ABSTRACT
The polysaccharide capsule of Streptococcus pneumoniae is the dominant surface structure of the organism and plays a critical role in virulence, principally by interfering with host opsonophagocytic clearance mechanisms. The capsule is the target of current pneumococcal vaccines, but there are 98 currently recognised polysaccharide serotypes and protection is strictly serotype-specific. Widespread use of these vaccines is driving changes in serotype prevalence in both carriage and disease. This chapter summarises current knowledge on the role of the capsule and its regulation in pathogenesis, the mechanisms of capsule synthesis, the genetic basis for serotype differences, and provides insights into how so many structurally distinct capsular serotypes have evolved. Such knowledge will inform ongoing refinement of pneumococcal vaccination strategies.
BACKGROUND
The presence of what is now recognized as the polysaccharide capsule on the surface of Streptococcus pneumoniae was noted by Pasteur in the first published description of the organism in 1880, and since that time it has been the direct or indirect focus of intensive investigation [reviewed by Austrian (1, 2)]. Studies during the first three decades of the 20th century demonstrated the existence of multiple capsular serotypes of S. pneumoniae and the fact that antibodies to the capsule conferred type-specific protection against challenge in laboratory animals. The capsular material itself was isolated by Dochez and Avery in 1917 (3), but the fact that it was immunogenic led them to believe that this “soluble substance of the pneumococcus” was proteinaceous in nature. It was not until 1925 that Avery and colleagues (4, 5) demonstrated that the pneumococcal capsule consisted of polysaccharide, the first nonprotein antigen to be recognized.
The capsule forms the outermost layer of encapsulated strains of S. pneumoniae. It can be up to approximately 400 nm thick, accounting for more than half of the pneumococcal volume (6), and for the vast majority of cases is covalently attached to the outer surface of the cell wall peptidoglycan (7). A total of 98 structurally and serologically distinct capsular polysaccharide (CPS) types have been recognized to date (8), and the chemical structures of the repeat units for approximately 70% of these have been determined (9). The simplest CPS types are linear polymers with repeat units comprising two or more monosaccharides. The more complicated structural types are branched polysaccharides with repeat unit backbones composed of one to six monosaccharides plus additional side chains. Two nomenclature systems for CPS serotypes have been developed, but the Danish system, which combines antigenically cross-reacting types into groups, is now preferred to the American system, which lists serotypes in chronological order of discovery. Examples of some of these structures are shown in Fig. 1.
FIGURE 1.

Comparison of the CPS biological repeat unit structures of S. pneumoniae serotypes 6A, 6B, 14, 15B, 15C 19F, 19A, 19B, and 19C. These are based on published chemical repeat unit structures (9), adjusting for the fact that Glc is the first sugar of the biological repeat unit.
The polysaccharide capsule has long been considered to be a sine qua non of pneumococcal virulence (2). The vast majority of fresh clinical isolates of S. pneumoniae are encapsulated, and spontaneous nonencapsulated derivatives of such strains are almost completely avirulent. Indeed, as early as 1931, Avery and Dubos (10) demonstrated that enzymic depolymerization of the CPS of a type 3 pneumococcus increased its 50% lethal dose more than 105-fold. Similar virulence defects were later reported for type 2 and 3 pneumococci with defined mutations in genes essential for CPS synthesis (11–14). However, notwithstanding the critical importance of the capsule for invasive disease, “nontypable” S. pneumoniae strains have been associated with outbreaks of conjunctivitis and have been isolated from the nasopharynx and middle ear. While some of these isolates may produce novel CPS serotypes that do not react with existing typing sera, most have genetic defects in the cps locus, which encodes CPS biosynthesis, or lack the cps locus altogether (8, 15, 16).
The clear morphological distinction between encapsulated (smooth) and nonencapsulated (rough) pneumococci, as well as the massive difference in systemic virulence, facilitated early studies of the phenomenon of capsular transformation. This was first demonstrated by Griffith (17), who found that a proportion of mice injected with a mixture of live rough and killed smooth pneumococci died. Smooth pneumococci expressing the same capsular serotype as the killed smooth strain were isolated from the blood of these mice. The transformation reaction was subsequently performed in vitro and led to the seminal discovery that the transforming principle, the carrier of genetic information, was in fact DNA (18).
ROLE IN PATHOGENESIS
The pneumococcal capsule is undoubtedly the most important defense against the host immune system and contributes to pathogenesis in multiple ways. The majority of CPS serotypes are highly charged at physiological pH, and this is thought to promote nasopharyngeal colonization by repelling sialic acid-rich mucopolysaccharides, thereby avoiding entrapment of S. pneumoniae in nasal mucus and facilitating access and attachment to the surface of epithelial cells (19). CPS inhibits both the classic and alternative complement pathways by limiting binding of immunoglobulins, complement components, and C-reactive protein to deeper bacterial surface structures. It reduces opsonization with C3b/iC3b and physically impairs interactions between bound C3b/iC3b or Fc regions of immunoglobulins with cognate receptors on phagocytic cells (20, 21). The CPS may also contribute to pathogenesis by inhibiting entrapment of pneumococci in neutrophil extracellular traps (22) and by impeding recognition of Toll-like receptor ligands on the bacterial surface, thereby impairing MyD88-mediated antibacterial defense (23).
Analysis of a series of mutants producing different amounts of CPS has demonstrated that within a given strain and serotype, the virulence of S. pneumoniae is directly related to capsular thickness (24). However, CPS serotypes differ in the efficacy with which they inhibit opsonophagocytosis in vitro and in their ability to elicit a humoral immune response (25), which largely correlates with differences between pneumococcal serotypes in their capacity to cause invasive disease (2). Otherwise isogenic pneumococci expressing different CPS serotypes exhibit marked differences in virulence for mice (26, 27), competitiveness in a nasopharyngeal carriage model (28), and efficiency of host-host transmission (29). Capsule-switch mutants expressing CPS serotypes associated with higher rates of invasive disease in humans also bound higher amounts of the alternative complement pathway inhibitor factor H in vitro (30). The difference in virulence between pneumococcal serotypes is clearly a function of the biological properties of the CPS itself and is not simply related to the thickness of capsule. For example, type 3 and type 37 pneumococci both produce very thick capsules, but only the former is of high virulence for humans or laboratory animals (2).
CPS-BASED VACCINES
In the immune host, binding of specific antibody to the CPS results in opsonization and rapid clearance of the invading pneumococci. For this reason, a polyvalent pneumococcal CPS vaccine was licensed in the late 1970s. The vaccine provides serotype-specific protection, and the current formulation contains CPS purified from 23 of the 98 recognized serotypes. However, the distribution of S. pneumoniae serotypes varies both temporally and geographically. The current vaccine covers approximately 85 to 90% of disease-causing serotypes in the United States and Europe, but in parts of Asia, coverage is <60% (31). Furthermore, serotype prevalence data are scanty for many developing countries, so vaccine coverage is uncertain. The CPS vaccine is protective in healthy adults (against invasive infections caused by those serotypes included in the formulation), but the efficacy is much lower in other groups at high risk of pneumococcal infection, such as the elderly; patients with underlying pulmonary, cardiac, or renal disease; and immunocompromised patients. Efficacy is poorest in young children, for whom the CPS vaccine has little or no demonstrable clinical benefit. Polysaccharides are T-cell-independent antigens and thus are poorly immunogenic in young children. This is particularly so for the five pneumococcal CPS types that were the most common causes of invasive disease in children (32). Poor immunogenicity of polysaccharide antigens can be overcome by conjugation to protein carriers, which converts them into T-cell-dependent antigens, resulting in immunoglobulin class switching, affinity maturation, and memory. A highly efficacious 7-valent pneumococcal PS-protein conjugate vaccine (PCV) was licensed in 2000, followed more recently by 10- and 13-valent formulations (9). These PCVs elicit strong, boostable antibody responses to the included serotypes and provide robust, albeit still strictly serotype-dependent, protection. An unexpected benefit of the high levels of serotype-specific immunoglobulin G generated by PCVs has been reduced acquisition of carriage among immunized children that has lowered overall rates of transmission, thereby protecting unimmunized populations against the included serotypes (“herd immunity”) (33, 34). However, the clinical benefit of PCVs is being offset by significant increases in both carriage and disease caused by nonvaccine serotypes (35, 36). These occupy the nasopharyngeal niche vacated by the vaccine types as a direct result of immune pressure from widespread use of PCVs (37, 38). Serotype replacement can result from two distinct scenarios. The first of these is by unmasking of strains belonging to nonvaccine serotypes already being carried in the population at low frequency. Elimination of more competitive serotypes covered by the PCV then allows the numbers of the nonvaccine strains to increase in the nasopharynx, increasing the likelihood of both transmission to others in the community and progression to localized or invasive disease. Alternatively, highly competitive strains whose CPS is included in the PCV may acquire the genes encoding synthesis of a nonvaccine serotype CPS by natural transformation with donor DNA from a nonvaccine strain simultaneously present in the nasopharynx. Such “capsule switching” is discussed in more detail later in this chapter. Regardless of the underlying mechanism, widespread use of PCVs is undoubtedly altering the serotype distribution of pneumococcal carriage and disease, which will necessitate ongoing surveillance with a view to periodic adjustment of PCV formulations.
