Abstract
Enteropathogenic Escherichia coli (EPEC) is a cause of prolonged watery diarrhea in children in developing countries. The ability of EPEC to kill host cells was investigated in vitro in assays using two human cultured cell lines, HeLa (cervical) and T84 (colonic). EPEC killed epithelial cells as assessed by permeability to the vital dyes trypan blue and propidium iodide. In addition, EPEC triggered changes in the host cell, suggesting apoptosis as the mode of death; such changes included early expression of phosphatidylserine on the host cell surface and internucleosomal cleavage of host cell DNA. Genistein, an inhibitor of tyrosine kinases, and wortmannin, an inhibitor of host phosphatidylinositol 3-kinase, markedly increased EPEC-induced cell death and enhanced the features of apoptosis. EPEC-induced cell death was contact dependent and required adherence of live bacteria to the host cell. A quantitative assay for EPEC-induced cell death was developed by using the propidium iodide uptake method adapted to a fluorescence plate reader. With EPEC, the rate and extent of host cell death were less that what has been reported for Salmonella, Shigella, and Yersinia, three other genera of enteric bacteria known to cause apoptosis. However, rapid apoptosis of the host cell may not favor the pathogenic strategy of EPEC, a mucosa-adhering, noninvasive pathogen.
Enteropathogenic Escherichia coli (EPEC) causes prolonged watery diarrhea in children in developing countries and is occasionally recognized as an agent of diarrhea in outbreaks among children and adults in developed countries. EPEC adheres intimately to intestinal epithelial cells, causes rearrangements of the cytoskeleton of the host cell, and activates several signalling pathways in the host cell, including protein kinases. While a great deal has been learned in recent years about EPEC adherence and early steps of EPEC-host cell interaction, the mechanism(s) by which EPEC actually causes diarrhea is still not well understood.
The ability of EPEC to damage and eventually kill host cells is one mechanism by which EPEC might cause disease. We became interested in these events while investigating the role of the host cell enzyme phosphatidylinositol 3-kinase (PI 3-kinase) in EPEC pathogenesis. Ireton et al. reported that wortmannin, an inhibitor of PI 3-kinase, blocked the ability of Listeria monocytogenes to invade cultured cells (21). Similarly, Wooldridge et al. showed that wortmannin inhibited the invasion of cultured cells by Campylobacter jejuni (45). While attempting to determine the effects of wortmannin on EPEC adherence and on EPEC invasion, we noted that while the cultured cell lines (HeLa, HEK-293, and T84) tolerated EPEC infection and wortmannin treatment separately, the combination of EPEC infection and wortmannin treatment caused massive cell detachment by 4 h. We sought to understand these events and to compare the mode of host cell death caused by EPEC with the cell death caused by other enteric bacteria, such as Salmonella, Shigella, and Yersinia species, which have been reported to trigger apoptosis (programmed cell death) in the host.
We found that EPEC alone causes host cells to become permeable to vital dyes in a manner dependent on contact with live bacteria. We also found that EPEC-induced cell death has features of apoptosis, including early expression of phosphatidylserine on the host cell surface and internucleosomal cleavage of DNA (the apoptotic DNA ladder). We were also able to develop a quantitative assay for EPEC-induced cell death based on uptake of propidium iodide or ethidium homodimer into cells in a multiwell plate format using a fluorescence plate reader.
MATERIALS AND METHODS
Materials.
Wortmannin and annexin V-fluorescein isothiocyanate (FITC) were from Alexis (San Diego, Calif.). Wortmannin was prepared as a 1 mM stock in dimethyl sulfoxide and stored at −20°C in individual aliquots for up to 2 months. Genistein, doxorubicin, propidium iodide, and ethidium bromide were obtained from Sigma (St. Louis, Mo.). Trypan blue, proteinase K, RNase A, tissue culture media, and animal sera were from Gibco/BRL (Gaithersburg, Md.). Lab-Tek chamber slides were from Nunc/Intermed (Napierville, Ill.). MG-132 (also known as Z-Leu-Leu-Leu-CHO), an inhibitor of proteosomal proteases, was obtained from Biomol (Plymouth Meeting, Pa.), prepared as a 2 mM stock solution in dimethyl sulfoxide or ethanol, and was stored at −70°C for up to 2 months.
Bacterial culture.
The E. coli strains used have been previously described (5, 8, 26). Briefly, strains E2348/69, E851/71, and B171-8 are wild-type EPEC. For brevity in figure labels and discussion, the name of strain E2348/69 is abbreviated as E2348. HB101 is a laboratory strain of E. coli, while HS is a normal colonic flora strain of E. coli. JPN15 is the plasmid-cured derivative of E2348 (27), and CVD206 has eae, the gene encoding intimin, deleted (10). Salmonella enterica var. enteritidis was a clinical isolate obtained from a patient at Erie County Medical Center, Buffalo, N.Y. For the experiments described here, bacteria were cultured overnight in Luria-Bertani (LB) broth supplemented with 10 g of mannose per liter and then subcultured at a ratio of 1:10 for 2 h at 37°C in eukaryotic tissue culture medium (Dulbecco modified Eagle medium [DMEM] or DMEM–F-12) supplemented with 10 g of mannose per liter, 15 mM HEPES, 18 mM NaHCO3, and 2% heat-inactivated newborn calf serum, which we refer to as EPEC adherence medium (EAM) (8). Subculturing in EAM induces the expression of the bundle-forming pilus and intimin, triggers spontaneous clumping of EPEC bacteria, and accelerates adhesion of EPEC to tissue cultured cells (43). Serum is not necessary for EPEC adherence but was included in the medium to reduce the possibility of host cell death due to serum withdrawal.
Culture of human cell lines.
Cell lines T84 (human colon carcinoma) and HeLa (human cervical cancer) were obtained from the American Type Culture Collection and maintained exactly as previously described (8).
Trypan blue uptake assay for microscopy.
Cells were grown in Lab-Tek chamber slides, subjected to various infection conditions and/or treatment with chemical reagents, rinsed twice gently with phosphate-buffered saline (PBS) to remove nonadhering bacteria, and then stained with 0.4% trypan blue at 37°C for 10 min. The trypan blue was rinsed off with two or three gentle washes with PBS, and then the cells were fixed with 0.1% cacodylate–2% glutaraldehyde in PBS for 10 min. After fixation, the upper buffer chamber of the Lab-Tek slide was snapped off, a drop of PBS was applied, a glass coverslip was placed over the slide, and the slide was examined by differential interference contrast microscopy.
Propidium iodide uptake assay.
