Abstract
Entry into S phase is dependent on the coordinated activation of CDK4,6 and CDK2 kinases. Once a cell commits to S phase, there must be a mechanism to ensure the irreversibility of this decision. The activity of these kinases is inhibited by their association with p27. In many cells, p27 plays a major role in the withdrawal from the cell cycle in response to environmental cues. Thus, it is likely that p27 is a target of the machinery required to ensure the irreversibility of S-phase entry. We have been interested in understanding the mechanisms regulating p27 at the G1/S transition. In this report, we define a cell-free degradation system which faithfully recapitulates the cell cycle phase-specific degradation of p27. We show that this reaction is dependent on active CDK2 activity, suggesting that CDK2 activity is directly required for p27 degradation. In addition to CDK2, other S-phase-specific factors are required for p27 degradation. At least some of these factors are ubiquitin and proteasome dependent. We discuss the relationships between CDK2 activity, ubiquitin-dependent, and possibly ubiquitin-independent proteasomal activities in S-phase extracts as related to p27.
Protein degradation is a key regulator of cell cycle transitions. Entry into S phase, separation of sister chromatids, and exit from mitosis are all dependent on the degradation of proteins by the proteasome (reviewed in references 26 and 30). Once a cell is committed to a transition by the action of cyclin-dependent kinases (CDKs), proteolytic events might act to ensure the irreversibility of this transition, thus maintaining order in the cell cycle.
Proteasomal ATP-dependent protein degradation is a mechanism to remove proteins that are either misfolded, not in appropriate complexes, or regulated during the cell cycle or by signal transduction pathways. Many of these proteins are targeted to the proteasome in a ubiquitin-dependent fashion (reviewed in references 10 and 30). This allows further regulation at the level of ubiquitin attachment and ubiquitin polymerization. Following ubiquitination, the protein is recognized by specific subunits on the 26S proteasome complex. There are a number of recognition proteins in 19S complexes, a subcomplex of the 26S particle (4, 45). This probably reflects the existence of multiple types of proteasomes, some nuclear and some cytosolic (1, 41, 65), as well as proteasomes with different substrate recognition properties, such as affinities for linkages through either K48 (8) or K63 (18) of ubiquitin and other potential lysines (2, 62). Ubiquitination is an ATP-dependent process. Ubiquitin is activated by the formation of a carboxyl-adenylate intermediate and conjugated to the E1 enzyme by a thioester bond. Next, it is trans-esterfied to a member of a family of ubiquitin-conjugating proteins called E2. Finally, it is transferred to a lysine residue on a target protein, either directly or with the aid of an E3 or ubiquitin-ligase complex. During the cell cycle, the specificity of the ubiquitination reaction, in both substrate choice and timing, is probably conferred by the E2 and E3 complexes (24).
The 26S proteasome is also ATP dependent. This might be due to the requirement for protein unfolding by the 19S complex (4). Thus, the ATP dependence of a proteasomal reaction is by no means a reflection of the need for ubiquitination, and protein degradation by the proteasome is not entirely dependent on ubiquitination. For example, ornithine decarboxylase is degraded by the proteasome following interaction with another factor, antizyme, neither of which has been reported to be ubiquitinated (16, 40). Furthermore, cyclin B1 degradation is initiated by a site-specific proteolytic cleavage activity which is ATP dependent and ubiquitin independent and copurifies with 26S proteasomes (59). Subsequently, the carboxyl fragment is likely to be ubiquitinated and degraded (19, 23, 58).
There are two paradigms for cell cycle-dependent proteolysis. One is exemplified by the cell cycle-dependent activity of the E3 complex. Cyclin B1 degradation is dependent on cell cycle changes in the anaphase-promoting complex (29, 58). The other regulates proteolysis by controlling the cell cycle-dependent phosphorylation of the substrate (11, 14, 21, 35, 63, 66, 67). A role for ubiquitin-dependent proteolysis at the G1/S transition in yeast has been demonstrated (reviewed in references 34 and 44). Entry into S phase requires the S-phase Clb5 and Clb6-associated kinases (50), which are held inactive by their association with the inhibitor p40sic1 (49). To enter S phase, p40sic1 has to by phosphorylated by G1-phase Cln-associated kinases (63) before being degraded (48, 61). Yeast with mutations in CLN or any of the SCFcdc4 components, CDC4, CDC34, CDC53, or SKP1, fails to degrade p40sic1 and cannot enter S phase and replicate DNA (3, 48, 49, 61, 63). cdc4 has a WD40 repeat domain which recognizes phosphorylated p40sic1 (55) and an F-box domain which interacts with the F-box domain of skp1 (3). In turn, skp1 has a binding domain to cdc53, which associates with cdc34, an E2 ubiquitin-conjugating enzyme (20, 37, 55). Phosphorylated p40sic1, through its interaction with cdc4, is recruited to this SCFcdc4 complex, where it is ubiquitinated. In vitro ubiquitination of phosphorylated p40sic1 had been established with reconstituted purified components of SCFcdc4 (17, 55).
In Xenopus, ubiquitin-dependent proteolysis is required for DNA replication. Extracts depleted of CDC34 cannot support DNA replication (68). Reconstitution of these with recombinant CDC34 leads to degradation of Xic1, a Xenopus homologue of mammalian p27kip1 (54, 57), and the ability to replicate DNA (68). This suggests that p27 might be a target of CDC34-mediated degradation in higher eukaryotes as well. Consistent with this possibility Pagano et al. have reported on the ubiquitin attachment to p27 in reticulocyte lysates supplemented with CDC34 (42). However, this system has not yet proven tractable for the identification of the enzymes and processes required for p27 degradation.
Recently, it was shown that p27 could be phosphorylated by CDK2 complexes in vitro (51), and in vivo this phosphorylation might depend on a transient association of p27 with the cyclin subunit (64). Increasing the abundance of p27 by expressing the cDNA from heterologous promoters leads to G1 arrest, which can be overcome by coexpression with cyclin E (51) or cyclin D (9, 51). Furthermore, the abundance of p27 protein often decreases at the G1/S boundary when CDK2 associated kinases become active (22, 39), although in some cell lines it does not (56). During cyclin E overexpression, the accumulation of p27 is reduced at the G1/S boundary (51). During cyclin D1 overexpression, the cell enters S phase but p27 is not degraded (9). Together, these data suggest that cyclin E and CDK2 might directly phosphorylate p27 at the G1/S transition, which may target p27 for ubiquitin-dependent proteolysis, analogous to how yeast CLN kinases phosphorylate p40sic1, targeting it for ubiquitination and subsequent proteolysis.
We have sought to define a cell-free degradation system that is cell cycle phase dependent and mimics the functions of CDK activity postulated for degradation. We hope to use this system eventually to entirely reconstitute the cell cycle phase dependence of p27 degradation. As a step toward this goal, we now present evidence demonstrating the relationship between CDK activity, the proteasome, and degradation of p27. We have shown that S-phase extracts are capable of degrading p27 in a CDK2-dependent manner and that mutation of threonine 187 to alanine can prevent this degradation. Furthermore, G1 extracts are incapable of degrading p27 even when supplemented with purified CDK2 kinases and are not able to inhibit degradation by S-phase extracts. This suggests that there are proteins in addition to the CDKs that are directly involved in the degradation pathway and might be regulated in a cell cycle phase-dependent manner. When S-phase extracts were depleted of p27T187A binding proteins and supplemented with CDK2 activity, the extracts could not degrade p27. Thus, we have established that CDK2 kinase can activate p27 degradation in a posttranscriptional manner in S-phase extracts and that degradation of p27 also requires S-phase-specific p27 binding proteins. Furthermore, p27 degradation was blocked in S extracts in the presence of K48R ubiquitin and in S extracts depleted of either ubiquitin binding proteins or the proteasome. These data suggested that p27 degradation was dependent on ubiquitin and proteasomal activity. In addition, we could detect a small amount of slower-migrating p27 following incubation of excess probe with extracts supplemented with K48R-ubiquitin and LLnL, an inhibitor of the proteasome. These forms were cell cycle phase dependent, being detected in S-phase extracts and not in G1 extracts, and could not be detected on the T187A mutant, which is not degraded in S phase. We discuss our findings in light of our current understanding of the cell cycle.
