Abstract
Identifying relationships between structural and functional networks is crucial for understanding the large-scale organization of the human brain. The potential contribution of emerging techniques like functional near-infrared spectroscopy to investigate the structure–functional relationship has yet to be explored. In our study, using simultaneous Electroencephalography (EEG) and Functional near-infrared spectroscopy (fNIRS) recordings from 18 subjects, we characterize global and local structure–function coupling using source-reconstructed EEG and fNIRS signals in both resting state and motor imagery tasks, as this relationship during task periods remains underexplored. Employing the mathematical framework of graph signal processing, we investigate how this relationship varies across electrical and hemodynamic networks and different brain states. Results show that fNIRS structure–function coupling resembles slower-frequency EEG coupling at rest, with variations across brain states and oscillations. Locally, the relationship is heterogeneous, with greater coupling in the sensory cortex and increased decoupling in the association cortex, following the unimodal to transmodal gradient. Discrepancies between EEG and fNIRS are noted, particularly in the frontoparietal network. Cross-band representations of neural activity revealed lower correspondence between electrical and hemodynamic activity in the transmodal cortex, irrespective of brain state while showing specificity for the somatomotor network during a motor imagery task. Overall, these findings initiate a multimodal comprehension of structure–function relationship and brain organization when using affordable functional brain imaging.
Keywords: Multimodal imaging, EEG, fNIRS, Brain structure–function relationship, Neural activity and Hemodynamic response
Subject terms: Neuroscience, Electroencephalography - EEG, Near-infrared spectroscopy, Network models
Introduction
The relationship between brain structure and function is a fundamental concept in neuroscience1,2. Clarifying the interplay or disturbances between structure and function is not only crucial for unveiling the neural mechanisms underlying behaviors, cognition, and disability3–9 but also holds significance in terms of early diagnosis and guiding therapeutic interventions10. Contemporary imaging technology enables the high-throughput reconstruction of neural circuits across various spatiotemporal scales. Anatomically, diffusion magnetic resonance imaging (dMRI) and diffusion tensor imaging (DTI) visualize physical connections and reconstruct nerve fibers, respectively11–13. Functionally, techniques like functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS) detect changes in hemoglobin oxygenation/deoxygenation, serving as indirect indicators of neuronal activity with high spatial resolution14–16. Magnetoencephalography (MEG) and electroencephalography (EEG), instead, measure magnetic and electrical activity generated by groups of neurons with high temporal resolution, respectively17–21. The relationship between functional activity and the underlying structural connectome is usually evaluated using whole-brain diffusion MRI-derived structural connectivity and functional connectivity at a desired temporal scale (either neural or hemodynamical). However, although the neurophysiological and the hemodynamic activity attempt to capture the same underlying biological process, they are sensitive to different physiological mechanisms at different timescales1,22,23 with hemodynamic activity indirectly reflecting the underlying patterns of neural activity due to slow neurovascular coupling. Neurovascular coupling describes a close temporal and regional linkage between neural activity and cerebral blood flow responses. When a specific region of the brain is active, it consumes more oxygen and nutrients, leading to an increase in blood flow to that area to meet the heightened demand24,25. Evidence indicates that the structure–function relationship is not uniform across brain regions, but is shaped based on the unimodal-transmodal organization of the brain. The unimodal-transmodal organization is a well-documented segregation of brain regions based on structure and function26–28. Unimodal areas refer to the areas that are associated with single sensory modality such as visual, auditory, and sensorimotor areas. In contrast, transmodal areas refer to regions associated with high-order or multiple functions such as temporoparietal junctions and frontal poles. Studies show a more robust coupling in the unimodal cortex, which is sensitive to immediate changes in the sensory environment, and a weaker association in the transmodal cortex, which is sensitive to prior context29–34. It is suggested that this regional heterogeneity may potentially reflect underlying molecular and cytoarchitectural gradients, highlighting a hierarchy of time scales of intrinsic fluctuations across the cortex2,35–39. The gradient concept in neuroscience describes the systematic and continuous change of characteristics in cortical organization across the entire cortex. Gradients in cortical structure and function are prevalent throughout the microstructural and functional areas of the mammalian neocortex40,41. Contemporary neuroanatomy studies suggest that cytoarchitectonic and laminar differentiation plays a fundamental role in the information processed by different cortical areas, shaping neural processing dynamics and corticocortical communication42.
Moreover, it is reported that the findings of the structure–function relationship can vary depending on the modality used, suggesting that functional connections estimated from longer time windows overlap significantly with the underlying structural connections. A mismatch may arise with shorter time windows due to distributed delays between neuronal populations, leading to transient phase (de-)synchronization43–45. This raises the possibility that the alignment between the relatively slower functional activity captured by the hemodynamic network and the faster functional activity captured by the electrical network may systematically vary across the cortex.
Evidence suggests that the functional information captured by EEG/MEG and fMRI are consistent early in the cortical hierarchy (the layered structure of the cortex), presumably reflecting activity related to instantaneous changes in the external environment. Conversely, as we move up the hierarchy, there is a gradual divergence between the information captured by these modalities, indicating that they are differently modulated by endogenous inputs and contextual information31,46,47.
However, it remains unclear how functional networks at different time scales relate to one another and their common structural substrate. Previous studies focused primarily on resting-state activity as inferred from EEG, MEG, and fMRI data, at both global and local levels48–52,52–55. However, the extent to which this relationship varies across resting and task-specific periods has not been fully explored. Additionally, while fNIRS recently demonstrated promise in probing resting-state functional connectivity47,56–60, its potential contribution to investigating the structure–functional relationship has yet to be explored.
This recent neuroimaging technique offers improved cost-effectiveness and portability compared to fMRI and provides a higher temporal resolution, typically ranging from 2 to 10 Hz59,61, than the typical fMRI whole-brain scan rate of 0.5 Hz.
Our study aims to investigate the relationship between functional patterns, captured by EEG and fNIRS techniques, and their common structural substrates. We hypothesize that the structure–function relationship varies between electrical (EEG) and hemodynamic (fNIRS) networks due to their different sensitivities to various physiological mechanisms at different timescales, as previously reported for EEG/MEG and fMRI1,22,23. Additionally, we explore how this relationship varies across cognitive states, which may reflect underlying organizational principles governing behavior. To our knowledge, this study represents the first investigation into the structure–functional relationship using fNIRS data alongside EEG. We use a group-consensus structural connectome from the ARCHI database and functional data from an open dataset comprising synchronous EEG and fNIRS recordings during resting-state and task conditions. To link the structural and functional data, we coregister EEG electrodes and fNIRS optodes by spatially aligning them to an anatomical MRI template using digitized positions relative to known scalp landmarks. Structural and functional data are mapped onto the Desikan-Killiany atlas to define regions of interest (ROIs). Our approach integrates structural and functional data within the same anatomical framework (Desikan-Killiany atlas) using the graph signal processing (GSP) tool. Although both data types are not derived from the same subjects, by projecting them onto the same brain atlas, we ensure that functional activity can be directly compared and correlated with the underlying structural connectivity. The introduction of GSP has offered a novel framework for combining structure–function analyses, allowing the extraction of harmonic basis functions from the structural connectivity (SC), which then serves to obtain a graph-spectral representation of the data29,62–66. In this context, Preti and Van De Ville (2019) introduced the structural-decoupling index (SDI)29, which quantifies the degree of structure–function dependency for each brain region. We use the SDI to quantify the global (cross-regional) and local (region-wise) (dis)alignment between the structural and functional networks within and between each modality in both resting state (RS) and task. Additionally, we investigate the degree to which regional patterns of structure–function coupling align with the canonical intrinsic functional networks (RSNs) to gain insights into how the underlying anatomy influences functional networks and supports various cognitive functions. This approach involves comparing hemodynamic and electrical activity in a common frame of reference which might shed light on the interaction between neural signaling and oxygenation demands during different brain conditions. The relevance of understanding the modality dependency of structure–function relationships might have implications for clinical applications, particularly in addressing questions such as identifying the most sensitive modality for detecting changes in functional networks in the presence of disease-specific structural network damage.
Methods
Data preprocessing
The functional data used in this study were acquired from 18 healthy subjects (28.5 ± 3.7 years) through an open dataset67 that has been extensively used in various studies related to BCI applications68–70, and complex analyses71–73. The dataset included synchronous EEG and fNIRS recordings during 1-minute resting state (RS) sessions and 30 trials of 10-second left and right-hand motor imagery tasks (MI) for each participant. EEG data were recorded with 30 electrodes placed according to the international 10-5 system (Fig. 1a) at a 1000 Hz sampling rate (down-sampled to 200 Hz). fNIRS data were collected by 36 channels (14 sources and 16 detectors with an inter-optode distance of 30 mm) (Fig. 1b), following the standardized 10-20 EEG system, at a 12.5 Hz sampling rate (down-sampled to 10 Hz). Two wavelengths at 760 nm and 850 nm were used to measure the changes in oxygenation levels.
Figure 1.
(a) EEG electrode locations. (b) NIRS optode locations. The red dots are the sources and the green dots are the detectors.
Resting-state and task functional fNIRS source timeseries
Resting-state and task fNIRS scans were processed using the MNE toolbox74 and Brainstorm software75. Optical density (OD) transformation was applied to the signals, and the scalp-coupled index (SCI) was used to assess signal quality76. Out of 29 subjects, we excluded those with more than 50% of channels displaying an SCI < 0.7, resulting in a final sample of 18 subjects. The signals were bandpass filtered with cutoff frequencies of 0.02-0.08 Hz using a finite impulse response (FIR) filter. Time segments with excessive head movements, identified through the global variance in temporal derivative (GVTD) metric77, were rejected. To address systemic physiological effects, principal component analysis (PCA) was applied, and components with the highest spatial uniformity value, indicative of superficial skin responses78, were removed. Task-related time epochs from 0 to 10 seconds relative to the stimulus onset were averaged across trials for each subject during both left and right MI. All the epochs had the equal length of 10 seconds. The diffuse optical tomography (DOT) method was implemented to reconstruct the signals in the source space for both the RS and task-related time epochs. A five-tissue segmentation of the Colin27 brain template was used to compute the forward model (sensitivity matrix). The fluences of light for each optode were estimated using Monte Carlo simulations with a number of photons equal to 10 and projecting the sensitivity values within each vertex of the cortex mesh. The forward model was computed from fluences by projecting the sensitivity values within each vertex of the cortex mesh using the Voronoi-based method79. The depth-weighted minimum norm estimate (depth-weighted MNE) method was utilized to estimate the sources. Parcellated time series were then estimated and mapped onto the three-dimensional space using a reduced set of 44 regions of interest (ROIs) from the Desikan-Killiany atlas instead of 82, due to the incomplete coverage of optodes across the scalp. The source time series were then converted into oxygenated hemoglobin (HbO) and deoxygenated hemoglobin (HbR) by applying the modified Beer-Lambert transformation.
and projecting the sensitivity values within each vertex of the cortex mesh. The forward model was computed from fluences by projecting the sensitivity values within each vertex of the cortex mesh using the Voronoi-based method79. The depth-weighted minimum norm estimate (depth-weighted MNE) method was utilized to estimate the sources. Parcellated time series were then estimated and mapped onto the three-dimensional space using a reduced set of 44 regions of interest (ROIs) from the Desikan-Killiany atlas instead of 82, due to the incomplete coverage of optodes across the scalp. The source time series were then converted into oxygenated hemoglobin (HbO) and deoxygenated hemoglobin (HbR) by applying the modified Beer-Lambert transformation.
