Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) are important pathogens in humans and certain animals. Molecular epidemiological analyses of ExPEC are based on structured observations of E. coli strains as they occur in the wild. By assessing real-world phenomena as they occur in authentic contexts and hosts, they provide an important complement to experimental assessment. Fundamental to the success of molecular epidemiological studies are the careful selection of subjects and the use of appropriate typing methods and statistical analysis. To date, molecular epidemiological studies have yielded numerous important insights into putative virulence factors, host-pathogen relationships, phylogenetic background, reservoirs, antimicrobial-resistant strains, clinical diagnostics, and transmission pathways of ExPEC, and have delineated areas in which further study is needed. The rapid pace of discovery of new putative virulence factors and the increasing awareness of the importance of virulence factor regulation, expression, and molecular variation should stimulate many future molecular epidemiological investigations. The growing sophistication and availability of molecular typing methodologies, and of the new computational and statistical approaches that are being developed to address the huge amounts of data that whole genome sequencing generates, provide improved tools for such studies and allow new questions to be addressed.
INTRODUCTION
Extraintestinal pathogenic Escherichia coli (ExPEC), the distinctive group of E. coli strains that possess an enhanced ability to overcome or subvert host defenses and cause extraintestinal disease in otherwise healthy hosts, are important pathogens in humans and certain animals (1, 2). Molecular epidemiological analyses contributed to the initial recognition of ExPEC strains as being distinct from other E. coli (including intestinal pathogenic and commensal variants), and have yielded insights into the ecology, evolution, reservoirs, transmission pathways, host-pathogen interactions, and pathogenic mechanisms of ExPEC, thereby providing an essential complement to experimental assessment of virulence mechanisms. This article first reviews the basic conceptual and methodological underpinnings of the molecular epidemiological approach, then summarizes the main aspects of ExPEC that have been investigated by using this approach.
The concept of ExPEC as representing a discrete microbiological subset of E. coli with special relevance to human and animal health has stimulated efforts to identify the bacterial correlates of extraintestinal virulence that determine, whether mechanistically or for classification purposes, a given strain’s likelihood of representing ExPEC. As discussed below in “Results of Molecular Epidemiological Studies,” such efforts are ongoing, and are increasingly informative thanks to the progressive advances in epidemiological methods and study design, typing technologies, and biostatistical approaches.
METHODOLOGICAL CONSIDERATIONS
Basic Principles of Molecular Epidemiology
Traditional versus molecular epidemiology
Epidemiology is the study of disease as it occurs in populations. In epidemiological studies, structured observations are used to identify host characteristics (that is, risk factors) that predict the occurrence, severity, or clinical manifestations of a particular illness (3–8). By analogy, molecular epidemiology is the study of a disease in relation to selected genetic characteristics of the host and/or the causative agent, such as an infectious disease and the causative microorganisms (9–14). In molecular epidemiological studies that focus on infectious diseases, structured observations are used to identify microbial traits (for example, specific genes, phylogenetic background, or clonal identity) that predict the occurrence, severity, or clinical manifestations of a particular infectious disease, or relevant characteristics of the affected hosts, including age, sex, and underlying predisposing conditions. Molecular epidemiological studies seek insights into the molecular basis for the virulence behavior and host predilections of the pathogen and to identify relevant reservoirs and transmission pathways. Such insights can be useful in developing strategies for managing and preventing infections caused by the particular pathogen.
Strengths and limitations of molecular epidemiology
By virtue of being observational rather than experimental, molecular epidemiological studies exhibit the strengths and limitations inherent to observational studies in general. Their main strength is that they examine “real world” phenomena, that is, wild-type microbes interacting with the natural host in a natural setting, rather than the artificially engineered host-pathogen interactions of experimental studies (15, 16), which can be of uncertain physiological relevance. In addition, molecular epidemiological studies can examine multiple predictor and outcome variables simultaneously, which can be challenging with experimental studies.
The main weakness of molecular epidemiological studies is that they allow the investigator no direct control over the variables analyzed. Consequently, a variable can be isolated only through careful selection of comparison groups so that the groups differ, to the extent possible, only according to the particular variable. Moreover, even with the most carefully selected comparison groups, associations that may emerge still are only that, associations. Although these may reflect causal relationships between the analyzed variables, they also may be due to confounding from other, unmeasured variables (in either the host or the pathogen). Consequently, the better characterized the source subjects and bacteria are, the greater the confidence with which associations between host variables and bacterial traits can be attributed to the particular variables themselves. In addition, because of the considerable variation within human and bacterial populations, large numbers of subjects are needed per group (relative to the number of replicate determinations needed to address variance in experimental studies), appropriate statistical tests are required to assess the significance of any observed differences between groups, and statistically significant findings require confirmation in different populations.
Finally, at least prior to the advent of whole-genome sequencing (WGS: discussed below) and transcriptome analysis, molecular epidemiological studies could assess only known microbial characteristics for which appropriate assays were available. That is, they required prior knowledge of the characteristics to be studied. Therefore, in contrast to exploratory methods such as signature-tagged mutagenesis (17), in vivo expression technology (18), and transcriptome analysis (19), they could not be used to discover new virulence factors. However, they provided an important complement to such discovery-based experimental approaches by assessing the epidemiological (that is, population-level) relevance of newly identified traits (20–23). This complementarity between epidemiology and experimentation is implicit in the molecular restatement of Koch’s postulates, the first of which is that the trait of interest must be epidemiologically associated with disease (24). Today, WGS makes it possible, in principle, to compare an extensive array of bacterial sequences, many of which may be of unknown significance or function, with epidemiological variables, thereby facilitating the unbiased discovery of novel bacterial correlates of virulence, which subsequently can be assessed experimentally.
Internal controls
Much stronger conclusions can be drawn from observed between-group differences in molecular characteristics if a study includes an internal control group that is tested in parallel with the clinical group of interest, in particular, if the controls are temporally, geographically, and demographically matched to the clinical isolates, thereby avoiding some of the problems associated with use of external (that is, historical) control groups (25, 26). Concurrent testing of cases and controls using the same methods and reagents, ideally by an operator who is unaware of sample identity, reduces the likelihood that cohort effects, technical factors, or subjective bias could influence the results.
Statistical considerations
Molecular epidemiological studies adhere to the same statistical principles and rely on the same statistical methods as conventional epidemiological studies (27, 28). Between-group comparisons are tested using standard statistical approaches such as a χ2 test, Fisher’s exact test, or univariate logistic regression analysis for dichotomous variables, and an unpaired t test or the Mann-Whitney U test for continuous variables. Multiple independent variables can be assessed simultaneously as predictors of an outcome (-dependent) variable by using appropriate multivariate methods. For comparisons involving multiple testing of a particular (bacterial or human) subject, whether for different traits as assessed at the same time or for a given trait as assessed at different times, appropriate tests for paired comparisons must be used, such as McNemar’s test for dichotomous variables and a paired t test or the Wilcoxon rank-sum test for continuous variables. Additionally, the ever-growing number of bacterial variables that can be determined and analyzed has led to increased use of multidimensional scaling methods, such as principle coordinates analysis and principle components analysis, which summarize the variance within complex data sets using a small number of derived variables, termed coordinates or components. Such methods determine and display graphically the overall extent of similarity or difference between source populations with respect to multiple variables when considered jointly, and the extent of correlation among different variables and between each variable and respective source populations. Other advanced statistical methods that can be used with complex data sets to identify bacterial correlates of epidemiological variables include discriminant analysis (29), classification and regression tree analysis, and machine learning (30).
Type I errors, which are the false conclusion of a difference when none actually exists, are a hazard of the use of multiple comparisons (since the probability of obtaining a “significant” P value is proportional to the number of comparisons) and the selective testing of associations suggested by post hoc data review (31). However, multiple comparisons are inevitable in molecular epidemiological studies that assess multiple bacterial traits, as increasingly is the practice, and are especially prominent with WGS analysis, which may generate thousands of candidate predictor variables. Statistical adjustment for multiple comparisons, and/or cautious interpretation of putatively significant associations, can be used to address this problem (32). Likewise, post hoc data review to discover new associations is an important means for generating new hypotheses. Recognition that such hypotheses require independent confirmation provides a helpful safeguard against false conclusions.
Type II errors, which are the false conclusion of absence of a difference when one actually exists, result from insufficient sample size, which limits statistical power for finding differences (33). However, the seemingly obvious remedy of studying large comparison groups may or may not be an option, depending on the context. This is because, unlike in experimental studies where the number of replicate determinations is largely a matter of investigator choice, in epidemiological studies clinical factors sometimes limit the number of subjects or isolates available for a particular group, thereby imposing insurmountable restrictions on sample size (34). As a consequence, conclusions may need to be tempered to reflect the inherent uncertainty resulting from limited power.
Typing Methods
The various bacterial traits analyzed in molecular epidemiological studies that attempt to define or characterize ExPEC represent a spectrum of levels of organization and complexity, ranging from subgenic DNA sequence (the most basic), through genes, operons, and pathogenicity islands (intermediate), through whole genomes and plasmids (complex), to clones, clonal groups, and phylogenetic groups (highly complex) (35, 36). Each level is important and informative, and each requires distinctive typing methods.
Sequence analysis
Historically, analysis of sequence diversity within virulence-associated genes or their flanking regions was done by using restriction fragment length polymorphism (RFLP) analysis (now little used) or direct Sanger sequencing of cloned DNA fragments or PCR products (37–44). The increasing accessibility and affordability of next-generation DNA-sequencing methods, e.g., Illumina (short reads) and PacBio (long reads, including for plasmid closure), now allows sequence analysis of whole chromosomes and plasmids (45, 46), providing an alternative or complement to conventional Sanger sequencing. Sequencing technologies, and the associated bioinformatics approaches needed to manipulate and interpret the resulting large-sequence files, are evolving rapidly, and may soon supplant conventional methods even for gene detection, let alone detailed sequence analysis.
Sequence analysis has multiple applications in molecular epidemiological studies (as discussed below). These include determination of the presence/absence of specific putative virulence genes or resistance genes and molecular variants thereof, identification of single-nucleotide polymorphisms in housekeeping genes that correspond with specific ExPEC-associated lineages, and clarification of the genetic architecture of mobile units (e.g., transposons and plasmids) that can transfer virulence and resistance genes between strains and lineages.
Gene detection
Detection of putative virulence markers, historically a mainstay of molecular epidemiology in E. coli, can be done using a variety of methods, with the method selected determining the nature of the results, which in turn shapes the conclusions that can be drawn. Conventional membrane or solid-support probe hybridization, which has largely been superseded by newer technologies, relies on complementarity between the probe, often several kilobases in length, and the target region (9, 37, 47). It can readily screen for broad genetic regions if large probes are used (40). Probes can be used either in solution for hybridization with target DNA that has been fixed to a solid substrate, or fixed to a solid substrate (using a macro- or microarray format) for hybridization with target DNA in solution (48).
PCR detection relies on precise matching between the primers and the target region, and usually is limited to relatively short targets, typically <2 kb (49). Thus, if the relevant sequence polymorphisms are known, PCR can differentiate between minor molecular variants of a particular gene (which may have different disease and/or host associations) and, if multiple primer pairs are used to map an operon, can identify suboperonic deletions (which also may have clinical correlates) (Fig. 1) (50). PCR can be done using either conventional or real-time methods, and with multiplexing of primers for simultaneous detection of multiple targets. Detection of conventional PCR products is usually done using agarose gel electrophoresis, fluorescence or luminescence-based real-time technology, or membrane-bound arrays (51).
Figure 1.
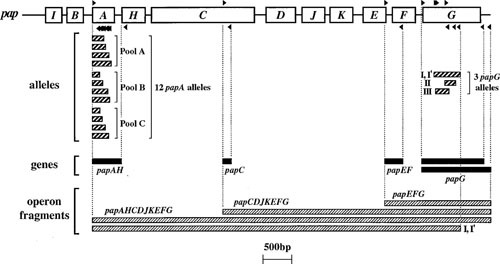
PCR analysis of pap operon. Open boxes represent genes within the pap operon (including papA, structural subunit; papC, usher; papEF, minor tip pilins; and papG, adhesin). Forward and reverse primers (right- and left-pointing black triangles, respectively, above and below the pap operon) are used in combinations as shown to yield the indicated PCR products (thin rectangles, below pap operon). Heavily striped rectangles, papA and papG allele PCR products. Solid black rectangles, pap gene PCR products. Finely striped rectangles, long PCR operon fragments (as generated using either flanking or internal allele-specific papG reverse primers, as illustrated for allele I-I′). Different papA and papG variants are associated with specific lineages, hosts, and clinical syndromes. Intraoperonic deletions that yield a null phenotype (which may be associated with compromised or asymptomatic hosts) can be detected as a truncated long-PCR product. Reprinted from reference 42, with permission.
WGS analysis is a rapidly emerging technology for detection of known virulence (and other) genes in E. coli, and may become the preferred genotyping method as sequencing prices drop and bioinformatics pipelines become increasingly robust, accessible, and user friendly (36, 45, 52–55). For this approach, enormous numbers of sequence reads of various lengths (depending on the particular sequencing method) are generated using one of several available massively parallel sequencing technologies.
Reads are analyzed by using (i) user-selected applications and a private sequence library; (ii) a web-based suite of applications, such as that hosted by the Center for Genomic Epidemiology (Danish Technical University, Copenhagen, DK: http://www.genomicepidemiology.org/); or (iii) a hybrid of these two approaches. Use of web-based platforms offers simplicity and speed, but limits the analysis to whatever markers the particular system happens to include. In contrast, manual analysis allows the user to search for any target sequence of interest, but is more technically demanding. Additionally, as mentioned in an earlier section, by using more sophisticated bioinformatics approaches, it is possible to identify and catalogue sequences of unknown identity that are present in an isolate, for comparison with other isolates from the same or different contexts. The huge amounts of data that WGS generates pose analytical challenges that oblige the development of new computational and statistical approaches (56, 57).
Clones, clonal groups, and phylogenetic groups
Clones and clonal groups (which are groups of closely related clones) can be identified at the molecular level by using typing methods that scan the entire genome. Of the historical “whole-genome” methods in general use, the most discriminating is pulsed-field gel electrophoresis (PFGE) analysis. This method involves electrophoretic separation of total bacterial DNA that has been digested using a restriction enzyme such as XbaI, which in E. coli recognizes a limited number of DNA sites (Fig. 2) (13, 58–60). Use of a second restriction enzyme can further enhance discrimination (61). Identity of two isolates by PFGE analysis implies that they represent the same strain or clone (62). However, PFGE is so discriminating that, beyond a limited level of divergence, it fails to perceive similarity between isolates, making it unreliable for identifying larger clonal groups. It also sometimes paradoxically calls as similar isolates that more precise genetic methods show are only distantly related. Additionally, it requires specialized equipment, is slow, has very limited capacity for data sharing, and is subject to multiple sources of error that limit its ability to reliably and consistently perceive genomic relationships, as compared with WGS-based phylogenetic analysis (see later section) (53, 63).
Figure 2.
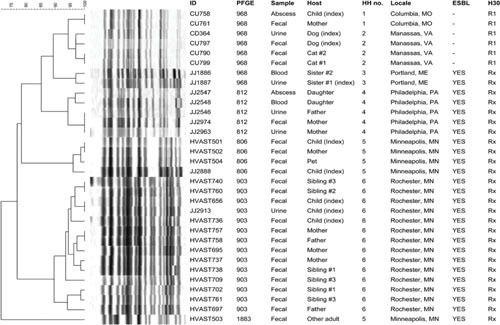
Dendrogram based on pulsed-field gel electrophoresis (PFGE) profiles of 33 clinical and fecal isolates of Escherichia coli sequence type 131 (ST131) from the members of 6 households. Profiles are diverse, despite all isolates deriving from the same ST, which reflects the superior resolving power of PFGE over MLST. Isolates from a given household cluster together, consistent with intrahousehold strain sharing. Scale is % profile similarity. All isolates were fluoroquinolone-resistant. H30, clonal subset within the ST131-H30 clade (R1 = H30R1, Rx = H30Rx). Abbreviations: ESBL, extended-spectrum β-lactamase production; HH, household; ID, identifier; PFGE, pulsotype. Reprinted from reference 63, with permission.
PCR-based whole-genome profiling methods are less discriminating than PFGE, but are simpler and faster, and can perceive broader clonal group relationships more effectively. Commonly used PCR-based genomic profiling methods include random amplified polymorphic DNA (RAPD) analysis, which uses random or arbitrary primers (Fig. 3) (12), and repetitive element PCR, which uses primers targeting various known chromosomal repeat elements (64). Both methods generate distinctive banding patterns that reflect the spacing of suitable primer sites in the isolate’s genome. In addition to allowing simple “same-versus-different” comparisons between isolates, PCR-generated genomic profiles can be subjected to cluster analysis to define quantitative profile similarity relationships, which provides a crude assessment of the underlying phylogenetic relationships (Fig. 4) (65, 66). Such profiling can be used to compare multiple colonies from a particular sample (e.g., feces) with one another to identify putative unique clones, or to compare epidemiologically unrelated clinical, fecal, or environmental isolates with selected reference isolates to allow classification of the unknown isolates as to putative clonal group, e.g., resistance-associated clonal group A (CGA; also known as ST69) by comparison (Fig. 4).
Figure 3.
