Abstract
Many bacteria metabolize ethanolamine as a nutrient source through cytoplasmic organelles named bacterial microcompartments (BMCs). Here we investigated the molecular assembly, regulation, and function of BMCs in Fusobacterium nucleatum – a Gram-negative oral pathobiont that is associated with adverse pregnancy outcomes. The F. nucleatum genome harbors a conserved ethanolamine utilization (eut) locus with 21 genes that encode several putative BMC shell proteins and a two-component signal transduction system (TCS), in addition to the enzymes for ethanolamine transport and catabolism. We show that the expression of most of these genes as well as BMC formation is highly increased in wild type fusobacteria when cultured in the presence of ethanolamine as a nutrient source. Deletion of the response regulator EutV eliminated this induction of eut mRNAs and BMCs, thus demonstrating that BMC formation is transcriptionally regulated by the TCS EutV-EutW in response to ethanolamine. Mass spectrometry of isolated BMCs unveiled the identity of the constituent proteins EutL, EutM1, EutM2, and EutN. Consistent with the role of these proteins in BMC assembly and metabolism, deletion of eutN, eutL/eutM1/eutM2, or eutL/eutM1/eutM2/eutN not only affected BMC formation, but also ethanolamine utilization, causing cell growth defects with ethanolamine as nutrient. BMCs also assembled in fusobacteria cultured with placental cells or the culture media, a process that is dependent on the BMC shell proteins. Significantly, we show that the eutN mutant is defective in inducing preterm birth in a mouse model. Together, these results establish that BMC-mediated metabolism of ethanolamine is critical for fusobacterial virulence.
Keywords: Fusobacterium nucleatum, bacterial microcompartment, ethanolamine utilization, preterm birth, electron microscopy
INTRODUCTION
Ethanolamine (EA) – a component of the membrane lipid phosphatidylethanolamine present in both eukaryotes and prokaryotes – is a notable nutrient source to gut bacteria, such as Escherichia, Salmonella, and Listeria (1, 2). Each of these microbes harbor a multi-gene locus, named eut for ethanolamine utilization, that encodes various proteins for EA transport and metabolism (1, 2). The eut locus also encodes several small structural proteins known as shell proteins (3). These shell proteins self-assemble to form polyhedral organelles called the bacterial microcompartments (BMCs) that encapsulate the EA catabolic enzymes and thereby empower bacteria with a highly efficient catabolic machine while also compartmentalizing and insulating bacterial cells from the toxic intermediates of EA catabolism (4, 5). In Escherichia coli, BMCs were shown to contain EutS, EutL, EutK, EutM, and EutN, with the first four representing the major shell constituents (6). Importantly, genetic studies in Salmonella enterica revealed that individual deletion mutants of eutK, eutL, eutM, or eutN fail to grow on EA as a carbon source at high pH (≥ 8), whereas mutant cells lacking eutS grow similarly as wild-type cells (7). Expression of the S. enterica shell proteins EutSMNLK in the heterologous host E. coli is sufficient to produce Salmonella BMCs, with increased BMC formation when EutQ is co-expressed (8). However, whether mutants lacking the aforementioned individual shell proteins are defective in BMC formation remained unclear.
Two distinct types of regulatory systems are known to be involved in controlling the formation of BMCs and the expression of eut genes in different organisms – the transcriptional regulator EutR and the two-component transduction system (TCS) EutV-EutW, with EutV acting as a response regulator and EutW as a sensor histidine kinase (1). BMCs are observed in wild-type Listeria monocytogenes grown in the EA-containing media but not in mutants lacking eutV (9). In contrast, EutR was shown to be a direct regulator of the eut locus in enterohemorrhagic E. coli (EHEC), as EutR binds to the promoter of eutS – the first gene of the eut locus, and eutR deletion drastically reduces eutS expression (10).
The multigene eut locus is present in the genomes of a wide range of Gram-positive and Gram-negative bacteria, including the subject of this study, Fusobacterium nucleatum, whose genome harbors three short operons of roughly 17 eut genes (3, 11, 12). F. nucleatum, a Gram-negative obligate anaerobe, is mainly found in the human oral cavity, but it has the ability to spread to distal organs such as colon and placenta (13–19), where the bacterium was found to promote colorectal cancer and adverse pregnancy outcomes including neonatal sepsis and preterm birth, respectively (20). The eut gene cluster is conserved in many other F. nucleatum subspecies such as F. nucleatum ssp. vincentii and F. nucleatum ssp. animalis (12, 21, 22). Consistent with the functionality of the eut gene locus in F. nucleatum, F. nucleatum ssp. animalis cells exposed to EA display a significant increase in expression of eut genes including those that encodes BMC proteins (22). This conservation of the EA utilization system and its regulation by EA and BMCs in a pathologically important fusobacterial clade underscores the importance of dissecting the genetic, molecular, and physiological determinants of BMC assembly and the role of BMCs in fusobacterial pathogenicity.
Here, using the F. nucleatum sub spp. nucleatum strain ATCC 23726, a clinical isolate that is amendable to robust genetic manipulation (23–28), we show that fusobacteria are responsive to EA as an environmental nutrient source that greatly increases the expression of eut genes and BMC formation. This EA response of fusobacteria requires the response regulator EutV, which appears to be an activator of eut genes and also a global transcriptional regulator of a large regulon under nutrient-rich conditions. We further show that F. nucleatum BMCs contain EutL, EutM1, EutM2, and EutN, and genetic disruption of eutN, eutLM1M2, or eutLM1M2N severely affects BMC formation in EA-supplementing conditions as well as in cocultures with placental cells. Most importantly, we show that the BMCs are essential for fusobacterial pathogenesis by demonstrating that the eutN mutant is significantly attenuated in virulence in a mouse model of preterm birth.
RESULTS
The conserved eut locus in F. nucleatum responds to environmental ethanolamine.
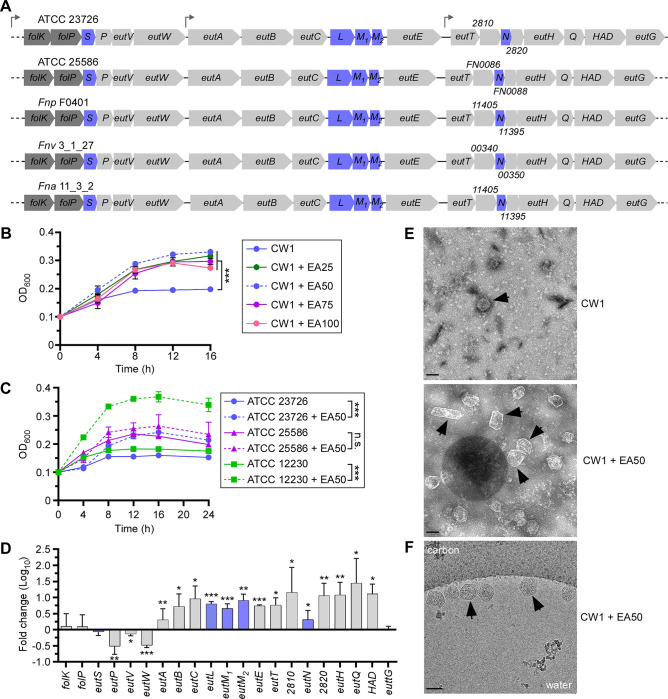
To elucidate the functionality of the eut gene cluster in F. nucleatum, we chose a genetically tractable clinical isolate of F. nucleatum ssp. nucleatum, ATCC 23726. The genome of this strain contains three short operons within a conserved gene cluster with several sets of genes that are characteristic of the eut locus: a two-component transduction system (TCS) (EutV-EutW), several putative bacterial microcompartment (BMC) proteins (EutS-EutL-EutM), an EA transporter (EutH), EA catabolic proteins (EutA/EutB/EutC) (Fig. 1A). The cluster also encodes several hypothetical proteins (RS02810 and RS02820), among many other predicted ethanolamine utilization proteins (EutP, EutE, EutT, EutQ, EutG, and HAD – haloacid dehalogenase-like hydrolase) (Fig. 1A). Another predicted gene, eutJ, coding for an EA utilization protein, is located elsewhere in the chromosome. Intriguingly, ATCC 23726 harbors 2 copies of EutM (M1 and M2) within the eut gene cluster, and it does not appear to encode a EutK homolog, (Fig. 1A & Fig. S1). Remarkably, this same gene arrangement was found in F. nucleatum ATCC 25586, as well as in many other F. nucleatum subspecies, including F. nucleatum ssp. polymorphum (Fnp F0401), F. nucleatum ssp. vincentii (Fnv 3_1_27), and F. nucleatum ssp. animalis (Fna 11_3_2) (Fig. 1A) (12), suggesting that EA utilization is a conserved feature in F. nucleatum.