CPS BIOSYNTHESIS GENES
The frequency of transformation of pneumococci from one CPS type to another observed during the classic studies of Griffith, Avery, and others suggested that at least those genes encoding the serotype-specific components of the CPS biosynthetic machinery were closely linked on the chromosome. Additional genetic and biochemical studies by Austrian, Bernheimer, and colleagues (39, 40) demonstrated that during transformation, CPS biosynthesis genes are transferred as a cassette. Advances in gene cloning and DNA sequencing technology in the 1990s resulted in publication of annotated sequences of the complete genetic loci encoding CPS biosynthesis for several S. pneumoniae serotypes and functional characterization of some of the genes therein (41–59). Further advances in high-throughput DNA sequencing enabled publication of annotated sequences for the remainder of the known serotypes by the mid 2000s (60). These studies have provided a sound basis for understanding how the pneumococcus acquired the capacity to synthesize such a vast array of CPS serotypes.
The type 3 locus (designated cps3 [61] or cap3 [62]) and the type 19F locus (designated cps19f [41]) were the first to be completely sequenced and were shown to be located at the same position in the chromosome, between dexB and aliA (41–44). The cps loci from all other serotypes except type 37 have also been localized to the same position on the chromosome. Type 37 has a cryptic copy of the type 33F cps locus located between dexB and aliA; it contains many deletions and point mutations and is not involved in type 37 CPS production. Instead, type 37 CPS biosynthesis is directed by one gene, tts, located elsewhere on the chromosome (54). The mechanism of CPS biosynthesis that occurs in type 37 is similar to that in type 3, and this mechanism is distinct from that in all other serotypes analyzed to date, as described below.
CPS Biosynthesis in Types 3 and 37
Type 3 CPS has a simple disaccharide repeat unit comprising glucose (Glc) and glucuronic acid (GlcA). The cps3/cap3 locus contains only three intact genes, which are transcribed as a single unit (42, 43). The first gene (cps3D or cap3A) encodes the UDP-Glc dehydrogenase required for the synthesis of UDP-GlcA (63). The second gene (cps3S or cap3B) encodes the type 3 synthase, a processive β-glycosyltransferase that links the alternating Glc and GlcA moieties via distinct glycosidic bonds (41, 42, 64). There is a significant degree of amino acid sequence similarity between Cps3S/Cap3B and other bacterial polysaccharide synthases, including HasA, which synthesizes the hyaluronic acid capsule of group A streptococci (65). These synthases have a common predicted architecture, with four transmembrane domains and a large central cytoplasmic domain. This latter region is believed to contain two distinct catalytic sites capable of forming the two different glycosidic linkages (66). The final complete gene in the cps3/cap3 locus (cps3U or cap3C) encodes a Glc-1-phosphate uridylyltransferase. However, Cps3U/Cap3C is not essential for CPS biosynthesis in type 3 pneumococci, because inactivation of the gene has no effect on CPS production (42). This enzyme is also encoded by the galU gene, which is located elsewhere in the chromosome, and its product (UDP-Glc) is required for CPS biosynthesis in all pneumococci, regardless of serotype (67).
Type 37 CPS is a branched polysaccharide with a linear backbone of Glc with a monosaccharide Glc side chain, and a single gene, tts, is responsible for its biosynthesis. Tts is a processive β-glucosyltransferase, which has sequence similarities to a group of plant and bacterial cellulose synthases, especially to the highly conserved motifs thought to be critical for catalysis and/or binding of UDP-Glc (54). The presence of the Cps3S/Cap3B and Tts synthases is sufficient for type 3 and type 37 CPS production, respectively. However, the expression of type 3 CPS also relies on the capacity of the cell to synthesize UDP-GlcA, which is a substrate for Cps3S/Cap3B (68, 69). CPS synthesized by processive transferases does not need a dedicated export system, because the C-terminal hydrophobic domains of these proteins have been predicted to form a pore in the membrane through which the growing polysaccharide chain is extruded as it is synthesized (66).
CPS Biosynthesis in Other Pneumococcal Serotypes
The CPS structures for most other serotypes are more complex than types 3 and 37, with an oligosaccharide repeat unit consisting of at least three sugars (9). These repeat units may also contain phosphodiester linkages, pyruvate, glycerol, phosphoryl choline, and/or ribitol. In some serotypes one or more sugars may also be acetylated (9). The respective cps loci are also much more complex than those of types 3 and 37, which reflects their more elaborate biosynthetic mechanism. The loci vary from approximately 13 to 30 kb in length and consist of from 10 to more than 20 genes that appear to be arranged as a single transcriptional unit (60). Functions for many of the gene products have been determined experimentally, with the remainder assigned on the basis of amino acid sequence similarities with known proteins and with reference to the structure of their CPS repeat unit. The genes in each cps locus are arranged in a common block-wise fashion, as shown for a selection of serotypes in Fig. 2. The first four genes (cpsABCD [also referred to as wzg, wzh, wzd, and wze, respectively (70)]) comprise the 5′ portion of each cps locus; their products have all been implicated in the regulation of CPS production and are discussed later. The central portion of each cps locus encodes the glycosyltransferases responsible for assembly of oligosaccharide repeat units, the repeat unit transporter (Wzx), and the polysaccharide polymerase (Wzy). The 3′ regions of the various cps loci encode enzymes for synthesis of those activated monosaccharide precursors required for CPS synthesis which are not ubiquitous in S. pneumoniae, acetyl transferases and sugar modifying enzymes (60).
FIGURE 2.
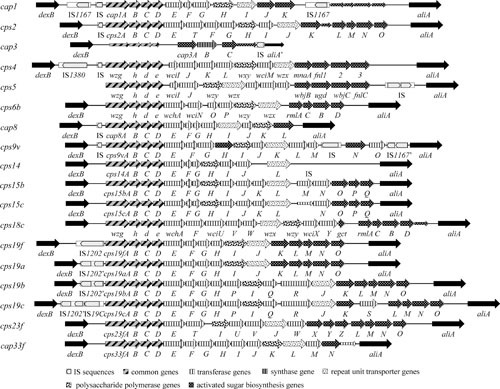
Organization of the cps loci from selected S. pneumoniae serotypes. Gene and locus designations are as published. Open reading frames (ORFs) within the DNA sequence are indicated by large boxed arrows. Highly conserved ORFs, or those encoding proteins belonging to a particular functional group, are identified as shown in the legend at the bottom of the figure. Assignment of an ORF to a given function-related group is based on the published information for each locus as well as on additional database comparisons for some of the ORFs. The short boxed arrows represent cryptic ORFs not required for CPS biosynthesis in the respective serotype.
