Abstract
Cancer and other chronic diseases are marked by alterations in the protein quality control system, affecting the posttranslational destiny of various proteins that regulate, structure, and catalyze cellular processes. Cellular chaperones, also known as heat shock proteins (HSPs), are pivotal in this system, performing protein triage that often determines the fate of proteins they bind to. Grasping the regulatory mechanisms of HSPs and their associated cofactors is crucial for understanding protein quality control in both healthy and diseased states. Recent research has shed light on the interactions within the protein quality control system and how post-translational modification govern protein interactions, function, and localization, which can drive or inhibit cell proliferation. This body of work encompasses critical elements of the heat shock response, including heat shock protein 70, heat shock protein 90, carboxyl-terminus of HSC70 interacting protein, and heat shock protein organizing protein. This review aims to synthesize these advancements, offering a holistic understanding of the system and its response when commandeered by diseases like cancer. We focus on the mechanistic shift in co-chaperone engagement—transitioning from heat shock protein organizing protein to carboxyl-terminus of HSC70 interacting protein in association with heat shock protein 70 and heat shock protein 90—which could influence cellular growth and survival pathways. A comprehensive examination of posttranslational modification–driven regulation within the protein quality control network is presented, highlighting the roles of activation factors, chaperones, and co-chaperones. Our insights aim to inform new strategies for therapeutically targeting diseases by considering the entire heat shock response system.
Keywords: heat shock proteins, cancer, posttranslational modifications, co-chaperones, protein quality control
Protein quality control and the heat shock response system
The cellular protein quality control (PQC) system is a complex network of enzymes and organelles crucial for maintaining cellular functions and responding to various stressors (1, 2). Within the PQC system, the heat shock response system (HSR) is critical in managing cellular stress, including heat, oxidative, metabolic, and hypoxia (3). The HSR operates through a sophisticated interplay of molecular chaperones, co-chaperones, transcription factors, and co-factors to ensure protein homeostasis and effectively respond to stressors (4). Central to the HSR are molecular chaperones like heat shock protein 70 (HSP70) and heat shock protein 90 (HSP90), which work in concert with co-chaperones to triage proteins for refolding or degradation (Fig. 1A) (5, 6, 7).
Figure 1.
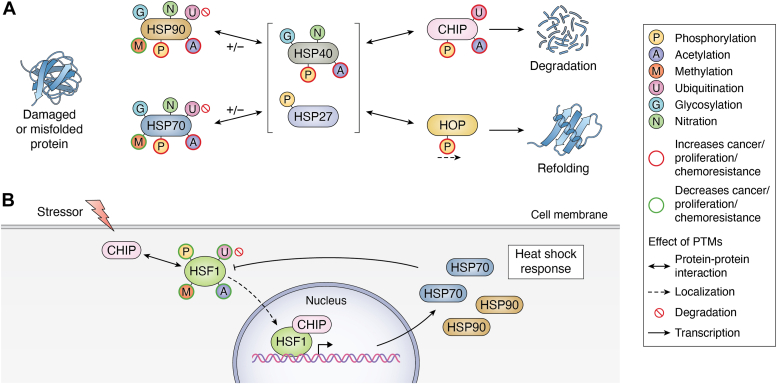
Mechanisms of the heat shock response and its implications in cancer.A, damaged or misfolded proteins are triaged by HSP70 or HSP90, with or without co-chaperones HSP40 or HSP27. The preference for CHIP versus HOP determines the fate of substrates, leading to either degradation or refolding. B, a stressor promotes the interaction of CHIP with HSF1 to initiate the HSR, leading to HSF1’s translocation to the nucleus. HSF1 then activates the transcription of HSR components. Posttranslational modifications (PTMs) on HSR components can influence protein–protein interactions, localization, degradation, and transcription. These PTMs can also affect cancer proliferation and chemoresistance.
The HSR is finely regulated by transcription factors such as heat shock factor 1 (HSF1), which orchestrates the transcriptional response to stress by upregulating various chaperones (8, 9). Upon stress induction, HSF1 changes protein–protein interactions and post-translational modifications (PTMs), leading to its activation and subsequent binding to DNA to promote the transcription of HSR proteins (Fig. 1B) (10). Notably, a negative feedback loop involving the chaperone HSP70 and co-chaperone heat shock protein 40 (HSP40, or J-domain proteins) regulates HSF1 activity, preventing excessive transcription of HSR genes (Fig. 1B) (11). Furthermore, PTMs play a significant role in modulating the HSR, with studies highlighting the impact of PTMs such as phosphorylation, acetylation, methylation, ubiquitination, glycosylation, and nitration on the system (Fig. 1) (12).
The HSR system operates through two primary chaperone complexes, anchored by HSP70 and HSP90, each utilizing a network of co-chaperones that influence whether proteins are directed toward refolding or degradation pathways (Fig. 1A) (13). In cases where degradation is necessary, the ubiquitin-proteasome system is the primary mechanism for disposing of damaged or misfolded proteins marked by ubiquitin (14, 15). The HSR system can also utilize chaperone-mediated autophagy and the lysosomal pathway to degrade larger aggregated substrates, further contributing to cellular proteostasis (14, 16). The balance between refolding and degradation of chaperone-bound substrates is critical for maintaining cellular PQC and overall cellular health (17).
The HSR is not confined to responding solely to heat stress but is also activated by various other stressors like infection or oxidants, showcasing its adaptability in combating different types of cellular stress (18). The intricate regulation of the HSR involves a network of molecular interactions and signaling cascades that ensure a timely and appropriate response to stress conditions (19, 20). Moreover, the involvement of co-chaperones like the carboxyl-terminus of HSP interacting protein (CHIP) and HSP organizing protein (HOP) in fine-tuning protein triage underscores the complexity of the HSR system and its role in maintaining cellular proteostasis (Fig. 1) (12, 21, 22, 23).
The HSR system represents a sophisticated cellular defense mechanism pivotal in managing PQC and responding to diverse stressors. Through the coordinated actions of molecular chaperones, co-chaperones, transcription factors, and PTMs, the HSR ensures the maintenance of proteostasis and cellular health in challenging environmental conditions. Understanding the intricate mechanisms underlying the HSR sheds light on fundamental cellular processes and holds promising implications for therapeutic interventions targeting PQC pathways in various diseases.
While previous reviews have provided valuable insights into the PTMs of individual HSR components (7, 8, 24, 25), our review aims to present a comprehensive mechanistic model that integrates these modifications within a broader system. Focusing on the pivotal molecular switch involving HSP70 and HSP90 with CHIP and HOP, we highlight the nuanced regulation of protein triage and the diverse PTMs that influence the HSR. Additionally, we explore the functional implications of HSR activity on cancer cell dynamics and discuss recent pharmacological advancements targeting HSR components and specific PTMs. This review identifies critical knowledge gaps (bold and italicized text) and emphasizes balancing CHIP- and HOP-containing chaperone complexes (underlined text).
Heat shock protein 70
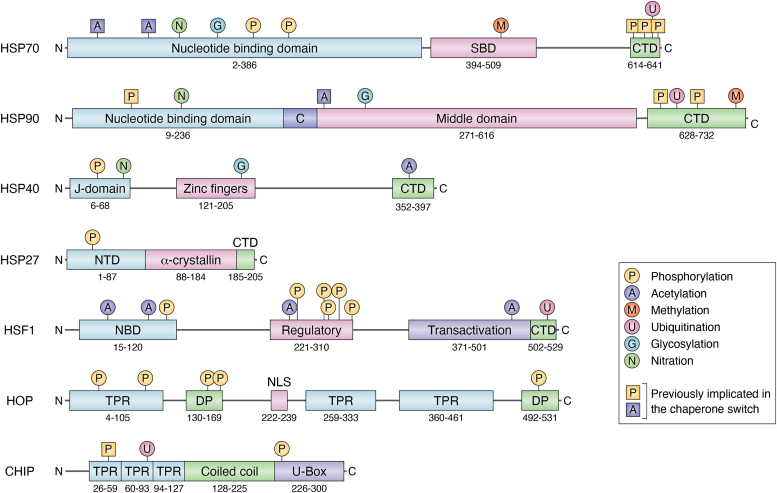
HSP70 is a crucial chaperone protein that plays a significant role in PQC within the cell. Structurally, HSP70 consists of a nucleotide-binding domain, a substrate-binding domain, and a C-terminal domain (CTD) that harbors essential regulatory elements like the lid domain and the EEVD motif for co-chaperone interactions (Fig. 2) (6). One of the primary functions of HSP70 is to recognize misfolded proteins by binding to exposed hydrophobic amino acid residues (4). Refolding of the substrate is catalyzed by repeated cycles of HSP70-substrate binding and release in an ATP-dependent manner (26). The energy released during ATP hydrolysis facilitates the refolding of the substrate, with the subsequent hydrolysis of ATP to ADP stabilizing the protein in place (27).
Figure 2.
Components of the heat shock response and posttranslational modifications. The figure illustrates various domains involved in the HSR, including the substrate-binding domain (SBD), C-terminal domain (CTD), charged domain (C), N-terminal domain (NTD), nucleotide-binding domain (NBD), tetratricopeptide repeat domains (TPR), Asp-Pro rich domain (DP), and nuclear localization sequence (NLS). Posttranslational modifications (PTMs) that impact the co-chaperone switch are indicated with squares. The domain boundaries for each protein are specified according to the following UniProt accessions: HSP70 (P11142 · HSP7C_HUMAN), HSP90 (P07900 · HS90A_HUMAN), HSP40 (P31689 · DNJA1_HUMAN), HSP27 (P04792 · HSPB1_HUMAN), HSF1 (Q00613 · HSF1_HUMAN), HOP (P31948 · STIP1_HUMAN), and CHIP (Q9UNE7 · CHIP_HUMAN).
Moreover, the hydrolysis of ATP by HSP70 is a relatively slow process, necessitating the involvement of co-chaperones like HOP and J-domain proteins to stimulate ATP hydrolysis by stabilizing the HSP70–substrate complex (10). Nucleotide exchange factors further promote nucleotide exchange, releasing both ADP and the substrate for subsequent cellular processes (27). This intricate mechanism highlights the dynamic nature of HSP70 in managing protein folding and quality control within the cell.
Heat shock protein 90
HSP90 represents another essential component of the HSR system, working alongside HSP70 to ensure proper protein folding and cellular homeostasis (28). HSP90 shares some primary structural similarities with HSP70, featuring a nucleotide-binding domain, a middle domain, and a CTD with an EEVD motif (Fig. 2) (29, 30). Like HSP70, HSP90 also binds ATP, albeit through different structural classes of binding sites, with HSP70 having a hexokinase fold and HSP90 a gyrase fold (31, 32).
One of HSP90's essential functions is stabilizing and activating a specific set of client proteins, thereby influencing various cellular processes (16, 24, 33). HSP90 interacts with co-chaperones to determine the fate of substrates, whether they undergo degradation, refolding, or modification, ultimately contributing to either pro-degradation or pro-folding cellular environments (34). Interestingly, HSP90 can complement the actions of HSP70 by participating in a secondary stage of the refolding process, enhancing protein folding and aiding in the final steps of protein maturation (33, 35).
HSP70 and HSP90, ying and yang
While HSP70 and HSP90 are critical players in the cellular PQC system, they exhibit distinct tertiary structural features and functional roles within the cell (19). HSP70 primarily recognizes and binds to misfolded proteins, initiating the refolding process through ATP-dependent mechanisms (27). In contrast, HSP90 stabilizes and activates specific client proteins, collaborating with co-chaperones to modulate substrate fate (36). Despite these functional differences, both chaperones share commonalities in their ATP-binding capabilities, highlighting the importance of nucleotide-dependent processes in protein folding and quality control (37).
Moreover, the interactions between HSP70 and HSP90, facilitated by co-chaperones like HOP and J-domain proteins, underscore the intricate network of molecular players involved in maintaining cellular proteostasis (38). The coordinated efforts of HSP70 and HSP90 and their associated co-chaperones ensure the proper folding, stabilization, and activation of diverse client proteins critical for cellular function (39). Understanding the distinct yet complementary roles of HSP70 and HSP90 sheds light on the sophisticated mechanisms governing PQC and cellular homeostasis in response to stress and environmental challenges (40).
Co-chaperones
CHIP comprises three main functional domains: an N-terminal tetratricopeptide repeat (TPR) domain, a coiled-coil domain, and a C-terminal U-box domain (Fig. 2) (41). CHIP functions as an E3 ligase, which mediates the transfer of ubiquitin molecules from E2-conjugating enzymes to substrates through CHIP’s U-box domain. This ubiquitination event can lead to degradation, transportation, or sequestration of substrates (42, 43, 44). Ubiquitin-marked substrates can be recognized by the proteasome or lysosome and degraded. Thus, HSP70 interaction with CHIP’s TPR domain leads to decreased ATPase activity of HSP70 and the potential for ubiquitination and subsequent degradation of the chaperone-bound substrate (45). A similar mechanism occurs with HSP90 substrates (46).
HOP contains three tetratricopeptide repeat domain regions, each contributing to binding to either HSP70 or HSP90, two Asp-Pro (DP) rich domains, and nuclear localization sequences (Fig. 2) (47, 48). HOP aids in the folding of newly synthesized proteins and the refolding of damaged or misfolded proteins (49). The three TPR domains of HOP with differential binding affinity to HSP70 and HSP90 also allow HOP to function as a bridge for substrates to be handed off between the chaperones, placing HOP as an essential player in proteostasis (21, 50, 51).
Co-chaperones such as the CHIP and HOP bind to the c-terminal tail of HSP70 and HSP90 (52, 53). HSP70 and HSP90’s interaction with CHIP leads to increased ubiquitination and subsequent degradation of the substrates (53, 54); conversely, the binding of HOP to HSP70 and HSP90 complexes leads to substrate refolding (50, 54). HOP stimulates the ATPase activity of HSP70 and HSP90, providing more energy to fold substrates (50, 55). Conversely, CHIP inhibits the ATPase activity of HSP70 and minimally the C-terminus domain of HSP90, providing more opportunity to ubiquitinate the substrate (23, 45, 56, 57).
HSP40 proteins, called J-domain proteins, form a large family with over fifty members. These proteins feature highly conserved J domains that bind to HSP70, enhancing its ATPase activity as a co-chaperone (Fig. 2), stabilizing complexes between HSP70 and non-native proteins (58, 59, 60). This cooperative stimulation by HSP40 proteins is a general feature, as they act on different reaction steps to enhance the ATPase activity of HSP70 severalfold (60). HSP40 co-chaperones can modulate various functions of HSP70, such as determining client specificity, enhancing disaggregase activity, and affecting localization (61). HSP40 proteins play a crucial role in contributing to protein folding, stress responses, and maintaining cellular proteostasis.
