Abstract
Background:
Esophageal squamous cell carcinoma (ESCC) comprises 90% of all esophageal cancer cases globally and is the most common histology in low-resource settings. Eastern Africa has a disproportionately high incidence of ESCC.
Methods:
We describe the genomic profiles of 61 ESCC cases from Tanzania and compare them to profiles from an existing cohort of ESCC cases from Malawi. We also provide a comparison to ESCC tumors in The Cancer Genome Atlas.
Results:
We observed substantial transcriptional overlap with other squamous histologies via comparison with The Cancer Genome Atlas (TCGA) PanCan dataset. DNA analysis revealed known mutational patterns, both genome-wide as well as in genes known to be commonly mutated in ESCC. TP53 mutations were the most common somatic mutation in tumors from both Tanzania and Malawi but were detected at lower frequencies than previously reported in ESCC cases from other settings. In a combined analysis, two unique transcriptional clusters were identified: a proliferative/epithelial cluster and an invasive/migrative/mesenchymal cluster. Mutational signature analysis of the Tanzanian cohort revealed common signatures associated with aging and cytidine deaminase activity (APOBEC) and an absence of signature 29, which was previously reported in the Malawi cohort.
Conclusion:
This study defines the molecular characteristics of ESCC in Tanzania, and enriches the Eastern African dataset, with findings of overall similarities but also some heterogeneity across two unique sites.
Impact:
Despite a high burden of ESCC in Eastern Africa, investigations into the genomics in this region are nascent. This represents the largest comprehensive genomic analysis ESCC from sub-Saharan Africa to date.
Keywords: esophageal squamous cell carcinoma, esophageal cancer, Africa, genomics
INTRODUCTION
Esophageal cancer is the sixth leading cause of cancer mortality worldwide (1). Adenocarcinoma and squamous cell carcinoma are two histologic subtypes of esophageal cancer, which have distinct biologic characteristics, geographical distributions, and risk factors (2). Esophageal squamous cell carcinoma (ESCC) is rare in the United States but comprises 90% of all esophageal cancer cases globally. There is significant geographic variation in the incidence of ESCC, with a majority of cases occurring in developing countries. Specific regions in Iran, central Asia, north-central China, southern Africa, East Africa, and southern South America, are impacted by a disproportionately high incidence of esophageal cancer (2).
The eastern coast of Africa, from Ethiopia to South Africa, has recently gained increased attention for its disproportionately high incidence of ESCC (3–6). Esophageal cancer is the leading cause of cancer mortality amongst males in Kenya, and the highest rates of ESCC mortality have been reported in Malawi (1). A disproportionately high number of patients younger than 40 at diagnosis has been described across a number of sites in East Africa (3,7–9). The high incidence of young-onset ESCC, as well as the geographic distribution along the eastern corridor of Africa, suggests plausible contribution of genetic susceptibility and/or a unique environmental or infectious risk factor(s).
Etiologic, genetic, and genomic studies of ESCC in Asian, European, and American populations have been extensive, but investigations into ESCC in sub-Saharan Africa are nascent by comparison. The few available studies on this topic have implicated possible contributions to the high incidence from thermal injury due to consumption of hot beverages, alcohol use, poor oral hygiene, low soil selenium levels, indoor air pollution from biomass burning and other environmental exposures, or possible infectious causes (4,10–17). Findings from a prior study conducted in Tanzania reported an association of low socioeconomic status with increased risk for ESCC, suggesting the contribution of some undetermined exposure or constellation of exposures (18).
Global collaborative studies in cancer genomics have potential to identify specific mutational signatures that may shed light on causal factors and opportunities for cancer prevention (19). While The Cancer Genome Atlas (TCGA) Network and International Cancer Genome Consortium (ICGC) comprehensively analyzed common malignancies in Western countries, ESCC is a relatively rare diagnosis in the United States and Europe. While earlier studies point to global heterogeneity in the genomic characteristics of ESCC, these have not included analyses of African ESCC tumors. Analyses from China and Japan have demonstrated frequent mutations in genes commonly aberrant in squamous cell cancers, including TP53, RB1, CDKN2A, PIK3CA, NOTCH1, and NFE2L2 (20–23). Mutation of the tumor suppressor gene TP53 is the most frequent genetic alteration in ESCC and esophageal adenocarcinoma alike, with higher rates in ESCC and mutation profiles known to vary widely across geographic areas (24–27).
A previous systematic review of studies on the genetics of ESCC in African populations included only 23 studies; almost all were candidate gene studies with only a single study from Malawi including whole-exome sequencing (28). The recent whole-exome sequencing and RNA transcriptomic analysis of 59 ESCC tumors in Malawi reported a high proportion of tumors without TP53 mutations and identified a unique tumor mutation signature interpreted as consistent with an unknown carcinogenic exposure (29). We hypothesized that detection of a novel mutational signature could be indicative of a carcinogenic exposure unique to eastern Africa, particularly if findings from Malawi were replicated in ESCC tumors from Tanzania.
The high societal burden in eastern Africa of this deadly and understudied disease emphasizes the need for a comprehensive molecular analysis of ESCC in this region, as identification of distinct mutational signatures (30) could point to potentially modifiable risk factors, including possible environmental exposures or infectious etiologies. Thus, we aimed to evaluate the somatic mutation rate, mutational patterns, copy number profiles, and recurrently mutated genes in tumor specimens obtained from ESCC patients in Tanzania. We aimed to compare the molecular characteristics of this disease from two representative sites in eastern Africa to molecular characteristics of ESCC tumors in TCGA, in effort to outline similarities and differences. Finally, in order to evaluate the possible heterogeneity of the disease within eastern African, we compared results from our Tanzanian cohort to those from the previously described cohort from Malawi.
MATERIALS AND METHODS
Study Design and Population
Muhimbili National Hospital (MNH) is the national referral and teaching hospital affiliated with Muhimbili University of Health and Allied Sciences (MUHAS) in Dar es Salaam, Tanzania. This prospective study recruited sequential patients with a suspected diagnosis of ESCC who sought care at MNH. From May 2016 to February 2018, all patients ≥18 years old who presented to MNH with symptoms of dysphagia who were planned to undergo an endoscopy due to concern for a diagnosis of esophageal cancer were considered eligible for participation. Non-permanent residents of Tanzania, and pregnant or lactating women were not eligible. Written informed consent was obtained from all participants prior to endoscopy. Informed consent was obtained in Swahili, the national language of Tanzania. Only those with endoscopic findings consistent with malignancy were considered for inclusion in the study protocol. Patients who were not found to have endoscopic findings consistent with malignancy at the time of endoscopy did not undergo biopsies for research purposes and were excluded from all subsequent study procedures. All biopsy specimens underwent pathologic review at the Central Pathology Lab at MNH, and a subset of cases underwent confirmatory pathologic review at the University of California, San Francisco (UCSF). The first 61 cases with biopsy confirmed ESCC histology were included in this analysis.
