Abstract
Genetic analysis has suggested that RAD17, RAD24, MEC3, and DDC1 play similar roles in the DNA damage checkpoint control in budding yeast. These genes are required for DNA damage-induced Rad53 phosphorylation and considered to function upstream of RAD53 in the DNA damage checkpoint pathway. Here we identify Mec3 as a protein that associates with Rad17 in a two-hybrid screen and demonstrate that Rad17 and Mec3 interact physically in vivo. The amino terminus of Rad17 is required for its interaction with Mec3, and the protein encoded by the rad17-1 allele, containing a missense mutation at the amino terminus, is defective for its interaction with Mec3 in vivo. Ddc1 interacts physically and cosediments with both Rad17 and Mec3, indicating that these three proteins form a complex. On the other hand, Rad24 is not found to associate with Rad17, Mec3, and Ddc1. DDC1 overexpression can partially suppress the phenotypes of the rad24Δ mutation: sensitivity to DNA damage, defect in the DNA damage checkpoint and decrease in DNA damage-induced phosphorylation of Rad53. Taken together, our results suggest that Rad17, Mec3, and Ddc1 form a complex which functions downstream of Rad24 in the DNA damage checkpoint pathway.
Eukaryotic cells employ a number of surveillance mechanisms to help ensure the orderly progression and completion of critical events such as chromosome replication and segregation during the cell division. The mechanisms that ensure the proper ordering of cell cycle events have been termed checkpoint controls (7). When DNA replication is delayed and DNA damage occurs, checkpoint controls activate cell cycle arrest, allowing DNA replication and repair (3, 22).
In the budding yeast Saccharomyces cerevisiae, checkpoint pathways induce cell cycle arrest in G1/S or G2/M and retard S-phase progression in response to DNA damage. Other checkpoints prevent cells with incompletely replicated DNA from exiting the S phase. Genetic studies have identified a number of genes that are involved in the DNA damage checkpoint and/or the replication block checkpoint (3, 22). These include RAD9, RAD17, RAD24, MEC3, DDC1, POL2, RFC5, MEC1/ESR1, and RAD53/SPK1/MEC2/SAD1. Among these genes, RAD9, RAD17, RAD24, MEC3, and DDC1 are involved not only in the G2/M-phase but also in the G1- and S-phase DNA damage checkpoints (12, 13, 21, 28–30, 40–42). POL2 (17, 18) and RFC5 (33, 35) are required for the checkpoints responding to replication block and DNA damage in the S phase. POL2 encodes a large subunit of DNA polymerase ɛ, and RFC5 encodes a small subunit of replication factor C (RFC). We have recently demonstrated that Rad24 interacts physically and cosediments with Rfc5, suggesting that Rad24 is an associated component of the RFC complex (27). MEC1 and RAD53 are necessary for the checkpoints operating in response to both DNA damage and incomplete DNA replication (1, 42). RAD53 encodes a dual-specificity protein kinase (32), and Mec1 belongs to the phosphatidylinositol kinase family that includes the human ATM proteins (9, 25). Rad53 is phosphorylated in response to DNA damage and DNA replication block in a MEC1-dependent manner (24, 36). DNA damage-induced Rad53 phosphorylation is also dependent on RAD9, RAD17, RAD24, RFC5, MEC3, and DDC1 (20, 27, 33, 36, 38).
The checkpoint genes RAD9 and RAD24 have been shown to affect the processing of single-stranded DNA (ssDNA), which accumulates in temperature-sensitive cdc13 mutants at the restrictive temperature (4, 15). Whereas cdc13 rad9 mutants accumulate ssDNA earlier than the wild-type cells, cdc13 rad24 mutants do not accumulate any measurable ssDNA. MEC3, RAD17, and RAD24 have been shown to belong to the same epistasis group (15). These observations have suggested that Mec3, Rad17, and Rad24 may function to promote degradation of ssDNA and Rad9 may inhibit the degradation. Consistent with this model, RAD17 encodes a protein with homology to a known 3′-5′ exonuclease encoded by the Ustilago maydis REC1 gene (15, 30, 37). DDC1 belongs to the MEC3 epistasis group and has a role in DNA damage checkpoints very similar to that of MEC3, RAD17, and RAD24 (13). Ddc1 is phosphorylated periodically during the cell cycle and hyperphosphorylated in response to DNA damage. Ddc1 phosphorylation depends on MEC3 function and DDC1 overexpression partially compensates the checkpoint defects of mec3 mutants, suggesting that Ddc1 may act downstream of Mec3 (13). Thus, Ddc1, Mec3, Rad17, and Rad24 appear to function in the same DNA damage checkpoint pathway, although how they interact in the pathway is obscure.
In this report, we describe the isolation of MEC3 on the basis of its interaction with Rad17 in the yeast two-hybrid system. We show that the amino-terminal region of Rad17 is required for its association with Mec3. We also present biochemical evidence showing that Rad17 and Mec3 form a complex with Ddc1 but not with Rad24. DDC1 overexpression partially suppresses the sensitivity of rad17Δ, mec3Δ, and rad24Δ mutants to DNA damage. These results suggest that the Rad17-Mec3-Ddc1 complex functions downstream of Rad24 in the DNA damage checkpoint pathway.
MATERIALS AND METHODS
Strains, media, and general methods.
Yeast strains used in this study (Table 1) are isogenic. DNA was manipulated by standard procedures (23). Standard genetic techniques were used for manipulating yeast strains (8). Synthetic complete (SC) medium containing 0.5% Casamino Acids and the appropriate supplements was used to maintain selection of TRP1 and URA3 plasmids. Galactose medium contained 2% galactose and 0.2% sucrose (for DDC1 overexpression) or 2% galactose and 2% sucrose (for RAD24 overexpression).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| KSC006 | MATa ade1 his2 trp1 ura3 leu2 |
| KSC973 | MATa rad17Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC975 | MATa mec3Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC980 | MATa rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1081 | MATa ddc1Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1085 | MATa rad17Δ::LEU2 mec3Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1086 | MATa rad17Δ::LEU2 ddc1Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1087 | MATa mec3Δ::LEU2 ddc1Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1088 | MATa rad17Δ::LEU2 rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1089 | MATa mec3Δ::LEU2 rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
Plasmids and gene disruptions.
