Abstract
Histidine phosphorylation (pHis) is a non-canonical post-translational modification (PTM) that is historically understudied due to a lack of robust reagents that are required for its investigation, such as high affinity pHis-specific antibodies. Engineering pHis-specific antibodies is very challenging due to the labile nature of the phosphoramidate (P-N) bond and the stringent requirements for selective recognition of the two isoforms, 1-phosphohistidine (1-pHis) and 3-phosphohistidine (3-pHis). Here, we present a strategy for in vitro engineering of antibodies for detection of native 3-pHis targets. Specifically, we humanized the rabbit SC44–8 anti-3-pTza (a stable 3-pHis mimetic) mAb into a scaffold (herein referred to as hSC44) that was suitable for phage display. We then constructed six unique Fab phage-displayed libraries using the hSC44 scaffold and selected high affinity 3-pHis binders. Our selection strategy was carefully designed to enrich antibodies that bound 3-pHis with high affinity and had specificity for 3-pHis versus 3-pTza. hSC44.20N32FL, the best engineered antibody, has an ~10-fold higher affinity for 3-pHis than the parental hSC44. Eleven new Fab structures, including the first reported antibody-pHis peptide structures were solved by X-ray crystallography. Structural and quantum mechanical calculations provided molecular insights into 3-pHis and 3-pTza discrimination by different hSC44 variants and their affinity increase obtained through in vitro engineering. Furthermore, we demonstrate the utility of these newly developed high-affinity 3-pHis-specific antibodies for recognition of pHis proteins in mammalian cells by immunoblotting and immunofluorescence staining. Overall, our work describes a general method for engineering PTM-specific antibodies and provides a set of novel antibodies for further investigations of the role of 3-pHis in cell biology.
Keywords: Antibody Engineering, Protein Engineering, Phosphohistidine, Post-translational Modifications, Phage-Display
Introduction
There are ~600 known PTMs that underlie almost every process in cell biology (1). Protein phosphorylation is the best characterized PTM, with phosphorylation of serine (Ser; pSer), threonine (Thr; pThr) and tyrosine (Tyr; pTyr) being the most abundant and best characterized (2). Given the importance of PTMs in signal transduction pathways, it is unsurprising that enzymes involved in post-translational modification of proteins are frequently mutated in many diseases, such as cancer (3), and therefore represent attractive drug targets (4–6). Rare and non-canonical PTMs are emerging as additional key regulators of cell biology and biochemistry (7). Of particular interest is the role of phosphohistidine (pHis) in mammalian cell biology (8). This non-canonical PTM was first described in 1962 (9) and synthesized in 1966 (10), but despite a role for His phosphorylation being well characterized in prokaryotic cells (11), it remains understudied in mammalian cells. One of the major challenges in the detection and analysis of pHis is the acid and heat labile nature of the phosphoramidate (P-N) bond in pHis (12) and the lack of suitable reagents to study this PTM. Despite these obstacles, the discovery of histidine kinases and phosphatases (13), as well as modified experimental pipelines (14) and phosphoproteomics workflows (15), have recently enabled the study of this PTM in mammalian cells at a systems level scale.
Advances in the development of non-hydrolyzable pHis mimetics led to their subsequent use for raising anti-pHis antibodies (16–22). Some of the pivotal anti-pHis antibodies are rabbit-derived monoclonal antibodies (mAbs) that were obtained by immunizing with non-hydrolyzable phosphoryl-triazolylalanine (pTza; a pHis mimetic)-containing peptides (23). Subsequent structure-function analysis has provided insights into how these antibodies discriminate between 1- and 3-pTza in a peptide context (24). This suite of anti-pHis antibodies has been used by many groups to investigate the role of pHis in bacterial and mammalian physiology (25–29). Of these antibodies, the rSC44–8 3-pHis mAb has been most widely used as it has relatively higher affinity towards 3-pHis proteins. However, rSC44–8 is more specific for the 3-pTza analogue (KD: 0.24 nM) than the natural 3-pHis, where the rSC44–8 affinity is ~75 fold lower (KD: 18 nM) (24).
The present study extends these advances by (i) humanizing the 3-pTza specific rabbit antibody, (ii) engineering the humanized antibody for improved affinity and specificity towards 3-pHis compared to 3-pTza, (iii) solving the first crystal structure of an antibody:3-pHis peptide complex, and (iv) demonstrating the use of the newly derived, high affinity anti-3-pHis antibodies to detect 3-pHis proteins in standard molecular and cell biology workflows. This advance was achieved by rational engineering of the human trastuzumab (4D5) antibody scaffold to accommodate rabbit complementarity determining regions (CDRs) by substituting specific variable region framework residues known as Vernier zone residues that modulate the conformation of CDRs (30, 31). We then used this humanized scaffold for constructing novel phage displayed antibody libraries and carried out affinity selections for both pTza- and pHis-containing peptides. Our engineering strategy allowed us to isolate new antibodies with increased affinity and specificity for 3-pTza- and 3-pHis-containing peptides. Furthermore, using novel, highly crystallizable Fab scaffolds (32–34), we solved 11 crystal structures of anti-3-pHis Fabs in complex with phosphorylated ligands, including the first antibody-pHis peptide structure. These structures in conjunction with quantum mechanical and molecular mechanical (QM/MM) studies provided insights into the recognition of 3-pTza versus 3-pHis by the humanized and engineered antibodies, explained the enhanced affinity of the engineered variant antibodies, and provide a basis for future engineering of antibodies towards specific pHis-peptides relevant to mammalian cell biology. Finally, we showed that the engineered 3-pHis specific antibodies identified biologically relevant pHis signals in mammalian cells using immunoblotting and immunofluorescence staining.
The significance of these studies is that they establish a generalizable strategy for engineering antibodies to recognize unstable non-canonical PTMs that can only be accomplished using in vitro evolution. Recent studies describing the engineering of a mouse monoclonal antibody for higher affinity recognition of pTyr highlight the importance of our approach (35). The enhanced affinity of anti-3-pHis antibodies can be used to elucidate the role of pHis in a variety of fields including cell biology, phosphoproteomics, signal transduction, and identifying novel drug targets (36).
Results
Rabbit Antibody Scaffold Engineering and Humanization.
To engineer 3-pTza specific rabbit antibodies for improved affinity and specificity for 3-pHis, we elected to use SC44–8 (hereafter referred to as rSC44), which is a 3-pHis specific rabbit mAb that has partial sequence selectivity for Gly-3-pHis-Ala sites and binds to an AGAG-3-pHis-AGAG peptide with ~20 nM affinity. We first assessed whether rSC44 could be produced as IgG and Fab proteins from mammalian cells and displayed as Fab and scFv versions on M13 bacteriophage. The rSC44 antibody could reliably be produced as IgG and Fab proteins, as previously described (24), but could not be displayed on phage in either Fab or scFv formats. This led to an extensive rational engineering campaign on the rSC44 scaffold to humanize it for directed evolution campaigns using phage display.
We first sought to fully humanize the rabbit antibody into the human 4D5 framework (trastuzumab), which displays efficiently on phage (37). Humanization of rabbit antibodies has traditionally been accomplished using CDR grafting and substitution of specific Vernier zone residues in the variable framework (38, 39). The sequences of rSC44 were aligned with those of the human 4D5 scaffold using Kabat (40) and IMGT nomenclatures (41) for both the variable light (VL) and variable heavy (VH) regions (Fig. S1), prior to loop grafting of all 6 rSC44 CDRs into the human 4D5 scaffold using both IMGT and Kabat boundaries. These humanized versions displayed on phage but did not bind the 3-pTza antigen. We then identified sets of Vernier zone residues that could be important for CDR orientation and therefore antigen binding. Several successive rounds of combinatorial back mutation were necessary to engineer a functional human scaffold for rSC44. Hereafter, we will refer to specific amino acid positions using Kabat nomenclature (Fig. S1C,D).
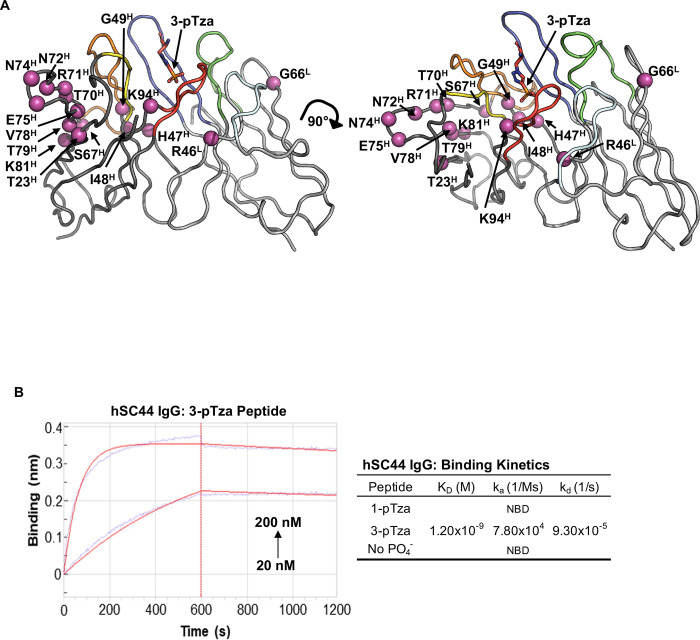
The humanized version of rSC44 (hSC44) had 2 and 14 Vernier zone residues in the VL and VH domains back mutated to rabbit sequences, respectively (Fig. 1A) and could be produced as IgG and Fab proteins (Fig. S1E). Furthermore, the IgG version of hSC44 bound the 3-pTza peptide with high affinity (KD = 1.2 nM) but exhibited no binding to the 1-pTza or the non-phosphorylated (No PO4−) version of the peptide as measured by biolayer interferometry (BLI; Fig. 1B). Finally, Fab and scFV versions of hSC44 displayed on phage could bind 3-pTza as measured by ELISA (Fig. S1F).
Fig. 1. Humanization of the rabbit (rSC44) antibody.
(A) Structure of rSC44 (PDB ID: 6X1V) in complex with a 3-pTza containing peptide (3-pTza shown as sticks colored in red) was used to guide humanization. The VH and VL domains are colored dark and light grey respectively. The CDRs are colored as follows: H3, red; H2, orange; H1, yellow; L3, blue; L2, cyan; L1, green. Residues in important framework positions, including known Vernier Zone residues are colored magenta and represented as spheres highlighting their Cα position. Residue numbering is in accordance with Kabat nomenclature. (B) Biolayer Interferometry (BLI) data for hSC44 IgG binding to 3-pTza, 1-pTza and a non-phosphorylated (no PO4−) peptide. The table shows the binding constants determined from BLI traces, and sample traces are shown for hSC44 binding to the immobilized 3-pTza peptide. NBD indicates no binding detected.
Phosphohistidine Peptide Immunoprecipitation.
To generate 1/3-pHis-containing peptides for antibody selection, we developed a custom immunoprecipitation workflow (Fig. S2). Biotinylated-His-peptides were incubated with potassium phosphoramidate for 16 h in 1x HEPES buffered saline (HBS; pH = 8.5) to generate pHis peptides. We then constructed a rProteinA-IgG resin using the rSC1 (1-pTza specific) and rSC44 (3-pTza specific) IgG’s described previously (23), in order to enrich either 1-pHis or 3-pHis peptides, respectively. Elution conditions using 2 M imidazole (pH 8.5) were optimized and eluates were analyzed using ELISA (see Materials and Methods for a detailed description).
Phage Displayed Library Construction and Affinity Selections.
In an attempt to both increase hSC44 affinity and improve specificity for 3-pHis versus 3-pTza peptides, we designed and constructed 6 unique phage displayed Fab libraries (Fig. S3–4). To aid library design, we analyzed the previously reported structure of rSC44 in complex with a 3-pTza peptide (PDB ID: 6X1V) and selected for diversification residues in both VL and VH regions. Libraries 1–5 (hSC44 1–5) diversified residues that are in direct contact with the 3-pTza side chain or have side chains oriented towards and proximal (within 10 Å) to the 3-pTza side chain (24). Library 6 diversified key Vernier zone and additional framework residues that were involved in CDR coordination and were important for creating a stable human scaffold. All libraries contained 109-1010 unique variants and were generated using a “soft randomization” strategy that favors wild-type (wt) sequence but allows for diversity across all positions sampled (Figs. S3 and S4, Table S1) (42).
To select for Fab variants with high affinity and specificity for 3-pHis peptides, but not the 1-pHis isoform, we devised a selection strategy in which 1-pTza and non-phosphorylated peptides (No PO4−) were used to deplete all 6 libraries of cross-reactive Fabs (pools a-d, Fig. 2A, Table S2). For example, pools a and b were positively selected for 3-pTza whereas pools c and d were positively selected for 3-pHis. Pools b and d were subjected to negative selections against 1-pTza. Selections were carried out with pre-clearing steps on BSA and streptavidin as well as stringent negative selections on non-phosphorylated peptides. All 6 hSC44 libraries were cycled through 5 rounds of affinity selections and yielded 32 unique sequences (Fig. 2B). No hSC44 template was detected across pools a-d, indicating the successful progression of affinity selections. We produced each unique variant as a phage displayed Fab as described previously (43), normalized their concentrations, and assessed binding to 1/3-pTza and 1/3-pHis peptides relative to hSC44 using high-throughput ELISAs. Thirty of the 32 unique sequences displayed increased binding to either 3-pTza or 3-pHis and no binding to 1-pTza or 1-pHis (Fig. 2B). Remarkably, we achieved high 3-pHis specificity in our selection campaigns, as all the enriched antibodies that were positively selected for 3-pHis showed preferable binding to 3-pHis over the phospho-mimetic 3-pTza embedded in the same peptide sequence.
Fig. 2. Outline of affinity selections protocol and selection of 3-pHis-specific hSC44 variants.
(A) Schema depicting affinity selection strategy. Libraries were combined on day 1 with unique outputs from different positive selections carried through the remainder of the selection process. The strategy resulted in 4 unique pools labelled a-d which represent unique positive selections. No PO4− is a non-phosphorylated version of the His-peptide. (B) hSC44 variants isolated after 5 rounds of affinity selections. The amino acid sequence (Kabat nomenclature) of hSC44 residues randomized in different libraries are shown at the top. Sequences in hSC44 variants that were the same as hSC44 are blank and differences are shown with the single-letter amino acid code. The pool, which each unique variant was identified in, is listed next to each sequence. Unique variants were reamplified as Fab-Phage, normalized for concentration and assayed by phage-ELISA for binding to non-phosphorylated (No PO4−), 1-pTza, 1-pHis, 3-pTza and 3-pHis peptides. Binding is shown as fold change in absorbance over hSC44.
