Abstract
Numerous recent developments in the biochemistry, molecular biology, and physiology of formate and H2 metabolism and of the [NiFe]-hydrogenase (Hyd) cofactor biosynthetic machinery are highlighted. Formate export and import by the aquaporin-like pentameric formate channel FocA is governed by interaction with pyruvate formate-lyase, the enzyme that generates formate. Formate is disproportionated by the reversible formate hydrogenlyase (FHL) complex, which has been isolated, allowing biochemical dissection of evolutionary parallels with complex I of the respiratory chain. A recently identified sulfido-ligand attached to Mo in the active site of formate dehydrogenases led to the proposal of a modified catalytic mechanism. Structural analysis of the homologous, H2-oxidizing Hyd-1 and Hyd-5 identified a novel proximal [4Fe-3S] cluster in the small subunit involved in conferring oxygen tolerance to the enzymes. Synthesis of Salmonella Typhimurium Hyd-5 occurs aerobically, which is novel for an enterobacterial Hyd. The O2-sensitive Hyd-2 enzyme has been shown to be reversible: it presumably acts as a conformational proton pump in the H2-oxidizing mode and is capable of coupling reverse electron transport to drive H2 release. The structural characterization of all the Hyp maturation proteins has given new impulse to studies on the biosynthesis of the Fe(CN)2CO moiety of the [NiFe] cofactor. It is synthesized on a Hyp-scaffold complex, mainly comprising HypC and HypD, before insertion into the apo-large subunit. Finally, clear evidence now exists indicating that Escherichia coli can mature Hyd enzymes differentially, depending on metal ion availability and the prevailing metabolic state. Notably, Hyd-3 of the FHL complex takes precedence over the H2-oxidizing enzymes.
INTRODUCTION
Formate and dihydrogen (H2) have negative redox potentials (CO2/formate: Eo′ = −432 mV; H+/H2: Eo′ = −414 mV) under standard conditions and therefore can be classified as high-energy compounds. Both serve as excellent energy sources for microorganisms. During fermentative growth, formate and H2 production provide a simple means for a microorganism to rid itself of excessive reducing equivalents. Enterobacteria such as E. coli and Salmonella enterica subsp. enterica serovar Typhimurium can produce, as well as consume, hydrogen and formate. Both compounds are also currently of considerable interest in applications such as metabolic engineering and biofuel production (1–4) and are therefore considered valuable bio-based feedstocks.
Two of the six carbon atoms of glucose are converted to formate during mixed acid fermentation (5). Each can contribute to H2 production by the cytoplasmic formate hydrogenlyase (FHL) complex, resulting in a theoretical maximum of 2 mol of H2 evolved per mol of glucose oxidized. This reaction serves to reduce cytotoxic levels of intracellular formate, but it is still unresolved as to whether it is directly involved in energy conservation. E. coli also encodes in its genome a second FHL-like complex; however, the physiological conditions under which it is synthesized remain to be determined. FHL and its “cryptic” homolog are structurally related to NADH:quinone oxidoreductase (complex I) of the respiratory chain (4, 6, 7). The FHL complex comprises the activities of a molybdenum/selenium-dependent formate dehydrogenase (FDH-H) and a [NiFe]-hydrogenase (Hyd), termed Hyd-3; Hyd-4 in the homologous FHL-2 complex. FDH-H and Hyd-3 are linked by three iron-sulfur (FeS) proteins and are attached to the membrane by two integral membrane proteins. Membrane association is essential for disproportionation of formate by the complex (2).
Because enterobacteria can switch between fermentative and respiratory growth modes, the fate of formate is often determined by the presence of exogenous electron acceptors (8). Thus, the FHL complex has to compete with two respiratory formate dehydrogenases (FDH-N and FDH-O) for its substrate. The genes coding for the FHL complex are only transcribed when a threshold concentration of formate has been reached in the cytoplasm, which is controlled in part by the formate-nitrate transport (FNT) membrane protein FocA. The FocA transporter is mainly responsible for formate export into the periplasm during the exponential growth phase and reimport into the cytoplasm during early stationary growth phase (Fig. 1A). The so-called formate regulon (8), in combination with nitrate- and oxygen-sensing regulators (9), presents a mechanism allowing the cell to adapt to prevailing metabolic conditions and to control transcription of genes required for formate oxidation. The H2 produced by the FHL complex is partially, or completely, reoxidized via the respiratory Hyds, the so-called H2-oxidizing Hyds, Hyd-1 and Hyd-2, in E. coli and Salmonella Typhimurium, and via a comparatively newly discovered Hyd-5 in Salmonella Typhimurium (10, 11). These enzymes face the periplasm and are electrochemically connected to the quinone pool. Biochemical data show that they fulfill different roles during H2 oxidation. Hyd-1 and its homolog Hyd-5 are oxygen-tolerant enzymes (12), while current evidence indicates that Hyd-2 is a H2-driven proton pump (13). Under conditions resulting in overreduction of the quinone pool, Hyd-2 can also serve as a “valve” to release reducing equivalents in the form of H2. These latter findings indicate that, while Hyd-2 has the primary function of a H2-oxidizing, or H2-uptake, hydrogenase, its function in vivo can be reversed so that it produces H2 by reducing protons. This facet provides the bacterium with added flexibility when switching rapidly between respiratory and fermentative metabolism, but it also raises interesting questions about the potential role of progenitors of Hyd-2-like enzymes in the early evolution of hydrogen-driven metabolism.
Figure 1.
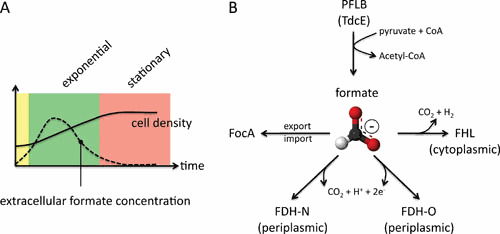
A, Schematic illustration of the increase in cell density (black line) during fermentative growth (green = exponential growth phase, red = stationary growth phase) and the formate concentration in the medium at the same time (dashed line). The curves are based on reference 299. B, Representation of the main metabolic pathways competing for the degradation of formate under different conditions. See the text for details.
All enzymes of formate and hydrogen metabolism have in common active sites that contain complex metal-based cofactors. Cofactor biosynthesis and insertion into the FDHs—selenocysteine (14), molybdenum cofactor generation (15), and FeS cluster insertion (16)—are topics covered elsewhere. However, in this updated review, we describe the most recent developments in the biosynthesis of the [NiFe] cofactor, which is involved in all H2 reactions in enterobacteria. Synthesis of this bimetallic cofactor is dependent on a set of accessory proteins, some of which are universal to all Hyds, while others are enzyme specific. These features of the maturation machinery allow the cell to selectively mature a preferred enzyme, which is determined in response to the metabolic status.
While based firmly on the original EcoSal Plus review by Sawers, Blokesch, and Böck (17), here we integrate the recent exciting developments in the physiology, biochemistry, and structural biology of formate and H2 metabolism in E. coli and Salmonella Typhimurium.
FORMATE METABOLISM AND HYDROGEN EVOLUTION
The Formate Hydrogenlyase System
The FHL pathway was first described by Stephenson and Stickland (18, 19), and it catalyzes the disproportionation of formate to H2 and carbon dioxide (CO2) (Fig. 1). Studies in the 1950s identified an absolute requirement of selenium and molybdenum for the synthesis of active formate dehydrogenase (FDH) and hydrogen gas production by E. coli (20), and it was established that an FDH, a Hyd, and three electron carriers constituted the FHL pathway (21, 22) (Table 1). An early genetic study resulted in the isolation of two mutants that were defective in FHL activity (23). One proved to carry a lesion in the fdhF gene, encoding the selenopolypeptide of FDH-H (which refers to the FDH component of FHL specifically involved in hydrogen production), while the other had an insertion element located within the second gene of the multicistronic hyc operon, encoding further structural components of the FHL complex (6, 23–26). FDH-H and Hyd-3 comprise the two catalytic components of FHL. Both of these enzymes are biochemically and genetically distinct from the other FDH and Hyd enzymes present in E. coli and Salmonella Typhimurium (reviewed in references 4 and 27). The FHL pathway constitutes a multiprotein complex located on the inner aspect of the cytoplasmic membrane and henceforth will be referred to as the FHL complex (Fig. 2A) (28, 29). The FHL complex shares structural features with the respiratory NADH:quinone oxidoreductase (complex I) (6, 28) (Table 2). After the crystal structure of complex I from Thermus thermophilus was solved (30), it became possible to predict the structure of the FHL complex and the biochemically related Hyd-4 complex and align these to complex I (31) (Fig. 2 and see below). As predicted, the FHL complex comprises a hydrophilic domain on the cytoplasmic side of the membrane with the selenium- and molybdenum-dependent FDH-H, the [NiFe]-cofactor-dependent Hyd-3 and the electron-transfer subunits HycB, HycF, and HycG. The cytoplasmic domain is attached to a cofactor-free membrane domain comprising two subunits, termed HycC and HycD. Because of the similarity with complex I, it was suggested that FHL has a role as an energy-conserving proton pump (reviewed in reference 32). However, recently conducted in vitro experiments using purified FHL complexes have so far been unable to verify this hypothesis (2, 29). Nevertheless, the homology between the complexes has been taken as evidence that complex I and components of the FHL complex diverged from a common ancestor (33, 34). The membrane subunit HycC of FHL complex is particularly striking in its homology to the proton translocation subunits of complex I (35, 36).
Table 1.
Comparison of the properties of the hydrogenase isoenzymes
| Enzyme | Subunit composition/electron pathway | Metals and cofactors | Function | Dye specificity | App. Km for H2 | References |
|---|---|---|---|---|---|---|
| Hydrogenase-1 | α2β2γ2 (HyaB-HyaA-HyaC)2 | [NiFe]; [4Fe-3S], [3Fe-4S], [4Fe-4S]; cyt b | H2 oxidation, O2 tolerance | Nitroblue tetrazolium, Benzyl viologen (BV) | 9 μM (PFEa)2 μM (BV) | (141, 154, 156) |
| Hydrogenase-2 | αβγδ (HybC-HybO-HybA-HybB) | [NiFe]; 2 [4Fe-4S], [3Fe-4S]; 4 [FeS] | H2 oxidation, energy conservation | Benzyl viologen | 17 μM (PFE)3.7 μM (BV) | (13, 144, 154, 158) |
| Hydrogenase-3 (FHL complex) | FdhH–HycB-G–HycE | Mo-bis-PGD,b Selenocysteine, [4Fe-4S]; 7 [4Fe-4S]; [NiFe] | Proton reduction | Benzyl viologen | 34 μM (PFE) | (28, 29) |
| Hydrogenase-4 (E. coli) | ?–HyfA-I | predicted FHL ortholog | Proton reduction? | Unknown | Unknown | (7) |
| Hydrogenase-5 (Salmonella Typhimurium) | α2β2γ2 (HydB-HydA-HydC)2 | [NiFe]; [4Fe-3S], [3Fe-4S], [4Fe-4S]; cyt b | H2 oxidation, O2 tolerance | Nitroblue tetrazolium, Benzyl viologen | 9 μM (PFE) | (11, 12) |
PFE, protein film electrochemistry.
Mo-bis-PGD, molybdopterin guanine dinucleotide.
Figure 2.
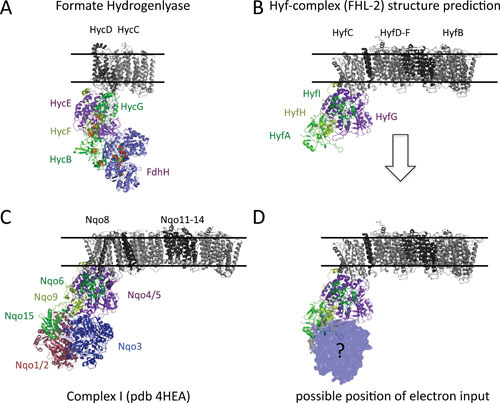
Structural similarity between FHL complexes and complex I. A structure prediction was carried out for the Hyc (A) and Hyf (B and D) subunits and together with the structure of FDH-H, which is shown in blue (Protein Data Bank [PDB]: 1AA6), aligned to the complex I structure (PDB: 4HEA; C). The membrane proteins are shown in black and gray, the small subunits in dark green, and the HycF electron transfer subunit in light green. The [NiFe] active site in FHL is located in the purple HycE subunit, and the diaphorase NADH oxidation site is shown in red in complex I only. The oxidation site for FHL-2 is unknown, and a model showing the possible location according to FHL is shown in D.
Table 2.
| FHL subunit | Molecular mass (kDa) | Function in FHL | Hyf subunit | Nuo complex I subunit (E. coli) | Thermus thermophilus |
|---|---|---|---|---|---|
| FDH-H (fdhF gene) | 79.12 | Formate oxidation | Unknown | NuoG (C terminus) | Nqo3 |
| HycA | 17.59 | Transcriptional regulator | |||
| HycB | 21.80 | FDH-H small subunit | HyfA | NuoG (N terminus) | Nqo3 |
| HycC | 63.98 | Transmembrane subunit (16 TMH)a | HyfB | NuoL | Nqo12 |
| Transmembrane subunit (14 TMH) | HyfD, HyfF | NuoM/N | Nqo13-14 | ||
| HycD | 32.97 | Transmembrane subunit (8 TMH), coupling site | HyfC | NuoH | Nqo8 |
| HycE (1–150 aa) | Hydrogenase large subunit | HyfG | NuoC | Nqo5 | |
| HycE (150–569 aa) | 64.89 (61.06)b | Hydrogenase large subunit, [NiFe] cofactor | HyfG | NuoD | Nqo4 |
| HycF | 20.27 | Electron transfer subunit | HyfH | NuoI | Nqo9 |
| HycG | 27.95 | Hydrogenase small subunit, FeS/N2-cluster | HyfI | NuoB | Nqo6 |
| HycH | 15.43 | Large subunit chaperone, not essential for FHL activity | HyfJ | ||
| HycI | 16.99 | Endoprotease | Unknown | Not required | Not required |
| Transmembrane protein | HyfE (C terminus) | NuoK | Nqo11 | ||
| Transcriptional regulator, similar to FhlA | HyfR | ||||
| Putative formate channel | FocB |
TMH, transmembrane α-helices.
Molecular mass after protein specific processing; mass given includes the first 150 amino acids (aa).
Hydrogenase 3
The existence of a third hydrogenase enzyme (Hyd-3; EC 1.12.99.-) was first established by performing immunoprecipitation studies with antibodies raised against the Hyd-1 and Hyd-2 enzymes (37). The nonimmunoprecipitable hydrogenase enzyme activity could be correlated with FHL complex synthesis. The functions of the hyc operon gene products in the FHL complex were established by molecular and biochemical methods (Table 2) (6, 28, 29, 38). The identification of the gene (hycE) encoding the large subunit of Hyd-3, which is located within the so-called hyc operon hycABCDEFGHI (see Fig. 3), facilitated subsequent characterization of the enzyme (6, 28). The HycE polypeptide has been purified and shown to contain up to 1 mol of nickel per mol of enzyme (39). Likewise, the entire FHL complex can be purified via affinity chromatography of a His-tagged HycE after dispersal of the membrane with a detergent cocktail (29). The Michaelis-Menten constant for H2 oxidation is 34 μM (pH 6), and it is interesting that the CO inhibition, which serves as an indicator of O2 tolerance, is in a similar range to that of Hyd-1 (29).
Figure 3.
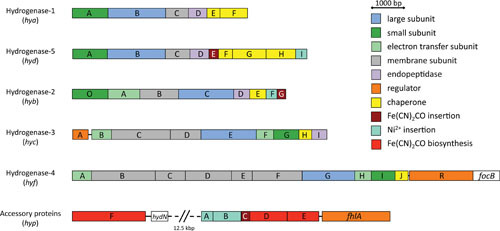
Organization of the “hydrogen metabolism” genes in E. coli and Hyd-5 operon in Salmonella Typhimurium. Genes whose products have a similar function or that have similar amino acid sequences are the same color, and a legend summarizes their function.
The small subunit of Hyd-3, HycG, carries a [4Fe-4S] cluster, which surprisingly has sequence features typically found in O2-sensitive Hyds (38). The corresponding [4Fe-4S] cluster in complex I is named N2 and transfers the electrons to the quinone pool. This quinone-binding site is presumably located where the [NiFe] cofactor in the active site of HycE resides. The HycE protein has two domains, whereby the N-terminal domain resembles the NuoC protein from complex I, while the C-terminal domain is more similar to Hyd (Table 2). The N-terminal domain of HycE harbors no cofactor but is essential for stability of the entire FHL complex (C. Pinske and F. Sargent, unpublished). The C-terminal domain has two CXXC motifs for binding of the [NiFe] cofactor, and, remarkably, it was reported that a truncated version lacking one of these motifs showed enhanced H2 production (40).
The proton-pumping mechanism in complex I is not fully understood but presumably involves a charge transfer between glutamate and lysine residues within the membrane subunits (41). These charged residues are likewise conserved in HycC and HycD; however, with the exception of being necessary to attach the cytoplasmic domain to the membrane, no requirement for the catalytic mechanism of FHL was apparent (2). Nevertheless, the presence of the membrane subunits is essential for FHL activity in vivo (2, 28).
The gene product of hycH is essential for full FHL activity, and a deletion reduces activity by over 70% (Pinske and Sargent, unpublished). Initially, a hycH deletion was characterized as being protease deficient, but this was because the downstream hycI gene was not originally noticed, and this led to polar effects of the hycH mutation, which prevented synthesis of the HycE-specific protease (28, 42). Although the function of HycH remains to be determined, the protein was found to interact tightly with HycE in the absence of the small subunit HycG (38).
The last gene in the operon, hycI, encodes the HycE-specific protease, which is required for C-terminal proteolytic processing after [NiFe]-cofactor insertion and which occurs prior to complex assembly (42). Finally, the first gene of the hyc operon, hycA, does not encode a structural component of the FHL complex. HycA is a regulator that interferes with the formate-sensing ability of formate hydrogenlyase activator (FHLA) (28, 43), although how this occurs mechanistically is unresolved.
Some pathogenic E. coli, Salmonella Typhimurium, and Shigella strains encode an additional open reading frame (E. coli Nissle GNBM-4002; Salmonella STM2844; Shigella flexneri SFV_2787) within the hyc operon that is located downstream of hycI and encodes a 30-kDa protein. The gene product has no assigned function, but it might be involved in controlling FHL activity or synthesis.
The requirement of the iron-sulfur cluster (ISC) insertion and biosynthetic machinery for Hyd activity was studied by phenotypic analysis of defined deletion strains (44–46). It could be shown that, of the two FeS-biosynthesis systems present in E. coli, only the ISC system, and not the SUF system (47), is involved in providing FeS clusters to these enzymes (45). The central scaffold protein IscU is essential for Hyd activity because ΔiscU mutants completely lack hydrogenase activity (45, 46). Notably, all three Hyd enzymes had no detectable small subunits. Deletion of genes coding for Fdx (ferredoxin) and the IscA and ErpA proteins, which are proposed to deliver FeS clusters to target proteins (16, 48), abolished the H2-oxidizing Hyd activities, while Hyd-3 retained some activity (44–46). FHL requires a total of eight [4Fe-4S] clusters (Table 1) (29), significantly more than the H2-oxidizing Hyd. Nevertheless, the FHL complex appears to be preferentially matured over the other Hyd enzymes (44–46, 49). It remains to be established how the ISC machinery controls the FeS cluster insertion process and how preference for particular target proteins is determined.
Formate Dehydrogenase H
FHL complex activity can be determined as the formate-dependent production of dihydrogen (37), or the activity of the formate dehydrogenase H (FDH-H) enzyme (EC 1.1.99.-) component can be determined in isolation by measuring the formate-dependent reduction of the one-electron, low-redox-potential dye benzyl viologen (BV) (21, 50–53). FDH-H is an 80-kDa selenopolypeptide and is encoded by the fdhF gene (24, 26, 51, 54, 55). Selenium, in the form of selenocysteine (SeCys), is located at amino acid position 140 in the FDH-H polypeptide chain (25, 55, 56). The FDH-H polypeptide was first purified in 1990 (54) and shown to contain 3.3 g atoms of iron and 1 g atom of molybdenum per mole of enzyme. These results suggested that the enzyme contains a single [4Fe-4S] cluster, and Heider and Böck (57) proposed that a conserved cysteine motif common to Mo-cofactor-dependent FDHs may be involved in forming a ligand to the cluster. Molybdenum was reported to be associated with the enzyme in the form of a molybdopterin guanine dinucleotide (Mo-bis-PGD) cofactor (54).
