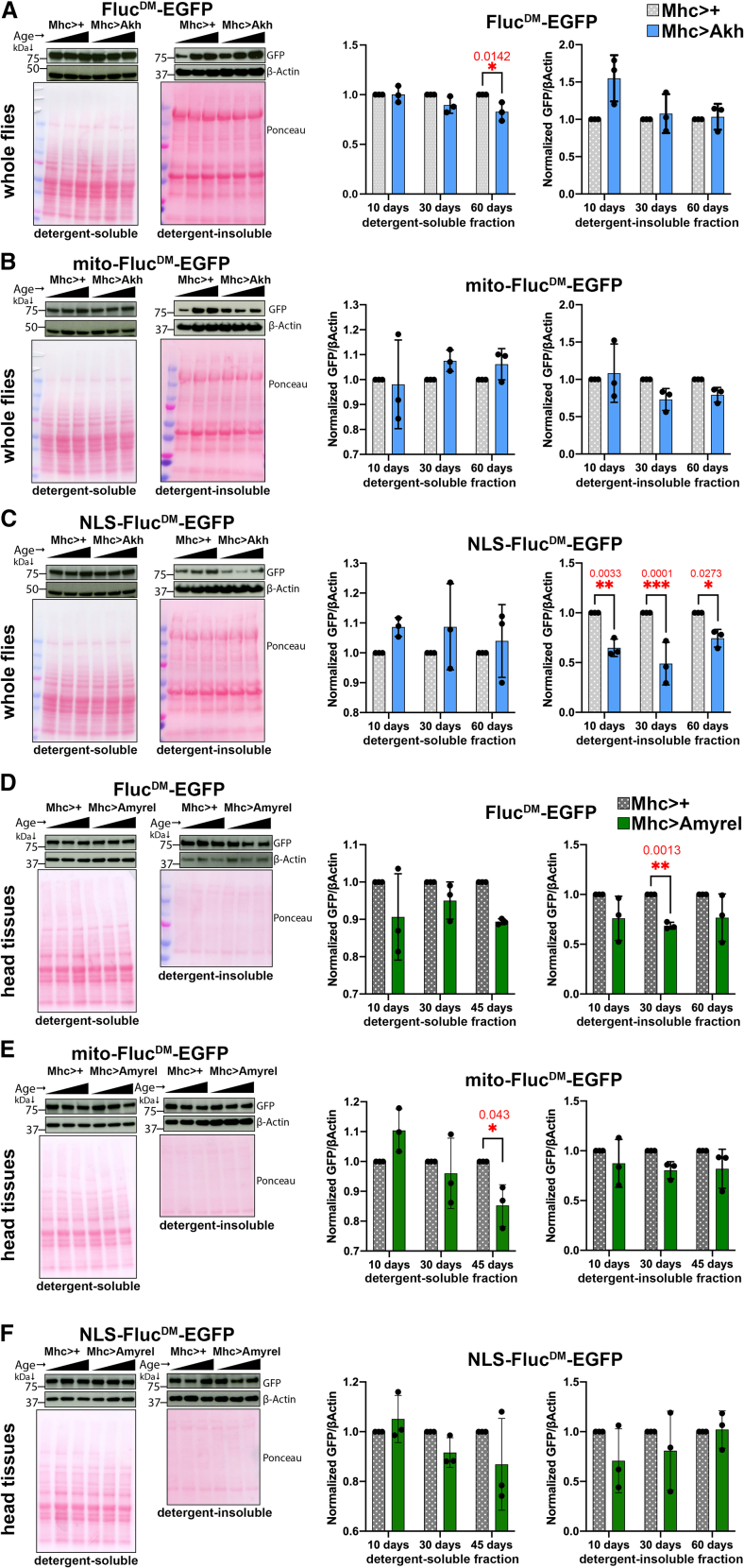
Aging is characterized by changes in multiple organ systems that collectively decrease the ability of the organism to maintain homeostasis.6,47,48,49 Previous studies have shown that inter-organ endocrine signaling regulates systemic aging and lifespan.50,51,52,53,54,55,56,57,58,59 Skeletal muscle has emerged as an important tissue in the systemic regulation of aging because of its capacity to secrete a variety of signaling factors (myokines) that contribute to inter-organ signaling in response to exercise, stress, and nutrient sensing.60,61,62,63,64,65,66,67,68 On this basis, we tested the function of the stress-induced amylase Amyrel, which regulates protein quality control in the central nervous system during aging via endocrine maltose/SLC45 signaling,68,69 and the adipokinetic hormone (Akh), a glucagon-like hormone that antagonizes insulin signaling and delays aging.70,71,72 In these studies, detergent-soluble and -insoluble fractions from whole flies were examined to determine the systemic outcome of muscle-expressed Akh. A ∼4-fold overexpression of Akh in muscle (Figure S1A) did not impact the detergent-soluble and -insoluble levels of FlucDM and mito-FlucDM (Figures 5A and 5B), apart from a significant reduction in the soluble levels of FlucDM in old age (Figure 5A). However, overexpression of Akh in muscle significantly reduced the detergent-insoluble levels of NLS-FlucDM across all ages (Figure 5C), indicating that muscle-derived Akh improves nuclear proteostasis. Altogether, these findings indicate that signaling by the glucagon-like hormone Akh has a compartment-specific effect on subcellular proteostasis during aging.
Figure 5.
Organelle-targeted sensors of proteostasis indicate that distinct endocrine signaling factors differentially impact subcellular proteostasis
(A–C) Western blot analyses of detergent-soluble and insoluble fractions from control whole flies (Mhc>+, gray) and isogenic flies with muscle-specific overexpression of Akh (Mhc>Akh, blue), the Drosophila functional homolog of glucagon and related peptides. Western blotting with anti-GFP antibodies indicates the levels of FlucDM-EGFP sensors, whereas Ponceau staining and β-actin are used as normalization controls. Akh differentially impacts the detergent-insoluble levels of FlucDM-EGFP variants targeted to distinct cell compartments during aging. Akh reduces the detergent-insoluble levels of nuclearly localized NLS-FlucDM at all ages (C), whereas it does not substantially impact the detergent-insoluble levels of cytoplasmic FlucDM (A) and mitochondrial mito-FlucDM (B). The ages analyzed for each genotype are 10, 30, and 60 days. n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired two-tailed t test).
(D–F) Western blot analysis of detergent-soluble and insoluble fractions from head extracts (enriched for the CNS) from control flies (Mhc>+, gray) and isogenic flies with muscle-specific overexpression of Amyrel (Mhc>Amyrel, green), which preserves proteostasis via maltose/SLC45 signaling. Amyrel reduces the detergent-insoluble levels of the untargeted (i.e., cytoplasmic) FlucDM (D) but not of mito-FlucDM (E) or NLS-FlucDM (F). Altogether, these findings indicate that muscle-derived Amyrel improves cytoplasmic proteostasis in the CNS. The ages analyzed for each genotype are 10, 30, and 45 days. n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05, ∗∗p < 0.01 (unpaired two-tailed t test).
We next tested whether another endocrine regulator of aging, Amyrel/maltose signaling,69 similarly impacts subcellular proteostasis. In these studies, detergent-soluble and -insoluble fractions from heads and thoraces were examined to determine the outcome of muscle-expressed Amyrel on the central nervous system (head extracts) or skeletal muscle (thoraces). Western blots of detergent-soluble and -insoluble fractions indicate that muscle-specific induction of Amyrel reduces the insoluble levels of the untargeted (i.e., cytoplasmic) FlucDM in heads (Figure 5D), whereas mito-FlucDM and NLS-FlucDM are not affected (Figures 5E and 5F). These findings, therefore, indicate that Amyrel promotes proteostasis in the cytoplasm but not in the mitochondria and nuclei, which is consistent with the previous finding that Amyrel induces the expression of the cytoplasmic chaperone Hsp23.69 Apart from the preservation of cytoplasmic proteostasis in head extracts (i.e., central nervous system), there was no effect of muscle-induced Amyrel on muscle proteostasis, as assessed from the analysis of detergent-soluble and insoluble levels of FlucDM, mito-FlucDM, and NLS-FlucDM in thoracic fractions (Figure S5). This is consistent with previous findings that have shown that overexpression of the maltose-producing enzyme Amyrel in skeletal muscle regulates protein quality control in the brain and retina but not in skeletal muscle during aging,69 presumably because of the high expression of SLC45 maltose transporters in the brain versus the skeletal muscle.69 Altogether, these findings indicate that anti-aging endocrine interventions may act by selectively improving proteostasis in specific subcellular compartments.
Divergent modulation of subcellular proteostasis by distinct aggregation-prone proteins
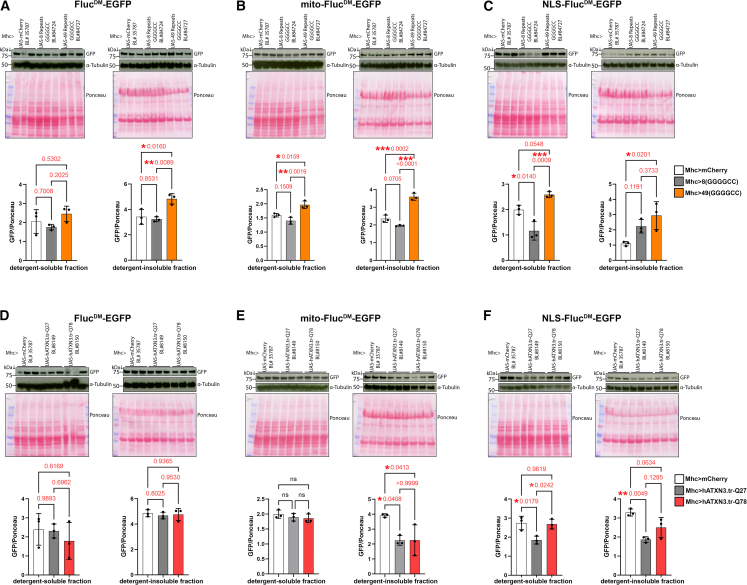
Aggregation-prone proteins are the culprits of several age-related diseases.23,73,74,75,76,77 Although they are primarily localized in a specific cell compartment, aggregation-prone proteins perturb the function of many organelles.1,2,3,14,78,79 However, it remains largely unknown whether such changes in cell function arise from corresponding changes in the protein quality control of subcellular compartments. To address this question, we monitored how the solubility of FlucDM, mito-FlucDM, and NLS-FlucDM is modulated by aggregation-prone proteins in skeletal muscle, an abundant tissue in which proteostasis is compromised with aging.8,32,33,63 In particular, we utilized flies that express a protein with 49(GGGGCC) repeats that model those found in human C9orf72 and that are associated with the etiology of amyotrophic lateral sclerosis and frontotemporal dementia.80 Such a protein with 49(GGGGCC) repeats forms toxic cytoplasmic and nuclear protein aggregates81 compared to a control non-toxic protein with 8(GGGGCC) repeats80 and to mCherry. Western blots of detergent-soluble and -insoluble fractions indicate that 49(GGGGCC) increases the insoluble levels of all FlucDM reporters (FlucDM, mito-FlucDM, and NLS-FlucDM) compared to the non-toxic controls, 8(GGGGCC)-containing proteins, and/or mCherry (Figures 6A–6C). Altogether, these findings indicate that a toxic protein with 49(GGGGCC) repeats compromises proteostasis widely across the cell in the cytoplasm, mitochondria, and the nucleus (Figures 6A–6C).
Figure 6.
Distinct aggregation-prone proteins differentially impact subcellular proteostasis
(A–C) Analyses of detergent-soluble and insoluble fractions from skeletal muscle from 10-day-old flies that express a toxic, aggregation-prone protein with 49(GGGGCC) repeats (orange) compared to non-toxic controls, 8(GGGGCC)-containing proteins (gray), and/or mCherry (white). Western blots with anti-GFP antibodies detect the levels of FlucDM-EGFP, whereas Ponceau staining and α-tubulin are used as normalization controls. The toxic 49(GGGGCC) protein increases the detergent-insoluble levels of FlucDM variants that localize to the cytoplasm (A), mitochondria (B), and the nucleus (C), indicating that aggregation-prone proteins with 49(GGGGCC) repeats generally disrupt proteostasis across multiple cell compartments. n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (one-way ANOVA).
