Summary
Annually, in India, 13% of all newborns are preterm, accounting for 23.4% of preterm birth (PTB) globally. The composition and diversity of the vaginal microbiome have a notable degree of ethnic inequality. For understanding differences in vaginal microbiome composition and functions between adverse and normal pregnancy, we have collected, processed and sequenced 600 high vaginal swab (HVS) samples across the three trimesters of pregnancy from 140 women who delivered at term and 60 women who delivered PTB, adopting a targeted metagenomics approach. The microbial signatures in HVS samples showed Lactobacillus genera to be highly abundant in term birth (TB), while in early pregnancy the abundances of Gardnerella, Atopobium, and Sneathia were found to be high in PTB. We further extended our analysis, identified specific microbial genomic signatures, and developed a dipstick assay for rapid identification of PTB-associated microbiota in HVS samples in low-resource settings.
Subject areas: Medicine, Medical science, Women’s health, Reproductive medicine, Diagnostics, Microbiology, Microbiome
Graphical abstract
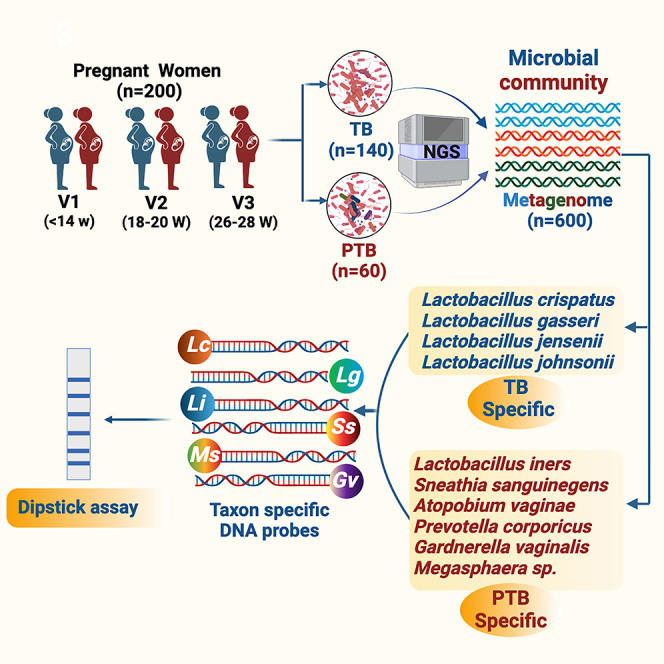
Highlights
-
•
Lactobacillus species in the vaginal microbiome lower the risk of preterm birth
-
•
Increased vaginal microbiome diversity is a significant risk factor for preterm birth
-
•
Higher abundance of Gardnerella, Atopobium, and Sneathia raise the risk of preterm birth
-
•
The dipstick assay quickly identifies PTB-associated microbiota in vaginal samples
Medicine; Medical science; Women’s health; Reproductive medicine; Diagnostics; Microbiology; Microbiome
Introduction
Preterm birth (PTB) is a global public health concern. An estimated 13.4 million babies were born preterm (before completing 37 weeks of gestation) in 2020 with the rate varying from 4 to 16% across countries.1 Children born preterm have increased risk of higher immediate mortality and long-term morbidity from respiratory, cardiac, and endocrine system disorders.2
Further, there is a higher risk of compromised growth and cognition. The complex pathophysiology and etiology of PTB are not well known and about 40–45% of these births happen due to unknown reasons. It is hypothesized that microbial invasion into the amniotic cavity is one of the most frequent causes of spontaneous PTB,3 and the most common route of microbial invasion is ascending from the lower genital tract.4,5,6 Microbial surface proteins are recognized by pattern-recognition receptors—toll-like receptors, which in turn elicit the release of inflammatory chemokines and cytokines. Microbial proinflammatory cytokines stimulate the production of prostaglandins and other inflammatory mediators. Prostaglandin stimulates uterine contractility and may facilitate early parturition of the baby from the mother’s womb. The microbes associated with bacterial vaginosis may ascend into the choriodecidual region leading to bacterial colonization thus triggering immunological response leading to uterine contractions and finally preterm labor.7 The vagina of reproductive-age women is mostly dominated by Lactobacillus species that are considered keystone members of the vagina.8 However, vaginal microbial communities significantly vary in different ethnic groups and based on high-throughput sequencing studies, the vaginal microbiome are classified into five community state types (CSTs) based on dominant Lactobacillus: CST-I to CST-V. CSTs I, II, III, and V are dominated by Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, and Lactobacillus jensenii, respectively, whereas the CST IV refers to high diversity of obligate anaerobic bacteria. Alteration of the normal bacterial flora can lead to bacterial vaginosis (BV), which has been identified as a possible risk factor for PTB.9,10 In a recent pilot study conducted as a cross sectional evaluation of pregnant women at the three trimesters, we showed that along with L. iners, several non-lactobacillus genera dominate the vaginal milieu of women delivering preterm babies compared to those delivering at term.9 To gain insights into the risk factors for PTB at its earliest stages, we investigated the vaginal microbiome of pregnant women. Using these data, we developed a dipstick-based assay for the rapid identification of microbiota linked to specific birth outcomes.
Results
Characteristics of the study participants
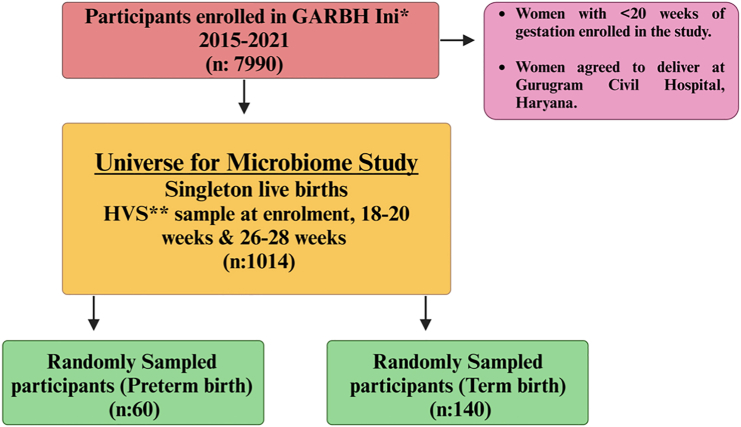
The participants for this case-control analysis were selected as described in STAR Methods and in Figure 1. The average age of women having term or preterm delivery was similar (mean 23.2 vs. 23.7 years). The mean gestational age at delivery was 40.2 weeks for term birth (TB) and 35.3 weeks for PTB. The proportions of normal vaginal deliveries were high for TB at 73.72%, compared to that for PTB at 61.29%. In case of Caesarean deliveries, TB had a lower rate at 26.28%, while PTB showed a higher percentage at 38.71% (Table 1).
Figure 1.
Infographic of participants enrolled in Interdisciplinary Group for Advanced Research on Birth Outcomes (GARBH-Ini)—DBT India Initiative cohort Gurugram, Haryana, India (∗Date: 18th May, 2015–8th August, 2020)
Out of 6,736 singleton live births, 834 women delivered preterm and 5,902 delivered at term. ∗∗High vaginal swabs (HVS) collected at all the three different time points (at enrollment, 18–20 weeks, 20–26 weeks of gestation) from preterm birth (PTB) delivery mothers (n = 124) and term birth delivery mothers (TB) (n = 890). In the present study, we have processed and sequenced 600 HVS samples across the three trimesters of pregnancy consisting of term (140) and preterm (60) delivering Indian women.
Table 1.
Demographic characteristic of the study participants
| Characteristic | Term (n = 140) |
Preterm (n = 60) |
|---|---|---|
| Mean (Min–Max) or n (%) | Mean (Min–Max) or n (%) | |
| Clinical and demographic | ||
| POG at delivery (weeks, days) | 40W 2D (37W 1D–42W 4D) | 35W 3D (27W 1D–36W 6D) |
| Maternal age (year) | 23.2 (18–35) | 23.7 (18–32) |
| Weight at enrollment (kg) | 49.3 (34.25–72.5) | 48.3 (34.3–76.4) |
| BMI (kg/m2) at enrollment | 20.72 (13.89–31.52) | 20.64 (14.90–31.15) |
| BMI (kg/m2) category | ||
| Underweight | 30 (22.14%) | 19 (31.67%) |
| Normal | 94 (67.14%) | 34 (56.67%) |
| Overweight | 13 (9.29%) | 6 (10.00%) |
| Obese | 2 (1.43%) | 1 (1.67%) |
| Gravidity | ||
| Primigravida | 61 (43.57%) | 20 (33.33%) |
| Multigravida | 79 (56.43%) | 40 (66.66%) |
| History of diarrhea | ||
| Present | 4 (2.86%) | 3 (5.00%) |
| Absent | 136 (97.14%) | 57 (95.00%) |
| Socioeconomic status/Household wealth (Kuppuswamy scale 2023, CPI-IW) a | n = 133 | n = 56 |
| Upper middle class (16–12) | 27 (30.30%) | 14 (25.00%) |
| Lower middle class (11–15) | 50 (37.59%) | 19 (33.93%) |
| Upper lower class (5–10) | 54 (40.60%) | 23 (41.07%) |
| Lower class (<5) | 2 (1.50%) | 0 (0.00%) |
| Education status | ||
| Illiterate | 21 (15.00%) | 11 (18.33%) |
| School | 92 (65.70%) | 44 (73.34%) |
| College | 27 (19.29%) | 5 (8.33%) |
| House type | ||
| Pucca house (concrete) | 135 (96.43%) | 58 (96.67%) |
| Kachha house (mud, bamboo, grass) | 5 (3.57%) | 2 (3.33%) |
| Laboratory characteristics | ||
| Mode of delivery | n = 137 | n = 59 |
| Normal vaginal | 101 (73.72%) | 38 (61.29%) |
| Caesarean | 36 (26.28%) | 24 (38.71%) |
| Nugent score | ||
| 0-3 (Negative for BV) | 20 (33.3%) | 41 (30.1%) |
| 4-6 (Intermediate for BV) | 32 (53.3%) | 87 (63.9%) |
| 7+ (Positive for BV) | 8 (13.3%) | 8 (5.8%) |
| Nugent score at enrollment | 3.9 (0–8) | 4.9 (0–10) |
| Nugent score at Visit 2 | 4.1 (0–8) | 4.2 (3–6) |
| Nugent score at Visit 3 | 3.5 (0–8) | 3.3 (0–7) |
| Vaginal pH at sampling |
n = 136 5.1 (4–6) |
n = 60 5.2 (5–6) |
| Antibiotic intake at sampling | 0 (0.00%) | 0 (0.00%) |
CPI-IW stand for Consumer Price Index Number for Industrial Workers, Labour Bureau, Ministry of Labour and Employment, Government of India; n represents numbers of study participants.
