Abstract
Background
Circulating tumour DNA (ctDNA) has emerged as a valuable liquid biopsy biomarker in the field of oncology, including head and neck squamous cell carcinomas (HNSCCs), offering potential insights into cancer diagnosis, progression, and prognosis. This review aims to comprehensively evaluate the utility of ctDNA as a prognostic biomarker in HNSCC.
Methods
PubMed and Ovid were searched as part of our review. Studies that investigated the relationship between ctDNA and prognosis in HNSCC patients were included. Outcomes extracted included basic characteristics, ctDNA details and survival data. Meta-analysis was performed on eligible studies to determine pooled progression-free/recurrence-free survival (RFS/PFS) and overall survival (OS).
Results
Twenty-two studies were included, involving 5062 HNSCC patients from 11 countries. The meta-analysis demonstrated that the positive ctDNA/methylation detection was associated with worse OS (HR = 2.00, 95% CI 1.35–2.96) and worse PFS/RFS (HR = 3.54, 95% CI 1.05–11.85). Positive ctEBV DNA was associated with poorer OS (HR = 2.86, 95% CI 1.84–4.45) and poorer PFS/RFS (HR = 1.93, 95% CI 1.74–2.13). Positive ctHPV DNA was associated with poorer OS (HR = 1.38, 95% CI 1.07–1.38) but not PFS/PFS (HR = 1.33, 95% CI 0.96–1.85).
Conclusion
Meta-analysis indicates that the status of ctDNA is significantly associated with the prognosis of HNSCC patients, with ctDNA/methylation-negative patients demonstrating better PFS/RFS and OS.
Keywords: Head and neck squamous cell carcinoma, Prognosis, Survival, ctDNA
Introduction
Head and neck squamous cell carcinomas (HNSCCs) develop from the mucosal epithelia of the oral cavity, pharynx, and larynx, and are the most common malignancies of the head and neck. Currently, HNSCC are typically treated by surgical resection, followed by adjuvant radiotherapy or chemotherapy plus radiotherapy depending on the stage of the disease [1]. Over years of research and clinical practice, the treatment of HNSCC has made considerable progress, but the therapeutic effect still needs to be improved [2]. Early detection and treatment are key to improving patient prognosis, but there is currently no effective screening strategy for HNSCC. Patients often come to consult when the disease has progressed to the late stage, which limits further treatment [3]. The current evaluation and dynamic monitoring of HNSCC treatment mainly rely on imaging techniques such as CT and MRI, which have low sensitivity and specificity for detecting small residual lesions, new detection methods are needed to further improve the monitoring of small lesions.
Liquid biopsies provide new insights into tumor treatment and prognosis evaluation [4, 5]. Among several types of liquid biopsies developed, circulating tumor DNA (ctDNA) has attracted increasing attention. CtDNA is cell-free DNA (cfDNA) carrying tumor-related mutations, and is believed to be closely related to tumor occurrence and development due to carrying tumor-related genes [6]. Monitoring ctDNA reflects the current state of the tumor to some extent. In clinical practice, ctDNA is receiving increasing attention as a means of disease monitoring. Comparing to traditional image monitoring, ctDNA detection can detect smaller lesions earlier, and its detection can help in tumor treatment dye to carrying information related to tumor tissue [7]. In terms of HNSCC, increasing studies have pointed to the role of ctDNA in patient diagnosis, treatment and prognosis, but different studies have inconsistent findings regarding the role of ctDNA in HNSCC. Some studies suggest that ctDNA can be used as a prognostic indicator while others indicate that it has no role in prognosis [8]. We now review and perform meta-analysis on existing studies to explore the relationship between ctDNA and the prognosis of HNSCC patients, aiming to provide guidance for clinical treatment.
Materials and methods
Literature search strategy
We conducted a systematic review and meta-analysis following the PRISMA reporting guidelines [9].
In April 2024, we conducted a systematic literature search to identify clinical studies describing the association between ctDNA detection and the prognosis of HNSCC patients. We used PubMed to search the MEDLINE database and Ovid to search the EMBASE database. The search terms used were: ctDNA, cfDNA, circulating tumor DNA, circulating free DNA, HNSCC, head and neck squamous cell carcinoma. Additionally, we manually searched the reference lists of these articles for other relevant studies.
Every retrieved abstract was independently reviewed by two authors to determine inclusion eligibility. A third person obtained full-text articles for further review. Any discrepancies were resolved through discussion among the three authors.
Study Selection Criteria
We included studies that investigated the relationship between ctDNA and prognosis in HNSCC patients. Included studies comprised patients diagnosed solely with HNSCC or those diagnosed with other diseases in addition to HNSCC. Studies involving both children and adults were included. Studies that included other tumors or concurrent diseases that could affect our analysis were excluded. Studies that did not provide essential information or had duplicate data reported multiple times were also excluded.
Data extraction
We used Overall Survival (OS), Progression-Free Survival (PFS) and Recurrence-Free Survival (RFS) as outcome measures in our analysis. Basic information about patients, treatment modalities used, cohort standardization, and time were also extracted. All data were verified by comparing with the results of the original publications, and any discrepancies were resolved through discussion or by contacting the authors if necessary.
Quality Assessment
Two authors independently assessed the methodological quality of each clinical trial, with involvement of a third author in case of disagreements. The Newcastle-Ottawa Scale (NOS) was used to assess quality on a ten-point scale [10]. Studies scoring below six were categorized as low quality, while those scoring six or above were considered high quality.
Statistical analysis
Analysis was performed using Review Manager software (RevMan 5.1) from the Cochrane Collaboration. We collected data on RFS, PFS, and OS of HNSCC patients with high vs. low ctDNA expression and analyzed the relationship between ctDNA and cancer prognosis. Hazard Ratios (HRs) with corresponding 95% confidence intervals (CIs) were collected from the included studies. Pooled HRs with their 95% CIs and p-values were calculated, where HR > 1 indicated worse prognosis and HR < 1 indicated better prognosis for cancer patients. I2 statistic was used to assess statistical heterogeneity among studies. A fixed-effects model was used to combine results if statistical homogeneity was observed (p > 0.05, I2 < 50%), while a random-effects model was used if statistical heterogeneity was present (p < 0.05, I2 > 50%). Efforts were made to identify the source of heterogeneity when found in included studies. Sensitivity analysis was conducted if the heterogeneity stemmed from low-quality studies or differing assessment methods. Otherwise, Engauge Digitizer version 12 was used to extract survival data not presented numerically from Kaplan-Meier curves, and HRs were calculated using Parmar and Tierney methods.
