Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive cancers, highly resistant to standard chemotherapy and immunotherapy. Regulatory T cells (Tregs) expressing tumor necrosis factor α receptor 2 (TNFR2) contribute to immunosuppression in PDAC. Treg infiltration correlates with poor survival and tumor progression in patients with PDAC. We hypothesized that TNFR2 inhibition using a blocking monoclonal antibody (mAb) could shift the Treg-effector T cell balance in PDAC, thus enhancing antitumoral responses.
Method
To support this hypothesis, we first described TNFR2 expression in a cohort of 24 patients with PDAC from publicly available single-cell analysis data. In orthotopic and immunocompetent mouse models of PDAC, we also described the immune environment of PDAC after immune cell sorting and single-cell analysis. The modifications of the immune environment before and after anti-TNFR2 mAb treatment were evaluated as well as the effect on tumor progression.
Results
Patients with PDAC exhibited elevated TNFR2 expression in Treg, myeloid cells and endothelial cells and lower level in tumor cells. By flow cytometry and single-cell RNA-seq analysis, we identified two Treg populations in orthotopic mouse models: Resting and activated Tregs. The anti-TNFR2 mAb selectively targeted activated tumor-infiltrating Tregs, reducing T cell exhaustion markers in CD8+ T cells. However, anti-TNFR2 treatment alone had limited efficacy in activating CD8+ T cells and only slightly reduced the tumor growth. The combination of the anti-TNFR2 mAb with agonistic anti-CD40 mAb promoted stronger T cell activation, tumor growth inhibition, and improved survival and immunological memory in PDAC-bearing mice.
Conclusion
Our data suggest that combining a CD40 agonist with a TNFR2 antagonist represents a promising therapeutic strategy for patients with PDAC.
Keywords: Immune modulatory, T regulatory cell - Treg, Tumor infiltrating lymphocyte - TIL, Tumor microenvironment - TME, Adenocarcinoma
WHAT IS ALREADY KNOWN ON THIS TOPIC.
WHAT THIS STUDY ADDS
We show that TNFR2 is mainly expressed by Tregs and myeloid cells in patients with PDAC. In an orthotopic mouse model of PDAC, administration of an anti-TNFR2 monoclonal antibody (mAb) induces a reduction of Tregs and T cell compartment with an exhausted phenotype. Combined with an agonistic CD40 mAb, the anti-TNFR2 treatment induces a potent antitumor effect associated with an immune memory establishment.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study highlights the role of TNFR2 in the antitumor immune response in PDAC and identifies TNFR2, alone or in combination therapy, as a new target in the therapeutic arsenal for the treatment of tumors refractory to immune checkpoint inhibitors.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the second leading gastrointestinal cancer in incidence in France. The overall 5-year survival rate for this malignancy is less than 12% and has not evolved in the last 10 years. In addition, the incidence of PDAC is consistently increasing and is expected to become the second leading cause of cancer death in the USA by 2030.1 Currently, there is no curative treatment for advanced PDAC. To date, surgical resection offers the only curative option, restricted to solely a small fraction of 20% of patients harboring a localized tumor at the time of diagnosis.2 For the vast majority of patients diagnosed with advanced or metastatic PDAC, chemotherapy protocols FOLFIRINOX or gemcitabine plus nab-paclitaxel association provide the only available options.3 PDAC treatment thus remains a true challenge in crucial need for new therapeutic development.4 In recent years, immunotherapy has emerged as a new tool in cancer treatment. Especially immune checkpoint inhibitors (ICIs) targeting programmed cell death protein-1 (PD-1)/programmed death-ligand 1 (PD-L1) axis and the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) molecule showed positive results in melanoma and non-small cell lung cancer. Unfortunately, such strategies have shown little to no efficacy in PDAC.5 The resistance of PDAC is partly due to the intrinsic resistance of cancer cells to therapy, their adaptive plasticity and the tumor microenvironment (TME).6 PDAC stroma is composed of a high proportion of cancer-associated fibroblasts and a fibrotic matrix that, together with tumor cells, promote tumor progression, chemoresistance and immunosuppressive signals.7 TME is characterized by poor infiltration of T lymphocytes.4 7 Moreover, the highly fibrotic and hypoxic stroma of PDAC creates a physical barrier to cytotoxic T cell infiltration8 and promotes immune suppressive cell accumulation such as tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs) thus contributing to immune evasion.7 9
An increased abundance of Tregs has been observed in PDAC tissues and is associated with poor prognosis and decreased survival.10,12 In particular, poor T cell infiltration and Treg enrichment have been described as an “immune escape” phenotype and can affect the prognostic of PDAC.13 14 Notably, Treg cell ablation in a mouse model of PDAC resulted in the control of tumor growth dependent on cytotoxic CD8+ T cells (CTL).15 Tregs highly express the tumor necrosis factor α (TNFα) receptor 2 (TNFR2), and the TNFα/TNFR2 axis was shown to play a crucial role in Treg stability, expansion and function in both mice and humans.16,20 Moreover, an accumulation of TNFR2+ Tregs with superior suppressive capacities have been found across a variety of cancers.21 Hence, blocking the TNFα/TNFR2 pathway has emerged as a new immunotherapy strategy to target Tregs in cancer.22 23 In addition, TNFR2 blockade was shown to not only inhibit Treg proliferation but also tumor cells expressing TNFR2 in ovarian cancer and Sezary syndrome.24 25
Anti-TNFR2 treatment in preclinical experiments has already been shown to decrease the frequency of tumor-infiltrated Tregs and promote CD8+ T cell infiltration and interferon (IFN)-γ expression in subcutaneous mouse models of colon and breast tumors.26 27 Here, we tested whether blockade of the TNFR2 pathway in PDAC could reverse the balance between Tregs and effector T cells and trigger an effective antitumor response.
Materials and methods
Cell culture
Murine pancreatic cancer cells mPDAC, were isolated and validated, as previously described28 from tumor-bearing p48cre, KrasLSL_G12D, p53R172H/+, Ink4a/Arfflox/+ FVB/n mice (kindly provided and validated by D HANAHAN, EPFL, Lausanne, Switzerland), and were cultured in DMEM (41965-039, Gibco) 10% fetal bovine serum (FBS). Murine pancreatic cancer cell line Panc02 (kindly provided by R Ronca (University of Brescia) in 2016) was cultured in RPMI 10% FBS and pyruvate 0,1%.
Tumor mouse model
Female FVB/n or C57BL/6j mice of 10–14 weeks of age were obtained from Janvier Labs (France). Male NSG (NOD/scid/IL-2Rγ−/−) mice were obtained from our own breeding. All in vivo experiments were carried out with the approval of the appropriate ethical committee (authorization number #24225–202001310859869, #29529–2020110222005935 and #11511–2017092610086943). All experiments were performed in the same animal facility (IMRB).
For tumor orthotopic mouse models
10–14 weeks of age FVB/n or NSG mice were injected orthotopically in the pancreas with mPDAC cells (103 cells/mouse in 50 µL) or C57BL/6j mice with Panc02 (105 cells/mouse in 50 µL), as previously described.28
For tumor ectopic model
10–14 weeks of age FVB/n mice were injected subcutaneously with mPDAC cells (105 cells/mouse in 100 µL) on the right flank.
Antibody treatments
Polyclonal Armenian hamster IgG (ref: BE0091), anti-mouse-TNFR2 (clone TR75-54.7, ref: BE0247), anti-mouse CD40 agonist (clone FGK4.5/FGK45, ref: BE0016-2), anti-OX40 (BX-BE0031) mAbs were purchased from Bio X Cell (Euromedex, France) and anti-CXC chemokine receptor 4 (CXCR4) from Abcam (ab120718). After orthotopic engraftment, FVB/n and C57Bl/6j mice received three intraperitoneal injections of anti-TNFR2 mAb (500 µg in 100 µL) on days 11, 13, and 15 or five injections on days 8, 11, 13, 15 and 18 respectively. Before treatments, animals were randomized in cages and the same investigator did the measures. For combination therapy, mice received three intraperitoneal injections of CD40 agonist mAb (100 µg in 100 µL) on days 11, 13, and 15. Control groups received either IgG control (500 µg), phosphate-buffered saline (PBS) or were untreated (as indicated on figures’ legends). Potential confounders as order administration of the therapeutic molecules were not evaluated. After subcutaneous engraftment, FVB/n mice received three intraperitoneal injections of anti-TNFR2 mAb or IgG control (500 µg in 100 µL) on days 8, 11, and 13. Tumor growth was monitored after reaching a volume of approximately 150–200 mm3. Mice were euthanized on day 18 or 21 after orthotopic engraftment (as indicated on figures’ legends) or day 20 after subcutaneous engraftment. At this time, total tumor burden was quantified as previously described,28 tumors and draining lymph nodes were collected for flow cytometry analysis.
For survival experiment, FVB/n (mPDAC) and C57Bl/6j (Panc02) mice received anti-TNFR2 and/or CD40 agonist on days 11, 13, 15, 20, 27 and 34 after orthotopic engraftment. Control groups received PBS. Mice were euthanized when the clinical score reached a limit established by a grid of symptoms (ventral swelling, slimming, anemia and cachexia). After 64 days, tumor presence in C57Bl/6j (Panc02) was assessed by echography. After 81 days, mice still alive and naïves mice were injected subcutaneously with mPDAC cells (105 cells/mouse in 100 µL) on the right flank and tumor growth was monitored.
Flow cytometry analysis
Lymph nodes were mechanistically dissociated, counted and passed through 70 µm cell strainers (Falcon 352350). To harvest infiltrated immune cells, tumors were minced with scissors and digested in RPMI-1640 2%-FBS, Collagenase IV (367.5 U/mL) (Worthington Biochemical, ref: WOLS04186) and DNase I (0.1 mg/mL) (Sigma, ref D5025) at 37°C for 60 min under rotation. Digested tumors were passed through a 70 µm cell strainer, and centrifuged at 400 g for 5 min. Then, the cell suspension was centrifuged in a gradient of Ficoll (GE Healthcare, ref: 17144003) for 40 min at 400 g. All samples were stained as previously described29 with the antibodies listed in supplementary table 1. Non-specific binding was blocked using anti-CD16/CD32 (Miltenyi, ref: 130-07-594). For cytokines staining, cells were stimulated in RPMI 10% FBS, PMA (1 µg/mL) and ionomycin (0.5 µg/mL) with GolgiPlug (1:1000) and GolgiStop (1:1000) solutions (Cytofix/Cytoperm Plus, BD Biosciences, ref: 555028) for 4–5 hour at 37°C to block Golgi’s exocytosis. For intracellular staining, cells were fixed and permeabilized with fixation/permeabilization buffer (Invitrogen, ref: 00-552300) according to the manufacturer’s instructions. Data were acquired with a BD Biosciences Canto II or a BD LSRFortessa flow cytometer, compensated and exported into FlowJo software (Tree Star) for analysis. After analysis, the data points<100 events were excluded. The complete list of Abs used can be found in supplementary table 1.
