Abstract
Cryptococcus neoformans is a pathogenic yeast that causes meningitis in immunocompromised patients. Because iron acquisition is critical for growth of a pathogen in a host, we studied the regulation of the ferric reductase and ferrous uptake system of this organism. We isolated 18 mutants, representing four independent loci, with dysregulated ferric reductase. The mutant strains had >10-fold higher than wild-type WT reductase activity in the presence of iron. Two of the strains also had >7-fold higher than WT iron uptake in the presence of iron but were not markedly iron sensitive. Both were sensitive to the oxidative stresses associated with superoxide and hydrogen peroxide. One strain exhibited only 23% of the WT level of iron uptake in the absence of iron and grew poorly without iron supplementation of the medium, phenotypes consistent with an iron transport deficiency; it was sensitive to superoxide but not to hydrogen peroxide. The fourth strain had high reductase activity but normal iron uptake; it was not very sensitive to oxidative stress. We also demonstrated that the ferric reductase was regulated by copper and could act as a cupric reductase. Sensitivity to oxidants may be related to iron acquisition by a variety of mechanisms and may model the interaction of the yeast with the immune system.
Cryptococcus neoformans is a pathogenic yeast that causes meningitis in immunosuppressed AIDS, cancer, and organ transplant patients. Like most organisms, it requires iron for housekeeping proteins such as ribonucleotide reductase, oxidases, and other cytochrome-containing proteins. In a host, however, little free iron is available: iron is generally bound to proteins or sequestered in cells, and serum iron may be lowered in response to infection (24). As demonstrated for Neisseria gonorrhoeae, Salmonella typhi, and Vibrio cholerae, pathogens deficient in iron acquisition are of diminished virulence (9, 17, 23). In addition, low iron may trigger the expression of other genes required for survival in a host: C. neoformans produces a polysaccharide capsule, a virulence factor that helps the cell escape phagocytosis (50).
Alteration of iron metabolism in the host may stimulate growth of pathogens. Some AIDS patients, instead of developing the anemia of infection, have iron overload in many organs and macrophages and elevated serum ferritin (reviewed in reference 5). In Africa, there is up to a 10% incidence of iron overload from dietary and genetic factors (39). Iron overload of macrophages decreases their microbicidal activity and may in fact stimulate the growth of microbes (38). This has been shown to increase risk of death from tuberculosis, which is the most common infection associated with human immunodeficiency virus in Africa (38). Iron overload has not yet been established as a risk factor for cryptococcal infections, yet such infections are common in African AIDS patients and may have heralded the emergence of the human immunodeficiency virus in the 1950s (37).
To obtain iron, C. neoformans expresses a ferric reductase, a plasma membrane enzyme which reduces and solubilizes Fe(III) (41). Expression of the reductase is down regulated over sevenfold by iron. The reductase is required to supply substrate to low- and high-affinity transmembrane transporters, which are specific for Fe(II). The low-affinity transporter was not saturable under the conditions used but appeared to be regulated by iron in the medium. The high-affinity transporter, with a Km for Fe(II) of 0.6 μM (27), was also down regulated ∼10-fold by iron in the medium. The high-affinity transporter requires copper to function. The combination of reductase and transporter is capable of removing and transporting iron chelated by deferoxamine, which is used clinically as an iron chelator.
Many of the components of iron reduction and uptake have been characterized at the molecular level in the model organism Saccharomyces cerevisiae. The FRE1 protein is the major ferric reductase (12). After reduction to Fe(II), the iron may be reoxidized by the FET3 oxidase concomitant with uptake into the cell through the FTR1 permease (46). FET3 assembly requires CCC2 and ATX1 (34, 54). Expression of the genes encoding these proteins is coordinately regulated by the transcription factor AFT1 (53). In addition to this high-affinity system, iron may be taken up by the low-affinity Fe(II) transporter FET4 (14).
In addition to reducing iron, the reductase has been shown to reduce copper in S. cerevisiae (22, 31), and its expression is regulated by copper. Copper is required for respiratory oxidases, the Cu/Zn superoxide dismutase (SOD), and laccase, an enzyme required for the production of the virulence factor melanin in C. neoformans. The FET3 oxidase contains copper, and so copper is required for iron uptake (2). The copper is transported by CCC2 across a post-Golgi compartment membrane for assembly with the FET3 apoprotein (54). In parallel with the iron uptake process, reduction is just the first step in the copper uptake process in S. cerevisiae. The copper ion is transported across the membrane by CTR1 (11). Expression of the cupric/ferric reductase and CTR1 is coordinately regulated by the transcription factor MAC1 (52).
Aberrant expression of any of these components could lead to dysregulated expression of the reductase. The S. cerevisiae mutant AFT1up has constitutive reductase and iron uptake in the presence of iron and so is markedly sensitive to high iron in the medium (51). The mutant MAC1up has constitutive reductase and copper uptake in the presence of copper and is sensitive to high copper in the medium; iron uptake is normal (22). In addition to mutations of the regulatory components, mutations in the structural components lead to reductase up regulation due to iron or copper deficiencies. High reductase activities were reported for a ctr1 mutant (13), for a fet3 fet4 double mutant (14), and for fet3 and ftr1 mutants (10a).
To study these components in C. neoformans, we isolated 18 mutants that are defective in their regulation of ferric reductase. The mutants represent four independent genetic loci. We examined the mutants’ reductase activities, iron uptake rates, growth physiology, and responses to oxidative stresses.
MATERIALS AND METHODS
Culture conditions and growth.
Cultures were stored on brain heart glucose agar slants (Scott Laboratories, Inc.), and liquid cultures were started in GYE (2% glucose–2% yeast extract broth; BBL, Becton, Dickinson and Co.). Limited iron medium (LIM) contained, per liter, 20 g of glucose, 5 g of asparagine, 400 mg of K2HPO4, 100 mg of MgSO4 · 7H2O, 50 mg of CaCl2 · 2H2O, 1 mg of thiamine, 57 μg of boric acid, 396 μg of CuSO4 · 5H2O, 72 μg of MnCl2 · 4H2O, 4.2 μg of ZnCl2, and 37 μg of (NH4)6Mo7O24 · 4H2O, buffered with 50 mM 2-(N-morpholino)ethanesulfonate acid-NaOH to pH 6.0, depleted of iron as described previously (41). For LIM plates, EDTA-purified Difco Bacto Agar (20 g/liter) was added. For iron repletion, 15 μM Fe(III) hydroxyethylenediaminetriacetate (FeHEDTA; Dow Chemicals) was added. All liquid cultures were grown with agitation at room temperature. Growth of cultures in exponential phase was measured by optical density at 700 nm, and generation times were determined by linear regression analysis of three independent optical density readings.