Results
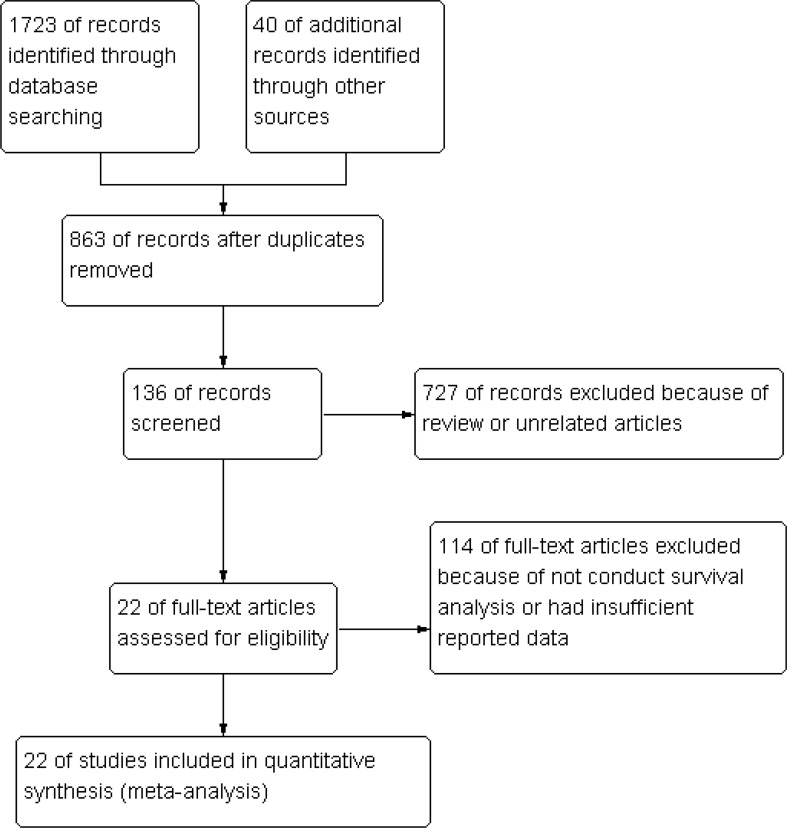
A total of 1763 articles were obtained through systematic literature search. After careful review of titles and abstracts, 1586 irrelevant papers and duplicates were excluded. Following the exclusion of articles that did not conduct survival analysis or had insufficient reported data, a total of 22 clinical studies evaluating the role of ctDNA in the prognosis of HNSCC patients were identified [11–32] (Fig. 1). The included studies were published between 2002 and 2024, involving 5062 HNSCC patients from 11 countries. The main clinical characteristics of the studies are presented in (Table 1). PCR (n = 16) and NGS (n = 6) were the main analytical methods used to detect peripheral blood circulating ctDNA. Eleven studies investigated the relationship between ctDNA levels from baseline blood samples and prognosis, while 13 studies examined the relationship between ctDNA from post-treatment blood samples and patient prognosis. The included studies explored the impact of peripheral blood ctDNA (n = 6), methylated ctDNA (n = 3), ctEBV DNA (n = 8), and ctHPV DNA (n = 5) on patient prognosis. Of the 21 studies included in the meta-analysis, survival outcomes were reported using various indicators, with 20 reporting OS, 2 reporting RFS, and 13 reporting PFS. RFS/PFS were analyzed as a single indicator. All studies had NOS scores greater than 7, indicating high-quality research (Table 2).
Fig. 1.
Flow diagram of the literature retrieval and selection for this study
Table 1.
Characteristics of studies
| study | location | Numbers of patients | Sampling time Baseline/ Post-treatment | Method of detection | Gene mutation detected | Sex male/female | Age median(range) |
|
|---|---|---|---|---|---|---|---|---|
| Chan 2002 [26] | China | 170 | Post-treatment | PCR based | ctEBV DNA | 143/27 | 46 (18–80) | |
| Lin 2004 [22] | China | 99 | Post-treatment | PCR based | ctEBV DNA | 73/26 | 47(24–74) | |
| Jiun 2012 [21] | Malaysia | 157 | Baseline | PCR based | ctEBV DNA | NA | NA | |
| Ahn 2014 [29] | Maryland | 53 | Post-treatment | PCR based | ctHPV DNA | 81/12 | NA | |
| Leung 2014 [25] | China | 107 | Post-treatment | PCR based | ctEBV DNA | NA | NA | |
| ZHAO 2015 [27] | China | 227 | Post-treatment | PCR based | ctEBV DNA | 477/160 | 46(14–80) | |
| Mydlarz 2015 [11] | USA | 100 | Baseline | PCR based | EDNRB, DCC, p16,ect. | 77/23 | 58.3 | |
| Schröck 2017[12] | Germany | 129 | Baseline | PCR based | SEPT9 | NA | NA | |
| Vos 2017 [13] | Germany | 137 | Baseline | PCR based | SEPT9,SHOX2 | 111/26 | 62(32–89) | |
| Chan 2018 [28] | China | 789 | Post-treatment | PCR based | ctEBV DNA | 593/196 | 51(19–81) | |
| Guo 2019 [24] | China | 1529 | Baseline | PCR based | ctEBV DNA | 1141/392 | NA | |
| Burgener 2021 [14] | Canada | 30 | Baseline | NGS based | TP53,PIK3CA, PCLO, ect. | NA | NA | |
| Wilson 2021 [15] | USA | 67 | Post-treatment | NGS based | TP53, CDKN2A, TERT, ect. | 52/23 | NA | |
| Burcher 2021 [16] | USA | 146 | Post-treatment | NGS based | BRCA1,BRCA2,ATM, ect. | 123/47 | NA | |
| Lv 2022 [23] | China | 821 | Post-treatment | PCR based | ctEBV DNA | 617/204 | NA | |
| Cao 2022 [30] | Michigan | 34 | Baseline | NGS based | ctHPV DNA | 31/3 | 64(47–79) | |
| Routman 2022 [31] | USA | 159 | Post-treatment | PCR based | ctHPV DNA | 141/18 | 58.6(35.7–77.6) | |
| Kogo 2022 [17] | Japan | 18 | Post-treatment | PCR based | TP53,CDKN2A, FAT1,ect. | 15/11 | 71 (42–86) | |
| Honoré 2023 [18] | Belgium | 53 | Post-treatment | NGS based | TP53,PIK3CA, LRP1B, ect. | 40/13 | 63.7 (41.1, 98.7) | |
| Economopoulou 2023 [19] | Greece | 46 | Baseline | NGS based | TP53, CDKN2A, HRAS, ect. | 46/16 | NA | |
| Adrian 2023 [32] | Sweden | 136 | Baseline | PCR based | ctHPV DNA | 110/82 | 60 (33–77) | |
| Hanna 2024 [20] | USA | 55 | Post-treatment | PCR based | BRCA1,BRCA2,ATM, ect. | 79/37 | 65.4 (21–94) | |
Table 2.