Ethics Statement
The study was approved by institutional review boards at UCSF (15–18275) and MUHAS (2018–08-22/AEC/Vol.XII/91, amendment of 2018–04-03/AEC/Vol.XII/84). A standardized Material Transfer Agreement, governing the transfer of tangible research materials between two organizations was ratified by UCSF and MUHAS prior to transfer of materials. Guidelines for shipment of Biologic Substances Category B (UN 3373) were adhered to. Human studies approval for the study conducted in Malawi was previously granted by the Malawi National Health Sciences Research Committee and the University of North Carolina Internal Review Board, granting permission for submission of data to dbGaP.
Data Collection
Demographic, clinical, and pathologic variables were abstracted from the medical records. In addition, each patient participated in an in-person questionnaire. Data were collected regarding educational level, occupational history, family history of illnesses, prior or ongoing tobacco and/or alcohol use, and environmental exposure.
Biospecimen Collection and Processing
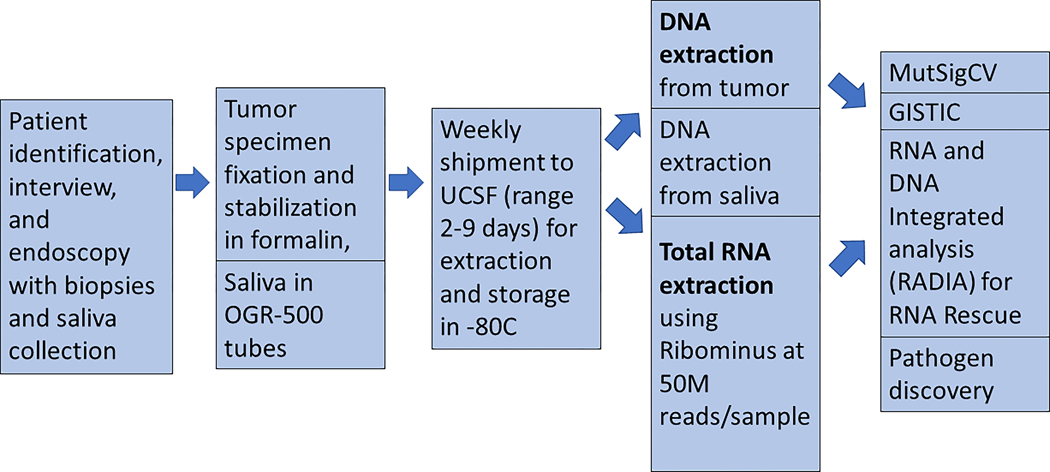
The specimen collection and processing workflow from Tanzania is summarized in Figure 1. If a tumor was visualized at the time of endoscopy, up to six core biopsies were obtained. Initially, we used both PAXgene Tissue Container (QIAGEN) and RNAlater (Thermo Fischer Scientific Inc.), both of which allowed for flexible transport of specimens from Tanzania and resulted in preserved genetic integrity and expression profiles in the first 10 samples (see Supplementary Figure S1). Based upon these results, RNAlater was used exclusively thereafter, and all specimens included in this analysis were preserved in RNAlater.
Figure 1. Workflow for ESCC specimens from Tanzania.
Stepwise progression from recruitment of participants, specimen fixation, shipment, DNA and RNA extraction, and molecular analyses.
Tumor specimens were immediately added to 10 ml of RNAlater at the time of collection. We processed all biopsy specimens to be <0.5 cm thick in order to ensure proper diffusion of RNAlater through the tissue. Each case was de-identified with a unique study identification number. Specimens were batched and shipped once per week at room temperature from Dar es Salaam, Tanzania to San Francisco, California using a commercial shipping service.
RNA and DNA extraction
Nucleic acids were extracted from tumor samples using the Qiagen AllPrep method and were quantified using Pico/RiboGreen. RNA integrity was evaluated using an Agilent bioanalyzer. DNA and RNA quantity were measured by nanodrop method, and DNA was further confirmed by picogreen method, yielding measures of total DNA and RNA acquired (ug). Saliva was collected in the Oragene DISCOVER OGR-500 tube as a source of germline DNA. Extraction of DNA from saliva samples was performed using the PrepIT L2P extraction kit from Oragene, and DNA was quantified using PicoGreen.
Whole genome sequencing
DNA sequencing libraries were prepared for both tumor and matched-normal samples with the KAPA Hyper prep kit. Sequencing was performed on the Illumina HiSeq or NovaSeq platform to a target depth of 60x coverage for tumor samples and 30x depth for normal samples. DNA sequencing reads were aligned to the UCSC hg19 build of GRCh37 using default parameters for “bwa mem -M” version 0.7.5, secondary alignments were removed with samtools, duplicates were marked with samblaster 0.1.21, and GATK version 2.3 was used for INDEL realignment and quality recalibration. Small variants were called with previously described methods (31).
RNA sequencing
RNA isolations with RIN>7 were incorporated into two independent libraries from each tumor sample using the KAPA Stranded RNA-seq with RiboErase kit, followed with sequencing on Illumina HiSeq or NovaSeq to a target of 200 million 150bp paired reads. RNA-seq read data were aligned to a RefSeq transcriptome using bowtie2 version 2.2.6 with options “-k 200 --dpad 0 --gbar 99999999 -mp 1,2 --np 1 --score-min L,0,0.1 --no-mixed --no-discordant --sensitive -I 1 -X 1000”. Gene level quantification of TPM was estimated with RSEM version 1.2.25. For assessing expression of variants found in DNA, local alignment was performed with bowtie2 with parameters “-z -k 5 --sensitive-local”.
We deployed the quantile normalization procedure (32) to bring various RNA-Seq datasets into a common expression space. We performed quantile normalization for each individual gene, excluding zero quantifications from both target and source distributions, adding them back in after the transformation. We transformed the source distribution’s data for each gene to a chosen target distribution by matching quantiles for each value. In the case of the samples from Tanzania, we used a set of clinical formalin-fixed, paraffin-embedded (FFPE) samples (n = 1,699) as the target distribution (33). In the case of Malawi samples, we used the Tanzanian samples (n = 61) as the target distribution.
RNA transcript quantification was computed as an average number of reads per base within each transcript. In order to assess gene expression, we obtained transcript per million (TPM) from RSEM output of RNA sequencing data. We summarized per-gene expression as a sum of TPM for all isoforms of a given gene. We quantified all genes that have at least one isoform that begins with NM_ (mRNA RefSeq category) to compose the final expression matrix of protein coding genes. For consistency in relative RNA quantification between samples, we applied rescaling to gene-wise TPM values in sample-wise manner, so each sample’s TPMs sum up to 1 million. This step allowed for a more uniform and interpretable comparison of expression levels across samples. The RNA sequencing coverage and quality statistics for each sample are summarized in Supplementary Table 1.