Figure 1: The eut locus of F. nucleatum and cellular response to ethanolamine.
(A) Shown are the eut loci in F. nucleatum strains ATCC 23726 and 25586, with genes coding for presumptive BMC shell proteins highlighted in blue. (B) Cells of CW1 (derivative of ATCC 23726) were grown at 37°C in minimal media supplemented with various concentrations of EA (0, 25, 50, 75, 100 mM). Cell growth was monitored by optical density at 600 nm (OD600). Error bars indicate means and standard deviations of three biological replicates; ***, P<0.001. (C) Cells of indicated strains were grown at 37°C in minimal media supplemented with 50 mM of EA, and growth was monitored by OD600. Error bars indicate means and standard deviations of three biological replicates; ***, P<0.001; n.s., not significant. (D) CW1 cells were grown at 37°C in minimal media supplemented with 50 mM of EA for 8 h and harvested for total RNA extraction. Expression of indicated genes was determined by qRT-PCR, with 16S RNA as control. Error bars indicate means and standard deviations of three biological replicates; *, P<0.05; **, P<0.01; ***, P<0.001. (E) BMCs of the parent strain (CW1) grown in conditions described in B in the absence or present of 50 mM EA were isolated and analyzed by electron microscopy; scale bars of 100 nm. (F) Aliquots of BMCs isolated in E were analyzed by cryo-electron microscopy at the nominal magnification of 22,000 x, showing BMCs clustered along the edge of a vitreous ice hole within the holey carbon grid; scale bars of 100 nm. Black arrows in panels E and F mark representative BMC structures.
To examine how F. nucleatum responds to EA, we grew cultures of F. nucleatum CW1, an isogenic ΔgalK derivative of ATCC 23726 that has been used for facile gene deletion (27), in minimal medium supplemented with increasing concentrations of EA, and monitored cell growth over time by measuring optical density (OD of 600 nm). F. nucleatum cells proliferated better and faster in the presence of EA, optimally at 50 mM (Fig. 1B; dashed blue line), compared to a culture without EA, whose growth arrested around 8 h in culture (Fig. 1B; solid blue line). To confirm that this growth phenotype is not strain-specific, we monitored the cell growth of strains F. nucleatum ATCC 23726, F. nucleatum ATCC 25586, and F. nucleatum ATCC 12230 in minimal medium with or without 50 mM EA. All three strains displayed a similar trend of increasing cell growth in the presence of EA, albeit no significant difference observed with ATCC 25586, with ATCC 12230 showing the largest increase (Fig. 1C). Next, we isolated total mRNA of CW1 grown in the same media in the presence or absence of 50 mM EA for 8 h for gene expression analysis by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Strikingly, while the expression of a few genes of the locus remained unchanged (eutG, eutS, eutV), and the expression of a few others decreased (eutP and eutW), a significant increase in expression was observed for a large number of genes including those predicted to encode the BMC shell proteins as well as EA transport and catabolic enzymes (Fig. 1D). Noticeably, this bimodal expression of eut genes in strain 23726 was also observed in strain 25586 (Fig. S2). It is noteworthy that a similar expression pattern for many eut genes in F. nucleatum ssp. animalis in response to EA was also observed (22).
To investigate whether the increased expression of predicted BMC proteins caused by EA led to increased BMC formation, we isolated BMCs from fusobacterial cells grown in two conditions as performed in the previous experiment (Fig. 1D) and quantified BMCs by electron microscopy (see Methods). Indeed, as shown in Fig. 1E, transmission electron microcopy (TEM) revealed a substantial increase in the number of polyhedral BMCs in the presence of EA. These polyhedral structures were also confirmed by cryogenic-electron microscopy (cryoEM) (Fig. 1F). Together, the results suggest that F. nucleatum harbors a functional ethanolamine utilization system involving BMCs that is responsive to environmental EA.
Modulation of eut gene expression by the response regulator EutV.
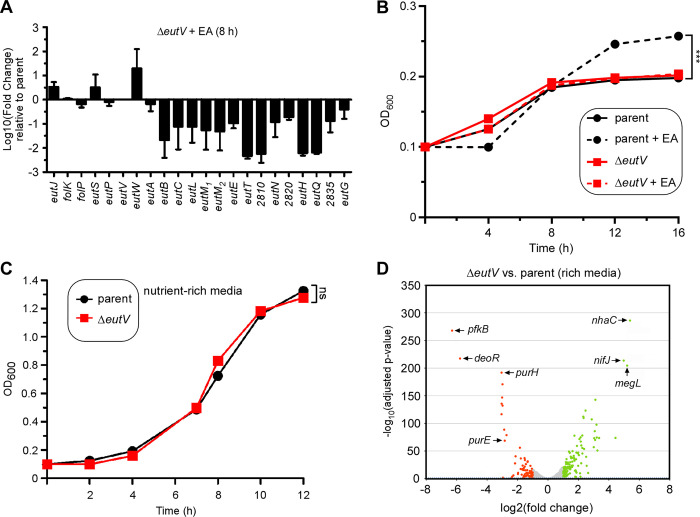
The bimodal expression of eut genes in the presence of EA, as demonstrated above, suggests that expression of eut genes is regulated, most likely by the TCS EutV-EutW given their close proximity with most other eut genes (Fig. 1A). To examine if this is the case, we generated an in-frame, non-polar deletion mutant devoid of the predicted response regulator EutV. Next, we isolated total mRNA of the parent and eutV mutant strains grown in minimal medium in the presence of 50 mM EA for 8 h and quantified gene expression by qRT-PCR. Significantly, mRNAs for all the genes whose expression was greatly elevated by EA were down-regulated in the eutV deletion mutant, while mRNAs for a few other genes (eutS, eutW, and eutJ) were up-regulated when eutV was absent (Fig. 2A), consistent with the bimodal expression of eut genes as observed above (Fig. 1D). Together these results support the view that the TCS gene products EutV-EutW responds to EA, and that EutV acts as an activator of a set of eut genes in response to EA.
Figure 2: Gene expression modulated by the response regulator EutV.
(A) Total RNA of the parent and eutV mutant strains grown at 37°C in minimal media supplemented with 50 mM of EA for 8 h was isolated for gene expression analysis by qRT-PCR. Error bars indicate means and standard deviations of three biological replicates. (B) Growth of indicated strains in minimal media supplemented with 50 mM of EA or without at 37°C was monitored by OD600. Error bars indicate means and standard deviations of three biological replicates; ***, P<0.001. (C) Total RNA of the parent and eutV mutant strains grown in rich media at 37°C to mid-log phase was isolated for RNA-seq analysis as previously described (25). The log2 (fold change) (LFC) was plotted against the statistical significance (−log10 of the adjusted p-value). All genes above a LFC of +1.0 and below 1.0 are shown in green and red, respectively. An insert shows a detailed portion of the volcano plot for a better presentation of the position of the adjusted p-value of 0.05 (dashed blue line). Some of the highly expressed genes are marked.