Direct involvement of the selenolate of SeCys in formate oxidation was demonstrated by comparing the enzymatic conversion of the selenocysteinyl enzyme with a cysteinyl-substituted derivative. The sulfur enzyme proved to be 20 times less active than its selenium-containing counterpart at physiological pH, thus emphasizing the advantage of the reactivity of the selenol over the thiol group in redox processes (50, 58). A subsequent electron paramagnetic resonance (EPR) spectroscopic analysis of 77Se-enriched FDH-H revealed that the selenolate of the SeCys residue is directly coordinated to the molybdenum, which was suggested to be in the Mo(V) species (59). A more-detailed EPR analysis using near-homogeneous enzyme revealed that the molybdenum in formate-reduced, crystalline FDH-H was in the Mo(IV) oxidation state and that the single [4Fe-4S] cluster was reduced (52). Oxidation of the enzyme with BV generated the Mo(VI) species and an oxidized [4Fe-4S] cluster. This study gave the first insights into the reaction mechanism and the possible intramolecular electron transfer route, which were substantiated by the determination of the crystal structure of FDH-H (55).
Formate Dehydrogenases: Structural and Mechanistic Insights Based on FDH-H
Apart from FDH-H, E. coli synthesizes two further phylogenetically highly similar FDH enzymes, termed FDH-N and FDH-O, so named to reflect under which respiratory (N for nitrate and O for oxygen) growth conditions they are optimally synthesized (20, 60, 61). Little is known about FDH-O apart from its ability to couple formate oxidation to oxygen reduction (61). Originally considered to be synthesized at a low level, more recent studies indicate that FDH-O is a comparatively abundant enzyme, even during growth with nitrate as exogenous electron acceptor (62, 63). Considerably more information is available concerning the FDH-N enzyme, however. The active site of FDH-N (64) is almost identical to that of FDH-H (55) and confirms that formate oxidation is performed at the SeCys-coordinated Mo-bis-PGD cofactor. Indeed, despite the α-subunit of FDH-N being substantially larger than FDH-H, the core structure of both proteins is superimposable (64).
All three FDH enzymes in E. coli belong to the dimethyl sulfoxide reductase family (65, 66). The organization of the Mo-bis-PGD cofactor is a general framework conserved in this class of redox enzymes, facilitating intramolecular electron transfer (67–69) and, recently, a common nomenclature, pyranopterin guanosine dinucleotide (PGD), has been proposed for these cofactors (65, 66). The Mo atom is hexa-coordinated to four sulfur atoms from two PGD molecules, one selenium atom from selenocysteine (SeCys), and one inorganic sulfur atom (Fig. 4) (70, 71). The reinterpretation of the sixth ligand as sulfur instead of an oxygen atom has implications for the proposed reaction mechanism (55, 66, 71, 72). The FdhD protein transfers this sulfur from the l-cysteine desulfurase IscS to the deeply buried molybdenum cofactor, possibly prior to insertion, a modification essential for activity (73, 74). A further FDH-H-specific protein is HydN, encoded adjacent to hypF (Fig. 3). A hydN deletion strain has reduced FDH-H activity, and the protein resembles other FDH small subunits like HycB (75). Therefore, it has been suggested that HydN associates with FDH-H to form an alternative pool for FDH-H when it is not incorporated into FHL (4).
Figure 4.
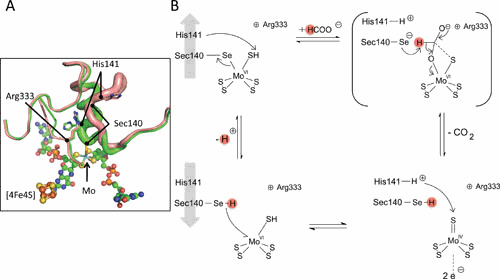
The reaction mechanism for formate oxidation has not been completely elucidated yet. A, The active site Mo-bis-PGD and the active site selenocysteine 140, His141, and Arg333 are shown. The red figure is based on PDB entry 2IV2 (71); the green figure is based on PDB entry 1AA6 (55). B, The proposed mechanism involves replacement of the SeCys ligand by formate, subsequent proton abstraction, possibly by the selenide, reduction of MoVI to MoIV during CO2 formation, and electron transfer to the [4Fe4S] cluster after which the proton is released. Here, the proton abstraction by selenide is shown, and alternative mechanisms are discussed in the text.
The FDH-H enzyme has a four-domain, αβ-structure in which domain I coordinates the [4Fe-4S] cluster, domains II and III coordinate the 2 Mo-bis-PGD in an αβα-sandwich, and the C-terminal domain IV forms a cap over the two PGD cofactors (55).
A further feature of the FDH-H active site revealed by the structure is that the ligand with the Mo also hydrogen bonds with the amide of His141 (55), although a reinterpretation of the data showed that, in the formate-bound state, the loop is too distant for bonding (71). Based on binding of the inhibitor nitrite, the substrate formate is suggested to be located between SeCys140 and His141, and upon formate binding the SeCys140 is displaced from the Mo (71). Arg333 is strictly conserved and initially orients the formate molecule and later forms an ionic interaction with free selenol (71). Experiments with 18O-labeled substrates have established that the FDH-H enzyme is not an oxotransferase, but instead dehydrogenates formate to form CO2 directly (53). Upon oxidation, the α-proton of formate was initially suggested to be transferred to His141 via the SeCys residue, which undergoes transient protonation (Fig. 4). This is also supported by EPR analysis (53). However, several different mechanisms for proton transfer have been proposed recently, including direct hydride transfer initially to the Mo atom with subsequent transfer of the proton to the selenol anion (76) or direct proton abstraction by the selenide (77). In these more recent models, the role of His141 has been suggested to involve positioning the negatively charged selenol anion optimally for deprotonation of the formate (77), which is supported by recent mutagenesis analysis of the FDH from the photosynthetic bacterium Rhodobacter capsulatus (78). The involvement of the sulfur ligand in the catalytic cycle has also been intensely debated (66). Therefore, the precise role of His141 in deprotonation events still needs to be unequivocally established.
Detailed kinetic studies using deuteroformate and proteoformate clearly demonstrated that the formate oxidation step is not rate limiting, but, rather, the subsequent one-electron transfer steps to BV in the in vitro analyses are rate determining (58). These findings have been substantiated by the structural data (55). The two electrons generated upon formate oxidation are transferred from the Mo(IV) to the [4Fe-4S] cluster, which is located just below the enzyme’s surface and transfers the electrons via HycB in the FHL complex. The electrons are transferred one at a time, generating the Mo(V) species observed by EPR (52, 53). Further oxidation of Mo(V) to Mo(VI) results in breaking of this SeCys-His141 hydrogen bond and release of the proton to the solvent.
While the reaction of the entire FHL complex is reversible (2, 79), isolated FdhF in solution was unable to reduce CO2 efficiently. The reaction of formate oxidation by isolated FDH-H was, however, shown to be bidirectional when the enzyme is attached to an electrode, a reaction that is of great interest for CO2 fixation (80).
The FHLA Transcriptional Activator
Transcription of the genes encoding the FHL complex occurs only during fermentative growth and is absolutely dependent on an acidic pH in the medium, formate, and the alternative σ54 (8, 81–85). Hence, the expression of the fdhF gene and the hyc and hyp operons is precisely coordinated. The isolation of trans-acting regulatory mutants identified the fhlA gene (Figs. 3 and 5), which encodes the transcriptional regulator that coordinates the expression of these genes in the presence of a critical threshold level of formate (86, 87). The FHLA protein is formally similar to regulators of two-component sensor-regulator pairs in that it has ATPase and DNA-binding domains characteristic of σ54-dependent transcriptional activators (86); however, it lacks the receiver domain typically found in two-component regulators. It has been shown to bind specifically to a cis-regulatory sequence located approximately 100 bp upstream of the fdhF gene (87), previously characterized by deletion analysis to be essential for the formate-dependent expression of fdhF-lacZ fusion (88). FHLA binds to two further cis-regulatory sequences; one sequence, termed IR1, is located between the hycA and hypA genes of the divergently oriented hyc and hyp operons, while the second binding site (IR2) is located between the hycA and hycB genes (87). A further transcriptional unit comprising hydN and hypF is also regulated by FHLA (75, 83) (see Fig. 5). The hydN promoter is further activated by the FlhDC transcriptional regulator that enables transition into the stationary phase (89).
Figure 5.
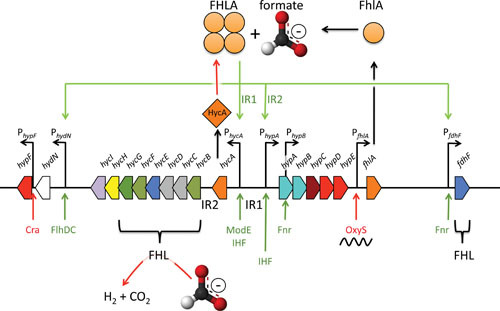
Organization of the FHLA-dependent formate regulon. The genes and distances are not to scale; the color scheme of genes is according to Fig. 3. The fhlA gene product oligomerizes as a homotetramer and is able to sense formate under fermentative growth conditions, whereupon it activates transcription (green arrows). The binding site for hyc activation is intergenic region 1 (IR1) and for hyp is IR2. FHLA autoactivates its own transcription via the hyp promoter; in the absence of formate, it is transcribed at a low constitutive level from its own promoter. The function of FHLA is antagonized by HycA, and by the small RNA OxyS, which binds to its mRNA, and the FHL complex removes the activating molecule formate (red arrows). Further transcriptional regulators to the respective promoters are shown in red (inhibiting) or green (activating).
Studies using an in vitro coupled transcription-translation system have demonstrated that IR1 is necessary for activation of hyc operon transcription and IR2 is required to activate transcription of the hyp operon (90). Formate probably interacts with the N-terminal domain of the FHLA protein to effect transcriptional activation (91, 92). Integration host factor (IHF) has also been shown to be required to optimize the expression from this complex regulatory region, and it has been proposed that one function may be to organize a supramolecular transcription complex (90). IHF is not involved in the transcriptional regulation of the fdhF gene.
The fhlA gene is transcribed at a low level from its own promoter (84, 85), and this level is enhanced anaerobically through the activity of the fumarate and nitrate reduction (FNR)-dependent promoter within the hypA gene (see Fig. 5) (8). Activation of the FHLA-dependent promoter in front of hypA further increases fhlA gene transcription. This scenario presents a novel positive-feedback mechanism for transcriptional control of a regulon (83). The HycA protein appears to antagonize the action of FHLA, thus preventing continuous activation of the formate regulon, but the exact mechanism of FHLA inhibition by HycA is still unclear (28, 43). Furthermore, FHLA synthesis is subject to translational control by OxyS, which is a 109-nucleotide-long, untranslated sRNA induced under oxidative stress (93). OxyS RNA inhibits translation by pairing with the fhlA mRNA covering the ribosome-binding site plus a small region of the coding segment and forming a stable mRNA-antisense complex (94). The interaction requires the binding of the RNA-binding chaperone Hfq (95). It can be speculated that oxidative stress, which is characteristic of the lifestyle of E. coli, can rapidly shut down translation of the components of the FHL system and thereby prevent wasteful synthesis of the oxygen-sensitive system.
Because the H2 production from FHL is an economically valuable reaction, current research aims at increasing its yield (for review, see reference 4). Although it was originally shown that hycA deletion mutants yield more H2 than the wild-type strain (28), other groups could not observe a significantly increased H2 production in hycA deletion strains (96, 97). However, the combination of a hycA deletion and FHLA overproduction allowed a 7-fold increase in hycE mRNA levels (98) and even an 80-fold increase in H2 production rates (3) after combination with genetic knockouts of genes whose products reduce cellular hydrogen or formate levels (99). Alternatively, the FHLA protein has been engineered to allow increased H2 production, and, through an E363G exchange, it was shown to be insensitive to HycA repression (100).
The Formate Regulon: fdhF, hyc, and hyp
In the absence of formate, no transcription of any FHLA-dependent promoter occurs (8). This indicates that there is an absolute requirement for formate to interact with FHLA to enable transcriptional control of the regulon. Molybdate also has an important subsidiary role in control of formate regulon expression (101, 102; see below). Control of intracellular formate levels, therefore, determines whether the formate regulon is activated or not, and this provides a simple mechanism to ensure that the FHL complex is only synthesized when it is required. Expression of the genes of the regulon is not activated when an alternative electron acceptor, such as nitrate, trimethylamine-N-oxide (TMAO), or oxygen, is present or when the pH of the medium is above 7.
A model for control of the formate regulon (reviewed in reference 85) is presented in Fig. 5. When pyruvate formate-lyase (PFLB) is activated under anaerobic conditions, formate, the product of the PFLB reaction, is excreted at neutral pH via the formate-specific channel, FocA (103). FocA belongs to a large and rapidly expanding class of pentameric anion channels with structural similarity to aquaporins (104–111). The specificity of transport is determined by a direct interaction of PFLB with FocA (112). In the presence of alternative electron acceptors such as TMAO or nitrate, formate is preferentially metabolized in the periplasm by FDH-O and FDH-N (8, 113) and so formate does not accumulate intracellularly (Fig. 1). One study has implicated the nitrate-responsive two-component NarXL component in mediating the nitrate effect by direct interaction of the NarL transcriptional regulator with the upstream regulatory elements of the FHLA-controlled genes (114). It should be pointed out, however, that, in the particular study mentioned, an appropriate control analysis in an fdn (encodes FDH-N) deletion mutant was not performed. Results of earlier studies demonstrated that preventing synthesis of alternative respiratory routes for formate metabolism, or introducing high levels of exogenous formate, relieve nitrate- and TMAO-dependent “repression” of the formate regulon (8, 113). If no exogenous electron acceptor is available, and as the external pH decreases, formate is transported back into the cell by FocA (Fig. 1A) and once the intracellular concentration increases above the threshold (Km of FHLA for formate is 5 mM) (115), the regulon becomes activated. Its main product is the formate-consuming FHL complex that counteracts the acidification of the growth medium (8, 116). These observations suggest that metabolic drainage of formate is the major mechanism underlying control of regulon induction and compartmentalization of formate determines whether the regulon is activated or not.
Additional support for such a model comes from the enzymatic properties of the formate dehydrogenases and of FHLA, which compete for their substrate, formate, under anaerobic conditions. FDH-H has an apparent Km for formate of about 26 mM (54); those of FDH-N (60) and FHLA (115) are 0.12 and 5 mM, respectively; the Km for formate of FDH-O is unknown but is assumed to be in a similar range to FDH-N. Induction of formate-dependent nitrate respiration by nitrate results in drainage of formate into the nitrate respiratory chain because of the lower Km of FDH-N compared with that of FHLA for the substrate formate (8, 85). The comparatively high levels of formate required to induce synthesis of the FHL complex are thus not attained during anaerobic respiration.
Effects of Metals on Regulation of FHL Genes
Iron, nickel, molybdenum, and selenium are essential for the assembly and maturation of a functional FHL complex. Selenium is an essential component of FDH-H and is incorporated as selenocysteine (14). However, there is no evidence that selenium affects transcription of the fdhF or sel (encoding the selenocysteine biosynthetic machinery) genes in E. coli (61).
A Ni2+-specific transport system is encoded by the nikABCDE operon (117), and its expression is regulated by FNR and the nickel-responsive regulator, NikR (118–120). Nickel is not known to regulate expression of the hyc or hyp genes directly.
Mutants unable to transport molybdate are impaired in expression of the fdhF and hyc genes (101, 102). This defect can be complemented phenotypically by addition of high levels of molybdate to the medium. A further mutational study identified the Mo-responsive transcriptional regulator ModE and the MoeA protein to be required for the effect of molybdate on hyc expression (121, 122). MoeA mediates molybdenum ligation from MogA into the molybdopterin cofactor precursor (123, 124) (reviewed in reference 125). It has been suggested that MoeA interacts with FHLA to enhance transcription (92, 126).
Mo-dependent binding of ModE to the hyc promoter-regulatory region has been demonstrated, and the ModE-Mo complex binds upstream of FHLA (121). Interestingly, the widely used E. coli protein production strain BL21(DE3) was shown to lack modE and some of the genes required for molybdenum transport, explaining, in part, the difficulties it has to synthesize the FHL complex (127). This led to the false conclusion that H2 can be produced from a heterologously expressed H2-oxidizing [NiFe]-Hyd (128); however, these findings nevertheless reveal that further regulatory mechanisms influence hyc expression.
Expression of the hyc operon is also influenced by iron homeostasis. Deletion of the fur gene encoding the ferric uptake regulator, Fur, causes reduced FHL activity, which is due to lower transcription of the fdhF and hyc genes (129). There is no obvious Fur-binding site in the upstream region of these genes, strongly suggesting that the effect of Fur could be indirect, possibly because of reduced formate levels. Lack of Fur releases repression of the small RNA RhyB, which, in turn, is a repressor of the iscSUA operon (130) and downregulates the pflA gene coding for pyruvate formate-lyase-activating enzyme (131).
The Fourth Hydrogenase, Hyf
DNA sequence analysis identified a 12-cistron operon, termed hyf (hydrogenase four), on the E. coli chromosome (7). This Hyd-4 enzyme complex is not encoded on the Salmonella Typhimurium chromosome. The operon (hyfABCDEFGHIRfocB; see Fig. 3 and Table 2) potentially codes for a hydrogenase complex comprising 10 subunits, resembling the FHL complex. Two further genes within the operon encode a σ54-dependent transcriptional regulator HyfR, which exhibits significant amino acid similarity to the formate-responsive transcriptional regulator FHLA (86), and a putative formate channel, FocB, which is similar to FocA (103). Seven of the putative proteins are orthologs of the Hyc proteins, while three membrane subunits (HyfD, HyfE, and HyfF) have no related subunits in the Hyc complex but are related to subunits of the proton-translocating complex I. Promising evidence for an involvement in proton translocation came from the concomitant overproduction of proteorhodopsin with the transcriptional activator HyfR that allowed an increase in H2 production by Hyd-4 and suggested a dependency upon ΔμH+ (132).
It has been proposed that the Hyf proteins, in particular HyfA, interact with FDH-H to form a novel proton-translocating complex, which has been termed FHL-2 (7). However, the electron input module, and thus the substrate for FHL-2, remains to be identified (Fig. 2). Alternatively, it was suggested that the FDH-H/HycB proteins interact with both FHL and FHL-2 (133), which seems unlikely because the hyf operon encodes its own HycB homolog, HyfA, and expression patterns of the hyc and hyf operons differ considerably. The uncharacterized FDH-H homolog YdeP has been suggested as a further potential interaction partner of the Hyf complex (4).
Unfortunately, expression of the hyf operon is very weak, and it has not yet been possible to characterize the gene products in wild-type E. coli cells (134, 135). However, peptides were identified by tandem affinity chromatography, and an interaction between the maturase HypC and the predicted large subunit HyfG has been shown (136). The hyf-operon does not encode a protease specific for a large subunit, despite HyfG carrying the C-terminal amino acid extension characteristic of cofactor-containing catalytic subunits capable of being processed (7). This might indicate that the predicted large subunit HyfG either is not processed, or it shares the HycI protease with HycE (Hyd-3).
It has been suggested that the Hyf complex is responsible for dihydrogen production at pH 7.5 and that activity of the complex requires F0F1-ATPase (133). Because this observation contradicts the increased expression levels of hyf genes at low pH, the H2 production under these conditions could possibly be attributable to Hyd-2 functioning in reverse (13, 137). In addition, clear evidence based on mutant analysis indicates that F0F1-ATPase is required for fermentative gas production in Salmonella Typhimurium, which has no hyf operon (138). Also the participation of hyf gene products in total H2 production seems to be higher when cells are growing by fermentation with low glucose concentrations, which indicates a complementary role to FHL (139, 140). Strains deleted for the genes encoding the large subunits of Hyd-1, Hyd-2, and Hyd-3 do not produce H2 from Hyd-4 (97). However, it has been suggested that this finding is due to a cross talk between, or codependence on, the other Hyd enzymes (32). Taken together, because the predicted FHL-2 complex is more closely related to complex I than the FHL complex, it might be involved in proton translocation (32, 35, 36).
Regulation of hyf Expression
Weak expression of a hyfA-lacZ fusion has been observed, and this was shown to be FHLA and σ54 dependent (134). Fermentative growth at low pH was required for expression, and formate was shown to induce expression. Expression was also maximal after cultures had exited the exponential phase of growth, which correlates with a pH reduction and intracellular formate accumulation.
