Abstract
Cyclic tetrapeptides (CTPs) are a diverse class of natural products with a broad range of biological activities. However, they are extremely challenging to synthesize due to the ring strain associated with their small ring size. While chemical methods have been developed to access CTPs, they generally require the presence of certain amino acids, limiting their substrate scopes. Herein, we report the first bioinformatics guided discovery of a thioesterase from a cryptic biosynthetic gene cluster for peptide cyclization. Specifically, we hypothesized that predicted Penicillin-binding type thioesterases (PBP-TEs) from cryptic nonribosomal peptide synthetase gene clusters containing four adenylation domains would catalyze tetrapeptide cyclization. We found that one of the predicted PBP-TEs, WP516, efficiently cyclizes a wide variety of tetrapeptide substrates. To date, it is only the second stand-alone enzyme capable of cyclizing tetrapeptides, and its substrate scope greatly surpasses that of the only other reported tetrapeptide cyclase Ulm16. AlphaFold modeling and covalent docking were used to rationalize the broad substrate scope of WP516 in comparison to other PBP-TEs. Overall, the bioinformatics guided workflow outlined in this paper, and the discovery of WP516, represent promising tools for the biocatalytic production of head-to-tail CTPs, as well as a more general strategy for discovery of enzymes for peptide cyclization.
Introduction
Peptide-based therapeutics have garnered significant interest in pharmaceutical development, resulting in hundreds of peptides currently undergoing clinical trials and a global market value exceeding tens of billions of dollars.1 Clinically relevant drugs exemplifying this success include the antibiotic murepavadin, the antifungal rezafungin, the immunosuppressant cyclosporine, and the anticancer agent octreotide.2 However, linear and unmodified peptides often display poor therapeutic properties due to their intrinsic instability and poor selectivity.3 Peptidyl natural products exhibit astounding structural diversity, which endows them with a remarkable range of biological activities.4 This diversity is generated by biosynthetic enzymes that construct core scaffolds and perform peripheral modifications, introducing pharmacophores, in stereoselective and regioselective fashion. These modifications can include methylation (N, O, C), halogenation, oxidation, and glycosylation.5 A particularly promising modification is head-to-tail macrocyclization, which enhances membrane permeability by eliminating the zwitterionic C and N termini, while also increasing potency, specificity, and proteolytic stability through defined secondary structures,6 enabling these peptides to occupy a ‘Goldilocks zone’ between small molecules and biologics.7 Despite these advantages, macrocyclization remains a synthetically challenging transformation. Epimerization at the α-carbon of the C-terminal residue, competing oligomerization, and conformational rigidity preclude remote residue coupling leading to poor yields and scalability issues.8,9 These problems are exacerbated as ring size decreases. Specifically, for tetrapeptides, the ground-state E geometry of the peptide bond impedes the adoption of a conformation favorable to cyclization, while the transannular ring strain further complicates the synthesis and derivatization of pharmaceutically and industrially relevant small cyclic peptides, at times rendering them inaccessible.9 This in turn severely limits our ability to access a class of cyclic peptides, cyclic tetrapeptides (CTPs), which show improved PK properties and oral bioavailability compared to their larger counterparts.10 CTP natural products have a variety of interesting bioactivities including inhibition of the chloroplast F1-ATPase (tentoxin), selective blockage of calcium channels by the cyclic peptide family oncychocins, antagonism of kappa opioid receptors by tetrapeptide CJ-15208 and inhibition of histone deacetylases (chlamydocin, trapoxin A, apicidin, microsporins and others), further motivating their investigation.11–14 While several synthetic strategies have been employed to gain access to CTPs such as ring contraction via Ser/Thr ligation mediated peptide cyclization15, on resin cyclization via anchoring of a Glu/Asp16, pseudo proline protecting groups17, and anion assisted cyclization18, they are either incompatible with Fmoc solid phase peptide synthesis (SPPS) or restrict amino acid sequence, often requiring a serine, threonine, or negatively charged amino acid (features not common in naturally occurring cyclic tetrapeptides).9
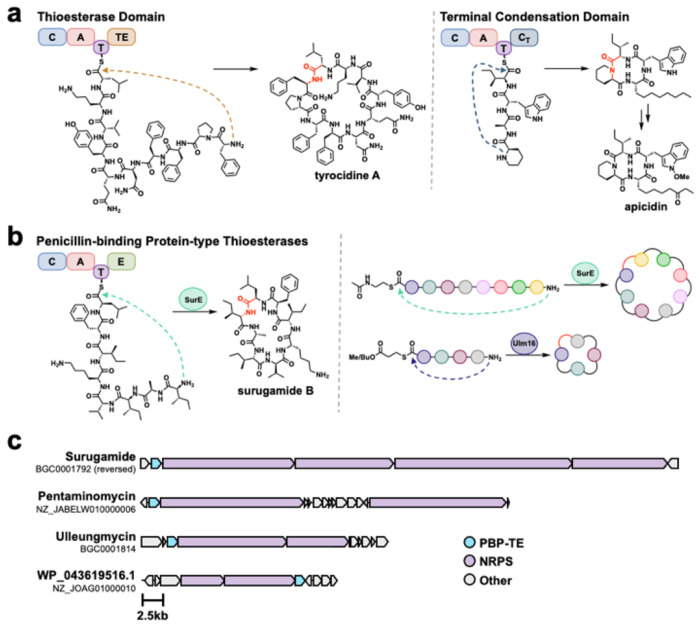
Recently, there has been increased interest in utilizing the biosynthetic machinery used to produce these cyclic peptides in vivo as biocatalysts for head-to-tail macrocyclization.19 Many peptidyl natural products from soil dwelling bacteria and fungi are produced by multimodular enzyme complexes known as Non-Ribosomal Peptide Synthases (NRPSs) and are frequently implicated in the production of small cyclic peptides including tetra- and pentapeptides.20–23 Offloading from the complex and cyclization are typically catalyzed by C-terminal domains which are in-cis (connected) to the complex, such as thioesterase domains (TE) in bacteria or condensation termination (CT) domains in fungi (Figure 1A).24 While these domains can efficiently catalyze the cyclization of strained cyclic peptides in vivo and have been studied as biocatalysts25, they have failed to reach widespread use due to their low substrate promiscuity,26 an inability to accurately predict cyclization modes (e.g. head-to-tail) via bioinformatics,27 and in the case of CT domains, the strict requirement for a peptidyl carrier protein (PCP) tethered substrate.24,28 However, a recently discovered thioesterase family sharing sequence similarity to the penicillin binding protein (PBP-TEs) have garnered significant attention for their use as biocatalysts for head-to-tail peptide macrocyclization as they act in trans (not connected) to the NRPS and have displayed unprecedented substrate promiscuity.29 The most extensively studied enzyme from this family is SurE from Streptomyces albidoflavus,30 which natively catalyzes the offloading and cyclization of two structurally unrelated octa- and decapeptides. SurE has demonstrated biocatalytic promise in vitro, catalyzing the cyclization of peptides ranging from 5-10 amino acids varying widely in sequence and peptide mimics with the equivalent length of 23 amino acids.31–35 Recently, we characterized Ulm16 from Streptomyces sp. KCB13F003, which natively catalyzes the offloading and cyclization of the hexapeptide antibiotics the ullugamycins (Figure 1B).36 While Ulm16 has a much more narrow substrate scope with respect to peptide length compared to SurE, it has demonstrated the unique ability to cyclize tetrapeptides in vitro with many substrates having cyclic:hydrolyzed ratios above 20:1 and high catalytic efficiencies up to 106 M−1s−1.37 However, its substrate scope was limited to the cyclization of DDLL and DDDL tetrapeptides (C to N-Terminus). For DLDL peptides, hydrolysis was favored, and Ulm16 was unable to produce any cyclic peptide with DLLL peptides, necessitating the need to find an alternative biocatalyst. Herein, we describe the bioinformatics identification of WP_043619516.1 (herein named WP516) a PBP-TE from a cryptic biosynthetic gene cluster (Figure 1C), which is a capable of cyclizing DLDL and DLLL peptides and has a greatly extended amino acid substrate scope compared to that of Ulm16.
