Abstract
Talin is a force-sensing multidomain protein and a major player in cellular mechanotransduction. Here, we use single-molecule magnetic tweezers to investigate the mechanical response of the R8 rod domain of talin. We find that under various force cycles, the R8 domain of talin can display a memory-dependent behavior: At the same low force (<10 pN), the same protein molecule shows vastly different unfolding kinetics. This history-dependent behavior indicates the evolution of a unique force-induced native state. We measure through mechanical unfolding that talin R8 domain binds one of its ligands, DLC1, with much higher affinity than previously reported. This strong interaction can explain the antitumor response of DLC1 by regulating inside-out activation of integrins. Together, our results paint a complex picture for the mechanical unfolding of talin in the physiological range and a new mechanism of function of DLC1 to regulate inside-out activation of integrins.
DLC1 locks talin R8 in a mechanically stable state, potentially preventing the inside-out activation of integrins.
INTRODUCTION
Since folding precedes the assembly of proteins into tethered structures, proteins operating under force in vivo may acquire new native structures during repeating unfolding-refolding cycles. It is currently well understood that mechanical forces alter the shape of the folding energy landscape of a protein (1, 2). However, this change in shape has not been associated with any change in function beyond flipping protein domains between folded and unfolded states. Structural and binding assays have been instrumental in determining binding partners for protein domains, but these experiments describe the natively folded domains that have never experienced mechanical unfolding-refolding cycles, contrary to their behavior in vivo. The advent of single-molecule techniques such as atomic force microscopy and optical or magnetic tweezers has opened the path to measure the nanomechanical heterogeneity of proteins under a force vector (3). Because of limitations related to instrumental drift and tethering stability, single proteins are typically tethered for short periods of time, typically up to several minutes, which does not allow the investigation of long-term force-induced effects on their energy landscape. The implementation of mechanically stable tethers has recently enabled magnetic tweezers to cross the day-long and even week-long measuring barriers for a single molecule and allows for the measurement of proteins over many unfolding-refolding cycles (4). Using these advancements, here we investigate the unfolding response of talin, the mechanical computer of cellular mechanotransduction, and report a unique force-induced behavior that triggers a change in the mechanical response of one of its rod domains through repeated force cycles.
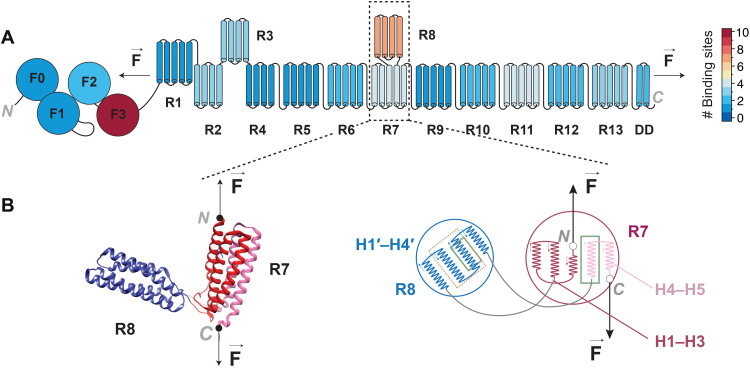
In vivo, talin is a force-sensing multidomain protein, which has evolved to operate under force and regulates the connections between the cellular cytoskeleton and its extracellular matrix (ECM) (5, 6). Talin is tethered between actin filaments and ECM-bound transmembrane integrins and acts as a mechanosensor by interacting with various ligands in a force-dependent manner. Its multidomain architecture allows it to respond to developing force vectors during mechanotransduction in a quantized fashion (Fig. 1A). Of the several ligands that bind to talin, vinculin can attach to mechanically unfolded rod (R) domains to consolidate focal adhesions (7–9). Many other ligands can bind to the same domains when they are in their folded state: Actin filaments and β-integrin provide the anchoring points for talin, while other ligands help localize talin at the plasma membrane or have other regulatory roles (5). Among the rod domains of talin, R8 has the highest number of known binding partners (Fig. 1A and table S1) and a unique molecular architecture (10): Talin R8 domain is inextricably linked to its R7 neighbor, which flanks it at both of its termini (Fig. 1B). This arrangement protects talin R8 domain from a direct force vector, as long as its flanking R7 domain remains folded, turning R7 into a molecular gatekeeper (Fig. 1). This unique geometry might explain the high number of binding ligands that this R8 domain has, since even when talin operates under force, R8 can be shielded from mechanical perturbations.
Fig. 1. Structure schematics and reactivity heatmap of talin.
(A) Schematics diagram of interaction heatmap of full-length talin based on the number of binding sites for each domain. (B) Left: Structure showing the α helices of talin domains R7R8, with R8 domain inserted inside R7 (Protein Data Bank: 4w8p). The direction of force applied to R7R8 from the N and C termini of R7 is shown with arrows. Right: Schematics of R7R8 showing R8 domain in between the third and fourth helices of R7 (H3 and H4). The helix numbers of each fragment of R7 and R8 are given. The helices of R8 inside the dotted gold rectangle show the binding site for DLC1, and the helices shown inside the solid green rectangle of R7 and R8 represent the vinculin binding sites.
Here, we take advantage of various protein constructs specifically engineered for single-molecule experiments to produce stable tethering and use magnetic tweezers to investigate the mechanical unfolding response of the R7R8 region, as well as R8 domain alone. Our approach samples the mechanical unfolding response of talin constructs at physiological forces (typically <10 pN) and for extended times of up to several hours. We find that R7R8 shows a broad range of previously unknown unfolding states, given by the partial or tandem denaturation of its component structures. When sampled in isolation, R8 domain displayed a rather puzzling behavior: not only that it is more mechanically weak than the other talin rod domains but also that it showed time-dependent unfolding-refolding transitions in the 4.5- to 9-pN range. This time-dependent behavior is indicative of an evolving energy landscape, where molecular memory can shift the binding affinity toward various ligands involved in mechanotransduction. We measure a stronger-than-expected binding of R8 to one of its ligands, deleted in liver cancer 1 (DLC1), in stark contrast to bulk measured binding of previously never unfolded states. On the basis of these findings, we hypothesize that some talin domains operate on a previously unknown biological mechanism, where mechanical memory triggered through unfolding-refolding of protein domains can produce novel binding interfaces, helping in the fine-tuning of focal adhesions.