(D–F) Western blots of detergent-soluble and -insoluble fractions from skeletal muscles of flies that express pathogenic ataxin-3 with poly-glutamine tract expansion (hATXN3.tr-Q78; red) versus a non-pathogenic ataxin-3 (hATXN3.tr-Q27, gray) and mCherry controls (white). There is no modulation of detergent-soluble and -insoluble FlucDM levels (D), whereas the effects of hATXN3.tr-Q78 on mito-FlucDM (E) and NLS-FlucDM (F) levels are inconsistent when compared to the hATXN3.tr-Q27 versus the mCherry control. n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05, ∗∗p < 0.01 (one-way ANOVA). Altogether, these findings indicate that distinct aggregation-prone proteins have strikingly different impacts on subcellular proteostasis.
In a second set of studies, we expressed a C-terminal fragment of the human Machado-Joseph disease/spinocerebellar ataxia type 3 protein with a 78-repeat polyglutamine tract expansion (hATXN3.tr-Q78),82 which forms toxic aggregates that are primarily located in the nucleus.83,84,85 As a control, hATXN3.tr-Q78 was compared to a non-toxic hATXN3 with only 27 repeats (hATXN3.tr-Q27)82 and to mCherry. These analyses indicate that hATXN3.tr-Q78 has no substantial effect on the solubility of the untargeted (i.e., cytoplasmic) FlucDM reporter (Figure 6D). There was minimal impact of hATXN3.tr-Q78 on the solubility of mito-FlucDM and NLS-FlucDM, and these effects were inconsistent when comparing hATXN3.tr-Q78 to distinct controls, hATXN3.tr-Q27 and mCherry (Figures 6E and 6F). Altogether, pathogenic ataxin-3 seems to minimally impact the folding status of FlucDM sensors located in distinct cell compartments, suggesting that pathogenic ataxin-3 may have a relatively limited impact on subcellular proteostasis.
In summary, the findings indicate that distinct pathogenic proteins have strikingly different impacts on subcellular protein quality control: while a toxic protein with 49(GGGGCC) repeats widely disrupts proteostasis in the cytoplasm, nucleus, and mitochondria, pathogenic ataxin-3 has minimal influence on the folding status of FlucDM proteins targeted to these compartments.
Compartment-specific effects of pathogenic tau on subcellar proteostasis during aging in Drosophila
Aggregation of MAPT deranges proteostasis, and it is a cause of Alzheimer’s disease and related dementias.86,87,88 While tau is a cytoplasmic protein, many other cell compartments, such as the nucleus and mitochondria, are perturbed by pathogenic tau.7,89,90,91,92,93 For example, tau deranges nucleocytoplasmic transport94 and nuclear architecture and chromatin organization,95,96,97 and tau nuclear aggregates alter the composition and organization of nuclear speckles.98 However, it remains largely unknown whether the effects of cytosolic tau on the nucleus and organelles derive from corresponding changes in the protein quality control of these cell compartments.
To address this question, we utilized knockin fly models99,100 in which the endogenous Drosophila tau was substituted by pathogenic human tauV337, which carries a mutation that causes frontotemporal dementia in humans.101,102,103 Previous studies have shown that these knockin Drosophila models develop progressive signs of tauopathy during aging when heterozygous, including neurodegeneration and behavioral deficits in memory, locomotion, and sleep.99,100 On this basis, we tested whether tauV337M impacts the detergent-soluble and -insoluble levels of FlucDM, mito-FlucDM, and NLS-FlucDM compared to isogenic controls. For these studies, flies with ubiquitous expression of tauV337M and of FlucDM variants were examined at 10, 30, and 60 days, which correspond to young, intermediate, and old ages, and fly heads (enriched for the central nervous system) and thoraces (enriched for skeletal muscles) were analyzed separately.
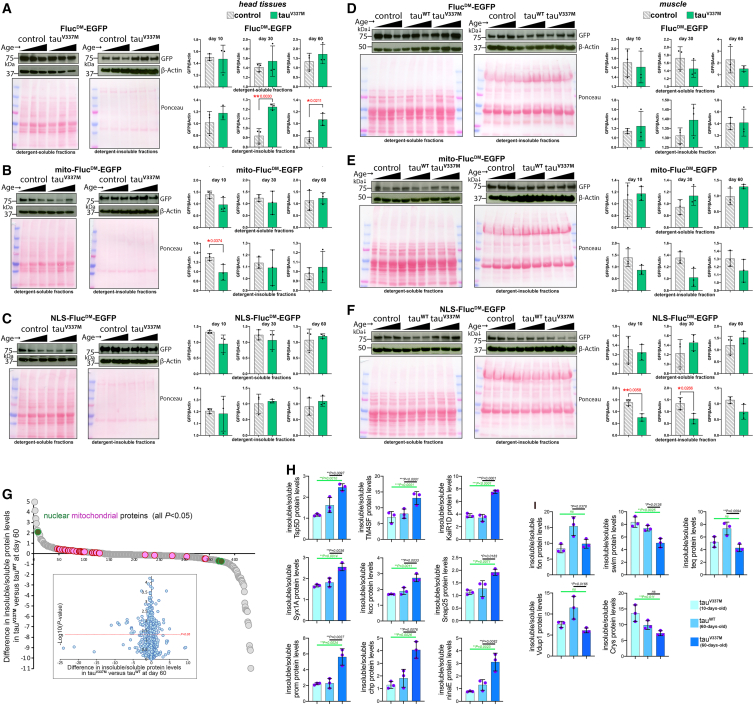
Western blots of detergent-soluble and -insoluble fractions from fly heads revealed that, as expected, tauV337M worsens cytosolic proteostasis, as indicated by a significant increase in the detergent-insoluble levels of the untargeted (∼cytosolic) FlucDM, whereas its soluble levels were not affected (Figure 7A). Interestingly, these effects were significant at day 30 and day 60 but not at day 10 (Figure 7A), consistent with the previous finding that the development of tauopathy is progressive and age dependent in heterozygous tauV337M flies.99,100 There were minimal effects of tauV337M on mitochondrial and nuclear proteostasis (Figures 7B and 7C), apart from a decrease in the insoluble mito-FlucDM levels at a young age (Figure 7B). Altogether, these analyses indicate that FlucDM insolubility is promoted by tauV337M primarily in the cytoplasm and in an age-dependent manner.
Figure 7.
Pathogenic tauV337M impacts subcellular proteostasis during aging in a contrasting manner
Western blots of detergent-soluble and -insoluble fractions from control flies (gray) and isogenic knockin flies that express human tauV337M (green), a mutant tau that causes frontotemporal dementia in humans. Western blotting with anti-GFP antibodies indicates the levels of FlucDM-EGFP sensors, whereas Ponceau staining and β-actin are used as normalization controls.
(A–C) Analysis of detergent-soluble and -insoluble fractions from head extracts (enriched for tissues of the CNS) indicates that heterozygous tauV337M mutations increase the detergent-insoluble levels of the cytoplasmic FlucDM reporter during aging (at 30 and 60 days) but not at a young age (10 days) compared to isogenic controls. There is little effect on the solubility of mito-FlucDM and NLS-FlucDM, indicating that tauV337M reduces cytoplasmic proteostasis but has no substantial effect on mitochondrial and nuclear proteostasis in the CNS.
(D–F) Western blots of detergent-soluble and insoluble fractions from skeletal muscle (thoracic fractions) of flies with heterozygous tauV337M mutations. There is no significant effect of pathogenic tau on FlucDM (D) and mito-FlucDM (E) levels, suggesting that the skeletal muscle may be resistant to tauV337M-induced defects in cytoplasmic proteostasis (D) compared to the CNS (A). However, heterozygous tauV337M mutations significantly reduce the detergent-insoluble levels of nuclearly localized NLS-FlucDM (F), suggesting that pathogenic tauV337M may derange protein import into the nuclei of skeletal muscle.
In (A)–(F), the ages analyzed for each genotype are 10, 30, and 60 days. n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05, ∗∗p < 0.01 (unpaired two-tailed t test).
(G–I) Ultra-deep-coverage TMT mass spectrometry indicates that pathogenic tauV337M divergently regulates the solubility of distinct protein sets. These proteomics analyses (18-plex TMT) are based on detergent-soluble and -insoluble fractions from isogenic knockin flies that express human tauWT and pathogenic human tauV337M at 10 and 60 days of age.
(G) The difference in the insoluble/soluble protein levels in tauV337M versus tauWT at approximately day 60. There are 432 proteins of 4,012 quantified proteins that display significantly (p < 0.05) increased (n = 140; n = 20 with log2R > 1) and decreased (n = 292, n = 43 with log2R < −1) insolubility in tauV337M versus tauWT at day 60. These include mitochondrial (pink) and nuclear (green) proteins.
(H and I) Examples of proteins with increased (H) and decreased (I) insolubility in response to tauV337M in old age compared to tauWT at the same age (black line of comparison) and compared to tauV337M at a young age (green line). n = 3 (biological replicates) with the mean ± SD indicated; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (one-way ANOVA).
Although pathogenic tau has been reported to impact skeletal muscle,104,105,106 our analyses revealed minimal effects of pathogenic tauV337M on cytosolic and mitochondrial proteostasis in this tissue, as assessed by monitoring the detergent-soluble and insoluble levels of the untargeted FlucDM (Figure 7D) and mito-FlucDM (Figure 7E) in muscle. Therefore, these findings suggest that skeletal muscle may be relatively resistant to derangement of cytoplasmic proteostasis by tauV337M compared to the central nervous system. There was, however, an unexpected decline in the insoluble levels of NLS-FlucDM in response to tauV337M, which may indicate derangement of nuclear protein import, whereas the soluble levels were unaffected (Figure 7F). Together, these findings indicate that pathogenic tau influences protein solubility in the nuclei of skeletal muscle cells (Figure 7F). Because such a reduction in insoluble NLS-FlucDM levels was not seen in fly head extracts (Figure 7C), which are enriched for the central nervous system, these findings suggest that the impact of pathogenic tau on subcellular proteostasis varies depending on the tissue and cell type.
Ultra-deep-coverage mass spectrometry indicates that pathogenic tau regulates, in opposite manners, the solubility of distinct protein sets during aging in Drosophila
To complement these analyses and determine how pathogenic tau impacts the solubility of the entire proteome, we next utilized ultra-deep-coverage tandem mass tag (TMT) mass spectrometry to analyze detergent-soluble and insoluble fractions. Also, in these studies, we utilized knockin tau models in which the endogenous Drosophila tau was replaced by wild-type human tau (tauWT) or by pathogenic human tauV337M.99,100 On this basis, an 18-plex TMT analysis was done from whole flies with homozygous tauV337M and control tauWT expression at day 60, which corresponds to an old age in which behavioral deficits are manifest in tauV337M versus tauWT fly strains.99,100 In addition, homozygous young (10-day-old) tauV337M flies were examined to determine the age-related changes in protein solubility that occur with aging. This TMT-based profiling indicates that the solubility of several proteins is reshaped by tauV337M compared to control, age-matched, isogenic tauWT flies at 60 days of age (Figures 7G–7I; Table S2).
Because many proteins are expressed at the threshold of their solubility,77,107,108,109,110,111,112,113,114 it has been hypothesized previously that aggregation-prone proteins generally tip the balance toward insolubility for other proteins. However, our TMT-based analyses indicate that pathogenic tauV337M has complex effects on protein solubility: while some proteins become more insoluble, others display increased solubility upon expression of tauV337M versus tauWT (Figures 7G–7I, Figures S6A, and S6B).
We next examined the proteins with a significant difference (p < 0.05) in protein solubility (insoluble/soluble levels) in tauV337M versus tauWT flies to determine whether any specific subcellular compartment is impacted. While several mitochondrial proteins were among the proteins with solubility regulated by tauV337M versus tauWT (with primarily increased insolubility in response to tauV337M), the magnitude of these changes, although significant (p < 0.05) was overall low (Figure 7G). Of the 3 significant proteins with nuclear localization, only one (nucleolar GTP-binding protein 1, CG8801) had increased insolubility in tauV337M versus tauWT (Figure 7G).