Results of the newcastle–ottawa scale (NOS) quality assessment
| Study | Selection | Comparability | Outcome | Total |
|---|---|---|---|---|
| Chan 2002 | **** | ** | *** | ********* |
| Lin 2004 | **** | ** | *** | ********* |
| Jiun 2012 | *** | ** | ** | ******* |
| Ahn 2014 | **** | ** | *** | ********* |
| Leung 2014 | *** | ** | *** | ******** |
| ZHAO 2015 | **** | ** | *** | ********* |
| Mydlarz 2015 | **** | ** | ** | ******* |
| Schröck 2017 | **** | ** | ** | ******* |
| Vos 2017 | **** | ** | *** | ******** |
| Chan 2018 | **** | ** | *** | ********* |
| Guo 2019 | *** | ** | *** | ******** |
| Burgener 2021 | *** | * | *** | ******* |
| Wilson 2021 | *** | ** | ** | ******* |
| Burcher 2021 | **** | ** | ** | ******** |
| Lv 2022 | *** | ** | *** | ******** |
| Cao 2022 | **** | ** | *** | ********* |
| Routman 2022 | **** | ** | ** | ******** |
| Kogo 2022 | **** | ** | *** | ********* |
| Honoré 2023 | **** | ** | *** | ******** |
| Economopoulou 2023 | *** | ** | *** | ******** |
| Adrian 2023 | **** | ** | *** | ********* |
| Hanna 2024 | **** | ** | *** | ********* |
Relationship between ctDNA/ methylation and prognosis in HNSCC patients
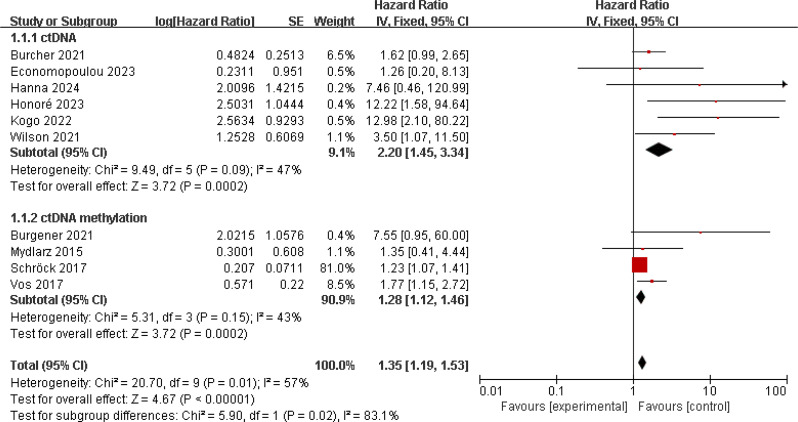
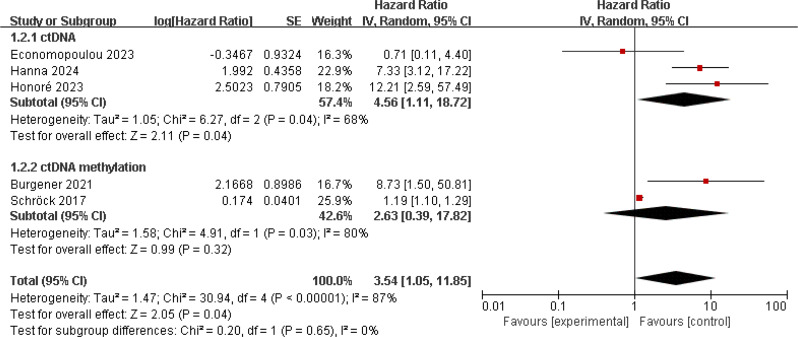
To analyze the correlation between ctDNA levels and patient prognosis in HNSCC, we first analyzed studies assessing OS. 10 studies reported on the impact of ctDNA and ctDNA methylation on prognosis, involving 781 HNSCC patients [11, 12, 14, 17–19, 33–36]. Studies on patient ctDNA methylation all utilized baseline samples, while one study used baseline samples for patient ctDNA research, and the rest used post-treatment samples. There was no significant heterogeneity among these studies (I2 = 57%; p = 0.01), so a random-effects model was used. The results showed that patients with positive ctDNA/methylation detection had worse OS (HR = 2.00, 95% CI 1.35–2.96, p = 0.0006). Subgroup analysis showed that patients positive for ctDNA (HR = 3.39, 95% CI 1.51–7.62, p = 0.003) or ctDNA methylation (HR = 1.47, 95% CI 1.03–2.09, p = 0.03) had worse OS (Fig. 2). We then analyzed studies involving patient PFS/RFS, totaling 5 studies, which showed significant heterogeneity (I2 = 87%; p < 0.001) [12, 18, 19, 33, 34]. A random-effects model was used, and the results indicated that patients with positive ctDNA/methylation detection had worse PFS/RFS (HR = 3.54, 95% CI 1.05–11.85, p = 0.04). Further research indicated an association between positive ctDNA and poorer PFS/RFS (HR = 4.56, 95% CI 1.11–18.72, p = 0.04) (Fig. 3).