MATERIALS AND METHODS
In vitro-translated probes.
The alanine-substituted mutant, p27T187A, was generated by PCR from the pCITE-p27 vector previously described (31). Oligonucleotides 5′-GTGGAGCAGGCGCCCAAGAAG-3′ and 5′-GATCAGCTAGCAATGGAAGCA-3′ were used for first-strand synthesis, and oligonucleotides 5′-CTTCTTGGGCGCCTGCTCCAC-3′ and 5′-GCCACGTTGTGAGTTGGATAG-3′ were used for second-strand synthesis, with pCITE-p27 as a template. The final product of the PCR was cut with XmaI and NcoI and subcloned directly into the pCite1 vector.
A cDNA encoding cyclin B1 was excised from pGEM4Z (46) by BamHI digestion. The digested DNA was made blunt ended with Klenow, cut with NcoI, and subcloned into NcoI- and StuI-digested pCITE1 vector.
[35S]methionine-labeled proteins were prepared by in vitro transcription with T7 polymerase and in vitro translation in nuclease-treated rabbit reticulocyte lysate as specified by the manufacturer (Promega). We calculated the amount of in vitro-translated p27 added to each reaction mixture by determining the counts per minute incorporated into trichloroacetic acid-precipitable product.
Recombinant proteins.
Human p27, the N terminus of p27 containing amino acids 1 to 86, and the C terminus of p27 containing amino acids 87 to 197, were cloned into pET21a (a gift of J. Massagué). These proteins were purified as described previously (36). Human p21 (15) was cloned into the pET21a vector, expressed in Escherichia coli, and purified on Ni-nitrilotriacetic acid with urea as a denaturant as specified by the manufacturer (Qiagen). All proteins were eluted with an imidazole gradient and dialyzed against 20 mM HEPES (pH 7.5)–50 mM KCl–1.5 mM MgCl2–1 mM dithiothreitol (DTT).
Lysates from Sf9 cells infected with baculovirus encoding cyclin E, cyclin A, CDK2, or CDK2K at a multiplicity of infection of 5 to 10 were prepared by the method of Desai et al. (12). The generation of complexes containing cyclin E and either CDK2 or CDK2K was described previously (33, 47).
Cyclin A and CDK2 were purified essentially as described previously (12). The cyclin A-CDK2 complex was purified by incubating equal molar amounts of purified cyclin A and CDK2 at 4°C overnight and then at room temperature for 1 h and applying the mixture to an ATP-agarose column. Purified cyclin A-CDK2 was incubated at 37°C for 30 min with purified cdk activating kinase (CAK; a kind gift from Robert Fisher) at an 8:1 ratio by mass in a buffer containing 3 mM ATP, 7 mM MgCl2, 0.5 mM DTT; 1 U is arbitrarily defined as 42 ng (or 483 fmol) of cyclin A-CDK2 with 5.2 ng of CAK.
Purified recombinant protein and ubiquitin (Sigma) were coupled to cyanogen bromide (CNBr)-activated Sepharose as recommended by the manufacturer. The efficiency of coupling was determined by Coomassie blue staining to be greater than 90%, which was approximately 10 mg/ml.
Cell cultures and extract preparation.
HeLa-S3 cells were grown in minimal essential medium-spinner supplemented with 10% enriched calf serum. For synchronization of cells in G1, the cells were incubated in 2 μM nocodozole (Sigma) for 12 h, thoroughly washed, and subsequently cultured in medium without nocodozole for another 5 h. For synchronization of cells in S phase, the cells were incubated in 2 mM hydroxyurea (Sigma) for 24 h, thoroughly washed, and subsequently cultured in medium without hydroxyurea for another 3 h. Extracts were prepared as described by Brandeis and Hunt (6) with minor modifications. Briefly, the cell pellet was washed twice with cold phosphate-buffered saline without magnesium or calcium and once with hypotonic buffer (20 mM HEPES [pH 7.5], 1.5 mM MgCl2, 5 mM KCl, 1 mM DTT) and resuspended in 0.75 volume of hypotonic buffer. The cell suspension was allowed to stand on ice for 30 min, and then the cells were lysed by Dounce homogenization. Subsequently, the crude cell lysate was clarified at 100,000 × g for 30 min at 4°C, and supernatants were collected, aliquoted, and stored at −80°C.
Extracts were depleted of p27 binding proteins as follows. A 1-ml volume of extract was incubated with 200 μl of affinity matrix for 45 min at 4°C with rotation. Supernatants were collected after a brief centrifugation and mixed with a fresh aliquot of matrix as above. The supernatant obtained after two sequential depletions was used in the assays described below.
To deplete the proteasome, extracts supplemented with rabbit reticulocyte lysate (RRL) were centrifuged for 6 h at 100,000 × g at 4°C and fractionated into supernatant and pellet. The pellets were subsequently resuspended in an equivalent volume of hypotonic buffer.
Degradation assay.
The degradation assay was performed essentially as described by Brandeis and Hunt (6) with some minor modifications. Unless otherwise specified for particular experiments, assay mixtures contained 200 μg of extract supplemented with an ATP-regenerating system (25 mM phosphocreatine, 10 μg of creatine kinase per ml), 1 mM ATP, and 1/15 volume of rabbit reticulolysate lysate (Promega) in a total volume of 20 μl with 0.1 μl of radiolabelled substrate. The reaction mixtures were incubated at 30°C for 2 h, and the reactions were stopped by the addition of sodium dodecyl sulfate (SDS) sample buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by autoradiography following amplification of the signal with 1 M sodium salicylate.
Addition of reagents to degradation assay mixtures.
For addition of purified CAK-activated cyclin A-CDK2, either 0.3 or 1.5 U of the activated kinase was added to extracts that had been preincubated with p27T187A for 15 min. The mixture was then incubated for another 15 min before the addition of radiolabeled p27.
Either 10 μM staurosporine (Sigma), 10 μM olomoucine (Research Biochemicals International), or apyrase (Sigma) was preincubated with extracts and the ATP-regenerating system for 15 min at room temperature before the addition of the radiolabelled substrate. N-Acetyl-l-leucinyl-l-leucinyl-l-norleucinol (LLnL; Sigma), MG132 (Calbiochem), and adenosine 5′-O-(3-thiotriphosphate) (ATPγS) (Sigma) were added directly to the degradation assay.
Ubiquitination assay.
In a total volume of 20 μl, 1 μl of radiolabelled p27 was incubated in 100 μg of G1- or S-phase extracts or hypotonic buffer supplemented with ATP regenerating system and 1 mM ATP, with or without the addition of 10 μg of K48R-ubiquitin or ubiquitin and in the presence of 100 μM LLnL, at 25°C for 30 min or for the times indicated in the figure legends. The reactions were stopped by the addition of SDS sample buffer.
Immunoprecipitation kinase assay.
One tenth of the degradation assay mixture was adjusted to 50 mM Tris (pH 7.4), 250 mM NaCl, 5 mM EDTA, and 0.5% Nonidet P-40 (NP-40 RIPA) and immunoprecipitated with 1 μg of antibody. The antibodies we used were CDK2-M2 (Santa Cruz), cyclin A-H432 (Santa Cruz), a rabbit anti-mouse antibody (Zymed), and 10 μl of a normal rabbit polyclonal serum or a rabbit polyclonal serum against cyclin E (31). Immune complexes were precipitated with protein A-Sepharose (Repligen) and washed twice with NP-40 RIPA buffer and four times with H1 kinase buffer (20 mM Tris [pH 7.4], 7.5 mM MgCl2, 1 mM dithiothreitol), and phosphorylation assays were performed as described by Koff et al. (32).
