Abstract
Oral squamous cell carcinoma (OSCC) is a common malignant tumor in the head and neck, associated with high recurrence and poor prognosis. We performed an integrated analysis of single-cell RNA and spatial transcriptomic data from cancerous and normal tissues to create a comprehensive atlas of epithelial cells and cancer-associated fibroblasts (CAFs). Our findings show that AKR1C3+ epithelial cells, located at the tumor’s stromal front, exhibit significant copy number variation and poor prognostic indicators, suggesting a role in tumor invasion. We also identified a distinct group of early-stage CAFs (named OSCC_Normal, characterized by ADH1B+, MFAP4+, and PLA2G2A+) that interact with AKR1C3+ cells, where OSCC_Normal may inhibit the FOXO1 redox switch in these epithelial cells via the IGF1/IGF1R pathway, causing oxidative stress overload. Conversely, AKR1C3+ cells use ITGA6/ITGB4 receptor to counteract the effects of OSCC_Normal, promoting cancer invasion. This study unveils complex interactions within the OSCC tumor microenvironment.
Subject terms: Oral cancer, High-throughput screening, Insulin signalling
Introduction
OSCC is the most common malignant tumor in the head and neck region, frequently found in the tongue, cheek, gums, floor of the mouth, palate, and lips1. OSCC is known for its high malignancy and aggressiveness, with significant rates of metastasis and recurrence. Currently, surgery is the primary treatment method, but the 5-year survival rate for patients is merely 64.4%2. Postoperative issues such as facial disfigurement, impaired speech, and difficulty in chewing severely impact the quality of life of the patients3.
The tumor microenvironment (TME) comprises cellular and non-cellular components, which are crucial in tumor initiation, progression, invasion, metastasis, and drug resistance. These components modulate the microenvironment to either promote or inhibit tumor development4. Fibroblasts, as a major component of the TME, participate in crosstalk with adjacent epithelial cells, influencing local epithelial stem cell behavior and coordinating immune system functions by producing chemokines and cytokines. Traditionally, cancer-associated fibroblasts (CAFs) have been recognized for their significant and diverse roles in supporting tumors5,6. Beyond directly interacting with malignant epithelial cells, CAFs help create a tumor-favorable TME by activating normal fibroblasts into CAFs, promoting angiogenesis in endothelial cells, and recruiting myeloid cells and T cells for immune suppression7–9. The advent of single-cell RNA sequencing (scRNA-seq) technology has unveiled previously unrecognized levels of CAF heterogeneity across various cancer types. This discovery is significant as it provides a potential explanation for the dual roles of CAFs in both tumor suppression and promotion10.
OSCC originates from epithelial cells, and epithelial–mesenchymal transition (EMT) signaling is pivotal in cancer metastasis and invasion11,12. During the local invasion, tumor cells initially detach from the primary tumor site, invade neighboring tissues, and breach the basement membrane. This transition from an epithelial to a mesenchymal state in tumor cells is continuous and varied, allowing tumor cells to maintain different transitional states with varying conversion rates and heterogeneity across different tumors and patients13–15. The epithelial state contributes to tumor mass increase, whereas the mesenchymal state enhances invasion and metastasis, with the mixed state exhibiting even more complex functions16. Oxidative stress plays a crucial role in EMT induction17. However, reactive oxygen species (ROS) concentration serves as a double-edged sword for cancer cells, with a critical mechanism being how cancer cells regulate their oxidative stress defense systems to maintain ROS levels, thereby acquiring high metabolic activity and an EMT-invasive phenotype.
Moreover, CAFs within the TME are also significant. scRNA-seq has revolutionized our understanding of the biological characteristics and dynamics within cancer lesions, offering high-resolution cellular transcriptomic maps and cell state transition trajectories of the OSCC TME, yet it does not indicate the spatial relationships of neighboring cells18,19. Spatial transcriptomics (ST) addresses this limitation by providing spatial distribution and positional information for each cell20–22. We employed high-resolution multi-omics analysis of single-cell and spatial transcriptomics to elucidate their roles in this process, aiming to uncover potential targeted therapeutic strategies.
Results
Integration of OSCC scRNA-seq Atlas
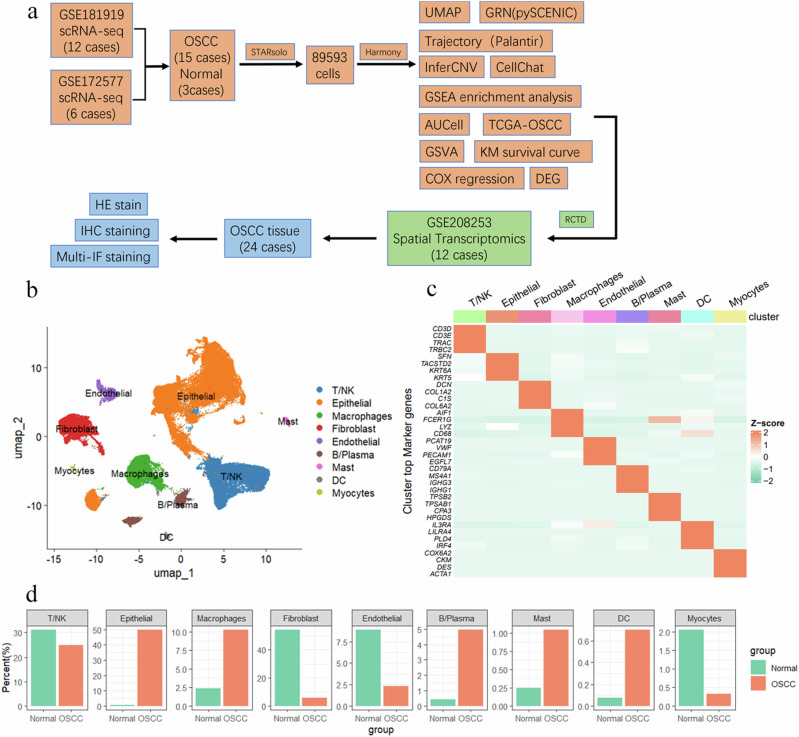
The study included a total of 18 scRNA-seq expression profiles from two datasets, comprising OSCC samples (n = 15) and normal tissues (n = 3). A workflow diagram (Fig. 1a) was created to illustrate this process. In addition to scRNA-seq data, we also incorporated spatial transcriptomics data for integrative multi-omics analysis, which was further validated using OSCC tissue samples. To achieve consistent gene alignment results and reduce batch effect impacts on downstream data analysis, we utilized STARsolo for uniform upstream data processing. After quality control, we included a total of 89,593 cells. Based on known cell marker genes, we identified nine cell types (Fig. 1b): T/NK cells (CD3D, CD3E, TRAC, TRBC2), epithelial cells (“SFN”, “TACSTD2”, “KRT6A”, “KRT5”), macrophages (“AIF1”, “IL1B”, “LYZ”, “CD68”), fibroblasts (“DCN”, “COL1A2”, “C1S”, “COL6A2”), endothelial cells (“PCAT19”, “VWF”, “PECAM1”, “EGFL7”), B/Plasma cells (“CD79A”, “IGLC2”, “IGHG3”, “IGHG1”), mast cells (“TPSB2”, “TPSAB1”, “CPA3”, “HPGDS”), DC cells (“IL3RA”, “LILRA4”, “PLD4”, “IRF4”), and myocytes (“COX6A2”, “CKM”, “DES”, “ACTA1”) (Fig. 1c). We analyzed the cell proportion changes between the OSCC and NORMAL groups. In the OSCC group, epithelial cells were predominant (49.89%), whereas fibroblasts were most abundant in the Normal group (54.28%). T/NK cells were more prevalent in the Normal group than in the OSCC group (31.20% vs. 24.90%), ranking second in both groups (Fig. 1d).
Fig. 1. Integration of OSCC scRNA-seq Atlas.
a The workflow diagram illustrates the data analysis process. The data were summarized and categorized into 15 OSCC samples and 3 normal samples. Initial data processing was conducted using the STARsolo software, resulting in a total of 89,593 cell data points. The Harmony algorithm was applied for batch effect correction. Various analytical methods were employed, including UMAP dimensionality reduction, GRN analysis (using pySCENIC), trajectory analysis (using Palantir), InferCNV, and CellChat. Single-cell annotations were mapped to spatial transcriptomics using RCTD, and further validation was conducted through histological staining. b UMAP plot showing nine major cell types: T/NK cells, epithelial cells, macrophages, fibroblasts, endothelial cells, B/plasma cells, mast cells, DC cells, and myocytes. c Heatmap showing the expression of representative marker genes for each cell type. Cell clusters are indicated on the x-axis, and gene names are on the y-axis. The color intensity represents the average gene expression level, with Z-score values standardized. d Proportion of cell types in OSCC patients; the y-axis represents the percentage of cell counts, and the x-axis represents the Normal and OSCC groups.
Identification of specific regulatory patterns in fibroblasts through gene regulatory network analysis
The heterogeneity of epithelial cells and fibroblasts is fundamental to the cellular heterogeneity of OSCC. Therefore, it is crucial to conduct a detailed differentiation of the cell states and types they encompass. However, the classic cell clustering annotation process is sensitive to resolution selection, which can lead to over-clustering and hinder the identification of cell states or subtypes. Gene regulatory network (GRN) analysis based on the SCENIC algorithm does not rely on cell classification labels. Instead, it calculates gene co-expression to abstract the single-cell gene expression matrix into co-expressed gene sets with regulatory relationships (i.e., regulons). Using this as a basis, we can employ algorithms such as AUCell to transform the single-cell gene expression matrix into an activity matrix reflecting the functional activity of the GRN, thereby distinguishing differences in cell states at the functional level of gene synergy. This approach precisely meets our needs for a deeper differentiation of OSCC cellular heterogeneity.
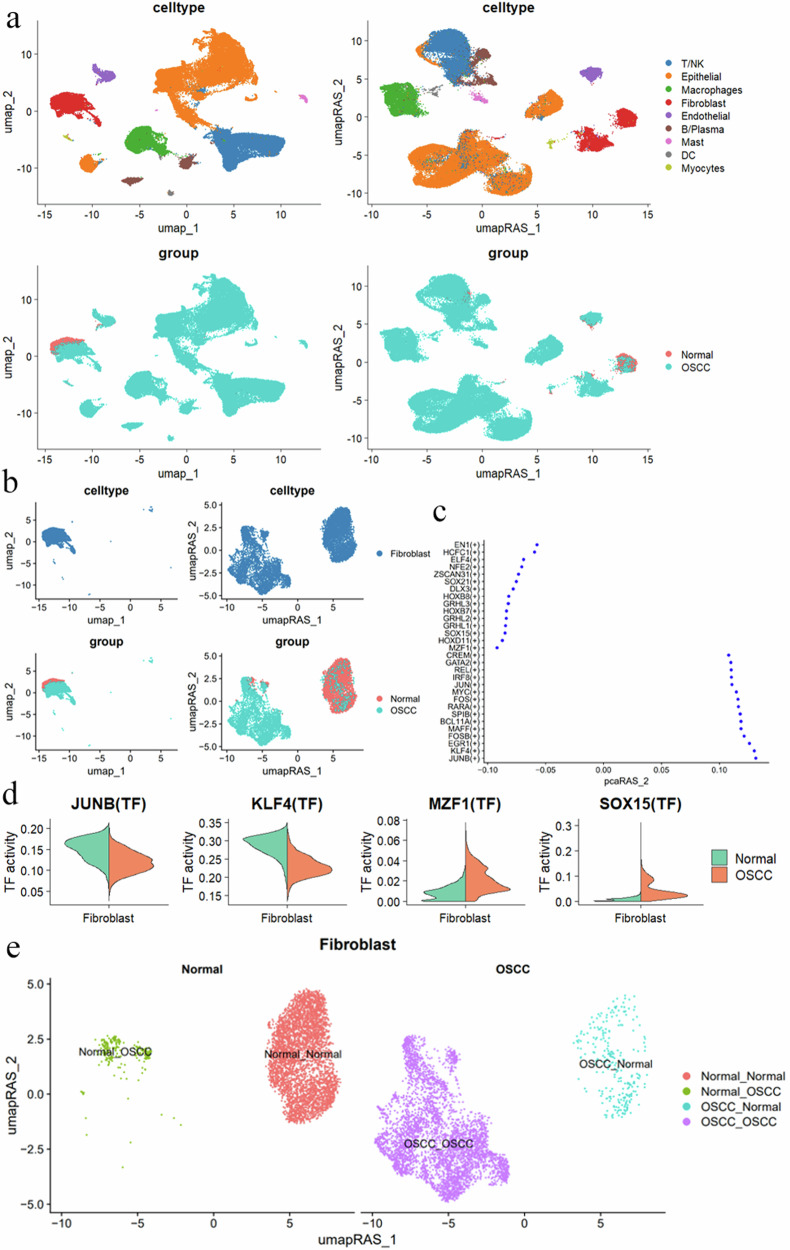
Initially, we used pySCENIC to establish the GRN of OSCC single cells23,24. Using AUCell, we calculated each cell’s regulon active score (RAS) to construct the GRN activity matrix. Through UMAP dimensionality reduction visualization (Fig. 2a), we unexpectedly found that fibroblasts clustered together in the gene expression matrix were clearly divided into two groups. This observation led us to realize that cells with high similarity at the shared gene expression level might exhibit significant differences in the activity levels of their GRNs (Fig. 2b, top left). This finding indicates that the above analytical strategy provides a reference anchor point for the subdivision of cell subtypes and states (Fig. 2b, top right).