Figure 1. Head-to-tail offloading mechanisms in non-ribosomal peptide synthesis.
A.) Thioesterase domain (TE) catalyzed cyclization and release25 and condensation Termination domain (CT) catalyzed cyclization and release from fungal NRPSs are common in peptide cyclization.20 Both act in-cis (connected) to the NRPS. B.) Penicillin-binding protein type thioesterase (PBP-TE) catalyze peptide cyclization and release in-trans (not connected) to the NRPS, with SurE and Ulm16 showing broad catalytic activity.32,36 C.) Biosynthetic gene clusters (BGCs) of known natural products produced by PBP-TEs along with the BGC of WP_04319516.1 a cryptic tetrapeptide BGC with a PBP-TE.
Identification and Initial Evaluation of PBP-TEs WP516 and SEC28031.1
To-date, no known cyclic tetrapeptide natural products have been associated with a PBP-TE; however, previous bioinformatics has identified multiple PBP-TEs colocalized with cryptic NRPS biosynthetic gene clusters (BGCs) that are predicted to produce a tetrapeptide (i.e. 4 adenylation, domains).38 While expression of traditional bioinformatically identified thioesterase (TE) domains is inherently risky due to their low substrate promiscuity (i.e. the natural products sequence must be known) and our inability to predict what reaction they will catalyze (hydrolysis, head-to-tail, or head-to-sidechain cyclization),27 PBP-TEs have displayed large substrate tolerance and, to date, only been shown to catalyze head-to-tail cyclization. Because of these characteristics, we chose to explore cryptic PBP-TEs as potential biocatalysts for tetrapeptide macrocyclization. To this end, we conducted an additional bioinformatics search utilizing BiG-SCAPE CORASON39 to better understand the genomic context of these predicted tetrapeptide cyclases (see SI Figure 1 for workflow and SI Figure 2 for results). From this, we identified 10 predicted tetrapeptide PBP-TEs. Two PBP-TEs were chosen for expression due to their predicted substrates: WP_031183424.1 (from Streptomyces seoulensis) and WP_043619516.1 (from Nonomuraea candida HMC10) (SI Figures 2–3). WP_031183424.1 was also further analyzed because it was hypothesized to cyclize a tetrapeptide with DLDL stereochemistry and clustered away from all other tetrapeptide BGCs. Instead, it clustered next to FlkO, a recently validated PBP-TE with a native hexapeptide substrate possessing DLDLDL stereochemistry.40 FlkO, despite sharing 70% amino acid identity with WP_031183424.1, was unable to cyclize a tetrapeptide with DLDL stereochemistry. Unfortunately, attempts to express this cyclase were unsuccessful as we were unable to obtain and solubilize the protein, preventing further investigation or characterization. We were, however, able to express WP_043619516.1 (SI Figure 4, here after referred to as WP516) which was predicted to natively cyclize a tetrapeptide with a C-terminal D-serine followed by D-enduracididine at the 2-position (a non-canonical amino acid whose biosynthetic precursor is arginine)41, L-phenylalanine at the 3-position, and an N-terminal L-valine. Additionally, we conducted a BlastP search of WP516 and identified SEC28301.1 (from Streptomyces sp. TLI_105) which, despite having the same predicted substrate as WP516, shared only 54% sequence identity (SI Figure 5). We hypothesized that this may cause them to possess differing substrate scopes from each other. In our initial evaluation of WP516 and SEC28301.1, we set out to confirm their activity as tetrapeptide cyclases and investigate their tolerance of DLDL stereochemistry, a characteristic that resulted in reduced cyclization activity and increased hydrolysis with Ulm16. Utilizing a substrate previously tested with Ulm16 (1), both WP516 and SEC28301.1 demonstrated the ability to cyclize a tetrapeptide possessing DLDL stereochemistry (Figure 2A, SI Figure 7). Due to SEC28301.1 having solubility issues and a narrower substrate scope with respect to some C-terminal residues, specifically polar residues (SI Figure 8), we chose to focus solely on WP516.
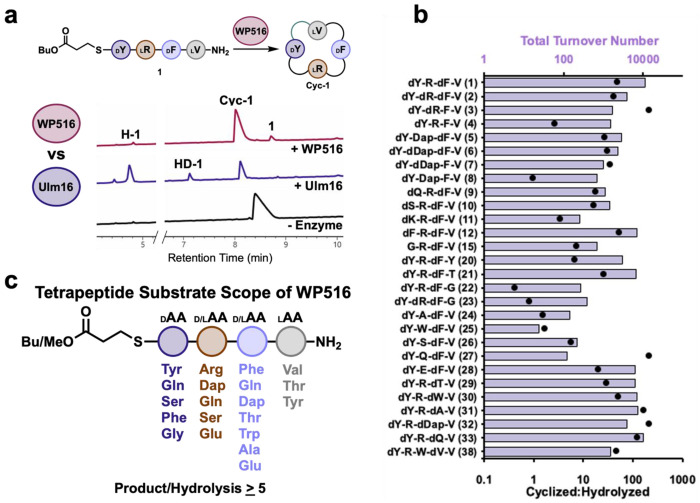
Figure 2. Substrate scope of WP516.