Biosynthesis of CPS in these serotypes occurs by a Wzx/Wzy-dependent polymerization pathway, analogous to group 1 CPS biosynthesis in Escherichia coli and O-antigen assembly in Gram-negative bacteria (71). The initial step involves transfer of a sugar-1-phosphate moiety to a lipid carrier (undecaprenyl-phosphate) on the cytoplasmic face of the cell membrane. In serotypes containing Glc, this step is carried out by the Glc-1-phosphate (Glc-1-P) transferase CpsE/WchA (60, 72). For those serotypes that lack Glc, the first sugar is linked to the lipid carrier by a cps locus transferase of the WciI, WcjG, or WcjH homology groups (60), except for serotype 1, where the function is provided by the glycosyltransferase that initiates teichoic acid biosynthesis (9). These glycosyltransferases are membrane-associated, which facilitates interaction with the lipid carrier (41). Other glycosyltransferases then catalyze the sequential transfer of the other component monosaccharide precursors (synthesized in the cytoplasm by the activated monosaccharide synthesis genes) to form the polysaccharide repeat unit. These lipid-linked repeat units are then translocated from the cytoplasmic to the extracellular side of the cell membrane by the repeat unit transporter (“flippase”) Wzx and polymerized in a block-wise fashion by the polysaccharide polymerase Wzy, extending the polysaccharide at the reducing terminus. The final step involves direct covalent linkage of the reducing end sugar (usually Glc) to the GlcNAc residue of the cell wall peptidoglycan via a 1,6 glycosidic bond (73). There is uncertainty regarding which enzyme is responsible for this linkage. CpsA, a LytR-CpsA-Psr protein family member, has been proposed to play a role (74), but CpsA deletion mutants are not defective in CPS attachment (75). A recent proposal is that Wzy may be a bifunctional glycosyltransferase capable of recognizing both peptidoglycan and CPS as acceptor substrates, thereby catalyzing both polymerization of CPS and its linkage to the cell wall (73).
COMPARISON OF CPS LOCI FROM DIFFERENT SEROTYPES
The common genomic location and block-wise arrangement of genes in the cps loci (see Fig. 2 for examples) has provided insights into the mechanism of generation of capsular diversity. The first five genes, cpsABCD and cpsE/wchA, are highly conserved in most cps loci, except in serotypes that do not contain Glc in their CPS. These serotypes contain cpsABCD but not cpsE/wchA, which encodes the initiating Glc-1-P transferase (48, 51, 60, 72). The cpsCDE genes exist as two distinct classes, suggesting that all pneumococcal serotypes have evolved from two distinct ancestral cps types (51, 60). Within one class, the genes share greater than 95% sequence identity but share only 70% identity with those from the other class. Some serotypes, such as type 4, contain a hybrid cps locus, where recombination between a class I and a class II locus has occurred. Additionally, the type 3 and type 37 cps loci, which do not require the cpsA-E gene products for CPS biosynthesis, still retain defective copies of these class I and class II sequences, respectively, suggesting that they also have a common ancestry with other pneumococci of the same class.
The central part of the cps locus is serotype-specific, because it encodes glycosyltransferases which exhibit a high degree of substrate specificity and form distinct glycosidic linkages. Accordingly, serotypes only have transferases in common if they have identical glycosidic linkages between identical sugar moieties in their CPS. For example, closely related glycosyltransferases present in the cps loci of types 19F, 9N, and 9V are predicted to join ManNAc via a β(1→4) linkage to Glc (44). This central region also encodes the repeat unit transporter/flippase Wzx and the polysaccharide polymerase Wzy. Closely related polymerase and transporter genes only occur between serotypes with near-identical repeat unit backbones, such as types 19F and 19A, types 19B and 19C, and types 14, 15B, and 15C (51, 52, 59). Recognition of the specificity of these genes laid the foundation for definitive determination of serotype by multiplex PCR rather than by classical serological methods (76), and this has been further facilitated by the availability of sequence data for all CPS serotypes.
Activated monosaccharide synthesis genes, located at the 3′ end of the cps locus, are more widely distributed among different S. pneumoniae serotypes than genes encoding specific glycosyltransferases. Even so, there is substantial deviation among functional homologues. For example, the genes encoding UDP-Glc dehydrogenases from serotypes 1, 2, and 3 exhibit 60 to 90% amino acid sequence identity to each other. Also, the type 19F and type 4 genes encoding UDP-N-acetylglucosamine-2-epimerases are only 66% identical. However, the four genes encoding synthesis of dTDP-rhamnose from Glc-1-P have been shown to be closely related with greater than 95% amino acid sequence identity in all serotypes tested that contain rhamnose in their CPS (44). Interestingly, defective copies of these genes have also been identified at the 3′ end of cps loci from serotypes not containing rhamnose in their CPS, such as type 1. Their presence suggests that the ancestor of type 1 pneumococci may have been a serotype containing rhamnose (48).
COMPARISON OF CPS LOCI FROM CLOSELY RELATED SEROTYPES
Comparison of cps loci of serotypes within group 19 enabled one of the earliest reconciliations of genetics with CPS structure. Serogroup 19 comprises serotypes 19F, 19A, 19B, and 19C, and the structures of their CPS repeat units are shown in Fig. 1. Types 19F and 19A have an identical trisaccharide repeat unit and differ only in the nature of the glycosidic linkage formed during polymerization. Both of these types are important causes of human disease, particularly in young children. Types 19B and 19C are only occasionally associated with human disease; they have more complicated CPS repeat units than types 19F and 19A but differ from each other only by one additional Glc side chain in type 19C. The cps loci of all four members of group 19 are closely related (41, 44, 45, 52). Thirteen genes (cps19A-H and K-O) are conserved in all four serogroup members. Nearly all of the common genes from types 19F, 19B, and 19C are >95% identical to each other, whereas those from the type 19A cps locus are more divergent. Although the 19A and 19F loci are identical in terms of the number and arrangement of the genes present, sequence homology between individual cps19a and cps19f genes varies from 70 to 99% (at both the DNA and the deduced amino acid sequence levels). This sequence divergence is surprising given that only an alteration in cps19fI/wzy, such that an α (1→3) rather than an α (1→2) linkage is formed during polymerization of the repeat units, is required to change a type 19F pneumococcus into type 19A (52). This suggests either that the type 19F and 19A loci diverged in the distant evolutionary past or that their component genes originated from different sources.
The type 19B and 19C cps loci are nearly identical and contain five genes between cps19H and cps19K unrelated to the genes in the type 19F and 19A cps loci (Fig. 2). These genes encode a repeat unit transporter and a polysaccharide polymerase that are specific for type 19B and 19C CPS, as well as two additional putative glycosyltransferases and an unknown protein that might possibly be involved in synthesis of an activated ribose precursor. These five genes encode all of the functions required to convert a type 19F pneumococcus to type 19B (45). The type 19C cps locus differs from the type 19B locus only in the insertion of a putative glucosyltransferase gene (cps19cS) between cps19cK and cps19cL (Fig. 2). The presence of this gene accounts for the additional Glc side chain in the otherwise identical repeat unit structures (Fig. 1).
The structures of the type 6A and 6B CPS repeat units differ only in the linkage between the rhamnose and ribitol moieties (Fig. 1), and as expected, the type 6A and 6B cps loci are very closely related (57, 60). The sequences for the genes encoding the rhamnosyltransferase differ only by a few bases and result in very few amino acid changes. However, these changes are clearly sufficient to alter the serotype-determining linkage. Genetic relationships of cps loci within serogroups 9, 12, and 15 and other closely related serotypes have been discussed by Bentley et al. (60).