Immunoblot assays.
Immunoblot assays were performed as described previously (56). Unless specified, 50-μg portions of extracts were used. The following antibodies were used: a 1:1,000 dilution of p27-C19 (Santa Cruz), a 1:1,000 dilution of p27-N20 (Santa Cruz), a 1:1,000 dilution of CDK2-M2 (Santa Cruz), a 1:2,000 dilution of cdc34 (Transduction), and 1:1,000 dilutions each of antibodies directed to the α and β subunits of the 20S proteasome (Calbiochem).
RESULTS
The degradation of p27 varies in a cell cycle phase-specific manner.
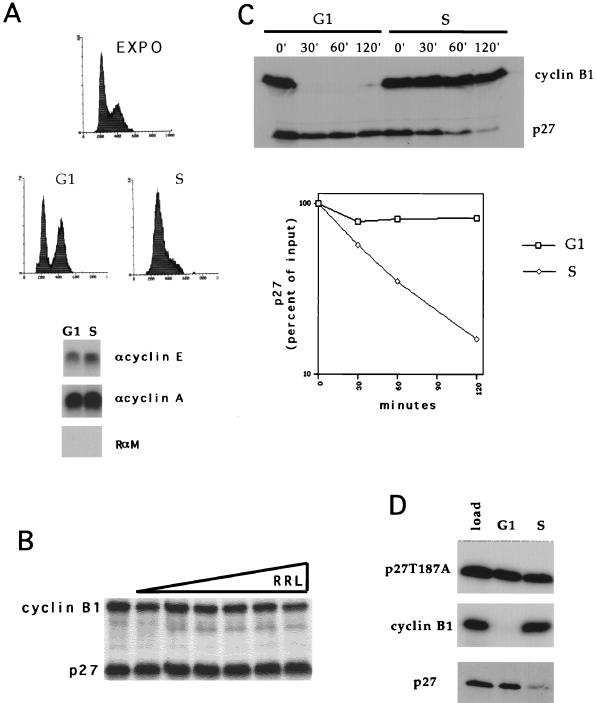
To identify proteins that regulate p27 stability during the cell cycle, we wanted to develop a cell-free system which would faithfully recapitulate the cell cycle phase-dependent changes in protein stability. To accomplish this, we either treated asynchronously growing HeLa cells with hydroxyurea for 24 h and released them from the drug for 3 h prior to harvest or treated them with nocadazole for 12 h and released them from the block for 5 h prior to harvest. This generated cells that were either 100% S phase or 60% G1 and 40% G2/M phase, respectively (Fig. 1A). Extracts were prepared from these cells and supplemented with an ATP-regenerating system and RRL. We examined the ability of these extracts to degrade tracer amounts of either cyclin B1, a protein unstable in G1 cell extract (6); p27, a protein unstable in S-phase cells (22, 39); or a mutant, p27T187A. We used only tracer amounts (0.3 fmol) of in vitro-translated protein because we did not remove endogenous proteins and suspected that stoichiometric elements in the proteolysis pathway might be limiting. RRL alone is incapable of degrading p27 under these conditions (Fig. 1B). We observed that these extracts faithfully mimic in vivo observations (Fig. 1C): p27 was degraded in S-phase extracts but not in G1 extracts, and this could be prevented by mutagenesis of the threonine at position 187 to alanine (Fig. 1D). Furthermore, degradation was dependent on elements in the C terminus, since deletion of amino acids 83 to 197 prevented degradation of the mutant proteins (data not shown). The stability of p27 in G1 extracts was not due to inactivation of proteolytic components during extract preparation, because cyclin B1 was degraded in these extracts.
FIG. 1.
Cell cycle phase-specific degradation of p27 can be recapitulated in a cell-free system. (A) Flow cytometry. Cells were treated with either nocadazole or hydroxyurea and then released to enter the G1 or S phase, respectively. The flow-cytometric profiles from these cultures are shown. DNA content and cell number are plotted on the ordinate and abscissa, respectively. Below the flow-cytometric profiles are the histone H1 kinase activities of cyclin E and cyclin A immunoprecipitates (antibodies are indicated on the right of each panel; RαM represents a rabbit-anti mouse control nonspecific antibody) from each extract (indicated at the top of each lane). (B) Cyclin B1 and p27 are stable in RRL supplemented with an ATP-regenerating system. Portions (0.3 fmol) of in vitro-translated cyclin B1 and p27 were incubated in reaction mixtures lacking extracts and containing increasing amounts of RRL (from left to right: 0.5, 1, 1.5, 2, and 2.5 μl). Neither protein was degraded under these conditions. (C) p27 is degraded in an S-phase extract-dependent manner. Portions (0.3 fmol) of each in vitro-translated cyclin B1 and p27 were combined with either 200 μg of G1-phase extract or S-phase extract, as indicated at the top of the panel, with an ATP-regenerating system at 30°C for the periods indicated above each lane. The amount of p27 at each point was determined by scanning densitometry and plotted on a semi-log scale. This experiment was repeated five times with similar results, and the autoradiogram and half-life plot are representative. (D) Mutation of threonine 187 to alanine stabilizes p27 in S-phase extracts. The in vitro-translated target proteins (indicated on the left of each autoradiogram) were added to extracts (indicated at the top of each lane), and the reaction was stopped 2 h later. p27T187A is a mutation of p27 where threonine-187 was mutated to alanine. This nomenclature is used throughout the figures.
The half-life of p27 was approximately 30 min in S phase and more than 2 h in G1 (Fig. 1C), values which were comparable with the in vivo half-life of p27 as reported by Hengst and Reed (22). Thus, the S-phase extracts faithfully recapitulate the stability and cis requirements of p27 degradation as reported in vivo.
CDK2 activity is necessary for p27 degradation in S-phase extracts.
Sheaff et al. (51) reported that threonine-187 of p27 was phosphorylated by the CDK2 kinase in vitro. Mutation of this residue to alanine resulted in G1 arrest and prevented p27 degradation when coexpressed with cyclin E (51). Because the in vitro system recapitulates the stability of p27T187A, it allows us to directly test, in a biochemical manner, the contribution of CDK2 associated kinase activity to p27 degradation postulated by the transfection studies (51). Because of the way the cells were synchronized, both G1 and S extract contained equivalent amounts of cyclin E- and cyclin A-associated histone H1 kinase activity (Fig. 1A), suggesting that CDK2 activity could not be sufficient for degradation but might be necessary.
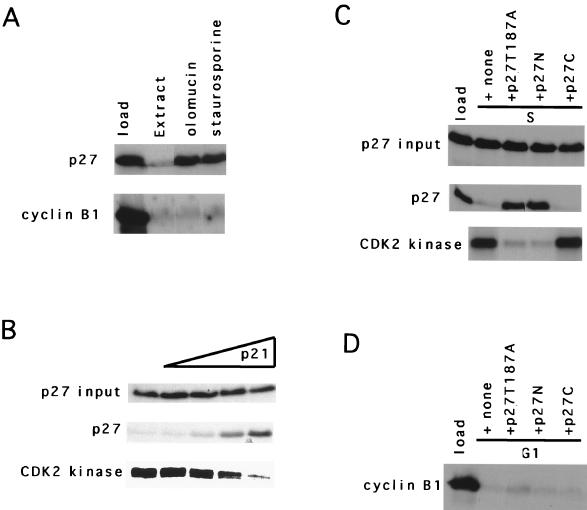
To address the requirement for CDK2 activity, we took many approaches. First, the CDK inhibitors staurosporine and olomoucine prevented degradation of p27 when added to S-phase extracts but did not affect cyclin B1 degradation when added to G1 extracts (Fig. 2A). At the concentrations used, these drugs should be inhibiting CDK2 activity and not protein kinase C, CDK4, or CDC2 activities (38); however, these chemical inhibitors might be targeting other pathways that have not yet been defined. Therefore, we explored the possibility of inhibiting CDKs by using a CDK inhibitor protein. We titrated bacterially produced p21Waf1/Cip1 into extracts and measured the effect on CDK2 kinase activity and p27 degradation. It was clear that the addition of p21 could prevent both CDK2-associated H1 kinase activity and p27 degradation (Fig. 2B). However, because p21 interacts with JNK/SAPK, at least at high stoichiometries (43, 53), we could not exclude these kinases from playing a role in degradation of p27.