A.) comparison between WP516 and Ulm16 on DLDL peptide substrate 1. Reaction conditions can be found in the methods section and SI Table 1. Cyc-1 is head-to-tail cyclic peptide of 1, H-1 represents hydrolyzed 1 and HD-1 represents 1 which has been dimerized and hydrolyzed. Traces were taken at a wavelength of 214 nm. Full UPLC traces can be found in SI Figure 7. B.) Substrate scope of WP516 featuring total turnover number (TTN, top, bars) and cyclized to hydrolyzed ratio (bottom, dots) for all substrates that were cyclized. For a full list of tested substrates see SI Table 1. C.) Representative substrate scope of WP516 featuring all amino acids and stereochemistries that yielded a cyclized:hydrolyzed ratio >5:1.
Substrate Scope Investigation
To fully explore the stereochemical tolerance of WP516, we synthesized diastereomers of 1 with DDDL (2), DDLL (3), or DLLL (4) stereochemistry. Interestingly, we found 1 with DLDL stereochemistry to be the preferred substrate compared to the other diastereomers with a TTN of 11219 (SI Table 1, Figure 2B). Modifying the stereochemistry from DLDL to DDDL (2) resulted in a roughly three-fold reduction in TTN (SI Figure 9 and 11). Surprisingly, we observed a six-fold reduction in TTN compared to 1 with the diastereomer possessing the stereochemistry of WP516’s predicted native substrate, DDLL (3) (SI Figure 12). Despite the differences in activity, high selectivity for the cyclized over hydrolyzed product was maintained with these diastereomers (>20:1 cyclized to hydrolyzed). With the DLLL diastereomer (4), WP516 displayed a greater propensity for hydrolysis (2.6:1 cyclized to hydrolyzed) and an approximately eight-fold reduction in TTN compared to 1 (SI Figure 13). Despite the reduction in activity, the ability of WP516 to cyclize 4 demonstrates unprecedented stereochemical promiscuity when compared to Ulm16, which was only capable of hydrolysis and dimerization with this substrate (Figure 3A, SI Figure S14). To further examine the ability of WP516 to cyclize substrates poorly tolerated by Ulm16, we investigated its activity toward a substrate with a 2,3-diaminopropionic acid (dap) at the 2 position and DLDL stereochemistry (5). Excitingly, 5 was well tolerated by WP516 (TTN: 2901, >20:1 cyclized to hydrolyzed) and exhibited a catalytic efficiency of 3.7 x 104 M−1s−1 (Figure 4A, SI Figure 15, SI Table 1). In the same manner as before, we synthesized diastereomers of 5 with DDDL (6), DDLL (7), and DLLL (9) stereochemistry then determined their TTNs (SI Table 1) and, if cyclization was favored, Michaelis-Menten kinetics (Figure 4). We observed a similar trend of stereochemical preference with the DDDL substrate (6) displaying a slightly reduced TTN and a two-fold reduction in catalytic efficiency compared to 5 (Figure 4B, SI Figure 16). Again, we observed a further decrease in catalytic activity with the DDLL diastereomer (7), demonstrated by a three-fold reduction in TTN and eight-fold reduction in catalytic efficiency (Figure 4C, SI Figure 17). While WP516 exhibited activity toward the DLLL substrate (8), it resulted in a four-fold reduction in TTN and complete abolishment of selectivity for cyclized over hydrolyzed products (0.9:1 cyclized to hydrolyzed, SI Figure 18).
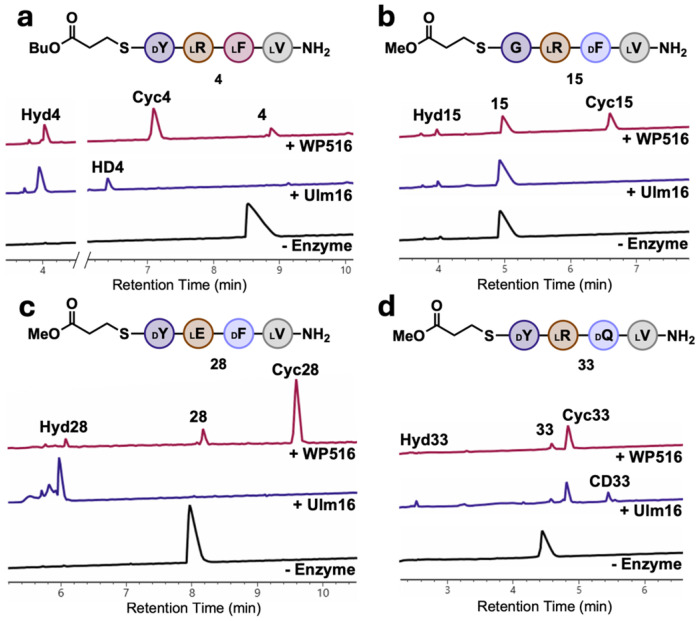
Figure 3. Comparison of WP516 and Ulm16 enzyme reactions with tetrapeptide substrates.
A.) Ulm16 and WP516 with peptide 4. B.) Ulm16 and WP516 with peptide 15. C.) Ulm16 and WP516 with peptide 28. D.) Ulm16 and WP516 with peptide 33. UV traces (214 nm) are from UPLC analysis. Full list of reaction conditions and traces can be found in the supplementary information. All reactions were run in triplicate with a representative 214 nm trace used for each reaction. CycX is head-to-tail cyclized peptide, HydX is hydrolyzed peptide, and CDx represents a peptide that has been dimerized then cyclized.
Figure 4. Michalis-Menten kinetics for WP516.

A.) peptide 5 B.) peptide 6 C.) peptide 7. Methods can be found in the supplementary information All reactions were run in triplicate. s.e.m., standard error of the mean.
