Abstract
Both acute and chronic stress have significant impact on brain functions. The amygdala is essential in mediating stress responses, but how its transcriptomic dynamics change under stress remains elusive. To overcome the difficulties in detecting subtle stress-induced changes by evaluating total RNA using classic RNA sequencing, we conducted thiol-labeled RNA sequencing (SLAM-seq). We injected 4-thiouridine (4sU) into mouse amygdala followed by SLAM-seq to detect nascent mRNA induced by acute and chronic restraint stress, and found that SLAM-seq could label actively transcribed genes in the major neuronal and glial subtypes. Using SLAM-seq, we found that chronic stress led to higher turnover of a group of genes associated with myelination, and this finding is confirmed by immunostaining which showed increased myelination in the chronically stressed amygdala. Additionally, genes detected by SLAM-seq and RNA-seq only partially overlapped, suggesting that SLAM-seq and RNA-seq are complementary in identifying stress-responsive genes. By applying SLAM-seq in vivo, we obtained a rich dataset of genes with higher turnover in the amygdala under stress.
Keywords: SLAM-seq, Amygdala, Acute stress, Chronic stress
1. Introduction
The impact of stress on the brain is profound and has been linked to an increased risk of mental illnesses, including depression and anxiety disorders (Caspi et al., 2003; Weger et al., 2018). Both acute and chronic stress can induce adaptive changes in various brain regions (de Kloet et al., 2005; McEwen, 2007; Xiao et al., 2021; Davis et al., 2010; Mah et al., 2016; Liu et al., 2020, 2021), including the amygdala, which plays a key role in processing physiological and behavioral responses to stress (Roozendaal et al., 2009; Zhang et al., 2019). While many studies have demonstrated that stress affects amygdala function, the underlying molecular mechanisms responsible for these changes remain elusive. To understand the stress-induced effects in the amygdala at the molecular level, it is essential to study how gene expression in amygdala is influenced by acute and chronic stress.
The thiol-labeled RNA sequencing (SLAM-seq) method specifically labels newly transcribed mRNAs (Herzog et al., 2017), and has been widely applied in cultured cells (Alalam et al., 2022; Muhar et al., 2018). Recently, SLAM-seq has also been extended to in vivo studies to explore genomic changes during zebrafish embryo development (Bhat et al., 2023), and to identify cell type-specific transcriptome in mouse tissues in a Cre-dependent manner (SLAM-ITseq (Matsushima et al., 2018)). Although SLAM-ITseq provides a solution to metabolically label RNA with 4-thiouracil (4-TU) in a specific cell type in vivo, this method requires usage of double-transgenic mouse lines to achieve cell type-specific expression of uracil phosphoribosyltransferase (UPRT), to transform 4-TU to 4-thiouridine (4sU) in Cre-expressing cells, limiting the applicability.
By conducting SLAM-seq with direct stereotactic injection of 4sU, we detected genes with higher turnover in the amygdala under acute and chronic stress. Bioinformatic analyses showed that the injected 4sU was integrated into the transcription of genes expressed in major brain cell types. In addition, chronic stress induced increased turnover of myelination-related genes, a result confirmed by immunostaining of myelin basic protein (MBP), which showed elevated myelination in the chronically stressed amygdala.
2. Results
2.1. Adapt SLAM-seq for the brain through stereotactic injection of 4sU
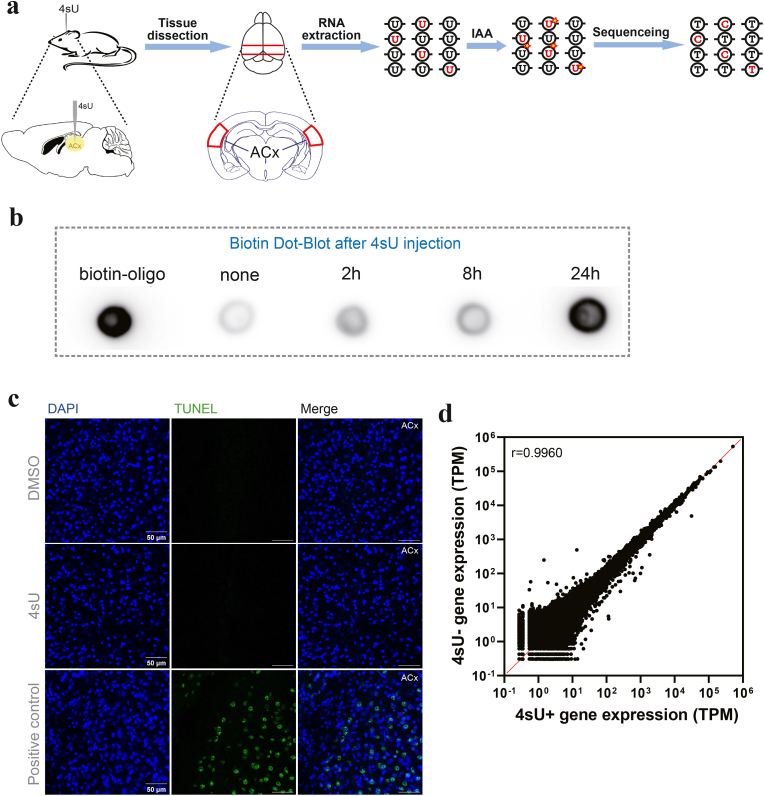
Neuronal activity is known to quickly trigger the transcription of many genes (Herdegen et al., 1998; O'Donovan et al., 1999; West et al., 2011). Seeking to detect gene transcription in response to diverse conditions of neuronal activity in different brain regions, we adapted the SLAM-seq protocol in vivo by injecting 4sU directly into the brain region of interest. The stereotactic injection method was used to ensure the target delivery of 4sU, a nucleotide analog that can be imported into animal cells by equilibrate nucleoside transporters (Yao et al., 2011), in selected brain regions (See Methods for details). To estimate the 4sU incorporation rate in brain cells, we first injected 4sU (100 mM, dissolved in 10% DMSO, 1uL) into the mouse auditory cortex (Fig. 1a, Supp. Fig. 1a), and performed biotin dot blot assay with mRNAs extracted from the cortex at 2 h, 8 h, and 24 h after 4sU injection (Radle et al., 2013). The labeling strength increased with labeling time (Fig. 1b), and at 24 h post injection, the labeling is highly efficient. Thus, we determined that 24 h is a sufficient duration to allow 4sU diffusion into brain cells to support SLAM-seq.
Fig. 1.
SLAM-seq for mouse brain cells by stereotactic injection of 4sU into the brain.
(a) Schematic showing SLAM-seq procedure, including stereotactic 4sU injection, RNA extraction, iodoacetamide (IAA) treatment, RNA sequencing and T-C conversion detection.
(b) Dot blot assay of RNA extracted from mouse brain labeled with 4sU for different durations. Left to right: biotin-oligo (positive control); RNA samples extracted from mouse auditory cortex with no 4sU injection (negative control); 2 h, 8 h, and 24 h after 4sU injection.
(c) TUNEL staining of brain slices containing auditory cortex (ACx). Top panels: ACx injected with DMSO. Middle panels: ACx injected with 4sU. Bottom panels: DNase I-treated ACx as the positive control. Scale bar, 50 μm.
(d) Correlation of RNA levels extracted from auditory cortex samples with (denoted as 4sU+, n = 3) or without (denoted as 4sU-, n = 3) 4sU injections. ρ, spearman correlation coefficient.
To evaluate the extent of tissue damage potentially caused by the injection of 4sU, we used TUNEL (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling) staining to assess cell death in the injected brain region (Gorczyca et al., 1993). We injected 4sU (100 mM, dissolved in 10% DMSO, 1uL) and DMSO (10%, 1uL) into the mouse auditory cortex and amygdala, and 24 h later, extracted the mouse brains for slicing and TUNEL staining (Methods). The glass pipettes used for injection were coated with DiI to mark the injection sites (Supp. Fig. 1a). No dead cells were observed in brain slices near the injection sites for both 4sU and DMSO injections, suggesting that neither 4sU nor DMSO induced detectable cell death or tissue damage in the injected brain regions (Fig. 1c, Supp. Fig. 1b).
