Abstract
While deep learning (DL) offers the compelling ability to detect details beyond human vision, its black-box nature makes it prone to misinterpretation. A key problem is algorithmic shortcutting, where DL models inform their predictions with patterns in the data that are easy to detect algorithmically but potentially misleading. Shortcutting makes it trivial to create models with surprisingly accurate predictions that lack all face validity. This case study shows how easily shortcut learning happens, its danger, how complex it can be, and how hard it is to counter. We use simple ResNet18 convolutional neural networks (CNN) to train models to do two things they should not be able to do: predict which patients avoid consuming refried beans or beer purely by examining their knee X-rays (AUC of 0.63 for refried beans and 0.73 for beer). We then show how these models’ abilities are tied to several confounding and latent variables in the image. Moreover, the image features the models use to shortcut cannot merely be removed or adjusted through pre-processing. The end result is that we must raise the threshold for evaluating research using CNNs to proclaim new medical attributes that are present in medical images.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79838-6.
Subject terms: Translational research, Predictive markers, Computational science, Medical imaging
Introduction
With the growth of artificial intelligence (AI) applications in medicine, concern over the accuracy of findings has also grown. Algorithmic shortcutting is a problem highlighted by multiple studies1–4, wherein deep learning (DL) models grasp superficial correlations in training data, potentially leading to biased or unreliable predictions. The risks of shortcutting are of particular concern in medical imaging (e.g., X-rays, CT scans, etc.), where machine learning is hoped to be deployed to improve the quality and efficiency of diagnosis and, hence, treatment5.
Within medical research, descriptions of algorithmic shortcutting belie its complexity. First, most studies of shortcutting have focused primarily on issues of fairness2,4,6–11. While fairness addresses biases that can lead to social inequities, algorithmic shortcutting within research can lead to biases that bend the truth. Second, proposed techniques to measure and/or address algorithmic shortcutting only reduce bias from a single confounding variable such as patient race, gender, or age2,12,13. This work aims to demonstrate that shortcutting goes far beyond fairness or single confounders.
This case study uses the Osteoarthritis Initiative (OAI) dataset14—an extensive 10-year dataset from the NIH’s longitudinal study documenting the natural history of knee osteoarthritis (OA). This widely known dataset has been used for hundreds of papers15, with dozens applying CNNs to the X-ray and MRI data included. Here, we use radiographs, but the principles apply to all medical images as the issue stems from deep learning and not specific image types1.
Our novel approach is to test whether a model can be trained to predict with reasonable confidence an outcome that lacks all face validity, for example, whether patients abstain from eating refried beans or avoid drinking beer solely from characteristics of their knee X-ray. As a test of robustness, we then remove the most obvious latent variable to test whether the deep learning algorithm continues to predict a nonsensical outcome. The goal is to highlight the perniciousness of shortcutting and the depth of care needed to surface valid relationships and not just mistake curious shortcuts as new findings.
Results
Predicting confounding and latent variables
This demonstration starts with an examination of how well several example confounding and latent variables can be detected in an X-ray. Table 1 shows the results from a series of models trained on PA fixed flexion knee views to predict the listed attribute. Sex was limited to male and female. Race was self-reported with options of White, Black, Asian, and other. Site was one of the five clinical sites where a patient initially signed up to join the study. Mfg covers seven manufacturers of X-ray machines used in this study. Year covers the years 2004–2014 when the X-rays were taken. Adjusted balanced accuracy is the macro-average of recall scores per class that is then rescaled so that the range 0–1 represents the expected accuracy from random guessing (0) to perfect accuracy (1). The adjusted balanced accuracy exceeded 0.90 in all cases except race (0.24) and year (0.55)).
Table 1.
