Abstract
In clinical practice, timely and accurate diagnosis can effectively reduce unnecessary treatment, avoid high medical costs, and prevent adverse prognoses. However, some patients with malignant tumors and those with infection often exhibit similar symptoms, which are difficult to distinguish, posing challenges in accurate clinical diagnosis. Metagenomic next-generation sequencing (mNGS) technology has been widely applied to confirm the source of infection. Recent studies have shown that for pathogen detection, mNGS technology can be used to perform chromosomal copy number variations (CNVs) analysis in two different analytical pipelines using the same wet test. mNGS technology has further demonstrated its utility in not only the determination of pathogenic microorganisms but also of CNVs, thereby facilitating early differential diagnosis for malignant tumors. In this review, we aim to analyze the diagnostic performance of mNGS technology in the simultaneous detection of pathogenic microorganisms and CNVs in current clinical practice and discuss the advantages and limitations of mNGS-CNV dual-omics detection technology. Our review highlights the need for more large-scale prospective research data on current mNGS-CNV dual-omics detection technology to provide more evidence-based results for researchers and clinicians and to promote the greater role of this technology in future clinical practice.
Keywords: mNGS, Pathogens, Tumor, Copy number variant, mNGS-CNV dual-omics
1. Introduction
With the continuous advancement of modern molecular biology techniques, a plethora of molecular diagnostic methods have emerged. These technological advancements have significantly enhanced the precision of clinical pathogen detection and played a pivotal role in diagnosing common infections. In contrast to traditional clinical microbiological diagnostics that typically target a limited number of pathogens and rely on subjective judgments by doctors, the widespread adoption of metagenomic next-generation sequencing (mNGS) technology in clinical practice can be attributed to its rapidity, high accuracy, and absence of bias. This technology has been widely applied in the identification of infectious pathogens and has been included in consensus literature by multiple experts [[1], [2], [3]]. mNGS is widely employed to detect pathogenic microorganisms, ascertaining their presence or absence. However, it may yield negative results, particularly in patients with potential malignant tumors but non-pathogenic microorganisms through mNGS, necessitating further testing to elucidate the underlying cause.
The occurrence of diseases may stem from infectious diseases, such as through external invasion, or non-infectious causes. For patients with unexplained fever, infection or tumors could be the culprit. Copy number variation (CNV) is a tumor marker referring to a class of structural variations characterized by alterations in copy numbers, typically involving DNA segments larger than 1 kb. Among these variations, the structural changes associated with CNV primarily encompass genomic acquisitions (such as repeats or transpositions), deletions, or complex rearrangements. CNVs have been extensively investigated using diverse methodologies, including whole-genome sequencing (WGS) and fluorescence in situ hybridization (FISH) [4,5]. Several studies have demonstrated the efficacy of mNGS in not only the accurate identification of infectious diseases but also in tumor detection through CNV analysis [6,7]. This analytical pipeline is based on the same wet experiment, where CNV analysis and pathogenic microorganism identification are conducted using sequencing data comprising host and microbial sequences in dry experiments. By fully leveraging and interpreting sequencing data, it yields results pertinent to infection and tumor diagnosis, facilitating the exploration of disease etiology and offering valuable insights into non-infectious diseases. This comprehensive approach provides reliable guidance for subsequent patient diagnosis and treatment strategies.
In this review, we present an overview of the technologies employed in pathogen detection and CNV analysis. We comprehensively evaluate the diagnostic performance of mNGS for the simultaneous detection of pathogens and CNVs in clinical practice. Furthermore, we discuss the technical advantages and limitations associated with mNGS-CNV dual-omics detection, aiming to enhance the understanding of clinicians and researchers.
2. mNGS pathogenic detection technology
The clinical application of metagenomics can be traced back to the early application of microarray technology in the early 2000s. Some early successful applications of this technology include the discovery of the SARS coronavirus, gene mapping of cancer mutations, and in-depth analysis of microbial communities in different parts of the human body [8]. In 2004, researchers completed human genome sequencing using Sanger sequencing technology, and the following need for human genome research drove the maturation of detection technologies, leading to the emergence of second-generation sequencing technologies [9,10]. mNGS uses high-throughput sequencing technology to perform unbiased sequencing of all nucleic acids in a sample and combined with medical databases and specific algorithms, it can detect the microbial spectrum and even the host's genome or transcriptome [8]. This method enables simultaneous sequencing and analysis of nearly all microbial nucleic acids in a sample, which can be used for pathogen characterization, population structure, and drug resistance studies, and is an important method for studying microbiomes in humans and various environments [11].
mNGS technology is a shotgun sequencing technique with sequencing depths of up to 10–20 million sequences per sample and is compatible with various clinical samples, including cerebrospinal fluid, plasma, respiratory secretions, urine, feces, and tissues, among others [8,12]. The currently used sequencing platforms include a series of platforms provided by Illumina (San Diego, CA) Scientific Equipment Co., Ltd. (iSeq, MiniSeq, MiSeq, HiSeq, NextSeq, and NovaSeq platforms), Ion Torrent platform provided by Thermo Fisher Scientific (USA), BGISEQ platform provided by BGI (China), and portable sequencers (MinION, GridION, and PromethION) provided by Oxford Nanopore Technologies (USA) [13]. The major operational processes include collection and processing of samples, extraction of nucleic acid, construction of libraries, sequencing, analysis of data, interpretation of results, and final clinical integration for diagnostic purposes. The primary clinical applications of mNGS technology lie in its ability to rapidly detect pathogenic microorganisms related to infectious diseases and assistance in disease diagnosis. mNGS can detect tens of thousands of pathogens in one test, including newly discovered, rare, cross-species transmitted, mixed infection pathogens, and culture-negative pathogens. It is particularly suitable for the diagnosis of challenging, critical, and novel infectious diseases [8,14].
