Abstract
The STATs (signal transducers and activators of transcription), latent cytoplasmic transcription factors, are activated by binding of extracellular polypeptides to cell surface receptors. Dimerization, accumulation in the nucleus, and transcriptional inductions of specific genes then occur. The COOH terminus of the STATs acts as a transcriptional activation domain (TAD). Stat1, one of seven mammalian STAT genes, forms a homodimer after activation by gamma interferon and induces transcription of a number of genes. These induced genes in turn produce the antiviral state. In the present experiments we used a Stat1-deficient cell line complemented with Stat1 or various fusion constructs in which the wild-type Stat1 TAD was replaced by other TADs to test the possibility that a specific activating domain was necessary for the induction of the antiviral response. We found that a wide variety of TADs with different activation potential appended to the Stat1 COOH terminus could substitute for the wild-type protein in inducing the antiviral state.
The inhibition by alpha interferon (IFN-α) and IFN-γ of viral infection depends upon the full transcriptional activation capacity of Stat1 and Stat2 proteins (9, 37). These transcription factors are latent in the cytoplasm until activated by tyrosine phosphorylation; dimerization, nuclear accumulation, and gene activation follow, with the result that the antiviral state becomes established (9, 37). A great deal has been learned about the functional anatomy of Stats 1 and 2 through mutagenesis of the coding sequences and introduction of mutants into cell lines deficient in one or the other of these proteins. The atomic structures of the core of Stats 1 and 3 and the highly conserved amino terminus have also been described (3, 8), and this information helps to guide such mutagenesis studies. These studies revealed that IFN-α treatment results in the activation of both Stat1 and Stat2, which form a heterodimer that interacts with a 48-kDa protein, p48, forming the interferon-stimulated gene factor 3 (ISGF3) DNA-binding complex (18, 31) that activates the IFN-α target genes (10, 37). In IFN-α-stimulated gene expression, the COOH domain of Stat2 is required, while the COOH terminus of Stat1 is not. IFN-γ treatment results in the activation of only Stat1, which forms a homodimer that activates target genes that contain gamma activation sequences (GAS) in their promoters (6, 10, 36).
The transcriptional activation of ISGF3 depends on the COOH-terminal segment of Stat2 but not that segment of Stat1. IFN-γ-dependent gene activation requires the COOH terminus of Stat1. Thus, at least one transcriptional activation function of the Stats is provided by the COOH terminus (16, 21, 24, 26, 38, 42). The C-terminal transcription activation domain (TAD) is known to interact with CBP-p300 (4, 14, 41) as well as with other proteins which contribute to transcriptional activation (41, 42). We have examined the requirement for and the specificity of the Stat1 COOH terminal domain in transcriptional activation by introducing a variety of recombinant Stat1 constructs transiently or into stable cell lines and correlated results from increasingly more specific assays for transcripitonal activation. The least specific assay was supplementing the yeast Gal4 DNA-binding domain (DBD) (32) with various transactivation domains. Next, the activation of endogenous genes by wild-type Stat1 and various Stat1 chimeric molecules was tested. Finally, induction of the antiviral state, an in vivo response presumably requiring the balanced activation of a set of genes to achieve a physiologic result (1, 27), were tested. We found that considerable variation exists in the ability of the Stat1 constructs with various COOH-terminal activation domains to drive transcription from synthetic promoters, several being more effective than the wild-type Stat1 COOH terminus. However, the Stat1 COOH terminus functions about as well as any other activation domain in stimulating endogenous genes or the antiviral state in response to IFN-γ. Activation domains other than the native Stat1 COOH domain can, however, support establishment of the antiviral state.
MATERIALS AND METHODS
Cell culture and transfection.
Human U3A cells (provided by George Stark, Cleveland Clinic Foundation Research Institute, Ohio) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% cosmic calf serum (Hyclone).
U3A cells stably transfected with Stat1N(1–716)-TADs were selected and maintained in G418 at 0.5 mg/ml (Gibco-BRL). The G418-resistant stable transfectants were directly lysed by sodium dodecyl sulfate (SDS) sample buffer and screened by Western blot with an antibody against the N-terminal domain of Stat1. Transient transfections of the Gal4-DBD fusion constructs were performed by the calcium phosphate method (Gibco-BRL). Altogether, 0.6 μg of Gal4DBD-TAD and 0.4 μg of the luciferase reporter construct with five copies of Gal4 binding sites (5×Gal4 DB) (42) were used in each transfection in a total DNA concentration of 3 μg per 24-well plate. Luciferase activity was assayed ∼40 h after transfection.