Inter individual microbiome diversity between term and preterm samples
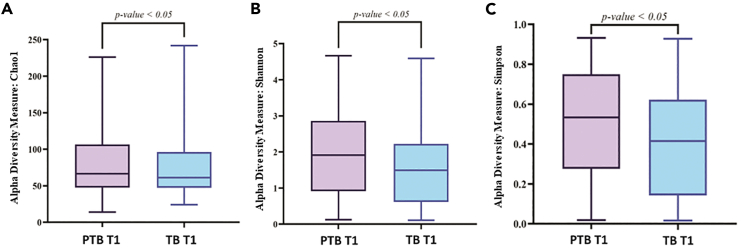
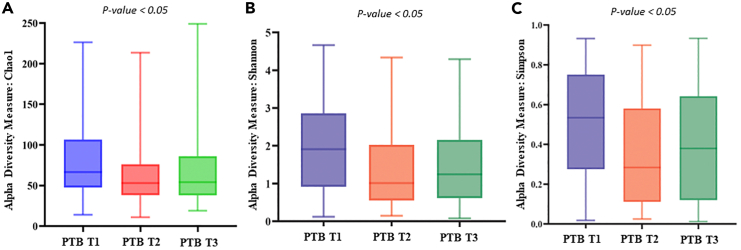
A total of 186.95 M (average 0.34 M reads/sample) raw reads were obtained from all the samples. After quality filtering and chimera removal, total of 163.06 M reads (average 0.30 M reads/samples) remained which were clustered to 8,834 amplicon sequence variant (ASV) sequences. At the end, total 8,324 ASVs remained for downstream analysis after singleton ASV removal and negative control adjustment. Rarefaction plot was generated to understand the minimum number of reads/samples required for estimating intra-individual diversity within that sample i.e., the alpha diversity index reached a plateau. In our data we observed that ∼5,000 reads/samples are sufficient to reach a plateau (Figure S2). From the alpha diversity analysis, it was found that, in the initial period of pregnancy or at the 1st trimester (T1), the Chao1 (PTB: 78.9 ± 41.6, TB: 74.8 ± 42.4; p value = 0.03), the Shannon (PTB: 2.0 ± 1.2, TB: 1.6 ± 1.1; p value = 0.003), and Simpson (PTB: 0.5 ± 0.3, TB: 0.4 ± 0.3; p value = 0.005) indices were significantly higher in PTB samples compared to TB samples (Figure 2). In the 2nd (T2) and 3rd (T3) trimesters the PTB samples showed a higher (not significantly different) alpha diversity indices compared to TB samples. Interestingly, it was also observed that alpha diversity measures were significantly altered within the PTB group (Chao1 p value = 0.02, Shannon p value = 0.004, Simpson p value = 0.005) across different trimesters (T1, T2, and T3) (Figure 3).
Figure 2.
Boxplots showing different alpha diversity measures between preterm (PTB) and term birth (TB) samples in 1st trimester of pregnancy (T1)
Alpha diversity was found to be significantly higher in PTB group compared to TB group for (A) Chao1 index (p value <0.05) (B) Shannon index (p value < 0.01), (C) Simpson index (p value < 0.01). The alpha diversity index was represented by box-and-whisker plot where the box indicates the interquartile range (IQR). The median value is represented as a line within the box and whiskers extend to the extreme (highest and lowest) value that is within 1.5∗IQR.
Figure 3.
Boxplots showing different alpha diversity measures within the preterm group (PTB) across different trimesters (T1, T2, and T3)
Alpha diversity measures were significantly altered in PTB group for (A) Chao 1 Index (p value = 0.024) (B) Shannon index (p value = 0.0045), and (C) Simpson index (p value = 0.0053). The alpha diversity index was represented by box-and-whisker plot where the box indicates the interquartile range (IQR). The median value is represented as a line within the box and whiskers extend to the extreme (highest and lowest) value that is within 1.5∗IQR.
Dynamics of vaginal microbiome composition in term and preterm samples during pregnancy
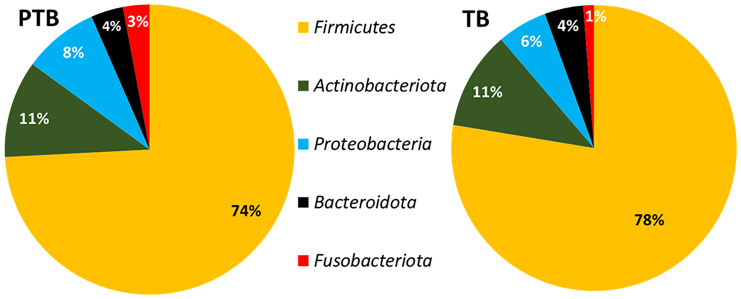
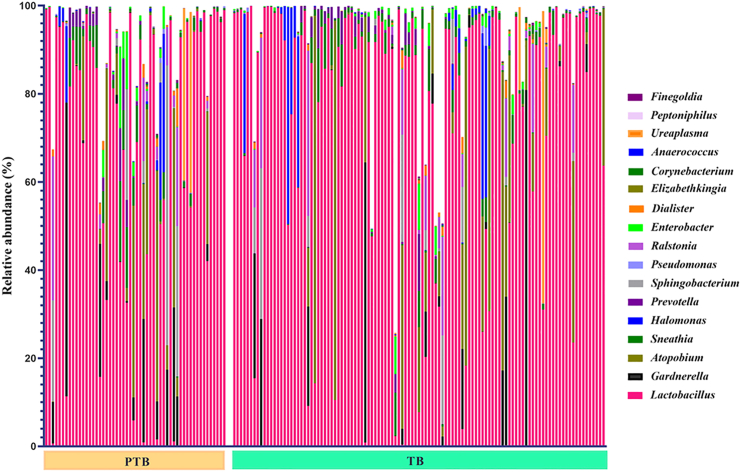
Total 25 microbial phyla and 618 genera were obtained after taxonomic assignment of ASV sequences. Core taxa were selected from the total number of taxa, on which the statistical comparisons were performed. Core taxa are those that are present in any of the two groups (i.e., TB or PTB) and at any of the three trimesters (T1/T2/T3): with (1) average relative abundance ≥0.1%, and (2) present in ≥50% of women in that group. Based on these criteria, a total of 5 phyla and 17 genera were selected as core taxa that comprised an average of 98% of the total relative abundance of the TB and PTB groups. At phylum level, it was found that Firmicutes was the most abundant phylum in both the groups (TB: 78%, PTB: 74%) followed by Actinobacteria (TB: 11%, PTB: 11%) and Proteobacteria (TB: 6%, PTB: 8%). The abundance of the phylum Fusobacteria was notably higher in PTB samples (3%) compared to TB samples (1%) (Figure 4). At phylum level, Fusobacteria was the only phylum found to be significantly high (p value = 0.001) in PTB compared to TB in the 1st trimester (T1) of pregnancy. At genera level, out of 17 core genera, Lactobacillus was the highest abundant taxa in both TB and PTB (TB: 74.16%, PTB: 68.63%, p value = 0.31) groups followed by Gardnerella (TB: 5.24%, PTB: 6.85%, p value = 0.24), Atopobium (TB: 2.98%, PTB: 4.05%, p value = 0.03), Sneathia (TB: 1.17%, PTB: 2.94%, p value = 0.005), Halomonas (TB: 1.87%, PTB: 2.83%, p value = 0.61), and Prevotella (TB: 1.93%, PTB: 1.82%, p value = 0.44) (Figure 5).
Figure 4.
Pie chart showing the core phyla with their relative abundances
Core phyla and their relative abundance (%) in preterm birth (PTB) group and core phyla and their relative abundance (%) in term birth (TB) samples. The colors in the pie chart represent different phyla, each indicating their mean relative abundance (%). Firmicutes was found to be the most abundant phylum in both PTB and TB (74% and 78%) followed by Actinobacteia (11% in both PTB and TB) and Proteobacteria (8% and 6%). The abundance of phylum Bacteroidota had a similar profile in both PTB and TB samples. Significant difference was seen for phylum Fusobacteria (p value = 0.001). The colors in the chart represent different phylum.
Figure 5.
Differential abundance of vaginal microbiome in preterm (PTB) and term birth (TB) delivering women from India
The bar plot showing the relative abundance (%) of seventeen different genera in PTB and TB women. Genus Lactobacillus was found to be the highest abundant in both PTB and TB delivering women. The bar plot illustrates individual-level data for the TB and PTB groups at the genus level. Each color in the bars corresponds to a different genus, while the height of the bars indicates the relative abundance of each genus within the groups.
