Abstract
Background
Zinc finger proteins (ZFPs) are important regulators in abiotic and biotic stress tolerance in plants. However, the role of the ZFPs in wheat responding to pathogen infection is poorly understood.
Results
In this study, we found TaZFP8-5B was down-regulated by Puccinia striiformis f. sp. tritici (Pst) infection. TaZFP8-5B possesses a single C2H2-type zinc finger domain with a plant-specific QALGGH motif, and an EAR motif (LxLxL) at the C-terminus. The EAR motif represses the trans-activation ability of TaZFP8-5B. Knocking down the expression of TaZFP8 by virus-induced gene silencing increased wheat resistance to Pst, whereas TaZFP8-5B-overexpressing reduced wheat resistance to stripe rust and rice resistance to Magnaporthe oryzae, suggesting that TaZFP8-5B plays a negative role in the modulation of plant immunity. Using bimolecular fluorescence complementation, split-luciferase, and yeast two-hybrid assays, we showed that TaZFP8-5B interacted with a wheat calmodulin-like protein TaCML21. Knock-down of TaCML21 reduced wheat resistance to Pst.
Conclusions
This study characterized the function of TaZFP8-5B and its interacting protein TaCML21. Our findings provide a new perspective on a regulatory module made up of TaCML21-TaZFP8-5B in plant immunity.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05843-6.
Keywords: Wheat, Cys2/His2-type transcription factor, Calmodulin-like protein, Plant immunity
Background
Plants suffer from possible invasion of pathogens throughout their life cycle. To defend pathogen attack, plants have developed two major innate immune systems: i.e. pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) [1, 2]. PTI is induced by several microbial patterns via cell surface-localized pattern-recognition receptors [3], and ETI is triggered by pathogen effectors via mostly intracellular receptors [4]. PTI has a role in the plant’s basal defense mechanism, triggering the production of reactive oxygen species (ROS), deposition of callose, and the activation of pathogenesis-related (PR) genes [5]. ETI confers strong resistance, leading to a hypersensitive response, which inhibits pathogen proliferation [6].
Transcriptional reprogramming is a major feature of plant immunity and is regulated by a series of transcription factors (TFs) and proteins associated with discrete transcriptional complexes [7]. TFs, such as WRKY, ethylene-responsive factor/APETALA2, basic-domain leucine-zipper, basic helix-loop-helix, and NAM/ATAF/CUC are involved in plant immunity [8–11]. Zinc finger proteins (ZFPs) are the largest family of transcription regulators in plants. ZFPs can be categorized into various subfamilies such as C2H2, C2HC, C2HC5, C3HC4, CCCH, C4, C4HC3, C6, and C8 types [12]. The C2H2-type ZFPs (C2H2-ZFPs) are known to have an important role in regulating the plant defense response to biotic and abiotic stresses [5, 13, 14]. For example, GmZFP03 modulates the expression of two superoxide dismutase1s to improve resistance against Phytophthora sojae in soybean [15]; Bsr-d1 regulates the expression of H2O2-degradation enzymes to accomplish rice blast resistance [5]; ZFP36 is a key regulator in ABA-induced antioxidant defense [16]; PeSTZ1 from Populus euphratica enhances freezing tolerance through the modulation of ROS scavenging by direct regulation of the PeAPX2 expression [17].
There are numerous C2H2-ZF genes in wheat (Triticum aestivum L.) and some of them are reported to be involved in the abiotic stress tolerance. For instance, Li et al. [18] identified 457 C2H2-ZFPs from the wheat genome and 18 C2H2-ZFPs were induced by heat and drought stresses; The overexpression of TaZFP1B or TaZFP13D significantly enhanced antioxidant enzyme activity and improved wheat tolerance to drought [19, 20]. TaZAT8 plays a significant role in regulating tolerance to the inorganic phosphate (Pi)-starvation [21]. However, the role of the C2H2-ZFPs in wheat responding to pathogen infection has been less reported.
Wheat stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most devastating fungal diseases of wheat. Improving host genetic resistance is a sustainable strategy to control this pathogen disease [22]. The plant defense response is determined by major resistance genes (R genes) and/or disease-resistant related genes that are involved in the defense responses [23]. Major R-genes are critical for the activation of hypersensitive response and often provide race-specific disease resistance [24]. The disease-resistant related genes are regulators in the pathogens’ defense pathway and their transcripts change are associated with defense response. Therefore, a continuous understanding of the regulation mechanisms of the defense-responsive genes in wheat against Pst infection are important to develop new disease resistance strategies.
In the present study, we have characterized the function of C2H2-ZF gene, TraesCS5B02G229800 (TaZFP8-5B hereafter), which was induced by Pst inoculation. Knock-down of TaZFP8 significantly improved wheat resistance against Pst, whereas the overexpression of TaZFP8-5B reduced the Pst resistance. Heterologous overexpression of TaZFP8-5B in rice enhanced plants susceptibility to Magnaporthe oryzae. The TaZFP8-5B interacted physically with a CaM-like protein TaCML21 which served as a positive regulator for stripe rust resistance. Our results demonstrate that the TaCML21-TaZFP8-5B complex is a potential key regulator of plant immunity.
Materials and methods
Plant materials and Pst inoculation
Two wheat cultivars, Shumai126 and Fielder were used in the current study. Shumai126 was used to amplify the cDNA sequences of TaZFP8-5B and TaCML21 and conduct BSMV-mediated gene silencing experiments. Fielder was used as receptor material to create TaZFP8-5B-overexpressed transgenic wheat plants. Shumai126 showed intermediate resistance to Pst race CYR34 at the seedling stage [25], while Fielder is susceptible to CYR34 and resistant to CYR23 [26]. The rice cultivar (Oryza sativa L. ssp. Japonica) Zhonghua11 (ZH11) was also transformed to generate TaZFP8-5B-overexpressed plants. Tobacco (Nicotiana benthamiana) was used for transiently expression and sub-cellular localization assays.
Wheat seedlings were grown in a growth chamber under a 16/8 h, 20/16 ◦C light/dark cycle. The Pst races CYR23 or CYR34 were used to inoculate the wheat plants as described by He et al. [27]. Seedlings sprayed with isododecane served as a mock control. Inoculated and mock-inoculated leaves were sampled at different days post inoculation (dpi) for expression analysis. Three biological replicates were conducted for each time point.
RNA extraction and expression analysis
Total RNA from the seedling leaves was isolated using TRNzol Universal Reagent (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. First-strand cDNA synthesis was performed using the Hifair® III 1st Strand cDNA Synthesis Super Mix kit (Shanghai Yeasen Biotechnology Co., Ltd., Shanghai, China). Quantitative real-time PCR (qRT-PCR) was performed in a 10.0 µL reaction volume including 1.0 µL diluted cDNA, 5.0 µL TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa), 400 nM of each primer. The Bio-Rad CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA) was used for amplification under the following program: 95 ◦C for 3 min, 40 cycles of 95 ◦C for 10 s, and 60 ◦C for 10 s. Primers were designed using the qPCR primer database (https://biodb.swu.edu.cn/qprimerdb/) (Supplementary Table S1). The wheat elongation factor TaEF-1α (GenBank accession no. Q03033), and the rice OsUbiquitin 5 (GenBank accession no. AK061988) were used as the internal references, respectively. The expression level was quantified using the 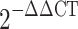 method [28].
method [28].
