Abstract
M is the most abundant structural membrane protein in coronaviruses and is essential for the formation of infectious virus particles. SARS-CoV-2 M adopts two conformations, Mshort and Mlong, and regulated transition between states is hypothesized to coordinate viral assembly and budding. However, the factors that regulate M conformation and roles for each state are unknown. Here, we discover a direct M-sphingolipid interaction that controls M conformational dynamics and virus assembly. We show M binds Golgi-enriched anionic lipids including ceramide-1-phosphate (C1P). Molecular dynamics simulations show C1P interaction promotes a long to short transition and energetically stabilizes Mshort. Cryo-EM structures show C1P specifically binds Mshort at a conserved site bridging transmembrane and cytoplasmic regions. Disrupting Mshort-C1P interaction alters M subcellular localization, reduces interaction with Spike and E, and impairs subsequent virus-like particle cell entry. Together, these results show endogenous signaling lipids regulate M structure and support a model in which Mshort is stabilized in the early endomembrane system to organize other structural proteins prior to viral budding.
Introduction
Coronaviruses are lipid envelope-encapsulated single-stranded RNA viruses with a genome encoding four structural proteins – spike (S), membrane (M), envelope (E), and nucleocapsid (N). The lipids that comprise the envelope are derived directly from host membranes at the site where viral budding occurs1. Viruses often utilize specific lipids to drive every step of the infection cycle – from viral entry2–5, to biosynthesis 6,7, assembly and egress8,9 – and have been shown to modulate the composition of cellular membranes in order to facilitate these processes10. Lipids are known to play multiple roles in betacoronavirus infection. Mouse hepatitis virus and SARS-CoV-2 target cholesterol- and ceramide-rich microdomains for viral entry5 and depletion of cholesterol reduces S receptor binding in cells11. Coronavirus genome amplification and biosynthesis are marked by the formation of double-membrane vesicles enriched in triacylglycerols and cholesterol6,12. Coronavirus N proteins, whether free or bound to viral RNA, associate with lipid membranes and specific interactions with anionic lipids may drive localization to sites of viral transcription and assembly13–15.
SARS-CoV-2 M is a highly conserved structural membrane protein that facilitates virus or virus-like particle assembly through direct protein-protein interactions with the other structural proteins S, E, and N16,17. Coronavirus M proteins adopt distinct conformations, Mshort and Mlong, in viral envelopes and cryo-EM structures of SARS-CoV-2 M have been captured in each state16,18,19. The transition between conformations has been speculated to be important for viral assembly or budding16,19. It is hypothesized that interactions between the M protein and specific lipids are critical for these conformational transitions. Recent AFM studies revealed a thinning of membrane thickness near the M protein dimer20. Despite the M protein’s critical roles in infectious viral particle assembly and release, and the growing evidence of the critical importance of lipids in these processes, potential roles for specific M-lipid interactions remain unstudied.
Here, we discover M directly interacts with sphingolipids including ceramide-1-phosphate (C1P) in a conformationally selective manner and show that the M-C1P interaction is critical for viral assembly.
Results
M binds anionic sphingolipids
Virus assembly and budding often require lipid-protein interactions for assembly site formation and budding and egress of virus particles. To determine if the M protein binds specific lipids, we screened purified M protein for binding to an array of sphingolipids. HA-tagged M protein robustly interacted with C1P as well as dihydro-C1P with appreciable binding to sphingosine-1-phosphate (So1P) and sphinganine-1-phosphate (S1P) (Fig. 1a). To control for construct-specific effects, we also tested a 50:50 mixture of EGFP-M:untagged M and His-SUMO-tagged M protein. Under all of these conditions, M displayed the most robust binding to C1P (Fig. 1a). M did not show appreciable binding to a host of other lipids tested (Supplementary Fig. 1a,b). M did not show appreciable binding to other lipids (Supplementary Fig. 1a,b). To confirm M-C1P binding in a complementary assay, we compared M binding to C1P-coated beads, anionic phosphatidic acid (PA)-coated beads, and control uncoated beads. M protein displayed appreciable binding to C1P-coated beads over control and PA-coated beads (Fig. 1b).
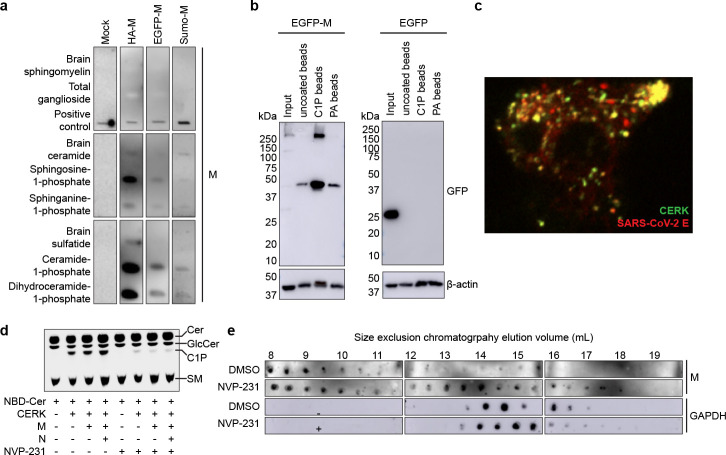
Figure 1. SARS-CoV-2 M protein interacts with C1P.
(a) Sphingolipid snooper assay for M-lipid interactions. Western blot of immobilized lipids probed with HA-tagged M, a 50:50 mixture of EGFP-tagged and untagged M, His- and Sumo-tagged M, or a control sample without M protein. All M constructs tested displayed strong binding to C1P and dihydroC1P as well as appreciable binding to anionic So1P and Sa1P. (b) Bead pull down assay of M-lipid interactions. Western blot of input EGFP-M (left) or EGFP (right) expressing cell lysate or eluant from control uncoated, C1P-coated, and PA-coated beads. EGFP-M protein was robustly pulled down by C1P-coated beads. (c) Colocalization of SARS-CoV-2 assembly sites with CERK. HEK293 cells expressing CERK and SARS-CoV-2 structural proteins were imaged. CERK displayed colocalization with fluorescently labeled E protein. (d) Assay for C1P production. Thin layer chromatography of samples from mock-, CERK-, CERK and M-, or CERK, M, and N-transfected HEK293 cells treated with 2.5 μM NBD-ceramide and either DMSO or 100 nM CERK inhibitor NVP-231. CERK increased C1P levels and was inhibited by NVP-231. (e) Assay for M oligomerization in cells. Western blot for M (upper) or GAPDH (lower) in size exclusion chromatography fractionated CERK, M, N, E, and S-expressing HEK293 cell lysate treated with DMSO or 100 nM NVP-231. Later eluting fractions correspond to smaller species. Lower C1P levels altered M protein oligomerization state with an increased proportion of smaller species detected.
Ceramide kinase (CERK) is the major producer of C1P in mammalian cells21 and to assess the role of CERK in M assembly, we imaged M and other structural proteins under different conditions (Supplementary Fig. 2). M protein colocalized with the S and E proteins. M protein also colocalized with markers of the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and Golgi, but not an ER marker (Supplementary Fig. 3a–c), and formed assembly sites similar in vesicle diameter (Supplementary Fig. 3d–f) to authentic SARS-CoV-222. A GFP-CERK construct displayed colocalization with SARS-CoV-2 assembly sites labeling the E protein (Fig. 1c). To assess the role of CERK in M protein’s cellular localization, we expressed CERK, CERK and M, or CERK, M, and N in HEK293 cells and assessed C1P content using thin layer chromatography. C1P production was increased upon CERK expression(Fig. 1d). Next, we treated cells with the CERK inhibitor NVP-231 to confirm reduction in C1P levels, which was similarly lowered under all conditions with CERK expression (Fig. 1d). We then expressed M, S, E, and N with CERK in cells and fractionated lysates using size exclusion chromatography (Figure 1e). Inhibition of CERK resulted in an increase in later-eluting, smaller M oligomers, demonstrating a reduction in higher order oligomerization of M protein when C1P was depleted.
C1P binds and stabilizes M in the short conformation
We used molecular dynamics (MD) simulations to better understand the nature and consequences of M-C1P interactions. Atomistic models of M were constructed from Mshort (7VGS) and Mlong (7VGR) structures embedded in lipid membranes19. Membrane composition mimicked the composition of the ERGIC (45% PC, 20% PI, 15% PE, 10% Cholesterol) with either 10% C1P or 10% PS as a control anionic lipid23,24. The conformation of M was monitored during the 4 μs simulations by two collective variables that distinguish Mshort from Mlong: (1) the angle between cytoplasmic C-terminal domains (CTDs) of each subunit and (2) the distance between hinge regions (Fig. 2a).