To fully elucidate the substrate scope of WP516, we used 1 as our model sequence, modifying each position and determining TTNs. WP516 was tolerant of a wide range of substitutions of the C-terminal residue as long as D-chirality was maintained (peptides 9-15, Figure 2B, SI Table 1). Polar uncharged residues at the C-terminus, such as D-glutamine (9) or D-serine (10), decreased catalytic activity compared to D-tyrosine (roughly ten- and seven-fold reduced TTN, respectively) but selectivity for cyclized over hydrolyzed products was maintained (>15:1 cyclized to hydrolyzed ratio, SI Figure 19 and 20). A positive charge at the C-terminus, such as D-lysine (11), was not well tolerated by WP516, resulting in a dramatically reduced TTN and greater propensity for hydrolysis (SI Figure 21). Substitution of D-tyrosine for D-phenylalanine (12) was very well tolerated by WP516 with good catalytic efficiency (kcat/KM = 2.7 x 104 M−1s−1) and little change in the ratio of cyclized to hydrolyzed products (SI Figure 22 and 23), however substitution for D-valine (13) resulted in a substrate not processed by WP516 (SI Figure 24), possibly due to the increase in steric bulk at the β-carbon. Negatively charged D-glutamate (14) at the C-terminus also resulted in a substrate that WP516 could not process (no cyclization or hydrolysis, SI Figure 25). Notably, WP516 was tolerant of glycine (15) at the C-terminus, maintaining selectivity for cyclized over hydrolyzed products with a ratio of 7:1 (Figure 3B, SI Figure 26). This is particularly noteworthy because many PBP-TEs are unable to tolerate a C-terminal glycine without further engineering.33 This is in contrast to Ulm16, which failed to cyclize a tetrapeptide with a C-terminal glycine residue (Figure 3B, SI Figure 27) despite previously showing the ability to cyclize a hexapeptide substrate bearing a C-terminal glycine.37 On the basis of this finding, we chose to probe the ability of WP516 to cyclize an all L tetrapeptide with glycine at the C terminus (GLLL, 16), but only hydrolysis was observed (SI Figure 28). Tolerance for substitutions at the N-terminus (peptides 17-24, Figure 2B, SI Table 1) was more limited, with WP516 displaying a preference for small, uncharged residues. Substituting L-valine for L-proline (17), which converted the N-terminus to a secondary amine, resulted exclusively in hydrolysis by WP516 (SI Figure 29). Similarly, a charged residue at the N-terminus, either L-histidine (18) or L-glutamate (19), yielded only the hydrolyzed product (SI Figures 30–31). Substitution for L-tyrosine (20) was better tolerated, although it still resulted in an approximately three-fold reduction in TTN and increased levels of hydrolyzed product (roughly 6:1 cyclized to hydrolyzed products, SI Figure 32). An N-terminal L-threonine (21) resulted in an approximately two-fold reduction in TTN compared to L-valine, although a high ratio of cyclized to hydrolyzed products (>20:1) was maintained (SI Figure 33). WP516 also exhibited cyclization activity toward substrates possessing an N-terminal glycine, with either D-D-D-G (22) or D-L-D-G (23) stereochemistry. However, this resulted in a 30-fold and 40-fold reduction in TTN, respectively, compared to L-valine and preference for hydrolysis over cyclization (SI Figures 34 and 35).
At the 2 position (Peptides 24-28, Figure 2B, SI Table 1), WP516 displays a preference for charged residues. Replacement of L-arginine with non-polar residues, such as L-alanine (24) or L-tryptophan (25), almost completely abolished cyclization activity and substantially increased hydrolysis with a ratio of cyclized to hydrolyzed products nearing 1:1 (SI Figures 36 and 37). Polar uncharged amino acids similarly reduced catalytic activity but maintained better cyclization to hydrolysis ratios. Specifically, a 5.5:1 ratio of cyclized to hydrolyzed products was observed with L-serine (26) (SI Figure 38) and for L-glutamine (27) complete selectivity for cyclized product was observed (SI Figure 39). Substitution for L-glutamate (28) at the 2 position was much better tolerated by WP516, maintaining high selectivity for the cyclized over hydrolyzed product (approximately 20:1) and retaining good catalytic activity with a less than two-fold reduction in TTN (SI Figure 40). In contrast, incubation of this substrate with Ulm16 resulted exclusively in hydrolysis (Figure 3C, SI Figure 41). Substitutions at the 3 position were very well tolerated by WP516 with a wide variety of residues accepted (Peptides 29-34). Replacement of D-phenylalanine with D-threonine (29), D-tryptophan (30), D-alanine (31), or D-diaminopropionic acid (D-dap, 32) yielded substrates that retained moderate to high TTNs with low levels of hydrolysis (SI Figures 42–45). Substitution for D-glutamine (33) was specifically well tolerated with little change in TTN compared to D-phenylalanine and high selectivity for cyclized over hydrolyzed products observed >100:1 (SI Figure 46). Again, these results strongly contrast Ulm16 which, when incubated with this substrate, produced cyclic dimer and increased levels of the hydrolyzed product alongside the cyclic peptide (Figure 3D, SI Figure 47). WP516 was also tolerant of D-glutamate (34) at the 3 position with minimal hydrolysis. Unfortunately, the cyclic peptide was too insoluble to determine TTN, though we still observed a ratio of cyclized to hydrolyzed products >20:1 (SI Figure 48).
Prompted by the previously observed ring size promiscuity of PBP-TEs, we explored the tolerance of WP516 to substrates ranging from 3 to 6 amino acids. Specifically, we investigated its ability to cyclize 3 tripeptide substrates with varying stereochemistry (35-37), a modified PenA substrate that is efficiently cyclized by Ulm16 (38), a derivative of Ulm16’s substrate (39), and a modified DsaJ substrate (40). While the tri- and hexapeptide substrates resulted exclusively in hydrolysis by WP516 (SI Figures 49–53), the PenA pentapeptide substrate was successfully cyclized (SI Figure 54), with a cyclized to hydrolyzed ratio >20:1. While WP516 was notably less efficient with this substrate when compared to its activity toward tetrapeptide substrates (>10-fold reduction in TTN and 102-fold reduction in catalytic efficiency, SI Figure 55), the minimal levels of hydrolysis highlight tolerance for pentapeptides in addition to tetrapeptides.
Rationalization of WP516’s Substrate Tolerance: Peptide Modeling
The distinct tetrapeptide substrate scope of WP516 compared to Ulm16 prompted us to investigate the basis of this unique substrate tolerance. Structurally, PBP-TEs have been shown to display two domains, a PBP domain containing an α-β-hydrolase fold and a lipocalin-like domain containing an eight-strand antiparallel β-barrel fold, connected by an unstructured loop. Previously, the crystal structure of Ulm16 (PDB: 8FEK) revealed that, while the PBP domains of Ulm16 and SurE (PDB: 6KSU and 6KSV) are highly similar, the orientation of the lipocalin domains differed between the two enzymes, suggesting that the lipocalin domain may play an important role in the enzymes ability to cyclize shorter peptides.37 Docking and subsequent mutagenesis studies with Ulm16 previously demonstrated that the lipocalin domain does play a key role in cyclization, with residue Arg431(Arg 438 of WP516) on the lipocalin domain being essential to cyclization.37 To investigate if a similar trend of lipocalin domain differences holds true for WP516, we attempted to solve its crystal structure. Unfortunately, crystallization efforts were unsuccessful due to insufficient protein solubility, prompting us to instead generate and compare its AlphaFold342 model to the crystal structures of SurE and Ulm16. Consistent with previous structural comparisons among this family of proteins, the α/β-hydrolase domains of Ulm16, SurE and WP516 are highly similar (Figure 5, SI Figure 56), sharing approximately 47% sequence identity and an RMSD of approximately 0.950 Å with each other (SI Figure 57). Interestingly, in the 70 amino acids (N to C terminus) leading into the unstructured loop between the two domains, the proteins exhibit close to 60% sequence identity, followed by a conserved XDVG motif (SI Figure 58A and C). However, after this motif, the proteins lose nearly all sequence identity, despite all ending all in a lipocalin domain (SI Figure 58 B and D). Of note, WP516 and Ulm16 are predicted to have similar lipocalin domain angles, yet only share 25% sequence identity. Looking at the lipocalin domain density between Ulm16’s crystal structure and the WP516 Alpha Fold 3 model revealed that WP516 has increased steric bulk from the addition of multiple Trp residues (W374, W394), resulting in a much shallower lipocalin domain, which possibly forces shorter peptides to turn sooner (SI Figure 59).