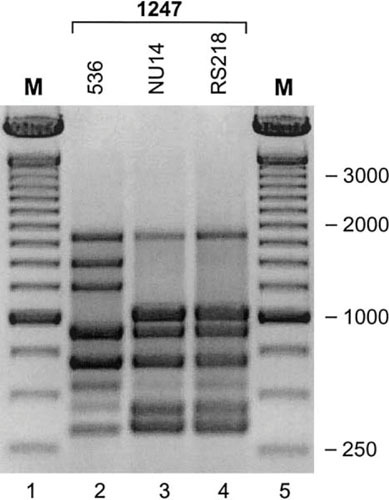
Random amplified polymorphic DNA (RAPD) analysis of E. coli strains 536, NU14, and RS218. RAPD profiles generated by using primer 1247 (12) show E. coli O18:K1:H7 strains NU14 (cystitis: lane 3) and RS218 (neonatal meningitis: lane 4) to be indistinguishable from one another, but distinct from strain 536 (O6:K15:H31, pyelonephritis: lane 2), illustrating both the broad syndrome capability of certain ExPEC lineages and the clonal diversity of urinary tract infection-causing ExPEC strains. M (lanes 1 and 5), 100-bp marker. Reprinted from reference 160, with permission.
Figure 4.
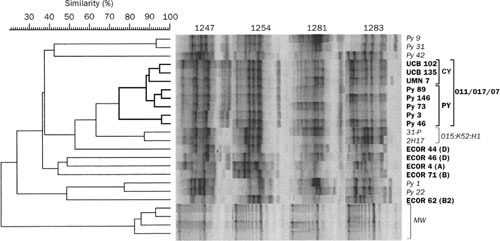
RAPD-based phylogenetic and clonal analysis of Escherichia coli isolates. Genomic profiles (shown in computer reconstruction), as generated for each isolate by using RAPD primers 1247, 1254, 1281, and 1283, were concatenated for cluster analysis. Pyelonephritis isolates (n = 10; “Py” strain designations) are labeled in bold if from E. coli clonal group A (CGA) (n = 5) and in lightface italic if non-CGA (n = 5). CGA isolates (bold) are bracketed and labeled as to syndrome (CY, cystitis; PY, pyelonephritis) and serogroup (O11/O17/O77) (right), with the corresponding cluster shown in bold (left). The two E. coli O15:K52:H1 control strains are bracketed and labeled by serotype. Reference strains from the E. coli Reference (ECOR) collection (bold) are identified as to phylogenetic group (right). The depth of the molecular weight ladder cluster (brackets; MW) reflects the intrinsic variability inherent in gel electrophoresis and image analysis, independent of amplification. The CGA isolates cluster together irrespective of clinical syndrome (pyelonephritis, cystitis) and geography (UCB: Berkeley, California; UMN: Minneapolis, MN; Py: multiple centers around the U.S.). Reprinted from reference 248, with permission.
Phylogenetic relationships and membership in specific clonal groups, which is relevant for studies involving potential ExPEC isolates because of the known phylogenetic distribution of extraintestinal virulence in E. coli (67, 68), can be resolved with greater accuracy by more focused methods, including PCR-based phylotyping, multilocus sequence typing (MLST), lineage-specific PCR assays, and WGS analysis. PCR-based phylotyping provides a rapid and simple, albeit approximate, method for classifying isolates into one of seven E. coli phylogenetic groups (A, B1, B2, C, D, E, F) or a cryptic clade (69). MLST involves DNA sequence analysis of multiple housekeeping genes (typically seven or more, depending on the particular MLST system) that are widely distributed around the chromosome, which enables assignment of an allele designation for the particular sequence variant found at each locus and assignment of a sequence type (ST) based on the isolate’s particular combination of alleles (https://enterobase.warwick.ac.uk/species/index/ecoli) (70–72). MLST is now used widely to define both the ST membership of individual isolates and the overall distribution of STs within a collection (i.e., bacterial population). Limitations include the need for a current, well-curated, universally accessible library of STs with reliable associated metadata, plus a system for translating STs into phylogenetic groups.
Lineage-specific PCR assays can detect either lineage-defining single-nucleotide polymorphisms within broadly conserved housekeeping genes (typically, those used for MLST) or lineage-specific accessory genes (i.e., genes variably present within the species). Currently, such assays are available for 12 broad subgroups within (ExPEC-associated) phylogenetic group B2 that correspond largely to ST complexes (73), and for specific STs and sub-ST clonal lineages within (ExPEC-associated) groups B2, D, and F (72, 74–81). Occasional misclassification results from genetic alteration of the targeted primer-binding sites.
WGS data, when limited to core genome sequences (i.e., those that are present in all members of the population), can be analyzed using appropriate phylogenetic algorithms to define the phylogenetic structure of a collection (45, 52). Inclusion of reference genomes that represent known phylogenetic or clonal groups allows such phylogenies to be referenced to the established E. coli population structure. Unlike the results of other genotyping methods, WGS-based phylogenies are informative across the entire spectrum of resolution within the phylogenetic tree, from the broadest (major phylogenetic groups) to the most resolved (minor sequence variants within individual sub-ST lineages, i.e., clones), as shown in Fig. 5 for the fine structure of the pandemic resistance-associated lineage ST131 (Fig. 5) (36, 63, 80, 82). They allow evolutionary time scale inferences, to clarify the chronology of emergence of epidemic ExPEC clones (36), and permit epidemiologically linked ExPEC isolates to be compared with one another and with unlinked isolates, to assess for clonal reservoirs and transmission (63). Notably, core genome phylogenies by definition ignore variation within the accessary genome (including plasmids, virulence genes, bacteriophages, and resistance genes), which may be important clinically or ecologically and require separate focused analysis.
Figure 5.
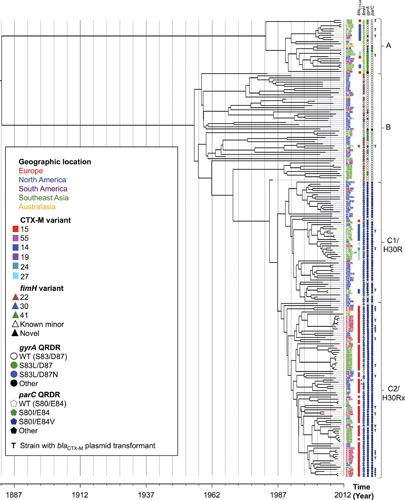
Time-scaled whole genome sequence (WGS)-based phylogeny of ST131 Escherichia coli (n = 215). Meta-data include allelic variants of blaCTX-M (extended-spectrum beta-lactamase gene) and fimH (type-1 fimbriae adhesin gene), plus mutations in the quinolone resistance-determining region (QRDR) of gyrA (WT, wild-type). Brackets identify defined ST131 clonal subsets. Branch tips are colored by geographic region, per the key. T, bla plasmid transformant generated for strain; *, cases with putative deletions in the assembled bla gene. Geographic clustering is evident, some of it linked with specific accessory gene variants; e.g., within the C1/H30R clonal subset, the Southeast Asian isolates (green) are largely confined to a specific clade that consists of two subclades, one characterized by blaCTX-M-14 and the other by blaCTX-M-27. Reprinted from reference 36, with permission.
RESULTS OF MOLECULAR EPIDEMIOLOGICAL STUDIES
Virulence Factors: Associations with Clinical Variables and Phylogenetic Background
Comparisons between clinical isolates and controls
E. coli isolates from the urine, blood, cerebrospinal fluid, etc., of patients with diverse extraintestinal infection syndromes typically exhibit a greater prevalence of specific molecular markers than do fecal isolates from uninfected hosts (9, 83–88). From an epidemiological perspective such virulence markers can be regarded as “virulence factors” (VFs), although this term must be understood as implying “factors associated with” rather than “contributing to” virulence, since epidemiological associations do not necessarily reflect causality (as discussed above).
These VFs can be grouped by functional category, for example, adhesins (fimbrial and nonfimbrial) (89–92), siderophore systems (23, 65, 93–97), toxins (20, 98–101), surface polysaccharides (102–105), invasins (50, 106, 107), serum resistance-associated traits (108, 109), and traits of miscellaneous or unknown function (22, 67) (Table 1, Fig. 6). Clinical isolates often contain multiple VFs from a particular functional category (110, 111); this conceivably may allow for redundancy, synergistic interactions among VFs, and/or adaptability to different environmental niches. Conversely, many seemingly virulent strains lack known representatives of one or more of these functional categories (110, 111). Whether such apparent deficits are compensated for by VFs from other functional categories, or these strains actually do contain unrecognized representatives of the “missing” VF categories, is unknown.
Table 1.
Virulence-associated traits of extraintestinal pathogenic Escherichia coli (ExPEC) by functional categorya
| Category | Gene(s) or operon | Comment | Evidence for trait as a virulence factor (reference[s]) | |
|---|---|---|---|---|
| Epidemiologicalb | Experimental (in vivo)c | |||
| Adhesinsd | afa/dra | Dr antigen-binding adhesins (AFA I-III, Dr, F1845) | Yes (176, 266, 267) | Yes (268) |
| afaE-8 | Afimbrial adhesin VIII | Yes (175) | No | |
| auf | Surface structure of unclear function | Yes (269) | No (269) | |
| bmaE | Blood group M-specific adhesin | Yes (270) | No | |
| clpG | CS31A adhesin (K88–related) | Yes (271) | No | |
| csgA | Curli | Yes (272) | Yes (272) | |
| ecp | Pilus | Yes (273) | No | |
| fim | D-mannose-specific adhesin, type-1 fimbriae | Yes (267, 274, 275) | Yes (89, 276) | |
| foc | F1C fimbriae | Yes (277) | No | |
| gafD | GlcNac-specific (G, F17c) fimbriae adhesin | Yes (270, 271) | No | |
| iha | Iron-regulated-gene-homologue adhesin | Yes (21, 111, 267) | Yes (96) | |
| nfa | NFA-1, -2, -3, -4 (nonfimbrial adhesins) | No (278–280) | No | |
| pap | Pilus associated with pyelonephritis (P fimbriae) | Yes (9, 151, 267) | Yes (281); no (282) | |
| pil | Type IV pilus | Yes (283) | Yes (284) | |
| sfa | S fimbriae (sialic acid-specific) | Yes (111) | Yes (285) | |
| sfa/foc | S and F1C fimbriae | Yes (286) | Yes (285) | |
| yad | Fimbria | Yes (283) | Yes (283) | |
| ygi | Fimbria | Yes (283) | Yes (283) | |
| yvc | Fimbria | Yes (283) | No | |
| Toxinsd | astA | EAST1, heat-stable enteroaggregative E. coli cytotoxin | Yes (79, 287) | No (189, 288) |
| cdtB | Cytolethal distending toxin, CDT | Yes (111) | No | |
| cnf1 | Cytotoxic necrotizing factor 1, CNF-1 | Yes (289) | Yes (98, 290, 291) | |
| hly | α-Hemolysin (Hly) | Yes (292) | Yes (100, 293) | |
| pic | Protein associated with intestinal colonization, PIC | Yes (294) | Yes (18, 284, 294, 295) | |
| sat | Secreted autotransporter toxin (serine protease) | Yes (20, 267) | Yes (296) | |
| tsh | Temperature-sensitive hemagglutinin, TSH | Yes (68, 294) | Yes (18, 294) | |
| upxA (tosA) | RTX toxin | Yes (297) | Yes (298) | |
| vat | Vacuolating autotransporter toxin | Yes (299, 300) | Yes (284) | |
| Nutrition | argC | Arginine synthesis | N/Ae | Yes (301) |
| aro | Shikimate synthesis | N/Ae | Yes (284) | |
| chuA | Heme receptor | Yes (268) | Yes (97) | |
| dppA-oppA-sapA | Uptake of short peptides and amino acids | Yes (284, 302) | ||
| dsd | Serine utilization | Yes (303) | No (304) | |
| entF | Enterobactin synthesis | No | Yes (65) | |
| fyuA, irp | Yersiniabactin (siderophore) receptor, synthesis | Yes (122, 267, 268) | Yes (305) | |
| guaA | Guanine synthesis | N/Ae | Yes (301) | |
| ireA | Iron-regulated element (catecholate siderophore) | Yes (23) | Yes (23) | |
| iroN | Salmochelin (siderophore) receptor | Yes (21, 306) | Yes (65, 95) | |
| iuc, iutA | Aerobactin (siderophore) synthesis, receptor | Yes (268, 307) | Yes (97) | |
| pckA | Gluconeogenesis | Yes (302) | ||
| sdhB | TCA cycle | Yes (302) | ||
| tonB | Siderophore uptake | N/Ae | Yes (65) | |
| Protectins | iss | Increased serum survival (outer membrane protein) | Yes (307, 308) | Yes (309) |
| kpsMT II | Group II capsule synthesis (e.g., K1, K5, K12) | Yes (112, 126, 267, 310) | Yes (311) | |
| kpsMT III | Group III capsule synthesis (e.g., K3, K10, K54) | Yes (112, 126, 310) | Yes (312) | |
| proP | Osmoprotection; proline permease | N/Ae | Yes (313) | |
| rfc | O4 lipopolysaccharide (LPS) synthesis | Yes (314, 315) | Yes (316) | |
| traT | Surface exclusion; serum resistance-associated | Yes (307) | No (317) | |
| TCSSf | barA-uvrY | Regulation of VFs, e.g., hemolysin, LPS | Yes (318) | |
| cpxAR | Regulation of VFs, e.g., hemolysin | Yes (319) | ||
| envZ-ompR | Regulation of VFs, e.g., osmoprotection | Yes (320) | ||
| kguRS | Regulation of VFs, e.g., nutrient acquisition | Yes (321) | Yes (321) | |
| qseBC | Regulation of VFs, e.g., adhesins, flagella | Yes (322) | ||
| Invasins | aslA | Cellular invasion (arylsulfatase-like gene) | N/Ae | Yes (323) |
| ibeA-C | Invasion of brain endothelium IbeA (Ibe10), B, and C | Yes (IbeA) (111) | Yes (323) | |
| nlpI | Invasion of brain endothelium, complement resistance | No | Yes (324, 325) | |
| ompA | Outer membrane protein A (cellular invasion) | N/Ae | Yes (323) | |
| traJ | Cellular invasion (F-like plasmid transfer region homologue) | No | Yes (323) | |
| Misc.g | cvaC | Microcin (colicin) V; on plasmids with traT, iss, iuc/iut | Yes (307) | Yes (326) |
| fliC | Flagella: motility, ascent to the kidneys | Yes (327) | ||
| malX | Pathogenicity island marker (from strain CFT073) | Yes (130, 132, 267) | No (139) | |
| ompT | Outer membrane protein T (protease) | Yes (328) | Yes (unpublished, JRJ) | |
| pga | Extracellular polysaccharide (poly-GlcNac), biofilm | Yes (284) | ||
| rfaH | Transcriptional antitermination: O antigen, capsule, Hly, ChuA | Yes (329) | ||
| usp | Uropathogenic-specific protein (bacteriocin) | Yes (22, 120, 267) | Yes (330) | |
| ydd-pqq | ABC transporter, inner-outer membrane proteins | Yes (284) | ||
Note: list is not comprehensive even for recognized markers, and additional markers remain to be identified and characterized. Conversely, not all the listed traits necessarily contribute to virulence; some are only epidemiologically linked with virulence or confer in vitro phenotypes that are suspected of promoting virulence. Additionally, some of the listed traits are prominent also among intestinal pathogenic E. coli, e.g., cytolethal distending toxin, certain Dr-binding adhesins, and EAST1 (astA).
Statistically associated with clinical isolates or specific host characteristics, or highly prevalent in a particular extraintestinal infection syndrome.
Based on animal model infection studies, not necessarily using isogenic strains or complemented mutants.
Certain adhesins and toxins function as invasins, e.g., type-1 fimbriae, some Dr-binding adhesins, and CNF-1 (115, 145, 197).
N/A, not applicable (ubiquitous within E. coli, which precludes epidemiological comparisons).
TCSS, two-component secretion system.
Misc., miscellaneous.
Figure 6.
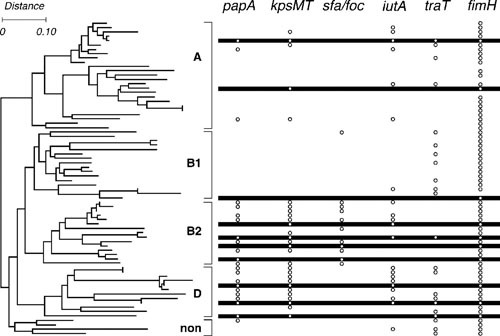
Phylogenetic distribution of extraintestinal pathogenic Escherichia coli (ExPEC)-associated virulence traits. Dendrogram at left depicts phylogenetic relationships for the 72 members of the E. coli Reference (ECOR) collection, as inferred based on multilocus enzyme electrophoresis (67). The four traditional major E. coli phylogenetic groups, i.e., A, B1, B2, and D (now split into groups D and F). The nonaligned (“non”) strains (now called group E) are bracketed and labeled. Bullets at right indicate presence of putative virulence genes (papA, P fimbriae; kpsMT, group II capsule synthesis; sfa/foc, S and F1C fimbriae; iutA, aerobactin system; traT, serum resistance; and fimH, type 1 fimbriae). Horizontal bars at right indicate the 10 ECOR strains isolated from humans with symptomatic UTI. The remaining strains, except for one asymptomatic bacteriuria isolate, are fecal isolates from healthy human or animal hosts. Note the concentration of (chromosomal) ExPEC-defining virulence genes papA, kpsMT, and sfa/foc within phylogenetic groups B2 and D, but their occasional joint appearance also in distant lineages, consistent with coordinate horizontal transfer, giving rise to ExPEC strains in historically non-ExPEC lineages. The more scattered phylogenetic distribution of iutA (ExPEC-defining) and traT is consistent with these two genes’ typically plasmid location, although iutA also can be chromosomal. fimH is nearly universally prevalent, consistent with its presence in other species of Enterobacteriaceae, presumably reflecting an origin in a shared enterobacterial ancestor. Note the concentration of UTI isolates within phylogenetic groups B2 and D and the concentration of virulence genes among UTI isolates. Note also the appreciable minority of fecal isolates with multiple virulence genes, reflecting a fecal reservoir of ExPEC. Reprinted from reference 67, with permission.