Mutagenesis.
We used serotype D strain B3501 from the National Institutes of Health collection as our wild-type (WT) strain (25). Cultures were treated with mutagenic UV light as previously described (25) in two independent treatments. Cells were harvested at 3-min intervals, and survival was studied. The time points selected in the first and second experiments gave 5 and 32% survival, respectively. The treated cells were grown in GYE to stationary phase and plated on LIM with 15 μM iron. When colonies reached 2-mm diameter, top agar containing bathophenanthroline disulfonate (BPDS; 1 mM), FeHEDTA (1 mM), and agar (Difco Bacto Agar; 1%) was adjusted to 45°C and poured over the colonies. Reduction of iron was indicated by a red color in and around the colonies. Red colonies were picked from a background population of white WT colonies and restreaked for further analysis.
Genetic analysis.
Crosses were made on hay infusion agar (44). Spores were collected with an inoculating loop, resuspended in 0.9% NaCl, and plated on Asp plates (3 g of glucose, 3 g of KHPO4, 1 g of asparagine, 0.5 MgSO4, 1 mg of thiamine, 20 g of Difco Bacto Agar) containing 15 μM FeHEDTA. The resulting colonies were overlaid with FeHEDTA-BPDS top agar and scored for iron reduction. Mutants were outcrossed to WT and a congenic MATa strain (15), and both MATα and MATa mutant progeny were collected.
Ferric reductase and cupric reductase assays.
Stationary-phase cultures grown in GYE were harvested, washed in LIM, and inoculated into 100 ml of LIM (15 μM CuSO4) with and without 15 μM FeHEDTA at a density of 106 cells/ml. Cultures were assayed every 2 h for 12 h and then every 12 h. For the assay (41), aliquots of cells were removed, mixed with FeHEDTA and BPDS (1 mM each), and incubated for 1 h prior to reading the A535 (ɛ = 22,140 M−1 cm−1 [10]).
Limited copper medium (LCM) contained 15 μM FeHEDTA and was supplemented where noted with 15 μM CuSO4. Cultures were grown as above, aliquots of cells with mixed with CuSO4 and bathocuproine disulfonate (BCDS) (1 mM each), and Cu reductase was measured by formation of the colored Cu(I)-BCDS complex at A478 (ɛ = 9,058 M−1 cm−1 [32]).
Statistical significance was determined by Student’s unpaired t test (Jandel Sigmaplot).
Ferrous iron uptake assays.
Cells grown in GYE were inoculated at a 1:1,000 dilution into LIM with or without 15 μM FeHEDTA, grown for 24 h, and washed twice in LIM. High-affinity ferrous uptake was measured with 55Fe in 1 μM carrier FeHEDTA in the presence of ascorbate and dithiothreitol (pH 6.0) (27).
Hyperbaric oxygen sensitivity.
Sensitivity to hyperbaric oxygen was determined as previously described (15). Strains grown to stationary phase in GYE were spotted on 1/3 brain heart infusion agar (13 g of brain heart infusion, 20 g of glucose, 20 g of Difco Bacto Agar) and grown for 24 h. These master plates were replica plated to two fresh plates and incubated for 6 h at 37°C either in ambient atmosphere or under 25 atm of 100% O2. The hyperbaric plates were removed from the high-pressure chamber and incubated at ambient conditions overnight prior to photography.
Oxidative stress assays.
Cultures were grown to late exponential phase in GYE, washed, and resuspended in 0.9% NaCl. Lawns were swabbed onto plates made from LICM (limited iron and copper medium, LIM without CuSO4) with additions of 500 μM BCDS, 15 μM CuSO4, 500 μM BPDS, 15 μM FeHEDTA, or 15 μM CuSO4 plus 15 μM FeHEDTA. Paper disks (0.25 in.; BBL) were inoculated with 10 μl of 30% H2O2 (Sigma) or 10 mM plumbagin (dissolved in 100% ethanol which was nonfungicidal; Sigma) and placed in the center of the plate (8). The diameters of the clear zones around the disks were measured after 3 days of growth at room temperature. Statistical significance was determined by Student’s unpaired t test (Jandel Sigmaplot).
RESULTS
Isolation of ferric reductase regulatory mutants.
C. neoformans reduces Fe(III) to Fe(II) at the plasma membrane prior to uptake. Iron in the medium down regulates this ferric reductase (41). To isolate regulatory mutants, we exposed WT cells to UV light and plated them on medium containing 15 μM iron. We then overlaid the colonies with top agar containing Fe(III) and the chromogenic Fe(II) chelator BPDS. Most colonies remained white; colonies developing a distinct red halo had dysregulated ferric reductase activity. We screened >20,000 colonies and found 1 red colony per ∼500 colonies. We picked and streak purified a total of 18 independent strains, but the mutagenesis may not have been saturating.
Genetic analysis of regulatory mutants.
Each strain was crossed to WT and was found to segregate in a 1:1 fashion (data not shown), as expected for a single mutation in a haploid genetic system. Progeny of both mating types were isolated. To determine the number of genetic loci represented by our mutants, we crossed each mutant to the others and screened progeny colonies for ferric reductase phenotypes. Crosses that yielded <2% WT progeny defined alleles of the same locus. Crosses that yielded ∼25% WT progeny defined segregation of two independent loci. By this means, we found that the 18 strains represented four independent loci (Table 1), named frr1 to frr4 (for ferric reductase regulatory mutant). Locus frr1 was represented by seven strains, frr2 was represented by eight strains, frr3 was represented by two strains, and frr4 was represented by one strain.
TABLE 1.
Percentages of WT colonies from genetic crosses of mutant frr strains
| Strain | % WT coloniesa
|
|||
|---|---|---|---|---|
| frr1606 | frr2608 | frr3632 | frr4616 | |
| frr1606 | <2 | 28 | 25 | 21 |
| frr2608 | <1 | 23 | 20 | |
| frr3632 | <1 | 24 | ||
| frr4616 | <2 | |||
<2, no WT in 40 to 100 colonies; <1%, no WT in 100 to 200 colonies.
Iron physiology of ferric reductase mutants.