Consequences of a dysfunctional HSR
Dysregulation and accumulation of misfolded proteins in the cell are hallmarks of many human diseases, including cancer, Alzheimer’s, and cardiac disease. For example, hallmarks of Alzheimer’s disease include tau aggregates, a microtubule-associated protein that escapes degradation, and accumulation of p53 correlates with a poor prognosis in lung cancer (62, 63). Additionally, proteotoxic stress, the accumulation of proteins in the cell, is a stress hallmark of cancer (64). The PQC system, and the HSR in particular, plays an essential role in the initiation and progression of these diseases. In normal healthy cells, the HSR system quenches the protein levels of oncogenic proteins such as c-myc, a proto-oncogene encoding a transcription factor, mutant p53, a cell cycle control protein, vascular endothelial growth factor, epidermal growth factor receptor (EGFR), and human epidermal growth factor receptor 2 (65, 66, 67, 68, 69). Cancer cells can hijack this system by targeting tumor suppressor proteins for degradation or buffering the HSR system to cope with the proteomic stress induced by rapid cancer cell proliferation. Mutant p53, for example, can induce HSF1 transcriptional activity to promote the transcription of other oncogenic proteins such as Her2 and EGFR (70). In addition, multiple PQC system proteins are upregulated in cancer cells and mediate chemoresistance to chemotherapies (71). Conversely, increasing CHIP expression in renal cancer cells decreased cancer proliferation, highlighting an opposing role the HSR system can play in cancer proliferation (22). We recently reviewed the pro-versus anti-proliferative role of the HSR in cancer (72), and given the complexities of the involvement of the HSR in cancer, understanding key regulatory events may reveal the molecular underpinnings of certain cancers and identify new pharmacological targets.
Co-chaperone expression is associated with cancer progression, and, in some cases, causality has been established; however, context is essential. For example, higher CHIP expression is associated with a better cancer prognosis, as seen in the better tumor, node, metastasis stage, a staging system used by doctors to classify cancer progression and survival rates in some types of cancer, including renal and breast (42, 73). Alternatively, higher CHIP expression is also associated with a worse cancer prognosis, with higher tumor, node, metastasis stage and lower survival rates in glioma and gallbladder cancer (74, 75). Increased CHIP protein expression inhibited both renal cancer cell growth and breast cancer tumor growth in mouse studies, while CHIP knockdown increased renal cancer cell growth and breast cancer tumor growth (73, 76). Interestingly, HOP is upregulated in glioblastomas, pancreatic cancer, and breast cancer tumors (77, 78, 79, 80). Knockdown of HOP in glioma cells reduced proliferation rate and invasion through alteration to the protein kinase B or serine/threonine kinase 1 (AKT)/tumor necrosis factor receptor protein 1 (TRAP1) signaling pathway (77). HOP knockdown through RNAi in pancreatic cancer cells decreased the invasiveness of the cancer cells (78). Similar results were documented in other cancer cell line studies (81).
A pivotal element to protein triage in diseases lies in the balance between the pro-folding (HOP-associated) versus pro-degradation (CHIP-associated) environment. This switch between CHIP and HOP mediates the degradation of mutated, unwanted, or misfolded proteins like mutant p53 and, therefore, the outcome of diseases, which can ultimately provide new therapeutic targets. Specifically, PTMs on HSP70, HSP90, CHIP, and HOP offer a new point of regulation and insight into the mechanisms of cancer.
Phosphorylation regulation of HSP70 and HSP90
Protein phosphorylation
Protein phosphorylation adds a phosphoryl group (PO3) to specific side chains that alter the protein’s charge (54). This reversible modification typically occurs on serine, threonine, tyrosine residues, and histidine residues to a lesser extent (82). Kinase enzymes catalyze the transfer of the gamma phosphoryl group from ATP to the hydroxyl group of the side chain residue, a process reversed by phosphatase enzymes (82). The charge of the phosphoryl group affects allosteric binding between proteins. Phosphorylation is crucial in cell signaling propagation, enzyme regulation, and protein–protein interactions (Table 1). For example, phosphorylation affects HSP70 function, including co-chaperone interactions and the triage outcome of bound substrates (25).
Table 1.
Key residues phosphorylated on HSP70 and HSP90 and the downstream effects of phosphorylation
| Protein & phosphorylated residue | Modifier | Downstream impact | Preferred Co-Chaperone |
|---|---|---|---|
| HSP70 | |||
| Ser 385 (83) | ERK (83) | Chemo-resistance, Refolding | HOP |
| Ser400 (83) | ERK (83) | Chemo-resistance, Refolding | HOP |
| Thr636 (22) | AKT (88, 96) | Chemo-resistance, Refolding, and Increased Proliferation | HOP |
| Ser634 (22) | Refolding | HOP | |
| Ser631 (88, 108) | AKT (88) | Decreased mitosis | HOP |
| Ser633 (108) | AKT (88) | Decreased mitosis | HOP |
| HSP90 | |||
| Thr725 (22) | Decreased proliferation | HOP | |
| Ser726 (22) | Decreased proliferation | HOP | |
| Thr90 (93) | PKA (93) | Increased degradation | CHIP |
| Thr5/Thr7 (111) | Increased chemo-response | CHIP | |
| Thr115/T425/Thr603 (94) | Increased cell migration | HOP | |
Numbers in parentheses refer to references for papers that describe the modifications. All modifiers have been referenced accordingly.
Phosphorylation of HSP70 in the NBD and linker domain
Phosphorylation of HSP70 at Ser385 and Ser400 by extracellular signal-related kinase (ERK) increased the refolding activity of HSP70 about 1.5-fold (83). Additionally, a phospho-null mutant of HSP70 at Ser385 and Ser400 sites showed increased ubiquitination of client proteins AKT and cyclin-dependent kinase 4. In contrast, a phospho-mimetic of those same sites had a decrease in the ubiquitination of client proteins (22, 83, 84). hYVH1 is a phosphatase that interacts with HSP70 and enhances cell survival after heat shock (85). Other kinases involved in HSP70 phosphorylation include polo-like kinase 1, PKA, and AKT (86, 87, 88).
Phosphorylation of HSP90 outside the CTD
Many studies have reported on HSP90 phosphorylation sites (89, 90, 91). PKA can phosphorylate HSP90 and allows for interaction with Cdc37, a cell cycle protein (92). However, phosphorylation via PKA at Thr90 increases the association with CHIP while decreasing the association with HOP (93). The Thr90 phosphorylation site also reduces the affinity for ATP to bind to HSP90⍺, decreasing the chaperone refolding cycling (93). Protein kinase Cγ (PKCγ) phosphorylates HSP90⍺ at multiple threonine residues Thr115/Thr425/Thr603, decreasing chaperone activity (94).
Phosphorylation of HSP70/90 tail influencing CHIP and HOP binding
Many studies have highlighted the C-terminus EEVD motif of HSP70, specifically PTMs on this motif, is vital in substrate fate outcome. The EEVD motif is where many co-chaperones interact with HSP70, making it a critical regulation point to determine co-chaperones associated with HSP70 (95). When unphosphorylated on the c-terminus domain at Thr636, HSP70 preferentially binds to CHIP, promoting the degradation of chaperone substrates, as seen in renal cancer (Fig. 2) (22). However, when phosphorylated at Thr636 and Ser634, HSP70 preferentially binds to HOP, decreasing the degradation of those downstream proteins in renal cancer (22, 96). Assimon et al. and Zemanovic et al. also highlighted the impact of phosphorylation on chaperone binding, specifically those that bind using a TPR domain, such as CHIP and HOP (88, 97). A pseudo-phosphorylated EEVD motif in the c-terminus domain of HSP70 altered the polarization binding between CHIP and the motif. It decreased the binding affinity constant, indicating a change in affinity for the pseudo-phosphorylated tail (97). Phosphorylation of Ser631, still on the tail of HSP70, impacts the association of CHIP, as seen in a decreased pull-down of CHIP in co-immunoprecipitation immunoblotting (88). Phosphorylation of HSP90 that occurs predominantly on its c-terminal tail affects its binding and interaction with co-chaperones (Fig. 2). Phosphorylation at Thr725 or Ser726 increases the affinity between HSP90⍺, one of four isoforms of HSP90, and HOP (22). In contrast, HSP90⍺ has a higher affinity for CHIP in its unphosphorylated form (22).
Phosphorylation regulation of CHIP and HOP
CHIP phosphorylation
Protein kinase G (PKG) phosphorylates CHIP at Ser19 and enhances its interaction with HSP70 via stabilizing CHIP’s half-life, improving its resistance to degradation (98). Ser19 phosphorylation also did not affect the ATPase activity of HSP70 (98), suggesting that Ser19 phosphorylation mainly functions through enhancing CHIP stability, supported by the finding that the phospho-null mutation at Ser19 decreased CHIP’s interaction with HSP70. Phosphorylated CHIP reduced proteotoxicity and more effectively cleared ubiquitinated proteins (98). CHIP can also be phosphorylated by cyclin-dependent kinase 5 at Ser19 (Ser20 in mice), causing a decrease in interaction between CHIP and truncated apoptosis-inducing factor in neuronal cells (99). This decreased interaction induces neuronal death, highlighting the essential role of CHIP in cell death regulation (99). Aurora kinase A phosphorylates CHIP at Ser273, enhancing ubiquitination by CHIP of androgen receptors (ARs), which are essential regulators involved in prostate cancer (100). A phospho-null CHIP Ser273 mutant attenuated AR degradation compared to WT CHIP, indicating that CHIP phosphorylation activates ubiquitin towards ARs (100). Since ARs are targets for prostate cancer treatment, increasing CHIP phosphorylation and, therefore, the HSP70-CHIP association could be a target for prostate cancer treatment. Current published data on CHIP phosphorylation (Table 2) focus on the interaction between HSP70 and CHIP, but gaps in knowledge remain regarding the regulatory impact on HSP90. However, based on the similarities in the interaction between HSP90 and HSP70 with CHIP, we predict that the phosphorylation of CHIP would have a similar effect on its association with HSP90.
Table 2.
Key residues phosphorylated on CHIP and HOP and the downstream effects of phosphorylation
| Phosphorylated residue | Modifier | Downstream impact |
|---|---|---|
| CHIP | ||
| Ser19 (98) | PKG (98) | Increased chaperone interaction |
| Ser19 (99) | CDK5 (99) | Increased neuronal death |
| Ser273 (100) | Aurora kinase A (100) | Increased ubiquitination activity |
| HOP | ||
| Ser189 (102) | Casein kinase II (101) | Nuclear translocation |
| Thr198 (102) | Cdk1 (101) | Nuclear exclusion |
| Thr37 (103) | Decreased activation activity | |
| Ser95 (103) | Decreased activation activity | |
| Tyr354 (104) | Decreased co-chaperone activity | |
Numbers in parentheses refer to references for papers that describe the modifications. All modifiers have been referenced accordingly.
HOP phosphorylation
On the competing side of the HSP70/90 co-chaperone switch, HOP can be phosphorylated by cyclin-dependent protein kinase 1 (Cdk1) and the protein kinase casein kinase II (CK2) at multiple sites that impact function (Table 2) (101). Ser189 phosphorylation causes a nuclear translocation of HOP and mediates the assembly of the HSP70 chaperone complex (102). However, phosphorylated HOP-Thr198, or a phosphomimetic HOP at Thr198, is restricted from the nucleus, compared to the phospho-null at Thr198, further demonstrating that PTM location dictates protein localization (102). Phosphorylation of the TPR1 domain of HOP on Thr37 and Ser95 decreased glucocorticoid receptor activation activity and reduced affinity for HSP70 (103), and the unphosphorylated Tyr354 of HOP is required for optimal HSP70 substrate accumulation and activity (104). Because it promotes protein folding and cell cycle progression, decreasing HOP expression is a target for cancer therapies. As mentioned above, HOP is highly expressed in tumors, including glioblastomas, pancreatic cancer, and breast cancer (77, 78, 79, 80). Pharmacologically altering the activity state of HOP through phosphorylation could decrease cancer proliferation and improve prognosis. Shifting the preference away from HOP toward CHIP–HSP70 and CHIP–HSP90 complexes may benefit when HOP is aberrantly expressed at high levels.
Impact of phosphorylation on cancer cell proliferation
Shifting co-chaperone binding
Depending on the co-chaperone complex, HSP70 and HSP90 can promote or prevent cancer cell proliferation. Promoting HSP70–CHIP interactions inhibited tumor necrosis factor-⍺–induced apoptosis by degrading apoptosis signal-regulating kinase 1 (105). Additional downstream substrate degradation mediated by CHIP–HSP70 interactions influences cancer progression. For example, c-myc, a common oncogenic protein mentioned above, is regulated by heat shock in cancer cells (65). Blocking CHIP ubiquitination of HSP70 through blocking the BCL2-associated athanogene 2–CHIP interaction induces apoptosis in gastric cancer, decreasing the CHIP–HSP70 interaction and allowing for a better prognosis in this cancer (106). A phospho-mimetic of HSP70 at Thr636 mimicking natural phosphorylation, reduced binding with CHIP, and increased cellular proliferation in HEK293 cell lines.
In contrast, a phospho-null mimetic has the opposite effect, increasing CHIP binding and decreasing cellular proliferation (22). Muller et al. found higher amounts of C-terminal tail phosphorylation on HSP70 (Thr636) in breast tumor samples compared to normal tissue that coincided with increased expression levels of HOP (22). Phosphorylation of HSP70 at Ser385 and Ser400 also increased the cell viability and resistance to HSP70 inhibitor–mediated cell death in HEK293T cells, another cancerous cell line (83). Phosphorylation of the Tyr288 site on HSP70 regulates the transportation of methotrexate, a chemotherapeutic agent, into leukemia cells, affecting these cancer cells’ resistance to methotrexate (107).
HSP70 function and activity
HSP70 and its PTMs play crucial roles in maintaining proteostasis by regulating various cellular functions. However, these mechanisms can become dysregulated during tumor progression. Phosphorylated HSP70 at Ser631 and Ser633 localizes to the centrosome during mitosis and induces mitotic spindle elongation and mitotic arrest (108). This phosphorylation prevents cells arrested in mitosis by arsenic trioxide from apoptosis (108). However, O’Regan et al. indicated that HSP72, a cytoplasmic isoform of HSP70, is phosphorylated by Nek6 at Thr66, and phosphorylated HSP72 promotes cell cycle progression, specifically highlighting the importance of HSP72 in regulating cancer cell division (109). Disruption of the cell cycle often leads to uncontrolled cell growth, a hallmark of cancer, highlighting PTMs of HSR as a critical point of regulation and control. Without HSP72, HeLa cancer cells could not align chromosomes and, therefore, properly divide, causing an increase in multinucleated cells and chromatin bridges, indicating mitotic progression breakdown (109). This data highlights a potential target to inhibit cancer cell cycle progression.
