Summary
The Gene Expression Omnibus (GEO) has millions of samples from thousands of studies. While users of GEO can search the metadata describing studies, there is a need for methods to search GEO at the data level. RummaGEO is a gene expression signature search engine for human and mouse RNA sequencing perturbation studies extracted from GEO. To develop RummaGEO, we automatically identified groups of samples and computed differential expressions to extract gene sets from each study. The contents of RummaGEO are served for gene set, PubMed, and metadata search. Next, we analyzed the contents of RummaGEO to identify patterns and perform global analyses. Overall, RummaGEO provides a resource that is enabling users to identify relevant GEO studies based on their own gene expression results. Users of RummaGEO can incorporate RummaGEO into their analysis workflows for integrative analyses and hypothesis generation.
Keywords: transcriptomics, gene expression, gene set enrichment analysis, data mining, data integration, signature search, LINCS, CFDE, ARCHS4, RNA sequencing
Highlights
-
•
RummaGEO is a search engine for gene expression signatures mined from GEO
-
•
Signatures were automatically computed by identifying groups of samples
-
•
The RummaGEO database contains 171,441 human and 195,265 mouse gene sets
-
•
The uniformly processed RNA-seq data from ARCHS4 was used to create the signatures
The bigger picture
Millions of samples of gene expression profiles have been deposited into the Gene Expression Omnibus (GEO) examining the effects in diseases, drugs, gene knockouts, and other perturbations. Although the GEO database can be queried by text prompting the indexed metadata, more methods to search GEO at the data level are needed. RummaGEO automatically partitions human and mouse GEO studies into groups of samples suitable for differential expression analysis, producing up- and down-regulated gene sets across most perturbation experiments in GEO. The RummaGEO search engine provides a user-friendly and fast search for these gene sets. RummaGEO promotes reuse, reanalysis, and integration of previously performed experiments, it can be accessed via API, and the gene sets are provided for download.
RummaGEO is a Gene Expression Omnibus (GEO) search engine that can be used to find matches for user-submitted gene sets or any metadata search term such as disease, phenotype, tissue, cell type, cell line, drug, or gene. To create RummaGEO, gene expression signatures were automatically computed by identifying groups of samples based on sample descriptions in GEO and the uniformly processed RNA-seq data from ARCHS4.
Introduction
The Gene Expression Omnibus (GEO) contains tens of thousands of transcriptomics studies and over 2 million genome-wide gene expression samples collected by RNA sequencing (RNA-seq).1 Such a massive transcriptomics profiling corpus covers many organisms, disease conditions, drug treatments, and genetic perturbations such as knockouts, knockdowns, and overexpression of genes across tissues, cell types, and cell lines. Although users of GEO can now download most RNA-seq datasets as gene expression count matrices, currently, it is difficult to search the GEO database at the data level. In addition, metadata about the conditions of each study and samples within each study have inconsistent formatting and follow different naming conventions.2 Multiple attempts have been made to make GEO studies better searchable by standardizing and restructuring the GEO metadata. For instance, GEOmetadb provides an R package together with an SQLite database to query GEO datasets locally, improving the speed of querying and the accessibility of the GEO metadata.3 Similarly, Restructured GEO (ReGEO) utilized natural language processing (NLP) to extract time points and disease terms from GEO metadata, enabling users to search by these attributes and many other biomedical terms embedded in the original GEO metadata.4 Another notable effort is MetaSRA.5 MetaSRA maps GEO metadata to ontologies and dictionaries. These mappings facilitate a metadata search engine that is better at identifying samples and studies. A more recent effort utilized multiple GPT4 “agents” to annotate and partition GEO studies, enabling improved on-the-fly labeling of control and perturbation conditions. The approach was used to create a drug-repurposing database from differential expression signatures identified with the method. However, the large language model (LLM)-powered pipeline was applied to only a subset of relevant GEO studies, and the annotations are not available for download and reuse.6
Although these efforts make GEO metadata more accessible and searchable, these resources do not enable users to search the GEO database at the data level, nor do they provide direct access to uniformly aligned samples and signatures from the processed GEO studies. Several efforts have aimed to uniformly align GEO RNA-seq transcriptomics samples and make these accessible to users for reuse. For example, Recount, in its third iteration called Recount3, uniformly aligned more than 750,000 human and mouse RNA-seq samples from GEO,1 GTEx,7 and The Cancer Genome Atlas,8 enabling users to more easily investigate and compare gene expression profiles across these resources.9 Recount3-processed datasets are served via an R Shiny web-based data explorer and an R package. The GEO RNA-Seq Experiments Interactive Navigator (GREIN) also uniformly aligned over 600,000 GEO RNA-seq samples from humans, mice, and rats. GREIN-processed data are served via an interactive R Shiny application that enables users to investigate these studies and request new GEO studies to be aligned and added to the database.10 We have developed the All RNA-seq and ChIP-seq (chromatin immunoprecipitation sequencing) sample and signature search (ARCHS4) resource.11 ARCHS4 provides access to over 2 million human and mouse uniformly aligned RNA-seq samples from GEO. Another, similar, effort called Digital Expression Explorer 2 provides uniformly aligned RNA-seq data for humans and mice and other species.12 These projects provide valuable uniformly processed data from GEO and other resources, making such data more accessible and reusable. However, none of these resources provide the uniformly aligned data for search at the signature level.
Differential gene expression signatures associated with these studies, however, still must be manually computed. This means users need to manually parse the metadata associated with each sample to determine proper groupings of samples, which can be a time-consuming process when attempted for thousands of studies. Various efforts have attempted to compute signatures automatically or manually from GEO studies. The Crowd Extracted Expression of Differential Signatures resource, for instance, provides manually curated and automatically generated gene, drug, and disease perturbation signatures extracted from GEO studies.13 These signatures were created via a crowdsourcing project that provided participants with access to the tool GEO2Enrichr.14 GEO2Enrichr enables users to extract differentially expressed genes from GEO studies using a browser extension. After identifying the control and perturbation samples, users can submit the computed signatures to Enrichr for reanalysis. A related project, GEN3VA,15 saves signatures extracted with GEO2Enrichr, and then makes these signatures available to the public as collections based on hashtags. The main limitation of GEO2Enrichr and GEN3VA is that they only work for processing data from microarray studies. There are many other projects that enable users to manually annotate GEO studies and their metadata in a user-friendly interface. For example, GEOMetaCuration provides users with an intuitive graphical user interface to label and submit relevant metadata and keywords associated with GEO studies.16 BioJupies17 enables users to select samples from GEO studies that were uniformly aligned by ARCHS4 to label control and perturbation conditions, compute differential expression, and perform a variety of analyses and visualizations using automatically executed Jupyter Notebook in the Cloud. In a similar vein, TidyGEO18 and iLINCS19 allow users to select GEO series and examine and label their metadata, as well as perform multiple data cleaning and filtering tasks, followed by standard downstream analyses such as differential expression and pathway analysis. These bioinformatics web applications, while useful, still require users to manually search and select their studies and conditions. To enable more efficient, automatic, and large-scale analyses, automated label extraction has attempted to label GEO metadata based on gene expression signatures using logistic regression, predicting the tissue, age, and gender of samples that do not have such annotations.20 While numerous projects aim to facilitate standardization, manipulation, and exploration of GEO studies and samples, there is a need for resources that enable searching GEO at the data level.
In the past, several efforts have been established to serve GEO data in a more digestible format. For example, ExpressionBlast used regular expressions to identify groups of samples and provided a search engine for normalized expression across studies for microarray data.21 Search-Based Exploration of Expression Compendium (SEEK) was developed to provide searching for gene or gene sets across a subset of human microarray and RNA-seq studies uniformly processed from GEO.22 Unfortunately, both ExpressionBlast and SEEK are no longer available or have not been updated since 2015, respectively. A more recent effort called GENe Expression Variance Analysis (GENEVA) leveraged the data from ARCHS4 to semi-automatically identify and serve groups of samples from human studies.23 The GENEVA website that hosted the data and the search engine to serve these processed datasets is also no longer publicly available. It should be noted that GENEVA and GEN3VA are two separate unrelated projects. To facilitate this type of search, here, we performed automatic identification and grouping of conditions of GEO samples from thousands of GEO studies, and then performed differential expression analysis producing hundreds of thousands of human and mouse signatures that are made available for search via a user-friendly web interface.