Vaginal microbiome composition in early pregnancy between term and preterm samples
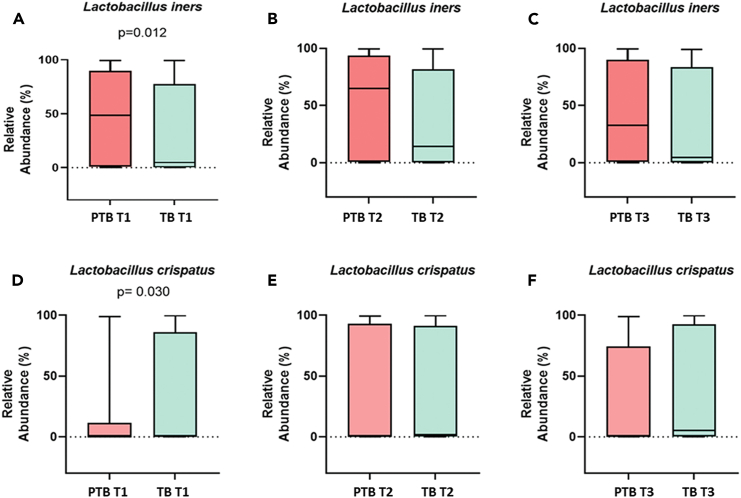
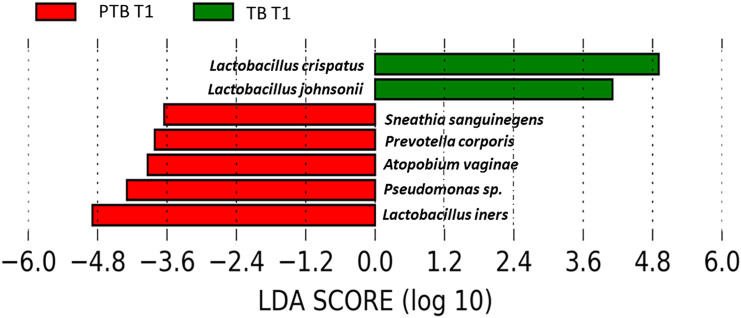
Lactobacillus is the highest abundant genus in both term and preterm delivering women. Species-level analysis revealed that a total of 26 species belonged to the Lactobacillus group. By comparing the relative abundance of each species of Lactobacillus, we have found that the higher abundance of L. crispatus was significantly associated with term delivery (TB: 32.23%, PTB: 14.83% in T1; p value = 0.03), whereas the higher abundance of L. iners was significantly associated with preterm delivery at 1st trimester of pregnancy (TB: 33.62%, PTB: 48.37% in T1; p value = 0.01).11,12 The higher abundance of L. crispatus in the early pregnancy (T1) are associated with a greater likelihood of delivering at term, while higher levels of L. iners are linked to an increased risk of preterm delivery. These associations are statistically significant, suggesting that the presence and abundance of these bacteria in early pregnancy (T1) could potentially influence the timing of delivery (Figure 6). By linear discriminant analysis (LDA) it was also observed L. crispatus is the most discriminant taxon in TB whereas L. iners is the most discriminant taxon in PTB in early pregnancy (T1) (Figure 7). Lactobacillus johnsonii was also found to be discriminant between TB and PTB in the 1st trimester. Some non-Lactobacillus vaginal flora, such as Atopobium vaginae (TB: 2.66%, PTB: 3.55%, p value = 0.009 in T1), Sneathia sanguinegens (TB: 0.62%, PTB: 1.24%, p value = 0.02 in T1), Prevotella corporis (TB: 0.002%, PTB: 0.1%; p value = 0.01 in T1), and Pseudomonas sp. identified in the 1st trimester (T1) vaginal fluid were found to be significantly higher in PTB samples compared to TB samples.
Figure 6.
Boxplots showing the relative abundance (%) of L. iners and L. crispatus in preterm (PTB) and term (TB) samples in three trimesters
(A–C) Represent relative abundance of L. iners and (D–F) represent relative abundance of L. crispatus.
(A) Relative abundance at 1st trimester (p value = 0.012).
(B) Relative abundance at 2nd trimester (p value > 0.05).
(C) Relative abundance at 3rd trimester (p value > 0.05) of L. iners in PTB and TB samples.
(D) Relative abundance at 1st trimester (p value = 0.03).
(E) Relative abundance at 2nd trimester (p value > 0.05).
(F) Relative abundance at 3rd trimester (p value > 0.05) of L. crispatus in PTB and TB samples. The box of box-and-whisker plot indicates the interquartile range (IQR). The median value is represented as a line within the box and whiskers extend to the extreme (highest and lowest) value that is within 1.5∗IQR.
Figure 7.
Differential abundance of vaginal microbiome in preterm (PTB) and term birth (TB) delivering women from India
Linear discriminant analysis size effect (LefSe) plot showing the most differentially abundant species (n = 7) among PTB and TB samples in the first trimester of pregnancy (T1). The x axis of the plot represents the LDA score on a log10 scale, while the y axis displays the discriminant taxa identified in the TB and PTB groups.
Community state type reveals vaginal microbial community structure
Interestingly, not all pregnant women are dominated by the same bacterial taxa in their vagina during pregnancy. We have categorized the pregnant women based on their vaginal microbiome composition; the community state types (CSTs) based on the relative abundance of the most dominant species present in the vagina.13 Each CST was named based on the most abundant taxa in that community. We obtained a total seven CSTs; CST-I was dominated by L. crispatus, CST-II by L. gasseri, CST-III by L. iners, and CST-IV by non-Lactobacillus taxa. Additionally, three Lactobacillus CSTs L. johnsonii, L. jensenii, and Lactobacillus mulieris were also observed. We found that nearly 36% of TB samples belonged to CST-I, compared to just 19% of PTB samples (L. crispatus CST) in 1st trimester (T1). In PTB, approximately 61% samples at T1, 54% at T2, and 52% at T3 belonged to the CST-III (L. iners CST) category, whereas in TB, only 41% at T1, 43% at T2, and 40% at T3 belonged to the CST-III category (Table 2). CST-I (L. crispatus CST) was found to be significantly higher in TB in the 1st trimesters (p value <0.05, equality of proportion test in R) whereas CST-III (L. iners CST) was found to be significantly higher in PTB in 1st and 3rd trimesters of pregnancy. L. johnsonii and L. jensenii dominant CSTs were completely absent in PTB samples at any of the trimester.
Table 2.
Community state type in term birth and preterm birth samples
| Community State Types (CSTs) | Dominant CST taxa | PTB-T1 (% of individuals) | TB-T1 (% of individuals) | p value | PTB-T2 (% of individuals) | TB-T2 (% of individuals) | p value | PTB-T3 (% of individuals) | TB-T3 (% of individuals) | p value |
|---|---|---|---|---|---|---|---|---|---|---|
| CST-I | Lactobacillus crispatus | 19% | 36% | 0.006 | 37% | 39% | 0.88 | 29% | 46% | 0.09 |
| CST-II | Lactobacillus gasseri | 2% | 3% | 0.999 | 2% | 2% | 0.924 | 3% | 2% | 0.693 |
| CST-III | Lactobacillus iners | 61% | 40% | 0.015 | 54% | 43% | 0.15 | 52% | 40% | 0.11 |
| CST-IV | Non-Lactobacillus | 16% | 14% | 0.88 | 4% | 7% | 0.53 | 14% | 6% | 0.009 |
| L. johnsonii CST | Lactobacillus johnsonii | 0% | 4% | 0.170 | 0% | 2% | 0.375 | 0% | 2% | 0.623 |
| L. jensenii CST | Lactobacillus jensenii | 0% | 1% | 1 | 0% | 3% | 0.225 | 0% | 2% | 0.356 |
| L. mulieris CST | Lactobacillus mulieris | 2% | 2% | 0.580 | 3% | 4% | 0.294 | 2% | 2% | 0.667 |
p value less than 0.05 is mentioned in bold font.
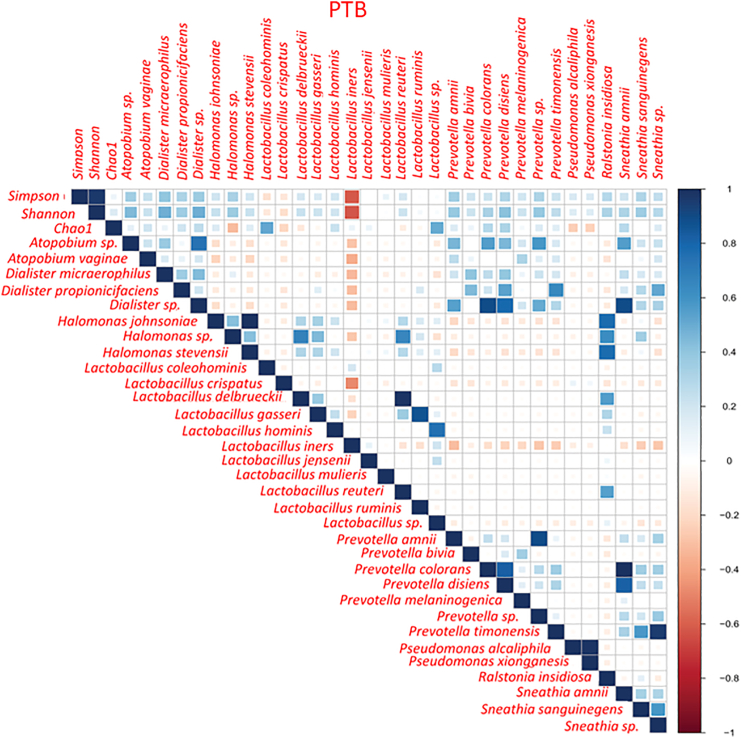
Correlation study reveals the positive and negative associations between microbial species in the vagina
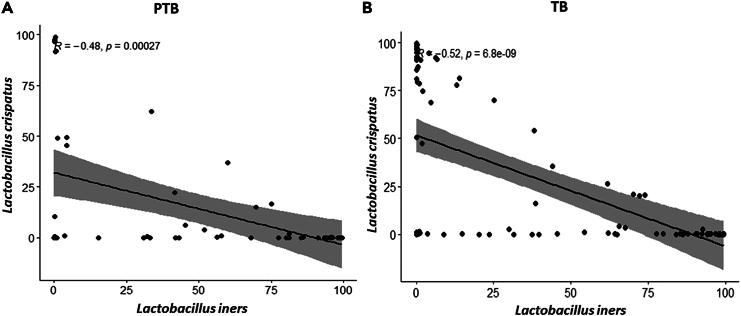
To understand the relation among all the species representing the core genera, Spearman rank correlation test was performed separately in term and preterm samples. We have found that L. crispatus and L. iners were negatively correlated (p value PTB = 2.7 × 10−4, p value TB = 6.8 × 10−9) to each other throughout the pregnancy in both the groups (Figure 8). L. crispatus was also found to be negatively correlated with other non-Lactobacillus taxa like A. vaginae, Dialister micraaerophilus, Halomonas sp. etc., in both TB and PTB samples (Figure 9; Figure S3). Correlation between vaginal microbial taxa and alpha diversity indices was also investigated in this current study. Non-parametric spearman rank correlation test revealed that the Shannon diversity index was positively correlated with relative abundance of non-Lactobacillus taxa but negatively correlated with L. crispatus, Lactobacillus coleohomonis, and L. iners in both the groups (Figure 9; Figure S3).
Figure 8.