Fig. 2. Single-cell transcriptional regulatory network analysis reveals regulatory patterns and cellular heterogeneity in fibroblastsm.
a UMAP and UMAP_RAS visualizations showing cell distribution based on different cell types and groups. The top left plot shows clustering by cell type, while the top right plot shows clustering by transcription factor regulatory patterns. The bottom left plot shows cell distribution in healthy and OSCC samples, while the bottom right plot shows the distribution between the two groups under transcription factor regulatory patterns. b Fibroblast distribution in UMAP and UMAP_RAS visualizations. The top left plot shows fibroblasts clustering together with similar gene expression. The top right plot reveals that fibroblasts are regulated by two distinct groups of regulons. The bottom left plot shows the distribution of healthy and OSCC fibroblasts in UMAP, while the bottom right plot shows the inter-group distribution characteristics of the two fibroblast regulon groups in UMAP_RAS. c The significantly different transcription factors (TFs) between normal and OSCC fibroblasts. Each point represents a transcription factor, with the Y-axis showing the transcription factor names and the X-axis showing the differential activity scores. d Comparison of transcription factor activities of JUNB, KLF4, MZF1, and SOX15 in normal and OSCC fibroblasts, with the Y-axis representing TF activity analysis and the X-axis representing cell types. e UMAP dimensionality reduction shows the heterogeneity of fibroblast populations and transcriptional regulatory differences. Different colors in the plot represent different cell states, such as Normal_Normal, Normal_OSCC, OSCC_Normal, and OSCC_OSCC. RAS regulon activity score.
Analysis of differential regulons in fibroblasts between OSCC and Normal tissues
We analyzed the differential regulons between fibroblasts from OSCC and Normal tissues (Fig. 2c and Supplementary Fig. 1d–f). The results showed that the activity of transcription factors JUNB and KLF4 was lower in OSCC fibroblasts, while MZF1 and SOX15 exhibited higher activity (Fig. 2d). Interestingly, we found that in the feature space of the GRN activity matrix, fibroblasts from the Normal group could be divided into two clusters: one projecting in the original Normal space and the other appearing in the OSCC space. We defined these as Normal–Normal and Normal–OSCC cell clusters, respectively. Similarly, fibroblasts from the OSCC group could also be divided into OSCC–Normal and OSCC–OSCC clusters (Fig. 2e). This indicates that a group of CAFs in OSCC (OSCC–Normal) possesses characteristics similar to normal cells at the GRN activity level. The function and role of these CAFs intrigued us. Furthermore, we found that OSCC–Normal cells still clustered together on the UMAP (Supplementary Fig. 1a), indicating consistent characteristics at the gene expression level.
However, when we attempted to cluster cell subgroups similar to OSCC–Normal directly at the gene expression matrix level using classic cell clustering strategies, specifically C35 (Supplementary Fig. 1b, c), we needed to set the resolution parameter of the clustering function to a very high level. This is extremely rare and difficult to achieve in typical analysis workflows. Moreover, high-resolution settings significantly increase the difficulty of cell annotation and the exploration of the biological functions of cell clusters. This indirectly proves the efficiency of our analytical strategy over classic approaches. Using GRN, we identified this special group of CAFs, OSCC–Normal, which retains the gene expression similarity of CAFs while exhibiting GRN activity characteristics similar to normal fibroblasts. This genetic profile may indicate that they are primitive cells among CAFs and have important functions in OSCC.
Functional typing and lineage analysis of CAF subgroups in OSCC
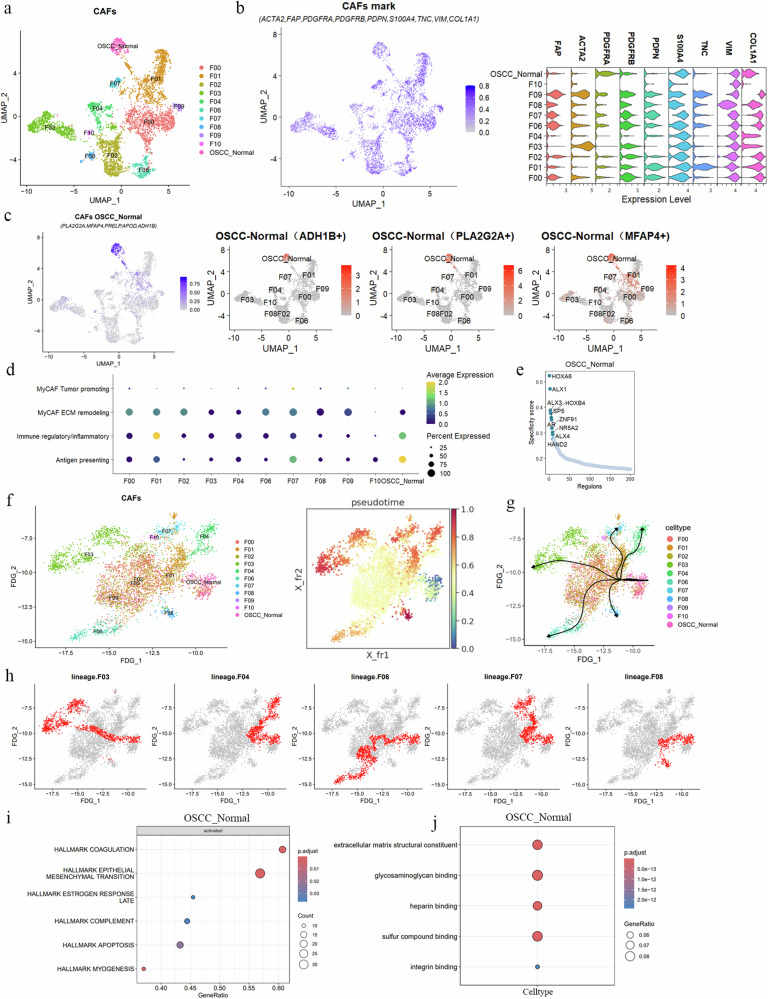
To better explore the heterogeneity of fibroblasts in OSCC, we divided fibroblasts in OSCC into 11 subgroups (named F00–F10) based on the resolution reference anchor points provided by the previous analysis (Table 1 and Fig. 3a). We used classical CAF surface markers to label our CAF cell groups (ACTA225, FAP26, PDGFRA26, PDGFRB26, PDPN27, S100A426, TNC28, VIM, and COL1A129). Additionally, we referenced the explorations of CAF markers by researchers such as Dwivedi30 and Lyu31. The results indicated that the expression of F10 is relatively weak, while the remaining fibroblast subtype exhibited widespread CAF marker expression (Fig. 3b). Additionally, we identified the top 5 characteristic markers of these CAF subgroups (Supplementary Fig. 2a), with F05 being the OSCC–Normal subgroup identified in the previous step. We labeled this subgroup with PLA2G2A, MFAP4, PRELP, APOD, and ADH1B, among which ADH1B, PLA2G2A, and MFAP4 were the most specific (Fig. 3c).
Table 1.
CAFs subtype mark
| CAFs subtype | Mark (top 3) |
|---|---|
| F00 | MMP11, ADAMTS2, TDO2 |
| F01 | IGHG3, IL24, TWIST2 |
| F02 | LUM, COL6A2, SFRP2 |
| F03 | PPP1R14A, PLN, MYH11 |
| F04 | CHRDL2, MYF5, DLK1 |
| F06 | MKI67, RRM2, BIRC5 |
| F07 | CD3E, CD2, CD3D |
| F08 | FGFBP1, LAD1, GJB6 |
| F09 | POSTN, LRRC17, COL11A1 |
| F10 | MLANA, TYR, FABP7 |
| OSCC–Normal | PLA2G2A, ADH1B, MFAP4 |
Fig. 3. Analysis of CAF subgroups in OSCC.
a UMAP analysis reveals the diversity of CAFs in OSCC, dividing them into 11 subgroups (F00–F10), with each color representing a subgroup, showing their distribution in the cell state space. b The left panel shows the expression levels of classical CAF markers in each CAF subgroup, with the bar chart on the right showing the expression intensity. The right panel displays a violin plot showing gene characteristic expression across clusters. c Expression intensity and location of marker genes in the OSCC_Normal specific subgroup, highlighting ADH1B+, PLA2G2A+, and MFAP4+ in red. d Dot plot of average expression of MyCAFs, immunoregulatory/inflammatory, and antigen-presenting genes across CAF subtypes (X-axis representing CAF clusters, Y-axis representing gene characteristics), with color indicating average expression intensity and dot size representing expression proportion. e Characteristic transcriptional regulators in the OSCC–Normal subgroup, showing the top 10 by specificity score. f Pseudotime trajectory reveals the dynamic process of fate determination in CAFs. The left panel shows the force-directed graph of FDG_1 dimensionality reduction, and the right panel shows the pseudotime scores of each CAF cluster, with darker colors indicating later cell fates. g, h Differentiation trajectories from the OSCC–Normal starting point to different terminal subgroups (F03, F04, F06, F07, F08), showing differentiation characteristics and molecular dynamics of specific subgroups, with each dot representing a cell in (h), and red cells indicating cells on the differentiation trajectory. i GSEA results show key biological processes activated in the OSCC–Normal subgroup, including epithelial–mesenchymal transition, coagulation, and others. j GO analysis results reveal the specific functions of the OSCC–Normal subgroup in ECM structure, actin binding, and other aspects, with dot size indicating gene count and color depth indicating statistical significance.
To further understand the biological functional characteristics of each OSCC fibroblast subgroup, we referred to the single-cell CAF annotations from pan-cancer tissues and Bienskowa’s32 classification of CAFs in HNSCC. Currently, the academic community generally agrees on three central CAF subtypes (myofibroblastic CAFs [MyCAFs], immunoregulatory/inflammatory CAFs, and antigen-presenting CAFs33). MyCAFs are further divided based on function into those promoting tumors and those involved in extracellular matrix (ECM) construction. Tumor-promoting MyCAFs highly accumulate IL4, IL13, and TGFB33,34, while ECM-constructing CAFs highly express POSTN, COL1A1, and MMP11 genes, participating in collagen formation and ECM remodeling29,35,36. Notably, the functions of MyCAFs are diverse and play different roles in various tissues and diseases. Immunoregulatory/inflammatory CAFs are primarily rich in various chemokines of the CXCL family and inflammatory factors such as IL-6 and IL-837,38. Additionally, antigen-presenting CAFs highly express MHC family genes, such as HLA-DRA and HLA-DQA39,40.
