Abstract
Background
Ancient Chang’an in the Tang Dynasty (618–907 AD) was one of the world’s largest and most populated cities and acted as the eastern end of the world-famous Silk Road. However, little is known about the genetics of Chang’an people and whether the Western Regions-related gene flows have been prevalent in this cosmopolitan city.
Results
Here, we present seven genomes from Xingfulindai (XFLD) sites dating to the Tang Dynasty in Chang’an. We observed that four of seven XFLD individuals (XFLD_1) were genetically homogenous with the Late Neolithic Wadian, Pingliangtai, and Haojiatai populations from the middle reaches of the Yellow River Basin (YR_LN), with no genetic influence from the Western Eurasian or other non-Yellow River-related lineages. The remaining three XFLD individuals were a mixture of YR_LN-related ancestry and ~ 3–15% Western Eurasian-related ancestry. Mixtures of XFLD_1 and Western Eurasian-related ancestry drove the main gradient of genetic variation in northern and central Shaanxi Province today.
Conclusions
Our study underlined the widespread distribution of the YR_LN-related ancestry alongside the Silk Road within the territory of China during the historical era and provided direct evidence of trans-Eurasian communication in Chang’an from a genetic perspective.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-024-02068-9.
Keywords: Ancient Chang’an, East Asia, Ancient DNA, Tang Dynasty, Silk Road, Population history, Neolithic middle Yellow River-related ancestry, Western-Eastern admixture
Background
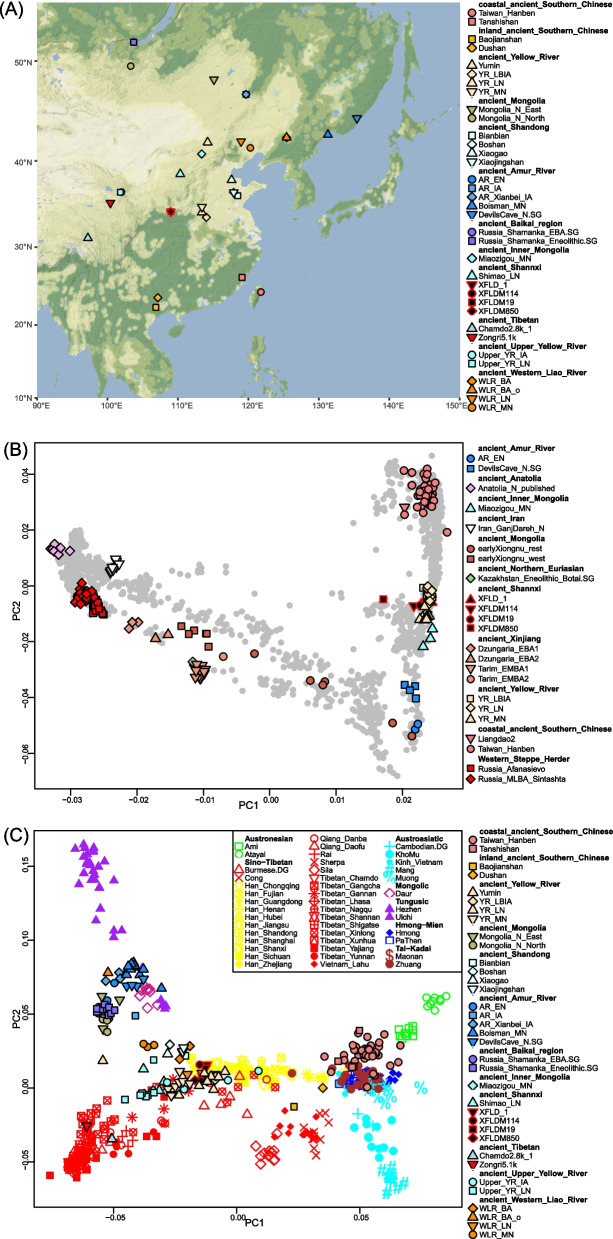
Xi’an (Fig. 1A), the capital of modern-day Shaanxi Province, is located in the central part of the Guanzhong Plain in the Yellow River Basin, adjacent to the Qinling Mountains to the south and the Wei River to the north. Thirteen feudal dynasties established the capitals in Xi’an (also known as Chang’an City) from the Zhou Dynasty (1111–221 BC) to the Tang Dynasty (618–907 AD). During the prosperous Tang Dynasty, Chang’an City grew into one of the largest urban centers of the ancient world. Its political and economic system greatly impacted later generations of China and even neighboring countries. Chang’an City in the Tang Dynasty also served as the eastern end of the Silk Road. This well-known ancient trade route linked China with Central Asia and Europe and also promoted the exchange of material culture and technology between the Tang Empire and the Western Regions.
Fig. 1.
Geographic location and population structure of newly sampled XFLD individuals. A The geographic location of newly sampled and published representative populations in East Asia. Our newly generated XFLD individuals were marked by black-filled shapes with red boundaries. B Principal components analysis (PCA) under Western and Eastern Eurasian backgrounds. The modern individuals are shown in light gray circles. See also Additional file 1: Fig. S2 for further details. C PCA under Eastern Eurasian background. The ancient individuals are projected on the PCs constructed by modern East Asian individuals, using the “lsqproject: YES” option
Recent ancient DNA studies documented that the expansion of Neolithic Yangshao culture-related ancestry from the middle reaches of the Yellow River (YR) left a genetic legacy to the ancients in the upper reaches of the Yellow River [1], Tibetan Plateau [2], and Xinjiang [3] to the west; the West Liao River basin, Inner Mongolia, and northern Shaanxi [1] to the north; inland southwest China and coastal southeast China to the south [4, 5]; and Japanese archipelago to the east [6]. However, the extent to which the expansion of middle YR-related ancestry influenced the genetic composition of Chang’an, the heart place of ancient China, remained unknown due to the lack of ancient genome data. Ancient DNA also revealed the genetic interaction between Western and Eastern Eurasian-related ancestry at Central and East Asian crossroads. In Xinjiang, the gene pool of Bronze Age people was formed by the different levels of admixture between local Xiaohe culture-related ancestry from Tarim Basin and non-local ancestries including Western Steppe herders, Chemurchek people with Bactria-Margiana Archaeology Complex (BMAC)-related ancestry, and Ancient Northeast Asian-related ancestry; the Iron Age and historical era Xinjiang people received additional gene flows from Central Asia and East Asia compared to the Bronze Age people [3]. In the westernmost part of Hexi Corridor, the passageway directly connected Central Plain and Xinjiang, all historical era individuals represented by Foyemiaowan_H were the direct descendants of the Late Neolithic middle YR-related ancestry (represented by YR_LN in [1]) except for two individuals dating to the Cao-Wei (220–266 AD) and Tang Dynasty (618–907 AD) with Eastern and Western Eurasian admixture profile (i.e., Foyemiaowan_Cao-Wei_o and Foyemiaowan_Tang_o) [7]. However, the genetic contribution of non-YR-related lineage was not detected in the central and eastern Hexi Corridor in the historical era (represented by Heishuiguo_H and Upper_YR_IA) [7]. Besides, historical records also indicated that non-Han people, such as Turkic, Xianbei, and Sogdian, lived in Chang’an during the Tang Dynasty [8]. There was an urgent need for ancient genome data to investigate the extent to which non-Han-related lineages had a genetic impact on the gene pool of the heart place of ancient China.