Correlation between L. crispatus and L. iners between preterm (PTB) and term birth (TB) samples using Spearman’s rank correlation test
The correlation plot between L. crispatus and L. iners in (A) PTB group and in (B) TB group. The relative abundances of both species were used for calculation of correlation coefficient value. The correlation coefficient and p value are mentioned in both the plots. The x axis of the plots denoted the relative abundance (%) of L. iners and y axis denoted the relative abundance (%) of L. crispatus. L. crispatus and L. iners were found to be negatively correlated with each other throughout the pregnancy in both the TB and PTB groups (PTB: R2 = −0.48, p value = 0.00027; TB: R2 = −0.52, p value = 6.8 × 10−9).
Figure 9.
Correlation between vaginal microbial species and Shannon diversity index using non-parametric Spearman’s rank correlation test in preterm birth (PTB) group
The x axis and y axis of the correlation plot represent the species and alpha diversity indices. Each box of the plot represents the correlation coefficient value between two variables. The color of each box represents either positive or negative correlation and the color intensity represents the strength of the correlation. Blue squares represent positive correlation, red squares represent negative correlation and white squares represent non-significant correlations.
To understand the relationship between the abundance of vaginal taxa and the most relevant clinical factor for the vaginal health, i.e., the Nugent score, the Spearman correlation was performed in TB and PTB samples separately. As a result, we have found that, the Nugent score is negatively correlated with L. crispatus abundance in both the groups (TB ρ: −0.16, p value T1 = 0.08; PTB ρ: −0.25, p value T1 = 0.06) but not significant (Figure S4). A positive correlation was observed between L. iners and the Nugent score in TB samples (ρ: 0.10, p value T1 = 0.29) only. Some significantly positive correlations were observed between Nugent score and non-Lactobacillus taxa such as Prevotella timonensis (ρ: 0.15, p value T1 = 0.007), Ralstonia sp. (ρ: 0.25, p value T1 = 0.0004) and Sneathia sp. (ρ: 0.25, p value T1 = 0.01) in TB samples only. A significantly negative correlation was also observed between Lactobacillus reuteri and the Nugent score (ρ: −0.17, p value T1 = 0.02) in term samples.
(Figure S4A). In preterm sample also L. coleohominis (ρ: −0.32, p value T1 = 0.02) was found to be significantly negatively correlated with Nugent score. Whereas, the Prevotella sp. (ρ: 0.23, p value T1 = 0.02) and A. vaginae (ρ: 0.13, p value T1 = 0.02) were found to be positively correlated with the Nugent score (Figure S4B).
Longitudinal trend of L. crispatus and L. iners in term and preterm samples
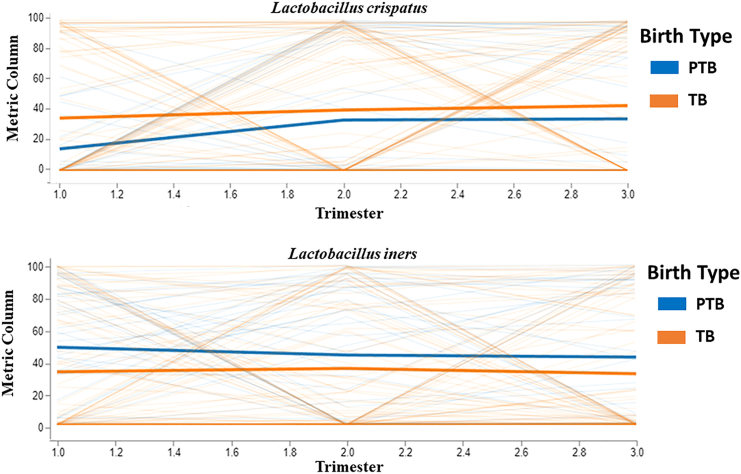
We investigated the fixed effects of birth type (preterm/term) and gestation time (first/second/third trimesters) on the relative abundance of taxa and their species level data, treating age of the mother as a random effect. The effects of both birth type (TB/PTB) and gestational time (trimesters) were significant for L. crispatus, L. jensenii, and L. iners longitudinally across all the trimesters. Longitudinal analysis showed that the relative abundance of L. crispatus and L. jensenii were higher in term samples and L. iners was higher in preterm samples throughout the pregnancy period (Figure 10) (Table S1).
Figure 10.
Linear mixed effect (LME) model-based analysis
The analysis is performed to study the fixed effect of birth type (preterm and term) and gestation time (1st, 2nd, and 3rd trimester) on taxa relative abundance using q2-longitudinal of Qiime2. The Volatility plot showing the relative abundance of L. crispatus and L. iners throughout the pregnancy in term and preterm samples. The plot features two colors, each representing the TB and PTB groups. The x axis indicates various time points during pregnancy, while the y axis shows the relative abundance (%) of species. Dark-colored lines illustrate the mean relative abundance of either L. crispatus or L. iners, whereas light-colored lines represent the distribution of individual-level data.
Predictive functional potency of the microbiome reveals several key pathways that are differentially enriched in preterm and term samples
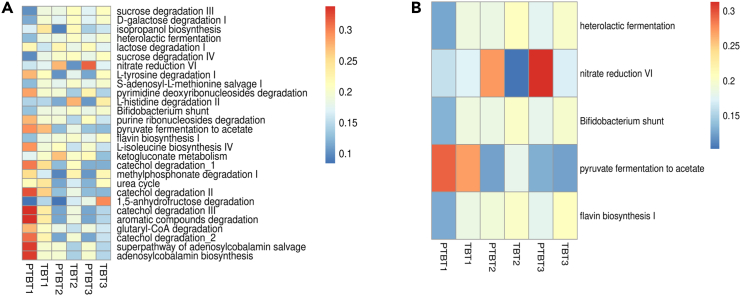
The metabolic capacity of the vaginal microbiome in term and preterm delivering women was inferred and this identified a total of 408 metabolic pathways across the samples. We found 28 metabolic pathways that were significantly altered (p value <0.05) between TB and PTB samples (Figure 11A; Table S2). However, in this study we will mainly focus on some specific pathways (n = 5) out of the 28 significant pathways that may have relevance with birth outcomes (Figure 11B). Heterolactic fermentation pathway that produces lactate along with carbon dioxide and ethanol as the end product was found to be highly enriched in less than 14 weeks (T1) of TB delivering mothers (mean relative abundance = 0.19) as compared to PTB delivering mothers (mean relative abundance = 0.13). In addition to acidifying the vaginal milieu and inhibiting the growth of pathogenic anaerobes, both the isomers of lactic acid (L and D) exhibit anti-inflammatory properties stimulating increased levels of IL-1RA without a concomitant increase in IL-1β, IL-6, IL-8, TNFα, RANTES, and MIP-3 α.ET.14 Furthermore, nitrate reduction VI (assimilatory) pathway showed a significant increase as the pregnancy progressed in mothers having preterm deliveries. Further, it was found to be highly enriched in the preterm delivering mothers in both T2, 18–20 weeks (mean in PTB = 0.27, mean in TB = 0.10) and T3, 26–28 weeks (mean in PTB = 0.31, mean in TB = 0.10) trimesters of pregnancy when compared with TB deliveries. The nitrite (NO2), which is produced as an intermediate compound, is previously reported to be associated with adverse gestational outcomes.15 Flavin biosynthesis I pathway was found to be highly enriched in the early pregnancy (T1) of TB (mean = 0.18) than in preterm deliveries (mean = 0.13). However, pathways (Bifidobacterium shunt) involved in the production of short-chain fatty acids, whose end product is lactate and acetate were found to be significantly altered in early pregnancy (T1) of TB (mean = 0.19) as compared to those being delivered preterm (mean = 0.13). Interestingly, the abundance of lactate and acetate gradually increased with increasing trimesters in mothers with PTB. Of note, another pathway producing butyrate and acetate as an intermediate product during pyruvate fermentation was found to be enriched in the mid trimester (T2) of mothers delivering at term (mean = 0.18) compared with those having PTB (mean = 0.12). However, abundance was found to be lower as the mothers proceeded toward the last trimester (T3) of pregnancy and this time it was found to be consistent with mothers delivering preterm. The functional potential of the targeted metagenomes was predicted based on the distribution of metabolic pathways in the core mycobiome; however, subsequent validation of the metabolic function is needed.
Figure 11.
Heatmap showing the significant functional profiles inferred by PICRUSt2
(A) Out of 408 metabolic pathways inferred through PICRUSt2, a total of 28 differentially abundant pathways (p value < 0.05) were identified among preterm (PTB) and term (TB) delivering mothers.
(B) Out of 28 significant metabolic pathways, 5 pathways that may have relevance with birth outcomes are represented here. The values represent the mean relative abundance in each trimester (T1, T2, and T3) among the two groups. The blue-red color in the scale represents lowest to highest mean relative abundance.