Resting-state and task functional EEG source timeseries
Resting-state and task EEG scans underwent processing using the MNE toolbox74 and Brainstorm software75. The preprocessing pipeline involved re-referencing with a common average reference and applying a second-order zero-phase Butterworth type FIR high-pass filter with a cutoff of 1 Hz. Independent component analysis (ICA) was performed using the Infomax algorithm for artifact removal. Artifacts were identified and eliminated through visual inspection of various diagnostic measures for each independent component (IC). Subsequently, the cleaned data were low-pass filtered (second-order zero-phase Butterworth type FIR filter) with a cutoff of 45 Hz. Task-related time epochs from 0 to 1.5 seconds relative to the stimulus onset were averaged across trials for each subject during left and right MI. Source estimation (ESI) for both the RS and task-related time epochs was performed using the standardized Low-Resolution brain Electromagnetic Tomography (sLORETA) method on an MRI template (Colin27). Data covariance regularization was applied using the median eigenvalue method to address variable source depth effects. Source orientations were constrained as normal to the 15000-vertex Colin27 surface, and parcellated time series were then estimated and mapped in the same 3D space of the Desikan-Killiany atlas as fNIRS for comparison. The source time series were then decomposed into the typical oscillatory activity by band-pass filtering: delta
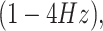 theta
theta
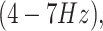 alpha
alpha
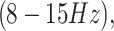 beta
beta
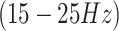 , and gamma
, and gamma
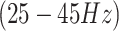 .
.
Structural and diffusion MRI
The group-consensus structural connectome was computed using the structural connectivity matrices derived from the CONNECT/ARCHI database. The dataset consisted of structural connectivity matrices and resting-state functional data (fMRI) obtained from 78 subjects in the ARCHI database. Individual structural connectivity matrices were constructed from diffusion MRI, with connection strength determined by fibers reconstructed through a tractography algorithm80–82. The connectivity data were then mapped onto the Desikan-Killiany atlas and subsequently reduced to 44 ROIs to align with the functional data. The group-consensus structural connectome, representing the entire population, was derived by averaging the structural connectivity (SC) values across subjects.
Structure–function coupling features
As an indicator of the structure–function relationship, the SDI was computed using the Graph Signal Processing (GSP) framework detailed in29. This analysis was performed for each subject, modality (EEG and fNIRS), and condition (resting state (RS), right and left motor imagery (MI) tasks). GSP provides a method for analyzing and representing graph signals in a spectral domain, utilizing the eigenvectors of the Laplacian matrix as fundamental basis functions. The core idea is that these fundamental basis functions can serve as ’building blocks’ for brain activity within the same domain. Each regional brain activity measurement s(t) at different time points t is considered as a graph signal, with each brain region represented as a node within the structural connectivity (SC) graph, described by the adjacency matrix 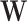 . The building blocks, referred to as harmonics, are obtained through an eigendecomposition of the symmetrically normalized Laplacian matrix
. The building blocks, referred to as harmonics, are obtained through an eigendecomposition of the symmetrically normalized Laplacian matrix 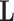 of
of 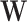 , denoted as:
, denoted as:
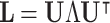 |
1 |
where 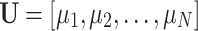 , is the matrix of eigenvectors
, is the matrix of eigenvectors 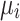 , and
, and 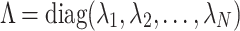 is the diagonal matrix of eigenvalues
is the diagonal matrix of eigenvalues 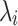 . Each eigenvalue
. Each eigenvalue 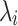 can be interpreted as the spatial frequency of the corresponding structural harmonic (eigenvector)
can be interpreted as the spatial frequency of the corresponding structural harmonic (eigenvector) 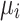 .
.
Subsequently, the graph Fourier transform (GFT) is utilized on the graph signal s(t) to extract their graph Fourier coefficients 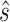 . Given the parcellated source reconstructed signal as an array of dimensionality
. Given the parcellated source reconstructed signal as an array of dimensionality 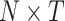 , where N is the number of brain regions (
, where N is the number of brain regions (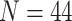 ) and T is the number of time points, each
) and T is the number of time points, each 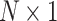 column of this array is an activation pattern s(t) indexed by time t and their graph Fourier coefficients
column of this array is an activation pattern s(t) indexed by time t and their graph Fourier coefficients 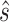 represented as:
represented as:
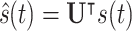 |
2 |
where UT is the transposed of the eigenvector matrix 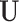 . Then, the filtering process, in the graph spectral domain, is applied to extract relevant frequency components, by using ideal low-pass
. Then, the filtering process, in the graph spectral domain, is applied to extract relevant frequency components, by using ideal low-pass 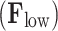 and high-pass
and high-pass 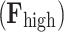 filters. These filters are represented as diagonal matrices with non-zero entries corresponding to the components to be retained. The cut-off frequency is determined based on a median split criterion, which ensures that the energy distribution in the retained frequency components is balanced between the lower and the upper portion of the signal spectrum. For each subject, EEG band, oxy- deoxyhemoglobin, and conditions (RS, left and right MI) we computed the power spectral density (PSD) using the Matlab pwelch function and identified the cut-off frequency that divided the averaged spectrum into two portions of approximately equal energy. The filtered graph signals are transformed back into the temporal domain by applying the inverse graph Fourier transform. This results in a low-frequency functional activity component
filters. These filters are represented as diagonal matrices with non-zero entries corresponding to the components to be retained. The cut-off frequency is determined based on a median split criterion, which ensures that the energy distribution in the retained frequency components is balanced between the lower and the upper portion of the signal spectrum. For each subject, EEG band, oxy- deoxyhemoglobin, and conditions (RS, left and right MI) we computed the power spectral density (PSD) using the Matlab pwelch function and identified the cut-off frequency that divided the averaged spectrum into two portions of approximately equal energy. The filtered graph signals are transformed back into the temporal domain by applying the inverse graph Fourier transform. This results in a low-frequency functional activity component 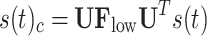 , which is coupled to the structure, and a high-frequency functional activity component
, which is coupled to the structure, and a high-frequency functional activity component 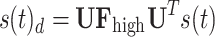 , more decoupled from the structure. The ratio between these two signal portions yields the SDI, quantified as:
, more decoupled from the structure. The ratio between these two signal portions yields the SDI, quantified as:
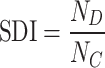 |
3 |
where 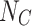 (coupling) and
(coupling) and 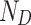 (decoupling) are the filtered spectral coefficients computed as the
(decoupling) are the filtered spectral coefficients computed as the 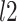 -norm across time of the coupled
-norm across time of the coupled 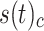 and decoupled
and decoupled 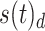 portions for each brain region. The SDI map for each subject is obtained for both the RS and the averaged SDI values of the left and right MI task (named task) for each EEG frequency band and fNIRS hemoglobin type.
portions for each brain region. The SDI map for each subject is obtained for both the RS and the averaged SDI values of the left and right MI task (named task) for each EEG frequency band and fNIRS hemoglobin type.
Statistical analyses significance testing
We elucidated how structural-function coupling systematically varies within and between modalities (EEG and fNIRS), globally across regions of interest (ROIs) and over the neocortex (for each ROI), from the perspective of topological, intrinsic functional organization during RS and task conditions. The statistical analysis was performed using the Matlab toolbox (version 2020a)83. We assessed the normality of data distribution (SDI across subjects and frequency bands for EEG and across subjects and hemoglobin types for fNIRS) by using the Skewness Matlab function. Given that the data did not exhibit a normal distribution (EEG: RS Skewness = − 0.6652, Task Skewness = − 0.6393; fNIRS: RS Skewness = − 0.5493, Task Skewness = − 0.5107), we opted for a non-parametric statistical tests.
False discovery rate (FDR) correction was applied to account for multiple comparisons, controlling the family-wise error rate at 0.05. Pearson’s correlation coefficient (r) was used to assess the similarity of SDI values across subjects and ROIs within (comparing the frequency bands and the hemoglobin types) and between modalities (comparing each frequency band with each hemoglobin type).
Global structure–function coupling
Our initial hypothesis aimed to determine significant differences in the overall distribution of SDI values between modalities for both RS and task conditions. To test this we used the Wilcoxon signed-rank test:
to compare EEG SDI values across ROIs, subjects, and frequency bands with fNIRS SDI values across ROIs, subjects, and hemoglobin types (HbO, HbR) for each condition.
to compare the SDI values within each modality for each condition. Specifically, we compared SDI values between each pair of frequency bands for EEG and between HbO and HbR for fNIRS for both RS and task.
to examine the role of different oscillations and hemoglobin types in modulating the dynamics of the structure–function relationship in response to the transition from RS to task, we compared SDI values within each modality between conditions, assessing each pair of frequency bands and HbO with HbR.
to compare the SDI values between modalities for each condition. Specifically, we compared each pair of EEG frequency bands with each HbO and HbR fNIRS.
Local structure–function coupling
Our second hypothesis aimed to determine how regional heterogeneity influences the structure–function relationships between modalities for both conditions. Regional variations in SDI values across subjects were used to determine the spatial distribution of structure–function coupling for each modality and condition. Then, to identify brain areas where the coupling/decoupling patterns differ between modalities, the Wilcoxon signed-rank test was employed to compare the regional SDI of each EEG band with HbO and HbR separately for both RS and task. False discovery rate (FDR) correction was applied to account for multiple comparisons, controlling the family-wise error rate at 0.05.
To shed light on how the underlying anatomy influences functional networks and supports various cognitive functions, we investigated how regional patterns of structure–function coupling align with the canonical intrinsic functional networks. We categorized regions based on their association with macro-scale intrinsic networks84 and computed the average SDI values across all regions (defined by the Desikan-Killiany atlas) belonging to each of the five functional networks: the default mode network (DMN), the attentional network (AN) which includes both dorsal (DAN) and ventral (VAN) attention network, the frontoparietal network (FPN), the visual network (VIS), and the somatomotor network (SMN)85,86. Subsequently, we performed comparisons between each EEG-band network and their corresponding HbO and HbR network separately, employing the Wilcoxon signed-rank test for both RS and task conditions. Additionally, we compared the cross-band EEG network with its corresponding HbO and HbR pairs separately. We averaged the SDI values across frequency bands and across all regions within each of the five functional networks, then compared them to the corresponding HbO and HbR pairs using the Wilcoxon signed-rank test for both resting state and task conditions.
