Abstract
Background
Glioblastoma is an aggressive cancer that originates from abnormal cell growth in the brain and requires metabolic reprogramming to support tumor growth. Metabolic reprogramming involves the upregulation of various metabolic pathways. Although the activation of specific metabolic pathways in glioblastoma cell lines has been documented, the comprehensive profile of metabolic reprogramming and the role of each pathway in glioblastoma tissues in patients remain elusive.
Methods
We analyzed 38 glioblastoma tissues. As a test set, we examined 20 tissues from Kyushu University Hospital, focusing on proteins related to several metabolic pathways, including glycolysis, the one-carbon cycle, glutaminolysis, and the mitochondrial tricarboxylic acid cycle. Subsequently, we analyzed an additional 18 glioblastoma tissues from Kagoshima University Hospital as a validation set. We also validated our findings using six cell lines, including U87, LN229, U373, T98G, and two patient-derived cells.
Results
The levels of mitochondria-related proteins (COX1, COX2, and DRP1) were correlated with each other and with glutaminolysis-related proteins (GLDH and GLS1). Conversely, their expression was inversely correlated with that of glycolytic proteins. Notably, inhibiting the glutaminolysis pathway in cell lines with high GLDH and GLS1 expression proved effective in suppressing tumor growth.
Conclusions
Our findings confirm that glioblastoma tissues can be categorized into glycolytic-dominant and mitochondrial-dominant types, as previously reported. The mitochondrial-dominant type is also glutaminolysis-dominant. Therefore, inhibiting the glutaminolysis pathway may be an effective treatment for mitochondrial-dominant glioblastoma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40170-024-00364-0.
Keywords: Glioblastoma, Glutaminolysis, Mitochondria, Metabolic changes
Background
Glioblastoma is a highly aggressive primary brain tumor with a poor prognosis [1]. Standard treatments for glioblastomas include surgery, radiotherapy, and chemotherapy [1]. However, there is currently no cure for this disease. Metabolic changes within glioma and overall cell metabolism represent potential therapeutic targets [2]. Specifically, mitochondria, key organelles in metabolism [3], play a critical role in oncogenesis and are promising targets for cancer treatment.
Cancer cells exhibit a highly proliferative capacity. Owing to limited metabolic resources, including nutrients, cancer cells undergo metabolic reprogramming to sustain growth [4, 5]. A defining hallmark of glioblastoma is its altered tumor metabolism [6]. Key metabolic activities, including aerobic glycolysis, mitochondrial oxidative phosphorylation, glutamine catabolism, macromolecular synthesis, and redox homeostasis, are essential for supporting the exponential growth of the tumor [4, 5]. Understating these processes could lead to the development of effective glioblastoma treatments. Notably, glycolysis and mitochondrial oxidative phosphorylation are the main metabolic pathways involved [6].
Two main types of brain tumors have been identified: glycolytic- and mitochondria-dominant gliomas [7]. Warburg et al. reported that cancer cells rely on glycolysis for energy even in the presence of oxygen [8]. Conversely, mitochondria generate energy via the tricarboxylic acid (TCA) cycle and are implicated in various diseases, including aging, diabetes, and cancer [9]. Notably, Hoang-Minh et al. [10] reported that slow-growing cells, which are prone to tumor invasion and chemoresistance, depend on mitochondrial activity [10–13]. Some mitochondrial pathways, such as one-carbon metabolism and glutaminolysis, are specific to cancer cells [14, 15]. Glutaminolysis not only generates energy and reactive oxygen species (ROS) [14, 16] but is also associated with resistance to radiotherapy and chemotherapy, making it a target for therapeutic strategies [14, 17]. Glutamate dehydrogenase (GLDH) converts glutamate to alpha-ketoglutarate (α-KG), which fuels the TCA cycle [18], and Zhang et al. reported that GLDH inhibitors decrease tumor proliferation [19]. One-carbon metabolism, critical for nucleic acid biosynthesis and linked with folate metabolism [20], is upregulated in cancer cells [15, 20, 21]. Kofuji et al. reported that the inactivation of IMPDH2, an enzyme vital for glioblastoma oncogenesis, is effective in treating glioblastoma [22]. However, a comprehensive understanding of aberrant metabolic pathways in glioblastoma remains lacking.
In the present study, we aimed to determine the pathways underlying glioblastoma development and explore the relationships between mitochondria-related and other pathway-related proteins using patient tumor specimens.
Methods
Cell culture
U87MG (U87), LN229, U373, and T98G cells were obtained from the American Type Culture Collection (Manassas, VA, USA) (certified by BEX [Japan]). The cells were cultured according to previously established protocols in Dulbecco’s modified Eagle’s medium (DMEM; Nacalai Tesque, Japan) containing 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin–streptomycin (Nacalai Tesque). The cells were cultured in a humidified incubator with 95% air and 5% CO2 at 37 °C. Additionally, two original patient-derived glioblastoma cell lines obtained from Kyushu University Brain Tumor Bank (KNS1435 and KNS1451) were suspended in DMEM/ham F12 (Nacalai Tesque) containing human FGF (R&D Systems, Minneapolis, MN, USA), EGF (R&D Systems), leukemia inhibitory factor (LIF; MilliporeSigma, Burlington, MA, USA), B27 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA USA), and penicillin/streptomycin and plated on a non-attached dish. Genetic analysis confirmed that KNS1451 harbors mutations in PTEN, TP53, NF1, and the TERT promoter C250T, whereas KNS1435 harbors mutations in PTEN, TP53, the TERT promoter C228T, PIK3R1, and NOTCH1 [23].
Brain tumor tissue
We analyzed samples collected from the tumor bank at Kyushu University Hospital and Kagoshima University Hospital. We randomly selected tumor samples histologically and genetically confirmed as glioblastoma, IDH-wild type, with sufficient specimen quantities. The cohort comprised 19 male patients and 18 female patients, with a mean age of 61.8 ± 4.2 years. Brain tumor samples were obtained during the surgery. A section of the tumor tissue was snap-frozen in liquid nitrogen and stored at -80 ℃. All study participants provided informed consent. This study was approved by a local ethics committee and conducted in accordance with the 1964 Declaration of Helsinki.
