Abstract
Background
Microglia play a crucial role in brain development and repair by facilitating processes such as synaptic pruning and debris clearance. They can be activated in response to various stimuli, leading to either pro-inflammatory or anti-inflammatory responses associated with specific metabolic alterations. The imbalances between microglia activation states contribute to chronic neuroinflammation, a hallmark of neurodegenerative diseases. N-acetylaspartate (NAA) is a brain metabolite predominantly produced by neurons and is crucial for central nervous system health. Alterations in NAA metabolism are observed in disorders such as Multiple Sclerosis and Canavan disease. While NAA’s role in oligodendrocytes and astrocytes has been investigated, its impact on microglial function remains less understood.
Methods
The murine BV2 microglial cell line and primary microglia were used as experimental models. Cells were treated with exogenous NAA and stimulated with LPS/IFN-γ to reproduce the pro-inflammatory phenomenon. HPLC and immunofluorescence analysis were used to study lipid metabolism following NAA treatment. Automated fluorescence microscopy was used to analyze phagocytic activity. The effects on the pro-inflammatory response were evaluated by analysis of protein/mRNA expression and ChIP assay of typical inflammatory markers.
Results
NAA treatment promotes an increase in both lipid synthesis and degradation, and enhances the phagocytic activity of BV2 cells, thus fostering surveillant microglia characteristics. Importantly, NAA decreases the pro-inflammatory state induced by LPS/IFN-γ via the activation of histone deacetylases (HDACs). These findings were validated in primary microglial cells, highlighting the impact on cellular metabolism and inflammatory responses.
Conclusions
The study highlighted the role of NAA in reinforcing the oxidative metabolism of surveillant microglial cells and, most importantly, in buffering the inflammatory processes characterizing reactive microglia. These results suggest that the decreased levels of NAA observed in neurodegenerative disorders can contribute to chronic neuroinflammation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01947-6.
Keywords: Microglial polarization, NAA, Anti-inflammatory response, Oxidative metabolism, Lipid turnover, Histone deacetylases
Background
Microglia are central nervous system (CNS)-resident immune cells involved in maintaining brain physiology and function. Surveillant microglia contribute to brain development and remodeling by promoting neuronal plasticity and tissue repair via synaptic pruning, cell debris scavenging, and trophic factor release [1]. Microglia become reactive upon the recognition of different stimuli, developing a wide range of different phenotypes and functions. Upon sensing pathogen- or damage-associated molecular patterns (PAMPs or DAMPs) they release pro-inflammatory cytokines (e.g., TNF-α, IL-6) and nitric oxide (NO) to accomplish CNS immunosurveillance. On the other hand, anti-inflammatory stimuli (e.g. IL-4, IL-10) can allow the resolution of inflammation and the repair of damaged sites [2]. The different reactive states of microglia are often associated with specific metabolic traits that influence their functional properties. Microglia responding to pro-inflammatory stimuli mainly rely on glucose metabolism boosting glycolysis for satisfying energetic demands and the pentose phosphate pathway for producing NADPH for NO synthesis by inducible NO synthase (iNOS). Surveillant as well as immunosuppressive microglia mainly exhibit an oxidative metabolism sustained by fatty acid oxidation and express arginase 1 (Arg-1) that ultimately contributes to polyamine synthesis involved in tissue recovery [3, 4]. The imbalance between pro-inflammatory and immunosuppressive properties of microglia may lead to chronic neuroinflammation [5–7], which represents one of the hallmarks of neurodegenerative diseases [6, 8].
The aberrant communication between neurons and glial cells, due to altered release of neurotransmitters, cytokines and metabolites, contributes to metabolic and inflammatory alterations of microglia in CNS disorders [9, 10]. N-acetylaspartate (NAA) is the second most abundant metabolite of the brain, where it is almost exclusively synthesized by neurons and continuously released in the microenvironment. NAA is selectively catabolized in acetate and aspartate by cells expressing the enzyme aspartoacylase (ASPA). Brain NAA concentration exhibits a very high intracellular/extracellular ratio reaching 10–15 mM in neurons and 100–200 µM in the extracellular space [11–13]. Along with the active efflux from neurons or the passive release after cell death, the extracellular level of NAA largely depends on the leakage from the blood-brain barrier and the uptake rate of neighboring cells. The effect of extracellular NAA in glial cells has been mainly delineated in oligodendroglia development and maturation. Oligodendrocytes catabolize NAA and use acetate as a relevant source of acetyl-CoA for lipid synthesis and thus myelin formation [14]. Moreover, NAA is involved in the regulation of protein acetylation process providing acetyl-CoA for acetyltransferases enzymes [15] and regulating the expression of histone deacetylases [16].
Alteration of NAA metabolism is typical of neurodegenerative disorders, particularly demyelinating diseases such as Multiple Sclerosis (MS), showing low levels of brain NAA, or the Canavan disease (CD), a genetic disorder with excessive NAA accumulation due to mutation in the ASPA gene [17–20]. NAA can also be transported in astrocytes [21], where its relevant role has been demonstrated by a gene therapy approach rescuing astrocyte-specific ASPA expression that was sufficient to normalize myelination in the CD mouse model [22]. Although ASPA expression has been also revealed in microglia [23] and neurodegenerative disorders are typically associated with both NAA dysmetabolism and neuroinflammatory processes [11, 24], no specific assessment of NAA involvement in microglial functions has been provided so far. In the present work, we demonstrate that exogenous NAA administration favors the oxidative metabolism of primary and immortalized microglia cells and restrains their reactivity in response to pro-inflammatory insults.
Methods
Cell lines and treatments
The murine microglia cell line BV2 and the murine macrophage cell line RAW 264.7, from the American Type Culture Collection (ATCC), were grown in DMEM containing 4.5 g/L of glucose supplemented with 10% fetal bovine serum, 10 U/ml penicillin/streptomycin and 2 mM l-glutamine. Cells were periodically tested for mycoplasma according to protocols from our laboratory. Cells were cultured at 37 °C in an atmosphere of 5% CO2 in air. RAW 264.7 cells were seeded at a density of 3 × 105 cells/ml whereas BV2 cells at a density of 1 × 105 cells/ml for all the experiments unless otherwise specified. N-Acetyl-L-aspartic acid was dissolved in de-ionized water, adjusting pH to 7.0 with NaOH, at a final concentration of 250 mM and used at concentrations indicated in the text. Lipopolysaccharide (LPS) was dissolved in de-ionized water at a final concentration of 1 mg/ml and used at a concentration of 100 ng/ml. IFN-γ was dissolved in de-ionized water containing 0.1% BSA at a final concentration of 0.25 mg/ml and used at a concentration of 20 ng/ml. The HDAC inhibitor Trichostatin A (TSA) was dissolved in DMSO at a final concentration of 50 µM and used at a concentration of 25 nM. The HDAC inhibitor Sodium butyrate (NaB) was dissolved in de-ionized water at a final concentration of 0.5 M and used at a concentration of 0.5 mM. Atglistatin was dissolved in DMSO at a final concentration of 25 mM and used at a concentration of 40 µM. N-methyl-D-aspartic acid (NMDA) was dissolved in de-ionized water at a final concentration of 250 mM and used at a concentration of 400 µM.
Primary microglia cell culture
Primary microglia cultures from the cortex were prepared as previously described [25]. Briefly, postnatal C57BL/6 mice (P0-P1) were sacrificed and, after removing the meninges, the cortex was digested with 0.01% trypsin (Sigma Aldrich) and 10 µg /ml DNase I (Sigma Aldrich). Following the process of dissociation and filtration through 70 μm filters, the cells were resuspended and placed in DMEM/F-12 media with GlutaMAX™ (Gibco, Invitrogen, UK). This media was supplemented with 10% FBS, 100 Units/ml of gentamicin, and 100 µg/ml of streptomycin/penicillin. The cells were then plated at a density of 6.25 × 104 cells/cm2. After approximately 15 days, mild trypsinization in DMEM/F-12 without FBS (0.08% trypsin in DMEM/F-12 without FBS) was performed for 40 min at 37 °C to remove non-microglial cells. The resultant adherent microglial cells were cultured in 50% mixed glial cells conditioned medium at 37 °C in a 5% CO2 and 95% air atmosphere for 48 h until use at the density of 5 × 104 cells/cm2.