Sequence analysis of the selected 3-pTza and 3-pHis antibodies revealed that most of the substitutions resided in CDR-H3, with almost all variants displaying S97TH and N99T/SH substitutions (Fig. 2B). These substitutions were correlated with increased binding to 3-pTza and/or 3-pHis. Furthermore, we observed that while N99H was substituted with D, H or G in the CDR-H3 of Fabs selected for 3-pTza-containing peptides, these substitutions were absent in Fabs positively selected for 3-pHis and correlated with a moderately increased affinity for 3-pTza and diminished or no binding to 3-pHis. Some variants had a V100aL/FH substitution in CDR-H3, but this substitution was less frequent and did not correlate with enhanced binding. Finally, the N32Y/FL substitution in CDR-L1 was observed only in variants selected for 3-pHis and conferred preferential binding to 3-pHis over 3-pTza (Fig. 2B right hand panels). Together, these results indicate that a small set of substitutions in CDR-H3 can increase affinity for both 3-pTza and 3-pHis, and that a single substitution in CDR-L1 contributes to enhanced binding of 3-pHis over 3-pTza.
hSC44 Variant Affinity, Specificity, and Developability.
To investigate the antibody variants in greater detail we expressed them as IgGs (Fig. S5A,B). Twenty-eight of the 32 variants had higher yields than hSC44, 17 of which were even greater than the high-yield trastuzumab benchmark. We selected 7 hSC44 variants that bound 3-pTza/3-pHis with high apparent affinity and had favorable IgG yields and measured their EC50 values for the 3-pHis peptide to estimate their affinities using ELISA (Fig. S5C). In agreement with the phage ELISA screening (Fig. 2B), all variants had EC50 values that were up to 2-fold lower than hSC44. We observed that all isolated antibodies displayed a substantial improvement in binding to 3-pHis relative to the parental hSC44, with hSC44.20 showing the highest apparent affinity (Fig. S5C).
We further investigated the affinity of 8 antibody variants with the greatest increase in apparent affinity for 3-pTza/3-pHis using biolayer interferometry (BLI, Table 1 and Fig. S6). BLI measurements demonstrated that hSC44 variants positively selected for the 3-pTza peptide displayed affinities in the 100–1000 pM range, with one variant (hSC44.20) reaching the detection limit of the instrument (<1×10−12 M) compared with 1.2 nM for parental hSC44. Variants that were positively selected for 3-pHis (hSC44.28 and hSC44.31) had a small increase or decrease in affinity for 3-pTza compared with hSC44, respectively.
Table 1.
Affinity measurements of hSC44 variants against 3-pTza peptides using bio-layer interferometry.
| Name | 3-pTza Peptide | ||
|---|---|---|---|
| KD (M) | ka (1/Ms) | kd (1/s) | |
|
| |||
| hSC44.1 | 5.88E−10 | 1.41E+05 | 8.32E−05 |
| hSC44.3 | 3.52E−10 | 4.07E+04 | 1.43E−05 |
| hSC44.8 | 1.85E−10 | 3.59E+05 | 6.66E−05 |
| hSC44.13 | 4.05E−10 | 1.82E+05 | 7.37E−05 |
| hSC44.20 | <1.0E−12 | 2.57E+05 | <1.0E−07 |
| hSC44.20.N32FL | 1.54E−09 | 3.73E+04 | 5.73E−05 |
| hSC44.20.N32YL | <1.0E−12 | 3.51E+04 | <1.0E−07 |
| hSC44.22 | 4.95E−10 | 9.06E+04 | 4.49E−05 |
| hSC44.28 | 1.21E−09 | 2.49E+04 | 3.00E−05 |
| hSC44.31 | 2.71E−09 | 1.47E+04 | 3.98E−05 |
Given that hSC44.20 had the highest affinity for 3-pTza and that all antibody variants isolated using 3-pHis as bait possessed a N32Y/FL in their CDR-L1, we decided to shuffle the light chain of hSC44.20 to obtain hSC44.20.N32YL and hSC44.20.N32FL. Affinity measurements for 3-pTza revealed that, while the N32YL substitution did not affect binding to 3-pTza (KD <1×10−12 M), the N32FL substitution substantially reduced 3-pTza affinity (KD <1.54×10−9 M, Table 1, Fig. S6). We then measured the binding affinity of hSC44.20.N32YL and hSC44.20.N32FL for 3-pHis and showed that both variants bound 3-pHis peptides with affinities higher (N32FL = 1.6 nM and N32YL = 9.5 nM) than the parental antibody (Table 2, and Fig S7). No binding to pTyr, pSer, pThr or the non-phosphorylated version of the peptide could be detected for any of the variants by ELISA (Fig. S5D).
Table 2.
Affinity measurements of hSC44 variants against 3-pHis peptides using bio-layer interferometry.
| Name | 3-pHis Peptide | ||
|---|---|---|---|
| KD (M) | ka (1/Ms) | kd (1/s) | |
|
| |||
| hSC44.20 | 2.10E−08 | 7.55E+04 | 1.60E−03 |
| hSC44.20.N32FL | 1.68E−09 | 1.39E+04 | 1.64E−05 |
| hSC44.20.N32YL | 9.50E−09 | 8.88E+03 | 2.61E−05 |
To assess the developability of the new antibody variants, we performed a specificity ELISA against known antigenic molecules, as cross-reactivity is indicative of poor developability (44) (Fig. S8). We did not detect binding to any of the molecules tested, indicating no cross-reactivity. Together these, results demonstrate that humanized hSC44 and its engineered variants provide a scaffold that can produce IgGs with high affinity for 3-pHis and are correlated with good developability for future engineering and use in research pipelines.
Antibody Structure Determination.
To better understand the structure-function relationships of antibody:pHis binding, we undertook an extensive crystallization campaign to solve the structures of Fab versions of hSC44, hSC44.20, and hSC44.20.N32FL. Initial crystallization attempts with hSC44 Fab failed to produce any crystals. To overcome this issue, we utilized a method that enhances the crystallization of Fabs by substituting several residues distal to the antibody paratope that favor crystal packing interfaces. This method was based on (i) altering the constant kappa (cκ) light chain β-sheet (34) along with two substitutions upstream of this site (32), which are collectively known as “S1C” (Fig. S9), and (ii) substituting the elbow region residues of the heavy chain (33) (Fig. S10; Table S3). The S1C substitutions were introduced either alone or in combination with the elbow substitutions (collectively called “S1CE” substitutions) into the scaffolds described above and used in crystallization trials to solve structures in the apo, 3-pTza or 3-pHis complexed forms. These substitutions enabled us to obtain dozens of new crystals and to solve 11 new structures, including three 3-pHis peptide/Fab co-crystal structures (Tables S4–5). Notably, the substitutions that enhanced crystallization did not affect the overall structure of the antibody or the epitope.
Effect of Vernier Zone Residue Back Mutation on hSC44 Structure.
Humanization of the rSC44 antibody required back mutation of 2 Vernier zone residues in the human light chain and 14 residues (Vernier plus additional framework residues) in the heavy chain (Fig 1A; Fig. S1). To evaluate the structural similarity of rSC44 and hSC44, we performed an alignment of the Cɑ coordinates of the VH region, which showed a root mean square deviation (RMSD) of 0.5 Å (Fig. S11A). Most of the Vernier zone residue changes are structurally justified as they are involved in positioning the CDR loops. For example, the L46RL back mutation in the light chain is important, as R46L interacts with the main chain of N99H and P100H on the CDR-H3 loop that is relevant for coordinating the phosphoryl group in 3-pTza (Fig. S11B). Most of the substituted Vernier residues in the heavy chain are located in framework regions 2 and 3 (Fig. 1A; Fig. S1). Residue F34H is involved in forming hydrophobic interactions with F27H, L4H, K94H and I102H. While these residues form the core of the antibody, K94H is involved in hydrogen bonding with the phosphoryl group (Fig. S11C). Substitutions from 47WVAS50 to 47HIGY50 in the framework region 2 and start of CDR-H2 of the heavy region are critical, as this loop is at the interface between the heavy chain and light chain variable domains. While H47H establishes hydrogen bond interactions, Y50H forms T-shaped pi-pi stacking with Y96L (distance 5.0 Å, angle 75°, and offset 1.5 Å) at the interface, which in turn interacts with the phosphoryl moiety (Fig. S11D). Residues 61SWAK64 in CDR-H2 of the heavy chain are directed towards the core of the antibody and structurally support the 47HIGY50 strand (Fig. S11D). Residues 70TRNTNENTV78 on framework region 3 of the heavy chain support the orientation of the CDR-H1 and CDR-H2 loops. Together, the Vernier zone substitutions uphold the structure of the paratope that interacts with either 3-pHis or 3-pTza containing epitopes.
Structural Analysis of 3-pHis Recognition by hSC44, hSC44.20, and hSC44.20.N32FL.
In order to understand the structural basis for recognition of 3-pHis versus 3-pTza by hSC44 variants, we co-crystallized them with 3-pTza and 3-pHis peptides. Though 3-pHis peptides are labile and undergo spontaneous hydrolysis, our previous study indicated that bound rSC44–8 shields the 3-pHis moiety and slows down its hydrolysis (24). Inclusion of the entire phosphorylation reaction together with the hSC44 variant proteins in the co-crystallization mix allowed the antibodies to bind to and protect 3-pHis peptides during crystallization leading to crystals containing intact 3-pHis peptides. We aligned co-crystal structures of hSC44, hSC44.20 and hSC44.20.N32FL in complex with a 3-pTza or 3-pHis peptide using the Cɑ coordinates of the VH domain and demonstrated that these structures are highly similar to one another with RMSD’s <1.0 Å (Fig. S12). Consequently, the peptide location in the CDR regions of the hSC44 variants are nearly identical to one another as well as in rSC44 (Fig. 3A–C and S11A). The phosphoryl moiety of 3-pHis forms a hydrogen bond network with K94H and the CDR-H3 residues 95–99 (LGSGNH in hSC44, LGTGSH in hSC44.20 and hSC44.20.N32FL) that form the canonical phosphoryl-recognizing GXGX motif (45) (Fig. 3D–F). The GXGX motif forms a loop around the phosphoryl group in the shape of a ‘nest’ and interacts through the main chain amides of G96H and G98H, and the side-chain hydroxyl of S/T97H and side-chain or backbone interactions with N/S99H. While N32L in CDR-L1 of hSC44.20 makes water mediated interactions with the phosphoryl group, the same interaction is absent in hSC44.20.N32FL due to the substituted Phe at this position (Fig. 3F). Additionally, the Y96L hydroxyl group from CDR-L3 in all variants also makes direct hydrogen bonds with the phosphoryl group. This interaction is further strengthened by water-mediated hydrogen bonds to G33H and Y50H.
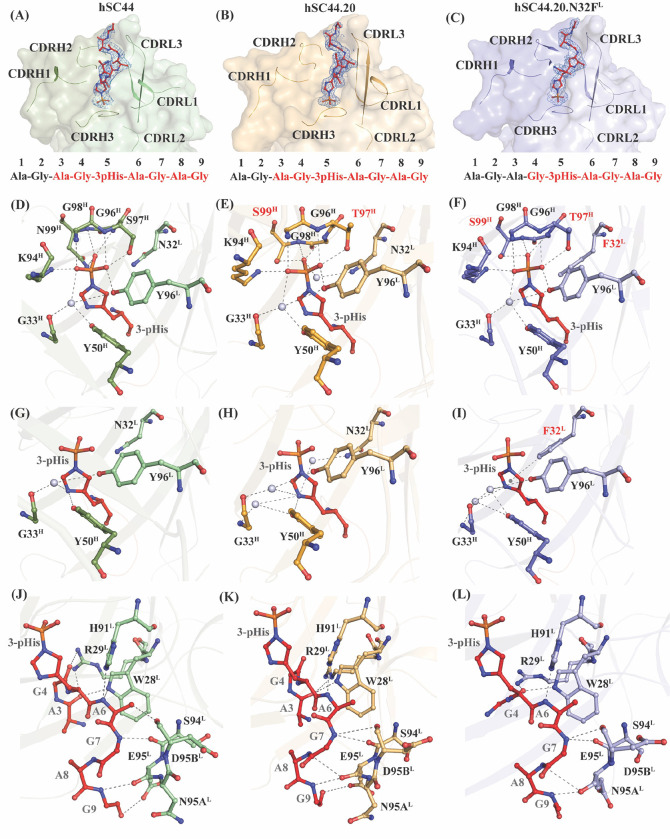
Fig. 3. Structural analysis of hSC44 variants bound to 3-pHis.
(A-C) Omit maps (FO-FC map) of (A) hSC44 Fab (green), (B) hSC44.20 Fab (orange), and (C) hSC44.20.N32FL Fab (blue) bound to a 3-pHis peptide (red). The peptide is contoured at 1.0 σ and the CDR loops are represented as cartoon under the surface. The sequence of the peptide used for crystallization is shown below and the residues in red font have interpretable electron density. (D-F) Hydrogen bond network of phosphoryl moiety contacts with CDR residues and coordinating water molecules (grey spheres) of (D) hSC44, (E) hSC44.20, and (F) hSC44.20.N32FL. Substituted residues in (E-F) that are different from hSC44 (D) are shown in red. (G-I) Hydrogen bond network of imidazole ring contacting residues and coordinating water molecules of (G) hSC44, (H) hSC44.20, and (I) hSC44.20.N32FL. (J-L) Hydrogen bond network of CDR residues contacting the ligand backbone and non-3-pHis side chain atoms in (J) hSC44, (K) hSC44.20, and (L) hSC44.20.N32FL.