Sequence analysis
The TaZFP8-5B and TaCML21 sequences were amplified from the cDNA library of Shumai126 seedling leaves inoculated with Pst race CYR34 using specific markers (Supplementary Table S1). The amino acid sequences and protein domains of TaZFP8 and TaCML21 were retrieved from the SMART website (http://smart.embl-heidelberg.de/). Multiple sequence alignments were carried out using MULTALIN (http://multalin.toulouse.inra.fr/multalin/multalin.html). The molecular sizes of TaZFP8 and TaCML21 were predicted using the Compute pI/Mw tool (https://web.expasy.org/compute_pi/). A phylogenetic tree was constructed with MEGA 11 software (http://www.megasoftware.net) using the neighbor-joining method with 1000 bootstrap replicates.
Plasmids construction
The coding sequences of TaZFP8-5B and TaCML21 were amplified from Shumai126 using primers containing BamHI and SalI restriction sites as overhangs. The PCR products were inserted into the plant binary expression vector pCAMBIA2300 with an enhanced green fluorescent protein (eGFP) tag. For barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) system, selected fragments of TaZFP8 and TaCML21 were amplified and inserted into the BSMV-γ vector [29]. The TaZFP8-5B and TaCML21 were sub-cloned into pSPYNE and pSPYCE with XbaI + KpnI and XbaI restriction sites to generate pSPYNE-TaZFP8-5B and pSPYCE-TaCML21 vectors for bimolecular fluorescence complementation (BiFC) assay. The TaZFP8-5B was linked to the N-terminal part of the luciferase reporter gene LUC to generate nLUC-TaZFP8-5B vector and TaCML21 was fused with the C-terminal part of LUC to construct TaCML21-cLUC for split-luciferase (LUC) assay. The primers used in this study are listed in Supplemental Table S1.
BSMV-mediated gene silencing
Different vectors were linearized using appropriate enzymes and transcribed to RNA in vitro. Transcripts of BSMV:α, β, γ (or γ-gene) were mixed in a 1:1:1 ratio and then added to the FES buffer. The third leaves of Shumai126 seedlings were infected with the virus mixture as described by Holzberg et al. [29]. The infected plants were then maintained in a 16/8 h, light/dark cycle at 28 ℃ for approximately 12 days. Following this, the fifth leaves were inoculated with uredospore of Pst race CYR34, and sampled at 1, 2 and 5 dpi for silencing efficiency estimation. The disease phenotype was recorded at 14 dpi.
Histological observation
Seedling leaves sampled at 1 and 5 dpi were observed for the hyphal development of Pst. Leaf segments were stained with wheat germ agglutinin (WGA) (Alexa-488; Thermo Fisher Scientific) conjugated with a fluorescent dye as described by Wang et al. [25] and were further observed using an BX-63 fluorescence microscope (Olympus Corp. Tokyo, Japan). The average value of hyphal length and infection areas from at least 30 infection sites of three independent biological repeats were calculated with ImageJ software.
Yeast two-hybrid assay
A high-quality Y2H library, previously constructed using Pst-infected wheat seedling leaves of Yr15 introgression line AVS + Yr15 (pedigree: V763/6*Avocet) [30], was screened using the Matchmaker GAL4 system (Clontech Laboratories) to identify candidate targets that interact with TaZFP8-5B. The pGBKT-TaZFP8-5B and pGAD-TaCML21 as well as other candidate targets were co-transformed into the yeast strain Y2HGold and grown on the selective medium (SD/-Trp/-Leu, SD/-Trp/-Leu/-His, and SD/-Trp/-Leu/-His/-Ade).
Agrobacterium tumefaciens infiltration assays
The Agrobacterium tumefaciens strains GV3101 (pSoup-p19, Weidi, Shanghai) were transformed with different recombinant vectors and then grown on LB medium supplemented with rifampicin (50 mg L− 1) and kanamycin (50 mg L− 1) at 28 ◦C for 24 h. The recombinant strains were adjusted to an optical density of 0.6 at 600 nm (OD600) and then infiltrated into 4-week-old N. benthamiana leaves. The transformed N. benthamiana plants were grown in a greenhouse at 20 ◦C with a 16 h/8 h light/dark photo-period for subsequent analysis.
The Agrobacterium strains carrying pCAMBIA35S-TaZFP8-5B-eGFP and pCAMBIA35S-TaCML21-eGFP were infiltrated into N. benthamiana leaves for sublocalization. The eGFP fluorescence signals were observed using a laser-scanning confocal microscope at 48 h post infiltration (hpi). The Agrobacterium strains carrying pSPYNE-TaZFP8-5B and pSPYCE-TaCML21 were co-infiltrated into N. benthamiana leaves for BiFC. Two days after infiltration, YFP fluorescence was captured by a Leica STELLARIS STED/EM CPD300 Confocal Laser Microscope with a 488 nm laser. Agrobacteria carrying nLUC-TaZFP8-5B and TaCML21-cLUC were co-infiltration into N. benthamiana leaves for split-LUC. 48 h after transfection, the 1-mM luciferin (Coolaber, China) was sprayed onto the inoculated leaves and the LUC activity was analyzed by a ChemiDoc imaging system (BIO-RAD, USA).
Overexpression of TaZFP8-5B in wheat and rice plants
The coding sequence (CDS) of TaZFP8-5B was fused into the downstream of the maize ubiquitin promoter to generate overexpression vector ProUbi: TaZFP8-5B. The vector was transformed into the wheat cultivar Fielder using the PureWheat technique developed by the Japan Tobacco Company. Positive TaZFP8-5B transgenic plants were identified by PCR using universal primers. The second leaves of the T1 transgenic lines were challenged with the avirulent Pst race CYR23. At 16 dpi, the disease phenotype was observed and the infected wheat leaves were sampled for fungal biomass analysis. The Pst biomass was calculated using the DNA amounts of fungal PstEF against the wheat TaEF-1α amounts by quantitative PCR [26].
The CDS of TaZFP8-5B was inserted into the pCAMBIA2300-CaMV35S-GFP vector to generate TaZFP8-5B-GFP. The construct was introduced into A. tumefaciens strain GV3101 and then transformed into rice cv. ZH11. Positive T0 transgenic plants were confirmed by PCR amplification using a gene-specific primer set (Table S1) and were further self-pollinated. T1 transgenic lines were detected by PCR and the expression of TaZFP8-5B was estimated by qRT-PCR using OsUbiquitin 5 as the endogenous control. Positive lines were selected to evaluate resistance to Magnaporthe oryzae strain GZ8 using a punch inoculation method as described by Li et al. [31]. After 7 dpi, the infected leaves were sampled for relative fungal biomass quantitation using the DNA amounts of M. oryzae Pot2 (Mopot2) against rice ubiquitin DNA amounts [32]. All experiments were repeated three times with consistent results.