Results
Descriptive statistics of the contents within the RummaGEO database
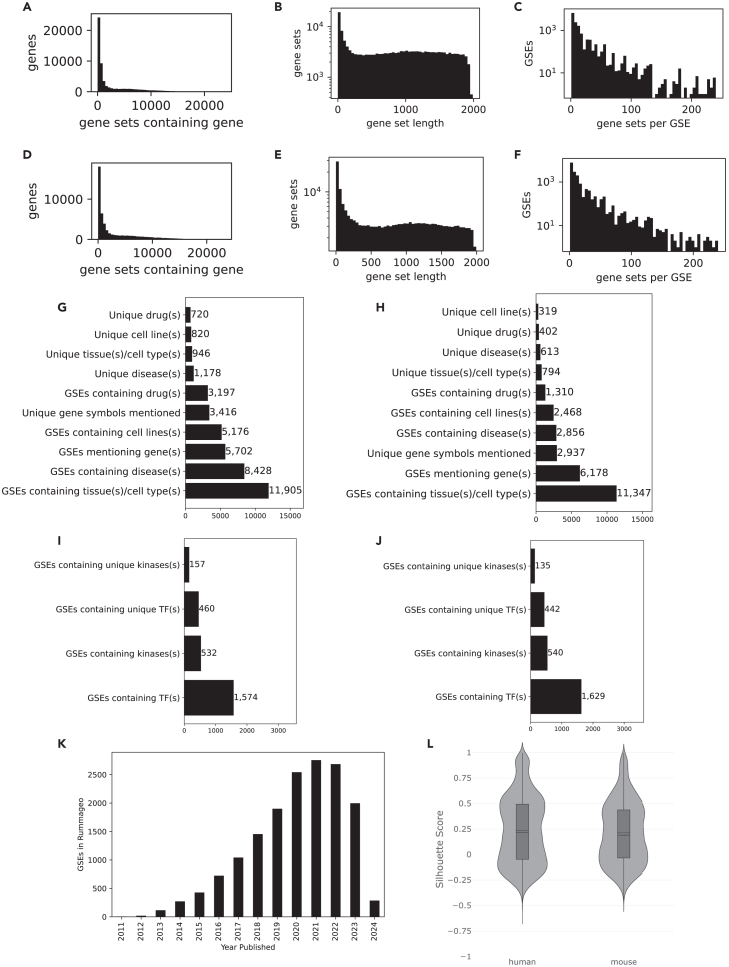
The current release of RummaGEO (https://rummageo.com/) contains 171,441 human and 195,265 mouse gene sets extracted from 29,294 GEO studies (Table S3). In general, most genes appear in only a small number of gene sets in both the collections of human sets (Figure 1A) and mouse sets (Figure 1D). There are many gene sets with less than 100 genes, while the remaining sets are relatively equally distributed for both human (Figure 1B) and mouse (Figure 1E). The maximum gene set size that we defined is 2,000. Additionally, while most of the studies contributed just a few gene sets, there are periodic peaks for studies that contributed more sets. This periodicity is a result of the possible combinations of conditions and groups with a bias toward having an even number of groups in the study design (Figures 1C–1F). By identifying functional terms in sample metadata, we found that a large majority of studies, GEO series (GSEs), contain the tissue or the cell type term that is often found in the source_ch1 metadata field. Diseases were also identified for over half of the human GSEs (8,428), but many fewer disease terms were identified for mouse GSEs (2,856). Additionally, smaller subsets of GSEs mentioned genes, drugs, and cell lines in the sample or the study metadata (Figures 1G and 1H). From the identified gene symbols, we also extracted the subsets of kinases and transcription factors (TFs; Figures 1I and 1J). When plotting the contribution of studies over time, we observe a linear increase in GSEs added to RummaGEO. The drop in GSEs that starts in 2022 is due to processing data from the 2023 release of ARCHS4 (Figure 1K). There is some delay in the process of GEO study availability, and thus we observe a decline in the number of studies from 2023, and to a lesser extent from 2022. Silhouette scores were computed on the dimensionality-reduced samples to examine whether the samples cluster in expression space as expected based on the groupings determined by the metadata. The silhouette scores for both human and mouse exhibit a bimodal distribution with one peak where the metadata and data levels are highly aligned (∼0.5) and another peak where there is less alignment between the data and metadata (∼−0.1) (Figure 1L).
Figure 1.
Distributions of genes and gene sets within the RummaGEO database
(A) Distribution of genes across the human gene sets.
(B) Distribution of gene set lengths across the human gene sets.
(C) Human gene sets per GSE.
(D) Distribution of genes across mouse gene sets.
(E) Distribution of gene set lengths across mouse gene sets.
(F) Mouse gene sets per GSE.
(G) GSEs per year.
(H) Silhouette scores across all human and mouse studies.
(I) Unique and non-unique diseases, drugs, cell lines, tissues, and cell types, and genes mentioned in GSE and GSM metadata for human studies.
(J) Unique and non-unique diseases, drugs, cell lines, tissues and cell types, and genes mentioned in GSE and GSM metadata for mouse studies.
(K and L) Unique and non-unique kinase and TFs identified from gene mentions in GSE and GSM metadata for human (K) and mouse (L) studies.
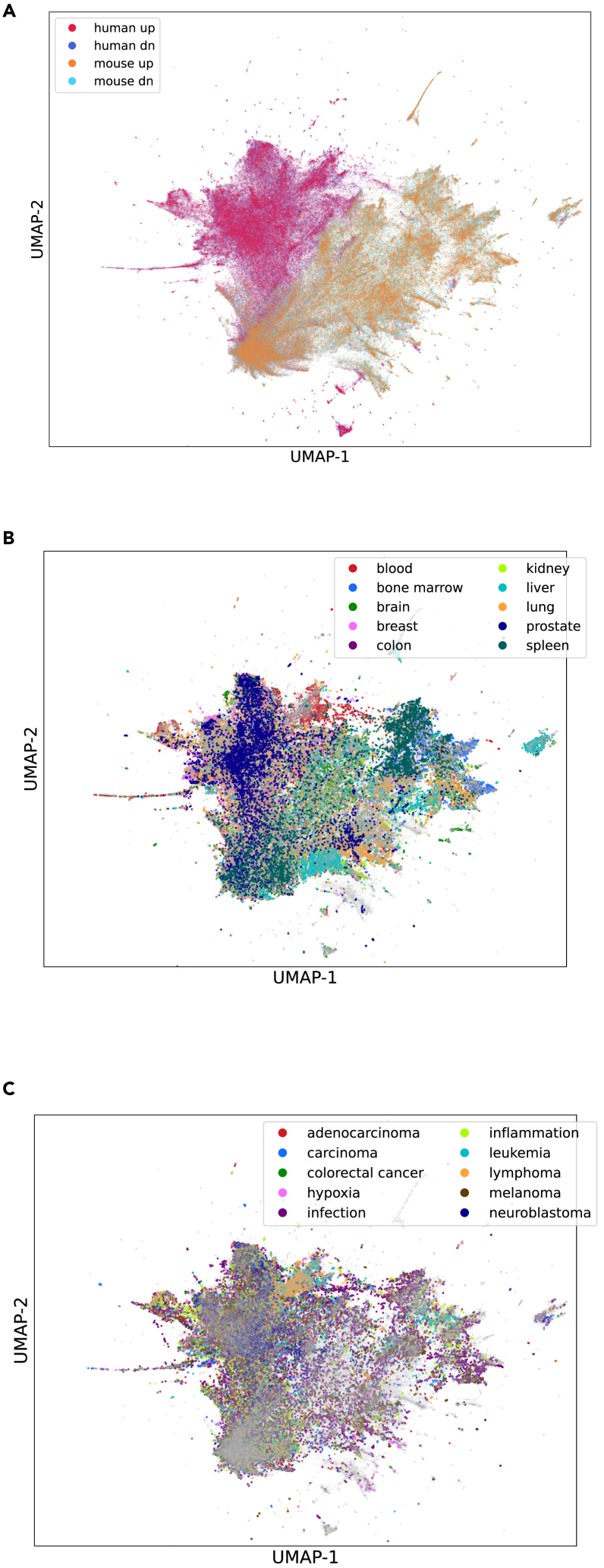
Global visualization of gene sets with UMAP
To visualize all of the gene sets within the RummaGEO database, gene sets were vectorized using inverse document frequency (IDF) followed by truncated singular value decomposition (SVD) and visualized as a uniform manifold approximation and projection (UMAP). SVD was utilized to compute the UMAP more efficiently on the large collection of vectors. Despite the harmonization of human and mouse gene symbols, we observe significant separation of the human and mouse gene sets in the global UMAP. Utilizing the species as the cluster label, a silhouette score was computed for the human and mouse gene sets (0.135) and for shuffled labels (−0.0065 ± 0.0038), indicating that there is species coherence (p = 1.456e−17) but not complete separation. However, the up and down signatures within each species are highly mixed (Figure 2A). Next, we used the metadata extracted from the GSEs and GEO samples (GSMs) to color the gene sets in the UMAP with the aim of elucidating additional patterns. When coloring by the most common tissues, we observe coherent groups of samples (Figure 2B), indicating that the gene sets in RummaGEO are significantly influenced by their tissue of origin. We performed Leiden clustering on the UMAP for gene sets with tissue labels and evaluated the association between each cluster (n = 258) and tissue type using a Monte Carlo chi-squared test (chi-squared statistic 1.35e6, p < 0.0001). Although less pronounced, the gene sets also appear to group by disease when visualizing the top 10 identified diseases (Figure 2C). This is particularly apparent for diseases such as leukemia and lymphoma, possibly due to their blood and bone marrow origins.
Figure 2.
UMAP projection of all human and mouse gene sets in the RummaGEO database
(A) Human up, human down, mouse up, and mouse down gene sets colored separately.
(B) The same UMAP is colored by the top 10 most mentioned tissues in the GSEs and GSMs metadata.
(C) The UMAP is colored by the top 10 most mentioned diseases in GSEs and GSMs metadata.
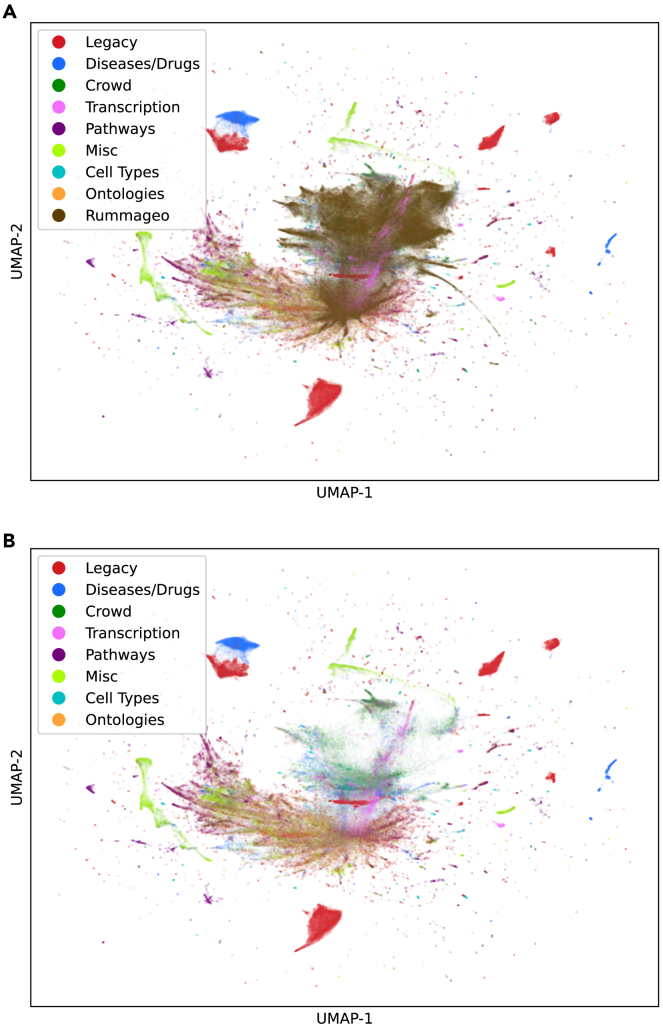
Comparison of the RummaGEO and Enrichr gene set spaces
To better understand the breadth of coverage of the RummaGEO gene sets, we compared it to the Enrichr24 gene set space. Enrichr has thousands of curated gene sets spanning several domains and categories, including transcription, pathways, ontologies, diseases and drugs, cell types and tissues, miscellaneous, and crowd generated. Enrichr gene sets mainly cluster by category (Figure 3A). Overlaying the RummaGEO gene sets on top of the Enrichr gene sets, we observe that most gene sets in RummaGEO fall in the “crowd generated” category. This is expected because the crowd generated category is also mainly composed of user-extracted gene sets from GEO studies (Figure 3B). There is also some overlap between Enrichr and RummaGEO gene sets in the center of the UMAP. However, these sets belong to several Enrichr categories, so it is difficult to discern a clear pattern from that region of the UMAP plot. To elucidate the similarity of the RummaGEO gene sets to the different Enrichr categories, we computed the Euclidean distance between each category centroid, wherein crowd generated was the closest by a large margin (0.165), followed by transcription (1.518), cell types (2.003), and diseases and drugs (2.417).