The mNGS pathogen detection technology plays a crucial role in infection diagnosis by enabling the fast and accurate identification of microbial pathogens responsible for infections or inflammation, thereby facilitating targeted treatment. According to a study by Duan et al., the sensitivity of mNGS pathogen detection (67.4 % vs 23.6 %, p < 0.001) is significantly higher than that of traditional culture methods, especially in samples such as bronchoalveolar lavage fluid (P = 0.002), blood (P < 0.001), and sputum (P = 0.037) [15]. Moreover, mNGS is highly efficient in that it has a short turnaround time, particularly in the detection of microorganisms such as strict anaerobes and intracellular bacteria. Data from Shi et al. demonstrated that mNGS can identify 67.23 % of infection cases within three days, whereas traditional culture methods for Mycobacterium tuberculosis, take approximately 90 days to detect and accurately identify 49.58 % of infection cases [16]. A study employed mNGS technology to examine blood samples from 14 children with unexplained hepatitis in the United States. The findings indicated that 13 children (93 %) tested positive for adeno-associated virus type 2 (AAV2) and suggested a potential association between the severity of symptoms in these children and co-infection of AAV2 and auxiliary viruses. This investigation highlights the utility of mNGS in diagnosing infectious cases in children with unexplained severe hepatitis [17]. In summary, mNGS holds promise in pathogen detection offering valuable insights for clinical practitioners and can provide references for clinical practitioners and future research [18].
The mNGS pathogen detection technology can also be used for the discovery of novel microbial pathogens. In recent years, mNGS has played a significant role in the discovery of novel coronaviruses, such as the SARS-CoV-2 virus that cause COVID-19 disease [19]. mNGS technology is primarily employed in the identification of novel pathogens and obtaining genomes of difficult-to-culture pathogens, which has implications for public health [20]. In 2011, China reported the use of mNGS technology to identify sequences resembling a suspected novel Bunyavirus from samples of patients with fever and thrombocytopenia syndrome. This enabled the isolation and identification of a new Bunyavirus, which was confirmed as the causative agent of the disease [21]. In late 2019, mNGS technology was used to identify sequences suspected to be from the SARS coronavirus in samples from patients with respiratory infections [19,22]. In the subsequent prevention and control of the COVID-19 pandemic, mNGS was also employed in transcriptome analysis of the host in COVID-19 patients, which helped establish a model for predicting the severity of COVID-19 [23].
mNGS can be used to study the microbial communities found in various samples, including those from humans. Through the analysis of microorganism genomes within these samples, it is possible to understand the composition and function of the microbial community. Mao et al. used the mNGS technology to analyze sputum samples from 29 patients with acute exacerbations of chronic obstructive pulmonary disease (AECOPD) of different phenotypes before treatment. The differences in the sputum microbiota between patients with emphysema phenotype (E), chronic bronchitis with emphysema phenotype (B + E), and asthma-COPD overlap phenotype (ACO) were analyzed, and the relationship between the sputum microbiota and clinical indicators during acute exacerbation was explored. This study found that inflammation caused by pathogenic bacterial colonization or dysbiosis plays a crucial role in the occurrence and development of COPD, guiding future research in changing patient prognosis by inhibiting specific bacterial groups associated with inflammation, thus guiding personalized treatment [24]. Yuan et al. performed mNGS analysis on BALF samples from 44 pneumoconiosis patients, comprehensively revealing the lung microbiota of patients with pneumoconiosis and comparing it with that of non-pneumoconiosis patients. The study confirmed a positive correlation between Mycobacterium avium complex and the duration of occupational exposure related to pneumoconiosis, preliminarily implicating microorganisms in pneumoconiosis. Additionally, the study identified different microbial species as potential biomarkers for pneumoconiosis, providing fundamental data for investigating the pathogenesis of pneumoconiosis [25].
3. Tumor CNV detection technology
CNVs are a type of structural variants (SV) within the genome, characterized by alterations in genomic material quantity resulting from either DNA loss or gain through deletion or duplication events, respectively [26,27]. The CNVs typically refers to genomic alterations that encompass a minimum of 100 basepairs and have been demonstrated to exert a significant impact on various human malignancies they are related to the development, invasion, and metastasis of tumors, such as breast, liver, kidney, and esophageal, cancers, among others [[28], [29], [30]]. Somatic CNVs are frequently acquired by tumor genomes during the development of cancer, and the amplification of oncogenes or the loss of tumor suppressor genes are commonly associated with pathogenicity, as a robust correlation exists between gene expression levels and their corresponding copy numbers [31]. CNV detection in cancer is significant in cancer diagnosis, prognosis, and treatment. According to the 2016 WHO classification system, the typical features of anaplastic oligodendroglioma are 1p/19q codeletion or IDH1/IDH2 mutation status [32]. The loss of TP gene copy number is recognized to be the marker of poor prognosis in various cancers [[33], [34], [35]]. Treatation-determining CNVs include ERBB2 amplification in breast cancer and MET amplification in lung adenocarcinoma [36]. The reliable and accurate detection technologies and characterization of CNVs in a given human genome are urgently needed to improve the efficiency of clinical diagnostics and treatment decision-making.
Several platforms for CNV detection in current clinical practice can be divided into cytogenetic detection and molecular detection. Cytogenetic detections include karyotype analysis and FISH [37,38]. Molecular detections include CNV microarrays (CMA) [39,40], NGS technologies, and droplet PCR (dPCR). Classical cytogenetic detections identify common and rare genome SV [41]. However, their low throughput and resolution limit their application to a few individuals and to particularly large structural differences (∼500 kb–5 mb) [42]. Therefore, more robust, high-resolution, and automatable methods are urgently needed.
In the early 2000s, microarray-based CNV analysis became the first-tier clinical method for diagnosing patients with cancer [43,44]. CMAs are typically used to identify small, unbalanced abnormalities or cryptic CNVs but detect neither mosaicism lower than 5–20 % or balanced chromosomal aberrations, nor decipher the orientation of duplicated segments or the location of inserted segments, and their resolution remains restricted to a few kilobases [45].
The advent of NGS technologies, which has brought revolutionary changes to the study of gene structure changes, has replaced microarray technology as a platform for SV discovery and genotyping [[46], [47], [48], [49], [50]]. NGS assays can cover both the entire genome and targeted regions of interest. The detection of CNVs utilizing NGS technology typically involves CNV-seq, WGS studies, and whole-exome sequencing (WES) studies. Four tools are frequently used for calling CNVs with NGS data: read-pair (RP), spilt-read (SR), read-depth (RD), and assembly (AS) [51]. NGS can be employed to comprehensively investigate genomic variations, encompassing single nucleotide variants (SNVs), small insertions and deletions (indels), structural alterations such as CNVs, gene fusions or chromosomal translocations, gene expression profiling, and DNA methylation analysis. Compared to clinical CNV detection, NGS methods are not limited or biased by a probe design and exhibit remarkably improved detection sensitivity and resolution of CNV. Despite these advantages, NGS presents a challenge for CNV calling due to their length, which, together with the complexity of the variant content and the lack of a gold standard for evaluating these algorithms, lead to wildly varying performance of callers [[52], [53], [54], [55], [56]].
