Abstract
Despite important advances in understanding the molecular basis of cancer, few treatments have been devised which rationally target known causal oncogenic defects. Selectively replicating viruses have a major advantage over nonreplicating viruses to target these defects because the therapeutic effect of the injected virus is augmented by virus produced within the tumor. To permit rational targeting of colon tumors, we have developed replicating adenoviruses that express the viral E1B and E2 genes from promoters controlled by the Tcf4 transcription factor. Tcf4 is constitutively activated by mutations in the adenomatous polyposis coli and β-catenin genes in virtually all colon tumors and is constitutively repressed by Groucho and CtBP in normal tissue. The Tcf-E2 and Tcf-E1B promoters are active in many, but not all, cell lines with activation of the wnt pathway. Viruses with Tcf regulation of E2 expression replicate normally in SW480 colon cancer cells but show a 50- to 100-fold decrease in replication in H1299 lung cancer cells and WI38 normal fibroblasts. Activation of wnt signaling by transduction of a stable β-catenin mutant into normal fibroblasts renders the cells permissive for virus replication. Insertion of Tcf4 sites in the E1B promoter has only small effects on replication in vitro but significantly reduces the inflammatory response in a rodent lung model in vivo. Replicating adenoviruses with Tcf regulation of both E1B and E2 transcription are potentially useful for the treatment of liver metastases from colorectal tumors, but additional changes will be required to produce a virus that can be used to treat all colon tumors.
Two strategies have been pursued to develop replicating adenoviruses that target tumor cells. The complementary defects approach, where cellular defects complement viral defects, requires a detailed understanding of the function of both the viral genes and the cellular pathways defective in cancer. The original virus of this type, dl1520, has a deletion of the E1B 55K gene. Since E1B 55K binds to and inhibits the p53 protein (33, 42), it was proposed that E1B 55K-deficient viruses would only replicate in p53 mutant cells (3). Subsequent analysis in a wider range of cells has found no clear correlation between p53 status and permissivity for dl1520 (14, 15, 17, 32). This may partly be explained by the loss of p53 function in cells with ostensibly wild-type p53, for example, through mutation of the p14-ARF gene (29). A more complex explanation is that E1B 55K has functions unrelated to p53. For the virus to replicate normally in p53-deficient tumor cells, all of the viral defects must be complemented by p53 loss. Since E1B 55K is required for late viral mRNA export (22), it is not surprising that p53 loss fails to restore viral replication in many cells, because p53 has no known role in viral mRNA export. A related tumor targeting strategy based on complementation of E1A defects by retinoblastoma pathway defects has recently been described (9, 19).
An alternative solution, which is less dependent on a complete understanding of virus biology, is to use transcriptional defects in tumors to regulate the expression of early viral proteins. Transcriptional defects are common in tumors and include activation of α-fetoprotein and prostate-specific antigen expression (16, 30, 43). Both the α-fetoprotein and prostate-specific antigen promoters have been used to regulate E1A expression in replicating adenoviruses, resulting in a 100-fold selectivity of virus replication for cells with the relevant defects (16, 30, 43). Many other genes are known from microarray and serial analysis of gene expression studies to be overexpressed in cancer (31, 37). In some cases, such as overexpression of E2F target genes, the link between mRNA overexpression and the underlying oncogenic defect is understood, but in most cases the basis for mRNA overexpression is unknown. In many cases the defect appears to be linked to cell type rather than transformation per se. Tissue targeting is acceptable provided the tissue of origin is nonessential, as is the case in breast and prostate cancer, but for tumors like those of the lung, brain, and colon, destruction of normal tissue would pose major problems.
To produce a replicating adenovirus targeting a known causal oncogenic defect, we have taken advantage of the constitutive activation of the wnt signaling pathway invariably seen in colon cancer (27). Activation of the pathway results mainly from mutations in the adenomatous polyposis coli (APC) and β-catenin genes, although mutations have also been described in the axin gene in hepatocellular carcinoma (27). In the absence of wnt signals, the APC-axin-GSK3β complex phosphorylates the amino terminus of β-catenin, resulting in proteasome-mediated β-catenin degradation. In response to wnt signals, GSK3β is inhibited and β-catenin is stabilized. β-Catenin then enters the nucleus, binds to Tcf/Lef family transcription factors, and activates transcription of wnt target genes, such as cyclin D, myc, and PPARδ (27). Activation of transcription reporters with Tcf binding sites is thus a constant feature of colon cancer cell lines (21).
To exploit the wnt signaling defect in colon tumors we have inserted Tcf binding sites in the E2 promoter of adenovirus type 5 (Ad5). To eliminate confounding regulation of E2 transcription by E1A, E4 orf6/7, and extraneous cellular transcription factors, we deleted the normal E2 control elements. The E2 transcription unit encodes the viral DNA polymerase, preterminal protein, and DNA binding protein (DBP). We chose to regulate E2 rather than other early genes because mutations elsewhere in the virus or cell cannot bypass the absolute requirement for E2 gene products in virus replication. To achieve tight E2 regulation we found it necessary to mutate the adjacent E3 promoter. Since E3 encodes immune-suppressant molecules (39), reduced E3 transcription could potentially augment undesirable inflammatory responses. In contrast, mutation of the E1B 55K gene decreases the inflammatory response, an effect independent of the reduction in viral replication caused by E1B mutation (12). To offset the potential increase in inflammatory response resulting from mutation of the E3 promoter, we have constructed viruses with combined transcriptional regulation of both the E2 and E1B promoters. These viruses replicate selectively in cells with activation of the wnt signaling pathway and provoke less inflammation than wild-type adenovirus.
MATERIALS AND METHODS
Adenovirus mutagenesis.
The adenovirus genome was modified by gap repair and two-step gene replacement as described by Gagnebin et al. (11). The wild-type Ad5 YAC/BAC (pMB20) was constructed by gap repair of pMB19 (11) with DNA prepared from Ad5 obtained from the American Type Culture Collection (ATCC) (VR5). The E1B, E2, and E3 promoter mutations were introduced sequentially in pMB20 using gene replacement vectors derived from pRS406 by selection for and against URA3.