Figure 5. WP516 Alpha Fold 3 model comparison with Ulm16 crystal structure (PDB: 8FEK).
highlighting the similarity between there α/β-hydrolase domain and lipocalin domain angle. RMSD 1.023
To further investigate the substrate tolerance and cyclization mechanism of WP516, we conducted covalent docking studies with its AlphaFold model. We began by docking peptide 5 to gain insight into unique ability of WP516 to cyclize peptides with a 2-position Dap and DLDL stereochemistry. Consistent with previous observations that linked variability in the docking outputs with hydrolysis,37 the top outputs when docking 5 with WP516 were highly uniform, with many taking a cyclic shape, while docking 5 with Ulm16 led to a peptide lacking uniformity and not taking a cyclic shape (SI Figure 60). Comparing the lowest MMGBSA scoring outputs for each protein (Figure 6 A and B) revealed that the (L372, L386, P387, Y428, S429, R431) Ulm16 pocket occupied by the 3 position Phe side chain contains two amino acid changes in WP516 (L372/ W374 Ulm16/WP516 and Y428/I435 Ulm16/WP516), forcing the 3rd amino acid to bind in a different lipocalin pocket (L345, W394, M415, Y418, and T436). Of note, L345 is from the unstructured loop and is positioned around the lipocalin domain due to a 5 amino acid insertion in WP516 that is not observed in SurE or Ulm16. M415 is part of a 5 amino acid insertion in the lipocalin domain that is likewise not found in Ulm16 (SI Figure 58B). This change in binding pocket might account for the ability of the enzyme to favorably cyclize multiple different tetrapeptides regardless of stereochemistry and amino acid at the 3rd position. We observed the conserved arginine in the lipocalin domain that was previously shown to be crucial for cyclization by Ulm16 (Arg438 in Ulm16; Arg431 in WP516), to be within hydrogen-bonding distance to two backbone carbonyls of the substrate. Both the side chain and amide N-H of Thr295 were also observed to be in hydrogen bonding distance of the substrate backbone. Interestingly, we found the orientation of the substrates in the active site to be flipped compared to what has been observed when docking tetrapeptides with Ulm16. However, this orientation was observed when Ulm16 was docked with a hexapeptide substrate bearing a C-Terminal Glycine (peptide 40 in this paper) which was cyclized without hydrolysis.37 Docking that same peptide into WP516 resulted in a mix of outputs with the peptide appearing to exit the lipocalin domain (SI Figure 61). This is consistent with the observation that this peptide was only hydrolyzed by WP516, likely due to the increased density of the lipocalin domain preventing the peptide from accessing the position and conformation necessary for cyclization. We do not believe that this was due to the C-Terminal glycine as tetrapeptide 15 bearing a C-terminal glycine was easily cyclized by WP516.
Figure 6. Covalent docking of peptide 5 with.
A.) WP516; W394 of the lipocalin domain has been omitted for clarity B.) Ulm16. Residues within hydrogen bonding distance or those that play a role in the lipocalin surface are highlighted.
Discussion
The chemical synthesis of cyclic tetrapeptides (CTPs) is a persistent challenge due to their high levels of ring strain. Current methods often severely limit the peptide sequence or are incompatible with Fmoc-based solid-phase peptide synthesis (Fmoc-SPPS), thereby restricting access to a class of highly valuable scaffolds with broad and diverse biological activities of interest across various industries. To address this, we explored a biocatalytic method to improve their accessibility. While excised thioesterases and ribosomally synthesized post-translationally modified peptide (RiPP) cyclases are efficient at cyclizing large peptides43,44 and even proteins45–47, none have demonstrated the ability to efficiently produce CTPs. We sought to expand upon the previous discovery of Ulm16, the first PBP-TE capable of efficiently producing CTPs, by conducting a bioinformatics search to identify a cyclase with a broader substrate scope. However, up to this point, the literature has lacked precedence on a workflow for the discovery, expression, and validation of TE domains belonging to cryptic BGCs as biocatalysts, despite this approach yielding many novel and synthetically useful enzymes of other classes.5,48 This is due, in part, to the poor substrate promiscuity of TE domains and lack of knowledge about the reaction they catalyze based on primary amino acid sequence.27 To circumvent these issues we aimed to exploit the broad substrate promiscuity, ease of bioinformatics identification, clustering based on peptide length, and native selectivity for head-to-tail macrocyclization of PBP-TEs towards the development of a workflow capable of identifying head-to-tail tetrapeptide cyclases.29,38 In doing so, we have successfully expanded the biocatalytic toolbox of PBP-TEs by demonstrating the utility of cryptic PBP-TEs, highlighted by the discovery of WP_043619516.1 (referred to as ‘WP516’). To the best of our knowledge, WP516 represents the first instance of a PBP-TE discovered entirely through bioinformatics, and one that is not associated with a known natural product. The substrate scope of WP516 surpassed all previously reported PBP-TEs with respect to tetrapeptides, favoring cyclization over hydrolysis for all stereochemistries, as long as heterochirality is maintained at the C- and N-termini. Interestingly, WP516 is one of the few PBP-TEs capable of utilizing a C-terminal glycine, accepting a GLDL substrate. The only other examples are Ulm16 and WolJ, both of which catalyze the cyclization of desotamides—a substrate WP516 does not accept. Additionally, WP516 tolerates a wide range of amino acid substitutions at the 2nd and 3rd positions, beyond what Ulm16 has previously been shown to accept.