Transient transfections of the Stat1N-TAD were performed using the Superfect reagent (Qiagen). Altogether, 0.3 μg of Stat1N-TAD and 0.3 μg of 3×Ly6E-GAS (39) were used in a total DNA concentration of 1 μg per 24-well plate of cells. Superfect-DNA complex incubated following the manufacturer's instruction was added to cells, and 3 h later the medium was replaced. At 24 h after transfection, cells were treated with IFN-γ (7.5 ng/ml) or left untreated for 6 h before harvesting for the luciferase assay. All transfection experiments were normalized to the activity of a cotransfected β-galactosidase expression construct. Recombinant human IFN-γ was a gift from Amgen.
Plasmid constructions.
Mammalian expression vectors Rc/CMV (Clontech), containing wild-type Stat1 or Stat1(S727A), and the 3×Ly6E-GAS luciferase reporter were described previously (39). Rc/CMV Stat1N-TAD fusions were constructed by replacing the XbaI-ApaI fragment at the COOH end of Stat1 with PCR-amplified TADs of Stats 2, 3, 4, 5a, and 6, VP16, and p53 (12, 20, 29, 43). The same TAD fragments were cloned into pSG424 (32) for generating Gal4 fusion proteins. The PCR regions of all constructs were confirmed by sequencing analysis. The Gal4-Stat1C(711–750) and the Gal4 luciferase reporter (5×Gal4 DB) were provided by J. Zhang (42).
RT-PCR.
Reverse transcriptase (RT)-PCR assays were performed on RNAs prepared from stably transfected cell lines with or without IFN-α or IFN-γ treatment as previously described with slight modifications (17). Briefly, total RNAs were isolated using Trizol reagent (Gibco BRL) from subconfluent cells treated with IFN-α or IFN-γ for 4.5 h or left untreated and digested with DNaseI (Promega), followed by reverse transcription with Moloney murine leukemia virus (MMLV) reverse transcriptase (Gibco-BRL) using random primers (Invitrogen). A mock transcription was carried out with no MMLV added (−RT). Typically, 5 μg of total RNA was used in each reverse transcription reaction, and 1/20 of the resulting cDNAs was then used as the template for 25 cycles of PCR amplification with radioactive deoxynucleotides using primers specific to the indicated genes. The products were resolved on a 5% polyacrylamide gel and detected by autoradiography. The primers for IRF-1, guanylate-binding proteins (GBP), ISG15, ISG54, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were as described (40). The primers for TAP1 were TAP1a (AACGGTTGGCTCCAAGAGC) and TAP1b (CGCACAGGGTTTCCAGAGC).
Antiviral CPE assay.
IFN-mediated antiviral response analyzed by a cytopathic effect (CPE) assay was performed as described (15) with modifications. Briefly, cells were plated on 96-well plates 1 day before the assay. Cells were pretreated with 1,000 IU of human IFN-α or 25 ng of IFN-γ per ml for 6 h or left untreated. Encephalomyocarditis virus (EMCV) was diluted in plain DMEM without serum to the desired concentration and added to the cells. After 24 h, the medium was removed, and cells were stained and visualized with 2% methylene blue in 50% ethanol. The absolute absorbance of the methylene blue staining was measured at 630 nm. The killing curve of each test was plotted, and the virus concentration required to kill 50% of the cells was calculated to evaluate the protection efficiency. EMCV was a gift from Robert H. Silverman and was produced and titrated on U3A cells.
RESULTS
Activity of the Gal4-TAD fusion proteins.
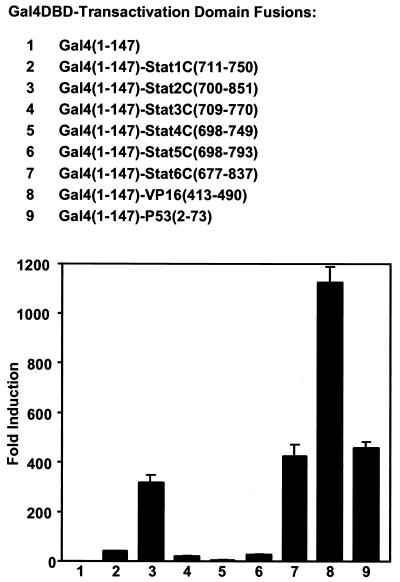
Recombinant DNA constructs encoding the COOH-terminal transactivation domains of Stat 1, 2, 3, 4, 5a, and 6 fused to the Gal4DBD were prepared to assess and compare their transactivation potential in parallel with TADs from two well-studied acidic activators, VP16 and P53 (5). All of these fusion constructs, when transiently transfected into cells, could activate transcription from a Gal4-luciferase reporter construct that has five Gal4 DNA-binding sites, indicating that all of the STAT carboxyl termini have transactivating capacity and thus constitute TADs (Fig. 1).