Results
Identification of TaZFP8
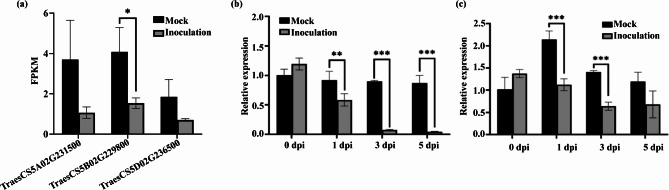
Utilizing our previous RNA-seq data from AVS + Yr15 (highly Pst-resistant), we found the expression levels of TraesCS5B02G229800 (TaZFP8-5B) were significantly reduced by Pst inoculation (Fig. 1a). qRT-PCR analysis confirmed that the expression levels of TraesCS5B02G229800 were significantly down-regulated by the Pst infection in wheat lines AVS + Yr15 and Shumai126 at different time points (Fig. 1b, c), implying the potential role of TraesCS5B02G229800 in wheat defense response.
Fig. 1.
The expression patterns of TaZFP8 upon Pst inoculation. (a) Expression levels of TraesCS5B02G229800 and its homologs in wheat leaves of AVS + Yr15 inoculated with Pst race CYR34 detected by RNA-seq. Expression levels of TaZFP8-5B in Pst-resistant wheat lines AVS + Yr15 (b) and Shumai126 (c) measured by qRT-PCR. Error bars represent SEM from three replications. Asterisks represent the level of significant differences. *p < 0.05, **p < 0.01, and ***p < 0.001
The TraesCS5B02G229800 was 1318 bp in length, with a predicted open reading frame (ORF) of 915 bp, encoded a 304 amino-acid protein with a molecular weight of 26.6 kDa and a pI of 12.67. The N-terminus of the predicted protein possesses a single C2H2-type zinc finger domain with a plant-specific QALGGH motif, and an ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif (LxLxL) at the C-terminus (Fig. S1a, b). The TaZFP8-5B sequence in AVS + Yr15 is identical to TraesCS5B02G229800, while two synonymous SNPs (C510G and G672A) were detected in Shumai126.
Phylogenetic analysis revealed that TaZFP8-5B was clustered with T. dicoccoides zinc finger protein 8-like protein (TdZFP8, accession number XP_037441652.1) and shared high similarity (> 96%) with Aegilops tauschii AetZFP8 (XP_020188179.1) and Hordeum vulgare HvZFP8 (XP_044946812.1) (Fig. S1c). Three homologs of TaZFP8 in Chinese Spring with 96–100% nucleotide sequence similarity were identified on chromosomes 5A, 5B, and 5D.
To investigate its localization in plants, we performed a subcellular localization experiment using N. benthamiana leaves. The fusion construct TaZFP8-5B-eGFP was transiently expressed in N. benthamiana. The fluorescence signal of the TaZFP8-5B-eGFP protein was only observed in the nucleus (Fig. S2), implying that TaZFP8-5B is located in the nucleus.
Knockdown of TaZFP8 enhances wheat resistance to stripe rust
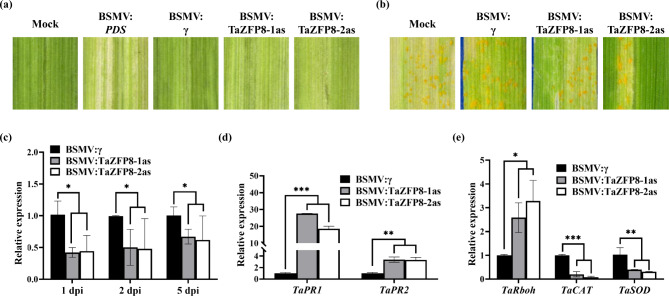
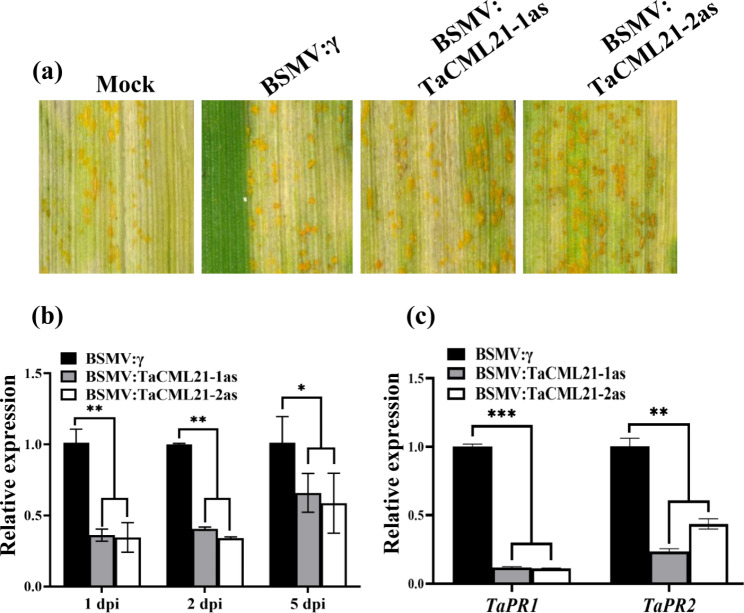
To determine the function of TaZFP8 in Pst resistance, two specific cDNA fragments were chosen to simultaneously silence three copies of TaZFP8 on wheat chromosome 5, due to the high similarity (95.53 to 96.73%) between the three copies. The BSMV:TaZFP8 and BSMV:γ leaves had chlorotic mosaic symptoms and no other evident defects. Photobleaching was observed in plants inoculated with BSMV:PDS (Fig. 2a), indicating that the silencing system was functioning correctly. After Pst inoculation, we observed less urediniospore production and necrosis on leaves treated with BSMV:TaZFP8 compared to the control (Fig. 2b). The transcript levels of TaZFP8 were decreased by 42-67% in BSMV:TaZFP8 leaves (Fig. 2c).
Fig. 2.
Silencing TaZFP8 increases wheat resistance to Pst. (a) The third leaves of wheat cultivar Shumai126 were inoculated with BSMV:γ, BSMV:PDS, BSMV:TaZFP8-1as and BSMV:TaZFP8-2as vectors. The virus symptoms (BSMV:γ and BSMV:TaZFP8) and photobleaching phenotypes (BSMV:PDS) on leaves were observed and photographed 10 days after infection. Mock, wheat leaves inoculated with FES buffer. (b) The resistance phenotype of the fifth leaves at 14 dpi, inoculated with Pst CYR34. (c-e) Relative expression of TaZFP8, TaPR1, TaPR2, TaRboh, TaCAT, and TaSOD in wheat leaves at 1 dpi. The transcript levels of the genes were detected by qRT-PCR. Error bars represent SEM from three independent biological replicates. Asterisks represent the level of significant differences. *p < 0.05, **p < 0.01, and ***p < 0.001
Histological analysis was performed to characterize the disease symptoms in TaZFP8-silenced leaves. Pst hyphal length was obviously decreased and Pst infection areas were significantly (p < 0.001) reduced in BSMV:TaZFP8 leaves compared to the controls (Fig. S3).