Materials
Cell culture medium and supplements are from EuroClone S.p.A. NAA (00920), NMDA (M3262), NaB (303410), TSA (T1952), LPS (L6529), EDTA (E6758), MgCl2 (A748033 012), nicotinamide adenine dinucleotide hydrate (NAD+) (N7004), fluorescent latex beads (L4655), oil red O (ORO) (O0625), formalin solution neutral buffered 10% (4% paraformaldehyde containing) (HT5012), TEMED (1.10732), sodium deoxycholate (D6750), sodium chloride (S9888), sodium fluoride (S7920), sodium orthovanadate (S6508), sodium pyrophosphate tetrabasic decahydrate (30411), 2-β-mercaptoetanol (M6250), blue of bromophenol (B5525), 2-propanol (33539-M), glycerol (G5516), trichloroacetic acid (TCA) (T6399) and Triton™ X-100 (T9284) were from Sigma-Aldrich. The 0.4% trypan blue solution (17-942E) was from Lonza. TRIS-base (1610716) and sodium dodecyl sulfate (SDS) (161–0300) were from Bio-Rad. Hoechst 33,342 (H3570) and phalloidin eFluor™ 570 (cat n. 41-6559-05) were from Thermo Fisher Scientific. l-lactate dehydrogenase (LDH) was obtained from Roche Applied Science. DTT (281) and protease inhibitors cocktail from AMRESCO. DAF-2DA was from Chemodex (D0085). Albumin (A1391), sucrose (A3935) and glycine (A1067) were from PanReac AppliChem. Hydrazinium sulphate (24696.186), sodium hydroxide (28244.295) and methanol (20864.320) were from VWR Chemicals. IFN-γ (cat n 50709) was from Sino Biological inc. ATGListatin (cat n. HY-15859) was from MedChem Express. IL-4 (cat n 214 − 14) (A3122) was from Preprotech.
Western blot analysis
Cell pellets were resuspended in a lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton™ X-100, 0.5% Sodium Deoxycholate, 0.1% SDS, 10 mM sodium fluoride, 5 mM Sodium Pyrophosphate, 2 mM Sodium Orthovanadate) supplemented with protease inhibitor cocktail (Sigma-Aldrich, PIC0002). The homogenates were sonicated and quantified by the Lowry method [26]. Lysates were added to sample buffer (0.125 M Tris-HCl pH 6.8, 4% SDS, 20% glycerol, 10% 2-β-mercaptoethanol, 0.004% blue of bromophenol), separated by SDS–PAGE and blotted into a nitrocellulose membrane (Bio-Rad). The following primary antibodies (dilution 1:1000) were used: ASPA (Novus, NBP1-31754), HK2 (Abnova, H00003099-M01), pPKM2 (Tyr105) (Cell Signaling Technology, #3827), PKM2 (Cell Signaling Technology, #4053), arginase 1, iNOS (Santa Cruz Biotechnology, sc-7271), STAT1 (Cell Signaling Technology), pNF-kB (Ser536), NF-kB, lamin A/C, tubulin (Santa Cruz Biotechnology, sc-5286), acetyl lysine (Santa Cruz Biotechnology, sc-32268), gp91phox (Santa Cruz Biotechnology, sc-130543) and β-actin (Sigma-Aldrich, A3853). The secondary antibodies used are Goat Anti-Rabbit IgG (H + L)–HRP Conjugate (Bio-Rad cat. #170–6515) and Goat Anti-Mouse IgG (H + L)-HRP Conjugate IgG (Bio-Rad, cat. #1706516). The signals derived from the incubation with Clarity Max Western ECL Substrate (Bio-Rad) were acquired using a Fluorchem imaging system (Alpha Innotech). ImageJ software was used to perform densitometry analyses.
Nuclear fraction isolation
Nuclear extraction was performed by incubation of cells for 30 min on ice with nuclei isolation buffer (10 mM Tris-HCl at pH 7.8, 10 mM MgCl2, 1 mM EDTA, 0.25 M sucrose, 1% Triton™ X-100, 50 mM sodium fluoride, 2 mM Sodium Pyrophosphate, 1 mM Sodium Orthovanadate, 0.5 mM DTT supplemented with protease inhibitor cocktail); after centrifugation at 600×g, the supernatant containing the cytosolic fraction was stored, while pelleted nuclei were washed with isolation buffer without Triton X-100 and centrifuged three times at 600×g.
Total cell extracts, cytosolic and nuclear fractions were analyzed by western blot for purity determination.
Phagocytosis assay
Microglial phagocytosis was evaluated by immunofluorescence analysis. BV2 cells were seeded at a density of 3 × 104 cells/ml on coverslips for 24 h before treatment. Two hours before the end of the treatment 2.25 µl/ml of fluorescence-labeled latex beads (starting from 2.5% solid content in the suspension) were added for 2 h at 37 °C. At the end of the time, the cell culture medium was removed, cells were washed three times with PBS to remove the non-phagocytized beads and then fixed with 4% paraformaldehyde for 30 min. After that, they were gently washed with PBS, permeabilized with 0.3% Triton™ X-100 for 20 min and blocked with 3% BSA for 1 h. The samples were incubated with Phalloidin (1:500) for 1 h. The coverslips were transferred onto glass slides after 10 min of staining with 1 µg/ml of Hoechst 33,342.
Automatized acquisition was performed using a Leica DMI6000 B epifluorescence inverted microscope (Leica Microsystems, Wetzlar, Germany), equipped with Matrix Screener software (version 3.0) at 20× magnification (HCX PL FLUOTAR 20 × NA 0.4). ScanR image analyses software (Ver.3.1; Olympus, Tokyo, Japan) was used to determine the number of fluorescent latex beads present inside NAA treated vs. NAA untreated cells. 25 images of three independent samples were analyzed.
Primary microglia cells were seeded into appropriate chambers on a standard microscope glass slide (Ibidi, USA) at the density of 3 × 104 cells/cm2. Fluorescent red latex beads (2 μm diameter, Sigma) were pre-opsonized in 50% horse serum (HS) at 37 °C for 1 h and incubated for 3 h to the cells at the final concentration of 1 × 106 beads/ml. Subsequently, the cells were washed with PBS to remove non-phagocytized beads and fixed in 4% paraformaldehyde for 5 min. Following phalloidin (1:200 Sigma Aldrich) and DAPI (1:1000 Thermo Fisher Scientific) counterstaining the number of beads ingested by the cells was performed using a Zeiss fluorescence microscope and analysis was done by ImageJ software.
Oil Red O staining
Oil Red O staining was performed to identify the content of lipid droplets inside the cells. BV2 cells were seeded at a density of 3 × 104 cells/ml and at the end of the treatment, the culture medium was removed and cells were washed with PBS. Cells were fixed with 4% paraformaldehyde for 30 min and then washed three times with PBS. Cells were incubated for 5 min with 60% isopropanol at room temperature and, after complete drying, incubated for 30 min with the Oil Red O working solution (filtered 6:4 dilution of a pre-filtered 0.35% Oil Red-O stock solution in 100% isopropanol with de-ionized water). Cells were washed with de-ionized water and incubated with 1 µg/ml of Hoechst 33,342 for 10 min. After Hoechst staining, cells were washed with PBS and then coverslips were transferred onto glass slides. Images were observed and captured under a Zeiss fluorescence microscope. The quantification of the red fluorescence was performed using ImageJ software.
HPLC
Metabolic analyses were performed after the deproteinization of cell samples according to a protocol suitable for obtaining protein-free extracts [27]. Briefly, cells were washed twice with ice-cold buffered PBS pH 7.4 and collected by centrifugation at 1860×g for 5 min at 4 °C. The cell pellets were deproteinized with the addition of a solution composed by 250 µl of ice-cold buffered PBS pH 7.40 and 750 µl of ice-cold HPLC-grade CH3CN acetonitrile. After vigorous vortexing for 60 s, samples were centrifuged at 20,690×g for 10 min at 4 °C. Supernatants were collected and subjected to two chloroform washings to eliminate the organic solvent. The upper aqueous phase, obtained by centrifugation under the same conditions, was then used for the HPLC analysis and injected into the HPLC to determine concentrations of NAA, acetyl-CoA, malonyl-CoA. Compounds were analyzed and quantified according to an ion-pairing HPLC method previously set up [28]. For the analyses, the HPLC equipment consisted of a Surveyor HPLC system (ThermoFisher Italia, Rodano, Milan, Italy) equipped with a highly-sensitive photodiode array detector provided with a 5 cm light path flow cell, and set up between 200 and 400 nm wavelength. Data acquisition and analyses were performed using the ChromQuest® software package provided by the HPLC manufacturer. Chromatographic separation of the various compounds was carried out using a Hypersil 250 × 4.6 mm, 5 μm particle-size column, provided with its own guard column (ThermoElectron Italia). Species identification and quantification in cell extracts was performed by matching retention times, peak areas and absorption spectra of those of freshly prepared ultrapure standards. The concentrations of acetyl-CoA and malonyl-CoA were calculated at 260 nm wavelength and those of NAA at 206 nm wavelength. Concentrations of compounds were normalized for the total cell number and expressed as nmol/106 cells.