Fig. 2.
Forest plot of OS in HNSCC patients with ctDNA-positive compared with ctDNA-negative
Fig. 3.
Forest plot of PFS/RFS in HNSCC patients with ctDNA-positive compared with ctDNA-negative
Relationship between ctEBV DNA and prognosis in HNSCC patients
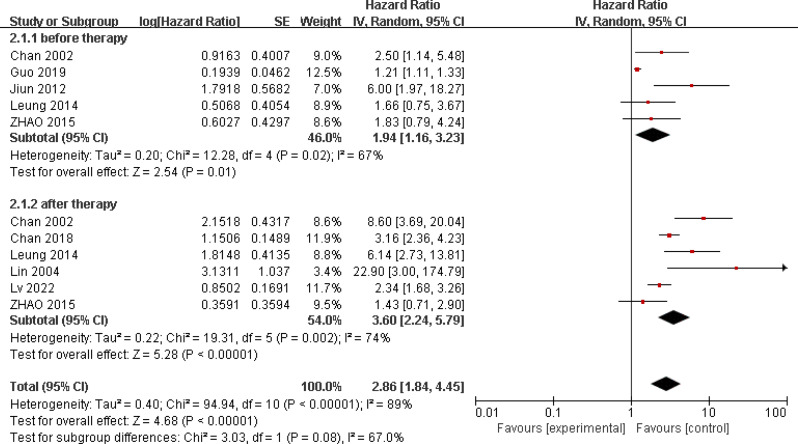
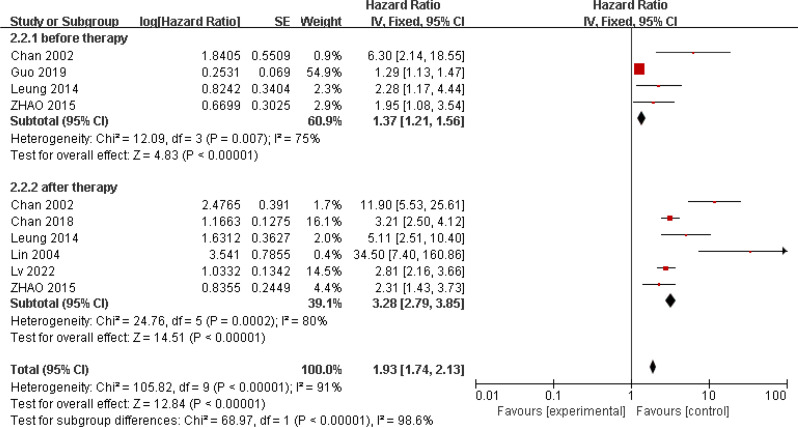
Eight studies reported on the relationship between ctEBV DNA and patient OS, involving 4313 patients [22, 23, 27, 37–41]. Five studies investigated the relationship between pre-treatment peripheral blood ctEBV DNA status and prognosis, while six studies examined the relationship between post-treatment peripheral blood EBV DNA status and prognosis. These studies showed significant heterogeneity (I2 = 89%; p < 0.001), so a random-effects model was used. The results showed an association between ctEBV DNA positivity and poorer OS (HR = 2.86, 95% CI 1.84–4.45, p < 0.001). Subgroup analysis showed that pre-treatment peripheral blood ctEBV DNA positivity (HR = 1.94, 95% CI 1.16–3.23, p = 0.01) and post-treatment peripheral blood ctEBV DNA positivity (HR = 3.6, 95% CI 2.24–5.79, p < 0.001) were both associated with poorer OS(Fig. 4). Seven studies reported on the relationship between ctEBV DNA and patient PFS, showing significant heterogeneity (I2 = 91%; p < 0.001). A random-effects model was used, and the results indicated an association between ctEBV DNA positivity and poorer PFS (HR = 1.93, 95% CI 1.74–2.13, p < 0.001). Subgroup analysis showed that pre-treatment peripheral blood ctEBV DNA positivity (n = 4, HR = 1.37, 95% CI 1.21–1.56, p < 0.001) and post-treatment ctEBV DNA positivity (n = 6, HR = 3.28, 95% CI 2.79–3.85, p < 0.001) were both associated with poorer PFS(Fig. 5).
Fig. 4.
Forest plot of OS in HNSCC patients with ctEBV DNA-positive compared with ctEBV DNA-negative
Fig. 5.
Forest plot of PFS/RFS in HNSCC patients with ctEBV DNA-positive compared with ctEBV DNA-negative
Relationship between ctHPV DNA and prognosis in HNSCC patients
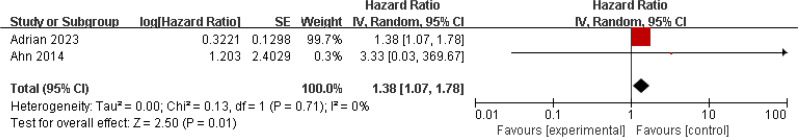
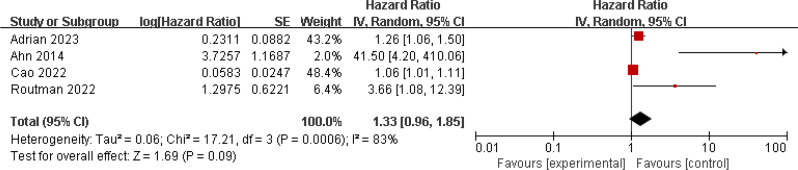
Three studies investigated the relationship between ctHPV DNA and patient OS, involving 469 patients [30, 32, 42]. There was no significant heterogeneity among the studies (I2 = 33%; p = 0.22), so a fixed-effects model was used. The results showed significant association between positive ctHPV DNA status and poorer OS (HR = 1.38, 95% CI 1.07–1.38, p = 0.01) (Fig. 6). Four studies examined the relationship between ctHPV DNA and patient PFS, showing significant heterogeneity (I2 = 83%; p < 0.001). A random-effects model was used, and the results did not suggest a significant association between ctHPV DNA status and patient PFS/RFS (HR = 1.33, 95% CI 0.96–1.85, p = 0.09) (Fig. 7).