Given that many EA metabolic and BMC-encoding genes are down-regulated in the absence of EutV, the eutV mutant might exhibit a defect in EA utilization and cell growth when cells are grown in media with EA as the major carbon and nitrogen source. Indeed, compared to the parent strain CW1, which was positively responsive to EA for rapid growth, the eutV mutant displayed significant growth reduction despite the presence of EA (Fig. 2B). In contrast, both parent and ΔeutV mutant cells grew at similar rate in nutrient-rich media (Fig. 2C), further supporting that the loss of this regulator impacts cellular response to environmental EA.
Considering EutV regulates the eutJ gene that is not located at the large eut gene cluster, we examine whether EutV regulates additional genes of the fusobacterial genome, we performed a comparative transcriptome analysis using RNA-sequencing (RNA-seq) with total RNA isolated from the parent and eutV mutant strains grown to mid-log phase in rich media as previously reported (25). When a twofold cutoff (log2 fold change ± 1) was set for comparison of RNA-seq reads, we found that mRNA counts for 121 genes were significantly elevated, while that for 83 genes were significantly reduced in the absence of eutV (Fig. 2C and Table S1). It is not surprising that eut genes are not found in the list of 204 differentially expressed genes as the culture condition used for this analysis did not require EA utilization (Table S1). Noticeably, many genes up-regulated in the absence of eutV encode proteins predicted to be involved in sugar/amino acid metabolism and transport (Table S1), with megL (> 36-fold increase) coding for the methionine gamma-lyase MegL that was previously shown to participate in cysteine metabolism in F. nucleatum (23). On the other hand, genes coding for glycolysis (PfkB), purine biosynthesis (PurE, PurH), and methionine metabolism/transport are among the highly down-regulated genes in the absence of eutV. Interestingly, many genes coding for multidrug transporters (RS07785, RS07780, RS01595, RS01290, and RS10825) are also significantly down-regulated in the eutV mutant (Table S1). Together, the transcriptome data combined with the quantitative RNA analysis demonstrates that EutV is a response regulator that not only modulates F. nucleatum eut genes in response to environmental EA but also has a broad physiological role under nutrient-rich conditions in regulating the expression of many metabolic genes outside of the eut gene locus.
Genetic determinants of BMC formation in F. nucleatum.
As we alluded in the introduction, the BMCs of Escherichia coli and Salmonella enterica are made of EutS, EutK, EutL, EutM, and EutN (1). By comparison, the eut gene loci in F. nucleatum strains ATCC 23726 and ATCC 25586 do not appear to encode EutK, but instead encode two copies of EutM, i.e. EutM1 and EutM2 (Fig. 1A). Because of amenability of in ATCC 23726 for genetic manipulation (25–27), we focused on this strain to examine whether these predicted genes participate in BMC formation by generating in-frame, non-polar deletion mutants according to our published protocol (26, 29). In this method, homologous exchange of the targeted genes with deletion alleles via sequential recombination at the flanking homology regions readily produce pairs of strains with either wild-type or mutant alleles (see Methods). While we succeeded generating individual mutants devoid of eutN, eutL/eutM1/eutM2, or eutL/eutM1/eutM2/eutN, we could not obtain any mutant alleles of eutS after several attempts, suggesting that the deletion of this gene might be lethal in F. nucleatum for unknown reasons.
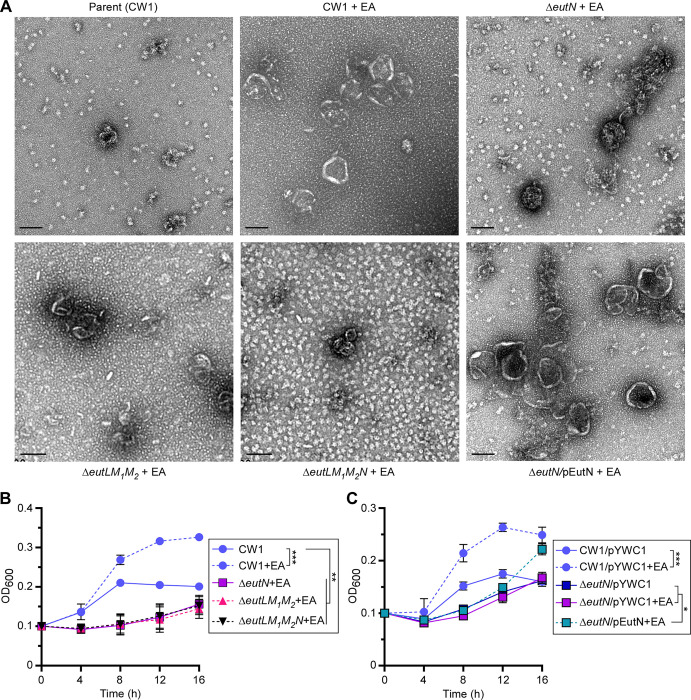
Next, to visualize BMCs by TEM, the parent and generated mutant strains were cultured in minimal media supplemented with 50 mM EA or without, and the harvested cells were used to isolate BMCs, which were analyzed by electron microscopy (see Methods). Compared to the parent strain grown in the presence of EA, deletion of eutN, eutL/eutM1/eutM2, or eutL/eutM1/eutM2/eutN significantly reduced BMC formation, and the resulting BMCs were visibly smaller in size (Fig. 3A). Importantly, the ectopic expression of eutN rescued the defect of the ΔeutN mutant (Fig. 3A and Fig. S3A). Furthermore, consistent with the role of BMCs in EA metabolism, all three shell mutant strains failed to respond to EA, unlike the parent strain (Fig. 3B). Note that complementation of the ΔeutN mutant slowly restored cell growth, which was more pronounced after 12 h (Fig. 3C).
Figure 3: Genetic determinants of BMC formation required for selective growth.
(A) BMCs of cells of the parent and indicated mutant strains grown in conditions described in 1E were isolated and analyzed by electron microscopy; scale bars of 100 nm. (B-C) Cell growth of indicated strains were monitored by OD600. Error bars indicate means and standard deviations of three biological replicates; *, P<0.05; **, P<0.01; ***, P<0.001.
To further confirm that EutL, EutM1, EutM2, and EutN are part of the F. nucleatum BMCs, we isolated BMCs from the parent strain grown in minimal media supplemented with EA for 8 h and performed mass spectrometry analysis for protein identification. As expected, we identified all aforementioned proteins among many other EA metabolic proteins, but surprisingly we did not find any signal for EutS (Table S2).