Functional typing and lineage analysis of CAF subgroups in OSCC
Building on this understanding, we categorized fibroblast subgroups in OSCC into functional types (Fig. 3d). The subgroups F00 (MMP11+), F02 (LUM+), F03 (PPP1R14A+), F06 (MKI67+, BIRC5+), and F09 (POSTN+) exhibit characteristics of MyCAFs, which are involved in ECM composition and remodeling (Supplementary Fig. 2b). Subgroups F01 (IGHG3+), OSCC–Normal (ADH1B+, PLA2G2A+, MFAP4), and F07 (VIM+, CD3E+) display characteristics of immunoregulatory, inflammatory, and antigen-presenting CAFs (Supplementary Fig. 2b). The remaining subgroups, F04, F08, and F10, do not correspond well to any known CAF subtypes. F04 is characterized by CHRDL2+, which has been reported to promote gastric cancer cell proliferation and is positively correlated with poor patient prognosis41 (Supplementary Fig. 2b). F08’s specific marker is FGFBP1, which enhances fibroblast growth factor (FGF) signaling, and is upregulated in various cancers, promoting cancer cell migration and invasion35,42–44. F10 specifically expresses MLANA, a marker often associated with melanocytes45–47.
The OSCC–Normal subgroup, characterized by ADH1B, PLA2G2A, and MFAP4, currently lacks related research on CAFs with this combination of markers. However, studies have shown that PLA2G2A is specifically expressed in inflammatory benign hepatocellular adenomas and detected in stromal cells around hepatocellular carcinoma tissues, without an association with poor prognosis in hepatocellular carcinoma48. A reduced number of ADH1B+ CAFs is linked to the invasive characteristics of lung adenocarcinoma. The ADH1B + CAF subgroup also exhibits high expression of CFD, DCN, SEPP1 (SELENOP), A2M, and MFAP4 genes, which in early-stage lung adenocarcinoma, are associated with better survival rates49. These findings suggest that OSCC–Normal CAFs might have potential anti-tumor functions. Lastly, we investigated CAF-specific transcription regulators (Supplementary Fig. 2d). The key transcription factors for OSCC–Normal include HOXA6, ALX1, ALX3, HOXB4, and SP5, which likely determine the state and identity of OSCC–Normal cells (Fig. 3e).
OSCC, as a chronic progressive disease, exhibits extensive changes in cell types and states, which can be captured by single-cell sequencing. Given that OSCC–Normal has GRN activity characteristics similar to normal fibroblasts, we believe that OSCC–Normal might act as a group of primitive CAFs that resist tumor progression, being at an earlier stage in the fibroblast state transition process in OSCC. Therefore, we used single-cell trajectory analysis tools to fit the lineage relationships and transcriptional fates among the 11 CAFs, starting from OSCC–Normal. The pseudotime trajectory originating from the OSCC–Normal cell cluster pointed to five major branches (F03, F04, F06, F07, and F08) (Supplementary Fig. 2c), while F00, F01, F02, F09, and F10 were considered intermediate states during CAF state transitions. The pseudotime map indicated the relative differentiation times of F03, F04, F06, F07, and F08 cells, with these five terminal CAF subgroups showing distinct cellular fates (Fig. 3f).
We explored the molecular dynamics of the five CAF groups, mapping the paths from OSCC–Normal to five different CAF states (Fig. 3g, h). We investigated the functions of the six different fate cell clusters, with GSEA results suggesting that EMT, complement processes, and others were activated in OSCC–Normal (Fig. 3I). The F03 cluster showed activated EMT (Supplementary Fig. 3a), the F04 cluster significantly activated hypoxia, complement, and NF-kB mediated TNF-α signaling processes (Supplementary Fig. 3b), the F06 cluster significantly activated angiogenesis, NF-kB mediated TNF-α signaling, inflammation, hypoxia, and EMT (Supplementary Fig. 3c), the F07 cluster significantly activated myogenesis, coagulation, and EMT (Supplementary Fig. 3d), while the F08 cluster only showed activation of the downregulated KRAS signaling pathway (Supplementary Fig. 3e).
GO analysis indicated that the specific gene functions of OSCC–Normal are involved in ECM composition (Fig. 3j). F03 genes mainly participate in actin binding (Supplementary Fig. 3f) and ECM composition, F04 genes are primarily involved in ribosomal structure components and rRNA binding, ECM binding (Supplementary Fig. 3g), F06 cluster genes participate in ECM structural components, cadherin binding, collagen binding, integrin binding (Supplementary Fig. 3h), F07 cluster genes are involved in ECM structural components, collagen binding, growth factor binding (Supplementary Fig. 3i), and F08 cluster genes are involved in cadherin binding, skin epidermis structural components, and cell–cell adhesion mediator activity (Supplementary Fig. 3j).
We analyzed the molecular characteristics and functional heterogeneity of various fibroblast subgroups (CAFs) in OSCC, revealing dynamic transitions and interactions of different CAF subgroups within the TME. This analysis mapped the lineage relationships and transcriptional fates of CAFs in OSCC. Particularly, the OSCC–Normal subgroup, which has GRN activity characteristics similar to normal fibroblasts and is upstream in the CAF state transition trajectory in OSCC, provides new perspectives for tumor research with its unique transcriptional regulators and gene function activity.
Analysis of heterogeneity and functional state evaluation of OSCC epithelial cells
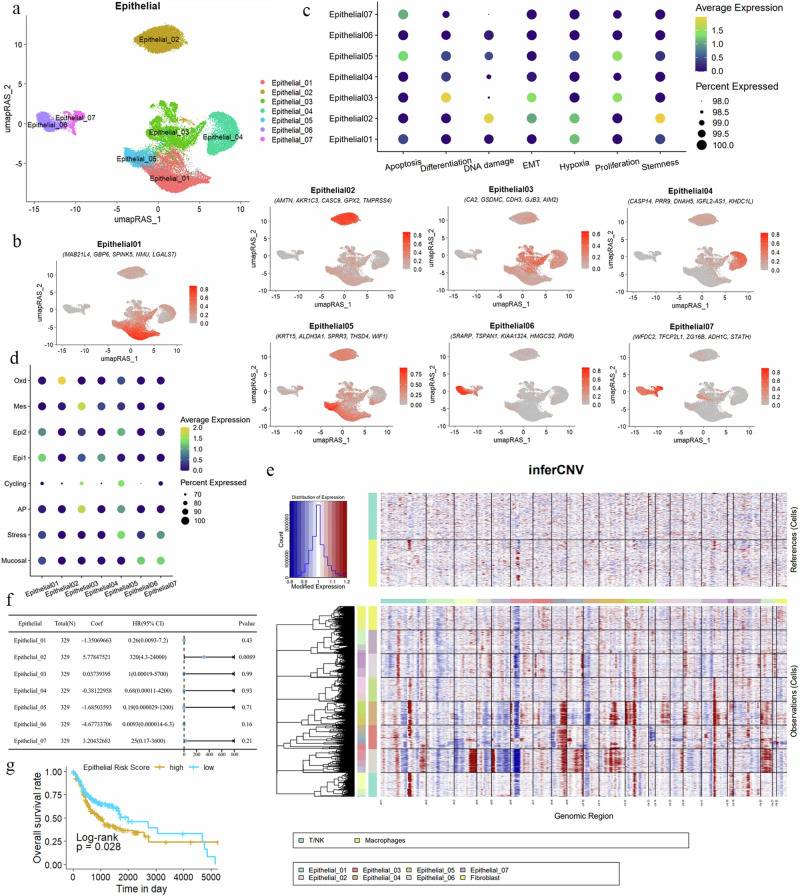
One of the main contributors to the heterogeneity and malignancy of OSCC is the OSCC epithelial cells. We applied the same strategy for the detailed identification of OSCC epithelial cells. The epithelial cells were divided into seven subgroups (Fig. 4a). We identified the characteristic genes of the epithelial cell subgroups and used the top 5 markers to indicate each epithelial subgroup. The specific markers for Epithelial01 are MAB21L4, GBP6, SPINK5, NMU, and LGALS7; for Epithelial02 they are AMTN, AKR1C3, CASC9, GPX2, and TMPRSS4; for Epithelial03 they are CA2, GSDMC, CDH3, GJB3, and AIM2; for Epithelial04 they are CASP14, PRR9, DNAH5, IGFL2-AS1, and KHDC1L; for Epithelial05 they are KRT15, ALDH3A1, SPRR3, THSD4, and WIF1; for Epithelial06 they are SRARP, TSPAN1, KIAA1324, HMGCS2, and PIGR; and for Epithelial07 they are WFDC2, TFCP2L1, ZG16B, ADH1C, and STATH (Table 2, Fig. 4b, and Supplementary Fig. 4b).
Fig. 4. Redefinition of OSCC malignant epithelial cells from the perspective of GRN.
a UMAP dimensionality reduction reveals that epithelial cells in OSCC are divided into seven distinct subgroups. b Specific marker genes for the epithelial cell subgroups are displayed, showing the five most significantly expressed genes in each subgroup. c A dot plot illustrates the gene expression related to processes such as apoptosis, proliferation, differentiation, DNA damage, hypoxia, stemness, and EMT in the seven epithelial cell subgroups. The color indicates the average expression intensity, while the size of the dots represents the proportion of expression. d Functions of epithelial cell subgroups based on specific biomarkers are depicted, highlighting characteristics such as mucosal properties, stress response, and antigen presentation. e A heatmap displays the CNV within the epithelial cell subgroups. Red indicates chromosomal amplification, blue indicates chromosomal deletion, with the X-axis representing chromosome numbers and the Y-axis representing the cell clusters included in the analysis. T/NK cells and macrophages are used as reference genomes. f Multivariate Cox regression analysis calculated the risk scores of epithelial cell subgroups for OSCC. For patients (n = 329), the coefficient (coef) represents the partial regression coefficient. A hazard ratio (HR) > 1 is considered a risk factor. g Kaplan–Meier curves show the relationship between the risk scores of epithelial subgroups and patient survival rates. A higher risk score correlates with poorer survival outcomes (P = 0.028). The Y-axis represents the probability of patient survival, and the X-axis represents patient survival time.
Table 2.
Epithelial cell subtype mark
| Epithelial subtype | Mark (top 5) |
|---|---|
| Epithelial01 | SPINK5, GBP6, NMU, MAB21L4, LGALS7 |
| Epithelial02 | AKRIC3, GPX2, AMTN, CASC9, TMPRSS4 |
| Epithelial03 | AIM2, CA2, GSDMC, CDH3, GJB3 |
| Epithelial04 | CASP14, PRR9, DNAH5, IGFL2-AS1, KHDC1L |
| Epithelial05 | KRT15, ALDH3A1, SPRR3, THSD4 |
| Epithelial06 | TSPAN1, KIAA1324, SRARP, HMGCS2, PIGR |
| Epithelial07 | WFDC2, ZG16B, STATH, ZG16B, ADH1C |
In the enrichment analysis of biological processes, we found that in Epithelial05, genes related to apoptosis and proliferation were activated (Fig. 4c). Epithelial03 showed activation of cell proliferation, differentiation, and EMT processes. In Epithelial02, DNA damage and hypoxia processes were observed, and stem cell genes were also activated. Additionally, we referenced epithelial cell types from esophageal squamous cell carcinoma (ESCC) since ESCC and OSCC are both epithelial-origin cancers and share common molecular and cellular biological characteristics50. By comparing the epithelial cell characteristics of these two cancers, we can gain a deeper understanding of the heterogeneity of epithelial cells in OSCC and their interactions with the TME.