Figure 3.
UMAP projection of the Enrichr and RummaGEO gene set spaces
(A) Enrichr gene set space, colored by Enrichr categories.
(B) RummaGEO human and mouse gene sets mapped to human protein coding genes overlaid onto the Enrichr gene set space.
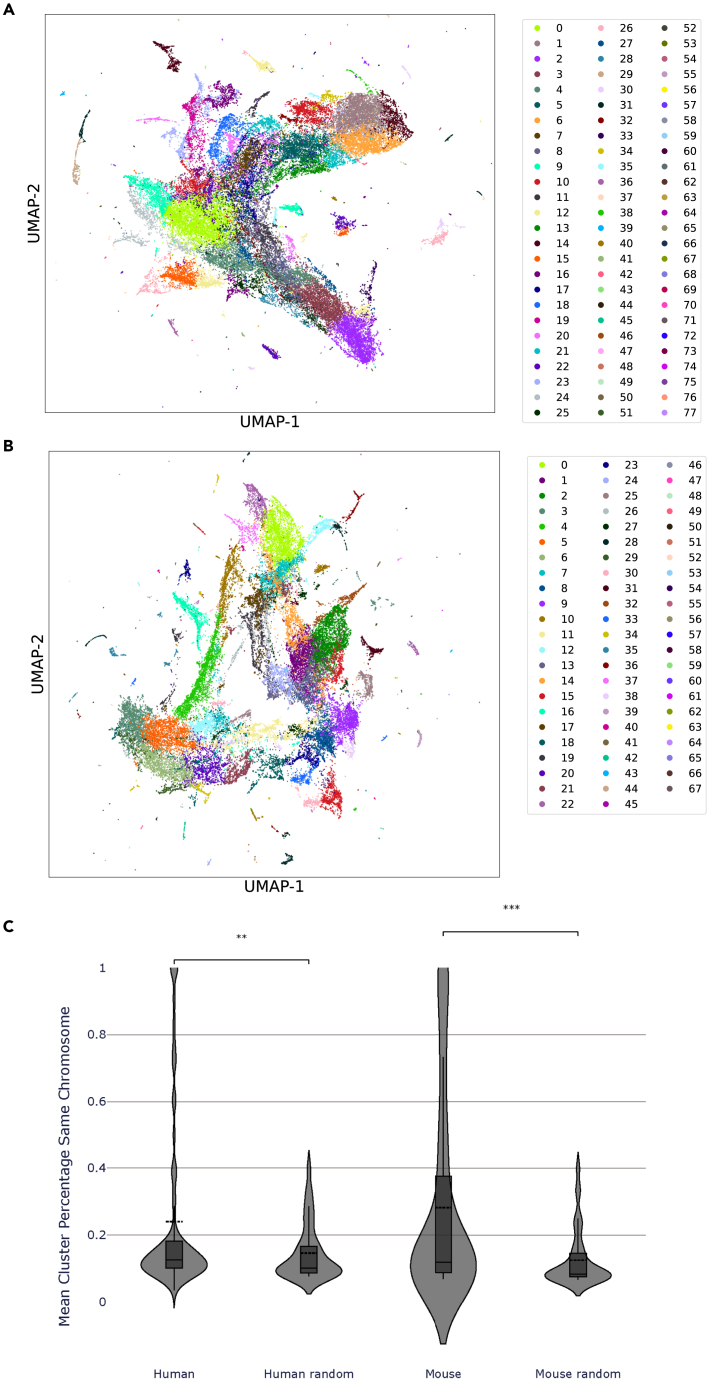
Global visualization of genes with UMAP
In addition to examining the RummaGEO gene set space, we can transpose the matrix to examine the gene similarity space. By plotting the gene vectors into two dimensions and then automatically identifying clusters of genes, we can identify functional clusters and see whether groups of genes are differentially expressed by their chromosomal locations. We automatically identified clusters using the Leiden algorithm.25 The algorithm identified 77 human clusters and 67 mouse clusters (Figures 4A and 4B). For more than two-thirds of the mouse clusters and for over half of human clusters, we identified consistent and clear statistically significant functional terms from the Enrichr24 libraries Gene Ontology (GO) Biological Processes,26 Kyoto Encyclopedia of Genes and Genomes (KEGG),27 Reactome,28 and WikiPathways29 (Table S1). Clusters of genes that co-occur in RummaGEO gene sets are organized into modules such as innate immune response, cytokine signaling, cell cycle, and regulation of autophagy (Figures 4A and 4B), while for other identified modules, there is less clear functional assignment. Additionally, to assess the influence of chromosome location on the formation of such modules, we computed the percentage of genes originating from the same chromosome in each cluster (Figure 4C). We observe that for both human and mouse clusters, there are some clusters that exhibit enrichment for specific chromosomes.
Figure 4.
UMAP projection of all human and mouse genes in the RummaGEO database
(A) UMAP projection of human genes clustered with the Leiden algorithm; clusters with consistent significant enrichment across multiple libraries and clusters with ≥33% membership to a single chromosome are labeled: 1 (organelle assembly); 3 (olfactory transduction); 5 (neuronal system); 6 (autophagy regulation); 7 (carbon dioxide transport); 9 (olfactory transduction); 12 (glutathione metabolism); 13 (extracellular matrix organization); 16 (antigen receptor signaling); 19 (inflammatory response); 23 (immune cytokine signaling); 24 (taste transduction); 26 (triglyceride biosynthesis); 27 (taste transduction); 32 (ciliopathies); 33 (sex differentiation); 34 (chr 19 72.8%); 35 (p53 signaling); 38 (Fanconi anemia pathway); 39 (cytoplasmic translation); 40 (melanin biosynthesis); 41 (tRNA aminoacylation); 42 (cholesterol biosynthesis); 43 (chr HG2023_PATCH 39.3%); 46 (histone demethylation); 47 (chr 1 100.0%); 49 (chr 7 76.9%); 51 (glycolysis); 52 (chr 17 90.9%); 53 (chr 21 40.0%); 57 (chr 15 37.5%); 61 (chr 21 71.4%); 63 (protein deubiquitination); 68 (chr Y 50.0%); 69 (chr 12 83.3%); and 75 (chr 1 60.0%); 76 (chr 9 100.0%).
(B) UMAP projection of mouse genes clustered with the Leiden algorithm; clusters with consistent significant enrichment across multiple libraries and clusters with ≥33% membership to a single chromosome are labeled: 2 (spliceosome); 6 (humoral immune response); 7 (sex determination); 8 (extracellular matrix organization); 9 (synaptic transmission); 11 (melanin biosynthesis); 13 (innate immune system); 14 (cilium assembly); 15 (metabolism); 17 (interferon signaling pathway); 19 (Th17 cell differentiation); 20 (female gonad development); 21 (meosis); 22 (phototransduction); 24 (cytokine signaling pathway); 25 (striated muscle contraction); 27 (lipolysis regulation); 29 (chr Y 48.6%); 30 (androgen biosynthesis); 31 (cilliopathies); 32 (cell cycle); 33 (ion channel transport); 34 (chr NA 37.8%); 37 (phototransduction); 40 (cytoplasmic translation); 41 (cellular respiration); 42 (chr 2 95.4%); 43 (chr 13 82.8%); 44 (chr 12 94.4%); 45 (chr 7 100.0%); 47 (chr 7 100.0%); 49 (chr X 55.0%); 50 (tRNA aminoacylation); 52 (chr Y 72.2%); 53 (chr Y 73.3%); 54 (chr Y 88.9%); 55 (chr Y 55.6%); 57 (chr 14 37.5%); 60 (chr X 71.4%); 61 (chr 4 50.0%); 63 (chr 10 33.3%); 64 (chr 4 33.3%); 65 (histone demethylation); and 66 (chr Y 100.0%).
(C) Fraction of genes from the same chromosome in human and mouse clusters and at random.