An Ad5 E2/E3 fragment (nucleotides [nt] 26688 to 27593) was amplified by PCR from VR5 DNA with primers TGCATTGGTACCGTCATCTCTA and GTTGCTCTGCCTCTCCACTT, cut with KpnI/SacI, and cloned into the KpnI/SacI sites in pRS406 to give pMB32. Four Tcf sites were inserted in the E2 promoter, and the normal sites were simultaneously deleted by inverse PCR with primers cAGATCAAAGGGattaAGATCAAAGGGccattatgagcaag and gatCCCTTTGATCTccaaCCCTTTGATCTagtccttaagagtc to give pMB69 (the Tcf sites in the primers are shown in capitals). The final sequence of the mutant region is gac tag ATCAAAGGGTTGGAGATCAAAGGGATCCAGATCAAAGGGATTA AGATCAAAGG gcc att atg, where the Ad5 sequence is in lowercase (the 33K stop codon and pVIII start codon are italicized).
The E3 mutations were introduced by two rounds of inverse PCR in pMB69. PCR with primers CTGCGCCCCGCTATTGGTCATCTGAACTTCGGCCTG and CTTGCGGGCGGCTTTAGACACAGGGTGCGGTC gave pMB46. PCR from pMB46 with primers AGCTGGGCTCTCTTGGTACACCAGTGCAGCGGGCCAACTA and CCCACCACTGTAGTGCTGCCAAGAGACGCCCAGGCCGAAGTT gave pMB49, which contains four Tcf sites in E2 and all of the desired mutations in E3. To facilitate gap repair, a 3′ extension was added as follows. An Ad5 VR5 PCR fragment (nt 27331 to 27689; primers ATGGCACAAACTCCTCAATAA and CCAAGACTACTCAACCCGAATA) was cut with EcoRI/PstI and cloned into the EcoRI/PstI sites in Bluescript to give pMB58. The EcoRI/PstI fragment from pMB58 was inserted into the EcoRI/SacI sites in pMB49 to give pMB63, the integrating vector with E2-Tcf mutations and a wild-type E3 region. The SacI/KpnI fragment of pMB49 was cloned into the SacI/KpnI sites in pMB63 to give pMB66, the integrating vector with E2-Tcf and E3 mutations. The final sequence of the mutant region is GCa CTG GTG TAC CAa GAg AGc CCa GCT CCC ACC ACT GTa GTg CTg CCa AGA GAC GCC CAG GCC GAA GTT CAG ATG ACc AAt agc GGG GCG CAG CTT GCG GGC GGC TTT aGa CAC, where the mutations are in lowercase. The E3 mutant retaining a wild-type E3 ATF site (the above sequence ending TTT CGT CAC) was obtained by two-step gene replacement with pMB66 because the ATF site is closest to the site of integration.
The SmaI Ad5 fragment (nt 1007 to 3940) containing the E1 region was cloned from VR5 DNA into Bluescript to give pMB22. Four Tcf sites were inserted in the E1B promoter and the Sp1 site was simultaneously deleted by inverse PCR from pMB22 with primers tCCCTTTGATCTccaaCCCTTTGATCT agtcctatataatgcgccgtg and tccAGATCAAAGGGattaAGATCAAAGGG atttaacacgccatgcaa to give pRDI-238. The pRDI-238 EcoRI/SacI fragment was then cloned into the EcoRI/SacI sites in pRS406 to give pRDI-239. The E1B-containing 2-kb SacI fragment from pMB22 was cloned into the SacI site in pRDI-239 to give pRDI-241, the E1B-Tcf integrating vector.
vMB12-14 viruses were made by transfection of PacI-digested YAC/BAC DNA into C7 cells (1). The E1B mutant viruses were made by transfection of PacI-digested DNA into 293 cells (ATCC CRL 1573) containing a plasmid expressing an amino-terminally truncated β-catenin mutant (36). The viruses were then plaque purified on SW480 cells, expanded on SW480, purified by CsCl banding, buffer exchanged using NAP25 columns into 1 M NaCl, 100 mM Tris-HCl (pH 8.0), and 10% glycerol, and stored frozen at −70°C. The identity of each batch was checked by restriction digestion and automated fluorescent sequencing on a Licor 4200L sequencer in the E1B (nt 1300 to 2300) and E2/E3 (nt 26700 to 27950) regions using primers IR 190 (E1B sense, TGT CTG AAC CTG AGC CTG AG), IR110 (E2/E3 sense, CAT CTC TAC AGC CCA TAC), and IF171 (E2/E3 antisense, AGT TGC TCT GCC TCT CCA C). Apart from the desired mutations, no differences were found between the sequences of VR5 and the Tcf viruses. Particle counts were based on the optical density at 260 nm (OD260) of virus in 0.1% sodium dodecyl sulfate, using the formula 1 OD260 = 1012 particles/ml.
Cell lines.
ISREC-01 (5), EB (4), SW480 (ATCC CCL-228), SW1116 (ATCC CCL-223), and LS513 (ATCC CRL-2134) were supplied by B. Sordat. H1299 cells were supplied by C. Prives (6). C7 cells were supplied by J. Chamberlain (1). HCT116 (ATCC CCL-247), LS174T (ATCC CL-188), HepG2 (ATCC HB-8065), HT29 (ATCC HTB-38), U2OS (ATCC HTB-96), WI38 (ATCC CCL-75), 293 (CRL-1573), and 293T were supplied by ATCC. HeLa (CCL-2) cells were supplied by Imperial Cancer Research Fund. To activate the wnt signaling pathway, WI38 cells were infected with vesicular stomatitis virus-G pseudotyped lentiviruses and selected for 2 days in puromycin, using virus prepared by transient transfection of 293T cells with gene transfer vector and packaging vectors pMD.G and pCMV ΔR8.91 as described previously (24). The self-inactivating lentiviral gene transfer vector is derived from pHR′ (24) and contains the SV40-puro cassette from pBabe-puro and myc-tagged ΔN-β-catenin (36) expressed from the cytomegalovirus promoter.
Luciferase assays.
The E2 luciferase reporters were constructed by cloning into pGL3-Basic an Eco47III/SacI fragment from pMB32 (wild-type Ad5 nt 26841 to 27594, E2-E3 promoter regions) and derivatives with the E2 and E3 mutations described above. SW480 cells were seeded at 2 × 105 cells per 35-mm well 24 h before transfection. Cells were lipofected (Life Technologies) for 18 h with 100 ng of reporter plasmid, 5 ng of control Renilla luciferase plasmid (Promega, Madison, Wis.), and 500 ng of pcDNA3 expressing E1A. Cells were harvested 48 h later and dual luciferase reporter assays were performed according to the manufacturer's instructions (Promega) using a Biocounter (Lumac bv, Landgraaf, The Netherlands). Each value is the mean of three independent experiments and transfection efficiency is normalized to the activity of the Renilla control.
Western blotting.