Propidium iodide uptake was determined by the method of Oberdoerster et al. (34). Propidium iodide is excluded from living, healthy cells and fluoresces much more intensely when bound to DNA than when free in solution. Since phenol red dye, present in most tissue culture media as a pH indicator, interferes with the emission of propidium iodide fluorescence, the propidium iodide uptake measurements were performed in optically clear medium consisting of phenol red-free RPMI 1640 (Gibco/BRL) supplemented with 25 mM HEPES (pH 7.4), 18 mM NaHCO3, bovine serum albumin (1 g/liter) and propidium iodide (2 μg/ml [3 μM]) but no serum. T84 cells were grown to confluency in Falcon 48-well tissue culture plates, with at least two wells left completely empty on each plate for use as blanks. On the day of the experiment, the medium was changed to EAM, inhibitors or reagents were added, and the cells were infected with bacteria at a multiplicity of infection (MOI) of 100:1 unless otherwise stated. Two hours after infection, the medium was removed and replaced with the phenol red-free, RPMI 1640-based medium mentioned above, with 2 μg of propidium iodide per ml. Fluorescence measurements were taken with a Cytofluor II fluorescence plate reader with an excitation wavelength of 530 ± 25 nm and an emission wavelength of 645 ± 40 nm. The readings from the two empty wells were averaged, and the mean blank value was subtracted from all remaining wells. Each 48-well plate was usually read at various time points over the course of an experiment (e.g., 3, 4, and 6 h) and was returned to the 37°C, 5% CO2 incubator between measurements. As a positive control for cell death, either 0.05% Triton X-100 or 0.1% H2O2 (final concentration) was added to some wells. In some experiments, ethidium homodimer was used instead of propidium iodide but at the same concentration (2 μg/ml).
For microscopy of propidium iodide-stained cells (Fig. 2), E. coli bacteria were subcultured in EAM supplemented with acridine orange (1 μg/ml) for 2 h in order to allow visualization of the bacteria. After 2 h of infection, the slides were rinsed in PBS, stained with propidium iodide (5 μg/ml) in RPMI 1640 medium at 37°C for 30 min, then rinsed again, allowed to dry, and examined under oil immersion in a fluorescence microscope using the blue filter cube, in which propidium iodide fluoresces red or reddish orange and acridine orange fluoresces yellow-green.
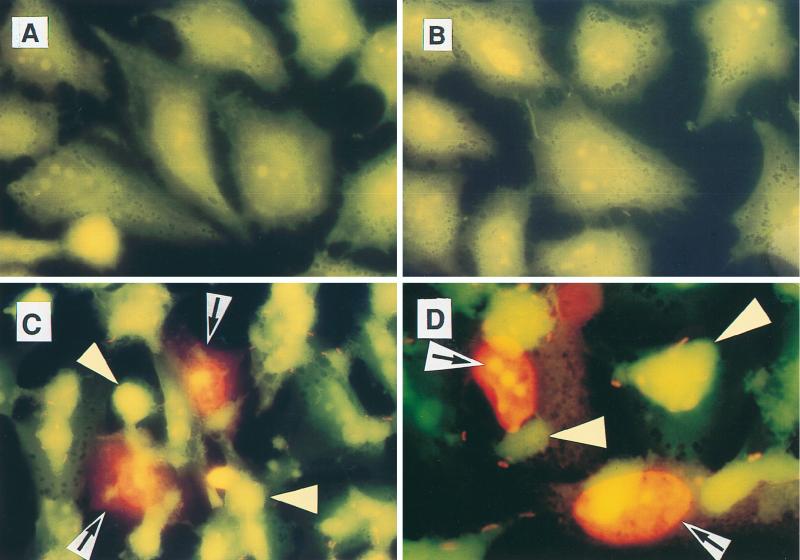
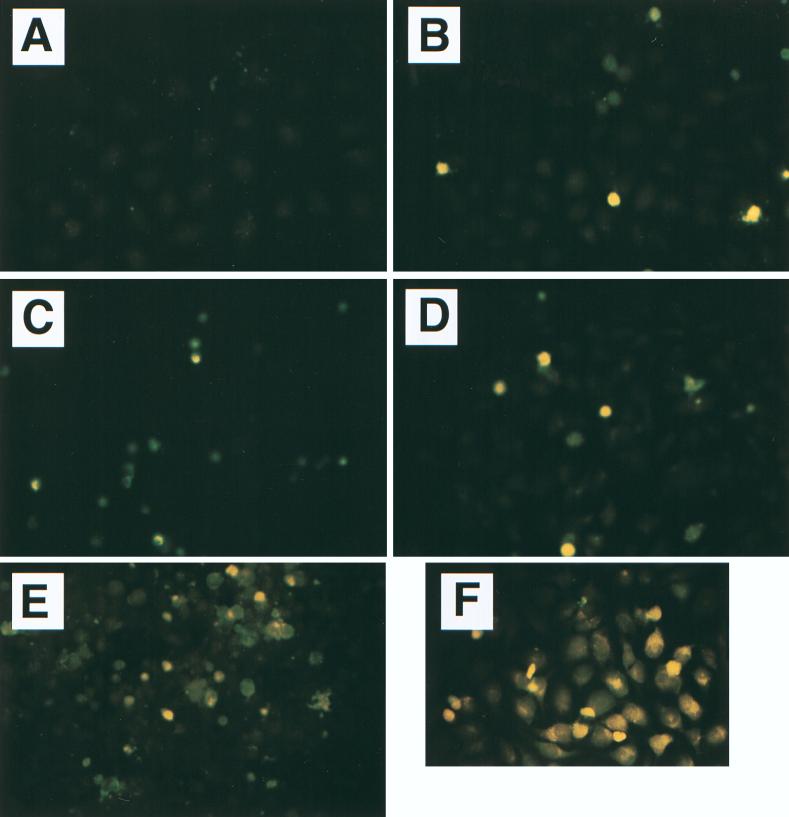
FIG. 2.
Colocalization of cell death with adherent EPEC. HeLa cells were left uninfected (A), infected with nonadherent strain E. coli HB101 (B), or infected with EPEC strain B171-8 (C and D) for 2 h. Bacteria had been fluorescently labeled by subculturing in the presence of acridine orange; after infection, HeLa cells were stained with propidium iodide for an additional 30 min as described in Materials and Methods. Original magnifications: A through C, ×600 (under oil); D, ×1,000. Propidium iodide-stained HeLa cell nuclei are 16 to 20 μm in size and fluoresce reddish orange (black arrows on white background), and clumps of adherent EPEC appear greenish yellow (yellow arrowheads). The apparent size of an individual bacterial cell (E. coli B171-8) is 1.8 μm (propidium iodide stains the central nucleoid, not the cell membrane or wall).
LDH release assay for cell death.
As a confirmation of the results obtained with the propidium iodide uptake method, cells were grown in multiwell plates and infected as described above for propidium iodide uptake except that serum-free adherence medium was used in order to avoid lactate dehydrogenase (LDH) activity present in serum. After 3 h of infection, the medium was aspirated and replaced with 0.25 ml of PBS; the plates were returned to the incubator for an additional 30 min. Once again, in some wells 0.5% Triton X-100 was added as a positive control (a higher concentration of Triton is used because in the LDH release method the intent is to lyse the cells completely rather than just permeabilize them in situ). The supernatant medium was collected and kept on ice; then the LDH was measured with a cytotoxicity detection kit from Boehringer Mannheim (now Roche Molecular Biochemicals, Indianapolis, Ind.) and read on a enzyme-linked immunosorbent assay reader at 492 nm. Raw absorbance readings were used to calculate the percent cell death compared to that released by Triton X-100 detergent, using the equation shown below (see “Data analysis and presentation”).
Assay for expression of phosphatidylserine on the surface of cells.