Analysis of a previously published dataset from ESCC cases in Malawi
Whole-exome DNA sequencing was previously performed on 59 untreated ESCC tumor specimens and paired normal DNA from Malawi, along with whole-transcriptome RNA sequencing at 100 million 48bp paired reads using previously described methods (29). We accessed publicly available data from dbGaP (study accession number phs001448).
t-Distributed Stochastic Neighbor Embedding (t-SNE) data projection and visualization
We utilized t-SNE methods (34) to project high-dimensional RNA-Seq data for the joint cohort of Tanzania, Malawi, TCGA and clinical FFPE samples into two-dimensional space. We first selected 3,000 most varying genes across the joint dataset and then applied Rtsne function from Rtsne R package with the following parameters: perplexity = 200, verbose = TRUE, eta = 500, check_duplicates = FALSE.
Mutation Detection using RNA
RNA and DNA Integrated Analysis (RADIA) is a published method to improve somatic mutation detection by analyzing the patient matched normal and tumor DNA along with the tumor RNA-Seq data to identify “RNA Rescue” mutations (31). Using patient-matched normal and tumor DNA along with tumor RNA, RADIA identified RNA editing events across the entire transcriptome.
MutSigCV
We next used MutSigCV (version 1.41) (35) with default coverage, covariate setting for background mutation adjustment to identify genes harboring more mutations than would be expected by the sample-specific background rate, in order to generate a list of significantly mutated genes. MAF files (read quality >=10) format from either the Tanzania and Malawi cohorts were used as input, the q<0.1 was considered statistically significant. Analyses were performed on the high-performance computing clustering at the Helen Diller Family Comprehensive Cancer Center at UCSF.
RNA-Seq cluster solution (k = 2)
In effort to further evaluate whether advanced molecular profiling could divide ESCC from East Africa into discrete subsets that are associated with biologic features, we performed a clustering analysis. We used Consensus Clustering R package (36), with hierarchical clustering, to scan a range of solutions from k = 2 to k = 10. Based on the average silhouette width for each cluster solution, k = 2 was the best solution, closely followed by k = 3 and k = 6 as the next best. This pattern remained irrespective of whether the whole transcriptome (WT) or top 3,000 (3k) most variable gene features were used to cluster samples. When comparing WT and 3k clusters for same-k solutions, almost all samples consistently fell into the same clusters, indicating high consistency in cluster solutions regardless of the feature space based on which the solutions are produced. We then compared k = 2 (average silhouette score = .87), k = 3 (average silhouette score = .83), and k = 6 (average silhouette score = .79) cluster solutions based on 3k. We found that two major clusters of samples (n = 94, out of total N = 120, 78%) persisted in every one of these solutions. In fact, these samples cluster together in every k = 2 through k = 10 solution. Using two-sided binomial test, we estimate that between 70% and 85% of samples in any sample size would cluster into these two clusters. Breaking k = 2 solution into more clusters only separates the remaining 26 samples into smaller and smaller groups, without affecting the two major groups of samples. Our final solution is k = 2 based on the 3,000 most variable genes.
Pathway enrichment analysis for gene clusters
As described in the previous section, our final RNA-Seq clustering solution is based on 3,000 most varying genes. Gene clustering was performed using hierarchical clustering method. Two major gene clusters were observed. Genes in each cluster were compared against MSigDB (37) and significant pathways were selected based on hypergeometric test significance (FDR <= 0.1). We computed per-gene t-test for RNA-Seq cluster 1 vs. 2 contrast. We then analyzed resulting per-gene t-statistic values using GSEA desktop application (38). Pathways with family-wise error rate (FWER) ≤0.05 were selected for plotting.
Statistical analysis
Statistical analyses were performed according to each bioinformatics method as described by relevant citations. Differences among clinical and environmental characteristics and subtypes were calculated using Chi-squared test or t-test. Age was considered as a continuous variable, using a t-test. All other variables were categorical variables, and Chi-squared test was used. A p-value of <0.05 was considered significant. Bonferroni multiple hypotheses correction was applied.
Data availability
Data generated for the Tanzania cohort in this study are publicly available in dbGaP at phs003217. Data generated for the Malawi cohort are publicly available in dbGaP at phs001448.
RESULTS
Patient Cohort
We recruited 200 patients with histologically confirmed ESCC into a parent study in Dar es Salaam, Tanzania. We performed whole-genome sequencing on the first 61 tumors with a histologic confirmation of ESCC, which met quality criteria. The median age was 49 (range 30–86). The majority were male (67%), and 15% were younger than 40 years old. Patient characteristics are presented in Table 1.
Table 1. Characteristics of patients with esophageal squamous cell carcinoma from Tanzania (n=61).
| N | % | |
|---|---|---|
| Gender | ||
| Male | 41 | 67.2 |
| Female | 20 | 32.8 |
|
| ||
| Age | ||
| <40 | 9 | 14.8 |
| >=40 | 52 | 85.2 |
|
| ||
| HIV status | ||
| Positive | 5 | 8.2 |
| Negative | 32 | 52.5 |
| Not reported | 24 | 39.3 |
|
| ||
| Smoking status | ||
| Current | 16 | 26.2 |
| Former | 19 | 31.1 |
| Never | 26 | 42.6 |
|
| ||
| Alcohol consumption | ||
| Yes | 31 | 50.8 |
| No | 30 | 49.2 |
|
| ||
| Cooking site | ||
| Indoors in a separate building | 26 | 42.6 |
| Indoors without chimney | 13 | 21.3 |
| Other | 1 | 1.6 |
| Outdoors | 21 | 34.4 |
|
| ||
| Burnt tongue or mouth in past year | ||
| Yes | 21 | 34.4 |
| No | 40 | 65.6 |
Mutation Detection and Copy Number Events in Tanzanian Tumors
We detected 4,159 somatic nonsynonymous mutations with a Q score >10 in 61 ESCC samples from Tanzania using Mutect. The median number of mutations was 68.18 (+/− 61.55) per sample. We found 3,876 single-nucleotide variants (missense, nonsense, silent) vs. 283 insertion-deletion (INDEL) mutations.