Analysis of single-cell RNA sequencing published data of patients with PDAC
The expression of TNFR2 in human patients with PDAC was analyzed by using public single-cell RNA-seq data.30 Data were analyzed from the raw counts matrix using standard Seurat workflow.31 Briefly, low-quality cells (<200 genes/cell, <3 cells/gene and>10% mitochondrial genes) were excluded. “NormalizeData” function with default parameters was applied to normalize the expression level of genes in each single cell. Then, 3000 highly variable genes were identified using the “FindVariableFeatures” function with “vst” method. All samples were processed independently and the data was then integrated using reciprocal Principal Component Analysis (PCA). The “ScaleData” function was used to scale and center gene expression matrices after regressing out heterogeneity associated with mitochondrial contamination. To perform clustering, the dimensionality of the data was determined by calculating relevant principal components (PC) using the ElbowPlot function. Relevant PC were selected to construct the shared nearest neighbor (SNN) graph with “FindNeighbors” function, and clusters were determined using the Louvain algorithm. The Uniform Manifold Approximation and Projection (UMAP) was finally applied based on the above described SNN graph to visualize the single-cell transcriptional profile in two-dimensional space. Annotation of the clusters was performed using marker genes and published gene signatures. Imputation of missing values in the count matrix was performed using adaptively thresholded low-rank approximation.32
Single-cell RNA sequencing of mouse models
Murine pancreatic cancer cells mPDAC were injected in syngeneic immunocompetent FVB/n mice as previously described.28 Mice were treated with anti-TNFR2 mAb or untreated. After 18 days, mice were euthanized, and tumors were processed with Collagenase IV and DNase I as previously described (flow cytometry). Cells were stained with DAPI, CD45, CD4, CD8, CD11c, Ly6G, Ly6C, CD19, NK1.1, F4/80 and T cells were sorted on a FACSAria III (BD) by gating on live cells, CD45+, lineage (CD11c, Ly6G, Ly6C, CD19, NK1.1, F4/80) negative cells, CD4+ and CD8+ cells. CD4 and CD8 isolated T cells were mixed. Sorted T cell samples were loaded on a 10x Chromium Controller (10x Genomics) according to the manufacturer’s protocol. Single-cell RNA-seq libraries were prepared using Chromium Single Cell 5′ v3 Reagent Kit (10x Genomics) according to the manufacturer’s protocol. Briefly, the initial step consisted of performing an emulsion where individual cells were isolated into droplets together with gel beads coated with unique primers bearing 10x cell barcodes, unique molecular identifiers, and poly (dT) sequences. Reverse transcription reactions were engaged to generate barcoded full-length complementary DNA (cDNA) followed by the disruption of emulsions. cDNA was then purified using DynaBeads MyOne Silane beads (Thermo Fisher Scientific). PCR amplification of 10x barcoded full-length cDNA was performed following manufacturer’s instructions. Finally, libraries were constructed following these steps: (1) fragmentation, end repair and A-tailing; (2) size selection with Solid Phase Reversible Immobilisation (SPRI) select beads; (3) adaptor ligation; (4) post-ligation cleanup with SPRI select beads; (5) sample index PCR and final cleanup with SPRI select beads. Library quantification and quality assessment were achieved by Qubit fluorometric assay (Invitrogen) using dsDNA HS (high sensitivity) Assay Kit and Bioanalyzer Agilent 2100 system using a high sensitivity DNA chip. Indexed libraries were tested for quality, equimolarly pooled and sequenced on an Illumina HiSeq2500 using paired-end 26×98 bp as sequencing mode. By using a full Rapid Flow Cell, coverage was around 100M reads per sample corresponding to 100,000 reads per cell.
After sequencing, single-cell expression was analyzed using the Cell Ranger single cell software suite (V.3.1.0) to perform quality control, sample de-multiplexing, barcode processing, and single-cell 5′ gene counting. Sequencing reads were aligned on 10x Genomics mm10-3.0.0 mouse genome reference using the Cell Ranger suite with default parameters. In this version of Cell Ranger, including EmptyDrops method, cells with low RNA content has been rescue. Downstream analyses were performed using Seurat (V.4.3.0) with R V.4.3.0. Some filters were applied for each sample imported into the seurat pipeline: Cells with fewer than 200 genes were removed to remove debris, dead cells and other cells with few genes. Uninformative cells and possible doublets cells were removed based on the percentage of mitochondrial genes (cells with a percentage of mitochondrial genes superior to 0,05 are filtered) and total counts of genes by cell (cells with a value of genes/cells inferior to 350 are filtered). Each sample were normalized separately using global-scaling normalization (using NormalizeData function with ‘‘LogNormalize’’ as normalization method) and the 2000 most highly variable genes was identified for each sample (using FindVariableFeatures with ‘‘vst’’ as a method, with low-cutoffs and high-cutoffs for feature dispersions fixed at 0.5 and Inf and with low-cutoffs and high-cutoffs for feature means fixed at –Inf and Inf ; in addition, Tra(vdj) and Trb(vdjc) genes are filtered from high variable genes). All the samples were merged by applying the integration process of Seurat based on the 2000 most variable features (using SelectIntegrationFeatures, FindIntegrationAnchors and IntegrateData with default settings). To reduce technical noise, PCA was performed to work on the most contributing PC. Graph-based clusterization was done at different resolutions (using FindNeighbors on the thirty first PCs and FindClusters for the resolution between 0 and 1 for each decimal) and visualized using Clustree V.0.5.0 (Zappia, Oshloack, 2018). UMAP reduction was done (using RunUMAP on the thirty first PCs) to visualize the cells in UMAP projection.
Clustering with resolution 0.2 was satisfying for the identification of contaminant cell based on the absence of expression of T cell markers (Cd3e, Cd3d, Cd4, Cd8) and expression of other immune cell population markers (Cd14). After the elimination of contaminant cells from the data of the different samples, the different steps described above (from normalization to UMAP reduction) were performed again.
Cluster identification: 12 clusters were initially defined at resolution 0.7 for all CD3+ cells and are visualized using UMAP. Clusters were assigned using “find all markers” analysis and known T cell markers. The expression of a selection of genes was then analyzed using Feature plots and Violin plots. For CD8 cells analysis the “add module score” function was used to compare an exhausted score using Tigit, Havcr2, Ctla4, Lag3 and Tox genes.
ScTCR-seq analysis
After sequencing, single-cell TCR data were analyzed using the Cell Ranger Single Cell Software suite (V.3.1.0) to perform quality control, sample de-multiplexing, barcode processing, and single-cell Variable, Diversity, Joining (VDJ) CDR3 counting. Sequencing reads were aligned on 10x Genomics vdj_GRCm38_alts_ensembl-3.1.0.gz-3.1.0 genome reference using the Cell Ranger suite with default parameters.
Filtered files produced by Cellranger were imported in R to study clonality. To improve the definition of clonotypes and considering that TRA and TRB chains are defined by combining CDR3 and V subunits, we considered for our analysis only those cells presenting only 1 TRA chain and 1 TRB chain to define clones. To estimate clonal diversity, we used the Gini-TCR Skewing Index.33
Statistical analysis
Statistical analyses were performed by using GraphPad Prism software. Unless indicated otherwise, bars represent mean±SE of mean (SEM). Two normality tests of sample data (the Kolmogorov-Smirnov and the Shapiro-Wilk test) were applied to determine if the data set had a normal distribution. Outliers were identified by the ROUT method and excluded from the data set. For statistical comparison of two independent samples, a parametric (Student’ t-test) or a non-parametric (Mann-Whitney). For statistical comparison of more than two independent groups, a parametric (one-way analysis of variance) or non-parametric (Kruskal-Wallis) tests were applied and the mean of each column compared with the mean of the other column with a Dunn or Tukey’s post-test. In the experiment comparing anti-TNFR2 to control mice, a Student’s t-test one-tailed was used to test if the tumor volume of the anti-TNFR2 was lower than control group. For Kaplan-Meier survival curves, groups were compared using the log-rank test. The χ² test was used to compare the proportion of mice with a tumor. Statistical significance is indicated as ns: non-significant *p<0.05, **p<0.01, ***p<0.001,****p<0.0001.
Results
TNFR2 emerges as a promising novel target in PDAC treatment
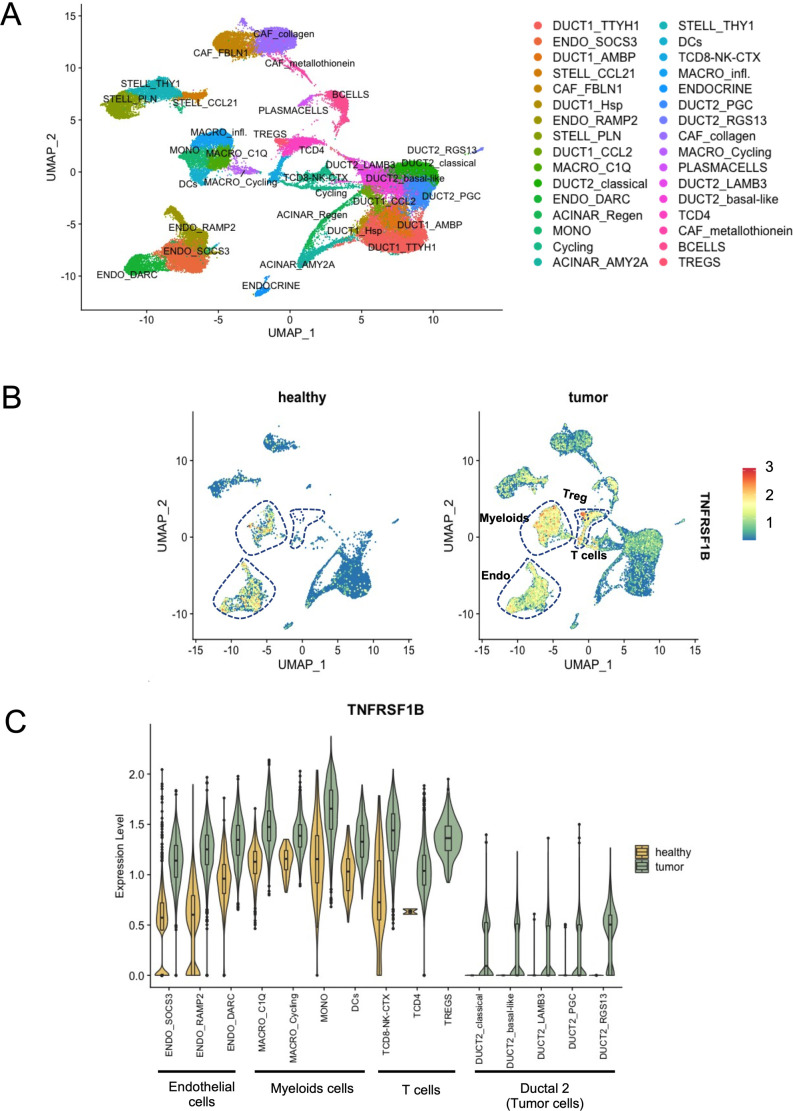
Targeting TNFR2 for antitumor purposes through the use of anti-TNFR2 mAb is based on the possibility to block local immunosuppression mediated by Tregs constitutively expressing TNFR2 and by the putative expression of this marker directly by certain tumor cells.24 34 We first took the opportunity of available public single-cell RNA-seq data30 to evaluate the expression of the TNFR2 gene (TNFRSF1B) in patients with PDAC. Clusters of the main populations present in PDAC tumors were assigned using module score of lists of differentially expressed genes published in Peng et al30 and visualized by UMAP (figure 1A). The level of expression of TNFRSF1B among the different main cell populations identified in PDAC was then analyzed. As expected, the main cell populations expressing TNFRSF1B in the tumors are Tregs, myeloid and endothelial cells (figure 1B). Importantly, after separating healthy (n=11) and tumor (n=24) single-cell RNA-seq data, the immune infiltration radically differed (figure 1B). Whereas conventional T cells and Tregs were not detected in healthy pancreas, they were present in tumor environment and they highly expressed TNFRSF1B (figure 1B, C). Among epithelial-derived cells, the ductal 2 cell clusters were present only in tumors and two clusters were characterized by markers for classical (TF1, TF2, CECAM6, REG4) and basal-like (KRT6A, KRT17, S100A2, S100A9, LY6D) PDAC cells.35 36 All tumor ductal 2 cell clusters expressed TNFRSF1B with a mean expression level<0.5 (figure 1A–C). Stromal and ductal 1 cells expressed also a low level of TNFRSF1B both in healthy and tumor tissues (figure 1A-C). These results support the relevance of TNFR2 as a potential target in immune cell infiltrate together with tumor cells of human PDAC.