Epithelial06 and Epithelial07 indicate mucosal characteristics (Fig. 4d), associated with innate immune response (S100P), mucosal defense mechanisms (CXCL17), and mucus production (e.g., AGR2 and MUC20). Epithelial05 indicates stress, composed of genes responding to a wide range of cellular stimuli (e.g., EGR1, JUN, and FOS). Epithelial03 may be related to antigen presentation (AP) processes, potentially indicating responsiveness to tumor neoantigens. The cell cycle (cycling) program is characterized by high expression of genes involved in cell proliferation (e.g., CENPW, CKS1B, and BIRC5), suggesting strong proliferative capacity in tumor cells in Epithelial05.
Reportedly, Epi1/2 are two highly co-existing epithelial cell groups. Epi1 is characterized by stress keratins (KRT6, KRT16, and KRT17), which may play a role in enhancing tumorigenesis and tumor growth. Epi2 features overexpressed genes associated with terminal differentiation, such as envelope proteins (SPRR1A/1B) and calgranulins (S100A8/9). Epithelial01 is enriched with Epi1 characteristics. Epithelial05 exhibits mesenchymal-like characteristics (Mes), composed of genes like VIM and SPARC, and shows activation of EMT and angiogenesis pathways. Finally, Epithelial02 shows oxidative stress (Oxd) characteristics, expressing various peroxidases and reductases (e.g., GPX2 and AKR1C1), involved in defending against oxidative damage.
Based on the characteristics of OSCC cases, epithelial cells in the samples are often regarded as malignant. To verify this assumption, we used normal cells, specifically macrophages and T/NK cells, as references and conducted a copy number variation (CNV) analysis. The results from inferCNV indicated that OSCC epithelial cells exhibit a significantly higher level of CNVs overall, highlighting their malignant features in comparison to normal cells. We also performed an inferCNV analysis on different epithelial cell subgroups (Supplementary Fig. 4c). The findings showed that the epithelial subgroups 02, 03, and 04 display extensive chromosomal copy number alterations, suggesting a higher degree of malignancy (Fig. 4e).
Building on this, we attempted to further validate the malignant characteristics and severity of the epithelial cell subgroups by conducting clinical prognosis modeling on 329 OSCC cases using the gene sets of characteristic genes from each subgroup. We performed lasso regression and multivariate Cox regression on the marker gene sets of the epithelial subgroups to calculate the Risk score for each subgroup and build clinical prognosis models. The results revealed that all epithelial subgroups in the TCGA–OSCC cohort were associated with lower survival rates. Notably, the multivariate Cox analysis indicated that Epithelial02 had a hazard ratio (HR) as high as 320 (p = 0.0089). Additionally, the prognosis nomogram, which included all epithelial subgroups, highlighted the prominence of the weight score for Epithelial02 (Fig. 4f).
Furthermore, the gene set variation analysis score constructed from the marker genes of epithelial subgroups created an overall risk score, and the Kaplan–Meier (KM) curve indicated that a higher score correlated with poorer patient prognosis (Fig. 4g). Collectively, these results consistently suggest that the Epithelial02 subgroup exhibits more pronounced malignant characteristics compared to other subgroups, drawing more attention to its role in cancer.
Spatial transcriptomics reveals CAF and tumor cell interactions
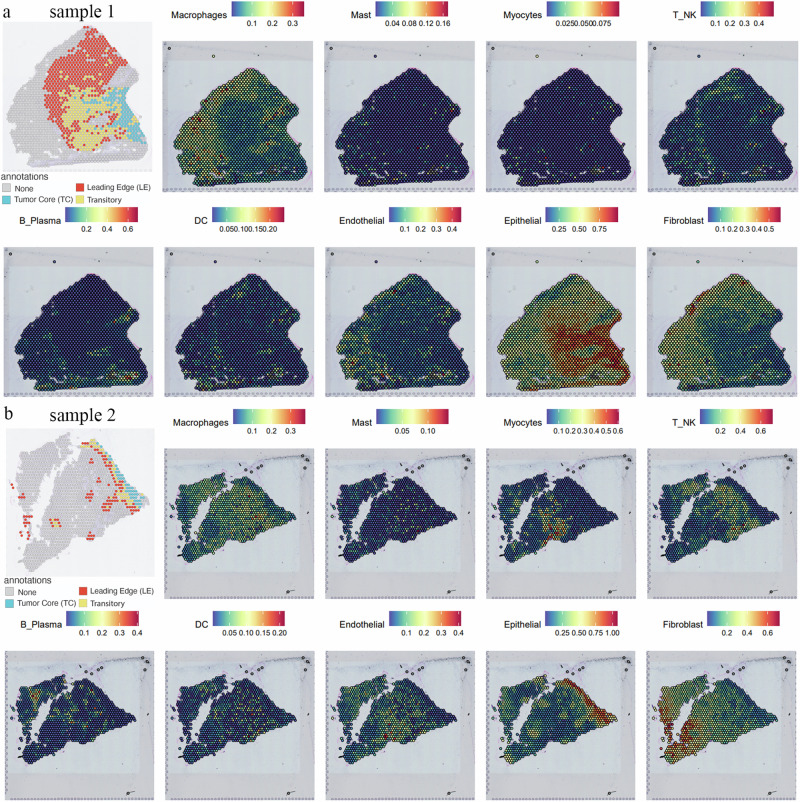
Current understanding highlights that interactions between tumor epithelial cells and the TME are critical mechanisms driving tumor development. The possibility and extent of these interactions are significantly influenced by the spatial distribution within the tissue. To comprehensively evaluate the spatial ecological domains of various cell subgroups, we integrated single-cell transcriptomics and spatial tissue information of OSCC (GSE20825351). Using robust cell type decomposition (RCTD), we spatially identified the characteristics of major cell groups and thoroughly analyzed the spatial distribution of all cell types present in OSCC (Supplementary Materials).
Arora51 classified the histological features of OSCC into “Tumor core (TC),” “Transition (TS),” “Leading edge (LE),” and “Non-cancer” regions. In the “TC” area, we observed a strong epithelial cell signal, which gradually transitioned toward the TS zone and stopped at the LE. In the LE area, we found a significant enrichment of T/NK cells, macrophages, and endothelial cells, while fibroblasts were located away from the tumor region (Fig. 5a). In sample 2, we observed that epithelial cells did not exhibit a noticeable invasion process, which might be due to the slice, but scattered epithelial budding clusters appeared in the stroma (Fig. 5b). Around these budding clusters, there were distributions of endothelial cells, macrophages, T/NK cells, B/plasma cells, and dendritic cells, consistent with the known tumor progression process.
Fig. 5. Spatial transcriptomics mapping of the OSCC tumor microenvironment cell Atlas.
a, b Spatial transcriptomics analysis of samples 1 and 2 shows the distribution of different cell types within the tumor microenvironment. Various cell types, including epithelial cells, macrophages, T/NK cells, and endothelial cells, are mapped. The top of the figure indicates the expression intensity of each cell cluster, with darker colors representing higher intensity.
This indicates that our integrated single-cell and spatial tissue maps of OSCC can accurately depict the dynamic process of tumor development. They clearly demonstrate how malignant epithelial cells invade the stroma, providing an important perspective for understanding cell interactions and tumor behavior within the TME.
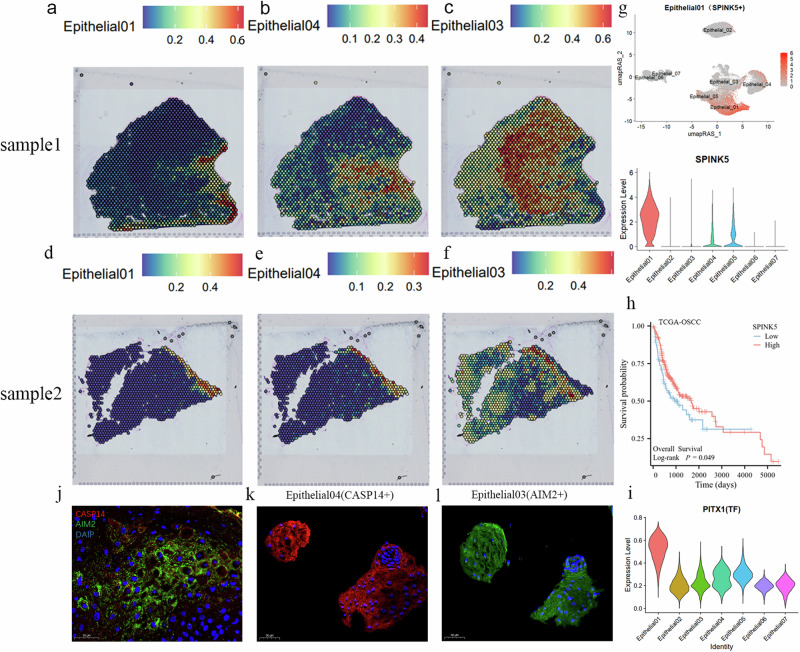
After projecting the characteristics of various cell subtypes into the spatial transcriptomics data space for joint analysis, we identified Epithelial01, 02, 03, and 04 as the most representative epithelial regions (Figs. 6a–f and 7a). The remaining epithelial subtype did not exhibit any discernible distribution patterns. Epithelial01, with its unique RNA expression profile, indicated the site of carcinoma in situ, appearing prominently in sample 02 only within the TC, and being most evident in sample 01, where the tumor invades the stroma after breaking through the basement membrane. During this process, Epithelial01 is most abundant at the primary site (Fig. 6a). The most specific marker gene for Epithelial01 is SPINK5 (Fig. 6g), which has been reported to inhibit the proliferation, migration, and invasion of esophageal cancer cells by suppressing the Wnt/β-catenin signaling pathway 52–54. In OSCC, SPINK5 acts as a tumor suppressor molecule and is also associated with a favorable prognosis (Fig. 6h) (P < 0.05). GO enrichment for Epithelial01-specific markers suggests a structural constituent of skin epidermis. Additionally, the transcription factor PITX (Fig. 6i), specific to Epithelial01, has been reported to act as a tumor suppressor gene in various human cancers, including OSCC55. We believe Epithelial01 may play a role in the early stages of cancer and remain fixed in the primary tumor niche in later stages, potentially aiding in locating the tumor’s origin.
Fig. 6. Tumor progression at spatial transcriptomics resolution.
a–f The spatial transcriptomics showcase the distribution of Epithelial01, 04, and 03 in OSCC samples. The top of each panel displays the expression intensity for each cell cluster, with darker colors indicating higher intensity. g The top UMAP plot shows the specific marker SPINK5 in Epithelial01. The bottom violin plot illustrates SPINK5 expression across different epithelial subtypes, with the Y-axis representing expression levels. h Survival analysis based on the TCGA–OSCC dataset is presented using a Kaplan–Meier curve, demonstrating that high SPINK5 expression is associated with better patient prognosis (P < 0.05). The Y-axis shows patient survival probability, and the X-axis shows patient survival time. i The violin plot depicts the expression of the transcription factor PITX1 across different epithelial subtypes, with the Y-axis indicating the expression levels. j–l Immunofluorescence staining validates the expression of AIM2 and CASP14 in tumor tissues. j The co-expression of AIM2 and CASP14 in tumor tissues, while (k) and (l) show the expression of these markers in tumor epithelial clusters.