Model accuracy learning to predict different confounding and latent variables.
| SEX | RACE | SITE | MFG | YEAR | |
|---|---|---|---|---|---|
| Accuracy | 0.987 | 0.921 | 0.982 | 0.999 | 0.644 |
| Adjusted balanced accuracy | 0.973 | 0.244 | 0.947 | 0.999 | 0.546 |
Predict patient diet through x-rays: beans and beer
Given how easily a CNN model can detect demographic and latent factors in an X-ray, it is simple to push the boundary of what predictions we can get from a model. To this end, a model was trained on the PA fixed flexion knee X-rays for all patients who answered the onboarding question of how often they ate refried beans in the past 12 months. These results were then binarized into patients who answered ‘Never’ and those who reported eating refried beans during that time. This simple setup reflects a common experiment design in many emerging CNN-based studies. While the effect is small (AUC 0.631), the results suggest that something within the knee X-ray is predictive of patients not liking refried beans.
A second model was made to confirm this ability to make implausible predictions, this time predicting whether a patient drinks beer less than once a year based on the same X-rays. Here, the effect is even more significant (AUC = 0.734). See Table 2 for full results and confusion matrix.
Table 2.
Model performances predicting patient avoidance of refried beans and beer. (a) Accuracy and AUC for each model against the test holdout set. (b) Confusion matrices for each model on the test holdout set.
| a) | ||||
|---|---|---|---|---|
| Refried beans | Beer | |||
| Accuracy | 0.600 | 0.702 | ||
| AUC | 0.631 | 0.734 | ||
| b) | ||||
|---|---|---|---|---|
| True answer | Predicted answer | |||
| Refried beans | Beer | |||
| Yes | No | Yes | No | |
| Yes | 1,258 | 686 | 1,876 | 610 |
| No | 839 | 1,031 | 529 | 810 |
Examining shortcutting sources
Table 3 provides preliminary evidence of prediction bias (aka differential prediction) across several confounding and latent variables. Differences between actual (P) and predicted prevalence (PP) across a variable’s sub-groups suggest which variables a model may be using for shortcutting. The results for clinical site serve as an example. Each clinical site has an actual prevalence, how those patients responded to the survey, and a predicted prevalence reflecting the models’ predictions for those patients. A well-calibrated model would have good parity between P and PP across sites. In this case, Site C has the fewest patients who avoid eating beans (P = 34.7), but an even lower predictive prevalence for the site (PP = 7.9). This suggests the model may detect the clinical site and generally predicting most patients from that site include beans in their diet.
Table 3.
Model performance across different subgroups within the test set.
| Refried beans | Beer | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % pop | % P | % PP | Acc, % | n | % pop | % P | % PP | Acc, % | ||
| Clinical site | A | 645 | 16.9 | 59.8 | 67.9 | 57.5 | 645 | 16.9 | 70.9 | 80.8 | 70.9 |
| B | 843 | 22.1 | 63.7 | 84.5 | 64.3 | 843 | 22.1 | 73.2 | 77.7 | 78.6 | |
| C | 1,030 | 27.0 | 34.7 | 7.9 | 65.0 | 1,030 | 27.0 | 57.8 | 62.3 | 67.7 | |
| D | 1,043 | 27.3 | 49.5 | 64.7 | 52.3 | 1,047 | 27.5 | 58.9 | 68.9 | 66.8 | |
| E | 253 | 6.3 | 58.5 | 76.3 | 63.2 | 260 | 6.8 | 76.9 | 88.5 | 80.0 | |
| Race | White | 3,164 | 83.0 | 47.1 | 51.4 | 59.7 | 3,175 | 83.2 | 62.0 | 69.9 | 70.0 |
| Black | 592 | 15.5 | 74.5 | 74.5 | 63.2 | 592 | 15.5 | 79.2 | 84.0 | 77.0 | |
| Asian | 7 | 0.2 | 85..7 | 42.9 | 57.1 | 7 | 0.2 | 100.0 | 85.7 | 85.7 | |
| Other | 51 | 1.3 | 74.5 | 54.9 | 41.2 | 51 | 1.3 | 84.3 | 92.2 | 76.5 | |
| Sex | Male | 1,512 | 39.6 | 44.7 | 36.4 | 57.5 | 1,512 | 39.6 | 41.5 | 34.3 | 58.2 |
| Female | 2,302 | 60.4 | 55.1 | 67.3 | 61.6 | 2,313 | 60.6 | 80.4 | 97.3 | 79.7 | |
n is the number of patients in a subgroup. % pop denotes the subgroup’s percentage of the test set population. %P is the percentage of the subgroup that answered positively about avoiding refried beans or beer. % PP is the model percentage of predicted positive answers for each subgroup. Acc, % is the model accuracy for each subgroup.