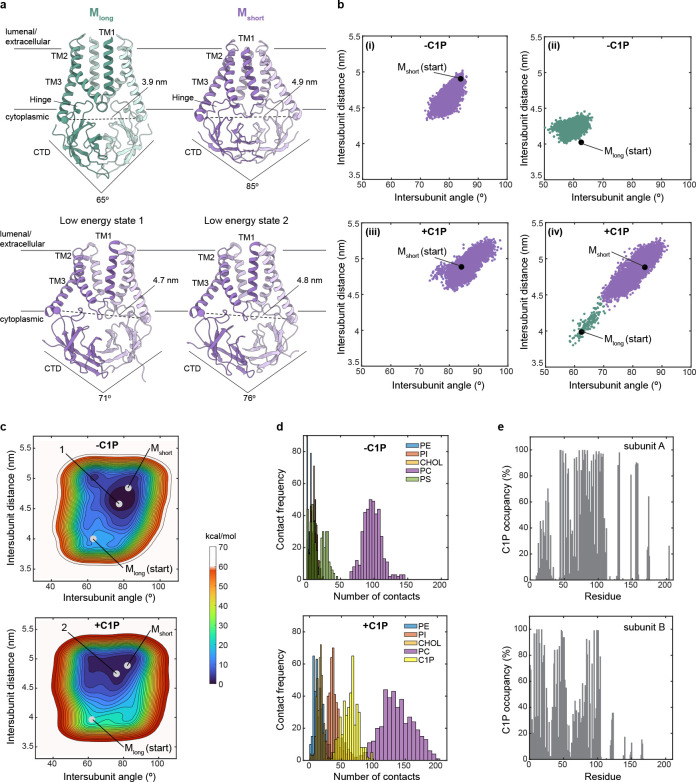
Figure 2. C1P energetically stabilizes Mshort.
(a) Structures of M protein viewed from the membrane plane. Collective variables that report on intersubunit distance and angle used to monitor conformational state during simulations are illustrated on the structures. (b) M protein conformational trajectory during molecular dynamics simulations. Intersubunit distance versus intersubunit angle of the structure at each time sample during 4 us simulations is plotted. Simulations were performed without (i,ii) or with C1P (iii, iv) and initiated from Mshort (i,iii) or Mlong (i, iv). Conformational switching was only observed from Mlong to Mshort in the presence (of C1P. (c) Free-energy landscape calculated from simulations without (above) or with C1P (below). Position of Mshort, Mlong, and a low energy state from each condition is indicated. (d) M-lipid contact frequency from simulations without (above) and with C1P (below). (e) M-C1P percentage occupancy by residue in subunit A (above) and subunit B (below).
Strikingly, MD simulations initiated from Mlong in the presence of C1P showed rapid conformational switching to Mshort-like conformations, which remained stable for the duration of the simulation (Fig. 2b). Conformational switching was dependent on C1P, as simulations initiated from Mlong in the absence of C1P did not exhibit a transition to the short state. Notably, switching was only observed from Mlong to Mshort; Mshort in the presence or absence of C1P remained in Mshort-like conformations throughout the simulations (Fig. 2b). This is consistent with the stability of Mshort we observed in prior simulations in the absence of C1P18.
These results suggest that C1P stabilizes Mshort. We computed the conformational free energy landscape using well-tempered (WT) metadynamics25, employing the same collective variables that utilize the intersubunit CTD angle and hinge distance as the pertinent coordinates (Fig. 2c). We reach two conclusions from this analysis. First, Mshort is a lower free energy state than Mlong, regardless of lipid composition. Minimum energy states in the absence (indicated by “1”) and presence of C1P (indicated by “2”) are similarly Mshort-like (Fig. 2a). Second, C1P markedly stabilizes Mshort. We calculate the magnitude of stabilization to be −6.99±1.00 kcal/mol (defined as ΔΔG = ΔG+C1P – ΔG−C1P = (−18.07±0.62)-(−11.08±0.79)} kcal/mol = −6.99±1.00 kcal/mol). Stabilization is primarily due to a deeper free energy well around Mshort-like conformations.
C1P stabilization could be explained by direct binding to Mshort. We observed more CTD-lipid headgroup contacts in MD simulations with C1P, with enrichment of C1P and PI contacts (Fig. 2d). C1P and PI lipids were localized around Mshort in visualizations of lipid position (Supplementary Fig. 4a). Analysis of membrane structure during the last 2 μs of the simulations with C1P shows M subtly thins and curves the membrane, particularly around the CTD-inner leaflet interface (Supplementary Fig. 4b–e). To identify candidate residues involved in M-C1P interactions, we calculated residence time (Supplementary Fig. 4f) and percentage occupancy (Fig. 2e) of C1P around M by amino acid. A continuous patch of residues at the TM-CTD interface showed both high C1P occupancy (>80%) and among the longest C1P residence times. The patch includes residues at the bottom of TM1 (R44, F45, and I48), bottom of TM3, hinge (A104, R107, S108, M109, and W110), proximal CTD (L129 and R131), and nearby charged residues in the CTD (H148, H155, and R158).
We next sought direct experimental structural insight into the molecular basis of M-C1P interaction. We prepared complexes of M with CTD-binding Fabs that stabilize either Mshort or Mlong conformations in the presence of 100 μM C1P. To facilitate comparisons to previously reported apo M structures, buffer and detergent conditions were otherwise identical. We determined cryo-EM structures of Mshort and Mlong in the presence of C1P to 3.0 Å overall resolution (Fig. 3a, Supplementary Fig. 5,6, Table 1).
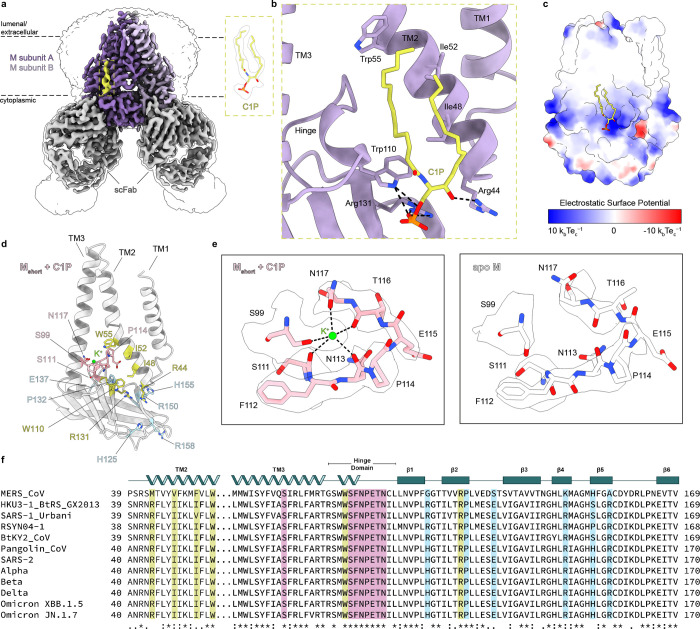
Figure 3. Cryo-EM structure of Mshort bound to C1P.
(a) 3.0 Å resolution cryo-EM map of SARS-CoV- 2 M in the short conformation viewed from the membrane. M subunits are purple, short conformation-selective Fabs (scFab) are gray, and C1P is yellow. Unsharpened map is shown transparent at low contour. (b) Zoomed in view of C1P binding site in one subunit. (c) Electrostatic surface view of M with C1P bound. (d) Single Mshort-C1P subunit with C1P binding residues yellow, K+ binding residues pink, and residues displaying associated conformational changes blue. (e) Zoomed in view of K+ binding site in Mshort-C1P (left) or apo Mshort (right). Cryo-EM density is shown as a transparent surface. (f) Sequence alignment of M across coronaviruses with residues colored as in (d).
Table 1 -.