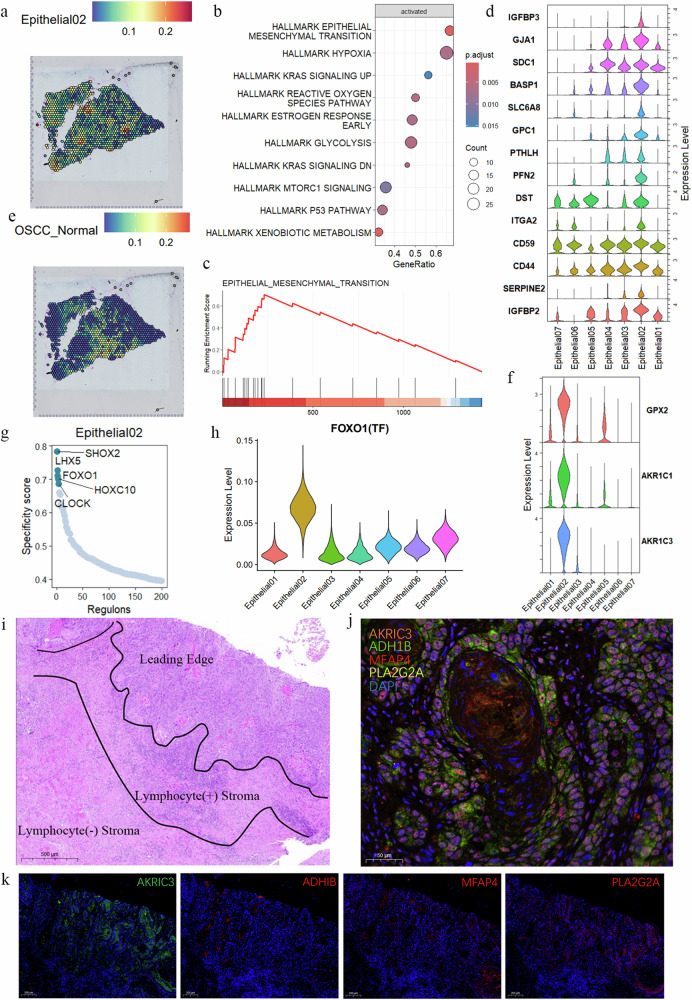
Fig. 7. Evidence of interaction between Epithelial02 tumor cell clusters and OSCC–Normal.
a, e The spatial transcriptomics display the distribution of Epithelial02 and OSCC_Normal in OSCC samples. The top of each panel shows the expression intensity for each cell cluster, with darker colors indicating higher intensity. b GSEA analysis results display the biological processes activated in Epithelial02. c GSEA results show the enrichment scores of EMT in Epithelial02. d The violin plot presents the expression of EMT genes in epithelial cells. f The violin plot shows the expression of GPX2, AKR1C1, and AKR1C3 in epithelial subtype cells. g Specific transcriptional regulators in Epithelial02. h The violin plot illustrates the expression of transcription factor FOXO1 in epithelial subtype cells. i HE-stained tumor sections, including the invasive front, lymphocyte-positive stromal region, and lymphocyte-negative stromal region. j, k Immunofluorescence staining highlights the specific genes in Epithelial02 and OSCC_Normal (AKR1C3, MFAP4, PLA2G2A, and ADH1B). The nuclei of the cells were stained blue with DAPI.
We also explored the positions and possible functions of high-CNV score epithelial subtype (02, 03, 04) based on spatial transcriptomics. Epithelial04 primarily represents the TC and TS of the tumor (Fig. 6b, e), while Epithelial03 is associated with the TS and the LE of the tumor (Fig. 6c, f). Interestingly, Epithelial03 and 04 exhibit relatively independent malignant expression profiles, residing in their respective epithelial spatial domains, and together forming a malignant epithelial region. AIM2 is the specific marker for Epithelial03, while CASP14 is specific to Epithelial04. We propose that Epithelial03 and 04 are independent yet interdependent populations in tumor development. Immunofluorescence using AIM+ to mark Epithelial03 and CASP14+ to mark Epithelial04 showed their co-occurrence in tumor nests (Fig. 6j). Both subtypes were observed in the center of tumor cell clusters, indicating their critical roles in tumor cell composition (Fig. 6k, l). Additionally, we found that CASP14+ Epithelial04 is closer to the granular and keratin layers, whereas AIM+ Epithelial03 is closer to the basal layer (Supplementary Fig. 4d). AIM2, an immune receptor, recognizes abnormal cytoplasmic double-stranded DNA to assemble the inflammasome56. Chronic inflammation is known to contribute to most stages of tumorigenesis, with inflammasome components promoting tumor cell proliferation, survival, immunosuppression, angiogenesis, and metastasis57. Studies have shown that AIM2 enhances the production of matrix metalloproteinases (MMPs) in skin squamous cell carcinoma (SCC), leading to increased invasion, possibly due to the indirect effects of cytokine responses to AIM2 inflammasome activation57. Caspases, including caspase-14, participate in apoptosis, necrosis, and inflammation signaling pathways58,59. Although the role of caspase-14 in cancer is unclear, its activation at the interface between the granular and keratin layers primarily functions within the keratin layer. Aberrant expression and localization of caspase-14 in epithelial tumors often result from altered transcriptional activity, potentially indicating early tumorigenesis60.
Characteristics and functional analysis of Epithelial02 in OSCC
Conversely, the Epithelial02, characterized by the highest inferCNV and malignant prognosis, is not located within the tumor epithelium but is widely distributed in the stromal region at the tumor’s invasive front (Fig. 7a). GSEA results for cluster 02 indicate that hypoxia and EMT are extensively activated in this subtype (Fig. 7b, c) and exhibit significantly higher expression levels compared to other subtype (Fig. 7d). Additionally, pathways related to ROS and glycolysis are also activated (Fig. 7b). During the EMT process of Epithelial02, OSCC_Normal CAFs were consistently present in the normal stromal tissue surrounding Epithelial02 (Fig. 7e), far from the malignant epithelial cell regions (Supplementary Fig. 5a–f).
AKR1C3 is specifically expressed in Epithelial02. Research indicates that AKR1C3 promotes liver cancer proliferation and metastasis through the activation of the NF-κB pathway, releasing pro-inflammatory cytokines and the IL-6/STAT3 pathway61. Another study identifies AKR1C3 as a novel EMT driver in prostate cancer metastasis through ERK signaling activation62. AKR1C3 expression is also positively correlated with poorer survival rates in oropharyngeal squamous cell carcinoma (OPSCC) cohorts63. GPX2, AKR1C1, and AKR1C3 are upregulated in four smoking-related cancers (OSCC, lung adenocarcinoma, bladder adenocarcinoma, and lung squamous cell carcinoma), all associated with the downregulation of arachidonic acid metabolism, an inflammatory pathway and a downstream target of the redox-sensitive Nrf2 transcription factor pathway64. GPX2 expression is negatively correlated with pro-inflammatory cytokines/chemokines and NF-kB activation, where GPX2 knockdown leads to increased secretion of prostaglandin E2 (PGE2) and interleukin-665. In HPV16-positive and HPV-negative OPSCC tumors, upregulated expression of AKR1C1 and AKR1C3 indicates poor prognosis66. In our study, GPX2, AKR1C1, and AKR1C3 are top markers for Epithelial02 (Fig. 7f). The oxidative stress or detoxification (Oxd) program involves various peroxidases and reductases (e.g., GPX2 and AKR1C1) in defense against oxidative damage50.
We analyzed the transcription factors with characteristic activity in Epithelial02, including SHOX2, LHX5, FOXO1, HOXC10, and CLOCK (Fig. 7g). Among these, FOXO1 shows significantly high expression in Epithelial02 (Fig. 7h). Studies have shown that FOXO1 plays crucial roles in cell proliferation, apoptosis, autophagy, metabolism, inflammation, and differentiation by activating or inhibiting downstream target genes and is associated with regulating oxidative stress67, functioning as a switch to modulate mitochondrial ROS production68. Epithelial02 is significantly enriched in oxidative stress and EMT characteristics, widely distributed in the stromal region at the tumor’s invasive front, suggesting its role as a pioneer in tumor invasion.
Given that Epithelial01 is only present in carcinoma in situ and might represent an early stage of the malignant epithelial lineage, we included malignant epithelial subtype from OSCC in trajectory analysis. The results show that malignant epithelial cells in OSCC progress from Epithelial01 as the starting point, with 02, 03, and 04 as trajectory endpoints. Epithelial03 and 04 indicate similar fate outcomes in pseudotime, while Epithelial02 represents a distinct progression direction, highlighting its potential significance (Supplementary Fig. 6a, b).
We further validated these findings with histological staining experiments on OSCC tissues. Tumors were categorized into invasive fronts, stroma enriched with lymphocytes, and stroma without lymphocytes based on histological evidence (Fig. 7i). Positive expression of Epithelial02 (AKRIC3+) was found at the tumor invasive front, in keratin pearls, and surrounding tumor cells, along with infiltrating OSCC_Normal cells (ADH1B+, MFAP4+, PLA2G2A+) (Fig. 7j and Supplementary Fig. 6c). Additionally, extensive positive signals for Epithelial02 were observed at the tumor invasive front. OSCC_Normal (ADH1B+, MFAP4+, PLA2G2A+) surrounded Epithelial02 but were absent from the invasive front, where Epithelial03 (AIM2+) and 04 (CASP14+) were not detected (Supplementary Fig. 6d). These results indicate that Epithelial02 is widely distributed in the stromal region at the tumor’s invasive front and may play a crucial role in guiding tumor invasion. OSCC_Normal CAFs are associated with Epithelial02, suggesting potential interactions between these populations.
Exploring cell communication and interactions
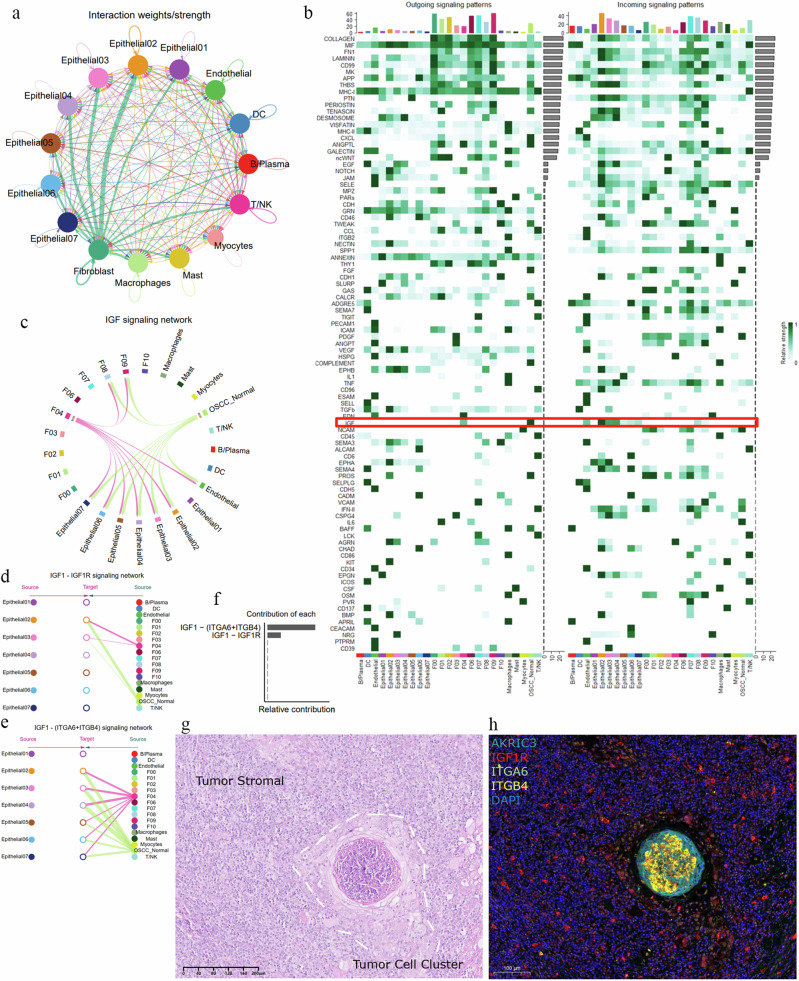
To further investigate the potential interactions between Epithelial02 and OSCC_Normal, we conducted a cell communication analysis. The results revealed that the communication signals between CAFs and Epithelial02 were the strongest (Fig. 8a), supporting our hypothesis that Epithelial02 acts as a pioneer in tumor invasion, which is consistent with its histological localization. Interestingly, OSCC_Normal also exhibited the highest communication intensity with Epithelial02 (Supplementary Fig. 7a). We further analyzed the cell–cell interactions between OSCC_Normal and Epithelial02 and identified 93 signaling interaction networks, including FGF, interleukins (IL-2, IL-6), chemokines (CCL, CXCL), and vascular endothelial growth factor (VEGF) (Fig. 8b).
