Abstract
Tissue biomarkers for immune checkpoint inhibitor (ICI) response are limited by tumor sample heterogeneity and availability. This study identifies clinically actionable pretreatment blood biomarkers that are associated with ICI treatment response and survival in recurrent/metastatic head and neck squamous cell carcinoma. A prospective multi-center study enrolled 100 patients before standard-of-care immunotherapy. Blood immune profiles, measured by methylation cytometry, were assessed alongside tumor mutational burden (TMB) and PD-L1 combined proportion score (CPS). TMB and PD-L1 CPS were available for 56 and 91 patients, respectively. High neutrophils, monocytes, and neutrophil-to-lymphocyte ratio were associated with worse survival, while high CD4T cells, especially naïve CD4T cells, and lymphocyte-to-monocyte ratio were associated with better survival. Significant interactions between TMB and peripheral immune profiles for both progression-free and overall survival were found. Clinically relevant pretreatment peripheral immune biomarkers were identified, demonstrating the potential of DNA-based immune profiling to predict ICI response before treatment.
Subject terms: Head and neck cancer, Tumour biomarkers, Prognostic markers
Introduction
An estimated 58,450 new cases of head and neck squamous cell carcinoma (HNSCC) will be diagnosed in 2024 in the US1. Late-stage distant HNSCC has a five-year survival rate between 23% and 41%2, and significant improvement in overall survival (OS) with immune checkpoint inhibitors (ICI) has been reported3–6. Programmed cell death ligand-1 (PD-L1) expression, microsatellite instability (MSI), and tumor mutational burden (TMB) are FDA-approved biomarkers for ICI response7. In recurrent/metastatic (R/M) HNSCC, PD-L1 expression is used to guide ICI treatment decisions based on the KEYNOTE-048 trial3, whereas TMB and MSI are not yet standardized for clinical decision-making. PD-L1 expression has shown inconsistent predictive value in HNSCC8, and higher levels of TMB and MSI have been associated with favorable survival outcomes5,9,10. All three markers require adequate archival tumor and/or invasive biopsy, and there remains a pressing need for noninvasive biomarkers that can be assessed before initiation of ICI therapy to predict response.
Abnormalities in peripheral blood immune cell counts have been associated with cancer treatment outcomes. In HNSCC managed with curative intent treatments (surgery and/or chemoradiation), posttreatment lymphopenia and increased neutrophil-to-lymphocyte ratio (NLR) have been associated with poor survival11,12. Lymphopenia is associated with lower response to concurrent definitive pembrolizumab and radiation therapy in locoregional disease13, as well as to Nivolumab in R/M HNSCC14. Low pretreatment absolute lymphocyte counts (<600 cells/μL) have also been reported to be associated with poor response to PD-1 inhibitors in R/M HNSCC15. These studies are consistent with reported associations of peripheral blood immune cell counts with ICI response in melanoma16, lung17, gastric18, and esophageal cancers19.
T-cell and monocyte subtypes have previously been associated with survival outcomes in patients with cancer receiving ICIs20,21. However, the depth of immune profiling has been limited, and with advances in high-resolution cell-type deconvolution based on DNA methylation in blood using methylation cytometry, we can now assess more detailed immune profiles22. Methylation cytometry has examined changes in cell composition across various diseases, including cancer23, hypertension24, and trisomy 2125. In clinical and large-scale research, traditional cell typing methods like flow cytometry need fresh biospecimens with intact cell membranes, limiting their use. These methods also face technical, logistical, and cost challenges, especially in population-scale studies where fresh samples are scarce. Variations in marker panels and subjective results further reduce reproducibility and rigor. Methylation cytometry, on the other hand, works with banked DNA samples and does not require intact cells. This makes it more scalable and cost-effective. It provides an objective and detailed analysis of immune cell types, subtypes, and activation states. These benefits make it ideal for clinical settings with both fresh and archived samples. We applied advances in methylation cytometry to interrogate a repertoire of 47 immune profile variables at baseline (before initial infusion) as biomarkers of ICI response and survival outcomes in patients with R/M HNSCC.
Methods
Study population
Patients with R/M HNSCC were consecutively recruited for an ongoing prospective multi-center study at the Dana-Farber Cancer Institute, Dartmouth Cancer Center, and Rhode Island Hospital. Eligible patients were at least 18 years old and were advised by their medical oncologists to initiate ICI-based therapy. There were no restrictions on prior lines of therapy. The majority of the patients in the cohort received standard-of-care chemotherapy and radiation therapy, with only six patients without a history of prior treatment. Patients with carcinoma in situ, cutaneous malignancies, and salivary gland cancers were not eligible for participation in the study. The institutional review boards approved the study protocol at each participating center.
Blood DNA methylation measurement
Pretreatment whole blood was collected via venipuncture on the same day but before the initiation of ICIs, immediately delivered to the clinical laboratory, and frozen at −80 °C. Complete blood counts (CBCs) were evaluated for all samples at their respective institutions. All blood samples were transported to the Laboratory for Molecular Medicine at Brown University, where DNA was uniformly processed. DNA was extracted and processed using the Zymo Quick DNA Miniprep Plus kit and quantified using a Qubit dsDNA Broad Range Assay kit (Thermo). DNA was sent to the Avera Institute for Human Genetics for DNA methylation quantification using the Illumina Infinium Methylation EPICv1 and EPICv2 array platforms. The ENmix and minfi packages in Bioconductor were implemented for data processing and quality control26,27. The normal-exponential out-of-band (Noob) method was used to generate DNA methylation beta values. The Noob preprocessing method (normal-out-of-band background correction plus linear dye bias correction) was chosen as it is recommended for methylation-based deconvolution22. Probes were filtered using probability Out-Of-Band probe-Hybridization Analysis (pOOBHA), which compares the intensity signals of the probes considering the expected target hybridization and the potential out-of-band hybridization. Probes with a pOOBHA > 0.05 were masked. Samples with >5% of probes masked, or probes across all samples with >5% masked content, would be flagged and removed. A sample would also be flagged if the average signal for the bisulfite conversion probes per sample exceeded 3 IQR. In this study, all samples passed these quality control criteria. A k-nearest neighbor beta value imputation was performed for those samples with partial probes masked. Per design, known polymorphic, X, Y, and mitochondrial probes are not used for cell deconvolution. As the EPICv2 array uses internal technical replicates, the rm.cgsuffix function was used for those persisting after quality control to average the beta values for EPIC v2 BeadChips for technical replicates into a single beta value.