Rapid detection of PTB associated bacterial taxa using dipstick-based chromatography
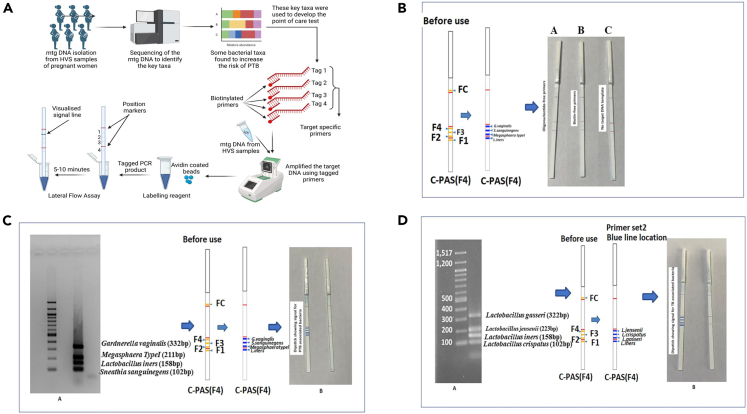
Improved diagnostic techniques, such as next-generation sequencing (NGS), have allowed for more accurate identification of microbial species present in the vaginal ecosystem. However, the use of specific approaches, such as deep sequencing of the 16S rRNA gene, is expensive, time-consuming, and hence not suitable in low-resource settings. To date, studies on the vaginal microbiome and PTB have been limited to metagenomic sequencing only. In the current study, we have translated our findings to the development of a rapid diagnostic point-of-care test that can detect four PTB and four TB-specific bacterial taxa at the early stage of gestation. Therefore, this can be a promising tool that can be used to screen Indian women at the community level for BV infection. Targeted metagenomics identified specific DNA sequences in the 16S rRNA gene of bacteria taxa that significantly increased the risk of PTB. In our previous study,9 we identified seven bacteria viz, L. crispatus, L. gasseri, and L. jensenii to be associated with TB, whereas S. sanguinegens, Gardnerella vaginalis, Megasphaera sp, and L. iners were found to be linked with PTB. We identified and used taxa specific DNA sequences for developing a potential point-of-care dipstick assay for rapid identification of bacterial taxa in the early pregnancy of women at high risk of PTB. The taxa specific oligonucleotides (n = 14) were tagged with biotin and the regions were amplified in a thermal cycler using genomic and metagenomic DNA isolated from high vaginal swab samples. Due to the high affinity between streptavidin and biotin, the biotin-labeled PCR amplicon quickly formed a hybrid with the streptavidin-coated blue latex particles in a post PCR reaction mixture. The PCR amplicon-blue latex hybrids moved up through the dipstick strip by capillary action when the tip of the strip was inserted into the mixture. Further, the amplicon was bound to the strip as it contains the primer complementary oligos. Hybridization of the primers causes accumulation of blue latex microspheres leading to a visible blue test line. The flow control line (FC) was embedded on the opposite tip of the strip to verify the proper functioning of the assay. With this approach, we detected seven bacterial taxa in two strips that are associated with term (n = 3) and PTB (n = 4) after 20 min of incubation at room temperature. The flow chart representing the multistep process is represented in Figure 12A.
Figure 12.
PCR-dipstick DNA chromatography
(A) Flowchart outlining the multistage process of dipstick DNA chromatography.
(B) Specificity of the PCR-Dipstick DNA chromatography: A: target amplicons tagged by oligonucleotides only. B: target amplicons tagged by biotin only. C: PCR performed with no target DNA templates.
(C) PCR-based dipstick DNA chromatography multiplex assay precisely identified the presence of Gardnerella, Sneathia, Megasphaera, and L. iners. A: gel image of multiplex PCR for preterm birth (PTB) associated bacteria. B: dipstick showing signal for PTB associated bacteria, i.e., G. vaginalis, Megasphaera type I, S. sanguinegens, and L. iners, along with the negative control.
(D) PCR-based dipstick DNA chromatography multiplex assay precisely identified the presence of L. gasseri, L. jensenii, L. iners, and L. crispatus. A: gel image of multiplex PCR for term birth (TB) associated bacteria. B: dipstick showing signal for TB associated bacteria, i.e., L. gasseri, L. jensenii, L. iners, and L. crispatus, along with the negative control.
Specificity and sensitivity of PCR-dipstick DNA chromatography
The specificity of PCR-dipstick DNA chromatography was confirmed under the following conditions: (1) oligonucleotide-free primers; (2) biotin-free primers; and (3) no target DNA template (Figure 12B). First, we optimized the amplification reactions in in vitro conditions utilizing the genome of seven representative bacteria including S. sanguinegens, G. vaginalis, Megasphaera sp, L. iners, L. crispatus, L. gasseri, and L. jensenii isolated from the vaginal milieu of women from the universe of microbiome study participants of GARBH-Ini cohort and further ruled out for any non-specific amplification due to cross-hybridization with other bacterial strains. Additionally, in-silico BLAST searches were also carried out using the National Center for Biotechnology Information (NCBI) Website to confirm the specificity of primers and further validated in vitro. Our PCR-based dipstick DNA chromatography multiplex assay precisely identified the presence of Gardnerella, Sneathia, Megasphaera, L. iners (Figure 12C), L. crispatus, L. gasseri, and L. jensenii (Figure 12D) in the high vaginal swab (HVS) samples of pregnant Indian women. The abundance profile of these 7 bacterial species was found to be consistent with our previous results, except Megasphaera (Table 3).
Table 3.
Abundance profile of bacterial species selected for dipstick assay (present study)
| Species | PTB-T1 | TB-T1 | p value | PTB-T2 | TB-T2 | p value | PTB-T3 | TB-T3 | p value |
|---|---|---|---|---|---|---|---|---|---|
| Lactobacillus iners | 0.58 | 0.42 | 0.03 | 0.53 | 0.45 | 0.2 | 0.53 | 0.42 | 0.1 |
| Gardnerella vaginalis | 0.1 | 0.12 | 0.2 | 0.06 | 0.08 | 0.6 | 0.09 | 0.07 | 0.88 |
| Megasphaera sp. | 0.05 | 0.02 | 0.6 | 0.02 | 0.01 | 0.6 | 0.04 | 0.01 | 0.1 |
| Sneathia sanguinegens | 0.04 | 0.02 | 0.02 | 0 | 0.01 | 0.16 | 0.01 | 0 | 0.53 |
| Lactobacillus crispatus | 0.2 | 0.37 | 0.02 | 0.35 | 0.4 | 0.5 | 0.3 | 0.45 | 0.53 |
| Lactobacillus gasseri | 0.04 | 0.03 | 0.2 | 0.03 | 0.02 | 0.66 | 0.03 | 0.02 | 0.45 |
| Lactobacillus jensenii | 0 | 0.01 | 0.4 | 0 | 0.03 | 0.03 | 0.01 | 0.03 | 0.9 |
We observed 67.1% specificity and 74.2% sensitivity of detection of bacteria that had shown significant association with either TB or PTB. The positive predictive value, or the probability likelihood that an individual with a positive screening test actually has the condition, is 62.9% for PTB associated bacteria and 56.9% for TB associated bacteria, respectively. The negative predictive value, i.e., the probability that the bacteria associated with PTB is absent in the sample when the test was negative is 79.4% for PTB and 77.8% for TB, respectively.
Discussion
PTB can lead to both short-term morbidity and mortality and long-term impact on childhood growth and neurodevelopment. To better understand environmental risk factors of PTB at its earliest stages we focused on the vaginal microbiome of pregnant women. Several studies have highlighted that genetic variations among individuals, which can correlate with race or ethnic background, significantly influence the composition and functional potency of the vaginal microbiome.13 Additionally, environmental factors such as diet, lifestyle, and geographic location play crucial roles in shaping microbiome composition and function. Previous studies have shown that the vaginal microbiome can vary based on the host’s race and ethnicity, revealing differences in microbial communities, particularly bacterial species and their relative abundances.9,16,17 Distinct genetic backgrounds among ethnic groups can predispose individuals to specific types of microbial colonization. Therefore, understanding the relationship between vaginal microbiome composition and ethnicity is essential for identifying individuals at risk of PTB and developing targeted interventions to reduce PTB rates. This study, which is pioneer of its kind in India with a substantial sample size of 600 high vaginal swabs, explores region-specific microbial taxa associated with PTB during the first trimester of pregnancy, offering valuable insights into ethnic-specific risk factors. Study subjects were selected from a large pregnancy cohort in India, GARBH-Ini, where 600 high vaginal swab samples were collected longitudinally across pregnancy from 200 pregnant women and evaluated the association of the vaginal microbiome composition with PTB in comparison to TB using a nested case-control study design. In our study, Lactobacillus was found to be the most dominant taxa in both the groups. However, species level investigation showed that distinct species of Lactobacillus are enriched in different study groups. We found L. crispatus has the highest abundance in the vaginal ecosystem of women who delivered at term whereas L. iners was mostly enriched in those who had a preterm baby. Longitudinal analyses showed that L. crispatus and L. iners were significantly higher across trimesters in term and preterm samples, respectively. Higher abundance of anaerobic non-Lactobacillus taxa during gestational period was found in women delivering preterm. The microbes associated with preterm and TB have distinct genomic signatures that helped us to develop a dipstick-based assay for rapid identification of the microbiota associated with specific birth outcomes.
It is a well-established fact that Lactobacillus sp. like L. crispatus, L. gasseri, L. jensenii and L. vaginalis are the hallmark of a healthy vagina that protects from the colonization of allochthonous microbiota and also maintains lower pH and hygiene.18 A number of previous studies reported the association between presence of allochthonous vaginal taxa and PTB.13,19 Our study corroborated these findings with distinct region-specific microbiota. More recently, increased abundance of allochthonous microbiota including G. vaginalis and other anaerobic microbes were associated with adverse pregnancy outcomes20 in Bangladesh. Other bacterial taxa associated with term (L. crispatus and Finegoldia) and preterm (Prevotella buccalis) birth outcomes have been seen in populations from Thailand-Myanmar.21 Further, BV associated non-Lactobacillus taxa were found to be linked with PTB in different ethnicities.16,22 In contrast, a few studies from different regions23,24 could not find any association with vaginal microbiome and PTB and TB. This suggests that microbial taxa associated with birth outcomes have region and population specific signatures.
Recent studies have shown that the BV-associated microbial taxa can mutually co-exist with L. iners and increases the risk of PTB in Caucasian and African-American ethnicity.17 In our cohort, we observed a strong negative correlation between L. crispatus and L. iners, indicating that there may be a possible negative correlation of PTB with other associated bacterial taxa. L. crispatus might play an important role to keep vagina less diverse and reduce the growth of facultative anaerobes in the vaginal milieu. Our results are further strengthened by reports showing strong association between L. iners and PTB in pregnant women from China.25 This emphasizes the potential of these bacteria as biomarkers for early prediction of bacterial dysbiosis that could be associated with PTB. L. crispatus and L. iners are the most dominant species in our study and are negatively correlated with Shannon diversity index that indicates that dominant Lactobacillus may inhibit the growth of other less abundant taxa in the vaginal cavity. The increased Shannon diversity in our preterm samples may be due to other non-Lactobacillus taxa that may explain the positive correlation between the Shannon index and non-lactobacillus taxa.