Extracellular lactate assay
Extracellular lactate assay was performed considering 500 µl of cell medium, that was precipitated with 1:2 volume of 30% trichloroacetic acid (TCA) and stored at − 20 °C for at least 1 h. Samples were centrifuged at 14,000×g for 20 min at 4 °C and then 10 µl of supernatants were incubated for 30 min at 37 °C in 290 µl of reaction buffer (0.2 M glycine, 0.2 M hydrazinium sulphate pH 9.2 with freshly added 0.6 mg/ml NAD+ and 17 U/ml LDH enzyme). After 30 min at 37 °C, NADH levels were evaluated by spectrophotometrically reading at 340 nm using a multi-plate reader (TECAN infinite 200 pro) and then converted to lactate concentration using the extinction coefficient of 6220 M− 1 cm− 1. Values were normalized on total proteins and expressed as µmol/µg of proteins.
Quantitative real-time PCR (RT-qPCR)
Cells were homogenized in TRItidy G (PanReact AppliChem, A4051) and then processed for RNA extraction according to the manufacturer’s instructions. Total RNA (1 µg) was used to synthesize cDNA using iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad) and RT-qPCR performed by using iTaq Universal SYBR® Green Supermix (Bio-Rad) on a QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific). Primers used in the study are as follows: CPT1α forward: 5′-CAGACTCGGTCACCACTCAAG-3′, reverse: 5′-GAGATCGATGCCATCAGGGG-3′; MCAD forward: 5′-AACACTTACTATGCCTCGATTGCA-3′, reverse: 5′-CCATAGCCTCCGAAAATCTGAA-3′; LCADH forward: 5′-GCATCAACATCGCAGAGAAA-3′, reverse: 5′-ACGCTTGCTCTTCCCAAGTA-3′; IL-6 forward: 5′-CTCTGCAAGAGACTTCCATCCA-3′, reverse: 5′-GACAGGTCTGTTGGGAGTGG-3′; TNFα forward: 5′-CCACCACGCTCTTCTGTCTA-3′, reverse: 5′-AGGGTCTGGGCCATAGAACT-3′; IGF-1 forward: 5′-CCGAGGGGCTTTTACTTCAACAA-3′, reverse: 5′-CGGAAGCAACACTCATCCACAA-3′; CD206 forward: 5′-GTTCACCTGGAGTGATGGTTCTC-3′, reverse: 5′-AGGACATGCCAGGGTCACCTTT-3′; HDAC1 forward: 5′-ACAGCAATAGGAGGCCAGTT-3′, reverse: 5′-TCCCTCCTTGCTTTCTCAGG-3′; HDAC3 forward: 5′-GCCGTGGTATTGGGAATGTC-3′, reverse: 5-GAGTGGGCACAAAAGGGAAG-3′; HDAC6 forward: 5′-GGAGACAACCCAGTACATGAATGAA-3′, reverse: 5′-CGGAGGACAGAGCCTGTAG-3′; HDAC11 forward: 5′-GGGGGATCTCAGTGATGGTA-3′, reverse: 5′-AAGAGAAGCTGCTGTCCGAT-3′; SIRT1 forward: 5′-GTAAGCGGCTTGAGGG-3′, reverse: 5′-TTCGGGCCTCTCCGTA-3′; ACTB forward: 5′-CACACCCGCCACCAGTTCGC-3′, reverse: 5′-TTGCACATGCCGGAGCCGTT-3′. Data were normalized to the internal standard ACTB and analyzed using the 2−ΔΔCt method. The fold changes refer to the control.
NO measurement
For nitric oxide detection DAF-2DA (4,5-diaminofluorescein diacetate) was used. DAF-2DA is a non-fluorescent cell permeable reagent that can measure intracellular NO. Once inside cells, it is deacetylated by intracellular esterases to become DAF-2. Production of NO in the cell, if any, converts the non-fluorescent dye, DAF-2, to its fluorescent triazole derivative, DAF-2 T. BV2 cells were seeded in 12-well plates. 30 min before the end of the treatment, cells were incubated with 10 µM DAF-2DA dissolved in DMEM at 37 °C. Cells were then washed with PBS and lysed in lysis buffer. Cell lysates were transferred in a 96 Well Black/Clear Bottom Plate (Greiner Black, 655090) and the fluorescence emission from each well was measured (λex 495 nm; λem 515 nm) with a multi-plate reader (TECAN infinite 200 pro). Results were expressed as DAF-2T fluorescent intensity.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation assay was performed on BV2 cells lysates. Briefly, cells were cross-linked for 12 min at room temperature with 1% final concentration of 37% formaldehyde and the reaction was stopped by 5 min incubation in 125 mM glycine. Cell monolayer was harvested by scraping in ice-cold PBS containing protease inhibitors and collected by centrifugation. Cell lysis was performed in 1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0, protease inhibitors and chromatin was sonicated using BRAISON SONIFIER 250 to High Power, 30 cycles for 10 s ON, 10 s OFF. Average size of sonicated DNA was around 500 bp, as measured by agarose gel electrophoresis. Soluble fraction of cells was isolated by centrifugation at 13.000×g for 15 min at 4 °C. Lysate - 300 µl per assay - was pre-cleared by addition of 2.7 ml of dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HC1 pH 8.0, 167 mM NaCl, protease inhibitors) and 70 µl of protein G Plus/Protein A Agarose suspension (Calbiochem) pre-blocked with salmon sperm DNA. Pre-cleared lysates were then incubated overnight with 3 µg of anti-acetyl-Histone H3 (Active Motif) antibody. Control immunoprecipitations with normal IgG were also performed. Immunocomplexes were recovered from lysates by incubation at 4 °C for 2 h with 65 µl of protein G Plus/Protein A Agarose suspension pre-blocked with salmon sperm DNA. Precipitates were successively washed (10 min each wash) with 1.0 ml of the following buffers: low salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 150 mM NaCI), high salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCI pH 8.0, 500 mM NaCl), LiCl (250 mM LiCl, 1% Nonidet P-40, 1% Na-deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0). All wash buffers had protease inhibitors added. Following two final washes in TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA), the immunocomplexes were finally eluted in 150 µl of TE/1% SDS buffer (10 mM Tris-HCl pH 8.0, 5 mM EDTA, 1% SDS) by incubation at 65 °C for 15 min. The formaldehyde cross-link was reversed by incubating the sample at 65 °C overnight. DNA fraction was recovered by proteinase K (Bioline) digestion followed by phenol-chloroform extraction and ethanol precipitation in the presence of glycogen. DNA pellets were resuspended in 40 µl of water. DNA amplification was performed using iTaq Universal SYBR® Green Supermix (Bio-Rad) on a QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific). All reactions were run as triplicates. Results were expressed as fold enrichment with respect to IgG controls. Primers used were the following: NOS2 forward: 5′-TAGTGGGGAAATGCTGGTCA-3′, reverse: 5′-ATATTCCAACACGCCCAGGA-3′.
Data analysis
Data analysis derived from at least 3 independent experiments. The results are presented as means ± SD. Statistical analysis was carried out through GraphPad Prism 7 software using Student’s t-test for comparisons of only two variables, one-way ANOVA with Tukey post hoc for multiple comparisons. Results were considered statistically significant at p ≤ 0.05.