Methylation cytometry
Methylation-based cell-type deconvolution, also known as methylation cytometry, was performed to estimate the immune cell profile of the samples22. The FlowSorted.BloodExtended.EPIC method was employed to infer 12 immune cell type proportions in whole blood samples22. The cell proportions were calibrated by incorporating the reported lower bound limit of detection, assuming that any values below the limit are unreliable28. Here, we used the imputeBDLs function from the robCompositions R package to account for the compositional data (cell proportions)29. We used partial least squares to predict missing values using 100 repetitions, a convergence criterion of 0.1, and a maximum number of iterations of 1000. Briefly, the algorithm iteratively imputes the parts of the composition below the limit of detection in the following steps: 1. compositional data are expressed in pivot coordinates using isometric log-ratio transformation; 2. Tobit regression is applied; 3. the values below the limit of detection are replaced by the expected values; 4. the corresponding inverse isometric log-ratio is applied. After all parts are imputed, the algorithm starts again until the imputations do not change using the convergence criterion. Additionally, absolute counts for the 12-cell types were calculated based on CBC data. Twenty-three-derived immune variables were calculated using the 12 immune cell proportions and counts. In total, a panel of 45 primary and derived immune variables were included in this study.
A comprehensive list of the immune variables, abbreviations, and their calculations is presented in Supplementary Table 1. A multidimensional scaling plot of baseline methylation cytometry immune profiles, colored by the three centers, was used to assess the inter-center effect across all subjects. The methylation cytometry algorithm is available at https://github.com/immunomethylomics/FlowSorted.BloodExtended.EPIC.
Reference group for the immune profiles
A previous study established reference levels of immune variables through methylation cytometry in post-menopausal women from the Women’s Health Initiative (WHI) cohort30. A comprehensive summary is included in Supplementary Table 2.
TMB and PD-L1 CPS measurements
TMB was obtained through clinically directed comprehensive genomic profiling on patients’ tumor samples. Commercially available from Foundation Medicine (FoundationOne CDx) or Oncopanel, the Dana-Farber/Brigham Cancer Center platform for genomic analysis was utilized. TMB was reported as the number of mutations per megabase harbored by tumor cells. PD-L1 CPS was measured by immunohistochemical quantitative analysis using Dako22C3 pharmDx™ companion diagnostic assay at Foundation Medicine or institutional assay at DFCI using E1L3N clone (Cell Signaling Technology). It was reported within a range of 0–100.
Patient outcomes
Progression-free survival (PFS) was defined as the time interval between the initiation of treatment and tumor progression, death, or censoring. Overall survival (OS) was defined as the time interval between the initiation of treatment and death or censoring. The durable benefit group regarding treatment with ICI was defined as patients who did not experience progression of disease or death within 365 days from the start of ICI-based treatment. The non-durable benefit group was defined as patients progressing or dying of the disease between 100 and 365 days after the start of ICI treatment. The non-benefit group was defined as patients experiencing progression of disease or death within 100 days after the start of treatment.
Statistical analyses
Multivariable linear regression models were fitted to assess the relationships between each pretreatment immune variable and the benefit outcomes. The models were adjusted for sex, age, and a blood methylation-based smoking score31. Similarly, TMB and PD-L1 CPS were investigated in relation to the benefit outcomes using multivariable linear regression models, adjusting for sex, age, and smoking. Cox proportional hazard models were fitted for survival outcomes to investigate the association between (i) pretreatment immune variables and PFS/OS, adjusting for sex, age, and smoking and (ii) TMB/PD-L1 CPS and PFS/OS, adjusting for sex, age, and smoking with pretreatment immune variables. A data-based statistical cut-point was optimized to dichotomize the variables using the surv_cutpoint() in the survminer R package. This outcome-oriented method provides a cut-point value corresponding to the most significant relationship with the outcome. The minimal proportion of observations per group was set to 20% of the total population. A sensitivity analysis was performed to examine the relationship between pretreatment immune variables and PFS/OS in patients who received anti-PD-1 monotherapy only at DFCI. Interaction effects of peripheral immune profiles and tumor-based biomarkers on survival outcomes were investigated. A series of Cox proportional hazard models were fitted with interaction terms for TMB and the 45 peripheral immune variables, adjusting for sex, age, and smoking. The same models were used for PDL1-CPS. A stratification analysis was conducted for immune variables that significantly interacted with TMB on survival outcomes. The subjects were only stratified by their median values for the significant immune variables. The log-rank test within each stratum was used to study the association of TMB with survival outcomes. Pearson’s correlation coefficient was used to evaluate the association between pretreatment TMB and immune variables. To assess the predictive value of methylation cytometry immune variables, TMB, PD-L1 CPS, sex, age, and smoking status on survival outcomes, the C-index in the Cox proportional hazard models for PFS and OS were calculated. The C-index for each variable model was calculated by removing the respective variable compared to the full model. Each Cox model’s proportional hazard assumption was checked using the cox.zph() function within the survival R package, and all models were adjusted to ensure the proportional hazard assumption was not violated32. Statistical significance was established for all models with a P-value < 0.05. The Benjamini–Hochberg false discovery rate (FDR)33 based on the P-values were generated by each model. All analyses were performed in R version 4.3.0.