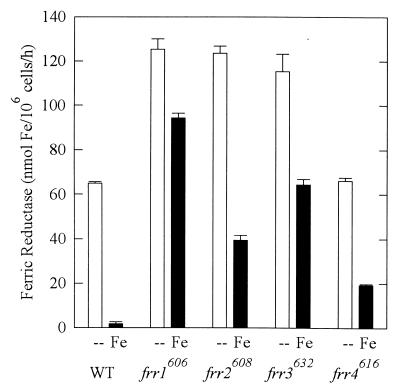
To confirm the phenotype of the mutant strains, we assayed the ferric reductase activities of WT and mutant strains in LIM with and without Fe(III). As shown in Fig. 1, WT cells produced substantial ferric reductase in the absence of iron. This activity peaked after several hours and then rapidly declined (41). Because the strains had different kinetics of ferric reductase induction, only the peak activity is reported. Strains frr1606, frr2608, and frr3632 expressed significantly higher than WT ferric reductase activity in the absence of iron (P < 0.05), while strain frr4616 produced a WT level of ferric reductase. When grown in the presence of iron, WT down regulated ferric reductase 30-fold, while the four mutant strains down regulated ferric reductase only 1.3-, 3.1-, 1.8-, and 3.4-fold. In this medium, all four mutant strains had significantly (57-, 24-, 39-, and 12-fold; P < 0.05) higher than WT ferric reductase activity.
FIG. 1.
Expression of ferric reductase in strains grown in LIM + 15 μM Cu(II) with no Fe (--) or with 15 μM Fe(III) (Fe). Ferric reductase was measured as the cell-mediated generation of the red Fe(II)-BPDS complex from Fe(III). Reductase was assayed every 2 h in exponential phase; each bar represents the peak of activity for each strain (mean ± standard deviation). The peaks occurred at the following time points in the absence and presence of Fe, respectively: WT, 12 and 0 h; frr1606 12 and 2 h; frr2608, 6 and 6 h; frr3632, 2 and 2 h; frr4616, 12 and 2 h.
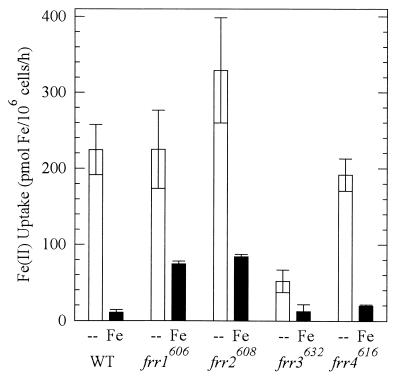
Uptake of iron across the plasma membrane of C. neoformans is also regulated by iron in the medium (27). To determine whether the mutant strains had dysregulated iron uptake, we measured high-affinity ferrous iron uptake of cells grown in LIM with and without Fe(III) (Fig. 2). In the absence of iron, WT took up Fe(II) at 200 pmol/106 cells/h; iron uptake rates of strains frr1606, frr2608, and frr4616 were not significantly different (P > 0.05). In contrast, the iron uptake rate of strain frr3632 was only 23% of the WT rate (P < 0.05). When grown in the presence of iron, WT down regulated iron uptake ∼20-fold, while the four mutant strains down regulated iron uptake only 3-, 4-, 4-, and 10-fold. When grown in iron, the residual rates of iron uptake by strains frr1606 and frr2608 were seven- and eightfold higher than the WT rate (P < 0.05). The rate of iron uptake by frr3632 was the same as the WT rate (P < 0.05). The rate for strain frr4616 was not statistically different from the WT rate.
FIG. 2.
Ferrous iron uptake of the strains grown in LIM (--) and LIM + 15 μM Fe(III) (Fe). Uptake was measured with 1 μM Fe, which represents the high-affinity system. All strains were assayed after 24 h of growth in defined medium, and values represent triplicate determinations (mean ± standard deviation). The experiment was repeated three times with identical relative uptake rates.
To determine the effects of the dysregulated ferric reductase and iron uptake phenotypes on growth of the mutants in low iron, we measured growth in medium containing the ferrous chelator BPDS. WT and strains frr1606, frr2608, and frr4616 grew with doubling times of 10 to 12 h (Table 2). Strain frr3632, however, grew with a doubling time of 27 h. To determine if high levels of iron were toxic in the face of high reductase, we measured the growth of the strains in medium supplemented with 5 mM FeHEDTA. Times for all strains varied only slightly from ∼5-h doubling times. To confirm the lack of iron sensitivity, the cells were grown on plates supplemented with disks containing 100 mM FeHEDTA or FeCl3. None of the strains exhibited clear zones—which would indicate inhibitory levels of iron—around the disks.
TABLE 2.
Generation times of WT and mutants in 500 μM BPDS and 5 mM Fe(III)
| Strain | Mean generation time (h) ± SD in:
|
|
|---|---|---|
| BPDS | 5 mM FeHEDTA | |
| WT | 10.1 ± 0.3 | 5.1 ± 0.2 |
| frr1606 | 11.9 ± 0.5 | 5.6 ± 0.4 |
| frr2608 | 10.3 ± 0.7 | 5.8 ± 0.7 |
| frr3632 | 27 ± 2 | 5.0 ± 0.1 |
| frr4616 | 11.9 ± 0.6 | 4.6 ± 0.0 |
Copper physiology.
Because the ferric reductase of S. cerevisiae has been reported to be a multifunctional enzyme with the ability to reduce Cu (22, 31), we tested the ability of C. neoformans cells to reduce Cu. When grown in the absence of Cu, the cells had cupric reductase activity (Table 3). This activity was down regulated 10-fold in the presence of 15 μM Cu. We then measured the ferric reductase activity of the copper-starved cells. Despite the fact that the cells had been grown in the presence of 15 μM Fe(III), the ferric reductase was active (Table 3). This activity was also down regulated by the addition of 15 μM Cu. The differences in the reduction rates of Fe(III) and Cu(II) substrates may be due to the difference in redox potential between Fe and Cu, or they may result from the use of HEDTA-chelated Fe versus unchelated Cu. Such cross-regulation supports the idea of a multifunctional reductase.
TABLE 3.
Cu regulation of Cu(II) and Fe(III) reductases in WT and frr1606
| Medium | Cu reductase (nmol of Cu/106 cells/h)
|
Fe reductase (nmol of Fe/106 cells/h)
|
||
|---|---|---|---|---|
| WT | frr1606 | WT | frr1606 | |
| No Cu + | 22.0 | 24.4 | 48.1 | 84.3 |
| 15 μM Fe | ||||
| 15 μM Cu + | 2.45 | 21.6 | 0.692 | 102.1 |
| 15 μM Fe | ||||
For additional support for this hypothesis, we examined the ferric reductase and cupric reductase activities of mutant strain frr1606. This mutant showed no down regulation of ferric reductase or cupric reductase by copper in the presence of iron (Table 3). This could indicate either that the ferric reductase and cupric reductases are distinct enzymes that are coregulated, with a regulatory mutation in strain frr1606, or that the reductase activities represent different activities of the same enzyme.