Transcription of the hyfR regulatory gene could not be detected in wild-type E. coli cells (134, 135). However, placing hyfR expression under the control of an inducible promoter revealed that HyfR could activate the hyf operon to significant levels. The same construct resulted in increased H2 yields in the presence of a proton-pumping proteorhodopsin (132). Furthermore, expression only occurred anaerobically and was formate-independent. HyfR was not able to activate the formate regulon, indicating that it has different properties to FHLA and thus probably senses different metabolites. This also correlates with its less conserved N-terminal domain compared with FHLA (7, 134, 135). Skibinski et al. (134) made the interesting observation that HyfR has a C-X6-H-C-X-C-P-X-C-X-P motif, which suggests that it might coordinate a metal center such as an FeS cluster. Notably, the FocB protein, which is encoded by the last gene of the hyf operon, fails to transport formate (Hunger and Sawers, unpublished), suggesting that its substrate is also different to that of the formate regulon.
The cAMP-CRP protein has also been shown to influence hyf expression, but this might be an indirect effect (135). It is necessary to identify the physiological conditions under which hyf is expressed, and these may well provide clues as to the function of the operon gene products in fermentative metabolism.
BIOCHEMISTRY AND PHYSIOLOGY OF H2-OXIDIZING HYDROGENASES
Hydrogenases 1 and 2
Hyd-1 and Hyd-2 are present at substantial levels when E. coli or Salmonella Typhimurium cells ferment hexoses (141–143), although it is still questionable whether they can be classified as true enzymes of fermentation, because they contribute directly to the establishment of a proton gradient. Both are mainly H2-oxidizing enzymes that are membrane associated, and each has its active site in the periplasm (Fig. 6). It is clear that Hyd-2 is the principle H2-oxidizing activity when E. coli cells grow on H2 and fumarate (37, 141, 142, 144), and the enzyme is probably proton translocating (13, 145). Hyd-1 and the Salmonella Typhimurium-specific homolog Hyd-5 have recently been characterized extensively because they have unique features in common with other O2-tolerant hydrogenases (reviewed in reference 146). The large subunit of the Hyd enzymes is subject to processing (see below) (147, 148), which has been shown to occur C-terminally and is part of the maturation process. The respective small subunits (HyaA and HybO) and an electron-transferring subunit specific to Hyd-2 (HybA) carry an N-terminal Tat (Twin arginine transport) signal peptide that is also proteolytically removed after membrane translocation (see below) (149, 150).
Figure 6.
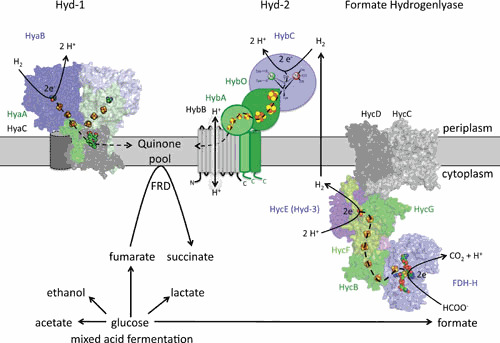
The hydrogenase-1 (Hyd-1), Hyd-2, and formate hydrogenlyase (FHL) complex in the cytoplasmic membrane of E. coli. The horizontal gray bar represents the cytoplasmic membrane. Components in each complex that have similar functions and exhibit amino acid sequence similarity share the same color. Large subunits are shown in blue tones, small subunits in greens, and integral membrane subunits in gray. The metal cofactors are shown as spheres with FeS clusters in brown/yellow, the [NiFe] cofactor in green/brown, and the molybdopterin guanine dinucleotide is shown as spheres in FDH-H. The Hyd-1 structure is based on PDB entry 4GD3 with one heme b molecule; an additional b-type cytochrome subunit has been added as a cylinder. A structure prediction based on complex I is shown for FDH-H and the Hyc components that form the FHL complex. Dashed arrows show the putative path of electron flow through each complex. The lower part of the panel shows the products of the mixed acid fermentation, of which succinate is generated by reduction of fumarate by fumarate reductase (FRD) using electrons derived from the quinone pool. The formate generated is the substrate for the FHL complex, yielding H2, which can be partially reoxidized by Hyd-1 and Hyd-2.
All H2-oxidizing Hyds (EC 1.12.1.-) have been purified (11, 141, 144, 151–154), and structural information is available for Hyd-1 and Hyd-5 (12, 155, 156). Summarized properties of E. coli and Salmonella Typhimurium Hyds are presented in Table 1. Hyd-2 can be purified either as a soluble, active, tryptic fragment that differs from the native membrane-bound enzyme only through the loss of a 5-kDa fragment from the small subunit (144) or by affinity chromatography after replacement of the transmembrane helix on the chromosomally encoded small subunit (HybO) by an affinity tag (154). This C-terminal hydrophobic helix on HybO anchors the catalytic core enzyme of large and small subunit (HybC-HybO) in the membrane, where it subsequently associates with the HybA-HybB dimer to transfer electrons into the quinone pool (157, 158). The HybA subunit is required to mediate bidirectional electron transport to and from the menaquinone pool. Hyd-2-dependent H2 production can be observed under glycerol-fermenting conditions, and this is dependent on the proton motive force (pmf), suggesting it might act as a “valve” to off-load excess reducing equivalents (13). Direct electron transfer via HybA to the quinone pool, the lack of a heme-binding site in the membrane anchor subunit, and the H2-driven proton-pumping activity of Hyd-2 suggest that a progenitor of this class of H2-oxidizing enzyme might have played an important role early in evolution by the coupling of H2-based metabolism to the generation of a transmembrane electrochemical proton gradient.
In contrast to Hyd-2, Hyd-1 is a heterotrimeric enzyme with an α-subunit (HyaB) and β-subunit (HyaA) that associate with a third membrane-bound cytochrome b γ-subunit (HyaC) after membrane translocation (Fig. 6, Table 1). Together, they form a dimer of trimers that cannot be released from the membrane fraction by proteolysis but can be readily solubilized through the action of detergents such as Triton X-100 (141, 152, 156, 158, 159).
General information about the Hyd catalytic mechanism has been garnered through examination of the available crystal structures. A conserved arginine directed toward the [NiFe] catalytic site was recently suggested to function as a general base in the catalytic mechanism, and its substitution with other amino acids resulted in virtually inactive enzyme (160). A recent crystal structure of Hyd-1 that includes HyaC, but that lacks a second heme b molecule, and has only one shared γ-subunit for the α2β2-complex, revealed the importance of the proximity of the two distal [4Fe-4S] clusters in HyaA for O2-tolerance of the enzyme because they can exchange electrons (156). It could be shown that the αβ-dimer is inactivated faster in the presence of 10% O2 than the α2β2-tetramer, but both forms similarly reactivate upon O2 removal, indicating that the active site only forms the so-called Ni-B state (161). The distal [4Fe-4S] cluster has also been suggested to be required for H2 production by Hyd-1, which so far could only be observed in vitro at a pH below 4 (162). Furthermore, the cluster proximal to the active site is a high-potential [4Fe-3S] cluster that provides the active site with two electrons, allowing rapid reactivation of the enzyme after O2 exposure (155). This [4Fe-3S] cluster is coordinated by 2 additional cysteinyl residues, a conserved feature in all O2-tolerant Hyds (163). Like Hyd-5 from Salmonella Typhimurium, Hyd-1 and other oxygen-tolerant Hyds have a conserved H229 in the large subunit that also interacts with the proximal [4Fe-3S] cluster of the small subunit and confers oxygen tolerance (12). Moreover, a structurally adjacent glutamic acid (E73) was shown to be necessary for the oxygen tolerance and catalysis of Hyd-5 (12).
It has been suggested that H2-dependent reduction of O2 catalyzed by Hyd-1 is of physiological relevance because it might afford protection against aerobic shock during passage through the intestine (156). Such a mechanism would require that electron transfer be directed toward the active site rather than into the quinone pool. The Hyd-1-specific coupling of H2 oxidation to O2 reduction was reported 15 years ago (164) and is also indirectly reflected by the specific interaction of Hyd-1 with nitroblue tetrazolium (Eh value of −80 mV) as electron acceptor (165). Although the ultimate physiological electron acceptor for Hyd-1 remains unknown, these findings suggest that Hyd-1 confers flexibility in electron transfer upon E. coli, allowing it to switch rapidly between anaerobiosis and aerobiosis via coupling H2 oxidation to the reduction of high-potential electron acceptors, for example, dimethyl sulfoxide, nitrate, or O2. In contrast, Hyd-2 is functional in the low redox potential range (164). Although Hyd-1 does not significantly contribute to H2 oxidation under fermentative conditions, its protein levels increase when glucose metabolism is restricted (166, 167).
In general, Hyd-2 couples H2 oxidation to reduction of electron acceptors with low midpoint potentials, while Hyd-1 is unable to couple electron transfer effectively to these acceptors (154, 168). Taken together, these findings suggest that Hyd-2 functions optimally in the redox range between −200 and −100 mV, while Hyd-1 is optimal in the range between +50 and +150 mV (154, 169). If substantiated, these results would suggest that Hyd-1 and Hyd-2 provide complementary H2-oxidation activities, covering a range of redox potentials to which facultative anaerobes such as E. coli might be exposed.
The finding that Hyd-1 and Hyd-2 respond differentially to external pH also indicates a complementary role of the two enzymes in anaerobic hydrogen metabolism (170) and is entirely consistent with the slightly alkaline pH optimum of Hyd-2 and the slightly acidic pH optimum of Hyd-1 (141, 144). Furthermore, the strong response of hya (encoding Hyd-1) expression to acidic pH and the stationary phase indicates a possible role for Hyd-1 in stress survival, perhaps through maintenance of the pmf via energy-conserving dihydrogen oxidation (170).
It has been proposed that either Hyd-1 or Hyd-2, or both, could serve the function of recycling the H2 evolved by the FHL complex during fermentation (141, 142). Such a H2-recycling mechanism could be useful in facilitating redox balance, for example, when particularly reducing substrates such as sugar alcohols are oxidized in the absence of exogenous electron acceptors (171). Supporting evidence for this has been obtained by growing E. coli on sorbitol, whereby cells produce excess ethanol and increased amounts of succinate relative to acetate and formate, which cannot be accounted for by calculating redox balance using standard fermentation pathways (172). One means of accounting for the excess ethanol production would be if the reducing equivalents from formate were recycled and channeled to fumarate via the quinone pool. This could be achieved by Hyd-1- or Hyd-2-dependent reoxidation of some of the H2 produced by the FHL complex. Indeed, Alam and Clark (172) could show that in a hypB mutant, which is incapable of synthesizing any active Hyd enzymes, the amount of ethanol and succinate produced was significantly decreased.
A much clearer distinction of the role of the various H2-oxidizing Hyd enzymes has been revealed by investigation of H2 metabolism in Salmonella Typhimurium (173). It could be shown that all three H2-uptake Hyds in Salmonella Typhimurium are required to show a pathogenic phenotype in a mouse model and strains deficient in Hyd-1, Hyd-2, and Hyd-5 are greatly attenuated (10, 174). The main contribution to survival and H2-dependent growth in the host is made by the Hyd-2 enzyme (175). Notably, however, the Hyd-1 enzyme is responsible for the oxidation of all H2 produced by the bacterium during fermentation and is the main enzyme that oxidizes exogenously added H2 (176). Because the hyd operon (encoding Hyd-5) is maximally expressed under aerobic conditions, as occurs in the liver and spleen, it has been suggested that Hyd-5 couples O2 reduction to the respiratory electron transport chain (10, 173). Indirect support for this proposal is provided by the fact that the hyd operon encodes its own homologs of HypA (HydI), HypC (HydE), and a scaffold protein known from Ralstonia eutropha HoxV (HydH) (Fig. 3, Table 3), which has features typical of accessory proteins required for enzyme maturation in the presence of O2 (11). When colonizing a host, the ability to oxidize H2 and couple this to respiration of different electron acceptors might contribute to bacterial survival; however, recycling of fermentative H2 is likely to be negligible because the majority of H2 is derived from the surrounding gut microbiota rather than from FHL (177). Attenuated strains of Salmonella Typhimurium were reported to have a defect in the rpoS gene, and consequently in Hyd synthesis (178), but our own single-nucleotide polymorphism analysis in comparison with strain LT2 revealed that the attenuated strain LT2a used in our laboratory carries a point mutation in HybO resulting in a G190D amino acid exchange. This amino acid is adjacent to Cys191, which coordinates the [4Fe-4S] cluster, and the variant lacks Hyd-2 activity, presumably due to the absence of the small subunit, despite having an intact rpoS gene (Pinske and Sargent, unpublished). Thus, the potential significance of RpoS deficiency in strain attenuation requires further study.
Table 3.
Function of the gene products of the hya, hyb, and hyd operons
| Gene product | Gene length (bp)/molecular mass (kDa) | Function/characteristics |
|---|---|---|
| HyaA | 1119/40.62 (35.64)a | Hyd-1 small subunit |
| HyaB | 1794/66.16 (64.59)a | Hyd-1 large subunit |
| HyaC | 708/27.55 | Hyd-1 b-type cytochrome membrane subunit |
| HyaD | 588/21.50 | Endopeptidase for HyaB, homology with HybD |
| HyaE | 399/14.85 | Chaperone, thioredoxin fold, interaction with signal peptide of small subunit |
| HyaF | 858/31.38 | Chaperone, possible interaction with small subunit precursor, redundancy with HyaE |
| HybO | 1119/39.59 (35.74)a | Hyd-2 small subunit |
| HybA | 328/35.91 (33.21)a | Hyd-2 ferredoxin-type subunit |
| HybB | 1179/43.53 | Hyd-2 integral membrane subunit |
| HybC | 1704/62.40 (60.88)a | Hyd-2 large subunit |
| HybD | 495/17.71 | Endopeptidase for HybC, homology with HyaD |
| HybE | 489/17.92 | Chaperone, recognizes HybO Tat signal and HybC C-terminal extension and coordinates dimer export |
| HybF | 342/12.66 | Nickel insertion in HybC and HyaB, homology with HypA |
| HybG | 249/8.78 | Delivery of Fe(CN)2CO cofactor to HybC and HyaB, homology with HypC |
| HydA (STM1539) | 1104/39.97 (34.86)a | Hyd-5 small subunit |
| HydB (STM1538) | 1803/66.65 (64.98)a | Hyd-5 large subunit |
| HydC (STM1537) | 744/29.33 | Hyd-5 b-type cytochrome membrane subunit |
| HydD (STM1536) | 609/22.10 | Endopeptidase for HydB |
| HydE (STM1535) | 300/10.74 | Delivery of Fe(CN)2CO cofactor to HydB, homology with HypC |
| HydF (STM1534) | 411/15.23 | HyaE-like chaperone |
| HydG (STM1533) | 1062/39.41 | HyaF homolog |
| HydH (STM1532) | 888/32.85 | HoxV (Ralstonia eutropha) homolog |
| HydI (STM1531) | 342/12.61 | Nickel insertion in HydB, Homology with HypA |
Molecular mass after protein specific processing.
Genetics of Hyd-1 and Hyd-2
The structural genes of Hyd-1 are encoded by the hya operon located between 1,031 and 1,036 kbp on the E. coli chromosome (179, 180). The hya operon includes six genes with hyaA-C encoding the structural subunits of the enzyme (Fig. 3, Table 3). The hyaD-F gene products are essential for synthesis of fully functional Hyd-1 (179, 181, 182). HyaD is an ortholog of HybD and is the specific protease required for maturation of the Hyd-1 large (α) subunit (148, 183). The roles of HyaE and HyaF are not entirely understood, except that they interact with the small subunit and with the Tat machinery. They might control small-subunit maturation, but both proteins exhibit redundancy in E. coli (158, 179, 182).
Mutants specifically defective in Hyd-2 biosynthesis have been isolated (184, 185). The mutations are likely to be located within the structural genes of the hyb operon, which encodes Hyd-2 (27, 186). The operon comprises eight genes located between 3,138 and 3,144 kbp on the E. coli chromosome (Fig. 3, Table 3). The first four genes, hybOABC, encode structural components of the enzyme (150, 186). The hybA gene was originally designated as the small subunit based on sequence homologies with the third subunit of the Hyd of Wolinella succinogenes (186, 187). Amino acid sequence analysis of the purified Hyd-2 small subunit revealed that it is encoded by an initially unidentified gene, termed hybO, and this gene is located immediately upstream of hybA (150). Transcriptional studies confirmed the reassigned operon structure (188). The HybO protein has a 37-amino-acid signal sequence that has the characteristic RRXFXK signature of the Sec-independent Tat pathway (149, 150). The fact that HybC lacks a signal sequence indicates that the HybC and HybO proteins fold and form a complex with bound cofactors in the cytoplasm prior to export to the periplasmic face of the membrane. Notably, the iron-sulfur (FeS) protein HybA also has a Tat-signal sequence and is transported independently of the HybO-HybC dimer to the periplasmic side of the membrane (158).
The hybD-G genes are essential for synthesis of a fully active Hyd-2 enzyme. HybD is the protease required for proteolytic processing of HybC after nickel insertion (183). HybE is a homolog of HyaE and interacts with the small subunit HybO (181). HybF is functionally related to HypA and is required for Ni2+ delivery to large subunits of Hyd-1 and Hyd-2 (189, 190). HybG exhibits amino acid similarity with HypC and is required for guiding the maturation machinery to the large subunits of Hyd-1 and Hyd-2 and insertion of the Fe(CN)2CO moiety ([191–193]; see also below).
The large and small subunits of both Hyd-1 and Hyd-2 share extensive similarities with the respective hydrogenase polypeptides from other organisms, and the various implications these homologies may have with regard to the structure and function of hydrogenases in general have been reviewed in detail (4, 194–200).
Regulation of hya and hyb Gene Expression
Regulation of hya and hyb operon expression has been examined and is complex (170, 188, 201, 202). Studies of enzyme levels indicate that Hyd-1 and Hyd-2 are anaerobically inducible (37, 159), and this has been confirmed to be due to transcriptional regulation by both transcript analysis (188) and through the use of lacZ fusions (170, 188, 201, 202). Expression of the hya operon is induced 50-fold during anaerobiosis, while hyb operon expression is induced 10-fold after anaerobic growth on glucose and 20-fold when E. coli is grown on glycerol and fumarate. Although an fnr mutation was shown to reduce significantly the levels of active Hyd-1 and Hyd-2 in anaerobically grown cells (37, 143), this was subsequently shown to be indirect and due to FNR-dependent control of nickel operon expression (203, 204). Consequently, fnr mutants, such as BL21(DE3) strains, can be phenotypically complemented by supplementation with exogenous nickel (127). Expression studies confirmed that FNR control of hya and hyb is indirect and that, in the case of hya, the ArcA two-component transcriptional regulator (205) and AppY control anaerobic induction (170, 188, 201, 202). AppY appears to be part of a regulatory cascade, and expression of the appY gene is negatively regulated by the two-component DpiAB system in aerobically grown cells (206). Furthermore, lacZ-fusions showed that IscR represses the expression of the hyaA and hybO promoters under aerobic conditions (130). IscR can bind a [2Fe-2S] cluster in response to FeS-cluster assembly, and both the apoprotein, lacking the [2Fe-2S] cluster, and the [2Fe-2S]-cluster-containing form of the protein control the expression of genes encoding FeS proteins, including those coding for the ISC pathway itself (130). Binding of IscR to the hya promoter is independent of its FeS cluster (207, 208). Instead, AppY and ArcA act as antirepressors and directly compete with IscR binding under anaerobic conditions, and therefore increase hya transcription (209).
Expression of hya also depends on the stationary phase sigma factor RpoS (170, 201), with operon expression being maximal in early-stationary phase. King and Pryzbyla (170) have also shown that hya is expressed maximally when the external pH of the growth medium is acidic and expression is abrogated under alkaline conditions. ArcA is required for pH regulation. Expression of hyb has the opposite response to external pH.
Precisely how hyb expression is controlled in response to anaerobic induction is unclear. Although anaerobic induction is reduced in an arcA fnr double null mutant, an approximate 5-fold anaerobic induction is still evident (188). AppY is not involved in controlling hyb expression; however, cAMP-CRP appears to have an indirect effect on expression. It is conceivable that cAMP-CRP controls expression of a regulatory gene whose product, in turn, controls hyb operon expression directly.