Ethics approval and consent to participate
This study involved human participants and was approved by the Institutional Review Boards of Dartmouth Cancer Center (approval number: STUDY02001227), Dana-Farber Cancer Institute (approval number: 18-548), and Brown University (approval number: 1901002321). Participants gave informed consent to participate in the study before taking part. Our research was conducted in full compliance with the ethical principles outlined in the Declaration of Helsinki.
Results
100 patients were recruited from August 2019 to January 2024 (Table 1). There were 75 males and 25 females, with an average age of 64. 78 patients received anti-PD-1 monotherapy, 21 patients received anti-PD-1 therapy with chemotherapy, while 1 patient received anti-PD-1 therapy with the EGFR inhibitor cetuximab. Four patients received manual CBCs and were excluded from the immune variable calculations.
Table 1.
Baseline characteristics of the study subjects
| Patients’ characteristics | N = 100 | % |
|---|---|---|
| Recruitment site | ||
| Dana-Farber Cancer Institute | 88 | 88 |
| Dartmouth Cancer Center | 10 | 10 |
| Rhode Island Hospital | 2 | 2 |
| Sex | ||
| Female | 25 | 25 |
| Male | 75 | 75 |
| Age, in years | ||
| Mean (standard deviation) | 64.4 (11.8) | |
| Location of primary tumor | ||
| Oral cavity | 44 | 44 |
| Oropharynx | 34 | 34 |
| Hypopharynx | 8 | 8 |
| Larynx | 7 | 7 |
| Paranasal sinuses and nasal cavity | 3 | 3 |
| Unknown | 4 | 4 |
| HPV status in oropharyngeal carcinomas (=34) | ||
| Positive | 25 | 73.5 |
| Negative | 6 | 17.6 |
| Missing | 3 | 8.8 |
| Smoking history | ||
| Current/former smoker | 51 | 51 |
| Never smoker | 40 | 40 |
| Missing | 9 | 9 |
| Initial treatment type | ||
| Immunotherapy only | 78 | 78 |
| Immunotherapy + Chemotherapy | 21 | 21 |
| Immunotherapy + EGFRi | 1 | 1 |
| Progression-free survival, in days | ||
| Median | 132 | |
| Overall survival, in days | ||
| Median | 538 | |
| Vital status | ||
| Alive | 63 | 63 |
| Deceased | 37 | 37 |
| Still continuing treatment among alive (=63) | ||
| Yes | 47 | 74.6 |
| No | 16 | 25.4 |
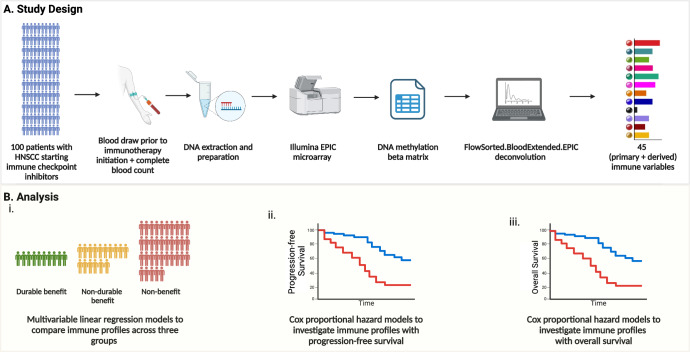
56 and 91 patients had available data for TMB (Supplementary Table 3) and PD-L1 CPS (Supplementary Table 4), respectively. The study design and analysis pipelines for peripheral immune profiling are shown in Fig. 1, for TMB in Supplementary Fig. 1, and for PD-L1 CPS in Supplementary Fig. 2. No inter-center effect was observed in the pretreatment immune profiles (Supplementary Fig. 3).
Fig. 1. Overview of study design and analysis pipeline.
A Schematic representation of the study design, including patient enrollment, sample collection, and methylation cytometry workflow. B Analysis pipeline showing statistical modeling to evaluate associations between immune profiles and clinical outcomes.
Comparison of immune profiles between pretreatment patients with HNSCC and a reference group
There were elevations in neutrophil and monocyte counts and proportions and reductions in lymphocyte counts and proportions among patients with HNSCC compared to reference levels in the WHI cohort. All lymphocyte subtypes exhibited reduced counts and proportions in patients compared to reference subjects in the WHI cohort (Supplementary Table 2).