Fig. 6.
Forest plot of OS in HNSCC patients with ctHPV DNA-positive compared with ctHPV DNA-negative
Fig. 7.
Forest plot of PFS/RFS in HNSCC patients with ctHPV DNA-positive compared with ctHPV DNA-negative
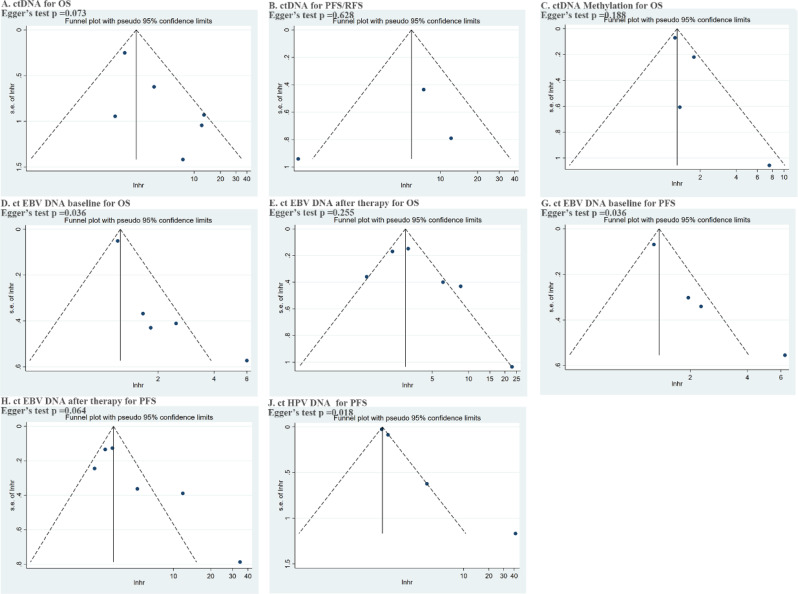
We summarized and visually estimated the risk of publication bias through funnel plots (Fig. 8). Signs of publication bias in each subgroup were assessed using Begg and Egger tests. The results indicated publication bias in terms of the impact of pre-treatment ctEBV DNA on OS or PFS, and ctHPV DNA on PFS.
Fig. 8.
Egger‘s test results
Discussion
Early detection of tumors and monitoring for recurrence are crucial for improving patient prognosis. Peripheral blood ctDNA testing provides a new approach for the diagnosis and treatment of HNSCC. Compared to traditional biochemical and imaging tests, ctDNA testing has its own unique advantages in both research and clinical practice. On one hand, unlike traditional imaging tests, ctDNA testing can detect tiny lesions within the body, thus enabling precise assessment of the patient’s disease status. On the other hand, ctDNA reflects the genetic information of tumor tissue specimens to some extent, which aids in tailoring tumor treatment [6, 43].In recent years, many studies have investigated the value of various liquid biopsy modes, including peripheral blood ctDNA testing, in screening, diagnosing, and predicting outcomes in HNSCC patients [44, 45]. We conducted a comprehensive meta-analysis for the first time, aimed at evaluating the role of ctDNA in HNSCC prognosis. We found an association between ctDNA and patient prognosis in HNSCC, where patients testing positive for peripheral blood ctDNA showed poorer PFS and OS. This suggests that ctDNA could serve as a potential biomarker for HNSCC prognosis, providing a reference for its clinical use.
CtDNA testing provides a new approach for the diagnosis and monitoring of tumors. However, the abundance of ctDNA in patient plasma is often low, and false positives in ctDNA testing due to factors like clonal hematopoiesis limit its effectiveness in cancer patient management, necessitating more stable indicators to increase reliability of test results [46].Methylation-based ctDNA analysis offers an alternative strategy for liquid biopsy. DNA methylation is a crucial mechanism regulating gene expression in cells, and cancer cells exhibit significant abnormalities in methylation patterns, such as global hypomethylation or specific hypermethylation on tumor suppressor gene promoters. This aberrant promoter hypermethylation has been proposed as a method to detect the status of solid tumors, including HNSCC [47].DNA methylation changes often occur at specific known sites, making them easier to target for sequencing. This lowers sequencing costs and improves the sensitivity and accuracy of detection [48]. Burgener et al. included 50 HNSCC patients and normal controls, using different methods for sequencing. Their results demonstrated high consistency between mutation-based and methylation-based analysis methods for detecting ctDNA and indicated both methods play a role in patient prognosis assessment. This suggests that methylation-based ctDNA testing contributes to biomarker discovery and clinical application in cases of low ctDNA abundance [14].Our subgroup analysis of related studies also found an association between methylation-positive ctDNA and poorer PFS/RFS and OS in HNSCC patients. This indicates that methylation-based ctDNA testing facilitates prognosis evaluation and serves as a potential biomarker for HNSCC.
The method of ctDNA detection impacts the test results. Currently, ctDNA detection methods mainly include PCR-based methods such as ddPCR and next-generation sequencing (NGS). ddPCR offers advantages of low cost and short detection time but is limited to detecting known mutations and analyzing a limited number of variants, thus restricting its further clinical use. NGS is a high-throughput technology capable of simultaneously detecting billions of DNA molecules and identifying previously unrecognized mutations, providing clinically relevant information with higher sequencing depth. NGS-based ctDNA detection methods such as TAm-Seq, eTAm-Seq, CAPP-Seq, and TEC-Seq are increasingly used in clinical practice, improving detection accuracy and providing more genetic information relevant to tumors [49, 50]. However, HNSCC encompasses tumors from various tissue sources such as the tongue, pharynx, and palate, leading to variability in mutation targets among different patients. For HNSCC patients, selecting an appropriate panel and conducting targeted NGS testing is crucial. Currently, there is no universally recognized specific mutation panel for HNSCC. TP53, PIK3CA, CDKN2A, among others, are considered potential targets, but extensive clinical trials are still needed to establish the optimal panel [43].