FIG. 2.
Degradation of p27 could be prevented by CDK2 inhibitors. (A) Pharamacologic inhibitors of CDK2 activity. Either 10 μM olomoucin or staurosporine, as indicated above each lane, was added to S-phase extracts (p27) or G1-phase extracts (cyclin B1). (B) p21. Increasing amounts of p21 (from left to right, 1.65, 3.3, 6.6, and 10.4 pmol) were added to S-phase extracts prior to addition of in vitro-translated p27. The amounts of p27 at the beginning and end of the reaction are shown on the left. The CDK2 kinase activity was determined at the end of the reaction by immunoprecipitation kinase assays as described (32) with a C-terminus-specific CDK2 antibody. (C) Degradation of p27 correlates with CDK2 activity. This is the same experiment as described in panel B but with 788 fmol of recombinant p27 T187A, an amino-terminal fragment of p27 encompassing residues 1 to 83 (p27N), or a carboxyl-terminal fragment encompassing amino acids 89 to 197 (p27C). (D) Degradation of cyclin B1 is resistant to inhibition of CDK2 kinase activity. This is the same as panel C except that G1 extracts and 35S-cyclin B1 were used.
We also examined the effect of three mutant versions of p27 on CDK2 activity and degradation: the nondegradable p27 (p27T187A), the CDK-inhibitory amino-terminal half of p27, and the carboxyl-terminal half of p27. We observed that a 2,000-fold molar excess (relative to the tracer) of either the bacterially produced amino-terminal half of p27 or p27T187A can inhibit immunoprecipitable CDK2 activity and the degradation of p27 (Fig. 2C). However, similar amounts of the carboxy-terminal half of p27 were unable to inhibit the CDK2 kinase and the degradation of p27 (Fig. 2C). Titration of p27T187A and p27N led to a sharp transition in immunoprecipitable CDK2-associated histone H1 kinase and degradation (data not shown). Additionally, none of these proteins could prevent G1 extract-dependent degradation of cyclin B1 (Fig. 2D). This established a correlation between kinase activity and p27 degradation.
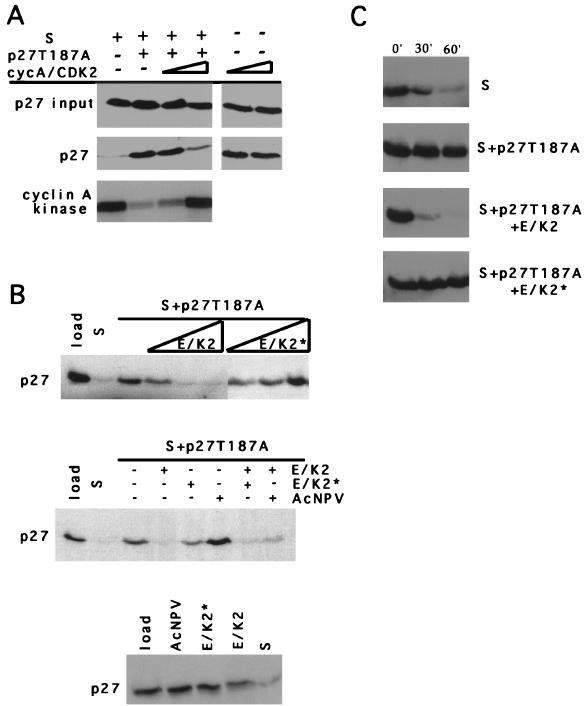
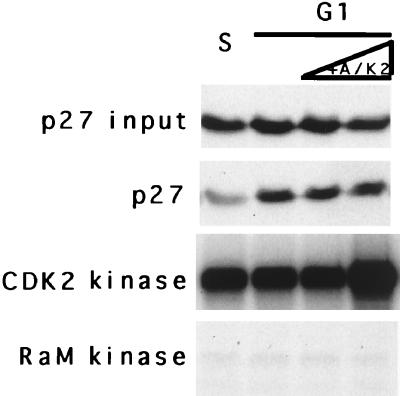
However, it was formally possible that this amount of p27T187A inhibitor might block degradation activity in a non-CDK-dependent fashion, i.e., by acting as a competitive substrate. To address this and to determine if the inhibitor acted by inhibiting CDK activity alone, we added either purified cyclin A-CDK2 (Fig. 3A), activated by recombinant CAK, or crude lysates from Sf9 cells coinfected with cyclin E- and CDK2-expressing baculovirus (Fig. 3B) to extracts preincubated with p27T187A for 15 min. This time is sufficient to inhibit all the CDK2 (data not shown). In either case, the reconstitution of CDK activity in the extract restored the ability of the extract to degrade p27, suggesting that the bacterial proteins did not act as suicide substrates but, rather, acted through another mechanism, presumably CDK2 inhibition. Furthermore, addition of cyclin A-CDK2 directly to G1 extracts did not stimulate degradation (Fig. 4), indicating that kinase is not sufficient and that S-phase extracts contain some functions in addition to CDK2 (see below).
FIG. 3.
p27 degradation is dependent on the histone H1 kinase activity of CDK2. (A) Purified cyclin A-CDK2 restores degradation activity to an S-phase extract to which p27T187A was added. As indicated at the top of the figure, S-phase extracts were preincubated for 15 mins with 788 fmol of p27T187A, and then purified CAK-activated cyclin A-CDK2 (from left to right, 144.8 and 724.5 fmol) was added. The amounts of p27 at the beginning and end of the reaction are shown (on the left). CDK2 kinase was determined at the end of the reaction by immunoprecipitation kinase assays with a cyclin A-specific antibody. The panels on the right demonstrate that the CAK-activated cyclin A-CDK2 was not contaminated with activities capable of degrading p27. (B) CDK2 catalytic activity is required for p27 degradation. In this experiment, S-phase extracts to which p27T187A was added were subsequently incubated with either cyclin E-CDK2 (E/K2) or a catalytically inactive mutant (E/K2*). The top panel shows titrations of increasing amounts of the lysate from Sf9 cells infected with the indicated viruses (from left to right, 0.1, 0.5, and 2 μl). The middle panel shows the effect of mixing 0.5 μl of the lysates indicated on the right. AcNPV represents lysates from Sf9 cells infected with parental baculovirus expressing the polyhedrin protein. The bottom panel shows the effect of mixing 0.3 fmol of in vitro-translated p27 with 5 μl of the lysates indicated above each lane. (C) Kinetic analysis of p27 degradation in reconstituted S-phase extracts. Aliquots from each reaction (indicated on the right) were collected at 30-min intervals (indicated at the top).
FIG. 4.
Supplementation of G1 extracts with cyclin A-CDK2 does not alter degradation. Extracts and addition of CAK-activated purified cyclin A-CDK2 are indicated above each lane. Increasing amounts of cyclin A-CDK2 were added to G1 extracts. At the beginning of the reaction, an aliquot was removed and the input p27 and immunoprecipitable CDK2 or rabbit anti-mouse (RaM) kinase activities were determined (left of each panel). At 2 h later, the reaction was stopped and the products were resolved on SDS-polyacrylamide gels.