Pretreatment peripheral blood immune profiles by benefit groups
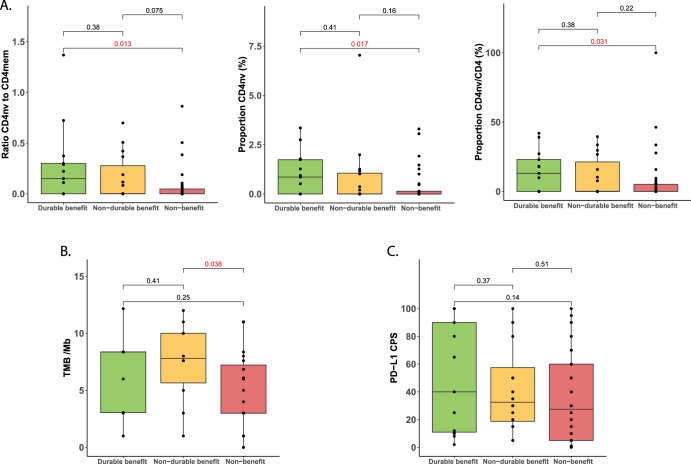
CD4nv cell levels exhibited significant differences between the durable (N = 13) and non-benefit groups (N = 35) in anti-PD-1 monotherapy patients. The durable benefit group showed a significantly higher CD4nv/CD4mem ratio (Δ = 0.22, P = 0.013), CD4nv proportion (Δ = 0.75%, P = 0.017), and CD4nv/CD4 proportion compared to the non-benefit group (Δ = 11.58%, P = 0.031) (Fig. 2A). A higher TMB was noted in the non-durable benefit (N = 10) group compared to the non-benefit group (N = 23) (Δ = 2.80/Mb, P = 0.036, Fig. 2B). The combined group showing any benefit (N = 19) had a significantly higher TMB compared to the non-benefit group (Δ = 2.20/Mb, P = 0.04, Supplementary Fig. 4A). While no significant differences were observed in PD-L1 CPS across the three groups, a monotonic downward trend of was observed from the durable (N = 13) to the non-durable (N = 16) to the non-benefit group (N = 32) (Fig. 2C). A similar trend was observed in the combined group showing any benefit (N = 29) compared to the non-benefit group (Δ = 11.03 CPS, P = 0.2, Supplementary Fig. 4B)
Fig. 2. Association of pretreatment peripheral immune profiles and tumor-based biomarkers with benefit to anti-PD-1 monotherapy.
A CD4nv-related immune variables, B TMB, and C PDL1-CPS.
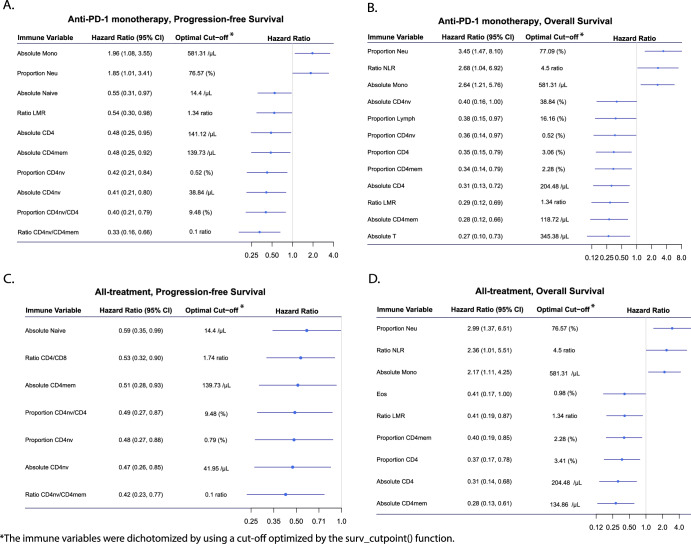
Pretreatment peripheral blood immune profiles by survival outcomes
The impact of pretreatment blood peripheral immune variables on survival outcomes was initially examined in 75 patients who received anti-PD-1 monotherapy exclusively. As binary variables, higher levels of neutrophil proportion and monocyte count were associated with shorter PFS (Fig. 3A). Similar trends were observed in OS with an additional NLR (Fig. 3B). Conversely, elevated levels of lymphocyte cell types, majorly including CD4T cell types, were associated with longer PFS and OS (Fig. 3A, B). As a continuous variable, a higher CD4mem count was significantly associated with longer OS (P = 0.04). Kaplan–Meier plots are shown for the significant binary immune variables in monotherapy patients, Supplementary Figs. 5 and 6. Analysis of all 96 patients yielded findings consistent with the monotherapy data with binary immune variables (Fig. 3C, D). As a continuous variable, a higher CD4mem count was associated with longer OS (P = 0.03). Kaplan–Meier plots are shown for the significant binary immune variables in all patients, Supplementary fig. 7-8. FDRs are shown to have significant associations between immune variables and survival outcomes (Supplementary Table 5). In the sensitivity analyses stratified to patients who received anti-PD-1 monotherapy at DFCI, 7 of the 10 significant immune variables for PFS (Supplementary Fig. 9A) and 8 of the 12 significant variables for OS from the full anti-PD-1 monotherapy patients were successfully replicated (Supplementary Fig. 9B). The C-index analysis showed that when removing methylation cytometry immune variables in the model, the C-index decreased by over 0.3, while removing TMB and smoking decreased the C-index by <0.01, with minimal contributions from other factors (Supplementary Fig. 10A). A similar result was found for overall survival, with methylation cytometry again boosting the C-index by over 0.3, while PD-L1 CPS contributed <0.01 (Supplementary Fig. 10B). These findings highlight the strong predictive value of pretreatment methylation cytometry immune variables for survival outcomes, beyond other demographic and clinical factors.
Fig. 3. Association of pretreatment immune variables with survival outcomes in HNSCC.
A PFS in the anti-PD-1 monotherapy subgroup. B OS in the anti-PD-1 monotherapy subgroup. C PFS in the all-treatment group. D OS in the all-treatment group.