To date, the history of Chang’an has been extensively studied, but genomic studies of ancient Chang’an have been limited. To fill the gap, we screened 24 specimens from Xingfulindai sites (幸福林带, hereafter called XFLD), an archaeological site dating to the Tang Dynasty located in Xi’an City, Shaanxi Province, northern China (Fig. 1A). Five individuals were directly radiocarbon dated to between 904 and 1286 calibrated years before the present (cal BP; “BP” is before 1950 AD), an interval that is chronological of the Tang Dynasty (1043–1332 BP). We successfully obtained genome-wide ancient DNA data from 7 individuals. We aim to investigate (1) the genetic relationships of the XFLD people with the Neolithic middle Yellow River-related ancestry, (2) whether Central Asian and Western Eurasian-related ancestries extended to Chang’an, the heart place of the Tang Empire, and (3) how Tang Dynasty XFLD people impacted present-day populations.
Results
Ancient DNA authenticity
Human endogenous content was estimated to range from 3.37 to 77.62%. The sequence data from each genome showed nucleotide misincorporation patterns, indicative of postmortem damage (Additional file 1: Fig. S1). All individuals showed low levels of modern human contamination rates. After masking 9 bp on both ends of each read to reduce the impact of postmortem DNA damage on genotyping, the average depth of autosomal coverage was estimated between 0.05- and 21.5732-fold coverage for the ~ 1.2 million single nucleotide polymorphisms (SNPs) and 63,463 to 1,228,428 at ~ 1.2 million SNPs covered by at least one reads (Additional file 2: Table S1A). Our kinship analysis confirmed that all pairs of individuals were unrelated (Additional file 2: Table S1B). For population genetic analyses, we merged genotype data of the XFLD individuals with the previously published 1240 k dataset and Human Origin dataset (Additional file 2: Table S1C and D).
Characterization of the genetic profile of XFLD individuals
To understand the genetic characteristics of our newly generated seven XFLD individuals, we performed principal components analysis (PCA) based on the Human Origin dataset. We projected ancient individuals onto the first two PCs calculated by present-day Western and Eastern Eurasians (Fig. 1B and Additional file 1: Fig. S2) and present-day Eastern Eurasians only (Fig. 1C). Our studied XFLD individuals fell within the genetic variation of present-day East Asians. Specifically, all XFLD individuals clustered with middle Yellow River-related populations (represented by Late Neolithic Longshan culture people from Wadian, Pingliangtai, and Haojiatai sites (i.e., YR_LN) and Late Bronze to Iron Ages people (i.e., YR_LBIA)). The strong affinity between middle YR populations and XFLD was also supported by outgroup-f3 statistics, providing direct genetic evidence that the expansion of middle YR-related ancestry arrived at Chang’an at least during the Tang Dynasty (Additional file 2: Table S2A and Additional file 1: Fig. S3). The mtDNA lineages of all XFLD individuals were commonly found in present-day northern and southern East Asians (Additional file 2: Table S1A). The F1a1a haplogroup carried by one XFLD individual was more common in southeast Asia than in East Asia.
We also noticed that one XFLD individual (i.e., XFLDM850) shifted along PC1 towards Western Eurasian populations compared to the rest of the XFLD individuals (Fig. 1B and Additional file 1: Fig. S2). Quantitatively, we performed f4 (Yoruba, X; each XFLD individuals, YR_LN/YR_LBIA) on the 1240 k panel to test whether any ancient Eurasian population shared more affinity to XFLD than middle YR-related ancestry. To capture any marginal but detectable genetic influence from non-Han-related ancestry to XFLD, we set Z-score = 2 as the cutoff to label a statistical significance. We observed that three XFLD individuals (i.e., XFLDM850, XFLDM114, and XFLDM19) shared more alleles with some ancient Central Asian or Western Eurasian populations compared with YR_LN/YR_LBIA, i.e., f4 (Yoruba, some Central Asian/Western Eurasian; XFLDM850/XFLDM114/XFLDM19, YR_LN/YR_LBIA) < 0 (Additional file 2: Table S2B). We also observed some Western Eurasian-related signals when using transversion-only SNPs to calculate f4 (Additional file 2: Table S2C). It should be noted that the overlapped transversion SNPs on four populations in the formular of all f4 statistics were less than 50,000 (relatively limited), not all Western Eurasian-related signals based on all mutations were observed on the transversion SNPs. As expected, pairwise qpWave modeling supported the genetic heterogeneity between XFLDM850/XFLDM114/XFLDM19 and YR_LN/YR_LBIA (Additional file 2: Table S2D). The other four XFLD individuals (i.e., XFLDM114, XFLDM635, XFLDM682, and XFLDM764) formed a clade with YR_LN/YR_LBIA in pairwise qpWave analysis (Additional file 2: Table S2D). These four individuals were, therefore, merged into a single main cluster as XFLD_1. The robustness of 1-way YR_LN/YR_LBIA qpWave modeling for XFLD_1 was confirmed by adding the populations identified from our f4 statistics as being significantly closer to either XFLD_1 or YR_LN/YR_LBIA (i.e., |Z-scores|> 2 in f4 statistics in the form of f4 (Yoruba, X; XFLD individual, YR_LN/YR_LBIA)) (Additional file 2: Table S2G). F4 statistics and pairwise qpWave analysis also supported the relatively genetic homogeny of population pairs between XFLD_1 and neighboring Xianyang people from the Tang Dynasty (represented by Xianyang_Tang), as well as historical era Hexi Corridor populations (represented by Heishuiguo_H, Foyemiaowan_H, and Upper_YR_IA) (Additional file 2: Table S2E–G).