The RAD17 open reading frame was cloned into the BamHI-SalI sites of plasmid pBTM116-8D (26, 39) by PCR using pDL179 (14) as the template. The 5′ primer for pBTM116-8D expressing the full-length Rad17 (pBTM-RAD17) and pBTM116-8D expressing the amino-terminal Rad17 [pBTM-RAD17(1-179)] was KS138 (5′-CTCGGATCCATGCGAATCAACAGTGAGC-3′). The 5′ primer for pBTM116-8D expressing the carboxyl-terminal Rad17 [pBTM-RAD17(177-401)] was KS211 (5′-CTAGGATCCATGGAGTGCTATGTATATGCAAAGAC-3′). The 3′ primer for pBTM-RAD17 and pBTM-RAD17(177-401) was KS139 (5′-CTCGTCGACTTAAAAAAATATAGGAATATCCTTTGTTGGAT-3′). The 3′ primer for pBTM-RAD17(1-179) was KS210 (5′-CTAGTCGACTTAATAGCACTCTTTGCATCCGATTTC-3′). To create pBTM-RAD17(E128K), the NheI-SphI fragment of pBTM-RAD17 was replaced by a 0.8-kb NheI-SphI fragment from pWSU171GR (30) that contains the rad17-1 mutation gene. The rad17-1 allele contains a single GC-AT transition resulting in a Glu-to-Lys amino acid change at position 128 of the Rad17 protein (31).
To create the mec3 disruption plasmid pDm3L, the amino- and carboxyl-terminal regions of the MEC3/PIP3 gene in pML46 (12) were amplified by PCR with the amino-terminal primers KS123 (5′-CTCACGCGTAGGGTTTACAAGCCCTTC-3′) and KS124 (5′-CTCGAATTCTGATACTAGCGGTGATACT-3′) or the carboxyl-terminal primers KS125 (5′-CTCAAGCTTAATCTGAATACATCATGAGGA-3′) and KS126 (5′-CTCACGCGTAAATGGTTGTGAAGCACCT-3′). The mec3 disruption plasmid was constructed by a three-part ligation of the EcoRI-MluI-treated PCR-amplified amino-terminal fragment and the HindIII-MluI-treated PCR-amplified carboxyl-terminal fragment with EcoRI-HindIII-linearized YIplac128 (5). To create the ddc1 disruption plasmid pDd1L, the amino- and carboxyl-terminal regions of the DDC1 gene were amplified by PCR with the amino-terminal primers KS237 (5′-CGTGGATCCTCAATGACAGCGATTACGTACGTAAACTAT-3′) and KS238 (5′-AAAAAGCTTCTGAAACCTAACCTGACAAAGAGTAGTATCTGT-3′) or the carboxyl-terminal primers KS239 (5′-TGCAAGGTCTGTTGAATTCCCAGAATGACACAAGT-3′) and KS241 (5′-CTCGGGATCCTTTCTTGGGATTTCTGTAGGCTCC-3′). The ddc1 disruption plasmid was constructed by a three-part ligation of the BamHI-HindIII-treated PCR-amplified amino-terminal fragment and the EcoRI-BamHI-treated PCR-amplified carboxyl-terminal fragment with EcoRI-HindIII-linearized YIplac128. To disrupt RAD17, MEC3, and DDC1, plasmids pDL183 (14), pDm3L, and pDd1L were cleaved by BamHI, MluI, and BamHI, respectively. The resulting DNA fragments were transformed into a diploid strain. The disruption of each gene was confirmed by PCR. The heterozygous diploids were then sporulated, and the tetrads were dissected. The disruption of RAD24 was described previously (27).
To create YCpT-RAD17, the BamHI-XbaI fragment from pDL179 was cloned into BamHI-XbaI-linearized YCplac22 (5). To tag the RAD17 gene, the DNA fragment encoding the epitope recognized by anti-Myc antibody was inserted into a NheI restriction site in YCpT-RAD17, creating YCpT-RAD17-myc. The EcoRI-XbaI fragment of YCpT-RAD17-myc was replaced by a 1.1-kb EcoRI-XbaI fragment from pWSU171GR that contains the rad17-1 mutation gene, creating YCpT-RAD17(E128K)-myc. The DNA sequences encoding the epitope recognized by anti-hemagglutinin (HA) or Myc antibody were attached in frame to the carboxyl-terminal end of MEC3 using PCR. The carboxyl-terminal MEC3 open reading frame was amplified by PCR with the 5′ primer KS140 (5′-CTCGGATCCATGAAATTAAAATTGATAGTAAATGGT-3′) and 3′ primer KS147 (5′-CTCGGATCCCCAAGCCCTTCGATCTTGCTATAT-3′). The MEC3-HA plasmid (YCpMEC3-HA) was constructed by ligation of the EcoRI-PvuI amino-terminal fragment of the MEC3 gene from pML46, the PvuI-BamHI-treated PCR-amplified carboxyl-terminal fragment, and a BamHI-SalI fragment containing DNA sequences encoding the HA epitope with EcoRI-SalI-linearized pRS316 (2). The EcoRI-BamHI fragment from YCpMEC3-HA and a BamHI-HindIII fragment containing DNA sequences encoding four Myc epitope tags were subcloned into EcoRI-HindIII-treated YCplac22, creating YCpT-MEC3-myc. The DDC1-HA plasmid (YCpDDC1-HA) was constructed by ligation of the BamHI-PstI fragment from pML89 (13) and the PstI-HindIII fragment of pML119 (13) with BamHI-HindIII-linearized YCplac33 (5). The tagged constructs (RAD17-myc, DDC1-HA, MEC3-HA, and MEC3-myc) expressed appropriate-sized proteins from their own promoters and complemented their null mutations with regard to sensitivity to DNA-damaging agents such as methyl methanesulfonate (MMS) and UV light.
To construct GAL1 promoter-fused DDC1, a fragment encoding DDC1 was amplified by PCR using the 5′ primer KS270 (5′-CGCGGATCCATGTCATTTAAGGCAACTATCACCGAGTC-3′) and 3′ primer KS280 (5′-CGCGTCGACTTAGTCAAATATACCCCTTGGCTTTTCTAC-3′), digested with BamHI and SalI, and cloned into BamHI-SalI-treated YCpG22 and YCpG33 (34), creating YCpG22-DDC1 and YCpG33-DDC1, respectively. Both plasmids fully complemented the ddc1Δ mutation. To construct a fusion of the GAL1 promoter to RAD24, a fragment encoding RAD24 was amplified by PCR using the 5′ primer KS130 (5′-CTCAGATCTATGGATAGTACGAATTTGAAC-3′) and 3′ primer KS132 (5′-CTCGTCGACGTTAGAGTATTTCCAGGTCTGAA-3′), digested with BglII and SalI, and cloned into BamHI-SalI-treated YCpG33, creating YCpG33-RAD24. The GAL1-RAD24 construct fully complemented the rad24Δ mutation. YCp-RAD53-HA and YCpRAD24-HA were described previously (27, 33). YCpT-RAD24-myc is a TRP1-marked version of YCpRAD24-myc (27).
