Abstract
The gammaherpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) (or human herpesvirus 8) is associated with the endothelial tumor Kaposi's sarcoma (KS) and lymphoproliferative disorders in immunocompromised individuals. Only a small number of viral proteins are expressed in B cells latently infected with KSHV; here we characterize the mechanism of expression of one of these, the viral FLICE inhibitory protein v-FLIP (K13, ORF71). The v-FLIP coding region is present in a bicistronic message, following the v-cyclin coding region. Using both in vitro translation and cell transfection assays, we have identified an internal ribosome entry site (IRES) preceding the v-FLIP start codon and overlapping the v-cyclin (ORF 72) coding region, which allows v-FLIP translation. Using an antibody against v-FLIP we have detected expression of the endogenous protein in latently infected KSHV-positive primary effusion lymphoma (PEL) cell lines. Induction of apoptosis by serum withdrawal from PEL cells results in a relative increase in v-FLIP synthesis, as previously described for some cellular proteins translated from IRES.
In 1994 Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) was first identified in an AIDS-KS patient (15). KSHV DNA is present in all epidemiological types of KS and can be detected in all fresh biopsies and most paraffin-embedded lesions (9, 33a). KSHV sequences are also present in hematopoietic tumors primary effusion lymphoma (PEL) and a subtype of multicentric Castleman's disease (MCD) (13, 20, 59). PEL represents the clonal expansion of virally infected B lymphocytes (36). MCD is a polyclonal proliferation of mantle zone B cells, with only a few cells within the lesion infected by KSHV (20, 21, 36, 59). Sequencing the KSHV genome revealed that the virus has acquired a large number of cellular genes which might affect cell proliferation, differentiation, or death (8, 45, 46, 54). Of these, the v-cyclin (ORF 72), the viral FLICE inhibitory protein (v-FLIP, K13, ORF71), v-interleukin-6 (v-IL-6), and one of the viral interferon response factors are transcribed in some latently infected PEL cell lines (45, 60), and the v-FLIP and v-cyclin are transcribed in latently infected KS spindle cells (60).
A single transcript containing the sequences of the viral latent nuclear antigen (LNA-1, ORF 73), the v-cyclin, and v-FLIP has been detected by in site reverse transcriptase PCR in latently infected KS spindle cells and B cells, as has a spliced derivative encoding v-cyclin and v-FLIP (17, 18, 37, 53, 56, 61, 65). The longer form is thought to express LNA-1, while v-cyclin and v-FLIP are believed to be coexpressed from the spliced bicistronic transcript. Northern hybridization analysis of RNA from PEL cell lines BC-1 (56) and BCP-1 (65) with a v-FLIP probe failed to detect a monocistronic v-FLIP transcript.
Expression of v-cyclin protein has been detected in PEL cell lines and primary isolates (12, 50). v-Cyclin is a D-type cyclin which binds to cellular cyclin-dependent kinase 6 (CDK6) to form an active kinase (15, 29, 42). LNA-1 maintains the viral episome and tethers KSHV DNA to chromatin during mitosis (3) and may contribute to cell transformation by inhibiting p53 and retinoblastoma protein function (25, 52). Cellular FLIPs and FLIPs encoded by other viruses block Fas-mediated apoptosis (34, 66), and expression of KSHV v-FLIP has been shown to enhance growth of a mouse tumor by blocking cytotoxic-T-cell lysis (19). v-FLIP has also been shown to activate NF-κB signaling (16).
As v-cyclin and v-FLIP coding sequences are present within a single transcript, in situ reverse transcriptase PCR results do not define how, or indeed whether, the proteins are coexpressed in cells latently infected with KSHV. For the majority of cellular messages the 5′ mRNA cap is essential for binding part of the translation initiation complex. A 43S preinitiation complex then scans the RNA until it reaches the first favorable initiation codon for translation, which is often the first AUG. The nucleotides surrounding the initiation codon are important; analysis of eukaryotic mRNAs has defined a consensus sequence for the initiation of translation (40). Following initiation, elongation occurs until a termination codon is reached. Occasionally, the 40S ribosome can stay bound to the RNA after the termination codon and continue scanning to the next favorable initiation codon to translate a further message. This process, known as reinitiation of translation, appears to be favored by a short first message (up to 30 codons) and an optimal intercistronic distance of about 80 nucleotides (nt) (41).
Cap-independent translation was first described for poliovirus, where a sequence in the 5′ untranslated region which could mediate internal initiation of translation when inserted into a bicistronic plasmid was identified (49). Such RNA regions, which can mediate cap-independent translation, have become known as internal ribosome entry sites (IRES) and contain conserved elements of secondary structure which allow direct binding of a translation initiation complex (reviewed in reference 35). IRES have now been identified in a number of RNA viruses with positive-sense, nonsegmented genomes (4–7, 27, 28, 67). Our aims were therefore to determine how the second, v-FLIP, reading frame is translated and to examine v-FLIP expression and its regulation in KSHV-infected B cells.
MATERIALS AND METHODS
IRES analysis.