We used XFLD_1 and YR_LBIA as the proximal source for XFLDM850/XFLDM114/XFLDM19 according to the high degree of shared genetic drift between XFLDM850/XFLDM114/XFLDM19 and middle YR-related ancestry inferred by outgroup-f3 statistics (Additional file 2: Table S2A and Additional file 1: Fig. S3). Populations that shared more alleles with XFLDM850/XFLDM114/XFLDM19 than with middle YR-related ancestry inferred from f4 (Yoruba, X; XFLDM850/XFLDM114/XFLDM19, YR_LN/YR_LBIA) < 0 (Additional file 2: Table S2B) were used as the second source for XFLDM850/XFLDM114/XFLDM19 in turn (Additional file 2: Table S2G). We found that two-source admixture modeling performed with qpAdm successfully modeled XFLDM850/XFLDM114/XFLDM19 as the mixture of XFLD_1/YR_LN/YR_LBIA and minor Western Eurasian-related ancestry component (ranging from ~ 3 to 15%) (Fig. 2B and Additional file 2: Table S2H). XFLDM850 who carried ~ 15% Western Eurasian-related ancestry on autosomes also carried the Y chromosomal lineage observed in the present-day Middle East (i.e., E-Z21014* (E1b1b1b2a1a1a1a1f1b)). The Y lineage E1b1b1b2a1a1 ~ was also observed in ancient individuals from the Near East [9]. We then applied DATES to clarify the admixture time for XFLDM850/XFLDM114/XFLDM19 (Additional file 1: Fig. S4 and Additional file 2: Table S2I). We found that 6 out of 8 results supported that the admixture was estimated to have occurred ~ 10 generations before the time of XFLDM850 (around 329–617 AD when assuming 29 years per generation), corresponding to the period between the Sixteen States period (304–439 AD) and the Sui Dynasty (589–618 AD). The other two results for XFLDM850 suggested the admixture might have occurred 40 or 60 generations earlier, corresponding to the Warring States period (403–221 BC) and Western Zhou Dynasty (1111–770 BC), respectively. The admixture dates for XFLDM19 ranged from ~ 15 to 300 generations earlier, corresponding to the Neolithic to Eastern Han Dynasty (25–220 AD). The admixture dates for XFLDM114 ranged from ~ 27 to 157 generations earlier, corresponding to the Neolithic to Western Han Dynasty (206 BC–8 AD).
Fig. 2.
Ancestry profile of ancient northern Chinese inferred by qpAdm-based modeling for A Neolithic, B historical era (see Additional file 2: Table S2B and D for further details), and C present-day (see Additional file 2: Table S3D). YR_LN denoted Late Neolithic Longshan culture people from Wadian, Pingliangtai, and Haojiatai sites published in ref [1]. Southern Chinese-related ancestry was represented by Taiwan_Hanben. ANA-related ancestry was represented by Devils Cave people. Western Eurasian-related ancestry was represented by Kazakhstan_Wusun
XFLD’s contribution to modern-day Shaanxi Han Chinese
We further quantified the genetic contribution of XFLD to modern Han Chinese from the northern, central, and southern Shaanxi Province published in ref [10] (Additional file 1: Fig. S5A). In outgroup-f3 statistics (Additional file 2: Table S3A), we observed that XFLD shared the strongest genetic affinity with Han Chinese from Central China (represented by Han_Zhejiang/Han_Hubei/Han_Jiangsu) and Korean instead of Han Chinese from Xi’an (denoted as Han_Shannxi_Xian). The symmetric f4 statistics in the form of f4 (Yoruba, XFLD_1; Han Chinese i, Han Chinese j) where i and j were any pairs of Han Chinese populations in China also suggested that XFLD_1 shared more alleles with Han Chinese from Central China than with local Han Chinese (i.e., Han_Shannxi_Xian) (Additional file 2: Table S3B). Next, we examined whether present-day Shaanxi populations were the direct descendants of XFLD. Firstly, admixture-f3 statistics in the form of f3 (source1, source2; target) (Additional file 2: Table S3C) were used to evaluate the admixture signals in the targets. The statistically significant negative f3 values with Z-scores less than − 2 suggested that the target population might be an admixture of source1 and source2-related populations. We observed that Han Chinese from northern and central Shaanxi (i.e., Yulin, Yan’an, Xiangyang, Xian, Weinan, and Baoji) could be the mixture between XFLD_1 and Western Eurasians; southern Shaanxi (i.e., Hanzhong and Ankang) showed the signal between XFLD_1 and Southern East Asians. We further applied qpAdm to explore the plausible admixture models for each Shaanxi Han Chinese population. As the results of admixture-f3 statistics suggested, we found that Han Chinese populations from northern and Central Shaanxi were best modeled as the mixture between XFLD_1 and ~ 2–5% Western Eurasian-related ancestry (Additional file 1: Fig. S5B and Additional file 2: Table S3D). The admixture proportions of northern and Central Shaanxi were similar to XFLDM114 and XFLDM19. Han Chinese from southern Shaanxi could be described as the mixture between XFLD_1 and southern East Asian-related ancestry represented by Taiwan_Hanben. ALDER produced consistent estimates of admixture time ~ 1000–1500 years ago for Han Chinese from northern (represented by Han_Shannxi_Yulin) and central Shaanxi (represented by Han_Shannxi_Xian) when using modern-day Western Eurasians and Han Chinese from Central China as sources (two reference Z-score > 3 and p < 0.001) (Additional file 1: Fig. S6 and Additional file 2: Table S3E).