Salmonella Typhimurium has the FHL pathway, as well as three H2-oxidizing Hyd (Hyd-1, Hyd-2, and Hyd-5) (143, 173, 210). As with the E. coli enzyme, synthesis of Hyd-1 is induced at acid pH in Salmonella Typhimurium (211). However, in contrast to hya in E. coli, expression of the hya operon in Salmonella Typhimurium is absolutely dependent on the cAMP-CRP complex. Surprisingly, expression of hya in Salmonella Typhimurium has an absolute requirement for the tyrosine-dependent regulator TyrR. No dependence of E. coli hya on tyrosine has been reported. In contrast to anaerobic expression of hya and hyb, the hyd operon (Hyd-5) is expressed maximally under aerobic conditions in Salmonella Typhimurium, and this is also consistent with the finding that the genes for maturation factors necessary for aerobic biosynthesis of the [NiFe] cofactor are encoded within the operon (173) (Fig. 3).
Anaerobic expression of both the hya and hyb operons in E. coli is reduced when nitrate is provided in the growth medium (188). The dual nitrate-responsive two-component systems NarXL and NarQP (9) are clearly involved in mediating nitrate repression. Nitrate repression of hyb operon expression can be accounted for solely through the NarXL and NarQP systems (188), although, surprisingly, anaerobic induction of hyb expression is abolished in a narP knockout mutant. This suggests that NarL, in the absence of NarP, represses operon expression.
Nitrate regulation of hya is more complex and, in the absence of both NarX and NarQ, nitrate repression is partially relieved but not to the extent that expression levels attain those observed in anaerobic, glucose-grown cultures. This suggests the involvement of an additional system in mediating nitrate repression of hya expression.
Additionally, posttranscriptional regulation of hya expression was suggested, adding another level of complexity to the control of Hyd-1 synthesis (166). Finally, posttranslational regulation is also exerted through the unusually long N-terminal domain of the HyaA Tat-signal peptide, which acts as a regulatory domain controlling membrane transport (212).
ASSEMBLY AND MATURATION OF THE HYDROGENASES
Structure of the Metal Center
The X-ray structures of [NiFe]-Hyds from various biological sources have been determined (Fig. 7) (for reviews, see references 199 and 213). The two subunits of the heterodimeric enzyme contact each other over an extraordinarily large interface, and the [NiFe] cofactor is located in the interior of the large subunit, close to the interface. The small subunit contains a specific number of FeS clusters that transfer the electrons to or from the catalytic [NiFe] metal center (Table 1) (155, 213). The bimetallic center is coordinated within the protein via four cysteinyl thiolates derived from the large subunit. Two of these cysteinyl residues function as bridging ligands between the Ni and Fe (Fig. 7, inset). One pair of cysteinyl residues is present in a strongly conserved motif located in the N-terminal domain of the large subunit, while the second pair is located close to the C terminus of the matured subunit. As disclosed by infrared spectroscopy (214–217) and X-ray analysis (218, 219), the Fe of the [NiFe] cofactor contains three diatomic ligands: two are cyano groups and the third is a carbonyl moiety (220).
Figure 7.
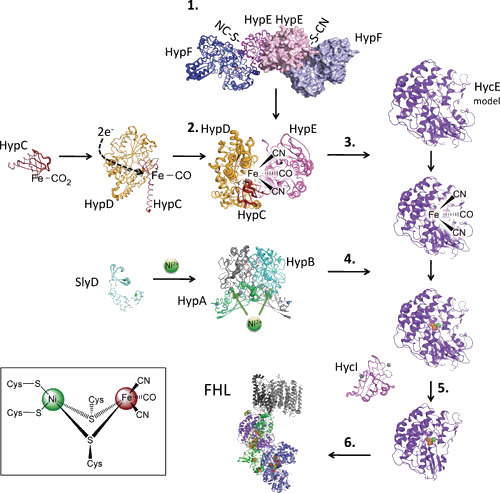
Postulated pathway of the maturation of hydrogenase 3 from E. coli. For details, see the text. The inset shows the [NiFe] cofactor. The proteins are represented as structures (not to scale) and are modified from PDB files: 5AUO (HypAB), 2Z1C (HypC), 3VYR (HypCD), 3VYS + 3VYU + 3WJQ (HypCDE), 3VTI (HypEF), 2E85 (HycI), 3CGM (SlyD), while HycE and FHL are models.
Genetic Analysis
Early work from a number of groups had shown that lesions in certain genes different from the structural genes block the generation of active Hyd enzymes. In E. coli, most of these genes were located in the 58- to 59-min region of the chromosome (185, 221–223). Determination of the nucleotide sequence of this region (6, 84) and systematic knockout of each gene by introducing an in-frame deletion revealed that this chromosome segment harbors the genes for the Hyd-3 component of the FHL complex (hyc operon) (28). Further DNA sequence analysis around the hyc operon identified a large locus (Fig. 3) required for synthesis of active Hyd enzymes (6, 84, 224). The genes were designated hyp, because inactivation of most of them (hypB, hypD, hypE, and hypF) affected Hyd formation pleiotropically (75, 224) (Table 4). Exceptions were hypA and hypC, because mutation of these genes mainly affected Hyd-3 (28); however, it was shown later that there are homologs of these genes (hybF and hybG, respectively) in the operon coding for Hyd-2 (Fig. 3, Table 3) (186, 224) and they fulfill the function of HypA and HypC in the formation of Hyd-1 and Hyd-2 (189, 190, 192). Immediately upon discovery of the diatomic cyanide (CN–) and carbon monoxide (CO) ligands attached to the iron of the active site cofactor (218, 220), it became obvious that the Hyp proteins must be involved in [NiFe]-cofactor synthesis, assembly, and insertion. In an early study, a screen for mutants of Salmonella Typhimurium lacking Hyd activity identified pyrA (= carAB) mutants (225). The carAB gene products are the subunits of carbamoyl phosphate synthetase, which generates carbamoyl phosphate, the metabolic precursor for CN– ligand synthesis (226). To date, no equivalent genes have been identified for the synthesis of the CO ligand.
Table 4.
Proteins involved in maturation of [NiFe] hydrogenases from E. coli
| Protein | Molecular mass (kDa) | Function |
|---|---|---|
| HypA/HybF | 13.14/12.66 | Nickel incorporation: delivery to apoenzyme |
| HypB | 31.51 | Nickel incorporation: binding, GTPase |
| HypC/HybG | 9.70/8.78 | CO-synthesis: CO2 delivery to HypCD complex, delivery of Fe(CN)2CO into apoenzyme; active residue Cys2 |
| HypD | 41.30 | CO-synthesis: scaffold protein for CO2 reduction and Fe(CN)2CO assembly, contains [4Fe-4S], active residue Cys41 |
| HypE | 35.00 | CN–-synthesis: ATP-dependent dehydration of the carbamate group to cyanate; active residue Cys336 |
| HypF | 81.95 | CN–-synthesis: produces carbamoyl adenylate from carbamoyl phosphate, transfer to HypE |
| CarAB | 41.35/117.69 | CN–-synthesis: carbamoyl phosphate synthetase from l-glutamine, HCO3–, 2 ATP, H2O |
Maturation involves six main steps: (i) the synthesis of the cyanide ligands from carbamoyl phosphate; (ii) assembly and attachment of all diatomic ligands to the iron atom; (iii) insertion of the iron moiety into the apoenzyme; (iv) nickel insertion; (v) endoproteolytic processing of the C-terminal peptide on the large subunit; (vi) assembly with the small subunit (Fig. 7); and (vii) in the case of periplasmically oriented enzymes, Tat-transport and association with other structural subunits. Therefore, some common phenotypic features of mutants blocked in one of the hyp genes are an accumulation of large subunit precursor (84, 224) and absence of iron (193) and/or nickel in the catalytic subunit (227, 228). Comprehensive reviews on diatomic ligand synthesis can be found here (229, 230).
Several of the hyp operon gene products are also required during respiratory growth conditions (see below). An FNR-dependent promoter is located within the hypA gene to ensure that sufficient levels of these gene products are available for the maturation of catalytically active Hyd enzymes, even in the absence of formate (Fig. 5) (84, 231). Although the original analysis of the hypA promoter suggested FNR-dependent activation occurs aerobically, a reassessment indicated that transcription of the hypA promoter occurs anaerobically and is FNR dependent (231). It is unclear why this discrepancy exists; however, the latter finding is in accord with the oxygen sensitivity of the maturation reaction in E. coli (232).
Biosynthesis of the cyanide ligands
Stimulated by the observation that HypF shares a sequence motif with O-carbamoyltransferases (233–235), it was discovered that carbamoyl phosphate is required for the formation of active hydrogenases (226). Mutants with a lesion in carAB were devoid of hydrogenase activity and the defect could be rescued, in part, by the inclusion of citrulline as a source of carbamoyl phosphate in the growth medium (226) (Fig. 8A). The response to citrulline was augmented when its conversion to arginine was blocked by a mutation in the argG gene and by concomitant overproduction of the ornithine transcarbamoylase protein to overcome the unfavorable equilibrium of the reaction (191). This approach further allowed the use of labeled citrulline to identify carbamoyl phosphate as the precursor for the CN– but not the CO ligand (236, 237), verifying and substantiating the original observations of Roseboom et al. (238).
Figure 8.
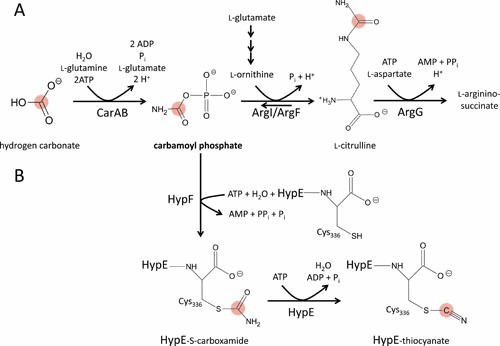
Reactions involving carbamoyl phosphate. The carbamoyl phosphate synthetase (CarAB) phosphorylates hydrogen carbonate in an ATP-dependent reaction. Carbamoyl phosphate serves as the substrate for the ornithine carbamoyltransferases (ArgF/ArgI) during arginine biosynthesis. HypF competes for the use of carbamoyl phosphate in a hydrolysis reaction coupled to the formation of carbamoyl adenylate that can be transferred to HypE. Variations of these pathways are explained in the text. The carbon atom derived from hydrogen carbonate is highlighted in red.
Purified HypF protein accepts carbamoyl phosphate as a substrate. Primary sequence analysis (239, 240) and structural insights from various organisms (234, 241–246) revealed 4 interconnected domains between the N and C terminus, including an acylphosphatase domain, a zinc finger domain, a YrdC-like domain, and finally a Kae1-like domain. The N-terminal acylphosphatase domain hydrolyzes carbamoyl phosphate and passes carbamate through the zinc finger domain to the ATP-binding YrdC domain. This reaction occurs in the absence of other substrates. A carbamoyl adenylate intermediate, which functions as an excellent leaving group for carbamate, is formed in the YrdC domain and is further transferred to HypE by the Kae1 carbamoyltransferase domain (234, 245). In the presence of ATP, HypF catalyzes the carbamoyl phosphate-dependent cleavage of ATP into AMP and pyrophosphate (233). The carbamoyl adenylate intermediate has also been observed in other HypF-related proteins (247). Inclusion of purified HypE in the reaction mixture showed that HypF carbamoylates the C-terminal cysteinyl (Cys336) residue of this protein, resulting in the generation of a protein-S-carboxamide (Fig. 8B) (248). Various structures of HypE exist, some of which show the carbamoylated and cyanated species (234, 244, 246, 249). Furthermore, the interaction of the HypE dimer showed that two HypF proteins interact at opposite ends of the complex (234) (Fig. 7). HypE activates the oxygen of the carboxamide by ATP-dependent phosphorylation followed by dephosphorylation converting the protein-S-carboxamide into the protein-thiocyanate (246, 248) or an isothiocyanate, as recently suggested by IR analysis (250). In summary, HypF functions as a carbamoyltransferase, while HypE catalyzes an ATP-dependent dehydratase reaction (248).
Assembly and attachment of CN– and CO ligands to the iron atom
HypC is required for the maturation of HycE (Hyd-3) and, to some extent, HyaB (Hyd-1), while HybG, which is encoded in the hyb operon, has a homologous role during the maturation of HyaB (Hyd-1) and HybC (Hyd-2) (192). HypC has an N-terminal β-barrel domain, where the terminal cysteinyl residue is exposed, and a C-terminal, flexible α-helix (251, 252) (Fig. 7). Mutants devoid of carbamoyl phosphate synthetase activity, and in which the genes for Hyp proteins were overexpressed, accumulate a HypC-HypD complex (253). When cells harboring this complex were provided with citrulline as a source of carbamoyl phosphate, the complex was converted to a more slowly migrating species, but only when cells were devoid of the large Hyd subunit (191). This was the first circumstantial evidence that at least a portion of the cofactor formed on a separate subcomplex, independent of the Hyd large subunit.
The HypC-HypD proteins probably share one or two iron atoms (254, 255) that serve as a scaffold for Fe-CO-(CN–)2 assembly (256). It was established that the CO ligand does not derive from carbamoyl phosphate (236–238), and different substrates were considered. For the aerobic maturation in Ralstonia, the HypX protein appears to convert the formyl group of N10-formyl-tetrahydrofolate to deliver the CO ligand (257); however, E. coli does not encode a HypX homolog. Instead, HypC enters the complex and has been suggested to deliver iron-bound CO2 (258). The CO2 must be from an intracellular source because exogenously supplied CO2 cannot be incorporated into the active site cofactor (236).
HypD carries a low redox potential [4Fe-4S] cluster that could provide reducing equivalents for the conversion of CO2 to CO (259), but the electron donor for that reaction is unknown. The [4Fe-4S] cluster is within electron-transferring distance to Cys41 on the opposite side of the protein, where the interaction with Cys2 of HypC takes place (251, 255). In this scenario the HypCD proteins carry iron-bound CO before the CN– ligands are attached (260), although the initial attachment of CN– ligands was suggested for the slightly different Ralstonia system (261).
The HypCD complex is also able to interact with HypE (253, 254). The crystal structures of HypF-HypE and the HypCDE complex (234, 254) show that HypE interacts with HypCD at the same interface as with HypF, suggesting a temporal order of events (Fig. 7). The HypE-(iso)thiocyanate transfers the CN– groups to the HypCD complex (253). It is still unclear how the distance of 34 Å between the (iso)thiocyanate on Cys336 in HypE and the Fe in the HypCD complex is overcome. It is thought that, rather than functioning in a sequential manner, the reactions take place in multiprotein complexes. Possibly, the presence of HypE as homodimer and HypCD as heterodimer explains why there are two CN– and one CO ligand attached to the iron. However, more research is required to identify the precise complex interactions and stoichiometries during maturation.
Fe-CO-(CN–)2 insertion into the apoenzyme
Cells in which hydrogenase maturation is blocked at nickel insertion or endoproteolytic processing lack the HypC-HypD complex but instead reveal HypC in a complex with the precursor of the large subunit (262, 263). The C termini of HypC and HybG are important determinants in the interaction with their respective large subunits (Thomas and Sawers, unpublished). A heterologously synthesized minimal version of HypC, together with its HypD counterpart, both derived from a deeply branching member of the Chloroflexi, is able to mature Hyd-1, Hyd-2, and Hyd-3 in an E. coli mutant lacking both endogenous hypC and hybG genes (264). The two homologous proteins HypC and HybG compete with each other during maturation. This can clearly be seen when one of the two partners is overproduced, resulting in a reduction in the maturation of the competing enzyme. Overproduction of HybG, for example, impairs the generation of mature Hyd-3 as it outcompetes HypC for the HypC-HypD complex and itself undergoes complex formation with HypD (191, 192). Under conditions where the FHL system and the hyp operon are fully induced by formate generated during fermentation, the increased amount of HypC formed recruits HypD to Hyd-3 maturation, thus reducing the availability of HypD to interact with HybG. Thus, the balance between HypC and HybG synthesis appears to determine when H2-oxidizing and H2-producing enzymes are synthesized.
In summary, the HypC-HypD complex assembles the Fe-CO-(CN–)2 cofactor and transfers it into the large subunit via HypC (Hyd-1/3) or HybG (Hyd-1/2).
Nickel insertion
Nickel insertion under physiological nickel concentrations in the medium requires the activity of two proteins, namely HypA and HypB, and is kinetically facilitated by the peptidyl prolyl cis-trans-isomerase SlyD (190, 224, 265). Maturation of Hyd-1 and Hyd-2 mainly relies on the HypA homolog HybF (189, 190).
One class of mutants blocked in Hyd maturation can be rescued by supplementation of the medium with nickel concentrations close to 1 mM (266). This indicated that the product, later on shown to be identical with HypB (227), is involved in nickel insertion. This also holds for strains with defective hypA, hybF, or slyD genes, as well as for a mutant lacking all three genes (189, 190, 265, 267, 268). There is convincing evidence that nickel insertion takes place at the precursor of the large hydrogenase subunit after iron incorporation. Arguments in favor of this are that the precursor present in cells grown under nickel limitation can be matured into active enzyme by the provision of nickel and the hydrogenase-specific protease (269). The successful suppression of the hypA, hypB, hybF, and slyD phenotypes by adding high concentrations of nickel to the medium indicates that these gene products are not absolutely required to complete maturation in vivo. Moreover, mutants with a deletion in any of the hyp genes contain a large subunit precursor devoid of nickel, whereas in a strain lacking the endopeptidase the large subunit contains the metal (227, 270).
HypB was first purified from E. coli (227), and later from other organisms (271–273). It was shown to be a GTP-binding and -hydrolyzing protein (228). Indeed, mutant HypB proteins with amino acid substitutions that interfere with hydrolysis of GTP are unable to incorporate nickel (at physiological concentrations) and to mature the large subunit (228, 273–275). HypB from E. coli, therefore, was the first GTPase discovered to have a role in metal insertion into a protein.
HypA, on the other hand, is a nickel-binding protein, as was initially demonstrated for the protein from Helicobacter pylori (267, 273) and subsequently also for HybF from E. coli (190). Apart from binding nickel, the HypA homolog HybF contains stably bound zinc, putatively coordinated in a classical zinc finger domain (190). The biochemical function of the zinc ion remains enigmatic.
Although a vast amount of biochemical data from the proteins involved exists, the mechanism of nickel insertion is not fully understood. HypB binds nickel in its N-terminal domain with high affinity (276). Furthermore, the HypB protein can dimerize upon binding of a nucleotide at the interface (227, 277), which is not essential for its GTPase activity and metal binding but is required for full Hyd activity (278). The nickel ion can be transferred from HypB to HypA (279) during a transient interaction (280, 281). HypA also forms homodimers, but the E. coli HypAB complex seems to include only one heterodimer (280). It was shown in Thermococcus kodakarensis that the interaction of HypA and HypB in a 2:2 complex induces a conformational change in HypA, which increases its nickel-binding affinity (282). However, GTP hydrolysis to GDP might be sufficient to transfer nickel from HypB to HypA in E. coli (279). In addition, SlyD interacts with HypB to facilitate nickel release (283, 284). Subsequently, both HypA and SlyD, but not HypB, were shown to interact directly with the large subunit to deliver nickel (285, 286). Nevertheless, an alternative nickel route from HypB to the large subunit HycE via SlyD has been proposed, where HypA would only mediate protein interaction (229). The nickel insertion complex of HypA, HypB, and SlyD forms also in the absence of Hyd enzymes (286), and the challenge will be to understand what the proteins do in this complex. A comprehensive review about nickel delivery with comparisons with other organisms can be found in reference 229.
Endoproteolytic maturation of the precursor
The last step in the maturation process involves the endoproteolytic removal of a short peptide from the C terminus of the hydrogenase large-subunit precursor (39, 287). The endopeptidases responsible are substrate specific and are encoded within the respective operon of the structural genes (Fig. 3). E. coli possesses at least three of these, namely HyaD, HybD, and HycI, with specificity for Hyd-1, Hyd-2, and Hyd-3, respectively (42, 179, 186). As discussed above, it is not known whether Hyd-4 requires a protease or shares HycI with HycE (7).
C-terminal processing only occurs after nickel has been inserted, indicating that the metal determines when cleavage occurs (288). The cleavage site is located three amino acid residues C-terminal to the fourth cysteinyl residue required for binding the [NiFe] cofactor. The peptide that is removed differs in length between Hyd enzymes and is 15 amino acids for Hyd-1 (HyaB) and Hyd-2 (HybC), but is 32 amino acids for Hyd-3 (HycE). It is similarly predicted to be 32 amino acids for Hyd-4 (HyfG).