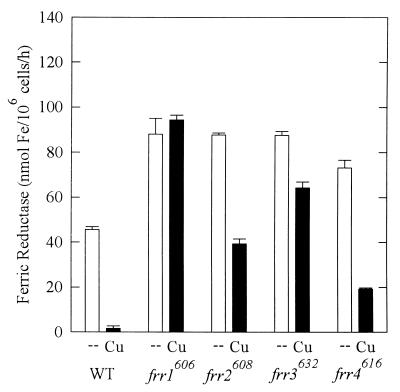
Ferric reductase activities, as a measure of the multifunctional reductase, were measured for all strains grown in LCM (15 μM Fe) with and without 15 μM CuSO4 (Fig. 3). The WT strain did not increase reductase expression in the absence of both copper and iron, which was presumably too deficient in metals to support maximal expression (data not shown). The mutant strains had significantly higher levels of reductase than WT in the absence of copper (P < 0.05). When grown in copper-containing media, WT cells down regulated the reductase 27-fold. Strain frr1606 maintained identical reductase levels in the absence and presence of copper. Strains frr2608 and frr4616 down regulated reductase 2.2- and 3.8-fold in the presence of copper. Strain frr3632 down regulated reductase only 1.4-fold. The four mutant strains had reductase activities 57-, 24-, 39-, and 12-fold greater than WT activity in the presence of copper (P < < 0.05).
FIG. 3.
Copper regulation of ferric reductase. Cells were grown in LIM + 15 μM Fe(III) with no copper (--) or with 15 μM CuSO4 (Cu). Ferric reductase was measured as described for Fig. 1. Reductase was assayed every 2 h in exponential phase; each bar represents the peak of activity for each strain (mean ± standard deviation). The peaks occurred at the following time points in the absence and presence of Cu, respectively: WT, 2 and 0 h; frr1606, 0 and 2 h; frr2608, 6 and 6 h; frr3632, 12 and 2 h; frr4616, 4 and 2 h.
To determine the effects of the reductase on copper physiology of the mutants, we measured the growth of the strains in medium rendered low in free copper by the addition of the Cu(I) chelator BCDS. WT and strains frr1606, frr2608, and frr4616 all grew with doubling times of 7 to 8 h; strain frr3632 grew slightly faster than WT, although statistical significance was borderline (P = 0.049) (Table 4). No strain grew significantly slower than WT. In this concentration of BCDS, all strains were equally affected and grew more slowly than in medium containing copper, demonstrating that copper limitation affected the cells. To stress the cells with copper, we grew the strains in 250 μM CuSO4. WT, frr3632, and frr4616 grew with doubling times of 4 to 5 h. The doubling time of strain frr2608 was slightly greater (7 h; P < 0.05), similar to its doubling time in BCDS. Strain frr1606, in contrast, required 13 h to double (P < 0.05); it seemed sensitive to high copper.
TABLE 4.
Generation times of WT and mutants in 500 μM BCDS and 250 μM Cu
| Strain | Mean generation time (h) ± SD in:
|
|
|---|---|---|
| BCDS | 250 μM CuSO4 | |
| WT | 7.8 ± 0.6 | 5.1 ± 0.2 |
| frr1606 | 8.5 ± 0.2 | 13 ± 3 |
| frr2608 | 8.3 ± 0.0 | 7.0 ± 0.2 |
| frr3632 | 6.7 ± 0.2 | 4.2 ± 0.3 |
| frr4616 | 8.2 ± 0.2 | 5.2 ± 0.2 |
Oxidative stress physiology.
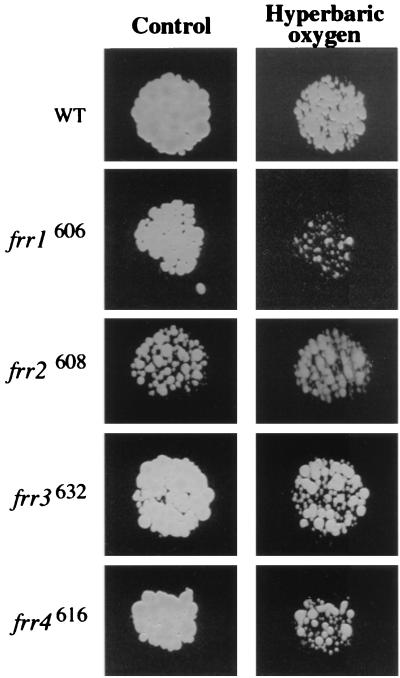
We reasoned that high reductase activity may lead to cell damage by reaction of reduced metals—either copper or iron—with oxidants to form highly damaging hydroxyl radicals (35). Since C. neoformans requires oxygen for respiration and cannot be grown anaerobically, we compared our mutants at ambient and hyperbaric oxygen (100% oxygen at 25 atm of pressure and 37°C [15]). Hyperbaric treatment is thought to produce superoxide radicals that dismutate to H2O2 and hydroxyl radical (42). For this experiment, the strains were grown on brain heart infusion agar (15). After 6 h of hyperbaric oxygen treatment, WT grew only slightly more slowly than its control (Fig. 4). In contrast, strains frr1606, frr3632, and frr4616 grew much more slowly after hyperbaric oxygen treatment than their controls or WT cells. This phenotype is similar to the Oxy− phenotypes of previously described mutant strains of C. neoformans (15). Strain frr2608, interestingly, grew more slowly than the other strains on the brain heart infusion agar but did not appear to be sensitive to hyperbaric oxygen.
FIG. 4.
Hyperbaric oxygen sensitivity of wild-type and regulatory mutants. Strains were grown on brain heart infusion agar overnight. Replicas were made, and the plates were incubated for 6 h in ambient atmosphere (control) or hyperbaric O2 (100% oxygen at 25 atm) and then grown for an additional 18 h in ambient atmosphere at 37°C.
