Abstract
Atrial fibrillation (AF) is the most common sustained arrhythmia in humans, yet the molecular basis of AF remains incompletely understood. To determine the cell type-specific transcriptional changes underlying AF, we perform single-nucleus RNA-seq (snRNA-seq) on left atrial (LA) samples from patients with AF and controls. From more than 175,000 nuclei we find that only cardiomyocytes (CMs) and macrophages (MΦs) have a significant number of differentially expressed genes in patients with AF. Attractin Like 1 (ATRNL1) was overexpressed in CMs among patients with AF and localized to the intercalated disks. Further, in both knockdown and overexpression experiments we identify a potent role for ATRNL1 in cell stress response, and in the modulation of the cardiac action potential. Finally, we detect an unexpected expression pattern for a leading AF candidate gene, KCNN3. In sum, we uncover a role for ATRNL1 which may serve as potential therapeutic target for this common arrhythmia.
Subject terms: Gene expression, Atrial fibrillation, Transcriptomics
Characterizing atrial fibrillation (AF) at the single cell level is challenging. Here, the authors perform snRNA-seq on 18 patients with AF to investigate the cell composition, and gene expression shifts associated with this common arrhythmia.
Introduction
AF is the most common arrhythmia in humans, which increases a patient’s risk for death, stroke, heart failure, and dementia. Clinical treatments for AF include stroke prevention, as well as rate and rhythm control, which can improve symptoms. AF has many known risk factors, including age, body mass index, height, hypertension, diabetes mellitus, sleep apnea, heart failure, smoking, alcohol use, as well as genetic predisposition1. While some risk factors for AF are modifiable, the strong genetic contribution to AF points to the need for personalized medicine to assess AF risk and severity more accurately. Moreover, the heritability also implicates several molecular pathways in AF onset and perpetuation.
The onset of AF results in both structural and electrical remodeling in the atria. Ion channels, gap junctions, and regulators of intracellular Ca2+ homeostasis are considered the principal molecular determinants of abnormal atrial electrical activity2,3. Indeed, mutations in ion channel genes, and gap-junctions4,5, have been found to alter cardiac conduction and promote AF. In addition to the molecular determinants of electrical remodeling, structural integrity of cardiac tissue also plays a major role in AF pathophysiology. Importantly, cardiac fibrosis presents a major impediment to physiological cardiac conduction and is also highly associated with AF. And while fibrosis and other structural remodeling-associated factors may not initiate AF, they are likely to contribute to the progression, persistence, and severity of the disease. An enhanced understanding of these molecular features, combined with clinical knowledge regarding modifiable risk factors have the potential to aid in the improvement of therapeutic efficacy for treating AF.
Single-nucleus RNA sequencing (snRNA-seq) has emerged as a powerful tool for characterizing cell diversity in preserved human tissue samples6,7. In addition to identifying cell types, snRNA-seq allows investigators to characterize tissue composition, sources of potential inflammation, cell status, and active gene regulatory networks. Recently, snRNA-seq has been applied to generate detailed cellular atlases of the healthy human heart8,9 and to large-scale studies investigating a broad range of cardiovascular diseases including hypertrophic-, dilated-, and ischemic-cardiomyopathies, as well as congenital heart disease(s)10–13. From these studies and others, cardiac fibroblasts (CFs) and macrophages appear to possess the potential to diversify during disease, particularly when compared to cardiomyocytes (CMs)11. However, a cell-type specific characterization of human AF has yet to be undertaken.
In this study, we perform large-scale snRNA-seq on LA samples from patients with and without AF. We generate over 170,000 snRNA transcriptomes allowing us to investigate the transcriptional and cellular responses associated with AF. We were able to identify disease-associated genes in cardiomyocytes and macrophages, such as ATRNL1, which were significantly dysregulated in AF. Genetic manipulation of ATRNL1 levels in hESC-aCMs allowed us to identify a role for ATRNL1 in the regulation of the cell stress response and cardiac conduction. Our findings have implications for the development of therapeutic strategies for treating AF and heart failure.
Results
snRNA-seq of human LA tissue from AF cases and controls
To characterize the cellular and molecular characteristics associated with AF, we performed snRNA-seq on samples from the LA of human patients with AF who were not in heart failure (n = 19) as well as non-AF controls (n = 17) (Supplementary Data 1, and Supplementary Data 2). We selected patients without heart failure to better control for the many diverse etiologies associated with this common arrythmia. Overall, our controls (CTRL) were 63% female, the mean age was 68 (SD = 7.5), 25% had a history of taking beta-blockers, and 6% had taken anticoagulants. The AF cases were 61% were female, the mean age was 66 (SD = 8.2), 56% had a history of taking beta-blockers, and 44% were being administered anticoagulants.
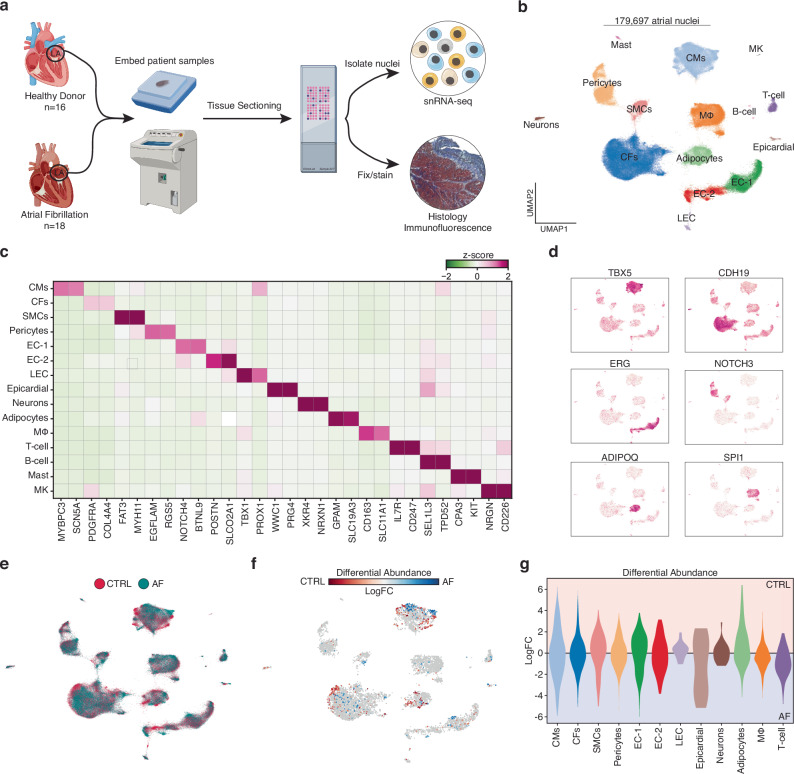
We carried out strict quality control, including sample sex check, unique molecular identifier (UMI) decay curve analysis, and a crosscheck of genotype fingerprinting with genome sequencing data (see Methods). A total of 2 samples failed our quality control standards and were removed from all subsequent analysis. Our final data set included 18 AF cases and 16 controls (Fig. 1a, Supplementary Data 1, and Supplementary Data 2). Next, batch correction was performed with the single-cell variational inference framework (scVI)14. Low-quality nuclei, doublets, and misclassified cells were then removed following clustering, leaving us with 179,697 nuclei that occupied a total of 15 transcriptionally distinct clusters (Fig. 1b, and Supplementary Fig. 1). Differential expression analysis was performed to determine the top representative genes for each cluster (Supplementary Data 3), and then clusters were annotated based on the expression of canonical cell markers (Fig. 1c). For example, TBX5 was enriched in cardiomyocytes (CMs), ERG in endothelial cell clusters (EC-1, EC-2, and LEC), and ADIPOQ was highly expressed in adipocytes (Fig. 1d).
Fig. 1. Cellular Composition in the LA from patients with and without atrial fibrillation.
a Schematic of experimental setup and tissue processing pipeline. Created partly in BioRender. Hill, M. (2021) BioRender.com/e34r263. b Uniform manifold approximation and projection (UMAP) representation of 179,697 left atrial cardiac nuclei isolated from a total of 30 donors. c Heatmap displaying the expression of marker genes for each cluster (cell type). Low median z-score of normalized expression highlighted in green. High expression showed in purple. d Feature plots of gene expression. High expression shown in purple. e UMAP displaying the clinical designation of each transcriptome. Healthy controls denoted by red, and AF patients are highlighted in blue. f Embedding of the Milo k-nearest neighbor differential abundance testing results for all cell types. Neighborhoods colored by log fold changes for Control (CTRL) versus AF. A negative (blue) log fold change indicates enrichment in AF, and a positive (red) log fold change represents an enrichment in CTRL. Nonsignificant neighborhoods (false discovery rate (FDR) > 5%) are shown in white. g Milo k-nearest neighbor differential abundance testing results scored by individual cell type. Cell types colored as in Fig. 1b.
Next, we sought to determine the changes in LA tissue composition that occur with AF, so we evaluated the proportions of cell-type annotated nuclei across all samples (Fig. 1e, and Supplementary Fig. 1b). Overall, control and AF samples contained similar percentages of all the major cardiac cell types and no clear trends were detectable. To formally determine which cell types were shifting in composition between AF and controls, we applied Milo, a statistical framework that avoids the resolution-associated pitfalls of clustering through performing differential abundance testing by assigning cells to neighborhoods on a k-nearest neighbor graph15. The differential abundance testing identified very few neighborhoods within the main cardiac cell types that shifted significantly between disease and controls (Fig. 1f, and Fig. 1g). To further evaluate changes in tissue composition with a different statistical approach we have decided to utilize a Bayesian model that we have successfully utilized and experimentally validated previously10,13 the single-cell compositional data analysis (scCODA) framework (Supplementary Fig. 2c)16. No statistically significant changes in nuclei abundance were detected between AF patients and control donors. To confirm these results, we next performed trichrome stained tissue sections collected directly proximal to those utilized for snRNA-seq to evaluate tissue composition (Supplementary Fig. 2d). The trichome stained sections contained similar tissue compositions between controls and AF patients, consistent with our snRNA-seq analysis. Overall, these results suggest that the tissue composition of the LA was not overly different in patients with AF compared to age matched controls.
