Abstract
The pathogenesis of head and neck squamous cell carcinoma (HNSCC) is notably complex. Early symptoms are often subtle, and effective early screening methods are currently lacking. The tumors associated with HNSCC develop rapidly, exhibit high aggressiveness, and respond poorly to existing treatments, leading to low survival rates and poor prognosis. Numerous studies have demonstrated that histone posttranslational modifications (HPTMs), including acetylation, methylation, phosphorylation, and ubiquitination, play a critical role in the occurrence and progression of HNSCC. Moreover, targeting histone posttranslationally modified molecules with specific drugs has shown potential in enhancing therapeutic outcomes and improving prognosis, underscoring their significant clinical value. This review aims to summarize the role of histone posttranslational modifications in the pathogenesis and progression of HNSCC and to discuss their clinical significance, thereby providing insights into novel therapeutic approaches and drug development for this malignancy.
Keywords: Head and neck squamous cell carcinoma, Epigenetics, Histone posttranslational modifications, Histone inhibitors
Introduction
Head and neck malignancies rank as the sixth most common type of cancer globally, with approximately 900,000 new cases and 500,000 deaths annually [1]. The incidence is expected to increase by 30% by 2030, reaching approximately 1.08 million new cases per year [2, 3]. Head and neck squamous cell carcinoma (HNSCC) arises from squamous epithelial cells in the oral cavity, nasopharynx, oropharynx, laryngopharynx, larynx, subglottic region, and nasal sinuses, making it the most common malignancy in this region. The pathogenesis of HNSCC is closely linked to HPV infection, tobacco use, and excessive alcohol consumption [4, 5]. This carcinoma is highly aggressive, with significant rates of recurrence and metastasis. Over 60% of patients exhibit no obvious symptoms or precancerous lesions in the early stages, often leading to a diagnosis of advanced HNSCC [6]. Current treatments for HNSCC include surgical resection, chemotherapy, radiotherapy, and multimodal combinations [7]. While these approaches can manage and alleviate the disease to some extent, they do not prevent the emergence of cancer cell resistance and can have significant side effects on the patient's body [8, 9]. This results in high recurrence rates (15–40%), high metastasis rates, a five-year survival rate of less than 50%, and poor prognosis [10]. Therefore, it is imperative to explore new diagnostic and therapeutic methods for HNSCC.
Epigenetic modifications are heritable changes in gene activity that co-regulate gene function, expression, and chromatin structure through covalent modifications of nucleic acids and histones, without altering the underlying DNA sequences. These modifications are reversible and dynamically regulated, occurring at various levels of gene expression. The primary mechanisms include chromatin remodeling (dynamic spatiotemporal localization of nucleosomes), DNA methylation, HPTMs, and noncoding RNA (ncRNA) regulation. Epigenetic mechanisms involve a diverse array of enzymes and protein domains, which fine-tune gene expression programs to control critical biological processes such as cell differentiation and embryogenesis. Epigenetic modifications can significantly influence gene expression activity, thereby driving tumor dynamic transcriptome heterogeneity. They have emerged as essential tools for tumor diagnosis and treatment, with their reversibility offering innovative strategies for tumor drug development [11].
HPTMs are multifunctional epigenetic markers that modulate chromatin conformation and the accessibility of transcription factors, coactivators, and repressors. They play pivotal roles in processes such as transcription, DNA damage repair, apoptosis, and cell cycle regulation [11, 12]. In the 1960s, Vincent Allfrey, a biologist from the USA, was the first to identify the association between histone acetylation and mammalian gene activity [13]. Since then, over ten covalent modifications have been identified, including lysine acetylation, arginine and lysine methylation, ubiquitination, ADP ribosylation, citrullination, serine and tyrosine phosphorylation, proline isomerization, acylation, carbonylation, and the debated biotinylation [14]. With the advancement of technology, the discovery of new histone modification sites and patterns continues to expand. Various pairs of histone-modifying enzymes antagonize the cis–trans regulation of chromatin, thereby influencing the physicochemical properties of histones and chromatin through distinct modification methods, resulting in a series of biological effects [12]. The miswriting, misinterpretation, and clearance of histone modifications are closely associated with tumorigenesis and cancer progression, as histone coding disorders lead to dysregulation of gene expression and cellular characteristics. Consequently, HPTMs are significant contributors to the initiation, progression, and metastasis of cancer [11]. Research has shown that histone modifications such as acetylation, methylation, phosphorylation, ubiquitination, acylation, ADP ribosylation, crotonylation, and 2-hydroxyisobutyrylation are involved in the development and progression of HNSCC. Enzymes associated with acetylation and methylation, in particular, represent important therapeutic targets for HNSCC, with substantial clinical value.
Histone posttranslational modifications in head and neck squamous carcinoma
Histone variants and modifications
Nucleosomes represent the fundamental units of chromatin, each comprising an octamer of histones—H3, H4, H2A, and H2B—around which a segment of DNA is wrapped. These histones form the core structure, providing scaffolds that encapsulate and compact DNA, thereby regulating chromatin organization and accessibility [12, 14]. Histone variants, such as H2A.Z, H3.3, and macroH2A, are nonallelic forms of the core histones that can substitute for their canonical counterparts within the nucleosome. These variants introduce diversity to chromatin structure and can lead to functional outcomes such as altering nucleosome stability, facilitating transcription factor access, and establishing distinct chromatin domains. For instance, H2A.Z deposition at gene promoters is often associated with active transcription [15], while macroH2A enrichment in heterochromatin regions contributes to gene silencing and chromosomal stability [16]. Moreover, histone modifications, including acetylation, methylation, and phosphorylation, dynamically regulate chromatin architecture by modulating the interaction between histones and DNA. These PTMs can either directly impact chromatin compaction or serve as recruitment platforms for chromatin-associated proteins, which in turn can lead to the formation of active or repressive chromatin states. For example, acetylation of histone H3 at lysine 27 (H3K27ac) is a mark of active enhancers and promoters, promoting a more open chromatin conformation that is permissive for transcription [17]. In contrast, trimethylation of histone H3 at lysine 27 (H3K27me3) is associated with Polycomb-mediated gene silencing and the establishment of repressive chromatin structures [18].
Histone acetylation
Histone acetylation is meticulously and dynamically regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), maintaining normal histone acetylation levels that control the initiation and termination of gene transcription. Changes in overall histone acetylation levels have been linked to cancer phenotypes in various cancers and can predict cancer patient prognosis. Elevated acetylation levels result in gene activation, while decreased acetylation levels, usually found at promoters and often occurring alongside DNA methylation, lead to gene silencing.
Histone acetylation by HATs
HATs utilizes acetyl coenzyme A as a cofactor to transfer the acetyl group to the ε-amino side chain of histone lysine residues. The mechanism by which HAT influences transcriptional activity operates through two primary pathways: (1) Acetylation neutralizes the positive charge of histones, thereby weakening the electrostatic interactions between the negatively charged DNA and histones. This weakening facilitates chromatin relaxation, increasing chromatin accessibility, and thereby allowing transcription factors to bind more readily. This posttranslational modification mechanism reduces DNA–histone interactions and promotes transcription factor binding. (2) Histone acetylation may influence intracellular pH levels, a hypothesis supported by evidence that many tumors exhibit lower pH levels alongside reduced histone acetylation. Additionally, low pH in tumors correlates with poor prognosis in cancer patients [19].
HATs are categorized into two types based on their intracellular localization: (1) Type A, which are situated in the nucleus. These enzymes add acetyl groups to lysine residues of nucleosome histones via bromodomain interactions. Type A HATs have been more thoroughly studied and are primarily classified into four groups in humans: [1] HAT1; [2] Gcn5/PCAF; [3] MYST: MOZ/YBF2/SAS2/TIP60; and [4] CBP/p300. (2) Type B, which are found in the cytoplasm. Unlike Type A, Type B HATs lack bromodomains and therefore acetylate newly synthesized, unacetylated histones before nucleosome assembly. Research on Type B HATs is relatively limited.
Research has demonstrated that head and neck squamous carcinoma cell lines can secrete endothelial secretory factors, which initiate histone acetylation. However, the acetylation levels in these carcinoma cell lines are significantly lower than those in normal mucosal cells [20, 21]. The co-expression of Vimentin, a marker of epithelial–mesenchymal transition (EMT), with acetylated histones has been detected in head and neck squamous carcinoma samples. This finding indicates a close relationship between histone acetylation and the EMT phenomenon, which further affects the invasive and metastatic capabilities of tumors [22–25]. Furthermore, a mouse implantation model of human-origin head and neck squamous carcinoma revealed that the combination of the epidermal growth factor receptor (EGFR) inhibitor cetuximab and the bromodomain inhibitor JQ1 delays tumor resistance, resulting in more effective treatment of head and neck squamous carcinoma. These observations underscore the clinical importance of HATs in influencing the therapeutic outcomes for head and neck squamous carcinoma [26].