Next-generation sequencing (NGS)
NGS was performed using an amplicon-based glioma-tailored gene panel as described previously [24, 25]. Amplicon sequences were aligned to the human reference genome GRCh37 (hg19) in the target region of the sequence. Data were analyzed using the QIAGEN Web Portal service (QIAGEN, Hilden, Germany; https://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/) and Mitsubishi Space Software (Amagasaki, Hyogo, Japan; https://www.mss.co.jp/business/life-science/). We identified the copy number variations and single nucleotide polymorphisms, including mutations in PDGFR, NF1, PTEN, Tp53, and EGFR. Samples were classified into three groups: wild type (WT), EGFR amplification, and PDGFR amplification.
Reagents
The reagents used in the study are listed in Table 1.
Table 1.
Reagents used in the study
| Antibodies | Catalog number | Source |
|---|---|---|
| anti-COX1(MTCO1) mouse mAb | #ab14705 | abcam |
| anti-COX2(MTCO2) mouse mAb | #ab110258 | abcam |
| anti-DRP1 mouse | 611112 | BD Biosciences |
| anti-HK2 rabbit mAb | #2867 | CST |
| anti-SHMT2 rabbit pAb | #ab224427 | abcam |
| anti-GLS1 rabbit pAb | 29519-1-AP | ProteinTech |
| anti-GLDH1/2 rabbit mAb | #12793 | CST |
| anti-rabbit IgG HRP-linked | #7074 | CST |
| anti-mouse IgG HRP-linked | #7076 | CST |
| anti-β-actin mouse mAb | #A5441 | Sigma-Aldrich |
| anti-TFAM rabbit mAb | Our laboratory | |
| Anti-MTHFD1 rabbit pAb | 10794-1-AP | ProteinTech |
| Chemicals | ||
| R162 (GLDH1 inhibitor) | E1170 | Selleck.co.jp |
| BPTES (GLS1 inhibitor) | S7753 | Selleck.co.jp |
Immunoblotting analysis
Immunoblotting was performed as previously described [26]. Cells were homogenized in a lysis buffer (20 mM Tris–HCl, 2 mM EDTA, 150 mM NaCl, and 1% NP40; pH 7.5) containing protease (161–26,021; Fujifilm Wako, Japan) and phosphatase inhibitors (4906837001; Sigma-Aldrich). After sonication, the cell lysates were centrifuged at 20,000 × g for 5 min at 4 ℃. The tumor sample was quickly frozen in liquid nitrogen in the operating room and stored at − 80 °C. For tumor samples, approximately 20 mg was processed by adding RIPA buffer, mashing, sonication, and resting for 30 min at 4 °C. Following sonication, the cell lysates were centrifuged at 20,000 × g for 30 min at 4 °C, and the supernatants were analyzed. Equal amounts of protein (5 μg) were separated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Immobilon-P transfer membranes (MilliporeSigma). The membranes were blocked using Blocking One (Nacalai Tesque) and probed overnight with primary antibodies. The membranes were incubated with secondary antibodies for 1 h at room temperature. Proteins were detected using enhanced chemiluminescence (GE Healthcare, Chicago, IL, USA). Chemiluminescence was recorded and quantified using a charge-coupled device camera (LAS1000 plus). The antibodies used in this study are listed in Table 1. Membranes were cut prior to antibody hybridization. The expression of each protein was compared to that of the internal reference β-actin.
Cell viability
Cells were seeded (1 × 104 cells/well in a 24-well dish) in triplicates and cultured in DMEM (containing glucose) with different drug concentrations for 72 h. Cells were counted using the Coulter counter (Beckman Coulter, Brea, CA, USA).
Metabolome assay
Glioblastoma tissue metabolites were analyzed using liquid chromatography-tandem mass spectrometry (LC–MS/MS) based on reverse-phase ion-pair chromatography and hydrophilic interaction chromatography coupled with a triple quadrupole mass spectrometer, LC-MS-8040 (Shimadzu, Kyoto, Japan), as previously described [27]. Data normalization was performed by autoscaling and analyzed using PCA and sPLA-DA methods, utilizing MetaboAnalyst 5.0.
Quantification and statistical analysis
Regarding the correlation between each pathway, western blot experiments were conducted simultaneously to detect the expression of each protein. The protein expression data were then normalized using the expression of the housekeeping protein β-actin. Subsequently, the normalized values for each sample and protein were plotted, and the correlation values were calculated using the Spearman method. Data are presented as the mean ± standard deviation. Significant differences between groups were examined using a one-way analysis of variance or Student’s t-test using GraphPad Prism version 9 (GraphPad Prism Software Inc.) and JMP 15 software (SAS Institute Inc., Cary, NC, USA). Tumor specimens were analyzed using nonparametric tests, while cell experiments were analyzed using parametric tests. All experiments were repeated at least three times.
Image generation
The image was created using bioRender (https://www.biorender.com/).