Cryo-EM data collection, refinement, and validation statistics
| M:scFab + C1P | M:lcFab + C1P | |
|---|---|---|
| PDB ID | 9CTU | 9CTW |
| EMDB ID | 45919 | 45921 |
| EMPIAR ID | xxx | xxx |
| Data collection | ||
| Total movies | 6,741 | 8,432 |
| Magnification | 105,000x | |
| Voltage (kV) | 300 | |
| Electron exposure (e−/Å2) | 50 | |
| Defocus range (um) | −0.5 to −1.8 | |
| Pixel size (Å2) | 0.4055 (super res) | 0.848 |
| Processing | ||
| Initial particle images (no.) | 351,588 | 561,632 |
| Final particle images (no.) | 44,736 | 190,251 |
| Map resolution Masked (Å, FSC = 0.143) | 3.03 | 3.01 |
| Symmetry imposed | C2 | C2 |
| Refinement | ||
| Model resolution (Å, FSC = 0.143 / 0.5) | 3.0/3.4 | 2.9/3.1 |
| Map-sharpening B factor (Å2) | −108.4 | −119.0 |
| Composition | ||
| Number of atoms | 9714 | 9754 |
| Number of protein residues | 1240 | 1256 |
| Number of ligands | 4 | 0 |
| RMS deviations | ||
| Bond lengths (Å) | 0.003 | 0.003 |
| Bond angles (Å) | 0.532 | 0.510 |
| Validation | ||
| MolProbity score | 1.63 | 1.25 |
| Clashscore | 9.58 | 2.07 |
| Ramachandran plot | ||
| Favored (%) | 97.39 | 96.06 |
| Allowed (%) | 2.61 | 3.78 |
| Disallowed (%) | 0 | 0.16 |
| Rotamer outliers (%) | 1.03 | 0.74 |
| Mean B factor (Å2) | ||
| Protein | 95.42 | 55.62 |
| Ligand | 73.27 | − |
The Mshort-C1P map shows lipid-like densities not visible in apo-Mshort maps. One density feature per subunit at the inner leaflet can be unambiguously modeled as C1P with well-defined phosphomonoester headgroup and acyl chains (Fig. 3a). C1P binds to a site formed by the bottom of TM1–3, hinge region, and membrane-proximal aspect of the CTD, corroborating our molecular dynamics simulations (Fig. 3b). The C1P head group fits into an electropositive pocket and forms polar interactions with R44, W110, and R131 (Fig. 3c). C1P acyl chains pack into a cleft between TM2 and TM3, forming hydrophobic interactions with F45, I48, L51, I52, W55, M109, W110, and P114 on TM2 (Fig. 3b,d). Density corresponding to the inner acyl chain is weakly visible in the apo Mshort map, suggesting partial occupancy by co-purified lipids or detergents in the absence of C1P.
In contrast, we see no evidence for specific C1P binding to Mlong (Supplementary Fig. 7a). Mlong+C1P and apo-Mlong structures are nearly indistinguishable (0.79 Å overall r.m.s.d.). Densities consistent with detergent or lipid acyl chains around the inner leaflet are similar in Mlong+C1P and apo-Mlong maps. The absence of C1P binding to Mlong is explained by reorganization of the Mshort-C1P binding site. In Mlong, the two halves of the C1P binding pocket (TM1/2 and TM3/hinge/CTD) spread apart, separating key residues R44 and R131 by > 7 Å (Supplementary Fig. 7b).
Comparing Mshort-C1P and apo Mshort structures reveals four consequences of C1P binding. First, C1P binding compacts M, particularly in the TM region with intersubunit TM2 and TM3 distances 1.5–3.5 Å shorter (Supplementary Fig. 8a). Second, C1P binding stabilizes Mshort. Mshort-C1P has more clearly resolved flexible regions (including in the hinge and TM linkers) and lower, less variable refined B-factors than apo Mshort (Supplementary Fig. 8c), despite similar particle numbers, processing routines, and overall resolutions. Third, C1P binding reveals an ion binding site bridging the hinge and bottom of TM3 (Fig. 3d,e). Electronegative groups, including S99 and N117 side chain hydroxyls and S111, N113, and T116 backbone carbonyls, surround a strong density feature not visible in the apo-Mshort map. We model a K+ in this site supported by buffer conditions (150 mM KCl), coordination geometry, and multiple validation tools. Fourth, a series of conformational changes propagate from the C1P binding site and alter the CTD surface character. Hinge residue W110 is shifted towards the CTD upon C1P binding, necessitating a shift in neighboring P132 on the β2 strand (Supplementary Fig. 8b). Charged residues including H125 on β1, R158 on β5, and E137, R150 and H155 at the intersubunit CTD interface change rotameric state, altering the electrostatic character of the CTD surface and potentially influencing M interactions with other structural proteins during virus assembly (Supplementary Fig. 8d). We conclude C1P specifically binds and stabilizes Mshort.
Notably, residues involved in C1P-binding and associated conformational changes have been recently implicated in the action of an anti-coronavirus small molecule. JNJ-9676 binds Mshort (but not Mlong) in a pocket above the hinge between TM2 and TM3, overlapping the potassium ion binding site identified here26. JNJ-9676 is reported to prevent virus assembly by trapping Mshort, suggesting an Mshort-Mlong transition is critical to the viral life cycle. Among JNJ-9676 escape mutants identified in M, two (P132S, S99A) are not readily explained by steric or chemical perturbation of the drug binding site. We reasoned these mutations could disrupt drug binding by promoting Mlong and that other residues implicated in C1P binding would have similar effects. We used AlphaFold3 to test this hypothesis. Wild-type M was predicted to adopt an Mshort-like conformation (Supplementary Fig. 9), consistent with our structural and MD results. In contrast, mutants at sites implicated in C1P binding or associated conformational changes (H155E, E137R, R150D, W110R, P132W, and S99D) are predicted to adopt increasingly Mlong-like conformations (Supplementary Fig. 9). Among these structures, Mshort-C1P is the most compacted in an MD trajectory from short to long conformations.
M-C1P binding is critical for virus assembly of the spike protein
Finally, we asked what role C1P binding has on M function in virus assembly. We prepared several mutations of M and assessed their functional consequences. A cationic patch in the M CTD implicated in MD and structural experiments in C1P interaction (residues 146-RGHLRIAGHH-155) was mutated to replace positively charged arginine and histidine residues with alanine. This mutation resulted in loss of M binding to C1P assessed with lipid-coated bead pull down and is referred to as MΔC1P (Fig. 4a). A second cationic rich region was also mutated (198-RYRIGNYK-205) as it was previously shown to be a trans Golgi retention signal for MERS M protein (MΔTGN)27. Similar to the previous report, mutation of the trans Golgi retention signal disrupted M localization (Supplementary Fig. 10) and colocalization with S and E. In a similar fashion, MΔC1P disrupted M localization (Fig. 4a,b) and colocalization with S and E (Figure 4b-d).
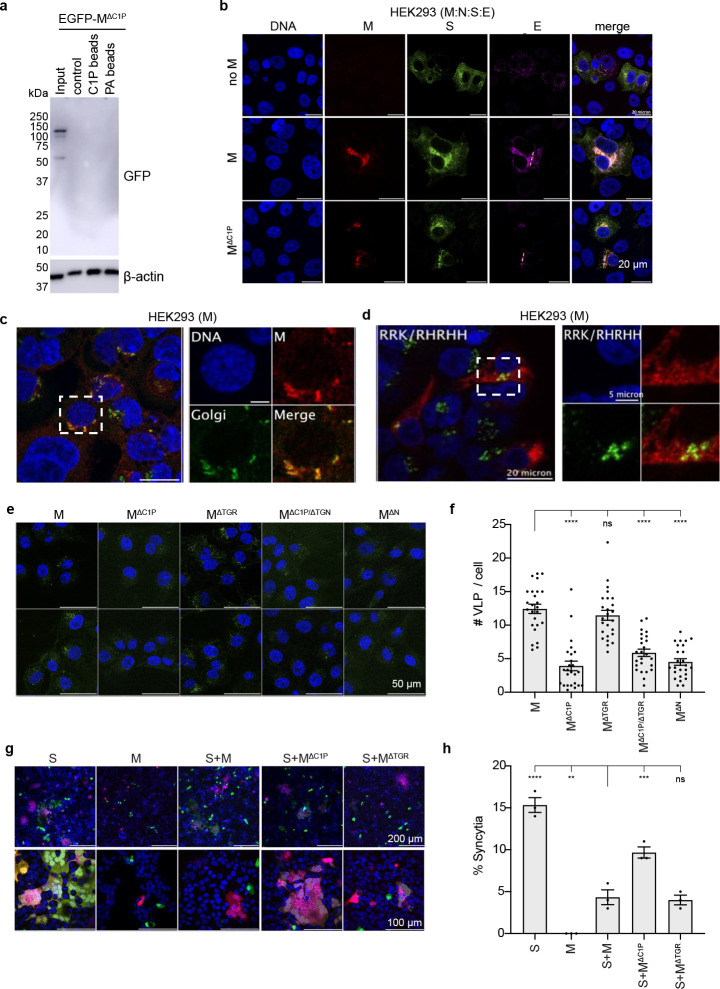
Figure 4. Functional assessment of M protein mutations.