Two-hybrid screening.
Screening of the pACT S. cerevisiae library (a gift from S. J. Elledge) with pBTM-RAD17 was carried out as described previously (26). After transformation with the pACT library, approximately 100 colonies of L40 cells carrying pBTM-RAD17 grew on selective medium containing 40 mM 3-aminotriazole (AT). A transformation efficiency test indicated that 2 × 106 Trp+ Leu+ transformants were obtained in this screening. pACT-cDNA plasmids that retested as positive were recovered from 36 of these transformants. Restriction analysis revealed that the plasmids were divided into two groups, one containing long inserts (about 2 kb) and the other containing short inserts (about 0.5 kb). Four out of those plasmids carrying long inserts were sequenced, and restriction and sequence analyses followed by DNA database search revealed that all of the plasmids contained the full-length MEC3 gene. Hereafter, the pACT plasmid carrying MEC3 is designated pACT-MEC3.
Immunoblotting.
Protein extracts for immunoblotting were prepared and resolved by electrophoresis on sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) as previously described (33). Proteins were then transferred to nylon membranes, subjected to immunoblot analysis with the monoclonal anti-HA (3F10, 12CA5, or 16B12) or anti-Myc (9E10) antibody or a rabbit polyclonal anti-HA or anti-Myc (MBL, Japan) antibody and then detected by ECL kit (Amersham).
Immunoprecipitation.
Yeast cells were grown in SC medium appropriate to select for TRP1 and/or URA3 plasmids. Cells were then diluted in YEPD and allowed to grow for 3 h. Cells were pelleted, washed, and resuspended in lysis buffer. An equal volume of glass beads was added, and the cells were lysed by vortexing. Extracts were clarified by 15 min of centrifugation at 4°C. The supernatant was diluted with lysis buffer and incubated at 4°C for 2 h with protein A-Sepharose beads bound with anti-HA or anti-Myc antibody. Protein concentration was determined by the Bio-Rad protein assay. Immunoprecipitates were washed four times with lysis buffer, washed twice with wash buffer (20 mM HEPES-Na [pH 7.5], 10 mM MgCl2), and boiled immediately in 1× SDS-PAGE sample buffer. The proteins were detected after immunoblotting with antibodies described above.
Sucrose density gradient centrifugation.
Cells were pelleted, washed, and resuspended in lysis buffer. An equal volume of glass beads was added, and the cells were lysed by vortexing. Extracts were clarified by 15 min of centrifugation at 4°C, and 150 μl of the supernatant was separated by sucrose density gradient sedimentation (4 ml of 10 to 40% sucrose gradient in lysis buffer centrifuged in an SW60 rotor at 40,000 rpm for 12 or 24 h at 4°C). The gradients were fractionated from the top (200 μl/fraction) and subjected to immunoblotting with antibodies described above.
MMS sensitivity and synchrony experiments.
To show colony formation in the presence of MMS, cultures of cells were serially diluted, spotted on the appropriate SC galactose plates with or without MMS, and incubated at 30°C. The MMS synchrony experiment was carried out at 30°C as described elsewhere (33). Cells were grown in the SC sucrose medium appropriate to select for plasmids and then in YEP galactose for 150 min and subsequently treated with α-factor (6 μg/ml) for 120 min. Cells synchronized with α-factor were released into YEP galactose containing 0.05% MMS. Viability in the MMS synchrony experiment was determined as described elsewhere (34).
RESULTS
Rad17 physically interacts with Mec3.
We used a LexA-based two-hybrid system (39) to screen budding yeast expression library for proteins that associate with Rad17. As baits, we used the full coding sequence of RAD17 fused to the DNA-binding domain of LexA. We isolated a full-length cDNA encoding MEC3 (see Materials and Methods). In the two-hybrid system, coexpression of the Rad17 and Mec3 fusion proteins specifically gave rise to histidine-prototrophic growth (Fig. 1A).
FIG. 1.
(A) Interaction between Rad17 and Mec3 in the two-hybrid assay. Strain L40 carrying pBTM-RAD17 was transformed with pACT-MEC3 or the vector. Transformants were streaked on a SC-Trp-Leu-His plate containing 40 mM AT. (B) Rad17 domain required for the interaction with Mec3. Strain L40 carrying pACT-MEC3 was transformed with pBTM-RAD17, pBTM-RAD17(1-179), pBTM-RAD17(177-401), or pBTM-RAD17(E128K). Transformants were streaked on an SC-Trp-Leu-His plate containing 40 mM AT. Interaction with Mec3 was assessed by growth of transformants.
To examine the physical interaction between Rad17 and Mec3 in vivo, epitope-tagged RAD17 and MEC3 were coexpressed and immunoprecipitation experiments were performed. Cells were cotransformed with YCpT-RAD17-myc, YCpMEC3-HA, and the control vectors. Extracts were prepared from the transformed cells and subjected to immunoprecipitation with anti-Myc antibody. The immunoprecipitates were then probed with antibodies against the Myc and HA epitopes. When probed with anti-Myc antibody, bands corresponding to Rad17 were detected in cells transformed with YCpT-RAD17-myc. When immunoblotted with anti-HA antibody, bands corresponding to Mec3-HA were detected only in cells transformed with both YCpT-RAD17-myc and YCpMEC3-HA (Fig. 2A). Extracts were also subjected to immunoprecipitation with anti-HA antibody. The immunoprecipitates were then analyzed by immunoblotting with antibodies against the HA and Myc epitopes. When probed with anti-Myc antibody, Rad17-myc was detected only in anti-HA immunoprecipitates from extracts of cells coexpressing Rad17-Myc and Mec3-HA (Fig. 2A). These observations demonstrate that Rad17 and Mec3 physically interact in vivo.
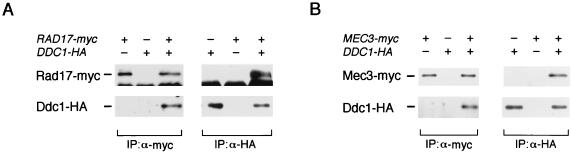
FIG. 2.