Results
Analysis of various pathways using tumor samples
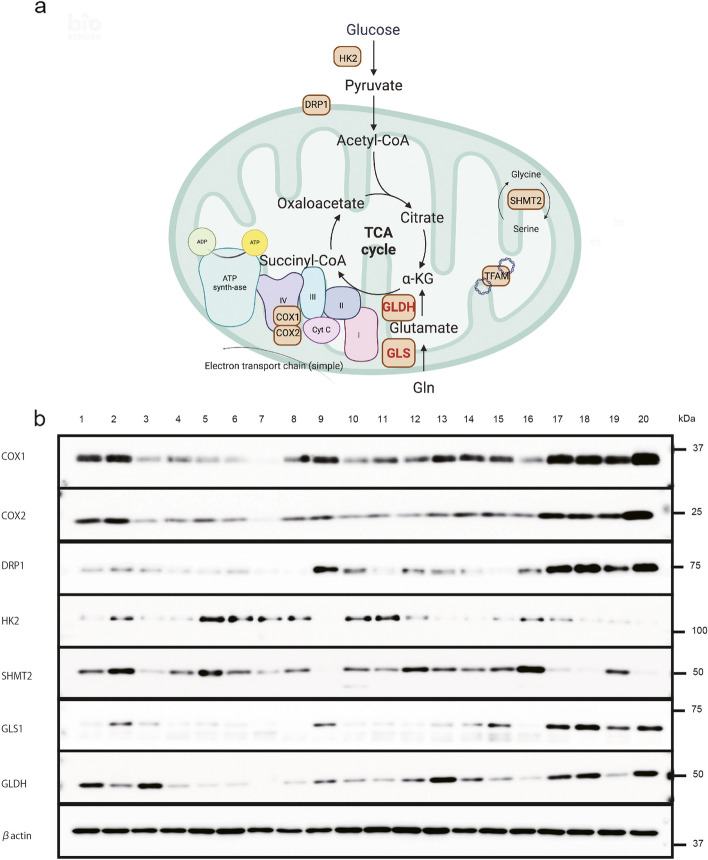
We initially selected 20 glioblastoma samples from Kyusyu University’s tumor bank as a test set, which included information on driver gene amplification, specifically EGFR and PDGFR. Patient characteristics are detailed in Additional file 1: Supplementary Table S2. The cohort comprised ten men and ten women, with a mean age of 58.8 ± 6.4 years. Among these patients, eight had PDGFR amplification (No. 2–9), five exhibited EGFR amplification (No. 1 and 10–12), and seven were WT (No. 13–16 and 18–20). We subsequently classified these samples into WT, EGFR amplification, and PDGFR amplification groups to examine the metabolic conditions of tumors relative to changes in driver gene amplifications. To further elucidate the metabolic changes in these glioma samples, we analyzed several metabolic pathways in the clinical glioma samples via western blotting (including the mitochondrial, glycolytic, one-carbon, and glutaminolytic pathways). The mitochondrial proteins analyzed included COX1, 2, DRP1, and TFAM; the glycolytic protein assessed was HK2; proteins related to the one-carbon pathway included serine hydroxymethyltransferase-2 (SHTM2) and methylenetetrahydrofolate dehydrogenase (MTHFD1); and proteins related to glutaminolysis included GLS1 and GLDH. Although no evident differences were observed between the driver gene amplifications, we noted that several metabolic proteins were correlated. Among these, mitochondrial proteins appeared particularly important. Thus, we redetected these proteins to investigate the correlation between different pathways. Sample No. 1 served as the control sample in both experiments, given its ample specimen availability. We determined the order of samples chronologically and by the expression level of COX1. We selected this order to illustrate a trend of increasing COX1 expression, facilitating a straightforward interpretation of the data. Additionally, we assessed the correlation of proteins from different pathways (Fig. 1; Additional file 1: Supplementary Fig. S1, and Additional file 2: Supplementary Table S1). All membranes were processed in the same experiments but are presented separately for clarity.
Fig. 1.
Protein expression of the metabolic pathway of tumor tissue. a Schematic representation of the study showing the relationship between each enzyme and mitochondria. b Expression of proteins of the metabolic pathway in tumor tissue, including COX1, COX2, DRP1, HK2, SHMT2, GLS1, GLDH, and β-actin
Interrelationships among mitochondria-related proteins
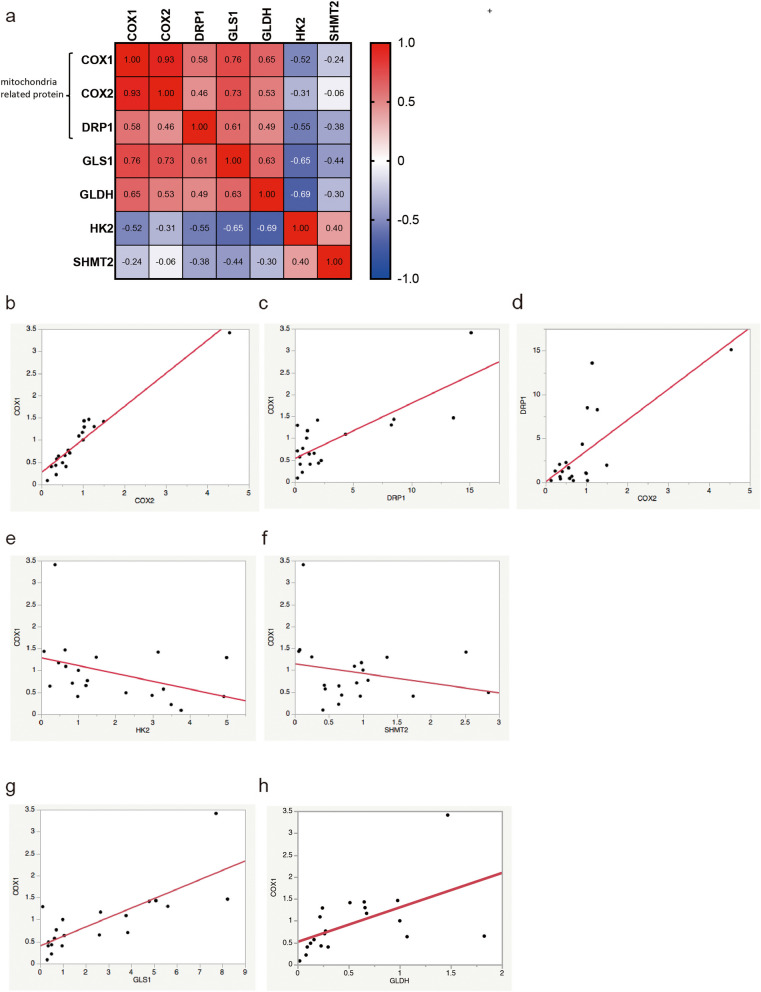
There are several types of proteins related to mitochondria, some of which are encoded by mitochondrial DNA, while others are encoded by nuclear DNA. COX1 and COX2 are encoded by mitochondrial DNA and are subunits of complex IV of the electron transport chain. DRP1, located in the outer mitochondrial membrane, plays a role in mitochondrial division. We plotted the normalized values for each sample and protein using β-actin as a reference and then calculated the correlation between the values of each protein using the Spearman method. The heatmap was generated using Prism software. We observed significant correlations among these proteins (COX1 and COX2, r = 0.93; COX1 and DRP1, r = 0.56; COX2 and DRP1, r = 0.46). Generally, |r| values exceeding 0.4 indicate modest to strong correlations [28]. Thus, the results suggest that the expression levels of COX1, COX2, and DRP1 are reliable indicators of overall mitochondria-related protein expression (Fig. 2a–d). However, TFAM, which protects mitochondrial DNA, did not show remarkable correlations with these mitochondrial proteins (Additional file 1: Supplementary Fig. S1a, b).
Fig. 2.