Results
NAA promotes lipid turnover and phagocytic activity in BV2 cells
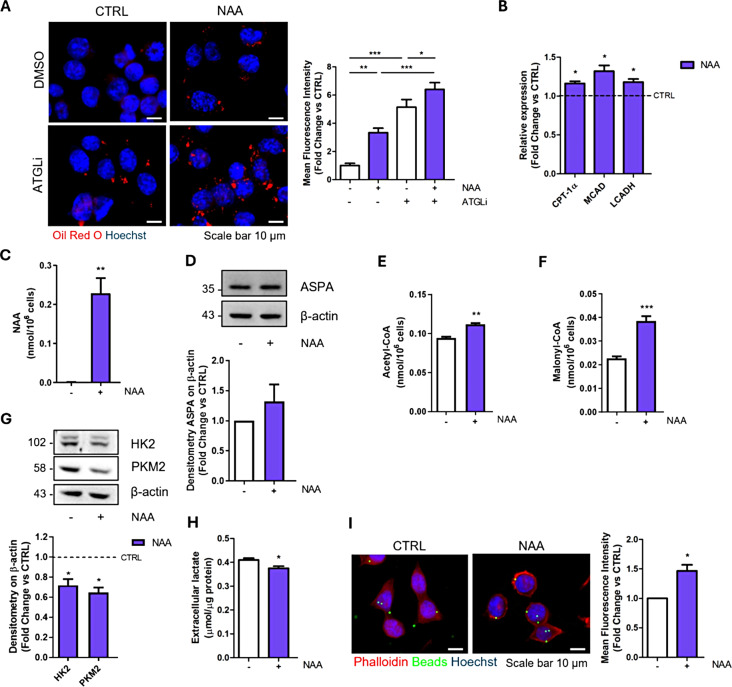
Considering the physiological release of NAA by neurons into the extracellular environment, where microglial cells are continuously exposed to this metabolite, we treated the BV2 microglial cell line for a period of 1 week with 200 µM NAA, a concentration comparable to the one present in brain extracellular fluid. We investigated neutral lipid content as a downstream product of acetate availability. The analysis of Oil Red O staining revealed that NAA-treated cells exhibit a higher number of intracellular lipid droplets (LDs) than control cells (Fig. 1A). The increase in LD content was more evident upon the addition of ATGListatin (ATGLi), an inhibitor of ATGL, the first and rate-limiting lipase of triacylglycerols degradation [29], by proving that NAA can boost lipogenesis and suggesting increased lipid turnover. The enhanced lipid catabolism was confirmed by the upregulation of β-oxidation genes - carnitine palmitoyltransferase 1α (CPT-1α), medium-chain acyl-coenzyme A dehydrogenase (MCAD) and long-chain 3-hydroxyacyl-CoA dehydrogenase (LCADH) (Fig. 1B).
Fig. 1.
NAA enhances lipid metabolism and phagocytic activity in BV2 cells treated for 1 week. (A) Representative images of immunofluorescence analysis of lipid droplet content after Oil red O staining in BV2 cells treated for 24 h with 40 µM ATGLi. Bar graph (right) refers to the immunofluorescence quantification (n = 3; *p < 0.05; **p < 0.01; ***p < 0.001 as indicated). (B) RT-qPCR analysis of genes involved in β-oxidation. ACTB was used as reference gene. Data are shown as fold change vs. CTRL, which was represented by a dashed line in the bar graph (n = 3; *p < 0.05 vs. CTRL). (C) Determination of NAA levels by HPLC analysis (n = 4; **p < 0.01 vs. CTRL). (D) Representative Western blot of ASPA levels. β-Actin was used as loading control. Bar graph (below) refers to the densitometry analysis (n = 3). Determination of Acetyl-CoA (E) and Malonyl-CoA (F) levels by HPLC analysis (n = 4; **p < 0.01; ***p < 0.001 vs. CTRL). (G) Representative Western blot of HK2 and PKM2 levels. β-Actin was used as loading control. Bar graph (below) refers to the densitometry analysis (n = 3; *p < 0.05 vs. CTRL). (H) Evaluation of extracellular lactate content normalized on total proteins (n = 3; * p < 0.05 vs. CTRL). (I) Representative images and quantification (right) of phagocytic activity using green fluorescent latex beads (n = 3; *p < 0.05 vs. CTRL)
Moreover, the detection of intracellular accumulation of NAA (Fig. 1C) by HPLC analysis, the presence of ASPA enzyme (Fig. 1D) and the increased concentration of NAA-derivative lipogenic precursors acetyl-CoA and malonyl-CoA (Fig. 1E-F) following NAA treatment indicate that the metabolite is internalized and catabolized by BV2 cells.
Along with the stimulation of lipid turnover, NAA treatment affects the glycolytic pathway as demonstrated by lower expression of two regulatory glycolytic enzymes, hexokinase 2 (HK2) and pyruvate kinase M2 (PKM2), and the reduced release of extracellular lactate (Fig. 1G-H). This metabolic setting was associated with an increased phagocytic capacity of cells as demonstrated by quantitative assessment of fluorescent latex beads uptake through automated fluorescence microscopy (Fig. 1I).
NAA treatment impacts the pro-inflammatory response of BV2 cells
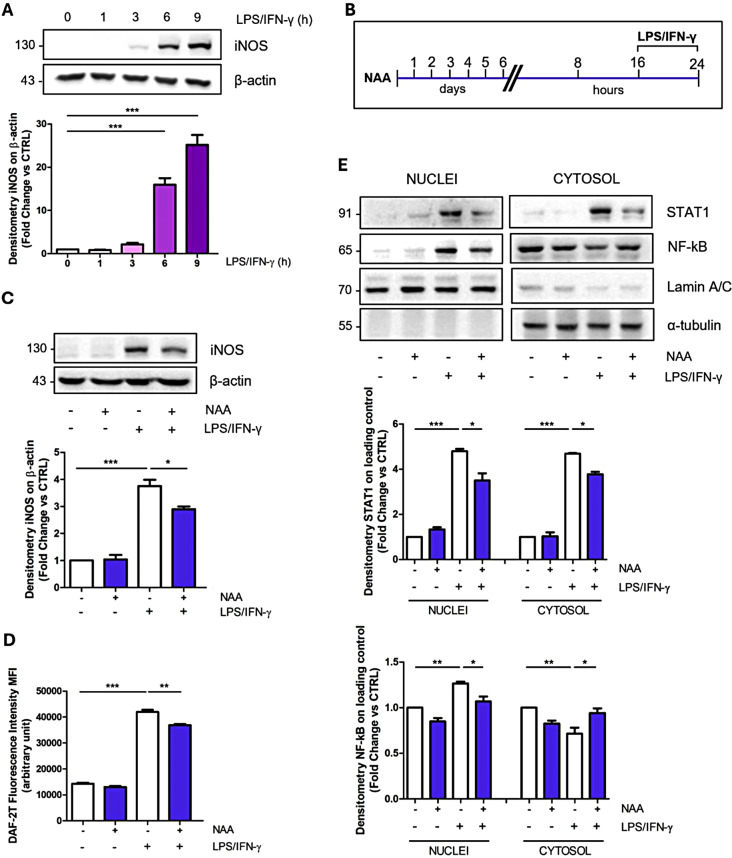
As the metabolic cell asset is fundamental for proper microglial function in inflammation [30], the results we obtained suggest that NAA could impact BV2 reactive phenotype. We challenged BV2 cells with pro- or anti-inflammatory stimuli in the presence of NAA, by using LPS/IFN-γ or IL-4, respectively. Time course experiments demonstrated that both the pro-inflammatory marker iNOS and the alternative activation marker Arg-1 were significantly upregulated starting from 6 h of LPS/IFN-γ or IL-4 treatment (Fig. 2A; Fig. S1A). The proper activation state of BV2 cells was confirmed by the upregulation of TNF-α and IL-6 genes following LPS/IFN-γ stimulation (Fig. S1B), and CD206 and IGF-1 following IL-4 treatment (Fig. S1C).
Fig. 2.