Associations among VFs
Certain VFs commonly occur together among clinical isolates in patterns suggesting either co-selection or direct genetic linkage (112, 113). Extensive genetic linkage of VFs has been demonstrated both within genomic islands, which have been called pathogenicity-associated islands (PAIs) and fitness islands, and can occur on either the chromosome or plasmids, and on plasmids, unassociated with PAIs (114–121). Certain VFs typically occur on the chromosome (for example, pap, sfa/foc, hly, cnf, and fyuA) (114, 122), others on plasmids (for example, iss, traT) (123), and some in either context (for example, afa/dra and iuc/iut) (118, 121). ExPEC strains often contain multiple PAIs, each with a distinctive combination of VFs, which sometimes results in a strain having multiple copies of a particular VF, for example, pap (Fig. 7) (114, 124, 125).
Figure 7.
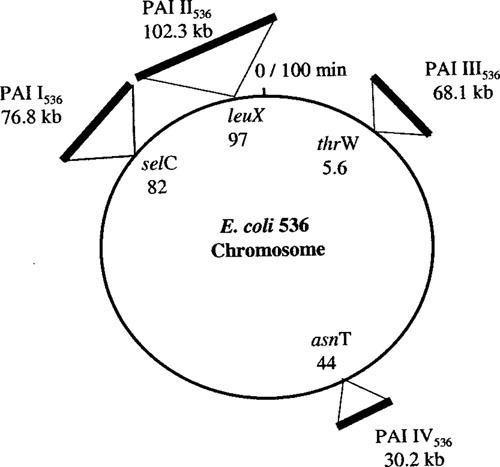
Genome map of ExPEC strain 536. The map is based on the chromosome of E. coli MG1655 (K-12). Pathogenicity islands (PAIs) are indicated according to their chromosomal insertion sites next to tRNA-encoding genes. Contents, by PAI, include: PAI I (α-hemolysin, F17-like fimbriae, CS12-like fimbriae); PAI II (α-hemolysin, P fimbriae with papG III); PAI III (S fimbriae, iro siderophore system, Tsh-like hemoglobin protease); PAI IV (yersiniabactin system). Many additional smaller DNA insertions compared to K-12 are present (not shown). Linkage of virulence genes in PAIs contributes to statistical associations between different virulence genes and between specific virulence genes and the lineages within which the corresponding PAIs tend to occur. Reprinted from reference 121, with permission.
This co-occurrence of VFs results in overlapping statistical associations of different VFs with clinical variables, leading to uncertainty as to which VF is primarily responsible for the association. Multivariable analysis can help in this situation, but is not definitive. Moreover, sequence analysis of PAIs, plasmids, and genomes invariably reveals genes of unknown function, some with homology to known VFs in other species (Fig. 8) (114, 119, 125). This suggests that the statistical associations of known VFs with virulence may be mediated through some of these as-yet-uncharacterized VFs; that is, the known VFs, although useful markers, may not themselves be the actual determinants of virulence. The genes of unknown function, once discovered, can become the focus of additional molecular epidemiological studies to clarify the genes’ associations with ecological source, clinical syndrome, host groups, lineages, etc.
Figure 8.
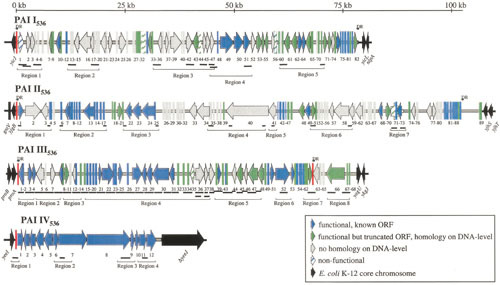
Maps of pathogenicity-associated islands (PAIs) from ExPEC strain 536. Known or putative open reading frames (ORFs) are grouped according to the following characteristics: blue, functional, known ORFs; green, truncated ORFs with a start codon and a stop codon; gray, as-yet-unidentified ORFs without homologues on the DNA level. Nonfunctional ORFs (e.g., due to internal stop codons or frameshifts) are indicated by hatched symbols. ORF numbers are indicated below the corresponding ORF symbols. Functional or truncated tRNA-encoding genes are marked in red. Direct repeat (DR) structures flanking PAIs are indicated. Thick black lines below the PAIs represent regions that were detected by PCR. Several PAI-specific PCRs were grouped into PAI regions. The molecular epidemiology of novel ORFs that are discovered through sequence analysis of PAIs can be investigated in subsequent studies. Reprinted from reference 121, with permission.
Phylogenetic group and clonal groups
According to molecular analyses, certain ExPEC clonal groups, as identified traditionally based on their O:K:H serotypes (for example, O18:K1:H7, O6:K2:H1, and O7:K1:H–) (126), which correspond closely with specific STs according to MLST, are disproportionately represented among clinical isolates in comparison with controls (110, 127, 128). These virulent clonal groups derive primarily from phylogenetic group B2 (e.g., STs 12, 14, 73, 95, 127, and 131), and to a lesser extent group D (e.g., STs 69, 394, and 405), which explains the observed predominance of groups B2 and D among clinical isolates (83, 110, 112, 129). Most of the traditionally recognized extraintestinal virulence markers (for example, pap, sfa/foc, hly, and kps) are typically concentrated within these virulent clonal groups and, hence, within phylogenetic groups B2 and/or D, whereas others (for example, afa/dra, iuc/iut, and traT) are more broadly and/or sporadically distributed across the species (Fig. 6) (35, 130). These divergent patterns of phylogenetic distribution correspond with vertical (within-lineage) versus horizontal (between-lineage) transmission, respectively, and reflect the typically chromosomal versus plasmid location of the respective sequences, as discussed above.
Considerable variation in VF profiles is evident at every level within the phylogenetic tree, including among the major phylogenetic groups, among the various clonal groups within these phylogenetic groups, and even among subclones within individual clonal groups. This is consistent with extensive ongoing remodeling of PAIs and/or virulence plasmids, in addition to acquisition and loss of entire PAIs or plasmids. Such evolutionary processes presumably result in the continuous emergence of new pathotypes upon which selective forces can act (110, 131).
Several studies have compared clinical isolates with fecal isolates from the same hosts, as opposed to fecal isolates from a separate control population (132–134). This strategy ensures a degree of matching for associated host characteristics greater than that provided by a traditional unpaired study design. The results of such studies, like those of most traditional comparison studies, suggest that special pathogenicity (as indicated by the presence of multiple VFs) rather than simple prevalence (that is, quantitative predominance in the fecal flora) is necessary for a fecal strain to cause urinary tract infection (UTI).
Phylogenetic background versus VFs
The overlapping associations of VFs and phylogenetic background with clinical virulence call into question which of these bacterial characteristics, VFs or phylogenetic background, more directly determines virulence, i.e., a strain’s ExPEC status. Several studies in which both phylogenetic group and VF profiles were analyzed have shown that VFs are statistically more closely associated with clinical virulence (132, 135, 136). However, phylogenetic group exhibits a residual association with virulence even after known VFs are accounted for (135). This is consistent with the existence of as-yet-undefined virulence-associated traits that are both phylogenetically distributed and incompletely linked with known VFs.
Comparisons among syndromes and host groups
Molecular epidemiological comparisons are not limited to E. coli populations from infected versus uninfected hosts. Comparisons also can be made between isolates (i) from patients with different clinical syndromes (to identify syndrome-specific, versus conserved, VFs or clonal groups) (113, 137, 138), (ii) from infected hosts who possess or lack particular predisposing conditions (to identify bacterial traits that may interact with specific host defense mechanisms or host receptors) (139–142), and (iii) from different host species (to identify species-specific, versus broad host range, VFs or pathogens) (106, 140–148).
The results of such studies support certain general conclusions regarding ExPEC strains, including which bacterial traits typify them, which host factors interact with these traits, and to what extent do these relationships vary with host species, sex, age, anatomical site of infection, and illness severity. First, invasive clinical syndromes, such as pyelonephritis, bacteremia, prostatitis, and meningitis, as compared with less invasive syndromes, such as cystitis and asymptomatic bacteriuria, on average usually involve strains with greater molecular virulence, as reflected in the number of VFs and a group B2 background. Second, various forms of host compromise significantly decrease the requirement for bacterial virulence within a defined clinical syndrome. This is exemplified by the reduced prevalence of pap among pyelonephritis isolates from patients with, versus those without, vesicoureteral reflux, that is, spontaneous retrograde flow of urine from the bladder back up to the kidneys (132, 149), and the reduced prevalence of pap and chromosomal aerobactin determinants among blood isolates from patients with urosepsis who have, versus those who lack, underlying anatomical or medical conditions predisposing to UTI (47). Third, although there is some syndrome and host specificity of VFs and clonal groups, there also is considerable commonality among syndromes and host groups, whereas tremendous diversity is apparent within each syndrome and host group. Examples of relative syndrome and host specificity that have been identified include the statistical association of sfaS (S fimbriae) with neonatal meningitis (83, 111); of pap with pyelonephritis (150, 151); of papG allele III, hly, and cnf with canine UTI (132, 143); and of the F11 variant of papA (P fimbriae structural subunit) with avian septicemic E. coli (152). However, each of these associations is incomplete, since the same VFs or clonal groups occur to various degrees also in other syndromes and host groups, as exemplified by the prominence of the O18:K1:H7 clonal group (ST95) in both neonatal meningitis (as traditionally recognized) and uncomplicated cystitis in women (as more recently appreciated) (Fig. 3) (1, 91, 119, 128, 153, 154). To what extent high-resolution phylogenetic analyses such as those based on WGS data will identify host or syndrome-specific subclades within certain clonal groups and STs that previously were regarded as unitary entities remains to be seen (53, 80).
Regulation of expression
The relative lack of syndrome-specific VFs does not necessarily negate the concept that selected VFs are critical for infection at selected sites. It may reflect instead the broad environmental flexibility of E. coli both within and outside the human host, which could be manifested by site-specific regulation and expression of genes or sets of genes. Therefore, in addition to the simple presence or absence of a particular gene, molecular epidemiological studies may also need to consider gene expression, since expression obviously is required if the genotype is to influence the virulence phenotype. Expression can be assessed through a variety of phenotypic tests, which moves beyond the realm of strict molecular epidemiology. However, in the instance of the fim operon, expression is regulated by an invertible switch element in the promoter region, the position of which (“on” versus “off”) can be defined for a bacterial population via a simple PCR assay (57, 124, 155, 156). Differences between UTI versus control isolates, and between cystitis versus pyelonephritis isolates, with respect to their fim switching bias can be demonstrated, supporting the concept that regulation of fim expression may influence not only overall pathogenicity but also anatomical site tropism (155, 156). Such a molecular epidemiological approach is particularly relevant for fim since, although expression of type 1 fimbriae appears from experimental studies to be quite important for UTI pathogenesis, the nearly uniform presence of fim in E. coli (and other enterobacteria) all but precludes demonstration of a virulence association for fim through conventional presence-absence comparisons between clinical isolates and controls (130).
Site-specific expression of VFs and core “housekeeping” genes directed by environmental cues and mediated by regulatory elements (e.g., two-component signal systems; Table 1) is a more global mechanism that enables growth/survival within a variety of challenging niches (157). Molecular epidemiological associations of regulatory elements may prove to be a fruitful area for additional study.
Molecular variants
Another potentially important consideration is the particular variant of a virulence gene present in an isolate. Molecular variation within a gene may produce pathogenetically important phenotypic alterations in the encoded peptide, such as the shifts in preferred receptor sugars or glycolipids that are associated with polymorphisms in fimH (type 1 fimbrial adhesin) and papG, respectively. Diverse single-nucleotide polymorphisms (SNPs) in fimH, which can be detected by sequence analysis or with SNP-specific PCR primers, cause single-amino-acid changes in the FimH peptide that produce a shift from a (commensal-associated) trimannose-binding phenotype to a (UTI-associated) monomannose-binding phenotype (158). Interestingly, the monomannose-binding variants, although at an advantage within the pathogenic niche (for example, because of their enhanced binding to bladder epithelium), also are more susceptible to inhibition by monomannose residues, such as are found in salivary glycoproteins; this presumably makes them less effective as gut colonization factors (158). An additional point mutation in a monomannose-binding fimH variant, resulting in a single FimH amino acid substitution (Ser-62-Ala), can further strengthen monomannose binding and also confer type IV collagen binding (Fig. 9), which may be important in the pathogenesis of neonatal meningitis (159, 160). Such mutations have been termed pathoadaptive, since they represent minor modifications of genes already present in nonpathogenic members of the species, with the mutations conferring enhanced fitness in the pathogenic niche (161).
Figure 9.
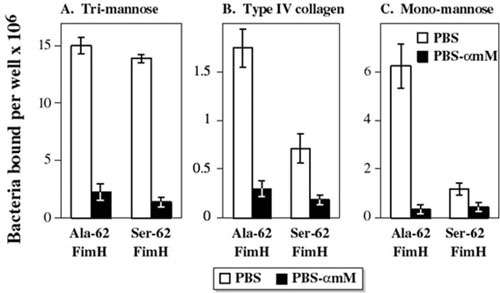
Receptor binding specificity of type 1 fimbrial adhesin (FimH) variants. In vitro binding of isogenic recombinant strains expressing the Ala-62 or Ser-62 FimH variants (from strains NU14 and 536, respectively) to (A) a trimannose substrate (bovine RNAse B), (B) human collagen type IV, and (C) a monomannose substrate (yeast mannan). Both variants bind equally well to trimannose, but the Ala-62 variant exhibits stronger type IV collagen and monomannose binding than does the Ser-62 variant. (Commensal-associated FimH variants exhibit equally strong trimannose binding but minimal binding to type IV collagen or monomannose [not shown].) Open columns, bacteria incubated without α-methyl mannoside (αmM); solid columns, bacteria incubated with 50 mM αmM. Data are mean + SEM (n = 4) of number of bacteria bound per well. Molecular epidemiological studies can be used to elucidate the likely clinical relevance of such genetic and phenotypic variation within different virulence factors. Adapted from reference 160, with permission.
More extensive alterations within papG, which can be detected by using specific primers or probes, or sequence analysis, are evident among the several known allelic variants of papG (42, 90, 162–164). The respective peptide products of the papG alleles bind preferentially to various forms of Gal(α1-4)Gal-containing glycolipids (165–168). Because of the varied distribution of these particular glycolipids by anatomical site and host species, the distinctive binding preference of the PapG variants may underlie their somewhat divergent associations with clinical syndromes and host groups, such as the associations of PapG II with pyelonephritis and of PapG III with cystitis, dogs and cats, and compromised hosts with urosepsis (84, 143, 168–173). Other examples of epidemiologically significant molecular variation within a particular VF, or VF family, include the sfa/foc (S and F1C fimbriae) family (174), the afa/dra (afimbrial and Dr-binding adhesins) family (92, 175, 176), the group 2 and group 3 kps (capsule) families (177, 178), and cnf1 and cnf2 (cytotoxic necrotizing factor I and II; associated with extraintestinal and intestinal virulence, respectively) (179). Likewise, papA (P fimbriae structural subunit) exhibits 13 known alleles (180, 181). These tend to be clonally distributed, such that they can be used, in conjunction with other VFs or seroantigens, as markers for particular clones (110, 181). They also exhibit somewhat distinctive associations with specific host groups or syndromes, for example, F10, F12, and F48 with canine UTI (132, 143, 182).
VFs as predictors of clinical outcomes
Apart from their role in pathogenesis, VFs also have been studied as potential clinical predictors that could be used to guide patient management. For example, P fimbriae (or pap) testing has been proposed as a way to identify boys at risk for renal scarring (183), adults with pyelonephritis or urosepsis who have unrecognized predisposing conditions (184), and patients whose household members should be screened for colonization with the patient’s UTI strain (185). Other proposed clinical applications of such testing include determining length of therapy for children with UTI (186) and identifying pregnant women at high risk for developing pyelonephritis (187). However, the true clinical utility and cost-effectiveness of such clinical applications of VF testing are unconfirmed (188), such that at present they cannot be recommended outside a research setting.