HSP90 function and activity
PTMs of HSP90 control its function in healthy cells, but these modifications also influence cancer cell proliferation and tumor progression. Phosphorylation of HSP90 is linked to enhanced chaperone function and many cancer-driving processes (24, 91). Phosphorylated HSP90⍺ at Thr725 and Ser726 decreased doubling time in HEK293 cells, while the phospho-null form of HSP90⍺ at those same sites increased the doubling time of the cells, indicating a decrease in proliferation (22). Additional immuno-blotting comparing breast cancer tissue samples with normal tissue samples showed an increase in phosphorylated HSP90α in the breast tissue (22). Regulation of these phosphorylation events impacts HSP90α′s role in cancer proliferation. In a reciprocal regulation loop, HSP90⍺ prevents PKCγ degradation and facilitates PKCγ translocation to the membrane, activating its kinase activity and promoting cell migration and proliferation (94). Inhibition of both phosphorylation of HSP90⍺ at Thr115/Thr425/Thr603 by PKCγ and PKCγ itself prevented cell migration and induced apoptosis in colon carcinoma cells (94). An upregulation of phosphorylated HSP90β on Ser254 was seen post-anticancer treatment with 5-fluorocytosine, a prodrug used in tumor gene therapy treatment, highlighting that HSP90β might contribute to cancer regression (110). Further studies with HSP90α showed the phosphorylated HSP90⍺ at Thr5/Thr7 correlated with a good response for patients to platinum-based chemotherapy. They highlighted a potential use of HSP90⍺ as a prognostic marker for this type of chemotherapy (111).
Altering the phosphorylation of HSP70 and HSP90 impacts many downstream processes (Table 1), including interactions with CHIP and HOP, ultimately influencing cancer prognosis. The data showcased in most cancer systems suggest the increased association of HSP70 and HSP90 with HOP is pro-proliferation; therefore, targeting a switch in preference to CHIP-containing chaperone complexes may have anticancer benefits, highlighting the role for balance between CHIP- versusHOP-containing chaperone complexes. Other research has focused on co-chaperones as a point of control and revealed how CHIP and HOP can also be phosphorylated, ultimately affecting their association with HSP70/90, a topic we explore below.
Impact of other PTMs and components
Additional PTMs of the HSP70/co-chaperone and HSP90/co-chaperone complex impacting protein–protein interactions and function include acetylation, methylation, ubiquitination, glycosylation, and nitration.
Acetylation
Protein acetylation is the addition of an acetyl group onto side chains, catalyzed by acetyltransferases (112). This modification can be reversible or irreversible, thus impacting the long-term and short-term functional consequences. Acetylation occurs predominantly on lysine residues and, to a lesser extent, on alanine, arginine, serine, and threonine residues (113).
Acetylation of HSP70 influences the binding preference of co-chaperones to HSP70 and can, therefore, dictate the result of protein triage. After the onset of stress, arrest-defective one protein (ARD1) acetylates HSP70 at Lys77, which pushes the preference of binding towards HOP, leading to the refolding of substrates (114). HSP70 is deacetylated in the later stages of the stress response, shifting the binding preference to CHIP (114). Without this acetylation, the cellular stress response is limited, and damaged cells do not undergo apoptosis, perhaps promoting cancerous cell formation. Acetylation at Lys126 also attenuates the chaperone activity of HSP70 by changing the physiological function of HSP70 and altering co-chaperone binding. This site increases HSP70’s association with CHIP over HOP, thus creating a pro-folding, pro-tumor environment (115). Acetylation of HSP70 at Lys126 inhibits MDA-MB-231 cancer cell proliferation, tumor cell invasion, and migration in breast cancer cells (115).
In addition to the HSR mechanism, HSP70 ensures cellular health by regulating chaperone-mediated autophagy and apoptosis. Yang et al. found that acetylated HSP70 is required to adequately form the autophagosomes that break down the cellular components for recycling (116). Specifically, acetylated HSP70 regulates Vps34, also known as the kinase PIK3C3, and its association with Beclin 1, a regulator of the autophagy pathway, marking a pivotal step in autophagosome formation (116). Acetylation of HSP70 at Lys126 increases the sequestering of Bcl2, a mitochondrial membrane protein that blocks cell death, for autophagosome formation (115). Additionally, this acetylation site prevents HSP70 from binding with apoptotic peptidase activating factor 1 and allograft inflammatory factor 1 and, therefore, mitigates the promotion of apoptosis (115). Without the ability to induce cell death, accumulating unhealthy cells could lead to cell transformation.
Acetylation of HSP90 also affects ATP binding, client binding, and co-chaperone activity (24). HSP90 is acetylated at multiple sites in the middle and CTDs, but the most influential factor affecting the co-chaperone role is at Lys294, between the charged linker region and the middle domain (Fig. 2) (117). When acetylated, HSP90 binds less CHIP, confirming that acetylation influences HSP90’s change in activity and interaction with co-chaperones (117). HSP90 is deacetylated by histone deacetylase 6 (HDAC6) (117, 118), and the inactivation of HDAC6 results in the hyperacetylation of HSP90, a loss of chaperone function, and dissociation with p23, a protein essential to glucocorticoid receptor maturation (118). Investigating this acetylation event in a cancer context could provide new insight and opportunities for treatment.
Methylation
Protein methylation by methyltransferases is a reversible PTM that occurs mainly in the nucleus, with histones being one of the most studied substrates for methylation and critical for transcriptional regulation of the genome. Methylation often occurs on lysine and arginine residues, significantly altering the target protein's structure and affecting its function (113). Methylation of HSP70 at Lys561 by HSPA- KMT, a lysine methyltransferase that methylates the specific residues in multiple HSP70 (HSPA) proteins, has a growth-promoting effect in HeLa cancer cells by enhancing Aurora kinase B activity (119, 120). Clinical tissue samples from bladder and liver tumors mirror this cancer influence where the dimethylation of Lys561 of HSP70 is significantly elevated in these cancer tissues (120). The methylation of HSP90 at Lys607 affects the closed confirmation of HSP90, changes ATP binding activity, and decreases Aha1 binding, a co-chaperone activator of HSP90 ATPase, ultimately altering client activation (121).
Ubiquitination, glycosylation, and nitration
There are limited reports on ubiquitination, glycosylation, and nitration, influencing co-chaperone association and subsequent cancer progression. Ubiquitination is the addition of ubiquitin, a small 8.5 kDa protein, onto a side chain of a protein, predominantly lysine residues, catalyzed by a cascade of three enzymes, E1, E2, and E3 (113). Events include single ubiquitination reactions (mono-ubiquitination) or polyubiquitination of any one of the seven lysine residues on ubiquitin, resulting in polyubiquitin chains. The result of ubiquitination alters the structure and interactions of the substrate protein. Ubiquitination is a critical signaling event in PQC. The 26S proteasome recognizes specific polyubiquitin chain topologies, and this interaction leads to the degradation of the ubiquitinated protein and recycling of the protein building blocks (122). CHIP can ubiquitinate HSP70 itself or substrates interacting with HSP70, leading to its degradation, but requires the HSP70-lid domain, positioned right before the EEVD motif in the CTD (Fig. 2), for proper ubiquitination of either (84, 123). Investigation of the functional effect of ubiquitinated HSP70 beyond degradation has not been reported (84, 124). Poly-ubiquitination of HSP90 at multiple sites (Lys6, Lys11, Lys48, and Lys63) leads to its degradation (125).
Although less studied, protein glycosylation and nitration can impact the function of proteins. In glycosylation, oligosaccharide chains are linked to residues through covalent bonds, affecting interactions structurally (113). The subsequent effect on protein–protein interaction depends on the specific type of glycosylation: O-linked, N-linked, or C-linked. However, it is known that HSP90 is glycosylated on the four ⍺-subunits and S434 and S461 of HSP70β, but few studies have explored the effect of glycosylation on the function of HSP90 (126). Nitration adds a nitro group to tyrosine residues of proteins and changes the proteins’ charge (127). The nitration of HSP90 at Tyr33 and Tyr56 induces cell death by activating the Fas pathway, a type of programmed cell death that activates apoptosis and is a toxic gain of function that turns HSP90 into a toxic protein (128). Triggering cell death via the Fas pathway by the nitration of HSP90 may lead to new cancer therapeutic approaches (128).
Despite many studies investigating the points of other PTMS, there is a lack of published studies on the effect of these PTMs on co-chaperone binding, highlighting a critical gap in knowledge. Based on the growth-promoting effects of these PTMs, it is predicted that the preference is for HOP in the cancer environment, highlighting a potential point of treatment.
Heat shock factor 1
HSFs include nucleotide-binding, regulatory, and CTDs (Fig. 2) (129). They regulate the transcriptional response to heat or stress (8). When faced with a stressor, HSFs dissociate from their inhibitory complex with HSP90 and bind to DNA to induce the transcription of numerous genes, including those that encode HSP70 and HSP90 (8). Early research on HSF1 implicated that CHIP is required for HSF1 dimerization and entry into the nucleus, promoting a feed-forward regulator mechanism to ramp up the HSR to alleviate proteostatic stress (130). PTMs of HSF1 alter its ability to bind and associate with DNA and other HSPs, modulating the cellular response (Table 3).
Table 3.
Key residues phosphorylated on HSF1 and the downstream effects of phosphorylation
| Phosphorylated residue | Modifier | Downstream impact |
|---|---|---|
| Ser326 (135) | AKT1 (135) | Increased transcriptional activity |
| Thr142 (135) | AKT1 (135) | Increased trimerization |
| Ser230 (135) | HSR activation | |
| Ser326 (135) | HSR activation | |
| Thr527 (135) | HSR activation | |
| Ser303/Ser307 (136) | Decreased transcriptional activity | |
| Ser216 (139) | PLK1 (139) | Cell cycle progression |
| Ser419 (138) | PLK1 (140) | Increases transcription |
| Thr120 (141) | Increases proteostasis |
Numbers in parentheses refer to references for papers that describe the modifications. All modifiers have been referenced accordingly.
Manipulating PTMs of HSF1 can inhibit cancer cell proliferation and susceptibility to chemotherapeutics. Multiple signaling pathways in cancer cells activate HSF1 to accommodate the rapidly changing tumor environment and stress of rapid cell proliferation (131). HSF1 is frequently overexpressed in cancer cells (132, 133). As mentioned above, HSF1 overexpression inhibited mitotic exit, causing an increase in aneuploidy and multinucleated cells, a hallmark of cancer (132). Additionally, HSF1-mediated aneuploidy was accelerated in p53-defective cells, a defect frequently seen in cancer cells (134). Therefore, decreasing HSF1 expression to lower HSP70 and its interaction with HOP would decrease cancer progression and thus make HSF1 a promising target for chemotherapy.
Phosphorylation
AKT phosphorylates and activates HSF1 (135). Specifically, AKT phosphorylation of Ser326 increases HSF1’s transcriptional activity, while phosphorylation at Thr142 is necessary for HSF1 trimerization (135). Other sites, such as Ser230, Ser326, and Thr527, are required for maximum activation of HSF1 during heat stress (135). Constitutive phosphorylation at Ser303 and Ser307 negatively regulates HSF1 transcription activity at physiological temperature, highlighting an essential step in HSF1 regulation (136). Cyclosporin A (CsA), an immune suppressant, enhances the phosphorylation of these sites (Ser303 and Ser307) and inhibits HSF1’s transcriptional activity and, thus, the ability to respond to heat shock (137). CsA also interferes with HSF1 complex formation, nuclear translocation, and binding to the HSP70 promoter (137). PLK1 also phosphorylates HSF1 at multiple sites, including Ser216 and Ser419 (138, 139, 140). Phosphorylation of HSF1 at Ser216 occurs during the early stages of mitosis and propels mitosis forward through interaction with Cdc20, a cell cycle regulatory protein (132, 139). Without the phosphorylation and subsequent binding to Cdc20, mitosis does not continue (132). However, too much HSF1 inhibits mitotic exit, leading to aneuploidy and multinucleated cells (132). The Ser419 phosphorylation site of HSF1 helps recruit the TRRAP–TIP60 acetyltransferase complex to promoters (138). This recruitment changes histone modifications, especially on the HSP72 promoter, and opens chromatin for transcription (138). Phosphorylation of HSF1 at Thr120 protects HSF1 from being ubiquitinated by F-box and WD repeat domain containing 7 (FBXW7), a tumor suppressor protein of the F-box protein family, and subsequently degraded (141, 142). This HSF1 Thr120 phosphorylation promotes proteostasis and binding to the programmed death ligand 1 (PD-L1) promoter (141).
Acetylation and ubiquitination
Although less reported than phosphorylation, acetylation and ubiquitination contribute to the post-translational landscape of HSF1 regulation. SIRT1, a well-characterized sirtuin family member, deacetylates HSF1 at Lys80 to induce the expression of other HSR proteins, such as HSP70 (143, 144). Acetylation negatively affects the DNA-binding ability of HSF1 (144). Acetylation by EP300, a transcriptional co-activator protein, at other sites, such as Lys118, Lys208, and Lys298, stabilizes the expression of HSF1 as it prevents ubiquitination and subsequent degradation (13, 145). FBXW7, a critical tumor suppressor, ubiquitinates HSF1 and leads to HSF1 degradation (142, 146). Without FBXW7, HSF1 accumulates and activates the metastatic pathway of melanoma, driving cancer survival and metastasis (142). Therapies could target these PTMs involved in HSF1 stability or activity.
Most work on HSF1's PTMs has focused on its nuclear activity as a transcription factor. However, HSF1 does have some role in the cytosol, where it interacts with CHIP. Investigating the roles of PTMs on HSF1 in the cytosol would provide a better understanding of HSF1’s role in HSR and cancer progression and reveal if there are cytosolic ramifications of HSF1 PTMs.
Heat shock protein 40
Another key player in the HSP70/co-chaperone complex is the HSP40 family of co-factor proteins, also known as J-domain proteins, that shares a conserved J domain on the N-terminus of the protein and includes cysteine string proteins and DNAJBs (Fig. 2) (147). Other domains in this protein include Zinc finger motifs and a CTD (Fig. 2) (148). HSP40s bind proteins and target them to HSP70 for folding and enhance the ATPase activity of HSP70s (147, 148). The primary functions of HSP40 are to increase the ATP hydrolysis rate of HSP70s, engage polypeptide substrates to HSP70s, and promote a profolding environment (149). CHIP can inhibit HSP40 by blocking the binding of HSP40 to HSP70 and, subsequently, the HSP40-stimulated activity of HSP70 (45). CHIP acts as a modulator of HSP40 to pause the ATPase activity of HSP70, promote substrate ubiquitination, and cause a shift to pro-degradation. HSP40 proteins, like the rest of the HSPs, undergo PTMs that contribute to the acute regulation of the heat shock mechanism and stress response. Manipulating HSP40s may impact the profolding versuspro-degradation associations of HSP70 and HSP90 with HOP versusCHIP, respectively.