Further analysis of over-represented protein categories (Table S3) indicates that transmembrane proteins (e.g., tetraspanins), proteins with disordered domains, and proteins that are components of cell projections (e.g., axons) are enriched among proteins with tauV337M-induced protein insolubility (p < 0.05 and Δ > 0.5 for insoluble/soluble levels in tauV337M versus tauWT). Curated analyses further indicate the presence of several proteins involved in synaptic transmission and brain function (e.g., the kainate-type ionotropic glutamate receptor subunit 1D, the potassium:chloride symporter Kazachoc, axotactin, syntaxin-1A, adenylate cyclase, and synaptosomal-associated protein 25) and proteins enriched in microvilli and rhabdomeres and, hence, required for photoreceptor function (e.g., prominin, chaoptin, and opsin Rh1) (Figures 7H and S6C). Together, these findings suggest that tauV337M may impact synaptic transmission88,115,116,117,118 and light detection by photoreceptors (as reported for human patients and mouse models of tauopathy119,120,121,122,123) by perturbing the solubility of specific components of the synapse and of the light-sensing apparatus. Altogether, organism-wide profiling of proteome solubility indicates that tauV337M particularly impacts the solubility of proteins involved in the function of the central nervous system. Conversely, proteins with tauV337M-induced upregulation of solubility (p < 0.05 and Δ < −0.5 for insoluble/soluble levels in tauV337M versus tauWT) are enriched in peptidases/proteases, glycoproteins, and proteins with disordered regions. Further refined analyses indicate that several secreted factors (e.g., Ance-3, Swim, teq, and fon) and the chaperone crystallin are among the proteins with improved solubility in tauV337M versus control tauWT flies (Figures 7I and S6C).
In summary, these proteomics analyses identify remarkably diverse impacts of pathogenic tauV337M on protein solubility and also highlight subcellular compartments (cell projections and plasma membrane) that are particularly affected.
Discussion
Eukaryotic cells have evolved complex functions via their compartmentalization into organelles and subcellular microenvironments.124 Such specialization of the cellular space has enabled optimal organelle function but also resulted in compartment-specific challenges to proteostasis.9,23,125 Consequently, organelle-specific stress responses and quality control mechanisms are in place to ensure proteostasis within each compartment.2,18,19,126 Although the loss of proteostasis is a key hallmark of aging,47 it remains largely unknown how subcellular proteostasis is impacted by environmental interventions and endocrine signaling factors that regulate aging. Moreover, while aggregation-prone proteins are the culprit of many age-related diseases,7 it remains undetermined whether they perturb proteostasis across the cell or only in specific subcellular compartments.
In this study, we have generated Drosophila strains that ubiquitously express misfolding-prone FlucDM protein variants that were engineered to selectively localize to the nucleus and mitochondria, along with an untargeted variant that localizes primarily to the cytoplasm (Figure 1). By using these tools, we demonstrate that FlucDM protein variants can be utilized to perturb proteostasis in a compartment-targeted manner in the cytoplasm, nucleus, and mitochondria when compared to isogenic flies with no FlucDM expression. Analysis of the organism-wide consequences of these ubiquitous subcellular perturbations indicates that mild stress induced by expression of compartment-targeted FlucDM variants extends lifespan, impedes age-associated neuromuscular aging, and promotes proteostasis (Figure 2). At the molecular level, there are both common and variant-specific transcriptional responses that are induced by FlucDM (Figure 3). Apart from a strong induction of the mitochondrial small heat shock protein Hsp22 by all FlucDM variants, chaperones were not an overrepresented category among the differentially regulated genes (Figure 3B), suggesting that the transcriptional stress response induced by FlucDM variants is different from the heat shock response (HSR).127,128 Likewise, apart from Hsp22, there was no upregulation of Hsp60 (Table S1; Figure S7), a mitochondrial chaperone that is stereotypically induced by the mitochondrial unfolded protein response (UPRmt).20,129 Altogether, these findings indicate that perturbation of subcellular proteostasis by FlucDM does not simply induce the HSR or the UPRmt (Figure 3). Previous studies in cultured cells engineered to express nuclear and cytoplasmic proteins with destabilizing domains similarly found that the transcriptional stress response induced by these misfolding-prone proteins is distinct from the HSR and the UPR.16 Altogether, these findings suggest that transcriptional responses different from the HSR and the UPR can be induced by misfolding-prone proteins targeted to specific subcellular compartments. Interestingly, Hsp22 levels have been identified previously as an important predictor of survival in Drosophila during aging,35,45 indicating a possible role of this chaperone as part of the stress response and lifespan extension induced by FlucDM (Figure 2). However, apart from Hsp22, chaperones do not appear to be a major category of genes transcriptionally induced by FlucDM (Figure S7). Other gene categories were rather induced by FlucDM, including lipases and proteases, which may, respectively, help maintain metabolic homeostasis and protein quality control in cooperation with the ubiquitin-proteasome system and autophagy.130
In addition to utilizing FlucDM variants as tools to perturb subcellular proteostasis (Figures 2 and 3), we found that they can be used as reporters of proteostasis. Specifically, by measuring the detergent-soluble and insoluble levels of each FlucDM, the impact of genetic and environmental interventions on subcellular proteostasis can be determined compared to controls. In particular, the detergent insolubility of the untargeted FlucDM, mito-FlucDM, and NLS-FlucDM sensors increases to different degrees upon heat shock (Figure 4A). Moreover, the beneficial effects of cold shock may derive from an improvement in cytoplasmic proteostasis, as indicated by a reduction in the detergent-insoluble levels of the untargeted FlucDM, whereas mito-FlucDM and NLS-FlucDM were not affected (Figure 4B). Different endocrine signals that regulate aging (the glucagon-like hormone Akh70,71,72 and Amyrel/maltose signaling69) also elicited remarkably distinct outcomes on subcellular proteostasis: while Akh improved nuclear proteostasis (by reducing the detergent-insoluble levels of NLS-FlucDM), Amyrel reduced the insoluble levels of the untargeted, largely cytoplasmic FlucDM. Because we induced Akh and Amyrel specifically in skeletal muscle and assessed the outcome systemically and/or in head extracts (which are enriched for the central nervous system), these findings suggest that endocrine signaling mediated by different muscle-secreted factors (myokines) may have remarkably distinct impacts on subcellular proteostasis. Physiologically, this may occur in response to muscle contraction because Amyrel is induced by cell stress,69 which is a component of exercise,131,132,133,134,135 and because glucagon-like peptide-1 (GLP-1, possibly related to Akh) is expressed by skeletal muscles in response to exercise in mice.136 Altogether, these findings indicate that compartment-targeted FlucDM can be utilized as a sensor to determine whether environmental and genetic interventions regulate subcellular proteostasis.
We also examined how distinct aggregation-prone proteins impact the solubility of FlucDM sensors during aging. Aggregation-prone proteins can interact with and disrupt the folding status of other native proteins and impede proteasome activity.7,23,74,76,125,137,138 On this basis, the toxicity of aggregation-prone proteins may derive from a widespread loss of protein quality control.7,23,74,76,125,137,138 To test this hypothesis, here, we estimated the impact of different aggregation-prone proteins on the detergent-insoluble levels of compartment-targeted FlucDM reporters of proteostasis (Figures 6 and 7). Our analyses indicate a range of diverse outcomes. A toxic protein with 49(GGGGCC) repeats, associated with amyotrophic lateral sclerosis and frontotemporal dementia,80 widely impaired proteostasis across cell compartments (cytoplasm, mitochondria, and nucleus). Conversely, pathogenic ataxin-3 (hATXN3.tr-Q78) associated with Machado-Joseph disease/spinocerebellar ataxia82 had minimal effects on the detergent-insoluble levels of all compartment-targeted FlucDM reporters (Figure 6).
In addition to 49(GGGGCC) and hATXN3.tr-Q78, we examined how a mutant form of MAPT90,139 impacts FlucDM solubility. Normally, tau is predominantly (>90%) attached to microtubules, but its mutation and/or hyperphosphorylation promote its aggregation and accumulation into cytoplasmic neurofibrillary tangles in Alzheimer’s disease and related dementias.7,90,101,139,140 Recent evidence indicates that cytoplasmic, soluble oligomers of tau, generated during tangle formation, are the most toxic tau species that cause neurodegeneration.7,89,90,91 Although the decline of proteostasis is a defining feature of tauopathies,7,89,90,91 it remains largely undetermined whether cytoplasmic mutant tau regulates protein quality control in other cell compartments and organelles apart from the cytosol. By utilizing untargeted (∼cytoplasmic) FlucDM, mito-FlucDM, and NLS-FlucDM, our analyses now indicate that pathogenic tauV337M impairs proteostasis in the cytoplasm but not in the nucleus and mitochondria (Figures 7A–7C) and that this occurs in the central nervous system but not in the skeletal muscle (Figures 7D–7F). Therefore, although tauV337M is known to derange the function of many cell compartments (e.g., mitochondria and the nucleus),90,139 this does not seem to occur via a general perturbation of proteostasis in these compartments (as inferred from the detergent-insoluble levels of mito-FlucDM and NLS-FlucDM) but only in the cytoplasm.
In summary, our analyses indicate that the impact of aggregation-prone proteins on subcellular proteostasis is widely diverse: there is an overt decline in cytoplasmic, nuclear, and mitochondrial proteostasis in response to 49(GGGGCC) expression (Figure 6), whereas tauV337M impedes only cytoplasmic protein quality control (Figure 7), and hATXN3.tr-Q78 has minimal impact (Figure 6). Altogether, these findings pinpoint that the toxicity of aggregation-prone proteins is not necessarily coupled to widespread disruption of proteostasis across cell compartments.
By analyzing the effects of pathogenic tauV337M on subcellular proteostasis in head extracts (consisting mostly of tissues from the central nervous system, CNS) versus thoraces (consisting of skeletal muscle), we found that tauV337M impedes cytoplasmic proteostasis in the CNS but not in muscle (Figure 7), consistent with the fact that tauopathies have primary manifestations in the brain and less so in peripheral tissues.86,87,88 Interestingly, tauV337M expression reduced the detergent-insoluble levels of NLS-FlucDM in muscle fractions (Figures 7D–7F), suggesting that pathogenic tauV337M may derange nuclear protein import in muscle cells.
To further explore how tauV337M impacts the solubility of the proteome, we utilized TMT mass spectrometry to profile the detergent-soluble and -insoluble levels of >4,000 proteins in whole flies that expressed endogenous levels of human tauV337M compared to tauWT. These analyses indicate that pathogenic tauV337M does not simply skew the proteome toward insolubility but, rather, remodels protein solubility in a balanced manner: ∼3.4% and ∼7.2% of detected proteins displayed significantly increased and decreased insolubility, respectively, in tauV337M versus tauWT (Figures 7G–7I). While proteins with tauV337M-modified solubility encompass several cell compartments, including the nucleus and mitochondria, there is an enrichment for transmembrane proteins and components of cell projections (e.g., axons). Functional categories of such proteins include synaptic function and light perception, suggesting that the solubility of proteins necessary for key functions of the CNS is prominently impacted by tauV337M. These studies therefore indicate a complex remodeling of proteome solubility by pathogenic tau.
Altogether, transgenic Drosophila strains that express compartment-targeted FlucDM sensors provide tools to survey subcellular proteostasis and guide subsequent analyses. Depending on the experimental design, compartment-targeted FlucDM variants can be utilized as reporters or as tools to perturb subcellular proteostasis. On this basis, transgenic FlucDM strains may be utilized to determine how protein quality control is regulated across cell compartments during the progression of age-related diseases and by interventions that delay aging.