(a) Bead pull down assay of MΔC1P mutant-lipid interactions. Western blot of input EGFP-MΔC1P expressing cell lysate and eluant from uncoated, C1P-coated, or PA-coated beads. (b) MΔC1P disrupts colocalization with S observed with wild-type M. (c,d) MΔC1P disrupts Golgi localization of wild-type M. (e,f) Virus-like particle (VLP) entry assay. MΔC1P, MΔC1P/ΔTGN, and MΔN (which reduces M:N-interaction) derived VLPs show reduced cell entry compared to VLPs formed with wild-type M or MΔC1P. (g,h) Cell syncytia formation assay. Cells expressing S alone, but not S and M, form robust syncytia. In contrast, co-expression with MΔC1P significantly increased syncytia formation. Data are mean ± sem with differences assessed with a one-way Annova with Dunnet correction for multiple comparisons (****p<0.0001, ***p<0.001, **p<0.01).
To test the hypothesis that the M-C1P interaction is an important factor for proper S incorporation into VLPs, we assessed VLP entry for WT and mutant M containing VLPs. WT M containing VLPs were able to undergo cell entry (quantified by number of GFP-positive puncta counted per cell) as were VLPs containing MΔTGN (Fig. 4e,f). In contrast, VLPs derived from M mutations that displayed altered S/M colocalization and reduced C1P binding (MΔC1P) had a significant reduction in cell entry (Fig. 4e,f). This underscores the importance of M-C1P interaction in proper S localization and incorporation into VLPs. To further confirm changes in S interaction and localization, we employed a syncytia formation assay which has been used to assess the trafficking of S to the plasma membrane of cells 28–30. S expression alone resulted in robust syncytia formation that was significantly reduced by co-expression with M or MΔTGN (Fig. 4g-h). In contrast, co-expression of MΔC1P with S resulted in increased syncytia formation consistent with disrupted S interaction(Fig. 4g-h).
Discussion
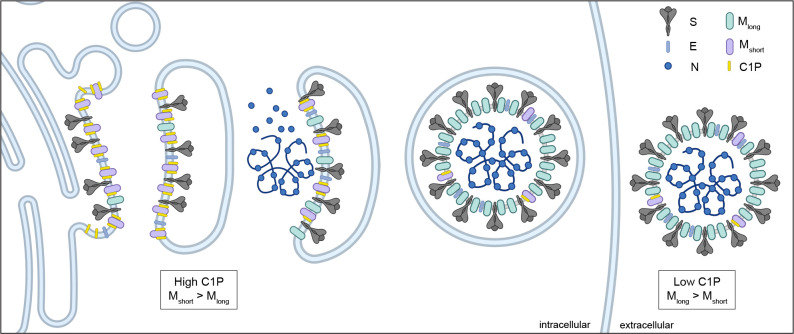
Together, the data presented here support a model for lipid-regulated coronavirus assembly through specific binding and conformational control of SARS-CoV-2 M protein (Fig. 5). We propose C1P binding enriches Mshort in ERGIC and Golgi membranes to facilitate its organization with other structural proteins S, E, and N in early steps of virus assembly. Transition to Mlong promotes higher order assembly and virus release and involves loss of C1P through dilution and/or additional promoting factors such as structural protein binding. Several lines of evidence support this model. M is in dynamic equilibrium Mshort and Mlong16,19, with Mshort energetically favored (Fig. 2)18. Specific binding of C1P further stabilizes and compacts Mshort (Figs. 1–3). C1P is synthesized and enriched in the ERGIC and Golgi membranes 31, where M organizes the other structural proteins S, E, and N 32 (Fig. 1). Depleting C1P results in mislocalization and higher order assembly of M (Fig. 1), while mutations that disrupt M-C1P interaction negatively impact structural protein binding and VLP entry (Fig. 4).
Figure 5. Model for lipid control of M conformational dynamics and coronavirus. assembly.
M protein adopts Mshort and Mlong conformations. Ceramide kinase localization to ERGIC membranes results in high C1P concentrations. C1P binds and stabilizes Mshort in the ERGIC to facilitate recruitment of E and S. Decreasing C1P along the secretory pathway and other potential factors trigger a conformational switch to Mlong that promotes higher membrane curvature and oligomerization necessary for virus assembly and budding.
This model provides insight into the mechanism of action of the recently reported small molecule antiviral JNJ-9676, the first that directly targets M protein26. JNJ-9676 mimics the effect of C1P, specifically binding to an Mshort-like conformation and disfavoring transition to Mlong. Unlike Mshort-C1P, Mshort-JNJ-9676 is inhibited from transitioning to Mlong, either because JNJ-9676 is not diluted during progression through the endomembrane systems or because the complex is insensitive to additional unknown Mlong-promoting factors. As a result, M is trapped in an early short-like conformation, preventing formation of mature virus particles. One prediction of this model is that additional Mshort-stabilizing molecules, perhaps additionally co-opting the C1P binding site, are likely to similarly prevent virus formation. A corollary is that Mlong stabilizing small molecules or antibodies, including those potentially patient-derived33–37, are predicted to inhibit virus disassembly and successful infection.
Lipid enveloped viruses hijack host membranes to form sites of virus assembly and give rise to new virus particles. Herein we demonstrate that SARS-CoV-2 M makes direct interactions with host cell C1P, which is critical for conformational stability of Mshort and correct assembly of S. This illustrates SARS-CoV-2 uses specific host lipid interactions to mediate formation of virus particles akin to other virus families (e.g., filoviruses, retroviruses, paramyxoviruses, flaviviruses, etc. 38–45. A number of studies have demonstrated increased cellular levels of ceramide during SARS-CoV-2 infection 46,47 where ceramide was shown to help facilitate viral entry 48,49. Further, drugs that disrupted ceramide synthesis (e.g., amitriptyline) reduced SARS-CoV-2 entry into cells 48,50 and reduced mortality in patients 51. While the details of C1P in this process have not been explored in great detail, ceramide is the direct substrate of C1P generation and increases in C1P have been shown in the sera of SARS-CoV-2 infected mice and in infected Vero E6 cells 52. Thus, more detailed studies examining ceramide/C1P ratio changes in SARS-CoV-2 infection and cellular models used herein are clearly warranted.
Methods
Molecular dynamics setup
The atomistic model of the M protein was built using the available cryo-EM structures of both the short (PDB ID: 7VGS) and long (PDB ID: 7VGR) conformations19. The CHARMM-GUI Membrane Builder 53–56 was employed to prepare initial MD systems, each consisting of a single M protein in either the short or long conformation embedded in a lipid bilayer. We utilized a 20×20 nm lipid bilayer membrane, which is sufficiently large to observe the membrane response of a single M protein. Based on biochemical experiments, we constructed two membrane models that mimic the composition of the endoplasmic reticulum-Golgi intermediate compartment 23,24. The first system is composed of a mixture of DYPC (45%), POPI (20%), POPE (15%), POPS (10%), and Cholesterol (10%). The second replaces POPS with C1P. We solvated the M/lipid-membrane system in a periodic rectangular box filled with TIP3P model water molecules, ensuring that the transmembrane region of the M protein was positioned close to the bilayer’s core. A multistep minimization and equilibration of the system was carried out following the protocols provided on CHARMM-GUI57. We used 0.15 M KCl to neutralize the system. We ran four conditions: Mshort with C1P, Mshort without C1P, M long with C1P, and Mlong without C1P. Each condition was run in triplicate for twelve total MD simulations.