Our study being a prospective longitudinal cohort was specifically placed to identify the dysbiotic pattern associated with PTB early in pregnancy. We identified L. iners to be significantly high in the first trimester in women who delivered preterm. In addition, a few other anaerobic bacteria like S. sanguinegens, Atopobium, and P. corporis were present in significantly high numbers in HVS of mothers with PTB in early pregnancy. Metagenomic sequencing is very sensitive and specific, but is expensive, time-consuming, and requires technical expertise to identify the microbiota. It is therefore difficult to implement this in low-resource settings as a diagnostic tool for identification of PTB associated microbiota in the vaginal milieu. Till now, people have identified the vaginal microbial taxa associated with TB and PTB mostly by exploring metagenomic sequences. Our study has translated the metagenomic findings into a diagnostic assay for rapid detection of specific bacterial taxa that could facilitate risk stratification of PTB in a minimal-resource health care facility. The nucleic acid sequence specific probes developed in this study allow for rapid identification of the seven major bacterial species found to be differentially abundant in the PTB and TB samples of Indian pregnant women using a highly sensitive metagenomic reads as the “gold standard.” Nucleic acid detection-based approaches eliminate the necessity for in vitro culture and reduce the time required to detect bacterial species. DNA biosensors provide a way to simplify post-PCR analysis, with much improved detectability, specificity, and reproducibility. This assay has the potential to screen women at the community level and provide an early indication of the risk of PTB. Early prediction of PTB-associated risk in low-resource settings is critical in reducing the complication associated with these adverse birth outcomes. The biophysical methods used to predict the risk of PTB involve uterine contractions (UC) and manual examination of the cervical length (CL) by endo-vaginal examination. In a recent study on the assessment of cervical length showed 50%, 50%, 53%, and 54% sensitivity at a 10% false predictive rate (FPR) at 20–23, 24–27, 28–31, and 32–35 weeks for predicting PTB, respectively.26,27 The addition of the dipstick assay to the other clinical and ultrasound markers could potentially improve the accuracy of the prediction. We plan to evaluate this in a large multicentric validation cohort study across India. Therefore, the early detection of PTB-associated bacteria in low resource settings is a promising tool for reducing complications associated with this complex syndrome that has immense public health implications.
In summary, the integration of metagenomic and dipstick assay helps us to identify microbial taxa that increase the risk of PTB in resource-limiting settings. The successful translation of knowledge of differences in vaginal microbiota derived from metagenomic study to simple scalable dipstick assay has the potential to positively impact public health.
Limitations of the study
Our study has a few limitations. The large focus is on the taxonomic dysbiosis of the vaginal microbiota. However, we are continuing to do a functional analysis for mechanistic understanding. While we have evaluated the predictive accuracy of the dipstick test against the metagenomic data as gold standard, the predictive ability for the risk of PTB needs a comprehensive evaluation. Such an evaluation is possible only in a large multicentric study that is currently being planned. Although the rapid dipstick assay identifies most of the bacterial taxa associated with birth outcomes some would be missed because of inter-oligo cross-reactivity.
Resource availability
Lead contact
Further information related to the manuscript, requests for any resources should be directed to the lead contact, Dr. Bhabatosh Das (bhabatosh@thsti.res.in).
Materials availability
Bacterial strains isolated from the high vaginal swab (HVS) samples are available with the lead contact.
Data and code availability
-
•
The raw 16S rRNA gene sequencing data generated in the context of present study have been submitted to the SRA National Center for Biotechnology Information (NCBI) databases. 16S rRNA gene sequences will be available in the BioProject: PRJNA1058804 (submission ID: SUB14115346), SRA:PRJNA1058804.
-
•
All original codes used in this study are available in the supplementary information (Data S1).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
The work was funded by the Department of Biotechnology (DBT), Govt. of India (No. BT/PR9983/MED/97/194/2013) and the Translational Research Program (TRP) (No. BT/PR30159/MED/15/188/2018) of BRIC-THSTI. We thank all the staff of GARBH-Ini cohort including research physicians, study nurses, clinical and laboratory technicians, field workers, internal quality improvement team, project and data management teams. We acknowledge the substantial support of Prof. Pramod K Garg to complete the study. Authors are grateful to Dr. A. Gambhir and N. Bansal from the Department of Biotechnology, Government of India, for their generous support. We acknowledge the support of administrative staff of all participating institutes. We wish to extend our thanks to the two hospitals (Gurugram Civil Hospital and Safdarjung Hospital) and their staff for facilitating the study. We thankfully acknowledge the Advanced Nucleotide Sequencing Facility at BRIC-THSTI. The funding agency had no role in study design, sample collection, analysis and interpretation of data, and writing the manuscript. D.T. and J.V. have received support from TRP, DBT, Govt. of India. We would like to acknowledge the National Genomics Core of BRIC-NIBMG, Kalyani for assisting in high throughput massively parallel 16S rRNA gene sequencing. We thank Roshan Kumar, Naveen Kumar, M. Rama Gowtham, Ridhima Mitra, Subhash Tanwar, and other members of the Functional Genomics Laboratory, BRIC-THSTI for technical support. M.S. is supported by CSIR-UGC-Senior Research Fellowship (UGC Reference No. 704). Memebrs of GARBH-Ini∗: BRIC-Translational Health Science and Technology Institute, Faridabad, Haryana, India (Shinjini Bhatnagar, Nitya Wadhwa, Bhabatosh Das, Pallavi S. Kshetrapal, Shailaja Sopory, Sumit Misra, Dharmendra Sharma, Kanika Sachdeva, Amanpreet Singh, G. Balakrish Nair, and Satyajit Rath); Gurugram Civil Hospital, Haryana, India (Alka Sharma, Sunita Sharma, Umesh Mehta, and Brahmdeep Sindhu); Vardhman Mahavir Medical College and Safdarjung Hospital, New Delhi, India (Pratima Mittal, Rekha Bharti, Harish Chellani, Rani Gera, Jyotsna Suri, Pradeep Debata, and Sugandha Arya); National Institute of Biomedical Genomics, Kalyani, West Bengal, India (Arindam Maitra and Souvik Mukherjee); Regional Centre for Biotechnology, Faridabad, Haryana, India (Tushar K. Maiti); International Centre for Genetic Engineering and Biotechnology, New Delhi, India (Dinakar M. Salunke); All India Institute of Medical Sciences, New Delhi, India (Nikhil Tandon, Yashdeep Gupta, Alpesh Goyal, Smriti Hari, Aparna Sharma K, and Anubhuti Rana); Maulana Azad Medical College, New Delhi, India (Siddarth Ramji and Anju Garg); The Ultrasound Lab, Defence Colony, New Delhi, India (Ashok Khurana); Sitaram Bhartia Institute of Science and Research, New Delhi, India (Reva Tripathi); President of India’s secretariat, Government of India (Rakesh Gupta); Indian Institute of Technology Madras, Chennai, Tamilnadu, India (Himanshu Sinha and Raghunathan Rengasamy), National Science Chair, Science and Engineering Board, Government of India (Partha P. Majumder), Indian Institute of Science Education and Research, Pune, Maharastra, India (Vineeta Bal), Amrita Institute of Medical Sciences & Research Centre, Faridabad, Haryana, India (Pratima Mittal); Center for Health Research and Development, Society for Applied Studies, New Delhi, India (Uma Chandra Mouli Natchu and Harish Chellani); and Pondicherry Institute of Medical Sciences, Puducherry, India (Ramachandran Thiruvengadam).
Author contributions
B.D., S. Bhatnagar, and G.B.N. conceived the idea and designed the experiments. S.B., N.W., R.T., and the members of GARBH-Ini conducted the clinical study, and collected HVS samples and relevant clinical information. D.T., M.S., T.A., S.K., D.D., S.N., P.J., O.M., A.K., J.R.Y., K.S., S. Bakshi, GARBH-Ini study group, S.M., and B.D. performed experiments. B.D., S.M., S. Bhatnagar, and P.K. contributed reagents. D.T., M.S., T.A., S.K., B.D., and S.M. performed data analysis. D.T., M.S., T.A., S.M., and B.D. wrote the manuscript. S. Bhatnagar, N.W., and R.T. edited the manuscript. All authors have read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human vaginal swab (HVS) samples | Indian women enrolled in the GARBH-Ini cohort | GARBH-Ini cohort study participants |
| Metagenomic DNA | High vaginal sample of Indian women enrolled in the GARBH-Ini cohort | Functional Genomics Lab., BRIC-THSTI |
| Lactobacillus crispatus genomic DNA | Vaginal swab samples of Indian women | Functional Genomics Lab., BRIC-THSTI |
| Lactobacillus gasseri genomic DNA | Vaginal swab samples of Indian women | Functional Genomics Lab., BRIC-THSTI |
| Lactobacillus jensenii genomic DNA | Vaginal swab samples of Indian women | Functional Genomics Lab., BRIC-THSTI |
| Gardnerella vaginalis ATCC strain | Human vaginal secretions | ATCC#14018 |
| Chemicals, peptides, and recombinant proteins | ||
| Mutanolysin | Sigma Aldrich, USA | Cat#M9901 |
| Lysostaphin | Sigma Aldrich, USA | Cat#L7386 |
| Lysozyme | Sigma Aldrich, USA | Cat# L6876 |
| Guanidine thiocyanate | Sigma Aldrich, USA | Cat#G9277 |
| N-Lauryl sarcosine | Sigma Aldrich, USA | Cat#L5777 |
| MRS agar | Sigma Aldrich, USA | Cat#69964 |
| MRS broth | Sigma Aldrich, USA | Cat#69966 |
| Trypticase soy broth | Sigma Aldrich, USA | Cat#22092 |
| Trypticase soy agar | Sigma Aldrich, USA | Cat#22091 |
| Defibrinated sheep blood | R-Biopharm Neugen Pvt. Ltd, India | Lot number – 260623 |
| BiomLife | Ruhvenile Biomedical Pvt. Ltd, India | Cat#RVN-BL-0.9-00 |
| Nextera XT DNA Library preparation kit | Illumina Inc., USA | Cat#FC-131-1096 |
| Q5 High Fidelity DNA Polymerase | New England biolabs, USA | M0491S |
| Deoxynucleotides | New England biolabs, USA | N0447S |
| Forward primer dry tube (dipsticks) | Tohoku bio-array Co., Ltd., Japan | HS Code#2934.99 |
| Reverse primer dry tube (dipsticks) | Tohoku bio-array Co., Ltd., Japan | HS Code#2934.99 |
| C-PAS Chromatography paper strip | Tohoku bio-array Co., Ltd., Japan | HS Code#4823.90 |