To confirm that CDK2 activity was required for degradation of p27, we compared the ability of catalytically active and inactive cyclin E-CDK2 to restore the degradative ability to the extracts containing p27T187A (Fig. 3B and C). The inactivating mutation does not affect cyclin E-CDK2 complex assembly, and we normalized the amounts added by quantitative immunoblotting against cyclin E and CDK2 (data not shown). Furthermore, when we incubated p27 with a 10-fold excess of the infected Sf9 lysates and an ATP-regenerating system under identical conditions, only a modest amount of degradation could be observed, and it was specific to lysates that contain the active form of cyclin E-CDK2 (Fig. 3B, bottom). Thus, we concluded that whereas the cell lysate from cyclin E-CDK2-infected Sf9 cells might contain some activities similar to the S-phase cell extract, they are not sufficient to explain the loss of p27 when combined with the S-phase extract. We observed that the catalytically inactive mutant could not reconstitute degradation activity (Fig. 3B). Furthermore, degradation could be restored by combining the catalytically active and inactive complexes, suggesting that the failure of the inactive form to restore degradation was due to the absence of CDK2 activity (Fig. 3B). Finally, to confirm that addition of cyclin E-CDK2 in baculovirus lysates reconstituted the degradation pathway, we compared the half-life of p27 in these experiments (Fig. 3C). The half-life of p27 in extracts containing p27T187A and cyclin E-CDK2 was approximately the same as observed in S-phase extracts alone. Together, these data supports the hypothesis that CDK2 activity is directly required for degradation and is not simply as an indirect effector promoting S-phase entry.
Mechanism of p27 degradation.
Pagano et al. (42) raised the possibility that p27 was degraded by the ubiquitin-dependent proteolysis pathway. They reported that RRL supplemented with cdc34 could ubiquitinate p27. In support of this hypothesis, inhibitors of the proteasome prevented p27 degradation in HL-60 cells (39) when added to culture medium. However, the HL-60 studies failed to demonstrate the formation of ubiquitin conjugates of p27, raising the possibility that degradation in this case was not due to direct ubiquitination of p27 but, rather, was due to an indirect mechanism perhaps involving CDK association and the proteasome.
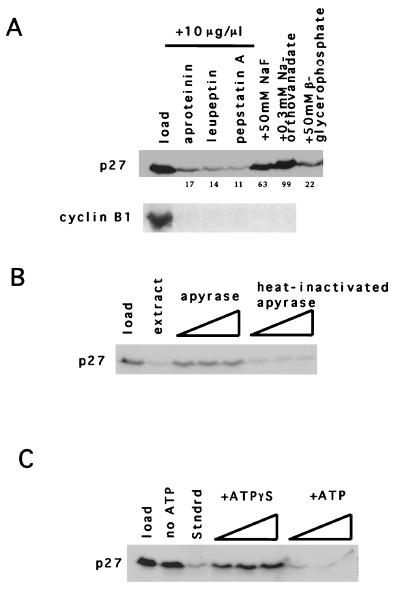
To gain further insight into the mechanism of p27 degradation, we examined the effects of various protease inhibitors on degradation in S-phase extracts. Inclusion of the microbial protease inhibitor leupeptin, aprotinin, or pepstatin A at 10 μg/μl did not prevent cyclin B1 or p27 degradation in G1- or S-phase extracts, respectively (Fig. 5A). The degradation of p27 could be inhibited by sodium fluoride and sodium orthovanadate but not by β-glycerolphosphate (Fig. 5A). Furthermore, degradation of p27 in S-phase extracts was dependent on ATP and could be prevented by either apyrase treatment (Fig. 5B) or inclusion of the ATP analogue ATPγS (Fig. 5C). Since both proteasomal degradation and CDK2 activity are ATP dependent, we next examined the sensitivity of the reaction to the proteasomal and calpain I inhibitors LLnL and MG132.
FIG. 5.
Degradation of p27 is ATPase dependent. (A) Microbial peptidase inhibitors. The inhibitors (indicated at the top of each lane) were added to S-phase extracts with in vitro-translated p27 or to G1 extracts with in vitro-translated cyclin B1. The numbers underneath the p27 autoradiogram represent the percentage of input p27 remaining after 2 h. (B) p27 degradation is sensitive to apyrase. p27 was added to S-phase extracts supplemented with an ATP-regenerating system and either increasing amounts of apyrase or heat-inactivated apyrase (0.02, 0.1, and 0.3 U). (C) ATPγS prevents p27 degradation. As in panel B, p27 was added to reaction mixtures containing either no ATP or an ATP-regenerating system with addition of increasing amounts of ATPγS (0.5, 1, and 1.75 mM).
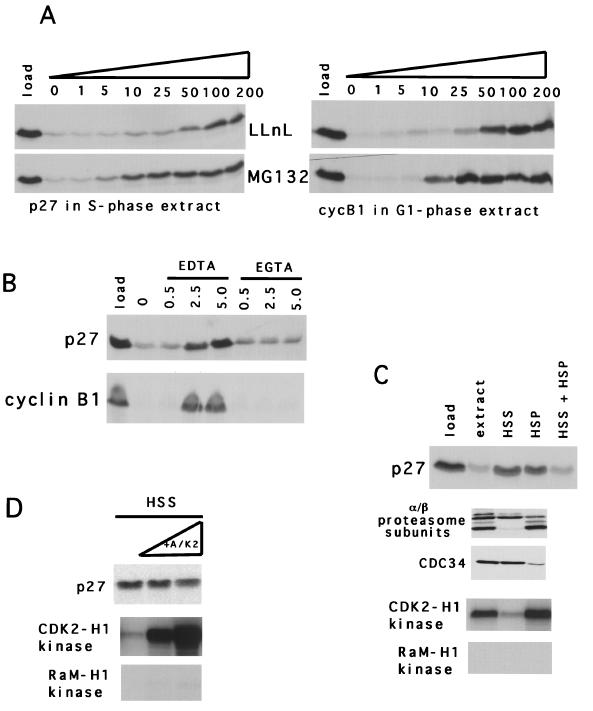
Degradation of p27 and cyclin B1 was sensitive to MG132 and LLnL, inhibitors of both calpain I and the proteasome (Fig. 6A). To discriminate between these two proteolytic pathways, we examined the sensitivity of the degradation reaction to EGTA and EDTA (Fig. 6B), since calpain I but not the proteasome is calcium dependent and thus would be inhibited by EGTA. The reaction was sensitive to EDTA but not EGTA, suggesting a non-calpain I cleavage pathway. Further evidence for a proteasome-dependent step in the degradation of p27 was suggested by examining proteolysis in crude fractions of the extract. When S-phase extracts supplemented with RRL were subjected to high-speed centrifugation, neither the pellet nor the supernatant fraction alone degraded p27 but when recombined they did (Fig. 6C). Immunoblotting the fractions showed that the α and β subunits of the proteasome were enriched in the pellet fraction that and CDC34, a putative E2 for G1/S control, was enriched in the supernatant fraction (Fig. 6C). Additionally, the CDK2-associated kinase activity largely fractionated to the pellet. Addition of CAK-activated and purified cyclin A-CDK2 to the supernatant fraction did not restore p27 degradation activity, suggesting that other factors in the pellet fraction, in addition to the CDK2 activity, were required for degradation (Fig. 6D).
FIG. 6.