Simulation details
All atomistic MD simulations were performed using GROMACS 2019 with CHARMM36m force field58,59. The simulations maintained a temperature of 310.15 K using a Nosé-Hoover thermostat 60,61 with a coupling time constant of 1.0 ps. During initial relaxation, the pressure was maintained at 1 Bar with a Berendsen barostat62. Periodic boundary conditions were applied with a time step of 2 fs. For the production run, the Parrinello-Rahman barostat was employed semi-isotropically due to the presence of a membrane63,64. A compressibility factor was set at 4.5×10−5 with a coupling time constant of 5.0 ps. Van der Waals interactions were computed using a switching function between 1.0 and 1.2 nm, and long-range electrostatics were calculated using the Particle Mesh Ewald (PME) method65. Hydrogen bonds were constrained using the LINCS algorithm66. We performed a 4 μs production run for each replica using Frontera (Texas Advanced Computing Center), and Midway2 (Research Computing Center at the University of Chicago) computing facilities.
Free energy calculations
We used metadynamics to explore the free energy landscape67. The Hamiltonian of the system includes a history-dependent Gaussian bias potential, which is a function of a finite set of collective variables (CVs). In this study, we employed the well-tempered metadynamics (WT-MetaD) 25 technique to investigate the switching between the short and long conformational states of the M protein. In WT-MetaD simulations, the Gaussian bias height is progressively lowered to avoid overfilling the free energy basins and to achieve convergence 68.
where is the Gaussian height, is the Gaussian width, and are the sequence of points in CV space at time . controls the rate of height decrease.
From manual inspection of superimposed Mshort and Mlong structures, we defined two collective variables (CVs) to monitor conformational switching during simulations (Fig. 2a). The first reports the angle between CTDs and is calculated between S108 (subunit A), S184 (subunit A), and S108 (subunit B). This angle in Mshort and Mlong structures is 85° and 65°, respectively. The second reports on intersubunit separation and is defined as the distance between R107 in subunit A and subunit B. This distance in Mshort and Mlong structures 4.9 nm and 3.9 nm, respectively (Fig. 2a). We selected the Cα atom of each residue for angle and distance calculations.
In this work, Gaussians were deposited every 500 steps with an initial height set to 0.3 kJ mol−1. A bias factor of 15 was used to reduce the Gaussian height. The σ was set to 0.1. We conducted WT-metaD simulations using PLUMED 2.8.2 patched with GROMACS 2019 for both the model-C1P and control systems, using the long conformation as the starting structure. The error bars were computed using block averaging, as implemented in PLUMED.
Analysis
Distances, angles and the number of contacts were calculated using the GROMACS module 58. Lipid distribution around the M protein and mean curvature were analyzed using the MDAnalysis Python package69. The residence time of interactions between lipid molecules and protein residues, as well as the occupancy of lipids around the protein, were determined using PyLipID70. Membrane thickness was calculated with VMD MEMBPLUGIN and Tcl scripts71.
Protein cloning, expression, and purification
The coding sequence for SARS-Cov-2 M protein (Uniprot P0DTC5) was synthesized (IDT, Newark, NJ) and cloned into a vector based on the pACEBAC1 backbone (MultiBac; Geneva Biotech, Geneva, Switzerland) with an added C-terminal PreScission protease (PPX) cleavage site, linker sequence, superfolder GFP (sfGFP) and 7xHis tag to generate the expression construct (SARS-CoV-2 M)-SNS-LEVLFQGP-SRGGSGAAAGSGSGS-sfGFP-GSS-7xHis18. Bacmid was generated by transposition in transformed DH10MultiBac cells according to the manufacturer’s instructions.
Sf9 cells were cultured in ESF 921 medium (Expression Systems, Davis, CA) and P1 virus was generated from cells transfected with Escort IV reagent (MillaporeSigma, Burlington, MA) according to manufacturer’s instructions. P2 virus was generated by infecting cells at a density of 2 million cells/mL with P1 virus at a MOI ~0.1. Infection was monitored by fluorescence and virus harvested at 72 hours. P3 virus was generated in a similar manner to expand the viral stock. The P2 or P3 viral stock was then used to infect Sf9 cells at 4 million cells/mL at a MOI ~2–5. At 72 hours, infected cells containing expressed M-sfGFP protein were harvested by centrifugation at 2500 × g for 10 minutes and frozen at −80 °C.
Infected Sf9 cells from 1 L of culture (~15 mL of cell pellet) were thawed in 100 mL of Lysis Buffer containing 50 mM HEPES pH 8.0, 150 mM KCl, 0.5 mM EDTA, 2 mM MgCl2, 2 mM DTT. Protease inhibitors (Final Concentrations: E64 (1 μM), pepstatin A (1 μg/mL), soy trypsin inhibitor (10 μg/mL), benzamidine (1 mM), aprotinin (1 μg/mL), leupeptin (1 μg/mL), AEBSF (1 mM), and PMSF (1 mM)) were added to the lysis buffer immediately before use. Benzonase (4 μl) was added after the cell pellet thawed. Cells were then lysed by sonication and centrifuged at 150,000 × g for 45 minutes. The supernatant was discarded, and residual nucleic acid was removed from the top of the membrane pellet using DPBS. Membrane pellets were resuspended in High Salt Lysis buffer (50 mM HEPES pH 8.0, 1 M KCl, 1 mM EDTA, 2 mM DTT) plus protease inhibitors, followed by ultracentrifugation at 150,000 × g for 45 minutes. The supernatant was discarded, membrane pellets were washed again with DPBS, then transferred into a Dounce homogenizer containing extraction buffer (50 mM HEPES, 150 mM KCl, 1 mM EDTA, 1% lauryl maltose neopentyl glycol (LMNG, Anatrace, Maumee, OH), 0.1% cholesteryl hemisuccinate Tris salt (CHS, Anatrace, Maumee, OH) pH 8). A stock solution of 10% LMNG, 1% CHS was dissolved and clarified by bath sonication in 200 mM HEPES pH 8 prior to addition to buffer to the indicated final concentration. Membrane pellets were homogenized in extraction buffer and this mixture (150 mL final volume) was gently stirred at 4°C for 1.5 hours. The extraction mixture was centrifuged at 33,000 × g for 45 minutes and the supernatant, containing solubilized membrane protein, was bound to 7.5 mL of Sepharose resin coupled to anti-GFP nanobody for 1.5 hours at 4°C. The resin was then collected in a column and washed with 100 mL of buffer 1 (20 mM HEPES pH 8.0, 150 mM KCl, 1 mM EDTA, 0.005% LMNG, 0.0005% CHS), 150 mL of buffer 2 (20 mM HEPES pH 8.0, 500 mM KCl, 1 mM EDTA, 0.005% LMNG, 0.0005% CHS), and 25 mL of buffer 1. The resin was then resuspended in 7.5 mL of buffer 1 with 0.5 mg of PPX protease and rocked gently in a capped column for 2 hours. M protein was then eluted with an additional 15 mL of buffer 1, spin concentrated to ~1 mL (Amicon Ultra 30 kDa cutoff, Millipore, Tullagreen, Ireland), and loaded onto a Superose 6 increase column (GE Healthcare, Chicago, IL) on an NGC system (Bio-Rad, Hercules, CA) equilibrated in buffer 1. Peak fractions containing M protein were stored overnight at 4 °C prior to preparation of Fab complexes.
Coding sequences for conformationally selective antibody fragments (lcFab and scFab)19 were synthesized and cloned in a pTwist CMV BetaGlobin WPRE Neo vector with C-terminal 7X-His tags on heavy chain (Twist Biosciences). Fabs were cotransfected in suspension HEK293T GNTI- cells for expression using PEI according to manufacturer’s instructions. Supernatants were thawed on ice, buffered with HEPES pH 8.0 at a final concentration of 100 mM, and centrifuged at 21,000 × g for 10 minutes to pellet any precipitated material. Clarified supernatant was incubated with 3 mL His-Pure Ni NTA resin (ThermoFisher Scientific, Rockford, IL) for 1.5 hours at 4°C. The Ni NTA resin was collected in a column and washed with 30 mL each of Cholate buffer (40 mM HEPES pH 8.0, 300 mM KCl, 50 mM Na-cholate, 10 mM Imidazole) and Imidazole Wash Buffer (40 mM HEPES pH 8.0, 300 mM KCl, 20 mM Imidazole). The Fabs were eluted in 4 column volumes of Ni NTA elution buffer (40 mM HEPES pH 8.0, 300 mM KCl, 200 mM Imidazole). The eluate was spin concentrated to ~1 mL with Amicon Ultra spin concentrator 30 kDa cutoff (Millipore, Tullagreen, Ireland) and loaded onto a Superdex 200 increase (S200) column (GE Healthcare, Chicago, IL) on an NGC system (Bio-Rad, Hercules, CA) equilibrated in SEC buffer (20 mM HEPES pH 8.0, 150 mM KCl, 1 mM EDTA). Fractions containing complete Fabs were pooled, concentrated, and stored at 4°C for future use.