Differentially expressed genes in AF
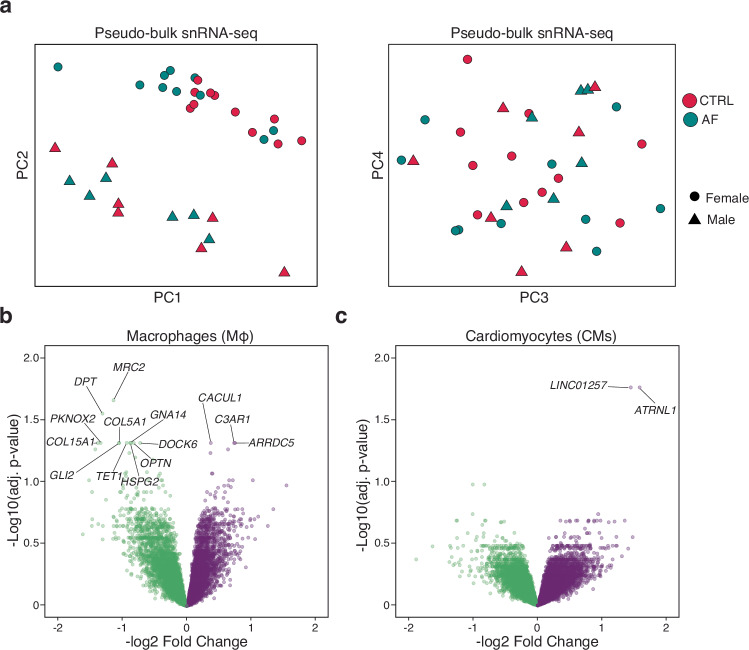
Next, we sought to identify individual transcripts that were associated with AF. We began by performing performed principal components analysis (PCA) across combined snRNA-seq data for each sample in a pseudo-bulk RNA-sequencing approach (Fig. 2a). Overall, we observed little to no transcriptional variation that could be explained by patient diagnosis. The greatest source of variation separating samples, regardless of disease status, was sex, as genes from X and Y chromosomes were included in our analysis. We next aimed to identify the differentially expressed genes between AF and control hearts for each individual cell type (Supplementary Data 4, see “Methods”). Very few substantial changes in transcription were identified after completing this comparison. However, we did observe significant transcriptional changes (adjusted p-value < 0.05) for 2 cell types, namely CMs and macrophages (Fig. 2e, f). In CMs, we observed a marked upregulation of LINC01257, a lncRNA associated with pediatric acute myeloid leukemia17, and ATRNL1.
Fig. 2. Differentially expressed genes in atrial fibrillation.
a PCA plots for pseudobulk RNA-seq analysis of samples by diagnosis. Red is control (CTRL) and blue is AF cases. Female samples are shaped as circular and male samples by triangles. b Volcano plot displaying the differentially expressed genes in macrophages between controls and AF cases. c Volcano plot displaying the differentially expressed genes in CMs between controls and AF cases. Volcano plots show log fold change and two-sided P-value expression changes between AF and CTRL for each gene tested using limma–voom differential expression analysis (“Methods”).
ATRNL1 localizes to the intercalated disk in cardiomyocytes
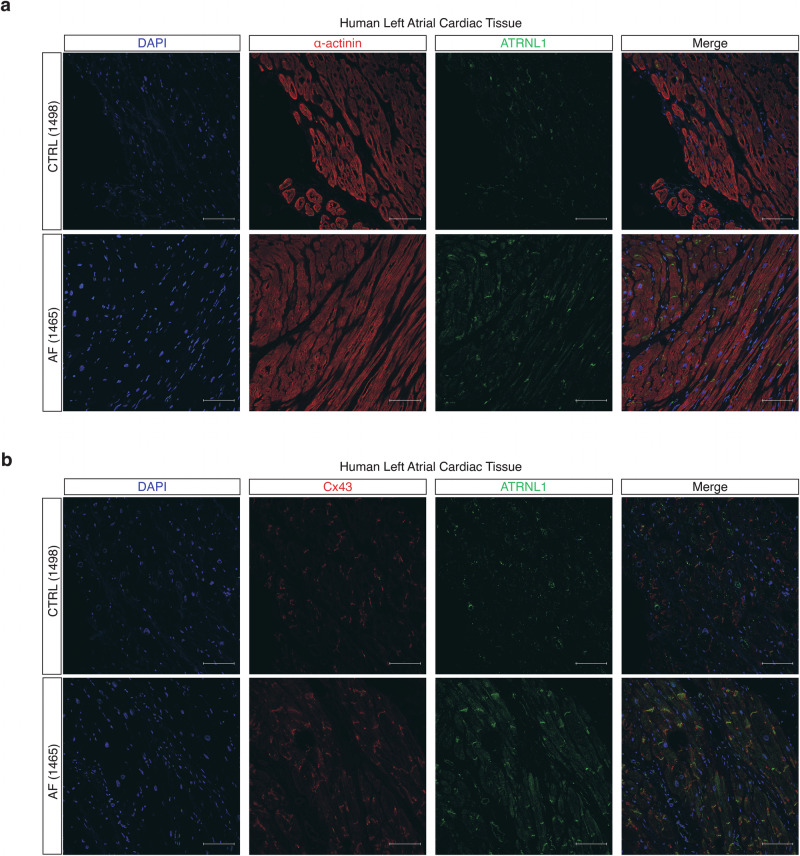
Attractin Like 1 (ATRNL1) is highly conserved transmembrane glycoprotein postulated to function in cell adhesion, cell signaling, and neuronal energy homeostasis. To our knowledge, only a single patient with an ATRNL1 mutation has been described, and this patient was heterozygous for a 325 kb microdeletion (10q25.3) that included ATRNL118. The patient presented with a dysmorphic facial appearance, ventricular septal defect, borderline microcephaly, global developmental delay with autistic features and mild ataxia, suggesting that the observed phenotype is the result of haploinsufficiency of ATRNL118. Interestingly, ATRNL1 knockout mice have no such phenotype19. Thus, a clear role for ATRNL1 in cardiac development and physiology has yet to be understood. We evaluated the localization of ATRNL1 with tissue immunofluorescence in fresh frozen LA samples obtained from patients included in our snRNA-seq analysis (Fig. 3a, and Supplementary Fig. 3a). We found that ATRNL1 was expressed primarily in α-actinin+ CMs. Interestingly, ATRNL1 localized strongly to the longitudinal borders of CMs at the intercalated disc. To confirm this subcellular localization, we co-stained the same LA tissues with the intercalated disc component connexin 43 (Cx43/GJA1) (Fig. 3b, Supplementary Fig. 3b) and found that ATRNL1 and Cx43 both localize at intercalated disks in human LA tissue samples (Pearson’s correlation coefficient = 0.87). Further, we confirmed the expression and co-localization of ATRNL1 with Cx43 in hESC-aCMs (Supplementary Fig. 3c). To assess the expression of ATRNL1 across different anatomical regions of the human heart we investigated the Human Heart Atlas20. We found that ATRNL1 was expressed in both ventricular and atrial cardiomyocytes (Supplementary Fig. 4a, b). Further, we found that ATRNL1 was expressed in pacemaker and epicardial cells (mesothelial). To confirm the presence of ATRNL1 in ventricular cardiomyocytes we performed immunofluorescence staining in non-failing LV samples and observed that ATRNL1 was localized at the intercalated disk in ventricular cardiomyocytes of the LV (Supplementary Fig. 4c).
Fig. 3. Subcellular localization of ATRNL1 in human left atrial tissue.
a Tissue immunohistochemistry for nuclei/DAPI (blue), α-actinin (red), and ATRNL1 (green) in CTRL (n = 3) and AF (n = 3) LA tissue. A total of 4 images were evaluated per tissue section. b Tissue immunohistochemistry for nuclei/DAPI (blue), Cx43 (red), and ATRNL1 (green) in CTRL (n = 3) and AF (n = 3) LA tissue. Scale bars = 100 μm. A total of 4 images were evaluated per tissue section.
Cardiomyocytes are dosage-sensitive to ATRNL1
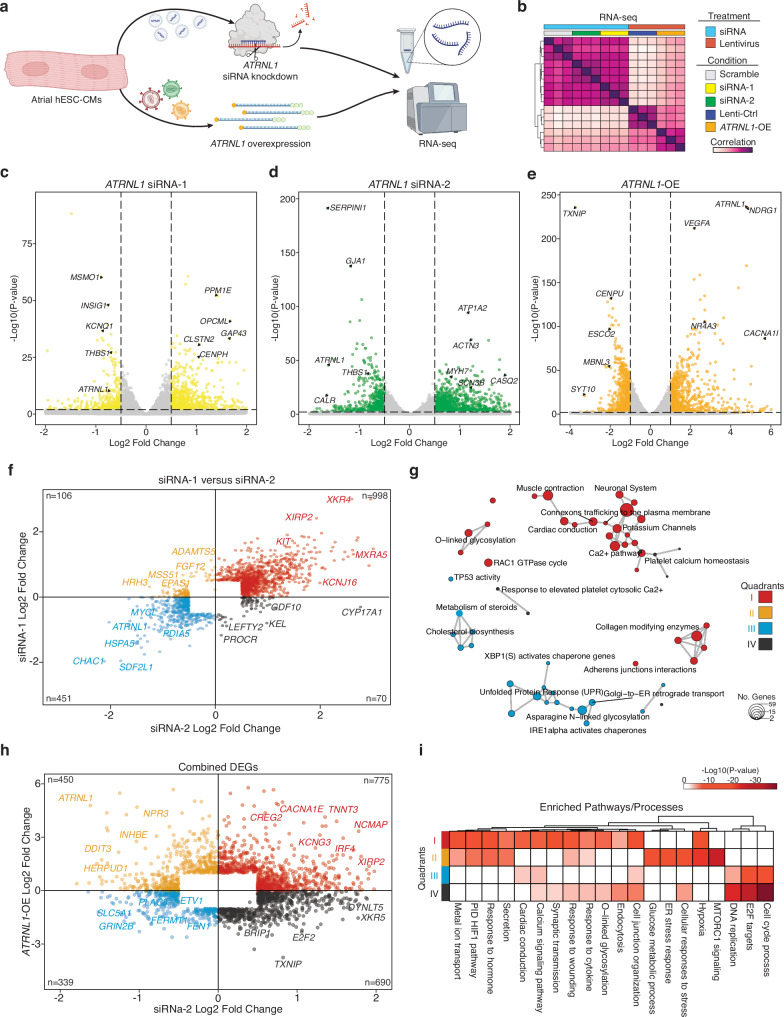
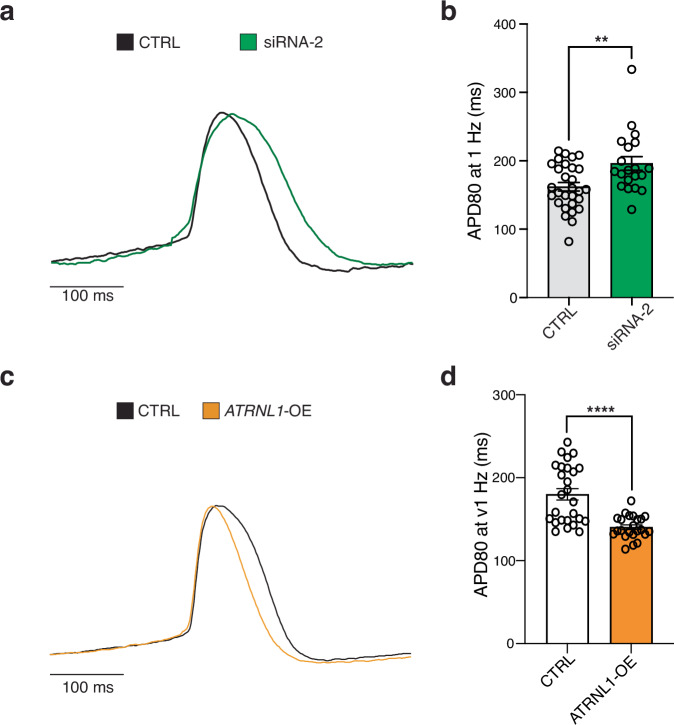
In AF patients we found that ATRNL1 was significantly higher expressed than in control LA tissues (Supplementary Fig. 5a). To comprehensively characterize the molecular role of ATRNL1 in CMs we elected to both knockdown its expression with siRNAs, as well as overexpress it with a lentiviral cDNA approach (Fig. 4a). While overexpression is more relevant for AF (Fig. 4d), the knockdown experiments were conducted to aid our analysis of the molecular function of ATRNL1 in CMs. First, we evaluated the expression of ATRNL1 in human tissues with data derived from the Genotype-Tissue Expression (GTEx) repository and identified two predominant isoforms of ATRNL1, a long isoform and a short isoform (Supplementary Fig. 5b). The predominant isoform in the brain is the long isoform, while the testis almost exclusively express the short isoform. Interestingly, the heart appears to express appreciable levels of both isoforms, though with a bias towards the short ATRNL1 transcript. We designed an siRNA to target the long isoform (siRNA-1), and another to target both the long and the short isoforms (siRNA-2).