In 3-pHis, the N3 nitrogen is phosphorylated, enabling the N1 nitrogen in the imidazole ring to interact with specific residues lining the binding pocket. The N1 nitrogen forms either direct or water-mediated hydrogen bond interactions with G33H, Y50H and Y96L in all hSC44 variants (Fig. 3G–I). The substitution N32FL in hSC44.20.N32FL increased the affinity of the antibody for the 3-pHis peptide by ~10-fold. This is likely due to the formation of T-shaped pi-pi stacking interactions with the imidazole side chain that are lacking in other variants (Fig. 3I). We further investigated the effect of N32FL substitution using Quantum Mechanics/Molecular Mechanics (QM/MM)-based contact analysis which revealed that though hSC44.20 and hSC44.20.N32FL have equal numbers of contacts with the 3-pHis peptide, the N32FL substitution introduced a new hydrophobic interaction (Fig. S13). Inspection of the structure following local alignment reveals a 1.2 Å shift of the imidazole ring closer to the N32FL substitution when 3-pHis is bound to hSC44.20.N32FL, relative to the position of the peptide when it is bound to hSC44.20 (Fig. S14A). A more detailed QM/MM analysis indicates that the side chain of the N32FL substitution establishes a T-shaped pi-pi interaction with the imidazole ring in pHis (Fig. S14B), which likely stabilizes the observed shift in peptide position when bound to hSC44.20.N32FL. The resulting stable π-π interaction explains the increased binding affinity of hSC44.20.N32FL to 3-pHis (Fig. S15 and Table S6).
Ordered electron density was found for the AG-3-pHis-AGAG peptide in the variants of hSC44, in keeping with the pseudo-sequence specificity or partial-sequence dependence property of rSC44 (24). CDR-L1 W28L and R29L interact with the N-terminal residues of the peptide (A3 and G4) while CDR-L3 residues (H91L, S94L, E95L, N95aL and D95bL) make hydrogen bond interactions with the peptide backbone at its C-terminus (G7-G9) (Fig. 3J–L). This network of hydrogen bonds is retained among hSC44 variants and is implicated in the higher affinity and specificity for the peptide ligand. This is further corroborated by thermal shift assays (Fig. S16A) and BLI assays (Fig. S16B) where the hSC44 variants show specificity for 3-pHis or 3-pTza-containing peptides of ACLYana (AGAG-pHis-AGAG), ACLY (VQFG-pHis-AGAC) and SCS (RRMG-pHis-AGAI), but do not bind other 3-pHis or 3-pTza peptides (Fig. S16).
In-Depth Structure-Function Analysis of hSC44.20.N32FL Substitutions.
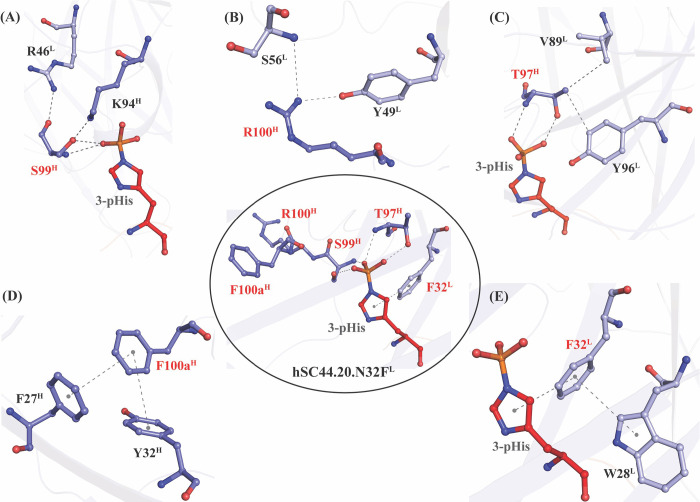
The high affinity variant hSC44.20.N32FL has four substitutions in CDR-H3 (S97TH, N99SH, P100RH and V100aFH) and one in CDR-L1 (N32FL) that differentiate it from the parental hSC44. These residues do not affect the conformations of their respective CDR-H3 loops and are essential for phosphoryl coordination of the pHis moiety, with T97H and S99H making hydrogen bonds with the phosphoryl group (Fig. 4 middle-inset). These residues also stabilize the CDR-H3 loop by forming intra- and inter-molecular networks with other CDRs as well as framework residues (Fig. 4A,C). S99H forms hydrogen bonds with K94H and R46L (Fig. 4A). The guanidino group of R100H in hSC44.20 forms hydrogen bond interactions with Y49L and S56L, which otherwise could not be formed with P100H of hSC44 (Fig. 4B). The methyl group on T97H in hSC44.20 is directed towards the core of the antibody and makes a hydrophobic interaction with V89L (distance 4.2 Å) and Y96L (distance 3.5 Å) (Fig. 4C). F100aH forms a T-type pi-pi stacking interaction with Y32H (distance 5.5 Å, angle 68.0°) and F27H (distance 4.4 Å, angle 86°) (Fig. 4D). The hSC44.20.N32FL variant is selective for 3-pHis while maintaining high affinity (Tables 1–2). This variant has an additional N32FL substitution in CDR-L1 that was specifically selected for in 3-pHis selections (Fig. 2B). This additional substitution forms T-shaped pi-pi stacking interactions with W28L (distance 5.9 Å, angle 81°) and the imidazole ring (distance 4.2 Å, angle 86°) of the 3-pHis peptide. These pi-pi interactions mediated by F32L may help to stabilize the CDR-L1 loop, resulting in selectivity for 3-pHis ligand (Fig. 4E).
Fig. 4. Structural analysis of effects of hSC44.20.N32FL substitutions relative to hSC44 on 3-pHis recognition.
The middle-inset shows the structure of hSC44.20.N32FL Fab and the 3-pHis side chain of the peptide ligand, and it highlights the CDR-H3 and CDR-L1 residues (red font) that differ from hSC44. (A) The sidechain of S99H makes hydrogen bonds with the phosphoryl group of 3-pHis. The sidechain and backbone residues interact with proximal framework residues R46L and K94H. (B) The sidechain of R100H points away from 3-pHis making contacts with framework residues S56L and Y49L. (C) The sidechain and backbone atoms of T97H make contact with the phosphoryl moiety of 3-pHis. The methyl group of the T97H sidechain makes hydrophobic contacts with framework residues V89L and Y96L. (D) The F100aH side chain makes a sandwiching T-shaped π-π interaction with the side chains of framework residues F27H and Y32H. (E) The F32L side chain also makes a sandwiching T-shaped π-π interaction with the imidazole ring of 3-pHis as well as the side chain of CDR-L1 residue W28L.
Analysis of Differences between 3-pTza and 3-pHis Coordination.
Although hSC44 antibody variants are structurally similar, their affinity towards the 3-pTza peptide is >1000 times more than 3-pHis peptide (Table 1 and 2). To understand these differences, we analyzed the structures of hSC44 variants bound to 3-pTza or 3-pHis in greater detail (Fig. S17). This analysis highlights the high structural similarity of hSC44 variants to one another (RMSD <1 Å between Cɑ atoms in the VH domain) when bound to either 3-pTza or 3-pHis. For simplicity, we further analyzed the hSC44.20 variant in complex with 3-pHis versus 3-pTza peptides (Fig. S18A,B). The CDR residues that interact with either 3-pTza or 3-pHis are similar in both structures (Fig. S18C–F). A water molecule buried at the interface between the peptide and CDR of the Fab, which mediate antibody-antigen interactions, is conserved between the 3-pTza and 3-pHis complexes (Fig. S18C,D). The anticipated difference between the 3-pTza and 3-pHis complexes mainly involves the triazolyl vs imidazole ring interactions. The triazolyl ring in the 3-pTza has three nitrogen groups (at ring positions 1, 2 and 5, with a phosphoryl group attached to the carbon at position 3) whereas 3-pHis has two nitrogens (at ring positions 1 and 3, with a phosphoryl group linked to the nitrogen at position 3 (Fig. S19A). The N1 atom of the 3-pHis sidechain ring and the N1 atom in the corresponding position of the pTza triazolyl ring both form water-mediated interactions with G33H and Y50H (Fig. S19B,C). The nitrogen atom at the triazolyl N2 position forms hydrogen bonds to K94H and S99H that are absent in the 3-pHis peptide, as it contains a carbon atom at this corresponding position (Fig. S19B). However, the >1000 fold difference in affinity observed for 3-pTza versus 3-pHis bound to hSC44 variants could not be explained by crystal structures. Hence, we carried out QM/MM calculations to rationalize this affinity difference between native and mimetic peptide. Molecular mechanics (MM)-based contact analysis reveals that, in comparison to 3-pHis, the 3-pTza moiety forms a greater number of contacts with the paratope of hSC44.20, and the stable contacts are predominantly associated with the phosphoryl moiety (Fig. S20). The chemical properties around the phosphoryl moiety differ between the two peptides. In 3-pHis, the phosphoryl moiety is attached via a nitrogen atom, whereas in 3-pTza, the phosphoryl moiety is attached via a carbon atom (Fig. S21A). Nitrogen, being more electronegative than carbon (46) (electronegativity 3.04 versus 2.55 for Nitrogen or Carbon, respectively), pulls more electron density from the phosphoryl moiety in 3-pHis (Fig. S21B–D), weakening its contacts with the surrounding environment. This accordingly leads to reduced binding affinity of 3-pHis to hSC44.20 in comparison to 3-pTza.
Apo Structures of hSC44 Variants.
The CDRs of the hSC44 variants are highly positively charged (24) and as a result, crystallization trials with hSC44 in the peptide-free state yielded structures with different small anionic groups bound in the paratope that were acquired during the purification process or from the crystallization buffers. These moieties include a citrate ion in hSC44, a phosphate ion or glutamic acid (from a symmetry-related Fab) in hSC44.20, and HEPES or a sulfate ion in hSC44.20.N32FL. Irrespective of the ligand bound to the paratope, the unliganded structures overlay on each other with RMSDs <1.0 Å on Cα carbons of the heavy-chain variable region (Fig. S22A). In addition, the apo forms of the antibodies overlay with 3-pTza and 3-pHis bound structures, with minimal deviations of their Cα carbons (Fig. S22B). As sulfate and phosphate ions share a tetrahedral geometry, their bonding interactions mimic that of the phosphoryl group from 3-pHis and 3-pTza peptides. Sulfate and phosphate ions make hydrogen bond interactions through the oxygen atoms with the GXGX residues (positions 96–99) in the CDR-H3 loop and Y96L or Y50H (Fig. S22C,D). The GXGX motif is also involved in forming hydrogen bonds with HEPES and citrate ions (Fig. 16E,F). Interestingly, the hSC44.20 GXGX motif in CDR-H3 is found to interact with E1H from the N-terminus of the adjacent Fab heavy chain during crystal packing (Fig. S23A). E1H also interacts with the GXGX motif in the CDR-H3 loop (Fig. S23B). Consequently, the CDR-H1 from the adjacent molecule comes into close contact with the CDR-L1 loop and is displaced from its usual position compared to other structures of hSC44 variants (Fig. S22A).
3-pHis Specific Antibodies Can Detect Biologically Relevant pHis modifications.
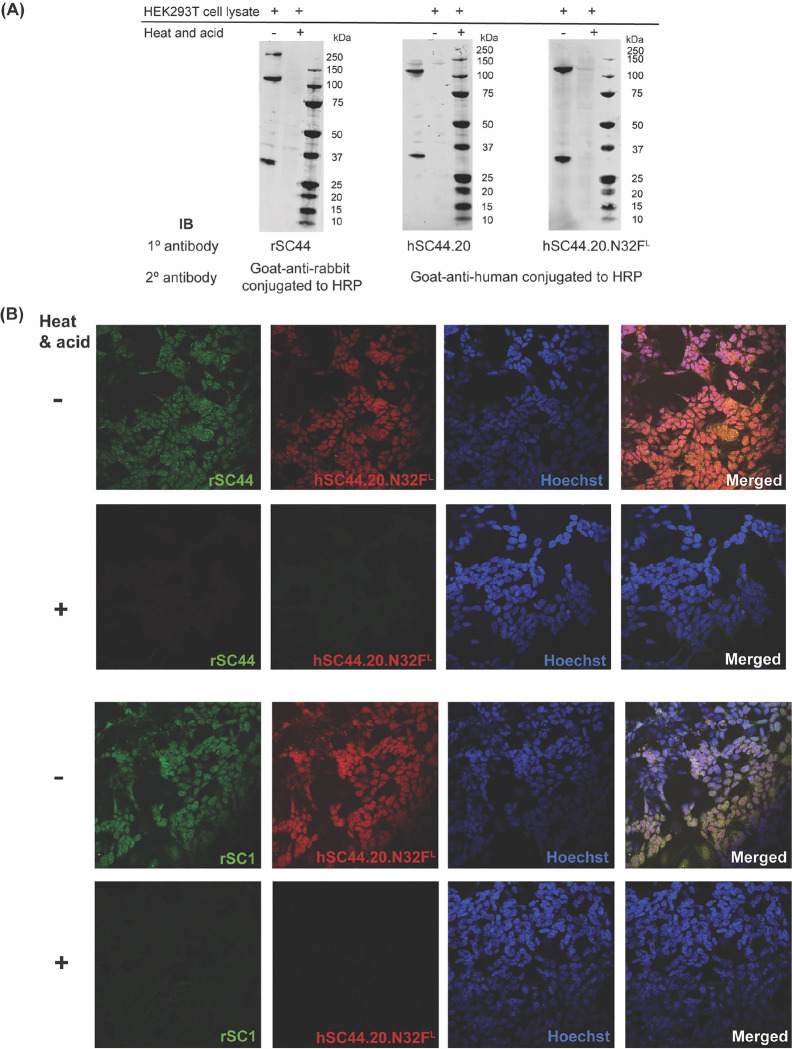
To evaluate the sensitivity and utility of the high-affinity, engineered 3-pHis specific antibodies for recognition of biologically relevant pHis proteins, we immunoblotted SDS PAGE-gel separated whole cell lysates from HEK293T cells with hSC44.20 or hSC44.20.N32FL IgGs. These immunoblots were compared with the banding pattern obtained with rSC44. All three antibodies generated similar banding patterns, indicating the presence of 3-pHis signals. This is further confirmed by loss of acid- and thermo-labile pHis signals upon incubation of lysates with acetic acid (pH = 5.5) at 95 °C prior to SDS-PAGE separation (Fig. 5A). As rSC44 and its humanized derivatives prefer a GpHAG motif, they predominantly recognize the 3-pHis forms of ACLY (120 kDa) and the α-subunit of SCS (30 kDa) proteins, as observed in the immunoblots with weaker protein signals in other molecular weight ranges (Fig. 5A). We then compared the performance of the hSC44.20.N32FL variant in immunofluorescence (IF) staining experiments by co-staining with either rSC44 or rSC1 (1-pTza/pHis specific) antibodies (Fig. 5B). hSC44.20.N32FL and rSC44 antibodies concomitantly recognized 3-pHis marks in the cytoplasm of SK-N-BE2 human neuroblastoma cells; as a control, acid-treatment of the slides with 90 °C PBS at pH 4.0 depleted the signals, confirming the specificity of these antibodies for 3-pHis. Notably, we observed 3-pHis localization mainly in the cytoplasm as well as the nucleus, consistent with our previous observations (14). Furthermore, when we co-stained with hSC44.20.N32FL and rSC1 in IF experiments (Fig. 5B), hSC44.20.N32FL successfully recognized 3-pHis residues in cells in the cytoplasm, whereas 1-pHis localization was observed primarily in the nucleus. Notably, these co-staining experiments take advantage of the Fc regions from different species (rabbit; rSC1 and human; hSC44.20.N32FL) allowing us to probe for 1-pHis and 3-pHis residues in cells simultaneously.