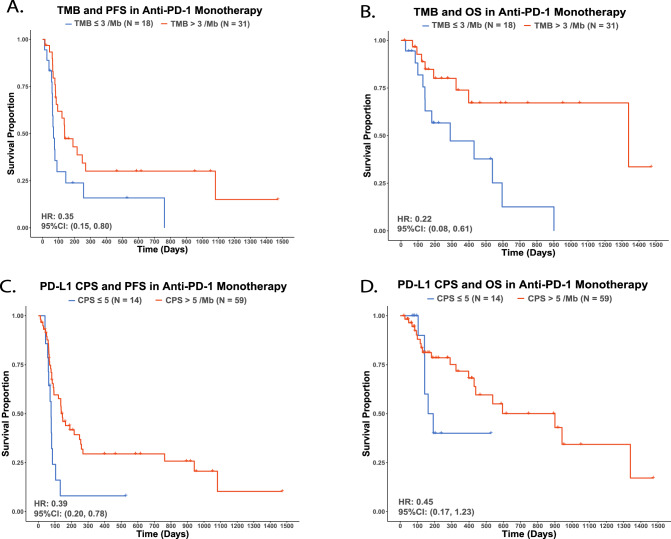
TMB and survival outcomes
Fifty-six out of 100 subjects (42: anti-PD-1 monotherapy, 5: along with chemotherapy, 1: along with cetuximab) with available TMB data were investigated for survival outcomes. TMB 3/Mb was selected as the optimized cut-off for both PFS and OS using the surv_cutpoint function. In anti-PD-1 monotherapy recipients, patients with high TMB had significantly longer PFS (HR for progression or death: 0.35, 95% CI: 0.15–0.80, Fig. 4A) and OS (HR for death: 0.22, 95% CI: 0.08–0.61, Fig. 4B) compared to patients with low TMB. Consistently, in all-treatment patients, those with TMB > 3/Mb also had significantly longer PFS (HR for progression or death: 0.34, 95% CI: 0.16–0.71, Supplementary Fig. 11A) and OS (HR for death: 0.22, 95% CI: 0.09–0.54) compared to patients with TMB ≤ 3/Mb (Supplementary Fig. 11B).
Fig. 4. Tumor-based biomarkers in anti-PD-1 monotherapy survival outcomes.
A TMB and PFS. B TMB and OS. C PDL1-CPS and PFS. D PDL1-CPS and OS.
TMB and peripheral immune profile interaction
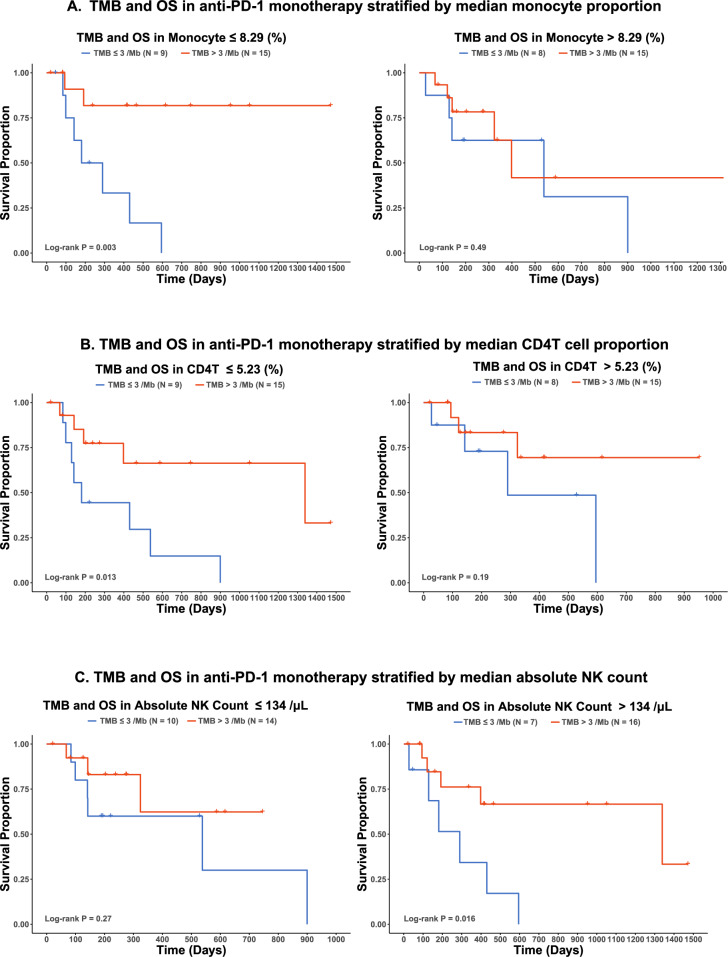
Since a high tumor mutation burden may generate numerous novel tumor antigens, we hypothesized that the immune profiles may differ in responders and non-responders by their TMB. In the 47 patients receiving anti-PD-1 monotherapy, peripheral Monocyte proportion, CD4T cell proportion, and NK count also had a significant interaction with TMB and impacted OS (Interaction term P < 0.05). When stratified by the median value, higher TMB was a better predictor of OS in the lower monocyte proportion group (Log-rank P: Lower Group 0.003 vs. Higher Group 0.49, Fig. 5A), lower CD4T cell proportion group (Log-rank P: Lower Group 0.013 vs. Higher Group 0.19, Fig. 5B), and higher NK count group (Log-rank P: Lower Group 0.27 vs. Higher Group 0.016, Fig. 5C). NK count and Neu count significantly interacted with TMB and affected PFS (Interaction term P < 0.05). Higher TMB was a better predictor of PFS in the higher NK group (Log-rank P: Lower Group 0.69 vs. Higher Group 2.4e−04, Supplementary Fig. 12A) and higher Neu count group (Log-rank P: Lower Group 0.62 vs. Higher Group 0.023, Supplementary Fig. 12B). FDRs are shown for significant immune variables and TMB interactions (Supplementary Table 6). Pearson’s correlation was used to assess the association between pretreatment TMB and immune variables. The top 5 most significant correlations, ranked by P-value, are shown in Supplementary Fig. 13A. Pretreatment NK cell proportion showed a statistically significant correlation with TMB (Pearson’s r = 0.28, P < 0.05), as illustrated in Supplementary Fig. 13B. NK cells and monocytes emerged as the top two cell types most strongly correlated with TMB, with both showing significant interactions in Fig. 5, though monocytes did not reach statistical significance in the correlation analysis.