To evaluate whether the expression level of defense-related genes in TaZFP8-silenced leaves was affected by Pst infection, transcripts of two wheat pathogenesis-related (PR) genes (TaPR1 and TaPR2) were examined by qRT-PCR. Transcript levels of TaPR1 and TaPR2 were significantly induced in TaZFP8-knockdown plants (Fig. 2d). We further assayed the expression of marker gene TaRboh which was reported to mediate H2O2 production [33], and a candidate catalase (i.e., TaCAT) as well as a superoxide dismutase (i.e., TaSOD) which are involved in ROS signaling. We found higher expression levels of TaRboh, whereas lower levels of TaCAT and TaSOD in BSMV:TaZFP8 plants compared to the control (Fig. 2e). These data suggest that simultaneous knockdown the three copies of TaZFP8 on chromosome 5 enhances wheat resistance to stripe rust pathogen.
Overexpression of TaZFP8-5B reduces plant disease resistance
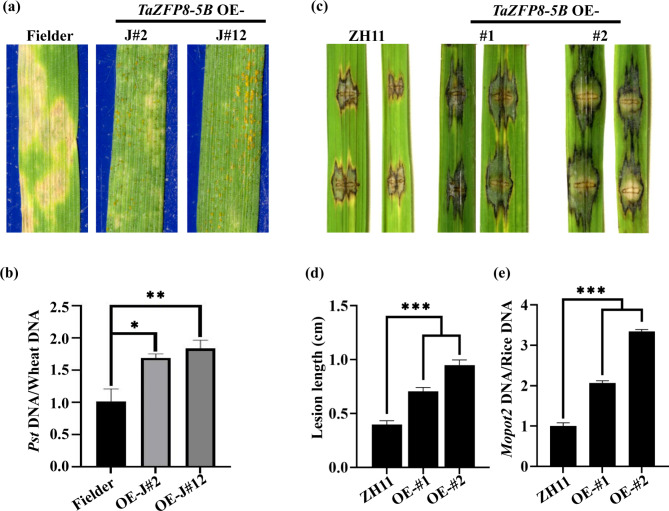
To specifically determine the role of TaZFP8-5B during the wheat-Pst interaction, we generated TaZFP8-5B overexpressing lines in the wheat cultivar Fielder. Two positive T0 transgenic plants TaZFP8-5BOE-J#2 and TaZFP8-5BOE-J#12 were obtained by PCR and qRT-PCR analysis (Fig. S4a). When inoculated with avirulent Pst race CYR23, Fielder leaves showed prominent cell death with no visible urediniospore pustules, whereas T1 plants overexpressing TaZFP8-5B produced pustules and had 67.1% and 81.3% increase in fungal biomass (Fig. 3a and b), indicating a negative role in wheat against stripe rust pathogen.
Fig. 3.
TaZFP8-5B negatively regulates plant disease resistance. (a and b) Overexpression of TaZFP8-5B increases wheat susceptibility to stripe rust. Fielder and TaZFP8-5B OE lines (TaZFP8-5B OE-J#2 and TaZFP8-5B OE-J#12) were inoculated with Pst CYR23, and disease symptoms were observed at 16 dpi (a), and fungal biomass in infected leaves was measured by qPCR (b). (c-e) Overexpression of TaZFP8-5B reduces rice resistance to Magnaporthe oryzae. (c) Disease symptoms of TaZFP8-5B OE lines (TaZFP8-5B OE-#1 and TaZFP8-5B OE-#2) and control. (d) Lesion length and (e) relative fungal growth were measured on leaves at 7 dpi. Error bars represent SEM from three independent biological replicates. Asterisks represent significant differences. ***p < 0.001
To further investigate the role of TaZFP8 in plant defense responses, we constructed rice lines heterologous expression of TaZFP8-5B. Five T1 transgenic lines (TaZFP8-5B OE-#1 to TaZFP8-5B OE-#5) expressing TaZFP8-5B were obtained and two of them (OE-#1 and OE-#2) with higher gene expression levels were selected for punch-inoculation (Fig. S4b). M. oryzae inoculation assay showed that the disease symptoms became more severe in TaZFP8-5B OE lines with significantly larger lesion sizes than ZH11 (Fig. 3c, d). Consistently, the fungal biomass in TaZFP8-5B OE lines was significantly higher than the control (Fig. 3e). To evaluate whether the expression of TaZFP8-5B affects the transcripts of ROS-related marker genes, we detected the expression levels of OsRbohA [5], OsCAT, and OsSOD [34] in TaZFP8-5B OE-#1 and -#2 transgenic lines (Fig. S4c). The results showed that the expression levels of OsCAT and OsSOD were significantly higher in OE lines than those in wild-type plants. The relative expression of OsRbohA was significantly increased in OE-#1 but not in OE-#2. Taken together, these results demonstrate that TaZFP8-5B functions as a negative regulator in plant disease resistance pathways.
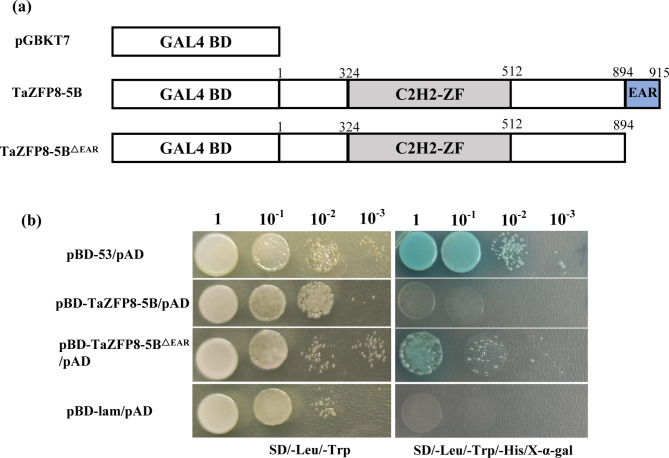
EAR motif represses the trans-activation ability of TaZFP8-5B in yeast
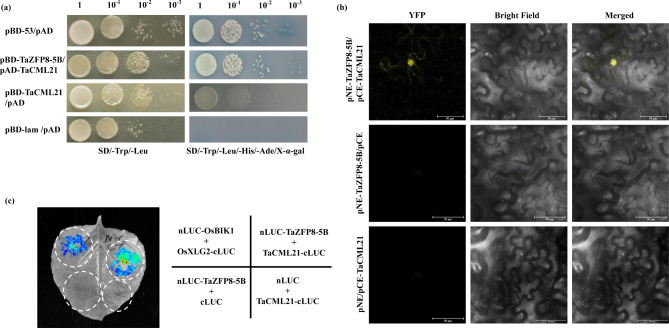
The EAR motif was found to be essential for transcriptional repression in plants [35, 36]. TaZFP8-5B contains an EAR motif at the C-terminus. To examine the transcriptional activity of TaZFP8-5B, fusion plasmids pBD-TaZFP8-5B (1–915 bp, full-length coding region), pBD-TaZFP8-5BΔEAR (1–894 bp, EAR deletion) (Fig. 4a), pBD-53 (positive control), and pBD-Lam (negative control) were separately co-transformed with pGADT7 into the yeast strain Y2HGold. The transformants harboring pBD-53 or pBD-TaZFP8-5BΔEAR grew well and colonies turned blue on SD/-Trp/-His/-Leu medium supplemented with X-α-gal, while transformants containing pBD-TaZFP8-5B or pBD-Lam did not (Fig. 4b). This results indicate that the EAR motif can repress the transactivation ability of TaZFP8-5B in yeast cells.