Compared with FISH, CAM, and NGS, dPCR offers a potential improvement over existing testing methods by providing absolute quantitation of both the target and reference sequences [57]. However, for each CNV, dPCR requires the design of a new specific hydrolysis probe, which limits its routine use as a complementary tool for CNV detection [58]. Nevertheless, the development of CNV analysis technology has remarkably improved a myriad of fields, as described in Table 1.
Table 1.
Summary of the features of currently available methods for CNV detection.
| Platform | resolution | TAT(days) | Alterations | Analytical method | Advantage | Disadvantage | Ref |
|---|---|---|---|---|---|---|---|
| Karyotyping | 5~10 Mb | 2~21 | CNV, SV | Qualitative | A gold standard | Highly dependent on the expertise of technicians and cytogeneticists | [59] |
| FISH | 80–200 kb | 3~5 | CNV, SV | Qualitative | A gold standard that serves as a means of verification of known tumor sites | Expensive, customization limited by the types of genomic variation, depending on subjective assessment | [37,38] |
| CMA | 20–100 kb | 3~14 | CNV, LOH | Quantitative | Cost-effective, significant advantages in detecting genomic variations greater than 100 Kb, | Limited by the types of genomic variation and the use of hybridization-based assays in repeat-rich and duplicated regions | [40,60,61] |
| CNV-seq | ~100 kb | 5~14 | CNV, SV, LOH, SNV, Indel | Quantitative | High throughput, excellent compatibility, enabling the detection of large fragments of CNVs at a whole-genome scale. | Cannot detect ROH | [62] |
| WGS | ≥1 bp | 5~14 | CNV, SV, LOH, SNV, Indel | Quantitative | High throughput, comprehensive evaluation of all tumor variation information | Expensive, long turnaround time, heavy computational requirements | [63] |
| WES | ≥1 bp | 5~14 | CNV, SV, LOH, SNV, Indel | Quantitative | High throughput, cost-efficient, and analytic-effective manner | Cannot detect pathogenic CNVs located within non-coding regions associated with human disease | [64] |
| dPCR | ≥1 bp | 2~5 | CNV, SV, LOH, SNV, Indel | Absolute quantification | Higher accuracy and reproducibility | Limited or biased by a probe design | [65,66] |
Abbreviations: CNV, copy number variations; SV, structural variants; FISH, fluorescence in situ hybridization; CMA, CNV microarrays; LOH, loss of heterozygosity; ROH, runs of homozygosity; SNV, single nucleotide variants; WGS,whole genomic sequencing; WES, whole exome sequencing; dRCR, droplet PCR.
4. Progress of mNGS for pathogens and CNV analysis
4.1. Bioinformatic analysis for CNV in mNGS pipeline
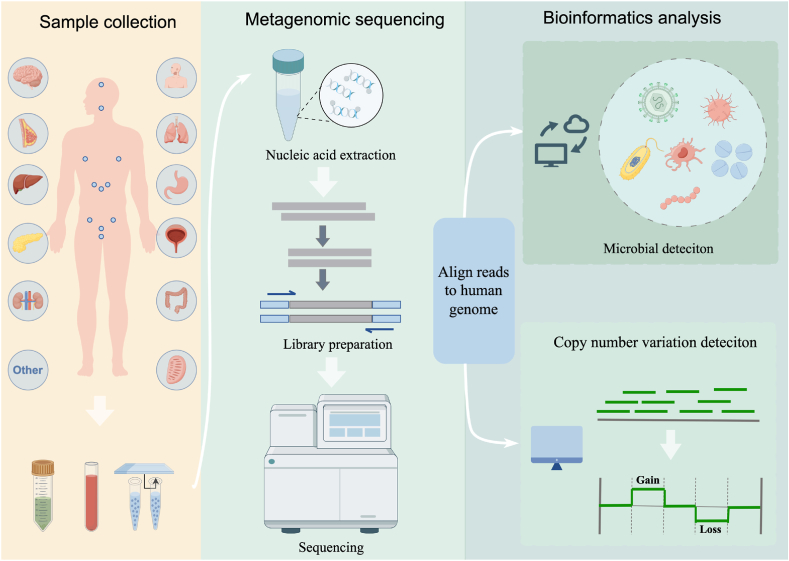
The workflow of mNGS used in clinical practice involves the wet-lab pipeline and dry-lab pipeline. The wet lab pipeline includes sample pretreatment, nucleic acid extraction, library construction, and sequencing. The process of dry-lab pipeline includes two components: pathogen detection pipeline and CNV detection pipeline. After sequencing, adapters and low-quality sequences are removed, and sequencing reads that are mapped to the human genome are used for CNV analysis, while the remaining reads that non-human reads are aligned to the microorganism genome database for pathogens identification. The basic procedure of mNGS is illustrated in Fig. 1.
Fig. 1.
Workflow of mNGS for pathogen detection and CNV analysis. Samples are subjected to DNA extraction, library preparation, and metagenomic sequencing. Following data quality control, they are aligned to the human genome for pathogens detection (upper) and CNV identification (lower).
The bioinformatic process for pathogens detection has been described in previous studies [[67], [68], [69]]. The obtained sequence data is proceeded using bioinformatics tools and algorithms. The sequence data is subjected to quality control, which involves trimming of reads to remove adaptors/adaptor trimming, quality filtering of reads, removal of low‐quality reads, removal of short reads, discarding reads shorter than 36 nucleotides, low‐complexity read filtering, and removal of duplicate reads. Subsequently, sequence alignments are referenced against the reference human reference genome (hg19, hg38), and only uniquely mapped sequencing reads are selected for subsequent CNV analysis, while other sequences are reserved for pathogen analysis. Unmapped human genome reads can be compared to various pathogen reference databases, including the National Center for Biotechnology Information (NCBI) nonredundant nucleotide sequence database (NT/NR), NCBI Reference Sequence (RefSeq) and genome database, NCBI GenBank database, and self-built microbial reference sequences or genome collections [70]. The approach of metagenomics classification, including the Kraken series represented by the k-mer algorithm includes Kraken, Bracken, KrakenUniq, Kraken 2, Centrifuge, and CLARK [[71], [72], [73]]. Finally, relevant potential pathogens are identified based on the clinical presentation and previous data results. Employing internal scripts for mapped data processing, conducting advanced data analysis including microbial diversity analysis, classification annotation, genome coverage/depth calculation, and abundance calculation.