We also assessed whether the presence of 4sU had any impact on the transcriptome by performing 150bp paired-end sequencing of total mRNA extracted from the isolated mouse auditory cortex and amygdala at 24 h after stereotactic 4sU or DMSO injections (Supp. Figs. 2a and 2b). By comparing the read counts for all genes (Herzog et al., 2017), we did not observe any significant difference in gene expression levels between 4sU (denoted as 4sU+) and DMSO (denoted as 4sU-) groups (Table 1, Table 2, Fig. 1d, Supp. Fig. 2c). These findings support that local injection of 4sU into specific brain regions enables efficient incorporation into transcription in brain cells, without obviously affecting the gene expression.
Table 1.
RNA sequencing data quality and yield.
| Brain Region | Stress Group | Labeling Group | Sample ID | A260/280 | concentration(ng/μL) | RIN | Yield(Gbases) |
|---|---|---|---|---|---|---|---|
| amygdala | Baseline | 4sU | PM17 | 2.15 | 134.1 | 8.9 | 6.622 |
| PM21 | 2.14 | 137.2 | 8.4 | 6.349 | |||
| PM22 | 2.09 | 254.5 | 8.7 | 6.55 | |||
| PM23 | 2.14 | 218.4 | 8.6 | 6.357 | |||
| PM24 | 2.14 | 109.6 | 8.8 | 6.367 | |||
| DMSO | D15 | 2.07 | 149.4 | 9.3 | 7.04017 | ||
| D16 | 2.08 | 168.2 | 9.1 | 7.2522 | |||
| D17 | 2.09 | 135.7 | 9.4 | 7.11745 | |||
| D18 | 2.09 | 150.9 | 9.5 | 6.6639 | |||
| Acute Stress | 4sU | PP3 | 2.17 | 98.1 | 9.1 | 7.2252 | |
| PP7 | 2.15 | 55 | 9 | 7.438 | |||
| PP14 | 2.12 | 176.9 | 8.8 | 7.99 | |||
| PP15 | 2.13 | 153.7 | 8.7 | 7.343 | |||
| PP16 | 2.13 | 89.6 | 8.8 | 6.935 | |||
| DMSO | AD3 | 2.09 | 161.5 | 9.1 | 7.23782 | ||
| AD4 | 2.05 | 134.9 | 9.1 | 6.68873 | |||
| AD5 | 2.03 | 85.7 | 9.2 | 6.83395 | |||
| AD6 | 2.04 | 82.6 | 9.2 | 7.09875 | |||
| AD8 | 2.1 | 154.3 | 9.1 | 7.281 | |||
| Chronic Stress | 4sU | U2 | 2.08 | 170 | 8.8 | 6.97511 | |
| U3 | 2.09 | 161.1 | 8.9 | 6.63578 | |||
| U4 | 2.11 | 148.5 | 9 | 6.97036 | |||
| U5 | 2.11 | 202.9 | 8.7 | 6.77345 | |||
| DMSO | CD1 | 2.09 | 146.2 | 9.5 | 6.81991 | ||
| CD3 | 2.11 | 154.3 | 9.5 | 7.21164 | |||
| CD5 | 2.11 | 156.5 | 9.2 | 7.06909 | |||
| auditory cortex | Baseline | 4sU | Z5L | 2.08 | 121.3 | 8.1 | 8.0123 |
| Z4L | 2.06 | 76.4 | 8.1 | 7.4506 | |||
| Z13L | 2.06 | 161.7 | 7.9 | 8.4367 | |||
| – | Z8L | 2.08 | 212.2 | 8.2 | 7.747 | ||
| S1 | 2.1 | 383 | 8.3 | 7.675 | |||
| S3 | 2.08 | 423 | 8.5 | 6.2753 |
Table 2.
Group comparisons.
| Figure ID | Group comparison |
|---|---|
| Fig. 1d | auditory cortex, baseline, 4sU + vs. DMSO |
| Supp Fig. 2c | amygdala, baseline, 4sU vs. DMSO |
| Fig. 2b, c, 2d | amygdala, baseline, 4sU vs. DMSO |
| Fig. 3b, Supp Fig. 4a | amygdala, acute stress, 4sU vs. DMSO |
| Fig. 3c | amygdala, acute stress (4sU) vs. baseline (4sU) |
| Fig. 3d | amygdala, acute stress (4sU-normalized) vs. baseline (4sU-normalized) |
| Supp Fig. 4c | amygdala, acute stress (DMSO) vs. baseline (DMSO) |
| Fig. 4b, Supp Fig. 5a | amygdala, chronic stress, 4sU vs. DMSO |
| Fig. 4c | amygdala, chronic stress (4sU) vs. baseline (4sU) |
| Fig. 4d | amygdala, chronic stress (4sU-normalized) vs. baseline (4sU-normalized) |
| Supp Fig. 5b | amygdala, chronic stress (DMSO) vs. baseline (DMSO) |
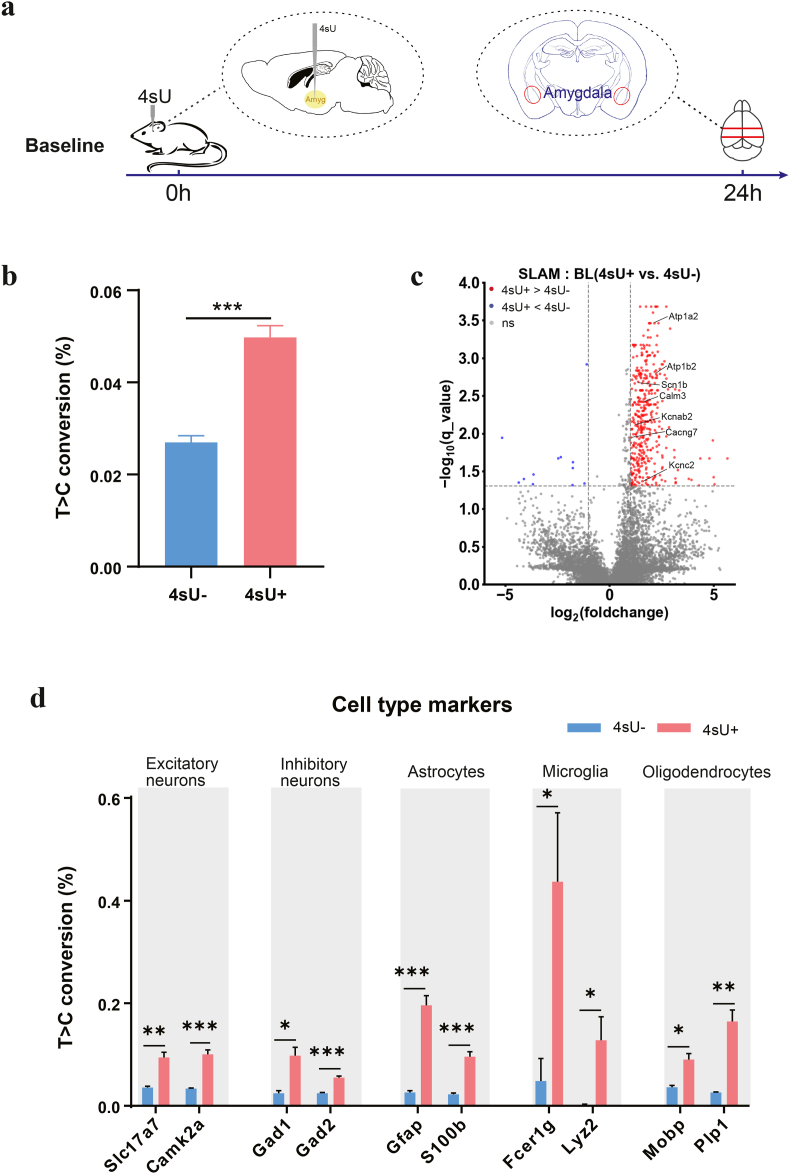
2.2. Identify 4sU-incorporated mRNA with SLAM-seq under basal conditions
Recently, in addition to cultured cells (Alalam et al., 2022; Muhar et al., 2018), SLAM-seq has also been applied in vivo by injecting 4sU into zebrafish embryo (Bhat et al., 2023), or injecting 4-thiouracil (4-TU) intraperitoneally into transgenic mice expressing UPRT in selected cell types (Matsushima et al., 2018). Given that local injection of 4sU into the mouse brain has never been reported, we needed to empirically determine whether the delivered 4sU can be incorporated into the nascently transcribed RNA molecules. We injected 4sU (100 mM, 1uL, dissolved in 10% DMSO) and DMSO (10%, 1uL) into the mouse amygdala 24 h prior to tissue dissection and RNA extraction (Fig. 2a), to establish a baseline (BL) for nascent RNA transcription in normal behaving mice. We then subjected the aforementioned total RNA extracts to iodoacetamide (IAA) treatment (Methods, Supp. Fig. 3a), to covalently attach a carboxyamidomethyl group to 4sU by nucleophilic substitution (Herzog et al., 2017), and then performed 150bp paired-end RNA-seq (Supp. Fig. 3b). During the reverse transcription step of RNA-seq library preparation, a guanine (G), instead of an adenine (A), is base paired to an alkylated 4sU, leading to the thymine (T) to cytosine (C) base conversion (T > C conversion) at the corresponding T position in mRNA (Herzog et al., 2017). To control for the background T > C conversion rate without 4sU, RNA obtained from DMSO-injected amygdala was subjected to the same procedures before RNA-seq.