HeLa or T84 cells were grown to confluency in Lab-Tek chamber slides, subjected to infection with bacteria or treatment with chemical reagents, and then rinsed twice with sterile PBS. Purified, human recombinant annexin V conjugated to FITC was used according to the manufacturer’s recommendations. Briefly, the annexin V was diluted 40-fold in binding buffer (140 mM NaCl, 10 mM HEPES [pH 7.4], 2.5 mM CaCl2), yielding a final annexin V-FITC concentration of 5 μg/ml; approximately 200 μl of this solution was placed in each chamber of a four-well Lab-Tek slide and allowed to incubate with the living cells at 37°C for 10 min. The cells were then rinsed twice more with PBS, the plastic chambers were snapped off, and the slide was covered with a coverslip and examined microscopically. In some experiments the cells were fixed with 0.1% cacodylate–2% glutaraldehyde prior to examination, a procedure which did not seem to affect the results. Fluorescence microscopy was performed with a Nikon Optiphot microscope using the blue filter set.
Extraction of apoptotic DNA from cells.
The procedure for extraction of apoptotic DNA was adapted from the phosphate-citric acid method of Gong et al. (18), modified at several steps in order to accommodate adherent rather than suspended cells according to Oberdoerster et al. (34) and our own pilot experiments. T84 cells were grown to confluency in six-well tissue culture plates (∼4 × 106 T84 cells per well at confluency). Antibiotic-containing medium was removed and replaced with 2 ml of EAM, and the cells were treated with inhibitor reagents and/or infected with 0.4 ml of bacterial subculture. The MOIs were 75:1 for the EPEC strain E2348 and 150:1 for nonadherent strain HS, which has a higher growth rate. After 2 h of infection, the medium was removed and replaced with 2 ml of fresh EAM. Six hours after infection, the medium was removed and centrifuged at 1,000 × g for 5 min to recover any detached cells; the resulting cell pellet, if any, was resuspended in 0.5 ml of 70% ethanol; 1.5 ml of 70% ethanol was added to each tissue culture well, and the plate and the resuspended cell pellets were kept at −20°C overnight. As a control, E. coli bacteria were also subjected to the DNA extraction method described for apoptotic host cells. Briefly, 0.5 ml of E. coli subculture (HB101 and E2348) was centrifuged at 14,000 × g for 5 min to pellet the bacteria, and then the bacterial pellet was subjected to the extraction steps described below for T84 cells.
As a positive control for internucleosomal DNA cleavage, H9 leukemia cells were obtained as a gift from E. James Bergey, Department of Oral Biology, State University of New York at Buffalo, and treated with doxorubicin (2 μg/ml) for 16 h. These cells (∼3 × 105) were recovered by centrifugation, kept at −20°C in 70% ethanol, and then processed in parallel with the T84 cells.
The next day, the adherent T84 cells were removed from the −20°C freezer, scraped from the wells with a Teflon cell scraper, and pooled in 12- by 75-mm test tubes with any detached cells from that well. The cells were collected by centrifugation at 1,000 × g at 4°C for 5 min, and the ethanol was decanted. The cell pellets were rinsed gently with 1.5 ml of Hanks’ balanced salt solution (Gibco/BRL) without breaking up or physically resuspending the pellet and again subjected to the same centrifugation; 0.4 ml of Hanks’ balanced salt solution was added to the cell pellet, and as gently as possible, the pellet was resuspended and transferred to a small Eppendorf microcentrifuge tube. The cell suspensions were centrifuged at 14,000 × g at 4°C for 5 min, and the supernatant was aspirated. The phosphate-citric acid extraction buffer was prepared freshly for each experiment by mixing 4.8 ml of 0.2 M Na2HPO4 with 0.2 ml of 0.1 M citric acid; 100 μl of the phosphate-citric acid solution was added to each cell pellet, and the material in the tube was mixed with a Nutator mixer (Becton Dickinson, Sparks, Md.) at room temperature for 40 min; vortexing was avoided. The Eppendorf tubes were again centrifuged at 14,000 × g at 4°C for 5 min, and the supernatant extracts were recovered and transferred to new small Eppendorf tubes. Each tube was treated with 10 μl of 0.25% Nonidet P-40 detergent and 5 μl of RNase A (10 mg/ml; Gibco/BRL) and incubated at 37°C for at least 30 min. Next, 5 μl of proteinase K (10 mg/ml; Gibco/BRL) was added, and the tubes were again incubated at 37°C for at least 30 min.
The DNA content of the extracts was estimated by UV spectrometry at 260 nm, and this value was averaged for several samples. The volume of extract needed to yield 3 μg of DNA was calculated, and this volume was then used for all samples. When possible, the extract was diluted with water to decrease the salt concentration and improve the electrophoresis.
Analysis of DNA fragments was performed with a 2% agarose gel in 1× Tris-borate-EDTA buffer; ethidium bromide (0.5 μg/ml) was added to the gel to allow visualization of DNA. Electrophoresis was carried out at 5 V/cm for ∼4 h. The gel was photographed under UV illumination with a Bio-Rad GelDoc 1000 camera. For ease of reproduction, the image of the gel was converted to a negative (inverted image).
Quantitative adherence assay using [3H]thymidine-labeled bacteria.
To reduce detachment of cells during the experiment, HeLa cells were grown in collagen-coated 24-well plates (Biocoat; Becton Dickinson, Bedford, Mass.). E. coli strains were labeled with [3H]thymidine by growth overnight in 0.5 ml of LB broth with 1% mannose and 4 μCi of [3H]thymidine. The next day, the labeled bacteria were subcultured 1:10 into EAM, and another 4 μCi of [3H]thymidine was added. After 2 h of incubation, the bacteria were pelleted by centrifugation at 1,000 × g at room temperature and resuspended in an equal volume of warm EAM. UV light irradiation of HeLa cells was carried out by placing a 24-well plate on a UV transilluminator (model TM-20; UVP, Inc., San Gabriel, Calif.) for 90 s according to the method of Kulik et al. (25); the UV exposure was performed 3 h prior to infection with EPEC. The resuspended, [3H]thymidine-labeled bacteria were used to infect control and UV-irradiated HeLa cells for 1.5 h; then the wells were rinsed twice with PBS and solubilized in 0.5% Triton X-100 in water; the solubilized material was transferred to scintillation vials, scintillant was added, and radioactivity was counted in a liquid scintillation counter.
Data analysis and presentation.
All numerical data presented are the means of at least triplicate determinations, and error bars shown are standard deviations. Each experiment shown was performed at least three times. Percent cell death (LDH release and propidium iodide uptake assays) was calculated as [(reading from experimental condition) − (reading from uninfected control)]/ [(reading from Triton-treated wells) − (reading from uninfected control)] × 100. Statistical analysis (Fig. 5 and 6) was by t test or one-way analysis of variance using Statview II for the Macintosh (Abacus Concepts, Berkeley, Calif.).
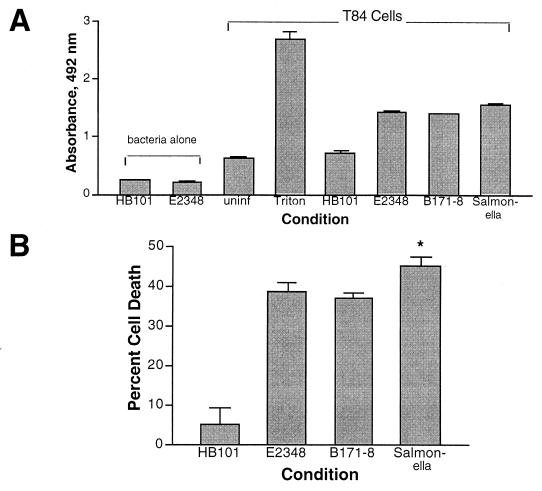
FIG. 5.