The v-cyclin/v-FLIP coding sequence of 1,423 nt, including both the initiation codon for v-cyclin and the termination codon for v-FLIP, was amplified by PCR from plasmid IgHP/E-73cycK13. The 5′ primer contained a Kozak consensus. This fragment was subcloned into the BamHI-XhoI sites of pcDNA 3.1(+) (Invitrogen) to give plasmid pcDNA3.1-cyclinFLIP-(wt) with the T7 bacteriophage promoter upstream of the v-cyclin/v-FLIP coding sequence. Mutations in the cyclin coding sequence were made by site-directed mutagenesis to give pcDNA3.1-cyclinFLIP-(frame shift), in which a base from the second codon for v-cyclin was deleted resulting in a 10-codon nonsense open reading frame and pcDNA3.1-cyclinFLIP-(truncation), in which codon 135 of v-cyclin was mutated to a stop. The monocistronic v-FLIP coding sequence was amplified by PCR from plasmid IgHP/E-73cycK13 with a 5′ primer that encoded an N-terminal HA epitope. The v-cyclin coding sequence was also amplified by PCR from plasmid IgHP/E-73cycK13 with a 5′ primer containing a Kozak consensus. Both were subcloned into pcDNA 3.1(+). pcDNA 3.1(+) was used as a vector control. The plasmids were linearized by digestion with XhoI prior to in vitro transcription.
PCR was used to amplify fragments corresponding to the 82, 363, and 655 nt proximal to the v-FLIP AUG from plasmid IgHP/E-73cycK13. PCR fragments were cloned into the ClaI-NcoI sites of plasmid pBSK-p35-p40 (31) between the coding sequences for the two subunits of IL-12 in which the NcoI site contains the initiation codon for p40. Plasmid pBSK-E12 (31), containing the IRES from encephalomyocarditis virus (EMCV) between the coding sequences for the IL-12 subunits was used as a positive control for IRES function. In each case cassette p35-IRES-p40 was excised with BamHI and inserted into pcDNA 3.1(+). Plasmids pcDNA-p40 and pcDNA-p35 contain the coding sequences for individual p35 and p40 subunits of IL-12 respectively. The above constructs and an empty pcDNA 3.1(+) control were linearized by digestion with NotI prior to in vitro transcription.
Plasmids were transcribed to produce capped RNA using the mMESSAGEmMACHINE kit (Ambion). RNA was precipitated by adding an equal volume of isopropanol and chilling at −20°C for 20 min. RNA size and concentration were determined by running an aliquot on a 1% agarose-formaldehyde gel. The translation reaction mixture was assembled from 5 μl of amino acid mixture, 5 μl of 0.2 M creatine phosphate, 5 μl of 2 M KCl–10 mM MgCl2, 5 μl of [35S]methionine-cysteine mixture (Amersham Pharmacia), and 80 μl of rabbit reticulocyte lysate. Equal amounts of capped, full-length RNA were added to 10 to 20 μl of translation mixture and incubated at 30°C for 90 min. Then 1 μl was removed and added to 9 μl of sodium dodecyl sulfate (SDS) sample buffer for resolution by SDS-polyacrylamide gel electrophoresis (PAGE).
293T-cell transfection.
Semiconfluent 10-cm-diameter dishes of 293T cells were transfected with 8 μg of plasmid using Lipofectamine. After 6 h of incubation at 37°C the cells were washed and incubated in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS) at 37°C for a further 48 h. Anti-v-FLIP polyclonal antibodies were raised in rabbits against a keyhole limpet hemocyanin-conjugated peptide corresponding to the N-terminal region of v-FLIP (TYEVLCEVARKLGT) (Severn Biotech). The immunization protocol was as described in reference 30. Anti-v-FLIP rat monoclonal antibody 6/14 was raised against full-length recombinant v-FLIP expressed in Escherichia coli. For immunodetection the polyclonal anti-v-FLIP rabbit serum was diluted 1:250 and detected by a goat anti-rabbit horseradish peroxidase (HRPO) antibody (1:2,000; Harlan) and enhanced chemiluminescence (Amersham) and the purified monoclonal anti-v-FLIP antibody was diluted 1:100 and detected by a goat anti-rat HRPO antibody (1:2,000; Harlan). The anti-v-cyclin was a rat monoclonal antibody (kind gift from Sybille Mittnacht, Institute of Cancer Research, London, United Kingdom) used at a dilution of 1:100 and detected with goat anti-rat HRPO (1:2,000; Harlan). Total cellular RNA was isolated using the RNAzol (Biogenesis) method, in accordance with the manufacturer's instructions. For RNA blot analysis, 10 μg of total RNA was loaded on a 1% agarose-formaldehyde gel and transferred to a nylon HybondN+ membrane (Amersham). Prehybridization, hybridization, and washes of the membrane were performed using the HybondN+ protocol. The v-FLIP and p40 probes were labeled with [α-32P]dCTP (Amersham) using a random-prime-labeling system (Promega). IL-12 was determined using a commercial sandwich enzyme-linked immunosorbent assay (ELISA) with antibodies which detect the p40 subunit; each supernatant was assayed in triplicate (Cytoscreen human IL-12; BioSource International Inc.).
v-FLIP expression in KSHV-infected cells.