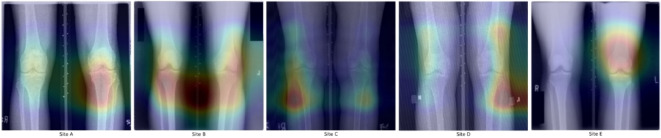
In this case, the source of the shortcutting may seem obvious to the human eye. The X-rays in this study often include laterality markers with fonts and placement unique to each clinical site (seen in Fig. 1). They also include patterns of blacked-out sections to obscure patient health indicators (PHI). However, when we look at what the saliency maps show as relevant to the model decisions, we quickly see this is not happening. Neither laterality markers nor PHI blackouts are consistently in the critical regions. This implies that something more subtle is being used to detect the clinical site.
Fig. 1.
Saliency maps from predictions of patient refried bean preferences. With an example from each clinical site, maps show the natural variance in model focus that spans all images. Note that despite the differences in laterality marker fonts (see site A vs D) across sites, the model only focuses on that in one image. The same goes for PHI blackout boxes (see upper corners of site B and lower left of site E).
Another thing we considered was X-ray manufacturer. We eliminated the most egregious differences between X-rays from different machines by normalizing all images to similar value ranges (see Methods). Even then, the CNN can still predict the X-ray manufacturer with high accuracy (Table 1). Yet, the X-ray machine manufacturer cannot be the sole source of determining the clinical site since clinical sites and X-ray machine manufacturers did not have a 1-to-1 mapping (see Supplementary Information Table S4).
Logically, we can see that the clinical site can’t be the only variable being used to make this prediction. When a model was explicitly trained to predict the clinical site (Table 1), it could not do so with perfect accuracy. Thus, the models must use more than indicators of clinical sites to predict patient consumption of beans. This is further confirmed by the model that predicts beer consumption, which Table 3 shows lacks a single site with less than a 50% positive response. In both cases, the results suggest a mix of biases across clinical sites and the other confounders.
Examining patterns learned by models
To better understand what kind of information correlates to the pixel patterns used by the prediction models, we transferred what each dietary model had learned to new models. Within each model, all but the final layers are convolutional neural network layers that learn useful pixel patterns. The final layer of the model uses a linear combination of the presence of different learned pixel patterns to predict patient preference for a given food. Three new models were created using a copy of the beans prediction model to predict sex, race, and site. These new models kept all layers of the bean prediction model trained to detect pixel patterns relevant to predicting bean consumption rates and only trained a final linear combination layer for their new task. The same was done with the beer model. The results in Table 4 show how well the pixel patterns, learned by the original dietary prediction models, contain information also needed to predict these confounding and latent variables. Both models seem to use pixel patterns that correlate heavily with clinical site and gender but much less so on race, despite significant differences in racial distribution across sites (Table 5).
Table 4.
Performance of the re-trained models.
| Refried beans | Beer | |||||
|---|---|---|---|---|---|---|
| SEX | RACE | SITE | SEX | RACE | SITE | |
| Accuracy | 0.847 | 0.849 | 0.833 | 0.905 | 0.848 | 0.829 |
| Adjusted balanced accuracy | 0.656 | 0.052 | 0.699 | 0.789 | 0.053 | 0.662 |
Using the weights from the CNN layers of the refried bean and beer avoidance prediction models were used to make six new models to predict confounding and latent variables.
Table 5.