Fig. 4.
Transcription activity assay of full-length or truncation of TaZFP8-5B in yeast. (a) Illustration of pGBKT7-TaZFP8-5B (full-length) and pGBKT7-TaZFP8-5BΔEAR (EAR motif deletion mutant), and pGBKT7 constructs. (b) The constructs were separately transformed into yeast strain Y2HGold and diluted yeast solutions were dropped onto SD/-Leu/-Trp, and then onto SD/-Leu/-Trp/-His/X-α-gal media, respectively. Photographs were recorded after 2-d of incubation. All experiments were repeated three times with consistent results
TaZFP8-5B interacts with a calmodulin-like protein, TaCML21
To identify the interacting proteins of TaZFP8-5B, we used TaZFP8-5B as a bait to screen the Y2H library of AVS + Yr15 which was constructed using RNA from Pst CYR34-infected wheat seedling leaves. A wheat calmodulin-like (CML) protein was identified as an interacting protein of TaZFP8-5B. The CML protein had the highest sequence similarity with CML21 in common wheat. Phylogenetic analysis showed its relatedness to other plant CML21 proteins, such as TdCML21, ZmCML21, and OsCML21 (Fig. S5). We therefore designated the CML protein as TaCML21.
We further analyzed the subcellular localization of TaCML21 in N. benthamiana leaf cells. When N. benthamiana leaves transformed with fusion construct TaCML21-eGFP, fluorescent signals were observed in both the cytoplasm and nucleus (Fig. S6).
We cloned TaCML21 into the pGADT7 and TaZFP8-5B into the pGBKT7. The growth of the Y2HGold strain co-transformed with AD-TaCML21 and BD-TaZFP8-5B on SD/-Trp/-Leu/-His/-Ade/X-α-gal medium, confirming the interaction between TaZFP8-5B and TaCML21 (Fig. 5a). The BiFC assay was used to further verify the interaction between TaZFP8-5B and TaCML21. Yellow fluorescent protein (YFP) signals were detected in the nuclei when TaZFP8-5B and TaCML21 were co-expressed in tobacco cells, whereas no signal was observed in the negative controls, indicating that TaZFP8-5B interacts with TaCML21 in the nucleus (Fig. 5b). Additionally, the TaZFP8-5B-TaCML21 interaction was further confirmed by LUC assay. Strong luminescence signals were observed in the area co-expressed TaZFP8-5B and TaCML21 proteins, while no signal was detected in the area expressed either nLUC-TaZFP8-5B + cLUC or nLUC + TaCML21-cLUC (Fig. 5c). These data therefore demonstrate the interaction between TaZFP8-5B and TaCML21.
Fig. 5.
TaZFP8-5B interacts with TaCML21. (a) Analysis of interaction between TaZFP8-5B and TaCML21 using yeast two-hybrid assay. Yeast cells of Y2HGold strains transformed with the recombinant constructs were assayed for growth on SD/-Leu/-Trp and SD/-Trp/-Leu/-His/-Ade/X-α-gal mediums. pBD-53/pAD and pBD-lam/pAD were positive and negative controls, respectively. (b) Confirmation of the interaction between TaZFP8-5B and TaCML21 using BiFC assay. pNE-TaZFP8-5B/pCE and pNE/pCE-TaCML21 were negative controls. Scale bars = 50 μm. (c) Detection of the TaZFP8-5B-TaCML21 interaction in N. benthamiana leaves transiently expressing the constructs by luciferase complementation imaging assay. OsBIK1 and OsXLG2 are the reported interacting proteins that were used as positive control. The LUC empty vector was used as the negative control
Silencing of TaCML21 reduces the wheat stripe rust resistance
We used VIGS system to test the effect of TaCML21 on Pst resistance. Two gene-specific fragments of TaCML21 (150-bp and 143-bp) were inserted into BSMV:γ plasmid. When the Pst CYR34 was inoculated on BSMV-infected leaves, uredinia in TaCML21-knockdown leaves were increased compared to BSMV:γ and mock controls at 14 dpi (Fig. 6a). The transcription of TaCML21 was reduced by 36–65% in TaCML21-silenced leaves from 1 to 5 dpi (Fig. 6b). Pst hyphal length and the infection areas of Pst were much larger in TaCML21-silenced leaves than those of controls (Fig. S7a-c). We further assessed the transcript levels of defense-related genes in TaCML21-knockdown leaves inoculated with CYR34. Transcript levels of TaPR1 and TaPR2 were significantly lower in TaCML21-knockdown leaves inoculated with CYR34 compared to that in the controls (Fig. 6c). These results indicate that silencing of TaCML21 reduced the wheat stripe rust resistance.
Fig. 6.
Knocking down TaCML21 reduces resistance to Pst. (a) The disease symptoms were photographed 14 days after Pst race CYR34 inoculation on the fifth leaf of BSMV-inoculated Shumai126. (b-c) Relative expression levels of TaCML21 and PR genes TaPR1 and TaPR2 in leaves inoculated with CYR34 were evaluated by qRT-PCR at 1 dpi. Error bars represent SEM from three independent biological replicates. Asterisks represent the level of significant differences. *p < 0.05, **p < 0.01, and ***p < 0.001
Discussion
Previous studies showed that ZFPs regulated wheat responses to abiotic stresses such as heat, drought, and inorganic phosphate (Pi)-starvation [19–21]. Here, we report a wheat C2H2-type ZFP (TaZFP8-5B) functions as a negative regulator in plant immunity, as demonstrated by VIGS assay and transgenic complementation. We show that TaZFP8-5B interacted with TaCML21, which is positively involved in wheat resistance against Pst.
TaZFP8-5B possesses a single ZF domain with a conserved QALGGH motif at the N-terminus, and an EAR motif at the C-terminus, which shares a similar structure to typical C2H2-ZFPs in plants [13, 14]. Previous studies showed that C2H2-ZFPs with a single zinc finger structure are mainly associated with plant growth and development [13, 14, 37]. A recent study shown that wheat C2H2-ZFPs with single zinc finger motif may have potential roles in responses to biotic stresses [38]. In the present study, we showed that the TaZFP8-5B with single zinc finger motif worked as a negative regulator for disease resistance, supporting the involvement of a single ZFP in biotic stress responses. The QALGGH motif is widely present in the ZFPs of both dicots and monocots plants [39, 40] and was considered as a necessary element for DNA-binding activity [41]. The function of the QALGGH motif in TaZFP8-5B needs to be further investigated. The EAR motif is the major form of transcriptional repression element in plants, which is known to function as negative regulators in a broader context of gene regulation [42]. In the present study, we showed that the EAR motif can inhibit the transactivation ability of TaZFP8-5B in yeast cells (Fig. 4). Further quantitative expression analysis revealed that TaPR1 and TaPR2 genes were up-regulated in TaZFP8-knockdown plants. Therefore, we speculate that the TaZFP8-5B may have a role in regulating the downstream gene expression negatively which is consistent with those of previous reports for C2H2-ZFPs [43, 44].