Results
Global modality-independent structure–function relationship
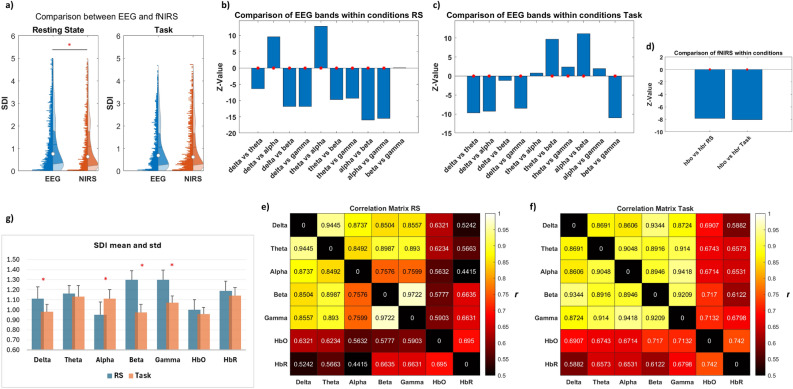
At the global level, our analysis revealed a notable overall difference in structure–function coupling between EEG and fNIRS for the RS. At the same time, no disparity was shown for the task condition (Fig. 2a). When investigating how the structure–function relationship changes across different EEG frequency bands and oxy- and deoxyhemoglobin in both conditions, we observed a general trend of increased coupling for both modalities during the task period, in contrast to the RS (Fig. 2g), alongside distinct patterns of coupling and decoupling. Specifically, in RS, the alpha oscillations demonstrated stronger coupling with the underlying structure than all other bands. However, the delta and theta bands exhibited higher coupling than the beta and gamma bands,
Figure 2.
Global structure–function coupling. (a) Overall assessment of SDI differences between EEG and fNIRS across ROIs, subjects, frequency bands, and hemoglobin types, in both RS and task conditions. The red asterisk highlights statistical significance (p < 0.05). (b,c) Bar plot illustrating the comparison of SDI between EEG bands, and (d) between HbO and HbR, during RS and task, represented as z-values. A higher absolute z-value indicates a more pronounced difference between the two groups. A positive z-value suggests that the first group exhibits higher values than the second group (indicating more decoupling), while a negative z-value implies lower values in the first group (indicating more coupling). The red asterisk denotes statistical significance (p < 0.05). (e,f). Evaluation of similarity within and between modalities (across each EEG frequency band and between HbO and HbR for fNIRS, as well as between each band and each hemoglobin) using Pearson’s correlation coefficient (r). (g) mean and std of SDI values, across ROIs and subjects, for each EEG band and fNIRS chromophore in RS and task. The red asterisk denotes statistical significance (p < 0.05).
while no significant differences were observed between the beta and gamma bands (Fig. 2b).
During the task, distinct patterns emerged, highlighting the interaction between structural and functional dynamics during cognitive engagement. The beta band demonstrated stronger coupling compared to all other bands,
while the delta band exhibited a higher degree of coupling than the theta, alpha, and gamma bands.
In contrast, theta and alpha oscillations showed increased decoupling compared to the delta, beta, and gamma bands (Fig. 2c).
For fNIRS, during both conditions, HbO displayed higher coupling than HbR (Fig. 2d).
When examining the similarities between EEG bands, frequency-wise correlations revealed higher similarities between delta and theta bands (r = 0.94, p < 0.001) and between beta and gamma bands (r = 0.97, p < 0.001) during resting state (RS), whereas alpha bands exhibited the lowest similarities (r = 0.75, p < 0.001). Conversely, in the task condition, the highest similarities were found between alpha and gamma bands (r = 0.94, p < 0.001) and delta and beta bands (r = 0.93, p < 0.001).
The similarities between HbO and HbR yielded correlation values of r = 0.69 (p < 0.001) for RS and r = 0.74 (p < 0.001) for the task (Fig. 2e,f).
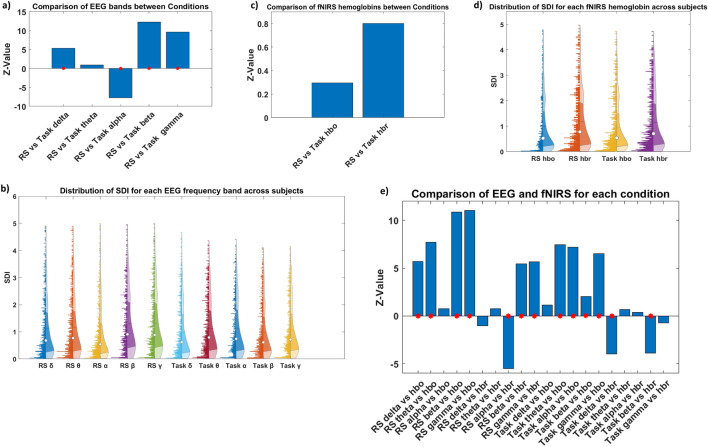
To give more insight into the role of the different oscillations and hemoglobins in modulating the dynamics of the structure–function relationship in response to the transition from RS to task, we compared each EEG frequency band and each oxy- and deoxyhemoglobin fNIRS between conditions. The observed patterns indicate that different EEG frequency bands exhibit varied dynamics of coupling/decoupling in the transition from RS to MI task. Specifically, higher coupling was observed within the alpha band during the RS accompanied by decoupling trends during the Task. Conversely, higher decoupling characterized the delta, beta, and gamma bands in RS, a trend that shifted towards higher coupling during the task. No notable differences were observed in the theta band (Fig. 3a,b).
Figure 3.
Global structure–function coupling. Bar plot comparing SDI values for each EEG band in (a) and for each HbO and HbR in (c) across conditions, displayed as z-values. A higher absolute z-value indicates a more significant difference between the two groups. A positive z-value suggests that the first group has higher values than the second group (indicating more decoupling), while a negative z-value implies lower values in the first group (indicating more coupling). The red asterisks indicate statistical significance (p < 0.05). (b–d) Illustration of SDI distribution across ROIs and subjects for each EEG band and each hemoglobin for fNIRS, respectively, in both resting state and task conditions. (e) Comparison between modalities (each band with each hemoglobin) for both resting state and task, represented as z-values.
These observations may reflect a task-specific modulation of neural activity for specific brain conditions.
On the contrary, no significant differences in the structure–function relationship were observed between RS and task for both HbO and HbR, suggesting an overall relatively stable relationship between oxygenated/deoxygenated hemoglobin levels and functional activity across both brain states (Fig. 3c,d).
Global modality-dependent structure–function relationship
Finally, we aimed to explore the interaction between the two neuroimaging modalities by comparing each EEG frequency band and fNIRS hemoglobin measures in both the RS and task conditions. We observed nuanced structure–functional patterns in limited-band EEG compared to oxy- and deoxyhemoglobin in both RS and task conditions. Specifically, HbO was more aligned with the alpha band, as no differences were highlighted between them, and demonstrated a higher coupling with the underlying structure compared to the other bands. This effect was particularly pronounced when compared to the faster bands (beta and gamma) during the RS. Contrarily, HbR more closely resembles the slower frequency bands (delta and theta) while showing higher decoupling than the alpha band.
During the task, HbO exhibited alignment with the delta band, demonstrating stronger coupling compared to all other rhythms, with less pronounced differences observed with the beta band. Conversely, HbR exhibited higher decoupling than the delta and beta bands while being more aligned with theta, alpha, and gamma oscillations (Fig. 3e).
When examining the similarities between EEG bands with oxy- and deoxyhemoglobin, a higher correlation between hemodynamic (HbO) and slower frequencies (delta r = 0.63, p < 0.001 and theta r = 0.62, p < 0.001) was displayed during the RS, and between HbO and faster frequencies (beta and gamma r = 0.663, p < 0.001) during the tasks. In contrast, HbR shows a higher correlation with faster frequencies in both RS (beta and gamma r = 0.71, p < 0.001) and Task (gamma r = 0.68, p < 0.001) conditions (Fig. 2e,f). The summary of statistical tests is reported in Tables S1 and S2 of the Supplementary Materials.
Local modality-independent structure–function relationship
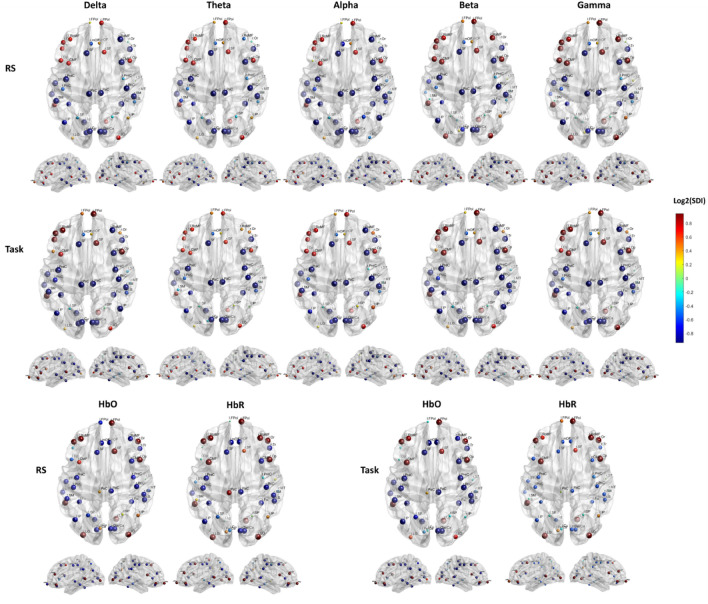
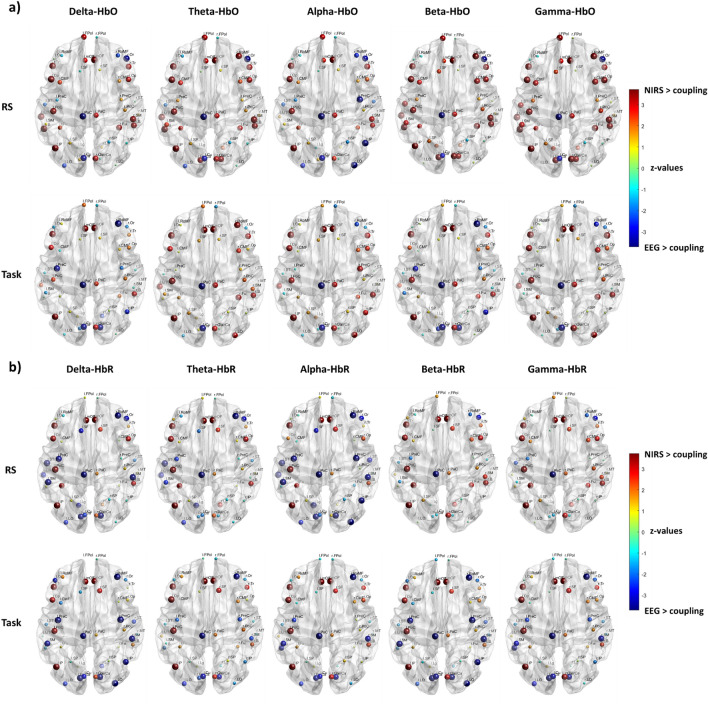
Our second hypothesis aimed to determine how regional heterogeneity influences the structure–function relationships between modalities and how this heterogeneous relationship evolves between RS and task conditions. We observed a similar spatial distribution of the structure–function patterns across different frequency bands and for both the neuroimaging modalities and conditions. This pattern was characterized by higher coupling in the sensory cortex and increased decoupling in the association cortex. Specifically, robust coupling (lower SDI) is evident in sensorimotor and visual areas, while robust decoupling (higher SDI) is observed in prefrontal and posterior areas, including superior and inferior parietal, and occipital association areas. However, we observed slightly divergent patterns across bands and hemoglobin types. In the RS, the alpha band demonstrates higher coupling in the right prefrontal and the right posterior parietal cortices compared to other frequency bands and hemoglobin types. Slower rhythms (delta and theta) exhibit increased coupling in the right prefrontal area, with HbO showing a similar trend. While faster rhythms (beta and gamma) display more decoupling in the right prefrontal area, with HbR following a similar pattern. During the task, there is a trend of increased coupling across the delta, beta, and gamma bands, as well as with HbO, in the right parietal regions compared to the RS. In contrast, an opposite pattern emerges for alpha oscillation and HbR, showing higher decoupling in the same regions (Fig. 4).