FIG. 1.
Comparison of the transactivation domains of the different STAT COOH termini by fusion to the DNA-binding domain of the yeast transcription factor Gal4. Gal4DBD-TAD fusions which comprise the DNA binding and dimerization domain of Gal4 (residues 1 to 147) and indicated TADs were constructed. These constructs were transiently transfected into U3A cells with a luciferase reporter (5×Gal4DB) with five copies of Gal4 binding sites. Luciferase activities were determined ∼40 h after transfection. Representative results of four experiments are shown with the standard error for triplicate samples.
Compared with the Gal4 DBD alone, Gal4-Stat1C(711–750), Gal4-Stat3C(709–770), and Gal4-Stat5C(698–793) activated transcription ∼40-, 19-, and 25-fold, respectively (Fig. 1, lanes 2, 4, and 6). Gal4-Stat4C (698–749) was the least active, giving only about fourfold activation in this test (Fig. 1, lane 5). Gal4-Stat2C(700–851) and Gal4-Stat6C(698–793) were the strongest activators among the Gal4-Stat TAD fusions (∼316 and 421-fold activation, respectively, Fig. 1, lanes 3 and 7), comparable to the strong activator Gal4-P53(2–73) (∼455-fold activation, Fig. 1, lane 9). (The strong activation capacity of the Stat6 TAD has been reported [24].) In these assays, as expected, VP16(413–490) is a very strong activator when fused to Gal4-DBD, giving over 1,000-fold activation under the conditions tested (Fig. 1, lane 8), consistent with earlier publications on activation in a Gal4-DBD-dependent system (5 and references therein).
The residues just after the phosphorylated tyrosine (Tyr-705) appear to be important for the transcriptional function of Stat3. Valine 713 and threonine 714 in Stat3 have been reported to be important for Stat3 dimerization (34). In transfection experiments, we found that the residues located in the carboxy-terminal domain just after phosphotyrosine 705 may also be important. The Gal4-Stat3 carboxyl terminus fusion Gal4-Stat3C(709–770) and two other fusion constructs, Gal4-Stat3(716–770) and Gal4-Stat3(713–770), were tested in the Gal4 system. Although only several residues shorter, the two shorter constructs both had only ∼60% of the activity of Gal4-Stat3C(709–770) (data not shown). The shorter Stat3 COOH termini were also somewhat less active when fused with Stat1 N-terminal domain and tested in the IFN-γ-dependent transient transfection assay discussed below.
Different transcriptional activity on a reporter with multiple Stat1 binding sites.
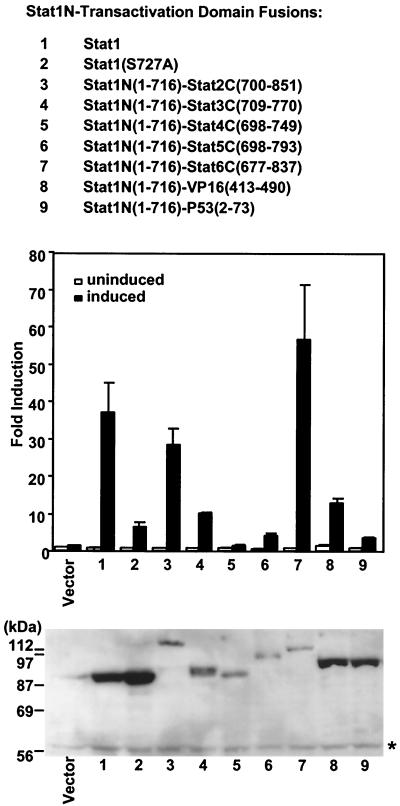
We next tested the various Stat1 COOH-terminal replacement constructs for their response to IFN-γ in transient transfections. The human cell line U3A lacks endogenous Stat1 (22, 25) and therefore can be used to assay IFN-γ-induced transcriptional activity after introduction of the various Stat1 constructs along with a cotransfected luciferase reporter with multiple Stat1 binding sites (39) (Fig. 2). The TADs assayed in the previous section were fused with the Stat1 N-terminal domain (residues 1 to 716, referred to as Stat1N hereafter), replacing the wild-type Stat1 COOH terminal TAD. These Stat1N-TAD chimeric proteins showed different activity in mediating IFN-γ activation of the reporter gene (see below), but all gave transcriptional activation.