The KSHV-transformed primary effusion lymphoma (PEL) cell lines BC3 and BCP1 (10, 26), Epstein-Barr virus (EBV)-transformed B-cell line B.45, and EBV-negative B-cell line DG75 were cultured in RPMI 1640 supplemented with 10% FCS, penicillin, and streptomycin at 37°C in 5% CO2. For immunostaining BC3 cells were fixed in 3% paraformaldehyde for 10 min and then washed in 0.2% Triton X-100 for 5 min. Cells were stained for v-FLIP with rat monoclonal antibody 6/14 (1/30 dilution), followed by a biotinylated rabbit anti-mouse antibody and peroxidase-conjugated avidin. The stained proteins were visualized with diaminobenzidine tetrahydrochloride and briefly counterstained with hematoxylin. For immunoprecipitation BC3 cells were washed twice with serum-free RPMI 1640 and incubated for 20 to 24 h in RPMI 1640 with or without serum. Cells were then incubated for 30 min in methionine- and cysteine-free RPMI1640 (Sigma) and then labeled for 2 h in the same medium supplemented with a mixture of [35S]methionine-cysteine (Redivue Pro-Mix; Amersham) and, where appropriate, with 10% FCS. After three washes in ice-cold phosphate-buffered saline (PSB), the cell pellets were lysed for 30 min on ice in 1 ml of radioimmunoprecipitation assay buffer (30) supplemented with protease inhibitors. Lysates were then centrifuged at 14,000 × g for 15 min at 4°C. Supernatant from the equivalent of 107 cells was precleared for 1 h with protein G-Sepharose (Sigma) for anti-LNA-1 (38) or protein A-sepharose (Sigma) for anti-v-FLIP and then immunoprecipitated for 2 h at 4°C with 2 μg of anti-LNA-1 or 10 μg of anti-v-FLIP antibodies. Immunoprecipitates were extensively washed with lysis buffer and then resuspended in sample buffer and loaded onto a 7% (for LNA-1) or 14% (for v-FLIP) SDS-polyacrylamide gel. Dried gels were exposed to a phosphorimager (Molecular Dynamics), and the relative levels of protein expression were determined using densitometry software (Imagequant; Molecular Dynamics). Apoptotic cells were quantitated by staining with ANNEXIN-V-Fluos (Boehringer). Cells were washed twice with ice-cold PBS and resuspended in 100 μl of annexin buffer (140 mM NaCl, 10 mM HEPES [pH 7.4] 5 mM CaCl2) containing 2 μl of annexin. The samples were incubated at room temperature in the dark for 15 min; at the end of the incubation 300 μl of annexin buffer was added to all samples, which were analyzed by FACScan.
RESULTS
An upstream RNA sequence allows v-FLIP translation in vitro.
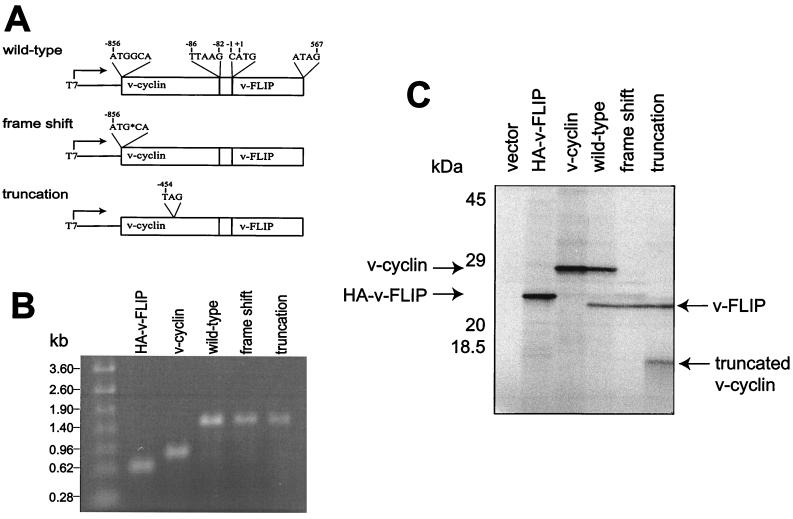
In the viral bicistronic transcript, the v-cyclin coding sequence is followed after 82 nt by the v-FLIP coding sequence. The constructs shown in Fig. 1A were designed to examine v-FLIP translation from this transcript in reticulocyte lysate. v-cyclin and v-FLIP contain similar numbers of methionine and cysteine residues for 35S incorporation (9 and 10, respectively). In vitro transcription of these constructs produced a single bicistronic RNA of approximately 1.5 kb, the predicted length, as shown in Fig. 1B. Translation of the bicistronic v-cyclin/v-FLIP transcript gave a strong v-cyclin band of 28 kDa and a slightly weaker 23-kDa v-FLIP band (Fig. 1C). v-FLIP expression was unlikely to be the result of cap-dependent translation, as the v-cyclin AUG lies within a strong Kozak consensus sequence (39) and a further five AUGs lie between the v-cyclin start codon and the v-FLIP start codon. Translation of v-FLIP was then examined in bicistronic constructs containing mutations in the v-cyclin coding region. In the first construct a guanosine residue in the second codon for v-cyclin was deleted, resulting in a change of frame to generate a predicted product of 10 amino acids, which was undetectable. When codon 135 for v-cyclin was changed to a termination codon, the predicted truncated protein of approximately 15 kDa was detected. v-FLIP translation was unaffected by both mutations (Fig. 1C). This suggested that reinitiation of translation was not responsible for v-FLIP expression because such reinitiation occurs efficiently only after a short intercistronic gap (40).