HSP40s can be phosphorylated by multiple kinases, including mitogen-activated protein kinase–activated protein kinase 5 (MK5) and CK2, or can even exist as phosphoproteins in the cell (147, 150, 151). When phosphorylated by MK5 at Ser149, Ser151, and Ser171, HSP40-mediated regulation of HSF1 activity increases, allowing for a more robust stress response (150). Phosphorylation on Ser10 of the cysteine string protein triggers a conformational switch in the protein’s orientation and activity and plays a role in presynaptic maintenance by preventing neurodegeneration, highlighting the crucial role of HSP40s in cellular viability (152, 153, 154).
The DNAJB-PRKACA gene fusion, created by the fusion of a subunit of the HSP40 complex (DNAJB) and the catalytic subunit of protein kinase A (PRKACA), creates a fusion kinase and drives fibrolamellar hepatocellular carcinoma, and PTMs of this kinase could impact cancer progression (155). DNAJB1 has four phosphorylation sites in its cytosolic domain that alter its nuclear transport (151) and directly affect DNAJB1’s role in cancer progression (156). DNAJB enhances the ubiquitination of MIG6, a tumor suppressor, and promotes EGFR signaling (157). When DNAJB was knocked down, lung cancer cells became sensitized to treatment with gefitinib, a common chemotherapeutic targeting tyrosine kinases (157). HEDJ, an HSP40 co-chaperone, is localized to the endoplasmic reticulum and can be glycosylated (158), although the potential implications of this glycosylation on cancer progression have not been reported.
Small HSPs
Small heat shock proteins (sHSPs) are molecular chaperones between 15 to 30 kDa and have a conserved sequence of 80 to 100 amino acids, essential for the function of sHSPs (Fig. 2) (159). sHSPs also have a conserved N-terminal region and a short sequence at the c-terminus (160). sHSPs include HSP beta-2, HSP17, HSP20, clusterin, and HSP17, all of which aid in the formation and folding of proteins by assisting chaperones (160, 161). However, only HSP27 is post-translationally modified, so we focus mainly on HSP27 (159). HSP27 can be phosphorylated on different regions by multiple kinases, including p38 mitogen-activated protein kinase (MAPK), MAPKAPK-2, another protein kinase in the MAPK family, AKT, and PKG (162). HSP27 is upregulated in many cancers, such as prostate, lung, and pancreatic (163, 164). Therefore, targeting HSP27, specifically phosphorylation of HSP27, has been promising for cancer therapies.
HSP27 phosphorylation is associated with cancer progression (165). Phosphorylation of HSP27 at Ser15, Ser78, or Ser82 initiates chemoresistance in prostate cancer cells (166). Overexpression of HSP27 led to increased survival rates of prostate cancer cells after docetaxel, a microtubule-targeted chemotherapy treatment, while the overexpression of the nonphosphorylatable HSP27 mutant decreased resistance to treatment (166). Similarly, overexpression of the same nonphosphorylatable HSP27 mutant with gemcitabine, a DNA replication–targeted chemotherapy treatment in pancreatic cancer cells, decreased cell proliferation (164, 167). Preventing phosphorylation of HSP27 by p38 MAPK and MAPKAPK-2 inhibition increased cell death of pancreatic cancer cells after treatment with gemcitabine (164, 167). Phosphorylated HSP27 at Ser15, Ser78, or Ser82 also contributes to the chemoresistance of adenocarcinoma cells (168). Overexpression of HSP27 phospho-mimetic mutants activated p53 and decreased cell death (168). Inhibition of this same phosphorylation in combination with doxorubicin decreased p53 induction and increased cell apoptosis (168). Because HSP27 promotes the folding and refolding of substrates, one would predict it preferentially associates with HOP. However, further studies would be needed to confirm that.
Advances in drug therapies
Our literature review suggests that the balance between a pro-degradation environment and a profolding environment can influence cancer cell proliferation, transformation, and resistance to chemotherapy. Consequently, developing drugs that can modulate the HSR and the balance between protein degradation and folding could be a promising approach for cancer therapy. This modulation could be achieved by directly targeting the proteins involved in the HSR or the upstream enzymes, such as kinases and acetyltransferases.
HSP70 and HSP90
Direct-acting drugs
HSP70 has already been shown to be a promising pharmacological target (169). Many studies investigated small molecules that target HSP70 function, summarized in a review by Nitzsche et al. (170). For example, the novel inhibitor for HSP70 named S1g-6 from Wang et al. showed selective cell death in cancer cells over normal cells (171). However, as mentioned above, PTM specificity determines the influence on cancer progression. For example, methylation of HSP70 has a growth-promoting effect, while acetylation inhibits cancer proliferation (115, 119). HSP70 plays a role in regulating apoptotic pathways promoting cancer progression, but it has also been shown to induce an immune response to target cancer cells, indicating a potential dual role in cancer progression (172). The duality of HSP70 is a crucial consideration for cancer therapies, and characterizing and specifying HSP70 substrates as pro-versus anti-cancer HSP70 is essential for developing cancer treatments.
Because of its prominent role in the HSR, HSP90 is another promising drug target for cancer treatment, with multiple drugs and compounds shown to decrease cancer cell proliferation that moved into clinical trials (34, 92, 173). One example is 17-AAG and its derivates, which showed anticancer activity in small-cell lung cancer, but the trials were stopped before reaching phase III due to safety concerned and limited efficacy (92). Another HSP90 inhibitor currently in clinical trials is ganetespib, a small-molecule inhibitor targeted at the ATP-binding site of HSP90 (174). In phase II trials, non-small cell lung cancer showed a response to ganetespib. However, no response was seen in refractory metastatic colorectal cancer, again highlighting a critical consideration when targeting the HSR system, which depends on the specific role and PTMs of the component that will determine the effectiveness of inhibitors (29). Further exploration of specific inhibitors for HSP90 is needed to ensure anti-cancer treatment rather than pro-cancer results.
Drugs targeted to the tail of HSP70/90 could significantly impact the pro-degradation and pro-folding environment mediated by CHIP and HOP occupancy. However, limited trials on compounds target the PTMs in the tail region of HSP70/90. Nadler et al. isolated a compound named DSG that targeted the EEVD motif of HPS70 and enhanced its steady-state ATPase activity (175). DGS showed antitumor properties but did not pass phase II trials (176, 177). HVH-2930 targets the CTD of HSP90, affecting the ATP-binding pocket of the HSP90 dimer and inhibiting its activity (178). Bhatia et al. also created a small-molecule inhibitor, 5b, that targeted the CTD of HSP90 and showed anti-leukemia properties (179). The impact of HVH-2930 and 5b on HSP90 PTMs and CHIP/HOP occupancy in these two molecules was not reported. However, targeting the CTD of HSP90 could shift CHIP- and HOP-binding dynamics and cancer progression.
Kinases and acetyltransferases
One approach to targeting PTMs is to drug the upstream enzymes, such as kinases and acetyltransferases. As mentioned above, ERK phosphorylates HSP70. Current progress on ERK inhibitors is limited, but a few are in clinical trials (180). Ulixertinib has shown promising anticancer effects in pancreatic cancer models (181). However, inhibiting these enzymes could be nonspecific to the HSR, as these enzymes target multiple other proteins and could have many adverse or opposite effects on cancer progression (182).
PKCγ is a kinase that targets HSP90 along with many other proteins. Although there are multiple places of inhibition on the PKC protein, most are targeted to the C3 domain, but some bind to the C1 domain. The compound UCN-01 showed promising results in multiple phase I drug combination therapies for cancer treatment, but few combinations made it through phase II (183). The upstream ORF peptide upstream of PKC inhibits the catalytic activity of PKCs and inhibits cancer cell survival, progression, and metastasis (184). Specifically, inhibiting PKC in non-small cell lung cancer has been a promising therapeutic focus to prevent resistance; however, many trials of PKC inhibitors have shown little clinical benefit (183, 185).
PKA is one of the kinases that phosphorylate HSP90. Endogenous PKA inhibitor (PKI) proteins such as PKIα, PKIβ, and PKIγ bind with PKA and inhibit its kinase activity. Prostate cancer cells show less migration and tumor growth when these inhibitor proteins are knocked down (186). However, like the rest of the HSR system, PKA signaling can be pro- and anti-tumor growth (187).
ARD1 and HDAC6 are the acetyltransferases responsible for acetylating HSP70. ARD1 genetic inhibition reduced the levels of NRF2, a critical oncogenic transcription factor protein in colon cancer cell lines, highlighting a protein role in cancer therapy (188). shRNA knockdown of ARD1 also inhibited prostate tumorigenesis (189); however, specific ARD1 inhibitors have yet to be available for clinical trials. HDAC6, on the other hand, has many promising inhibitors that enhance the effectiveness of different chemical therapies (190). The small molecule inhibitor ACY1215 targeted to HDAC6 reduced tumor growth rate in non-small lung and gallbladder cancer cells (191, 192). Therefore, HDAC6 could be therapeutically beneficial for cancer treatment.
CHIP and HOP
Direct-acting drugs
Although there are no current drug therapies in clinical trials that target CHIP currently, many studies have alluded to inhibiting CHIP as a mechanism of action to prevent cancer cell proliferation. Inhibiting CHIP in both colorectal and prostate cancer, where CHIP acts as a tumor promoter, decreases cell growth, migration, and invasion (193, 194). However, as highlighted in previous reviews, CHIP plays a dual role as a tumor suppressor and oncogenic protein (42, 72, 195, 196). Overexpression of CHIP decreased renal cancer cell growth, and the suppression of CHIP promoted ovarian cancer metastasis, both instances where CHIP acts as a tumor suppressor (73, 197). Therefore, a better understanding of its role in different cancer types is needed before any drugs are targeted to CHIP.
The HSP70 inhibitor VER155008 suppresses the expression of HOP, which decreases prostate cancer growth (198). HOP was implicated as a prognostic biomarker in ovarian cancer and breast cancer, and when knocked down, pancreatic, colorectal, and breast cancer cell progression decreased, highlighting how targeting HOP and likely shifting the binding occupancy of HSP70/90 to HOP and CHIP could be beneficial for cancer treatment (78, 80, 199, 200). A compound generated by Pimienta et al. inhibited the HOP–HSP90 complex and demonstrated anticancer effects in breast cancer cell lines (201). However, further studies on this compound or others targeted towards HOP have yet to be published.
Kinases and acetyltransferases
Activation of PKG, the kinase that phosphorylates CHIP, is implicated in multiple cancer studies as a target to decrease cancer cell progression (202, 203). However, other studies have indicated that the nitric oxide pathway, containing PKG, protects against apoptosis in ovarian cancer cells and increases the stemness and metastasis in lung and breast cancer cells (204, 205). Compound KT5823 inhibits protein kinase activity in vitro but did not show any effects in human platelets or rat mesangial cells (206, 207). As indicated above, CHIP also has a complicated, dual role in cancer progression; therefore, the dual role of PKG does correlate with its influence on CHIP.
Aurora kinase A (AURKA) is another kinase that phosphorylates CHIP. It has been implicated as a synthetic lethal target for many other tumor suppressors. Targeting AURKA and other tumor suppressors results in cancer cell death, highlighting a promising target for cancer treatment (208). Clinical trials of AURKA molecular inhibitors are currently underway (209).
Targeting the kinases and acetyltransferases that modify HOP to decrease its association with HSP70/90 is another promising strategy to reduce cancer progression. CDK1 is the kinase that phosphorylates HOP. Many inhibitors targeted to CDK1 have shown promising results in decreasing cancer cell progression and have entered clinical trials (210). Specifically, a few novel CDK1 inhibitors from Akl et al. had antiproliferative activity in a panel of pancreatic cancer cell lines (211). Other reviews have highlighted the impact of CDK1 inhibitors in breast cancer, emphasizing that inhibiting HOP phosphorylation would be promising to decrease cancer cell progression (210, 212). CK2 is another kinase that can target HOP for phosphorylation. Inhibitors targeted towards this kinase have been implicated in treating myeloid leukemia and triple-negative breast cancer, although there have been limited clinical effects in leukemia treatment (213, 214, 215).
Heat shock factor 1
Recent studies detail HSF1 pharmacology, especially regarding its phosphorylation status. AKT phosphorylates HSF1 independent of heat stress in breast cancer cells (216). Inhibition of AKT and, therefore, inhibition of HSF1 phosphorylation reduced the proliferation of PIK3CA mutated cells, a common mutation in the gene that makes the PI3K enzyme (216). Biochemical inhibition or siRNA silencing of HSF1 increased benzimidazole carbamate drug potency in colorectal cancer cells (217). Further experiments found that ERK-1/2 activates HSF1 through phosphorylation of S326 and promotes chemotherapeutic resistance in colorectal cancer cells (217). Bortezomib, a proteasome inhibitor for treating multiple myeloma and mantle cell lymphoma, induces cancer cell apoptosis and cell cycle arrest by blocking the 26S proteasome, which also disrupts DNA replication and repair (218). Bortezomib-induced phosphorylation of HSF1 on Ser326 contributes to multiple myeloma resistance to treatment (219). Silencing of HSF1 sensitized multiple myeloma cells to bortezomib treatment (219). Phosphorylation of HSF1 on Thr120 promotes breast cancer tumorigenesis compared to an overexpression of WT HSF1 (141). Another phosphorylation site on S419 was found at higher levels on melanoma cell lines, and the substitution of HSF1-Ser419 reduced the proliferation of these melanoma cell lines (138).
CsA, an immunosuppressant, enhances the phosphorylation of the negative activator site on HSF1, causing the death of HeLa cancer cells (137). Thus, CsA, via the inhibition of HSF1 through phosphorylation, could be used as chemotherapy to decrease tumor progression. HSF1 modulates the effects of cisplatin, an alkylating chemotherapy agent for oral squamous cell carcinoma, acting on PD-L1 (220). Additionally, cisplatin activates HSF1 by phosphorylating S326, leading to an upregulation of PD-L1 surface expression (220). Therefore, modulating HSF1 phosphorylation, especially of S326, is a promising chemotherapy target. One caveat to specifically targeting HSF1 PTMs is that some PTMs increase transcriptional activity while other PTM sites decrease activity, requiring specific PTM inhibitors to ensure a pro-cancer environment is not promoted with drug treatment (133). Because of HSF1’s role in promoting HSP70 and HSP90 refolding chaperone activity, inhibiting HSF1 would likely shift the preference of HSP70 and HSP90 towards CHIP and substrate degradation, thus providing a promising cancer treatment strategy.
Heat shock protein 40
Targeting HSP40s would affect the refolding chaperone activity of HSP40 and ultimately decrease cancer cell proliferation. For example, inhibition of HSP40 results in a depletion of mutant p53 and reduced cancer cell proliferation in a panel of cancer cells (221). HSP40 controls the fate of the misfolded mutant p53, promoting the accumulation of this protein (222). Therefore, blocking the refolding activity of HSP40 would shift the chaperone preference towards CHIP and result in the degradation of cancer-promoting substrates. Inhibiting the interaction of the HSP70–HSP40 complex and its substrates decreased cancer stem cell growth, again highlighting how a shift towards CHIP and degradation may be necessary for cancer cell progression (223).