The CNV analysis pipeline is well-established, leveraging data obtained from comparative analysis with the human genome sequence for CNV detection and interpretation. The commonly employed analytical approach involves utilizing the sliding window method to partition the genome into equidistant windows, wherein the sequencing depth is computed for each window, enabling the assessment of CNVs across chromosomal regions based on the distribution of sequencing depths. Adjusting the original sequencing depth requires the utilization of the GC content [74]. When employing a reading algorithm to screen for >10M CNVs, these chromosomal variants can be regarded as potential initiators of neoplasms [75].
4.2. Applications of mNGS for CNV analysis
With the growing incidence of human tumors, the search for accurate diagnostic methods and treatment strategies is crucial. Chromosomal instability is one of the characteristics of malignant tumors, and CNV is one of the manifestations of chromosomal instability, including amplifications or deletions of chromosomal segments. CNV plays a crucial role in the occurrence and development of tumors. mNGS technology provides a new approach to CNV detection. We searched reports with the keywords “mNGS and CNV, mNGS and neoplasms, mNGS and cancer, mNGS and chromosome” up to February 2024. The final review included 14 clinically relevant research articles. Of these, case reports (6/14) and observational studies (8/14) were predominant. We summarized these research findings, focusing on the application of mNGS technology in CNV detection in different types of tumors to evaluate the potential application of mNGS technology in CNV detection (Table 2).
Table 2.
Summary of research articles employing mNGS for CNV detection.
| Research type | Sample type | Cancer type | Number of participants for CNV analysis | Gold standard | mNGS-CNV Sensitivity | mNGS-CNV Specificity | Published data & Ref. |
|---|---|---|---|---|---|---|---|
| Retrospective | BALF, sputum, tissue, blood | Lung cancer | 23 | Clinical diagnosis/pathology | BALF 38.5 % Tissue 100 % Blood 100 % |
– | 2023 [77] |
| Retrospective | Pleural effusion | Malignant pleural effusion | 113 | Clinical diagnosis | 54.1 % | 80.8 % | 2023 [7] |
| Retrospective | Blood | Malignant diseases with EBV-positivity (lymphoma/nasopharyngeal carcinoma) | 29 | Clinical diagnosis | 90 % | 89.5 % | 2023 [78] |
| Retrospective (case-control study) | Pleural fluid/Peritoneal fluid/BALF/other body fluids | Clinical diagnosis of either malignancy | 124 | Cytology and/or flow cytometry testing | 87 % for cytology/cytometry-positive case 68% for cytology/cytometry-negative cases |
100 % for negative controls | 2021 [75] |
| Prospective (verification cohort) | Pleural/Peritoneal/BALF/Pericardial/fine needle aspirate | / | 81 | Cytology and/or flow cytometry testing | |||
| Prospective | Peripheral blood/BALF/CSF/others (e.g., sputum, pleural fluid, ascites, and pericardial effusion | Lung adenocarcinomas/hematological tumor/ | 140 | clinical diagnosis | 66.7 | 98.3 % | 2022 [76] |
| Retrospective (Test performance case-control study) | CSF | CNS malignant neoplasm | / | Clinical criterion standard (flow cytometry and/or cytologic testing) | 64 % | 100 % | 2021 [81] |
| Prospective (Neuroinflammatorydisease case-control study) | CSF | CNS malignant neoplasm | 29 | Clinical criterion standard (flow cytometry and/or cytologic testing) | 55 % | 100 % | |
| Retrospective | BALF | Lung cancer | 61.22 %(30/49) | 99.65 % | 2023 [80] | ||
| Descriptive study | CSF | Meningeal carcinomatosis | 10 | Cytology | 80 %(8/10) | / | 2023 [83] |
| Case report | Blood/CSF/biopsy/BLAF | Various cancers | 2024 [79] | ||||
| Case report | BALF | Lung squamous cell carcinoma | / | H&E staining and immunohistochemistry of lung biopsy | / | / | 2022 [84] |
| Case report | pericardial drainage fluid & pleural drainage fluid | T-lymphoblastic lymphoma | / | Bone marrow biopsy, immunohistochemistry, flow cytometry, PCR testing, and FISH | / | / | 2023 [85] |
| Case report | BALF | Pneumonic-type lung cancer | / | CT scans and pathology examinations | / | / | 2022 [86] |
| Case report | Plasma | Diffuse large B-cell lymphoma | / | Mandibular biopsy | / | / | 2021 [87] |
| Case report | Blood | Liver disease | / | Imaging and biochemical examination | / | / | 2023 [88] |
Abbreviations: mNGS, metagenomic next-generation sequencing; CNV, copy number variants; BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; EBV, Epstein-Barr virus; H&E staining, hematoxylin-eosin staining; PCR, polymerase chain reaction; FISH, Fluorescence in situ hybridization; CT, Computed Tomography.
Existing clinical results suggest that mNGS technology has been successfully applied to various types of samples (BALF fluid, blood, CSF, pleural effusion, tissue samples, etc.) for simultaneous pathogen and human tumor CNV detection. Su et al. conducted a multicenter, prospective case study in four hospitals in Shanghai, China, selecting patients with suspected infections between July 2019 and January 2020 [76]. Based on mNGS technology and CNV analysis, a new method called Onco-mNGS was developed as a diagnostic tool for the identification of both pathogens and malignant tumors in patient samples. Of the 140 patients tested by Onco-mNGS detection, 115 were diagnosed with infections, and 17 exhibited significantly aberrant CNV signals, indicative of the presence of malignant tumors in patients based on clinical confirmation. This preliminary evidence further substantiates the viability and clinical significance of Onco-mNGS technology in concurrently detecting potential pathogens and malignant tumors CNV among patients with suspected infections. Huang et al. conducted mNGS pathogen and CNV detection on 188 patients admitted between January 2020 and September 2022, including samples from BALF, sputum, lung biopsy tissues, peripheral blood, etc [77]. mNGS had higher sensitivity and accuracy in diagnosing pathogens than traditional culture and CMT. Additionally, 8 % (15/188) of patients were CNV positive. Of these 15 cases, 10 were initially misdiagnosed with non-neoplastic diseases, with a misdiagnosis rate of 66.7 % (10/15), indicating that CNV detection is an important auxiliary diagnostic tool for cancer, especially suitable for screening occult tumors. Moreover, mNGS has shown potential in the diagnosis of tuberculous pleurisy and malignant pleural effusion, differentiation between benign and malignant diseases in EBV-positive patients, and identification of infection and tumors in patients with unexplained fever [7,78,79].