RESULTS
Mechanical clamp obtained during unfolding of talin rod domains
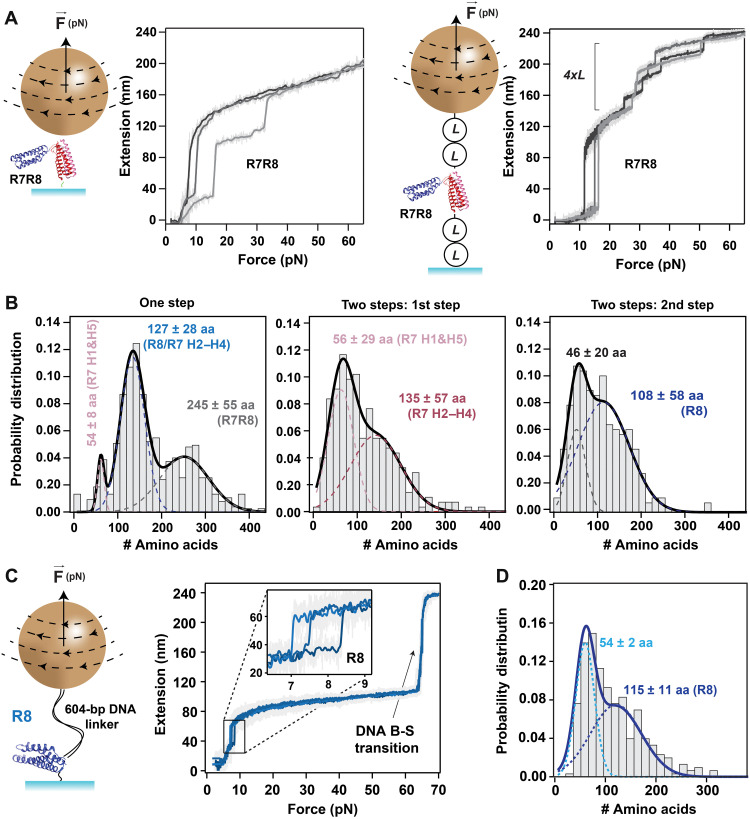
To study the unfolding behavior of talin R7R8 and R8 rod domains, we first engineered two simple constructs with a HaloTag at the N terminus and AviTag at the C terminus (schematics of the R7R8 construct is shown in Fig. 2A, left). These proteins were attached to the bottom glass surface of the fluid chamber using a covalently imbedded chloroalkane ligand (4) and pulled via streptavidin-coated paramagnetic beads (Fig. 2A, left). Unexpectedly, a variety of unfolding steps was seen, pointing to a much more complex unfolding behavior than anticipated. This behavior was later confirmed with protein constructs that have a more complex unfolding fingerprint. In this case, we sandwiched the R7R8 fragment in between two protein L domains on each side and replaced the C terminus with a SpyTag, following a procedure that we recently developed (Fig. 2A, right) (11). For these experiments, the surface was coated with the complementary SpyCatcher molecules, and the paramagnetic beads with the chloroalkane ligand that binds HaloTag. Unfolding traces obtained from this latter construct also showed the unfolding fingerprint originating from protein L, in the form of four equal steps of ~15 nm (Fig. 2A, right). As the results obtained from both constructs were similar, we combined them into a single histogram (Fig. 2B). Our data show that R7R8 can unfold either as a single step, or as two steps, and, in some rare occasions, as three steps. The number of amino acids comprising the mechanical clamp was then calculated from the measured extension and force using a worm-like chain model with the characteristic persistence length for polypeptides of 0.58 nm (12) and a size per amino acid of 0.4 nm (13). For traces with one unfolding step, three peaks were seen with sizes of 54 ± 8 amino acids, 127 ± 28 amino acids, and 245 ± 55 amino acids (Fig. 2B, left). These peaks can be assigned to a part of R7 (H1 and H5), to R8/R7 (H2 to H4), and R7R8, respectively (see also Fig. 1B and table S2). Similarly, the histogram of the number of amino acids obtained from the two-step traces showed a bimodal distribution (Fig. 2B, center and right). From these fits, the number of amino acids was determined as follows: for the first step, 56 ± 29 amino acids for the first peak and 135 ± 57 amino acids for the second peak, and for the second step, 46 ± 20 amino acids and 108 ± 58 amino acids from the first and second peaks, respectively. An in-depth molecular dynamics simulation study of the response of the talin R7R8 fragment to force was reported by Hytonen, del Rio Hernandez, and collaborators (14). These authors found that a stable three-helix intermediate can form between the second and fourth α helices of R7 (H2 to H4), which encompasses a total of 118 amino acids (see fig. S1). In the histogram made from traces with a single step, the most prevalent peak corresponds to this value, within our measuring error. However, R8 unfolding is also expected to produce a step corresponding to 118 amino acids. This coincidence might explain why the step that can be assigned to complete unfolding of the R7R8 domain does not have the highest probability. For the histogram made from traces that showed two steps, the first step had a peak that can be assigned to the unfolding of the first and fifth helices (H1 and H5), while the second peak corresponds to the three-helix structure of R7. The second step had an intermediate at around 46 amino acids, which can be assigned to the unfolding of H4′ or H1′&H4′ from R8, followed by full-length unfolding of R8. In the case of talin R8, we also covalently cross-linked a 604–base pair (bp) double-stranded DNA fragment, terminated with a biotin group (Fig. 2C). The advantage of having a DNA fragment in series with our protein is that the trace also displays a well-characterized overstretching transition from the B to a stretched (S) state (B-S transition) at exactly 65 pN, constituting both a molecular fingerprint and force reporter (4). Similar to the histogram in Fig. 2B (right), the unfolding histogram of R8 showed a bimodal distribution with the second peak at 115 ± 11 amino acids, which is at the expected length when all four α helices denature (Fig. 2D). The first peak of 54 ± 2 amino acids is equivalent to the unfolding of H1′ and H4′ of the four helices.
Fig. 2. Mechanical unfolding of talin domains R7R8 and R8.
(A) Left: Schematics of R7R8 construct attached between a paramagnetic bead and the glass surface and representative traces of extension versus pulling force showing the unfolding of R7R8 as single step or two steps at a pulling rate of 15.5 pN/s. Right: Schematics of the same protein construct R7R8 flanked by two protein L on each side (L2-R7R8-L2) attached between a paramagnetic bead and the glass surface and the corresponding traces of unfolding. Unfolding traces of extension versus pulling force from construct L2-R7R8-L2 show the unfolding of R7R8 followed by the steps of unfolding of four protein L at a pulling rate of 0.33 pN/s. (B) Histograms showing the probability distribution of number of amino acids: (left) from R7R8 when both domains were unfolded as a single step (N = 746) and (middle) from first step and (right) from second step, when R7R8 displayed two unfolding steps (N = 274). aa, amino acids. (C) Left: Schematics of protein construct R8 attached in series to a 604-bp DNA linker, tethered in between the glass surface and paramagnetic bead. Right: Representative traces of extension versus pulling force at 0.33 pN/s show the unfolding of R8 and the B-S overstretching transition of DNA at 65 pN. The inset shows the single-step unfolding of R8 domain. (D) Histogram showing the probability distribution of the number of amino acids unfolded from R8 construct (N = 301). The dotted lines on histogram represent the individual fits for Gaussian distribution, and the continuous lines represent their sum. The bin size of the histograms is 15. Errors are SDs.
Talin R8 domain shows time- and force-dependent unfolding-refolding transitions in the physiological range
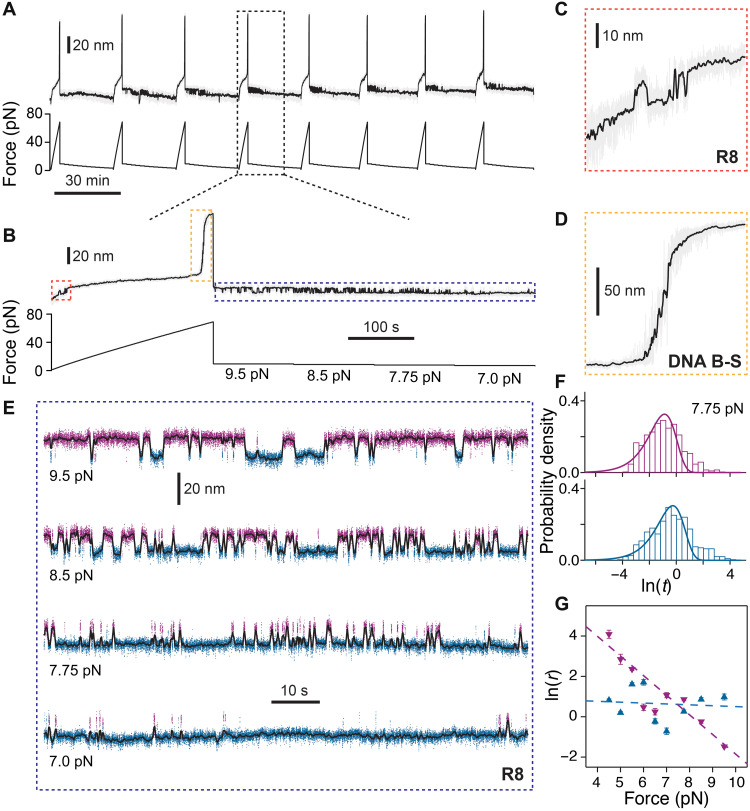
Magnetic tweezers allows for sampling of the mechanical response of the same protein molecule over several minutes and over repeating force-changing protocols. For example, we exposed molecules made from talin R8 domain in series with a covalently linked DNA fragment to repeating cycles (Fig. 3A). These cycles consisted of a linear increase in the applied force up to ~75 pN, followed by quenching the force in small increments from 9.5 to 4 pN in intervals of 2 min (Fig. 3B). The first part of the pulse shows the unfolding of R8 domain (Fig. 3C), followed by the overstretching DNA B-S transition at 65 pN (Fig. 3D). The third part of the pulse takes place at low force, where R8 domain can unfold and refold in a force-dependent manner (Fig. 3E). The times spent in the unfolded or folded states at a given force (tu/f) were then used to calculate their transition rates, ru/f = 1/τu/f, where τu/f is the mean transition time. In this regard, we made histograms of the natural logarithm of the measured dwell times and fitted the rates using exp[x − x0 − exp (x − x0)], where x = ln [t] and x0 = ln [τ] (15) (Fig. 3F and fig. S2). The folding transition had a regular dependency with force: the higher the applied force, the lower the refolding rate. The measured distance to transition state of 3.99 nm is similar to that reported for the R3 domain of talin (16). However, the unfolding rates in this low force range did not show a clear dependency on force. These trends indicate that while unfolding is dominated by entropy and breaking of hydrogen bonds, the refolding depends on the entropic collapse of the unfolded chain (17). Furthermore, the unfolding transitions seemed to appear in a sequence of pulses, with many fast folded-unfolding transitions followed by slow ones (see, for example, the 8.5-pN trace in Fig. 3E). This behavior is not captured by the histograms of the dwell times, as the two rates are too close to be resolved (Fig. 3F). The lack of force dependency of unfolding rate at low force and the inhomogeneous unfolding dwell times are indicative of a more complex energy landscape, where several native states might coexist.