Figure 4.
Local structure–function coupling. Spatial distribution of the structure–function coupling across frequency bands (delta, theta, alpha, beta, and gamma) and hemoglobin (HbO and HbR), in RS and task. The blue dots highlight regions more coupled (lower SDI) while the red dots the regions more decoupled (higher SDI).
Local modality-dependent structure–function relationship
We then quantitatively assessed the degree of alignment between structural connectivity and electrical activity versus hemodynamic activity to discern whether different modalities exhibit similar or divergent patterns of regional structure–function coupling for both RS and task conditions.
We found that the correspondence between HbO and HbR with the EEG bands converged in the profile of the regional structure–function coupling. This convergence manifested as consistent patterns of coupling or decoupling within the same ROI, albeit varying in intensity, indicating a predominant coupling of HbO over the different EEG bands than HbR (Fig. 5a). This discrepancy differed between the RS and task conditions, highlighting a higher level of alignment during the task between HbO and EEG, more pronounced for higher frequencies (beta and gamma), as evidenced by fewer ROIs displaying significant disparities (Table S3 of the Supplementary Materials). Conversely, differences between RS and task conditions were also noted for HbR, but they appeared to be less prominent (a similar number of ROIs displayed significant disparities in both conditions). These differences were marked by a tendency towards greater coupling for electrical activity, particularly evident in the lower frequencies (delta, theta, and alpha) during RS, while across all bands during the Task (Fig. 5b).
Figure 5.
Local structure–function coupling. Spatial distribution of the cross-modal differences in structure–function coupling between (a) EEG and HbO, and (b) EEG and HbR, in RS and task conditions, represented as z-values. A higher absolute z-value indicates a more significant difference between the two groups, with dot size indicating the absolute z-value. A positive z-value indicates higher coupling for fNIRS, while a negative z-value indicates higher coupling for EEG. Only significant regions of interest (ROIs) are shown (p < 0.05), FDR corrected.
Network modality-independent structure–function relationship
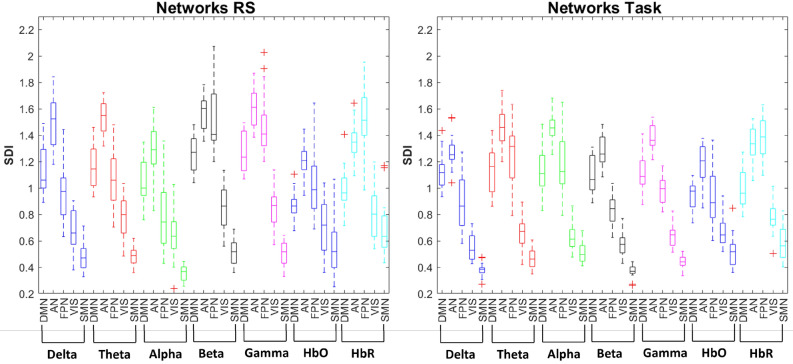
To further explore how this distinctive pattern of structure–function coupling observed between electrical and hemodynamic activity is related to functional networks, we categorized the ROIs based on their association with macro-scale intrinsic networks. We observed higher structure–function coupling in the VIS and SMN networks (unimodal cortex) and lower structure–function coupling in the DMN and AN (transmodal cortex) across all EEG frequency bands and fNIRS hemoglobin types. In contrast, the extent of structure–function coupling in the FPN varied depending on the frequency bands and conditions. A pattern of higher coupling in lower frequency bands (delta, theta, and alpha) and greater decoupling in higher-frequency rhythms (beta and gamma), as well as for HbO during the resting state, was observed. However, higher coupling was found in delta, beta, and HbO during the task. On the other hand, a consistent profile of structure–function relationship in the FPN was observed for deoxyhemoglobin across conditions, characterized by stronger decoupling (Fig. 6). The summary of statistics is reported in Table S4 of the Supplementary Materials.
Figure 6.
Network structure–function coupling. Boxplot illustrating the distribution of the SDI values across the macro-scale intrinsic networks (DMN, AN, FPN, VIS, and SMN) for each EEG frequency band (Delta, Theta, Alpha, Beta, and Gamma), oxyhemoglobin (HbO), and deoxyhemoglobin (HbR) in resting state (RS) and motor imagery task.
Network modality-dependent structure–function relationship
Differences emerged when comparing each EEG band with the corresponding HbO and HbR functional networks. We observed, in RS, greater convergence between EEG and HbO in the primary sensory and motor cortices (VIS and SMN), as well as in the FPN for slower bands (delta, theta), and in the SMN for faster rhythms (beta and gamma). However, the differences in VIS were marginally significant (p = 0.04 for beta and p = 0.03 for gamma band). Greater divergence between modalities was found in the DMN across all frequency bands and in the AN, except for the alpha band. When comparing HbR with EEG, convergences were observed in the DMN and AN for the alpha band, as well as in the FPN and VIS for faster oscillations (beta and gamma).
Under the task condition, the differences between EEG and fNIRS shifted slightly compared to the resting state. In the DMN divergent EEG-HbO pattern was significant across all bands. Greater convergence was observed in the VIS and SMN within the theta and alpha bands, and for VIS also within the gamma band, although in SMN, the difference was only marginally significant (p = 0.04). Convergences were seen in the AN and FPN within the delta and beta bands, extending to the gamma band in FPN. When comparing to HbR, convergent patterns between modalities were highlighted in the AN within the delta, beta, and gamma bands, in the DMN for alpha and beta bands, in the FPN for theta, and the SMN for alpha rhythms. The summary of statistical tests is reported in Table 1.
Table 1.
Network structure–function coupling: summary of statistical tests comparing band-limited EEG and fNIRS measures across different conditions.
| Network | p_delta_HbO | p_theta_HbO | p_alpha_HbO | p_beta_HbO | p_gamma_HbO | p_delta_HbR | p_theta_HbR | p_alpha_HbR | p_beta_HbR | p_gamma_HbR |
|---|---|---|---|---|---|---|---|---|---|---|
| RS | ||||||||||
| DMN | 0.0004 | 0.0002 | 0.0007 | 0.0002 | 0.0002 | 0.0108 | 0.0084 | 0.2145 | 0.0003 | 0.0003 |
| AN | 0.0005 | 0.0004 | 0.157 | 0.0003 | 0.0002 | 0.0279 | 0.0038 | 0.6475 | 0.001 | 0.0014 |
| FPN | 0.3061 | 0.3271 | 0.0249 | 0.0006 | 0.0007 | 0.0002 | 0.0002 | 0.0002 | 0.9133 | 0.7112 |
| VIS | 0.5862 | 0.3271 | 0.3061 | 0.0429 | 0.0347 | 0.0156 | 0.7771 | 0.0096 | 0.3958 | 0.4204 |
| SMN | 0.3491 | 0.3958 | 0.0009 | 0.7439 | 0.5566 | 0.0043 | 0.0043 | 0.0002 | 0.0096 | 0.0084 |
| Network | z-stat_delta_HbO | z-stat_theta_HbO | z-stat_alpha_HbO | z-stat_beta_HbO | z-stat_gamma_HbO | z-stat_delta_HbR | z-stat_theta_HbR | z-stat_alpha_HbR | z-stat_beta_HbR | z-stat_gamma_HbR |
|---|---|---|---|---|---|---|---|---|---|---|
| RS | ||||||||||
| DMN | 3.5494 | 3.68 | 3.3752 | 3.7236 | 3.7236 | 2.5477 | 2.6348 | 1.2412 | 3.5929 | 3.6365 |
| AN | 3.4623 | 3.5494 | 1.4154 | 3.6365 | 3.68 | 2.1993 | 2.8961 | − 0.4573 | 3.2881 | 3.201 |
| FPN | − 1.0234 | 0.9799 | − 2.2428 | 3.4187 | 3.3752 | − 3.68 | − 3.68 | − 3.7236 | − 0.1089 | − 0.3702 |
| VIS | − 0.5444 | 0.9799 | − 1.0234 | 2.0251 | 2.1122 | − 2.417 | − 0.2831 | − 2.5912 | 0.8492 | 0.8057 |
| SMN | − 0.9363 | − 0.8492 | − 3.3316 | − 0.3266 | − 0.5879 | − 2.8525 | − 2.8525 | − 3.68 | − 2.5912 | − 2.6348 |
| Network | p_delta_HbO | p_theta_HbO | p_alpha_HbO | p_beta_HbO | p_gamma_HbO | p_delta_HbR | p_theta_HbR | p_alpha_HbR | p_beta_HbR | p_gamma_HbR |
|---|---|---|---|---|---|---|---|---|---|---|
| Task | ||||||||||
| DMN | 0.0002 | 0.0006 | 0.0006 | 0.0009 | 0.0005 | 0.0198 | 0.0347 | 0.0778 | 0.1221 | 0.0386 |
| AN | 0.1119 | 0.0004 | 0.0005 | 0.1446 | 0.0009 | 0.1841 | 0.0429 | 0.0108 | 0.3061 | 0.3491 |
| FPN | 0.8789 | 0.0029 | 0.005 | 0.4204 | 0.1841 | 0.0002 | 0.0582 | 0.0084 | 0.0002 | 0.0002 |
| VIS | 0.0057 | 0.5566 | 0.5277 | 0.0074 | 0.4204 | 0.0002 | 0.0108 | 0.0016 | 0.0002 | 0.0038 |
| SMN | 0.0007 | 0.157 | 0.9826 | 0.0005 | 0.0386 | 0.0002 | 0.0043 | 0.0854 | 0.0002 | 0.0016 |
| Network | z-stat_delta_HbO | z-stat_theta_HbO | z-stat_alpha_HbO | z-stat_beta_HbO | z-stat_gamma_HbO | z-stat_delta_HbR | z-stat_theta_HbR | z-stat_alpha_HbR | z-stat_beta_HbR | z-stat_gamma_HbR |
|---|---|---|---|---|---|---|---|---|---|---|
| Task | ||||||||||
| DMN | 3.7236 | 3.4187 | 3.4187 | 3.3316 | 3.4623 | 2.3299 | 2.1122 | 1.7638 | 1.546 | 2.0686 |
| AN | 1.5896 | 3.5494 | 3.5058 | 1.4589 | 3.3316 | − 1.3283 | 2.0251 | 2.5477 | − 1.0234 | 0.9363 |
| FPN | − 0.1524 | 2.9832 | 2.809 | − 0.8057 | 1.3283 | − 3.68 | − 1.8944 | − 2.6348 | − 3.7236 | − 3.7236 |
| VIS | − 2.7654 | − 0.5879 | − 0.6315 | − 2.6783 | − 0.8057 | − 3.7236 | − 2.5477 | − 3.1574 | − 3.68 | − 2.8961 |
| SMN | − 3.3752 | − 1.4154 | − 0.0218 | − 3.4623 | − 2.0686 | − 3.7236 | − 2.8525 | − 1.7202 | − 3.7236 | − 3.1574 |
P-values and z-values are reported for each comparison, indicating the significance of observed differences (in italics) or similarities between the measures.