Fig. 2.
Identifying 4sU-labeled genes with SLAM-seq and match to different cell types
(a) Time line showing stereotactic 4sU injection and brain extraction under baseline (BL) condition.
(b) T > C conversion rates of total mRNA extracted from DMSO (n = 5) and 4sU (n = 4) injected amygdala under baseline condition. Student's t-test, ∗∗∗p < 0.001.
(c) Volcano plots showing T > C conversion rates of transcripts under baseline, with or without 4sU injection. Red dots represent transcripts with significantly higher T > C conversion rates in the 4sU + group (FDR <0.05 and log2FC > 1); blue dots represent transcripts with significantly lower T > C conversion rates in the 4sU + group (FDR <0.05 and log2FC < −1).
(d) T>C Conversion rates of marker genes for multiple neuron and glial cell types. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We calculated the T > C conversion rates for each gene in both the 4sU-injected and DMSO-injected samples using SLAM-DUNK software (Neumann et al., 2019). At the whole transcriptome level, the average T > C conversion rate of the 4sU-injected group was significantly higher than that of DMSO-injected group (0.027 ± 0.0014% for DMSO, 0.050 ± 0.0025% for 4sU, Table 1, Table 2, Fig. 2b), indicating that the injected 4sU was incorporated into transcription. We identified 413 genes with significantly higher T > C conversion rates in the 4sU group than in the DMSO group (beta-binomial test, Fig. 2c), which can be plausibly interpreted as genes with higher turnover. Note that these mice were not subjected to any behavioral or physiological perturbations, therefore, the 413 differentially 4sU-labeled genes had higher turnover rates under normal baseline conditions. These genes included sodium channels (Scn1b), potassium channels (Kcnab2, Kcnc2), cation transport ATPases (Atp1a2, Atp1b2), calcium channels (Cacng7) and Calmodulin (Calm3), all of which are essential for maintaining basic neuronal functions (Goaillard et al., 2021).
2.3. Determine the 4sU labeling efficiency in distinct cell types
Identification of genes encoding neuron-specific ion channels indicated that 4sU was successfully incorporated into transcription of genes in the amygdala neurons. To additionally evaluate the efficiency of 4sU incorporation into the transcription of other cell types in amygdala, we cross-referenced the 413 differentially 4sU-labeled genes with known marker genes of major cell types, as reported in single-cell RNA sequencing studies (Hochgerner et al., 2023; Yao et al., 2023). We found that the 413 differentially labeled genes under baseline conditions covered marker genes of excitatory (Slc17a7, Camk2a) and inhibitory neurons (Gad1, Gad2), astrocytes (Gfap, S100b), microglia (Fcer1g, Lyz2) and oligodendrocytes (Mobp, Plp1). The T > C conversion rates of these genes were significantly higher in 4sU-injected than DMSO-injected amygdala (Fig. 2d). These results indicate that 4sU has been imported into major neuronal and glial cell types of the amygdala, and could be effectively incorporated into the transcription of these cells to signal RNA turnover.
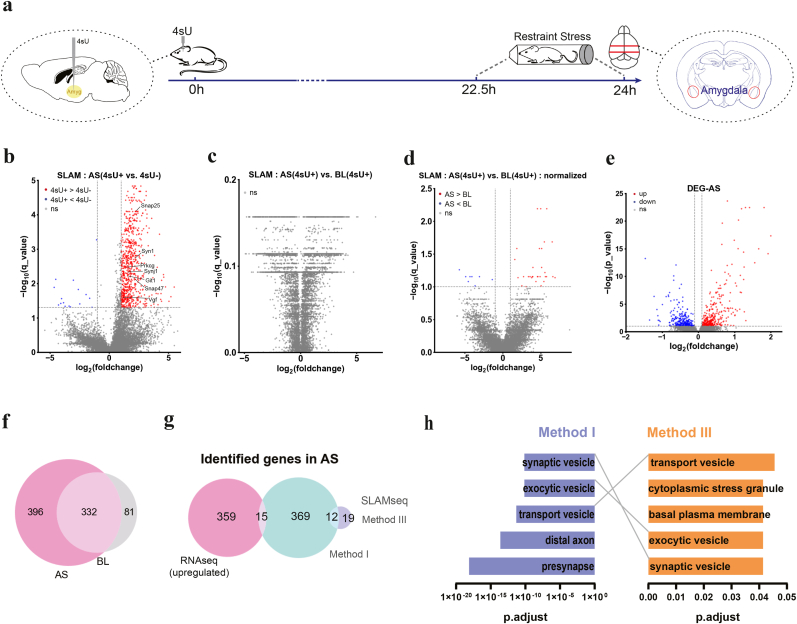
2.4. Transcriptome dynamics of the amygdala under acute stress identified by SLAM-seq
The amygdala has been extensively implicated in stress responses (Roozendaal et al., 2009; Zhang et al., 2019). To identify genes with higher turnover under acute stress (AS), we subjected 4sU- and DMSO-injected mice to 1.5 h of restraint stress, as 1 h and 2 h of body restraint was commonly used for induction of acute stress in rodents (Li et al., 2012; Satoh et al., 2006; Chen et al., 2024; Kuti et al., 2022). Immediately after the restraint, we isolated the amygdala and extracted RNAs (Methods, Fig. 3a), followed by IAA treatment and subsequent 150bp paired-end RNA-seq.
Fig. 3.
Transcriptome dynamics of mouse amygdala under acute stress.
(a) Time line of acute stress experiment, including time points for 4sU injection and brain extraction.
(b) Volcano plots showing T > C conversion rates of transcripts under AS, 4sU-injected (4sU+) vs. DMSO-injected (4sU-), related to Method I. Red dots represent transcripts with significant higher T > C conversion rates in the 4sU + group (FDR <0.05 and log2FC > 1); blue dots represent transcripts with significant lower T > C conversion rates in the 4sU + group (FDR <0.05 and log2FC < −1).
(c) Volcano plots showing T > C conversion rates of transcripts, AS 4sU + vs. BL 4sU+, Method II. No significant gene was detected.
(d) Volcano plots showing T > C conversion rates of transcripts, normalized AS 4sU + vs. normalized BL 4sU+, Method III. Red dots represent transcripts with significant higher T > C conversion rates in the AS group (FDR <0.1 and log2FC > 1); blue dots represent transcripts with significant lower T > C conversion rates in the AS group (FDR <0.1 and log2FC < −1).