Distribution of patient racial identification across clinical sites.
| A (%) | B (%) | C (%) | D (%) | E (%) | |
|---|---|---|---|---|---|
| White or caucasian | 50.7 | 95.1 | 91.4 | 84.2 | 57.0 |
| Black/African American | 47.1 | 1.7 | 7.0 | 13.7 | 38.8 |
| Other/non-white | 1.3 | 2.1 | 1.2 | 0.9 | 2.8 |
| Asian | 0.6 | 0.9 | 0.4 | 1.2 | 1.2 |
Blinding models to a latent variable
To show how the gains from shortcuts on the original dietary models are not linear, we blind the models to one of those variables. For this demonstration, we blind the model to the clinical site. By using k-fold cross-validation across clinical sites, we make five models, each trained on the data of four sites but tested on the data of the remaining site. In the case of refried beans, this results in an accuracy of 0.536 and an AUC of 0.607 (a drop of only 0.02). In the case of beer, the accuracy becomes 0.641, and the AUC becomes 0.690 (a drop of only 0.04).
Discussion
We demonstrate how convolutional neural networks can be trained on properly pre-processed medical imaging studies, yet make surprisingly accurate predictions that lack all face validity (i.e. they suggest relationships that have no known/plausible medical explanation). In this case study, we present a model that can accurately predict a person’s affection for eating refried beans solely from a radiograph of the knee. We repeat this parlor trick by training a model to predict beer consumption from knee radiographs. The models are not uncovering a hidden truth about beans or beer hidden within our knees, nor are the accuracies attributable to mere chance. Rather, the models are “cheating” by shortcut learning –a situation where a model learns to achieve its objective by exploiting unintended or simpler patterns in the data rather than learning the more complex, underlying relationships it was intended to learn. Shortcut learning is a widely known phenomenon in natural images but is known to a lesser degree in medical imaging studies. The eagerness with which these models learn to shortcut is important to recognize. Equally critical is how difficult it is to remove these effects. Understanding the full extent of how shortcutting happens on medical images is critical to researchers, reviewers, and readers alike. Our series of examples walk through different facets of this problem.
Earlier work has detected known confounders: race, age, and gender from chest X-rays6,7,13,16; age, gender, and smoking status from retinal fundus images17; age from dermatology images2. We confirm these findings on knee X-rays, with notably high accuracies. We build upon these findings by showing how well we can detect latent variables, such as the manufacturer of an X-ray machine and even the clinical site from which the X-ray was obtained. Yet merely using the X-ray machine manufacturer isn’t enough to explain the high accuracy in predicting a patient’s clinical site. The model must be using additional subtle clues (possibly machine settings and subtle differences in protocol between sites). Then, to highlight how the model can learn variables that aren’t immediately obvious, we show that the model can learn and predict the calendar year during which an X-ray was obtained. These examples show the potential and range of what shortcutting can pick up. Note that just because a model can learn to spot these variables does not mean it necessarily does when trained for another task7; this merely establishes potential.
Given the risks posed by shortcutting, should we be using these techniques? Deep learning algorithms are not hypothesis tests. Yet, CNNs can see what the human eye can’t. This is both the reason to use them and the reason their use requires extraordinary scrutiny18. Medical images contain a tremendous amount of information, and there is good reason to be excited about the potential for these algorithms to find information within medical images for scientific discovery. Traditional methods largely limit us to visual features humans are already aware of. Handing this feature engineering over to the algorithm in its entirety poses something of a double-edged sword: CNNs automatically detect features, features we know we wanted as well as the ones we didn’t know we needed, but it also means we get the features we never wanted and shouldn’t have. Despite awareness of deep learning’s black-box nature, even smart audiences can be fooled into believing that a CNN is uncovering enigmatic relationships within medical imaging studies. By feeding the model images of knees, it is tempting to assume it is learning a hidden truth about beans and beer in knee anatomy. As Geirhos points out1, human thinking is biased to assume that if a CNN successfully recognizes an object, it seems natural to assume that they are using object properties like shape and color the way humans do (but better). They aren’t19,20. We want to think of confounding and latent variables in a traditional sense –a concrete measure. Internally, these models don’t capture gender or race specifically. It is critical to remember that the “features” these models may learn aren’t clearly defined things; instead, they learn statistical pixel patterns21,22. These can be amorphous, beyond simple reasoning, and it may not even be a cause-and-effect relationship – merely an indescribable association.