Fig. 4. ATRNL1 is part of the CM cell stress response.
a Experimental overview for RNA-seq. Created partly in BioRender. Hill, M. (2021) BioRender.com/e34r263. b Correlation heatmap of RNA-seq sample distances. Euclidean distance between samples based on DESeq2 normalized gene expression (“Methods”). Individual libraries are annotated type of treatment and condition. We profiled 3 replicates for each condition (total n = 15). c Volcano plot displaying the differentially expressed genes in hESC-aCMs between siRNA-1 treated cells and Scramble siRNA control treated cells. Genes highlighted in yellow are significantly differentially expressed (adjusted p-value < 0.05, and Log2 Fold Change > 0.5). d Volcano plot displaying the differentially expressed genes in hESC-aCMs between siRNA-2 treated cells and Scramble siRNA control treated cells. Genes highlighted in green are significantly differentially expressed (adjusted p-value < 0.05, and Log2 Fold Change > 0.5). e Volcano plot displaying the differentially expressed genes in hESC-aCMs between ATRNL1-OE treated cells and control GFP lentiviral treated cells. Genes highlighted in orange are significantly differentially expressed (adjusted p-value < 0.01, and Log2 Fold Change > 1.0). f Scatterplot of all combined significantly differentially expressed genes compared across siRNA-1 treated cells and siRNA-2 treated cells. Individual genes are colored by their location on the Cartesian coordinate plane, separating them into quadrants. Representative genes are highlighted. g Enrichment map for gene pathway over-representation analysis colored by quadrant from Fig. 5f. The size of each dot represents the number of genes in that pathway category. h Scatterplot of all combined significantly differentially expressed genes compared across siRNA-2 treated cells and ATRNL1-OE cells. Individual genes are colored by their location on the Cartesian coordinate plane, separating them into quadrants. Representative genes are highlighted. i Heatmap for pathway enrichment analysis colored by significance of enrichment (2-sided P-value). Quadrant are annotated and colored according to Fig. 5h. Volcano plots show log fold change and two-sided P-value expression changes between AF and CTRL for each gene tested using DEseq2 differential expression analysis (“Methods”).
For the overexpression we chose to focus on the short isoform (ATRNL1-OE), due to the size restrictions of lentiviral vectors. We manipulated ATRNL1 expression with these reagents in hESC-aCMs and then performed total RNA-seq (Fig. 4b). Next, we performed differential expression analysis and found that for both siRNAs we achieved a significant knockdown of ATRNL1 expression (adjusted p-value < 0.05, log2 fold change cutoff = 0.5)(Fig. 4c, d, and Supplementary Data 5–6). Further, both siRNAs lead to decreased expression of similar genes including, THBS1, a stress- and injury-induced matricellular protein known to mediate cellular attachment21. Overexpression of ATRNL1 in hESC-aCMs caused the downregulation of proliferation-related genes like CENPU, and the upregulation of VEGFA, and the voltage gated calcium channel gene CACNA1I (adjusted p-value < 0.01, log2 fold change cutoff = 0.5)(Fig. 4e, and Supplementary Data 7).
To determine if the rarer long isoform had a distinct function in hESC-aCMs, we examined the effect of siRNA-1 (targeting the long-isoform) and siRNA-2 (targeting the short and long isoforms) (Fig. 5f). We selected all genes that were significantly differentially expressed in at least one siRNA treatment condition, and then compared the directionality of expression across both experiments. Many genes displayed concordant directionality for both siRNAs, with 451 genes being downregulated, and 998 being upregulated in both conditions (Supplementary Data 8). We explored the transcriptional characteristics of ATRNL1 deficient iPS-CMS by performing pathway enrichment analysis on these differentially expressed genes (Fig. 4g). The genes that are upregulated upon depletion of ATRNL1 (quadrant I) are associated most with cardiac conduction, connexons trafficking, potassium channels, calcium signaling, adherens junctions, and glycosylation. Genes that are down regulated when ATRNL1 was knocked down (quadrant III) are associated with cholesterol biosynthesis, the unfolded protein response (UPR), endoplasmic reticulum (ER) stress, and protein transport. Overall, these results suggest that, at least in hESC-aCMs, both isoforms function similarly to actively reduce cell stress and regulate levels of key conduction-related pathways in CMs.
Fig. 5. Modulation of ATRNL1 alters the atrial action potential duration.
a Representative action potentials recorded by optical mapping of ArcLight fluorescence in hESC-aCMs transfected with scrambled siRNA (CTRL) or siRNA-2 targeting ATRNL1. b Bar plot displaying the mean ± SEM of action potential durations at 80% repolarization (APD80, ms) in CTRL and ATRNL1 siRNA-2 treated hESC-aCMs. N = 29 in CTRL and n = 20 in ATRNL1 siRNA-2, N is the total number of wells from three independent biological replicates. **p = 0.0092, CTRL vs. siRNA-2, two-tailed Mann-Whitney test. c Representative tracings of action potentials in hESC-aCMs transduced with lentivirus carrying an empty vector backbone (CTRL), or an ATRNL1 overexpression construct (ATRNL1-OE). d Bar plot displaying the mean ± SEM of APD80 in hESC-aCMs overexpressing the short isoform of ATRNL1 or CTRL. N = 26 in CTRL and N = 23 in ATRNL1-OE from two biological replicates. ****p < 0.0001, CTRL vs. ATRNL1-OE, two-tailed Mann-Whitney test.
We next integrated the results of the knockdown experiments with the ATRNL1-OE dataset (Fig. 4h, and Supplementary Fig. 5c). We selected all genes that were significantly differentially expressed in at least one condition, the siRNA-2 and/or ATRNL1-OE experiment, and then compared the directionality of expression across both experiments (Supplementary Data 9). First, we wanted to identify genes that displayed concordance between experiments. Genes and pathways that are activated by ATRNL1, directly or indirectly, should be decreased in the knockdown and upregulated with the overexpression of ATRNL1 (quadrant II). We identified 450 such genes that fit this description. For genes/pathways negatively impacted or inhibited by ATRNL1, they should be upregulated after its depletion and strongly downregulated with ATRNL1 overexpression (quadrant IV). There were 690 genes negatively impacted by ATRNL1 expression. The discordantly regulated genes in quadrants I and III were abundant, with 775 and 339 gene members, respectively. Such large numbers highlight the dose sensitivity of CMs to ATRNL1 levels and may also be an indication of its role within an intricate feedback loop or represent noise within our experimental system. Pathway enrichment analysis was next performed on all concordant and discordant quadrants (Fig. 4i). We found that ATRNL1 positively regulates, directly or indirectly, pathways and gene sets related to MTORC1 signaling, hypoxia, cell stress, ER stress, glucose metabolism, protein secretion/transport, and ion transport (quadrant II). Conversely, ATRNL1 inhibits, directly or indirectly, genes and pathways related to cell cycle progression, cell junction organization, endocytosis, glycosylation, calcium signaling, and synaptic transmission (quadrant IV).
The large number of gene expression changes observed following the manipulation of ATRNL1 levels prompted us to look at more diverse forms of gene regulation. Interestingly, a circular form of the ATRNL1 transcript has been previously described in several cancer studies22–25. We re-analyzed our total RNA-seq data from hESC-aCMs to detect circular RNAs. We identified several circular RNA isoforms of ATRNL1 (circATRNL1) (Supplementary Fig. 6a). The most reliably detectable circATRNL1 species spanned from exon 2 through exon 8 (Supplementary Fig. 6b). This circular RNA was not detectable in 2 out of our 3 siRNA-2 libraries, suggesting that the siRNA is also capable of targeting this molecule. In cancer cicrATRNL1 is thought to act as a microRNA (miRNA) sponge25. So, we next scanned the circATRNL1 transcript for putative miRNA binding sites. We restricted this analysis to miRNAs conserved across mammals, giving us a total of 231 miRNAs to analyze. We filtered all hits by imposing a dissociation constant (log(Kd)) cutoff of -4, and were left with 20 putative miRNAs (Supplementary Data 10). Next, we sought to determine the gene targets of these microRNAs. For miRNA target-enrichment we incorporated the results from 2 well-known predictive models, TargetScan26 and miRTarBase (Supplementary Fig. 6c)27,28. We then intersected these results with our combined differentially expressed genes (Fig. 4h) and identified a total of 98 genes (Supplementary Fig. 6d, and Supplementary Data 11). Next, we wanted to determine which other putative gene networks could be regulated by circATRNL1 in CMs, so we performed pathway enrichment analysis on the 98 genes that intersected our differential expression data (Supplementary Fig. 6e). Consistent with circATRNL1s suggested role as a tumor suppressor, cell cycle was among the most enriched pathways. Further, we found genes enriched in pathways/processes related to MTORC1 signaling, hypoxia, cell-cell adhesion, TGFB-, and WNT-signaling.