Fig. 8. The potential role of the IGF1–IGF1R/(ITGA6 + ITGB4) signaling network in OSCC progression.
a Signal interaction strengths between different cell types, with circles representing cell clusters and lines representing communication signals. The thicker the line, the stronger the interaction signal. b Heatmap displaying all cell signaling pathways occurring in OSCC, totaling 93 cell–cell signaling pathways. The red box highlights the IGF pathway, and darker colors within the heatmap indicate higher communication flow intensity. c Analysis of the insulin-like growth factor (IGF) signaling network. d, e The interactions within the IGF signaling network are revealed, including the mutual signals between IGF–IGFR1 and IGF–(ITGA6 + ITGB4) receptor pairs. “Source” denotes the signaling cell subpopulation, while “Target” indicates the receiving cell. The color of the circles represents different cell types. f Analysis of the relative contribution of the IGF–IGFR1 and IGF–(ITGA6 + ITGB4) ligand–receptor pairs to the overall IGF signaling pathway communication. This is shown as the ratio of each ligand–receptor pair’s inferred network communication probability to the total pathway communication probability. g HE staining of OSCC tissue is shown, with white dashed lines indicating tumor cell aggregates surrounded by stromal tissue. h Multiplex immunofluorescence staining marked Epithelial02 and the receptors (AKR1C3, IGF1R, ITGB4, ITGA6), with blue DAPI staining indicating cell nuclei.
Among these, the insulin-like growth factor (IGF) signaling network displayed the most specific and intense communication between OSCC_Normal and Epithelial02 (Fig. 8b). We detailed the interactions within the IGF signaling network, showing that ligands were primarily emitted by OSCC_Normal and F04, with OSCC_Normal having a significantly higher signal intensity than F04 (Supplementary Fig. 7b). Meanwhile, Epithelial02 served as the strongest signal receptor (Fig. 8c and Supplementary Fig. 7b). This signaling network includes two pairs of ligand–receptor interactions: IGF–IGFR1 and IGF–(ITGA6 + ITGB4) (Fig. 8d, e). Comparing the weights of these interactions in epithelial cells and CAFs revealed that IGF–(ITGA6 + ITGB4) was dominant (Fig. 8f). The IGF1 ligand originating from CAFs is expressed exclusively in OSCC_Normal and F04. Similarly, the receptors IGFR1 and the ITGA6 + ITGB4 complex are primarily found in Epithelial02 (Fig. S7c). The cell communication results indicate that OSCC_Normal exerts its effects primarily through the release of IGF1 ligands binding to receptors on Epithelial02.
Studies have reported that insulin-like growth factor receptor (IGF1R) can inactivate FOXO1 through phosphorylation67,69, whereas FOXO1 regulates mitochondrial ROS production as a switch for oxidative stress. Inactivation of FOXO1 leads to uncontrolled oxidative stress, adversely affecting cells. This IGF1–IGF1R inhibitory pathway mediated by OSCC_Normal is primarily observed in Epithelial02 and 03, with a stronger presence in subtype 02, indicating its role at the tumor invasion front (Fig. 8d and Supplementary Fig. 7b). The results of the histological staining of OSCC indicate that tumor cell aggregates in the stroma exhibit strong positivity for P63 and P40, which are markers of squamous cell-derived tumors (Fig. 8g and Supplementary Fig. 7d). Additionally, these invasive tumor cell aggregates show positivity for Epithelial02 (AKR1C3), while positive expression of IGF1R, ITGB4, and ITGA6 is observed surrounding the tumor cell aggregates (Fig. 8h). This finding suggests that the potential intercellular interaction relationships identified through CellChat are indeed present during tumor progression and imply a possible regulatory mechanism.
Interestingly, the communication strength of IGF1–(ITGA6 + ITGB4) is higher than IGF1–IGF1R, with OSCC_Normal activating integrin α6β4 (ITGA6 + ITGB4) across all epithelial subtype except 01. This integrin expression promotes various aspects of tumor progression, including proliferation signaling, invasion and metastasis, apoptosis evasion, and angiogenesis70. ITGA6 can accelerate Keap1 proteasomal degradation, leading to increased Nrf2 stability and activation71. Nrf2 is known as the “master regulator” of the oxidative stress response, providing inducible defense against oxidative damage72. Additionally, the upregulation of ITGB4 in airway epithelial cells enhances wound healing and antioxidative capabilities, which are blocked when ITGB4 is silenced73.
Some studies suggest that ITGA6 + ITGB4 can downregulate oxidative stress through genes like Nrf2, protecting cells from ROS. The competitive binding of IGF1R and ITGA6 + ITGB4 to the same ligand IGF1 suggests that Epithelial02 may actively regulate the expression of ITGA6 + ITGB4 on the membrane surface. This regulation competes for IGF1 released by OSCC_Normal to prevent the adverse effects of the IGF1–IGF1R axis, such as FOXO1 inactivation and ROS overload in tumor cells. Concurrently, ITGA6 + ITGB4 can activate oxidative stress defense and downregulate ROS. Tumor cells use this mechanism to maintain redox homeostasis, ensuring that the pro-tumor effects of ROS are not quenched while avoiding senescence or apoptosis. ITGA6 + ITGB4 is widely expressed in Epithelial02–06, with the highest expression in 02, 03, and 04, suggesting that this process is a tumor cell-initiated response to host defense mechanisms, promoting their growth (Fig. 8e). Furthermore, we found that ITGA6 and ITGB4 are adverse prognostic factors for OSCC (Supplementary Fig. 7e). This also highlights the critical role of ITGA6 and ITGB4 in the invasion process of OSCC; however, further investigation is needed.
Discussion
Thanks to the development of single-cell omics technology, researchers have significantly elucidated the progression of OSCC tumors and the landscape of the TME. Kim et al. used single-cell analysis to explore the heterogeneity of normal tissue, precancerous leukoplakia, primary, and metastatic HNSCC, finding that regulatory T cells expressing LAIR2 promote tumor growth74. Sun et al. established single-cell transcriptomic and spatial transcriptomic maps, observing unique immunosuppressive monocyte subtypes and spatial transitions of VEGF signaling around precancerous lesions75. He et al. discovered significant infiltration changes in SPP1+ macrophages and POSTN+ fibroblasts and their strong interactions with tumor cells during HNSCC progression76.
In our study, we focused on the highly heterogeneous tumor epithelial cells and CAFs in OSCC. First, we identified a group of CAFs regulated by normal fibroblast transcription factors: OSCC_Normal (ADH1B+, MFAP4+, and PLA2G2A+), potentially representing an early stage in OSCC CAFs. Second, we finely characterized the tumor cells, discovering that Epithelial01 (SPINK5+), Epithelial03 (AIM2+), and Epithelial04 (CASP14+) possess specific identities during tumor progression, while Epithelial02 (AKR1C3+) is highly invasive, indicating poor prognosis and closely associated with biological processes such as hypoxia, ROS, and EMT. Third, we found strong interactions between OSCC_Normal and Epithelial02 through spatial transcriptomics and tissue multiplex fluorescence localization. We hypothesize that OSCC_Normal may resist tumor development by regulating oxidative stress in Epithelial02, although the tumor appears to counteract this inhibitory effect. Overall, we enriched the map of tumor cells and CAFs in OSCC, defined different tumor cell states during progression, and discovered the potential inhibitory role of ADH1B+, MFAP4+, and PLA2G2A+ CAFs. This detailed study of the cellular landscape and potential complex interactions in OSCC can aid in developing precise therapeutic strategies for the host cells’ active resistance against tumor progression in the TME.
Fibroblasts are the main components of the TME across all types of stromal cells, engaging in crosstalk with adjacent epithelial cells, affecting local epithelial stem cell behavior77, and coordinating immune system functions by producing chemokines and cytokines. Different fibroblasts exhibit heterogeneity in the cytokines they produce78–80. In many cases, ECM expansion caused by fibroblast proliferation occurs concurrently with tumor development, often limiting early or precancerous lesions81,82. Experimental studies and observations of early lesions surrounded by fibroblasts support the idea that initial fibroblast responses can inhibit tumors83,84. The high heterogeneity within CAFs results in various subgroups that exhibit both tumor-suppressing and tumor-promoting functions.
Identifying whether a CAF promotes or inhibits tumor function remains challenging, requiring multiple lines of evidence to demonstrate simultaneously. Our transcriptional regulatory network identified a CAF subtype in OSCC, OSCC_Normal, whose gene expression matrix resembles other CAF subtypes in OSCC but is regulated similarly to normal fibroblasts, hence named as such. We hypothesize that OSCC_Normal CAFs combat epithelial invasion, as normal fibroblasts have been shown to inhibit cancer progression85. However, finding a subgroup of CAFs with inhibitory functions within the tumor ECM is challenging. We identified transcription factors like HOXA4, ALX1, ALX3, ALX4, and HOXB4 that determine the identity of OSCC_Normal cells. The proportion of OSCC_Normal accounts for ~5% of the entire CAF population (254/4576). However, in scRNA data, cellular function, and proportion are not linearly related. For example, during tooth development, many early differentiated progenitor cells are present in low proportions. Similarly, in single-cell data for sepsis, IL1R2+ immature neutrophils constitute less than 0.2% of all neutrophils, yet they play a crucial role in regulating sepsis86. We analyzed the trajectory fate of CAFs in OSCC, which shows a continuous state of change, with OSCC_Normal indeed being at an initial stage. We hypothesize that if other CAFs could be reverted to the OSCC_Normal state through intervention, it could be decisive in inhibiting tumor invasion. Currently, some researchers have successfully targeted CAFs to inhibit tumors. For instance, in rectal cancer, neoadjuvant chemotherapy has been used to remodel CAF populations, regulating the TME through spatial recruitment and crosstalk, activating immunity, and inhibiting tumor progression via various cytokines, including CXCL12, SLIT2, and DCN87.
Additionally, we observed high specificity within epithelial cells, leveraging spatial transcriptomics to model the potential roles of epithelial subtype during tumor invasion. Our primary focus was on Epithelial02, which exhibited the highest CNV variation and clinical risk, characterized by activated EMT signals, oxidative stress states, and unique spatial localization. This subtype potentially represents the core driving cell cluster of tumor invasion. Research reports indicate that the marker AKR1C3 in Epithelial02 promotes proliferation and metastasis in hepatocellular carcinoma88 and prostate cancer61. Additionally, the most specific transcription factor in Epithelial02, SHOX2, has been extensively reported to promote tumor invasion and metastasis in various cancers, such as prostate cancer89, breast cancer90, and lung cancer91. Throughout tumor development, cancer cells must endure oxidative stress, including during tumorigenesis, EMT, metastasis, and disease recurrence. Tumor cells employ various adaptive strategies to mitigate the cytotoxicity of high ROS levels, maintaining ROS within a dynamic range to preserve high proliferative activity and metabolic levels while avoiding cell death from oxidative stress overload.