Fig. 3. Unfolding and refolding kinetics of talin R8 domain as a function of force.
(A) Representative trace of unfolding and refolding of R8-DNA. A repeating force protocol is used: First, a force ramp is applied at a pulling force of 0.33 pN/s to obtain the unfolding of R8 and the B-S transition of DNA as the molecular fingerprint, and then subsequently, the force is quenched in the range of 9.5 to 4.5 pN for 2 min at each force. (B) Enlarged view of a single force cycle. (C) Zoom-in of the unfolding and refolding steps of R8 during the force-ramp part of the pulse. (D) B-S transition of 604-bp DNA linker at 65 pN. (E) Representative traces of unfolding and refolding transition of R8 at several forces of 9.5, 8.5, 7.75, and 7 pN. Magenta and blue colors indicate the protein in the unfolded and folded states, respectively, assigned using a hidden Markov model. (F) Histograms showing the probability density of the natural logarithm of the measured dwell times of R8 for the refolding (magenta) and unfolding (blue) states at 7.75 pN. The continuous lines represent the fit using single exponential law. (G) Graph of natural logarithm of refolding (magenta points) and unfolding (blue points) rates of R8 domain as a function of force from N = 8 different molecules. The dotted lines represent the fits using the Bell model, showing a distance to transition state of ∆xf→TS = − 3.99 ± 0.08 nm for the folded state and ∆xu→TS = − 0.18 ± 0.08 nm for the unfolded state. Error bars are SDs of the fit.
When measuring the low-force behavior of R8 refolding, an unexpected observation comes from the heterogeneity of these transitions at a given force. In some cases, molecules that showed the correct unfolding fingerprint in the force-ramp part of the pulse (unfolding step of R8 and the B-S overstretching of DNA transition at 65 pN) did not show any unfolding-refolding transitions in the low-force part of the pulse, while others started showing folding-unfolding transitions only after several cycles (Fig. 3A). Hence, the folding-unfolding equilibrium force varied from molecule-to-molecule in the 5.5- to 7.5-pN range, as reflected by the average rates (Fig. 3G). However, these rates represent the average behavior and do not capture the history dependency of R8 transitions. The time-dependent behavior was further confirmed when analyzing pulses from the same molecule. In this case, the bead heterogeneity or small force calibration errors do not affect our measurements, as all traces in this figure are from the same molecule. As shown in fig. S3, the same protein molecule can show a broad spectrum of unfolding-refolding distributions. In this case, at every cycle, the protein was first unfolded completely (Fig. 3A). During the low-force part of the pulse, the same molecule exposed to the same low forces showed notably varied unfolding-refolding kinetics with each cycle (fig. S3). For a given force, while the size of the steps remained constant between cycles, the probability of folding varied notably with each cycle. As expected, there were no significant changes in the measured position of the overstretching DNA transition between cycles (fig. S4). This heterogeneous behavior might be responsible for how talin R8 domain interacts with its binding partners, such as with deleted in liver cancer (DLC1), while being in its folded state.
Interaction of talin R8 domain with its binding partner DLC1
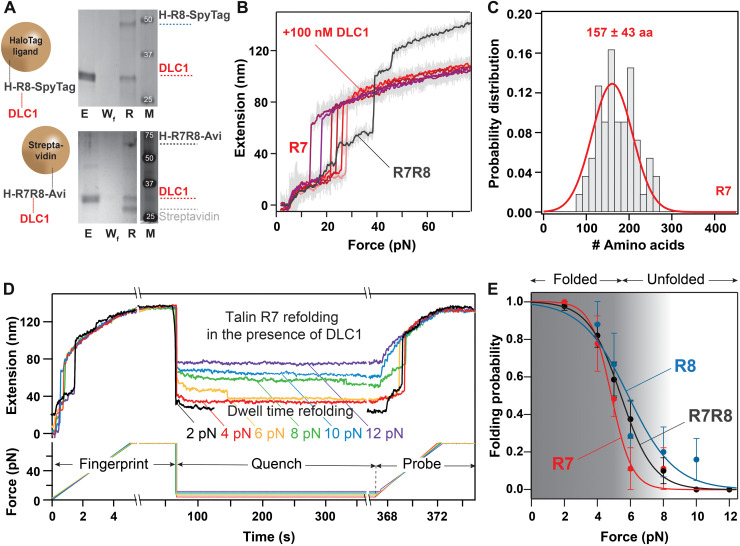
DLC1 was shown to be an important regulator for focal adhesions in vivo (18). It has a single known binding site to talin, at rod domain R8. Here, we investigated the interaction between talin R8 and DLC1 using the exceptional capability of magnetic tweezers to study the same protein molecule in the absence and presence of ligands. First, we confirmed that DLC1 binds in solution to our constructs, which were specifically engineered for magnetic tweezers (Fig. 4A). The same R8 or R7R8 constructs used in our single-molecule experiments were here adsorbed on the same type of paramagnetic beads, terminated with either HaloTag or streptavidin. Following washing, the beads were reacted with 10 μM DLC1, then washed extensively, and eluted in the presence of Coomassie blue and SDS. The collected solution (R-lane) was run on an SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel, together with the eluted fraction (E-lane) and the final wash after the DLC1 reaction (Wf lane). The E-lane contained the nonadsorbed DLC1; the Wf lane did not show any nonspecifically bound DLC1 ligand, indicative of complete washing of nonspecifically adsorbed proteins, while the R-lane of the boiled beads showed both the DLC1 and the talin signals (Fig. 4A). In our single-molecule experiments, we exposed talin R7R8 and R8 domains to initial force-ramp pulses, without any DLC1 present (Fig. 4B). Once we measured the unfolding fingerprint of talin molecules, we added DLC1 at concentrations as low as 10 nM and measured the exact same protein molecule using the same force-ramp pulse. While we used a concentration well below the reported dissociation constant Kd = 48 μM (19), starting from the very first pulse in the presence of DLC1, only a single step was seen for the R7R8 construct (Fig. 4B), and no unfolding step was measured for the R8 construct (fig. S5). As previously reported for other protein systems, binding of ligands is expected to induce a mechanical stabilization of proteins (11, 20). However, in the case of talin R8, we could not force the ligand to detach, even when applying ~90 pN, which is the upper boundary for our measuring approach. This unexpected behavior has, however, allowed us to measure the unfolding behavior of R7 while its R8 neighbor stayed in the folded state, resembling its mode of operation in vivo (Fig. 4C). The step histogram for R7 showed a peak of 157 ± 43 amino acids, indicating a mechanical clamp for the full-length structure. This result indicates that, when R8 stays folded because of ligand binding, R7 only unfolds in an all-or-none configuration. Similarly, we used this binding-induced inactivation of R8 to measure the refolding of R7 and compare it with that of R8 alone and of R7R8 (Fig. 4D). To measure the refolding probability of a protein as a function of force, we perform fingerprint-quench-probe experiments (12). In these experiments, a molecule was first exposed to a fingerprint pulse, where the force was ramped, to assess that the protein folded from the previous pulse. Then, the force was quenched to a low value (between 2 and 12 pN), allowing the protein to refold (quench pulse). Last, the folding at low force was probed by again increasing the force (probe pulse) and unfolding any refolded domains from the quench pulse (Fig. 4D). The folding probability for a given force applied during the quench part was then computed as the ratio of the number of unfolding steps in the probe pulse to the number of domains in the fingerprint pulse. This approach also allows us to measure folding probabilities at low forces, where the refolding steps can be too small to be detected with our technique and represents how likely an unfolded protein will refold at a set constant force (12). It also eliminates false positives in the event of mechanical aging of the protein (21), which would become registered in the fingerprint pulse. As seen in Fig. 4E, the measured folding probability for the R7R8 construct lies between the individual folding probability of R7 and R8. These data suggest that R8 is the first to refold under a force vector (half force of F1/2 = 5.9 ± 0.5 pN), followed by R7 (F1/2 = 5.5 ± 0.2 pN for R7R8 construct), while R7 refolding when its neighboring R8 domain stays folded had a F1/2 = 4.9 ± 0.5 pN.