Fig. 5. TMB and OS in anti-PD-1 monotherapy subgroup according to immune variables stratified by median.
A monocyte proportion, B CD4T proportion, and C absolute NK count.
PD-L1 CPS and survival outcomes
Ninety out of 100 subjects (53: anti-PD-1 monotherapy, 6: along with chemotherapy, 1: along with cetuximab) had available PD-L1 CPS. PD-L1 CPS at 5 was selected, using the surv_cutpoint function, as the optimized cut-off for PFS and OS. In anti-PD-1 monotherapy recipients, patients with PD-L1 CPS > 5 had a significantly longer PFS compared to patients with PD-L1 CPS ≤ 5 (HR: 0.39, 95% CI: 0.20–0.78) (Fig. 5C). A similar trend was observed with OS but not statistically significant (HR: 0.45, 95% CI: 0.17–1.23) (Fig. 5D). In all treatment patients, patients with PD-L1 CPS > 5 had a significantly better PFS (HR: 0.53, 95% CI: 0.30–0.95, Supplementary Fig. 11C) and OS compared to patients with PD-L1 CPS ≤ 5 (HR: 0.41, 95% CI: 0.19–0.88, Supplementary Fig. 11D). No significant interactions were observed between the peripheral immune profile and PD-L1 CPS.
Discussion
We utilized methylation cytometry in peripheral blood samples to identify key pretreatment immune variables linked to survival outcomes in patients with HNSCC undergoing palliative intent ICI therapy. We also found an interaction between peripheral immune profiles and TMB, perhaps arising as a result of the action of specific immune subtypes on novel tumor antigens. While further research is warranted to develop sophisticated models with aggregated immune variables for ICI response and prognosis prediction in patients with HNSCC, our findings establish significant associations between specific immune variables and survival outcomes: elevated levels of neutrophil- and monocyte-related immune variables were correlated with poorer prognosis, whereas higher levels of lymphocyte cell types, especially CD4T cells, were associated with better prognosis.
These results are consistent with previous findings associating pretreatment absolute lymphocyte counts of <600 cells/μL with poor response to ICI in a smaller study15. Methylation cytometry provided us with more granular details of the immune profile (e.g., delineating lymphocyte subtypes), revealing that CD4T cells were primarily responsible for the prognostic benefit. While memory T cells are recognized for their propensity to mount a robust immune response to ICIs due to prior antigen exposure, our findings highlight the significant contribution of naïve CD4T cells to prognostic outcomes in HNSCC, consistent with previous findings in lung and bladder cancers34,35. This underscores their pivotal role in initiating and augmenting the anti-tumor immune response upon PD-1 pathway blockade36. It is well-recognized that standard chemoradiation can induce lymphopenia11,37. This may partially explain the poor prognostic outcomes for patients with the development of recurrent/metastatic disease within 6 months of the end of curative intent chemoradiation therapy6.
Regarding myeloid cells, previous studies have associated elevated blood neutrophil and monocyte levels with worse survival outcomes in other cancers38–42, consistent with our observations in R/M HNSCC. Neutrophils and monocytes play a key role in cancer development and progression43,44. Neutrophils suppress the immune response against cancer cells, allowing them to evade destruction by the immune system and limit the efficacy of immunotherapy, while circulating monocytes can differentiate into macrophages upon entering the tumor microenvironment, promoting chronic inflammation, angiogenesis, and extracellular matrix remodeling. Although we observed higher monocyte count associated with worse prognosis, future studies are needed to distinguish the precise nature of the immune modulation associated with changes in monocyte subsets.
Two variables that we found consistently associated with prognosis are NLR and LMR. Elevated pretreatment NLR levels were associated with poorer survival outcomes, aligning with prior research findings across other cancer types, including lung cancer45, melanoma46, and kidney cancer47. This correlation underscores NLR’s role in reflecting the balance between pro-inflammatory neutrophils and anti-tumor lymphocytes, pivotal components of the immune response against cancer. Conversely, higher LMR levels were associated with improved survival outcomes in our HNSCC cohort receiving ICIs, congruent with previous research findings48,49. This relationship underscores LMR’s utility in gauging the balance between anti-tumor lymphocytes and pro-inflammatory monocytes in response to immunotherapy.
Despite TMB and PD-L1 CPS being commonly used, FDA-approved, tumor tissue-based markers, their interactive effects with peripheral blood immune profiles on the prognosis of patients with HNSCC receiving ICIs have not been previously explored. Here, we identified several immune cell types that strongly interacted with TMB in predicting prognosis. High-level TMB in patients with lower levels of peripheral monocyte and CD4T cell proportions, and higher levels of peripheral NK cell counts is a stronger predictor of better OS. Tumors with a higher TMB tend to have more neoantigens, potentially providing the immune system with a broader range of targets in response to immunotherapy50. Prior research has also indicated the potential of NK cells to contribute to the effectiveness of cancer immunotherapy by providing a targeted and memory-based immune response against cancer cells51. Our data suggests that a low level of peripheral monocytes and a high level of NK cells can assist in cancer cell elimination through TMB-associated enhanced neoantigen recognition through immunotherapies. Moreover, the favorable impact of CD4 T cells on OS appears to mitigate the adverse prognostic effect of low TMB, thereby attenuating the predictive capacity of TMB for OS in the high CD4 T cell group.
Our study has some limitations. Firstly, the sample size may limit the power to detect significant associations. While we calculated the FDR, we set the statistical significance at the P-value cut-off of 0.05 without adjusting for the family-wise error rate due to limited power. Future studies with larger sample sizes must train, test, and validate these biomarkers. Secondly, although 12 cell types can be deconvolved with the current technology, an even higher resolution deconvolution approach is necessary for more granular monocyte and effector/memory cell subtypes. For example, the association between peripheral monocytes, worse survival rates, and their interaction with TMB may be primarily attributed to mMDSCs suppressing the immune response to cancer. Additionally, studies have shown differential roles of central and effector memory T cells in the immunotherapy response52. Finally, validation of the markers in larger studies in other cancer types will help us understand the universal tumor-agnostic implications of our findings.