During the early stages of cancer initiation, cancer cells typically experience high and sustained ROS loads, necessitating upregulation of their antioxidant defense systems. Notably, different ROS concentrations impact tumor cells differently. Acute, high ROS levels induce apoptosis or necrosis by oxidatively damaging proteins, lipids, and even DNA. Moderate ROS levels can temporarily or permanently arrest cells at specific cell cycle stages, eventually leading to differentiation through physiological responses. Chronic, low ROS levels promote cell proliferation and increase genomic instability in newly formed cells, fostering tumorigenesis.
Cells adapt to oxidative stress in the short term through metabolic reprogramming and in the long term through genetic reprogramming92. Glucose-6-phosphate dehydrogenase (G6PD)-produced NADPH plays a crucial role in alleviating oxidative stress during acute stimulation, as NADPH depletion occurs due to ROS-induced stress93,94. Metabolic reprogramming within a short to medium term involves the abundance and/or subcellular distribution of hypoxia-inducible factor 1α (HIF-1α). This process includes hypoxia and oxidation of Cys-326 in PHD2, stabilizing HIF-1α, leading to transcriptional changes, and shifting from glucose oxidation to glycolysis95. Prolonged oxidative stimulation may cause cancer cells to open redox switches to ensure their survival and proliferation. Several transcription factors, including NRF2, P53, HSF1, NF-KB, FOXO, and PGC-1α, regulate intracellular redox states96–98.
Epithelial02 exhibited invasive capabilities under abnormal ROS conditions. We discovered that the antioxidant transcription factor FOXO1 mediated oxidative stress adaptation in Epithelial02. Activation of the IGF1R leads to phosphorylation and activation of PI3K and AKT. AKT then phosphorylates FOXO, causing it to exit the nucleus and become transcriptionally inactive67,69,99. Through cell communication analysis, we found that OSCC_Normal CAFs inactivate the FOXO1 redox switch in Epithelial02 via their IGF1/IGF1R/PI3K/AKT signaling axis, disrupting the redox homeostasis in invasive Epithelial02. Interestingly, Epithelial02 exploits this resistance mechanism of OSCC_Normal CAFs by consuming IGF1 ligands through (ITGA6 + ITGB4) to reduce oxidative stress interference from host cells.
The IGF1/(ITGA6 + ITGB4) signaling axis also promotes cancer cell invasion and resistance to oxidative stress. Epithelial cells connect to the basement membrane via focal adhesions and hemidesmosomes. Integrin α6β4, as a cell adhesion molecule, anchors cells to the basement membrane through hemidesmosomes, crucial for maintaining the basal-lateral polarity and tissue homeostasis of epithelial cells. During cancer progression, integrin α6β4 is released from hemidesmosomes, altering signaling pathways and stimulating tumor progression through multiple avenues, including sustained proliferative signaling, invasion and metastasis, apoptosis evasion, and angiogenesis100–103. Studies have shown that ITGA6 and ITGB4 can resist intracellular oxidative stress levels71–73, but further research is needed to explore the regulatory mechanisms involved.
In conclusion, this study provides a multi-omics perspective on the ecological map of tumor cells and CAFs. We explored the potential interactions between OSCC_Normal and Epithelial02, contributing to a comprehensive understanding of cell components during OSCC progression and offering molecular intervention strategies for cell interactions in OSCC treatment.
Methods
Data preprocessing and integration
We downloaded two scRNA-seq datasets, GSE172577104 and GSE18191974, from the GEO database. The inclusion criteria were sequencing samples originating from the oral cavity that had not been processed. The raw data were processed using STARsolo’s upstream analysis, resulting in 18 raw sparse matrices from single-cell sequencing. During the single-cell analysis, we used Seurat version 4.4 to merge all data and further screened low-quality cells with thresholds of nFeature >200, <6000, and mitochondrial transcript percentage <25%. We selected 2000 highly variable genes and used Harmony for batch correction and data integration, ensuring the reliability of the analysis results.
Gene regulatory network (GRN) analysis
We employed pySCENIC105 to explore cell state transitions and GRNs. This method leverages transcription factors and cis-regulatory sequences to guide cell state analysis in single-cell RNA-seq data. We established GRNs in OSCC and identified different cell states, quantifying each regulon’s activity using the AUCell algorithm. We marked characteristic regulons between groups and cell types through variance decomposition to further investigate complex regulatory relationships in OSCC.
Cell trajectory analysis
We used Palantir to simulate the trajectory of epithelial cells and fibroblasts in OSCC and measured cell plasticity along the trajectory using entropy. Based on pseudo-time ordering, we assigned each cell state the probability of differentiating into each terminal state106 (https://github.com/JarningGau/scutilsR).
InferCNV for inferring copy number variations
We used the R package inferCNV to study large-scale chromosomal copy number changes in OSCC somatic cells. Using default parameters, we analyzed T/NK cells and macrophages as reference groups to explore gene expression intensities at different genomic locations, inferring chromosomal changes and displaying the results in a heatmap.
Clinical prognostic analysis
We calculated the marker genes for each epithelial subtype and initially screened characteristic genes for each subtype using a threshold of P < 10−6 and FC > 2. Based on the RNA expression matrix and clinical information from TCGA–OSCC, sourced from TCGA_HNSCC datasets, we selected samples from the oral cavity. Prognostic analysis was conducted using KM survival analysis with log-rank tests for statistical significance. We incorporated subtype characteristic genes into Lasso regression, performing multivariate Cox regression analysis on genes appearing more than 45 times, and calculated HR and risk scores for each subtype.
Spatial transcriptomics analysis
We applied the RCTD107 method, a supervised learning approach, to map the spatial information of cell subtype defined in single-cell data onto spatial transcriptomics data. The data were sourced from GSE208253, and we referenced the annotation results from the authors’ data exploration platform.
Cell–cell communication
CellChat108, a tool for quantitatively inferring and analyzing cell–cell communication networks from scRNA-seq data, was used to construct communication patterns and ligand–receptor networks among OSCC. We used the “cellchatDB” ligand–receptor database with default parameters to analyze and create heatmaps displaying signal strength and calculate the relative weight of each ligand–receptor pair within each signal.
Clinical sample collection
Tumor tissues and adjacent normal tissues were collected from OSCC patients, with approval from the Medical Ethics Committee of the Affiliated Stomatology Hospital of Kunming Medical University. All patients signed informed consent forms.
Ethics approval
The study was conducted in accordance with the Declaration of Helsinki and under the approval of the Ethics Committee of Stomatology Hospital Affiliated to Kunming Medical University (Approval Number: KYKQ2024MEC0066). The patients signed written informed consent, agreeing to the anonymized use of their clinical and molecular data for research purposes.
Immunofluorescence staining
Tissue samples were obtained from OSCC surgery patients at the Affiliated Stomatology Hospital of Kunming Medical University. Tissues were fixed in 4% paraformaldehyde for 24 h, dehydrated, embedded, and sectioned into 4 μm slices for staining. After blocking with 3% BSA, sections were incubated overnight at 4 °C with primary antibodies AKRIC3 (Bioss, 11401R, 1:300), ADH1B (Bioss, 10591R, 1:300), MFAP4 (Bioss, 18824R, 1:300), PLA2G2A (Bioss, 20483R, 1:300), AIM2 (Bioss, 5986R, 1:300), and CASP14 (Bioss, 10136R, 1:300). After washing with TBST, sections were incubated at room temperature for 2 h with goat anti-rabbit secondary antibodies and imaged using a fluorescence microscope.
Supplementary information
Acknowledgements
The authors would like to express their sincere gratitude to the Affiliated Stomatology Hospital of Kunming Medical University and the Yunnan Key Laboratory of Stomatology for their generous support and collaboration throughout this project. This research was supported by grants from the National Natural Science Foundation of China (No. 82101004), the Science and Technology Project of Yunnan Province (No. 202101AY070001-189), the Medicine Leading Talent of Yunnan Province Health Care Committee (No. L-2018010), and Yunnan Province Xing Dian Talent (No. XDYC-MY-2022-0052).
Author contributions
H.R.Z. and Y.Z. performed data analyses. Y.M.Q., S.Y.S., and X.Z. performed experiments. H.R.Z. and Y.Z. acquired tissues and data. Z.Y.S. and W.H.W. developed the study design, interpreted data, and supervised the study. All the authors contributed to writing the manuscript.
Data availability
The datasets used in this paper are available online, as described in the “Methods” section.
Code availability
The underlying code for this study is available in GitHub and can be accessed via this link https://github.com/Zhang-hr666/OSCC_code. No new algorithms were developed for this article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hongrong Zhang, Yemei Qian, Yang Zhang.
Contributor Information
Zheyi Sun, Email: sunzheyi@kmmu.edu.cn.
Weihong Wang, Email: wangweihong@kmmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00764-x.