Degradation of p27 is dependent on the proteasome. (A) Pharmcological inhibitors. LLnL or MG132, as indicated between the panels, was added to p27 or cyclin B1 degradation reaction mixtures in either S-phase or G1-phase extracts, respectively, as indicated below each panel, at the concentrations indicated above each lane. (B) Divalent cation requirement. EDTA or EGTA was added at the millimolar concentrations indicated above each lane to p27 or cyclin B1 degradation reaction mixtures in either S-phase or G1-phase extracts, respectively (indicated to the left of each panel). (C) High-speed fractionation of the inhibitory activity. S-phase extracts were subjected to centrifugation and fractionated into a supernatant (HSS) and pellet (HSP). These fractions, either alone or combined, were assessed for degradation activity (top). The presence of proteasome subunits and CDC34 was determined by immunoblot analysis, and the histone H1 kinase activity of anti-CDK2 or rabbit anti-mouse immunoprecipitates was determined (indicated to the left of each panel). (D) The pellet contains a component(s) other than CDK2 activity that is required for restoration of p27 degradation. The stability of p27 was determined in the supernatant (HSS) extracts supplemented with increasing amounts of purified active cyclin A-CDK2 kinase. The presence of p27 and the amount of kinase activity were determined 2 h after incubation (indicated to the left of each panel).
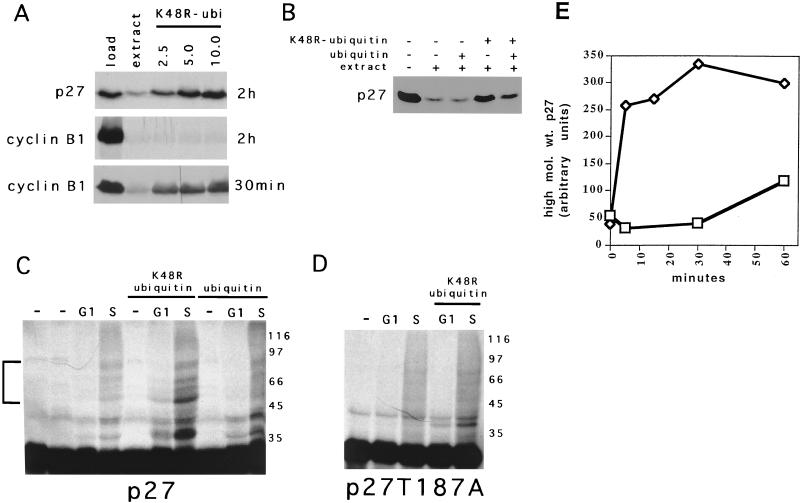
Thus, we next asked if p27 degradation was dependent on ubiquitin. To address this, we titrated a modified ubiquitin, K48R, into S-phase extracts and measured its effect on p27 degradation (Fig. 7A). We observed that this modified ubiquitin could stabilize p27 in S-phase extracts in a dose-dependent manner. Similar results were obtained with methylated ubiquitin and ubiquitin-aldehyde (data not shown). Furthermore, including unmodified ubiquitin with K48R could reverse this stabilization, consistent with the possibility that a ubiquitin-dependent event is involved in degradation of p27 (Fig. 7B). We could also block cyclin B1 degradation in G1 extracts with this mutant ubiquitin, but this reaction was reversible by continued incubation, suggesting that deubiquitinases are active in these extracts (Fig. 7A, compare the 30-min and 2-h time points).
FIG. 7.
Degradation of p27 is ubiquitin dependent. (A) A mutant ubiquitin, K48R, inhibits p27 degradation. The amount of K48R ubiquitin (in micrograms) added to lysates is indicated at the top of each lane. Its effect on cyclin B1 was determined in G1-phase extracts, and its effect on p27 was determined in S-phase extract. The time for which the reaction was incubated is indicated to the right of each panel. (B) K48R is a competitive inhibitor of degradation. Either 5 μg of K48R-ubiquitin or 10 μg of ubiquitin was added to reaction mixtures as indicated at the top of each lane. (C) Slower-migrating forms of p27 are detected in S-phase extracts supplemented with K48R-ubiquitin and LLnL. The extract indicated at the top of each lane was incubated with p27, LLnL, and the ubiquitin species indicated at the top of the gel. After 30 min, the reaction was stopped and the products were resolved by SDS-PAGE. The mobility of prestained protein molecular weight standards is indicated to the right of the gel, in thousands. (D) High-molecular-weight species cannot form on a T187A mutant. The experiment is the same as described in panel C. (E). The high-molecular-weight species forms rapidly in a cell cycle phase-dependent manner. S-phase (◊) and G1-phase (□) extracts were combined with K48R-ubiquitin, LLnL, and p27, and samples were removed at 0, 5, 15, 30, and 60 min. Following resolution on SDS-polyacrylamide gels and autoradiography, the signal in the regions marked in panel C on the left side of the gel were quantitated by scanning densitometry with an alpha-Innotech 1000 gel documentation system. The total area under the curve was then plotted on the y axis, and the time is given on the x axis.
Our ability to block p27 degradation in S-phase extracts with K48R ubiquitin raised the possibility that we could identify higher-molecular-weight conjugates of p27, presumably covalent linkages of ubiquitin to p27. To examine this, we increased the amount of target in the reaction 10-fold and added K48R-ubiquitin with LLnL. Importantly, these conditions did not inhibit the CDK2 activity of the extract (data not shown). After 30 min, we observed slower-mobility species of p27 specifically enriched in S-phase extracts and dependent on the inclusion of K48R-ubiquitin (Fig. 7C and E). These species did not form if p27T187A was used as the target (Fig. 7D). Consequently, formation of these species can be correlated with the degradation of p27, as reported in the previous experiments. Together, these data suggest that these “bands” might represent ubiquitinated precursors which are ultimately degraded in the S-phase extract. The cell cycle phase dependence of this reaction suggests that we are not simply observing the ubiquitination of a fraction of target that is either unfolded or misfolded. Therefore, we conclude that the pathway leading to p27 degradation at the G1/S boundary is regulated in a ubiquitin-dependent, proteasome-dependent manner and that at least some fraction of p27 is itself ubiquitinated.
A p27 binding protein is required for degradation in addition to cyclin-CDK2 activity.
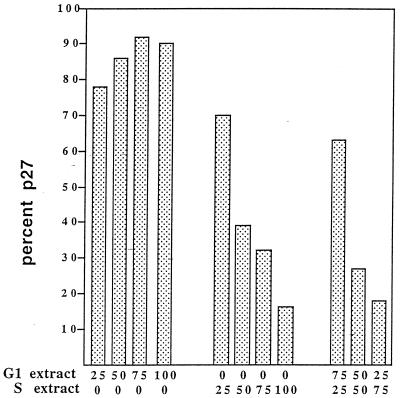
G1 extracts containing cyclin E- and cyclin A-associated kinase activity (Fig. 1A) were unable to promote p27 degradation. In addition, increasing the amount of cyclin A-CDK2 activity in these extracts by adding purified kinase did not stimulate degradation in G1 extracts, indicating that the amount of this kinase was not a limiting factor (Fig. 4). These observations suggested either that the G1 extracts lack the activities required for p27 degradation or that there was a molecular mechanism capable of suppressing these activities. Another possibility is that the S-phase extracts contain an inhibitor of a G1 inhibitor and such relationships can continue endlessly. To begin to distinguish between these possibilities, we first mixed variable amounts of G1- and S-phase extracts and examined the half-life of p27. We observed that the extent of p27 degradation was dependent on the proportion of S-phase extract in the reaction (Fig. 8). This suggests that G1 extracts probably do not contain suppressors of p27 degradation but that if they do, they must be balanced by the activating proteins in S-phase extracts.
FIG. 8.
G1 extracts lack the activities required for p27 degradation. To determine if G1 extracts contained factors that could inhibit S-phase-specific degradation, extracts were mixed at the percentages indicated and the amount of p27 was determined by scanning densitometry and plotted as percent input. This experiment was performed five times with three different sets of extracts. The data shown is representative of one experiment.