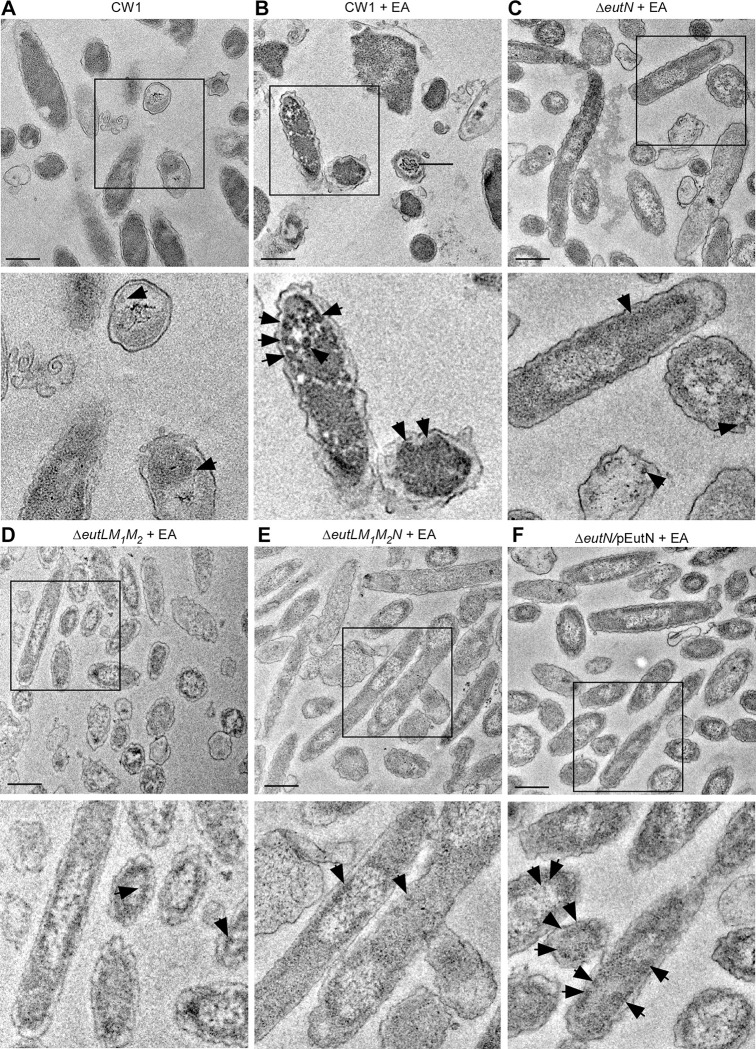
We next examined BMCs within fusobacterial cells by thin-section TEM, whereby F. nucleatum cells grown in the same conditions as described in Fig. 3A were harvested for preparation of thin-sections, which were stained with 1% uranyl acetate prior to TEM observation (see Methods). Consistent with the above results, the parent cells (CW1) produced a higher number of BMCs in the presence of 50 mM EA than without it (Fig. 4A–4B). Significantly, all three shell protein mutants (single, triple, and quadruple) produced less BMCs than the parent strain (Fig. 4C–4E), whereas complementation of the ΔeutN mutant rescued this defect (Fig. 4F and Fig. S3B).
Figure 4: BMC formation analyzed by thin-section electron microscopy.
(A-F) Cells of the parent and indicated mutant strains grown in conditions described in 1E were harvested for thin-section electron microscopy; scale bars of 200 nm. Enlargement of boxed areas is shown in below panels. Black arrows mark representative BMC structures.
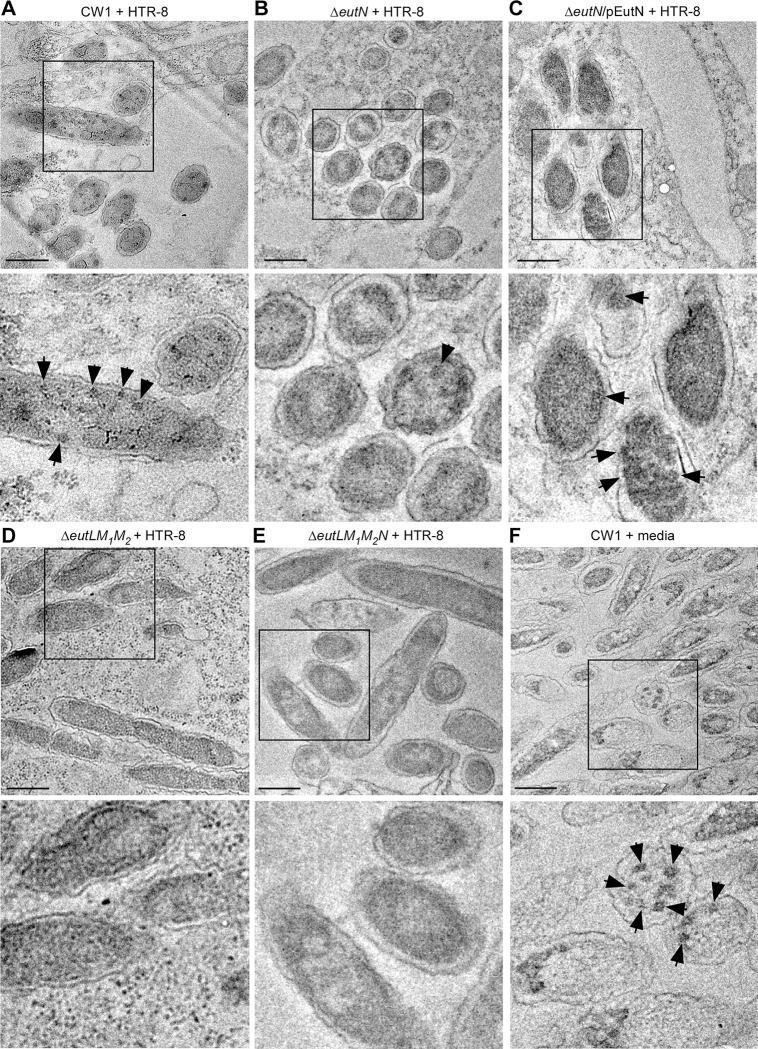
Finally, we examined whether F. nucleatum forms BMCs in culture in the presence of placental cells. We co-cultured fusobacterial cells with HTR8/SVneo placental cells for 4.5 h prior to embedding them in resin for thin-sectioning. Compared to the parent strain, which formed BMCs in the presence of HTR8, the ΔeutN mutant produced significantly less BMCs (Fig. 5A–5B). This defect was recused by ectopic expression of eutN (Fig. 5C). Notably, BMCs were rarely observed in the triple and quadruple mutants (Fig. 5D–5E). Interestingly, the parent CW1 cells grown in media used for growing HTR8 cells (RPMI+ 5% FBS) produced a high number of BMCs (Fig. 5F and Fig. S3C), suggesting these media contain EA and/or some other inducer that triggered BMC formation.
Figure 5: BMC formation in placental cells.
(A-F) HTR-8/SVneo cells seeded at 3 × 106 cells/ml were infected with F. nucleatum CW1 at the multiplicity of infection (MOI) of 450 for 4.5 h then embedded into resin before sectioning; scale bars of 200 nm. Black arrows in all images mark BMC structures. Images are representative of observed phenotypes.
Altogether, the results demonstrate that EutL, EutM1, EutM2, and EutN of F. nucleatum serve as a molecular determinant of leading to the EA-induced assembly of BMCs that are critical for bacterial growth under conditions whereby fusobacteria must utilize EA as a carbon and nitrogen source.
BMC formation is critical for F. nucleatum virulence.
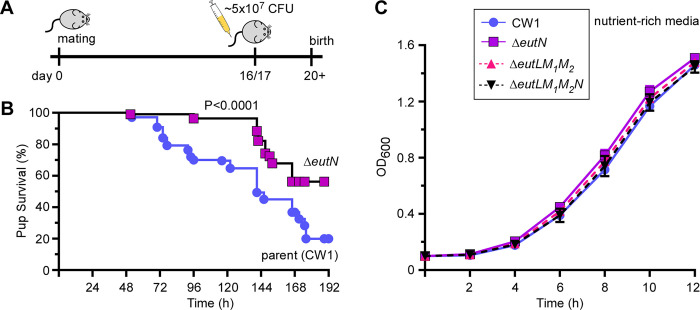
Since deletion of either eutN, eutL/eutM1/eutM2, or eutL/eutM1/eutM2/eutN severely affects BMC formation, we next proceeded to determine whether the ΔeutN mutant has a virulence phenotype in a mouse model of preterm birth, using an experimental setup we tested out previously for examining other potential virulence factors (26, 29). We utilized groups of five pregnant CF-1 mice that were infected via tail vein injection on day 16/17 of gestation with ~5.0 ×107 CFU of the parent or ΔeutN mutant strain and monitored pup recovery and pup survival over time (Fig. 6A). Remarkably, the ΔeutN mutant showed a significant attenuation in virulence, with roughly 60% of pups being born alive by the end of experimentation, in contrast to the parent strain (CW1) which caused only 20% of pup survival (P < 0.001). Of note, no significant defects in growth and propagation of fusobacteria were observed in the ΔeutN mutant, as compared to the parent strain when fusobacterial cells were grown in rich media (Fig. 6C), a condition that was used to prepare bacterial cultures for infection. We conclude that BMC formation is critical for fusobacterial virulence in the mouse host.