Histone deacetylation by HDACs
HDACs facilitate the removal of acetyl groups from histones, thereby restoring the positive charge of lysine residues. This restoration strengthens the DNA–histone interaction, reduces chromatin accessibility, and inhibits chromatin transcription. Based on their enzymatic activity, structural characteristics, and cofactor requirements, HDACs are classified into four categories: (1) Class I, comprising HDAC1, HDAC2, HDAC3, and HDAC8; (2) Class II, consisting of HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10; (3) Class III, which includes the sirtuin proteins (SIRT1-7); and (4) Class IV, which solely includes HDAC11. Notably, Class III enzymes utilize NAD+ as a cofactor, whereas the other classes employ Zn2+ as a cofactor. Due to their relatively low substrate specificity, each HDAC can deacetylate multiple histone sites, and each lysine residue can be targeted by more than one HDAC. HDACs play a crucial role in regulating tumor development by activating oncogenic cell signaling pathways and repressing oncogenes. HDACs are frequently overexpressed in cancer patients. Notably, HDAC2 can disrupt apoptotic mechanisms and regulate the cell cycle through the deacetylation of p53 and cyclin-dependent kinase inhibitors 1B, 1C, and 2A, thereby facilitating tumor cell activation [27]. Numerous studies have demonstrated that HDACs, including HDAC1, HDAC2, HDAC6, and HDAC9, are upregulated in oral squamous carcinoma and nasal squamous carcinoma tissues, contributing to the progression of these cancers. Among these, the expression of HDAC6 is notably elevated in oral squamous carcinoma tissues, indicating that histone deacetylation is associated with EMT, which further enhances tumor invasiveness [28]. In addition, research has found that HDAC inhibitors can suppress the invasiveness of HNSCC by blocking the EGFR-Arf1 axis [29].
Histone methylation
Histone methylation is a posttranslational modification occurring on histone lysine or arginine residues, regulated synergistically by histone methyltransferases (HMTs) and histone demethylases (HDMTs). This modification is integral to the regulation of chromatin structure and genetic alterations, without altering the histone's charge [30]. The epigenetic impact of histone methylation is contingent on the methylation state and specific location. Lysine residue methylation is linked to chromatin transcription and DNA repair, whereas arginine residue methylation exclusively affects chromatin transcription [31, 32]. Histone H3 possesses several lysine methylation sites, including K4, K9, K27, K36, and K79. The methylation of H3K4 results in the opening of chromatin structures, whereas the methylation of H3K9 leads to chromatin condensation [30]. Furthermore, H3K4me3, H3K36me3, and H3K79me3 are indicative of transcriptionally active genes, whereas H3K27me3, H3K9me2, and H3K9me3 are markers of gene silencing [33, 34].
Histone methylation by HMTs
HMTs are predominantly classified into histone lysine methyltransferases and histone arginine methyltransferases. These enzymes catalyze the transfer of 1–3 methyl groups to the ε-amino or ω-guanidino groups of lysine and arginine residues, respectively [30]. Histone lysine methyltransferases exhibit high specificity; following monomethylation of lysine, a distinct aromatic residue within their catalytic site determines whether further dimethylation or trimethylation occurs, allowing lysine residues to exist in monomethylated, dimethylated, or trimethylated states [35, 36]. Arginine methyltransferases are categorized into type I and type II; both types catalyze monomethylation of arginine, with type I facilitating asymmetric dimethylation and type II facilitating symmetric dimethylation, thus resulting in monomethylated or dimethylated (symmetric or asymmetric) arginine [37]. Among histone lysine methyltransferases, EHMT2/G9a, EZH2, NSD1, NSD2, NSD3, SMYD2, and DOT1L are notable. Research has identified five of these—NSD1, NSD2, NSD3, EZH2, and EHMT2—as particularly relevant to head and neck squamous carcinoma. The NSD family of enzymes, which catalyze H3K36me and H3K36me2, may enhance transcriptional activity, with each member playing a critical role in the pathogenesis and progression of head and neck squamous carcinoma. EHMT2/G9a is a histone lysine methyltransferase that induces H3K9me and H3K9me2, leading to the transcriptional silencing of target genes [38]. Furthermore, G9a (H3K9 methyltransferase) has been implicated in promoting malignant progression in head and neck squamous carcinoma, correlating with a poorer prognosis for patients [39]. EZH2, primarily responsible for catalyzing H3K27me3 methylation, functions as an oncogene in various cancers. It independently predicts poor patient survival by facilitating cell cycle progression, EMT, chemotherapy resistance, and dedifferentiation in head and neck squamous carcinoma. As a potential drug target, EZH2 may serve as a cisplatin sensitizer or be utilized for secondary prevention after curative radiotherapy, offering significant clinical research value [40]. Analysis of oral cancer cases in Taiwan demonstrated that overexpression of EZH2 and SUV39H1 (H3K9 methyltransferase) in the nucleus correlates with lymph node metastasis and tumor stage, respectively, while cytoplasmic staining intensity of G9a is associated with histological typing related to differentiation [41]. The latest research has found that EZH1 and EZH2, as key components of PRC2, compete with each other and can affect the development of oral squamous cell carcinoma through H3K27 methylation [42]. Research has demonstrated that the levels of H3K4 methylation in head and neck squamous carcinoma cells differ significantly from those in normal cells, with H3K4me3 being underexpressed and H3K4me2 being overexpressed in oral squamous carcinoma tissues. Furthermore, EZH2 (H3K27 methyltransferase), responsible for methylating the H3K27, is highly expressed in oral squamous carcinoma cell lines, along with H3K27me3. The expression levels of both EZH2 and H3K27me3 correlate with tumor progression (tumor T and N stages), disease-free survival, and cancer-specific survival, indicating their potential as prognostic markers for patients with oral squamous carcinoma [30, 43]. Consequently, histone methylation is intricately associated with the development of head and neck squamous carcinoma.
Histone arginine methyltransferases include PRMT1, PRMT4, PRMT5, and PRMT6; however, only PRMT1 and PRMT5 have been implicated in head and neck squamous carcinoma according to the literature. PRMT1 can monomethylate and asymmetrically dimethylate a range of histone substrates, whereas PRMT5 is involved in monomethylation and symmetric dimethylation of histone proteins and has been identified as an oncogene in various cancers [39]. Yang et al. reported that elevated PRMT5 expression is significantly correlated with advanced lymph node staging and decreased overall survival in patients with head and neck squamous carcinoma [44].
Histone demethylation by HDMTs
HDMTs, analogous to HMTs, are divided into histone lysine demethylases and histone arginine demethylases. These enzymes catalyze the removal of methyl groups from lysine and arginine residues, respectively, which results in a range of biological effects.
Lysine-specific histone demethylases (KDMs) are aerobic enzymes and are classified into two primary groups based on their catalytic mechanisms and cofactor utilization: FAD-dependent KDMs and KDMs of the JumonjiC (JmjC) family. The latter group, which contains a Fe2+ ion in its active site, requires 2-oxoglutarate for activation. JmjC family KDMs are further subdivided into seven subfamilies—KDM2, KDM3, KDM4, KDM5, KDM6, KDM7, and KDM8—according to their substrate specificity [45, 46].
KDM1A/LSD1, a component of the transcriptional co-repressor complex, specifically removes methyl groups from H3K4me1 or H3K4me2, thereby mediating gene silencing [47]. Evidence from in vitro and in vivo preclinical studies underscores LSD1's significant role in enhancing proliferation and viability while inhibiting apoptosis in head and neck squamous carcinoma cells. KDM2 is categorized into two types, KDM2A and KDM2B, both targeting H3K36me1 and H3K36me2 [48, 49]. The KDM3/JMJD1 subfamily, comprising KDM3A, KDM3B, and KDM3C, removes methyl groups from H3K9me2 and activates target gene expression. Nevertheless, the exact role of KDM3 in head and neck squamous carcinoma is not fully elucidated and requires further research [50]. The KDM4/JMJD2 subfamily, including KDM4A, KDM4B, KDM4C, and KDM4D, facilitates the demethylation of H3K9me2/3. Additionally, KDM4A-C can demethylate H3K36me3. Given that H3K9me2/3 functions as a transcriptional repressor marker associated with heterochromatin formation, and H3K36me3 is generally linked to transcriptional repression at the transcription start site but promotes elongation, KDM4 is proposed to act as a transcriptional activator [51, 52]. KDM5 catalyzes the demethylation of H3K4me2/3, with KDM5A being identified in various head and neck squamous carcinomas [53] and other malignant tumors. KDM6A/UTX and KDM6B/JMJD3 demethylate H3K27me2/3, leading to the activation of target genes, and both exhibit elevated expression in head and neck squamous carcinomas. KDM7A and KDM7B/PHF8 preferentially target mono- and dimethylation repressor markers, such as H3K9me1/2, H3K27me2, and H4K20me1, thereby promoting transcriptional activation. They can also bind to H3K4me3, enhancing its demethylase activity toward H3K27/KDM7A or H3K9/KDM7B. Additionally, KDM8 impacts multiple histone tails, including the demethylation of H3K36me3 [54].