A multicenter retrospective study using mNGS to detect chromosomal instability in lung cancer confirmed malignancy among 30 BALF samples with positive chromosomal instability, according to pathological results. The sensitivity, specificity, and accuracy of CNV detection for lung cancer by mNGS were 61.22 %, 99.65 %, and 83.17 %, respectively [80]. GUO et al. studied the clinical application of mNGS for the detection of chromosomal instability in lung biopsy tissue based on the principle that mNGS can identify host chromosomal instability and achieved a sensitivity, specificity, and accuracy of mNGS of 83.7 %, 97.6 %, and 92.9 %, respectively. This demonstrates the use of mNGS for tumor detection in lung biopsy tissue specimens for accurate and timely tumor diagnosis [6]. Further, in 2021, Gu et al. reported the application of mNGS-CNV technology in the diagnosis of neurogenic tumors and meningiomas, yielding a sensitivity of 75 % and specificity of 100 % in identifying neurogenic tumors. This suggests that mNGS-CNV can provide genetic evidence for central nervous system malignant tumors in patients with negative results and a low risk of false positives in CSF cytology and/or flow cytometry testing [81]. Similarly, other studies have confirmed the high positive rate (80 %, 8/10) of mNGS-CNV in the diagnosis of meningiomas, indicating that this method can serve as a diagnostic biomarker for meningiomas. In addition, mNGS technology can be combined with other methods such as CNV technology and ctDNA technology. By analyzing the genomic sequences in the samples using mNGS, CNVs can be accurately detected, providing a new non-invasive tumor diagnosis method for lung cancer brain metastasis patients [82].
In conclusion, the potential application of mNGS technology in tumor CNV detection has been preliminarily validated in different sample types. Through the analysis of genomic sequences in samples, mNGS demonstrates accurate detection of CNVs in chromosomes, offering significant support for tumor diagnosis and treatment.
4.3. Challenges in CNV analysis using mNGS
Aneuploidy and other large CNVs are prevalent in primary and metastatic tumors, with approximately 90 % of all malignant tumors harboring an abnormal number of chromosomes [89]. However, the most prevalent CNVs are between 1 kb and a few kb in size [90,91], whereas larger fragments of CNVs (>100 kb) are relatively rare. The highest sensitivity and resolution in CNV detection is achieved through deep-coverage paired-end WGS [40,92]. Several studies have also shown that short-insert (standard 350 bp) WGS can detect approximately 5-fold more CNVs (>1 kb) by ≥ 50 % reciprocal overlap, with lower sensitivity than mate-pair WGS [63].
Sequencing depth utilized for infection diagnosis reportedly varies from 2 to 25 million reads [[93], [94], [95]]. Despite the fact that human DNA reads constitute more than 90 % of the initial NGS reads, the data for CNV analysis is still relatively low. mNGS generates short fragments (50–120 bp), which decrease the sensitivity and resolution of assays for CNVs and result in false negatives for CNVs with small fragment variants. High-depth analysis results in higher rates of CNV detection with increasing coverage but increased costs. Emphasizing the importance of prospective clinical studies and economic data is essential to substantiate the cost-effectiveness of mNGS in CNV analysis and pathogen detection.
Notably, the detection of CNVs via mNGS merely implies the existence of genetic alterations associated with tumors in the specimen and cannot serve as diagnostic evidence for neoplasms; it only indicates potential malignancies. Moreover, this approach does not provide precise tumor localization. Therefore, further investigations are imperative to gain deeper insights into these aspects.
5. Conclusion
mNGS successfully facilitated the diagnosis of neuroleptospirosis in a 14-year-old boy, representing a significant milestone in the application of mNGS within the field of microbial detection [96]. mNGS is a high-throughput nucleic acid sequencing method that has been extensively validated for its rapid turnaround time, exceptional sensitivity, and remarkable specificity. Currently, mNGS is increasingly employed for unbiased pathogen detection in clinical samples from patients with diverse infectious diseases [[97], [98], [99]].
Chromosomal instability is a prevalent phenomenon in tumors, leading to SV during division. Various conventional testing methods, including karyotype analysis and flow cytometry, are employed in clinical practice to detect SVs; however, these techniques rely on the integrity of tumor cells, which limits their applicability. CNVs in tumors are characterized by the duplication or deletion of large segments or entire chromosomes. The presence of large CNVs (>10M) involving several chromosomes is indicative of somatic events within tumor cells rather than hereditary diseases [75]. When detecting pathogenic microorganisms in the samples of patients, it is necessary to pre-filter the host nucleic acid, resulting in the wastage of valuable genetic material. However, the host nucleic acid harbors significant information pertaining to non-infectious diseases. Under such circumstances, leveraging CNVs identified through human-derived data in mNGS can offer more comprehensive insights for clinical research on malignant diseases.
Clinicians often encounter challenges when diagnosing infections and tumors, as these cases frequently present with overlapping symptoms such as fever [100]. Additionally, some infection patients may exhibit elevated tumor markers, making the identification of the underlying cause of illness challenging. In the absence of timely and effective pathogen diagnosis, even cancer patients may receive empirical antibiotic treatment despite the absence of microbial infection, leading to inappropriate antibiotic usage [84]. Currently, a range of diagnostic methods are used in clinical practice for distinguishing between infections and tumors. However, these individual methods have limitations as they can only detect either infection or tumor. For instance, FISH can detect chromosomal aberrations but not the presence of infection-causing pathogens. Moreover, these approaches may be associated with high costs and long turnaround times [101]. In contrast, advanced diagnostic tools such as mNGS offer simultaneous detection of pathogens and tumors while providing rapid results within a few hours(<24 h) [102].
mNGS possesses the unique advantage of providing both pathogens and CNVs information in a single test. Although the results of mNGS detection for chromosomal CNVs cannot be directly used for definitive cancer diagnosis, they can prompt clinicians to conduct more targeted tumor diagnostic tests and increase the likelihood of tumor detection. In comparison to tumor-associated NGS testing, mNGS presents a potential practical tool with lower requirements for sample DNA input and faster turnaround time. Moreover, mNGS enables simultaneous pathogenic microbial analysis and CNV analysis without increasing costs or requiring additional sampling, thus alleviating the economic burden on patients.