Benchmarking TF and kinase libraries created from the RummaGEO database
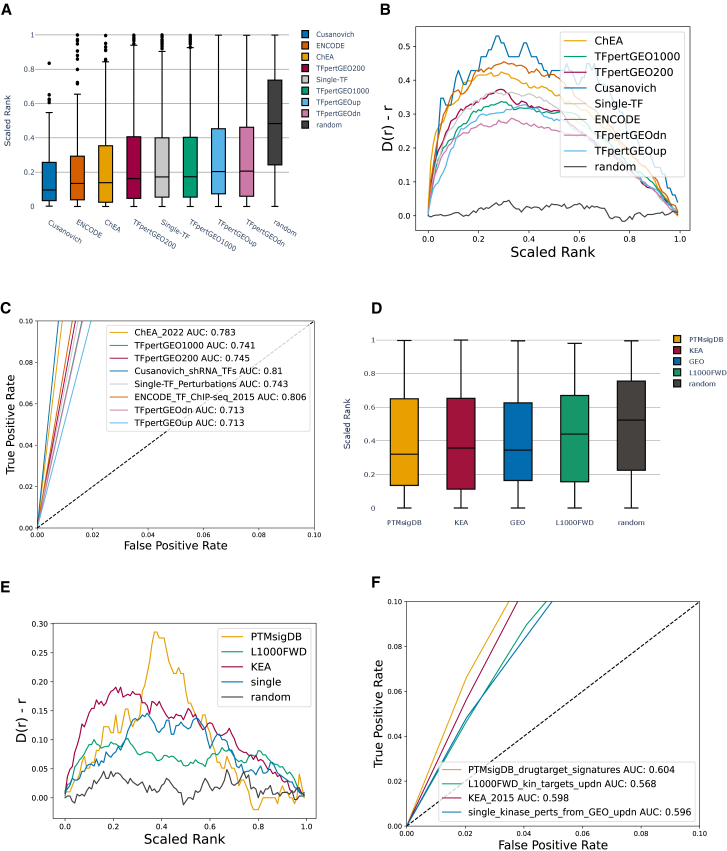
To assess the ability of RummaGEO signatures to recover known TF targets and kinase substrates, we created TF and kinase gene set libraries from the RummaGEO gene-gene co-occurrence matrix for coding genes. Transcription factors are direct regulators of differential gene expression, whereas kinases are key cell-signaling molecules that control the activity of TFs and, in turn, the regulation of gene expression. Kinase activity is used to regulate cell proliferation, cell growth, immune response, and many other key biological processes. In addition, kinases serve as drug targets. Hence, understanding their function, interactions, and activity is critical to understanding the molecular networks that control cellular phenotypes. To benchmark these libraries, we utilized ChIP-X Enrichment Analysis 3 (ChEA3)30 and Kinase Enrichment Analysis 3 (KEA3)31 and the benchmarking libraries these resources previously collected. Enrichment analysis was performed using the RummaGEO TFs and kinases libraries, and the rank of a given TF or a kinase from the benchmarking datasets was determined by the significance of the overlap based on the p value from Fisher’s exact test. For TFs, the Cusanovich short hairpin RNA (shRNA) TFs32 library showed the most accurate recovery of TFs (area under the receiver operating characteristic [ROC] curve [AUC]: 0.81) (Figures 5A–5C). All other TFs benchmarking libraries showed greater than 0.70 AUC. For the kinase libraries, the Post-Translational Modification Signatures Database (PTMsigDB33) drug signatures showed the highest AUC of 0.604 (Figures 5D–5F). The lower performance for kinases is expected because the RummaGEO gene-gene co-expression matrix is based on mRNA expression, while kinase phosphorylation events happen at the proteome and phosphoproteome levels. Signatures from TF and kinase perturbations extracted from GEO did not show the highest recovery. This is less expected because the RummaGEO kinase and TF libraries originate from the same source. Comparing the performance of the RummaGEO TF and kinase libraries to those created from Rummagene,34 the RummaGEO library performs similarly, with TF recovery being slightly better (mean AUC: 0.757 vs. 0.708) and kinase recovery slightly worse (mean AUC: 0.592 vs. 0.640). This is expected because the gene sets of RummaGEO are exclusively from transcriptomics and the gene sets from Rummagene are from multiple sources, including transcriptomics, proteomics, and the literature. Additionally, we created a partial intersection gene set library from signatures mentioning a TF or a kinase in their study metadata; however, this approach performed worse than the gene set libraries created based on co-occurrence (TF mean AUC: 0.625, kinase mean AUC: 0.593) (Figure S2).
Figure 5.
RummaGEO kinase and TF libraries benchmarking
(A) Scaled rank (0 highest rank, 1 lowest rank) for TF benchmarking libraries as computed by Fisher’s exact test.
(B) Deviation of the cumulative distribution for scaled ranks of each TF from uniform distribution (Kolmogorov-Smirnov test for goodness of fit compared to uniform distribution: ChEA [ChEA_2022] p = 2.20E−88; TFpertGEO1000 p = 4.40E−115; TFpertGEO200 p = 6.84E−119; Cusanovich_shRNA_TFs p = 2.05E−12; Single-TF_Perturbations p = 2.98E−117; ENCODE_TF_ChIP-seq_2015 p = 1.14E−110; TFpertGEOdn p = 1.35E−89; TFpertGEOup p = 3.32E−92.
(C) A total of 5,000 bootstrapped curves with downsampled negative class were generated to compute mean ROC curves and mean AUC for TFs. Only the leading edge is visualized.
(D) Scaled rank (0 highest rank, 1 lowest rank) for kinase benchmarking libraries as computed by Fisher’s exact test.
(E) Deviation of the cumulative distribution for scaled ranks of each kinase from uniform distribution (Kolmogorov-Smirnov test for goodness of fit compared to uniform distribution: PTMsigDB_drugtarget_signatures p = 1.66E−6; L1000FWD_kin_targets_updn p = 1.09E−8; KEA_2015 p = 9.39E−10; single_kinase_perts_from_GEO_updn (single) p = 5.94E−8.
(F) A total of 5,000 bootstrapped curves with downsampled negative class generated to compute mean ROC curves and mean AUC for kinases. Only the leading edge is visualized.
Benchmarking sample partitions
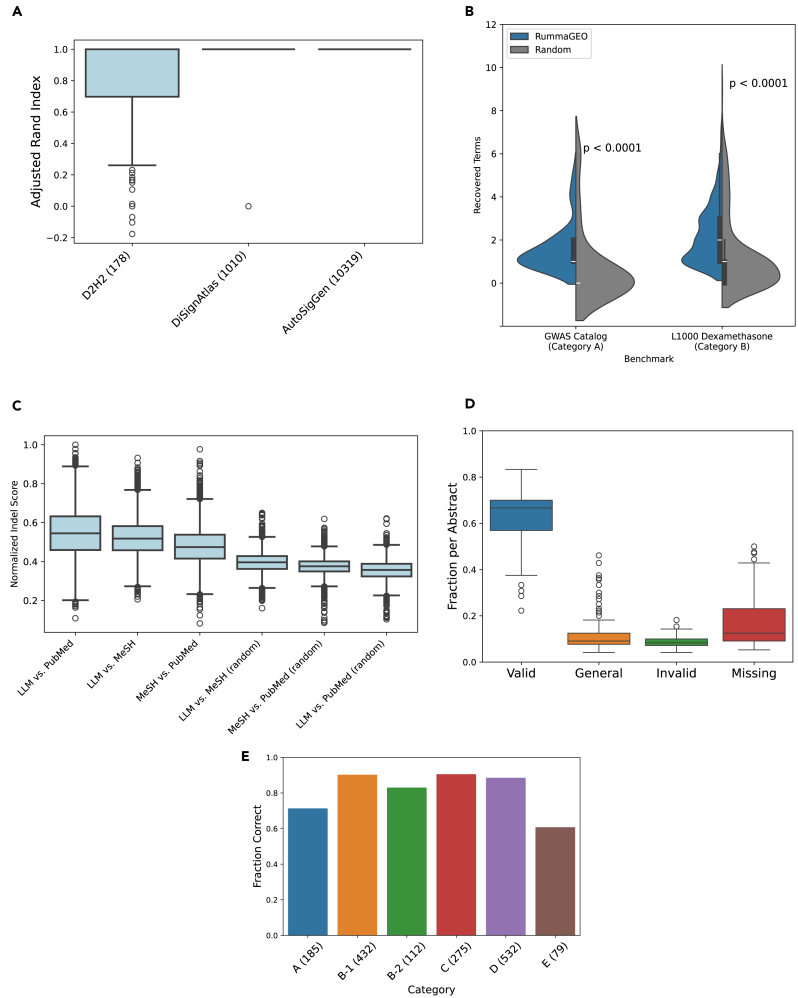
To assess the accuracy of the automated sample partitions into groups by the RummaGEO resource, we utilized several resources that manually identified groups of samples. The Diabetes and Data Hypothesis Hub (D2H2)35 resource contains hundreds of RNA-seq and microarray gene expression signatures manually extracted from GEO studies related to diabetes and other metabolic disorders. A total of 178 of these studies overlap with the RummaGEO database. These studies often contain multiple conditions and groups, making them more complex to partition. All studies contained within the D2H2 resource are manually partitioned according to the GEO metadata and series description. Comparing the groupings from D2H2 using the Adjusted Rand Index (ARI), RummaGEO partitioning performs similarly to the resource for ∼124 studies (∼70%, ARI >0.8), demonstrating the ability of the automated method to effectively partition complex perturbation experiments (Figure 6A). Further inspection of cases where the ARI is low (<0.5) reveals that some of these cases relate to time-series conditions. In these cases, the partitions are not separated by time points but instead by groups. Manual inspection of other studies revealed manual curation errors, as well as cases where some experimental conditions were grouped by a different criterion—for example, young vs. old instead of healthy vs. disease. Overall, although a high ARI gives confidence that a study is partitioned correctly, a lower ARI does not always indicate incorrect partitioning for valuable signature computation. For more basic studies containing one perturbation, or one disease group, and one control group, the RummaGEO method partitions are perfectly aligned with DiSignAtlas,36 except one case (n = 1,010). DiSignAtlas contains manually partitioned experiments from GEO. Additionally, comparing the RummaGEO partitions to a previous automated approach we applied to create the GEO Reverse Search Appyter,37 which grouped studies more stringently, the 10,000 studies that were compared grouped exactly the same.
Figure 6.
Benchmarking sample partitioning and keyword extraction
(A) Comparing sample partitions by ARI from RummaGEO to the D2H2 manual partitions (n = 178), DiSignAtlas (n = 1,010), and AutoSigGen, which is another automatic resource used for the Gene Centric GEO Reverse Search Appyter (n = 10,319).
(B) Recovery of related disease/phenotype terms (category A) from GWAS Catalog gene sets (n = 1,000) and dexamethasone-related biomolecules (category B) from up- and down- L1000 dexamethasone signatures (n = 934), compared to random GWAS gene sets and random L1000 signatures.
(C) Mean normalized string similarity (indel) of LLM, PubMed, and MeSH terms compared to random.
(D) Manually evaluated and categorized LLM extracted keywords (n = 200 abstracts).