ATRNL1 levels effect atrial cardiomyocyte electrophysiology
Our transcriptomics data indicated that elevated levels of ATRNL1 promotes the expression of many genes with roles in cardiac conduction and calcium signaling (Fig. 4). Changes in expression of such genes could lead to changes of the electrophysiological properties of CMs and promote arrhythmias like AF. Further, reduced expression of intercalated disc components can lead to the misexpression of others, which can lead to diminished cell-cell coupling and arrhythmias. Previously, it was found that with reduced N-cadherin levels (an important intercalated disc protein component) you also observe reduced Cx43/GJA1 expression which predisposes animals to arrhythmias29. Interestingly, we also observed significant changes in Cx43/GJA1 expression in our ATRNL1 knockdown experiments (Fig. 4d). Thus, we next wanted to determine if ATRNL1 plays a functional role in CM electrophysiology. For these experiments we wanted to phenotype hESC-aCMs through measuring their action potentials. The cardiac action potential duration (APD) determines the refractory period of the heart and if it becomes too short or too long, then the result is an arrhythmia30. Importantly, APD is focused on so heavily because this parameter is essential for modeling inherited arrhythmogenic syndromes (short and long QT syndromes) and for the pharmacologic field of cardiac safety31. We transduced hESC-aCMs with a lentivirus carrying a voltage-sensitive fluorescent protein, ArchLight32. We then manipulated the levels of ATRNL1 expression in these cells (Supplementary Fig. 7a, b), recorded their action potentials with a high-speed fluorescence imager, and finally calculated their APD at 80% repolarization (APD80). Knocking down ATRNL1 with siRNA-2 significantly prolonged the APD (Fig. 5a, b). Conversely, overexpressing ATRNL1 significantly shortened the APD of hESC-aCMs (Fig. 5c, d). Overall, these results suggest that the manipulation of ATRNL1 in atrial CMs is arrhythmogenic.
snRNA-seq and AF GWAS uncover unexpected expression of KCNN3
Finally, we sought to use our large scale snRNAseq dataset from the human LA to prioritize genes at the known AF genetic loci. To date, genome-wide association studies (GWAS) have identified more than 150 genetic loci associated with AF. However, a limitation of GWAS is that they implicate a genetic locus rather than directly a gene. The term effector gene is typically used to refer to the causal gene at a GWAS locus, that is driving the common variant signal observed in the study. Finding the effector gene at each locus remains challenging. Typically, gene prioritization approaches are applied to identify a subset of candidate effector genes for follow-up. One available gene prioritization method is the intersection with gene expression in relevant tissues and cell types. Since the LA is a highly relevant tissue for AF, our dataset serves as a rich resource to perform such analyzes.
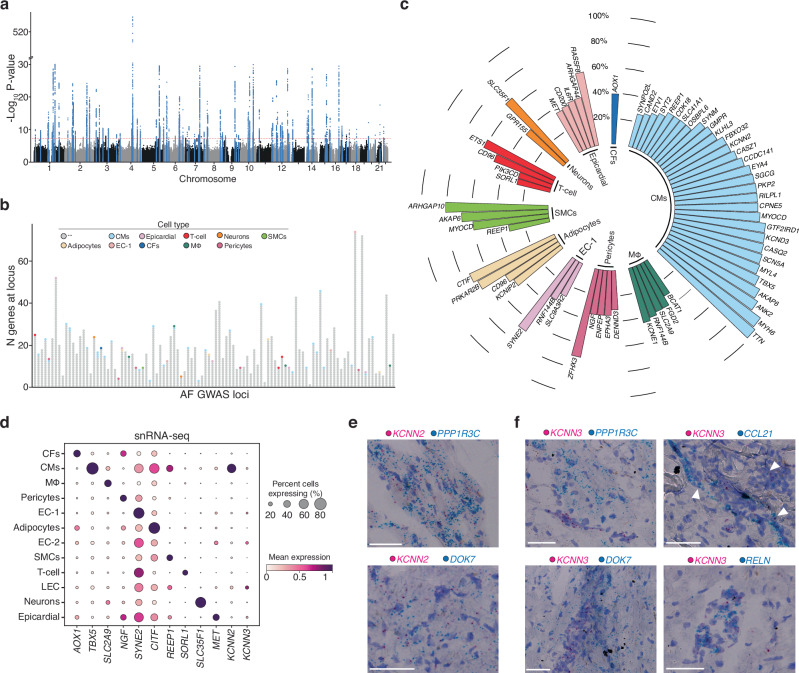
Overall, we identified 1379 marker genes from our snRNA-seq dataset (Supplementary Data 3) with roughly a quarter of the marker genes (24.3%) arising from CMs. We then sought to intersect these markers with the genes at the genetic loci of a previously published AF GWAS study33 (Fig. 6a). At each GWAS locus (defined as 1 mega base (Mb) around the sentinel variant) there can be many effector gene candidates. And, at the 104 AF GWAS loci we found a total of 1793 candidate genes. Importantly, some of these loci contained over 60 candidate genes, emphasizing the pressing need for prioritization (Fig. 6b). By intersecting the GWAS candidate genes with the marker genes for each cell type of our dataset, we were able to highlight 1 gene, in most cases. The rare cases included 6 loci each with 2 prioritized candidates, and 3 additional loci each with 3 gene hits. In sum, we found at least 1 candidate effector gene using this approach for 52 of the 104 AF GWAS loci. The majority of the 59 candidate effector genes are CM marker genes (N = 31), and up to 5 each are coming from the other cell types (Fig. 6c). A few genes are marker genes in multiple cell types, namely REEP1 (CMs and smooth muscle cells(SMCs)), CD96 (Adipocytes, and T-cells), RNF144B (endothelial cells (EC-1, EC2, and macrophages (MΦ)), AKAP6 (CMs, and SMCs), and MYOCD (CMs, and SMCs). We also identified several known candidates for AF that have been previously described, including TTN34, ZFHX335, TBX536 and KCNIP2, which supports the validity of our approach. Next, we evaluated the expression of all candidate AF loci across the cell states found in the Human Heart Cell Atlas (Supplementary Fig. 8a)20. Our data matched the Heart Cell Atlas cell state annotations well. The CM AF loci were expressed across the different subtypes of atrial and ventricular CMs, as well as pacemaker cells, and Purkinje cells. Some candidate genes did display differential expression among the CM clusters, including known atrial-enriched transcripts like TBX5 and MYL437. Further, with the large enrichment of immune cells in the Human Heart Atlas dataset we found improved resolution among our immune cell-enriched candidate AF loci. For example, the macrophage marker gene RNF144B exhibited high expression in neutrophils. RNF144B is an E3 ubiquitin ligase known to promote lipopolysaccharide-inducible IL-1b expression and inflammasome priming in human macrophages. The interconnection of inflammasome activation and AF have been recently established38. Overall, the Human Heart Atlas data agrees with our cell type annotations and provides improved cell-type-specific expression information for the identified AF GWAS loci. Additionally, the gene KCNN2 emerged as a candidate effector gene at the genetic locus 5q22.3 (sentinel variant rs716845).
Fig. 6. Integration of AF snRNA-seq data with a multi-ancestry genome-wide association study for AF.
a Manhattan plot for Roselli et al. AF GWAS main results17. The dotted line marks the cutoff for genome-wide significance (P-value < 5 × 10-8, that accounts for multiple testing across the genome). Highlighted in blue are the genetic loci that reached genome-wide significance, which were used to select the region (1 mega base (MB) around sentinel variants) and identify overlapping genes for the integration with snRNA-seq data. The y-axis of the plot has a break between -log10(P-value) of 30 and 510 to for visibility. The significance level accounts for multiple testing of independent variants with using a Bonferroni correction. P values (two-sided) were derived from a meta-analysis using a fixed-effects model with an inverse-variance weighted approach. b Stacked dotplot with one column for each genetic locus examined. Each dot represents a gene at the genetic locus, showing the range of number of genes within 1 MB of sentinel variants. Some loci including more than 60 genes close to the sentinel variant. Genes that overlap with the marker genes for the snRNA-seq cell types are labeled in cell-type specific colors as in Fig. 1b. Gray dots indicate genes that did not overlap. The cell-types color scheme is shown in the legend. c Circular plot of all marker genes that intersected with the AF GWAS loci, sorted by percent of cells expressing the gene and cell-type. Cell types are labeled in cell-type specific colors as in Fig. 1b. d Dot plot displaying expression for each indicated cell population. Size represents the percent of cells expressing each marker. e RNA-ISH images of LA tissue displaying expression of KCNN2 (shown in pink) and the CM markers PPP1R3C (top, shown in blue) and DOK7 (bottom, shown in blue). Scale bars = 50 μm. f RNA-ISH images of LA tissue displaying expression of KCNN3 (shown in pink) and the CM markers PPP1R3C (top left, shown in blue) and DOK7 (bottom left, shown in blue). Expression of KCNN3 (shown in pink) and the LEC markers CCL21 (top right, shown in blue) and RELN (bottom right, shown in blue). Scale bars = 50 μm.
KCNN2 and KCNN3 encode small conductance calcium-activated potassium channels (SK). The channels are known to regulate cardiac excitability and are gated by changes in intracellular Ca2+ derived from the sarcoplasmic reticulum39. Importantly, SK channels are known to contribute to arrythmias, as it has been found that both gain- and loss-of-function of SK channels can increase AF susceptibility39–42. In the search for novel antiarrhythmics, inhibition of small conductance calcium-activated potassium, or SK, channels has recently been proposed as a therapeutic approach. SK channels are encoded by the KCNN1 (KCa2.1; SK1), KCNN2 (KCa2.2; SK2), and KCNN3 (KCa2.3; SK3) genes43. Both KCNN2 and KCNN3 have been independently associated with AF in genome-wide association studies33,44–46.
To further characterize the expression patterns of KCNN2 and KCNN3 we evaluated their transcript abundance across all snRNA-seq clusters in comparison to other cell type-specific GWAS loci mentioned above (Fig. 2d). Interestingly, while KCNN2 was expressed highly by CMs, KCNN3 levels were surprisingly low in CMs. Rather, KCNN3 was expressed by lymphatic endothelial cells (LECs), a finding that was confirmed by RNA in situ hybridization (RNA-ISH) of fresh frozen LA tissue as follows. We began by identifying markers from the snRNA-seq data that were: a) specific for the cell populations of interest, CMs and LECs, and b) expressed at levels comparable to KCNN2 and KCNN3. The genes DOK7 and PPP1R3C fit these criteria and were therefore used to identify CMs in LA tissue. RELN and CCL21 were selected as appropriate markers for the LEC population. Our RNA-ISH results indicated that KCNN2 expression is widespread in atrial tissue and often observed in cells simultaneously expressing the CM markers DOK7 and PP1R3C (Fig. 6e). These results support our snRNA-seq data and existing literature that shows widespread KCNN2 expression in the human heart, specifically within CMs. When KCNN3 expression was similarly assessed by RNA-ISH, we observed sporadic groups of cells positive for KCNN3, and in many cases, these KCNN3+ cells were also co-expressing the LEC-specific marker genes, RELN and CCL21 (Fig. 6f). Performing RNA-ISH with probes targeting KCNN3 conjointly with the CM markers DOK7 and PPP1R3C failed to provide any evidence of co-expression; rather, there was clear spatial separation of cells expressing KCNN3 and these CM markers (Fig. 6f). Together, these results demonstrated that KCNN3 is expressed in LECs, and not in CMs as was previously expected.
Discussion
We present a comprehensive snRNA-seq analysis of LA tissue from patients with and without AF. In total, our dataset includes 34 individuals, and we were able to generate an embedding of over 170,000 nuclei from LA tissue. We identified a gene significantly upregulated in AF patients, ATRNL1. We performed immunofluorescence analysis of human LA tissue and hESC derived cardiomyocytes, transcriptomic profiling of siRNA and overexpression cells, and electrophysiological phenotyping to characterize the role of ATRNL1 in AF and cardiomyocyte physiology. Finally, we integrated our large-scale transcriptomics data with high-quality AF GWAS results to prioritize effector genes at single-cell resolution.