To rationalize the broad tetrapeptide substrate scope of WP516, we performed covalent docking studies on its AlphaFold model with two substrates that displayed varying degrees of cyclization efficiency. Despite the lipocalin domain angle being highly similar between Ulm16 and WP516, we observed that the steric bulk imposed by multiple residue substitutions in the lipocalin domain of WP516 likely forces the peptide to turn earlier, facilitating cyclization, thereby increasing its tetrapeptide substrate promiscuity. Similar observations have been made with models of SurE, which can cyclize peptides as small as 5 amino acids but has a more limited substrate scope compared to 8-amino-acid peptides that are predicted to preorganize into a cyclic conformation.49 Additionally, Ulm16L300G, which possesses an active site mimicking that of WP516 more closely, shows a slight preference for cyclization over hydrolysis of DLDL tetrapeptides, further highlighting how subtle changes in the α/β-hydrolase domain can influence an enzyme’s ability to cyclize peptides with differing stereochemistries.37 Taken together, our covalent docking and AlphaFold modeling studies suggest a molecular basis for WP516’s ability to cyclize tetrapeptides with a broader substrate range than Ulm16. To date, Ulm16 is the only other enzyme known to catalyze CTP formation. More generally, the workflow described here is applicable to other PBP-TEs and will facilitate the discovery of additional PBP-TE biocatalysts for peptides of various lengths and sequences. These findings contribute to our understanding of enzymatic mechanisms in peptide cyclization and provide a foundation for developing novel biocatalytic methods for highly efficient CTP cyclization.
Supplementary Material
Acknowledgements
We wish to thank Prof. Chittaranjan Das (Purdue University) and his students Rishi Patel and Gunaratne (Jayamini) Dhanapala for attempts to obtain a crystal structure of WP516, and Dr. César Aguilar for his help with bioinformatics. This work was funded by the National Institutes of Health (grant no. 1R35GM138002 to E.I.P.). Z.L.B. acknowledges the National Science Foundation for support under the Graduate Research Fellowship Program under grant no. DGE-1842166. We acknowledge the support from the Purdue Center for Cancer Research, NIH grant no. P30 CA023168.
Footnotes
Ethics declarations
The authors declare no competing interests
Supplementary Information
Materials and methods, along with supplementary figures, tables, NMRs, and mass spectra can be found in the Supplementary Information.
Data Availability
Detailed experimental procedures, characterization data, NMR spectra of compounds, detailed computational results and calculated structures are available within the Supplementary Information. Protein structures used in this paper are available at the PDB (SurE: 6KSU and Ulm16: 8FEK). Protein sequences used in this study are from the NCBI database: Ulm16 (accession ATU31793.1), SurE (BBZ90014.1), FlkO (AGI87381.1), WP516 (WP_04319516.1), WP_031183424.1, and SEC28301.1.
References
- (1).Wang L.; Wang N.; Zhang W.; Cheng X.; Yan Z.; Shao G.; Wang X.; Wang R.; Fu C. Therapeutic Peptides: Current Applications and Future Directions. Signal Transduct. Target. Ther. 2022, 7 (1), 48. 10.1038/s41392-022-00904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zorzi A.; Deyle K.; Heinis C. Cyclic Peptide Therapeutics: Past, Present and Future. Curr. Opin. Chem. Biol. 2017, 38, 24–29. 10.1016/j.cbpa.2017.02.006. [DOI] [PubMed] [Google Scholar]
- (3).Tsomaia N. Peptide Therapeutics: Targeting the Undruggable Space. Eur. J. Med. Chem. 2015, 94, 459–470. 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- (4).Ribeiro R.; Pinto E.; Fernandes C.; Sousa E. Marine Cyclic Peptides: Antimicrobial Activity and Synthetic Strategies. Mar. Drugs 2022, 20 (6), 397. 10.3390/md20060397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Scott T. A.; Piel J. The Hidden Enzymology of Bacterial Natural Product Biosynthesis. Nat. Rev. Chem. 2019, 3 (7), 404–425. 10.1038/s41570-019-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Malde A. K.; Hill T. A.; Iyer A.; Fairlie D. P. Crystal Structures of Protein-Bound Cyclic Peptides. Chem. Rev. 2019, 119 (17), 9861–9914. 10.1021/acs.chemrev.8b00807. [DOI] [PubMed] [Google Scholar]
- (7).Morrison C. Constrained Peptides' Time to Shine? Nat. Rev. Drug Discov. 2018, 17 (8), 531–533. 10.1038/nrd.2018.125. [DOI] [PubMed] [Google Scholar]
- (8).White C. J.; Yudin A. K. Contemporary Strategies for Peptide Macrocyclization. Nat. Chem. 2011, 3 (7), 509–524. 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- (9).Sarojini V.; Cameron A. J.; Varnava K. G.; Denny W. A.; Sanjayan G. Cyclic Tetrapeptides from Nature and Design: A Review of Synthetic Methodologies, Structure, and Function. Chem. Rev. 2019, 119 (17), 10318–10359. 10.1021/acs.chemrev.8b00737. [DOI] [PubMed] [Google Scholar]
- (10).Nielsen D. S.; Shepherd N. E.; Xu W.; Lucke A. J.; Stoermer M. J.; Fairlie D. P. Orally Absorbed Cyclic Peptides. Chem. Rev. 2017, 117 (12), 8094–8128. 10.1021/acs.chemrev.6b00838. [DOI] [PubMed] [Google Scholar]
- (11).Du L.; Risinger A. L.; King J. B.; Powell D. R.; Cichewicz R. H. A Potent HDAC Inhibitor, 1-Alaninechlamydocin, from a Tolypocladium Sp. Induces G2/M Cell Cycle Arrest and Apoptosis in MIA PaCa-2 Cells. J. Nat. Prod. 2014, 77 (7), 1753–1757. 10.1021/np500387h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chloroplast coupling factor 1: A species-specific receptor for tentoxin. 10.1073/pnas.737.2245. [DOI] [PMC free article] [PubMed]