(e) Volcano plots showing differentially expressed genes (DEGs) under AS compared to BL. Red dots represent significantly AS-upregulated genes (FDR <0.1 and log2FC > 0.1); blue dots represent significantly AS-downregulated genes (FDR <0.1 and log2FC < −0.1). ns, no significant difference; up, significantly upregulated; down, significantly downregulated.
(f) Venn diagram showing intersections of 4sU-labeled genes identified by SLAM-seq under AS (AS 4sU + vs. AS 4sU-) and BL (BL 4sU + vs. BL 4sU-) conditions.
(g) Venn diagram showing intersection of genes identified by SLAM-seq and RNA-seq. (h) GO term analysis for genes detected by SLAM-seq Method I and Method III. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To identify genes with higher turnover under AS, we used three analysis methods. Method I, we compared T > C conversion rates between 4sU and DMSO-injected amygdala samples after AS treatment (Table 1, Table 2, Supp. Fig. 4a). We identified 728 differentially 4sU-labeled genes with significantly higher T > C conversion rates in the 4sU-injected group (FDR <0.05, log2FC > 1, Table 1, Table 2, Fig. 3b. FDR: false discovery rate. FC: fold change). Nearly half of the AS 4sU-enriched genes (332 out of 728) were also identified at BL condition (Fig. 3f), including known housekeeping genes such as Sdha, Ywhaz, Gfap (Kosuth et al., 2019). These genes had high turnover rates regardless of stress conditions, indicating their important roles in maintaining normal cellular functions. We performed a gene ontology (GO) analysis on the remaining 396 AS-specific genes, and found enrichment for terms closely associated with synaptic functions (Fig. 3h, Supp. Fig. 4b).
As identifying genes by intersecting lists of significant genes may induce artifacts, we performed a second analysis (Method II) by directly comparing T > C conversion rates for each gene between the 4sU-injected AS and 4sU-injected BL samples (Methods). We found no significant genes using Method II (Fig. 3c). By examining the T > C conversion rates in the DMSO-injected samples, we found that the T > C conversion rates for each gene show poor correlation in BL vs AS groups (Supp Fig. 4c). This result suggested that the baseline (DMSO-injected) T > C conversion rates of many genes are influenced by AS, likely attributed to altered expression levels, RNA turnover, endogenous RNA modifications, and spontaneous T > C mutation rates. To correct for the baseline T > C conversion rates under different stress conditions, we performed a third analysis (Method III), in which we normalized the T > C conversion rate of each gene in the 4sU-injected samples by its corresponding mean T > C conversion rate in the DMSO-injected samples, before conducting the AS vs. BL comparison. This analysis yielded 31 genes with significantly higher T > C conversion rates in 4sU-injected AS samples compared to 4sU-injected BL samples (Fig. 3d). We performed a GO analysis on these genes, and again found enrichment for terms associated with synapses (Fig. 3h, Supp. Fig. 4e), supporting the notion that AS had a strong influence on synaptic functions.
2.5. Differentially expressed genes under acute stress identified by RNA-seq
By conducting a bulk RNA-seq analysis using DESeq2 software package (Love et al., 2014), we identified differentially expressed genes (DEGs) in AS condition compared to BL (Table 2). In total, 374 significantly upregulated and 279 downregulated genes were identified (FDR <0.1, Log2FC > 0.1 or Log2FC < −0.1, Table 1, Table 2, Fig. 3e). Only 15 overlapping genes were identified by intersecting these AS-upregulated genes with genes identified using SLAM-seq analysis method I, and none with method II or III (Fig. 3g, Supp. Fig. 4d). The overlapping genes include known stress-related genes such as Cldn11 and Etnppl (Flati et al., 2020; Kolbasi et al., 2021). These results suggest that SLAM-seq and RNA-seq are highly complementary in identifying genes responsive to acute stress.
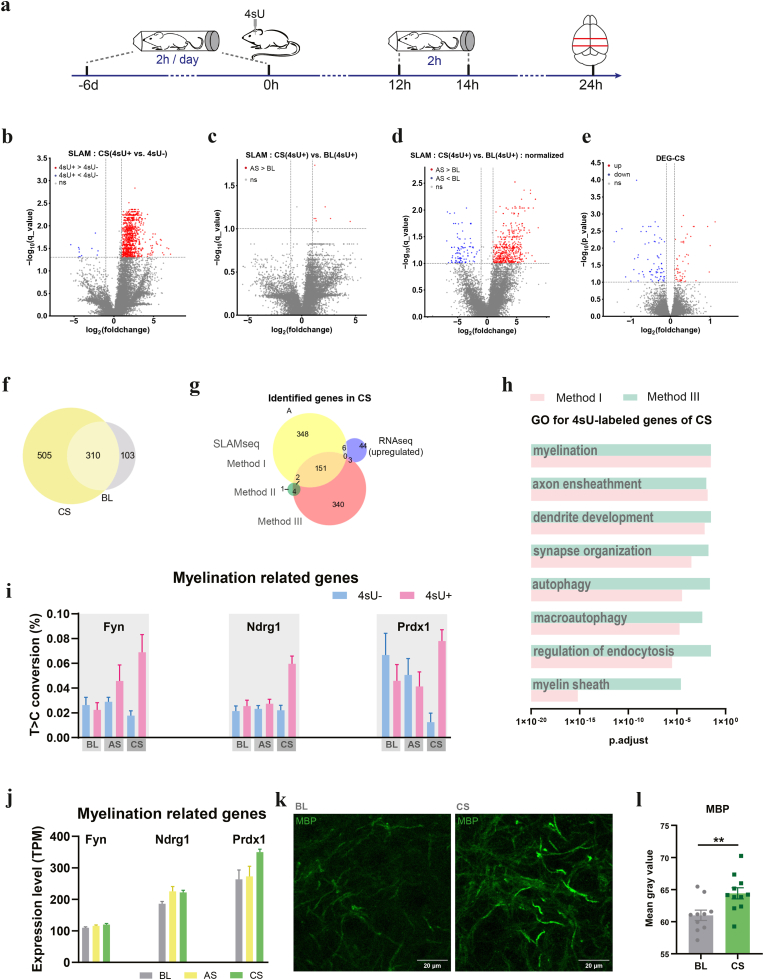
2.6. Transcriptome dynamics of amygdala under chronic stress identified by SLAM-seq
Chronic stress (CS) poses prolonged impact in the brain (McEwen, 2017). For chronic stress experiments, we subjected mice to 7 days of body restraint, 2 h each day (Mozhui et al., 2010; Chen et al., 2018) (Methods, Fig. 4a). To examine the long-lasting transcriptomic changes under CS, we extracted the amygdala at 10 h after the final restraint stress session on day 7. We reasoned that by shifting 10 h, we reduced the possibility of detecting acute changes induced by the last session of restraint stress. 4sU was injected into the amygdala 24 h before brain extraction, same as in BL and AS experiments.
Fig. 4.
Transcriptome dynamics of mouse amygdala under chronic stress.
(a) Time line of chronic stress experiment, including time points for 4sU injection and brain extraction.
(b) Volcano plots showing T > C conversion rates of transcripts, CS 4sU + vs. CS 4sU-, related to Method I. Red dots represent transcripts with significant higher T > C conversion rates in the 4sU + group (FDR <0.05 and log2FC > 1); blue dots represent transcripts with significant lower T > C conversion rates in the 4sU + group (FDR <0.05 and log2FC < −1).
(c) Volcano plots showing T > C conversion rates of transcripts, AS 4sU + vs. BL 4sU+, Method II. Red dots represent transcripts with significant higher T > C conversion rates in the 4sU + group (FDR <0.1 and log2FC > 1); blue dots represent transcripts with significant lower T > C conversion rates in the 4sU + group (FDR <0.1 and log2FC < −1).
(d) Volcano plots showing T > C conversion rates of transcripts, normalized AS 4sU + vs. normalized BL 4sU+, Method III. Red dots represent transcripts with significant higher T > C conversion rates in the CS group (FDR <0.1 and log2FC > 1); blue dots represent transcripts with significant lower T > C conversion rates in the CS group (FDR <0.1 and log2FC < −1).