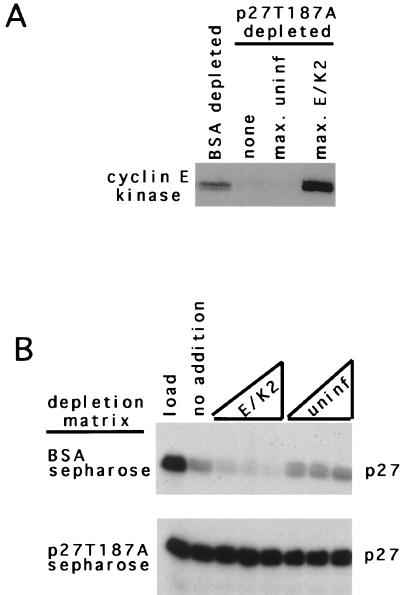
Thus, we wanted to determine if these S-phase-specific factors could interact with p27 or its associated proteins. To accomplish this, we covalently linked p27T187A or bovine serum albumin (BSA) to Sepharose beads and, following incubation with S-phase extracts, assayed the supernatants for CDK2 activity and p27 degradation activity. As expected, the extracts incubated with BSA retained CDK2 kinase activity whereas the extracts incubated with p27T187A did not (Fig. 9A). Furthermore, p27 degradation activity correlated with the presence of CDK2 activity. Extracts depleted of BSA binding proteins were capable of degradation, but those depleted of p27 binding proteins were not (Fig. 9B). We next added lysates from infected Sf9 cells expressing cyclin E-CDK2 or noninfected cells to the extracts depleted of p27T187A binding proteins and assayed them for degradation. Addition of the kinase was not capable of restoring p27 degradation activity to the S-phase extract (Fig. 9B), although there was clearly an increase in the amount of CDK2 activity in the extract (Fig. 9A).
FIG. 9.
A non-cyclin-CDK2 factor is required for p27 degradation and can be depleted from S-phase extracts. (A) Kinase assay. The immunoprecipitable cyclin E kinase activity for each condition is shown above each lane. (B) Degradation reaction. Extracts depleted of either BSA binding proteins or p27 binding proteins (indicated on the left) were incubated with 0.3 fmol of in vitro-translated p27 and either increasing amounts of lysates (from left to right, 0.1, 0.5, and 2 μl) from Sf9 cells infected with cyclin E- and CDK2-expressing baculoviruses or uninfected (uninf) lysates.
This data and the results of the experiments in Fig. 3 are consistent with the following model. When p27T187A is added to extracts, it binds both cyclin-CDK complexes and another factor, SPC (S-phase proteolytic complex). Depending on the equilibrium constants of the various partners, addition of cyclin-CDK complexes might displace SPC from p27. It is unlikely that the recombinant complex can displace the endogenous complex due to the strong binding affinity (consistent with our observation that catalytically inactive cyclin-CDK complexes could not restore degradation activity [Fig. 3B]). On the other hand, when all p27 binding proteins are depleted from extracts, addition of active cyclin-dependent kinases would not be able to support degradation, since supernatant is depleted of SPC. This is consistent with the interpretation that degradation of p27 occurs by a multimolecular process, one event being directly CDK2 dependent and the other requiring an S-phase-specific protein(s) that binds to p27.
DISCUSSION
From the phenotypes of mice deficient for each cdk inhibitor and from studies on p27-deficient oligodendrocytes (7) and luteal cells (60), it appears that p27 plays a major role in the regulation of proliferation in response to environmental signals. Thus, one way to ensure that cells would not be able to respond to antimitogenic environmental signals following commitment would be to have the same mechanism promoting S phase and eliminating p27. This would be enforced if p27 degradation was a direct consequence of CDK2-mediated phosphorylation and other mitogen-induced factors.
In this report, we demonstrate that p27 is targeted for degradation in a pathway that involves cyclin-CDK2 activity and S-phase-specific functions. Because depletion of cyclin-CDK2 activity from S-phase extracts prevents p27 degradation, its role in degradation cannot be attributed solely to its ability to promote S phase. Furthermore, it is not the presence of this complex but, rather, its activity that is required for degradation. Because catalytically inactive cyclin-CDK complexes do not stimulate degradation, it is unlikely that degradation is due to a “bystander” effect of cyclin E-CDK2 association. The observation that threonine-187 is phosphorylated by CDK2 in vitro (51) and the inability of p27T187A to be degraded in S-phase extract (as reported here) are consistent with a direct role of CDK2 phosphorylation.
How this phosphorylation occurs is not clear—does the binding of p27 to cyclin-CDK unmask the carboxyl terminus of the inhibitor, or does it increase the concentration of inhibitor around the kinase? p27 binds to both cyclin D-CDK4,6 and cyclin E-CDK2 complexes (52). In Rat1 cells, mutants of p27 deficient for cyclin E-CDK2 binding appear to be less efficiently degraded (64), and we have shown that the cyclin-CDK binding domain is not efficiently degraded (unpublished data). These data suggest that if cyclin E-CDK2-p27 complex formation is an important component of the degradation pathway, it is not sufficient for degradation in the absence of the carboxyl domains of the protein and presumably the T187 residue. Whereas T187 is phosphorylated by cyclin E-CDK2, it is not appreciably phosphorylated by cyclin D-CDK complexes (51). In addition, p27 can associate with cyclin D-CDK complexes in both inhibitory (52) and noninhibitory fashions (5, 56). The association of p27 with cyclin D1-CDK even stabilizes the protein in S phase (9). It is possible that this association does not expose the carboxyl portions of the inhibitor to the appropriate kinase or that it prevents p27 phosphorylation by cyclin E-CDK2 complexes. These observations suggest that cells can regulate p27 activity by two distinct mechanisms at the G1/S transition: they can either degrade the protein in a cyclin-CDK2-dependent manner or sequester it in a cyclin D-CDK4-dependent manner. The mechanism that is used may depend on the cell type or particular environmental signals.
It is also interesting that the G1 extracts we used have abundant CDK2 kinase activity, due to contaminating G2 cells, but do not promote p27 degradation. We also found that elutriated G1 and G2 cells lack the p27 degradation activity present in S-phase extracts (unpublished data). It is not yet clear if S-phase-specific degradation is due to cell cycle phase-specific alteration in the activity of a constitutively expressed protein or if proteins which recognize phosphorylated p27 and target it for degradation are regulated at the level of expression during the cell cycle. It is formally possible that p27 is not phosphorylated in G1 extracts, either because of an increased phosphatase activity, an inability of CDK2 to phosphorylate p27 because of interactions with other G1-specific proteins (either the cyclin-CDK2 complex or p27), or the lack of a required activity (another cell cycle-regulated factor). Our data from mixing experiments and from the depletion of p27 binding proteins suggests that there is an S-phase-specific factor that is essential for degradation of p27. Purification of this factor is required before its nature can be discussed further.
Degradation of p27 involves the proteasome. The degradation of p27 is sensitive to proteasomal inhibitors both in vivo and in S-phase extracts. In vitro, degradation is correlated with the presence of the proteasome, CDC34, CDK2 activity, and other unidentified S-phase-specific factors. Our data strongly suggests that the addition of proteasomal inhibitors to cells directly prevents p27 degradation and does not act in an indirect manner. Furthermore, we can demonstrate that the degradation reaction is sensitive to inclusion of an arginine-substituted mutant ubiquitin, K48R, which prevents side chain elongation by a subset of ubiquitin ligases (17). This data is consistent with a role for ubiquitin-dependent proteasomal degradation in the degradation of p27.