FIG. 1.
In vitro translation of the the v-cyclin/v-FLIP cassette. The bicistronic plasmids (A), with monocistronic controls and empty vector, were transcribed in vitro. RNA was analyzed on a 1% agarose-formaldehyde gel (B) and translated in reticulocyte lysate as described in Materials and Methods. (C) Labeled proteins resolved by SDS–15% PAGE. HA, hemagglutinin.
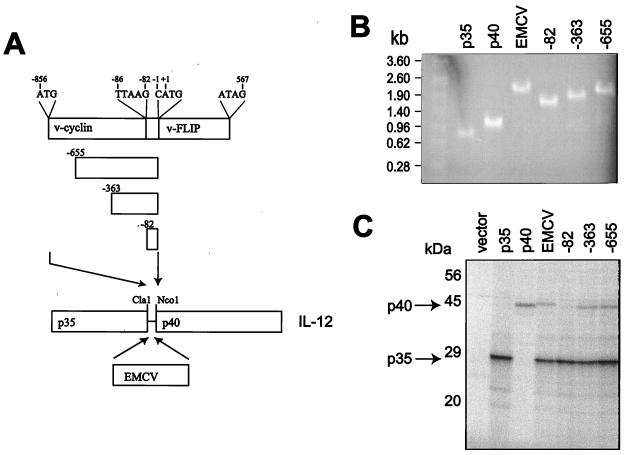
To identify the region of the bicistronic transcript that allows v-FLIP translation, three regions of the transcript were cloned into a bicistronic vector between the subunits of the heterodimeric cytokine IL-12, with the p40 initiation codon in the position of that for v-FLIP (Fig. 2A). Again in vitro transcription produced bicistronic RNAs of the expected lengths (Fig. 2B), which were added to reticulocyte lysate to analyze translation. When the intercistronic 82-nt region between the coding sequences for v-cyclin and v-FLIP was inserted between the coding sequences for the two IL-12 subunits a p35 signal but no p40 signal was seen, whereas the 363- and 655-nt KSHV sequences allowed p40 translation. The p40 band densities seen with the two KSHV constructs were similar to that seen with an encephalomyocarditis virus (EMCV) IRES (Fig. 2C). p35 contains 18 potential methionine or cysteine residues compared to 14 such residues in p40, which may account for the stronger intensity of the p35 signal. The size of p35 is about 30 kDa, as p35 is extensively glycosylated in vivo. Again cap-dependent translation or reinitiation of translation was unlikely to explain p40 expression, as this was not observed with the intercistronic 82-nt region. Therefore we concluded that an IRES was present upstream of the FLIP start codon and overlapping the v-cyclin coding sequence. It should be noted that translation of v-cyclin does not inhibit the activity of this IRES, as demonstrated in Fig. 1C.
FIG. 2.
Mapping a region upstream of v-FLIP which allows translation. The portions of the v-cyclin/v-FLIP cassette (A) and a previously characterized IRES from EMCV (25) were cloned between the coding sequences for the subunits of heterodimeric cytokine IL-12. The bicistronic plasmids, with monocistronic controls and empty vector, were transcribed in vitro. RNA was analyzed on a 1% agarose-formaldehyde gel (B) and translated in reticulocyte lysate as described in Materials and Methods. (C) Labeled proteins resolved by SDS–12.5% PAGE.
Function of the IRES in mammalian cells.
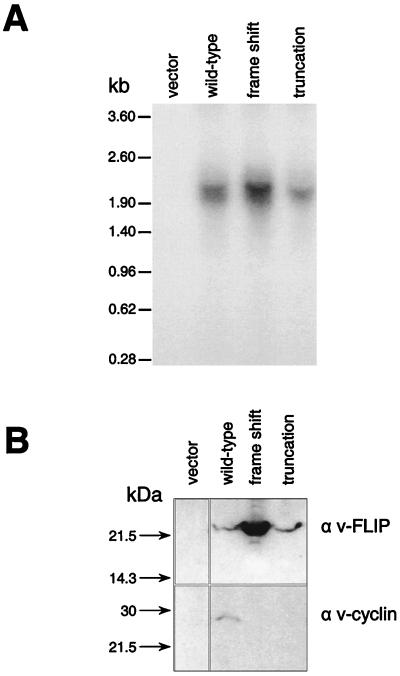
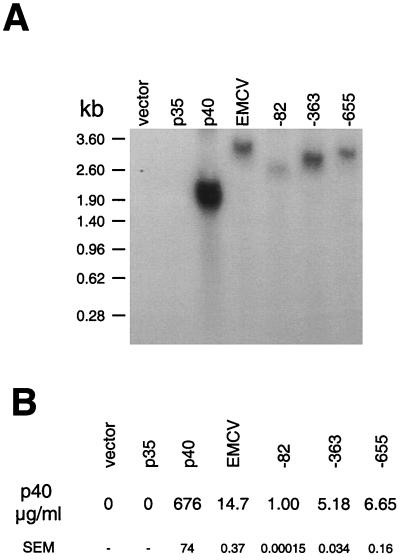
The expression of v-FLIP was also examined in mammalian cells where the cytomegalovirus promoter drives expression of the bicistronic v-cyclin/v-FLIP transcripts. Figure 3A shows that the predicted 1.9-kb bicistronic transcript was detected in transfected 293T cells, with no shorter v-FLIP transcripts being detected even on overexposure of the blot (data not shown). To detect v-FLIP expression, we raised a polyclonal antiserum against a peptide corresponding to the 14 N-terminal amino acids. Figure 3B shows that both v-cyclin and v-FLIP were expressed from the bicistronic message. p40 expression from the bicistronic IL-12 cassettes was also examined following mammalian