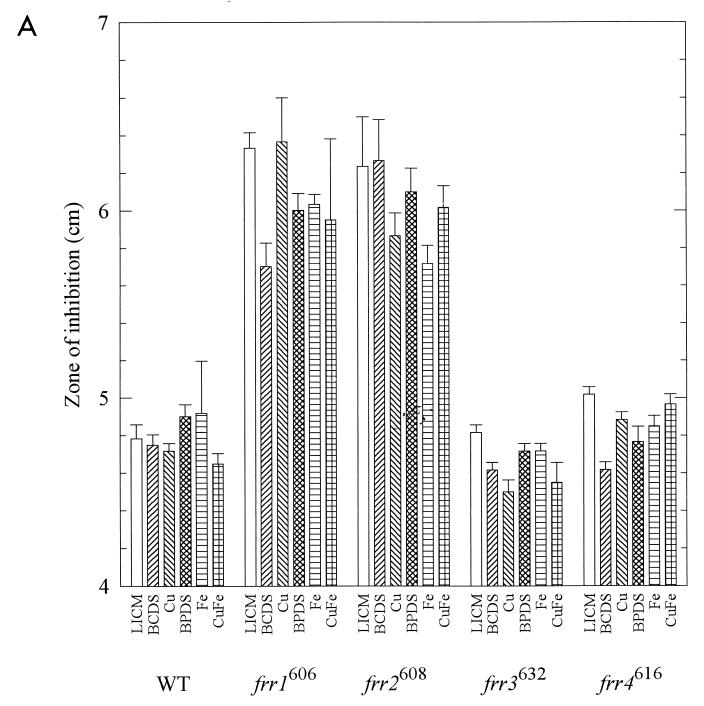
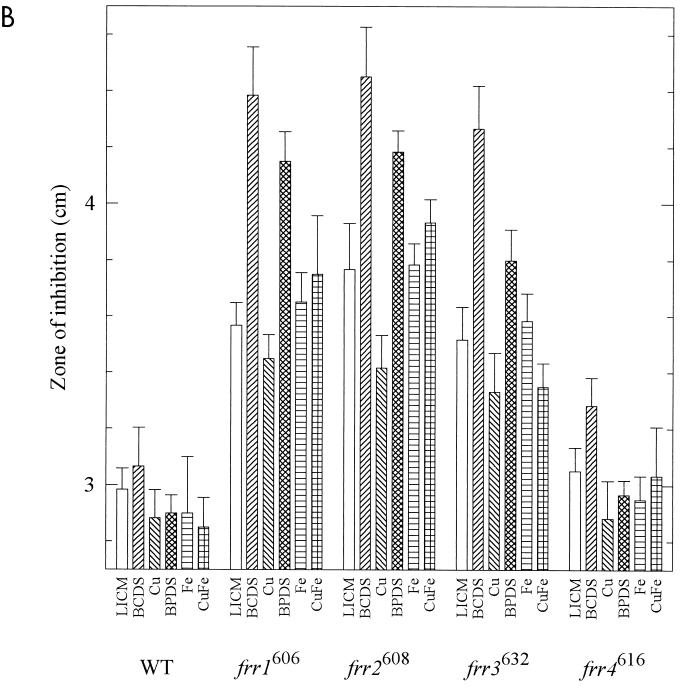
We next tested the sensitivity of the strains to oxidants on defined media in the ambient atmosphere. To evaluate sensitivities under many metal conditions, we used disk diffusion assays (8). Strains were grown in GYE medium. Lawns were streaked on LICM with or without BCDS, BPDS, Cu, and Fe, and disks containing the oxidants were applied. As the lawns of cells grew up, clear zones, indicating inhibitory levels of oxidant, were measured (Fig. 5). Small differences in the diameters of the clear zones (on the order of millimeters) were statistically significant in some cases, but the physiological significance of this is unknown. Since the largest deviation from the mean was 0.43 cm, we considered a difference of 0.5 cm between means significant.
FIG. 5.
Sensitivity of strains to oxidants. Cultures grown in GYE were swabbed on LICM with various additions: 500 μM BCDS, 15 μM Cu, 500 μM BPDS, 15 μM Fe, and 15 μM Cu plus 15 μM Fe. Disks containing 10 μl of 30% H2O2 (A) or 15 μl of 10 mM plumbagin (B) were applied to the center of each plate. Three measurements of the diameter of the clear zones were made after 3 days of growth at room temperature (mean ± standard deviation).
H2O2 can react with reduced metals to generate hydroxyl radicals. We found that frr1606 was more sensitive than WT to H2O2 on all media but was less sensitive when grown on BCDS than on LICM. Strain frr2608 was more sensitive than WT to H2O2 on all media but was less sensitive when grown on 15 μM Fe than on LICM. Strains frr3632 and frr4616 were similar to WT (Fig. 5A) and had similar sensitivities on all media. Addition of 15 μM Cu, Fe, or both did not increase the sensitivity of any strain to H2O2. Thus metals, albeit at modest levels, did not always potentiate oxidative stress.
The redox-cycling agent plumbagin is thought to promote generation of superoxide (48). We found that frr1606 was more sensitive than WT to plumbagin on all media (Fig. 5B). When comparing the different media for each strain, we found that strain frr1606 was more sensitive to plumbagin on BCDS and BPDS than on LICM. Strain frr2608 was more sensitive than WT to plumbagin on all media. Growth on BCDS increased its sensitivity relative to LICM. In a complex pattern, strain frr3632 was more sensitive than WT to plumbagin on LICM, BCDS, BPDS, and Fe-containing media but not on Cu medium and was at the borderline of sensitivity for Cu-Fe medium. It was significantly more sensitive on BCDS than on LICM. Strain frr4616 was not significantly more sensitive than WT to plumbagin on any of the media; it appeared slightly more sensitive on BCDS, but the zone size did not reach our level of significance.
Genetic linkage of reductase and oxidant-sensitive phenotypes.