CAPSULAR TRANSFORMATION IN VIVO
There is growing evidence that the phenomenon of capsular transformation first observed by Griffith (17) is a common event in vivo, even before the widespread introduction of PCVs which provide additional immune selection. Indeed, the cps locus has recently been identified as an evolutionary hot spot with elevated rates of recombination and substitution (77). Even before the advent of high-throughput genome sequencing, the application of molecular typing techniques resulted in the detection of otherwise genetically indistinguishable pneumococci expressing different capsular types. This was particularly evident in clonal groups that are resistant to multiple antibiotics. Indeed, derivatives of a highly successful, multiply resistant type 23F clone (which originated in Spain) expressing types 3, 9N, 14, 19A, and 19F capsules were isolated well before the introduction of PCVs (27, 78–80). Examination of sequence polymorphisms in the conserved regions of the cps loci for the parental multiresistant type 23F strain and eight otherwise identical type 19F clinical isolates indicated that in each case, the 5′ recombination occurred upstream of dexB. In six of the eight type 19F strains, the 3′ crossover point was downstream of aliA. However, in the other two, a recombination crossover point between the introduced type 19F sequences and the type 23F chromosome was identified; this was in cpsM in one strain and cpsN in the other (81). Thus, capsule switching involves the exchange of very large DNA fragments, ranging from at least 15 kb to over 22.5 kb. The existence of multiple crossover points as well as additional minor polymorphisms within the type 19F-derived cps genes also indicated that the eight multiply resistant type 19F strains that were studied arose as a consequence of a minimum of four independent transformation events involving different type 19F donors. It therefore appears that these capsule switching events may be relatively common among pneumococci in nature (81). Multiple serotypes of S. pneumoniae are frequently carried concurrently in the human nasopharynx (2), providing ample opportunity for exchange of DNA between types. In addition to enhancing the spread of drug resistance among diverse capsular types, these exchanges provide a mechanism for evasion of serotype-specific host immune defenses, such as those resulting from immunization with PCVs, which provide cover against a limited range of serotypes.
Complete or partial insertion sequences have been located adjacent to many of the cps loci, and in some serotypes, the locus is flanked at both the 5′ and 3′ ends by such elements. The cps-flanking regions appear to be common targets for insertion elements, and this has led to the suggestion that they may play a role in horizontal transfer of cps genes (47, 48). There are precedents for this in other bacteria; the H. influenzae type b capsule genes, for example, are located on a 17-kb compound transposon (82). However, the identification of crossover points within the cps loci of the type 19F derivatives of the multiresistant type 23F S. pneumoniae clone referred to above confirms that at least in these cases, capsular exchange occurred as a consequence of homologous recombination rather than by transposition. Nevertheless, Muñoz et al. (48) demonstrated that IS1167 sequences flanking the cap1 locus could direct ectopic integration of these genes into copies of IS1167 located elsewhere in the pneumococcal chromosome, resulting in genetically binary strains.
The contribution of the CPS itself to in vivo recombination is somewhat paradoxical. On one hand, dense genomic sampling of carriage isolates in a refugee camp indicated that the highest frequencies of donation and receipt of recombined DNA fragments occurred in nonencapsulated lineages of S. pneumoniae, consistent with the capsule acting as a physical barrier to DNA exchange (83). On the other hand, among encapsulated lineages, recombination was positively associated with serotypes expressing larger capsules and extended nasopharyngeal carriage duration (84). The nasopharyngeal environment is particularly conducive to genetic exchange because not only are multiple S. pneumoniae strains carried simultaneously, colocalizing donors and recipients, but transformation frequency itself is enhanced in nasopharyngeal biofilms wherein competence genes are upregulated and capsule thickness is reduced (85). However, the complexity and diversity of pneumococcal cps loci can be only partially explained by genetic recombination within the S. pneumoniae species. Recent studies have demonstrated the presence of intact cps loci genes in a wide range of commensal streptococci. The degree of sequence homology indicates that many pneumococcal CPS serotypes have evolved by import of cps fragments from commensal Streptococcus species, resulting in a mosaic of genes of different origins (86). Such interspecies recombination is now recognized as having an important role in the evolution of pneumococcal cps loci and the emergence of new CPS serotypes (77).
REGULATION OF CPS PRODUCTION
Colonization of the nasopharyngeal mucosa is an essential first step in the pathogenesis of pneumococcal disease and involves direct interaction between pneumococcal adhesins and specific receptors on host epithelial cells. In some cases, asymptomatic carriage progresses to invasive disease, and although the events involved are not fully understood, it is clearly a watershed in the bacteria-host relationship. This transition involves a major switch in the expression of important virulence determinants as the pneumococcus adapts to the altered microenvironment (87, 88). Maximal expression of capsule is clearly important for systemic virulence, but the degree of exposure of other important pneumococcal surface structures, such as phosphoryl choline moieties on cell wall teichoic acid and various surface proteins involved in adherence to host cells, may also be influenced by capsular thickness. Nonencapsulated pneumococci exhibit higher adherence to, and invasion of, human respiratory epithelial cells in vitro than otherwise isogenic derivatives expressing either type 3 or type 19F capsules (89). Thus, the very feature (encapsulation) that is most important for systemic virulence of S. pneumoniae could be disadvantageous during the colonization or cellular invasion phase. Thus, regulation of CPS production is crucial for survival of the pneumococcus in different host environments.
Role of cpsABCD
As mentioned previously, the first four genes of the cps locus, cpsABCD (wzg, wzh, wzd, and wze) are common to all pneumococcal serotypes, except types 3 and 37, and deletion of any one of these genes has been shown to affect the level of CPS expression (90). Homologues of cpsB-D are found in most Gram-positive capsule loci, but cpsA homologues have only been identified in the cps loci of other members of the genus Streptococcus. Interestingly, although CpsA resembles a Bacillus subtilis transcriptional attenuator, its homologue in group B streptococci appears to function as a transcriptional enhancer (91). To date, cpsA has not been shown to affect cps transcription in S. pneumoniae. Type 19F and type 2 mutants in which the cpsA gene had been deleted produced reduced levels of CPS (approximately two-thirds that of the respective wild-type strains) (14, 90, 92). However, there were no obvious differences in the amounts of CpsB and CpsD proteins detected by Western blotting in any of the strains (90, 93). Nor is there convincing evidence that pneumococcal CpsA catalyzes the linkage of CPS to peptidoglycan, as discussed above. Thus, the precise function of CpsA in pneumococcal CPS biosynthesis or regulation remains uncertain.
CpsB, CpsC, and CpsD collectively constitute a phosphorelay system implicated in modulation of CPS production at a posttranscriptional level. CpsC and CpsD belong to the PCP2b family of polysaccharide copolymerases and were predicted to function together in polymerization and export of CPS in a fashion similar to PCP2a proteins, such as Wzc and ExoP, in the production of CPS and exopolysaccharide in Gram-negative bacteria (41, 71, 94, 95). CpsD has similarities to the C-terminal domain of PCP2a proteins and is an autophosphorylating protein-tyrosine kinase (90, 95). These kinases contain Walker A and B ATP-binding motifs and a tyrosine-rich region at the C-terminus which becomes phosphorylated at multiple tyrosine residues, initially via the ATP-binding domain and also via transphosphorylation which occurs independently of the ATP-binding domain (11, 90, 92). In S. pneumoniae, the tyrosine-rich region of CpsD is arranged as an ordered (YGX) motif varying from two to four repeats in different serotypes, with four being the most prevalent. CpsC is a membrane protein with two membrane-spanning hydrophobic domains; the N and C termini are located in the cytoplasm, while the central portion is exposed on the external side of the cell membrane. It has similarities to PCP1/Wzz proteins associated with polymerization of O-antigen in Gram-negative bacteria and to the N-terminal domain of PCP2a proteins (95). CpsC is required for initial CpsD tyrosine-autophosphorylation but is not required for transphosphorylation between CpsD proteins (90). CpsB has been identified as a manganese-dependent phosphotyrosine-protein phosphatase belonging to the PHP (polymerase and histidinol phosphatase) family of phosphoesterases and is required to dephosphorylate CpsD (96). CpsB is also thought to be able to bind CpsD and prevent transphosphorylation between CpsD proteins (11).