NAA reduces iNOS and STAT1 levels and modulates NF-kB nuclear translocation in BV2 cells. (A) Representative Western blot of iNOS levels in BV2 cells treated with LPS/IFN-γ for 1, 3, 6 and 9 h. β-Actin was used as loading control. Bar graph (below) refers to the densitometry analysis (n = 3; ***p < 0.001 as indicated). (B) Schematic of BV2 cells pre-treated with 200 µM NAA for 1 week and then treated with LPS/IFN-γ for 8 h. (C) Representative Western blot of iNOS levels. β-Actin was used as loading control. Bar graph (below) refers to the densitometry analysis (n = 3; *p < 0.05; ***p < 0.001 as indicated). (D) Determination of intracellular NO levels. Thirty minutes before the end of the experimental time, cells were incubated with 10 µM DAF-2DA and the fluorescent intensity of DAF-2T was measured by a fluorometer (n = 3; **p < 0.01; ***p < 0.001 as indicated). (E) Representative Western blot of cytosolic and nuclear fractions of STAT1 and NF-kB levels in BV2 cells. Lamin A/C and α-tubulin were used as nuclear and cytosolic loading control, respectively. Bar graphs (below) refer to the densitometry analysis (n = 3; *p < 0.05; **p < 0.01; ***p < 0.001 as indicated)
When BV2 cells pre-treated with NAA were subjected to LPS/IFN-γ stimulation (Fig. 2B), iNOS levels were significantly decreased (Fig. 2C). This result was also observed upon extensive washout of NAA from the medium before LPS/INF-γ stimulation (Fig. S1D). The NAA effect on pro-inflammatory markers was corroborated by lower intracellular NO levels and the decreased expression of TNF-α and IL-6 genes (Fig. 2D; Fig. S1E-F). Moreover, NAA was able to reduce gp91phox levels, the plasma membrane subunit of NADPH oxidase (NOX), involved in the production of reactive oxygen species (ROS) during inflammation (Fig. S1G).
NAA also abrogated the increase in extracellular lactate levels typically elicited by pro-inflammatory stimuli (Fig. S2A). Notably, the application of a NAA concentration 10-fold higher (2 mM) than 200 µM treatment resulted in a comparable iNOS downregulation, whereas a 10-fold lower concentration (20 µM) was largely ineffective (Fig. S2B). On the other hand, no modulation in iNOS levels was observed after LPS/IFN-γ stimulation in cells treated with different NAA concentrations for only 24 h (Fig. S2C). Remarkably, when cells pre-treated for a long-term period with NAA were stimulated with IL-4 (Fig. S2D), no significant modulation of Arg-1 and IGF-1 expression was observed (Fig. S2E-F).
To dissect the effect of NAA on cell signaling events following LPS/IFN-γ stimulation we monitored the expression and nuclear translocation of NF-kB and STAT1, key transcriptional factors downstream of LPS/Toll-like receptor 4 (TLR4) and INF-γ receptor (IFNGR) signal transduction pathways, respectively. By analyzing total cell lysates, we observed a reduction in STAT1 levels following NAA treatment (Fig. S2G), while no significant alteration in NF-kB levels was detected (Fig. S2H). However, nuclear/cytosolic fractionation in NAA-treated cells highlighted the reduction in NF-kB nuclear accumulation following LPS/IFN-γ activation and confirmed the decreased levels of STAT1 protein in both compartments (Fig. 2E).
NAA effect on pro-inflammatory markers is not dependent on NMDA receptor in BV2 cells
N-methyl-D-aspartate receptor (NMDAR) activity is known to promote pro-inflammatory response in microglia and it was recently shown to be affected by NAA in macrophages [31]. To test the reliance of NAA on this receptor, we used the selective agonist NMDA. In particular, we monitored pro-inflammatory effects elicited by LPS/INF-γ stimulation in presence of 400 µM NMDA in BV2 cells exposed to short-term and prolonged NAA pre-treatment. The short-term pre-incubation of BV2 cells with 2 mM NAA (a five-fold higher concentration than NMDA) for only one hour was used to test the direct effect of NAA on pro-inflammatory receptors. In this condition, NAA did not abrogate the iNOS upregulation elicited by LPS/INF-γ with/without NMDA (Fig. S3A). Nevertheless, when murine macrophages RAW 264.7 were pre-treated with different doses of NAA for one hour before LPS stimulation, we observed that NAA efficiently inhibited iNOS expression in this cell type (Fig. S3B). On the other hand, prolonged NAA pre-treatment of BV2 cells inhibits iNOS upregulation also in the presence of NMDA (Fig. S3C). These results suggest that NAA exerts its effect in microglia independently of the inhibition of pro-inflammatory receptors (e.g. NMDAR).
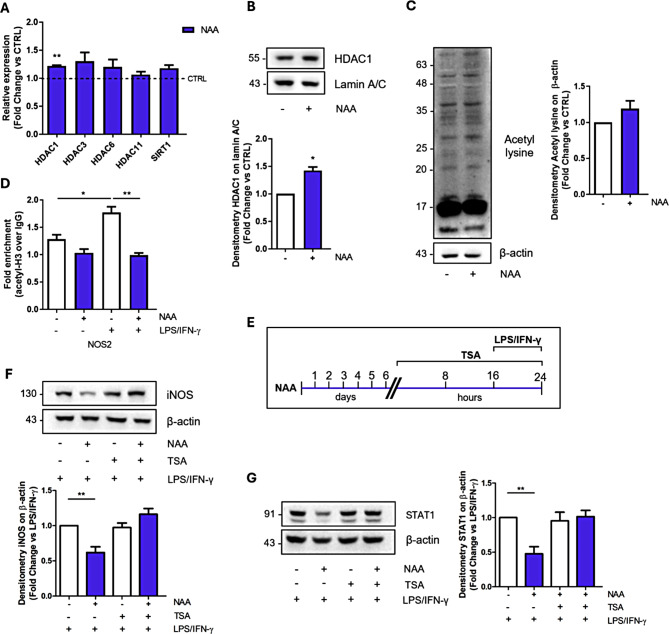
HDAC activity mediates NAA effects on pro-inflammatory markers
NAA was recently demonstrated to foster the expression of histone deacetylase (HDAC) in other glial cells [16] and can provide acetyl moieties not only for lipid biosynthesis but also for protein acetylation processes. Based on this, the inhibition of BV2 pro-inflammatory response achieved by protracted NAA treatment may be mediated by long-term epigenetic effects. Consistently, NAA-treated BV2 cells exhibit a significant upregulation of HDAC1, an HDAC class I enzyme (Fig. 3A-B). Since we observed no significant change in the total lysine acetylation levels after NAA treatment (Fig. 3C), we focused on the locus-specific epigenetic analysis of the NOS2 gene (i.e. iNOS) by ChIP assay. In this context, we demonstrated that the upregulation of iNOS after LPS/INF-γ stimulation is coupled with increased occupancy of acetylated histone H3 at the promoter region (Fig. 3D). Interestingly, NAA was able to counteract that increase in H3 acetylation levels (Fig. 3D), which is consistent with the upregulation of HDAC expression/activity in NAA-treated samples (Fig. 3A-B). To verify the actual involvement of HDAC enzymes downstream of NAA treatment, we utilized the class I/II HDAC inhibitor Trichostatin A (TSA) (Fig. 3E). After validation of TSA efficacy by monitoring the increase in histone 3 and 4 acetylation levels (Fig. S4A), we focused on its effects on pro-inflammatory markers in combination with NAA. We demonstrated that TSA treatment abrogated NAA inhibitory action on LPS/INF-γ mediated upregulation of iNOS and STAT1 (Fig. 3F-G). The same effect on iNOS and STAT1 levels was elicited when we treated BV2 cells with Sodium butyrate (NaB), another class I/IIa HDAC inhibitor, (Fig. S4B-C) implying an active role of HDACs in the NAA-mediated effects.
Fig. 3.