(e) Volcano plots showing T > C conversion rates of transcripts under CS, with or without 4sU injection. Red dots represent transcripts with significant higher T > C conversion rates in the CS group (FDR <0.1 and log2FC > 1); blue dots represent transcripts with significant lower T > C conversion rates in the CS group (FDR <0.1 and log2FC < −1).
(f) Venn diagram showing intersections of 4sU-labeled genes identified by SLAM-seq under AS (AS 4sU + vs. AS 4sU-) and BL (BL 4sU + vs. BL 4sU-) conditions.
(g) Venn diagram showing intersection of genes identified by SLAM-seq and RNA-seq.
(h) GO term analysis for genes detected by SLAM-seq Method I and Method III.
(i) T > C conversion rates for myelination related genes detected by SLAM-seq under BL, AS and CS.
(j) Gene expression levels for the myelination related genes in (i).
(k) Example images showing immunostaining of myelin basic protein (MBP) in the amygdala of control (BL) and chronically stressed (CS) mice. Scale bar, 20 μm.
(l) Increased MBP signal intensity in CS mice (n = 5) compared to BL mice (n = 5), measured by mean grey value. Student's t-test, ∗∗p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We again used Method I, II, and III (same as in the AS sections) to identify genes with higher turnover under CS. Using Method I, we identified 815 differentially 4sU-labeled genes under CS (Table 1, Table 2, Fig. 4b, Supp. Fig. 5a), of which 310 overlapped with BL (Fig. 4f). Using Method II, by directly comparing T > C conversion rates for each gene between the 4sU-injected CS and 4sU-injected BL samples, we found only 7 significant genes (Fig. 4c). As the T > C conversion rates for each gene are poorly correlated in BL vs CS groups (Supp Fig. 5b), we then used Method III, normalizing the T > C conversion rate of each gene in the 4sU-injected CS samples before comparing CS and BL. This analysis identified 494 genes with significantly higher T > C conversion rates in 4sU-injected CS samples compared to 4sU-injected BL samples (Fig. 4d). DESeq2 analysis detected 53 significantly up-regulated and 11 down-regulated genes for CS, compared to BL (FDR <0.1, Log2FC > 0.1 or Log2FC < −0.1, Table 2, Fig. 4e). Intersection of genes identified by RNA-seq and SLAM-seq again revealed very few overlapping genes (Fig. 4g, Supp. Fig. 5c).
We performed two separate GO analyses on the 505 genes identified using SLAM-seq Method I, and 494 genes identified using SLAM-seq Method III. The two GO analyses yielded highly consistent results. In addition to terms associated with neuronal functions such as “synapse organization”, there is also enrichment for terms closely linked to glial cells, especially oligodendrocytes and microglia (Fig. 4h). These terms include “myelination” and “axon ensheathment”, which are tightly linked to myelination (Supp. Fig. 5d), suggesting that CS leads to increased myelination in the amygdala.
2.7. Validation of SLAM-seq results by quantitative immunofluoresence
Recent studies suggested that elevated neuronal and axonal activity could trigger myelination (Mount et al., 2017; Fields, 2015), a phenomenon known as “activity-dependent myelination”. SLAM-seq detected an increased turnover of glutamate and GABA receptor genes (Supp. Fig. 6), indicating altered neuronal activity of excitatory and inhibitory neurons in the stressed amygdala (Zhang et al., 2018). In line with these results, the enriched GO terms for CS condition detected by SLAM-seq included “myelination” and “axon ensheathment”. Myelination-related genes, such as Fyn, Ndrg1, and Prdx1, were identified by SLAM-seq (Fig. 4i), These genes were not detected by RNA-seq, but exhibited a trend of elevated transcription levels under CS (Fig. 4j). To examine whether the increased turnover of these genes is indicative of increased myelination in the amygdala, we stained brain slices of CS mice and control mice with a myelin marker, the myelin basic protein (MBP), and compared the immunofluorescence signal intensity between the two groups (Fig. 4k). We found that MBP signal was indeed significantly stronger in the chronically stressed amygdala than the control (p < 0.01, Student's t-test, Fig. 4l), validating the SLAM-seq results that chronic stress led to increased myelination in the mouse amygdala.
3. Discussion
By directly injecting 4- thiouridine (4sU) into the amygdala followed by SLAM-seq, we were able to identify newly transcribed genes under both acute and chronic stress. By cross-referencing these genes with known cell type-specific marker genes, we found that the injected 4sU was integrated into the transcription of major subtypes of neurons and glial cells in the amygdala. Analyses revealed that chronic stress had a strong impact on transcription genes associated with myelination, in line with heightened activity of the amygdala (Zhang et al., 2018). Thus, by applying SLAM-seq to the amygdala, we generated a rich dataset which can be used in future studies to dissect the functions of genes related to stress responses.
The transcriptional networks in neurons are dynamically modulated by neuronal activities (Frey et al., 1997; Rangaraju et al., 2017), which may lead to changes in RNA turnover, without altering the total mRNA of these genes (Lee et al., 2021). SLAM-seq, a metabolic RNA labeling method, is an approach to gain insights into the dynamic transcriptional regulation (Alalam et al., 2022; Muhar et al., 2018; Bhat et al., 2023). A previously published method, SLAM-ITseq, extended SLAM-seq to mouse tissues by crossing Cre-dependent UPRT mice with Cre lines (Matsushima et al., 2018). This method requires usage of transgenic animals, and the cell types that can be investigated are restricted by available Cre lines. Instead of relying on Cre-dependent UPRT expression for cell-specific labeling, we used stereotactic 4sU injection to target all cell types in selected brain regions. Our data showed that 4sU can be effectively imported into major cell types in the brain (Yao et al., 2011), and incorporated into the transcription, without causing detectable tissue damage or cell death. This is a simplified method that can be applied to any brain region in mice and possibly in other species, without the need for transgenic animals.
We used this method to investigate the RNA dynamics of a key brain region involved in stress conditions, the amygdala, under acute and chronic restraint stress. We found partial overlap in genes detected by SLAM-seq and RNA-seq, and most of the overlapped genes identified under CS condition had been implicated in chronic stress by previous studies (Gautam et al., 2015; Kuehner et al., 2023; Mingardi et al., 2023; Issler et al., 2014; Jaffe et al., 2022; Ochi et al., 2023; McMurray et al., 2018). Notably, SLAM-seq identified an ensemble of genes related to myelination, which were not detected by RNA-seq. Immunostaining of myelin-specific protein revealed increased myelination following CS treatment, confirming the SLAM-seq results, suggesting that SLAM-seq is a complementary method to RNA-seq.
Alternative methods, such as nuclear RNA sequencing (nuRNA-seq) and ribosome-bound RNA sequencing (riboRNAseq), had also been used to profile activity-driven transcriptome dynamics in neurons (Fernandez-Albert et al., 2019). These methods specialized on detecting nuclear RNAs or RNAs undergoing translation, while SLAM-seq could also capture cytoplasmic RNAs. For the same reason, SLAM-seq lacks these specificities of nuRNA-seq and riboRNA-seq. Along with the development of single-cell RNA sequencing, single-cell SLAM-seq (scSLAM-seq) had been developed to probe transcription activity in single cells in vitro (Erhard et al., 2019). It is possible to combine scSLAM-seq with in vivo stereotaxic injection of 4sU to investigate single-cell resolution transcriptome dynamics, providing insights into the heterogeneity of RNA dynamics in individual cells, which the current method is unable to capture.
In summary, we provided a dataset of stress-responsive genes, and confirmed that CS can induce increased myelination in the amygdala. Future studies will be needed for uncovering the functions of other genes identified in this study.
4. Materials and methods
4.1. Animals
C57BL/6 mice were purchased from Slac Laboratory Animals. Mice were housed and bred in a 12 h light-dark cycle (7 a.m. - 7 p.m. light) in the animal facility of ShanghaiTech University. Eight-to ten-week-old male mice were used for the experiments. They were kept 5 per cage, always with free access to water and food. All procedures were approved by the Animal Committee of ShanghaiTech University.