Discussion
To date, the ancient DNA study associated with Xi’an on Guanzhong Plain is limited to the DNA fragments of mitochondrial hypervariable control region from two individuals from the Neolithic Yangshao culture-related Banpo site [11]. However, limited by reference populations and methodological framework, the demographic history of Neolithic Xi’an remained unclear. Previous ancient genomic studies revealed distinguishable genetic profiles of Neolithic people neighboring Xi’an, i.e., northern Shaanxi in the north (represented by Shimao_LN), Henan in the east (represented by YR_LN and YR_Yangshaocun_Longshan), and the eastern area of Hexi Corridor in the west (represented by Upper_YR_LN) [1] (Fig. 2A). In northern Shaanxi and the eastern area of Hexi Corridor, the Late Neolithic of Shimao sites (labeled as Shimao_LN) and Qijia culture-related sites (labeled as Upper_YR_LN) were a mixture of Ancient Northeast Asian (labeled as ANA) and YR_MN [1]. In Henan, the Late Neolithic Longshan culture-related populations (labeled as YR_LN) from Pingliangtai, Wadian, and Haojiatai sites received additional Southern East Asian-related ancestry when compared to its preceding Middle Neolithic Yangshao populations from Xiaowu and Wanggou sites (labeled as YR_MN) [1]. Recently published Late Neolithic Longshan culture-related Yangshaocun genomes from Sanmenxia City in Henan (adjacent to the Xi’an City) displayed the Shimao_LN-related genetic profile [12]. We could assume Yangshaocun_Longshan people as the genetic profile of Late Neolithic Xi’an people. Yangshaocun_Longshan was not genetically homogenous with YR_LN: all Yangshaocun_Longshan individuals did not harbor additional Southern East Asian-related ancestry compared with YR_MN as YR_LN did.
However, in our study, we observed that the YR_LN-related genetic profile was widely distributed alongside the Silk Road within the territory of China during the historical era (Fig. 2B). Four of seven Tang Dynasty XFLD individuals (labeled as XFLD_1) could be modeled as the direct descendants of YR_LN/YR_LBIA without a significant contribution from non-YR lineages, including Xiongnu, Xianbei, and Western Regions-related lineages, who once resided in the heart place of ancient China according to the historical records [13–15]. The historical era populations from Xian’yang [16] and Hexi Corridor (represented by Heshuiguo_H, Foyemiaowan_H, and Upper_YR_IA) [7] were also genetically homogenous with XFLD_1 and YR_LN/YR_LBIA. These results suggested the strong westward expansion of YR_LN-related ancestry led to the genetic turnover in Shaanxi and Hexi Corridor, i.e., from Neolithic Shimao_LN/Yangshaocun_Longshan/Upper_YR_LN-related to historical era YR_LN-related ancestry.
We also detected three of seven XFLD individuals with marginal but detectable Western Eurasian-related ancestry (~ 3–15%) (Fig. 2B). It should be noted that numerous genetically distinguishable groups could be used as Western Eurasian-related sources for XFLDM850/XFLDM114/XFLDM19 in the qpAdm analysis. The limited genetic legacy of Western Eurasian-related ancestry in XFLDM850/XFLDM114/XFLDM19 made it more challenging to determine which kinds of Western Eurasian-related ancestry or population directly contributed to XFLDM850/XFLDM114/XFLDM19. The admixture time for XFLDM114/XFLDM19 was estimated to have occurred at the latest in the Han Dynasty (206 BC–220 AD). The admixture time for XFLDM850 was estimated to have occurred at the latest in the Sui Dynasty (589–618 AD). These results were consistent with the historical records about the marriage between Han and non-Han people in ancient China [13–15]. It should be noted that DATES software only considered a single pulse-like admixture. The actual population admixture may have been a continuous process. Therefore, the initiation of the period of frequent Western and Eastern Eurasian contacts in Chang’an may precede the admixture date estimated by DATES software. This is unsurprising because transcontinental material and cultural dissemination occurred early in the Bronze Age [17].
Present-day northern Han Chinese, such as Han_Shandong, Han_Henan, and Han_Shanxi, were also the direct descendants of XFLD_1 or YR_LN-related populations. However, the Han Chinese from Shaanxi had prevalently received low levels of Western Eurasian-related ancestry compared to XFLD_1 (Fig. 2C). The date of admixture in northern and central Shaanxi Han Chinese populations was estimated at 1000–1500 years ago by ALDER software, corresponding to the Sui and Tang Dynasties (589–907 AD) which were the heyday of the Silk Road trade exchange and had the frequent intermarriage between Han and non-Han [15]. Like DATES software, ALDER assumed a single-pulse admixture model. Therefore, the admixture event in northern and central Shaanxi Han Chinese populations occurred at the latest during the Sui and Tang Dynasties. It matched the observation that three Tang Dynasty XFLD individuals had already obtained a genetic profile similar to present-day central Shaanxi Han Chinese. Southern Shaanxi Han Chinese people received more Southern East Asian-related ancestry compared with XFLD_1, with no trace of Western Eurasian-related ancestry (Fig. 2C). Historically, today’s southern Shaanxi was not included in the jurisdiction of Shaanxi Province until the Yuan Dynasty (1279–1368 AD). Qinling Mountains, which stretch 400–500 km from east to west and 100–150 km from north to south, serve as the boundary between central and southern Shaanxi. The Qinling Mountains are also the natural boundary between the geology, geography, ecology, environment, climate, and even culture of the north and south of China. Therefore, it is unsurprising that Southern Shaanxi Han Chinese groups showed similar genetic profiles with Han Chinese from the Yangtze River delta, such as Han_Jiangsu and Han_Shanghai. The Qinling Mountains could be considered as a geographical barrier to gene flows of Western Eurasian-related ancestry southward and Southern East Asian-related ancestry northward.
We noted that our study was based on only a single site and seven ancient genomes and thus may not represent the full genetic diversity in Chang’an. Additional ancient genomic data from XFLD or other sites is needed to comprehensively clarify the genetic history of Chang’an City during the Tang Dynasty. Future archaeogenetic studies on human remains from earlier period sites in and around Shaanxi will contribute to a more comprehensive understanding of the interplay between ancient China and non-Han ancestry.