In our snRNA-seq analyzes, we did not identify large changes in cellular composition in the LA from patients with AF compared to control donors. Interestingly, it’s generally acknowledged that fibrotic replacement of the atrial myocardium is the basis of atrial pathology in patients with AF47. However, we did not observe any obvious shifts in cardiac fibroblast (CF) composition or cellular status. Further, there is a high correlation of AF with increased amounts of epicardial adipose tissue (EAT) and subsequent infiltration of that adipose tissue into the myocardium48,49. Adipose and lipid accumulation in the atrial myocardium is also thought to promote fibrosis and immune cell infiltration. Increased inflammation and fibro-fatty tissue accumulation are believed to strongly contribute to the development and maintenance of substrates for arrhythmias. Yet, from our study presented here, we did not see a dramatic change in tissue composition after comparing between control hearts and patients with AF. A potential limitation of our study would that the observed tissue compositional variability among the patients profiled here may be difficult to properly account for even with modern analytical frameworks (e.g., Milo and scCODA) and could require larger sample sizes with greater statistical power to determine the distinctions more accurately between patient groups. Moreover, profiling more patients at a greater depth (nuclei per sample) could improve our statistical power to accurately identify differentially expressed genes across more cell types50. Further studies on a broader range of AF cases classified from mild to severe (e.g., AF with heart failure) could help identify subtle changes in intercellular signaling and tissue composition that accompany AF.
Interestingly, a recent scRNA-seq study conducted on patients undergoing open heart surgery with AF and mitral valve regurgitation found evidence of macrophage expansion and a commensurate decrease in endothelial and mural cell composition in the left atrial appendage compared to controls51. The flow cytometry-based cellular enrichment approach taken in that study may be an ideal approach for characterizing the changes in all non-cardiomyocytes from living tissue, especially rare immune cells that our nuclei-based profiling approach may not properly account for. Importantly, here we only focused on the LA, an important tissue for evaluating AF. In AF the right atrium (RA) is also affected, however, the cellular and molecular changes to the RA in AF are not well understood. The pulmonary vein (PV) is known to play a role in the pathogenesis of AF, and catheter ablation of foci in the PV is an effective treatment for arrhythmias52,53. Future studies interrogating the single cell characteristics of the PV-LA junction, and the RA are also warranted.
Although there has been growing interest in SK channels as therapeutic targets for the treatment of AF, the close homology between the SK channels and the lack of isoform-specific tools have presented considerable challenges to establishing the role of SK2 and SK3 in the heart at steady-state and during AF. Previous studies of SK channel expression in the heart have relied on qRT-PCR, Western Blot, and immunofluorescence analysis of isolated CM or cardiac tissue lysates54–60. Our goal was to use snRNAseq to obtain additional information about the expression of KCNN2 and KCNN3 at the level of individual cell populations to help clarify the role of these channels in the adult human heart. Our snRNAseq studies demonstrated that while KCNN2 was readily identifiable in the CMs of the heart, KCNN3 expression was negligible within the CM compartment. Rather, KCNN3 was expressed by cells falling into what we annotated as LECs. These results were validated using RNA-ISH where again, KCNN2 was easily detectable in cells bearing common CM markers, while KCNN3 was found in cells co-expressing markers specific to LECs. Recently it was discovered that in human small resistance KCNN3 colocalizes with TRPV4 channels in endothelial caveolae61,62. The interaction of these proteins allows Ca2+ entry through TRPV4 channels, initiating the endothelium-derived hyperpolarizing factor response and contributes to vasodilation and regulation of blood flow and blood pressure63. This interaction of KCNN3 and TRPV4 was found to be impaired in the small resistance arteries of patients with hypertension and in a mouse model of hypertension63. Importantly, among patients with persistent AF, hypertension is present in about 60% to 80% of individuals64. Future studies will determine if KCNN3 plays a role in the development of AF and may help shed light on the well-known epidemiological association between AF and hypertension.
A major finding of our study was the differential expression of ATRNL1 in CMs from patients with AF. The role of ATRNL1 in cardiovascular biology is unknown. However, a patient heterozygous for a deletion of ATRNL1 has been identified with a heart murmur and a partial ventricular septal defect18. Interestingly, a Lebanese patient with a 64 kb heterozygous deletion of exons 9-13 of ATRNL1 has recently been identified and this patient didn’t present any abnormal cardiac phenotype65. This may suggest that only the short ATRNL1 isoform is required for proper human cardiac development, as exon 9 is the first exon distinguishing the long isoform from the short, which is only 8 exons long. When we selectively knocked down the long isoform of ATRNL1, we overall observe a similar transcriptional profile to when we knocked down both the long and the short isoforms. The long isoform is distinguished from the short at the protein level via the following additional features; a C-type lectin-like domain, several plexin-semaphorin-integrin (PSI) domains, a lamin-type EGF domain, a transmembrane helix, and a small domain predicted to be localized in the cytoplasm. The short isoform lacks the predicted transmembrane helix domain, but contains features typically associated with extracellular or secreted proteins like EGF domains, a CUB domain, and a lectin domain. Our immunofluorescence analysis indicated that ATRNL1 localizes to the intercalated disk and may also be localized in the cytoplasm. The antibody used in this study recognizes an antigen only found in the long ATRNL1 isoform. Interestingly, in other cell types ATRNL1 has been found to be localized to the nucleoplasm and mitochondria (proteinatlas.org)66. Future studies dissecting the functional roles of these two isoforms, including the protein-protein interactions and subcellular localization will be invaluable for deciphering the role of ATRNL1.
We found that knockdown of ATRNL1 led to an upregulation of genes involved in cardiac conduction, and a downregulation of genes associated with the UPR, protein glycosylation, cholesterol biosynthesis. While a role has been proposed for ATRNL1 in energy homeostasis previously19, no other clear molecular functions have been ascribed. Our analysis indicated a broader function for ATRNL1 and implicated it as a regulator and/or sensor of various pathways associated with the cellular response to stress. ATRNL1 appears to repress cell cycle associated pathways, cell junction organization, calcium signaling, and genes involved in synaptic transmission. Given the highly pleiotropic nature of the phenotype caused by a heterozygous ATRNL1 deletion in humans, it could be anticipated that ATRNL1 may have several molecular functions. ATRNL1 appears to positively impact the expression of Natriuretic Peptide Receptor 3 (NPR3), which is a clearance receptor for natriuretic peptides and has been shown to have an anti-apoptotic role in CMs67. The ER-associated degradation (ERAD) complex member HERPUD1, was similarly affected by ATRNl1, and HERPUD1 also appears to play a protective role in CMs as it’s been identified as an inhibitor of pathologic cardiac hypertrophy68. We also observed a strong enrichment of genes related to MTORC1 signaling. While MTORC1 signaling is required for heart development, some studies suggest that pharmacologically inhibiting MTORC1 signaling is cardioprotective69. Further, we found that ATRNL1 also activates the expression of MYC. In CMs it has been found that MYC regulates metabolism and mitochondrial biogenesis in response to pathologic stress and enhances the heart’s ability to respond to ischemic insults70. Similarly, ATRNL1 levels had an inhibitory impact on the expression of the thioredoxin interacting protein TXNIP, and mouse studies have found that knockout of TXNIP Deletion of thioredoxin-interacting protein in mice impairs mitochondrial function but protects the heart from ischemia-reperfusion injury71. Together, these findings suggest a cardioprotective role for ATRNL1. A limitation of our study is the use of hESC-aCMs, which are not as mature as adult human CMs in respect to features like sarcomere structure, Ca2+ kinetics, and ion channel density. Recent developments in 3D cardiac tissue technologies and methods have led to improvements in CM maturation and cardiac tissue modeling, including engineered heart tissue (EHT)72,73. Future work evaluating the physiological role of ATRNL1 in CMs will need to be conducted in EHTs or other 3D cardiac tissue models containing more mature and therapeutically relevant cell populations.
Altogether, our study has identified several findings regarding the cellular and molecular mechanisms underlying AF. First, we found that there were no significant changes in cellular composition in the LA from patients with AF compared to control donors. This suggests that the development of AF may not be due to a single cell type or population, but rather a complex interplay between multiple cell types, and potentially even cardiac regions. Second, we found that the expression of the gene ATRNL1 was significantly upregulated in CMs from AF patients. Finally, we found that knockdown of ATRNL1 in hESC-aCMs causes an upregulation of genes involved in cardiac conduction, and a downregulation of cell stress response genes and pathways. These findings suggest that ATRNL1 may have a cardioprotective role. However, we also found that elevated ATRNL1 levels cause a shortening of the APD, and thus may be arrhythmogenic. The dosage sensitivity of CMs to ATRNL1 highlights its important biological role in humans. In total, our findings extend our understanding of the transcriptional, cellular, and molecular basis of arrhythmias, results that will launch new areas of investigation and opportunities for the therapeutic development of improved AF treatments.
Methods
Human tissue samples
Written informed consent for research use of donated tissue was obtained from either the patient providing tissue or the next-of-kin in the case of deceased organ donors. Research use of tissues were approved by the relevant institutional review boards at the Gift-of-Life Donor Program, the University of Pennsylvania, Massachusetts General Hospital, and the Broad Institute (IRB-5999). All samples originated from deceased donors with non-failing hearts that were rejected for organ donation. Adult left atrial tissue samples were collected from organ donors by Myocardial Applied Genetics Network (MAG-Net;) as previously described10,13. Control samples were collected from organ donors with no previous history of heart disease or atrial fibrillation AF samples were collected from patients that had a history of atrial fibrillation but no history of other heart disease. Briefly, hearts were arrested in situ with at least 1 L ice-cold crystalloid cardioplegia solution and were transported to the on ice until cryopreservation (always less than 4 h). Tissue from the left atria was collected, flash frozen and stored at -80 until used.
Statistics and Reproducibility
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Trichrome staining of cardiac tissue
Trichrome staining was carried out on 10 um thick fresh frozen sections of LA tissue following manufacturer’s protocol (Abcam ab150686).
Immunofluorescence staining of cardiac tissue
Immunofluorescence staining was carried out on 10 um thick fresh frozen sections of LA tissue. In brief, samples were fixed in 4% PFA for 15 mins and then permeabilized with 0.1% TX-100 for 5 mins. Following 3x PBS washes samples were blocked in 7% donkey serum for 1 h (h) at room temperature (RT). Sections were incubated with primary antibodies overnight at 4 degrees C. Sections were washed 3 x in PBS and then incubated with secondary antibodies and hoescht for 1 h at RT. Sections were washed 3 x in PBS and then mounted with prolong gold mounting media (Invitrogen). Images were taken at 20x on a Leica SP8 microscope. Antibodies used in this study; ATRNL1, rabbit, Thermo fisher PA5-58234 (1:100), Cx43, mouse, Thermofisher 14-4759-82 (1:100), α-actinin: mouse: abcam ab9465 (1:200). Secondary antibodies: donkey anti-rabbit 488 thermo, A-21206 (1:200); donkey anti-mouse 568 thermo, A10037 (1:200). Pearson’s correlation coefficient measurements were carried out on images co-stained for ATNRL1 and Cx43 using the Image J plug in Just Another Colocalization Plugin74. Images were converted to 16 bit and ATRNL1 and Cx43 images were analyzed. The average of all analyzed images was reported in the results section.