Interaction between mitochondria and other pathway-related proteins. Mitochondria-related protein expression levels are correlated. a Correlation coefficient between each protein. The correlation values (r) are calculated using the Spearman method using GraphPad Prism version 9. b COX1 expression correlates with COX2 expression (r = 0.93). c COX1 expression correlates with DRP1 expression (r = 0.56). d COX2 expression correlates with DRP1 expression (r = 0.46). Mitochondria-related protein expression levels are inversely correlated with glycolytic protein expression levels. e COX1 expression tends to be inversely correlated with that of HK2 (r = -0.52). Mitochondria-related protein expression is not correlated with that of one-carbon metabolism proteins. f COX1 expression is not correlated with that of SHMT2 (r = -0.24). Mitochondria-related protein expression is correlated with that of glutaminolysis-related proteins. g COX1 expression is correlated with GLS1 (r = 0.76). h COX1 expression is correlated with GLDH (r = 0.65)
Mitochondrial-related protein levels are inversely correlated with glycolytic protein levels
A low expression of glycolytic proteins was observed among samples exhibiting high levels of mitochondrial proteins. Therefore, we speculated that the levels of mitochondrial proteins, especially mitochondrial DNA-coded proteins, may be associated with the activity of the TCA pathway. Specifically, an increase in the levels of these mitochondria-related proteins may be related to a decrease in the glycolytic pathway. As expected, the expression of HK2 expression, a representative protein of the glycolytic pathway, tended to be inversely correlated with mitochondrial proteins (COX1 and HK2, r = -0.52) (Fig. 2a, e; and Additional file 1: Supplementary Fig. S2a).
Mitochondrial proteins are not associated with one-carbon metabolism
Several pathways, including those involved in one-carbon metabolism and glutaminolysis, are specific to cancer cells. Therefore, we examined the relationship between mitochondria and one-carbon metabolism. Two of the primary enzymes involved in carbon metabolism are SHMT2 and MTHFD1. Our results revealed no correlation between the expression of mitochondrial-related proteins and that of one-carbon-related proteins (Fig. 2a, f; and Additional file 1: Supplementary Fig. S1c).
Mitochondrial proteins are associated with glutaminolysis
The glutaminolysis pathway can produce α-KG, which is a metabolite of the TCA cycle. Glutaminolysis plays an essential role in energy production [14, 16], and the main enzymes involved are GLDH and GLS-1 [29]. Hence, we examined the correlation between mitochondria-related proteins and glutaminolysis-related proteins. Notably, the levels of mitochondria-related proteins were correlated with glutaminolysis (Fig. 2g, h; and Additional file 1: Supplementary Fig. S2b, c).
Validation of results using a validation sample set from other institutions
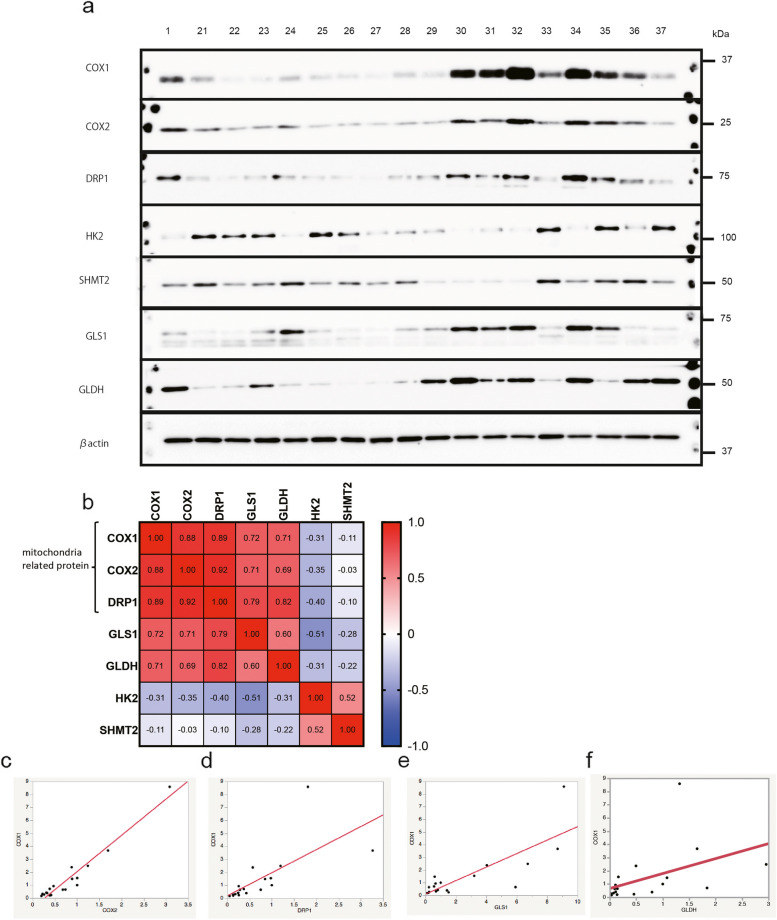
We conducted additional experiments on specimens obtained as a validation set from the Kagoshima University tumor bank. This group consisted of eight men and nine women, with a mean age of 65 ± 10.2 years. Among them, six had PDGFR amplification (No. 21–24, 30, and 35), five had EGFR amplification (No. 25, 26, 31, 32, and 36), and six were WT (No. 27–29, 33, 34, and 37). Sample No. 1 presented in Fig. 3a is the same control sample from Kyushu University, which was used to compare both test and validation sets. The sample sequence was organized chronologically and by the expression level of COX1, ensuring a clear trend of increasing COX1 expression. The results corroborated our initial findings, revealing correlations among the levels of mitochondria-related proteins and the glutaminolysis pathway (Fig. 3a–f; and Additional file 2: Supplementary Tables S3, 4b).
Fig. 3.