Small HSPs
Specific drug targets are needed to ensure effective anticancer therapies because of the broad range of cellular roles for sHSPs. Selective inhibitors targeted towards other sHSPs have been challenging to create; thus, most research has focused on HSP27 as a target for drug therapies to reduce expression or inhibit action (224). Because of its role in cell death and PQC, HSP27 is a promising drug target for chemotherapies (163, 165). However, HSP27 lacks an ATP-binding domain; therefore, targeted chemotherapies likely need to focus on altering HSP27 function, such as targeting phosphorylation or dimerization (225). Small molecule inhibitors, peptides, and antisense oligonucleotides targeted to HSP27 mRNA have effectively decreased cancer proliferation. However, only the antisense oligonucleotide treatment is currently in clinical trials (226). Small molecule inhibitors such as quercetin bind directly to the sHSPs and inhibit their function. Quercetin works by inhibiting HSP27 phosphorylation and, therefore, its expression (227). It has also shown promising results in ovarian and breast cancer (228, 229). Peptides are designed to bind to specific protein domains, but little preclinical work has explored using peptide targets to sHSPs (224). sHSPs play a role in the formation, folding, and refolding of proteins. Therefore, chemically shifting the activity towards CHIP to promote degradation through sHSP inhibition would benefit cancer treatment.
Conclusions
The HSR is modulated by various PTMs that affect protein interactions, outcomes on substrates, and disease progression. These proteins and their alterations are identified as potential targets for cancer therapy. A deeper understanding of this aspect of cellular PQC could lead to more effective pharmacological strategies. One area of further exploration is the HSP110 family of chaperones protein, which is not highlighted in this review but is still an essential system component. Although large HSPs were discovered in the 1990s, little research has investigated the PTMs on these proteins, highlighting a gap in knowledge in our understanding of the complete HSR system (230). HSP110 is the only protein in the HSP family with a mutation linked to the progression of colorectal cancer (231). Therefore, targeting this protein could be beneficial in treating colorectal cancer harboring this mutation. Research on large HSPs in cancer, specifically their role in cancer immunotherapy, demonstrates they are essential and influence cancer progression, emphasizing a key area of further study.
Current research on the HSR often focuses on isolated components. However, this review underscores the complex interaction among all molecular entities involved and the importance of understanding the entire system for developing drug therapies. It is noted that encouraging the transition from HSP70/90 binding with HOP to CHIP could be advantageous in most cancer treatments. Nonetheless, modifying one element of the system may impact the overall response and significantly alter the therapeutic effects of drugs. Therefore, further research is necessary to grasp the dynamics between HSR components. Our review synthesizes critical elements of the HSR and how their alterations influence interactions, which could pave the way for novel therapeutic strategies against diseases like cancer. Future studies should consider the entire HSR network to fully comprehend the mechanisms of therapies that lead to successful treatment outcomes.
Conflict of interests
The authors declare that they have no conflicts of interests with the contents of this article.
Acknowledgments
We thank Becky Sanchez for reviewing the manuscript, all the members of the Schisler Laboratory, the administration teams at the McAllister Heart Institute, and the Department of Pharmacology. Figures were created using BioRender.com.
Author contributions
M. S. writing–original draft; M. S. and J. C. S. conceptualization; J. C. S. writing–review and editing; J. C. S. funding acquisition.
Funding and additional information
This research was funded by the National Institute on Aging, grant numbers R01AG066710 and R01AG061188, to J. C. S., and a Graduate Research Fellowship Program award from the National Science Foundation to M. S.
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
References
- 1.Kaufman R.J. Molecular chaperones and the heat shock response. Biochim. Biophys. Acta (BBA) - Rev. Cancer. 1999;1423:R13–R27. doi: 10.1016/s0304-419x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A., Kwon Y.T. Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 2017;11:185. doi: 10.3389/fnins.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang B.J., Guerrero M.E., Prince T.L., Okusha Y., Bonorino C., Calderwood S.K. The functions and regulation of heat shock proteins; key orchestrators of proteostasis and the heat shock response. Arch. Toxicol. 2021;95:1943–1970. doi: 10.1007/s00204-021-03070-8. [DOI] [PubMed] [Google Scholar]
- 4.Whitesell L., Lindquist S. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 5.Mallouk Y., Vayssier-Taussat M., Bonventre J.V., Polla B.S. Heat shock protein 70 and ATP as partners in cell homeostasis (Review) Int. J. Mol. Med. 1999;4:463–537. doi: 10.3892/ijmm.4.5.463. [DOI] [PubMed] [Google Scholar]
- 6.Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019;20:665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Pastor R., Burchfiel E.T., Thiele D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y.-M., Huang D.-Y., Chiu J.-F., Lau A.T.Y. Post-translational modification of human heat shock factors and their functions: a recent update by proteomic approach. J. Proteome Res. 2012;11:2625–2634. doi: 10.1021/pr201151a. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S. Dynamic M6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prodromou C. Mechanisms of Hsp90 regulation. Biochem. J. 2016;473:2439–2452. doi: 10.1042/BCJ20160005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guisbert E., Yura T., Rhodius V.A., Gross C.A. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol. Mol. Biol. Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian S.-B., McDonough H., Boellmann F., Cyr D.M., Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raychaudhuri S., Loew C., Körner R., Pinkert S., Theis M., Hayer-Hartl M., et al. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 2014;156:975–985. doi: 10.1016/j.cell.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 14.Korolchuk V.I., Menzies F.M., Rubinsztein D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 15.Ohnishi K., Ohkura S., Nakahata E., Ishisaka A., Kawai Y., Terao J., et al. Non-specific protein modifications by a phytochemical induce heat shock response for self-defense. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vihervaara A., Sergelius C., Vasara J., Blom M.A., Elsing A.N., Roos-Mattjus P., et al. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc. Natl. Acad. Sci. 2013;110:E3388–E3397. doi: 10.1073/pnas.1305275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berwin B., Nicchitta C.V. To find the road traveled to tumor immunity: the trafficking itineraries of molecular chaperones in antigen-presenting cells. Traffic. 2001;2:690–697. doi: 10.1034/j.1600-0854.2001.21003.x. [DOI] [PubMed] [Google Scholar]
- 18.Jacob P., Hirt H., Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017;15:405–414. doi: 10.1111/pbi.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panda A.K. Editorial: influence of mutations, stresses and post-translational modifications on the structure and function of heat shock proteins and their relationship with various diseases. Front. Mol. Biosciences. 2023;10 doi: 10.3389/fmolb.2023.1310272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirkkala L., Nykänen P., Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya K., Weidenauer L., Luengo T.M., Pieters E.C., Echeverría P.C., Bernasconi L., et al. The Hsp70-Hsp90 co-chaperone Hop/Stip1 shifts the proteostatic balance from folding towards degradation. Nat. Commun. 2020;11:5975. doi: 10.1038/s41467-020-19783-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller P., Ruckova E., Halada P., Coates P.J., Hrstka R., Lane D.P., et al. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32:3101–3110. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- 23.Kampinga H.H., Kanon B., Salomons F.A., Kabakov A.E., Patterson C. Overexpression of the cochaperone CHIP enhances hsp70-dependent folding activity in mammalian cells. Mol. Cell Biol. 2003;23:4948–4958. doi: 10.1128/MCB.23.14.4948-4958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backe S.J., Sager R.A., Woodford M.R., Makedon A.M., Mollapour M. Post-translational modifications of Hsp90 and translating the chaperone code. J. Biol. Chem. 2020;295:11099–11117. doi: 10.1074/jbc.REV120.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitika, Porter C.M., Truman A.W., Truttmann M.C. Post-translational modifications of Hsp70 family proteins: expanding the chaperone code. J. Biol. Chem. 2020;295:10689–10708. doi: 10.1074/jbc.REV120.011666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma D., Masison D.C. Hsp70 structure, function, regulation and influence on yeast prions. Protein Pept. Lett. 2009;16:571–581. doi: 10.2174/092986609788490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Qian S.-B., McDonough H., Boellmann F., Cyr D.M., Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson S.E. In: Molecular Chaperones. 240. Jackson S., editor. Springer; Berlin, Heidelberg: 2013. Hsp90: structure and function; p. 155. (Topics in Current Chemistry). [Google Scholar]
- 30.Hoter A., El-Sabban M.E., Naim H.Y. The HSP90 family: structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 2018;19:2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo H.-J., Jiang J., Lafer E.M., Sousa R. ATP induced conformational changes in Hsp70: molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry. 2009;48:11470–11477. doi: 10.1021/bi901256y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prodromou C., Roe S.M., O’Brien R., Ladbury J.E., Piper P.W., Pearl L.H. Identification and structural characterization of the ATP/ADP-Binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 33.Albakova Z., Mangasarova Y., Albakov A., Gorenkova L. HSP70 and HSP90 in cancer: cytosolic, endoplasmic reticulum and mitochondrial chaperones of tumorigenesis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.829520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birbo B., Madu E.E., Madu C.O., Jain A., Lu Y. Role of HSP90 in cancer. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luengo T.M., Mayer M.P., Rüdiger S.G.D. The hsp70–hsp90 chaperone cascade in protein folding. Trends Cell Biol. 2019;29:164–177. doi: 10.1016/j.tcb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Peng H.M., Morishima Y., Clapp K.M., Lau M., Pratt W.B., Osawa Y. Dynamic cycling with Hsp90 stabilizes neuronal nitric oxide synthase through calmodulin-dependent inhibition of ubiquitination. Biochemistry. 2009;48:8483–8490. doi: 10.1021/bi901058g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cartledge K., Elsegood C.L., Roiniotis J., Hamilton J.A., Scholz G.M. Importance of the C-terminal domain of harc for binding to Hsp70 and hop as well as its response to heat shock. Biochemistry. 2007;46:15144–15152. doi: 10.1021/bi701041p. [DOI] [PubMed] [Google Scholar]
- 38.Peng G., Zhao W., Zhen-guang S., Chen H., Liu Y., Wei J., et al. Cloning HSP70 and HSP90 genes of kaluga (Huso dauricus) and the effects of temperature and salinity stress on their gene expression. Cell Stress and Chaperones. 2016;21:349–359. doi: 10.1007/s12192-015-0665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirigin E., Matandirotya L., Ruck J.-L., Weaver F., Jackson Z., Chakraborty A., et al. 2024. HOP/STIP1 is required for KSHV lytic replication. (bioRxiv). [preprint] [DOI] [Google Scholar]
- 40.Brychzy A., Rein T., Winklhofer K.F., Hartl F.U., Young J.C., Obermann W.M.J. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the hsp70/hsp90 chaperone system. Embo J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonough H., Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C., Li H.-L., Shi M.-L., Liu Q.-H., Bai J., Zheng J.-N. Diverse roles of C-terminal Hsp70-interacting protein (CHIP) in tumorigenesis. J. Cancer Res. Clin. Oncol. 2014;140:189–197. doi: 10.1007/s00432-013-1571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty S., Karmakar S., Basu M., Ghosh M.K. E3 ubiquitin ligase CHIP drives monoubiquitination-mediated nuclear import of tumor suppressor PTEN. J Cell Sci. 2023;136 doi: 10.1242/jcs.260950. [DOI] [PubMed] [Google Scholar]
- 44.Hu C., Yang J., Qi Z., Wu H., Wang B., Zou F., et al. Heat shock proteins: biological functions, pathological roles, and therapeutic opportunities. MedComm. 2022;3:e161. doi: 10.1002/mco2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballinger C.A., Connell P., Wu Y., Hu Z., Thompson L.J., Yin L.-Y., et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quintana-Gallardo L., Martín-Benito J., Marcilla M., Espadas G., Sabidó E., Valpuesta J.M. The cochaperone CHIP marks Hsp70- and Hsp90-bound substrates for degradation through a very flexible mechanism. Sci. Rep. 2019;9:5102. doi: 10.1038/s41598-019-41060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Xu X., Jiang Z., You Q. Modulation of protein fate decision by small molecules: targeting molecular chaperone machinery. Acta Pharmaceutica Sinica B. 2020;10:1904–1925. doi: 10.1016/j.apsb.2020.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odunuga O.O., Longshaw V.M., Blatch G.L. Hop: more than an Hsp70/Hsp90 adaptor protein. BioEssays. 2004;26:1058–1068. doi: 10.1002/bies.20107. [DOI] [PubMed] [Google Scholar]
- 49.Baindur-Hudson S., Edkins A.L., Blatch G.L. In: The Networking of Chaperones by Co-chaperones: Control of Cellular Protein Homeostasis. Blatch G.L., Edkins A.L., editors. Springer International Publishing; Cham: 2015. Hsp70/Hsp90 organising protein (hop): beyond interactions with chaperones and prion proteins; pp. 69–90. (Subcellular Biochemistry). [Google Scholar]