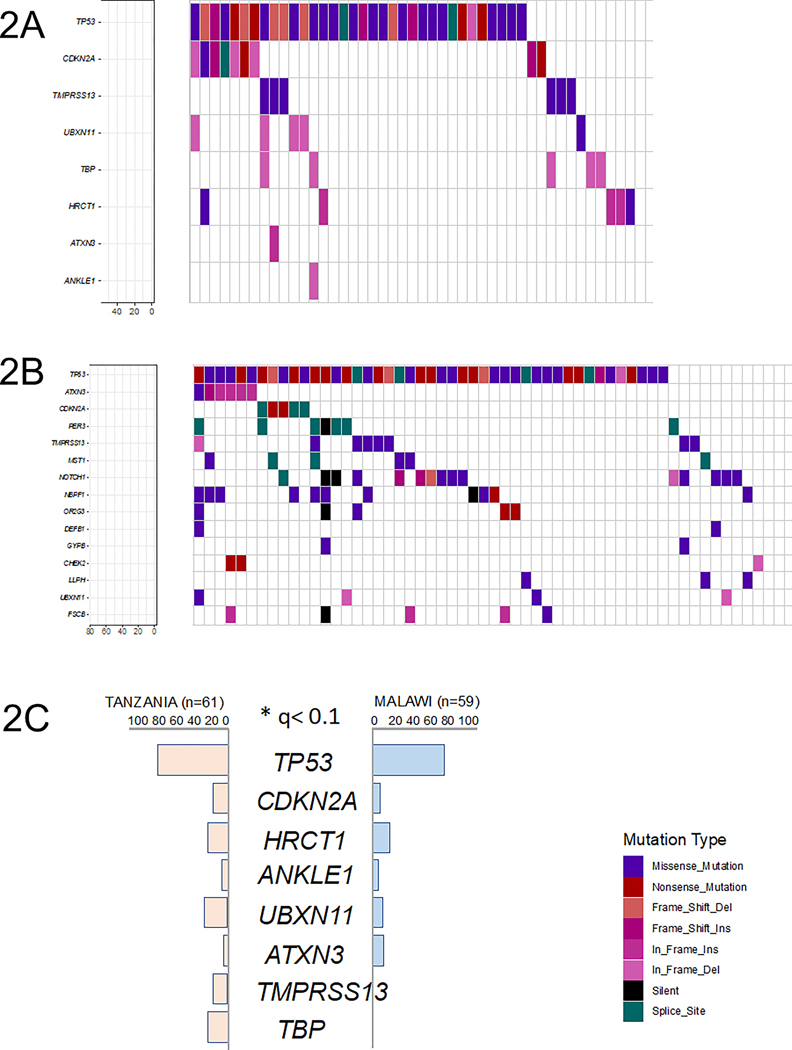
Genes mutated in >5% of Tanzanian samples appear in Figure 2A. We identified nine genes as significantly mutated using MutSigCV: TP53, CDKN2A, HRCT1, ANKLE1, UBXN11, ATXN3, TMPRSS13, and TBP (q<0.1). A total of 15 genes were significantly mutated in samples from Malawi with this q-value cut-off (see Figure 2B). To remove potential false positives, we applied MutSigCV to merged Mutation Annotation Format (.MAF) files from the two populations and identified TP53, CDKN2A, HRCT1, ANKLE1, UBXN11, ATXN3, TMPRSS13, and TBP as significantly mutated in ESCC. The differential patterns between the Tanzanian and Malawian datasets of mutations occurring in ≥5 samples in each are shown in Figure 2C. To further characterize biological function of these genes and to evaluate the capability of RADIA to rescue low-confidence DNA calls, RNA expression levels of these genes were evaluated in both cohorts and appear in Supplementary Figure S2.
Figure 2. Summary of recurrently mutated genes.
Each column represents one patient sample, and columns are ordered according to gene mutation frequency. The overall frequencies of synonymous and non-synonymous mutations are summarized at the top of the figure. Percentage of the cohort harboring a mutation(s) in a given gene is depicted at the bottom left of each figure.
(2A) Somatic point mutations in the Tanzania cohort (n=61).
(2B) Somatic point mutations in the Malawi cohort (n=59).
(2C) Significantly mutated genes in the Tanzania and Malawi cohorts.
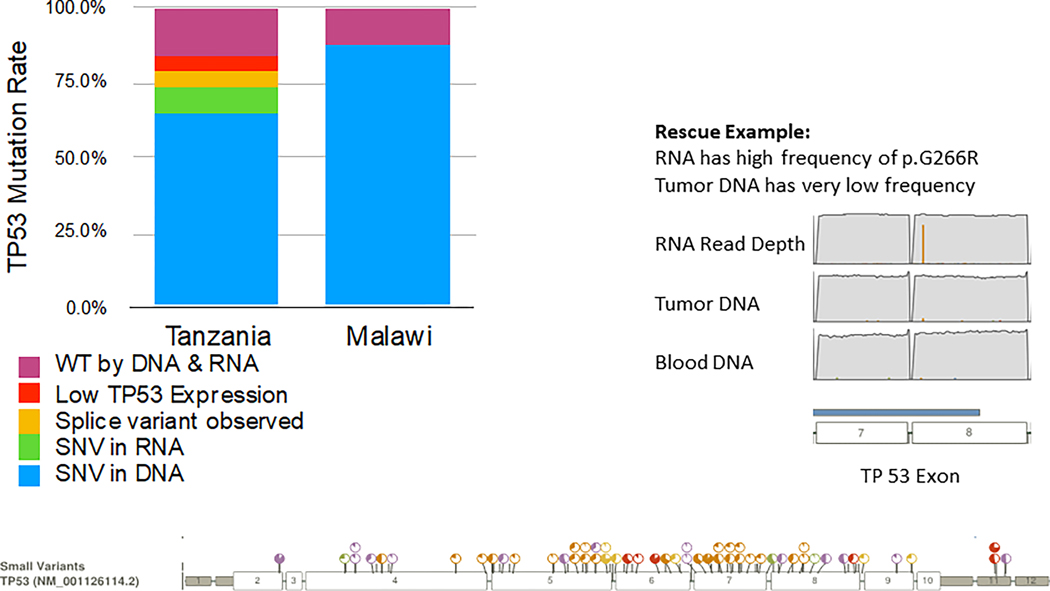
TP53 Mutation Detection and Rescue
The TP53 mutation rate originally reported in the Malawi cohort was 78% (46 out of 59 samples) (29). We initially detected TP53 mutation in 62% of samples from Tanzania, based on DNA calls alone (38 out of 61 samples). However, RNA rescue added eight mutation calls in seven unique samples. One of those calls occurred in a sample that already had a distinct TP53 mutation call, based on DNA, and two calls occurred in the same sample. Therefore, use of RNA sequencing (RNAseq) reads resulted in detection of TP53 mutations in an additional six samples in which mutations were not initially detected using DNA calls alone. Malawi samples were not analyzed in this manner; thus, a direct comparison is not available. The overall TP53 mutation rate and rescue in the Tanzanian samples is summarized in Figure 3.
Figure 3. TP53 mutation rate and rescue in Tanzania and Malawi cases.
For cases from Tanzania, RNA and DNA Integrated Analysis (RADIA) was used to improve somatic mutation detection by analyzing the patient matched normal and tumor DNA along with the tumor RNA-Seq data to identify “RNA Rescue” mutations. The plot along the bottom of the figure shows all positions at which somatic variants in TP53 were detected.