M-Fab complex formation and purification
The peak fraction from size exclusion chromatography containing pure homodimeric M protein was collected, incubated with purified lcFab or scFab at a 1:2.2 M monomer:Fab molar ratio for 45 minutes on ice, and run on a Superose 6 increase column in Buffer 1 to separate excess Fabs. Pooled peak fractions containing M:Fab complexes were spin concentrated with an Amicon Ultra 50 kDa cutoff spin concentrator to 2.2 mg/mL (scFab) and 2.7 mg/mL (lcFab). Concentrated complex samples were cleared by a 10-minute 21,000 × g spin at 4 °C, then d18:1/16:0 ceramide-1-phoshate (C1P; N-palmitoyl-ceramide-1-phosphate ammonium salt, (Avanti Polar Lipids, Alabaster, AL)) was added to each sample to a final concentration of 100 μM from a 1 mM stock in 1:1 MeOH:CHCl3. Samples were then incubated on ice for 30 minutes prior to grid preparation.
Cryo-EM sample preparation and data collection
3.4 μl samples of M:lcFab or M:scFab in LMNG/CHS and C1P were applied to freshly glow discharged Holey Carbon, 300 mesh R 1.2/1.3 gold grids (Quantifoil, Großlöbichau, Germany) (M:scFab) or C-Flat Au 300 mesh 1.2/1.3 (Electron Microscopy Sciences, Hatfield, PA) (M:lcFab) then plunge frozen in liquid ethane using a FEI Vitrobot Mark IV (ThermoFisher Scientific) with a ~5 second wait time set to 4°C, 100% humidity, blot force 1, and 3 second blot time.
Grids were clipped and cryo-EM micrographs were collected on a Titan Krios G3i microscope operated at 300 kV. Fifty frame videos with a total electron dose of 50 e-/Å2 were recorded on a Gatan K3 Summit direct electron detector. The M:scFab dataset was collected in super resolution mode with a pixel size of 0.4055 Å (0.8110 Å physical pixel size), and the M:lcFab dataset was collected in counting mode with a pixel size of 0.848 Å. Videos were recorded around a central hole position via image shift in an 11×11 hole pattern with two targets per hole. Target defocus range was −0.5 to −1.8 μm. See Supplemental Table 4.1 for detailed data collection statistics.
Cryo-EM data processing
Data processing for both samples were done in cryoSPARC v4.4.172. For M:scFab in LMNG/CHS/C1P 6,741 movie stacks were collected, motion-corrected, binned to 0.8110 Å pixel−1 using Patch Motion Correction, and CTF-corrected using Patch CTF Estimation (Supplementary Fig. 5). Micrographs with an estimated CTF resolution below 7 Å were discarded, leaving 5,105 for further processing. Initial particle picking was performed using cryoSPARC’s reference free blob picker and curated from 404,953 particles down to 29,857 ‘good’ particles by 2D classification.
Topaz training, picking and extraction yielded 351,588 particles which were then iteratively 2D classified, resulting in a set of 163,911 ‘good’ particles. A two-class ab-initio reconstruction was performed to further sort out 44,905 particles that did not contribute to a high-resolution 3D map. Subsequent non-uniform refinement (C1, 2 extra passes, 14 Å initial resolution) produced a map with an overall 2.94 Å resolution showing clear lipid densities on both leaflets. This map and corresponding 119,006 particles were subjected to local CTF refinement, then global CTF refinement, followed by polishing via Reference Based Motion Correction which further improved the resolution to 2.7 Å with improved lipid densities. A round of 3D classification (2 classes, 3 Å target resolution, 10 O-EM epochs, 0.95 class similarity) and subsequent C2 non-uniform refinement on the class with the clearest lipid densities produced a final 3.03 Å resolution map (44,726 particles) where a C1P molecule could be unambiguously modeled (Supplementary Fig. 5, Supplementary Table 1). The same non-uniform refinement with C1 symmetry produced a 3.26 Å resolution map with no apparent differences between subunits or lipid binding, so the C2 map was chosen for building an atomic model as the C1P densities were more complete and clearly defined.
The M:lcFab LMNG/CHS/C1P dataset was processed in a similar manner, with 9,910 initial micrographs culled to 8,432 using a 5 Å cutoff. Topaz was trained on 136,478 particles yielding a stack of 561,632 particles, which was cleaned, refined and polished as described for the scFab dataset, yielding 301,406 ‘shiny’ particles. A round of 3D classification yielded a class of 190,251 particles with 3.01 Å final resolution after non-uniform refinement with C2 symmetry (Supplementary Fig. 6, Supplementary Table 1). Further attempts at 3D classification and heterogenous refinement failed to separate these particles into subsets with unique characteristics or improved map quality.
Modeling, refinement, and analysis
Modeling of the M:scFab complex in LMNG/CHS/C1P was performed in Coot 73 using cryoSPARC sharpened cryo-EM maps, the SARS-CoV-2 M protein short conformation structure (PDB ID: 8CTK), and an AlphaFold374 model of the scFab variable domains rigid body fitted into the map in Coot as initial models. The C1P ligands (3 letter code 1PX) were added in Coot. The model was real space refined in Phenix 75 and validated using Molprobity76. Mutant models were generated with AlphaFold3.
We assessed putative metal coordination using several computational tools. First, AlphaFold374 structure predictions of M dimers with two cations (potassium, sodium, calcium, and magnesium) all show ion occupancy of the putative binding site with minimal distortions to the overall fold. Second, Metric Ion Classification (a deep learning tool for assigning ions in cryo-EM maps77) and CheckMyMetal (CMM, an ion validation tool that accounts for ligand chemistry, geometry, and refined B-factor statistics78) both predict metal over water binding to this site. We therefore separately modeled each ion in the structure, refined the model in Phenix 75 and compared validation statistics in CMM. The site was predicted in all cases to be most likely occupied by potassium and K+ is included in the final models.
Lipid binding assays
HEK293 cells were used to express and purify M protein for lipid binding overlay assays. Eight μg of DNA was diluted into Opti-MEM, mixed with PEI in the ratio of 3:1 (PEI:DNA), and incubated for 10 minutes at room temperature. The PEI-DNA mixture was added to cells plated in a 60 mm dish at 80% confluency. 5 hours post-transfection the media was changed to DMEM + 10 % FBS. The cells were then incubated for 24 hours at 37°C and 5% CO2. Media was then removed from the dish and cells were washed twice with PBS and lysed with lysis buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.25% Igepal CA-630, 5 μM PMSF, 5 mM EDTA, and Halt protease inhibitor cocktail). Cells were incubated in lysis buffer at 4°C for 10 min. Cells were then scraped from the dish and transferred to a prechilled centrifuge tube. The cell lysate was incubated for 30 min at 4°C with frequent 30–40 sec vortexing. The cell lysate was then centrifuged at 16,000 × g for 10 min at 4°C to separate the soluble and insoluble fractions. The supernatant was transferred to a microcentrifuge tube and small aliquots were used to determine the protein concentration with the BCA method. SDS-PAGE followed by Western blot was performed on the purified cell lysates to confirm M protein expression and check expression levels for different conditions/replicates. An anti-M antibody was used for M protein detection and an anti-actin antibody was used for control blots on cell lysates.
Lipid strips or sphingolipid snoopers were used to detect binding of M protein to different lipids. 0.5 μg of cell lysate was added as a control to one corner of each membrane to ensure M detection in each blot. The membrane was allowed to fully dry after cell lysate blotting. Once dry, the membrane was saturated with 1% milk in TBS-T for 1 hr at room temperature. After 1 hour, the solution was replaced with 4 μg/ml of total cell lysate in 1% milk in TBS-T and incubated overnight at 4°C. The membrane was then washed three times for 10 min each with TBS-T at room temperature. The membrane was incubated with anti-SARS-COV-2 M protein antibody diluted 1:5000 in 1% milk in TBS-T for 1 hr at room temperature. The membrane was then washed three times for 10 min each with TBS-T at room temperature. The membrane was then incubated for 1 hour at room temperature with secondary antibody (goat anti-rabbit) diluted at 1:5000 in 1% milk in TBS-T for 1 hr at room temperature. M protein interactions with different lipid classes was resolved using ECL Clarity reagents and an imager (Amersham Imager 600) to visualize M protein location on each membrane.