There is some suggestion that p27 might be degraded following ubiquitination. Ubiquitination of p27 is supported by a number of observations and analogies. The strongest analogy comes from the comparison of p27 with the yeast CDK inhibitors p40sic1 and Far1 and the Xenopus CDK inhibitor Xic1. p40 and Far1 degradation depends on CLN and CDC34 activity (21, 48, 49, 61, 63). Xic1 degradation requires CDK2 activity as well as the presence of CDC34 (68). The strongest evidence that p27 is ubiquitinated comes from the direct demonstration of ubiquitination in crude whole-cell extract systems supplemented with additional CDC34 and RRL (42). We have documented the formation of S-phase- and K48R-ubiquitin-dependent slower-migrating species of p27, suggesting that ubiquitin-p27 intermediates form in this system in a cell cycle-regulated manner. However, only a small amount of protein can be demonstrably ubiquitinated. This may be due to the inefficiency of the reaction, the presence of deubiquitinating enzymes, or another unidentified process. Regardless, the correlation between degradation and formation of these higher-molecular-weight species argues against any model, such as misfolded substrate, that fails to invoke some aspect of cell cycle dependence.
Ultimately, the fractionation of the S-phase extract and purification of the factors required for degradation will be the final arbiter of the relationship between CDK2 activity, proteasome activity, and ubiquitin in this process.
ACKNOWLEDGMENTS
We thank Joan Massagué (The Howard Hughes Medical Institute, MSKCC), Robert Fisher (MSKCC), and Ray Deshaies (California Institute of Technology) for providing reagents crucial to these studies. In addition, we thank Jim Roberts for critically evaluating the manuscript.
Work in our laboratory is supported by funds from the NIH (GM52597), the Memorial Sloan-Kettering Cancer Center NCI Core Grant (CA08748), the SPORE program of NCI (CA68425), and the Koch fund of CAPcure. A.K. is supported by a Pew Scholarship in Biomedical Science and an Irma T. Hirschl Scholarship and is the incumbent of the Frederick R. Adler Chair for Junior Faculty.
REFERENCES
- 1.Amsterdam A, Pitzer F, Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc Natl Acad Sci USA. 1993;90:99–103. doi: 10.1073/pnas.90.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baboshina O V, Haas A L. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2 and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271:2823–2831. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- 3.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 4.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 5.Blain S W, Montalvo E, Massagué J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 6.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 7.Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Jr, Chao M V, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavshky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 9.Cheng M, Sexl V, Sherr C J, Roussel M F. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1) Proc Natl Acad Sci USA. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 11.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Turnover of cyclin E by the ubiquitin-dependent proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 12.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diehl J A, Zindy F, Sherr C J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 16.Elias S, Bercovich B, Kahana C, Coffino P, Fischer M, Hilt W, Wolf D H, Ciechanover A. Degradation of ornithine decarboxylase by the mammalian and yeast 26S proteasome complexes requires all the components of the protease. Eur J Biochem. 1995;229:276–283. [PubMed] [Google Scholar]
- 17.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 18.Galan J-M, Haguenauer-Tsapis R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 20.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;212:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 21.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengst L, Reed S I. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 23.Hershko A, Ganoth D, Pehrson J, Palazzo R E, Cohen L H. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J Biol Chem. 1991;266:16376–16379. [PubMed] [Google Scholar]
- 24.Hilt W, Wolf D H. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 25.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 26.Hoyt M A. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 27.Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 28.Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 29.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 30.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 31.Kiyokawa H, Kineman R D, Manova-Todorova K O, Soares V C, Hoffman E S, Ono M, Khanam D, Hayday A C, Frohman L A, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27Kip1. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 32.Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, Philippe M, Roberts J M. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991;66:1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- 33.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Formation and activation of a cyclin E-cdk2 complex during G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 34.Krek W. Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 35.Lanker S, Vladivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 36.Luo Y, Hurwitz J, Massagué J. Cell-cycle inhibition by independent CDK and PCNA binding domains of p21Cip1. Nature. 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- 37.Mathias N, Johnson S L, Wilney M, Adams A E, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53 acts in concert with Cdc4 and Cdc34 to control the G1 to S phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meijer L. Chemical inhibitors of cyclin-dependent kinases. In: Meijer L, Guidet S, Tung H Y L, editors. Progress in cell cycle research. Vol. 1. New York, N.Y: Plenum Press; 1995. pp. 351–363. [DOI] [PubMed] [Google Scholar]
- 39.Millard S S, Yan J S, Nguyen H, Pagano M, Kiyokawa H, Koff A. Enhanced ribosomal association of p27Kip1 mRNA is a mechanism contributing to accumulation during growth arrest. J Biol Chem. 1997;272:7093–7098. doi: 10.1074/jbc.272.11.7093. [DOI] [PubMed] [Google Scholar]
- 40.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is a degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 41.Nederlof P M, Wang H-R, Baumeister W. Nuclear localization signals of human and Thermoplasma proteasomal subunits are functional in vitro. Proc Natl Acad Sci USA. 1995;92:12060–12064. doi: 10.1073/pnas.92.26.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 43.Patel R, Bartosch B, Blank J L. p21WAF1 is dynamically associated with JNK in human T-lymphocytes during cell cycle progression. J Cell Sci. 1998;111:2247–2255. doi: 10.1242/jcs.111.15.2247. [DOI] [PubMed] [Google Scholar]
- 44.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 45.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 46.Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 47.Polyak K, Kato J-Y, Solomon M J, Sherr C J, Massagué J, Roberts J M, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 48.Schneider B L, Yang Q H, Futcher A B. Linkage of replication to Start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 49.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40sic1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 50.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in S phase and mitotic spindle formation in S. cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 51.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 52.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 53.Shim J, Lee H, Park J, Kim H, Choi E J. A non-enzymatic p21 protein inhibitor of stress-activated protein kinases. Nature. 1996;381:804–806. doi: 10.1038/381804a0. [DOI] [PubMed] [Google Scholar]
- 54.Shou W, Dunphy W G. Cell cycle control by Xenopus p28Kix1, a developmentally regulated inhibitor of cyclin-dependent kinases. Mol Biol Cell. 1996;7:457–469. doi: 10.1091/mbc.7.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 56.Soos T J, Kiyokawa H, Yan J S, Rubin M S, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A. Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ. 1996;7:135–146. [PubMed] [Google Scholar]
- 57.Su Y, Rempel R E, Erikson E, Maller J L. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27xic1. Proc Natl Acad Sci USA. 1995;92:10187–10191. doi: 10.1073/pnas.92.22.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tokumoto T, Yamashita M, Tokumoto M, Katsu Y, Horiguchi R, Kajiura H, Nagahama Y. Initiation of cyclin B degradation by the 26S proteasome upon egg activation. J Cell Biol. 1997;138:1313–1322. doi: 10.1083/jcb.138.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tong W, Kiyokawa H, Soos T J, Park M S, Soares V C, Manova K, Pollard J W, Koff A. The absence of p27Kip1, an inhibitor of G1 cyclin-dependent kinases, uncouples differentiation and growth arrest during the granulosa→luteal transition. Cell Growth Differ. 1998;9:787–794. [PubMed] [Google Scholar]
- 61.Tyers M. The cyclin-dependent kinase inhibitor p40sic1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 63.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 64.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H-R, Kania M, Baumeister W, Nederlof P M. Import of human and thermoplasma 20S proteasomes into nuclei of HeLa cells requires functional NLS sequences. Eur J Cell Biol. 1997;73:105–113. [PubMed] [Google Scholar]
- 66.Won K-A, Reed S I. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 67.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yew P R, Kirschner M W. Proteolysis and DNA replication: the CDC34 requirement in the Xenopus egg cell cycle. Science. 1997;277:1672–1676. doi: 10.1126/science.277.5332.1672. [DOI] [PubMed] [Google Scholar]