(A) Physical interaction between Rad17 and Mec3 in vivo. Extracts were prepared from rad17Δ mec3Δ (KSC1085) cells transformed with YCpT-RAD17-myc, YCpMEC3-HA, or the vectors and subjected to immunoprecipitation (IP) with anti-Myc (α-myc) or anti-HA (α-HA) antibody. The immunoprecipitates were separated by SDS-PAGE and subjected to immunoblotting analysis with anti-Myc or anti-HA antibody. (B) Failure of Rad17E128K to interact physically with Mec3. rad17Δ mec3Δ (KSC1085) cells carrying YCpMEC3-HA were transformed with YCpT-RAD17-myc or YCpT-RAD17(E128K)-myc. Extracts were prepared from the transformants and subjected to immunoprecipitation with anti-Myc antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-HA or anti-Myc antibody.
The amino terminus of Rad17 is required for its interaction with Mec3.
To define the region of Rad17 that interacts with Mec3, we constructed LexA fusions containing either the amino-terminal [Rad17(1-179)] or carboxyl-terminal [Rad17(177-401)] domain of Rad17 and used the two-hybrid system to test for their interaction with Mec3. Mec3 interacted with the amino-terminal but not the carboxyl-terminal domain of Rad17 (Fig. 1B). This result suggests that the interaction with Mec3 involves the amino terminus of Rad17.
The rad17-1 mutant allele encodes a protein containing an amino acid change in the amino-terminal part of Rad17 (Glu to Lys at position 128) (31). The rad17-1 mutation confers DNA damage sensitivity similar to that caused by the rad17 disruption (30). To examine the possibility that the protein encoded by the rad17-1 gene is defective for its interaction with Mec3, we constructed a LexA fusion of Rad17E128K and examined its interaction with Mec3 in the two-hybrid assay. As shown in Fig. 1B, the Rad17E128K fusion failed to interact with Mec3. To confirm that Rad17E128K loses the ability to interact with Mec3 in vivo, we prepared extracts from rad17Δ mec3Δ cells coexpressing Mec3-HA and Rad17-Myc or Rad17E128K-Myc, subjected them to immunoprecipitation with anti-Myc antibody, and then probed the immunoprecipitates with antibodies against the HA and Myc epitopes. We found that the wild-type Rad17 protein coimmunoprecipitated with Mec3 but the Rad17E128K mutant did not (Fig. 2B). Immunoblotting analysis showed that the rad17-1 mutation did not affect the expression level of either Mec3 or Rad17 (Fig. 2B). Thus, Rad17E128K does not interact physically with Mec3 in vivo. Taken together, these results suggest that the Rad17-Mec3 interaction requires the amino-terminal region of Rad17 and may be crucial for the DNA damage checkpoint.
Ddc1, but not Rad24, interacts physically with Rad17 and Mec3.
Genetic analyses of checkpoint genes have demonstrated that RAD17, RAD24, MEC3, and DDC1 are in the same epistasis group, as determined by the sensitivity of the double- and/or triple-disruption mutants to DNA-damaging agents (13, 15, 21). We thus addressed the possibility that the Rad17-Mec3 complex associates with Rad24 and/or Ddc1. To examine whether Ddc1 interacts with Rad17, we prepared extracts from strains expressing Rad17-Myc and Ddc1-HA, subjected them to immunoprecipitation with anti-Myc antibody, and then probed the immunoprecipitates with antibodies against the Myc and HA epitopes. When immunoblotted with anti-HA antibody, bands corresponding to Ddc1-HA were detected only in cells expressing both Rad17-Myc and Ddc1-HA (Fig. 3A). Subsequently, extracts were subjected to immunoprecipitation with anti-HA antibody and immunoblotting analysis. Rad17-Myc was detected in anti-HA immunoprecipitates from extracts of cells coexpressing Rad17-Myc and Ddc1-HA (Fig. 3A). To examine the physical interaction between Mec3 and Ddc1, we prepared extracts from cells coexpressing Mec3-myc and Ddc1-HA, subjected them to immunoprecipitation with anti-Myc or anti-HA antibody, and then probed the immunoprecipitates with antibodies against the Myc and HA epitopes. Immunoblotting analysis showed that Mec3 and Ddc1 were found to coprecipitate only in extracts prepared from cells coexpressing Mec3-myc and Ddc1-HA (Fig. 3B). These results indicate that both Rad17 and Mec3 physically interact with Ddc1. Very recently, Paciotti et al. also showed that Mec3 and Ddc1 form a stable complex in vivo (20). The interactions described above raise the possibility that Rad17, Mec3, and Ddc1 form a multiprotein complex.
FIG. 3.
Rad17 and Mec3 physically interact with Ddc1. (A) Physical interaction between Rad17 and Ddc1 in vivo. Extracts were prepared from rad17Δ ddc1Δ (KSC1086) cells carrying YCpT-RAD17-myc, YCpDDC1-HA, or the vectors and subjected to immunoprecipitation (IP) with anti-Myc (α-myc) or anti-HA (α-HA) antibody. The immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-Myc or anti-HA antibody. (B) Physical interaction between Mec3 and Ddc1 in vivo. Extracts were prepared from mec3Δ ddc1Δ (KSC1087) cells carrying YCpT-MEC3-myc, YCpDDC1-HA, or the vector and subjected to immunoprecipitation with anti-Myc or anti-HA antibody. The immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-Myc or anti-HA antibody.
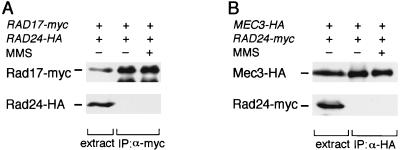
We next examined the physical interaction of Rad24 with Rad17 and Mec3. Extracts were prepared from cells expressing Rad17-Myc and Rad24-HA and subjected to immunoprecipitation with anti-myc antibody (Fig. 4A). Extracts were also prepared from cells expressing Mec3-HA and Rad24-Myc and subjected to immunoprecipitation with anti-HA antibody (Fig. 4B). The immunoprecipitates were then analyzed by immunoblotting to detect Rad24-HA or Rad24-Myc. As shown in Fig. 4, neither Rad17 nor Mec3 was found to coimmunoprecipitate with Rad24 from cells treated or not treated with MMS (Fig. 4). Similarly, Ddc1 failed to coprecipitate with Rad24 (data not shown). These results suggest that Rad24 cannot physically interact with Rad17, Mec3, or Ddc1.