HPV is considered to be associated with the occurrence and development of HNSCC, serving as a potential biomarker for disease prediction and treatment monitoring in HNSCC patients [51]. The majority of HPV infections are asymptomatic, and most patients do not develop tumors. In some cases, HPV DNA from the virus integrates into the human keratinocyte nuclear DNA, expressing oncogenic proteins E6 and E7. This integration process, particularly the expression of E7, leads to uncontrolled cell proliferation, while increased p16 expression often serves as a marker for HPV-associated HNSCC [52]. HPV DNA status is believed to be associated with the prognosis of head and neck cancer patients. Shi et al. conducted a meta-analysis of 18 studies and found that HPV DNA positivity in tissue is associated with better OS in hypopharyngeal carcinoma patients (HR = 0.61, 95% CI 0.54–0.69) [53]. Apart from tissue HPV DNA, circulating tumor HPV DNA (ctHPV DNA) has been extensively studied in the diagnosis and treatment of HNSCC. Tanaka et al. examined ctHPV DNA in 35 HPV16-related HNSCC patients and observed that detection of ctHPV16 DNA after treatment correlates with increased likelihood of treatment failure [54]. Research by Cao et al. involving 34 p16 + Oropharyngeal squamous cell carcinoma (OPSCC) patients indicated that early changes in ctHPV DNA levels correlate with sensitivity to radiochemotherapy [30]. Abbas et al.’s meta-analysis of 6 studies suggested a higher recurrence rate (HR = 12.25) in OPSCC patients with ctHPV DNA positivity [55].While these studies highlight the role of ctHPV DNA in HNSCC diagnosis and treatment, our analysis of included studies indicates that the plasma ctHPV DNA status is not significantly correlated with patients PFS, but correlated with patients OS. Further research is necessary to clarify the impact of ctHPV DNA on patient outcomes.
Similar to HPV, EBV infection plays a significant role in the development of HNSCC, particularly in nasopharyngeal carcinoma (NPC). In China, EBV-associated NPC is prevalent in the southern regions and Southeast Asia, where nearly every cancer cell contains the EBV genome [56]. In the human body, circulating EBV DNA is generally rapidly cleared, resulting in low levels of EBV DNA in circulation from asymptomatic carriers through cell death release. However, in EBV-related NPC, the situation changes due to virus reactivation and rapid turnover of tumor cells, leading to increased release of EBV DNA into circulation and detectability [57]. Therefore, elevated levels of circulating EBV DNA are associated with the disease itself in patients and are considered a potential tumor marker, playing a role in the diagnosis and treatment of NPC. Chan et al. screened 20,174 subjects and found that compared to endoscopy and MRI, early detection of NPC patients using ctEBV DNA testing showed significantly higher proportions (71% vs. 20%) and improved 3-year progression-free survival (HR 0.10; 95% CI 0.05–0.18), indicating that EBV DNA testing can effectively identify early-stage NPC patients and improve patient prognosis [58]. Guo et al. included 1141 EBV-related NPC patients, detected ctEBV DNA before treatment, and developed a new analysis system. Results showed that incorporating ctEBV DNA improved hazard consistency (1.81 vs. 4.97), discrimination (0.50 vs. 0.56), outcome prediction (27.8 vs. 26.4), and sample balance (0.48 vs. 0.53) compared to the eighth edition TNM staging system, indicating enhancements to the existing staging system [24]. These studies demonstrate the role of pre-treatment ctEBV DNA testing in patient diagnosis and treatment. Subgroup analysis exploring the relationship between ctEBV DNA and patient prognosis showed that EBV DNA status effectively predicts outcomes, with EBV DNA-negative patients exhibiting better PFS and OS. Furthermore, our analysis indicates that both post-treatment and pre-treatment EBV DNA testing play a role in predicting the prognosis of NPC patients.
This meta-analysis represents the first study of ctDNA in HNSCC, including the largest number of studies and subgroup analyses based on detection timing and target points. However, there are several limitations to this research. Firstly, the included studies exhibit variability in both the methods of ctDNA detection and the interpretation of ctDNA test results, which may contribute to heterogeneity. Some studies categorize patients into ctDNA detected and ctDNA undetected groups, while others establish different cutoff values based on clinical circumstances and classify patients into ctDNA high and ctDNA low groups. We uniformly use ctDNA positive to refer to ctDNA detected and ctDNA high in the studies, and ctDNA negative to refer to ctDNA undetected and ctDNA low, which are significant sources of heterogeneity in the analysis. Secondly, the scope of ctDNA detection varied among studies, and some results may not fully represent the overall ctDNA mutation status of patients, thus contributing to further heterogeneity. Lastly, despite comprehensive and systematic literature searches, publication bias remains unavoidable. Future studies should focus on large-scale, multi-center, prospective clinical research to validate the findings obtained from current analyses.
Conclusion
Our meta-analysis indicates that the status of ctDNA is significantly associated with the prognosis of HNSCC patients, with ctDNA/methylation-negative patients demonstrating better PFS/RFS and OS.
Acknowledgements
This article was supported by the Chengdu international science and technology cooperation project [2019-GH02-00003-HZ], Sichuan Provincial Science and Technology Department[2022JDRC0142] and Sichuan Health Commission of Sichuan Province Medical Science and Technology Program [24LCYJZD06], [24LCYJPT06].
Author contributions
R.Y. wrote the main manuscript text, prepare Figs. 1, 2, 3, 4, 5, 6 and 7 and approved the submitted version. T.L. interpretation of data and approved the submitted version. S.Z. acquisition, analysis and approved the submitted version. C.S. analysis and approved the submitted version. H.M.prepare Table 1, and 2 and approved the submitted version. C.L. have drafted the work or substantively revised text and approved the submitted version. All authors reviewed the manuscript.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hong Ma, Email: mahong@gmc.edu.cn.