Protein expression of the metabolic pathway of tumor tissues (used as a validation set). Expression of mitochondria-related proteins is correlated with each other and with that of glutaminolysis proteins. a Proteins associated with metabolic pathways in tumor tissues include COX1, COX2, DRP1, HK2, SHMT2, GLS1, GLDH, and actin. b Correlation coefficient between each protein. The correlation values (r) are calculated using the Spearman method using GraphPad Prism version 9. c COX1 expression correlates with COX2 expression (r = 0.88). d COX1 expression correlates with DRP1 expression (r = 0.89). e COX1 expression correlates with GLS1 (r = 0.72). f COX1 expression tends to correlate with GLDH (r = 0.71)
Mitochondrial-dominant tissues rely on the TCA cycle
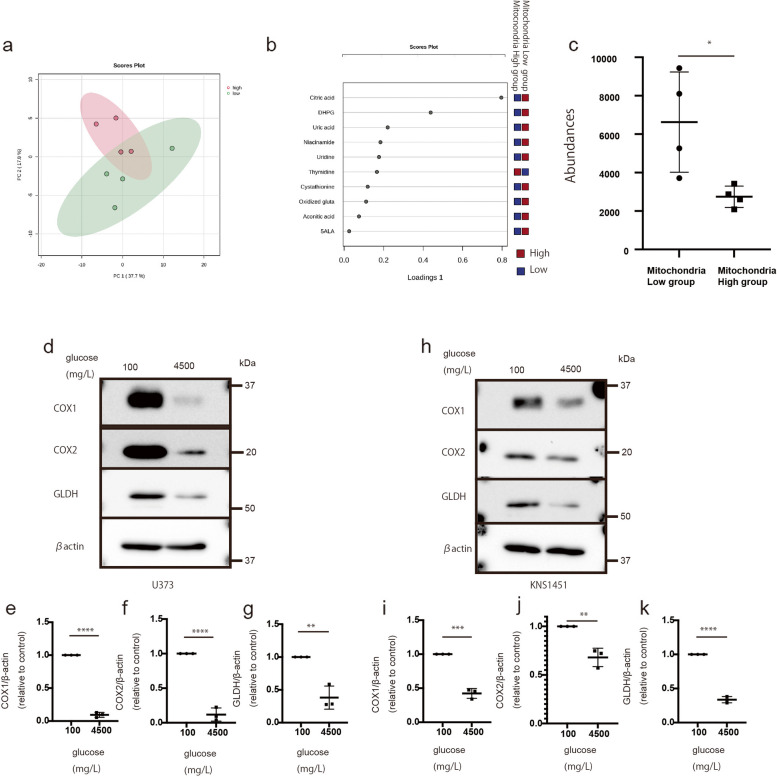
There are two types of tumor cells: glycolytic and mitochondrial [7, 10]. Cells with active mitochondrial function require glutamine metabolism [18]. We hypothesized that mitochondrial-dominant cells are dependent on the TCA cycle and utilize nutrients from glutaminolysis. To confirm our hypothesis, we use MS analysis to compare samples with high expression of mitochondria-related proteins (mitochondria-high expression group; No. 3, 4, 10, and 11) and samples with low expression of mitochondria-related proteins (mitochondria-low expression group) (No. 16–20) (Additional file 2: Supplementary Table S4a.) The data were analyzed using the PCA and sPLA-DA methods with MetaboAnalyst 5.0. PCA method revealed differences between the two groups, while the sPLA-DA method identified citric acid as one of the most distinguishing factors between them (Fig. 4a and b). Notably, citric acid levels were low in the samples with high expression of mitochondria-related proteins, suggesting that citric acid is consumed by these tissues (Fig. 4c). These results suggest that tumor tissues with high expression of mitochondria-related proteins are dependent on TCA cycle activity for growth.
Fig. 4.
Mass spectrometry analysis of the tumor sample comparing the correlation of glycolytic- and mitochondria-type tissue proteins with glutaminolysis protein levels under glucose starvation. a Schema of the differences between the sample with high expression of mitochondria-related proteins and the sample with low expression of mitochondria-related proteins using the PCA method. b One of the main metabolome compounds distinguishing between glycolytic- and mitochondria-type tissue using the sPLS-DA method. c Quantitative value of citric acid levels in each group (P < 0.05). Values are presented as the mean ± SD. The Mann–Whitney test was performed for comparison between the low group vs. the high group (*P < 0.05). d–k Expression of mitochondrial and glutaminolysis proteins under different glucose concentrations. d expression of COX1, COX2, GLDH, and actin in U373. e–g Quantification of COX1 (e), COX2 (f), and GLDH (g), normalized to the expression of β-actin (h). Expression of COX1, COX2, GLDH, and actin in KNS1451. i–k Quantification of COX1 (i), COX2 (j), and GLDH (k), normalized to the expression of β-actin. Values are presented as the mean ± SD. A Student’s t-test was performed to compare conditions of 100 vs. 4,500 mg/L of glucose. **P < 0.01; ***P < 0.001 ****P < 0.0001
Mitochondrial proteins are associated with glutaminolysis in cell lines
We further explored whether tissues with high expression of mitochondria-related proteins rely on mitochondrial energy production. We investigated this hypothesis using traditional cell lines, including U87 and U373, and patient-derived stem-like cells, including KNS1435 and KNS1451. We examined the change in glutaminolysis-related protein expression under glucose-starved conditions because mitochondrial activity and mitochondrial translation-related proteins typically increase under glucose-starved conditions [26]. As expected, we observed elevated levels of mitochondrial proteins, specifically COX1 and 2, across all cells under glucose-starved conditions (Fig. 4d-k; and Additional file 1: Supplementary Fig. S3a-g). Notably, under the same conditions, all cells exhibited elevated levels of GLDH, a protein related to glutaminolysis.
Patient characteristics and glutaminolysis protein expression
We investigated the relationship between the expression of glutaminolysis-related proteins and patient characteristics, including sex, age, gene amplification, and OS. However, given the limited number of patients, we could not identify the specific characteristics of patients with high expression of glutaminolysis-related proteins.