Limitations of the study
This study examined how FlucDM-based sensors of subcellular proteostasis can be utilized to either monitor or perturb subcellular proteostasis (depending on the experimental design) in the context of aging and age-associated conditions. To perturb subcellular proteostasis, FlucDM variants targeted to different subcellular compartments are compared to isogenic controls with no FlucDM to detect responses that are triggered by compartment-targeted FlucDM. However, we did not systematically examine the proteomic alterations resulting from FlucDM and how these compare to other established proteostatic challenges occurring during normal aging or experimental interventions. In a second application, FlucDM variants can be utilized to monitor the impact of an experimental intervention (genetic, pharmacologic, etc.) on subcellular proteostasis by comparing such interventions to controls that also express the same FlucDM. For this application, the measurement of luciferase activity does not appear to be a convenient readout for the mitochondrial and nuclear luciferases (mito-FlucDM-EGFP and NLS-FlucDM-EGFP) because of the low levels detected, which are presumably due to the low luciferase activity and/or low stability of these FlucDM variants. Because mitochondrial and nuclear proteins are translated in the cytoplasm and then imported into mitochondria and nuclei, the insolubility of mito-FlucDM-GFP and NLS-FlucDM-GFP may also reflect defects in protein import into these cellular compartments. Another limitation is that the FlucDM sensors indicate the general status of proteostasis but not necessarily that of individual proteins or specific protein categories. Our mass spectrometry data indeed indicate that pathogenic tauV337M differentially remodels the solubility of distinct protein subsets (Figure 7).
Resource availability
Lead contact
Requests for further information, resources, and reagents should be directed to and will be fulfilled by the lead contact, Fabio Demontis (Fabio.Demontis@stjude.org).
Materials availability
There are no restrictions on the availability of the data and tools generated by this study.
Data and code availability
-
•
The data supporting the findings of this study are available within the paper (Figures 1, 2, 3, 4, 5, 6, 7 and S1–S7 and Table S1. Transcriptional stress responses induced by compartment-targeted misfolding-prone FlucDM proteins in Drosophila, related to Figure 3, Table S2. Protein solubility changes induced by pathogenic human tau/MAPT during aging in Drosophila, related to Figure 7, Table S3. GO term analysis of protein categories that display tauV337M-induced changes in solubility during aging, related to Figure 7, Table S4. Source data file, related to all figures and supplemental figures, Table S5. Full scans of western blots, related to Figures 4–7, S1–S3, and S5). The TMT mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository and are accessible with the dataset identifier PRIDE: PXD045325. The RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) with the identifier GEO: GSE243224.
-
•
This paper does not report original code.
-
•
Any other information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank Dr. Doris Kretzschmar for the tau knockin fly stocks and Dr. Ulrich Hartl for the pCI-FlucDM-EGFP plasmid. We also thank the Light Microscopy facility and the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital and the Bloomington Drosophila stock center. Schematics were drawn with BioRender. This project was supported by the National Institute on Aging of the NIH (R21AG079267). The Demontis lab is also supported by the Alzheimer's Association (AARG-NTF-22-973220) and by the National Institute on Aging (R01AG075869). The Peng lab is supported by the NIH (RF1AG068581). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Research at St. Jude Children’s Research Hospital is supported by ALSAC.
Author contributions
M.R. generated the FlucDM transgenic sensors. M.C. did most of the experiments with the FlucDM sensors together with M.R., C.-L.C., A.S., Z.C., and M.R.-M. V.P. did the TMT mass spectrometry and corresponding data analyses. Y.-D.W. analyzed the RNA-seq data. J.P. supervised the mass spectrometry studies. F.D. supervised the project and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-alpha-tubulin (11H10) | Cell Signaling Technologies | 2125; RRID:AB_2619646 |
| Rabbit anti-beta-actin | Cell Signaling Technologies | 8457; RRID:AB_10950489 |
| Rabbit anti-GFP | Cell Signaling Technologies | 2956; RRID:AB_1196615 |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Technologies | 7074; RRID:AB_2099233 |
| Chicken anti-GFP | Aves | GFP-1010; RRID:AB_2307313 |
| AlexaFluor488-conjugated anti-chicken | ThermoFisher | A-11039; RRID:AB_2534096 |
| Mouse anti-ATP5a | Abcam | ab14748; RRID:AB_301447 |
| AlexaFluor555-conjugated anti-mouse | LifeTechnologies | A28180; RRID:AB_2536164 |
| Chemicals, peptides, and recombinant proteins | ||
| Alexa Fluor 635 Phalloidin | LifeTechnologies | A22284 |
| DAPI | ThermoFisher | D1306 |
| SlowFade Gold antifade | Invitrogen | S36937 |
| IQ Sybr Green supermix | Bio-Rad | 170–8885 |
| 96-well PCR plates | Bio-Rad | HSP9601 |
| PBS | Gibco | 10010023 |
| Blue loading buffer pack | Cell Signaling Technologies | 7722 |
| Precision Plus protein standard | Bio-Rad | 1610374 |
| 4–20% Mini-PROTEAN TGX pre-cast gels | Bio-Rad | 4561096 |
| Immobilon-P PVDF membrane | Millipore | IPVH00010 |
| Ponceau S | ThermoFisher | A40000279 |
| 16% Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| NP40 cell lysis buffer | Invitrogen | FNN0021 |
| TRIzol | Ambion | 15596018 |
| Critical commercial assays | ||
| Pierce BCA protein assay kit | ThermoScientific | 23225 |
| Dual-Glo luciferase assay system | Promega | E2920 |
| iScript reverse transcriptase | Bio-Rad | 1708840 |
| Deposited data | ||
| TMT mass spectrometry proteomics data of solubility changes induced by tau | This study | ProteomeXchange Consortium, PRIDE: PXD045325 |
| RNA-seq data of gene expression changes induced by FlucDM | This study | Gene Expression Omnibus, GEO: GSE243224 |
| Experimental models: Organisms/strains | ||
| Drosophila: Mhc-Gal4 | Demontis lab collection | Demontis et al., 2014141 |
| Drosophila: GMR-Gal4 | Demontis lab collection | Jiao et al., 202333 |
| Drosophila: UAS-mCherry | Bloomington stock center | #35787 |
| Drosophila: UAS-8(GGGGCC) | Bloomington stock center | #84724 |
| Drosophila: UAS-49(GGGGCC) | Bloomington stock center | #84727 |
| Drosophila: UAS-hATXN3.tr-Q27 | Bloomington stock center | #8149 |
| Drosophila: UAS-Htt-Q72-GFP | Gift of Dr. Sheng Zhang | Zhang et al., 201031 |
| Drosophila: knock-in tauV337M | Gift of Dr. Doris Kretzschmar | Cassar et al., 202099 |
| Drosophila: knock-in tauWT | Gift of Dr. Doris Kretzschmar | Cassar et al., 202099 |
| Drosophila: UAS-mCherryRNAi | Bloomington stock center | #35785 |
| Drosophila: UAS-Hsp70RNAi | Bloomington stock center | #33948 |
| Drosophila: UAS-Hsp22 | Bloomington stock center | #20055 |
| Drosophila: UAS-Akh | Bloomington stock center | #27343 |
| Drosophila: UAS-Amyrel | Demontis lab collection | Rai et al., 202169 |
| Drosophila: tubulin_FlucDM-EGFP | This study | N/A |
| Drosophila: tubulin_mito-FlucDM-EGFP | This study | N/A |
| Drosophila: tubulin_NLS-FlucDM-EGFP | This study | N/A |
| Oligonucleotides | ||
| Primers: Hsp22 forward: 5′-CTTACCGATGTTTTGGCGCA-3′ |
This study | N/A |
| Primers: Hsp22 reverse: 5′-TCGTGGAAGAAGGCGTGAAA-3′ |
This study | N/A |
| Primers: Hsp70Bb forward: 5′-AAATCGGATGGAGAGTTGGC-3′ |
This study | N/A |
| Primers: Hsp70Bb reverse: 5′-TGTAGGCGGGTTTTTGTTT-3′ |
This study | N/A |
| Primers: Akh forward: 5′-AAGCACCGCGAGTAGATAGC-3′ |
This study | N/A |
| Primers: Akh reverse: 5′-TGTGTGTGCGTGCTAGACAT-3′ |
This study | N/A |
| Primers: Tub84B forward: 5′-GCTGTTCCACCCCGAGCAGCTGATC-3′ |
This study | N/A |
| Primers: Tub84B reverse: 5′-GGCGAACTCCAGCTTGGACTTCTTGC-3′ |
This study | N/A |
| Primers: Fluc (firefly luciferase) forward: 5′-CCCTGGTTCCTGGAACAATTGC-3′ |
This study | N/A |
| Primers: Fluc (firefly luciferase) reverse: 5′-AAGAATTGAAGAGAGTTTTCACTGC-3′ |
This study | N/A |
| Software and algorithms | ||
| Cell Profiler 3.0.0 | Cell Profiler | https://www.cellprofiler.org |
| GraphPad Prism | GraphPad Prism | https://www.graphpad.com/ |
| Photoshop 2023 | Adobe | https://www.adobe.com/products/photoshop.html |
| Excel | Microsoft | https://www.microsoft.com |
| BioRender | BioRender | https://www.biorender.com |
Experimental model and subject details
Drosophila husbandry
Flies were kept (∼30 flies/tube) at 25°C, 60% humidity, and a 12h/12h light-dark cycle in tubes containing cornmeal/soy flour/yeast fly food. The fly food was changed regularly every 2–3 days. Flies were aged to 10, 30, and 60 days at 25°C (unless otherwise indicated). Similar procedures as previously reported were utilized for measuring survival during aging and for negative geotaxis.141,142,143,144 All experiments were done with male flies.
Method details
Drosophila stocks
The following fly stocks were utilized in this study: Mhc-Gal4,141,142,143,144 GMR-Gal4,33 UAS-mCherry (Bloomington stock center #35787), UAS-8(GGGGCC) repeats (non-toxic control; #84724), UAS-49(GGGGCC) repeats (toxic poly-GA proteins, as found in C9orf72 associated with ALS and FTD; #84727),80 UAS-hATXN3.tr-Q27 (non-toxic control; #8149), UAS-hATXN3.tr-Q78 (expresses a toxic C-terminal fragment of the human Machado-Joseph Disease/Spinocerebellar Ataxia 3 protein with a 78 repeat polyglutamine tract; #8150), UAS-Htt-Q72-GFP,31,32,33 and knock-in tau models in which the endogenous Drosophila tau has been replaced by wild-type human tau (tauWT) and by pathogenic human tauV337M.99,100 These additional fly stocks were also utilized: UAS-mCherryRNAi (#35785), UAS-Hsp70RNAi (#33948), and UAS-Hsp22 (#20055). FlucDM stocks (generated as indicated below), UAS-Akh (#27343), and UAS-Amyrel69 were backcrossed against the w1118 background and compared to their respective litter-mate isogenic controls with no transgene expression.
Cloning and establishment of compartment-targeted FlucDM transgenic sensors in Drosophila
A pCaSpeR5-attB-tubulin promoter vector was generated by PCR-mediated amplification and cloning of a tubulin promoter and attB sequences by using KpnI with EcoRI-HF, and EcoRI-HF, respectively. To express a general, untargeted version of a misfolding-prone luciferase (FlucDM),10 FlucDM fused with EGFP was PCR-amplified from the pCI-FlucDM-EGFP plasmid10 and cloned into the pCaSpeR5-attB-tubulin promoter vector with KpnI-HF and XbaI. A similar procedure was used to clone a mitochondrially-targeted version of FlucDM-EGFP (mito-FlucDM), which consists of a mitochondrial target sequence from human COX VIII (MRLRLTVFCGVLRAIMSVLTPLLLRGLTGSARRLPVPRAK, previously utilized for establishing the mito-GFP Drosophila stock, Bloomington #8442),24,25 a linker (RSS), and FlucDM-EGFP. For establishing FlucDM targeted to the nucleus (NLS-FlucDM), the canonical PAAKRVKLD nuclear localization signal26 was added by PCR to the N-terminus of FlucDM-EGFP. These plasmids were injected into w1118;attP40 embryos and tubulin-FlucDM-EGFP transgenic Drosophila stocks were established by phiC31-mediated site integration.145
Immunostaining and laser scanning confocal microscopy of Drosophila tissues
Drosophila brains and guts from FlucDM transgenic stocks, established as described above, were immunostained according to standard procedures.32,144,146 Specifically, brains and guts were dissected and immediately fixed with 4% paraformaldehyde and 0.2% Triton X-100 for 30 min, washed, and then incubated with 1:200 chicken anti-GFP (Aves #GFP-1010) primary antibodies, followed by AlexaFluor488-conjugated anti-chicken secondary antibodies (ThermoFisher #A11039) together with DAPI (ThermoFisher #D1306) and AlexaFluor635-conjugated phalloidin (Life Technologies #A22284). Co-staining with mouse anti-ATP5a (abcam #14748) was done to assess the degree of mito-FlucDM localization to mitochondria, identified by the mitochondrial marker ATP5a. The samples were mounted on glass slides with SlowFade Gold antifade reagent (Invitrogen #S36937) and #1.5 coverslips and imaged by using a Zeiss LSM780 confocal microscope. Enterocytes of the midgut and brain cells of the antennal lobes are shown in the representative images in Figures 1B–1D.