A comparison of cleavage sites from various precursors reveals a striking conservation of amino acids. The amino acid at position −1 is usually a His or an Arg, and at position +1 a nonpolar residue is found (Val, Met). Surprisingly, however, certain amino acid substitutions introduced into the precursor of Hyd-3 reveal a remarkable tolerance. Thus, exchange of the Arg by other basic or nonpolar residues did not influence cleavage; acidic and large nonpolar residues, however, yielded an enzymatically inactive, yet nevertheless processed subunit (148, 289). Replacement of the +1 Met residue by acidic residues abolished processing, whereas substitution by other nonpolar amino acids was tolerated. Furthermore, the replacement by polar residues greatly destabilized the whole protein (290) (Theodoratou and Böck, unpublished results, cited in reference 148).
Extension of the peptide of both HyaB (Hyd-1) and HybC (Hyd-2) resulted in a lack of nickel insertion and the inability of the precursor to be processed (291). Truncations down to a critical lower limit in size, on the other hand, were better tolerated with regard to processing, but the overall stability of the total precursor was dramatically reduced. This indicates that the C-terminal extension interacts with the main body of the molecule, stabilizing it in a particular conformation (290, 292).
HybC was used to show that the C-terminal extension prevents premature folding of the protein when the cofactor is not inserted (193). Similarly, it was shown that the precursor of the large subunit undergoes a dramatic change in its electrophoretic mobility in nondenaturing polyacrylamide gels after cleavage (270). The C-terminal extension does not appear to interact directly with the maturation machinery (193) but nevertheless possibly modulates the interaction of the Hyp complex with the apoenzyme (Thomas and Sawers, unpublished).
The proteases HybD and HycI have been purified, and the proteolytic activity of HycI has been demonstrated in vitro (42, 183, 269). HybD could be crystallized with cadmium in the crystallization buffer. In the crystals of the protein, which has an α/β-structure, one cadmium ion is bound by HybD within a cleft, possibly representing the active site (270). The cadmium is penta-coordinated by the carboxylate oxygens of a Glu and an Asp, the imidazole nitrogen of a His side chain, and a water molecule (183). Replacement of these residues by nonsimilar amino acids completely abolishes enzymatic activity, whereas mutant proteins with chemically similar amino acid substitutions retain residual activity. The nuclear magnetic resonance and crystal structures of HycI revealed a congruent fold, and the nickel-binding site identified through cadmium binding is coordinated by Asp16, Asp62, and His90, as predicted (270, 293, 294). In the presence of calcium, two further metal-binding sites were identified in the structure (293). As purified HybD and HycI proteins are devoid of metal and also do not bind nickel tightly in vitro, it was concluded that the nickel present in the precursor of the large subunit is used as a recognition motif for the endopeptidase (148, 270, 295). Indeed, whereas proteolytic processing of a nickel-free precursor of the large subunit is inhibited by addition of a nickel-complexing agent, this was not observed when nickel had already been incorporated (270).
Altogether, the C-terminal extension appears to function in “freezing” the polypeptide in an open conformation, allowing metal cofactor insertion, rather like a baited mousetrap. Cleavage of the C-terminal peptide springs the trap and results in the final, active conformation of the protein.
Protein assembly
Differences also exist between the further maturation process of the Hyd-3 enzyme and that of Hyd-1 and Hyd-2. Hyd-3 is attached to the inner side of the cytoplasmic membrane (28), and maturation can take place in the absence of the other subunits. However, assembly of the core complex is primed only after the proteolytic cleavage of HycE (Pinske and Sargent, unpublished).
Tat transport
In contrast to Hyd-3, Hyd-1 and Hyd-2 are transported to the periplasmic side of the membrane as fully matured and assembled heterodimers (157). It is currently unclear why the small subunits for the Tat-dependent hydrogenases are encoded upstream of the large subunits while the FHL small subunit is encoded downstream of HycE and whether this has implications for FeS insertion and complex assembly. Current evidence suggests, however, that maturation of the large and small subunits of the Tat-dependent Hyds co-occurs in a large assembly complex (Braussemann and Sawers, unpublished). The small subunits of Hyd-1 and Hyd-2 carry an N-terminal Tat signal sequence and transfer the mature large subunits “piggyback-style” through the membrane (296, 297). Genetic removal of the 15 amino acid C-terminal extension from the large subunit of Hyd-2 resulted in Tat-dependent translocation of a HybO-HybC heterodimer that was inactive because the [NiFe] cofactor was missing (193). Therefore, it seems that the large-subunit and the small-subunit heterodimer complex, as is found in the final cofactor-containing enzyme, is prevented from forming because of the incorrect conformation adopted by the large-subunit precursor. Only after successful cofactor insertion of the large subunit has occurred, and the conformation of the mature polypeptide has been adopted, is a tight interaction with the small subunit possible. Premature export of the small subunit is prevented by degradation of the small subunit in the absence of the correctly matured and folded large subunit. This latter process is controlled further by enzyme-specific chaperones (see reference 298).
ACKNOWLEDGMENTS
Prof. Frank Sargent is thanked for his valuable discussions. R.G.S. and C.P. receive support from the DFG (projects SA 494/3, SA 494/7, and PI 1252/2). August Böck and Melanie Blokesch are thanked for their invaluable contributions to a previous version of this review.
Conflict of interest: The authors declare no conflicts.
REFERENCES
- 1.Schuchmann K, Müller V. 2013. Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342:1382–1385. [PubMed] 10.1126/science.1244758 [DOI] [PubMed] [Google Scholar]
- 2.Pinske C, Sargent F. 2 May 2016. Exploring the directionality of Escherichia coli formate hydrogenlyase: a membrane-bound enzyme capable of fixing carbon dioxide to organic acid. Microbiologyopen 10.1002/mbo3.365. 10.1002/mbo3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda T, Sanchez-Torres V, Wood TK. 2008. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol 1:30–39. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sargent F. 2016. The Model [NiFe]-Hydrogenases of Escherichia coli. Adv Microb Physiol 68:433–507. [PubMed] 10.1016/bs.ampbs.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 5.Sawers RG, Clark DP. 2004. Fermentative pyruvate and acetyl-coenzyme a metabolism. Ecosal Plus 10.1128/ecosalplus.3.5.3. [PubMed] 10.1128/ecosalplus.3.5.3 [DOI] [PubMed] [Google Scholar]
- 6.Böhm R, Sauter M, Böck A. 1990. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol 4:231–243. [PubMed] 10.1111/j.1365-2958.1990.tb00590.x [DOI] [PubMed] [Google Scholar]
- 7.Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P, Guest JR. 1997. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 143:3633–3647. [PubMed] 10.1099/00221287-143-11-3633 [DOI] [PubMed] [Google Scholar]
- 8.Rossmann R, Sawers G, Böck A. 1991. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol 5:2807–2814. [PubMed] 10.1111/j.1365-2958.1991.tb01989.x [DOI] [PubMed] [Google Scholar]
- 9.Stewart V. 2003. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem Soc Trans 31:1–10. [PubMed] 10.1042/bst0310001 [DOI] [PubMed] [Google Scholar]
- 10.Zbell AL, Maier SE, Maier RJ. 2008. Salmonella enterica serovar Typhimurium NiFe uptake-type hydrogenases are differentially expressed in vivo. Infect Immun 76:4445–4454. [PubMed] 10.1128/IAI.00741-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkin A, Bowman L, Roessler MM, Davies RA, Palmer T, Armstrong FA, Sargent F. 2012. How Salmonella oxidises H2 under aerobic conditions. FEBS Lett 586:536–544. [PubMed] 10.1016/j.febslet.2011.07.044 [DOI] [PubMed] [Google Scholar]
- 12.Bowman L, Flanagan L, Fyfe PK, Parkin A, Hunter WN, Sargent F. 2014. How the structure of the large subunit controls function in an oxygen-tolerant [NiFe]-hydrogenase. Biochem J 458:449–458. [PubMed] 10.1042/BJ20131520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinske C, Jaroschinsky M, Linek S, Kelly CL, Sargent F, Sawers RG. 2015. Physiology and bioenergetics of [NiFe]-hydrogenase 2-catalyzed H2-consuming and H2-producing reactions in Escherichia coli. J Bacteriol 197:296–306. [PubMed] 10.1128/JB.02335-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Böck A, Thanbichler M. 2004. Selenocysteine. EcoSal Plus 10.1128/ecosalplus.3.6.1.1. [PubMed] 10.1128/ecosalplus.3.6.1.1 [DOI] [PubMed] [Google Scholar]
- 15.Magalon A, Mendel RR. 2015. Biosynthesis and insertion of the molybdenum cofactor. EcoSal Plus 10.1128/ecosalplus.ESP-0006-2013. [PubMed] 10.1128/ecosalplus.ESP-0006-2013 [DOI] [PubMed] [Google Scholar]
- 16.Fontecave M, Py B, Ollagnier-de Choudens S, Barras F. 2008. From iron and cysteine to iron-sulfur clusters: the biogenesis protein machineries. EcoSal Plus 10.1128/ecosalplus.3.6.3.14. 10.1128/ecosalplus.3.6.3.14 [DOI] [PubMed] [Google Scholar]
- 17.Sawers RG, Blokesch M, Böck A. 2004. Anaerobic formate and hydrogen metabolism. EcoSal Plus 10.1128/ecosalplus.3.5.4. [PubMed] 10.1128/ecosalplus.3.5.4 [DOI] [PubMed] [Google Scholar]
- 18.Stephenson M, Stickland LH. 1931. Hydrogenase: a bacterial enzyme activating molecular hydrogen: the properties of the enzyme. Biochem J 25:205–214. [PubMed] 10.1042/bj0250205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephenson M, Stickland LH. 1932. Hydrogenlyases: bacterial enzymes liberating molecular hydrogen. Biochem J 26:712–724. [PubMed] 10.1042/bj0260712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinsent J. 1954. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem J 57:10–16. [PubMed] 10.1042/bj0570010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peck HD Jr, Gest H. 1957. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol 73:706–721. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gest H, Peck HD Jr. 1955. A study of the hydrogenlyase reaction with systems derived from normal and anaerogenic coli-aerogenes bacteria. J Bacteriol 70:326–334. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pecher A, Zinoni F, Jatisatienr C, Wirth R, Hennecke H, Böck A. 1983. On the redox control of synthesis of anaerobically induced enzymes in enterobacteriaceae. Arch Microbiol 136:131–136. [PubMed] 10.1007/BF00404787 [DOI] [PubMed] [Google Scholar]
- 24.Pecher A, Zinoni F, Böck A. 1985. The seleno-polypeptide of formic dehydrogenase (formate hydrogen-lyase linked) from Escherichia coli: genetic analysis. Arch Microbiol 141:359–363. [PubMed] 10.1007/BF00428850 [DOI] [PubMed] [Google Scholar]
- 25.Zinoni F, Birkmann A, Stadtman TC, Böck A. 1986. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA 83:4650–4654. [PubMed] 10.1073/pnas.83.13.4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zinoni F, Birkmann A, Leinfelder W, Böck A. 1987. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci USA 84:3156–3160. [PubMed] 10.1073/pnas.84.10.3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawers G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie van Leeuwenhoek 66:57–88. [PubMed] 10.1007/BF00871633 [DOI] [PubMed] [Google Scholar]
- 28.Sauter M, Böhm R, Böck A. 1992. Mutational analysis of the operon (hyc ) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol 6:1523–1532. [PubMed] 10.1111/j.1365-2958.1992.tb00873.x [DOI] [PubMed] [Google Scholar]
- 29.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. 2014. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci USA 111:E3948–E3956. [PubMed] 10.1073/pnas.1407927111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. 2013. Crystal structure of the entire respiratory complex I. Nature 494:443–448. [PubMed] 10.1038/nature11871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. [PubMed] 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 32.Trchounian A, Gary Sawers R. 2014. Novel insights into the bioenergetics of mixed-acid fermentation: can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life 66:1–7. [PubMed] 10.1002/iub.1236 [DOI] [PubMed] [Google Scholar]
- 33.Friedrich T, Scheide D. 2000. The respiratory complex I of bacteria, archaea, and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett 479:1–5. [PubMed] 10.1016/S0014-5793(00)01867-6 [DOI] [PubMed] [Google Scholar]
- 34.Albracht SP. 1993. Intimate relationships of the large and the small subunits of all nickel hydrogenases with two nuclear-encoded subunits of mitochondrial NADH: ubiquinone oxidoreductase. Biochim Biophys Acta 1144:221–224. [PubMed] 10.1016/0005-2728(93)90176-G [DOI] [PubMed] [Google Scholar]
- 35.Batista AP, Marreiros BC, Pereira MM. 2013. The antiporter-like subunit constituent of the universal adaptor of complex I, group 4 membrane-bound [NiFe]-hydrogenases and related complexes. Biol Chem 394:659–666. [PubMed] 10.1515/hsz-2012-0342 [DOI] [PubMed] [Google Scholar]
- 36.Marreiros BC, Batista AP, Duarte AMS, Pereira MM. 2013. A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. Biochim Biophys Acta 1827:198–209. [PubMed] 10.1016/j.bbabio.2012.09.012 [DOI] [PubMed] [Google Scholar]
- 37.Sawers RG, Ballantine SP, Boxer DH. 1985. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J Bacteriol 164:1324–1331. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDowall JS, Hjersing MC, Palmer T, Sargent F. 2015. Dissection and engineering of the Escherichia coli formate hydrogenlyase complex. FEBS Lett 589(20 Pt B):3141–3147. [PubMed] 10.1016/j.febslet.2015.08.043 [DOI] [PubMed] [Google Scholar]
- 39.Rossmann R, Sauter M, Lottspeich F, Böck A. 1994. Maturation of the large subunit (HYCE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg537. Eur J Biochem 220:377–384. [PubMed] 10.1111/j.1432-1033.1994.tb18634.x [DOI] [PubMed] [Google Scholar]
- 40.Maeda T, Sanchez-Torres V, Wood TK. 2008. Protein engineering of hydrogenase 3 to enhance hydrogen production. Appl Microbiol Biotechnol 79:77–86. [PubMed] 10.1007/s00253-008-1416-3 [DOI] [PubMed] [Google Scholar]
- 41.Friedrich T. 2014. On the mechanism of respiratory complex I. J Bioenerg Biomembr 46:255–268. [PubMed] 10.1007/s10863-014-9566-8 [DOI] [PubMed] [Google Scholar]
- 42.Rossmann R, Maier T, Lottspeich F, Böck A. 1995. Characterisation of a protease from Escherichia coli involved in hydrogenase maturation. Eur J Biochem 227:545–550. [PubMed] 10.1111/j.1432-1033.1995.tb20422.x [DOI] [PubMed] [Google Scholar]
- 43.Leonhartsberger S, Ehrenreich A, Böck A. 2000. Analysis of the domain structure and the DNA binding site of the transcriptional activator FhlA. Eur J Biochem 267:3672–3684. [PubMed] 10.1046/j.1432-1327.2000.01399.x [DOI] [PubMed] [Google Scholar]
- 44.Jaroschinsky M, Sawers RG. 2014. Ferredoxin has a pivotal role in the biosynthesis of the hydrogen-oxidizing hydrogenases in Escherichia coli. Int J Hydrogen Energy 39:18533–18542. 10.1016/j.ijhydene.2014.03.124 [DOI] [Google Scholar]
- 45.Pinske C, Sawers RG. 2012. Delivery of iron-sulfur clusters to the hydrogen-oxidizing [NiFe]-hydrogenases in Escherichia coli requires the A-type carrier proteins ErpA and IscA. PLoS One 7:e31755. [PubMed] 10.1371/journal.pone.0031755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinske C, Jaroschinsky M, Sawers RG. 2013. Levels of control exerted by the Isc iron-sulfur cluster system on biosynthesis of the formate hydrogenlyase complex. Microbiology 159:1179–1189. [PubMed] 10.1099/mic.0.066142-0 [DOI] [PubMed] [Google Scholar]
- 47.Takahashi Y, Tokumoto U. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem 277:28380–28383. [PubMed] 10.1074/jbc.C200365200 [DOI] [PubMed] [Google Scholar]
- 48.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. 2013. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827:455–469. [PubMed] 10.1016/j.bbabio.2012.12.010 [DOI] [PubMed] [Google Scholar]
- 49.Pinske C, Sawers G. 2011. Iron restriction induces preferential down-regulation of H2-consuming over H2-evolving reactions during fermentative growth of Escherichia coli. BMC Microbiol 11:196. 10.1186/1471-2180-11-196. 10.1186/1471-2180-11-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Axley MJ, Grahame DA. 1991. Kinetics for formate dehydrogenase of Escherichia coli formate-hydrogenlyase. J Biol Chem 266:13731–13736. [PubMed] [PubMed] [Google Scholar]
- 51.Cox JC, Edwards ES, DeMoss JA. 1981. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J Bacteriol 145:1317–1324. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gladyshev VN, Boyington JC, Khangulov SV, Grahame DA, Stadtman TC, Sun PD. 1996. Characterization of crystalline formate dehydrogenase H from Escherichia coli. Stabilization, EPR spectroscopy, and preliminary crystallographic analysis. J Biol Chem 271:8095–8100. [PubMed] 10.1074/jbc.271.14.8095 [DOI] [PubMed] [Google Scholar]
- 53.Khangulov SV, Gladyshev VN, Dismukes GC, Stadtman TC. 1998. Selenium-containing formate dehydrogenase H from Escherichia coli: a molybdopterin enzyme that catalyzes formate oxidation without oxygen transfer. Biochemistry 37:3518–3528. [PubMed] 10.1021/bi972177k [DOI] [PubMed] [Google Scholar]
- 54.Axley MJ, Grahame DA, Stadtman TC. 1990. Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem 265:18213–18218. [PubMed] [PubMed] [Google Scholar]
- 55.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. 1997. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275:1305–1308. [PubMed] 10.1126/science.275.5304.1305 [DOI] [PubMed] [Google Scholar]
- 56.Stadtman TC. 1991. Biosynthesis and function of selenocysteine-containing enzymes. J Biol Chem 266:16257–16260. [PubMed] [PubMed] [Google Scholar]
- 57.Heider J, Böck A.1993. Selenium metabolism in microorganisms. Adv Microb Physiol 35:71–109. [PubMed] 10.1016/S0065-2911(08)60097-1 [DOI] [PubMed] [Google Scholar]
- 58.Axley MJ, Böck A, Stadtman TC. 1991. Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc Natl Acad Sci USA 88:8450–8454. [PubMed] 10.1073/pnas.88.19.8450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gladyshev VN, Khangulov SV, Axley MJ, Stadtman TC. 1994. Coordination of selenium to molybdenum in formate dehydrogenase H from Escherichia coli. Proc Natl Acad Sci USA 91:7708–7711. [PubMed] 10.1073/pnas.91.16.7708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enoch HG, Lester RL. 1975. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem 250:6693–6705. [PubMed] [PubMed] [Google Scholar]