Although CpsB, CpsC, and CpsD clearly have a role in modulation of CPS production, the precise manner in which this is achieved remains uncertain, and the situation has been complicated by phenotypic differences between mutants constructed in different strain backgrounds. Indeed, CpsD phosphorylation has been reported to have opposite effects on total CPS production in some studies (90, 93). The capacity of CpsD to cycle between the phosphorylated and dephosphorylated state may facilitate interactions with the polymerization machinery that modulate CPS chain length and cell wall ligation (14, 75, 90, 92, 93). Regardless of its actual role, loss of CPS regulation via phosphorylation of tyrosine in CpsD affects the ability of S. pneumoniae to translocate from the lungs into the bloodstream in a murine intranasal challenge model (14). Interestingly, CpsD phosphorylation has been reported to be suppressed by high oxygen concentrations via modulation of CpsB phosphatase activity, which may imply a role in the adjustment of CPS expression between host sites that differ in oxygen availability (97, 98).
Recent advances in cell imaging technologies have provided novel insights into the collective function of the CpsBCD phosphorelay system. CpsC and CpsD have been shown to localize to the division septum in growing pneumococcal cells, and this depends on a functional ATP binding domain in CpsD. In the absence of either CpsC or CpsD, CPS was still produced and linked to the cell surface, but it was absent from the division septum, suggesting that the two proteins are spatial regulators of CPS synthesis (99). Further studies showed that the cytoplasmic C-terminal portion of CpsC required for CpsD autophosphorylation was essential for localization at mid-cell and that the CpsCD complex captures the polymerase Wzy at the division site. Furthermore, dephosphorylation of CpsD impaired both CPS synthesis and cell division itself (100). Collectively, these studies support a model whereby the CpsBCD system ensures that CPS and new cell wall synthesis are colocalized, thereby enabling the newly synthesized cell wall to be promptly masked by CPS to prevent complement deposition at the site (99, 100).
Interestingly, the CpsBCD phosphorelay system has recently been shown to also be capable of phosphorylating and dephosphorylating the major pneumococcal autolysin LytA. LytA amidase activity was stimulated in the phosphorylated state, suggesting a regulatory link between CPS synthesis and LytA-dependent cellular autolysis (101). As mentioned previously, the presence of the pneumococcal capsule significantly impairs both adherence to, and invasion of, epithelial cells in vitro (89). However, electron microscopy studies have shown that pneumococci are capable of shedding their capsules at the point of epithelial cell invasion (102). Such capsule shedding is dependent on LytA and protects pneumococci from attack by cationic antimicrobial peptides released in the immediate vicinity by host cells (103). Thus, it may not be a coincidence that the LytA-dependent capsule shedding process is modulated by the CpsBCD phosphorelay (103).
Transcriptional Regulation of the cps Locus
The first direct evidence for transcriptional regulation of the cps locus in the host environment used quantitative reverse transcriptase PCR (RT-PCR) to show that the level of cps mRNA relative to 16S rRNA in pneumococci isolated from the blood of infected mice was approximately 4-fold higher compared to that in pneumococci grown in vitro (104). RegM, a homologue of the staphylococcal catabolite control protein CcpA involved in the regulation of sugar metabolism pathways, was also shown to affect transcription of the cps locus, suggesting that the carbon source may also influence capsular expression (105).
The cps loci are arranged as single transcriptional units in all Wzy-dependent pneumococcal serotypes. The σ70 promoter (cpsp) is highly conserved, and the transcriptional start site is located about 20 to 30 nucleotides upstream of the initiation codon of the first cps gene, cpsA (48). The core promoter incorporating the -10 and -35 sequences is essential for cps transcription, but sequences immediately upstream also impact the level of transcript (106, 107). The upstream region presumably enables fine-tuning of cps transcription, which is necessary for optimal survival of the pneumococcus in diverse host niches (107). Furthermore, sequence polymorphisms in this upstream region result in differences between strains in cps transcript levels that correlate with the virulence profile in vivo (108). A recent study identified a GntR family transcriptional regulator CpsR (cps repressor), which is encoded elsewhere on the chromosome but directly binds to a region of DNA approximately 100 nucleotides upstream of the core promoter and represses cps transcription (109). A cpsR deletion mutant exhibited enhanced CPS production and systemic virulence in mice relative to the wild type but was attenuated in a nasopharyngeal colonization model (109). Interestingly, in vitro binding of CpsR to cpsp was competitively inhibited by Glc, leading to increased total CPS expression (109). This provides a mechanism whereby CPS expression could be increased in vivo when pneumococci enter the bloodstream, where Glc concentrations are maintained at high levels. The competence response regulator ComE has also been shown to be capable of binding to cpsp in its phosphorylated form in vitro, and a comE deletion mutant exhibited increased CPS production (110). Like CpsR, ComE repression of cps transcription was alleviated by Glc, but this effect was mediated by repression of comE transcription rather than by interfering with the binding of ComE to cpsp (110).
Phase Variation and Quorum Sensing
Pneumococci have been shown to undergo bidirectional phase variation between two distinct colonial morphologies, described as “opaque” and “transparent.” The transparent phenotype exhibits increased in vitro adherence to buccal epithelial cells and cytokine-activated A549 cells relative to opaque variants of the same strain, as well as an enhanced capacity to colonize the nasopharynx of infant rats (111, 112). On the other hand, the opaque form is associated with massively increased virulence in animal models of systemic disease, and this correlates with increased production of CPS relative to cell wall teichoic acid compared with the transparent phenotype. Phase variation also correlated with alteration in the levels of several surface proteins (113). A recent study has shown that the underlying mechanism involves a type I restriction-modification system (SpnIII) with a genetic locus containing inverted repeats that enable spontaneous rearrangement of alternative specificity domain genes. This generates six different SpnIII target specificities, each with distinct genome-wide DNA methylation patterns, gene expression profiles, and virulence phenotypes (114). Moreover, pneumococci were shown to readily switch between SpnIII alleles during progression of disease in a murine model. Differentially expressed genes included the CPS biosynthesis locus cps, various sugar transporters, the Mn transporter psaBCA, and luxS (114). The luxS gene is of particular interest because it encodes synthesis of the ubiquitous quorum sensing molecule autoinducer 2 (AI-2), which is known to be an important regulator of biofilm formation and virulence in pneumococci (115). Recent studies showed that AI-2 accumulating in the extracellular compartment is sensed by the pneumococcal fructose-specific phosphotransferase system component FruA, leading to upregulation of the galactose (Gal) ABC transporter and the Leloir pathway, presumably by phosphorylation of the regulator GalR (116). This entire process is repressed in the presence of Glc. However, Gal is an important carbon source for S. pneumoniae in the respiratory tract, where Glc is very scarce, and AI-2-mediated quorum sensing is essential for Gal uptake and metabolism. The Leloir pathway converts Gal to Gal-1-P and then to Glc-1-P, the precursor for all the activated nucleotide sugars required for CPS synthesis. In the absence of Glc, Leloir pathway upregulation increases total CPS production and confers a hypervirulent phenotype in vivo (116). Two other proteins involved in sugar metabolism have been shown to affect CPS production. PGM is the phosphoglucomutase, which catalyzes the conversion of Glc-6-P (derived from imported Glc when it is available) to Glc-1-P, and GalU is a Glc-1-P uridylyltransferase that catalyzes the formation of UDP-Glc from Glc-1-P. Pneumococcal mutants in which either the galU or the pgm genes were disrupted produced almost no CPS and exhibited growth defects (12, 67). Glc-1-P is also a precursor for synthesis of other cellular structures, such as teichoic acid, so limiting the supply of this precursor would be expected to impact heavily on CPS production in the pneumococcus. Indirect modulation of CPS production by controlled availability of precursors or cofactors may be one of the regulatory mechanisms used by the pneumococcus.