NAA-induced epigenetic modulation via HDAC upregulation mediates its effects on pro-inflammatory response in BV2 cells. (A) RT-qPCR analysis of genes involved in deacetylation processes. ACTB was used as reference gene. Data are shown as fold change vs. CTRL (n = 3; **p < 0.01 vs. CTRL). (B) Representative Western blot of HDAC1 levels in the nuclear fraction. Lamin A/C was used as loading control. Bar graph (below) refers to the densitometry analysis (n = 3; *p < 0.05 vs. CTRL). (C) Representative Western blot of total acetyl lysine levels in BV2 cells treated with 200 µM NAA for 1 week. β-Actin was used as loading control. Bar graph (right) refers to the densitometry analysis (n = 3). (D) ChIP analysis to reveal H3 acetylation levels on NOS2 promoter region. Data are expressed as fold enrichment relative to the sample immunoprecipitated using normal purified IgG (n = 3, *p < 0.05; **p < 0.01 as indicated). (E) Schematic of BV2 cells pre-treated with 200 µM NAA for 1 week and then treated with TSA 25 nM for 24 h and with LPS/IFN-γ for 8 h. Representative Western blot of iNOS (F) and STAT1 (G) levels in BV2 cells. β-Actin was used as loading control. Bar graphs (below and right) refer to the densitometry analysis (n = 3; **p < 0.01 as indicated)
NAA dampens pro-inflammatory activation in primary microglia
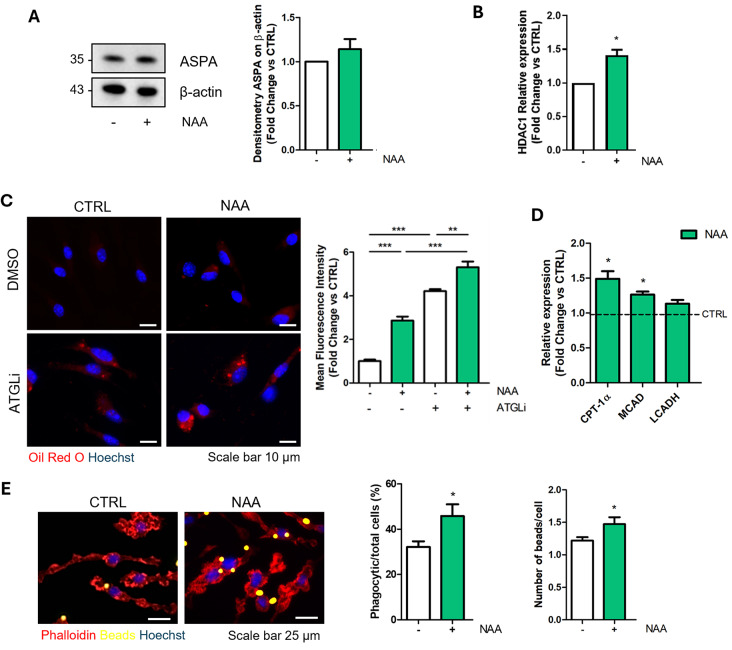
To corroborate the role of NAA in the regulation of microglia reactivity upon pro-inflammatory stimuli, we replicated key experiments on primary microglial cells exposed to NAA treatment. After confirmation that primary microglia express ASPA enzyme and upregulate HDAC1 expression upon NAA treatment (Fig. 4A-B), we assessed whether the metabolic and functional features induced by NAA in BV2 cells also occurred in primary microglial cells. We observed an increase in lipid turnover in primary microglia treated with NAA as demonstrated by the accumulation of Oil Red O staining with/without ATGLi and the upregulation of β-oxidation genes (Fig. 4C-D). This metabolic trait was associated with an enhanced ability to engulf fluorescent latex beads (Fig. 4E).
Fig. 4.
NAA enhances lipid turnover and phagocytic activity in primary microglial cells treated for 1 week. (A) Representative Western blot of ASPA levels. β-Actin was used as loading control. Bar graph (right) refers to the densitometry analysis (n = 3). (B) RT-qPCR analysis of HDAC1 mRNA. ACTB was used as reference gene. Data are shown as fold change vs. CTRL (n = 3; *p < 0.05 vs. CTRL). (C) Representative images of immunofluorescence analysis of lipid droplet content after Oil red O staining in microglial cells pre-treated with 200 µM NAA for 1 week and 40 µM ATGLi for 24 h. Bar graph (right) refers to the immunofluorescence quantification (n = 3; **p < 0.01; ***p < 0.001 as indicated). (D) RT-qPCR analysis of genes involved in β-oxidation. ACTB was used as reference gene. Data are shown as fold change vs. CTRL, which was represented by a dashed line in the bar graph (n = 3; *p < 0.05 vs. CTRL). (E) Representative images and quantification (right) of phagocytic activity tested by using fluorescent latex beads (n = 3; *p < 0.05 vs. CTRL)
Regarding the response of primary microglia to LPS/IFN-γ activation (Fig. 5A), we observed that NAA impeded the efflux of lactate triggered by the pro-inflammatory stimuli (Fig. 5B). Moreover, we confirmed that NAA dampens the upregulation of STAT1 and phosphorylated active form of NF-κB (p-NF-κB) as well as their targets iNOS, TNF-α and IL-6 (Fig. 5C-E). Finally, NAA impeded the LPS/IFN-γ-mediated increased phagocytic activity in primary microglia (Fig. 5F).
Fig. 5.
NAA modulates pro-inflammatory response in primary microglial cells. (A) Schematic of primary microglial cells pre-treated with 200 µM NAA for 1 week and then treated with LPS/IFN-γ for 8 h. (B) Evaluation of extracellular lactate content (n = 6; ***p < 0.001 as indicated). (C) Representative Western blot of STAT1, p-NF-kB, NF-kB and iNOS levels. β-Actin was used as loading control. Bar graphs (below) refer to the densitometry analysis (n = 3; *p < 0.05; **p < 0.01; ***p < 0.001 as indicated). RT-qPCR analysis of TNF-α (D) and IL-6 (E) mRNA. ACTB was used as reference gene. Data are shown as fold change vs. CTRL (n = 3; *p < 0.05; ***p < 0.001 as indicated). (F) Representative images and quantification (right) of phagocytic activitytested by using fluorescent latex beads (n = 3; *p < 0.05 vs. LPS/IFN-γ)
Discussion
In this study we demonstrated that NAA promotes oxidative metabolism in microglial cells and negatively impacts pro-inflammatory stimulation. The main metabolic change is the increase in lipid turnover characterized by augmentation of both triacylglycerol synthesis and degradation. The same phenomenon has been already described in brown adipocytes producing high levels of NAA following NAT8L overexpression [32]. The increase in lipogenesis can be ascribed to ASPA-mediated production of acetate from NAA, as suggested by the increase in intracellular NAA content and consequently acetyl-CoA and malonyl-CoA levels. Consistently, acetate administration alone was previously shown to promote fatty acid synthesis in BV2 cells [33]. The concomitant activation of lipid catabolism may represent an adaptive response of microglia to harmful LD overaccumulation. In fact, excessive LD content is a trait of microglial cells during aging and leads to reduced phagocytosis, oxidative stress and inflammation [34]. The enhanced utilization of lipids as energetic sources following NAA treatment was coupled with a reduction of glycolytic flux. Thus, NAA favors an oxidative phenotype in microglial cells, which is the typical metabolic setting of surveillant microglia. The increase in the phagocytic activity without the activation of proinflammatory markers further indicates that NAA treatment supports microglial homeostatic functions such as debris clearance. These results are in agreement with the physiological role of a metabolite like NAA that is highly produced by neurons and released in the extracellular space.
Based on this, the ability of NAA to mitigate the inflammatory response following LPS/IFN-γ stimulation may represent a way to limit the harmful effects of protracted inflammatory events on neurons. This possibility is supported by the fact that even when NAA concentrations higher than 200 µM were used, the effects were maintained but not intensified, while at low concentrations (20 µM), no anti-inflammatory response can be appreciated. Interestingly, a decrease in brain NAA content is typical of neurodegenerative disorders and thus conditions associated with chronic neuroinflammation [35]. Since no effect was obtained treating cells for a few hours even at high concentrations, the role of NAA in the modulation of the inflammatory response is unlikely due to any inhibition of pro-inflammatory receptors. This behavior is opposed to what was observed in macrophages suggesting that microglia do not respond to acute fluctuations in the concentration of the metabolite but rather they are physiologically primed to adapt to NAA presence in the brain extracellular milieu. As NAA exerts anti-inflammatory activity also on astroglia cells [36] and it was shown to promote the differentiation process of oligodendrocytes [16], a more general contribution of NAA in preserving glial functions in the brain is conceivable.