Figure 1. TNFR2 expression in human PDAC. (A) UMAP plot of main populations present in PDAC tumors from Peng et al.30 (B) Expression of TNFRSF1B split between tumor and healthy samples. (C) Violin plot displaying the expression of TNFRSF1B across the cell identified in PDAC. Adaptive thresholded low rank approximation-imputed data are depicted. PDAC, pancreatic ductal adenocarcinoma; TNFR2, tumor necrosis factor α receptor 2; Treg, regulatory T cells; UMAP, Uniform Manifold Approximation and Projection.
TNFR2 blockade decreases tumor-infiltrating Treg proportions in PDAC tumors
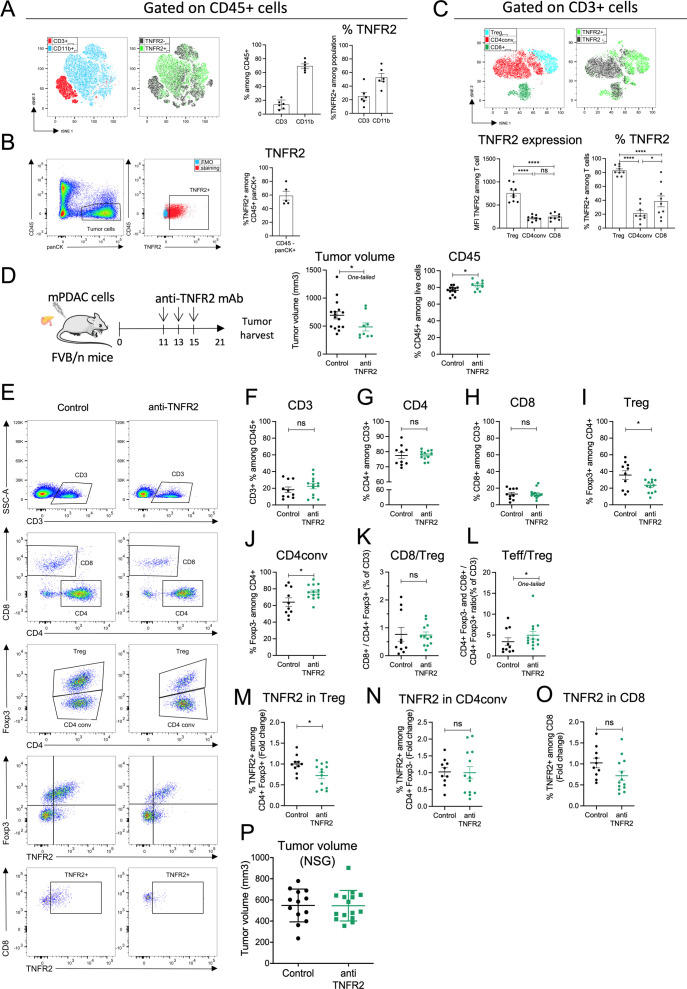
We sought to study the expression of TNFR2 by tumor infiltrated immune cells in orthotopic and immunocompetent mouse models of PDAC obtained by using mPDAC and Panc02 cell lines previously described.28 We initially observed that mPDAC tumors have an immune-escape TME infiltrated by CD45+ immune cells, highly enriched in CD4+Foxp3+ Tregs and characterized by low CD8+ T cell infiltration.29 Here, tumor infiltrated lymphocytes (TILs) were isolated from mPDAC tumors and analyzed by flow cytometry: 14% (±3) of CD45+ infiltrated immune cells were CD3+ T cells and 69% (±3) were CD11b+ myeloid cells (figure 2A). TNFR2 was detected in CD3+ T cells, CD11b+ myeloid cells (figure 2A) and in tumor cells (figure 2B). Among T cells, the intensity of TNFR2 expression was significantly higher in tumor-infiltrated CD4+Foxp3+ Tregs than in CD4 conventional (CD4conv) CD4+Foxp3− T cells or CD8+ T cells (figure 2C). The vast majority of Tregs infiltrating the mPDAC tumor express TNFR2 but a lower proportion of CD8+ (38%) and CD4+Foxp3− T cells (21%) expressed this receptor (figure 2C). Comparable results of higher TNFR2 expression in tumor infiltrated CD4+Foxp3+ Tregs compared with CD4+Foxp3− T cells were observed in a second model of orthotopic PDAC tumors obtained by injecting murine Panc02 cells (online supplemental figure 1A-C).
Figure 2. TNFR2 blockade decreases tumor volume and tumor-infiltrating Treg proportions in mouse models of PDAC. FvB/n immunocompetent mice (A–O) or NSG mice (P) were grafted with mPDAC cells into the pancreas. After 21 days, tumors were harvested for flow cytometry analysis. (A) Cell clustering using a t-distributed stochastic neighbor embedding (t-SNE) algorithm performed on CD45+ FVS− (Fixable Viability Stain) gate. CD3+ and CD11b+ cell clusters localization and TNFR2+ cell localization among the above-mentioned populations are indicated. Histograms show the proportion of intratumoral CD3+ and CD11b+ cells among CD45+ FVS− cells and of TNFR2+ cells among CD3+ or CD11b+ cells. (B) Gating strategy of tumor cells (CD45− panCK+) and TNFR2+ cells among tumor cells and proportion of TNFR2+ cells among CD45−panCK+ cells. (C) Cell clustering using a t-SNE algorithm performed on CD3+ gate. Treg (CD4+Foxp3+), CD4conv (CD4+Foxp3−) and CD8+ cell clusters, and TNFR2+ cell cluster localization are indicated. Histograms show TNFR2 expression (MFI TNFR2) and proportion in intratumoral Treg (CD4+Foxp3+), CD4conv (CD4+Foxp3-) and CD8+ cells. (D) Schemas of the experiment: mice were treated with anti-TNFR2 mAb, IgG control or PBS at day 11, 13 and 15 or were untreated. Tumor volume at day 21 was measured and infiltrated CD45+ cells among FVS− cells analyzed without previous separation on density gradient (n=16). (E) Gating strategy of infiltrated T lymphocytes analyzed by flow cytometry. Representative scatter plots of intratumoral (F–H) CD3+, CD4+ and CD8+; (I, J) Treg (CD4+Foxp3+) and CD4conv (CD4+Foxp3−); (K–L) CD8+/Treg (CD4+Foxp3+) ratio, and Teff (CD4+Foxp3− and CD8+)/Treg(CD4+Foxp3+) ratio. (M–O) TNFR2 proportion among Treg (CD4+Foxp3+), CD4conv (CD4+Foxp3−) and CD8+ cells is represented as fold change (calculated by reporting each point to the mean of the control group). For (F–O) (n=20 including two experiments, representative of three experiments). Data are plotted as the mean±SEM. Statistical significance of (C) between population in control was determined using ANOVA multiple comparison test. Statistical significance of anti-TNFR2 treated groups from controls was determined using a Mann-Whitney test. Ns: non-significant, p>0.05, *p<0.05, **p<0.01, ***p<0.001. ANOVA, analysis of variance; IgG, immunoglobulin G; mAb, monoclonal antibody; PBS, phosphate-buffered saline; PDAC, pancreatic ductal adenocarcinoma; TNFR2, tumor necrosis factor α receptor 2; Treg, regulatory T cells.
Since disruption of the TNFα/TNFR2 pathway was shown to decrease tumor growth and metastasis progression in different experimental models of cancer21, we sought to test the effect of TNFR2 blockade on PDAC progression and on the regulation of the immune microenvironment in the mPDAC and Panc02 orthotopic or ectopic tumors. We previously observed a Treg enrichment between day 7 and day 17 in mPDAC tumor-bearing mice.29 To target CD4+Foxp3+ Tregs expressing TNFR2, mice were treated with three injections of a blocking anti-TNFR2 mAb at day 11, 13 and 15 for mPDAC or KPC orthotopic tumors (figure 2D and online supplemental figure 1O) and at day 8, 11, 13, 15 and 18 for Panc02 tumors (online supplemental figure 1D). 21 days after mPDAC or Panc02 cell injection, mice were euthanized and their tumor volumes were measured. The mPDAC, Panc02 and KPC tumor volumes statistically decreased in mice treated with the anti-TNFR2 (figure 2D and online supplemental figure 1E and O). TNFR2 blockade did not have any effect on the progression of mPDAC ectopic tumors (online supplemental figure 1K-M). We analyzed the TILs after anti-TNFR2 mAb administration by flow cytometry. The frequency of CD45+ among total number of cells isolated from tumor digestion increased in anti-TNFR2 treated mice (figure 2D). The proportion of CD3+, CD4+ and CD8+ cells among CD3+ cells was not modified (figure 2E–H and online supplemental figure 1F). However, the frequency of CD4+Foxp3+ Tregs cells significantly decreased by 1.5-fold both in mPDAC and Panc02 tumor-bearing mice (figure 2I and online supplemental figure 1G) whereas the CD4+Foxp3− (CD4conv) increased (figure 2J). While the CD8+/Treg ratio did not change (figure 2K and online supplemental figure 1I), the ratio between all effector T cells (CD4+Foxp3− and CD8+/CD4+Foxp3+) and Treg (Teff/Treg) increased (figure 2L and online supplemental figure 1J) in anti-TNFR2 treated mice compared with untreated mice. Also, the frequency of TNFR2 among CD4+Foxp3+ cells but not of CD4+Foxp3− or CD8+ T cells was lower in anti-TNFR2 treated mice compared with untreated m
ice (figure 2M–O). In tumor-draining lymph nodes, no major modifications were observed (online supplemental figure 2). We did not observe any impact of the treatment on the immune infiltrate of mPDAC when tumors were ectopically injected (online supplemental figure 1K-N), in accordance with the absence of clinical impact of anti-TNFR2 treatment in the last setting. The involvement of the immune system in the observed antitumor clinical effect was also tested by grafting PDAC tumor cells into the pancreas of NSG immunodeficient mice. Injection of an anti-TNFR2 treatment in these mice had no effect on tumor growth (figure 2P).