Figure 6: Virulence attenuation of the eutN mutant.
(A-B) A group of 5 pregnant mice was infected with ~5 × 107 CFU of the parent or ΔeutN strain via the tail vein on day 16 or 17 of gestation, and pup survival was recorded. This experimented was repeated independently 3x. The statistical differences were analyzed by Mantel-Cox (*** P<0.001). (C) Cell growth of indicated strains in rich media were monitored by OD600.
DISCUSSION
F. nucleatum is one of the key inhabitants of the human oral cavity that can migrate to and colonize multiple distal organs, promoting the development of several extra-oral diseases, most notably colorectal cancer and preterm birth (20). Our previous studies of fusobacterial pathogenesis employing targeted gene deletion mutants suggest that the metabolic versatility of F. nucleatum may contribute to its adaptability to different environments within its animal hosts (29). This is the trait that must be essential for a commensal organism to cause diseases in multiple organs that present unique challenges in varied cellular environments. Here, we present an important advance in support of this hypothesis by demonstrating that fusobacteria form specific metabolic organelles, BMCs, in response to environmental ethanolamine and that these BMCs serve as a major determinant of this pathobiont to maintain its virulent potential within its host.
Like many gastrointestinal bacteria, F. nucleatum harbors a large eut gene cluster, coding for 17 putative EA utilization (Eut) proteins and two hypothetical proteins, with the adjoining folK and folP genes seemingly a part of the eut transcriptional units (Fig 1A). We demonstrated that when EA is provided as a major carbon and nitrogen source, expression of many eut genes is significantly elevated, concomitant with increased BMC formation (Fig. 1). Interestingly, expression of the eut gene cluster appears to be bimodal, with eutS, eutP, eutV, and eutW genes down-regulated in the EA-conditioned media and the remaining genes starting from eutA highly expressed, including the genes coding for BMC shell proteins (eutLM1M2N), (Fig. 1D). This bimodal pattern of expression of eut genes is also mirrored in the gene expression analysis of the eutV mutant (Fig. 2A). The differential expression of eutS and eutLM1M2N suggests that EutS might not be part of the BMC structures in F. nucleatum under the tested experimental conditions. Consistent with this, mass spectrometry did not identify EutS among proteins associated with isolated BMCs (Table S2). Nonetheless, we cannot rule out a role of EutS in the process of BMC formation since we were unable thus far to obtain an eutS mutant via our conventional gene deletion method.
A key question our study addressed is the identity of the molecular components of F. nucleatum BMCs. Our biochemical and genetic analyses so far suggest that EutL, EutM1, EutM2, and EutN make up the shell of F. nucleatum BMCs: they are present as components enriched in the isolated BMCs, and the genetic disruption of EutLM1M2N severely affects BMC formation (Figures 3–5; Table S2). It is intriguing that F. nucleatum encodes two copies of EutM (EutM1 and EutM2), which are strikingly similar to E. coli EutM (30), except that EutM1 has an extra α-helix at the C-terminus as predicted by AlphaFold (31, 32) (Fig. S1). Moreover, while no EutK homologs are found in F. nucleatum, its eut gene cluster encodes two hypothetical proteins (RS2810 and RS2820). Whether these hypothetical proteins participate in BMC formation and if so, in what capacity remains to be investigated.
Significantly, we also demonstrated that BMCs are formed when fusobacteria are exposed to human placental cells in culture, an experimental condition that leads to the attachment and invasion of placental cells by fusobacteria (Fig. 5). In this context, it is important to note that fusobacterial BMCs are also observed when F. nucleatum was grown in Roswell Park Memorial Institute (RPMI) media supplemented with 5% fetal bovine serum (FBS) – the medium used for culturing the placental cells (Fig. 5F). The efficient assembly of fusobacterial BMCs observed in the RPMI/FBS culture medium alone might be mediated by EA and phosphatidyl-EA present in FBS (33) or some other yet unknown component that acts as an inducer of the eut genes and BMCs. Whatever the mechanism might be, we infer that fusobacteria are capable of assembling BMCs under conditions that mimic an environment pertinent to fusobacterial encounter of one of its target host tissue, the placenta, where it induces adverse pregnancy outcomes.
The induction of BMC formation and eut gene expression in response to EA suggested that these processes are transcriptionally regulated. Indeed, we showed that the response regulator EutV of the EutV-EutW pair constituting an EA-responsive TCS that is encoded within the eut locus modulates not only transcription of the eut locus in response to EA (Fig. 7A) but also the transcription of many additional genes outside of the eut gene cluster under a nutrient-rich condition (Fig. 2 and Fig. 7B). F. nucleatum EutV is annotated as a Fis family transcriptional regulator (biocyc.org), with Fis (factor for inversion stimulation) previously shown to be a regulator of metabolism in E. coli (34). Interestingly, as predicted by AlphaFold, EutV harbors an ANTAR domain (ANTAR for AmiR and NasR transcription antitermination regulators), which is an RNA-binding domain identified in many response regulators of the two-component systems (35). Notably, as a representative member of the ANTAR-containing response regulators, the E. faecalis EutV regulates expression of eut genes (36), via its interaction with terminator elements located in the untranslated regions preceding eutP, eutG, eutS, and eutA (37). Whether or not a similar regulatory mechanism is operative in the case of the ANTAR domain in F. nucleatum EutV remains to be examined. It is worth mentioning that expression of eut genes appears to be cross-regulated by some other two-component systems and transcriptional regulators at certain conditions and cell growth. When cells are grown to mid-log phase in a nutrient-rich medium, deletion of either carR, coding for the response regulator CarR of the TCS CarRS or modR, encoding the response regulator ModR of the TCS ModRS, enhances expression of many eut genes (24, 25). In the early stationary phase, in mutants devoid of either the RNA-binding protein KhpA or KhpB, expression of 13 eut genes is down-regulated (38).
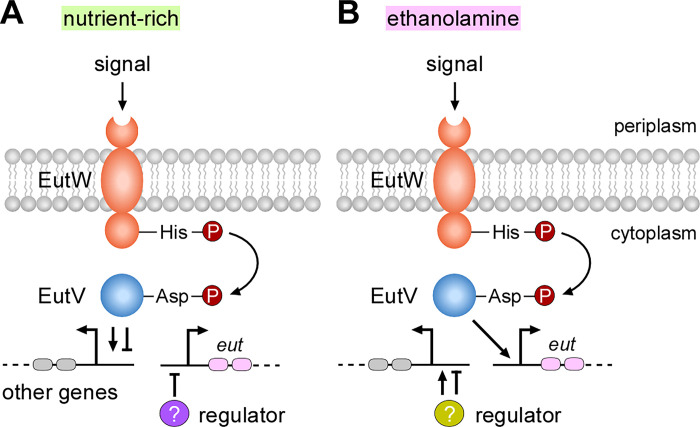
Figure 7: A working model of gene expression regulated by the two-component transduction system EutVW in F. nucleatum.
(A) In nutrient-rich conditions, a specific signal(s) is recognized by the sensor kinase EutW, leading to autophosphorylation at a conserved histidine residue. The phosphate is transferred to a conserved aspartate residue of the response regulator EutV. Phosphorylation of EutV triggers the transcriptional regulation response of EutV, resulting in the activation or repression of various genes. It is unclear if EutV also regulates expression of a regulator that represses expression of eut genes in these conditions. (B) In the presence of environmental ethanolamine, which may act as a signal that activates EutW, the response regulator EutV becomes activated, leading to the upregulation of eut genes. In this condition, it is unclear if EutV and/or other regulators controls expression of other genes.