Antimicrobial resistance and virulence
Acquired resistance to therapeutically important antimicrobial agents is increasingly prevalent among clinical E. coli isolates, making management of E. coli infections more difficult and costly (189, 190). The relationship between resistance and virulence or phylogenetic background has been explored in multiple molecular epidemiological studies. Older data indicated that among E. coli isolates from patients with urosepsis, resistance to historical antimicrobial agents such as ampicillin, sulfonamides, tetracycline, and streptomycin is negatively associated with virulence and a group B2 phylogenetic background, but is positively associated with host compromise (191). This is consistent with a scenario wherein resistance provides a greater fitness advantage than do traditional VFs or a group B2 background for infections in compromised hosts, who have weakened defenses but are frequently exposed to antimicrobial agents.
Subsequent studies regarding fluoroquinolone resistance that were done prior to the emergence of ST131-H30, the currently dominant fluoroquinolone-resistant E. coli lineage (which is from group B2), demonstrated similar negative associations between resistance and VFs or a B2 phylogenetic background (192–195). Although these were interpreted as suggesting that VFs may be lost concomitant with mutation to fluoroquinolone resistance (195), that explanation did not account for the concomitant resistance-associated phylogenetic shifts away from group B2, which suggested instead that resistant isolates derive predominantly from distinct, less virulent bacterial populations (192, 193). Therefore, selection for antimicrobial resistance within different host and bacterial populations, rather than loss of virulence genes in exchange for resistance, may have produced the observed VF differences between susceptibility groups. Indeed, fluoroquinolone resistance has been shown to be associated with host compromise. Thus, among clinical isolates selection factors similar to those that historically produced statistical associations between low virulence and resistance to older antimicrobial agents may be operative also with fluoroquinolones. Notably, the current prominence of ST131-H30, with its broad virulence gene repertoire despite extensive multidrug resistance, has largely erased the historical negative associations of resistance with virulence genes and group B2 (79, 196).
Other findings confirm that antimicrobial resistance does not necessarily equate with reduced virulence. For example, among fecal E. coli from diseased cattle and swine, and from retail meat products (which strains likely derive from the source animal hosts), settings in which most of the organisms are nonpathogens, resistance to extended-spectrum cephalosporins or fluoroquinolones is associated with minimal shifts in VF profile (192, 197–204). Likewise, two notable epidemic multidrug-resistant clonal groups that preceded ST131-H30 by several decades, i.e., E. coli “clonal group A” (ST69) and the O15:K52:H1 clonal group (STc31), are replete with VFs, which presumably contributed to these clonal groups’ success as pathogens among otherwise healthy hosts. Resistance and virulence presumably are uncoupled for the animal-source isolates by the absence of selection for virulence, and for human clinical isolates from clonal group A and the O15:K52:H1 clonal group (as for those from ST131-H30) by the uniform requirement for virulence, irrespective of resistance. Further studies clearly are needed to clarify the relationship between virulence and resistance, taking into account ecological source and relevant selection factors.
VFs versus colonization factors
Paradoxically, certain molecular epidemiological (205–207) and experimental (208–212) data suggest that at least some of what traditionally have been regarded as VFs in ExPEC may also promote intestinal survival and colonization, and that virulence may even be a by-product of commensalism (213). This hypothesis provides a more plausible mechanism for the evolution of these traits than does the enhanced pathogenicity the traits confer, since the ability to persist and flourish within the host intestinal tract represents a more obvious survival advantage than does the ability to cause sporadic and usually self-limited or even fatal disease.
Moreover, although many of the traditionally recognized extraintestinal VFs are encoded on what have been designated PAIs, which implies that pathogenicity is their raison d’être, this terminology is evolving. The newer, more inclusive designation “fitness island” reflects the recognition that similar accretions of genes encoding related functions, with associated insertion sequences and other mobility-promoting elements, occur in nonpathogens, including even environmental (non-host-associated) organisms (214), and that even in ExPEC strains PAIs may function as colonization factors (215). However, the hypothesis that VFs have evolved primarily as colonization factors does not explain why ExPEC are not the dominant clone(s) within the intestinal tract in most human hosts, as would be expected if they truly have a fitness advantage in this niche (113, 216). Additional epidemiological and experimental studies are needed to clarify the relationship between specific bacterial traits, including recently discovered putative VFs, and intestinal colonizing ability.
Genomic Profiling
In most of the studies discussed above, E. coli isolates were analyzed in the aggregate, with conclusions being based on comparisons between groups of isolates, without particular regard to the constituent clones. In contrast, genomic profiling (also called molecular fingerprinting or clonal typing) allows individual clones or strains to be resolved and analyzed. This approach underlies a distinct branch of molecular epidemiology, one that focuses on the individual clone rather than on the group. Topics that have been addressed using this approach include the reservoirs, colonization patterns, transmission pathways, and clinical diagnostics of ExPEC.
Fecal-vaginal-urethral hypothesis
According to the fecal-vaginal-urethral hypothesis, E. coli strains causing UTI usually derive immediately from the host’s own fecal and perineal flora. This model, which was first suggested based on O serogroup data (217), is now supported also by molecular data showing that, in most episodes of acute cystitis or pyelonephritis in women, prostatitis in men, or UTI in dogs, the urine organism is also the host’s predominant fecal strain (60, 63, 132, 218, 219). This concept was also demonstrated by a study that used daily sampling to obtain high-resolution tracking of colonization patterns leading up to an acute UTI episode (220). Over the week prior to the UTI episode, the causative UTI strain’s prevalence in the vaginal reservoir rose rapidly, preceding the strain’s appearance in urine cultures, which in turn preceded the onset of UTI symptoms. This is relevant to prevention efforts, since it suggests that fecal (and vaginal) colonization with a urovirulent organism is a potentially modifiable risk factor for subsequent UTI. This provides a rationale for studying the determinants of intestinal (and vaginal) colonization with particular E. coli strains and for searching for external reservoirs of virulent E. coli that might be acquired by the host as intestinal (and vaginal) colonizers.
Same- versus different-strain recurrent UTI
Molecular fingerprinting also has been used to assess the same- versus different-strain nature of recurrent UTI isolates, in comparison with index isolates. The findings have been quite variable, with same-strain episodes accounting for from 25 to 100% of recurrences in different studies (60, 63, 64, 221–224), with differences in selection criteria and patient populations contributing to the variability. In one study, 30/44 (68%) of recurrent UTIs were caused by a strain previously identified in that person (60). This percentage rose with the number of recurrences per person-year, from 55% (6/11) among patients with two recurrences, through 72% (17/24) among those with four or five recurrences, to 78% (7/9) among those with six or more recurrences. Analysis of a subset of subjects established that most recurrent UTIs were due to same-strain reinfection, not overt persistence within the urinary tract, and suggested that the colonic flora was the reservoir for these reinfecting strains (although intracellular persistence within the urinary tract, as discussed below, could not be ruled out). As observed in this study and others, some patients experience multiple same-strain recurrences, some of which can occur months or years after the initial episode, occasionally with intervening UTI episodes due to unrelated strains (60, 63, 64, 221, 224, 225).
The biological relevance of this sort of analysis is that different-strain recurrence implies an independent infection episode whereas same-strain recurrence implies either relapse from a persisting endogenous focus or reintroduction of the strain from a persisting external reservoir in the host or the environment. This distinction may have clinical relevance for prevention and treatment efforts, since the occurrence of multiple independent infection episodes suggests an underlying host predisposition to infection (which may be amenable to intervention, for example, through a change in contraceptive method) (66), whereas the presence of a persisting reservoir (whether external or internal) suggests a need to identify and eradicate the reservoir.
Potential endogenous reservoirs include the long-term intracellular persistence of a strain within the bladder epithelium that seems to underlie the intermittent episodes of recurrent bacteriuria that occur in experimentally infected mice following apparent resolution of the initial infection (226). Limited clinical evidence supports that this phenomenon occurs also in humans, although its proportional contribution to same-strain recurrent UTI is undefined (227). For external reservoirs, the host may be persistently colonized with a strain in the intestine and/or vagina or may repeatedly reacquire it from the (animate or inanimate) external environment (221). In either situation, efforts to identify and eliminate the external reservoir conceivably could be protective.
Strain sharing between associated hosts
Environmental sources of uropathogenic or antimicrobial-resistant E. coli have been investigated by using genomic profiling, with or without VF detection and phylogenetic analysis. Within-household strain sharing has been demonstrated by PCR-based genomic profiling, PFGE, and whole-genome analysis between adult sex partners (in some instances, accompanied by symptomatic UTI in one or both individuals) and between parents and children, siblings, and even humans and pets (Figs. 2, 10, and 11) (63, 148, 224, 228–237). Likewise, several hospital-based pyelonephritis outbreaks have been documented in which health care workers seemingly transmitted a virulent strain to patients (238–240). Interestingly, some evidence suggests that what classically have been regarded as VFs also predict co-colonization of epidemiologically associated hosts, implying that these traits may promote person-to-person transmission as well as infection (224, 233, 234, 237, 241, 242).
Figure 10.
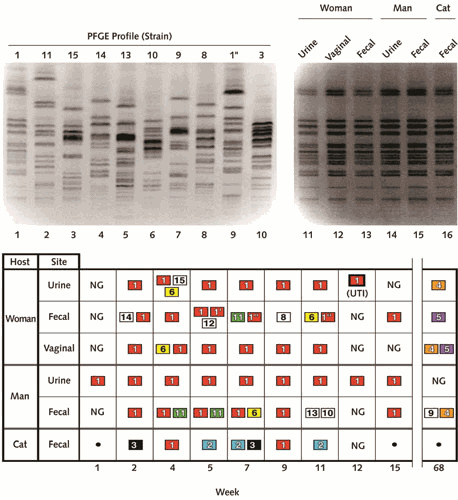
Pulsed-field gel electrophoresis (PFGE) profiles and colonization patterns of Escherichia coli isolates from three household members (man, woman, and pet cat). (Top panel) PFGE profiles. Lane numbers are shown below gel images. Lanes 1 through 10, profiles of nine of the unique strains, with strain designations shown above gel lanes, plus subtype 1″) (lane 9). Lanes 11 through 16, profiles of independent isolates of strain 1, as recovered from various anatomical sites from the woman (lanes 11–13), man (lanes 14 and 15), and cat (lane 16). (Bottom panel) Distribution of 14 unique E. coli strains over time (week of sampling shown below grid), as recovered from various anatomical sites from the three household members. NG, no growth; •, no sample. Strains isolated more than once appear in colored boxes, with a unique color for each strain. Strains isolated only once appear in colorless boxes. Week 12, which coincided with symptoms of acute urinary tract infection (UTI) in the woman, yielded strain 1 from the woman’s urine specimen (boldface box). There is no strain 7. Strain 1, the woman’s UTI strain, was the most extensively shared and persistent strain, and had the most virulence genes of the 14 strains. Reprinted from reference 242, with permission.
Figure 11.
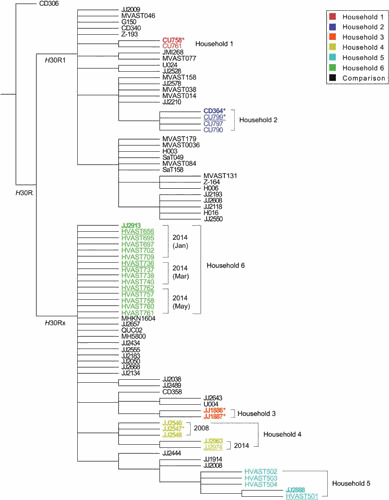
Bootstrapped consensus core genome phylogeny for 33 household isolates and 47 comparison isolates of Escherichia coli sequence type 131 (ST131). The tree was based on 1000 bootstrapped maximum likelihood trees, retaining only those nodes that appeared in >70% of trees, and was rooted with strain CD306. Branch lengths are meaningless. Household isolates (as in Fig. 2) are color-coded by household (1–6); comparison isolates are shown in black. For the household isolates: boldface indicates clinical isolates; regular font indicates fecal isolates; underlining indicates fecal isolates from a clinical isolate’s source host; and asterisks indicate the 6 household isolates that were included in Price et al (11), i.e., JJ1886, JJ1887, JJ2547, CU758, CU799, and CD364 (which in Price et al was labeled as CD449). Dates are shown for the 2 households that underwent serial sampling (households 4 and 6). Clustering by household supports within-household transmission (strain sharing); near identify of clinical and fecal isolates within each household supports the fecal reservoir as the source for infection-causing strains; and variation in a given strain within its source household over time (households 4 and 5) suggests microevolution during long-term host colonization. Reprinted from reference 63, with permission.
However, that person-to-person transmission actually occurs is inferential except in the hospital setting: the underlying mechanism for the observed within-household strain sharing remains to be defined. Sexual transmission, a favored hypothesis, is supported by epidemiological evidence associating certain sexual practices with co-colonization of adult sex partners and by anatomical coincidences such as colonization of the male partner’s urethra or glans penis with a strain also found in the female partner’s vagina and urine (229, 241, 243). However, sexual transmission is unlikely to explain co-colonization involving children, pets, and nonsexually associated adults (63, 148, 230, 236, 237, 242, 244); other modes of host-to-host transmission (whether direct or indirect) must be considered, as must be possible coordinate acquisition from a common external source, such as the food supply.
Community-wide dissemination of ExPEC
Transmission pathways of ExPEC within the larger community are relevant to the dissemination of virulent clonal groups within the human population. Older examples of this phenomenon include the O15:K52:H1 clonal group, which (as mentioned above) first came to attention when it caused the mysterious South London outbreak of 1986 and 1987 (245–247), and E. coli clonal group A, which (as also mentioned above) has emerged as a prominent cause of drug-resistant UTI across the United States (10, 61, 248) and appears to have undergone point-source spread within at least one community (61). The most recent and dramatic example is ST131-H30, which first emerged around 2000 and expanded rapidly thereafter to become, by 2010, the leading cause of fluoroquinolone-resistant and ESBL-associated E. coli infections worldwide (249, 250). Several other resistance-associated ExPEC clonal groups likewise have recently emerged and disseminated (78, 251, 252).
The seeming point-source outbreak behavior of clonal group A, mentioned above, prompted the suggestion that, by analogy to E. coli O157:H7, Campylobacter, and Salmonella, contaminated food products might serve as a transmission vehicle also for ExPEC (61). Indeed, retail foods are commonly contaminated with antibiotic-resistant E. coli and ExPEC, along a descending prevalence gradient from poultry (highest) through other meats (beef and pork) to produce and miscellaneous foods (lowest), and a subset of these isolates resembles human clinical isolates according to virulence genes and genomic profiles (192, 197–204). The human health implications of such contamination remain to be fully defined. Insights into this question conceivably could derive from higher-resolution molecular epidemiological comparisons of ExPEC isolates from retail foods versus colonized and infected humans to ascertain the extent of commonality between these populations. Indeed, preliminary results from a WGS-based comparison of temporally and geographically matched E. coli isolates from retail meat products and patients with UTI in a relatively isolated community (Flagstaff, Arizona) suggest that 11 to 20% of UTI isolates derive from genome clades that are dominated by meat-source isolates, implying a food animal source (L. B. Price, personal communication). Such cross-sectional ecological studies could be complemented by longitudinal molecular epidemiological surveillance of individuals and the foods they consume, thereby identifying temporal patterns of genomic commonality of E. coli strains suggesting food-borne transmission.
Within-host epidemiology in a single sample or across samples
Various molecular typing methods have been used to investigate the clonal composition of a single fecal or clinical sample or of concurrent paired samples (e.g., fecal or blood versus urine) from a given subject, and to track this over time by analyzing serial samples, or to compare same-clone and different-clone isolates from a given host. Such studies have shown that what conventionally would be regarded as monoclonal infections may actually involve multiple clones (253–255) or a single clone that has developed microheterogeneity (including for antimicrobial resistance gene content) in vivo, likely during adaptation to the infection niche (255). Plasmid segregation can occur in vivo or in the clinical laboratory, producing genetic variants with discordant resistance profiles that could conceal the underlying clonal commonality and, thus, potentially obscure the likely source of bacteremia (256). Subtle genetic differences between concurrent or sequential same-clone isolates from different niches or disease contexts (e.g., feces, cystitis, bacteremia) in a single host, or closely associated hosts, may (257, 258) or may not (255, 259) have apparent virulence-related implications, so may or may not be responsible for the isolates’ divergent clinical behaviors. Finally, at the time of an acute urinary tract infection the disease-causing clone often not only predominates in the host’s feces, consistent with both the “fecal-urethral” and “prevalence” hypotheses, but also exhibits more virulence-associated traits (e.g., VFs and group B2 background) than do the host’s concurrent “fecal-only” clones, consistent with the “special pathogenicity” hypothesis (132, 134, 260, 261).
Clonal diagnostics
The well-established associations of specific E. coli lineages (as defined at the level of ST complexes, STs, and sub-ST clades, e.g., ST131-H30) with antimicrobial resistance, clinical manifestations, and host characteristics, together with the increasing availability of rapid molecular detection methods, allow for the development of clone-specific diagnostic tests to predict, in real time, both antimicrobial resistance and clinical phenotype (262–265). Emerging data suggest that if such tests were to be used clinically to inform empirical treatment choices, this could reduce significantly the frequency of “drug-bug” (i.e., antimicrobial versus organism) mismatch and use of overly broad-spectrum therapy, thereby improving both clinical outcomes and antimicrobial stewardship (264). Clinical studies are needed to determine the real-world effectiveness of such tests.