Since, TNFR2 was also expressed by CD11b+ infiltrating the tumors (figure 2A), TNFR2 expression and blocking effect were analyzed in different types of myeloid cells (online supplemental figure S3A). TNFR2 was highly expressed by the TAMs and the polarized macrophages M1 and M2 CD206+-cells (online supplemental figure 3B-D). Also, it was shown that TNFα drives the accumulation of peripheral Myeloid-Derived Suppressor Cells (MDSCs) via TNFR2 signaling, regulating MDSC survival and helping tumor cells evade the immune system.37 In mPDAC mouse models, infiltrated CD11b+ suppressive cells constituted more than 50% of the CD45+ cell populations as in the human pathology.29 Polymorphonuclear MDSCs (PMN-MDSCs) directly inhibit T cell proliferation, express PDL-1 and immunosuppressive cytokines and mobilize Tregs.38 Indeed, PMN-MDSCs are the dominant source of TNFα leading to stromal inflammation and immune tolerance to promote therapeutic resistance in PDAC.39 This was observed by using the etanercept, a soluble form of TNFR2, to block TNFα. However, etanercept inhibits both TNFR1 and TNFR2 signaling by targeting TNFα. In mice treated with anti-TNFR2, the proportion of the CD11b+ myeloid cells was not modified (online supplemental figure 3E) whereas the frequency of PMN-MDSCs but not the monocytic MDSCs significantly decreased (online supplemental figure 3F, G). No impact on the percentage of TAMs, M1, M2 CD206+ or M2 CD206− (online supplemental figure 3H-J) or dendritic cells (DCs) (online supplemental figure 3K-M) or CD80 and CD86 expression by DC (online supplemental figure 3N-P) was observed. These results indicate that despite a strong expression of TNFR2 on tumor-infiltrated myeloid cells, anti-TNFR2 mAb treatment only impact PMN-MDSCs in mPDAC tumors.
Altogether, these results suggest that anti-TNFR2 mAb treatment mainly targeted Tregs and PMN-MDSCs in the pancreatic TME thus promoting higher infiltration of immune cells leading to a more favorable balance between effector T cells and Tregs.
TNFR2 blockade decreases Treg activation and effector T cell exhaustion
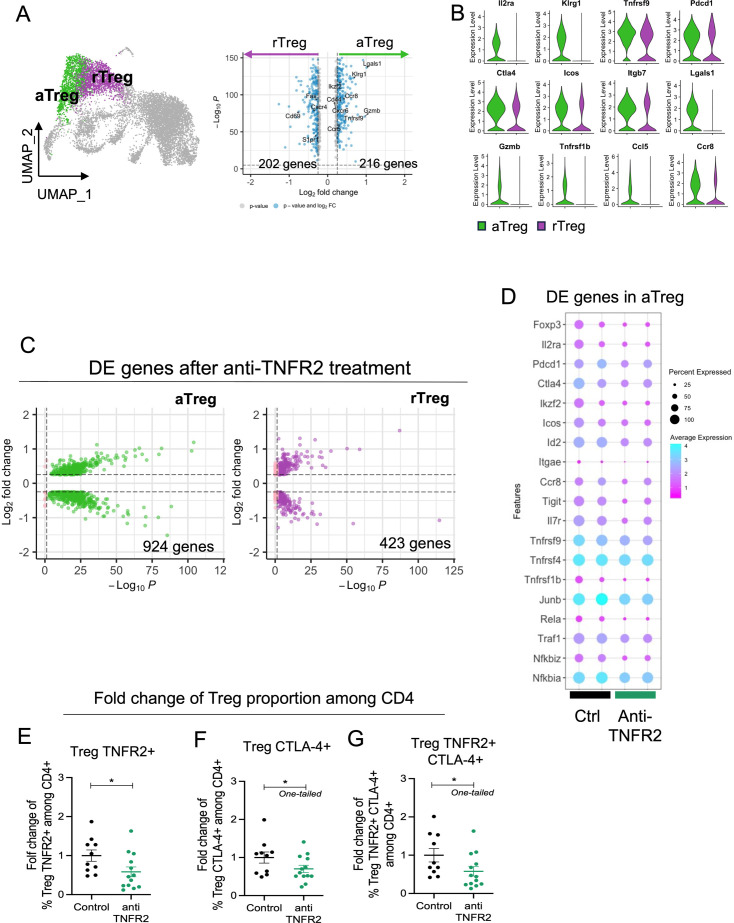
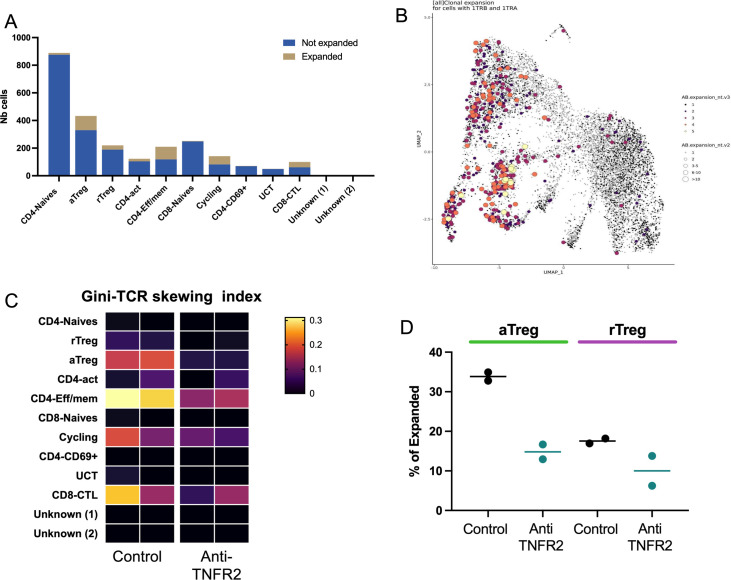
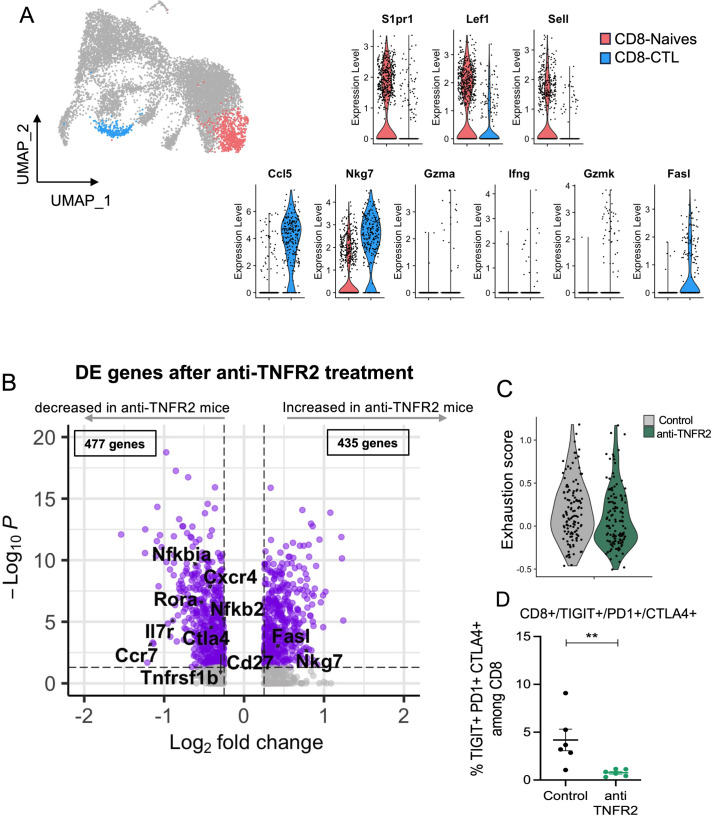
To go further inside the mechanisms of regulation due to anti-TNFR2 mAb treatment, we next analyzed mPDAC TILs by single-cell RNA-seq analysis. For this, mice were euthanized at day 18 to be closer to the end of the anti-TNFR2 mAb treatment, and thus to increase the possibility to detect any direct effect. We isolated CD45+ cells from tumor-control mice and anti-TNFR2-treated mice at day 18. CD3+ cells were sorted and analyzed using single-cell RNA-seq. After data normalization and using known markers of T cells, cells were clustered in two dimensions using the UMAP dimensionality reduction technique (online supplemental figure 4). Two clusters of Tregs (activated, (a)Treg and resting (r)Treg) were observed and cell clusters of CD4+ effector/memory T cells (Cd44, Cd40L, Cxcr6, Icos), CD8 effector/memory T cells (Cd8a, Pdcd1, Gzmb FasL), naives CD8+ and CD4+ T cells (S1pr1, TCF7, Lef1, Sell) and cycling T cells (Mki67) were identified (online supplemental figure 4). Focusing on the Treg clusters, rTreg and aTreg clusters were separated by differently expressed genes such as Il2ra, Klrg1, Lgals1, Gzmb, Ccr8 (figure 3A). The activation markers Pdcd1, Ctla4, Icos, Itgb7 and Tnfrsf1b were more expressed in aTreg than in rTreg (figure 3B). Anti-TNFR2 treatment induced deregulation of a greater number of genes in the aTreg (924 genes) cluster than in the rTreg one (423 genes) (figure 3C). In particular, it induced a decreased expression of several genes of activation and NF-kβ signaling pathway (figure 3D). Importantly, we observed that anti-TNFR2 mAb treatment reduced the expression level of Tnfrsf1b and Ctla4 (figure 3D). We validated these anti-TNFR2 effects by flow cytometry. The frequency of CD4+FoxP3+ Tregs expressing TNFR2, CTLA-4 or both significantly decreased under anti-TNFR2 mAb treatment (figure 3E–G). TCR sequencing revealed that among infiltrated T lymphocytes, cell populations displaying high numbers of expanded clones were identified in aTregs, CD4-Eff/mem, cycling and CD8/CTL clusters (figure 4A, B). We next calculated the Gini index which captures the inequality in clonotype size across the population (figure 4C). First, its global value was limited (less than 0.3) suggesting a low clonal expansion in the PDAC tumors. As expected, activated aTreg, CD4-eff/mem and CD8 CTL clusters had the highest Gini index, reflecting their higher expansion. Interestingly, anti-TNFR2 mAb treatment induced a Gini index reduction for these three clusters, with a less pronounced effect for the CD8 CTL cluster (figure 4C). Importantly, anti-TNFR2 treatment specifically targets aTregs, as evidenced by the strong reduction of the percentage of cells with expanded clones, likely revealing a less immunosuppressive environment in the tumor of treated mice (figure 4D).