To date, only three histone arginine demethylases—PAD4, JMJD6, and JMJD1B—have been documented [55, 56]. PAD4 regulates arginine methylation and gene expression by removing methyl groups from residues H3R2, H3R8, H3R17, and H3R26 and converting arginine to citrulline [57]. A reduction in PAD4 activity is associated with increased expression of mesenchymal markers, which plays a crucial role in inhibiting tumor growth and metastasis. Furthermore, PAD4 knockdown induces cellular autophagy and apoptosis, consequently impairing cell proliferation [58]. JMJD6, an iron-containing, 2-oxoglutarate-dependent dioxygenase from the JmjC family, functions as an arginine demethylase, removing methyl groups from H3R2me2 and H4R3me2. JMJD1B is involved in the demethylation of H3K9me2 and, indirectly, the demethylation of H4R3me2/me1 [56]. As of now, no studies have explored the relationship between histone arginine demethylases and head and neck squamous carcinoma (Tables 1, 2).
Table 1.
Histone posttranslational modifications in head and neck squamous cell carcinoma
| PTMs | Sites | Specific functions | Effect (↑↓) | References |
|---|---|---|---|---|
| Acetylation | Lysine residues ε-amino side chains |
1. Promotes chromatin release, thereby increasing chromatin accessibility 2. Exhibits a lower pH with reduced levels of histone acetylation |
↑ | [79] |
| ↓ | ||||
| Deacetylation | — |
1. Upregulated in oral squamous cell carcinoma promotes the development of cancer 2. Deacetylation is associated with epithelial–mesenchymal transition, affecting the aggressiveness of tumors |
↑ | [80] |
| [30] | ||||
|
Methylation Demethylation |
H3K36me |
1. Induces transcriptional activity, EHMT2/G9a can silence target gene transcription 2. G9a can promote the malignant progression of HNSCC |
↓ | [38] |
| H3K36me2 | ↑ | [81] | ||
| H3K27me3 | An oncogene for many types of cancer and can be used as a drug target | ↑ | [40] | |
| H3K27 |
1. Associated with lymph node metastasis and the stage of the tumor 2. Associated with histological typology of differentiation |
↑ | [41] | |
| H3K9 | ||||
| H3K79me3 | ||||
| H3K4me1 | KDM1A/LSD1 can selectively remove its methyl markers and mediate gene silencing | ↓ | [47] | |
| H3K4me2 | ||||
| H3K36me1 | Targets of KDM2 KDM3/JMJD1 can remove methyl groups in H3K9me2 and activate target gene expression | ↓ |
[48] [49] |
|
| H3K36me2 | ||||
| H3K36me3 | KDM4A-C also demethylates H3K36me3, KDM8 has a demethylating effect on histone tails | ↓ | [52] | |
| [82] | ||||
| H3K9me1/2 | KDM7A and KDM7B/PHF8 act on monomethylation and dimethylation. Repressive markers lead to transcriptional activation | ↓ | [82] | |
| H3K27Me2 | ||||
| H4K20Me1 | ||||
| H3K36me3 | KDM4A-C also demethylates H3K36me3 | ↓ | [52] | |
| H3R2me2 | JMJD6 removes methyl groups as arginine demethylase | ↓ | [56] | |
| H4R3me2 | ||||
| Phosphorylation | Histone residues of serine threonine tyrosine |
1. Upregulation in the nucleus of ARK2 cells is associated with poor clinical outcomes in patients, and overexpression in the cytoplasm is associated with tumor size and cancer stage 2. Associated with histological differentiation, cell proliferation, and metastasis of oral cancer |
↑ | [41] |
| [59] | ||||
| Acylation | Histone lysine residues | Semps can reverse SUMOization, SNEP5 expression is associated with the differentiation of oral squamous cell carcinoma, SUMO-1 overexpression is associated with tumor cell proliferation | ↑ | [30] |
| ADP ribosylation | — |
Affects biological functions such as DNA damage repair, transcription, and apoptosis An increase in the degree of malignancy of oral tissues, an increase in the degree of ADP ribosylation |
↑ | [71] |
| Kcr | — |
1. Involved in cell metabolism, cell cycle and other processes 2. The expression of KCR regulators is associated with the occurrence and development of HNSCC |
↑ | [72] |
| Khib | — |
1. Chromatin is involved in the regulation of cellular function 2. It plays an important role in the occurrence and development of pancreatic cancer |
↑ | [75] |
| Lactylation | H3K18 |
1. Stabilize key regulators within cells 2. Activates relevant transcriptional and signaling pathways 3. It plays an important role in a variety of tumors such as cervical cancer |
↑ | (78) |
Table 2.
Potential clinical significance of histone posttranslational modifications in HNSCC
| PTMs | Inhibitor | Function | References | |
|---|---|---|---|---|
| Acetylation | HATs | CCS1477 | Phase I/II clinical studies for the treatment of hematologic malignancies, prostate cancer, breast cancer and non-small cell lung cancer have been initiated | [83] |
| EP31670 | Phase I clinical trial of safety and maximum tolerated dose in advanced solid tumors | |||
| HDACs | SAHA |
1. Treatment of cutaneous T-cell lymphoma, laryngeal squamous cell carcinoma 2. Inhibits cell proliferation, induces cell cycle arrest and apoptosis 3. The combination with pembrolizumab has stronger therapeutic activity against HNSCC |
[86] [88] [92] |
|
| Romidepsin |
1. Treatment of cutaneous T-cell lymphoma 2. Inhibits tumor-associated histone deacetylase |
[83] | ||
| Valproic acid |
1. Increase cell arrest and promote apoptosis 2. It has strong efficacy in combination with cisplatin |
[93] | ||
| TSA |
1. By regulating cell cycle mediators 2. Inhibits cell proliferation and induces G2/M phase arrest in tumor cells 3. TSA-induced H3K9ac can ameliorate the chemoresistance of HNSCC to cisplatin under active NF-κB signaling pathway |
[94] [95] |
||
| PBA |
1. Damage to the EMT process 2. It increased p21 and p27, decreased the levels of G1/S molecules CDK6, cyclin D1 and phosphorylated kinase, and inhibited the growth of oral squamous cell carcinoma |
[96] | ||
| [97] | ||||
| Methylation | HMTs | Chaetocin | It plays an important role in the combination of drugs in cancer treatment | [83] |
|
BIX-01294 CUDC-101 |
Inhibits H3K9me2 expression Phage I study on intermediate or high-risk HNSCC patients |
[103] [113] |
||
| GSK-343 |
1. The tumor growth of tongue squamous cell carcinoma was significantly inhibited after DZNeP administration 2. DZNeP is effective against a variety of cancers, including HNSCC, either alone or in combination 3. Associated with HPV status |
[104] [105] [106] |
||
| DZNeP | ||||
| EPZ-568 | ||||
| HNMTs | TCP | Most are associated with hematologic disorders | [83] | |
| ORY-1001 | ||||
| INCB059872 | ||||
| IMG-7289 | ||||
| TAK-418 | ||||
| GSK-LSD1 |
1. Blocks EGF-induced proliferation 2. Weakens genes involved in carcinogenic properties Inhibits the growth of oral squamous cell carcinoma |
[112] | ||
Histone phosphorylation
Histone phosphorylation occurs at serine, threonine, and tyrosine residues across all histones, with a notable prevalence on histone H4 [30]. This modification is subject to reversible regulation by kinases and phosphatases. Phosphorylation decreases the positive charge of histones, potentially altering chromatin structure. A study of oral cancer cases within a Taiwanese cohort identified overexpression of the threonine protein kinase ARK2 (ARAF-related kinase 2). This upregulation in the nucleus was associated with adverse clinical outcomes, whereas cytoplasmic overexpression correlated with tumor size and cancer stage [41]. Consequently, ARK2 may serve as a valuable prognostic biomarker [41]. Additionally, research by Qi et al. established that ARK2 is overexpressed in head and neck squamous carcinoma patients, with elevated enzyme levels correlating with histological differentiation, cell proliferation, and metastasis of oral cancer. This highlights ARK2's critical role in the progression of oral squamous cell carcinoma [59]. Studies have shown that the EGFR-specific inhibitor cetuximab (Erbitux) can cause abnormal phosphorylation of tyrosine 1173 in HNSCC cell lines [60].
Histone ubiquitination and histone SUMOylation
Ubiquitin is a small regulatory protein comprising 76 amino acids, present in nearly all cell types. Histone ubiquitination represents a multi-step posttranslational modification process wherein ubiquitin attaches to a substrate protein. This process involves three critical stages: activation, binding, and attachment. Regulation of histone ubiquitination is mediated by three primary types of enzymes: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s). Ubiquitinizing, deubiquitinizing, and ubiquitin-modifying enzymes are integral to transcriptional regulation and DNA repair [61–63]. Evidence suggests that various histone ubiquitination-modifying enzymes exert either oncogenic or tumor-suppressive effects, indicating a significant connection between histone ubiquitination and cancer. For example, ubiquitin-specific protease 14 (USP14) is overexpressed in tongue cancer relative to adjacent noncancerous tissues and enhances proliferation and invasion of HNSCC in both in vivo and in vitro settings [64]. USP4, which facilitates TNF-α-induced apoptosis, also functions as an oncogene in head and neck squamous carcinoma [65]. Moreover, survival analyses have revealed that elevated expression of USP7 correlates with poor prognosis in head and neck squamous carcinoma [66]. PiR-has-23533 can bind to USP7 and promote its expression, thereby affecting the proliferation and apoptosis of HNSCC cells [66].