In the present study, we found TaCML21 is a direct partner of TaZFP8-5B (Fig. 5). CML proteins were known to function as Ca2+ sensors and transducers in plant-pathogen signaling [45–48]. Increasing evidence shows that reduction of CML expression or loss of CML function in plants strongly affects immune responses [49]. For example, silencing of CML24 in Arabidopsis impaired the defense response to bacterial strains of Pseudomonas syringae [50]. NtCaM13-knockout tobacco plants reduces basal resistance against pathogens [51]. In the current study, TaCML21-silenced wheat leaves display enhanced plant susceptibility to stripe rust and the deregulation of PR gene expression, suggesting that TaCML21 also acts as a positive regulator in plant immunity.
In plants, the CaM/CML family can bind various proteins including diverse TFs [52]. Recent reports demonstrated that the CaM-TF complex functions as a repressor in regulation of gene expression [53, 54]. Upon pathogen attack, the elevation of nuclear Ca2+ signal interacts with CaM-TF which relieves transcriptional repression conferred by the CaM-TF complex and leads to de-repress the expression of the immune system [53]. In the current study, we showed that TaCML21 interacts with TaZFP8-5B. TaCML21 functions as a positive regulator, whereas TaZFP8-5B is a negative regulator in plant immune responses. We hypothesize that the possible mechanism of TaCML21-TaZFP8-5B in defense responses may fit the transcription repression and de-repression model [53]. In this scenario, the TaCML21-TaZFP8-5B complex may act as a repressor of the plant immune response by their interactions with promoters of target gene(s) in the absence of pathogen attack; upon pathogen infection, the increased Ca2+ signal could interact with TaCML21-TaZFP8-5B complex and affects the protein conformation, causing the abrogation of transcriptional repression and activation of plant defense responses (Fig. S8). Further study is required to clarify the regulation mechanism of the TaCML21-TaZFP8-5B complex in plant immunity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1: Sequence analysis of TaZFP8. (a) Schematic diagram of TaZFP8 protein structure. (b) Sequence alignment of TaZFP8 with other C2H2-type ZFPs. The sequences were aligned with DNAMAN software. C2H2-ZF domain (gray), QALGGH conserved motif (green), and EAR motif (blue). (c) Phylogenetic analysis of TaZFP8. Different C2H2-ZFs from T. aestivum (Ta), T. dicoccoides (Td), Hordeum vulgare (Hv), Zea mays (Zm), Oryza sativa (Os), Setaria italica (Si), Aegilops tauschii (Att), Arabidopsis thaliana (At), Solanum tuberosum (St), Nicotiana benthamiana (Nb) and Vitis vinifera (Vv) were used for the phylogenetic analyses using MEGA11
Supplementary Figure S2: Subcellular localization of TaZFP8-5B protein. TaZFP8-5B-eGFP fusion protein and eGFP were separately mixed with NLS-mRFP (nuclear localization marker protein) and expressed in the leaves of N. benthamiana
Supplementary Figure S3: Fungal development in TaZFP8-silenced leaves during initial stages of Pst inoculation. (a) Microscopic observation of pathogen development in TaZFP8-silenced leaves at different time points after Pst inoculation. (b) Hyphal length and (c) infection area of Pst in TaZFP8-silenced leaves after being infected with CYR34. Wheat leaves were sampled at 1 and 5 dpi and observed microscopically after stained with WGA. SV, substomatal vesicle; H, haustoria; IH, infection hypha; HMC, haustorial mother cell; and SH, secondary hyphae. Bars = 100 μm. Asterisks represent significant differences. ** p < 0.01, ***p < 0.001
Supplementary Figure S4: Transcript levels of TaZFP8-5B and ROS-related genes in transgenic plants. (a) Transcript levels in Fielder and TaZFP8-5BOE plants. (b) Transcript levels of TaZFP8-5B in the rice cultivar ZH11 background. TaZFP-5BOE-#1 to -#5, overexpression transgenic events. (c) Relative expression of OsRbohA, OsCAT, and OsSOD in rice leaves at 1 dpi. The transcript levels of the genes in TaZFP8-5BOE-#1 and TaZFP8-5B#2 leaves were detected by qRT-PCR. Error bars represent SEM from three replicates. Asterisks represent significant differences. *p < 0.05, ***p < 0.001
Supplementary Figure S5: Phylogenetic tree of TaCML21 and CML variants from various species. T. aestivum (Ta), T. dicoccoides (Td), T. urartu (Tu), Z. mays (Zm), O. sativa (Os), A. thaliana (At), S. tuberosum (St), Capsicum annuum (Ca), Glycine max (Gm)
Supplementary Figure S6: Subcellular localization of TaCML21 protein. TaCML21-eGFP and eGFP were separately mixed with mRFP (NLS-mRFP, nuclear localization protein) and expressed in the leaves of N. benthamiana
Supplementary Figure S7: Fungal development in TaCML21-silenced leaves during initial stages of Pst inoculation. (a) Microscopic observation of Pst growth in TaCML21-silenced leaves at 1 and 5 dpi. (b) Hyphal length and (c) infection area of Pst in gene silenced leaves after infected with CYR34. Wheat leaves were sampled at different time points and observed microscopically after stained with WGA. SV, substomatal vesicle; H, haustoria; IH, infection hypha; HMC, haustorial mother cell; and SH, secondary hyphae. Bars = 100 μm. * p < 0.05,** p < 0.01, ***p < 0.001
Supplementary Figure S8: Possible working model of TaCML21-TaZFP-5B complex in plant defense responses. The TaCML21-TaZFP8-5B complex represses the expression of target gene(s) that are involved in regulation of plant defense; upon pathogen infection, the generated Ca2+ signal could interact with TaCML21-TaZFP8-5B complex that leads to the complex degradation and transcriptional de-repression and activation of plant defense responses. Dash lines represent hypothetical situations. Red spheres denote Ca2+ ions. The number of red spheres represent the level of concentration
Supplementary Table S1: List of primers used in this study
Author contributions
LH, BHW, and DCL conceived and designed the research. LH, RJX, YLH, LLD, XEZ, FW, XJC, MH, LHF, ZZW conducted experiments. RJX, YYH, QX and LQZ analyzed data. LH, RJX, and DCL wrote the manuscript. All authors read and approved the final manuscript.
Funding
The work was supported by the National Natural Science Foundation of China (No. 32341035, 32272068), Sichuan Science and Technology Program (2024YFNH0024, 2021YFYZ0002), Sichuan Provincial Agricultural Department Innovative Research Team (SCCXTD-2024-11), and State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China (SKL-KF202322).