LA sample processing and snRNAseq
Nuclei were isolated from fresh frozen LA tissue as previously described10,13. In brief, LA tissue was sectioned into 100 um thick slices using a cryostat. Nuclei were liberated by douncing in NIM2 buffer (250 mM sucrose, 25 mM KCl, 0.05% IGEPAL-630, 3 mM MgCl2, 1 μM DTT, 10 mM Tris pH 8.0), and debris was pelleted at 400 × g for 2 mins at 4 degrees C. Nuclei were filtered through 40 and 10 um filters and then washed with PBS wash buffer (0.01% BSA, 5 mM MgCl2, PBS). Nuclei were pelleted by centrifugation at 550 × g for 5 min at 4 degrees C, washed with 6 mL of PBS wash buffer and centrifuged at 550 × g for 5 min at 4 degrees C. Nuclei were resuspended in Nuclei Suspension buffer (Nuclear wash buffer + 0.4 U μl − 1 murine RNAse inhibitor) and then counted and 6000 nuclei were loaded into the 10x genomics microfluidic device (10x genomics Single cell 3′ solution v 3). Libraries were constructed using manufacturer’s protocol with a few modifications. First, to promote nuclei lysis, GEMS were incubated at 4 °C for 15 min after emulsion generation. Second, the reverse transcription protocol was lengthened to be 42 °C for 20 min then 53 °C for 120 min. Libraries were QCed and quantified using BioAnalyzer and qPCR. Libraries were pooled and sequenced on Illumina Novaseq S1 flow cells, at an average of 16 samples per flow cell.
snRNA-seq data pre-processing
The processing pipeline implemented here followed similar steps than previously described10. In brief, the raw files of 36 samples were processed on the TERRA platform (https://app.terra.bio/). The raw base call files (BCLs) were demultiplexed into FASTQ files using the mkfastq workflow from the 10x Genomics CellRanger v 4.0.0 tool. Homopolymer repeats (A30, C30, T30 and G30, settings: max_error_rate = 0.1; min_overlap = 20) and the template-switch oligonucleotide and its reverse complement (AAGCAGTGGTATCAACGCAGAGTACATGGG and CCCATGTACTCTGCGTTGATACCACTGCTT, settings: max_error_rate = 0.07 and min_overlap = 10) were trimmed using cutadapt v1.18 in Python v3.7.9. Alignment of trimmed reads was performed with CellRanger count workflow, to the GRCh38 pre-mRNA human reference (v2020-A), using the parameter expect-cells = 5000 and otherwise default values.
The standard quality control output metrics from the CellRanger count workflow were inspected regarding percent of reads in cells, percent of reads confidently mapped to transcriptome, percent valid barcodes, and Q30 for reads (Supplementary Data 12). To identify low quality samples, we performed visual inspection of the unique molecular identifier (UMI) curves, the automatically created graph-based t-distributed stochastic neighbor embedding (t-SNE) projection, and the Y-chromosome count distribution in comparison to the reported sex. One sample (LA_1512) was removed during this procedure, as it presented as an outlier on the metrics distribution and had an atypical UMI curve with insufficient ambient plateau. As an additional quality control step, we fingerprinted the samples against whole-genome sequencing (WGS) data (available for 33 of the 36 samples) to identify mismatches using the CrosscheckFingerprints function from the Picard tool. The generation and processing of the WGS data has been previously described10. In brief, Whole-genome sequencing was performed targeting 30x coverage at the Broad Institute of Harvard and the Massachusetts Institute of Technology using the Illumina NovaSeq platform. Reads were aligned to the GRCh38 reference using BWA-MEM75. Variants were called using GATK HaplotypeCaller (v3.5.054)76. We removed one sample (LA_1344) with a mismatch between the WGS data and the genotypes extracted from the single nuclei RNA-sequencing. The baseline data for the 34 samples included in the final analysis can be found in Supplementary Data 1.
For a sensitivity analysis all samples that passed QC were processed with the remove-background workflow from the CellBender v0.2 tool77. The following parameters were used: --expected-cells 5000, --z-dim 100, --total-droplets-included 25000, --epochs 150, --learning_rate 1e-4, and --fpr 0.01. For the sample LA_1440 the parameters were adjusted to achieve training convergence: --learning_rate 2e-5, --expected_cells 10000, --total_droplets_included 30000.
Construction of nuclei map
We generated a nuclei map using scanpy 1.7.0. We log-normalized the count data using scanpy.pp.normalize_per_cell(1e4) and scanpy.pp.log1p() and calculated highly variable genes with scanpy.pp.highly_variable_genes. Afterwards scVI14 was applied for sample alignment by including the patient of origin as a batch indicator, and a nuclei map was constructed using scanpy.pp.neighbors(n_neighbors=10, n_pcs=50, use_rep = ‘X_scvi’,metric = ‘cosine’). Then clustering was performed with scanpy.tl.leiden(resolution=0.75).
The following 7 quality control (QC) metrics were calculated for each nucleus. We used the tool scanpy 1.7.0 to calculate several QC metrics unless otherwise indicated. 1) exon_ratio: the tool scR-Invex (https://github.com/getzlab/scrinvex) was used to assign reads to exons, introns, or both. The total proportion of exonic reads was then calculated for each droplet. A larger proportion of exonic reads could be indicative of cytoplasmic contamination in the droplet. 2) percent_mito: the percentage of mitochondrial reads out of all reads was calculated. 3) doublet_scores: The tool Scrublet78 was used to calculate a double score that can detect doublet nuclei in the data. 4) log_n_genes: The number of unique genes per nucleus were computed with scanpy.pp.filter_cells(min_genes=0) and logarithmized with numpy.log1p(). 5) log_ncount: The number of UMI counts per nucleus was calculated and logarithmized with numpy.log1p(). 6) entropy: The entropy per nucleus based on CellRanger counts was calculated with Bayesian entropy estimation (https://pypi.org/project/ndd/1.6.3/). 7) loggene_entropy: The entropy value was multiplied by the logarithmized number of genes. We first examined each cluster for outliers. One cluster was identified as highly enriched with contaminated nuclei based on a high proportion of reads mapping to exons and high mitochondrial gene counts, a second cluster was identified as enriched for doublets though high doublet score and high number of genes. All nuclei (N = 28,040) from both clusters were removed from the dataset (Supplemental Fig. 1). For all remaining clusters, nuclei were removed if a QC value was an outlier as defined by the interquartile range (IQR) = 1.5*IQR. Cutoffs were determined by cluster to account for inherent differences in transcriptional complexity between cell types. Then we reran scanpy.tl.leiden (resolution=0.5) and performed additional nuclei QC within the cell type clusters obtained at this resolution. To identify low quality or misclassified nuclei, the 7 nuclei QC metrics listed above were inspected, along with a marker gene-based score for each major cell type, to identify misclassifications. These marker gene-based scores were generated by performing a differential expression test based on the log normalized expression matrix across the clusters using scanpy.tl.rank_genes_groups(method = ‘wilcoxon’). Then the area under the curve (AUC) per gene and cluster was calculated. The top 50 differentially expressed genes per cluster (by highest AUC) were selected to generate a cell type score for each nucleus using scanpy.tl.score_genes(). To identify subpopulations that may represent misclassified or low quality nuclei that were not removed in our previous QC, we iteratively subclustered each cluster, stepwise increasing the resolution by 0.1 until no more subcluster with marker genes with an AUC greater than 0.6 emerged. Then the distribution of all the nuclei QC metrics was inspected visually (via boxplots) and outlier sub-clusters were identified. A total of 8059 nuclei were marked for exclusion during this QC step. The final nuclei map contained 179,697 nuclei. Uniform Manifold Approximation and Projection (UMAP) plots of the nuclei map were created with scanpy.tl.umap (min_dist=0.2).
Identification of cell-types and marker genes
Marker genes for the top 13 most abundant cell types were calculated with the differential expression method limma-voom using the R-package limma v3.40.6. First, the expression counts were summarized by cluster and sample, including each observation with at least 50 nuclei per sample/cluster combination. For each cluster, genes were filtered using the algorithm implemented in filterByExpr from the R-package edgeR v3.26.879 to retain genes with sufficiently large counts with default settings. The data was then normalized using DESeq2 v1.24.080 normalization. Differential expression was then calculated using the limma-voom pipeline81 with the model ‘~0 + cluster + individual’, followed by extraction of the contrast comparing expression in each cluster versus all other clusters. Marker genes were selected if the AUC was > 0.6, the log fold change > 2, the adjusted P-value (by Benjamini Hochberg) was < 0.01 and the percentage of nuclei expressing the gene in the given cell-type was > 25%. For the 2 smallest clusters there was not enough power to calculate differential expression with the limma-voom pipeline. Instead, the marker genes were calculated using the function scanpy.tl.rank_genes_groups(method = “Wilcoxon”) on the log normalized expression matrix. The marker genes were selected using the same criteria as listed above but based on the Wilcoxon test results. The final set of marker genes for each cell-type can be found in Supplementary Data 3. The marker genes were then used to perform pathway enrichment analyzes to confirm the cell-type of each cluster. The web-based tool g:Profiler82 was used to perform gene set enrichment analyzes based on Gene Ontology (GO) gene sets.
Differential expression analysis
A formal differential expression model controlling for the correlation amongst nuclei from the same individual was performed by summing gene counts across all nuclei in a cluster within an individual patient and treating the data as a bulk RNA sequencing experiment before running limma-voom in a similar manner as mentioned above. The differential expression analysis across cases and controls was performed with the method limma-voom using the R-package limma v3.40.6. First, the expression counts were summarized by cluster and sample, including each observation with at least 20 nuclei per sample/cluster combination. For each cluster, genes were filtered using the algorithm implemented in filterByExpr from the R-package edgeR v3.26.879 to retain genes with sufficiently large counts with default settings. The data was then normalized using DESeq2 v1.24.080 normalization. Differential expression was then calculated using the limma-voom pipeline81 with the model ‘~af + sex’, followed by extraction of the contrast comparing expression in cases versus controls. Genes were considered significant in the differential expression analyzes when the adjusted P-value (by Benjamini Hochberg) was < 0.05. For visualization of the differential expression results in violin plots, the filtered and normalized count data was converted to the unit counts per million (CPM) as implemented in the cpm function from the edgeR R-package. Since there can be notable background contamination present in single nuclei RNA-sequencing experiments we generated a flag that would identify genes that have a high probability of coming from the background. We followed a procedure to calculate the flag based on CellRanger and also CellBender counts.