Catalog of Somatic Mutations in Cancer (COSMIC) Signatures
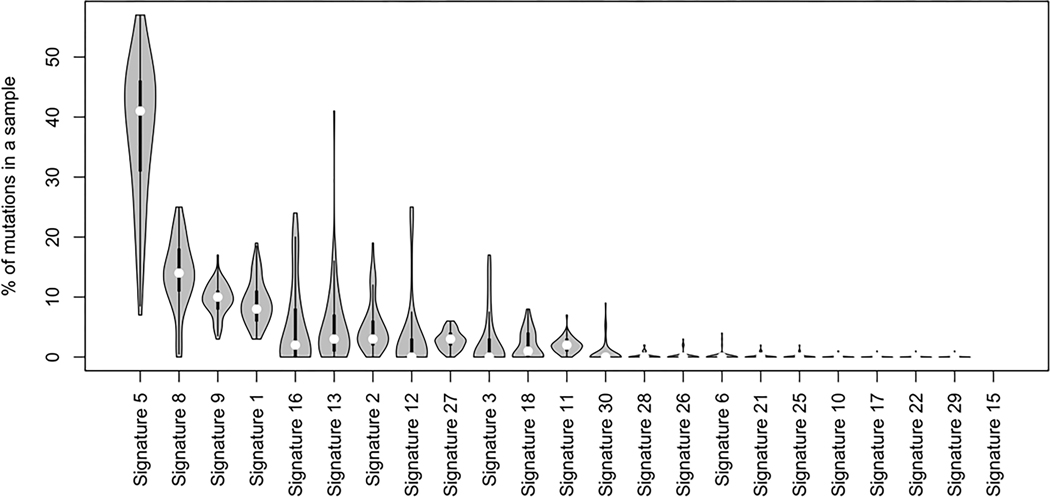
Mutational signatures offer clues to the oncogenic process(es) shaping tumor initiation. We used WGS data to assign COSMIC mutational signatures to each tumor. Signature frequencies in specimens from Tanzania appear in Figure 4. COSMIC mutational signature 5 was the most common, consistent with other reports in squamous cell cancers.
Figure 4. COSMIC Signatures in the Tanzania cohort.
Mutational signature frequencies in specimens from Tanzania, ordered by frequency. COSMIC mutational signature 5 was the most common, consistent with other reports in squamous cell cancers.
RNA Expression
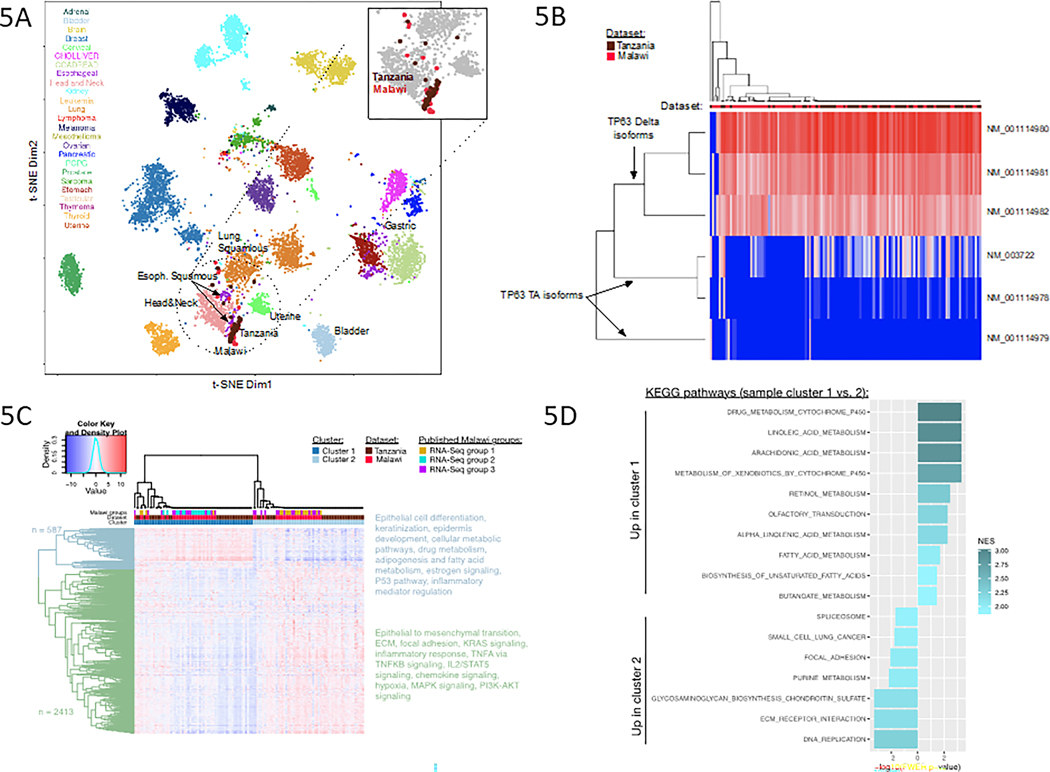
Figure 5A presents a scatterplot of t-Distributed Stochastic Neighbor Embedding (t-SNE) projection of pan-cancer RNAseq expression data composed of clinical (n=1,699) (33) and TCGA (n = 10,471) samples. Each point represents a single sample. The colors reflect the sample’s annotated cancer type. Tanzania and Malawi datasets cluster with ESCC and other squamous tumors in this background cohort, indicating that Tanzania and Malawi tumors are most similar to other malignancies with squamous histology and most similar to other esophageal tumors.
Figure 5. Transcriptomic analysis of esophageal squamous cell carcinoma from Tanzania and Malawi.
(5A.) t-SNE projection of pan-cancer RNA-Seq data, showing that both the Tanzania and Malawi datasets cluster with other squamous tumors from a combined cohort of clinical and TCGA.
(5B.) Heatmap of expression levels of six different TP63 isoforms, three Delta and three TA, in which columns represent individual samples in Tanzania and Malawi datasets and rows represent individual isoforms.
(5C.) Final clustering solution (k=2), based on expression of the 3,000 most varying genes across combined Tanzania and Malawi datasets.
(5D.) KEGG pathways from Gene Set Enrichment Analysis results for cluster 1 versus cluster 2 contrast, based on whole transcriptome.
Alternative splicing of TP63 into the Delta isoforms define many squamous cell types and is common ubiquitous in ESCC. We examined alternative splicing of TP63 using RNAseq data. Figure 5B confirms TP63 Delta isoforms are highly expressed across most samples, consistent with previously described analysis of Malawi cohort RNAseq data. This robust expression of TP63 is also consistent with findings from TCGA.