The function of NAA on microglia reactivity entails the regulation of the acetylation/deacetylation machinery. Acetate may be a driver of the observed response as its administration in rat models of neuroinflammation and in BV2 cells leads to histone acetylation changes diminishing inflammation [37, 38]. Nevertheless, the increase in acetylation levels upon NAA administration is not significantly consistent; this event can be ascribed to the upregulation of the HDAC1 enzyme aimed at preventing excessive acetylation. In fact, histone hyperacetylation at gene promoters is associated with high basal/LPS-mediated activation of inflammatory genes [39] and the acetylation of mitochondrial proteins generally causes enzyme activity inhibition and mitochondrial dysfunction [40]. In our context, HDACs attenuate microglia response since their activity is required for NAA anti-inflammatory effects. However, HDAC inhibitors have been largely demonstrated to abrogate neuroinflammation in different disease states [41–43], where the protracted inflammatory conditions and the complexity of pathogenic events may largely subvert homeostatic metabolism affecting, among others, NAA concentration and functions. Overall, NAA emerges as a key neuronal metabolite that is released in the extracellular space to fine-tune basal and adaptive responses to inflammation in microglial cells exerting epigenetic and metabolic effects.
Conclusions
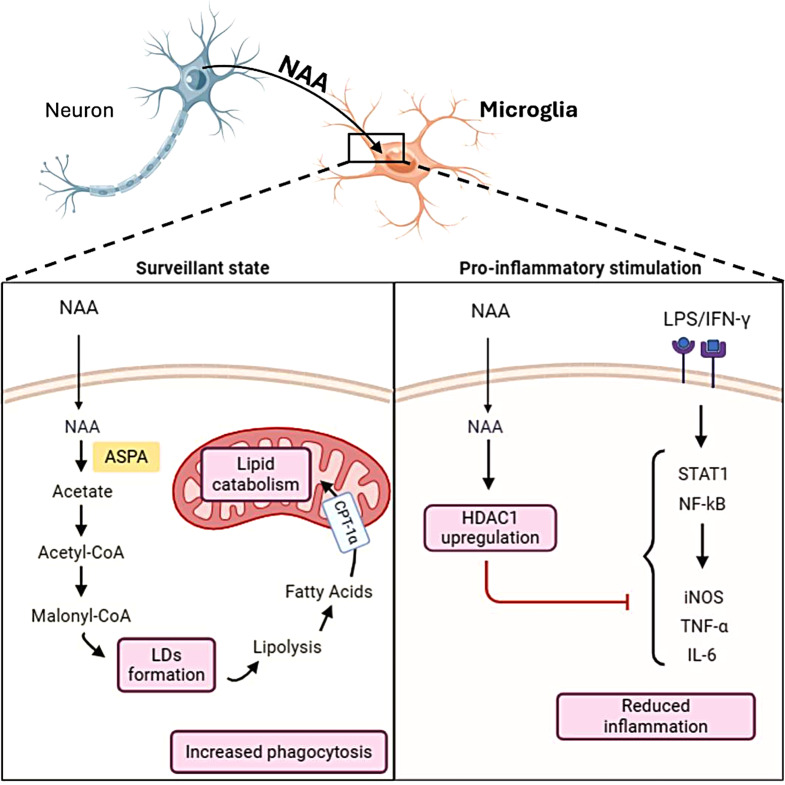
Our data demonstrated that NAA promotes oxidative metabolism in microglial cells and reduces inflammation (Fig. 6). It enhances lipid turnover and phagocytic activity while decreasing glycolysis and inflammatory responses. NAA’s effects are likely mediated through epigenetic mechanisms involving histone deacetylation. These findings suggest NAA regulates microglial functions and mitigates chronic neuroinflammation, playing a crucial role in maintaining brain health.
Fig. 6.
Schematic model illustrating the effects of NAA on microglial cell metabolism and inflammation. This figure illustrates the role of NAA in regulating microglial functions in both surveillant (left panel) and inflammatory (right panel) states. Once inside the cell, NAA is converted to acetate by ASPA enzyme. Acetate is activated to Acetyl-CoA and then undergoes carboxylation to Malonyl-CoA leading to fatty acids synthesis and storage in lipid droplets (LDs). This phenomenon is coupled with enhanced lipolysis fueling lipid catabolism in the mitochondria. Functionally, this condition is associated with increased phagocytic activity in microglial cells. NAA also upregulates HDAC1 expression, which inhibits inflammatory signaling pathways and mediators induced by LPS/IFN-γ
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 2: The online version contains supplementary figures 1-4.
Acknowledgements
Federica Felice is enrolled in PhD Program in Cellular and Molecular Biology, Department of Biology, University of Rome “Tor Vergata”, Rome, Italy.
Abbreviations
- ARG-1
Arginase-1
- ASPA
Aspartoacylase
- ATGL
Adipose triglyceride lipase
- ATGLi
Atglistatin
- CD
Canavan disease
- CD206
Mannose receptor Cluster of Differentiation 206
- ChIP
Chromatin Immunoprecipitation
- CNS
Central nervous system
- CPT-1α
Carnitine palmitoyltransferase-1α
- DAMPs
Damage-associated molecular patterns
- HDAC
Histone deacetylase
- HK2
Hexokinase 2
- IFN-γ
Interferon-γ
- IFNGR
Interferon-gamma receptor
- IGF-1
Insuline-like growth factor-1
- IL-4
Interleukin-4
- IL-6
Interleukin-6
- IL-10
Interleukin-10
- iNOS
Inducible nitric oxide synthase
- LCADH
Long-chain 3-hydroxyacyl-CoA dehydrogenase
- LDs
Lipid droplets
- LPS
Lipopolysaccharide
- MCAD
Medium-chain acyl-coenzyme A dehydrogenase
- MS
Multiple sclerosis
- NAA
N-acetylaspartate
- NaB
Sodium butyrate
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NAT8L
N-acetyltransferase 8-like
- NF-kB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NMDA
N-methyl-D-aspartate
- NMDAR
N-methyl-D-aspartate receptor
- NO
Nitric oxide
- NOX
NADPH oxidase
- PAMPs
Pathogen-associated molecular patterns
- PKM2
Pyruvate kinase isozyme M2
- ROS
Reactive oxygen species
- STAT1
Signal transducer and activator of transcription 1
- TLR4
Toll-like receptor 4
- TNF-α
Tumor necrosis factorα
- TSA
Trichostatin A
Author contributions
F.F. designed and managed the experiments, performed the statistical analysis, conceptualized data and wrote the original draft; P.D.F. and S.C. provided methodological support; G.L. performed HPLC analysis; N.D.A and M.M. performed experiments on primary microglia; A.R.W. analyzed immunofluorescent experiments of phagocytosis; F.C. and M.R.C. performed study concept and design, edited and reviewed the manuscript; M.R.C. and F.C. acquired funding. F.C. and M.R.C. equally contributed as co-last authors. All authors read and approved the final manuscript.
Funding
This work was partially supported by: Ricerca Scientifica d’Ateneo 2021 (E83C22000160005 to FC) University of Rome “Tor Vergata”; Italian Ministry of Health - Ricerca Corrente; #NEXTGENERATIONEU (NGEU) funded by the Ministry of University and Research (MUR), National Recovery and Resilience Plan (NRRP), project MNESYS (PE0000006) – (DN. 1553 11.10.2022).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors are aware of their work and approve of the content of the article and the fact that they are listed as authors of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fabio Ciccarone and Maria Rosa Ciriolo contributed equally to this work as co-last authors.