RNA sequencing and hierarchical clustering
Samples for RNA sequencing were prepared with TRIzol (Ambion #15596018) from whole flies and the RNA was extracted by isopropanol precipitation from the aqueous phase.147,148,149 RNA sequencing libraries for each sample were prepared from 1 μg total RNA by using the Illumina TruSeq RNA Sample Prep v2 Kit per the manufacturer’s instructions, and sequencing was completed on the Illumina NovaSeq 6000. The 100-bp paired-end reads were trimmed, filtered against quality (Phred-like Q20 or greater) and length (50-bp or longer), and aligned to the Drosophila melanogaster reference genome (BDGP6/dm6) by using CLC Genomics Workbench v12.0.1 (Qiagen). For gene expression comparisons, we obtained the TPM (transcripts per million) counts from the CLC RNA-Seq Analysis tool. The differential gene expression analysis was performed by applying the non-parametric ANOVA using Kruskal-Wallis and Dunn’s tests on log-transformed TPM, implemented in the Partek Genomics Suite v7.0 software (Partek Inc.). Hierarchical clustering of RNA-seq data into heatmaps was done by using the UPGMA clustering method (unweighted pair group method with arithmetic mean) with similarity measure of correlation, implemented in Spotfire v7.5.0 software (TIBCO). The RNA-seq data discussed in this publication are available in Table S1 and have been deposited in the NCBI’s Gene Expression Omnibus (GEO) with accession number GEO: GSE243224. GO terms analysis was done with DAVID.150
qRT-PCR
For qRT-PCR, cDNAs were reverse transcribed with the iScript cDNA synthesis kit (Bio-Rad #1708840) from 1 μg total RNA. qRT-PCR was performed by using the IQ Sybr Green supermix (Bio-Rad #170–8885). Tub84B was utilized as normalization. The following qRT-PCR oligos were used:
Hsp22: 5′-CTTACCGATGTTTTGGCGCA-3′ and 5′-TCGTGGAAGAAGGCGTGAAA-3’
Hsp70Bb: 5′-AAATCGGATGGAGAGTTGGC-3′ and 5′-TGTAGGCGGGTTTTTGTTT-3’
Akh: 5′-AAGCACCGCGAGTAGATAGC-3′ and 5′-TGTGTGTGCGTGCTAGACAT-3’
Tub84B: 5′-GCTGTTCCACCCCGAGCAGCTGATC-3′ and 5′-GGCGAACTCCAGCTTGGACTTCTTGC-3’
Fluc (firefly luciferase): 5′-CCCTGGTTCCTGGAACAATTGC-3′ and 5′-AAGAATTGAAGAGAGTTTTCACTGC-3’
Analysis of aggregates of pathogenic huntingtin
Pathogenic huntingtin-polyQ72-GFP protein aggregates were imaged as previously done33 by using a ZEISS SteREO Discovery.V12 epifluorescence microscope. Subsequently, the images were analyzed using Cell Profiler 3.0.0 (cellprofiler.org) to quantify the total area of protein aggregates (Huntingtin-polyQ72-GFP speckles) normalized by the retinal tissue area. This analysis was done with male flies after aging them at 25°C for 30 days.
Heat shock and cold shock
For the analysis of FlucDM solubility upon thermal stress, 14-days-old wild-type flies were heat shocked at 36°C for 3 h. For cold shock, 14-days-old flies were housed at 4°C for 2 h. Only flies that were alive after the heat and cold shocks were utilized for subsequent analyses.
Western blots of detergent-soluble and insoluble protein fractions
Western blots of detergent-soluble and insoluble fractions were obtained substantially as described previously.142,151 Specifically, whole bodies, heads, or thoraces were homogenized in 60 μL ice-cold Triton X-100 buffer (1% Triton X-100 in PBS containing protease inhibitors and phosphatase inhibitors) for 5 min at the highest speed. Homogenates were centrifuged at 14,000 rpm at 4°C for 10 min and the supernatant was collected (Triton X-100 soluble fraction). The remaining pellet was washed in 400 μL Triton X-100 buffer and centrifuged twice at 14,000 rpm for 5 min at 4°C. The pellet was then resuspended at room temperature in 60 μL RIPA buffer containing 8M urea and 5% SDS, centrifuged at 14,000 rpm at 4°C for 10 min, and the supernatant collected (Triton X-100 insoluble fraction). 8 μL of soluble/insoluble protein extracts were boiled with sample buffer containing DTT and used for SDS-PAGE. Detergent-soluble and insoluble fractions were analyzed on 4–20% SDS-PAGE with anti-GFP antibodies (1:500, dissolved in TBST; Cell Signaling Technologies, #2956). Ponceau S staining (ThermoFisher #A40000279) and anti-β-actin and/or anti-α-tubulin antibodies (Cell Signaling Technologies, #8457 and #2125) were used as loading controls. The western blots with head samples were typically normalized by β-actin whereas the blots with thoracic samples were normalized by Ponceau staining or α-tubulin.
Luciferase assays
For luciferase assays, 10 thoraces/replicate were homogenized in 170 μL ice-cold homogenization buffer (250 mM sucrose, 50 mM Tris-HCl, 5 mM MgCl2, and protease inhibitors) in a NextAdvance bullet blender at 4°C. Subsequently, 90 μL of the homogenate was transferred to a new tube and gently mixed with an equal volume of Dual-Glo luciferase reagent (Promega).141,142,152 After a 10-min incubation, the luminescence was read with a Tecan Infinite 200 Pro. Luciferase readings were normalized by the protein content, as estimated with the Pierce BCA protein assay kit (Thermo Scientific).
Preparation of detergent-soluble and insoluble fractions, protein digestion, and peptide isobaric labeling by tandem mass tags
For TMT of Drosophila samples, 25 male flies per sample (n = 3 biological replicates per condition) were collected at day 10 and day ∼60 from eclosion and homogenized in 200 μL NP40 cell lysis buffer (Invitrogen #FNN0021) with protease and phosphatase inhibitors and 0.5-mm zirconium beads in a NextAdvance bullet blender.153,154 After homogenization, the samples were centrifuged at maximum speed for 10 min, and the supernatant was collected (corresponding to the detergent-soluble fraction, typically consisting of >100 μg). The remaining pellet was washed 3x in PBS and homogenized in insoluble lysis buffer containing 8M urea, PBS, and 1% SDS (with protease and phosphatase inhibitors). The resulting supernatant was collected (corresponding to the detergent-insoluble fraction, typically consisting of >10 μg). All the samples were then prepared for subsequent analyses by adding SDS-blue loading buffer and DTT (Cell Signaling Technologies #7722), and by heating them for 5 min at 95°C. The protein concentration of the lysates was determined by Coomassie-stained short gels using bovine serum albumin (BSA) as standard.155 Specifically, the samples were run on 7.5% SDS-page gels at 60V for 10–15 min, or until all the protein was through the initial lane. BSA was used as a standard at 0.2, 1, and 5 mg. The gels were washed in distilled water and stained with Coomassie blue for 1–4 h with gentle shaking at room temperature, shielded from light. The gels were then de-stained with distilled water overnight at 4°C with gentle shaking. The bands were excised from the gels, cut into several pieces, collected into 1.5 mL-eppendorf tubes on ice, and stored at −80°C.
In preparation for TMT,156 the short gel bands were washed twice with 50% acetonitrile and dried. The dried gel bands were then incubated with trypsin at an enzyme-to-substrate ratio of 1:10 (w/w) for overnight digestion. Following the overnight digestion, the peptide solution from the short gel bands was extracted, and the peptides were reduced by adding 1 mM DTT for 30 min at room temperature followed by alkylation with 10 mM iodoacetamide (IAA) for 30 min in the dark at room temperature. The unreacted IAA was quenched with 30 mM DTT for 30 min. Finally, the digestion was terminated and acidified by adding trifluoroacetic acid (TFA) to 1%, desalted using C18 cartridges (Harvard Apparatus), and dried by speed vac. The peptide mixture was resuspended in 50 mM HEPES (pH 8.5) and labeled with 18-plex Tandem Mass Tag (TMT) reagents (ThermoScientific) following the manufacturer’s recommendations.
Two-dimensional HPLC and mass spectrometry
The TMT-labeled samples were mixed equally, desalted, and fractionated on an offline HPLC (Agilent 1220) using basic pH reverse-phase liquid chromatography (pH 8.0, XBridge C18 column, 4.6 mm × 25 cm, 3.5 μm particle size, Waters). The fractions were dried and resuspended in 5% formic acid and analyzed by acidic pH reverse phase LC-MS/MS analysis. The peptide samples were loaded on a nanoscale capillary reverse phase C18 column (New objective, 75 μm ID × ∼25 cm, 1.9 μm C18 resin from Dr. Maisch GmbH) by an HPLC system (Thermo Ultimate 3000) and eluted by a 60-min gradient. The eluted peptides were ionized by electrospray ionization and detected by an inline Orbitrap Fusion mass spectrometer (ThermoScientific). The mass spectrometer is operated in data-dependent mode with a survey scan in Orbitrap (60,000 resolution, 1 × 106 AGC target and 50 m maximal ion time) and MS/MS high-resolution scans (60,000 resolution, 2 × 105 AGC target, 120 m maximal ion time, 32 HCD normalized collision energy, 1 m/z isolation window, and 15 s dynamic exclusion).157
MS data analysis
The MS/MS raw files were processed by the tag-based hybrid search engine JUMP.158 The raw data were searched against the UniProt Drosophila database concatenated with a reversed decoy database for evaluating false discovery rates. Searches were performed using a 15-ppm mass tolerance for both precursor and product ions, fully tryptic restriction with two maximal missed cleavages, three maximal modification sites, and the assignment of a, b, and y ions. TMT tags on Lys and N-termini (+304.20715 Da) were used for static modifications and Met oxidation (+15.99492 Da) was considered as a dynamic modification. Matched MS/MS spectra were filtered by mass accuracy and matching scores to reduce the protein false discovery rate to ∼1%. Proteins were quantified by summing reporter ion intensities across all matched PSMs using the JUMP software suite.159 Categories enriched in protein sets were identified with DAVID.150
The TMT mass spectrometry proteomics data are reported in Table S2 and have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD045325.
Quantification and statistical analysis
Data organization, scientific graphing, and statistical analyses were done with Microsoft Excel (version 14.7.3) and GraphPad Prism (version 8). The unpaired two-tailed Student’s t test was used to compare the means of two independent groups to each other. One-way ANOVA was used for multiple comparisons of more than two groups of normally distributed data. Survival and climbing data were analyzed with the log-rank test. The n for each experiment can be found in the figures and represents independently generated samples. Bar graphs represent the mean ± SD or the mean ± SEM, as specified in the figure legend. A significant result was defined as p < 0.05. Throughout the figures, asterisks indicate the significance of p values: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Published: October 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2024.100875.