Consensus Clustering of samples based on RNA transcription examines intrinsic subtypes in larger sample collections (36) revealed two major RNAseq subtypes. To avoid noise affecting clustering solution, we performed our clustering analysis based on gene expression data for the 3,000 genes with highest variance across the combined Tanzania and Malawi datasets. Final RNAseq clustering solution is shown in Figure 4C; both Tanzania and Malawi samples were used, annotated by the dataset and cluster color tracks. We re-clustered the data using whole transcriptome profiles and also examined alternative clustering approaches, to evaluate robustness of our clustering solution. The two major sample clusters in the final solution were consistently identified across choices of k (k=2:10). Using sample proportion estimates from two-sided binomial test we estimate that 78% (95% confidence interval: 70–85%) of samples in any sample size fall into these two major clusters under any solution. Our final solution is k = 2, based on the silhouette score and the sample composition analysis. Two major gene clusters are annotated by different dendrogram colors (blue vs. green), and the list of pathways enriched in these gene groups are listed on the right side of Figure 4C.
Figure 4D depicts the differential pathway enrichment analysis results for the two sample clusters in the RNAseq solution, using Gene Set Enrichment Analysis (GSEA) method on the whole transcriptome (38). Subtype 1 (proliferative/epithelial cluster) is characterized by epithelial cell differentiation, keratinization, epidermal development, cellular metabolic pathways, drug metabolism, adipogenesis and fatty acid metabolism, estrogen signaling, the p53 pathway, and inflammatory mediator regulation. Subtype 2 (invasive/migrative/mesenchymal cluster) was characterized by epithelial to mesenchymal transition, extracellular matrix, focal adhesion, KRAS signaling, inflammatory response, TNFA vs. TNFKB signaling, IL2/STAT5 signaling, chemokine signaling, hypoxia, MAPK signaling, and PI3K signaling.
To determine significant associations of clinical features with the RNAseq subtypes, we tested associations of clinical and environmental variables with the two subtypes. These variables included age (as a continuous variable), gender, smoking history, pesticide exposure, HIV status, tumor purity, and tumor ploidy. No statistically significant associations of any of these factors with the individual clusters were identified. We also tested for statistically significant associations of molecular markers in Tanzania cohort with these two clusters and found that two RNA editing events in TGM5 and GBP6 genes are highly enriched in cluster 1 (p-value <e-16). TGM5 is involved in maintenance of epidermis and GbP6 has previously been lined to aggressiveness of tumorigenesis in head and neck squamous tumors (39).
DISCUSSION
This work provides an unprecedented and comprehensive genomic profiling of ESCC tumors from two countries in eastern Africa. The primary analysis was conducted using whole-genome sequencing of 61 histologically confirmed ESCC tumors from Tanzania to determine molecular signatures, RNA expression profiles, and RNA editing events. We also performed a secondary analysis, comparing data from Tanzanian ESCC tumors to an existing dataset of 59 ESCC Malawian patients for whom whole-exome sequencing and RNAseq data was publicly available. Comparing ESCC cases from sub-Saharan Africa with other squamous cell carcinomas, including ESCC cases from TCGA, demonstrated substantial transcriptional overlap between squamous histologies.
ESCC tumors included in TCGA originated from North America, Eastern Europe, Vietnam, and Brazil. In this sample of ninety tumors, significantly mutated genes included TP53, NFE2L2, MLL2, ZNF750, NOTCH1 and TGFBR2 (40). In TCGA, ESCC tumors resembled squamous cell carcinomas from other organs, and molecular features were notably distinct from those of the esophageal adenocarcinoma tumors. Similarly, ESCC tumors from both Tanzania and Kenya clustered with ESCC tumors from TCGA and squamous cell carcinomas from other origins, including head and neck, lung, and uterus. Moreover, our work demonstrated that the molecular characteristics of ESCC tumors from Tanzania and Malawi are similar to ESCC tumors from Japan and China (20–23).
DNA analysis revealed known mutational patterns, both genome-wide as well as in known, commonly mutated genes. TP53 is the most commonly mutated gene in squamous cell cancers from many tissues of origin. Our analysis found TP53 mutations to be the most common somatic mutation in ESCC tumors from both sites in eastern Africa. Based upon the subset of tumors without TP53 mutations reported in the original publication of the cohort from Malawi, we initially hypothesized that TP53 mutations may have been under-represented due to inability to detect somatic mutations missed by conventional DNA mutation callers (29). To address this, we utilized an iterative “rescue” process, RADIA (31). RNA rescue mutations have low support in the DNA but strong support in the RNA and are typically missed by traditional DNA mutation calling algorithms that only examine the DNA. The inclusion of the RNA increases the power to detect somatic mutations at low DNA allelic frequencies (31) or when tumor purity is low (41). RNA rescue identified TP53 mutations in six additional cases from Tanzania, resulting in an overall mutation rate of 72%. RNA editing events, including nonsynonymous events that have been shown to have significant differential editing in cancer specific subtypes, stages, and overall survival were detected (42). RNA editing of MDM2 3’UTRs in the miRNA target sites essential for binding was common (43) and we speculate this could be a contributing factor to a decrease in TP53 gene expression.
Nonetheless, our final frequencies of detection of TP53 mutations in the cohorts from Tanzania and Malawi were strikingly similar (72% vs. 78%), even despite methodologic differences. Even with employment of the RADIA technique to improve somatic mutation detection, this study corroborated prior findings from Malawi that a subset of ESCC tumors in Eastern Africa are notable for the absence of the TP53 mutation. By comparison, the TP53 mutation is nearly ubiquitous in ESCC tumors from Western countries, Japan, and China. (24,25) Variations in mutation frequency according to ethnicity have been previously reported in ESCC, with TP53 mutations occurring more frequently in Asian ESCC patients than in Caucasians (44). In a small cohort of African-American ESCC cases (n = 10), TP53 was found to be mutated in only 50% of samples (45). Further investigation, with expanded sample sizes, is warranted to determine the true prevalence of the TP53 mutation in ESCC cases from sub-Saharan Africa and in individuals of African descent.
In the combined analysis, two unique clusters were identified: a proliferative/epithelial cluster and an invasive/migrative/mesenchymal cluster. We were only able to recapitulate, and not fully, the subtypes reported by Liu et al. when clustering on whole transcriptome (29); however, when clustering was performed on 3,000 most varying genes, the third group did not persist as a separate subtype. When combining the Tanzania and Malawi data sets, clusters associated with the Malawi RNA-seq subtype 2 is comprised only of Malawi samples. Our results support a finding of two subtypes that are molecularly different. These two subtypes likely reflect the relative differentiation states of squamous epithelium, distinct cells of origin, variations in contaminating non-neoplastic epithelium. Alternatively, differences in immune infiltration could also account for these clusters. Single cell analyses of sorted populations may bring further insights into these alternative potential explanations.