References
- 1.Colonna M, Butovsky O. Microglia function in the Central Nervous System during Health and Neurodegeneration. Annu Rev Immunol. 2017;35:441–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo S, Wang H, Yin Y. Microglia polarization from M1 to M2 in neurodegenerative diseases. Front Aging Neurosci. 2022;14:815347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orihuela R, McPherson CA, Harry GJ. Microglial M1/M2 polarization and metabolic states. Br J Pharmacol. 2016;173(4):649–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauro C, Limatola C. Metabolic reprograming of Microglia in the regulation of the Innate Inflammatory Response. Front Immunol. 2020;11:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa Y, Chiba K. Role of microglial M1/M2 polarization in Relapse and Remission of Psychiatric disorders and diseases. Pharmaceuticals. 2014;7(12):1028–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon HS, Koh SH. Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl Neurodegener. 2020;9(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuntzel T, Bagnard D. Manipulating Macrophage/Microglia polarization to Treat Glioblastoma or multiple sclerosis. Pharmaceutics. 2022;14(2):344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Xiao D, Mao Q, Xia H. Role of neuroinflammation in neurodegeneration development. Signal Transduct Target Ther. 2023;8(1):267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haidar MA, Ibeh S, Shakkour Z, Reslan MA, Nwaiwu J, Moqidem YA, et al. Crosstalk between Microglia and neurons in Neurotrauma: an overview of the underlying mechanisms. Curr Neuropharmacol. 2022;20(11):2050–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavda V, Singh K, Patel V, Mishra M, Mishra AK. Neuronal glial crosstalk: Specific and Shared mechanisms in Alzheimer’s Disease. Brain Sci. 2022;12(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AM. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol. 2007;81(2):89–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rebelos E, Daniele G, Campi B, Saba A, Koskensalo K, Ihalainen J, et al. Circulating N-Acetylaspartate does not track brain NAA concentrations, cognitive function or features of small vessel disease in humans. Sci Rep. 2022;12(1):11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baslow MH. N-Acetylaspartate in the Vertebrate Brain: metabolism and function. Neurochem Res. 2003;28(6):941–53. [DOI] [PubMed] [Google Scholar]
- 14.Nordengen K, Heuser C, Rinholm JE, Matalon R, Gundersen V. Localisation of N-acetylaspartate in oligodendrocytes/myelin. Brain Struct Funct. 2015;220(2):899–917. [DOI] [PubMed] [Google Scholar]
- 15.Prokesch A, Pelzmann HJ, Pessentheiner AR, Huber K, Madreiter-Sokolowski CT, Drougard A, et al. N-acetylaspartate catabolism determines cytosolic acetyl-CoA levels and histone acetylation in brown adipocytes. Sci Rep. 2016;6(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominicis A, Del Giovane A, Torreggiani M, Recchia AD, Ciccarone F, Ciriolo MR, et al. N-Acetylaspartate drives oligodendroglial differentiation via histone deacetylase activation. Cells. 2023;12(14):1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai G, Coyle JT. N-acetylaspartate in neuropsychiatric disorders. Prog Neurobiol. 1995;46(5):531–40. [DOI] [PubMed] [Google Scholar]
- 18.Namboodiri AMA, Peethambaran A, Mathew R, Sambhu PA, Hershfield J, Moffett JR, et al. Canavan disease and the role of N-acetylaspartate in myelin synthesis. Mol Cell Endocrinol. 2006;252(1):216–23. [DOI] [PubMed] [Google Scholar]
- 19.von Jonquieres G, Spencer ZH, Rowlands BD, Klugmann CB, Bongers A, Harasta AE, et al. Uncoupling N-acetylaspartate from brain pathology: implications for Canavan disease gene therapy. Acta Neuropathol (Berl). 2018;135(1):95–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Song H, Yang Y, Zhang K, Gao X, Li X, et al. Metabolic changes in normal appearing white matter in multiple sclerosis patients using multivoxel magnetic resonance spectroscopy imaging. Med (Baltim). 2017;96(14):e6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sager TN, Thomsen C, Valsborg JS, Laursen H, Hansen AJ. Astroglia contain a specific transport mechanism for N-acetyl-L-aspartate. J Neurochem. 1999;73(2):807–11. [DOI] [PubMed] [Google Scholar]
- 22.Gessler DJ, Li D, Xu H, Su Q, Sanmiguel J, Tuncer S, et al. Redirecting N-acetylaspartate metabolism in the central nervous system normalizes myelination and rescues Canavan disease. JCI Insight. 2017;2(3):e90807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffett JR, Arun P, Ariyannur PS, Garbern JY, Jacobowitz DM, Namboodiri AMA. Extensive aspartoacylase expression in the rat central nervous system. Glia. 2011;59(10):1414–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song T, Song X, Zhu C, Patrick R, Skurla M, Santangelo I, et al. Mitochondrial dysfunction, oxidative stress, neuroinflammation, and metabolic alterations in the progression of Alzheimer’s disease: a meta-analysis of in vivo magnetic resonance spectroscopy studies. Ageing Res Rev. 2021;72:101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Apolloni S, Parisi C, Pesaresi MG, Rossi S, Carrì MT, Cozzolino M et al. The NADPH oxidase pathway is dysregulated by the P2X7 receptor in the SOD1-G93A microglia model of amyotrophic lateral sclerosis. J Immunol Baltim Md. 1950. 2013;190(10):5187–95. [DOI] [PubMed]
- 26.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–75. [PubMed] [Google Scholar]
- 27.Lazzarino G, Amorini AM, Fazzina G, Vagnozzi R, Signoretti S, Donzelli S, et al. Single-sample preparation for simultaneous cellular redox and energy state determination. Anal Biochem. 2003;322(1):51–9. [DOI] [PubMed] [Google Scholar]
- 28.Tavazzi B, Lazzarino G, Leone P, Amorini AM, Bellia F, Janson CG, et al. Simultaneous high performance liquid chromatographic separation of purines, pyrimidines, N-acetylated amino acids, and dicarboxylic acids for the chemical diagnosis of inborn errors of metabolism. Clin Biochem. 2005;38(11):997–1008. [DOI] [PubMed] [Google Scholar]
- 29.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis – a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50(1–4):14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, Qin C, Hu ZW, Zhou LQ, Yu HH, Chen M, et al. Microglia reprogram metabolic profiles for phenotype and function changes in central nervous system. Neurobiol Dis. 2021;152:105290. [DOI] [PubMed] [Google Scholar]
- 31.Menga A, Favia M, Spera I, Vegliante MC, Gissi R, De Grassi A et al. N-acetylaspartate release by glutaminolytic ovarian cancer cells sustains protumoral macrophages. EMBO Rep. 2021;e51981. [DOI] [PMC free article] [PubMed]
- 32.Pessentheiner AR, Pelzmann HJ, Walenta E, Schweiger M, Groschner LN, Graier WF, et al. NAT8L (N-Acetyltransferase 8-Like) accelerates lipid turnover and increases Energy Expenditure in Brown adipocytes *. J Biol Chem. 2013;288(50):36040–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatt DP, Rosenberger TA. Acetate treatment increases fatty acid content in LPS-Stimulated BV2 microglia. Lipids. 2014;49(7):621–31. [DOI] [PubMed] [Google Scholar]
- 34.Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, et al. Lipid droplet accumulating microglia represent a dysfunctional and pro-inflammatory state in the aging brain. Nat Neurosci. 2020;23(2):194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuff N, Meyerhoff DJ, Mueller S, Chao L, Sacrey DT, Laxer K, et al. N-acetylaspartate as a marker of neuronal injury in neurodegenerative disease. Adv Exp Med Biol. 2006;576:241–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rael LT, Thomas GW, Bar-Or R, Craun ML, Bar-Or D. An anti-inflammatory role for N-acetyl aspartate in stimulated human astroglial cells. Biochem Biophys Res Commun. 2004;319(3):847–53. [DOI] [PubMed] [Google Scholar]
- 37.Soliman ML, Puig KL, Combs CK, Rosenberger TA. Acetate reduces microglia inflammatory signaling in vitro. J Neurochem. 2012;123(4):555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soliman ML, Smith MD, Houdek HM, Rosenberger TA. Acetate supplementation modulates brain histone acetylation and decreases interleukin-1β expression in a rat model of neuroinflammation. J Neuroinflammation. 2012;9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meleady L, Towriss M, Kim J, Bacarac V, Dang V, Rowland ME, et al. Histone deacetylase 3 regulates microglial function through histone deacetylation. Epigenetics. 2023;18(1):2241008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson KA, Hirschey MD. Mitochondrial protein acetylation regulates metabolism. Essays Biochem. 2012;52:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G, Shi Y, Jiang X, Leak RK, Hu X, Wu Y, et al. HDAC inhibition prevents white matter injury by modulating microglia/macrophage polarization through the GSK3β/PTEN/Akt axis. Proc Natl Acad Sci U S A. 2015;112(9):2853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai Y, Wei T, Shen Z, Bei Y, Lin H, Dai H. Classical HDACs in the regulation of neuroinflammation. Neurochem Int. 2021;150:105182. [DOI] [PubMed] [Google Scholar]
- 43.Wang C, Shen D, Hu Y, Chen J, Liu J, Huang Y, et al. Selective targeting of Class I HDAC reduces microglial inflammation in the Entorhinal Cortex of Young APP/PS1 mice. Int J Mol Sci. 2023;24(5):4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 2: The online version contains supplementary figures 1-4.
Data Availability Statement
No datasets were generated or analysed during the current study.