Supplemental information
RNA-sequencing data from Drosophila melanogaster strains (whole flies, males, 10-days-old) reared at 18°C, 25°C, and 29°C and that express misfolding-prone proteins that localize primarily to the cytoplasm (FlucDM), the mitochondria (mito-FlucDM), and the nucleus (NLS-FlucDM), compared to isogenic control flies (w1118) that do not express FlucDM. For mito-FlucDM and NLS-FlucDM, homozygous and heterozygous strains that carry respectively 2 copies and 1 copy of the FlucDM transgenes were analyzed. For each condition, 3 biological replicates were analyzed. Tab 1: gene expression changes induced by FlucDM variants compared to w1118 controls at 18°C, 25°C, and 29°C. Tab 2: TPM (transcripts per million) values, indicating the mRNA levels for each gene across all samples. Tab 3: FlucDM cross-comparison: gene expression changes induced by mito-FlucDM and NLS-FlucDM compared to the untargeted (∼cytoplasmic) FlucDM at 18°C, 25°C, and 29°C. Tab 4: no meaningful upregulation of Hsp60 mRNA levels, a marker of the UPRmt
Ultra-deep-coverage proteomics (18-plex TMT) of detergent-soluble and insoluble fractions from isogenic knock-in male flies that express wild-type human tauWT and pathogenic human tauV337M at 10 and 60 days of age
Protein categories that display change in detergent-solubility in tauV337M flies at 60 versus 10 days, and in tauV337M versus tauWT flies at 60 days of age
References
- 1.Dillin A., Gottschling D.E., Nyström T. The good and the bad of being connected: the integrons of aging. Curr. Opin. Cell Biol. 2014;26:107–112. doi: 10.1016/j.ceb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottschling D.E., Nyström T. The Upsides and Downsides of Organelle Interconnectivity. Cell. 2017;169:24–34. doi: 10.1016/j.cell.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouska M., Huang K., Kang P., Bai H. Organelle aging: Lessons from model organisms. J Genet Genomics. 2019;46:171–185. doi: 10.1016/j.jgg.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang K., Miao T., Chang K., Kim J., Kang P., Jiang Q., Simmonds A.J., Di Cara F., Bai H. Impaired peroxisomal import in Drosophila oenocytes causes cardiac dysfunction by inducing upd3 as a peroxikine. Nat. Commun. 2020;11:2943. doi: 10.1038/s41467-020-16781-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J., Bai H. Peroxisomal Stress Response and Inter-Organelle Communication in Cellular Homeostasis and Aging. Antioxidants. 2022;11:192. doi: 10.3390/antiox11020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiLoreto R., Murphy C.T. The cell biology of aging. Mol. Biol. Cell. 2015;26:4524–4531. doi: 10.1091/mbc.E14-06-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas P.M., Dillin A. Protein homeostasis and aging in neurodegeneration. J. Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demontis F., Piccirillo R., Goldberg A.L., Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis. Model. Mech. 2013;6:1339–1352. doi: 10.1242/dmm.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirstein J., Morito D., Kakihana T., Sugihara M., Minnen A., Hipp M.S., Nussbaum-Krammer C., Kasturi P., Hartl F.U., Nagata K., Morimoto R.I. Proteotoxic stress and ageing triggers the loss of redox homeostasis across cellular compartments. EMBO J. 2015;34:2334–2349. doi: 10.15252/embj.201591711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta R., Kasturi P., Bracher A., Loew C., Zheng M., Villella A., Garza D., Hartl F.U., Raychaudhuri S. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods. 2011;8:879–884. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]
- 11.Samant R.S., Livingston C.M., Sontag E.M., Frydman J. Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature. 2018;563:407–411. doi: 10.1038/s41586-018-0678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson K.A., Hughes A.L., Gottschling D.E. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife. 2014;3:e03504. doi: 10.7554/eLife.03504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veatch J.R., McMurray M.A., Nelson Z.W., Gottschling D.E. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peric M., Bou Dib P., Dennerlein S., Musa M., Rudan M., Lovric A., Nikolic A., Saric A., Sobocanec S., Macak Z., et al. Crosstalk between cellular compartments protects against proteotoxicity and extends lifespan. Sci. Rep. 2016;6:28751. doi: 10.1038/srep28751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Oosten-Hawle P., Porter R.S., Morimoto R.I. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell. 2013;153:1366–1378. doi: 10.1016/j.cell.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyazaki Y., Chen L.C., Chu B.W., Swigut T., Wandless T.J. Distinct transcriptional responses elicited by unfolded nuclear or cytoplasmic protein in mammalian cells. Elife. 2015;4:e07687. doi: 10.7554/eLife.07687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shpilka T., Du Y., Yang Q., Melber A., Uma Naresh N., Lavelle J., Kim S., Liu P., Weidberg H., Li R., et al. UPR(mt) scales mitochondrial network expansion with protein synthesis via mitochondrial import in Caenorhabditis elegans. Nat. Commun. 2021;12:479. doi: 10.1038/s41467-020-20784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovaisaite V., Mouchiroud L., Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marciniak S.J., Ron D. Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 20.Shpilka T., Haynes C.M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 21.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenstock S., Schulz-Trieglaff E.K., Voelkl K., Bolender A.L., Lapios P., Lindner J., Hipp M.S., Hartl F.U., Klein R., Dudanova I. Fluc-EGFP reporter mice reveal differential alterations of neuronal proteostasis in aging and disease. The EMBO journal. 2021;40:e107260. doi: 10.15252/embj.2020107260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labbadia J., Morimoto R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox R.T., Spradling A.C. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130:1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- 25.Rizzuto R., Brini M., Pizzo P., Murgia M., Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr. Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 26.Lu J., Wu T., Zhang B., Liu S., Song W., Qiao J., Ruan H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021;19:60. doi: 10.1186/s12964-021-00741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gargano J.W., Martin I., Bhandari P., Grotewiel M.S. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 2005;40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Rhodenizer D., Martin I., Bhandari P., Pletcher S.D., Grotewiel M. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Exp. Gerontol. 2008;43:739–748. doi: 10.1016/j.exger.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sujkowski A., Bazzell B., Carpenter K., Arking R., Wessells R.J. Endurance exercise and selective breeding for longevity extend Drosophila healthspan by overlapping mechanisms. Aging (Albany NY) 2015;7:535–552. doi: 10.18632/aging.100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S., Binari R., Zhou R., Perrimon N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics. 2010;184:1165–1179. doi: 10.1534/genetics.109.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunt L.C., Schadeberg B., Stover J., Haugen B., Pagala V., Wang Y.D., Puglise J., Barton E.R., Peng J., Demontis F. Antagonistic control of myofiber size and muscle protein quality control by the ubiquitin ligase UBR4 during aging. Nat. Commun. 2021;12:1418. doi: 10.1038/s41467-021-21738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao J., Curley M., Graca F.A., Robles-Murguia M., Shirinifard A., Finkelstein D., Xu B., Fan Y., Demontis F. Modulation of protease expression by the transcription factor Ptx1/PITX regulates protein quality control during aging. Cell Rep. 2023;42:111970. doi: 10.1016/j.celrep.2022.111970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z., Tito A.J., Rui Y.N., Zhang S. Studying polyglutamine diseases in Drosophila. Exp. Neurol. 2015;274:25–41. doi: 10.1016/j.expneurol.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrow G., Samson M., Michaud S., Tanguay R.M. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 36.Leuenberger P., Ganscha S., Kahraman A., Cappelletti V., Boersema P.J., von Mering C., Claassen M., Picotti P. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355:eaai7825. doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 37.Mateus A., Määttä T.A., Savitski M.M. Thermal proteome profiling: unbiased assessment of protein state through heat-induced stability changes. Proteome Sci. 2016;15:13. doi: 10.1186/s12953-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen V.T., Morange M., Bensaude O. Protein denaturation during heat shock and related stress. Escherichia coli beta-galactosidase and Photinus pyralis luciferase inactivation in mouse cells. J. Biol. Chem. 1989;264:10487–10492. [PubMed] [Google Scholar]
- 39.Lindquist J.A., Mertens P.R. Cold shock proteins: from cellular mechanisms to pathophysiology and disease. Cell Commun. Signal. 2018;16:63. doi: 10.1186/s12964-018-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H.J., Alirzayeva H., Koyuncu S., Rueber A., Noormohammadi A., Vilchez D. Cold temperature extends longevity and prevents disease-related protein aggregation through PA28gamma-induced proteasomes. Nat. Aging. 2023;3:546–566. doi: 10.1038/s43587-023-00383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvalho G.B., Drago I., Hoxha S., Yamada R., Mahneva O., Bruce K.D., Soto Obando A., Conti B., Ja W.W. The 4E-BP growth pathway regulates the effect of ambient temperature on Drosophila metabolism and lifespan. Proc. Natl. Acad. Sci. USA. 2017;114:9737–9742. doi: 10.1073/pnas.1618994114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudkevich R., Koh J.H., Beaudoin-Chabot C., Celik C., Lebenthal-Loinger I., Karako-Lampert S., Ahmad-Albukhari S., Thibault G., Henis-Korenblit S. Neuronal IRE-1 coordinates an organism-wide cold stress response by regulating fat metabolism. Cell Rep. 2022;41:111739. doi: 10.1016/j.celrep.2022.111739. [DOI] [PubMed] [Google Scholar]
- 43.Al-Fageeh M.B., Smales C.M. Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Biochem. J. 2006;397:247–259. doi: 10.1042/BJ20060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., Tower J. Expression of hsp22 and hsp70 transgenes is partially predictive of drosophila survival under normal and stress conditions. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64:828–838. doi: 10.1093/gerona/glp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. Hallmarks of aging: An expanding universe. Cell. 2022;186:243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Otin C., Galluzzi L., Freije J.M.P., Madeo F., Kroemer G. Metabolic Control of Longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 50.Panowski S.H., Dillin A. Signals of youth: endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009;20:259–264. doi: 10.1016/j.tem.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Droujinine I.A., Perrimon N. Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annu. Rev. Genet. 2016;50:539–570. doi: 10.1146/annurev-genet-121415-122024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neufer P.D., Bamman M.M., Muoio D.M., Bouchard C., Cooper D.M., Goodpaster B.H., Booth F.W., Kohrt W.M., Gerszten R.E., Mattson M.P., et al. Understanding the Cellular and Molecular Mechanisms of Physical Activity-Induced Health Benefits. Cell Metab. 2015;22:4–11. doi: 10.1016/j.cmet.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 53.Murphy R.M., Watt M.J., Febbraio M.A. Metabolic communication during exercise. Nat. Metab. 2020;2:805–816. doi: 10.1038/s42255-020-0258-x. [DOI] [PubMed] [Google Scholar]
- 54.Russell S.J., Kahn C.R. Endocrine regulation of ageing. Nat. Rev. Mol. Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 55.Jones C.M., Boelaert K. The Endocrinology of Ageing: A Mini-Review. Gerontology. 2015;61:291–300. doi: 10.1159/000367692. [DOI] [PubMed] [Google Scholar]
- 56.Smith H.J., Sharma A., Mair W.B. Metabolic Communication and Healthy Aging: Where Should We Focus Our Energy? Dev. Cell. 2020;54:196–211. doi: 10.1016/j.devcel.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antebi A. Inside insulin signaling, communication is key to long life. Sci. Aging Knowledge Environ. 2004;2004:pe25. doi: 10.1126/sageke.2004.23.pe25. [DOI] [PubMed] [Google Scholar]
- 58.Galluzzi L., Yamazaki T., Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:731–745. doi: 10.1038/s41580-018-0068-0. [DOI] [PubMed] [Google Scholar]