Figure 3. TNFR2 blockade selectively targets activated Tregs over resting Tregs in a PDAC mouse model. FvB/n immunocompetent mice were grafted with mPDAC cells into the pancreas. Mice were treated with anti-TNFR2 mAb and tumors were harvest and intratumoral CD4+ and CD8+ were isolated for single-cell RNA sequencing. (A) UMAP plot with showing the two identified Treg clusters and violin plot comparing expression of different genes between aTreg (green) and rTreg (purple). (B) Volcano plot of differential gene expression analysis between aTreg and rTreg. (C) Volcano Plot of differential gene expression analysis between control and anti-TNFR2 treated mice for aTreg (Left) and rTreg (right) clusters. (D) Dot plot highlighting anti-TNFR2 treatment effect on gene expression level on aTreg. (E–G) Scatter plots of fold change of (E) Foxp3+ TNFR2+ (Treg TNFR2+), (F) Foxp3+ CTLA-4+ (Treg CTLA-4+) and (G) Foxp3+ TNFR2+ CTLA-4+ (Treg TNFR2+ CTLA-4+) proportions among CD4. Data are plotted as the mean±SEM. Statistical significance from controls was determined using a Mann-Whitney test two-tailed or one-tailed (F–G), *p<0.05. aTreg, activated regulatory T cell; CTLA-4, cytotoxic T-lymphocytes-associated protein 4; mAb, monoclonal antibody; PDAC, pancreatic ductal adenocarcinoma; rTreg, resting regulatory T cell; TNFR2, tumor necrosis factor α receptor 2; UMAP, Uniform Manifold Approximation and Projection.
Figure 4. TNFR2 blockade specifically reduces clonally expanded aTreg in a mouse model of PDAC. FvB/n immunocompetent mice were grafted with mPDAC cells into the pancreas. Mice were treated with anti-TNFR2 mAb and tumors were harvest and intratumoral CD4+ were isolated for single-cell RNA and TCR sequencing. (A) Distribution of cells with expanded clones among the different identified clusters, the clusters with more expanded cells are Treg, CD4-EFF/MEM and CD8 CTL (B) UMAP representation of expanded clones (same conclusion of A) (C) Gini index (explaining the diversity) of all cluster from all mice (near 1=expansion of clones, near 0=no expansion). (D) Specific effect of treatment on cells with expanded clones inside each Tregs cluster. aTreg, activated regulatory T cell; mAb, monoclonal antibody; PDAC, pancreatic ductal adenocarcinoma; rTreg, resting regulatory T cell; TNFR2, tumor necrosis factor α receptor 2; UMAP, Uniform Manifold Approximation and Projection.
We then turned our analysis on the effect of the anti-TNFR2 mAb on CD8+ T cells. We focused on the cytotoxic CD8+ T cells (CD8 CTL) and the naive CD8+ T cells (figure 5A). Anti-TNFR2 treatment induced deregulation of 912 genes in CD8 CTL (figure 5B). A significant decrease in the expression of the markers of exhaustion (Ctl4, Tigit, Tim3, Entpd1, havcr2) in the CD8 CTL cluster was individually observed and the exhaustion score taking into account all these markers was lower in anti-TNFR2 mAb treated tumors (figure 5C). Importantly, the proportion of CD8+ T cells positive for the exhaustion markers CTLA-4, PD-1 and T cells with Immunoreceptor with Ig and ITIM domains (TIGIT) was shown to be reduced by flow cytometry analysis in the tumors from mice treated with the anti-TNFR2 compared with those from untreated mice (figure 5D).
Figure 5. TNFR2 blockade decreases the exhausted profile of CD8 T cells. FvB/n immunocompetent mice were grafted with mPDAC cells into the pancreas. Mice were treated with anti-TNFR2 mAb and tumors were harvest and intratumoral CD8+ were isolated for single-cell RNA and TCR sequencing. (A) UMAP plot showing CD8 clusters differentiated by color (CD8 CTL in blue and CD8-Naives in coral) and violin plot comparing expression of different genes between CD8 CTL and CD8-Naives. (B) Volcano plots of differential gene expression analysis between control and anti-TNFR2 treated mice within CD8 CTL cluster. (C) Violin plot showing expression level of CD8 exhaustion score (genes=Tigit, Havcr2, Ctla4, Lag3 and Tox) for each condition (gray: control group, green: anti-TNFR2 treated group). (D) Scatter plot of TIGIT+ CTLA-4+ PD-1+ cells proportion among intratumoral CD8+ cells analyzed by flow cytometry. Data are plotted as the mean±SEM. Statistical significance from controls was determined using a Mann-Whitney test, **p<0.01. CTLA-4, cytotoxic T-lymphocytes-associated protein 4; mAb, monoclonal antibody; PDAC, pancreatic ductal adenocarcinoma; PD-1, programmed cell death protein-1; TNFR2, tumor necrosis factor α receptor 2; UMAP, Uniform Manifold Approximation and Projection.
Combination therapy with blocking anti-TNFR2 and agonistic anti-CD40 mAbs has synergistic effect on PDAC antitumor response
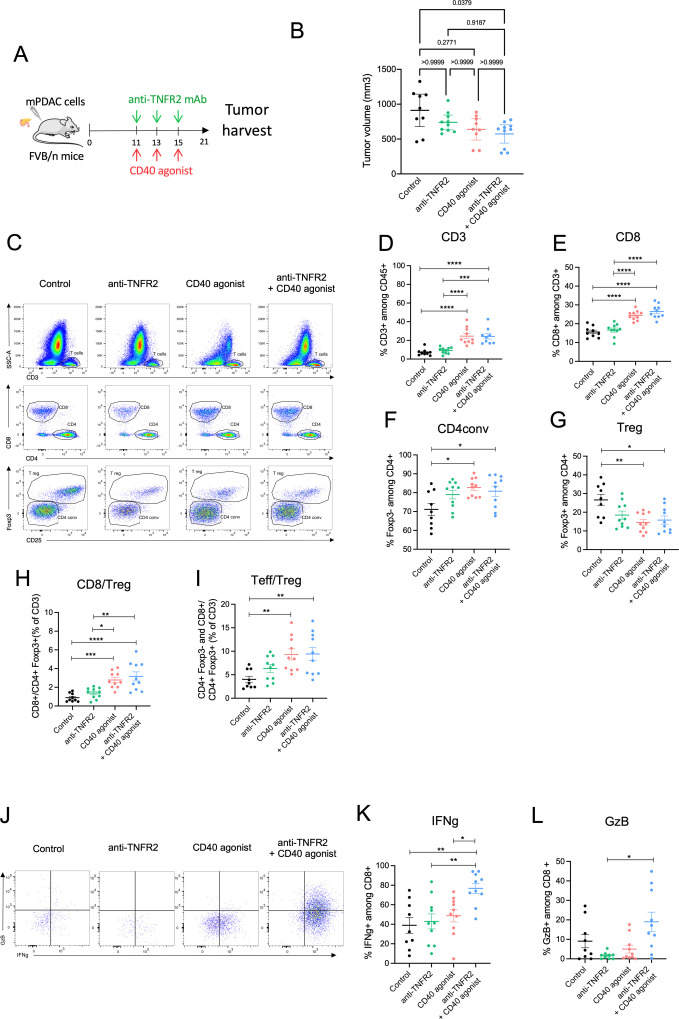
The single-cell RNA-seq data from mPDAC clearly indicate that TNFR2 blockade inhibited immunosuppressive cells and T cell exhaustion but is not sufficient to induce enough antitumoral cytotoxic T cells. We sought to improve the antitumoral response induced by the anti-TNFR2 mAb by designing a combined treatment with immunotherapy that could boost T-cell activation. Agonist mAbs that target costimulatory pathways such as CD40 and OX40 have been shown to successfully promote antigen-specific T-cell expansion, and CXCR4 blockade to promote T cell tumor infiltration and activation in PDAC,40,42 CD40, TNFRSF4 (OX40) and CXCR4 gene expression was analyzed in public human single-cell RNA-seq data from figure 1, showing that all genes were highly expressed by tumor infiltrated immune cells in patients with PDAC and were potential target for combination together with TNFR2 blockade (online supplemental figure 5). mPDAC-carrying mice were treated with a blocking anti-TNFR2 mAb together with agonists of CD40 or OX40 or antagonist of CXCR4 using mAbs administered at day 11, 13, 15. 21 days after mPDAC cell injection, mice were euthanized and the tumor volumes were measured (online supplemental figure 6). Only the anti-TNFR2 mAb combined with an agonistic anti-CD40 mAb, but not with an agonistic anti-OX40 or anti-CXCR4 mAbs, significantly reduced tumor growth (online supplemental figure 6). Tumor volume and infiltrated T cell proportion of the anti-TNFR2 mAb combined with an agonistic anti-CD40 mAb group were compared with the administration of an anti-TNFR2 mAb or an anti-CD40 agonist alone (figure 6). Whereas the anti-TNFR2 mAb or an anti-CD40 agonist alone tended to reduce tumor progression only the anti-TNFR2 mAb combined with an agonistic anti-CD40 mAb slightly reduced tumor growth in a statistical analysis of the comparison between the groups (figure 6A, B). Whenever agonist anti-CD40 mAb was administered, alone or in the presence of anti-TNFR2 mAb, an increase in CD3+, CD8+, CD4conv cells (figure 6C–F), and a decrease in Tregs (figure 6G) were observed, in line with an improved antitumor immune response. Importantly, only the combined treatment induced a higher frequency of IFN-γ or granzyme B expressing T cells compared with anti-TNFR2 mAb alone (figure 6J–L). However, in a dedicated experiment, we did not observed improved survival of mice whatever the treatment used compared with untreated mice (online supplemental figure 7).
Figure 6. Combination of blocking anti-TNFR2 and agonistic anti-CD40 mAbs increases effector T cell activation. (A) Schemas of the experiment: FvB/n immunocompetent mice were grafted with mPDAC cells into the pancreas. Mice were treated with either anti-TNFR2 mAb or CD40 agonist or both or received PBS at day 11, 13, 15 (n=10). After 21 days, tumors were harvest for flow cytometry analysis (B) Scatter plot of tumor volume at day 21 are plotted as the mean with 95% CI. (C–G) Scatter plots of intratumoral CD3+, CD8+, Treg (CD4+ Foxp3+) and CD4conv (CD4+Foxp3−) cells proportion with representative dot plot (C). (H–I) Scatter plots of CD8+/Treg(CD4+Foxp3+) ratio and Teff (CD4+Foxp3− and CD8+)/Treg (CD4+ Foxp3+) ratio. (J) Representative dot plot of IFN-γ+, and GzB+ (granzyme B) cells proportion in intratumoral CD8. (K–L) Scatter plots of IFN-γ+ and GzB+ proportion in CD8 cells. Data (B, D–I, K, L) are plotted as the mean±SEM. Statistical significance of the difference between groups was determined using the Kruskal-Wallis test (in B p=0.0446 in L p=0.0160) or One-way ANOVA (in D, H and E p<0.0001, in G p=0.0033, in I p=0.0031, in K p=0.018) (depending if data follow a normal distribution). The mean of each column was compared with the mean of the other columns with a Dunn (B and L) or Tukey’s (D–K) test: non-significant (ns) p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. All groups without asterisks in panels B, D–I, and K, L are ns. ANOVA, analysis of variance; IFN, interferon; mAb, monoclonal antibody; PBS, phosphate-buffered saline; PDAC, pancreatic ductal adenocarcinoma; TNFR2, tumor necrosis factor α receptor 2; Treg, regulatory T cell.