In summary, this study emphasizes the remarkable sensitivity and efficiency of mNGS in detecting CNVs within clinical samples. Its ability to simultaneously identify pathogens and oncogenic CNVs underscores mNGS's transformative potential in the clinical diagnositc. By streamlining the diagnostic workflow and reducing the need for multiple tests, mNGS not only enhances the quality of clinical decision-making but also promises significant cost savings. Future research should focus on optimizing the application of mNGS technology in infectious and oncological diseases to further solidify its role in personalized medicine.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
This work was financially supported by the Key Research and Development Plan of Science and Technology Department of Jiangsu Provice(Project No. BE2023704) and the Project of MOE Key Laboratory of Geriatric Diseases and Immunology of Soochow University(Project No. KJS2403).
CRediT authorship contribution statement
Xiaofang Xie: Writing – original draft, Conceptualization. Xiaotong Xi: Writing – original draft, Conceptualization. Dan Zhao: Writing – original draft, Data curation. Yingyue Zhao: Writing – original draft, Data curation. Tiantian Yi: Project administration. Dongsheng Chen: Project administration. Rui Liu: Project administration. Lin Qi: Project administration. Zhen Pan: Validation. Hongqiu Wang: Validation. Haifang Zhang: Validation. Ran Ding: Writing – review & editing, Conceptualization. Hong Du: Writing – review & editing, Resources, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Ran Ding, Email: ran.ding@simceredx.com.
Hong Du, Email: hong_du@126.com.
References
- 1.Timsit Jean-François, et al. Bloodstream infections in critically ill patients: an expert statement. Intensive Care Med. 2020;46(2):266–284. doi: 10.1007/s00134-020-05950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.2 Consensus Group Of Experts On Application Of Metagenomic Next Generation Sequencing In The Pathogen Diagnosis In Clinical Moderate And Severe Infections et al. Zhonghua wei zhong bing ji jiu yi xue. 2020;32(5):531–536. doi: 10.3760/cma.j.cn121430-20200228-00095. [DOI] [PubMed] [Google Scholar]
- 3.Chinese Society of Infectious Diseases. Cytology Cerebrospinal Fluid. Expert consensus on clinical application of metagenomic next-generation sequencing of cerebrospinal fluid in the diagnosis of infectious diseases of the central nervous system. Chin. J. Neurol. 2021;54(12):1234–1240. doi: 10.3760/cma.j.cn113694-20210730-00532. [DOI] [Google Scholar]
- 4.Hanahan Douglas, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Torres-Ruiz Raul, et al. Detection of chromosome instability by interphase FISH in mouse and human tissues. STAR protocols. 28 Jun. 2021;2 doi: 10.1016/j.xpro.2021.100631. 3 100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Yifan, et al. Metagenomic next-generation sequencing to identify pathogens and cancer in lung biopsy tissue. EBioMedicine. 2021;73 doi: 10.1016/j.ebiom.2021.103639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Fudong, et al. Simultaneous diagnosis of tuberculous pleurisy and malignant pleural effusion using metagenomic next-generation sequencing (mNGS) J. Transl. Med. 2023;21(1):680. doi: 10.1186/s12967-023-04492-x. 30 Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu Charles Y., Steven A Miller. Clinical metagenomics. Nat. Rev. Genet. 2019;20(6):341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 10.Besser J., et al. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2018;24(4):335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virgin Herbert W., Todd John A. Metagenomics and personalized medicine. Cell. 2011;147(1):44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Donglai, et al. Challenges and considerations on quality control and evaluation of pathogen metagenomic next-generation sequencing. Chinese journal of biotechnology. 2020;36(12):2598–2609. doi: 10.13345/j.cjb.20037. [DOI] [PubMed] [Google Scholar]
- 13.Gu Wei, et al. Clinical metagenomic next-generation sequencing for pathogen detection. Annual review of pathology. 2019;14:319–338. doi: 10.1146/annurev-pathmechdis-012418-012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consensus Group Of Experts On Application Of Metagenomic Next Generation Sequencing In The Pathogen Diagnosis In Clinical Moderate And Severe Infections et al. Zhonghua wei zhong bing ji jiu yi xue. 2020;32(5):531–536. doi: 10.3760/cma.j.cn121430-20200228-00095. [DOI] [PubMed] [Google Scholar]
- 15.Duan Hongxia, et al. The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases. BMC Infect. Dis. 2021;21(1 62) doi: 10.1186/s12879-020-05746-5. 13 Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Cui-Lin, et al. Clinical metagenomic sequencing for diagnosis of pulmonary tuberculosis. J. Infect. 2020;81(4):567–574. doi: 10.1016/j.jinf.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Servellita Venice, et al. Adeno-associated virus type 2 in US children with acute severe hepatitis. Nature. 2023;617(7961):574–580. doi: 10.1038/s41586-023-05949-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Sike, et al. The application of metagenomic next-generation sequencing in pathogen diagnosis: a bibliometric analysis based on Web of Science. Front. Cell. Infect. Microbiol. 2023;13(3 Aug) doi: 10.3389/fcimb.2023.1112229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren Li-Li, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qasem Ahmad, et al. Coronavirus disease 2019 (COVID-19) diagnostic tools: a focus on detection technologies and limitations. Curr. Issues Mol. Biol. 20 Jul. 2021;43(2):728–748. doi: 10.3390/cimb43020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Bianli, et al. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog. 2011;7(11) doi: 10.1371/journal.ppat.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Fan, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Haocheng, et al. Metatranscriptomic characterization of coronavirus disease 2019 identified a host transcriptional classifier associated with immune signaling. Clin. Infect. Dis. : an official publication of the Infectious Diseases Society of America. 2021;73(3):376–385. doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Xiaoyan, et al. Analysis of sputum microbial flora in chronic obstructive pulmonary disease patients with different phenotypes during acute exacerbations. Microb. Pathog. 2023;184 doi: 10.1016/j.micpath.2023.106335. [DOI] [PubMed] [Google Scholar]
- 25.Yuan Xingya, et al. Application of mNGS in the study of pulmonary microbiome in pneumoconiosis complicated with pulmonary infection patients and exploration of potential biomarkers. Front. Cell. Infect. Microbiol. 2023;13(21 Jul) doi: 10.3389/fcimb.2023.1200157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stankiewicz Paweł, Lupski James R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Gao J., Ling C. Deletion genotype calling on the basis of sequence visualisation and image classification. Int. J. Data Min. Bioinf. 2018;20(2) doi: 10.1504/IJDMB.2018.093682. [DOI] [Google Scholar]
- 28.Park Richard W., et al. Identification of rare germline copy number variations over-represented in five human cancer types. Mol. Cancer. 2015;14 25(3 Feb) doi: 10.1186/s12943-015-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Yulin, et al. ABCC4 copy number variation is associated with susceptibility to esophageal squamous cell carcinoma. Carcinogenesis. 2014;35(9):1941–1950. doi: 10.1093/carcin/bgu043. [DOI] [PubMed] [Google Scholar]