Histone SUMOylation represents a posttranslational modification analogous to histone ubiquitination, involving three principal enzymes: E1-activating enzyme, E2-binding enzyme, and E3 ligase. These enzymes facilitate the addition of small ubiquitin-like modifiers (SUMOs) to histone lysine residues, while SUMO-specific proteases (SENPs) can reverse this modification. Among the seven known SENPs in humans, SENP5 has been identified as particularly relevant to head and neck squamous carcinoma. Research indicates that SENP5 is overexpressed in oral squamous carcinoma specimens relative to normal epithelial cells, suggesting a correlation between SENP5 expression and the differentiation of oral squamous carcinoma [67, 68]. Furthermore, SUMO-1 has been shown to be overexpressed in human oral squamous carcinoma cell lines and patient tissues, potentially contributing to tumor cell proliferation [30].
Histone ADP ribosylation
ADP ribosylation of lysine residues represents a relatively infrequent histone modification, occurring in approximately 1% of histones, yet it is significantly pronounced in response to single DNA strand breaks. This modification impacts crucial biological processes such as DNA damage repair, transcription, and apoptosis [69]. The enzymes responsible for ADP ribosylation, known as poly(ADP-ribose) polymerases (PARPs), utilize nicotinamide adenine dinucleotide (NAD) to synthesize poly(ADP-ribose) (PAR) [70]. Studies have demonstrated that the extent of ADP ribosylation correlates with the level of malignancy in oral tissues [71]. In recent years, there has been an increasing focus on targeting ADP ribosylation in clinical trials for various cancers, including head and neck squamous carcinoma.
Histone crotonylation
Lysine crotonylation (Kcr) represents a novel posttranslational modification of histone proteins, which is reversibly regulated by protein crotonyltransferases and decrotonylases. This modification is implicated in various cellular processes, including metabolism and the cell cycle [72]. Studies have demonstrated that the expression of lysine crotonylation regulatory factors is associated with the progression of head and neck squamous carcinoma. Many of these factors are aberrantly expressed in HNSCC, potentially providing insights into the prognostic characteristics of the disease [73].
Histone 2-hydroxyisobutyrylation
Lysine 2-hydroxyisobutyrylation (Khib) is a recently identified posttranslational modification discovered via mass spectrometry (MS). This modification is implicated in the regulation of chromatin and various cellular processes [74]. Although the role of histone lysine 2-hydroxyisobutyrylation has been elucidated in the context of pancreatic cancer development [75], there is a lack of literature addressing Lysine 2-hydroxyisobutyrylation in head and neck squamous carcinoma. Further in-depth research is required to explore its potential relevance in this context.
Histone lactylation
Histone lactylation represents a novel histone modification characterized by the addition of lactoyl groups to lysine residues. This modification not only alters nucleosome structure but also profoundly affects chromatin dynamics and gene expression, playing a significant role in cellular metabolism, inflammatory responses, and embryonic development [76, 77]. Studies have demonstrated that lactate, following its entry into cancer cells through monocarboxylic acid transporter protein 1, facilitates the lactylation of histone H3K18. This modification stabilizes key intracellular regulators, such as hypoxia-inducible factor 1α, and activates associated transcriptional and signaling pathways, thus playing a critical role in various cancers, including cervical cancer [78]. Despite these findings, histone lactylation in head and neck squamous carcinoma remains underexplored and requires further comprehensive investigation.
Clinical significance of histone posttranslational modifications in HNSCC
In recent years, research into novel drug therapies for HNSCC has largely failed to produce effective new treatments. As a result, current management strategies remain limited to surgery, radiotherapy, and platinum-based chemotherapy, which are considered the standard first-line interventions [7]. While these treatments can provide some degree of relief, they do not entirely prevent the emergence of cancer cell resistance or mitigate the associated side effects. This scenario leads to high rates of recurrence and metastasis, a low five-year survival rate, and a generally poor prognosis [10]. Thus, there is an urgent need to explore innovative diagnostic and therapeutic approaches. Epigenetic modifications, due to their reversible nature, present promising new opportunities for drug development in the context of HNSCC [12].
Histone acetyltransferase inhibitors
Numerous experimental molecules have been developed as inhibitors of HATs. These include PU139, a broad-spectrum inhibitor; PCAF and p300 inhibitors such as isothiazolone, Garcinol, and hydroxybenzoquinone-based compounds like Embelin; the p300-specific inhibitor PU141 and the small molecule pyrazolinone C646; as well as the MYST family HAT Tip60 inhibitor TH1834, and anaxin acid and its derivative, 6-alkyl salicylate. Despite these developments, few histone acetyltransferase inhibitors have advanced to clinical trials. For instance, the CBP/p300 inhibitor CCS1477 is undergoing Phase I/II clinical trials for hematologic malignancies, prostate cancer, breast cancer, and non-small cell lung cancer. Additionally, a Phase I clinical trial evaluating the safety and maximum tolerated dose of the dual BET and CBP/p300 inhibitor EP31670 in advanced solid tumors was initiated in 2022 [83]. Nonetheless, there are currently no reported studies on histone acetyltransferase inhibitors specifically targeting HNSCC.
Histone deacetylase inhibitors
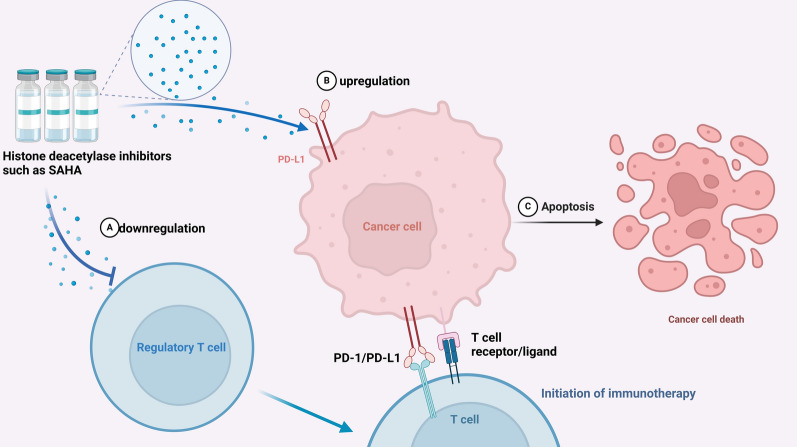
Factors within the tumor microenvironment, such as those secreted by endothelial cells, can induce acetylation in HNSCC cells. Nevertheless, the acetylation levels in these cancer cells remain comparatively low relative to those in normal oral mucosal cells [25]. These findings indicate that histone deacetylase inhibitors may serve as a novel target for anticancer therapy. As the development and application of histone deacetylase inhibitors advance, their therapeutic efficacy in various solid tumors increasingly depends on combination regimens [84]. Furthermore, histone deacetylase inhibitors have been shown to function as modulators of immunotherapy initiation by upregulating PD‐L1 expression and decreasing the population of regulatory T cells [85] (Fig. 1).
Fig. 1.
Modulation of Immunotherapy by Histone Deacetylase Inhibitors: Histone deacetylase inhibitors such as suberoylanilide hydroxamic acid downregulate regulatory T cells (A) and upregulate PD-L1 expression on cancer cells (B), enhancing the immunotherapy response. This leads to apoptosis (C) and cancer cell death, improving immunotherapy efficacy.
Among the first-generation histone deacetylase inhibitors, suberoylanilide hydroxamic acid (SAHA) was approved by the US Food and Drug Administration in 2006 for the treatment of cutaneous T-cell lymphoma, and research into its efficacy across various cancer types commenced subsequently [86]. A Phase II clinical trial found that vorinostat did not achieve a significant tumor response in patients with recurrent and metastatic HNSCC [87]. Nevertheless, vorinostat demonstrated notable therapeutic potential when utilized in combination therapies. For instance, the combination of vorinostat with cisplatin has shown considerable therapeutic promise in laryngeal squamous cell carcinoma [88]. Besides its role in inhibiting cell proliferation, inducing cell cycle arrest, and promoting apoptosis, vorinostat also enhances the sensitivity of squamous cell carcinoma cells to cisplatin [89]. Additionally, vorinostat has proven effective when combined with concurrent chemoradiotherapy [90] and has displayed positive results when used in conjunction with the EGFR inhibitor gefitinib [91]. Furthermore, a Phase II clinical trial assessing the combination of vorinostat and the anti-PD-1 agent pembrolizumab has revealed superior therapeutic activity in HNSCC compared to pembrolizumab monotherapy [92].