Cells were infected with either 10,000 (WI38) or 1,000 (other cell lines) viral particles per cell. Two hours after infection, the medium was replaced. Cells were harvested 24 h later. E1B 55K and DBP were detected with 2A6 (34) and B6 (28) monoclonal antibodies, respectively. The myc-tagged β-catenin was detected with 9E10 monoclonal antibody.
Quantitative PCR assays.
Cells were infected with either 300 (SW480 and H1299) or 1,000 (W138) viral particles per cell. Two hours after infection, the medium was replaced. Hydroxyurea (10 mM) was added to cultures destined for RNA extraction. Twenty-four hours later, RNA and DNA extractions were performed with RNeasy and DNeasy kits (Qiagen) according to the manufacturer's instructions. Reverse transcription (RT) was performed using 1 μg of total RNA and Moloney murine leukemia virus Superscript Core Reagents (LifeTechnologies) in a 20-μl reaction volume. TaqMan PCRs were performed using a TaqMan Universal PCR Master Mix kit (Perkin-Elmer), a 900 nM concentration of primers (Microsynth and Eurogentec), and 500 nM TaqMan probe (Eurogentec). Five microliters of RT reaction product was used for RT-PCR. Sybr green PCRs were performed using the Sybr green Universal PCR Master Mix kit (Perkin-Elmer) and a 900 nM concentration of fiber gene primers (Eurogentec). Results for DNA were normalized to the OD260, and results for RNA were normalized to ribosomal RNA (Ribosomal RNA Control; Perkin-Elmer). The primers and probes used for quantitative PCR were as follows: E2 early forward primer, TTCGCTTTTGTGATACAGGCA; E2 early reverse primer, GTCTTGGACGCGACGAGAAG; E2 probe, CGGAGCGTTTGCCGCGC; E3 forward primer, AGCTCGGAGAGGTTCTCTCGTAG; E3 reverse primer, AACACCTGGTCCACTGTCGC; E3 probe, CCGCGACTCCGTTTCAACCCAGA; E1B-55K forward primer, TGCTTCCATCAAACGAGTTGG; E1B-55K reverse primer, GCGCTGAGTTTGGCTCTAGC; E1B-55K probe, CGGCGGCTGCTCAATCTGTATCTTCA; fiber forward primer, TGATGTTTGACGCTACAGCCATA; and fiber reverse primer, GGGATTTGTGTTTGGTGCATTAG.
Virus replication assay.
Cells in six-well plates were infected with either 300 (SW480 and H1299) or 1,000 (W138) viral particles per cell. Two hours after infection, the medium was replaced. Cells were harvested 48 h later and lysed by three cycles of freeze-thawing. The supernatant was tested for virus production by counting plaques formed on SW480 cells after 10 days under 0.9% agarose in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Each bar in the figures represents the mean ± standard deviation of duplicate infections tested in triplicate plaque assays.
Cotton rats.
Animal experiments were performed in accordance with local legislation (cantonal authorization number VD1276). Cotton rats were infected intranasally with 3 × 1010 particles of each virus in 50 μl of buffer. Three days later the animals were killed with CO2-isoflurane, and four pieces of lung were taken from each animal. Each sample was divided in two. DNA was extracted from one part and viral DNA content was measured by Sybr green quantitative PCR using fiber primers as described above. The remainder of the sample was fixed in neutral buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. The severity of infection was scored using the following scale: 0, normal histology; 1, mild changes present in <50% of the sample; 2, mild changes in >50% or severe changes in <50%; 3, severe changes in >50% but <100%; 4, severe changes throughout the lung. Mean scores (four samples per animal) are given for infection of five animals per virus, normalized so that wild-type virus gives a score of 1.
RESULTS
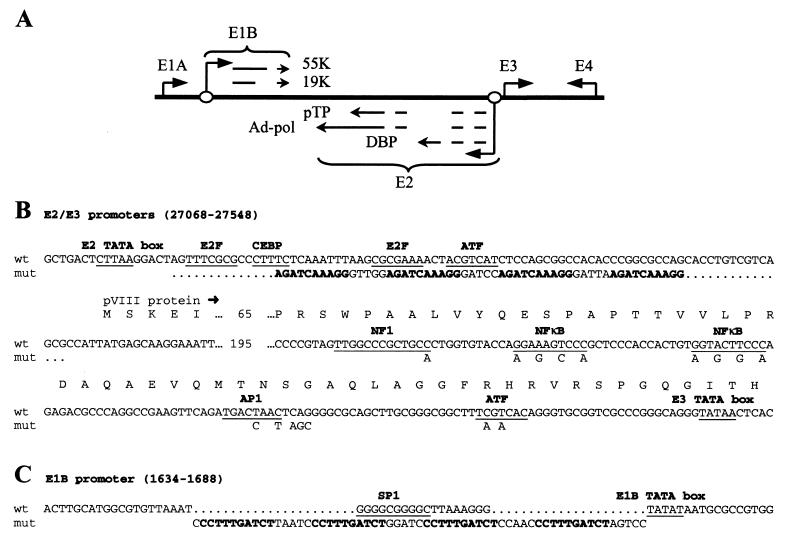
To produce a virus in which E2 expression would respond selectively to activation of the wnt signaling pathway, we removed all of the existing transcription factor binding sites in the E2 promoter and replaced them with multiple copies of a known binding site for Tcf4 (Fig. 1A and B) (21). Since the E2 and E3 promoters are contiguous in the adenovirus genome (Fig. 1A), we suspected that E3 activity might interfere with tight regulation of the E2 promoter. To test the potential for cross talk between the two promoters, we performed transcription assays with luciferase reporters containing the full E2 and E3 promoter region. To inactivate the E3 promoter, mutations were introduced in the E3 NF1, NFκB, AP1, and ATF sites (Fig. 1B). The luciferase gene was inserted in the E2 5′ untranslated region. SW480 colon carcinoma cells were tested because they contain a truncated APC gene, resulting in strong activation of the wnt signaling pathway. The E3 mutations had no effect on the luciferase activity of the wild-type E2 promoter construct but markedly reduced transcription from the Tcf-E2 constructs (Fig. 2). This demonstrates that the E3 enhancer can transactivate the mutant Tcf-E2 promoter. Luciferase activity increased progressively as the number of Tcf binding sites was increased, with four Tcf sites giving near wild-type levels of E2 activity (Fig. 2). We therefore constructed a set of viruses with four Tcf sites in the E2 promoter and wild-type or mutant E3 promoters (Table 1). In addition to the virus with all of the E3 mutations shown in Fig. 1B (vMB14), we constructed an E3 mutant virus retaining the wild-type E3 ATF binding site (vMB13).