For the differential abundance analysis with Milo, we used the python version, MiloPy (0.1.1)15. We selected neighbors with a k = 50 for our differential abundance testing analysis. Compositional analysis with scCODA was carried out in python with the PertPy package (version 0.7.0)16. For pseudo-bulk RNA-seq analysis the python apdbulk (https://github.com/noamteyssier/adpbulk) module was utilized to generate pseudo-bulk counts, and the resulting count table was then analyzed with DEseq2 (1.30.1)80.
Integration of snRNA-seq with AF GWAS results
The marker genes for each cell type were intersected with results from an AF genome-wide association study (GWAS)33. We intersected the 104 genetic loci (sentinel variant +/- 500 kb) with the human gene reference GRCh38.p13 exported from BioMart to identify all genes at the genetic loci from the GWAS. That gene list was then intersected with the marker genes from this analysis to 1) prioritize genes at GWAS loci and 2) highlight the most relevant cell-types for AF.
hPSC culture and ACM differentiation
Human embryonic stem cells (hESCs, WiCell) were cultured in feeder-free culture on Geltrex (Invitrogen) coated flasks in a modified Essential 8 medium containing DMEM/F12 with HEPES (Thermo Fisher, 1133057), 540 mg/L sodium bicarbonate (Thermo Fisher, 25080094), 1% MEM Non-Essential Amino Acids (Invitrogen 11140) and 1x Pen-Strep (ThermoFisher 15140122), 8.5 mM NaCl (Sigma, S5150), 20 μg/mL insulin (Thermo Fisher, 12585014), 200 μM ascorbic acid (Sigma, A8960), 10 μg/mL holo-transferrin (Sigma, T0665), 1 μg/mL heparin (Sigma, H3149), 14 ng/mL sodium selenite (Sigma, S9133), 20 ng/mL bFGF (Thermo Fisher, 13256027), 10 ng/mL activin A (R&D Systems, 338-AC-01M), and 10 ng/mL Neuregulin-beta 1 (R&D Systems, 396-HB-050). Cells were incubated at 5% O2 and 10% CO2 at 37 °C.
For atrial cardiomyocyte (ACM) differentiation, dissociated single cell PSC suspension was incubated overnight with continuous shaking at 98 RPM to form spheroid clusters (day -1). Spheroids were collected (day 0) and resuspended in Basal Differentiation Media (BDM) containing DMEM/F12 with HEPES, 540 mg/L sodium bicarbonate, 1% non-essential amino acids (Thermo Fisher, 11140), 10 ug/mL holo-transferrin, 200 uM ascorbic acid, 63 ng/mL progesterone (Sigma, P8783), and 14 ng/mL sodium selenite, supplemented with 50 ng/mL activin A, 10 ng/mL BMP4 (R&D Systems, 314-BP-050), 20 ng/mL bFGF (Shenandoah, 100-28-500UG), with 2 μM CHIR-99021 (Selleckchem, S2924). Spheroids were replenished with fresh BDM to remove CHIR-99021 on day 1 and treated with 10 uM XAV-939 (APExBio, A1877), 1 uM IWR1-Endo (APExBio, B2306) and 1 uM IWP-2 (APExBio, A3512) on day 2. To promote atrial induction, 1 nM TTNPB (Tocris, 0761) was applied along with 100 nM retinoic acid, 1 uM SAG (Selleckchem, S7779) and 10 uM XAV-939 on day 3. TTNPB and RA were removed on days 4–5. On day 6 and onward, spheroids were maintained in BDM supplemented with 4 μg/ml insulin. On day 17, spheroids were collected, washed and dissociated with 1 mg/ml collagenase II (Thermo Fisher, 117010015) for one hour at 37 °C. Dissociated cardiomyocytes were seeded in fibronectin-coated plates and cultured in Lactate Media (DMEM without glucose (Thermo Fisher, 11966025), 540 mg/L sodium bicarbonate, 4 mM lactate (Sigma L6402), 200 uM ascorbic acid, and 14 ng/mL sodium selenite) for 4 days to remove non-cardiomyocytes. Cardiomyocytes were then kept in Cardiomyocyte Maintenance Media (DMEM/F12, 10% FBS and 1% Pen/Strep) until day 30.
Cellular electrophysiology phenotyping
Beating atrial cardiomyocytes at day 30 were dissociated as described above and seeded onto a fibronectin-coated 96-well (Greiner Bio-One) at 55,000 cells per well densities. Cardiomyocytes were infected with lentiviruses carrying a genetically encoded voltage sensor – Arclight. After 72 h of infection, ACMs were transfected with a scrambled siRNA control (Life Technologies, 4390843) or a ATRNL1 siRNA using Lipofectamine RNAiMAX (Invitrogen 13778) according to the manufacturer’s instructions. After 7 days of transfection, Arclight fluorescence of ACM was captured at 500 frames/sec on a Vala Sciences IC200-KIC Imager with electrical pacing at 1 Hz. Cardiomyocytes were maintained in a heating chamber at 37 °C with 10% CO2 throughout the recordings. The raw data were exported as sequences of TIFF files and transferred to an internal cloud database for further analysis. MATLAB (R2019b) was used to calculate durations of action potentials at 80% recovery to determine APD8083.
RNA extraction and qPCR
Total RNAs from ACMs were extracted with the QIAGEN RNeasy Mini Kit (74106) according to the manufacturer’s instructions. cDNA was synthesized with the iScript kit (Bio-Rad 1708891). qPCR was performed with the SsoAdvanced Universal probes supermix (Bio-Rad 1725284) on a Bio-rad CFX384 Real-time system. TaqMan probes (Life Technologies 4331182) for ATRNL1 and HPRT1 (Hs02800695_m1) were used for gene expression assay. Analysis was performed using the delta CT method normalized to HPRT1.
Immunohistochemistry of hESC-aCMs
HESC derived ACMs seeded on fibronectin-coated 96-well plate (Perkin Elmer, 6055300) were fixed in 4% paraformaldehyde for 15 min followed by 3 times washing with PBS. Cells were permeabilized with 0.2% Triton-X100 for 15 min and blocked with 2% BSA in PBS for 1 h at RT, followed by incubation with primary antibodies ATRNL1 and MYOM1 (DSHB, B4) overnight at 4 °C. After washing three times with PBS, cells were incubated with fluorescent secondary antibodies donkey anti-rabbit IgG alexa 488 (Invitrogen A32790), donkey anti-mouse IgG alexa 568 (Invitrogen A10037) and DAPI (Thermo Fisher, D1306) for 1 h at RT. Images were captured with Perkin Elmer Opera Phenix Imaging system and processed with Harmony. Human left atrial tissue from normal donor was sectioned at 10 μm and fixed in 4% PFA for 5 min, followed by washing with PBS, permeabilization in 0.1% Triton-X100 for 30 min and blocking in 3% BSA for an hour at RT. Tissue section was stained with primary antibodies against ATRNL1 and MYOM1 at 4 °C overnight, followed by washing and incubation with fluorescent secondary antibodies in 7 % donkey serum as described above, and mounted with Prolong Gold Antifade Mountant (Life Technologies, P36934). Fluorescent images were captured with a Leica SP8 confocal microscope. Adult human myocardial samples were collected and research use of tissues was approved by the relevant institutional review boards at the Gift-of-Life Donor Program, the University of Pennsylvania, Massachusetts General Hospital, and the Broad Institute.
RNA-seq and RNA-seq analysis
All hESC-aCMs were transfected with ATRNL1 siRNAs as described above. For the overexpression of ATRNL1 we packaged the short isoform (ENST00000609571.5) into lentiviral particles via transfection of HEK293T cells according to the manufacturers protocol (Dharmacon lentiviral packaging kit, Horizon Discovery, TLP5913). The overexpression lentiviral vector was obtained from GeneCopoeia (EX-E1118-Lv242). All hESC-aCMs were transduced with lentiviral particles and selected for overexpression with puromycin. Total RNA was extracted from atrial iPS-CMs using the Direct-zol RNA Miniprep kit (Zymo Research, R2051). RNA-seq libraries were generated with a ribosomal depletion protocol (KAPA RNA HyperPrep with RiboErase, KK8560). All RNA-seq libraries were sequenced on an Illumina NovaSeq. Sequenced reads were aligned to the human genome (GRCh38) using STAR (2.7.9a)84. Differential expression analysis was performed with DESeq2 (1.30.1)80. Pathway enrichment analysis was carried out with Metascape85 and ClusterProfiler (version 4.6.2)86. For the detection of circular RNA, we utilized the CircRNA_Finder scripts with STAR (2.7.9a)87. We scanned the circATRNL1 for miRNAs with ScanMiR (1.5.2)88. And the miRNA target enrichment analysis was carried out with MIENTURNet28.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Some clip artwork used in this manuscript were generated via BioRender.com. Dr. Ellinor is supported by grants from the National Institutes of Health (RO1HL092577, RO1HL157635), from the American Heart Association (18SFRN34230127, 961045), and from the European Union (MAESTRIA 965286). This research was also supported by the Carol and Roch Hillenbrand and the George L. Nardi, MD, funds at Massachusetts General Hospital. Dr. Xiao was supported by an American Heart Association Career Development Award (20CDA35260081). Dr. Tucker is supported by the National Institutes of Health (R01HL170051). Matthew C. Hill is supported by NIH grant 5T32HL007208. We thank the Gift-of-Life Donor Program, Philadelphia, PA, who helped provide nonfailing heart tissue from unused donor hearts for this research.
Author contributions
M.C.H., B.S., C.R., N.R.T., C.A.K., and P.T.E. conceived and designed the study. B.S. generated snRNA-seq and bulk RNA-seq data. M.C.H., C.R., and M.C. analyzed the snRNA-seq data. M.C.H., B.S., C.R., M.C., C.H, and P.T.E. interpreted snRNA-seq analyzes. B.S., O.A., H.B., and Y.G. performed histology and immunofluorescence. M.C.H. performed bulk RNA-seq analysis. C.H. performed RNA in situ hybridization. L.X. and H.M. performed all hESC-aCM work. L.X. carried out and analyzed all electrophysiology. K.C.B. and K.B.M. acquired LA and LV samples for this study. M.C.H., B.S., C.R., and P.T.E. drafted the manuscript, which was revised by all authors.
Peer review
Peer review information
Nature Communications thanks Kory Lavine, Arnela Saljic and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Processed single-nucleus transcriptomic data are available through the Broad Institute’s Single Cell Portal under project ID SCP2489. Raw and processed next-generation sequencing data have been deposited at the NCBI Gene Expression Omnibus with accession number GSE255992. Requests for Source Data can be made directly to the corresponding author. Source data are provided with this paper.