Chao Li, Email: lichao@scszlyy.org.cn.
References
- 1.Johnson D E Burtnessb, Leemans C R, et al. Head and neck squamous cell carcinoma [J]. Nat Reviews Disease Primers. 2020;6(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung K-W, Won Y-J, Kong H-J, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2015 [J]. Cancer Res Treatment: Official J Korean Cancer Association. 2018;50(2):303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamoli A, Gosavi A S, Shirwadkar U P, et al. Overview of oral cavity squamous cell carcinoma: risk factors, mechanisms, and diagnostics [J]. Oral Oncol. 2021;121:105451. [DOI] [PubMed] [Google Scholar]
- 4.Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer [J]. Nature. 2019;570(7761):385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbosh C, Birkbak N J, Wilson G A, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution [J]. Nature. 2017;545(7655):446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management [J]. Comput Struct Biotechnol J. 2018;16:370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pessoa L S, Heringer M, Ferrer V P. ctDNA as a cancer biomarker: a broad overview [J]. Crit Rev Oncol/Hematol. 2020;155:103109. [DOI] [PubMed] [Google Scholar]
- 8.Huang X, Duijf P H, Sriram S, et al. Circulating tumour DNA alterations: emerging biomarker in head and neck squamous cell carcinoma [J]. J Biomed Sci. 2023;30(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page M J, Mckenzie J E, Bossuyt P M, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews [J]. BMJ. 2021;372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses [J]. Eur J Epidemiol. 2010;25(9):603–5. [DOI] [PubMed] [Google Scholar]
- 11.Mydlarz W K, Hennessey P T, WANG H, et al. Serum biomarkers for detection of head and neck squamous cell carcinoma [J]. Head Neck. 2016;38(1):9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schröck A, Leisse A, de Vos L, et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study [J]. Clin Chem. 2017;63(7):1288–96. [DOI] [PubMed] [Google Scholar]
- 13.de Vos L, Gevensleben H, Schröck A, et al. Comparison of quantification algorithms for circulating cell-free DNA methylation biomarkers in blood plasma from cancer patients [J]. Clin Epigenetics. 2017;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgener JM, Zou J, Zhao Z, et al. Tumor-naïve multimodal profiling of circulating tumor DNA in head and neck squamous cell carcinoma [J]. Clin Cancer Res. 2021;27(15):4230–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson HL, D’agostino JR R B, Meegalla N, et al. The prognostic and therapeutic value of the mutational profile of blood and tumor tissue in head and neck squamous cell carcinoma [J]. Oncologist. 2021;26(2):e279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burcher K M, Faucheux A T, Lantz J W, et al. Prevalence of DNA repair gene mutations in blood and tumor tissue and impact on prognosis and treatment in HNSCC [J]. Cancers. 2021;13(13):3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogo R, Manako T, Iwaya T, et al. Individualized circulating tumor DNA monitoring in head and neck squamous cell carcinoma [J]. Cancer Med. 2022;11(21):3960–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honoré N, Van Marcke C, Galot R, et al. Tumor-agnostic plasma assay for circulating tumor DNA detects minimal residual disease and predicts outcome in locally advanced squamous cell carcinoma of the head and neck [J]. Ann Oncol. 2023;34(12):1175–86. [DOI] [PubMed] [Google Scholar]
- 19.Economopoulou P, Spathis A, Kotsantis I, et al. Next-generation sequencing (NGS) profiling of matched tumor and circulating tumor DNA (ctDNA) in head and neck squamous cell carcinoma (HNSCC) [J]. Oral Oncol. 2023;139:106358. [DOI] [PubMed] [Google Scholar]
- 20.Hanna G J, Dennis M J, Scarfo N et al. Personalized circulating tumor DNA for monitoring disease status in head and neck squamous cell carcinoma [J]. Clin Cancer Res, 2024. [DOI] [PMC free article] [PubMed]
- 21.Pua K C Saleha, Yap Y Y, et al. Clinical significance of plasma Epstein–Barr Virus DNA loads in a large cohort of Malaysian patients with nasopharyngeal carcinoma [J]. J Clin Virol. 2012;55(1):34–9. [DOI] [PubMed] [Google Scholar]
- 22.Lin J-C, Wang W-Y, Chen K Y, et al. Quantification of plasma Epstein–Barr virus DNA in patients with advanced nasopharyngeal carcinoma [J]. N Engl J Med. 2004;350(24):2461–70. [DOI] [PubMed] [Google Scholar]
- 23.LV J, WU C, LI J, et al. Improving on-treatment risk stratification of cancer patients with refined response classification and integration of circulating tumor DNA kinetics [J]. BMC Med. 2022;20(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo R, Tang L L, Mao Y P, et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein‐Barr virus‐related nasopharyngeal carcinoma [J]. Cancer. 2019;125(1):79–89. [DOI] [PubMed] [Google Scholar]
- 25.Leung SF, Chan K, MA B, et al. Plasma Epstein–Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma [J]. Ann Oncol. 2014;25(6):1204–8. [DOI] [PubMed] [Google Scholar]
- 26.Chan A T, LO Y D Zeeb, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma [J]. J Natl Cancer Inst. 2002;94(21):1614–9. [DOI] [PubMed] [Google Scholar]
- 27.Zhao F P, Liu X, Chen X M, et al. Levels of plasma Epstein-Barr virus DNA prior and subsequent to treatment predicts the prognosis of nasopharyngeal carcinoma [J]. Oncol Lett. 2015;10(5):2888–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan A T, Hui E P, Ngan R K, et al. Analysis of plasma Epstein-Barr virus DNA in nasopharyngeal cancer after chemoradiation to identify high-risk patients for adjuvant chemotherapy: a randomized controlled trial [J]. J Clin Oncol. 2018;36(31):3091–100. [DOI] [PubMed] [Google Scholar]