Romidesine, a bicyclic peptide isolated from Purple Bacillus, is an additional histone deacetylase inhibitor that has been approved by the US Food and Drug Administration for the treatment of cutaneous T-cell lymphoma, following the earlier approval of vorinostat. In a Phase II clinical trial focusing on patients with recurrent or metastatic HNSCC, romidesine effectively inhibited tumor-associated histone deacetylase. Nevertheless, it did not exhibit significant efficacy as a monotherapy and demonstrated greater clinical potential when combined with other therapeutic agents [21].
Valproic acid, traditionally used as an anticonvulsant for neurological conditions such as epilepsy, also functions as a Class I and II histone deacetylase inhibitor. It promotes cell cycle arrest and apoptosis. In vitro studies have shown that valproic acid is effective in treating HNSCC, especially when combined with cisplatin [93]. However, a Phase II clinical trial assessing the combination of valproic acid with standard platinum-based chemoradiotherapy was terminated due to considerable toxicity and severe side effects [80].
Trichostatin A (TSA) is a natural inhibitor of Class I and II HDACs, known for its ability to inhibit cell proliferation and induce G2/M phase arrest in tumor cells. This effect is mediated through modulation of cell cycle mediators, including the upregulation of p21 expression, reduction of cyclin B1, and alteration of cyclin E and cyclin A levels. Additionally, TSA decreases transcription factors E2F-1 and E2F-4 and reduces the hyperphosphorylation of retinoblastoma tumor suppressor proteins [94]. TSA-induced acetylation of H3K9 also mitigates chemoresistance to cisplatin in HNSCC cells, especially in the presence of active NF-κB signaling [95] 0.4-Phenylbutyric acid (PBA), initially introduced as an ammonia scavenger for urea cycle disorders, has been shown to induce apoptosis, differentiation, and cell cycle arrest. Sodium phenylbutyrate exhibits antitumor effects and can interfere with the EMT process [96]. When used as an adjunct to radiation therapy, PBA reduces oxidative stress, TNF-α levels, and oral mucositis, thereby promoting DNA repair and cell survival. The phenylbutyric acid-derived histone deacetylase inhibitor HDAC42 demonstrates superior antiproliferative activity relative to vorinostat and inhibits the growth of oral squamous cell carcinoma by increasing p21 and p27 levels, while decreasing G1/S phase molecules, including CDK6, cyclin D1, and phosphorylated kinases [97].
Experimental studies have demonstrated that the histone deacetylase inhibitor apxidin can effectively inhibit cell proliferation, impede the G2/M transition through the upregulation of p21, and induce autophagy, thereby exerting its tumor-suppressive effects. However, the role of apxidin in the context of HNSCC remains to be investigated [98].
Histone methyltransferase inhibitors
Chaetocin was the first histone lysine methyltransferase inhibitor identified, specifically targeting Drosophila 3–9 HMTs [99]. Since then, a range of inhibitors has been developed for HMTs, and numerous phase I/II clinical trials have been conducted across various cancers [100]. Importantly, the integration of histone methyltransferase inhibitors with other therapeutic modalities has garnered substantial attention in these clinical trials, underscoring their potential significance in cancer treatment [83].
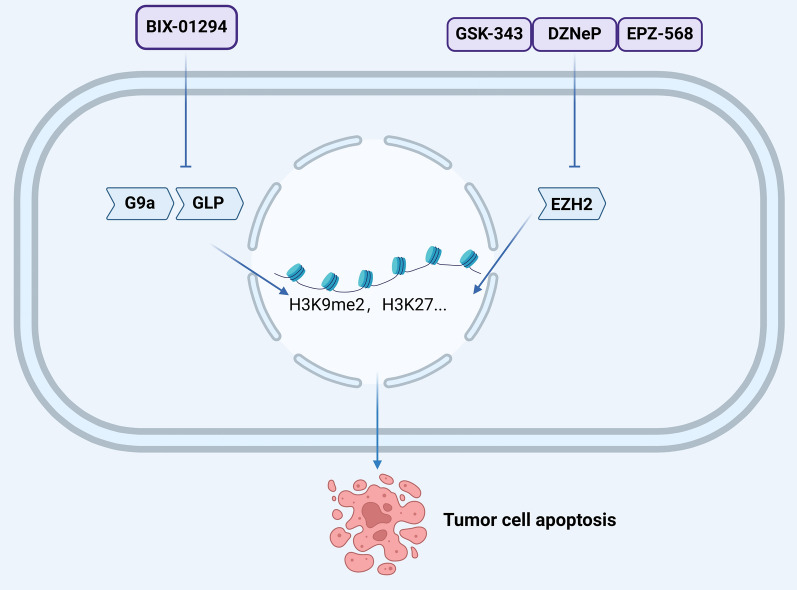
Numerous lysine and arginine methyltransferase inhibitors have been identified, investigated, and utilized, including those targeting G9a and EZH2 [101, 102]. BIX-01294, a selective inhibitor of G9a and GLP, impedes H3K9me2 expression and has demonstrated efficacy in reducing cell growth and proliferation, promoting cell phagocytosis, inhibiting colony formation, and suppressing tumor growth in mouse xenograft models [103]. EZH2, which is implicated in the trimethylation of H3K27, has several inhibitors, including GSK-343, DZNeP, and EPZ-568. Experimental data reveal that DZNeP significantly inhibits tumor growth in a tongue squamous cell carcinoma transplant model, induces targeted apoptosis, and enhances the epithelial phenotype [104]. DZNeP, whether used alone or in combination, has shown effectiveness across various cancers, including HNSCC [105]. GSK-343 effectively inhibits H3K27me3 in both HPV-positive and HPV-negative oral squamous cell carcinoma cell lines, DZNeP is effective against HPV-negative cell lines, whereas EPZ-568 exhibits no significant impact. These findings suggest that the antitumor efficacy of histone methyltransferase inhibitors may be influenced by HPV status [106] (Fig. 2). HPV is classified into low-risk and high-risk types based on their carcinogenic potential, primarily driven by two viral oncoproteins: E6 and E7 [107]. These proteins, through interactions with cellular partners, engage in the mutually dependent viral and cellular cycles within stratified squamous epithelia, concurrently inducing epigenetic alterations in both infected and malignant transforming cells. Oncoproteins E6 and E7 interact with and/or regulate the expression of numerous proteins involved in epigenetic regulation, including DNA methyltransferases, histone-modifying enzymes, and subunits of chromatin remodeling complexes, thereby affecting host cell transcriptional programs [108, 109] Upregulation of HPV oncoproteins promotes DNA methylation, further repressing the transcription of tumor suppressor genes. These epigenetic mechanisms play a crucial role in the regulation of both free and integrated HPV DNA expression, potentially driving early tumor progression [110, 111].
Fig. 2.
Impact of Histone Methyltransferase Inhibitors on Tumor Cell Apoptosis: BIX-01294, an inhibitor of H3K9 methyltransferase and GLP, reduces H3K9me2 expression, decreasing cell growth and tumor proliferation. H3K27 methyltransferase inhibitors like GSK-343, DZNeP, and EPZ-568 block H3K27 trimethylation, suppressing oncogenic gene expression and inducing tumor cell apoptosis.