Efforts are ongoing to identify an unidentified carcinogen, or a constellation of carcinogens, that are contributing to the high incidence of ESCC affecting the eastern corridor of Africa. Risk factors traditionally associated with this disease in western populations, including tobacco and alcohol use, do not seem to be solely responsible for the unusually high incidence in this population, with a significant proportion of patients identifying as never-smokers. Our use of whole-genome sequencing empowered exhaustive profiling of underlying mutational processes. However, mutational signature analysis from the Tanzanian cohort did not detect any specific mutagenic insult beyond the aging and cytidine deaminase activity (APOBEC) pathway. While a unique signature 29 was identified in a small number of tumors from Malawi, this finding was not replicated in the cohort from Tanzania. Additionally, we did not observe a significant frequency of signature 17 as previously reported in the Malawi cohort. These differences could possibly be due to technical differences in the input data from the two unique sites.
Several limitations of this study must be mentioned. The two cohorts are notable for significant methodologic differences in specimen collection and processing. The Malawi specimens were collected as frozen tumors while Tanzania tumors were collected and processed in RNAlater medium (Thermo Fischer Scientific Inc.) to facilitate transport at room temperature. The Malawi specimens were selected for polyadenylation, while the Tanzania tumors were processed using ribosomal depletion, designed to enrich the whole spectrum of RNA transcripts, irrespective of polyadenylation status. Thus, our comparison of these two groups may be subject to some technical artifacts specific to one technique vs. the other; nonetheless, our findings in clustering based on RNAseq and overall TP53 mutation rate were comparable between the two cohorts, supporting the overall similarity of the samples on a larger scale. In addition, we did not evaluate whether there was any association of the two unique transcriptional clusters that were identified in the Tanzanian cohort with clinical outcomes Because >90 percent of patients present with very advanced disease in this context and available therapies were limited in Tanzania during the study period, this modest sample size would like have been inadequately powered to detect a meaningful difference in survival.
Substantial inequities exist in the access to genomic sequencing in resource-constrained settings (19). Specifically, clinical applications of these endeavors are not routinely available. Moreover, genomic analyses are costly in the face of myriad needs affecting cancer patients in resource-constrained settings and also rely heavily on transport of specimens to distant laboratories equipped with the analytic resources. Therefore, it is particularly important that this study is interpreted as part of a broader research portfolio and efforts which collectively aim to understand the etiology of ESCC in sub-Saharan African and to identify risk factor(s) which can be targeted by prevention and early detection efforts. In a low-resource setting, ‘translational genomic studies’ must be accompanied by and interpreted along with data from traditional epidemiologic studies and in the context of a broader public health perspective.
In conclusion, this combined analysis is a first report of a multi-site genomic analysis of ESCC from sub-Saharan Africa. Parallel studies were undertaken in Malawi and Tanzania with a goal to enhance the understanding of the molecular profiles of ESCC tumors in Eastern Africa. The findings from the current study did not fully recapitulate the finding of three subtypes previously reported from Malawi (29) but rather we identified two molecularly unique subtypes which were in part consistent with the findings previously published by Liu et al. While the unique findings from these two cohorts may suggest that ESCC is a heterogeneous disease, even within a circumscribed geographic region, this result also highlights the importance of collaborative studies to enrich both numbers and diversity from within the African continent.
We sought to identify factors causal for the high incidence of this disease in eastern Africa in order to inform development of preventive and early detection interventions. Our results further highlight the need for collaborative investigation; thus, future studies must include efforts to corroborate these findings in ESCC tumors from other sites in eastern Africa that are similarly impacted by a high incidence of this disease. Our future work will include a pathogen-finding analysis using the RNA sequencing data from Tanzanian specimens. A multi-site genome-wide association study to evaluate the possible contribution of genetic susceptibility that could explain the geographic clustering of this disease predominantly along the eastern corridor of the African continent is also underway.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients in both Tanzania and Malawi and their family members for their participation in this research study. We are grateful to each participating institution and to the ward and endoscopy nurses at MNH for assistance with case ascertainment. We acknowledge the members of the African Esophageal Cancer Consortium (AfrECC) for their ongoing scientific collaboration in East Africa.
FINANCIAL SUPPORT:
K. Van Loon, E.J. Mmbaga, B.P. Mushi, S. Msiba, L. Akoko, and A. Mwanga received support from the National Institutes of Health, National Cancer Institute Cancer Center Administrative Supplement to Promote Cancer Prevention and Control Research in Low and Middle Income Countries, A119617, [CA0082629]. E.A. Collisson was supported by R01 [CA178015, CA222862, CA227807, CA239604, CA230263], and U24 [CA210974] grants. Content does not reflect the views of the National Cancer Institute or National Institutes of Health.
LIST OF ABBREVIATIONS:
- APOBEC
aging and cytidine deaminase activity
- dbGaP
database of Genotypes and Phenotypes
- ESCC
esophageal squamous cell carcinoma
- FFPE
formalin-fixed, paraffin-embedded
- ICGC
International Cancer Genome Consortium
- INDEL
insertion-deletion
- MNH
Muhimbili National Hospital
- MUHAS
Muhimbili University of Health and Allied Sciences
- RADIA
RNA and DNA Integrated Analysis
- TCGA
The Cancer Genome Atlas
- TPM
transcript per million
- t-SNE
t-Distributed Stochastic Neighbor Embedding
- UCSF
University of California, San Francisco
- WT
whole transcriptome
Footnotes
CONFLICT OF INTERESTS: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Wild CP, Weiderpass E SB. Esophageal Cancer: A Tale of Two Malignancies. In: World Cancer Report: Cancer Research for Cancer Prevention. 2020. [PubMed] [Google Scholar]