FIG. 1.
(A) Schematic diagram showing the positions of the adenovirus early promoters. Open circles represent Tcf binding sites. (B and C) Annotated sequence of the E2/E3 and E1B promoter regions. Changes in Tcf viruses are shown below the wild-type sequence. Gaps are indicated by dots in the sequence (the Tcf insert is not the same length as the deleted wild-type sequence). The Tcf consensus sites are shown in bold. The E3 mutations do not change the encoded pVIII protein sequence (note that to meet this requirement the NF1 site mutation affects only a poorly conserved residue and may not influence NF1 binding).
FIG. 2.
Luciferase assays in SW480 colon tumor cells showing that the E3 promoter transactivates the Tcf-E2 promoter. + E3, wild-type E3 promoter; − E3, mutant E3 promoter; wt E2, wild-type E2 promoter; 2, 3, or 4 Tcf E2, E2 promoter containing 2, 3, or 4 Tcf sites. The assays were performed in the presence of cotransfected E1A.
TABLE 1.
Mutant adenoviruses used in this studyd
| Virus | E1B promoter | E2 promoter | E3 promoter |
|---|---|---|---|
| vMB12 | wt | Tcfa | wt |
| vMB13 | wt | Tcf | mut + ATFb |
| vMB14 | wt | Tcf | mut − ATFc |
| vMB23 | Tcf | wt | wt |
| vMB27 | Tcf | Tcf | wt |
| vMB31 | Tcf | Tcf | mut + ATFb |
| vMB19 | Tcf | Tcf | mut − ATFc |
Replacement of normal binding sites by four Tcf binding sites.
Mutation of the NF1, NFκB, and AP1 sites in the E3 promoter.
Mutation of the NF1, NFκB, AP1, and ATF sites in the E3 promoter.
wt, wild type; mut, mutant.
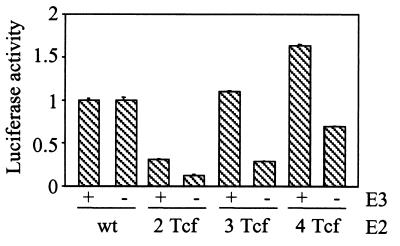
SW480 colon carcinoma cells, H1299 lung carcinoma cells, and WI38 normal fibroblasts were infected with the new viruses, and E2 promoter activity was assessed by Western blotting for DBP. H1299 and WI38 were used as negative controls because the wnt pathway is inactive in these cells. Compared to wild-type adenovirus, DBP expression from the Tcf-E2 viruses was normal in SW480 cells but reduced in H1299 and WI38 cells (Fig. 3A). This is the expected result if the mutant E2 promoter is responsive to activation of the wnt pathway. To determine more exactly the effect of the E3 mutations, the activity of both the E2 and E3 promoters was tested by quantitative RT-PCR using Taqman probes spanning splice sites in the E2 and E3 mRNAs (Fig. 3B). To avoid changes in viral copy number the experiments were performed in the presence of hydroxyurea to block virus replication. All three Tcf-E2 viruses showed wild-type levels of E2 and E3 transcription in SW480 cells (Fig. 3C). As expected, the Tcf viruses showed reduced E2 activity in H1299 cells, in which the wnt pathway is inactive, and the E3 ATF site contributed to both E2 and E3 activation in these cells (compare vMB13 with vMB14). To determine whether the difference in E2 transcription translates into an effect on virus replication, viral DNA content was measured 24 h after infection in the absence of hydroxyurea (Fig. 3C). As for transcription, there was a 100-fold decrease in DNA replication of the most attenuated virus (vMB14) in nonpermissive cells. Finally, to confirm that the difference in DNA replication translates into a difference in virus production, burst assays were performed (Fig. 3D). Compared to SW480, the amount of vMB14 virus produced by normal fibroblasts was reduced over 100-fold. We conclude that tight regulation of E2 transcription by insertion of Tcf sites in the E2 promoter and mutation of the E3 promoter permits normal virus replication in SW480 but significantly impairs replication in H1299 and normal fibroblasts.
FIG. 3.
Activity of viruses with Tcf sites in the E2 promoter. (A) Western blot showing DBP expression 24 h after infection of SW480, H1299, and WI38 with the indicated viruses. (B) E2 and E3 exon structure showing the position of the RT-PCR primers and Taqman probes. (C) PCR quantitation of adenoviral E2 and E3 mRNA by Taqman assay (upper two panels) and adenoviral genomic DNA by Sybr green assay (lower panel) 24 h after infection of SW480 and H1299 with the indicated viruses. (D) Burst assay for virus production by SW480, H1299, and WI38 48 h after infection with the indicated viruses. The viral titer was measured by plaque assay on SW480, which is permissive for all the viruses.
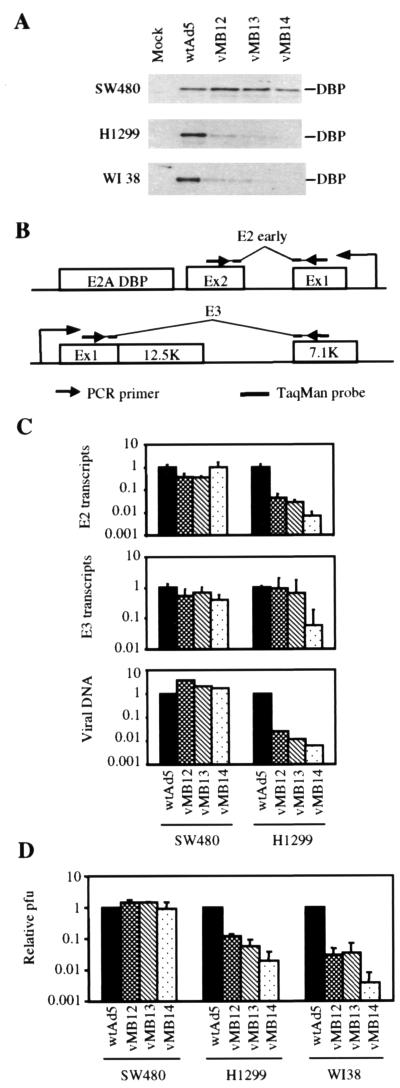
Preliminary experiments showed that deletion of E1B 55K in combination with Tcf regulation of E2 produced a virus that was Tcf specific but severely attenuated in some cell lines, with active wnt signaling and mutant p53 (data not shown). To avoid the adverse effects of E1B 55K deletion in target cells while retaining the beneficial effects of E1B deletion in normal cells, we attempted to regulate E1B transcription by insertion of Tcf sites in the E1B promoter. To determine whether this could increase the selectivity of the Tcf-E2 viruses described above, a set of viruses with Tcf sites in the E1B and E2 promoters was constructed (Fig. 1A and C; Table 1). The Tcf sites replace the Sp1 site in the E1B promoter, which is the only major regulatory site other than the TATA box (40). In addition to the reduction in DBP expression seen with Tcf-E2 regulation alone, the Tcf-E1B viruses showed strongly reduced E1B 55K expression in H1299 and W138 cells by Western blotting (Fig. 4A). Despite the large differences seen by Western blotting, quantitative RT-PCR using a Taqman probe spanning a splice site in the E1B mRNA (Fig. 4B) showed that the reduction in E1B expression at the mRNA level was only 10-fold (Fig. 4C). Taqman assays for E2 expression and viral DNA replication showed only slight improvements in selectivity relative to the parental Tcf-E2 viruses (Fig. 4C). Transcription assays were performed in the presence of hydroxyurea and replication assays were done in the absence of hydroxyurea. Loss of E1B 55K function is known to have much larger effects on virus production than on viral DNA replication, but examination of virus production by burst assay showed that the viruses with combined Tcf-E1B and Tcf-E2 regulation suffered similar reductions in virus production in lung cells and normal fibroblasts as did the parental Tcf-E2 viruses (Fig. 4D). We conclude that replacement of the Sp1 site with four Tcf sites in the E1B promoter leads to a reduction in E1B expression that is too small to significantly restrict viral replication in vitro.