Fig. 5. Engineered hSC44 antibodies in typical molecular biology experiments.
(A) Immunoblot of SDS PAGE-gel separated HEK293T cell lysates probed with rSC44, hSC44.20 and hSC44.20.N32FL IgGs. HRP-conjugated secondary antibodies against rabbit and human IgG were used for detection. Heat and acid treated cell lysates were used as a negative control for pHis signals. (B) Immunofluorescence staining of SK-N-BE human neuroblastoma cells probed with rSC44, rSC1 and hSC44.20.N32FL IgGs. Alexa fluor conjugated goat anti-rabbit or goat anti-human antibody was used for immunofluorescence staining. Nuclei were stained with Hoechst and acquired images were merged. Heat and acid treated cells represent negative controls for pHis signals.
Discussion
The study of pHis in mammalian cells has been historically difficult due to a paucity of tools capable of sensitive detection and quantitative analysis of pHis proteins. The development and use of rabbit antibodies capable of selectively binding 1- and 3-pHis mimetics has enabled new approaches towards understanding the role of pHis in mammalian cell biology (23). Using advanced antibody engineering methods, we were able to solve many of the challenges affecting the development of pHis-specific antibodies from immunized animals. The acid labile and unstable nature of the phosphoramidate bond means that pHis modifications on peptides and proteins are short-lived in animals, and therefore not amenable for use as antigens for the generation of pHis specific antibodies by animal immunization. In these studies, we used prior sequence and structural information of anti-3-pTza rabbit monoclonal antibodies isolated via rabbit immunizations with a 3-pTza peptide to create a humanized version (hSC44) of rSC44 that was amenable to additional engineering. We then constructed phage displayed antibody libraries to engineer hSC44 for improved pHis recognition. Phage display was essential to our strategy, as it allowed us to: (i) control buffer formulations in a pH range that is favorable for pHis stability, (ii) establish a reliable method of 1- and 3-pHis peptide synthesis and isolation, and (iii) implement a carefully controlled selection procedure allowing us to isolate pHis-specific amino acid substitutions in the hSC44 scaffold using a tailored negative and positive selection scheme. Together, this approach coupled with rational engineering of the light chain of the selected variants generated humanized antibodies with favorable developability properties and unparalleled affinity and specificity for 3-pHis. More remarkably, none of the engineered antibodies bound to other phosphorylated amino acids under the conditions used, confirming the 3-pHis specificity of the engineered antibodies. Given the difficulty in generating other phospho-specific antibodies, our work represents a testament to the power of phage display to select antibodies with atomic-level precision.
To complement this approach, we sought to further understand the sets of amino acid substitutions in the hSC44 CDRs that governed high affinity and selectivity for 3-pHis. To do this, we undertook an extensive crystallization campaign and now report for the first time several structures of antibodies complexed with pHis peptides. To aid our structural studies, we used novel technologies that favored antibody crystallization by promoting interactions at interfaces away from the paratope (32–34). These substitutions are located in the elbow region of the heavy chain as well as the constant region of the kappa light chain and enabled us to obtain crystals that lead to 11 structures consisting of hSC44, hSC44.20 and hSC44.20.N32FL in the apo form as well as in complex with 3-pTza and 3-pHis peptides. This large dataset revealed the molecular determinants governing high affinity interactions with 3-pTza/3-pHis peptides in hSC44 variants. Additionally, QM/MM analysis offers a molecular-level insight into the effect of the mutation (N32FL), which leads to a newly established pi-pi interaction with pHis, resulting in improved binding affinity to pHis by hSC44.20.N32FL in comparison to hSC44.20.
The high affinity of isolated antibodies for 3-pHis was likely crucial for the successful crystallization of antibodies in complex with a 3-pHis phosphorylated peptide, possibly by preventing the spontaneous and rapid hydrolysis of the pHis moiety (24). The comparison between 3-pTza and 3-pHis peptide-bound hSC44 variants demonstrated that CDRs of these variants interact with the modifications in a similar way. However, the large affinity difference between the 3-pTza versus 3-pHis bound antibodies (>1000 fold) could not be explained by the crystal structures. Hence, we resorted to QM/MM analysis of the structures which revealed that the imidazole ring makes the phosphoryl moiety less electronegative compared to the triazolyl ring by drawing electrons from the phosphoryl moiety, thereby decreasing the number of interactions with the paratope. Another positive feature of hSC44 is its absolute isomer specificity, as we did not observe any interaction with 1-pTza or 1-pHis peptides using BLI. This property was further confirmed during crystallization, as the native peptides were chemically phosphorylated using a strategy that results in a mixture of unphosphorylated, 1-pHis, 3-pHis and 1,3-di-pHis peptides (see Materials and Methods for a detailed description). We subjected this mixture to co-crystallization with hSC44 variants without peptide purification, and in every case, we obtained crystals of antibodies in complex with 3-pHis peptides. We failed to obtain a purely apo structure of any of the hSC44 variants, as the positive electrostatic potential of its CDRs facilitated interactions with small electronegative anions from either the purification buffers, crystallization buffers, or neighboring Fabs. However, this is likely an artifact of crystallization and the high concentration of anions, as we did not observe binding to other negatively charged molecules (i.e., ssDNA, dsDNA) in our specificity ELISA (Fig. S8) nor to other phosphoryl-based PTMs such as pSer, pThr, pTyr by ELISA (Fig S5D) or the 1-pHis isoform (Tables 1–2), as measured by BLI. Overall, our structural analysis of these Fabs reveals that humanization and engineering did not alter the hallmark properties of the original rabbit rSC44–8 antibody. These antibodies recognize the phosphoryl moiety through a GXGX motif (‘nest’ motif) that was retained during engineering. They displayed properties of isomer specificity and structures also reveal that they retained the property of sequence dependence towards GpHAGA motif in the epitope. However, individual changes at the single residue level in hSC44.20.N32FL contribute to its higher affinity binding to 3-pHis.
There are several important advances made by the work presented here. First, we developed a workflow that allows the use of unstable pHis for in vitro engineering of anti-3-pHis antibodies. This in vitro selection protocol can be expanded to other non-canonical and unstable PTMs. There are 9 amino acids with modifiable side chains and six, like pHis, are understudied due to a lack of reagents for their analysis (23). We anticipate that the methods developed in this study can be adapted for isolating antibodies recognizing additional non-canonical PTMs. Second, the high affinity 3-pHis specific antibodies described here can be used to probe the pHis phosphoproteome in greater depth using immunoblotting, immunofluorescence staining, and phosphoproteomics. To complement these tools, we have generated an unprecedented amount of structural data on pHis recognition. These data, and particularly the N32F substitution, can be exploited to construct next-generation phage-displayed libraries of pHis specific Fabs and to rationally engineer sequence-specific pHis antibodies. In conclusion, our work demonstrates how in vitro evolution of PTM-specific antibodies can be used to generate a novel set of tools for investigating the biology of pHis.
Materials and Methods
IgG/Fab Protein Expression and Purification.
IgG/Fab protein was expressed as previously described (47) with some modifications. Briefly, plasmids designed to express the heavy and light chains were co-transfected into Expi293 cells (ThermoFisher Scientific; A14527) using the FectoPRO® transfection reagent kit (Polyplus) according to manufacturer’s instructions. After 5 days of expression, cell culture media was harvested and applied to CaptoL affinity column (Cytiva; 17547806). IgG/Fab protein was eluted using IgG elution buffer (Pierce™; 21009) and neutralized with 1 M Tris-HCl (pH = 8.0). Fractions containing IgG/Fab protein were combined, concentrated, and buffer exchanged into 1x HBS (50mM HEPES (pH 8.5), 150 mM NaCl) using Amicon Ultra-15 centrifugal units (50 kDa/10 kDa respectively, Millipore Sigma; UFC9010). IgG/Fab protein was characterized for purity by SDS-PAGE with concentrations being determined by spectrophotometry (280 nm).
For crystallization, Fabs were expressed as described, but were affinity purified from the cell culture media with CaptureSelect CH1-XL resin (Thermo-Fisher) followed by a wash with PBS and elution with 50 mM acetate, pH = 4.0, and neutralization with 1 M Tris-HCl (pH = 9.0).
Size Exclusion Chromatography.
IgG/Fab protein was purified by size exclusion chromatography (SEC) using an ÄKTA system (GE Healthcare) equipped with a Hi-Load 16/60 Superdex 75 gel filtration column (GE Healthcare Life Sciences). SEC columns were pre-equilibrated and run with 1x HBS with a flow rate of 1 mL/min. Fractions containing IgG/Fab protein were combined and concentrated using Amicon Ultra-15 centrifugal units (50 kDa/10 kDa, respectively, Millipore Sigma; UFC9010). IgG/Fab protein was characterized for purity by SDS-PAGE with concentrations being determined using spectrophotometry (280 nm). Fabs for crystallization were purified by SEC with running buffer consisting of 20 mM Tris and 150 mM NaCl.
Peptide Synthesis.
Phosphohistidine mimetic (1/3-pTza) peptides (Biotin-AminocaproicAcid-A.G.A.G.1-pTza.A.G.A.G; 1-pTza peptide and Biotin-AminocaproicAcid-A.G.A.G.3-pTza.A.G.A.G; 3-pTza peptide) and histidine containing peptide (Biotin-AminocaproicAcid-A.G.A.G.H.A.G.A.G; His-peptide) were synthesized as previously described (23).
Peptides ACLYana-3-pTza and ACLYana used in the crystallization experiments were synthesized by the Salk Peptide Synthesis Core. Both peptides have the same sequence Ala-Gly-Ala-Gly-X-Ala-Gly-Ala-Gly except for one residue. X is 3-pTza (3-phosphotriazolyl alanine) in ACLYana-3-pTza peptide and histidine in ACLYana peptide. These peptides have an acetyl group at the N-terminus and an amide at the C-terminus. They were synthesized using standard Fmoc chemistry on a Gyros Protein Technologies Tribute peptide synthesizer with UV monitoring. After deprotection, peptides were analyzed by mass spectrometry and analytical HPLC by the Proteomics Core at the Salk Research Institute. The peptides were purified by HPLC to 99% purity by 21st Century Biochemicals, Inc. The Fmoc-pTza molecules were synthesized at 10 g scale for incorporation into the peptides.
Preparation of rProtein A-IgG Matrix.
A 100 μL aliquot of rProtein A Sepharose® beads (Cytiva; 17–1279-01) was washed with 1 mL dH2O three times and equilibrated in 1 mL 1x HBS. 100 μg rSC1 (1-pTza/pHis) IgG (24) was added to one-half of equilibrated rProtein A beads and 100 μg of rSC44 (3-pTza/pHis) (24) IgG to the other half of equilibrated rProtein A beads, to make two separate resins. IgG binding to rProtein A beads was carried out at 4 °C for 1 h with end-over-end rotation. rProtein A-IgG resin mixture was then spun down at 1000 rpm for 1 min, supernatant was removed, and the beads were washed with 1 mL 1x HBS to remove unbound IgG. This was repeated three times. IgGs were then crosslinked to rProtein A resin by adding BS3 (Thermo Scientific™; 21580; resuspended in 20 mM HEPES, pH = 8.5) to a final concentration of 1 mM. The IgG-rProtein A-BS3 mixture was then incubated at room temperature for 1 h with end-over-end rotation. The reaction was quenched by adding an equal volume 1M Tris-HCl (pH 8.5) and incubated at room temperature for 10 mins with end-over-end rotation. The resin was spun down at 1000 rpm for 1 min, supernatant was removed, and the resin was washed with 1 mL 1x HBS to remove unbound BS3. This was repeated 3 times and resin was stored in 300 μL 1x HBS with 0.05% NaN3 at 4°C.
Phosphoramidate Reaction and Phosphohistidine Peptide Production.
The synthesis of potassium phosphoramidate and the phosphorylation of His side-chains using potassium phosphoramidate have been described previously (48). Briefly, 10 mg of potassium phosphoramidate was resuspended in 1 mL of 1x HBS (10 μg/μL). 100 μg of His-peptide (biotin-aminocaproic-acid.A.G.A.G.H.A.G.A.G) resuspended in 1x HBS was mixed with 1 mg phosphoramidate (100 μL, 1:10 ratio). Reaction was incubated overnight at room temperature for 16 h and subsequently quenched with an equal volume of 1 M Tris-HCl (pH 8.5).
Phosphohistidine Peptide Immunoprecipitation.
Preparation of rProtein A-IgG resins. 1-pHis and 3-pHis resins were washed with 1 mL 1x HBS three times to remove NaN3. Half of the His-peptide:potassium phosphoramidate reaction was added to anti-1-pHis mAb (SC1–1) resin and the other half to anti-3-pHis (SC44–8) mAb resin and incubated at room temperature for 1 h with end-over-end rotation. Resins were washed thrice with 500 μL 1x HBS by centrifugation at 1000 RPM. Bound pHis peptides were eluted by adding 500 μL 2 M Imidazole (re-suspended in 1x HBS, pH 8.5) to resin and incubating at room temperature with end-over-end rotation for 10 min. This was repeated once more for a total elution volume of 1 mL. Elution volume can change depending on desired use or concentration of peptide. Typical yields using this protocol were ~500 ng of phosphopeptide and can be scaled up as needed. Peptide concentrations were estimated using a quantitative colorimetric peptide assay kit (Pierce™; 23275) according to manufacturer’s instructions. We also developed a quantitative ELISA assay using 3-pTza peptides and r/hSC44 to generate standard curves for the estimation of pHis peptide concentrations.