- 3.Cheng ML, Zhang L, Borok M, Chokunonga E, Dzamamala C, Korir A, et al. The incidence of oesophageal cancer in Eastern Africa: Identification of a new geographic hot spot? Cancer Epidemiol. 2015;39(2):143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormack VA, Menya D, Munishi MO, Dzamalala C, Gasmelseed N, Leon Roux M, et al. Informing etiologic research priorities for squamous cell esophageal cancer in Africa: A review of setting-specific exposures to known and putative risk factors. Int J Cancer. 2017;140(2):259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Loon K, Mwachiro MM, Abnet CC, Akoko L, Assefa M, Burgert SL, et al. The African Esophageal Cancer Consortium: A Call to Action. J Glob Oncol. 2018;(4):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy G, McCormack V, Abedi-Ardekani B, Arnold M, Camargo MC, Dar NA, et al. International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann Oncol. 2017;28(9):2086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mmbaga EJ, Deardorff KV, Mushi B, Mgisha W, Merritt M, Hiatt RA, et al. Characteristics of Esophageal Cancer Cases in Tanzania. J Global Oncol. 2018;154(2):360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker RK, Dawsey SM, Abnet CC, White RE. Frequent occurrence of esophageal cancer in young people in western Kenya. Dis Esophagus. 2010; 23(2):128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel K, Wakhisi J, Mining S, Mwangi A, Patel R. Esophageal Cancer, the Topmost Cancer at MTRH in the Rift Valley, Kenya, and Its Potential Risk Factors. ISRN Oncol. 2013;2013:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Middleton DRS, Menya D, Kigen N, Oduor M, Maina SK, Some F, et al. Hot beverages and oesophageal cancer risk in western Kenya: Findings from the ESCCAPE case–control study. Int J Cancer. 2019;144(11):2669–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munishi MO, Hanisch R, Mapunda O, Ndyetabura T, Ndaro A, Schüz J, et al. Africa’s oesophageal cancer corridor: Do hot beverages contribute? Cancer Causes Control. 2015; 26(10):1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mlombe YB, Rosenberg NE, Wolf LL, Dzamalala CP, Chalulu K, Chisi J, et al. Environmental risk factors for oesophageal cancer in Malawi: A case-control study. Malawi Med J. 2015;27(3):88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menya D, Kigen N, Oduor M, Maina SK, Some F, Chumba D, et al. Traditional and commercial alcohols and esophageal cancer risk in Kenya. Int J Cancer. 2019;144(3):459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chetwood JD, Garg P, Finch P, Gordon M. Systematic review: the etiology of esophageal squamous cell carcinoma in low-income settings. Expert Rev Gastroenterol Hepatol. 2019;13(1):71–88. [DOI] [PubMed] [Google Scholar]
- 15.Okello S, Akello SJ, Dwomoh E, Byaruhanga E, Opio CK, Zhang R, et al. Biomass fuel as a risk factor for esophageal squamous cell carcinoma: a systematic review and meta-analysis. Environ Heal. 2019;18(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okello S, Churchill C, Owori R, Nasasira B, Tumuhimbise C, Abonga CL, et al. Population attributable fraction of Esophageal squamous cell carcinoma due to smoking and alcohol in Uganda. BMC Cancer. 2016. Dec 11;16(1):446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menya D, Maina SK, Kibosia C, Kigen N, Oduor M, Some F, et al. Dental fluorosis and oral health in the African Esophageal Cancer Corridor: Findings from the Kenya ESCCAPE case–control study and a pan-African perspective. Int J Cancer. 2019;145(1):99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mmbaga EJ, Mushi BP, Deardorff K, Mgisha W, Akoko LO, Paciorek A, et al. A Case-Control Study to Evaluate Environmental and Lifestyle Risk Factors for Esophageal Cancer in Tanzania. Cancer Epidemiol Biomarkers Prev. 2021; 30(2):305–316. [DOI] [PubMed] [Google Scholar]
- 19.Ginsburg O, Ashton-Prolla P, Cantor A, Mariosa D, Brennan P. The role of genomics in global cancer prevention. Nat Rev Clin Oncol. 2021;18(2):116–128. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y-B, Chen Z-L, Li J-G, Hu X-D, Shi X-J, Sun Z-M, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46(10):1097–102. [DOI] [PubMed] [Google Scholar]
- 21.Lin D-C, Hao J-J, Nagata Y, Xu L, Shang L, Meng X, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014; 46(5):467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–5. [DOI] [PubMed] [Google Scholar]
- 23.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology. 2016;150(5):1171–82. [DOI] [PubMed] [Google Scholar]
- 24.Harris CC, Peri L, Mandard AM, Welsh JA, Montesano R, Metcalf RA, et al. Genetic Analysis of Human Esophageal Tumors from Two High Incidence Geographic Areas: Frequent p53 Base Substitutions and Absence of ras Mutations. Cancer Res. 1991;51(15):4102–6. [PubMed] [Google Scholar]
- 25.Montesano R, Hollstein M, Hainaut P. Generic alterations in esophageal cancer and their relevance to etiology and pathogenesis: A review. Int J Cancer. 1996;69(3):225–35. [DOI] [PubMed] [Google Scholar]
- 26.Tanière P, Martel-Planche G, Puttawibul P, Casson A, Montesano R, Chanvitan A, et al. TP53 mutations andMDM2 gene amplification in squamous-cell carcinomas of the esophagus in South Thailand. Int J Cancer. 2000;88(2):223–7. [DOI] [PubMed] [Google Scholar]
- 27.Alaouna M, Hull R, Penny C, Dlamini Z. Esophageal cancer genetics in South Africa. Clin Exp Gastroenterol. 2019; 12:157–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simba H, Kuivaniemi H, Lutje V, Tromp G, Sewram V. Systematic Review of Genetic Factors in the Etiology of Esophageal Squamous Cell Carcinoma in African Populations. Front Genet. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Snell JM, Jeck WR, Hoadley KA, Wilkerson MD, Parker JS, et al. Subtyping sub-Saharan esophageal squamous cell carcinoma by comprehensive molecular analysis. JCI Insight. 2016;1(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500(7463):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radenbaugh AJ, Ma S, Ewing A, Stuart JM, Collisson EA, Zhu J, et al. RADIA: RNA and DNA Integrated Analysis for Somatic Mutation Detection. Chellappan SP, editor. PLoS One. 2014; 9(11):e111516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010; 11(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton Y, Sedgewick AJ, Cisneros L, Golovato J, Johnson M, Szeto CW, et al. Large scale, robust, and accurate whole transcriptome profiling from clinical formalin-fixed paraffin-embedded samples. Sci Rep. 2020;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurens van der Maaten and Geoffrey Hinton. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9(86):2579–605. [Google Scholar]
- 35.Lawrence MS, Stojanov P, Polak P, Kryukov GV., Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monti S, Tamayo P, Mesirov J, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach Learn. 2003; 52(1):91–118. [Google Scholar]
- 37.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu ZH, Cai F, Zhong Y. Comprehensive Analysis of the Expression and Prognosis for GBPs in Head and neck squamous cell carcinoma. Sci Rep. 2020;10(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, Cherniack AD, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkerson MD, Cabanski CR, Sun W, Hoadley KA, Walter V, Mose LE, et al. Integrated RNA and DNA sequencing improves mutation detection in low purity tumors. Nucleic Acids Res. 2014; 42(13):e107–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell. 2015; 28(4):515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanbin Z, Jing Z. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem Biophys Res Commun. 2019; 516(1):102–11. [DOI] [PubMed] [Google Scholar]
- 44.Deng J, Chen H, Zhou D, Zhang J, Chen Y, Liu Q, et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat Commun. 2017;8(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erkizan HV, Sukhadia S, Natarajan TG, Marino G, Notario V, Lichy JH, et al. Exome sequencing identifies novel somatic variants in African American esophageal squamous cell carcinoma. Sci Rep. 2021;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for the Tanzania cohort in this study are publicly available in dbGaP at phs003217. Data generated for the Malawi cohort are publicly available in dbGaP at phs001448.