FIG. 2.
Transcription activity of various TADs when fused to Stat1 N-terminal domain. Various TADs as indicated were fused to Stat1N (residues 1 to 716). These constructs were transiently transfected into U3A cells with a luciferase reporter (3×Ly6E-Gas) with three copies of Stat1 binding sites. At 24 h after transfection, cells were treated or left untreated with IFN-γ for 6 h and harvested for luciferase assays. The experiments were performed five times, each time with triplicate or quadruple samples. A representative experiment is shown, with the mean and standard deviation of triplicate samples. Shown at the bottom is the Western blot analysis with an antibody reactive with Stat1N on equal amounts of cell extracts from the IFN-γ treated samples. The faint signal in the vector lane at the STAT1 position was due to contamination from the leak of lane 1. ∗, nonspecific protein band. The untreated samples showed similar protein expression levels (data not shown).
Western analysis using an antibody reactive with the Stat1 N-terminal domain showed that the chimeric proteins were expressed at or accumulated to different levels (Fig. 2, bottom panel). These experiments were repeated several times, with similar relative expression levels obtained each time. Obviously, the differences in the transcriptional activity of the various Stat1N-TADs did not correlate directly with protein expression levels. Stat1β, lacking the C-terminal 38 residues, is incompetent in transcription (25, 35), and both Stat1β and Stat1N(1–716) were inactive in this assay (not shown). Wild-type Stat1 was expressed well and gave ∼40-fold activation of the reporter gene upon IFN-γ treatment (Fig. 2, lane 1). Stat1(S727A), carrying a mutation in a residue, S727A, that is known to be required for full Stat1-driven transcription, was expressed well and showed ∼20% of the activity of wild-type Stat1, as reported previously (39, 42). This mutation impairs the interaction of Stat1 with a possible coactivator, MCM5 (42). The Stat3 C-terminal TAD does not interact with MCM5 (42). The Stat3 COOH-terminal construct showed slightly decreased activity compared to wild-type Stat1, but better than Stat1(S727A) (Fig. 2, compare lane 4 with lanes 1 and 2). Stat1N-Stat4C(698–749), which had the weakest activity among the STAT TADs in the Gal4 assay, showed almost no activity when fused with Stat1N (Fig. 2, lane 5). Stat1N-Stat5C(698–793) gave about 50% of the activity of Stat1N-Stat3C(709–770) (Fig. 2, lanes 4 and 6), although Gal4-Stat5C(698–793) showed a slightly higher (∼30%) activity than the Stat3 COOH terminus construct in the Gal4 assay (Fig. 1. lanes 6 and 4). Both Stat1N-Stat6C(677–837) and Stat1N-Stat2C(700–851) fusions, although expressed at low levels, showed strong stimulating activity compared to wild-type Stat1 (Fig. 2, compare lane 1 with lanes 3 and 7), in accord with their strong activities in the Gal4 assay. Thus, the strongest TADs among the STATs come from Stats 2 and 6, which have longer COOH-terminal TADs than the other STATs. The potent TADs of VP16 and P53 showed weaker activity than wild-type Stat1 when fused with Stat1N, despite a level of expression comparable to Stat1 (Fig. 2, compare lanes 8 and 9 with lane 1), differing from the results in the Gal4 assay.
Thus, there is not a consistent correlation between activation by the Stat1 recombinants as full-length molecules driving a synthetic promoter with Stat1 binding sites and the activity of TADs when fused with Gal4-DBD. However, all TADs except the weak Stat4 TAD did give some transcriptional response to IFN-γ when fused with Stat1N; the most potent TADs in the Gal4 fusion experiments, the TADs of VP16 and p53, showed weak activity for inducing IFN-γ-dependent transcription from a synthetic promoter when fused with Stat1N.
Stat1N-TAD fusion proteins activate endogenous IFN target genes.