Perhaps more importantly, we demonstrated here that the ability of F. nucleatum to form BMCs is critical to its virulence potential in its host by showing that the eutN mutant that is defective in BMC formation is attenuated in the mouse model of fusobacterial infection causing preterm birth (Fig. 6). In this context, it is prudent to recall the striking clinical observation that the level of EA increases in the homogenates of human placental villi sampled during the third trimester, in parallel with a corresponding decrease in phosphor-ethanolamine, a precursor of EA (39). These observations lead us to posit that the ability of F. nucleatum to utilize environmental EA provides fusobacteria a physiologically significant advantage to adapt to specific niches such as the placenta and the colon, in spite of the changing and challenging environments that fusobacteria encounter along their long journey from the oral cavity to those distal organs.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Media.
Bacterial strains and plasmids used in this study are listed in Supplemental Material (SM) Table S3. F. nucleatum cells were grown in tryptic soy broth supplemented with 1% Bacto Peptone plus 0.05% cysteine (TSPC) or TSPC agar plates under anaerobic conditions (10% CO2, 10% H2, and 80% N2). Escherichia coli strains were grown in Luria broth (LB). When necessary, chloramphenicol and thiamphenicol were added to culture media at a concentration of 15 μg/mL and 5 μg/ml (Fisher Scientific), respectively. All reagents were purchased from Sigma-Aldrich and unless noted otherwise.
Genetic manipulation in F. nucleatum.
(i). Generation of non-polar, in-frame deletion mutants –
Generation of single (ΔeutV or ΔeutN) and triple (ΔeutLM1M2) deletion mutants was performed according to a published protocol (26, 29). Briefly, 1-kb flanking regions upstream and downstream of eutV, eutN or eutLM1M2 were PCR-amplified with appropriate primers (Table S4) and cloned into the deletion vector pCM-GalK (27), generating the corresponding deletion vectors pΔeutV, pΔeutN, and pΔeutLM1M2 (Table S3). The generated plasmids were electroporated into F. nucleatum CW1 cells, plasmid integration into the bacterial chromosome and producing integrant strains that were selected by thiamphenicol-containing TSPC agar plates. The integrant strains were grown in media without antibiotics to allow the second homologous recombination event. This led to plasmid excision that formed wild-type or mutant alleles, which were then selected on agar plates containing 2-deoxy-D-galactose and verified by PCR and DNA sequencing.
To create a quadruple mutant lacking eutLM1M2N (ΔeutLM1M2N), the ΔeutLM1M2 deletion mutant and the deletion vector pΔeutN (Table S3) were used in the deletion procedure mentioned above.
(ii). Generation of complementing vectors –
To generate pYWC1, the 321-bp promoter region of the ribosomal RNA small subunit methyltransferase A (rsmA) in F. nucleatum ATCC23726 was PCR-amplified using appropriate primers (Table S4). The amplicon was treated with SacI and NdeI restriction enzymes prior to be ligated overnight at 16°C into pCWU6 precut with the same enzymes. The generated plasmid was introduced into E. coli DH5α for preparation of plasmid DNA, which was used for confirmation by DNA sequencing.
To generate pEutN, primers com-eutN-F and com-eutN-R were used to PCR-amplify the eutN coding sequence, with chromosomal DNA of F. nucleatum ATCC 23726 as a template, while appending BamHI/KpnI sites for cloning into pYWC1 precut with restriction enzymes BamHI/KpnI. The resulting vector, confirmed by DNA sequencing, was electroporated into the ΔeutN mutant strain.
Bacterial growth assay.
Overnight cultures of various F. nucleatum strains grown in TSPC were used to inoculate fresh cultures in minimal media with a starting optical density at 600 nm (OD600) of 0.1. Cell growth in an anaerobic chamber was monitored by OD600. The minimal media was made of 5x M9 salts (64 g of Na2HPO4•7H2O, 15 g of KH2PO4, 2.5 g of NaCl, and 5 g of NH4Cl in 1L (40), 1% Bacto peptone, 0.5 M MgSO4, 0.1 mM CaCl2, and 0.05% cysteine. Various concentrations of ethanolamine (Fisher Scientific) were added when necessary.
RNA extraction.
Total RNA of various F. nucleatum strains was extracted utilizing RNeasy Mini Kits (Qiagen) in accordance with the manufacturer’s protocol, with some modifications. Briefly, cell pellets harvested from 3-ml bacterial cultures were suspend in 200 μl of RNA-free 10mM Tris-EDTA, and the suspensions were transferred to a chilled Fast Protein Tube (Qbiogene) containing 700 μl of RNeasy lysis buffer (RNeasy Mini Kit, Qiagen) and 7 μL of β-mercaptoethanol (Thermo Fisher Scientific). Cells were lysed via Ribolyser (Hybaid), and supernatants were collected via centrifugation at 13,000 × g for 5 min at 4°C. RNA was then purified from the supernatants accordingly, and purified RNA was treated with deoxyribonuclease (DNase; Qiagen) and cleaned by using a RNeasy clean up kit (Qiagen).
Gene expression analysis.
For qRT-PCR, ~100 ng of extracted RNA was used for complementary DNA (cDNA) synthesis using iScript Supermix (Bio-Rad) according to the manufacturer’s protocol. qRT-PCR was performed using a CFX96 Real-Time System (Bio-Rad), with reactions prepared using a SYBR Green PCR Master Mix (Bio-Rad) with appropriate primers (Table S4). Changes in gene expression were calculated as previously described (25).
For RNA-seq analysis, cDNA libraries were prepared, and sequencing was performed as previously reported (25). Genes with a log2 (fold change) above +1.0 or below −1.0 were considered to be differentially transcribed under the examined conditions.
BMC isolation.
Overnight cultures of various F. nucleatum strains in TSPC were used to inoculate fresh cultures (1:10 dilution), which were grown to mid-log phase. 8-ml aliquots of these cultures were transferred to 400 ml of minimal media supplemented with 50 mM of ethanolamine or without. The resulting cultures were grown overnight (14–16 h), and BMCs were isolated according to published protocols (8, 41). Briefly, harvested cells were washed in 20 ml of Buffer A (50 mM Tris, 500 mM KCl,12.5 mM MgSO4), followed by centrifugation at 5,000 × g for 15 min. Washed cells were resuspended in 10 ml of 60% bacterial protein extraction reagent (BPER) in Buffer A supplemented with 1 mM PMSF, 1 mg/ml lysozyme, and 10 μg/ml DNase and rocked at 30°C for 1 h. Supernatants were separated from cells by centrifugation at 12,000 × g for 8 min, and the supernatants were spun at 20,000 × g for 40 min to harvest BMCs. Crude BMCs were resuspended in 50 μl of chilled Buffer B (50mM Tris, 50mM KCl, 5mM MgCl2). BMC suspensions were centrifuged at 12,000 × g to remove contaminants.
Mass spectrometry analysis of BMCs.
Obtained BMC samples in lysis buffer (100 μl, 12 mM sodium lauroyl sarcosine, 0.5% sodium deoxycholate, 50 mM triethylammonium bicarbonate (TEAB)) were heated for 5 min. The samples were treated with tris(2-carboxyethyl) phosphine (10 μl, 55 mM in 50 mM TEAB, 30 min, 37°C), followed by treatment with chloroacetamide (10 μl, 120 mM in 50 mM TEAB, 30 min., 25°C in the dark). They were then diluted 5-fold with aqueous 50 mM TEAB and incubated overnight with Sequencing Grade Modified Trypsin (1 μg in 10 μl of 50 mM TEAB; Promega, Madison, WI) following which an equal volume of ethyl acetate/trifluoroacetic acid (TFA, 100/1, v/v) was added. After vigorous mixing (5 min) and centrifugation (13,000 x g; 5 min), the supernatants were discarded, and the lower phases were dried in a centrifugal vacuum concentrator. The samples were then dissolved in acetonitrile/water/TFA (solvent A, 100 μL, 2/98/0.1, v/v/v) and loaded onto a small portion of a C18-silica disk (3M, Maplewood, MN) placed in a 200 μl pipette tip. Prior to sample loading, the C18 disk was prepared by sequential treatment with methanol (20 μl), acetonitrile/water/TFA (solvent B, 20 μl, 80/20/0.1, v/v/v) and finally with solvent A (20 μl). After loading the sample, the disc was washed with solvent A (20 μl, eluent discarded) and eluted with solvent B (40 μl) (42). The collected eluent was dried in a centrifugal vacuum concentrator.