cell transfection with expression vectors. Figure 4A shows that only the predicted bicistronic transcripts encoding p40 were detected in the transfected cells. Levels of p40, measured using an ELISA which detects p40 whether present as a homodimer or heterodimer, are shown in Fig. 4B. A low level of p40 was detected following the transfection of the vector containing the 82-nt intercistronic v-cyclin/v-FLIP coding sequence region. The vector containing the 655-nt v-cyclin/v-FLIP coding sequence region produced more p40 than that the 363-nt region. The low activity of the 82-nt sequence is in contrast to its inactivity in vitro. Furthermore, while a sequence of 363 nt can function optimally in vitro, full activity in cells occurs with the longer 655-nt sequence. This could imply that the interaction of cellular proteins with sequences proximal to the 363-nt sequence is required for translation or alternatively could suggest that the longer sequence is required for optimum stability of an IRES structure in cells.
FIG. 3.
Function of the v-cyclin/v-FLIP transcript in mammalian cells. 293T cells were transiently transfected with parental pcDNA 3.1(+) vector or with vectors containing the coding sequence for bicistronic v-cyclin/v-FLIP. After 48 h, half the cells were used to prepare RNA as described in Materials and Methods and half were lysed in radioimmunoprecipitation assay buffer for SDS-PAGE. (A) RNA probed with a v-FLIP probe. (B) v-cyclin and v-FLIP detected on a Western blot as described in Materials and Methods.
FIG. 4.
Mapping a region upstream of v-FLIP which allows translation in mammalian cells. 293T cells were transiently transfected with parental pcDNA 3.1(+) vector or with vectors containing sequences from the v-cyclin/v-FLIP-coding region of KSHV between the coding sequences for the subunits of the heterodimeric cytokine IL-12. The same plasmid containing the IRES from EMCV was used for comparison. After 48 h, cell supernatant was collected and the cells were used to prepare RNA as described in Materials and Methods. (A) RNA probed with a p40 probe. (B) Level of p40 protein in supernatant detected by ELISA, as described in Materials and Methods. p40 levels are shown as the means of triplicate determinations with standard errors of the means (SEM).
v-FLIP expression in KSHV-transformed PEL cells.
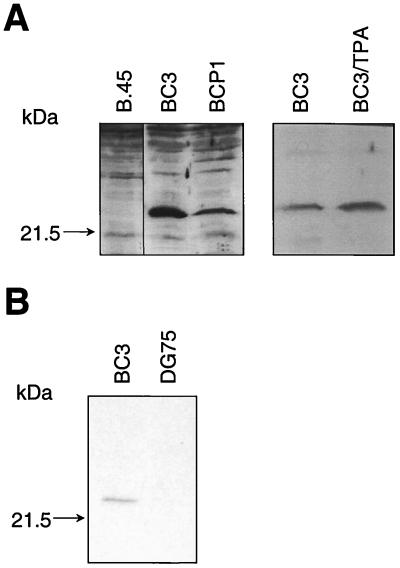
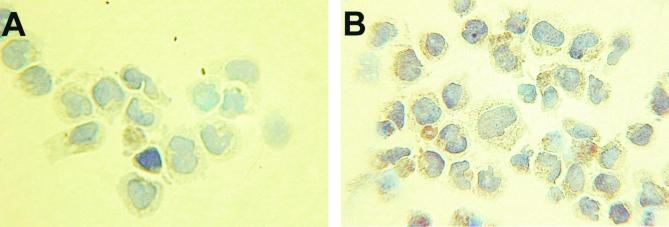
To demonstrate that v-FLIP is expressed from the bicistronic message detected in latently infected cells, we used the polyclonal antiserum which recognizes v-FLIP. Figure 5A shows that this antiserum recognized a protein of approximately 23 kDa in lysates from the EBV-negative PEL cell lines BC3 and BCP1, but not the EBV-transformed cell line B.45. Induction of KSHV replication by tetradecanoyl phorbol acetate in BC3 cells (25) caused only a small increase in v-FLIP expression (Fig. 5A), demonstrating that it is a latently expressed protein. Anti-v-FLIP rat monoclonal antibody 6/14 was raised against full-length recombinant v-FLIP expressed in E. coli. This monoclonal antibody also recognized a protein of approximately 23 kDa in BC3 cell lysate (Fig. 5B) and stained the cytoplasm of BC3 cells (Fig. 6).