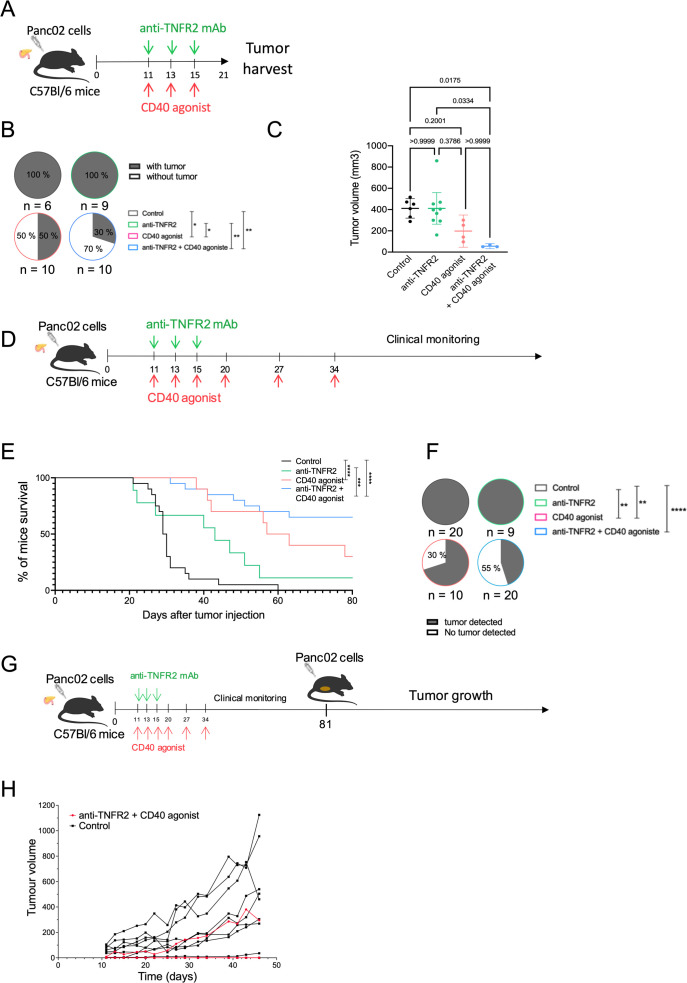
In order to confirm our biological and clinical observations obtained with the combined treatment, we reproduced these experiments using the Panc02-derived orthotopic tumors (figure 7A). The treatments between mPDAC and Panc02 tumors for evaluating tumor volume were similar (figures6A 7A). After 21 days, the mice were euthanized. As observed for mPDAC, the combination treatment had a statistically significant impact on the tumor incidence compared with control and anti-TNFR2-treated mice (figure 7B). Remarkably, 70% of mice treated by the combination treatment versus 50% treated by anti-CD40 did not develop tumors (figure 7B). The tumor mass of mice treated with the combination treatment that still developed tumors was very low compared with control, anti-TNFR2 or anti-CD40 treated mice (figure 7C). We previously described that mPDAC-bearing mice die between the third and the fourth week after cancer cell injection into the pancreas.28 In a second set of experiments, we assessed whether the observed antitumor effects of anti-TNFR2, anti-CD40 or the co-administration of both mAbs would also enhance the survival of Panc02-bearing mice (figure 7D). Whereas Panc02 control mice died after the fourth week, the anti-CD40 and the anti-TNFR2 mAbs induced a statistically significant improved survival. The better survival was obtained in mice receiving the combined treatment (figure 7E). Strikingly after 64 days, 55% of mice treated with anti-TNFR2 and anti-CD40 mAbs were still alive and tumor-free compared with 20% mice with the sole anti-CD40 (figure 7F). In protected mice, we evaluated the possible presence of subclinical tumors by ultrasound. Of the 13 mice treated with the Ab combination, tumors were detected in 2 of them. One-third of the anti-CD40-protected mice had tumors.
Figure 7. Combination of blocking anti-TNFR2 and agonistic anti-CD40 mAbs induces immunological T memory and improves survival of mice. (A) Schemas of the experiment: C56Bl/6j immunocompetent mice were grafted with Panc02 cells into the pancreas. Mice were treated with either anti-TNFR2 mAb or CD40 agonist or both or received PBS at day 11, 13 and 15 (n=10). Mice were euthanized at day 21. (B) Proportion of tumor incidence in control and treated groups represented in pie charts (χ², *p<0.05, **p<0.01). (C) Scatter plot of tumor volume at day 21 are plotted as the mean with 95% CI. Data are plotted as the mean±SEM. Statistical significance of the difference between groups was determined using the Kruskal-Wallis test (p=0.0062) and with a Dunn test for multiple comparison between all groups. ns: non-significant, *p<0.05. All groups without asterisks in panel C are ns. (D) Schemas of the experiment: C56Bl/6j immunocompetent mice were grafted with Panc02 cells into the pancreas. Mice were treated with either anti-TNFR2 mAb or CD40 agonist or both or received PBS at day 11, 13 and 15, 20, 27 and 34 and were clinically monitored. (E) Kaplan-Meier survival curve of the following groups: control (n=20), anti-TNFR2 (n=9), CD40 agonist (n=10) and anti-TNFR2+CD40 agonist (n=20). Mice are euthanized when the limit of the clinical score (established by a grid of symptoms) is reached. (Kaplan-Meier test, ***p<0.001, ****p<0.0001). (F) Presence of tumor (primary and/or metastatic) is challenged by echography at day 64. (χ², **p<0.01, ***p<0.001, ****p<0.0001). (G) Schemas of the experiment: mice still alive from anti-TNFR2 + CD40 agonist group and C57Bl/6 naive mice received subcutaneous injection of Panc02 cells at day 81, and monitored for tumor growth. Tumor growth for individual mice is shown in (H). mAbs, monoclonal antibody; PBS, phosphate-buffered saline; TNFR2, tumor necrosis factor α receptor 2.
Then, we rechallenged the alive mice from the combination group with Panc02 cells injected subcutaneously (figure 7G). Control mice developed subcutaneous tumors as in online supplemental figure 1 while the anti-TNFR2/anti CD40 mice did not have visible tumors growing suggesting that these mice rejected Panc02 cells due to acquired immunological memory (figure 7H).
Discussion
In this study, we evaluated the relevance of inhibiting the immunosuppressive effect of Tregs by blocking the TNF/TNFR2 pathway in PDAC. We first validated TNFR2 as a relevant target by showing that, both in humans and in mice, tumor-infiltrating Tregs express TNFR2 more intensively than other TME TNFR2+ cell populations. In mice, treatment with anti-TNFR2 reduced the percentage of Tregs in the TME of PDACs, thereby reducing tumor growth, paralleled with a reduction in the percentage of CD8+ T cells displaying an exhausted phenotype. Moreover, the combination of the anti-TNFR2 mAb with agonistic anti-CD40 mAbs improved survival in PDAC-bearing mice and promoted immunological memory.
This study corresponds to the first accurate description of TNFR2 expression in patients with PDAC by single-cell analysis. Using published dataset of single-cell RNA-seq of 24 tumors from patients with PDAC compared with 11 non-PDAC pancreas biopsies,30 we showed that the main cell populations expressing TNFRSF1B at the expression level>0.5 in patients with PDAC are T cells, macrophages and endothelial cells. The TME is also characterized by the presence of immunosuppressive cells such as Tregs and MDSCs that both require TNFR2 for suppressive functions.43 Importantly, the cell population that expresses TNFRSF1B at the highest level in human PDAC turned out to be Treg while TNFRSF1B expression in tumor cells seems to be at a lower level. In human PDAC, we thus validated that TNFR2 is a relevant target whose blockade could directly impact Tregs and tumor cells accordingly the level of expression. Previous studies already described TNFR2 expression in PDAC tumor cells34 44 while transcriptomic data of TNFRSF1B expression in human stromal PDAC cells should be confirmed by TNFR2 protein expression. However, due to low immune infiltrate and the great heterogeneity between patients, this study is very challenging and would require a very large number of patient biopsies before drawing any conclusion. We then focused the rest of our study on evaluating the impact of a TNFR2-blocking treatment on tumor growth and modulation of the immune environment in mouse models of PDAC.
We used our previously described murine model of orthotopic PDAC, in which we had initially observed that the immune TME was similar to that of human PDAC, characterized by an immune-escape profile, enriched in Tregs and with a paucity of CD8+ T cells.29 Here, deeper analysis revealed that TNFR2 was expressed by T cells, myeloid cells and to a lesser level by tumor cells. Among T cells, the intensity of TNFR2 expression in Tregs was 3.5-fold higher than in CD4+FoxP3− T cells. We therefore have a relevant experimental model to test TNFR2-based immunotherapies due to similarities with what is observed in human PDACs.