- 50.Dahiya V., Rutz D.A., Moessmer P., Mühlhofer M., Lawatscheck J., Rief M., et al. The switch from client holding to folding in the Hsp70/Hsp90 chaperone machineries is regulated by a direct interplay between co-chaperones. Mol. Cell. 2022;82:1543–1556.e6. doi: 10.1016/j.molcel.2022.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Chen S., Smith D.F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and Hsp90 chaperone machinery. J. Biol. Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 52.Morimoto R.I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 53.Schopf F.H., Biebl M.M., Buchner J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 54.Griffith A.A., Holmes W. Fine tuning: effects of post-translational modification on Hsp70 chaperones. Int. J. Mol. Sci. 2019;20:4207. doi: 10.3390/ijms20174207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto S., Subedi G.P., Hanashima S., Satoh T., Otaka M., Wakui H., et al. ATPase activity and ATP-dependent conformational change in the co-chaperone HSP70/HSP90-organizing protein (HOP) J. Biol. Chem. 2014;289:9880–9886. doi: 10.1074/jbc.M114.553255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murata S., Minami Y., Minami M., Chiba T., Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stankiewicz M., Nikolay R., Rybin V., Mayer M.P. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 2010;277:3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- 58.Needham P.G., Patel H.J., Chiosis G., Thibodeau P.H., Brodsky J.L. Mutations in the yeast Hsp70, Ssa1, at P417 alter ATP cycling, interdomain coupling, and specific chaperone functions. J. Mol. Biol. 2015;427:2948–2965. doi: 10.1016/j.jmb.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaida A., Iwakuma T. Regulation of P53 and cancer signaling by heat shock protein 40/J-domain protein family members. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222413527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cyr D.M., Ramos C.H. In: The Networking of Chaperones by Co-Chaperones: Control of Cellular Protein Homeostasis. Blatch G.L., Edkins A.L., editors. Springer International Publishing; Cham: 2015. Specification of Hsp70 Function by Type I and Type II Hsp40; pp. 91–102. [Google Scholar]
- 61.Chen K.-C., Qu S., Chowdhury S., Noxon I.C., Schonhoft J., Plate L., et al. The endoplasmic reticulum HSP40 Co-chaperone ERdj3/DNAJB11 assembles and functions as a tetramer. Embo J. 2017;36:2296–2309. doi: 10.15252/embj.201695616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mito R., Matsubara E., Komohara Y., Shinchi Y., Sato K., Yoshii D., et al. Clinical impact of TROP2 in non-small lung cancers and its correlation with abnormal p53 nuclear accumulation. Pathol. Int. 2020;70:287–294. doi: 10.1111/pin.12911. [DOI] [PubMed] [Google Scholar]
- 63.Quinlan D.C., Davidson A.G., Summers C.L., Warden H.E., Doshi H.M. Accumulation of p53 protein correlates with a poor prognosis in human lung cancer. Cancer Res. 1992;52:4828–4831. [PubMed] [Google Scholar]
- 64.Ho Zhi Guang M., Kavanagh E.L., Dunne L.P., Dowling P., Zhang L., Lindsay S., et al. Targeting proteotoxic stress in cancer: a review of the role that protein quality control pathways play in oncogenesis. Cancers (Basel) 2019;11:66. doi: 10.3390/cancers11010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bukh A., Martinez-Valdez H., Freedman S.J., Freedman M.H., Cohen A. The expression of c-fos, c-jun, and c-myc genes is regulated by heat shock in human lymphoid cells. J. Immunol. 1990;144:4835–4840. [PubMed] [Google Scholar]
- 66.Jolly C., Morimoto R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000;92:1564–1572. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- 67.Li D., Marchenko N.D., Schulz R., Fischer V., Velasco-Hernandez T., Talos F., et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruns A.F., Yuldasheva N., Latham A.M., Bao L., Pellet-Many C., Frankel P., et al. A heat-shock protein Axis regulates VEGFR2 proteolysis, blood vessel development and repair. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patani N., Jiang W., Newbold R., Mokbel K. Prognostic implications of carboxyl-terminus of Hsc70 interacting protein and lysyl-oxidase expression in human breast cancer. J. Carcinog. 2010;9:9. doi: 10.4103/1477-3163.72505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexandrova E.M., Marchenko N.D. Mutant p53 – heat shock response oncogenic cooperation: a new mechanism of cancer cell survival. Front. Endocrinol. 2015;6:53. doi: 10.3389/fendo.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cyran A.M., Zhitkovich A. Heat shock proteins and HSF1 in cancer. Front. Oncol. 2022;12:860320. doi: 10.3389/fonc.2022.860320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altinok S., Sanchez-Hodge R., Stewart M., Smith K., Schisler J.C. With or without you: Co-chaperones mediate health and disease by modifying chaperone function and protein triage. Cells. 2021;10:3121. doi: 10.3390/cells10113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Min B., Park H., Lee S., Li Y., Choi J.-M., Lee J.Y., et al. CHIP-mediated degradation of transglutaminase 2 negatively regulates tumor growth and angiogenesis in renal cancer. Oncogene. 2016;35:3718–3728. doi: 10.1038/onc.2015.439. [DOI] [PubMed] [Google Scholar]
- 74.Liang Z.L., Kim M., Huang S.M., Lee H.J., Kim J.-M. Expression of carboxyl terminus of Hsp70-interacting protein (CHIP) indicates poor prognosis in human gallbladder carcinoma. Oncol. Lett. 2013;5:813–818. doi: 10.3892/ol.2013.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu T., Zhou Q., Zhou J., Huang Y., Yan Y., Li W., et al. Carboxyl terminus of Hsp70-interacting protein (CHIP) contributes to human glioma oncogenesis. Cancer Sci. 2011;102:959–966. doi: 10.1111/j.1349-7006.2011.01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kajiro M., Hirota R., Nakajima Y., Kawanowa K., So-ma K., Ito I., et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat. Cell Biol. 2009;11:312–319. doi: 10.1038/ncb1839. [DOI] [PubMed] [Google Scholar]
- 77.Yin H., Deng Z., Li X., Li Y., Yin W., Zhao G., et al. Down-regulation of STIP1 regulate apoptosis and invasion of glioma cells via TRAP1/AKT signaling pathway. Cancer Genet. 2019;237:1–9. doi: 10.1016/j.cancergen.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 78.Walsh N., Larkin A., Swan N., Conlon K., Dowling P., McDermott R., et al. RNAi knockdown of Hop (Hsp70/Hsp90 organising protein) decreases invasion via MMP-2 down regulation. Cancer Lett. 2011;306:180–189. doi: 10.1016/j.canlet.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Wu R., Liu F., Peng P., Qiu H., Xiong H., Yu S., et al. Tumor stress-induced phosphoprotein 1 as a prognostic biomarker for breast cancer. Ann. Transl Med. 2018;6:302. doi: 10.21037/atm.2018.06.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin L., Wen J., Lin B., Xia E., Zheng C., Ye L., et al. Stress-induced phosphoprotein 1 facilitates breast cancer cell progression and indicates poor prognosis for breast cancer patients. Hum. Cell. 2021;34:901–917. doi: 10.1007/s13577-021-00507-1. [DOI] [PubMed] [Google Scholar]
- 81.Schwarz K., Baindur-Hudson S., Blatch G.L., Edkins A.L. Hsp70/Hsp90 organising protein (hop): coordinating much more than chaperones. Subcell Biochem. 2023;101:81–125. doi: 10.1007/978-3-031-14740-1_3. [DOI] [PubMed] [Google Scholar]
- 82.Panni S. Phospho-peptide binding domains in S. cerevisiae model organism. Biochimie. 2019;163:117–127. doi: 10.1016/j.biochi.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 83.Lim S., Kim D.G., Kim S. ERK-dependent phosphorylation of the linker and substrate-binding domain of HSP70 increases folding activity and cell proliferation. Exp. Mol. Med. 2019;51:112. doi: 10.1038/s12276-019-0317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang H., Amick J., Chakravarti R., Santarriaga S., Schlanger S., McGlone C., et al. A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure. 2015;23:472–482. doi: 10.1016/j.str.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sharda P.R., Bonham C.A., Mucaki E.J., Butt Z., Vacratsis P.O. The dual-specificity phosphatase hYVH1 interacts with Hsp70 and prevents heat-shock-induced cell death. Biochem. J. 2009;418:391–401. doi: 10.1042/BJ20081484. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y.-J., Lai K.-C., Kuo H.-H., Chow L.-P., Yih L.-H., Lee T.-C. HSP70 colocalizes with PLK1 at the centrosome and disturbs spindle dynamics in cells arrested in mitosis by arsenic trioxide. Arch. Toxicol. 2014;88:1711–1723. doi: 10.1007/s00204-014-1222-x. [DOI] [PubMed] [Google Scholar]
- 87.Ding C.-L., Xu G., Tang H.-L., Zhu S.-Y., Zhao L.-J., Ren H., et al. Anchoring of both PKA-RIIα and 14-3-3θ regulates retinoic acid induced 16 mediated phosphorylation of heat shock protein 70. Oncotarget. 2015;6:15540–15550. doi: 10.18632/oncotarget.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zemanovic S., Ivanov M.V., Ivanova L.V., Bhatnagar A., Michalkiewicz T., Teng R.-J., et al. Dynamic phosphorylation of the C terminus of Hsp70 regulates the mitochondrial import of SOD2 and redox balance. Cell Rep. 2018;25:2605–2616.e7. doi: 10.1016/j.celrep.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mollapour M., Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mimnaugh E.G., Worland P.J., Whitesell L., Neckers L.M. Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the hsp90 stress protein and the pp60v-src tyrosine kinase (∗) J. Biol. Chem. 1995;270:28654–28659. doi: 10.1074/jbc.270.48.28654. [DOI] [PubMed] [Google Scholar]
- 91.Zhao Y.-G., Gilmore R., Leone G., Coffey M.C., Weber B., Lee P.W.K. Hsp90 phosphorylation is linked to its chaperoning function: assembly of the reovirus cell attachment protein. J. Biol. Chem. 2001;276:32822–32827. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
- 92.Li L., Wang L., You Q.-D., Xu X.-L. Heat shock protein 90 inhibitors: an update on achievements, challenges, and future directions. J. Med. Chem. 2020;63:1798–1822. doi: 10.1021/acs.jmedchem.9b00940. [DOI] [PubMed] [Google Scholar]
- 93.Wang X., Lu X., Song X., Zhuo W., Jia L., Jiang Y., et al. Thr90 phosphorylation of Hsp90α by protein kinase A regulates its chaperone machinery. Biochem. J. 2012;441:387–397. doi: 10.1042/BJ20110855. [DOI] [PubMed] [Google Scholar]
- 94.Lu X., Wang X., Zhuo W., Jia L., Jiang Y., Fu Y., et al. The regulatory mechanism of a client kinase controlling its own release from Hsp90 chaperone machinery through phosphorylation. Biochem. J. 2014;457:171–183. doi: 10.1042/BJ20130963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson O.T., Nadel C.M., Carroll E.C., Arhar T., Gestwicki J.E. Two distinct classes of co-chaperones compete for the EEVD motif in heat shock protein 70 (Hsp70) to tune its chaperone activities. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barati M.T., Rane M.J., Klein J.B., McLeish K.R. A proteomic screen identified stress-induced chaperone proteins as targets of akt phosphorylation in mesangial cells. J. Proteome Res. 2006;5:1636–1646. doi: 10.1021/pr0502469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Assimon V.A., Southworth D.R., Gestwicki J.E. Specific binding of tetratricopeptide repeat proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) is regulated by affinity and phosphorylation. Biochemistry. 2015;54:7120–7131. doi: 10.1021/acs.biochem.5b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ranek M.J., Oeing C., Sanchez-Hodge R., Kokkonen-Simon K.M., Dillard D., Aslam M.I., et al. CHIP phosphorylation by protein kinase G enhances protein quality control and attenuates cardiac ischemic injury. Nat. Commun. 2020;11:5237. doi: 10.1038/s41467-020-18980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim C., Yun N., Lee J., Youdim M.B.H., Ju C., Kim W.-K., et al. Phosphorylation of CHIP at Ser20 by Cdk5 promotes tAIF-mediated neuronal death. Cell Death Differ. 2016;23:333–346. doi: 10.1038/cdd.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sarkar S., Brautigan D.L., Larner J.M. Aurora kinase A promotes AR degradation via the E3 ligase CHIP. Mol. Cancer Res. 2017;15:1063–1072. doi: 10.1158/1541-7786.MCR-17-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Longshaw V.M., Dirr H.W., Blatch G.L., Lässle M. The in vitro phosphorylation of the co-chaperone mSTI1 by cell cycle kinases substantiates a predicted casein kinase II-p34cdc2-NLS (CcN) motif. Biol. Chem. 2000;381:1133–1138. doi: 10.1515/BC.2000.139. [DOI] [PubMed] [Google Scholar]
- 102.Longshaw V.M., Chapple J.P., Balda M.S., Cheetham M.E., Blatch G.L. Nuclear translocation of the Hsp70/Hsp90 organizing protein mSTI1 is regulated by cell cycle kinases. J. Cell Sci. 2004;117:701–710. doi: 10.1242/jcs.00905. [DOI] [PubMed] [Google Scholar]
- 103.Röhl A., Tippel F., Bender E., Schmid A.B., Richter K., Madl T., et al. Hop/Sti1 phosphorylation inhibits its co-chaperone function. EMBO Rep. 2015;16:240–249. doi: 10.15252/embr.201439198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Castelli M., Bhattacharya K., Abboud E., Serapian S.A., Picard D., Colombo G. Phosphorylation of the Hsp90 Co-chaperone hop changes its conformational dynamics and biological function. J. Mol. Biol. 2023;435 doi: 10.1016/j.jmb.2022.167931. [DOI] [PubMed] [Google Scholar]
- 105.Gao Y., Han C., Huang H., Xin Y., Xu Y., Luo L., et al. Heat shock protein 70 together with its co-chaperone CHIP inhibits TNF-α induced apoptosis by promoting proteasomal degradation of apoptosis signal-regulating kinase1. Apoptosis. 2010;15:822–833. doi: 10.1007/s10495-010-0495-7. [DOI] [PubMed] [Google Scholar]
- 106.Liu Q., Wei H., Tang B., Tian B., Ma Z., Gu Q., et al. Blockage of BAG2-CHIP Axis Combats Gastric Cancer by Inducing HSP70 Ubiquitination-Mediated Apoptosis. 2024. [Google Scholar]
- 107.Liu T., Singh R., Rios Z., Bhushan A., Li M., Sheridan P.P., et al. Tyrosine phosphorylation of HSC70 and its interaction with RFC mediates methotrexate resistance in murine L1210 leukemia cells. Cancer Lett. 2015;357:231–241. doi: 10.1016/j.canlet.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Y.-J., Lai K.-C., Kuo H.-H., Chow L.-P., Yih L.-H., Lee T.-C. HSP70 colocalizes with PLK1 at the centrosome and disturbs spindle dynamics in cells arrested in mitosis by arsenic trioxide. Arch. Toxicol. 2014;88:1711–1723. doi: 10.1007/s00204-014-1222-x. [DOI] [PubMed] [Google Scholar]
- 109.O’Regan L., Sampson J., Richards M.W., Knebel A., Roth D., Hood F.E., et al. Hsp72 is targeted to the mitotic spindle by Nek6 to promote K-fiber assembly and mitotic progression. J. Cell Biol. 2015;209:349–358. doi: 10.1083/jcb.201409151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Negroni L., Samson M., Guigonis J.-M., Rossi B., Pierrefite-Carle V., Baudoin C. Treatment of colon cancer cells using the cytosine deaminase/5-fluorocytosine suicide system induces apoptosis, modulation of the proteome, and Hsp90β phosphorylation. Mol. Cancer Ther. 2007;6:2747–2756. doi: 10.1158/1535-7163.MCT-07-0040. [DOI] [PubMed] [Google Scholar]