Fig. 4. Interaction of talin domains R7R8 to DLC1.
(A) SDS-PAGE analysis showing the binding of R7R8 to DLC1. The E-lane contained the nonadsorbed DLC1; the Wf lane confirms the wash procedure, while the R-lane of the boiled beads showed both the DLC1 and the talin signals. M, molecular weight standards. (Top) Binding of DLC1 to R8 separated from R7, and (bottom) binding of DLC1 to R8 using the R7R8 construct. (B) Representative trace of the extension of the same molecule of R7R8 at increasing force of 15.5 pN/s: Before adding DLC1, unfolding of R7R8 shows three steps (gray trace) and, after adding 100 nM DLC1, shows the unfolding of only R7 domain having a single step of ~40 nm and a shorter final extension (red and purple traces). (C) Histogram showing the probability distribution of the number of amino acids unfolding from R7 on a single step (N = 55). The average number of amino acids unfolded is obtained from Gaussian fit. (D) Traces showing the folding probability of talin R7 domain in the presence of R8-DLC1 complex. A first fingerprint pulse shows unfolding of R7 as a ~40-nm step. Force is then quenched to various low forces, where refolding could take place. Last, in the probe pulse, force is increased and any refolded domains in the quench pulse unfold once more. (E) Comparison of folding probability as a function of refolding forces for R7R8, R8, and R7 as shown by black, blue, and red points, respectively. The half force of refolding corresponding to R7R8, R8, and R7 are 5.5 ± 0.2 pN, 5.9 ± 0.5 pN, and 4.9 ± 0.5 pN, respectively. The solid lines are sigmoidal fit to the data, and error bars represent SEs.
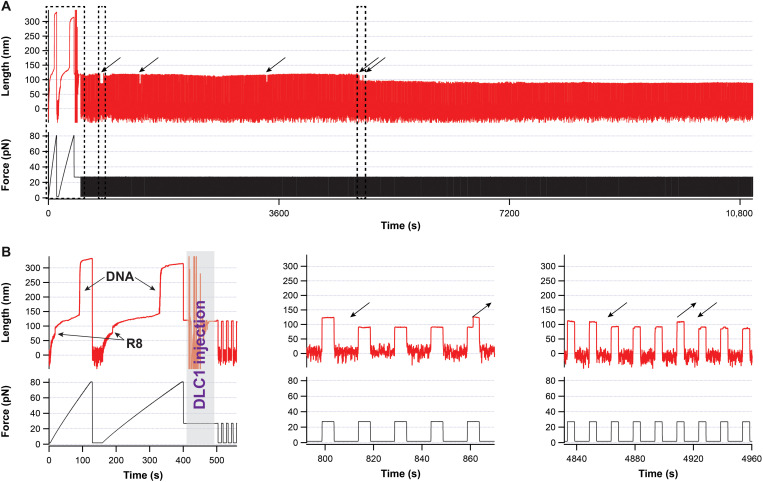
To further investigate this unexpected binding-induced stabilization behavior, we designed our experiments to maintain talin R8 domain unfolded while adding the DLC1 ligand by applying a force of 27 pN (Fig. 5). We then cycled the force between 2 and 27 pN, in 10-s intervals. This pulse is sufficient to refold R8 at low force in ~74% of the cases and to unfold DLC1-free talin in the high-force part in 95.2% of the pulses. During our cycling force protocol, when DLC1 binds, the measured extension at 27 pN decreases to the same value expected for the molecule having R8 folded (Fig. 5). In the first part of some of these experiments, we did see unbinding, followed by rebinding, with both transitions taking place relatively fast (<1 min, with the longest binding time of ~30 min; see table S3 for rates and Fig. 5 for the longest measured binding time, measured from N = 118). Most of the traces showed a final binding event, which, in our longest occurrence, was stable for more than 7 hours, before we stopped the experiment (Fig. 5 shows the first part of this event). However, unlike the force-ramp protocols, where the tether broke before DLC1 detached, using the pulse approach, we measured concentration-dependent unbinding rates as well, with the longest DLC1 detachment time being 78.5 min. This recovery suggests that the connection between DLC1 and R8 is not covalent.
Fig. 5. Binding of DLC1 to talin domain R8.
(A) First 3 hours of a trace lasting over 30,000-s and monitoring the binding of DLC1. Arrows represent binding events, and dotted boxes mark chosen zoom-ins. (B) Left: Zoom-in of the fingerprint pulse, where the force is ramped to ~80 pN, showing the unfolding of R8 followed by the DNA overstretching transition. After two force-ramp pulses, a solution of DLC1 is injected, while a force of 27 pN is maintained to prevent talin R8 from refolding. The noise in length during injection is an artifact from the bead responding to flow. Middle: A zoom-in showing a temporary binding event lasting three force-step changes. Right: A magnified view showing two binding events. No other unbinding events were measured in this trace after time t = 4920 s, in the remainder of ~7 hours.
DISCUSSION
Talin was deemed to be the most important protein involved in the formation of focal adhesions, as it generates several possible responses under force while reacting with a variety of ligands (22). Talin was shown to operate at forces between 5 and 10 pN per molecule (23) and extend inside cells between 100 and 350 nm (24), which can result in the mechanical unfolding of all its 13 rod domains (25). Previously, we made the analogy that talin functions as a biomechanical computer, as its response resembles that of a complex computer program (26, 27), and if talin is the mechanical computer of cellular adhesion, then R8 represents the central processing unit, as it is capable of binding to the highest number of known ligands among rod domains and has a complex molecular architecture (Fig. 1). Among the binding partners of talin, the interaction with vinculin is best understood. Talin-vinculin interaction has been extensively studied with single-molecule techniques, as its response requires some partial unfolding of the talin rod domain to expose vinculin binding sites on talin (7). Another intriguing talin binding partner is DLC1, a protein first discovered as lacking in patients with liver cancer (28) but later reported to be lacking in many other types of cancers (29). DLC1 is thought to act as a tumor suppressor by interacting with Rho–guanosine triphosphate (GTP) and was shown to inhibit the activation of Rho–guanosine triphosphatase (GTPase) by accelerating its intrinsic GTPase activity (18). DLC1 was also reported to bind to talin, and its only known binding location is at the R8 site (19). Here, we report that the complex molecular architecture of talin R7R8 results in an intricate response to force and find a stronger-than-expected binding of R8 to its DLC1 partner.
The number of amino acids comprising the mechanical clamp of a protein can be calculated from its contour length. The extension of a protein during unfolding depends on the applied force and the number of amino acids contained inside the unfolding structure and can be directly used to estimate the contour length of the molecular assembly opposing the force vector. We find an unexpected variation in the measured unfolding structures for the talin R7R8 construct (Fig. 2). As magnetic tweezers applies force to all the molecules inside the fluid chamber at once, it is worth noting that our data represent snapshots of how proteins unfold repeatedly, similar to their mode of operation in vivo. The unfolding histograms presented in Fig. 2 show steps that can be attributed to the synchronous unfolding of both R7 and R8 domains, as well as their full or partial asynchronous denaturation. Using molecular dynamics simulations, a three-helix stable structure was predicted for the unfolding of R7, which was confirmed by our measurements (Fig. 2B middle peak in the left and center histogram) (14). Unexpectedly, our data showed that this structure is more prevalent than the unfolding of all five helices of R7 in one step. The same study predicted that R7 helix H1 could form an intermediate structure with R8 domain during unfolding, which would produce a stable structure from H2 to H5 helix of R7, with an expected step of 147 amino acids (table S2). However, our data did not have the necessary resolution to distinguish between structures with three and four helices.