SUMMARY AND CONCLUSIONS
Molecular epidemiological analyses of ExPEC, which are based on structured observations of E. coli strains as they occur in the wild, provide an important complement to experimental assessment. Fundamental to the success of molecular epidemiological studies are the careful selection of subjects and the use of appropriate methods for genotyping and statistical analysis. To date, molecular epidemiological studies have yielded numerous important insights into host-pathogen relationships, phylogenetic background, reservoirs, clinical diagnostics, and transmission pathways of ExPEC, including antimicrobial-resistant strains, and have delineated areas in which further study is needed. The rapid pace of discovery of new putative VFs and the increasing awareness of the importance of VF regulation, expression, and molecular variation should stimulate many future molecular epidemiological investigations. The ever-increasing sophistication and availability of molecular typing methodologies, and the new computational and statistical approaches that are being developed to address the huge amounts of data that WGS generates, provide improved tools for such studies and allow entirely new questions to be addressed.
ACKNOWLEDGMENTS
This material is based in part on work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs (grants # 2I01CX000920-04 to J.R.J. and # IBX000984A to T.A.R.).
The authors declare a conflict of interest. JRJ has received research grants from Allergan, Merck, and Tetraphase, is a coinvestigator on an NIH grant to IDGenomics, is a paid consultant to Crucell/Janssen, and has patent applications for tests to detect specific E. coli strains. TAR lists no financial conflicts of interest.
REFERENCES
- 1.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis 181:1753–1754. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect 5:449–456. [DOI] [PubMed] [Google Scholar]
- 3.Lipsky BA. 1989. Urinary tract infections in men. Epidemiology, pathophysiology, diagnosis, and treatment. Ann Intern Med 110:138–150. [DOI] [PubMed] [Google Scholar]
- 4.Andriole VT, Patterson TF. 1991. Epidemiology, natural history, and management of urinary tract infections in pregnancy. Med Clin North Am 75:359–373. [DOI] [PubMed] [Google Scholar]
- 5.Rothman KJ, Greenland S. 1998. The Emergence of Modern Epidemiology, 2nd ed. Lippincott-Raven, Philadelphia. [Google Scholar]
- 6.Senay H, Goetz MB. 1991. Epidemiology of bacteremic urinary tract infections in chronically hospitalized elderly men. J Urol 145:1201–1204. [DOI] [PubMed] [Google Scholar]
- 7.Stull TL, LiPuma JJ. 1991. Epidemiology and natural history of urinary tract infections in children. Med Clin North Am 75:287–297. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Warren JW. 1996. Clinical Presentations and Epidemiology of Urinary Tract Infections. ASM Press, Washington, DC. [Google Scholar]
- 9.Arthur M, Johnson CE, Rubin RH, Arbeit RD, Campanelli C, Kim C, Steinbach S, Agarwal M, Wilkinson R, Goldstein R. 1989. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli. Infect Immun 57:303–313. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burman WJ, Breese PE, Murray BE, Singh KV, Batal HA, MacKenzie TD, Ogle JW, Wilson ML, Reves RR, Mehler PS. 2003. Conventional and molecular epidemiology of trimethoprim-sulfamethoxazole resistance among urinary Escherichia coli isolates. Am J Med 115:358–364. [DOI] [PubMed] [Google Scholar]
- 11.Eisenstein BI. 1990. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis 161:595–602. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Lupski JR. 1993. Molecular epidemiology and its clinical application. JAMA 270:1363–1364. [PubMed] [PubMed] [Google Scholar]
- 13.Maslow JN, Slutsky AM, Arbeit RD (ed). 1993. The Application of Pulsed Field Gel Electrophoresis to Molecular Epidemiology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 14.Wachsmuth K. 1986. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis 8:682–692. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Edén C. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagberg L, Hull R, Hull S, Falkow S, Freter R, Svanborg Edén C. 1983. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect Immun 40:265–272. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badger JL, Wass CA, Weissman SJ, Kim KS. 2000. Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infect Immun 68:5056–5061. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dozois CM, Daigle F, Curtiss R III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA 100:247–252. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HLT. 2014. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci USA 111:18327–18332. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guyer DM, Henderson IR, Nataro JP, Mobley HLT. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol 38:53–66. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Johnson JR, Russo TA, Tarr PI, Carlino U, Bilge SS, Vary JCJ Jr, Stell AL. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN(E. coli), among Escherichia coli isolates from patients with urosepsis. Infect Immun 68:3040–3047. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurazono H, Yamamoto S, Nakano M, Nair GB, Terai A, Chaicumpa W, Hayashi H. 2000. Characterization of a putative virulence island in the chromosome of uropathogenic Escherichia coli possessing a gene encoding a uropathogenic-specific protein. Microb Pathog 28:183–189. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Russo TA, Carlino UB, Johnson JR. 2001. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun 69:6209–6216. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falkow S. 1988. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis 10(Suppl 2):S274–S276. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Bailar JC III, Louis TA, Lavori PW, Polansky M. 1984. Studies without internal controls. N Engl J Med 311:156–162. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Sartwell PE. 1974. Retrospective studies. A review for the clinician. Ann Intern Med 81:381–386. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Moses LE. 1985. Statistical concepts fundamental to investigations. N Engl J Med 312:890–897. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Rothman KJ. 1977. Epidemiologic methods in clinical trials. Cancer 39(Suppl):1771–1775. [DOI] [PubMed] [Google Scholar]
- 29.Kluytmans JA, Overdevest IT, Willemsen I, Kluytmans-van den Bergh MF, van der Zwaluw K, Heck M, Rijnsburger M, Vandenbroucke-Grauls CM, Savelkoul PH, Johnston BD, Gordon D, Johnson JR. 2013. Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis 56:478–487. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Bakhtiarizadeh MR, Moradi-Shahrbabak M, Ebrahimi M, Ebrahimie E. 2014. Neural network and SVM classifiers accurately predict lipid binding proteins, irrespective of sequence homology. J Theor Biol 356:213–222. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Louria DB, Skurnick J, Holland B. 1990. Listeria “epidemic” overinterpreted. J Infect Dis 162:274–276. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Schlesselman JJ. 1982. Limitations of Statistics. Oxford University Press, New York. [Google Scholar]
- 33.Freiman JA, Chalmers TC, Smith H Jr, Kuebler RR. 1978. The importance of beta, the type II error and sample size in the design and interpretation of the randomized control trial. Survey of 71 “negative” trials. N Engl J Med 299:690–694. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Johnson JR, Gajewski A, Lesse AJ, Russo TA. 2003. Extraintestinal pathogenic Escherichia coli as a cause of invasive nonurinary infections. J Clin Microbiol 41:5798–5802. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyd EF, Hartl DL. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol 180:1159–1165. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TE, Johnson JR, Didelot X, Walker AS, Crook DW, Modernizing Medical Microbiology Informatics Group (MMMIG). 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 7:e02162. 10.1128/mBio.02162-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arthur M, Campanelli C, Arbeit RD, Kim C, Steinbach S, Johnson CE, Rubin RH, Goldstein R. 1989. Structure and copy number of gene clusters related to the pap P-adhesin operon of uropathogenic Escherichia coli. Infect Immun 57:314–321. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JR, Stell AL. 2001. PCR for specific detection of H7 flagellar variant of fliC among extraintestinal pathogenic Escherichia coli. J Clin Microbiol 39:3712–3717. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hull SI, Bieler S, Hull RA. 1988. Restriction fragment length polymorphism and multiple copies of DNA sequences homologous with probes for P-fimbriae and hemolysin genes among uropathogenic Escherichia coli. Can J Microbiol 34:307–311. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Plos K, Carter T, Hull S, Hull R, Svanborg Edén C. 1990. Frequency and organization of pap homologous DNA in relation to clinical origin of uropathogenic Escherichia coli. J Infect Dis 161:518–524. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Boyd EF, Hartl DL. 1998. Diversifying selection governs sequence polymorphism in the major adhesin proteins fimA, papA, and sfaA of Escherichia coli. J Mol Evol 47:258–267. [DOI] [PubMed] [Google Scholar]
- 42.Johnson JR, Stell AL, Kaster N, Fasching C, O’Bryan TT. 2001. Novel molecular variants of allele I of the Escherichia coli P fimbrial adhesin gene papG . Infect Immun 69:2318–2327. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid SD, Selander RK, Whittam TS. 1999. Sequence diversity of flagellin (fliC ) alleles in pathogenic Escherichia coli. J Bacteriol 181:153–160. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokurenko EV, Courtney HS, Ohman DE, Klemm P, Hasty DL. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J Bacteriol 176:748–755. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. 2015. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson TJ, Danzeisen JL, Youmans B, Kase K, Llop K, Munoz-Aquayo J, Flores-Figueroa C, Aziz M, Stoesser N, Sokurenko E, Price LB, Johnson JR. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere 1:e00121-16. 10.1128/mSphere.00121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JR, Moseley SL, Roberts PL, Stamm WE. 1988. Aerobactin and other virulence factor genes among strains of Escherichia coli causing urosepsis: association with patient characteristics. Infect Immun 56:405–412. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bekal S, Brousseau R, Masson L, Prefontaine G, Fairbrother J, Harel J. 2003. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. J Clin Microbiol 41:2113–2125. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson JR. 2000. Development of polymerase chain reaction-based assays for bacterial gene detection. J Microbiol Methods 41:201–209. [DOI] [PubMed] [Google Scholar]
- 50.Huang S-H, Chen Y-H, Fu Q, Stins M, Wang Y, Wass C, Kim KS. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB , required for penetration of brain microvascular endothelial cells. Infect Immun 67:2103–2109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kudinha T, Kong F, Johnson JR, Andrew SD, Anderson P, Gilbert GL. 2012. Multiplex PCR-based reverse line blot assay for simultaneous detection of 22 virulence genes in uropathogenic Escherichia coli. Appl Environ Microbiol 78:1198–1202. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanamori H, Parobek CM, Juliano JJ, Johnson JR, Johnston BD, Johnson TJ, Weber DJ, Rutala WA, Anderson DJ. 2017. Genomic analysis of multidrug-resistant Escherichia coli from North Carolina community hospitals: ongoing circulation of CTX-M-producing ST131-H30Rx and ST131-H30R1 strains. Antimicrob Agents Chemother 61:e00912–e00917. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of ESBL-producing Escherichia coli ST131 is driven by a single highly virulent subclone, H30-Rx. MBio 6:e00377-13. 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNally A, Oren Y, Kelly D, Pascoe B, Dunn S, Sreecharan T, Vehkala M, Välimäki N, Prentice MB, Ashour A, Avram O, Pupko T, Dobrindt U, Literak I, Guenther S, Schaufler K, Wieler LH, Zhiyong Z, Sheppard SK, McInerney JO, Corander J. 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12:e1006280. 10.1371/journal.pgen.1006280. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. MBio 7:e00347-16. 10.1128/mBio.00347-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Earle SG, Wu C-H, Charlesworth J, Stoesser N, Gordon NC, Walker TM, Spencer CCA, Iqbal Z, Clifton DA, Hopkins KL, Woodford N, Smith EG, Ismail N, Llewelyn MJ, Peto TE, Crook DW, McVean G, Walker AS, Wilson DJ. 2016. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol 1:16041. 10.1038/nmicrobiol.2016.41. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lees JA, Vehkala M, Välimäki N, Harris SR, Chewapreecha C, Croucher NJ, Marttinen P, Davies MR, Steer AC, Tong SYC, Honkela A, Parkhill J, Bentley SD, Corander J. 2016. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat Commun 7:12797. 10.1038/ncomms12797. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arbeit RD, Arthur M, Dunn R, Kim C, Selander RK, Goldstein R. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J Infect Dis 161:230–235. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.Ott M, Bender L, Blum G, Schmittroth M, Achtman M, Tschäpe H, Hacker J. 1991. Virulence patterns and long-range genetic mapping of extraintestinal Escherichia coli K1, K5, and K100 isolates: use of pulsed-field gel electrophoresis. Infect Immun 59:2664–2672. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russo TA, Stapleton A, Wenderoth S, Hooton TM, Stamm WE. 1995. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J Infect Dis 172:440–445. [PubMed] [DOI] [PubMed] [Google Scholar]
- 61.Manges AR, Johnson JR, Foxman B, O’Bryan TT, Fullerton KE, Riley LW. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med 345:1007–1013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson JR, Davis G, Clabots C, Johnston BD, Porter S, DebRoy C, Pomputius W, Ender PT, Cooperstock M, Slater BS, Banerjee R, Miller S, Kisiela D, Sokurenko EV, Aziz M, Price LB. 2016. Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole genome sequence analysis. Open Forum Infect Dis 3:ofw129. 10.1093/ofid/ofw129 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1996. Persistent bacteriuria caused by uropathogenic Escherichia coli. Urol Int 57:89–92. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Nègre VL, Bonacorsi S, Schubert S, Bidet P, Nassif X, Bingen E. 2004. The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect Immun 72:1216–1220. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stapleton A, Stamm WE. 1997. Prevention of urinary tract infection. Infect Dis Clin North Am 11:719–733. [DOI] [PubMed] [Google Scholar]
- 67.Johnson JR. 2003. Microbial virulence determinants and the pathogenesis of urinary tract infection. Infect Dis Clin North Am 17:261–278, viii. [DOI] [PubMed] [Google Scholar]
- 68.Dale AP, Woodford N. 2015. Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J Infect 71:615–626. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Enright MC, Spratt BG. 1999. Multilocus sequence typing. Trends Microbiol 7:482–487. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95:3140–3145. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52:1358–1365. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clermont O, Christenson JK, Daubié AS, Gordon DM, Denamur E. 2014. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J Microbiol Methods 101:24–27. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppé E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother 64:274–277. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Doumith M, Day M, Ciesielczuk H, Hope R, Underwood A, Reynolds R, Wain J, Livermore DM, Woodford N. 2015. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J Clin Microbiol 53:160–166. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson JR, Owens K, Sabate M, Prats G, Prats G. 2004. Rapid and specific detection of the O15:K52:H1 clonal group of Escherichia coli by gene-specific PCR. J Clin Microbiol 42:3841–3843. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson JR, Owens K, Manges AR, Riley LW. 2004. Rapid and specific detection of Escherichia coli clonal group A by gene-specific PCR. J Clin Microbiol 42:2618–2622. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson JR, Johnston BD, Gordon DM. Rapid and specific detection of the Escherichia coli sequence type 648 complex (STc648) within phylogroup F. J Clin Microbiol 55:1116–1121. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson JR, Porter S, Thuras P, Castanheira M. 2017. The pandemic H30 subclone of sequence type 131 (ST131) is the leading cause of multidrug-resistant Escherichia coli infections in the United States (2011–2012). Open Forum Infect Dis 4:ofx089. 10.1093/ofid/ofx089. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gordon DM, Geyik S, Clermont O, O’Brien CL, Huang S, Abayasekara C, Rajesh A, Kennedy K, Collignon P, Pavli P, Rodriguez C, Johnston BD, Johnson JR, Decousser J-W, Denamur E. 2017. Fine-scale structure analysis shows epidemic patterns of clonal complex 95, a cosmopolitan Escherichia coli lineage responsible for extraintestinal infection. MSphere 2:e00168-17. 10.1128/mSphere.00168-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsumura Y, Pitout J, Peirano G, DeVinney R, Noguchi T, Yamamoto M, Gomi R, Matsuda T, Nakano S, Nagao M, Tanaka M, Ichiyama S. Rapid identification of different Escherichia coli ST131 clades. Antimicrob Agents Chemother 61:e00179-17. 10.1128/AAC.00179-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsumura Y, Pitout JD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradford PA, Motyl MR, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bingen E, Bonacorsi S, Brahimi N, Denamur E, Elion J. 1997. Virulence patterns of Escherichia coli K1 strains associated with neonatal meningitis. J Clin Microbiol 35:2981–2982. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johanson I-M, Plos K, Marklund B-I, Svanborg C. 1993. Pap, papG and prsG DNA sequences in Escherichia coli from the fecal flora and the urinary tract. Microb Pathog 15:121–129. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Mitsumori K, Terai A, Yamamoto S, Ishitoya S, Yoshida O. 1999. Virulence characteristics of Escherichia coli in acute bacterial prostatitis. J Infect Dis 180:1378–1381. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Sannes MR, Kuskowski MA, Owens K, Gajewski A, Johnson JR. 2004. Virulence factor profiles and phylogenetic background of Escherichia coli isolates from veterans with bacteremia and uninfected control subjects. J Infect Dis 190:2121–2128. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin Microbiol Infect 19:E173–E180. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr Infect Dis J 32:543–548. [PubMed] [DOI] [PubMed] [Google Scholar]