Some groups demonstrated that Treg depletion elicits effective antitumor immunity in mouse PDAC and supports the efficacity of a potential therapeutic strategy targeting Tregs.15 45 Different strategies of Treg targeting during PDAC progression have been tested: total Treg ablation in Foxp3DTR mice was sufficient to evoke effective antitumor response in orthotopically implanted KPC cells (Kras G12D and p53 R172H mutated cells). This was associated with an induction of cytotoxic CD8+ T cells activated by CD11c+ DCs.15 On opposite, anti-CTLA-4 or anti-CD25 blocking Abs had no clinical effect in mPDAC5 45 nor in a transgenic KCiMist1 mouse model spontaneously developing PDAC tumors,46 suggesting that total Treg ablation was more effective than treatment by anti-CTLA-4 or anti-CD25. Since we observed that TNFR2 was highly expressed by Tregs in human and mouse models of PDAC, blocking TNFR2 was tested as an alternative approach for Treg targeting in a clinically compatible approach using anti-TNFR2 mAb treatment. In two immunocompetent mouse models of orthotopically implanted PDAC, we demonstrated that blocking TNFR2 with the TR75-54.7 mAb was able to reduce tumor-infiltrating Tregs and promote a more favorable balance between CD4conv/Treg and globally between Teff/Treg cells for an antitumor response. This result was important if we consider that anti-CTLA-4 or anti-CD25 antibodies did not show a significant change in CD4+Foxp3+ Tregs in the PDAC tumor.45 Moreover, compared with anti-CTLA-4 or anti-CD25 targeting treatments,45 Tregs and T cells in lymph nodes (LNs) did not vary under anti-TNFR2, suggesting that the TNFR2 blockade in our models mainly targets TME-resident Tregs. This could be an important advantage to limit off-target effects and potential systemic Treg depletion. These results were reproducible in three different models of orthotopic PDAC, supporting that anti-TNFR2 is generally applicable and has a relevant immunomodulatory effect in PDAC. In a recently published study using the same anti-TNFR2 mAb, the depletion of TNFR2 in PDAC murine cells reduced the tumor growth of orthotopic KPC-induced tumors both in immunocompetent or immunodeficient mice, suggesting that TNFR2 expression in tumor cells directly participate to tumor cell growth.44 They very interestingly hypothesized that the expression of TNFR2 by tumor cells promotes tumorigenesis by inhibiting cancer immunogenicity that would occur through the PD-1/PD-L1 axis. Their experiments of anti-TNFR2 pre-incubation of tumor cells or TNFR2 invalidation in tumor cells, both subcutaneously administered, seem to support this hypothesis and the clinical antitumor effect of a treatment combining anti-TNFR2 and anti-PDL1 administration is convincing. However, they are by no means conclusive regarding the absence of effect in the immune system. To establish this, it would have been necessary to compare the observed effects in these two models with those observed in additional groups of mice treated with anti-TNFR2. In our hands and using two different PDAC models, the pancreatic immune TME was sensitive to the treatment and at the center of the clinical effect observed. In our study, in mPDAC and Panc02 orthotopic tumors, TNFR2 blockade mainly impacts tumor growth by acting on the pancreatic TME as suggested by the fact that we did not observe any antitumor response nor immunomodulatory effect in ectopic compared with orthotopic tumors generated by the same PDAC cells. The crucial involvement of the immune system in the observed antitumor response was confirmed by the complete absence of a clinical effect when the anti-TNFR2 treatment was administered to NSG immunodeficient mice orthotopically grafted with PDAC. It is also possible that expression of TNFR2 on the tumor cells results in immunosuppression and impairment of the CD8 T cell response, independent of the role of Tregs and their expression level of TNFR2. Our and previous results together strongly support that TNFR2 is a multicellular target in PDAC, mainly immune and tumor cells, and that the impact of its blockade may be due to different effects in PDAC TME. Consequently, the potential response of patients with PDAC to the anti-TNFR2 would probably depend both on the expression of TNFR2 in the different cell types and the characteristics of the immune cell infiltrate.
We then tried to better define the mechanism of TNFR2 inhibition by single-cell transcriptomic analysis of T cells infiltrated in the TME in untreated and treated mice. We found that Tregs within mPDAC TME were clustered in two groups: aTregs and rTregs. These clusters were identified based on a similar gene expression signature as the one found in physiological condition.47 Interestingly, the signature found in the aTreg cluster from the pathological murine PDAC TME includes genes hindering tumor immunity, which were previously identified in Treg from patients with PDAC.48 Indeed, highly immunosuppressive states of Tregs in patients with PDAC were associated with high Tigit, Icos and Cd39 expression. The anti-TNFR2 mAb treatment strongly impacted the cluster of aTregs compared with rTregs by decreasing specific markers. In particular, a signature of genes associated with the activated Treg clusters Tnfrsf1b, Ctla4, Icos, Cd39, Il2ra, Tigit, Tnfrsf4, Tnfrsf9, Ccr8, Pdcd1 significantly decreased in the PDAC TME of mice treated with anti-TNFR2 antibody. Using flow cytometry analysis, the frequency of Treg expressing TNFR2, CTLA-4 or TNFR2 and CTLA-4 was significantly reduced after anti-TNFR2 treatment, thus validating the transcriptomic data.
CD8+ T cells in human PDAC have an exhausted and senescent profile48 and phenotypes of exhaustion were clearly identified in the TME of our mouse models of PDAC. TNFR2 blockade did not change the phenotype of naïve CD8+ T cell cluster or the CD8 CTL cluster, but impacted the expression of exhaustion markers of CD8-Eff/mem T cells. A transcriptomic exhaustion score calculated by the expression of Tigit, Lag3, Pdcd1, Entpd1, Ctla4 and Havcr2, and the frequency of CD8+ T cells expressing TIGIT, PD-1 and CTLA-4 were significantly decreased on anti-TNFR2 treatment. TNFR2 inhibition is therefore able to affect Treg activation and CD8+ T cell exhaustion in the TME. Moreover, TCR sequencing revealed that TNFR2 blockade decreased the frequency of expanded aTreg clones.
Finally, TNFR2 blockade alone was not sufficient to decisively boost the clinical antitumoral response. However, the strong restriction of activated Tregs and the increase of the ratio between Teffs and Tregs suggested that a combination therapy with immunotherapy boosting CD8+ T cell activation and infiltration could promote more efficient T cell responses against PDAC.
Importantly, we showed that the combined treatment associating anti-TNFR2 and an agonistic anti-CD40 mAbs was able to increase CD3+ and CD8+ T cell frequency, as well as the expansion of IFN-γ-producing and granzyme B-producing CD8+ T cells. This combined treatment impacted tumor growth in mPDAC and Panc02 mouse models compared with control mice and to monotherapies with the anti-TNFR2 or anti-CD40 mainly in Panc02 model. Moreover, the survival of mice bearing Panc02-derived tumors was improved compared with control mice and 55% of anti-TNFR2 plus anti-CD40-treated mice were tumor free. This could reflect differences in the frequency of CD8+ T cell infiltration among CD45+ cells that in our hands is 10-fold higher in Panc02 than in PDAC. In this setting, it is not surprizing that a therapeutic strategy targeting the immune system has little effect in a poorly infiltrated model, while it has a significant clinical effect in a higher infiltrated tumor model. These observations are in line with a study in humans showing significant variation in the T cell infiltrate in PDAC, and that the size of the infiltrate correlates with prolonged survival.49 Interestingly, Panc02 treated mice developed a memory response against tumor re-challenge in the absence of any additional treatment.
Targeting immune checkpoints that suppress antitumor immune responses, mainly by using anti-CTLA-4 and anti-PD-1/PD-L1 mAbs, has demonstrated robust clinical activity in several cancers, but not in PDAC.5 Potential antitumoral activity of three types of immunotherapy in the ICI-refractory PDAC mouse models has been demonstrated, the agonist CD40, the antagonist CXCR4 and agonist OX40 by inducing T-cell priming and T cell infiltration.40,42 Only the combination of the blockade of TNFR2 together with the agonist anti-CD40 was able to impact tumor growth of PDAC mouse models. CD40 agonist have met a significant challenge in clinical development due to systemic toxicity. However, novel approaches are designed to circumvent the systemic toxicity associated with CD40 agonism.50 51 In an upcoming clinical trial, combining an anti-TNFR2 antibody with a modified CD40 agonist molecule would be the preferable choice. Recently, the CD40 agonist antibody tested in a phase II clinical trial did not show benefit.52 However, despite the failure of this immunotherapy, some patients had increased intratumoral T cell infiltration. Here, we provide a proof of principle in mice that treatment combining a CD40 agonist and a TNFR2 antagonist represents a novel promising immunotherapy approach that deserves to be tested to effectively treat patients with PDAC.
supplementary material
Acknowledgements
We are grateful to the IMRB for providing access to their animal facility team and the flow cytometry platform team for their help. We thank Virginie Reynal, Benoit Albaud, Laura Baudrin, and Patricia Legoix at the CurieCoreTech Next Generation Sequencing (ICGex) platform at Institut Curie. High throughput sequencing was performed by the ICGex NGS platform. Data management, quality control and primary analysis were performed by the Bioinformatics platform of the Institut Curie. We acknowledge Coralie Guerin, Anna Chipont, Annick Viguier, and Lea Guyonnetin at CurieCoreTech Cytometry platform at Institut Curie. The authors are grateful to Dr Sebastien Lemoine for his decisive help with the analysis in humans in sRNA-seq data.
Footnotes
Funding: This work was supported by INCA PRT-K 18-022 “IMPROVE ICI”, ANR-Theranuc from the Agence Nationale de la Recherche and the French charitable organisation “Ligue National contre le Cancer” “Combination of tumor vessel normalization strategy and immune checkpoint therapies for digestive cancers”, the LabEx DCBIOL (ANR-10-IDEX-0001-02 PSL; ANR-11-LABX-0043); and Center of Clinical Investigation (CIC IGR- Curie 1428). Institut Curie NGS analysis was supported by the grants ANR-10-EQPX-03 (Equipex) and ANR-10-INBS-09-08 (France Génomique Consortium) from the Agence Nationale de la Recherche ("Investissements d’Avenir" program), by the ITMO-Cancer Aviesan (Plan Cancer III) and by the SiRIC-Curie program (SiRIC Grant INCa-DGOS-465 and INCa-DGOSInserm_12554) and has received support under the program France 2030 launched by the French Government. AD received a PhD grant from the Université Paris-Est-Créteil (UPEC). SM was supported by grants from the National Institute for Cancer Research (INCA PRT-K 18-022 “IMPROVE ICI”).
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: Not applicable.
Data availability free text: Single-cell RNA sequencing raw datasets are available on the GEO database (accession number: GSE280808).
Contributor Information
Anais Debesset, Email: anais.debesset@gmail.com.
Caroline Pilon, Email: caroline.pilon@inserm.fr.
Sylvain Meunier, Email: sylvain.meunier@inserm.fr.
Orianne Cuelenaere-Bonizec, Email: orianne.bonizec@inserm.fr.
Wilfrid Richer, Email: Wilfrid.Richer@curie.fr.
Allan Thiolat, Email: allan.thiolat@inserm.fr.
Claire Houppe, Email: claire.justine@u-pec.fr.
Matteo Ponzo, Email: matteo.ponzo@ircc.it.
Jeanne Magnan, Email: jeanne.magnan@inserm.fr.
Jonathan Caron, Email: jonathan.caron@inserm.fr.
Pamela Caudana, Email: pamela.caudana@curie.fr.
Jimena Tosello Boari, Email: jimena.tosello@curie.fr.
Sylvain Baulande, Email: sylvain.baulande@curie.fr.
Nhu Han To, Email: nhuhanh.to@aphp.fr.
Benoit Laurent Salomon, Email: benoit.salomon@inserm.fr.
Eliane Piaggio, Email: Eliane.Piaggio@curie.fr.
Ilaria Cascone, Email: ilaria.cascone@inserm.fr.
José Laurent Cohen, Email: jose.cohen@inserm.fr.