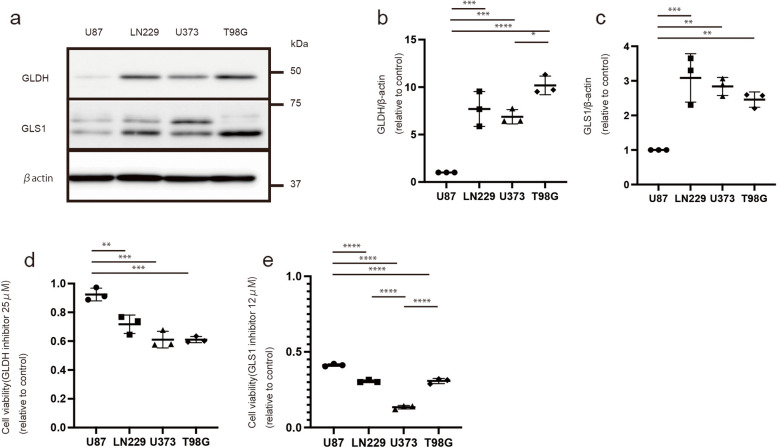
A GLDH inhibitor is effective against cells with high GLDH expression
We also examined the expression of mitochondria-related and glutaminolysis-related proteins in various cell lines, including U87, LN229, U373, and T98G cells. Although batch-to-batch differences in the expression of COX1 and COX2 were observed as cells were harvested in separate dishes, the expression levels of GLDH and GLS1 increased in LN229, U373, and T98G cells with high reproducibility (Fig. 5a–c, Additional file 1: Supplementary Fig. S4a–d; these experiments were simultaneously conducted). Subsequently, we tested the anti-tumor efficacy of treatment with 25 μM GLDH (R162) and 12 μM GLS1 inhibitors (BPTES) for 72 h. These inhibitors demonstrated more pronounced effects in LN229, U373, and T98G cells than in U87 cells (Fig. 5d, e). These results indicate that GLDH and GLS1 inhibitors may be effective for treating tumors with elevated glutaminolysis.
Fig. 5.
Expression of glutaminolysis proteins (GLDH and GLS1) and the effectiveness of their inhibitors (GLDH and GLS1 inhibitors) in each cell line. a GLDH and GLS1 expressions in U87, LN229, U373, and T98G. b Quantification of GLDH expression, normalized to the expression of β-actin. c Quantification of GLS1 expression, normalized to the expression of β-actin. d Cell number (relative to control) following the administration of the 25 μM GLDH(R162) inhibitor for 72 h. e Cell number (relative to control) following the administration of the 12 μM GLS1(BPTES) inhibitor for 72 h. Values are presented as the mean ± SD. One-way ANOVA and Tukey’s multiple comparison tests were performed for comparisons between U87 vs. LN229, U373, and T98G. *P < 0.05, **P < 0.01; ***P < 0.001 ****P < 0.0001
Glioblastoma genotype is not correlated with mitochondrial or glutaminolysis pathways
Glioblastoma is classified into four cellular states characterized by copy number amplification of the CDK4, EGFR, and PDGFRA loci and by mutations in the NF1 locus [30]. Among these changes, EGFR and PDGFR amplifications are representative of genetic changes related to metabolic conditions [30–33]. Therefore, we analyzed and genotyped tissue samples sourced from Kagoshima University using a custom NGS panel [24]. We classified the samples into the WT, EGFR, and PDGFR amplification groups (Additional file 2: Supplementary Table S4b). We also assessed the relationship between genetic change and metabolic protein expression. Although the sample size was limited, we observed no significant difference in protein expression between the groups (Additional file 1: Supplementary Fig. S4a–g).
Discussion
In this study, we found that the expressions of mitochondria-related proteins, including COX1, COX2, and DRP1, are interrelated and related to the glutaminolysis pathway. Conversely, they exhibit an inverse correlation with the glycolytic pathway. Therefore, mitochondria-related protein levels can reflect certain aspects of the metabolic condition of tumor tissue, including both glycolysis and glutaminolysis.
Mitochondrial DNA-encoded proteins are important for assessing mitochondrial activity because they reflect mitochondrial translation. Therefore, we focused on the expression of COX1, a subunit of complex IV, which plays a pivotal role in understanding tumor metabolism. Although we observed correlations among the expression of various mitochondria-related proteins, TFAM did not exhibit any correlation. As TFAM is an important protein for protecting mitochondrial DNA but is not encoded by mitochondrial DNA [34], the lack of correlation between TFAM and COX1 or COX2 is expected.
Cancer cells were initially reported to be dependent on glycolysis, a phenomenon known as the Warburg effect [8]. However, recent studies have demonstrated that under low-nutrient conditions, cancer cells are dependent on mitochondrial activity [26, 35]. Moreover, there are two main energy pathways: glycolysis and mitochondria [7]. Our results indicated an inverse correlation between mitochondrial-related protein and glycolytic protein expression, which is a reasonable finding. Furthermore, we examined the relationships between mitochondria-related pathways and tumor-specific metabolic pathways, including glutaminolysis and one-carbon metabolism [14]. We observed no correlation between mitochondria-related proteins and one-carbon pathway-related proteins. However, there was a considerable correlation between mitochondrial-related proteins and the glutaminolysis pathways. Therefore, we further examined the association between mitochondria-related proteins and glutaminolysis-related proteins. Oizel et al. reported that tumor cells with high glutamine levels exhibit increased mitochondrial respiration in the presence of glutamine [36]. However, in some cases, inhibition of glutaminolysis can restore mitochondrial function in a contrasting phenomenon. For example, consistent with our results, Rupprecht et al. reported that during a glucose shortage, glutamine utilization is facilitated [37, 38]. Therefore, increased expression of glutaminolysis-related proteins may imply a low-nutrient and glucose-starved microenvironment or a dependence on the TCA cycle. Further supporting this notion, we observed that under glucose-starvation conditions, both mitochondria and glutaminolysis-related protein expression increased in glioblastoma cell lines [26].
In these experiments presented study, as presented in Fig. 4d-k, under glucose-starvation conditions, the expression of mitochondria and glutaminolysis-related proteins increased. Moreover, our MS analysis identified citric acid as a crucial metabolite for distinguishing between glycolytic-dominant and mitochondria-dominant tissues. Notably, we utilized frozen samples to accurately represent intracellular conditions. Our findings reveal that citric acid levels are significantly reduced in mitochondria-dominant tissues. This decrease in intracellular citrate levels may imply an increase in extracellular citrate. Parkinson et al. reported that the presence of extracellular citrate leads to a reduction in intracellular amino acid levels [39] and that changes in intracellular citrate levels could reflect the effects of therapeutic interventions [40]. Moreover, the enzymes of the TCA pathway are sensitive to ROS, and in the presence of ROS, these enzymes may not effectively function [41]. Therefore, cells would be unable to utilize the TCA cycle, potentially leading to the accumulation of citrate acid. Collectively, these results suggest cells in mitochondrial-dominant tissue may experience low-nutrient conditions and that glutaminolysis or mitochondria-dominant tissue may rely on the TCA cycle to produce energy.