References
- 1.Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Zanoni, D. K. et al. Survival outcomes after treatment of cancer of the oral cavity (1985-2015). Oral. Oncol.90, 115–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bschorer, M. et al. Quality of life and survival rate after primary surgical treatment of oral squamous cell carcinoma: a retrospective study with 18 years of follow-up. J. Craniomaxillofac. Surg.50, 170–177 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer16, 582–598 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Kalluri, R. & Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer6, 392–401 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Fukumura, D. et al. Tumor induction of VEGF promoter activity in stromal cells. Cell94, 715–725 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Kumar, V. et al. Cancer-associated fibroblasts neutralize the anti-tumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell32, 654–668.e655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearon, D. T. The carcinoma-associated fibroblast expressing fibroblast activation protein and escape from immune surveillance. Cancer Immunol. Res.2, 187–193 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Biffi, G. & Tuveson, D. A. Diversity and biology of cancer-associated fibroblasts. Physiol. Rev.101, 147–176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heerboth, S. et al. EMT and tumor metastasis. Clin. Transl. Med.4, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakir, B., Chiarella, A. M., Pitarresi, J. R. & Rustgi, A. K. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol.30, 764–776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramón, Y. C. S. et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J. Mol. Med.98, 161–177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, R. et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat. Med.27, 141–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzywa, T. M., Paskal, W. & Włodarski, P. K. Intratumor and intertumor heterogeneity in melanoma. Transl. Oncol.10, 956–975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhmetkaliyev, A., Alibrahim, N., Shafiee, D. & Tulchinsky, E. EMT/MET plasticity in cancer and Go-or-Grow decisions in quiescence: the two sides of the same coin? Mol. Cancer22, 90 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farahzadi, R. et al. Oxidative stress regulation and related metabolic pathways in epithelial-mesenchymal transition of breast cancer stem cells. Stem Cell Res. Ther.14, 342 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C. & Teichmann, S. A. The technology and biology of single-cell RNA sequencing. Mol. Cell58, 610–620 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Lei, Y. et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J. Hematol. Oncol.14, 91 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet.22, 627–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao, A., Barkley, D., França, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature596, 211–220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Q. et al. Spatially resolved transcriptomics technology facilitates cancer research. Adv. Sci.10, e2302558 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bravo González-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods20, 1355–1367 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoto, K., Ito, K. & Aoki, S. Complex formation between platelet-derived growth factor receptor β and transforming growth factor β receptor regulates the differentiation of mesenchymal stem cells into cancer-associated fibroblasts. Oncotarget9, 34090–34102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahai, E. et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer20, 174–186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman, G. et al. Cancer-associated fibroblast compositions change with breast cancer progression linking the ratio of S100A4(+) and PDPN(+) CAFs to clinical outcome. Nat. Cancer1, 692–708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon, H. et al. TGF-β1-mediated transition of resident fibroblasts to cancer-associated fibroblasts promotes cancer metastasis in gastrointestinal stromal tumor. Oncogenesis10, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov.9, 1102–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi, N., Shukla, N., Prathima, K. M., Das, M. & Dhar, S. K. Novel CAF-identifiers via transcriptomic and protein level analysis in HNSC patients. Sci. Rep.13, 13899 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyu, S. I. et al. Co-expression patterns of cancer associated fibroblast markers reveal distinct subgroups related to patient survival in oropharyngeal squamous cell carcinoma. Front. Cell Dev. Biol.12, 1337361 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bienkowska, K. J., Hanley, C. J. & Thomas, G. J. Cancer-associated fibroblasts in oral cancer: a current perspective on function and potential for therapeutic targeting. Front. Oral Health2, 686337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavie, D., Ben-Shmuel, A., Erez, N. & Scherz-Shouval, R. Cancer-associated fibroblasts in the single-cell era. Nat. Cancer3, 793–807 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebastian, A. et al. Single-cell transcriptomic analysis of tumor-derived fibroblasts and normal tissue-resident fibroblasts reveals fibroblast heterogeneity in breast cancer. Cancers1210.3390/cancers12051307 (2020). [DOI] [PMC free article] [PubMed]
- 35.Wang, Y. et al. Single-cell analysis of pancreatic ductal adenocarcinoma identifies a novel fibroblast subtype associated with poor prognosis but better immunotherapy response. Cell Discov.7, 36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharjee, S. et al. Tumor restriction by type I collagen opposes tumor-promoting effects of cancer-associated fibroblasts. J. Clin. Investig.13110.1172/jci146987 (2021). [DOI] [PMC free article] [PubMed]
- 37.Wu, S. Z. et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J.39, e104063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartoschek, M. et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun.9, 5150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyazaki, Y. et al. Adipose-derived mesenchymal stem cells differentiate into heterogeneous cancer-associated fibroblasts in a stroma-rich xenograft model. Sci. Rep.11, 4690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, M. et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol.73, 1118–1130 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Wang, L. et al. CHRDL2 promotes cell proliferation by activating the YAP/TAZ signaling pathway in gastric cancer. Free Radic. Biol. Med.193, 158–170 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, M. O. et al. The role of fibroblast growth factor-binding protein 1 in skin carcinogenesis and inflammation. J. Investig. Dermatol.138, 179–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang, W. et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology61, 1920–1933 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Han, X. et al. Construction of a human cell landscape at single-cell level. Nature581, 303–309 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues-Junior, D. M. et al. High expression of MLANA in the plasma of patients with head and neck squamous cell carcinoma as a predictor of tumor progression. Head Neck41, 1199–1205 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol.39, 1375–1384 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busam, K. J. & Jungbluth, A. A. Melan-A, a new melanocytic differentiation marker. Adv. Anat. Pathol.6, 12–18 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Iwabuchi, S. et al. Phospholipase A2 group IIA is associated with inflammatory hepatocellular adenoma. Cancers1610.3390/cancers16010159 (2023). [DOI] [PMC free article] [PubMed]
- 49.Vasiukov, G. et al. Cancer-associated fibroblasts in early-stage lung adenocarcinoma correlate with tumor aggressiveness. Sci. Rep.13, 17604 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X. et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun.12, 5291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arora, R. et al. Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response. Nat. Commun.14, 5029 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Q. et al. A novel tumor suppressor SPINK5 targets Wnt/β-catenin signaling pathway in esophageal cancer. Cancer Med.8, 2360–2371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, S., Su, G. & Zheng, X. Inhibition of the tumor suppressor gene SPINK5 via EHMT2 induces the oral squamous cell carcinoma development. Mol. Biotechnol.66, 208–221 (2024). [DOI] [PubMed] [Google Scholar]
- 54.Ye, H. et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics9, 69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, J. & Xu, Y. PITX1 plays essential functions in cancer. Front. Oncol.13, 1253238 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma, B. R., Karki, R. & Kanneganti, T. D. Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur. J. Immunol.49, 1998–2011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karki, R. & Kanneganti, T. D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer19, 197–214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholson, D. W. & Thornberry, N. A. Caspases: killer proteases. Trends Biochem. Sci.22, 299–306 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Markiewicz, A., Sigorski, D., Markiewicz, M., Owczarczyk-Saczonek, A. & Placek, W. Caspase-14—from biomolecular basics to clinical approach. A review of available data. Int. J. Mol. Sci.2210.3390/ijms22115575 (2021). [DOI] [PMC free article] [PubMed]
- 60.Denecker, G., Ovaere, P., Vandenabeele, P. & Declercq, W. Caspase-14 reveals its secrets. J. Cell Biol.180, 451–458 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, Q. et al. A positive feedback loop of AKR1C3-mediated activation of NF-κB and STAT3 facilitates proliferation and metastasis in hepatocellular carcinoma. Cancer Res.81, 1361–1374 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Wang, B. et al. AKR1C3, a crucial androgenic enzyme in prostate cancer, promotes epithelial-mesenchymal transition and metastasis through activating ERK signaling. Urol. Oncol.36, 472.e411–472.e420 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Peraldo-Neia, C. et al. AKR1C3 is a biomarker and druggable target for oropharyngeal tumors. Cell. Oncol.44, 357–372 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Ahmed, K. M. et al. Glutathione peroxidase 2 is a metabolic driver of the tumor immune microenvironment and immune checkpoint inhibitor response. J. Immunother. Cancer1010.1136/jitc-2022-004752 (2022). [DOI] [PMC free article] [PubMed]
- 65.Chun, J. Y. et al. Interleukin-6 regulates androgen synthesis in prostate cancer cells. Clin. Cancer Res.15, 4815–4822 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huebbers, C. U. et al. Upregulation of AKR1C1 and AKR1C3 expression in OPSCC with integrated HPV16 and HPV-negative tumors is an indicator of poor prognosis. Int. J. Cancer144, 2465–2477 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y., Zhou, Y. & Graves, D. T. FOXO transcription factors: their clinical significance and regulation. BioMed. Res. Int.2014, 925350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim, S. W. et al. Klotho enhances FoxO3-mediated manganese superoxide dismutase expression by negatively regulating PI3K/AKT pathway during tacrolimus-induced oxidative stress. Cell Death Dis.8, e2972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hay, N. Interplay between FOXO, TOR, and Akt. Biochim. Biophys. Acta1813, 1965–1970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stewart, R. L. & O’Connor, K. L. Clinical significance of the integrin α6β4 in human malignancies. Lab. Investig.95, 976–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, K. et al. m6A/HOXA10-AS/ITGA6 axis aggravates oxidative resistance and malignant progression of laryngeal squamous cell carcinoma through regulating Notch and Keap1/Nrf2 pathways. Cancer Lett.587, 216735 (2024). [DOI] [PubMed] [Google Scholar]
- 72.Hybertson, B. M., Gao, B., Bose, S. K. & McCord, J. M. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol. Asp. Med.32, 234–246 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Liu, C. et al. Wound repair and anti-oxidative capacity is regulated by ITGB4 in airway epithelial cells. Mol. Cell. Biochem.341, 259–269 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Choi, J. H. et al. Single-cell transcriptome profiling of the stepwise progression of head and neck cancer. Nat. Commun.14, 1055 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun, L. et al. Single-cell and spatial dissection of precancerous lesions underlying the initiation process of oral squamous cell carcinoma. Cell Discov.9, 28 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu, Z. L. et al. Single cell deciphering of progression trajectories of the tumor ecosystem in head and neck cancer. Nat. Commun.15, 2595 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong, H. et al. Tumor microenvironment as niche constructed by cancer stem cells: Breaking the ecosystem to combat cancer. J. Adv. Res.10.1016/j.jare.2024.06.014 (2024). [DOI] [PubMed]
- 78.Philippeos, C. et al. Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations. J. Investig. Dermatol.138, 811–825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, L. C. et al. Targeting fibroblast activation protein in tumor stroma with chimeric antigen receptor T cells can inhibit tumor growth and augment host immunity without severe toxicity. Cancer Immunol. Res.2, 154–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buechler, M. B. & Turley, S. J. A short field guide to fibroblast function in immunity. Semin. Immunol.35, 48–58 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Abbas, O. & Mahalingam, M. Desmoplasia: not always a bad thing. Histopathology58, 643–659 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Beacham, D. A. & Cukierman, E. Stromagenesis: the changing face of fibroblastic microenvironments during tumor progression. Semin. Cancer Biol.15, 329–341 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Rhim, A. D. et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell25, 735–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Özdemir, B. C. et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell25, 719–734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trimboli, A. J. et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature461, 1084–1091 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwok, A. J. et al. Neutrophils and emergency granulopoiesis drive immune suppression and an extreme response endotype during sepsis. Nat. Immunol.24, 767–779 (2023). [DOI] [PubMed] [Google Scholar]
- 87.Qin, P. et al. Cancer-associated fibroblasts undergoing neoadjuvant chemotherapy suppress rectal cancer revealed by single-cell and spatial transcriptomics. Cell Rep. Med.4, 101231 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cui, X. et al. Establishing a proteomics-based signature of AKR1C3-related genes for predicting the prognosis of prostate cancer. Int. J. Mol. Sci.2410.3390/ijms24054513 (2023). [DOI] [PMC free article] [PubMed]
- 89.Yang, W. et al. SHOX2 promotes prostate cancer proliferation and metastasis through disruption of the Hippo-YAP pathway. iScience26, 107617 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teng, Y. et al. SHOX2 cooperates with STAT3 to promote breast cancer metastasis through the transcriptional activation of WASF3. J. Exp. Clin. Cancer Res.40, 274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phuong, N. A. et al. Novel semi-nested real-time PCR assay leveraging extendable blocking probes for improved SHOX2 methylation analysis in lung cancer. Biomolecules1410.3390/biom14060729 (2024). [DOI] [PMC free article] [PubMed]
- 92.Hayes, J. D., Dinkova-Kostova, A. T. & Tew, K. D. Oxidative stress in cancer. Cancer Cell38, 167–197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dick, T. P. & Ralser, M. Metabolic remodeling in times of stress: who shoots faster than his shadow? Mol. Cell59, 519–521 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Kuehne, A. et al. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell59, 359–371 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Lee, G. et al. Oxidative dimerization of PHD2 is responsible for its inactivation and contributes to metabolic reprogramming via HIF-1α activation. Sci. Rep.6, 18928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marinho, H. S., Real, C., Cyrne, L., Soares, H. & Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol.2, 535–562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown, A. K. & Webb, A. E. Regulation of FOXO factors in mammalian cells. Curr. Top. Dev. Biol.127, 165–192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo, X. et al. The AMPK-PGC-1α signaling axis regulates the astrocyte glutathione system to protect against oxidative and metabolic injury. Neurobiol. Dis.113, 59–69 (2018). [DOI] [PubMed] [Google Scholar]
- 99.O’Neill, B. T. et al. Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J. Clin. Investig.126, 3433–3446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo, W. & Giancotti, F. G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol.5, 816–826 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Lipscomb, E. A. & Mercurio, A. M. Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev.24, 413–423 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Nikolopoulos, S. N., Blaikie, P., Yoshioka, T., Guo, W. & Giancotti, F. G. Integrin beta4 signaling promotes tumor angiogenesis. Cancer Cell6, 471–483 (2004). [DOI] [PubMed] [Google Scholar]
- 103.Rabinovitz, I., Toker, A. & Mercurio, A. M. Protein kinase C-dependent mobilization of the alpha6beta4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol.146, 1147–1160 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng, Y. et al. Single-cell profiling of tumor-infiltrating TCF1/TCF7(+) T cells reveals a T lymphocyte subset associated with tertiary lymphoid structures/organs and a superior prognosis in oral cancer. Oral. Oncol.119, 105348 (2021). [DOI] [PubMed] [Google Scholar]
- 105.Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc.15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 106.Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol.37, 451–460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol.40, 517–526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this paper are available online, as described in the “Methods” section.
The underlying code for this study is available in GitHub and can be accessed via this link https://github.com/Zhang-hr666/OSCC_code. No new algorithms were developed for this article.