FIG. 4.
Activity of viruses with Tcf sites in the E1B and E2 promoters. (A) Western blot showing DBP and E1B 55K expression 24 h after infection of SW480, H1299, and WI38 with the indicated viruses. (B) E1B exon structure showing the position of the RT-PCR primers and Taqman probe. (C) PCR quantitation of adenoviral E1B and E2 mRNA (upper two panels) and adenoviral genomic DNA (lower panel) 24 h after infection of SW480 and H1299 with the indicated viruses. (D) Burst assay for virus production by SW480, H1299, and WI38 48 h after infection with the indicated viruses. The viral titer was measured by plaque assay on SW480.
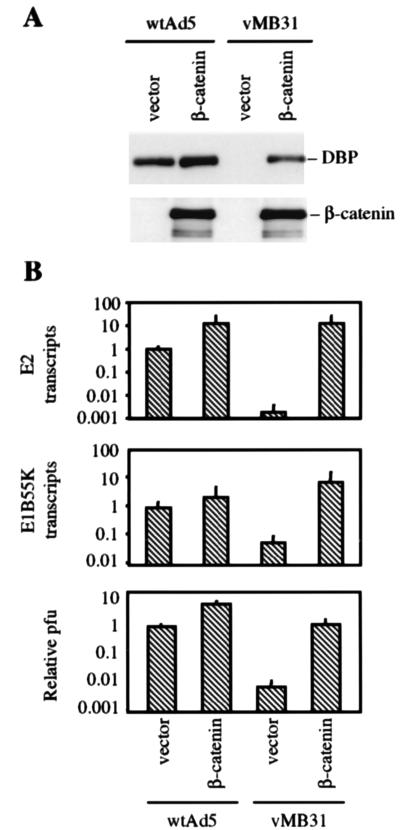
To prove that the difference in virus replication in SW480, H1299, and W138 cells was due to the difference in wnt pathway activity and not some other difference between the cell lines, the wnt signaling pathway was artificially activated in W138 cells by infection with a lentivirus expressing a stable β-catenin mutant. Lentivirus-infected cells were then superinfected with wild-type adenovirus or a Tcf-E2/Tcf-E1B virus (vMB31). Western blotting showed that DBP protein expression was induced in vMB31-infected cells by transduction of the β-catenin mutant (Fig. 5A). Activation of E1B and E2 expression by β-catenin was confirmed by quantitative RT-PCR for E1B and DBP mRNA (Fig. 5B, upper panels). Finally, burst assays showed that activation of the wnt pathway by the β-catenin mutant resulted in a 100-fold increase in vMB31 virus production (Fig. 5B, lower panel). In each case there was a small increase in wild-type virus activity, possibly due to the general transforming effect of the oncogenic β-catenin mutant. We conclude that the Tcf-E1B and Tcf-E2 promoters in the mutant viruses are able to respond selectively to artificial activation of the wnt signaling pathway and that additional genetic alterations are not required.
FIG. 5.
Activation of the wnt signaling pathway renders normal fibroblasts permissive for Tcf virus replication. WI38 cells were first infected with empty vector or Δ-N β-catenin-expressing lentiviruses and then infected with adenoviruses. (A) Western blot showing DBP expression 24 h after infection with wild-type Ad5 and vMB31. (B) PCR quantitation of adenoviral E1B and E2 mRNA 24 h after infection with wild-type Ad5 and vMB31 (upper two panels), and burst assay for virus production 48 h after infection with wild-type Ad5 and vMB31 (lower panel). The viral titer was measured by plaque assay on SW480.
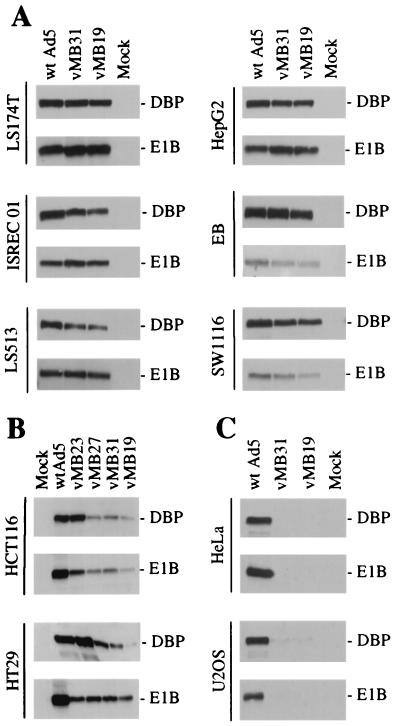
To determine whether the Tcf-E1B and Tcf-E2 promoters are active in all cells with mutations in the wnt signaling pathway, we tested a panel of cell lines by Western blotting for E1B and DBP (Fig. 6). As expected, relative to wild-type virus, the Tcf viruses gave substantially reduced E1B and DBP expression in control cell lines in which the wnt pathway is inactive (HeLa and U2OS; Fig. 6C). In LS174T, ISREC01, LS513, EB, and SW1116 colon cancer cells and HepG2 hepatocellular carcinoma cells, in which the wnt pathway is active, E1B and DBP were expressed at near-wild-type levels (Fig. 6A). In contrast, both proteins were poorly expressed in HCT116 and HT29 colon cancer cells (Fig. 6B). Poor expression of both E1B 55K and DBP suggests that the problem is at the level of Tcf activation rather than differences in the E1B and E2 basal promoters or upstream sites. In conclusion, the Tcf viruses are likely to be effective in many but not all tumors with oncogenic activation of the wnt signaling pathway. Possible reasons for the poor expression of E1B and DBP from the Tcf viruses in some colon tumor cells are discussed below.