Glutaminolysis plays a dual role in energy production and ROS generation [14, 16]. In cancer cells, targeting glutamine metabolism is a promising therapeutic strategy, as evidenced by the efficacy of GLDH inhibitors in reducing tumor proliferation [14, 17, 19]. Regarding glioblastoma, GLDH is particularly exploitable; inhibition of glycolysis increases GLDH activity, and glutamine addition is a metabolic adaptation that supplements oxidative glycolysis in the unfavorable condition of hypoxia [42, 43]. Specifically, GLDH inhibitors (R162) and glutaminase inhibitors are effective for glioma cells [19, 44]. In our study, inhibitor experiments demonstrated the effectiveness of the GLDH inhibitor in cell lines with high GLDH expression, including LN229, U373, and T98G cells. Therefore, inhibition of glutaminolysis may serve as an effective treatment strategy for tissues with high glutaminolysis activity. However, it is important to note that the GLDH antibody used in this study recognizes both GLDH1 and 2, whereas R162 only inhibits GLDH1. While GLDH1 inhibition has been proven effective in tumor treatment [45], the effectiveness of GLDH2 inhibition remains unclear. Combined inhibition of both GLDH1 and 2 may offer a more effective treatment strategy, as evidenced by the reduction in tumor volume following GLDH1/2 shRNA treatment [46]. According to our results, among all samples, samples No. 17, 18, and 20 exhibited high expression of both mitochondria-related and glutaminolysis-related proteins, including GLDH and GLS1. Therefore, the use of inhibitors targeting GLDH and GLS1 may offer enhanced therapeutic efficacy for patients with these characteristics.
Finally, we examined the relationship between driver gene amplification, including EGFR and PDGFR, and metabolic changes, considering their relation to metabolism [30, 33]. The tyrosine kinase receptor is involved in the PI3K and MAPK pathways [31, 32]. However, EGFR and PDGFR amplifications did not affect glutaminolysis or mitochondrial protein expression in this study. Further investigations with higher sample sizes and explorations of other metabolic pathways are required to validate these results.
In this study, we assessed protein expression in tissue samples, acknowledging that metabolic activities may vary within the same samples based on the location. When comparing expression differences among genotypes, it is crucial to consider factors such as tumor heterogeneity and tumor necrosis. Therefore, in the future, experiments utilizing the spatial transcriptome or metabolomics must be performed to clarify the relationship between genotypes and metabolic conditions. Plasticity is another crucial characteristic of glioblastomas [30]. Specimens can reflect metabolic conditions at a specific point in time, representing a snapshot of the metabolic state, which may diverge significantly from other time points. Our results provide insights for elucidating the relationship between each metabolic pathway. For example, if tumors transition to being primarily reliant on mitochondrial metabolism, they may concurrently increase their dependence on glutaminolysis while reducing their reliance on glycolysis. Therefore, our findings can guide the development of novel treatments that accommodate tumor plasticity.
This study had some limitations, including the methodological approach used to determine if tissues are mitochondria-dominant, relying on western blot and MS analysis. It is important to note that tumor samples may contain substances other than tumor cells, potentially affecting the accuracy of the analysis. There remains a possibility that high expression of mitochondria-related proteins leads to the accumulation of nonfunctional proteins. Therefore, OXPHOS activity or mitochondrial fraction experiments using fresh tumor samples should be assessed in the future. Nevertheless, this research established the different characteristics between high and low expression of mitochondria-related proteins and that there are two types of tissue present in glioblastoma. Finally, to confirm the effectiveness of glutaminolysis inhibition, we should use mouse models or patient organoids in the future. Despite these limitations, we believe that our results are novel and serve as a basis for the development of new glioblastoma treatments.
Conclusion
Our study revealed that mitochondrial proteins are interrelated with glutaminolysis. Moreover, we identified two types of tumor tissues: one with a high expression of mitochondria and glutaminolysis-related proteins and another with a high expression of glycolytic proteins. Inhibition of GLDH or GLS1 may be effective in cases with high glutaminolysis activity. Our findings can be used in future studies aiming to develop medical treatments for glioblastoma targeting cancer metabolism and can lead to effective treatment for refractory malignant brain tumors.
Supplementary Information
Additional file 1: Supplementary Fig. S1–5.
Additional file 2: Supplementary Tables S1–S4.
Abbreviations
- α-KG
Alpha-ketoglutarate
- DMEM
Dulbecco’s modified Eagle’s medium
- GLDH
Glutamine dehydrogenase
- LC–MS/MS
Liquid chromatography-tandem mass spectrometry
- MS
Mass spectrometry
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SHMT2
Serine hydroxymethyltransferase-2
- MTHFD1
Methylenetetrahydrofolate dehydrogenase
- TCA
Tricarboxylic acid
Authors’ contributions
KM conducted the investigation, experiments, funding, wrote the manuscript, and analyzed the data. DS conducted MS experiments and supervision. RO, TM, KG, YF, YS, DK, NH, TT, YH, TA, AT, and RH contributed to patient enrollment and collection of tumor samples. MY, TU and YK supervised the study. KY and RH analyzed the data and supervised the study and funding. All the authors read and approved the final version of the manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number JP19K17673 and JP23H03021, the Clinical Research Foundation, and the Fukuoka Public Health Promotion Organization Cancer Research Fund.
Data availability
Data is provided within the manuscript of supplementary information files.