- 61.Sawers G, Heider J, Zehelein E, Böck A. 1991. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J Bacteriol 173:4983–4993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soboh B, Pinske C, Kuhns M, Waclawek M, Ihling C, Trchounian K, Trchounian A, Sinz A, Sawers G. 2011. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen: benzyl viologen oxidoreductase activity. BMC Microbiol 11:173. 10.1186/1471-2180-11-173. 10.1186/1471-2180-11-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zorn M, Ihling CH, Golbik R, Sawers RG, Sinz A. 2014. Mapping cell envelope and periplasm protein interactions of Escherichia coli respiratory formate dehydrogenases by chemical cross-linking and mass spectrometry. J Proteome Res 13:5524–5535. [PubMed] 10.1021/pr5004906 [DOI] [PubMed] [Google Scholar]
- 64.Jormakka M, Törnroth S, Byrne B, Iwata S. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295:1863–1868. [PubMed] 10.1126/science.1068186 [DOI] [PubMed] [Google Scholar]
- 65.Magalon A, Fedor JG, Walburger A, Weiner JH. 2011. Molybdenum enzymes in bacteria and their maturation. Coord Chem Rev 255:1159–1178. 10.1016/j.ccr.2010.12.031 [DOI] [Google Scholar]
- 66.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. 2013. The prokaryotic Mo/W-bisPGD enzymes family: a catalytic workhorse in bioenergetic. Biochim Biophys Acta 1827:1048–1085. [PubMed] 10.1016/j.bbabio.2013.01.011 [DOI] [PubMed] [Google Scholar]
- 67.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NCJ. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol 10:681–687. [PubMed] 10.1038/nsb969 [DOI] [PubMed] [Google Scholar]
- 68.Jormakka M, Richardson D, Byrne B, Iwata S. 2004. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 12:95–104. [PubMed] 10.1016/j.str.2003.11.020 [DOI] [PubMed] [Google Scholar]
- 69.Schindelin H, Kisker C, Hilton J, Rajagopalan KV, Rees DC. 1996. Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination. Science 272:1615–1621. [PubMed] 10.1126/science.272.5268.1615 [DOI] [PubMed] [Google Scholar]
- 70.Najmudin S, González PJ, Trincão J, Coelho C, Mukhopadhyay A, Cerqueira NM, Romão CC, Moura I, Moura JJ, Brondino CD, Romão MJ. 2008. Periplasmic nitrate reductase revisited: a sulfur atom completes the sixth coordination of the catalytic molybdenum. J Biol Inorg Chem 13:737–753. [PubMed] 10.1007/s00775-008-0359-6 [DOI] [PubMed] [Google Scholar]
- 71.Raaijmakers HCA, Romão MJ. 2006. Formate-reduced E. coli formate dehydrogenase H: the reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem 11:849–854. [PubMed] 10.1007/s00775-006-0129-2 [DOI] [PubMed] [Google Scholar]
- 72.Leopoldini M, Chiodo SG, Toscano M, Russo N. 2008. Reaction mechanism of molybdoenzyme formate dehydrogenase. Chemistry 14:8674–8681. [PubMed] 10.1002/chem.200800906 [DOI] [PubMed] [Google Scholar]
- 73.Thomé R, Gust A, Toci R, Mendel R, Bittner F, Magalon A, Walburger A. 2012. A sulfurtransferase is essential for activity of formate dehydrogenases in Escherichia coli. J Biol Chem 287:4671–4678. [PubMed] 10.1074/jbc.M111.327122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arnoux P, Ruppelt C, Oudouhou F, Lavergne J, Siponen MI, Toci R, Mendel RR, Bittner F, Pignol D, Magalon A, Walburger A. 2015. Sulphur shuttling across a chaperone during molybdenum cofactor maturation. Nat Commun 6:6148. 10.1038/ncomms7148. [PubMed] 10.1038/ncomms7148 [DOI] [PubMed] [Google Scholar]
- 75.Maier T, Binder U, Böck A. 1996. Analysis of the hydA locus of Escherichia coli: two genes (hydN and hypF ) involved in formate and hydrogen metabolism. Arch Microbiol 165:333–341. [PubMed] 10.1007/s002030050335 [DOI] [PubMed] [Google Scholar]
- 76.Tiberti M, Papaleo E, Russo N, De Gioia L, Zampella G. 2012. Evidence for the formation of a Mo-H intermediate in the catalytic cycle of formate dehydrogenase. Inorg Chem 51:8331–8339. [PubMed] 10.1021/ic300863d [DOI] [PubMed] [Google Scholar]
- 77.Cerqueira NMFSA, González PJ, Fernandes PA, Moura JJG, Ramos MJ. 2015. Periplasmic nitrate reductase and formate dehydrogenase: similar molecular architectures with very different enzymatic activities. Acc Chem Res 48:2875–2884. [PubMed] 10.1021/acs.accounts.5b00333 [DOI] [PubMed] [Google Scholar]
- 78.Hartmann T, Schrapers P, Utesch T, Nimtz M, Rippers Y, Dau H, Mroginski M-A, Haumann M, Leimkühler S. 2016. The molybdenum active site of formate dehydrogenase is capable of catalyzing C-H bond cleavage and oxygen atom transfer reactions. Biochemistry 55:2381–2389. [PubMed] 10.1021/acs.biochem.6b00002 [DOI] [PubMed] [Google Scholar]
- 79.Woods DD. 1936. Hydrogenlyases: the synthesis of formic acid by bacteria. Biochem J 30:515–527. [PubMed] 10.1042/bj0300515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bassegoda A, Madden C, Wakerley DW, Reisner E, Hirst J. 2014. Reversible interconversion of CO2 and formate by a molybdenum-containing formate dehydrogenase. J Am Chem Soc 136:15473–15476. [PubMed] 10.1021/ja508647u [DOI] [PubMed] [Google Scholar]
- 81.Birkmann A, Sawers RG, Böck A. 1987. Involvement of the ntrA gene product in the anaerobic metabolism of Escherichia coli. Mol Gen Genet 210:535–542. [PubMed] 10.1007/BF00327209 [DOI] [PubMed] [Google Scholar]
- 82.Birkmann A, Zinoni F, Sawers G, Böck A. 1987. Factors affecting transcriptional regulation of the formate-hydrogen-lyase pathway of Escherichia coli. Arch Microbiol 148:44–51. [PubMed] 10.1007/BF00429646 [DOI] [PubMed] [Google Scholar]
- 83.Lutz S, Böhm R, Beier A, Böck A. 1990. Characterization of divergent NtrA-dependent promoters in the anaerobically expressed gene cluster coding for hydrogenase 3 components of Escherichia coli. Mol Microbiol 4:13–20. [PubMed] 10.1111/j.1365-2958.1990.tb02010.x [DOI] [PubMed] [Google Scholar]
- 84.Lutz S, Jacobi A, Schlensog V, Böhm R, Sawers G, Böck A. 1991. Molecular characterization of an operon (hyp ) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol 5:123–135. [PubMed] 10.1111/j.1365-2958.1991.tb01833.x [DOI] [PubMed] [Google Scholar]
- 85.Leonhartsberger S, Korsa I, Böck A. 2002. The molecular biology of formate metabolism in enterobacteria. J Mol Microbiol Biotechnol 4:269–276. [PubMed] [PubMed] [Google Scholar]
- 86.Schlensog V, Böck A. 1990. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol Microbiol 4:1319–1327. [PubMed] 10.1111/j.1365-2958.1990.tb00711.x [DOI] [PubMed] [Google Scholar]
- 87.Schlensog V, Lutz S, Böck A. 1994. Purification and DNA-binding properties of FHLA, the transcriptional activator of the formate hydrogenlyase system from Escherichia coli. J Biol Chem 269:19590–19596. [PubMed] [PubMed] [Google Scholar]
- 88.Birkmann A, Böck A. 1989. Characterization of a cis regulatory DNA element necessary for formate induction of the formate dehydrogenase gene (fdhF ) of Escherichia coli. Mol Microbiol 3:187–195. [PubMed] 10.1111/j.1365-2958.1989.tb01807.x [DOI] [PubMed] [Google Scholar]
- 89.Prüss BM, Liu X, Hendrickson W, Matsumura P. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol Lett 197:91–97. [PubMed] 10.1016/S0378-1097(01)00092-1 [DOI] [PubMed] [Google Scholar]
- 90.Hopper S, Babst M, Schlensog V, Fischer HM, Hennecke H, Böck A. 1994. Regulated expression in vitro of genes coding for formate hydrogenlyase components of Escherichia coli. J Biol Chem 269:19597–19604. [PubMed] [PubMed] [Google Scholar]
- 91.Korsa I, Böck A. 1997. Characterization of fhlA mutations resulting in ligand-independent transcriptional activation and ATP hydrolysis. J Bacteriol 179:41–45. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Self WT, Hasona A, Shanmugam KT. 2001. N-terminal truncations in the FhlA protein result in formate- and MoeA-independent expression of the hyc (formate hydrogenlyase) operon of Escherichia coli. Microbiology 147:3093–3104. [PubMed] 10.1099/00221287-147-11-3093 [DOI] [PubMed] [Google Scholar]
- 93.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 17:6069–6075. [PubMed] 10.1093/emboj/17.20.6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Argaman L, Altuvia S. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol 300:1101–1112. [PubMed] 10.1006/jmbi.2000.3942 [DOI] [PubMed] [Google Scholar]
- 95.Salim NN, Feig AL. 2010. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS One 5:e13028. 10.1371/journal.pone.0013028. [PubMed] 10.1371/journal.pone.0013028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mathews J, Li Q, Wang G. 2010. Characterization of hydrogen production by engineered Escherichia coli strains using rich defined media. Biotechnol Bioprocess Eng 15:686–695. 10.1007/s12257-009-3139-4 [DOI] [Google Scholar]
- 97.Redwood MD, Mikheenko IP, Sargent F, Macaskie LE. 2008. Dissecting the roles of Escherichia coli hydrogenases in biohydrogen production. FEMS Microbiol Lett 278:48–55. [PubMed] 10.1111/j.1574-6968.2007.00966.x [DOI] [PubMed] [Google Scholar]
- 98.Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. 2005. Enhanced hydrogen production from formic acid by formate hydrogen lyase-overexpressing Escherichia coli strains. Appl Environ Microbiol 71:6762–6768. [PubMed] 10.1128/AEM.71.11.6762-6768.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maeda T, Sanchez-Torres V, Wood TK. 2007. Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 77:879–890. [PubMed] 10.1007/s00253-007-1217-0 [DOI] [PubMed] [Google Scholar]
- 100.Sanchez-Torres V, Maeda T, Wood TK. 2009. Protein engineering of the transcriptional activator FhlA To enhance hydrogen production in Escherichia coli. Appl Environ Microbiol 75:5639–5646. [PubMed] 10.1128/AEM.00638-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rosentel JK, Healy F, Maupin-Furlow JA, Lee JH, Shanmugam KT. 1995. Molybdate and regulation of mod (molybdate transport), fdhF , and hyc (formate hydrogenlyase) operons in Escherichia coli. J Bacteriol 177:4857–4864. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schlensog V, Birkmann A, Böck A. 1989. Mutations in trans which affect the anaerobic expression of a formate dehydrogenase (fdhF ) structural gene. Arch Microbiol 152:83–89. [PubMed] 10.1007/BF00447016 [DOI] [PubMed] [Google Scholar]
- 103.Suppmann B, Sawers G. 1994. Isolation and characterization of hypophosphite--resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol 11:965–982. [PubMed] 10.1111/j.1365-2958.1994.tb00375.x [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Huang Y, Wang J, Cheng C, Huang W, Lu P, Xu Y-N, Wang P, Yan N, Shi Y. 2009. Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel. Nature 462:467–472. [PubMed] 10.1038/nature08610 [DOI] [PubMed] [Google Scholar]
- 105.Waight AB, Love J, Wang D-N. 2010. Structure and mechanism of a pentameric formate channel. Nat Struct Mol Biol 17:31–37. [PubMed] 10.1038/nsmb.1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lü W, Du J, Wacker T, Gerbig-Smentek E, Andrade SLA, Einsle O. 2011. pH-dependent gating in a FocA formate channel. Science 332:352–354. [PubMed] 10.1126/science.1199098 [DOI] [PubMed] [Google Scholar]
- 107.Lü W, Du J, Schwarzer NJ, Gerbig-Smentek E, Einsle O, Andrade SLA. 2012. The formate channel FocA exports the products of mixed-acid fermentation. Proc Natl Acad Sci USA 109:13254–13259. [PubMed] 10.1073/pnas.1204201109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Waight AB, Czyzewski BK, Wang D-N. 2013. Ion selectivity and gating mechanisms of FNT channels. Curr Opin Struct Biol 23:499–506. [PubMed] 10.1016/j.sbi.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lü W, Schwarzer NJ, Du J, Gerbig-Smentek E, Andrade SLA, Einsle O. 2012. Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium. Proc Natl Acad Sci USA 109:18395–18400. [PubMed] 10.1073/pnas.1210793109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Czyzewski BK, Wang D-N. 2012. Identification and characterization of a bacterial hydrosulphide ion channel. Nature 483:494–497. [PubMed] 10.1038/nature10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beckham KSH, Potter JA, Unkles SE. 2010. Formate-nitrite transporters: optimisation of expression, purification and analysis of prokaryotic and eukaryotic representatives. Protein Expr Purif 71:184–189. [PubMed] 10.1016/j.pep.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 112.Doberenz C, Zorn M, Falke D, Nannemann D, Hunger D, Beyer L, Ihling CH, Meiler J, Sinz A, Sawers RG. 2014. Pyruvate formate-lyase interacts directly with the formate channel FocA to regulate formate translocation. J Mol Biol 426:2827–2839. [PubMed] 10.1016/j.jmb.2014.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abaibou H, Giordano G, Mandrand-Berthelot MA. 1997. Suppression of Escherichia coli formate hydrogenlyase activity by trimethylamine N-oxide is due to drainage of the inducer formate. Microbiology 143:2657–2664. [PubMed] 10.1099/00221287-143-8-2657 [DOI] [PubMed] [Google Scholar]
- 114.Wang H, Gunsalus RP. 2003. Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J Bacteriol 185:5076–5085. [PubMed] 10.1128/JB.185.17.5076-5085.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hopper S, Böck A. 1995. Effector-mediated stimulation of ATPase activity by the sigma 54-dependent transcriptional activator FHLA from Escherichia coli. J Bacteriol 177:2798–2803. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vivijs B, Haberbeck LU, Baiye Mfortaw Mbong V, Bernaerts K, Geeraerd AH, Aertsen A, Michiels CW. 2015. Formate hydrogen lyase mediates stationary-phase deacidification and increases survival during sugar fermentation in acetoin-producing enterobacteria. Front Microbiol 6:150. 10.3389/fmicb.2015.00150. 10.3389/fmicb.2015.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Navarro C, Wu LF, Mandrand-Berthelot MA. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol 9:1181–1191. [PubMed] 10.1111/j.1365-2958.1993.tb01247.x [DOI] [PubMed] [Google Scholar]
- 118.De Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. 1999. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J Bacteriol 181:670–674. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu LF, Mandrand-Berthelot MA. 1986. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie 68:167–179. [PubMed] 10.1016/S0300-9084(86)81081-1 [DOI] [PubMed] [Google Scholar]
- 120.Wang SC, Dias AV, Zamble DB. 2009. The “metallo-specific” response of proteins: a perspective based on the Escherichia coli transcriptional regulator NikR. Dalton Trans (14):2459–2466. [PubMed] 10.1039/b818167p [DOI] [PubMed] [Google Scholar]
- 121.Self WT, Grunden AM, Hasona A, Shanmugam KT. 1999. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41–55. [PubMed] 10.1099/13500872-145-1-41 [DOI] [PubMed] [Google Scholar]
- 122.Hasona A, Self WT, Ray RM, Shanmugam KT. 1998. Molybdate-dependent transcription of hyc and nar operons of Escherichia coli requires MoeA protein and ModE-molybdate. FEMS Microbiol Lett 169:111–116. [PubMed] 10.1111/j.1574-6968.1998.tb13306.x [DOI] [PubMed] [Google Scholar]
- 123.Nichols J, Rajagopalan KV. 2002. Escherichia coli MoeA and MogA. Function in metal incorporation step of molybdenum cofactor biosynthesis. J Biol Chem 277:24995–25000. [PubMed] 10.1074/jbc.M203238200 [DOI] [PubMed] [Google Scholar]
- 124.Nichols JD, Rajagopalan KV. 2005. In vitro molybdenum ligation to molybdopterin using purified components. J Biol Chem 280:7817–7822. [PubMed] 10.1074/jbc.M413783200 [DOI] [PubMed] [Google Scholar]
- 125.Yokoyama K, Leimkühler S. 2015. The role of FeS clusters for molybdenum cofactor biosynthesis and molybdoenzymes in bacteria. Biochim Biophys Acta 1853:1335–1349. [PubMed] 10.1016/j.bbamcr.2014.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Self WT, Shanmugam KT. 2000. Isolation and characterization of mutated FhlA proteins which activate transcription of the hyc operon (formate hydrogenlyase) of Escherichia coli in the absence of molybdate(1). FEMS Microbiol Lett 184:47–52. [PubMed] 10.1111/j.1574-6968.2000.tb08988.x [DOI] [PubMed] [Google Scholar]
- 127.Pinske C, Bönn M, Krüger S, Lindenstrauss U, Sawers RG. 2011. Metabolic deficiencies revealed in the biotechnologically important model bacterium Escherichia coli BL21(DE3). PLoS One 6:e22830. 10.1371/journal.pone.0022830. 10.1371/journal.pone.0022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jo BH, Cha HJ. 2015. Activation of formate hydrogen-lyase via expression of uptake [NiFe]-hydrogenase in Escherichia coli BL21(DE3). Microb Cell Fact 14:151. 10.1186/s12934-015-0343-0. 10.1186/s12934-015-0343-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pinske C, Sawers RG. 2010. The role of the ferric-uptake regulator Fur and iron homeostasis in controlling levels of the [NiFe]-hydrogenases in Escherichia coli. Int J Hydrogen Energy 35:8938–8944. 10.1016/j.ijhydene.2010.06.030 [DOI] [Google Scholar]
- 130.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol 60:1058–1075. [PubMed] 10.1111/j.1365-2958.2006.05160.x [DOI] [PubMed] [Google Scholar]
- 131.Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. [PubMed] 10.1128/JB.187.20.6962-6971.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kuniyoshi TM, Balan A, Schenberg ACG, Severino D, Hallenbeck PC. 2015. Heterologous expression of proteorhodopsin enhances H2 production in Escherichia coli when endogenous Hyd-4 is overexpressed. J Biotechnol 206:52–57. [PubMed] 10.1016/j.jbiotec.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 133.Bagramyan K, Mnatsakanyan N, Poladian A, Vassilian A, Trchounian A. 2002. The roles of hydrogenases 3 and 4, and the F0F1-ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett 516:172–178. [PubMed] 10.1016/S0014-5793(02)02555-3 [DOI] [PubMed] [Google Scholar]
- 134.Skibinski DAG, Golby P, Chang Y-S, Sargent F, Hoffman R, Harper R, Guest JR, Attwood MM, Berks BC, Andrews SC. 2002. Regulation of the hydrogenase-4 operon of Escherichia coli by the sigma(54)-dependent transcriptional activators FhlA and HyfR. J Bacteriol 184:6642–6653. [PubMed] 10.1128/JB.184.23.6642-6653.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Self WT, Hasona A, Shanmugam KT. 2004. Expression and regulation of a silent operon, hyf, coding for hydrogenase 4 isoenzyme in Escherichia coli. J Bacteriol 186:580–587. [PubMed] 10.1128/JB.186.2.580-587.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537. [PubMed] 10.1038/nature03239 [DOI] [PubMed] [Google Scholar]
- 137.Trchounian K, Soboh B, Sawers RG, Trchounian A. 2013. Contribution of hydrogenase 2 to stationary phase H2 production by Escherichia coli during fermentation of glycerol. Cell Biochem Biophys 66:103–108. [PubMed] 10.1007/s12013-012-9458-7 [DOI] [PubMed] [Google Scholar]
- 138.Sasahara KC, Heinzinger NK, Barrett EL. 1997. Hydrogen sulfide production and fermentative gas production by Salmonella typhimurium require F0F1 ATP synthase activity. J Bacteriol 179:6736–6740. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Trchounian K, Trchounian A. 2014. Hydrogen producing activity by Escherichia coli hydrogenase 4 (hyf) depends on glucose concentration. Int J Hydrogen Energy VL 39:16914–16918. 10.1016/j.ijhydene.2014.08.059 [DOI] [Google Scholar]
- 140.Mnatsakanyan N, Bagramyan K, Trchounian A. 2004. Hydrogenase 3 but not hydrogenase 4 is major in hydrogen gas production by Escherichia coli formate hydrogenlyase at acidic pH and in the presence of external formate. Cell Biochem Biophys 41:357–366. [PubMed] 10.1385/CBB:41:3:357 [DOI] [PubMed] [Google Scholar]