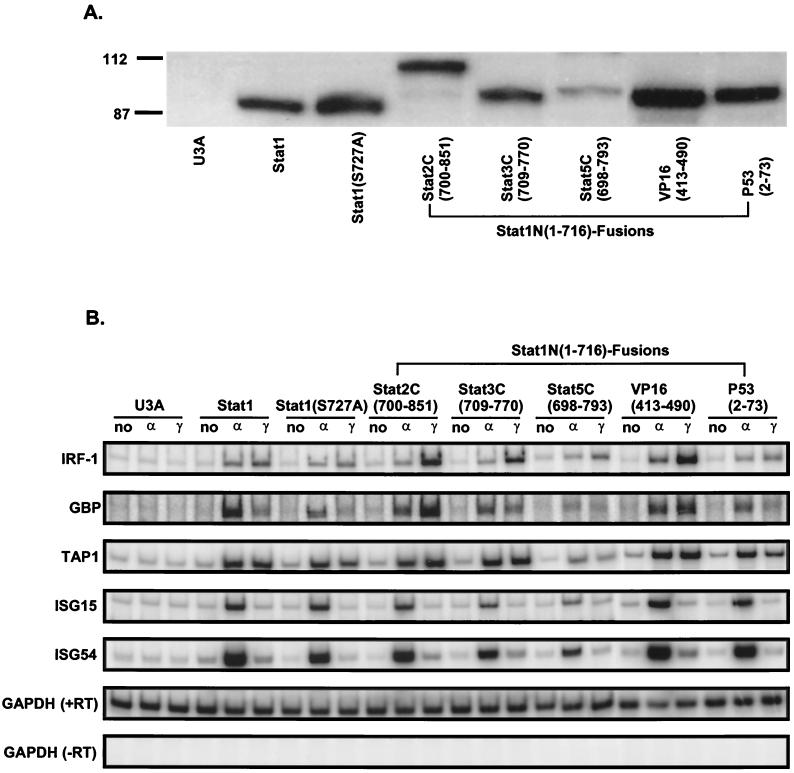
To further examine the requirement and specificity of the TADs in supporting Stat1 transcriptional activity, activation of IFN-γ-inducible target genes in the chromosome was tested (Fig. 3). Expression vectors encoding the Stat1N-TAD chimeric proteins were permanently transfected into U3A cells. Individual cell lines were selected for expression of the Stat1 COOH-terminal fusion proteins (Fig. 3A). The Stat1N-Stat4C(698–749) cell line was not constructed, since the Stat4 COOH-terminal domain showed very low activity in both of the above assays. To determine how the Stat1 fusions function in mediating IFN-α and IFN-γ activation, a semiquantitative RT-PCR analysis (17) was used to assay activation of several target genes. Representative results are shown in Fig. 3. IRF-1, GBP, and TAP1 (23) can be activated to various degrees by both IFN-α and IFN-γ, whereas ISG15 and ISG54 are activated by IFN-α but not IFN-γ (6).
FIG. 3.
Activation of IFN-responsive genes by Stat1N-TAD fusions. U3A stable transfectants expressing wild-type Stat1 or various Stat1N-TAD fusions were selected. (A) Western blot analysis using an antibody against Stat1 N-terminal domain. (B) RT-PCR analysis as detailed in Materials and Methods was performed on the indicated endogenous genes from the Stat1N-TAD stable cell lines treated with IFN-α or IFN-γ or left untreated. In the samples of GAPDH (−RT), reverse transcriptase was left out in the reverse transcription.
As expected, all the Stat1N-TAD fusion proteins tested were approximately equally active in response to IFN-α where Stat1 is part of the IFN-α-induced ISGF-3 but Stat2 supplies the functional TAD (30).
When the ability of the fusion proteins to activate chromosomal IFN-γ target genes was assayed, activities different from those in transient-transfection assays were found (compare Fig. 2 and 3). Stat1N-Stat2C(700–851) and Stat1N-VP16(413–490) showed the strongest activation of the IFN-γ-responsive genes IRF-1, GBP, and TAP1. Stat1N-Stat3C(709–770) showed activity similar to that of the wild-type Stat1. Stat1N-P53(2–73) showed a similar or slightly reduced IFN-γ response of the IFN-γ target genes compared with wild-type Stat1. Stat1N-Stat5C(698–793) was expressed at a lower level and showed weak activation of the IFN-γ-responsive genes.