| Dilution Buffer | Tohoku bio-array Co., Ltd., Japan | HS Code#3822.00 |
| Streptavidin coated latex beads | Tohoku bio-array Co., Ltd., Japan | HS Code#3822.00 |
| Nextera XT DNA Library preparation kit | Illumina Inc., USA | Cat#FC-131-1096 |
| Oligonucleotides | ||
| L. gasseri specific | Sigma Aldrich, USA | F- 5` TGGAAACAGRTGCTAATACCG 3′ R- 5` CAGTTACTACCTCTATCTTTCTTCACTAC 3′ |
| Megasphaera type 1 specific | Sigma Aldrich, USA | F-5` GATGCCAACAGTATCCGTCCG 3′ R- 5` CCTCTCCGACACTCAAGTTCGA 3′ |
| L. jensenii specific | Sigma Aldrich, USA | F- 5` ATGCTTGCGCTTATCCTT 3′ R-5` GTTTAGCGATGTCAGGATC 3′ |
| L. crispatus specific | Sigma Aldrich, USA | F-5` TCCCCAATTAGATCCTGCATC 3′ R-5` AGAAGTGCTTGCTGGAGGTG 3′ |
| S. sanguinegens specific | Sigma Aldrich, USA | F-5` AATTATTGGGCTTAAAGGGCATC 3′ R-5` AGTACTCTAGTTATACAGTTTTGTAG 3′ |
| G. vaginalis specific | Sigma Aldrich, USA | F-5` TTACTGGTGTATCACTGTAA 3′ R-5` CCGTCACAGGCTGAACAGT 3′ |
| L. iners specific | Sigma Aldrich, USA | F-5` GTCTGCCTTGAAGATCGG 3′ R-5` ACAGTTGATAGGCATCATC 3′ |
| Software and algorithms | ||
| DADA2 package (Version: 1 · 16) | Callahan et al.28 | http://benjjneb.github.io/dada2/ |
| SILVA 138 | Quast et al.29 | https://www.arb-silva.de |
| PICRUSt2 | Douglas et al.30 | https://github.com/picrust/picrust2/wiki |
| q2-longitudinal of Qiime2 | Bokulich et al.31 | https://github.com/qiime2/q2-longitudinal.git |
Experimental model and study participant details
Study design and participants
Our study is a nested case-control study embedded in the GARBH-Ini (Interdisciplinary Group for Advanced Research on Birth Outcomes - A DBT India Initiative) pregnancy cohort conducted between 2015 and 2021. The details of the cohort are described elsewhere.10,28 Briefly, pregnant women visiting the antenatal clinic at Gurugram Civil Hospital (GCH) before completion of 20-weeks period of gestation (POG), as assessed by ultrasound dating scan, were enrolled in the GARBH-Ini cohort. These women were followed up at least 4 times during pregnancy till delivery. After enrollment, each subject was monitored until childbirth to collect information on the period of gestation (POG) at delivery. Based on this POG data, subjects were classified into either the term or preterm group. In total, the study included 60 women who delivered preterm and 140 women who delivered at term. Serial clinical information and biospecimens, including samples of high vaginal swabs (HVS) were collected and stored in the biorepository situated at THSTI. Among women enrolled in the GARBH-Ini cohort those who had singleton babies without congenital abnormalities by spontaneous delivery and HVS at all three (T1 swab: <14 weeks), 2nd (T2 swab: 18–20 weeks) and 3rd (T3 swab: 26–28 weeks) trimesters and had no history of antibiotic usage in the 7 days prior to sampling, or complications during pregnancy, such as pregnancy with preeclampsia, or any placental abnormalities constituted the universe of study participants (n = 1014) for the current microbiome study (Figure 1). From this universe, 60 women who delivered preterm were chosen randomly as cases and 140 women with term birth (TB) were selected as controls. The subcategories of Preterm birth of the study participants based on gestational age are as follows; Extreme preterm (<28 weeks): 1.6%, very preterm (28–32 weeks): 10% and moderate preterm (32–34 weeks):15%. Information on abortion history was available only for 16 PTB and 36 TB out of 60 PTB and 140 TB delivering women. Total, 10 PTB (16%) and 25 TB (18%) participants had a previous history of abortion. Conversely, 6 PTB (10%) and 11 TB (8%) participants reported no history of abortion. High vaginal swabs taken at three time points for the selected participants were retrieved from the biorepository. Microbiome DNA isolation and massively parallel sequencing were performed.
Method details
High vaginal swab sample collection procedure
The study participants were positioned in the lithotomy position in the procedure room at Gurugram Civil Hospital, Haryana. Collection of high vaginal swab samples were done aseptically from the midpoint of the vagina with the help of a Cusco’s speculum and four sterile Catch-All sample collection swabs that were then gently rubbed against the mid vaginal wall for ∼20 s. Microcentrifuge tube prefilled with 0.5 mL of sterile 50 mM Tris-1 mM EDTA buffer (pH 8.0) supplemented with nuclease inhibitors and protein-denaturing agents (Guanidinium thiocyanate) was used for placing the HVS swabs. Further, vortexing was done in order to remove microbial cells. These tubes were further transferred to Functional Genomics Laboratory (FGL), BRIC, Translational Health Science and Technology Institute (THSTI) for carrying out the microbiome study in freezing conditions (−192°C) within 12 h of sample collection.
Amplicon sequencing of 16S rRNA gene and bioinformatics analysis
After microbiome DNA isolation, the V3-V4 hyper-variable region of 16S rRNA gene was amplified using universal primer pairs: 175F (5′-CCTACGGGNGGCWGCAG-3′) and 512R (5′-GACTACHVGGGTATCTAATCC-3′). Amplified products were purified using Agencourt AMPure- XP (Beckman Coulter) paramagnetic beads and viewed by 1% agarose gel electrophoresis. Sample indexing was done by Nextera XT Index Kit (Illumina) and quantification of DNA library was performed by Qubit Flurometer using Qubit dsDNA HS Assay Kit (Invitrogen) and amplicon length was checked using 2100 TapeStation (Agilent 4200) instrument. Then the final libraries were pooled and sequenced using HiSeq2500 platform following massively parallel 2×250 paired-end chemistry.
After sequencing the sequence data were processed using DADA2 package (Version: 1 · 16) in R (V 4 · 1·1).28 The quality profile of demultiplexed raw reads were checked using the “plotQualityProfile '' function of DADA2 R package. No reads were removed as Phred QV was ≥30 for all paired-end reads. Using “filterAndTrim'' function in DADA2, the primer sequences at the 5′ end of the reads were trimmed and those reads with (a) read length <200 bp and >600 bp and (b) ambiguous bases (N) > 0 were discarded. The filtered paired-end reads were de-replicated and processed for error rate estimation for each base position using dada and learnErrors function. The de-replicated unique paired-end reads were joined only when at least 12 bases overlapped (mergepairs function) and merged reads were subsequently binned into Amplicon Sequence Variants (ASVs) based on 100% sequence similarity (makeSequenceTable function). Removal of chimeric as well as singleton ASVs were then performed. The codes used in this study are mentioned in supplemental file Data S1. Adjustment for negative control was performed by removing those ASVs from all the samples having at least 1% abundance in negative controls. Rarefaction analysis and alpha (Shannon, Simpson and Chao1) diversity estimation were done using vegan package. Finally, taxonomy assignment till the genus level was done by aligning the representative sequences of each ASVs to the SILVA Reference database (Version 138).29 For species level classification, the NCBI’s 16S Microbial database was used and alignment was performed by BLASTn.32,33 Only those representative sequences with ≥98% sequence identity in BLAST were selected for species-level annotation.
Predictive functional analysis by PICRUSt2
Functional predictions of the bacterial communities from the vaginal microbiome were computed through the latest PICRUSt2 v2·4 · 2 software described by Douglas et al.,2020.30 The unique amplicon sequence variants (ASVs) and a Biome file were given as input for the analysis. HMMER (http://www.hmmer.org/) was used for the multiple assignments of the exact sequence variants (ESVs) in the first step followed by evolutionary placement-ng (EPA-ng) and Genesis Applications for Phylogenetic Placement Analysis (gappa) omics.34,35 Finally, prediction of pathway-level abundances was done using the pathway_pipeline.py function that assigns EC numbers to Meta Cyc reactions and KO abundances in KEGG pathways.36 The code used for the PICRUSt2 based analysis is mentioned in supplemental file Data S1.
Samples and bacterial strains used for the dipstick assay
Seven bacterial species, Lactobacillus crispatus, Lactobacillus jensenii, Lactobacillus gasseri, Lactobacillus iners, Sneathia sanguinegens, Gardnerella vaginalis, and Megasphaera sp. that were identified from our previous study9 were selected for the development of the dipstick assay. In the previous study, L. iners (<14 weeks, 18–20 weeks and 26–28 weeks, p-value <0·02), Megasphaera sp. (<14 weeks, p-value <0·05), G. vaginalis (18–20 weeks, p-value = 0·01) and S. sanguinegens (18–20 weeks, p-value <0·0001) were found to be significantly associated with preterm deliveries whereas the higher abundance of L. gasseri (26–28 weeks, p-value = 0·010) was observed in term deliveries.
We developed the dipstick assay using a two-step process. A total of 220 samples were selected from the universe of our microbiome study participants (n = 1014). Among 220 samples, 82 were chosen from our earlier case control study (n = 110) and 51 were selected from this study. Since, metagenomic sequencing data were available for 133 out of the total 220, these samples were used as gold standard for calculating the sensitivity and specificity. The remaining 87 samples for which the final birth outcome was known, were tested for the assay.
Dipstick DNA chromatography
Dipstick strips and the reagents for this assay were obtained from the Tohoku bio-array (TBA) Co., Sendai, Japan (Figure S1). As shown in the above table, for each pair of primers, the 5` terminus of the forwarding primer was tagged with different oligonucleotides and the 5′ terminus of the reverse primer was biotinylated. Blue-colored latex particles coated with streptavidin were linked to biotinylated terminal amplicons through a streptavidin-biotin interaction. Dipstick strips were manufactured by immobilizing four complementary oligonucleotides to specifically recognize the PCR amplicons through hybridization with 5′ terminus tags. On the strip, 4 test lines in total are available. A flow control line was also set up at the end of the strip. One PCR-amplified product was diluted with H2O into a total volume of 10 μL and mixed with 10 μL of developing buffer (containing salts solution) and 1 · 5 μL of streptavidin-coated blue latex suspension (both were supplied by TBA Co.). A DNA strip was then inserted into the mixture. The appearance of a blue test line at the position where the complementary oligonucleotide is immobilized indicates the presence of the target DNA sequence.