- 89.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA 93:9827–9832. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marklund BI, Tennent JM, Garcia E, Hamers A, Båga M, Lindberg F, Gaastra W, Normark S. 1992. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol 6:2225–2242. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Mitsumori K, Terai A, Yamamoto S, Yoshida O. 1998. Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol Med Microbiol 21:261–268. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Nowicki B, Labigne A, Moseley S, Hull R, Hull S, Moulds J. 1990. The Dr hemagglutinin, afimbrial adhesins AFA-I and AFA-III, and F1845 fimbriae of uropathogenic and diarrhea-associated Escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect Immun 58:279–281. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Opal SM, Cross AS, Gemski P, Lyhte LW. 1990. Aerobactin and α-hemolysin as virulence determinants in Escherichia coli isolated from human blood, urine, and stool. J Infect Dis 161:794–796. [PubMed] [DOI] [PubMed] [Google Scholar]
- 94.Schubert S, Cuenca S, Fischer D, Heesemann J. 2000. High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J Infect Dis 182:1268–1271. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Barnard TJ, Johnson JR. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun 70:7156–7160 . [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HLT, Tarr PI. 2005. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect Immun 73:965–971. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69:6179–6185. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rippere-Lampe KE, Lang M, Ceri H, Olson M, Lockman HA, O’Brien AD. 2001. Cytotoxic necrotizing factor type 1-positive Escherichia coli causes increased inflammation and tissue damage to the prostate in a rat prostatitis model. Infect Immun 69:6515–6519. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scott DA, Kaper JB. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect Immun 62:244–251. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Welch RA, Dellinger EP, Minshew B, Falkow S. 1981. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature 294:665–667. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Yamamoto T, Echeverria P. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect Immun 64:1441–1445. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jann K, Jann B. 1992. Capsules of Escherichia coli, expression and biological significance. Can J Microbiol 38:705–710. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Russo TA, Davidson BA, Carlino-MacDonald UB, Helinski JD, Priore RL, Knight PR III, McFadden CD, Beanan JM, Barnard TJ, Johnson JR, Carlino UB, Jodush ST, Brown JJ, Liang Y, Cross AS. 2003. The effects of Escherichia coli capsule, O-antigen, host neutrophils, and complement in a rat model of Gram-negative pneumonia. FEMS Microbiol Lett 226:355–361. [DOI] [PubMed] [Google Scholar]
- 104.Kusecek B, Wloch H, Mercer A, Vaisänen V, Pluschke G, Korhonen T, Achtman M. 1984. Lipopolysaccharide, capsule, and fimbriae as virulence factors among O1, O7, O16, O18, or O75 and K1, K5, or K100 Escherichia coli. Infect Immun 43:368–379. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cross AS, Kim KS, Wright DC, Sadoff JC, Gemski P. 1986. Role of lipopolysaccharide and capsule in the serum resistance of bacteremic strains of Escherichia coli. J Infect Dis 154:497–503. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.He XL, Wang Q, Peng L, Qu Y-R, Puthiyakunnon S, Liu X-L, Hui C-Y, Boddu S, Cao H, Huang S-H. 2015. Role of uropathogenic Escherichia coli outer membrane protein T in pathogenesis of urinary tract infection. Pathog Dis 73:ftv006. 10.1093/femspd/ftv006. [DOI] [PubMed] [Google Scholar]
- 107.Huang S-H, Wan Z-S, Chen Y-H, Jong AY, Kim KS. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J Infect Dis 183:1071–1078. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Kanukollu U, Bieler S, Hull S, Hull R. 1985. Contribution of the traT gene to serum resistance among clinical isolates of Enterobacteriaceae. J Med Microbiol 19:61–67. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Johnson TJ, Giddings CW, Horne SM, Gibbs PS, Wooley RE, Skyberg J, Olah P, Kercher R, Sherwood JS, Foley SL, Nolan LK. 2002. Location of increased serum survival gene and selected virulence traits on a conjugative R plasmid in an avian Escherichia coli isolate. Avian Dis 46:342–352. [DOI] [PubMed] [Google Scholar]
- 110.Johnson JR, O’Bryan TT, Kuskowski M, Maslow JN. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect Immun 69:5363–5374. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnson JR, Oswald E, O’Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J Infect Dis 185:774–784. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Kanamaru S, Kurazono H, Ishitoya S, Terai A, Habuchi T, Nakano M, Ogawa O, Yamamoto S. 2003. Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J Urol 170:2490–2493. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Dobrindt U, Blum-Oehler G, Hartsch T, Gottschalk G, Ron EZ, Fünfstück R, Hacker J. 2001. S-Fimbria-encoding determinant sfa(I) is located on pathogenicity island III(536) of uropathogenic Escherichia coli strain 536. Infect Immun 69:4248–4256. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guyer DM, Kao J-S, Mobley HLT. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and Catheter-associated bacteriuria and from fecal samples. Infect Immun 66:4411–4417. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Le Bouguenec C, Garcia MI, Ouin V, Desperrier J-M, Gounon P, Labigne A. 1993. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun 61:5106–5114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Low D, David V, Lark D, Schoolnik G, Falkow S. 1984. Gene clusters governing the production of hemolysin and mannose-resistant hemagglutination are closely linked in Escherichia coli serotype O4 and O6 isolates from urinary tract infections. Infect Immun 43:353–358. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Swenson DL, Bukanov NO, Berg DE, Welch RA. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun 64:3736–3743. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Valvano MA, Silver RP, Crosa JH. 1986. Occurrence of chromosome- or plasmid-mediated aerobactin iron transport systems and hemolysin production among clonal groups of human invasive strains of Escherichia coli K1. Infect Immun 52:192–199. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun 70:6365–6372. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun 66:480–485. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Binns MM, Mayden J, Levine RP. 1982. Further characterization of complement resistance conferred on Escherichia coli by the plasmid genes traT of R100 and iss of ColV,I-K94. Infect Immun 35:654–659. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bloch CA, Rode CK. 1996. Pathogenicity island evaluation in Escherichia coli K1 by crossing with laboratory strain K-12. Infect Immun 64:3218–3223. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Welch RA, Burland V, Plunkett G III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA 99:17020–17024. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Orskov I, Orskov F, Birch-Andersen A, Kanamori M, Svanborg-Edén C. 1982. O, K, H and fimbrial antigens in Escherichia coli serotypes associated with pyelonephritis and cystitis. Scand J Infect Dis Suppl 33:18–25. [PubMed] [PubMed] [Google Scholar]
- 127.Orskov F, Orskov I. 1983. From the national institutes of health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis 148:346–357. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Johnson JR, Delavari P, O’Bryan TT. 2001. Escherichia coli O18:K1:H7 isolates from patients with acute cystitis and neonatal meningitis exhibit common phylogenetic origins and virulence factor profiles. J Infect Dis 183:425–434. [PubMed] [DOI] [PubMed] [Google Scholar]
- 129.Picard B, Journet-Mancy C, Picard-Pasquier N, Goullet P. 1993. Genetic structures of the B2 and B1 Escherichia coli strains responsible for extra-intestinal infections. J Gen Microbiol 139:3079–3088. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Johnson JR, Delavari P, Kuskowski M, Stell AL. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183:78–88. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Bingen-Bidois M, Clermont O, Bonacorsi S, Terki M, Brahimi N, Loukil C, Barraud D, Bingen E. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect Immun 70:3216–3226. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Johnson JR, Kaster N, Kuskowski MA, Ling GV. 2003. Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. J Clin Microbiol 41:337–345. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Plos K, Connell H, Jodal U, Marklund BI, Mårild S, Wettergren B, Svanborg C. 1995. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. J Infect Dis 171:625–631. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol 46:2529–2534. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Johnson JR, Kuskowski M, Denamur E, Elion J, Picard B. 2000. Clonal origin, virulence factors, and virulence. Infect Immun 68:424–425. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blanco M, Blanco JE, Alonso MP, Blanco J. 1996. Virulence factors and O groups of Escherichia coli isolates from patients with acute pyelonephritis, cystitis and asymptomatic bacteriuria. Eur J Epidemiol 12:191–198. [PubMed] [DOI] [PubMed] [Google Scholar]
- 138.Ruiz J, Simon K, Horcajada JP, Velasco M, Barranco M, Roig G, Moreno-Martínez A, Martínez JA, Jiménez de Anta T, Mensa J, Vila J. 2002. Differences in virulence factors among clinical isolates of Escherichia coli causing cystitis and pyelonephritis in women and prostatitis in men. J Clin Microbiol 40:4445–4449. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johnson JR, Kuskowski MA, O’Bryan TT, Maslow JN. 2002. Epidemiological correlates of virulence genotype and phylogenetic background among Escherichia coli blood isolates from adults with diverse-source bacteremia. J Infect Dis 185:1439–1447. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Maslow JN, Mulligan ME, Adams KS, Justis JC, Arbeit RD. 1993. Bacterial adhesins and host factors: role in the development and outcome of Escherichia coli bacteremia. Clin Infect Dis 17:89–97. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Drekonja DM, Kuskowski MA, Anway R, Johnston BD, Johnson JR. 2016. The niche for Escherichia coli sequence type 131 among veterans: urinary tract abnormalities and long-term care facilities. Open Forum Infect Dis 3:ofw138. 10.1093/ofid/ofw138. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnson JR, Thuras P, Johnston BD, Weissman SJ, Limaye AP, Riddell K, Scholes D, Tchesnokova V, Sokurenko E. 2016. The pandemic H30 subclone of Escherichia coli sequence type 131 is associated with persistent infections and adverse outcomes independent from its multidrug resistance and associations with compromised hosts. Clin Infect Dis 62:1529–1536. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Johnson JR, Stell AL, Delavari P, Murray AC, Kuskowski M, Gaastra W. 2001. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J Infect Dis 183:897–906. [PubMed] [DOI] [PubMed] [Google Scholar]
- 144.Johnson JR, Johnston B, Clabots CR, Kuskowski MA, Roberts E, DebRoy C. 2008. Virulence genotypes and phylogenetic background of Escherichia coli serogroup O6 isolates from humans, dogs, and cats. J Clin Microbiol 46:417–422. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother 55:3782–3787. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo S, Wakeham D, Brouwers H, Cobbold RN, Abraham S, Platell J, Johnson JR, Chapman T, Gordon D, Barrs V, Trott D. 2015. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including O1-ST38/ST648, ST354, O7-ST457, and O15:K52:H1-ST393, isolated from canine faeces and extraintestinal infections in Australia. Microbes Infect 17:266–274. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, Kim KS, Spanjaard L, Nolan LK. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol 74:7043–7050. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Caugant DA, Levin BR, Selander RK. 1984. Distribution of multilocus genotypes of Escherichia coli within and between host families. J Hyg (Lond) 92:377–384. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Majd M, Rushton HG, Jantausch B, Wiedermann BL. 1991. Relationship among vesicoureteral reflux, P-fimbriated Escherichia coli, and acute pyelonephritis in children with febrile urinary tract infection. J Pediatr 119:578–585. [DOI] [PubMed] [Google Scholar]
- 150.Hull RA, Hull SI, Falkow S. 1984. Frequency of gene sequences necessary for pyelonephritis-associated pili expression among isolates of Enterobacteriaceae from human extraintestinal infections. Infect Immun 43:1064–1067. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.O’Hanley P, Low D, Romero I, Lark D, Vosti K, Falkow S, Schoolnik G. 1985. Gal-Gal binding and hemolysin phenotypes and genotypes associated with uropathogenic Escherichia coli. N Engl J Med 313:414–420. [PubMed] [DOI] [PubMed] [Google Scholar]
- 152.Dozois CM, Harel J, Fairbrother JM. 1996. P-fimbriae-producing septicaemic Escherichia coli from poultry possess fel-related gene clusters whereas pap-hybridizing P-fimbriae-negative strains have partial or divergent P fimbrial gene clusters. Microbiology 142:2759–2766. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun 39:315–335. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kunin CM, Hua TH, Krishnan C, Van Arsdale White L, Hacker J. 1993. Isolation of a nicotinamide-requiring clone of Escherichia coli O18:K1:H7 from women with acute cystitis: resemblance to strains found in neonatal meningitis. Clin Infect Dis 16:412–416. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Gunther NW IV, Lockatell V, Johnson DE, Mobley HL. 2001. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect Immun 69:2838–2846. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lim JK, Gunther NW IV, Zhao H, Johnson DE, Keay SK, Mobley HL. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect Immun 66:3303–3310. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Breland EJ, Eberly AR, Hadjifrangiskou M. 2017. An overview of two-component signal transduction systems implicated in extra-intestinal pathogenic E. coli infections. Front Cell Infect Microbiol 7:162. 10.3389/fcimb.2017.00162. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sokurenko EV, Chesnokova V, Doyle RJ, Hasty DL. 1997. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J Biol Chem 272:17880–17886. [PubMed] [DOI] [PubMed] [Google Scholar]
- 159.Pouttu R, Puustinen T, Virkola R, Hacker J, Klemm P, Korhonen TK. 1999. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol Microbiol 31:1747–1757. [PubMed] [DOI] [PubMed] [Google Scholar]
- 160.Johnson JR, Weissman SJ, Stell AL, Trintchina E, Dykhuizen DE, Sokurenko EV. 2001. Clonal and pathotypic analysis of archetypal Escherichia coli cystitis isolate NU14. J Infect Dis 184:1556–1565. [PubMed] [DOI] [PubMed] [Google Scholar]
- 161.Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu X-R, Krogfelt KA, Struve C, Schembri MA, Hasty DL. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA 95:8922–8926. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jacobson SH, Kühn I, Brauner A. 1992. Biochemical fingerprinting of urinary Escherichia coli causing recurrent infections in women with pyelonephritic renal scarring. Scand J Urol Nephrol 26:373–377. [PubMed] [DOI] [PubMed] [Google Scholar]
- 163.Johnson JR, Brown JJ. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(α 1-4)Gal-binding PapG adhesins of Escherichia coli. J Infect Dis 173:920–926. [DOI] [PubMed] [Google Scholar]
- 164.Karkkainen U-M, Kauppinin J, Ikaheimo R, Katila M-L, Siitonen A. 1998. Rapid and specific detection of three different G adhesin classes of P-fimbriae in uropathogenic Escherichia coli by polymerase chain reaction. J Microbiol Methods 34:23–29. [Google Scholar]
- 165.Senior D, Baker N, Cedergren B, Falk P, Larson G, Lindstedt R, Edén CS. 1988. Globo-A--a new receptor specificity for attaching Escherichia coli. FEBS Lett 237:123–127. [DOI] [PubMed] [Google Scholar]
- 166.Stapleton AE, Stroud MR, Hakomori SI, Stamm WE. 1998. The globoseries glycosphingolipid sialosyl galactosyl globoside is found in urinary tract tissues and is a preferred binding receptor In vitro for uropathogenic Escherichia coli expressing pap-encoded adhesins. Infect Immun 66:3856–3861. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stroud MR, Stapleton AE, Levery SB. 1998. The P histo-blood group-related glycosphingolipid sialosyl galactosyl globoside as a preferred binding receptor for uropathogenic Escherichia coli: isolation and structural characterization from human kidney. Biochemistry 37:17420–17428. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Strömberg N, Marklund BI, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson KA, Normark S. 1990. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Gal α 1-4Gal-containing isoreceptors. EMBO J 9:2001–2010. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jantunen ME, Siitonen A, Koskimies O, Wikström S, Kärkkäinen U, Salo E, Saxén H. 2000. Predominance of class II papG allele of Escherichia coli in pyelonephritis in infants with normal urinary tract anatomy. J Infect Dis 181:1822–1824. [PubMed] [DOI] [PubMed] [Google Scholar]