Regeneration of rProtein A-IgG resins. Used resins were washed with 1 mL dH2O three times. To strip any 1/3-pHis peptide still bound to the resin, 1 mL 50 mM sodium acetate, pH = 4.0 was added and incubated at room temperature for 10 min with end-over-end rotation. This was repeated two times. Resins were then washed with 1 mL 1x HBS three times and resuspended in 300 μL 1x HBS with 0.05% NaN3 and stored at 4 °C.
ELISA.
A Nunc® MAXISORP™ 384-well ELISA plate (Millipore Sigma; P6366) was coated with 25 μL/well of 2 μg/μL streptavidin (New England Biolabs; N7021S) diluted in 1x HBS and incubated overnight at 4 °C. For experiments involving phage-ELISAs, anti-FLAG M2 mouse derived IgG (MilliporeSigma; F3165) was diluted 1/5000 in 1x HBS and 25 μL/well was dispensed to appropriate wells and incubated overnight at 4 °C. The following day, plates were washed four times with 1x HBS-T (1x HBS, 0.05% Tween 20) with 90 μL/well. Plates were then blocked with 75 μL/well 1x HBS, 0.5% BSA for 1 h at room temperature with shaking, followed by plate washing. Biotinylated peptides were added to the plate at various concentrations at 25 μL/well, incubated at room temperature for 30 mins with shaking, followed by plate washing. IgG was added at various concentrations at 25 μL/well and incubated at room temperature for 1 h with shaking, followed by plate washing. For the detection of rabbit IgG (rSC44), HRP conjugated goat anti-rabbit IgG (Thermo Scientific™/Invitrogen; A16110) diluted 1/5000 in 1x HBS-T, 0.05% BSA was added at 25 μL/well and incubated at room temperature for 30 mins with shaking, followed by plate washing. For humanized IgGs (hSC44 variants), HRP-conjugated goat anti-Human Kappa (SouthernBiotech; 2060–05) was used with identical conditions. For Fab-phage ELISA (hSC44 variants), HRP conjugated mouse anti-M13 (Sino Biological; 11973-MM05T-H) was used at a 1:12000 dilution. Binding was detected using 25 μL/well 3,3′,5,5′-tetramethylbenzidine (TMB) (Thermo Scientific™; 34021) chromogenic substrate and reactions were quenched using 25 μL/well 1 M phosphoric acid. Plate absorbance values (450nm) were read using a PowerWave XS microplate reader (BioTek).
For quantitative ELISA to estimate 3-pHis peptide concentration, a 2-fold serial dilution (12 dilutions total) of 1/3-pTza peptides starting at ~3.5 mM (~4 mg/mL) was added to streptavidin coated plates. A 10-fold serial dilution (6 dilutions total) of purified biotinylated 3-pHis peptide was also added to separate wells on streptavidin coated plates. Wells were probed with 100 nM rSC44 and ELISA was carried out as previously described. Standard curves corresponding to linear detection range of 3-pTza peptides were generated using Prism 6 (GraphPad Prism) and concentrations of 3-pHis with absorbance values falling within the linear range were solved. Concentrations of 3-pHis peptides were extrapolated back to undiluted sample and averaged. Three technical replicates were conducted for each measurement.
For EC50 measurements using hSC44 variant IgGs, 100 nM peptide was added to each streptavidin coated well. A 10-fold serial dilution (7 dilutions total) of IgG starting at 1 μM was added to the plate. Streptavidin and 1x HBS were used as negative controls and rSC44 was used as a positive control. ELISA was carried out as previously described and EC50 curves were calculated (Graphpad Prism). Experiments were performed with 4 technical replicates and 3 experimental replicates.
For phage ELISA using hSC44 variant Fab displayed on phage after selections, 1 μM peptide was added to each streptavidin coated wells. While streptavidin and 1x HBS were used as negative controls and rSC44 was used as a positive control. hSC44 phage pools were first normalized based on FLAG expression using anti-FLAG M2 ELISA. Briefly, a 2-fold serial dilution (7 dilutions total) of overnight phage pools was added to the wells of a plate coated with anti-FLAG M2 antibody (Sigma). hSC44 variant concentrations were then normalized based on FLAG display and used to probe peptide containing plates. Experiments were performed with 4 technical replicates and 3 experimental replicates.
For cross-reactive ELISAs, 25 μL/well of select macromolecules were added to wells at final concentrations of cardiolipin (50 μg/mL; C0563; Sigma), keyhole limpet hemocyanin (KLH; 5 μg/mL, H8283; Sigma), lipopolysaccharide (LPS; 10 μg/mL; tlrl-eblps; InvivoGen), single-stranded DNA (ssDNA; 1 μg/mL; D8899; Sigma), double-stranded DNA (dsDNA; 1 μg/mL; D4522; Sigma), insulin (1 μg/mL; 91077C; Millipore Sigma), BSA (1% (w/v)), streptavidin (10 μg/mL) and goat anti-human Fc (1 μg/mL; 109–005-098; Jackson). IgGs being tested for cross-reactivity were added at 100 nM in a volume of 25 μL/well. ELISAs were then carried out as previously described.
Construction of Phage-Displayed Libraries.
Phage-displayed library construction methods fusing Fab to phage coat protein III (pIII) of M13 bacteriophage have been previously described (49). Here we use some minor modifications for obtaining hSC44 phage-displayed libraries. Briefly, residues that were oriented towards, proximal to (within 10 Å) or directly contacting the 3-pTza peptide ligand in a crystal structure (PDB ID: 6X1V) were selected for mutagenesis for libraries hSC44 1–5 (Fig S2–3). Key framework residues required for CDR orientation were selected for mutagenesis for library hSC44 6. Appropriately designed “stop template” versions of each proposed library were synthesized by introducing stop codons into regions intended to be mutagenized. Mutagenic oligonucleotides targeting these positions while also repairing stop codons were synthesized (Integrated DNA Technologies) and used to construct libraries (Table S1). Mutagenic oligonucleotides were synthesized to adjust the nucleotide ratio of diversified positions to 70% of the wild-type nucleotide and 10% of each of the other nucleotides retaining the wild-type amino acid roughly 50% of the time (42). The libraries diversities ranged from 109-1010 unique variants.
Affinity Selections.
Libraries hSC44 1–6 were pooled and cycled through five rounds of affinity selections on 96-well Maxisorp Immunoplates (Nunc™ Thermo Scientific™; 12–565-135) as previously described (25), with some modifications. Selections were carried out as outlined in Fig 2A. 3-pHis peptides used in selections were generated fresh each day (see Phosphoramidate Reaction and Phosphohistidine Peptide Production section of Materials and Methods). Plates were coated with 100 μL/well of reagent indicated in Table S2 overnight at 4 °C and blocked the following day with 1x HBS containing 0.5% BSA. Phage pools were cycled through negative and positive selections by incubation at room temperature for 1 hour with shaking for each condition. Washing steps after positive selection used 1x HBS-T, 0.05% Tween-20 with 2–5 min incubations with shaking at each washing step. Phage were eluted from positive selection using 100 μL/well 100 mM HCl with incubation for 10 min with shaking at room temperature, eluted phage pools were neutralized with 1 M Tris-HCl (pH 11) and used to infect OmniMax™ (Invitrogen™; C854003) Escherichia coli (E. coli) cells at OD = 0.6–0.8 for overnight amplification of phage pools.
Biolayer Interferometry.
Binding kinetics and estimation of affinity (KD) measurements were carried out using an Octet HTX instrument (FortèBio) as previously described (50) with some minor modifications. Biotinylated 1/3-pTza/pHis peptides and non-phosphorylated His-peptides were captured on streptavidin biosensors at 20 nM. Assay buffer used was 1x HBS-T, 1% BSA. IgGs were used at various concentrations (20–200 nM).
Data Analysis.
ELISA data were analyzed using Prism 6 software (GraphPad Software Inc). Structural alignments were processed and visualized in PyMol (v.2.5.2). Library design and DNA sequencing was analyzed using Geneious v.10.0.09 (https://www.geneious.com). Biolayer interferometry data were analyzed using Octet Systems Software v.9.0 (FortèBio).
Crystallization and Structure Solution.
ACLYana-pHis peptides were prepared by incubating 50 mM of unphosphorylated ACLYana peptide with 250 mM of phosphoramidate in HBS for 12 h at room temperature. The resulting reaction has mixed species of unphosphorylated peptide, phosphorylated peptide with 1-pHis modification and 3-pHis modification. The whole reaction was used for co-crystallization. Fab versions of hSC44, hSC44.20 and hSC44.20.N32FL antibodies described in Table S4–5 were pre-incubated with either ACLYana-3-pTza or ACLYana-pHis peptides at a molar ratio of 1:5 (Fab:peptide). hSC44 Fab variants (hSC44, hSC44.20 and hSC44.20.N32FL) complexed with ACLYana-3-pTza peptides as well as unliganded hSC44 proteins were screened for crystallization using our Rigaku CrystalMation system at Scripps Research, with JCSG Screens 1–4 at 4 °C and 20 °C. The crystallization conditions used are reported in Table S5. Data were collected at either APS beamline 23-ID-D, SSRL beamline 12–2 or ALS beamline 5.0.3 at 100 K. The diffraction images were indexed, integrated, and scaled using either HKL-2000 (51) or XDS (52). The rSC44 Fab (PDB ID: 6X1V) (24) was used as search model for molecular replacement with PHASER-MR (53) to obtain initial phase information. Refinement was carried out by alternating rounds of Phenix.refine (54) and model building software Coot (55). The final models were validated using MolProbity (56) and figures were prepared using PyMOL (Schrodinger LLC) and UCSF Chimera (57). Buried molecular surfaces were analyzed with the MS program (58) using a 1.7 Å probe radius. The pi-pi interaction distances and angles were measured using either BINANA 2.0 or PLIP (59). The What If server was used to measure torsion angles (60). The x-ray data collection, refinement statistics and the model validation values are reported in Table S4.
Quantum Mechanics/Molecular Mechanics (QM/MM).
The experimentally resolved structures (hSC44.20:ACLYana-3-pTza, hSC44.20:ACLYana-3-pHis and hSC44.20.N32FL:ACLYana-3-pHis) underwent QM/MM minimizations to achieve geometry-optimized structures of the active site at the ab initio DFT level, allowing assessment of the quantitative response of the ligand-bound structures to mutations. The MM region was modelled using the OPLS force field [3], while the active site was treated with the LACVP* basis sets, combining the successful 6–31G basis set with the LANL2DZ effective core basis set (61, 62) as implemented in the Schrodinger Qsite tool (63–65). QM/MM calculations were performed in a non-periodic manner, with atoms more than 30 Å from the ligand (antibody: pHis/pTza) excluded, and those between 20 and 30 Å fixed. Population and Orbital analyses for the QM region were conducted using the Jaguar tool (66) as implemented in Schrodinger 2023–2. Additionally, QM analysis was also performed on the antibodies (pHis and pTza) using the Jaguar tool (66), as implemented in Schrodinger-2023. The peptides are locally aligned to analyze the shift in the antibody near the Fab region caused by the N32F mutations using UCSF Chimera (57). For graphics preparation and visualization, we used UCSF Chimera and Maestro-2023 from the Schrodinger suite tool.
Molecular Dynamics and Binding Affinity (FEP) Simulation Details.
For molecular dynamics-based contact analysis and FEP simulations, the starting model was the experimentally resolved structures (hSC44.20:ACLYana-3-pTza, hSC44.20:ACLYana-3-pHis and hSC44.20.N32FL:ACLYana-3-pHis).
Contact Analysis.
MD-based contact analysis was performed on three experimentally resolved structures (hSC44.20:ACLYana-3-pTza, hSC44.20:ACLYana-3-pHis and hSC44.20.N32FL:ACLYana-3-pHis) using the Maestro-Desmond Interoperability Tools (67) in Schrodinger. Each system was solvated with ~40,000 TIP3P water molecules, depending on the antibody and mutation. Water molecules directly involved in metal coordination were retained from the original starting structure. All three systems were prepared using the Protein Preparation Wizard in Schrodinger. Minimization was conducted using a steepest descent algorithm for 100,000 steps, followed by a 20 ns equilibration with restrained heavy atoms (backbone heavy atoms of the protein and nucleic acids with an isotropic force of 1000 kJ mol−1nm−1) under constant number, pressure, and temperature (NPT) and constant number, volume, and temperature (NVT) conditions up to 1 ns at 300 K using the standard MD procedure in the Desmond tool of Schrodinger 2023.
Criteria of Contact Analysis:
The contacts presented here are chosen based on these criteria.
H-bonds:
The current geometric criteria for protein-ligand H-bond are: (1) distance of 2.5 Å between the donor and acceptor atoms. (2) (D—H···A); A donor angle of 120°–180° between the donor-hydrogen-acceptor atoms (D—H···A). (3) An acceptor angle of 120°–180° between the hydrogen-acceptor-bonded atom atoms (H···A—X).
Water Bridge:
Hydrogen-bonded protein-ligand interactions mediated by a water molecule. The current geometric criteria for a protein-water or water-ligand H-bond are: (1) A distance of 2.8 Å between the donor and acceptor atoms (D—H···A); a donor angle of 120°–180° between the donor-hydrogen-acceptor atoms (D—H···A); (2) An acceptor angle of 120°–180° between the hydrogen-acceptor-bonded atom atoms (H···A—X).
Hydrophobic Contacts:
The current geometric criteria for hydrophobic interactions are as follows: (1) π-Cation — Aromatic and charged groups within 4.5 Å. (2) π-π — Two aromatic groups stacked face-to-face or face-to-edge within 3.5–5 Å.
Binding Affinity (FEP) Simulations.