- 59.Rajan A., Perrimon N. Drosophila as a model for interorgan communication: lessons from studies on energy homeostasis. Dev. Cell. 2011;21:29–31. doi: 10.1016/j.devcel.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chow L.S., Gerszten R.E., Taylor J.M., Pedersen B.K., van Praag H., Trappe S., Febbraio M.A., Galis Z.S., Gao Y., Haus J.M., et al. Exerkines in health, resilience and disease. Nat. Rev. Endocrinol. 2022;18:273–289. doi: 10.1038/s41574-022-00641-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demontis F., Piccirillo R., Goldberg A.L., Perrimon N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell. 2013;12:943–949. doi: 10.1111/acel.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta R., Khan R., Cortes C.J. Forgot to Exercise? Exercise Derived Circulating Myokines in Alzheimer's Disease: A Perspective. Front. Neurol. 2021;12:649452. doi: 10.3389/fneur.2021.649452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiao J., Demontis F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr. Opin. Pharmacol. 2017;34:1–6. doi: 10.1016/j.coph.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 64.Pedersen B.K. Physical activity and muscle-brain crosstalk. Nat. Rev. Endocrinol. 2019;15:383–392. doi: 10.1038/s41574-019-0174-x. [DOI] [PubMed] [Google Scholar]
- 65.Pedersen B.K., Akerström T.C.A., Nielsen A.R., Fischer C.P. Role of myokines in exercise and metabolism. J. Appl. Physiol. 2007;103:1093–1098. doi: 10.1152/japplphysiol.00080.2007. [DOI] [PubMed] [Google Scholar]
- 66.Pedersen B.K., Febbraio M.A. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol. Rev. 2008;88:1379–1406. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- 67.Rai M., Demontis F. Systemic Nutrient and Stress Signaling via Myokines and Myometabolites. Annu. Rev. Physiol. 2016;78:85–107. doi: 10.1146/annurev-physiol-021115-105305. [DOI] [PubMed] [Google Scholar]
- 68.Rai M., Demontis F. Muscle-to-Brain Signaling Via Myokines and Myometabolites. Brain Plast. 2022;8:43–63. doi: 10.3233/BPL-210133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rai M., Coleman Z., Curley M., Nityanandam A., Platt A., Robles-Murguia M., Jiao J., Finkelstein D., Wang Y.D., Xu B., et al. Proteasome stress in skeletal muscle mounts a long-range protective response that delays retinal and brain aging. Cell Metab. 2021;33:1137–1154.e9. doi: 10.1016/j.cmet.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Post S., Liao S., Yamamoto R., Veenstra J.A., Nässel D.R., Tatar M. Drosophila insulin-like peptide dilp1 increases lifespan and glucagon-like Akh expression epistatic to dilp2. Aging Cell. 2019;18:e12863. doi: 10.1111/acel.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katewa S.D., Demontis F., Kolipinski M., Hubbard A., Gill M.S., Perrimon N., Melov S., Kapahi P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waterson M.J., Chung B.Y., Harvanek Z.M., Ostojic I., Alcedo J., Pletcher S.D. Water sensor ppk28 modulates Drosophila lifespan and physiology through AKH signaling. Proc. Natl. Acad. Sci. USA. 2014;111:8137–8142. doi: 10.1073/pnas.1315461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eisele Y.S., Monteiro C., Fearns C., Encalada S.E., Wiseman R.L., Powers E.T., Kelly J.W. Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 2015;14:759–780. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klaips C.L., Jayaraj G.G., Hartl F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 76.Scior A., Juenemann K., Kirstein J. Cellular strategies to cope with protein aggregation. Essays Biochem. 2016;60:153–161. doi: 10.1042/EBC20160002. [DOI] [PubMed] [Google Scholar]
- 77.Vendruscolo M. Proteome folding and aggregation. Curr. Opin. Struct. Biol. 2012;22:138–143. doi: 10.1016/j.sbi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Wilson D.M., 3rd, Cookson M.R., Van Den Bosch L., Zetterberg H., Holtzman D.M., Dewachter I. Hallmarks of neurodegenerative diseases. Cell. 2023;186:693–714. doi: 10.1016/j.cell.2022.12.032. [DOI] [PubMed] [Google Scholar]
- 79.Stamer K., Vogel R., Thies E., Mandelkow E., Mandelkow E.M. Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J. Cell Biol. 2002;156:1051–1063. doi: 10.1083/jcb.200108057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goodman L.D., Prudencio M., Kramer N.J., Martinez-Ramirez L.F., Srinivasan A.R., Lan M., Parisi M.J., Zhu Y., Chew J., Cook C.N., et al. Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat. Neurosci. 2019;22:863–874. doi: 10.1038/s41593-019-0396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frottin F., Perez-Berlanga M., Hartl F.U., Hipp M.S. Multiple pathways of toxicity induced by C9orf72 dipeptide repeat aggregates and G(4)C(2) RNA in a cellular model. Elife. 2021;10:e62718. doi: 10.7554/eLife.62718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsou W.L., Burr A.A., Ouyang M., Blount J.R., Scaglione K.M., Todi S.V. Ubiquitination regulates the neuroprotective function of the deubiquitinase ataxin-3 in vivo. J. Biol. Chem. 2013;288:34460–34469. doi: 10.1074/jbc.M113.513903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raj K., Akundi R.S. Mutant Ataxin-3-Containing Aggregates (MATAGGs) in Spinocerebellar Ataxia Type 3: Dynamics of the Disorder. Mol. Neurobiol. 2021;58:3095–3118. doi: 10.1007/s12035-021-02314-z. [DOI] [PubMed] [Google Scholar]
- 84.Johnson S.L., Ranxhi B., Libohova K., Tsou W.L., Todi S.V. Ubiquitin-interacting motifs of ataxin-3 regulate its polyglutamine toxicity through Hsc70-4-dependent aggregation. Elife. 2020;9:e60742. doi: 10.7554/eLife.60742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee D., Lee Y.I., Lee Y.S., Lee S.B. The Mechanisms of Nuclear Proteotoxicity in Polyglutamine Spinocerebellar Ataxias. Front. Neurosci. 2020;14:489. doi: 10.3389/fnins.2020.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y., Mandelkow E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 87.Strang K.H., Golde T.E., Giasson B.I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Invest. 2019;99:912–928. doi: 10.1038/s41374-019-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ballatore C., Lee V.M.Y., Trojanowski J.Q. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 89.Cowan C.M., Mudher A. Are tau aggregates toxic or protective in tauopathies? Front. Neurol. 2013;4:114. doi: 10.3389/fneur.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Polanco J.C., Li C., Bodea L.G., Martinez-Marmol R., Meunier F.A., Götz J. Amyloid-beta and tau complexity - towards improved biomarkers and targeted therapies. Nat. Rev. Neurol. 2018;14:22–39. doi: 10.1038/nrneurol.2017.162. [DOI] [PubMed] [Google Scholar]
- 91.Szabo L., Eckert A., Grimm A. Insights into Disease-Associated Tau Impact on Mitochondria. Int. J. Mol. Sci. 2020;21:6344. doi: 10.3390/ijms21176344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lasagna-Reeves C.A., Castillo-Carranza D.L., Sengupta U., Clos A.L., Jackson G.R., Kayed R. Tau oligomers impair memory and induce synaptic and mitochondrial dysfunction in wild-type mice. Mol. Neurodegener. 2011;6:39. doi: 10.1186/1750-1326-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shafiei S.S., Guerrero-Muñoz M.J., Castillo-Carranza D.L. Tau Oligomers: Cytotoxicity, Propagation, and Mitochondrial Damage. Front. Aging Neurosci. 2017;9:83. doi: 10.3389/fnagi.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eftekharzadeh B., Daigle J.G., Kapinos L.E., Coyne A., Schiantarelli J., Carlomagno Y., Cook C., Miller S.J., Dujardin S., Amaral A.S., et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer's Disease. Neuron. 2018;99:925–940.e7. doi: 10.1016/j.neuron.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cornelison G.L., Levy S.A., Jenson T., Frost B. Tau-induced nuclear envelope invagination causes a toxic accumulation of mRNA in Drosophila. Aging Cell. 2019;18:e12847. doi: 10.1111/acel.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mahoney R., Ochoa Thomas E., Ramirez P., Miller H.E., Beckmann A., Zuniga G., Dobrowolski R., Frost B. Pathogenic Tau Causes a Toxic Depletion of Nuclear Calcium. Cell Rep. 2020;32:107900. doi: 10.1016/j.celrep.2020.107900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frost B., Hemberg M., Lewis J., Feany M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014;17:357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lester E., Ooi F.K., Bakkar N., Ayers J., Woerman A.L., Wheeler J., Bowser R., Carlson G.A., Prusiner S.B., Parker R. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron. 2021;109:1675–1691.e9. doi: 10.1016/j.neuron.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cassar M., Law A.D., Chow E.S., Giebultowicz J.M., Kretzschmar D. Disease-Associated Mutant Tau Prevents Circadian Changes in the Cytoskeleton of Central Pacemaker Neurons. Front. Neurosci. 2020;14:232. doi: 10.3389/fnins.2020.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Law A.D., Cassar M., Long D.M., Chow E.S., Giebultowicz J.M., Venkataramanan A., Strauss R., Kretzschmar D. FTD-associated mutations in Tau result in a combination of dominant and recessive phenotypes. Neurobiol. Dis. 2022;170:105770. doi: 10.1016/j.nbd.2022.105770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gistelinck M., Lambert J.C., Callaerts P., Dermaut B., Dourlen P. Drosophila models of tauopathies: what have we learned? Int. J. Alzheimer's Dis. 2012;2012:970980. doi: 10.1155/2012/970980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spillantini M.G., Crowther R.A., Kamphorst W., Heutink P., van Swieten J.C. Tau pathology in two Dutch families with mutations in the microtubule-binding region of tau. Am. J. Pathol. 1998;153:1359–1363. doi: 10.1016/S0002-9440(10)65721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tanemura K., Murayama M., Akagi T., Hashikawa T., Tominaga T., Ichikawa M., Yamaguchi H., Takashima A. Neurodegeneration with tau accumulation in a transgenic mouse expressing V337M human tau. J. Neurosci. 2002;22:133–141. doi: 10.1523/JNEUROSCI.22-01-00133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X., Miller N.M., Afghah Z., Geiger J.D. Development of AD-Like Pathology in Skeletal Muscle. J. Parkinson’s Dis. Alzheimer's Dis. 2019;6 doi: 10.13188/2376-922x.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kitazawa M., Trinh D.N., LaFerla F.M. Inflammation induces tau pathology in inclusion body myositis model via glycogen synthase kinase-3beta. Ann. Neurol. 2008;64:15–24. doi: 10.1002/ana.21325. [DOI] [PubMed] [Google Scholar]
- 106.Wang J., Gu B.J., Masters C.L., Wang Y.J. A systemic view of Alzheimer disease - insights from amyloid-beta metabolism beyond the brain. Nat. Rev. Neurol. 2017;13:612–623. doi: 10.1038/nrneurol.2017.111. [DOI] [PubMed] [Google Scholar]
- 107.Ciryam P., Antalek M., Cid F., Tartaglia G.G., Dobson C.M., Guettsches A.K., Eggers B., Vorgerd M., Marcus K., Kley R.A., et al. A metastable subproteome underlies inclusion formation in muscle proteinopathies. Acta Neuropathol. Commun. 2019;7:197. doi: 10.1186/s40478-019-0853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ciryam P., Kundra R., Morimoto R.I., Dobson C.M., Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ciryam P., Tartaglia G.G., Morimoto R.I., Dobson C.M., Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sui X., Pires D.E.V., Ormsby A.R., Cox D., Nie S., Vecchi G., Vendruscolo M., Ascher D.B., Reid G.E., Hatters D.M. Widespread remodeling of proteome solubility in response to different protein homeostasis stresses. Proc. Natl. Acad. Sci. USA. 2020;117:2422–2431. doi: 10.1073/pnas.1912897117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tartaglia G.G., Vendruscolo M. Correlation between mRNA expression levels and protein aggregation propensities in subcellular localisations. Mol. Biosyst. 2009;5:1873–1876. doi: 10.1039/b913099n. [DOI] [PubMed] [Google Scholar]
- 112.Vecchi G., Sormanni P., Mannini B., Vandelli A., Tartaglia G.G., Dobson C.M., Hartl F.U., Vendruscolo M. Proteome-wide observation of the phenomenon of life on the edge of solubility. Proc. Natl. Acad. Sci. USA. 2020;117:1015–1020. doi: 10.1073/pnas.1910444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vendruscolo M., Knowles T.P.J., Dobson C.M. Protein solubility and protein homeostasis: a generic view of protein misfolding disorders. Cold Spring Harb. Perspect. Biol. 2011;3:a010454. doi: 10.1101/cshperspect.a010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walther D.M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R.I., Dobson C.M., Vendruscolo M., Mann M., Hartl F.U. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell. 2015;161:919–932. doi: 10.1016/j.cell.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sydow A., Van der Jeugd A., Zheng F., Ahmed T., Balschun D., Petrova O., Drexler D., Zhou L., Rune G., Mandelkow E., et al. Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic Tau mutant. J. Neurosci. 2011;31:2511–2525. doi: 10.1523/JNEUROSCI.5245-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu T.H., Lu Y.N., Chuang C.L., Wu C.L., Chiang A.S., Krantz D.E., Chang H.Y. Loss of vesicular dopamine release precedes tauopathy in degenerative dopaminergic neurons in a Drosophila model expressing human tau. Acta Neuropathol. 2013;125:711–725. doi: 10.1007/s00401-013-1105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yoshiyama Y., Higuchi M., Zhang B., Huang S.M., Iwata N., Saido T.C., Maeda J., Suhara T., Trojanowski J.Q., Lee V.M.Y. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 118.Mandelkow E.M., Mandelkow E. Tau in Alzheimer's disease. Trends Cell Biol. 1998;8:425–427. doi: 10.1016/s0962-8924(98)01368-3. [DOI] [PubMed] [Google Scholar]