Histone demethylase inhibitors
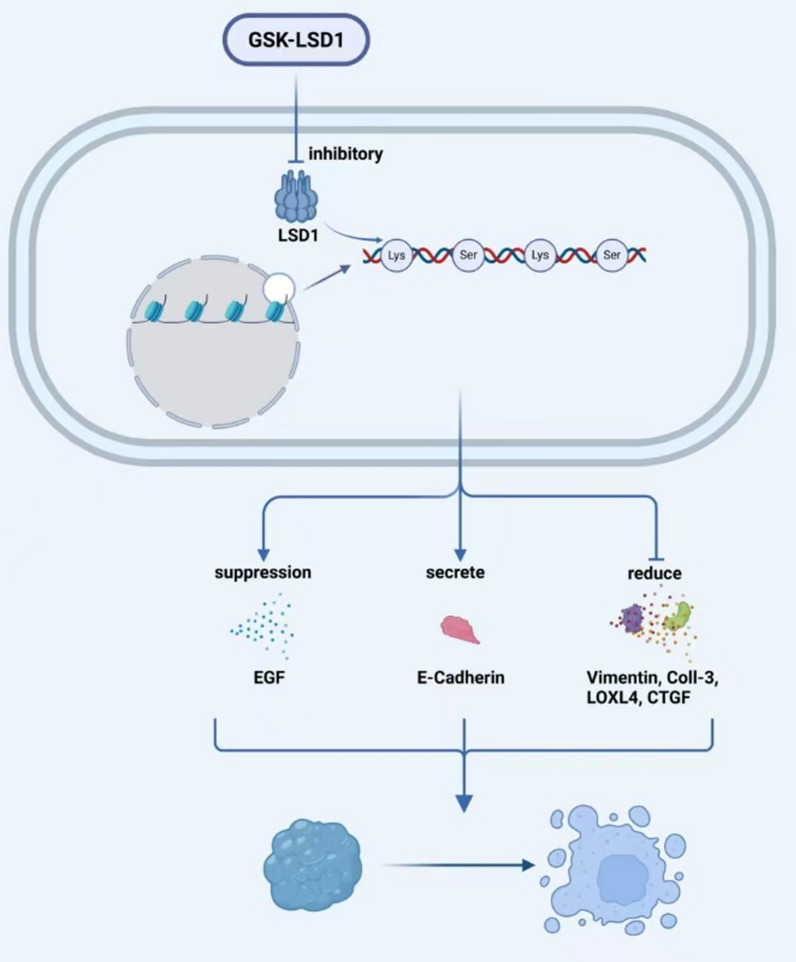
Among demethylases, KDM1, KDM4, KDM5, and KDM6 proteins are regarded as the most promising prognostic and therapeutic targets for head and neck cancer. Currently, a range of histone demethylase inhibitors targeting LSD1 has been developed, with several progressing to clinical trials to evaluate their efficacy and safety. These include the irreversible LSD1 inhibitor TCP, the highly selective covalent LSD1 inhibitor ORY-1001, the novel LSD1 inhibitors INCB059872 and IMG-7289, and the small molecule LSD1 inhibitor TAK-418. Nevertheless, most of these inhibitors are primarily associated with hematologic diseases, and there is a paucity of research concerning their application in HNSCC [83]. In contrast, inhibitors targeting other demethylases have rarely advanced to clinical trials when compared to LSD1 inhibitors. Notably, among histone demethylase inhibitors, only the LSD1 inhibitor GSK-LSD1 has been identified as capable of blocking EGF-induced proliferation, modulating oncogenic gene expression, and inhibiting oral squamous cell carcinoma growth by enhancing E-cadherin expression and reducing levels of vimentin, collagenase-3, lysine oxidase-like 4, and connective tissue growth factor [112] (Fig. 3).
Fig. 3.
Effect of LSD1 Inhibitor GSK-LSD1 on EGF-Induced Proliferation: GSK-LSD1, an LSD1 inhibitor, blocks EGF-induced proliferation and modulates oncogenic gene expression, reducing oral squamous cell carcinoma growth. It enhances E-cadherin expression and reduces vimentin, collagenase-3, LOXL4, and CTGF levels, thereby suppressing tumor growth.
Discussion and conclusion
This review provides a comprehensive overview of the relationship between HPTMs and HNSCC, addressing modifications such as acetylation, methylation, phosphorylation, ubiquitination, vanoylation, ADP ribosylation, crotonylation, 2-hydroxyisobutyrylation, and lactylation. The discussion then shifts to the advancements in utilizing these modifications for the treatment of HNSCC, with a particular focus on histone deacetylase inhibitors. The reversible nature of epigenetic modifications renders them promising therapeutic targets for cancer, offering potential benefits in overcoming the limitations of traditional treatment methods or enhancing their efficacy, thereby holding considerable clinical research potential. Nonetheless, it is important to acknowledge that current research on histone PTMs in HNSCC is still in its nascent stages. The exploration of certain novel modification methods such as lysine 2-hydroxyisobutyrylation and lactylation and their associations with HNSCC remains relatively limited, and many underlying mechanisms and targets are not yet fully elucidated. It cannot be denied that in the future, there will be more research on epigenetics in HNSCC, which will provide new ideas for the treatment of HNSCC and bring more effective and less toxic treatment methods for HNSCC patients.
Abbreviations
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial–mesenchymal transition
- HNSCC
Head and neck squamous cell carcinoma
- HATs
Histone acetyltransferases
- HDACs
Histone deacetylases
- HDMTs
Histone demethylases
- HMTs
Histone methyltransferases
- MS
Mass spectrometry
- NAD
Nicotinamide adenine dinucleotide
- ncRNA
Noncoding RNA
- PBA
4-Phenylbutyric acid
- PTM
Posttranslational modification
- SUMOs
Small ubiquitin-like modifiers
- SAHA
Suberoylanilide hydroxamic acid
- TSA
Trichostatin A
- USP14
Ubiquitin-specific protease 14
Author contributions
YX, XW, SY and YQ designed this study. YX, YZ, YH, JT and YQ reviewed and revised the manuscript. YX and JT drafted the original manuscript. YX and YZ designed and completed the figures. All authors approved the final manuscript submission and agreed to be responsible for all aspects of the work.
Funding
This research was funded by the National Natural Science Foundation of China (No.82303133); China Postdoctoral Science Foundation (No. 2022M723559); Natural Science Foundation of Hunan Province (No. 2022JJ30963, NO.2021JJ40951); Project of Hunan Health Commission (No. B202307017799, No. B202309018525).
Data availability
No datasets were generated or analyzed during the current study.
Declarations
Ethics approval and consent to participation
Not applicable.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xingwei Wang, Email: 371156510@qq.com.
Yuexiang Qin, Email: 602466@csu.edu.cn.
References:
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–53. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 4.Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48(11):3282–7. [PubMed] [Google Scholar]
- 5.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: An emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110(7):1429–35. [DOI] [PubMed] [Google Scholar]
- 6.Braakhuis BJ, Brakenhoff RH, Leemans CR. Treatment choice for locally advanced head and neck cancers on the basis of risk factors: biological risk factors. Ann Oncol. 2012;23(Suppl 10):x173–7. [DOI] [PubMed] [Google Scholar]
- 7.Pfister DG, Spencer S, Adelstein D, Adkins D, Anzai Y, Brizel DM, et al. Head and neck cancers, Version 22020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2020;18(7):873–98. [DOI] [PubMed] [Google Scholar]
- 8.Cui L, Lu Y, Zheng J, Guo B, Zhao X. ACTN1 promotes HNSCC tumorigenesis and cisplatin resistance by enhancing MYH9-dependent degradation of GSK-3β and integrin β1-mediated phosphorylation of FAK. J Exp Clin Cancer Res. 2023;42(1):335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Guo B, Sun W, Yu J, Cui L. Targeting squalene epoxidase confers metabolic vulnerability and overcomes chemoresistance in HNSCC. Adv Sci (Weinh). 2023;10(27): e2206878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow LQM. Head and neck cancer. N Engl J Med. 2020;382(1):60–72. [DOI] [PubMed] [Google Scholar]
- 11.Davalos V, Esteller M. Cancer epigenetics in clinical practice. CA Cancer J Clin. 2023;73(4):376–424. [DOI] [PubMed] [Google Scholar]
- 12.Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer. 2021;21(7):413–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51(5):786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Zhang Q, Zhang Y, Han J. The role of histone modification in dna replication-coupled nucleosome assembly and cancer. Int J Mol Sci. 2023;24(5). [DOI] [PMC free article] [PubMed]
- 15.Ibarra-Morales D, Rauer M, Quarato P, Rabbani L, Zenk F, Schulte-Sasse M, et al. Histone variant H2A.Z regulates zygotic genome activation. Nat Commun. 2021;12(1):7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393(6685):599–601. [DOI] [PubMed] [Google Scholar]
- 17.Qian H, Zhu M, Tan X, Zhang Y, Liu X, Yang L. Super-enhancers and the super-enhancer reader BRD4: tumorigenic factors and therapeutic targets. Cell Death Discov. 2023;9(1):470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu SJ, Furlan SN, Mihalas AB, Kaya-Okur HS, Feroze AH, Emerson SN, et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat Biotechnol. 2021;39(7):819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, et al. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49(2):310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida TFA, Oliveira SR, Mayra da Silva J, Fernandes de Oliveira AL, de Lourdes CZ, Menezes HC, et al. Effects of high-dose bisphenol A on the mouse oral mucosa: a possible link with oral cancers. Environ Pollut. 2021;286:117296. [DOI] [PubMed] [Google Scholar]
- 21.Haigentz M Jr, Kim M, Sarta C, Lin J, Keresztes RS, Culliney B, et al. Phase II trial of the histone deacetylase inhibitor romidepsin in patients with recurrent/metastatic head and neck cancer. Oral Oncol. 2012;48(12):1281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang JQ, Yan FQ, Wang LH, Yin WJ, Chang TY, Liu JP, et al. Identification of new hypoxia-regulated epithelial-mesenchymal transition marker genes labeled by H3K4 acetylation. Genes Chromosom Cancer. 2020;59(2):73–83. [DOI] [PubMed] [Google Scholar]
- 23.Caponigro F, Di Gennaro E, Ionna F, Longo F, Aversa C, Pavone E, et al. Phase II clinical study of valproic acid plus cisplatin and cetuximab in recurrent and/or metastatic squamous cell carcinoma of Head and Neck-V-CHANCE trial. BMC Cancer. 2016;16(1):918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohd-Sarip A, Teeuwssen M, Bot AG, De Herdt MJ, Willems SM, Baatenburg de Jong RJ, et al. DOC1-dependent recruitment of NURD reveals antagonism with SWI/SNF during epithelial-mesenchymal transition in oral cancer cells. Cell Rep. 2017;20(1):61–75. [DOI] [PubMed] [Google Scholar]