To determine the ability of M protein to interact with lipid coated beads, a pull down assay was performed using control beads, C1P beads, and PA beads (Echelon Biosciences, Inc., Salt Lake City, UT) and adapted from a published lipid coated bead binding protocol (https://bio-protocol.org/en/bpdetail?id=1039&type=0). M protein was expressed in HEK293 cells for 24 hours and cells were collected and lysed, which was followed by low speed centrifugation to remove cellular debris. Beads were incubated with cell lysates for 30 minutes at room temperature and the lipid bound and unbound fractions were separated centrifugation. SDS-PAGE analysis was performed on the bead pull down samples, which was followed by Western blot to detect the M protein in each sample.
C1P cellular assays
TLC plates were prepared with 20 cm height and width and incubated with 50 of 1:1 chloroform:methanol (v/v) in a glass tank. The TLC plate was secured in a plate holder for stability and the tank lid was closed until the solution ascended to 0.5 cm from the top of the plate (usually 2 to 3 hrs depending on room temperature). The TLC plate was allowed to dry for 5 min in a fume hood. The TLC plate was then activated for 30 minutes in an oven at 100°C. Lipid samples were prepared in chloroform (5–10 μl), sonicated, spun down, and loaded (0.5 to 2 μl of each sample) ~1 cm from the bottom of the TLC plate. Spots were dried prior to loading the plate in a saturated tank. Plates were sprayed with primuline (0.005% (w/v) in 4:1 acetone:water (v/v)), dried in the fume hood, and imaged at 345 nm (Amersham Imager 600). To assess M protein chromatography profiles with and without inhibition of CERK, cells were treated with vehicle (DMSO) or 100 nM NVP-231 and lysed and for analysis of M protein size using a Superdex™ 200 Increase small-scale SEC column (Cytiva Life Sciences, Marlborough, MA). Fractions were collected every 1 mL and a portion of each fraction was spotted onto nitrocellulose to determine M protein content in each fraction via detection of M using Western blot.
Cellular imaging and fractionation assays
HEK293, COS-7, or Vero E6 cells were used for SARS-CoV-2 protein expression and detection with confocal microscopy. Transfections were performed for M, N, S, and Flag-E alone or in combinations to assess M protein localization and colocalization with S or E. Imaging analysis was performed 16–24 hours post-transfection. Images were analyzed in Image J for localization and co-localization analysis. The cellular synctia assay was adapted from Zhang et al. 79. 24 hours post-transfection of S, S and M, or S with different M mutations, were imaged post nuclei staining and percent synctia formation was determined as the percentage of nuclei with fused cells.
VLP budding and entry assays
VLP production was performed and modified from Plescia et al. 17. Transfections of plasmid DNAs of M:N:S:Flag-E at a 6:3:8:8 ratio in 50 μl Opti-MEM, mixed with PEI in a 3:1 ratio (PEI:DNA), and incubated for 10–15 min at room temperature, and added to cells. Cells were placed at 37°C and 5% CO2 and five to six hours post-transfection, the media was changed to DMEM + 10 % FBS. Transfected cells were incubated at 37°C and 5% CO2. 48 hours post transfection, supernatants were collected in a chilled falcon tube with 1x Halt™ protease inhibitor cocktail. Cells were washed with 1x PBS and centrifuged 10 min 4° C at 1000 × g. The supernatant was transferred to a new falcon and centrifuged 10 min, 4° C at 1000 × g and filtered through 0.45 micron filter into a new tube. Supernatants were deposited on a 20% sucrose cushion. VLPs were centrifuged at 28,000 rpm for 3 hours at 4°C. 4. The resulting VLPs were used for entry assays and quantified for protein content using a BCA assay. VLP entry assays were performed using equal amounts of VLPs for each condition. Cells subjected to VLPs were spinoculated for 45 min at 4°C, and allowed to incubate for 1 hour at 37°C. Plates were then rinsed with PBS and nuclei were stained with Hoechst 33342 for 15 min at 37 °C prior to fixation with 4% paraformaldehyde (PFA) and confocal imaging. Quantification of VLP entry signal was performed by calculating the total number of VLPs (green puncta)/the total number of cells.
Figures were prepared using VMD, PyMOL, ChimeraX80, Prism, Python, Matlab, and Adobe Illustrator software.
Supplementary Material
Acknowledgements
This work was funded by the National Institutes of Allergy and Infectious Diseases grant R01-AI169896. Computational resources were provided by the Frontera Supercomputer at the Texas Advanced Computer Center (TACC) and the Research Computing Center (RCC) at the University of Chicago.
Footnotes
Competing interest statement
The authors declare no competing interests.
References
- 1.Navaratnarajah C. K., Warrier R. & Kuhn R. J. Encyclopedia of Virology (Third Edition). Artic. Titles: A 193–200 (2008) doi: 10.1016/b978-012374410-4.00667-1. [DOI] [Google Scholar]
- 2.Mazzon M. & Mercer J. Lipid interactions during virus entry and infection. Cell. Microbiol. 16, 1493–1502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meisen I. et al. The human H3N2 influenza viruses A/Victoria/3/75 and A/Hiroshima/52/2005 preferentially bind to α2–3-sialylated monosialogangliosides with fucosylated poly-N-acetyllactosaminyl chains. Glycobiology 22, 1055–1076 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Roth S. L. & Whittaker G. R. Promotion of vesicular stomatitis virus fusion by the endosome-specific phospholipid bis(monoacylglycero)phosphate (BMP). FEBS Lett. 585, 865–869 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Niort K. et al. Cholesterol and Ceramide Facilitate Membrane Fusion Mediated by the Fusion Peptide of the SARS-CoV-2 Spike Protein.ACS Omega 8, 32729–32739 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roingeard P. et al. The double-membrane vesicle (DMV): a virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses. Cell. Mol. Life Sci. 79, 425 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabata K. et al. Convergent use of phosphatidic acid for hepatitis C virus and SARS-CoV-2 replication organelle formation. Nat. Commun. 12, 7276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motsa B. B. & Stahelin R. V. Lipid–protein interactions in virus assembly and budding from the host cell plasma membrane. Biochem. Soc. Trans. 49, 1633–1641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckmann N. & Becker K. A. Ceramide and Related Molecules in Viral Infections. Int. J. Mol. Sci. 22, 5676 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meer-Janssen Y. P. M. van der, Galen J. van, Batenburg J. J. & Helms J. B. Lipids in host–pathogen interactions: Pathogens exploit the complexity of the host cell lipidome. Prog. Lipid Res. 49, 1–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baglivo M. et al. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Bio Med. : Atenei Parm. 91, 161–164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding C., Chen Y., Miao G. & Qi Z. Research Advances on the Role of Lipids in the Life Cycle of Human Coronaviruses. Microorganisms 12, 63 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong F. & Anderson R. Coronaviruses, Molecular Biology and Virus-Host Interactions. Adv. Exp. Med. Biol. 342, 203–208 (2011). [Google Scholar]
- 14.Anderson R. & Wong F. Membrane and Phospholipid Binding by Murine Coronaviral Nucleocapsid N Protein. Virology 194, 224–232 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta M., Su Y., Plescia C. B., Voth G. A. & Stahelin R. V. The SARS-CoV-2 nucleoprotein associates with anionic lipid membranes. J. Biol. Chem. 300, 107456 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuman B. W. et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol 174, 11–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plescia C. B. et al. SARS-CoV-2 viral budding and entry can be modeled using BSL-2 level virus-like particles. J Biological Chem 296, 100103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolan K. A. et al. Structure of SARS-CoV-2 M protein in lipid nanodiscs. Elife 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z. et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat Commun 13, 4399 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. et al. Synthesis, insertion, and characterization of SARS-CoV-2 membrane protein within lipid bilayers. Sci. Adv. 10, eadm7030 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornancin F. Ceramide kinase: The first decade. Cell. Signal. 23, 999–1008 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Mendonça L. et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 12, 4629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor K., Chen T. & Tajkhorshid E. Posttranslational modifications optimize the ability of SARS-CoV-2 spike for effective interaction with host cell receptors. Proc. Natl. Acad. Sci. 119, e2119761119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweizer A., Clausen H., Meer G. van & Hauri H. P. Localization of O-glycan initiation, sphingomyelin synthesis, and glucosylceramide synthesis in Vero cells with respect to the endoplasmic reticulum-Golgi intermediate compartment. J. Biol. Chem. 269, 4035–4041 (1994). [PubMed] [Google Scholar]