Data availability
The data and plant materials generated and analyzed supporting the findings of the current work are available within the manuscript and its supplementary information files or are available from the corresponding author upon request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lin Huang and Ruijie Xie contributed equally to this work.
Contributor Information
Lin Huang, Email: lhuang@sicau.edu.cn.
Dengcai Liu, Email: dcliu7@sicau.edu.cn.
References
- 1.Ngou BPM, Ahn H, Ding P, Jones JDG. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 2021;592(7852):110–5. [DOI] [PubMed] [Google Scholar]
- 2.Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, Karelina D, Hua C, Frohlich K, Wan WL, et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature. 2021;598(7881):495–9. [DOI] [PubMed] [Google Scholar]
- 3.Couto D, Zipfel. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16(9):537–52. [DOI] [PubMed] [Google Scholar]
- 4.Monteiro F, Nishimura MT. Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu Rew Phytopathol. 2018;56(1):243–67. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170(1):114–26. [DOI] [PubMed] [Google Scholar]
- 6.Balint-Kurti P. The plant hypersensitive response: concepts, control and consequences. Mol Plant Pathol. 2019;20(8):1163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206(3):932–47. [DOI] [PubMed] [Google Scholar]
- 8.Seo E, Choi D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief Funct Genomics. 2015;14(4):260–7. [DOI] [PubMed] [Google Scholar]
- 9.Amorim LLB, Santos RDFD, Neto JAOP, Guida-Santos M, Crovella S, Benko-Iseppon AM. Transcription factors involved in plant resistance to pathogens. Curr Protein Pept Sc. 2017;18 4:335–51. [DOI] [PubMed] [Google Scholar]
- 10.Burke R, Schwarze J, Sherwood OL, Jnaid Y, McCabe PF, Kacprzyk J. Stressed to death: the role of transcription factors in plant programmed cell death induced by abiotic and biotic stimuli. Front Plant Sci. 2020;11:1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valandro F, Menguer PK, Cabreira-Cagliari C, Margis-Pinheiro M, Cagliari A. Programmed cell death (PCD) control in plants: new insights from the Arabidopsis thaliana deathosome. Plant Sci. 2020;299:110603. [DOI] [PubMed] [Google Scholar]
- 12.Li W, He M, Wang J, Wang Y. Zinc finger protein (ZFP) in plants-a review. Plant Omics. 2013;6:474–80. [Google Scholar]
- 13.Wang K, Ding Y, Cai C, Chen Z, Zhu C. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol Plant. 2019;165(4):690–700. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Khan AR, Gan Y. C2H2 zinc finger proteins response to abiotic stress in plants. Int J Mol Sci. 2022;23(5):2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Zheng X, Cheng R, Zhong C, Zhao J, Liu TH, Yi T, Zhu Z, Xu J, Meksem K, et al. Soybean ZINC FINGER PROTEIN03 targets two SUPEROXIDE DISMUTASE1s and confers resistance to Phytophthora sojae. Plant Physiol. 2023;192(1):633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Liu Y, Wen F, Yao D, Wang L, Guo J, Ni L, Zhang A, Tan M, Jiang M. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J Exp Bot. 2014;65(20):5795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He F, Li HG, Wang JJ, Su Y, Wang HL, Feng CH, et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol J. 2019;17(11):2169–83. [DOI] [PMC free article] [PubMed]
- 18.Li Y, Sun A, Wu Q, Zou X, Chen F, Cai R, Xie H, Zhang M, Guo X. Comprehensive genomic survey, structural classification and expression analysis of C(2)H(2)-type zinc finger factor in wheat (Triticum aestivum L). BMC Plant Biol. 2021;21(1):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheuk A, Ouellet F, Houde M. The barley stripe mosaic virus expression system reveals the wheat C2H2 zinc finger protein TaZFP1B as a key regulator of drought tolerance. BMC Plant Biol. 2020;20(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouard W, Houde M. The C2H2 zinc finger protein TaZFP13D increases drought stress tolerance in wheat. Plant Stress. 2022;6:100119. [Google Scholar]
- 21.Ding W, Wang Y, Fang W, Gao S, Li X, Xiao K. TaZAT8, a C2H2-ZFP type transcription factor gene in wheat, plays critical roles in mediating tolerance to Pi deprivation through regulating P acquisition, ROS homeostasis and root system establishment. Physiol Plant. 2016;158(3):297–311. [DOI] [PubMed]
- 22.Zeng QD, Zhao J, Wu J, Zhan G, Han D, Kang Z. Wheat stripe rust and integration of sustainable control strategies in China. Front Agric Sci Eng. 2022;9:37. [Google Scholar]
- 23.Singh KP, Kumari P, Rai PK. Current status of the disease-resistant gene(s)/QTLs, and strategies for improvement in Brassica juncea. Front Plant Sci. 2021;12:617405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Hu Y, Gong F, Jin Y, Xia Y, He Y, Jiang Y, Zhou Q, He J, Feng L, et al. Identification and mapping of QTL for stripe rust resistance in the Chinese wheat cultivar Shumai126. Plant Dis. 2022;106(4):1278–85. [DOI] [PubMed] [Google Scholar]
- 26.Wang N, Tang C, Fan X, He M, Gan P, Zhang S, Hu Z, Wang X, Yan T, Shu W, et al. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell. 2022;185(16):2961–74. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Feng L, Jiang Y, Zhang L, Yan J, Zhao G, Wang J, Chen G, Wu B, Liu D, et al. Distribution and nucleotide diversity of Yr15 in wild emmer populations and Chinese wheat germplasm. Pathogens. 2020;9(3):212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 29.Holzberg S, Brosio P, Gross C, Pogue GP. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002;30(3):315–27. [DOI] [PubMed] [Google Scholar]
- 30.Yaniv E, Raats D, Ronin Y, Korol AB, Grama A, Bariana H, Dubcovsky J, Schulman AH, Fahima T. Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol Breed. 2015;35:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XP, Ma XC, Wang H, Zhu Y, Liu XX, Li TT, Zheng YP, Zhao JQ, Zhang JW, Huang YY, et al. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and yield. Rice. 2020;13(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24(11):4748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, et al. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell. 2003;15(3):706–18. [DOI] [PMC free article] [PubMed]
- 34.Chen Z, Fei X, Sun F, Cui X. Effects of saline-alkali stress on activities and gene expression of antioxidant enzymes of transgenic Lc-CDPK rice. J Northwest Sci-Tech Univ Agric for. 2019;47(05):15–22. [Google Scholar]
- 35.Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics-Us. 2011;6(2):141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow V, Kirzinger MW, Kagale S. Lend me your EARs: a systematic review of the broad functions of EAR motif-containing transcriptional repressors in plants. Genes. 2023;14(2):270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X, Wu Y, Dai T, Gu Y, Wang L, Qin X, Xu Y, Chen F. JcZFP8, a C2H2 zinc finger protein gene from Jatropha curcas, influences plant development in transgenic tobacco. Electron J Biotechn. 2018;34:76–82. [Google Scholar]