The eluants were reconstituted in water/acetonitrile/FA (solvent E, 10 μl, 98/2/0.1, v/v/v), and aliquots (5 μl) were injected onto a reverse phase nanobore HPLC column (AcuTech Scientific, C18, 1.8 μm particle size, 360 μm × 20 cm, 150 μm ID), equilibrated in solvent E and eluted (500 μl/min.) with an increasing concentration of solvent F (acetonitrile/water/FA, 98/2/0.1, v/v/v: min./% F; 0/0, 5/3, 18/7, 74/12, 144/24, 153/27, 162/40, 164/80, 174/80, 176/0, 180/0) using an Eksigent NanoLC-2D system (Sciex (Framingham, MA)). The effluent from the column was directed to a nanospray ionization source connected to a high-resolution orbitrap mass spectrometer (Q Exactive Plus, Thermo Fisher Scientific) acquiring mass spectra in a data-dependent mode alternating between a full scan (m/z 350–1700, automated gain control (AGC) target 3 × 106, 50 ms maximum injection time, FWHM resolution 70,000 at m/z 200) and up to 10 MS/MS scans (quadrupole isolation of charge states ≥ 2, isolation width 1.2 Th) with previously optimized fragmentation conditions (normalized collision energy of 32, dynamic exclusion of 30 s, AGC target 1 × 105, 100 ms maximum injection time, FWHM resolution 35,000 at m/z 200). The raw data was analyzed in Proteome Discoverer 2.4 with appropriate chemical modifications.
Placental cells co-culturing with F. nucleatum.
Placental HTR-8/SVneo cells were grown in Roswell Park Memorial Institute (RPMI) supplemented with 5% fetal bovine serum (FBS) to ~90% confluence in an incubator maintained at 5% CO2 and infected with individual F. nucleatum strains, grown to mid-log phase in TSPC, at a MOI of 450 for 4.5 h. Cells were gently washed with ice-cold phosphate-buffered saline (PBS) to remove unattached cells prior to being subjected to the embedding procedure described below.
Electron microscopy.
BMCs were analyzed by transmission electron microscopy (TEM), adapted from a published protocol (27). Briefly, a drop of isolated BMCs in suspension was placed onto carbon-coated TEM grids and wicked off with filtered papers after 10 min. BMCs on grids were stained twice with 2% uranyl acetate prior to electron microscopic analysis (FEI Tecnai 20 iCorr TEM).
For thin-section TEM of fusobacteria alone or fusobacteria cocultured with placental cells, samples were prepared according to a published protocol with some minor modifications (43). For cocultured samples, cells were fixed overnight in 4% paraformaldehyde in 0.2 M HEPES at 4°C and washed with 0.1 M HEPES for 15 min. Cells were then fixed with 2% OsO4 and 1.5% potassium ferricyanide in 0.1M phosphate buffer at 4°C and dehydrated in increasing concentrations of pure-grade ethanol (50–100%) for 15 min with centrifugation at 13,523 × g for 3 min followed by 100% acetone for 7 min. Fixed cells were embedded in 33% Spurr’s resin in acetone for 1 h, 66% Spurr’s Resin in acetone overnight, and 100% Spurr’s Resin for ~6 h and polymerized for ~72 h. For bacterial cells alone, sample preparation was performed similarly except that cells were fixed in 4% paraformaldehyde in 0.2 M HEPES at room temperature for 2 h and washed in 0.1 M phosphate buffer, followed by post-fixation in 1% OsO4 in 0.1 M phosphate buffer and washed again. All samples were thin sectioned using an UCT Ultramicrotome (Leica) and collected on Formvar-coated nickel grids. Bacterial samples were post-stained with 2% uranyl acetate, while cocultured samples were post-stained with 3% lead citrate prior to analysis using a T20 iCorr TEM.
For cryoEM, 3 μL aliquots of BMC samples were applied to glow-discharged Quantifoil R2/1 holey carbon grids (300 mesh, Ted Pella). After 3-s incubation, grids were plunge-frozen into liquid ethane using a Vitrobot Mark IV vitrification system (Thermo-Fisher Scientific). During freezing condition optimization, cryo-EM grids were screened on an FEI Technai F20 electron microscope operated at 200 kV. Optimal conditions were achieved with a 90-s glow-discharge using a PELCO easiGlow™ (air as gas source, target vacuum of 0.37 mbar, target current of 15 mA) and Vitrobot settings at 4°C, 100% humidity, 5.5 s blotting time, blotting force 2, and 0 s drain time. CryoEM grids were imaged on the same instrument as used during screening, with a TIETZ F415MP 16-megapixel CCD camera at a nominal magnification of 25,000 × (pixel size 4.3 Å/pixel).
Mouse model of preterm birth.
To compare the infectivity of ΔeutN with that of the parent strain, we employed a mouse model of preterm birth as previously reported (26, 29). Briefly, groups of six CF-1 (Charles River Laboratories) pregnant mice were infected with ~5×107 CFU of the parent or ΔeutN strain at day 16 or 17 of gestation via tail vein injection. Pup survival was recorded for the next 8 days. Statistical analysis was determined via the Mantel-Cox text, using GraphPad Prism.
IMPORTANCE.
The oral anaerobe Fusobacterium nucleatum can spread to distal internal organs, such as the colon and placenta, and thereby promote the development of colorectal cancer and induce preterm birth, respectively. Yet, how this opportunistic pathogen adapts to the various metabolically distinct host cellular niches remains poorly understood. We demonstrated here that this microbe assembles specialized metabolic organelles, termed bacterial microcompartments (BMCs), to utilize environmental ethanolamine (EA) as a key environmental nutrient source. The formation of F. nucleatum BMCs, containing BMC shell proteins EutLM1M2N, is controlled by a two-component system, EutV-EutW, responsive to EA. Significantly, this ability of F. nucleatum to form BMCs in response to EA is crucial for its pathogenicity evidenced by the fact that the genetic disruption of BMC formation reduces fusobacterial virulence in a mouse model of preterm birth.
ACKNOWLEDGMENTS
We thank Drs. Chenggang Wu, Ju Huck Lee, and Luong T. Truc (UT McGovern Medical School) for providing reagents, Lucy Gao and Andrew Goring (UCLA) for technical assistance, and the Ton-That lab members for their critical input and discussion. Research reported in this publication was supported by the National Institute of Dental & Craniofacial Research of the National Institutes of Health under the Award Numbers DE032906 and DE026758 (to H.T.-T). Dana Franklin was supported by the Ruth L. Kirschstein National Research Service Award T32AI007323. Angela Agnew was supported by the UCLA Dentist-Scientist and Oral Health-Researcher Training Program, NIDCR grant T90DE030860. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests
DATA AVAILABILITY
All data generated in this study are provided in this manuscript, including the Supplemental Material. The RNA-seq data were deposited in the NCBI Gene Expression Omnibus (GEO) database with the accession number of GSE280934.