- 25.Barducci A., Bussi G. & Parrinello M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 100, 020603 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Loock M. V. et al. A small-molecule SARS-CoV-2 inhibitor targeting the membrane protein. (2024) doi: 10.21203/rs.3.rs-3975125/v1. [DOI] [Google Scholar]
- 27.Perrier A. et al. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J. Biol. Chem. 294, 14406–14421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boson B. et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 296, 100111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang Q. et al. SARS-CoV-2 Membrane protein regulates the function of Spike by inhibiting its plasma membrane localization and enzymatic activity of Furin. Microbiol. Res. 282, 127659 (2024). [DOI] [PubMed] [Google Scholar]
- 30.Cattin-Ortolá J. et al. Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation. Nat. Commun. 12, 5333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boath A. et al. Regulation and Traffic of Ceramide 1-Phosphate Produced by Ceramide Kinase COMPARATIVE ANALYSIS TO GLUCOSYLCERAMIDE AND SPHINGOMYELIN*. J. Biol. Chem. 283, 8517–8526 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Scherer K. M. et al. SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress. Sci. Adv. 8, eabl4895 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y., Zhou Y., Siddiqui P., Niu J. & Jiang S. Identification of Immunodominant Epitopes on the Membrane Protein of the Severe Acute Respiratory Syndrome-Associated Coronavirus. J Clin Microbiol 43, 3718–3726 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotop S.-K. et al. Peptide microarrays coupled to machine learning reveal individual epitopes from human antibody responses with neutralizing capabilities against SARS-CoV-2. Emerg Microbes Infec 11, 1037–1048 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heffron A. S. et al. The landscape of antibody binding in SARS-CoV-2 infection. Plos Biol 19, e3001265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin S. et al. SARS-CoV-2 integral membrane proteins shape the serological responses of COVID-19 patients. Iscience 24, 103185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jörrißen P. et al. Antibody Response to SARS-CoV-2 Membrane Protein in Patients of the Acute and Convalescent Phase of COVID-19. Front Immunol 12, 679841 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson K. A. et al. PI(4,5)P2 binding sites in the Ebola virus matrix protein VP40 modulate assembly and budding. J. Lipid Res. 65, 100512 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goellner S. et al. Zika virus prM protein contains cholesterol binding motifs required for virus entry and assembly. Nat. Commun. 14, 7344 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Husby M. L. et al. Phosphatidylserine clustering by the Ebola virus matrix protein is a critical step in viral budding. EMBO Rep. 23, e51709 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martín-Acebes M. A. et al. The Composition of West Nile Virus Lipid Envelope Unveils a Role of Sphingolipid Metabolism in Flavivirus Biogenesis. J. Virol. 88, 12041–12054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chukkapalli V., Hogue I. B., Boyko V., Hu W.-S. & Ono A. Interaction between the Human Immunodeficiency Virus Type 1 Gag Matrix Domain and Phosphatidylinositol-(4,5)-Bisphosphate Is Essential for Efficient Gag Membrane Binding. J. Virol. 82, 2405–2417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dick R. A. & Vogt V. M. Membrane interaction of retroviral Gag proteins. Front. Microbiol. 5, 187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inlora J., Collins D. R., Trubin M. E., Chung J. Y. J. & Ono A. Membrane Binding and Subcellular Localization of Retroviral Gag Proteins Are Differentially Regulated by MA Interactions with Phosphatidylinositol-(4,5)-Bisphosphate and RNA. mBio 5, 10.1128/mbio.02202-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norris M. J. et al. Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization. Sci. Adv. 8, eabn1440 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel-Megied A. M. et al. Characterization of the cellular lipid composition during SARS-CoV-2 infection. Anal. Bioanal. Chem. 415, 5269–5279 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abusukhun M. et al. Activation of Sphingomyelinase-Ceramide-Pathway in COVID-19 Purposes Its Inhibition for Therapeutic Strategies. Front. Immunol. 12, 784989 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moseley K. R., Knapp R. C. & Haisma H. J. An assay for the detection of human anti-murine immunoglubulins in the presence of CA125 antigen. J. Immunol. Methods 106, 1–6 (1988). [DOI] [PubMed] [Google Scholar]
- 49.Pauletto P. J. T., Delgado C. P. & Rocha J. B. T. da. Acid sphingomyelinase (ASM) and COVID-19: A review of the potential use of ASM inhibitors against SARS-CoV-2. Cell Biochem. Funct. 41, 284–295 (2023). [DOI] [PubMed] [Google Scholar]
- 50.D’Avila H. et al. Lipid Metabolism Modulation during SARS-CoV-2 Infection: A Spotlight on Extracellular Vesicles and Therapeutic Prospects. Int. J. Mol. Sci. 25, 640 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoertel N. et al. Medications Modulating the Acid Sphingomyelinase/Ceramide System and 28-Day Mortality among Patients with SARS-CoV-2: An Observational Study. Pharmaceuticals 16, 1107 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vitner E. B., Avraham R., Politi B., Melamed S. & Israely T. Elevation in sphingolipid upon SARS-CoV-2 infection: possible implications for COVID-19 pathology. Life Sci. Alliance 5, e202101168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jo S., Kim T., Iyer V. G. & Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Jo S., Kim T. & Im W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE 2, e880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brooks B. R. et al. CHARMM: The biomolecular simulation program. Journal of Computational Chemistry 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J. et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S., Lim J. B., Klauda J. B. & Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys J 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abraham M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softwarex 1, 19–25 (2015). [Google Scholar]
- 59.Huang J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoover W. G. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 61.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol Phys 52, 255–268 (2006). [Google Scholar]
- 62.Berendsen H. J. C., Postma J. P. M., Gunsteren W. F. van, DiNola A. & Haak J. R. Molecular dynamics with coupling to an external bath. J Chem Phys 81, 3684–3690 (1984). [Google Scholar]
- 63.Parrinello M. & Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys 52, 7182–7190 (1981). [Google Scholar]
- 64.Nosé S. & Klein M. L. Constant pressure molecular dynamics for molecular systems. Mol Phys 50, 1055–1076 (2006). [Google Scholar]
- 65.Darden T., York D. & Pedersen L. Particle mesh Ewald: An N ⋅log( N ) method for Ewald sums in large systems. J Chem Phys 98, 10089–10092 (1993). [Google Scholar]
- 66.Hess B., Bekker H., Berendsen H. J. C. & Fraaije J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J Comput Chem 18, 1463–1472 (1997). [Google Scholar]
- 67.Laio A. & Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. 99, 12562–12566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dama J. F., Parrinello M. & Voth G. A. Well-Tempered Metadynamics Converges Asymptotically. Phys. Rev. Lett. 112, 240602 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Michaud-Agrawal N., Denning E. J., Woolf T. B. & Beckstein O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J Comput Chem 32, 2319–2327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song W. et al. PyLipID: A Python Package for Analysis of Protein–Lipid Interactions from Molecular Dynamics Simulations. J. Chem. Theory Comput. 18, 1188–1201 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Humphrey W., Dalke A. & Schulten K. VMD: Visual molecular dynamics. J Mol Graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 72.Punjani A., Rubinstein J. L., Fleet D. J. & Brubaker M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 1–11 (2021) doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Afonine P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta crystallographica. Section D, Structural biology 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein science : a publication of the Protein Society 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shub L., Liu W., Skiniotis G., Keiser M. J. & Robertson M. J. Metric Ion Classification (MIC): A deep learning tool for assigning ions and waters in cryo-EM and x-ray crystallography structures. bioRxiv 2024.03.18.585639 (2024) doi: 10.1101/2024.03.18.585639. [DOI] [Google Scholar]
- 78.Gucwa M. et al. CMM—An enhanced platform for interactive validation of metal binding sites. Protein Sci. 32, e4525 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Z. et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ. 28, 2765–2777 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goddard T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein science : a publication of the Protein Society 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.