Quantitation of EPEC-induced cell death by LDH release. (A) LDH activity was measured as described in Materials and Methods in E. coli bacteria alone and in T84 cells treated as indicated for 3 h. uninf, uninfected control. (B) The data in panel A expressed as percent cell death. *, statistically increased in Salmonella-infected compared to both EPEC-infected conditions (P < 0.05).
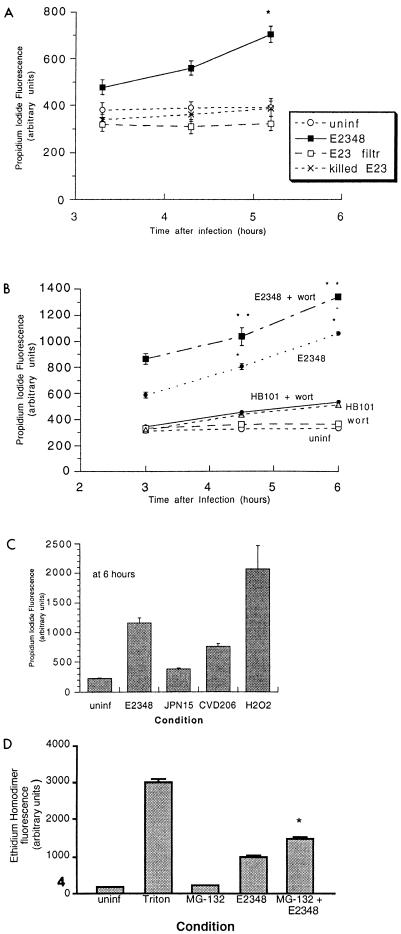
FIG. 6.
Quantitation of EPEC-induced cell death by propidium iodide uptake. T84 cells grown to confluency in 48-well plates were treated with or without inhibitors or infected with E. coli in adherence medium for 2 h, and then the medium was replaced with phenol red-free medium containing propidium iodide (2 μg/ml) as described in Materials and Methods; measurements were taken at various times afterward, as indicated on the graph. Abbreviations: uninf, uninfected control; E2348, infected with E2348 alone; E23 filtr, cells treated with a sterile filtrate of EPEC subculture; killed E23, E2348 bacteria killed by incubation with ciprofloxacin (15 μg/ml) for 5 min prior to addition to the T84 cells; wort, treated with 100 nM wortmannin; JPN15 and CVD206, the plasmid-cured and eae-deleted derivatives of E2348, respectively; H2O2, cells treated with 0.1% H2O2 as a positive control. In panel B, the single asterisk indicates statistical significance at P < 0.05 compared to HB101 plus wortmannin and the double asterisk indicates statistical significance (P < 0.05) compared to E2348 alone. In panel C, the three infected conditions gave results which were significantly different from each other (P < 0.05). In panel D, ethidium homodimer was used instead of propidium iodide; the concentration of MG-132 used was 40 μM, and the data shown are at 6 h after infection; the asterisk indicates statistical significance (P < 0.05) compared to E2348 alone.
RESULTS
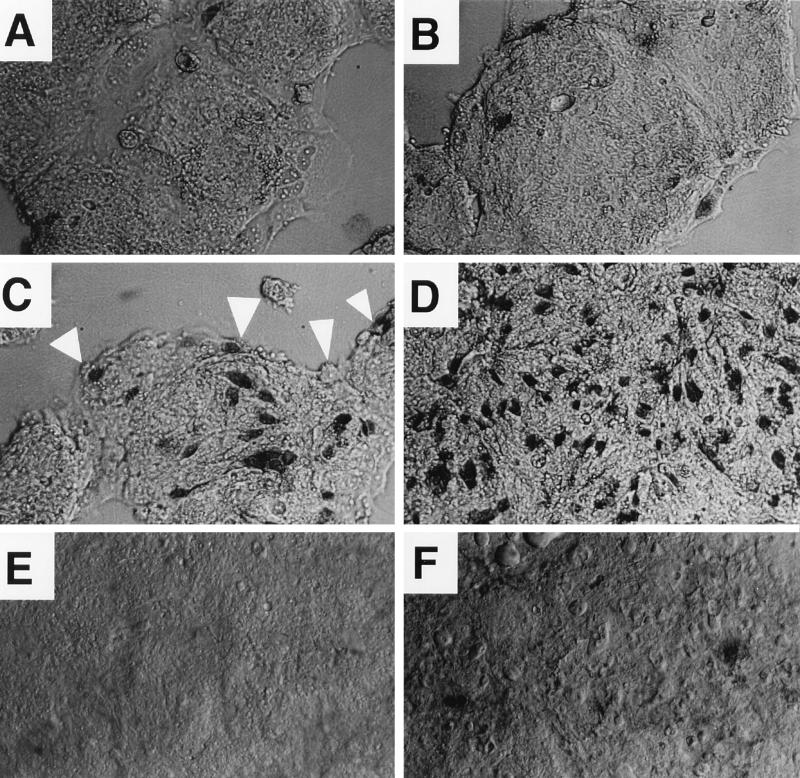
While attempting to use the inhibitor wortmannin to investigate the role of PI 3-kinase, a host cell enzyme, in EPEC signalling, we observed marked cytotoxicity with simultaneous wortmannin treatment and EPEC infection on three different cell lines (T84, HeLa, and HEK-293). To investigate this phenomenon further, we examined cells for trypan blue uptake at somewhat earlier times after EPEC infection, wortmannin treatment, or both. The results of a trypan blue uptake assay using T84 cells after 2 h of various treatments showed that 100 nM wortmannin alone had little effect on T84 cell viability (Fig. 1B), whereas infection with EPEC strain E851/71 caused a noticeable increase in trypan blue-positive cells (Fig. 1C). Trypan blue-positive cells were almost invariably in direct contact with adherent clumps of EPEC bacteria. The converse was not true, however; often EPEC clumps were seen adhering to cells which still excluded the trypan blue dye (Fig. 1C). EPEC infection in the presence of wortmannin resulted in about twice as many blue cells per field as seen with EPEC alone (Fig. 1D). In contrast, the nonadherent E. coli strain HB101 did not cause significant cell death, even in the presence of wortmannin (Fig. 1E and F). Results very similar to those in Fig. 1 were obtained with HeLa cells (not shown).
FIG. 1.
EPEC-induced cell killing by trypan blue uptake. T84 cells were grown in Lab-Tek chamber slides, changed to antibiotic-free adherence medium, treated with inhibitors and/or infected with an E. coli strain for 2 h, and then stained with trypan blue as described in Materials and Methods. Cells were viewed by differential interference contrast microscopy at a magnification of ×200. (A) Uninfected control; (B) cells treated with 100 nM wortmannin alone; (C) cells infected with EPEC strain E851/71; (D) cells infected with E851/71 in the presence of 100 nM wortmannin; (E) cells infected with laboratory strain HB101; (F) cells infected with HB101 in the presence of wortmannin.