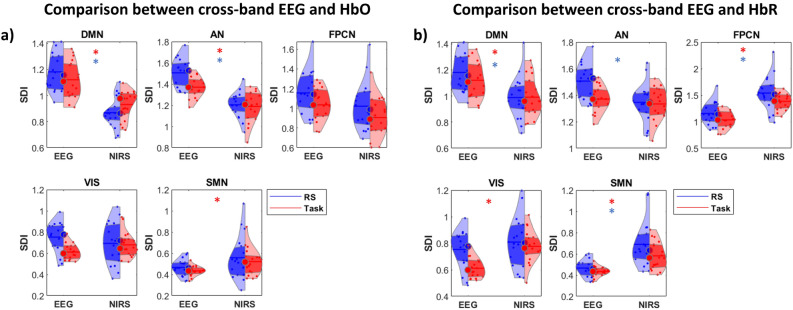
We then performed comparisons between cross-band EEG with HbO and HbR functional networks, highlighting differences in structure–function coupling that emerged in the DMN and AN during both resting state (RS) and task conditions. Notably, these differences were reflected in greater decoupling for EEG in both conditions. During the task, these disparities extended to the SMN, reflecting greater coupling for the electrical network (Fig. 7a). Interestingly, the convergent pattern in the FPN was marginally not significant in both RS and task, with (p = 0.064) and (p = 0.078) respectively. When comparing cross-band EEG with HbR, only the VIS network in RS and the AN in the task displayed convergence between modalities. Greater decoupling was observed within the DMN and AN, while higher coupling was found in the FPN, VIS, and SMN for EEG in both RS and task conditions (Fig. 7b). The summary of statistical tests is reported in Table S5 of the Supplementary Materials.
Figure 7.
Network structure–function coupling. Violin plot illustrating the differences between (a) cross-band EEG and HbO network in RS and task and (b) cross-band EEG and HbR network in RS and task. Colored asterisks indicate significant differences: blue asterisks for RS and red asterisks for task.
Discussion
This study assesses the patterns of structure–function coupling throughout the neocortex by analyzing synchronized EEG and fNIRS data during resting state and motor imagery tasks.
We identify five primary findings. First, the fNIRS structure–function coupling typically resembles slower-frequency EEG structure–function coupling. Second, the structure–function coupling varies across different oscillations between the resting and task periods, alongside the associated hemodynamic response. Third, the structure–function relationship of the hemodynamic activity is specific for oxy- and deoxyhemoglobin in response to neuronal activity. Fourth, local structure–function relationships exhibit heterogeneity but follow a systematic pattern across different modalities and conditions, organized along the sensorimotor association axis. However, discrepancies in specific cortical regions between the two modalities depend on brain state and frequency. Fifth, cross-band representations of neural activity reveal lower correspondence between electrical and hemodynamic activity in the transmodal cortex regardless of brain state while showing specificity for the somatomotor network during a motor imagery task.
Correlation between modalities
At the global level, our results show that fNIRS structure–function coupling typically resembles that of slower-frequency EEG. However, inter-modality differences, as well as variations among different EEG rhythms and conditions, are also evident. As previously reported, slower and intermediate neural rhythms (delta, theta, and alpha bands) and hemodynamic activity (oxyhemoglobin) are more coupled with the underlying structure than faster neural oscillations (beta and gamma) and deoxyhemoglobin31,87. These findings can be explained by the time-scale dependency of structural-functional overlap. Functional connections estimated from larger time windows strongly overlap with the underlying structural connections, while smaller time windows can exhibit a structural-functional network discrepancy due to distributed delays between neuronal populations, causing transient phase (de-)synchronization45,88. When examining the correlation between EEG bands and oxy- and deoxyhemoglobin, we observed a higher correlation between HbO and slower frequencies (delta and theta) during the resting state, and between HbO and faster frequencies (beta and gamma) during tasks. The slower oscillations reflect states of neural inhibition, such as mind-wandering or low cognitive engagement, which align with reduced metabolic demands and oxygenation levels captured by HbO. In contrast, beta and gamma rhythms are typically involved in active cognitive processing, sensory-motor integration, and higher-order functions that require increased neuronal activity and oxygen consumption. HbR, on the other hand, shows a consistent relationship with faster frequencies, suggesting that these faster neural oscillations drive oxygen consumption regardless of brain state due to their higher metabolic cost. Slower frequencies are associated with long-range, large-scale brain networks that reflect the brain’s structural architecture and operate over extended time windows. In contrast, faster frequencies are linked to shorter time windows and more transient, task-specific processes, facilitating rapid, localized communication necessary for cognitive and motor functions. The shift in HbO correlation from slower frequencies during rest to faster frequencies during tasks can be explained by the fact that, despite fNIRS having slower temporal resolution compared to EEG, it still captures, to some extent, the neural synchronization reflected by these faster oscillations.
Task-specific modulation of structure–function coupling
A general trend of increased coupling for both modalities, with no significant difference between the two techniques was observed during the task period, in contrast to the RS. This implies that the modalities exhibit more similar patterns of structure–function relationship when the brain is engaged in a specific cognitive task. This phenomenon may indicate the dynamic integration of information across distant brain regions to facilitate task-relevant processes and cognitive functions. It is reported that a higher resemblance to the structural connectome is evident when the functional network is in a highly integrated state89. However, increased modular segregation might be manifested, reflecting flexibility away from the structural connectome65,89. In the case of fNIRS, hemodynamic activity exhibits an overall relatively stable structure–function relationship across both brain states for both hemoglobin types with HbO consistently demonstrated higher coupling than HbR during both conditions, suggesting that the relationship between structural and functional dynamics is closely linked to the demand for oxygenation. It was reported that oscillation-based networks are stable over long periods, and as in the case of fNIRS, their organization is largely invariant to changing cognitive demands1.
Variations in coupling guided by oscillations
We note variations in structure–function coupling among different oscillations between the resting and task periods, alongside the associated hemodynamic response. Specifically, EEG activity in the alpha band demonstrates a stronger coupling relative to other oscillations at rest. Contrarily, during the task, the delta and beta bands display a more pronounced coupling pattern. This aligns with the well-known notion that alpha oscillations are linked to an idling or inhibitory state90, while delta and beta bands are indicative of task-specific modulation of neural interactions during motor imagery processing. It can be argued that the specific patterns of structure–function coupling in EEG observed in RS and task conditions might align with the concept of nested oscillations and cross-spectral coupling theory91. Indeed, connectome harmonics have been theoretically proposed as a mechanism for macroscopic brain activity, enabling nested functional segregation and integration across multiple spatiotemporal scales92. Oxyhemoglobin activity shows a greater alignment with the alpha band in the resting state and the delta band during the task. It is reported that alpha power shows a close fit between the observed and predicted hemodynamic responses as measured by fNIRS93 and that the modulation of hemodynamics fNIRS response by EEG power is not limited to the alpha band94–97. Moreover, there is some consensus that the best agreement between resting-state fMRI and MEEG/EEG signals is in the alpha and beta bands98,99, both in empirical and simulated MEG connectivity based on coupled oscillators with parameters derived from structural networks52. Interestingly, HbR shows a higher decoupling compared to the alpha rhythm during the RS and to delta and beta oscillations during the task period, indicating the distinct roles of the two hemoglobin types in relation to electrical activity. These findings are in line with concurrent fNIRS-fMRI studies, which have demonstrated a strong correlation between the spatiotemporal characteristics of HbR and the BOLD signal across various experimental conditions100–105.
Regional heterogeneity in electrical and hemodynamic networks
At the local level, we observe a heterogeneous but systematic structure–function relationship across different modalities and conditions. The cross-modal correspondence between modalities is manifested as consistent patterns of coupling or decoupling within the same ROI, albeit varying in intensity and indicating a predominant coupling of HbO over the different EEG bands than HbR. This discrepancy differed between the RS and task conditions, highlighting a higher level of alignment during the task between HbO and EEG, more pronounced for higher frequencies (beta and gamma), as evidenced by fewer ROIs displaying significant disparities. This pattern revealed a greater coupling in the sensory cortex and increased decoupling in the association cortex, following the unimodal to transmodal gradient as previously reported for EEG and fMRI3,31,32. This supports the idea that different brain regions exhibit unique patterns of structural-functional interaction and that the spatial organization of EEG structure–function coupling shares similarities with fNIRS but also displays unique characteristics, varying depending on the specific band and condition and indicating differential sensitivity of the modalities to underlying neural processes.
Frequency-specific patterns of regional specialization
Differences in key regions related to motor imagery tasks can be observed when comparing electrical and hemodynamic activity, which are condition- and frequency-specific. Higher coupling in the dorsolateral prefrontal cortex, premotor cortex, and posterior parietal regions is emphasized for EEG over fNIRS in the alpha band during the resting state and in delta and beta bands during the task. At rest, slower rhythms (delta and theta) also show increased coupling in the same regions with HbO reflecting a similar trend. In contrast, faster rhythms (beta and gamma) demonstrate more decoupling, aligned with the pattern observed in HbR. During the task, these areas showed increased coupling across the delta, beta, and gamma bands, along with HbO, compared to RS. Conversely, alpha oscillations and HbR show higher decoupling in this region. These findings might agree with the idea that the cortical regions operate at different timescales based on their functional specialization within the cortical hierarchy, suggesting that slower rhythms, primarily localized in deeper layers, are associated with higher-order cognitive processing and integration of sensory information, while faster rhythms, primarily localized in superficial layers, are involved in processing more specific or localized information. Previous studies have reported that the correspondence between hemodynamic and electromagnetic connectivity is linked to the underlying cytoarchitectural variation across the cortex, with regions with a higher density of granular cells (unimodal cortex) exhibiting higher cross-modal correspondence and vice versa (transmodal cortex)31,106–108. Indeed, patterns of neurovascular coupling display a topography with the greatest vascularization density in layer IV31,109,110 which encompasses primary sensory cortices and receives the majority of feedforward input111–113. This suggests that areas with richer vasculature exhibit a greater ability to demonstrate strong neurovascular coupling, as measured by both fMRI and fNIRS. Thus, the correspondence between EEG and fNIRS in these regions may reflect the underlying structural and functional architecture, similar to the correspondence observed between EEG and fMRI. However, variability between modalities is dependent on brain state and frequency. Considering the hierarchical model that delineates separate superficial and deep layers, it is plausible that this architecture could exhibit more intricate spectral characteristics114,115. These characteristics may encompass a broader range of temporal dynamics, a complexity that may be challenging to capture with fNIRS due to its lower temporal resolution but can be addressed by EEG.