FIG. 4.
Failure of Rad24 to coprecipitate with Rad17 and Mec3. (A) Interaction between Rad17 and Rad24. Extracts were prepared from rad17Δ rad24Δ (KSC1088) cells carrying YCpT-RAD17-myc and YCpRAD24-HA with (+) or without (−) MMS treatment and subjected to immunoprecipitation (IP) with anti-Myc (α-myc) antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-Myc or anti-HA antibody. (B) Interaction between Mec3 and Rad24. Extracts were prepared from mec3Δ rad24Δ (KSC1089) cells carrying YCpT-RAD24-myc and YCpMEC3-HA with (+) or without (−) MMS treatment and subjected to immunoprecipitation with anti-HA (α-HA) antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-HA or anti-Myc antibody.
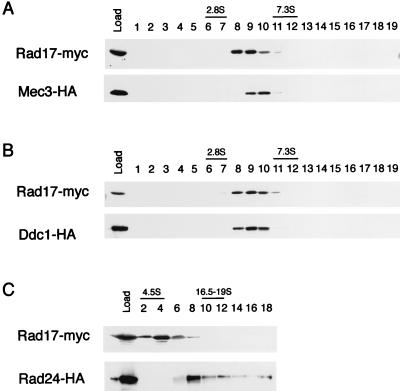
To further examine the possibility that Rad24 cannot interact physically with Rad17, Mec3, and Ddc1, extracts from strains expressing epitope-tagged proteins were fractionated by sucrose density gradient centrifugation and subjected to immunoblotting analysis. Mec3-HA and Ddc1-HA cosedimented with Rad17-Myc at 5S, while Rad24-HA sedimented separately as a 10S particle (Fig. 5). MMS treatment did not affect sedimentation of Rad17, Mec3, or Ddc1 (data not shown). These results strongly suggest that Rad17, Mec3, and Ddc1 are not associated with Rad24. Cosedimentation of Rad17, Mec3, and Ddc1 is consistent with the possibility that these three proteins form a multiprotein complex. However, mec3 disruption did not significantly alter the sedimentation profile of Rad17 (data not shown). Thus, it is not clear whether a large proportion of the proteins in the 5S fraction exists in a complex, because sedimentation analysis did not give separate fractions corresponding to the complex and free monomeric proteins.
FIG. 5.
Sedimentation of Rad17, Mec3, Ddc1, and Rad24 in sucrose density gradient centrifugation. Extracts were prepared from rad17Δ mec3Δ (KSC1085) cells carrying YCpT-RAD17-myc and YCpMEC3-HA (A), rad17Δ ddc1Δ (KSC1086) cells carrying YCpT-RAD17-myc and YCpDDC1-HA (B), or rad17Δ rad24Δ (KSC1087) cells carrying YCpT-RAD17-myc and YCpRAD24-HA (C). Extracts were separated by running in a 10 to 40% sucrose gradient for 24 h (A and B) or 12 h (C), and fractions (removed from the top of the gradient) were analyzed by immunoblotting using anti-HA and anti-Myc antibodies. DNase I (2.8S), bovine serum albumin (4.5S), immunoglobulin G (7.3S), and thyroglobulin (16.5 to 19S) were separated simultaneously in an independent gradient as markers.
Interactions among Rad17, Mec3, and Ddc1.
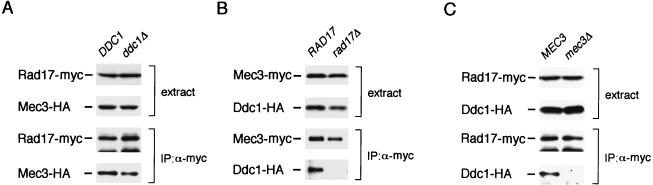
To further investigate interactions among Rad17, Mec3, and Ddc1, we performed coimmunoprecipitation using cells disrupted for each gene. To examine whether Rad17 and Mec3 form a complex in the absence of Ddc1, extracts were prepared from wild-type and ddc1Δ mutant cells carrying YCpT-RAD17-myc and YCpMEC3-HA and then subjected to immunoprecipitation with anti-Myc antibody and immunoblotting with anti-HA antibody. Mec3-HA was found to coimmunoprecipitate with Rad17-Myc in both wild-type and ddc1Δ mutant cells (Fig. 6A). Therefore, the Rad17-Mec3 interaction is not dependent on Ddc1. To examine whether Rad17 is required for the interaction between Mec3 and Ddc1, extracts were prepared from wild-type and rad17Δ mutant cells carrying YCpT-MEC3-myc and YCpDDC1-HA and subjected to immunoprecipitation with anti-Myc antibody. The immunoprecipitates were then analyzed by immunoblotting with anti-Myc or anti-HA antibody. As shown in Fig. 6B, Mec3-Myc failed to coimmunoprecipitate with Ddc1-HA in rad17Δ mutants. To test the dependence of the Rad17-Ddc1 interaction on Mec3, we subjected extracts from wild-type and mec3Δ mutant cells carrying YCpT-RAD17-myc and YCpDDC1-HA to immunoprecipitation with anti-Myc antibody and then analyzed the immunoprecipitates by immunoblotting with anti-Myc or anti-HA antibody. Ddc1-HA was not observed to coimmunoprecipitate with Rad17-Myc in mec3Δ mutants (Fig. 6C). The levels of the tagged Mec3, Rad17, and Ddc1 proteins were not changed in rad17Δ and mec3Δ mutants (Fig. 6). In contrast to the Rad17-Mec3 interaction, the Mec3-Ddc1 and Rad17-Ddc1 interactions are dependent on the presence of Rad17 and Mec3, respectively. These observations further suggest that Rad17, Mec3, and Ddc1 form a complex. It is possible that Rad17 and Mec3 form the core for the Rad17-Mec3-Ddc1 complex.
FIG. 6.