(E) Manually evaluated category designations of keywords. A, disease/phenotype; B1, genes/proteins; B2, other biomolecules; C, tissue/cell/organ/organism/model; D, pathway/biological process; E, assay; and other.
Benchmarking functional term extraction
To benchmark functional term extraction from RummaGEO studies, we randomly selected 1,000 disease/phenotype gene sets from the GWAS (genome-wide association study) Catalog38 and performed enrichment analysis with the extracted terms in category A (disease, phenotype). A p value threshold was applied such that an average of 25 enriched terms were identified for each gene set, ignoring nonspecific terms enriched in >50% of the gene sets. RummaGEO predicted significantly more terms closely related to the disease/phenotype than expected by chance (Mann-Whitney p < 0.0001) (Figure 6B). Relevant terms were identified by literature-based similarity39 (>0.95 to the GWAS term). Similarly, we submitted 467 up and down L1000 dexamethasone perturbation signatures40 to RummaGEO and performed enrichment analysis with the extracted terms in category B (biomolecules). Compared to results for submitting random L1000 perturbation signatures, RummaGEO recovered more terms specific to dexamethasone targets (NR3C1, NR0B1, NR1I2) and related biomolecules (e.g., glucocorticoids) from the dexamethasone signatures (Mann-Whitney p < 0.0001) (Figure 6B). We also compared LLM extracted keywords to those manually determined by the authors or journal (PubMed key terms) and MeSH (Medical Subject Headings) terms manually assigned by librarians at the National Library of Medicine (NLM) (Figure 6C). Comparing the similarity of the terms in each set shows there are clearly many terms matching those that were manually extracted. The similarity of the LLM terms to MeSH and PubMed terms compared to the similarity of the MeSH and PubMed terms to each other show that the LLM terms have slightly higher mean similarity, suggesting a greater breadth of coverage compared with the manually curated keyword resources. Additionally, we manually evaluated the LLM keywords to assess the level of “noise” added through this methodology. Terms in each abstract were sorted into four categories: valid, valid but too general, invalid, and “missing” keywords (Figure 6D; Table S4). In general, most terms (∼65%) extracted from each abstract were determined to be accurate and specific, while a smaller portion (∼10%) were either invalid or general. Compared to a manual curation of the terms, ∼15%–25% were missing by the LLM extracted keywords but were manually identifiable. When manually evaluating the category designations of terms (disease/phenotype, molecule/gene/protein/drug, tissue/cell/organ/organism, pathway/biological process, and assay/method) we observe an average accuracy of ∼75% (Figure 6E; Table S5). In general, categories A (disease/phenotype; ∼71.3%) and E (assay; ∼60.8%) were the least accurate, and categories B (biomolecules; ∼88.8%), C (tissue/cell type; ∼90.5%), and D (pathway/biological process; ∼88.5%) were the most accurate. Overall, although not perfect, the LLM can extract the functional term from abstracts. This feature of RummaGEO provides useful information concerning the relevance of biomedical terms of enriched gene sets, helping users to summarize results when large collections of gene sets match their input signature.
Generating hypotheses with GPT-4
After submitting a gene set for analysis with RummaGEO and examining the top matching results, the user can provide a textual description of the gene set they submitted, and then generate hypotheses with GPT-4, an LLM model developed by OpenAI. Once we obtain the descriptions of the query gene set, the matching gene set from the RummaGEO database, and significantly enriched terms from their overlapping genes from Enrichr,24 we can prompt an LLM to produce plausible reasoning for the observed highly significant set overlap using these data as the input. Although caution should be exercised in interpreting the hypotheses generated by the LLM, these hypotheses can provide valuable insight and assist with the initial reasoning for explaining the observed overlap between the two sets. We outline several use cases below to demonstrate this feature of RummaGEO.
Use case 1: Investigating dysregulated mechanisms in Alzheimer disease (rummagene PMC5534941-tp2017110x1.docx-6-Alzheimer_s_disease; n = 42)
Searching “Alzheimer’s” in Rummagene34 provides numerous gene sets extracted from supporting materials of articles deposited in PubMed Central (PMC) related to this disease. One such gene set examines the genetic risk factors shared between coronary artery disease and Alzheimer disease (AD). In Table S3, the authors list a set of genes known to have an association with AD based on published genetics studies (https://rummagene.com/term-search?page=1&q=PMC5534941-tp2017110x1.docx-6-Alzheimer_s_disease; Figure S3). Submitting this gene set to RummaGEO results in the most significantly overlapping gene set to come from a study that inhibits STAT5 in acute myeloid leukemia (AML) (https://rummageo.com/enrich?dataset=eb388fc4-6ead-46d5-93c1-28c56548ee5d; Figure S4).
To further investigate why these two gene sets might be related, we generated a hypothesis using the abstract of the PMC article from which the gene set was sourced as the description. From the descriptions of both gene sets and the significant Enrichr terms from their overlapping genes, GPT-4 produces a plausible explanation for the highly significant overlap: “The high overlap between the user-submitted gene set and the GEO gene set could be due to the shared involvement of these genes in lipid metabolism and cholesterol transport, as well as their association with disease states such as acute myeloid leukemia (AML) and Alzheimer’s disease (AD).” The hypothesis describes both gene sets utilizing the provided descriptions: “The GEO gene set is derived from a study investigating the role of STAT5 in AML, particularly its activation by FLT3-ITD, a constitutively active tyrosine kinase. The study also explores the potential of a novel inhibitor, AC-4-130, in disrupting STAT5 activation and thereby impairing the proliferation and growth of AML cells. On the other hand, the user-submitted gene set is based on a study examining the genetic overlap between coronary artery disease (CAD) and AD, as well as the shared risk factors between these two diseases. The study found that genetic susceptibility to CAD modifies the association between cardiovascular disease (CVD) and dementia, likely through associations with shared risk factors.”
Then, the hypothesis describes how the enrichment of their overlapping genes supports the shared mechanism related to lipid metabolism and cholesterol transport: “The enriched terms from the overlapping genes of the two sets further support this hypothesis. The terms Statin Inhibition of Cholesterol Production WP430, Fatty Acids And Lipoproteins Transport In Hepatocytes WP5323, and Cholesterol Metabolism WP5304 from WikiPathway_2023_Human suggest a shared involvement in lipid metabolism and cholesterol transport. This is further supported by the GO_Biological_Process_2023 terms Phospholipid Efflux (GO:0033700), Cholesterol Efflux (GO:0033344), and Cholesterol Transport (GO:0030301).”
Several studies in the literature, including the selected study, already support the hypothesis that there is accumulation of cholesterol41 and dysregulation of lipid metabolism42 in AD, and cholesterol metabolism reprogramming43 and lipid metabolism reprogramming44 are also happening in AML. The high overlap between the AD genes and genes from AML also further support the link between AD and inflammation. Thus, through this approach, we identify a dysregulated mechanism shared between AD and AML pointing to the key regulator STAT5. STAT5 activation has been reported to be protective in a mouse model of AD.45
Use case 2: Relation of senescence related genes and Ewing’s sarcoma tumors
To identify GEO studies related to cellular senescence, a set of 301 genes related to senescence was sourced from SenoRanger.46 The 301 senescence-related genes were identified via a consensus analysis applied to six independent transcriptomics studies where fibroblast cells were induced to undergo senescence in vitro. The genes that are differentially highly expressed in senescent cells were compared to gene expression levels in healthy normal human cells and tissues to identify genes uniquely expressed at high levels only in senescence cells. Results for submitting the 301 genes for enrichment analysis against the RummaGEO database can be found at https://rummageo.com/enrich?dataset=cee77176-0611-40b8-8a97-26c870d5c363 (Figure S5).
The most significantly overlapping signature identified by RummaGEO is related to the analysis of Ewing’s sarcoma family of tumors (ESFT) cell lines and the dysregulation of EWSR1 and BRCA1 genes.47 The hypothesis is that there are shared biological pathways between ES and senescence, particularly those related to extracellular matrix organization, and the structure and strength of connective tissue, as provided by GPT-4: “The terms ‘Extracellular Matrix Organization (GO:0030198),’ ‘Extracellular Structure Organization (GO:0043062),’ and ‘External Encapsulating Structure Organization (GO:0045229)’ from GO_Biological_Process_2023 suggest that both senescent cells and ESFT cells may undergo changes in their extracellular matrix and structure, possibly as a response to stress or as a mechanism to evade immune surveillance. Finally, the terms ‘abnormal cutaneous collagen fibril morphology MP:0008438,’ ‘decreased skin tensile strength MP:0003089,’ and ‘abnormal tendon morphology MP:0005503’ from MGI_Mammalian_Phenotype_Level_4_2021 suggest that both senescence and ESFT may affect the structure and function of connective tissues, possibly due to alterations in extracellular matrix organization and structure.”
Senescence and its role and relation to extracellular matrix organization are actively being investigated.48,49 Interestingly, the ES gene Ews has been observed to be essential in modulating senescence in hematopoietic stem cells.50 Given the relation of these sets, and the enriched terms related to their overlap, the hypothesis identifies ECM organization as the central theme of the apparent overlap. It should be noted that SenoRanger was created by extracting gene sets from several published studies that induced fibroblast cell lines to undergo senescence. Hence, the enrichment for ECM organization when combining ES disease signatures with fibroblasts undergoing senescence may provide actionable clues toward the identification of therapeutic strategies for ES.
The RummaGEO search engine
The RummaGEO website supports four main components of functionality and search. The first is gene set enrichment that utilizes an in-memory algorithm51 to calculate Fisher’s exact test results quickly. Enriched signatures can be filtered by a term and by a silhouette score threshold. For each significantly enriched signature, users may also generate a hypothesis with GPT-4, as described above. In addition to returning significantly overlapping gene sets, RummaGEO provides term enrichment from three sources: MeSH terms,52 PubMed key terms, and terms extracted by an LLM. Once a gene set has been enriched, the enriched terms are available as an additional tab, in which users are provided with a bar chart, a table, and a word cloud created from the enriched terms. The RummaGEO database can be queried in conjunction with PubMed to find signatures from GEO studies associated with any publications returned from a PubMed search. Users can additionally search RummaGEO through the GEO metadata to find signatures associated with any search term. Furthermore, the human and mouse gene set libraries and accompanying metadata are available to download from the RummaGEO website. Additionally, users can learn more about how to use RummaGEO and the available API from the “About” and “Documentation” pages.