- 30.Cai Lei, et al. DeepSV: accurate calling of genomic deletions from high-throughput sequencing data using deep convolutional neural network. BMC Bioinf. 2019;20(1 665) doi: 10.1186/s12859-019-3299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis Christina, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:7403 346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis David N., et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 33.Shah Vallari, et al. Subclonal TP53 copy number is associated with prognosis in multiple myeloma. Blood. 2018;132(23):2465–2469. doi: 10.1182/blood-2018-06-857250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Qian, et al. Proteogenomic characterization of small cell lung cancer identifies biological insights and subtype-specific therapeutic strategies. Cell. 2024;187(1):184–203.e28. doi: 10.1016/j.cell.2023.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Stengel A., et al. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31(3):705–711. doi: 10.1038/leu.2016.263. [DOI] [PubMed] [Google Scholar]
- 36.Dumbrava Ecaterina E Ileana, et al. Targeting ERBB2 (HER2) amplification identified by next-generation sequencing in patients with advanced or metastatic solid tumors beyond conventional indications. JCO precision oncology. 2019;3(18) doi: 10.1200/PO.18.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speicher Michael R., Carter Nigel P. The new cytogenetics: blurring the boundaries with molecular biology. Nat. Rev. Genet. 2005;6(10):782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 38.Schaaf Christian P., et al. Copy number and SNP arrays in clinical diagnostics. Annu. Rev. Genom. Hum. Genet. 2011;12:25–51. doi: 10.1146/annurev-genom-092010-110715. [DOI] [PubMed] [Google Scholar]
- 39.Kallioniemi A., et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science (New York, N.Y.) 1992;258(5083):818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 40.Alkan Can, et al. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trask B.J., et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum. Mol. Genet. 1998;7(13):2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- 42.Levy Brynn, et al. Optical genome mapping in acute myeloid leukemia: a multicenter evaluation. Blood advances. 2023;7(7):1297–1307. doi: 10.1182/bloodadvances.2022007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iafrate A John, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 44.McCarroll Steven A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40(10):1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 45.Duncavage Eric J., et al. Genomic profiling for clinical decision making in myeloid neoplasms and acute leukemia. Blood. 2022;140(21):2228–2247. doi: 10.1182/blood.2022015853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler David A., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 47.Bentley David R., et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKernan Kevin Judd, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19(9):1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills Ryan E., et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudmant Peter H., et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pirooznia Mehdi, et al. Whole-genome CNV analysis: advances in computational approaches. Front. Genet. 2015;6(13 Apr):138. doi: 10.3389/fgene.2015.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duan Junbo, et al. Comparative studies of copy number variation detection methods for next-generation sequencing technologies. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Legault Marc-André, et al. Comparison of sequencing based CNV discovery methods using monozygotic twin quartets. PLoS One. 2015;10(26 Mar) doi: 10.1371/journal.pone.0122287. 3 e0122287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kosugi Shunichi, et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome biology. 3 Jun. 2019;20(1):117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cameron Daniel L., et al. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 19 Jul. 2019;10(1):3240. doi: 10.1038/s41467-019-11146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Le, et al. Comprehensively benchmarking applications for detecting copy number variation. PLoS Comput. Biol. 28 May. 2019;15(5) doi: 10.1371/journal.pcbi.1007069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogelstein B., Kinzler K.W. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999;96(16):9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cassinari Kévin, et al. A simple, universal, and cost-efficient digital PCR method for the targeted analysis of copy number variations. Clin. Chem. 2019;65(9):1153–1160. doi: 10.1373/clinchem.2019.304246. [DOI] [PubMed] [Google Scholar]
- 59.Mantere Tuomo, et al. Optical genome mapping enables constitutional chromosomal aberration detection. Am. J. Hum. Genet. 2021;108(8):1409–1422. doi: 10.1016/j.ajhg.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michels Evi, et al. Detection of DNA copy number alterations in cancer by array comparative genomic hybridization. Genet. Med. : official journal of the American College of Medical Genetics. 2007;9(9):574–584. doi: 10.1097/gim.0b013e318145b25b. [DOI] [PubMed] [Google Scholar]
- 61.Lavrichenko Ksenia, et al. Comprehensive characterization of copy number variation (CNV) called from array, long- and short-read data. BMC Genom. 2021;22(1 826) doi: 10.1186/s12864-021-08082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Chao, Tammi Martti T. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinf. 2009;10(6 Mar):80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Bo, et al. Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. J. Med. Genet. 2018;55(11):735–743. doi: 10.1136/jmedgenet-2018-105272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyatake Satoko, et al. Detecting copy-number variations in whole-exome sequencing data using the eXome Hidden Markov Model: an 'exome-first' approach. J. Hum. Genet. 2015;60(4):175–182. doi: 10.1038/jhg.2014.124. [DOI] [PubMed] [Google Scholar]
- 65.Beck Julia, et al. Genome aberrations in canine mammary carcinomas and their detection in cell-free plasma DNA. PLoS One. 30 Sep. 2013;8(9) doi: 10.1371/journal.pone.0075485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Härmälä Suvi K., et al. Copy number variation analysis by droplet digital PCR. Methods Mol. Biol. 2017;1654:135–149. doi: 10.1007/978-1-4939-7231-9_9. [DOI] [PubMed] [Google Scholar]
- 67.Chinese Society of Laboratory Medicine Expert consensus on the standardized management of bioinformatics analysis for the detection of pathogenic microorganisms in mNGS. Chin J Lab Med. 2021;44(9):799–807. doi: 10.3760/cma.j.cn114452-20210322-00178. [DOI] [Google Scholar]