CONCLUDING REMARKS
In this article we have summarized the current state of knowledge of the CPS of S. pneumoniae, with particular reference to the genes encoding biosynthesis of this most important of all pneumococcal surface antigens. Notwithstanding the insights gained from the elegant classical genetic studies of CPS production carried out before the early 1970s, research in the past 2 decades has been revolutionized by the advent of modern methods for gene cloning, DNA amplification, and high-throughput sequence analysis. The complete nucleotide sequences of the cps loci of all of the known serotypes are available, and although the functions of many of the individual genes in these loci await confirmation by conventional biochemical and genetic analysis, access to the enormous body of information on sequence databases now available, combined with knowledge of the chemical structures of many of the CPS repeat units, has enabled accurate predictions of function for a significant proportion of these genes. It has also been possible to predict the mechanisms of CPS biosynthesis in pneumococci by analogy with those operating in Gram-negative bacteria. The existence of two distinct mechanisms of CPS biosynthesis in S. pneumoniae has already been recognized. However, much remains to be learned about the precise molecular events involved in both of these processes and about how CPS production is regulated in response to the microenvironmental conditions to which pneumococci are exposed in vivo. Given the importance of capsules to the virulence of S. pneumoniae and several other Gram-positive pathogens, such conserved components of the CPS biosynthesis machinery may prove to be useful targets for novel antimicrobial strategies. Furthermore, the availability of thousands of complete pneumococcal genome sequences in recent years is providing insights into the continuing evolution of diversity of pneumococcal CPS serotypes, particularly in the face of strong immune selection provided by PCVs. A better understanding of these processes will undoubtedly inform ongoing refinement of pneumococcal vaccination strategies.
ACKNOWLEDGMENTS
Research in the authors’ laboratory is supported by program grant 1071659 from the National Health and Medical Research Council of Australia (NHMRC) (to J.C.P.).
J.C.P. is an NHMRC senior principal research fellow; C.T. holds a University of Adelaide Beacon Fellowship.
REFERENCES
- 1.Austrian R. 1981. Pneumococcus: the first one hundred years. Rev Infect Dis 3:183–189 10.1093/clinids/3.2.183. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Austrian R. 1981. Some observations on the pneumococcus and on the current status of pneumococcal disease and its prevention. Rev Infect Dis 3(Suppl):S1–S17 10.1093/clinids/3.Supplement_1.S1. [DOI] [PubMed] [Google Scholar]
- 3.Dochez AR, Avery OT. 1917. The elaboration of specific soluble substance by pneumococcus during growth. J Exp Med 26:477–493 10.1084/jem.26.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avery OT, Heidelberger M. 1925. Immunological relationships of cell constituents of pneumococcus. J Exp Med 42:367–376 10.1084/jem.42.3.367. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery OT, Morgan HJ. 1925. Immunological reactions of the isolated carbohydrate and protein of pneumococcus. J Exp Med 42:347–353 10.1084/jem.42.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skov Sørensen UB, Blom J, Birch-Andersen A, Henrichsen J. 1988. Ultrastructural localization of capsules, cell wall polysaccharide, cell wall proteins, and F antigen in pneumococci. Infect Immun 56:1890–1896. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sørensen UBS, Henrichsen J, Chen HC, Szu SC. 1990. Covalent linkage between the capsular polysaccharide and the cell wall peptidoglycan of Streptococcus pneumoniae revealed by immunochemical methods. Microb Pathog 8:325–334 10.1016/0882-4010(90)90091-4. [DOI] [PubMed] [Google Scholar]
- 8.Geno KA, Saad JS, Nahm MH. 2017. Discovery of novel pneumococcal serotype 35D, a natural WciG-deficient variant of serotype 35B. J Clin Microbiol 55:1416–1425 10.1128/JCM.00054-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avery OT, Dubos R. 1931. The protective action of a specific enzyme against type III pneumococcus infections in mice. J Exp Med 54:73–89 10.1084/jem.54.1.73. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender MH, Yother J. 2001. CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J Biol Chem 276:47966–47974 10.1074/jbc.M105448200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Hardy GG, Magee AD, Ventura CL, Caimano MJ, Yother J. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect Immun 69:2309–2317 10.1128/IAI.69.4.2309-2317.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun 69:3755–3761 10.1128/IAI.69.6.3755-3761.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morona JK, Miller DC, Morona R, Paton JC. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J Infect Dis 189:1905–1913 10.1086/383352. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Marsh R, Smith-Vaughan H, Hare KM, Binks M, Kong F, Warning J, Gilbert GL, Morris P, Leach AJ. 2010. The nonserotypeable pneumococcus: phenotypic dynamics in the era of anticapsular vaccines. J Clin Microbiol 48:831–835 10.1128/JCM.01701-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller LE, Robinson DA, McDaniel LS. 2016. Nonencapsulated Streptococcus pneumoniae: emergence and pathogenesis. MBio 7:e01792 10.1128/mBio.01792-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159 10.1017/S0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158 10.1084/jem.79.2.137. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect Immun 75:83–90 10.1128/IAI.01475-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. 2010. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun 78:704–715 10.1128/IAI.00881-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeyta M, Hardy GG, Yother J. 2003. Genetic alteration of capsule type but not PspA type affects accessibility of surface-bound complement and surface antigens of Streptococcus pneumoniae. Infect Immun 71:218–225 10.1128/IAI.71.1.218-225.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wartha F, Beiter K, Albiger B, Fernebro J, Zychlinsky A, Normark S, Henriques-Normark B. 2007. Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol 9:1162–1171 10.1111/j.1462-5822.2006.00857.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.de Vos AF, Dessing MC, Lammers AJ, de Porto AP, Florquin S, de Boer OJ, de Beer R, Terpstra S, Bootsma HJ, Hermans PW, van ’t Veer C, van der Poll T. 2015. The polysaccharide capsule of Streptococcus pneumonia partially impedes MyD88-mediated immunity during pneumonia in mice. PLoS One 10:e0118181 10.1371/journal.pone.0118181. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLEOD CM, Kraus MR. 1950. Relation of virulence of pneumococcal strains for mice to the quantity of capsular polysaccharide formed in vitro. J Exp Med 92:1–9 10.1084/jem.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyams C, Yuste J, Bax K, Camberlein E, Weiser JN, Brown JS. 2010. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect Immun 78:716–725 10.1128/IAI.01056-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly T, Dillard JP, Yother J. 1994. Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infect Immun 62:1813–1819. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nesin M, Ramirez M, Tomasz A. 1998. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis 177:707–713 10.1086/514242. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Trzciński K, Li Y, Weinberger DM, Thompson CM, Cordy D, Bessolo A, Malley R, Lipsitch M. 2015. Effect of serotype on pneumococcal competition in a mouse colonization model. MBio 6:e00902-15 10.1128/mBio.00902-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zafar MA, Hamaguchi S, Zangari T, Cammer M, Weiser JN. 2017. Capsule type and amount affect shedding and transmission of Streptococcus pneumoniae. MBio 8:e00989-17 10.1128/mBio.00989-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyams C, Trzcinski K, Camberlein E, Weinberger DM, Chimalapati S, Noursadeghi M, Lipsitch M, Brown JS. 2013. Streptococcus pneumoniae capsular serotype invasiveness correlates with the degree of factor H binding and opsonization with C3b/iC3b. Infect Immun 81:354–363 10.1128/IAI.00862-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CJ, Banks SD, Li JP. 1991. Virulence, immunity, and vaccine related to Streptococcus pneumoniae. Crit Rev Microbiol 18:89–114 10.3109/10408419109113510. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Douglas RM, Paton JC, Duncan SJ, Hansman DJ. l983. Antibody response to pneumococcal vaccination in children younger than five years of age. J Infect Dis 48:131–137. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien KL, Millar EV, Zell ER, Bronsdon M, Weatherholtz R, Reid R, Becenti J, Kvamme S, Whitney CG, Santosham M. 2007. Effect of pneumococcal conjugate vaccine on nasopharyngeal colonization among immunized and unimmunized children in a community-randomized trial. J Infect Dis 196:1211–1220 10.1086/521833. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention (CDC). 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease--United States, 1998-2003. MMWR Morb Mortal Wkly Rep 54:893–897. [PubMed] [PubMed] [Google Scholar]