- 111.Sottile M.L., Cuello-Carrión F.D., Gómez L.C., Semino S., Ibarra J., García M.B., et al. DNA damage repair proteins, HSP27, and phosphorylated-HSP90α as predictive/prognostic biomarkers of platinum-based cancer chemotherapy: an exploratory study. Appl. Immunohistochem. Mol. Morphol. 2022;30:425–434. doi: 10.1097/PAI.0000000000001037. [DOI] [PubMed] [Google Scholar]
- 112.Yang X.-J., Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramazi S., Zahiri J. Post-translational modifications in proteins: resources, tools and prediction methods. Database (Oxford). 2021;2021 doi: 10.1093/database/baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seo J.H., Park J.-H., Lee E.J., Vo T.T.L., Choi H., Kim J.Y., et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat. Commun. 2016;7 doi: 10.1038/ncomms12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun F., Jiang X., Wang X., Bao Y., Feng G., Liu H., et al. Vincristine ablation of Sirt2 induces cell apoptosis and mitophagy via Hsp70 acetylation in MDA-MB-231 cells. Biochem. Pharmacol. 2019;162:142–153. doi: 10.1016/j.bcp.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 116.Yang C., Huntoon K., Ksendzovsky A., Zhuang Z., Lonser R.R. Proteostasis modulators prolong missense VHL protein activity and halt tumor progression. Cell Rep. 2013;3:52–59. doi: 10.1016/j.celrep.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scroggins B.T., Robzyk K., Wang D., Marcu M.G., Tsutsumi S., Beebe K., et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kovacs J.J., Murphy P.J.M., Gaillard S., Zhao X., Wu J.-T., Nicchitta C.V., et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 119.Jakobsson M.E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., et al. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 2013;288:27752–27763. doi: 10.1074/jbc.M113.483248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cho H.-S., Shimazu T., Toyokawa G., Daigo Y., Maehara Y., Hayami S., et al. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat. Commun. 2012;3:1072. doi: 10.1038/ncomms2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rehn A., Lawatscheck J., Jokisch M.-L., Mader S.L., Luo Q., Tippel F., et al. A methylated lysine is a switch point for conformational communication in the chaperone Hsp90. Nat. Commun. 2020;11:1219. doi: 10.1038/s41467-020-15048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park J., Cho J., Song E.J. Ubiquitin–proteasome system (UPS) as a target for anticancer treatment. Arch. Pharm. Res. 2020;43:1144–1161. doi: 10.1007/s12272-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kundrat L., Regan L. Identification of residues on Hsp70 and Hsp90 ubiquitinated by the co-chaperone CHIP. J. Mol. Biol. 2010;395:587–594. doi: 10.1016/j.jmb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gomes R.A., Miranda H.V., Silva M.S., Graça G., Coelho A.V., Ferreira A.E., et al. Yeast protein glycation in vivo by methylglyoxal. FEBS J. 2006;273:5273–5287. doi: 10.1111/j.1742-4658.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- 125.Kundrat L., Regan L. Identification of residues on Hsp70 and Hsp90 ubiquitinated by the cochaperone CHIP. J. Mol. Biol. 2010;395:587–594. doi: 10.1016/j.jmb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Overath T., Kuckelkorn U., Henklein P., Strehl B., Bonar D., Kloss A., et al. Mapping of O-GlcNAc sites of 20 S proteasome subunits and hsp90 by a novel biotin-cystamine tag. Mol. Cell Proteomics. 2012;11:467–477. doi: 10.1074/mcp.M111.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophysical Res. Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 128.Franco M.C., Ye Y., Refakis C.A., Feldman J.L., Stokes A.L., Basso M., et al. Nitration of Hsp90 induces cell death. Proc. Natl. Acad Sci. U. S. A. 2013;110:E1102–E1111. doi: 10.1073/pnas.1215177110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vihervaara A., Sistonen L. HSF1 at a glance. J. Cell Sci. 2014;127:261–266. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 130.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R.A., Godfrey V., et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gomez-Pastor R., Burchfiel E.T., Thiele D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee Y.J., Lee H.J., Lee J.S., Jeoung D., Kang C.M., Bae S., et al. A novel function for HSF1-induced mitotic exit failure and genomic instability through direct interaction between HSF1 and Cdc20. Oncogene. 2008;27:2999–3009. doi: 10.1038/sj.onc.1210966. [DOI] [PubMed] [Google Scholar]
- 133.Chin Y., Gumilar K.E., Li X.-G., Tjokroprawiro B.A., Lu C.-H., Lu J., et al. Targeting HSF1 for cancer treatment: mechanisms and inhibitor development. Theranostics. 2023;13:2281–2300. doi: 10.7150/thno.82431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim E.-H., Lee Y.-J., Bae S., Lee J.S., Kim J., Lee Y.-S. Heat shock factor 1–mediated aneuploidy requires a defective function of p53. Cancer Res. 2009;69:9404–9412. doi: 10.1158/0008-5472.CAN-09-1411. [DOI] [PubMed] [Google Scholar]
- 135.Lu W.-C., Omari R., Ray H., Wang J., Williams I., Jacobs C., et al. AKT1 mediates multiple phosphorylation events that functionally promote HSF1 activation. Febs J. 2022;289:3876. doi: 10.1111/febs.16375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kline M.P., Morimoto R.I. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol. Cell Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shao J., Han B., Cao P., Zhang B., Liu M., Li D., et al. HSF1 phosphorylation by cyclosporin A confers hyperthermia sensitivity through suppression of HSP expression. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 2019;1862:846–857. doi: 10.1016/j.bbagrm.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 138.Fujimoto M., Takii R., Matsumoto M., Okada M., Nakayama K.I., Nakato R., et al. HSF1 phosphorylation establishes an active chromatin state via the TRRAP–TIP60 complex and promotes tumorigenesis. Nat. Commun. 2022;13:4355. doi: 10.1038/s41467-022-32034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee Y.-J., Kim E.-H., Lee J.S., Jeoung D., Bae S., Kwon S.H., et al. HSF1 as a mitotic regulator: phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 2008;68:7550–7560. doi: 10.1158/0008-5472.CAN-08-0129. [DOI] [PubMed] [Google Scholar]
- 140.Kim S.-A., Yoon J.-H., Lee S.-H., Ahn S.-G. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. J. Biol. Chem. 2005;280:12653–12657. doi: 10.1074/jbc.M411908200. [DOI] [PubMed] [Google Scholar]
- 141.Yang T., Ren C., Lu C., Qiao P., Han X., Wang L., et al. Phosphorylation of HSF1 by PIM2 induces PD-L1 expression and promotes tumor growth in breast cancer. Cancer Res. 2019;79:5233–5244. doi: 10.1158/0008-5472.CAN-19-0063. [DOI] [PubMed] [Google Scholar]
- 142.Kourtis N., Moubarak R.S., Aranda-Orgilles B., Lui K., Aydin I.T., Trimarchi T., et al. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell Biol. 2015;17:322–332. doi: 10.1038/ncb3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Westerheide S.D., Anckar J., Stevens S.M., Sistonen L., Morimoto R.I. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Raynes R., Pombier K.M., Nguyen K., Brunquell J., Mendez J.E., Westerheide S.D. The SIRT1 modulators AROS and DBC1 regulate HSF1 activity and the heat shock response. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huang C., Wu J., Xu L., Wang J., Chen Z., Yang R. Regulation of HSF1 protein stabilization: an updated review. Eur. J. Pharmacol. 2018;822:69–77. doi: 10.1016/j.ejphar.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 146.Yeh C.-H., Bellon M., Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol. Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Velasco L., Dublang L., Moro F., Muga A. The complex phosphorylation patterns that regulate the activity of Hsp70 and its cochaperones. Int. J. Mol. Sci. 2019;20:4122. doi: 10.3390/ijms20174122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Faust O., Abayev-Avraham M., Wentink A.S., Maurer M., Nillegoda N.B., London N., et al. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature. 2020;587:489–494. doi: 10.1038/s41586-020-2906-4. [DOI] [PubMed] [Google Scholar]
- 149.Liu Q., Liang C., Zhou L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020;29:378–390. doi: 10.1002/pro.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kostenko S., Jensen K.L., Moens U. Phosphorylation of heat shock protein 40 (Hsp40/DnaJB1) by mitogen-activated protein kinase-activated protein kinase 5 (MK5/PRAK) Int. J. Biochem. Cell Biol. 2014;47:29–37. doi: 10.1016/j.biocel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 151.Poon I.K.H., Jans D.A. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 152.Patel P., Prescott G.R., Burgoyne R.D., Lian L.-Y., Morgan A. Phosphorylation of cysteine string protein triggers a major conformational switch. Structure. 2016;24:1380–1386. doi: 10.1016/j.str.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shirafuji T., Ueyama T., Adachi N., Yoshino K.-I., Sotomaru Y., Uwada J., et al. The role of cysteine string protein α phosphorylation at serine 10 and 34 by protein kinase Cγ for presynaptic maintenance. J. Neurosci. 2018;38:278–290. doi: 10.1523/JNEUROSCI.1649-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Burgoyne R.D., Morgan A. Cysteine string protein (CSP) and its role in preventing neurodegeneration. Semin. Cell Dev. Biol. 2015;40:153–159. doi: 10.1016/j.semcdb.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kastenhuber E.R., Lalazar G., Houlihan S.L., Tschaharganeh D.F., Baslan T., Chen C.-C., et al. DNAJB1–PRKACA fusion kinase interacts with β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. 2017;114:13076–13084. doi: 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim H.-Y., Hong S. Multi-faceted roles of DNAJB protein in cancer metastasis and clinical implications. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232314970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Park S.-Y., Choi H.-K., Seo J.S., Yoo J.-Y., Jeong J.-W., Choi Y., et al. DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 2015;1853:2722–2730. doi: 10.1016/j.bbamcr.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 158.Yu M., Haslam R.H.A., Haslam D.B. HEDJ, an Hsp40 Co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 159.Zoubeidi A., Gleave M. Small heat shock proteins in cancer therapy and prognosis. Int. J. Biochem. Cell Biol. 2012;44:1646–1656. doi: 10.1016/j.biocel.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 160.Lelj-Garolla B., Mauk A.G. Roles of the N- and C-terminal sequences in Hsp27 self-association and chaperone activity. Protein Sci. : A Publ. Protein Soc. 2012;21:122. doi: 10.1002/pro.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jakob U., Gaestel M., Engel K., Buchner J. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 162.Kostenko S., Moens U. Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell. Mol. Life Sci. 2009;66:3289–3307. doi: 10.1007/s00018-009-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Choi S.-K., Kam H., Kim K.-Y., Park S.I., Lee Y.-S. Targeting heat shock protein 27 in cancer: a druggable target for cancer treatment? Cancers. 2019;11:1195. doi: 10.3390/cancers11081195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nakashima M., Adachi S., Yasuda I., Yamauchi T., Kawaguchi J., Itani M., et al. Phosphorylation status of heat shock protein 27 plays a key role in gemcitabine-induced apoptosis of pancreatic cancer cells. Cancer Lett. 2011;313:218–225. doi: 10.1016/j.canlet.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 165.Katsogiannou M., Andrieu C., Rocchi P. Heat shock protein 27 phosphorylation state is associated with cancer progression. Front. Genet. 2014;5:346. doi: 10.3389/fgene.2014.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stope M.B., Weiss M., Preuss M., Streitbörger A., Ritter C.A., Zimmermann U., et al. Immediate and transient phosphorylation of the heat shock protein 27 initiates chemoresistance in prostate cancer cells. Oncol. Rep. 2014;32:2380–2386. doi: 10.3892/or.2014.3492. [DOI] [PubMed] [Google Scholar]
- 167.Taba K., Kuramitsu Y., Ryozawa S., Yoshida K., Tanaka T., Maehara S.-I., et al. Heat-shock protein 27 is phosphorylated in gemcitabine-resistant pancreatic cancer cells. Anticancer Res. 2010;30:2539–2543. [PubMed] [Google Scholar]
- 168.Xu Y., Diao Y., Qi S., Pan X., Wang Q., Xin Y., et al. Phosphorylated Hsp27 activates ATM-dependent p53 signaling and mediates the resistance of MCF-7 cells to doxorubicin-induced apoptosis. Cell Signal. 2013;25:1176–1185. doi: 10.1016/j.cellsig.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 169.Zhao K., Zhou G., Liu Y., Zhang J., Chen Y., Liu L., et al. HSP70 family in cancer: signaling mechanisms and therapeutic advances. Biomolecules. 2023;13:601. doi: 10.3390/biom13040601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nitzsche B., Höpfner M., Biersack B. Synthetic small molecule modulators of Hsp70 and Hsp40 chaperones as promising anticancer agents. Int. J. Mol. Sci. 2023;24:4083. doi: 10.3390/ijms24044083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang Z., Song T., Guo Z., Uwituze L.B., Guo Y., Zhang H., et al. A novel Hsp70 inhibitor specifically targeting the cancer-related Hsp70-Bim protein-protein interaction. Eur. J. Med. Chem. 2021;220 doi: 10.1016/j.ejmech.2021.113452. [DOI] [PubMed] [Google Scholar]
- 172.Sha G., Jiang Z., Zhang W., Jiang C., Wang D., Tang D. The multifunction of HSP70 in cancer: guardian or traitor to the survival of tumor cells and the next potential therapeutic target. Int. Immunopharmacology. 2023;122 doi: 10.1016/j.intimp.2023.110492. [DOI] [PubMed] [Google Scholar]
- 173.Li Z.-N., Luo Y. HSP90 inhibitors and cancer: prospects for use in targeted therapies (Review) Oncol. Rep. 2023;49:1–13. doi: 10.3892/or.2022.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Youssef M.E., Cavalu S., Hasan A.M., Yahya G., Abd-Eldayem M.A., Saber S. Role of ganetespib, an HSP90 inhibitor, in Cancer Therapy: From Molecular Mechanisms to Clinical Practice. Int. J. Mol. Sci. 2023;24:5014. doi: 10.3390/ijms24055014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nadler S.G., Tepper M.A., Schacter B., Mazzucco C.E. Interaction of the immunosuppressant deoxyspergualin with a member of the Hsp70 family of heat shock proteins. Science. 1992;258:484–486. doi: 10.1126/science.1411548. [DOI] [PubMed] [Google Scholar]
- 176.Plowman J., Harrison S.D., Jr., Trader M.W., Griswold D.P., Jr., Chadwick M., McComish M.F., et al. Preclinical antitumor activity and pharmacological properties of Deoxyspergualin1. Cancer Res. 1987;47:685–689. [PubMed] [Google Scholar]
- 177.Dhingra K., Valero V., Gutierrez L., Theriault R., Booser D., Holmes F., et al. Phase II study of deoxyspergualin in metastatic breast cancer. Invest. New Drugs. 1994;12:235–241. doi: 10.1007/BF00873965. [DOI] [PubMed] [Google Scholar]