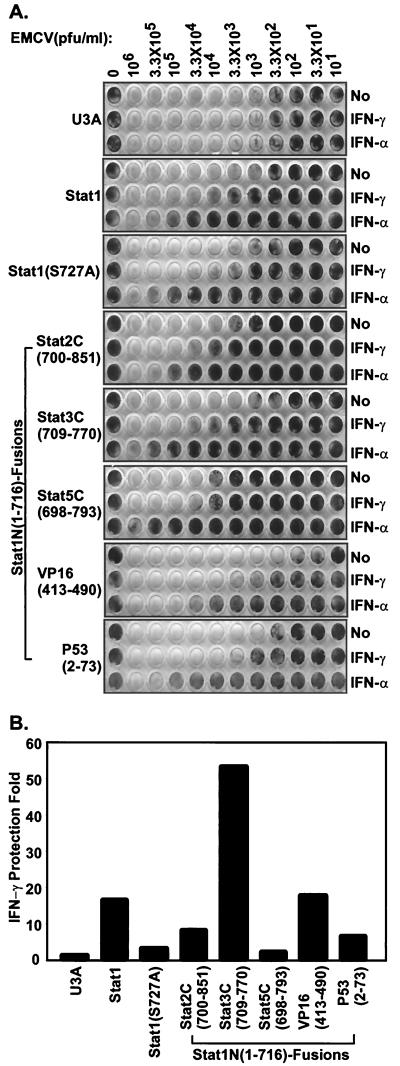
Stat1N-TAD fusion proteins induce antiviral state.
Presumably through activation of a large number of different genes, IFN can induce an antiviral state (37). Using a standard CPE assay of EMCV infection, we next assayed the effectiveness of the various Stat1 chimeras in establishing the IFN-α- and IFN-γ-induced antiviral state (33, 44). Antiviral response to both IFN-α and IFN-γ can be reconstituted in U3A cells by permanent transfection of Stat1 (25). Monolayer U3A cells or U3A cells permanently transfected with various Stat1 fusion constructs were treated with a maximally protective dose of either IFN-α or IFN-γ, followed by infection with serially diluted EMCV (Fig. 4A). Protection by IFN-γ was quantitated by comparing the virus concentrations required to generate 50% of the cell killing as measured by staining of remaining cells (Fig. 4B). All assays were performed on at least two cell lines derived from each construct, and Fig. 4 shows the results from one cell line which were reproduced in several other tests. Without IFN treatment, different cell lines showed slightly different basal levels of susceptibility to EMCV infection, and the maximal protection generated by the IFN-γ antiviral effect was between ∼15- and 50-fold. (It is known that IFN-γ induces less protection than does IFN-α [1], as is evident in Fig. 4A.) The Stat1 fusion proteins protected cells from viral infection in response to IFN-α to a similar level as did wild-type Stat1, consistent with the dominant role of Stat2 in mediating the IFN-α response (30). Stat1N-Stat3C(709–770) and Stat1N-VP16(413–490) were more effective than or as effective as wild-type Stat1 in mediating the IFN-γ-induced antiviral state (Fig. 4A and B, ∼53- and ∼17-fold protection, respectively, compared with ∼16-fold by Stat1). The Stat1 COOH-terminal mutant Stat1(S727A) was less effective in this assay (Fig. 4A and B, ∼3-fold, compared with ∼16-fold by Stat1).
FIG. 4.
Antiviral responses of U3A cells complemented with various Stat1N-TADs. (A) The indicated cell lines were treated with IFN-α or IFN-γ for 6 h or left untreated. The indicated amount of EMCV was added to the corresponding wells and left on the cells for 24 h. The viable cells left in the wells were visualized by methylene blue staining. Similar results were obtained from several experiments performed in duplicate and cells plated at different densities on at least two cell lines from each construct. (B) The above results were quantitated by measuring the absorbance of methylene blue staining of remaining cells at 630 nm. The IFN-γ protection efficiency was evaluated by comparing the virus concentrations required to kill 50% of the nontreated cells versus 50% of the IFN-γ-treated cells.
The concentration of IRF-1 was reported to be very important in establishing IFN-γ-induced antiviral resistence to EMCV (19). Surprisingly, Stat1N-Stat2C(700–851), which activates IRF-1 more strongly than wild-type Stat1 (Fig. 3), induced only (modest about eightfold) IFN-γ antiviral protection (Fig. 4A and B). The Stat1N-P53(2–73) clone, which expressed the fusion protein well (Fig. 3A), also only induced modest (about sixfold) protection (Fig. 4A and B) after IFN-γ treatment. Stat1N-Stat5C(698–793) gave only a marginal level (about twofold) of protection (Fig. 4A and B).