Convergence and divergence in intrinsic functional networks
When comparing the intrinsic functional networks between EEG and fNIRS, we find a convergent pattern between modalities characterized by greater structure–function coupling in the visual and somatomotor networks and lower coupling in the default mode and attention network. However, nuances arise in the frontoparietal network, where the structure–function coupling is influenced by frequency bands, hemoglobin types, and brain states. Specifically, greater convergence in the FPN is observed between HbO and slower frequency bands (delta, theta) in the resting state, while between HbO and delta, beta, and gamma bands during the task. On the contrary, higher convergence is displayed between HbR and beta and gamma bands at rest, while between HbR and theta band during the task. These findings suggest that the spatial organization of fNIRS structure–function coupling is reminiscent of but distinct from EEG. This resemblance is dependent on the hemoglobin types, the rhythm being considered, and the brain state, implying potentially different modes of intrinsic functional organization in EEG116,117, and aligns with existing research on the distinct functional roles of oxyhemoglobin and deoxyhemoglobin118. Indeed, specific frequency bands may not always directly correspond to fNIRS functional activity, but different canonical EEG bands and coupling modes can influence hemodynamic responses via neurovascular coupling. This has been previously reported in fMRI studies, which demonstrate that synchronized oscillations across multiple frequency bands best explain the well-established fMRI functional networks46. Therefore, we incorporate the cross-band representations of the electrical networks to gain further insights into the convergence/divergence of EEG and fNIRS structure–function relationship within the intrinsic functional networks. Differences between modalities emerge in the DMN and AN in both the resting and task conditions, confirming a lower correspondence between electrical and hemodynamic activity in the transmodal cortex. These differences extend to the SMN during the task period, indicating a broader pattern of discordance between EEG and HbO structure–function coupling, during an active cognitive engagement. These variations are characterized by greater decoupling in the DMN and AN, alongside increased coupling in the SMN for EEG activity.
Influence of long- and short-range connections on modality differences
The transmodal cortex integrates information from multiple sensory modalities and higher-order cognitive processes through long-range connections65,119. Different studies provide evidence for the functional role of long-range neuronal coupling in integrating distributed information in the human brain and demonstrate that inter-areal synchronization predicts behavioral performance, illustrating the functional relevance of large-scale coupling for cognitive processing120–122. Long-range connections may not exhibit strong coupling with specific structural motifs or local cortical architecture, resulting in a weaker association between functional activity and underlying structural features123. These long-range integrative functions of transmodal regions, rely heavily on vascular resources and neurovascular coupling124 and, thus are better captured by fNIRS. This complex processing may involve a higher degree of functional segregation, leading to weaker coupling in EEG measurements but more robust long-range coupling in fNIRS. Conversely, higher coupling in the somatomotor network could be attributed to its strong and well-defined structural connections that support the rapid exchange of information related to motor control and sensory response. These connections are often shorter-range and tied to faster processes, making EEG, with its high temporal resolution, particularly effective in detecting the fast, localized neural dynamics of the SMN, leading to stronger coupling with the underlying structural connectome. Hemodynamic activity measured by fNIRS may not fully capture the rapid dynamics of neural activity in the SMN. Additionally, it may be influenced by factors such as vascular reactivity and metabolic demands, which could introduce additional variability and lead to weaker coupling with the underlying structural connectivity in the SMN compared to EEG93,125. Furthermore, structural measures are identified, delineating differences across cognitive states, with interhemispheric and local dense intrahemispheric connectivity supporting resting-state function and long-range intrahemispheric connectivity supporting task-driven function122. This distinction may explain the greater convergence between EEG and fNIRS during task conditions, as the task-driven state engages more widespread, long-range intrahemispheric networks, which both modalities are capable of detecting despite their differences in temporal resolution. In contrast, the resting-state involves more localized, dense connectivity patterns that may not be as well captured by the slower hemodynamic responses measured by fNIRS, resulting in less convergence between modalities during this state. Our findings are consistent with prior research revealing that connectivity within the beta band becomes prominent during task engagement, confirming the significant role of this frequency range in facilitating interaction between distant brain regions126. The crucial involvement of the beta band in long-range connectivity aligns with predictions from computational models127,128, as well as evidence from invasive recordings in monkeys129–131, and clinical studies involving various patient cohorts132–135.
Our study highlights that structure–function coupling captures characteristic features of the functional organization of the human brain, with these relationships being modality and state-dependent. Specifically, our findings underscore the differential impacts of underlying brain structure on resting and task-driven cognitive states, with clearer frequency effects observed for modalities operating on faster time scales. Additionally, the dynamic nature of the structure–function relationship in the human brain across modalities, brain regions, frequency bands, and brain states emphasizes the need to consider multiple modalities and conditions when investigating mechanisms underlying brain function. These findings set the stage for further exploration into the mechanisms that govern brain function and dysfunction in future research endeavors.
It is essential to acknowledge three key methodological limitations. Firstly, our assessment of structural connectivity relies on diffusion MRI and a constructed group consensus structural connectivity. Although we utilize data from synchronous EEG-fNIRS recordings, our study is constrained by a limited sample size. Future investigations aiming to compare multimodal structure–function relationships could benefit from larger sample sizes and the use of precision imaging techniques that integrate multiple modalities within individual subjects136,137. Secondly, we utilize data with a scan duration of 1 minute for the resting-state condition. While for the task, we captured both electrical and hemodynamic activity at temporal scales able to detect the typical event-related responses (ERPs), for the resting state, this short duration of data, unlike EEG, may not be sufficient for fNIRS due to the slow temporal dynamics of the hemodynamic response to stabilize. However, it has been demonstrated that a 1-minute scan is adequate for obtaining functional connectivity and network metrics stabilization and reproducibility138. Future studies aiming to compare multimodal structure–function relationships could explore longer scan durations. Finally, we used a reduced number of regions of interest derived from the Desikan-Killiany atlas due to the limited coverage of fNIRS optodes. This constraint may have resulted in an incomplete representation of cortical regions, potentially overlooking important functional connections between brain areas. While this limitation suggests that future studies could benefit from the use of more comprehensive brain atlases or parcellation schemes with higher spatial resolution to better align with the imaging modalities used139,140, it is important to recognize that the regions and connections we investigated still provide valuable insights into the structure–function relationship, ensuring meaningful coverage of key brain areas.
Supplementary Information
Acknowledgements
The publication was created within the project of the Minister of Science and Higher Education “Support for the activity of Centers of Excellence established in Poland under Horizon 2020” on the basis of the contract number MEiN/2023/DIR/3796. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 857533. This publication is supported by Sano project carried out within the International Research Agendas programme of the Foundation for Polish Science, co-financed by the European Union under the European Regional Development Fund. Sano Centre for Computational Medicine, Kraków, Poland ( https://sano.science/). MGP was supported by the CIBM Center for Biomedical Imaging, a Swiss research center of excellence founded and supported by Lausanne University Hospital (CHUV), University of Lausanne (UNIL), Ecole polytechnique fédérale de Lausanne (EPFL), University of Geneva (UNIGE) and Geneva University Hospitals (HUG).
Author contributions
R.B.: Conceptualization, Methodology, Data Analysis, Writing - Original draft preparation, Writing, Visualization. M.G.P.: Methodology, Data curation, Writing. C.K.: Methodology, Data curation, Writing. D.V.D.V.: Supervision. A.C.: Supervision. All authors reviewed the manuscript.
Data availibility
Data used in preparation of this article were obtained from the Open access dataset for simultaneous EEG and NIRS brain-computer interface (BCI) https://doc.ml.tu-berlin.de/hBCI/contactthanks.php and the Structural connectivity data of the ARCHI database in the Desikan atlas. Human Brain Project Neuroinformatics Platform https://doi.org/10.25493/91BN-SZ9. The supporting code will be available on GitHub https://github.com/RosmaryBlancoSano/Structure-Function-Relationship-EEG-and-fNIRS.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79817-x.