- 35.Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog 3:e168 10.1371/journal.ppat.0030168. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, Jackson D, Thomas A, Beall B, Lynfield R, Reingold A, Farley MM, Whitney CG. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J Infect Dis 196:1346–1354 10.1086/521626. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Klugman KP. 2009. The significance of serotype replacement for pneumococcal disease and antibiotic resistance. Adv Exp Med Biol 634:121–128 10.1007/978-0-387-79838-7_11. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.von Gottberg A, de Gouveia L, Tempia S, Quan V, Meiring S, von Mollendorf C, Madhi SA, Zell ER, Verani JR, O’Brien KL, Whitney CG, Klugman KP, Cohen C, GERMS-SA Investigators. 2014. Effects of vaccination on invasive pneumococcal disease in South Africa. N Engl J Med 371:1889–1899 10.1056/NEJMoa1401914. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Austrian R, Bernheimer HP, Smith EEB, Mills GT. 1959. Simultaneous production of two capsular polysaccharides by pneumococcus. II. The genetic and biochemical bases of binary capsulation. J Exp Med 110:585–602 10.1084/jem.110.4.585. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernheimer HP, Wermundsen IE, Austrian R. 1967. Qualitative differences in the behavior of pneumoncoccal deoxyribonucleic acids transforming to the same capsular type. J Bacteriol 93:320–333. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guidolin A, Morona JK, Morona R, Hansman D, Paton JC. 1994. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun 62:5384–5396. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dillard JP, Vandersea MW, Yother J. 1995. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med 181:973–983 10.1084/jem.181.3.973. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrecubieta C, García E, López R. 1995. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene 167:1–7 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 44.Morona JK, Morona R, Paton JC. 1997. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol 23:751–763 10.1046/j.1365-2958.1997.2551624.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Morona JK, Morona R, Paton JC. 1997. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol 179:4953–4958 10.1128/jb.179.15.4953-4958.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolkman MAB, van der Zeijst BAM, Nuijten PJM. 1997. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem 272:19502–19508 10.1074/jbc.272.31.19502. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Kolkman MAB, Wakarchuk W, Nuijten PJM, van der Zeijst BAM. 1997. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol 26:197–208 10.1046/j.1365-2958.1997.5791940.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Muñoz R, Mollerach M, López R, García E. 1997. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol 25:79–92 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 49.Llull D, López R, García E, Muñoz R. 1998. Molecular structure of the gene cluster responsible for the synthesis of the polysaccharide capsule of Streptococcus pneumoniae type 33F. Biochim Biophys Acta 1443:217–224 10.1016/S0167-4781(98)00213-9. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez M, Tomasz A. 1998. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol 180:5273–5278. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morona JK, Morona R, Paton JC. 1999. Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J Bacteriol 181:3599–3605. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morona JK, Morona R, Paton JC. 1999. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J Bacteriol 181:5355–5364. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morona JK, Miller DC, Coffey TJ, Vindurampulle CJ, Spratt BG, Morona R, Paton JC. 1999. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 23F. Microbiology 145:781–789 10.1099/13500872-145-4-781. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Llull D, Muñoz R, López R, García E. 1999. A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide. Type 37 pneumococci are natural, genetically binary strains. J Exp Med 190:241–251 10.1084/jem.190.2.241. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muñoz R, Mollerach M, López R, García E. 1999. Characterization of the type 8 capsular gene cluster of Streptococcus pneumoniae. J Bacteriol 181:6214–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iannelli F, Pearce BJ, Pozzi G. 1999. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol 181:2652–2654. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang SM, Wang L, Reeves PR. 2001. Molecular characterization of Streptococcus pneumoniae type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect Immun 69:1244–1255 10.1128/IAI.69.3.1244-1255.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Selm S, Kolkman MA, van der Zeijst BA, Zwaagstra KA, Gaastra W, van Putten JP. 2002. Organization and characterization of the capsule biosynthesis locus of Streptococcus pneumoniae serotype 9V. Microbiology 148:1747–1755 10.1099/00221287-148-6-1747. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.van Selm S, van Cann LM, Kolkman MA, van der Zeijst BA, van Putten JP. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect Immun 71:6192–6198 10.1128/IAI.71.11.6192-6198.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31 10.1371/journal.pgen.0020031. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dillard JP, Yother J. 1994. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol Microbiol 12:959–972 10.1111/j.1365-2958.1994.tb01084.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.García E, García P, López R. 1993. Cloning and sequencing of a gene involved in the synthesis of the capsular polysaccharide of Streptococcus pneumoniae type 3. Mol Gen Genet 239:188–195. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Arrecubieta C, López R, García E. 1994. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J Bacteriol 176:6375–6383 10.1128/jb.176.20.6375-6383.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arrecubieta C, López R, García E. 1996. Type 3-specific synthase of Streptococcus pneumoniae (Cap3B) directs type 3 polysaccharide biosynthesis in Escherichia coli and in pneumococcal strains of different serotypes. J Exp Med 184:449–455 10.1084/jem.184.2.449. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeAngelis PL, Papaconstantinou J, Weigel PH. 1993. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem 268:19181–19184. [PubMed] [PubMed] [Google Scholar]
- 66.Keenleyside WJ, Whitfield C. 1996. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem 271:28581–28592 10.1074/jbc.271.45.28581. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Mollerach M, López R, García E. 1998. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J Exp Med 188:2047–2056 10.1084/jem.188.11.2047. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cartee RT, Forsee WT, Jensen JW, Yother J. 2001. Expression of the Streptococcus pneumoniae type 3 synthase in Escherichia coli. Assembly of type 3 polysaccharide on a lipid primer. J Biol Chem 276:48831–48839 10.1074/jbc.M106481200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Llull D, García E, López R. 2001. Tts, a processive beta-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in Pneumococcus and other gram-positive species. J Biol Chem 276:21053–21061 10.1074/jbc.M010287200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Yother J. 2011. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65:563–581 10.1146/annurev.micro.62.081307.162944. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Whitfield C, Paiment A. 2003. Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr Res 338:2491–2502 10.1016/j.carres.2003.08.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Kolkman MAB, Morrison DA, Van Der Zeijst BAM, Nuijten PJM. 1996. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol 178:3736–3741 10.1128/jb.178.13.3736-3741.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larson TR, Yother J. 2017. Streptococcus pneumoniae capsular polysaccharide is linked to peptidoglycan via a direct glycosidic bond to β-d-N-acetylglucosamine. Proc Natl Acad Sci U S A 114:5695–5700 10.1073/pnas.1620431114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eberhardt A, Hoyland CN, Vollmer D, Bisle S, Cleverley RM, Johnsborg O, Håvarstein LS, Lewis RJ, Vollmer W. 2012. Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 18:240–255 10.1089/mdr.2011.0232. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Morona JK, Morona R, Paton JC. 2006. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci U S A 103:8505–8510 10.1073/pnas.0602148103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawrence ER, Griffiths DB, Martin SA, George RC, Hall LM. 2003. Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J Clin Microbiol 41:601–607 10.1128/JCM.41.2.601-607.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mostowy RJ, Croucher NJ, De Maio N, Chewapreecha C, Salter SJ, Turner P, Aanensen DM, Bentley SD, Didelot X, Fraser C. 2017. Pneumococcal capsule synthesis locus cps as evolutionary hotspot with potential to generate novel serotypes by recombination. Mol Biol Evol 34:2537–2554 10.1093/molbev/msx173. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barnes DM, Whittier S, Gilligan PH, Soares S, Tomasz A, Henderson FW. 1995. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis 171:890–896 10.1093/infdis/171.4.890. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Coffey TJ, Dowson CG, Daniels M, Zhou J, Martin C, Spratt BG, Musser JM. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol 5:2255–2260 10.1111/j.1365-2958.1991.tb02155.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Coffey TJ, Enright MC, Daniels M, Wilkinson P, Berrón S, Fenoll A, Spratt BG. 1998. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist 4:51–55 10.1089/mdr.1998.4.51. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Coffey TJ, Enright MC, Daniels M, Morona JK, Morona R, Hryniewicz W, Paton JC, Spratt BG. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol 27:73–83 10.1046/j.1365-2958.1998.00658.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Kroll JS, Loynds BM, Moxon ER. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol 5:1549–1560 10.1111/j.1365-2958.1991.tb00802.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309 10.1038/ng.2895. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaguza C, Andam CP, Harris SR, Cornick JE, Yang M, Bricio-Moreno L, Kamng’ona AW, Parkhill J, French N, Heyderman RS, Kadioglu A, Everett DB, Bentley SD, Hanage WP. 2016. Recombination in Streptococcus pneumoniae lineages increase with carriage duration and size of the polysaccharide capsule. MBio 7:e01053-16 10.1128/mBio.01053-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marks LR, Reddinger RM, Hakansson AP. 2012. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. MBio 3:e00200-12 10.1128/mBio.00200-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skov Sørensen UB, Yao K, Yang Y, Tettelin H, Kilian M. 2016. Capsular polysaccharide expression in commensal Streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. MBio 7:e01844-16 10.1128/mBio.01844-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun 72:5582–5596 10.1128/IAI.72.10.5582-5596.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ogunniyi AD, Mahdi LK, Trappetti C, Verhoeven N, Mermans D, Van der Hoek MB, Plumptre CD, Paton JC. 2012. Identification of genes that contribute to the pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect Immun 80:3268–3278 10.1128/IAI.00295-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Talbot UM, Paton AW, Paton JC. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect Immun 64:3772–3777. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morona JK, Paton JC, Miller DC, Morona R. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol Microbiol 35:1431–1442 10.1046/j.1365-2958.2000.01808.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J Biol Chem 276:139–146 10.1074/jbc.M005702200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Morona JK, Morona R, Miller DC, Paton JC. 2003. Mutational analysis of the carboxy-terminal (YGX)4 repeat domain of CpsD, an autophosphorylating tyrosine kinase required for capsule biosynthesis in Streptococcus pneumoniae. J Bacteriol 185:3009–3019 10.1128/JB.185.10.3009-3019.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bender MH, Cartee RT, Yother J. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J Bacteriol 185:6057–6066 10.1128/JB.185.20.6057-6066.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Glucksmann MA, Reuber TL, Walker GC. 1993. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol 175:7045–7055 10.1128/jb.175.21.7045-7055.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morona R, Van Den Bosch L, Daniels C. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146:1–4 10.1099/00221287-146-1-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Morona JK, Morona R, Miller DC, Paton JC. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J Bacteriol 184:577–583 10.1128/JB.184.2.577-583.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weiser JN, Bae D, Epino H, Gordon SB, Kapoor M, Zenewicz LA, Shchepetov M. 2001. Changes in availability of oxygen accentuate differences in capsular polysaccharide expression by phenotypic variants and clinical isolates of Streptococcus pneumoniae. Infect Immun 69:5430–5439 10.1128/IAI.69.9.5430-5439.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geno KA, Hauser JR, Gupta K, Yother J. 2014. Streptococcus pneumoniae phosphotyrosine phosphatase CpsB and alterations in capsule production resulting from changes in oxygen availability. J Bacteriol 196:1992–2003 10.1128/JB.01545-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Henriques MX, Rodrigues T, Carido M, Ferreira L, Filipe SR. 2011. Synthesis of capsular polysaccharide at the division septum of Streptococcus pneumoniae is dependent on a bacterial tyrosine kinase. Mol Microbiol 82:515–534 10.1111/j.1365-2958.2011.07828.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Nourikyan J, Kjos M, Mercy C, Cluzel C, Morlot C, Noirot-Gros MF, Guiral S, Lavergne JP, Veening JW, Grangeasse C. 2015. Autophosphorylation of the bacterial tyrosine-kinase CpsD connects capsule synthesis with the cell cycle in Streptococcus pneumoniae. PLoS Genet 11:e1005518 10.1371/journal.pgen.1005518. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Standish AJ, Whittall JJ, Morona R. 2014. Tyrosine phosphorylation enhances activity of pneumococcal autolysin LytA. Microbiology 160:2745–2754 10.1099/mic.0.080747-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Müller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun 73:4653–4667 10.1128/IAI.73.8.4653-4667.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kietzman CC, Gao G, Mann B, Myers L, Tuomanen EI. 2016. Dynamic capsule restructuring by the main pneumococcal autolysin LytA in response to the epithelium. Nat Commun 7:10859 10.1038/ncomms10859. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ogunniyi AD, Giammarinaro P, Paton JC. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045–2053 10.1099/00221287-148-7-2045. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Giammarinaro P, Paton JC. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect Immun 70:5454–5461 10.1128/IAI.70.10.5454-5461.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wen Z, Sertil O, Cheng Y, Zhang S, Liu X, Wang WC, Zhang JR. 2015. Sequence elements upstream of the core promoter are necessary for full transcription of the capsule gene operon in Streptococcus pneumoniae strain D39. Infect Immun 83:1957–1972 10.1128/IAI.02944-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shainheit MG, Mulé M, Camilli A. 2014. The core promoter of the capsule operon of Streptococcus pneumoniae is necessary for colonization and invasive disease. Infect Immun 82:694–705 10.1128/IAI.01289-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wen Z, Liu Y, Qu F, Zhang JR. 2016. Allelic variation of the capsule promoter diversifies encapsulation and virulence in Streptococcus pneumoniae. Sci Rep 6:30176 10.1038/srep30176. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu K, Xu H, Zheng Y, Wang L, Zhang X, Yin Y. 2016. CpsR, a GntR family regulator, transcriptionally regulates capsular polysaccharide biosynthesis and governs bacterial virulence in Streptococcus pneumoniae. Sci Rep 6:29255 10.1038/srep29255. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng Y, Zhang X, Wang X, Wang L, Zhang J, Yin Y. 2017. ComE, an essential response regulator, negatively regulates the expression of the capsular polysaccharide locus and attenuates the bacterial virulence in Streptococcus pneumoniae. Front Microbiol 8:277 10.3389/fmicb.2017.00277. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62:2582–2589. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cundell DR, Weiser JN, Shen J, Young A, Tuomanen EI. 1995. Relationship between colonial morphology and adherence of Streptococcus pneumoniae. Infect Immun 63:757–761. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis 177:368–377 10.1086/514205. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055 10.1038/ncomms6055. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trappetti C, Potter AJ, Paton AW, Oggioni MR, Paton JC. 2011. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect Immun 79:4550–4558 10.1128/IAI.05644-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trappetti C, McAllister LJ, Chen A, Wang H, Paton AW, Oggioni MR, McDevitt CA, Paton JC. 2017. Autoinducer 2 signaling via the phosphotransferase FruA drives galactose utilization by Streptococcus pneumoniae, resulting in hypervirulence. MBio 8:e02269-16 10.1128/mBio.02269-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