- (13).Isolation and Structural Elucidation of Cyclic Tetrapeptides from Onychocola sclerotica. 10.1021/np3000987. [DOI] [PubMed]
- (14).CJ-15, 208, a Novel Kappa Opioid Receptor Antagonist from a Fungus, Ctenomyces serratus ATCC15502. https://www.jstage.jst.go.jp/article/antibiotics1968/55/10/55_10_847/_article (accessed 2023-08-24). [DOI] [PubMed]
- (15).Wong C. T. T.; Lam H. Y.; Song T.; Chen G.; Li X. Synthesis of Constrained Head-to-Tail Cyclic Tetrapeptides by an Imine-Induced Ring-Closing/Contraction Strategy. Angew. Chem. Int. Ed. 2013, 52 (39), 10212–10215. 10.1002/anie.201304773. [DOI] [PubMed] [Google Scholar]
- (16).Alcaro M. C.; Sabatino G.; Uziel J.; Chelli M.; Ginanneschi M.; Rovero P.; Papini A. M. On-Resin Head-to-Tail Cyclization of Cyclotetrapeptides: Optimization of Crucial Parameters. J. Pept. Sci. 2004, 10 (4), 218–228. 10.1002/psc.512. [DOI] [PubMed] [Google Scholar]
- (17).Skropeta D.; Jolliffe K. A.; Turner P. Pseudoprolines as Removable Turn Inducers: Tools for the Cyclization of Small Peptides. J. Org. Chem. 2004, 69 (25), 8804–8809. 10.1021/jo0484732. [DOI] [PubMed] [Google Scholar]
- (18).Vidović N.; Horvat G.; Riva D.; Rinkovec T.; Cindro N.; Tomišić V.; Speranza G. Chloride-Assisted Peptide Macrocyclization. Org. Lett. 2020, 22 (6), 2129–2134. 10.1021/acs.orglett.0c00036. [DOI] [PubMed] [Google Scholar]
- (19).Liu D.; Rubin G. M.; Dhakal D.; Chen M.; Ding Y. Biocatalytic Synthesis of Peptidic Natural Products and Related Analogues. iScience 2021, 24 (5), 102512. 10.1016/j.isci.2021.102512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Li Y.-H.; Han W.-J.; Gui X.-W.; Wei T.; Tang S.-Y.; Jin J.-M. Putative Nonribosomal Peptide Synthetase and Cytochrome P450 Genes Responsible for Tentoxin Biosynthesis in Alternaria Alternata ZJ33. Toxins 2016, 8 (8), 234. 10.3390/toxins8080234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Witte T. E.; Villeneuve N.; Boddy C. N.; Overy D. P. Accessory Chromosome-Acquired Secondary Metabolism in Plant Pathogenic Fungi: The Evolution of Biotrophs Into Host-Specific Pathogens. Front. Microbiol. 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Xu H.-M.; Yi H.; Zhou W.-B.; He W.-J.; Zeng G.-Z.; Xu W.-Y.; Tan N.-H. Tataricins A and B, Two Novel Cyclotetrapeptides from Aster Tataricus, and Their Absolute Configuration Assignment. Tetrahedron Lett. 2013, 54 (11), 1380–1383. 10.1016/j.tetlet.2012.12.111. [DOI] [Google Scholar]
- (23).Ma G.-L.; Candra H.; Pang L. M.; Xiong J.; Ding Y.; Tran H. T.; Low Z. J.; Ye H.; Liu M.; Zheng J.; Fang M.; Cao B.; Liang Z.-X. Biosynthesis of Tasikamides via Pathway Coupling and Diazonium-Mediated Hydrazone Formation. J. Am. Chem. Soc. 2022, 144 (4), 1622–1633. 10.1021/jacs.1c10369. [DOI] [PubMed] [Google Scholar]
- (24).Gao X.; Haynes S. W.; Ames B. D.; Wang P.; Vien L. P.; Walsh C. T.; Tang Y. Cyclization of Fungal Nonribosomal Peptides by a Terminal Condensation-like Domain. Nat. Chem. Biol. 2012, 8 (10), 823–830. 10.1038/nchembio.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kohli R. M.; Walsh C. T.; Burkart M. D. Biomimetic Synthesis and Optimization of Cyclic Peptide Antibiotics. Nature 2002, 418 (6898), 658–661. 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- (26).Trauger J. W.; Kohli R. M.; Mootz H. D.; Marahiel M. A.; Walsh C. T. Peptide Cyclization Catalysed by the Thioesterase Domain of Tyrocidine Synthetase. Nature 2000, 407 (6801), 215–218. 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- (27).Horsman M. E.; Hari T. P. A.; Boddy C. N. Polyketide Synthase and Non-Ribosomal Peptide Synthetase Thioesterase Selectivity: Logic Gate or a Victim of Fate? Nat. Prod. Rep. 2016, 33 (2), 183–202. 10.1039/C4NP00148F. [DOI] [PubMed] [Google Scholar]
- (28).Hoyer K. M.; Mahlert C.; Marahiel M. A. The Iterative Gramicidin S Thioesterase Catalyzes Peptide Ligation and Cyclization. Chem. Biol. 2007, 14 (1), 13–22. 10.1016/j.chembiol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Matsuda K.; Wakimoto T. Penicillin-Binding Protein-Type Thioesterases: An Emerging Family of Non-Ribosomal Peptide Cyclases with Biocatalytic Potentials. Curr. Opin. Chem. Biol. 2024, 80, 102465. 10.1016/j.cbpa.2024.102465. [DOI] [PubMed] [Google Scholar]
- (30).Kuranaga T.; Matsuda K.; Sano A.; Kobayashi M.; Ninomiya A.; Takada K.; Matsunaga S.; Wakimoto T. Total Synthesis of the Nonribosomal Peptide Surugamide B and Identification of a New Offloading Cyclase Family. Angew. Chem. 2018, 130 (30), 9591–9595. 10.1002/ange.201805541. [DOI] [PubMed] [Google Scholar]
- (31).Matsuda K.; Kobayashi M.; Kuranaga T.; Takada K.; Ikeda H.; Matsunaga S.; Wakimoto T. SurE Is a Trans -Acting Thioesterase Cyclizing Two Distinct Non-Ribosomal Peptides. Org. Biomol. Chem. 2019, 17 (5), 1058–1061. 10.1039/C8OB02867B. [DOI] [PubMed] [Google Scholar]
- (32).Matsuda K.; Fujita K.; Wakimoto T. PenA, a Penicillin-Binding Protein-Type Thioesterase Specialized for Small Peptide Cyclization. J. Ind. Microbiol. Biotechnol. 2021, 48 (3–4), kuab023. 10.1093/jimb/kuab023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kobayashi M.; Fujita K.; Matsuda K.; Wakimoto T. Streamlined Chemoenzymatic Synthesis of Cyclic Peptides by Non-Ribosomal Peptide Cyclases. J. Am. Chem. Soc. 2023, 145 (6), 3270–3275. 10.1021/jacs.2c11082. [DOI] [PubMed] [Google Scholar]