- 25.Giudice FS, Pinto DS Jr, Nör JE, Squarize CH, Castilho RM. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE. 2013;8(3): e58672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard B, Brand TM, O’Keefe RA, Lee ED, Zeng Y, Kemmer JD, et al. BET inhibition overcomes receptor tyrosine kinase-mediated cetuximab resistance in HNSCC. Cancer Res. 2018;78(15):4331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryu HW, Shin DH, Lee DH, Choi J, Han G, Lee KY, et al. HDAC6 deacetylates p53 at lysines 381/382 and differentially coordinates p53-induced apoptosis. Cancer Lett. 2017;391:162–71. [DOI] [PubMed] [Google Scholar]
- 28.Sakuma T, Uzawa K, Onda T, Shiiba M, Yokoe H, Shibahara T, et al. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int J Oncol. 2006;29(1):117–24. [PubMed] [Google Scholar]
- 29.He L, Gao L, Shay C, Lang L, Lv F, Teng Y. Histone deacetylase inhibitors suppress aggressiveness of head and neck squamous cell carcinoma via histone acetylation-independent blockade of the EGFR-Arf1 axis. J Exp Clin Cancer Res. 2019;38(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaździcka J, Gołąbek K, Strzelczyk JK, Ostrowska Z. Epigenetic modifications in head and neck cancer. Biochem Genet. 2020;58(2):213–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang K, Prasse C, Greenberg MM. Effect of histone lysine methylation on DNA lesion reactivity in nucleosome core particles. Chem Res Toxicol. 2019;32(5):910–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SH, Kim O, Kim HJ, Hwangbo C, Lee JH. Epigenetic regulation of TGF-β-induced EMT by JMJD3/KDM6B histone H3K27 demethylase. Oncogenesis. 2021;10(2):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Li W, Zang W, Liu Z, Xu X, Yu H, et al. JMJD2B promotes epithelial-mesenchymal transition by cooperating with β-catenin and enhances gastric cancer metastasis. Clin Cancer Res. 2013;19(23):6419–29. [DOI] [PubMed] [Google Scholar]
- 35.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107(3):323–37. [DOI] [PubMed] [Google Scholar]
- 36.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HW, Kim S, Paik WK. S-adenosylmethionine: protein-arginine methyltransferase. Purification and mechanism of the enzyme. Biochemistry. 1977;16(1):78–85. [DOI] [PubMed] [Google Scholar]
- 38.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A. 2011;108(14):5718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li KC, Hua KT, Lin YS, Su CY, Ko JY, Hsiao M, et al. Inhibition of G9a induces DUSP4-dependent autophagic cell death in head and neck squamous cell carcinoma. Mol Cancer. 2014;13:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He C, Sun J, Liu C, Jiang Y, Hao Y. Elevated H3K27me3 levels sensitize osteosarcoma to cisplatin. Clin Epigenet. 2019;11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JH, Yeh KT, Yang YM, Chang JG, Lee HE, Hung SY. High expressions of histone methylation- and phosphorylation-related proteins are associated with prognosis of oral squamous cell carcinoma in male population of Taiwan. Med Oncol. 2013;30(2):513. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Tang S, Zheng Q, Li J, Jiang H, Lu H, et al. The competitive mechanism of EZH1 and EZH2 in promoting oral squamous cell carcinoma. Exp Cell Res. 2024;436(1): 113957. [DOI] [PubMed] [Google Scholar]
- 43.Mancuso M, Matassa DS, Conte M, Colella G, Rana G, Fucci L, et al. H3K4 histone methylation in oral squamous cell carcinoma. Acta Biochim Pol. 2009;56(3):405–10. [PubMed] [Google Scholar]
- 44.Yang D, Liang T, Gu Y, Zhao Y, Shi Y, Zuo X, et al. Protein N-arginine methyltransferase 5 promotes the tumor progression and radioresistance of nasopharyngeal carcinoma. Oncol Rep. 2016;35(3):1703–10. [DOI] [PubMed] [Google Scholar]
- 45.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 2014;5(19):8906–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleszcz R, Skalski M, Krajka-Kuźniak V, Paluszczak J. The inhibitors of KDM4 and KDM6 histone lysine demethylases enhance the anti-growth effects of erlotinib and HS-173 in head and neck cancer cells. Eur J Pharm Sci. 2021;166: 105961. [DOI] [PubMed] [Google Scholar]
- 47.Narayanan SP, Singh S, Gupta A, Yadav S, Singh SR, Shukla S. Integrated genomic analyses identify KDM1A’s role in cell proliferation via modulating E2F signaling activity and associate with poor clinical outcome in oral cancer. Cancer Lett. 2015;367(2):162–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ai Y, Wu S, Zou C, Wei H. Circular RNA circFOXO3 regulates KDM2A by targeting miR-214 to promote tumor growth and metastasis in oral squamous cell carcinoma. J Cell Mol Med. 2022;26(6):1842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peta E, Sinigaglia A, Masi G, Di Camillo B, Grassi A, Trevisan M, et al. HPV16 E6 and E7 upregulate the histone lysine demethylase KDM2B through the c-MYC/miR-146a-5p axys. Oncogene. 2018;37(12):1654–68. [DOI] [PubMed] [Google Scholar]
- 50.Brauchle M, Yao Z, Arora R, Thigale S, Clay I, Inverardi B, et al. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS ONE. 2013;8(4): e60549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125(4):691–702. [DOI] [PubMed] [Google Scholar]
- 52.Krishnan S, Trievel RC. Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure. 2013;21(1):98–108. [DOI] [PubMed] [Google Scholar]
- 53.Hu D, Jablonowski C, Cheng PH, AlTahan A, Li C, Wang Y, et al. KDM5A regulates a translational program that controls p53 protein expression. iScience. 2018;9:84–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams ST, Walport LJ, Hopkinson RJ, Madden SK, Chowdhury R, Schofield CJ, et al. Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics. 2014;9(12):1596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–7. [DOI] [PubMed] [Google Scholar]
- 56.Li S, Ali S, Duan X, Liu S, Du J, Liu C, et al. JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 2018;23(2):389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mondal S, Thompson PR. Protein Arginine deiminases (PADs): biochemistry and chemical biology of protein citrullination. Acc Chem Res. 2019;52(3):818–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, Wei L, Luo M, Wang X, Zhu C, Huang H, et al. LINC00324 suppresses apoptosis and autophagy in nasopharyngeal carcinoma through upregulation of PAD4 and activation of the PI3K/AKT signaling pathway. Cell Biol Toxicol. 2022;38(6):995–1011. [DOI] [PubMed] [Google Scholar]
- 59.Qi G, Ogawa I, Kudo Y, Miyauchi M, Siriwardena BS, Shimamoto F, et al. Aurora-B expression and its correlation with cell proliferation and metastasis in oral cancer. Virchows Arch. 2007;450(3):297–302. [DOI] [PubMed] [Google Scholar]
- 60.Mandic R, Rodgarkia-Dara CJ, Zhu L, Folz BJ, Bette M, Weihe E, et al. Treatment of HNSCC cell lines with the EGFR-specific inhibitor cetuximab (Erbitux) results in paradox phosphorylation of tyrosine 1173 in the receptor. FEBS Lett. 2006;580(20):4793–800. [DOI] [PubMed] [Google Scholar]
- 61.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10(5):319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17(10):626–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10(11):755–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X, Wu J, Chen Y, Ye D, Lei H, Xu H, et al. Ubiquitin-specific protease 14 regulates cell proliferation and apoptosis in oral squamous cell carcinoma. Int J Biochem Cell Biol. 2016;79:350–9. [DOI] [PubMed] [Google Scholar]
- 65.Hou X, Wang L, Zhang L, Pan X, Zhao W. Ubiquitin-specific protease 4 promotes TNF-α-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Lett. 2013;587(4):311–6. [DOI] [PubMed] [Google Scholar]
- 66.Hu H, Lu J, Xu M, Wang J, Zhang Y, Yang S, et al. PiR-hsa-23533 promotes malignancy in head and neck squamous cell carcinoma via USP7. Transl Oncol. 2024;45: 101990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng Y, Li X. Expression and prognosis analysis of SUMOylation regulators in oral squamous cell carcinoma based on high-throughput sequencing. Front Genet. 2021;12: 671392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding X, Sun J, Wang L, Li G, Shen Y, Zhou X, et al. Overexpression of SENP5 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. 2008;20(5):1041–5. [PubMed] [Google Scholar]
- 69.Gupte R, Nandu T, Kraus WL. Nuclear ADP-ribosylation drives IFNγ-dependent STAT1α enhancer formation in macrophages. Nat Commun. 2021;12(1):3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das BR. Increased ADP-ribosylation of histones in oral cancer. Cancer Lett. 1993;73(1):29–34. [DOI] [PubMed] [Google Scholar]