The FEP+ program (68, 69) from the Schrödinger suite 2023–2 was used to determine the relative binding free energies of pHis between the wild-type and mutant N32F, evaluating the effect of the N32F mutation on pHis binding. The wild-type and mutant complexes were prepared using the Protein Preparation Wizard. For each wild-type complex and its corresponding mutant, the FEP Protein Mutation for Ligand Selectivity GUI was used to build a perturbation map. The OPLS4 force field modelled proteins and ligands (70) and torsion parameters for all ligand fragments were checked using a Force Field Builder. A 10 Å cubic box filled with approximately ~ 40,000 TIP3P water molecules (71) was used for the complex and solvent perturbation leg (wild-type and mutant systems). The number of alchemical λ windows was set to 12 by default to connect the wild-type and mutant states. For each λ window, production MD was run for 20 ns in the NPT ensemble. The Bennett acceptance ratio method (72) (BAR) was used to calculate the free energy differences. Mutational free energy calculations were performed in triplicate, initializing the MD with different random seeds. The reported relative binding free energies were calculated as the average of the three ΔΔG values from independent simulations.
Immunoblotting.
The protocol used here was described previously (73). To maintain pHis signals, cell lysates were processed at 4 °C and alkaline buffer conditions were used throughout the experiment. HEK293T cells were grown in DMEM medium in a 10-cm dish for 3 days before processing. Cells were washed twice with ice-cold 1x PBS buffer, detached from the dish surface using a cell scraper and lysed with 400 μL of lysis buffer (25 mM Tris pH 8.5, 140 mM NaCl, 0.1 % Tween-20, 1 mM PMSF, 1x protease inhibitor tablet, and 1x PhosStop tablet. Cells were lysed by sonicating for 1 min using a 10 sec on and 10 sec off cycle at 40% amplitude. Lysates were clarified by centrifugation at 12,000 rpm at 4 °C and 30 μg of supernatant was loaded onto an alkaline 12 % SDS-PAGE gel followed by alkaline immunoblotting as previously described (Methods Kalagiri Ref). Electrophoresis and immunoblotting were carried out 4 °C. Lysates treated with acid (pH 5.5) and heat (95 °C for 15 min) were used as negative controls. Membranes were blocked with blocking buffer (5 mM Tris pH 8.5, 30 mM NaCl, 0.1 % Tween-20, and 0.1 % casein) and then incubated with 0.5 μg/mL of primary antibodies (rSC44, hSC44.20, or hSC44.20.N32FL) diluted in blocking buffer for 1 h at room temperature. Membranes were washed three times with wash buffer (25 mM Tris pH 8.5, 140 mM NaCl, 0.1 % Tween-20) with 10 min incubation times. The blot was then incubated for 1 h at room temperature with 0.5 μg/mL of goat anti-human kappa antibody conjugated to HRP (31460, SouthernBiotech) if hSC44.20, or hSC44.20.N32FL were used as primary antibodies and goat anti-rabbit antibody conjugated to HRP (31460, Thermo Fisher) if rSC44 was used as a primary antibody. Membranes were washed with wash buffer three times and developed using luminol reagent which is a substrate for HRP. The resultant chemiluminescence was detected using BioRad ChemiDoc MP Touch Imaging System.
Immunofluorescence.
SKN-BE(2) cells were fixed with 4% paraformaldehyde (pH 8.5) for 15 min at room temperature and washed three times with 4 °C PBS (pH 8.5) (control slides and coverslips were washed three times with 90 °C PBS at pH 4.0). Then, the cells were permeabilized with 0.2% Triton X-100 for 15 min at room temperature and washed three times with 4 °C PBS (pH 8.5) (control slides and coverslips were washed with 90 °C PBS at pH 4.0). Cells were blocked in 10% BSA in TBS-T (pH = 8.5) at room temperature for 30 min, before incubating with primary antibody diluted in 1% BSA in TBS-T (pH 8.5) at 4 °C overnight. The dilutions were used as follows: 2 μg/mL for rSC44–8 and rSC1–1 and 8 μg/mL for hSC44.20.N32FL. The day after, cells were washed 3 times for 5 minutes with PBS at room temperature. Cells were then incubated with secondary antibodies diluted in 1% BSA in TBS-T (pH 8.5) for one hour as follows: 1/200 for secondary goat anti-rabbit IgG antibody Alexa Fluor® 488 (A-11034, Invitrogen, Carlsbad, CA, USA), 1/20 for secondary mouse anti-human Ig light chain K antibody Alexa Fluor® 647 (316514, BioLegend®, San Diego, CA, USA) and mixed with Hoescht 33342 (H3570, Invitrogen, Carlsbad, CA, USA). Finally, the cells were washed 3 times with PBS (pH 8.5) at room temperature and then mounted with Prolong Gold Antifade (P36930, Invitrogen, Carlsbad, CA, USA). Cell imaging was performed with an SP8 confocal microscope (Leica) using a 63×/NA 1.4 Plan Neofluor objective lens. Each channel was imaged sequentially using the multitrack recording module before merging in order to prevent contamination between fluorochromes.
Supplementary Material
Fig. S1. Kabat numbering of rSC44 and hSC44 antibodies with associated protein expression and phage-display data. (A-B) IMGT nomenclature for (A) the VL domain and (B) the VH domain. (C-D) Kabat nomenclature for (C) the VL domain and (D) the VH domain. CDR positions and important framework positions necessary for humanization are colored as in Fig. 1. Positions in hSC44 that are the same as in rSC44 are left blank and differences are shown with the single-letter amino acid code. (E) Non-reducing SDS-PAGE of hSC44 and Trastuzumab Fab and IgG protein. (F) Phage-ELISA was used to confirm hSC44 Fab (left) and scFv (right) displayed on phage as well as binding to 3-pTza. Phage were purified and concentrated to 1×1012 cfu/mL and serially diluted. FLAG indicates display levels of hSC44 Fab and scFv.
Fig. S2. Schematic of the workflow for generation and enrichment of pHis containing peptides. A degenerate histidine containing peptide (Biotin-AGAGHAGAG) was reacted with potassium phosphoramidate. The reaction yielded products: non-phosphorylated, 1-pHis, 3-pHis and 1/3 di-pHis. The reaction was quenched and custom rSC1 and rSC44 antibody resins were used to preferentially enrich 1-pHis or 3-pHis containing peptides, respectively. See Materials and Methods for a more detailed description of the protocol.
Fig. S3. Structure guided design of phage-displayed hSC44 libraries. The structure of rSC44 in complex with a 3-pTza peptide (PDB ID: 6X1V) was used to rationalize randomization strategies. Residue side chains are shown as sticks and colored as in Fig 1. Residues that were proximal (within 10 Å) to the 3-pTza moiety were selected for randomization (A-E) Different residues in CDRs L1, L3, H1, H2, and H3 were randomized to maximize diversity and likelihood of identifying new combinations of specificity determining residues contacting 3-pHis. (F) Vernier zone and additional framework residues that were necessary for humanization of rSC44 were selected for randomization.
Fig. S4. Residues selected for randomization in hSC44 libraries. The amino acid sequences of rSC44 and hSC44 are shown using Kabat nomenclature. CDRs and Vernier Zone residues are highlighted and colored as in Fig. 1. Residues that were selected for randomization in each unique library are shown in bold underneath the sequence of hSC44. (A) Sequence of VL domain. (B) Sequence of VH domain.
Fig. S5. Characterization of hSC44 variants isolated from selections. (A) Variants of hSC44 were reformatted as IgG1 and expressed in Expi293 cells. The total protein yields were determined for each antibody with trastuzumab (black bar) and hSC44 (white bar) used as benchmarks. Variants with higher or lower yields than hSC44 are depicted with red or grey bars, respectively. (B) Non-reducing SDS-PAGE of purified hSC44 variant IgGs. Each variant IgG was loaded at 2.5 μg/well. (C) Lead hSC44 variants positively selected against 3-pTza or 3-pHis were assayed for binding to a 3-pHis peptide using ELISA. Data shown are an average of 4 replicates +/− SEM and displayed as fold change over hSC44 with hSC44 normalized to 1. (E) Lead hSC44 variants tested for binding to 3-pTza. 3-pHis, pTyr, pSer, and pSer peptides (biotin-Aminocaproic Acid-AGAGXAGAG; where “X” is 3-pTza, 3-pHis, pTyr, pSer or pThr) using ELISA. Each hSC44 variant IgG was used at 50 nM with varying concentrations of indicated peptides.
Fig. S6. BLI traces of hSC44 variants assayed for binding to 3-pTza peptides.
Fig. S7. BLI traces of hSC44 variants assayed for binding to 3-pHis peptides.
Fig. S8. Specificity ELISA for hSC44 variants. hSC44 variant IgGs were tested for off-target binding against known antigenic molecules typically associated with poor antibody developability. See Materials and Methods for an in-depth description of the methodology.
Fig. S9. Effect of S1C substitutions on crystal packing. Replacement of the FG β-strand (198–204 Kabat numbering) of the light chain of the humanized antibody with residues from rabbit antibody (198–202 Kabat numbering) was used to enhance crystallization. The FG β-strand of the light chain formed hydrogen bond interactions with the constant domain of the heavy chain (206–210 Kabat numbering) from an adjacent protomer. These interactions propagated across protomers and mediated effective crystal packing.
Fig. S10. Effect of S1CE substitutions in the heavy chain elbow region on the elbow angle. Replacing SSASTKG with FNQIKG residues in the elbow region of the heavy chain shifted the elbow angles of Fabs to >160° (red) while Fabs without elbow mutations (green) stayed below this angle (Table S3). hSC44 Fabs are overlaid on their Cα carbons of the heavy chain variable region.
Fig. S11. Structural analysis of Vernier zone residue in hSC44. (A) Rabbit rSC44 (salmon red) and hSC44 (green) are overlaid on each other with a Cα RMSD of 0.75 Å2. (B) Vernier residue R46L interacts with N99H and P100H residues on CDR-H3 loop which in turn interacts with phosphoryl group of 3-pTza. Note that Vernier zone residues are shown in green text. (C) F34H is oriented towards the core of the antibody and forms hydrophobic interactions with residues from several framework regions of the heavy chain and K94H from CDR-H3 that interacts with phosphoryl group. (D) Two patches of substitutions in the heavy chain framework region enhanced the structure and function of the hSC44 Fab. H47H and Y50H at the interface of heavy and light chains interact with Y96L, which is essential for phosphoryl recognition. The 61SWAK64 further stabilized the 47HIGY50 region in CDR-H2 and the heavy framework region 3.
Fig. S12. Structural overlay of hSC44 variants. Fabs are overlaid based on the Cα carbons of their heavy chain variable region. Superposed hSC44 (green), hSC44.20 (orange) and hSC44.20.N32FL (blue) have Cα RMSDs less than <1 Å in complex with (A) 3-pTza (magenta) or (B) 3-pHis (red) peptides.
Fig. S13. Contact analysis between WT and N32F hSC44.20 and 3-pHis. (A) The time-dependent variation in the total number of contacts for 3-pHis with (Orange) WT and (Blue) mutant (N32F) hSC44.20 in their respective environments. The black dashed line represents the average number of contacts between 3-pHis and the Fab region of the antibody over time. (B) Analysis of the types of contacts and their interaction fractions for (left) hSC44.20 with 3-pHis and (right) hSC44.20.N32FL with 3-pHis. The interactions presented at figure B are those that are found >50% of the time during the 20 ns simulations. The π-π interaction involving the mutation (N32FL) with 3-pHis are outlined with a black dotted line in Figure 1B (bottom). This stable interaction is present ~75% of the time during the 20 ns simulation, but it is not found in the structure of WT hSC44.20 with 3-pHis (left). In the Y-axis, “H” refers to the heavy-chain and “L” refers to the light-chain. Superscripts “A” and “B” refer to different chains in the protein structure.
Fig. S14. (A) Effect of N32F mutation on 3-pHis. (B) Electrostatic potential of 3-pHis and Phe32 showing the T-shaped π-π interactions. The color bar shows the change in charges across electron density. The black dashed line shows the T-shaped π-π interactions in each plot.
Fig. S15. Thermodynamic cycles employed for MD-based FEP simulations were utilized to compute the changes in binding free energy resulting from residue mutations in the antibody binding process. We examined the changes in the binding free energy caused by the N32F mutation in the binding of 3-pHis by using FEP (free energy perturbation) to calculate the relative binding free energy for 3-pHis. In the thermodynamic cycle, the relative binding free energy can be obtained by the two vertical legs, instead of simulating the physical binding process.
Fig. S16. Sequence specificity assays of rSC44–8 and hSC44 variants. Thermal shift assay of rSC44–8 (A), hSC44.20 (B) and hSC44.20.N32FL (C) with 3-pTza peptides. Biolayer interferometry assay of rSC44–8 (D), hSC44.20 (E) and hSC44.20.N32FL (F) variants with pHis peptides. Biotinylated peptides from pHis substrate proteins were phosphorylated by phosphoramidate and immobilized on the streptavidin biosensor tip.
Fig. S17. Differences in CDR loop orientations of hSC44 variants complexed with 3-pTza and 3-pHis peptides. Fabs are overlaid on the basis of the Cα carbons of their heavy chain variable regions. The 3-pTza (magenta) and 3-pHis (red) peptides bound to (A) hSC44 CDRs (green), (B) hSC44.20 CDRs (orange), and (C) hSC44.20.N32FL CDRs (blue).
Fig. S18. hSC44.20.N32FL in complex with 3-pTza and 3-pHis peptides. (A-B) Omit maps (FO-FC map) of (A) hSC44.20.N32FL Fab (blue) in complex with 3-pTza peptide (magenta) and (B) hSC44.20.N32L (blue) in complex with 3-pHis peptide (red). The bound peptide is contoured at 1.0 sigma and the CDR loops are represented as cartoon. The sequence of the peptide used for crystallization is shown below and the residues in red font have interpretable electron density. (C-D) Residues from CDRs H1, H2, H3, L1 and L3 of hSC44.20.N32FL are making direct and water-mediated hydrogen bond interactions with the phosphoryl group of the 3-pTza peptide (C) or the 3-pHis peptide (D). A coordinated water molecule (grey sphere) is conserved in both complexes. Residues depicted in red font are substitutions that are different from hSC44. (E-F) hSC44.20.N32FL interactions with the peptide backbone of either (E) 3-pTza peptide or (F) 3-pHis peptide.