- 170.Johnson JR, Brown JJ, Ahmed P. 1998. Diversity of hemagglutination phenotypes among P-fimbriated wild-type strains of Escherichia coli in relation to papG allele repertoire. Clin Diagn Lab Immunol 5:160–170. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Johnson JR, Russo TA, Brown JJ, Stapleton A. 1998. papG alleles of Escherichia coli strains causing first-episode or recurrent acute cystitis in adult women. J Infect Dis 177:97–101. [PubMed] [DOI] [PubMed] [Google Scholar]
- 172.Otto G, Sandberg T, Marklund BI, Ulleryd P, Svanborg C. 1993. Virulence factors and pap genotype in Escherichia coli isolates from women with acute pyelonephritis, with or without bacteremia. Clin Infect Dis 17:448–456. [PubMed] [DOI] [PubMed] [Google Scholar]
- 173.Senior DF, deMan P, Svanborg C. 1992. Serotype, hemolysin production, and adherence characteristics of strains of Escherichia coli causing urinary tract infection in dogs. Am J Vet Res 53:494–498. [PubMed] [PubMed] [Google Scholar]
- 174.Ott M, Hoschützky H, Jann K, Van Die I, Hacker J. 1988. Gene clusters for S fimbrial adhesin (sfa) and F1C fimbriae (foc) of Escherichia coli: comparative aspects of structure and function. J Bacteriol 170:3983–3990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Le Bouguénec C, Lalioui L, du Merle L, Jouve M, Courcoux P, Bouzari S, Selvarangan R, Nowicki BJ, Germani Y, Andremont A, Gounon P, Garcia MI. 2001. Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coli isolates: PCR assays for detection of Afa adhesins that do or do not recognize Dr blood group antigens. J Clin Microbiol 39:1738–1745. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang L, Foxman B, Tallman P, Cladera E, Le Bouguenec C, Marrs CF. 1997. Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coli isolates and identification of new subtypes. Infect Immun 65:2011–2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Roberts I, Mountford R, High N, Bitter-Suermann D, Jann K, Timmis K, Boulnois G. 1986. Molecular cloning and analysis of genes for production of K5, K7, K12, and K92 capsular polysaccharides in Escherichia coli. J Bacteriol 168:1228–1233. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Russo TA, Wenderoth S, Carlino UB, Merrick JM, Lesse AJ. 1998. Identification, genomic organization, and analysis of the group III capsular polysaccharide genes kpsD, kpsM, kpsT, and kpsE from an extraintestinal isolate of Escherichia coli (CP9, O4/K54/H5). J Bacteriol 180:338–349. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Blanco J, Blanco M, Alonso MP, Blanco JE, Garabal JI, González EA. 1992. Serogroups of Escherichia coli strains producing cytotoxic necrotizing factors CNF1 and CNF2. FEMS Microbiol Lett 75:155–159. [PubMed] [DOI] [PubMed] [Google Scholar]
- 180.Johnson JR, Stell AL, Delavari P. 2001. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect Immun 69:1306–1314. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Johnson JR, Stell AL, Scheutz F, O’Bryan TT, Russo TA, Carlino UB, Fasching C, Kavle J, Van Dijk L, Gaastra W. 2000. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun 68:1587–1599. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Garcia E, Bergmans HEN, Van den Bosch JF, Orskov I, Van der Zeijst BAM, Gaastra W. 1988. Isolation and characterisation of dog uropathogenic Escherichia coli strains and their fimbriae. Antonie van Leeuwenhoek 54:149–163. [PubMed] [DOI] [PubMed] [Google Scholar]
- 183.de Man P, Cläeson I, Johanson IM, Jodal U, Svanborg Edén C. 1989. Bacterial attachment as a predictor of renal abnormalities in boys with urinary tract infection. J Pediatr 115:915–922. [DOI] [PubMed] [Google Scholar]
- 184.Dowling KJ, Roberts JA, Kaack MB. 1987. P-fimbriated Escherichia coli urinary tract infection: a clinical correlation. South Med J 80:1533–1536. [PubMed] [DOI] [PubMed] [Google Scholar]
- 185.Roberts JA. 1986. Pyelonephritis, cortical abscess, and perinephric abscess. Urol Clin North Am 13:637–645. [PubMed] [PubMed] [Google Scholar]
- 186.Tambic T, Oberiter V, Delmis J, Tambic A. 1992. Diagnostic value of a P-fimbriation test in determining duration of therapy in children with urinary tract infections. Clin Ther 14:667–671. [PubMed] [PubMed] [Google Scholar]
- 187.Stenqvist K, Lidin-Janson G, Sandberg T, Edén CS. 1989. Bacterial adhesion as an indicator of renal involvement in bacteriuria of pregnancy. Scand J Infect Dis 21:193–199. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Johnson JR. 1995. Epidemiological Considerations in Studies of Adherence, vol 253. Academic Press, Orlando, FL. [PubMed] [Google Scholar]
- 189.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE, Infectious Diseases Society of America, European Society for Microbiology and Infectious Diseases. 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52:e103–e120. [PubMed] [DOI] [PubMed] [Google Scholar]
- 190.Talan DA, Takhar SS, Krishnadasan A, Abrahamian FM, Mower WR, Moran GJ, EMERGEncy ID Net Study Group. 2016. Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli infections in patients with pyelonephritis, United States. Emerg Infect Dis 22:1594–1603. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Johnson JR, Goullet P, Picard B, Moseley SL, Roberts PL, Stamm WE. 1991. Association of carboxylesterase B electrophoretic pattern with presence and expression of urovirulence factor determinants and antimicrobial resistance among strains of Escherichia coli that cause urosepsis. Infect Immun 59:2311–2315. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Johnson JR, Kuskowski MA, Owens K, Gajewski A, Winokur PL. 2003. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J Infect Dis 188:759–768. [PubMed] [DOI] [PubMed] [Google Scholar]
- 193.Johnson JR, van der Schee C, Kuskowski MA, Goessens W, van Belkum A. 2002. Phylogenetic background and virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from the Netherlands. J Infect Dis 186:1852–1856. [PubMed] [DOI] [PubMed] [Google Scholar]
- 194.Velasco M, Horcajada JP, Mensa J, Moreno-Martinez A, Vila J, Martinez JA, Ruiz J, Barranco M, Roig G, Soriano E. 2001. Decreased invasive capacity of quinolone-resistant Escherichia coli in patients with urinary tract infections. Clin Infect Dis 33:1682–1686. [PubMed] [DOI] [PubMed] [Google Scholar]
- 195.Vila J, Simon K, Ruiz J, Horcajada JP, Velasco M, Barranco M, Moreno A, Mensa J. 2002. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J Infect Dis 186:1039–1042. [PubMed] [DOI] [PubMed] [Google Scholar]
- 196.Johnson JR, Porter S, Thuras P, Castanheira M. 2017. Epidemic emergence in the United States of Escherichia coli sequence type 131-H30, 2000-2009. Antimicrob Agents Chemother 61:e00732-17. 10.1128/AAC.00732-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xia X, Meng J, Zhao S, Bodeis-Jones S, Gaines SA, Ayers SL, McDermott PF. 2011. Identification and antimicrobial resistance of extraintestinal pathogenic Escherichia coli from retail meats. J Food Prot 74:38–44. [PubMed] [DOI] [PubMed] [Google Scholar]
- 198.Johnson JR, McCabe JS, White DG, Johnston B, Kuskowski MA, McDermott P. 2009. Molecular Analysis of Escherichia coli from retail meats (2002–2004) from the United States National Antimicrobial Resistance Monitoring System. Clin Infect Dis 49:195–201. [PubMed] [DOI] [PubMed] [Google Scholar]
- 199.Johnson JR, Delavari P, O’Bryan TT, Smith KE, Tatini S. 2005. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis 2:38–49. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Johnson JR, Kuskowski MA, Smith K, O’Bryan TT, Tatini S. 2005. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis 191:1040–1049. [PubMed] [DOI] [PubMed] [Google Scholar]
- 201.Johnson JR, Sannes MR, Croy C, Johnston B, Clabots C, Kuskowski MA, Bender J, Smith KE, Winokur PL, Belongia EA. 2007. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002–2004. Emerg Infect Dis 13:838–846. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Johnson JR, Porter SB, Johnston B, Thuras P, Clock S, Crupain M, Rangan U. 2017. Extraintestinal pathogenic and antimicrobial resistant Escherichia coli, including sequence type 131 (ST131), from retail chicken breasts: United States, 2013. Appl Environ Microbiol 83:e02956-16. 10.1128/AEM.02956-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hannah EL, Angulo FJ, Johnson JR, Haddadin B, Williamson J, Samore MH. 2005. Drug-resistant E. coli, rural Idaho. Emerg Infect Dis 11:1614–1617. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nowrouzian F, Adlerberth I, Wold AE. 2001. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol Infect 126:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nowrouzian F, Hesselmar B, Saalman R, Strannegard I-L, Aberg N, Wold AE, Adlerberth I. 2003. Escherichia coli in infants’ intestinal microflora: colonization rate, strain turnover, and virulence gene carriage. Pediatr Res 54:8–14. [PubMed] [DOI] [PubMed] [Google Scholar]
- 207.Wold AE, Caugant DA, Lidin-Janson G, de Man P, Svanborg C. 1992. Resident colonic Escherichia coli strains frequently display uropathogenic characteristics. J Infect Dis 165:46–52. [PubMed] [DOI] [PubMed] [Google Scholar]
- 208.Wold AE, Thorssén M, Hull S, Edén CS. 1988. Attachment of Escherichia coli via mannose- or Gal α 1----4Gal β-containing receptors to human colonic epithelial cells. Infect Immun 56:2531–2537. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Adlerberth I, Hanson LA, Svanborg C, Svennerholm A-M, Nordgren S, Wold AE. 1995. Adhesins of Escherichia coli associated with extra-intestinal pathogenicity confer binding to colonic epithelial cells. Microb Pathog 18:373–385. [PubMed] [DOI] [PubMed] [Google Scholar]
- 210.Herías MV, Midtvedt T, Hanson LÅ, Wold AE. 1995. Role of Escherichia coli P fimbriae in intestinal colonization in gnotobiotic rats. Infect Immun 63:4781–4789. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hérias MV, Midtvedt T, Hanson LA, Wold AE. 1997. Escherichia coli K5 capsule expression enhances colonization of the large intestine in the gnotobiotic rat. Infect Immun 65:531–536. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mahmood A, Engle MJ, Hultgren SJ, Goetz GS, Dodson K.Alpers DH. 2000. Role of intestinal surfactant-like particles as a potential reservoir of uropathogenic Escherichia coli. Biochim Biophys Acta 1523:49–55. [PubMed] [DOI] [PubMed] [Google Scholar]
- 213.Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, Tenaillon O. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol Biol Evol 24:2373–2384. [PubMed] [DOI] [PubMed] [Google Scholar]
- 214.Hacker J, Carniel E. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep 2:376–381. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Diard M, Garry L, Selva M, Mosser T, Denamur E, Matic I. 2010. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J Bacteriol 192:4885–4893. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Siitonen A. 1992. Escherichia coli in fecal flora of healthy adults: serotypes, P and type 1C fimbriae, non-P mannose-resistant adhesins, and hemolytic activity. J Infect Dis 166:1058–1065. [PubMed] [DOI] [PubMed] [Google Scholar]
- 217.Grüneberg RN. 1969. Relationship of infecting urinary organism to the faecal flora in patients with symptomatic urinary infection. Lancet 2:766–768. [DOI] [PubMed] [Google Scholar]
- 218.Terai A, Ishitoya S, Mitsumori K, Ogawa O. 2000. Molecular epidemiological evidence for ascending urethral infection in acute bacterial prostatitis. J Urol 164:1945–1947. [PubMed] [Google Scholar]
- 219.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 157:1127–1129. [PubMed] [Google Scholar]
- 220.Czaja CA, Stamm WE, Stapleton AE, Roberts PL, Hawn TR, Scholes D, Samadpour M, Hultgren SJ, Hooton TM. 2009. Prospective cohort study of microbial and inflammatory events immediately preceding Escherichia coli recurrent urinary tract infection in women. J Infect Dis 200:528–536. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ikäheimo R, Siitonen A, Heiskanen T, Kärkkäinen U, Kuosmanen P, Lipponen P, Mäkelä PH. 1996. Recurrence of urinary tract infection in a primary care setting: analysis of a 1-year follow-up of 179 women. Clin Infect Dis 22:91–99. [PubMed] [DOI] [PubMed] [Google Scholar]
- 222.Kärkkäinen UM, Ikäheimo R, Katila ML, Siitonen A, Ulla-Maija Kärkkäinen, Risto Ikähei. 2000. Recurrence of urinary tract infections in adult patients with community-acquired pyelonephritis caused by E. coli: a 1-year follow-up. Scand J Infect Dis 32:495–499. [PubMed] [DOI] [PubMed] [Google Scholar]
- 223.Jantunen ME, Saxén H, Salo E, Siitonen A. 2002. Recurrent urinary tract infections in infancy: relapses or reinfections? J Infect Dis 185:375–379. [PubMed] [DOI] [PubMed] [Google Scholar]
- 224.Ulleryd P, Sandberg T, Scheutz F, Clabots C, Johnston BD, Thuras P, Johnson JR. 2015. Colonization with Escherichia coli strains among female sex partners of men with febrile urinary tract infection. J Clin Microbiol 53:1947–1950. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Brauner A, Jacobson SH, Kühn I. 1992. Urinary Escherichia coli causing recurrent infections--a prospective follow-up of biochemical phenotypes. Clin Nephrol 38:318–323. [PubMed] [PubMed] [Google Scholar]
- 226.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. 2007. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med 4:e329. 10.1371/journal.pmed.0040329 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bailey RR, Peddie BA, Swainson CP, Kirkpatrick D. 1986. Sexual acquisition of urinary tract infection in a man. Nephron 44:217–218. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.Foxman B, Zhang L, Tallman P, Andree BC, Geiger AM, Koopman JS, Gillespie BW, Palin KA, Sobel JD, Rode CK, Bloch CA, Marrs CF. 1997. Transmission of uropathogens between sex partners. J Infect Dis 175:989–992. [PubMed] [DOI] [PubMed] [Google Scholar]
- 230.Johnson JR, Brown JJ, Carlino UB, Russo TA. 1998. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J Infect Dis 177:1120–1124. [PubMed] [DOI] [PubMed] [Google Scholar]
- 231.Johnson JR, Delavari P. 2002. Concurrent fecal colonization with extraintestinal pathogenic Escherichia coli in a homosexual man with recurrent urinary tract infection and in his male sex partner. Clin Infect Dis 35:E65–E68. [PubMed] [DOI] [PubMed] [Google Scholar]
- 232.Wong ES, Stamm WE. 1983. Sexual acquisition of urinary tract infection in a man. JAMA 250:3087–3088. [PubMed] [PubMed] [Google Scholar]
- 233.Johnson JR, Clabots C, Kuskowski MA. 2008. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. J Clin Microbiol 46:4078–4082. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Johnson JR, Owens K, Gajewski A, Clabots C. 2008. Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. J Infect Dis 197:218–224. [PubMed] [DOI] [PubMed] [Google Scholar]
- 235.Johnson JR, Miller S, Johnston B, Clabots C, Debroy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and Urovirulent E. coli strains among dogs and cats within a household. J Clin Microbiol 47:3721–3725. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Johnson JR, Anderson JT, Clabots C, Johnston B, Cooperstock M. 2010. Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr Infect Dis J 29:473–475. [PubMed] [DOI] [PubMed] [Google Scholar]
- 237.Madigan T, Johnson JR, Clabots C, Johnston BD, Porter SB, Slater BS, Banerjee R. 2015. Extensive household outbreak of urinary tract infection and intestinal colonization due to extended-spectrum beta lactamase (ESBL)-producing Escherichia coli sequence type 131 (ST131). Clin Infect Dis 61:e5–e12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kenny JF, Medearis DN Jr, Klein SW, Drachman RH, Gibson LE. 1966. An outbreak of urinary tract infections and septicemia due to Escherichia coli in male infants. J Pediatr 68:530–541. [Google Scholar]
- 239.Sweet AY, Wolinsky E. 1964. An outbreak of urinary tract and other infections due to E. coli. Pediatrics 33:865–871. [PubMed] [PubMed] [Google Scholar]
- 240.Tullus K, Hörlin K, Svenson SB, Källenius G. 1984. Epidemic outbreaks of acute pyelonephritis caused by nosocomial spread of P fimbriated Escherichia coli in children. J Infect Dis 150:728–736. [PubMed] [DOI] [PubMed] [Google Scholar]
- 241.Foxman B, Manning SD, Tallman P, Bauer R, Zhang L, Koopman JS, Gillespie B, Sobel JD, Marrs CF. 2002. Uropathogenic Escherichia coli are more likely than commensal E. coli to be shared between heterosexual sex partners. Am J Epidemiol 156:1133–1140. [PubMed] [DOI] [PubMed] [Google Scholar]
- 242.Murray AC, Kuskowski MA, Johnson JR. 2004. Virulence factors predict Escherichia coli colonization patterns among human and animal household members. Ann Intern Med 140:848–849. [PubMed] [DOI] [PubMed] [Google Scholar]
- 243.Stamey TA, Sexton CC. 1975. The role of vaginal colonization with enterobacteriaceae in recurrent urinary infections. J Urol 113:214–217. [DOI] [PubMed] [Google Scholar]
- 244.Ender PT, Gajanana D, Johnston B, Clabots C, Tamarkin FJ, Johnson JR. 2009. Transmission of an extended-spectrum-beta-lactamase-producing Escherichia coli (sequence type ST131) strain between a father and daughter resulting in septic shock and Emphysematous pyelonephritis. J Clin Microbiol 47:3780–3782. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Eykyn SJ, Phillips I. 1986. Community outbreak of multiresistant invasive Escherichia coli infection. Lancet 2:1454. 10.1016/S0140-6736(86)92756-X. [DOI] [PubMed] [Google Scholar]
- 246.O’Neill PM, Talboys CA, Roberts AP, Azadian BS. 1990. The rise and fall of Escherichia coli O15 in a London teaching hospital. J Med Microbiol 33:23–27. [PubMed] [DOI] [PubMed] [Google Scholar]
- 247.Phillips I, Eykyn S, King A, Gransden WR, Rowe B, Frost JA, Gross RJ. 1988. Epidemic multiresistant Escherichia coli infection in West Lambeth health district. Lancet 1:1038–1041. [DOI] [PubMed] [Google Scholar]
- 248.Johnson JR, Manges AR, O’Bryan TT, Riley LW. 2002. A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 359:2249–2251. [DOI] [PubMed] [Google Scholar]
- 249.Nicolas-Chanoine MH, Bertrand X, Madec J-Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. [PubMed] [DOI] [PubMed] [Google Scholar]
- 252.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-β-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 67:2612–2620. [PubMed] [DOI] [PubMed] [Google Scholar]