FIG. 5.
v-FLIP protein expression in KSHV-infected B cells. (A) Western blot of 70 μg of protein from the EBV-transformed B.45 cells and the KSHV-transformed EBV-negative PEL BC3 and BCP1 cells and protein from BC3 cells uninduced or induced for 48 h with 200 ng of tetradecanoyl phorbol acetate (TPA)/ml and separated by SDS–14% PAGE. Immunoblotting with a polyclonal anti-v-FLIP antiserum was performed as described in Materials and Methods. (B) Western blot of 70 μg of protein from EBV-negative B-cell line DG75 and the KSHV-transformed PEL BC3 cells separated by SDS–14% PAGE. Immunoblotting with monclonal anti-v-FLIP antibody 6/14 was performed as described in Materials and Methods.
FIG. 6.
BC3 cells (×500 magnification) stained with a second-layer antibody only (A) or monoclonal anti v-FLIP antibody 6/14, where a brown stain in the cytoplasm was detected (B) as described in Materials and Methods.
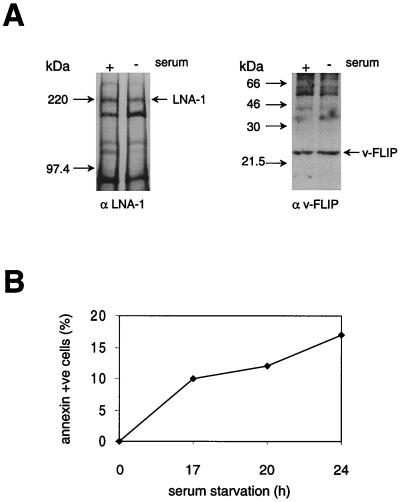
During apoptosis cap-dependent protein translation is inhibited due to cleavage of eIF4G by caspase 3 (11, 44). Recently, the translation of cellular proteins involved in apotosis induction, myc (63) and apoptosis inhibition, XIAP (32), was shown to be initiated from an IRES and therefore maintained during apoptosis. The rate of v-FLIP synthesis was therefore examined during apoptosis of PEL cell line BC3 induced by serum withdrawal, which caused eIF4G cleavage (data not shown) and induced apoptosis as detected by ANNEXIN-V-Fluos staining (Fig. 7B). For comparison the rate of LNA-1 synthesis was measured, since this requires cap-dependent protein translation and since v-cyclin could not be immunoprecipitated with the available antibody. Figure 7A shows that the rate of synthesis of LNA-1 decreased in serum-deprived cells whereas that of v-FLIP was maintained. Quantitation of the relative amounts of newly synthesized LNA-1 and v-FLIP demonstrated a threefold increase in the ratio of v-FLIP synthesis to LNA-1 synthesis during apoptosis (Fig. 7). This is a change similar to the previously reported relative up-regulation of myc (63) or XIAP (32) synthesis during apoptosis. Clearly v-FLIP synthesis may be relatively higher in individual apoptotic cells, as not all the cells are apoptotic at any one time (Fig. 7B).
FIG. 7.
Synthesis of LNA-1 and v-FLIP during apoptosis of PEL cells. BC3 cells were serum starved for 20 h to induce apoptosis, which was detected by ANNEXIN-V Fluos staining (B) as described in Materials and Methods. Viable cells (107) were then pulsed for 2 h with labeled methionine and cysteine, after which LNA-1 and v-FLIP were immunoprecipitated as described in Materials and Methods and the immunoprecipitates were separated by SDS–7 or 14% PAGE, respectively (A). The relative levels of labeled protein were determined using densitometry software on a phosphorimager as described in Materials and Methods. The FLIP/LNA-1 labeled-protein ratio was normalized to 1 in cells with serum; the relative FLIP/LNA-1 labeled-protein ratio in cells without serum was 3.25. In a second experiment cells were serum starved for 24 h and the relative FLIP/LNA-1 labeled-protein ratio increased from 1 to 3.1 on serum starvation.
DISCUSSION
We have demonstrated that a sequence upstream of the v-FLIP coding sequence functions to allow translation by acting as an IRES. IRES have previously been identified in RNA viruses with a positive-sense, nonsegmented genome (4–7, 27, 28, 67); they possess certain conserved RNA secondary structural features (35, 62). In such viruses the RNA must serve both as the genome and as an mRNA for translation of viral proteins; thus IRES have been thought to be crucial for this dual usage.
The putative IRES upstream of the v-FLIP coding sequence is the first to be identified in a DNA virus; its presence and the unusual overlap of the IRES with the v-cyclin coding sequence suggest that expression of v-cyclin and v-FLIP needs to be tightly coordinated. Three other gammaherpesviruses closely related to KSHV (1) are known to contain adjacent v-cyclin and v-FLIP reading frames. In ateline herpesvirus 3 (unpublished) (GenBank accession no. AF083424) and herpesvirus saimiri (2) a single base pair separates the v-cyclin and v-FLIP reading frames. Macaca mulatta rhadinovirus 17577 shows spacing similar to that for KSHV, with 69 bp between the v-cyclin and v-FLIP reading frames (57). Whether any of these viruses employ a bicistronic transcript to express v-cyclin and v-FLIP remains to be determined. The nearest relative to this group, the gammaherpesvirus murine herpesvirus 68, encodes v-cyclin alone (1, 68), suggesting that v-FLIP was a later acquisition from cellular DNA. A second bicistronic transcript from KSHV may also use an IRES to allow expression of the G-protein-coupled receptor (v-GPCR) during lytic replication (39).