Fig. S19. Differences in triazolyl and imidazole ring interactions in hSC44.20.N32FL. (A) Structures of 3-pTza and 3-pHis molecules. The nitrogen atoms that undergo phosphorylation in 3-pHis and the corresponding atoms in the triazolyl ring are marked in red. (B) hSC44.20.N32FL makes direct and water-mediated (grey sphere) interactions with nitrogen 2 and nitrogen 3 atoms of the triazolyl moiety of the peptide through residues in CDRs H1, H2, H3, L1, and L3. (C) hSC44.20.N32FL makes direct and water-mediated interactions with the imidazole moiety of 3-pHis through residues in CDRs H1, H2, L1, and L3. Interactions from CDR-H3 are absent due to the presence of a carbon atom in the imidazole ring position corresponding to position of nitrogen 3 in the triazolyl ring.
Fig. S20. Time-dependent variation in the total number of contacts for 3-pHis (Orange) and 3-pTza (Green) with their surroundings (criteria detailed in the computational methods section). The black dashed line represents the average number of contacts over time.
Fig. S21. QM/MM analysis of 3-pHis and 3-pTza. (A) Key atomic level structural differences near PO3− between (left) 3-pHis (Backbone carbon atoms indicated with green) and (right) 3-pTza (Backbone carbon atoms indicated with white) antigens. (B) Electrostatic potential surface of (left) 3-pHis and (right) 3-pTza, showing Mulliken charges [18] on the O atom of PO3− from QM analysis (charges are in atomic units (au); the color bar indicates charge variations across the electron density. (C) Highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) (iso-surface value 0.05 au) and corresponding energies of (left) 3-pHis and (right) 3-pTza from natural bond orbital (NBO) population analysis using QM; a lower HOMO-LUMO gap indicates that 3-pTza is more chemically active towards binding than 3-pHis. (D) Electrostatic potential surface of (left) 3-pHis and (right) 3-pTza with their surrounding environment from QM/MM analysis; the color bar shows charge variations across the electron density. The antigen with N-terminal histidine phosphorylation (N-PO3−, 3-pHis) and C-terminal phosphohistidine analog (C-PO3−, 3-pTza), facilitating binding to the Fab region, is shown with black dotted circles throughout the figure.
Fig. S22. Unliganded structures of hSC44 variants. (A) Differences in CDR loop orientation across unliganded structures of hSC44 (green), hSC44.20 (orange) and hSC44.20.N32FL (blue) Fabs. (B) Comparison of CDR loop orientation in hSC44.20.N32FL (blue) in complex with 3-pTza peptide (magenta) or 3-pHis peptide (red) or in unliganded state. (C-F) Hydrogen bond interactions of (C) sulfate ion, (D) phosphate ion, (E) HEPES, and (F) citrate ion bound to hSC44.20.N32FL, hSC44.20, hSC44.20.N32FL, or hSC44 Fabs, respectively.
Fig. S23. Structure of unliganded hSC44.20 Fab. (A) hSC44.20 Fab (orange) makes crystal contacts with an adjacent protomer (gray). The CDR-L2 loop and the epitope-binding region are marked with red and grey circles respectively. (B) The CDRs of unliganded hSC44.20 Fab bound the sidechain of residue E1H of an adjacent protomer.
Significance Statement.
Histidine phosphorylation is an elusive PTM whose role in mammalian cell biology is largely unknown due to the lack of robust tools and methods for its analysis. Here we report the development of antibodies with unprecedented affinity and specificity towards 3-pHis and present the first crystal structures of a pHis peptide in complex with an antibody. Finally, we show how these antibodies can be used in standard molecular biology workflows to investigate pHis-dependent biology.
Acknowledgments
GM/CA@APS has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). This research used resources of the Advanced Photon Source; a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. The Berkeley Center for Structural Biology is supported in part by the Howard Hughes Medical Institute. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231. The Pilatus detector on 5.0.1. was funded under NIH grant S10OD021832. The ALS-ENABLE beamlines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169. R.M.L was supported by NIH R35 GM132090 and U01 AI136680. D.L was supported by the Hearst Foundations grant U01 AI136680. A.B. would like to acknowledge support from the Eric and Wendy Schmidt AI in Science Postdoctoral Fellowship, a program of Schmidt Sciences. The Salk Peptide Synthesis and Proteomics Cores are supported by an NCI-designated Cancer Center Support Grant P30 CA014195. T.H. is a Frank and Else Schilling American Cancer Society Professor and holds the Renato Dulbecco Chair in Cancer Research. T.H. is supported by an NIH National Cancer Institute R35 (5 R35 CA242443) award, R.K. was supported by a Salk Innovation Award and Salk Alumni Fellowship, and M.S. was supported by a Hewitt Foundation Fellowship. The structure work was supported in part by the Skaggs Institute of Chemical Biology (I.A.W).
Footnotes
Competing Interest Statement: Authors declare no competing interests.
Data deposition
The X-ray coordinates and structure factors of the pHis Fab:peptide complexes and pHis Fabs have been deposited in the Protein Data Bank with accession codes- 8UJI (hSC44.S1C:AGAG-3pTza-AGAG); 8UIT (hSC44.S1C:AGAG-3pHis-AGAG); 8UIO (hSC44.S1C); 8UIH (hSC44.S1C.20:AGAG-3pTza-AGAG); 8UIG (hSC44.S1C.20:AGAG-3pHis-AGAG); 8UHT (hSC44.S1C.20); 8UHS (hSC44.S1C.20); 8UHP (hSC44.S1C.20.N32FL:AGAG-3pTza-AGAG); 8UHN (hSC44.S1C.20.N32FL:AGAG-3pHis-AGAG); 8UHJ (hSC44.S1C.20.N32FL); and 8UHH (hSC44.S1C.20.N32FL).
References
- 1.Aebersold R., et al. , How many human proteoforms are there? Nat Chem Biol 14, 206–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoury G. A., Baliban R. C., Floudas C. A., Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci Rep 1, 1–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L., et al. , The human phosphotyrosine signaling network: Evolution and hotspots of hijacking in cancer. Genome Res 22, 1222–1230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Page B. D. G., et al. , Small molecule STAT5-SH2 domain inhibitors exhibit potent antileukemia activity. J Med Chem 55, 1047–1055 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Imran M., et al. , Innovations and patent trends in the development of USFDA approved protein Kinase inhibitors in the last two decades. Pharmaceuticals 14, 1–59 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attwood M. M., Fabbro D., Sokolov A. V., Knapp S., Schiöth H. B., Trends in kinase drug discovery: targets, indications and inhibitor design. Nat Rev Drug Discov 20, 839–861 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Ke M., et al. , Identification, quantification, and site localization of protein posttranslational modifications via mass spectrometry-based proteomics. Adv Exp Med Biol 919, 345–382 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Adam K., Hunter T., Histidine kinases and the missing phosphoproteome from prokaryotes to eukaryotes. Laboratory Investigation 98, 233–247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer P. D., Ebner K. E., Hultquist D. E., Peter J. B., Identification of phospohistidine in digests from a probable intermediate of oxidative phosphorylation. J Biol Chem 237, PC3306–PC3308 (1962). [PubMed] [Google Scholar]
- 10.Hultquist D. E., Moyer R. W., Boyer P. D., The preparation and characterization of 1-phosphohistidine and 3-phosphohistidine. Biochemistry 5, 322–331 (1966). [DOI] [PubMed] [Google Scholar]
- 11.Capra E. J., Laub M. T., Evolution of two-component signal transduction systems. Annu Rev Microbiol 66, 325–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. R. Fuhs, T. Hunter, pHisphorylation: the emergence of histidine phosphorylation as a reversible regulatory modification. Curr Opin Cell Biol 45, 8–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panda S., et al. , Identification of PGAM5 as a mammalian protein histidine phosphatase that plays a central role to negatively regulate CD4+ T cells. Mol Cell 63, 457–469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oslund R. C., et al. , A phosphohistidine proteomics strategy based on elucidation of a unique gas-phase phosphopeptide fragmentation mechanism. J Am Chem Soc 136, 12899–12911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardman G., et al. , Strong anion exchange-mediated phosphoproteomics reveals extensive human non-canonical phosphorylation. EMBO J 38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilley M., Mambwe B., Thompson M. J., Jackson R. F. W., Muimo R., 4-Phosphopyrazol-2-yl alanine: a non-hydrolysable analogue of phosphohistidine. Chemical Communications 51, 7305–7308 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Kee J.-M., Villani B., Carpenter L. R., Muir T. W., Development of Stable Phosphohistidine Analogues. J Am Chem Soc 132, 14327–14329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kee J.-M., Oslund R. C., Perlman D. H., Muir T. W., A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol 9, 416–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schenkels C., Erni B., Reymond J.-L., Phosphofurylalanine, a stable analog of phosphohistidine. Bioorg Med Chem Lett 9, 1443–1446 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Makwana M. V., Muimo R., Jackson R. F. W., Advances in development of new tools for the study of phosphohistidine. Laboratory Investigation 98, 291–303 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Makwana M. V., et al. , Chemical Tools for Studying Phosphohistidine: Generation of Selective τ-Phosphohistidine and π-Phosphohistidine Antibodies**. ChemBioChem 24 (2023). [DOI] [PubMed] [Google Scholar]
- 22.Kee J.-M., Oslund R. C., Couvillon A. D., Muir T. W., A Second-Generation Phosphohistidine Analog for Production of Phosphohistidine Antibodies. Org Lett 17, 187–189 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Fuhs S. R., et al. , Monoclonal 1- and 3-phosphohistidine antibodies: New tools to study histidine phosphorylation. Cell 162, 198–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalagiri R., et al. , Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies. Proceedings of the National Academy of Sciences 118, e2010644118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava S., et al. , Histidine phosphorylation relieves copper inhibition in the mammalian potassium channel KCa3.1. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindupur S. K., et al. , The protein histidine phosphatase LHPP is a tumour suppressor. Nature 555, 678–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Affandi T., McEvoy M. M., Mechanism of metal ion-induced activation of a two-component sensor kinase. Biochemical Journal 476, 115–135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulte J. E., Roggiani M., Shi H., Zhu J., Goulian M., The phosphohistidine phosphatase SixA dephosphorylates the phosphocarrier NPr. Journal of Biological Chemistry 296, 100090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai Y., Hanafusa H., Hisamoto N., Matsumoto K., Histidine dephosphorylation of the Gβ protein GBP-1 promotes axon regeneration in C. elegans. EMBO Rep 23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foote J., Winter G., Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol 224, 487–499 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Jones P. T., Dear P. H., Foote J., Neuberger M. S., Winter G., Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321, 522–525 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Bruce H. A., et al. , Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes. Protein Science 33 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey L. J., et al. , Locking the elbow: Improved antibody Fab fragments as chaperones for structure determination. J Mol Biol 430, 337–347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieu R., et al. , Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing. PLoS One [Preprint] (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mou Y., et al. , Engineering improved antiphosphotyrosine antibodies based on an immunoconvergent binding motif. J Am Chem Soc 140, 16615–16624 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Makwana M. V., Williamson M. P., Jackson R. F. W., Muimo R., Quantitation of phosphohistidine in proteins in a mammalian cell line by 31P NMR. PLoS One 17, e0273797 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fellouse F. A., et al. , High-throughput Generation of Synthetic Antibodies from Highly Functional Minimalist Phage-displayed Libraries. J Mol Biol 373, 924–940 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Rader C., et al. , The rabbit antibody repertoire as a novel source for the generation of therapeutic human antibodies. Journal of Biological Chemistry 275, 13668–13676 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Liang W. C., et al. , Dramatic activation of an antibody by a single amino acid change in framework. Sci Rep 11, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabat E. A., Wu T. T., Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol 147, 1709–19 (1991). [PubMed] [Google Scholar]
- 41.Ehrenmann F., Kaas Q., Lefranc M. P., IMGT/3Dstructure-DB and IMGT/DomainGapAlign: A database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MHcSF. Nucleic Acids Res 38, D301–D307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidhu S. S., Lowman H. B., Cunningham B. C., Wells J. A., “Phage display for selection of novel binding peptides” in Methods in Enzymology, (2000), pp. 333–363. [DOI] [PubMed] [Google Scholar]
- 43.Sidhu S. S., et al. , Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol 338, 299–310 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Avery L. B., et al. , Establishing in vitro in vivo correlations to screen monoclonal antibodies for physiochemical properties related to favorable human pharmacokinetics. MAbs 10, 244–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinoshita K., Sadanami K., Kidera A., Go N., Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Eng 12, 11–14 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Pauling Linus, The Nature of the Chemical Bond - An Introduction to Modern Structural Chemistry, 3rd Ed. (Cornell University Press, 1960). [Google Scholar]
- 47.Kelil A., Gallo E., Banerjee S., Adams J. J., Sidhu S. S., CellectSeq: In silico discovery of antibodies targeting integral membrane proteins combining in situ selections and next-generation sequencing. Commun Biol 4, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei Y.-F., Mathews H. R., “Identification of Phosphohistidine in Proteins and Purification of Protein-Histidine Kinases” in Methods in Enzymology, (1991), pp. 388–414. [DOI] [PubMed] [Google Scholar]
- 49.Chen G., Sidhu S. S., “Design and generation of synthetic antibody libraries for phage display” in Methods in Molecular Biology, (2014), pp. 113–131. [DOI] [PubMed] [Google Scholar]
- 50.Miersch S., et al. , Tetravalent SARS-CoV-2 neutralizing antibodies show enhanced potency and resistance to escape mutations. J Mol Biol 433, 167177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z., Minor W., “Processing of X-ray diffraction data collected in oscillation mode” in Methods in Enzymology, (1997), pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 52.Kabsch W., XDS. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy A. J., et al. , Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr D60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr D66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Connolly M. L., Analytical molecular surface calculation. J Appl Crystallogr 16, 548–558 (1983). [Google Scholar]
- 59.Adasme M. F., et al. , PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res 49, W530–W534 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vriend G., WHAT IF: A molecular modeling and drug design program. J Mol Graph 8, 52–56 (1990). [DOI] [PubMed] [Google Scholar]
- 61.Dolg M., Stoll H., Preuss H., A combination of quasirelativistic pseudopotential and ligand field calculations for lanthanoid compounds. Theor Chim Acta 85, 441–450 (1993). [Google Scholar]
- 62.Dolg M., Stoll H., Savin A., Preuss H., Energy-adjusted pseudopotentials for the rare earth elements. Theor Chim Acta 75, 173–194 (1989). [Google Scholar]
- 63.Philipp D. M., Friesner R. A., Mixedab initio QM/MM modeling using frozen orbitals and tests with alanine dipeptide and tetrapeptide. J Comput Chem 20, 1468–1494 (1999). [Google Scholar]
- 64.Tian L., Friesner R. A., QM/MM Simulation on P450 BM3 Enzyme Catalysis Mechanism. J Chem Theory Comput 5, 1421–1431 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murphy R. B., Philipp D. M., Friesner R. A., A mixed quantum mechanics/molecular mechanics (QM/MM) method for large-scale modeling of chemistry in protein environments. J Comput Chem 21, 1442–1457 (2000). [Google Scholar]
- 66.Bochevarov A. D., et al. , Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int J Quantum Chem 113, 2110–2142 (2013). [Google Scholar]
- 67.Bowers K. J., Proceedings of the 2006 ACM/IEEE conference on Supercomputing (SC06) in (Association for Computing Machinery, 2006), pp. 11–17. [Google Scholar]
- 68.Chen W., et al. , Enhancing Hit Discovery in Virtual Screening through Absolute Protein–Ligand Binding Free-Energy Calculations. J Chem Inf Model 63, 3171–3185 (2023). [DOI] [PubMed] [Google Scholar]
- 69.Ross G. A., et al. , The maximal and current accuracy of rigorous protein-ligand binding free energy calculations. Commun Chem 6, 222 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu C., et al. , OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J Chem Theory Comput 17, 4291–4300 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926–935 (1983). [Google Scholar]
- 72.Bennett C. H., Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys 22, 245–268 (1976). [Google Scholar]
- 73.Kalagiri R., Adam K., Hunter T., “Empirical evidence of cellular histidine phosphorylation by immunoblotting using pHis mAbs” in Histidine Phosphorylation: Methods and Protocols , Eyers C. E., Ed. (Springer US, 2020), pp. 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Kabat numbering of rSC44 and hSC44 antibodies with associated protein expression and phage-display data. (A-B) IMGT nomenclature for (A) the VL domain and (B) the VH domain. (C-D) Kabat nomenclature for (C) the VL domain and (D) the VH domain. CDR positions and important framework positions necessary for humanization are colored as in Fig. 1. Positions in hSC44 that are the same as in rSC44 are left blank and differences are shown with the single-letter amino acid code. (E) Non-reducing SDS-PAGE of hSC44 and Trastuzumab Fab and IgG protein. (F) Phage-ELISA was used to confirm hSC44 Fab (left) and scFv (right) displayed on phage as well as binding to 3-pTza. Phage were purified and concentrated to 1×1012 cfu/mL and serially diluted. FLAG indicates display levels of hSC44 Fab and scFv.