Another unexpected result came when measuring talin R8 domain, engineered separately from its neighboring R7 domain. This domain showed folding-unfolding transitions in the 4- to 10-pN force range. Several initial experiments done solely with R8 tethered between a glass surface and paramagnetic bead using HaloTag-SpyTag chemistry showed such unfolding-refolding transitions, but these measurements puzzled us in the beginning, as other molecules with the correct unfolding fingerprint at high force did not have such transitions at low force. We initially attributed these transitions to the extremely unlikely event of having two molecules tethered perfectly in parallel (as any misalignment would produce two steps). By improving our molecular fingerprint with the addition of a DNA linker, covalently cross-linked to the HaloTag-R8 molecule via cysteine chemistry, and using AviTag-streptavidin at the opposite end, we were able to exclude the possibility that the fluctuations come from a perfectly parallel double tether. A double-tether scenario would result in a change in the measured force for the DNA overstretching transition, as the tension would be distributed across two molecules in parallel. This was not the case, as the DNA overstretching transitions appeared at the expected magnet position for 65 pN (~0.99 mm) (4). The downside of the protein-DNA linkers comes from using noncovalent AviTag-streptavidin attachment, which rarely produces the long-lasting tethers at forces over 65 pN, needed to measure the overstretching transition. This noncovalent interaction was shown to have a possible mechanically stable state when the domain of the tetrameric streptavidin that attaches to AviTag is also directly connected to the bead (30, 31). Such a long-lasting AviTag-streptavidin tether is shown in Fig. 3. Here, the molecule was first exposed to a force ramp increasing up to ~75 pN, where both the unfolding of R8 and the overstretching DNA transition appeared. When cycling between various low forces in the physiological range, we saw no folding-unfolding transitions in the first few cycled. However, as the molecule cycled through, we measured in the low-force part folding-unfolding transitions for the R8 domain, starting after a few cycles. We excluded instrumental error in the applied force since the measured overstretching transition showed no dependency with magnet position between cycles (figs. S4 and S6). Notably, these transitions show a memory dependency, as, at the same force, the same molecule shows slightly different kinetics (fig. S3). So, what could produce such a memory effect in the folding behavior of talin R8 in its physiological force range? We speculate that repeated unfolding-refolding cycles might lead to different native structures of the R8 domain, which can result in changes in its mechanical response. The changes affecting this process can be physical or chemical. Several cases were reported where repeated unfolding induced interdomain misfolded structures in other proteins through energetic dependencies among its constituent domains (32, 33). This possible mechanism would involve kinetic trapping in a stable structure (34). Using molecular dynamics simulations, it was proposed that, if unfolded, then half of the cytosolic Escherichia coli proteins could exhibit subpopulations of misfolded conformations, which would be kinetically trapped, but without propensity to aggregate (35). These conformations are rarely encountered in vivo for freshly expressed proteins, where the cotranslational folding restricts the folding kinetics (36). Kinetic trapping was also corroborated in an experimental study, which reports on different unfolding behaviors for different molecules of the same protein (37). Furthermore, in its unfolded state, a protein has all of its amino acids exposed to the solution environment. Some amino acids that typically are buried inside the protein structure are more prone to posttranslational modifications, such as oxidation, which would chemically change the folding energy landscape of a protein (21). However, these transitions did not showed a shortening specific for disulfide bond formation (38) or lack of folding, specific to oxidation (21), but rather a strengthening of the folded state. While the oxidation effect reported in (21) seems to be universal, appearing with various probabilities in all measured proteins, including talin, it manifests through the absence of refolding, as the molecule extends to the same length as when first unfolded. Both the unfolding/refolding transitions and the molecules with bound DLC1 showed shorter overall extensions between cycles, representative of a different mechanism (see also Fig. 5 and fig. S5).
Another important finding was that the interaction of talin R8 domain with its ligand DLC1 locks R8 into a mechanically stable state, which can withstand forces over 90 pN for long times (Figs. 4 and 5 and fig. S5). Using calorimetry, the dissociation constant of never-unfolded R8 with its DLC1 ligand was measured to be Kd = 48 μM (19). Hence, our expectation was that when exposed to a solution containing DLC1 in concentrations well below Kd, binding will take place only after several cycles. Again, it is worth mentioning here that we are measuring the exact same protein molecule without and with its ligand present in solution using an approach as previously described (11). When DLC1 was introduced into the measuring chamber while the bead was maintained at 2 pN, R8 stopped unfolding from the initial cycle, and the ligand could not be detached, even at high forces. Molecular dynamics simulations predicted no change in the mechanical stability of the R8 upon DLC1 binding (14). However, these simulations used the structure reported from crystallography studies, where the talin domains were not previously unfolded mechanically.
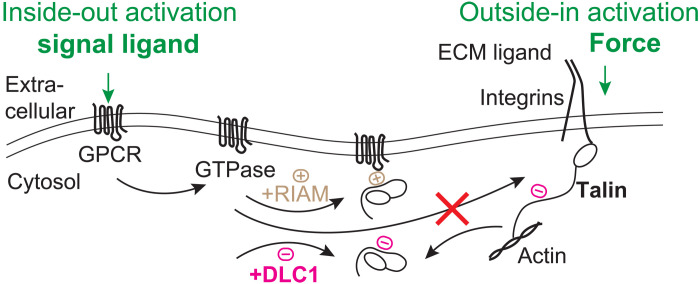
What is the biological interpretation of this extreme mechanical stability of R8 upon DLC1 binding? Here, we propose that a mechanism of action for the potential tumor suppressor activity of DLC1 comes from its competing interaction against RIAM (Rap1-GTP–interacting adaptor molecule) for talin R8 (Fig. 6). DLC1 was studied as a tumor suppressor in the context of its interaction with Rho-GTP and was shown to inhibit the activation of Rho-GTPase by accelerating its intrinsic GTPase activity (18, 39). However, tumor-associated loss-of-function mutants showed normal Rho-GTPase activating protein activity (40), suggesting that a different mechanism makes DLC1 act as a tumor inhibitor. A landmark study identified DLC1 as a ligand for talin R8 domain (19). The same R8 site is used by GTPases to bring talin at the plasma membrane through RIAM (Fig. 6). Once at the plasma membrane, two-dimensional (2D) versus 3D diffusion facilitates better binding of talin to integrins, leading to a 1000- to 10,000-fold increased affinity for ECM ligands (41). However, in calorimetry experiments, RIAM was shown to bind more strongly to talin R8 than DLC1 (Kd = 3.5 μM for RIAM-R8 and Kd = 48 μM for DLC1-R8), which would make DLC1 a poor regulator (19). These findings are in contradiction with experiments done with cells, where, during the formation of focal adhesions, RIAM/talin complexes were undetectable when DLC1 was present (19). Hence, the extreme binding between DLC1 to talin R8 domain measured here supports R8 as a strong regulator for inside-out activation of integrins. Furthermore, this mechanical locking of R8 will affect the (un)folding behavior of its neighbor R7. The half-force for refolding of R7 within 5 min is 4.9 pN, smaller than for the refolding of R8 alone, or for R7R8. This half-force indicates that R7 will be the last protein domain to fold in the R7R8 complex and that a folded R8 will favor R7 refolding.
Fig. 6. Schematics of the interaction between talin and DLC1 in the context of integrin activation.
Inside-out activation of integrins is triggered through binding of an extracellular ligand to a G protein–coupled receptor (GPCR), which later activates a GTPase. Adapter molecule Rap1-interacting molecule (RIAM) can link the GTPase to talin when binding via the R8 domain and can maintain talin at the plasma membrane, increasing the chances of activation of integrins and binding the ECM. The strong binding of DLC1 can compete with RIAM, regulating activation, or can prevent membrane-bound RIAM from recruiting talin during focal adhesion disassembly.