To demonstrate that the oxidant-sensitive phenotypes are genetically linked to the reductase phenotypes, we crossed each mutant to a WT MATa strain and isolated approximately equal numbers of white progeny with WT reductase activities and red progeny with mutant reductase activities. Strain frr1606 progeny were tested for hyperbaric oxygen sensitivity; the 50 WT progeny were all resistant to hyperbaric oxygen, while 43 red mutant progeny were all sensitive. As a check, they were also tested for H2O2 sensitivity on medium containing Fe. The white WT progeny had inhibition zones of 5.0 ± 0.17 cm, while the red mutant progeny had zones of 6.2 ± 0.18 cm (P < 0.05). These values are identical to those obtained for WT and parental strain frr1606 as shown in Fig. 5A. Strain frr2608 progeny were tested for sensitivity to H2O2 on LICM medium. The 51 white WT progeny had inhibition zones of 4.8 ± 0.21 cm, while the 39 red mutant progeny had zones of 5.4 ± 0.26 cm. While the value for the WT progeny is identical to the value shown for WT in Fig. 5A, the red progeny were slightly less sensitive in this experiment than parental strain frr2608 shown in Fig. 5A but were still significantly different from the WT progeny (P < 0.05). Strain frr3632 progeny were tested for hyperbaric oxygen sensitivity; 45 white WT progeny were resistant and 42 red mutant progeny were sensitive. They were also tested for plumbagin sensitivity on medium containing 500 μM BCDS. The white WT progeny gave inhibition zones of 3.2 ± 0.26 cm, while the red mutant progeny gave zones of 4.2 ± 0.22 cm (P < 0.05). These values are identical to those obtained for WT and parental strain frr3632 as shown in Fig. 5B. Strain frr4616 progeny were tested by hyperbaric oxygen sensitivity. The 50 white WT progeny were all resistant to hyperbaric oxygen, while the 46 red mutant progeny were sensitive. The results showed that only the progeny with mutant reductase activities were oxidant sensitive, thereby demonstrating genetic linkage of these two phenotypes.
DISCUSSION
In this report, we describe strains with mutations at four genetic loci (frr1 to -4). All of the mutant strains had significantly higher than WT ferric reductase activity in the presence of iron, and three (frr1606, frr2608, and frr3632) also had higher than WT ferric reductase activities in the absence of iron. Thus, these mutants were always more derepressed than WT, similar to the fur regulatory mutants of Escherichia coli (21). A second possibility is that the mutants were expressing a second reductase; S. cerevisiae does have ferric reductase FRE2 in addition to FRE1 (18). Since many of the steps in iron uptake have been characterized genetically in S. cerevisiae, we will compare the phenotypes of mutants defined in that model organism to our four mutants. We recognize that, unlike S. cerevisiae, C. neoformans must always obtain sufficient copper and iron to carry out respiration and that S. cerevisiae is an ascomycete whereas C. neoformans is an evolutionarily distant basidiomycete.
Analysis of the mutants leads us to the conclusion that the ferric reductase of C. neoformans is bifunctional, using both Fe(III) and Cu(II) as substrates, as in S. cerevisiae (22, 31). We present three lines of evidence. First, the ferric reductase was active in the presence of iron if copper is absent from the medium. Second, this ferric reductase activity was down regulated by copper, and a copper reductase activity was regulated in parallel. Third, one of the ferric reductase mutants (selected at random) had high activity in the presence of both iron and copper. Its copper reductase was similarly unregulated by copper. Thus, we infer that the reductase is regulated by both copper and iron levels in a complex synergy.
Strain frr1606 appears to be a regulatory mutant. While its elevated levels of reductase and iron uptake in the presence of iron are similar to those for the constitutive mutant AFT1up of S. cerevisiae (51), growth of our strain was not affected by 5 mM Fe, whereas AFT1up was sensitive to 500 μM Fe. Strain frr1606 was capable of some down regulation of iron uptake; perhaps this degree of down regulation is sufficient to protect cells from iron overload, or perhaps efficient intracellular sequestration and damage control prevented measurable effects of iron overload. The elevated reductase and growth sensitivity to Cu are phenotypes similar to those of the copper uptake regulatory mutant of S. cerevisiae, MAC1up (29). Although the copper uptake rate of frr1606 has not been determined, this hypothesis does not explain why this strain had dysregulated iron uptake; perhaps the Cu sensitivity stemmed from the high iron uptake. The sensitivity of the strain to hyperbaric oxygen, superoxide-generating plumbagin, and hydrogen peroxide could be due to either Fe or Cu accumulation. We predicted that the strain would be more sensitive to oxidants in the presence of iron since it has elevated iron uptake. However, the addition of iron had no effect on the H2O2 or superoxide sensitivity of frr1606, but the iron chelator BPDS increased the sensitivity of frr1606 to superoxide. This is similar to the finding that in E. coli SOD-deficient strains, iron supplementation decreased superoxide sensitivity (4). Similarly, the addition of copper did not increase oxidant sensitivities; the concentration of copper may have been too low for such an effect to be observed. We did find that the copper chelator BCDS increased the sensitivity of frr1606 to superoxide, perhaps by sequestering copper required for SOD. The same chelator decreased sensitivity of the strain to H2O2, perhaps by decreasing intracellular levels of reactive copper.
Strain frr2608, similarly to frr1606, showed little down regulation of ferric reductase by Fe or Cu and little regulation of ferrous uptake by Fe, and growth was sensitive neither to high iron nor to high copper. In contrast to the other strains, frr2608 was not sensitive to hyperbaric oxygen at 37°C but was sensitive to H2O2 and superoxide on all defined media at room temperature. BCDS increased the sensitivity of frr2608 to superoxide, perhaps by sequestering copper required for SOD. Although this strain also has high iron uptake, the addition of iron decreased sensitivity of frr2608 to H2O2, perhaps by increasing levels of iron-requiring catalase. The addition of iron had no effect on its superoxide sensitivity. While these phenotypes are consistent with mutations in AFT1 or MAC1, they are also consistent with the phenotypes of mutations in YFH1 and SSC2 (3, 30), in which reductase and iron uptake are increased because intracellular iron is sequestered in the mitochondrion. Thus incubation at 37°C in combination with excess iron could cause sufficient oxidative stress to damage mitochondrial proteins and DNA. The slow growth may reflect reduced electron flow in mitochondria that would prevent the formation of oxygen free radicals under hyperbaric oxygen conditions. In this hypothesis, supplementation with iron could reduce sensitivity to H2O2 by increasing cytoplasmic iron for catalase.
Strain frr3632 is clearly an iron uptake mutant, transporting iron at only 23% of the WT rate and growing more slowly than WT in medium containing the ferrous iron chelator BPDS. Supplementation with Fe corrected its deficiency, perhaps allowing it to acquire iron through the low-affinity uptake system (27). However, the reductase activity was only slightly decreased by growth with Fe and Cu. These phenotypes are similar to those of the fet3 and ftr1 mutants of S. cerevisiae (46), with high reductase expression due to poor iron uptake. Strain frr3632 does not seem to be a copper transport mutant, homologous to ctr1, atx1, or ccc2 mutants (13, 11, 34, 54), since it grew in low copper, and addition of copper did not correct the iron uptake defect (data not shown). Strain frr3632 was sensitive to hyperbaric oxygen, and to superoxide on most media, suggesting that iron is required for detoxification of superoxide, although iron supplementation (to 15 μM) did not correct the sensitivity to superoxide. While an Fe-containing SOD has not been found in fungi, iron supplementation increases the superoxide resistance of superoxide dismutase mutants of E. coli (4). BCDS increased sensitivity of frr3632 to superoxide, but Cu decreased sensitivity to a level not significantly different from the WT level. Strikingly, this strain was resistant to H2O2 on all media, which is puzzling since detoxification of this molecule requires iron-containing catalase. The level of catalase in this strain has not been determined.