- 141.Sawers RG, Boxer DH. 1986. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K12. Eur J Biochem 156:265–275. [PubMed] 10.1111/j.1432-1033.1986.tb09577.x [DOI] [PubMed] [Google Scholar]
- 142.Sawers RG, Jamieson DJ, Higgins CF, Boxer DH. 1986. Characterization and physiological roles of membrane-bound hydrogenase isoenzymes from Salmonella typhimurium. J Bacteriol 168:398–404. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jamieson DJ, Sawers RG, Rugman PA, Boxer DH, Higgins CF. 1986. Effects of anaerobic regulatory mutations and catabolite repression on regulation of hydrogen metabolism and hydrogenase isoenzyme composition in Salmonella typhimurium. J Bacteriol 168:405–411. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ballantine SP, Boxer DH. 1986. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur J Biochem 156:277–284. [PubMed] 10.1111/j.1432-1033.1986.tb09578.x [DOI] [PubMed] [Google Scholar]
- 145.Jones RW. 1980. The role of the membrane-bound hydrogenase in the energy-conserving oxidation of molecular hydrogen by Escherichia coli. Biochem J 188:345–350. [PubMed] 10.1042/bj1880345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fritsch J, Lenz O, Friedrich B. 2013. Structure, function and biosynthesis of O2-tolerant hydrogenases. Nat Rev Microbiol 11:106–114. [PubMed] 10.1038/nrmicro2940 [DOI] [PubMed] [Google Scholar]
- 147.Forzi L, Sawers RG. 2007. Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 20:565–578. [PubMed] 10.1007/s10534-006-9048-5 [DOI] [PubMed] [Google Scholar]
- 148.Böck A, King PW, Blokesch M, Posewitz MC. 2006. Maturation of hydrogenases. Adv Microb Physiol 51:1–71. [PubMed] 10.1016/S0065-2911(06)51001-X [DOI] [PubMed] [Google Scholar]
- 149.Palmer T, Sargent F, Berks BC. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol 13:175–180. [PubMed] 10.1016/j.tim.2005.02.002 [DOI] [PubMed] [Google Scholar]
- 150.Sargent F, Ballantine SP, Rugman PA, Palmer T, Boxer DH. 1998. Reassignment of the gene encoding the Escherichia coli hydrogenase 2 small subunit--identification of a soluble precursor of the small subunit in a hypB mutant. Eur J Biochem 255:746–754. [PubMed] 10.1046/j.1432-1327.1998.2550746.x [DOI] [PubMed] [Google Scholar]
- 151.Adams MW, Hall DO. 1979. Purification of the membrane-bound hydrogenase of Escherichia coli. Biochem J 183:11–22. [PubMed] 10.1042/bj1830011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.DerVartanian ME, Menon NK, Przybyla AE, Peck HD Jr, DerVartanian DV. 1996. Electron paramagnetic resonance (EPR) studies on hydrogenase-1 (HYD1) purified from a mutant strain (AP6) of Escherichia coli enhanced in HYD1. Biochem Biophys Res Commun 227:211–215. [PubMed] 10.1006/bbrc.1996.1491 [DOI] [PubMed] [Google Scholar]
- 153.Francis K, Patel P, Wendt JC, Shanmugam KT. 1990. Purification and characterization of two forms of hydrogenase isoenzyme 1 from Escherichia coli. J Bacteriol 172:5750–5757. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lukey MJ, Parkin A, Roessler MM, Murphy BJ, Harmer J, Palmer T, Sargent F, Armstrong FA. 2010. How Escherichia coli is equipped to oxidize hydrogen under different redox conditions. J Biol Chem 285:3928–3938. [PubMed] 10.1074/jbc.M109.067751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Volbeda A, Amara P, Darnault C, Mouesca J-M, Parkin A, Roessler MM, Armstrong FA, Fontecilla-Camps JC. 2012. X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli. Proc Natl Acad Sci USA 109:5305–5310. [PubMed] 10.1073/pnas.1119806109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Volbeda A, Darnault C, Parkin A, Sargent F, Armstrong FA, Fontecilla-Camps JC. 2013. Crystal structure of the O2-tolerant membrane-bound hydrogenase 1 from Escherichia coli in complex with its cognate cytochrome b. Structure 21:184–190. [PubMed] 10.1016/j.str.2012.11.010 [DOI] [PubMed] [Google Scholar]
- 157.Rodrigue A, Chanal A, Beck K, Müller M, Wu LF. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J Biol Chem 274:13223–13228. [PubMed] 10.1074/jbc.274.19.13223 [DOI] [PubMed] [Google Scholar]
- 158.Dubini A, Pye R, Jack R, Palmer T, Sargent F. 2002. How bacteria get energy from hydrogen: a genetic analysis of periplasmic hydrogen oxidation in Escherichia coli. Int J Hydrogen Energy 27:1413–1420. 10.1016/S0360-3199(02)00112-X [DOI] [Google Scholar]
- 159.Ballantine SP, Boxer DH. 1985. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol 163:454–459. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Evans RM, Brooke EJ, Wehlin SAM, Nomerotskaia E, Sargent F, Carr SB, Phillips SEV, Armstrong FA. 2016. Mechanism of hydrogen activation by [NiFe] hydrogenases. Nat Chem Biol 12:46–50. [PubMed] 10.1038/nchembio.1976 [DOI] [PubMed] [Google Scholar]
- 161.Wulff P, Day CC, Sargent F, Armstrong FA. 2014. How oxygen reacts with oxygen-tolerant respiratory [NiFe]-hydrogenases. Proc Natl Acad Sci USA 111:6606–6611. [PubMed] 10.1073/pnas.1322393111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murphy BJ, Sargent F, Armstrong FA. 2014. Transforming an oxygen-tolerant [NiFe] uptake hydrogenase into a proficient, reversible hydrogen producer. Energy Environ Sci 7:1426–1433. 10.1039/c3ee43652g [DOI] [Google Scholar]
- 163.Lukey MJ, Roessler MM, Parkin A, Evans RM, Davies RA, Lenz O, Friedrich B, Sargent F, Armstrong FA. 2011. Oxygen-tolerant [NiFe]-hydrogenases: the individual and collective importance of supernumerary cysteines at the proximal Fe-S cluster. J Am Chem Soc 133:16881–16892. [PubMed] 10.1021/ja205393w [DOI] [PubMed] [Google Scholar]
- 164.Laurinavichene TV, Tsygankov AA. 2001. H2 consumption by Escherichia coli coupled via hydrogenase 1 or hydrogenase 2 to different terminal electron acceptors. FEMS Microbiol Lett 202:121–124. [PubMed] 10.1111/j.1574-6968.2001.tb10790.x [DOI] [PubMed] [Google Scholar]
- 165.Pinske C, Jaroschinsky M, Sargent F, Sawers G. 2012. Zymographic differentiation of [NiFe]-hydrogenases 1, 2, and 3 of Escherichia coli K-12. BMC Microbiol 12:134. [PubMed] 10.1186/1471-2180-12-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pinske C, McDowall JS, Sargent F, Sawers RG. 2012. Analysis of hydrogenase 1 levels reveals an intimate link between carbon and hydrogen metabolism in Escherichia coli K-12. Microbiology 158:856–868. [PubMed] 10.1099/mic.0.056622-0 [DOI] [PubMed] [Google Scholar]
- 167.Pinske C, Krüger S, Soboh B, Ihling C, Kuhns M, Braussemann M, Jaroschinsky M, Sauer C, Sargent F, Sinz A, Sawers RG. 2011. Efficient electron transfer from hydrogen to benzyl viologen by the [NiFe]-hydrogenases of Escherichia coli is dependent on the coexpression of the iron-sulfur cluster-containing small subunit. Arch Microbiol 193:893–903. [PubMed] 10.1007/s00203-011-0726-5 [DOI] [PubMed] [Google Scholar]
- 168.Laurinavichene TV, Zorin NA, Tsygankov AA. 2002. Effect of redox potential on activity of hydrogenase 1 and hydrogenase 2 in Escherichia coli. Arch Microbiol 178:437–442. [PubMed] 10.1007/s00203-002-0471-x [DOI] [PubMed] [Google Scholar]
- 169.Flanagan LA, Parkin A. 2016. Electrochemical insights into the mechanism of NiFe membrane-bound hydrogenases. Biochem Soc Trans 44:315–328. [PubMed] 10.1042/BST20150201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.King PW, Przybyla AE. 1999. Response of hya expression to external pH in Escherichia coli. J Bacteriol 181:5250–5256. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Clark DP. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev 5:223–234. [PubMed] 10.1016/0168-6445(89)90033-8 [DOI] [PubMed] [Google Scholar]
- 172.Alam KY, Clark DP. 1989. Anaerobic fermentation balance of Escherichia coli as observed by in vivo nuclear magnetic resonance spectroscopy. J Bacteriol 171:6213–6217. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zbell AL, Benoit SL, Maier RJ. 2007. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology 153:3508–3516. [PubMed] 10.1099/mic.0.2007/009027-0 [DOI] [PubMed] [Google Scholar]
- 174.Maier RJ, Olczak A, Maier S, Soni S, Gunn J. 2004. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect Immun 72:6294–6299. [PubMed] 10.1128/IAI.72.11.6294-6299.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lamichhane-Khadka R, Kwiatkowski A, Maier RJ. 2010. The Hyb hydrogenase permits hydrogen-dependent respiratory growth of Salmonella enterica serovar Typhimurium. MBio 1:e00284-10. 10.1128/mBio.00284-10. 10.1128/mBio.00284-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zbell AL, Maier RJ. 2009. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 75:1456–1459. [PubMed] 10.1128/AEM.02064-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lamichhane-Khadka R, Benoit SL, Miller-Parks EF, Maier RJ. 2015. Host hydrogen rather than that produced by the pathogen is important for Salmonella enterica serovar Typhimurium virulence. Infect Immun 83:311–316. [PubMed] 10.1128/IAI.02611-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Swords WE, Cannon BM, Benjamin WH Jr. 1997. Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun 65:2451–2453. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Menon NK, Robbins J, Wendt JC, Shanmugam KT, Przybyla AE. 1991. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol 173:4851–4861. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Menon NK, Robbins J, Peck HD Jr, Chatelus CY, Choi ES, Przybyla AE. 1990. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol 172:1969–1977. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dubini A, Sargent F. 2003. Assembly of Tat-dependent [NiFe] hydrogenases: identification of precursor-binding accessory proteins. FEBS Lett 549:141–146. [PubMed] 10.1016/S0014-5793(03)00802-0 [DOI] [PubMed] [Google Scholar]
- 182.Pinske C, Sawers RG. 2014. The importance of iron in the biosynthesis and assembly of [NiFe]-hydrogenases. Biomol Concepts 5:55–70. [PubMed] 10.1515/bmc-2014-0001 [DOI] [PubMed] [Google Scholar]
- 183.Fritsche E, Paschos A, Beisel HG, Böck A, Huber R. 1999. Crystal structure of the hydrogenase maturating endopeptidase HYBD from Escherichia coli. J Mol Biol 288:989–998. [PubMed] 10.1006/jmbi.1999.2719 [DOI] [PubMed] [Google Scholar]
- 184.Stoker K, Oltmann LF, Stouthamer AH. 1989. Randomly induced Escherichia coli K-12 Tn5 insertion mutants defective in hydrogenase activity. J Bacteriol 171:831–836. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee JH, Patel P, Sankar P, Shanmugam KT. 1985. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J Bacteriol 162:344–352. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Menon NK, Chatelus CY, Dervartanian M, Wendt JC, Shanmugam KT, Peck HD Jr, Przybyla AE. 1994. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J Bacteriol 176:4416–4423. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dross F, Geisler V, Lenger R, Theis F, Krafft T, Fahrenholz F, Kojro E, Duchene A, Tripier D, Juvenal K, Kroger A. 1992. The quinone-reactive Ni/Fe-hydrogenase of Wolinella succinogenes. Eur J Biochem 206:93–102. [PubMed] 10.1111/j.1432-1033.1992.tb16905.x [DOI] [PubMed] [Google Scholar]
- 188.Richard DJ, Sawers G, Sargent F, McWalter L, Boxer DH. 1999. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology 145:2903–2912. [PubMed] 10.1099/00221287-145-10-2903 [DOI] [PubMed] [Google Scholar]
- 189.Hube M, Blokesch M, Böck A. 2002. Network of hydrogenase maturation in Escherichia coli: role of accessory proteins HypA and HybF. J Bacteriol 184:3879–3885. [PubMed] 10.1128/JB.184.14.3879-3885.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Blokesch M, Rohrmoser M, Rode S, Böck A. 2004. HybF, a zinc-containing protein involved in NiFe hydrogenase maturation. J Bacteriol 186:2603–2611. [PubMed] 10.1128/JB.186.9.2603-2611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Blokesch M, Böck A. 2002. Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J Mol Biol 324:287–296. [PubMed] 10.1016/S0022-2836(02)01070-7 [DOI] [PubMed] [Google Scholar]
- 192.Blokesch M, Magalon A, Böck A. 2001. Interplay between the specific chaperone-like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J Bacteriol 183:2817–2822. [PubMed] 10.1128/JB.183.9.2817-2822.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Thomas C, Muhr E, Sawers RG. 2015. Coordination of synthesis and assembly of a modular membrane-associated [NiFe]-hydrogenase is determined by cleavage of the C-terminal peptide. J Bacteriol 197:2989–2998. [PubMed] 10.1128/JB.00437-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Frey M, Fontecilla-Camps JC, Volbeda A. 2011. Nickel-Iron Hydrogenases. John Wiley & Sons, Ltd, Chichester, UK. 10.1002/9781119951438.eibc0590 [DOI] [Google Scholar]
- 195.Przybyla AE, Robbins J, Menon N, Peck HD Jr. 1992. Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev 8:109–135. [PubMed] 10.1111/j.1574-6968.1992.tb04960.x [DOI] [PubMed] [Google Scholar]
- 196.Vignais PM, Billoud B, Meyer J. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev 25:455–501. [PubMed] 10.1111/j.1574-6976.2001.tb00587.x [DOI] [PubMed] [Google Scholar]
- 197.Vignais PM, Colbeau A. 2004. Molecular biology of microbial hydrogenases. Curr Issues Mol Biol 6:159–188. [PubMed] [PubMed] [Google Scholar]
- 198.Wu LF, Mandrand MA. 1993. Microbial hydrogenases: primary structure, classification, signatures, and phylogeny. FEMS Microbiol Rev 10:243–269. [PubMed] 10.1111/j.1574-6968.1993.tb05870.x [DOI] [PubMed] [Google Scholar]
- 199.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MWW. 2015. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta 1853:1350–1369. [PubMed] 10.1016/j.bbamcr.2014.11.021 [DOI] [PubMed] [Google Scholar]
- 200.Lubitz W, Ogata H, Rüdiger O, Reijerse E. 2014. Hydrogenases. Chem Rev 114:4081–4148. [PubMed] 10.1021/cr4005814 [DOI] [PubMed] [Google Scholar]
- 201.Atlung T, Knudsen K, Heerfordt L, Brøndsted L. 1997. Effects of sigmaS and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J Bacteriol 179:2141–2146. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Brøndsted L, Atlung T. 1994. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J Bacteriol 176:5423–5428. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rodrigue A, Boxer DH, Mandrand-Berthelot MA, Wu LF. 1996. Requirement for nickel of the transmembrane translocation of NiFe-hydrogenase 2 in Escherichia coli. FEBS Lett 392:81–86. [PubMed] 10.1016/0014-5793(96)00788-0 [DOI] [PubMed] [Google Scholar]
- 204.Wu LF, Mandrand-Berthelot MA, Waugh R, Edmonds CJ, Holt SE, Boxer DH. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol Microbiol 3:1709–1718. [PubMed] 10.1111/j.1365-2958.1989.tb00156.x [DOI] [PubMed] [Google Scholar]
- 205.Sawers G. 1999. The aerobic/anaerobic interface. Curr Opin Microbiol 2:181–187. [PubMed] 10.1016/S1369-5274(99)80032-0 [DOI] [PubMed] [Google Scholar]
- 206.Ingmer H, Miller CA, Cohen SN. 1998. Destabilized inheritance of pSC101 and other Escherichia coli plasmids by DpiA, a novel two-component system regulator. Mol Microbiol 29:49–59. [PubMed] 10.1046/j.1365-2958.1998.00895.x [DOI] [PubMed] [Google Scholar]
- 207.Nesbit AD, Giel JL, Rose JC, Kiley PJ. 2009. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol 387:28–41. [PubMed] 10.1016/j.jmb.2009.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rajagopalan S, Teter SJ, Zwart PH, Brennan RG, Phillips KJ, Kiley PJ. 2013. Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat Struct Mol Biol 20:740–747. [PubMed] 10.1038/nsmb.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nesbit AD, Fleischhacker AS, Teter SJ, Kiley PJ. 2012. ArcA and AppY antagonize IscR repression of hydrogenase-1 expression under anaerobic conditions, revealing a novel mode of O2 regulation of gene expression in Escherichia coli. J Bacteriol 194:6892–6899. [PubMed] 10.1128/JB.01757-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jamieson DJ, Higgins CF. 1986. Two genetically distinct pathways for transcriptional regulation of anaerobic gene expression in Salmonella typhimurium. J Bacteriol 168:389–397. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Park KR, Giard JC, Eom JH, Bearson S, Foster JW. 1999. Cyclic AMP receptor protein and TyrR are required for acid pH and anaerobic induction of hyaB and aniC in Salmonella typhimurium. J Bacteriol 181:689–694. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bowman L, Palmer T, Sargent F. 2013. A regulatory domain controls the transport activity of a twin-arginine signal peptide. FEBS Lett 587:3365–3370. [PubMed] 10.1016/j.febslet.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 213.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. 2007. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem Rev 107:4273–4303. [PubMed] 10.1021/cr050195z [DOI] [PubMed] [Google Scholar]
- 214.Bagley KA, Duin EC, Roseboom W, Albracht SP, Woodruff WH. 1995. Infrared-detectable groups sense changes in charge density on the nickel center in hydrogenase from Chromatium vinosum. Biochemistry 34:5527–5535. [PubMed] 10.1021/bi00016a026 [DOI] [PubMed] [Google Scholar]
- 215.Happe RP, Roseboom W, Pierik AJ, Albracht SP, Bagley KA. 1997. Biological activation of hydrogen. Nature 385:126. [PubMed] 10.1038/385126a0 [DOI] [PubMed] [Google Scholar]
- 216.van der Spek TM, Arendsen AF, Happe RP, Yun S, Bagley KA, Stufkens DJ, Hagen WR, Albracht SP. 1996. Similarities in the architecture of the active sites of Ni-hydrogenases and Fe-hydrogenases detected by means of infrared spectroscopy. Eur J Biochem 237:629–634. [PubMed] 10.1111/j.1432-1033.1996.0629p.x [DOI] [PubMed] [Google Scholar]
- 217.Bagley KA, Van Garderen CJ, Chen M, Duin EC, Albracht SP, Woodruff WH. 1994. Infrared studies on the interaction of carbon monoxide with divalent nickel in hydrogenase from Chromatium vinosum. Biochemistry 33:9229–9236. [PubMed] 10.1021/bi00197a026 [DOI] [PubMed] [Google Scholar]
- 218.Volbeda A, Garcin E, Piras C, De Lacey AL, Fernandez VM, Hatchikian EC, Frey M, Fontecilla-Camps JC. 1996. Structure of the [NiFe] hydrogenase active site: evidence for biologically uncommon Fe ligands. J Am Chem Soc 118:12989–12996. 10.1021/ja962270g [DOI] [Google Scholar]
- 219.Volbeda A, Charon MH, Piras C, Hatchikian EC, Frey M, Fontecilla-Camps JC. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580–587. [PubMed] 10.1038/373580a0 [DOI] [PubMed] [Google Scholar]
- 220.Pierik AJ, Roseboom W, Happe RP, Bagley KA, Albracht SP. 1999. Carbon monoxide and cyanide as intrinsic ligands to iron in the active site of [NiFe]-hydrogenases. NiFe(CN)2CO, Biology’s way to activate H2. J Biol Chem 274:3331–3337. [PubMed] 10.1074/jbc.274.6.3331 [DOI] [PubMed] [Google Scholar]