DISCUSSION
We have tested the requirement for and specificity of the Stat1 C-terminal domain in mediating its transcriptional activation function. The first major conclusion from these experiments is that the IFN-γ-mediated protection against virus infection does not specifically require the natural Stat1 COOH-terminal sequences, i.e., other activator sequences can suffice. Similar results have been reported in other systems whereby heterologous activator sequences fused to the DNA-binding domain can mediate in vivo biological responses. For example, VP16 can turn ZEBRA into a more powerful activator in vivo when fused with ZEBRA, although part of the ZEBRA activation domain needs to be present for the fusion protein to work (2). Likewise, the mutant bicoid (Bcd−) phenotype (11) could be rescued by injection of Bcd− mutant embryos with mRNAs encoding fusion proteins consisting of the DNA-binding domain of Bcd attached to several heterologous acidic activating sequences, including acidic regions derived from yeast GAL4- and Escherichia coli-derived sequences. However, when the Bcd DNA-binding domain was fused to the most potent activator VP16, its mRNA had a deleterious toxic effect even when injected at a low concentration.
Second, it is clear that all of the COOH-terminal domains of the STATs have demonstrable transactivation potential that varies both among the different proteins and according to the assay used in assessing transcription. Perhaps the least specific assay, the ability of Gal4DBD-TADs to activate transcription, is the least specific guide to physiologic function. Significant variation was found when the activities of the TADs as Gal4-DBD fusions and Stat1N-TAD fusions were compared. Stat6C(677–837), either as a Gal4 fusion product or when fused to Stat1N(1–716), seems to be more effective than wild-type Stat1 in activating the reporter constructs in transient-transfection assays, in accord with earlier reports (13, 24). In contrast, the other much stronger TADs determined by Gal4 reporter assay, Stat2C(700–851), VP16(413–490), and P53(2–73), showed activity weaker than wild-type Stat1 when fused with Stat1N instead (Fig. 2). These results suggest caution in use of synthetic promoters to derive physiologic conclusions.
Likewise, differences between the effectiveness of each TAD exist when comparing transient and permanent transfections. Of course, in these cases there is also a difference in the promoters, synthetic promoters being used in the transient transfections and endogenous promoters in the permanent transfections. Perhaps not surprisingly, these results make a case for scoring endogenous gene activation when attempting to determine the possible contribution of a transcription factor to a physiologic decision.
When a more stringent and perhaps physiologically relevant comparison was made, namely, for the capacity to induce the Stat1-mediated antiviral response involving activation of a dozen(s) genes organized in the in vivo chromosomal context (6, 37), again a discrepancy with the transient assay was found. Unrelated TADs from VP16 and P53 could also function in this assay, with VP16-TAD being very effective. There was a reasonable correspondence between induction of several endogenous genes and IFN responsiveness; Stat1 induced both, as did Stat3C(709–770), the closest in sequence to Stat1-TAD. This was the case in spite of their relatively weak activation potential, as assayed by Gal4 fusion. It also appears that different promoters may have different requirements. For example, the TAP1 promoter is less sensitive to the difference in the Stat1N-TAD fusions than IRF1 or GBP [Fig. 4B, compare wild-type Stat1 and Stat1N-Stat2C(700–851)]. Such results may reflect the use of different auxilliary transcriptional activators in the enhanceosomal complexes (7, 28). Nevertheless, no specific Stat1 C-terminal TAD appears to be required to bring about the antiviral state.
ACKNOWLEDGMENTS
We thank B. Groner for the Stat5a cDNA clone, J. Ihle for the Stat6 cDNA clone, R. Roeder for the VP16 and P53 clones, and G. Stark for the U3A cells. We thank Michael Carey and Darnell laboratory members for discussions. We are grateful to Stas Mamonov for the StatW-Stat2C(700–851)-complemented U3A cell line and Lois Cousseau for preparation of the manuscript.
This work was supported by NIH grant AI32489 to J.E.D. Y.S. was partially supported by a Leukemia Research Foundation postdoctoral fellowship.