Conclusions
In our study, we investigated the demographic history of Capital Chang’an of the Tang Dynasty by analyzing seven ancient genomes from XFLD archaeological sites dating to the Tang Dynasty. We observed that XFLD individuals were categorized into two groups: one was the direct descendant of Neolithic middle YR-related people represented by Wadian, Pingliangtai, and Haojiatai (YR_LN), and another was the mixture between YR_LN and ~ 3–15% Western Eurasians. The present-day Han Chinese population residing in Chang’an tends to exhibit a predominantly middle YR-related ancestry, with approximately 3% of the genetic makeup being of Western Eurasian origin. These results provide new insights into the demography of the ancient Chang’an population and extend our understanding of the exchange and integration of genes and cultures between Eastern and Western Eurasians.
Methods and material
Archaeological information for Xingfulindai (幸福林带)
The specimens for this study are from the Xingfu Forest Belt Site in the eastern suburbs of Xi’an City, Shaanxi Province (Fig. 1A). In August 2017, the Xi’an Institute of Cultural Relics Protection and Archaeology conducted a salvage excavation of the site to cooperate with the construction project of the Xingfu Forest Belt in the eastern suburbs of Xi’an City. Through typological research on unearthed burial objects and epitaph records, it is reflected that the majority of this site was used by the Chang’an people in the mid-Tang Dynasty.
◾ XFLDM695: 1294–1223 cal BP
◾ XFLDM84: 982–904 cal BP
◾ XFLDM318: 1286–1180 cal BP
◾ XFLDM785: 1188–1063 cal BP
◾ XFLDM339: 1184–1052 cal BP
Ancient DNA extraction and sequencing
A total of 24 skeletal specimens were subjected to experimental analysis in this study. All experimental procedures were conducted in the specially designed ancient DNA cleanroom laboratory at Xiamen University [18–20]. We utilized 75% ethanol and 10% sodium hypochlorite (NaClO) to eliminate external contaminants. After surface abrasion and ultraviolet (UV) radiation treatment, we employed a drill bit to obtain inner core bone powder ranging from 130 to 230 mg. For each 100 mg of bone powder, we added 1 ml of 0.5 M EDTA and 1 μl of 20 mg/ml proteinase K, followed by incubation at 37 °C on a shaker at 300 rpm for 20 h. We used the MinElute System (Qiagen, Germany) to obtain concentrated DNA fragments. We prepared double-stranded libraries using the NEBNext® Ultra™ II DNA Library Prep Kit, paired with specially in-house designed sticky-end adaptors. We then sequenced the libraries on the DNBSEQ-T7 platform.
For samples XFLDM114 and XFLDM764, we followed the protocol from David Reich’s lab, employing Twist capture reagents for hybridization capture to enhance the acquisition of additional SNP loci [21]. Post-PCR clean-up throughout the entire experimental procedure was achieved using 1.8X Ampure beads (Beckman Coulter, USA) to eliminate non-target DNA fragments below 100 bp. We then sequenced the libraries on the Illumina Novaseq platform.
DNA sequence data processing
Using AdapterRemoval (version 2.3.2) [22], we removed adapters from both read pairs, trimmed bases at 5′/3′ termini with quality scores ≤ 20 and ambiguous bases (N) (–trimns –trimqualities –minquality 20), and collapsed forward and reverse reads (–collapse), and discarded reads shorter than 30 bp. Collapsed reads were aligned against the human reference genome hs37d5 (GRCh37 with decoy sequences) using the aln and samse modules in the Burrows-Wheeler Aligner (BWA) program (version 0.7.17-r1188) [23], with parameters “-l 1024” (seeding disabled) and “-n 0.01” (additional mismatches allowed). BAM files were sorted and indexed using Samtools (version 1.7) [24] before being utilized to remove PCR duplicates using dedup (version 0.12.8) [25]. BAM data were filtered with Samtools (version 1.7) [24] for a minimum Phred-scaled mapping quality score of 30.
Data quality control
We used a variety of methods to assess the ancient DNA’s authenticity. First, we used mapDamage (version 2.2.1) [26] to compute the postmortem damage pattern. We determined if each library had > 10% C-to-T misincorporations at 5′ termini and > 10% G-to-A misincorporations at 3′ termini, as is expected for double-stranded libraries. Second, we estimated the contamination rate. To calculate the mtDNA contamination rate, (1) we used the schmutzi.pl module in schmutzi software [27] and the contaminant database developed for Eurasian samples from share/schmutzi/alleleFreqMT/eurasian/freqs, and (2) we followed the pipeline posted in https://github.com/mnievesc/Ancient_mtDNA_Pipeline/blob/master/MT_DataAnalysis_Pipeline.sh to conduct contamMix [28]-related analysis. The X chromosomal contamination rate was calculated for males using the contamination module in ANGSD software (version 0.910) [29]. We used the HapMap resources for CHB (Han Chinese in Beijing) provided by ANGSD software to define X chromosomal polymorphic sites. We focused our investigation on the non-recombining part of the X chromosome (X:5,000,000–154,900,000). The library was treated as contaminated when the mtDNA contamination was greater than 3%, except when nuclear DNA contamination was assessed by the X chromosome < 3% for males.
Biological sex determination
An individual with one X chromosome and one Y chromosome was designated as “male,” whereas an individual with two X chromosomes was designated as “female.” Rx and Ry statistics [30, 31] were computed to estimate the biological sex of the samples based on shotgun data. The biological sex of samples based on 1240 K capture data was determined by the method described in reference [32]. This method calculated the ratio of the alignments to chromosome Y to chromosome autosomes, divided by the expected value of the quantity based on the number of SNPs in the relevant target set. The depth of coverage was calculated by the depth module of Samtools (version 1.7) [24]. is predicted to be approximately 0.5 for males and ~ 0 for females. Individuals with an observed value of > 0.3 are classified as males, whereas value of < 0.1 are classified as females, as described in ref [33].
Genotyping
We used the trimBam module in BamUtil (version 1.0.15) [34] to mask 9 bp from both ends to reduce the bias caused by ancient DNA deamination. We randomly picked one high-quality base (–q 30 and –Q 30) as a pseudohaploid using pileupcaller (https://github.com/stschiff/sequenceTools).
Estimation of genetic relatedness
We used the default parameters of the READ software [35] to estimate biological relatedness between each pair of individuals of XFLD.