High-resolution DNA methylation-based immune cell deconvolution in pretreatment peripheral blood identified immune variables associated with survival outcomes in patients with R/M HNSCC receiving ICIs. Furthermore, peripheral immune profiles interacted with TMB on survival outcomes in these patients. Taken together, our results demonstrate the potential of detailed immune profiles to predict immunotherapy response and survival outcomes prior to the start of treatment.
Supplementary information
Acknowledgements
This work was supported by R01CA253976, R01CA216265, R01CA258375, P30CA023108, W81XWH-20-1-0778, P20GM104416, P20GM130454, P20GM130423, P30CA168524, P20GM103418, P50CA097257, R01CA207360, and the Robert Magnin Newman Endowed Chair in Neuro-oncology. Administrative work from Sean Brian Donnelly and Julianna Miller is gratefully acknowledged.
Author contributions
Conceptualization: B.C.C., K.T.K., R.H., D.C.K., L.A.S., J.K.W., K.Sehgal., K.Shirai.; Methodology: Z.Z., K.Sehgal, B.C.C., D.C.K., L.A.S., J.K.W., A.M.M., R.B., Formal analysis: Z.Z., G.R., M.K.L., H.G.S.; Original draft preparation: Z.Z., K.Sehgal.; Review and editing: B.C.C., K.T.K., R.H., R.B., D.C.K., L.A.S., J.K.W., K.Sehgal., K.Shirai., Z.Z., G.R., M.K.L., H.G.S., A.M.M.; Supervision: B.C.C., K.T.K., R.H.; Project administration: B.C.C., K.T.K., R.H., D.C.K., L.A.S., J.K.W., K.Sehgal., K.Shirai., R.B.
Data availability
All data used in this study are publicly available on Gene Expression Omnibus with the accession number GSE277573.
Code availability
The methylation cytometry algorithm is available under a free license for academic purposes. Licensing algorithm information is available at https://github.com/immunomethylomics/FlowSorted.BloodExtended.EPIC and the code used to generate the figures in the manuscript can be accessed at https://github.com/zzhang23/HNSCC_ICI_Methylation_Cytometry.
Consent for publication
The final version of the manuscript has been reviewed and approved by all authors.
Competing interests
K.T.K. and J.K.W. are founders of Cellintec, which had no role in this research. Dr. K.S. reported receiving consulting fees from Scholar Rock and Equinox Group, Inc.; receiving personal fees for consulting or advisory board participation from Exelexis Inc. and MedScape; receiving research funding to the institution from Merck; and travel honorarium from Merck; none of which conflict with research conducted on this study. B.C.C. is an advisor to Guardant Health, which had no role in this research.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ze Zhang, Kartik Sehgal.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00759-8.
References
- 1.Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024). [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2023. (Atlanta, Ga, American Cancer Society, 2023).
- 3.Burtness, B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet394, 1915–28 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Cohen, E. E. W. et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet393, 156–67 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Poulose, J. V. & Kainickal, C. T. Immune checkpoint inhibitors in head and neck squamous cell carcinoma: a systematic review of phase-3 clinical trials. World J. Clin. Oncol.13, 388–411 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris, R. L. et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med.375, 1856–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, Y. et al. FDA-approved and emerging next generation predictive biomarkers for immune checkpoint inhibitors in cancer patients. Front. Oncol.11, 683419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park, J. C., Krishnakumar, H. N. & Saladi, S. V. Current and future biomarkers for immune checkpoint inhibitors in head and neck squamous cell carcinoma. Curr. Oncol.29, 4185–98 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan, L., Wang, Z., Xue, J., Yang, H. & Zhu, Y. Tumor mutation burden predicts response and survival to immune checkpoint inhibitors: a meta-analysis. Transl. Cancer Res.9, 5437–49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haddad, R. I. et al. Influence of tumor mutational burden, inflammatory gene expression profile, and PD-L1 expression on response to pembrolizumab in head and neck squamous cell carcinoma. J. Immunother. Cancer10, e003026 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campian, J. L., Sarai, G., Ye, X., Marur, S. & Grossman, S. A. Association between severe treatment-related lymphopenia and progression-free survival in patients with newly diagnosed squamous cell head and neck cancer. Head Neck36, 1747–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, A. J. et al. Association of posttreatment lymphopenia and elevated neutrophil-to-lymphocyte ratio with poor clinical outcomes in patients with human papillomavirus-negative oropharyngeal cancers. JAMA Otolaryngol. Head Neck Surg.145, 413–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss, J. et al. Concurrent definitive immunoradiotherapy for patients with stage III–IV head and neck cancer and cisplatin contraindication. Clin. Cancer Res.26, 4260–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesaire, M. et al. Impact of lymphopenia on efficacy of nivolumab in head and neck cancer patients. Eur. Arch. Otorhinolaryngol.280, 2453–61 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Ho, W. J. et al. Association between pretreatment lymphocyte count and response to PD1 inhibitors in head and neck squamous cell carcinomas. J. Immunother. Cancer6, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, A. C. et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature545, 60–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabet, B. Y. et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell183, 363–76 e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon, M. et al. Determinants of response and intrinsic resistance to PD-1 blockade in microsatellite instability-high gastric cancer. Cancer Discov.11, 2168–85 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Kato, R. et al. Increased Tim-3(+) T cells in PBMCs during nivolumab therapy correlate with responses and prognosis of advanced esophageal squamous cell carcinoma patients. Cancer Immunol. Immunother.67, 1673–83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An, H. J., Chon, H. J. & Kim, C. Peripheral blood-based biomarkers for immune checkpoint inhibitors. Int. J. Mol. Sci.22, 9414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez, C. et al. Systemic blood immune cell populations as biomarkers for the outcome of immune checkpoint inhibitor therapies. Int. J. Mol. Sci.21, 2411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salas, L. A. et al. Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling. Nat. Commun.13, 761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, J. Q. et al. Genome-scale methylation analysis identifies immune profiles and age acceleration associations with bladder cancer outcomes. Cancer Epidemiol. Biomark. Prev.32, 1328–37 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kresovich, J. K. et al. Peripheral immune cell composition is altered in women before and after a hypertension diagnosis. Hypertension80, 43–53 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Z., Stolrow, H. G., Christensen, B. C. & Salas, L. A. Down syndrome altered cell composition in blood, brain, and buccal swab samples profiled by DNA-methylation-based cell-type deconvolution. Cells12, 1168 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu, Z., Niu, L., Li, L. & Taylor, J. A. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res.44, e20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics30, 1363–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell-Glenn, S. et al. Calculating detection limits and uncertainty of reference-based deconvolution of whole-blood DNA methylation data. Epigenomics15, 435–51 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Templ, M., Hron, K., Filzmoser, P. & Gardlo, A. Imputation of rounded zeros for high-dimensional compositional data. Chemom. Intell. Lab155, 183–90 (2016). [Google Scholar]
- 30.Nissen, E. et al. Assessment of immune cell profiles among post-menopausal women in the Women’s Health Initiative using DNA methylation-based methods. Clin. Epigenet.15, 69 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCartney, D. L. et al. Epigenetic prediction of complex traits and death. Genome Biol.19, 136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grambsch, P. M. & Therneau, T. M. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika81, 515–26 (1994). [Google Scholar]
- 33.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B57, 289–300 (1995). [Google Scholar]
- 34.Xia, L. et al. Peripheral CD4(+) T cell signatures in predicting the responses to anti-PD-1/PD-L1 monotherapy for Chinese advanced non-small cell lung cancer. Sci. China Life Sci.64, 1590–601 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Joshi, M. et al. Concurrent durvalumab and radiation therapy (DUART) followed by adjuvant durvalumab in patients with localized urothelial cancer of bladder: results from phase II study, BTCRC-GU15-023. J. Immunother. Cancer11, e006551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou, J., Yang, X., Xie, S., Zhu, B. & Zha, H. Circulating T cells: a promising biomarker of anti-PD-(L)1 therapy. Front. Immunol.15, 1371559 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byun, H. K. et al. Clinical predictors of radiation-induced lymphopenia in patients receiving chemoradiation for glioblastoma: clinical usefulness of intensity-modulated radiotherapy in the immuno-oncology era. Radiat. Oncol.14, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bahig, H. et al. Neutrophil count is associated with survival in localized prostate cancer. BMC Cancer15, 594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meunier, S. et al. Elevated Baseline neutrophil count correlates with worse outcomes in patients with muscle-invasive bladder cancer treated with chemoradiation. Cancers (Basel)15, 1886 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, W. et al. Neutrophil count and percentage: potential independent prognostic indicators for advanced cancer patients in a palliative care setting. Oncotarget8, 64499–508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanford, D. E. et al. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin. Cancer Res.19, 3404–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibutani, M. et al. The peripheral monocyte count is associated with the density of tumor-associated macrophages in the tumor microenvironment of colorectal cancer: a retrospective study. BMC Cancer17, 404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahid, K. R. et al. Neutrophils: musketeers against immunotherapy. Front. Oncol.12, 975981 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards, D. M., Hettinger, J. & Feuerer, M. Monocytes and macrophages in cancer: development and functions. Cancer Microenviron.6, 179–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romano, F. J. et al. Neutrophil-to-lymphocyte ratio is a major prognostic factor in non-small cell lung carcinoma patients undergoing first line immunotherapy with pembrolizumab. Cancer Diagn. Progn.3, 44–52 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartlett, E. K. et al. High neutrophil-to-lymphocyte ratio (NLR) is associated with treatment failure and death in patients who have melanoma treated with PD-1 inhibitor monotherapy. Cancer126, 76–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, X., Meng, F. & Jiang, R. Neutrophil-to-lymphocyte ratio as a prognostic biomarker for patients with metastatic renal cell carcinoma treated with immune checkpoint inhibitors: a systematic review and meta-analysis. Front. Oncol.11, 746976 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan, L., Wu, C., Luo, S. & Xie, X. Prognostic value of lymphocyte-to-monocyte ratio (LMR) in cancer patients undergoing immune checkpoint inhibitors. Dis. Markers2022, 3610038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, Y. et al. Association of lymphocyte-to-monocyte ratio with survival in advanced gastric cancer patients treated with immune checkpoint inhibitor. Front. Oncol.11, 589022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jardim, D. L., Goodman, A., de Melo Gagliato, D. & Kurzrock, R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell39, 154–73 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimasaki, N., Jain, A. & Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov.19, 200–18 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Liu, Q., Sun, Z. & Chen, L. Memory T cells: strategies for optimizing tumor immunotherapy. Protein Cell11, 549–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are publicly available on Gene Expression Omnibus with the accession number GSE277573.
The methylation cytometry algorithm is available under a free license for academic purposes. Licensing algorithm information is available at https://github.com/immunomethylomics/FlowSorted.BloodExtended.EPIC and the code used to generate the figures in the manuscript can be accessed at https://github.com/zzhang23/HNSCC_ICI_Methylation_Cytometry.