- 38.Manser B, Zbinden H, Herren G, Steger J, Isaksson J, Bräunlich S, Wicker T, Keller B. Wheat zinc finger protein TaZF interacts with both the powdery mildew AvrPm2 protein and the corresponding wheat Pm2a immune receptor. Plant Commun. 2023;5:100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheuk A, Houde M. Genome wide identification of C1-2i zinc finger proteins and their response to abiotic stress in hexaploid wheat. Mol Genet Genomics. 2016;291(2):873–90. [DOI] [PubMed] [Google Scholar]
- 40.Gourcilleau D, Lenne C, Armenise C, Moulia B, Julien JL, Bronner G, Leblanc-Fournier N. Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic, cold and mechanical stresses. DNA Res. 2011;18(2):77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol. 1999;39(6):1073–8. [DOI] [PubMed] [Google Scholar]
- 42.Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11(3):109–12. [DOI] [PubMed] [Google Scholar]
- 43.Ciftci-Yilmaz S, Morsy M, Song L, Coutu A, Krizek B, Lewis M, Warren D, Cushman J, Connolly E, Mittler R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem. 2007;282:9260–8. [DOI] [PubMed] [Google Scholar]
- 44.Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Bioph Res. 2004;321(1):172–8. [DOI] [PubMed] [Google Scholar]
- 45.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–8. [DOI] [PubMed] [Google Scholar]
- 46.Bender KW, Snedden WA. Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 2013;163(2):486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Liu Z, Han S, Liu P, Sadeghnezhad E, Liu M. Growth or survival. What is the role of calmodulin-like proteins in plant? Int J Biol Macromol. 2023;242:124733. [DOI] [PubMed] [Google Scholar]
- 48.Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah BW. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015;6:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheval C, Aldon D, Galaud JP, Ranty B. Calcium/calmodulin-mediated regulation of plant immunity. Biochim Biophys Acta. 2013;1833(7):1766–71. [DOI] [PubMed] [Google Scholar]
- 50.Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 2008;148(2):818–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takabatake R, Karita E, Seo S, Mitsuhara I, Kuchitsu K, Ohashi Y. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 2007;48(3):414–23. [DOI] [PubMed] [Google Scholar]
- 52.Galon Y, Finkler A, Fromm H. Calcium-regulated transcription in plants. Mol Plant. 2010;3(4):653–69. [DOI] [PubMed] [Google Scholar]
- 53.Fromm H, Finkler A. Repression and de-repression of gene expression in the plant immune response: the complexity of modulation by Ca2+ and calmodulin. Mol Plant. 2015;8(5):671–3. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Du L, Shen C, Yang Y, Poovaiah BW. Regulation of plant immunity through ubiquitin-mediated modulation of Ca2+–calmodulin–AtSR1/CAMTA3 signaling. Plant J. 2014;78(2):269–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Sequence analysis of TaZFP8. (a) Schematic diagram of TaZFP8 protein structure. (b) Sequence alignment of TaZFP8 with other C2H2-type ZFPs. The sequences were aligned with DNAMAN software. C2H2-ZF domain (gray), QALGGH conserved motif (green), and EAR motif (blue). (c) Phylogenetic analysis of TaZFP8. Different C2H2-ZFs from T. aestivum (Ta), T. dicoccoides (Td), Hordeum vulgare (Hv), Zea mays (Zm), Oryza sativa (Os), Setaria italica (Si), Aegilops tauschii (Att), Arabidopsis thaliana (At), Solanum tuberosum (St), Nicotiana benthamiana (Nb) and Vitis vinifera (Vv) were used for the phylogenetic analyses using MEGA11
Supplementary Figure S2: Subcellular localization of TaZFP8-5B protein. TaZFP8-5B-eGFP fusion protein and eGFP were separately mixed with NLS-mRFP (nuclear localization marker protein) and expressed in the leaves of N. benthamiana
Supplementary Figure S3: Fungal development in TaZFP8-silenced leaves during initial stages of Pst inoculation. (a) Microscopic observation of pathogen development in TaZFP8-silenced leaves at different time points after Pst inoculation. (b) Hyphal length and (c) infection area of Pst in TaZFP8-silenced leaves after being infected with CYR34. Wheat leaves were sampled at 1 and 5 dpi and observed microscopically after stained with WGA. SV, substomatal vesicle; H, haustoria; IH, infection hypha; HMC, haustorial mother cell; and SH, secondary hyphae. Bars = 100 μm. Asterisks represent significant differences. ** p < 0.01, ***p < 0.001
Supplementary Figure S4: Transcript levels of TaZFP8-5B and ROS-related genes in transgenic plants. (a) Transcript levels in Fielder and TaZFP8-5BOE plants. (b) Transcript levels of TaZFP8-5B in the rice cultivar ZH11 background. TaZFP-5BOE-#1 to -#5, overexpression transgenic events. (c) Relative expression of OsRbohA, OsCAT, and OsSOD in rice leaves at 1 dpi. The transcript levels of the genes in TaZFP8-5BOE-#1 and TaZFP8-5B#2 leaves were detected by qRT-PCR. Error bars represent SEM from three replicates. Asterisks represent significant differences. *p < 0.05, ***p < 0.001
Supplementary Figure S5: Phylogenetic tree of TaCML21 and CML variants from various species. T. aestivum (Ta), T. dicoccoides (Td), T. urartu (Tu), Z. mays (Zm), O. sativa (Os), A. thaliana (At), S. tuberosum (St), Capsicum annuum (Ca), Glycine max (Gm)
Supplementary Figure S6: Subcellular localization of TaCML21 protein. TaCML21-eGFP and eGFP were separately mixed with mRFP (NLS-mRFP, nuclear localization protein) and expressed in the leaves of N. benthamiana
Supplementary Figure S7: Fungal development in TaCML21-silenced leaves during initial stages of Pst inoculation. (a) Microscopic observation of Pst growth in TaCML21-silenced leaves at 1 and 5 dpi. (b) Hyphal length and (c) infection area of Pst in gene silenced leaves after infected with CYR34. Wheat leaves were sampled at different time points and observed microscopically after stained with WGA. SV, substomatal vesicle; H, haustoria; IH, infection hypha; HMC, haustorial mother cell; and SH, secondary hyphae. Bars = 100 μm. * p < 0.05,** p < 0.01, ***p < 0.001
Supplementary Figure S8: Possible working model of TaCML21-TaZFP-5B complex in plant defense responses. The TaCML21-TaZFP8-5B complex represses the expression of target gene(s) that are involved in regulation of plant defense; upon pathogen infection, the generated Ca2+ signal could interact with TaCML21-TaZFP8-5B complex that leads to the complex degradation and transcriptional de-repression and activation of plant defense responses. Dash lines represent hypothetical situations. Red spheres denote Ca2+ ions. The number of red spheres represent the level of concentration
Supplementary Table S1: List of primers used in this study
Data Availability Statement
The data and plant materials generated and analyzed supporting the findings of the current work are available within the manuscript and its supplementary information files or are available from the corresponding author upon request.