- 29.Ahn S M, Chan J Y, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction–based detection and surveillance of human papillomavirus–related head and neck cancer [J]. Jama otolaryngology–head neck Surg. 2014;140(9):846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y, Haring C T, Brummel C, et al. Early HPV ctDNA kinetics and imaging biomarkers predict therapeutic response in p16 + oropharyngeal squamous cell carcinoma [J]. Clin Cancer Res. 2022;28(2):350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Routman D M Kumars, Chera B S, et al. Detectable postoperative circulating tumor human papillomavirus DNA and association with recurrence in patients with HPV-associated oropharyngeal squamous cell carcinoma [J]. Int J Radiation Oncology* Biology* Phys. 2022;113(3):530–8. [DOI] [PubMed] [Google Scholar]
- 32.Adrian G, Forslund O. Circulating tumour HPV16 DNA quantification–A prognostic tool for progression-free survival in patients with HPV-related oropharyngeal carcinoma receiving curative chemoradiotherapy [J]. Radiother Oncol. 2023;186:109773. [DOI] [PubMed] [Google Scholar]
- 33.Burcher K M, Faucheux A T, Lantz J W et al. Prevalence of DNA Repair Gene Mutations in Blood and Tumor Tissue and Impact on Prognosis and Treatment in HNSCC [J]. Cancers (Basel), 2021, 13(13). [DOI] [PMC free article] [PubMed]
- 34.Hanna G J, Dennis M J, Scarfo N, et al. Personalized ctDNA for monitoring Disease Status in Head and Neck squamous cell carcinoma [J]. Clin Cancer Res. 2024;30(15):3329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson HL, D’agostino R B JR. The prognostic and therapeutic value of the Mutational Profile of Blood and Tumor tissue in Head and Neck squamous cell carcinoma [J]. Oncologist. 2021;26(2):e279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Vos L, Gevensleben H, Schröck A, et al. Comparison of quantification algorithms for circulating cell-free DNA methylation biomarkers in blood plasma from cancer patients [J]. Clin Epigenetics. 2017;9:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo R, Tang L L, Mao Y P, et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma [J]. Cancer. 2019;125(1):79–89. [DOI] [PubMed] [Google Scholar]
- 38.Chan A T, Lo Y M Zeeb, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma [J]. J Natl Cancer Inst. 2002;94(21):1614–9. [DOI] [PubMed] [Google Scholar]
- 39.Chai SJ, Pua K C, Saleh A, et al. Clinical significance of plasma Epstein-Barr Virus DNA loads in a large cohort of Malaysian patients with nasopharyngeal carcinoma [J]. J Clin Virol. 2012;55(1):34–9. [DOI] [PubMed] [Google Scholar]
- 40.Leung S F, Chan K C, MA B B, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma [J]. Ann Oncol. 2014;25(6):1204–8. [DOI] [PubMed] [Google Scholar]
- 41.Chan A T C, Hui E P, Ngan R K C et al. Analysis of plasma Epstein-Barr Virus DNA in Nasopharyngeal Cancer after Chemoradiation to identify high-risk patients for adjuvant chemotherapy: a randomized controlled trial [J]. J Clin Oncol, 2018: Jco2018777847. [DOI] [PubMed]
- 42.Ahn S M, Chan J Y, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer [J]. JAMA Otolaryngol Head Neck Surg. 2014;140(9):846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X, Leo P. Jones L, A comparison between mutational profiles in tumour tissue DNA and circulating tumour DNA in head and neck squamous cell carcinoma—A systematic review [J]. Mutat Research/Reviews Mutat Res, 2023: 108477. [DOI] [PubMed]
- 44.Rapado-González Ó, Rodríguez-Ces A M, LóPez-López R, et al. Liquid biopsies based on cell-free DNA as a potential biomarker in head and neck cancer [J]. Japanese Dent Sci Rev. 2023;59:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aulakh SS, Silverman D A, Young K, et al. The promise of circulating tumor DNA in head and neck cancer [J]. Cancers. 2022;14(12):2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fiala C, Diamandis E P. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection [J]. BMC Med. 2018;16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allis C D, Jenuwein T. The molecular hallmarks of epigenetic control [J]. Nat Rev Genet. 2016;17(8):487–500. [DOI] [PubMed] [Google Scholar]
- 48.Liang N, LI B, Jia Z, et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning [J]. Nat Biomedical Eng. 2021;5(6):586–99. [DOI] [PubMed] [Google Scholar]
- 49.Newman A M, Bratman S V Toj, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage [J]. Nat Med. 2014;20(5):548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin C, Liu X. Liquid biopsy, ctDNA diagnosis through NGS [J]. Life. 2021;11(9):890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabatini ME. Human papillomavirus as a driver of head and neck cancers [J]. Br J Cancer. 2020;122(3):306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pytynia K B, Dahlstrom K R, Sturgis EM. Epidemiology of HPV-associated oropharyngeal cancer [J]. Oral Oncol. 2014;50(5):380–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi J, Wang L, Yao N, et al. The effect of HPV DNA and p16 status on the prognosis of patients with hypopharyngeal carcinoma: a meta-analysis [J]. BMC Cancer. 2022;22(1):658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka H, Takemoto N, Horie M, et al. Circulating tumor HPV DNA complements PET-CT in guiding management after radiotherapy in HPV‐related squamous cell carcinoma of the head and neck [J]. Int J Cancer. 2021;148(4):995–1005. [DOI] [PubMed] [Google Scholar]
- 55.Karimi A, Jafari-Koshki T, Zehtabi M, et al. Predictive impact of human papillomavirus circulating tumor DNA in treatment response monitoring of HPV‐associated cancers; a meta‐analysis on recurrent event endpoints [J]. Cancer Med. 2023;12(17):17592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang E T Yew, Zeng Y-X, et al. The evolving epidemiology of nasopharyngeal carcinoma [J]. Cancer Epidemiol Biomarkers Prev. 2021;30(6):1035–47. [DOI] [PubMed] [Google Scholar]
- 57.Lam W K J, Chan K C A, Lo Y M. D. plasma Epstein-Barr virus DNA as an archetypal circulating tumour DNA marker [J]. J Pathol. 2019;247(5):641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan K A, Woo J K, King A, et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer [J]. N Engl J Med. 2017;377(6):513–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is provided within the manuscript or supplementary information files.