Discussion
By automatically identifying conditions from uniformly aligned studies from GEO and computing differential gene signatures, we were able to produce 171,441 human and 195,265 mouse gene sets extracted from 29,294 GEO studies. These sets provide differential expression knowledge across a wide array of experimental conditions. It should be noted that the identified gene sets come in pairs of up and down sets for each condition. Hence, if we term the paired up and down sets as signatures, then there are ∼85,000 human and ∼97,000 mouse signatures in the RummaGEO database. We also make a substantial effort to annotate these signatures by parsing GEO metadata, categorized functional terms extracted by an LLM, and terms from selected Enrichr libraries. We serve these annotations alongside the gene set search results. This enables users to gain a broader perspective about the top matching gene sets. The additional metadata assists users with filtering the returned matching gene sets and identifying common themes within the top results. We also provide users with the ability to generate hypotheses for overlapping gene sets utilizing abstracts and summaries of the GEO studies and user-submitted descriptions of their gene set. We demonstrate how this feature can uncover pathways, targets, and shared molecular mechanisms across diseases and conditions. We also show how the data within RummaGEO can be used for applications such as TF and kinase enrichment analyses. Transposing the RummaGEO gene sets into a matrix that defines similarity between genes presents the opportunity to identify gene modules and predict gene function for understudied genes. There are numerous other applications that can be enabled by reusing the RummaGEO database, for example, creating a cell type-maker library for single cell identification or developing dynamical models for cell phenotype trajectory analysis. In addition, by crossing the gene sets within RummaGEO with other large sources of gene sets such as Rummagene34 and Enrichr24 we can further discover connections between biological processes and disease mechanisms. The RummaGEO resource has some limitations that should be considered. Although RummaGEO effectively groups samples from each study by condition, for some studies, especially for those with a larger number of conditions, the partitioning can be improved. Additionally, given the large collection of signatures in RummaGEO, most searches return many significant matches, making it difficult to prioritize or sort through all the results. Moreover, RummaGEO provides the search at the gene level. Transcript level search may be more specific and accurate. Currently, RummaGEO covers only humans and mice and is geared toward bulk RNA-seq. Adding more organisms and supporting other types of assays such as microarrays and single-cell RNA-seq could extend the breadth and depth of the resource. Overall, RummaGEO presents an unprecedented resource for the community to query, analyze, and generate hypotheses with gene expression signatures massively mined from GEO.
Experimental procedures
Identifying conditions and computing signatures
All the human and mouse GEO studies aligned by ARCHS4 (version 2.4) with at least three samples per condition and at least six samples in total for a specific study were considered for inclusion in the RummaGEO database. Studies with more than 50 samples were discarded because such studies typically contain expression data collected from large patient cohorts, and this is not amenable for simple signature computation that compares two or more conditions. We also discarded groups of samples that only have one sample and studies with only one identified condition. Samples were grouped using the metadata provided by each study. Specifically, k-means clustering of the embeddings of the concatenated sample_title, characteristic_ch1, and source_ch1 fields were used to classify the conditions. First, the text that describes each sample was converted into an embedding vector of 768 dimensions. We begin with assuming that there are three samples per condition, so the total number of clusters is n-samples divided by three. We then perform k-means clustering and compute the between-clusters and within-cluster distances as the objective function. The k is then decreased to allow 4, 5, 6, or more samples per condition, depending on the similarity of the condition strings in the embedding space. The silhouette score is used to evaluate the quality of the clusters at each step. For string embeddings, we use the SentenceTransformer53 Python module, which utilizes the all-mpnet-base-v2 model enabling embedding of sentences or paragraphs as 768-dimension vectors. If no control conditions are identified, then each condition is compared to every other condition. To create condition titles, common words across all samples for each condition were retained. The limma-voom R package54 was used to compute differential expression signatures for each condition against all other conditions within each study. Additionally, we attempted to first identify any control conditions based on the metadata associated with each sample. To achieve this, a set of keywords that describe control conditions was compiled. The set of such terms contain, for example, “wildtype,” “ctrl,” and “DMSO.” If such terms were identified, then they were used to compare the samples labeled with such terms to all other condition groups. Up and down gene sets were extracted from each signature for genes with an adjusted p < 0.05. If fewer than five genes met this threshold, the gene set was discarded. If more than 2,000 genes met this threshold, then the threshold was lowered incrementally from 0.05 to 0.01, then 0.005, and lastly 0.001, until fewer than 2,000 genes were retained. A total of 2,000 genes was chosen as the cutoff by computing the number of significantly enriched terms for RummaGEO signatures based on cutoffs ranging from 5 to 5,000 submitted for enrichment against multiple Enrichr libraries. We observe that a cutoff of more than 2,000 genes results in little or no gain or lower number of significantly enriched terms (Figure S1).
Data-level silhouette scores
For each study with identified conditions based on metadata clustering, a silhouette score was computed to determine the data-level adherence to the assigned groupings based on the metadata. All aligned counts were extracted from ARCHS411 and normalized by the number of reads aligned, followed by log2 transformation and quantile and Z score normalization. Principal-component analysis (PCA) was then performed on the normalized data, and the silhouette scores were computed from the distance between the samples in each condition in the two-dimensional PCA space. Silhouette scores range from −1 to 1, where a value of 1 would indicate perfect clustering and −1 would indicate fully disjointed clusters.
Search engine implementation
Given the large number of gene sets contained within the RummaGEO database, we employ a fast search engine strategy. This strategy is outlined in more detail in a recent publication that describes the Rummagene resource, a web-server application that hosts over 700,000 gene sets extracted from the supplemental materials of publications listed on PMC.34 Briefly, the enrichment analysis is performed by a Rust-powered API whereby gene set overlaps are computed on bit vectors stored in random access memory.
Identifying functional terms from sample and study metadata
Functional terms were extracted from both the GEO sample (GSM) and the GEO series (GSE) metadata. These functional terms include tissues, cell types, and cell lines (these terms were associated with the BRENDA Tissue Ontology55); diseases and phenotypes (these terms were sourced from DisGeNET56); drugs and small molecules (these terms were associated with International Chemical Identifiers keys); and genes and proteins (these terms were associated with NCBI57 gene symbols). Synonyms and official terms were retained and associated with each study (GSE). Exact matches of these various functional terms were searched for in the GSE summary as well as in the GSM metadata columns used to partition the samples: sample_title, characteristic_ch1, and source_ch1.
Co-occurrence gene-gene similarity matrix
Gene-gene co-occurrence matrices were computed for human and mouse coding genes (19,484 for human and 22,350 for mouse) and non-coding genes (41,366 for human and 29,143 for mouse) using 50,000 randomly selected RummaGEO gene sets. The co-occurrence probabilities for any two genes were computed as previously described by Ma’ayan and Clark.58 For each pair of genes , the co-occurrence count is divided by the total number of occurrences in the matrix. Using the co-occurrence matrix of human coding genes, we then computed the cosine similarity, Jaccard similarity, and normalized pointwise mutual information (NPWMI) between all pairs of genes as follows:
Benchmarking TF and kinase enrichment analyses
The co-occurrence matrices from the coding genes were used to create TF and kinase gene sets libraries by taking the 200 most co-occurring genes with each TF or kinase. The benchmarking datasets previously employed for the ChEA330 and KEA331 resources were used to assess the quality of the TFs and kinases RummaGEO-generated gene set libraries in regard to their ability to identify the “correct” TFs and kinases given sets of differentially expression genes or differentially phosphorylated proteins, respectively. Benchmarking gene sets for TFs are sourced from single TF knockouts extracted from GEO,11,13,59 shRNA knockdowns in a B cell cell line,32 and the ChEA3 gene set library. The kinase benchmarking gene sets are derived from kinase perturbation experiments extracted from GEO,13 LINCS L1000 kinase inhibitor perturbation signatures based on the signatures computed for L1000FWD,60 phosphoproteomics signatures from PTMsigDB,61 and the KEA3 gene set library. Fisher’s exact test was applied to perform the enrichment analysis to rank TFs or kinases by p value in each benchmarking dataset. The input for this test is the gene sets associated with TFs or kinases from the benchmarking dataset. The output is the rank of the TFs or the kinases (sorted by p value) in the RummaGEO TF/kinase gene set library. When generating the receiver operating characteristic (ROC) curves, the positive class is the rank of the correct TF or the kinase, and the negative class is the ranks of all other TFs or kinases. Since there is a large class imbalance, the negative class was downsampled to an equal size as the positive class. Downsampling was randomly performed over 5,000 iterations, and the mean ROC curves and AUCs were reported. To generate the composite ROC curves for each benchmarking library, the numpy interp function was utilized, enabling linear interpolation of the generated points from the 5,000 ROC curves.
Extracting key terms from abstracts
To enrich the metadata of RummaGEO gene sets with descriptive terms from biomedical text, the human and mouse studies (GSEs) included in RummaGEO were annotated with three different types of key terms based on the published paper associated with each study. The GEO DataSets database was first queried for the earliest PubMed ID linked to each study, and the corresponding article information was then retrieved from the PubMed database using the NCBI’s E-utilities Esearch function. In total, 13,427 human studies were annotated with data from 8,804 unique articles, and 15,478 mouse studies were annotated with data from 10,269 unique articles. For 53.5% of the human studies and for the 54.0% of the mouse studies, the PubMed metadata included key terms provided by the authors and/or the publishing journals. Another source of key terms is the MeSH thesaurus, a controlled vocabulary produced by the NLM. MeSH headings were found for 87.7% of the human studies and 92.0% of the mouse studies. A third set of key terms was generated for all studies in RummaGEO by submitting the abstracts from each article associated with each study to the LLM Mistral-7B-Instruct-v0.2 LLM,62 accessed via the HuggingFace API. The LLM was presented with the abstract text and prompted to return up to 10 of the most relevant biomedical key terms. Without processing any terms, the collection of human key terms included 16,488 unique PubMed key terms, 7,935 unique MeSH headings, and 48,462 unique LLM-generated key terms. The mouse key terms comprised 17,742 unique PubMed key terms, 8,212 unique MeSH headings, and 52,152 unique LLM-generated key terms.