Figure 2 shows that EPEC infection also triggered host cell permeability to propidium iodide in a contact-dependent manner. In this experiment, E. coli bacteria had been labeled by subculturing in the presence of acridine orange (1 μg/ml) to allow their visualization. No uptake of propidium iodide was observed in normal, control cells (Fig. 2A) or cells infected with E. coli HB101 (Fig. 2B). However, after 2.5 h of infection with wild-type EPEC strain B171-8, vivid staining of nuclei is seen in host cells directly adhered to by EPEC (Fig. 2C and D). EPEC strains adhere as large clumps of 100 or more bacteria. Only a few EPEC bacteria at the edges of each clump take up the propidium iodide stain, which is seen better at high power (Fig. 2D) and provides an indication of the size of an individual bacterium.
To determine if EPEC-induced cell death showed features of apoptosis, we looked for distinctive morphological changes, expression of abnormal phospholipids on the host cell surface, and evidence of internucleosomal cleavage of DNA. In the cell lines that we examined (T84 and HeLa), only rarely did EPEC-infected cells show distinct morphological changes of apoptosis such as cell shrinkage, membrane blebbing, nuclear condensation, or nuclear fragmentation. However, morphological changes indicating death by necrosis, such as cell swelling and decreased retention of histological stains, were also not seen.
The phospholipids phosphatidylserine and phosphatidylethanolamine are highly asymmetrically distributed in the plasma membrane of normal cells, with at least 99% of these lipids found in the inner, cytoplasmic leaflet of the lipid bilayer. An early marker of apoptosis is the surface expression of these lipids (47). We labeled HeLa cells with FITC-tagged annexin V, a protein which binds to phosphatidylserine, and examined them by fluorescence microscopy. Normal, control HeLa cells showed only faint, background fluorescence (Fig. 3A). Wortmannin alone for 3 h triggered the appearance of a few cells exhibiting bright yellow fluorescence typical of fluorescein (Fig. 3B). EPEC infection caused a gradual increase in the number of bright yellow cells with increasing duration of infection (Fig. 3C [1 h] and D [3 h]). The combination of wortmannin treatment and EPEC infection again triggered a large increase in the number of brightly fluorescing cells (Fig. 3E), an increase which appeared to be supra-additive compared to either treatment applied alone. In fact, the combination of EPEC plus wortmannin was almost as effective in triggering phosphatidylserine expression as a 20-h exposure to doxorubicin, a known inducer of apoptosis, at a concentration of 1 μg/ml (Fig. 3F). Results very similar to those in Fig. 3 were obtained with T84 cells.
FIG. 3.
Early expression of phosphatidylserine on the host cell surface after EPEC infection. HeLa cells were grown to confluency on Lab-Tek slides, infected with EPEC strain E2348 in the presence or absence of wortmannin, and then incubated with FITC-labeled annexin V to reveal phosphatidylserine on the cell surface as described in Materials and Methods. (A) Normal, uninfected control HeLa cells; (B) cells treated with 100 nM wortmannin for 3 h; (C) cells infected with E2348 for 1 h; (D) cells infected with E2348 for 3 h; (E) cells infected with E2348 in the presence of 100 nM wortmannin for 3 h; (F) cells treated for 20 h with doxorubicin (1 μg/ml) as a positive control. Magnifications: A through E, ×200; F, ×400.
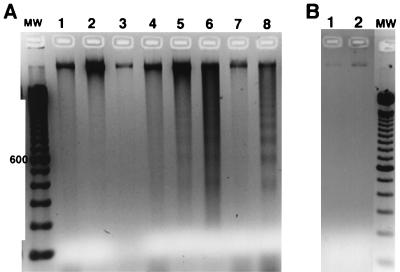
Internucleosomal cleavage of DNA is one of the most distinctive features of apoptosis. To determine if EPEC-induced cell death showed this pattern, we compared several different methods for extraction of DNA from cells. Of these various techniques, the method of Gong et al. (18) using a phosphate-citric acid extraction step, as modified to accommodate adherent rather that suspended cells, gave the best results. Figure 4A shows the result of agarose gel electrophoresis of the DNA extracted from T84 cells after they were subjected to various treatment conditions. No DNA cleavage was observed in control cells, in cells treated with wortmannin alone or genistein alone, or in cells infected with normal flora E. coli strain HS for 6 h (Fig. 4A, lanes 1, 2, 3, and 7). EPEC infection alone for 6 h caused a small amount of DNA breakdown with faint apoptotic banding (Fig. 4A, lane 4). The combination of EPEC plus wortmannin (lane 5) or EPEC plus genistein (lane 6) generated an apoptotic DNA ladder from T84 cells which was similar in intensity but not quite as clear-cut as that generated from H9 leukemia cells treated with doxorubicin (lane 8). HeLa cells seemed to give results similar to those shown for T84 cells, except that HeLa cells were more likely to detach toward the end of the infection period or during the final rinses (results not shown). Other investigators have noted that epithelial cell lines may be defective in the ability to carry out internucleosomal cleavage of DNA (35). In other experiments, we observed that the protein kinase inhibitor staurosporine also potentiated EPEC-induced DNA breakdown (data not shown). Apoptosis is not part of the biochemical repertoire of prokaryotes, but as a control, E. coli bacteria were subjected to the phosphate-citric acid extraction procedure used for infected host cells. In Fig. 4B, no low-molecular-weight DNA was detectable from strain HS (Fig. 4B, lane 1) or EPEC strain E2348 (lane 2) even though the number of bacteria used in the extraction procedure was about 5 times the number which adhered in Fig. 4A.
FIG. 4.
Internucleosomal breakdown of host cell DNA after EPEC infection. T84 cells were treated with inhibitors or infected with E. coli for 6 h; medium was replenished once at 2 h, at which time wortmannin was readded. Cells were recovered in 70% ethanol and subjected to extraction with phosphate-citric acid as described in Materials and Methods and then analyzed by agarose gel electrophoresis. A negative image of the ethidium bromide-stained gel is shown. (A) Extracts from human cells; (B) extracts from E. coli alone; MW, 100-bp DNA molecular size markers, with the 600-bp marker indicated. (A) Lane 1, uninfected, control T84 cells; lane 2, cells treated with 100 nM wortmannin alone; lane 3, cells treated with 200 μM genistein alone; lane 4, cells infected with EPEC strain E2348; lane 5, cells treated with EPEC plus wortmannin; lane 6, cells treated with E2348 plus genistein; lane 7, cells infected with strain HS alone; lane 8, H9 leukemia cells treated with doxorubicin (2 μg/ml) for 16 h as a positive control. (B) DNA extracts from E. coli HS (lane 1) and EPEC strain E2348 (lane 2). In panel B, the number of bacteria subjected to the extraction was about five times greater than that calculated to be present in panel A.
To compare EPEC mutants for the ability to trigger host cell death, a quantitative assay was needed. Counting trypan blue-positive cells under the microscope was the first method tried, but this is very tedious, time-consuming, and susceptible to observer bias. Therefore, we invested effort in comparing two quantitative methods, the LDH release assay and propidium iodide uptake method. The propidium iodide uptake method has been used extensively with nonadherent cells by flow cytometry (46) but has been adapted for adherent cells by Oberdoerster et al. for use with a fluorescence plate reader (34). This method was rapid and nondestructive, meaning that a single multiwell plate could be read and reread at multiple time points after infection. Results obtained with the LDH release assay are shown in Fig. 5, and those obtained with the propidium iodide method are shown in Fig. 6.