- 178.Park M., Jung E., Park J.M., Park S., Ko D., Seo J., et al. The HSP90 inhibitor HVH-2930 exhibits potent efficacy against trastuzumab-resistant HER2-positive breast cancer. Theranostics. 2024;14:2442–2463. doi: 10.7150/thno.93236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bhatia S., Spanier L., Bickel D., Dienstbier N., Woloschin V., Vogt M., et al. Development of a first-in-class small-molecule inhibitor of the C-terminal Hsp90 dimerization. ACS Cent. Sci. 2022;8:636–655. doi: 10.1021/acscentsci.2c00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Song Y., Bi Z., Liu Y., Qin F., Wei Y., Wei X. Targeting RAS–RAF–MEK–ERK signaling pathway in human cancer: current status in clinical trials. Genes Dis. 2023;10:76–88. doi: 10.1016/j.gendis.2022.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jiang H., Xu M., Li L., Grierson P., Dodhiawala P., Highkin M., et al. Concurrent HER or PI3K inhibition potentiates the antitumor effect of the ERK inhibitor ulixertinib in preclinical pancreatic cancer models. Mol. Cancer Ther. 2018;17:2144–2155. doi: 10.1158/1535-7163.MCT-17-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sugiura R., Satoh R., Takasaki T. ERK: a double-edged sword in cancer. ERK-dependent apoptosis as a potential therapeutic strategy for cancer. Cells. 2021;10:2509. doi: 10.3390/cells10102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kawano T., Inokuchi J., Eto M., Murata M., Kang J.-H. Activators and inhibitors of protein kinase C (PKC): their applications in clinical trials. Pharmaceutics. 2021;13:1748. doi: 10.3390/pharmaceutics13111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jayaram D.R., Frost S., Argov C., Liju V.B., Anto N.P., Muraleedharan A., et al. Unraveling the hidden role of a uORF-encoded peptide as a kinase inhibitor of PKCs. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2018899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sadeghi M.M., Salama M.F., Hannun Y.A. Protein kinase C as a therapeutic target in non-small cell lung cancer. Int. J. Mol. Sci. 2021;22:5527. doi: 10.3390/ijms22115527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hoy J.J., Parra N.S., Park J., Kuhn S., Iglesias-Bartolome R. Protein kinase A inhibitor proteins (PKIs) divert GPCR-Gαs-cAMP signaling towards EPAC and ERK activation and are involved in tumor growth. FASEB J. 2020;34:13900–13917. doi: 10.1096/fj.202001515R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang H., Kong Q., Wang J., Jiang Y., Hua H. Complex roles of cAMP–PKA–CREB signaling in cancer. Exp. Hematol. Oncol. 2020;9:32. doi: 10.1186/s40164-020-00191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fang X., Lee Y.-H., Jang J.-H., Kim S.-J., Kim S.H., Kim D.-H., et al. ARD1 stabilizes NRF2 through direct interaction and promotes colon cancer progression. Life Sci. 2023;313 doi: 10.1016/j.lfs.2022.121217. [DOI] [PubMed] [Google Scholar]
- 189.Wang Z., Wang Z., Guo J., Li Y., Bavarva J.H., Qian C., et al. Inactivation of androgen-induced regulator ARD1 inhibits androgen receptor acetylation and prostate tumorigenesis. Proc. Natl. Acad. Sci. 2012;109:3053–3058. doi: 10.1073/pnas.1113356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kaur S., Rajoria P., Chopra M. HDAC6: a unique HDAC family member as a cancer target. Cell Oncol. 2022;45:779–829. doi: 10.1007/s13402-022-00704-6. [DOI] [PubMed] [Google Scholar]
- 191.Deskin B., Yin Q., Zhuang Y., Saito S., Shan B., Lasky J.A. Inhibition of HDAC6 attenuates tumor growth of non-small cell lung cancer. Translational Oncol. 2020;13:135–145. doi: 10.1016/j.tranon.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ruan Y., Wang L., Lu Y. HDAC6 inhibitor, ACY1215 suppress the proliferation and induce apoptosis of gallbladder cancer cells and increased the chemotherapy effect of gemcitabine and oxaliplatin. Drug Dev. Res. 2021;82:598–604. doi: 10.1002/ddr.21780. [DOI] [PubMed] [Google Scholar]
- 193.Xu J., Zhou J., Dai H., Liu F., Li W., Wang W., et al. CHIP functions as an oncogene by promoting colorectal cancer metastasis via activation of MAPK and AKT signaling and suppression of E-cadherin. J Transl Med. 2018;16:169. doi: 10.1186/s12967-018-1540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cheng L., Zang J., Dai H.-J., Li F., Guo F. Ubiquitin ligase CHIP functions as an oncogene and activates the AKT signaling pathway in prostate cancer. Int. J. Oncol. 2018;53:203–214. doi: 10.3892/ijo.2018.4377. [DOI] [PubMed] [Google Scholar]
- 195.Seo J., Han S.Y., Seong D., Han H.-J., Song J. Multifaceted C-terminus of HSP70-interacting protein regulates tumorigenesis via protein quality control. Arch. Pharm. Res. 2019;42:63–75. doi: 10.1007/s12272-018-1101-8. [DOI] [PubMed] [Google Scholar]
- 196.Kumar S., Basu M., Ghosh M.K. Chaperone-assisted E3 ligase CHIP: a double agent in cancer. Genes Dis. 2021;9:1521–1555. doi: 10.1016/j.gendis.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Park S., Park S., Ryu K., Kim I., Han H., Kim H., et al. Downregulation of CHIP promotes ovarian cancer metastasis by inducing Snail-mediated epithelial–mesenchymal transition. Mol. Oncol. 2019;13:1280–1295. doi: 10.1002/1878-0261.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Brünnert D., Langer C., Zimmermann L., Bargou R.C., Burchardt M., Chatterjee M., et al. The heat shock protein 70 inhibitor VER155008 suppresses the expression of HSP27, HOP and HSP90β and the androgen receptor, induces apoptosis, and attenuates prostate cancer cell growth. J. Cell Biochem. 2020;121:407–417. doi: 10.1002/jcb.29195. [DOI] [PubMed] [Google Scholar]
- 199.Chao A., Lai C.-H., Tsai C.-L., Hsueh S., Hsueh C., Lin C.-Y., et al. Tumor stress-induced Phosphoprotein1 (STIP1) as a prognostic biomarker in ovarian cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xia Y., Chen J., Liu G., Huang W., Wei X., Wei Z., et al. STIP1 knockdown suppresses colorectal cancer cell proliferation, migration and invasion by inhibiting STAT3 pathway. Chemico-Biological Interactions. 2021;341 doi: 10.1016/j.cbi.2021.109446. [DOI] [PubMed] [Google Scholar]
- 201.Pimienta G., Herbert K.M., Regan L. A compound that inhibits the HOP–Hsp90 complex formation and has unique killing effects in breast cancer cell lines. Mol. Pharmaceutics. 2011;8:2252–2261. doi: 10.1021/mp200346y. [DOI] [PubMed] [Google Scholar]
- 202.Deguchi A., Thompson W.J., Weinstein I.B. Activation of protein kinase G is sufficient to induce apoptosis and inhibit cell migration in colon cancer cells. Cancer Res. 2004;64:3966–3973. doi: 10.1158/0008-5472.CAN-03-3740. [DOI] [PubMed] [Google Scholar]
- 203.Fallahian F., Karami-Tehrani F., Salami S., Aghaei M. Cyclic GMP induced apoptosis via protein kinase G in oestrogen receptor-positive and -negative breast cancer cell lines. FEBS J. 2011;278:3360–3369. doi: 10.1111/j.1742-4658.2011.08260.x. [DOI] [PubMed] [Google Scholar]
- 204.Leung E.L., Wong J.C., Johlfs M.G., Tsang B.K., Fiscus R.R. Protein kinase G type iα activity in human ovarian cancer cells significantly contributes to enhanced src activation and DNA synthesis/cell proliferation. Mol. Cancer Res. 2010;8:578–591. doi: 10.1158/1541-7786.MCR-09-0178. [DOI] [PubMed] [Google Scholar]
- 205.Lv Y., Wang X., Li X., Xu G., Bai Y., Wu J., et al. Nucleotide de novo synthesis increases breast cancer stemness and metastasis via cGMP-PKG-MAPK signaling pathway. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chan S.L., Fiscus R.R. Guanylyl cyclase inhibitors NS2028 and ODQ and protein kinase G (PKG) inhibitor KT5823 trigger apoptotic DNA fragmentation in immortalized uterine epithelial cells: anti-apoptotic effects of basal cGMP/PKG. Mol. Hum. Reprod. 2003;9:775–783. doi: 10.1093/molehr/gag094. [DOI] [PubMed] [Google Scholar]
- 207.Burkhardt M., Glazova M., Gambaryan S., Vollkommer T., Butt E., Bader B., et al. KT5823 inhibits cGMP-dependent protein kinase activity in vitro but not in intact human platelets and rat mesangial cells. J. Biol. Chem. 2000;275:33536–33541. doi: 10.1074/jbc.M005670200. [DOI] [PubMed] [Google Scholar]
- 208.Mou P.K., Yang E.J., Shi C., Ren G., Tao S., Shim J.S. Aurora kinase A, a synthetic lethal target for precision cancer medicine. Exp. Mol. Med. 2021;53:835–847. doi: 10.1038/s12276-021-00635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lin X., Xiang X., Hao L., Wang T., Lai Y., Abudoureyimu M., et al. The role of Aurora-A in human cancers and future therapeutics. Am. J. Cancer Res. 2020;10:2705–2729. [PMC free article] [PubMed] [Google Scholar]
- 210.Wang Q., Bode A.M., Zhang T. Targeting CDK1 in cancer: mechanisms and implications. NPJ Precis. Onc. 2023;7:1–14. doi: 10.1038/s41698-023-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Akl L., Abd El-Hafeez A.A., Ibrahim T.M., Salem R., Marzouk H.M.M., El-Domany R.A., et al. Identification of novel piperazine-tethered phthalazines as selective CDK1 inhibitors endowed with in vitro anticancer activity toward the pancreatic cancer. Eur. J. Med. Chem. 2022;243 doi: 10.1016/j.ejmech.2022.114704. [DOI] [PubMed] [Google Scholar]
- 212.Izadi S., Nikkhoo A., Hojjat-Farsangi M., Namdar A., Azizi G., Mohammadi H., et al. CDK1 in breast cancer: implications for theranostic potential. Anti-Cancer Agents Med. Chem. 2020;20:758–767. doi: 10.2174/1871520620666200203125712. [DOI] [PubMed] [Google Scholar]
- 213.Bruserud Ø., Reikvam H. Casein kinase 2 (CK2): a possible therapeutic target in acute myeloid leukemia. Cancers. 2023;15:3711. doi: 10.3390/cancers15143711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Richter A., Sender S., Lenz A., Schwarz R., Hinz B., Knuebel G., et al. Influence of Casein kinase II inhibitor CX-4945 on BCL6-mediated apoptotic signaling in B-ALL in vitro and in vivo. BMC Cancer. 2020;20:184. doi: 10.1186/s12885-020-6650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Shoaib T.H., Ibraheem W., Abdelrahman M., Osman W., Sherif A.E., Ashour A., et al. Exploring the potential of approved drugs for triple-negative breast cancer treatment by targeting casein kinase 2: insights from computational studies. PLoS One. 2023;18 doi: 10.1371/journal.pone.0289887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Da Costa R., De Almeida S., Chevarin M., Hadj-Rabia S., Leclerc-Mercier S., Thauvin-Robinet C., et al. Neutralization of HSF1 in cells from PIK3CA-related overgrowth spectrum patients blocks abnormal proliferation. Biochem. Biophysical Res. Commun. 2020;530:520–526. doi: 10.1016/j.bbrc.2020.04.146. [DOI] [PubMed] [Google Scholar]
- 217.Wales C.T.K., Taylor F.R., Higa A.T., McAllister H.A., Jacobs A.T. ERK-dependent phosphorylation of HSF1 mediates chemotherapeutic resistance to benzimidazole carbamates in colorectal cancer cells. Anti-Cancer Drugs. 2015;26:657–666. doi: 10.1097/CAD.0000000000000231. [DOI] [PubMed] [Google Scholar]
- 218.Alwahsh M., Farhat J., Talhouni S., Hamadneh L., Hergenröder R. Bortezomib advanced mechanisms of action in multiple myeloma, solid and liquid tumors along with its novel therapeutic applications. EXCLI J. 2023;22:146–168. doi: 10.17179/excli2022-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shah S.P., Nooka A.K., Jaye D.L., Bahlis N.J., Lonial S., Boise L.H. Bortezomib-induced heat shock response protects multiple myeloma cells and is activated by heat shock factor 1 serine 326 phosphorylation. Oncotarget. 2016;7:59727–59741. doi: 10.18632/oncotarget.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sasaya T., Kubo T., Murata K., Mizue Y., Sasaki K., Yanagawa J., et al. Cisplatin-induced HSF1-HSP90 axis enhances the expression of functional PD-L1 in oral squamous cell carcinoma. Cancer Med. 2023;12:4605–4615. doi: 10.1002/cam4.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Alalem M., Bhosale M., Ranjan A., Yamamoto S., Kaida A., Nishikawa S., et al. Mutant p53 depletion by novel inhibitors for HSP40/J-domain proteins derived from the natural compound plumbagin. Cancers. 2022;14:4187. doi: 10.3390/cancers14174187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Parrales A., Ranjan A., Iyer S.V., Padhye S., Weir S.J., Roy A., et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat. Cell Biol. 2016;18:1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Suzuki M., Yamamoto Y., Nishijima-Matsunobu A., Kawasaki Y., Shibata H., Omori Y. A curcumin analogue GO-Y030 depletes cancer stem cells by inhibiting the interaction between the HSP70/HSP40 complex and its substrates. FEBS Open Bio. 2023;13:434–446. doi: 10.1002/2211-5463.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xiong J., Li Y., Tan X., Fu L. Small heat shock proteins in cancers: functions and therapeutic potential for cancer therapy. Int. J. Mol. Sci. 2020;21:6611. doi: 10.3390/ijms21186611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hoter A., Rizk S., Naim H.Y. The multiple roles and therapeutic potential of molecular chaperones in prostate cancer. Cancers. 2019;11:1194. doi: 10.3390/cancers11081194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Spigel D.R., Shipley D.L., Waterhouse D.M., Jones S.F., Ward P.J., Shih K.C., et al. A randomized, double-blinded, phase II trial of carboplatin and pemetrexed with or without apatorsen (OGX-427) in patients with previously untreated stage IV non-squamous-non-small-cell lung cancer: the SPRUCE trial. The Oncologist. 2019;24:e1409–e1416. doi: 10.1634/theoncologist.2018-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chen S.-F., Nieh S., Jao S.-W., Liu C.-L., Wu C.-H., Chang Y.-C., et al. Quercetin suppresses drug-resistant spheres via the p38 MAPK–hsp27 apoptotic pathway in oral cancer cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vafadar A., Shabaninejad Z., Movahedpour A., Fallahi F., Taghavipour M., Ghasemi Y., et al. Quercetin and cancer: new insights into its therapeutic effects on ovarian cancer cells. Cell Biosci. 2020;10:32. doi: 10.1186/s13578-020-00397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang R., Yang L., Li S., Ye D., Yang L., Liu Q., et al. Quercetin inhibits breast cancer stem cells via downregulation of aldehyde dehydrogenase 1A1 (ALDH1A1), chemokine receptor type 4 (CXCR4), mucin 1 (MUC1), and epithelial cell adhesion molecule (EpCAM) Med. Sci. Monit. 2018;24:412–420. doi: 10.12659/MSM.908022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Easton D.P., Kaneko Y., Subjeck J.R. The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gozzi G.J., Gonzalez D., Boudesco C., Dias A.M.M., Gotthard G., Uyanik B., et al. Selecting the first chemical molecule inhibitor of HSP110 for colorectal cancer therapy. Cell Death Differ. 2020;27:117–129. doi: 10.1038/s41418-019-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]