Every term was first normalized to a standard capitalization, punctuation, and grammatical form. Due to the unstructured nature of the PubMed key terms and LLM-generated key terms, these two collections were further processed to consolidate key terms that are semantically synonymous or similar. To organize synonymous terms without introducing new terminology, a stemming-like procedure was implemented to identify small clusters of terms (<5) with high textual overlap. Each term in the cluster was replaced with the most frequent term, and these substitutions were assessed manually to ensure semantic equivalency. Terms that are overly general were then filtered out manually. The final processed collection of human key terms includes 10,713 unique PubMed key terms, 6,506 unique MeSH headings, and 31,210 unique LLM-generated key terms. The mouse processed key term collection contains 11,587 unique PubMed key terms, 7,140 unique MeSH headings, and 33,302 unique LLM-generated key terms.
Term categorization and enrichment analysis
To enable domain-specific enrichment analyses, RummaGEO key terms were sorted into four categories using the LLM model Mistral-7B-Instruct-v0.2.62 The categories were defined as follows: category A: disease, phenotype; category B: gene, metabolite, protein, drug, lipid, RNA, variant, receptor; category C: organism, organ, tissue, cell line, cell type, organelle; category D: pathway, biological process, family of genes, chromosome. The LLM was provided with a description of each of these categories and was asked to select the most appropriate choice for each key term. For cases where the LLM responses were uncertain, the term was categorized manually. Out of all human and mouse key terms, 5.4% were categorized as A, 33.3% as B, 19.5% as C, 32.5% as D, and 9.3% as “other” (Table S2). Term enrichment is implemented using the LLM-extracted functional terms for the four categories described above. Since functional terms are extracted per study (GSE), we compute term significance utilizing Fisher’s exact test on categorized terms from the first 5,000 unique GSEs returned from the RummaGEO gene set search, and additionally adjusted p values are computed with the Benjamini-Hochberg method.
Assigning enrichment terms to gene sets with Enrichr
To provide additional metadata and functional terms for RummaGEO gene sets, enrichment analysis is precomputed for each gene set for seven Enrichr63 libraries: ChEA 2022,30 KEGG 2021 Human,27 WikiPathway 2023 Human,29 GO Biological Process 2023,26 MGI Mammalian Phenotype Level 4 2021,64 Human Phenotype Ontology,65 and GWAS Catalog 2023.38 Significance is assessed using Fisher’s exact test, and adjusted p values are computed with the Benjamini-Hochberg method.66 Only significant terms with an adjusted p < 0.05 were retained. To assess the significance of the Enrichr terms’ appearance in the RummaGEO gene set search page, the Kolmogorov-Smirnov test is utilized, comparing the distribution of sets that are significantly enriched for that term compared to a uniform distribution.
Global visualization of signatures with UMAP
To integrate the human and mouse gene sets from GEO, all genes were mapped to uppercase and only protein-coding genes were retained. Gene sets were then converted to one-hot vectors for each set using the Scikit-learn package.67 We utilized truncated SVD68 to reduce the dimensionality of the IDF vectors to the largest 50 singular values. Then, to convert the vectors into two-dimensional space, UMAP69 was applied with the default parameters.
Hypothesis generation with GPT
To generate hypotheses relevant to the user-inputted gene set, we utilize the OpenAI chat completion API (endpoint:/v1/chat/completions, model: gpt-4o). The user is required to submit a textual description of their submitted gene set in the form of a summary or an abstract. RummaGEO takes this description together with the matching RummaGEO gene set study abstract and the top three significantly enriched terms from the overlapping genes from the Enrichr24 libraries WikiPathway 2023 Human, GWAS Catalog 2023, GO Biological Process 2023, and MGI Mammalian Phenotype Level 4 2021. The prompt additionally instructs the LLM to reference all the provided descriptions and contexts of the gene sets, as well as the highly enriched terms from Enrichr. Hypotheses are then parsed to find references for any of the enriched terms and insert the enrichment statistics as part of the hypothesis description output.
Resource availability
Lead contact
Avi Ma’ayan, PhD (avi.maayan@mssm.edu).
Materials availability
The study did not generate any new unique reagents.
Data and code availability
The RummaGEO search engine is available from https://rummageo.com/.
The RummaGEO data, metadata, and correlation matrices are available from the RummaGEO download page at https://rummageo.com/download.
The RummaGEO source code is available from https://github.com/MaayanLab/rummageo.70
The code used to generate the figures for the paper is available at https://github.com/MaayanLab/rummageo/tree/rummageo/figures.
A snapshot of the code was deposited into Zenodo and received the following https://doi.org/10.5281/zenodo.13358070.
The source code and processed datasets produced for the project are protected under the BSD Source Code Attribution license.
Acknowledgments
The authors would like to acknowledge the members of the Ma’ayan Lab for their help in manually curating abstracts for the identification of keywords. This study was supported by NIH grants R01DK131525, OT2OD036435, OT2OD030160, U24CA264250, U24CA271114, and RC2DK131995.
Author contributions
G.B.M. performed the analysis, generated the figures and tables, wrote the paper, and developed the web-based software resource. D.J.B.C. developed the web-based software resource. A.L. contributed to the analysis and updated the ARCHS4 resource. E.Z.D. performed the analysis and generated the figures and tables. A.M. initiated the study, managed the project, provided funding, and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: October 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2024.101072.
Supplemental information
The highlighted terms are organized into four categories by GSE: “valid”, “valid but too general”, “invalid”, and “missing” (n = 200 abstracts).
Terms (n = 1734) from 200 abstracts were classified into the following categories include: (A) Disease/Phenotype, (B1) Genes/Proteins, (B2) Other Biomolecules, (C) Tissue/Cell/Organ/Organism/Model, (D) Pathway/Biological Process, (E) Assay, and Other.
References
- 1.Clough E., Barrett T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z., Lachmann A., Ma’ayan A. Mining data and metadata from the gene expression omnibus. Biophys. Rev. 2019;11:103–110. doi: 10.1007/s12551-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y., Davis S., Stephens R., Meltzer P.S., Chen Y. GEOmetadb: powerful alternative search engine for the Gene Expression Omnibus. Bioinformatics. 2008;24:2798–2800. doi: 10.1093/bioinformatics/btn520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G., Ramírez J.C., Deng N., Qiu X., Wu C., Zheng W.J., Wu H. Restructured GEO: restructuring Gene Expression Omnibus metadata for genome dynamics analysis. Database. 2019;2019 doi: 10.1093/database/bay145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein M.N., Doan A., Dewey C.N. MetaSRA: normalized human sample-specific metadata for the Sequence Read Archive. Bioinformatics. 2017;33:2914–2923. doi: 10.1093/bioinformatics/btx334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H., Zhang S., Zhang L., Geng J., Lu J., Hou C., He P., Lu X. Multi role ChatGPT framework for transforming medical data analysis. Sci. Rep. 2024;14 doi: 10.1038/s41598-024-64585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilks C., Zheng S.C., Chen F.Y., Charles R., Solomon B., Ling J.P., Imada E.L., Zhang D., Joseph L., Leek J.T., et al. recount3: summaries and queries for large-scale RNA-seq expression and splicing. Genome Biol. 2021;22:323. doi: 10.1186/s13059-021-02533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahi N.A., Najafabadi M.F., Pilarczyk M., Kouril M., Medvedovic M. GREIN: An Interactive Web Platform for Re-analyzing GEO RNA-seq Data. Sci. Rep. 2019;9:7580. doi: 10.1038/s41598-019-43935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma’ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziemann M., Kaspi A., El-Osta A. Digital expression explorer 2: a repository of uniformly processed RNA sequencing data. GigaScience. 2019;8 doi: 10.1093/gigascience/giz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Monteiro C.D., Jagodnik K.M., Fernandez N.F., Gundersen G.W., Rouillard A.D., Jenkins S.L., Feldmann A.S., Hu K.S., McDermott M.G., et al. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat. Commun. 2016;7:12846. doi: 10.1038/ncomms12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gundersen G.W., Jones M.R., Rouillard A.D., Kou Y., Monteiro C.D., Feldmann A.S., Hu K.S., Ma’ayan A. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics. 2015;31:3060–3062. doi: 10.1093/bioinformatics/btv297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gundersen G.W., Jagodnik K.M., Woodland H., Fernandez N.F., Sani K., Dohlman A.B., Ung P.M.-U., Monteiro C.D., Schlessinger A., Ma’ayan A. GEN3VA: aggregation and analysis of gene expression signatures from related studies. BMC Bioinf. 2016;17:461. doi: 10.1186/s12859-016-1321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Li J., Yu P. GEOMetaCuration: a web-based application for accurate manual curation of Gene Expression Omnibus metadata. Database. 2018;2018 doi: 10.1093/database/bay019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torre D., Lachmann A., Ma’ayan A. BioJupies: Automated Generation of Interactive Notebooks for RNA-Seq Data Analysis in the Cloud. Cell Syst. 2018;7:556–561.e3. doi: 10.1016/j.cels.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mecham A., Stephenson A., Quinteros B.I., Brown G.S., Piccolo S.R. TidyGEO: preparing analysis-ready datasets from Gene Expression Omnibus. J. Integr. Bioinform. 2024;21 doi: 10.1515/jib-2023-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilarczyk M., Fazel-Najafabadi M., Kouril M., Shamsaei B., Vasiliauskas J., Niu W., Mahi N., Zhang L., Clark N.A., Ren Y., et al. Connecting omics signatures and revealing biological mechanisms with iLINCS. Nat. Commun. 2022;13:4678. doi: 10.1038/s41467-022-32205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giles C.B., Brown C.A., Ripperger M., Dennis Z., Roopnarinesingh X., Porter H., Perz A., Wren J.D. ALE: automated label extraction from GEO metadata. BMC Bioinf. 2017;18:509. doi: 10.1186/s12859-017-1888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinman G.E., Naiman S., Kanfi Y., Cohen H., Bar-Joseph Z. ExpressionBlast: mining large, unstructured expression databases. Nat. Methods. 2013;10:925–926. doi: 10.1038/nmeth.2630. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Q., Wong A.K., Krishnan A., Aure M.R., Tadych A., Zhang R., Corney D.C., Greene C.S., Bongo L.A., Kristensen V.N., et al. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat. Methods. 2015;12:211–214. doi: 10.1038/nmeth.3249. 3 p following 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur N., Oskotsky B., Butte A.J., Hu Z. Systematic identification of ACE2 expression modulators reveals cardiomyopathy as a risk factor for mortality in COVID-19 patients. Genome Biol. 2022;23:15. doi: 10.1186/s13059-021-02589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gene Ontology Consortium. Aleksander S.A., Balhoff J., Carbon S., Cherry J.M., Drabkin H.J., Ebert D., Feuermann M., Gaudet P., Harris N.L., et al. The Gene Ontology knowledgebase in 2023. Genetics. 2023;224 doi: 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M. The KEGG database. Novartis Found. Symp. 2002;247:91–252. discussion 101-3, 119–128, 244–252. [PubMed] [Google Scholar]