REFERENCES
- 1.Baron S, Coppenhaver D H, Dianzani F, Fleischmann W R, Jr, Hughes T K, Jr, Klimpel G R, Niesel D W, Stanton G J, Tyring S K. Interferon: principles and medical applications. Galveston, Tex: The University of Texas Medical Branch at Galveston; 1992. [Google Scholar]
- 2.Baumann R, Grogan E, Ptashne M, Miller G. Changing Epstein-Barr viral ZEBRA protein into a more powerful activator enhances its capacity to disrupt latency. Proc Natl Acad Sci USA. 1993;90:4436–4440. doi: 10.1073/pnas.90.10.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker S, Groner B, Muller C W. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 5.Blau J, Xiao H, McCracken S, O'Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehm U, Klamp T, Groot M, Howard J C. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 7.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 9.Darnell J E. Stats and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 10.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.Driever W, Ma J, Nusslein-Volhard C, Ptashne M. Rescue of bicoid mutant Drosophila embryos by bicoid fusion proteins containing heterologous activating sequences. Nature. 1989;342:149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- 12.Fu X Y, Schindler C, Improta T, Aebersold R, Darnell J E., Jr The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoey T, Schindler U. STAT structure and function in signaling. Curr Opin Genet Dev. 1998;8:582–587. doi: 10.1016/s0959-437x(98)80015-4. [DOI] [PubMed] [Google Scholar]
- 14.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of Jak/Stat and Ras/Ap-1 signaling by Cbp and P300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath C M, Darnell J E., Jr The antiviral state induced by alpha interferon and gamma interferon requires transcriptionally active Stat1 protein. J Virol. 1996;70:647–650. doi: 10.1128/jvi.70.1.647-650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath C M, Darnell J E. The state of the STATs: recent developments in the study of signal transduction to the nucleus. Curr Opin Cell Biol. 1997;9:233–239. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 17.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Improta T, Schindler C, Horvath C M, Kerr I M, Stark G R, Darnell J E., Jr Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci USA. 1994;91:4776–4780. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak T W, et al. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu B, Reichel M, Fisher D A, Smith J F, Rothman P. Identification of a STAT6 domain required for IL-4-induced activation of transcription. J Immunol. 1997;159:1255–1264. [PubMed] [Google Scholar]
- 22.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min W, Pober J S, Johnson D R. Kinetically coordinated induction of TAP1 and HLA class I by IFN-gamma: the rapid induction of TAP1 by IFN-gamma is mediated by Stat1 alpha. J Immunol. 1996;156:3174–3183. [PubMed] [Google Scholar]
- 24.Moriggl R, Berchtold S, Friedrich K, Standke G J R, Kammer W, Heim M, Wissler M, Stocklin E, Gouilleux F, Groner B. Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells. Mol Cell Biol. 1997;17:3663–3678. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulson M, Pisharody S, Pan L, Guadagno S, Mui A L, Levy D E. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 27.Pestka S, Langer J A, Zoon K C, Samuel C E. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 28.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 29.Quelle F W, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben S M, Cleveland J L, Pierce J H, Keegan A D, Nelms K, et al. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol Cell Biol. 1995;15:3336–3343. doi: 10.1128/mcb.15.6.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qureshi S A, Leung S, Kerr I M, Stark G R, Darnell J E., Jr Function of Stat2 protein in transcriptional activation by alpha interferon. Mol Cell Biol. 1996;16:288–93. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qureshi S A, Salditt-Georgieff M, Darnell J E., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci USA. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel C E. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 34.Sasse J, Hemmann U, Schwartz C, Schniertshauer U, Heesel B, Landgraf C, Schneidermergener J, Heinrich P C, Horn F. Mutational analysis of acute-phase response factor stat3 activation and dimerization. Mol Cell Biol. 1997;17:4677–4686. doi: 10.1128/mcb.17.8.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindler C, Fu X Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuai K, Horvath C M, Huang L H, Qureshi S A, Cowburn D, Darnell J E., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 37.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 38.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 40.Yang E, Wen Z, Haspel R L, Zhang J J, Darnell J E., Jr The linker domain of Stat1 is required for gamma interferon-driven transcription. Mol Cell Biol. 1999;19:5106–5112. doi: 10.1128/mcb.19.7.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J J, Zhao Y, Chait B T, Lathem W W, Ritzi M, Knippers R, Darnell J E., Jr Ser727-dependent recruitment of MCM5 by Stat1 alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17:6963–69671. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong Z, Wen Z, Darnell J E., Jr Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci USA. 1994;91:4806–4810. doi: 10.1073/pnas.91.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou A, Paranjape J M, Der S D, Williams B R, Silverman R H. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]