We have also shown for the first time that v-FLIP is expressed in latently infected PEL cell lines. These lines express Fas but are resistant to Fas-mediated killing (data not shown). It is likely that this v-FLIP inhibition of Fas-mediated cell killing is one mechanism by which KSHV evades anti-viral cytotoxic T-lymphocyte (CTL) responses. This may occur during persistent KSHV infection of immunocompetent individuals, in whom KSHV-specific CTL have been detected (48). However, v-FLIP expression continues when tumors arise in immunosuppressed individuals, whose anti-KSHV CTL responses are likely to be compromised. This suggests that v-FLIP also directly affects the survival of the tumor cells.
The v-cyclin–CDK6 complex is resistant to inhibition by p16 and p21 (64), and its expression causes degradation of the p27 inhibitor (22, 43). It can therefore override normal controls to induce G1/S-phase transition. However, constitutive expression of molecules which potentiate G1/S transition such as c-myc (23), E2F (51), and cyclin D1 (58) in the absence of optimal growth factors or v-cyclin in the presence of a high level of CDK6 (47) has been shown to cause apoptosis. It therefore seems an attractive hypothesis that v-cyclin may cooperate with v-FLIP to transform endothelial cells and/or B cells, which could explain the need to link their expression. Indeed, death receptor signaling is known to be involved in apoptosis of myc-expressing cells following serum removal (33) or of epithelial cells following substrate detachment (55), and v-FLIP has been shown to activate NF-κB signaling (16). However, the fact that v-FLIP synthesis can be up-regulated when apoptosis is triggered may also provide a survival advantage for KSHV-infected cells under some circumstances and could be another reason for an IRES-dependent translation of v-FLIP.
ACKNOWLEDGMENTS
W. Low and M. Harries contributed equally to this work.
BC3 was kindly provided by Ethel Ceserman.
This work was supported by the Clinical Research and Development Committee of the Special Trustees of the Middlesex Hospital and a Cancer Research Campaign Fellowship for a Clinician awarded to M.H.
REFERENCES
- 1.Alba, M. M., R. Das, C. Orengo, and P. Kellam. Genome wide function conservation and phylogeny in the Herpesviridae. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittman S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Belsham G J, Brangwyn J K. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–5395. doi: 10.1128/jvi.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlioz C, Darlix J-L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienkowska-Szewczyk K, Ehrenfeld E. An internal 5′-noncoding region required for translation of poliovirus RNA in vitro. J Virol. 1988;62:3068–3072. doi: 10.1128/jvi.62.8.3068-3072.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman A, Jackson R J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff C. Coupling herpesvirus to angiogenesis: viral pirates on a cellular sea. Nature. 1998;391:24–25. doi: 10.1038/34054. [DOI] [PubMed] [Google Scholar]
- 9.Boshoff C. Kaposi's sarcoma-associated herpesvirus. In: Newton R, Beral V, Weiss R A, editors. Cancer surveys: infections and human cancer. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1999. pp. 157–190. [Google Scholar]
- 10.Boshoff C, Gao S-J, Healy L E, Matthews S, Thomas A J, Coignet L, Warnke R A, Strauchen J A, Matutes E, Kamel O W, Moore P S, Weiss R A, Chang Y. Establishment of a KSHV positive cell line (BCP-1) from peripheral blood and characterizing its growth in vivo. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 11.Bushell M, McKendrick L, Janicke R U, Clemens M J, Morley S J. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 1999;451:332–336. doi: 10.1016/s0014-5793(99)00614-6. [DOI] [PubMed] [Google Scholar]
- 12.Carbone A, Gloghini A, Bontempo D, Monini P, Tirelli U, Volpe R, Browning P J, Gaidano G. Proliferation in HHV-8-positive primary effusion lymphomas is associated with expression of HHV-8 cyclin but independent of p27(kip1) Am J Pathol. 2000;156:1209–1215. doi: 10.1016/S0002-9440(10)64991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden K, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhary P M, Jasmin A, Eby M, Hood L. Modulation of the NF-kappaB pathway by virally encoded death effector domain-containing proteins. Oncogene. 1999;18:5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 17.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1829–1831. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 18.Dittmer D, Lagunoff M, Renne R, Stastus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djerbi M, Screpanti V, Catrina A I, Bogen B, Biberfeld P, Grandien A. The inhibitor of death receptor signaling. FLICE-inhibitory protein, defines a new class of tumor progression factors. J Exp Med. 1999;190:1025–1031. doi: 10.1084/jem.190.7.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupin N, Diss T, Kellam P, Tulliez M, Du M-Q, Weiss R A, Isaacson P G, Boshoff C. HHV-8 is associated with a plasmablastic variant of Castleman's disease that is linked to HHV-8 positive plasmablastic lymphoma. Blood. 2000;95:1406–1412. [PubMed] [Google Scholar]