Figure 5A shows that spontaneous release of LDH from normal, control T84 cells was substantial. In addition, E. coli bacteria alone possessed detectable LDH activity, unlike what has been reported for Yersinia and Salmonella grown aerobically (31). Cell death induced by EPEC strains E2348 and B171-8 was essentially equal and slightly less than that induced by S. enterica var. enteritidis. The relatively low cell death seen with Salmonella may be due in part to the cell line used, T84. Elsinghorst reported that Salmonella typhi invasion of T84 cells was the lowest of any of eight cell lines tested (11). In Fig. 5B, the data in Fig. 5A are expressed as percent cell death, where the LDH release in the presence of Triton X-100 is taken as 100%.
Figure 6A shows that while live EPEC caused death of the T84 host cells, a sterile filtrate of EPEC medium, which contains EPEC-secreted proteins, did not induce death. In addition, EPEC killed by incubation with the antibiotic ciprofloxacin failed to kill T84 cells. These results, taken together with the results of Fig. 1 and 2, indicate the requirement for live EPEC to be in contact with the host cell to cause the host cell death.
Figure 6B demonstrates quantitatively what was observed visually in Fig. 1 with trypan blue, that wortmannin potentiated the cell death caused by EPEC (P < 0.05 at all time points). Wortmannin alone, in contrast, was nontoxic and did not potentiate the effects of nonadherent strain HB101.
In Fig. 6C, the effects of two EPEC mutants and the wild type are compared. In this experiment H2O2 was used instead of Triton X-100 detergent as the positive control for cell death. Infection with wild-type EPEC strain E2348 resulted in 42% cell death at 6 h. Strain JPN15, the plasmid-cured derivative of E2348 which adheres poorly, was almost completely attenuated in its ability to cause cell death (6% cell death). Strain CVD206, the eae mutant which shows nonintimate adherence, gave 24% cell death at 6 h, or about half the wild-type level.
EPEC activates the transcription factor NF-κB (41), and NF-κB activation inhibits apoptosis in response to a wide variety of cell death stimuli (4, 42, 46), including rickettsial infection (7). Therefore, we tested whether an inhibitor of NF-κB activation would alter cellular sensitivity to EPEC infection. A cell-permeant peptide inhibitor of NF-κB activation is MG-132, which blocks proteosomal degradation of IκB-α. Figure 6D shows that 40 μM MG-132 did significantly potentiate EPEC-induced cell death in a way similar to that observed by Clifton et al. with Rickettsia rickettsii (7).
Last, although it seemed intuitively obvious how invasive enteric bacteria such as Salmonella and Shigella could benefit from inducing apoptosis in the host cell, it was not clear how triggering apoptosis could fit the pathogenesis strategy of EPEC, a noninvasive, mucosa-adhering pathogen. We tested whether inducing apoptosis in host cells could increase the ability of EPEC to adhere. We induced apoptosis in HeLa cells by exposure to UV light. After 3 h, when the HeLa cells were beginning to show morphological changes of apoptosis, such as membrane blebbing, but before wholesale cell detachment occurred, we measured the adherence of 3H-labeled EPEC. Adherence of EPEC strain E2348 was increased about 40% in UV-exposed HeLa cells (4,553 ± 329 cpm/well [17.9% ± 1.3% of the initial inoculum) compared to control cells (3,224 ± 225 cpm/well [12.7% ± 0.9% of the initial inoculum]), a finding that reached statistical significance (P < 0.001). Barnett-Foster et al. have shown that enterohemorrhagic E. coli and EPEC can adhere to phosphatidylethanolamine, a lipid normally not exposed on the surface of cells but which, like phosphatidylserine, is exposed on the cell surface during apoptosis (3). Thus, is it possible that induction of apoptosis in the host cell benefits EPEC in part by increasing the availability of receptors for bacterial adhesins.
To summarize these findings, cell death caused by EPEC has striking features of apoptosis, including early surface expression of phosphatidylserine and internucleosomal DNA cleavage. However, the early cell permeability to vital dyes is not typical of a purely apoptotic mode of cell death (23, 36, 47). In addition, the tyrosine kinase inhibitor genistein, the PI 3-kinase inhibitor wortmannin, and the NF-κB inhibitor MG-132 increased EPEC-induced cell death. Host cell death required contact with live EPEC.
DISCUSSION
The ability of EPEC to kill host cells in vitro was observed previously by the trypan blue uptake method (2), but the apoptotic features of cell death were not noted. In human EPEC cases examined pathologically, evidence of intestinal cell damage was noted but the specific features of apoptosis were not reported (13, 38, 39). This is not surprising, however, since in our in vitro work, EPEC infection alone did not induce morphological changes typical of apoptosis (Fig. 1 and 2), although these changes were clearly visible in cells irradiated with UV light (data not shown). Furthermore, in infected animals, recognizing apoptosis in intestinal epithelial cells presents a special additional challenge since the majority of apoptotic cells are shed into the lumen (19). Therefore, it seems likely that EPEC do induce intestinal cell damage and eventually cell death in vivo, but future studies are needed which specifically look for features of apoptosis in EPEC-infected human or animal tissues.
Recently, species of several genera within the family Enterobacteriaceae, including Salmonella, Shigella, and Yersinia, have been found to trigger apoptosis in host cells in vitro or in vivo (31, 32, 49, 50). In addition, Helicobacter pylori, which shares with EPEC the ability to adhere intimately and to cause tyrosine phosphorylation in the host cell, causes apoptosis of gastric cells (33).
The EPEC-induced permeability of host cells to vital dyes, which occurs early after infection, is not a feature of pure apoptosis as originally defined (23). However, Salmonella-, Shigella-, and Yersinia-induced cell death is also accompanied by this type of membrane damage in the host cell, as measured by the early release of chromium-51 or of LDH (31, 48). While it may be tempting to use the fashionable term “apoptosis” to refer to all forms of cell death induced by bacteria, enteric bacterial pathogens actually trigger host cell death with some features of necrosis (15, 28). Based on the work presented here, EPEC appears to be included in this group of pathogens which induce host cell death with mixed features of apoptosis and necrosis.
EPEC-induced cell death appears to differ from the cell death resulting from the invasive enteric pathogens in some important ways. First, compared to Salmonella- and Shigella-mediated killing of macrophages, EPEC is much weaker in its ability to cause cell death. For example, Monack et al. showed that Salmonella typhimurium was able to kill between 70 and 90% of target macrophages within 2 h, depending on which assay was used (31, 32). In contrast, EPEC strains killed only 50% of host cells at 6 h, despite a higher MOI (100:1). Second, the host range for EPEC-induced cell death differs from that of Salmonella and Shigella; the latter are highly efficient at killing lymphocytes and macrophages but less able or unable to kill intestinal epithelial cells (Fig. 5). Moreover, invasion and induction of apoptosis are separable processes, since Shigella flexneri can invade HeLa cells (11) but does not induce apoptosis in HeLa cells (47a).
In contrast, EPEC is able to kill all of the epithelial cell lines we have studied (HeLa, T84, and HEK-293) and appears to be able to do so while remaining in an extracellular location (Fig. 1C). We did not examine whether EPEC could kill macrophages, but other authors have reported that it does not (15).