4.2. 4sU injection
The 4sU powder (APExBIO, C3722) was dissolved in dimethyl sulfoxide (DMSO) to reach a concentration of 1 M, and the solution was aliquoted, frozen, and stored at −20 °C. Before injection, the solution was 1:10 diluted with phosphate-buffered saline (PBS). For injection, mice were anesthetized with isoflurane (1.5–2 %) and positioned in a stereotaxic frame (RWD Life Science Co.). Body temperature was maintained at 37 °C using a heating pad. 4sU solution (1 μL) was injected using a glass micropipette with a tip diameter of 15–20 μm, through a small skull opening (<0.5 mm2), with a microinjector (Nanoject III), at a rate of 1 nL/s. The following coordinates were used (in mm relative to Bregma): −1.1 AP, 3.25 ML, −3.95 DV and −2.4 AP, 3.2 ML, −4.1 DV for BLA; −0.85 AP, 2.8 ML, −4.1 DV for CEA; −2.2 AP, 4.4 ML, −1.1 DV for auditory cortex. In baseline, acute stress and chronic stress experiments, 4sU was injected into bilateral amygdala 24 h before brain extraction. After injection, mice were kept on the heating pad until they recovered from the anesthesia, and then they were returned to the home cage.
4.3. TUNEL staining
The TUNEL assay was performed following the one-step method outlined in the Promega TUNEL Cell Apoptosis Detection Kit (meilunbio, MA0223). Mice were intracranially injected with 1 μL of 100 mM 4sU into the auditory cortex (−2.2 AP, 4.4 ML, −1.1 DV) or amygdala (−1.1 AP, 3.25 ML, −3.95 DV), as described above. Before injection, the glass micropipette was dipped into the DiI (Beyotime, C1036) solution to mark the injection site. After 24 h, mice were deeply anesthetized and transcardially perfused with PBS and paraformaldehyde (PFA), followed by brain extraction. The extracted brains were postfixed in PFA at 4 °C for 12 h and embedded in agarose for vibratome sectioning at 70 μm per slice. Brain slices with DiI signal were collected for TUNEL staining, in which terminal deoxynucleotidyl transferase (TdT) labels DNA strand breaks with fluorophore-labeled nucleotides. The brain slices were fixed in 4% paraformaldehyde at room temperature for 30 min and then washed twice with PBS. Then, 20 μg/ml proteinase K was used for permeabilization for 10 min and then washed 3 times with PBS. For the positive control, the brain slices were incubated in 1 × DNase I Buffer and a buffer containing 5 μg/ml DNase I for 5 min and 10 min respectively, washed with PBS. The brain slices were incubated in TUNEL detection solution (TdT Enzyme (10 × ): FITC-12-dUTP Labeling Mix = 1:9) in the dark for 60 min and then washed 3 times with PBS. Stained brain slices were mounted on glass slides and imaged using confocal fluorescence microscopy (Leica).
4.4. Tissue RNA extraction
Mice were deeply anesthetized and transcardially perfused with cold PBS. Each mouse was then quickly decapitated, and the head was immersed in ice-cold PBS for rapid cooling for 10 s. The mouse brain was then carefully extracted using forceps, placed in a pre-chilled mouse brain mold (RWD Life Science Inc.), and a 2 mm-thick brain tissue block containing the target region (auditory cortex or amygdala) was sliced using a razor blade. The brain slice was then placed on a 6 mm Petri dish and dropped into liquid nitrogen briefly for 7–8 s. Then, the auditory cortex or amygdala was cut out using a 15 G needle, and promptly transferred to cryotubes for rapid freezing in liquid nitrogen, and subsequent storage at −80 °C.
RNA extraction from mouse brain tissue was performed using the Trizol method. The frozen tissue in cryotubes was thawed, and 1 mL of Trizol (MagZol Reagent, R4801-02) was added to each tube. Tissue homogenization was performed using an electric homogenizer, with a 10-s pause every 10 s to prevent overheating. The mixture was then left at room temperature for 5 min for tissue digestion, and the liquid was transferred to 1.5 mL centrifuge tubes. To each tube, 200 μL of chloroform (one-fifth of the Trizol volume) was added, and after thorough vortex mixing, the mixture was left at room temperature for 15 min. The mixture was then centrifuged at 12,000 rpm for 15 min at 4 °C. Then, the supernatant containing RNA was carefully aspirated into new tubes, an equal volume of pre-cooled isopropanol was added, along with 1/100 volume of 10 mM DTT (final concentration 0.1 mM) and 1 μL glycogen (20 mg/mL). After thorough mixing, the mixture was left at −20 °C for 1 h for RNA precipitation. Subsequently, it was centrifuged at 12,000 rpm for 30 min at 4 °C, resulting in a white gel-like RNA substance at the bottom. The RNA pellet was washed once with pre-cooled 75% ethanol (containing 5 μL 10 mM DTT), and after removing the supernatant, the RNA pellet was air-dried for approximately 3–5 min. Finally, the RNA was dissolved in 20 μL RNase-free water, and the concentration was determined using Nanodrop 2000.
4.5. Dot blot assay
Four RNA samples with different 4sU injection conditions were used in this experiment: 1) RNA extracted from cortex with no 4sU injection, 2) RNA extracted from cortex dissected 2 h after 4sU injection, 3) RNA extracted from cortex dissected 8 h after 4sU injection, 4) RNA extracted from cortex dissected 24 h after 4sU injection.
10 μL 10 × BB buffer (100 mM Tris–HCl, pH 7.4, 10 mM EDTA), 1 μg RNA, and 20 μl 1 mg/ml Biotin-HPDP buffer (Thermo, 21341) were mixed, and incubated at room temperature in the dark for 1.5 h. RNA was purified using the Easy Pure RNA Purification Kit (TransGen, ER701-01). RNA was diluted to 200 ng/μL RNA using ice-cold dot blot binding buffer (10 mM NaOH, 1 mM EDTA) and 5 μL was loaded per dot on the pre-incubated nylon membrane. The membrane was dried at room temperature and irradiated with 254 nm ultraviolet wavelength at a distance of about 5–10 cm from the membrane for 5 min. The samples were processed according to the Chemiluminescent Biotin-labeled Nucleic Acid Detection Kit (Beyotime, D3308) and photographed.
4.6. Restraint stress experiment
Mice were acclimated in the animal facility for at least one week before the initiation of restraint stress experiments.
4.6.1. Acute stress group
Each mouse was carefully removed from its home cage and put in a Falcon 50 mL centrifuge tube, with 8 small ventilation holes drilled on the tube walls. Once the mouse entered the tube, the rear was sealed with a clean tissue, and the tube was capped with the lid, preventing forward or backward movements. Following the 1.5-h restraint, the mouse was perfused and the brain was extracted.
4.6.2. Chronic stress group
Each mouse was carefully removed from its home cage and put in a 50 mL tube, as described in the acute stress experiment. Daily restraint sessions lasting 2 h (from 10:00 a.m. to 12:00 p.m.) were conducted for one week. On the 7th day, 10 h after the final restraint session ended, the mouse was perfused and the brain was extracted.
4.6.3. Baseline group
Mice did not undergo any restraint stress, and remained in their home cages until they were perfused.
4.7. IAA treatment
Extracted RNA was reacted with IAA to alkylate the thiol group following (Herzog et al., 2017). For a 50 μL reaction: 2–3 μg RNA, 25 μL DMSO, 5 μL 100 mM iodoacetamide (IAA), and 5 μL 500 mM sodium phosphate (pH = 8), and RNase-free water. IAA was freshly prepared and diluted with anhydrous ethanol. The prepared mixture was incubated at 50 °C for 15 min, followed by the addition of 1 M dithiothreitol (DTT) to terminate the reaction. Then, 125 μL anhydrous ethanol, 1 μL glycogen (20 mg/mL), and 5 μL sodium acetate (3 M, pH = 5.2) were added to the reaction system. After thorough mixing, the solution was left at −80 °C for 30 min, followed by centrifugation at 12,000 rpm for 30 min at 4 °C, resulting in a white gel-like RNA substance at the bottom. The RNA pellet was washed once with pre-cooled 75% ethanol. After removing the supernatant, the RNA pellet was air-dried for approximately 3–5 min, and dissolved in 20 μL RNase-free water.