How would a memory-dependent force fluctuation have a physiological role in the functioning of the talin R7R8 domains? While R8 was predicted to be mechanically weak, similar to the R3 domain (14), currently, the only talin domain known to show folding-unfolding transitions in the physiological force range is R3 (16, 42, 43). The R1-R4 region was shown to recruit vinculin without being exposed to force (44). These reported behaviors make R3 the likely place for the initiation of focal adhesion, where vinculin can be easily recruited. Because of its complex architecture, R8 can only undergo folding transitions while R7 is locked into an unfolded conformation, when bound to vinculin (42). Hence, we speculate that a potential role for the folding transitions seen for R8 is in terminating focal adhesions. In such a role, R8 folding-unfolding transition would act as a probe to determine whether vinculin, or other binding partners such as DLC1, have higher concentrations, as vinculin would lock R8 in an unfolded state, while other ligands would maintain it folded. In its folded state, R8 would favor R7 refolding and, hence, vinculin detachment. How would such a strong interaction with DLC1 affect the function of R7R8? First, such an interaction would prevent recruitment of RIAM, a linker protein that keeps talin at the plasma membrane during inside-out activation (45). Furthermore, detachment of talin with R8-bound DLC1 from integrins would prevent interaction with RIAM and attachment to membrane proteins that use the same site (Fig. 6). Another possible scenario involves the attachment of talin to actin filaments and microtubules. Attachment to actin is accomplished though vinculin via R7R8 unfolding, while attachment to microtubules is accomplished via adaptor protein KN motif and ankyrin repeat domains 1 (KANK1) when R7 stays folded (46). One would expect that the folded R8 would stabilize the folded state of R7 and favor binding of talin to microtubules over binding to actin.
In conclusion, repeated mechanical unfolding of talin R8 domain via magnetic tweezers showed a history-dependent mechanism, where exposure to unfolding transitions of a protein domain determines its future behaviors. Furthermore, an unusually strong binding interaction of talin R8 with DLC1 ligand was measured and can explain the regulatory potential of this interaction for integrin activation and focal adhesion formation. While we currently do not fully understand the exact mechanism for the history dependence of unfolding and how it relates to DLC1 binding, we envision that future studies will elucidate how common such a history-dependent response occurs in the proteome and how it influences the binding to proteins involved in mechanotransduction.
MATERIALS AND METHODS
Protein engineering, expression, and purification
If not otherwise mentioned, then all chemicals were purchased from Sigma-Aldrich. Talin rod domains R7 and R8 were cloned from human TLN1 gene and were engineered as inserts (residues 1351 to 1656) for R7-R8, inserts (residues 1453 to 1582) for R8, and (L)2-R7R8-(L)2 (L denotes protein L) into several pFN18A expression vectors that were modified to add HaloTag at the N terminus, and either AviTag or SpyTag with a terminal cysteine at the C terminus. The pFN18A plasmids also had a (Histidine)6 tag at the C terminus, used for purification. They were inserted into E. coli BLR(DE3) cells, which were grown in Luria Broth (LB) in the presence of carbenicillin (50 μg/ml) at 37°C until OD600nm (optical density at 600 nm) reached 0.6 to 0.8. Then, overexpression was induced overnight at 25°C with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were then pelleted and resuspended in E/W buffer [50 mM Na2HPO4/NaH2PO4, 300 mM NaCl, 1 mM dithiothreitol, and 5% (v/v) glycerol (pH 7.0)] and lysed with lysozyme, 1% Triton X-100, deoxyribonuclease (DNase), and ribonuclease (RNase), in the presence of protease inhibitors, followed by sonication. Following cell lysis, the soluble protein fraction was separated from the insoluble fraction using high-speed centrifugation or filtration and passed through a chemical affinity purification Ni-NTA (nitrilotriacetic acid) column. Washing of the Ni-NTA column with the adsorbed protein of interest was done with E/W buffer containing also 7 mM imidazole, while elution was achieved with E/W buffer containing 250 mM imidazole. Following affinity purification, the protein solution was injected into a size exclusion chromatography column (S-300, ÄKTA GE) and eluted with a Hepes buffer [50 mM Hepes, 150 mM NaCl, and 5% (v/v) glycerol (pH 7.2) buffer]. DLC1 with a sequence of NEDIFPELDDILYHVKGMQRIVNQWSE (which was shown to bind R8 domain) (19) fragment was ordered from Integrated DNA Technologies and was inserted in a pQ80E expression vector, which also contained mVenus protein and the (Histidine)6 tag at the N terminus. The used concentrations for DLC1 in our experiments were 10 nM, 100 nM, 1 μM, and 14 μM. The purification procedure was similar to the one used for HaloTag-Talin constructs, with the exception that the competent cells used for protein overexpression were E. coli C41, and the buffers used during purification did not contain glycerol. The purified proteins with the AviTag construct were concentrated to ~100 μM and biotinylated using a BirA biotin-protein ligase standard reaction kit (Avidity), following the manufacturer’s protocol. An up-to-date protocol of our protein purification is maintained and available on our website at https://popalab.uwm.edu/.
Attachment chemistry
An experimental fluid chamber was used to measure the immobilized molecules. Fluid chambers were assembled by sandwiching two parafilm strips between the cleaned glass top surface (260140, Ted Pella) and the silanized bottom surface or by using a 3D-printed scaffold (with PLA using a Prusa Research 3D printer). The assembled fluid chambers were incubated with a mixture of 1% (v/v) glutaraldehyde in phosphate-buffered saline (PBS) buffer at pH 7.2 and 0.05% (w/v) amine-terminated polystyrene nonmagnetic (reference) beads of diameter 2.6 μm for 1 hour (Spherotech). The nonadsorbed glutaraldehyde and reference beads were then washed with PBS buffer [50 mM Na2HPO4/NaH2PO4 and 150 mM KCl (pH 7.2)] and incubated for 4 hours at room temperature with amine-terminated chloroalkane ligand (10 μg/ml; HaloTag Ligand, Promega), also dissolved in PBS buffer. Last, the chambers were washed and passivated with bovine serum albumin (BSA) for 12 hours [1% (w/v) sulfhydryl blocked BSA, 20 mM tris, and 150 mM KCl (pH 7.4)].
Dynabeads M-270 (Thermo Fisher Scientific) with functional amine or streptavidin groups were used to apply force to biomolecules. The amine beads were used for the HaloTag-SpyTag constructs and were functionalized with chloroalkane ligand in a similar way as the surfaces. The streptavidin beads were washed three times with PBS. Before experiments, beads were also passivated for more than 1 hour in bead-blocking solution (S-4023, TriLink BioTechnologies) or with a casein solution [1.5% (g/v) in PBS; Fisher Scientific] at 4°C. The excess bead-blocking solution was washed three times, and beads were incubated in PBS buffer to use.
Protein-DNA constructs were synthetized as described in (4) using the same DNA sequence, terminated with a di-biotin and amine. Briefly, cloning of the 604-bp lambda-DNA was done via polymerase chain reaction in the presence of the biotin and amine primers. The product was then cleaned with a clean-up column (Macherey-Nagel NucleoSpin from Fisher Scientific) and reacted for 30 min at room temperature with a sulfosuccinimidyl-trans-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC) bifunctional ligand (EMD Millipore). Following a second cleanup to remove unreacted Sulfo-SMCC reagent, the DNA was reacted to the cysteine-terminated protein overnight at 4°C. An up-to-date protocol of our surface chemistry approaches is maintained and available on our website at https://popalab.uwm.edu/.