Strain frr4616 down regulated ferric reductase only three- to fourfold in response to Fe and Cu and exhibited normal iron uptake and normal growth in high Cu and Fe. The reason for its high reductase activity is not known; perhaps it has an up mutation in the promoter of the reductase gene. It was sensitive neither to hydrogen peroxide nor to superoxide. We conclude that the activity of the extracellular reductase is not sufficient to promote oxidant sensitivity: intracellular metabolic pathways must also be perturbed.
Why are four genes involved in the regulation of the reductase? First, because the reductase is bifunctional for copper and iron reduction, both iron-responsive and copper-responsive regulatory genes may be required. Second, in order for iron uptake to occur, copper must be assembled into FET3, a multicopper oxidase, and in order for the reductase to reduce copper, iron must be assembled into the heme cofactor (45). Third, there may be additional regulators of reductase, such as a global regulator of both iron acquisition and oxidative stress responses. All of these requirements ensure that the expression of reductase will be complex. Correlating cryptococcal genes with homologues in S. cerevisiae will require molecular characterization; we are currently transforming the mutant strains with a wild-type genomic DNA library and screening for complementation of the mutant phenotype.
Paradoxically, both metal deficiency and metal overload have been shown to promote oxidative stress. Iron-deficient E. coli strains are oxidant sensitive due to insufficient FeSOD (20). The atx1 mutant of S. cerevisiae is oxidant sensitive due to its inability to deliver copper to cytoplasmic targets (33). On the other hand, metal overload may stress cells when metals react with oxidants to generate toxic free radicals that cause lipid peroxidation and DNA damage (47); thus, organisms also have regulatory strategies to minimize metal-induced oxidative stress. In humans with hemochromatosis, overexpression of the intestinal ferric reductase (43) and dysregulation of transferrin uptake (16) cause iron overload and multiorgan disease, apparently through oxidative damage (19). Normally, mammalian iron metabolism is posttranscriptionally regulated by oxidative stress to decrease iron uptake (6). In bacteria, constitutive Δfur (ferric uptake regulator) mutants have iron overload and increased sensitivity to oxidative stress (49). The bacterial iron uptake regulatory protein Fur also regulates SOD gene expression at the level of transcription (40).
The study of oxidative stress responses in these mutants of iron metabolism is important, as C. neoformans is subject to oxidative attack by neutrophils (7) and certain macrophages (32). It may also be subject to nitrogen-based oxidants, secreted by certain macrophages, that specifically react with iron (1). On that basis, the overlap of oxidant sensitivity and the various frr-associated phenotypes provides a model for fungal interaction with immune effectors in vivo. Indeed, the finding that frr1606 is allelic to the hypovirulent, oxidant-sensitive oxy1 strain (28) suggests that regulation of the iron acquisition system is critical in the host-pathogen interaction.
ACKNOWLEDGMENTS
The Department of Veterans Affairs supported this research.
We are grateful to Amy T. Wilborn for development of the mutant screen.
REFERENCES
- 1.Alspaugh J A, Granger D L. Inhibition of Cryptococcus neoformans replication by nitrogen oxides supports the role of these molecules as effectors of macrophage-mediated cytostasis. Infect Immun. 1991;59:2291–2296. doi: 10.1128/iai.59.7.2291-2296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askwith C, Eide D, Van Ho A, Bernard P S, Li L, Davis-Kaplan S, Sipe D M, Kaplan J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 1994;76:403–410. doi: 10.1016/0092-8674(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 3.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 4.Benov L, Fridovich I. Growth in iron-enriched medium partially compensates Escherichia coli for the lack of manganese and iron superoxide dismutase. J Biol Chem. 1998;273:10313–10316. doi: 10.1074/jbc.273.17.10313. [DOI] [PubMed] [Google Scholar]
- 5.Boelaert J R, Weinberg G A, Weinberg E D. Altered iron metabolism in HIV infection: mechanisms, possible consequences, and proposals for management. Infect Agents Dis. 1996;5:36–46. [PubMed] [Google Scholar]
- 6.Cairo G, Castrusini E, Minotti G, Bernelli-Zazzera A. Superoxide and hydrogen peroxide-dependent inhibition of iron regulatory protein activity: a protective stratagem against oxidative injury. FASEB J. 1996;10:1326–1335. doi: 10.1096/fasebj.10.11.8836047. [DOI] [PubMed] [Google Scholar]
- 7.Chaturvedi V, Wong B, Newman S L. Oxidative killing of Cryptococcus neoformans by human neutrophils. J Immunol. 1996;156:3836–3840. [PubMed] [Google Scholar]
- 8.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen C N, Kelley M, Hobbs M M, Anderson J E, Cannon J G, Cohen M S, Sparling P F. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol Microbiol. 1998;27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 10.Cox C D. Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol. 1994;235:315–329. doi: 10.1016/0076-6879(94)35150-3. [DOI] [PubMed] [Google Scholar]
- 10a.Dancis, A. Personal communication.