- 21.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J-P, Weiss R A, Alitalo K, Boshoff C. Distribution of HHV-8 positive cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis M, Chew Y P, Fallis L, Freddersdorf S, Boshoff C, Weiss R A, Lu X, Mittnacht S. Degradation of p27KIP cdk inhibitor triggered by Kaposi's sarcoma virus cyclin-cdk6 complex. EMBO J. 1999;18:644–653. doi: 10.1093/emboj/18.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters L M, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 24.Flore O, Gao S J. Effect of DNA synthesis inhibitors on Kaposi's sarcoma-associated herpesvirus cyclin and major capsid protein gene expression. AIDS Res Hum Retroviruses. 1997;13:1229–1233. doi: 10.1089/aid.1997.13.1229. [DOI] [PubMed] [Google Scholar]
- 25.Friborg J, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 26.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 27.Ghattas I R, Sanes J R, Majors J E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glass M J, Jia X Y, Summers D F. Identification of the hepatitis A virus internal ribosome entry site: in vivo and in vitro analysis of bicistronic RNAs containing the HAV 5′ noncoding region. Virology. 1993;193:842–852. doi: 10.1006/viro.1993.1193. [DOI] [PubMed] [Google Scholar]
- 29.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus (KSHV) stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harlow E, Lane D. Antibodies: a laboratory manual. New York, N.Y: CSHL Press; 1988. [Google Scholar]
- 31.Harries M, Phillipps N, Anderson R, Prentice G, Collins M. Comparison of bicistronic retroviral vectors containing IRES using expression of human IL-12 as a readout. J Gene Med. 2000;2:243–249. doi: 10.1002/1521-2254(200007/08)2:4<243::AID-JGM115>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 32.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 33.Hueber A O, Zornig M, Lyon D, Suda T, Nagata S, Evan G I. Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- 33a.IARC Working Group. Epstein-Barr virus and Kaposi's sarcoma herpesvirus/human herpesvirus 8. IARC Monogr Eval Carcinog Risks Hum. 1997;70:1–492. [PMC free article] [PubMed] [Google Scholar]
- 34.Irmler M, Thome M, Hahne M, Schneider P, Hofman K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, Rimoldi D, French L E, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 35.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 36.Judde J G, Lacoste V, Briere J, Kassa-Kelembho E, Clyti E, Couppie P, Buchrieser C, Tulliez M, Morvan J, Gessain A. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi's sarcoma and other diseases. J Natl Cancer Inst. 2000;92:729–736. doi: 10.1093/jnci/92.9.729. [DOI] [PubMed] [Google Scholar]
- 37.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen (LNA-1) in the human herpesvirus 8 (HHV-8) genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 38.Kellam P, Bourboulia D, Dupin N, Talbot S, Boshoff C, Weiss R A. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J Virol. 1999;73:5149–5155. doi: 10.1128/jvi.73.6.5149-5155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirshner J R, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann D J, Child E S, Swanton C, Laman H, Jones N. Modulation of p27(Kip1) levels by the cyclin encoded by Kaposi's sarcoma-associated herpesvirus. EMBO J. 1999;18:654–663. doi: 10.1093/emboj/18.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marissen W E, Lloyd R E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 46.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ojala P M, Tiainen M, Salven P, Veikkola T, Castanos-Velez E, Sarid R, Biberfeld P, Makela T P. Kaposi's sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res. 1999;59:4984–4989. [PubMed] [Google Scholar]
- 48.Osman M, Kubo T, Gill Y, Neipel F, Smith J, Weiss R A, Gazzard B, Boshoff C, Gotch F. Identification of human herpesvirus-8-specific cytotoxic T-cell responses. J Virol. 1999;73:6136–6140. doi: 10.1128/jvi.73.7.6136-6140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 50.Platt G M, Cannell E, Cuomo M E, Singh S, Mittnacht S. Detection of the human herpesvirus 8-encoded cyclin protein in primary effusion lymphoma-derived cell lines. Virology. 2000;272:257–266. doi: 10.1006/viro.2000.0343. [DOI] [PubMed] [Google Scholar]
- 51.Qin X Q, Livingston D M, Kaelin W G, Jr, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radkov S A, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 53.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rytomaa M, Downward J. Involvement of FADD and caspase-8 signalling in detachment-induced apoptosis. Curr Biol. 1999;9:1043–1046. doi: 10.1016/s0960-9822(99)80454-0. [DOI] [PubMed] [Google Scholar]
- 56.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sofer-Levi Y, Resnitzky D. Apoptosis induced by ectopic expression of cyclin D1 but not cyclin E. Oncogene. 1996;13:2431–2437. [PubMed] [Google Scholar]
- 59.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 60.Staskus K A, Sun R, Miller G, Racz P, Jaslowski A, Metroka C, Brett-Smith H, Haase A T. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J Virol. 1999;73:4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart S R, Semler B L. RNA determinants of picornavirus cap-independent translation initiation. Semin Virol. 1997;8:242–255. [Google Scholar]
- 63.Stoneley M, Chappell S A, Jopling C L, Dickens M, MacFarlane M, Willis A E. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Herpesviral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 65.Talbot S, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 (HHV-8) open reading frames 71, 72, 73, K14 and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 66.Thome M, Schneider P, Hofmann K, Fickenscher H, Meini E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidl C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 67.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genome analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]