Single-molecule magnetic tweezer measurements
Our magnetic tweezer instrument is built on top of an inverted Olympus IX71 microscope and was described in detail in (11). When measuring SpyTag-terminated proteins, the chloroalkane functionalized chamber was first incubated for 10 min with 5 μM HaloTag-SpyCatcher, washed with PBS buffer, and incubated for 30 more minutes with the protein of interest (diluted to ~100 nM in PBS). All other proteins were added directly to the chloroalkane functionalized chamber for 10 min, at the same concentration. Then, the fluid chamber was washed with PBS buffer to remove nonadsorbed protein, and the paramagnetic beads were added and left to sediment for ~1 min while the chamber was mounted on the inverted microscope. Then, the permanent magnets were approached to 5 mm to attract nonbound paramagnetic beads. Regions of interest (ROIs) of 128 × 128 pixels encompassing a tethered paramagnetic bead and a surface-glued nonmagnetic reference bead were selected. Using the selected two beads, a stack profile was obtained by moving the objective, with the help of a piezo actuator, in steps of 20 nm before the start of the experiment. During the experiment, this stack library was used to determine the relative position of the paramagnetic bead in respect to the reference bead, hence yielding the extension of the tethered molecule. The live-image processing was done using Igor Pro (WaveMetrics) by computing the 2D fast Fourier transform of the ROI, followed by a radial profile around the center (4). Last, the correlation between the radial profile obtained during measurement and the radial profiles stored in the stack library was fitted using a Gaussian law, to determine the absolute position of a bead.
Binding experiments were conducted as described in (11). Briefly, after finding a tether displaying the expected molecular fingerprint, the solution inside the chamber was changed to contain the ligand (in this case DLC1) while the bead was maintained either under a small force (~2 pN) to prevent binding to glass during generated flow, or at a force of 27-pN to prevent R8 refolding. Then, the same molecule was exposed to the desired force pulse, and its response was measured. As single molecules behave in a probabilistic manner, we designed our pulse to maximize folding probability and minimize the low-force exposure time. Folding probability of R8 at 2 pN for 10 s was determined to be 0.74 ± 0.02. In control experiments, when cycled between 2 and 27 pN for 10 s each, without added DLC1, R8 reached unfolding extension in 95.3% of the pulses. As this number was still less than 100%, we assigned binding events only where two or more pulses had R8 folded, while single folded pulses were considered as having DLC1 nonbound. The dwell times required for DLC1 to bind were determined from the low-force times, when R8 refolded. Obviously, the rates shown in table S3 are an underestimate, as they consider the folding of R8 to be immediate at 2 pN.
SDS-PAGE binding assay
SDS-PAGE (Bio-Rad Laboratories) was also used to analyze the binding of DLC1 to the folded R8 and R7R8 constructs. In this process, the purified protein was incubated with paramagnetic beads for an hour in Hepes buffer at 4°C on the rotor. The excess nonadsorbed protein was washed three times with Hepes buffer, obtained as supernatant by the process of sedimentation using a strong magnet. Following the wash, the paramagnetic beads with a protein attached to them were incubated with mVenus-DLC1 at 4°C overnight on the rotor. Following the incubation process, the excess nonadsorbed DLC1 was again washed four times by sedimentation using a strong magnet, and the nonadsorbed and final wash supernatant was collected. The sedimented beads with bound protein and ligand were incubated in 10 μl of Hepes buffer. The nonadsorbed supernatant was diluted to 2 μM with Hepes buffer. Ten microliters from each sample was mixed with 4 μl of buffer [50 μl of 4× Laemmli sample buffer (Bio-Rad) and 5.5 μl of 2-mercaptoethanol (Amresco) as a reducing agent], and the samples were further denatured by heating for 10 min at 100°C. The samples were then loaded onto a 4 to 15% polyacrylamide gel (Bio-Rad) and run at a constant voltage of 200 V for 30 min using 1× tris-glycine SDS buffer (Amresco Inc.). Following three washes with DDi H2O and Coomassie staining (Bio-Rad), the protein bands on the gels were visualized using a destaining solution (20% methanol, 10% acetic acid, and 70% H2O).
Data analysis and errors estimation
Data analysis was done using Igor Pro (WaveMetrics). To estimate the structures resisting mechanical force, step sizes from the single-molecule data were measured as changes in the extension of the tethered molecule. The measured extensions were then used to estimate the number of amino acids forming the mechanical clamp that generated them. For this calculation, we used the worm-like chain model of polymer elasticity, assuming a persistence length of 0.58 nm (12) and the size of an amino acid under force of 0.4 nm (13). The data were then aggregated into histograms and fitted with a multi-Gaussian model using the built-in multipeak fitting procedure from Igor Pro to determine the mean and SD. The number of peaks estimated from histograms was verified by using the minimization of the fitting residual method: , where A is the fitted amplitude, x is the calculated number of amino acids, x0 is the position of the peak, SD is the standard deviation, and the number of degrees of freedom f = N − nf, with N being the number of bins and nf being the number of fitting parameters (47).
To determine the dwell times for the unfolding and refolding transitions from the equilibrium traces measured for R8, we used a hidden Markov analysis procedure, as described in (48). Briefly, the data were first resampled to contain equidistant points in time, then fitted using the code provided in the reference above, which was initialized from Igor Pro and uses MATLAB. Once the transition and emission matrices were calculated, a Viterbi algorithm was used to calculate the most probable path. Transitions having less than 30 points were ignored. Last, the traces were analyzed by hand for quality control. The dwell times were then aggregated into histograms by plotting the natural log and fitted to determine the rates with exp[x − x0 − exp (x − x0)], where x = ln [t] and x0 = ln [τ] (15). The measured rates were plotted as a function of force, and the expected rate at zero force r0 and distance to transition state ∆x were determined assuming a Bell model: , where kT is the thermal energy term.
The folding probability was determined from the ratio of the number of domains unfolding in the probe pulse over the number of domains unfolding in the fingerprint pulse, as the molecule was exposed to various low forces for 300 s. The folding probability was then fitted with a sigmoid function to determine the half-force: . Errors for single molecule statistics were evaluated using boot-strapping analysis, as described in (49).
Acknowledgments
We thank A. del Rio Hernandez for sending us the talin R7R8 gene, Y. Zhang for sharing the hidden Markov data analysis code, and V. Raicu for suggestions on improving our data analysis.
Funding: This work was funded by the NSF (grant numbers MCB-1846143 and DBI-1919670) and the Greater Milwaukee Foundation (Shaw Award to I.P.).