- 28.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A., Mélius J., Waagmeester A., Sinha S.R., Miller R., et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keenan A.B., Torre D., Lachmann A., Leong A.K., Wojciechowicz M.L., Utti V., Jagodnik K.M., Kropiwnicki E., Wang Z., Ma’ayan A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47:W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov M.V., Xie Z., London A.B.K., Yang J., Evangelista J.E., Lachmann A., Shu I., Torre D., Ma’ayan A. KEA3: improved kinase enrichment analysis via data integration. Nucleic Acids Res. 2021;49:W304–W316. doi: 10.1093/nar/gkab359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cusanovich D.A., Pavlovic B., Pritchard J.K., Gilad Y. The functional consequences of variation in transcription factor binding. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krug K., Mertins P., Zhang B., Hornbeck P., Raju R., Ahmad R., Szucs M., Mundt F., Forestier D., Jane-Valbuena J., et al. A curated resource for phosphosite-specific signature analysis. Mol. Cell. Proteomics. 2019;18:576–593. doi: 10.1074/mcp.TIR118.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke D.J.B., Marino G.B., Deng E.Z., Xie Z., Evangelista J.E., Ma’ayan A. Rummagene: massive mining of gene sets from supporting materials of biomedical research publications. Commun. Biol. 2024;7:482. doi: 10.1038/s42003-024-06177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marino G.B., Ahmed N., Xie Z., Jagodnik K.M., Han J., Clarke D.J.B., Lachmann A., Keller M.P., Attie A.D., Ma’ayan A. D2H2: diabetes data and hypothesis hub. Bioinform. Adv. 2023;3 doi: 10.1093/bioadv/vbad178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai Z., Lin Z., Meng X., Zheng X., Du Y., Li Z., Zhang X., Liu C., Zhou L., Zhang X., et al. DiSignAtlas: an atlas of human and mouse disease signatures based on bulk and single-cell transcriptomics. Nucleic Acids Res. 2024;52:D1236–D1245. doi: 10.1093/nar/gkad961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke D.J.B., Jeon M., Stein D.J., Moiseyev N., Kropiwnicki E., Dai C., Xie Z., Wojciechowicz M.L., Litz S., Hom J., et al. Appyters: Turning Jupyter Notebooks into data-driven web apps. Patterns (N Y) 2021;2 doi: 10.1016/j.patter.2021.100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Y., Tinn R., Cheng H., Lucas M., Usuyama N., Liu X., Naumann T., Gao J., Poon H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. arXiv. 2020 doi: 10.48550/arXiv.2007.15779. [DOI] [Google Scholar]
- 40.Xie Z., Chen C., Ma’ayan A. Dex-Benchmark: datasets and code to evaluate algorithms for transcriptomics data analysis. PeerJ. 2023;11 doi: 10.7717/peerj.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feringa F.M., van der Kant R. Cholesterol and Alzheimer’s Disease; From Risk Genes to Pathological Effects. Front. Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.690372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin F. Lipid metabolism and Alzheimer’s disease: clinical evidence, mechanistic link and therapeutic promise. FEBS J. 2023;290:1420–1453. doi: 10.1111/febs.16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao L., Zhan H., Jiang X., Li Y., Zeng H. The role of cholesterol metabolism in leukemia. Blood Sci. 2019;1:44–49. doi: 10.1097/BS9.0000000000000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D., Liang J., Yang W., Guo W., Song W., Zhang W., Wu X., He B. A distinct lipid metabolism signature of acute myeloid leukemia with prognostic value. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.876981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X., Shen Q., Chang H., Li J., Xing D. Promoted CD4+ T cell-derived IFN-γ/IL-10 by photobiomodulation therapy modulates neurogenesis to ameliorate cognitive deficits in APP/PS1 and 3xTg-AD mice. J. Neuroinflammation. 2022;19:253. doi: 10.1186/s12974-022-02617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng E.Z., Fleishman R.H., Xie Z., Marino G.B., Clarke D.J.B., Ma’ayan A. Computational screen to identify potential targets for immunotherapeutic identification and removal of senescence cells. Aging Cell. 2023;22 doi: 10.1111/acel.13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorthi A., Romero J.C., Loranc E., Cao L., Lawrence L.A., Goodale E., Iniguez A.B., Bernard X., Masamsetti V.P., Roston S., et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018;555:387–391. doi: 10.1038/nature25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mavrogonatou E., Pratsinis H., Papadopoulou A., Karamanos N.K., Kletsas D. Extracellular matrix alterations in senescent cells and their significance in tissue homeostasis. Matrix Biol. 2019;75–76:27–42. doi: 10.1016/j.matbio.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Brauer E., Lange T., Keller D., Görlitz S., Cho S., Keye J., Gossen M., Petersen A., Kornak U. Dissecting the influence of cellular senescence on cell mechanics and extracellular matrix formation in vitro. Aging Cell. 2023;22 doi: 10.1111/acel.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho J., Shen H., Yu H., Li H., Cheng T., Lee S.B., Lee B.C. Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence. Blood. 2011;117:1156–1166. doi: 10.1182/blood-2010-04-279349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clarke D.J.B., Marino G.B., Deng E.Z., Xie Z., Evangelista J.E., Ma’ayan A. Rummagene: Mining Gene Sets from Supporting Materials of PMC Publications. bioRxiv. 2023 doi: 10.1101/2023.10.03.560783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dhammi I.K., Kumar S. Medical subject headings (MeSH) terms. Indian J. Orthop. 2014;48:443–444. doi: 10.4103/0019-5413.139827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimers N., Gurevych I. Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing. EMNLP-IJCNLP) (Association for Computational Linguistics; 2019. Sentence-BERT: Sentence embeddings using Siamese BERT-networks; pp. 3980–3990. [DOI] [Google Scholar]
- 54.Law C.W., Chen Y., Shi W., Smyth G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gremse M., Chang A., Schomburg I., Grote A., Scheer M., Ebeling C., Schomburg D. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., García-García J., Sanz F., Furlong L.I. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma’ayan A., Clark N.R. Large Collection of Diverse Gene Set Search Queries Recapitulate Known Protein-Protein Interactions and Gene-Gene Functional Associations. arXiv. 2016 doi: 10.48550/arXiv.1601.01653. [DOI] [Google Scholar]
- 59.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z., Lachmann A., Keenan A.B., Ma’ayan A. L1000FWD: fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics. 2018;34:2150–2152. doi: 10.1093/bioinformatics/bty060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krug K., Mertins P., Zhang B., Hornbeck P., Raju R., Ahmad R., Szucs M., Mundt F., Forestier D., Jane-Valbuena J., et al. A curated resource for phosphosite-specific signature analysis. Mol. Cell. Proteomics. 2019;18:576–593. doi: 10.1074/mcp.TIR118.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang A.Q., Sablayrolles A., Mensch A., Bamford C., Chaplot D.S., de las Casas D., Bressand F., Lengyel G., Lample G., Saulnier L., et al. Mistral 7B. arXiv. 2023 [Google Scholar]
- 63.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eppig J.T. Mouse Genome Informatics (MGI) Resource: Genetic, Genomic, and Biological Knowledgebase for the Laboratory Mouse. ILAR J. 2017;58:17–41. doi: 10.1093/ilar/ilx013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gargano M.A., Matentzoglu N., Coleman B., Addo-Lartey E.B., Anagnostopoulos A.V., Anderton J., Avillach P., Bagley A.M., Bakštein E., Balhoff J.P., et al. The Human Phenotype Ontology in 2024: phenotypes around the world. Nucleic Acids Res. 2024;52:D1333–D1346. doi: 10.1093/nar/gkad1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 67.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 68.Chicco D., Masseroli M. Software Suite for Gene and Protein Annotation Prediction and Similarity Search. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;12:837–843. doi: 10.1109/TCBB.2014.2382127. [DOI] [PubMed] [Google Scholar]
- 69.Van Der Maaten L., Postma E.O., van den Herik H.J. Dimensionality reduction: A comparative review. J. Mach. Learn. Res. 2009;10:13. [Google Scholar]
- 70.Giacomo M., Clarke D., Deng E., Ma’ayan A. Zenodo; 2024. RummaGEO Source Code Snapshot from 08222024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The highlighted terms are organized into four categories by GSE: “valid”, “valid but too general”, “invalid”, and “missing” (n = 200 abstracts).
Terms (n = 1734) from 200 abstracts were classified into the following categories include: (A) Disease/Phenotype, (B1) Genes/Proteins, (B2) Other Biomolecules, (C) Tissue/Cell/Organ/Organism/Model, (D) Pathway/Biological Process, (E) Assay, and Other.
Data Availability Statement
The RummaGEO search engine is available from https://rummageo.com/.
The RummaGEO data, metadata, and correlation matrices are available from the RummaGEO download page at https://rummageo.com/download.
The RummaGEO source code is available from https://github.com/MaayanLab/rummageo.70
The code used to generate the figures for the paper is available at https://github.com/MaayanLab/rummageo/tree/rummageo/figures.
A snapshot of the code was deposited into Zenodo and received the following https://doi.org/10.5281/zenodo.13358070.
The source code and processed datasets produced for the project are protected under the BSD Source Code Attribution license.