4.8. RNA sequencing
The IAA-methylated RNA was stored in a −80 °C freezer, and library construction and sequencing were conducted by Suzhou Geneseeq Technology Inc. The libraries were sequenced using the Illumina NovaSeq for 150-base pair paired-end sequencing, generating approximately 6 GB of sequencing data for each RNA sample.
4.9. Bioinformatics analysis
4.9.1. Preprocessing of RNA-seq data
Raw data obtained from RNA-Seq was preprocessed using Cutadapt v3.1. Adapter sequences in paired-end sequencing were eliminated, and sequences shorter than 15 nucleotides were discarded. The preprocessed data was then aligned to the NCBI reference mouse genome (mm10) using Hisat2 v2.1.0. The resulting alignment data in SAM format was converted to BAM format using Samtools v1.9.
4.9.2. SLAM-seq analysis
The software SLAM-DUNK v0.2.4 (tneumann.github.io/slamdunk/), designed for SLAM-Seq analysis, was used to compute the T > C mutation counts for each sample (Neumann et al., 2019). Annotation data for 3′ UTR regions were obtained from the RefSeq and Ensembl databases.
To identify significantly 4sU-labeled transcripts in BL, AS and CS: Firstly, genes with zero T read counts were excluded. Secondly, only genes with read counts >5 in all samples were kept for further analyses. Statistical analysis was performed on 4sU + vs. 4sU- (BL 4sU + vs. BL 4sU-; AS 4sU + vs. AS 4sU-; CS 4sU + vs. CS 4sU-), using the beta-binomial test (ibb R package, version 13.06), as described in (Pham et al., 2010), following the protocol of SLAM-ITseq (Matsushima et al., 2018, 2019). In short, To control for the false discovery rate (FDR), adjusted P-values were calculated using the Benjamini-Hochberg method.
To identify genes with significantly higher turnover under AS/CS.
Method I: Intersect significantly 4sU-labeled transcripts under AS/CS with significantly 4sU-labeled transcripts under BL to identify AS/CS-specific genes.
Method II: Perform beta-binomial test on AS/CS 4sU + vs. BL 4sU+.
Method III: Firstly, normalize T > C conversion rates of each gene in AS/CS/BL 4sU + dataset by subtracting the mean T > C conversion rates of each gene in the corresponding AS/CS/BL 4sU- dataset, and then convert the conversion rates back to T and T > C counts for beta-binomial test. Secondly, Perform beta-binomial test on the normalized AS/CS 4sU + vs. BL 4sU+.
4.9.3. Differential gene expression (DEG) analysis
Quantification of gene expression was performed using FeatureCounts (v2.0.1). The Transcripts Per Million (TPM) values for each gene were calculated using the formula:
| TPM = RPK/∑RPK×106 |
Here, RPK (Reads Per Kilobase) represents the reads per thousand base pairs, and ∑RPK is the sum of RPK values for all genes in the sample. Differential expression analysis was performed using the DESeq2 package in R 3.6.1. The standard DESeq2 workflow was followed for data normalization, variance estimation, and statistical modeling. Identification of significantly differentially expressed genes was based on the criterion of adjusted P-values <0.05 or <0.1.
4.10. GO term enrichment analysis
List of genes was used as input for metascape (metascape.org/) to perform KEGG pathway enrichment analysis. Gene Ontology (GO) terms for biological processes, cellular components and molecular functions were analyzed with the clusterProfiler package (v4.2.2) in R (v4.1.0). Only gene sets with P < 0.05 and q < 0.05 were considered as significantly enriched.
5. Immunofluorescence
Chronic stressed mice (n = 5) were perfused 2 weeks after the final restraint session ended, with 0.01M PBS and 4% PFA. Control (n = 5) mice were perfused without any stress treatment. Brains were sliced (at 50 μm) using a vibratome (Leica, VT1200S) after post-fixed for 5 d in 4% PFA at 4 °C. Brain slices including the amygdala were permeabilized with 0.4% Triton X-100 in PBS for 30 min, blocked with 5% bovine serum albumin (BSA) in PBS for 1 h at RT, and incubated with rat anti-MBP (1:200; Serotec, MCA409S) in 0.1% Triton X-100 and 1% BSA in PBS for 40 h at 4 °C, then washed with PBS 3 × 10 min. Sections were incubated with DAPI (1:2500) and fluorophore-conjugated secondary antibodies (1:500; abcam, AB150165) in 0.1% Triton X-100 and 1% BSA in PBS for 2h at RT, then washed with PBS 3 × 10 min. Images were acquired at a Leica STELLARIS 8 FALCON, with a HC FLUOTAR L VISIR 25x/0.95 water objective; pinhole was set to 1 AU, pixel size to 113.55 nm × 113.55 nm and z-stack interval to 1 μm.
Image processing and quantification were performed using FIJI. For the quantitative immunofluorescence analysis, the mean grey value of each field of view was first thresholded for identification of myelin structures, and then calculated for 11 optical planes of acquired z-stacks (step 1 μm). For each mouse, the mean grey value is averaged across two sections containing the amygdala, one from each hemisphere.
6. Statistics
The statistical details of experiments including statistical tests used, number of replicates and the number of data points used (n) are denoted in the relevant figures and listed in Table 1. All statistical tests and correlations were analyzed using GraphPad Prism 8.0. All data were presented as mean ± SEM. A p value or FDR of <0.05 was considered statistically significant unless indicated otherwise.
CRediT authorship contribution statement
Dan Zhao: Writing – original draft, Visualization, Methodology, Data curation. Lu Zhang: Writing – original draft, Visualization, Data curation. Yang Yang: Writing – original draft, Supervision, Funding acquisition, Conceptualization.
Data availability
Sequencing data generated for this study and software parameters used for data analysis have been deposited at GEO (GSE274148) and are publicly available as of Aug. 14th, 2024.
Any additional information related to sequencing data used in this paper is available from the corresponding author upon request.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank Drs. Margaret Ho and Dan-Qian Liu for comments on the manuscript. We thank Dr. Hao Wu and Ms. Liang He for assistance in data analysis and experiments. We thank the animal facility of ShanghaiTech for their excellent care of mice. This work was supported by grants from the Ministry of Science and Technology of China (2022ZD0204900), Central Guidance on Local Science and Technology Development Fund (YDZX20233100001002), and ShanghaiTech University Startup Fund to Y.Y.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2024.100688.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data is deposited at GEO (GSE274148) and are publicly available as of Aug. 14th, 2024.