References
- 1.Sadaghiani, S. & Wirsich, J. Intrinsic connectome organization across temporal scales: New insights from cross-modal approaches. Netw. Neurosci.4, 1–29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suárez, L. E., Markello, R. D., Betzel, R. F. & Misic, B. Linking structure and function in macroscale brain networks. Trends Cogn. Sci.24, 302–315 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Baum, G. L. et al. Development of structure-function coupling in human brain networks during youth. Proc. Natl. Acad. Sci.117, 771–778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, R. et al. Abnormal anatomical rich-club organization and structural-functional coupling in mild cognitive impairment and alzheimer’s disease. Front. Neurol.11, 53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocchi, L. et al. Disruption of structure–function coupling in the schizophrenia connectome. NeuroImage: Clin.4, 779–787 (2014). [DOI] [PMC free article] [PubMed]
- 6.Raj, A. & Powell, F. Models of network spread and network degeneration in brain disorders. Biol. Psychiatry Cogn. Neurosci. Neuroimaging3, 788–797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Heuvel, M. P. & Sporns, O. A cross-disorder connectome landscape of brain dysconnectivity. Nat. Rev. Neurosci.20, 435–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, M. et al. Neuroimaging of brain structure-function coupling mechanism in neuropsychiatric disorders. Front. Neurosci.17, 1270645 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H. et al. Alterations of brain network topology and structural connectivity-functional connectivity coupling in capsular versus pontine stroke. Eur. J. Neurol.28, 1967–1976 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Horn, A. & Fox, M. D. Opportunities of connectomic neuromodulation. Neuroimage221, 117180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, L. et al. Diffusion models: A comprehensive survey of methods and applications. ACM Comput. Surv.56, 1–39 (2023). [Google Scholar]
- 12.Nucifora, P. G., Verma, R., Lee, S.-K. & Melhem, E. R. Diffusion-tensor MR imaging and tractography: exploring brain microstructure and connectivity. Radiology245, 367–384 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Zhang, L. et al. Predicting brain structural network using functional connectivity. Med. Image Anal.79, 102463 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, Z., Zhang, Q., Tu, W. & Zhang, N. Gaining insight into the neural basis of resting-state fMRI signal. Neuroimage250, 118960 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friston, K. J. Functional and effective connectivity: a review. Brain Connect.1, 13–36 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Rahman, M. A., Siddik, A. B., Ghosh, T. K., Khanam, F. & Ahmad, M. A narrative review on clinical applications of fNIRS. J. Digit. Imaging33, 1167–1184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fred, A. L. et al. A brief introduction to magnetoencephalography (MEG) and its clinical applications. Brain Sci.12, 788 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorina-Careta, N., Kurkela, J. L., Hämäläinen, J., Astikainen, P. & Escera, C. Neural generators of the frequency-following response elicited to stimuli of low and high frequency: A magnetoencephalographic (MEG) study. Neuroimage231, 117866 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Cao, J. et al. Brain functional and effective connectivity based on electroencephalography recordings: A review. Hum. Brain Mapp.43, 860–879 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, A. F. & Bolger, D. J. The neurophysiological bases of eeg and eeg measurement: A review for the rest of us. Psychophysiology51, 1061–1071 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Babiloni, C. et al. International federation of clinical neurophysiology (ifcn)–eeg research workgroup: Recommendations on frequency and topographic analysis of resting state eeg rhythms. part 1: Applications in clinical research studies. Clin. Neurophysiol.131, 285–307 (2020). [DOI] [PubMed]
- 22.Hall, E. L., Robson, S. E., Morris, P. G. & Brookes, M. J. The relationship between MEG and fMRI. Neuroimage102, 80–91 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Hari, R. & Parkkonen, L. The brain timewise: how timing shapes and supports brain function. Philos. Trans. R. Soc. B Biol. Sci.370, 20140170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips, A. A., Chan, F. H., Zheng, M. M. Z., Krassioukov, A. V. & Ainslie, P. N. Neurovascular coupling in humans: physiology, methodological advances and clinical implications. J. Cereb. Blood Flow Metab.36, 647–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cauli, B. & Hamel, E. Revisiting the role of neurons in neurovascular coupling. Front. Neuroenerget.2, 1661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huntenburg, J. M., Bazin, P.-L. & Margulies, D. S. Large-scale gradients in human cortical organization. Trends Cogn. Sci.22, 21–31 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Bassett, D. S. et al. Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci.28, 9239–9248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vézquez-Rodríguez, B., Liu, Z.-Q., Hagmann, P. & Misic, B. Signal propagation via cortical hierarchies. Netw. Neurosci.4, 1072–1090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preti, M. G. & Van De Ville, D. Decoupling of brain function from structure reveals regional behavioral specialization in humans. Nat. Commun.10, 4747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffa, A., Amico, E., Liégeois, R., Van De Ville, D. & Preti, M. G. Brain structure-function coupling provides signatures for task decoding and individual fingerprinting. Neuroimage250, 118970 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Liu, Z.-Q., Shafiei, G., Baillet, S. & Misic, B. Spatially heterogeneous structure-function coupling in haemodynamic and electromagnetic brain networks. Neuroimage278, 120276 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Vázquez-Rodríguez, B. et al. Gradients of structure-function tethering across neocortex. Proc. Natl. Acad. Sci.116, 21219–21227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y. et al. Enhanced brain structure-function tethering in transmodal cortex revealed by high-frequency eigenmodes. bioRxiv 2022–07 (2022). [DOI] [PMC free article] [PubMed]
- 34.Zamani Esfahlani, F., Faskowitz, J., Slack, J., Mišić, B. & Betzel, R. F. Local structure-function relationships in human brain networks across the lifespan. Nat. Commun.13, 2053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shafiei, G. et al. Topographic gradients of intrinsic dynamics across neocortex. elife9, e62116 (2020). [DOI] [PMC free article] [PubMed]
- 36.Bazinet, V. et al. Assortative mixing in micro-architecturally annotated brain connectomes. Nat. Commun.14, 2850 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fotiadis, P. et al. Myelination and excitation-inhibition balance synergistically shape structure-function coupling across the human cortex. Nat. Commun.14, 6115 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raut, R. V., Snyder, A. Z. & Raichle, M. E. Hierarchical dynamics as a macroscopic organizing principle of the human brain. Proc. Natl. Acad. Sci.117, 20890–20897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao, R., van den Brink, R. L., Pfeffer, T. & Voytek, B. Neuronal timescales are functionally dynamic and shaped by cortical microarchitecture. Elife9, e61277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Heuvel, M. P., Scholtens, L. H., Barrett, L. F., Hilgetag, C. C. & de Reus, M. A. Bridging cytoarchitectonics and connectomics in human cerebral cortex. J. Neurosci.35, 13943–13948 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haueis, P. Multiscale modeling of cortical gradients: the role of mesoscale circuits for linking macro-and microscale gradients of cortical organization and hierarchical information processing. Neuroimage232, 117846 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Zilles, K. & Amunts, K. Anatomical basis for functional specialization. fMRI: From nuclear spins to brain functions 27–66 (2015).
- 43.Meier, J. et al. A mapping between structural and functional brain networks. Brain Connect.6, 298–311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ton, R., Deco, G. & Daffertshofer, A. Structure-function discrepancy: inhomogeneity and delays in synchronized neural networks. PLoS Comput. Biol.10, e1003736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honey, C. J., Kötter, R., Breakspear, M. & Sporns, O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl. Acad. Sci.104, 10240–10245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shafiei, G., Baillet, S. & Misic, B. Human electromagnetic and haemodynamic networks systematically converge in unimodal cortex and diverge in transmodal cortex. PLoS Biol.20, e3001735 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanco, R., Koba, C. & Crimi, A. Investigating the interaction between EEG and fNIRS: A multimodal network analysis of brain connectivity. J. Comput. Sci.82 (2024).
- 48.Deco, G. et al. Resting-state functional connectivity emerges from structurally and dynamically shaped slow linear fluctuations. J. Neurosci.33, 11239–11252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdelnour, F., Dayan, M., Devinsky, O., Thesen, T. & Raj, A. Functional brain connectivity is predictable from anatomic network’s Laplacian eigen-structure. Neuroimage172, 728–739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atasoy, S., Donnelly, I. & Pearson, J. Human brain networks function in connectome-specific harmonic waves. Nat. Commun.7, 10340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raj, A., Verma, P. & Nagarajan, S. Structure-function models of temporal, spatial, and spectral characteristics of non-invasive whole brain functional imaging. Front. Neurosci.16, 959557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabral, J., Kringelbach, M. L. & Deco, G. Exploring the network dynamics underlying brain activity during rest. Prog. Neurobiol.114, 102–131 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Tewarie, P. et al. Tracking dynamic brain networks using high temporal resolution MEG measures of functional connectivity. Neuroimage200, 38–50 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Glomb, K. et al. Connectome spectral analysis to track eeg task dynamics on a subsecond scale. Neuroimage221, 117137 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Wirsich, J. et al. Complementary contributions of concurrent EEG and fMRI connectivity for predicting structural connectivity. Neuroimage161, 251–260 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Zhang, F., Khan, A. F., Ding, L. & Yuan, H. Network organization of resting-state cerebral hemodynamics and their aliasing contributions measured by functional near-infrared spectroscopy. J. Neural Eng.20, 016012 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, H. et al. Functional connectivity as revealed by independent component analysis of resting-state fNIRS measurements. Neuroimage51, 1150–1161 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Lu, C.-M. et al. Use of fNIRS to assess resting state functional connectivity. J. Neurosci. Methods186, 242–249 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Chen, W.-L. et al. Functional near-infrared spectroscopy and its clinical application in the field of neuroscience: advances and future directions. Front. Neurosci.14, 724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y. & Zhu, C. Assessing brain networks by resting-state dynamic functional connectivity: an fNIRS-EEG study. Front. Neurosci.13, 1430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, F. et al. Correcting physiological noise in whole-head functional near-infrared spectroscopy. J. Neurosci. Methods360, 109262 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Rigoni, I. et al. Structure-function coupling increases during interictal spikes in temporal lobe epilepsy: A graph signal processing study. Clin. Neurophysiol.153, 1–10. 10.1016/j.clinph.2023.05.012 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Huang, W. et al. A graph signal processing perspective on functional brain imaging. Proc. IEEE106, 868–885 (2018). [Google Scholar]
- 64.Lioi, G., Gripon, V., Brahim, A., Rousseau, F. & Farrugia, N. Gradients of connectivity as graph fourier bases of brain activity. Netw. Neurosci.5, 322–336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medaglia, J. D. et al. Functional alignment with anatomical networks is associated with cognitive flexibility. Nat. Hum. Behav.2, 156–164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shuman, D. I., Narang, S. K., Frossard, P., Ortega, A. & Vandergheynst, P. The emerging field of signal processing on graphs: Extending high-dimensional data analysis to networks and other irregular domains. IEEE Signal Process. Mag.30, 83–98 (2013). [Google Scholar]
- 67.Shin, J. et al. Open access dataset for EEG + NIRS single-trial classification. IEEE TNSRE25, 1735–1745 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Abdalmalak, A. et al. Assessing time-resolved fnirs for brain-computer interface applications of mental communication. Front. Neurosci.14, 105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jayaram, V. & Barachant, A. Moabb: trustworthy algorithm benchmarking for bcis. J. Neural Eng.15, 066011 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Gao, Y., Jia, B., Houston, M. & Zhang, Y. Hybrid eeg-fnirs brain computer interface based on common spatial pattern by using eeg-informed general linear model. IEEE Trans. Instrum. Meas.72, 1–10 (2023).37323850 [Google Scholar]
- 71.Maghsoudi, A. & Shalbaf, A. Mental arithmetic task recognition using effective connectivity and hierarchical feature selection from eeg signals. Basic Clin. Neurosci.12, 817 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghouse, A., Faes, L. & Valenza, G. Inferring directionality of coupled dynamical systems using gaussian process priors: Application on neurovascular systems. Phys. Rev. E104, 064208 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Chiarelli, A. M., Croce, P., Merla, A. & Zappasodi, F. Deep learning for hybrid eeg-fnirs brain-computer interface: application to motor imagery classification. J. Neural Eng.15, 036028 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Gramfort, A. et al. MNE software for processing MEG and EEG data. neuroimage86, 446–460 (2014). [DOI] [PMC free article] [PubMed]
- 75.Tadel, F. et al. Brainstorm: a user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. (2011). [DOI] [PMC free article] [PubMed]
- 76.Hernandez, S. M. & Pollonini, L. Nirsplot: a tool for quality assessment of fnirs scans. In Optics and the Brain, BM2C–5 (Optica Publishing Group, 2020).
- 77.Sherafati, A. et al. Global motion detection and censoring in high-density diffuse optical tomography. Hum. Brain Mapp.41, 4093–4112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kohno, S. et al. Removal of the skin blood flow artifact in functional near-infrared spectroscopic imaging data through independent component analysis. J. Biomed. Opt.12, 062111–062111 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Hiyoshi, H. & Sugihara, K. Voronoi-based interpolation with higher continuity. In Proceedings of the sixteenth annual symposium on Computational geometry, 242–250 (2000).
- 80.Rivière, D., Lefranc, S., Poupon, C., Duclap, D. & Mangin, J. F. Structural and functional connectivity data of the archi database in the desikan atlas, 10.25493/91BN-SZ9 (2019).
- 81.Assaf, Y. et al. The connect project: combining macro-and micro-structure. Neuroimage80, 273–282 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Lefranc, S. et al. Groupwise connectivity-based parcellation of the whole human cortical surface using watershed-driven dimension reduction. Med. Image Anal.30, 11–29 (2016). [DOI] [PubMed] [Google Scholar]
- 83.MATLAB, M. Version r2020b. Natick, Massachusetts: The MathWorks Inc (2020).