- 72.Liu S, Xue C, Fang Y, Chen G, Peng X, Zhou Y, et al. Global involvement of lysine crotonylation in protein modification and transcription regulation in rice. Mol Cell Proteomics. 2018;17(10):1922–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang L, Yin X, Zhang H, Zhang X, Cao Z, Zhou M, et al. Development and validation of a prognostic signature based on the lysine crotonylation regulators in head and neck squamous cell carcinoma. Biomed Res Int. 2023;2023:4444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang H, Luo Z, Qi S, Huang J, Xu P, Wang X, et al. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 2018;28(1):111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu Y, Li X, Zhao K, Qiu P, Deng Z, Yao W, et al. Global landscape of 2-hydroxyisobutyrylation in human pancreatic cancer. Front Oncol. 2022;12:1001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell. 2016;62(2):181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meng Q, Sun H, Zhang Y, Yang X, Hao S, Liu B, et al. Lactylation stabilizes DCBLD1 activating the pentose phosphate pathway to promote cervical cancer progression. J Exp Clin Cancer Res. 2024;43(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le JM, Squarize CH, Castilho RM. Histone modifications: Targeting head and neck cancer stem cells. World J Stem Cells. 2014;6(5):511–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suchanti S, Stephen BJ, Awasthi S, Awasthi SK, Singh G, Singh A, et al. Harnessing the role of epigenetic histone modification in targeting head and neck squamous cell carcinoma. Epigenomics. 2022;14(5):279–93. [DOI] [PubMed] [Google Scholar]
- 81.Liu S, Ye D, Guo W, Yu W, He Y, Hu J, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6(9):6887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dorna D, Paluszczak J. The emerging significance of histone lysine demethylases as prognostic markers and therapeutic targets in head and neck cancers. Cells. 2022;11(6). [DOI] [PMC free article] [PubMed]
- 83.Wang N, Ma T, Yu B. Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct Target Ther. 2023;8(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng F, Zheng B, Wang J, Zhao G, Yao Z, Niu Z, et al. Comprehensive analysis of a new prognosis signature based on histone deacetylases in clear cell renal cell carcinoma. Cancer Med. 2021;10(18):6503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen L, Ciesielski M, Ramakrishnan S, Miles KM, Ellis L, Sotomayor P, et al. Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLoS ONE. 2012;7(1): e30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12(10):1247–52. [DOI] [PubMed] [Google Scholar]
- 87.Blumenschein GR Jr, Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL, et al. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest New Drugs. 2008;26(1):81–7. [DOI] [PubMed] [Google Scholar]
- 88.Grabarska A, Łuszczki JJ, Nowosadzka E, Gumbarewicz E, Jeleniewicz W, Dmoszyńska-Graniczka M, et al. Histone deacetylase inhibitor SAHA as potential targeted therapy agent for larynx cancer cells. J Cancer. 2017;8(1):19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pan CH, Chang YF, Lee MS, Wen BC, Ko JC, Liang SK, et al. Vorinostat enhances the cisplatin-mediated anticancer effects in small cell lung cancer cells. BMC Cancer. 2016;16(1):857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teknos TN, Grecula J, Agrawal A, Old MO, Ozer E, Carrau R, et al. A phase 1 trial of Vorinostat in combination with concurrent chemoradiation therapy in the treatment of advanced staged head and neck squamous cell carcinoma. Invest New Drugs. 2019;37(4):702–10. [DOI] [PubMed] [Google Scholar]
- 91.Citro S, Bellini A, Miccolo C, Ghiani L, Carey TE, Chiocca S. Synergistic antitumour activity of HDAC inhibitor SAHA and EGFR inhibitor gefitinib in head and neck cancer: a key role for ΔNp63α. Br J Cancer. 2019;120(6):658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rodriguez CP, Wu QV, Voutsinas J, Fromm JR, Jiang X, Pillarisetty VG, et al. A phase II trial of pembrolizumab and vorinostat in recurrent metastatic head and neck squamous cell carcinomas and salivary gland cancer. Clin Cancer Res. 2020;26(4):837–45. [DOI] [PubMed] [Google Scholar]
- 93.Erlich RB, Rickwood D, Coman WB, Saunders NA, Guminski A. Valproic acid as a therapeutic agent for head and neck squamous cell carcinomas. Cancer Chemother Pharmacol. 2009;63(3):381–9. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki T, Yokozaki H, Kuniyasu H, Hayashi K, Naka K, Ono S, et al. Effect of trichostatin A on cell growth and expression of cell cycle- and apoptosis-related molecules in human gastric and oral carcinoma cell lines. Int J Cancer. 2000;88(6):992–7. [DOI] [PubMed] [Google Scholar]
- 95.Almeida LO, Abrahao AC, Rosselli-Murai LK, Giudice FS, Zagni C, Leopoldino AM, et al. NFκB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC). FEBS Open Bio. 2014;4:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qian K, Sun L, Zhou G, Ge H, Meng Y, Li J, et al. Sodium phenylbutyrate inhibits tumor growth and the epithelial-mesenchymal transition of oral squamous cell carcinoma in vitro and in vivo. Cancer Biother Radiopharm. 2018;33(4):139–45. [DOI] [PubMed] [Google Scholar]
- 97.Bai LY, Chiu CF, Pan SL, Sargeant AM, Shieh TM, Wang YC, et al. Antitumor activity of a novel histone deacetylase inhibitor (S)-HDAC42 in oral squamous cell carcinoma. Oral Oncol. 2011;47(12):1127–33. [DOI] [PubMed] [Google Scholar]
- 98.Ahn MY, Ahn SG, Yoon JH. Apicidin, a histone deaceylase inhibitor, induces both apoptosis and autophagy in human oral squamous carcinoma cells. Oral Oncol. 2011;47(11):1032–8. [DOI] [PubMed] [Google Scholar]
- 99.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005;1(3):143–5. [DOI] [PubMed] [Google Scholar]
- 100.Morschhauser F, Tilly H, Chaidos A, McKay P, Phillips T, Assouline S, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020;21(11):1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Piunti A, Meghani K, Yu Y, Robertson AG, Podojil JR, McLaughlin KA, et al. Immune activation is essential for the antitumor activity of EZH2 inhibition in urothelial carcinoma. Sci Adv. 2022;8(40):eabo8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25(3):473–81. [DOI] [PubMed] [Google Scholar]
- 103.Ren A, Qiu Y, Cui H, Fu G. Inhibition of H3K9 methyltransferase G9a induces autophagy and apoptosis in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2015;459(1):10–7. [DOI] [PubMed] [Google Scholar]
- 104.Gannon OM, Merida de Long L, Endo-Munoz L, Hazar-Rethinam M, Saunders NA. Dysregulation of the repressive H3K27 trimethylation mark in head and neck squamous cell carcinoma contributes to dysregulated squamous differentiation. Clin Cancer Res. 2013;19(2):428–41. [DOI] [PubMed] [Google Scholar]
- 105.Bourguignon LY, Wong G, Shiina M. Up-regulation of histone methyltransferase, DOT1L, by matrix hyaluronan promotes microRNA-10 expression leading to tumor cell invasion and chemoresistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2016;291(20):10571–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lindsay CD, Kostiuk MA, Harris J, O’Connell DA, Seikaly H, Biron VL. Efficacy of EZH2 inhibitory drugs in human papillomavirus-positive and human papillomavirus-negative oropharyngeal squamous cell carcinomas. Clin Epigenet. 2017;9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schwarz E, Freese UK, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, et al. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314(6006):111–4. [DOI] [PubMed] [Google Scholar]
- 108.Durzynska J, Lesniewicz K, Poreba E. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res. 2017;772:36–50. [DOI] [PubMed] [Google Scholar]
- 109.Kim MJ, Lee HJ, Choi MY, Kang SS, Kim YS, Shin JK, et al. UHRF1 induces methylation of the TXNIP promoter and down-regulates gene expression in cervical cancer. Mol Cells. 2021;44(3):146–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun Y, et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m(6)A-MYC expression. Int J Biol Sci. 2022;18(2):507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zygouras I, Leventakou D, Pouliakis A, Panagiotou S, Tsakogiannis D, Konstantopoulos G, et al. Human papillomavirus 16 DNA methylation patterns and investigation of integration status in head and neck cancer cases. Int J Mol Sci. 2023;24(19). [DOI] [PMC free article] [PubMed]
- 112.Alsaqer SF, Tashkandi MM, Kartha VK, Yang YT, Alkheriji Y, Salama A, et al. Inhibition of LSD1 epigenetically attenuates oral cancer growth and metastasis. Oncotarget. 2017;8(43):73372–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Galloway TJ, Wirth LJ, Colevas AD, Gilbert J, Bauman JE, Saba NF, et al. A phase I study of CUDC-101, a multitarget inhibitor of HDACs, EGFR, and HER2, in combination with chemoradiation in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2015;21(7):1566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analyzed during the current study.