While the ability to rapidly induce apoptosis in host cells would intuitively appear to be an advantage for invasive pathogens such as Salmonella and Shigella, there appears to be no obvious benefit to EPEC from host cell killing, since EPEC is an adherent, noninvasive pathogen. On the contrary, rapid death of intestinal epithelial cells would result in loss of sites of adherence before the organism had a chance to complete the attaching and effacing lesion, replicate, and infect other nearby cells. Moreover, in comparing recent findings on EPEC signalling with the literature on apoptosis, one is struck by the fact that EPEC adherence is able to strongly activate at least three separate antiapoptotic pathways within the host cell. Stimulation of tyrosine kinases, protein kinase C, and the transcription factor NF-κB all suppress cell death (9, 12, 20, 22, 30, 40, 44); EPEC potently stimulates all of these signalling pathways (8, 37, 41). The ability of EPEC to stimulate these known, antiapoptotic pathways, the slower host cell killing that we observed with EPEC, and the apparent disadvantage to EPEC of rapid intestinal cell death in the pathogen’s life cycle have led us to speculate that EPEC has developed strategies to slow rather than stimulate apoptosis (Fig. 7). Such an antiapoptotic mechanism has been demonstrated for the obligate intracellular bacterium R. rickettsii, which inhibits host cell apoptosis by activation of NF-κB (7). Other examples of microbes which manipulate the host cell to inhibit apoptosis are found among plant pathogens, chlamydiae, viruses, and protozoa (1, 14, 16).
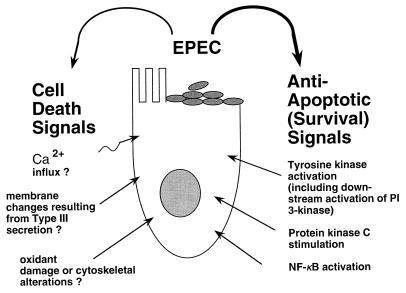
FIG. 7.
Scheme for understanding the slow host cell killing by EPEC and enhanced killing in the presence of inhibitors. When antiapoptotic signals are blocked by genistein, wortmannin, staurosporine, or the NF-κB inhibitor MG-132, death-promoting pathways act unopposed and killing is enhanced.
While antiapoptotic signals from EPEC to host are apparent, the nature of the cell death-inducing stimulus (or stimuli) from EPEC is unclear (Fig. 7, left side). In Shigella, introduction of the IpaB protein into the host cell cytoplasm is necessary and sufficient to induce apoptosis. IpaB enters the cytoplasm and binds to and directly activates ICE (caspase-1, an apoptotic protease) (6). However, the mechanism of EPEC-induced cell death may be different from that of cell death induced by Shigella, based on the difference in host range and on the failure of the cell-permeant caspase-1 inhibitor Z-Val-Ala-Asp-fluoromethylketone to protect cells from EPEC-induced death (our unpublished data). Death-inducing signals from EPEC could include increases in intracellular calcium, cytoskeletal rearrangements, oxidant damage, and membrane changes resulting from introduction of bacterial proteins into the host cell via the type III secretion pathway (29). It is possible that type III-mediated secretion into the host cell is itself responsible for the breakdown of membrane integrity observed in response to EPEC, Salmonella, Shigella, and Yersinia infection. Indeed, Frankel and colleagues have recently proposed an model for EPEC secretion in which filaments formed by the EPEC-secreted protein EspA interact with EspB to form a pore in the host cell membrane (17, 24). Homologs of EspA and EspB exist in Salmonella and Yersinia (17). A pore sufficiently large to allow the insertion of EPEC proteins such as Tir into the host cell could also result in the permeability to vital dyes and release of LDH which we observed.
If EPEC does have mechanisms to suppress apoptosis in host cells, it may be possible to isolate EPEC mutants which show enhanced killing of cultured cells (hyperlethal mutants) as well as additional, more instructive mutants which are attenuated in cell death (Fig. 6C). However, addressing the issue of whether apoptosis is protective or deleterious to the host following EPEC infection will probably not be possible with in vitro systems and may require animal models to assess.
ACKNOWLEDGMENTS
This work was supported by NIH grant R29 DK49410 (to J.K.C.).
We thank Richard Rabin and Jan Oberdoerster, Department of Pharmacology, SUNY at Buffalo, for help in optimizing DNA extraction methods and in setting up the propidium iodide uptake assay.
REFERENCES
- 1.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin T, Lee-Delaunay M, Knutton S, Williams P. Calcium-calmodulin dependence of actin accretion and lethality in cultured HEp-2 cells infected with enteropathogenic Escherichia coli. Infect Immun. 1993;61:760–763. doi: 10.1128/iai.61.2.760-763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett-Foster, D., D. Philpott, M. Huesca, P. Sherman, and C. Lingwood. Comparison of enteropathogenic E. coli and enterohemorrhagic E. coli binding to lipid receptors. Gastroenterology, in press. [DOI] [PubMed]
- 4.Beg A, Baltimore D. An essential role for NF-κB in preventing TNF-α induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S, Wu C-Y, Murray W, Tobe T, Fernandez R, Schoolnik G. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Smith M, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by directly binding ICE. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 7.Clifton D, Goss R, Sahni S, van Antwerp D, Baggs R, Marder V, Silverman D, Sporn L. NF-κB dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc Natl Acad Sci USA. 1998;95:4646–4651. doi: 10.1073/pnas.95.8.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crane J K, Oh J S. Activation of host cell protein kinase C by enteropathogenic Escherichia coli. Infect Immun. 1997;65:3277–3285. doi: 10.1128/iai.65.8.3277-3285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLuca C, Kwon H, Pelletier N, Wainberg M A, Hiscott J. NF-kappaB protects HIV-1-infected myeloid cells from apoptosis. Virology. 1998;244:27–38. doi: 10.1006/viro.1998.9085. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg M, Kaper J. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsinghorst E. Measurement of invasion by gentamicin resistance. Methods Enzymol. 1994;236:405–420. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 12.Evans C A, Lord J M, Owen-Lynch P J, Johnson G, Dive C, Whetton A D. Suppression of apoptosis by v-ABL protein tyrosine kinase is associated with nuclear translocation and activation of protein kinase C in an interleukin-3-dependent haemopoietic cell line. J Cell Sci. 1995;108:2591–2598. doi: 10.1242/jcs.108.7.2591. [DOI] [PubMed] [Google Scholar]
- 13.Fagundes-Neto U, Freymuller E, Schimitz L, Scaletsky I. Nutritional impact and ultrastructural intestinal alterations in severe infections due to enteropathogenic Escherichia coli strains in infants. J Am Coll Nutr. 1996;15:180–185. doi: 10.1080/07315724.1996.10718586. [DOI] [PubMed] [Google Scholar]