- 84.Yeo, B. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol.106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest. Neuroimage31, 968–980 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Seitzman, B. A., Snyder, A. Z., Leuthardt, E. C. & Shimony, J. S. The state of resting state networks. Top. Magn. Reson. Imaging TMRI28, 189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deligianni, F., Centeno, M., Carmichael, D. W. & Clayden, J. D. Relating resting-state fMRI and EEG whole-brain connectomes across frequency bands. Front. Neurosci.8, 258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Messé, A., Rudrauf, D., Benali, H. & Marrelec, G. Relating structure and function in the human brain: relative contributions of anatomy, stationary dynamics, and non-stationarities. PLoS Comput. Biol.10, e1003530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukushima, M. et al. Structure-function relationships during segregated and integrated network states of human brain functional connectivity. Brain Struct. Funct.223, 1091–1106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
-
90.Sadaghiani, S. & Kleinschmidt, A. Brain networks and
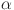 -oscillations: structural and functional foundations of cognitive control. Trends Cogn. Sci.20, 805–817 (2016).
[DOI] [PubMed] [Google Scholar]
-oscillations: structural and functional foundations of cognitive control. Trends Cogn. Sci.20, 805–817 (2016).
[DOI] [PubMed] [Google Scholar] - 91.Sorrentino, P. et al. The structural connectome constrains fast brain dynamics. Elife10, e67400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, R. et al. Hierarchical connectome modes and critical state jointly maximize human brain functional diversity. Phys. Rev. Lett.123, 038301 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Keles, H. O., Barbour, R. L. & Omurtag, A. Hemodynamic correlates of spontaneous neural activity measured by human whole-head resting state EEG+ fNIRS. Neuroimage138, 76–87 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Roche-Labarbe, N. et al. NIRS-measured oxy-and deoxyhemoglobin changes associated with eeg spike-and-wave discharges in children. Epilepsia49, 1871–1880 (2008). [DOI] [PubMed] [Google Scholar]
- 95.Trambaiolli, L. R., Cassani, R. & Falk, T. H. EEG spectro-temporal amplitude modulation as a measurement of cortical hemodynamics: An EEG-fNIRS study. In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 3481–3484 (IEEE, 2020). [DOI] [PubMed]
- 96.Grippa, E. et al. Hemodynamic responses (fNIRS) and EEG modulation of prefrontal cortex during emotion processing. Neuropsychol. Trends 97–98 (2014).
- 97.Chen, L.-C., Sandmann, P., Thorne, J. D., Herrmann, C. S. & Debener, S. Association of concurrent fNIRS and EEG signatures in response to auditory and visual stimuli. Brain Topogr.28, 710–725 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Finger, H. et al. Modeling of large-scale functional brain networks based on structural connectivity from dti: comparison with eeg derived phase coupling networks and evaluation of alternative methods along the modeling path. PLoS Comput. Biol.12, e1005025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brookes, M. J. et al. Measuring functional connectivity using MEG: methodology and comparison with fcMRI. Neuroimage56, 1082–1104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toronov, V. et al. The roles of changes in deoxyhemoglobin concentration and regional cerebral blood volume in the fMRI BOLD signal. Neuroimage19, 1521–1531 (2003). [DOI] [PubMed] [Google Scholar]
- 101.Mehagnoul-Schipper, D. J. et al. Simultaneous measurements of cerebral oxygenation changes during brain activation by near-infrared spectroscopy and functional magnetic resonance imaging in healthy young and elderly subjects. Hum. Brain Mapp.16, 14–23 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huppert, T. J., Hoge, R. D., Diamond, S. G., Franceschini, M. A. & Boas, D. A. A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. Neuroimage29, 368–382 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pouratian, N. et al. Spatial/temporal correlation of BOLD and optical intrinsic signals in humans. Magn. Reson. Med.47, 766–776 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Steinbrink, J. et al. Illuminating the BOLD signal: combined fMRI-fNIRS studies. Magn. Reson. Imaging24, 495–505 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Schroeter, M. L., Kupka, T., Mildner, T., Uludağ, K. & von Cramon, D. Y. Investigating the post-stimulus undershoot of the BOLD signal-a simultaneous fMRI and fNIRS study. Neuroimage30, 349–358 (2006). [DOI] [PubMed] [Google Scholar]
- 106.Shafiei, G., Baillet, S. & Misic, B. Hierarchical decoupling of electromagnetic and haemodynamic cortical networks. bioRxiv 2021–09 (2021).
- 107.Collins, C. E. Variability in neuron densities across the cortical sheet in primates. Brain Behav. Evol.78, 37–50 (2011). [DOI] [PubMed] [Google Scholar]
- 108.Wang, P. et al. Inversion of a large-scale circuit model reveals a cortical hierarchy in the dynamic resting human brain. Sci. Adv.5, eaat7854 (2019). [DOI] [PMC free article] [PubMed]
- 109.Drew, P. J., Mateo, C., Turner, K. L., Yu, X. & Kleinfeld, D. Ultra-slow oscillations in fMRI and resting-state connectivity: neuronal and vascular contributions and technical confounds. Neuron107, 782–804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scheeringa, R. & Fries, P. Cortical layers, rhythms and BOLD signals. Neuroimage197, 689–698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vezoli, J. et al. Cortical hierarchy, dual counterstream architecture and the importance of top-down generative networks. Neuroimage225, 117479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roelfsema, P. R. & Holtmaat, A. Reply to ‘can neocortical feedback alter the sign of plasticity?’. Nat. Rev. Neurosci.19, 637–638 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Markov, N. T. et al. The role of long-range connections on the specificity of the macaque interareal cortical network. Proc. Natl. Acad. Sci.110, 5187–5192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mejias, J. F., Murray, J. D., Kennedy, H. & Wang, X.-J. Feedforward and feedback frequency-dependent interactions in a large-scale laminar network of the primate cortex. Sci. Adv.2, e1601335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Demirtaş, M. et al. Hierarchical heterogeneity across human cortex shapes large-scale neural dynamics. Neuron101, 1181–1194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baillet, S. Magnetoencephalography for brain electrophysiology and imaging. Nat. Neurosci.20, 327–339 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Sadaghiani, S., Brookes, M. J. & Baillet, S. Connectomics of human electrophysiology. Neuroimage247, 118788 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jensen, F. B. The dual roles of red blood cells in tissue oxygen delivery: oxygen carriers and regulators of local blood flow. J. Exp. Biol.212, 3387–3393 (2009). [DOI] [PubMed] [Google Scholar]
- 119.Wang, Y. et al. Long-range functional connections mirror and link microarchitectural and cognitive hierarchies in the human brain. Cereb. Cortex33, 1782–1798 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang, P., Göschl, F., Friese, U., König, P. & Engel, A. K. Long-range functional coupling predicts performance: Oscillatory eeg networks in multisensory processing. Neuroimage196, 114–125 (2019). [DOI] [PubMed] [Google Scholar]
- 121.Knösche, T. R. & Tittgemeyer, M. The role of long-range connectivity for the characterization of the functional-anatomical organization of the cortex. Front. Syst. Neurosci.5, 58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hermundstad, A. M. et al. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc. Natl. Acad. Sci.110, 6169–6174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park, B.-Y. et al. Signal diffusion along connectome gradients and inter-hub routing differentially contribute to dynamic human brain function. Neuroimage224, 117429 (2021). [DOI] [PubMed] [Google Scholar]
- 124.Braga, R. M. & Leech, R. Echoes of the brain: local-scale representation of whole-brain functional networks within transmodal cortex. Neuroscientist21, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schumacher, F. K. et al. The impact of physiological noise on hemodynamic-derived estimates of directed functional connectivity. Brain Struct. Funct.224, 3145–3157 (2019). [DOI] [PubMed] [Google Scholar]
- 126.Womelsdorf, T. et al. Modulation of neuronal interactions through neuronal synchronization. Science316, 1609–1612 (2007). [DOI] [PubMed]
- 127.Lee, J. H., Whittington, M. A. & Kopell, N. J. Top-down beta rhythms support selective attention via interlaminar interaction: a model. PLoS Comput. Biol.9, e1003164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kopell, N., Ermentrout, G., Whittington, M. A. & Traub, R. D. Gamma rhythms and beta rhythms have different synchronization properties. Proc. Natl. Acad. Sci.97, 1867–1872 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pesaran, B., Nelson, M. J. & Andersen, R. A. Free choice activates a decision circuit between frontal and parietal cortex. Nature453, 406–409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buschman, T. J. & Miller, E. K. Top-down versus bottom-up control of attention in the prefrontal and posterior parietal cortices. Science315, 1860–1862 (2007). [DOI] [PubMed]
- 131.Saalmann, Y. B., Pigarev, I. N. & Vidyasagar, T. R. Neural mechanisms of visual attention: how top-down feedback highlights relevant locations. Science316, 1612–1615 (2007). [DOI] [PubMed] [Google Scholar]
- 132.Sun, L. et al. Evidence for dysregulated high-frequency oscillations during sensory processing in medication-naïve, first episode schizophrenia. Schizophr. Res.150, 519–525 (2013). [DOI] [PubMed] [Google Scholar]
- 133.Bangel, K. A. et al. Reduced beta band connectivity during number estimation in autism. NeuroImage: Clin.6, 202–213 (2014). [DOI] [PMC free article] [PubMed]
- 134.Lange, J., Christian, N. & Schnitzler, A. Audio-visual congruency alters power and coherence of oscillatory activity within and between cortical areas. Neuroimage79, 111–120 (2013). [DOI] [PubMed] [Google Scholar]
- 135.Uhlhaas, P. J. & Singer, W. Neural synchrony in brain disorders: relevance for cognitive dysfunctions and pathophysiology. Neuron52, 155–168 (2006). [DOI] [PubMed]
- 136.Naselaris, T., Allen, E. & Kay, K. Extensive sampling for complete models of individual brains. Curr. Opin. Behav. Sci.40, 45–51 (2021). [Google Scholar]
- 137.Groves, A. R. et al. Benefits of multi-modal fusion analysis on a large-scale dataset: life-span patterns of inter-subject variability in cortical morphometry and white matter microstructure. Neuroimage63, 365–380 (2012). [DOI] [PubMed] [Google Scholar]
- 138.Geng, S. & Niu, H. Effect of resting-state fnirs scanning duration on functional brain connectivity and graph theory metrics of brain network. Front. Neurosci.11, 237179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Michel, C. M. et al. Eeg source imaging. Clin. Neurophysiol.115, 2195–2222 (2004). [DOI] [PubMed] [Google Scholar]
- 140.Hiwa, S., Miki, M. & Hiroyasu, T. Validity of decision mode analysis on an roi determination problem in multichannel fNIRS data. Artif. Life Robot.22, 336–345 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in preparation of this article were obtained from the Open access dataset for simultaneous EEG and NIRS brain-computer interface (BCI) https://doc.ml.tu-berlin.de/hBCI/contactthanks.php and the Structural connectivity data of the ARCHI database in the Desikan atlas. Human Brain Project Neuroinformatics Platform https://doi.org/10.25493/91BN-SZ9. The supporting code will be available on GitHub https://github.com/RosmaryBlancoSano/Structure-Function-Relationship-EEG-and-fNIRS.