- 253.Clermont O, Glodt J, Burdet C, Pognard D, Lefort A, Branger C, Denamur E, COLIBAFI Group Members. 2013. Complexity of Escherichia coli bacteremia pathophysiology evidenced by comparison of isolates from blood and portal of entry within single patients. Int J Med Microbiol 303:529–532. [PubMed] [DOI] [PubMed] [Google Scholar]
- 254.Levert M, Zamfir O, Clermont O, Bouvet O, Lespinats S, Hipeaux MC, Branger C, Picard B, Saint-Ruf C, Norel F, Balliau T, Zivy M, Le Nagard H, Cruveiller S, Chane-Woon-Ming B, Nilsson S, Gudelj I, Phan K, Ferenci T, Tenaillon O, Denamur E. 2010. Molecular and evolutionary bases of within-patient genotypic and phenotypic diversity in Escherichia coli extraintestinal infections. PLoS Pathog 6:e1001125. 10.1371/journal.ppat.1001125. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.McNally A, Alhashash F, Collins M, Alqasim A, Paszckiewicz K, Weston V, Diggle M. 2013. Genomic analysis of extra-intestinal pathogenic Escherichia coli urosepsis. Clin Microbiol Infect 19:E328–E334. [PubMed] [DOI] [PubMed] [Google Scholar]
- 256.Johnson JR, Moseley SL, Coyle MB, Stamm WE. 1992. Success of DNA fingerprinting after failure of biotyping, antimicrobial susceptibility testing, and plasmid analysis to reveal clonality of multiple blood and urine isolates from a patient with Escherichia coli urosepsis. Diagn Microbiol Infect Dis 15:399–405. [PubMed] [DOI] [PubMed] [Google Scholar]
- 257.Kisiela DI, Radey M, Paul S, Porter S, Polukhina K, Tchesnokova V, Shevchenko S, Chan D, Aziz M, TJ J, Price LB, Johnson J, Sokurenko E. 2017. Inactivation of transcriptional regulators during within-household evolution of Escherichia coli. J Bacteriol 199:e00036-17. 10.1128/JB.00036-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Weissman SJ, Beskhlebnaya V, Chesnokova V, Chattopadhyay S, Stamm WE, Hooton TM, Sokurenko EV. 2007. Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect Immun 75:3548–3555. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, Clabots C, Lan R, Johnson JR, Wang L. 2011. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6:e26907. 10.1371/journal.pone.0026907. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Johnson JR, Scheutz F, Ulleryd P, Kuskowski MA, O’Bryan TT, Sandberg T. 2005. Phylogenetic and pathotypic comparison of concurrent urine and rectal Escherichia coli isolates from men with febrile urinary tract infection. J Clin Microbiol 43:3895–3900. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Johnson JR, Scheutz F, Ulleryd P, Kuskowski MA, O’Bryan TT, Sandberg T. 2005. Host-pathogen relationships among Escherichia coli isolates recovered from men with febrile urinary tract infection. Clin Infect Dis 40:813–822. [PubMed] [DOI] [PubMed] [Google Scholar]
- 262.Tchesnokova V, Billig M, Chattopadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova V, Johnston B, Gileva A, Igusheva I, Toland A, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. 2013. Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J Clin Microbiol 51:2991–2999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tchesnokova V, Avagyan H, Billig M, Chattopadhyay S, Aprikian P, Chan D, Pseunova J, Rechkina E, Riddell K, Scholes D, Fang FC, Johnson JR, Sokurenko EV. 2016. A novel 7-single nucleotide polymorphism-based clonotyping test allows rapid prediction of antimicrobial susceptibility of extraintestinal Escherichia coli directly from urine specimens. Open Forum Infect Dis 3:ofw002. 10.1093/ofid/ofw002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tchesnokova V, Avagyan H, Rechkina E, Chan D, Muradova M, Haile HG, Radey M, Weissman S, Riddell K, Scholes D, Johnson JR, Sokurenko EV. 2017. Bacterial clonal diagnostics as a tool for evidence-based empiric antibiotic selection. PLoS One 12:e0174132. 10.1371/journal.pone.0174132. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, Khan AM, Woodford N, Saunders NJ, Wain J, O’Grady J, Livermore DM. 2017. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 72:104–114. [PubMed] [DOI] [PubMed] [Google Scholar]
- 266.Nowicki B, Svanborg-Edén C, Hull R, Hull S. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect Immun 57:446–451. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Olesen B, Hansen DS, Nilsson F, Frimodt-Møller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol 51:1779–1785. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Clermont O, Bonacorsi S, Bingen E. 2001. The Yersinia high-pathogenicity island is highly predominant in virulence-associated phylogenetic groups of Escherichia coli. FEMS Microbiol Lett 196:153–157. [PubMed] [DOI] [PubMed] [Google Scholar]
- 269.Buckles EL, Bahrani-Mougeot FK, Molina A, Lockatell CV, Johnson DE, Drachenberg CB, Burland V, Blattner FR, Donnenberg MS. 2004. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect Immun 72:3890–3901. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Johnson JR, Kuskowski MA, Owens K, Soto S, Horcajada JP, Jimenez de Anta MT, Vila J. 2005. Extended virulence genotypes of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J Infect Dis 191:46–50. [PubMed] [DOI] [PubMed] [Google Scholar]
- 271.Bertin Y, Girardeau J-P, Darfeuille-Michaud A, Contrepois M. 1996. Characterization of 20K fimbria, a new adhesin of septicemic and diarrhea-associated Escherichia coli strains, that belongs to a family of adhesins with N-acetyl-D-glucosamine recognition. Infect Immun 64:332–342. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bian Z, Brauner A, Li Y, Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis 181:602–612. [PubMed] [DOI] [PubMed] [Google Scholar]
- 273.Saldaña Z, De la Cruz MA, Carrillo-Casas EM, Durán L, Zhang Y, Hernández-Castro R, Puente JL, Daaka Y, Girón JA. 2014. Production of the Escherichia coli common pilus by uropathogenic E. coli is associated with adherence to HeLa and HTB-4 cells and invasion of mouse bladder urothelium. PLoS One 9:e101200. 10.1371/journal.pone.0101200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mobley HLT, Chippendale GR, Tenney JH, Hull RA, Warren JW. 1987. Expression of type 1 fimbriae may be required for persistence of Escherichia coli in the catheterized urinary tract. J Clin Microbiol 25:2253–2257. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sokurenko EV, Courtney HS, Maslow J, Siitonen A, Hasty DL. 1995. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol 177:3680–3686. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner JS, Burlein J, Barren P, Koenig S, Leath S, Jones CH, Hultgren SJ. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276:607–611. [PubMed] [DOI] [PubMed] [Google Scholar]
- 277.Pere A, Leinonen M, Väisänen-Rhen V, Rhen M, Korhonen TK. 1985. Occurrence of type-1C fimbriae on Escherichia coli strains isolated from human extraintestinal infections. J Gen Microbiol 131:1705–1711. [DOI] [PubMed] [Google Scholar]
- 278.Goldhar J, Perry R, Golecki JR, Hoschutzky H, Jann B, Jann K. 1987. Nonfimbrial, mannose-resistant adhesins from uropathogenic Escherichia coli O83:K1:H4 and O14:K?:H11. Infect Immun 55:1837–1842. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Grünberg J, Perry R, Hoschützky H, Jann B, Jann K, Goldhar J. 1988. Nonfimbrial blood group N-specific adhesin (NFA-3) from Escherichia coli O20:KX104:H-, causing systemic infection. Fed Eur Microbiol Soc Microbiol Lett 56:241–246. [Google Scholar]
- 280.Hoschützky H, Nimmich W, Lottspeich F, Jann K. 1989. Isolation and characterization of the non-fimbrial adhesin NFA-4 from uropathogenic Escherichia coli O7:K98:H6. Microb Pathog 6:351–359. [DOI] [PubMed] [Google Scholar]
- 281.Roberts JA, Marklund B-I, Ilver D, Haslam D, Kaack MB, Baskin G, Louis M, Möllby R, Winberg J, Normark S. 1994. The Gal(α 1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc Natl Acad Sci USA 91:11889–11893. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mobley HLT, Jarvis KG, Elwood JP, Whittle DI, Lockatell CV, Russell RG, Johnson DE, Donnenberg MS, Warren JW. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol Microbiol 10:143–155. [PubMed] [DOI] [PubMed] [Google Scholar]
- 283.Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HL. 2011. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of ygi and yad fimbriae. Infect Immun 79:4753–4763. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HL. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. 10.1371/journal.ppat.1003788. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Marre R, Hacker J. 1987. Role of S- and common-type I-fimbriae of Escherichia coli in experimental upper and lower urinary tract infection. Microb Pathog 2:223–226. [DOI] [PubMed] [Google Scholar]
- 286.Schönian G, Sokolowska-Köhler W, Bollmann R, Schubert A, Gräser Y, Presber W. 1992. Determination of S fimbriae among Escherichia coli strains from extraintestinal infections by colony hybridization and dot enzyme immunoassay. Zentralbl Bakteriol 276:273–279. [DOI] [PubMed] [Google Scholar]
- 287.Janben T, Schwarz C, Preikschat P, Voss M, Philipp HC, Wieler LH. 2001. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int J Med Microbiol 291:371–378. [PubMed] [DOI] [PubMed] [Google Scholar]
- 288.Savarino SJ, Fasano A, Watson J, Martin BM, Levine MM, Guandalini S, Guerry P. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci USA 90:3093–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Caprioli A, Falbo V, Ruggeri FM, Baldassarri L, Bisicchia R, Ippolito G, Romoli E, Donelli G. 1987. Cytotoxic necrotizing factor production by hemolytic strains of Escherichia coli causing extraintestinal infections. J Clin Microbiol 25:146–149. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Khan NA, Wang Y, Kim KJ, Chung JW, Wass CA, Kim KS. 2002. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J Biol Chem 277:15607–15612. [PubMed] [DOI] [PubMed] [Google Scholar]
- 291.Rippere-Lampe KE, O’Brien AD, Conran R, Lockman HA. 2001. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf(1)) attenuates the virulence of uropathogenic Escherichia coli. Infect Immun 69:3954–3964. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Brooks HJL, O’Grady F, McSherry MA, Cattell WR. 1980. Uropathogenic properties of Escherichia coli in recurrent urinary-tract infection. J Med Microbiol 13:57–68. [PubMed] [DOI] [PubMed] [Google Scholar]
- 293.O’Hanley P, Lalonde G, Ji G. 1991. α-hemolysin contributes to the pathogenicity of piliated digalactoside-binding Escherichia coli in the kidney: efficacy of an α-hemolysin vaccine in preventing renal injury in the BALB/c mouse model of pyelonephritis. Infect Immun 59:1153–1161. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Heimer SR, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. 2004. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect Immun 72:593–597. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lloyd AL, Henderson TA, Vigil PD, Mobley HL. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol 191:3469–3481. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Guyer DM, Radulovic S, Jones F-E, Mobley HLT. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect Immun 70:4539–4546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Vigil PD, Stapleton AE, Johnson JR, Hooton TM, Hodges AP, He Y, Mobley HL. 2011. Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. MBio 2:e00066-11. 10.1128/mBio.00066-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Vigil PD, Wiles TJ, Engstrom MD, Prasov L, Mulvey MA, Mobley HL. 2012. The repeat-in-toxin family member TosA mediates adherence of uropathogenic Escherichia coli and survival during bacteremia. Infect Immun 80:493–505. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Restieri C, Garriss G, Locas MC, Dozois CM. 2007. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol 73:1553–1562. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Parham NJ, Pollard SJ, Desvaux M, Scott-Tucker A, Liu C, Fivian A, Henderson IR. 2005. Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol 43:4076–4082. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Russo TA, Jodush ST, Brown JJ, Johnson JR. 1996. Identification of two previously unrecognized genes (guaA and argC) important for uropathogenesis. Mol Microbiol 22:217–229. [PubMed] [DOI] [PubMed] [Google Scholar]
- 302.Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5:e1000448. 10.1371/journal.ppat.1000448. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Moritz RL, Welch RA. 2006. The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA. J Clin Microbiol 44:4038–4048. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hryckowian AJ, Baisa GA, Schwartz KJ, Welch RA. 2015. dsdA does not affect colonization of the murine urinary tract by Escherichia coli CFT073. PLoS One 10:e0138121. 10.1371/journal.pone.0138121. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect Immun 70:5335–5337. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Bauer RJ, Zhang L, Foxman B, Siitonen A, Jantunen ME, Saxen H, Marrs CF. 2002. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection-usp, iha, and iroN(E. coli). J Infect Dis 185:1521–1524. [PubMed] [DOI] [PubMed] [Google Scholar]
- 307.Fernandez-Beros ME, Kissel V, Lior H, Cabello FC. 1990. Virulence-related genes in ColV plasmids of Escherichia coli isolated from human blood and intestines. J Clin Microbiol 28:742–746. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Delicato ER, de Brito BG, Gaziri LC, Vidotto MC. 2003. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet Microbiol 94:97–103. [DOI] [PubMed] [Google Scholar]
- 309.Binns MM, Davies DL, Hardy KG. 1979. Cloned fragments of the plasmid ColV,I-K94 specifying virulence and serum resistance. Nature 279:778–781. [PubMed] [DOI] [PubMed] [Google Scholar]
- 310.Cross AS, Gemski P, Sadoff JC, Orskov F, Orskov I. 1984. The importance of the K1 capsule in invasive infections caused by Escherichia coli. J Infect Dis 149:184–193. [PubMed] [DOI] [PubMed] [Google Scholar]
- 311.Kim KS, Itabashi H, Gemski P, Sadoff J, Warren RL, Cross AS. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Invest 90:897–905. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Russo TA, Liang Y, Cross AS. 1994. The presence of K54 capsular polysaccharide increases the pathogenicity of Escherichia coli in vivo. J Infect Dis 169:112–118. [PubMed] [DOI] [PubMed] [Google Scholar]
- 313.Culham DE, Dalgado C, Gyles CL, Mamelak D, MacLellan S, Wood JM. 1998. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology 144:91–102. [PubMed] [DOI] [PubMed] [Google Scholar]
- 314.Väisänen V, Elo J, Tallgren LG, Siitonen A, Mäkelä PH, Svanborg-Edén C, Källenius G, Svenson SB, Hultberg H, Korhonen T. 1981. Mannose-resistant haemagglutination and P antigen recognition are characteristic of Escherichia coli causing primary pyelonephritis. Lancet 2:1366–1369. [DOI] [PubMed] [Google Scholar]
- 315.Opal SM, Cross A, Gemski P, Lyhte LW. 1988. Survey of purported virulence factors of Escherichia coli isolated from blood, urine and stool. Eur J Clin Microbiol Infect Dis 7:425–427. [PubMed] [DOI] [PubMed] [Google Scholar]
- 316.Russo T, Brown JJ, Jodush ST, Johnson JR. 1996. The O4 specific antigen moiety of lipopolysaccharide but not the K54 group 2 capsule is important for urovirulence of an extraintestinal isolate of Escherichia coli. Infect Immun 64:2343–2348. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Montenegro MA, Bitter-Suermann D, Timmis JK, Agüero ME, Cabello FC, Sanyal SC, Timmis KN. 1985. traT gene sequences, serum resistance and pathogenicity-related factors in clinical isolates of Escherichia coli and other gram-negative bacteria. J Gen Microbiol 131:1511–1521. [DOI] [PubMed] [Google Scholar]
- 318.Palaniyandi S, Mitra A, Herren CD, Lockatell CV, Johnson DE, Zhu X, Mukhopadhyay S. 2012. BarA-UvrY two-component system regulates virulence of uropathogenic E. coli CFT073. PLoS One 7:e31348. 10.1371/journal.pone.0031348. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Debnath I, Norton JP, Barber AE, Ott EM, Dhakal BK, Kulesus RR, Mulvey MA. 2013. The Cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect Immun 81:1450–1459. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Schwan WR. 2009. Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155:1832–1839. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Cai W, Wannemuehler Y, Dell’anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli’s ability to exploit abundant host metabolites. PLoS Pathog 9:e1003428. 10.1371/journal.ppat.1003428. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. 2009. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol 73:1020–1031. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Kim KS. 2001. Escherichia coli translocation at the blood-brain barrier. Infect Immun 69:5217–5222. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wang X, Maruvada R, Morris AJ, Liu JO, Wolfgang MJ, Baek DJ, Bittman R, Kim KS. 2016. Sphingosine 1-phosphate activation of EGFR as a novel target for meningitic Escherichia coli penetration of the blood-brain barrier. PLoS Pathog 12:e1005926. 10.1371/journal.ppat.1005926. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Tseng YT, Wang SW, Kim KS, Wang YH, Yao Y, Chen CC, Chiang CW, Hsieh PC, Teng CH. 2012. NlpI facilitates deposition of C4bp on Escherichia coli by blocking classical complement-mediated killing, which results in high-level bacteremia. Infect Immun 80:3669–3678. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Aguero ME, de la Fuente G, Vivaldi E, Cabello F. 1989. ColV increases the virulence of Escherichia coli K1 strains in animal models of neonatal meningitis and urinary infection. Med Microbiol Immunol 178:211–216. [PubMed] [DOI] [PubMed] [Google Scholar]
- 327.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci USA 104:16669–16674. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lundrigan MD, Webb RM. 1992. Prevalence of ompT among Escherichia coli isolates of human origin. FEMS Microbiol Lett 76:51–56. [PubMed] [DOI] [PubMed] [Google Scholar]
- 329.Nagy G, Dobrindt U, Schneider G, Khan AS, Hacker J, Emödy L. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect Immun 70:4406–4413. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Yamamoto S, Nakano M, Terai A, Yuri K, Nakata K, Nair GB, Kurazono H, Ogawa O. 2001. The presence of the virulence island containing the usp gene in uropathogenic Escherichia coli is associated with urinary tract infection in an experimental mouse model. J Urol 165:1347–1351. [PubMed] [Google Scholar]