Author contributions: N.D., S.S., and B.P. performed the measurements. N.D., S.S., and I.P. analyzed the data. A.E. engineered the protein constructs; N.D., S.S., and A.E. expressed and purified the proteins used in this study. I.P. designed the research, supervised the project, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S6
References
REFERENCES AND NOTES
- 1.Mora M., Stannard A., Garcia-Manyes S., The nanomechanics of individual proteins. Chem. Soc. Rev. 49, 6816–6832 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Berkovich R., Fernandez V. I., Stirnemann G., Valle-Orero J., Fernandez J. M., Segmentation and the entropic elasticity of modular proteins. J. Phys. Chem. Lett. 9, 4707–4713 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Neuman K. C., Nagy A., Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popa I., Rivas-Pardo J. A., Eckels E. C., Echelman D. J., Badilla C. L., Valle-Orero J., Fernandez J. M., A halotag anchored ruler for week-long studies of protein dynamics. J. Am. Chem. Soc. 138, 10546–10553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haining A. W., Lieberthal T. J., Del Rio Hernandez A., Talin: A mechanosensitive molecule in health and disease. FASEB J. 30, 2073–2085 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Yao M., Goult B. T., Klapholz B., Hu X., Toseland C. P., Guo Y., Cong P., Sheetz M. P., Yan J., The mechanical response of talin. Nat. Commun. 7, 11966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P., Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tapia-Rojo R., Alonso-Caballero A., Fernandez J. M., Direct observation of a coil-to-helix contraction triggered by vinculin binding to talin. Sci. Adv. 6, eaaz4707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluger C., Braun L., Sedlak S. M., Pippig D. A., Bauer M. S., Miller K., Milles L. F., Gaub H. E., Vogel V., Different vinculin binding sites use the same mechanism to regulate directional force transduction. Biophys. J. 118, 1344–1356 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingras A. R., Bate N., Goult B. T., Patel B., Kopp P. M., Emsley J., Barsukov I. L., Roberts G. C. K., Critchley D. R., Central region of talin has a unique fold that binds vinculin and actin. J. Biol. Chem. 285, 29577–29587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahal N., Nowitzke J., Eis A., Popa I., Binding-induced stabilization measured on the same molecular protein substrate using single-molecule magnetic tweezers and heterocovalent attachments. J. Phys. Chem. B 124, 3283–3290 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Valle-Orero J., Rivas-Pardo J. A., Popa I., Multidomain proteins under force. Nanotechnology 28, 174003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ainavarapu S. R., Brujic J., Huang H. H., Wiita A. P., Lu H., Li L., Walther K. A., Carrion-Vazquez M., Li H., Fernandez J. M., Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J. 92, 225–233 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haining A. W. M., Rahikainen R., Cortes E., Lachowski D., Rice A., von Essen M., Hytonen V. P., Hernandez A. D., Mechanotransduction in talin through the interaction of the R8 domain with DLC1. PLOS Biol. 16, e2005599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tapia-Rojo R., Eckels E. C., Fernandez J. M., Ephemeral states in protein folding under force captured with a magnetic tweezers design. Proc. Natl. Acad. Sci. U.S.A. 116, 7873–7878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapia-Rojo R., Alonso-Caballero A., Fernandez J. M., Talin folding as the tuning fork of cellular mechanotransduction. Proc. Natl. Acad. Sci. U.S.A. 117, 21346–21353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berkovich R., Garcia-Manyes S., Urbakh M., Klafter J., Fernandez J. M., Collapse dynamics of single proteins extended by force. Biophys. J. 98, 2692–2701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim T. Y., Healy K. D., Der C. J., Sciaky N., Bang Y. J., Juliano R. L., Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J. Biol. Chem. 283, 32762–32770 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zacharchenko T., Qian X., Goult B. T., Jethwa D., Almeida T. B., Ballestrem C., Critchley D. R., Lowy D. R., Barsukov I. L., LD motif recognition by talin: Structure of the talin-DLC1 complex. Structure 24, 1130–1141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y., Balamurali M. M., Sharma D., Li H., A functional single-molecule binding assay via force spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 15677–15681 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valle-Orero J., Rivas-Pardo J. A., Tapia-Rojo R., Popa I., Echelman D. J., Haldar S., Fernandez J. M., Mechanical deformation accelerates protein ageing. Angew. Chem. Int. Ed. Engl. 56, 9741–9746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klapholz B., Brown N. H., Talin—The master of integrin adhesions. J. Cell Sci. 130, 2435–2446 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Austen K., Ringer P., Mehlich A., Chrostek-Grashoff A., Kluger C., Klingner C., Sabass B., Zent R., Rief M., Grashoff C., Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margadant F., Chew L. L., Hu X., Yu H., Bate N., Zhang X., Sheetz M., Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 9, e1001223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haining A. W. M., von Essen M., Attwood S. J., Hytonen V. P., Hernandez A. D., All subdomains of the talin rod are mechanically vulnerable and may contribute to cellular mechanosensing. ACS Nano 10, 6648–6658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popa I., Gutzman J. H., The extracellular matrix–myosin pathway in mechanotransduction: From molecule to tissue. Emerg. Top. Life Sci. 2, 727–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S., Subramani S., Popa I., Does protein unfolding play a functional role in vivo? FEBS J. 288, 1742–1758 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Wong C. M., Lee J. M., Ching Y. P., Jin D. Y., Ng I. O., Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 63, 7646–7651 (2003). [PubMed] [Google Scholar]
- 29.Ullmannova V., Popescu N. C., Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int. J. Oncol. 29, 1127–1132 (2006). [PubMed] [Google Scholar]
- 30.Sedlak S. M., Schendel L. C., Gaub H. E., Bernardi R. C., Streptavidin/biotin: Tethering geometry defines unbinding mechanics. Sci. Adv. 6, eaay5999 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruber S., Lof A., Sedlak S. M., Benoit M., Gaub H. E., Lipfert J., Designed anchoring geometries determine lifetimes of biotin-streptavidin bonds under constant load and enable ultra-stable coupling. Nanoscale 12, 21131–21137 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Scholl Z. N., Yang W., Marszalek P. E., Chaperones rescue luciferase folding by separating its domains. J. Biol. Chem. 289, 28607–28618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu K., Chen X., Kaiser C. M., Energetic dependencies dictate folding mechanism in a complex protein. Proc. Natl. Acad. Sci. U.S.A. 116, 25641–25648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baiesi M., Orlandini E., Seno F., Trovato A., Sequence and structural patterns detected in entangled proteins reveal the importance of co-translational folding. Sci. Rep. 9, 8426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nissley D. A., Jiang Y., Trovato F., Sitarik I., Narayan K. B., Yo E. P., Xia P., Fried S. D., Universal protein misfolding intermediates can bypass the proteostasis network and remain soluble and non-functional. Nat. Commun. 13, 3081 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waudby C. A., Dobson C. M., Christodoulou J., Nature and regulation of protein folding on the ribosome. Trends Biochem. Sci. 44, 914–926 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tapia-Rojo R., Alonso-Caballero A., Badilla C. L., Fernandez J. M., Identical sequences, different behaviors: Protein diversity captured at the single-molecule level. bioRxiv, 2021.02.24.432730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Manyes S., Liang J., Szoszkiewicz R., Kuo T. L., Fernandez J. M., Force-activated reactivity switch in a bimolecular chemical reaction. Nat. Chem. 1, 236–242 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Durkin M. E., Avner M. R., Huh C. G., Yuan B. Z., Thorgeirsson S. S., Popescu N. C., DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 579, 1191–1196 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Liao Y. C., Shih Y. P., Lo S. H., Mutations in the focal adhesion targeting region of deleted in liver cancer-1 attenuate their expression and function. Cancer Res. 68, 7718–7722 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Springer T. A., Dustin M. L., Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24, 107–115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao M., Goult B. T., Chen H., Cong P., Sheetz M. P., Yan J., Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4, 4610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stannard A., Mora M., Beedle A. E. M., Castro-Lopez M., Board S., Garcia-Manyes S., Molecular fluctuations as a ruler of force-induced protein conformations. Nano Lett. 21, 2953–2961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dedden D., Schumacher S., Kelley C. F., Zacharias M., Biertumpfel C., Fassler R., Mizuno N., The architecture of Talin1 reveals an autoinhibition mechanism. Cell 179, 120–131.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y. C., Zhang H., Franco-Barraza J., Brennan M. L., Patel T., Cukierman E., Wu J., Structural and mechanistic insights into the recruitment of talin by RIAM in integrin signaling. Structure 22, 1810–1820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouchet B. P., Gough R. E., Ammon Y. C., van de Willige D., Post H., Jacquemet G., Altelaar A. M., Heck A. J., Goult B. T., Akhmanova A., Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife 5, e18124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoneman M. R., Biener G., Ward R. J., Pediani J. D., Badu D., Eis A., Popa I., Milligan G., Raicu V., A general method to quantify ligand-driven oligomerization from fluorescence-based images. Nat. Methods 16, 493–496 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y., Jiao J., Rebane A. A., Hidden Markov modeling with detailed balance and its application to single protein folding. Biophys. J. 111, 2110–2124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiita A. P., Perez-Jimenez R., Walther K. A., Grater F., Berne B. J., Holmgren A., Sanchez-Ruiz J. M., Fernandez J. M., Probing the chemistry of thioredoxin catalysis with force. Nature 450, 124–127 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan R. B., Goult B. T., Adhesions assemble!-autoinhibition as a major regulatory mechanism of integrin-mediated adhesion. Front. Mol. Biosci. 6, 144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bass M. D., Patel B., Barsukov I. G., Fillingham I. J., Mason R., Smith B. J., Bagshaw C. R., Critchley D. R., Further characterization of the interaction between the cytoskeletal proteins talin and vinculin. Biochem. J. 362, 761–768 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H., Choudhury D. M., Craig S. W., Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 281, 40389–40398 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Miller P. W., Pokutta S., Mitchell J. M., Chodaparambil J. V., Clarke D. N., Nelson W. J., Weis W. I., Nichols S. A., Analysis of a vinculin homolog in a sponge (phylum Porifera) reveals that vertebrate-like cell adhesions emerged early in animal evolution. J. Biol. Chem. 293, 11674–11686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goult B. T., Zacharchenko T., Bate N., Tsang R., Hey F., Gingras A. R., Elliott P. R., Roberts G. C. K., Ballestrem C., Critchley D. R., Barsukov I. L., RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J. Biol. Chem. 288, 8238–8249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S6
References