REFERENCES
- 1.Kaval KG, Garsin DA. 2018. Ethanolamine Utilization in Bacteria. mBio 9. 10.1128/mBio.00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luzader DH, Kendall MM. 2016. Commensal ‘trail of bread crumbs’ provide pathogens with a map to the intestinal landscape. Curr Opin Microbiol 29:68–73. 10.1016/j.mib.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsoy O, Ravcheev D, Mushegian A. 2009. Comparative genomics of ethanolamine utilization. J Bacteriol 191:7157–64. 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobik TA, Lehman BP, Yeates TO. 2015. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol 98:193–207. 10.1111/mmi.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravcheev DA, Moussu L, Smajic S, Thiele I. 2019. Comparative Genomic Analysis Reveals Novel Microcompartment-Associated Metabolic Pathways in the Human Gut Microbiome. Front Genet 10:636. 10.3389/fgene.2019.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka S, Sawaya MR, Yeates TO. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81–4. 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- 7.Penrod JT, Roth JR. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol 188:2865–74. 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Held M, Kolb A, Perdue S, Hsu SY, Bloch SE, Quin MB, Schmidt-Dannert C. 2016. Engineering formation of multiple recombinant Eut protein nanocompartments in E. coli. Sci Rep 6:24359. 10.1038/srep24359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee A, Kaval KG, Garsin DA. 2024. Role of ethanolamine utilization and bacterial microcompartment formation in Listeria monocytogenes intracellular infection. Infect Immun 92:e0016224. 10.1128/iai.00162-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luzader DH, Clark DE, Gonyar LA, Kendall MM. 2013. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195:4947–53. 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D’Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184:2005–18. 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponath F, Tawk C, Zhu Y, Barquist L, Faber F, Vogel J. 2021. RNA landscape of the emerging cancer-associated microbe Fusobacterium nucleatum. Nat Microbiol 6:1007–1020. 10.1038/s41564-021-00927-7. [DOI] [PubMed] [Google Scholar]
- 13.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. 2009. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol 47:38–47. 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Buhimschi CS, Temoin S, Bhandari V, Han YW, Buhimschi IA. 2013. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One 8:e56131. 10.1371/journal.pone.0056131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaim W, Mazor M. 1992. Intraamniotic infection with fusobacteria. Arch Gynecol Obstet 251:1–7. 10.1007/BF02718272. [DOI] [PubMed] [Google Scholar]
- 16.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Liu F, Ling Z, Tong X, Xiang C. 2012. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7:e39743. 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. 2014. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33:1381–90. 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 19.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–8. 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han YW. 2015. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol 23:141–7. 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapatral V, Ivanova N, Anderson I, Reznik G, Bhattacharyya A, Gardner WL, Mikhailova N, Lapidus A, Larsen N, D’Souza M, Walunas T, Haselkorn R, Overbeek R, Kyrpides N. 2003. Genome analysis of F. nucleatum sub spp vincentii and its comparison with the genome of F. nucleatum ATCC 25586. Genome Res 13:1180–9. 10.1101/gr.566003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zepeda-Rivera M, Minot SS, Bouzek H, Wu H, Blanco-Miguez A, Manghi P, Jones DS, LaCourse KD, Wu Y, McMahon EF, Park SN, Lim YK, Kempchinsky AG, Willis AD, Cotton SL, Yost SC, Sicinska E, Kook JK, Dewhirst FE, Segata N, Bullman S, Johnston CD. 2024. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature 628:424–432. 10.1038/s41586-024-07182-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YW, Camacho MI, Chen Y, Bhat AH, Chang C, Peluso EA, Wu C, Das A, Ton-That H. 2022. Genetic Determinants of Hydrogen Sulfide Biosynthesis in Fusobacterium nucleatum Are Required for Bacterial Fitness, Antibiotic Sensitivity, and Virulence. mBio 13:e0193622. 10.1128/mbio.01936-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheible M, Nguyen CT, Luong TT, Lee JH, Chen YW, Chang C, Wittchen M, Camacho MI, Tiner BL, Wu C, Tauch A, Das A, Ton-That H. 2022. The Fused Methionine Sulfoxide Reductase MsrAB Promotes Oxidative Stress Defense and Bacterial Virulence in Fusobacterium nucleatum. mBio 13:e0302221. 10.1128/mbio.03022-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, Chen YW, Scheible M, Chang C, Wittchen M, Lee JH, Luong TT, Tiner BL, Tauch A, Das A, Ton-That H. 2021. Genetic and molecular determinants of polymicrobial interactions in Fusobacterium nucleatum. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2006482118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peluso EA, Scheible M, Ton-That H, Wu C. 2020. Genetic Manipulation and Virulence Assessment of Fusobacterium nucleatum. Curr Protoc Microbiol 57:e104. 10.1002/cpmc.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Al Mamun AAM, Luong TT, Hu B, Gu J, Lee JH, D’Amore M, Das A, Ton-That H. 2018. Forward Genetic Dissection of Biofilm Development by Fusobacterium nucleatum: Novel Functions of Cell Division Proteins FtsX and EnvC. mBio 9. 10.1128/mBio.00360-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponath F, Zhu Y, Cosi V, Vogel J. 2022. Expanding the genetic toolkit helps dissect a global stress response in the early-branching species Fusobacterium nucleatum. Proc Natl Acad Sci U S A 119:e2201460119. 10.1073/pnas.2201460119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Britton TA, Wu C, Chen YW, Franklin D, Chen Y, Camacho MI, Luong TT, Das A, Ton-That H. 2024. The respiratory enzyme complex Rnf is vital for metabolic adaptation and virulence in Fusobacterium nucleatum. mBio 15:e0175123. 10.1128/mbio.01751-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takenoya M, Nikolakakis K, Sagermann M. 2010. Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli. J Bacteriol 192:6056–63. 10.1128/JB.00652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varadi M, Bertoni D, Magana P, Paramval U, Pidruchna I, Radhakrishnan M, Tsenkov M, Nair S, Mirdita M, Yeo J, Kovalevskiy O, Tunyasuvunakool K, Laydon A, Zidek A, Tomlinson H, Hariharan D, Abrahamson J, Green T, Jumper J, Birney E, Steinegger M, Hassabis D, Velankar S. 2024. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res 52:D368–D375. 10.1093/nar/gkad1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee DY, Lee SY, Yun SH, Jeong JW, Kim JH, Kim HW, Choi JS, Kim GD, Joo ST, Choi I, Hur SJ. 2022. Review of the Current Research on Fetal Bovine Serum and the Development of Cultured Meat. Food Sci Anim Resour 42:775–799. 10.5851/kosfa.2022.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Gil G, Bringmann P, Kahmann R. 1996. FIS is a regulator of metabolism in Escherichia coli. Mol Microbiol 22:21–9. 10.1111/j.1365-2958.1996.tb02652.x. [DOI] [PubMed] [Google Scholar]
- 35.Shu CJ, Zhulin IB. 2002. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci 27:3–5. 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 36.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A 106:4435–40. 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. 2012. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet 8:e1002666. 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Ponath F, Cosi V, Vogel J. 2024. A global survey of small RNA interactors identifies KhpA and KhpB as major RNA-binding proteins in Fusobacterium nucleatum. Nucleic Acids Res 52:3950–3970. 10.1093/nar/gkae010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sooranna SR, Burston D, Ramsay B, Steer PJ. 1994. Free amino acid concentrations in human first and third trimester placental villi. Placenta 15:747–51. 10.1016/0143-4004(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 40.Buda R, Liu Y, Yang J, Hegde S, Stevenson K, Bai F, Pilizota T. 2016. Dynamics of Escherichia coli’s passive response to a sudden decrease in external osmolarity. Proc Natl Acad Sci U S A 113:E5838–E5846. 10.1073/pnas.1522185113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochoa JM, Mijares O, Acosta AA, Escoto X, Leon-Rivera N, Marshall JD, Sawaya MR, Yeates TO. 2021. Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments. Acta Crystallogr F Struct Biol Commun 77:275–285. 10.1107/S2053230X21007470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 43.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 2009. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol 452:143–64. 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated in this study are provided in this manuscript, including the Supplemental Material. The RNA-seq data were deposited in the NCBI Gene Expression Omnibus (GEO) database with the accession number of GSE280934.