- 14.Fan T, Lu H, Hu H, Shi L, McLarty G, Nance D, Greenberg A, Zhong G. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med. 1998;187:487–496. doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Prada C, Tall B, Elliott S, Hoover D, Nataro J, Venkatesan M. Hemolysin-positive enteroaggregative and cell-detaching Escherichia coli strains cause oncosis of human monocyte-derived macrophages and apoptosis of murine J774 cells. Infect Immun. 1998;66:3918–3924. doi: 10.1128/iai.66.8.3918-3924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox J. Current topics: some microbes elicit, others block apoptosis. ASM News. 1997;63:412–413. [Google Scholar]
- 17.Frankel G, Phillips A, Rosenshine I, Dougan G, Kaper J, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 18.Gong J, Traganos F, Darzynkiewicz Z. A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry. Anal Biochem. 1994;218:314–319. doi: 10.1006/abio.1994.1184. [DOI] [PubMed] [Google Scholar]
- 19.Hall P, Coates P J, Ansari B, Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J Cell Sci. 1994;107:3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 20.Iimuro Y, Nishiura T, Hellerbrand C, Behrns K E, Schoonhoven R, Grisham J W, Brenner D A. NFkappaB prevents apoptosis and liver dysfunction during liver regeneration. J Clin Investig. 1998;101:802–811. doi: 10.1172/JCI483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ireton K, Payrastre B, Chap H, Ogawa W, Sakaue H, Kasuga M, Cossart P. A role for phosphoinositide 3-kinase in bacterial invasion. Science. 1996;274:780–782. doi: 10.1126/science.274.5288.780. [DOI] [PubMed] [Google Scholar]
- 22.Karnes W E, Jr, Weller S G, Adjei P N, Kottke T J, Glenn K S, Gores G J, Kaufmann S H. Inhibition of epidermal growth factor receptor kinase induces protease-dependent apoptosis in human colon cancer cells. Gastroenterology. 1998;114:930–939. doi: 10.1016/s0016-5085(98)70312-9. [DOI] [PubMed] [Google Scholar]
- 23.Kerr J, Wyllie A, Currie A. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel Espa-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulik G, Klippel A, Weber M. Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine M, Nalin D, Hornick R, Bergquist E, Waterman D, Young C, Sotman S, Rowe B. Escherichia coli strains that cause diarrhea but do not produce heat-labile or heat-stable enterotoxins and are not invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 27.Levine M, Nataro J, Karch H, Baldini M, Kaper J, Black R, Clements M, O’Brien A. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 28.Lindgren S, Heffron F. To sting or be stung: bacteria-induced apoptosis. Trends Microbiol. 1997;5:263–264. doi: 10.1016/S0966-842X(97)88832-4. [DOI] [PubMed] [Google Scholar]
- 29.Mannick E E, Bravo L E, Zarama G, Realpe J L, Zhang X J, Ruiz B, Fontham E T, Mera R, Miller M J, Correa P. Inducible nitric oxide synthase, nitrotyrosine, and apoptosis in Helicobacter pylori gastritis: effect of antibiotics and antioxidants. Cancer Res. 1996;56:3238–3243. [PubMed] [Google Scholar]
- 30.Messmer U K, Lapetina E G, Brune B. Nitric oxide-induced apoptosis in RAW 264.7 macrophages is antagonized by protein kinase C- and protein kinase A-activating compounds. Mol Pharmacol. 1995;47:757–765. [PubMed] [Google Scholar]
- 31.Monack D, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss S F, Calam J, Agarwal B, Wang S, Holt P R. Induction of gastric epithelial apoptosis by Helicobacter pylori. Gut. 1996;38:498–501. doi: 10.1136/gut.38.4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberdoerster J, Kamer A, Rabin R. Differential effect of ethanol on PC12 cell death. J Pharmacol Exp Ther. 1998;287:359–365. [PubMed] [Google Scholar]
- 35.Oberhammer F, Wilson J, Dive C, Morris I, Hickman J, Wakeling A, Walker P, Sikorska M. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993;12:3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry S, Epstein L, Gelbard H. Simultaneous in situ detection of apoptosis and necrosis in monolayer cultures by TUNEL and trypan blue staining. BioTechniques. 1997;22:1102–1106. doi: 10.2144/97226st01. [DOI] [PubMed] [Google Scholar]
- 37.Rosenshine I, Donnenberg M, Kaper J, Finlay B. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 1992;11:3551–3560. doi: 10.1002/j.1460-2075.1992.tb05438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothbaum R, McAdams A, Giannella R, Partin J. A clinicopathologic study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology. 1982;83:441–454. [PubMed] [Google Scholar]
- 39.Rothbaum R, Partin J, Saalfield K, McAdams A. An ultrastructural study of enteropathogenic Escherichia coli infection in human infants. Ultrastruct Pathol. 1983;4:291–304. doi: 10.3109/01913128309140582. [DOI] [PubMed] [Google Scholar]
- 40.Ruckdeschel K, Harb S, Roggenkamp A, Hornef M, Zumbihl R, Kohler S, Heesemann J, Rouot B. Yersinia enterocolitica impairs activation of transcription factor NF-kappaB: involvement in the induction of programmed cell death and in the suppression of the macrophage tumor necrosis factor alpha production. J Exp Med. 1998;187:1069–1079. doi: 10.1084/jem.187.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savkovic S D, Koutsouris A, Hecht G. Activation of NF-kappa-B in intestinal epithelial cells by enteropathogenic Escherichia coli. Am J Physiol. 1997;42:C1160–C1167. doi: 10.1152/ajpcell.1997.273.4.C1160. [DOI] [PubMed] [Google Scholar]
- 42.Van Antwerp D, Martin S, Kafri T, Green D, Verma I. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 43.Vuopio-Varkila J, Schoolnik G. Localized adherence by enteropathogenic Escherichia coli is an inducible phenotype associated with the expression of new outer membrane proteins. J Exp Med. 1991;174:1167–1177. doi: 10.1084/jem.174.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whelan R D, Parker P J. Loss of protein kinase C function induces an apoptotic response. Oncogene. 1998;16:1939–1944. doi: 10.1038/sj.onc.1201725. [DOI] [PubMed] [Google Scholar]
- 45.Wooldridge K G, Williams P H, Ketley J M. Host signal transduction and endocytosis of Campylobacter jejuni. Microb Pathog. 1996;21:299–305. doi: 10.1006/mpat.1996.0063. [DOI] [PubMed] [Google Scholar]
- 46.Wu M, Ao Z, Prasad K, Wu R, Schlossman S. IEX-1L, an apoptosis inhibitor involved in NF-κB mediated cell survival. Science. 1998;281:998–1001. doi: 10.1126/science.281.5379.998. [DOI] [PubMed] [Google Scholar]
- 47.Zhang G, Gurtu V, Kain S, Yan G. Early detection of apoptosis using a fluorescent conjugate of Annexin V. BioTechniques. 1997;23:525–532. doi: 10.2144/97233pf01. [DOI] [PubMed] [Google Scholar]
- 47a.Zychlinsky, A., and J. Moss. Personal communication.
- 48.Zychlinsky A, Prevost M, Sansonetti P. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–169. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]
- 49.Zychlinsky A, Sansonetti P J. Apoptosis as a proinflammatory event: what can we learn from bacteria-induced cell death? Trends Microbiol. 1997;5:201–204. doi: 10.1016/S0966-842X(97)01044-5. [DOI] [PubMed] [Google Scholar]
- 50.Zychlinsky A, Thirumalai K, Arondel J, Cantey J R, Aliprantis A O, Sansonetti P J. In vivo apoptosis in Shigella flexneri infections. Infect Immun. 1996;64:5357–5365. doi: 10.1128/iai.64.12.5357-5365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]