Declarations
Ethics approval and consent to participate
The use of glioblastoma tissues for this study was approved by the Ethics Committee of the Graduate School of Medical Sciences, Kyushu University, and Kagoshima University. Written informed consent was obtained by all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 2.Bi J, Chowdhry S, Wu S, Zhang W, Masui K, Mischel PS. Altered cellular metabolism in gliomas - an emerging landscape of actionable co-dependency targets. Nat Rev Cancer. 2020;20:57–70. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Sun Y, Guo Y, Shi X, Chen X, Feng W, et al. An overview: the diversified role of mitochondria in cancer metabolism. Int J Biol Sci. 2023;19:897–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab. 2022;34:355–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18:160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibao S, Minami N, Koike N, Fukui N, Yoshida K, Saya H, et al. Metabolic heterogeneity and plasticity of glioma stem cells in a mouse glioblastoma model. Neuro Oncol. 2018;20:343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburg O. The metabolism of carcinoma cells. J Cancer Res. 1925;9:148–63. [Google Scholar]
- 9.Bhatti JS, Bhatti GK, Reddy PH. Mitochondrial dysfunction and oxidative stress in metabolic disorders - a step towards mitochondria based therapeutic strategies. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoang-Minh LB, Siebzehnrubl FA, Yang C, Suzuki-Hatano S, Dajac K, Loche T, et al. Infiltrative and drug-resistant slow-cycling cells support metabolic heterogeneity in glioblastoma. EMBO J. 2018;37:e98772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iranmanesh Y, Jiang B, Favour OC, Dou Z, Wu J, Li J, et al. Mitochondria’s role in the maintenance of cancer stem cells in glioblastoma. Front Oncol. 2021;11:582694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talasila KM, Rosland GV, Hagland HR, Eskilsson E, Flones IH, Fritah S, et al. The angiogenic switch leads to a metabolic shift in human glioblastoma. Neuro Oncol. 2017;19:383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson DC, Bayik D, Storevik S, Moreino SS, Sprowls SA, Han J, et al. GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat Cancer. 2023;4:648–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernhard C, Reita D, Martin S, Entz-Werle N, Dontenwill M. Glioblastoma metabolism: insights and therapeutic strategies. Int J Mol Sci. 2023;24:9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghannad-Zadeh K, Das S. One-carbon metabolism associated vulnerabilities in glioblastoma: a review. Cancers (Basel). 2021;13:3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natarajan SK, Venneti S. Glutamine metabolism in brain tumors. Cancers (Basel). 2019;11:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucharzewska P, Christianson HC, Belting M. Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. PLoS One. 2015;10:e0116740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Wang G, Mao Q, Li S, Xiong W, Lin Y, et al. Glutamate dehydrogenase (GDH) regulates bioenergetics and redox homeostasis in human glioma. Oncotarget. 2014;5. Retrieved from https://www.oncotarget.com/article/7657/text/.
- 20.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kofuji S, Hirayama A, Eberhardt AO, Kawaguchi R, Sugiura Y, Sampetrean O, et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol. 2019;21:1003–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amemiya T, Hata N, Mizoguchi M, Yokokawa R, Kawamura Y, Hatae R, et al. Mesenchymal glioblastoma-induced mature de-novo vessel formation of vascular endothelial cells in a microfluidic device. Mol Biol Rep. 2021;48:395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higa N, Akahane T, Yokoyama S, Yonezawa H, Uchida H, Takajo T, et al. A tailored next-generation sequencing panel identified distinct subtypes of wildtype IDH and TERT promoter glioblastomas. Cancer Sci. 2020;111:3902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higa N, Akahane T, Yokoyama S, Yonezawa H, Uchida H, Fujio S, et al. Molecular genetic profile of 300 Japanese patients with diffuse gliomas using a glioma-tailored gene panel. Neurol Med Chir (Tokyo). 2022;62:391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miki K, Yagi M, Yoshimoto K, Kang D, Uchiumi T. Mitochondrial dysfunction and impaired growth of glioblastoma cell lines caused by antimicrobial agents inducing ferroptosis under glucose starvation. Oncogenesis. 2022;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito T, Uchiumi T, Yagi M, Amamoto R, Setoyama D, Matsushima Y, et al. Cardiomyocyte-specific loss of mitochondrial p32/C1qbp causes cardiomyopathy and activates stress responses. Cardiovasc Res. 2017;113:1173–85. [DOI] [PubMed] [Google Scholar]
- 28.Akoglu H. User’s guide to correlation coefficients. Turk J Emerg Med. 2018;18:91–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villar VH, Merhi F, Djavaheri-Mergny M, Duran RV. Glutaminolysis and autophagy in cancer. Autophagy. 2015;11:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–49 e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Khayari A, Bouchmaa N, Taib B, Wei Z, Zeng A, El Fatimy R. Metabolic rewiring in glioblastoma cancer: EGFR, IDH and beyond. Front Oncol. 2022;12:901951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westermark B. Platelet-derived growth factor in glioblastoma-driver or biomarker? Ups J Med Sci. 2014;119:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. [DOI] [PubMed] [Google Scholar]
- 36.Oizel K, Chauvin C, Oliver L, Gratas C, Geraldo F, Jarry U, et al. Efficient mitochondrial glutamine targeting prevails over glioblastoma metabolic plasticity. Clin Cancer Res. 2017;23:6292–304. [DOI] [PubMed] [Google Scholar]
- 37.Choudhury D, Rong N, Ikhapoh I, Rajabian N, Tseropoulos G, Wu Y, et al. Inhibition of glutaminolysis restores mitochondrial function in senescent stem cells. Cell Rep. 2022;41:111744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rupprecht A, Moldzio R, Modl B, Pohl EE. Glutamine regulates mitochondrial uncoupling protein 2 to promote glutaminolysis in neuroblastoma cells. Biochim Biophys Acta Bioenerg. 2019;1860:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson EK, Adamski J, Zahn G, Gaumann A, Flores-Borja F, Ziegler C, et al. Extracellular citrate and metabolic adaptations of cancer cells. Cancer Metastasis Rev. 2021;40:1073–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordan K, Stanton EH, Milenkovic VM, Federlin M, Drexler K, Buchalla W, et al. Potential involvement of extracellular citrate in brain tumor progression. Curr Mol Med. 2022;22:506–13. [DOI] [PubMed] [Google Scholar]
- 41.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci U S A. 1997;94:11168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obara-Michlewska M, Szeliga M. Targeting glutamine addiction in gliomas. Cancers (Basel). 2020;12:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Restall IJ, Cseh O, Richards LM, Pugh TJ, Luchman HA, Weiss S. Brain tumor stem cell dependence on glutaminase reveals a metabolic vulnerability through the amino acid deprivation response pathway. Cancer Res. 2020;80:5478–90. [DOI] [PubMed] [Google Scholar]
- 45.Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R, Nishimura MC, Kharbanda S, Peale F, Deng Y, Daemen A, et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci U S A. 2014;111:14217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Fig. S1–5.
Additional file 2: Supplementary Tables S1–S4.
Data Availability Statement
Data is provided within the manuscript of supplementary information files.