FIG. 6.
Western blots for DBP and E1B 55K 24 h after infection of cell lines with wild-type Ad5 and Tcf viruses. (A) Permissive cell lines (HepG2 is a hepatocellular carcinoma cell line, and the rest are colon cancer cell lines). (B) Nonpermissive colon cancer cell lines (vMB23 expresses DBP because it has a wild-type E2 promoter). (C) Cell lines in which the wnt pathway is inactive.
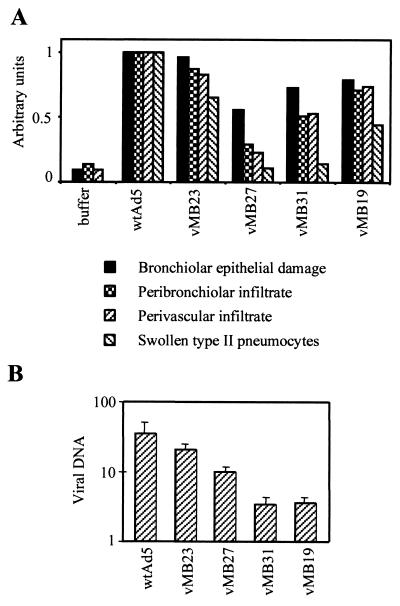
Inflammatory reactions are a source of concern in gene therapy protocols using adenoviruses. Despite the modest effects of Tcf-E1B regulation in vitro, we considered the possibility that the reduction in E1B 55K expression in normal tissue might still have a useful effect in vivo, because E1B mutation is known to reduce the inflammatory reaction to adenoviruses in rodent lung models (12). To test this, we performed intranasal infections of cotton rats, which are permissive for human adenovirus replication. Inflammatory response was scored on a semiquantitative histological scale normalized to the effect of wild-type virus. Each virus was tested on five animals, which were sacrificed 3 days after infection with 3 × 1010 particles of virus. This is approximately 10-fold less than the 50% lethal dose for wild-type virus (13). The viruses with combined Tcf-E1B and Tcf-E2 regulation provoked substantially less inflammatory reaction than wild-type virus (Fig. 7A). The strongest reduction was seen with vMB27, which has combined Tcf-E1B and Tcf-E2 regulation but a normal E3 promoter. The progressive increase in inflammatory response with vMB31 and vMB19, relative to vMB27, is expected because the E3 region encodes immune-suppressant proteins (39) whose expression should be progressively reduced by attenuation of the E3 promoter in vMB31 and, particularly, vMB19. The increase in inflammatory response with vMB31 and vMB19 was accompanied by a decrease in the amount of viral DNA that could be detected by quantitative PCR (Fig. 7B), suggesting that the E3 promoter changes reduce viral replication in normal tissue in vivo.
FIG. 7.
Cotton rat lungs were tested 3 days after intranasal instillation of the indicated adenoviruses. (A) Histological assessment of bronchiolar epithelial damage, peribronchiolar inflammatory infiltrate, perivascular inflammatory infiltrate, and the presence of swollen type II pneumocytes. (B) Viral DNA content measured by quantitative PCR.
DISCUSSION
We have developed selectively replicating adenoviruses capable of targeting cells with activation of the wnt pathway. These viruses could potentially be used for treatment of liver metastases from colorectal tumors. Since the Tcf-E2 and Tcf-E1B promoters are not activated to wild-type levels in some colon cancer cell lines (Fig. 6B), the Tcf viruses are likely to replicate poorly in some colon tumors in vivo, despite the near universality of wnt pathway activation in colon cancer. The reason for the failure of activation of the viral Tcf promoters in some colon cell lines is unknown. Globally reduced activation of the wnt pathway is one possibility. If that is the explanation, it should be possible to develop a clinical test to identify susceptible tumors, perhaps based on microarray analysis or immunohistochemical staining for overexpression of β-catenin or wnt target genes. Such a test would also permit use of the viruses to treat tumor types in which wnt pathway activation is less frequent than in colon cancer. A more specific explanation is that E1A may inhibit E2 promoter transactivation by β-catenin–Tcf4. Sequestration of the histone acetyltransferase p300 by E1A is a plausible mechanism for this, because p300 is a coactivator for β-catenin–Tcf4 (18, 35). Mutation of the p300 binding site in E1A partially relieves repression of the Tcf-E2 promoter in transient-transfection luciferase assays (C. Furer and R. Iggo, unpublished data), and we are currently making viruses with Tcf binding sites in the early promoters combined with mutations in the E1A gene to test this model. An alternative explanation is that the viruses may have acquired specific adaptations favoring growth on SW480 cells, in addition to their requirement for wnt pathway activation. The viruses were produced by transfection of plasmid DNA into packaging cells derived from 293 and HER911, followed by plaque purification and expansion on SW480. We can rule out adaptation by mutation of the Tcf-E1B and Tcf-E2 promoters because we sequenced 370 bp upstream of the E1B TATA box and the entire region between the E2 and E3 TATA boxes and found no mutations. There could be mutations elsewhere, but we saw no evidence of progressive adaptation during early passages on SW480. Indeed, the reason for expanding the viruses on SW480 was precisely to avoid selection of revertants adapted to growth in normal packaging cells. Furthermore, the fact that the Tcf-E2 promoter is active in many cells with wnt pathway activation but not in control cells (Fig. 3, 4, and 6) and activation of the pathway renders normal fibroblasts permissive for Tcf virus replication (Fig. 5) suggests that the host cell restriction of the Tcf viruses is due to the promoter changes we have introduced rather than coincidental spontaneous mutations.