Interactions among Rad17, Mec3, and Ddc1. (A) Physical interaction between Rad17 and Mec3 in ddc1Δ mutants. Wild-type (KSC006) and ddc1Δ (KSC1081) cells were transformed with YCpT-RAD17-myc and YCpMEC3-HA. Extracts prepared from the transformants were subjected to immunoprecipitation (IP) with anti-Myc (α-myc) antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-HA or anti-Myc antibody. (B) Physical interaction between Mec3 and Ddc1 in rad17Δ mutants. Wild-type (KSC006) and rad17Δ (KSC973) cells were transformed with YCpT-MEC3-myc and YCpDDC1-HA. Extracts prepared from the transformants were subjected to immunoprecipitation with anti-Myc antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-HA or anti-Myc antibody. (C) Physical interaction between Rad17 and Ddc1 in mec3Δ mutants. Wild-type (KSC006) and mec3Δ (KSC975) cells were transformed with YCpT-RAD17-myc and YCpDDC1-HA. Extracts prepared from the transformants were subjected to immunoprecipitation with anti-Myc antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-HA or anti-Myc antibody.
Effects of DDC1 overexpression in rad24 mutants.
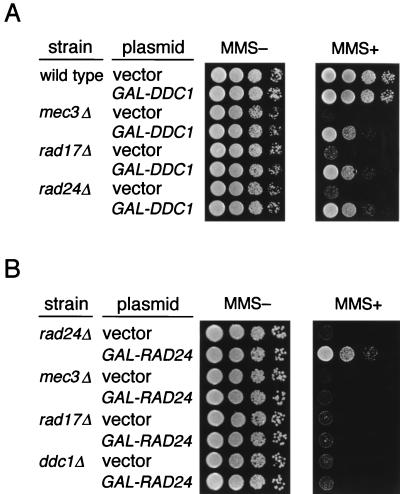
It has been shown that DDC1 overexpression from the GAL1 promoter can partially suppress the sensitivity to MMS in mec3Δ mutants (13). To explore the relationship between the Rad17-Mec3-Ddc1 complex and Rad24, we examined whether DDC1 overexpression suppresses the sensitivity to MMS in rad17Δ and rad24Δ mutants. Wild-type, mec3Δ, rad17Δ, and rad24Δ mutant cells were transformed with YCpG33-DDC1 (GAL1p-DDC1 marked with URA3) and the vector and spotted on galactose medium without or with MMS (0.01%). As previously observed for mec3Δ mutants (13), DDC1 overexpression partially suppressed the sensitivity to MMS in rad17Δ and rad24Δ mutants (Fig. 7A). On the other hand, DDC1 overexpression did not suppress the sensitivity to MMS in mec1-1 (42) and rad53 (spk1-101 [34]) mutants (data not shown). In experiments carried out to examine the effects of RAD24 overexpression in rad17Δ, mec3Δ, and ddc1Δ mutant cells, its overexpression from YCpG33-RAD24 (GAL1p-RAD24 marked with URA3) did not suppress the MMS sensitivity of the rad17Δ, mec3Δ, or ddc1Δ strains (Fig. 7B).
FIG. 7.
Effect of DDC1 or RAD24 overexpression on sensitivity to MMS in ddc1Δ, mec3Δ, rad17Δ, and rad24Δ mutants. Wild-type (KSC006), ddc1Δ (KSC1081), mec3Δ (KSC975), rad17Δ (KSC973), and rad24Δ (KSC980) cells were transformed with YCpG33-DDC1 (A), YCpG33-RAD24 (B), or the vector. Serial dilutions of cultures of the transformants were spotted on galactose-containing SC medium without or with 0.01% MMS.
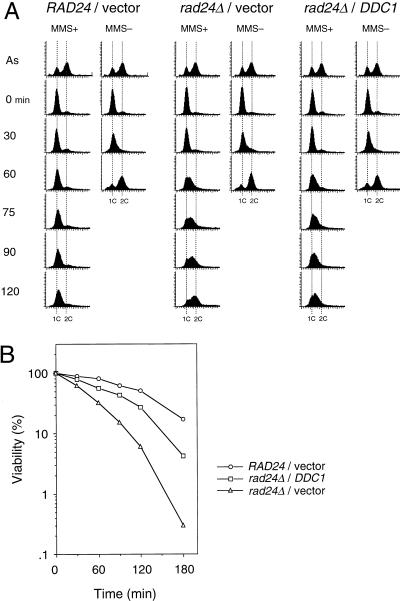
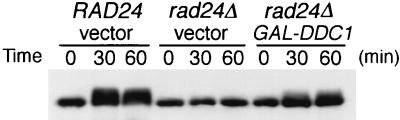
Since overexpression of DDC1 suppressed the DNA damage sensitivity in rad24Δ mutants, we examined the effect of DDC1 overexpression on the DNA damage checkpoint. Wild-type and rad24Δ mutants carrying YCpG22-DDC1 (GAL1p-DDC1 marked with TRP1) or the vector were synchronized with α-factor and then released from G1 arrest into YEP galactose medium containing MMS. As shown in Fig. 8A, rad24Δ mutants carrying YCpG22-DDC1 progressed through S phase more slowly than rad24Δ mutants carrying the control vector. Furthermore, DDC1 overexpression resulted in increased cell survival of rad24Δ mutants following MMS treatment (Fig. 8B). DDC1 overexpression did not cause any significant effect on the cell cycle kinetics or the cell cycle distribution of the cells in the absence of MMS (Fig. 8A). These results demonstrate that the rad24Δ checkpoint defect is partially restored by DDC1 overexpression. It has been shown that RAD24 and DDC1 are required for DNA damage-induced Rad53 phosphorylation (20, 27, 36, 38). The observation that overexpression of DDC1 can suppress the rad24Δ checkpoint defect raises the possibility that its overexpression would also have an effect on the Rad53 phosphorylation in rad24Δ mutants. To test this hypothesis, the phosphorylation state of Rad53 was examined in vivo by immunoblotting analysis. As previously demonstrated, rad24Δ mutants were defective for Rad53 phosphorylation in response to MMS treatment compared to the wild-type cells (Fig. 9). However, the DNA damage-induced phosphorylation of Rad53 was partially restored in rad24Δ mutants by overexpression of DDC1, as evidenced by the appearance of shifted bands corresponding to Rad53 (Fig. 9). Thus, DDC1 overexpression suppresses the defect for the S-phase DNA damage checkpoint and the DNA damage-induced Rad53 phosphorylation in rad24Δ mutants. Together with the observation that Rad17, Mec3, and Ddc1 form a complex, these results suggest that the Rad17-Mec3-Ddc1 complex may function downstream of Rad24 in the DNA damage checkpoint pathway.
FIG. 8.