Uniparental haplogroup assignment
We produced mitochondrial consensus sequences of quality ≥ 20 using the log2fasta tool in schmutzi software [27]. Y-chromosomal and mitochondrial haplogroups were assigned by Yleaf (version 3.1) [36] with “-r 1 -q 20 -b 90” option and Haplogrep (version 3) software [37], respectively.
Data merging
We merged XFLD samples with published genome-wide SNP data of present-day and ancient Eurasians using the mergeit function in EIGENSOFT software [38]. The information on co-analyzed ancient and modern genomes is listed in Additional file 2: Table S1C and D. Two datasets were used in our analysis: (1) We merged XFLD data with a 1240 k dataset curated by The Allen Ancient DNA Resource (AADR) [39], which covered ancient DNA data with maximum SNP sites (1,135,618). (2) We merged XFLD data with the Human Origin (HO) dataset curated by The Allen Ancient DNA Resource (AADR) [39], which covered ancient DNA data with overlapped SNP sites between 1240 k and HO SNP panel (593,120), as well as present-day Eurasians genotyped on Human Origin SNP chip. (3) We merged the published Shaanxi Han Chinese [10] genotyped on Illumina Bead Chip with the above 1240 k and HO datasets, resulting in 176,837 and 70,969 SNP sites, respectively. These datasets were denoted as the “1240 k-Illumina dataset” and “HO-Illumina dataset” only for testing the relationship between XFLD and present-day Shaanxi Han Chinese.
Principal components analysis (PCA)
We ran PCA on the HO dataset using the smartpca (version 16,000) algorithm in EIGENSOFT software [40] with the options “lsqproject: YES” and “numoutlieriter:0,” and “shrinkmode: YES.” We employed the “lsqproject: YES” option to project all ancient genomes onto the PC spaces calculated with the modern genomes.
F statistics
We applied qp3pop (version 651) from the ADMIXTOOLS software [38] with the option “inbred: YES” to calculate outgroup f3 in the form of f3 (A, B; Yoruba) and admixture-f3 (source1, source2; target). The outgroup f3 was used to quantify the shared genetic drift between A and B. The admixture-f3 was used to evaluate the admixture signals in the targets. We used qpDstat (version 980) from the ADMIXTOOLS software [38] with the option “f4mode: YES” to calculate f4 statistics in the form of f4 (Yoruba, A; B, C). The standard error (Std.err) was calculated with 5 cM block jackknifing implemented in the ADMIXTOOLS software [38].
Pairwise qpWave
We conducted pairwise qpWave analysis using the qpWave program (version 600) in ADMIXTOOLS software [38]. We chose the following populations as the base outgroups: Mbuti.DG, Loschbour.DG, Onge.DG, Iran_GanjDareh_N, Kazakhstan_Eneolithic_Botai.SG, Shandong_EN (pooled by Boshan, Xiaogao, Xiaojingshan, and Bianbian), Mongolia_N_East, Russia_Shamanka_Eneolithic.SG, Japan_Jomon, Fujian_LN (pooled by Tanshishan and Xitoucun), Anatolia_N_published, Upper_YR_LN. Rank = 0 models with p value > 0.01 suggested that the left populations were genetically homogeneous compared to the base outgroups; in other words, one stream of ancestry from a set of outgroup populations could explain the ancestry of left populations.
Admixture modeling using qpAdm
We modeled our genomes as an admixture of two source populations and estimated the proportions of ancestry using qpAdm (version 1000) in ADMIXTOOLS software [38], with settings “details: YES” and “allsnps: YES.” We chose the following populations as the base outgroups: Mbuti.DG, Loschbour.DG, Onge.DG, Iran_GanjDareh_N, Kazakhstan_Eneolithic_Botai.SG, Shandong_EN (pooled by Boshan, Xiaogao, Xiaojingshan, and Bianbian), Mongolia_N_East, Russia_Shamanka_Eneolithic.SG, Japan_Jomon, Fujian_LN (pooled by Tanshishan and Xitoucun), Anatolia_N_published. All the conditions listed below should be satisfied: (1) coefficient > 0 and coefficient ± Std.err > 0; (2) p value > 0.01; (3) p value for the nested model < 0.05.
Estimating the admixture dates
We applied the DATES algorithm (version: 753) [41] under a single pulse admixture model to date the admixture for a single ancient genome. We used the following parameters: “binsize: 0.001,” “maxdis: 1,” “runmode: 1,” “mincunt: 1,” and “lovaifit: 0.45.” We used ALDER [42] (version: 1.03) to date admixture events with default parameters for present-day groups. We assumed 29 years per generation [38].
Supplementary Information
Additional file 1: Fig. S1 The plot of terminal damage pattern for all the double-stranded libraries with no UDG treatment. All samples displayed the typical damage pattern at both ends was observed (5′ C > T and 3′ G > A). Fig. S2 Principal components analysis (PCA) of ancient and present-day Eurasians. We calculated PCs using present-day populations and projected ancient individuals onto the top two PCs. Present-day individuals were color-coded based on the language family they belonged to. Ancient individuals were marked by color-filled shapes with black boundaries. Our newly generated XFLD individuals were marked by black-filled shapes with red boundaries. Fig. S3 The genetic affinity of XFLD and reference populations measured by outgroup-f3 statistics. Here, we list the top 30 populations that share the highest amount of genetic drift with XFLD based on (A) the 1240 k dataset for ancient Eurasian reference populations, (B) the Human Origin dataset, and (C) “HO-Illumina” dataset for present-day Eurasian reference populations. Error bars show one standard deviation. The raw f3 results are listed in Additional file 2: Tables S2A and S3A. See “Data merging” in the“Methods and material” section for the details of the “1240 k dataset,”“Human Origin dataset,” and “HO-Illumina” dataset used in this study. Fig. S4 Genetic admixture dates for XFLDM850/XFLDM114/XFLDM19 estimated via DATES software. We report the results with Z-scores > 2. The error bars represented the ± 1 standard error calculated by the leave-one-chromosome-out jackknifing method. Fig. S5 The genetic contribution of XFLD for present-day Shaanxi Han Chinese. (A) Geographic distribution of previously published Han Chinese [9] from the northern Shaanxi (Yulin and Yan’an), central Shaanxi (Xiangyang, Xian, Weinan, and Baoji), and southern Shaanxi (Hanzhong and Ankang); (B) the qpAdm-based autosome modeling for each Shaanxi Han Chinese population. The error bars represented the ± 1 standard errors (SE) of estimated ancestry proportions. The raw qpAdm results are provided in Additional file 2: Table S3D. Fig. S6 ALDER estimated the dates of admixture events between Western Eurasian-related ancestry (represented by present-day Europeans/Central Asians) and Northern Han Chinese-related ancestry (present-day Han Chinese) for present-day central Shaanxi Han Chinese (represented by Han_Shannxi_Xian) and northern Shaanxi (represented by Han_Shannxi_Yulin). The estimated date in generation was converted into years with the assumption of 29 years per generation. Details are provided in Additional file 2: Table S3E.