Data availability statement
Data are available upon reasonable request. Data are available in a public, open access repository.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Słodkowski M, Wroński M, Karkocha D. Current approaches for the curative-intent surgical treatment of pancreatic ductal adenocarcinoma. Cancers (Basel) 2023;15:2584. doi: 10.3390/cancers15092584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. Lancet. 2020;395:2008–20. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 4.Halbrook CJ, Lyssiotis CA, Pasca di Magliano M, et al. Pancreatic cancer: advances and challenges. Cell. 2023;186:1729–54. doi: 10.1016/j.cell.2023.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong J, Hendifar A, Tuli R, et al. Combination systemic therapies with immune checkpoint inhibitors in pancreatic cancer: overcoming resistance to single-agent checkpoint blockade. Clin Transl Med. 2018;7:32. doi: 10.1186/s40169-018-0210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat Rev Clin Oncol. 2020;17:527–40. doi: 10.1038/s41571-020-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bärthel S, Falcomatà C, Rad R, et al. Single-cell profiling to explore pancreatic cancer heterogeneity, plasticity and response to therapy. Nat Cancer . 2023;4:454–67. doi: 10.1038/s43018-023-00526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolas-Boluda A, Vaquero J, Vimeux L, et al. Tumor stiffening reversion through collagen crosslinking inhibition improves T cell migration and anti-PD-1 treatment. Elife. 2021;10:e58688. doi: 10.7554/eLife.58688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samain R, Brunel A, Douché T, et al. Pharmacologic normalization of pancreatic cancer-associated fibroblast secretome impairs prometastatic cross-talk with macrophages. Cell Mol Gastroenterol Hepatol. 2021;11:1405–36. doi: 10.1016/j.jcmgh.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiraoka N, Onozato K, Kosuge T, et al. Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res. 2006;12:5423–34. doi: 10.1158/1078-0432.CCR-06-0369. [DOI] [PubMed] [Google Scholar]
- 11.Ino Y, Yamazaki-Itoh R, Shimada K, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 2013;108:914–23. doi: 10.1038/bjc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Zhao G, Wu W, et al. Low intratumoral regulatory T cells and high peritumoral CD8(+) T cells relate to long-term survival in patients with pancreatic ductal adenocarcinoma after pancreatectomy. Cancer Immunol Immunother. 2016;65:73–82. doi: 10.1007/s00262-015-1775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wartenberg M, Zlobec I, Perren A, et al. Accumulation of FOXP3+T-cells in the tumor microenvironment is associated with an epithelial-mesenchymal-transition-type tumor budding phenotype and is an independent prognostic factor in surgically resected pancreatic ductal adenocarcinoma. Oncotarget. 2015;6:4190–201. doi: 10.18632/oncotarget.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wartenberg M, Cibin S, Zlobec I, et al. Integrated genomic and immunophenotypic classification of pancreatic cancer reveals three distinct subtypes with prognostic/predictive significance. Clin Cancer Res. 2018;24:4444–54. doi: 10.1158/1078-0432.CCR-17-3401. [DOI] [PubMed] [Google Scholar]
- 15.Jang J-E, Hajdu CH, Liot C, et al. Crosstalk between regulatory T cells and tumor-associated dendritic cells negates anti-tumor immunity in pancreatic cancer. Cell Rep. 2017;20:558–71. doi: 10.1016/j.celrep.2017.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Subleski JJ, Hamano R, et al. Co-expression of TNFR2 and CD25 identifies more of the functional CD4+FOXP3+ regulatory T cells in human peripheral blood. Eur J Immunol. 2010;40:1099–106. doi: 10.1002/eji.200940022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Subleski JJ, Kopf H, et al. Cutting edge: expression of TNFR2 defines a maximally suppressive subset of mouse CD4+CD25+FoxP3+ T regulatory cells: applicability to tumor-infiltrating T regulatory cells. J Immunol. 2008;180:6467–71. doi: 10.4049/jimmunol.180.10.6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Bäumel M, Männel DN, et al. Interaction of TNF with TNF receptor type 2 promotes expansion and function of mouse CD4+CD25+ T regulatory cells. J Immunol. 2007;179:154–61. doi: 10.4049/jimmunol.179.1.154. [DOI] [PubMed] [Google Scholar]
- 19.Okubo Y, Mera T, Wang L, et al. Homogeneous expansion of human T-regulatory cells via tumor necrosis factor receptor 2. Sci Rep. 2013;3:3153. doi: 10.1038/srep03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaragoza B, Chen X, Oppenheim JJ, et al. Suppressive activity of human regulatory T cells is maintained in the presence of TNF. Nat Med. 2016;22:16–7. doi: 10.1038/nm.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moatti A, Cohen JL. The TNF-α/TNFR2 pathway: targeting a brake to release the anti-tumor immune response. Front Cell Dev Biol. 2021;9:725473. doi: 10.3389/fcell.2021.725473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moatti A, Debesset A, Pilon C, et al. TNFR2 blockade of regulatory T cells unleashes an antitumor immune response after hematopoietic stem-cell transplantation. J Immunother Cancer. 2022;10:e003508. doi: 10.1136/jitc-2021-003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Oppenheim JJ. Targeting TNFR2, an immune checkpoint stimulator and oncoprotein, is a promising treatment for cancer. Sci Signal. 2017;10:eaal2328. doi: 10.1126/scisignal.aal2328. [DOI] [PubMed] [Google Scholar]
- 24.Torrey H, Butterworth J, Mera T, et al. Targeting TNFR2 with antagonistic antibodies inhibits proliferation of ovarian cancer cells and tumor-associated Tregs. Sci Signal. 2017;10:eaaf8608. doi: 10.1126/scisignal.aaf8608. [DOI] [PubMed] [Google Scholar]
- 25.Torrey H, Khodadoust M, Tran L, et al. Targeted killing of TNFR2-expressing tumor cells and Tregs by TNFR2 antagonistic antibodies in advanced Sézary syndrome. Leukemia. 2019;33:1206–18. doi: 10.1038/s41375-018-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie Y, He J, Shirota H, et al. Blockade of TNFR2 signaling enhances the immunotherapeutic effect of CpG ODN in a mouse model of colon cancer. Sci Signal. 2018;11:eaan0790. doi: 10.1126/scisignal.aan0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q, Shen Q, Tong J, et al. Anti-tumor necrosis factor receptor 2 antibody combined with anti-PD-L1 therapy exerts robust antitumor effects in breast cancer. Front Cell Dev Biol. 2021;9:720472. doi: 10.3389/fcell.2021.720472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilles M-E, Maione F, Cossutta M. Nucleolin targeting impairs the progression of pancreatic cancer and promotes the normalization of tumor vasculature. Cancer Res. 2016;76:7181–93. doi: 10.1158/0008-5472.CAN-16-0300. [DOI] [PubMed] [Google Scholar]
- 29.Ponzo M, Debesset A, Cossutta M, et al. Nucleolin therapeutic targeting decreases pancreatic cancer immunosuppression. Cancers (Basel) 2022;14:4265. doi: 10.3390/cancers14174265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J, Sun B-F, Chen C-Y, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–38. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linderman GC, Zhao J, Roulis M, et al. Zero-preserving imputation of single-cell RNA-seq data. Nat Commun. 2022;13:192. doi: 10.1038/s41467-021-27729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Geest KSM, Abdulahad WH, Horst G, et al. Quantifying distribution of flow cytometric TCR-Vβ usage with economic statistics. PLoS ONE. 2015;10:e0125373. doi: 10.1371/journal.pone.0125373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, Zhang Q, Chen H, et al. TNFR2 promotes pancreatic cancer proliferation, migration, and invasion via the NF-κB signaling pathway. Aging (Milano) 2023;15:8013–25. doi: 10.18632/aging.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan-Seng-Yue M, Kim JC, Wilson GW, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–40. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, Rong L, Zhao X, et al. TNF signaling drives myeloid-derived suppressor cell accumulation. J Clin Invest. 2012;122:4094–104. doi: 10.1172/JCI64115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pergamo M, Miller G. Myeloid-derived suppressor cells and their role in pancreatic cancer. Cancer Gene Ther. 2017;24:100–5. doi: 10.1038/cgt.2016.65. [DOI] [PubMed] [Google Scholar]
- 39.Bianchi A, De Castro Silva I, Deshpande NU, et al. Cell-autonomous Cxcl1 sustains tolerogenic circuitries and stromal inflammation via neutrophil-derived TNF in pancreatic cancer. Cancer Discov. 2023;13:1428–53. doi: 10.1158/2159-8290.CD-22-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winograd R, Byrne KT, Evans RA, et al. Induction of t-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol Res. 2015;3:399–411. doi: 10.1158/2326-6066.CIR-14-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y, Li J, Wang H, et al. Combination of PD-1 inhibitor and OX40 agonist induces tumor rejection and immune memory in mouse models of pancreatic cancer. Gastroenterology. 2020;159:306–19. doi: 10.1053/j.gastro.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bockorny B, Semenisty V, Macarulla T, et al. BL-8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: the COMBAT trial. Nat Med. 2020;26:878–85. doi: 10.1038/s41591-020-0880-x. [DOI] [PubMed] [Google Scholar]
- 43.Polz J, Remke A, Weber S, et al. Myeloid suppressor cells require membrane TNFR2 expression for suppressive activity. Immun Inflamm Dis. 2014;2:121–30. doi: 10.1002/iid3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Lao M, Xu J, et al. Combination cancer immunotherapy targeting TNFR2 and PD-1/PD-L1 signaling reduces immunosuppressive effects in the microenvironment of pancreatic tumors. J Immunother Cancer. 2022;10:e003982. doi: 10.1136/jitc-2021-003982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengsch F, Knoblock DM, Liu A, et al. CTLA-4/CD80 pathway regulates T cell infiltration into pancreatic cancer. Cancer Immunol Immunother. 2017;66:1609–17. doi: 10.1007/s00262-017-2053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feig C, Jones JO, Kraman M, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20212–7. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zemmour D, Zilionis R, Kiner E, et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19:291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivakumar S, Abu-Shah E, Ahern DJ, et al. Activated regulatory T-cells, dysfunctional and senescent T-cells hinder the immunity in pancreatic cancer. Cancers (Basel) 2021;13:1776. doi: 10.3390/cancers13081776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orhan A, Vogelsang RP, Andersen MB. The prognostic value of tumour-infiltrating lymphocytes in pancreatic cancer: a systematic review and meta-analysis. Eur J Cancer. 2020;132:71–84. doi: 10.1016/j.ejca.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Sum E, Rapp M, Fröbel P, et al. Fibroblast activation protein α-targeted CD40 agonism abrogates systemic toxicity and enables administration of high doses to induce effective antitumor immunity. Clin Cancer Res. 2021;27:4036–53. doi: 10.1158/1078-0432.CCR-20-4001. [DOI] [PubMed] [Google Scholar]
- 51.Salomon R, Dahan R. Next generation CD40 agonistic antibodies for cancer immunotherapy. Front Immunol. 2022;13:940674. doi: 10.3389/fimmu.2022.940674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Hara MH, O’Reilly EM, Varadhachary G, et al. CD40 agonistic monoclonal antibody APX005M (sotigalimab) and chemotherapy, with or without nivolumab, for the treatment of metastatic pancreatic adenocarcinoma: an open-label, multicentre, phase 1b study. Lancet Oncol. 2021;22:118–31. doi: 10.1016/S1470-2045(20)30532-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request. Data are available in a public, open access repository.