Effect of DDC1 overexpression on S-phase progression of MMS-treated rad24Δ mutants. (A) RAD24 (KSC006) and rad24Δ (KSC980) cells carrying YCpG22-DDC1 or YCpG22 (vector) were synchronized in G1 by α-factor and released in YEP galactose with or without 0.05% MMS as described in Materials and Methods. Aliquots of cells were collected at the indicated times after release from α-factor treatment and examined for DNA content by flow cytometry. Dotted lines indicate the DNA content of 1C and 2C cells. The top panels represent asynchronous cells grown in galactose medium without MMS and are included as a reference. (B) The viability of cells was measured at the indicated times after release from α-factor treatment into MMS as described in Materials and Methods.
FIG. 9.
Effect of DDC1 overexpression on modification of Rad53 in rad24Δ mutants. RAD24 (KSC006) and rad24Δ (KSC980) cells carrying YCp-RAD53-HA were transformed with YCpG22-DDC1 or YCpG22 (vector). Cells were grown in YEP galactose for 4 h and incubated with 0.06% MMS for the indicated time. Cells were then subjected to immunoblotting analysis as described in Materials and Methods.
DISCUSSION
Genetic analysis has identified numerous genes required for DNA damage checkpoint in budding yeast, including RAD17, RAD24, MEC3, and DDC1. Since RAD17, RAD24, MEC3, and DDC1 belong to the same epistasis group (13, 15, 21) and are required for the DNA damage-induced Rad53 phosphorylation (20, 27, 36, 38), it has been proposed that they function upstream of RAD53 in the DNA damage checkpoint pathway. It is, however, unclear how these gene products function in the pathway. In this study, we characterized the relationships among Rad17, Rad24, Mec3, and Ddc1 in the DNA damage checkpoint.
To investigate the interactions among the DNA damage checkpoint genes, we screened for proteins that associate with Rad17 in a two-hybrid system and isolated Mec3. We showed that Rad17 and Mec3 physically interact in vivo. We also demonstrated that Ddc1 interacts physically and cosediments with Rad17 and Mec3. On the other hand, Rad24 did not interact physically with Rad17, Mec3, or Ddc1 or cosediment with Rad17. Thus, Rad17, Mec3, and Ddc1 appear to form a complex that does not comprise Rad24. To address the epistatic relationships among RAD17, MEC3, DDC1, and RAD24, we examined the effects of DDC1 overexpression in checkpoint mutants. DDC1 overexpression suppressed the DNA damage sensitivity of rad17Δ, rad24Δ, and mec3Δ mutants. Furthermore, DDC1 overexpression suppressed defects in the DNA damage checkpoint and DNA damage-induced Rad53 phosphorylation in rad24Δ mutants. Together with the observation that Rad17, Mec3, and Ddc1 form a complex, these genetic results suggest that the Rad17-Mec3-Ddc1 complex functions downstream of Rad24 in the DNA damage checkpoint pathway. Consistent with this possibility, RAD24 overexpression failed to suppress the DNA damage sensitivity of rad17Δ, mec3Δ, or ddc1Δ mutants. The finding that RAD24 overexpression fails to suppress the rad17Δ and mec3Δ mutations differs from that recently reported by Torre-Ruiz et al. (38). This discrepancy could be due to the difference of strains used in the studies. Consistent with this possibility, Lydall and Weinert (14) observed that phenotypes caused by RAD24 overexpression are affected by the strain background.
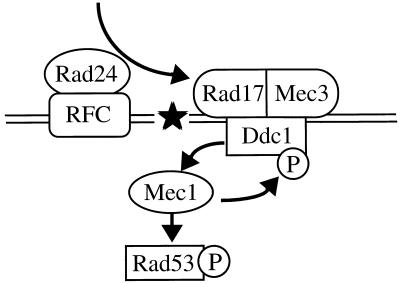
If Rad24 functions upstream of the Rad17-Mec3-Ddc1 complex in the DNA damage checkpoint pathway, one might imagine that Rad24 is required for assembly of the complex. However, the physical interactions among Rad17, Mec3, and Ddc1 are affected neither in rad24Δ mutants nor in response to DNA damage (data not shown). Therefore, Rad24 appears to regulate other properties of the Rad17-Mec3-Ddc1 complex. The RAD24 gene encodes a protein which is structurally related to subunits of the RFC complex. We have recently demonstrated that Rad24 interacts physically with RFC subunits Rfc2 and Rfc5 and cosediments with Rfc5, suggesting that Rad24 is an associated component of the RFC complex (27). Recently, it was shown that RFC is required for the activation of flap endonuclease 1 (FEN1) to excise a 5′-incised apurinic/apyrimidinic (AP) site in a proliferating cell nuclear antigen (PCNA)-dependent manner in vitro (10, 16). In this reaction, RFC and PCNA appear to recognize an AP site and load FEN1 on the AP site to effect base excision DNA repair. FEN1 carries several distinct nuclease activities on specific-structured DNA substrates. RAD17 encodes a protein homologous to U. maydis Rec1 that is shown to possess a 3′-5′ exonuclease activity (15, 30). It is possible that Rad24 is required for recruitment of the Rad17-Mec3-Ddc1 complex to damaged DNA (Fig. 10).
FIG. 10.
Model of the DNA damage checkpoint pathway in budding yeast. The Rad17-Mec3-Ddc1 complex may be recruited to DNA damage by Rad24 and may increase the Mec1 activity through an interaction between Ddc1 and Mec1. Rad24 interacts physically with RFC. Mec1 is required for phosphorylation and activation of Rad53. The phosphorylation of Ddc1 is dependent on Mec1, although the role of the Ddc1 phosphorylation is not known. The star indicates damage to DNA. See text for details.
We show that the Rad17-Mec3 interaction is dependent on the amino-terminal region of Rad17. The rad17-1 allele has a mutation within this region resulting in the expression of the protein Rad17E128K. The rad17-1 mutation confers DNA damage sensitivity indistinguishable from that of the rad17Δ mutation. To test whether the rad17-1 defect is associated with the failure of Rad17E128K to interact with Mec3, we investigated the interaction between Rad17E128K and Mec3. Two-hybrid assay and immunoprecipitation experiments showed that Rad17E128K does not interact with Mec3. These results confirm that the Rad17-Mec3 interaction is dependent on the amino terminus of Rad17 and suggest that the Rad17-Mec3 interaction is essential for the DNA damage checkpoint control. The amino terminus of the Rad17 protein shows high similarity to U. maydis Rec1. Interestingly, this homologous region of U. maydis Rec1 is essential for the exonuclease activity (19, 37). It will be interesting to see whether Rad17 possesses an exonuclease activity and whether its activity is affected by its association with Mec3.