The decrease in E2 promoter activity documented by Western blotting and quantitative PCR was similar in magnitude to the decrease in virus production measured by plaque assay (Fig. 3 and 4). Given the dependence of the latter on the former, it is perhaps surprising that a larger decrease in virus production was not seen. Since the virus has an absolute requirement for E2 gene products for replication, it is unlikely that cell-specific factors can account for the disparity. The simplest explanation is that E2 gene products may only become limiting when expressed at extremely low levels. Since the expression assays were performed at 24 h but the replication assays were at 48 h, it is also possible that a threshold for initiation of replication is reached in non-colon cells between 24 and 48 h. A delay of 24 h in initiating replication could still have an important effect in vivo, where reinfection is blocked by the production of neutralizing antibody, because the entire replicating virus strategy is based on the assumption that the virus will undergo multiple rounds of replication and spread within a tumor.
Replacement of the E1B promoter with a prostate-specific promoter has been shown to confer a 100-fold decrease in virus production in non-prostate cells (43). Deletion of the E1B 55K gene likewise confers a 100-fold reduction in virus yield relative to wild-type virus in many cell types (32). Replacement of the Sp1 site in the E1B promoter with four Tcf sites clearly does not achieve a comparable level of restriction of virus production in putative nonpermissive cells (Fig. 4). There are several possible explanations for this. In the absence of β-catenin, Tcf factors normally repress transcription through recruitment of Groucho and CtBP. To achieve tight promoter regulation this repression must overcome basal promoter activity. The poor selectivity of the Tcf-E1B viruses may reflect high basal promoter activity, mediated either by direct E1A-dependent transactivation via the TATA box (41) or through recruitment of cellular transactivators to distant upstream sites located within the E1A coding region (25). These upstream sites are generally not considered to play an important part in regulating the normal E1B promoter (41) but could play a greater role in the context of a Tcf-E1B promoter. A further possibility is that read-through transcription from the E1A promoter is contributing to E1B expression (10, 23). The effect of E1B 55K on virus production is largely mediated at the level of late mRNA export, acting after viral DNA replication (22). One motivation in regulating E1B expression in our viruses was the possibility that leakiness of E2 expression was due to E2 late promoter activity. Unlike the E2 early promoter, which lies in a noncoding sequence, the E2 late promoter lies in a coding sequence and the normal transcription factor binding sites cannot easily be replaced with Tcf sites (2). Regulation of E1B 55K expression provides an alternative means to regulate E2 late expression. In practice, we could see no effect on DBP protein level of Tcf-E1B regulation, suggesting either that the level of E1B expression was too high or that E2 late activity was not contributing significantly to DBP expression. Despite these largely negative results, it is interesting that the Tcf-E1B viruses did induce less inflammatory damage in vivo. This could be due to more efficient regulation in vivo than in vitro. Alternatively, the inflammatory response may depend in a more quantitative way on E1B 55K expression than does viral replication.
To achieve tight regulation of E2 expression it was necessary to mutate the E3 promoter, including mutation of the NFκB sites. Since NFκB is activated during liver regeneration (20), mutation of these sites may reduce the potential for virus replication in regenerating liver after successful treatment. Mutation of these sites may also reduce persistence of the virus in lymphocytes (38). Cross talk between the E2 and E3 promoters means that these viruses will be relatively protected from immune attack in tumors, which should favor virus replication, but more sensitive to immune clearance from normal tissue. Although on superficial examination preservation of immune responses in normal tissue may appear undesirable, the real danger with replicating viruses designed to kill human cells is that of creating new pathogens. Achieving the correct balance between causing harm to the patient and guaranteeing public safety is a delicate issue which will require careful consideration as the field evolves towards the creation of more potent viruses capable of producing substantial tumor responses. The viruses we have developed show how the extent of the inflammatory response can be deliberately manipulated to balance the conflicting needs of patients and society. Even within an individual tumor there is a balance to be struck between viral killing and immune killing. Although suppression of the immune response within the tumor is essential to permit multiple rounds of virus replication early after infection, because this is a prerequisite for infecting many tumor cells, at late stages a strong immune response is probably desirable to enhance the killing of tumor cells expressing viral antigens. There is unfortunately no easy way to select the optimal virus which balances all of these conflicting aims, because there is no immune-competent, replication-competent animal model for colon cancer (for example, cotton rat colon cancer cell lines that can be grafted into isogenic inbred cotton rats).
Despite the widespread involvement of wnt factors in development, there are very few sites in adult organisms where the wnt pathway is active. These tissues are sites of potential adverse effects and include hair follicles (8) and probably early T cells and colon crypt stem cells. Injection of virus into the hepatic artery to treat liver metastases would limit the exposure of these tissues to virus because high-level systemic infection with adenovirus is difficult to achieve even by deliberate systemic vascular administration (7). If hematological or gastrointestinal side effects were to prove limiting in animal studies, an obvious solution would be to introduce selectivity for additional oncogenic defects, for example, by driving E1A expression from the E2F promoter (26) or mutating E1A (9).
In conclusion, we have shown that adenoviral replication can be restricted to cells with activation of the wnt signaling pathway by placing Tcf sites in the E2 promoter, that tight regulation requires concomitant inactivation of the E3 promoter, and that combined Tcf regulation of the E2 and E1B promoters reduces the inflammatory response to adenovirus infection in cotton rat lungs.
ACKNOWLEDGMENTS
We thank B. Sordat for advice on histopathology, E. Lurati for technical assistance, B. Amati, J. Chamberlain, H. Clevers, O. Hagenbuechle, A. J. Levine, C. Prives, B. Sordat, and D. Trono for supplying reagents, and P. Beard and M. Peter for critical reading of the manuscript.
We thank the Swiss National Science Foundation and Swiss Cancer League for financial support. H. Kashiwazaki received a research fellowship from the Japanese Society for the Promotion of Science.