- 221.Sankar P, Lee JH, Shanmugam KT. 1985. Cloning of hydrogenase genes and fine structure analysis of an operon essential for H2 metabolism in Escherichia coli. J Bacteriol 162:353–360. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sankar P, Shanmugam KT. 1988. Biochemical and genetic analysis of hydrogen metabolism in Escherichia coli: the hydB gene. J Bacteriol 170:5433–5439. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sankar P, Shanmugam KT. 1988. Hydrogen metabolism in Escherichia coli: biochemical and genetic evidence for a hydF gene. J Bacteriol 170:5446–5451. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jacobi A, Rossmann R, Böck A. 1992. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol 158:444–451. [PubMed] 10.1007/BF00276307 [DOI] [PubMed] [Google Scholar]
- 225.Barrett EL, Kwan HS, Macy J. 1984. Anaerobiosis, formate, nitrate, and pyrA are involved in the regulation of formate hydrogenlyase in Salmonella typhimurium. J Bacteriol 158:972–977. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Paschos A, Glass RS, Böck A. 2001. Carbamoylphosphate requirement for synthesis of the active center of [NiFe]-hydrogenases. FEBS Lett 488:9–12. [PubMed] 10.1016/S0014-5793(00)02408-X [DOI] [PubMed] [Google Scholar]
- 227.Maier T, Jacobi A, Sauter M, Böck A. 1993. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol 175:630–635. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maier T, Lottspeich F, Böck A. 1995. GTP hydrolysis by HypB is essential for nickel insertion into hydrogenases of Escherichia coli. Eur J Biochem 230:133–138. [PubMed] 10.1111/j.1432-1033.1995.0133i.x [DOI] [PubMed] [Google Scholar]
- 229.Lacasse MJ, Zamble DB. 2016. [NiFe]-Hydrogenase maturation. Biochemistry 55:1689–1701. [PubMed] 10.1021/acs.biochem.5b01328 [DOI] [PubMed] [Google Scholar]
- 230.Soboh B, Sawers RG. 2013. [NiFe]-hydrogenase cofactor assembly, p 1–9. In Encyclopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Ltd, Chichester, UK. [Google Scholar]
- 231.Messenger SL, Green J. 2003. FNR-mediated regulation of hyp expression in Escherichia coli. FEMS Microbiol Lett 228:81–86. [PubMed] 10.1016/S0378-1097(03)00726-2 [DOI] [PubMed] [Google Scholar]
- 232.Soboh B, Krüger S, Kuhns M, Pinske C, Lehmann A, Sawers RG. 2010. Development of a cell-free system reveals an oxygen-labile step in the maturation of [NiFe]-hydrogenase 2 of Escherichia coli. FEBS Lett 584:4109–4114. [PubMed] 10.1016/j.febslet.2010.08.037 [DOI] [PubMed] [Google Scholar]
- 233.Paschos A, Bauer A, Zimmermann A, Zehelein E, Böck A. 2002. HypF, a carbamoyl phosphate-converting enzyme involved in [NiFe] hydrogenase maturation. J Biol Chem 277:49945–49951. [PubMed] 10.1074/jbc.M204601200 [DOI] [PubMed] [Google Scholar]
- 234.Shomura Y, Higuchi Y. 2012. Structural basis for the reaction mechanism of S-carbamoylation of HypE by HypF in the maturation of [NiFe]-hydrogenases. J Biol Chem 287:28409–28419. [PubMed] 10.1074/jbc.M112.387134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Blokesch M, Paschos A, Bauer A, Reissmann S, Drapal N, Böck A. 2004. Analysis of the transcarbamoylation-dehydration reaction catalyzed by the hydrogenase maturation proteins HypF and HypE. Eur J Biochem 271:3428–3436. [PubMed] 10.1111/j.1432-1033.2004.04280.x [DOI] [PubMed] [Google Scholar]
- 236.Lenz O, Zebger I, Hamann J, Hildebrandt P, Friedrich B. 2007. Carbamoylphosphate serves as the source of CN(-), but not of the intrinsic CO in the active site of the regulatory [NiFe]-hydrogenase from Ralstonia eutropha. FEBS Lett 581:3322–3326. [PubMed] 10.1016/j.febslet.2007.06.027 [DOI] [PubMed] [Google Scholar]
- 237.Forzi L, Hellwig P, Thauer RK, Sawers RG. 2007. The CO and CN(-) ligands to the active site Fe in [NiFe]-hydrogenase of Escherichia coli have different metabolic origins. FEBS Lett 581:3317–3321. [PubMed] 10.1016/j.febslet.2007.06.028 [DOI] [PubMed] [Google Scholar]
- 238.Roseboom W, Blokesch M, Böck A, Albracht SPJ. 2005. The biosynthetic routes for carbon monoxide and cyanide in the Ni-Fe active site of hydrogenases are different. FEBS Lett 579:469–472. [PubMed] 10.1016/j.febslet.2004.12.013 [DOI] [PubMed] [Google Scholar]
- 239.Stefani M, Taddei N, Ramponi G. 1997. Insights into acylphosphatase structure and catalytic mechanism. Cell Mol Life Sci 53:141–151. [PubMed] 10.1007/PL00000585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wolf I, Buhrke T, Dernedde J, Pohlmann A, Friedrich B. 1998. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol 170:451–459. [PubMed] 10.1007/s002030050666 [DOI] [PubMed] [Google Scholar]
- 241.Rosano C, Zuccotti S, Stefani M, Bucciantini M, Ramponi G, Bolognesi M. 2002. Crystallization and preliminary X-ray characterization of the acylphosphatase-like domain from the Escherichia coli hydrogenase maturation factor HypF. Acta Crystallogr D Biol Crystallogr 58:524–525. [PubMed] 10.1107/S0907444901021874 [DOI] [PubMed] [Google Scholar]
- 242.Tominaga T, Watanabe S, Matsumi R, Atomi H, Imanaka T, Miki K. 2012. Structure of the [NiFe]-hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1. Acta Crystallogr Sect F Struct Biol Cryst Commun 68:1153–1157. [PubMed] 10.1107/S1744309112036421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Winter G, Dökel S, Jones AK, Scheerer P, Krauss N, Höhne W, Friedrich B. 2010. Crystallization and preliminary X-ray crystallographic analysis of the [NiFe]-hydrogenase maturation factor HypF1 from Ralstonia eutropha H16. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:452–455. [PubMed] 10.1107/S1744309110006196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Watanabe S, Sasaki D, Tominaga T, Miki K. 2012. Structural basis of [NiFe] hydrogenase maturation by Hyp proteins. Biol Chem 393:1089–1100. [PubMed] 10.1515/hsz-2012-0197 [DOI] [PubMed] [Google Scholar]
- 245.Petkun S, Shi R, Li Y, Asinas A, Munger C, Zhang L, Waclawek M, Soboh B, Sawers RG, Cygler M. 2011. Structure of hydrogenase maturation protein HypF with reaction intermediates shows two active sites. Structure 19:1773–1783. [PubMed] 10.1016/j.str.2011.09.023 [DOI] [PubMed] [Google Scholar]
- 246.Rangarajan ES, Asinas A, Proteau A, Munger C, Baardsnes J, Iannuzzi P, Matte A, Cygler M. 2008. Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF. J Bacteriol 190:1447–1458. [PubMed] 10.1128/JB.01610-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Parthier C, Görlich S, Jaenecke F, Breithaupt C, Bräuer U, Fandrich U, Clausnitzer D, Wehmeier UF, Böttcher C, Scheel D, Stubbs MT. 2012. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew Chem Int Ed Engl 51:4046–4052. [PubMed] 10.1002/anie.201108896 [DOI] [PubMed] [Google Scholar]
- 248.Reissmann S, Hochleitner E, Wang H, Paschos A, Lottspeich F, Glass RS, Böck A. 2003. Taming of a poison: biosynthesis of the NiFe-hydrogenase cyanide ligands. Science 299:1067–1070. [PubMed] 10.1126/science.1080972 [DOI] [PubMed] [Google Scholar]
- 249.Tominaga T, Watanabe S, Matsumi R, Atomi H, Imanaka T, Miki K. 2013. Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation. Proc Natl Acad Sci USA 110:20485–20490. [PubMed] 10.1073/pnas.1313620110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Stripp ST, Lindenstrauss U, Sawers RG, Soboh B. 2015. Identification of an isothiocyanate on the HypEF complex suggests a route for efficient cyanyl-group channeling during [NiFe]-hydrogenase cofactor generation. PLoS One 10:e0133118. 10.1371/journal.pone.0133118. 10.1371/journal.pone.0133118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Watanabe S, Matsumi R, Arai T, Atomi H, Imanaka T, Miki K. 2007. Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling. Mol Cell 27:29–40. [PubMed] 10.1016/j.molcel.2007.05.039 [DOI] [PubMed] [Google Scholar]
- 252.Wang L, Xia B, Jin C. 2007. Solution structure of Escherichia coli HypC. Biochem Biophys Res Commun 361:665–669. [PubMed] 10.1016/j.bbrc.2007.07.094 [DOI] [PubMed] [Google Scholar]
- 253.Blokesch M, Albracht SPJ, Matzanke BF, Drapal NM, Jacobi A, Böck A. 2004. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J Mol Biol 344:155–167. [PubMed] 10.1016/j.jmb.2004.09.040 [DOI] [PubMed] [Google Scholar]
- 254.Watanabe S, Matsumi R, Atomi H, Imanaka T, Miki K. 2012. Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation. Structure 20:2124–2137. [PubMed] 10.1016/j.str.2012.09.018 [DOI] [PubMed] [Google Scholar]
- 255.Soboh B, Stripp ST, Muhr E, Granich C, Braussemann M, Herzberg M, Heberle J, Gary Sawers R. 2012. [NiFe]-hydrogenase maturation: isolation of a HypC-HypD complex carrying diatomic CO and CN- ligands. FEBS Lett 586:3882–3887. [PubMed] 10.1016/j.febslet.2012.09.019 [DOI] [PubMed] [Google Scholar]
- 256.Stripp ST, Soboh B, Lindenstrauss U, Braussemann M, Herzberg M, Nies DH, Sawers RG, Heberle J. 2013. HypD is the scaffold protein for Fe-(CN)2CO cofactor assembly in [NiFe]-hydrogenase maturation. Biochemistry 52:3289–3296. [PubMed] 10.1021/bi400302v [DOI] [PubMed] [Google Scholar]
- 257.Bürstel I. 2013. Die pleiotrope Maturation der sauerstofftoleranten [NiFe]-Hydrogenasen aus Ralstonia eutropha. Humboldt-Universität zu Berlin. [Google Scholar]
- 258.Soboh B, Stripp ST, Bielak C, Lindenstrauß U, Braussemann M, Javaid M, Hallensleben M, Granich C, Herzberg M, Heberle J, Gary Sawers R. 2013. The [NiFe]-hydrogenase accessory chaperones HypC and HybG of Escherichia coli are iron- and carbon dioxide-binding proteins. FEBS Lett 587:2512–2516. [PubMed] 10.1016/j.febslet.2013.06.055 [DOI] [PubMed] [Google Scholar]
- 259.Blokesch M, Böck A. 2006. Properties of the [NiFe]-hydrogenase maturation protein HypD. FEBS Lett 580:4065–4068. [PubMed] 10.1016/j.febslet.2006.06.045 [DOI] [PubMed] [Google Scholar]
- 260.Stripp ST, Lindenstrauss U, Granich C, Sawers RG, Soboh B. 2014. The influence of oxygen on [NiFe]-hydrogenase cofactor biosynthesis and how ligation of carbon monoxide precedes cyanation. PLoS One 9:e107488. 10.1371/journal.pone.0107488. [PubMed] 10.1371/journal.pone.0107488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bürstel I, Siebert E, Winter G, Hummel P, Zebger I, Friedrich B, Lenz O. 2012. A universal scaffold for synthesis of the Fe(CN)2(CO) moiety of [NiFe] hydrogenase. J Biol Chem 287:38845–38853. [PubMed] 10.1074/jbc.M112.376947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Drapal N, Böck A. 1998. Interaction of the hydrogenase accessory protein HypC with HycE, the large subunit of Escherichia coli hydrogenase 3 during enzyme maturation. Biochemistry 37:2941–2948. [PubMed] 10.1021/bi9720078 [DOI] [PubMed] [Google Scholar]
- 263.Magalon A, Böck A. 2000. Analysis of the HypC-hycE complex, a key intermediate in the assembly of the metal center of the Escherichia coli hydrogenase 3. J Biol Chem 275:21114–21120. [PubMed] 10.1074/jbc.M000987200 [DOI] [PubMed] [Google Scholar]
- 264.Hartwig S, Thomas C, Krumova N, Quitzke V, Türkowsky D, Jehmlich N, Adrian L, Sawers RG. 2015. Heterologous complementation studies in Escherichia coli with the Hyp accessory protein machinery from Chloroflexi provide insight into [NiFe]-hydrogenase large subunit recognition by the HypC protein family. Microbiology 161:2204–2219. [PubMed] 10.1099/mic.0.000177 [DOI] [PubMed] [Google Scholar]
- 265.Zhang JW, Butland G, Greenblatt JF, Emili A, Zamble DB. 2005. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J Biol Chem 280:4360–4366. [PubMed] 10.1074/jbc.M411799200 [DOI] [PubMed] [Google Scholar]
- 266.Waugh R, Boxer DH. 1986. Pleiotropic hydrogenase mutants of Escherichia coli K12: growth in the presence of nickel can restore hydrogenase activity. Biochimie 68:157–166. [PubMed] 10.1016/S0300-9084(86)81080-X [DOI] [PubMed] [Google Scholar]
- 267.Olson JW, Mehta NS, Maier RJ. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol 39:176–182. [PubMed] 10.1046/j.1365-2958.2001.02244.x [DOI] [PubMed] [Google Scholar]
- 268.Pinske C, Sargent F, Sawers RG. 2015. SlyD-dependent nickel delivery limits maturation of [NiFe]-hydrogenases in late-stationary phase Escherichia coli cells. Metallomics 7:683–690. [PubMed] 10.1039/C5MT00019J [DOI] [PubMed] [Google Scholar]
- 269.Maier T, Böck A. 1996. Generation of active [NiFe] hydrogenase in vitro from a nickel-free precursor form. Biochemistry 35:10089–10093. [PubMed] 10.1021/bi960567l [DOI] [PubMed] [Google Scholar]
- 270.Theodoratou E, Paschos A, Magalon A, Fritsche E, Huber R, Böck A. 2000. Nickel serves as a substrate recognition motif for the endopeptidase involved in hydrogenase maturation. Eur J Biochem 267:1995–1999. [PubMed] 10.1046/j.1432-1327.2000.01202.x [DOI] [PubMed] [Google Scholar]
- 271.Fu C, Olson JW, Maier RJ. 1995. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc Natl Acad Sci USA 92:2333–2337. [PubMed] 10.1073/pnas.92.6.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Rey L, Imperial J, Palacios JM, Ruiz-Argüeso T. 1994. Purification of Rhizobium leguminosarum HypB, a nickel-binding protein required for hydrogenase synthesis. J Bacteriol 176:6066–6073. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Mehta N, Olson JW, Maier RJ. 2003. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol 185:726–734. [PubMed] 10.1128/JB.185.3.726-734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mehta N, Benoit S, Maier RJ. 2003. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb Pathog 35:229–234. [PubMed] 10.1016/S0882-4010(03)00151-7 [DOI] [PubMed] [Google Scholar]
- 275.Olson JW, Maier RJ. 2000. Dual roles of Bradyrhizobium japonicum nickelin protein in nickel storage and GTP-dependent Ni mobilization. J Bacteriol 182:1702–1705. [PubMed] 10.1128/JB.182.6.1702-1705.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Leach MR, Sandal S, Sun H, Zamble DB. 2005. Metal binding activity of the Escherichia coli hydrogenase maturation factor HypB. Biochemistry 44:12229–12238. [PubMed] 10.1021/bi050993j [DOI] [PubMed] [Google Scholar]
- 277.Gasper R, Scrima A, Wittinghofer A. 2006. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem 281:27492–27502. [PubMed] 10.1074/jbc.M600809200 [DOI] [PubMed] [Google Scholar]
- 278.Cai F, Ngu TT, Kaluarachchi H, Zamble DB. 2011. Relationship between the GTPase, metal-binding, and dimerization activities of E. coli HypB. J Biol Inorg Chem 16:857–868. [PubMed] 10.1007/s00775-011-0782-y [DOI] [PubMed] [Google Scholar]
- 279.Douglas CD, Ngu TT, Kaluarachchi H, Zamble DB. 2013. Metal transfer within the Escherichia coli HypB-HypA complex of hydrogenase accessory proteins. Biochemistry 52:6030–6039. [PubMed] 10.1021/bi400812r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Atanassova A, Zamble DB. 2005. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J Bacteriol 187:4689–4697. [PubMed] 10.1128/JB.187.14.4689-4697.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chan K-H, Lee K-M, Wong K-B. 2012. Interaction between hydrogenase maturation factors HypA and HypB is required for [NiFe]-hydrogenase maturation. PLoS One 7:e32592. 10.1371/journal.pone.0032592. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Watanabe S, Kawashima T, Nishitani Y, Kanai T, Wada T, Inaba K, Atomi H, Imanaka T, Miki K. 2015. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc Natl Acad Sci USA 112:7701–7706. [PubMed] 10.1073/pnas.1503102112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Leach MR, Zhang JW, Zamble DB. 2007. The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J Biol Chem 282:16177–16186. [PubMed] 10.1074/jbc.M610834200 [DOI] [PubMed] [Google Scholar]
- 284.Kaluarachchi H, Zhang JW, Zamble DB. 2011. Escherichia coli SlyD, more than a Ni(II) reservoir. Biochemistry 50:10761–10763. [PubMed] 10.1021/bi201590d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Chung KC, Zamble DB. 2011. The Escherichia coli metal-binding chaperone SlyD interacts with the large subunit of [NiFe]-hydrogenase 3. FEBS Lett 585:291–294. [PubMed] 10.1016/j.febslet.2010.12.024 [DOI] [PubMed] [Google Scholar]
- 286.Chan Chung KC, Zamble DB. 2011. Protein interactions and localization of the Escherichia coli accessory protein HypA during nickel insertion to [NiFe] hydrogenase. J Biol Chem 286:43081–43090. [PubMed] 10.1074/jbc.M111.290726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Gollin DJ, Mortenson LE, Robson RL. 1992. Carboxyl-terminal processing may be essential for production of active NiFe hydrogenase in Azotobacter vinelandii. FEBS Lett 309:371–375. [PubMed] 10.1016/0014-5793(92)80809-U [DOI] [PubMed] [Google Scholar]
- 288.Magalon A, Böck A. 2000. Dissection of the maturation reactions of the [NiFe] hydrogenase 3 from Escherichia coli taking place after nickel incorporation. FEBS Lett 473:254–258. [PubMed] 10.1016/S0014-5793(00)01542-8 [DOI] [PubMed] [Google Scholar]
- 289.Theodoratou E, Huber R, Böck A. 2005. [NiFe]-Hydrogenase maturation endopeptidase: structure and function. Biochem Soc Trans 33:108–111. [PubMed] 10.1042/BST0330108 [DOI] [PubMed] [Google Scholar]
- 290.Theodoratou E, Paschos A, Mintz-Weber, Böck A. 2000. Analysis of the cleavage site specificity of the endopeptidase involved in the maturation of the large subunit of hydrogenase 3 from Escherichia coli. Arch Microbiol 173:110–116. [PubMed] 10.1007/s002039900116 [DOI] [PubMed] [Google Scholar]
- 291.Soboh B, Kuhns M, Braussemann M, Waclawek M, Muhr E, Pierik AJ, Sawers RG. 2012. Evidence for an oxygen-sensitive iron-sulfur cluster in an immature large subunit species of Escherichia coli [NiFe]-hydrogenase 2. Biochem Biophys Res Commun 424:158–163. [PubMed] 10.1016/j.bbrc.2012.06.096 [DOI] [PubMed] [Google Scholar]
- 292.Binder U, Maier T, Böck A. 1996. Nickel incorporation into hydrogenase 3 from Escherichia coli requires the precursor form of the large subunit. Arch Microbiol 165:69–72. [PubMed] 10.1007/s002030050299 [DOI] [PubMed] [Google Scholar]
- 293.Kumarevel T, Tanaka T, Bessho Y, Shinkai A, Yokoyama S. 2009. Crystal structure of hydrogenase maturating endopeptidase HycI from Escherichia coli. Biochem Biophys Res Commun 389:310–314. [PubMed] 10.1016/j.bbrc.2009.08.135 [DOI] [PubMed] [Google Scholar]
- 294.Yang F, Hu W, Xu H, Li C, Xia B, Jin C. 2007. Solution structure and backbone dynamics of an endopeptidase HycI from Escherichia coli: implications for mechanism of the [NiFe] hydrogenase maturation. J Biol Chem 282:3856–3863. [PubMed] 10.1074/jbc.M609263200 [DOI] [PubMed] [Google Scholar]
- 295.Magalon A, Blokesch M, Zehelein E, Böck A. 2001. Fidelity of metal insertion into hydrogenases. FEBS Lett 499:73–76. [PubMed] 10.1016/S0014-5793(01)02525-X [DOI] [PubMed] [Google Scholar]
- 296.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10:483–496. [PubMed] 10.1038/nrmicro2814 [DOI] [PubMed] [Google Scholar]
- 297.Berks BC. 2015. The twin-arginine protein translocation pathway. Annu Rev Biochem 84:843–864. [PubMed] 10.1146/annurev-biochem-060614-034251 [DOI] [PubMed] [Google Scholar]
- 298.Palmer T, Sargent F, Berks BC. 2010. The Tat Protein Export Pathway. Ecosal Plus 10.1128/ecosalplus.4.3.2. [PubMed] 10.1128/ecosalplus.4.3.2 [DOI] [PubMed] [Google Scholar]
- 299.Beyer L, Doberenz C, Falke D, Hunger D, Suppmann B, Sawers RG. 2013. Coordination of FocA and pyruvate formate-lyase synthesis in Escherichia coli demonstrates preferential translocation of formate over other mixed-acid fermentation products. J Bacteriol 195:1428–1435. [PubMed] 10.1128/JB.02166-12 [DOI] [PMC free article] [PubMed] [Google Scholar]