References
- Alalam H., Zepeda-Martinez J.A., Sunnerhagen P. Global SLAM-seq for accurate mRNA decay determination and identification of NMD targets. RNA. 2022;28(6):905–915. doi: 10.1261/rna.079077.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P., et al. SLAMseq resolves the kinetics of maternal and zygotic gene expression during early zebrafish embryogenesis. Cell Rep. 2023;42(2) doi: 10.1016/j.celrep.2023.112070. [DOI] [PubMed] [Google Scholar]
- Caspi A., et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chen C.C., et al. Selective activation of parvalbumin interneurons prevents stress-induced synapse loss and perceptual defects. Mol Psychiatry. 2018;23(7):1614–1625. doi: 10.1038/mp.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., et al. Microglia govern the extinction of acute stress-induced anxiety-like behaviors in male mice. Nat. Commun. 2024;15(1):449. doi: 10.1038/s41467-024-44704-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M., et al. Phasic vs sustained fear in rats and humans: role of the extended amygdala in fear vs anxiety. Neuropsychopharmacology. 2010;35(1):105–135. doi: 10.1038/npp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet E.R., Joëls M., Holsboer F. Stress and the brain: from adaptation to disease. Nat. Rev. Neurosci. 2005;6(6):463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- Erhard F., et al. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature. 2019;571(7765):419–423. doi: 10.1038/s41586-019-1369-y. [DOI] [PubMed] [Google Scholar]
- Fernandez-Albert J., et al. Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 2019;22(10):1718–1730. doi: 10.1038/s41593-019-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields R.D. A new mechanism of nervous system plasticity: activity-dependent myelination. Nat. Rev. Neurosci. 2015;16(12):756–767. doi: 10.1038/nrn4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flati T., et al. A gene expression atlas for different kinds of stress in the mouse brain. Sci. Data. 2020;7(1):437. doi: 10.1038/s41597-020-00772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U., Morris R.G. Synaptic tagging and long-term potentiation. Nature. 1997;385(6616):533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- Gautam A., et al. Acute and chronic plasma metabolomic and liver transcriptomic stress effects in a mouse model with features of post-traumatic stress disorder. PLoS One. 2015;10(1) doi: 10.1371/journal.pone.0117092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goaillard J.M., Marder E. Ion Channel degeneracy, variability, and covariation in neuron and circuit resilience. Annu. Rev. Neurosci. 2021;44:335–357. doi: 10.1146/annurev-neuro-092920-121538. [DOI] [PubMed] [Google Scholar]
- Gorczyca W., et al. Presence of DNA strand breaks and increased sensitivity of DNA in situ to denaturation in abnormal human sperm cells: analogy to apoptosis of somatic cells. Exp. Cell Res. 1993;207(1):202–205. doi: 10.1006/excr.1993.1182. [DOI] [PubMed] [Google Scholar]
- Herdegen T., Leah J.D. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev. 1998;28(3):370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- Herzog V.A., et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods. 2017;14(12):1198–1204. doi: 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgerner H., et al. Neuronal types in the mouse amygdala and their transcriptional response to fear conditioning. Nat. Neurosci. 2023;26(12):2237–2249. doi: 10.1038/s41593-023-01469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O., et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83(2):344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- Jaffe A.E., et al. Decoding shared versus divergent transcriptomic signatures across cortico-amygdala circuitry in PTSD and depressive disorders. Am J Psychiatry. 2022;179(9):673–686. doi: 10.1176/appi.ajp.21020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbasi B., et al. Chronic unpredictable stress disturbs the blood-testis barrier affecting sperm parameters in mice. Reprod. Biomed. Online. 2021;42(5):983–995. doi: 10.1016/j.rbmo.2020.12.007. [DOI] [PubMed] [Google Scholar]
- Kosuth J., et al. Selection of reliable reference genes for analysis of gene expression in spinal cord during rat postnatal development and after injury. Brain Sci. 2019;10(1) doi: 10.3390/brainsci10010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner J.N., et al. Social defeat stress induces genome-wide 5mC and 5hmC alterations in the mouse brain. G3 (Bethesda) 2023;13(8) doi: 10.1093/g3journal/jkad114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuti D., et al. The metabolic stress response: adaptation to acute-, repeated- and chronic challenges in mice. iScience. 2022;25(8) doi: 10.1016/j.isci.2022.104693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.R., Fields R.D. Activity-dependent gene expression in neurons. Neuroscientist. 2021;27(4):355–366. doi: 10.1177/1073858420943515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. Effects of acute restraint stress on different components of memory as assessed by object-recognition and object-location tasks in mice. Behav. Brain Res. 2012;227(1):199–207. doi: 10.1016/j.bbr.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Liu W.Z., et al. Identification of a prefrontal cortex-to-amygdala pathway for chronic stress-induced anxiety. Nat. Commun. 2020;11(1):2221. doi: 10.1038/s41467-020-15920-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., et al. Stress induces microglia-associated synaptic circuit alterations in the dorsomedial prefrontal cortex. Neurobiol Stress. 2021;15 doi: 10.1016/j.ynstr.2021.100342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah L., Szabuniewicz C., Fiocco A.J. Can anxiety damage the brain? Curr Opin Psychiatry. 2016;29(1):56–63. doi: 10.1097/YCO.0000000000000223. [DOI] [PubMed] [Google Scholar]
- Matsushima W., et al. SLAM-ITseq: sequencing cell type-specific transcriptomes without cell sorting. Development. 2018;145(13) doi: 10.1242/dev.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima W., et al. Sequencing cell-type-specific transcriptomes with SLAM-ITseq. Nat. Protoc. 2019;14(8):2261–2278. doi: 10.1038/s41596-019-0179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.S. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol. Rev. 2007;87(3):873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- McEwen B.S. vol. 1. Thousand Oaks); 2017. Neurobiological and systemic effects of chronic stress. (Chronic Stress). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray K.M.J., et al. Identification of a novel, fast-acting GABAergic antidepressant. Mol Psychiatry. 2018;23(2):384–391. doi: 10.1038/mp.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingardi J., et al. Involvement of miR-135a-5p downregulation in acute and chronic stress response in the prefrontal cortex of rats. Int. J. Mol. Sci. 2023;24(2) doi: 10.3390/ijms24021552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount C.W., Monje M. Wrapped to adapt: experience-dependent myelination. Neuron. 2017;95(4):743–756. doi: 10.1016/j.neuron.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K., et al. Strain differences in stress responsivity are associated with divergent amygdala gene expression and glutamate-mediated neuronal excitability. J. Neurosci. 2010;30(15):5357–5367. doi: 10.1523/JNEUROSCI.5017-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhar M., et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360(6390):800–805. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann T., et al. Quantification of experimentally induced nucleotide conversions in high-throughput sequencing datasets. BMC Bioinf. 2019;20(1):258. doi: 10.1186/s12859-019-2849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan K.J., et al. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22(4):167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- Ochi S., Dwivedi Y. Dissecting early life stress-induced adolescent depression through epigenomic approach. Mol Psychiatry. 2023;28(1):141–153. doi: 10.1038/s41380-022-01907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.V., et al. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 2010;26(3):363–369. doi: 10.1093/bioinformatics/btp677. [DOI] [PubMed] [Google Scholar]
- Radle B., et al. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J. Vis. Exp. 2013;78 doi: 10.3791/50195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V., Tom Dieck S., Schuman E.M. Local translation in neuronal compartments: how local is local? EMBO Rep. 2017;18(5):693–711. doi: 10.15252/embr.201744045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B., McEwen B.S., Chattarji S. Stress, memory and the amygdala. Nat. Rev. Neurosci. 2009;10(6):423–433. doi: 10.1038/nrn2651. [DOI] [PubMed] [Google Scholar]
- Satoh E., Edamatsu H., Omata Y. Acute restraint stress enhances calcium mobilization and proliferative response in splenic lymphocytes from mice. Stress. 2006;9(4):223–230. doi: 10.1080/10253890601095794. [DOI] [PubMed] [Google Scholar]
- Weger M., Sandi C. High anxiety trait: a vulnerable phenotype for stress-induced depression. Neurosci. Biobehav. Rev. 2018;87:27–37. doi: 10.1016/j.neubiorev.2018.01.012. [DOI] [PubMed] [Google Scholar]
- West A.E., Greenberg M.E. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3(6) doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., et al. A new GABAergic somatostatin projection from the BNST onto accumbal parvalbumin neurons controls anxiety. Mol Psychiatry. 2021;26(9):4719–4741. doi: 10.1038/s41380-020-0816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S.Y., et al. Nucleobase transport by human equilibrative nucleoside transporter 1 (hENT1) J. Biol. Chem. 2011;286(37):32552–32562. doi: 10.1074/jbc.M111.236117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature. 2023;624(7991):317–332. doi: 10.1038/s41586-023-06812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., et al. Stress-induced functional alterations in amygdala: implications for neuropsychiatric diseases. Front. Neurosci. 2018;12:367. doi: 10.3389/fnins.2018.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., et al. Chronic stress remodels synapses in an amygdala circuit-specific manner. Biol Psychiatry. 2019;85(3):189–201. doi: 10.1016/j.biopsych.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated for this study and software parameters used for data analysis have been deposited at GEO (GSE274148) and are publicly available as of Aug. 14th, 2024.
Any additional information related to sequencing data used in this paper is available from the corresponding author upon request.
Data is deposited at GEO (GSE274148) and are publicly available as of Aug. 14th, 2024.