- 119.Ashok A., Singh N., Chaudhary S., Bellamkonda V., Kritikos A.E., Wise A.S., Rana N., McDonald D., Ayyagari R. Retinal Degeneration and Alzheimer's Disease: An Evolving Link. Int. J. Mol. Sci. 2020;21:7290. doi: 10.3390/ijms21197290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chiasseu M., Alarcon-Martinez L., Belforte N., Quintero H., Dotigny F., Destroismaisons L., Vande Velde C., Panayi F., Louis C., Di Polo A. Tau accumulation in the retina promotes early neuronal dysfunction and precedes brain pathology in a mouse model of Alzheimer's disease. Mol. Neurodegener. 2017;12:58. doi: 10.1186/s13024-017-0199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hart N.J., Koronyo Y., Black K.L., Koronyo-Hamaoui M. Ocular indicators of Alzheimer's: exploring disease in the retina. Acta Neuropathol. 2016;132:767–787. doi: 10.1007/s00401-016-1613-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ho W.L., Leung Y., Cheng S.S.Y., Lok C.K.M., Ho Y.S., Baum L., Yang X., Chiu K., Chang R.C.C. Investigating degeneration of the retina in young and aged tau P301L mice. Life Sci. 2015;124:16–23. doi: 10.1016/j.lfs.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 123.Lacomme M., Hales S.C., Brown T.W., Stevanovic K., Jolicoeur C., Cai J., Bois T., Desrosiers M., Dalkara D., Cayouette M. Numb regulates Tau levels and prevents neurodegeneration in tauopathy mouse models. Sci. Adv. 2022;8:eabm4295. doi: 10.1126/sciadv.abm4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gabaldon T. Origin and Early Evolution of the Eukaryotic Cell. Annu. Rev. Microbiol. 2021;75:631–647. doi: 10.1146/annurev-micro-090817-062213. [DOI] [PubMed] [Google Scholar]
- 125.Morimoto R.I., Cuervo A.M. Proteostasis and the aging proteome in health and disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014;69:S33–S38. doi: 10.1093/gerona/glu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Richter K., Haslbeck M., Buchner J. The heat shock response: life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 128.Gomez-Pastor R., Burchfiel E.T., Thiele D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Owusu-Ansah E., Song W., Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rai M., Curley M., Coleman Z., Demontis F. Contribution of proteases to the hallmarks of aging and to age-related neurodegeneration. Aging Cell. 2022;21:e13603. doi: 10.1111/acel.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cordeiro A.V., Peruca G.F., Braga R.R., Brícola R.S., Lenhare L., Silva V.R.R., Anaruma C.P., Katashima C.K., Crisol B.M., Barbosa L.T., et al. High-intensity exercise training induces mitonuclear imbalance and activates the mitochondrial unfolded protein response in the skeletal muscle of aged mice. Geroscience. 2021;43:1513–1518. doi: 10.1007/s11357-020-00246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu J., Ruas J.L., Estall J.L., Rasbach K.A., Choi J.H., Ye L., Boström P., Tyra H.M., Crawford R.W., Campbell K.P., et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13:160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hart C.R., Ryan Z.C., Pfaffenbach K.T., Dasari S., Parvizi M., Lalia A.Z., Lanza I.R. Attenuated activation of the unfolded protein response following exercise in skeletal muscle of older adults. Aging (Albany NY) 2019;11:7587–7604. doi: 10.18632/aging.102273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hentila J., Ahtiainen J.P., Paulsen G., Raastad T., Hakkinen K., Mero A.A., Hulmi J.J. Autophagy is induced by resistance exercise in young men, but unfolded protein response is induced regardless of age. Acta Physiol. 2018;224:e13069. doi: 10.1111/apha.13069. [DOI] [PubMed] [Google Scholar]
- 135.Ogborn D.I., McKay B.R., Crane J.D., Parise G., Tarnopolsky M.A. The unfolded protein response is triggered following a single, unaccustomed resistance-exercise bout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R664–R669. doi: 10.1152/ajpregu.00511.2013. [DOI] [PubMed] [Google Scholar]
- 136.Wu L., Zhou M., Li T., Dong N., Yi L., Zhang Q., Mi M. GLP-1 regulates exercise endurance and skeletal muscle remodeling via GLP-1R/AMPK pathway. Biochim. Biophys. Acta. Mol. Cell Res. 2022;1869:119300. doi: 10.1016/j.bbamcr.2022.119300. [DOI] [PubMed] [Google Scholar]
- 137.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 138.Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 139.Masters C.L., Bateman R., Blennow K., Rowe C.C., Sperling R.A., Cummings J.L. Alzheimer's disease. Nat. Rev. Dis. Primers. 2015;1:15056. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 140.Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S., Laskowski P.R., MacKenzie D., Kamath T., Commins C., et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37:e98049. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Demontis F., Patel V.K., Swindell W.R., Perrimon N. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep. 2014;7:1481–1494. doi: 10.1016/j.celrep.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Demontis F., Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hunt L.C., Demontis F. Age-Related Increase in Lactate Dehydrogenase Activity in Skeletal Muscle Reduces Life Span in Drosophila. J. Gerontol. A Biol. Sci. Med. Sci. 2022;77:259–267. doi: 10.1093/gerona/glab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hunt L.C., Jiao J., Wang Y.D., Finkelstein D., Rao D., Curley M., Robles-Murguia M., Shirinifard A., Pagala V.R., Peng J., et al. Circadian gene variants and the skeletal muscle circadian clock contribute to the evolutionary divergence in longevity across Drosophila populations. Genome Res. 2019;29:1262–1276. doi: 10.1101/gr.246884.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Keravala A., Calos M.P. Site-specific chromosomal integration mediated by phiC31 integrase. Methods Mol. Biol. 2008;435:165–173. doi: 10.1007/978-1-59745-232-8_12. [DOI] [PubMed] [Google Scholar]
- 146.Hunt L.C., Demontis F. Whole-mount immunostaining of Drosophila skeletal muscle. Nat. Protoc. 2013;8:2496–2501. doi: 10.1038/nprot.2013.156. [DOI] [PubMed] [Google Scholar]
- 147.Graca F.A., Stephan A., Wang Y.D., Shirinifard A., Jiao J., Vogel P., Labelle M., Demontis F. Progressive development of melanoma-induced cachexia differentially impacts organ systems in mice. Cell Rep. 2023;42:111934. doi: 10.1016/j.celrep.2022.111934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Graca F.A., Rai M., Hunt L.C., Stephan A., Wang Y.-D., Gordon B., Wang R., Quarato G., Xu B., Fan Y., et al. The myokine Fibcd1 is an endogenous determinant of myofiber size and mitigates cancer-induced myofiber atrophy. Nat. Commun. 2022;13:2370. doi: 10.1038/s41467-022-30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hunt L.C., Pagala V., Stephan A., Xie B., Kodali K., Kavdia K., Wang Y.D., Shirinifard A., Curley M., Graca F.A., et al. An adaptive stress response that confers cellular resilience to decreased ubiquitination. Nat. Commun. 2023;14:7348. doi: 10.1038/s41467-023-43262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rai M., Curley M., Coleman Z., Nityanandam A., Jiao J., Graca F.A., Hunt L.C., Demontis F. Analysis of proteostasis during aging with western blot of detergent-soluble and insoluble protein fractions. STAR Protoc. 2021;2:100628. doi: 10.1016/j.xpro.2021.100628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Robles-Murguia M., Rao D., Finkelstein D., Xu B., Fan Y., Demontis F. Muscle-derived Dpp regulates feeding initiation via endocrine modulation of brain dopamine biosynthesis. Genes Dev. 2020;34:37–52. doi: 10.1101/gad.329110.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jiao J., Kavdia K., Pagala V., Palmer L., Finkelstein D., Fan Y., Peng J., Demontis F. An age-downregulated ribosomal RpS28 protein variant regulates the muscle proteome. G3 (Bethesda) G3 (Bethesda). 2021;11:jkab165. doi: 10.1093/g3journal/jkab165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hunt L.C., Stover J., Haugen B., Shaw T.I., Li Y., Pagala V.R., Finkelstein D., Barton E.R., Fan Y., Labelle M., et al. A Key Role for the Ubiquitin Ligase UBR4 in Myofiber Hypertrophy in Drosophila and Mice. Cell Rep. 2019;28:1268–1281.e6. doi: 10.1016/j.celrep.2019.06.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xu P., Duong D.M., Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J. Proteome Res. 2009;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zaman M., Fu Y., Chen P.C., Sun H., Yang S., Wu Z., Wang Z., Poudel S., Serrano G.E., Beach T.G., et al. Dissecting Detergent-Insoluble Proteome in Alzheimer's Disease by TMTc-Corrected Quantitative Mass Spectrometry. Mol. Cell. Proteomics. 2023;22:100608. doi: 10.1016/j.mcpro.2023.100608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bai B., Tan H., Pagala V.R., High A.A., Ichhaporia V.P., Hendershot L., Peng J. Deep Profiling of Proteome and Phosphoproteome by Isobaric Labeling, Extensive Liquid Chromatography, and Mass Spectrometry. Methods Enzymol. 2017;585:377–395. doi: 10.1016/bs.mie.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang X., Li Y., Wu Z., Wang H., Tan H., Peng J. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Mol. Cell. Proteomics. 2014;13:3663–3673. doi: 10.1074/mcp.O114.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pagala V.R., High A.A., Wang X., Tan H., Kodali K., Mishra A., Kavdia K., Xu Y., Wu Z., Peng J. Quantitative protein analysis by mass spectrometry. Methods Mol. Biol. 2015;1278:281–305. doi: 10.1007/978-1-4939-2425-7_17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-sequencing data from Drosophila melanogaster strains (whole flies, males, 10-days-old) reared at 18°C, 25°C, and 29°C and that express misfolding-prone proteins that localize primarily to the cytoplasm (FlucDM), the mitochondria (mito-FlucDM), and the nucleus (NLS-FlucDM), compared to isogenic control flies (w1118) that do not express FlucDM. For mito-FlucDM and NLS-FlucDM, homozygous and heterozygous strains that carry respectively 2 copies and 1 copy of the FlucDM transgenes were analyzed. For each condition, 3 biological replicates were analyzed. Tab 1: gene expression changes induced by FlucDM variants compared to w1118 controls at 18°C, 25°C, and 29°C. Tab 2: TPM (transcripts per million) values, indicating the mRNA levels for each gene across all samples. Tab 3: FlucDM cross-comparison: gene expression changes induced by mito-FlucDM and NLS-FlucDM compared to the untargeted (∼cytoplasmic) FlucDM at 18°C, 25°C, and 29°C. Tab 4: no meaningful upregulation of Hsp60 mRNA levels, a marker of the UPRmt
Ultra-deep-coverage proteomics (18-plex TMT) of detergent-soluble and insoluble fractions from isogenic knock-in male flies that express wild-type human tauWT and pathogenic human tauV337M at 10 and 60 days of age
Protein categories that display change in detergent-solubility in tauV337M flies at 60 versus 10 days, and in tauV337M versus tauWT flies at 60 days of age
Data Availability Statement
-
•
The data supporting the findings of this study are available within the paper (Figures 1, 2, 3, 4, 5, 6, 7 and S1–S7 and Table S1. Transcriptional stress responses induced by compartment-targeted misfolding-prone FlucDM proteins in Drosophila, related to Figure 3, Table S2. Protein solubility changes induced by pathogenic human tau/MAPT during aging in Drosophila, related to Figure 7, Table S3. GO term analysis of protein categories that display tauV337M-induced changes in solubility during aging, related to Figure 7, Table S4. Source data file, related to all figures and supplemental figures, Table S5. Full scans of western blots, related to Figures 4–7, S1–S3, and S5). The TMT mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository and are accessible with the dataset identifier PRIDE: PXD045325. The RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) with the identifier GEO: GSE243224.
-
•
This paper does not report original code.
-
•
Any other information required to reanalyze the data reported in this paper is available from the lead contact upon request.