Additional file 2: Tables S1, S2, and S3.
Acknowledgements
We sincerely thank the editors and reviewers for their contributions and suggestions. SF and ZX from the Information and Network Center of Xiamen University are acknowledged for their help with high-performance computing.
Abbreviations
- XFLD
Xingfulindai
- SNPs
Single nucleotide polymorphisms
- HO
Human Origin
- PCA
Principal components analysis
- N
Neolithic
- EN
Early Neolithic
- MN
Middle Neolithic
- LN
Late Neolithic
- BA
Bronze Age
- IA
Iron Age
- EMBA
Early and Middle Bronze Age
- LBIA
Late Bronze and Iron Ages
- SEA
Southeast Asian
- SC
Southern Chinese
- EA
East Asian
- YR
Yellow River
- WLR
West Liao River
- ANA
Ancient Northeast Asian
- cal BP
Calibrated years before present
- BMAC
Bactria-Margiana Archaeology Complex
- ANE
Ancient Northern Eurasian
Authors’ contribution
C.C.W. conceived and supervised the project. M.L., Y.Z., H.L., and X.Z. provided the materials and resources. H.M., W.Z., Y.Z., Z.Q., H.Z., Y.L., Y.L., L.T., and H.H. performed the wet laboratory work. R.W., X.Y., and K.Z. performed the genetic data analysis and prepared the figures. Y.Z. and H.M. performed the radiocarbon dating of ancient samples. R.W. and C.C.W. wrote and edited the manuscript. All authors contributed to the article and approved the final version for submission.
Funding
The work was funded by the National Natural Science Foundation of China (T2425014 and 32270667), the Natural Science Foundation of Fujian Province of China (2023J06013), the Major Project of the National Social Science Foundation of China (21&ZD285), Open Research Fund of State Key Laboratory of Genetic Engineering at Fudan University (SKLGE-2310), Open Research Fund of Forensic Genetics Key Laboratory of the Ministry of Public Security (2023FGKFKT07), and National Key Research and Development Program of China (2023YFC3303701-02).
Data availability
The BAM files reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA008730).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Minglei Lv, Hao Ma and Rui Wang contributed equally to this work.
Contributor Information
Rui Wang, Email: 17786126601@163.com.
Yawei Zhou, Email: zhouyawei469@163.com.
Chuan-Chao Wang, Email: wang@xmu.edu.cn.
References
- 1.Ning C, Li T, Wang K, Zhang F, Li T, Wu X, et al. Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat Commun. 2020;11:2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Yang MA, Wangdue S, Lu H, Chen H, Li L, et al. Human genetic history on the Tibetan Plateau in the past 5100 years. Sci Adv. 2023;9:eadd5582. [DOI] [PMC free article] [PubMed]
- 3.Kumar V, Wang W, Zhang J, Wang Y, Ruan Q, Yu J, et al. Bronze and Iron Age population movements underlie Xinjiang population history. Science. 2022;376:62–9. [DOI] [PubMed] [Google Scholar]
- 4.Yang MA, Fan X, Sun B, Chen C, Lang J, Ko Y-C, et al. Ancient DNA indicates human population shifts and admixture in northern and southern China. Science. 2020;369:282–8. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, Wang W, Xie G, Li Z, Fan X, Yang Q, et al. Human population history at the crossroads of East and Southeast Asia since 11,000 year ago. Cell. 2021;184:3829-41.e21. [DOI] [PubMed] [Google Scholar]
- 6.Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, et al. Ancient genomics reveals tripartite origins of Japanese populations. Sci Adv. 2021;7:eabh2419. [DOI] [PMC free article] [PubMed]
- 7.Xiong J, Wang R, Chen G, Yang Y, Du P, Meng H, et al. Inferring the demographic history of Hexi Corridor over the past two millennia from ancient genomes. Science Bulletin. 2024;69:606–11. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y. Tang Dynasty Chang’an and the Silk Road. Journal of Northwest University (Philosophy and Social Sciences Edition). 2015;1:30–2 (in Chinese). [Google Scholar]
- 9.Skourtanioti E, Erdal YS, Frangipane M, Balossi Restelli F, Yener KA, Pinnock F, et al. Genomic history of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell. 2020;181:1158-1175.e28. [DOI] [PubMed] [Google Scholar]
- 10.He G, Wang M, Li Y, Zou X, Yeh H, Tang R, et al. Fine-scale north-to-south genetic admixture profile in Shaanxi Han Chinese revealed by genome-wide demographic history reconstruction. J of Sytematics Evolution. 2022;60:955–72. [Google Scholar]
- 11.Lai X, Yang S, Tang X, Shi S, Li R, Yang H, et al. Preliminary study on ancient DNA of human remains from Yangshao culture. Earth Science-Journal of China University of Geoscience. 2004;29:15–9 (in Chinese). [Google Scholar]
- 12.Li S, Wang R, Ma H, Tu Z, Qiu L, et al. Ancient genomic time transect unravels the population dynamics of Neolithic middle Yellow River farmers. Science Bulletin. 2024. 10.1016/j.scib.2024.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Mao Y. Hu people from the Western Regions in the Yellow River Basin during the Northern Dynasties to the Sui and Tang Dynasties. Root exploration. 2006;2:35–41 (in Chinese) [Google Scholar]
- 14.Ma Y. Examination of Central Asians coming to China in the Late Eastern Han Dynasty. Journal of Xinjiang University (Philosophy and Social Sciences Edition). 1984;2:18–28 (in Chinese). [Google Scholar]
- 15.Fu Y. On the integration and complementarity between the Hu and Han ethnic groups in the Tang Dynasty. Journal of Shandong University (Philosophy and Social Sciences Edition). 1992;3:55–64 (in Chinese). [Google Scholar]
- 16.Zhao D, Chen Y, Xie G, Ma P, Wen Y, Zhang F, et al. A multidisciplinary study on the social customs of the Tang Empire in the Medieval Ages. PLoS ONE. 2023;18:e0288128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong G, Yang yishi, Liu X, Li H, Yifu C, Wang H, et al. Prehistoric trans-continental cultural exchange in the Hexi Corridor, northwest China. The Holocene. 2017;28.