REFERENCES
- 1.Amalfitano A, Chamberlain J S. Isolation and characterization of packaging cell lines that coexpress the adenovirus E1, DNA polymerase, and preterminal proteins: implications for gene therapy. Gene Ther. 1997;4:258–263. doi: 10.1038/sj.gt.3300378. [DOI] [PubMed] [Google Scholar]
- 2.Bhat G, SivaRaman L, Murthy S, Domer P, Thimmappaya B. In vivo identification of multiple promoter domains of adenovirus EIIA-late promoter. EMBO J. 1987;6:2045–2052. doi: 10.1002/j.1460-2075.1987.tb02469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 4.Brattain M G, Brattain D E, Fine W D, Khaled F M, Marks M E, Kimball P M, Arcolano L A, Danbury B H. Initiation and characterization of cultures of human colonic carcinoma with different biological characteristics utilizing feeder layers of confluent fibroblasts. Oncodev Biol Med. 1981;2:355–366. [PubMed] [Google Scholar]
- 5.Cajot J F, Sordat I, Silvestre T, Sordat B. Differential display cloning identifies motility-related protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res. 1997;57:2593–2597. [PubMed] [Google Scholar]
- 6.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Yu D C, Charlton D, Henderson D R. Pre-existent adenovirus antibody inhibits systemic toxicity and antitumor activity of CN706 in the nude mouse LNCaP xenograft model: implications and proposals for human therapy. Hum Gene Ther. 2000;11:1553–1567. doi: 10.1089/10430340050083289. [DOI] [PubMed] [Google Scholar]
- 8.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 9.Doronin K, Toth K, Kuppuswamy M, Ward P, Tollefson A, Wold W. Tumor-specific, replication-competent adenovirus vectors overexpressing the adenovirus death protein. J Virol. 2000;74:6147–6155. doi: 10.1128/jvi.74.13.6147-6155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falck-Pedersen E, Logan J, Shenk T, Darnell J E., Jr Transcription termination within the E1A gene of adenovirus induced by insertion of the mouse beta-major globin terminator element. Cell. 1985;40:897–905. doi: 10.1016/0092-8674(85)90349-6. [DOI] [PubMed] [Google Scholar]
- 11.Gagnebin J, Brunori M, Otter M, Juillerat-Jeaneret L, Monnier P, Iggo R. A photosensitising adenovirus for photodynamic therapy. Gene Ther. 1999;6:1742–1750. doi: 10.1038/sj.gt.3300992. [DOI] [PubMed] [Google Scholar]
- 12.Ginsberg H S, Moldawer L L, Prince G A. Role of the type 5 adenovirus gene encoding the early region 1B 55-kDa protein in pulmonary pathogenesis. Proc Natl Acad Sci USA. 1999;96:10409–10411. doi: 10.1073/pnas.96.18.10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsberg H S, Prince G A. The molecular basis of adenovirus pathogenesis. Infect Agents Dis. 1994;3:1–8. [PubMed] [Google Scholar]
- 14.Goodrum F D, Ornelles D A. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrum F D, Ornelles D A. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J Virol. 1998;72:9479–9490. doi: 10.1128/jvi.72.12.9479-9490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallenbeck P L, Chang Y N, Hay C, Golightly D, Stewart D, Lin J, Phipps S, Chiang Y L. A novel tumor-specific replication-restricted adenoviral vector for gene therapy of hepatocellular carcinoma. Hum Gene Ther. 1999;10:1721–1733. doi: 10.1089/10430349950017725. [DOI] [PubMed] [Google Scholar]
- 17.Harada J-N, Berk A J. p53-independent and -dependent requirements for E1B–55K in adenovirus type 5 replication. J Virol. 1999;73:5333–5344. doi: 10.1128/jvi.73.7.5333-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht A, Vleminckx K, Stemmler M P, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes A, Williams A, Hawkins L, Kirn D. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 2000;6:1134–1139. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 20.Iimuro Y, Nishiura T, Hellerbrand C, Behrns K E, Schoonhoven R, Grisham J W, Brenner D A. NFkappaB prevents apoptosis and liver dysfunction during liver regeneration. J Clin Investig. 1998;101:802–811. doi: 10.1172/JCI483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 22.Leppard K N. Selective effects on adenovirus late gene expression of deleting the E1b 55K protein. J Gen Virol. 1993;74:575–582. doi: 10.1099/0022-1317-74-4-575. [DOI] [PubMed] [Google Scholar]
- 23.Maxfield L F, Spector D J. Readthrough activation of early adenovirus E1b gene transcription. J Virol. 1997;71:8321–8329. doi: 10.1128/jvi.71.11.8321-8329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 25.Parks C L, Banerjee S, Spector D J. Organization of the transcriptional control region of the E1b gene of adenovirus type 5. J Virol. 1988;62:54–67. doi: 10.1128/jvi.62.1.54-67.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parr M J, Manome Y, Tanaka T, Wen P, Kufe D W, Kaelin W G, Fine H A. Tumor-selective transgene expression in vivo mediated by an E2f-responsive adenoviral vector. Nat Med. 1997;3:1145–1149. doi: 10.1038/nm1097-1145. [DOI] [PubMed] [Google Scholar]
- 27.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 28.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 29.Ries S J, Brandts C H, Chung A S, Biederer C H, Hann B C, Lipner E M, McCormick F, Korn W M. Loss of p14ARF in tumor cells facilitates replication of the adenovirus mutant dl1520. Nat Med. 2000;6:1128–1133. doi: 10.1038/80466. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez R, Schuur E R, Lim H Y, Henderson G A, Simons J W, Henderson D R. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 1997;57:2559–2563. [PubMed] [Google Scholar]
- 31.Ross D T, Scherf U, Eisen M B, Perou C M, Rees C, Spellman P, Iyer V, Jeffrey S S, Van de Rijn M, Waltham M, Pergamenschikov A, Lee J C, Lashkari D, Shalon D, Myers T G, Weinstein J N, Botstein D, Brown P O. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 32.Rothmann T, Hengstermann A, Whitaker N J, Scheffner M, zur Hausen H. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b–58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 34.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b–58Kd tumor antigen: characterization of the E1b–58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 35.Takemaru K I, Moon R T. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 37.Velculescu V E, Vogelstein B. Analysis of human transcriptomes. Nat Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 38.Williams J L, Garcia J, Harrich D, Pearson L, Wu F, Gaynor R. Lymphoid specific gene expression of the adenovirus early region 3 promoter is mediated by NF-kappa B binding motifs. EMBO J. 1990;9:4435–4442. doi: 10.1002/j.1460-2075.1990.tb07894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wold W S, Doronin K, Toth K, Kuppuswamy M, Lichtenstein D L, Tollefson A E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr Opin Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Berk A. Constraints on spacing between transcription factor binding sites in a simple adenovirus promoter. Genes Dev. 1988;2:403–411. doi: 10.1101/gad.2.4.403. [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Rosser D S, Schmidt M C, Berk A. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature. 1987;326:512–515. doi: 10.1038/326512a0. [DOI] [PubMed] [Google Scholar]
- 42.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 43.Yu D C, Sakamoto G T, Henderson D R. Identification of the transcriptional regulatory sequences of human kallikrein 2 and their use in the construction of calydon virus 764, an attenuated replication competent adenovirus for prostate cancer therapy. Cancer Res. 1999;59:1498–1504. [PubMed] [Google Scholar]