- 68.Yang Aimei, et al. Application of metagenomic next-generation sequencing (mNGS) using bronchoalveolar lavage fluid (BALF) in diagnosing pneumonia of children. Microbiol. Spectr. 2022;10(5) doi: 10.1128/spectrum.01488-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miao Qing, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin. Infect. Dis. : an official publication of the Infectious Diseases Society of America. 2018;67(suppl_2):S231–S240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 70.de Vries, Jutte J.C., et al. Recommendations for the introduction of metagenomic next-generation sequencing in clinical virology, part II: bioinformatic analysis and reporting. J. Clin. Virol. : the official publication of the Pan American Society for Clinical Virology. 2021;138 doi: 10.1016/j.jcv.2021.104812. [DOI] [PubMed] [Google Scholar]
- 71.Kim Daehwan, et al. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood Derrick E., Salzberg Steven L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome biology. 2014;15(3):R46. 3 Mar. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ounit Rachid, et al. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genom. 25 Mar. 2015;16(1 236) doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Biao, et al. Computational methods for detecting copy number variations in cancer genome using next generation sequencing: principles and challenges. Oncotarget. 2013;4(11):1868–1881. doi: 10.18632/oncotarget.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu Wei, et al. Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids. Genome Med. 2021;13(1 98. 1 Jun) doi: 10.1186/s13073-021-00912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su Jiachun, et al. Simultaneous detection of pathogens and tumors in patients with suspected infections by next-generation sequencing. Front. Cell. Infect. Microbiol. 2022;12(9 Jun) doi: 10.3389/fcimb.2022.892087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang Jinbao, et al. Clinical application and evaluation of metagenomic next-generation sequencing for lower respiratory tract infections and human tumor screening. Int. J. Gen. Med. 2023;16(7 Dec):5763–5777. doi: 10.2147/IJGM.S437800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song Jieyu, et al. Utility of clinical metagenomics in diagnosing malignancies in a cohort of patients with Epstein-Barr virus positivity. Front. Cell. Infect. Microbiol. 2023;13(22 Aug) doi: 10.3389/fcimb.2023.1211732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qin Feng, et al. Utility of metagenomic Next-Generation Sequencing for simultaneously detecting pathogens and neoplasms. Heliyon. 2024;10(10 Jan) doi: 10.1016/j.heliyon.2024.e24399. 2 e24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin Ping, et al. A multicenter-retrospective cohort study of chromosome instability in lung cancer: clinical characteristics and prognosis of patients harboring chromosomal instability detected by metagenomic next-generation sequencing. J. Thorac. Dis. 2023;15(1):112–122. doi: 10.21037/jtd-22-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu Wei, et al. Detection of neoplasms by metagenomic next-generation sequencing of cerebrospinal fluid. JAMA Neurol. 2021;78(11):1355–1366. doi: 10.1001/jamaneurol.2021.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hai tao Ren, Ke chi Fang, Fan Si yuan, et al. Diagnosis of meningeal carcinomatosis by copy number variation analysis based on cerebrospinal fluid metagenome next-generation sequencing. Chinese Medical Case Repository. 2022;4(1) doi: 10.3760/cma.j.cmcr.2022.e01291. [DOI] [Google Scholar]
- 83.Ren Hai tao. Detection of meningeal carcinomatosis by metagenomic next-generation sequencing and copy number variation analysis of cerebrospinal fluid. China Med. Abstr. 2023;40(3):190. [Google Scholar]
- 84.Wei Ping, et al. Diagnosis of lung squamous cell carcinoma based on metagenomic Next-Generation Sequencing. BMC Pulm. Med. 27 Mar. 2022;22(1):108. doi: 10.1186/s12890-022-01894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mo Lianqin, et al. T-lymphoblastic lymphoma in a child diagnosed by metagenomic sequencing: a case report. Oncol. Lett. 2023;26(1):289. doi: 10.3892/ol.2023.13875. 22 May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shui Yuexiang, Wang Huabin. Metagenomic next-generation sequencing as an unconventional approach to warn of tumor cells in a patients with non-mucinous pneumonic-type lung adenocarcinoma: case report. Medicine. 2022;101(51) doi: 10.1097/MD.0000000000032448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Kaili, et al. Diffuse large B-cell lymphoma of the mandible diagnosed by metagenomic sequencing: a case report. Front. Med. 2021;8(23 Dec) doi: 10.3389/fmed.2021.752523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Liuqing, et al. Case Report: Mycobacterium kansasii causing infective endocarditis explored by metagenomic next-generation sequencing. Front. Cell. Infect. Microbiol. 2023;13(22 Aug) doi: 10.3389/fcimb.2023.1227537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taylor Alison M., et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33(4):676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martínez-Jiménez Francisco, et al. Pan-cancer whole-genome comparison of primary and metastatic solid tumours. Nature. 2023;618(7964):333–341. doi: 10.1038/s41586-023-06054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beroukhim Rameen, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Hongbin, et al. Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin. Infect. Dis. : an official publication of the Infectious Diseases Society of America. 2020;71(Suppl 4):S416–S426. doi: 10.1093/cid/ciaa1516. [DOI] [PubMed] [Google Scholar]
- 94.Zinter Matt S., et al. Pulmonary metagenomic sequencing suggests missed infections in immunocompromised children. Clin. Infect. Dis. : an official publication of the Infectious Diseases Society of America. 2019;68(11):1847–1855. doi: 10.1093/cid/ciy802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takeuchi Suguru, et al. Metagenomic analysis using next-generation sequencing of pathogens in bronchoalveolar lavage fluid from pediatric patients with respiratory failure. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-49372-x. 9 Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilson Michael R., et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mei Jian, et al. Diagnostic role of mNGS in polymicrobial periprosthetic joint infection. J. Clin. Med. 24 Feb. 2023;12(5):1838. doi: 10.3390/jcm12051838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun Limin, et al. Clinical application and influencing factor analysis of metagenomic next-generation sequencing (mNGS) in ICU patients with sepsis. Front. Cell. Infect. Microbiol. 2022;12(13 Jul) doi: 10.3389/fcimb.2022.905132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qian Yi-Yi, et al. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front. Cell. Infect. Microbiol. 2021;10(26 Jan) doi: 10.3389/fcimb.2020.567615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Foggo Vanessa, Cavenagh Jamie. Malignant causes of fever of unknown origin. Clinical medicine (London, England) 2015;15(3):292–294. doi: 10.7861/clinmedicine.15-3-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fan H Christina, et al. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. U.S.A. 2008;105(42):16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han Dongsheng, et al. mNGS in clinical microbiology laboratories: on the road to maturity. Crit. Rev. Microbiol. 2019;45(5–6):668–685. doi: 10.1080/1040841X.2019.1681933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.