Fig. S2. Schematic of the workflow for generation and enrichment of pHis containing peptides. A degenerate histidine containing peptide (Biotin-AGAGHAGAG) was reacted with potassium phosphoramidate. The reaction yielded products: non-phosphorylated, 1-pHis, 3-pHis and 1/3 di-pHis. The reaction was quenched and custom rSC1 and rSC44 antibody resins were used to preferentially enrich 1-pHis or 3-pHis containing peptides, respectively. See Materials and Methods for a more detailed description of the protocol.
Fig. S3. Structure guided design of phage-displayed hSC44 libraries. The structure of rSC44 in complex with a 3-pTza peptide (PDB ID: 6X1V) was used to rationalize randomization strategies. Residue side chains are shown as sticks and colored as in Fig 1. Residues that were proximal (within 10 Å) to the 3-pTza moiety were selected for randomization (A-E) Different residues in CDRs L1, L3, H1, H2, and H3 were randomized to maximize diversity and likelihood of identifying new combinations of specificity determining residues contacting 3-pHis. (F) Vernier zone and additional framework residues that were necessary for humanization of rSC44 were selected for randomization.
Fig. S4. Residues selected for randomization in hSC44 libraries. The amino acid sequences of rSC44 and hSC44 are shown using Kabat nomenclature. CDRs and Vernier Zone residues are highlighted and colored as in Fig. 1. Residues that were selected for randomization in each unique library are shown in bold underneath the sequence of hSC44. (A) Sequence of VL domain. (B) Sequence of VH domain.
Fig. S5. Characterization of hSC44 variants isolated from selections. (A) Variants of hSC44 were reformatted as IgG1 and expressed in Expi293 cells. The total protein yields were determined for each antibody with trastuzumab (black bar) and hSC44 (white bar) used as benchmarks. Variants with higher or lower yields than hSC44 are depicted with red or grey bars, respectively. (B) Non-reducing SDS-PAGE of purified hSC44 variant IgGs. Each variant IgG was loaded at 2.5 μg/well. (C) Lead hSC44 variants positively selected against 3-pTza or 3-pHis were assayed for binding to a 3-pHis peptide using ELISA. Data shown are an average of 4 replicates +/− SEM and displayed as fold change over hSC44 with hSC44 normalized to 1. (E) Lead hSC44 variants tested for binding to 3-pTza. 3-pHis, pTyr, pSer, and pSer peptides (biotin-Aminocaproic Acid-AGAGXAGAG; where “X” is 3-pTza, 3-pHis, pTyr, pSer or pThr) using ELISA. Each hSC44 variant IgG was used at 50 nM with varying concentrations of indicated peptides.
Fig. S6. BLI traces of hSC44 variants assayed for binding to 3-pTza peptides.
Fig. S7. BLI traces of hSC44 variants assayed for binding to 3-pHis peptides.
Fig. S8. Specificity ELISA for hSC44 variants. hSC44 variant IgGs were tested for off-target binding against known antigenic molecules typically associated with poor antibody developability. See Materials and Methods for an in-depth description of the methodology.
Fig. S9. Effect of S1C substitutions on crystal packing. Replacement of the FG β-strand (198–204 Kabat numbering) of the light chain of the humanized antibody with residues from rabbit antibody (198–202 Kabat numbering) was used to enhance crystallization. The FG β-strand of the light chain formed hydrogen bond interactions with the constant domain of the heavy chain (206–210 Kabat numbering) from an adjacent protomer. These interactions propagated across protomers and mediated effective crystal packing.
Fig. S10. Effect of S1CE substitutions in the heavy chain elbow region on the elbow angle. Replacing SSASTKG with FNQIKG residues in the elbow region of the heavy chain shifted the elbow angles of Fabs to >160° (red) while Fabs without elbow mutations (green) stayed below this angle (Table S3). hSC44 Fabs are overlaid on their Cα carbons of the heavy chain variable region.
Fig. S11. Structural analysis of Vernier zone residue in hSC44. (A) Rabbit rSC44 (salmon red) and hSC44 (green) are overlaid on each other with a Cα RMSD of 0.75 Å2. (B) Vernier residue R46L interacts with N99H and P100H residues on CDR-H3 loop which in turn interacts with phosphoryl group of 3-pTza. Note that Vernier zone residues are shown in green text. (C) F34H is oriented towards the core of the antibody and forms hydrophobic interactions with residues from several framework regions of the heavy chain and K94H from CDR-H3 that interacts with phosphoryl group. (D) Two patches of substitutions in the heavy chain framework region enhanced the structure and function of the hSC44 Fab. H47H and Y50H at the interface of heavy and light chains interact with Y96L, which is essential for phosphoryl recognition. The 61SWAK64 further stabilized the 47HIGY50 region in CDR-H2 and the heavy framework region 3.
Fig. S12. Structural overlay of hSC44 variants. Fabs are overlaid based on the Cα carbons of their heavy chain variable region. Superposed hSC44 (green), hSC44.20 (orange) and hSC44.20.N32FL (blue) have Cα RMSDs less than <1 Å in complex with (A) 3-pTza (magenta) or (B) 3-pHis (red) peptides.
Fig. S13. Contact analysis between WT and N32F hSC44.20 and 3-pHis. (A) The time-dependent variation in the total number of contacts for 3-pHis with (Orange) WT and (Blue) mutant (N32F) hSC44.20 in their respective environments. The black dashed line represents the average number of contacts between 3-pHis and the Fab region of the antibody over time. (B) Analysis of the types of contacts and their interaction fractions for (left) hSC44.20 with 3-pHis and (right) hSC44.20.N32FL with 3-pHis. The interactions presented at figure B are those that are found >50% of the time during the 20 ns simulations. The π-π interaction involving the mutation (N32FL) with 3-pHis are outlined with a black dotted line in Figure 1B (bottom). This stable interaction is present ~75% of the time during the 20 ns simulation, but it is not found in the structure of WT hSC44.20 with 3-pHis (left). In the Y-axis, “H” refers to the heavy-chain and “L” refers to the light-chain. Superscripts “A” and “B” refer to different chains in the protein structure.
Fig. S14. (A) Effect of N32F mutation on 3-pHis. (B) Electrostatic potential of 3-pHis and Phe32 showing the T-shaped π-π interactions. The color bar shows the change in charges across electron density. The black dashed line shows the T-shaped π-π interactions in each plot.
Fig. S15. Thermodynamic cycles employed for MD-based FEP simulations were utilized to compute the changes in binding free energy resulting from residue mutations in the antibody binding process. We examined the changes in the binding free energy caused by the N32F mutation in the binding of 3-pHis by using FEP (free energy perturbation) to calculate the relative binding free energy for 3-pHis. In the thermodynamic cycle, the relative binding free energy can be obtained by the two vertical legs, instead of simulating the physical binding process.
Fig. S16. Sequence specificity assays of rSC44–8 and hSC44 variants. Thermal shift assay of rSC44–8 (A), hSC44.20 (B) and hSC44.20.N32FL (C) with 3-pTza peptides. Biolayer interferometry assay of rSC44–8 (D), hSC44.20 (E) and hSC44.20.N32FL (F) variants with pHis peptides. Biotinylated peptides from pHis substrate proteins were phosphorylated by phosphoramidate and immobilized on the streptavidin biosensor tip.
Fig. S17. Differences in CDR loop orientations of hSC44 variants complexed with 3-pTza and 3-pHis peptides. Fabs are overlaid on the basis of the Cα carbons of their heavy chain variable regions. The 3-pTza (magenta) and 3-pHis (red) peptides bound to (A) hSC44 CDRs (green), (B) hSC44.20 CDRs (orange), and (C) hSC44.20.N32FL CDRs (blue).
Fig. S18. hSC44.20.N32FL in complex with 3-pTza and 3-pHis peptides. (A-B) Omit maps (FO-FC map) of (A) hSC44.20.N32FL Fab (blue) in complex with 3-pTza peptide (magenta) and (B) hSC44.20.N32L (blue) in complex with 3-pHis peptide (red). The bound peptide is contoured at 1.0 sigma and the CDR loops are represented as cartoon. The sequence of the peptide used for crystallization is shown below and the residues in red font have interpretable electron density. (C-D) Residues from CDRs H1, H2, H3, L1 and L3 of hSC44.20.N32FL are making direct and water-mediated hydrogen bond interactions with the phosphoryl group of the 3-pTza peptide (C) or the 3-pHis peptide (D). A coordinated water molecule (grey sphere) is conserved in both complexes. Residues depicted in red font are substitutions that are different from hSC44. (E-F) hSC44.20.N32FL interactions with the peptide backbone of either (E) 3-pTza peptide or (F) 3-pHis peptide.
Fig. S19. Differences in triazolyl and imidazole ring interactions in hSC44.20.N32FL. (A) Structures of 3-pTza and 3-pHis molecules. The nitrogen atoms that undergo phosphorylation in 3-pHis and the corresponding atoms in the triazolyl ring are marked in red. (B) hSC44.20.N32FL makes direct and water-mediated (grey sphere) interactions with nitrogen 2 and nitrogen 3 atoms of the triazolyl moiety of the peptide through residues in CDRs H1, H2, H3, L1, and L3. (C) hSC44.20.N32FL makes direct and water-mediated interactions with the imidazole moiety of 3-pHis through residues in CDRs H1, H2, L1, and L3. Interactions from CDR-H3 are absent due to the presence of a carbon atom in the imidazole ring position corresponding to position of nitrogen 3 in the triazolyl ring.
Fig. S20. Time-dependent variation in the total number of contacts for 3-pHis (Orange) and 3-pTza (Green) with their surroundings (criteria detailed in the computational methods section). The black dashed line represents the average number of contacts over time.
Fig. S21. QM/MM analysis of 3-pHis and 3-pTza. (A) Key atomic level structural differences near PO3− between (left) 3-pHis (Backbone carbon atoms indicated with green) and (right) 3-pTza (Backbone carbon atoms indicated with white) antigens. (B) Electrostatic potential surface of (left) 3-pHis and (right) 3-pTza, showing Mulliken charges [18] on the O atom of PO3− from QM analysis (charges are in atomic units (au); the color bar indicates charge variations across the electron density. (C) Highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) (iso-surface value 0.05 au) and corresponding energies of (left) 3-pHis and (right) 3-pTza from natural bond orbital (NBO) population analysis using QM; a lower HOMO-LUMO gap indicates that 3-pTza is more chemically active towards binding than 3-pHis. (D) Electrostatic potential surface of (left) 3-pHis and (right) 3-pTza with their surrounding environment from QM/MM analysis; the color bar shows charge variations across the electron density. The antigen with N-terminal histidine phosphorylation (N-PO3−, 3-pHis) and C-terminal phosphohistidine analog (C-PO3−, 3-pTza), facilitating binding to the Fab region, is shown with black dotted circles throughout the figure.
Fig. S22. Unliganded structures of hSC44 variants. (A) Differences in CDR loop orientation across unliganded structures of hSC44 (green), hSC44.20 (orange) and hSC44.20.N32FL (blue) Fabs. (B) Comparison of CDR loop orientation in hSC44.20.N32FL (blue) in complex with 3-pTza peptide (magenta) or 3-pHis peptide (red) or in unliganded state. (C-F) Hydrogen bond interactions of (C) sulfate ion, (D) phosphate ion, (E) HEPES, and (F) citrate ion bound to hSC44.20.N32FL, hSC44.20, hSC44.20.N32FL, or hSC44 Fabs, respectively.
Fig. S23. Structure of unliganded hSC44.20 Fab. (A) hSC44.20 Fab (orange) makes crystal contacts with an adjacent protomer (gray). The CDR-L2 loop and the epitope-binding region are marked with red and grey circles respectively. (B) The CDRs of unliganded hSC44.20 Fab bound the sidechain of residue E1H of an adjacent protomer.