We also examined the protein interactions among Rad17, Mec3, and Ddc1 in vivo. The Rad17-Ddc1 and Mec3-Ddc1 interactions are dependent on Mec3 and Rad17, respectively. These results further support the possibility that Rad17, Mec3, and Ddc1 form a complex. In contrast, Ddc1 is not required for the Rad17-Mec3 interaction. These results suggest that Rad17 and Mec3 may form an intermediate complex which is subsequently associated with Ddc1.
DDC1 overexpression failed to suppress the DNA damage sensitivity of mec1 and rad53 mutants, suggesting that DDC1 may function upstream of MEC1 and RAD53. This possibility is consistent with the observation that DDC1 is required for DNA damage-induced Rad53 phosphorylation. Ddc1 is hyperphosphorylated in response to DNA damage, although the function of this phosphorylation has not been elucidated. Paciotti et al. (20) have recently shown that DNA damage-induced phosphorylation of Ddc1 is abolished in mec1 but not in rad53 mutants, raising the possibility that Mec1 interacts with and phosphorylates Ddc1. Ddc1 phosphorylation is greatly reduced but not completely abolished in rad17Δ, mec3Δ, and rad24Δ mutants (20). Since Ddc1 is not assembled into the complex in rad17Δ or mec3Δ mutants, it is possible that Ddc1 alone interacts with Mec1 and becomes partially phosphorylated in response to DNA damage. DDC1 overexpression might suppress the rad17Δ, mec3Δ, and rad24Δ mutations by increasing Mec1 activity through a direct interaction between Ddc1 and Mec1 (Fig. 10).
The function of checkpoint genes may be conserved since in many cases there are related genes in eukaryotes. The S. cerevisiae RAD24 gene has a fission yeast Schizosaccharomyces pombe homolog rad17+ (6, 14). The S. cerevisiae RAD17 gene product is structurally related to Rad1 of S. pombe (15, 30). The Ddc1 amino acid sequence shows some, although weak, homology with the product of the S. pombe rad9+ gene (13). Recently, Kostrub et al. (11) showed that S. pombe Rad1 and Hus1 form a complex in a Rad9-dependent manner, suggesting that fission yeast Rad1, Hus1, and Rad9 may exist in a discrete complex. Although the budding yeast Mec3 and the fission yeast Hus1 are not structurally related, it is suggested that Rad1, Rad9, and Hus1 in fission yeast form a complex similar to the Rad17-Ddc1-Mec3 complex in budding yeast.
In summary, our data indicate that Rad17, Mec3, and Ddc1 form a complex which may participate at an early step in the DNA damage response. Our results also suggest that the Rad17-Mec3-Ddc1 complex functions downstream of Rad24 in the DNA damage checkpoint pathway. However, it remains to be determined exactly how these checkpoint proteins are involved in the DNA damage checkpoint. Future work will focus on elucidating the biochemical properties by which these proteins recognize DNA damage and activate the checkpoint pathway. These analyses should further our understanding of the checkpoint control in eukaryotes.
ACKNOWLEDGMENTS
We thank S. J. Elledge, M. P. Longhese, G. Lucchini, D. Lydall, W. Siede, and T. Weinert for materials; we thank S. Ando, T. Enoch, M. Lamphier, and T. Shimomura for helpful discussions and suggestions. We also thank M. P. Longhese, G. Lucchini, and Y. Matsumoto for helpful discussions and sharing unpublished information. K.S. is especially indebted to Kay Sullivan for encouragement and advice.
This work was supported by Grant-in-Aid for Scientific Research on Priority Areas and General Research from the Ministry of Education, Science, Sports and Culture of Japan (K.M. and K.S.).
REFERENCES
- 1.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 2.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 3.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 4.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths D J F, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17; a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:229–234. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 9.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair, and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8843. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 11.Kostrub C F, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longhese M P, Fraschini R, Plevani P, Lucchini G. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow down DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol Cell Biol. 1996;16:3235–3244. doi: 10.1128/mcb.16.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longhese M P, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;17:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 15.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto, Y. Personal communication.
- 17.Navas T A, Sanchez Y, Elledge S J. RAD9 and DNA polymerase ɛ form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- 18.Navas T A, Zhou Z, Elledge S J. DNA polymerase ɛ links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 19.Onel K, Thelen M P, Ferguson D O, Bennett R L, Holloman W K. Mutation avoidance and DNA repair proficiency in Ustilago maydis are differentially lost with progressive truncation of the REC1 gene product. Mol Cell Biol. 1995;15:5329–5338. doi: 10.1128/mcb.15.10.5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulovich A G, Toczyski D P, Harwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 25.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanajali S R, Simmons A, Clines G A, Sartiel A, Gatti R A, Chessa L, Sanal O, Lavin M F, Jaspers N G J, Taylor A M R, Arlett C F, Miki T, Weissman S M, Lovett M, Collins F S, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 26.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. TAB1: an activator of the TAK1 MAPKKK in TGFβ-signal transduction. Science. 1996;272:1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 27.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siede W, Friedberg A S, Dianova I, Friedberg E C. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agent. Genetics. 1994;138:271–281. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siede W, Friedberg A S, Friedberg E C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siede W, Nusspaumer G, Portillo V, Rodriguez R, Friedberg E C. Cloning and characterization of RAD17, a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:1669–1675. doi: 10.1093/nar/24.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siede, W., G. Nusspaumer, V. Portillo, R. Rodriguez, and E. C. Friedberg. DNA database accession no. P48581. [DOI] [PMC free article] [PubMed]
- 32.Stern D F, Zheng P, Beidler D R, Zerillo C. Spk1, a new kinase from Saccharomyces cerevisiae, phosphorylates proteins on serine, threonine, and tyrosine. Mol Cell Biol. 1991;13:3744–3755. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto K, Sakamoto Y, Takahashi O, Matsumoto K. HYS2, an essential gene required for DNA replication in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:3493–3500. doi: 10.1093/nar/23.17.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 37.Thelen M P, Onel K, Holloman W K. The REC1 gene of Ustilago maydis involved in the cellular response to DNA damage encodes an exonuclease. J Biol Chem. 1994;269:747–754. [PubMed] [Google Scholar]
- 38.Torre-Ruiz M-A, Green C M, Lowndes N F. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 40.Weinert T A, Hartwell L H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 42.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]