Quantification and statistical analysis
Differences in the alpha diversities and vaginal microbiome composition were determined by non-parametric Wilcoxon rank-sum test (two-tailed) using the R command (wilcox.test, paired = FALSE) (Figure 2; Figure 6). Multivariate analysis was performed using Linear Discriminant Analysis (LDA) Effect Size (LEfSe) (Figure 7). Analysis of Variance (ANOVA) tests were performed for the three groups (T1, T2, T3) comparison (Figure 3). Correlations between vaginal taxa were evaluated using non-parametric Spearman’s rank correlation test in R (cor.test, method = spearman) (Figures 8, 9, and S4). Linear Mixed Effects (LME) model-based analysis was performed using q2-longitudinal of Qiime231 to understand longitudinal shift of microbial species during pregnancy in the term birth (TB) and preterm birth (PTB) samples. Birth type (TB/PTB) and gestation time (first/second/third trimesters) were used as fixed effects and age of the mother was used as a random effect in this LME model (Figure 10).
Since the dipstick assay involves a parallel testing approach, we first tested for the TB associated microbes followed by testing for the PTB associated microbes, as described earlier. Therefore, the combined sensitivity of the assay was calculated using formula (sensitivity of first test × sensitivity of second test), and the specificity was calculated using (specificity of first test + specificity of second test – [specificity of first test × specificity of second test]).
Additional resources
This study is reviewed and approved by Translational Health Science and Technology Institute Human Ethics Committee. [Ref.# THS 1.8.1/(30) dated 11th Feb 2015].
Published: October 23, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111238.
Contributor Information
Shinjini Bhatnagar, Email: shinjini.bhatnagar@thsti.res.in.
Souvik Mukherjee, Email: sm2@nibmg.ac.in.
Bhabatosh Das, Email: bhabatosh@thsti.res.in.
Supplemental information
References
- 1.Preterm birth. 2023. https://www.who.int/news-room/fact-sheets/detail/preterm-birth
- 2.Pravia C.I., Benny M. Long-term consequences of prematurity. Cleve Clin. J. Med. 2020;87:759–767. doi: 10.3949/ccjm.87a.19108. [DOI] [PubMed] [Google Scholar]
- 3.Han Y.W., Shen T., Chung P., Buhimschi I.A., Buhimschi C.S. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J. Clin. Microbiol. 2009;47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiGiulio D.B., Romero R., Kusanovic J.P., Gómez R., Kim C.J., Seok K.S., Gotsch F., Mazaki-Tovi S., Vaisbuch E., Sanders K., et al. Prevalence and Diversity of Microbes in the Amniotic Fluid, the Fetal Inflammatory Response, and Pregnancy Outcome in Women with Preterm Pre-Labor Rupture of Membranes. Am. J. Reprod. Immunol. 2010;64:38–57. doi: 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero R., Gomez-Lopez N., Winters A.D., Jung E., Shaman M., Bieda J., Panaitescu B., Pacora P., Erez O., Greenberg J.M., et al. Evidence that intra-amniotic infections are often the result of an ascending invasion - a molecular microbiological study. J. Perinatal Med. 2019;47:915–931. doi: 10.1515/jpm-2019-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillier S.L., Nugent R.P., Eschenbach D.A., Krohn M.A., Gibbs R.S., Martin D.H., Cotch M.F., Edelman R., Pastorek J.G., Rao A.V., McNellis D. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N. Engl. J. Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 7.Leitich H., Bodner-Adler B., Brunbauer M., Kaider A., Egarter C., Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: A meta-analysis. Am. J. Obstet. Gynecol. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 8.Chee W.J.Y., Chew S.Y., Than L.T.L. Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb. Cell Factories. 2020;19 doi: 10.1186/s12934-020-01464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Kumari N., Talukdar D., Kothidar A., Sarkar M., Mehta O., Kshetrapal P., Wadhwa N., Thiruvengadam R., Desiraju B.K., et al. The Vaginal Microbial Signatures of Preterm Birth Delivery in Indian Women. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.622474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatnagar S., Majumder P.P., Salunke D.M., Interdisciplinary Group for Advanced Research on Birth Outcomes—DBT India Initiative GARBH-Ini A Pregnancy Cohort to Study Multidimensional Correlates of Preterm Birth in India: Study Design, Implementation, and Baseline Characteristics of the Participants. Am. J. Epidemiol. 2019;188:621. doi: 10.1093/aje/kwy284. [DOI] [PubMed] [Google Scholar]
- 11.Kindinger L.M., Bennett P.R., Lee Y.S., Marchesi J.R., Smith A., Cacciatore S., Holmes E., Nicholson J.K., Teoh T.G., MacIntyre D.A. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5:6. doi: 10.1186/s40168-016-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan D., Bennett P.R., Lee Y.S., Kundu S., Teoh T.G., Adan M., Ahmed S., Brown R.G., David A.L., Lewis H.V., et al. Microbial-driven preterm labour involves crosstalk between the innate and adaptive immune response. Nat. Commun. 2022;13:975. doi: 10.1038/s41467-022-28620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fettweis J.M., Serrano M.G., Brooks J.P., Edwards D.J., Girerd P.H., Parikh H.I., Huang B., Arodz T.J., Edupuganti L., Glascock A.L., et al. The vaginal microbiome and preterm birth. Nat. Med. 2019;25:1012. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hearps A.C., Tyssen D., Srbinovski D., Bayigga L., Diaz D.J.D., Aldunate M., Cone R.A., Gugasyan R., Anderson D.J., Tachedjian G. Vaginal lactic acid elicits an anti-inflammatory response from human cervicovaginal epithelial cells and inhibits production of pro-inflammatory mediators associated with HIV acquisition. Mucosal Immunol. 2017;10:1480. doi: 10.1038/mi.2017.27. [DOI] [PubMed] [Google Scholar]
- 15.Estarlich M., Ballester F., Davdand P., Llop S., Esplugues A., Fernández-Somoano A., Lertxundi A., Guxens M., Basterrechea M., Tardón A., et al. Exposure to ambient air pollution during pregnancy and preterm birth: A Spanish multicenter birth cohort study. Environ. Res. 2016;147:50. doi: 10.1016/j.envres.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 16.Serrano M.G., Parikh H.I., Brooks J.P., Edwards D.J., Arodz T.J., Edupuganti L., Huang B., Girerd P.H., Bokhari Y.A., Bradley S.P., et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019;25:1001–1011. doi: 10.1038/s41591-019-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan B.J., DiGiulio D.B., Goltsman D.S.A., Sun C.L., Costello E.K., Jeganathan P., Biggio J.R., Wong R.J., Druzin M.L., Shaw G.M., et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl. Acad. Sci. USA. 2017;114:9966–9971. doi: 10.1073/pnas.1705899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amabebe E., Anumba D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018;5:181. doi: 10.3389/fmed.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenberg R.L., Culhane J.F., Iams J.D., Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baqui A.H., Lee A.C.C., Koffi A.K., Khanam R., Mitra D.K., Dasgupta S.K., Uddin J., Ahmed P., Rafiqullah I., Rahman M., et al. Prevalence of and risk factors for abnormal vaginal flora and its association with adverse pregnancy outcomes in a rural district in north-east Bangladesh. Acta Obstet. Gynecol. Scand. 2019;98:309–319. doi: 10.1111/aogs.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar M., Murugesan S., Singh P., Saadaoui M., Elhag D.A., Terranegra A., Kabeer B.S.A., Marr A.K., Kino T., Brummaier T., et al. Vaginal Microbiota and Cytokine Levels Predict Preterm Delivery in Asian Women. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.639665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donders G.G., Van Calsteren K., Bellen G., Reybrouck R., Van den Bosch T., Riphagen I., Van Lierde S. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. Br. J. Obstet. Gynaecol. 2009;116:1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]
- 23.Ng S., Chen M., Kundu S., Wang X., Zhou Z., Zheng Z., Qing W., Sheng H., Wang Y., He Y., et al. Large-scale characterisation of the pregnancy vaginal microbiome and sialidase activity in a low-risk Chinese population. NPJ Biofilms Microbiomes. 2021;7:89. doi: 10.1038/s41522-021-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero R., Hassan S.S., Gajer P., Tarca A.L., Fadrosh D.W., Bieda J., Chaemsaithong P., Miranda J., Chaiworapongsa T., Ravel J. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng N., Guo R., Yao Y., Jin M., Cheng Y., Ling Z. Lactobacillus iners Is Associated with Vaginal Dysbiosis in Healthy Pregnant Women: A Preliminary Study. BioMed Res. Int. 2019;2019 doi: 10.1155/2019/6079734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elovitz M.A., Gajer P., Riis V., Brown A.G., Humphrys M.S., Holm J.B., Ravel J. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-09285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudicha D.W., Romero R., Kabiri D., Hernandez-Andrade E., Pacora P., Erez O., Kusanovic J.P., Jung E., Paredes C., Berry S.M., et al. Personalized assessment of cervical length improves prediction of spontaneous preterm birth: a standard and a percentile calculator. Am. J. Obstet. Gynecol. 2021;224:288.e1. doi: 10.1016/j.ajog.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bokulich N.A., Dillon M.R., Zhang Y., Rideout J.R., Bolyen E., Li H., Albert P.S., Caporaso J.G. q2-longitudinal: Longitudinal and Paired-Sample Analyses of Microbiome Data. mSystems. 2018;3 doi: 10.1128/msystems.00219-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbera P., Kozlov A.M., Czech L., Morel B., Darriba D., Flouri T., Stamatakis A. EPA-ng: Massively Parallel Evolutionary Placement of Genetic Sequences. Syst. Biol. 2019;68:365. doi: 10.1093/sysbio/syy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czech L., Stamatakis A. Scalable methods for analyzing and visualizing phylogenetic placement of metagenomic samples. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Y., Doak T.G. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw 16S rRNA gene sequencing data generated in the context of present study have been submitted to the SRA National Center for Biotechnology Information (NCBI) databases. 16S rRNA gene sequences will be available in the BioProject: PRJNA1058804 (submission ID: SUB14115346), SRA:PRJNA1058804.
-
•
All original codes used in this study are available in the supplementary information (Data S1).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.