As we demonstrated, tracking accuracy imbalances across patient sub-groups is not proof of “cheating.” In reality, the models merely learn patterns that happen to correlate to some degree with each confounding or latent variable, making it hard to say that the model does or doesn’t use a specific variable. It is extremely difficult to separate how much those correlations are shortcuts and how much are mere coincidences. When we take the ‘visual’ layers of the models for predicting beans and beer and train a new final layer that uses the original model’s ‘knowledge’, we can see how the learned pixel patterns also correlate to predict patient race, gender, and clinical site. While accuracy imbalances would suggest the model had learned race, this final step shows that the imbalance was more likely coming from clinical site acting as a proxy for race. Further, these variables are not fully separable. When we blind the models to the clinical site, one might assume that AUC scores would drop dramatically. Instead, the resulting drop is minor.
With a dataset of over 25,000 images, this study exceeds the size of most studies, and yet shortcutting still persists. In fact, earlier work has shown that greater data volumes also don’t eliminate shortcutting1. It is also important to note that these shortcutting patterns were found using extremely low-resolution versions of these images (224 × 224). Higher-resolution versions have more information providing more subtle statistical patterns within the pixels for shortcutting. Finally, as Brown et al. points out, not all shortcutting is harmful2. Certain diseases aren’t uniform across patient demographics and models may be justified in leveraging this for more accurate diagnosis.
All this is to say that it is extremely difficult to address the effects of shortcutting and that no complete solution exists. The problem of shortcutting goes far beyond simple contextual clues like identifying a cow based on a green grassy background versus a camel on a tan sandy background23, healthy patients getting chest X-rays in an upright position and sicker patients in a reclined position24, pen marks in skin lesion classification25 and chest drains in pneumothorax classification26. This demonstration shows that the confounders and latent variables can be thoroughly entangled and smeared across the image. Several papers have focused on ways to measure or prevent a model from shortcutting on a single variable, such as class balancing. While class balancing a cohort is possible for a single variable, we’ve established that shortcutting isn’t using a single variable. Furthermore, it’s important to note that the models weren’t using laterality markers to predict the clinical site, even though they could, and has happened in prior studies27. By blinding a model to one latent or confounder in the training set, a model may just learn others or rely on existing ones more. Biases from shortcutting are pernicious, pervasive, and exceedingly difficult to correct.
There are several policy implications. First is the realization that, while critical, preprocessing and data augmentation will not solve the problem of shortcutting. For example, we found that obvious site identifiers on images (laterality markers, PHI blackouts) were not the pathway through which models identified clinical sites. As such, preprocessing is necessary but by no means sufficient. Second, while tools and methods exist to limit the effect of single confounding and latent variables, in many cases this may not be enough. Deep learning was designed for prediction, not hypothesis testing28. This means that discovery through a black-box tool like CNNs demands far greater proof than simply showing a model found correlations in the sea of data within an image.
Methods
Dataset
In this work, we utilized the OAI dataset, an extensive 10-year dataset from the NIH’s longitudinal study documenting the natural history of knee osteoarthritis (OA). This study examined the Bilateral PA Fixed Flexion X-rays collected over five clinical sites anonymized in the data as sites A-E. This study’s cohort only included those patients who self-identified their race during the study on-boarding. This resulted in 26,495 X-rays across 4,789 patients.
Image preprocessing
To preprocess the images, initially, all images were transformed to set the minimum pixel value as the darkest. This corrective measure eliminates differences based on how the image data is stored. Then, the range of original pixel values of each image was scaled from 0–1. This eliminates differences in range sizes between different source X-ray machines. The images were then reduced to 224 × 224 pixels. Finally, using the population mean and standard deviation pixel values across all training images, the pixels of each image were Z-score normalized. This linear transform is standard practice pre-processing within deep learning to facilitate better convergence during model training and ultimately improve the overall performance of the CNN. These transformations are the standard preparation of images for use by a CNN. See Supplementary Information for an exploration of further normalization.
Convolutional neural network architecture
In this study, we utilized the ResNet18 CNN architecture, which is known for its efficacy in image classification tasks29. The model consisted of 18 layers, utilizing residual connections to address the vanishing gradient problem during training and enhance its ability to learn from complex data. All models were initialized via transfer learning with weights learned from training on the ImageNet dataset30.