- (34).Fazal A.; Wheeler J.; Webb M. E.; Seipke R. F. The N-Terminal Substrate Specificity of the SurE Peptide Cyclase. Org. Biomol. Chem. 2022, 20 (36), 7232–7235. 10.1039/D2OB01061E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kobayashi M.; Onozawa N.; Matsuda K.; Wakimoto T. Chemoenzymatic Tandem Cyclization for the Facile Synthesis of Bicyclic Peptides. Commun. Chem. 2024, 7 (1), 67. 10.1038/s42004-024-01147-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Son S.; Hong Y.-S.; Jang M.; Heo K. T.; Lee B.; Jang J.-P.; Kim J.-W.; Ryoo I.-J.; Kim W.-G.; Ko S.-K.; Kim B. Y.; Jang J.-H.; Ahn J. S. Genomics-Driven Discovery of Chlorinated Cyclic Hexapeptides Ulleungmycins A and B from a Streptomyces Species. J. Nat. Prod. 2017, 80 (11), 3025–3031. 10.1021/acs.jnatprod.7b00660. [DOI] [PubMed] [Google Scholar]
- (37).Budimir Z. L.; Patel R. S.; Eggly A.; Evans C. N.; Rondon-Cordero H. M.; Adams J. J.; Das C.; Parkinson E. I. Biocatalytic Cyclization of Small Macrolactams by a Penicillin-Binding Protein-Type Thioesterase. Nat. Chem. Biol. 2024, 20 (1), 120–128. 10.1038/s41589-023-01495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hostetler M. A.; Smith C.; Nelson S.; Budimir Z.; Modi R.; Woolsey I.; Frerk A.; Baker B.; Gantt J.; Parkinson E. I. Synthetic Natural Product Inspired Cyclic Peptides. ACS Chem. Biol. 2021, 16 (11), 2604–2611. 10.1021/acschembio.1c00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Navarro-Muñoz J. C.; Selem-Mojica N.; Mullowney M. W.; Kautsar S. A.; Tryon J. H.; Parkinson E. I.; De Los Santos E. L. C.; Yeong M.; Cruz-Morales P.; Abubucker S.; Roeters A.; Lokhorst W.; Fernandez-Guerra A.; Cappelini L. T. D.; Goering A. W.; Thomson R. J.; Metcalf W. W.; Kelleher N. L.; Barona-Gomez F.; Medema M. H. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 2020, 16 (1), 60–68. 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Matsuda K.; Ichihara R.; Wakimoto T. FlkO, a Penicillin-Binding Protein-Type Thioesterase in Cyclofaulknamycin Biosynthesis. Org. Biomol. Chem. 2024, 22 (33), 6713–6717. 10.1039/D4OB00907J. [DOI] [PubMed] [Google Scholar]
- (41).Atkinson D. J.; Naysmith B. J.; Furkert D. P.; Brimble M. A. Enduracididine, a Rare Amino Acid Component of Peptide Antibiotics: Natural Products and Synthesis. Beilstein J. Org. Chem. 2016, 12, 2325–2342. 10.3762/bjoc.12.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Abramson J.; Adler J.; Dunger J.; Evans R.; Green T.; Pritzel A.; Ronneberger O.; Willmore L.; Ballard A. J.; Bambrick J.; Bodenstein S. W.; Evans D. A.; Hung C.-C.; O’Neill M.; Reiman D.; Tunyasuvunakool K.; Wu Z.; Žemgulytė A.; Arvaniti E.; Beattie C.; Bertolli O.; Bridgland A.; Cherepanov A.; Congreve M.; Cowen-Rivers A. I.; Cowie A.; Figurnov M.; Fuchs F. B.; Gladman H.; Jain R.; Khan Y. A.; Low C. M. R.; Perlin K.; Potapenko A.; Savy P.; Singh S.; Stecula A.; Thillaisundaram A.; Tong C.; Yakneen S.; Zhong E. D.; Zielinski M.; Žídek A.; Bapst V.; Kohli P.; Jaderberg M.; Hassabis D.; Jumper J. M. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630 (8016), 493–500. 10.1038/s41586-024-07487-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).McIntosh J. A.; Robertson C. R.; Agarwal V.; Nair S. K.; Bulaj G. W.; Schmidt E. W. Circular Logic: Nonribosomal Peptide-like Macrocyclization with a Ribosomal Peptide Catalyst. J. Am. Chem. Soc. 2010, 132 (44), 15499–15501. 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ludewig H.; Czekster C. M.; Oueis E.; Munday E. S.; Arshad M.; Synowsky S. A.; Bent A. F.; Naismith J. H. Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates. ACS Chem. Biol. 2018, 13 (3), 801–811. 10.1021/acschembio.8b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Nguyen G. K. T.; Qiu Y.; Cao Y.; Hemu X.; Liu C.-F.; Tam J. P. Butelase-Mediated Cyclization and Ligation of Peptides and Proteins. Nat. Protoc. 2016, 11 (10), 1977–1988. 10.1038/nprot.2016.118. [DOI] [PubMed] [Google Scholar]
- (46).Hemu X.; Qiu Y.; Nguyen G. K. T.; Tam J. P. Total Synthesis of Circular Bacteriocins by Butelase 1. J. Am. Chem. Soc. 2016, 138 (22), 6968–6971. 10.1021/jacs.6b04310. [DOI] [PubMed] [Google Scholar]
- (47).Nguyen G. K. T.; Kam A.; Loo S.; Jansson A. E.; Pan L. X.; Tam J. P. Butelase 1: A Versatile Ligase for Peptide and Protein Macrocyclization. J. Am. Chem. Soc. 2015, 137 (49), 15398–15401. 10.1021/jacs.5b11014. [DOI] [PubMed] [Google Scholar]
- (48).Xu G.; Torri D.; Cuesta-Hoyos S.; Panda D.; Yates L. R. L.; Zallot R.; Bian K.; Jia D.; Iorgu A. I.; Levy C.; Shepherd S. A.; Micklefield J. Cryptic Enzymatic Assembly of Peptides Armed with β-Lactone Warheads. Nat. Chem. Biol. 2024, 20 (10), 1371–1379. 10.1038/s41589-024-01657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Du Z.; Ma Y.; Shen Y.; Jiang X.; Zhou Y.; Shi T. Exploring the Substrate Stereoselectivity and Catalytic Mechanism of Nonribosomal Peptide Macrocyclization in Surugamides Biosynthesis. iScience 2024, 27 (2), 108876. 10.1016/j.isci.2024.108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed experimental procedures, characterization data, NMR spectra of compounds, detailed computational results and calculated structures are available within the Supplementary Information. Protein structures used in this paper are available at the PDB (SurE: 6KSU and Ulm16: 8FEK). Protein sequences used in this study are from the NCBI database: Ulm16 (accession ATU31793.1), SurE (BBZ90014.1), FlkO (AGI87381.1), WP516 (WP_04319516.1), WP_031183424.1, and SEC28301.1.