- 11.Dancis A, Haile D, Yuan D S, Klausner R D. The Saccharomyces cerevisiae copper transport protein (Ctr1p) J Biol Chem. 1994;269:25660–25667. [PubMed] [Google Scholar]
- 12.Dancis A, Roman D G, Anderson G J, Hinnebusch A G, Klausner R D. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc Natl Acad Sci USA. 1992;89:3869–3873. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dancis A, Yuan D S, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner R D. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- 14.Dix D R, Bridgham J T, Broderius M A, Byersdorfer C A, Eide D J. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 15.Emery H S, Shelburne C P, Bowman J P, Fallon P G, Schulz C A, Jacobson E S. Genetic study of oxygen resistance and melanization in Cryptococcus neoformans. Infect Immun. 1994;62:5694–5697. doi: 10.1128/iai.62.12.5694-5697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feder J N, Penny D M, Irrinki A, Lee V K, Lebron J A, Watson N, Tsuchihashi Z, Sigal E, Bjorkman P J, Schatzman R C. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA. 1998;95:1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman M, Fica A, Saxena M, Di Fabio J L, Cabello F C. Salmonella typhi iron uptake mutants are attenuated in mice. Infect Immun. 1994;62:4091–4094. doi: 10.1128/iai.62.9.4091-4094.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgatsou E, Mavrogiannis L A, Fragiadakis G S, Alexandraki D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J Biol Chem. 1997;272:13786–13792. doi: 10.1074/jbc.272.21.13786. [DOI] [PubMed] [Google Scholar]
- 19.Gutteridge J M C, Rowley D A, Griffiths E, Halliwell B. Low-molecular-weight iron complexes and oxygen radical reactions in idiopathic haemochromatosis. Clin Sci. 1985;68:463–467. doi: 10.1042/cs0680463. [DOI] [PubMed] [Google Scholar]
- 20.Haas A, Goebel W. Microbial strategies to prevent oxygen-dependent killing by phagocytes. Free Radical Res Commun. 1992;16:137–157. doi: 10.3109/10715769209049167. [DOI] [PubMed] [Google Scholar]
- 21.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 22.Hassett R, Kosman D J. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J Biol Chem. 1995;270:128–134. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 23.Henderson D P, Payne S M. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun. 1994;62:5120–5125. doi: 10.1128/iai.62.11.5120-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershko C. Iron, infection and immune function. Proc Nutr Soc. 1993;52:165–174. doi: 10.1079/pns19930048. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson E S, Ayers D J, Harrell A C, Nicholas C C. Genetic and phenotypic characterization of capsule mutants of Cryptococcus neoformans. J Bacteriol. 1982;25:415–418. doi: 10.1128/jb.150.3.1292-1296.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson E S, Emery H S. Temperature regulation of the cryptococcal phenoloxidase. J Med Vet Mycol. 1991;29:121–124. doi: 10.1080/02681219180000201. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson E S, Goodner A P, Nyhus K J. Ferrous iron uptake in Cryptococcus neoformans. Infect Immun. 1998;66:4169–4175. doi: 10.1128/iai.66.9.4169-4175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson E S, Nyhus K J. Abstracts of the IDSA 36th Annual Meeting 1998. Chicago, Ill: Infectious Diseases Society of America; 1998. Iron phenotypes of cryptococcal Oxy mutants, abstr. 333; p. 983. [Google Scholar]
- 29.Jungmann J, Reins H-A, Lee J, Romeo A, Hassett R, Kosman D, Jentsch S. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993;12:5051–5056. doi: 10.1002/j.1460-2075.1993.tb06198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight S A B, Sepuri N B V, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 31.Lesuisse E, Labbe P. Reductive iron assimilation in Saccharomyces cerevisiae. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 149–178. [Google Scholar]
- 32.Levitz S M. Macrophage-Cryptococcus interactions. In: Zwilling B S, Eisenstein T K, editors. Macrophage-pathogen interactions. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 533–543. [Google Scholar]
- 33.Lin S, Culotta V C. The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci USA. 1995;92:3784–3788. doi: 10.1073/pnas.92.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S, Pufahl R A, Dancis A, O’Halloran T V, Culotta V C. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem. 1997;272:9215–9220. [PubMed] [Google Scholar]
- 35.Lubec G. The hydroxyl radical: from chemistry to human disease. J Investig Med. 1996;44:324–346. [PubMed] [Google Scholar]
- 36.Lynch S M, Frei B. Reduction of copper, but not iron, by human low density lipoprotein (LDL) J Biol Chem. 1995;270:55158–5163. doi: 10.1074/jbc.270.10.5158. [DOI] [PubMed] [Google Scholar]
- 37.Molez J F. The historical question of acquired immunodeficiency syndrome in the 1960s in the Congo River basin area in relation to cryptococcal meningitis. Am J Trop Med Hyg. 1998;58:273–276. doi: 10.4269/ajtmh.1998.58.273. [DOI] [PubMed] [Google Scholar]
- 38.Moyo V M, Gangaidzo I T, Gordeuk V R, Kiire C F, Macphail A P. Tuberculosis and iron overload in Africa: a review. Cent Afr J Med. 1997;43:334–339. [PubMed] [Google Scholar]
- 39.Moyo V M, Mandishona E, Hasstedt S J, Gangaidzo I T, Gomo Z A, Khumalo H, Saungweme T, Kiire C F, Paterson A C, Bloom P, Macphail A P, Rouault T, Gordeuk V R. Evidence of genetic transmission in African iron overload. Blood. 1998;91:1076–1082. [PubMed] [Google Scholar]
- 40.Niederhoffer E C, Naranjo C M, Bradley K L, Fee J A. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyhus K J, Wilborn A T, Jacobson E S. Ferric iron reduction by Cryptococcus neoformans. Infect Immun. 1997;65:434–438. doi: 10.1128/iai.65.2.434-438.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park M K, Myers R A M, Marzella L. Oxygen tensions and infections: modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin Infect Dis. 1992;14:720–740. doi: 10.1093/clinids/14.3.720. [DOI] [PubMed] [Google Scholar]
- 43.Raja K B, Pountney D, Bomford A, Przemioslo R, Sherman D, Simpson R J, Williams R, Peters T J. A duodenal mucosal abnormality in the reduction of Fe(III) in patients with genetic haemochromatosis. Gut. 1996;38:765–769. doi: 10.1136/gut.38.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raper K B. Growth and development of Dictyostelium discoideum with different bacterial associates. J Agric Res. 1937;55:289–316. [Google Scholar]
- 45.Shatwell K P, Dancis A, Cross A R, Klausner R D, Segal A W. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J Biol Chem. 1996;271:14240–14244. doi: 10.1074/jbc.271.24.14240. [DOI] [PubMed] [Google Scholar]
- 46.Stearman R, Yuan D S, Yamaguchi-Iwai Y, Klausner R D, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 47.Stohs S J, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radical Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 48.Storz G, Toledano M B. Regulation of bacterial gene expression in response to oxidative stress. Methods Enzymol. 1994;236:196–207. doi: 10.1016/0076-6879(94)36017-0. [DOI] [PubMed] [Google Scholar]
- 49.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vartivarian S E, Anaissie E J, Cowart R E, Sprigg H A, Tingler M J, Jacobson E S. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi-Iwai Y, Dancis A, Klausner R D. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi-Iwai Y, Serpe M, Haile D, Yang W, Kosman D J, Klausner R D, Dancis A. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J Biol Chem. 1997;272:17711–17718. doi: 10.1074/jbc.272.28.17711. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner R D. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan D S, Stearman R, Dancis A, Dunn T, Beeler T, Klausner R D. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc Natl Acad Sci USA. 1995;92:2632–2636. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]