Predicting confounding and latent variables
In this stage, five models were created, one for each target variable of patient gender, self-reported race, clinical site, X-ray manufacturer, and X-ray year. To prevent the models from learning to identify individual patients in an X-ray, we partitioned the datasets by patient X-ray sets. This data was split 70/15/15% (training/validation/test) across the 4,695 patients, with the exception of the X-ray manufacturer, which was only available for 24,761 X-rays (4,751 patients). The performance of each model was measured through accuracy and adjusted balanced accuracy, along with providing the resulting confusion matrix. See Supplementary Information for a comparison against similar models built using raw ImageNet-based weights.
Score-CAM images31 were generated to act as saliency maps to elucidate which regions of an image played a significant role in a model’s prediction. Score-CAM (Score-weighted Class Activation Mapping) was chosen because it frequently gives more detailed activation maps than other techniques.
Predict patient diet through x-rays
In this stage, two models were created, one for each target variable of a patient’s self-reported preference for beans and beer during the study enrollment. Again, we partitioned the datasets by patient, with a data split 70/15/15% (training/validation/test). Only 4,648 patients answered the survey about how frequently they ate refried beans, leaving a dataset of 25,743 X-rays. For beer, 4,655 patients answered, giving 25,776 X-rays for model building and testing.
Examining patterns learned by models
The weights from the CNN layers of the two models created in the prior stage were used to initialize six new models. Three models were created to predict gender, race, and clinical site from each of the prior dietary models. These weights were frozen during training so that only the final layer was allowed to change. This transfer learning step allows us to see how much the patterns learned by each dietary model reflect patterns that also predict confounding and latent variables. The dataset sizes were the same as when originally predicting confounding and latent variables.
Blinding models to a latent variable
To prevent a model from being able to leverage what patterns it learned that imply a clinical site, we retrained the model using k-folds32 (k = 5). Each fold used the data from four clinical sites as training data and data from the remaining site as test data. The reported performance metrics of accuracy and AUC are based on the combination of test predictions from all five folds. Both the beans and beer models were retrained using this technique. In this experiment, model training can benefit from learning patterns that predict clinical site, but none of the accuracy in the test datasets will come from this knowledge. In the case of refried beans, the sizes of the clinical site folds are 4,000/5,718/7,564/6,280/2,181 (A-E, respectively). For beer, the sizes are 4,013/5,707/7,567/6,284/2,205.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
B.H. and P.S. wrote the main manuscript. F.K. contributed to an early draft and prepared Fig. 1. All authors reviewed the manuscript.
Data availability
X-ray images used in this study were part of the publicly available NIH-funded OsteoArthritis Initiative (OAI) dataset (https://nda.nih.gov/oai).
Code availability
Code and scripts to reproduce this work can be found at: https://github.com/cairo-lab/xray_fingerprints
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Geirhos, R. et al. Shortcut learning in deep neural networks. Nature Machine Intelligence2, 665–673 (2020). [Google Scholar]
- 2.Brown, A. et al. Detecting shortcut learning for fair medical AI using shortcut testing. Nat. Commun.14, 4314 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, I. et al. “Shortcuts” causing bias in radiology artificial intelligence: Causes, evaluation, and mitigation. J. Am. Coll. Radiol.20, 842–851 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, R. J. et al. Algorithmic fairness in artificial intelligence for medicine and healthcare. Nat Biomed Eng7, 719–742 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajpurkar, P., Chen, E., Banerjee, O. & Topol, E. J. AI in health and medicine. Nat. Med.28, 31–38 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Gichoya, J. W. et al. AI recognition of patient race in medical imaging: A modelling study. Lancet Digit Health10.1016/S2589-7500(22)00063-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glocker, B., Jones, C., Bernhardt, M. & Winzeck, S. Algorithmic encoding of protected characteristics in chest X-ray disease detection models. EBioMedicine89, 104467 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celis, L. E., Huang, L., Keswani, V. & Vishnoi, N. K. Classification with Fairness Constraints: A Meta-Algorithm with Provable Guarantees. In Proceedings of the Conference on Fairness, Accountability, and Transparency 319–328 (Association for Computing Machinery, New York, NY, USA, 2019).