- 18.Knapp M, Clarke AC, Horsburgh KA, Matisoo-Smith EA. Setting the stage – building and working in an ancient DNA laboratory. Annals of Anatomy - Anatomischer Anzeiger. 2012;194:3–6. [DOI] [PubMed] [Google Scholar]
- 19.Llamas B, Valverde G, Fehren-Schmitz L, Weyrich LS, Cooper A, Haak W. From the field to the laboratory: controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. STAR: Science & Technology of Archaeological Research. 2017;3:1–14.
- 20.Zhu K, He H, Tao L, Ma H, Yang X, Wang R, et al. Protocol for a comprehensive pipeline to study ancient human genomes. STAR Protoc. 2024;5:102985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohland N, Mallick S, Mah M, Maier R, Patterson N, Reich D. Three assays for in-solution enrichment of ancient human DNA at more than a million SNPs. Genome Res. 2022;32:2068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltzer A, Jäger G, Herbig A, Seitz A, Kniep C, Krause J, et al. EAGER: efficient ancient genome reconstruction. Genome Biol. 2016;17:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginolhac A, Rasmussen M, Gilbert MTP, Willerslev E, Orlando L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–5. [DOI] [PubMed] [Google Scholar]
- 27.Renaud G, Slon V, Duggan AT, Kelso J. Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Q, Mittnik A, Johnson PLF, Bos K, Lari M, Bollongino R, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23:553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci. 2013;40:4477–82. [Google Scholar]
- 31.Mittnik A, Wang C-C, Svoboda J, Krause J. A molecular approach to the sexing of the triple burial at the Upper Paleolithic site of Dolní Věstonice. PLoS ONE. 2016;11:e0163019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, et al. Genetic population structure of the Xiongnu Empire at imperial and local scales. Sci Adv. 2023;9:eadf3904. [DOI] [PMC free article] [PubMed]
- 34.Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monroy Kuhn JM, Jakobsson M, Günther T. Estimating genetic kin relationships in prehistoric populations. PLoS ONE. 2018;13: e0195491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralf A, Montiel González D, Zhong K, Kayser M. Yleaf: software for human Y-chromosomal haplogroup inference from next-generation sequencing data. Mol Biol Evol. 2018;35:1291–4. [DOI] [PubMed] [Google Scholar]
- 37.Schönherr S, Weissensteiner H, Kronenberg F, Forer L. Haplogrep 3 - an interactive haplogroup classification and analysis platform. Nucleic Acids Res. 2023;51:W263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallick S, Micco A, Mah M, Ringbauer H, Lazaridis I, Olalde I, et al. The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes. Sci Data. 2024;11:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chintalapati M, Patterson N, Moorjani P. The spatiotemporal patterns of major human admixture events during the European Holocene. eLife. 2022;11:e77625. [DOI] [PMC free article] [PubMed]
- 42.Loh P-R, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1 The plot of terminal damage pattern for all the double-stranded libraries with no UDG treatment. All samples displayed the typical damage pattern at both ends was observed (5′ C > T and 3′ G > A). Fig. S2 Principal components analysis (PCA) of ancient and present-day Eurasians. We calculated PCs using present-day populations and projected ancient individuals onto the top two PCs. Present-day individuals were color-coded based on the language family they belonged to. Ancient individuals were marked by color-filled shapes with black boundaries. Our newly generated XFLD individuals were marked by black-filled shapes with red boundaries. Fig. S3 The genetic affinity of XFLD and reference populations measured by outgroup-f3 statistics. Here, we list the top 30 populations that share the highest amount of genetic drift with XFLD based on (A) the 1240 k dataset for ancient Eurasian reference populations, (B) the Human Origin dataset, and (C) “HO-Illumina” dataset for present-day Eurasian reference populations. Error bars show one standard deviation. The raw f3 results are listed in Additional file 2: Tables S2A and S3A. See “Data merging” in the“Methods and material” section for the details of the “1240 k dataset,”“Human Origin dataset,” and “HO-Illumina” dataset used in this study. Fig. S4 Genetic admixture dates for XFLDM850/XFLDM114/XFLDM19 estimated via DATES software. We report the results with Z-scores > 2. The error bars represented the ± 1 standard error calculated by the leave-one-chromosome-out jackknifing method. Fig. S5 The genetic contribution of XFLD for present-day Shaanxi Han Chinese. (A) Geographic distribution of previously published Han Chinese [9] from the northern Shaanxi (Yulin and Yan’an), central Shaanxi (Xiangyang, Xian, Weinan, and Baoji), and southern Shaanxi (Hanzhong and Ankang); (B) the qpAdm-based autosome modeling for each Shaanxi Han Chinese population. The error bars represented the ± 1 standard errors (SE) of estimated ancestry proportions. The raw qpAdm results are provided in Additional file 2: Table S3D. Fig. S6 ALDER estimated the dates of admixture events between Western Eurasian-related ancestry (represented by present-day Europeans/Central Asians) and Northern Han Chinese-related ancestry (present-day Han Chinese) for present-day central Shaanxi Han Chinese (represented by Han_Shannxi_Xian) and northern Shaanxi (represented by Han_Shannxi_Yulin). The estimated date in generation was converted into years with the assumption of 29 years per generation. Details are provided in Additional file 2: Table S3E.
Additional file 2: Tables S1, S2, and S3.
Data Availability Statement
The BAM files reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA008730).