Competing interests
C.A.K. is an employee of Bayer US LLC (a subsidiary of Bayer AG) and may own stock in Bayer. The Precision Cardiology Laboratory is joint effort between the Broad Institute and Bayer AG. P.T.E. has received sponsored research support from Bayer AG, Novo-Nordisk, Bristol Myers Squibb and Pfizer; he has also served on advisory boards or consulted for Bayer AG. C.R. is a full-time employee at GSK as of July 2024. All remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Matthew C. Hill, Bridget Simonson, Carolina Roselli.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54296-w.
References
- 1.Nattel, S., Heijman, J., Zhou, L. & Dobrev, D. Molecular basis of atrial fibrillation pathophysiology and therapy: a translational perspective. Circ. Res.127, 51–72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah, M., Akar, F. G. & Tomaselli, G. F. Molecular basis of arrhythmias. Circulation112, 2517–2529 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Dobrev, D. Electrical remodeling in atrial fibrillation. Herz31, 108–112 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y.-H. et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science299, 251–254 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Kato, T., Iwasaki, Y. & Nattel, S. Connexins and atrial fibrillation: filling in the gaps. Circulation125, 203–206 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slyper, M. et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med.26, 792–802 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker, N. R. et al. Transcriptional and cellular diversity of the human heart. Circulation142, 466–482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litviňuková, M. et al. Cells of the adult human heart. Nature588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaffin, M. et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature608, 174–180 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Koenig, A. L. et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat. Cardiovasc. Res.1, 263–280 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, M. C. et al. Integrated multi-omic characterization of congenital heart disease. Nature608, 181–191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonson, B. et al. Single-nucleus RNA sequencing in ischemic cardiomyopathy reveals common transcriptional profile underlying end-stage heart failure. Cell Rep.42, 112086 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol.40, 245–253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Büttner, M., Ostner, J., Müller, C. L., Theis, F. J. & Schubert, B. scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun.12, 6876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connerty, P. et al. Development of siRNA-loaded lipid nanoparticles targeting long non-coding RNA LINC01257 as a novel and safe therapeutic approach for t(8;21) pediatric acute myeloid leukemia. Pharmaceutics13, 1681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark, Z., Bruno, D. L., Mountford, H., Lockhart, P. J. & Amor, D. J. De novo 325 kb microdeletion in chromosome band 10q25.3 including ATRNL1 in a boy with cognitive impairment, autism and dysmorphic features. Eur. J. Med. Genet.53, 337–339 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Walker, W. P. et al. Genetic analysis of attractin homologs. Genes. N. Y. N. 200045, 744–756 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature619, 801–810 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanhoutte, D. et al. Thbs1 induces lethal cardiac atrophy through PERK-ATF4 regulated autophagy. Nat. Commun.12, 3928 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, G. et al. Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol. Ther. Nucleic Acids19, 961–973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song, X. et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res44, e87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hj, F. et al. Circular RNA CpG island hypermethylation-associated silencing in human cancer. Oncotarget9, 29208–29219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyu, M. et al. CircATRNL1 and circZNF608 inhibit ovarian cancer by sequestering miR-152-5p and encoding protein. Front. Genet.13, 784089 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.V, A., Gw, B., Jw, N. & Dp, B. Predicting effective microRNA target sites in mammalian mRNAs. eLife4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hy, H. et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res50, D222–D230 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licursi, V., Conte, F., Fiscon, G. & Paci, P. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinforma.20, 545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J. et al. N-cadherin haploinsufficiency affects cardiac gap junctions and arrhythmic susceptibility. J. Mol. Cell. Cardiol.44, 597–606 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Da, E., Km, D. & Aw, T. The mechanism and significance of the slow changes of ventricular action potential duration following a change of heart rate. Exp. Physiol.94, 520–528 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Shinnawi, R. et al. Monitoring human-induced pluripotent stem cell-derived cardiomyocytes with genetically encoded calcium and voltage fluorescent reporters. Stem Cell Rep.5, 582–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin, L. et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron75, 779–785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roselli, C. et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet.50, 1225–1233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi, S. H. et al. Association between titin loss-of-function variants and early-onset atrial fibrillation. JAMA320, 2354–2364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudbjartsson, D. F. et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat. Genet.41, 876–878 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postma, A. V. et al. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ. Res.102, 1433–1442 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Wang, T. Y. et al. Human cardiac myosin light chain 4 (MYL4) mosaic expression patterns vary by sex. Sci. Rep.9, 12681 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ariffin, J. K. et al. The E3 ubiquitin ligase RNF144B is LPS-inducible in human, but not mouse, macrophages and promotes inducible IL-1β expression. J. Leukoc. Biol.100, 155–161 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Zhang, X.-D., Thai, P. N., Lieu, D. K. & Chiamvimonvat, N. Cardiac small-conductance calcium-activated potassium channels in health and disease. Pflug. Arch.473, 477–489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahida, S. et al. Overexpression of KCNN3 results in sudden cardiac death. Cardiovasc. Res.101, 326–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, N. et al. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. J. Physiol.587, 1087–1100 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozgen, N. et al. Early electrical remodeling in rabbit pulmonary vein results from trafficking of intracellular SK2 channels to membrane sites. Cardiovasc. Res.75, 758–769 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köhler, M. et al. Small-conductance, calcium-activated potassium channels from mammalian brain. Science273, 1709–1714 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Ellinor, P. T. et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat. Genet.42, 240–244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olesen, M. S. et al. Screening of KCNN3 in patients with early-onset lone atrial fibrillation. Eur. Eur. Pacing Arrhythm. Card. Electrophysiol. J. Work. Groups Card. Pacing Arrhythm. Card. Cell. Electrophysiol. Eur. Soc. Cardiol.13, 963–967 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Chang, S.-H. et al. Significant association of rs13376333 in KCNN3 on chromosome 1q21 with atrial fibrillation in a Taiwanese population. Circ. J. J. Jpn. Circ. Soc.76, 184–188 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Pg, P. Atrial fibrosis: an obligatory component of arrhythmia mechanisms in atrial fibrillation? J. Geriatr. Cardiol. JGC14, 233–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iacobellis, G. Epicardial adipose tissue in contemporary cardiology. Nat. Rev. Cardiol.19, 593–606 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suffee, N. et al. Reactivation of the epicardium at the origin of myocardial fibro-fatty infiltration during the atrial cardiomyopathy. Circ. Res.126, 1330–1342 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Schmid, K. T. et al. scPower accelerates and optimizes the design of multi-sample single cell transcriptomic studies. Nat. Commun.12, 6625 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hulsmans, M. et al. Recruited macrophages elicit atrial fibrillation. Science381, 231–239 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haïssaguerre, M. et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N. Engl. J. Med.339, 659–666 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Mahida, S. et al. Science linking pulmonary veins and atrial fibrillation. Arrhythmia Electrophysiol. Rev.4, 40–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kokubun, S., Nishimura, M., Noma, A. & Irisawa, H. Membrane currents in the rabbit atrioventricular node cell. Pflug. Arch.393, 15–22 (1982). [DOI] [PubMed] [Google Scholar]
- 55.Mangoni, M. E. et al. Functional role of L-type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc. Natl Acad. Sci. Usa.100, 5543–5548 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mangoni, M. E. et al. Bradycardia and slowing of the atrioventricular conduction in mice lacking CaV3.1/alpha1G T-type calcium channels. Circ. Res.98, 1422–1430 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Marionneau, C. et al. Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J. Physiol.562, 223–234 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Y. et al. Molecular identification and functional roles of a Ca(2+)-activated K+ channel in human and mouse hearts. J. Biol. Chem.278, 49085–49094 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Tuteja, D. et al. Differential expression of small-conductance Ca2+-activated K+ channels SK1, SK2, and SK3 in mouse atrial and ventricular myocytes. Am. J. Physiol. Heart Circ. Physiol.289, H2714–H2723 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Tuteja, D. et al. Cardiac small conductance Ca2+-activated K+ channel subunits form heteromultimers via the coiled-coil domains in the C termini of the channels. Circ. Res.107, 851–859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Absi, M. et al. Effects of methyl beta-cyclodextrin on EDHF responses in pig and rat arteries; association between SK(Ca) channels and caveolin-rich domains. Br. J. Pharmacol.151, 332–340 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saliez, J. et al. Role of caveolar compartmentation in endothelium-derived hyperpolarizing factor-mediated relaxation: Ca2+ signals and gap junction function are regulated by caveolin in endothelial cells. Circulation117, 1065–1074 (2008). [DOI] [PubMed] [Google Scholar]
- 63.He, D. et al. Treatment of hypertension by increasing impaired endothelial TRPV4-KCa2.3 interaction. EMBO Mol. Med.9, 1491–1503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verdecchia, P., Angeli, F. & Reboldi, G. Hypertension and atrial fibrillation: doubts and certainties from basic and clinical studies. Circ. Res.122, 352–368 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Choucair, N. et al. Contribution of copy number variants (CNVs) to congenital, unexplained intellectual and developmental disabilities in Lebanese patients. Mol. Cytogenet.8, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thul, P. J. et al. A subcellular map of the human proteome. Science356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Lin, D., Chai, Y., Izadpanah, R., Braun, S. E. & Alt, E. NPR3 protects cardiomyocytes from apoptosis through inhibition of cytosolic BRCA1 and TNF-α. Cell Cycle15, 2414–2419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Torrealba, N. et al. Herpud1 negatively regulates pathological cardiac hypertrophy by inducing IP3 receptor degradation. Sci. Rep.7, 13402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sciarretta, S., Volpe, M. & Sadoshima, J. mTOR signaling in cardiac physiology and disease. Circ. Res.114, 549–564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahuja, P. et al. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J. Clin. Invest.120, 1494–1505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshioka, J. et al. Deletion of thioredoxin-interacting protein in mice impairs mitochondrial function but protects the myocardium from ischemia-reperfusion injury. J. Clin. Invest.122, 267–279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tani, H. & Tohyama, S. Human engineered heart tissue models for disease modeling and drug discovery. Front. Cell Dev. Biol.10, 855763 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen, A. et al. Development of a drug screening platform based on engineered heart tissue. Circ. Res.107, 35–44 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Bolte, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc.224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at 10.48550/arXiv.1303.3997 (2013).
- 76.van der Auwera, G. & O’Connor, B. D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. (O’Reilly Media, Incorporated, 2020).
- 77.Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods20, 1323–1335 (2023). [DOI] [PubMed] [Google Scholar]
- 78.Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst.8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen, Y., Lun, A. T. L. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research5, 1438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol.15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leyton-Mange, J. S. et al. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep.2, 163–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl.29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov. Camb. Mass2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Westholm, J. O. et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep.9, 1966–1980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soutschek, M., Gross, F., Schratt, G. & Germain, P.-L. scanMiR: a biochemically based toolkit for versatile and efficient microRNA target prediction. Bioinforma. Oxf. Engl.38, 2466–2473 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Processed single-nucleus transcriptomic data are available through the Broad Institute’s Single Cell Portal under project ID SCP2489. Raw and processed next-generation sequencing data have been deposited at the NCBI Gene Expression Omnibus with accession number GSE255992. Requests for Source Data can be made directly to the corresponding author. Source data are provided with this paper.