- 9.Kamishima, T., Akaho, S., Asoh, H. & Sakuma, J. Fairness-Aware Classifier with Prejudice Remover Regularizer. In Machine Learning and Knowledge Discovery in Databases 35–50 (Springer Berlin Heidelberg, 2012).
- 10.Wang, R. et al. Drop the shortcuts: image augmentation improves fairness and decreases AI detection of race and other demographics from medical images. EBioMedicine102, 105047 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, H. et al. Improving the Fairness of Chest X-ray Classifiers. In Conference on Health, Inference, and Learning 204–233 (PMLR, 2022).
- 12.Jiménez-Sánchez, A., Juodelyte, D., Chamberlain, B. & Cheplygina, V. Detecting Shortcuts in Medical Images -- A Case Study in Chest X-rays. arXiv [cs.CV] (2022).
- 13.Jabbour, S., Fouhey, D., Kazerooni, E., Sjoding, M. W. & Wiens, J. Deep Learning applied to chest x-rays: Exploiting and preventing shortcuts. Proceedings of Machine Learning Research126, 750–782 (2020). [Google Scholar]
- 14.Michael C. Nevitt, David T. Felson, Gayle Lester. StudyDesignProtocolAndAppendices.Pdf. https://nda.nih.gov/static/docs/StudyDesignProtocolAndAppendices.pdf (2006).
- 15.NIMH Data Archive - Publications Using OAI Data by Year. https://nda.nih.gov/oai/publications.
- 16.Banerjee, I. et al. Reading race: AI Recognises patient’s racial identity in medical images. arXiv [cs.CV] (2021).
- 17.Poplin, R. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng2, 158–164 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Doersch, C., Gupta, A. & Efros, A. A. Unsupervised Visual Representation Learning by Context Prediction. In 2015 IEEE International Conference on Computer Vision (ICCV) 1422–1430 (IEEE, 2015).
- 19.Baker, N., Lu, H., Erlikhman, G. & Kellman, P. J. Deep convolutional networks do not classify based on global object shape. PLoS Comput. Biol.14, e1006613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geirhos, R. et al. ImageNet-trained CNNs are biased towards texture; increasing shape bias improves accuracy and robustness. International Conference on Learning Representations (2018).
- 21.Jo, J. & Bengio, Y. Measuring the tendency of CNNs to Learn Surface Statistical Regularities. arXiv [cs.LG] (2017).
- 22.Ilyas, A. et al. Adversarial examples are not bugs, They Are Features. arXiv [stat.ML] (2019).
- 23.Beery, S., Van Horn, G. & Perona, P. Recognition in terra incognita. In Computer Vision – ECCV 2018 472–489 (Springer International Publishing, 2018).
- 24.DeGrave, A. J., Janizek, J. D. & Lee, S.-I. AI for radiographic COVID-19 detection selects shortcuts over signal. medRxiv (2020) 10.1101/2020.09.13.20193565.
- 25.Winkler, J. K. et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol.155, 1135–1141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakden-Rayner, L., Dunnmon, J., Carneiro, G. & Ré, C. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging. Proc ACM Conf Health Inference Learn2020(2020), 151–159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zech, J. R. et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med.15, e1002683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, J. J. & Tong, X. Statistical hypothesis testing versus machine learning binary classification: Distinctions and guidelines. Patterns (N Y)1, 100115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. arXiv [cs.CV] (2015).
- 30.Deng, J. et al. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (2009).
- 31.Wang, H. et al. Score-CAM: score-weighted visual explanations for convolutional neural networks. arXiv [cs.CV] (2019).
- 32.Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th international joint conference on Artificial intelligence - Volume 2 1137–1143 (Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, 1995).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray images used in this study were part of the publicly available NIH-funded OsteoArthritis Initiative (OAI) dataset (https://nda.nih.gov/oai).
Code and scripts to reproduce this work can be found at: https://github.com/cairo-lab/xray_fingerprints